Feedback loops arising from crosstalk between early endocytic trafficking and receptor signaling can be co-opted or amplified in cancer cells to enhance their metastatic abilities. Lakoduk et al. reveal that mutant p53 upregulates dynamin-1 expression and recruitment of the APPL1 signaling scaffold to a spatially localized subpopulation of endosomes to increase receptor recycling and cell migration.
Abstract
Multiple mechanisms contribute to cancer cell progression and metastatic activity, including changes in endocytic trafficking and signaling of cell surface receptors downstream of gain-of-function (GOF) mutant p53. We report that dynamin-1 (Dyn1) is up-regulated at both the mRNA and protein levels in a manner dependent on expression of GOF mutant p53. Dyn1 is required for the recruitment and accumulation of the signaling scaffold, APPL1, to a spatially localized subpopulation of endosomes at the cell perimeter. We developed new tools to quantify peripherally localized early endosomes and measure the rapid recycling of integrins. We report that these perimeter APPL1 endosomes modulate Akt signaling and activate Dyn1 to create a positive feedback loop required for rapid recycling of EGFR and β1 integrins, increased focal adhesion turnover, and cell migration. Thus, Dyn1- and Akt-dependent perimeter APPL1 endosomes function as a nexus that integrates signaling and receptor trafficking, which can be co-opted and amplified in mutant p53–driven cancer cells to increase migration and invasion.
Introduction
Cancer cell invasion and metastasis are mechanistically heterogeneous and adaptive processes contributing to the majority of cancer-related deaths (Bacac and Stamenkovic, 2008; Friedl and Alexander, 2011). While driver gene mutations and epigenetic alterations can increase cancer cell proliferation, survival, invasion, and migration, they cannot account for all of the metastatic traits acquired through evolution of aggressive cancer cells (Schmid, 2017). The underlying mechanisms governing the transition from primary to aggressive tumors, during which cancer cells acquire their adaptive metastatic abilities, are heterogeneous and remain largely unknown. Understanding the possible mechanisms leading to cancer metastasis is crucial for successful cancer treatment.
Signaling receptors, including receptor tyrosine kinases and integrins, control many aspects of cell physiology and behavior and are frequently dysregulated in and associated with the initiation and progression of cancer (Sever and Brugge, 2015). Their signaling activities are, in turn, modulated by endocytic trafficking (Mellman and Yarden, 2013). Indeed, expression of gain-of-function (GOF) p53 mutants, which contribute to a more invasive phenotype in multiple cancers (Lang et al., 2004; Olive et al., 2004), results in increased recycling of EGF receptor (EGFR), cMET, and β1 integrins (Muller et al., 2009, 2013; Lanzetti and Di Fiore, 2017). This leads to increased invasion and migration. The mechanisms responsible for GOF mutant p53–dependent changes in endocytic trafficking remain incompletely understood.
Endocytic trafficking of signaling receptors, which are internalized primarily via clathrin-mediated endocytosis (CME), involves delivery through distinct early endosomal compartments marked by the scaffold proteins APPL1 (adaptor protein, phosphotyrosine interacting with PH domain and leucine zipper 1) or EEA1 (early endosome antigen 1; Zoncu et al., 2009; Kalaidzidis et al., 2015). Receptors can be recycled back to the cell surface along either fast (i.e., directly from early endosomes) or slow (i.e., via perinuclear recycling endosomes) pathways. Alternatively, receptors can be packaged in intralumenal vesicles and delivered to lysosomes for degradation (Kalaidzidis et al., 2015). Difficulties in quantitatively measuring fast recycling render this the least mechanistically understood of these trafficking pathways.
GOF p53-dependent increases in receptor recycling require the Rab11 effector, Rab-coupling protein (RCP; Muller et al., 2009, 2013). However, RCP expression levels are not regulated by p53; thus the mechanisms by which mutant p53 regulates components of the endocytic machinery to alter endocytic trafficking remain unknown. Also unknown are the identities of the endosomal compartments from which this recycling occurs, although Rab11 is associated with recycling endosomes and the slow recycling pathway (Wandinger-Ness and Zerial, 2014).
The temporal and functional relationships between the early APPL1 and EEA1 endosomes also remain incompletely defined (Zoncu et al., 2009; Kalaidzidis et al., 2015). One study suggested that APPL1 endosomes are intermediates along a maturation pathway from nascent endocytic vesicles to EEA1-positive early endosomes (Zoncu et al., 2009), while a second study suggested that they function as distinct, albeit dynamically interacting, sorting stations (Kalaidzidis et al., 2015). APPL1-positive endosomes are often referred to as “signaling endosomes” because APPL1, through its scaffolding properties, regulates many signaling events including Akt/GSK3β activity (Schenck et al., 2008; Ding et al., 2016; Diggins and Webb, 2017). In addition, APPL1 endosomes have been linked to the regulation of cell migration (Tan et al., 2010; Broussard et al., 2012; Ding et al., 2016) and to recycling of some G protein–coupled receptors (GPCRs; Jean-Alphonse et al., 2014; Sposini et al., 2017). APPL1 endosomes have been reported to be regulated by PKA signaling downstream of GPCRs (Sposini et al., 2017) and by CME itself (Zoncu et al., 2009, but see Kalaidzidis et al., 2015). Thus, while still poorly defined, APPL1 endosomes are emerging as important integrators of signaling and endocytic trafficking.
The large GTPase dynamin plays an important role in endocytosis. Vertebrates encode three differentially expressed isoforms: of these, dynamin 2 (Dyn2) is uniformly expressed, Dyn1 is highly enriched in neurons, and Dyn3 is primarily detected in neurons, testes, and lung. We recently reported that neuronally enriched and typically quiescent Dyn1 is specifically up-regulated and/or activated in many non–small cell lung cancer (NSCLC) cell lines (Reis et al., 2017; Schmid, 2017). Indeed, Dyn1 has emerged as a nexus in regulating signaling and endocytic trafficking in cancer cells (Reis et al., 2015; Chen et al., 2017; Srinivasan et al., 2018). Its activation leads to altered EGFR trafficking, increased Akt signaling, and the accumulation of peripheral APPL1-positive endosomes (Chen et al., 2017). Together, these effects are associated with increased metastatic activity of H1299 NSCLC cells in a mouse model for lung metastasis (Chen et al., 2017). Whether this pathway is also linked to GOF p53-dependent changes in endocytic trafficking is unknown.
We have developed quantitative approaches to study rapid recycling from early endosomes and APPL1 endosome regulation. Applying these tools to study mutant p53–driven alterations in endocytic membrane trafficking and cell migration, we find that both Dyn1 and myosin VI (Myo6) are up-regulated in cells expressing mutant p53 and are required for enhanced recruitment of APPL1 to a spatially restricted subpopulation of endosomes that is also dependent on EGFR and Akt signaling. This EGFR/Akt/Dyn1–dependent subpopulation of APPL1-positive endosomes functions as a hub to modulate signaling and fast recycling of β1 integrins and enhance the migratory and invasive phenotypes in cancer cells. This study reveals mechanisms that govern complex reciprocal regulation between endocytic trafficking and receptor signaling, which can be co-opted and/or amplified in cancer cells to acquire properties that could enhance their metastatic activities during cancer progression.
Results
EGFR/Akt signaling regulates a subpopulation of peripheral APPL1 endosomes in p53-driven cancer cells
The underlying mechanisms and pathways by which the p53 GOF mutant, R273H, drives alterations in the recycling and signaling of surface receptors (Muller et al., 2009) remain incompletely defined. Given the recently discovered link between EGFR/Akt signaling, Dyn1 activation, and the accumulation of APPL1 endosomes (Reis et al., 2015; Chen et al., 2017), we tested whether these factors might also be required for mutant p53–driven changes in early endocytic trafficking and cancer invasion.
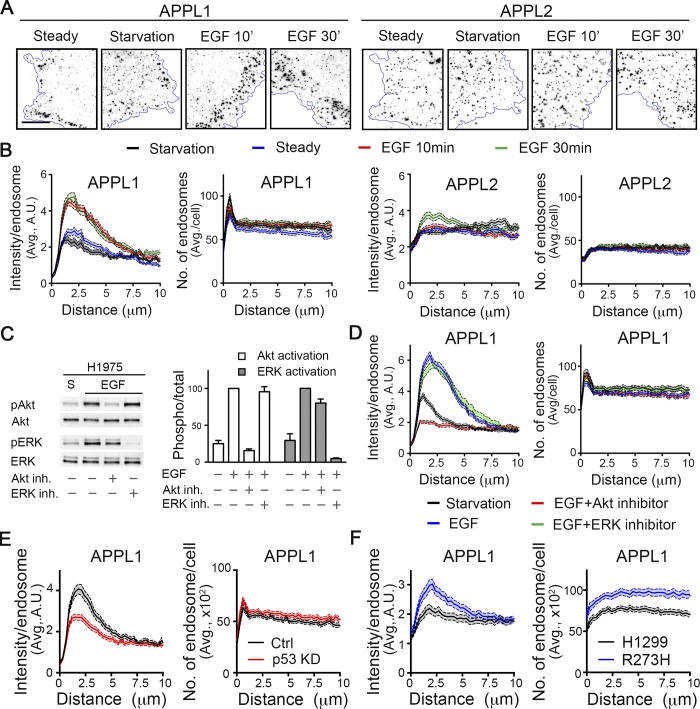
H1975 NSCLC cells, which carry a homozygous R273H GOF p53 mutation, were serum starved and then stimulated with 20 ng/ml of EGF. The distribution of APPL1-positive endosomes was then determined by immunofluorescence. APPL2, its closely related isoform, and EEA1 (Fig. S1, A and B) were also examined to determine the specificity of these effects (Fig. 1 A). To reduce noise from out-of-focus planes and to limit our analyses to peripherally localized early endosomes, total internal reflection fluorescence (TIRF) microscopy was used after adjusting the angle of illumination to generate a theoretical penetration depth of ∼200 nm (“thick TIRF”).
Figure 1.
EGFR/Akt signaling regulates perimeter APPL1 endosomes in p53-driven cancer cells. (A) Representative thick-TIRF immunofluorescence images of APPL1 or APPL2 endosomes in H1975 cells at steady state or after starvation, and stimulation with EGF (20 ng/ml) for 10 or 30 min as indicated. Scale bar, 10 µm. (B) Accompanying quantification of fluorescence intensity per endosome (left) and number (right) of APPL1- or APPL2-positive endosomes relative to their distance from the cell edge. Errors bars along each curve represent 95% confidence intervals. (C) Representative Western blot and accompanying quantification (n = 3) of phosphorylated Akt (Ser 473) and phospho-ERK in starved or EGF-stimulated treated with either Akt or ERK inhibitors. (D) Quantification of intensity/endosome or number of APPL1-positive endosomes in starved or EGF-treated H1975 cells, with or without 30-min pretreatment with 10 µM of either Akt (Akt Inhibitor X) or ERK (SCH772984) inhibitors. (E and F) Quantification of intensity and number of APPL1-positive endosomes in control and p53 siRNA-treated H1975 cells (E) or parental and p53 R273H-expressing H1299 cells (F).
