Th17 cells provide a protective immunity against extracellular bacterial and fungal pathogens. Kotov et al. identify and characterize a mechanism by which BCOR promotes Th17 formation after Streptococcus pyogenes infection by repressing genes that inhibit the Th17 lineage.
Abstract
CD4+ T helper 17 (Th17) cells protect vertebrate hosts from extracellular pathogens at mucosal surfaces. Th17 cells form from naive precursors when signals from the T cell antigen receptor (TCR) and certain cytokine receptors induce the expression of the RORγt transcription factor, which activates a set of Th17-specific genes. Using T cell–specific loss-of-function experiments, we find that two components of the Polycomb repressive complex 1.1 (PRC1.1), BCL6 corepressor (BCOR) and KDM2B, which helps target the complex to unmethylated CpG DNA islands, are required for optimal Th17 cell formation in mice after Streptococcus pyogenes infection. Genome-wide expression and BCOR chromatin immunoprecipitation studies revealed that BCOR directly represses Lef1, Runx2, and Dusp4, whose products inhibit Th17 differentiation. Together, the results suggest that the PRC1.1 components BCOR and KDM2B work together to enhance Th17 cell formation by repressing Th17 fate suppressors.
Introduction
CD4+ T cells participate in host immunity by using TCRs to recognize MHCII-bound microbial peptides on host cells. TCR signaling causes naive CD4+ T cells to proliferate and differentiate into specialized T helper 1 (Th1), Th2, Th9, Th17, T follicular helper (Tfh), or induced regulatory T (T reg) cells depending on signals from cytokine receptors. Th17 cells are generated from naive precursors that proliferate in the context of signals from receptors for innate immune system cytokines IL-6 and TGF-β (Bettelli et al., 2006; Veldhoen et al., 2006). These receptors activate the transcription factors STAT3 and SMAD3 (Zhong et al., 1994; Derynck and Zhang, 2003), which activate transcription of Rorc, the gene encoding RORγt, the master transcription factor for Th17 cells (Ivanov et al., 2006). RORγt binds to the promoters of Th17-specific genes such as Il17a, Il17f, Il22, and Ccr6 and stimulates their expression (Skepner et al., 2014; Xiao et al., 2014). CCR6 directs Th17 cells to effector mucosal sites (Sinclair et al., 2008; Yamazaki et al., 2008), where they secrete cytokines, including IL-17A, IL-17F, and IL-22. These cytokines act on other immune cells, including neutrophils, to promote clearance of extracellular bacteria, such as Streptococcus pyogenes (Sp), Klebsiella pneumoniae, and Bordetella pertussis (Higgins et al., 2006; Liang et al., 2006, 2007; Aujla et al., 2008; Dileepan et al., 2011; Fan et al., 2014).
Repression of genes that dictate other fates is another important component of Th differentiation. For example, RORγt promotes Th17 differentiation by inhibiting expression of Tbx21 and Foxp3 (Xiao et al., 2014; Fang and Zhu, 2017), which encode proteins that promote Th1 or T reg cell formation, respectively (Szabo et al., 2000; Fontenot et al., 2005). Similarly, the transcription factor BCL6 promotes the Tfh fate by repressing Tbx21 and Rorc to suppress the Th1 and Th17 fates, respectively (Yu et al., 2009). Previous work from our laboratory and others suggests that BCL6 represses genes and promotes the germinal center subset of Tfh cells by recruiting the BCL6 corepressor (BCOR), a component of a variant Polycomb repressive complex 1.1 (PRC1.1; Nance et al., 2015; Yang et al., 2015). BCOR-mediated repression is required for orchestrating many aspects of cellular differentiation (Ng et al., 2004; Wamstad et al., 2008), and although originally named for its interaction with BCL6 (Huynh et al., 2000), BCOR can be recruited independently of BCL6 by other components of PRC1.1 such as KDM2B (Farcas et al., 2012; Wang et al., 2018).
Here, we show that BCOR-mediated repression also facilitates the formation of Th17 cells. We found that the loss of BCOR or KDM2B, but not BCL6, led to a reduction in the formation of Th17 cells after Sp infection. Chromatin immunoprecipitation sequencing (ChIP-seq) and RNA expression analysis revealed that BCOR was bound to and repressed the Lef1, runt-related transcription factor 2 (Runx2), and dual-specificity phosphatase 4 (Dusp4) genes, which encoded proteins that suppress the Th17 cell fate, thereby enhancing Th17 development.
Results
BCOR is required for optimal Th17 differentiation after Sp infection
We previously found that T cell BCOR mutant mice produce fewer of the germinal center subset of Tfh cells and more Th1 cells than WT T cells during an immune response to Listeria monocytogenes (Yang et al., 2015). We compared T cell responses of WT and BCOR mutant T cells to a Th17-inducing pathogen to determine whether BCOR also influences Th17 differentiation. As in our previous study (Yang et al., 2015), we used a Bcor conditional allele, Bcorfl, with Lck-cre to mutate Bcor in T cells. Cre-mediated deletion of this allele removes exons 9 and 10 and results in a premature stop codon. The resulting truncated protein product, if stable, is incapable of incorporation into PRC1.1. We refer to Bcorfl/fl;Lck-cre mice as “BCOR deficient” throughout the paper.
We used Sp infection to generate a robust Th17 response (Dileepan et al., 2011; Ruiz-Romeu et al., 2016). Our studies relied on an engineered Sp strain expressing a model antigenic peptide called 2W (Sp-2W; Rees et al., 1999; Dileepan et al., 2011). Both WT and BCOR-deficient mice are on a C57BL/6 (B6) background and express I-Ab MHCII molecules. The Sp-2W strain was used so that a fluorochrome-labeled 2W:I-Ab tetramer and flow cytometry could be used to monitor the fate of a Th population specific for an epitope from the microbe, because no natural I-Ab–binding Sp epitopes have been discovered.
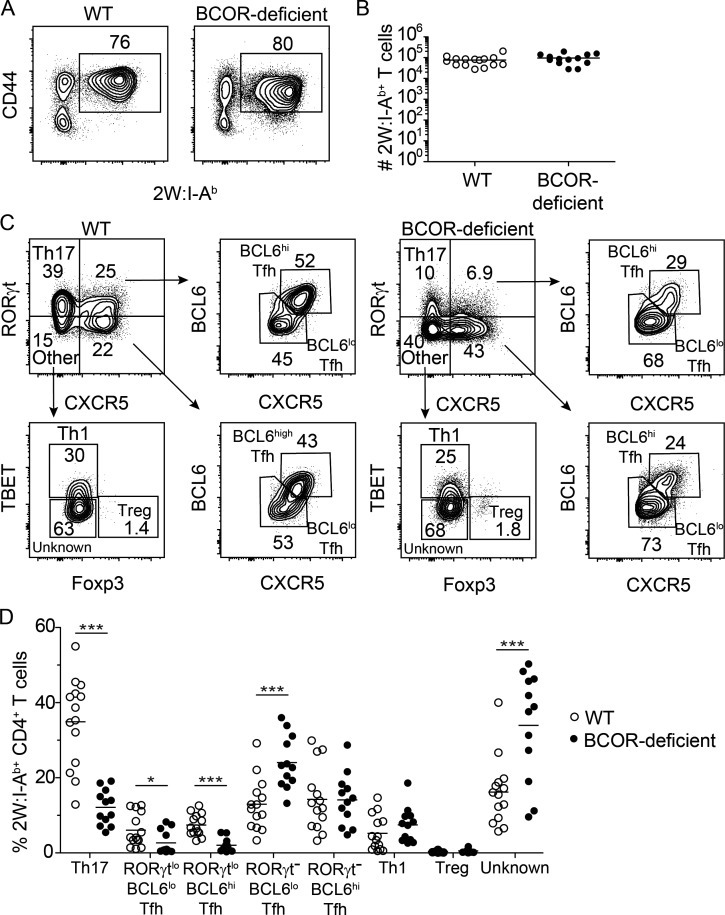
We first determined whether BCOR deficiency affected the clonal expansion of 2W:I-Ab-specific CD4+ T cells. 2W:I-Ab tetramer–based cell enrichment (Moon et al., 2007) was performed to identify 2W:I-Ab–specific CD4+ T cells in spleen and lymph node samples on day 7 after Sp-2W infection. The ∼300 2W:I-Ab–specific CD4+ T cells in uninfected mice (Moon et al., 2007) underwent extensive and similar clonal expansion in WT and T cell BCOR-deficient mice (Fig. 1, A and B).
Figure 1.
BCOR is required for optimal Th17 cell differentiation after Sp infection. WT and Bcorfl/fl;Lck-cre+ (T cell BCOR-deficient) mice were infected with Sp-2W bacteria. After 7 d, 2W:I-Ab–specific CD4+ T cells were enriched from spleen and cervical lymph nodes using 2W:I-Ab tetramer and a magnetic based bead enrichment strategy. (A) B220− CD11b− CD11c− CD4+ T cells from 2W:I-Ab tetramer–enriched samples with gates on CD44+ 2W:I-Ab tetramer+ cells. (B) Numbers of 2W:I-Ab–specific cells in individual WT and Bcorfl/fl;Lck-cre+ mice with horizontal bars at the means. (C) Identification of 2W:I-Ab–specific (from gate in A) RORγt+ CXCR5− Th17, RORγtlo BCL6lo Tfh, RORγtlo BCL6hi Tfh, RORγt− BCL6lo Tfh, RORγt− BCL6hi Tfh, TBET+ Th1, and FOXP3+ T reg cells. (D) Percentages of each subset among 2W:I-Ab–specific cells. Numbers in flow cytometry plots indicate percentages of gated populations. Data shown in all panels are representative of two independent experiments (n = 6–11 mice per group). Student’s t test; *, P < 0.05; ***, P < 0.001.
We then examined CD4+ T cell subsets within the 2W:I-Ab–specific population by staining for the lineage-defining markers RORγt (Th17), CXCR5 (Tfh), BCL6 (Tfh), TBET (Th1), and FOXP3 (T reg; Szabo et al., 2000; Fontenot et al., 2005; Ivanov et al., 2006; Crotty, 2011). Approximately half of the 2W:I-Ab–specific T cells in WT mice did not express CXCR5, and approximately two thirds of these cells were RORγt+ Th17 cells (Fig. 1 C). The CXCR5− cells that lacked RORγt contained some TBET+ Th1 cells, other cells of unknown lineage, and a few FOXP3+ T reg cells. The 2W:I-Ab–specific population in WT mice also contained CXCR5+ Tfh cells, some of which expressed low amounts of RORγt. The RORγtlo and RORγt– Tfh populations contained BCL6lo and BCL6hi subsets (Fig. 1 C) that expressed more BCL6 than CXCR5− cells (data not shown), likely corresponding to germinal center and non-germinal Tfh cells described in other systems (Johnston et al., 2009; Pepper et al., 2011; Liu et al., 2012). In total, the 2W:I-Ab–specific population in WT mice was composed of 35% Th17, 6% RORγtlo BCL6lo Tfh, 8% RORγtlo BCL6hi Tfh, 14% RORγt− BCL6lo Tfh, 15% RORγt− BCL6hi Tfh, 5% Th1, 0.4% T reg cells, and 16% cells of unknown lineage (Fig. 1 D).
