Nowak et al. show that loss of the AKT-inactivating phosphatase PHLPP2 paradoxically blocks prostate tumor growth and metastasis. PHLPP2, they find, is critical for MYC stability, suggesting that PHLPP2 inhibitors may present a therapeutic opportunity to target MYC.
Abstract
Metastatic prostate cancer commonly presents with targeted, bi-allelic mutations of the PTEN and TP53 tumor suppressor genes. In contrast, however, most candidate tumor suppressors are part of large recurrent hemizygous deletions, such as the common chromosome 16q deletion, which involves the AKT-suppressing phosphatase PHLPP2. Using RapidCaP, a genetically engineered mouse model of Pten/Trp53 mutant metastatic prostate cancer, we found that complete loss of Phlpp2 paradoxically blocks prostate tumor growth and disease progression. Surprisingly, we find that Phlpp2 is essential for supporting Myc, a key driver of lethal prostate cancer. Phlpp2 dephosphorylates threonine-58 of Myc, which renders it a limiting positive regulator of Myc stability. Furthermore, we show that small-molecule inhibitors of PHLPP2 can suppress MYC and kill PTEN mutant cells. Our findings reveal that the frequent hemizygous deletions on chromosome 16q present a druggable vulnerability for targeting MYC protein through PHLPP2 phosphatase inhibitors.
Introduction
Prostate cancer (PC) is one of the most prevalent cancers among men, causing almost 30,000 deaths in the United States alone. Death is mainly due to metastasis, as the 5-yr survival rate of metastatic PC is only 28%. In contrast, the 5-yr survival rate of organ-confined disease is almost 99% (National Cancer Institute, 2016). Understanding how genetic alterations are linked to cancer progression can help explain how tumor cells escape from focal disease sites to distant metastatic sites. However, there is a scarcity of human prostate metastatic samples for research purposes because invasive biopsies at metastatic sites can be dangerous and offer uncertain clinical benefit to patients.
Large-scale genomics efforts on both primary and metastatic PC have transformed our basic understanding of the genetics behind patient progression to metastatic disease. Two major lessons learned from these collaborative studies can be summarized as follows. First, PC has a low DNA missense mutation rate (Lawrence et al., 2013), resulting in only a few recurrent mutations (Barbieri et al., 2012) that show no increase in metastatic sample analysis (Robinson et al., 2015). In contrast, DNA repair–associated mutations may offer new therapeutic opportunities (Grasso et al., 2012; Cancer Genome Atlas Research Network, 2015; Mateo et al., 2015, 2017), but at this point they cannot help to identify the bulk of men who are at risk of progression. Second, metastatic patient samples reveal a sharp increase in the number of recurrent DNA copy number alterations (CNAs). These cover known drivers of disease, including PTEN, TP53, MYC, RB1, and amplification of AR, which is induced by hormone ablation therapy (Taylor et al., 2010; Cancer Genome Atlas Research Network, 2015; Robinson et al., 2015). However, most regions that suffer recurrent CNAs are still awaiting rigorous validation of the candidate cancer genes that they contain.
We, and others, use genetically engineered mice (GEMs) to functionally test the contribution of individual gene alterations to the disease (Irshad and Abate-Shen, 2013). Modeling of primary PC has demonstrated the pivotal role of Pten as an essential suppressor of PI 3-kinase/Akt signaling in controlling onset, progression, and outcome of the disease, either alone (Podsypanina et al., 1999; Kwabi-Addo et al., 2001; Trotman et al., 2003) or when combined with other insults (Di Cristofano et al., 2001; Chen et al., 2005; Trotman et al., 2006; Ding et al., 2011). Of particular relevance to this study, we have previously shown that loss of the phospho-Akt inhibiting Phlpp1 phosphatase triggers prostatic neoplasia on its own, and when combined with hemizygous loss of Pten results in highly penetrant prostate carcinoma (Chen et al., 2011). These results were consistent with the notion that the degree of PI 3-kinase/Akt pathway activation dictates disease course (Trotman et al., 2003), a notion that long served as the blueprint for target therapy of PC (Majumder and Sellers, 2005).
To now explore the mechanisms behind metastasis, we have recently developed RapidCaP (Cho et al., 2014). In this GEM model for analysis and therapy of endogenous metastatic PC, we are using somatic gene transfer to trigger loss of Pten and Trp53 in prostate, two alterations that have emerged as a hallmark of the human metastatic PC genome (Armenia et al., 2018). The analysis of primary lesions and visceral metastases revealed a surprise: in contrast to primary PC, suppression of Akt was seen in metastasis (Cho et al., 2014; Nowak et al., 2015). Mechanistically, we showed that inactivation of phospho-Akt was mediated by its phosphatase, Phlpp2, consistent with high Phlpp2 expression in the phospho-Akt–negative metastatic lesions from multiple histological sites (Nowak et al., 2015).
PHLPP2 and the closely related paralog PHLPP1 are members of the protein phosphatase 2C (PP2C) family of Mg2+/Mn2+-dependent phosphatases, which are insensitive to most common phosphatase inhibitors, including okadaic acid (OA; Brognard et al., 2007). They can inactivate signaling of their targets AKT and PKC by dephosphorylation of the C-terminal hydrophobic phosphorylation motifs (Brognard and Newton, 2008; Gao et al., 2008).
Since Phlpp1 loss triggers PC initiation by activation of Akt (Chen et al., 2011), but in metastasis we found that Akt is suppressed by a mechanism that requires Phlpp2, it has become unclear if Phlpp2 promotes or prevents the disease. Human PC genomics does not provide strong clues, as PHLPP2 is part of a recurrent broad hemizygous deletion in primary and metastatic disease. Therefore, we used genetics to directly test the role of Phlpp2 in vivo using the RapidCaP system as done previously for other candidate cancer genes (Cho et al., 2015; Chen et al., 2017). Our results show that despite its ability to suppress Akt kinase, Phlpp2 is required for PC and its progression because it can dephosphorylate and stabilize the Myc oncogene. The frequent hemizygous PHLPP2 deletions therefore make it an attractive drug target.
Results
PHLPP2 maintains MYC levels and cell proliferation
To dissect mechanistic connections between genes of interest, we first used in vitro recombination of mouse-derived primary cells, as published recently (Nowak et al., 2015). This approach allows us to dissect immediate mechanistic consequences of gene deactivation(s) from adaptive long-term responses in tissue culture (Fig. 1, A and B; and Fig. S1 A). Pten/Trp53 co-deletion results in increased levels of total Myc and Phlpp2, consistent with our previously published results (Nowak et al., 2015; Fig. 1 A and Fig. S1 B). In contrast to Phlpp2, Phlpp1 was not increased in the PtenΔ/Δ;Trp53Δ/Δ double mutants relative to the Trp53Δ/Δ single mutant, a consistent observation that was in line with our earlier findings suggesting that cells modulate Phlpp2 rather than Phlpp1 to compensate for Pten loss (Fig. 1 A; Chen et al., 2011).
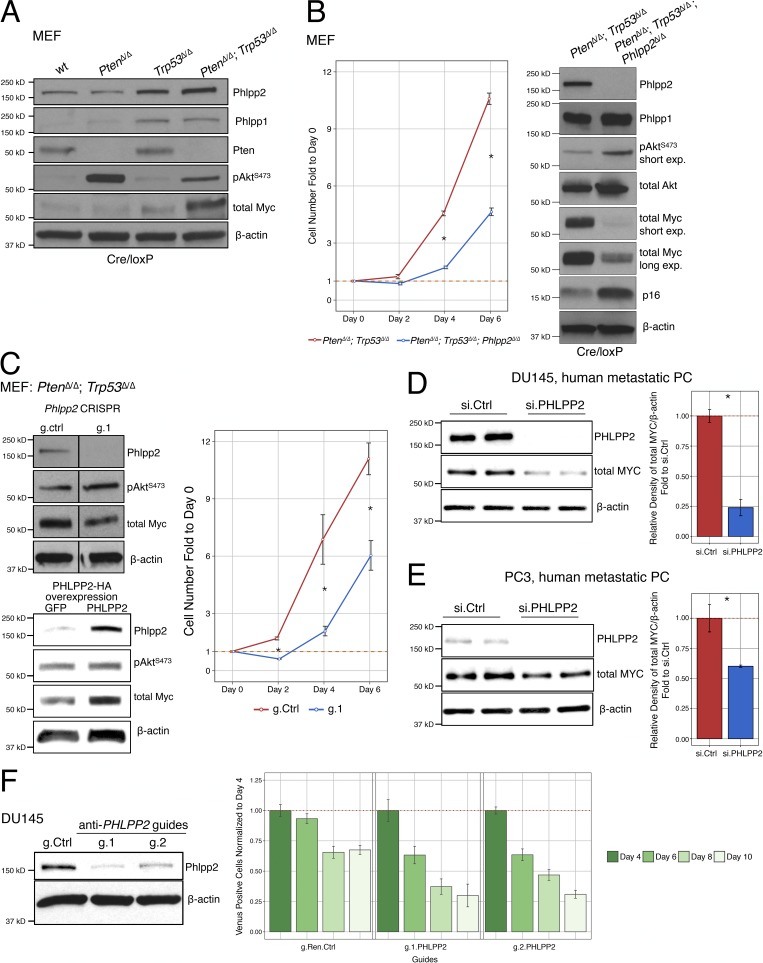
Figure 1.
Deletion of Phlpp2 suppresses proliferation of primary and cancer cells. (A) Analysis of signaling pathways by Western blot analysis of cell lysates from different genotypes of primary MEFs. (B) Loss of the Phlpp2 gene causes primary MEFs to proliferate less aggressively when compared with the Pten/Trp53-null cells (left panel). Error bars, SD; n ≥ 3; *, P < 0.05 with Student’s t test at days 4 and 6. Combined loss of the Pten/Trp53/Phlpp2 genes decreases expression of Myc and increases p16 expression (right panel). (C) Western blot analysis confirms efficient knockout of Phlpp2 with the CRISPR/Cas9 system and a decrease in activation of Myc expression (left upper panel, lines indicate splicing of two irrelevant lanes). Guide 1 anti-Phlpp2 (g.1) suppresses proliferation of Pten/Trp53–negative cells (right panel). Overexpression of PHLPP2 leads to increased Myc expression (lower left panel). Error bars, SD; n ≥ 3; *, P < 0.05 with Student’s t test at days 2, 4, and 6. (D) Western blot analysis confirms knockdown of PHLPP2 in DU145-human PC cells with siRNA (left panel), reaching almost 75% efficiency after 72 h, as quantified by densitometric analysis (right panel). Error bars, SD; n = 2; *, P < 0.05 with Student’s t test. (E) Western blot analysis confirms knockdown of PHLPP2 in PC3 human PC cells with siRNA (left panel), reaching almost 40% efficiency after 72 h, as quantified by densitometric analysis (right panel). Error bars, SD; n = 2; *, P < 0.05 with Student’s t test. (F) Western blot analysis shows knockdown of PHLPP2 in DU145 cells with two different guides (left panel). Depletion of the PHLPP2 mutant cells as seen by depletion of Venus-positive cells compared with g.Ren.Ctrl, normalized at day 4. Error bars, SD; n = 4; ANOVA, Dunnett’s post hoc test; P < 0.05 versus g.Ren.Ctrl at each respective day. wt, wild-type.
