Medina et al. demonstrate that Kit oncogene activity in gastrointestinal stromal tumor modulates the CD103+CD11b− dendritic cell (DC) lineage. The antitumor efficacy of oncogene inhibition initially depends on preexisting immunity orchestrated by CD103+CD11b− DCs, but subsequently is limited by a decrease in DC lineage maturation.
Abstract
Gastrointestinal stromal tumor (GIST) is driven by an activating mutation in the KIT proto-oncogene. Using a mouse model of GIST and human specimens, we show that intratumoral murine CD103+CD11b− dendritic cells (DCs) and human CD141+ DCs are associated with CD8+ T cell infiltration and differentiation. In mice, the antitumor effect of the Kit inhibitor imatinib is partially mediated by CD103+CD11b− DCs, and effector CD8+ T cells initially proliferate. However, in both mice and humans, chronic imatinib therapy decreases intratumoral DCs and effector CD8+ T cells. The mechanism in our mouse model depends on Kit inhibition, which reduces intratumoral GM-CSF, leading to the accumulation of Batf3-lineage DC progenitors. GM-CSF is produced by γδ T cells via macrophage IL-1β. Stimulants that expand and mature DCs during imatinib treatment improve antitumor immunity. Our findings identify the importance of tumor cell oncogene activity in modulating the Batf3-dependent DC lineage and reveal therapeutic limitations for combined checkpoint blockade and tyrosine kinase inhibition.
Introduction
Antitumor immune responses are driven in large part by the interplay between T cells and antigen-presenting cells. The Batf3-dependent dendritic cell (DC) lineage mediates the cross-priming of CD8+ T cells (Hildner et al., 2008). In mice, lymphoid resident Batf3-dependent DCs are characterized by expression of CD8α, whereas Batf3-dependent tissue–resident and migratory lymphoid DCs are instead CD103+CD11b− (Edelson et al., 2010). Homologous DCs in human tissue are characterized by expression of CD141 (Bachem et al., 2010; Jongbloed et al., 2010; Haniffa et al., 2012). Studies in Batf3−/− mice have identified a crucial role for CD103+CD11b− DCs in the cross-presentation of tumor-associated antigens and the antitumor response to checkpoint blockade (Salmon et al., 2016; Sánchez-Paulete et al., 2016).
Gastrointestinal stromal tumor (GIST) is the most common human sarcoma and is often driven by a single activating mutation in the KIT proto-oncogene (Hirota et al., 1998; Joensuu and DeMatteo, 2012). Imatinib mesylate is a small molecule inhibitor of KIT and improves median overall survival in metastatic disease from 9 mo to >5 yr (Demetri et al., 2002; Blanke et al., 2008). The antitumor effect of imatinib is partially mediated by CD8+ T cells through inhibition of the immunosuppressive enzyme indoleamine 2,3-dioxygenase (IDO), which is produced by tumor cells as a product of constitutive Kit signaling (Balachandran et al., 2011). Crucially, the checkpoint inhibitors anti–PD-1 and anti–PD-L1 require imatinib to demonstrate antitumor efficacy, establishing GIST as a model of combined targeted molecular and immunotherapy (Seifert et al., 2017). However, despite T cell activation in the context of both imatinib and checkpoint blockade, the antitumor benefit of adding immunotherapy is modest, suggesting the development of immune evasion.
In this study, we examined CD103+CD11b− and CD141+ DCs in murine and human GIST. We identified their role in tumor growth and defined their reciprocal interaction with imatinib. In mice, tumor cell oncogene activity modulates the Batf3-dependent DC lineage, resulting in divergent CD8+ T cell responses depending on the duration of imatinib treatment. In patients treated with tyrosine kinase inhibitors, the antitumor effect of checkpoint blockade may be limited in the absence of strategies to maintain Batf3-dependent DCs.
Results
CD103+CD11b− DCs are essential for CD8+ T cell immunosurveillance and partially mediate the antitumor effects of imatinib in GIST
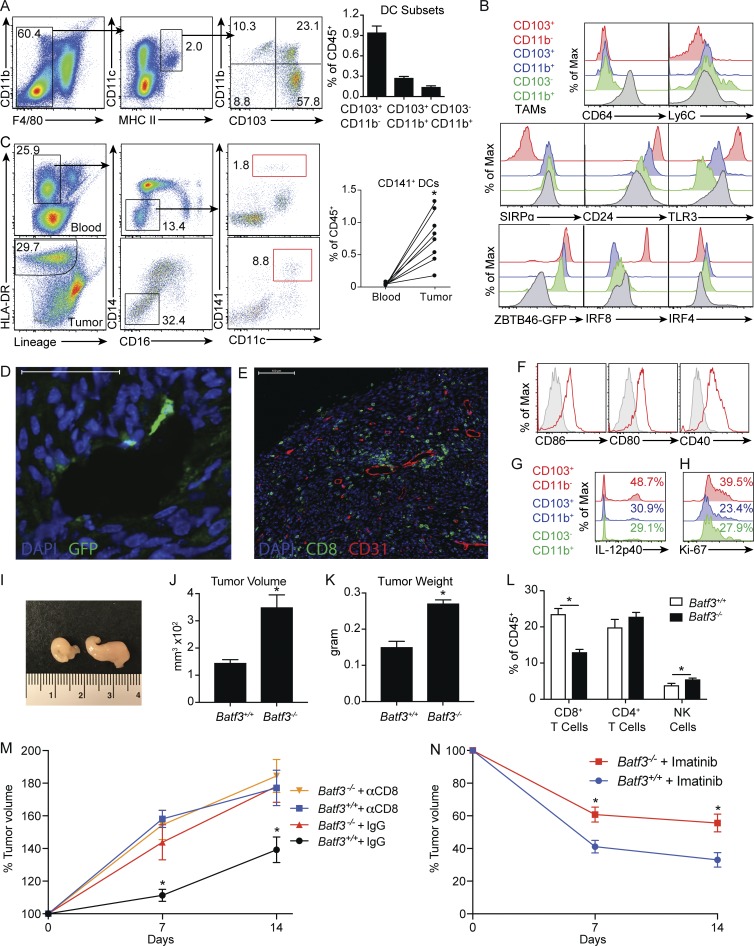
To characterize the role of CD103+CD11b− DCs in GIST, we used a murine model containing a single deletion in exon 11 of Kit, the most frequently affected site in human GIST. KitV558Δ/+ mice develop a single imatinib-sensitive GIST in the cecum with 100% penetrance, and untreated mice have a median lifespan of ∼6 mo secondary to progressive bowel obstruction (Sommer et al., 2003). Our previous work identified a large number of tumor-associated macrophages (TAMs) based on expression of F4/80 that harbored a uniquely inflammatory phenotype with near-uniform expression of CD11c and MHC II (Cavnar et al., 2013). Therefore, we defined DCs as F4/80−MHC II+CD11c+ to distinguish them from TAMs. Three DC populations infiltrated murine GIST based on differential expression of CD103 and CD11b (Fig. 1 A). CD103+CD11b− DCs were the most frequent DC subset. We characterized them further by examining other established DC and TAM markers. Only TAMs expressed CD64, whereas both TAMs and CD103−CD11b+ DCs expressed low levels of Ly6C, reflecting their monocytic origin (Fig. 1 B; Bogunovic et al., 2009). CD103+CD11b− DCs did not express SIRPα, unlike DCs and TAMs of monocytic origin. In contrast, CD103+CD11b− DCs expressed high levels of CD24 and Toll-like receptor 3 (TLR3). The transcription factor Zbtb46 is a marker of classic DCs and their progenitors (Satpathy et al., 2012). In tumors of KitV558Δ/+;Zbtb46GFP/+ mice, DCs expressed high levels of GFP, but TAMs did not. An alternate gating strategy based on CD45 and Zbtb46-GFP expression, instead of F4/80 and CD11b, demonstrated similar intratumoral DC subset composition (Fig. S1 A). In KitV558Δ/+ tumors, CD103+CD11b− DCs expressed high levels of the transcription factor Irf8 and lower levels of Irf4 compared with both CD11b+ DC subsets. In seven matched peripheral blood and tumor samples from surgically resected untreated human GIST specimens, CD141+ DCs were expanded in tumor relative to blood in all patients and, as in KitV558Δ/+ mice, comprised ∼1% of all immune cells (Fig. 1 C).
Figure 1.
CD103+CD11b− DCs are essential for CD8+ T cell immunosurveillance and partially mediate the antitumor effects of imatinib in GIST. (A) DC subsets in tumors of KitV558Δ/+ mice were identified by flow cytometry (four mice/group). (B) DCs and TAMs in KitV558Δ/+ mice and KitV558Δ/+;ZBTB46GFP/+ mice were analyzed for surface markers and transcription factors. A and B are representative of two independent experiments. (C) Matched peripheral blood and untreated human GIST specimens (seven specimens from four patients) were analyzed by flow cytometry for CD45+ lineage (CD3, CD19, CD56)−CD14−CD16−CD11c+CD141+ DCs. (D) Tumors of untreated KitV558Δ/+;ZBTB46GFP/+ mice were stained for GFP and examined by immunofluorescence. Bar represents 50 µm. (E) Tumors of KitV558Δ/+ mice were stained for CD8 and CD31 and examined by immunofluorescence. Bar represents 100 µm. D and E are representative of three mice. (F) CD103+CD11b− DCs in tumors of KitV558Δ/+ mice were analyzed for expression of CD86, CD80, and CD40. (G) Untreated KitV558Δ/+ mice were injected i.v. with brefeldin A to measure IL-12p40 production by DCs. (H) Untreated KitV558Δ/+ mice were stained for intracellular Ki67 in DCs. F–H are representative of three mice. (I) Gross appearance of tumors in KitV558Δ/+;Batf3+/+ (left) and KitV558Δ/+;Batf3−/− (right) mice. Representative of six mice. (J) Untreated 8-wk-old KitV558Δ/+;Batf3+/+ and KitV558Δ/+;Batf3−/− mice underwent MRI, and tumor volume was measured (six mice/group, representative of two independent experiments). (K) Tumors in untreated 8-wk-old KitV558Δ/+;Batf3+/+ and KitV558Δ/+;Batf3−/− mice were weighed (three mice/group, representative of two independent experiments). (L) Tumors of KitV558Δ/+;Batf3+/+ and KitV558Δ/+;Batf3−/− mice were analyzed for frequency of CD8+ T cells, CD4+ T cells, and natural killer (NK) cells (five mice/group, representative of two independent experiments). (M) KitV558Δ/+;Batf3+/+ and KitV558Δ/+;Batf3−/− mice were treated with anti-CD8 antibody or isotype control on days 0 and 1 and every 3–4 d thereafter. MRI was performed on days 0, 7, and 14. Graph represents tumor volume as a percent of control (three mice/group, each mouse recruited and analyzed independently after reaching 8 wk of age). (N) KitV558Δ/+;Batf3+/+ and KitV558Δ/+;Batf3−/− mice were treated with vehicle or imatinib, and MRI was performed on days 0, 7, and 14. Graph represents tumor volume as a percentage of control (five mice/group, each mouse recruited and analyzed independently after reaching 8 wk of age). Data represent mean ± SEM; P values were calculated using a Student’s t test; *, P < 0.05.
To spatially locate DCs in murine GIST tumors, we performed immunofluorescence staining of untreated tumors from KitV558Δ/+;Zbtb46GFP/+ mice. Rare GFP+ cells with cytoplasmic projections resembling DCs were located in the vicinity of vascular structures (Fig. 1 D). Similarly, immunofluorescence staining of CD8 and CD31 in untreated tumors of KitV558Δ/+ mice demonstrated heterogeneous distribution of CD8+ cells with clusters surrounding CD31+ vessels in the periphery (Fig. 1 E). CD103+CD11b− DCs expressed the costimulatory markers CD86, CD80, and CD40 (Fig. 1 F). CD141+ DCs in human GIST also had a mature phenotype with higher levels of CD86, CD80, and CD40 in the tumor relative to the blood (Fig. S1 B). Murine CD103+CD11b− DCs produced more IL-12p40, which is known to enhance CD8+ T cell proliferation and effector function, and expressed more Ki67 relative to other DC subsets, suggesting a high degree of activation (Fig. 1, G and H; and Fig. S1 C).