The 2W:I-Ab–specific population in T cell BCOR-deficient mice contained significantly smaller fractions of Th17, RORγtlo BCL6lo Tfh, and RORγtlo BCL6hi Tfh cells, a significantly larger fraction of RORγt– BCL6lo Tfh cells and cells of unknown lineage, and the same fraction of RORγt− BCL6hi Tfh, Th1, and T reg cells as WT mice. T cell BCOR deficiency resulted in a more significant reduction in RORγtlo BCL6hi than in RORγtlo BCL6lo Tfh cells. Thus, BCOR was required for maximal formation of Th17 cells and RORγtlo Tfh cells, especially the BCL6hi subset (Fig. 1 D).
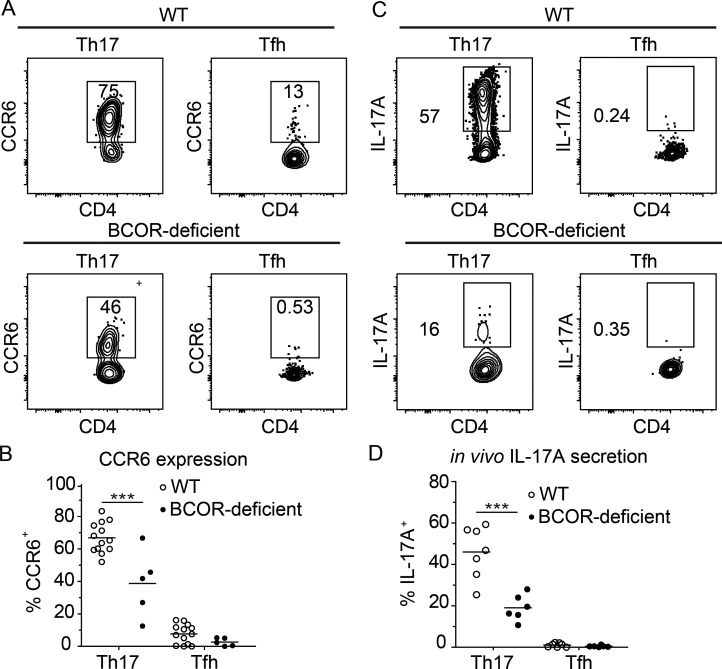
We also determined whether BCOR was required for expression of CCR6 and IL-17, two proteins that provide canonical functions of Th17 cells. Most of the 2W:I-Ab–specific RORγt+ CXCR5− Th17 cells in WT mice on day 7 after Sp-2W infection expressed CCR6 and produced IL-17A 2 h after intravenous injection of 2W peptide (Fig. 2, A–D). BCOR deficiency led to significant reductions in CCR6+ Th17 cells and peptide-induced IL-17A production by Th17 cells. The total CXCR5+ Tfh population containing all the subsets shown in Fig. 1 C was also examined. This population contained very few CCR6+ or IL-17A–producing cells in WT or BCOR-deficient mice. In summary, these data suggest that BCOR is important for Th17 cell formation and function.
Figure 2.
BCOR is important for the induction of canonical Th17 cell–associated chemokine receptor and cytokine expression. WT and BCOR-deficient mice were infected with Sp-2W bacteria, and 2W:I-Ab-specific CD4+ T cells from spleen and cervical lymph nodes were analyzed 7 d later. IL-17A production was assessed 2 h following 2W peptide injection. (A) CCR6 expression by 2W:I-Ab–specific Th17 (RORγt+ CXCR5−) and Tfh (RORγt− CXCR5+) cells. (B) CCR6 percentage of each subset from individual mice identified as in A with horizontal bars at the means. (C) IL-17A secretion by 2W:I-Ab–specific subsets. (D) IL-17A percentage of each subset from individual mice with horizontal bars at the means. Numbers in flow cytometry plots indicate percentages of gated populations. Data shown in all panels are representative of two independent experiments (n = 2–9 mice per group). Student’s t test; ***, P < 0.001.
KDM2B, but not BCL6, is required for optimal Th17 differentiation
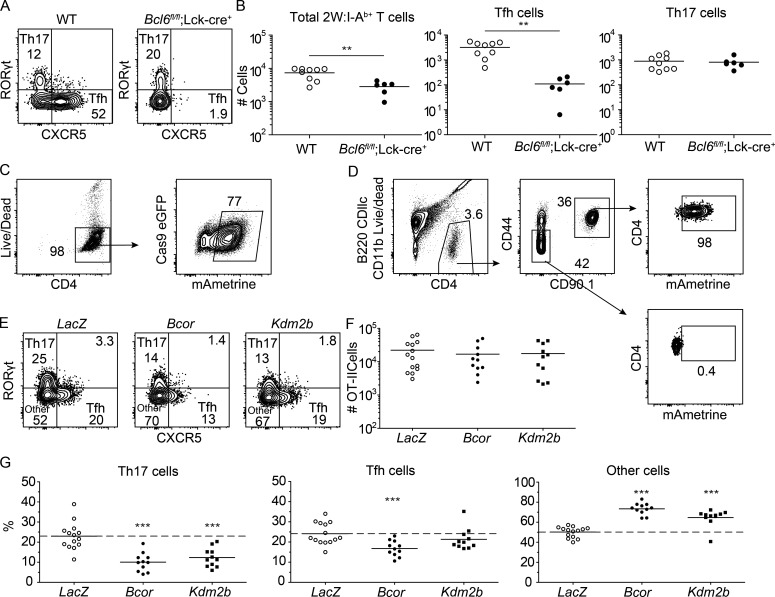
We asked whether BCL6, which is known to directly interact with BCOR (Huynh et al., 2000), promotes Th17 cell differentiation. This possibility was tested by immunizing WT and Bcl6fl/fl;Lck-cre mice, which have a Bcl6 mutation in T cells, with a 2W peptide–phycoerythrin (2W-PE) conjugate in CFA. This immunization strategy was used so that effects on antigen-specific germinal center B cells could be monitored. In WT mice, the 2W-PE immunogen induced 2W:I-Ab–specific Th17 and Tfh cells (Fig. 3 A). The mutation of Bcl6 in T cells led to reduced expansion of 2W:I-Ab–specific CD4+ T cells (Fig. 3 B) and significant defects in the formation of all CXCR5+ Tfh (Fig. 3, A and B) and PE-specific germinal center B cells (data not shown). In contrast, WT and Bcl6fl/fl;Lck-cre mice contained the same number of Th17 cells (Fig. 3 B), indicating that BCOR does not cooperate with BCL6 to promote Th17 differentiation.
Figure 3.
KDM2B, but not BCL6, is required for optimal Th17 cell differentiation. WT and Bcl6fl/fl;Lck-cre+ mice were immunized with 2W-PE in CFA. After 9 d, 2W:I-Ab–specific CD4+ T cells were enriched from spleen and cervical lymph nodes using 2W:I-Ab tetramer and a magnetic based bead strategy. (A) B220− CD11b− CD11c− CD4+ CD44+ 2W:I-Ab–specific T cells from tetramer-enriched samples with gates on RORγt+ Th17 and CXCR5+ Tfh cells. (B) Numbers of total 2W:I-Ab–specific cells, Tfh, and Th17 cells in WT and Bcl6fl/fl;Lck-cre+ mice based on gates in A, with horizontal bars at the means. Cas9-expressing CD90.1+ OT-II transgenic CD4+ T cells were stimulated in vitro and transduced with retroviruses encoding gRNAs and mAmetrine. (C) Transduced CD4+ T cells were sorted based on mAmetrine expression, and 10,000 were transferred into naive B6 mice. After 4 d of in vivo rest, mice were immunized with heat-killed Sp expressing OVA bacteria, and spleen and cervical lymph nodes were taken 6 d later to assess OT-II differentiation. Transduced OT-II cells were enriched from spleen and cervical lymph nodes with CD90.1 antibody. (D) Gating strategy used to identify B220− CD11b− CD11c− CD4+ T cells, CD90.1+ OT-II cells, and CD90.1– CD4+ T cells of the recipient and their expression of mAmetrine. (E and F) Identification of Th17, Tfh, and other cells in LacZ-, Bcor-, or Kdm2b-targeted OT-II cell populations (E) and their numbers (F), with horizontal bars at the means. (G) Percentages of each subset among OT-II cells based on gates in E, with horizontal bars at the means. Numbers in flow cytometry plots indicate percentages of gated populations. Data shown in A and B are representative of two independent experiments (n = 2–6 mice per group). Data shown in C–G are representative of three independent experiments (n = 2–5 mice per group). Student’s t test; **, P < 0.01; ***, P < 0.001.
To confirm the importance of the PRC1.1 complex in Th17 differentiation, we investigated the role of KDM2B, a second unique component of PRC1.1. KDM2B is a histone demethylase and has a CXXC motif that can recruit BCOR/PRC1.1 to nonmethylated CpG islands in other cell types (Farcas et al., 2012; Wang et al., 2018). We adopted an in vitro clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9–based gene targeting approach to test the role of KDM2B in Th17 differentiation in vivo. OT-II TCR transgenic CD4+ T cells expressing Cas9 and a chicken OVA peptide:I-Ab–specific TCR (Robertson et al., 2000) were transduced in vitro with retroviruses encoding the mAmetrine fluorescent protein and guide RNAs (gRNAs) targeting genes of interest. The Il2ra (encoding CD25) and Cd27 genes, which are highly expressed during the in vitro stimulation protocol, were targeted to assess the efficiency of this approach. mAmetrine+ OT-II cells transduced with Il2ra or Cd27 gRNAs had 93% and 83% fewer CD25+ or CD27+ cells, respectively, than mAmetrine+ OT-II cells transduced with irrelevant LacZ gRNAs (data not shown) indicating that this targeting system was very efficient.
This approach was then used to determine the role of KDM2B in Th17 differentiation in vivo. mAmetrine+ Cas9-expressing CD90.1+ OT-II cells were sorted after transduction with retroviruses encoding gRNAs targeting LacZ, Bcor, or Kdm2b and then transferred into naive CD90.2+ B6 mice (Fig. 3 C). The recipients were inoculated intranasally with heat-killed Sp expressing the chicken OVA peptide (Sp-OVA; Park et al., 2004) 4 d after transfer. 6 d later, the OT-II cells were enriched from spleen and cervical lymph node cells after staining with CD90.1 antibody. Heat-killed Sp bacteria were used for these experiments to eliminate the possibility that mice lacking critical gene products in T cells might develop different levels of infection, which could influence Th cell differentiation.
The results of these experiments are shown Fig. 3, D–G. The LacZ-targeted OT-II cells maintained mAmetrine expression (Fig. 3 D) and proliferated and differentiated into RORγt+ Th17 cells, Tfh cells, or uncommitted cells but did not become RORγtlo Tfh cells, as in the case of live Sp infection (Fig. 3 E). Deletion of Kdm2b or Bcor did not affect OT-II cell proliferation (Fig. 3 F). The Bcor-targeted OT-II cell population, however, contained a smaller fraction of Th17 cells than the LacZ-targeted population (Fig. 3 G). Bcor deletion also led to a reduction in Tfh cells, which was only observed for the RORγtlo Tfh subsets during live Sp-2W infection (Fig. 1 D). The Kdm2b-targeted OT-II cell population exhibited reduced Th17 formation with no significant alteration in Tfh differentiation as compared with the LacZ control (Fig. 3, E–G), confirming the importance of the PRC1.1 complex in Th17 differentiation and suggesting that KDM2B may contribute to the recruitment of BCOR to DNA in Th17, but not Tfh, cells. Both Bcor- and Kdm2b-targeted OT-II cell populations had increased fractions of RORγt− CXCR5− other cells (Fig. 3 G), which primarily consists of uncommitted cells (Fig. 1 C). These results indicate that BCOR and KDM2B drive uncommitted cells to become Th17 cells.