To assess the impact of Phlpp2 loss in double-mutant cells, we generated and tested triple-mutant PtenΔ/Δ;Trp53Δ/Δ;Phlpp2Δ/Δ mouse embryonic fibroblasts (MEFs). First, we noted a significant drop in proliferation of these cells compared with the double-mutant PtenΔ/Δ;Trp53Δ/Δ cells (Fig. 1 B, left panel). Protein analysis revealed increases in Akt activation upon Phlpp2 loss, as expected. To our surprise, however, we found that loss of Phlpp2 caused a sharp reduction in the total levels of Myc (Fig. 1 B, right panel). MYC is a driver oncogene of many cancer types, including human PC and classic GEMs of the disease (Ellwood-Yen et al., 2003; Farrell and Sears, 2014; McKeown and Bradner, 2014; Fernandez-Salas et al., 2016). Notably, we found that Myc is a spontaneously activated, essential driver of metastasis and resistance to castration therapy in RapidCaP (Cho et al., 2014; Nowak et al., 2015). Activation of the p16 tumor suppressor was also seen in these cells (Fig. 1 B, right panel), a response that can be overcome by Myc to cause immortalization (Gil et al., 2005). In addition, Phlpp2 loss did not affect levels of Phlpp1, showing lack of general compensation between these two proteins (Fig. 1 B, right panel).
To explore the result on MYC suppression in more depth, we next turned from simultaneous deletion of the three cancer genes to spontaneous loss of PHLPP2 in a stable PTEN/TP53 mutant (cancer) cell background using CRISPR/Cas9 and siRNA. Using the latter approach first in PtenΔ/Δ;Trp53Δ/Δ MEFs, we confirmed suppression of Myc upon Phlpp2 knockout together with increased phosphorylation of Akt (Fig. 1 C, upper left panel). Functional analysis of these cells also showed decreased proliferation after Phlpp2 deletion (Fig. 1 C, right panel). Conversely, overexpression of PHLPP2 in the double-mutant cells caused an increase in Myc protein, confirming the notion that PHLPP2 regulates Myc levels (Fig. 1 C, lower left panel). Next, we knocked down PHLPP2 in DU145 cells, a human metastatic PC cell line. As shown in Fig. 1 D (left panel), PHLPP2 siRNA caused an ∼75% reduction in MYC protein, as quantified by densitometric analysis (Fig. 1 D, right panel). PHLPP2-siRNA knockdown in PC3, another PC metastatic cell line (Fig. 1 E, left panel), also caused a reduction in MYC protein levels (by ∼40%; Fig. 1 E, right panel).
Finally, we tested if genetic targeting of PHLPP2 can be used to deplete DU145 cells upon stable infection with our anti–PHLPP2-CRISPR-Cas9-P2A-VenusFP viral vector system (ECPV; Chen et al., 2017; Senturk et al., 2017). Fig. 1 F shows how targeting of PHLPP2 with two different guides efficiently depleted the fluorescence-positive cells relative to the control. In contrast, the nonmetastatic cell line 22Rv1 showed no difference after Phlpp2 targeting (Fig. 1 F and Fig. S1 C). Collectively, our results suggested that PHLPP2 both controls and is required to maintain MYC levels and proliferation of cells, possibly more efficiently in metastatic cell lines.
PHLPP2 controls MYC stability
MYC up-regulation in human cancers occurs via a multitude of mechanisms (see Discussion); we observed gene amplification in metastatic and castration-resistant RapidCaP lesions (Cho et al., 2014). MYC stability is regulated by an intriguing phosphorylation system involving two conserved residues. Phosphorylation of Serine 62 (S62) by the kinases CDK2 (Hydbring et al., 2010), extracellular signal–related kinase (ERK; Sears et al., 2000), or CAMKIIγ (Gu et al., 2017) increases MYC stability (Fig. 2 A). In contrast, phosphorylation of threonine-58 (T58) by GSK3β triggers dephosphorylation of phospho-S62 by PP2A (Yeh et al., 2004). This is followed by ubiquitination of MYC by the SCF-Fbw7 E3 ligase and proteasomal degradation (Welcker et al., 2004). MYC degradation takes place in the nucleolus because, in the nucleoplasm, ubiquitination is counteracted by USP28, an ubiquitin-specific protease. USP28 binds to MYC via an interaction with FBW7α (Popov et al., 2007). We therefore tested if PHLPP2 affects phosphorylation of MYC and its stability as outlined in Fig. 2 A.
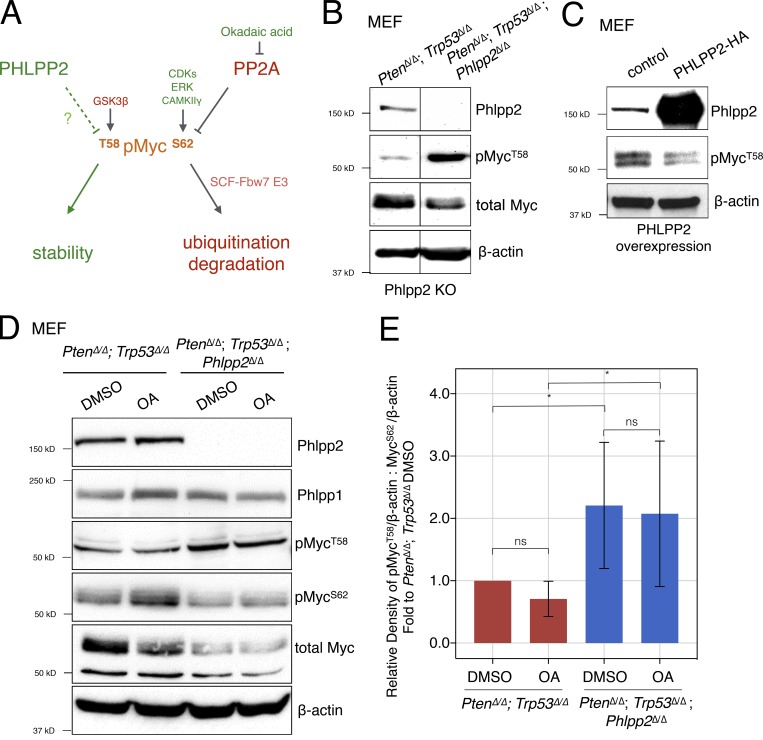
Figure 2.
Phlpp2 status controls phosphorylation of Myc. (A) Scheme representing regulators of MYC protein through T58 and S62 phosphorylation sites. These sites can be phosphorylated by GSK3β and CDK1/ERK/CAMKIIγ, respectively. Note that decreased ratios of T58 to S62 phosphorylation and T58 mutation to non–phospho-mimetic residues (e.g., T58A) are recurrent in human cancer. (B) Phlpp2 knockout in Pten/Trp53-null MEFs increases phosphorylation at the T58 site. Lines indicate splicing of three irrelevant lanes. (C) Overexpression of PHLPP2 leads to decreased T58 phosphorylation. (D) Treatment of Pten/Trp53-null MEFs with OA results in an increase in Myc phosphorylation on S62, but not T58. Total levels of Myc were reduced upon deletion of Phlpp2. (E) Densitometric analysis shows that the phosphorylation ratio of T58 to S62 is significantly increased in the triple-deleted MEFs. Data are presented as the densitometric measurements of pMycT58 and MycS62 normalized to β-actin and then these ratios of pMycT58 to MycS62 were used in graph as fold to PtenΔ/Δ;Trp53Δ/Δ DMSO. Error bars, SD; n = 5; one-way ANOVA, P values corrected for multicomparison testing using the two-stage linear step-up procedure of Benjamini, Krieger, and Yekutieli, with a false discovery rate at 10%. *, P < 0.05. KO, knockout; ns, not significant.
Deletion of Phlpp2 clearly increased the steady-state phosphorylation of Myc at T58 (Fig. 2 B), and PHLPP2 overexpression led to decreased T58 phosphorylation (Fig. 2 C). OA is an inhibitor of PP2A to which the PP2C-type phosphatases, such as PHLPP2, are insensitive (Gao et al., 2008). First, we observed that the Phlpp2 knockout boosted T58 phosphorylation, but not that of S62 (Fig. 2, D and E, lane 1 vs. 3). Second, Phlpp2 loss lowered Myc total levels (Fig. 2 D, lane 1 vs. lane 3). Furthermore, we saw that treatment with OA boosts Myc phosphorylation on S62, but not T58 (Fig. 2, D and E; lane 1 vs. lane 2). These results confirmed the previously proposed dominant role of T58 phosphorylation in dictating Myc levels (Sears, 2004; Farrell and Sears, 2014) and introduced Phlpp2 as a key regulator.
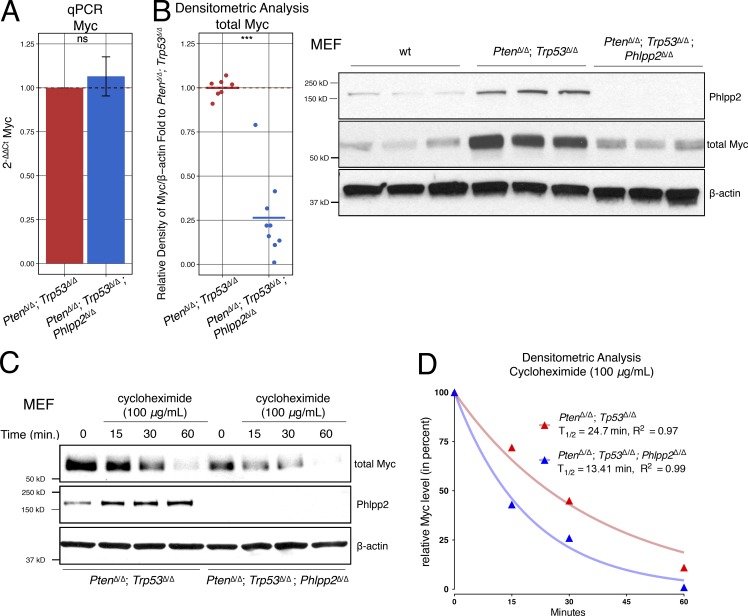
Based on our quantitative PCR results, we could exclude transcriptional suppression of Myc upon Phlpp2 knockout (Fig. 3 A). Instead, Myc protein was decreased by ∼75% on average (Fig. 3 B). Note that, for example, p16/Ink4a was in contrast increased at both protein and mRNA levels (Fig. 1 B and Fig. S2 A). Next, we studied Myc protein turnover as a function of Phlpp2 status using cycloheximide. As shown in Fig. 3, C and D, the Phlpp2-deficient MEFs consistently showed a reduced half-life of Myc protein (see Fig. S2, B and C, for replicates at various cycloheximide concentrations). Taken together, our results introduced Phlpp2 as a regulator of Myc stability via regulation of T58 phosphorylation.
