Peroxisomes import proteins with a C-terminal SKL sequence by a poorly understood mechanism. Romano et al. use Xenopus egg extracts to study peroxisome import in vitro. The novel assay recapitulates import in vivo and provides mechanistic insights.
Abstract
Peroxisomes import their luminal proteins from the cytosol. Most substrates contain a C-terminal Ser-Lys-Leu (SKL) sequence that is recognized by the receptor Pex5. Pex5 binds to peroxisomes via a docking complex containing Pex14, and recycles back into the cytosol following its mono-ubiquitination at a conserved Cys residue. The mechanism of peroxisome protein import remains incompletely understood. Here, we developed an in vitro import system based on Xenopus egg extracts. Import is dependent on the SKL motif in the substrate and on the presence of Pex5 and Pex14, and is sustained by ATP hydrolysis. A protein lacking an SKL sequence can be coimported, providing strong evidence for import of a folded protein. The conserved cysteine in Pex5 is not essential for import or to clear import sites for subsequent rounds of translocation. This new in vitro assay will be useful for further dissecting the mechanism of peroxisome protein import.
Introduction
Peroxisomes house diverse metabolic functions, notably those involved in lipid metabolism and reactive oxygen detoxification (Braverman and Moser, 2012; Smith and Aitchison, 2013; Wanders, 2014). In humans, defects in peroxisome biogenesis cause neurological diseases, such as Zellweger syndrome (Braverman et al., 2013; Fujiki, 2016; Waterham et al., 2016). While peroxisome membrane proteins are probably derived from the ER, matrix proteins are synthesized in the cytosol and must then be transported across the peroxisome membrane (Hettema et al., 2014; Agrawal and Subramani, 2016). Most matrix proteins use a C-terminal Ser-Lys-Leu (SKL) sequence as an import signal, otherwise known as the peroxisome targeting signal 1 (PTS1; Gould et al., 1989). This motif is recognized by the import receptor Pex5 through its C-terminal tetratricopeptide repeat domain (McCollum et al., 1993; Van der Leij et al., 1993; Brocard et al., 1994). Pex5 uses an N-terminal domain to bind to a docking complex on the peroxisome membrane, which contains Pex13 and Pex14 as conserved subunits (Erdmann and Blobel, 1996; Gould et al., 1996; Albertini et al., 1997). The PTS1 cargo is then translocated across the peroxisome membrane by mechanisms that have not been fully elucidated. The current evidence also suggests that Pex5 is mono-ubiquitinated at a conserved Cys residue close to its N terminus (Carvalho et al., 2007; Williams et al., 2007). Pex5 is subsequently returned to the cytosol by an ATPase complex and can start a new translocation cycle (Platta et al., 2005). Despite progress over several decades, important aspects of peroxisome import remain unclear. A particularly mysterious point is the reported import of folded proteins and oligomeric assemblies (Léon et al., 2006).
Further progress on the mechanism of peroxisome protein import critically depends on in vitro experiments in which components can be depleted and manipulated. Several in vitro import assays have been reported, either based on permeabilized cells or fractionated extracts (Fujiki and Lazarow, 1985; Wendland and Subramani, 1993; Rodrigues et al., 2016; Okumoto et al., 2017). However, none of these systems has been used extensively, probably because they have to be prepared freshly, or are difficult to reproduce, or import is of low efficiency and hard to quantify. A confounding problem with in vitro systems is the fragility of peroxisomes, which makes the use of purified organelles difficult. Here, we describe a reliable and quantifiable in vitro system based on Xenopus laevis egg extracts, which recapitulates peroxisome protein import. We use this assay to investigate several aspects of the mechanism of peroxisome protein import.
Results
An in vitro system for peroxisome protein import
Xenopus egg extracts have been used extensively to reproduce various biological processes, as they contain all cellular components at physiological concentrations. We therefore decided to test this system for peroxisome protein import. Eggs from the frog X. laevis were centrifuged in the absence of the Ca2+-chelator EGTA to move the extract into interphase of the cell cycle (Wang et al., 2019). The resulting crude extract was subjected to ultra-centrifugation in the absence of the actin-depolymerizing reagent cytochalasin D. The gel-like cytosolic fraction and membranes contained in it were collected and frozen in aliquots (Fig. S1 A). Glycogen and a large portion of the membranes sedimented to the bottom of the tube and were discarded. The isolated membrane/cytosol material, called “cleared Xenopus extract,” is active in peroxisome protein transport after thawing (see below). The extract contains peroxisomes (see below), as well as ER and mitochondria (Fig. S1 B), and it maintains microtubule and ER dynamics even after thawing (Fig. S1 C). Extracts generated with the normal procedure, which involves the addition of cytochalasin D before sedimentation of the membranes, also showed peroxisome protein import, but they lost activity for import and microtubule dynamics after freeze-thawing (data not shown). The generation of cleared Xenopus extract may provide a facile alternative to a recently reported procedure to generate frozen extracts for the study of other biological processes (Takagi and Shimamoto, 2017).
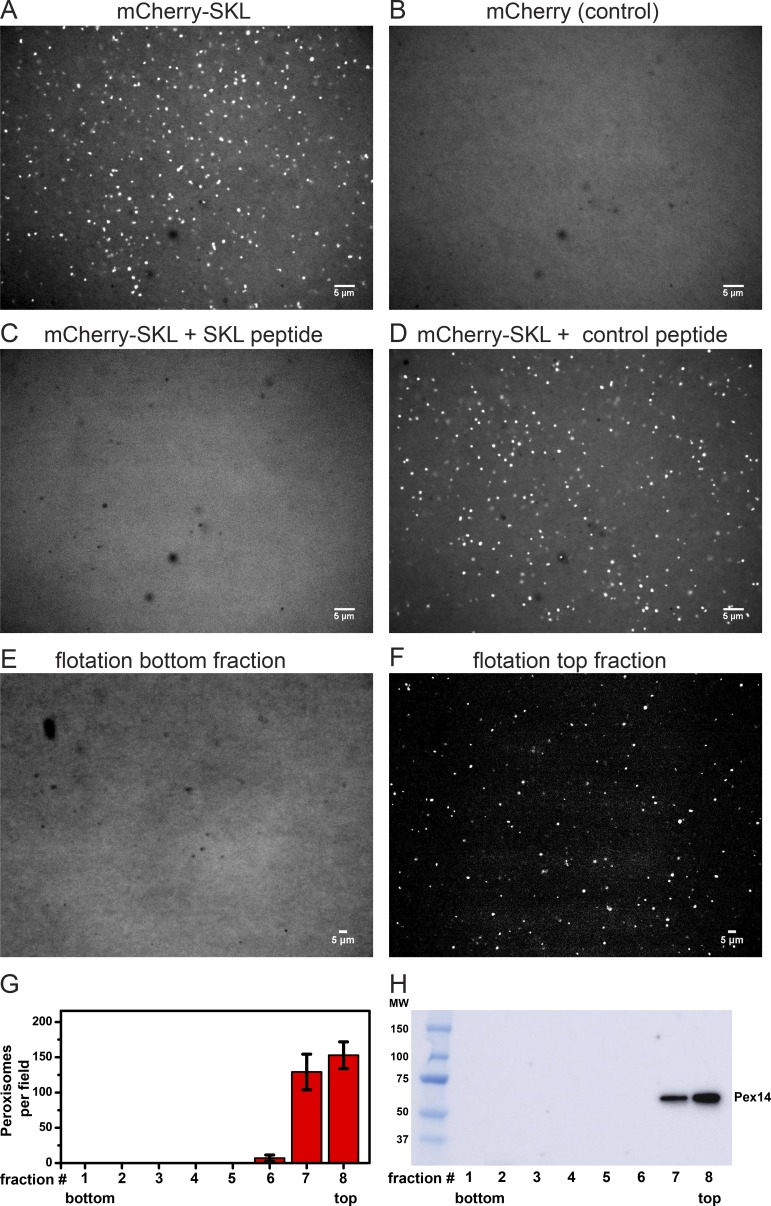
To test for peroxisome protein transport, we fused a C-terminal SKL sequence to a fluorescent protein, either superfolder GFP, mCherry, or mScarlet. These proteins were expressed in Escherichia coli and purified using N-terminal His tags, followed by gel filtration (Fig. S2). After incubation of the purified fusion proteins with cleared Xenopus extract, bright foci were observed (shown for mCherry in Fig. 1 A). In contrast, no foci were seen with fluorescent proteins lacking the SKL sequence (Fig. 1 B). Foci formation was prevented when the incubation of mCherry-SKL was performed in the presence of an excess of a synthetic peptide that contains the SKL sequence at its C terminus (Fig. 1 C). In contrast, foci formation was not perturbed by a control peptide that contains the sequence KLS at its C terminus (Fig. 1 D), a sequence that does not function as a PTS1 signal (Neuberger et al., 2003). These results suggest that the foci correspond to peroxisomes that bound or imported proteins containing a PTS1 signal. It should be noted that the extract was not fixed, allowing the foci to move freely (Video 1).
Figure 1.
Peroxisome targeting of SKL-containing fluorescent proteins in Xenopus egg extracts. (A) Cleared egg extract was incubated with 0.9 µM purified mCherry-SKL for 1 h at 18°C. The formation of bright puncta was visualized with a spinning-disk confocal microscope. (B) As in A, but with mCherry lacking the SKL targeting sequence. (C) As in A, but in the presence of a synthetic peptide (300 µM) with a C-terminal SKL sequence. (D) As in C, but with a peptide containing a scrambled import signal at its C terminus (KLS). (E) Cleared egg extract was incubated with 0.6 µM purified mScarlet-SKL for 5 h at 18°C. The sample was subjected to flotation in a discontinuous sucrose gradient. Fractions were collected and analyzed with a fluorescence microscope. Shown is the bottom fraction containing nonimported substrate. (F) As in E, but for the top fraction, containing peroxisome-associated substrate. (G) Quantification of the number of fluorescent peroxisome foci in the different fractions of the sucrose gradient. Shown are the mean and standard deviation from >40 images of two different experiments. (H) All fractions of the sucrose gradient were analyzed by SDS-PAGE, followed by immunoblotting with antibodies against the peroxisome membrane protein Pex14. Bars, 5 µm.