Expression profiling discriminates T cell subsets in WT and BCOR-deficient CD4+ T cells
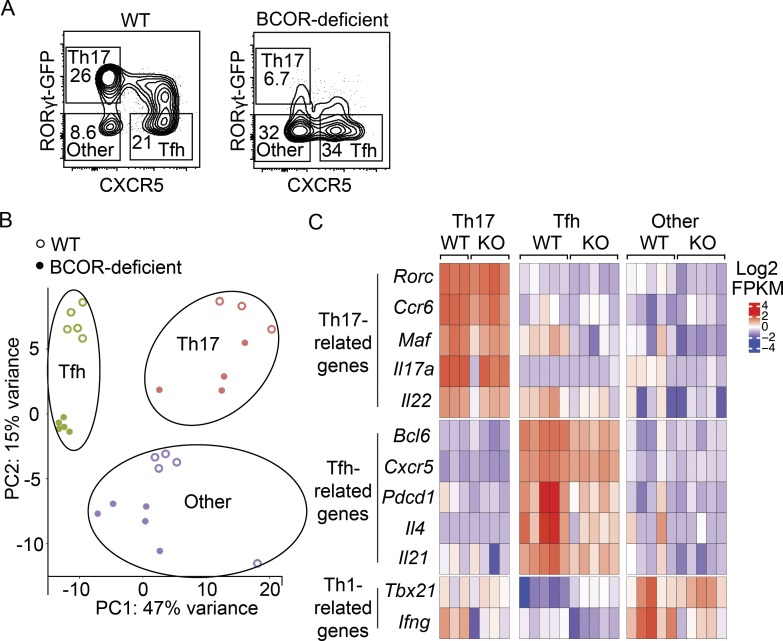
We used genome-wide expression profiling to further characterize the T cell subsets generated upon Sp infection in both WT and BCOR-deficient populations. WT RorcGFP or Bcorfl/fl;Lck-cre+;RorcGFP mice were infected with the Sp-2W bacterial strain, and 7 d later, cells were harvested from the spleen and cervical lymph nodes. 2W:I-Ab tetramer-binding Th17, Tfh, and RORγt− CXCR5− other cells were sorted based on RORγt and CXCR5 expression (Fig. 4 A). The RorcGFP reporter-based strategy led to better resolution of the RORγtlo Tfh cells than RORγt antibody–based intracellular staining used in other experiments. As noted in Fig. 1, however, very few of these cells were present in BCOR-deficient mice, precluding their inclusion in this analysis. Libraries for RNA sequencing were generated, and gene expression values were used for principal component analysis. As shown in Fig. 4 B, WT Th17 cells, Tfh cells, and other subsets formed distinct clusters, and BCOR-deficient Th17 cells, Tfh cells, and other populations formed clusters that were related to but distinct from those formed by the WT populations. The first principal component along the horizontal axis separated the Tfh and Th17 cell subsets and was driven by variation in expression of Ccr2, Adam8, Cxcr5, Nhsl2, and Il17re. The second principal component separated the WT and BCOR-deficient samples within each subset and was driven by variation in Spock2, Ccr6, Il17re, Nkg7, and Rorc (encoding RORγt). The analysis revealed that Th17 cells highly expressed Th17-related genes such as Rorc, Ccr6, and Maf, while Tfh cells highly expressed Tfh-related genes such as Bcl6, Cxcr5, and Pdcd1 (encoding PD-1). The other population expressed higher levels of Tbx21 and Ifng than the Th17 and Tfh populations (Fig. 4 C), consistent with the observation that the other population contains a Th1 subset (Fig. 1, C and D). Taken together, the expression profiles recapitulate the known characteristics of Th17, Tfh, and Th1 cells.
Figure 4.
RNA sequencing of WT and BCOR-deficient CD4+ T cells following Sp infection. WT (RorcGFP) or BCOR-deficient (Bcorfl/fl;Lck-cre+;RorcGFP) mice were infected with Sp-2W bacteria. After 7 d, 2W:I-Ab–specific CD4+ T cell subsets from spleen and cervical lymph nodes were sorted for RNA sequencing analysis. (A) Flow cytometry plots displaying B220− CD11b− CD11c− CD4+ CD44+ 2W:I-Ab–specific T cells from tetramer-enriched samples with gates used to identify Th17, Tfh, and RORγt− CXCR5− other cells. (B) Principal component (PC) analysis of WT (open circles) and BCOR-deficient (closed circles) CD4+ T cell subsets. (C) Centered and scaled Log2 FPKM expression values of canonical markers used to identify Th cell subsets. Numbers in flow cytometry plots indicate percentages of gated populations. Data shown in all panels are representative of two independent experiments (n = 2–3 mice per data point and n = 1–3 data points per group in each experiment).
Identification of genes directly regulated by BCOR
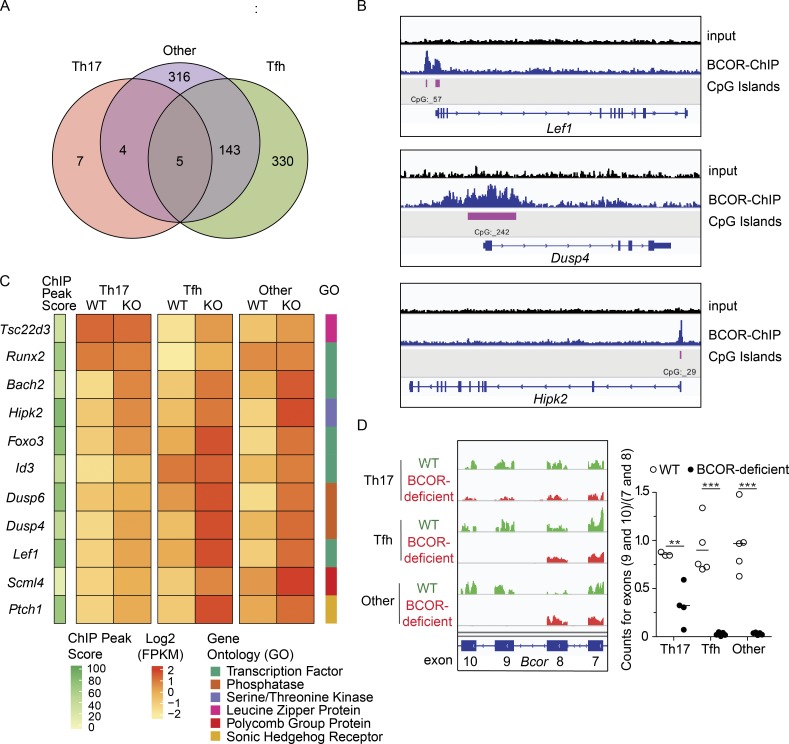
We sought to identify BCOR-regulated genes given the distinct nature of the BCOR-deficient cells in the principal component analysis. We found only 16 differentially expressed genes for the Th17 cell subset, while 478 and 468 genes were identified in the Tfh and RORγt− CXCR5− other populations, respectively (twofold, up or down, and Benjamini–Hochberg adjusted P value < 0.05; Fig. 5 A). Because of the possibility that BCOR enhances Th17 cell formation by acting as a repressor, we focused on the 157 genes that were expressed more highly in BCOR-deficient than WT cells for one or more subsets (twofold up and Benjamini-Hochberg adjusted P value < 0.05). We then used BCOR-specific ChIP-seq to determine which of these genes were direct BCOR targets. WT CD4+ T cells were cultured in vitro for 3 d under Th17-inducing conditions to mimic a situation whereby BCOR represses gene targets in the context of Th17 differentiation. We identified 10,761 BCOR peaks above threshold levels (q-value < 0.05), 82% of which were found at promoters. CpG islands were observed at 83% of the peaks, consistent with the CXXC domain of KDM2B contributing to the recruitment of BCOR. Of the 157 genes that were more highly expressed in BCOR-deficient cells, 123 were directly bound by BCOR in Th17 cells (Table S1).
Figure 5.
RNA expression analysis and BCOR ChIP-seq reveal genes directly regulated by BCOR. (A) Venn diagram displaying the number of BCOR-regulated genes identified by RNA sequencing in the sorted Th subsets shown in Fig. 4 A (twofold, up or down, and Benjamini–Hochberg–adjusted P value < 0.05). (B) Input and BCOR-enriched ChIP tracks for select BCOR-bound genes in WT CD4+ T cells cultured in vitro with Th17-polarizing conditions for 3 d. (C) BCOR-repressed genes identified by RNA sequencing were cross-referenced with BCOR-specific binding peaks from ChIP-seq data, and 11 genes were chosen for further analysis based on expression level (mean counts of BCOR-deficient cells > 150) and potential of the encoded proteins to suppress differentiation by regulating transcription or phosphatase activity. ChIP peak score (10 × −log10 q-value), scaled log2 FPKM values, and gene ontology terms are displayed for each sample. (D) Representative RNA sequencing tracks for BCOR exons 7–10 (left) and splicing efficiency of exons 9–10 (right) with horizontal bars at the means. RNA sequencing data shown in A, C, and D are representative of two independent experiments (n = 2–3 mice per data point and n = 1–3 data points per group). Statistical significance testing to determine the number of differentially expressed genes shown in A was performed using the Wald test in DESeq2, and P values were calculated using the Benjamini–Hochberg correction for multiple testing. ChIP-seq data shown in B and C are representative of two independent experiments (n = 3 mice per experiment). The q-values for each ChIP peak were determined using MACS software. Statistical significance for D was determined using Student’s t test. **, P < 0.01; ***; P < 0.001.
11 of the 123 BCOR-repressed direct target genes (Tsc22d3, Runx2, Id3, Bach2, Scml4, Foxo3, Hipk2, Dusp6, Dusp4, Ptch1, and Lef1) were chosen for further study based on expression level (mean counts in BCOR-deficient cells >150) and because they encoded proteins with the potential to suppress differentiation by regulating transcription or phosphatase activity. BCOR was found at promoters and CpG islands for 10 of the 11 target genes (examples for Lef1, Dusp4, and Hipk2 are shown in Fig. 5 B). BCOR-deficient Tfh and RORγt− CXCR5− other cells expressed these genes to a higher level than the comparable WT subsets (Fig. 5 C). BCOR deficiency led to more modest increases in the expression of these genes in the Th17 cells that formed in the absence of BCOR (Fig. 5 C). This observation led us to investigate the efficiency of Cre-mediated deletion of exons 9 and 10 of Bcor in each subset, since inefficient deletion would result in expression of intact BCOR protein. Indeed, quantification of RNA sequence reads mapping to exons 9 and 10 indicated that the residual Th17 cells in Bcorfl/fl;Lck-cre+ mice retained these exons more often than Tfh or RORγt− CXCR5− other cells (Fig. 5 D). The presence of intact Bcor alleles in some Th17 cells is consistent with the small numbers of significant gene expression changes observed for this subset (Fig. 5 A). This result suggests that many of the Th17 cells that developed in Bcorfl/fl;Lck-cre+ mice arose from precursors that expressed intact BCOR. The enrichment of cells that failed to delete Bcor in the Th17 subset is strong evidence that BCOR is beneficial to their development.
