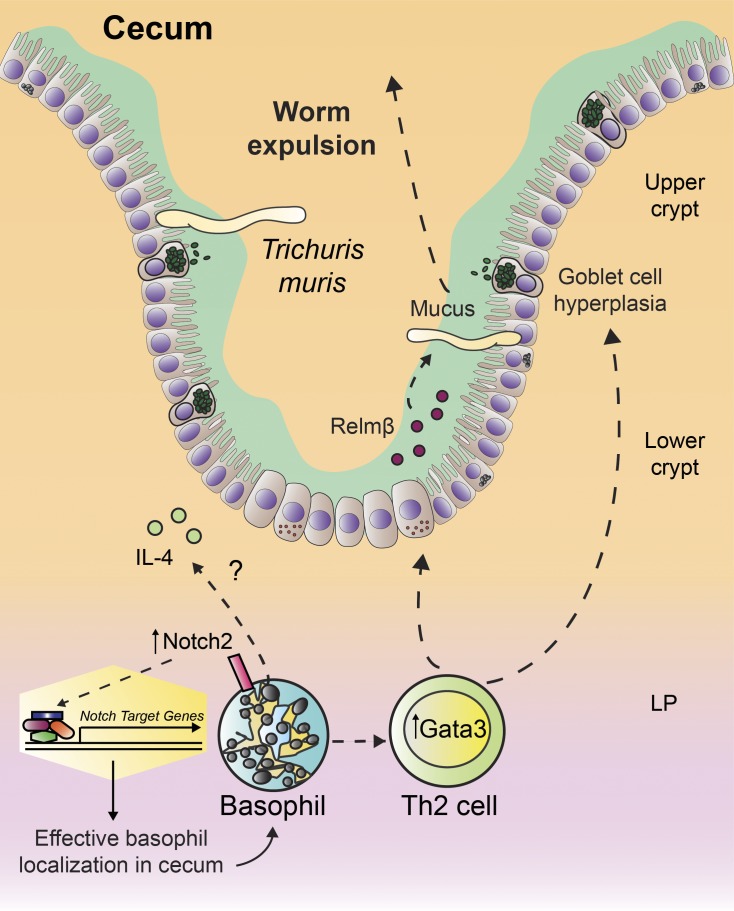
Basophils promote type 2 inflammation that mediates worm clearance during murine infection with the gastrointestinal helminth parasite Trichuris muris. Webb et al. show for the first time that basophil–intrinsic Notch signaling is required for basophil gene expression and a functional program that support helminth expulsion.
Abstract
Type 2 inflammation drives the clearance of gastrointestinal helminth parasites, which infect over two billion people worldwide. Basophils are innate immune cells that support host-protective type 2 inflammation during murine infection with the helminth Trichuris muris. However, the mechanisms required for basophil function and gene expression regulation in this context remain unclear. We show that during T. muris infection, basophils localized to the intestine and up-regulated Notch receptor expression, rendering them sensitive to Notch signals that rapidly regulate gene expression programs. In vitro, Notch inhibition limited basophil cytokine production in response to cytokine stimulation. Basophil-intrinsic Notch signaling was required for T. muris–elicited changes in genome-wide basophil transcriptional programs. Mice lacking basophil-intrinsic functional Notch signaling had impaired worm clearance, decreased intestinal type 2 inflammation, altered basophil localization in the intestine, and decreased CD4+ T helper 2 cell responses following infection. These findings demonstrate that Notch is required for basophil gene expression and effector function associated with helminth expulsion during type 2 inflammation.
Graphical Abstract
Introduction
Type 2 inflammation characterizes infection with gastrointestinal helminth parasites, which afflict over two billion people worldwide, causing significant morbidity and economic loss (Bethony et al., 2006; Harhay et al., 2010). Type 2 inflammation promotes helminth expulsion and tissue repair (Gause et al., 2013; Allen and Sutherland, 2014) and is initiated when helminths and their products cause intestinal epithelial cells (IECs) to release thymic stromal lymphopoietin (Tslp), IL-25, and IL-33. These cytokines and helminth antigens activate innate immune cells, including basophils, group 2 innate lymphoid cells (ILC2s), and APCs that support the polarization of CD4+ T helper 2 (Th2) cells (Allen and Maizels, 2011; Pulendran and Artis, 2012; Van Dyken et al., 2016). Innate and Th2 cells produce the type 2 cytokines IL-4, IL-5, IL-9, and IL-13, some of which, notably IL-13, act back on IECs to induce antiworm responses, including goblet cell hyperplasia, mucus production, increased cellular turnover, and tuft cell activation (Artis and Grencis, 2008; Gerbe et al., 2016; von Moltke et al., 2016).
Studies have shown that basophils, which are rare, short-lived innate granulocytes, can support host-protective type 2 inflammation during infection with some helminth species (Perrigoue et al., 2009; Siracusa et al., 2011; Eberle and Voehringer, 2016; Schwartz et al., 2016). During infection, basophils accumulate, interact with other immune cells, and are a source of IL-4 (Min et al., 2004; Gessner et al., 2005; Perrigoue et al., 2009; Siracusa et al., 2011). Basophils release IL-4 and other effector molecules following FcεRIα cross-linking and in response to Tslp, IL-3, IL-33, or helminth proteases (Phillips et al., 2003; Shen et al., 2008; Siracusa et al., 2011; Cassard et al., 2012). Critically, during primary infection with the murine intestinal parasite Trichuris muris, the absence of Tslp-elicited basophils was associated with impaired worm clearance that could be reversed by transfer of wild-type basophils (Siracusa et al., 2011; Rivera et al., 2016). However, the molecular mechanisms required for basophil gene expression programs and effector functions in the tissue that promote helminth expulsion remain poorly understood.
Notch signaling orchestrates organismal development, lymphopoiesis, and immune cell activation via regulation of Notch-dependent gene expression. Signaling is initiated when a ligand binds a cell surface Notch receptor (Notch1–4). A γ secretase enzyme releases the Notch receptor intracellular domain (Radtke et al., 2013), which translocates to the nucleus and forms a complex with the transcription factor Rbpj and a Mastermind-like protein that recruits critical coactivators. The complex is then able to bind to Notch target genes and regulate their expression (Kopan and Ilagan, 2009). During T. muris infection, Notch signaling in T cells is required for worm clearance and Th2 cell differentiation by driving or stabilizing Gata3 and Il4 expression (Tu et al., 2005; Fang et al., 2007; Bailis et al., 2013). Notch signaling can also promote mast cell differentiation, localization, and function (Sakata-Yanagimoto et al., 2008; Nakano et al., 2015; Jiang et al., 2018), and the pro-allergic functions of bone marrow–derived basophils in vitro and after transfer in vivo (Qu et al., 2017a,b). Together, these data suggest a role for Notch in promoting changes in basophil gene expression programs and functions in vivo during helminth infection.
Here, we show for the first time that basophils up-regulated expression of Notch receptors during T. muris infection, rendering them sensitive to Notch-mediated signals. In vitro, Notch inhibition was associated with impaired basophil IL-4 and IL-6 production in response to IL-33, and Il4 and Il6 were direct Notch targets. Genetic inhibition of Notch signaling specifically in basophils during T. muris infection resulted in an aberrant basophil gene expression program, delayed worm clearance, decreased intestinal type 2 effector responses, and altered patterns of basophil localization in the cecum that were associated with impaired accumulation of Th2 cells in the tissue. These results represent the first description of a noncytokine, non-antibody–mediated molecular pathway that is required for primary basophil function and gene expression programs in the intestine that support helminth expulsion.
Results and discussion
Basophils up-regulate the Notch2 receptor during T. muris infection
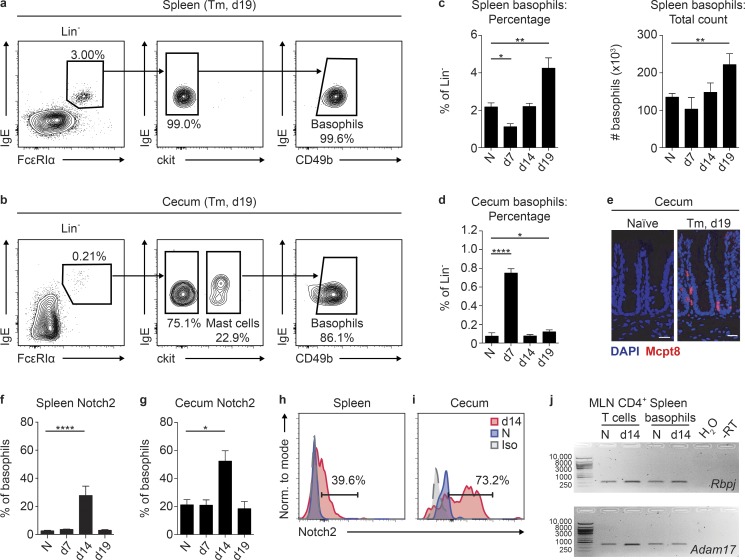
To investigate the regulation of basophil responses during T. muris infection, we first assessed basophil population dynamics throughout infection in resistant C57BL/6 mice in the spleen and in the cecum, where T. muris resides. Basophils were identified by flow cytometry as Lineage−IgE+FceRIα+CD49b+ckit− cells (Fig. 1, a and b) that also expressed the IL-33 receptor, ST2 (Fig. S1, a and b). In the spleen, basophil frequencies and total numbers declined from steady state levels at day 7 post-infection (p.i.) and increased at day 19 (Fig. 1 c) after the emergence of the Gata3-expressing Th2 cell response at day 14 p.i. (Fig. S1, c and d) and just before full worm clearance at day 21 in C57BL/6 mice (D’Elia et al., 2009; Klementowicz et al., 2012).