To test whether the fluorescent puncta correspond to peroxisomes, we performed an import reaction with mScarlet-SKL and then subjected the mixture to flotation in a discontinuous sucrose gradient. Fractions were analyzed by fluorescence imaging (bottom and top fractions are shown in Fig. 1, E and F, respectively) and the number of foci was quantified using an automated script (Fig. 1 G). Essentially all fluorescent foci floated to the top of the gradient. Immunoblotting with an antibody raised against a N-terminal domain of the peroxisomal membrane protein Pex14 showed that these fractions also contained endogenous peroxisomes (Fig. 1 H). Non-imported mScarlet-SKL substrate stayed at the bottom of the gradient and showed diffuse fluorescence (Fig. 1 E). These results provide evidence that the foci correspond to peroxisomes and are not caused by protein aggregation.
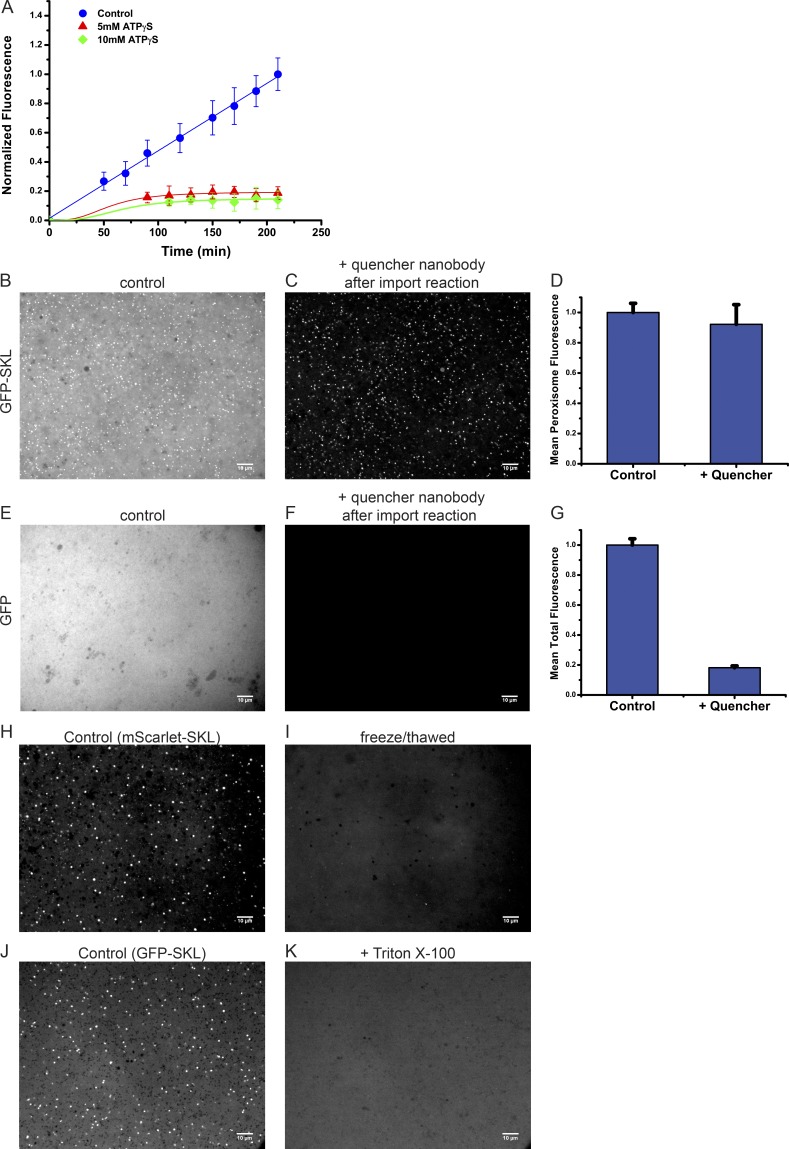
To distinguish between substrate binding to peroxisomes and actual import into these organelles, we followed the accumulation of mScarlet-SKL at peroxisomes over time. Given that substrate was added in excess over endogenous Pex5 and docking protein complex (Wühr et al., 2014), we reasoned that the fluorescence of each peroxisome would plateau rapidly if the substrate was only binding, whereas it would increase linearly if the substrate was imported. To follow substrate over time, we placed the reaction mix into a small microscope chamber and imaged it repeatedly. An automated script was developed to identify peroxisomes in the images and calculate their average fluorescence. At early time points, the detection of peroxisomes was difficult because of their weak fluorescence, but imaging at later time points showed that their average fluorescence increased linearly over several hours (Fig. 2 A). These data are consistent with real protein import into peroxisomes. The addition of the nonhydrolysable ATP analogue ATPγS drastically reduced import, although import was still detectable at the beginning of the reaction (Fig. 2 A), perhaps because a single round of peroxisome protein import can occur in the absence of ATP hydrolysis. Depletion of ATP from crude extracts by blocking ATP production by both mitochondrial respiration and glycolysis also inhibited peroxisome protein import (Fig. S3).
Figure 2.
Peroxisome protein import in Xenopus egg extract. (A) mScarlet-SKL (0.6 µM) was added at time point zero to cleared Xenopus egg extract. Where indicated, 5 or 10 mM ATPγS was added 20 min before substrate. The samples were immediately mounted on PEG-passivated glass coverslips and imaged at the indicated time points, using a spinning-disk confocal microscope. The mean fluorescence of peroxisomes was determined from >10 images using an automated image analysis script. Shown are the combined data of two different experiments (>20 images per time point), each normalized to the final time point of the control. For each time point, the mean and the standard deviation of the mean are given. (B) Cleared extract was incubated with 0.5 µM GFP-SKL for 1 h at 20–23°C and imaged with a spinning-disk confocal microscope. (C) As in B but at the end of the incubation, 0.75 µM GFP-fluorescence quenching nanobodies (Kirchhofer et al., 2010) were added for ∼15 min before imaging. (D) Quantification of the mean peroxisome fluorescence in B and C of two separate import reactions. Shown are the mean and standard deviation of >20 images. (E) As in B, but with GFP lacking the SKL sequence. (F) As in C, but with GFP lacking the SKL sequence. (G) Quantification of the total fluorescence in E and F of two separate import reactions. Shown are the mean and standard deviation of >20 images. (H) mScarlet-SKL (0.6 µM) was incubated with cleared egg extract for 1 h at 18°C. The membranes were sedimented twice by centrifugation and resuspension in buffer. The sample was imaged directly in a confocal microscope. (I) As in H, but the sample was subjected to three freeze–thaw cycles before imaging. (J) GFP-SKL (0.4 µM) was incubated with crude extract for 1 h and imaged. (K) As in J, but 0.1% Triton X-100 was added before imaging. Bars, 10 µM.
To directly test for substrate translocation, we used GFP-quenching nanobodies (Kirchhofer et al., 2010) and asked whether the peroxisome membrane would prevent quenching of previously imported GFP-SKL. When the nanobodies were added after incubation of the extract with GFP-SKL, the foci remained unquenched, whereas residual, nonimported substrate was quenched, as seen by a decrease of the background fluorescence (Fig. 2 C versus Fig. 2 B). Quantification confirmed that the fluorescence of the peroxisomes was unaffected by the quenching nanobodies, whereas the unimported material was quenched (Fig. 2 D). These data suggest that the substrate is inside the peroxisomes and therefore inaccessible to the nanobodies. No foci were seen with GFP lacking SKL, and ∼80% of the total fluorescence was quenched by the nanobodies (Fig. 2, E–G).
Additional support for real substrate import comes from experiments in which we isolated peroxisomes after import of mScarlet-SKL. More than 80% of the fluorescent foci could be recovered after two rounds of sedimentation and resuspension. However, when the resuspended sample was subjected to three freeze–thaw cycles, all foci were lost, as expected from such treatment disrupting the peroxisome membrane (Fig. 2 H). Addition of detergent had the same effect (Fig. 2 I). Taken together, these experiments provide strong evidence that the Xenopus system recapitulates actual protein import into peroxisomes.