In vivo CRISPR/Cas9 screen reveals novel Th17 regulators
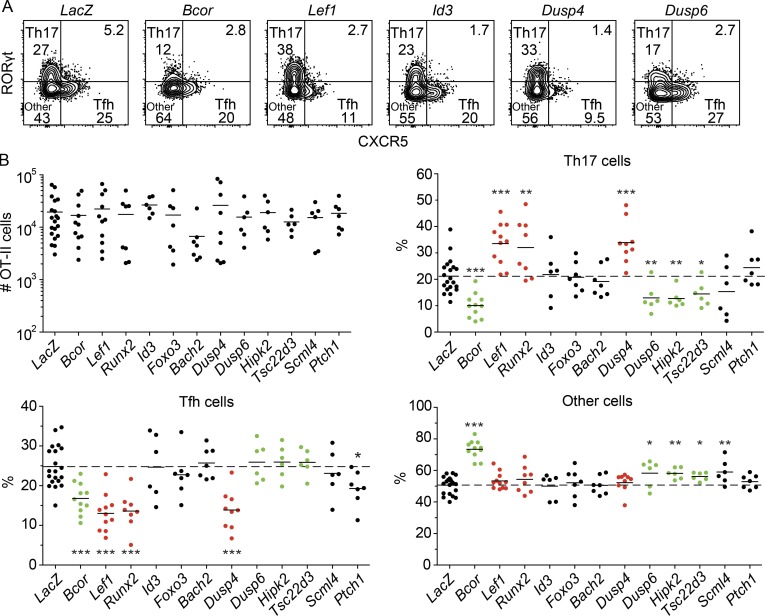
As described above, we found genes that were significantly up-regulated in Tfh and RORγt− CXCR5− cells upon BCOR depletion (Fig. 5 C). We tested the hypothesis that products of these genes suppress the Th17 fate during normal Th cell differentiation. Cas9+ OT-II cells were transduced in vitro with retroviruses expressing mAmetrine and gRNAs targeting each of the 11 genes. mAmetrine+ cells were transferred into recipient mice, which were immunized intranasally with heat-killed Sp-OVA bacteria 4 d after transfer. The transferred OT-II cells were enriched from spleen and cervical lymph node cells after staining with fluorochrome-conjugated CD90.1 antibody and fluorochrome antibody-conjugated magnetic beads. Targeting the BCOR-repressed genes did not result in significant alterations to OT-II cell proliferation when compared with a LacZ control (Fig. 6 B). Ablation of Id3, Foxo3, Bach2, Scml4, and Ptch1 had little to no effect on the differentiation of the OT-II cells. In contrast, disruption of Lef1, Runx2, and Dusp4 enhanced Th17 differentiation, reduced Tfh differentiation, and had no effect on other cells (Fig. 6, A and B). These results indicate that the products of these genes suppress the Th17 fate. BCOR repression of these genes in WT cells would therefore be predicted to enhance Th17 differentiation. Surprisingly, disruption of Dusp6, Hipk2, and Tsc22d3 reduced the number of Th17 cells and increased the differentiation of uncommitted cells (Fig. 6, A and B), indicating that the products of these genes enhance Th17 differentiation in WT cells. These results suggest that BCOR is targeted to some genes that enhance Th17 differentiation and others that suppress it. Enhancement must dominate suppression, however, since the net effect of BCOR deficiency is a reduction in Th17 formation.
Figure 6.
In vivo CRISPR/Cas9 screen reveals novel Th17 regulators. Cas9-expressing OT-II transgenic CD4+ T cells were in vitro stimulated and retrovirally transduced with guides targeting LacZ or the genes listed in Fig 5 C. 10,000 mAmetrine+ transduced CD90.1+ OT-II cells were adoptively transferred into naive B6 mice and immunized with heat-killed Sp-OVA bacteria 4 d later. Spleen and cervical lymph nodes were harvested 6 d after immunization to assess OT-II differentiation. (A) Flow cytometry plots displaying B220− CD11b− CD11c− CD4+ CD44+ CD90.1+ OT-II T cells and Th17 cells, Tfh cells, and other subsets from CD90.1-enriched samples. (B) Numbers of OT-II CD4+ T cells and percentages of each subset in the population, with horizontal bars at the means. Numbers in flow cytometry plots indicate percentages of gated populations. Data shown in all panels are representative of two to four independent experiments (n = 2–5 mice/group). Student’s t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
The CD25–STAT5–BLIMP1 pathway promotes Th17 differentiation
The mechanism of action of DUSP4, the protein product of the Dusp4 gene, during Th17 formation was explored in more detail. DUSP4 is a phosphatase that inhibits the phosphorylation and action of the STAT5 transcription factor (Huang et al., 2012; Hsiao et al., 2015). During Th1 priming conditions, STAT5 phosphorylation is triggered by TCR signaling and induces Il2ra encoding CD25, a component of the IL-2 receptor, which promotes the Th1 fate (Nakajima et al., 1997; Welte et al., 1999; Liao et al., 2011; Johnston et al., 2012). It was therefore possible that STAT5 signaling is also required to promote the Th17 fate during Th17 priming conditions and that BCOR promotes this process by preventing DUSP4 from dephosphorylating STAT5.
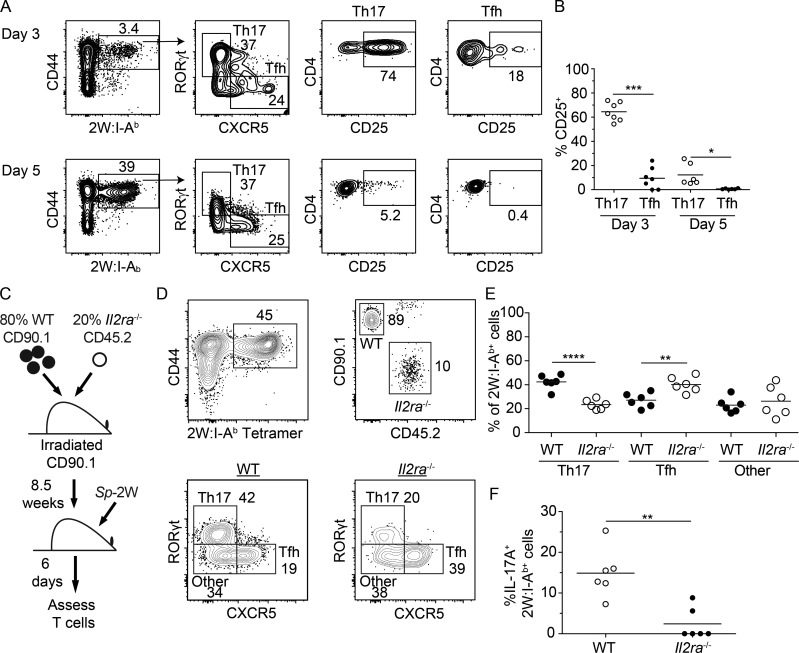
We assessed the plausibility of this model by examining the pattern of CD25 expression at early times after Sp infection. RORγt− CXCR5+ and RORγt+ CXCR5− 2W:I-Ab–specific T cells were already present on day 3 after intranasal infection with live Sp-2W bacteria (Fig. 7 A). Approximately 60% of the RORγt+ CXCR5− cells, but only ∼10% of the RORγt− CXCR5+ cells, expressed CD25 (Fig. 7 B). Although CD25 expression fell by day 5, more RORγt+ CXCR5− than RORγt− CXCR5+ cells retained this molecule. Thus, a much larger fraction of the early Th17 population expressed CD25 than did the early Tfh population.
Figure 7.
CD25 is selectively expressed by Th17 cells early after Sp infection and is important for Th17 cell differentiation. WT mice were infected with Sp-2W bacteria, and 2W:I-Ab–specific CD4+ T cells were enriched from spleen and cervical lymph nodes 3 or 5 d later. (A) Flow cytometry gates used to identify 2W:I-Ab–specific CD4+ T cells, 2W:I-Ab–specific Th17 and Tfh cells, and CD25 expression by 2W:I-Ab–specific Th17 or Tfh cells (right). (B) Percentages of CD25+ 2W:I-Ab–specific Th17 or Tfh cells, with horizontal bars at the means. (C) Diagram of the WT plus Il2ra−/− mixed bone marrow chimera experiment. (D) Flow cytometry gates used to identify 2W:I-Ab–specific WT or Il2ra−/− CD4+ T cells and their differentiation into Th17, Tfh, or other cells 6 d after administration of heat-killed Sp-2W bacteria. (E) Percentages of the 2W:I-Ab–specific WT or Il2ra−/− subsets, with horizontal bars at the means. (F) WT and Il2ra−/− mixed bone marrow chimeras were infected with Sp-2W bacteria for 7 d and challenged with heat-killed Sp-2W bacteria for 3 h before IL-17A expression analysis. Numbers in flow cytometry plots indicate percentages of gated populations. Data shown in all panels are representative of two independent experiments (n = 2–8 mice per group). Student’s t test; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
We then determined whether CD25 was required for Th17 differentiation using a radiation chimera-based approach. A mixture of 80% WT CD45.1+ and 20% Il2ra−/− CD45.2+ bone marrow cells was transplanted into lethally irradiated CD90.1+ B6 mice (Fig. 7 C). This strategy was used to prevent the autoimmunity that occurs in intact Il2ra−/− mice due to T reg cell insufficiency (Sadlack et al., 1995). After reconstitution, these mice were immunized with heat-killed Sp-2W bacteria, and CD44high 2W:I-Ab+ CD4+ T cells were identified 6 d later. WT or Il2ra−/− 2W:I-Ab–specific CD4+ T cells were identified based on congenic markers and classified as Th17, Tfh, or other cells based on the expression of RORγt, CXCR5, or neither (Fig. 7 D). CD25 deficiency reduced Th17 cell differentiation, enhanced Tfh differentiation, and had no effect on the other population (Fig. 7, D and E). Furthermore, CD25 expression was critical for IL-17A production, a function of Th17 cells (Fig. 7 F). These results demonstrate that CD25 expression is beneficial for Th17 differentiation and detrimental for Tfh differentiation after inoculation of the heat-killed Sp bacteria.
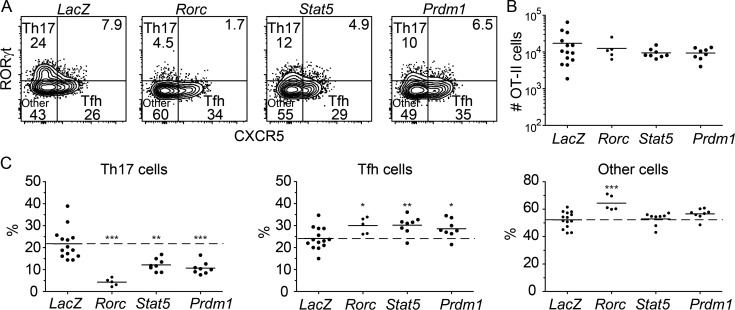
The CRISPR/Cas9-based approach described above was then used to determine whether CD25 and STAT5 enhance Th17 formation in part by preventing the Tfh fate via activation of Prdm1 (BLIMP1) as observed for Th1 cells (Johnston et al., 2012; Nurieva et al., 2012). Rorc, which encodes RORγt, the master transcription factor for Th17 cells (Ivanov et al., 2006), was also targeted to serve as a control. As expected, Rorc-targeted OT-II cells exhibited reduced Th17 differentiation and enhanced differentiation of Tfh and RORγt− CXCR5− other cells (Fig. 8, A–C). Ablation of Stat5 or Prdm1 had similar effects. These results demonstrate that STAT5 and BLIMP1 are required for optimal Th17 cell differentiation and raise the possibility that DUSP4 acts as a Th17 fate suppressor by inhibiting the CD25–STAT5–BLIMP1 signaling pathway.