To accurately quantify the spatial distribution of APPL endosomes, we adapted our cmeAnalysis platform (Aguet et al., 2013) for high-throughput endosome detection and implemented distance analysis to assess endosome distribution relative to the cell edge (see Materials and methods). Using these tools, we measured both the average intensity of individually detected endosomes and the total number of detected endosomes relative to the distance they reside from the cell edge (Fig. 1 B).
Starvation triggered a significant reduction in the fluorescence intensity of the subpopulation of APPL1 endosomes localized within ∼2.5 µm from the cell edge (hereafter referred to as “perimeter endosomes”), without a corresponding change in the number of these peripherally localized structures (Fig. 1, A and B). In contrast, the intensities of APPL2 endosomes did not change, and they remained uniformly distributed throughout the cell periphery (Fig. 1, A and B). Stimulation with EGF, at either 20 ng/ml (Figs. 1, A and B) or 1 ng/ml (Fig. S1 A), resulted in a rapid (within 10 min) and dramatic accumulation of perimeter-localized, high-intensity APPL1 endosomes. The EGF-stimulated increase in perimeter APPL2 endosomes occurred at later time points (30 min) and to a much lesser extent (Fig. 1 B), indicating a specificity for the APPL1 isoform. Additionally, the distribution of EEA1-positive endosomes was unaffected (Fig. S1 A), and EEA1-positive endosomes were spatially segregated from APPL1-positive endosomes (Fig. S1 B) under all conditions. Interestingly, in isolated migrating cells (Fig. S1 C) or within cell clusters (Fig. S1 D), the EGF-stimulated APPL1-positive perimeter endosomes appeared to accumulate toward the outside or leading edge of the cell.
The observed EGF-dependent increase in the intensity of perimeter APPL1 endosomes was not accompanied by an increase in their numbers (Fig. 1 B). This suggests that EGF treatment either increased the expression of APPL1 and/or its recruitment to and accumulation at the membrane. We did not detect any EGF-dependent increase in total APPL1 but were able to detect EGF-dependent increases in the recruitment of APPL1 and its interactor Akt to cellular membranes (Fig. S2). Thus, we conclude that EGFR signaling enhances the selective recruitment of APPL1 to a subpopulation of perimeter endosomes.
To identify which of the two major signaling pathways downstream of EGFR, phosphatidylinositol 3-kinase/Akt or Ras/MAPK/extracellular signal–regulated kinase (ERK), was required for the selective accumulation of APPL1 on perimeter endosomes, cells were treated with either Akt inhibitor X or the ERK1/2 inhibitor SCH772984. Control experiments established that both compounds selectively inhibited their respective target kinases (Fig. 1 C). The EGF-stimulated accumulation of perimeter APPL1 endosomes selectively required Akt but not ERK1/2 activity (Fig. 1 D). Interestingly, even the residual subpopulation of perimeter APPL1 endosomes in starved cells were sensitive to Akt inhibition (Fig. 1 D), suggesting a direct link between Akt signaling and APPL1 recruitment. Taken together, our data establish that the accumulation of APPL1 on perimeter endosomes is dependent on EGFR and/or Akt signaling.
Given that Akt is activated by a GOF mutant p53 downstream of EGFR (Muller et al., 2009), we examined the relationship between mutant p53 expression and perimeter APPL1 endosomes. Knockdown of the R273H mutant p53 expressed in H1975 cells resulted in a decrease in the intensity of perimeter APPL1 endosomes (Fig. 1 E). Correspondingly, ectopic overexpression of R273H mutant p53 in a p53-null NSCLC cell line, H1299, resulted in an increase in both the intensity and number of perimeter APPL1-positive endosomes (Fig. 1 F). These data link the increased APPL1 recruitment and accumulation on perimeter endosomes to mutant p53 expression.
Mutant p53 expression up-regulates Dyn1
Intriguingly, the reported effects of mutant p53 expression on EGFR recycling (Muller et al., 2009, 2013; Mellman and Yarden, 2013), and our finding that p53-expression increases the accumulation of APPL1 on endosomes (Fig. 1, E and F), mirror effects previously seen following activation of Dyn1 (Chen et al., 2017). These observations prompted us to investigate a possible role for Dyn1 in mediating mutant p53–driven accumulation of perimeter APPL1 endosomes.
Strikingly, ectopic expression of GOF mutant p53 R273H in p53-null H1299 cells increased both mRNA, as detected by RT-PCR relative to actin controls (see Materials and methods; Fig. 2 A) and protein (Fig. 2 B) levels of Dyn1. siRNA-mediated knockdown of p53 R273H in H1975 cells resulted in a corresponding decrease in both Dyn1 mRNA (Fig. 2 C) and protein (Fig. 2 D) levels. Thus, the expression of GOF R273H mutant p53 directly or, more likely, indirectly regulates Dyn1 expression. APPL1 protein levels were unaffected by mutant p53 expression (Fig. 2, B and D). Thus, as occurred upon EGF stimulation, the mutant p53–dependent increase in perimeter APPL1 endosomes likely reflects increased recruitment of APPL1 to nascent endosomal structures.
Figure 2.
Dyn1 is up-regulated downstream of mutant p53. (A) Quantification and representative RT-PCR of Dyn1 mRNA expression in parental and p53 R273H-expressing H1299 cells. (B) Quantification of Dyn1 expression and representative Western blots of the indicated proteins in parental and p53 R273H-expressing H1299 cells. (C) Quantification and representative RT-PCR of Dyn1 mRNA in parental and p53 KD H1975 cells. (D) Quantification of Dyn1 expression and representative Western blots of the indicated proteins in parental and p53 KD H1975 cells. All quantifications are from three or more independent experiments. Unpaired t tests were used to assess statistical significance; **, P < 0.01; ***, P < 0.001. (E) Box plot of The Cancer Genome Atlas RNA expression profiles of DNM1 and DNM2 in human NSCLC tumors with WT (n = 358) or any p53 mutation (i.e., deletions and/or mutations; n = 733). Mann–Whitney U test was performed to compute statistical significance; *, P < 0.05; ***, P < 0.001.
To connect these observations with human cancers, we mined NSCLC patient data in The Cancer Genome Atlas from Genome Data Commons Data Portal and examined the relationship of Dyn1 mRNA expression and p53 mutation status. Downloaded patient data were split into two groups: patients with WT p53 and those with any p53 mutations. The numbers were insufficient to further parse the data between GOF and p53-null genotypes. Nonetheless, and consistent with our observations, the expression of Dyn1 mRNA in tumors with p53 mutations was significantly higher than in those expressing WT p53 (Fig. 2 E). In contrast, tumors with p53 mutations exhibited a slight, but significant, reduction in the expression of Dyn2 mRNA relative to those expressing WT p53. The above data suggest that early endocytic trafficking is altered in GOF mutant p53–expressing cancer cells, in part through up-regulation of Dyn1 and an increased accumulation of APPL1 on perimeter endosomes.
Dyn1 isoform-specific regulation of perimeter APPL1 endosomes
There have been conflicting reports as to the role of CME in regulating the accumulation of APPL1 endosomes (Zoncu et al., 2009; Kalaidzidis et al., 2015). Given the differential regulation of CME by dynamin isoforms (Reis et al., 2015; Srinivasan et al., 2018), we next tested their roles in regulating the accumulation of APPL1 on perimeter endosomes. Despite that fact that siRNA-mediated knockdown of Dyn2 more potently inhibits CME in NSCLC cells than does Dyn1 knockdown (not depicted; but see Reis et al., 2015; Srinivasan et al., 2018), only depletion of Dyn1 abrogated the accumulation of perimeter APPL1 endosomes in response to EGF signaling. Neither Dyn2 nor Dyn3 loss had any significant effect (Fig. S3). As for EGFR stimulation, the effect of Dyn1 depletion was specific to APPL1-positive endosomes, as neither the distribution nor number of early EEA1-positive endosomes was affected (Fig. S4).
To determine whether the role of Dyn1 in the EGF-induced accumulation of perimeter APPL1 endosomes was more general and/or restricted to cells expressing mutant p53, we evaluated the influence of depletion of each dynamin isoform on APPL1 and EEA1 endosomes in various cell lines that express WT or mutant p53. These included two noncancerous cell lines, ARPE-19 (human retinal pigment epithelial) and HBEC-3KT (human bronchial epithelial) that both express WT p53, as well as a panel of cancerous cells, including NSCLC A549 cells that express WT p53; MDA-MB-231 breast cancer cells that express homozygous p53 R280K; A375 and MV3 melanoma cells that both express WT p53; and DU145 and PC3 prostate cancer cells that express heterozygous P223L and V274F p53 (DU145) and 138del p53 (PC3). The number of APPL1 and EEA1 endosomes did not significantly change in any dynamin knockdown condition or cell line (not depicted). However, siRNA-mediated knockdown of Dyn1, but not that of Dyn2 or Dyn3, selectively reduced the intensity of perimeter APPL1 endosomes in all cell lines tested (Fig. S3), without affecting the distribution of EEA1 endosomes (Fig. S4). Knockdown efficiencies for Dyn1 and Dyn2 are shown in Fig. S5. Dyn3 is expressed at low, undetectable levels in the cells we have studied (not depicted). In some cells, knockdown of Dyn2 and/or Dyn3 resulted in increased intensity of perimeter APPL1 endosomes (Fig. S3). Although not investigated further, this could reflect a compensatory up-regulation of Dyn1. Again, in all cases, EEA1-positive endosomes were unaffected (Fig. S4). Notably, overexpression of Dyn1-eGFP alone was insufficient to trigger the redistribution of APPL1 endosomes in p53-null H1299 cells, suggesting that other factors cooperate with Dyn1 to contribute to these effects (not depicted). Together, these data establish that the EGF-dependent accumulation of APPL1 perimeter endosomes selectively requires Dyn1 in both noncancerous and cancer cells. Thus, expression of GOF p53 mutations appears to amplify a Dyn1/APPL1 endosome nexus that normally functions in many cell types.