Peroxisome protein import depends on Pex5 and its membrane receptor Pex14
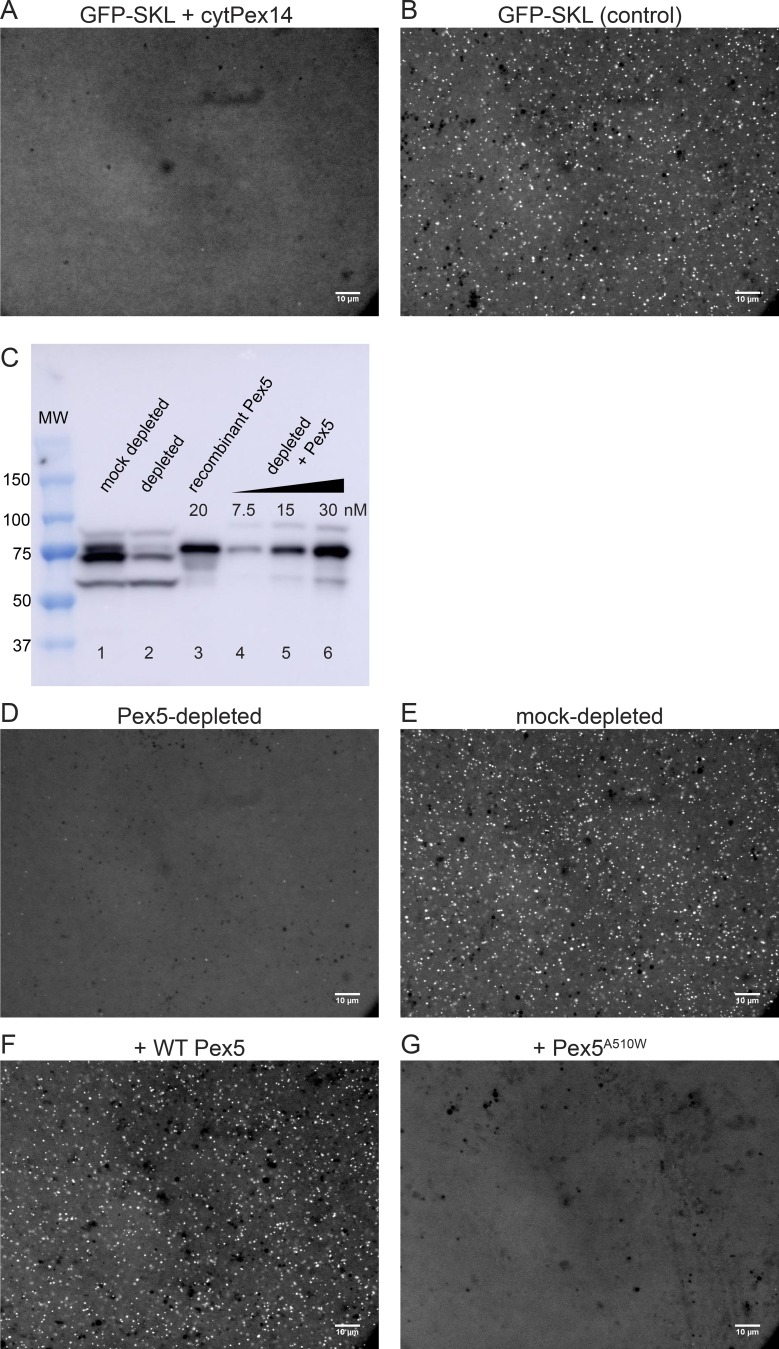
Next, we tested whether peroxisome foci formation depends on the known import components Pex5 and Pex14. To this end, we purified a Pex5-interacting cytosolic domain of the docking protein Pex14 (amino acids 17–78 in Xenopus Pex14; Neufeld et al., 2009). The protein was expressed in E. coli as a glutathione S-transferase (GST) fusion, and purified after proteolytic removal of the GST tag (Fig. S2). Addition of this cytosolic Pex14 fragment to cleared Xenopus extract completely inhibited foci formation by GFP-SKL (Fig. 3 A; parallel control shown in Fig. 3 B) or mCherry-SKL (data not shown). Next, we coupled the cytosolic Pex14 fragment to a resin and used these beads to deplete the cleared extract of Pex5. The mock-depleted extract contained two major bands detected by immunoblotting with an antibody raised against Xenopus Pex5 (Fig. 3 C, lane 1), possibly corresponding to the short and long isoforms of Pex5 reported in other species (Braverman et al., 1998; Otera et al., 1998). The intensity of both bands was reduced by ∼80% after incubation with Pex14 fragment–containing resin (Fig. 3 C, lane 2). The depleted extract was almost completely inactive in foci formation with GFP-SKL (Fig. 3 D; control with a mock-depleted extract shown in Fig. 3 E). To test whether Pex5 is the only essential component missing in the depleted extract, we performed rescue experiments. Full-length Xenopus Pex5, corresponding to the long isoform of Pex5 in other species, was expressed in E. coli as a GST fusion and purified after removal of the GST tag by gel filtration (Fig. S2). This protein migrated at the same position as the higher molecular weight band detected by immunoblotting in Xenopus extracts (Fig. 3 C, lane 3 versus lane 1). Addition of the recombinant Pex5 protein restored foci formation in a depleted extract (Fig. 3 F). As a control, we purified a Pex5 mutant in which a conserved Ala (Ala510) in the SKL-binding pocket of the tetratricopeptide repeat domain (Gatto et al., 2000) was mutated to Trp. Gel filtration experiments confirmed that the Pex5 Ala510Trp mutant bound mScarlet-SKL significantly more weakly than WT Pex5 (Fig. S4). Pex5 Ala510Trp did not restore foci formation in the depleted extract (Fig. 3 G). Taken together, these experiments suggest that our system reproduces Pex5- and Pex14-dependent peroxisome protein import.
Figure 3.
Protein targeting to peroxisomes depends on Pex5 and Pex14. (A) Cleared Xenopus egg extract was incubated for 1 h at 18°C with 0.5 µM GFP-SKL in the presence of 6 µM of a cytosolic fragment of the peroxisome docking protein Pex14 (cytPex14). The sample was imaged with a spinning-disk confocal microscope. (B) As in A, but without adding cytPex14. (C) Egg extract was incubated with beads containing immobilized cytPex14 and subjected to SDS-PAGE, followed by immunoblotting with Pex5 antibodies (lane 2). A control was done with beads lacking cytPex14 (lane 1; mock depletion). Purified, recombinant Pex5 was analyzed either without added extract (lane 3) or after addition of different amounts to depleted extract (lanes 4–6). MW, molecular weight. (D) Pex5-depleted extract was incubated for 1 h at 18°C with 0.5 µM GFP-SKL and imaged with a spinning-disk confocal microscope. (E) As in D, but with mock-depleted extract. (F) As in D, but in the presence of 1 µM purified Pex5. (G) As in F, but with 1 µM purified Pex5A510W, a Pex5 mutant defective in SKL binding. All experiments were performed at least three times. Bars, 10 µm.
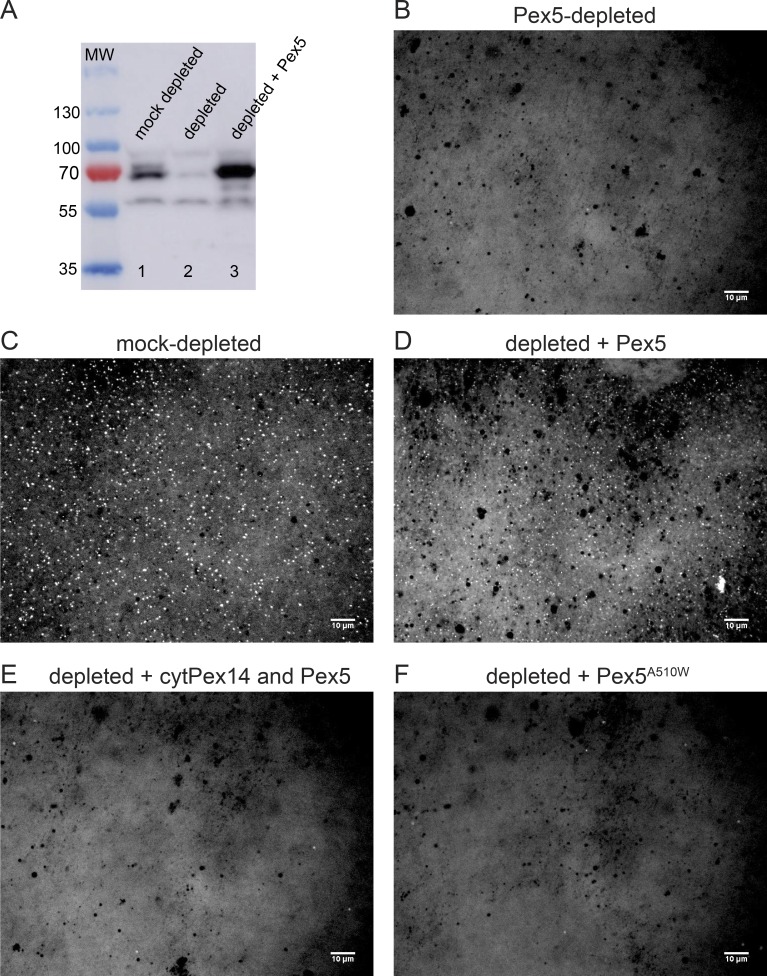
To further test this conclusion, we used cleared Xenopus extracts that were depleted of Pex5 with antibodies. Pex5 antibodies were isolated from crude antiserum with beads containing recombinant Pex5. After elution from the beads, the antibodies were coupled to a resin and then used to deplete cleared extract. Pex5 depletion was almost complete (Fig. 4 A). The depleted extract was entirely inactive (Fig. 4 B versus Fig. 4 C), but activity could be restored by addition of recombinant Pex5 (Fig. 4 D). When the Pex5-interacting cytosolic Pex14 fragment was also added, no foci were observed (Fig. 4 E). As before, addition of the SKL-binding mutant Ala510Trp did not restore foci formation (Fig. 4 F). These results further indicate that our Xenopus system recapitulates Pex5-dependent protein import into peroxisomes.
Figure 4.
Peroxisome targeting in Pex5-depleted and -replenished egg extract. (A) Cleared Xenopus egg extract was incubated with beads containing immobilized, affinity-purified antibodies to Pex5. The depleted extract was subjected to SDS-PAGE, followed by immunoblotting with Pex5 antibodies (lane 2). A control was done with beads lacking Pex5 antibodies (lane 1; mock depletion). In lane 3, 30 nM purified, recombinant Pex5 was added to the depleted extract. (B) Pex5-depleted extract was incubated with 0.6 µM mScarlet-SKL for 1 h at 18°C and imaged with a spinning-disk confocal microscope. (C) As in B, but with mock-depleted extract. (D) As in B, but in the presence of 0.5 µM purified Pex5. (E) As in D, but with additionally added 6 µM cytPex14. (F) As in D, but with 0.5 µM purified Pex5A510W, a Pex5 mutant defective in SKL binding. These experiments were performed twice. Bars, 10 µm.