Figure 1.
Basophils up-regulate Notch signaling machinery during T. muris infection. (a and b) Representative plots of spleen (a) and cecum (b) basophils (CD45+Lineage[Lin]−FcεRIα+IgE+CD49b+ckit−) in T. muris (Tm)–infected C57BL/6 mice on day (d) 19 p.i. (c and d) Spleen basophil percentage (c) of Lin− cells and total numbers and cecum basophil percentage (d) of Lin− cells in naive (N) and T. muris–infected C57BL/6 mice on day 7, 14, and 19 p.i. (e) Immunofluorescent staining for Mcpt8+ (red) basophils in the cecum of N and day 19 T. muris–infected C57BL/6 mice (bar, 50 µm). (f–i) Percentage of spleen (f) and cecum (g) basophils that expressed Notch2 in N and T. muris–infected C57BL/6 mice on day 7, 14, and 19 p.i. with representative histograms showing Notch2 staining on splenic (h) and cecum (i) basophils (gray, isotype; blue, N; red, day 14 T. muris–infected). (j) Gel image of quantitative real-time PCR products for Rbpj and Adam17 reactions performed on mRNA isolated from sort-purified MLN CD4+ T cells or spleen basophils from N or day 14 T. muris-infected C57BL/6 mice (RT, reverse transcription). Mean ± SEM; *, P < 0.05; **, P < 0.01; ****, P < 0.0001; analyzed using (c, d, f, and g) a linear fixed-effect model with pairwise comparison; (a and b) representative of three experiments; (c and d) three experiments pooled (N, n = 18; Tm, n = 7–13/time point); (e) representative of three experiments; (f and g) three experiments pooled (N, n = 18; Tm, n = 7–13/time point); (h and i) representative of three experiments; and (j) CD4+ T cells from three mice pooled and basophils from 12 mice pooled.
Strikingly, in the cecum, we observed an inverse pattern with a significant increase in basophil frequencies at day 7 p.i. (Fig. 1 d) along with a large increase in frequencies of IgE+FceR1α+ cells that did not express CD49b or ckit (Fig. S1 e). This was followed by a contraction of the basophil population with maintenance of slightly elevated basophil frequencies at days 14 and 19 p.i. over those observed in naive mice (Fig. 1 d). Immunofluorescent staining for the basophil-specific marker Mcpt8 (Schwartz et al., 2014) confirmed infection-induced cecum basophil accumulation on day 19 p.i. (Fig. 1 e). We did not detect an appreciable basophil population in the draining mesenteric lymph node (MLN) in the steady state or early during infection (Fig. S1 f), supporting the notion that basophils may not be key APCs during helminth infection (Voehringer, 2017). These data suggest that there are temporal alterations in the size of the basophil population in the spleen and inflamed cecum during T. muris infection.
To examine if basophils were responsive to Notch signals during infection, we tracked surface expression of Notch receptors by spleen and cecum basophils throughout infection using flow cytometry. Spleen and cecum basophils from naive mice expressed low levels of all Notch receptors and a small percentage of spleen or cecum basophils up-regulated Notch1, 3, or 4 at various time points p.i. (Fig. 1, f–i; and Fig. S1, g–i). Strikingly, basophil Notch2 expression was significantly up-regulated in the spleen and cecum at day 14 p.i., with more than half of all basophils in the cecum expressing Notch2 at this time point (Fig. 1, f–i). Splenic basophils sort-purified from naive or day 14 p.i. T. muris–infected mice also had detectable expression of the transcription factor Rbpj and Adam17, a γ secretase enzyme (Kopan and Ilagan, 2009; Fig. 1 h). Together, these data show that basophils in the spleen and cecum are competent to receive Notch signals during T. muris infection.
Notch signaling is required for optimal IL-33–elicited basophil cytokine production
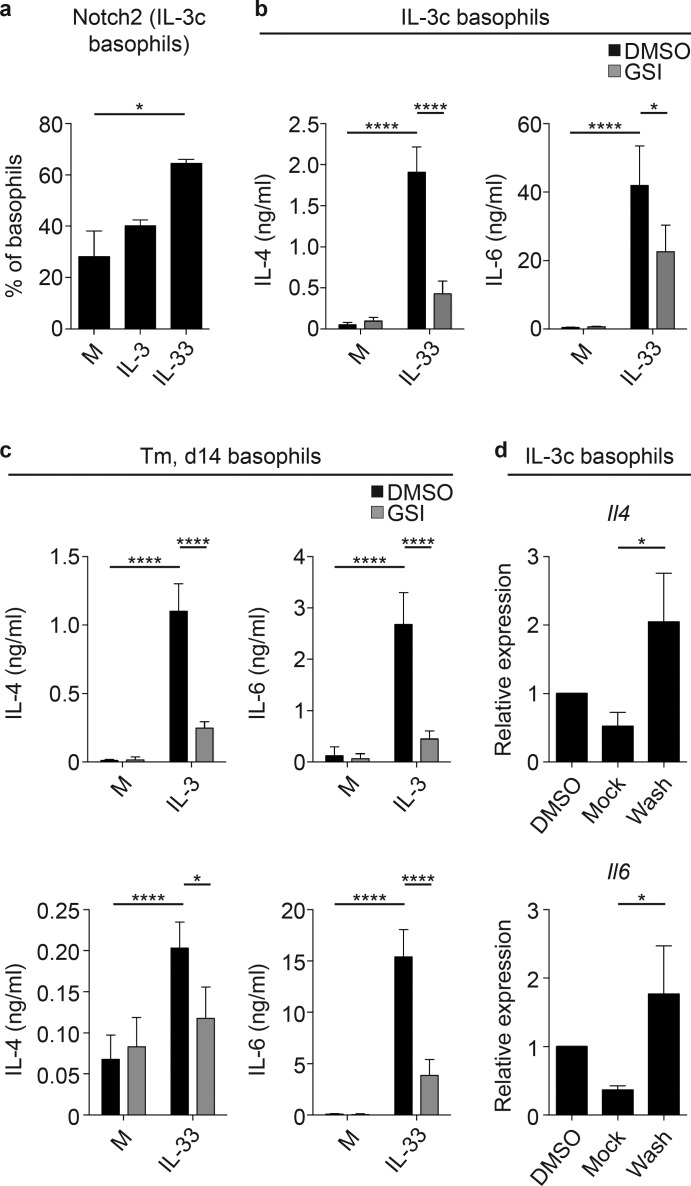
Next, we hypothesized that Notch signaling promotes basophil effector function in response to helminth infection–associated cytokines such as IL-3 and IL-33 (Webb and Tait Wojno, 2017). Thus, we first examined how IL-3 and IL-33 affected basophil cytokine production and Notch receptor expression in vitro. C57BL/6 mice were treated with IL-3 complexes (IL-3c) to expand the basophil population, enabling sort-purification of primary splenic basophils for in vitro assays (Ohmori et al., 2009; average purity 96.1%; Fig. S2 a). Basophils cultured for 18 h with 10 ng/ml recombinant murine (rm)IL-3 (Ohmori et al., 2009; Siracusa et al., 2011) or rmIL-33 (Pecaric-Petkovic et al., 2009; Venturelli et al., 2016) produced IL-4 and IL-6 as detected by intracellular cytokine staining (Fig. S2, b and c). Basophils from IL-3c–treated mice expressed Notch2 (Fig. S2 d), and rmIL-33 exposure in vitro further increased Notch2 expression (Fig. 2 a), suggesting that IL-3 and IL-33 promote basophil cytokine production and sensitivity to Notch signaling.
Figure 2.
Basophil IL-4 and IL-6 production is partially Notch signaling dependent. (a) Percentage of basophils from IL-3c–treated mice that expressed Notch2 following 18 h culture in media alone or with 10 ng/ml rmIL-33 or rmIL-3. (b and c) IL-4 and IL-6 levels in the cell-free supernatant measured by ELISA from in vitro basophils sort-purified from (b) IL-3c–treated mice and (c) mice on day (d) 14 after T. muris (Tm) infection, cultured in media alone or with 10 ng/ml rmIL-3 or rmIL-33 in the presence of DAPT (GSI) or a vehicle control (DMSO) for 5 d. (d) Il4 and Il6 expression (relative to Actβ and normalized to DMSO) measured using quantitative real-time PCR in basophils from IL-3c–treated mice subjected to the GSI washout assay. Mean ± SEM; *, P < 0.05; ****, P < 0.0001; analyzed using (a–c) a linear fixed-effect model with pairwise comparison or (d) the Wilcoxon rank sum test with χ2 test; (a) two experiments pooled (six to seven wells/condition total); (b and c) three to five experiments pooled (five to ten wells/condition total); and (d) three experiments pooled (three wells/condition total).
We next tested how Notch inhibition affects basophil IL-4 and IL-6 release after IL-3 or IL-33 stimulation (Gessner et al., 2005; Schwartz et al., 2014; Qu et al., 2017a). Sort-purified splenic basophils from C57BL/6 IL-3c–treated mice were cultured for 5 d with vehicle (DMSO) or the γ secretase inhibitor (GSI) N-[N-(3,5-Difluorophenacetyl)-l-alanyl]-S-phenylglycine t-butyl ester (DAPT) in media with or without rmIL-33. Basophils secreted high levels of IL-4 and IL-6 in response to IL-33 as measured by ELISA, and this response was significantly decreased in the presence of GSI (Fig. 2 b), independent of any effects on basophil expression of the IL-3 receptor CD123 or ST2 (Fig. S2 e) or viability (Fig. S2 f). Similarly, splenic basophils sort-purified from T. muris–infected mice secreted significantly less IL-4 and IL-6 in response to IL-3 and IL-33 when Notch signaling was inhibited (Fig. 2 c), suggesting that Notch can promote basophil IL-4 and IL-6 production in the context of IL-3c treatment and T. muris infection. Importantly, GSI did not completely abolish IL-3– and IL-33–induced basophil cytokine production (Fig. 2, b and c), and IL-4+ IL-3c–elicited basophils were not more likely to be Notch2+ than IL-4− basophils (Fig. S2 g), suggesting that Notch is not the only pathway that promotes basophil IL-4 and IL-6 secretion.
