Abstract
Although accumulating evidence suggests that long non-coding RNAs (lncRNAs) are critical determinants of ovarian cancer development and progression, reports of metastasis-associated lncRNAs are limited. Here, we focused on NONHSAT076754 and explored its expression level, clinical value, biological behavior and molecular basis in epithelial ovarian cancer (EOC) metastasis. The results showed that NONHSAT076754 expression was increased in EOC tissues and cell lines and that this expression was closely related with FIGO stage, high tumor grade and lymph node metastasis. Furthermore, NONHSAT076754 knockdown markedly inhibited EOC cell migration and invasion in vitro. Consistently, the in vivo data from both the bioluminescence imaging and tumor dissection revealed that depletion of NONHSAT076754 reduced EOC metastasis. Mechanically, the pro-metastatic activities of NONHSAT076754 were partially regulated by PTEN and HTATIP2. Further rescue assays validated that knockdown of HTATIP2 remarkably reversed NONHSAT076754 silencer-induced inhibition of EOC cell metastasis. These data indicate that NONHSAT076754 is a vital regulator of EOC metastasis, laying the foundation for lncRNA-based clinical management of EOC aggressiveness and metastasis.
Keywords: long non-coding RNA, NONHSAT076754, invasion, metastasis, ovarian cancer
Introduction
Epithelial ovarian cancer (EOC) is among the most common gynecological malignancies and causes the most deaths in women with these malignancies. The high mortality is attributed to the frequent spread of metastases at time of diagnosis in patients with generally limited symptoms 1, 2. Despite progresses in clinical management with effective surgery and chemotherapy, overall survival remains at an uncomfortable rate of 30% largely because of abdominal or other systemic metastases 3. Hence, the identification of an available metastasis-associated biomarker and exploration of its molecular mechanism will be great beneficial for the clinical management of ovarian cancer (OC).
Recently, high-throughput sequencing efforts have revealed that long non-coding RNAs (lncRNAs), which are longer than 200 nucleotides in length, account for the majority of the human genome rather than “transcription noise” 4, 5. Growing evidence has showed that lncRNAs are abnormally expressed in various cancers and participate in multiple biological processes, including apoptosis, proliferation, cell migration, and invasion 6. Furthermore, several lncRNAs, such as MALAT1 7, H19 8, HOTAIR 9, AFAP1-AS1 10, CCAT2 11, lncRNA- ATB 12 and lincRNA-RoR 13 have emerged as promising candidates for gene regulation during tumor progression. In the case of EOC, existing evidence, including our previous findings, have unveiled a handful of lncRNAs are implicated in EOC metastasis. For example, the long non-coding RNA ANRIL exhibited great clinical significance in predicting the aggressive prognosis of OC and stimulated the invasion and metastasis of serous ovarian cancer 14. In another study, we revealed that HOTAIR also acted as a pro-metastasis lncRNA in EOC, and its overexpression was highly connected with poor prognosis in patients with EOC 15. However, despite these findings, the essential roles and mechanisms of metastasis-associated lncRNAs in EOC are unclear. Based on the current situation on metastasis-associated lncRNAs research, it is necessary to identify novel lncRNAs involved in metastasis process and to further explore their molecular mechanisms.
In the present study, the expression of four metastasis-associated lncRNAs (NONHSAT076754, HEIRCC, lncTCF7 and ZEB2NAT) was examined, and the novel OC-related lncRNA NONHSAT076754 captured our attention due to its remarkably consistently high expression in EOC 16-21. NONHSAT076754, which is located on chromosome 2, is 695 bp in length and transcribed from fibronectin 1, which is closely associated with cells' adhesive and migrative capabilities 22-24. NONHSAT076754 was originally found to promote migration and invasiveness in papillary thyroid cancer (PTC) and could be a valuable biomarker and predictor of lymph node metastasis 16. However, other than these studies, no data regarding the role of NONHSAT076754 in the invasion and metastasis of EOC are currently available.
The present investigation aimed to illustrate the involvement of NONHSAT076754 in EOC metastasis. Thus, we first detected the expression level of NONHSAT076754 in EOC clinical samples and cell lines and analysed the association between NONHSAT076754 expression and EOC clinicopathological characteristics. Additionally, NONHSAT076754 was silenced to evaluate its effect on invasion and metastasis both in vitro and vivo. Moreover, the mechanism by which NONHSAT076754 modulated EOC metastasis was explored. The major objective of the present study is to highlight the instructive significance of NONHSAT076754 in EOC metastasis.
Materials and Methods
Patients and tissue samples
In total, 70 EOC tissue samples were gained from the Tissue Bank of the Obstetrics and Gynecology Hospital of Fudan University. Inclusion criteria were as follows: (1) Patients were diagnosed with first-time EOC and samples were confirmed by postoperative pathology; (2) patients underwent surgery between August 2013 and September 2016. Exclusion criteria were as follows: (1) Patients had two or more different malignancies; (2) patients had received preoperative chemotherapy, radiotherapy, or hormonal therapy; (3) patients had coexisting autoimmune diseases or evidence of active infection; (4) there were metastatic or non-epithelial tumors in ovary. Ten normal ovarian epithelial tissues were collected from the uterine fibroids participants scheduled to experience hysterectomy and oophorectomy. Inclusion criteria were as follows: All samples were confirmed by postoperative pathology as uterine leiomyoma with no abnormality in adnexa. Exclusion criteria were the same as EOC group apart from those patients who had previous ovarian pathology or surgery and who had history of malignancy. This study was approved by the Ethics Committee of the Obstetrics and Gynecology Hospital of Fudan University (No. [2017]82).
Cell lines and transfection
The human EOC cell lines (SKOV3, OVCAR5, OVCAR3, OVCAR8, HO8910) and a normal human ovarian epithelial cell line (HOSEPIC) were offered by the University of Texas MD Anderson Cancer Center (Houston, TX, USA). The cells were grown in RPMI-1640 medium (Genom, Hangzhou, China) supplemented with 10% fetal bovine serum (FBS) (Gibco, Grand, Island, NY, USA) and 100 u/ml penicillin streptomycin in a humidified 5% carbon dioxide incubator at 37 °C.