Figure 8.
The STAT5–BLIMP1 pathway promotes Th17 differentiation. Cas9-expressing CD90.1+ OT-II transgenic CD4+ T cells were transduced in vitro with gRNAs targeting LacZ, Rorc, Stat5, or Prdm1 before in vivo transfer to assess differentiation in response to heat-killed Sp-OVA bacteria as in Fig. 6. (A) Flow cytometry plots used to identify B220− CD11b− CD11c− CD4+ CD44+ CD90.1+ Th17, Tfh, and other OT-II T cells from CD90.1-enriched samples. (B) Numbers of OT-II CD4+ T cells, with horizontal bars at the means. (C) Percentages of OT-II cells in each subset, with horizontal bars at the means. Numbers in flow cytometry plots indicate percentages of gated populations. Data shown in all panels are representative of two independent experiments (n = 2–8 mice per group). Student’s t test; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Discussion
We found that Sp infection causes Sp epitope–specific naive CD4+ T cells to differentiate into CXCR5− non-Tfh effector cells as observed in Listeria monocytes, lymphocytic choriomeningitis virus, and influenza A virus infections (Johnston et al., 2009; Pepper et al., 2011; Ballesteros-Tato et al., 2012). The non-Tfh cells induced by the L. monocytes infection, however, are TBET+ Th1 cells (Pepper et al., 2011), whereas the Sp-induced non-Tfh cells are RORγt-dependent Th17 cells. This difference is probably related to the fact that Sp epitope–specific naive CD4+ T cells are stimulated by IL-6– and TGF-β–producing dendritic cells in nasal-associated lymph nodes, whereas naive T cells are stimulated by IL-12–producing dendritic cells in the spleen in the other infections (Linehan et al., 2015). Despite this difference, the non-Tfh cells induced in all these infections depend on CD25. Thus, epitope-specific Th cells that express the largest amounts of CD25 may experience IL-2 receptor–induced phosphorylation of STAT5, which induces BLIMP1, a repressor of the Tfh pathway (Johnston et al., 2009). CD25high cells that are denied the Tfh option would then be free to become Th17 or Th1 cells depending on which innate immune cytokines are present. The CD25 dependence of Th2 cells induced by house dust mite inhalation (Hondowicz et al., 2016) indicates this model also applies to this Th cell subset. It may, however, not apply to the in vitro situation, since STAT5 signaling inhibits Th17 cell formation (Laurence et al., 2007) in a cell culture system.
Some Sp epitope–specific Th cells, presumably those that expressed lesser amounts of CD25 early after infection, became CXCR5+ Tfh cells. The Tfh population consisted of BCL6hi germinal center and BCL6lo subsets, as observed during other infections (Johnston et al., 2009; Pepper et al., 2011; Liu et al., 2012). Unlike infections that also induce Th1 cells, however, some of the Sp-induced Tfh cells expressed RORγt, although in lower amounts than Th17 cells. Sp-infected lymphoid tissues may contain microenvironments in which IL-6, TGF-β, and Tfh-inducing cytokines are all produced, thereby causing some Sp epitope–specific Th cells to acquire characteristics of Th17 and Tfh cells. This finding, together with reports on hybrid Tfh cells in other systems (Crotty, 2014), suggests that the lines of Tfh lineage commitment can be blurry.
We found that BCOR is essential for the optimal differentiation of Sp-induced Th17 cells. Unlike RORγt, which primarily binds to and activates the genes that define the Th17 lineage, BCOR promoted the Th17 fate by repressing genes that encode proteins that promote the differentiation of Tfh cells and other subsets. The fact that T cell BCOR deficiency causes increases in Tfh cells, as Th17 cells are lost, supports this contention. The significant occupancy of BCOR at CpG islands in Th17 cells suggests that BCOR recruitment to DNA is based on its interaction with KDM2B, which contains a CXXC DNA-binding domain that specifically targets unmethylated CpG nucleotides (Farcas et al., 2012). This result, and the fact that ablation of Kdm2b reduced Th17 formation, suggest that KDM2B recruits BCOR to unmethylated CpG islands in Th17 cells to repress genes encoding proteins that promote other fates. BCOR–KDM2B complexes may be mainly responsible for alternate fate suppression in Th17 cells, because the relevant target genes become unmethyated under Th17 polarizing conditions, and Th17 cells do not express the other BCOR partner, BCL6.
Maximal formation of RORγtlo Tfh cells, and especially RORγtlo BCL6hi Tfh cells, was also dependent on BCOR. Differential BCOR dependence of Tfh subsets was previously observed with L. monocytogenes infection, which generates BCL6lo and BCL6hi Tfh that express small amounts of TBET instead of RORγt. The BCL6hi, but not the BCL6lo, Tfh cells induced by L. monocytogenes infection are dependent on BCOR (Yang et al., 2015). The RORγtlo BCL6hi Tfh cells in Sp infection and the TBETlo BCL6hi Tfh cells in L. monocytogenes infection may depend on BCOR–BCL6, BCOR–KDM2B, or both complexes. In contrast, the RORγt− Tfh cells in Sp infection and BCL6lo Tfh cells in L. monocytogenes infection appear to depend only on BCOR–BCL6 complexes, perhaps because the requisite target genes are methylated and thus unavailable to KDM2B.
ChIP-seq and CRISPR/Cas9 targeting studies suggest that BCOR is targeted by KDM2B to unmethylated CpG islands in the Lef1, Runx2, and Dusp4 genes, the products of which promote Th17 formation by suppressing Th17 inhibitors. The fact that the Lef1, Runx2, and Dusp4 gene products have different functions indicates that BCOR-mediated repression facilitates Th17 cell formation via multiple pathways. For example RUNX2, which promotes the Tfh fate in the context of lymphocytic choriomeningitis virus infection (Choi et al., 2015), could activate transcription of genes encoding proteins that repress Th17 cell differentiation. In contrast, BCOR-mediated repression of Dusp4 could enhance Th17 cell formation by reducing DUSP4 and dephosphorylation of STAT5, thereby increasing BLIMP1 and preventing pre-Th17 cells from becoming Tfh cells.
Although we found that BCOR enhances Th17 cell formation by repressing genes that suppress the Th17 cell fate, we also identified BCOR target genes such as Dusp6, Hipk2, and Tsc22d3, which encoded proteins that enhanced Th17 formation. The role of DUSP6 in promoting the Th17 fate is consistent with previous findings showing that Dusp6−/− CD4+ T cells have reduced production of the Th17 cytokine IL-17A after in vitro stimulation (Bertin et al., 2015). The roles of homeodomain-interacting protein kinase 2 (HIPK2) and TSC22 domain family member 3 (TSC22D3) will require additional studies to understand their importance in the context of Th17 differentiation. In any case, although BCOR represses genes encoding proteins that promote and suppress the Th17 cell fate, the fact that BCOR is required for optimal Th17 cell formation suggests that its dominant activity in Th cell differentiation is to repress genes that inhibit the Th17 cell fate.
Materials and methods
Mice
Mice with a conditional Bcor allele (Bcorfl) were generated by homologous recombination (unpublished data). Bcorfl mice contain LoxP sites flanking Bcor exons 9 and 10 on the X chromosome, and Cre-mediated deletion causes a premature stop codon and a null allele. Bcorfl/+ mice were fully backcrossed with B6 mice (NCI Frederick) for over 10 generations. Bcorfl/+ mice were bred with Lck-cre mice (B6.Cg-Tg(Lck-icre)3779Nik/J; stock 012837; The Jackson Laboratory; Wang et al., 2001) to generate WT (Bcorfl/fl;Lck-Cre− or Bcorfl/Y;Lck-Cre−) and BCOR T cell–deficient (Bcorfl/fl;Lck-Cre+ or Bcorfl/Y;Lck-Cre+) mice. Bcorfl/fl;Lck-Cre+/− mice were bred with RorcGFP reporter mice (B6.129P2(Cg)-Rorctm2Litt/J; stock 007572; The Jackson Laboratory; Eberl et al., 2004) to generate Bcorfl/fl;Lck-cre−;RorcGFP or Bcorfl/fl;Lck-cre+;RorcGFP reporter mice for cell sorting and RNA sequencing analysis. Bcl6fl/fl mice (B6.129S(FVB)-Bcl6tm1.1Dent/J; stock 023727; The Jackson Laboratory; Hollister et al., 2013) were bred with Lck-cre mice described above to generate Bcl6fl/fl;Lck-Cre+ mice. Cas9 eGFP+ mice (B6(C)-Gt(ROSA)26Sorem1.1(CAG-cas9*,-EGFP)Rsky/J; stock 028555; The Jackson Laboratory; Chu et al., 2016) were crossed with CD90.1+ OVA peptide 323–339:I-Ab–specific (OT-II) Rag1−/− TCR transgenic mice (Barnden et al., 1998) to generate CD90.1+ Cas9 eGFP+ OT-II Rag1−/− mice for CRISPR experiments. CD45.1 (B6.SJL-PtprcaPepcb/BoyJ; stock 002014; The Jackson Laboratory), CD90.1 (B6.PL-Thy1a/CyJ; stock 000406; The Jackson Laboratory), and Il2ra−/− mice (B6;129S4-Il2ratm1Dw/J; stock 002462; The Jackson Laboratory; Willerford et al., 1995) were used for bone marrow chimera experiments. All mice were housed in specific pathogen–free conditions at the University of Minnesota. All experimental protocols were performed in accordance with guidelines of the University of Minnesota Institutional Animal Care and Use Committee and National Institutes of Health.
Bone marrow chimera
Bones from CD45.1+ WT and CD45.2+ Il2ra−/− mice were crushed separately with a mortar and pestle in PBS to prepare bone marrow cells. Bone marrow cells were resuspended in 10% DMSO in FBS (Omega), aliquoted, and frozen. Frozen aliquots were thawed, spun down in DMEM containing 10% FBS, Pen Strep (Gibco), and Glutamax (Gibco), and resuspended in PBS. For the experiment shown in Fig. 7 C, recipient CD90.1+ mice, ≥8 wk of age, were irradiated with 2 × 500 rad with 6 h between doses. After the second irradiation, each CD90.1+ recipient was retro-orbitally injected with 4 × 106 CD45.1+ WT and 106 Il2ra−/− CD45.2+ bone marrow cells. Alternatively, 1:1 WT and Il2ra−/− mixed bone marrow chimeras were made for the experiment shown in for Fig. 7 F. After reconstitution, mice were infected with Sp-2W bacteria and challenged with heat-killed Sp-2W bacteria for 3 h before analysis of IL-17A expression.
Infections and immunizations
Male or female mice 6–12 wk of age were infected intranasally with 10 μl of 2 × 108 CFUs per mouse of live Sp expressing 2W (Sp-2W) peptide in PBS (Dileepan et al., 2011). Alternatively, mice were immunized intranasally with 15 μl of 3 × 108 CFUs of heat-killed Sp-2W bacteria (Dileepan et al., 2011) or Sp expressing OVA323–339 peptide (Sp-OVA) in PBS (Park et al., 2004).