Given potential off-target effects of the siRNA experiments, we confirmed these findings using inducible CRISPR knockout technology (Senturk et al., 2017) in H1975 cells. The advantage of inducible CRISPR over constitutive CRISPR is that it does not require clonal selection and expansion of knockout cells, which are susceptible to the induction/selection of compensatory mechanisms and clonal variation, which is especially problematic with heterogeneous and genomically unstable cancer cell lines. We focused on Dyn1 and Dyn2, given the low levels of Dyn3 expression and the lack of effect in siRNA knockdown experiments (Fig. S3).
Consistent with the siRNA experiments, conditional knockout (cKO) of Dyn1, but not Dyn2, suppressed the accumulation of APPL1 on perimeter endosomes compared with control H1975 cells expressing DD-Cas9 (Cas9 fused to a destabilizing domain) without targeting sequences (Fig. 3). Together, these results reveal a specific and isoform-selective role for the neuronally enriched isoform, Dyn1, in regulating the accumulation of an EGF-dependent and/or mutant p53–driven spatially restricted subpopulation of APPL1 signaling endosomes at the cell edge.
Figure 3.
Dyn1 isoform-specific regulation of perimeter APPL1 endosomes. Representative immunofluorescence images and accompanying quantification of intensity or number of APPL1-positive endosomes in control (cells expressing DD-Cas9 without guide sequence), DD-Cas9-Dyn1 (Dyn1 cKO), or DD-Cas9-Dyn2 (Dyn2 cKO) H1975 cells after 4 d of Shield-1 treatment. The representative Western blot demonstrates Dyn1 and Dyn2 cKO efficiency. Scale bar, 10 µm.
The above studies also reinforce the distinct responses of APPL1- versus EEA1-positive endosomes to changes in expression of Dyn1 or GOF mutant p53. Indeed, in H1975 cells (Fig. S1 B), and all other cell lines tested (note that the images in Figs. S3 and S4 are derived from dual-channel images of the same cells), we detected little or no overlap between APPL1- and EEA1-positive endosomes, while the latter are often excluded from the cell perimeter.
Myo6 regulates the localization of perimeter APPL1 endosomes
A recent study showed that Myo6, a minus end–directed actin motor, is required for the peripheral localization of APPL1 endosomes and their role in Akt activation (Masters et al., 2017). Therefore, we next tested whether Myo6 might contribute to the mutant p53–dependent effects on APPL1 endosomes in H1975 cells. Interestingly, Myo6 has also been reported as a transcriptional target of WT and the R294S GOF mutant p53 (Jung et al., 2006). We extended these results, showing that depletion of the R273H mutant p53 by siRNA in H1975 cells also decreased Myo6 protein expression (Fig. 4 A). Importantly, siRNA-mediated depletion of Myo6 suppressed the EGF-dependent accumulation of APPL1 endosomes at the cell perimeter (Fig. 4 B) and correspondingly attenuated EGF-stimulated Akt signaling in H1975 cells (Fig. 4 C). Together, these data suggest that Myo6 expression is an additional mechanism downstream of mutant p53 required to modulate the accumulation of APPL1 endosomes at the cell perimeter and their Akt signaling activity.
Figure 4.

Myo6 is involved in mutant p53–driven perimeter APPL1 endosome accumulation and activity. (A) Representative Western blots and quantification of Myo6 and p53 levels in control and p53 KD H1975 cells. (B) Quantification of fluorescence intensity per endosome (top) or number (middle) of APPL1-positive endosomes relative to their distance from the cell edge in control or Myo6 KD H1975 cells. Errors bars along each curve represent 95% confidence intervals. n = 60 cells/condition. Representative Western blot showing knockdown efficiency of Myo6 (bottom). (C) Representative Western blot and accompanying quantification of Akt activity in control and Myo6 KD H1975 cells. Cells were serum-starved and treated with EGF (20 ng/ml) for the indicated times before cell lysis. Akt activity was calculated as the ratio of phospho-Akt/total Akt for each time point, and then normalized to the control (Ctrl) 10-min EGF-treated sample. Unpaired t tests were used to assess statistical significance; *, P < 0.05; **, P < 0.01; ***, P < 0.001. All quantifications are from three or more independent experiments.
Dyn1 and APPL1 depletion reveal the plasticity of mechanisms regulating EGFR recycling
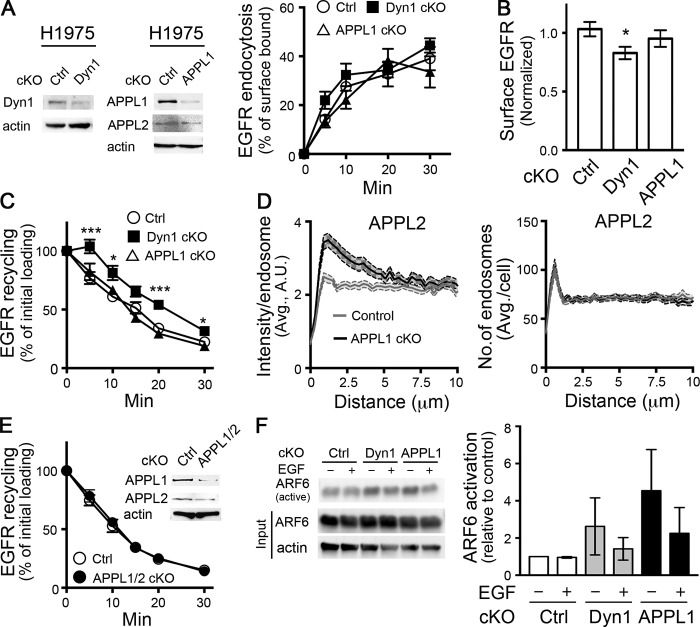
We next tested for the function of Dyn1 and APPL1 in EGFR endocytic trafficking. Conditional depletion of neither Dyn1 nor APPL1 by DD-Cas9 altered the rate of EGFR endocytosis (Fig. 5 A). However, cKO of Dyn1, but not APPL1, induced a slight reduction of surface EGFR levels (Fig. 5 B), suggesting a defect in receptor recycling.
Figure 5.
Dyn1 and APPL1 depletion reveal the plasticity of mechanisms regulating EGFR recycling. (A) Representative Western blots showing efficiency of Dyn1 and APPL1 cKO and the effect of their cKO on EGFR endocytosis. Percentage of internalized biotinylated EGF was calculated relative to the initial surface bound. (B) Surface expression of EGFR in control, Dyn1, or APPL1 cKO H1975 cells. (C) Effects of Dyn1 or APPL1 cKO on rapid recycling of EGFR. Cells were pulsed for 5 min with 20 ng/ml biotinylated EGF, stripped, and reincubated at 37°C for the indicated times before measuring the remaining intracellular EGF. Percentage of recycled biotinylated EGF was calculated relative to the initial loading. (D) Quantification of the intensity and distribution of APPL2 endosomes in APPL1 cKO H1975 cells. (E) EGFR recycling in control, APPL1/2 double-cKO cells. Inset shows cKO efficiency. (F) Representative Western blot and quantification showing compensatory up-regulation of Arf6⋅GTP (i.e., active Arf6) in Dyn1 and APPL1 cKO cells. Error bars represent SEM (n ≥ 3 independent experiments). Unpaired t tests were used to assess statistical significance; *, P < 0.05; ***, P < 0.001.
To directly measure the potential short-circuited, rapid recycling of EGFR from perimeter endosomes, we developed a sensitive in-cell ELISA assay to follow the fate of a brief, 5-min pulse of internalized biotinylated EGF (see Materials and methods). As expected from the observed decrease in surface EGFR, cKO of Dyn1 resulted in an ∼5 min lag before we could detect EGFR recycling (Fig. 5 C), suggesting progression of the EGF pulse deeper along the endosomal pathway before returning to the cell surface.
Given that APPL1 endosome distribution is dependent on Dyn1, we were surprised that APPL1 cKO did not alter the kinetics of EGFR recycling (Fig. 5 C). Therefore, we looked for compensatory mechanisms that might account for the unexpected differential effects of Dyn1 and APPL1 knockout on EGFR recycling. Indeed, we found that APPL2 accumulated on perimeter endosomes upon APPL1 depletion (compare Figs. 1 B and 5 D). Depletion of APPL1 did not alter the number of APPL2 endosomes or APPL2 protein levels (Fig. 5, A and D). These data suggested that APPL2 might be recruited to perimeter endosomes in compensation for the loss of APPL1. To test this, we generated double APPL1/APPL2 DD-Cas9 H1975 cells and found, surprisingly, that EGFR recycling was not affected, even by simultaneous depletion of both APPL isoforms (Fig. 5 E). We conclude that either Dyn1 and APPLs have differential effects on EGFR recycling, or that yet other compensatory mechanisms exist.
The small GTPase Arf6 regulates receptor recycling through both APPL1- and EEA1-positive endosomes (Chen et al., 2014) To determine whether changes in Arf6 activity might constitute another compensatory mechanism for EGFR recycling, we measured Arf6 activity in control and cKO cells using an Arf6⋅GTP pull-down assay (Cohen and Donaldson, 2010). Interestingly, steady-state Arf6 activity was elevated upon cKO of either Dyn1 or APPL1 compared with control (Fig. 5 F). The greater degree of activation upon APPL1 knockdown may compensate for the loss of APPL1 in these cells, perhaps by accelerating recycling through EEA1-positive endosomes (Chen et al., 2014). Interestingly, EGF treatment decreased Arf6 activity in both cKO cell lines relative to corresponding steady-state levels, suggesting other links between EGFR signaling and endocytic trafficking. Further studies are needed to test and characterize the role of Arf6 in compensating EGFR recycling in APPL1 knockout cells. Regardless, together these data suggest a high degree of plasticity in early endocytic trafficking and recycling of activated EGFR, as has also been observed for its uptake via CME (Goh et al., 2010). That these compensatory mechanisms are induced in response to depletion of Dyn1 and/or APPL1 is consistent with their role, and presumably that of perimeter APPL1 endosomes, in regulating the rapid recycling of EGFR.