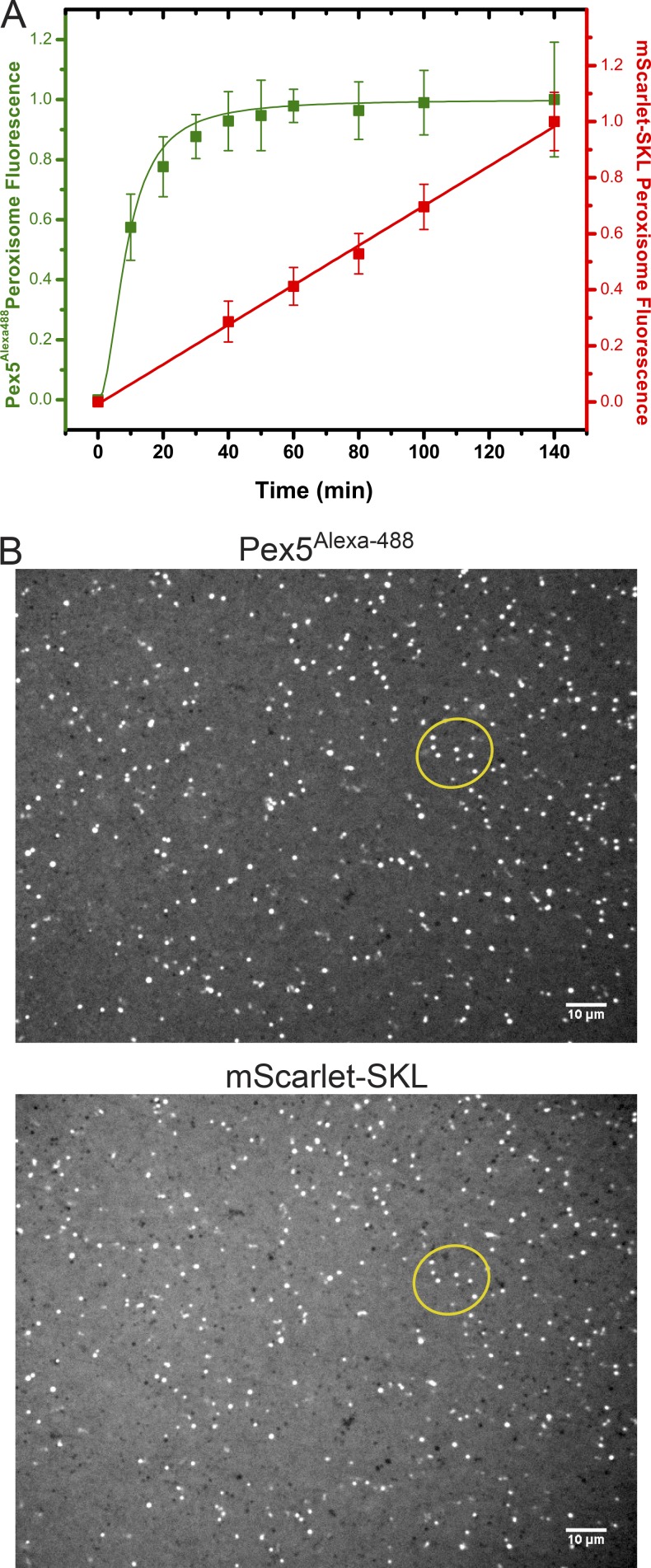
Next we tested the binding of Pex5 to peroxisomes. An 11-residue ybbR tag (Yin et al., 2006) was added at the C terminus of Pex5, and the purified protein (Fig. S2) was labeled with Alexa Fluor 488. Labeled Pex5 was then added to the cleared Xenopus extract and its binding to peroxisomes followed over time in a fluorescent microscope (Fig. 5 A). Pex5 binding reached a plateau after 20 min, whereas substrate import in the same experiment continued for several hours (Fig. 5 A). Because of the mobility of the peroxisomes, the puncta recorded in the two fluorescent channels do not show exact colocalization, and the fluorescence ratio varies, but the pattern is similar enough to conclude that they correspond to the same puncta (Fig. 5 B). These experiments are consistent with the idea that Pex5 rapidly reaches a steady state of binding to and releasing from peroxisomes, while the substrate continuously accumulates inside the organelle.
Figure 5.
Kinetics of Pex5 binding and peroxisome protein import. (A) mScarlet-SKL (0.6 µM) and fluorescently labeled Pex5 (Pex5Alexa488; 0.15 µM) were added at time point zero to cleared Xenopus egg extract. The sample was mounted on a PEG-passivated glass chamber and imaged over time using a spinning-disk confocal microscope. The red (mScarlet-SKL) and green (Pex5Alexa-488) emission channels were imaged simultaneously. Shown are the combined data of two different experiments (>20 images per time point), each normalized to the final time point of the control. For each time point, the mean and the standard deviation of the mean are given. (B) Images taken at 140 min sequentially in the two channels. The foci partially overlap (compare foci inside the oval). Overlap is not perfect because the peroxisomes are moving. Bars, 10 µm.
Testing the role of the conserved Cys residue of Pex5
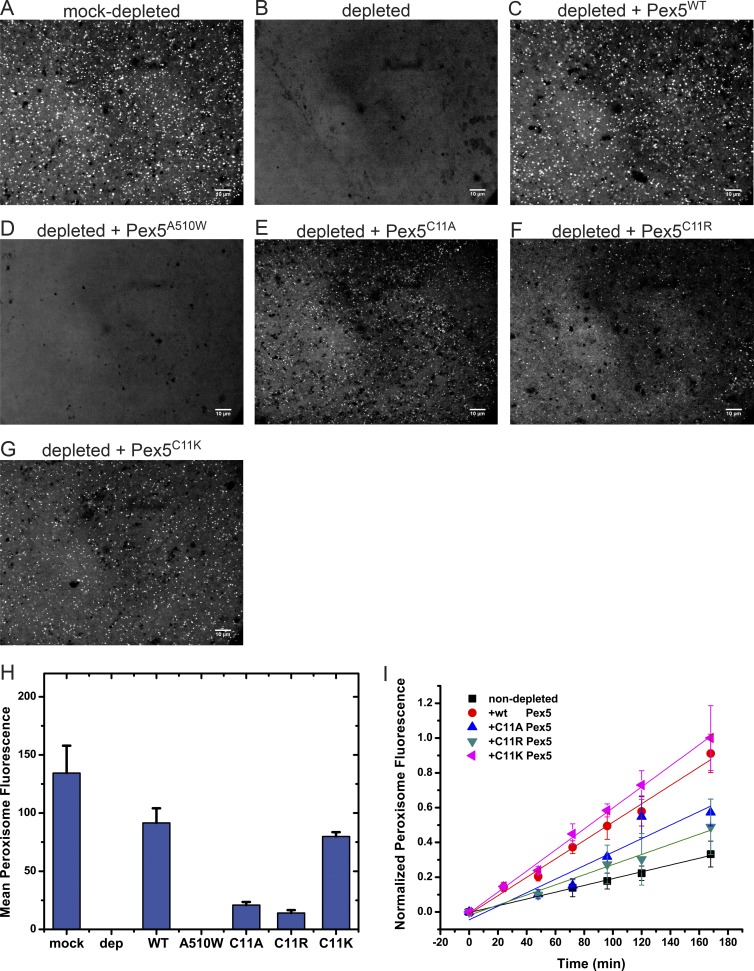
In all species, Pex5 has a conserved Cys residue close to its N terminus (Cys11 in Xenopus Pex5), which seems to be mono-ubiquitinated, a modification that is thought to be required for the recycling of Pex5 from the peroxisome membrane back into the cytosol (Braverman and Moser, 2012; Smith and Aitchison, 2013; Wanders, 2014). We used our depletion/add-back Xenopus system to test various Cys mutants. The Cys11Lys mutant was almost as active as the WT Pex5 protein in restoring peroxisome import in Pex5 antibody-depleted extracts (Fig. 6 E versus Fig. 6 C; controls in Fig. 6, A and B; and quantification in Fig. 6 I). Replacement of Cys11 with either Ala or Arg drastically reduced the activity of Pex5, but did not completely abolish it (Fig. 6, F and G). These mutants retained 10–20% of WT activity (Fig. 6 I), in contrast to the SKL-binding mutant A510W, which was completely inactive. These results are consistent with the idea that modification of Cys11 is only required for the recycling of Pex5, not for a single round of protein import. The Cys11Ser mutant did not support import (data not shown), but the protein may not have been correctly folded, as it was expressed in E. coli at lower levels than WT Pex5 or all other mutants and was prone to aggregation and proteolysis during purification.
Figure 6.
Peroxisome protein import with Cys11 mutations in Pex5. (A) Xenopus cleared egg extract was mock-depleted with beads, as in Fig. 3, and incubated with 0.6 µM mScarlet-SKL for 1 h at 20–23°C. The sample was imaged with a spinning-disk confocal microscope. (B) As in A, but with an extract depleted of Pex5 with beads containing affinity-purified Pex5 antibodies (see Fig. 3). (C) As in B, but with 1 µM WT Pex5 added. (D) As in B, but with 1 µM Pex5A510W, defective in SKL binding. (E) As in B, but with 1 µM Pex5C11A. (F) As in B, but with 1 µM Pex5C11R. (G) As in B, but with 1 µM Pex5C11K. (H) Quantification of the end-point fluorescence in peroxisomes, using automated image analysis. Shown are the combined data of two parallel experiments (>20 images). The mean and standard deviation of the mean are given. (I) WT Pex5 or the indicated Cys11 mutants were added at 1 µM to cleared, un-depleted extract. The samples were imaged at the indicated time points with a spinning-disk confocal microscope, and the mean fluorescence per peroxisome was determined by automated image analysis. Shown are the combined data of two experiments done on different days (>20 images per time point), each normalized to the final time point of the brightest sample. For each time point, the mean and the standard deviation of the mean are given. Bars, 10 µm.
To test whether the Pex5 mutants are dominant-negative, we added them to nondepleted cleared extracts and followed peroxisome import of mScarlet-SKL over time (Fig. 6 J). Although WT Pex5 and the Cys11Lys mutant had the greatest effect, even the Cys11Ala and Cys11Arg mutants stimulated import. Thus, these mutants are not dominant-negative, suggesting that any modification at Cys11 is not necessary to clear import sites for subsequent rounds of translocation.