To delineate the role of CD103+CD11b− DCs in murine GIST, we generated KitV558Δ/+;Batf3−/− mice (Batf3−/− mice). CD103+CD11b− DCs were largely depleted in tumors of Batf3−/− mice (Fig. S1 D). At 8 wk of age, tumors in Batf3−/− mice were strikingly larger than tumors in KitV558Δ/+;Batf3+/+ mice (Batf3+/+ mice; Fig. 1, I–K). There was a 50% reduction in intratumoral CD8+ T cells in Batf3−/− mice (Fig. 1 L). To further evaluate the relationship between CD103+CD11b− DCs, CD8+ T cells, and tumor growth, we performed serial magnetic resonance imaging (MRI) of Batf3+/+ and Batf3−/− mice treated with a depleting anti-CD8 antibody or isotype control. Tumors in Batf3+/+ mice grew more slowly compared with Batf3−/− mice, but CD8+ T cell depletion abrogated this difference (Fig. 1 M). Meanwhile, CD8+ T cell depletion did not affect tumor growth in Batf3−/− mice. Therefore, CD103+CD11b− DCs were required for CD8+ T cell–mediated immunosurveillance in murine GIST tumors. We then performed serial MRI of Batf3+/+ and Batf3−/− mice treated with imatinib and found an attenuated response in Batf3−/− mice, indicating that the effect of imatinib in GIST is partially dependent on CD103+CD11b− DCs (Fig. 1 N).
Batf3 dependence distinguishes resident memory from antitumor effector CD8+ T cells
Following immunological challenge, antigen-specific effector CD8+ T cells develop from naive circulating precursors into effector T cells, which are characterized by down-regulation of CD62L, up-regulation of CD44, KLRG1, and the transcription factor Tbet, and secretion of the effector cytokines IFN-γ and TNF (Intlekofer et al., 2005; Sarkar et al., 2008). In the case of tumors and chronic viral infections, persistent antigen exposure results in up-regulation of inhibitory receptors such as PD-1, TIM3, and LAG3 (Pauken and Wherry, 2015). Indeed, PD-1 is a marker demonstrated to identify T cells with antitumor specificity (Gros et al., 2014). In contrast, T cells that occupy tissues without recirculating are characterized as tissue-resident memory cells, which have a CD44+CD62L− memory phenotype and variably express CD103, CD69, or both (Casey et al., 2012; Mackay et al., 2012).
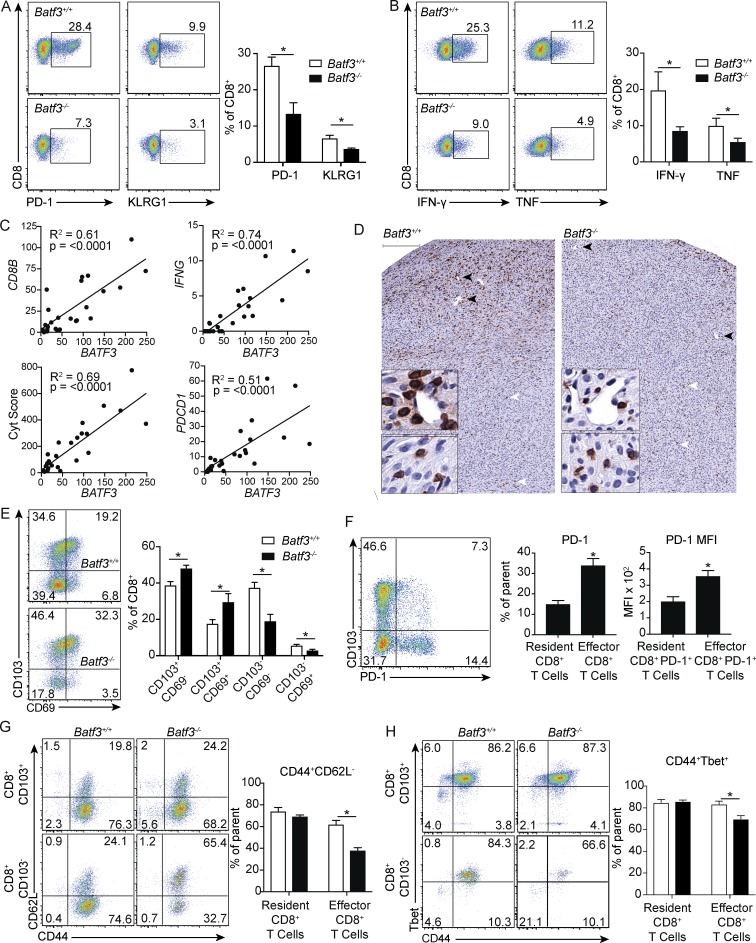
While CD103+CD11b− DCs were essential for CD8+ T cell–mediated immunosurveillance in GIST, a substantial portion of CD8+ T cells were still present in tumors of Batf3−/− mice, suggesting potential heterogeneity in the bulk CD8+ T cell population. We reasoned that dependence on CD103+CD11b− DCs could be used to further phenotype intratumoral CD8+ T cells. CD8+ T cells in Batf3−/− mice had no difference in the frequency of CD44+CD62L− or CD44+Tbet+ cells, but they had decreased expression of PD-1 and KLRG1 and lacked TIM3 and LAG3 (Fig. 2 A and data not shown). CD8+ T cells in Batf3−/− mice also produced less IFN-γ and TNF upon restimulation (Fig. 2 B). We then compared our mouse findings to RNA sequencing (RNaseq) expression data in 25 untreated human GIST specimens. BATF3 was variably expressed between patients but correlated strongly with CD8B, IFNG, PDCD1, and cytolytic score, suggesting that the presence of CD141+ DCs was associated with infiltration and differentiation of CD8+ T cells with an effector phenotype (Fig. 2 C). In contrast, BATF3 expression was not associated with tumor size, mitotic rate, or location (Fig. S2, A and B).
Figure 2.
Batf3 dependence distinguishes resident memory from antitumor effector CD8+ T cells. (A) Flow cytometry and frequency of PD-1 and KLRG1 on CD8+ T cells from tumors of untreated KitV558Δ/+;Batf3+/+ and KitV558Δ/+;Batf3−/− mice (six mice/group pooled from two experiments). (B) Tumor cell suspensions from KitV558Δ/+;Batf3+/+ and KitV558Δ/+;Batf3−/− mice were stimulated with PMA and ionomycin in the presence of brefeldin A followed by intracellular analysis of IFN-γ and TNF in CD8+ T cells (five mice/group, representative of two independent experiments). (C) Normalized gene expression of BATF3 was plotted against CD8B, IFNG, and the cytolytic score from 26 untreated human GIST specimens. (D) Untreated tumors in KitV558Δ/+;Batf3+/+ and KitV558Δ/+;Batf3−/− mice were stained for CD8. Top bar represents 200 µm. Black arrowheads point to staining in periphery and correspond to top inset. White arrowheads point to staining in central core and correspond to lower inset. Inset bar represents 50 µm (representative of three mice/group). (E) Flow cytometry and frequency of CD103 and CD69 on CD8+ T cells from untreated tumors of KitV558Δ/+;Batf3+/+ and KitV558Δ/+;Batf3−/− mice (six mice/group pooled from two experiments). (F) Flow cytometry and frequency of PD-1 on CD103+ resident and CD103− effector CD8+ T cells and PD-1 MFI of PD-1+ effector and PD-1+ resident CD8+ T cells from tumors of KitV558Δ/+;Batf3+/+ mice (five mice/group, representative of two independent experiments). (G) Flow cytometry and frequency of CD44 and CD62L on resident CD8+CD103+ and effector CD8+CD103− T cells from tumors of KitV558Δ/+;Batf3+/+ and KitV558Δ/+;Batf3−/− mice (five mice/group, representative of two independent experiments). (H) Flow cytometry and frequency of CD44 and Tbet on resident and effector CD8+ T cells from tumors of KitV558Δ/+;Batf3+/+ and KitV558Δ/+;Batf3−/− mice (four mice/group, pooled from two experiments). Data represent mean ± SEM; P values were calculated using a Student’s t test; *, P < 0.05. Correlation is shown using R2, and significance was determined using Spearman correlation.
We then performed immunohistochemistry to characterize the location of the relevant CD8+ cells. In Batf3+/+ mice, CD8+ cells were again heterogeneously distributed, with some cells clustering around vascular structures at the tumor periphery and others located within the tumor core (Fig. 2 D). Both peripheral and central CD8+ cells were round with prominent nuclei, but peripheral CD8+ cells were larger, consistent with an activated phenotype. Batf3−/− mice had fewer peripheral, perivascular CD8+ cells, while centrally located CD8+ cells appeared unchanged (Fig. S2 C). Together, these findings suggested that effector CD8+ T cells responsible for immunosurveillance were recruited by CD103+CD11b− DCs and clustered at the periphery.
Given that CD8+ cells in Batf3−/− mice were centrally located away from tumor vasculature, but still expressed high levels of CD44 and Tbet, we hypothesized that these might represent resident memory T cells. Consistent with this, we found an increased frequency of CD8+CD103+ T cells in Batf3−/− mice (Fig. 2 E). To confirm CD103 as a marker of tissue residence in GIST, we performed intravascular staining as previously described using peripheral blood as a positive control (Anderson et al., 2014). CD8+ T cells within tumor vasculature represented a small fraction of the overall population; however, these did not express CD103, whereas bulk tumor CD8+ T cells contained a distinct CD103+ population (Fig. S2 D). These data suggest that the antitumor effector CD8+ T cell subset in GIST does not express CD103 and is derived from the circulation. In contrast, resident CD8+ T cells express CD103 but are dispensable for immunosurveillance. Effector CD8+CD103− T cells were further distinguished by increased PD-1 expression in Batf3+/+ mice, and these PD-1+ cells had increased PD-1 mean fluorescence intensity (MFI) relative to the resident memory subset, consistent with persistent antigen exposure (Fig. 2 F; Paley et al., 2012). While there were no significant differences in bulk memory CD44+CD62L− cells between Batf3+/+and Batf3−/− mice, when we analyzed CD8+ T cells in tissue-resident CD103+ and antitumor effector CD103− populations, we found a decrease in the percentage of CD44+CD62L− cells only in the effector population (Fig. 2 G). Similarly, the percentage of CD44+Tbet+ cells was decreased only in the effector population (Fig. 2 H). Overall, despite being driven by a single oncogenic driver mutation, GIST contains CD8+ T cells with diverse specificity, not all of which are antitumoral. CD103+CD11b− DCs were responsible for both the recruitment and differentiation of effector CD8+CD103− T cells with antitumor activity.
Short- and long-term imatinib have divergent effects on effector CD8+ T cells
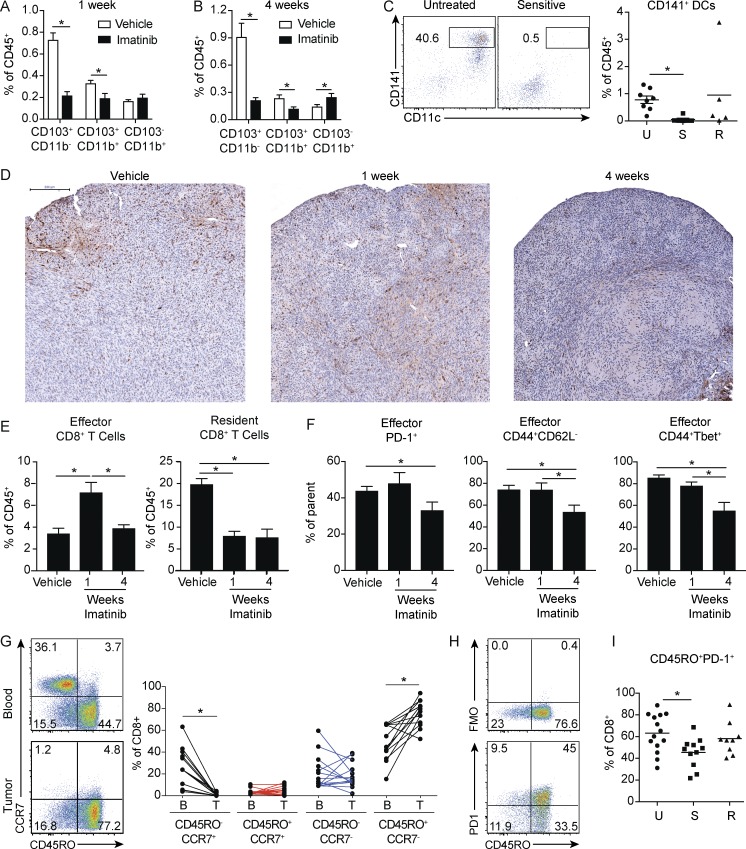
Given that the effects of imatinib partially depended on CD103+CD11b− DCs, we sought to examine changes in this population with treatment. Strikingly, both tumor CD103+ DC populations were dramatically decreased after just 1 wk, with the most pronounced effect on the CD103+CD11b− DCs (Fig. 3 A). This effect persisted at 4 wk of treatment (Fig. 3 B). Similarly, CD141+ DCs were nearly depleted in imatinib-sensitive human GIST tumors (Fig. 3 C).