A primary feature of Notch signaling is the rapid regulation of direct Notch target genes (Radtke et al., 2013). To test whether Il4 and Il6 are direct Notch targets in basophils, we performed an in vitro Notch target identification assay in which Notch target gene expression in a cell type of interest is decreased following GSI treatment but recovers immediately upon GSI washout (with cycloheximide to limit translation; Bailis et al., 2013). Quantitative real-time PCR analysis showed that Il4 was a direct Notch target, as Il4 expression increased significantly in cells that were subjected to GSI but then washed (Wash) compared with cells left in GSI (Mock; Fig. 2 d), similar to observations in CD4+ T cells (Bailis et al., 2013). Il6 was also a direct Notch target in basophils (Fig. 2 d). Taken together, these data suggest that Notch provides one signal that can promote IL-3– and IL-33–elicited IL-4 and IL-6 production from primary basophils in vitro and that Notch can directly target the Il4 and Il6 genes.
Notch regulates helminth infection–induced basophil gene expression programs
These data prompted us to test the hypothesis that Notch signaling is required for T. muris–elicited basophil gene expression programs using mice with basophil-specific Notch inhibition. These mice express Cre recombinase-YFP under the control of the basophil-specific Mcpt8 gene (Sullivan et al., 2011) and a construct containing a floxed stop cassette that prevents transcription of a dominant-negative Mastermind-like protein (DNMAML)–GFP in cells that lack Cre (Tu et al., 2005; Fig. S2 h). In basophils alone, Cre excises the stop codon, allowing expression of DNMAML that binds the transcriptional activating complex yet fails to recruit necessary coactivators, rendering the complex nonfunctional (Tu et al., 2005; Maillard et al., 2006) and inhibiting Notch signaling.
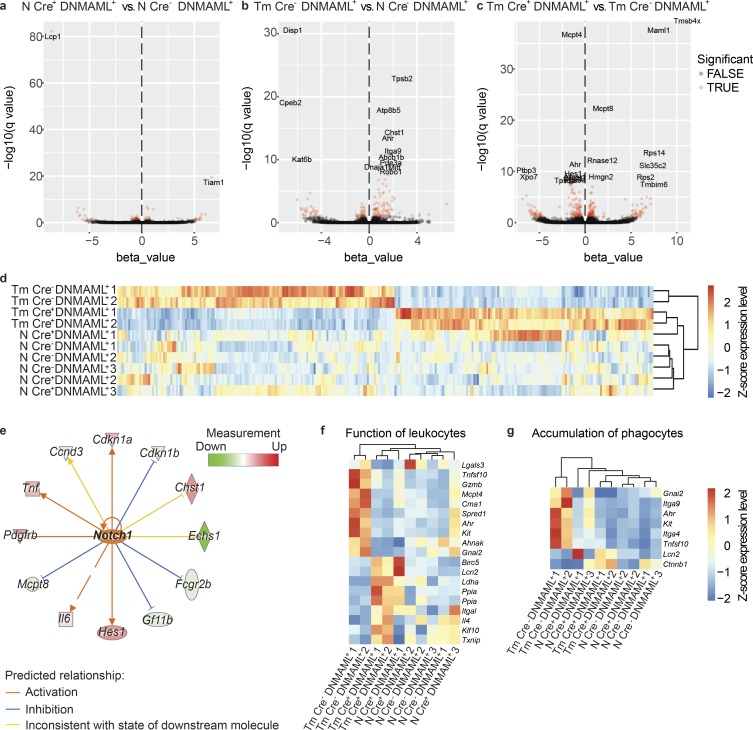
Primary splenic Notch signaling–sufficient (Mcpt8Cre− DNMAML+) and Notch signaling–deficient (Mcpt8Cre+ DNMAML+) basophils were sort-purified from naive and day 14 T. muris–infected mice treated with IL-3c to expand the basophil population and were assessed for gene expression by using RNA sequencing (RNaseq) and Kallisto/Sleuth analysis software (Bray et al., 2016; Pimentel et al., 2017). There were few significant changes in gene expression between basophils from naive, IL-3c–treated Mcpt8Cre− DNMAML+ and Mcpt8Cre+ DNMAML+ mice (Fig. 3 a), but 151 transcripts were differentially expressed in Mcpt8Cre− DNMAML+ basophils from IL-3c–treated, T. muris–infected versus naive mice (Fig. 3 b). Critically, 293 transcripts were differentially expressed between Mcpt8Cre+ DNMAML+ and Mcpt8Cre− DNMAML+ basophils from infected, IL-3c–treated mice (Fig. 3 c and Table S1) with the gene expression profile of Mcpt8Cre+ DNMAML+ basophils from infected, IL-3c–treated mice clustering distinctly from that of Mcpt8Cre− DNMAML+ basophils from infected, IL-3c–treated mice (Fig. 3 d).
Figure 3.
Notch signaling promotes helminth infection–induced changes in effector gene expression in primary basophils. (a–c) Volcano plots of differentially expressed genes between splenic basophils from (a) naive, IL-3c–treated (N), Mcpt8Cre+ DNMAML+ versus Mcpt8Cre− DNMAML+ mice, (b) day 14 T. muris (Tm)–infected, IL-3c–treated versus N Mcpt8Cre− DNMAML+ mice, or (c) day 14 T. muris–infected Mcpt8Cre+ DNMAML+ vs. Mcpt8Cre− DNMAML+ mice. Differentially expressed genes with −log10(q value) > ∼1.3 shown in red with gene names labeled when –log10(q value) > ∼8. (d) Heatmap of differentially expressed genes between basophils from day 14 T. muris–infected Mcpt8Cre+ DNMAML+ versus Mcpt8Cre− DNMAML+ mice (−log10(q value) > ∼1.3), reported by z-score with samples organized by hierarchal clustering. (e) Network depicting genes downstream of Notch1 activation in splenic basophils from day 14 T. muris–infected versus N Mcpt8Cre− DNMAML+ mice. (f and g) Heatmaps showing expression of genes depicted as z-score associated with IPA-designated groups of genes associated with (f) function of leukocytes and (g) accumulation of phagocytes in basophils from N or day 14 T. muris–infected Mcpt8Cre+ DNMAML+ and Mcpt8Cre− DNMAML+ mice (genes filtered on differences between day 14 T. muris–infected Mcpt8Cre+ DNMAML+ and Mcpt8Cre− DNMAML+ basophils (–log10(q value) > 1), samples arranged by hierarchal clustering). Samples included cells from three to four pooled mice.
Ingenuity Pathway Analysis (IPA) of differentially expressed genes identified Notch signaling (called Notch1 but consistent with pan-Notch activation; Kopan and Ilagan, 2009) as a likely upstream regulator of changes in gene expression observed between Mcpt8Cre− DNMAML+ but not Mcpt8Cre+ DNMAML+ basophils from IL-3c–treated, infected versus naive mice (Fig. 3 e), validating Notch inhibition in Mcpt8Cre+ DNMAML+ basophils. IPA also identified transcripts associated with the function of leukocytes (Fig. 3 f), accumulation of immune cells (Fig. 3 g), and lipid synthesis (Fig. S2 i) that were significantly up-regulated in Mcpt8Cre− DNMAML+ but not Mcpt8Cre+ DNMAML+ basophils from IL-3c–treated, infected versus naive mice. These transcripts included genes involved in MAPK signaling, which is active downstream of Notch in rat basophilic leukemia cells (Jiang et al., 2018), and genes that encode for integrins associated with intestinal cellular positioning such as Itga4 and Itga9 (Rivera-Nieves et al., 2005; Agace, 2008). Other transcripts encode for proteins that could allow basophils to promote accumulation, survival, or function of other cells, such as Tnfsf10, which encodes TNF-related apoptosis-inducing ligand, which induces apoptosis but can also promote tissue Th2 cell accumulation (Weckmann et al., 2007), and Hpgds, which encodes the synthesis enzyme for prostaglandin D2, an attractant and activator of Th2 cells, ILC2s, and eosinophils (Hirai et al., 2001).
Notably, there were some up-regulated genes in Mcpt8Cre+ DNMAML+ basophils versus Mcpt8Cre− DNMAML+ basophils in infected, IL-3c–treated mice (Fig. 3, d, f, and g; and Fig. S2 i), including Il4. This finding further suggests that, while Notch promotes basophil IL-4 secretion in response to IL-33 in vitro (Fig. 2), direct Notch targeting of Il4 transcription is not the only factor required for basophil IL-4 production during infection. This is pertinent considering that while Il4 is transcribed in steady state basophils, Il4 transcription and IL-4 protein production are not always equivalent (Mohrs et al., 2005). Overall, while changes in gene expression detected in this assay may be due to pathways downstream of Notch signaling in basophils (indirect effects; Radtke et al., 2013) and may be influenced by IL-3c treatment, these data suggest that basophil-intrinsic Notch signaling is required for the emergence of a basophil gene expression program that promotes basophil activation, localization, and production of effector molecules during T. muris infection.