Import of a folded protein into peroxisomes
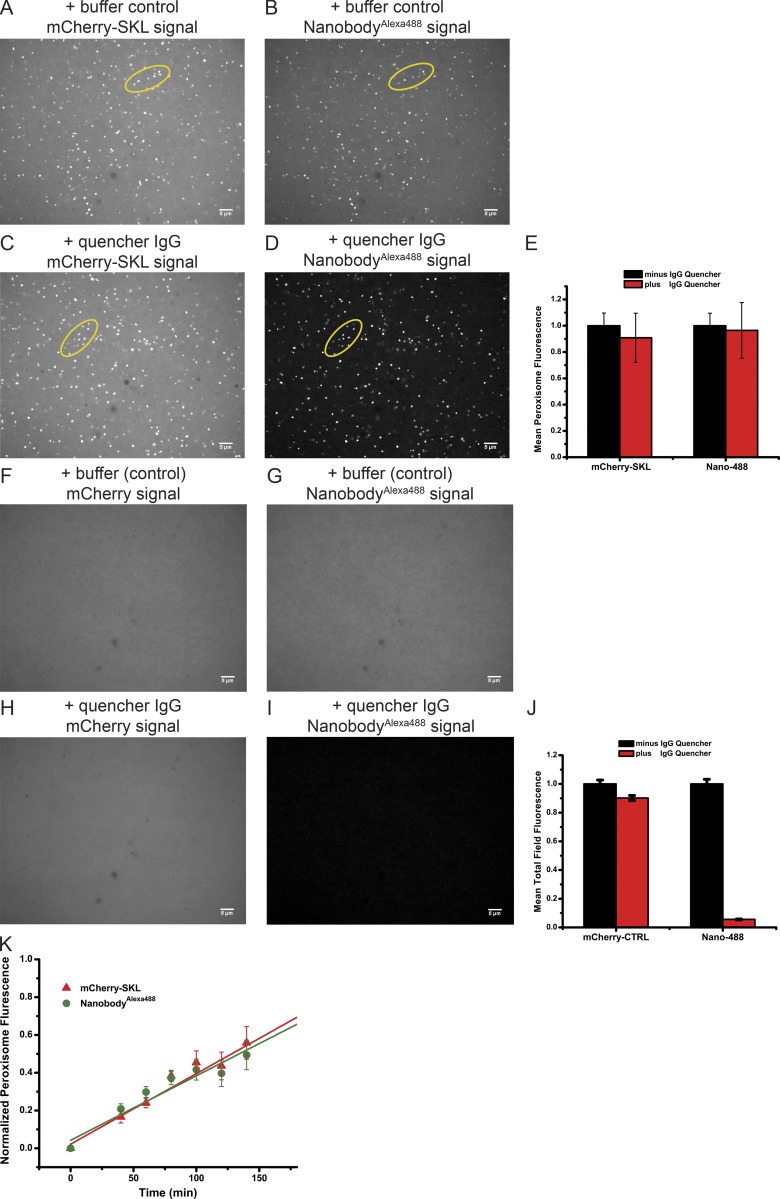
Next we used the established in vitro system to test whether a folded protein can be imported into peroxisomes. We added mCherry-SKL to an interacting, fluorescently labeled nanobody (Alexa Fluor 488 nanobody) that lacks a SKL sequence, and then incubated the mixture with cleared Xenopus extract. Peroxisomes could be detected both by mCherry and Alexa Fluor 488 fluorescence (Fig. 7, A and B), indicating colocalization of the two interacting proteins. Import of the nanobody into peroxisomes was tested by adding after the import reaction IgG from monoclonal antibodies that quench Alexa Fluor 488 fluorescence (Baldridge and Rapoport, 2016). Addition of IgG had no effect on the fluorescence of peroxisomes labeled with GFP or Alexa Fluor 488 (Fig. 7, C and D; quantification shown in Fig. 7 E). However, nonimported Alexa Fluor 488 nanobody was quenched, as seen by the reduction of background fluorescence (Fig. 7 D). As expected, no labeled peroxisomes were visible in either the mCherry or the Alexa Fluor 488 channel when mCherry lacked the SKL sequence (Fig. 7, F–I; quantification shown in Fig. 7 J). The addition of IgG reduced the total fluorescence only in the Alexa Fluor 488 channel (Fig. 7 I). Consistent with the coimport of mCherry-SKL and Alexa Fluor 488 nanobody into peroxisomes, the average fluorescence of peroxisomes increased with the same kinetics in both channels (Fig. 7 K). Taken together, these results indicate that the nanobody lacking an SKL sequence can be transported “piggyback” together with mCherry-SKL into peroxisomes. We also performed the reverse experiment with crude Xenopus extract: GFP lacking an SKL sequence could be imported into peroxisomes with a nanobody containing a C-terminal SKL sequence (Fig. S5). Given that the nanobody can only bind to folded fluorescent proteins, the complex is likely transported without prior dissociation.
Figure 7.
Transport of a folded protein into peroxisomes. (A) mCherry-SKL (40 µM) was preincubated with mCherry-nanobodies labeled with Alexa Fluor 488 (nanobodyAlexa488; 40 µM) for 20 min at 20–23°C. The complex was added at 0.9 µM final concentration to cleared Xenopus egg extract. The sample was incubated for 2 h at 18°C and imaged with a spinning-disk confocal microscope for mCherry fluorescence. (B) The same field shown in A was imaged for nanobodyAlexa488 fluorescence. The foci in A and B overlap (compare foci in ovals). (C) As in A, but Alexa Fluor 488-fluorescence–quenching antibodies (0.5 µM) were added after the import reaction. (D) The same field as in C was analyzed for Alexa Fluor 488 fluorescence. The foci in C and D overlap (compare foci in ovals). (E) Quantification of the experiments in A–D. Shown are the combined data of two parallel experiments in the absence or presence of the quenching antibodies (>20 images per condition). The fluorescence in each channel observed in the presence of quencher was normalized relative to the mean fluorescence in the absence of quencher. The mean and standard deviation of the mean are given. (F) As in A, but with mCherry lacking SKL. (G) The same field shown in F was imaged for nanobodyAlexa488 fluorescence. (H) As in F, but Alexa Fluor 488-fluorescence–quenching antibodies (0.5 µM) were added after the import reaction. (I) The same field as in H was analyzed for Alexa Fluor 488 fluorescence. (J) Quantification of the experiments in F–I. Shown are the combined data of two parallel experiments in the absence or presence of the quenching antibodies (>20 images per condition). The fluorescence in each channel observed in the presence of quencher was normalized relative to the mean fluorescence in the absence of quencher. The mean and standard deviation of the mean are given. (K) Kinetics of the accumulation of mCherry-SKL and nanobodyAlexa488 in peroxisomes. The fluorescence in both channels was quantitated at the indicated time points by automated image analysis. Shown are the mean and standard deviation of the combined data of two parallel experiments (>20 images per time point). Bars, 5 µm.
Discussion
We have developed a new in vitro assay based on Xenopus egg extracts that recapitulates peroxisome protein import. In this system, fluorescent proteins that carry a SKL sequence at their C terminus are imported into peroxisomes in a process that is dependent on the import receptor Pex5 and its docking complex component Pex14. Sustained import is dependent on ATP hydrolysis and can occur with folded proteins.
Our in vitro system has several advantages over others reported before: import can be followed over time without fixation and is quantifiable, highly reproducible, and easily performed, as the extract can be frozen and thawed. Our “cleared extract” retains not only peroxisome protein import but also other activities seen with crude Xenopus extracts. The protocol for extract preparation is much simpler than a method recently reported for the study of spindle mechanics (Takagi and Shimamoto, 2017), and it might generally replace crude extracts, which cannot be subjected to a freeze–thaw cycle and therefore suffer from reproducibility.
Multiple pieces of evidence suggest that the system recapitulates actual protein import into peroxisomes, rather than mere binding to the surface. First, imported fluorescent substrate is inaccessible to quenching nanobodies or antibodies. Second, the substrate accumulates linearly over an extended time period, while Pex5 binding rapidly reaches a plateau. Third, substrate is released after freeze–thaw cycles or detergent addition. However, we found that only a small percentage of imported substrate is protected when reisolated peroxisomes are treated with protease (data not shown). One possibility is that the peroxisome membrane becomes fragile during proteolysis, but perhaps some portion of the imported substrate remained exposed to the cytosol.
We provide strong evidence that a folded protein lacking a SKL sequence can be piggyback imported into peroxisomes. Previous experiments also reached the conclusion that folded or oligomeric proteins can be imported (Brul et al., 1988; Walton et al., 1992, 1995), but caveats can be raised for each reported study. In several cases, it could not be excluded that proteins are first unfolded and then refolded inside peroxisomes. In other cases, import of folded proteins was not unambiguously demonstrated or its efficiency was not established. For example, microinjection of gold nanoparticles conjugated to a peroxisome targeting signal showed their import into peroxisomes, but many particles could be detected outside the peroxisomes, and the extent of enrichment in peroxisomes remained unclear (Walton et al., 1995). Most experiments assessed peroxisome localization by differential centrifugation, in which peroxisome content tends to distribute across many fractions. Perhaps the best data in the field come from in vivo piggyback experiments (Glover et al., 1994; McNew and Goodman, 1994; Elgersma et al., 1996), but it remained unclear how much of the substrate was really imported, rather than only associated with peroxisomes. In contrast, our experiments show that a prefolded SKL-containing fluorescent protein accumulates with the same kinetics as the associated nanobody in peroxisomes, and that the piggyback imported nanobody is completely shielded from quenching antibodies. In principle, it is conceivable that the protein complex dissociates after binding to peroxisomes and that actual membrane translocation of the two binding partners occurs sequentially, but their identical import kinetics would be difficult to explain, as the SKL-containing protein has an affinity for the docking complex, whereas the dissociated binding partner would be expected to frequently move back into the cytosol.
The mechanism by which folded proteins move across the peroxisome membrane remains unclear. However, in the only other known system in which folded proteins cross a membrane, the Tat secretion system in bacteria, two components distort the bilayer: the single-spanning TatA component appears to thin the bilayer through its short trans-membrane segment (Rodriguez et al., 2013), and the multi-spanning TatC protein has a deep cytosolic hydrophilic cavity (Rollauer et al., 2012; Ramasamy et al., 2013). Perhaps translocation across the peroxisome membrane also occurs through a protein-distorted lipid bilayer.
Our Pex5 depletion and rescue experiments show that the conserved Cys residue at the N terminus of Pex5 (Cys11 in mammals) can be replaced by a Lys residue, just as in experiments in yeast (Platta et al., 2007; Williams et al., 2007). According to the prevailing model of import, Pex5 would be ubiquitinated at the Cys or Lys and then be returned into the cytosol by the Pex1/Pex6 ATPase to start a new targeting cycle (Carvalho et al., 2007; Platta et al., 2007; Williams et al., 2007; Okumoto et al., 2011). Indeed, in our experiments, ATPγS inhibited import with some delay, as would be expected if the recycling of Pex5 was affected. Our results show that replacement of the Cys residue by Ala or Arg does not completely abolish import, which again would be consistent with this model if one assumes that these mutants can only perform one round of import. Surprisingly, however, the Ala and Arg mutants stimulate import when added to an extract that has not been depleted of Pex5, rather than being dominant-negative (Fig. 6 I). These results suggest that modification of the Cys residue of Pex5 is not absolutely required to clear the import sites on peroxisomes for the next round of translocation. The reported dominant-negative effect of the Cys11Ala mutant in mammalian tissue culture cells might be caused by its extended expression for 48 h (Okumoto et al., 2011).
