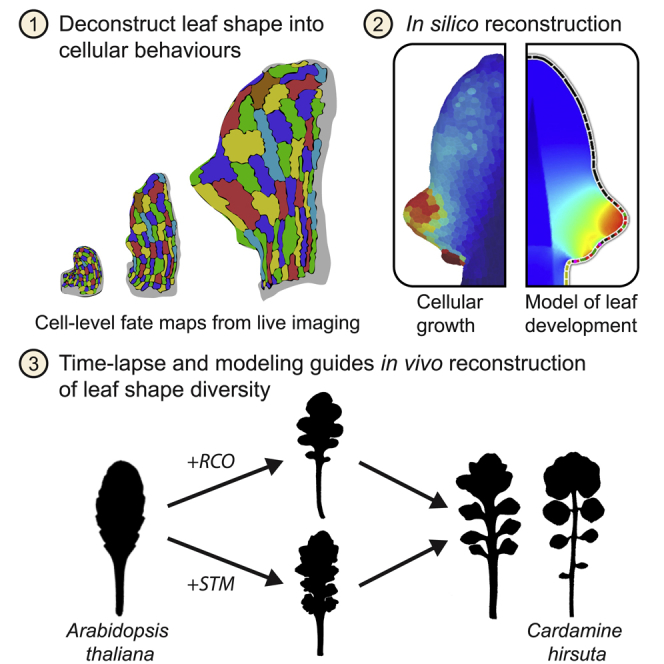
Summary
How do genes modify cellular growth to create morphological diversity? We study this problem in two related plants with differently shaped leaves: Arabidopsis thaliana (simple leaf shape) and Cardamine hirsuta (complex shape with leaflets). We use live imaging, modeling, and genetics to deconstruct these organ-level differences into their cell-level constituents: growth amount, direction, and differentiation. We show that leaf shape depends on the interplay of two growth modes: a conserved organ-wide growth mode that reflects differentiation; and a local, directional mode that involves the patterning of growth foci along the leaf edge. Shape diversity results from the distinct effects of two homeobox genes on these growth modes: SHOOTMERISTEMLESS broadens organ-wide growth relative to edge-patterning, enabling leaflet emergence, while REDUCED COMPLEXITY inhibits growth locally around emerging leaflets, accentuating shape differences created by patterning. We demonstrate the predictivity of our findings by reconstructing key features of C. hirsuta leaf morphology in A. thaliana.
Video Abstract
Keywords: leaf development, growth and patterning, live-imaging, organ shape, Cardamine hirsuta, Arabidopsis thaliana, KNOX, RCO, computational modelling, morphogenesis
Graphical Abstract
Highlights
-
•
Complete growth and fate maps are made for C. hirsuta and A. thaliana leaf surfaces
-
•
Patterning of the leaf margin modifies organ-wide growth pattern to produce leaf shape
-
•
Altering growth relative to patterning generates leaf shape diversity
-
•
Reconstructing dissected leaf shape by combining STM and RCO in A. thaliana leaves
By understanding the impact of growth and patterning on leaf shape diversity, it is possible to convert the leaf morphology of A. thaliana to that of another plant species.
Introduction
How gene activity translates into distinct organ morphologies remains poorly understood (e.g., Runions and Tsiantis, 2017, Zuniga, 2015). To understand the complex interactions that link gene action to tissue form, we need quantitative data on cellular growth at high spatial and temporal resolution, which are technically challenging to acquire (Etournay et al., 2016, Fox et al., 2018). Such data are also difficult to interpret because local gene activity can have complex non-local effects on tissue deformations, which arise from interactions between genetically specified growth and tissue mechanics (Coen and Rebocho, 2016). We also do not yet understand how evolutionary changes in gene activity alter how growth, patterning, and differentiation interact to produce diverse organ forms.
Plant leaves are an attractive system in which to address these questions as they grow from almost indistinguishable primordia into shapes that vary tremendously among species. Leaves can be simple, with smooth undivided margins (the botanical term for leaf edge), or complex, with protrusions of different size and geometry. The leaves of A. thaliana are simple and bear small marginal protrusions called serrations, while those of its relative, C. hirsuta, are dissected into distinct leaflets that each resemble a simple leaf (Figures 1A and 1B). Genetic studies have identified KNOX (Knotted1-like homeobox) and RCO (REDUCED COMPLEXITY) homeobox genes as important molecular regulators of leaf complexity that are involved in the evolutionary diversification of leaf form (Bharathan et al., 2002, Hareven et al., 1996, Hay and Tsiantis, 2010, Vlad et al., 2014). While target genes are known for some KNOX proteins (Bolduc et al., 2012), we still do not understand how KNOX and RCO affect cell- and tissue-level growth during leaf primordium development. Furthermore, we lack information on the growth of morphogenetically important domains at the margin and base of developing leaves, and cell-level fate maps for leaf primordia do not exist. It thus remains unclear how local growth regulation at the margin integrates with global patterns of growth, proliferation, and differentiation to produce divergent leaf forms (Bar and Ori, 2014, Bilsborough et al., 2011, Alvarez et al., 2016, Donnelly et al., 1999, Fox et al., 2018, Kuchen et al., 2012, Poethig, 1987). For example, current evidence indicates that growth polarity is vital for leaf geometry, yet the degree to which this polarity is a local or global feature of organ development and how it shapes leaf form remain unclear (Bringmann and Bergmann, 2017, Kuchen et al., 2012, Mansfield et al., 2018). Computational modeling offers one way to address these questions (Ali et al., 2014), by enabling us to investigate how multiple processes interact to create geometry in a growing tissue.
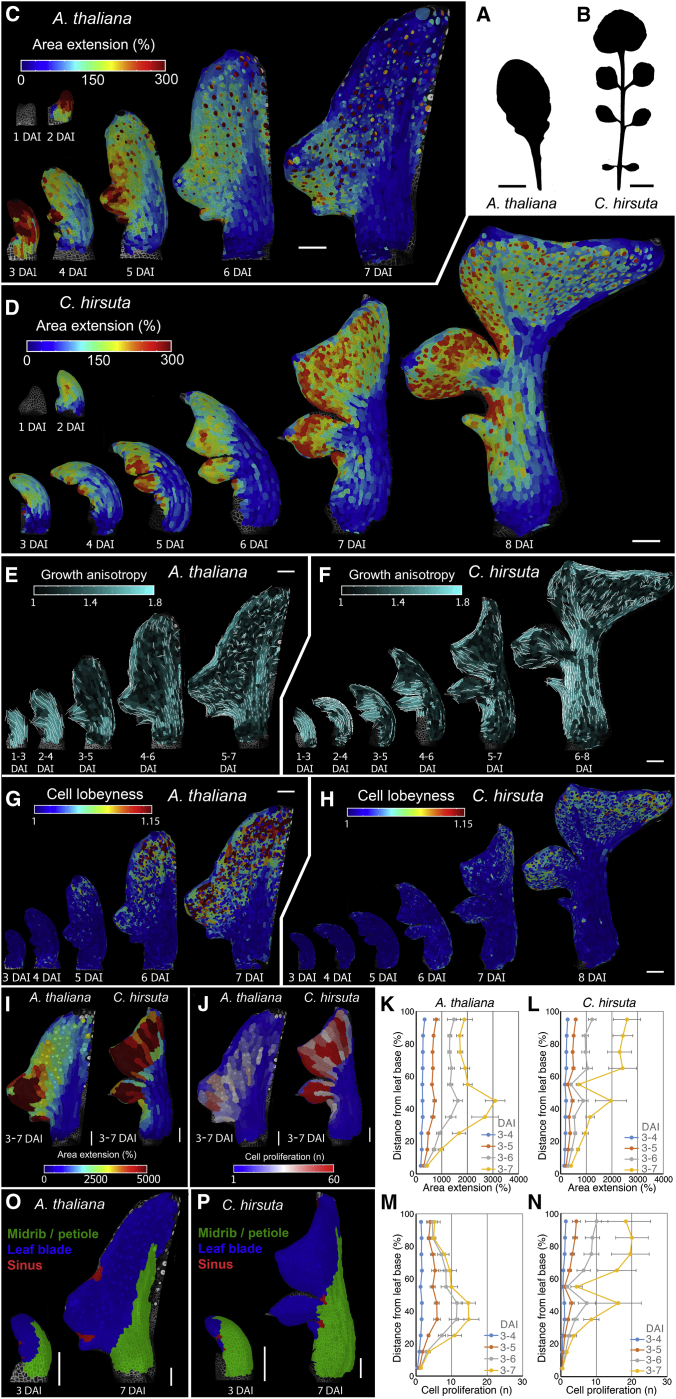
Figure 1.
Conserved versus Divergent Growth Patterns in A. thaliana and C. hirsuta Leaves
(A and B) Eighth rosette leaf of A. thaliana (A) and C. hirsuta (B).
(C–H) Heat-maps of area extension (C and D), growth anisotropy (E and F), and cell lobeyness (G and H) for A. thaliana (C, E, and G) and C. hirsuta (D, F, and H) leaves. Lobeyness measures pavement cell undulation (Sapala et al., 2018, see STAR Methods). White lines in (E) and (F) indicate cell-growth orientation where anisotropy > 40%. In both species, proliferation and growth increased at protrusions and decreased in adjacent sinuses (Figure S2). At the leaf margin, a basipetal transition from dynamic growth to tissue-dependent patterning occurs, coinciding with differentiation progression (C–H). See also Figures S1A–S1J.
(I and J) Heat-maps of area extension (I) and cell proliferation (J), 3–7 DAI for A. thaliana (left) and C. hirsuta (right).
(K–N) Growth alignment graph of mean area extension (K and L), and cell proliferation (M and N), from 3–7 DAI as a function of distance from leaf base in A. thaliana (K and M) and C. hirsuta (L and N). Error bars, SEM (n = 5–28, K and M; n = 11–27, L and N).
(O and P) Lineage tracing of leaf blades (blue), petiole and midrib (green), and sinuses (red) in A. thaliana (O) and C. hirsuta (P).
DAI indicates days after primordium initiation. Scale bars, 1 cm in (A) and (B), 100 μm in (C)–(J), (O), and (P). See also Figure S1 and S2 and Video S1. Replication for imaging data is reported for all figures in STAR Methods.
Here, we identify differences in growth patterns that yield simple, elliptical A. thaliana leaves versus dissected C. hirsuta leaves with a broad terminal leaflet. We use live imaging and genetics alongside computational modeling and analyses of growth to deconstruct these two divergent leaf forms into their cell-level constituent elements: the amount and direction of growth and the rate of differentiation. Surprisingly, we find that key features of growth are conserved between these two leaf forms. Differences in leaf geometry originate from two distinct processes that act in C. hirsuta, but not in A. thaliana leaves. In the first process, which requires the KNOX gene SHOOTMERISTEMLESS (STM), delayed differentiation and slower but prolonged growth throughout the leaf primordium increase the size and number of protrusions initiated by a conserved auxin-based pattern-generating mechanism. This process also allows protrusions to grow for longer in a polarized fashion. In the second process, local growth inhibition, mediated by the RCO gene, accentuates growth differences created by marginal patterning. We demonstrate the predictive power of our approach by experimentally reconstructing key aspects of the dissected C. hirsuta leaf form in A. thaliana.
Results
Development of Simple versus Dissected Leaf Primordia
We developed an imaging protocol (see STAR Methods) to understand how the balance of conserved versus diverged cellular growth patterns produce the simple leaf forms of A. thaliana and the dissected leaves of C. hirsuta (Figures 1A and 1B). We measured leaf primordium growth at cellular resolution, from its emergence until 7–8 days after initiation (DAI), when shape divergence between the two species is established. Our measurements included the leaf margin where serrations and leaflets form (Figure 1; Video S1). We computed complete lineage maps from these data (Barbier de Reuille et al., 2015) to understand how cells in the early primordium contribute to the development of the mature leaf form. Using the lineage maps, we quantified cell growth parameters that affect form (see STAR Methods), including growth amount (rate of cell-area increase), directionality (anisotropy, the ratio of expansion in the max. and min. principal directions of growth), and cell proliferation. To assess the progression of tissue differentiation, we also measured cell size, stomatal density, and pavement cell geometry.
1
We observed that despite their different final forms, leaves from these two species shared three commonalities in their growth patterns (Figures 1C–1H and S1A–S1S). (1) During leaf initiation (1–3 DAI), primordium growth is uniform and anisotropic. (2) After the initiation phase (∼3–4 DAI), dynamic marginal growth patterns are established in lateral regions, which are defined by changing patterns of proliferation, growth, and anisotropy that accompany protrusion outgrowth (Figures 1C–1F and S2A–S2F). (3) Later, in association with the basipetal (i.e., tip-to-base) progression of differentiation, tissue-dependent growth patterns emerge, with mainly isotropic growth occurring in the blade and anisotropic growth occurring along the margin circumference, midrib, and petiole (Figures 1E, 1H and S1L–S1S). Notably, in both plants, fronts of differentiation initiated at the leaf tip and each protrusion tip, rather than as a single front, and progressed toward the leaf base (Figures 1C–1D, 1G, 1H, S1A–S1D, and S2A–S2F). Proliferation decreased abruptly in the midrib/petiole region at ∼4 DAI (Figures S1A–S1D) with differentiation onset, while in the rest of the blade, proliferation decreased gradually, coinciding with the basipetal progression of differentiation. These results indicate that species-specific differences in leaf form emerge in the context of these three conserved growth patterns.
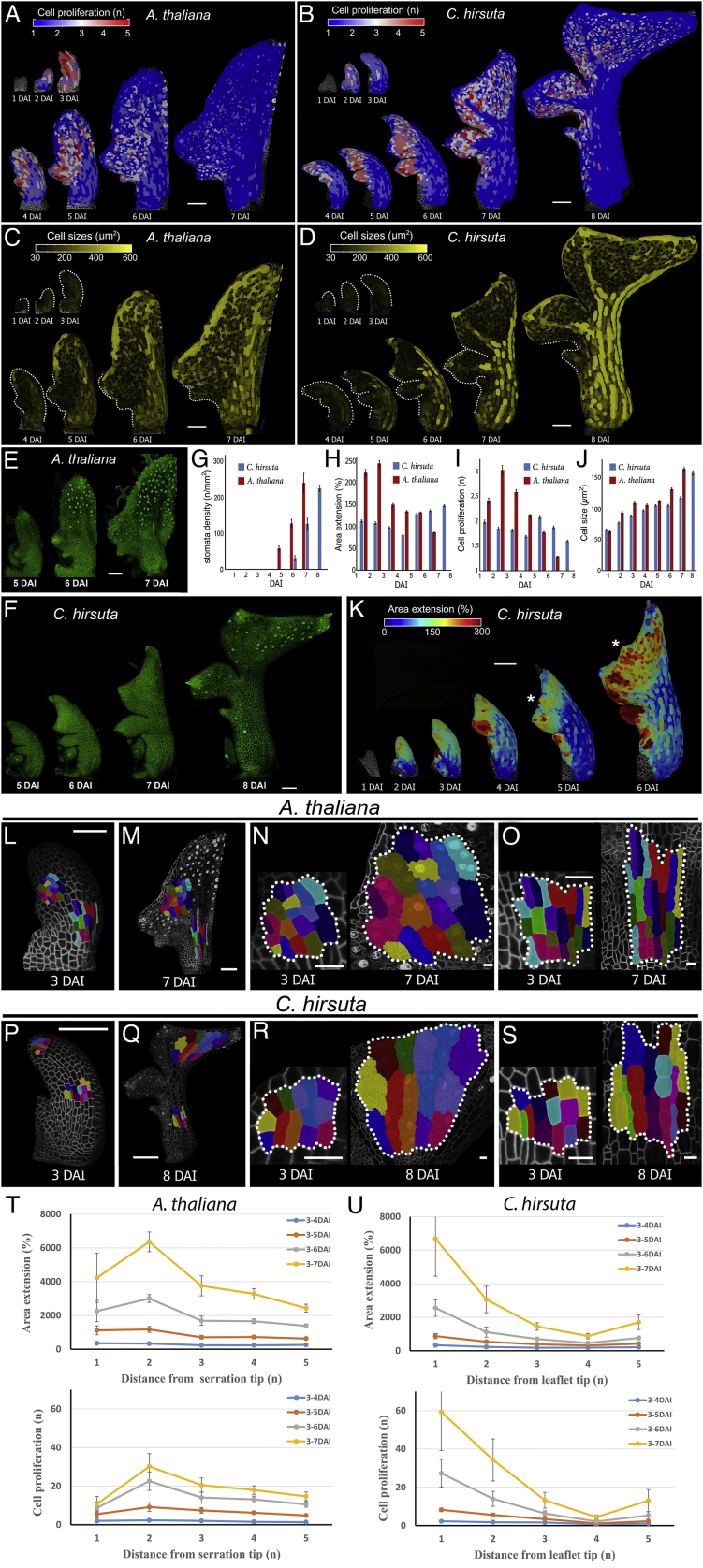
Figure S1.