The control small interfering RNA (siRNA) and siRNAs targeting NONHSAT076754 and HTATIP2 (GenePharma, Shanghai, China) were applied in combination with Lipofectamine 3000 transfection reagent (Invitrogen, Carlsbad, CA, USA) to SKOV3, OVCAR5 cells based on the manufacturer's protocol. The target sequences of the NONHSAT076754 siRNAs were 5'-UGAUGUGGUGGUCUGGUCUC-3' (siRNA-1), 5'-GACCAGACCACCACAUCAUTT-3' (siRNA-2) and 5'-GCAUACUGUAACCUCUGCATT-3' (siRNA-3). The target sequences of the HTATIP2 siRNAs were 5'-GGAGGGAUUUGUUCGUGUUTT-3' (siRNA-1), 5'-GCAGAAUAAAUCCGUCUUUTT-3' (siRNA-2) and 5'-CCAGGUGAAUGGCUGGUUATT-3' (siRNA-3). The sequence of the negative control (NC) was 5'-UUCUCCGAACGUGUCACGUTT-3. After 48 h, the knockdown efficiency was examined using quantitative real-time polymerase chain reaction (qRT-PCR).
Establishment of stable NONHSAT076754-knockdown tumor cells
Stable knockdown tumor cells were established by using lentiviral vectors carrying NONHSAT076754-siRNA-1 and a luciferase sequence (KD-luc-SKOV3). Accordingly, the negative control lentivirus (NC-luc- SKOV3) (Genechem, Shanghai, China) were prepared. At 72h post-infection, luciferin (0.15mg/ml) was added, and cells were imaged by Bioluminescence Imaging Facility to confirm luciferase expression. Subsequently, puromycin (4μg/ml) was applied for approximately 12 days to isolate the puromycin-resistant cell clones.
RNA extraction and qRT-PCR
The total RNA was isolated using TRIZOL reagent (TAKARA, Code No.9109, Dalian, China) and the cDNA was synthesized using the Prime-Script™ RT Master Mix (TAKARA, RR036A, Dalian, China). qRT-PCR was performed using the SYBR Green qPCR (TAKARA, RR820A, Dalian, China). All steps were conducted according to the manufacturer's instructions. The relative quantification results were displayed as fold change using 2-ΔΔCt. The primer sequences are listed in Table 1.
Table 1.
The primers used in the study.
| Gene | Primer | Sequence (5'-3') |
|---|---|---|
| GAPDH | Forward | GTCATCAATGGAAATCCCATCA |
| Reverse | CCAG TGGACTCCACGACGTAC | |
| NONHSAT076754 | Forward | AAGTTTCTCACTCACCCACCTG |
| Reverse | GAAGCATGTACAGTTCAGCATGTG | |
| HEIRCC | Forward | GCTGCTATTCTGGTGCCC |
| Reverse | TCAACTCCGATAAACAGGTGA | |
| lncTCF7 | Forward | AGGAGTCCTTGGACCTGAGC |
| Reverse | AGTGGCTGGCATATAACCAACA | |
| ZEB2NAT | Forward | GAG AGA CGA GAG ACC CTG AA |
| Reverse | TGC ACA CAC CCT AAT ACA CAT | |
| HPSE | Forward | TTCTCACAGCAAACCTCTCC |
| Reverse | AACAACAACGAGGTTCCTAT | |
| HRAS | Forward | GGCTTCCTGTGTGTGTTTGC |
| Reverse | AGTACAGGGAGCAGATCAAA | |
| PTEN | Forward | CTGGTTCACATCCTACCCCT |
| Reverse | CGAAGGGTTTTGCTACATTC | |
| FGFR4 | Forward | TCAAAGACAACGCCTCTGAC |
| Reverse | GAGATGGAGGTGATGAAGCT | |
| HTATIP2 | Forward | GTTTTCCAAAGTCACGCTCA |
| Reverse | GTTGGATTCTGTTGCCTGGG | |
| NR4A3 | Forward | GTCTGAAAGGGAGGAGAGGT |
| Reverse | ACCTTCTCAGCCCTCTCCAC |
Scratch assay
The cells were seeded onto 6-cell plates. Once approximately 80% confluency was reached, the cells were scraped with a sterile pipette tip, washed with phosphate-buffer-solution (PBS) and maintained in serum-free media at 37°C and 5% CO2. The scratch width was photographed at 0 h, 24h and 48 h at the same positions.
Cell migration assay
The cell migration assay was conducted by using transwell chambers with a diameter of 8 μm (Millipore, Billerica, MA). The cells were seeded onto the upper chamber with 200 μL of serum-free RPMI-1640. Then, 600 μL of RPMI-1640 containing 10% FBS were added to the bottom transwell chamber as a chemo- inducer. After 24 h, the cells in the bottom layer were fixed and stained. Further analyses were conducted through imaging and counting the number of cells in five random regions using inverted microscopy.
Invasion assay
The invasion assay was performed similar to the migration assay; however, the transwell membranes were coated with Matrigel (BD Biosciences, Franklin Lakes, NJ, USA) before the cell inoculation.
In vivo intraperitoneal metastasis model
All animal protocols were approved by the Institutional Animal Care and Use Committee of the Fudan University. Female 5- to 6-week-old Balb/c nude mice were obtained from Slac Laboratory Animal Co. Ltd. (Shanghai, China), maintained in a specific-pathogen free (SPF) environment and randomly assigned to the following two groups: the control group and the experimental knockdown group (six mice per group), which were intraperitoneally (i.p.) injected with NC-luc-SKOV3 and KD-luc-SKOV3 cells (5× 106/0.2 ml), respectively. Then, the mice were analyzed weekly at the Small Animal Bioluminescence Imaging Facility, and luciferin(150mg/kg) i.p. injected into each mouse before imaging. After 4 weeks, all mice were sacrificed, and the tumors were dissected for analysis.
Tumour metastasis real-time PCR array
The total RNA extraction was conducted on SKOV3-NONHSAT076754-siRNA1 cells and SKOV3- NC-siRNA cells with TRIZOL reagent (TAKARA, Code No.9109, Dalian, China). Then, the RNA was reverse transcribed into cDNA using an RT2 First Strand kit (Qiagen, Mississauga, ON, Canada). Following the protocol provided by the manufacturer, a cDNA library was produced and the cDNA was applied to the Human Tumour Metastasis RT2 Profiler™ PCR array (Qiagen, Mississauga, ON, Canada), which assessed 84 genes related to tumour metastatic activity. The 2-ΔΔCt method was used to calculate the expression fold-changes.
Western blot (WB) analysis
WB was conducted as described previously 15. The primary antibodies used were rabbit anti- GAPDH (Cell Signaling Technology, #5174, USA), anti-PTEN (Abcam, ab32199, Cambridge, UK) and anti-HTATIP2 (Abcam, ab177961, Cambridge, UK).