Figure 3.
Phlpp2 status controls Myc stability. (A) Myc transcript levels measured by quantitative PCR is not affected by genotype. Error bars, SD; n = 4; P > 0.05 with Student’s t test. (B) Myc protein is decreased in triple-mutant cells by almost 75%. Error bars, SD; n ≥ 7; ***, P < 0.001 with Student’s t test (left panel). Right panel: Myc and Phlpp2 protein levels from WT, Pten/Trp53-, and Pten/Trp53/Phlpp2-negative cells. (C) Myc half-life as measured in MEFs of both genotypes after treatment with 100 µg/ml of cycloheximide. (D) Quantification of Western blot shows significantly shortened half-life of Myc in triple-mutant cells compared with the Phlpp2-positive cells. This graph represents densitometric analysis of Myc normalized with β-actin from the corresponding Western blot in Fig. 3 C. Lines (blue: PtenΔ/Δ;Trp53Δ/Δ;Phlpp2Δ/Δ; and red: PtenΔ/Δ;Trp53Δ/Δ) represent the nonlinear regression model, using the one-phase exponential decay curves. Myc half-life T1/2 was calculated based on this model, n = 1. Note that independent biological replicates are shown and quantified in Fig. S2, B and C. ns, not significant; qPCR, quantitative PCR; wt, wild-type.
PHLPP2 directly dephosphorylates Myc
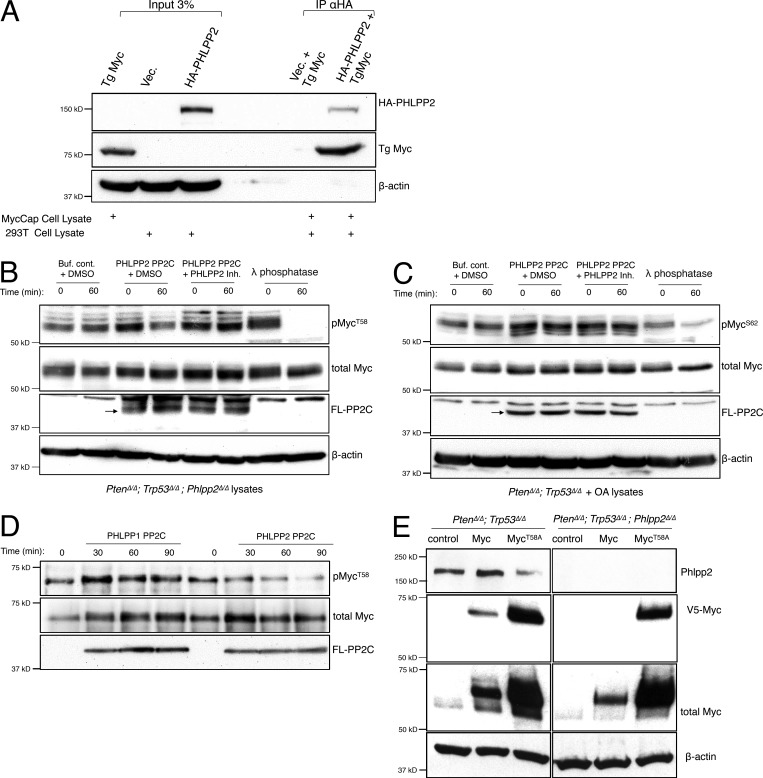
To address whether PHLPP2 targets Myc directly, we first expressed HA-PHLPP2 in 293T cells. These lysates were then mixed with lysates from the murine PC-derived MycCaP cells, which express elevated levels of transgenic Myc (Watson et al., 2005). Co-immunoprecipitation revealed that overexpressed HA-PHLPP2 specifically pulled down the transgenic Myc, consistent with a direct interaction (Fig. 4 A). We next examined if PHLPP2 directly and selectively dephosphorylates the T58 residue on Myc. The purified FLAG-tagged PP2C domain of PHLPP2 (Sierecki et al., 2010) was incubated with cell lysates derived from Pten/Trp53/Phlpp2-negative cells, which present well-detected levels of phosphorylated Myc at T58. As shown (Fig. 4 B), this incubation resulted in strong dephosphorylation of T58 (lane 3 vs. lane 4) that was effectively blocked by a PHLPP2 inhibitor (lane 5 vs. lane 6). Similar results were obtained when testing our positive control, the known regulation of PKC by PHLPP2 (Fig. 4 B and Fig. S3 A). To test if PHLPP2 also affects S62 of Myc, the purified FLAG-tagged PP2C domain of PHLPP2 was incubated with cell lysates derived from OA-treated, Pten/p53-deficient cells generating visibly phosphorylated Myc-S62 because of the PP2A inhibition. In contrast to T58, however, this incubation did not result in dephosphorylation of S62 (Fig. 4 C, lane 3 vs. lane 4), consistent with specific dephosphorylation of pMYCT58 by PHLPP2. Lambda phosphatase, a broadly active serine–threonine phosphatase, was used as a positive control for dephosphorylation in these assays (Fig. 4, B and C, lane 7 vs. lane 8). The PP2C domain is mediating PHLPP1/2 phosphatase activity, but the similarity between the paralogs is only 57% at the amino acid level (Fig. 4 D and Fig. S3 B). We thus also tested the kinetics of dephosphorylation by the PP2C domain of PHLPP1 and found that it did not cause dephosphorylation of pMYC at T58 (Fig. 4 D). Collectively, these results strongly suggested that PHLPP2 directly dephosphorylates the T58 site of MYC, but not S62.
Figure 4.
Phlpp2 dephosphorylates Myc directly. (A) Co-immunoprecipitation experiments revealed that overexpressed HA-tagged PHLPP2 pulled-down transgenic (Tg) Myc (from MycCaP cells), consistent with a direct interaction. (B) PHLPP2 directly dephosphorylates T58 on Myc. Incubation with the FLAG-tagged PHLPP2 PP2C domain (arrow) resulted in a decrease in T58 phosphorylation after 60 min of incubation (lanes 3 and 4). Dephosphorylation was blocked by a small-molecule inhibitor of PHLPP2 (lanes 5 and 6). (C) PHLPP2 does not dephosphorylate S62 on Myc. Incubation with the FLAG-PP2C domain of PHLPP2 (arrow) did not affect S62 phosphorylation after 60 min of incubation (lanes 3 and 4). Dephosphorylation was not affected by a small-molecule inhibitor of PHLPP2 (lanes 5 and 6). (D) The purified FLAG-tagged PP2C domain of PHLPP2 was incubated with phosphorylated purified Myc protein for different time points. Incubation with the PP2C domain of PHLPP1 did not affect T58 even after 90 min. In contrast, incubation with the PP2C domain of PHLPP2 resulted in a decrease in T58 phosphorylation that was most significant after 90 min. (E) MycT58A levels increase compared with WT MYC regardless of Phlpp2 demonstrating that this site is key for Phlpp2-mediated Myc regulation. wt, wild-type.
Phosphorylation on T58 is a signal for MYC degradation (Sears et al., 2000; Welcker et al., 2004), and the lymphoma-associated T58A mutation of MYC has therefore enhanced oncogenicity (Hemann et al., 2005). Based on our findings, MycT58A protein levels should thus not be affected by Phlpp2 deletion. Indeed, as shown in Fig. 4 E, we found that MycT58A levels were insensitive to Phlpp2 deletion as we observed strong MycT58A expression under conditions where we could not detect MycWT expression in the Phlpp2-deficient cells (Fig. 4 E, see V5-Myc, right panels).
Pharmacological inhibition of Phlpp2 suppresses Myc and proliferation
Based on the above findings, we next tested the intriguing hypothesis that an inhibitor developed to target the PHLPP2 PP2C domain could have an anticancer effect by suppressing MYC protein, a key oncogene that cannot yet be pharmacologically controlled. Various cell types were therefore treated with this inhibitor (NCI45586; see Sierecki et al., 2010). First, we tested two GEM-derived cell lines, known to drive PC through Myc (Watson et al., 2005; Cho et al., 2014). As expected, the Phlpp2 inhibitor induced a dose-dependent increase in phospho-S473 of Akt in the PtenΔ/Δ;Trp53Δ/Δ cells (Fig. 5 A). Importantly, this was accompanied by a dose-dependent reduction in total Myc protein, entirely consistent with our observations above. The murine PC-derived MycCaP cell line showed the same opposite effects on Myc and Akt (Fig. 5 B) as well as an increase in cell death (Fig. S3 C). We confirmed that the inhibitor also caused cell death in our RapidCaP PC-derived RCaP cells (Naguib et al., 2018; see Fig. S3 D) and that genetic ablation of Phlpp2 increased cell death in MEFs (Fig. S3 E). These results strongly indicated that Myc levels, rather than Akt activation, drive survival and proliferation in the cells, a hierarchical relation that is consistent with our previously published results in cells (Nowak et al., 2015) and in mouse primary and metastatic PC (Cho et al., 2014).
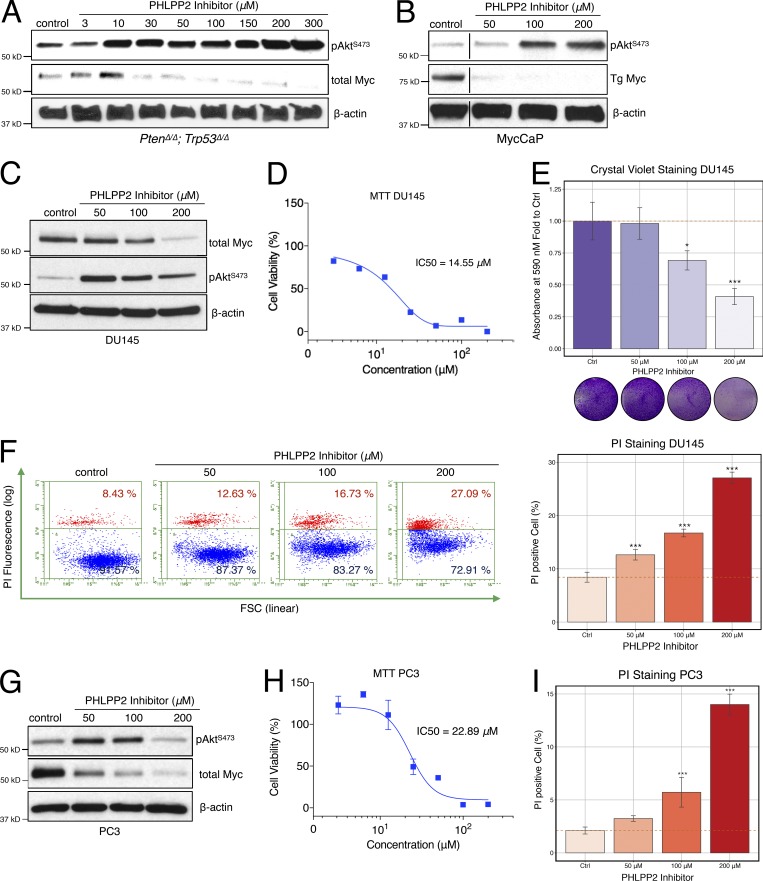
Figure 5.