Analysis of the Conserved versus Divergent Aspects of Growth of A. thaliana and C. hirsuta Leaves, Related to Figure 1
(A-D) Heat-maps of cell proliferation (A-B) and cell size (C-D) for A. thaliana (A,C), and C. hirsuta (B,D) leaves.
(E-F) Confocal time-lapse series of developing A. thaliana (E) and C. hirsuta (F) leaves with plasma membrane marker in green. Note the earlier appearance of stomata (dots marked with stronger YFP signal) in A. thaliana.
(G) Bar-graph representing stomata density in relation to DAI (error bars indicate SE). n = 2-7 (A. thaliana), n = 3-6 (C. hirsuta).
(H-J) Quantifications of mean area extension (H), cell proliferation (I), and cell size (J) for A. thaliana and C. hirsuta leaves (error bars indicate SE). A. thaliana, n > 100 (3 independent time-lapse series); C. hirsuta, n > 350 (4 independent time-lapse series).
(K) Heat-maps of area extension for a C. hirsuta leaf. Asterisk indicates additional protrusion emerging from the leaf margin of the terminal leaflet.
(L-S) Cell lineage tracing analysis in the leaf blade (N and R) or the petiole/midrib (O and S) for A. thaliana (L-O) and C. hirsuta (P-S) leaves. Location of the sectors at the beginning (L and P) and end (M and Q) of observations. Colors show the correspondence between cells at 3 DAI and their clonal sectors at 7 DAI.
(T-U) Growth alignment graphs showing mean area extension and cell proliferation from 3-7 DAI as a function of distance from the tip of the first serration in A. thaliana (T) and leaflet in C. hirsuta (U). Distance is measured in cell-number. Error bars indicate SEM (n = 5-28, T; n = 3-24, U).
DAI indicates days after primordia initiation. Scale bars, 100 μm in (A-F, K-M, P-Q), and 20 μm in (N-O and R-S).
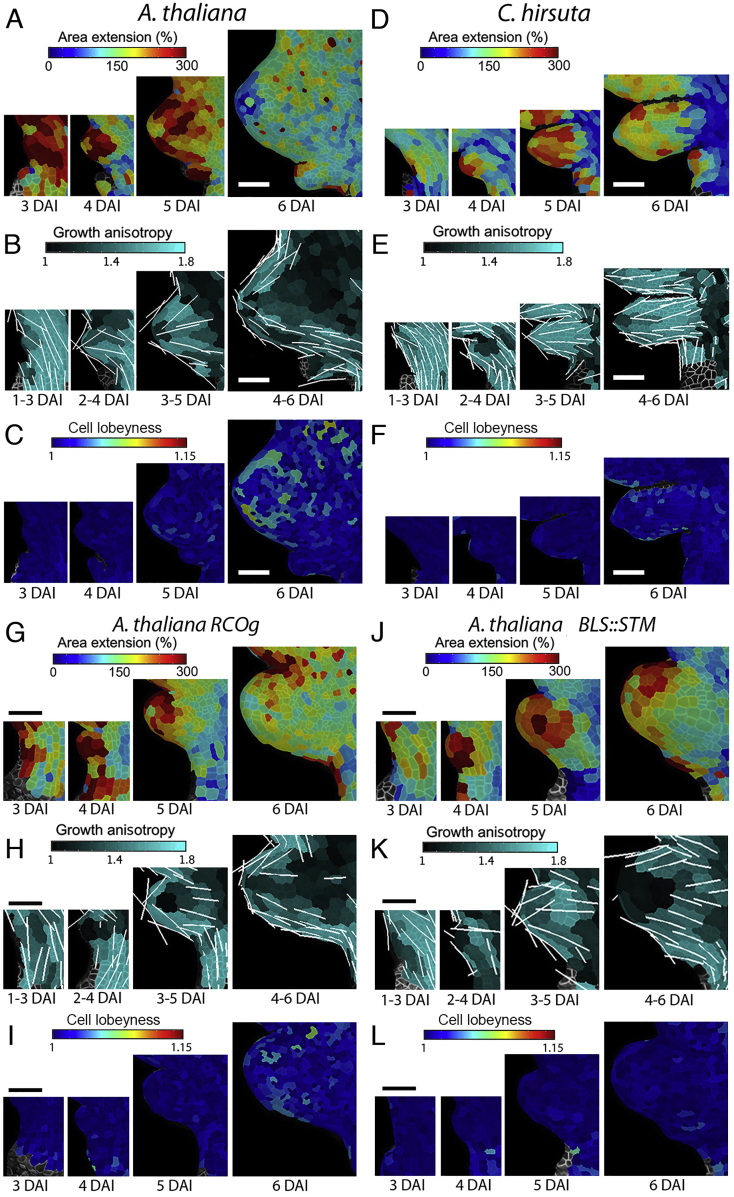
Figure S2.
Growth Patterns of Margin Protrusions, Related to Figure 1, Figure 2, Figure 3, Figure 4
(A-C) Heat-maps of area extension (A), growth anisotropy (B), and cell lobeyness (C) for A. thaliana WT serration.
(D-F) Heat-maps of area extension (D), growth anisotropy (E), and cell lobeyness (F) for C. hirsuta WT lateral leaflet.
(G-I) Heat-maps of area extension (G), growth anisotropy (H), and cell lobeyness (I) for A. thaliana RCOg lobe.
(J-L) Heat-maps of area extension (J), growth anisotropy (K), and cell lobeyness (L) for A. thaliana BLS::STM lobe.
White lines in (B, E, H and K) indicate the orientation of cell growth where anisotropy is higher than 40%. Scale bars 50 μm.
To identify in an unbiased manner the differences in cellular growth distribution that yield these distinct leaf forms, we quantified and compared the growth and proliferation of clonal lineages of each species from 3–7 DAI (Figures 1I and 1J). In this context, we also computed growth alignment graphs by mapping growth and proliferation, according to the location of cells along a leaf’s Proximal-Distal axis (P-D axis, location assessed at 3 DAI, Figures 1K–1N). This approach provides a developmental biology equivalent of a sequence alignment, and we used it to determine the balance of conservation versus divergence in cell-level growth properties at a given developmental stage in diverse genotypes. Although primordium length was comparable between species at both 3 and 7 DAI (∼150 and ∼800 μm, respectively), the P-D distributions of growth and proliferation differed (Figures 1I–1N). Growth and proliferation were restricted to proximal regions in A. thaliana but were broadly distributed in the C. hirsuta blade (Figures 1I and 1J), increasing the contribution of the distal primordium to the leaf surface (Figures 1K–1N) and correlating with the emergence of a broad terminal leaflet. Increased distal growth in C. hirsuta, compared to A. thaliana, coincided with the delayed onset of tissue-dependent growth patterns (Figures 1C–1H), delayed differentiation (Figures S1E–S1J), and extended dynamic marginal growth (as indicated by the emergence of additional protrusions in the terminal leaflet, Figure S1K). Thus, the distribution of growth and proliferation in C. hirsuta leaves is shifted distally along the P-D axis relative to A. thaliana. This shift is associated with prolonged marginal patterning and a global delay in differentiation.
We next investigated the local growth features that underlie the different leaf protrusion shapes of each species. Protrusion shape is influenced by the growth differential between rapidly growing protrusion tips and slow-growing adjacent tissues (Bilsborough et al., 2011, Nikovics et al., 2006, Vlad et al., 2014). We observed large differences in this growth differential between species (Figures S1T and S1U). Compared to serrations, the duration of anisotropic growth was extended in leaflets (2–6 DAI in leaflets versus 2–5 DAI in serrations; Figures 1E–1F, S2B, and S2E), as was the duration of reduced growth and proliferation in the sinus regions between leaflets (3–7 DAI in leaflets versus 3–5 DAI in serrations, Figure 1C, 1D, S1A, S1B, S2A, and S2D). Consequently, in C. hirsuta, both increased protrusion outgrowth and reduced growth at the protrusion base contributed to leaflet formation (compare Figure S1T and S1U). Reduced growth at the protrusion base likely reflects local growth inhibition (Vlad et al., 2014) but may also result from global differences in the growth of primordia (C. hirsuta growth is almost half that of A. thaliana over 1–3 DAI, Figures S1H and S1I). Thus, reduced growth at the protrusion base distinguishes leaflets in C. hirsuta from serrations in A. thaliana and likely involves both local and global growth regulation.
The hypothesis that both global and local aspects of growth contribute to the increased growth inhibition between protrusions in C. hirsuta leaves raised the question of how these two different growth modes interact to shape leaf geometry. To investigate this, we examined the origin of protrusions and sinuses relative to the global (organ-wide) growth patterns of the leaf blade, petiole, and midrib. Based on patterns of growth, proliferation, and cell morphology (Figures 1C–1J and S1A–S1D), we divided 3 and 7 DAI primordia into three regions: (1) blade, (2) petiole and midrib, and (3) the slowly growing sinus cells that lie adjacent to emerging protrusions (Figures 1O–1P, see STAR Methods). We found that in A. thaliana, sinuses initiated in the leaf blade and were thus always surrounded by rapidly growing blade cells (Figure 1O). By contrast, at 3 DAI in C. hirsuta, growth-repressed zones appeared directly adjacent to the midrib (Figure 1P), preventing blade establishment between protrusions and enabling the formation of a dissected leaf. Thus, sinus establishment in the blade is associated with a simple leaf, and in the midrib, with a dissected leaf.
These results suggest that three key differences distinguish dissected leaves from simple ones: (1) the local context in which protrusions initiate (midrib versus blade); (2) decreased growth at the base of initiating marginal protrusions; and (3) a global change in growth pattern, coinciding with delayed differentiation and marked by increased lateral and distal growth and by reduced growth in proximal and medial regions. Thus, both global and local factors influence growth to shape simple and dissected leaves.
Relationship between Marginal Patterning and Leaf Growth
Our data suggest that differences in tissue growth during the patterning of marginal protrusions help to create distinct leaf forms. The placement and growth of marginal protrusions requires polar auxin transport, which is controlled by the auxin efflux carrier PIN-FORMED1 (PIN1, Figures 2A and 2B) and is influenced by the transcription factor CUP-SHAPED COTYLEDON2 (CUC2) (Barkoulas et al., 2008, Bilsborough et al., 2011, Hay et al., 2006, Nikovics et al., 2006, Rast-Somssich et al., 2015). To investigate how components of the auxin-PIN1-CUC2 module influence leaf growth and differentiation, we first examined the role of auxin. Activity of this hormone, as reported by the DR5 auxin activity sensor, marks outgrowing protrusions in both A. thaliana and C. hirsuta leaves (Barkoulas et al., 2008, Hay et al., 2006). Two lines of evidence indicated that auxin activity maxima influence the rate and direction of growth. First, we found that initial growth in protrusions is rapid and anisotropic (Figures 2D, 2F, S2A, S2B, S2D, and S2E). Second, these points of rapid anisotropic growth are absent in the leaves of C. hirsuta and A. thaliana plants treated with the polar auxin-transport inhibitor, 1-N-naphthylphthalamic acid (NPA) (Figures S3A–S3J), which lack auxin maxima (Hay et al., 2006, Barkoulas et al., 2008). NPA treatment generated simple leaves in both species, and a growth pattern also observed in the leaves of pin1 and cuc2 A. thaliana mutants (Figures 2G–2I and S4A–S4G; Video S2), which also lack discrete marginal auxin maxima (Bilsborough et al., 2011, Hay et al., 2006). By contrast, in addition to its role in patterning auxin maxima, CUC2 appears to inhibit growth. Following the establishment of auxin maxima, CUC2 is expressed primarily in the sinuses of A. thaliana leaves (Nikovics et al., 2006, Bilsborough et al., 2011), where growth and proliferation are reduced (Figures 1C, 1I, 1J, S1A, and S1T), and this growth inhibition is lost in cuc2 mutants (Figures 2G and S4B). Thus, auxin and CUC2 shape the leaf margin by locally increasing growth at protrusions and decreasing growth at their flanks, respectively.
Figure 2.
Relationship between Marginal Patterning and Growth
(A and B) Localization of pPIN1::PIN1:GFP (green) in A. thaliana (A) and C. hirsuta (B) leaves at consecutive developmental stages.
(C–F) pPIN1::PIN1:GFP signal distribution near cell border (C and E) and growth anisotropy (D and F) in A. thaliana (C and D) and C. hirsuta (E and F) emerging protrusions.
(G–I) Heat-maps of area extension (G), growth anisotropy (H), and cell lobeyness (I) for cuc2-3 A. thaliana mutant.
(J and K) Heat-maps of area extension (J) and cell lobeyness (K) for 5 μm IAA treated A. thaliana leaves.
Yellow lines in (C) and (E) indicate cellular orientation of PIN1 polarization. White lines in (D), (F), and (H) indicate cell growth orientation when anisotropy > 20% (D and F) or 40% (H). Scale bars, 100 μm (A and B, G–K), 20 μm (C–F).
See also Figure S2, Figure S3, Figure S4, Figure S5 and Video S2.
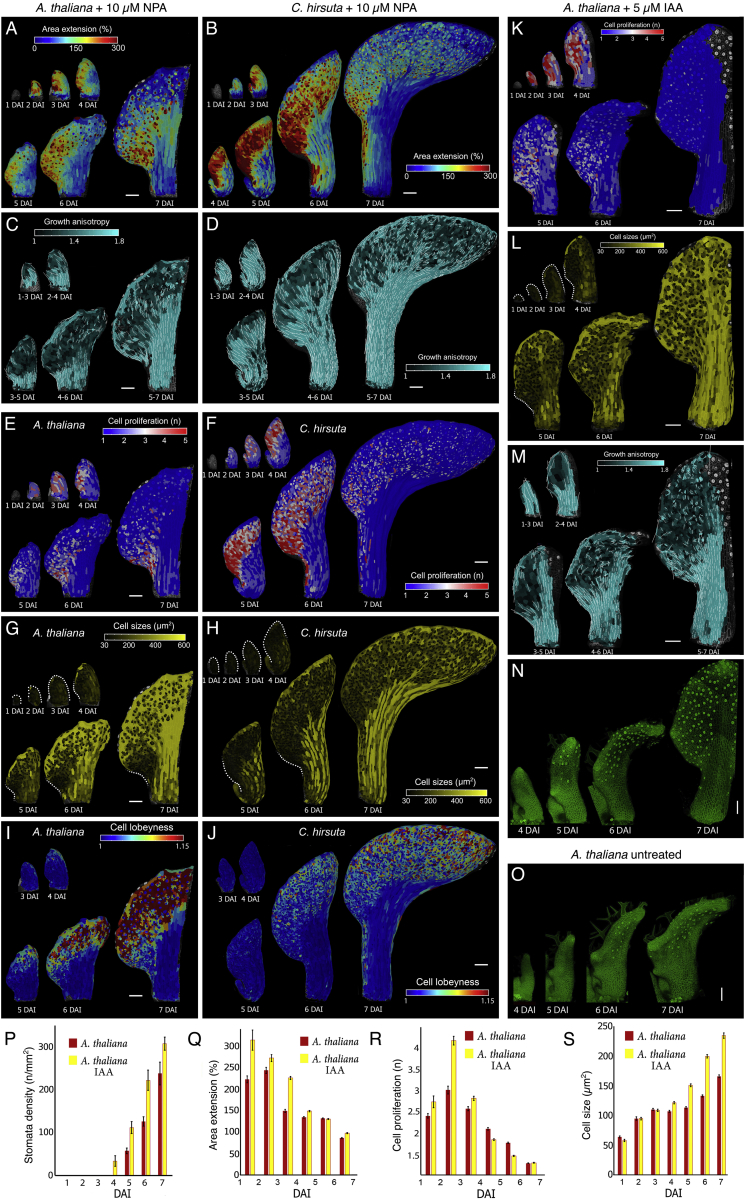
Figure S3.
IAA and NPA Treated Wild-Type A. thaliana Leaves, Related to Figure 2
(A-J) Heat-maps of area extension (A,B), growth anisotropy (C,D), proliferation (E,F), cell size (G,H) and lobeyness (I,J) for NPA-treated A. thaliana (A,C,E,G,I), and C. hirsuta (B,D,F,H,J) leaves.
(K-M) Heat-maps of cell proliferation (K), cell size (L), and growth anisotropy (M) for a A. thaliana wild-type leaf treated with 5 μM IAA.
(N-O) Confocal time-lapse series of the developing A. thaliana leaf treated with 5 μM IAA (N) as compared to the control (O).
(P-S) Quantifications of stomatal density (P), mean area extension (Q), cell proliferation (R), and cell size (S) for A. thaliana WT leaves treated with 5 μM IAA as compared to control (error bars indicate SEM). A. thaliana leaf treated with 5 μM IAA, n = 3-7 (P) and n > 80 (Q-S, 4 independent time-lapse series).
White lines in (C-D, M) indicate cell growth orientation where anisotropy > 40%. Scale bars, 100 μm. Dotted lines in (L) indicate leaf outlines.
Figure S4.