Figure 3.
Short- and long-term imatinib have divergent effects on antitumor effector CD8+ T cells. (A) Frequency of DC subsets in tumors of KitV558Δ/+ mice treated with vehicle or imatinib for 1 wk (four mice/group). (B) Frequency of DC subsets in tumors of KitV558Δ/+ mice treated with vehicle or imatinib for 4 wk (four mice/group). A and B are representative of two independent experiments. (C) Human GIST specimens characterized by sensitivity to imatinib treatment (19 specimens from 14 patients) were analyzed for CD141+ DCs. Untreated tumors (U) were from patients who never received a tyrosine kinase inhibitor before surgery. Sensitive tumors (S) were responding to imatinib or another tyrosine kinase inhibitor at the time of surgery. Resistant tumors (R) were progressing despite treatment with imatinib or another tyrosine kinase inhibitor. (D) KitV558Δ/+ mice were treated with vehicle or imatinib for 1 or 4 wk and stained for CD8. Bar represents 200 µm. Representative of three mice/group. (E) Frequency of effector CD8+CD103− and resident CD8+CD103+ T cells from tumors of KitV558Δ/+ mice treated with vehicle or imatinib for 1 or 4 wk (four mice/group). (F) Frequency of PD-1+, CD44+CD62L−, and CD44+ Tbet+ effector CD8+CD103− T cells from tumors of KitV558Δ/+ mice treated with vehicle or imatinib for 1 or 4 wk (four mice/group). E and F represent two pooled experiments. (G) Matched peripheral blood and untreated human GIST specimens (14 tumor specimens from 10 patients) were analyzed for frequency of CD8+ T cell memory subsets. (H) Representative flow cytometry of CD45RO and fluorescence minus one (FMO) or PD-1 in CD8+ T cells from untreated human GIST specimens. (I) 38 human GIST specimens from 28 patients were characterized by sensitivity to imatinib treatment and analyzed for CD8+CD45RO+PD1+ T cells. Data represent mean ± SEM; P values were calculated using a Student’s t test; *, P < 0.05.
In light of the above findings, we sought to evaluate T cell dynamics after short and long treatment. At 1 wk of imatinib treatment, CD8+ cells were decreased in the perivascular regions at the tumor margin, while infiltration in the tumor core was increased (Figs. 3 D and S2 E). At 4 wk of treatment, there was substantial loss of CD8+ cells in both regions. However, by flow cytometry, effector CD8+CD103− T cells were increased at 1 wk while resident CD8+CD103+ T cells were decreased, suggesting that the centrally located CD8+ cells were no longer dominated by the resident subtype at this time point (Fig. 3 E). In contrast, both effector and resident subsets decreased between 1 and 4 wk. Moreover, the frequency of PD-1+, CD44+CD62L−, and CD44+Tbet+ effector cells was also decreased after 4 wk, but not 1 wk, of treatment, indicating decreased differentiation and antigen stimulation at this time point (Fig. 3 F). We then sought to correlate our findings to human GIST. In untreated specimens, effector memory CD8+CD45RO+CCR7− T cells were expanded in tumor compared with matched peripheral blood specimens (Fig. 3 G). Moreover, the CD45RO+ population was enriched for PD-1 expression, suggesting this was the tumor-specific phenotype (Fig. 3 H). Similar to our findings in mice, CD45RO+PD-1+ cells were decreased in imatinib-sensitive, but not imatinib-resistant, human GIST specimens (Fig. 3 I). Overall, these results indicate that during the course of imatinib treatment, early mobilization of a preexisting immune response depended on CD103+CD11b− DCs. At the same time, imatinib decreased the frequency of CD103+CD11b− DCs. Consequently, the infiltration and differentiation of effector CD8+ T cells at later time points was decreased, likely leading to diminished antitumor immunity.
CD103+CD11b− DC progenitors accumulate after Kit oncogene inhibition
We investigated whether the effect of imatinib on CD103+CD11b− DCs was tumor specific or affected the DC lineage globally. DC development in mice begins in the bone marrow and proceeds in a stepwise fashion from a distinct hematopoietic lineage. The macrophage and DC precursor (MDP) is the earliest myeloid-committed precursor in bone marrow (Fogg et al., 2006). MDPs give rise to monocytes and the common DC precursor (CDP), which itself gives rise to plasmacytoid DCs and preDCs (Naik et al., 2007; Onai et al., 2007). PreDCs then circulate to peripheral tissues and sequentially up-regulate MHC II, CD103 or CD11b, and costimulation markers to give rise to mature tissue-resident DCs (Ginhoux et al., 2009; Liu et al., 2009).
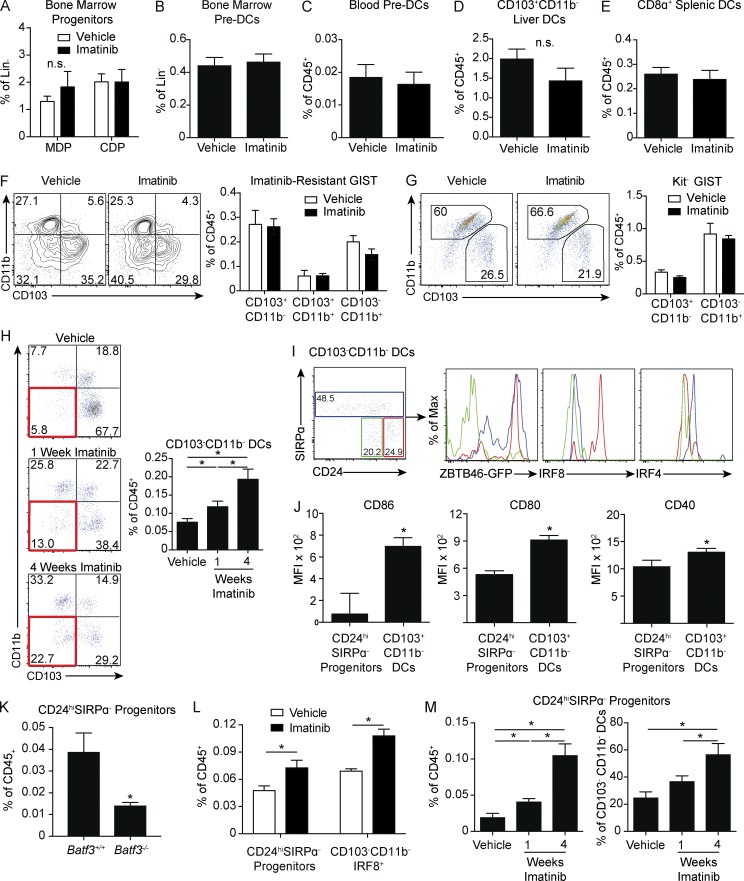
MDPs and CDPs are defined in part by expression of Kit, but we found no difference in the frequencies of either subset in imatinib-treated mice (Figs. 4 A and S3 A). There were also no differences in bone marrow or circulating preDCs, resident CD103+CD11b− DCs in the liver, or the developmentally related CD8α+ DCs in the spleen (Fig. 4, B–E; and Fig. S3 B). To account for the fact that constitutive Kit signaling might alter bone marrow DC development in KitV558Δ/+ mice, we also treated non–tumor-bearing wild-type mice with imatinib but found no difference in MDP or CDP frequency (Fig. S3 C). Similarly, cultures of wild-type bone marrow in the presence of FMS-like tyrosine kinase 3 ligand (Flt3L) and GM-CSF demonstrated no significant difference in the yield of CD103+ DCs at either low or high concentrations of imatinib (Fig. S3 D). Altogether, these data indicate that the inhibition of CD103+CD11b− DCs in GIST is strictly a tumor-related phenomenon.
Figure 4.
Kit oncogene inhibition leads to the accumulation of CD103+CD11b− DC progenitors. (A–E) KitV558Δ/+ mice were treated with vehicle or imatinib for 1 wk and analyzed for (A) frequency of Lin−CSF1R+CD117+MHC II−CD11c−CD135+ MDPs and lin−CSF1R+CD117intMHC II−CD11c−CD135+ CDPs in the bone marrow (five mice/group); (B) Lin−CD11c+MHC II−CD135+SIRPαint preDCs in the bone marrow (four mice/group); (C) Lin−CD11c+MHC II−CD135+SIRPαint preDCs in the peripheral blood (four mice/group); (D) Siglec F−F4/80−CD11c+MHC II+CD103+CD11b− DCs in the liver (five mice/group); and (E) CD11c+MHC II+CD8α+ DCs in the spleen (five mice/group). A–E are representative of two independent experiments. (F) Flow cytometry and frequency of DC subsets in tumors were analyzed in KitV558Δ;T669I/+ mice treated with vehicle or imatinib for 1 wk (six mice/group pooled from two independent experiments). (G) S2 cells were injected into the flanks of wild-type mice. After 3 wk of growth, imatinib was administered for 1 wk. Flow cytometry and frequency of DC subsets were analyzed (five mice/group, representative of two independent experiments). (H) Tumors of KitV558Δ/+ mice treated with vehicle or imatinib for 1 or 4 wk were analyzed by flow cytometry for DC populations. Red box identifies CD103−CD11b− DCs. Graph represents frequency of CD103−CD11b− DCs as a percentage of CD45+ cells (four to five mice/group, representative of two independent experiments). (I) Flow cytometry of CD103−CD11b− DCs distinguished by CD24 and SIRPα expression in tumors of untreated KitV558−/+ mice. Histograms represent transcription factor expression within these subsets. Histogram color matches gating within the CD103−CD11b− DC compartment (five mice/group). (J) CD86, CD80, and CD40 MFI in CD24hiSIRPα− progenitors and CD103+CD11b− DCs in tumors of untreated KitV558Δ/+ mice (five mice/group). I and J are representative of two independent experiments. (K) Untreated tumors in KitV558Δ/+;Batf3+/+ and KitV558Δ/+;Batf3−/− mice were analyzed for frequency of CD24hiSIRPα− progenitors (three mice/group, representative of two independent experiments). (L) Frequency of CD24hiSIRPα− progenitors and IRF8+ progenitors in tumors of KitV558Δ/+ mice treated with vehicle or imatinib for 1 wk (four mice/group, representative of two independent experiments). (M) Frequency of CD24hiSIRPα− progenitors as a percentage of CD45+ or CD103−CD11b− DCs in tumors of KitV558Δ/+ mice treated with vehicle or imatinib for 1 or 4 wk (four to five mice/group, similar results obtained in experiments separately analyzing 1- and 4-wk treatment durations). Data represent mean ± SEM; P values were calculated using a Student’s t test; *, P < 0.05. n.s., not significant.
To determine whether the decrease in CD103+CD11b− DCs was due to inhibition of Kit or an off-target effect, we used a model of imatinib-resistant GIST. KitV558Δ/+;T669I/+ mice contain an additional Kit mutation in exon 14, which inhibits imatinib binding (Bosbach et al., 2012). We found no changes in any of the DC populations after 1 wk of imatinib (Fig. 4 F). To determine if the decrease in CD103+CD11b− DCs in KitV558Δ/+ mice required inhibition of Kit on tumor cells specifically, we used the S2 GIST cell line that was derived from a KitV558Δ/+ mouse tumor, lost Kit expression in vitro, and is therefore resistant to imatinib. There was no difference in the frequency of CD103+CD11b− DCs in either vehicle- or imatinib-treated mice, indicating that the effect on DCs in KitV558Δ/+ mice required inhibition of Kit specifically on tumor cells (Fig. 4 G).