Data analysis
The data analysis was done through the SPSS version 20.0 software (SPSS, Inc., Chicago, IL, USA) and GraphPad Prism (Prism 5.0, GraphPad Software, San Diego, CA, USA). Continuous data were processed by Student's t-test (2 groups) or one-way ANOVA (>2 groups), while categorical data were analyzed by chi-square test or Fisher's exact test. P <0.05 were defined statistically significant.
Results
NONHSAT076754 expression is elevated in ovarian cancer cells
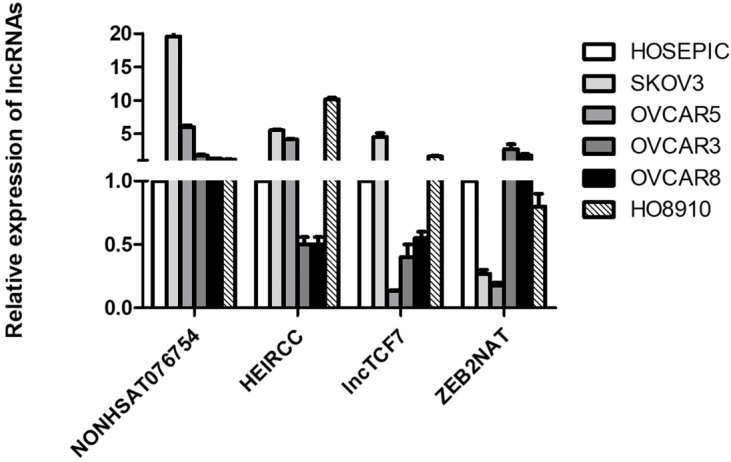
Based on existing studies 16-21, four metastasis-associated lncRNAs (NONHSAT076754, HEIRCC, lncTCF7 and ZEB2NAT) were selected and their expression levels were measured in five EOC cell lines (SKOV3, OVCAR5, OVCAR3, OVCAR8 and HO8910) and a normal human ovarian epithelial cell line (HOSEPIC). The results showed that NONHSAT076754 expression exhibited optimal consistency and that compared to its expression in HOSEPIC, NONHSAT076754 expression in ovarian cancer cells was notably higher (Fig. 1). Thus, NONHSAT076754 was selected for further clinical, functional and mechanical investigations.
Figure 1.
Expression analysis of four metastasis-associated lncRNAs in EOC cell lines (SKOV3, OVCAR5, OVCAR3, OVCAR8 and HO8910) and a normal human ovarian epithelial cell line (HOSEPIC).
Overexpression of NONHSAT076754 is associated with advanced and metastatic ovarian cancer clinical phenotype
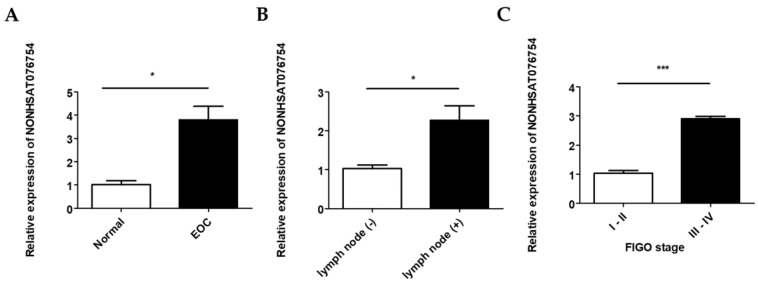
The NONHSAT076754 expression levels in 70 EOC and 10 normal ovarian surface epithelial tissues were determined by qRT-PCR. NONHSAT076754 expression in the EOC tissue was significantly higher than that in the normal tissues (Fig. 2A). In addition, there were significant differences in NONHSAT076754 expression levels between the lymph node-positive group (with lymph node metastasis) and the lymph node-negative group (without detectable lymph node metastasis) (Fig. 2B). Additionally, EOC samples from patients with advanced FIGO stage (III-IV) cancer showed higher NONHSAT076754 expression levels than those from early FIGO stage (I-II) patients (Fig. 2C). For the clinicopathological correlation analysis, 70 EOC patients were divided into two groups, i.e., high NONHSAT076754 group (n = 35) and low NONHSAT076754 group (n = 35), according to the median relative NONHSAT076754 expression. Consequently, high NONHSAT076754 expression levels were closely associated with FIGO stage, tumor grade and lymph node metastasis while no significant differences in age, histological subtype, residual tumor diameter, CA125, ascites and distant metastasis were found (Table 2). These results illustrated that up-regulation of NONHSAT076754 might have an essential impact on EOC progression.
Figure 2.
Relative NONHSAT076754 expression levels in EOC. A. Expression of NONHSAT076754 in EOC and normal ovarian tissues. B. Expression of NONHSAT076754 in lymph node-positive and lymph node-negative EOC samples. C. Expression of NONHSAT076754 in samples from EOC patients with advanced and early FIGO stage cancers. Data are shown as the mean± SD. * p < 0.05, *** p < 0.001 vs. NC groups.
Table 2.
Association between NONHSAT076754 expression and clinicopathological factors in 70 EOC patients.