Leaf Growth Characteristics Converge in the Absence of Marginal Patterning, Related to Figures 2 and 3
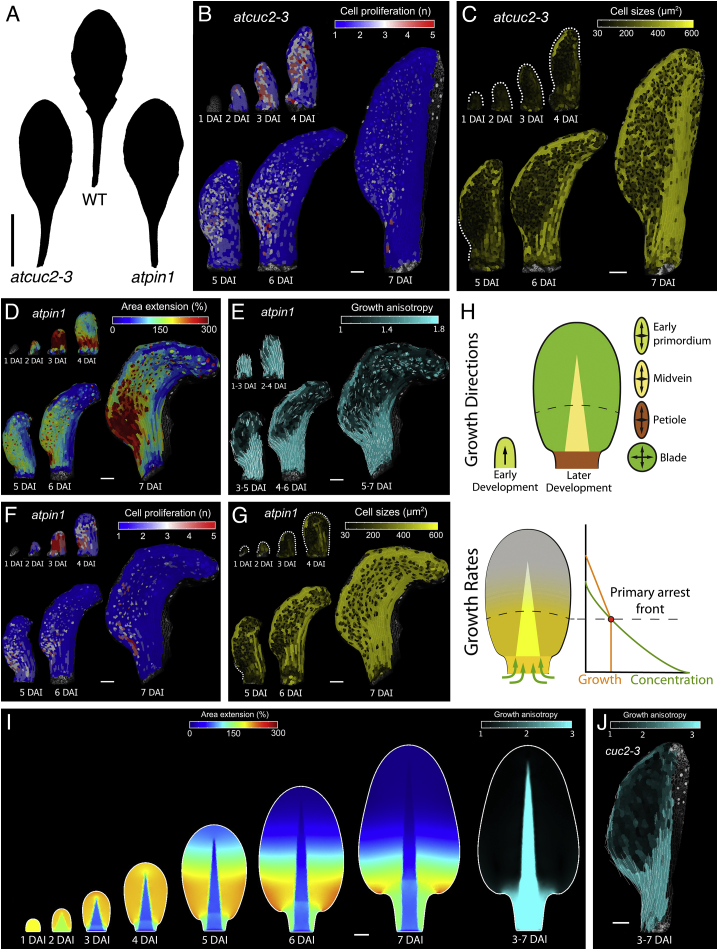
(A) Silhouettes of A. thaliana wild-type (middle), cuc2-3 (left), and pin1 (right) mutant leaves.
(B-C) Heat-maps of cell proliferation (B), and cell size (C) for A. thaliana cuc2-3 leaf.
(D-G) Heat-maps of area extension (D), growth anisotropy (E), cell proliferation (F), and cell size (G) for the A. thaliana pin1 mutant leaf.
(H-J) Model of leaf growth in the absence of marginal patterning. (H) Specified growth is homogeneous and anisotropic during early development (initiation, 1-1.85 DAI), and follow indicated tissue-dependent directions thereafter (anisotropic in the midrib/petiole, isotropic in the blade). Specified growth rates depend on tissue type and differentiation. Differentiation begins when a compound diffusing from the leaf base (green curve, graph) falls below a threshold value (dotted line). Following differentiation, growth (orange curve) decreases. (I-J) Resultant shape and distribution of growth rates and anisotropy of the model (I), compared to growth anisotropy in cuc2-3 mutant (J). Note that growth rates decrease from the leaf tip to the base, mirroring the progression of differentiation. See also Video S3.
White lines in (E,J) indicate the orientation of cell growth where anisotropy is higher than 40%. Dotted lines in (C, G) indicate leaf outlines. Scale bars, 1 cm in (A), 100 μm in (B-G and I-J).
S3
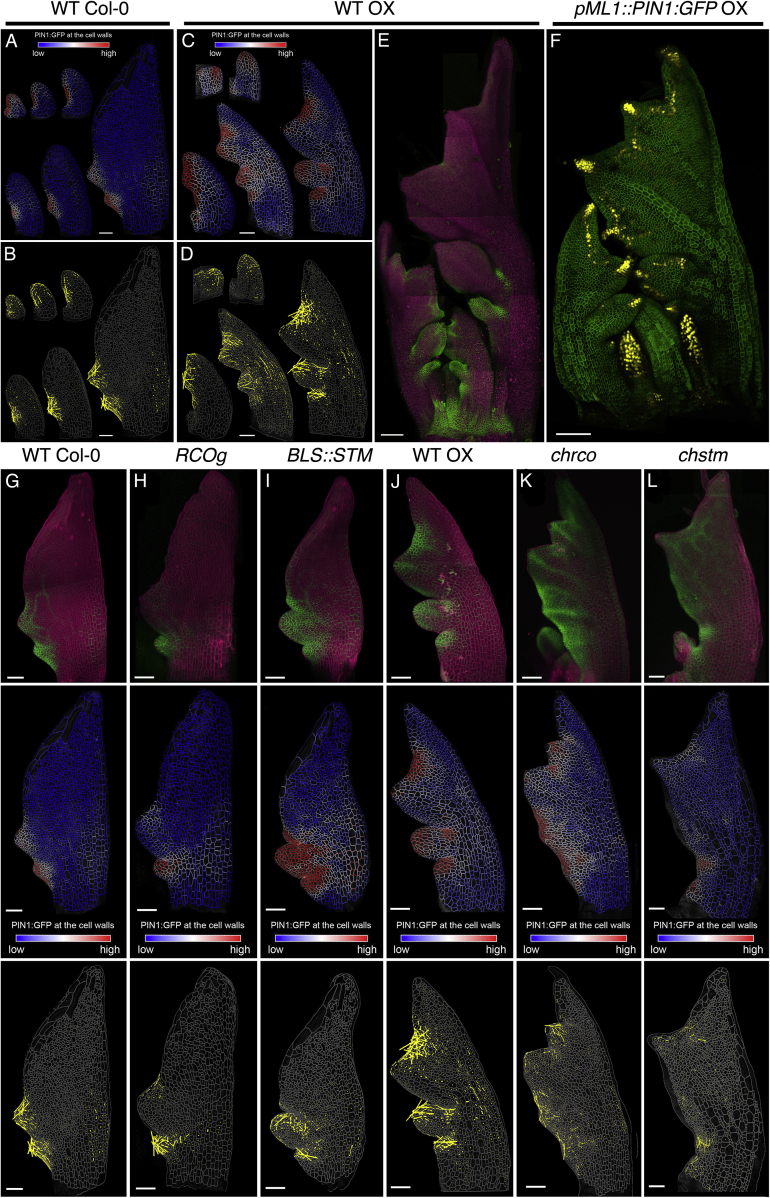
To assess the role of PIN1 in tissue growth polarity in leaves (Bilsborough et al., 2011, Hay et al., 2006), we monitored its distribution during leaf development in both species. During leaf initiation, PIN1 was uniformly expressed, with its expression later coinciding with regions of active marginal patterning (Figures 2A–2B and S5A–S5E). PIN1 polarities pointed toward protrusion tips, mirroring the directions of growth at emerging protrusions (Figures 2C–2F, S5B, and S5D). We did not detect PIN1 expression in the remainder of the blade, where we observed isotropic growth in time-lapse samples (Figures 1E and 1F). In C. hirsuta primordia, prolonged PIN1 expression coincided with delayed differentiation (Figures 2B and S5C) and with the late emergence of protrusions in the terminal leaflet (Figure S1K). To test whether PIN1 expression can influence growth direction following differentiation, we expressed PIN1 throughout the L1 layer in C. hirsuta, including in the distal leaf blade where it is typically not expressed (Figure S5F). This had no apparent effect on patterning or leaf shape, consistent with previous results in A. thaliana (Bilsborough et al., 2011). Thus, our results indicate that PIN1 localization influences growth polarities but that its action is local rather than global and lost following differentiation.
Figure S5.
Patterns of PIN1 Expression and Polarization in Developing Leaves, Related to Figure 2, Figure 3, Figure 4
(A-B) Expression of pAtPIN1::PIN1:GFP in A. thaliana leaves. Quantification of PIN1:GFP signal at cell membranes (A) and the orientation of PIN1 polarization (B) in the epidermis.
(C-E) Expression patterns of pAtPIN1::PIN1:GFP in C. hirsuta leaves. Quantification of PIN1:GFP signal at cell borders (C) and the orientation of PIN1 polarization (D) in the epidermis. (E) Confocal stacks of PIN1:GFP signal in green and autofluorescence in magenta at a later stage of development. PIN1 expression is maintained for longer in C. hirsuta leaves compared to A. thaliana. Note high PIN1 expression at early stages of leaf development and at protrusions. Color scales in (A and C) represent the intensity of GFP fluorescence. Cell outlines in (B and D) are marked in gray. Yellow lines in (B and D) indicate the orientation of PIN1 polarization in each cell (length of the lines is proportional to the strength of PIN1 polarization). Note that cells with strongly polarized PIN1 are mainly located at the tip of the early leaf primordia or at emerging protrusions.
(F) The expression pattern of pAtML1::PIN1:GFP in C. hirsuta leaves with GFP signal in green and pDR5::VENUS signal in yellow.
(G-L) The expression patterns of pAtPIN1::PIN1:GFP (top row), quantification of PIN1:GFP expression at cell borders (middle row) and the orientation of PIN1 polarization in the epidermis (bottom row) of A. thaliana WT (G), RCOg (H), and BLS::STM (I) as compared to C. hirsuta WT (J), rco (K) and stm (L) mutant leaves. The duration of PIN1 expression is extended in C. hirsuta WT leaves. Confocal stacks with PIN1:GFP signal in green and propidium iodide in magenta (top row). Color scales in the middle row represent intensity of GFP fluorescence. Cell outlines in bottom row are marked in gray, with yellow lines indicating the orientation of PIN1 polarization in each cell (length of the lines is proportional to the strength of PIN1 polarization).
Scale bars 50 μm in A-D and G-L, and 100 μm in E-F.
We next assessed how marginal patterning relates to differentiation. Differentiation fronts were initiated at marginal protrusions (Figures 1C–1D, S1A–S1D, and S2A–S2F) and their formation depended on the auxin-PIN1-CUC2 module (Figures 2G–2I, S3A–S3J, and S4B–S4G). We therefore reasoned that auxin maxima may direct the formation of these differentiation fronts. To test this, we treated A. thaliana leaves with auxin and observed accelerated growth and differentiation (Figures 2J–2K and S3K–S3S), indicating that auxin can promote both growth and differentiation and that auxin maxima help to establish multiple differentiation fronts.
A Computational Model of Marginal Patterning and Leaf Growth
We developed a physically based computational model using the finite element method (FEM) to further investigate how the auxin-PIN1-CUC2 module controls the dynamic marginal growth patterns that underlie leaf shape and to understand how margin growth is integrated with overall leaf blade growth (Figure 3). We first modeled leaf growth when margin patterning was disrupted. Our results indicate that, in this case, leaf development converges on a common growth pattern characterizing pin1 and cuc2 A. thaliana mutants (Figures 2G–2I and S4B–S4G) and development of both species following NPA treatment (Figures S3A–S3J). Each of these perturbations generates a simple leaf with smooth margins. We thus used this common growth pattern to characterize the bulk growth of the leaf (Figures S4H and S4I and Methods S1). We assumed that growth of the leaf depends on tissue type (blade versus midrib/petiole) and growth rates decrease with the basipetal progression of differentiation. This model reproduced the convergent development and form observed when margin patterning is perturbed, including the emergence of a radiating pattern of anisotropy at the blade-petiole junction (Figure S4I and S4J).
Figure 3.
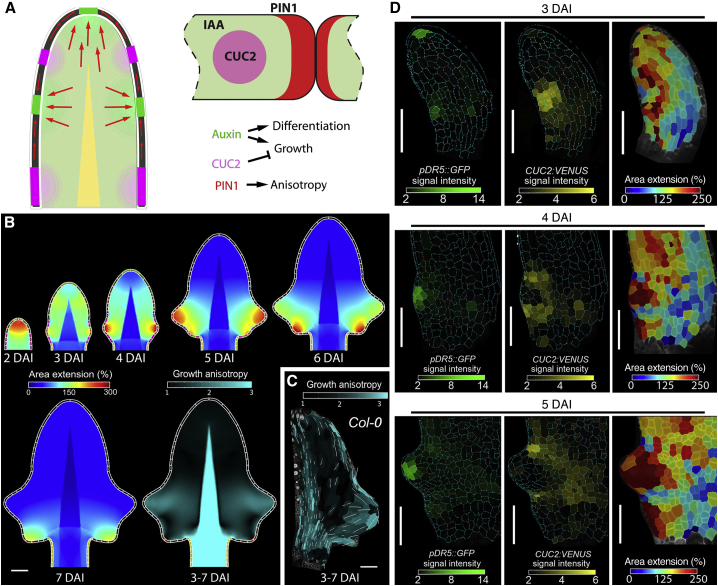
Model of Marginal Patterning and Leaf Growth
(A–C) Model of A. thaliana wild-type leaf. (A) Margin cells at the leaf edge simulate a feedback between auxin (IAA), CUC2 and PIN1. PIN1-dependent auxin maxima in the margin promote anisotropic growth, while CUC2 inhibits growth to influence nearby blade growth. (B) Resulting leaf shape, area extension, and anisotropy distribution from the model (C) as compared to growth anisotropy in an A. thaliana leaf.
(D) Heat-maps of pDR5::GFP (left panel) and pCUC2::CUC2:VENUS (middle panel) signal intensity, and area extension (right panel) in 3–5 DAI A. thaliana leaf epidermis. The time intervals for area extension heat-maps are: 3–4 DAI (3 DAI), 4–5 DAI (4 DAI and 5 DAI).
White lines in (C) indicate cell growth orientation when anisotropy > 40%. Scale bars, 100 μm (B and C), 50 μm (D).
We then extended this model to incorporate both molecular regulation at the leaf margin (Figure 3A and Methods S1) (Bilsborough et al., 2011) and bulk interior growth. For margin growth regulation, we assumed that (1) PIN1 locally regulates growth polarities by organizing auxin maxima; (2) auxin locally increases growth rates, accelerates differentiation, and negatively regulates CUC2; and (3) CUC2 locally inhibits growth and permits the organization of auxin maxima by PIN1. The resulting model reproduces the growth patterns and leaf shapes observed during A. thaliana development (Figures 3B and 3C; Video S3). Thus, by combining time-lapse imaging with simulations of growth and patterning in a physically connected leaf blade, we obtained an integrated model of auxin-PIN1-CUC2-mediated growth regulation that conceptualizes how marginal and non-marginal tissue growth may interact to create a simple serrated leaf form.
The model of the default growth pattern observed when marginal patterning at the leaf margin is absent, followed by the model of wild type A. thaliana leaf development.
Time-Lapse Imaging of Marginal Patterning Tests Computational Model
In our simulation, we observed the emergence of an interspersed distribution of auxin activity maxima and CUC2, as has been observed in confocal micrograph snapshots (Bilsborough et al., 2011, Maugarny-Calès et al., 2019). However, it is possible that the mechanism generating this interspersed distribution cannot be inferred from snapshots. For example, an oscillatory mechanism (as in somitogenesis; Cotterell et al., 2015) could sequentially establish auxin maxima and CUC2 domains. Or distal CUC2 expression could organize the neighboring auxin maximum (as in ectopic abaxial outgrowths of kanadi1;2 mutants; Abley et al., 2016). Both mechanisms could generate auxin maxima together with a strong CUC2 expression domain on the proximal or distal side of a protrusion. If CUC2 inhibits growth, then the asymmetric distribution of CUC2 should cause the protrusion to be asymmetric from emergence. By contrast, if a protrusion is flanked on both sides by CUC2, our model predicts it to be symmetric at initiation and to only later become asymmetric due to the proximodistal distribution of growth rates.
To investigate these possibilities, we live-imaged CUC2 and DR5 expression during serration initiation in A. thaliana leaves. Consistent with our model’s predictions, we observed that the auxin maximum that patterns a serration is organized in a CUC2-expression domain, such that DR5 expression precedes the elimination of CUC2 at this site (Figure 3D). As in our model, we found that high growth at the auxin maxima, juxtaposed against slow growth in adjacent CUC2-expressing sinuses, created a growth differential that leads to the emergence of a symmetric protrusion. Thus, our current model captures how local growth regulation by marginal patterning involving auxin, PIN1 and CUC2 influences leaf shape in A. thaliana. It also links gene activities, as studied in real time, to specific aspects of growth amount and direction.
In Silico Exploration of Quantitative Differences in Development
We next set out to explore how both global and local growth inputs are integrated to generate leaflets in C. hirsuta (Figure 1). To do so, we constructed a geometric model of margin development (Figure 4A) (see Methods S1; Runions et al., 2017) to examine how growth activators, repressors, and differentiation interact to form marginal protrusions. In this model, the local growth of lateral protrusions in the context of global leaf blade extension generates periodic outgrowths, with forms dependent on the model’s parameter values (Figure 4B).
Figure 4.