In probing the origin of our findings in KitV558Δ/+ mice, we found a progressive increase in a CD103−CD11b− subset within the F4/80−CD11c+MHC II+ tumor compartment at 1 and 4 wk of treatment with imatinib (Fig. 4 H). We sought to further investigate this compartment based on alternative myeloid and DC markers (Fig. 1 B). Within the CD103−CD11b−compartment, there were three populations based on differential expression of CD24 and SIRPα (Fig. 4 I). CD103−CD11b−CD24hiSIRPα− cells expressed high levels of ZBTB46-GFP, indicating that these were DCs. CD24hiSIRPα− cells also expressed high levels of IRF8 and lower levels of IRF4, suggesting that these were a CD103+CD11b− DC progenitor subset. Furthermore, expression of CD86, CD80, and CD40 was decreased in CD24hiSIRPα− cells compared with CD103+CD11b− DCs, consistent with immaturity (Fig. 4 J). CD24hiSIRPα− cells were also reduced in Batf3−/− mice to a similar extent as CD103+CD11b− DCs, confirming that these cells were progenitors in the Batf3-dependent DC lineage (Fig. 4 K). After 1 wk of imatinib treatment, we found an increase in CD24hiSIRPα− progenitors as well as an increase in the overall percentage CD103−CD11b−IRF8+ cells (Fig. 4 L). Longer treatment resulted in progressive accumulation of CD24hiSIRPα− progenitors, which were the dominant CD103−CD11b− population after 4 wk of imatinib (Fig. 4 M). Overall, these results indicate that Kit oncogene inhibition reduces CD103+CD11b− DCs and increases their CD24hiSIRPα− progenitors.
GM-CSF is required for CD103+CD11b− DCs and is decreased by imatinib
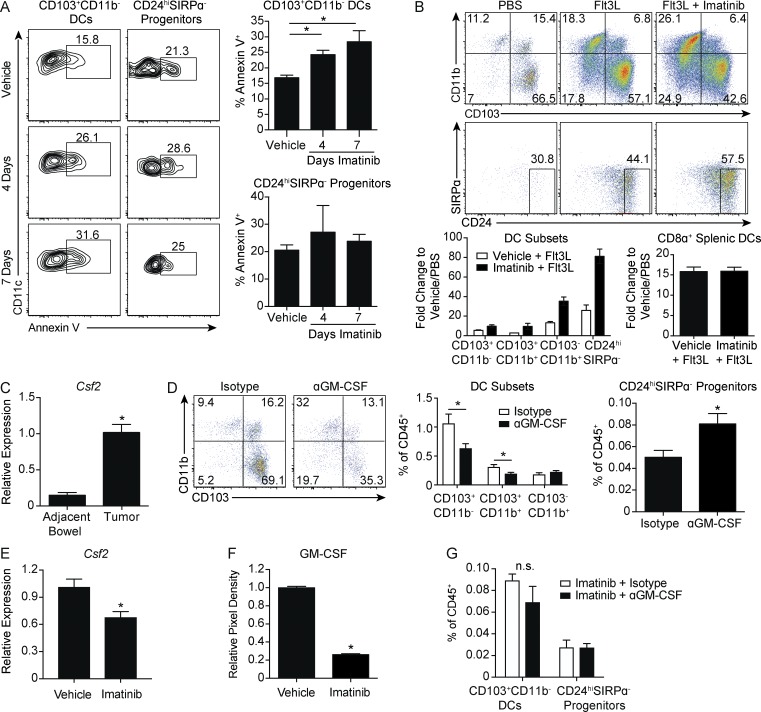
The selective intratumoral accumulation of CD24hiSIRPα− progenitors suggested that local factors responsible for CD103+CD11b− DC maintenance might be decreased during imatinib treatment. Consistent with this, apoptosis measured by Annexin V staining progressively increased in the CD103+CD11b−DCs, but not their CD24hiSIRPα− progenitors, at 4 and 7 d of imatinib (Fig. 5 A). In vivo expansion of DCs with the growth factor Flt3L, which promotes hematopoietic progenitor commitment to the DC lineage (Maraskovsky et al., 1996), led to far greater expansion of CD24hiSIRPα− progenitors in the presence of imatinib, compared with vehicle-treated mice (Fig. 5 B). In contrast, CD8α+ splenic DCs expanded similarly regardless of exposure to imatinib.
Figure 5.
GM-CSF is required for CD103+CD11b− DCs and is decreased by imatinib. (A) Tumors of KitV558Δ/+ mice treated with vehicle or imatinib for 4 or 7 d were analyzed for apoptosis by Annexin V in CD103+CD11b− DCs or CD24hiSIRPα− progenitors (four mice/group, similar results obtained with a 4-wk treatment duration). (B) KitV558Δ/+ mice were treated with nine daily injections of PBS or Flt3L and vehicle or imatinib in the drinking water. Flow cytometry of DC subsets in tumors. Graph represents fold change of DC subsets in tumor and CD8α+ DCs in spleen with respect to PBS plus vehicle–treated mice (three mice/group, representative of two independent experiments). (C) Quantitative RT-PCR of Csf2 mRNA expression in tumor and adjacent bowel of untreated KitV558Δ/+ mice (two mice/group plated and analyzed in triplicate). (D) KitV558Δ/+ mice were treated with 1 mg of isotype or anti-GM-CSF antibody on days 0, 3, and 6. Mice were sacrificed on day 7. Flow cytometry and frequency of DC subsets (eight mice/group, pooled from two experiments). (E) Quantitative RT-PCR of Csf2 mRNA expression in tumors of KitV558Δ/+ mice treated with vehicle or imatinib for 1 wk (three mice/group, representative of two independent experiments). (F) Cytokine array against GM-CSF protein in tumors of KitV558Δ/+ mice treated with vehicle or imatinib for 1 wk. Graph represents relative pixel density (two mice/group, two biological replicates). (G) Mice treated as in D except that imatinib was present in the drinking water for both groups. Graph of frequency of CD24hiSIRPα− progenitors and CD103+CD11b− DCs (three mice/group). Data represent mean ± SEM; P values were calculated using a Student’s t test; *, P < 0.05. n.s., not significant.
GM-CSF (Csf2) is a hematopoietic growth factor that controls the differentiation of the myeloid lineage. GM-CSF is critical for the development of resident CD103+ DCs in the steady state in several tissues, including the bowel, and is additionally required for cross-priming of CD8+ T cells (King et al., 2010; Greter et al., 2012). We found that Csf2 expression was increased in the tumor relative to adjacent bowel (Fig. 5 C). 1 wk of antibody-mediated depletion of GM-CSF decreased CD103+CD11b− DCs and increased CD24hiSIRPα− progenitors, recapitulating our findings with imatinib treatment (Fig. 5 D). Csf2 mRNA expression and, to a greater extent, GM-CSF protein were decreased in tumors after 1 wk of imatinib treatment (Fig. 5, E and F). There were no changes in the frequency of CD103+CD11b− DCs or CD24hiSIRPα− progenitors when imatinib was added to isotype or anti–GM-CSF, suggesting overlapping mechanisms (Fig. 5 G).
TAMs promote maturation of CD103+CD11b− DCs through GM-CSF+ γδ T cells via IL-1β
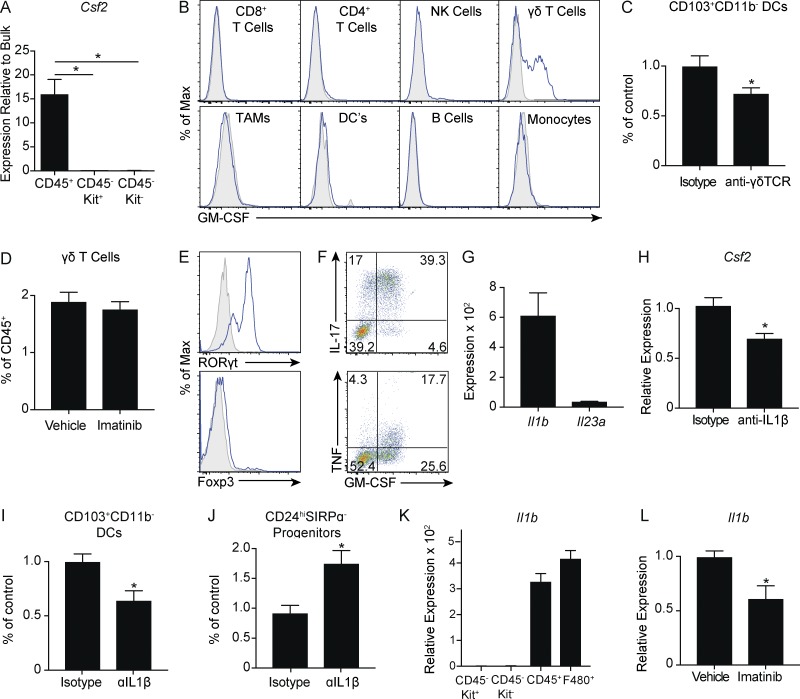
We next sought to determine the source of GM-CSF in GIST tumors. Expression of Csf2 was greatest in isolated CD45+ immune cells compared with CD45−Kit+ tumor cells and CD45− Kit− stromal cells (Fig. 6 A). Surprisingly, on restimulation of bulk tumor cell suspensions, γδ T cells were the only immune subset to produce GM-CSF (Fig. 6 B). Consistent with this, antibody-mediated depletion of γδ T cells resulted in fewer CD103+CD11b− DCs, indicating partial dependence on γδ T cells (Fig. 6 C). We did not observe a decrease in the frequency of γδ T cells after 1 wk of imatinib treatment despite a marked decrease in GM-CSF protein, suggesting that imatinib altered their function (Fig. 6 D). γδ T cells expressed high levels of the transcription factor RORγt and little to no Foxp3 (Fig. 6 E). Consistent with this inflammatory phenotype, GM-CSF+ γδ T cells secreted high levels of IL-17A and TNF (Fig. 6 F).
Figure 6.
Macrophages promote maturation of CD103+CD11b− DCs through GM-CSF+ γδ T cells via IL-1β. (A) CD45+ immune cells, CD45−Kit+ tumor cells, and CD45−Kit− stromal cells in untreated tumors of KitV558−/+ mice were isolated by magnetic beads. Graph represents quantitative RT-PCR of Csf2 mRNA expression relative to Kit+ tumor cells (three mice analyzed in duplicate). (B) Single-cell suspensions of tumors from KitV558Δ/+ mice were stimulated with PMA and ionomycin in the presence of brefeldin A and analyzed for intracellular GM-CSF in immune cells. (C) Tumors of KitV558Δ/+ mice were treated with 500 µg of isotype or anti-γδTCR antibody on day 0 and 250 µg every other day thereafter. Mice were sacrificed on day 14. Frequency of CD103+CD11b− DCs (seven mice/group pooled from two experiments). (D) Frequency of γδ T cells in tumors of KitV558Δ/+ mice treated with vehicle or imatinib for 1 wk (five mice/group, representative of two independent experiments). (E) γδ T cells in untreated KitV558Δ/+ tumors were analyzed for transcription factors by flow cytometry. (F) Single-cell suspensions of tumors from untreated KitV558Δ/+ mice were stimulated with PMA and ionomycin in the presence of brefeldin A and analyzed for intracellular cytokines in γδ T cells. (G) Intratumoral mRNA expression in untreated KitV558Δ/+ mice by Affymetrix array (three mice/group). (H) KitV558Δ/+ mice were treated every 3 d with 200 µg of isotype or anti–IL-1β and sacrificed on day 14. Quantitative RT-PCR of tumor Csf2 mRNA expression was performed (six to eight mice/group, pooled from two experiments). (I and J) Frequency of CD103+CD11b− DCs and CD24hiSIRPα− progenitors in mice treated as in H. (K) CD45+ immune cells, CD45−Kit+ tumor cells, CD45−Kit− stromal cells, and F4/80+ macrophages in untreated tumors of KitV558−/+ mice were isolated by magnetic beads. Quantitative RT-PCR of Il1b mRNA expression was performed relative to CD45−Kit+ tumor cells (four mice/group analyzed in triplicate). (L) Quantitative RT-PCR of Il1b mRNA expression in tumors of KitV558Δ/+ mice treated with vehicle or imatinib for 1 wk (three to four mice/group, representative of two independent experiments). Data represent mean ± SEM; P values were calculated using a Student’s t test; *, P < 0.05.
