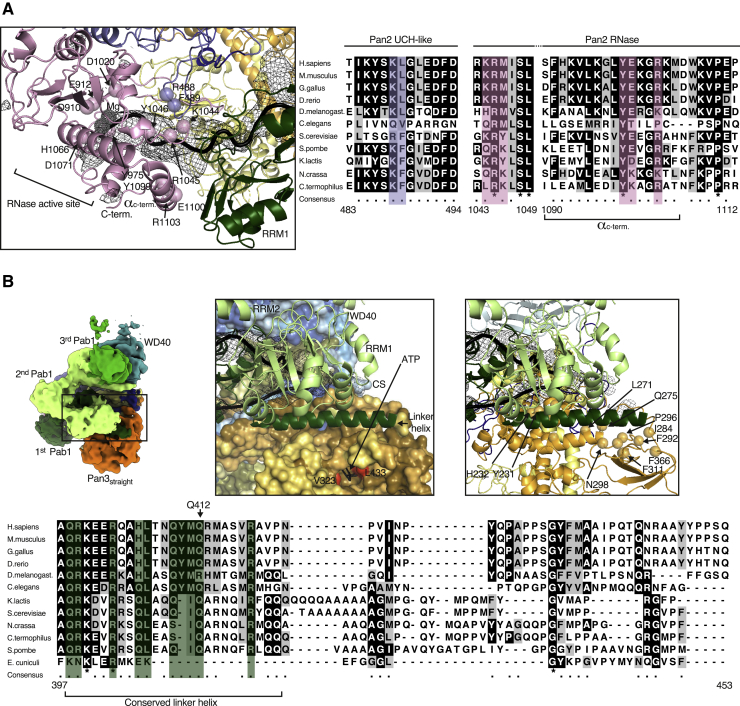
Figure S5.
Evolutionarily Conserved Interactions between Pan2-Pan3 and Pab1, Related to Figure 4
(A) Details of the interactions at the 3′ end of the 90A RNP. On the left is a panel corresponding to Figure 4A, with the position of conserved residues from the Pan2 UCH-like domain (violet) and RNase (pink) indicated by spheres. On the right are sequence alignments of the corresponding regions of Pan2 (with conservation shown in blue for the UCH-like domain and pink for the RNase domain). The strongly conserved catalytic residues of the catalytic DEDDh motif around D1020 and D910 are highlighted as is residue Y975 which interacts with the base moiety of AMP in a previously described UCH-like–RNase crystal structure (Schäfer et al., 2014).
(B) Details of the interactions at the first Pab1-Pab1 oligomerization interface of the 90A RNP. The panel on the left highlights the overall position of the interface in the context of the reconstruction (corresponding to Figure 4B, after a counterclockwise rotation of 90º). The panel in the middle shows the position of ATP bound to the pseudokinase domain of Pan3straight (from PDB:4bwp, Pan3s shown in surface representation; Christie et al., 2013). In red are evolutionarily conserved ATP-binding residues as reference. The panel on the right lays out the RNP-interacting structural elements of Pan3straight. Spheres indicate the position of conserved residues on Pan3. The lower panel shows the evolutionary conservation of the Pab1 helical segment downstream of RRM4 (the linker helix) and of parts of the (less conserved) rest of the linker.