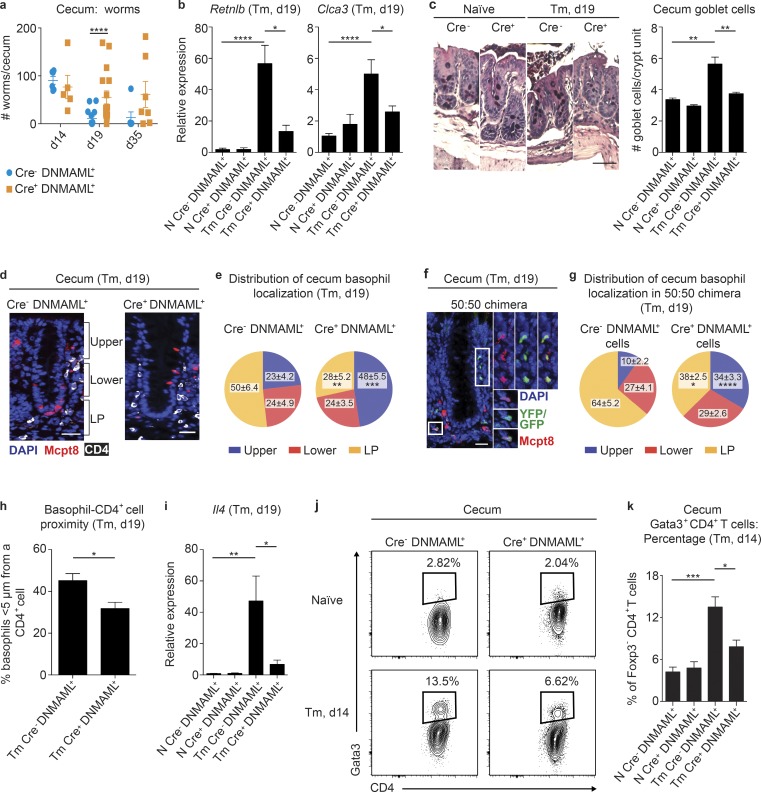
Basophil-intrinsic Notch signaling promotes optimal worm clearance, type 2 inflammation, and basophil positioning in the cecum during T. muris infection
Our in vitro and transcriptional data prompted us to investigate whether basophil-intrinsic Notch signaling was important in promoting helminth expulsion and type 2 inflammatory responses during T. muris infection. We infected Mcpt8Cre− DNMAML+ and Mcpt8Cre+ DNMAML+ mice with T. muris and assessed their response to infection. While Mcpt8Cre− DNMAML+ and Mcpt8Cre+ DNMAML+ mice had similar worm burdens on day 14 p.i., strikingly, Mcpt8Cre− DNMAML+ but not Mcp8Cre+ DNMAML+ mice had cleared most of their worms by day 19, and some Mcpt8Cre+ DNMAML+ mice retained significant numbers of adult parasites out to day 35 p.i. (Fig. 4 a). Increased susceptibility to infection in Mcpt8Cre+ DNMAML+ mice at day 19 p.i. was associated with decreased infection-induced IEC responses that have been associated with worm expulsion (Harris and Loke, 2017), including colonic expression of Retnlb, the gene that encodes for Relm-β (Herbert et al., 2009), and Clca3 (Gob5), a mediator of mucin up-regulation (Nakanishi et al., 2001; Fig. 4 b), and cecum goblet cell hyperplasia (Fig. 4 c). These data show that a loss of functional Notch signaling in basophils led to suboptimal helminth-elicited IEC responses and worm clearance.
Figure 4.
Notch signaling in basophils is required for optimal worm clearance and intestinal type 2 inflammation following T. muris infection. (a) Worm burden in the cecum. d, day. (b) Retnlb and Clca3 gene expression (relative to Actβ and normalized to Cre− DNMAML+ naive [N]) measured by real-time PCR in colon homogenates. Tm, T. muris. (c) Representative images (bar, 50 µm) and average number cecum goblet cells per crypt unit. (d) Immunofluorescence staining for Mcpt8+ (red) basophils and CD4+ (white) cells in the cecum (bar, 50 µm). (e) Percentage of basophils that localized in the upper and lower intraepithelial regions of the crypt and the lamina propria (LP) in Mcpt8Cre− DNMAML+ and Mcpt8Cre+ DNMAML+ mice on day 19 after T. muris infection. (f and g) Immunofluorescent staining for Mcpt8Cre− DNMAML+ (Mcpt8 stained, red) and Mcpt8Cre+ DNMAML+ (YFP/GFP+ and Mcpt8 stained, green/red [insets]) basophils in the cecum (f; bar, 50 µm), and percentage of basophils (g) that localized in the upper and lower intraepithelial regions of the crypt and the LP in Mcpt8Cre− DNMAML+:Mcpt8Cre+ DNMAML+ 50:50 bone marrow chimeric mice on day 19 after T. muris infection. (h) Percentage of basophils that were <5 µm from a CD4+ cell quantified from d. (i) Il4 gene expression (relative to Actβ and normalized to Cre− DNMAML+ N) measured by real-time PCR in colon homogenates in Mcpt8Cre− DNMAML+ and Mcpt8Cre+ DNMAML+ mice that were N or on day 19 after T. muris infection. (j and k) Representative plots (j) and percentage (k) of TCRβ+CD4+CD8−Foxp3− cells that expressed Gata3 in the cecum in Mcpt8Cre− DNMAML+ and Mcpt8Cre+ DNMAML+ mice that were N or on day 14 after T. muris infection. Mean ± SEM; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; (a–c, e, g–i, and k) analyzed by using a linear fixed-effect model with pairwise comparison; (a) six experiments pooled (n for all groups displayed on graph); (b) three (Retnlb) or four (Clca3) experiments pooled (Retnlb: N, n = 6; Tm, n = 12–13; Clca3: N, n = 7–8; Tm, n = 16–17); (c) two experiments pooled (N, n = 3; Tm, n = 7–8); (d) representative of two experiments; (e and h) two experiments pooled (n = 8); (f) representative of two experiments; (g) two experiments pooled (n = 7); (i) five experiments pooled (N, n = 9; Tm, n = 20–21); (j) representative of three experiments; and (k) three experiments pooled (N, n = 6; Tm, n = 6).
We next tested if these defects in Mcpt8Cre+ DNMAML+ mice were associated with impaired basophil responses. Notch signaling was not required for infection-induced spleen or cecum basophil population dynamics (Fig. S3, a and b) or accumulation of Mcpt8+ cells in the cecum at day 19 p.i. counted in immunofluorescent images (Fig. S3 c). However, imaging analysis revealed that basophil-intrinsic Notch signaling was required for normal basophil localization in the cecum during infection because basophils were primarily located in the lamina propria near the crypt base in Mcpt8Cre− DNMAML+ mice but were more likely to be in the intraepithelial region and upper crypt unit portion in Mcpt8Cre+ DNMAML+ mice (Fig. 4, d and e). This pattern was also observed in mixed–bone marrow chimeric mice that had equal populations of Mcpt8Cre− DNMAML+ and Mcpt8Cre+ DNMAML+ basophils (Fig. 4, f and g; and Fig. S3 d). Strikingly, the altered localization pattern of Mcpt8Cre+ DNMAML+ basophils was associated with a significant decrease in the percentage of basophils in close proximity to a CD4+ cell (Fig. 4 h). These data are consistent with RNaseq data showing that Notch signaling is required for basophil gene expression that may determine basophil localization in the cecum during infection (Fig. 3 g). These findings also suggest that basophil-intrinsic Notch signaling plays a key role in orienting basophils in the inflamed tissue to allow them access to CD4+ T cells.
These data led us to hypothesize that Notch programs or positions basophils to support IL-4–producing CD4+ Th2 cells in the cecum during T. muris infection that are critical for worm expulsion (Finkelman et al., 1997). Consistent with this idea, Mcpt8Cre+ DNMAML+ mice had impaired infection-induced colonic Il4 upregulation compared with Mcpt8Cre− DNMAML+ mice, measured by real-time PCR at day 19 p.i. (Fig. 4 i). CD4+ cells were the major cecum IL-4 source as detected by intracellular cytokine staining at day 14 p.i. (Fig. S3, e and f) when infection-induced colonic Il4 expression was highest (Fig. S3 g) and the cecum Th2 cell response emerged (Fig. S1 d). Thus, we tested if Mcpt8Cre+ DNMAML+ mice had a deficient Th2 cell response at this time point. Indeed, despite similar infection-induced Th2 cell frequencies in the MLN of Mcpt8Cre− DNMAML+ and Mcpt8Cre+ DNMAML+ mice (Fig. S3 h), Mcpt8Cre+ DNMAML+ mice had lower frequencies of cecum Gata3+ Th2 cells compared with Mcpt8Cre+ DNMAML+ mice at day 14 p.i. (Fig. 4, j and k).
Together, our findings show that Notch signaling in primary basophils is required for optimal T. muris infection–induced basophil gene expression and effector function, allowing basophils to support Th2 cell responses in the tissue that promote worm expulsion. However, a number of outstanding questions remain. For example, where and when basophils encounter Notch ligands is unknown, with endothelial cells (Butler et al., 2010; Gamrekelashvili et al., 2016), fibroblastic stromal cells (Fasnacht et al., 2014; Chung et al., 2017), immune cells, and IECs (Radtke et al., 2013; Demitrack and Samuelson, 2016) all potential ligand sources. In addition, how basophils integrate Notch signals and other cues to regulate gene expression and function is unclear. We did observe variance in the ability of Mcpt8Cre+ DNMAML+ mice to expel T. muris (Fig. 4 a), which could be due to transgene penetrance but could also result from variation in how basophils in each animal integrate stimuli from cytokines, FcεRIα cross-linking, and other pathways in the absence of functional Notch. Since Notch was clearly not the only factor that drove basophil Il4 expression and IL-4 production (Fig. 2, b and c; Fig. 3; and Fig. S2 g), it seems likely that basophils must integrate signals from multiple sources to fully acquire their full complement of normal functions that support helminth clearance.