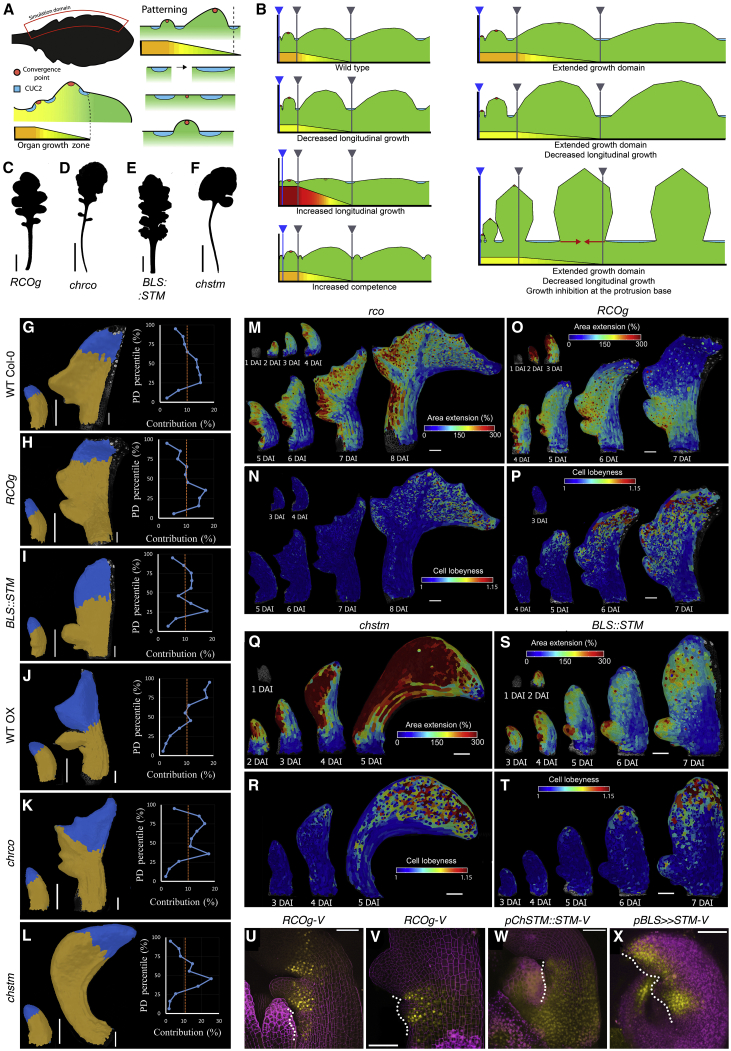
RCO and STM Together Shape Leaf Growth
(A and B) Geometric model of protrusion development. (A) Principle of the simulation. Red inset indicates simulated margin area. Blade extension drives patterning. CUC2 intervals (blue) that exceed a threshold length are broken by a convergence point (red). Distal portions of the margin differentiate at the black dotted line. Growth has a basipetal gradient (red-green scale, with red denoting regions of highest growth). (B) Effects of varying simulation parameters on protrusion form, with WT simulation acting as reference. Loss of competence to produce protrusions (blue bars, arrowheads), start and end of decreasing growth zone (gray bars, arrowheads), and growth restriction at the protrusion base (red arrows) are indicated.
(C–F) Leaf silhouettes of A. thaliana RCOg (C) and BLS::STM (E) transgenic lines; C. hirsuta rco (D), and stm (F) mutants.
(G–L) Left panels, lineage tracing of distal quartile (blue) of early primordia (3–7 DAI, G–K; or 2 DAI until end of the time-lapse, L), for A. thaliana WT (G), RCOg (H), BLS::STM (I), and C. hirsuta WT (J, OX), rco (K), and stm (L). Right panels, growth alignment graphs of estimated cellular contribution at final time-point (%, x axis) of cells along PD-axis at 3 DAI (y axis, 10 bins, each containing 10% of cells at 3 DAI).
(M–T) Heat-maps of area extension (M, O, Q, and S) and cell lobeyness (N, P, R, and T) for C. hirsuta rco (M and N), and stm (Q and R) mutant leaves and A. thaliana RCOg (O and P) and BLS::STM (S and T) leaves.
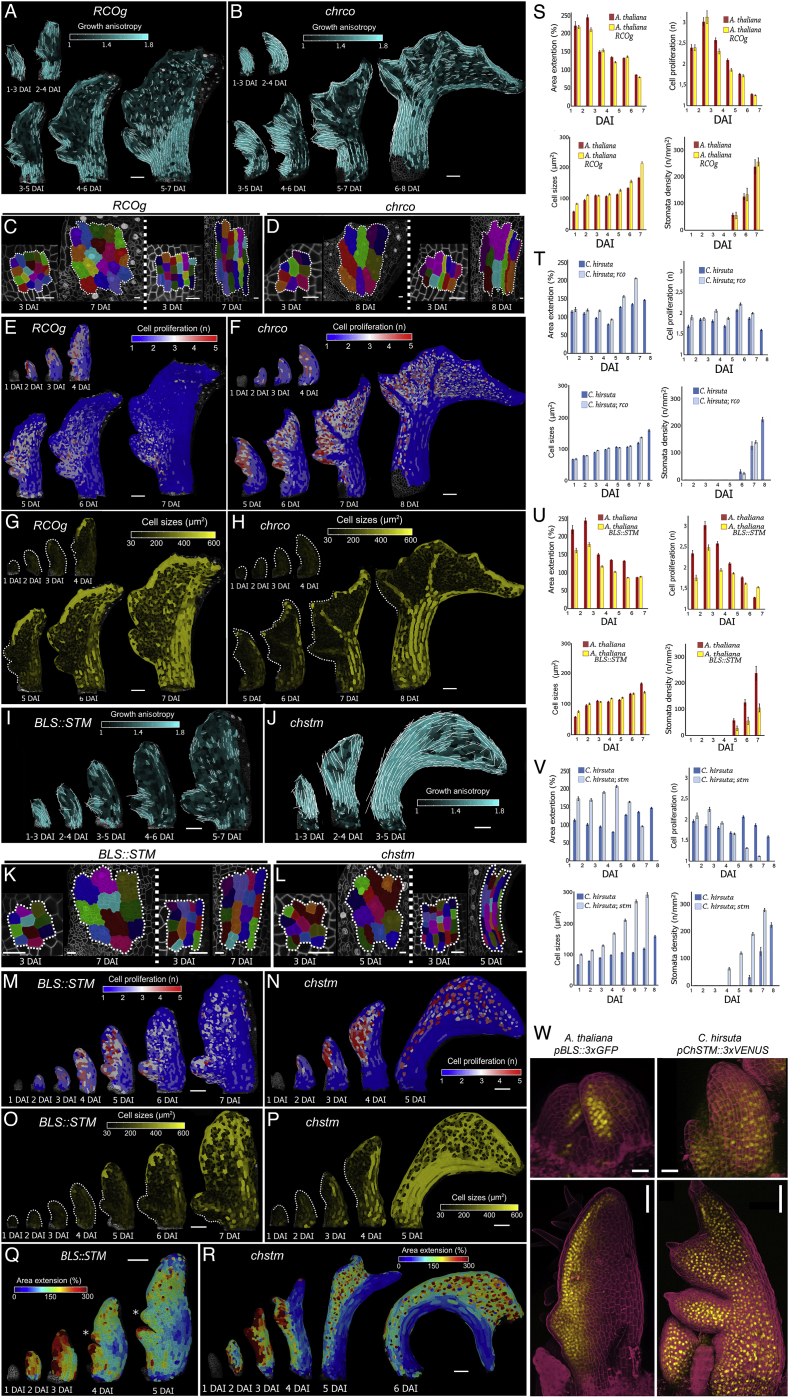
(U–X) Localization of RCO:VENUS (yellow) in C. hirsuta (U) and A. thaliana (V), pChSTM::STM:VENUS in C. hirsuta (W) and BLS≫STM:VENUS in A. thaliana (X) leaves; dotted-lines mark the leaf margin. Scale bars, 1 cm (C–F), and 100 μm (G–X). See also Figure S2, Figure S5, Figure S6 and Videos S4 and S5.
We explored the parameters that affect growth at the protrusion base to evaluate their impact on margin form (Figure 4B and Video S4). In the model, decreased longitudinal growth produced more focused outgrowths. By contrast, increased longitudinal growth impeded the formation of focused protrusions by smoothing outgrowths. Increasing the duration of marginal patterning led to the initiation of intercalary protrusions but did not affect protrusion form. Extending the growth domain increased protrusion size without otherwise affecting their form. Protrusion size was further increased by both slowing longitudinal growth and by extending the growth domain. These effects coincide with the suggested role of the KNOX gene STM in retarding growth and differentiation in C. hirsuta leaves (Hay et al., 2006). However, protrusions still lack the narrow base found in leaflets. By introducing a domain at the protrusion base that resists longitudinal extension, the model generates outgrowths with a narrow base (Figure 4B and Video S4), thereby eliminating the characteristic asymmetric shape of serrations in A. thaliana. Local growth inhibition in this region is consistent with the proposed function of the RCO gene at the base of leaflets in C. hirsuta (Vlad et al., 2014). In summary, our model predicts three parameters, which together contribute to the contrasting leaf morphologies of C. hirsuta and A. thaliana: (1) a global decrease in longitudinal growth, (2) an extended duration of growth, and (3) growth inhibition at leaflet bases. We hypothesized that these parameters can be mapped to the actions of STM and RCO, with STM acting to globally reduce growth and delay differentiation (1 and 2) and RCO acting to locally inhibit growth at the base of emerging protrusions (3).
A wild-type simulation is shown followed by simulations where parameters affecting protrusion form are varied, including: the rate of longitudinal growth, the region of competence for protrusion generation or the length of the growth domain.
RCO Acts Locally, and STM Acts Broadly, to Increase Leaf Complexity
To test predictions from our geometric model in the context of mechanically connected tissues, we analyzed the roles of RCO and STM genes in leaf development. These genes are active in leaf primordia of C. hirsuta, but not A. thaliana. STM is expressed in the pluripotent shoot apical meristem of both species and excluded from A. thaliana leaves, while RCO is absent from the A. thaliana genome (Long et al., 1996, Vlad et al., 2014). To assess how these genes influence global differences in primordium growth, we compared fate maps of wild-type leaf cells at 3 DAI to those of plants with modified STM and RCO expression (Figures 4C–4L). C. hirsuta leaves can be distinguished from A. thaliana’s by the increased contribution of distal cells to the leaf blade (Figures 4G and 4J). This feature is not accounted for by RCO, as expressing RCO in A. thaliana leaves from its own regulatory sequence—or eliminating it in the C. hirsuta rco mutant—did not alter this pattern of distal cell contribution (Figures 4H and 4K). However, this feature is lost in the simple leaves of C. hirsuta loss-of-function stm mutants (Figure 4L) and appears in A. thaliana when STM expression is driven from a leaf margin promoter that is active from 2 DAI onward (Figures 4I and 4W–4X; BLS, Figure S6W; Shani et al., 2009). These results show that RCO and STM expression during leaf development increases leaf complexity (Figures 4C–4F) (Vlad et al., 2014, Shani et al., 2009) but that they differ in their effects on the contribution of cells to the primordium.
Figure S6.
The Influence of RCO and STM on Leaf Growth, Related to Figure 4
(A-B) Heat-maps of growth anisotropy for A. thaliana RCOg (A), and C. hirsuta rco (B) leaves (white lines indicate the orientation of cell growth where anisotropy is higher than 40%).
(C-D) Cell lineage tracing analysis in the leaf blade (left panels) or petiole/midrib (right panels) for A. thaliana RCOg (C) and C. hirsuta rco (D) leaves from 3-7 DAI. Colors show the correspondence between cells at 3 DAI and their clonal sectors at 7 DAI.
(E-F) Heat-maps of cell proliferation for A. thaliana RCOg (E) and C. hirsuta rco (F) leaves.
(G-H) Heat-maps of cell size for A. thaliana RCOg (G) and C. hirsuta rco (H) leaves.
(I-J) Heat-maps of growth anisotropy for A. thaliana BLS::STM (I) and C. hirsuta stm (J) leaves (white lines indicate the orientation of cell growth where anisotropy is higher than 40%). As the BLS::STM sample used for 3 DAI in (M) and (N) was not captured at 1 DAI; the heat-map of anisotropy over 1-3 DAI in (I) was obtained from an independent time-lapse series.
(K-L) Cell lineage tracing analysis in the leaf blade (left panel) or petiole/midrib (right panel) for A. thaliana BLS::STM (K) and C. hirsuta stm (L) leaves from 3-7 DAI. Colors show the correspondence between cells at 3 DAI and their clonal sectors at 7 DAI.
(M-N) Heat-maps of cell proliferation for A. thaliana BLS::STM (M) and C. hirsuta stm (N) leaves.
(O-P) Heat-maps of cell size for A. thaliana BLS::STM (O) and C. hirsuta stm (P) leaves.
(Q-R) Heat-maps of area extension in an A. thaliana BLS::STM transgenic leaf (Q) and a C. hirsuta stm mutant (R). Asterisk indicates additional protrusion emerging from the leaf margin in (Q).
(S-V) Quantifications of mean area extension, cell proliferation, cell size, and stomata density for A. thaliana RCOg (S), C. hirsuta rco (T), A. thaliana BLS::STM (U) and C. hirsuta stm (V) compared to WT leaves (error bars indicate SEM). (S) A. thaliana RCOg, n = 2-4 (stomata density) and n > 150 (remaining plots, 4 independent time-lapse series). (T) C. hirsuta rco, n = 3 (stomata density) and n > 80 (remaining plots, 3 independent time-lapse series). (U) A. thaliana BLS::STM, n = 3-4 (stomata density) and n > 100 (remaining plots, 4 independent time-lapse series). (V) C. hirsuta stm, n = 2-4 (stomata density) and n > 300 (remaining plots, 4 independent time-lapse series).
(W) pAtBLS::3xGFP expression pattern in A. thaliana (left column) and pChSTM::3xVENUS expression pattern in C. hirsuta (right column). Confocal images with GFP or VENUS signal in yellow and propidium iodide staining in magenta.
Note that tissue-dependent growth patterns are still observed in leaf primordia of A. thaliana RCOg (C) and BLS::STM (K) plants, as well as C. hirsuta rco (D) and stm (L) mutants.
DAI indicates days after primordia initiation. Scale bars, 100 μm in (A-B, E-J, M-R, and W bottom panels), 20 μm in (C-D, K-L, and W top panels).
We then examined how the action of each gene translated into cellular behaviors. As RCO does not affect the contribution of distal cells to the primordium, we reasoned that it might affect leaf development locally, consistent with its proposed role as a local growth inhibitor (Vlad et al., 2014). Loss of RCO activity in C. hirsuta and introduction of RCO activity in A. thaliana did not affect the global distribution of growth (Figure 4M, 4O, and S6A–S6D and Video S5) and differentiation (Figures 4N, 4P, S6E–S6H, S6S, and S6T; for wild type see: Figures 1C and 1G, A. thaliana; Figure 1D and 1H, C. hirsuta), or marginal patterning (PIN1 expression and the order of protrusion initiation, compare Figure S5G and S5J to S5H and S5K; Figure 1C to Figure 4O; and Figure 1D to Figure 4M). Instead, when expressed in either species, RCO inhibited local growth (Figure 4M, 4O, and S2G compared to Figures 1C and 1D; S2A) within its restricted expression domain at the base of emerging protrusions (Figures 4U and 4V). Thus, RCO is required for local growth repression at the base of protrusions but has no apparent global effects on leaf development.
4
Our fate maps show that STM increases the relative contribution of distal cells to the primordium (Figures 4G–4L). We thus reasoned that it might broadly influence leaf development. Time-lapse imaging of C. hirsuta stm mutants and A. thaliana BLS::STM leaves confirmed that STM broadly affects patterns of growth, proliferation, and differentiation. Growth, proliferation, and differentiation were strongly accelerated in C. hirsuta stm mutant leaves (Figures 4Q, 4R, S6J, S6N, S6P, S6R, and S6V; Video S5), concomitant with an early decrease in PIN1 expression (Figures S5J and S5L) and a reduction in the lateral growth of emerging leaflets (Figure 4Q). Conversely, compared to A. thaliana wild-type leaf primordia (Figures 1C and S1A), BLS::STM primordia showed significantly reduced growth and proliferation (Figures 4S, S6I, and S6U and Video S5), delayed differentiation (Figures 4T and 1G), a broader domain of PIN1 expression and prolonged margin patterning (Figure S5G, S5I, and S6Q). Growth reduction in BLS::STM leaves was progressively restricted to proximal regions of the primordium (Figures 4S, S6M, and S6O) and was most apparent adjacent to emerging protrusions, where differentiation was particularly protracted (compare Figure 1G and S2C with Figures 4T and S2L). Nevertheless, increased growth at protrusion tips was comparable to WT but was maintained for longer (Figure 4S, S2J, and S2K). Thus, expressing STM in A. thaliana leaves increases the contribution of cells in distal and lateral regions to the early primordium (Figures 4G and 4I), shifting cellular growth patterns toward those of C. hirsuta (compare Figures 1C–1H, 4S–4T, S1A, S1B, S6I and S6M). Our observations suggest that STM retards early growth while delaying tissue maturation, thus prolonging the duration of growth and patterning. Notably, this STM-dependent redistribution of leaf growth might account both for the formation of lateral leaflets and the broad terminal leaflet that distinguish C. hirsuta from A. thaliana, consistent with the loss of both these features in C. hirsuta stm mutants (Figures 4G, 4I, 4J, and 4L).