CRISPR/Cas9 and retroviral transductions
Interpro was used to identify functional motifs within target genes, and two or three gRNAs targeting the functional motifs for each gene were designed using Benchling (Finn et al., 2017). Primers encoding the gRNA were generated using the Voytas laboratory toolkit, and the cloning of the transfer RNA (tRNA)–gRNA array was performed as described previously (Čermák et al., 2017), with modifications as follows. An MSCV-based gamma retroviral vector was created for generating gRNA delivery. This vector involves modification of the LMP-Amt vector (Chen et al., 2014), a gift from S. Crotty (La Jolla Institute, La Jolla, CA). The SapI cut site in the vector was disrupted by mutation. Then, the shRNA-encoding segment was replaced with cytotoxic protein CCDB bacterial gene flanked by SapI cut sites using In-Fusion cloning (Takara Bio). The CCDB gene derived from the pMOD_B2303 plasmid from D. Voytas (University of Minnesota, Minneapolis, MN). The PGK promoter present in the LMP-Amt vector was replaced with an internal ribosome entry site (plasmid 52109; Addgene) to generate the pMCIA vector. A golden gate reaction was performed to generate the tRNA–gRNA arrays (Xie et al., 2015; Dong et al., 2017; Xu et al., 2017) using AarI (Thermo Fisher Scientific) and T4 DNA ligase (New England Biolabs). The product was purified with a MinElute PCR Purification Kit (Qiagen), and a second golden gate reaction was performed using SapI (New England Biolabs) and T4 DNA ligase (New England Biolabs) to replace the CCDB gene in the pMCIA vector with the tRNA–gRNA arrays made from the first golden gate reaction.
Retroviral supernatant was prepared as previously described (Choi et al., 2013), with some alterations. Briefly, Platinum-E cells (Cell Biolabs) were plated in 6-well plates the evening before transfection. Platinum-E cells were grown in D10 media containing DMEM media (Gibco) supplemented with 10% FBS (Omega), penicillin/streptomycin (Life Technologies), Glutamax (Life Technologies), β-mercaptoethanol (Sigma), Hepes (Life Technologies), sodium pyruvate (Life Technologies), MEM nonessential amino acids (Life Technologies), and Plasmocin prophylactic (Invivogen). The following morning, media was replaced and after 1–2 h, a transfection solution was added dropwise to the cells. The transfection solution contained DMEM media (Gibco) with polyethylenimine (linear, mol wt 25,000) for transfection (Polysciences), pCL-Eco, and plasmid DNA containing gRNA. After 5–6 h, the media was aspirated and D10 media supplemented with ViralBoost reagent (Alstem Cell Advancements) and 30 μM water-soluble cholesterol (Sigma) was added to the cells. After 24 and 48 h, viral supernatant was collected, combined, filtered through a 0.45-µm nylon 25-mm syringe filter (Thermo Fisher Scientific), and frozen at −80°C for retroviral transductions.
Retroviral transduction of CD4+ T cells was performed as previously described (Choi et al., 2013), with some alterations. Cas9 eGFP+ CD90.1+ OT-II Rag1−/− CD4+ T cells were isolated using a negative CD4+ T cell isolation kit (Stemcell Technologies) and incubated in a 96-well plate coated with anti-CD3 (8 µg/ml; 2C11; Bio X Cell), anti-CD28 (8 µg/ml; 37.51; Bio X Cell), and IL-7 (2 ng/ml; Tonbo Biosciences). Cells were grown in I10 media containing IMDM (Sigma) supplemented with 10% FBS (Omega), penicillin/streptomycin (Life Technologies), Glutamax (Life Technologies), β-mercaptoethanol (Sigma), Hepes (Life Technologies), sodium pyruvate (Life Technologies), MEM nonessential amino acids (Life Technologies), and Plasmocin prophylactic (Invivogen). At ∼24 and 40 h after stimulation, two spin transductions were conducted by removing the media, adding retroviral supernatant, and spinning the cells at 1,500 rpm for 2 h at 37°C. After transductions, retroviral supernatants were replaced with I10 media containing IL-2 (10 ng/ml; Peprotech). Approximately 24 h after the second transduction, cells were removed from the anti-CD3– and anti-CD28–stimulating conditions and split into a 24-well plate. The next day, additional IL-2–containing media was added, and the day after that, the cells are washed out of the IL-2–containing media and resuspended in I10 media containing IL-7 (2 ng/ml; Tonbo Biosciences). After 24 h, OT-II cells were cell sorted for guide-mAmetrine expression using a FACS Aria (BD). Then 10,000 mAmetrine+ OT-II cells were transferred into B6 mice. After 4 d of in vivo rest, the mice were immunized with heat-killed Sp-2W bacteria, and organs were harvested 6 d later for processing as described below.
Cell enrichment and flow cytometry
Spleen and cervical lymph node cells were stained with CXCR5 antibody (L138D7; BioLegend) and fluorochrome-labeled I-Ab tetramers containing 2W (2W:I-Ab) for 1 h at room temperature (Moon et al., 2007). For the detection of CD90.1+ OT-II CD4+ T cells, suspensions were incubated with CD90.1 antibody (OX-7; BioLegend) for 8 min at room temperature and then CXCR5 antibody for 1 h at room temperature. Cell suspensions were enriched for 2W:I-Ab–specific or CD90.1+ CD4+ T cells using the EasySep Mouse fluorochrome positive selection kit (Stemcell Technologies). Briefly, cells were incubated with 12.5 μl of EasySep fluorochrome selection cocktail per sample for 15 min at room temperature and 12.5 μl of EasySep magnetic particles per sample for 10 min at room temperature. The magnetically bound cells were isolated using magnets (Stemcell Technologies) according to the manufacturer’s protocol. The positively enriched CD4+ T cells were surface stained with fluorochrome-labeled antibodies specific for B220 (RA3-6B2; all antibodies are from eBioscience unless otherwise indicated), CD11b (M1/70), CD11c (N418), CD44 (IM7), CD27 (LG.3A10; BD), and CD4 (GK1.5; BD). For intracellular staining, cells were incubated in intracellular fixation and permeabilization buffer (eBioscience) for 1 h at room temperature and stained with fluorochrome-labeled RORγt (Q31-378; BD), BCL6 (K112-91; BD), TBET (4B10; BioLegend), and FOXP3 (FJK-16s) antibodies in permeabilization buffer (Tonbo Biosciences) overnight.
For in vivo cytokine secretion analysis, mice were injected intravenously with 100 μg of 2W peptide or heat-killed Sp-2W bacteria. Cells from the spleen and cervical lymph nodes were harvested 2 or 3 h later and processed in Brefeldin A (eBioscience)–containing FACS media for all steps before fixation with Cytofix/Cytoperm (BD) for 15 min at 4°C and staining with fluorochrome-labeled anti–IL-17A antibody (TC11-18H10; BD) overnight. Cells were analyzed on a Fortessa (Becton Dickinson) flow cytometer and analyzed with FlowJo (TreeStar).
RNA sequencing
Bcorfl/fl;Lck-cre−;RorcGFP or Bcorfl/fl;Lck-cre+;RorcGFP reporter mice were intranasally infected with Sp-2W bacteria, and 2W:I-Ab–specific CD4+ T cells were isolated 7 d later and surface stained as described above. Th17 (RORγt+ CXCR5−), Tfh (RORγt− CXCR5+), and RORγt− CXCR5− 2W:I-Ab–specific CD4+ T cells were individually sorted with a FACS Aria (BD) into tubes containing Trizol (Invitrogen). Total RNA was isolated from the Trizol samples and treated with DNase using an RNA isolation kit (Qiagen). Sequencing libraries were made from PolyA+ mRNA using the KAPA mRNA Hyper Prep kit per the manufacturer’s instructions (KAPA Biosystems). The libraries were submitted to the University of Minnesota Genomics Center for 2 × 50-bp sequencing using the High-Output HiSeq 2500 (Illumina). Demultiplexed reads were trimmed with Trimmomatic (v0.32, parameters: ILLUMINACLIP:TruSeq3-PE.fa:2:30:10 LEADING:3 TRAILING:3 SLIDINGWINDOW:4:5 MINLEN:25). Trimmed reads were mapped to the GRCm38 version of the mouse genome with STAR (v2.4.2a_modified) and indexed and sorted with samtools (v1.0). PCR duplicates were removed using Picard MarkDuplicates (2.17.10-SNAPSHOT). Reads that overlap features in the Gencode M16 annotation were enumerated with Rsubread (1.22.3). Differential expression analysis was performed with R (v3.5.1) and DESeq2 (v1.20.0) using custom scripts (available at https://github.com/micahgearhart/th17). Raw sequence data, count tables, and normalized fragments per kilobase of transcript per million (FPKM) values are available in the Gene Expression Omnibus under accession number GSE121766.
ChIP-seq of in vitro Th17 cells
CD4+ T cells were isolated from B6 mice using a CD4 T cell positive isolation kit (Stemcell) and stimulated with Th17 polarizing conditions as follows: plate-coated CD3 (4 µg/ml; 2C11; Bio X Cell) and CD28 (4 µg/ml; 37.51; Bio X Cell) antibodies, IL-6 (60 ng/ml; Tonbo Biosciences), TGF-β1 (0.8 ng/ml; Tonbo Biosciences), IL-23 (20 ng/ml; Miltenyi), IL-1β (20 ng/ml; Tonbo Biosciences), IFN-γ (10 µg/ml; XMG1.2), anti–IL-12 (10 µg/ml; C17.8; Tonbo Biosciences), and anti–IL-4 (10 µg/ml; 11B11; Tonbo Biosciences) antibodies. Cells were grown in IMDM (Sigma) supplemented with 10% FBS (Omega), penicillin/streptomycin (Life Technologies), Glutamax (Life Technologies), β-mercaptoethanol (Sigma), Hepes (Life Technologies), sodium pyruvate (Life Technologies), and MEM nonessential amino acids (Life Technologies). After 3 d, the cells were washed with PBS two times and resuspended in PBS at a concentration of 10 million cells per milliliter. DNA and proteins were cross-linked by incubating cells with 1.5 mM ethylene glycol bis(succinimidyl succinate) (Thermo Fisher Scientific) for 30 min at room temperature on a rotator. Paraformaldehyde was added at a final concentration of 1% during the last 10 min. The cross-linking reaction was quenched by adding 2.5 M glycine at a 1:20 dilution and washed two times with PBS. After the last spin, media was completely removed, and the cell pellet was frozen using dry ice and isopropanol and kept at −80°C until shearing. The truChIP Chromatin Shearing Reagent kit (Covaris) was applied to lyse the cells, isolate nuclei, and shear chromatin with the AFA Focused-ultrasonicator per the manufacturer’s instructions. An aliquot was taken for the input. The rest of the sample was enriched for BCOR-bound DNA by incubating sheared chromatin overnight with Protein A Dynabeads (Invitrogen) bound to rabbit polyclonal mouse BCOR antibody (RRID: AB_2750631). The sample was washed using the DynaMag-2 Magnet (Invitrogen) and buffers in the following order: low-salt buffer (0.1% SDS, 1.0% Triton X-100, 2 mM EDTA, 20 mM Tris-HCL, pH 8.0, and 150 mM NaCl), high-salt buffer (0.1% SDS, 1.0% Triton X-100, 2 mM EDTA, 20 mM Tris-HCL, pH 8.0, and 500 mM NaCl), lithium chloride buffer (0.25 M lithium chloride, 1% NP-40 or IGEPAL-CA630 1% deoxycholate (DOC), 1 mM EDTA, and 10 mM Tris, pH 8.0), Morohashi RIPA buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 5 mM EDTA, 0.5% NP-40, and 0.1% SDS), DOC/Triton buffer (25 mM Tris pH 7.5, 150 mM NaCl, 5 mM EDTA, 1% Triton-X-100, and 0.5% DOC), and TE buffer (10 mM Tris-HCL, pH 8.0, and 1 mM EDTA). DNA was eluted off the magnetic beads using elution buffer (1% SDS, 100 mM NaHCO3). Cross-links were reversed for the BCOR-enriched DNA and input samples by incubating with 200 mM NaCl at 55°C overnight. DNA was purified using a PCR purification kit (Qiagen), and cDNA sequencing libraries were made using the KAPA Hyper Prep kit per the manufacturer’s instructions (KAPA Biosystems). The libraries were submitted to the University of Minnesota Genomics Center for 1 × 50-bp sequencing using the High-Output HiSeq 2500 (Illumina). Reads were trimmed (Trimmomatic 0.32), mapped to the GRCm38 reference genome (Burrows-Wheeler Aligner mem 0.7.12-r1039), and sorted and indexed (Samtools 1.0). Peaks were identified using MACS version 2.1.1.20160309 using the --broad --broad-cutoff 0.1 options. BCOR peak locations were analyzed in relationship to the Gencode M16 reference annotation and the University of California–Santa Cruz custom-defined CpG islands using custom R scripts, which are available at https://github.com/micahgearhart/th17. Raw sequence data, coverage tracks, and peak lists are available in the Gene Expression Omnibus under accession number GSE121766.