Small-molecule PHLPP2 inhibitors suppress MYC and trigger cell death. (A) Treatment with a PHLPP2 inhibitor decreases Myc expression and increases pAktS473 activation in PtenΔ/Δ;Trp53Δ/Δ cells, compared with control DMSO treatment. (B) PHLPP2 inhibitor treatment results in decreased Myc expression and increased Akt activation in the MycCaP murine PC cell line. Lines indicate splicing of three irrelevant lanes. (C) Treatment with the PHLPP2 inhibitor decreases Myc expression and increases pAktS473 activation in DU145 cells, compared with control DMSO treatment. (D) Relative cell viability of DU145 cells upon PHLPP2 inhibitor treatment. The mean with error bars, SD (n = 6), and dose–response curves are plotted. IC50 values were derived from curves fitted using a nonlinear regression model. (E) Treatment with the PHLPP2 inhibitor decreases cell numbers measured with crystal violet assays in DU145 cells, compared with control DMSO. ANOVA, Dunnett’s post hoc test; *, P < 0.05; ***, P < 0.001 versus control; error bars, SD; n = 4. (F) Treatment with the PHLPP2 inhibitor increases cell death as shown by PI staining. ANOVA, Dunnett’s post hoc test; ***, P < 0.001 versus control; error bars, SD; n = 4. (G) PHLPP2 inhibitor treatment suppresses Myc and activates Akt in the PC3 human metastatic PC cell line. (H) Relative cell viability of PC3 cells upon PHLPP2 inhibitor treatment. The mean ± SD (n = 3) and dose–response curves are plotted. IC50 values were derived from curves fitted using a nonlinear regression model using. (I) Treatment with the PHLPP2 inhibitor increases cell death as shown by the PI-positive cell fraction. ANOVA, Dunnett’s post hoc test; ***, P < 0.001 versus control; error bars, SD; n = 4.
We next tested human PC cell lines. In the metastasis-derived DU145 cells, NCI45586 also showed suppression of MYC and some activation of AKT (Fig. 5 C; see Fig. S3 F for replicates). Treatment with inhibitor led to decreased cell viability and proliferation (Fig. 5 E), and it increased cell death (Fig. 5 F). Similarly, in the bone metastasis–derived PC3 cells, the inhibitor caused suppression of MYC and a decrease in cell viability, despite activation of AKT (Fig. 5, G–I; and Fig. S3 G). Our results with the NCI45586 tool compound closely mirrored those from genetic ablation of the PHLPP2 gene, a proof of concept that could lead to development of pharmacologically relevant drugs. In summary, our results showed that the PHLPP2 function of protecting MYC can be targeted to kill PTEN mutant cells.
Deletion of Phlpp2 slows down primary tumor growth and prevents progression to metastasis
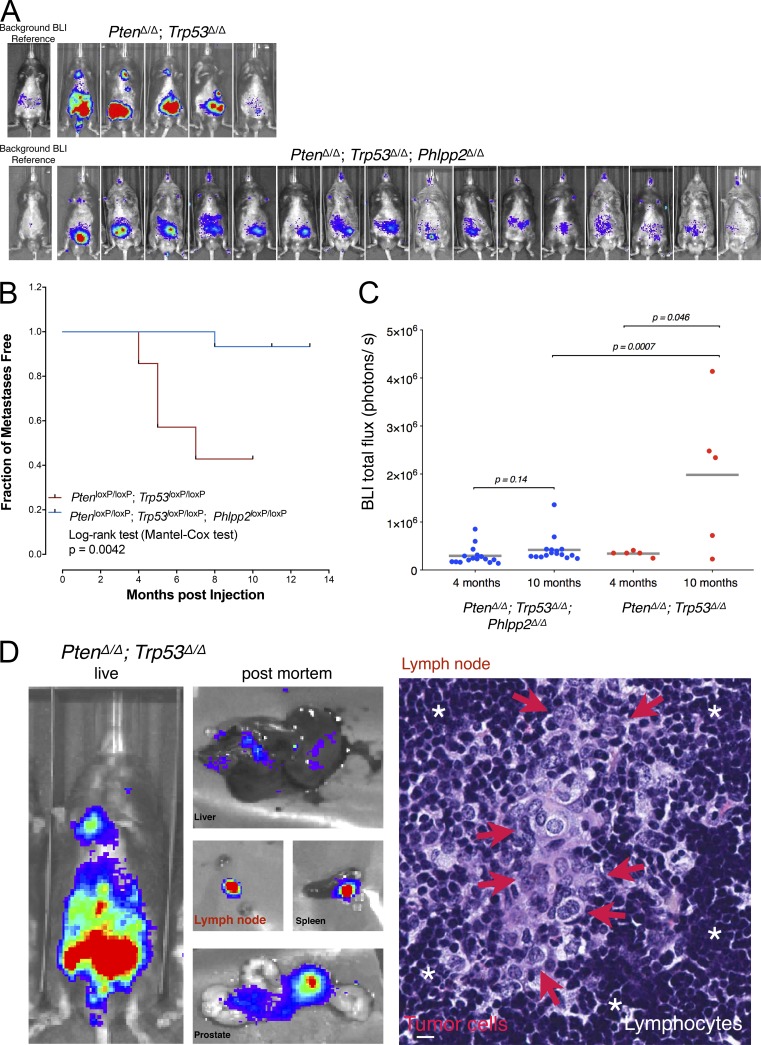
We have recently developed RapidCaP, a GEM model for analysis and therapy of endogenous metastatic PC (Cho et al., 2014). Of note, we could show that metastasis to multiple organs can be selectively targeted using JQ1 to transcriptionally suppress Myc. Thus, we now used RapidCaP to test how ablation of Phlpp2 would affect disease (note that NCI45586 cannot be used in mice). We generated conditional knockout mice (Phlpp2loxP/loxP) and crossed them to the PtenloxP/loxP;Trp53loxP/loxP mice on which RapidCaP is based to generate the novel triple-mutant PtenloxP/loxP;Trp53loxP/loxP;Phlpp2loxP/loxP mice (Fig. S4, A and B). Next, we used the RapidCaP system of intraprostatic Cre-Luciferase virus delivery to generate a trial cohort of PtenΔ/Δ;Trp53Δ/Δ;Phlpp2Δ/Δ mice and compared their disease progression and outcome to that of the PtenΔ/Δ;Trp53Δ/Δ RapidCaP mice. As shown in Fig. 6 A, four out of five Pten/Trp53 double-mutant mice presented with strong disease signals over a period of 10 mo. They also showed bioluminescent signal derived from metastases that was clearly distant and distinct from the prostate-derived signal seen in the lower abdomen. Kaplan-Meier analysis of metastasis onset and outcome in the double-mutant mice (Fig. 6, A and B) showed overall kinetics that were very similar to what we originally published (Cho et al., 2014). In contrast, the cohort with Phlpp2 deletion showed significantly reduced overall disease burden after a 10-mo follow up (Fig. 6, A–C; PtenΔ/Δ;Trp53Δ/Δ;Phlpp2Δ/Δ). Yet, as seen in Fig. 6 C, quantification showed an insignificant difference between the two cohorts at 4 mo (P = 0.6, two-tailed Fisher’s exact test), demonstrating the lack of metastatic progression potential of the Phlpp2 mutant prostate tumors. Postmortem analysis of the disease (Fig. 6 D) indicated metastasis to lymph nodes, liver, and spleen, as previously described for RapidCaP (Cho et al., 2014), but not in the triple-mutant animals (Fig. S4 C). Histological examination of luciferase-positive lymph nodes allowed us to identify the nests of large metastatic tumor cells with prominent nucleoli and pale pleomorphic nuclei (Fig. 6 D, right panel, red arrows) embedded within the lymphocytes. Note that total sectioning of lymph nodes from the triple-mutant cohort did not reveal any metastatic cells (data not shown).
Figure 6.
Phlpp2is required for progression of Pten mutant PC. (A) Comparison of PC progression to metastasis in double- and triple-mutant RapidCaP mice as determined by bioluminescent (BLI) imaging. In the PtenΔ/Δ;Trp53Δ/Δ system, four out of five animals present with primary and metastatic signal within 10 mo, while little to insignificant disease is imaged in PtenΔ/Δ;Trp53Δ/Δ;Phlpp2Δ/Δ. (B) Kaplan–Meier analysis shows the early onset of metastasis in PtenΔΔ/Δ;Trp53Δ/Δ compared with PtenΔ/Δ;Trp53Δ/Δ;Phlpp2Δ/Δ mice as determined by monthly luciferase imaging. P = 0.0042 with log-rank (Mantel-Cox) test; PtenΔ/Δ;Trp53Δ/Δ: n = 5; PtenΔ/Δ;Trp53Δ/Δ;Phlpp2Δ/Δ: n = 16. (C) Bioluminescent signals are significantly lowered in PtenΔ/Δ;Trp53Δ/Δ;Phlpp2Δ/Δ animals compared with PtenΔ/Δ;Trp53Δ/Δ mice at 10 mo but not at 4 mo (P = 0.6, two-tailed Student’s t test). Each dot represents bioluminescent (BLI) signal from one animal, lines are means, and indicated P values are for two-tailed Student’s t tests. PtenΔ/Δ;Trp53Δ/Δ: n = 5; PtenΔ/Δ;Trp53Δ/Δ;Phlpp2Δ/Δ: n = 16. (D) Postmortem luciferase analysis confirms metastasis to secondary organs in PtenΔ/Δ;Trp53Δ/Δ (left panels). The right panel shows a typical nest of prostate tumor cells having metastasized to the luciferase-positive lymph node shown in the left panel. White asterisks indicate normal lymphocytes, and red arrows indicate metastatic prostate tumor cells. Scale bar, 20 µm.
Taken together, our in vivo genetic validation data demonstrated that Phlpp2 is required for the tumor and metastasis formation seen in the Pten/Trp53-mutant RapidCaP model.