Secretion of GM-CSF and IL-17 and expression of RORγt can be regulated by IL-1β or IL-23p19 (Codarri et al., 2011; Lukens et al., 2012; Wu et al., 2016). We hypothesized that one or both of these might contribute to GM-CSF production. Using a previously published gene expression array, we found that Il1b was expressed in untreated tumors with comparatively minimal Il23a expression (Fig. 6 G; Balachandran et al., 2011). Moreover, antibody-mediated depletion of IL-1β decreased expression of Csf2 (Fig. 6 H). More importantly, depletion of IL-1β reduced the frequency of CD103+CD11b− DCs and increased CD24hiSIRPα− progenitors (Fig. 6, I and J), recapitulating our findings with GM-CSF depletion. In probing the source of IL-1β, we found that Il1b expression was greatest in isolated CD45+ immune cells compared with CD45−Kit+ tumor cells and CD45− Kit− stromal cells and that expression was enriched in F4/80+ TAMs (Fig. 6 K). Finally, imatinib treatment reduced expression of Il1b in bulk tumor by RT-PCR (Fig. 6 L). Altogether, our results link TAM activity via IL-1β to the maturation of CD103+CD11b− DCs through GM-CSF+ γδ T cells.
TAMs produce DC chemoattractants that are reduced by imatinib
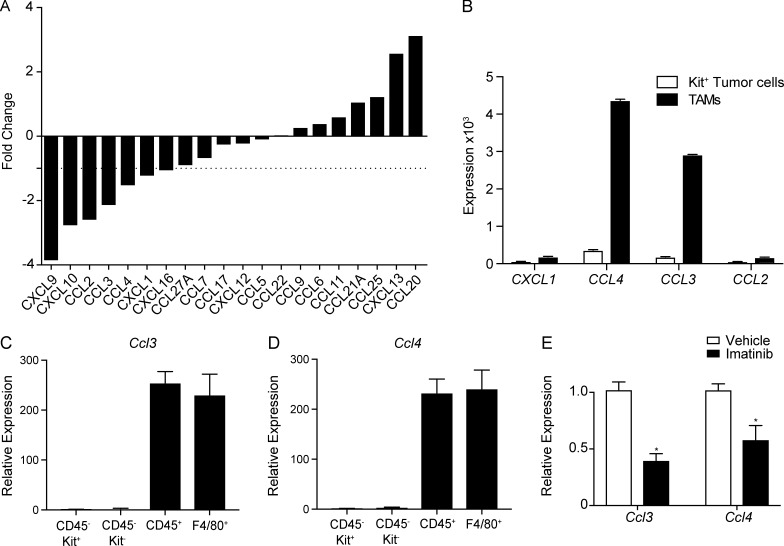
While depletion of either GM-CSF or IL-1β decreased CD103+CD11b− DCs and increased CD24hiSIRPα− progenitors, neither fully accounted for the magnitude of the effect observed with imatinib. We sought to examine expression of various chemokines in KitV558Δ/+ mice given their potential role in DC migration and localization. RNaseq was performed on bulk KitV558Δ/+ tumors and revealed a number of chemokines reduced by ≥50% in imatinib-treated specimens (Fig. 7 A). The most dramatically decreased were CXCL9 and CXCL10, which have been implicated in CD103+CD11b− DC–mediated effector T cell recruitment (Spranger et al., 2017). Of the remaining candidate chemokines, CXCL1, CCL4, CCL3, and CCL2 have all been implicated in recruitment of various DC subsets. We compared their expression between isolated tumor cells and TAMs using a previously published array (Fig. 7 B; Cavnar et al., 2013). Expression was greater in TAMs than tumor cells for all of the chemokines. Moreover, expression of CCL3 and CCL4 was particularly high. CCL3 is known to promote preDC migration into tumors (Diao et al., 2010). In contrast, CCL4 has been shown to promote intratumoral accumulation of CD103+CD11b− DCs specifically (Spranger et al., 2015). To further clarify the source of each chemokine, we again compared isolated CD45+ immune cells with CD45−Kit+ tumor cells and CD45− Kit− stromal cells as well as F4/80+ TAMs. Ccl3 and Ccl4 expression was highest in the CD45+ immune compartment and highly expressed by TAMs (Fig. 7, C and D). We then confirmed that expression of both Ccl3 and Ccl4 was decreased after imatinib treatment by RT-PCR (Fig. 7 E). Thus, in addition to the indirect effects on DC maturation observed with IL-1β, TAMs in GIST may directly enhance CD103+CD11b− DC migration through the production of chemokines.
Figure 7.
Macrophages produce DC chemoattractants that are reduced by imatinib. (A) RNaseq was performed in bulk tumors of KitV558Δ/+ mice treated with vehicle or imatinib for 3 wk. Graph shows expression of various chemokines (three mice/group). (B) Quantitative RT-PCR for chemokine mRNA expression in tumors of KitV558Δ/+ mice treated with imatinib (three to four mice/group). (C and D) CD45+ immune cells, CD45−Kit+ tumor cells, CD45−Kit− stromal cells, and F4/80+ macrophages in untreated tumors of KitV558Δ/+ mice were isolated by magnetic beads. Quantitative RT-PCR of Ccl3 and Ccl4 mRNA expression was performed relative to Kit+ tumor cells (three mice/group). (E) Quantitative RT-PCR of Ccl3 and Ccl4 mRNA expression in tumors of KitV558Δ/+ mice treated with vehicle or imatinib for 2 wk (three to four mice/group, representative of two independent experiments). Data represent mean ± SEM; P values were calculated using a Student’s t test; *, P < 0.05.
Targeting Batf3-dependent DCs in the presence of imatinib increases antitumor immunity
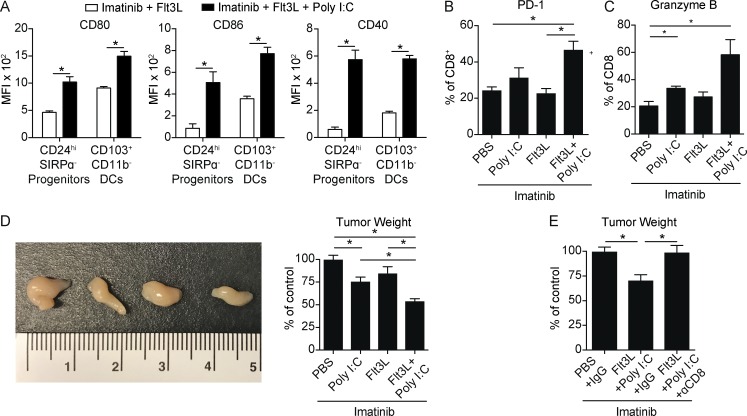
Immature Batf3-dependent CD103−CD11b−CD24hiSIRPα− DC progenitors accumulated in the tumor after imatinib therapy. Polyinosinic:polycytidylic acid (poly I:C) is a synthetic analogue of double-stranded RNA and induces DC maturation through recognition by TLR3, which is highly expressed by the Batf3-dependent DC lineage (Fig. 1 B; Longhi et al., 2009). We hypothesized that in vivo expansion of Batf3-dependent DCs with Flt3L beginning before imatinib treatment and poly I:C–mediated maturation during imatinib treatment would result in enhanced CD8+ T cell–mediated immunity (Fig. S4 A). Indeed, after Flt3L expansion in the presence of imatinib, poly I:C massively up-regulated the costimulation markers CD80, CD86, and CD40 on both CD103+CD11b− DCs and their CD24hiSIRPα− progenitors, while having minimal effect on TAMs (Figs. 8 A and S4 B). The combination therapy also increased the frequency of PD-1+ and granzyme B+ CD8+ T cells (Fig. 8, B and C). The frequencies of CD44+CD62L− and CD44+Tbet+ CD8+ T cells were also increased, altogether indicating an increase in effector differentiation and activity (Fig. S4 C). Accordingly, tumors of mice treated with the combination appeared smaller and weighed 50% less than tumors of mice treated with imatinib and PBS, poly I:C, or Flt3L (Fig. 8 D). Moreover, the effect was completely abrogated in the absence of CD8+ T cells (Fig. 8 E).
Figure 8.
Targeting Batf3-dependent DCs in presence of imatinib increases antitumor immunity. (A) KitV558Δ/+ mice treated with imatinib and Flt3L with or without Poly I:C as in Fig. S4 A and sacrificed on day 5 with respect to start of imatinib. Graph of CD80, CD86, and CD40 MFI in CD103+CD11b− DCs and CD24hiSIRPα− progenitors (five mice/group, representative of two independent experiments). (B and C) Tumors of KitV558Δ/+ mice treated as in Fig. S4 A and sacrificed on day 7 with respect to start of imatinib. Graph of PD-1 expression in CD8+ T cells (three mice/group, representative of two independent experiments). (D) Gross appearance (in the order of the accompanying graph) and tumor weight in KitV558Δ/+ mice treated as in B (pooled from two experiments, six mice/group). (E) Tumor weight from KitV558Δ/+ mice were treated as in B with antibody against CD8 or its isotype given on days −4, −2, 0, 2, 4, and 6 with respect to imatinib treatment (three mice/group). Data represent mean ± SEM; P values were calculated using a Student’s t test; *, P < 0.05.
Discussion
In the present study, we found that CD103+CD11b− DCs and CD141+ DCs infiltrated murine and human GIST tumors, respectively. These DCs appeared phenotypically and functionally identical to the DC subset highlighted by others for its ability to capture tumor antigens and cross-prime CD8+ T cells (Engelhardt et al., 2012; Roberts et al., 2016). Consistent with this, CD8+ T cell–mediated immunosurveillance in murine GIST depended on CD103+CD11b− DCs. Similar to other studies showing that these DCs promote effector T cell trafficking to tumors, it seems likely that CD103+CD11b− DCs also attract T cells in GIST, given the spatial correlation of DCs with CD8+ and CD31+ cells and the reduced CD8+ T cell infiltrate in Batf3−/− mice (Broz et al., 2014; Spranger et al., 2017).
The surprising finding that Batf3−/− mice harbored a residual CD8+ T cell population presented an opportunity to deduce the relevant effector phenotype in GIST using our genetically engineered mouse model of GIST. In contrast to CD8+ cells that depended on CD103+CD11b− DCs, CD8+ cells in Batf3−/− mice were situated throughout the tumor and expressed high levels of CD44 and Tbet. These findings supported our hypothesis that these were likely tissue-resident memory cells and may explain why the marker CD103 was highly expressed in this population. In contrast, CD8+CD103− T cells had a higher frequency of and expressed more PD-1, a marker demonstrated to identify T cells with antitumor specificity (Gros et al., 2014). Moreover, effector memory markers were reduced only in the CD8+CD103− T cell fraction of Batf3−/− mice. Overall, then, we established CD103 as a marker to distinguish effector T cells dependent on CD103+CD11b− DCs from resident memory CD8+ T cells in GIST.
It is unclear what role resident memory T cells play in modulating tumor growth. Several recent studies have demonstrated enhanced antitumor immunity when resident memory T cells are induced by tumor vaccination (Enamorado et al., 2017; Nizard et al., 2017). This is consistent with the proposed function of resident memory T cells as a noncirculating subset specialized in rapid response to immunological challenge (Schenkel and Masopust, 2014). Despite this, tumor growth was not altered in Batf3−/− mice when comparing antibody-mediated depletion of CD8+ T cells to isotype-treated controls. The resident memory subset may then be agnostic with respect to the tumor, responding instead to inflammatory cues from the gut in the context of small bowel obstruction from progressing tumors. It is possible that resident memory T cells were passively immunosuppressive through the consumption of cytokines and growth factors that would otherwise promote expansion of the effector subset. TGFβ has been implicated in resident memory T cell maintenance and is expressed in GIST and reduced by imatinib treatment (Seifert et al., 2017), raising the possibility that tumor cells themselves may maintain the resident memory subset for this purpose. It will be important to determine the precise specificity and function of this subset, given their high frequency in tumors, to potentially target them to promote effector T cell proliferation.
While murine GIST is infiltrated by CD103+CD11b− DCs and is subject to CD8+ T cell immunosurveillance, this net response is functionally restrained given the persistence and growth of tumors. In support of this, our previous work established IDO expression in tumor cells as a product of constitutive Kit signaling and a mechanism of adaptive immune resistance in GIST (Balachandran et al., 2011). This, coupled with the ubiquity of PD-L1 expression on tumor and other stromal cells, may explain why effector T cells were confined to the tumor periphery. Whereas we previously showed that the effects of imatinib are partially mediated by CD8+ T cells due to inhibition of IDO, we now show that imatinib is partially dependent on CD103+CD11b− DCs, likely through effector CD8+ T cells. Consistent with this, we found increased proliferation and activation of effector CD8+ T cells after 1 wk of imatinib treatment. We additionally observed decreased marginalization of CD8+ cells at the tumor periphery, persistent infiltration of the core, and decreased frequency of the resident CD8+ T cell subset. Our data imply that imatinib treatment resulted in the translocation of the effector subset from the periphery to the core, where it exerted antitumor activity. This finding is consistent with the observation that tumor regression in response to checkpoint blockade is associated with proliferation of CD8+ T cells that localize from tumor margins to the tumor (Tumeh et al., 2014). We attribute this to the on-target effect of imatinib with respect to inhibition of IDO as well as down-regulation of PD-L1 on tumor cells.