In summary, this novel in vitro assay recapitulates peroxisome protein import, provides mechanistic insight, and will be useful for further dissecting the molecular mechanism of the process.
Materials and methods
Plasmids
Xenopus tropicalis Pex5 cDNA coding for the long isoform (GenBank accession no. BC088562.1) was purchased from GE Healthcare. The open reading frame of Pex5 was amplified by PCR and cloned into the expression vector pGEX-6p-3 (GE Healthcare) immediately downstream of the 3C protease cleavage site using the Gibson Assembly (NEB). The open reading frame of Xenopus Pex14 was synthesized (Genscript) based on a Pex14 protein sequence obtained from mass spectrometry data (Wühr et al., 2014). The N-terminal, Pex5-interacting domain of X. laevis Pex14 was predicted based on the homology to human Pex14 (PDB code 2W84). Pex14 residues 19 to 78 (cytPex14) were cloned into the expression vector pGEX-6p-3, as described for Pex5. Anti-GFP minimizer nanobody and anti-mCherry nanobody plasmids were purchased from Addgene (61838 and 70696, respectively). The open reading frames of the nanobodies were cloned into the vector pGEX-6p-3, as described for Pex5. Except for GFP, SKL-tagged and control fluorescent proteins constructs were made as described for Pex5, but SKL-tagged constructs contained the DNA sequence 5′-TCTAAACTG-3′ before the stop codon. GFP-SKL was purified via an N-terminal His tag on a Ni-NTA resin. cytATL (residues 1–462 of X. laevis atlastin) was synthetized by Genscript and cloned as described for Pex5. A ybbR-tagged Pex5 construct (Pex5-ybbR; Yin et al., 2006) was created by inserting the tag sequence at the 3′-end of a C-terminally extended isoform of X. tropicalis Pex5 (X1 Pex5; NCBI accession no. XP_012821977.1), using PCR of the vector followed by DNA phosphorylation and plasmid recircularization by ligation. Point mutations in the respective constructs were generated using the QuickChange Site-Directed Mutagenesis kit (Agilent Technologies). All plasmid inserts were confirmed by DNA sequencing.
Protein expression and purification
All recombinant proteins were expressed in BL21 (DE3) Rosetta (Novagen) E. coli strains (EMD Millipore). Bacterial cultures were grown at 37°C until OD600nm reached 0.5. Then 0.5 mM isopropyl β-D-1-thiogalactopyranoside was added to the culture, the temperature was reduced to 18°C, and incubation was continued for an additional 16 h. Harvested bacteria were lysed using a microfluidizer. GST-tagged protein was isolated using glutathione Sepharose 4B resin (GE Healthcare). The GST tag was removed by on-column digestion with GST-tagged 3C protease (BPS Bioscience) overnight at 4°C. The protein was further purified by gel filtration (Superdex 200; GE Healthcare). The samples were concentrated and subjected to centrifugation at 20,000 × g for 10 min at 4°C before snap-freezing in liquid nitrogen using 0.2-ml PCR tubes. The final storage buffer for all proteins was 20 mM Hepes, pH 7.8, 150 mM KCl, 250 mM sucrose, and 1 mM DTT.
Peptides
A nine-residue peptide corresponding to the C-terminal sequence of the peroxisome matrix protein malate dehydrogenase 3 from Saccharomyces cerevisiae (N-EFILDSSKL-C) was synthesized by Genscript. A control peptide was synthesized in which the C-terminal SKL sequence was changed to KLS. These peptides contained acetylated N termini and unmodified C termini. The lyophilized peptides were resuspended at 10 mM in 100 mM Hepes, pH 7.8, and 100 mM KCl.
Fluorescent labeling of proteins
The anti-mCherry nanobody was labeled at Lys residues using N-hydroxysuccinimide-ester Alexa Fluor 488 (Thermo Fisher Scientific) as follows. 120 µM nanobody was incubated with 240 µM of the dye in reaction buffer (50 mM Hepes, pH 7.8, and 100 mM KCl) for 2 h at 23°C with gentle agitation. The reaction was stopped by addition of 100 mM Tris buffer, pH 7.8, and unreacted dye was removed by gel filtration on a Superdex 200 10/30 column (GE Healthcare), using 30 mM Hepes, pH 7.8, and 150 mM KCl as a running buffer. The efficiency of the labeling reaction was determined to be 30%, using extinction coefficients of 23,000 M−1cm−1 at 280 nm for the nanobody and 73.000 M−1cm−1 at 490 nm for the Alexa Fluor 488 dye. To label the Pex5-ybbR protein, reduced coenzyme A (Sigma-Aldrich) was incubated with Alexa Fluor 488–maleimide (Thermo Fisher Scientific) as follows. 2 mM coenzyme A was combined with 3 mM Alexa Fluor 488–maleimide in reaction buffer (50 mM Hepes, pH 7.8, and 100 mM KCl). The sample was incubated at 23°C with gentle agitation for 2.5 h, and the reaction was terminated by addition of 10 mM DTT. Next, 40 µM of labeled coenzyme A was mixed with 15 µM Pex5-ybbR and 6 µM Bacillus subtilis 4′-phosphopantetheinyl transferase (SFP synthase) in 50 mM Hepes, pH 7.8, and 10 mM MgCl2. The reaction was incubated at 37°C for 1 h, and then the labeled protein was purified on a Superdex 200 10/300 column, using 30 mM Hepes, pH 7.8, and 150 mM KCl as a running buffer. The efficiency of the labeling reaction was determined to be 49%, using extinction coefficients of 98.890 M−1cm−1 at 280 nm for Pex5-ybbR and 73.000 M−1cm−1 at 495 nm for the Alexa Fluor 488 dye. Enhancer nanobody against GFP was labeled with Dylight-650 maleimide (Life Technologies) at a 2:1 ratio for several hours. The reaction was quenched with 10 mM β-mercaptoethanol, and excess dye was removed by gel filtration.
Antibodies to Pex5
Antibodies against purified Pex5 were raised in rabbits by Thermo Fisher Scientific. The terminal bleeds were used for affinity purification of the antibodies. 4 ml of packed Affigel-15 beads (BioRad) was incubated with 24 mg of purified Pex5 in 30 mM Hepes, pH 7.8, and 100 mM KCl overnight at 4°C. The beads were washed and incubated overnight at 4°C with 12 ml of serum in the presence of protease inhibitors. After washing, the antibodies were eluted with 100 mM glycine HCl, pH 2.4, and 150 mM KCl. The eluate was collected in one tenth the volume of 1 M Hepes, pH 7.8. The buffer was exchanged to 30 mM Hepes, pH 7.8, 100 KCl, and 0.25 M sucrose, and aliquots of the sample were frozen in liquid nitrogen.
Antibodies to Pex14
Antibodies to Pex14 were raised in rabbits against a GST fusion to Pex14 residues 19 to 78 (GST-cytPex14). Affinity purification was done as described for Pex5 antibodies, using cytPex14 coupled to beads.
Depletion of Pex5 from Xenopus extracts by immobilized Pex5 antibodies or Pex14 fragment
The purified Pex5 antibodies (600 µl at 1.5 mg/ml) were coupled to Affigel-15 resin (400 µl) by incubation overnight at 4°C. The beads were washed with buffer and used for depletion of Pex5 from Xenopus extracts. The purified Pex14 fragment (6 mg) was incubated with 1 ml of Affigel-15 beads overnight at 4°C. The beads were washed with buffer and used for depletion of Pex5 from Xenopus extracts. Cleared extract (300 µl) was incubated with 30 µl of packed beads for 20 min at room temperature on a rotator. To remove the beads, the samples were centrifuged on a Micro-spin column (Pierce) at 300 × g.
Preparation of Xenopus cleared egg extracts
Interphase crude extracts from X. laevis eggs were prepared as described previously (Desai et al., 1999), with the following modifications. All steps were performed at 18°C. Eggs were de-jellied using XB buffer (10 mM Hepes, pH 7.7, 0.1 M KCl, 1 mM MgCl2, 0.1 mM CaCl2, and 50 mM sucrose) supplemented with 2.4% wt/vol cysteine (Sigma-Aldrich) adjusted to pH 7.8. De-jellied eggs were washed several times with XB buffer and packed in a centrifuge tube (344057; Beckman) by spinning at 300 × g for 1 min, followed by 900 × g for 20 s. Excess buffer was removed, and after addition of protease inhibitors (10 µg/ml leupeptin, 10 µg/ml pepstatin, and 10 µg/ml chymostatin; all from Sigma-Aldrich), the eggs were subjected to a crush spin at 12,100 × g (10,000 rpm in a SW55 Ti Beckman rotor) for 15 min at 16°C. Note that no cytochalasin B, cycloheximide, or energy-regenerating system was added. From here on, the crude extract was kept on ice at all times. The crude extract was carefully collected from tubes by side-puncture using a syringe with an 18-gauge needle and supplemented again with protease inhibitors. The crude extract was placed in centrifuge tubes (347356; Beckman) and spun at 166,000 × gav (50,000 rpm in a TLS-55 Beckman rotor) for 50 min at 4°C. The layer immediately above the light membrane fraction (the cleared extract) was carefully collected from the top of the tube using a wide-bore pipette tip. After mixing, the cleared extract was divided into 50-µl aliquots in PCR tubes, snap-frozen in liquid nitrogen, and stored at −80°C.
Peroxisome import assay using cleared extract from Xenopus eggs
Cleared extract aliquots were thawed and maintained on ice at all times. Typical import reactions were prepared in 0.5-ml tubes by pipetting 20 µl cleared extract followed by small molecules or proteins being tested, other than the substrates. When ATPγS was included, the reaction mix was incubated for <20 min at 23°C before addition of the fluorescent protein. The import reaction was started by addition of SKL-tagged fluorescent protein (or a control fluorescent protein lacking SKL), and the incubation was continued for >1 h at 18°C before imaging. 8 µl of the reaction mix was mounted between two polyethylene glycol (PEG)–passivated square glass coverslips (22 × 22 mm), sealed with VALAP (vaseline, lanolin, and paraffin mixed at 1:1:1 weight ratio), and imaged on a spinning-disk fluorescence confocal microscope. When the import reaction was followed in real time, a 20-µl reaction volume was mounted as described and maintained on the microscope stage. The incubation temperature was 20–23°C. At least 10 randomly selected microscope fields were captured at each indicated time point. Photobleaching before image acquisition was minimized by using the Perfect Focus System (Nikon), where the microscope objective lens is positioned on a previously unexposed field and the image is acquired without preexposure to excitation light.