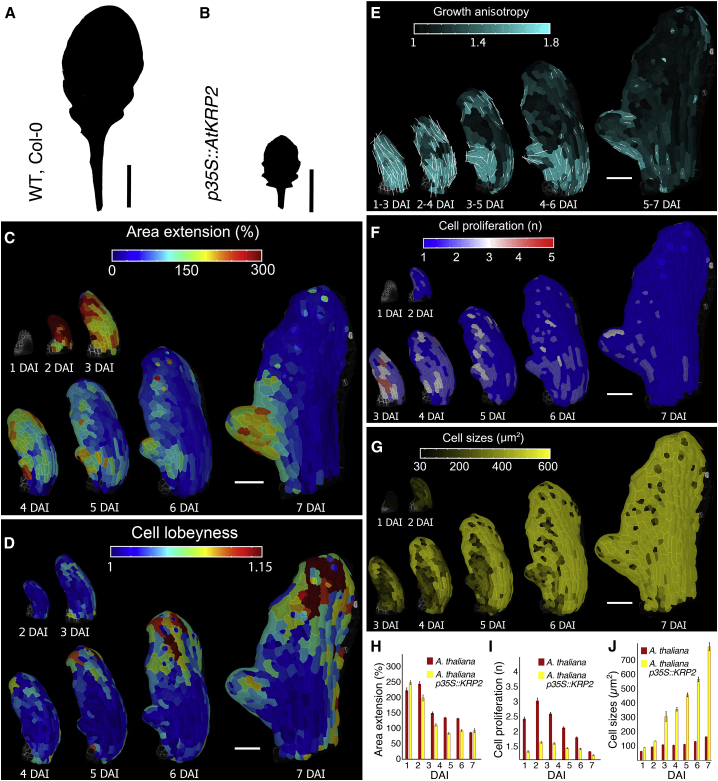
We observe that STM increases leaf complexity by delaying differentiation. However, accelerated differentiation can also increase leaf complexity, as seen in A. thaliana plants overexpressing KIP-RELATED PROTEIN2 (KRP2) under the 35S promoter (Figures 5A and 5B; De Veylder et al., 2001). Thus, both delayed and accelerated differentiation can increase leaf complexity. We investigated this contradiction by recording time-lapse images of p35S::KRP2 leaves (Figure 5C–5J), which confirmed the early onset of differentiation, relative to wild-type (∼3 DAI versus ∼5 DAI in wild type; compare Figures 5C–5J with Figures 1C, 1G, S1A, and S1C). p35S::KRP2 leaves initiated slow-growing symmetric serrations and sharp sinuses that were maintained over time, unlike in wild-type leaves (Figure 5C). Thus, a common feature of BLS::STM, RCOg and p35S::KRP2 leaves was decreased growth in the regions surrounding initiating protrusions. These observations indicate that a slow growth context enables the maintenance of more prominent protrusions, regardless of the differentiation status of surrounding tissue, as also suggested by our geometric model (Figure 4B).
Figure 5.
Increased Complexity of A. thaliana p35S::KRP2 Leaves
(A and B) Silhouettes of A. thaliana WT (A), and p35S::KRP2 transgenic line (B) leaves.
(C–G) Heat-map of area extension (C), cell lobeyness (D), growth anisotropy (E), cell proliferation (F), and cell size (G) for the A. thaliana p35S::KRP2 leaf.
(H–J) Quantifications of mean area extension (H), cell proliferation (I), and cell size (J) for A. thaliana p35S::KRP2 leaves compared to WT leaves (error bars indicate SEM). A. thaliana p35S::KRP2, n = 135-979 (H, 5 independent time-lapse series), n = 123–1,225 (I, 5 independent time-lapse series), n = 135–1,322 (J, 5 independent time-lapse series).
White lines in (E) indicate cell growth orientation where anisotropy is higher than 40%. Scale bars, 1 cm (A and B), and 100 μm (C–G)
Modulating Both Local and Global Growth Produces a Dissected Leaf
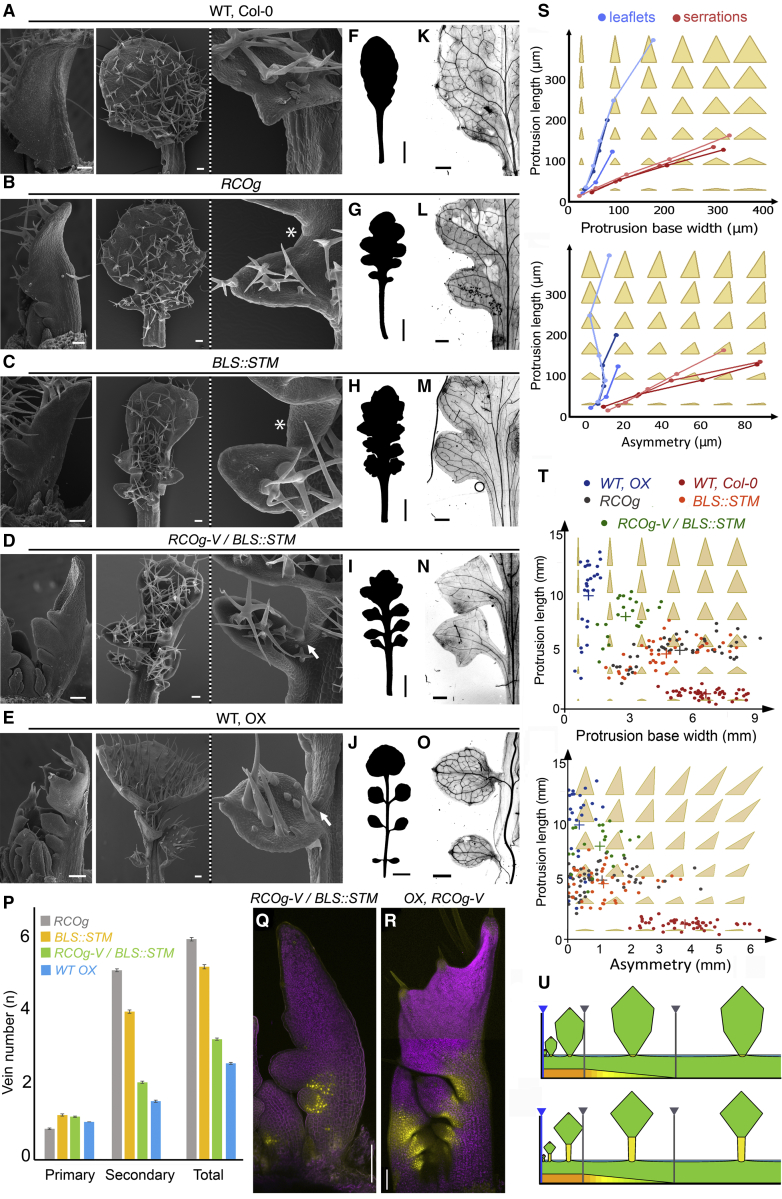
Our observations indicate that differences in A. thaliana and C. hirsuta leaf development include the local modification of growth during patterning and a global redistribution of growth that results from delayed differentiation (Figures 1, 2, and 3). Alone, neither of these features generated a dissected leaf (Figures 4 and 5), leading us to hypothesize that they act together to produce leaflets. In agreement with this hypothesis, the combined expression of RCO and STM in A. thaliana double transgenic plants reproduced key aspects of the C. hirsuta dissected leaf form (Figures 6A–6J), including a broad terminal leaflet. In A. thaliana plants expressing RCOg-VENUS (RCOg-V) and BLS::STM transgenes, protrusions were completely separated from each other and supported by a narrow base (Figure 6I). RCO expression localized to the petiolules of both terminal and lateral leaflets (Figures 6Q and 6R), and the vascular architecture of RCOg-V; BLS::STM leaves resembled that of C. hirsuta rather than A. thaliana leaves (Figures 6K–6P).
Figure 6.
Reconstructing Dissected Leaf Shape by Combining RCO and STM in A. thaliana Leaves
(A–E) Scanning electron micrographs of A. thaliana WT (A), RCOg (B), BLS::STM (C), RCOg-V; BLS::STM (D), and C. hirsuta WT (OX) (E) leaves. Insets: close-up views of marginal protrusions. Leaf blade between protrusions (white stars) and absence of blade between leaflets (white arrow) are shown.
(F–J) Silhouettes of fully developed eighth rosette leaves of A. thaliana WT (F), RCOg (G), BLS::STM (H), RCOg-V; BLS::STM (I), and C. hirsuta WT (J).
(K–P) Vascular architecture in protrusions of A. thaliana WT (K), RCOg (L), BLS::STM (M), RCOg-V; BLS::STM (N), C. hirsuta WT (O) leaves, and (P) vein number at protrusion bases (n = 21). Error bars indicate SE.
(Q and R) Expression of pRCO::RCO:VENUS fusion protein (yellow) in RCOg-V; BLS::STM A. thaliana (Q) and C. hirsuta (R) leaves.
(S and T) Analysis of protrusion geometry in time-lapse series (S) and mature leaves (T) of C. hirsuta (blue), and A. thaliana (red), RCOg-V (gray), BLS::STM (orange), and RCOg; BLS::STM (green) for protrusion length versus width (top), and protrusion length versus asymmetry (bottom); crosses indicate the mean for each background. Triangle shape captures the base to protrusion width (distance between sinuses), the height to protrusion length, and asymmetry to the length difference between left and right triangle edges.
(U) Geometric model extension to account for leaflets. Yellow at leaflet base indicates zone of RCO action. (Top) Strong RCO repression at leaflet rachis junction yields sessile leaflets. (Bottom) Extending zone of RCO action into the leaflet base produces petiolate leaflets. Scale bars, 100 μm in (A-E and Q-R), 1 cm in (F-J), and 1 mm in (K–O). See also Video S6.
These results suggest that growth inhibition at the base of emerging protrusions, combined with an increase in their outgrowth, can convert a serration into a leaflet. To test this idea, we quantified the overall form of emerging protrusions in our time-lapse series by using a triangle to approximate protrusion shape (Figure 6S, Methods S1). Our analysis showed that during serration development, the protrusion base quickly increased in width and gradually increased in length. By contrast, in leaflets, the protrusion base remained narrow while the protrusion elongated substantially. Notably, leaflets remained symmetric, whereas serrations adopted an asymmetric form during development (Figure 6S). Measurements of mature leaves are consistent (Figure 6T) and, thus, indicate that leaflets result from the combined effects of BLS::STM and RCOg on shape. Qualitatively, the changes in serration and leaflet shapes (Figures 6S and 6T) were consistent with the changes in growth predicted by the geometric model to increase the prominence and symmetry of protrusions (Figure 4B), specifically: the local effects of RCO inhibiting growth at the protrusion base, and the global effects of STM in slowing the rate of growth and differentiation, while prolonging growth duration. However, the simulations shown in Figure 4B evidently do not reproduce the narrow stalk observed in leaflets of A. thaliana RCOg-V; BLS::STM (Figure 6I and 6N) and C. hirsuta (Figure 6J and 6O).
To conceptualize the combined effects of RCO and STM on leaf form, we thus returned to the geometric model of A. thaliana margins. Inspired by our biological data (Figure 6I, 6N, and 6Q), we explored the effects of manipulating the amount and domain of growth repression at protrusion bases (Figure 6U and Video S6). Increasing growth inhibition at the protrusion base transformed lobes into leaflet shapes but did not produce a stalked base. Extending the inhibition zone at the protrusion base to match RCO expression (Figures 6Q and 6R) yielded a leaflet supported by a narrow stalk. Taken together, our data clarify how the combined action of RCO and STM during leaf development is sufficient to account for key aspects of simple versus dissected leaf development despite the many multi-scale interactions that influence leaf shape. Both genes contribute to slow growth at the protrusion base, which enables the lateral growth anisotropy of protrusions to generate discrete leaflets (Figures 1C–1F and 4). However, STM also contributes to the sustained longitudinal extension that allows leaflets to separate. Thus, growth inhibition both shapes the outgrowths driven by anisotropic lateral growth (2–6 DAI) and enables the anisotropic extension of the central leaf stalk (termed rachis) to separate leaflets (7–8 DAI) (Figures 1D and 1F). The relatively mild phenotypes that distinguish A. thaliana RCOg-V; BLS::STM plants from C. hirsuta might reflect other differences in gene expression between the leaf primordia of the two species, including differential expression of several meristem transcription factors (Gan et al., 2016).
The lateral extent of growth repression in the protrusion base is varied between simulations.
Discussion
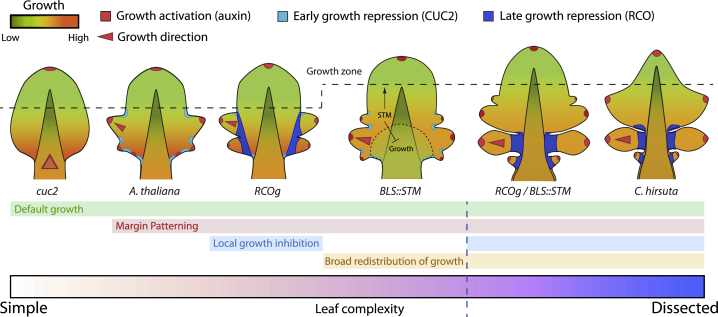
We investigated how key regulators of organogenesis influence the rate and orientation of growth, cell proliferation, and the timing of cellular differentiation to generate different leaf forms. To do so, we acquired real-time, cellular-level growth data at an unprecedented resolution. This information, coupled with theoretical analysis, guided the in vivo reconstruction of a complex morphological trait from its cell-level constituent elements (Figure 7).
Figure 7.
How Genetic Factors Influence Growth to Shape Divergent Leaf Forms
Schematic representation of A. thaliana and C. hirsuta leaf shapes; green-orange gradients represent growth rates, dashed line represents distal boundary of growth zone. Without auxin-based patterning, species have a common growth pattern, characterized by a basal growth zone and tissue-dependent growth polarities (midrib/petiole, anisotropic; blade, isotropic), producing simple leaves with a smooth margin (as seen in A. thaliana cuc2-3 mutants, where it may represent a default growth pattern). Auxin, PIN1 and CUC2 pattern alternate regions of growth repression and anisotropic growth activation along the leaf margin. Margin patterning creates outgrowths (serrations in A. thaliana). Local growth inhibition by RCO accentuates the growth differences created by margin patterning, generating more pronounced outgrowths (as in A. thaliana RCOg). STM functions to broadly redistribute growth by extending the growth zone, which increases leaf complexity. STM slows differentiation and growth in median and proximal regions, and prolongs growth and patterning in distal and lateral parts of leaf primordia (as in A. thaliana BLS::STM). This has two consequences: (1) it provides a second mode of growth repression, which creates leaflets when RCO is present. (2) It increases the relative contribution of lateral and distal regions to final leaf form, creating a broader leaf tip. Together, RCO and KNOX actions account for key differences in growth between C. hirsuta and A. thaliana leaves, as demonstrated by reconstructing a dissected leaf form in A. thaliana RCOg-V; BLS::STM.
At the single-gene level, we resolved how CUC2 can both stimulate and repress growth (Bilsborough et al., 2011, Blein et al., 2008, Maugarny-Calès et al., 2019); first, it triggers auxin activity maxima within its own domain; then, it represses cell growth as a separate effect in the region flanking these maxima (Figure 3). We also show how the KNOX gene, STM, influences leaf form by slowing growth and delaying differentiation in the proximal domain where marginal patterning occurs. These effects enable the auxin-PIN1-CUC2 patterning module to increase both the number and prominence of marginal protrusions. These interactions also concur with previous findings on how KNOX genes act, including their possible repression by auxin (Barkoulas et al., 2008, Blein et al., 2008, Bolduc et al., 2012, Hay et al., 2006, Heisler et al., 2005, Richardson et al., 2016). By explicitly considering dynamic cell-level growth, we extend our understanding of how these molecular regulators create simple versus complex leaves beyond the view that they mainly influence local tissue identities in the leaf (Bar and Ori, 2014, Hay and Tsiantis, 2010). A future challenge is to resolve how other regulators affect cell growth, such as NGATHA and CINCINNATA genes, which appear to prevent marginal outgrowth independently of CUC2 (Alvarez et al., 2016).
At the organ level, we show how local foci of genetically controlled growth anisotropy at the leaf margin interact with a broadly isotropic, organ-wide growth pattern to produce leaf form. In this context, the generation of leaflets versus serrations involves both the global modification of growth by STM and local growth inhibition conferred by RCO. These effects accentuate the non-uniform growth created by the patterning of leaf margins, thus shaping form (Figure 7). Notably, the growth repressive effects of RCO do not propagate broadly, despite leaf cells being mechanically interconnected by their walls. It will be interesting to explore the biophysical basis of this restricted propagation of RCO-dependent growth inhibition, which may also limit the potentially pleiotropic effects of evolutionary changes to RCO expression (Vlad et al., 2014).