Discussion
By activating MYC and suppressing AKT, PHLPP2 exhibits a dual nature in cancer. This may be reflected in its cancer-associated alteration patterns, which are dictated by broad chromosomal 16q hemizygous deletions and some amplifications, as shown in Fig. S5 A. Also, in PC, the vast majority of PHLPP2 mutations are broad 16q deletions (Fig. S5 B), which may result in some AKT activation, as we have shown for Phlpp1 loss (Chen et al., 2011). Yet, the remaining allele is widely retained in human PC metastasis (Fig. S5 C) and clearly necessary to obtain metastasis in RapidCaP, as shown above. This requirement for the retention of one allele has been recognized as a druggable vulnerability of cancer (“Cyclops” genes; Nijhawan et al., 2012). Indeed, an unbiased analysis of such Cyclops gene candidates from human cancer data did include PHLPP2 (see Table S3 in Nijhawan et al., 2012). Thus, PHLPP2 could fall into a new type of category of cancer genes. Its hemizygous loss can promote cancer through AKT activation. The remaining allele, however, is protected because it mediates survival through MYC. Thus, PHLPP2 is in a unique position to regulate the signaling plasticity of a cell and its switching between AKT (epithelial) and MYC (mesenchymal) phenotypes. This switch can be observed in primary PC, and it can drive Pten-deficient metastasis (Cho et al., 2014; Nowak et al., 2015). It is intriguing to speculate that these opposing activities may be achieved by regulation of PHLPP2 pools in the nucleus (MYC) and the cytoplasm (AKT).
Several independent studies have now confirmed the existence of a Pten-null Akt-off state in PC (Ku et al., 2017; Zou et al., 2017). In humans, the modeling of “AR indifference” by pharmaco-genetic AR suppression of the PTEN-deficient LNCaP cell line (Bluemn et al., 2017) revealed that the derived AR-negative, ADT (androgen deprivation therapy)-resistant cells no longer showed detectable AKT pathway activity despite PTEN deficiency, closely mirroring our findings in RapidCaP mice, especially after castration. Collectively, these results highlight the importance of understanding AKT-negative, PTEN-deficient disease, as it appears to be among the drivers of resistance to the latest generation of androgen deprivation therapy.
It is estimated that 450,000 Americans are diagnosed each year with a cancer that is driven by MYC (McKeown and Bradner, 2014). MYC is a validated driver of PC in GEM models of primary disease and metastasis (Ellwood-Yen et al., 2003; Cho et al., 2014). To date, there is no drug that directly targets MYC because inhibitor design is limited by the lack of good target motifs or clefts (McKeown and Bradner, 2014). Yet, several indirect strategies have shown promise. Foremost among them is inhibition at the level of transcription using inhibitors of BRD4, such as the small molecule JQ1 (Delmore et al., 2011). This approach has shown much promise in GEM models of, for example, leukemia (Zuber et al., 2011), metastatic PC (Cho et al., 2014), lung cancer, and neuroblastoma (Puissant et al., 2013; Shimamura et al., 2013). This strategy is now the basis for several clinical trials (Liu et al., 2017). Other approaches target upstream activators of MYC, such as NOTCH1 (Weng et al., 2006), or interfere with MYC stability through indirect activation of PP2A. Activation of PP2A, however, presents a formidable challenge because it (1) requires indirect suppression of its endogenous inhibitors (Janghorban et al., 2014; Kaur and Westermarck, 2016), (2) forms over 70 active holoenzyme complexes, and thus (3) is known to control many proteins and processes (Sangodkar et al., 2016).
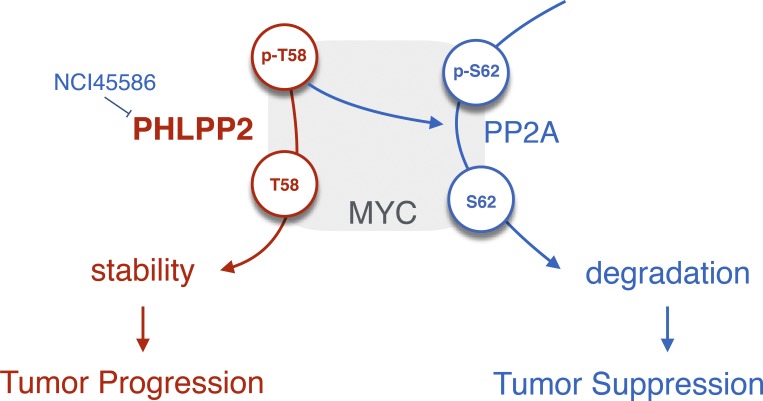
We show that PHLPP2 protects MYC from degradation. PHLPP2 dephosphorylates the T58 site of MYC, thus directly increasing its stability (see model in Fig. 7). This residue suffers recurrent mutation (T58A) in patients with Burkitt’s lymphoma, and it was shown to cause increased transformation in vitro and lymphoma in vivo (Chang et al., 2000; Hemann et al., 2005). The T58A mutant is constitutively dephosphorylated, mimicking constant PHLPP2 activity. Thus, its natural selection in MYC-driven human lymphoma impressively demonstrates the oncogenic power of PHLPP2 action on MYC.
Figure 7.
Proposed model for PHLPP2. PHLPP2 protects MYC from degradation resulting in PC progression. PHLPP2 dephosphorylates the T58 site of MYC, thus directly increasing its stability. Small-molecule inhibitors of PHLPP2 have been discovered (Sierecki et al., 2010). PHLPP2 thus has opposing roles in PC: while suppressing AKT activation, it stabilizes MYC progression. Our data suggest that the latter function is critical for PC progression.
Small-molecule inhibitors of PHLPP2 have been discovered (Sierecki et al., 2010). We show that they can be used to suppress MYC and cause cell death. The compounds were screened from the First Diversity Set of the National Cancer Institute, which comprises 1990 small molecules. At this stage, they should be considered proof of principle compounds, rather than drugs. This suggests, however, that targeted efforts for designing PHLPP2 inhibitors could result in very efficient new drugs that suppress MYC-driven cancer (see proposed model, Fig. 7).
The PHLPP2 phosphatase is an attractive drug target for several reasons. First, we show that Pten-deficient metastatic PC cells fully depend on it. In contrast, normal cells do not, as demonstrated by the normal viability of the Phlpp2 knockout mice (Wen et al., 2015). Next, general advances in structural understanding of phosphatases have led to a “renaissance” of inhibitor design (Krishnan et al., 2014; Chen et al., 2016). By looking beyond the active site, novel allosteric inhibitors can now shut down phosphatases that used to be considered “undruggable.” This could also apply to the PP2C-type active site of PHLPP2. Thus, our work introduces the PHLPP2 phosphatase as an unexpected, yet druggable, driver of MYC-driven PC and its progression.
Materials and methods
Mice
PtenloxP/loxP;Trp53loxP/loxP and PtenloxP/loxP;Trp53loxP/loxP;Phlpp2loxP/loxP were used in this study. All protocols for mouse experiments were done in accordance with the institutional guidelines and approved by Institutional Animal Care and Use Committee. For the creating of Phlpp2loxP/loxP animals, we used embryonic stem cells that were received from the Mouse Phenotype Consortium (Phlpp2-IKMC Project: 47191). We used promoter-driven targeting cassettes for the generation of a “knockout-first allele” in C57BL/6N mice. First, mice were bred to a flipase (Flp) mouse to remove lacZ/loxP/neo cassette that gave a rise to loxP conditional mice. Following removal of the floxed region using Cre, our expression analysis confirmed knockouts of Phlpp2 at the protein level. PtenloxP/loxP;Trp53loxP/loxP, PtenloxP/loxP;Trp53loxP/loxP, and Phlpp2loxP/loxP transgenic mice were generated by crossing PtenloxP/loxP;Trp53loxP/loxP and Phlpp2loxP/loxP. For genotyping, PCR of tail DNA was used with the following primers: for PtenloxP/loxP, primer 1 (5′-TGTTTTTGACCAATTAAAGTAGGCTGTG-3′) and primer 2 (5′-AAAAGTTCCCCTGCTGATGATTTGT-3′) were used; for Trp53loxP/loxP, primer 1 (5′-CACAAAAACAGGTTAAACCCAG-3′) and primer 2 (5′-AGCACATAGGAGGCAGAGAC-3′) were used; and for Phlpp2loxP/loxP, primer 1 (5′-GATGCTCTGCTTCTGCTCCTGTGC-3′) and primer 2 (5′-GATATGAGGAACGAAGCAATATGGG-3′) were used.
MEFs
Animals with different combinations of loxP alleles were used. At 13.5 d, embryos were harvested by sacrificing a female using CO2. Embryos were minced and trypsinized for 15 min in a 37°C water bath. Cells were plated on a 10-cm dish and cultured in 10% DMEM and 1× penicillin–streptomycin. 72 h after plating, cells were split into a 10-cm dish at 900,000 cells per dish per 15-ml volume of 10% DMEM and 1× penicillin–streptomycin. Cells were cultured for the next 3 d, collected, frozen in 20% dimethyl sulfoxide/80% FBS, and stored in batches of 1,000,000 cells per cryovial in liquid nitrogen. All protocols for mouse experiments were conducted in accordance with the institutional guidelines and were approved by the Institutional Animal Care and Use Committee at Cold Spring Harbor Laboratory (protocol number 562709, “Tumor suppressor mutation in mouse models for cancer”).
Cancer cell lines
Cell lines were used and cultured in 10% DMEM (MycCaP) or RPMI (PC3, DU45) medium and 1× penicillin–streptomycin.
Drug treatments on 2D cell lines and propidium iodide analysis
For PHLPP2 inhibitor experiments, the NCI45586 compound was used to assess effects of PHLPP2 on pAkt, Myc signaling, and cell death measured with propidium iodide (PI) using Guava easyCyte 8HT Benchtop Flow Cytometer (Millipore Sigma). The PHLPP2 inhibitor was resuspended in DMSO and used as previously described (Sierecki et al., 2010).
Drug treatment on 3D cultured RapidCaP-derived prostate epithelial cells
3D cultures of a RapidCaP-derived prostate cell line was performed as per the protocol in Drost et al. (2016). Matrigel was thawed overnight at 4°C and placed on ice during the experiment. Prechilled glass-bottom 6-well plates were coated with a thin layer of Matrigel (5 µl) and incubated for 30 min at 37°C to allow the Matrigel to set. Cells (80% confluent) were harvested to a single-cell suspension by trypsinization. Approximately 15,000 cells were aliquoted into a 1.5-ml microcentrifuge tube, pelleted, and resuspended in 100 µl of Matrigel. The mixture of cells and Matrigel was gently mixed. Three 30-µl aliquots of the mixture was carefully overlaid onto the precoated surface of the glass plate and allowed to set for 15 min at 37°C. Once the Matrigel had set, cell culture media was carefully added to the wells. The culture was maintained for 5 d before drug treatment and media changed every 2 d. For drug treatment, 1 ml of fresh basal media with vehicle or drug was added to overnight serum-starved cells. 72 h after treatment, media were removed and cells were extracted from Matrigel and dissociated for cell viability analysis using trypan blue dye.