Dyn1 and APPL1 are required for β1 integrin recycling
EGFR trafficking can involve multiple sorting signals (Goh et al., 2010) and pathways, especially in cancer cells (Tomas et al., 2014). Given this complexity, we next studied β1 integrin trafficking, as these adhesion receptors are critical for mutant p53–dependent changes in cancer cell migration and invasion (Muller et al., 2013, 2014). However, to accurately measure rapid, early recycling of β1 integrins required the development of more sensitive assays than those previously used. For example, in studies using surface biotinylation, cells were incubated for ≥30 min to accumulate sufficient internalized signal for subsequent recycling measurements (Roberts et al., 2001; Caswell et al., 2008; Muller et al., 2009, 2013, 2014; Arjonen et al., 2012). Other studies used surface bound mAbs to follow β1 integrin trafficking, which can inhibit rapid recycling due to clustering of the bivalent Ab-bound receptor (Weissman et al., 1986). To circumvent these problems, we developed a single-chain antibody variable fragment (scFv) derived from mAb K20, a well-characterized, nonfunction perturbing monoclonal anti–β1 integrin antibody (Amiot et al., 1986; Takada and Puzon, 1993; Byron et al., 2009) that can be used to measure β1 integrin uptake and recycling (unpublished data). A comparison of internalization rates of the β1 integrin scFv with its parent mAb revealed that uptake of mAb K20 continued linearly for ≥30 min, whereas the scFv is internalized faster and reached equilibrium after 15 min (Fig. 6 A). These data are consistent with rapid and efficient recycling of scFv.
Figure 6.
APPL1 and Dyn1 regulate β1 integrin recycling. (A) Endocytosis of biotinylated β1 integrin scFv (5 µg/ml) or mAb K20 IgG (1 µg/ml) in H1975 cells. Percentage of internalized antibody was calculated relative to the initial surface bound. (B) Endocytosis of biotinylated β1 integrin scFv (5 µg/ml) was measured in control (Ctrl), Dyn1 cKO, and APPL1 cKO H1975 cells. Shown is the percentage of internalized scFv after 10 min at 37°C. (C) Recycling of biotinylated β1 integrin scFv (5 µg/ml) in control, Dyn1 cKO, and APPL1 cKO H1975 cells. Percentage of recycled β1 integrin scFv was calculated relative to the initial loading (10 min). (D) Surface levels of β1 integrin in Dyn1 and APPL1 cKO cells normalized to control. Error bars represent SEM (n ≥ 4 independent experiments). Unpaired t tests were used to assess statistical significance; **, P < 0.01; ***, P < 0.001.
In contrast to EGFR, conditional depletion of Dyn1 selectively inhibited β1 integrin internalization (Fig. 6 B). A cargo-selective role for Dyn1 in CME has been previously reported (Reis et al., 2017). More importantly, the rapid recycling of β1 integrins, which we could measure directly after a 10-min internalization pulse of scFv, was equally inhibited following either Dyn1 or APPL1 depletion (Fig. 6 C), both of which resulted in a 5-min lag before recycling could be detected. In combination, these two effects resulted in unchanged levels of surface β1 integrin (Fig. 6 D). Together, these data indicate that both Dyn1 and APPL1 play obligatory roles in rapid β1 integrin recycling.
Dyn1 and APPL1 modulate focal adhesion (FA) turnover, migration, and invasion
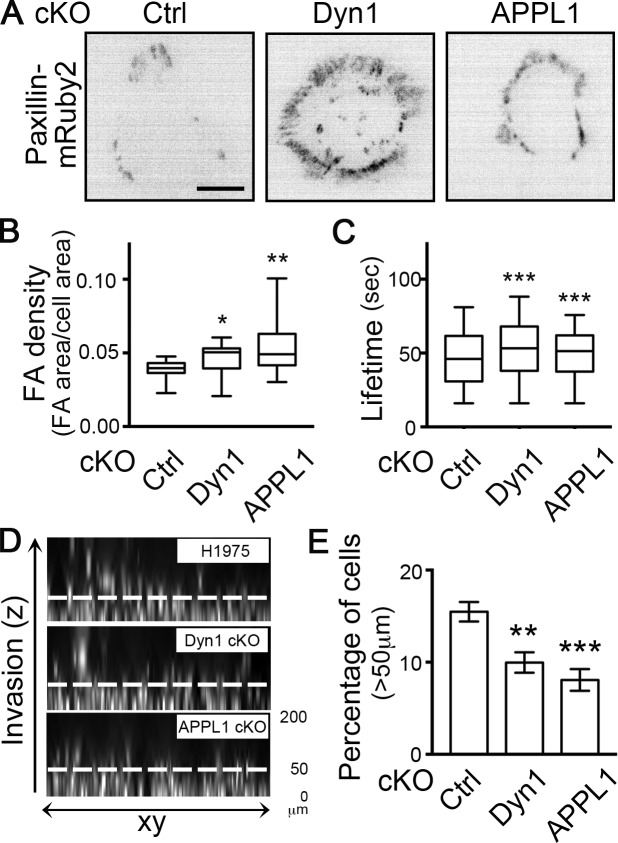
Mutant p53 expression has been shown to increase cancer cell invasion in vitro and in vivo (Muller et al., 2009, 2014). Therefore, we next tested whether these GOF p53-dependent changes in Dyn1 expression and perimeter APPL1 endosome accumulation, and their effects on rapid integrin recycling, can alter cancer cell invasion. The dynamic turnover of cell surface integrins regulates FAs that form at the edges of cells and are critical for cell migration. Therefore, we first used paxillin immunofluorescence in fixed cells to assess the density of FAs (FA area/cell area) at steady state in GOF mutant p53–expressing H1975 cells. cKO of Dyn1 and APPL1 resulted in increased FA density compared with control (Fig. 7 B), especially at cell edges (Fig. 7 A), from where perimeter APPL endosomes are lost. Depletion of Dyn1 and APPL1 also resulted in significantly increased FA lifetimes, as measured by expressing low levels of mRuby2-tagged paxillin (Fig. 7, A and C). Together, our results demonstrate a role for Dyn1 and perimeter APPL1 endosomes in regulating integrin and adhesion turnover in cancer cells.
Figure 7.
Dyn1 and APPL1 modulate FA turnover, cell migration, and invasion. (A) Representative inverted images of H1975 cells expressing Paxillin-mRuby2 in control, Dyn1, and APPL1 cKO conditions. Scale bar, 10 µm. (B) FA density measured by immunofluorescence in control (Ctrl), Dyn1, and APPL1 cKO H1975 cells. Box-and-whisker plots represent the 5th to 95th percentile, mean, and outliers of the data from three independent experiments (n > 49 images with more than one cell per image). (C) FA lifetimes in control (Ctrl), Dyn1, and APPL1 cKO H1975 cells. Data from three independent experiments (n > 11 videos). Mann–Whitney U test was performed to compute significance: *, P < 0.05; **, P < 0.01; ***, P < 0.001. (D and E) Representative Hoechst-stained images (D) and quantification (E) of migration and invasion of control, Dyn1 cKO, APPL1 cKO H1975 cells evaluated in an inverted 3D collagen matrix overlaid with media containing 80 ng/ml EGF. Values are mean ± SEM; n = 3. Statistical significance was analyzed by Student’s t test; **, P < 0.01; ***, P < 0.001.
Finally, we investigated the effect of depletion of Dyn1 and APPL1 on cell migration/invasion using an inverted invasion assay in which cells migrate upwards through a collagen/fibronectin layer toward a chemotactic EGF signal. cKO of either Dyn1 or APPL1 significantly inhibited cell invasion beyond 50 µm (Fig. 7, D and E). Together, our data establish that Dyn1-regulated perimeter APPL1 endosomes modulate cancer cell migration and invasion through alterations in early endocytic trafficking of EGFR and β1 integrin.
Discussion
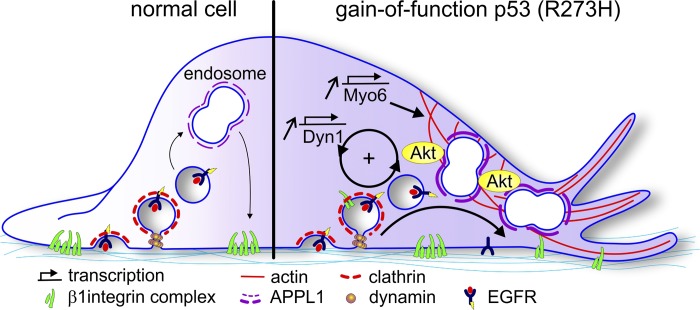
Solid tumors, initially triggered by oncogenic transformation, require numerous adaptations, often acquired over many years, to progress to the aggressive metastatic state. Among these are alterations in receptor signaling and endocytic membrane trafficking that can lead to enhanced proliferation, survival, and invasive properties of the evolved cancer cell (Lanzetti and Di Fiore, 2017; Schmid, 2017). Previous studies have shown that GOF p53 mutations linked to numerous cancers can alter endocytic trafficking of receptor tyrosine kinases and integrins, resulting in increased cell migration and invasion (Muller et al., 2009, 2013). Here, we identify additional molecular mechanisms underlying these effects. Our data establish that Dyn1 and Myo6 are up-regulated in association with GOF mutant p53 expression, resulting in the increased recruitment and accumulation of APPL1 on a spatially restricted endosome subpopulation. Increased membrane association of the APPL1 scaffold accounts for increased Akt signaling previously described in mutant p53 cells (Muller et al., 2009), and Akt, in turn, activates Dyn1 (Reis et al., 2015; Chen et al., 2017), which results in further accumulation of perimeter APPL1 endosomes. Thus, together these activities create a positive feedback loop, which is amplified upon GOF mutant p53 expression, that leads to enhanced migration and invasion in cancer cells. Our findings exemplify how crosstalk between receptor signaling and endocytic trafficking, mediated by a Dyn1/APPL1 endosome/Akt nexus, can contribute to aggressive cancer cell behaviors (Fig. 8).
Figure 8.
A Dyn1 and APPL1 endosome nexus regulates β1 integrin recycling downstream of EGFR, Akt, and GOF mutant p53. Dyn1 and Myo6, whose expression levels are increased in cells expressing GOF mutant p53, regulate the increased recruitment and accumulation of APPL1 “signaling” endosomes at the cell perimeter to accelerate EGFR and β1 integrin recycling. A positive feedback loop involving APPL1-dependent Akt signaling and activation of Dyn1 is established. Together, these activities, which can be co-opted and amplified in GOF p53-driven cancer cells, alter endocytic membrane trafficking to enhance migration and invasion.
We have previously reported that APPL1 endosome-dependent Akt/GSK3β signaling activates normally quiescent Dyn1 in nonneuronal cells to increase rates of clathrin-coated pit initiation and maturation (Reis et al., 2015; Chen et al., 2017; Srinivasan et al., 2018). Here we show that both Akt activity and Dyn1 are also required for the increased recruitment and accumulation of APPL1 on perimeter endosomes. Future work is necessary to define the mechanisms underlying the reciprocal regulation of Dyn1 and APPL1 and their roles in rapid recycling.