Critically, though our data suggest that Notch programs basophil positioning in the cecum and their ability to support Th2 cell responses during T. muris infection, the exact Notch signaling–dependent basophil effector mechanisms that promote resistance to infection have yet to be identified. Future studies are required to assess how Notch programs basophils to position in the cecum and the functional significance of positioning-related genes identified by RNaseq as differentially regulated between Notch signaling–deficient and –sufficient basophils during infection. Further, how basophil-intrinsic Notch signaling programs basophils to influence Th2 cell responses, whether via IL-4 production or otherwise, and the identity of the critical cellular sources of IL-4 throughout T. muris infection remain unclear. The significance of basophil IL-6 production and of IL-6 generally during helminth infection is also unclear. Despite remaining questions, our data show for the first time that basophil-intrinsic Notch is required for basophil gene expression and functions during helminth infection that promote worm expulsion. This is an important finding for our understanding of innate immune cell biology in type 2 inflammation in the intestine and has implications for the development of effective therapeutics targeting type 2 immune pathways.
Materials and methods
Mice
Male and female C57BL/6 wild-type and CD45.1 mice were purchased from The Jackson Laboratory or bred in-house. DNMAML+ C57BL/6 mice (Tu et al., 2005; DNMAML+ denotes DNMAML+/+ throughout) were provided by Warren Pear (University of Pennsylvania, Philadelphia, PA) and were crossed to Mcpt8Cre+/− mice (The Jackson Laboratory) provided by David Artis (Weill Cornell Medicine, New York, NY). Of note, the DNMAML transgene is fused to GFP (Tu et al., 2005), while the Mcpt8Cre is YFP tagged (Sullivan et al., 2011). Cre+ DNMAML+ mice have only one copy of Cre in all cases, allowing for normal Mcpt8 expression from the other allele. Mice were used at 6–16 wk of age, and all experiments used age- and sex-matched controls. Animals were housed in specific pathogen–free conditions at Cornell University in accordance with protocols approved by the Cornell University Institutional Animal Care and Use Committee. Mcpt8Cre− DNMAML+ and Mcpt8Cre+ DNMAML+ mice were cohoused.
In vivo treatments and ex vivo tissue preparation
T. muris eggs were generated in-house and tested for infectivity by infecting Rag−/− mice with 200 T. muris eggs. Briefly, worms collected from the cecum of Rag−/− mice at 6 wk p.i. were cultured overnight at 37°C and 5% CO2. Eggs were collected and stored in ultrapure water at room temperature for 7 wk to allow for embryonation (Klementowicz et al., 2012). Experimental mice were infected with 200 T. muris eggs by oral gavage and sacrificed on days 7, 14, or 19 p.i. for analysis. At various time points p.i., ceca were harvested, and the number of adult worms in each cecum was ascertained microscopically.
For harvesting large numbers of splenic basophils from naive or infected mice, some mice were treated twice, 3 d apart, intraperitoneally with IL-3c (1 µg rmIL-3 [R&D Systems] with 0.5 µg anti-IL-3 [MP2-8F8; BioLegend] per mouse per dose) and euthanized on day 5 following initiation of treatment.
To make Mcpt8Cre− DNMAML+:Mcpt8Cre+ DNMAML+ 50:50 bone marrow chimeras, CD45.1 female mice were lethally irradiated with 1,000 rad and reconstituted with a 50:50 mix of 106 Mcpt8Cre− DNMAML+ and 106 Mcpt8Cre+ DNMAML+ bone marrow cells from female mice, as previously described (Tait Wojno et al., 2015). Recipients were treated for 3 wk after transfer with sulfamethoxozole/trimethoprim (STI Pharma LLC) and reconstituted for 6–8 wk before T. muris infection.
Spleens and MLNs were digested with 1 U/ml Liberase TL (Roche) and 20 µg/ml DNase (Sigma-Aldrich) for 15 min at 37°C. Single-cell suspensions were prepared by disrupting the tissue through a 70-µm strainer. Spleens were red blood cell lysed with ammonium-chloride-potassium lysis buffer (Lonza). For analysis of cecum immune cell populations, ceca were collected directly into ice-cold PBS, cut open longitudinally, cleaned of fecal contents, and washed twice in ice-cold HBSS (Thermo Fisher Scientific) with 10% FBS (Denville Scientific). Tissues were chopped roughly into 1-cm pieces in HBSS/FBS, stored on ice, and then shaken by hand vigorously for 10 s before being strained and incubated in HBSS plus 2 mM EDTA (Thermo Fisher Scientific) for 15 min at 37°C in a shaking incubator. Samples were shaken by hand again, strained, and incubated for 25 min in fresh HBSS/EDTA at 37°C in a shaking incubator. After further shaking, samples were incubated in 1 U/ml Liberase TL and 20 µg/ml DNase for 35 min at 37°C in a shaking incubator and further vigorous manual shaking every 7 min. Samples were washed through 40-µm strainers to collect single-cell suspensions.
Flow cytometry and cell sorting
Single-cell suspensions were incubated with Aqua LIVE/DEAD Fixable Dye (Life Technologies) and fluorochrome-conjugated monoclonal antibodies against mouse CD3ε (145-2C11), CD4 (GK1.5), CD5 (53–7.3), CD8 (53–6.7), CD11b (M1/70), CD11c (N418), CD19 (eBio1D3), CD44 (1M7), CD45 (30-F11), CD45.1 (A20), CD45.2 (104), CD49b (DX5), CD62L (MEL-14), CD64 (X54-5/7.1), CD123 (5B11), CD200R (OX-110), ckit (2B8), FcεRIα (MAR-1), IgE (R35-72), NK1.1 (PK136), Notch1 (22E6), Notch2 (16F11), Notch3 (HMN3-133), Notch4 (HMN4-14), SiglecF (E50-2440), ST2 (RMST2-2), TCRβ (H57-597), or TCRγδ (eBioGL3) from Thermo Fisher Scientific, BD Biosciences, or BioLegend. Surface staining was performed in the presence of 10% rat serum (Jackson ImmunoResearch) and 1 µg/ml FcR block (anti-CD16/32, 2.4G2; Bio X Cell). For intracellular cytokine staining, cells were fixed in 2% paraformaldehyde (PFA) and incubated in permeabilization buffer (BD Biosciences) before staining with rat serum and FcR block with antibodies against IL-4 (11B11) and IL-6 (MP5-20F3) from Thermo Fisher Scientific. For nuclear staining, cells were fixed with Foxp3 fixation/permeabilization buffer and permeabilized by using Foxp3 permeabilization buffer (Thermo Fisher Scientific) and stained for Foxp3 (FJK-16s) and Gata3 (TWAJ) from Thermo Fisher Scientific. Samples were run on a three-laser LSR II (BD Biosciences), a four-laser Attune NxT (Thermo Fisher Scientific), or a Gallios (Beckman Coulter) flow cytometer, and FlowJo 10 (Tree Star) was used to analyze data. Gates were set by using fluorescence minus one or isotype controls. For cell sorting, single-cell suspensions were sorted by using a four-laser FACS Fusion (BD Biosciences) with an 85-µm nozzle. All leukocyte populations were first gated as singlet, live (LIVE/DEAD Aqua−), leukocytes (by forward scatter area/side scatter area). Basophils were gated as CD45+Lin−IgE+FcεRIα+CD49b+c-kit−. For sorting, basophils were sorted as CD45+Lin−IgE+FcεRIα+, CD45+Lin−CD200R+CD49b+, or CD45+Lin−CD123+CD49b+. The lineage gate (Lin) included CD3, CD5, CD11c, CD19, CD64, TCRγδ, NK1.1 and, in some instances for sorting to exclude mast cells, ckit.
RNaseq
Groups of three or four Mcpt8Cre− DNMAML+ and Mcpt8Cre+ DNMAML+ female mice were either naive or infected on day 0 with 200 T. muris eggs. On day 9 and 12 of the infection time course, naive and infected mice were treated with IL-3c as described above. On day 14 p.i., spleen basophils were sort-purified from single-cell suspensions as described above, resuspended in TRIzol (Invitrogen), and snap frozen. Total RNA was purified by using TRIzol according to the manufacturer’s protocol, with an additional chloroform extraction step of the aqueous layer in Phase Lock Gel heavy tubes (Quantabio) after the first-phase separation, addition of 1 µl GlycoBlue (Thermo Fisher Scientific) immediately before isopropanol precipitation, and use of two washes of the RNA pellet with 75% ethanol. RNA concentration and integrity were determined by using a Qubit 3 (RNA HS kit; Thermo Fisher Scientific) and with a Fragment Analyzer (Advanced Analytical). PolyA+ RNA was isolated with the NEBNext Poly(A) mRNA Magnetic Isolation Module (New England Biolabs). To prepare libraries, TruSeq-barcoded RNaseq libraries were generated with the NEBNext Ultra Directional RNA Library Prep Kit (New England Biolabs). Each library was quantified with a Qubit 2.0 (dsDNA HS kit; Thermo Fisher Scientific), and the size distribution was determined with a Fragment Analyzer before pooling. Libraries were sequenced on a NextSeq 500 instrument (Illumina). At least 20 M single-end 75-bp reads were generated per library. RNaseq data are available in the Gene Expression Omnibus database with accession no. GSE129056.