RNA isolation and quantitative RT-PCR
Isolation of RNA was performed using TRIzol (Invitrogen) and purified by precipitation with isopropanol. Total RNA was reverse transcribed using the High Capacity cDNA Reverse Transcription Kit (Life Technologies) according to the manufacturer’s indications. 2 μl of this cDNA was then amplified by real-time PCR in a final volume of 10 μl per reaction on a StepOnePlus System thermocycler using the respective assays (Life Technologies). All reactions were performed in triplicate. The relative expression of Myc, p16, and Phlpp2 was determined using the mean value of the control samples (WT) as calibrator and following the 2-ΔΔCt method.
Western blotting
To asses protein expression, we used SDS-PAGE (8% and 10% reducing gels, 5% 2-β mercaptoethanol). Gels were loaded with 20–40 μg protein per well (Bradford assay). The following antibodies were used: β-actin (1:30,000; Sigma-Aldrich), pAKT (S473; 193H12, 1:3,000; Cell Signaling Technology), total AKT (40D4, 1:3,000; Cell Signaling Technology), c-Myc (ab32072; 1:3,000; Abcam), pMYC (T58; ab28842, 1:1,000; Abcam), pMYC (S62; E1J4K, 1:1,000; Cell Signaling Technology), proliferating cell nuclear antigen (PCNA) (PC10, 1:6,000; Santa Cruz Biotechnology), PTEN (6H2.1; 1:1,000; Cascade Bioscience), p16 (M-156; 1:1,000; Santa Cruz Biotechnology), PHLPP2 (A300-661A; 1:1,000), and V5-tag (#13202, 1:1,000; Cell Signaling Technology). ECL was used with Amersham Hyperfilm ECL (Amersham Bioscience) combined with signal intensity analysis using the ImageJ 1.38× software. Infrared-based recording and quantification were done using the LiCor Odyssey Classic infrared scanner and Image Studio Lite software version 5.2. Histogram analysis was done to prevent pixel saturation and clipping.
MTT assays
10,000 cells/well were seeded in full media (RPMI, 10% FBS) on a 96-well plate and left to adhere overnight. The cells were serum starved (RPMI, 0% FBS) for 24 h. After serum starvation, the cells were treated with an eight-point twofold dilution of the Phlpp2 inhibitor (compound #13) in 2% FBS-RPMI for 96 h. After treatment, the medium containing the compound was removed and MTT solution was added. After labeling cells with MTT, the absorbance was read at 570 nM.
Crystal violet assay
Cell proliferation was also measured using the crystal violet method as previously described (Nowak et al., 2015). Cells were fixed in 10% formalin for 15 min, washed with sterile water, and stained with 0.1% crystal violet solution. Solutions were aspirated and washed with water, and then plates were air dried overnight. After this, 10% acetic acid was added and mixed with water in a 1:3 ratio. Absorbance was measured at 590 nm using a plate reader (Synergy H4 Hybrid Multi-Mode Microplate Reader; Biotek).
Myc mutant transfection
PtenloxP/loxP;Trp53loxP/loxP and PtenloxP/loxP;Trp53loxP/loxP;Phlpp2loxP/loxP cells previously infected with adenovirus carrying Cre recombinase were seeded in the full media (DMEM, 10% FBS) on a 6-well plate and left to adhere overnight. Cells then were transfected with plasmids: pD40-His/V5-c-Myc (Addgene; #45597) and pD40-His/V5-c-MycT58A (Addgene; #45598; Yeh et al., 2004) using Lipofectamine 2000 following the manufacturer’s protocol (Thermo Fisher Scientific; 11668027). 16 h after transfection, cells were analyzed using Western blots.
siRNAi transfection targeting PHLPP2
PC3 and DU145 cells were transfected with siRNAi control (ON-TARGETplus Non-targeting Pool; Darmacon) and siRNAi targeting PHLPP2 (ON-TARGETplus Human PHLPP2; Darmacon) using DharmaFECT following the manufacturer’s protocol. 48–72 h after transfection, cells were analyzed using Western blots.
Immunoprecipitation
DNA was transfected into HEK-293T cells using FuGene6 (Roche). Cells were collected 24 h after transfection and then lysed in a buffer containing 50 mM Na2HPO4 (pH 7.5), 1 mM sodium pyrophosphate, 20 mM NaF, 2 mM EDTA, 2 mM EGTA, 1% SDS, 1 mM DTT, 1 µM microcystin, 20 µM bezamidine, 40 µg/ml leupeptin, and 1 mM PMSF and then were sonicated briefly. For co-immunoprecipitation, cells were lysed and the detergent-solubilized cell lysates were incubated with an anti-HA antibody (BioLegend; #901503) at 4°C overnight. The next day, the samples were incubated with protein A/G PLUS-Agarose (Santa Cruz Biotechnology; #sc-2003) for 1 h at 4°C and then washed three times in lysis buffer containing 0.05 M NaCl and 0.25% Triton-X. Bound proteins and lysates were separated by SDS-PAGE gel and analyzed by Western blotting.
Phosphatase assays
A 3XFLAG-tagged protein PHLPP2-PP2C was expressed in HEK-293T cells and purified using anti-FLAG M2 affinity gel (Sigma-Aldrich; A2220). The FLAG-PP2C protein was eluted from the beads in elution buffer containing 50 mM Hepes, pH 7.4, 100 mM NaCl, 1 mM DTT, 0.1 mM Na3VO4, 0.01% NP-40, 10% glycerol, and 0.5 mg/ml 3XFLAG peptide (Sigma-Aldrich; F4799). Recombinant human Myc protein was purchased from Abcam (ab169901). To obtain phosphorylated Myc as substrate, in vitro kinase assays were performed by incubating cMyc (0.30 µM) with recombinant human GSK3 β (54.79 nM; ab60863; Abcam) at 30°C for 15 min. Reactions were stopped using 40 µM staurosporine (Calbiochem). The dephosphorylation reactions were performed with 3XFLAG-PP2C (0.76 µM) and phosphorylated cMyc (0.30 µM) as a substrate in a reaction buffer containing 50 mM Tris (pH 7.4), 1 mM DTT, and 5 mM MnCl2 at 30°C for 0–90 min.
Lentivirus production
293FT cells were plated at a density of 400,000 cells/ml in the volume of 27 ml per 15-cm dish. The next day, target DNA (27 μg) was combined with packaging constructs, 8 μg of pMD2.G and 20 µg of psPAX2, and 2 mol/liter calcium chloride. The mixture was then added dropwise to cells that were treated with 6.7 μl 100 mmol/liter chloroquine. Medium was changed after 14 h, and viral supernatants were collected 72 h after transfection. Supernatants were then centrifuged and filtered with a 0.45-μm filter before infection of cells. The 293FT cells were bought from Thermo Fisher Scientific (R70007).
Lentivirus CRISPR/Cas9 experiments
gRNA were designed using the R package CRISPRseek (Zhu et al., 2014). The highest-ranking gRNAs were chosen. The gRNA oligonucleotides were purchased from Sigma-Aldrich and phosphorylated and annealed according to previously published protocols (Chen et al., 2017). The duplex oligonucleotides were subsequently cloned into the lentiCRISPR_V2 plasmid (Addgene Plasmid #52961) or ECPV vector (Senturk et al., 2017), replacing the 2-kb filler.
Virus for mouse injections
The Luc.Cre lentiviral plasmid for mice injections was purchased from Tyler Jacks (Koch Institute for Integrative Cancer Research, MIT; Addgene plasmid #20905). The lentivirus was packaged and purchased from the University of Iowa’s Viral Vector Core.
Intraprostate injection
Prior to surgery, the lower half of the mouse abdomen was shaved. The mouse was given 2% isoflurane for the duration of the 15-min procedure. Once deep anesthesia was determined, the abdomen was cleaned with betadine and ethanol twice before the incision was made. A 1.5-cm incision was made through the skin and the peritoneum to the right of the center abdomen. The seminal vesicle and the anterior prostate were removed and injected with 20 μl of lentivirus using a 30-gauge insulin syringe. The peritoneum incision was sutured with dissolvable sutures, and the skin incision was closed with steel EZ clips. The mouse was laid on a warmed cage, allowed to fully recover, and given DietGel. Lidocaine and buprenorphine were provided for pain management.
Bioluminescence imaging and tissue histology
For all bioluminescent imaging, a Xenogen IVIS Spectrum was used. For live imaging, animals were injected intraperitoneally with 200 mg/kg of luciferin (D-luciferin, potassium salt; Gold Biotechnology). 3 min after injection, animals were put under 2% isoflurane. At 10 min after luciferin administration, the animals were transferred to the warmed camera stage and placed in a nose cone with continuous 2% isoflurane. Animals were scanned for 3 min and then removed from the camera and recovered in cages. For postmortem imaging, animals were humanely sacrificed using CO2. The body of the animal was opened up and luciferin was dropped throughout the body and scanned for 3 min in the IVIS Spectrum. After all organs of interest were also removed, luciferin was applied to the organs and scanned for 3 min. For histology analysis, tissue was dissected from the animal and placed in 10% buffered formalin. Tissues were fixed for 24 h, followed by gentle wash and transfer to PBS. After paraffin embedding the tissue was total sectioned at 5-µm thickness and placed on charged glass slides. Every 10th section was stained with hematoxylin and eosin to locate tumor cells. Histology imaging was done at RT on an Axioplan microscope (Zeiss) using a Zeiss 40× A-plan objective (Air; NA = 0.65) and recorded on a Spot Flex 15.2, 64-Mp Shifting Pixel camera and the Spot software version 5.2.
Data analysis
Graph analyses were performed using the R programming language and ggplot2 package on Apple Macintosh computers. Values are expressed as the means with SDs unless otherwise stated. To compare two groups, Student’s t test was used, and P < 0.05 was considered statistically significant. For multiple comparisons, statistical analyses were performed on raw data using the one-way ANOVA test (Dunnett’s post-test) or the one-way ANOVA, with P values corrected for multicomparison testing using the two-stage linear step-up procedure of Benjamini, Krieger, and Yekutieli, with a false discovery rate at 10% using GraphPad Prism 7. P < 0.05 was considered statistically significant. IC50 values were derived from curves fitted using a nonlinear regression model using GraphPad Prism 7. See Table S1 for a list of key resources.