Sedimentation of peroxisomes after protein import
A 100-µl import reaction with 0.6 µM mScarlet-SKL was performed for 2 h at 18°C. The sample was centrifuged in a Beckman polypropylene tube (342630) in a TLS-55 swinging bucket rotor at 50,000 rpm for 25 min at 4°C using Teflon tube adapters. The membrane pellet was resuspended in 190 μl of 30 mM Hepes, pH 7.8, 100 mM KCl, 2 mM DTT, protease inhibitors, and 250 mM sucrose using a wide-bore pipette tip. The sample was centrifuged at 30,000 rpm for 25 min at 4°C in a TLS-55 swinging bucket rotor. Resuspension and sedimentation were repeated, and the sample was finally resuspended in 40 µl buffer (final volume ∼60–80 µl).
Flotation of peroxisomes after protein import
A 75-µl import reaction with 0.6 µM mScarlet-SKL was performed for 5 h at 18°C and placed in a TLS-55 centrifuge tube (347356; Beckman). A total of 225 µl of 70% sucrose in 30 mM Hepes, pH 7.8, 100 mM KCl, and 2 mM DTT was added in 20-µl increments with mixing in between. The sample was overlaid with 1 ml of 50% sucrose and with 100 µl of 8.5% sucrose and then centrifuged at 4°C in a TLS-55 swinging bucket rotor for 1.5 h at 50,000 rpm. Fractions (200 µl) were taken from the bottom and analyzed by fluorescence imaging and by SDS-PAGE, followed by immunoblotting with affinity-purified Pex14 antibodies.
Fluorescent staining of ER networks in cleared egg extract
Staining of the ER network was performed as previously described for crude extracts (Wang et al., 2019). 30 µl of extract was incubated with 100 µg/ml DiI-C18 for 45 min at 18°C and diluted 1:10 with fresh extract. After 15 min, the sample was imaged in a fluorescence microscope.
Glass coverslip passivation
Square glass coverslips (48366–227; VWR) were placed in metal racks; rinsed sequentially with pure water, ethanol, and pure water; and then air-dried. Coverslips were treated with an oxygen-plasma etcher (500-II Plasma System; Technics) for 4 min at 200-W power. Immediately thereafter, one side of the glass coverslip was layered with a 35 mg/ml solution of 20,000 molecular weight methoxy-polyethylene glycol-silane (JenKem Technology USA), dissolved in 95% ethanol/4.9% water/0.1% glacial acetic acid, and incubated at 80°C for >5 h. Coverslips were stored in a sealed container at room temperature. Prior to use, they were rinsed with ethanol and pure water, and then air dried.
Fluorescence microscopy
All samples were visualized using a spinning-disk confocal head (CSU-X1; Yokogawa) with Borealis modification (Spectral Applied Research) and a quad bandpass 405/491/561/642 dichroic mirror (Semrock). The confocal was mounted on a Ti inverted microscope (Nikon) equipped with a 60× Plan Apo NA 1.4 oil immersion objective or 60× CFI Plan Apochromat NA 1.2 water immersion objective and the Perfect Focus System for continuous maintenance of focus (Nikon). Green fluorescence images were collected using a 491-nm solid-state laser controlled with an acousto-optic tunable filter (AOTF; Spectral Applied Research) and ET525/50 emission filter (Chroma Technology). Red fluorescence images were collected using a 561-nm solid-state laser controlled with an AOTF and ET620/60 emission filter (Chroma Technology). All images were acquired with a cooled charge coupled device camera (ORCA AG; Hamamatsu Photonics) controlled with MetaMorph software (version 7.0; Molecular Devices) and archived using ImageJ (National Institutes of Health). In some cases, linear adjustments were applied to enhance the contrast of images using levels in the image adjustments function of ImageJ.
Quantitative data analysis
An automated ImageJ macro script was written to segment and quantify the fluorescence in peroxisomes. The initialization parameters of the automated script were determined for each time course as follows. First, the standard deviation of the background fluorescence was determined in randomly selected regions that did not contain peroxisomes. Second, single foci were randomly selected, and the fluorescence profile on a line across the center of the particle was fit to a Gaussian distribution. This resulted in estimates of particle size and its standard deviation. These parameters were then used in an automated script to segment individual peroxisomes and calculate their mean fluorescence intensity in all images. Specifically, for each raw image, the background was subtracted using a rolling-ball procedure (Sternberg, 1983) with a ball diameter twice the preestimated particle size. Then, the image was subjected to a Laplacian filter to enhance the contrast of the particles regardless of their fluorescence intensity. The filter used as a parameter the previously calculated particle size standard deviation. The resulting images were then inverted, as they originally show the particles as dark objects on a bright background. The images were then segmented using a standard automated segmentation routine based on statistical region-merging (Robust Automatic Threshold Selection ImageJ plugin; Wilkinson and Schut, 1998). During this analysis, the pre-estimated standard deviation of background pixel values was used as a parameter. The resulting image was converted to a binary image, where the positive values indicate the position, size, and shape of the peroxisomes. These images were then used in the standard ImageJ “analyze-particles” function to generate a list of regions of interest (ROIs). The ROIs were then transferred to the background-subtracted initial unprocessed image and used to determine the mean fluorescence per peroxisome. For each time point, >10 images were analyzed, giving 15,000–20,000 particles. The number of ROIs gives the number of peroxisomes.
Online supplemental material
Fig. S1 shows the preparation and characteristics of cleared Xenopus egg extract. Fig. S2 shows purified proteins analyzed by SDS-PAGE. Fig. S3 shows that ATP depletion blocks peroxisome protein import. Fig. S4 compares the affinity of WT and mutant Pex5 for import substrate. Fig. S5 shows that GFP can be piggyback imported by nanobody-SKL. Video 1 shows the mobility of puncta labeled with mScarlet-SKL followed in real time.
Supplementary Material
Acknowledgments
We thank the Nikon Imaging Center at Harvard Medical School, in particular Jennifer Waters, and Clarence Yapp from the Image and Data Analysis Core and Marco Catipovic for advice with automated image analysis.
T.A. Rapoport is a Howard Hughes Medical Institute investigator.
The authors declare no competing financial interests.
Author contributions: F.B. Romano and N.B. Blok performed all experiments. T.A. Rapoport wrote the paper together with F.B. Romano and N.B. Blok.
References
- Agrawal G., and Subramani S.. 2016. De novo peroxisome biogenesis: Evolving concepts and conundrums. Biochim. Biophys. Acta. 1863:892–901. 10.1016/j.bbamcr.2015.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertini M., Rehling P., Erdmann R., Girzalsky W., Kiel J.A., Veenhuis M., and Kunau W.H.. 1997. Pex14p, a peroxisomal membrane protein binding both receptors of the two PTS-dependent import pathways. Cell. 89:83–92. 10.1016/S0092-8674(00)80185-3 [DOI] [PubMed] [Google Scholar]
- Baldridge R.D., and Rapoport T.A.. 2016. Autoubiquitination of the Hrd1 ligase triggers protein retrotranslocation in ERAD. Cell. 166:394–407. 10.1016/j.cell.2016.05.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braverman N.E., and Moser A.B.. 2012. Functions of plasmalogen lipids in health and disease. Biochim. Biophys. Acta. 1822:1442–1452. 10.1016/j.bbadis.2012.05.008 [DOI] [PubMed] [Google Scholar]
- Braverman N., Dodt G., Gould S.J., and Valle D.. 1998. An isoform of pex5p, the human PTS1 receptor, is required for the import of PTS2 proteins into peroxisomes. Hum. Mol. Genet. 7:1195–1205. 10.1093/hmg/7.8.1195 [DOI] [PubMed] [Google Scholar]
- Braverman N.E., D’Agostino M.D., and Maclean G.E.. 2013. Peroxisome biogenesis disorders: Biological, clinical and pathophysiological perspectives. Dev. Disabil. Res. Rev. 17:187–196. 10.1002/ddrr.1113 [DOI] [PubMed] [Google Scholar]
- Brocard C., Kragler F., Simon M.M., Schuster T., and Hartig A.. 1994. The tetratricopeptide repeat-domain of the PAS10 protein of Saccharomyces cerevisiae is essential for binding the peroxisomal targeting signal-SKL. Biochem. Biophys. Res. Commun. 204:1016–1022. 10.1006/bbrc.1994.2564 [DOI] [PubMed] [Google Scholar]
- Brul S., Wiemer E.A., Westerveld A., Strijland A., Wanders R.J., Schram A.W., Heymans H.S., Schutgens R.B., Van den Bosch H., and Tager J.M.. 1988. Kinetics of the assembly of peroxisomes after fusion of complementary cell lines from patients with the cerebro-hepato-renal (Zellweger) syndrome and related disorders. Biochem. Biophys. Res. Commun. 152:1083–1089. 10.1016/S0006-291X(88)80395-4 [DOI] [PubMed] [Google Scholar]
- Carvalho A.F., Pinto M.P., Grou C.P., Alencastre I.S., Fransen M., Sá-Miranda C., and Azevedo J.E.. 2007. Ubiquitination of mammalian Pex5p, the peroxisomal import receptor. J. Biol. Chem. 282:31267–31272. 10.1074/jbc.M706325200 [DOI] [PubMed] [Google Scholar]
- Desai A., Murray A., Mitchison T.J., and Walczak C.E.. 1999. The use of Xenopus egg extracts to study mitotic spindle assembly and function in vitro. Methods Cell Biol. 61:385–412. 10.1016/S0091-679X(08)61991-3 [DOI] [PubMed] [Google Scholar]
- Elgersma Y., Vos A., van den Berg M., van Roermund C.W., van der Sluijs P., Distel B., and Tabak H.F.. 1996. Analysis of the carboxyl-terminal peroxisomal targeting signal 1 in a homologous context in Saccharomyces cerevisiae. J. Biol. Chem. 271:26375–26382. 10.1074/jbc.271.42.26375 [DOI] [PubMed] [Google Scholar]