We used Kallisto (Bray et al., 2016) to estimate the abundance of annotated transcripts on reference GRCm38 (mm10) release 79 from RNaseq data. We specified the estimated fragment length (-l = 300), SD (-s = 100), and 100 bootstrap iterations. We also specified strand-specific reads with the first read on the reverse strand (--rf-stranded). Differentially expressed transcripts were identified by using the Wald test in Sleuth (Pimentel et al., 2017) and filtered for a false discovery rate of 10%. Heatmaps were generated in R after centering transcript abundance estimates by the mean and scaling to the SD. Functional analysis of gene lists containing significantly differentially expressed genes (−log10[q value] > 1) was done with QIAGEN’s IPA. IPA utilizes a right-tailed Fisher’s exact test to determine if pathways are significantly altered between conditions (−log10[q value] > ∼1.3). An activation z-score >2 was considered significant.
In vitro cultures and GSI washout assay
For analysis of sorted splenic basophil cytokine production by intracellular cytokine staining, 15 × 104 basophils per well from mice treated with IL-3c were cultured at 37°C and 5% CO2 in complete Roswell Park Memorial Institute medium (RPMI; Corning) with 10% FBS (Denville Scientific), 1% l-glutamine (Corning), and 1% penicillin/streptomycin (Corning), plus 55 µM β-mercaptoethanol (Thermo Fisher Scientific) in the presence or absence of 10 ng/ml rmIL-3 or rmIL-33 (R&D Systems) for 18 h. For the final 5 h, Brefeldin A (Sigma-Aldrich) was spiked into the cultures according to manufacturers’ instruction, and the cells were then stained as described above.
For analysis of cytokine production following Notch inhibition, 50 × 104 sorted splenic basophils per well from IL-3c mice or 104 basophils per well from the spleens of T. muris–infected mice were cultured at 37°C and 5% CO2 in complete RPMI with or without rmIL-3 or rmIL-33 and with or without the addition of 1 µM of the GSI DAPT (Sigma-Aldrich) or vehicle (DMSO). On day 3 of culture, fresh cytokines were spiked into cultures. Cell-free supernatants were collected for ELISA on day 5 of culture. For analysis of the impact of GSI on cell surface receptor expression, sorted basophils were cultured in the presence or absence of GSI for 18 h before flow analysis.
To demonstrate induction of DNMAML-GFP expression following exposure to Tat-Cre, 5 × 104 splenic basophils from IL-3c–treated Mcpt8Cre− DNMAML+ mice were cultured with or without 100 µg/ml Tat-Cre (Excellgen) in Opti-MEM (Thermo Fisher Scientific) for 45 min at 37°C and 5% CO2. Cells were washed repeatedly in PBS to remove excess Cre and then cultured for 18 h in complete RPMI plus 55 µM β-mercaptoethanol and 25 mM Hepes buffer (Thermo Fisher Scientific) at 37°C and 5% CO2.
For analysis of IL-4 production in the cecum, 3 × 105 cecum cells were cultured with Brefeldin A (Sigma-Aldrich) according to the manufacturers’ instruction in complete RPMI plus 55 µM β-mercaptoethanol and 25 mM Hepes buffer at 37°C and 5% CO2 for 5 h. Cells were then collected for intracellular cytokine staining as described above.
For the GSI washout assay, sorted splenic basophils from mice treated with IL-3c were cultured for 18 h at 37°C and 5% CO2 in complete RPMI with 1 µM DAPT or DMSO. Cells were then washed twice in PBS before being cultured for 4 h with DAPT or DMSO in the presence of 20 µg/ml cycloheximide and then collected for real-time PCR analysis.
ELISA and real-time PCR
Cytokine levels in cell-free supernatants were assessed by using standard sandwich ELISA for IL-4 and IL-6 (Thermo Fisher Scientific). For real-time PCR, RNA was isolated from intestinal lysates by using TRIzol extraction according to the manufacturers’ instruction. Real-time PCR was performed on cDNA generated by using a Superscript II reverse transcriptase kit (Life Technologies) with SYBR Green master mix (Applied Biosystems) and commercially available primer sets (QIAGEN QuantiTect). Samples were run on the ABI 7500 Real-Time PCR System (Life Technologies). PCR products for Notch pathway–associated genes were visualized by using gel electrophoresis.
Histology and immunofluorescence
At necropsy, a section of cecum was collected and stored directly in 10% buffered formalin. Tissues were paraffin-embedded, and 5-mm sections were stained with periodic acid–Schiff/Alcian blue. Image acquisition was performed by using a Zeiss Observer Z1 microscope, a Zeiss Axiocam MR3 camera, and AxioVision version 4.8 acquisition software (Zeiss). Adobe Photoshop (Adobe Systems Inc.) was used to adjust brightness and contrast (changes were applied to all images). The average number of goblet cells (periodic acid–Schiff/Alcian blue positive staining) per crypt unit was enumerated for all intact crypt units that could be visualized in sections from each mouse. A researcher blinded to sample and group identity performed the enumerations at 10× magnification, with representative images at 20×.
For immunofluorescence in Mcpt8Cre− DNMAML+ and Mcpt8Cre+ DNMAML+ mice, 5-µm paraffin-embedded cecum sections were washed in xylene three times for 5 min before being washed down in an ethanol gradient (100% to 50%). Slides were washed in water and boiled in sodium citrate buffer for 15 min. For 50:50 Mcpt8Cre− DNMAML+:Mcpt8Cre+ DNMAML+ chimeras, cecal samples were fixed in 4% PFA (Sigma-Aldrich) at 4°C, sequentially incubated in 15% and 30% sucrose (Sigma-Aldrich) solutions overnight to quench residual PFA, mounted in optimal cutting temperature embedding medium (Sakura), frozen, and sectioned. 25-μm cryosections were blocked in 2% normal goat serum (Jackson ImmunoResearch), 0.05% Tween (Thermo Fisher Scientific), and 0.1% Triton (Sigma-Aldrich) for 1 h at room temperature, incubated with primary antibodies against Mcpt8 (TUG8; BioLegend) and CD4 (EPR19514; Abcam) or isotype controls overnight at 4°C, washed, and incubated with Alexa Fluor 488–conjugated donkey anti–rabbit IgG and/or Alexa Fluor 647–conjugated goat anti–rat IgG (Jackson ImmunoResearch) for 1 h at room temperature. Slides were washed before the addition of mounting medium containing DAPI (Vector Laboratories) and imaged directly on a TCS SP8 Leica confocal microscope with laser lines with emission wavelengths of 488, 633, and 405 (DAPI) at room temperature under the 40× objective with oil immersion (Leica). Images were rendered in Imaris software for analysis.
The average number of basophils, defined based on punctate Mcpt8 staining surrounding the multilobular nucleus, per high-powered field (10 fields total) was calculated for each sample in Mcpt8Cre− DNMAML+ and Mcpt8Cre+ DNMAML+ mice. The location of at least 25 basophils (lamina propria, lower intraepithelial crypt portion [lower 50% of the total length of the crypt unit from base to longest point at the tip, measured in micrometers] or upper intraepithelial crypt portion [upper 50%]) was recorded for Mcpt8Cre− DNMAML+, Mcpt8Cre+ DNMAML+, and chimeric 50:50 Mcpt8Cre−DNMAML+:Mcpt8Cre+ DNMAML+ mice. Whether a basophil was in proximity to a CD4-expressing cell was determined by assessing if the leading edge of the basophil was <5 µm from the nearest leading edge of a CD4 cell, and the average frequency of basophils that were in proximity to a CD4 cell per field (10 fields total) was calculated for each sample. A researcher blinded to sample and group identity of each image performed the enumerations at 20× magnification. Adobe Photoshop was used to adjust brightness and contrast of representative images (changes were applied to all images).
Statistics
Statistical analysis was performed by using JMP software (SAS). Data were analyzed by using linear mixed-effects models with a fixed effect of experimental group and a random effect of experiment day. Model assumptions of normality and homogeneous variance were assessed by a visual analysis of the raw data and the model residuals. Right-skewed data were log or square root transformed as indicated by the residuals. Experimental group was considered statistically significant if the fixed-effect F test P value was ≤0.05, with results in graphs shown as mean ± SEM. Post hoc pairwise comparisons between experimental groups were made using a Student’s t test for two groups or Tukey’s honestly significant difference multiple-comparison test for more than two groups. Statistical outliers were identified by using the extreme Studentized deviate method and were omitted before performing the mixed-model analysis. For the GSI washout assay, where sample size was low, mock and wash conditions were compared by using a Wilcoxon rank sum test, which does not assume normal distribution of data. Significant differences between these two groups were calculated using a χ2 test, with results ≤0.05 judged to be significant, with results shown in graphs as mean ± SEM. For analysis of basophil localization within the cecum, a linear model with fixed effects for genotype, location, and experiment was employed, initially with all two-way/three-way interactions between effects included and any interactions that were not significant excluded from the final model. Post hoc comparisons between group and location were made using Tukey’s method to correct for multiple comparisons. Results ≤0.05 were judged to be significant, with results shown in graphs as mean ± SEM.
Online supplemental material
Fig. S1 provides additional characterization of basophils during T. muris infection in the spleen, cecum, and MLN and the kinetics of the CD4+ Gata3+ T cell response in the MLN and cecum. Fig. S2 highlights the cytokine response and phenotype of sorted splenic basophils in the presence or absence of Notch inhibition by GSI in vitro; the ability of exogenous Cre to induce DNMAML-GFP in Mcpt8Cre− DNMAML+ cells in vitro; and a heatmap to show basophil-intrinsic, Notch-dependent differentially expressed genes associated with lipid synthesis in basophils from IL-3c–treated naive versus T. muris–infected mice. Fig. S3 provides characterization of basophils in naive and T. muris–infected Mcpt8Cre− DNMAML+ and Mcpt8Cre+ DNMAML+ mice; chimerism levels in basophils from 50:50 Mcpt8Cre− DNMAML+:Mcpt8Cre+ DNMAML+ bone marrow chimeric mice; IL-4 characterization in the cecum by flow cytometry; Il4 expression in colon homogenates; and MLN Gata3+ CD4+ T cell percentages in T. muris–infected Mcpt8Cre− DNMAML+ and Mcpt8Cre+ DNMAML+ mice. Table S1 includes gene information for all differentially expressed genes (with q value <0.1, β value >|1.5|) between Mcpt8Cre− DNMAML+ and Mcpt8Cre+ DNMAML+ basophils from mice on day 14 of T. muris infection following IL-3c treatment.