Given the partial dependence of the antitumor effect of imatinib on CD103+CD11b− DCs, we were surprised to find that imatinib greatly reduced this subset and the homologous CD141+ DCs in mouse and human GIST, respectively. Although imatinib initially increased CD8+ T cell infiltration and activation in mice, longer treatment decreased CD8+ T cell infiltration and differentiation, which correlated with findings in imatinib-sensitive human GIST. Our results indicate that while the short-term effect of imatinib is to unleash preexisting CD8+ T cell immunity, long-term immunity is diminished along with a decrease in CD103+CD11b− DCs, which are required to recruit and prime CD8+ T cells to maintain this response.
Several studies have explored the effect of imatinib treatment on the generation of DCs in vitro as well as the expansion of DCs in vivo (Appel et al., 2004; Taïeb et al., 2004). However, those studies predate our current knowledge of the various DC lineages and the heterogeneity of DC subsets in peripheral tissues. Consequently, it is unclear if the findings apply to the Batf3-dependent lineage. We employed in vitro studies and explored multiple tissue compartments in mice to identify DC progenitors and resident DCs in mice, none of which were affected by imatinib treatment in either KitV558Δ/+ or wild-type mice. Using two models of imatinib-resistant GIST, we demonstrated that the inhibition of CD103+CD11b− DCs could not be recapitulated without inhibiting Kit on tumor cells. In contrast, treatment of KitV558Δ/+ mice with imatinib was associated with the progressive accumulation of CD24hiSIRPα progenitors, which did not express CD103, but otherwise resembled CD103+CD11b− DCs transcriptionally and were similarly Batf3 dependent. GM-CSF dependence of CD103+CD11b− DCs is consistent with established steady state requirements. In several studies of pancreatic ductal adenocarcinoma, GM-CSF is produced as a byproduct of oncogenic KRAS and recruits myeloid-derived suppressor cells and CD11b+ DCs (Bayne et al., 2012; Pylayeva-Gupta et al., 2012; Kenkel et al., 2017). In KitV558Δ/+ mice, we discovered that GM-CSF is produced by γδT cells. The net effect of GM-CSF then may simply be one of quantity, where high levels promote the proliferation of immunosuppressive myeloid cells while lower levels maintain the expanded population of CD103+CD11b− DCs relative to the steady state.
While several recent studies have targeted CD103+CD11b− DCs in vivo using Flt3L and poly I:C, this strategy in GIST was dictated by the variable immune effects of imatinib (Salmon et al., 2016; Sánchez-Paulete et al., 2016). In our mouse model, we exploited the on-target inhibition of IDO to as a mechanism to enable immunotherapy. DCs expanded with Flt3L in the presence of imatinib necessitated the use of poly I:C as a means to activate the substantial proportion of immature CD24hiSIRPα progenitors. While this approach led to an increased antitumor effect compared with imatinib alone, it is unclear whether targeting DCs in this way represents the optimal approach to immunotherapy in GIST. Given the immune activating and suppressive mechanisms associated with imatinib, it may be that alternative regimens to continuous imatinib therapy would result in a more ideal immune environment for combination strategies.
Overall, our findings identified the inherent limitations of Kit tyrosine kinase inhibitors on antitumor immunity in GIST and highlight the dynamic, complex, intratumoral immune network. In our mouse model, effector CD8+ T cells were dependent on CD103+CD11b− DCs, which required GM-CSF that was produced only by γδ T cells. The secretion of GM-CSF was itself regulated by IL-1β, which was produced by TAMs. Accordingly, the frequency of CD103+CD11b− DCs was decreased when GM-CSF, γδ T cells, or IL-1β were depleted.
There are noteworthy parallels between the present findings and our previous investigation of TAMs in murine and human GIST (Cavnar et al., 2013). TAMs in GIST are inflammatory and antitumoral, and CSF1R inhibition increased tumor growth. TAMs became functionally suppressive with oncogene inhibition yet reverted back to an antitumor phenotype in the face of imatinib resistance. We identified IL-1β as an indirect mechanism by which CD103+CD11b− DCs depend on the inflammatory activity of TAMs in murine GIST. Moreover, it is likely that TAMS directly enhance pre-DC and CD103+CD11b− DC migration into tumors through production of CCL3 and CCL4 respectively. In contrast to other models where tumor cell–intrinsic mechanisms have been associated with altered lineage DC development and exclusion of CD103+CD11b− DCs from tumors, our results clearly implicated oncogene activity in the maintenance of CD103+CD11b− DCs and adaptive immunity (Spranger et al., 2015; Böttcher et al., 2018; Meyer et al., 2018). Indeed, reduced expression of IL-1β, CCL3, and CCL4 after treatment with imatinib is consistent with our prior work connecting inflammatory TAMs to intact oncogenic signaling. In concert with our previous work, the present findings underscore the central importance of oncogene activity in modulating the immune signature of GIST, characterized by an inflammatory innate response limited by mechanisms of adaptive immune resistance. Kit oncogene inhibition flips this dynamic, limiting antitumoral innate immunity, but also promoting adaptive immune suppression.
Materials and methods
Mice
8–12-wk-old KitV558Δ/+ and KitV558Δ;T669I/+ mice (Sommer et al., 2003; Bosbach et al., 2012) and Batf3−/−, Zbtb46GFP/+, and C57BL/6J mice (B6; The Jackson Laboratory), all on a B6 background, were maintained in a specific pathogen–free animal facility and age- and sex-matched for experiments. The animal experiments were approved by the Institutional Animal Care and Use Committee at Memorial Sloan-Kettering and the University of Pennsylvania.
Implantable flank tumors
106 S2 cells in PBS were mixed 1:1 with growth factor–reduced Matrigel (BD) and injected subcutaneously in the right flank of B6 mice. Flank tumor volume was calculated using the ellipse formula (1/2 length × width × height as measured with calipers).
Imaging
MRI was used to measure tumors in some mice. In brief, mice were anesthetized with 2% isofluorane, and images were obtained on a Bruker 4.7T 40-cm bore magnet with a commercial 7-cm inner diameter birdcage coil in the institutional Animal Imaging MRCore Facility. Low-resolution axial scout images were obtained initially, followed by high-spatial-resolution T2-weighted axial images. Volumetric analysis was performed using ImageJ software (National Institutes of Health [NIH]).
Treatments
Imatinib was obtained from Novartis and dissolved in the drinking water at 600 mg/liter. For in vivo DC expansion, Flt3L (30 µg in 100 µl PBS; generously provided by Celldex) or control PBS was injected i.p. (nine daily injections). At the indicated time points, the following were administered i.p.: high molecular weight poly I:C (50 µg in 100 µl; InvivoGen) or control PBS; 250 µg anti-CD8 (2.43; Bio X Cell) or rat IgG2b (LTF-2); 1 mg anti–GM-CSF (MP1-22E9; Bio X Cell) or rat IgG2a (2A3); 100 µg anti-IL-1β (B122) or Armenian Hamster IgG (BE0091); or 500 µg loading dose and 250 µg maintenance dose anti-γδTCR (UC7-13D5) or Armenian Hamster IgG (BE0091).
Preparation of cell suspensions
Excised tumors were minced and incubated in Liberase TL (Roche) in serum-free medium for 30 min while shaking at 37°C, quenched with FBS, and washed through 100- and 40-µm nylon cell strainers (Falcon; BD Biosciences) in PBS with 1% FBS. Lymph nodes and spleens were incubated in 0.5 mg/ml collagenase IV (Sigma) for 25 min at 37°C, quenched with EDTA, mechanically dissociated with the flat portion of a plunger from a 3-ml syringe, and sequentially washed through 70- and 40-µm nylon cell strainers. In the case of spleens, red blood cells were lysed with ammonium-chloride-potassium (ACK) lysing buffer (Life Technologies) for 1 min and washed in PBS. Bone marrow was harvested from femurs and tibias of mice after flushing with 3 ml PBS with 1% FBS. Red blood cells were lysed in ACK lysing buffer. Suspensions were washed through 40-µm nylon cell strainers. Blood was obtained by direct cardiac puncture. Red blood cells were lysed with ACK lysing buffer. Suspensions were washed through 40-µm nylon cell strainers. After procurement, all specimens were immediately analyzed with flow cytometry.
Flow cytometry
Flow cytometry was performed using a LSRFortessa (BD). Fc receptors were blocked using anti-CD16/32 (Bio X Cell). Mouse-specific antibodies conjugated to various fluorochromes were purchased from BioLegend (CD45, 30-F11; Tbet, 4B10; CD64, X54-5/7.1; and TLR3, 11F8), BD Biosciences (CD11c, HL3; CD80, 16-10A1; CD86, Gl1; PDL1, MIH5; CD45RA, 14.8; CD44, IM7; IFNγ, XMG1.2; CD117, 2B8; CD62L, MEL14; CD11b, M1/70; NK1.1, PK136; IL12-p40, C15.6; TNF, MP6-XT22; Ly6C, AL-21; CD4, GK1.5; CD3, 145-2C11; and GM-CSF, MP1-22E9), eBioscience (CD8, 53-6.7; F4/80, BM8; CD40, 1C10; Ki67, SolA15; CD172a, SIRPα, P84; CD24, M1/69; PD1, J43; CD115, AFS98; CD135, A2F10; MHC II, M5/114.15.2; CD103, 2E7, IRF8, V3GYWCH; and IRF4, 3E4), and Invitrogen (Granzyme B, GB11). Human-specific antibodies conjugated to various fluorochromes were purchased from BioLegend (CD45, HI30; CD45RO, UCHL1; and CD11b, ICRF44), BD Biosciences (PD1, MIH4; CD45RA, HI100; CD80, L307; CD86, 2331; CD40, 5C3; CD14, M5E2; CD11c, B-ly6; CD3, SK7; CD56, B159; CD19, HIB19; HLA-DR, TU36; CD4, RPA-T4; and CD8, RPA-T8), eBioscience (CD16, CB16), and Miltenyi Biotec (CD141, AD5-14H12). Annexin V staining was performed using the BD Bioscience Annexin V staining kit as directed. Transcription factor staining was performed using the eBioscience Fixation and Permeabilization Buffer Kit. Intracellular cytokine staining was performed with the BD Cytofix/Cytoperm Kit.
Cytokine detection and intravascular staining
For intracellular IFN-γ, TNF, and GM-CSF, cells were stimulated with PMA (50 ng/ml) and ionomycin (750 ng/ml) for 4 h at 37°C, 5% CO2 in the presence of 10 µg/ml brefeldin A (Sigma-Aldrich). For IL-12p40, 250 µg brefeldin A was injected i.v., the animals were sacrificed 6 h later, and tumors were processed as described above. For GM-CSF protein, whole protein from frozen tumor tissues was lysed, and total protein was tested for expression using a Proteome Profiler Array (R&D Systems). Densitometry was conducted on blots using ImageJ (NIH). For intravascular staining, 3 µg of anti-CD8a-Alexa Fluor 700 or anti-CD8b-PE (clone: 53-6.7 from BD) were injected i.v. as described. The animals were sacrificed 3 min later, and organs were processed as described above.
Cell isolation
Tumor single-cell suspensions were incubated with anti-mouse CD45 microbeads (Miltenyi Biotec) and run through two sequential LS columns per 3 × 107 cells, saving the positive fraction. Kit+ tumor cell selection was performed by incubating the CD45− fraction with CD117 microbeads (Miltenyi Biotec) and collecting the positive fraction. CD45−Kit− cells were collected from the remaining negative fraction. For TAM isolation, single-cell suspensions were incubated with anti-mouse F4/80 microbeads (Miltenyi Biotec) and run through two sequential LS columns per 3 × 107 cells. Purity was >90% by flow cytometry.