Online supplemental material
Fig. S1 shows the mechanism of interaction between Akt and Myc. Fig. S2 shows how Phlpp2 status affects Myc stability. Fig. S3 presents an in vitro analysis of Phlpp2 activity and stability. Fig. S4 shows modeling of Phlpp2 deletion in RapidCaP. Fig. S5 shows that hemizygous, not complete loss of PHLPP2, is common in advanced PC patients. Table S1 shows a list of key resources.
Supplementary Material
Acknowledgments
The authors thank Drs. Kuhulika Bhalla, Danielle Ramazzotti, and Serif Senturk and members of the Trotman laboratory for valuable discussions; the Cold Spring Harbor Laboratory Animal Resources team, Lisa Bianco, Jodi Coblentz, and Michael Cahn for help with animal work; Pamela Moody for cell sorting and FACS advice; and Dorothy Tsang for help with the article. The authors are also very thankful for the generous donations from the local foundation Glen Cove Cares.
L.C. Trotman is a Research Scholar of the American Cancer Society and is supported by the Pershing Square Sohn Foundation. This work was directly supported by grants to L.C. Trotman from the National Institutes of Health (R01 CA137050), the U.S. Department of Defense (DoD W81XWH-14-1-0247), the Pershing Square Sohn Cancer Research Alliance, and the American Cancer Society (RSG-14-069-01-TBE); by grants from the National Institutes of Health to A.C. Newton (R35 GM122523) and K.C. Katsenelson (T32 CA009523); and by grants to D.G. Nowak from the National Institutes of Health (Career Enhancement Program, Weill Cornell Medicine SPORE in Prostate Cancer; P50 CA211024). This research was furthermore supported by the Robertson Research Fund of Cold Spring Harbor Laboratory and by the Cold Spring Harbor Laboratory Cancer Center Support Grant from the National Institutes of Health (5P30CA045508) to support the flow cytometry, sequencing, gene targeting, animal husbandry, and histology analysis of this project.
The authors declare no competing financial interests.
Author contributions: D.G. Nowak, K.C. Katsenelson, K. Watrud, M. Chen, G. Mathew, V.D. D’Andrea, M.F. Lee, M.M. Swamynathan, I. Casanova-Salas, M.C. Jibilian, C.L. Buckholtz, A.J. Ambrico, and C.-H. Pan designed and performed experiments and interpreted the data; J.E. Wilkinson designed animal-related experiments and interpreted data; A.C. Newton and L.C. Trotman guided and provided funding for the research, designed experiments, and interpreted data; and D.G. Nowak, K.C. Katsenelson, A.C. Newton, and L.C. Trotman wrote the manuscript.
References
- Armenia J., Wankowicz S.A.M., Liu D., Gao J., Kundra R., Reznik E., Chatila W.K., Chakravarty D., Han G.C., Coleman I., et al. PCF/SU2C International Prostate Cancer Dream Team . 2018. The long tail of oncogenic drivers in prostate cancer. Nat. Genet. 50:645–651. 10.1038/s41588-018-0078-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri C.E., Baca S.C., Lawrence M.S., Demichelis F., Blattner M., Theurillat J.P., White T.A., Stojanov P., Van Allen E., Stransky N., et al. . 2012. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat. Genet. 44:685–689. 10.1038/ng.2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bluemn E.G., Coleman I.M., Lucas J.M., Coleman R.T., Hernandez-Lopez S., Tharakan R., Bianchi-Frias D., Dumpit R.F., Kaipainen A., Corella A.N., et al. . 2017. Androgen receptor pathway-independent prostate cancer is sustained through FGF signaling. Cancer Cell. 32:474–489.e6. 10.1016/j.ccell.2017.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brognard J., and Newton A.C.. 2008. PHLiPPing the switch on Akt and protein kinase C signaling. Trends Endocrinol. Metab. 19:223–230. 10.1016/j.tem.2008.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brognard J., Sierecki E., Gao T., and Newton A.C.. 2007. PHLPP and a second isoform, PHLPP2, differentially attenuate the amplitude of Akt signaling by regulating distinct Akt isoforms. Mol. Cell. 25:917–931. 10.1016/j.molcel.2007.02.017 [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network 2015. The molecular taxonomy of primary prostate cancer. Cell. 163:1011–1025. 10.1016/j.cell.2015.10.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D.W., Claassen G.F., Hann S.R., and Cole M.D.. 2000. The c-Myc transactivation domain is a direct modulator of apoptotic versus proliferative signals. Mol. Cell. Biol. 20:4309–4319. 10.1128/MCB.20.12.4309-4319.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Pratt C.P., Zeeman M.E., Schultz N., Taylor B.S., O’Neill A., Castillo-Martin M., Nowak D.G., Naguib A., Grace D.M., et al. . 2011. Identification of PHLPP1 as a tumor suppressor reveals the role of feedback activation in PTEN-mutant prostate cancer progression. Cancer Cell. 20:173–186. 10.1016/j.ccr.2011.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Nowak D.G., Narula N., Robinson B., Watrud K., Ambrico A., Herzka T.M., Zeeman M.E., Minderer M., Zheng W., et al. . 2017. The nuclear transport receptor Importin-11 is a tumor suppressor that maintains PTEN protein. J. Cell Biol. 216:641–656. 10.1083/jcb.201604025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.N., LaMarche M.J., Chan H.M., Fekkes P., Garcia-Fortanet J., Acker M.G., Antonakos B., Chen C.H., Chen Z., Cooke V.G., et al. . 2016. Allosteric inhibition of SHP2 phosphatase inhibits cancers driven by receptor tyrosine kinases. Nature. 535:148–152. 10.1038/nature18621 [DOI] [PubMed] [Google Scholar]
- Chen Z., Trotman L.C., Shaffer D., Lin H.K., Dotan Z.A., Niki M., Koutcher J.A., Scher H.I., Ludwig T., Gerald W., et al. . 2005. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 436:725–730. 10.1038/nature03918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H., Herzka T., Zheng W., Qi J., Wilkinson J.E., Bradner J.E., Robinson B.D., Castillo-Martin M., Cordon-Cardo C., and Trotman L.C.. 2014. RapidCaP, a novel GEM model for metastatic prostate cancer analysis and therapy, reveals myc as a driver of Pten-mutant metastasis. Cancer Discov. 4:318–333. 10.1158/2159-8290.CD-13-0346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H., Herzka T., Stahlhut C., Watrud K., Robinson B.D., and Trotman L.C.. 2015. Rapid in vivo validation of candidate drivers derived from the PTEN-mutant prostate metastasis genome. Methods. 77-78:197–204. 10.1016/j.ymeth.2014.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmore J.E., Issa G.C., Lemieux M.E., Rahl P.B., Shi J., Jacobs H.M., Kastritis E., Gilpatrick T., Paranal R.M., Qi J., et al. . 2011. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 146:904–917. 10.1016/j.cell.2011.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Cristofano A., De Acetis M., Koff A., Cordon-Cardo C., and Pandolfi P.P.. 2001. Pten and p27KIP1 cooperate in prostate cancer tumor suppression in the mouse. Nat. Genet. 27:222–224. 10.1038/84879 [DOI] [PubMed] [Google Scholar]
- Ding Z., Wu C.J., Chu G.C., Xiao Y., Ho D., Zhang J., Perry S.R., Labrot E.S., Wu X., Lis R., et al. . 2011. SMAD4-dependent barrier constrains prostate cancer growth and metastatic progression. Nature. 470:269–273. 10.1038/nature09677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drost J., Karthaus W.R., Gao D., Driehuis E., Sawyers C.L., Chen Y., and Clevers H.. 2016. Organoid culture systems for prostate epithelial and cancer tissue. Nat. Protoc. 11:347–358. 10.1038/nprot.2016.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellwood-Yen K., Graeber T.G., Wongvipat J., Iruela-Arispe M.L., Zhang J., Matusik R., Thomas G.V., and Sawyers C.L.. 2003. Myc-driven murine prostate cancer shares molecular features with human prostate tumors. Cancer Cell. 4:223–238. 10.1016/S1535-6108(03)00197-1 [DOI] [PubMed] [Google Scholar]
- Farrell A.S., and Sears R.C.. 2014. MYC degradation. Cold Spring Harb. Perspect. Med. 4:a014365 10.1101/cshperspect.a014365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Salas E., Wang S., and Chinnaiyan A.M.. 2016. Role of BET proteins in castration-resistant prostate cancer. Drug Discov. Today. Technol. 19:29–38. 10.1016/j.ddtec.2016.07.001 [DOI] [PubMed] [Google Scholar]
- Gao T., Brognard J., and Newton A.C.. 2008. The phosphatase PHLPP controls the cellular levels of protein kinase C. J. Biol. Chem. 283:6300–6311. 10.1074/jbc.M707319200 [DOI] [PubMed] [Google Scholar]
- Gil J., Kerai P., Lleonart M., Bernard D., Cigudosa J.C., Peters G., Carnero A., and Beach D.. 2005. Immortalization of primary human prostate epithelial cells by c-Myc. Cancer Res. 65:2179–2185. 10.1158/0008-5472.CAN-03-4030 [DOI] [PubMed] [Google Scholar]
- Grasso C.S., Wu Y.M., Robinson D.R., Cao X., Dhanasekaran S.M., Khan A.P., Quist M.J., Jing X., Lonigro R.J., Brenner J.C., et al. . 2012. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 487:239–243. 10.1038/nature11125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y., Zhang J., Ma X., Kim B.W., Wang H., Li J., Pan Y., Xu Y., Ding L., Yang L., et al. . 2017. Stabilization of the c-Myc protein by CAMKIIγ promotes T cell lymphoma. Cancer Cell. 32:115–128.e7. 10.1016/j.ccell.2017.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemann M.T., Bric A., Teruya-Feldstein J., Herbst A., Nilsson J.A., Cordon-Cardo C., Cleveland J.L., Tansey W.P., and Lowe S.W.. 2005. Evasion of the p53 tumour surveillance network by tumour-derived MYC mutants. Nature. 436:807–811. 10.1038/nature03845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hydbring P., Bahram F., Su Y., Tronnersjö S., Högstrand K., von der Lehr N., Sharifi H.R., Lilischkis R., Hein N., Wu S., et al. . 2010. Phosphorylation by Cdk2 is required for Myc to repress Ras-induced senescence in cotransformation. Proc. Natl. Acad. Sci. USA. 107:58–63. 10.1073/pnas.0900121106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irshad S., and Abate-Shen C.. 2013. Modeling prostate cancer in mice: something old, something new, something premalignant, something metastatic. Cancer Metastasis Rev. 32:109–122. 10.1007/s10555-012-9409-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janghorban M., Farrell A.S., Allen-Petersen B.L., Pelz C., Daniel C.J., Oddo J., Langer E.M., Christensen D.J., and Sears R.C.. 2014. Targeting c-MYC by antagonizing PP2A inhibitors in breast cancer. Proc. Natl. Acad. Sci. USA. 111:9157–9162. 10.1073/pnas.1317630111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur A., and Westermarck J.. 2016. Regulation of protein phosphatase 2A (PP2A) tumor suppressor function by PME-1. Biochem. Soc. Trans. 44:1683–1693. 10.1042/BST20160161 [DOI] [PubMed] [Google Scholar]