Finally, our work sheds new light on the regulatory logic that connects cell- and tissue-scale effects of molecular regulators. Specifically, we found that auxin activity maxima increase growth while accelerating differentiation, while STM proteins have the opposite effect on both cellular growth and differentiation. Consequently, unlike growth regulators that predominantly influence the amount of growth (e.g., RCO), STM and auxin maxima strongly influence both growth amount and its duration, where duration depends on the proliferative status of cells. In this way, the opposing effects of auxin and STM are both self-limiting. Auxin activity increases growth, but its potential to alter form is limited by the induction of differentiation. STM delays differentiation, prolonging proliferation and patterning, but its potential is limited by its capacity to retard growth. The logic of these dual activities is similar to an incoherent feed-forward loop (Alon, 2006), a well-known gene-regulatory architecture in which a single, upstream regulator both inhibits and promotes the activity of a downstream target. These networks can provide temporally limited activation (i.e., a pulse of activity) in response to activation by the upstream regulator. In growing tissues, the opposing regulation of growth and differentiation affects the spatial-temporal distribution of growth by acting inversely on growth duration and amount. This regulatory logic might be well-suited for controlling the shape of determinate organs, by allowing fine-grained geometric changes to occur during development without dramatic changes to organ size.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, Peptides, and Recombinant Proteins | ||
| Propidium iodide (PI) | Sigma-Aldrich | 87-51-4 |
| Indole-3-Acetic Acid (IAA) | Sigma-Aldrich | I0901.0100 |
| Plant Preservative Mixture (PPM) | Plant Cell Technology | 250 |
| 1-N-naphthylphthalamic acid (NPA) | Sigma-Aldrich | 132-66-1 |
| Roti Histol | Roth | 6640 |
| Experimental Models: Organisms/Strains | ||
| A. thaliana: pin1 (RCR-genotyped to be insertion GABI-Kat -051A10) | Kierzkowski et al., 2013 | Cris Kuhlemeier, UniBe |
| A. thaliana: cuc2-3 | Nikovics et al., 2006 | Patrick Laufs, Institut Jean-Pierre Bourgin |
| A. thaliana: pUBQ10::acyl:TDT | Segonzac et al., 2012 | Elliot Meyerowitz, Caltech |
| A. thaliana: pUBQ10::acyl:YFP | Willis et al., 2016 | Raymond Wightman, University of Cambridge |
| A. thaliana: pin1 x pUBQ10::acyl:YFP | This study | N/A |
| A. thaliana: cuc2-3 x pUBQ10::acyl:YFP | This study | N/A |
| A. thaliana: pBLS::3xGFP | This study | N/A |
| A. thaliana: pDR5::GFP | Friml et al., 2003 | Gerd Jurgens, Max Planck Institute for Developmental Biology |
| A. thaliana: pUBQ10::acyl:TDT x pCUC2::CUC2:VENUS x pDR5::GFP | This study | N/A |
| A. thaliana: pBLS≫STM:VENUS | This study | N/A |
| A. thaliana: pPIN1::PIN1:GFP | Heisler et al., 2005 | N/A |
| A. thaliana: p35S::KRP2 | This study | NA |
| A. thaliana: RCOg:VENUS (translational fusion of VENUS to RCO in genomic context in pMLBART) | Vuolo et al., 2016 | Miltos Tsiantis, MPIPZ |
| A. thaliana: pBLS::STM | Shani et al., 2009 | Yuval Eshed, Weizmann Institute Naomi Ori, Hebrew University |
| A. thaliana: pBLS::STM x pUBQ10::acyl:YFP | This paper | N/A |
| A. thaliana: RCOg | Vlad et al., 2014 | Miltos Tsiantis, MPIPZ |
| A. thaliana: RCOg x pUBQ10::acyl:YFP | This study | N/A |
| A. thaliana: p35S::KRP2 x pUBQ10::acyl:YFP | This study | N/A |
| A. thaliana: pBLS::STM x pAtPIN1::PIN1:GFP | This study | N/A |
| A. thaliana: RCOg x pPIN1::PIN1:GFP | This study | N/A |
| A. thaliana: RCOg:VENUS x pBLS::STM | This study | N/A |
| C. hirsuta: rco | Vlad et al., 2014 | Miltos Tsiantis, MPIPZ |
| C. hirsuta: stm | Rast-Somssich et al., 2015 | Miltos Tsiantis, MPIPZ |
| C. hirsuta: stm x pUBQ10::acyl:YFP | This study | N/A |
| C. hirsuta: rco x pUBQ10::acyl:YFP | This study | N/A |
| C. hirsuta: pChSTM::3xNLS-VENUS | This study | N/A |
| C. hirsuta: pUBQ10::acyl:YFP | This study | N/A |
| C. hirsuta: pPIN1::PIN1:GFP | Barkoulas et al., 2008 | Miltos Tsiantis, MPIPZ |
| C. hirsuta: pML1::PIN1:GFP x DR5::VENUS | This study | N/A |
| C. hirsuta: stm x pPIN1::PIN1:GFP | This study | N/A |
| C. hirsuta: rco x pPIN1::PIN1:GFP | This study | N/A |
| C. hirsuta: pChSTM::ChSTM:VENUS | Rast-Somssich et al., 2015 | Miltos Tsiantis, MPIPZ |
| Recombinant DNA | ||
| pUBQ10::acyl:YFP | Willis et al., 2016 | Raymond Wightman, University of Cambridge |
| pBLS::STM; pBLS::LhG4 | Shani et al., 2009 | Yuval Eshed, Weizmann Institute Naomi Ori, Hebrew University |
| pPIN1::PIN1:GFP | Heisler et al., 2005 | N/A |
| pML1::PIN1:GFP | Bilsborough et al., 2011 | Miltos Tsiantis, MPIPZ |
| pBLS::3xGFP | This paper | N/A |
| pChSTM::3xNLS-VENUS | This paper | N/A |
| OP::STM:VENUS | This paper | N/A |
| pMLBART- STM:VENUS | Heisler et al., 2005 | N/A |
| pVTOp | Hay et al., 2006 and references therein | Miltos Tsiantis, MPIPZ |
| Software and Algorithms | ||
| Fiji | NIH | https://fiji.sc/ |
| Physically based simulations (C++/VVe) | This paper | Available upon request |
| Geometric models of margin development (L+C/lpfg) | This paper | Available upon request |
| MorphoGraphX | Barbier de Reuille et al., 2015 | http://www.mpipz.mpg.de/MorphoGraphX/ |
| VVe | Smith et al., 2004 | http://www.algorithmicbotany.org |
| LPFG | Karwowski and Prusinkiewicz, 2003 | http://www.algorithmicbotany.org |
| Lineage tracing script (Python) | Barbier de Reuille et al., 2015 | http://www.mpipz.mpg.de/MorphoGraphX |
| FantaMoprh | Abrosoft | http://www.fantamorph.com/ |
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Miltos Tsiantis (tsiantis@mpipz.mpg.de).
Experimental Model and Subject Details
Arabidopsis thaliana and Cardamine hirsuta
All transgenic plants and mutants were generated in the Col-0 background for A. thaliana and in the Ox background for C. hirsuta (Hay et al., 2006).
Growth Conditions
Plants were grown on soil in a growth chamber under long day conditions (16 h of illumination, 110 μEm-2s-1) at 20 ± 2°C, with 65 ± 10% relative humidity. For time-lapse experiments, soil grown plants were transferred at 11 days after germination to 1/2 MS medium including vitamins (Duchefa Biochem, M0222.0050) supplemented with 1% sucrose, 0.1% PPM and grown in long day conditions.
Method Details
Construction of transgenes
All transgenes were constructed using standard cloning techniques. All finished constructs were verified by sequencing. BLS::3xGFP: BLS promoter (7.1kb upstream of BLS) in pDRIVE vector (gift from Yuval Eshed lab). BLS was excised by PstI and BamHI and inserted upstream of 3xNLS-GFP in pBJ36. BLS::3xGFP was excised by NotI and inserted into pMLBART. pChSTM::3xNLS-VENUS:ChSTM promoter in pGEM-T-EASY as described before (Rast-Somssich et al., 2015). The pChSTM fragment was excised by SalI and BamHI and inserted into pBJ97 upstream of 3xNLS-VENUS. pChSTM::3xNLS-VENUS was then excised by NotI and inserted into pMLBART. STM:VENUS in pMLBART was a gift from Markus Heisler lab (Heisler et al., 2005). To construct OP::STM:VENUS, STM:VENUS was inserted in pVTOp (Hay et al., 2006 and references therein). We used the BLS promoter (Shani et al., 2009) to express STM in a minimal domain that is sufficient for leaflet formation in A. thaliana. The genomic locus of C. hirsuta STM was not sufficient for this purpose because gChSTM expression is active but not sustained in A. thaliana leaves and does not recreate the endogenous pattern observed in C. hirsuta (Rast-Somssich et al., 2015). For the AtML1::PIN1:GFP construct in C. hirsuta, we confirmed that this construct complements when introduced in a C. hirsuta pin1 background.
Plant transformations were performed using the floral dip method, using Agrobacterium tumefaciens strain GV3101. For each construct, a minimum of 10 independent lines were self-pollinated to obtain T2 seeds. Plants carrying two or more transgenes were produced by crossing and analyzed in the F2 or F3 generation.
Chemical treatments
For 1-N-naphthylphthalamic acid (NPA) treatment, ½ MS medium supplemented with 1% sucrose was supplemented with 10μM NPA. The same concentration was added to the water used to submerge plants during confocal observations. For auxin treatment, 1μM indole-3-acetic acid (IAA, Fluka) was used as described for NPA.
PI staining
Seedlings were submerged in PI solution (10 mg/mL) for 5-10 min and then imaged immediately using a Leica SP8 up-right confocal microscope.
Phenotypic and histological analysis
To obtain leaf silhouettes and quantify leaf margin protrusions, the 8th leaf of 5-weeks-old plants was removed, flattened onto clear adhesive on paper and scanned. Protrusions were measured in Fiji and analyzed as described in Methods S1.
To visualize vascular architecture, chlorophyll was eliminated using a graded ethanol series from 50% to 100% (v/v), and subsequent clearing with 50% to 100% (v/v) Roti Histol, followed by incubation with 50% glycerol overnight. Images were taken using a Nikon SMZ 1500 microscope equipped with a Nikon DS-Fi2 camera.
Microscopy and image analysis
All confocal imaging was performed using a SP8 upright confocal microscope equipped with a long working-distance water immersion objective (AP 20x/0.8 or AP 40x/0.8; Leica). Excitation was performed using an argon laser with 488 nm for GFP and PI, 514 nm for VENUS, YFP and PI. Images were collected at 529-545 nm for VENUS and YFP, 499-526 nm for GFP, 600-660 nm for PI and 660-749 nm for chlorophyll auto-fluorescence. For reporter gene analysis, cell walls were visualized after staining with 0.1% propidium iodide. For scanning electron microscopy, samples were prepared as described in Bilsborough et al. (2011). Imaging was performed using a JSM-5510 microscope (Joel). Images were processed and analyzed using Photoshop (Adobe) and MorphGraphX software (Barbier de Reuille et al., 2015). Quantification of fluorescence signals for each cell in the L1 layer (1-6 μm from the surface) was performed as described previously (Barbier de Reuille et al., 2015). Signal intensity of CUC2::CUC2:VENUS and pDR5::VENUS was quantified for the entire cell. pPIN1::PIN1:GFP signal intensity was quantified for each cell membrane based on the 1 μm region adjacent to the cell wall. PIN1:GFP orientations were assessed for each cell using the polarization plugin in MorphoGraphX (see Methods S1).
Growth tracking experiments
For growth analysis, cotyledons and older leaf primordia were removed from 11-days-old soil-grown plants to expose the apex for imaging. Only plants with undamaged roots, hypocotyls and emerging leaf primordia were used for time-lapse experiments. Dissected plants were then transferred into Ø60 mm Petri dishes filled with ½ MS medium including vitamins (Duchefa Biochem, M0222.0050) supplemented with 1.5% plant agar, 1% sucrose and 0.1% plant protective medium (Plant Cell Technology). To visualize cell outlines pUBQ10::acyl:YFP (Willis et al., 2016) or propidium iodide was used. During each experiment, at least half of the abaxial epidermis of the leaf was imaged at 24 h intervals using a Leica SP8 confocal microscope. Adult leaves 8 ± 1 were imaged for all genotypes except for C. hirsuta stm mutants which produce variable and reduced leaf number (Rast-Somssich et al., 2015) and where primordia as equivalently staged as possible to wild-type were observed. Between imaging, samples were transferred to a growth chamber and cultured in vitro under standard long-day conditions. Confocal stacks were acquired at 512x512 resolution, with 0.5-0.8 μm distance in Z-dimension, and no averaging to minimalize imaging stress. The resulting confocal time-lapse series were then analyzed using MorphoGraphX as described in Vlad et al. (2014). For samples that were larger than the scanning area, acquired stacks were stitched using MorphoGraphX. After cells were segmented, parent relations between successive days were determined manually in MorphoGraphX. These were used to calculate cell area, area extension, cell proliferation and growth anisotropy for each cell. The extent of lobing in pavement cells (lobeyness) was calculated as in Sapala et al. (2018) using a MorphoGraphX plugin and taking the ratio of each cell’s perimeter to that of its convex-hull (the smallest convex shape containing the cell). Lineage tracing analysis was performed automatically in MorphoGraphX. To compute corresponding cell lineages over multiple observations (i.e., multiple days), a custom python script was used that linked the parent relations between successive days to provide lineage relations over larger time-periods. We used lineages computed as described above to analyze protrusion shape and compute growth alignment graphs (see Methods S1), and the blade, petiole/midrib and slow growing sinus cells adjacent to emerging protrusions in A. thaliana and C. hirsuta (Figure 1 O,P). The blade, petiole/midrib and sinus cells in 3 and 7 DAI WT leaf primordia were identified by: 1) labeling the blade and petiole/midrib regions at 7 DAI based on cell shape, growth and proliferation, 2) mapping these regions to 3 DAI using lineage tracing, 3) marking the slow growing sinuses between protrusions, and 4) mapping the marked sinuses to 7 DAI.
Heat-maps for area extension, anisotropy and proliferation between two time-points are displayed on the later time-point (e.g., Leaf area extension for 3-4 DAI is visualized on the leaf at 4 DAI), with the exception of Figure 3L. In Figure 3L, area extension is visualized on the first time-point for 3 and 4 DAI, to make relations between pDR5::GFP and CUC2::Venus signal intensity and growth more apparent. Unless otherwise indicated, the time interval visualized for area extension, anisotropy and proliferation heat-maps is 1 day. Representative time-lapse series shown in main and supplemental figures were obtained from a single time-lapse experiment, with the exception of wild-type C. hirsuta and chrco leaves, as well as A. thaliana BLS::STM, 35S::KRP2 and RCOg leaves, where the first days of observation (1-2 or 1-3 DAI) were obtained from other time-lapse series. A minimum of 3 time-lapse series were performed for each genotype. These covered a period from leaf primordium emergence until up to 7-8 days after primordium initiation (DAI).
Movies
Movies of time-lapse sequences (Movies S1-2, S5) were created using images visualizing the average growth of each cell and its neighbors. Images were morphed using FantaMorph software (Abrosoft).
Modeling
Detailed description of all models can be found in Methods S1.
Quantification and Statistical Analysis
Statistical analysis
Statistical analysis was performed using Microsoft Excel. Error bars in figures represent standard errors of the mean (SEM).