| Variable | Low NONHSAT076754 expression (n=35) | High NONHSAT076754 expression (n=35) | P-value | |
|---|---|---|---|---|
| n(%) | n(%) | |||
| Age (years) | ||||
| <50 | 18 (51.4) | 11 (31.4) | ||
| ≥50 | 17 (48.6) | 24 (68.6) | 0.090 | |
| Histological subtype | ||||
| Serous | 28 (80.0) | 22 (62.9) | ||
| Others | 7 (20.0) | 13 (37.1) | 0.112 | |
| Tumor grade | ||||
| Low | 13 (37.1) | 5 (14.3) | ||
| High | 22 (62.9) | 30 (85.7) | 0.029 | |
| FIGO Stage | ||||
| I-II | 12 (34.3) | 4 (11.4) | ||
| III-IV | 23 (65.7) | 31 (88.6) | 0.023 | |
| Residual tumor diameter (cm) | ||||
| <1 | 24 (68.6) | 21 (60.0) | ||
| ≥1 | 11 (31.4) | 14 (40.0) | 0.454 | |
| Lymph node metastasis | ||||
| Absent | 21 (60.0) | 7 (20.0) | ||
| Present | 14 (40.0) | 28 (80.0) | <0.001 | |
| CA125 level (U/ml) | ||||
| <600 | 16 (45.7) | 14 (40.0) | ||
| ≥600 | 19 (54.3) | 21 (60.0) | 0.629 | |
| Ascites | ||||
| <100 | 18 (51.4) | 21 (60.0) | ||
| ≥100 | 17 (48.6) | 14 (40.0) | 0.470 | |
| Distant metastasis | ||||
| Absent | 33 (94.3) | 28 (80.0) | ||
| Present | 2 (5.7) | 7 (20.0) | 0.074 | |
NONHSAT076754 silencing represses EOC cell metastasis in vitro
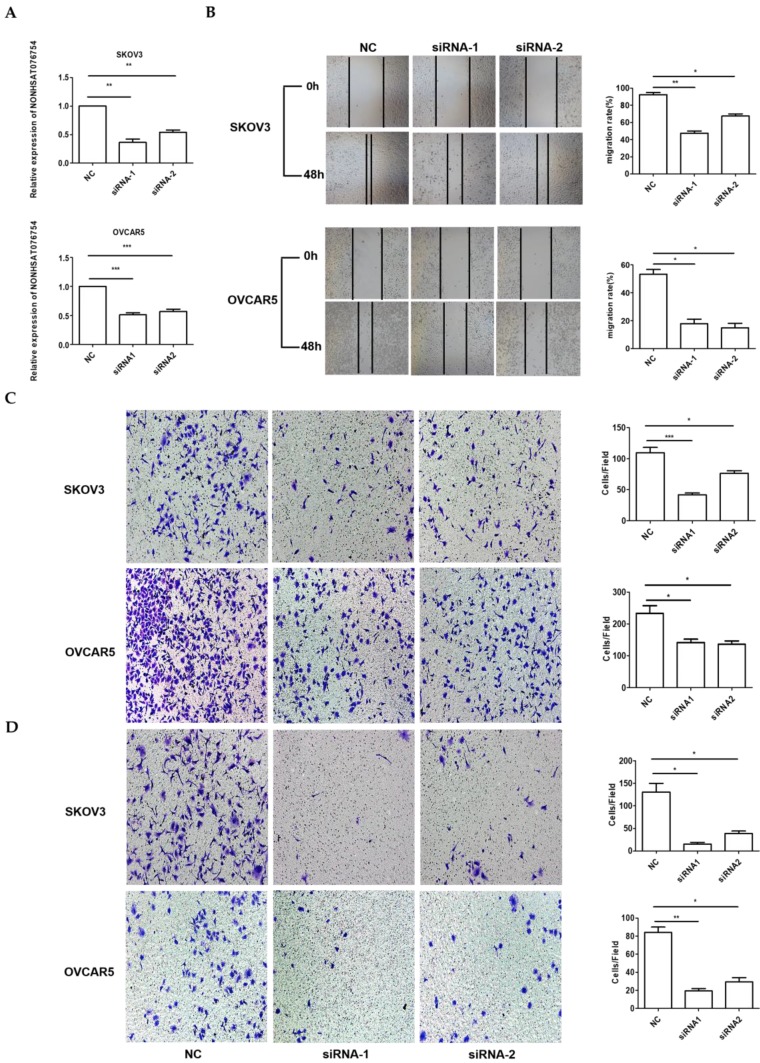
Because the highest NONHSAT076754 expression was observed in the SKOV3 and OVCAR5 cells (Fig. 1), the loss-of-function experiments were performed using these two EOC cell lines. Firstly, we designed three siRNAs to silence NONHSAT076754 in the SKOV3 and OVCAR5 cells. Consequently, siRNA1 and siRNA2 showed a higher knockdown efficiency, as determined by qRT-PCR (Fig. 3A). Subsequently, to explore the role of NONHSAT076754 in cell migration and invasion, we preformed wound-healing and transwell assays using NONHSAT076754-siRNA-transfected cells. The results showed that the NONHSAT076754 reduction significantly attenuated SKOV3 and OVCAR5 metastasis in vitro (Fig. 3B-D).
Figure 3.
Knockdown of NONHSAT076754 hinders migration and invasion in SKOV3 and OVCAR5 cells. A. Efficiency of NONHSAT076754 silencing in SKOV3 and OVCAR5 cells transfected with siRNAs was confirmed by qRT-PCR. B. Migration of NONHSAT076754-knockdown cells was evaluated by wound healing assays. C. Transwell migration assays were conducted to confirm the cell migration activity. D. Invasion of NONHSAT076754-knockdown cells was detected by transwell invasion assays. Data are shown as the mean± SD. * p < 0.05, ** p < 0.01, *** p < 0.001 vs. NC groups.
NONHSAT076754 silencing represses EOC cell metastasis in vivo
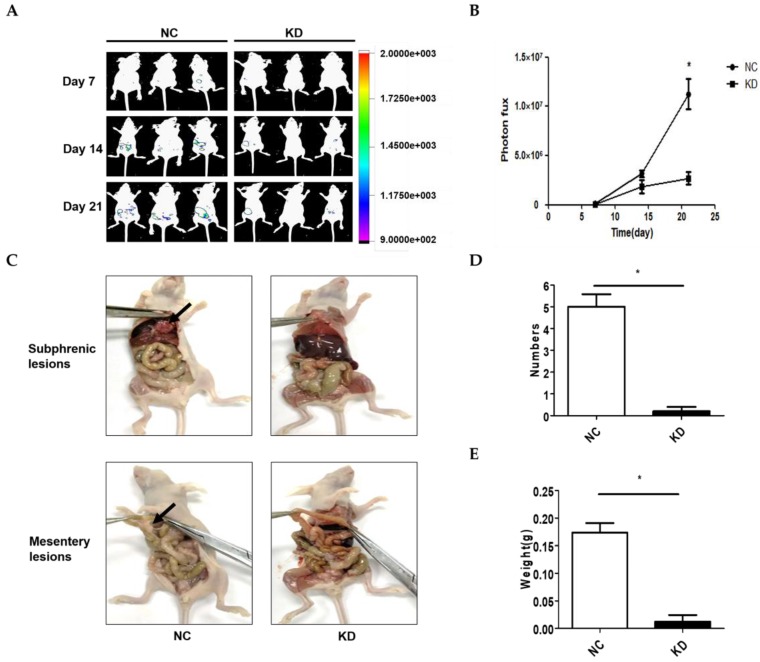
A model of abdominal metastasis was used to assess the metastatic role of NONHSAT076754 in vivo. Firstly, we established stable NONHSAT076754- knockdown 1 SKOV3 cells expressing luciferase (KD-luc-SKOV3) and the corresponding controls (NC-luc-SKOV3). Then, nude mice were i.p. injected with the above cells. The bioluminescence imaging showed that the mice injected with the KD-luc-SKOV3 cells exhibited an apparent decrease in the bioluminescence signal compared with those injected with NC-luc-SKOV3 cells (Fig. 4A and 4B). After four weeks, all mice were sacrificed, and the tumors were dissected for analysis. Consequently, the mice injected with the NC-luc-SKOV3 cells displayed multiple metastatic nodules in the abdominal cavity primarily visible on the subphrenic and mesentery spaces adjacent to the bowel. Additionally, several smaller metastatic foci were seeded throughout the abdominal cavity. In comparison, the mice injected with the KD-luc-SKOV3 cells exhibited either no or little tumour formation limited to the injection site of abdominal wall at four weeks (Fig. 4C). Statistically, the mice injected with the NC-luc-SKOV3 cells developed more and larger metastatic nodules than those injected with the KD-luc-SKOV3 (Fig. 4D and 4E). Collectively, NONHSAT076754 silencing could repress EOC cell metastasis in vivo.