Statistical analysis
Statistical tests were performed using Prism (GraphPad) software, and P values were obtained using two-tailed unpaired t tests with a 95% confidence interval. Statistical significance testing for RNA sequencing data were performed using the Wald test in DESeq2, and P values were calculated using the Benjamini–Hochberg correction for multiple testing. For the ChIP-seq data, q-values for each peak were determined using MACS software.
Online supplemental material
Table S1 lists the direct BCOR-repressed genes from RNA sequencing and ChIP-seq coanalysis.
Supplementary Material
Acknowledgments
The authors thank Jennifer Walter and Charles Ellwood for technical assistance and all members of the Jenkins lab for helpful discussions. The authors also thank Jason Motl at the University of Minnesota Flow Cytometry Facility for cell sorting and the University of Minnesota Genomics Center for sequencing RNA and ChIP libraries.
This work was supported by the National Institutes of Health (grants R01 AI039614, R01 AI103760, and R37 AI027998 to M.K. Jenkins; grants T32 AI007313 and F31 AI133716 to J.A. Kotov; grants T32 AI83196 and T32 AI007313 to D.I. Kotov; and grants R01 CA071540 and R01 HD084459 to V.J. Bardwell). This work was also supported by the University of Minnesota Microbiology and Immunology Department (Dennis W. Watson Fellowship to J.A. Kotov).
The authors declare no competing financial interests.
Author contributions: J.A. Kotov designed and performed all of the experiments, conducted bioinformatics analyses, and wrote the paper. D.I. Kotov provided expertise for CRISPR/Cas9 knockout experiments. J.L. Linehan conducted the bone marrow chimera experiment in Fig. 7 E, bottom panel. V.J. Bardwell and M.D. Gearhart oversaw and provided expertise for the RNA and ChIP sequencing experiments. M.D. Gearhart performed and oversaw bioinformatics analyses. M.K. Jenkins conceptualized and oversaw all experiments. All authors listed reviewed and provided edits to the manuscript.
References
- Aujla S.J., Chan Y.R., Zheng M., Fei M., Askew D.J., Pociask D.A., Reinhart T.A., McAllister F., Edeal J., Gaus K., et al. . 2008. IL-22 mediates mucosal host defense against Gram-negative bacterial pneumonia. Nat. Med. 14:275–281. 10.1038/nm1710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballesteros-Tato A., León B., Graf B.A., Moquin A., Adams P.S., Lund F.E., and Randall T.D.. 2012. Interleukin-2 inhibits germinal center formation by limiting T follicular helper cell differentiation. Immunity. 36:847–856. 10.1016/j.immuni.2012.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnden M.J., Allison J., Heath W.R., and Carbone F.R.. 1998. Defective TCR expression in transgenic mice constructed using cDNA-based alpha- and beta-chain genes under the control of heterologous regulatory elements. Immunol. Cell Biol. 76:34–40. 10.1046/j.1440-1711.1998.00709.x [DOI] [PubMed] [Google Scholar]
- Bertin S., Lozano-Ruiz B., Bachiller V., García-Martínez I., Herdman S., Zapater P., Francés R., Such J., Lee J., Raz E., and González-Navajas J.M.. 2015. Dual-specificity phosphatase 6 regulates CD4+ T-cell functions and restrains spontaneous colitis in IL-10-deficient mice. Mucosal Immunol. 8:505–515. 10.1038/mi.2014.84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettelli E., Carrier Y., Gao W., Korn T., Strom T.B., Oukka M., Weiner H.L., and Kuchroo V.K.. 2006. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 441:235–238. 10.1038/nature04753 [DOI] [PubMed] [Google Scholar]
- Čermák T., Curtin S.J., Gil-Humanes J., Čegan R., Kono T.J.Y., Konečná E., Belanto J.J., Starker C.G., Mathre J.W., Greenstein R.L., and Voytas D.F.. 2017. A multipurpose toolkit to enable advanced genome engineering in plants. Plant Cell. 29:1196–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R., Bélanger S., Frederick M.A., Li B., Johnston R.J., Xiao N., Liu Y.C., Sharma S., Peters B., Rao A., et al. . 2014. In vivo RNA interference screens identify regulators of antiviral CD4(+) and CD8(+) T cell differentiation. Immunity. 41:325–338. 10.1016/j.immuni.2014.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y.S., Yang J.A., Yusuf I., Johnston R.J., Greenbaum J., Peters B., and Crotty S.. 2013. Bcl6 expressing follicular helper CD4 T cells are fate committed early and have the capacity to form memory. J. Immunol. 190:4014–4026. 10.4049/jimmunol.1202963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y.S., Gullicksrud J.A., Xing S., Zeng Z., Shan Q., Li F., Love P.E., Peng W., Xue H.H., and Crotty S.. 2015. LEF-1 and TCF-1 orchestrate T(FH) differentiation by regulating differentiation circuits upstream of the transcriptional repressor Bcl6. Nat. Immunol. 16:980–990. 10.1038/ni.3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu V.T., Weber T., Graf R., Sommermann T., Petsch K., Sack U., Volchkov P., Rajewsky K., and Kühn R.. 2016. Efficient generation of Rosa26 knock-in mice using CRISPR/Cas9 in C57BL/6 zygotes. BMC Biotechnol. 16:4 10.1186/s12896-016-0234-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty S. 2011. Follicular helper CD4 T cells (TFH). Annu. Rev. Immunol. 29:621–663. 10.1146/annurev-immunol-031210-101400 [DOI] [PubMed] [Google Scholar]
- Crotty S. 2014. T follicular helper cell differentiation, function, and roles in disease. Immunity. 41:529–542. 10.1016/j.immuni.2014.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derynck R., and Zhang Y.E.. 2003. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 425:577–584. 10.1038/nature02006 [DOI] [PubMed] [Google Scholar]
- Dileepan T., Linehan J.L., Moon J.J., Pepper M., Jenkins M.K., and Cleary P.P.. 2011. Robust antigen specific th17 T cell response to group A Streptococcus is dependent on IL-6 and intranasal route of infection. PLoS Pathog. 7:e1002252 10.1371/journal.ppat.1002252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong F., Xie K., Chen Y., Yang Y., and Mao Y.. 2017. Polycistronic tRNA and CRISPR guide-RNA enables highly efficient multiplexed genome engineering in human cells. Biochem. Biophys. Res. Commun. 482:889–895. 10.1016/j.bbrc.2016.11.129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberl G., Marmon S., Sunshine M.J., Rennert P.D., Choi Y., and Littman D.R.. 2004. An essential function for the nuclear receptor RORgamma(t) in the generation of fetal lymphoid tissue inducer cells. Nat. Immunol. 5:64–73. 10.1038/ni1022 [DOI] [PubMed] [Google Scholar]
- Fan X., Wang X., Li N., Cui H., Hou B., Gao B., Cleary P.P., and Wang B.. 2014. Sortase A induces Th17-mediated and antibody-independent immunity to heterologous serotypes of group A streptococci. PLoS One. 9:e107638 10.1371/journal.pone.0107638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang D., and Zhu J.. 2017. Dynamic balance between master transcription factors determines the fates and functions of CD4 T cell and innate lymphoid cell subsets. J. Exp. Med. 214:1861–1876. 10.1084/jem.20170494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farcas A.M., Blackledge N.P., Sudbery I., Long H.K., McGouran J.F., Rose N.R., Lee S., Sims D., Cerase A., Sheahan T.W., et al. . 2012. KDM2B links the Polycomb Repressive Complex 1 (PRC1) to recognition of CpG islands. eLife. 1:e00205 10.7554/eLife.00205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R.D., Attwood T.K., Babbitt P.C., Bateman A., Bork P., Bridge A.J., Chang H.Y., Dosztányi Z., El-Gebali S., Fraser M., et al. . 2017. InterPro in 2017-beyond protein family and domain annotations. Nucleic Acids Res. 45(D1):D190–D199. 10.1093/nar/gkw1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontenot J.D., Rasmussen J.P., Williams L.M., Dooley J.L., Farr A.G., and Rudensky A.Y.. 2005. Regulatory T cell lineage specification by the forkhead transcription factor foxp3. Immunity. 22:329–341. 10.1016/j.immuni.2005.01.016 [DOI] [PubMed] [Google Scholar]
- Higgins S.C., Jarnicki A.G., Lavelle E.C., and Mills K.H.. 2006. TLR4 mediates vaccine-induced protective cellular immunity to Bordetella pertussis: role of IL-17-producing T cells. J. Immunol. 177:7980–7989. 10.4049/jimmunol.177.11.7980 [DOI] [PubMed] [Google Scholar]
- Hollister K., Kusam S., Wu H., Clegg N., Mondal A., Sawant D.V., and Dent A.L.. 2013. Insights into the role of Bcl6 in follicular Th cells using a new conditional mutant mouse model. J. Immunol. 191:3705–3711. 10.4049/jimmunol.1300378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hondowicz B.D., An D., Schenkel J.M., Kim K.S., Steach H.R., Krishnamurty A.T., Keitany G.J., Garza E.N., Fraser K.A., Moon J.J., et al. . 2016. Interleukin-2-dependent allergen-specific tissue-resident memory cells drive asthma. Immunity. 44:155–166. 10.1016/j.immuni.2015.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao W.Y., Lin Y.C., Liao F.H., Chan Y.C., and Huang C.Y.. 2015. Dual-specificity phosphatase 4 regulates STAT5 protein stability and helper T cell polarization. PLoS One. 10:e0145880 10.1371/journal.pone.0145880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C.Y., Lin Y.C., Hsiao W.Y., Liao F.H., Huang P.Y., and Tan T.H.. 2012. DUSP4 deficiency enhances CD25 expression and CD4+ T-cell proliferation without impeding T-cell development. Eur. J. Immunol. 42:476–488. 10.1002/eji.201041295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh K.D., Fischle W., Verdin E., and Bardwell V.J.. 2000. BCoR, a novel corepressor involved in BCL-6 repression. Genes Dev. 14:1810–1823. [PMC free article] [PubMed] [Google Scholar]
- Ivanov I.I., McKenzie B.S., Zhou L., Tadokoro C.E., Lepelley A., Lafaille J.J., Cua D.J., and Littman D.R.. 2006. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 126:1121–1133. 10.1016/j.cell.2006.07.035 [DOI] [PubMed] [Google Scholar]