Supplementary Material
Acknowledgments
We thank members of the Tait Wojno and Rudd laboratories for their feedback during manuscript development; Adam Wojno, Carol Bayles, Rebecca Williams, and Chris Donohue and the Cornell University Biomedical Sciences Flow Cytometry Core (which acknowledges support from the New York State Stem Cell Science, National Institutes of Health, and Cornell University College of Veterinary Medicine) and the Biotechnology Resource Center Imaging Core for cell sorting and assistance with flow cytometry; Christine Butler and the Cornell University College of Veterinary Medicine RNA Sequencing Core for assistance with RNA isolation and RNaseq services; Lynn Johnson from the Cornell Statistical Consulting Unit for advice on statistical analysis; Roy Cohen for assistance with confocal microscopy; Will Bailis for advice on the design of the Tat-Cre assay; and Warren Pear and David Artis for provision of the DNMAML and Mcpt8Cre mice.
This work was supported by the National Institutes of Health National Institute of Allergy and Infectious Diseases (K22 AI116729 and R01 AI132708 to E.D. Tait Wojno), National Human Genome Research Institute (R01 HG009309 to C.G. Danko), National Institute of Neurological Disorders and Stroke (R01 NS091067 to T.H. Harris), and Cornell University start-up funds (to E.D. Tait Wojno). The content of this manuscript is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The authors declare no competing financial interests.
Author contributions: L.M. Webb, O.O. Oyesola, S.P. Früh, and E.D. Tait Wojno designed the project, carried out experimental work, analyzed data, and wrote the manuscript, with valuable input from all authors. E. Kamynina and K.M. Still carried out experimental work and analyzed data. S.A. Peng and R.L. Cubitt carried out experimental work. J.K. Grenier carried out experimental work and analyzed RNaseq data. R.K. Patel, A. Grimson, and C.G. Danko assisted in RNaseq analysis. T.H. Harris assisted in microscopy. E.D. Tait Wojno conceived the project and supervised the research.
References
- Agace W.W. 2008. T-cell recruitment to the intestinal mucosa. Trends Immunol. 29:514–522. 10.1016/j.it.2008.08.003 [DOI] [PubMed] [Google Scholar]
- Allen J.E., and Maizels R.M.. 2011. Diversity and dialogue in immunity to helminths. Nat. Rev. Immunol. 11:375–388. 10.1038/nri2992 [DOI] [PubMed] [Google Scholar]
- Allen J.E., and Sutherland T.E.. 2014. Host protective roles of type 2 immunity: parasite killing and tissue repair, flip sides of the same coin. Semin. Immunol. 26:329–340. 10.1016/j.smim.2014.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artis D., and Grencis R.K.. 2008. The intestinal epithelium: sensors to effectors in nematode infection. Mucosal Immunol. 1:252–264. 10.1038/mi.2008.21 [DOI] [PubMed] [Google Scholar]
- Bailis W., Yashiro-Ohtani Y., Fang T.C., Hatton R.D., Weaver C.T., Artis D., and Pear W.S.. 2013. Notch simultaneously orchestrates multiple helper T cell programs independently of cytokine signals. Immunity. 39:148–159. 10.1016/j.immuni.2013.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethony J., Brooker S., Albonico M., Geiger S.M., Loukas A., Diemert D., and Hotez P.J.. 2006. Soil-transmitted helminth infections: ascariasis, trichuriasis, and hookworm. Lancet. 367:1521–1532. 10.1016/S0140-6736(06)68653-4 [DOI] [PubMed] [Google Scholar]
- Bray N.L., Pimentel H., Melsted P., and Pachter L.. 2016. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34:525–527. 10.1038/nbt.3519 [DOI] [PubMed] [Google Scholar]
- Butler J.M., Nolan D.J., Vertes E.L., Varnum-Finney B., Kobayashi H., Hooper A.T., Seandel M., Shido K., White I.A., Kobayashi M., et al. 2010. Endothelial cells are essential for the self-renewal and repopulation of Notch-dependent hematopoietic stem cells. Cell Stem Cell. 6:251–264. 10.1016/j.stem.2010.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassard L., Jönsson F., Arnaud S., and Daëron M.. 2012. Fcγ receptors inhibit mouse and human basophil activation. J. Immunol. 189:2995–3006. 10.4049/jimmunol.1200968 [DOI] [PubMed] [Google Scholar]
- Chung J., Ebens C.L., Perkey E., Radojcic V., Koch U., Scarpellino L., Tong A., Allen F., Wood S., Feng J., et al. 2017. Fibroblastic niches prime T cell alloimmunity through Delta-like Notch ligands. J. Clin. Invest. 127:1574–1588. 10.1172/JCI89535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Elia R., Behnke J.M., Bradley J.E., and Else K.J.. 2009. Regulatory T cells: a role in the control of helminth-driven intestinal pathology and worm survival. J. Immunol. 182:2340–2348. 10.4049/jimmunol.0802767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demitrack E.S., and Samuelson L.C.. 2016. Notch regulation of gastrointestinal stem cells. J. Physiol. 594:4791–4803. 10.1113/JP271667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberle J.U., and Voehringer D.. 2016. Role of basophils in protective immunity to parasitic infections. Semin. Immunopathol. 38:605–613. 10.1007/s00281-016-0563-3 [DOI] [PubMed] [Google Scholar]
- Fang T.C., Yashiro-Ohtani Y., Del Bianco C., Knoblock D.M., Blacklow S.C., and Pear W.S.. 2007. Notch directly regulates Gata3 expression during T helper 2 cell differentiation. Immunity. 27:100–110. 10.1016/j.immuni.2007.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasnacht N., Huang H.Y., Koch U., Favre S., Auderset F., Chai Q., Onder L., Kallert S., Pinschewer D.D., MacDonald H.R., et al. 2014. Specific fibroblastic niches in secondary lymphoid organs orchestrate distinct Notch-regulated immune responses. J. Exp. Med. 211:2265–2279. 10.1084/jem.20132528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelman F.D., Shea-Donohue T., Goldhill J., Sullivan C.A., Morris S.C., Madden K.B., Gause W.C., and Urban J.F. Jr. 1997. Cytokine regulation of host defense against parasitic gastrointestinal nematodes: lessons from studies with rodent models. Annu. Rev. Immunol. 15:505–533. 10.1146/annurev.immunol.15.1.505 [DOI] [PubMed] [Google Scholar]
- Gamrekelashvili J., Giagnorio R., Jussofie J., Soehnlein O., Duchene J., Briseño C.G., Ramasamy S.K., Krishnasamy K., Limbourg A., Häger C., et al. 2016. Regulation of monocyte cell fate by blood vessels mediated by Notch signalling. Nat. Commun. 7:12597 10.1038/ncomms12597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gause W.C., Wynn T.A., and Allen J.E.. 2013. Type 2 immunity and wound healing: evolutionary refinement of adaptive immunity by helminths. Nat. Rev. Immunol. 13:607–614. 10.1038/nri3476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbe F., Sidot E., Smyth D.J., Ohmoto M., Matsumoto I., Dardalhon V., Cesses P., Garnier L., Pouzolles M., Brulin B., et al. 2016. Intestinal epithelial tuft cells initiate type 2 mucosal immunity to helminth parasites. Nature. 529:226–230. 10.1038/nature16527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gessner A., Mohrs K., and Mohrs M.. 2005. Mast cells, basophils, and eosinophils acquire constitutive IL-4 and IL-13 transcripts during lineage differentiation that are sufficient for rapid cytokine production. J. Immunol. 174:1063–1072. 10.4049/jimmunol.174.2.1063 [DOI] [PubMed] [Google Scholar]
- Harhay M.O., Horton J., and Olliaro P.L.. 2010. Epidemiology and control of human gastrointestinal parasites in children. Expert Rev. Anti Infect. Ther. 8:219–234. 10.1586/eri.09.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris N.L., and Loke P.. 2017. Recent Advances in Type-2-Cell-Mediated Immunity: Insights from Helminth Infection. Immunity. 47:1024–1036. 10.1016/j.immuni.2017.11.015 [DOI] [PubMed] [Google Scholar]
- Herbert D.R., Yang J.-Q., Hogan S.P., Groschwitz K., Khodoun M., Munitz A., Orekov T., Perkins C., Wang Q., Brombacher F., et al. 2009. Intestinal epithelial cell secretion of RELM-beta protects against gastrointestinal worm infection. J. Exp. Med. 206:2947–2957. 10.1084/jem.20091268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai H., Tanaka K., Yoshie O., Ogawa K., Kenmotsu K., Takamori Y., Ichimasa M., Sugamura K., Nakamura M., Takano S., and Nagata K.. 2001. Prostaglandin D2 selectively induces chemotaxis in T helper type 2 cells, eosinophils, and basophils via seven-transmembrane receptor CRTH2. J. Exp. Med. 193:255–261. 10.1084/jem.193.2.255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S., Da Y., Han S., He Y., and Che H.. 2018. Notch ligand Delta-like1 enhances degranulation and cytokine production through a novel Notch/Dok-1/MAPKs pathway in vitro. Immunol. Res. 66:87–96. 10.1007/s12026-017-8977-0 [DOI] [PubMed] [Google Scholar]