Figure 4.
NONHSAT076754 silencing suppresses metastasis in an SKOV3-luc ovarian cancer intraperitoneal metastasis model. A. Representative images of luciferase signals in the KD-luc-SKOV3 groups compared with those in the NC-luc-SKOV3. B. Quantification of the photon flux from the metastatic lesions. C. Representative views of visible metastasis in the peritoneal cavity. D. Analysis of the numbers of dissected tumors. E. Analysis of the weight of dissected tumors. Data are shown as mean± SD. * p < 0.05 vs. NC groups.
NONHSAT076754 affects EOC metastasis via PTEN and HTATIP2
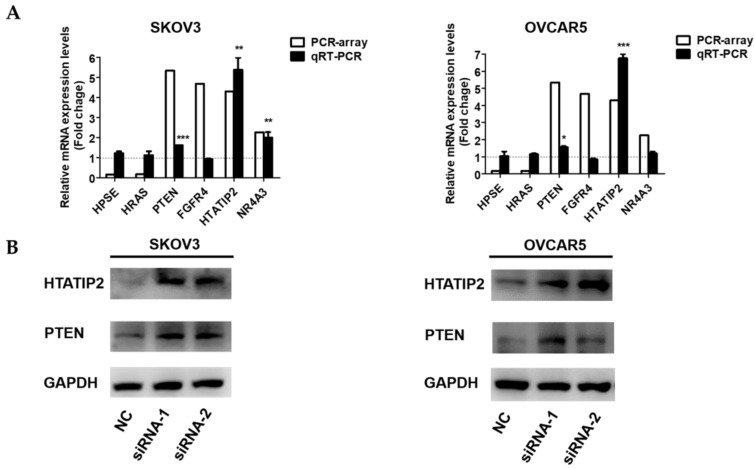
To further investigate the potential mechanisms underlying the pro-metastatic role of NONHSAT076754 in EOC, a tumour metastasis real-time PCR array was applied to compare the metastasis-related gene alterations in SKOV3-NC with those in SKOV3- siRNA1. Consequently, six genes were tremendously dysregulated (>2-fold) after the NONHSAT076754- knockdown in the SKOV3 cells; among these genes, four genes (PTEN, FGFR4, HTATIP2, and NR4A3) were upregulated and two genes (HPSE and HRAS) were downregulated (Table 3). Subsequently, qRT- PCR and WB assays were conducted for further validation of these differentially expressed genes. Consistent with the results derived from the array, the up-regulation of PTEN and HTATIP2 was observed at both the mRNA and protein levels in the SKOV3 and OVCAR5 cells transfected with the two siRNAs (Fig. 5A and 5B). Collectively, these data indicated that the two metastasis-related genes PTEN and HTATIP2 might participate in NONHSAT076754-mediated pro-metastatic effect.
Table 3.
Differential expressed genes (>2-fold) after NONHSAT076754-knockdown in SKOV3 cells as indicated by an array analysis.
| Gene name | GeneBank ID | Description | Function | Fold change |
|---|---|---|---|---|
| HPSE | NM_006665 | Heparanase | Remodeling of the extracellular matrix | 0.16 |
| HRAS | NM_005343 | HRas proto-oncogene, GTPase | HRas proto-oncogene | 0.17 |
| PTEN | NM_000314 | Phosphatase and tensin homolog | Tumor suppressor | 5.34 |
| FGFR4 | NM_002011 | Fibroblast growth factor receptor 4 | Cell proliferation, cell differentiation, cell migration | 4.68 |
| HTATIP2 | NM_006410 | HIV-1 Tat interactive protein 2 | Tumor suppressor | 4.3 |
| NR4A3 | NM_006981 | Nuclear receptor subfamily 4, group A, member 3 | Tumor suppressor | 2.26 |
Figure 5.
NONHSAT076754 regulates EOC metastasis via the downstream metastasis-related genes PTEN and HTATIP2. A. qRT-PCR examination of relative PTEN and HTATIP2 expression in SKOV3 and OVCAR5 transfected with NC or NONHSAT076754-siRNA. B. WB verification of PTEN and HTATIP2 expression at the protein level in SKOV3 and OVCAR5 cells transfected with NC or NONHSAT076754-siRNA. Data are shown as the mean± SD. * p < 0.05, ** p < 0.01, *** p < 0.001 vs. NC groups.