- Johnston R.J., Poholek A.C., DiToro D., Yusuf I., Eto D., Barnett B., Dent A.L., Craft J., and Crotty S.. 2009. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 325:1006–1010. 10.1126/science.1175870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston R.J., Choi Y.S., Diamond J.A., Yang J.A., and Crotty S.. 2012. STAT5 is a potent negative regulator of TFH cell differentiation. J. Exp. Med. 209:243–250. 10.1084/jem.20111174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurence A., Tato C.M., Davidson T.S., Kanno Y., Chen Z., Yao Z., Blank R.B., Meylan F., Siegel R., Hennighausen L., et al. . 2007. Interleukin-2 signaling via STAT5 constrains T helper 17 cell generation. Immunity. 26:371–381. 10.1016/j.immuni.2007.02.009 [DOI] [PubMed] [Google Scholar]
- Liang S.C., Tan X.Y., Luxenberg D.P., Karim R., Dunussi-Joannopoulos K., Collins M., and Fouser L.A.. 2006. Interleukin (IL)-22 and IL-17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. J. Exp. Med. 203:2271–2279. 10.1084/jem.20061308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang S.C., Long A.J., Bennett F., Whitters M.J., Karim R., Collins M., Goldman S.J., Dunussi-Joannopoulos K., Williams C.M., Wright J.F., and Fouser L.A.. 2007. An IL-17F/A heterodimer protein is produced by mouse Th17 cells and induces airway neutrophil recruitment. J. Immunol. 179:7791–7799. 10.4049/jimmunol.179.11.7791 [DOI] [PubMed] [Google Scholar]
- Liao W., Lin J.X., Wang L., Li P., and Leonard W.J.. 2011. Modulation of cytokine receptors by IL-2 broadly regulates differentiation into helper T cell lineages. Nat. Immunol. 12:551–559. 10.1038/ni.2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linehan J.L., Dileepan T., Kashem S.W., Kaplan D.H., Cleary P., and Jenkins M.K.. 2015. Generation of Th17 cells in response to intranasal infection requires TGF-β1 from dendritic cells and IL-6 from CD301b+ dendritic cells. Proc. Natl. Acad. Sci. USA. 112:12782–12787. 10.1073/pnas.1513532112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Yan X., Zhong B., Nurieva R.I., Wang A., Wang X., Martin-Orozco N., Wang Y., Chang S.H., Esplugues E., et al. . 2012. Bcl6 expression specifies the T follicular helper cell program in vivo. J. Exp. Med. 209:1841–1852: S1–S24. 10.1084/jem.20120219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J.J., Chu H.H., Pepper M., McSorley S.J., Jameson S.C., Kedl R.M., and Jenkins M.K.. 2007. Naive CD4(+) T cell frequency varies for different epitopes and predicts repertoire diversity and response magnitude. Immunity. 27:203–213. 10.1016/j.immuni.2007.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima H., Liu X.W., Wynshaw-Boris A., Rosenthal L.A., Imada K., Finbloom D.S., Hennighausen L., and Leonard W.J.. 1997. An indirect effect of Stat5a in IL-2-induced proliferation: a critical role for Stat5a in IL-2-mediated IL-2 receptor alpha chain induction. Immunity. 7:691–701. 10.1016/S1074-7613(00)80389-1 [DOI] [PubMed] [Google Scholar]
- Nance J.P., Bélanger S., Johnston R.J., Takemori T., and Crotty S.. 2015. Cutting edge: T follicular helper cell differentiation is defective in the absence of Bcl6 BTB repressor domain function. J. Immunol. 194:5599–5603. 10.4049/jimmunol.1500200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng D., Thakker N., Corcoran C.M., Donnai D., Perveen R., Schneider A., Hadley D.W., Tifft C., Zhang L., Wilkie A.O., et al. . 2004. Oculofaciocardiodental and Lenz microphthalmia syndromes result from distinct classes of mutations in BCOR. Nat. Genet. 36:411–416. 10.1038/ng1321 [DOI] [PubMed] [Google Scholar]
- Nurieva R.I., Podd A., Chen Y., Alekseev A.M., Yu M., Qi X., Huang H., Wen R., Wang J., Li H.S., et al. . 2012. STAT5 protein negatively regulates T follicular helper (Tfh) cell generation and function. J. Biol. Chem. 287:11234–11239. 10.1074/jbc.M111.324046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H.S., Costalonga M., Reinhardt R.L., Dombek P.E., Jenkins M.K., and Cleary P.P.. 2004. Primary induction of CD4 T cell responses in nasal associated lymphoid tissue during group A streptococcal infection. Eur. J. Immunol. 34:2843–2853. 10.1002/eji.200425242 [DOI] [PubMed] [Google Scholar]
- Pepper M., Pagán A.J., Igyártó B.Z., Taylor J.J., and Jenkins M.K.. 2011. Opposing signals from the Bcl6 transcription factor and the interleukin-2 receptor generate T helper 1 central and effector memory cells. Immunity. 35:583–595. 10.1016/j.immuni.2011.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees W., Bender J., Teague T.K., Kedl R.M., Crawford F., Marrack P., and Kappler J.. 1999. An inverse relationship between T cell receptor affinity and antigen dose during CD4(+) T cell responses in vivo and in vitro. Proc. Natl. Acad. Sci. USA. 96:9781–9786. 10.1073/pnas.96.17.9781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson J.M., Jensen P.E., and Evavold B.D.. 2000. DO11.10 and OT-II T cells recognize a C-terminal ovalbumin 323-339 epitope. J. Immunol. 164:4706–4712. 10.4049/jimmunol.164.9.4706 [DOI] [PubMed] [Google Scholar]
- Ruiz-Romeu E., Ferran M., Sagristà M., Gómez J., Giménez-Arnau A., Herszenyi K., Hóllo P., Celada A., Pujol R., and Santamaria-Babí L.F.. 2016. Streptococcus pyogenes-induced cutaneous lymphocyte antigen-positive T cell-dependent epidermal cell activation triggers TH17 responses in patients with guttate psoriasis. J. Allergy Clin. Immunol. 138:491–499.e6. 10.1016/j.jaci.2016.02.008 [DOI] [PubMed] [Google Scholar]
- Sadlack B., Löhler J., Schorle H., Klebb G., Haber H., Sickel E., Noelle R.J., and Horak I.. 1995. Generalized autoimmune disease in interleukin-2-deficient mice is triggered by an uncontrolled activation and proliferation of CD4+ T cells. Eur. J. Immunol. 25:3053–3059. 10.1002/eji.1830251111 [DOI] [PubMed] [Google Scholar]
- Sinclair L.V., Finlay D., Feijoo C., Cornish G.H., Gray A., Ager A., Okkenhaug K., Hagenbeek T.J., Spits H., and Cantrell D.A.. 2008. Phosphatidylinositol-3-OH kinase and nutrient-sensing mTOR pathways control T lymphocyte trafficking. Nat. Immunol. 9:513–521. 10.1038/ni.1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skepner J., Ramesh R., Trocha M., Schmidt D., Baloglu E., Lobera M., Carlson T., Hill J., Orband-Miller L.A., Barnes A., et al. . 2014. Pharmacologic inhibition of RORγt regulates Th17 signature gene expression and suppresses cutaneous inflammation in vivo. J. Immunol. 192:2564–2575. 10.4049/jimmunol.1302190 [DOI] [PubMed] [Google Scholar]
- Szabo S.J., Kim S.T., Costa G.L., Zhang X., Fathman C.G., and Glimcher L.H.. 2000. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 100:655–669. 10.1016/S0092-8674(00)80702-3 [DOI] [PubMed] [Google Scholar]
- Veldhoen M., Hocking R.J., Atkins C.J., Locksley R.M., and Stockinger B.. 2006. TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity. 24:179–189. 10.1016/j.immuni.2006.01.001 [DOI] [PubMed] [Google Scholar]
- Wamstad J.A., Corcoran C.M., Keating A.M., and Bardwell V.J.. 2008. Role of the transcriptional corepressor Bcor in embryonic stem cell differentiation and early embryonic development. PLoS One. 3:e2814 10.1371/journal.pone.0002814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Strong J., and Killeen N.. 2001. Homeostatic competition among T cells revealed by conditional inactivation of the mouse Cd4 gene. J. Exp. Med. 194:1721–1730. 10.1084/jem.194.12.1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Gearhart M.D., Lee Y.W., Kumar I., Ramazanov B., Zhang Y., Hernandez C., Lu A.Y., Neuenkirchen N., Deng J., et al. . 2018. A Non-canonical BCOR-PRC1.1 complex represses differentiation programs in human ESCs. Cell Stem Cell. 22:235–251.e9. 10.1016/j.stem.2017.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welte T., Leitenberg D., Dittel B.N., al-Ramadi B.K., Xie B., Chin Y.E., Janeway C.A. Jr., Bothwell A.L., Bottomly K., and Fu X.Y.. 1999. STAT5 interaction with the T cell receptor complex and stimulation of T cell proliferation. Science. 283:222–225. 10.1126/science.283.5399.222 [DOI] [PubMed] [Google Scholar]
- Willerford D.M., Chen J., Ferry J.A., Davidson L., Ma A., and Alt F.W.. 1995. Interleukin-2 receptor alpha chain regulates the size and content of the peripheral lymphoid compartment. Immunity. 3:521–530. 10.1016/1074-7613(95)90180-9 [DOI] [PubMed] [Google Scholar]
- Xiao S., Yosef N., Yang J., Wang Y., Zhou L., Zhu C., Wu C., Baloglu E., Schmidt D., Ramesh R., et al. . 2014. Small-molecule RORγt antagonists inhibit T helper 17 cell transcriptional network by divergent mechanisms. Immunity. 40:477–489. 10.1016/j.immuni.2014.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie K., Minkenberg B., and Yang Y.. 2015. Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc. Natl. Acad. Sci. USA. 112:3570–3575. 10.1073/pnas.1420294112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Zhao L., Gao Y., Xu J., and Han R.. 2017. Empower multiplex cell and tissue-specific CRISPR-mediated gene manipulation with self-cleaving ribozymes and tRNA. Nucleic Acids Res. 45:e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki T., Yang X.O., Chung Y., Fukunaga A., Nurieva R., Pappu B., Martin-Orozco N., Kang H.S., Ma L., Panopoulos A.D., et al. . 2008. CCR6 regulates the migration of inflammatory and regulatory T cells. J. Immunol. 181:8391–8401. 10.4049/jimmunol.181.12.8391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J.A., Tubo N.J., Gearhart M.D., Bardwell V.J., and Jenkins M.K.. 2015. Cutting edge: Bcl6-interacting corepressor contributes to germinal center T follicular helper cell formation and B cell helper function. J. Immunol. 194:5604–5608. 10.4049/jimmunol.1500201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D., Rao S., Tsai L.M., Lee S.K., He Y., Sutcliffe E.L., Srivastava M., Linterman M., Zheng L., Simpson N., et al. . 2009. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity. 31:457–468. 10.1016/j.immuni.2009.07.002 [DOI] [PubMed] [Google Scholar]
- Zhong Z., Wen Z., and Darnell J.E. Jr. 1994. Stat3: a STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science. 264:95–98. 10.1126/science.8140422 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.