Replication for imaging data
Replication for imaging data shown in main and supplementary figures is provided below.
| Replication of imaging data | ||
|---|---|---|
| Figure panels | Replication | |
| A. thaliana: WT | ||
| time-lapse | Fig. 1 C, E, G, I, J; 2D; 3C; S1 A, C, E, L-O; S3 O | 5 |
| time-lapse protrusion | Fig. S2 A-C | 5 |
| Sinus initiation in the blade (time-lapse) | Fig. 1 O | 5 |
| Growth/Proliferation vs PD-position (growth alignment map) | Fig. 1 K, M | 3 |
| Protrusion growth alignment map | Fig. S1 T | 3 |
| Primordium contribution (growth alignment map) | Fig. 4 G | 5 |
| pPIN1::PIN1:GFP expression | Fig. 2 A, C; S5 A, B, G | 10 |
| pDR5::GFP/CUC2:VENUS time-lapse and expression | Fig. 3D | 3 |
| BLS::3xGFP expression | Fig. S6 W | 7 |
| A. thaliana: NPA treatment (10 uM) | ||
| time-lapse | Fig. S3 A, C, E, G, I | 3 |
| A. thaliana: IAA treatment (5 um) | ||
| time-lapse | Fig. 2J,K; S3 K-N | 4 |
| A. thaliana: cuc2-3 mutant | ||
| time-lapse | Fig. 2G-I; S4 B, C, J | 3 |
| A. thaliana: pin1 mutant | ||
| time-lapse | Fig. S4 D-G | 4 |
| A. thaliana: RCOg | ||
| time-lapse | Fig. 4 O,P; S6 A, C, E, G | 3 |
| time-lapse: protrusion | Fig. S2 G-I | 3 |
| RCOg-VENUS expression | Fig. 4 V | 5 |
| pPIN1::PIN1:GFP expression | Fig. S5 H | 3 |
| Primordia contribution (Growth alignment map) | Fig. 4 H | 5 |
| A. thaliana: BLS::STM | ||
| time-lapse | Fig. 4 S,T; S6 I,K,M,O | 5 |
| protrusion time-lapse | Fig. S2 J-L | 5 |
| extra protrusion (compared to WT) | Fig. S6 Q | 5/8 |
| pPIN1::PIN1:GFP expression | Fig. S5 I | 8 |
| BLS≫STM-VENUS expression | Fig. 4 X | 7 |
| Primordia contribution (Growth alignment map) | Fig. 4 I | 3 |
| A. thaliana: p35S::AtKRP2 | ||
| time-lapse | Fig. 5 C-G | 4 |
| A. thaliana: RCOg-V/BLS::STM | ||
| RCOg-VENUS expression | Fig. 6 Q | 4 |
| C. hirsuta: WT | ||
| time-lapse | Fig. 1 D, F, H-J; 2F; S1 B, D, F, P-S | 3 |
| time-lapse protrusion | Fig. S2 D-F | 3 |
| Sinus initiation adjacent to the midrib (time-lapse) | Fig. 1 P | 3 |
| Growth/Proliferation vs PD-position (growth alignment map) | Fig. 1 L, N | 3 |
| Additional protrusion in terminal leaflet | Fig. S1 K | 3 |
| Protrusion growth alignment map | Fig. S1 U | 3 |
| Primordium contribution (growth alignment map) | Fig. 4 J | 3 |
| pPIN1::PIN1:GFP expression | Fig. 2 B, E; S5 C, D, E, J | 6 |
| RCOg-VENUS expression | Fig. 4 U; 6 R | 6 |
| pCHSTM::STM-VENUS expression | Fig. 4 W | 7 |
| pChSTM::3xVENUS expression | Fig. S6 W | 5 |
| C. hirsuta: NPA treatment (10 uM) | ||
| time-lapse | Fig. S3 B, D, F, H, J | 3 |
| C. hirsuta: pML1::PIN1:GFP | ||
| pDR5::VENUS expression and leaf morphology | Fig. S5 F | 5 |
| C. hirsuta: rco mutant | ||
| time-lapse | Fig. 4 M, N; S6 B, D, F, H | 3 |
| pPIN1::PIN1:GFP expression | Fig. S5 K | 7 |
| Primordia contribution (Growth alignment map) | Fig. 4 K | 3 |
| C. hirsuta: stm mutant | ||
| time-lapse | Fig. 4 Q,R; S6 J, L, N, P, R | 5 |
| pPIN1::PIN1:GFP expression | Fig. S5 L | 6 |
| Primordia contribution (Growth alignment map) | Fig. 4 L | 3 |
| A. thaliana vs C. hirsuta | ||
| Stomata appear earlier in A. thaliana vs C. hirsuta | Fig. S1 E, F | 4 from each species |
Acknowledgments
We thank A. Hay for valuable suggestions on the manuscript, Tsiantis lab members for comments, P. Prusinkiewicz and M. Heisler for discussions, Y. Eshed, N. Ori, P. Piazza, R. Wightman, and M. Barkoulas for materials, and I. Karakasilioti for assistance. The work was supported by DFG awards SFB 680 (M.T.) and FOR2581 (M.T. and R.S.S.), BBSRC grants BB/F012934/1 and BB/H011455/1 (M.T.), the Gatsby Charitable Foundation (M.T.), a Max Planck Society core grant (M.T.), and a Marie Skłodowska-Curie IF (Horizon 2020, 703886) (A.R.). M.T. also acknowledges the support of CEPLAS.
Author Contributions
D.K.: time-lapse imaging, method development, quantification; A.R.: modeling, method development, quantification including growth alignments; F.V.: genetics, SEM; R.L., S.S., and P.H.: data extraction, visualization; S.S. and R.S.S.: modeling, quantification, method development; R.D.I., C.C., and Z.Z.: material generation; H.J., C.G., A.-L.R.-K., and G.M.: method development; D.W.-S.: quantification, imaging. Written and revised by M.T. and A.R. with input from D.K., who co-wrote the first draft. M.T. designed and directed the study.
Declaration of Interests
The authors declare no competing interests.
Published: May 23, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.cell.2019.05.011.
A video abstract is available at https://doi.org/10.1016/j.cell.2019.05.011#mmc8.
Supporting Citations
The following references appear in the Supplemental Information: Bassel et al., 2014, Crane et al., 2013, Hejnowicz and Romberger, 1984, Rivara and Inostroza, 1997, Rubio-Somoza et al., 2014, Scarpella et al., 2006, Smith et al., 2006.
Supplemental Information
Descriptions of PIN quantification plug-in, regional primordium analysis and triangle-based protrusion morphospaces, as well as all computational models.
References
- Abley K., Sauret-Güeto S., Marée A.F., Coen E. Formation of polarity convergences underlying shoot outgrowths. eLife. 2016;5:e18165. doi: 10.7554/eLife.18165. [DOI] [PMC free article] [PubMed] [Google Scholar]; Abley, K., Sauret-Gueto, S., Maree, A.F., and Coen, E. (2016). Formation of polarity convergences underlying shoot outgrowths. eLife 5, e18165. [DOI] [PMC free article] [PubMed]
- Ali O., Mirabet V., Godin C., Traas J. Physical models of plant development. Annu. Rev. Cell Dev. Biol. 2014;30:59–78. doi: 10.1146/annurev-cellbio-101512-122410. [DOI] [PubMed] [Google Scholar]; Ali, O., Mirabet, V., Godin, C., and Traas, J. (2014). Physical models of plant development. Annu. Rev. Cell Dev. Biol. 30, 59-78. [DOI] [PubMed]
- Alon U. Chapman & Hall/CRC; 2006. An Introduction to Systems Biology: Design Principles of Biological Circuits. [Google Scholar]; Alon, U. (2006). An Introduction to Systems Biology: Design Principles of Biological Circuits (Chapman & Hall/CRC).
- Alvarez J.P., Furumizu C., Efroni I., Eshed Y., Bowman J.L. Active suppression of a leaf meristem orchestrates determinate leaf growth. eLife. 2016;5:e15023. doi: 10.7554/eLife.15023. [DOI] [PMC free article] [PubMed] [Google Scholar]; Alvarez, J.P., Furumizu, C., Efroni, I., Eshed, Y., and Bowman, J.L. (2016). Active suppression of a leaf meristem orchestrates determinate leaf growth. eLife 5, e15023. [DOI] [PMC free article] [PubMed]
- Bar M., Ori N. Leaf development and morphogenesis. Development. 2014;141:4219–4230. doi: 10.1242/dev.106195. [DOI] [PubMed] [Google Scholar]; Bar, M., and Ori, N. (2014). Leaf development and morphogenesis. Development 141, 4219-4230. [DOI] [PubMed]
- Barbier de Reuille P., Routier-Kierzkowska A.L., Kierzkowski D., Bassel G.W., Schüpbach T., Tauriello G., Bajpai N., Strauss S., Weber A., Kiss A. MorphoGraphX: A platform for quantifying morphogenesis in 4D. eLife. 2015;4:05864. doi: 10.7554/eLife.05864. [DOI] [PMC free article] [PubMed] [Google Scholar]; Barbier de Reuille, P., Routier-Kierzkowska, A.L., Kierzkowski, D., Bassel, G.W., Schupbach, T., Tauriello, G., Bajpai, N., Strauss, S., Weber, A., Kiss, A., et al. (2015). MorphoGraphX: A platform for quantifying morphogenesis in 4D. eLife 4, 05864. [DOI] [PMC free article] [PubMed]
- Barkoulas M., Hay A., Kougioumoutzi E., Tsiantis M. A developmental framework for dissected leaf formation in the Arabidopsis relative Cardamine hirsuta. Nat. Genet. 2008;40:1136–1141. doi: 10.1038/ng.189. [DOI] [PubMed] [Google Scholar]; Barkoulas, M., Hay, A., Kougioumoutzi, E., and Tsiantis, M. (2008). A developmental framework for dissected leaf formation in the Arabidopsis relative Cardamine hirsuta. Nat. Genet. 40, 1136-1141. [DOI] [PubMed]
- Bassel G.W., Stamm P., Mosca G., Barbier de Reuille P., Gibbs D.J., Winter R., Janka A., Holdsworth M.J., Smith R.S. Mechanical constraints imposed by 3D cellular geometry and arrangement modulate growth patterns in the Arabidopsis embryo. Proc. Natl. Acad. Sci. USA. 2014;111:8685–8690. doi: 10.1073/pnas.1404616111. [DOI] [PMC free article] [PubMed] [Google Scholar]; Bassel, G.W., Stamm, P., Mosca, G., Barbier de Reuille, P., Gibbs, D.J., Winter, R., Janka, A., Holdsworth, M.J., and Smith, R.S. (2014). Mechanical constraints imposed by 3D cellular geometry and arrangement modulate growth patterns in the Arabidopsis embryo. Proc. Natl. Acad. Sci. USA 111, 8685-8690. [DOI] [PMC free article] [PubMed]
- Bharathan G., Goliber T.E., Moore C., Kessler S., Pham T., Sinha N.R. Homologies in leaf form inferred from KNOXI gene expression during development. Science. 2002;296:1858–1860. doi: 10.1126/science.1070343. [DOI] [PubMed] [Google Scholar]; Bharathan, G., Goliber, T.E., Moore, C., Kessler, S., Pham, T., and Sinha, N.R. (2002). Homologies in leaf form inferred from KNOXI gene expression during development. Science 296, 1858-1860. [DOI] [PubMed]
- Bilsborough G.D., Runions A., Barkoulas M., Jenkins H.W., Hasson A., Galinha C., Laufs P., Hay A., Prusinkiewicz P., Tsiantis M. Model for the regulation of Arabidopsis thaliana leaf margin development. Proc. Natl. Acad. Sci. USA. 2011;108:3424–3429. doi: 10.1073/pnas.1015162108. [DOI] [PMC free article] [PubMed] [Google Scholar]; Bilsborough, G.D., Runions, A., Barkoulas, M., Jenkins, H.W., Hasson, A., Galinha, C., Laufs, P., Hay, A., Prusinkiewicz, P., and Tsiantis, M. (2011). Model for the regulation of Arabidopsis thaliana leaf margin development. Proc. Natl. Acad. Sci. USA 108, 3424-3429. [DOI] [PMC free article] [PubMed]
- Blein T., Pulido A., Vialette-Guiraud A., Nikovics K., Morin H., Hay A., Johansen I.E., Tsiantis M., Laufs P. A conserved molecular framework for compound leaf development. Science. 2008;322:1835–1839. doi: 10.1126/science.1166168. [DOI] [PubMed] [Google Scholar]; Blein, T., Pulido, A., Vialette-Guiraud, A., Nikovics, K., Morin, H., Hay, A., Johansen, I.E., Tsiantis, M., and Laufs, P. (2008). A conserved molecular framework for compound leaf development. Science 322, 1835-1839. [DOI] [PubMed]
- Bolduc N., Yilmaz A., Mejia-Guerra M.K., Morohashi K., O’Connor D., Grotewold E., Hake S. Unraveling the KNOTTED1 regulatory network in maize meristems. Genes Dev. 2012;26:1685–1690. doi: 10.1101/gad.193433.112. [DOI] [PMC free article] [PubMed] [Google Scholar]; Bolduc, N., Yilmaz, A., Mejia-Guerra, M.K., Morohashi, K., O’Connor, D., Grotewold, E., and Hake, S. (2012). Unraveling the KNOTTED1 regulatory network in maize meristems. Genes Dev. 26, 1685-1690. [DOI] [PMC free article] [PubMed]
- Bringmann M., Bergmann D.C. Tissue-wide Mechanical Forces Influence the Polarity of Stomatal Stem Cells in Arabidopsis. Curr. Biol. 2017;27:877–883. doi: 10.1016/j.cub.2017.01.059. [DOI] [PubMed] [Google Scholar]; Bringmann, M., and Bergmann, D.C. (2017). Tissue-wide Mechanical Forces Influence the Polarity of Stomatal Stem Cells in Arabidopsis. Curr. Biol. 27, 877-883. [DOI] [PubMed]
- Coen E., Rebocho A.B. Resolving Conflicts: Modeling Genetic Control of Plant Morphogenesis. Dev. Cell. 2016;38:579–583. doi: 10.1016/j.devcel.2016.09.006. [DOI] [PubMed] [Google Scholar]; Coen, E., and Rebocho, A.B. (2016). Resolving Conflicts: Modeling Genetic Control of Plant Morphogenesis. Dev. Cell 38, 579-583. [DOI] [PubMed]
- Cotterell J., Robert-Moreno A., Sharpe J. A Local, Self-Organizing Reaction-Diffusion Model Can Explain Somite Patterning in Embryos. Cell Syst. 2015;1:257–269. doi: 10.1016/j.cels.2015.10.002. [DOI] [PubMed] [Google Scholar]; Cotterell, J., Robert-Moreno, A., and Sharpe, J. (2015). A Local, Self-Organizing Reaction-Diffusion Model Can Explain Somite Patterning in Embryos. Cell Syst. 1, 257-269. [DOI] [PubMed]
- Crane K., Weischedel C., Wardetzky M. Geodesics in heat: A new approach to computing distance based on heat flow. ACM Trans. Graph. 2013;32:152. [Google Scholar]; Crane, K., Weischedel, C., and Wardetzky, M. (2013). Geodesics in heat: A new approach to computing distance based on heat flow. ACM Trans. Graph. 32, 152.