Knockdown of HTATIP2 significantly rescues the repressive effects of silencing NONHAT076754 on EOC metastasis
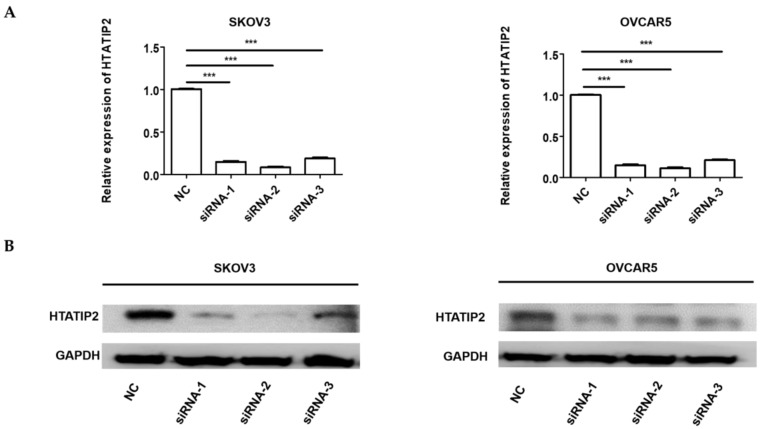
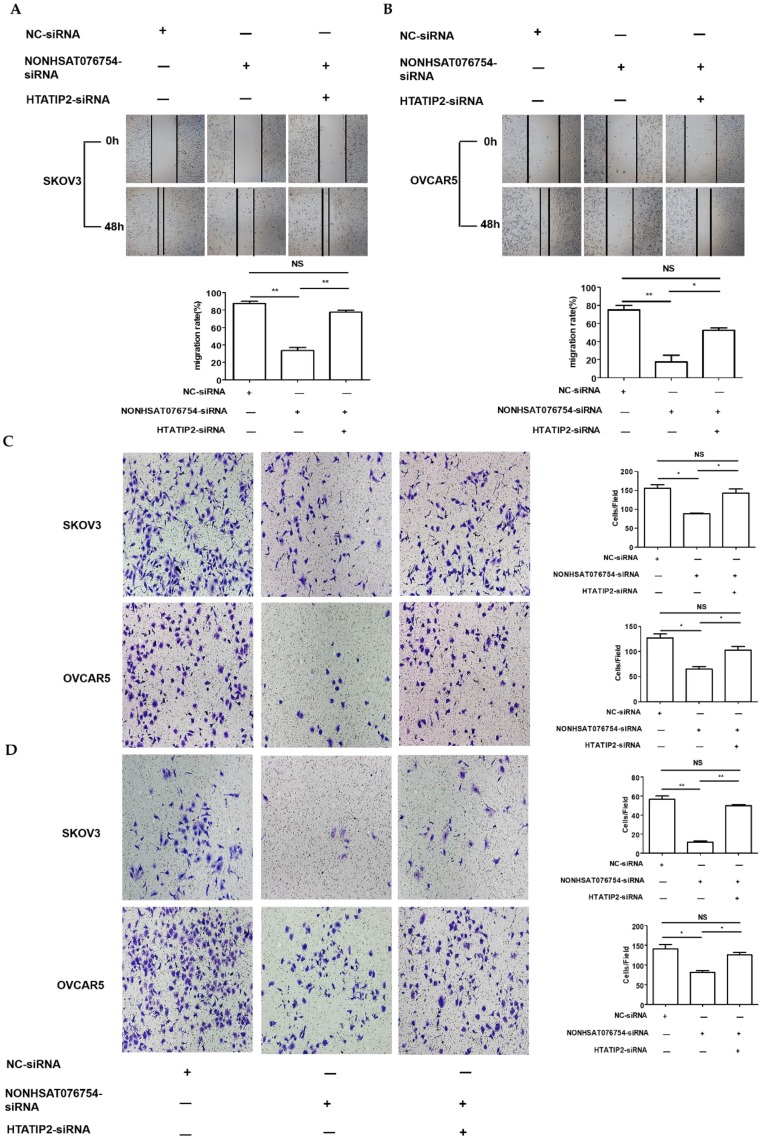
Since HTATIP2 was the most markedly altered genes when NONHAT076754 was silenced in both two EOC cells (SKOV3 and OVCAR5), we further attempted to ascertain whether NONHAT076754 acted its pro-metastatic effect partly through its target HTATIP2. Firstly, three siRNAs (siRNA-1, siRNA-2 and siRNA-3) targeting HTATIP2 were synthesized, among which, siRNA-2 acquired the most significant knockdown efficiency in SKOV3 and OVCAR5 cells at both mRNA and protein levels (Fig. 6A and 6B). Therefore, siRNA-2 was used for further co-transfection assays. Secondly, two EOC cells were co-transfected NONHAT076754-siRNA or NC with HTATIP2-siRNA. Consequently, the results showed that knockdown of HTATIP2 partially reversed the inhibitory effect of silencing NONHAT076754 on EOC migration (Fig. 7A-C), and invasion (Fig. 7D). These data further determined that NONHAT076754 might alternate cell migration, and invasion partly through its modulation on HTATIP2.
Figure 6.
Expression of HTATIP2 is silenced by siRNAs in SKOV3 and OVCAR5 cells. A. The mRNA expression levels of HTATIP2 in SKOV3 and OVCAR5 cells transfected with siRNAs were examined by qRT-PCR. B. The protein expression levels of HTATIP2 in SKOV3 and OVCAR5 cells after transfection of siRNAs were determined by WB assays. The results are shown as the mean± SD. *** p < 0.001 vs. NC groups.
Figure 7.
Knockdown of HTATIP2 partially reverses NONHAT076754 silencer-induced repressive effects on EOC metastasis. A. Knockdown of HTATIP2 partially reversed NONHAT076754 silencer-induced inhibition of migration in SKOV3 detected by wound healing assays. B. Knockdown of HTATIP2 partially reversed NONHAT076754 silencer-induced inhibition of migration in OVCAR5 cells detected by wound healing assays. C. Transwell migration assays confirmed the rescue action of HTATIP2 interference on altered migration activity caused by NONHAT076754-siRNA. D. Knockdown of HTATIP2 partially reversed NONHAT076754 silencer-generated anti-invasion in SKOV3 and OVCAR5 cells determined by transwell invasion assays. Data are shown as the mean± SD. * p < 0.05, ** p < 0.01 vs. NC groups.
Discussion
Ovarian cancer ranks among the most common and lethal malignant diseases worldwide. The poor prognosis of ovarian cancer is primarily linked to the occurrence of metastasis, which is a complex process that is poorly understood 2. Recently, accumulating evidence has indicated that lncRNAs play prominent roles in tumour metastasis 25. Although various lncRNAs have been reported to act as crucial determinants of cancer metastasis, knowledge regarding the metastasis-associated lncRNAs involved in EOC is still limited. Therefore, further studies investigating lncRNAs in EOC metastasis are imperative for improving the clinical diagnosis and treatment of EOC.
In this investigation, we identified four metastasis-associated lncRNAs including NONHSAT076754, HEIRCC, lncTCF7 and ZEB2NAT, whose roles in EOC have not been explored. Among these lncRNAs, NONHSAT076754 attracted much interest as the most significantly differentially expressed lncRNA in EOC cell lines and tissues. NONHSAT076754, which is a newly functionally characterized lncRNA, was initially implicated in PTC and elevation in NONHSAT076754 led to PTC progression 16. To the best of our knowledge, this study is the first to evaluate the contribution of NONHSAT076754 to EOC metastasis.