- Klementowicz J.E., Travis M.A., and Grencis R.K.. 2012. Trichuris muris: a model of gastrointestinal parasite infection. Semin. Immunopathol. 34:815–828. 10.1007/s00281-012-0348-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopan R., and Ilagan M.X.G.. 2009. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 137:216–233. 10.1016/j.cell.2009.03.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillard I., Tu L., Sambandam A., Yashiro-Ohtani Y., Millholland J., Keeshan K., Shestova O., Xu L., Bhandoola A., and Pear W.S.. 2006. The requirement for Notch signaling at the β-selection checkpoint in vivo is absolute and independent of the pre-T cell receptor. J. Exp. Med. 203:2239–2245. 10.1084/jem.20061020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min B., Prout M., Hu-Li J., Zhu J., Jankovic D., Morgan E.S., Urban J.F. Jr., Dvorak A.M., Finkelman F.D., LeGros G., and Paul W.E.. 2004. Basophils produce IL-4 and accumulate in tissues after infection with a Th2-inducing parasite. J. Exp. Med. 200:507–517. 10.1084/jem.20040590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohrs K., Wakil A.E., Killeen N., Locksley R.M., and Mohrs M.. 2005. A two-step process for cytokine production revealed by IL-4 dual-reporter mice. Immunity. 23:419–429. 10.1016/j.immuni.2005.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi A., Morita S., Iwashita H., Sagiya Y., Ashida Y., Shirafuji H., Fujisawa Y., Nishimura O., and Fujino M.. 2001. Role of gob-5 in mucus overproduction and airway hyperresponsiveness in asthma. Proc. Natl. Acad. Sci. USA. 98:5175–5180. 10.1073/pnas.081510898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano N., Nishiyama C., Yagita H., Hara M., Motomura Y., Kubo M., Okumura K., and Ogawa H.. 2015. Notch signaling enhances FcεRI-mediated cytokine production by mast cells through direct and indirect mechanisms. J. Immunol. 194:4535–4544. 10.4049/jimmunol.1301850 [DOI] [PubMed] [Google Scholar]
- Ohmori K., Luo Y., Jia Y., Nishida J., Wang Z., Bunting K.D., Wang D., and Huang H.. 2009. IL-3 induces basophil expansion in vivo by directing granulocyte-monocyte progenitors to differentiate into basophil lineage-restricted progenitors in the bone marrow and by increasing the number of basophil/mast cell progenitors in the spleen. J. Immunol. 182:2835–2841. 10.4049/jimmunol.0802870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecaric-Petkovic T., Didichenko S.A., Kaempfer S., Spiegl N., and Dahinden C.A.. 2009. Human basophils and eosinophils are the direct target leukocytes of the novel IL-1 family member IL-33. Blood. 113:1526–1534. 10.1182/blood-2008-05-157818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrigoue J.G., Saenz S.A., Siracusa M.C., Allenspach E.J., Taylor B.C., Giacomin P.R., Nair M.G., Du Y., Zaph C., van Rooijen N., et al. 2009. MHC class II-dependent basophil-CD4+ T cell interactions promote T(H)2 cytokine-dependent immunity. Nat. Immunol. 10:697–705. 10.1038/ni.1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips C., Coward W.R., Pritchard D.I., and Hewitt C.R.A.. 2003. Basophils express a type 2 cytokine profile on exposure to proteases from helminths and house dust mites. J. Leukoc. Biol. 73:165–171. 10.1189/jlb.0702356 [DOI] [PubMed] [Google Scholar]
- Pimentel H., Bray N.L., Puente S., Melsted P., and Pachter L.. 2017. Differential analysis of RNA-seq incorporating quantification uncertainty. Nat. Methods. 14:687–690. 10.1038/nmeth.4324 [DOI] [PubMed] [Google Scholar]
- Pulendran B., and Artis D.. 2012. New paradigms in type 2 immunity. Science. 337:431–435. 10.1126/science.1221064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu S.-Y., He Y.-L., Zhang J., and Wu C.-G.. 2017b Transcription factor RBP-J-mediated signalling regulates basophil immunoregulatory function in mouse asthma model. Immunology. 152:115–124. 10.1111/imm.12753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu S.-Y., Lin J.-J., Zhang J., Song L.-Q., Yang X.-M., and Wu C.-G.. 2017a Notch signaling pathway regulates the growth and the expression of inflammatory cytokines in mouse basophils. Cell. Immunol. 318:29–34. 10.1016/j.cellimm.2017.05.005 [DOI] [PubMed] [Google Scholar]
- Radtke F., MacDonald H.R., and Tacchini-Cottier F.. 2013. Regulation of innate and adaptive immunity by Notch. Nat. Rev. Immunol. 13:427–437. 10.1038/nri3445 [DOI] [PubMed] [Google Scholar]
- Rivera A., Siracusa M.C., Yap G.S., and Gause W.C.. 2016. Innate cell communication kick-starts pathogen-specific immunity. Nat. Immunol. 17:356–363. 10.1038/ni.3375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Nieves J., Olson T., Bamias G., Bruce A., Solga M., Knight R.F., Hoang S., Cominelli F., and Ley K.. 2005. L-selectin, α 4 β 1, and α 4 β 7 integrins participate in CD4+ T cell recruitment to chronically inflamed small intestine. J. Immunol. 174:2343–2352. 10.4049/jimmunol.174.4.2343 [DOI] [PubMed] [Google Scholar]
- Sakata-Yanagimoto M., Nakagami-Yamaguchi E., Saito T., Kumano K., Yasutomo K., Ogawa S., Kurokawa M., and Chiba S.. 2008. Coordinated regulation of transcription factors through Notch2 is an important mediator of mast cell fate. Proc. Natl. Acad. Sci. USA. 105:7839–7844. 10.1073/pnas.0801074105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz C., Turqueti-Neves A., Hartmann S., Yu P., Nimmerjahn F., and Voehringer D.. 2014. Basophil-mediated protection against gastrointestinal helminths requires IgE-induced cytokine secretion. Proc. Natl. Acad. Sci. USA. 111:E5169–E5177. 10.1073/pnas.1412663111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz C., Eberle J.U., and Voehringer D.. 2016. Basophils in inflammation. Eur. J. Pharmacol. 778:90–95. 10.1016/j.ejphar.2015.04.049 [DOI] [PubMed] [Google Scholar]
- Shen T., Kim S., Do J.-S., Wang L., Lantz C., Urban J.F., Le Gros G., and Min B.. 2008. T cell-derived IL-3 plays key role in parasite infection-induced basophil production but is dispensable for in vivo basophil survival. Int. Immunol. 20:1201–1209. 10.1093/intimm/dxn077 [DOI] [PubMed] [Google Scholar]
- Siracusa M.C., Saenz S.A., Hill D.A., Kim B.S., Headley M.B., Doering T.A., Wherry E.J., Jessup H.K., Siegel L.A., Kambayashi T., et al. 2011. TSLP promotes interleukin-3-independent basophil haematopoiesis and type 2 inflammation. Nature. 477:229–233. 10.1038/nature10329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan B.M., Liang H.-E., Bando J.K., Wu D., Cheng L.E., McKerrow J.K., Allen C.D.C., and Locksley R.M.. 2011. Genetic analysis of basophil function in vivo. Nat. Immunol. 12:527–535. 10.1038/ni.2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tait Wojno E.D., Monticelli L.A., Tran S.V., Alenghat T., Osborne L.C., Thome J.J., Willis C., Budelsky A., Farber D.L., and Artis D.. 2015. The prostaglandin D2 receptor CRTH2 regulates accumulation of group 2 innate lymphoid cells in the inflamed lung. Mucosal Immunol. 8:1313–1323. 10.1038/mi.2015.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu L., Fang T.C., Artis D., Shestova O., Pross S.E., Maillard I., and Pear W.S.. 2005. Notch signaling is an important regulator of type 2 immunity. J. Exp. Med. 202:1037–1042. 10.1084/jem.20050923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyken S.J., Nussbaum J.C., Lee J., Molofsky A.B., Liang H.-E., Pollack J.L., Gate R.E., Haliburton G.E., Ye C.J., Marson A., et al. 2016. A tissue checkpoint regulates type 2 immunity. Nat. Immunol. 17:1381–1387. 10.1038/ni.3582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venturelli N., Lexmond W.S., Ohsaki A., Nurko S., Karasuyama H., Fiebiger E., and Oyoshi M.K.. 2016. Allergic skin sensitization promotes eosinophilic esophagitis through the IL-33-basophil axis in mice. J. Allergy Clin. Immunol. 138:1367–1380.e5. 10.1016/j.jaci.2016.02.034 [DOI] [PubMed] [Google Scholar]
- Voehringer D. 2017. Recent advances in understanding basophil functions in vivo. F1000 Res. 6:1464 10.12688/f1000research.11697.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Moltke J., Ji M., Liang H.E., and Locksley R.M.. 2016. Tuft-cell-derived IL-25 regulates an intestinal ILC2-epithelial response circuit. Nature. 529:221–225. 10.1038/nature16161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb L.M., and Tait Wojno E.D.. 2017. The role of rare innate immune cells in Type 2 immune activation against parasitic helminths. Parasitology. 144:1288–1301. 10.1017/S0031182017000488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weckmann M., Collison A., Simpson J.L., Kopp M.V., Wark P.A.B., Smyth M.J., Yagita H., Matthaei K.I., Hansbro N., Whitehead B., et al. 2007. Critical link between TRAIL and CCL20 for the activation of TH2 cells and the expression of allergic airway disease. Nat. Med. 13:1308–1315. 10.1038/nm1660 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.