- Krishnan N., Koveal D., Miller D.H., Xue B., Akshinthala S.D., Kragelj J., Jensen M.R., Gauss C.M., Page R., Blackledge M., et al. . 2014. Targeting the disordered C terminus of PTP1B with an allosteric inhibitor. Nat. Chem. Biol. 10:558–566. 10.1038/nchembio.1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku S.Y., Rosario S., Wang Y., Mu P., Seshadri M., Goodrich Z.W., Goodrich M.M., Labbé D.P., Gomez E.C., Wang J., et al. . 2017. Rb1 and Trp53 cooperate to suppress prostate cancer lineage plasticity, metastasis, and antiandrogen resistance. Science. 355:78–83. 10.1126/science.aah4199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwabi-Addo B., Giri D., Schmidt K., Podsypanina K., Parsons R., Greenberg N., and Ittmann M.. 2001. Haploinsufficiency of the Pten tumor suppressor gene promotes prostate cancer progression. Proc. Natl. Acad. Sci. USA. 98:11563–11568. 10.1073/pnas.201167798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M.S., Stojanov P., Polak P., Kryukov G.V., Cibulskis K., Sivachenko A., Carter S.L., Stewart C., Mermel C.H., Roberts S.A., et al. . 2013. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 499:214–218. 10.1038/nature12213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Wang P., Chen H., Wold E.A., Tian B., Brasier A.R., and Zhou J.. 2017. Drug discovery targeting bromodomain-containing protein 4. J. Med. Chem. 60:4533–4558. 10.1021/acs.jmedchem.6b01761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumder P.K., and Sellers W.R.. 2005. Akt-regulated pathways in prostate cancer. Oncogene. 24:7465–7474. 10.1038/sj.onc.1209096 [DOI] [PubMed] [Google Scholar]
- Mateo J., Carreira S., Sandhu S., Miranda S., Mossop H., Perez-Lopez R., Nava Rodrigues D., Robinson D., Omlin A., Tunariu N., et al. . 2015. DNA-repair defects and Olaparib in metastatic prostate cancer. N. Engl. J. Med. 373:1697–1708. 10.1056/NEJMoa1506859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateo J., Boysen G., Barbieri C.E., Bryant H.E., Castro E., Nelson P.S., Olmos D., Pritchard C.C., Rubin M.A., and de Bono J.S.. 2017. DNA Repair in Prostate Cancer: Biology and Clinical Implications. Eur. Urol. 71:417–425. 10.1016/j.eururo.2016.08.037 [DOI] [PubMed] [Google Scholar]
- McKeown M.R., and Bradner J.E.. 2014. Therapeutic strategies to inhibit MYC. Cold Spring Harb. Perspect. Med. 4:a014266 10.1101/cshperspect.a014266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naguib A., Mathew G., Reczek C.R., Watrud K., Ambrico A., Herzka T., Salas I.C., Lee M.F., El-Amine N., Zheng W., et al. . 2018. Mitochondrial complex I inhibitors expose a vulnerability for selective killing of Pten-null cells. Cell Reports. 23:58–67. 10.1016/j.celrep.2018.03.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Cancer Institute 2016. Cancer Stat Facts. Surveillance, Epidemiology, and End Results Program (SEER). https://seer.cancer.gov/statfacts/html/prost.html
- Nijhawan D., Zack T.I., Ren Y., Strickland M.R., Lamothe R., Schumacher S.E., Tsherniak A., Besche H.C., Rosenbluh J., Shehata S., et al. . 2012. Cancer vulnerabilities unveiled by genomic loss. Cell. 150:842–854. 10.1016/j.cell.2012.07.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak D.G., Cho H., Herzka T., Watrud K., DeMarco D.V., Wang V.M., Senturk S., Fellmann C., Ding D., Beinortas T., et al. . 2015. MYC drives Pten/Trp53-deficient proliferation and metastasis due to IL6 secretion and AKT suppression via PHLPP2. Cancer Discov. 5:636–651. 10.1158/2159-8290.CD-14-1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podsypanina K., Ellenson L.H., Nemes A., Gu J., Tamura M., Yamada K.M., Cordon-Cardo C., Catoretti G., Fisher P.E., and Parsons R.. 1999. Mutation of Pten/Mmac1 in mice causes neoplasia in multiple organ systems. Proc. Natl. Acad. Sci. USA. 96:1563–1568. 10.1073/pnas.96.4.1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popov N., Wanzel M., Madiredjo M., Zhang D., Beijersbergen R., Bernards R., Moll R., Elledge S.J., and Eilers M.. 2007. The ubiquitin-specific protease USP28 is required for MYC stability. Nat. Cell Biol. 9:765–774. 10.1038/ncb1601 [DOI] [PubMed] [Google Scholar]
- Puissant A., Frumm S.M., Alexe G., Bassil C.F., Qi J., Chanthery Y.H., Nekritz E.A., Zeid R., Gustafson W.C., Greninger P., et al. . 2013. Targeting MYCN in neuroblastoma by BET bromodomain inhibition. Cancer Discov. 3:308–323. 10.1158/2159-8290.CD-12-0418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson D., Van Allen E.M., Wu Y.M., Schultz N., Lonigro R.J., Mosquera J.M., Montgomery B., Taplin M.E., Pritchard C.C., Attard G., et al. . 2015. Integrative clinical genomics of advanced prostate cancer. Cell. 161:1215–1228. 10.1016/j.cell.2015.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangodkar J., Farrington C.C., McClinch K., Galsky M.D., Kastrinsky D.B., and Narla G.. 2016. All roads lead to PP2A: exploiting the therapeutic potential of this phosphatase. FEBS J. 283:1004–1024. 10.1111/febs.13573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears R.C. 2004. The life cycle of C-myc: from synthesis to degradation. Cell Cycle. 3:1131–1135. 10.4161/cc.3.9.1145 [DOI] [PubMed] [Google Scholar]
- Sears R., Nuckolls F., Haura E., Taya Y., Tamai K., and Nevins J.R.. 2000. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev. 14:2501–2514. 10.1101/gad.836800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senturk S., Shirole N.H., Nowak D.G., Corbo V., Pal D., Vaughan A., Tuveson D.A., Trotman L.C., Kinney J.B., and Sordella R.. 2017. Rapid and tunable method to temporally control gene editing based on conditional Cas9 stabilization. Nat. Commun. 8:14370 10.1038/ncomms14370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimamura T., Chen Z., Soucheray M., Carretero J., Kikuchi E., Tchaicha J.H., Gao Y., Cheng K.A., Cohoon T.J., Qi J., et al. . 2013. Efficacy of BET bromodomain inhibition in Kras-mutant non-small cell lung cancer. Clin. Cancer Res. 19:6183–6192. 10.1158/1078-0432.CCR-12-3904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierecki E., Sinko W., McCammon J.A., and Newton A.C.. 2010. Discovery of small molecule inhibitors of the PH domain leucine-rich repeat protein phosphatase (PHLPP) by chemical and virtual screening. J. Med. Chem. 53:6899–6911. 10.1021/jm100331d [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor B.S., Schultz N., Hieronymus H., Gopalan A., Xiao Y., Carver B.S., Arora V.K., Kaushik P., Cerami E., Reva B., et al. . 2010. Integrative genomic profiling of human prostate cancer. Cancer Cell. 18:11–22. 10.1016/j.ccr.2010.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotman L.C., Niki M., Dotan Z.A., Koutcher J.A., Di Cristofano A., Xiao A., Khoo A.S., Roy-Burman P., Greenberg N.M., Van Dyke T., et al. . 2003. Pten dose dictates cancer progression in the prostate. PLoS Biol. 1:E59 10.1371/journal.pbio.0000059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotman L.C., Alimonti A., Scaglioni P.P., Koutcher J.A., Cordon-Cardo C., and Pandolfi P.P.. 2006. Identification of a tumour suppressor network opposing nuclear Akt function. Nature. 441:523–527. 10.1038/nature04809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson P.A., Ellwood-Yen K., King J.C., Wongvipat J., Lebeau M.M., and Sawyers C.L.. 2005. Context-dependent hormone-refractory progression revealed through characterization of a novel murine prostate cancer cell line. Cancer Res. 65:11565–11571. 10.1158/0008-5472.CAN-05-3441 [DOI] [PubMed] [Google Scholar]
- Welcker M., Orian A., Jin J., Grim J.E., Harper J.W., Eisenman R.N., and Clurman B.E.. 2004. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc. Natl. Acad. Sci. USA. 101:9085–9090. 10.1073/pnas.0402770101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Y.A., Li X., Goretsky T., Weiss H.L., Barrett T.A., and Gao T.. 2015. Loss of PHLPP protects against colitis by inhibiting intestinal epithelial cell apoptosis. Biochim. Biophys. Acta. 1852:2013–2023. 10.1016/j.bbadis.2015.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng A.P., Millholland J.M., Yashiro-Ohtani Y., Arcangeli M.L., Lau A., Wai C., Del Bianco C., Rodriguez C.G., Sai H., Tobias J., et al. . 2006. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 20:2096–2109. 10.1101/gad.1450406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E., Cunningham M., Arnold H., Chasse D., Monteith T., Ivaldi G., Hahn W.C., Stukenberg P.T., Shenolikar S., Uchida T., et al. . 2004. A signalling pathway controlling c-Myc degradation that impacts oncogenic transformation of human cells. Nat. Cell Biol. 6:308–318. 10.1038/ncb1110 [DOI] [PubMed] [Google Scholar]
- Zhu L.J., Holmes B.R., Aronin N., and Brodsky M.H.. 2014. CRISPRseek: a bioconductor package to identify target-specific guide RNAs for CRISPR-Cas9 genome-editing systems. PLoS One. 9:e108424 10.1371/journal.pone.0108424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou M., Toivanen R., Mitrofanova A., Floch N., Hayati S., Sun Y., Le Magnen C., Chester D., Mostaghel E.A., Califano A., et al. . 2017. Transdifferentiation as a mechanism of treatment resistance in a mouse model of castration-resistant prostate cancer. Cancer Discov. 7:736–749. 10.1158/2159-8290.CD-16-1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber J., Shi J., Wang E., Rappaport A.R., Herrmann H., Sison E.A., Magoon D., Qi J., Blatt K., Wunderlich M., et al. . 2011. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 478:524–528. 10.1038/nature10334 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.