Here, we confirmed that high NONHSAT076754 expression was linked to aggressive clinicopathological variables including advanced tumor stage, high tumor grade and lymph node metastasis. Interestingly, there was no significant difference (P-value 0.074) between NONHSAT076754 expression and distant metastasis, which might due to the limitation of tissue bank-derived clinical samples and small sample size. Therefore, clinical observation with larger sample size was essential to ascertain the real relevance in future. Furthermore, the primary tumors with lymph node metastasis expressed higher NONHSAT076754 levels than those in the non-lymph node metastasis groups, suggesting that this lncRNA is highly related to the metastatic potential of EOC. Based on the expression and clinical features of NONHSAT076754, we speculated that this molecule might perform a critical function in the biological process of EOC metastasis.
To verify our hypothesis, we subsequently performed a set of in vitro and in vivo assays. The cell function experiments investigating migration and invasion showed that the depletion of NONHSAT076754 impaired the metastatic and invasive potential in vitro. Consistently, by using an animal model of abdominal metastasis, the results from both the bioluminescence imaging and tumor dissection showed that NONHSAT076754 silencing in nude mice resulted in a remarkable reduction in the number, weight and range of the tumor. Taken together, these results suggest that NONHSAT076754 has a pro-metastasis effect on EOC.
Since the metastatic value of NONHSAT076754 in EOC was determined, we further supposed whether certain metastasis-related downstream genes were affected by NONHSAT076754. To date, the mechanisms underlying NONHSAT076754-mediated pro-metastasis behavior are unclear. Metastasis is a complex process when multiple events occur and plenty of genes are dysregulated 26. To systematically explore the relevant molecular regulation of NONHSAT076754, a tumour metastasis real-time PCR array containing 84 well-known genes that are well known to be associated with tumour metastasis was applied. The subsequent validation of the array data confirmed that the expression of PTEN and HTATIP2, two well-known tumor suppressors 27-31, was altered at both mRNA and protein levels after the knockdown of NONHSAT076754. Further rescue assays determined that the repressive effects of silencing NONHAT076754 on metastasis could be reversed by co-transfection with HTATIP2-siRNA. Collectively, this study implied that NONHSAT076754 facilitated EOC aggressiveness and metastasis partially by modulating HTATIP2. Notably, future efforts are needed to fully understand the direct or indirect regulatory relationship between NONHSAT076754 and downstream genes. Moreover, pertaining to clinical application, more clinical data, including blood specimen and prognosis of patients inaccessible in this investigation for limitation in tissue bank-derived samples, are necessary to assess the effectiveness of NONHSAT076754 in clinical management.
Conclusion
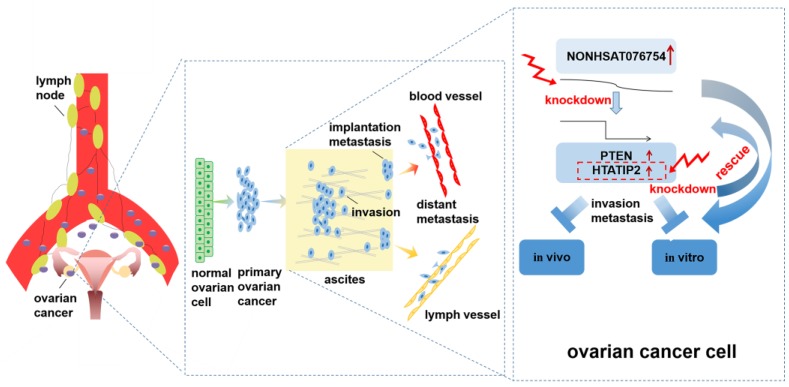
In conclusion, the present study provides the first evidence that the overexpression of NONHSAT076754 is associated with advanced tumor stage, high tumor grade and lymph node metastasis in EOC. Furthermore, NONHSAT076754 plays a pro- metastatic role in EOC by regulating certain metastasis-associated genes. Particularly, HTATIP2 might be the target downstream gene of NONHAT076754 due to rescue action of HTATIP2 interference on altered migration and invasion activity caused by NONHAT076754-siRNA (Fig. 8). This data highlight the value of NONHSAT076754 in EOC progression and imply that this molecule is a potentially crucial predictor of EOC metastasis.
Figure 8.
Schematic diagram of the mechanism proposed in this research. NONHSAT076754 was overexpressed in EOC. NONHAT076754 knockdown markedly inhibited EOC invasion and metastasis in vitro and in vivo. Mechanically, two metastasis-related genes PTEN and HTATIP2 might participate in NONHSAT076754-mediated pro-metastatic effect. Further rescue assays validated that knockdown of HTATIP2 remarkably reversed NONHAT076754 silencer-induced inhibition of EOC cell metastasis.
Acknowledgments
This research was supported by the National Natural Science Foundation of China grant 81370689 and 81571404 (to Ke-Qin Hua), the Shanghai Science and Technology Innovation Foundation (16411950500; to Ke-qin Hua), the National Natural Science Foundation for Young Scholars of China grant 81502240 (to Jun-jun Qiu), and the Shanghai Science and Technology Development Funds for the Talents grant 15YF1401400 (to Jun-jun Qiu).