Recent studies have shown that APPL1 endosomes function in the rapid recycling of a subset of GPCRs (Jean-Alphonse et al., 2014; Sposini et al., 2017) and that this recycling pathway is inhibited by a negative feedback loop driven by cAMP-dependent activation of protein kinase A. Together with our findings, these results suggest that APPL1 endosomes are central players in the crosstalk between specific signaling receptors and their endocytic trafficking. That spatially restricted subpopulations of APPL1 endosomes are differentially regulated adds another layer of complexity to this crosstalk. The assays we have developed to measure rapid recycling and peripheral endosome distribution should help to unravel this complexity.
The distribution of EEA1 endosomes appears not to be altered by EGFR signaling or mutant p53 expression. This observation is consistent with an independent role for perimeter APPL1 endosomes in localized, rapid recycling, as has been suggested (Kalaidzidis et al., 2015). However, dynamic measurements of integrin trafficking and endosomal maturation at the cell periphery would be required to confidently define this rapid recycling pathway and the relationship between EEA1 and APPL1 endosomes.
Altered β1 integrin recycling is associated with promoting cancer cell dissemination (Jones et al., 2006; Muller et al., 2009; Paul et al., 2015). Canonically, β1 integrin can recycle back to the plasma membrane through either a short-loop (fast) or a perinuclear long-loop (slow) pathway (Caswell et al., 2009; Ivaska and Heino, 2011; Paul et al., 2015). The former is selectively regulated by growth factor receptor signaling (Fang et al., 2010; Arjonen et al., 2012; Onodera et al., 2012), although the mechanisms for this regulation were unknown. Previous studies have shown the polarized redistribution of RCP-positive recycling endosomes at leading extensions of cells migrating in 3D microenvironments toward a chemotactic signal (Caswell et al., 2008). Thus, it is possible that the recruitment of APPL1 to endosomes at the cell edge we observe in response to global EGF stimulation might be more polarized in response to directional chemotactic signals. Indeed, we observe a polarized distribution toward the outer edges of cell clusters (Fig. S1, C and D). Together, our findings support a molecular mechanism for the necessary role of growth factors or serum in regulating a spatially restricted subpopulation of endosomes involved in the fast recycling of β1 integrin.
APPL1 was initially identified as an Akt interacting protein (Mitsuuchi et al., 1999) involved in cell survival and proliferation (Schenck et al., 2008; Ding et al., 2016). However, the role of APPL1 in regulating cell migration is still controversial. Recent studies suggest a negative role of APPL1 in cell migration and Akt signaling in Ras-driven HT1080 fibrosarcoma cells (Broussard et al., 2012; Diggins et al., 2018). In contrast, other reports support a role for APPL1 in increasing cell migration in leptin-stimulated cancer cells (Ding et al., 2016) and in HGF-stimulated mouse embryo fibroblasts (Tan et al., 2016). Our results support a positive role for a specific subpopulation of APPL1 endosomes in cell migration in EGF-stimulated, mutant p53–driven H1975 cells. Differences in cell types used and the signaling pathways studied may account for these different findings. More importantly, these differences reflect the heterogeneity and plasticity in mechanisms driving cancer cell progression.
Cancer cells exhibit significant heterogeneity, and there are no doubt multiple pathways by which they can adapt their early endocytic trafficking during tumor progression. Indeed, the plasticity of the mechanisms governing early endocytic trafficking in cancer cells was evident in the induction of compensatory mechanisms governing EGFR recycling upon conditional CRISPR-mediated knockout of Dyn1 or APPL1 (e.g., ARF6 activation in Dyn1 and APPL1 cKO cells, and APPL2 redistribution in APPL1 cKO cells). Interestingly, integrin recycling was not restored by these compensatory mechanisms, perhaps reflecting the plasticity and complexity of EGFR trafficking and signaling (Goh et al., 2010), compared with integrins. These observations suggest a flexibility of endocytic and signaling systems that can lead to increased plasticity and adaptation in cancer cells.
Both factors, the induction of compensatory mechanisms and heterogeneity, can complicate mechanistic analyses of cancer cells. State-of-the-art CRISPR-based DNA editing has greatly advanced our understanding of gene functions and improves off-target issues associated with siRNA experiments (Hsu et al., 2014; Setodji et al., 2017). However, most constitutive CRISPR knockout studies require single-cell clonal expansion and are therefore susceptible to inherent problems of clonal variation and compensation arising from prolonged cell culture. We adopted a conditional CRISPR knockout system to circumvent these issues, but still observed that cancer cells adapt to maintain early endocytic trafficking. Given the rapid kinetics of endocytic trafficking relative to the 4 d required to fully deplete endogenous proteins, coupled to the ability to regulate these pathways through altered signaling (Srinivasan et al., 2018; Xiao et al., 2018) and/or activation of small G proteins (Fig. 5), researchers should be alert to the induction of compensatory mechanisms that can mask phenotypes.
The extent of our current knowledge of the molecular mechanisms governing early endocytic trafficking provides a foundation for identifying and testing possible compensatory mechanisms. As we have demonstrated, applying this growing knowledge base regarding membrane trafficking and its regulation will improve our understanding of cancer cell plasticity and heterogeneity. Reciprocally, our studies on the regulation of early endocytic trafficking in cancer cells have revealed added complexity with regard to the spatial organization and functional diversity of early endosomes.
Together, our findings suggest that tunable crosstalk between endosomal recycling and receptor signaling networks, in combination with feedback loops, can be activated or amplified in cancer cells to enhance their metastatic traits. Future studies will be needed to test whether these altered in vitro activities manifest in enhanced metastatic activity in vivo. However, that the components of the endocytic machinery that regulate APPL1 signaling endosomes and rapid recycling are targeted by GOF p53 mutations frequently associated with cancer speaks to the physiological/pathological significance of manipulating this pathway. More generally, our data provide mechanistic insight into how selective activation of endocytic isoforms can alter endosomal recycling and receptor signaling to promote the adaptation required for aggressive phenotypes in cancers.
Materials and methods
Cell culture
H1299, H1975, and A549 cells were kindly provided by Dr. John Minna (Hamon Center for Therapeutic Oncology, University of Texas Southwestern [UTSW] Medical Center, Dallas, TX); DU145 and PC3 cells were kindly provided Dr. Jer-Tsong Hsieh (UTSW Medical Center); and A375 and MV3 cells were kindly provided by Dr. Gaudenz Danuser (UTSW Medical Center). A549, DU145, and PC3 cells were maintained in RPMI 1640 (Gibco) supplemented with 10% FBS. HBEC-3KT cells were maintained in keratinocyte-SFM medium (Thermo Fisher Scientific) supplemented with bovine pituitary extract (Thermo Fisher Scientific). ARPE-19 cells were maintained in F12/DMEM (Thermo Fisher Scientific) supplemented with 10% FBS. MDA-MB-231, A375, and MV3 cells were maintained in DMEM supplemented with 10% FBS. H1975 and derived cell lines were maintained in biotin-free RPMI 1640 (USBiological) supplemented with 10% FBS to reduce biotin background for biochemical assays.
Cell engineering
p53 R273 reconstituted H1299 cells were generated by transfecting with pCMV-Neo-Bam p53 R273H plasmid (Addgene; plasmid 16439) and selected in complete medium containing G418 (1 mg/ml; Thermo Fisher Scientific). Conditional knockout cell lines were generated by infection with the lentiviral vector EDCPV, which encodes DD-Cas9, targeting sequences (or not for control), and Venus for cell sorting (Addgene; plasmid 90085; Senturk et al., 2017). Specifically, isogenic inducible H1975 derived DD-Cas9-Dyn1, DD-Cas9-Dyn2, DD-Cas9-APPL1, and DD-Cas9-APPL2 cells were generated by expansion of Venus-positive cells by FACS. Targeting sequences were as follows: human Dyn1 guide sequence (positive strand, 5′-GCAGGTCGAGGTCCGCGTTC-3′; negative strand, 5′-GAACGCGGACCTCGACCTGC-3′), human Dyn2 guide sequence (positive strand, 5′-CCCGCTGGTCAACAAACTGC-3′; negative strand, 5′-GCAGTTTGTTGACCAGCGGG-3′), human APPL1 guide sequence (positive strand, 5′-CTTGTCGATCCCCGGCATCG-3′; negative strand, 5′CGATGCCGGGGATCGACAAG-3′), or human APPL2 guide sequence (positive strand, 5′-TGTCCACGGCGGGCATGGTG-3′; negative strand, 5′-CACCATGCCCGCCGTGGACA-3′).
Immunofluorescence analysis
Briefly, 22 × 22-mm glass coverslips were coated with 0.01% poly-l-lysine for 10 min at RT, washed with PBS, and coated with 0.02% gelatin (in PBS containing 2% sucrose) at 37°C for 30 min, followed by cross-linking with 1% PFA for 15 min at RT. Gelatin-coated coverslips were completely washed with PBS and incubated in complete medium at 4°C overnight. 3 × 105 cells were seeded on gelatin-coated coverslips overnight, washed with PBS, and starved in serum-free medium for 1 h. After that, 20 ng/ml EGF was added to cells for 10 min, washed once with PBS, fixed in 4% PFA for 30 min at 37°C, quenched in 100 mM glycine for 5 min, washed with PBS, permeabilized in 0.2% Triton X-100/PBS for 10 min at RT, and then blocked with 5% BSA/PBS for 1 h at RT. Cells were then incubated overnight at 4°C with primary antibodies (1:250 dilution in Q-PBS, which contains 0.2% BSA, 0.001% saponin, and 0.01% glycine). After three PBS washes, the cells were incubated with Alexa Fluor–conjugated secondary antibodies (1:1,000 dilution in Q-PBS) at 37°C for 1 h, washed three times in PBS, mounted, and imaged by TIRF microscopy.
siRNA and plasmid transfection
Plasmids and siRNA were transfected according to the manufacturer’s recommendations using Lipofectamine 2000 and Lipofectamine RNAiMAX, respectively (Invitrogen). Briefly, 110 pmol of the indicated siRNA and 6.5 µl of Lipofectamine RNAiMAX reagent were diluted in 100 µl of OptiMEM (Thermo Fisher Scientific) and incubated for 5 min at RT. The mixed siRNA-lipid complex was added to each well of a six-well plate containing cells for 4 h. For plasmid transfections, 1 µg of the indicated plasmids and 10 µl of Lipofectamine 2000 reagent were diluted in 100 µl of OptiMEM, incubated for 5 min at RT, and then added to each well of a six-well plate containing cells for 4 h. The cells were replaced in fresh complete medium after 4-h transfection.