Bone marrow–derived DCs
For the generation of CD103+ DCs, 106 bone marrow cells were maintained in RPMI 1640 medium supplemented with 10% FBS, 2 mmol/liter l-glutamine, 50 U/ml penicillin-streptomycin, 0.1% 2-mercaptoethanol, and 10 mmol/liter Hepes supplemented with 200 ng/ml recombinant human Flt3L (Peprotech) for 8 d at 37°C in 5% CO2. Recombinant murine GM-CSF (2 ng/ml, Peprotech) was added for the final 2 d of culture. Imatinib was dissolved in PBS at the indicated concentrations and was present at the start of the culture.
Quantitative real-time PCR
Total RNA was extracted from tumor tissues or cells, reverse transcribed, and amplified with PCR TaqMan probes for murine Csf2, Il1b, Ccl3, Ccl4, and Gapdh (Applied Biosystems). Quantitative PCR was performed using a ViiA 7 real-time PCR system (Applied Biosystems). Data were calculated by the 2−ΔΔCt method as described in the manufacturer’s instructions and expressed as fold increase over control.
Microscopy
Formalin-fixed and paraffin-embedded specimens were sectioned at 5-µm thickness and mounted on glass slides. Antigen retrieval was achieved with citrate buffer. Anti-CD8 (4SM16; eBioscience, 5 µg/ml) was applied on tissue sections overnight at 4°C. After washing with TBS (0.05 mol/liter Tris base and 0.9% NaCl, pH 8.4), tissue sections were incubated with biotinylated anti-rat IgG (1:100; BA-2001; Vector Laboratories), followed by incubation with Elite ABC Kit for 30 min at room temperature. Antibody binding was detected with the TSA Biotin System (NEL700A001KT; PerkinElmer) and DAB (K3468; Dako). Staining was quantified by counting the positive staining in a 40× field. Immunofluorescence staining against GFP, CD8, and CD31 was performed by our institutional Molecular Cytology Core. Slides were scanned with MIRAX scan (Zeiss) and analyzed with Pannoramic Viewer.
Bioinformatics
Next-generation RNaseq of fresh, frozen tumor specimens treated with either vehicle or imatinib was performed by the institutional Integrated Genomics Operations core facility using an Illumina HiSeq 2500 platform. We obtained a minimum of 40–50 million reads per sample, with a read length of 50 bp with paired ends. Sequencing reads were quantified and aligned.
Patient samples
Tumor specimens and matched peripheral blood were obtained from patients with GIST who underwent surgery at our institution and consented to a protocol approved by the Institutional Review Board at Memorial Sloan-Kettering. Blood was drawn before surgical incision, and peripheral blood mononuclear cells were isolated by density centrifugation over Ficoll-Plaque PLUS (GE Healthcare). Tumor tissue was sectioned and digested in 5 mg/ml collagenase IV (Sigma-Aldrich) and DNase I (0.5 mg/ml; Roche Diagnostics) in HBSS for 30 min while shaking at 37°C. After procurement, all processed cells were immediately analyzed with flow cytometry.
Protein chunks from human GIST tumors were flash frozen in liquid nitrogen and stored at −80°C until use. Total RNA was isolated using the RNAeasy Plus Mini Kit (Qiagen). Quality of RNA was ensured before labeling by analyzing 20 ng of each sample using the RNA 6000 NanoAssay and a Bioanalyzer 2100 (Agilent). High-throughput RNaseq was performed by the Integrated Genomics Core laboratory of the Sloan Kettering Institute using the Illumina platform and normalized using the software package DESeq. Cytolytic score was calculated as the square root of the product of granzyme A (GZMA) and perforin (PRF1) expression.
Statistical analysis
Unpaired two-tailed Student’s t test was performed on datasets using Graph Pad Prism 6.0 (Graph Pad Software). A P value <0.05 was considered significant. Data are shown as mean ± SEM or median. Spearman correlation was performed where applicable.
Online supplemental material
Fig. S1 demonstrates DC frequencies using two unique gating strategies, increased activation of DCs in tumor relative to blood in human GIST patients, in vivo brefeldin staining, and depletion of CD103+CD11b− DCs in KitV558Δ/+;Batf3−/− mice. Fig. S2 compares characteristics of human GIST specimens with BATF3 expression, quantifies immunohistochemistry in the periphery and core of tumors in KitV558Δ/+;Batf3−/− mice and KitV558Δ/+ mice treated with imatinib, and demonstrates CD103 staining in intravascular CD8+ T cells. Fig. S3 shows the gating strategy for CD103+CD11b− DC precursors and lineage-related DCs in multiple tissue compartments and in vitro generation of CD103+CD11b− DCs in the presence of imatinib. Fig. S4 outlines the regimen for mice treated with imatinib, Flt3L, and poly I:C.
Supplementary Material
Acknowledgments
We are grateful to the Tissue Procurement Service for assistance in the acquisition of human tumor specimens. We thank members of the Sloan Kettering Institute Laboratory of Comparative Pathology, Molecular Cytology, Flow Cytometry, Integrated Genomics, and Pathology Core Facilities, Colony Management Group, and Research Animal Resource Center. We thank Russell Holmes for logistical and administrative support.
The investigators were supported by NIH grants R01 CA102613 and T32 CA09501, Betsy Levine-Brown and Marc Brown, David and Monica Gorin, The David Foundation, the GIST Cancer Research Fund (R.P. DeMatteo), NIH grant F32 CA186534 (J.Q. Zhang), and NIH grant L30 TR002111 (G.A. Vitiello). The Flow Cytometry and Molecular Cytology Core Facilities were supported by National Cancer Institute Cancer Center Support Grant P30 CA008748.
The authors declare no competing financial interests.
All authors contributed to the experimental design. B.D. Medina, M. Liu, G.A. Vitiello, A.M. Seifert, and S. Zeng performed the experiments. All authors participated in data analysis. B.D. Medina and R.P. DeMatteo wrote the manuscript with critical comments from all authors.
References
- Anderson K.G., Mayer-Barber K., Sung H., Beura L., James B.R., Taylor J.J., Qunaj L., Griffith T.S., Vezys V., Barber D.L., and Masopust D.. 2014. Intravascular staining for discrimination of vascular and tissue leukocytes. Nat. Protoc. 9:209–222. 10.1038/nprot.2014.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appel S., Boehmler A.M., Grünebach F., Müller M.R., Rupf A., Weck M.M., Hartmann U., Reichardt V.L., Kanz L., Brümmendorf T.H., and Brossart P.. 2004. Imatinib mesylate affects the development and function of dendritic cells generated from CD34+ peripheral blood progenitor cells. Blood. 103:538–544. 10.1182/blood-2003-03-0975 [DOI] [PubMed] [Google Scholar]
- Bachem A., Güttler S., Hartung E., Ebstein F., Schaefer M., Tannert A., Salama A., Movassaghi K., Opitz C., Mages H.W., et al. 2010. Superior antigen cross-presentation and XCR1 expression define human CD11c+CD141+ cells as homologues of mouse CD8+ dendritic cells. J. Exp. Med. 207:1273–1281. 10.1084/jem.20100348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balachandran V.P., Cavnar M.J., Zeng S., Bamboat Z.M., Ocuin L.M., Obaid H., Sorenson E.C., Popow R., Ariyan C., Rossi F., et al. 2011. Imatinib potentiates antitumor T cell responses in gastrointestinal stromal tumor through the inhibition of Ido. Nat. Med. 17:1094–1100. 10.1038/nm.2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayne L.J., Beatty G.L., Jhala N., Clark C.E., Rhim A.D., Stanger B.Z., and Vonderheide R.H.. 2012. Tumor-derived granulocyte-macrophage colony-stimulating factor regulates myeloid inflammation and T cell immunity in pancreatic cancer. Cancer Cell. 21:822–835. 10.1016/j.ccr.2012.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanke C.D., Rankin C., Demetri G.D., Ryan C.W., von Mehren M., Benjamin R.S., Raymond A.K., Bramwell V.H., Baker L.H., Maki R.G., et al. 2008. Phase III randomized, intergroup trial assessing imatinib mesylate at two dose levels in patients with unresectable or metastatic gastrointestinal stromal tumors expressing the kit receptor tyrosine kinase: S0033. J. Clin. Oncol. 26:626–632. 10.1200/JCO.2007.13.4452 [DOI] [PubMed] [Google Scholar]
- Bogunovic M., Ginhoux F., Helft J., Shang L., Hashimoto D., Greter M., Liu K., Jakubzick C., Ingersoll M.A., Leboeuf M., et al. 2009. Origin of the lamina propria dendritic cell network. Immunity. 31:513–525. 10.1016/j.immuni.2009.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosbach B., Deshpande S., Rossi F., Shieh J.H., Sommer G., de Stanchina E., Veach D.R., Scandura J.M., Manova-Todorova K., Moore M.A., et al. 2012. Imatinib resistance and microcytic erythrocytosis in a KitV558Δ;T669I/+ gatekeeper-mutant mouse model of gastrointestinal stromal tumor. Proc. Natl. Acad. Sci. USA. 109:E2276–E2283. 10.1073/pnas.1115240109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttcher J.P., Bonavita E., Chakravarty P., Blees H., Cabeza-Cabrerizo M., Sammicheli S., Rogers N.C., Sahai E., Zelenay S., and Reis e Sousa C.. 2018. NK Cells Stimulate Recruitment of cDC1 into the Tumor Microenvironment Promoting Cancer Immune Control. Cell. 172:1022–1037.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broz M.L., Binnewies M., Boldajipour B., Nelson A.E., Pollack J.L., Erle D.J., Barczak A., Rosenblum M.D., Daud A., Barber D.L., et al. 2014. Dissecting the Tumor Myeloid Compartment Reveals Rare Activating Antigen-Presenting Cells Critical for T Cell Immunity. Cancer Cell. 26:938 10.1016/j.ccell.2014.11.010 [DOI] [PubMed] [Google Scholar]
- Casey K.A., Fraser K.A., Schenkel J.M., Moran A., Abt M.C., Beura L.K., Lucas P.J., Artis D., Wherry E.J., Hogquist K., et al. 2012. Antigen-independent differentiation and maintenance of effector-like resident memory T cells in tissues. J. Immunol. 188:4866–4875. 10.4049/jimmunol.1200402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavnar M.J., Zeng S., Kim T.S., Sorenson E.C., Ocuin L.M., Balachandran V.P., Seifert A.M., Greer J.B., Popow R., Crawley M.H., et al. 2013. KIT oncogene inhibition drives intratumoral macrophage M2 polarization. J. Exp. Med. 210:2873–2886. 10.1084/jem.20130875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codarri L., Gyülvészi G., Tosevski V., Hesske L., Fontana A., Magnenat L., Suter T., and Becher B.. 2011. RORγt drives production of the cytokine GM-CSF in helper T cells, which is essential for the effector phase of autoimmune neuroinflammation. Nat. Immunol. 12:560–567. 10.1038/ni.2027 [DOI] [PubMed] [Google Scholar]
- Demetri G.D., von Mehren M., Blanke C.D., Van den Abbeele A.D., Eisenberg B., Roberts P.J., Heinrich M.C., Tuveson D.A., Singer S., Janicek M., et al. 2002. Efficacy and safety of imatinib mesylate in advanced gastrointestinal stromal tumors. N. Engl. J. Med. 347:472–480. 10.1056/NEJMoa020461 [DOI] [PubMed] [Google Scholar]
- Diao J., Zhao J., Winter E., and Cattral M.S.. 2010. Recruitment and differentiation of conventional dendritic cell precursors in tumors. J. Immunol. 184:1261–1267. 10.4049/jimmunol.0903050 [DOI] [PubMed] [Google Scholar]
- Edelson B.T., Kc W., Juang R., Kohyama M., Benoit L.A., Klekotka P.A., Moon C., Albring J.C., Ise W., Michael D.G., et al. 2010. Peripheral CD103+ dendritic cells form a unified subset developmentally related to CD8alpha+ conventional dendritic cells. J. Exp. Med. 207:823–836. 10.1084/jem.20091627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enamorado M., Iborra S., Priego E., Cueto F.J., Quintana J.A., Martínez-Cano S., Mejías-Pérez E., Esteban M., Melero I., Hidalgo A., and Sancho D.. 2017. Enhanced anti-tumour immunity requires the interplay between resident and circulating memory CD8+ T cells. Nat. Commun. 8:16073 10.1038/ncomms16073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelhardt J.J., Boldajipour B., Beemiller P., Pandurangi P., Sorensen C., Werb Z., Egeblad M., and Krummel M.F.. 2012. Marginating dendritic cells of the tumor microenvironment cross-present tumor antigens and stably engage tumor-specific T cells. Cancer Cell. 21:402–417. 10.1016/j.ccr.2012.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogg D.K., Sibon C., Miled C., Jung S., Aucouturier P., Littman D.R., Cumano A., and Geissmann F.. 2006. A clonogenic bone marrow progenitor specific for macrophages and dendritic cells. Science. 311:83–87. 10.1126/science.1117729 [DOI] [PubMed] [Google Scholar]