- Erdmann R., and Blobel G.. 1996. Identification of Pex13p a peroxisomal membrane receptor for the PTS1 recognition factor. J. Cell Biol. 135:111–121. 10.1083/jcb.135.1.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki Y. 2016. Peroxisome biogenesis and human peroxisome-deficiency disorders. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 92:463–477. 10.2183/pjab.92.463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki Y., and Lazarow P.B.. 1985. Post-translational import of fatty acyl-CoA oxidase and catalase into peroxisomes of rat liver in vitro. J. Biol. Chem. 260:5603–5609. [PubMed] [Google Scholar]
- Gatto G.J.J. Jr., Geisbrecht B.V., Gould S.J., and Berg J.M.. 2000. Peroxisomal targeting signal-1 recognition by the TPR domains of human PEX5. Nat. Struct. Biol. 7:1091–1095. 10.1038/81930 [DOI] [PubMed] [Google Scholar]
- Glover J.R., Andrews D.W., and Rachubinski R.A.. 1994. Saccharomyces cerevisiae peroxisomal thiolase is imported as a dimer. Proc. Natl. Acad. Sci. USA. 91:10541–10545. 10.1073/pnas.91.22.10541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould S.J., Keller G.-A., Hosken N., Wilkinson J., and Subramani S.. 1989. A conserved tripeptide sorts proteins to peroxisomes. J. Cell Biol. 108:1657–1664. 10.1083/jcb.108.5.1657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould S.J., Kalish J.E., Morrell J.C., Bjorkman J., Urquhart A.J., and Crane D.I.. 1996. Pex13p is an SH3 protein of the peroxisome membrane and a docking factor for the predominantly cytoplasmic PTs1 receptor. J. Cell Biol. 135:85–95. 10.1083/jcb.135.1.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettema E.H., Erdmann R., van der Klei I., and Veenhuis M.. 2014. Evolving models for peroxisome biogenesis. Curr. Opin. Cell Biol. 29:25–30. 10.1016/j.ceb.2014.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhofer A., Helma J., Schmidthals K., Frauer C., Cui S., Karcher A., Pellis M., Muyldermans S., Casas-Delucchi C.S., Cardoso M.C., et al. . 2010. Modulation of protein properties in living cells using nanobodies. Nat. Struct. Mol. Biol. 17:133–138. 10.1038/nsmb.1727 [DOI] [PubMed] [Google Scholar]
- Léon S., Goodman J.M., and Subramani S.. 2006. Uniqueness of the mechanism of protein import into the peroxisome matrix: transport of folded, co-factor-bound and oligomeric proteins by shuttling receptors. Biochim. Biophys. Acta. 1763:1552–1564. 10.1016/j.bbamcr.2006.08.037 [DOI] [PubMed] [Google Scholar]
- McCollum D., Monosov E., and Subramani S.. 1993. The pas8 mutant of Pichia pastoris exhibits the peroxisomal protein import deficiencies of Zellweger syndrome cells--the PAS8 protein binds to the COOH-terminal tripeptide peroxisomal targeting signal, and is a member of the TPR protein family. J. Cell Biol. 121:761–774. 10.1083/jcb.121.4.761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNew J.A., and Goodman J.M.. 1994. An oligomeric protein is imported into peroxisomes in vivo. J. Cell Biol. 127:1245–1257. 10.1083/jcb.127.5.1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuberger G., Maurer-Stroh S., Eisenhaber B., Hartig A., and Eisenhaber F.. 2003. Motif refinement of the peroxisomal targeting signal 1 and evaluation of taxon-specific differences. J. Mol. Biol. 328:567–579. 10.1016/S0022-2836(03)00318-8 [DOI] [PubMed] [Google Scholar]
- Neufeld C., Filipp F.V., Simon B., Neuhaus A., Schüller N., David C., Kooshapur H., Madl T., Erdmann R., Schliebs W., et al. . 2009. Structural basis for competitive interactions of Pex14 with the import receptors Pex5 and Pex19. EMBO J. 28:745–754. 10.1038/emboj.2009.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumoto K., Misono S., Miyata N., Matsumoto Y., Mukai S., and Fujiki Y.. 2011. Cysteine ubiquitination of PTS1 receptor Pex5p regulates Pex5p recycling. Traffic. 12:1067–1083. 10.1111/j.1600-0854.2011.01217.x [DOI] [PubMed] [Google Scholar]
- Okumoto K., Honsho M., Liu Y., and Fujiki Y.. 2017. Peroxisomal Membrane and Matrix Protein Import Using a Semi-Intact Mammalian Cell System. Methods Mol. Biol. 1595:213–219. 10.1007/978-1-4939-6937-1_20 [DOI] [PubMed] [Google Scholar]
- Otera H., Okumoto K., Tateishi K., Ikoma Y., Matsuda E., Nishimura M., Tsukamoto T., Osumi T., Ohashi K., Higuchi O., and Fujiki Y.. 1998. Peroxisome targeting signal type 1 (PTS1) receptor is involved in import of both PTS1 and PTS2: studies with PEX5-defective CHO cell mutants. Mol. Cell. Biol. 18:388–399. 10.1128/MCB.18.1.388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platta H.W., Grunau S., Rosenkranz K., Girzalsky W., and Erdmann R.. 2005. Functional role of the AAA peroxins in dislocation of the cycling PTS1 receptor back to the cytosol. Nat. Cell Biol. 7:817–822. 10.1038/ncb1281 [DOI] [PubMed] [Google Scholar]
- Platta H.W., El Magraoui F., Schlee D., Grunau S., Girzalsky W., and Erdmann R.. 2007. Ubiquitination of the peroxisomal import receptor Pex5p is required for its recycling. J. Cell Biol. 177:197–204. 10.1083/jcb.200611012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramasamy S., Abrol R., Suloway C.J.M., and Clemons W.M. Jr. 2013. The glove-like structure of the conserved membrane protein TatC provides insight into signal sequence recognition in twin-arginine translocation. Structure. 21:777–788. 10.1016/j.str.2013.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues T.A., Francisco T., Dias A.F., Pedrosa A.G., Grou C.P., and Azevedo J.E.. 2016. A cell-free organelle-based in vitro system for studying the peroxisomal protein import machinery. Nat. Protoc. 11:2454–2469. 10.1038/nprot.2016.147 [DOI] [PubMed] [Google Scholar]
- Rodriguez F., Rouse S.L., Tait C.E., Harmer J., De Riso A., Timmel C.R., Sansom M.S.P., Berks B.C., and Schnell J.R.. 2013. Structural model for the protein-translocating element of the twin-arginine transport system. Proc. Natl. Acad. Sci. USA. 110:E1092–E1101. 10.1073/pnas.1219486110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollauer S.E., Tarry M.J., Graham J.E., Jääskeläinen M., Jäger F., Johnson S., Krehenbrink M., Liu S.-M., Lukey M.J., Marcoux J., et al. . 2012. Structure of the TatC core of the twin-arginine protein transport system. Nature. 492:210–214. 10.1038/nature11683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J.J., and Aitchison J.D.. 2013. Peroxisomes take shape. Nat. Rev. Mol. Cell Biol. 14:803–817. 10.1038/nrm3700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg S.R. 1983. Biomedical Image Processing. IEEE Computer. 16:22–34. 10.1109/MC.1983.1654163 [DOI] [Google Scholar]
- Takagi J., and Shimamoto Y.. 2017. High-quality frozen extracts of Xenopus laevis eggs reveal size-dependent control of metaphase spindle micromechanics. Mol. Biol. Cell. 28:2170–2177. 10.1091/mbc.e17-03-0174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Leij I., Franse M.M., Elgersma Y., Distel B., and Tabak H.F.. 1993. PAS10 is a tetratricopeptide-repeat protein that is essential for the import of most matrix proteins into peroxisomes of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 90:11782–11786. 10.1073/pnas.90.24.11782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton P.A., Gould S.J., Feramisco J.R., and Subramani S.. 1992. Transport of microinjected proteins into peroxisomes of mammalian cells: inability of Zellweger cell lines to import proteins with the SKL tripeptide peroxisomal targeting signal. Mol. Cell. Biol. 12:531–541. 10.1128/MCB.12.2.531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton P.A., Hill P.E., and Subramani S.. 1995. Import of stably folded proteins into peroxisomes. Mol. Biol. Cell. 6:675–683. 10.1091/mbc.6.6.675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanders R.J.A. 2014. Metabolic functions of peroxisomes in health and disease. Biochimie. 98:36–44. 10.1016/j.biochi.2013.08.022 [DOI] [PubMed] [Google Scholar]
- Wang S., Romano F.B., and Rapoport T.A.. 2019. Endoplasmic Reticulum Network Formation with Xenopus Egg Extracts. Cold Spring Harb. Protoc. 2019:pdb.prot097204 10.1101/pdb.prot097204 [DOI] [PubMed] [Google Scholar]
- Waterham H.R., Ferdinandusse S., and Wanders R.J.A.. 2016. Human disorders of peroxisome metabolism and biogenesis. Biochim. Biophys. Acta. 1863:922–933. 10.1016/j.bbamcr.2015.11.015 [DOI] [PubMed] [Google Scholar]
- Wendland M., and Subramani S.. 1993. Cytosol-dependent peroxisomal protein import in a permeabilized cell system. J. Cell Biol. 120:675–685. 10.1083/jcb.120.3.675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson M.H.F., and Schut F.. 1998. Digital image analysis: imaging, morphometry, fluorometry and mitlity techniques and applications. Wiley, New York. [Google Scholar]
- Williams C., van den Berg M., Sprenger R.R., and Distel B.. 2007. A conserved cysteine is essential for Pex4p-dependent ubiquitination of the peroxisomal import receptor Pex5p. J. Biol. Chem. 282:22534–22543. 10.1074/jbc.M702038200 [DOI] [PubMed] [Google Scholar]
- Wühr M., Freeman R.M. Jr., Presler M., Horb M.E., Peshkin L., Gygi S., and Kirschner M.W.. 2014. Deep proteomics of the Xenopus laevis egg using an mRNA-derived reference database. Curr. Biol. 24:1467–1475. 10.1016/j.cub.2014.05.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J., Lin A.J., Golan D.E., and Walsh C.T.. 2006. Site-specific protein labeling by Sfp phosphopantetheinyl transferase. Nat. Protoc. 1:280–285. 10.1038/nprot.2006.43 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.