Quantification of endosomal imaging data
An automatic pipeline for the analysis of endosomal distribution in two-channel TIRF microscopy imaging data was adapted from our previous work (Reis et al., 2015) and optimized for high-throughput analysis. This pipeline is released as an open-source software package named codistirf, available online at https://github.com/proudot/codistirf. The detection algorithm reuses the method described in Aguet et al. (2013). Briefly, the detection algorithm uses a locally adaptive thresholding approach to obtain a mask of candidate endosome signal followed by subpixellic fitting of a Gaussian function with a scale fixed to 1.5 pixels. Every location presenting intensity significantly above the background signal (P < 0.05) was then classified as an endosome. The cell boundary was estimated on the EGFP channel after Gaussian smoothing (scale set to 5 pixels). The cell mask is defined as the largest connected component in the set of pixels above a segmentation threshold. The threshold value was estimated for each image using the least probable intensity value lying between the two modes of the histogram of the smoothed image: the first mode describes the background pixel, and the second mode describes the cytosolic locations (bin number set to 100). For each cell, the average count- and intensity-measured functions of the membrane distance were computed and exported to Prism (GraphPad) software for flexible representations. The pipeline has been optimized and parallelized to allow for the processing of hundreds of cell images on the day of acquisition. A summarized view of each video was produced for a systematic review of quantification quality and cellular heterogeneity.
Thick-TIRF microscopy
To selectively image endosomal compartments close to the cell periphery, we adjusted the illumination of TIRF evanescent field to a theoretical penetration depth of ∼200 nm (thick TIRF). Briefly, fixed cells were mounted in PBS and imaged using a 60×, 1.49 NA APO TIRF objective (Nikon) mounted on a fully motorized Nikon Ti-Eclipse inverted microscope with Perfect Focus System and coupled to an Andor Diskovery TIRF/Borealis widefield illuminator equipped with an additional 1.8× tube lens (yielding a final magnification of 108×). TIRF illumination was achieved using a Diskovery Platform (Andor Technology). During imaging, cells were maintained at 37°C. Image sequences were acquired using a scientific CMOS camera with 6.5-µm pixel size (pco.edge).
RNA isolation and RT-PCR
RNA was isolated from 3 × 106 cells grown on 6-cm dishes using TRIzol (Thermo Fisher Scientific). cDNA was synthesized using SuperScript IV Reverse transcription kit (Thermo Fisher Scientific) and oligo dT primers (Promega) from 5 µg of total mRNA, following the kit instructions. Gene-specific primers were used in subsequent PCR reactions to amplify DNM1 and actin, and products were visualized after agarose gel electrophoresis. Results are presented as fold-change DNM1 over actin controls.
Purification and biotinylation of β1 integrin scFv
A scFv targeting β1 integrin (unpublished data) was biotinylated for use in biochemical endocytosis and recycling assays. HiFive insect cells obtained from Vincent Tagliabracci (UTSW Medical Center) were infected for 48 h with P2 baculovirus encoding for the expression of a cell-secreted His6-tagged and maltose binding protein–fused β1 integrin scFv (unpublished data). Insect cell culture supernatant was harvested and purified using a 1-ml HisTrapExcel column (GE Healthcare) by fast protein liquid chromatography. Pooled His-purified scFv were further purified via size exclusion chromatography on a Superdex200 Increase column (GE Healthcare). Purified recombinant scFv was biotinylated at a 5:1 molar ratio with EZ-Link Sulfo-NHS-SS-Biotin (Thermo Fisher Scientific) for 2 h at 4°C. Free biotin was removed using Zeba spin desalting columns (Thermo Fisher Scientific). Biotinylated scFv (in PBS + 5% glycerol) was flash frozen in liquid nitrogen (LN2) in single-use aliquots and stored at −80°C.
Cell fractionation
For simple cell fractionation experiments, 107 cells were seeded overnight in duplicate 10-cm dishes. After 16 h, cells were washed three times with PBS and starved for 1 h in serum-free biotin-free RPMI. EGF-stimulated samples were treated with 20 ng/ml EGF for 10 min at 37°C/5% CO2. Starved and EGF-stimulated cells were then washed on ice with cold PBS containing 1 mM sodium orthovanadate and lysed directly in ice-cold cell fractionation (CF) buffer (250 mM sucrose, 20 mM Hepes, pH 7.5, 1 mM EGTA, 2 mM MgCl2, cOmplete protease inhibitor tablet [Roche], PhosSTOP phosphatase inhibitor tablet [Roche], 1 mM 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride, and 1 mM sodium orthovanadate). Cells were ruptured with three cycles of freeze-thaw in LN2 and thawed on ice for 10 min. Nuclei and cell debris were cleared by centrifugation at 5,000 rpm for 5 min at 4°C. Postnuclear supernatant was then used as input to separate membrane fractions from the soluble cytoplasm by ultracentrifugation (100,000 g for 1 h at 4°C). Pelleted fractions were washed in 1× CF buffer and repelleted at 100,000 g for 45 min at 4°C. Pellets were resuspended in CF buffer to be 10× more concentrated than the input samples. 5× Laemmli buffer was added to input, soluble supernatant, and membrane/pellet fractions to be used for Western blotting.
Dyn1 pull-down
Endogenous Dyn1 protein levels in H1299 and H1299 cells expressing mutant p53 (R273H) were assayed via pull-down using GST-AMPHIISH3. Approximately 107 cells were lysed in 10-cm dishes on ice in pull-down buffer (20 mM Hepes, pH 7.4, 150 mM KCl, 2 mM MgCl2, 0.2% Triton X-100, cOmplete tablets, and PhosSTOP) via cell scraping. Cell lysates were collected in Eppendorf tubes and incubated on ice for 30 min, followed by centrifugation at 4°C for 10 min at 14,000 rpm. Clarified lysate was used to measure protein concentrations (A280). Lysates were precleared with prewashed glutathione-Sepharose beads (GE Healthcare) for 30 min at 4°C. Approximately 1 mg of precleared lysate was incubated with GST-AMPHIISH3 bead slurry for 1 h at 4°C. Beads were washed three times in pull-down buffer at 4°C. Bound endogenous Dyn1 was eluted directly from beads in 2× Laemmli buffer (Bio-Rad) and used for Western blotting.
Arf6 activation assay
Endogenous Arf6 activation was assessed through a pull-down assay (Cohen and Donaldson, 2010) using the construct pGST-GGA3VHS-GAT, which encodes the VHS and ARF-binding domains of GGA3 fused to GST, obtained from Juan Bonifacino (National Institutes of Health, Bethesda, MD). Briefly, 8 × 106 cells were seeded in duplicate 10-cm dishes overnight (16 h) in biotin-free RPMI. The next day, cells were washed three times with PBS and starved for 1 h in serum-free biotin-free RPMI. Starved samples were then washed once with cold PBS and lysed directly on ice in ice-cold assay buffer (50 mM Tris, pH 7.5, 100 mM NaCl, 2 mM MgCl2, 1% Triton X-100, 10% glycerol, cOmplete tablets, and PhosSTOP). EGF-stimulated samples were incubated with 20 ng/ml EGF for 10 min at 37°C/5% CO2, and cells were then washed once with cold PBS and lysed on ice in assay buffer. Lysates were immediately subjected to centrifugation at 13,000 g for 5 min at 4°C. Clarified lysates were incubated with GST-GGA3VHS-GAT beads for 30 min at 4°C. Beads were washed on ice three times in assay buffer. Bound proteins were eluted directly in 2× Laemmli buffer and used for Western blotting.
Endocytosis assay
Cells were seeded for 6 h in 96-well plates at a density of 3.5 × 104 cells/well and incubated with 20 ng/ml of biotinylated EGF (Invitrogen), 5 µg/ml of biotinylated β1 integrin scFv, or 1 µg/ml of K20 β1 integrin antibody in assay buffer (PBS4+: PBS supplemented with 1 mM MgCl2, 1 mM CaCl2, 5 mM glucose, and 1% BSA) at 37°C for the indicated times. Cells were then immediately cooled to 4°C to stop internalization. The remaining surface-bound biotinylated EGF, biotinylated β1 integrin scFv, or K20 β1 integrin antibody was removed from the cells by an acid wash step (0.2 M acetic acid and 0.2 M NaCl, pH 2.5). Cells were washed with cold PBS and fixed in 4% PFA (Electron Microscopy Sciences) in PBS for 30 min and then permeabilized with 0.2% Triton X-100/PBS for 10 min. Internalized K20 β1 integrin antibody was assessed using a goat anti-mouse HRP-conjugated antibody (Life Technologies), and internalized biotinylated EGF or biotinylated β1 integrin scFv was assessed by streptavidin-POD (Roche). The reaction was developed with o-phenylenediamine (P1536; Sigma-Aldrich) and then stopped with 5 M H2SO4. The absorbance was read at 490 nm (Biotek Synergy H1 Hybrid Reader). Internalized ligand was expressed as the percentage of the total surface-bound ligand at 4°C (i.e., without acid wash step), measured in parallel (Reis et al., 2015).
Recycling assay
Cells were seeded for 6 h in 96-well plates at a density of 3 × 104 cells/well and pulsed with 20 ng/ml of biotinylated EGF or 5 µg/ml of biotinylated β1 integrin scFv in PBS4+ buffer at 37°C for 5 min (for biotinylated EGF) or 10 min (for biotinylated scFv). Cells were then immediately cooled to 4°C to stop internalization. The remaining surface-bound biotinylated EGF or biotinylated scFv was removed by acid stripping at 4°C (Chen et al., 2017) or cleaved by incubation with 10 mM Tris(2-carboxyethyl)phosphine (TCEP) for 30 s at RT, respectively. Cells were washed with cold PBS4+ buffer and then incubated in PBS4+ containing 20 ng/ml of EGF and 10 mM of TCEP at 37°C for the indicated times. Cells were then washed with 0.2 M acetic acid/0.2 M NaCl (pH 2.5) and PBS, and then fixed in 4% PFA in PBS for 30 min and permeabilized with 0.2% Triton X-100/PBS for 10 min. Remaining intracellular biotinylated ligand was assessed by streptavidin-POD (1:10,000 dilution in Q-PBS [Roche]) and assayed as described above for internalization. The decrease in intracellular biotinylated EGF or biotinylated scFv (recycling) was calculated relative to the total internal pool of ligand internalized.
FA analysis
FAs were analyzed by both fixed-cell immunofluorescence and live-cell microscopy. For fixed-cell immunofluorescence, 3 × 105 cells were seeded overnight on 22 × 22-mm glass coverslips coated with gelatin (0.02%) and fibronectin (25 µg/ml). After 16 h, cells were washed three times with PBS and fixed with simultaneous cytoplasm washout in 2% PFA/0.5% Triton X-100 for 2 min at RT. Cells were then fixed in 4% PFA for 30 min at RT. To block nonspecific antibody binding, cells were incubated in Q-PBS for 30 min at RT. Cells were incubated for 1 h at RT with mouse monoclonal anti-paxillin antibody (clone 177; BD Biosciences) diluted 1:250 in Q-PBS, washed three times with PBS, and then incubated with Alexa Fluor–conjugated secondary antibodies (1:1,000 dilution in Q-PBS) at 37°C for 1 h. Alexa Fluor–conjugated phalloidin was added during the secondary antibody incubation step and used to detect cell boundaries in adhesion quantification analysis. After three PBS washes, the cells were mounted and imaged by TIRF microscopy.