- Ginhoux F., Liu K., Helft J., Bogunovic M., Greter M., Hashimoto D., Price J., Yin N., Bromberg J., Lira S.A., et al. 2009. The origin and development of nonlymphoid tissue CD103+ DCs. J. Exp. Med. 206:3115–3130. 10.1084/jem.20091756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greter M., Helft J., Chow A., Hashimoto D., Mortha A., Agudo-Cantero J., Bogunovic M., Gautier E.L., Miller J., Leboeuf M., et al. 2012. GM-CSF controls nonlymphoid tissue dendritic cell homeostasis but is dispensable for the differentiation of inflammatory dendritic cells. Immunity. 36:1031–1046. 10.1016/j.immuni.2012.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gros A., Robbins P.F., Yao X., Li Y.F., Turcotte S., Tran E., Wunderlich J.R., Mixon A., Farid S., Dudley M.E., et al. 2014. PD-1 identifies the patient-specific CD8+ tumor-reactive repertoire infiltrating human tumors. J. Clin. Invest. 124:2246–2259. 10.1172/JCI73639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haniffa M., Shin A., Bigley V., McGovern N., Teo P., See P., Wasan P.S., Wang X.N., Malinarich F., Malleret B., et al. 2012. Human tissues contain CD141hi cross-presenting dendritic cells with functional homology to mouse CD103+ nonlymphoid dendritic cells. Immunity. 37:60–73. 10.1016/j.immuni.2012.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildner K., Edelson B.T., Purtha W.E., Diamond M., Matsushita H., Kohyama M., Calderon B., Schraml B.U., Unanue E.R., Diamond M.S., et al. 2008. Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science. 322:1097–1100. 10.1126/science.1164206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota S., Isozaki K., Moriyama Y., Hashimoto K., Nishida T., Ishiguro S., Kawano K., Hanada M., Kurata A., Takeda M., et al. 1998. Gain-of-function mutations of c-kit in human gastrointestinal stromal tumors. Science. 279:577–580. 10.1126/science.279.5350.577 [DOI] [PubMed] [Google Scholar]
- Intlekofer A.M., Takemoto N., Wherry E.J., Longworth S.A., Northrup J.T., Palanivel V.R., Mullen A.C., Gasink C.R., Kaech S.M., Miller J.D., et al. 2005. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat. Immunol. 6:1236–1244. 10.1038/ni1268 [DOI] [PubMed] [Google Scholar]
- Joensuu H., and DeMatteo R.P.. 2012. The management of gastrointestinal stromal tumors: a model for targeted and multidisciplinary therapy of malignancy. Annu. Rev. Med. 63:247–258. 10.1146/annurev-med-043010-091813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongbloed S.L., Kassianos A.J., McDonald K.J., Clark G.J., Ju X., Angel C.E., Chen C.J., Dunbar P.R., Wadley R.B., Jeet V., et al. 2010. Human CD141+ (BDCA-3)+ dendritic cells (DCs) represent a unique myeloid DC subset that cross-presents necrotic cell antigens. J. Exp. Med. 207:1247–1260. 10.1084/jem.20092140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenkel J.A., Tseng W.W., Davidson M.G., Tolentino L.L., Choi O., Bhattacharya N., Seeley E.S., Winer D.A., Reticker-Flynn N.E., and Engleman E.G.. 2017. An Immunosuppressive Dendritic Cell Subset Accumulates at Secondary Sites and Promotes Metastasis in Pancreatic Cancer. Cancer Res. 77:4158–4170. 10.1158/0008-5472.CAN-16-2212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King I.L., Kroenke M.A., and Segal B.M.. 2010. GM-CSF-dependent, CD103+ dermal dendritic cells play a critical role in Th effector cell differentiation after subcutaneous immunization. J. Exp. Med. 207:953–961. 10.1084/jem.20091844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K., Victora G.D., Schwickert T.A., Guermonprez P., Meredith M.M., Yao K., Chu F.F., Randolph G.J., Rudensky A.Y., and Nussenzweig M.. 2009. In vivo analysis of dendritic cell development and homeostasis. Science. 324:392–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longhi M.P., Trumpfheller C., Idoyaga J., Caskey M., Matos I., Kluger C., Salazar A.M., Colonna M., and Steinman R.M.. 2009. Dendritic cells require a systemic type I interferon response to mature and induce CD4+ Th1 immunity with poly IC as adjuvant. J. Exp. Med. 206:1589–1602. 10.1084/jem.20090247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukens J.R., Barr M.J., Chaplin D.D., Chi H., and Kanneganti T.D.. 2012. Inflammasome-derived IL-1β regulates the production of GM-CSF by CD4(+) T cells and γδ T cells. J. Immunol. 188:3107–3115. 10.4049/jimmunol.1103308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay L.K., Stock A.T., Ma J.Z., Jones C.M., Kent S.J., Mueller S.N., Heath W.R., Carbone F.R., and Gebhardt T.. 2012. Long-lived epithelial immunity by tissue-resident memory T (TRM) cells in the absence of persisting local antigen presentation. Proc. Natl. Acad. Sci. USA. 109:7037–7042. 10.1073/pnas.1202288109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maraskovsky E., Brasel K., Teepe M., Roux E.R., Lyman S.D., Shortman K., and McKenna H.J.. 1996. Dramatic increase in the numbers of functionally mature dendritic cells in Flt3 ligand-treated mice: multiple dendritic cell subpopulations identified. J. Exp. Med. 184:1953–1962. 10.1084/jem.184.5.1953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M.A., Baer J.M., Knolhoff B.L., Nywening T.M., Panni R.Z., Su X., Weilbaecher K.N., Hawkins W.G., Ma C., Fields R.C., et al. 2018. Breast and pancreatic cancer interrupt IRF8-dependent dendritic cell development to overcome immune surveillance. Nat. Commun. 9:1250 10.1038/s41467-018-03600-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik S.H., Sathe P., Park H.Y., Metcalf D., Proietto A.I., Dakic A., Carotta S., O’Keeffe M., Bahlo M., Papenfuss A., et al. 2007. Development of plasmacytoid and conventional dendritic cell subtypes from single precursor cells derived in vitro and in vivo. Nat. Immunol. 8:1217–1226. 10.1038/ni1522 [DOI] [PubMed] [Google Scholar]
- Nizard M., Roussel H., Diniz M.O., Karaki S., Tran T., Voron T., Dransart E., Sandoval F., Riquet M., Rance B., et al. 2017. Induction of resident memory T cells enhances the efficacy of cancer vaccine. Nat. Commun. 8:15221 10.1038/ncomms15221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onai N., Obata-Onai A., Schmid M.A., Ohteki T., Jarrossay D., and Manz M.G.. 2007. Identification of clonogenic common Flt3+M-CSFR+ plasmacytoid and conventional dendritic cell progenitors in mouse bone marrow. Nat. Immunol. 8:1207–1216. 10.1038/ni1518 [DOI] [PubMed] [Google Scholar]
- Paley M.A., Kroy D.C., Odorizzi P.M., Johnnidis J.B., Dolfi D.V., Barnett B.E., Bikoff E.K., Robertson E.J., Lauer G.M., Reiner S.L., and Wherry E.J.. 2012. Progenitor and terminal subsets of CD8+ T cells cooperate to contain chronic viral infection. Science. 338:1220–1225. 10.1126/science.1229620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauken K.E., and Wherry E.J.. 2015. SnapShot: T Cell Exhaustion. Cell. 163:1038–1038.e1. [DOI] [PubMed] [Google Scholar]
- Pylayeva-Gupta Y., Lee K.E., Hajdu C.H., Miller G., and Bar-Sagi D.. 2012. Oncogenic Kras-induced GM-CSF production promotes the development of pancreatic neoplasia. Cancer Cell. 21:836–847. 10.1016/j.ccr.2012.04.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts E.W., Broz M.L., Binnewies M., Headley M.B., Nelson A.E., Wolf D.M., Kaisho T., Bogunovic D., Bhardwaj N., and Krummel M.F.. 2016. Critical Role for CD103(+)/CD141(+) Dendritic Cells Bearing CCR7 for Tumor Antigen Trafficking and Priming of T Cell Immunity in Melanoma. Cancer Cell. 30:324–336. 10.1016/j.ccell.2016.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon H., Idoyaga J., Rahman A., Leboeuf M., Remark R., Jordan S., Casanova-Acebes M., Khudoynazarova M., Agudo J., Tung N., et al. 2016. Expansion and Activation of CD103(+) Dendritic Cell Progenitors at the Tumor Site Enhances Tumor Responses to Therapeutic PD-L1 and BRAF Inhibition. Immunity. 44:924–938. 10.1016/j.immuni.2016.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Paulete A.R., Cueto F.J., Martínez-López M., Labiano S., Morales-Kastresana A., Rodríguez-Ruiz M.E., Jure-Kunkel M., Azpilikueta A., Aznar M.A., Quetglas J.I., et al. 2016. Cancer Immunotherapy with Immunomodulatory Anti-CD137 and Anti-PD-1 Monoclonal Antibodies Requires BATF3-Dependent Dendritic Cells. Cancer Discov. 6:71–79. 10.1158/2159-8290.CD-15-0510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S., Kalia V., Haining W.N., Konieczny B.T., Subramaniam S., and Ahmed R.. 2008. Functional and genomic profiling of effector CD8 T cell subsets with distinct memory fates. J. Exp. Med. 205:625–640. 10.1084/jem.20071641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satpathy A.T., Kc W., Albring J.C., Edelson B.T., Kretzer N.M., Bhattacharya D., Murphy T.L., and Murphy K.M.. 2012. Zbtb46 expression distinguishes classical dendritic cells and their committed progenitors from other immune lineages. J. Exp. Med. 209:1135–1152. 10.1084/jem.20120030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenkel J.M., and Masopust D.. 2014. Tissue-resident memory T cells. Immunity. 41:886–897. 10.1016/j.immuni.2014.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert A.M., Zeng S., Zhang J.Q., Kim T.S., Cohen N.A., Beckman M.J., Medina B.D., Maltbaek J.H., Loo J.K., Crawley M.H., et al. 2017. PD-1/PD-L1 Blockade Enhances T-cell Activity and Antitumor Efficacy of Imatinib in Gastrointestinal Stromal Tumors. Clin. Cancer Res. 23:454–465. 10.1158/1078-0432.CCR-16-1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer G., Agosti V., Ehlers I., Rossi F., Corbacioglu S., Farkas J., Moore M., Manova K., Antonescu C.R., and Besmer P.. 2003. Gastrointestinal stromal tumors in a mouse model by targeted mutation of the Kit receptor tyrosine kinase. Proc. Natl. Acad. Sci. USA. 100:6706–6711. 10.1073/pnas.1037763100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spranger S., Bao R., and Gajewski T.F.. 2015. Melanoma-intrinsic β-catenin signalling prevents anti-tumour immunity. Nature. 523:231–235. 10.1038/nature14404 [DOI] [PubMed] [Google Scholar]
- Spranger S., Dai D., Horton B., and Gajewski T.F.. 2017. Tumor-Residing Batf3 Dendritic Cells Are Required for Effector T Cell Trafficking and Adoptive T Cell Therapy. Cancer Cell. 31:711–723.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taïeb J., Maruyama K., Borg C., Terme M., and Zitvogel L.. 2004. Imatinib mesylate impairs Flt3L-mediated dendritic cell expansion and antitumor effects in vivo. Blood. 103:1966–1967, author reply :1967. 10.1182/blood-2003-10-3475 [DOI] [PubMed] [Google Scholar]
- Tumeh P.C., Harview C.L., Yearley J.H., Shintaku I.P., Taylor E.J., Robert L., Chmielowski B., Spasic M., Henry G., Ciobanu V., et al. 2014. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 515:568–571. 10.1038/nature13954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Diny N.L., Ong S., Barin J.G., Hou X., Rose N.R., Talor M.V., and Čiháková D.. 2016. Pathogenic IL-23 signaling is required to initiate GM-CSF-driven autoimmune myocarditis in mice. Eur. J. Immunol. 46:582–592. 10.1002/eji.201545924 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.