Abbreviations
- EOC
epithelial ovarian cancer
- OC
ovarian cancer
- lncRNAs
long non-coding RNAs
- PTC
papillary thyroid cancer
- FBS
fetal bovine serum
- siRNA
small interfering RNA
- NC
negative control
- qRT-PCR
quantitative real-time polymerase chain reaction
- PBS
phosphate-buffer-solution
- SPF
specific-pathogen free
- WB
western blot
References
- 1.Siegel R, Naishadham D, Jemal A. CA Cancer J Clin. Cancer statistics, 2012. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Pradeep S, Kim SW, Wu SY. et al. Hematogenous metastasis of ovarian cancer: rethinking mode of spread. Cancer Cell. 2014;26:77–91. doi: 10.1016/j.ccr.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goff BA, Mandel L, Muntz HG. et al. Ovarian carcinoma diagnosis - Results of a National Ovarian Cancer Survey. Cancer. 2000;89:2068–75. doi: 10.1002/1097-0142(20001115)89:10<2068::aid-cncr6>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 4.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–9. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 5.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanchez Calle A, Kawamura Y, Yamamoto Y. et al. Emerging roles of long non-coding RNA in cancer. Cancer Sci. 2018;109:2093–100. doi: 10.1111/cas.13642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang MH, Hu ZY, Xu C. et al. MALAT1 promotes colorectal cancer cell proliferation/migration/invasion via PRKA kinase anchor protein 9. Biochim Biophys Acta. 2015;1852:166–74. doi: 10.1016/j.bbadis.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raveh E, Matouk IJ, Gilon M. et al. The H19 Long non-coding RNA in cancer initiation, progression and metastasis - a proposed unifying theory. Mol Cancer. 2015;14:184. doi: 10.1186/s12943-015-0458-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang J, Zhang P, Wang L. et al. Long non-coding RNA HOTAIR in carcinogenesis and metastasis. Acta Biochim Biophys Sin. 2014;46:1–5. doi: 10.1093/abbs/gmt117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deng J, Liang Y, Liu C. et al. The up-regulation of long non-coding RNA AFAP1-AS1 is associated with the poor prognosis of NSCLC patients. Biomed Pharmacother. 2015;75:8–11. doi: 10.1016/j.biopha.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 11.Qiu M, Xu Y, Yang X. et al. CCAT2 is a lung adenocarcinoma-specific long non-coding RNA and promotes invasion of non-small cell lung cancer. Tumour Biol. 2014;35:5375–80. doi: 10.1007/s13277-014-1700-z. [DOI] [PubMed] [Google Scholar]
- 12.Yuan JH, Yang F, Wang F. et al. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25:666–81. doi: 10.1016/j.ccr.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 13.Eades G, Wolfson B, Zhang Y. et al. lincRNA-RoR and miR-145 regulate invasion in triple-negative breast cancer via targeting ARF6. Mol Cancer Res. 2015;13:330–8. doi: 10.1158/1541-7786.MCR-14-0251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qiu JJ, Lin YY, Ding JX. et al. Long non-coding RNA ANRIL predicts poor prognosis and promotes invasion/metastasis in serous ovarian cancer. Int J Oncol. 2015;46:2497–505. doi: 10.3892/ijo.2015.2943. [DOI] [PubMed] [Google Scholar]
- 15.Qiu JJ, Lin YY, Ye LC. et al. Overexpression of long non-coding RNA HOTAIR predicts poor patient prognosis and promotes tumor metastasis in epithelial ovarian cancer. Gynecol Oncol. 2014;134:121–8. doi: 10.1016/j.ygyno.2014.03.556. [DOI] [PubMed] [Google Scholar]
- 16.Xia SJ, Wang CD, Ni XF. et al. NONHSAT076754 aids ultrasonography in predicting lymph node metastasis and promotes migration and invasion of papillary thyroid cancer cells. Oncotarget. 2017;8:2293–306. doi: 10.18632/oncotarget.13725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lan T, Chang L, Wu L. et al. Downregulation of ZEB2-AS1 decreased tumor growth and metastasis in hepatocellular carcinoma. Mol Med Rep. 2016;14:4606–12. doi: 10.3892/mmr.2016.5836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu J, Wang D. Long noncoding RNA TCF7 promotes invasiveness and self-renewal of human non-small cell lung cancer cells. Hum Cell. 2017;30:23–9. doi: 10.1007/s13577-016-0147-5. [DOI] [PubMed] [Google Scholar]
- 19.Wu J, Zhang J, Shen B. et al. Long noncoding RNA lncTCF7, induced by IL-6/STAT3 transactivation, promotes hepatocellular carcinoma aggressiveness through epithelial-mesenchymal transition. J Exp Clin Cancer Res. 2015;34:116. doi: 10.1186/s13046-015-0229-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiong J, Liu Y, Luo S. et al. High expression of the long non-coding RNA HEIRCC promotes Renal Cell Carcinoma metastasis by inducing epithelial-mesenchymal transition. Oncotarget. 2017;8:6555–63. doi: 10.18632/oncotarget.14149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhuang J, Lu Q, Shen B. et al. TGFbeta1 secreted by cancer-associated fibroblasts induces epithelial-mesenchymal transition of bladder cancer cells through lncRNA-ZEB2NAT. Sci Rep. 2015;5:11924. doi: 10.1038/srep11924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li-Korotky HS, Hebda PA, Lo CY. et al. Age-dependent differential expression of fibronectin variants in skin and airway mucosal wounds. Arch Otolaryngol. 2007;133:919–24. doi: 10.1001/archotol.133.9.919. [DOI] [PubMed] [Google Scholar]
- 23.Wang YM, Reheman A, Spring CM. et al. Plasma fibronectin supports hemostasis and regulates thrombosis. J Clin Invest. 2014;124:4281–93. doi: 10.1172/JCI74630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang C, Wu MZ, Zhang LZ. et al. Fibrotic microenvironment promotes the metastatic seeding of tumor cells via activating the fibronectin 1/secreted phosphoprotein 1-integrin signaling. Oncotarget. 2016;7:45702–14. doi: 10.18632/oncotarget.10157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen XH, Qi P, Du X. Long non-coding RNAs in cancer invasion and metastasis. Mod Pathol. 2015;28:4–13. doi: 10.1038/modpathol.2014.75. [DOI] [PubMed] [Google Scholar]
- 26.Vanharanta S, Massague J. Origins of metastatic traits. Cancer Cell. 2013;24:410–21. doi: 10.1016/j.ccr.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simpson L, Parsons R. PTEN: life as a tumor suppressor. Exp Cell Res. 2001;264:29–41. doi: 10.1006/excr.2000.5130. [DOI] [PubMed] [Google Scholar]
- 28.Yang H, Kong W, He L. et al. MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res. 2008;68:425–33. doi: 10.1158/0008-5472.CAN-07-2488. [DOI] [PubMed] [Google Scholar]
- 29.Shtivelman E. A link between metastasis and resistance to apoptosis of variant small cell lung carcinoma. Oncogene. 1997;14:2167–73. doi: 10.1038/sj.onc.1201059. [DOI] [PubMed] [Google Scholar]
- 30.Ito M, Jiang C, Krumm K. et al. TIP30 deficiency increases susceptibility to tumorigenesis. Cancer Res. 2003;63:8763–7. [PubMed] [Google Scholar]
- 31.Xiao H, Palhan V, Yang Y. et al. TIP30 has an intrinsic kinase activity required for up-regulation of a subset of apoptotic genes. EMBO J. 2000;19:956–63. doi: 10.1093/emboj/19.5.956. [DOI] [PMC free article] [PubMed] [Google Scholar]