For live-cell adhesion analyses, cells were infected with a lentivirus encoding an mRuby2-Paxillin fusion protein under the control of a crippled CMV promoter. This construct was generated by substituting mRuby2 fluorescent protein for the mNeonGreen (mNG) in pLVX-CMV100-mNG-Paxillin plasmid (Dean et al., 2017) obtained from Dr. Kevin Dean (UTSW Medical Center). H1975 and all engineered cKO cells stably infected with mRuby2-Paxillin were sorted by FACS for very low expression and were used in all live-cell adhesion imaging experiments. 3 × 105 cells were seeded on 22 × 22-mm glass coverslips coated with gelatin (0.02%) and fibronectin (25 µg/ml) for 6 h. 7-min videos were acquired by TIRF microscopy with 100-ms exposure, at a frame rate of one frame/2 s.
FA analysis was performed using a previously published Focal Adhesion Analysis Package (Han et al., 2015). Fixed-cell paxillin immunofluorescence images were used to quantify total cellular adhesion density. Live-cell paxillin videos were used to determine adhesion dynamics and lifetimes. The analysis software is available online at https://git.biohpc.swmed.edu/danuser/applications/pipelines/1944.
Inverted invasion assay
Invasion assays were performed in 96-well dishes (PerkinElmer) as previously described (Bendris et al., 2016). In brief, 5 × 104 cells/ml were suspended in collagen (1.5 mg/ml; Nutacon; 5409) supplemented with 25 µg/ml of human fibronectin (Sigma-Aldrich). 100-µl aliquots of collagen/cells were dispensed into plates. Plates were centrifuged at 1,000 rpm at 4°C and then incubated in a 37°C/5% CO2 tissue culture incubator for 1 h for polymerization. 30 µl of serum-free medium containing 80 ng/ml EGF was added on top of the collagen plug. After 48 h, cells were fixed/stained with 4% formaldehyde and 10 µg/ml Hoechst 33342. For quantification, 15 adjacent images were acquired in each well, yielding ∼90% field of view for each well. Nuclei labeled with Hoechst 33342 from 0 µm (bottom of the plate) to 200 µm into the collagen plug, with 50-µm steps, were detected with the object counts feature of Nikon Elements. The invasion ratio was calculated as the sum of cell counts at 50, 100, 150, and 200 µm over cell counts at 0 µm. Results were obtained from at least three independent experiments including four replicates on each day. Bar charts are plotted as means of all experiments ± SEM.
The Cancer Genome Atlas analysis
mRNA expression level datasets of lung cancer (lung adenocarcinoma and squamous cancer carcinoma) were downloaded from the National Cancer Institute Genomic Data Commons Data Portal (https://portal.gdc.cancer.gov/, data release 10.1, February 15, 2018). We grouped tumor patients into WT p53 and mutant p53 groups based on p53 status. Dyn1 RNA expression value readout fragments per kilobase of transcript per million mapped reads–upper quartile normalization were used.
Online supplemental material
Fig. S1 shows quantitation (A) and representative immunofluorescence (B) of APPL1 and EEA1 endosome redistribution in response to EGF stimulation (1 or 20 ng/ml, respectively). Fig. S1 (C and D) shows examples of the polarized distribution of APPL1 endosomes within cells. Fig. S2 shows the EGF-dependent recruitment of APPL1 to cellular membranes. Fig. S3 shows the Dyn1 isoform-specific dependence of APPL1 endosome redistribution in a panel of cancer and noncancerous cell lines. This Dyn1 isoform-specific effect is specific to APPL1, and not EEA1-positive early endosomes (shown in Fig. S4). Fig. S5 shows knockdown efficiencies of Dyn1 and Dyn2 quantified by Western blot.
Supplementary Material
Acknowledgments
We are grateful to Dr. Szu-Chin Fu (University of Texas Southwestern [UTSW] Medical Center, Dallas, TX) for help in downloading, analysis, and discussion of the Cancer Genome Atlas from Genome Data Commons and Dr. Bo-Jui Chang (UTSW Medical Center) for image analysis of inverted invasion assay. We also appreciate Dr. Sangyoon J. Han (Michigan Technological University, Houghton, MI) and Dr. Andrew Jamieson (UTSW Medical Center) for helpful discussion regarding the FA analysis software, as well as additional help from Schmid laboratory members.
This research was supported by National Institutes of Health grants GM73165 to S.L. Schmid and G. Danuser and GM42455 to S.L. Schmid. P. Roudot was partly supported by the Human Frontier Science Program under grant LT000954/2015. P.-H. Chen was partly supported by Taiwan National Science Council grant 103-2917-I-564-029.
The authors declare no competing financial interests.
Author contributions: A.M. Lakoduk and P.-H. Chen designed the project and performed the experiments. A.M. Lakoduk, P.-H. Chen, and S.L. Schmid discussed and analyzed the results. P. Roudot generated new computational analysis software for endosome immunofluorescence experiments. M. Mettlen assisted with experiments and analyses. H.M. Grossman assisted with virus preparation and cell line generation. A.M. Lakoduk, P.-H. Chen, and S.L. Schmid wrote the manuscript with input from all authors.
References
- Aguet F., Antonescu C.N., Mettlen M., Schmid S.L., and Danuser G.. 2013. Advances in analysis of low signal-to-noise images link dynamin and AP2 to the functions of an endocytic checkpoint. Dev. Cell. 26:279–291. 10.1016/j.devcel.2013.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amiot M., Bernard A., Tran H.C., Leca G., Kanellopoulos J.M., and Boumsell L.. 1986. The human cell surface glycoprotein complex (gp 120,200) recognized by monoclonal antibody K20 is a component binding to phytohaemagglutinin on T cells. Scand. J. Immunol. 23:109–118. 10.1111/j.1365-3083.1986.tb01948.x [DOI] [PubMed] [Google Scholar]
- Arjonen A., Alanko J., Veltel S., and Ivaska J.. 2012. Distinct recycling of active and inactive β1 integrins. Traffic. 13:610–625. 10.1111/j.1600-0854.2012.01327.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacac M., and Stamenkovic I.. 2008. Metastatic cancer cell. Annu. Rev. Pathol. 3:221–247. 10.1146/annurev.pathmechdis.3.121806.151523 [DOI] [PubMed] [Google Scholar]
- Bendris N., Williams K.C., Reis C.R., Welf E.S., Chen P.H., Lemmers B., Hahne M., Leong H.S., and Schmid S.L.. 2016. SNX9 promotes metastasis by enhancing cancer cell invasion via differential regulation of RhoGTPases. Mol. Biol. Cell. 27:1409–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broussard J.A., Lin W.H., Majumdar D., Anderson B., Eason B., Brown C.M., and Webb D.J.. 2012. The endosomal adaptor protein APPL1 impairs the turnover of leading edge adhesions to regulate cell migration. Mol. Biol. Cell. 23:1486–1499. 10.1091/mbc.e11-02-0124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byron A., Humphries J.D., Askari J.A., Craig S.E., Mould A.P., and Humphries M.J.. 2009. Anti-integrin monoclonal antibodies. J. Cell Sci. 122:4009–4011. 10.1242/jcs.056770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caswell P.T., Chan M., Lindsay A.J., McCaffrey M.W., Boettiger D., and Norman J.C.. 2008. Rab-coupling protein coordinates recycling of alpha5beta1 integrin and EGFR1 to promote cell migration in 3D microenvironments. J. Cell Biol. 183:143–155. 10.1083/jcb.200804140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caswell P.T., Vadrevu S., and Norman J.C.. 2009. Integrins: masters and slaves of endocytic transport. Nat. Rev. Mol. Cell Biol. 10:843–853. 10.1038/nrm2799 [DOI] [PubMed] [Google Scholar]
- Chen P.W., Luo R., Jian X., and Randazzo P.A.. 2014. The Arf6 GTPase-activating proteins ARAP2 and ACAP1 define distinct endosomal compartments that regulate integrin α5β1 traffic. J. Biol. Chem. 289:30237–30248. 10.1074/jbc.M114.596155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P.H., Bendris N., Hsiao Y.J., Reis C.R., Mettlen M., Chen H.Y., Yu S.L., and Schmid S.L.. 2017. Crosstalk between CLCb/Dyn1-Mediated Adaptive Clathrin-Mediated Endocytosis and Epidermal Growth Factor Receptor Signaling Increases Metastasis. Dev. Cell. 40:278–288.e275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen L.A., and Donaldson J.G.. 2010. Analysis of Arf GTP-binding protein function in cells. Curr. Protoc. Cell Biol. Chapter 3:Unit 14.12.1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean K.M., Roudot P., Welf E.S., Pohlkamp T., Garrelts G., Herz J., and Fiolka R.. 2017. Imaging Subcellular Dynamics with Fast and Light-Efficient Volumetrically Parallelized Microscopy. Optica. 4:263–271. 10.1364/OPTICA.4.000263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diggins N.L., and Webb D.J.. 2017. APPL1 is a multifunctional endosomal signaling adaptor protein. Biochem. Soc. Trans. 45:771–779. 10.1042/BST20160191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diggins N.L., Kang H., Weaver A., and Webb D.J.. 2018. α5β1 integrin trafficking and Rac activation are regulated by APPL1 in a Rab5-dependent manner to inhibit cell migration. J. Cell Sci. 131:jcs207019 10.1242/jcs.207019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y., Cao Y., Wang B., Wang L., Zhang Y., Zhang D., Chen X., Li M., and Wang C.. 2016. APPL1-Mediating Leptin Signaling Contributes to Proliferation and Migration of Cancer Cells. PLoS One. 11:e0166172 10.1371/journal.pone.0166172 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Fang Z., Takizawa N., Wilson K.A., Smith T.C., Delprato A., Davidson M.W., Lambright D.G., and Luna E.J.. 2010. The membrane-associated protein, supervillin, accelerates F-actin-dependent rapid integrin recycling and cell motility. Traffic. 11:782–799. 10.1111/j.1600-0854.2010.01062.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl P., and Alexander S.. 2011. Cancer invasion and the microenvironment: plasticity and reciprocity. Cell. 147:992–1009. 10.1016/j.cell.2011.11.016 [DOI] [PubMed] [Google Scholar]