- De Veylder L., Beeckman T., Beemster G.T., Krols L., Terras F., Landrieu I., van der Schueren E., Maes S., Naudts M., Inzé D. Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell. 2001;13:1653–1668. doi: 10.1105/TPC.010087. [DOI] [PMC free article] [PubMed] [Google Scholar]; De Veylder, L., Beeckman, T., Beemster, G.T., Krols, L., Terras, F., Landrieu, I., van der Schueren, E., Maes, S., Naudts, M., and Inze, D. (2001). Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell 13, 1653-1668. [DOI] [PMC free article] [PubMed]
- Donnelly P.M., Bonetta D., Tsukaya H., Dengler R.E., Dengler N.G. Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev. Biol. 1999;215:407–419. doi: 10.1006/dbio.1999.9443. [DOI] [PubMed] [Google Scholar]; Donnelly, P.M., Bonetta, D., Tsukaya, H., Dengler, R.E., and Dengler, N.G. (1999). Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev. Biol. 215, 407-419. [DOI] [PubMed]
- Etournay R., Merkel M., Popović M., Brandl H., Dye N.A., Aigouy B., Salbreux G., Eaton S., Jülicher F. TissueMiner: A multiscale analysis toolkit to quantify how cellular processes create tissue dynamics. eLife. 2016;5:e14334. doi: 10.7554/eLife.14334. [DOI] [PMC free article] [PubMed] [Google Scholar]; Etournay, R., Merkel, M., Popović, M., Brandl, H., Dye, N.A., Aigouy, B., Salbreux, G., Eaton, S., and Julicher, F. (2016). TissueMiner: A multiscale analysis toolkit to quantify how cellular processes create tissue dynamics. eLife 5, e14334. [DOI] [PMC free article] [PubMed]
- Fox S., Southam P., Pantin F., Kennaway R., Robinson S., Castorina G., Sánchez-Corrales Y.E., Sablowski R., Chan J., Grieneisen V. Spatiotemporal coordination of cell division and growth during organ morphogenesis. PLoS Biol. 2018;16:e2005952. doi: 10.1371/journal.pbio.2005952. [DOI] [PMC free article] [PubMed] [Google Scholar]; Fox, S., Southam, P., Pantin, F., Kennaway, R., Robinson, S., Castorina, G., Sanchez-Corrales, Y.E., Sablowski, R., Chan, J., Grieneisen, V., et al. (2018). Spatiotemporal coordination of cell division and growth during organ morphogenesis. PLoS Biol. 16, e2005952. [DOI] [PMC free article] [PubMed]
- Friml J., Vieten A., Sauer M., Weijers D., Schwarz H., Hamann T., Offringa R., Jürgens G. Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature. 2003;426:147–153. doi: 10.1038/nature02085. [DOI] [PubMed] [Google Scholar]; Friml, J., Vieten, A., Sauer, M., Weijers, D., Schwarz, H., Hamann, T., Offringa, R., and Jurgens, G. (2003). Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426, 147-153. [DOI] [PubMed]
- Gan X., Hay A., Kwantes M., Haberer G., Hallab A., Ioio R.D., Hofhuis H., Pieper B., Cartolano M., Neumann U. The Cardamine hirsuta genome offers insight into the evolution of morphological diversity. Nat. Plants. 2016;2:16167. doi: 10.1038/nplants.2016.167. [DOI] [PMC free article] [PubMed] [Google Scholar]; Gan, X., Hay, A., Kwantes, M., Haberer, G., Hallab, A., Ioio, R.D., Hofhuis, H., Pieper, B., Cartolano, M., Neumann, U., et al. (2016). The Cardamine hirsuta genome offers insight into the evolution of morphological diversity. Nat. Plants 2, 16167. [DOI] [PMC free article] [PubMed]
- Hareven D., Gutfinger T., Parnis A., Eshed Y., Lifschitz E. The making of a compound leaf: genetic manipulation of leaf architecture in tomato. Cell. 1996;84:735–744. doi: 10.1016/s0092-8674(00)81051-x. [DOI] [PubMed] [Google Scholar]; Hareven, D., Gutfinger, T., Parnis, A., Eshed, Y., and Lifschitz, E. (1996). The making of a compound leaf: genetic manipulation of leaf architecture in tomato. Cell 84, 735-744. [DOI] [PubMed]
- Hay A., Tsiantis M. KNOX genes: versatile regulators of plant development and diversity. Development. 2010;137:3153–3165. doi: 10.1242/dev.030049. [DOI] [PubMed] [Google Scholar]; Hay, A., and Tsiantis, M. (2010). KNOX genes: versatile regulators of plant development and diversity. Development 137, 3153-3165. [DOI] [PubMed]
- Hay A., Barkoulas M., Tsiantis M. ASYMMETRIC LEAVES1 and auxin activities converge to repress BREVIPEDICELLUS expression and promote leaf development in Arabidopsis. Development. 2006;133:3955–3961. doi: 10.1242/dev.02545. [DOI] [PubMed] [Google Scholar]; Hay, A., Barkoulas, M., and Tsiantis, M. (2006). ASYMMETRIC LEAVES1 and auxin activities converge to repress BREVIPEDICELLUS expression and promote leaf development in Arabidopsis. Development 133, 3955-3961. [DOI] [PubMed]
- Heisler M.G., Ohno C., Das P., Sieber P., Reddy G.V., Long J.A., Meyerowitz E.M. Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr. Biol. 2005;15:1899–1911. doi: 10.1016/j.cub.2005.09.052. [DOI] [PubMed] [Google Scholar]; Heisler, M.G., Ohno, C., Das, P., Sieber, P., Reddy, G.V., Long, J.A., and Meyerowitz, E.M. (2005). Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr. Biol. 15, 1899-1911. [DOI] [PubMed]
- Hejnowicz Z., Romberger J.A. Growth tensor of plant organs. J. Theor. Biol. 1984;110:93–114. [Google Scholar]; Hejnowicz, Z., and Romberger, J.A. (1984). Growth tensor of plant organs. J. Theor. Biol. 110, 93-114.
- Karwowski R., Prusinkiewicz P. Design and Implementation of the L+C Modeling Language. Electron. Notes Theor. Comput. Sci. 2003;86:134–152. [Google Scholar]; Karwowski, R., and Prusinkiewicz, P. (2003). Design and Implementation of the L+C Modeling Language. Electron. Notes Theor. Comput. Sci. 86, 134-152.
- Kierzkowski D., Lenhard M., Smith R., Kuhlemeier C. Interaction between meristem tissue layers controls phyllotaxis. Dev. Cell. 2013;26:616–628. doi: 10.1016/j.devcel.2013.08.017. [DOI] [PubMed] [Google Scholar]; Kierzkowski, D., Lenhard, M., Smith, R., and Kuhlemeier, C. (2013). Interaction between meristem tissue layers controls phyllotaxis. Dev. Cell 26, 616-628. [DOI] [PubMed]
- Kuchen E.E., Fox S., de Reuille P.B., Kennaway R., Bensmihen S., Avondo J., Calder G.M., Southam P., Robinson S., Bangham A., Coen E. Generation of leaf shape through early patterns of growth and tissue polarity. Science. 2012;335:1092–1096. doi: 10.1126/science.1214678. [DOI] [PubMed] [Google Scholar]; Kuchen, E.E., Fox, S., de Reuille, P.B., Kennaway, R., Bensmihen, S., Avondo, J., Calder, G.M., Southam, P., Robinson, S., Bangham, A., and Coen, E. (2012). Generation of leaf shape through early patterns of growth and tissue polarity. Science 335, 1092-1096. [DOI] [PubMed]
- Long J.A., Moan E.I., Medford J.I., Barton M.K. A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature. 1996;379:66–69. doi: 10.1038/379066a0. [DOI] [PubMed] [Google Scholar]; Long, J.A., Moan, E.I., Medford, J.I., and Barton, M.K. (1996). A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379, 66-69. [DOI] [PubMed]
- Mansfield C., Newman J.L., Olsson T.S.G., Hartley M., Chan J., Coen E. Ectopic BASL Reveals Tissue Cell Polarity throughout Leaf Development in Arabidopsis thaliana. Curr. Biol. 2018;28:2638–2646.e4. doi: 10.1016/j.cub.2018.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]; Mansfield, C., Newman, J.L., Olsson, T.S.G., Hartley, M., Chan, J., and Coen, E. (2018). Ectopic BASL Reveals Tissue Cell Polarity throughout Leaf Development in Arabidopsis thaliana. Curr. Biol. 28, 2638-2646.e4. [DOI] [PMC free article] [PubMed]
- Maugarny-Calès A., Cortizo M., Adroher B., Borrega N., Gonçalves B., Brunoud G., Vernoux T., Arnaud N., Laufs P. Dissecting the pathways coordinating patterning and growth by plant boundary domains. PLoS Genet. 2019;15:e1007913. doi: 10.1371/journal.pgen.1007913. [DOI] [PMC free article] [PubMed] [Google Scholar]; Maugarny-Cales, A., Cortizo, M., Adroher, B., Borrega, N., Gonçalves, B., Brunoud, G., Vernoux, T., Arnaud, N., and Laufs, P. (2019). Dissecting the pathways coordinating patterning and growth by plant boundary domains. PLoS Genet. 15, e1007913. [DOI] [PMC free article] [PubMed]
- Nikovics K., Blein T., Peaucelle A., Ishida T., Morin H., Aida M., Laufs P. The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell. 2006;18:2929–2945. doi: 10.1105/tpc.106.045617. [DOI] [PMC free article] [PubMed] [Google Scholar]; Nikovics, K., Blein, T., Peaucelle, A., Ishida, T., Morin, H., Aida, M., and Laufs, P. (2006). The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell 18, 2929-2945. [DOI] [PMC free article] [PubMed]
- Poethig R.S. Clonal analysis of cell lineage patterns in plant development. Am. J. Bot. 1987;74:581–594. [Google Scholar]; Poethig, R.S. (1987). Clonal analysis of cell lineage patterns in plant development. Am. J. Bot. 74, 581-594.
- Rast-Somssich M.I., Broholm S., Jenkins H., Canales C., Vlad D., Kwantes M., Bilsborough G., Dello Ioio R., Ewing R.M., Laufs P. Alternate wiring of a KNOXI genetic network underlies differences in leaf development of A. thaliana and C. hirsuta. Genes Dev. 2015;29:2391–2404. doi: 10.1101/gad.269050.115. [DOI] [PMC free article] [PubMed] [Google Scholar]; Rast-Somssich, M.I., Broholm, S., Jenkins, H., Canales, C., Vlad, D., Kwantes, M., Bilsborough, G., Dello Ioio, R., Ewing, R.M., Laufs, P., et al. (2015). Alternate wiring of a KNOXI genetic network underlies differences in leaf development of A. thaliana and C. hirsuta. Genes Dev. 29, 2391-2404. [DOI] [PMC free article] [PubMed]
- Richardson A., Rebocho A.B., Coen E. Ectopic KNOX Expression Affects Plant Development by Altering Tissue Cell Polarity and Identity. Plant Cell. 2016;28:2079–2096. doi: 10.1105/tpc.16.00284. [DOI] [PMC free article] [PubMed] [Google Scholar]; Richardson, A., Rebocho, A.B., and Coen, E. (2016). Ectopic KNOX Expression Affects Plant Development by Altering Tissue Cell Polarity and Identity. Plant Cell 28, 2079-2096. [DOI] [PMC free article] [PubMed]
- Rivara M., Inostroza P. Using longest-side bisection techniques for the automatic refinement of delaunay triangulations. Int. J. Numer. Methods Eng. 1997;40:581–597. [Google Scholar]; Rivara, M., and Inostroza, P. (1997). Using longest-side bisection techniques for the automatic refinement of delaunay triangulations. Int. J. Numer. Methods Eng. 40, 581-597.
- Rubio-Somoza I., Zhou C.M., Confraria A., Martinho C., von Born P., Baena-Gonzalez E., Wang J.W., Weigel D. Temporal control of leaf complexity by miRNA-regulated licensing of protein complexes. Curr. Biol. 2014;24:2714–2719. doi: 10.1016/j.cub.2014.09.058. [DOI] [PubMed] [Google Scholar]; Rubio-Somoza, I., Zhou, C.M., Confraria, A., Martinho, C., von Born, P., Baena-Gonzalez, E., Wang, J.W., and Weigel, D. (2014). Temporal control of leaf complexity by miRNA-regulated licensing of protein complexes. Curr. Biol. 24, 2714-2719. [DOI] [PubMed]
- Runions A., Tsiantis M. The shape of things to come: From typology to predictive models for leaf diversity. Am. J. Bot. 2017;104:1437–1441. doi: 10.3732/ajb.1700251. [DOI] [PubMed] [Google Scholar]; Runions, A., and Tsiantis, M. (2017). The shape of things to come: From typology to predictive models for leaf diversity. Am. J. Bot. 104, 1437-1441. [DOI] [PubMed]
- Runions A., Tsiantis M., Prusinkiewicz P. A common developmental program can produce diverse leaf shapes. New Phytol. 2017;216:401–418. doi: 10.1111/nph.14449. [DOI] [PMC free article] [PubMed] [Google Scholar]; Runions, A., Tsiantis, M., and Prusinkiewicz, P. (2017). A common developmental program can produce diverse leaf shapes. New Phytol. 216, 401-418. [DOI] [PMC free article] [PubMed]
- Sapala A., Runions A., Routier-Kierzkowska A.L., Das Gupta M., Hong L., Hofhuis H., Verger S., Mosca G., Li C.B., Hay A. Why plants make puzzle cells, and how their shape emerges. eLife. 2018;7:e32794. doi: 10.7554/eLife.32794. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sapala, A., Runions, A., Routier-Kierzkowska, A.L., Das Gupta, M., Hong, L., Hofhuis, H., Verger, S., Mosca, G., Li, C.B., Hay, A., et al. (2018). Why plants make puzzle cells, and how their shape emerges. eLife 7, e32794. [DOI] [PMC free article] [PubMed]
- Scarpella E., Marcos D., Friml J., Berleth T. Control of leaf vascular patterning by polar auxin transport. Genes Dev. 2006;20:1015–1027. doi: 10.1101/gad.1402406. [DOI] [PMC free article] [PubMed] [Google Scholar]; Scarpella, E., Marcos, D., Friml, J., and Berleth, T. (2006). Control of leaf vascular patterning by polar auxin transport. Genes Dev. 20, 1015-1027. [DOI] [PMC free article] [PubMed]
- Segonzac C., Nimchuk Z.L., Beck M., Tarr P.T., Robatzek S., Meyerowitz E.M., Zipfel C. The shoot apical meristem regulatory peptide CLV3 does not activate innate immunity. Plant Cell. 2012;24:3186–3192. doi: 10.1105/tpc.111.091264. [DOI] [PMC free article] [PubMed] [Google Scholar]; Segonzac, C., Nimchuk, Z.L., Beck, M., Tarr, P.T., Robatzek, S., Meyerowitz, E.M., and Zipfel, C. (2012). The shoot apical meristem regulatory peptide CLV3 does not activate innate immunity. Plant Cell 24, 3186-3192. [DOI] [PMC free article] [PubMed]
- Shani E., Burko Y., Ben-Yaakov L., Berger Y., Amsellem Z., Goldshmidt A., Sharon E., Ori N. Stage-specific regulation of Solanum lycopersicum leaf maturation by class 1 KNOTTED1-LIKE HOMEOBOX proteins. Plant Cell. 2009;21:3078–3092. doi: 10.1105/tpc.109.068148. [DOI] [PMC free article] [PubMed] [Google Scholar]; Shani, E., Burko, Y., Ben-Yaakov, L., Berger, Y., Amsellem, Z., Goldshmidt, A., Sharon, E., and Ori, N. (2009). Stage-specific regulation of Solanum lycopersicum leaf maturation by class 1 KNOTTED1-LIKE HOMEOBOX proteins. Plant Cell 21, 3078-3092. [DOI] [PMC free article] [PubMed]
- Smith C., Prusinkiewicz P., Samavati F. Local Specification of Surface Subdivision Algorithms. Lect. Notes Comput. Sci. 2004;3062:313–327. [Google Scholar]; Smith, C., Prusinkiewicz, P., and Samavati, F. (2004). Local Specification of Surface Subdivision Algorithms. Lect. Notes Comput. Sci. 3062, 313-327.
- Smith R.S., Guyomarc’h S., Mandel T., Reinhardt D., Kuhlemeier C., Prusinkiewicz P. A plausible model of phyllotaxis. Proc. Natl. Acad. Sci. USA. 2006;103:1301–1306. doi: 10.1073/pnas.0510457103. [DOI] [PMC free article] [PubMed] [Google Scholar]; Smith, R.S., Guyomarc’h, S., Mandel, T., Reinhardt, D., Kuhlemeier, C., and Prusinkiewicz, P. (2006). A plausible model of phyllotaxis. Proc. Natl. Acad. Sci. USA 103, 1301-1306. [DOI] [PMC free article] [PubMed]
- Vlad D., Kierzkowski D., Rast M.I., Vuolo F., Dello Ioio R., Galinha C., Gan X., Hajheidari M., Hay A., Smith R.S. Leaf shape evolution through duplication, regulatory diversification, and loss of a homeobox gene. Science. 2014;343:780–783. doi: 10.1126/science.1248384. [DOI] [PubMed] [Google Scholar]; Vlad, D., Kierzkowski, D., Rast, M.I., Vuolo, F., Dello Ioio, R., Galinha, C., Gan, X., Hajheidari, M., Hay, A., Smith, R.S., et al. (2014). Leaf shape evolution through duplication, regulatory diversification, and loss of a homeobox gene. Science 343, 780-783. [DOI] [PubMed]
- Vuolo F., Mentink R.A., Hajheidari M., Bailey C.D., Filatov D.A., Tsiantis M. Coupled enhancer and coding sequence evolution of a homeobox gene shaped leaf diversity. Genes Dev. 2016;30:2370–2375. doi: 10.1101/gad.290684.116. [DOI] [PMC free article] [PubMed] [Google Scholar]; Vuolo, F., Mentink, R.A., Hajheidari, M., Bailey, C.D., Filatov, D.A., and Tsiantis, M. (2016). Coupled enhancer and coding sequence evolution of a homeobox gene shaped leaf diversity. Genes Dev. 30, 2370-2375. [DOI] [PMC free article] [PubMed]
- Willis L., Refahi Y., Wightman R., Landrein B., Teles J., Huang K.C., Meyerowitz E.M., Jönsson H. Cell size and growth regulation in the Arabidopsis thaliana apical stem cell niche. Proc. Natl. Acad. Sci. USA. 2016;113:E8238–E8246. doi: 10.1073/pnas.1616768113. [DOI] [PMC free article] [PubMed] [Google Scholar]; Willis, L., Refahi, Y., Wightman, R., Landrein, B., Teles, J., Huang, K.C., Meyerowitz, E.M., and Jonsson, H. (2016). Cell size and growth regulation in the Arabidopsis thaliana apical stem cell niche. Proc. Natl. Acad. Sci. USA 113, E8238-E8246. [DOI] [PMC free article] [PubMed]
- Zuniga A. Next generation limb development and evolution: old questions, new perspectives. Development. 2015;142:3810–3820. doi: 10.1242/dev.125757. [DOI] [PubMed] [Google Scholar]; Zuniga, A. (2015). Next generation limb development and evolution: old questions, new perspectives. Development 142, 3810-3820. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
S3
The model of the default growth pattern observed when marginal patterning at the leaf margin is absent, followed by the model of wild type A. thaliana leaf development.
A wild-type simulation is shown followed by simulations where parameters affecting protrusion form are varied, including: the rate of longitudinal growth, the region of competence for protrusion generation or the length of the growth domain.
4
The lateral extent of growth repression in the protrusion base is varied between simulations.
Descriptions of PIN quantification plug-in, regional primordium analysis and triangle-based protrusion morphospaces, as well as all computational models.