- Goh L.K., Huang F., Kim W., Gygi S., and Sorkin A.. 2010. Multiple mechanisms collectively regulate clathrin-mediated endocytosis of the epidermal growth factor receptor. J. Cell Biol. 189:871–883. 10.1083/jcb.201001008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S.J., Oak Y., Groisman A., and Danuser G.. 2015. Traction microscopy to identify force modulation in subresolution adhesions. Nat. Methods. 12:653–656. 10.1038/nmeth.3430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P.D., Lander E.S., and Zhang F.. 2014. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 157:1262–1278. 10.1016/j.cell.2014.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivaska J., and Heino J.. 2011. Cooperation between integrins and growth factor receptors in signaling and endocytosis. Annu. Rev. Cell Dev. Biol. 27:291–320. 10.1146/annurev-cellbio-092910-154017 [DOI] [PubMed] [Google Scholar]
- Jean-Alphonse F., Bowersox S., Chen S., Beard G., Puthenveedu M.A., and Hanyaloglu A.C.. 2014. Spatially restricted G protein-coupled receptor activity via divergent endocytic compartments. J. Biol. Chem. 289:3960–3977. 10.1074/jbc.M113.526350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M.C., Caswell P.T., and Norman J.C.. 2006. Endocytic recycling pathways: emerging regulators of cell migration. Curr. Opin. Cell Biol. 18:549–557. 10.1016/j.ceb.2006.08.003 [DOI] [PubMed] [Google Scholar]
- Jung E.J., Liu G., Zhou W., and Chen X.. 2006. Myosin VI is a mediator of the p53-dependent cell survival pathway. Mol. Cell. Biol. 26:2175–2186. 10.1128/MCB.26.6.2175-2186.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalaidzidis I., Miaczynska M., Brewińska-Olchowik M., Hupalowska A., Ferguson C., Parton R.G., Kalaidzidis Y., and Zerial M.. 2015. APPL endosomes are not obligatory endocytic intermediates but act as stable cargo-sorting compartments. J. Cell Biol. 211:123–144. 10.1083/jcb.201311117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang G.A., Iwakuma T., Suh Y.A., Liu G., Rao V.A., Parant J.M., Valentin-Vega Y.A., Terzian T., Caldwell L.C., Strong L.C., et al. . 2004. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell. 119:861–872. 10.1016/j.cell.2004.11.006 [DOI] [PubMed] [Google Scholar]
- Lanzetti L., and Di Fiore P.P.. 2017. Behind the Scenes: Endo/Exocytosis in the Acquisition of Metastatic Traits. Cancer Res. 77:1813–1817. 10.1158/0008-5472.CAN-16-3403 [DOI] [PubMed] [Google Scholar]
- Masters T.A., Tumbarello D.A., Chibalina M.V., and Buss F.. 2017. MYO6 Regulates Spatial Organization of Signaling Endosomes Driving AKT Activation and Actin Dynamics. Cell Reports. 19:2088–2101. 10.1016/j.celrep.2017.05.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellman I., and Yarden Y.. 2013. Endocytosis and cancer. Cold Spring Harb. Perspect. Biol. 5:a016949 10.1101/cshperspect.a016949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuuchi Y., Johnson S.W., Sonoda G., Tanno S., Golemis E.A., and Testa J.R.. 1999. Identification of a chromosome 3p14.3-21.1 gene, APPL, encoding an adaptor molecule that interacts with the oncoprotein-serine/threonine kinase AKT2. Oncogene. 18:4891–4898. 10.1038/sj.onc.1203080 [DOI] [PubMed] [Google Scholar]
- Muller P.A., Caswell P.T., Doyle B., Iwanicki M.P., Tan E.H., Karim S., Lukashchuk N., Gillespie D.A., Ludwig R.L., Gosselin P., et al. . 2009. Mutant p53 drives invasion by promoting integrin recycling. Cell. 139:1327–1341. 10.1016/j.cell.2009.11.026 [DOI] [PubMed] [Google Scholar]
- Muller P.A., Trinidad A.G., Timpson P., Morton J.P., Zanivan S., van den Berghe P.V., Nixon C., Karim S.A., Caswell P.T., Noll J.E., et al. . 2013. Mutant p53 enhances MET trafficking and signalling to drive cell scattering and invasion. Oncogene. 32:1252–1265. 10.1038/onc.2012.148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller P.A., Trinidad A.G., Caswell P.T., Norman J.C., and Vousden K.H.. 2014. Mutant p53 regulates Dicer through p63-dependent and -independent mechanisms to promote an invasive phenotype. J. Biol. Chem. 289:122–132. 10.1074/jbc.M113.502138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olive K.P., Tuveson D.A., Ruhe Z.C., Yin B., Willis N.A., Bronson R.T., Crowley D., and Jacks T.. 2004. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell. 119:847–860. 10.1016/j.cell.2004.11.004 [DOI] [PubMed] [Google Scholar]
- Onodera Y., Nam J.M., Hashimoto A., Norman J.C., Shirato H., Hashimoto S., and Sabe H.. 2012. Rab5c promotes AMAP1-PRKD2 complex formation to enhance β1 integrin recycling in EGF-induced cancer invasion. J. Cell Biol. 197:983–996. 10.1083/jcb.201201065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul N.R., Jacquemet G., and Caswell P.T.. 2015. Endocytic Trafficking of Integrins in Cell Migration. Curr. Biol. 25:R1092–R1105. 10.1016/j.cub.2015.09.049 [DOI] [PubMed] [Google Scholar]
- Reis C.R., Chen P.H., Srinivasan S., Aguet F., Mettlen M., and Schmid S.L.. 2015. Crosstalk between Akt/GSK3β signaling and dynamin-1 regulates clathrin-mediated endocytosis. EMBO J. 34:2132–2146. 10.15252/embj.201591518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis C.R., Chen P.H., Bendris N., and Schmid S.L.. 2017. TRAIL-death receptor endocytosis and apoptosis are selectively regulated by dynamin-1 activation. Proc. Natl. Acad. Sci. USA. 114:504–509. 10.1073/pnas.1615072114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts M., Barry S., Woods A., van der Sluijs P., and Norman J.. 2001. PDGF-regulated rab4-dependent recycling of alphavbeta3 integrin from early endosomes is necessary for cell adhesion and spreading. Curr. Biol. 11:1392–1402. 10.1016/S0960-9822(01)00442-0 [DOI] [PubMed] [Google Scholar]
- Schenck A., Goto-Silva L., Collinet C., Rhinn M., Giner A., Habermann B., Brand M., and Zerial M.. 2008. The endosomal protein Appl1 mediates Akt substrate specificity and cell survival in vertebrate development. Cell. 133:486–497. 10.1016/j.cell.2008.02.044 [DOI] [PubMed] [Google Scholar]
- Schmid S.L. 2017. Reciprocal regulation of signaling and endocytosis: Implications for the evolving cancer cell. J. Cell Biol. 216:2623–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senturk S., Shirole N.H., Nowak D.G., Corbo V., Pal D., Vaughan A., Tuveson D.A., Trotman L.C., Kinney J.B., and Sordella R.. 2017. Rapid and tunable method to temporally control gene editing based on conditional Cas9 stabilization. Nat. Commun. 8:14370 10.1038/ncomms14370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setodji C.M., McCaffrey D.F., Burgette L.F., Almirall D., and Griffin B.A.. 2017. The Right Tool for the Job: Choosing Between Covariate-balancing and Generalized Boosted Model Propensity Scores. Epidemiology. 28:802–811. 10.1097/EDE.0000000000000734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sever R., and Brugge J.S.. 2015. Signal transduction in cancer. Cold Spring Harb. Perspect. Med. 5:a006098 10.1101/cshperspect.a006098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sposini S., Jean-Alphonse F.G., Ayoub M.A., Oqua A., West C., Lavery S., Brosens J.J., Reiter E., and Hanyaloglu A.C.. 2017. Integration of GPCR Signaling and Sorting from Very Early Endosomes via Opposing APPL1 Mechanisms. Cell Reports. 21:2855–2867. 10.1016/j.celrep.2017.11.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan S., Burckhardt C.J., Bhave M., Chen Z., Chen P.H., Wang X., Danuser G., and Schmid S.L.. 2018. A noncanonical role for dynamin-1 in regulating early stages of clathrin-mediated endocytosis in non-neuronal cells. PLoS Biol. 16:e2005377 10.1371/journal.pbio.2005377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada Y., and Puzon W.. 1993. Identification of a regulatory region of integrin beta 1 subunit using activating and inhibiting antibodies. J. Biol. Chem. 268:17597–17601. [PubMed] [Google Scholar]
- Tan Y., You H., Wu C., Altomare D.A., and Testa J.R.. 2010. Appl1 is dispensable for mouse development, and loss of Appl1 has growth factor-selective effects on Akt signaling in murine embryonic fibroblasts. J. Biol. Chem. 285:6377–6389. 10.1074/jbc.M109.068452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y., Xin X., Coffey F.J., Wiest D.L., Dong L.Q., and Testa J.R.. 2016. Appl1 and Appl2 are Expendable for Mouse Development But Are Essential for HGF-Induced Akt Activation and Migration in Mouse Embryonic Fibroblasts. J. Cell. Physiol. 231:1142–1150. 10.1002/jcp.25211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomas A., Futter C.E., and Eden E.R.. 2014. EGF receptor trafficking: consequences for signaling and cancer. Trends Cell Biol. 24:26–34. 10.1016/j.tcb.2013.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandinger-Ness A., and Zerial M.. 2014. Rab proteins and the compartmentalization of the endosomal system. Cold Spring Harb. Perspect. Biol. 6:a022616 10.1101/cshperspect.a022616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman A.M., Klausner R.D., Rao K., and Harford J.B.. 1986. Exposure of K562 cells to anti-receptor monoclonal antibody OKT9 results in rapid redistribution and enhanced degradation of the transferrin receptor. J. Cell Biol. 102:951–958. 10.1083/jcb.102.3.951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao G.Y., Mohanakrishnan A., and Schmid S.L.. 2018. Role for ERK1/2-dependent activation of FCHSD2 in cancer cell-selective regulation of clathrin-mediated endocytosis. Proc. Natl. Acad. Sci. USA. 115:E9570–E9579. 10.1073/pnas.1810209115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R., Perera R.M., Balkin D.M., Pirruccello M., Toomre D., and De Camilli P.. 2009. A phosphoinositide switch controls the maturation and signaling properties of APPL endosomes. Cell. 136:1110–1121. 10.1016/j.cell.2009.01.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.