Tunneling nanotubes (TNT) are membranous tubes that connect two cells, but their functional roles and mechanism of biogenesis remain obscure. Sharma and Subramaniam demonstrate that Rhes, a brain-enriched protein, increases biogenesis of TNT-like cellular protrusions or “Rhes tunnels” through which Rhes travels from cell to cell and transports Huntington disease (HD) protein.
Abstract
Tunneling nanotubes (TNT) are thin, membranous, tunnel-like cell-to-cell connections, but the mechanisms underlying their biogenesis or functional role remains obscure. Here, we report, Rhes, a brain-enriched GTPase/SUMO E3-like protein, induces the biogenesis of TNT-like cellular protrusions, “Rhes tunnels,” through which Rhes moves from cell to cell and transports Huntington disease (HD) protein, the poly-Q expanded mutant Huntingtin (mHTT). The formation of TNT-like Rhes tunnels requires the Rhes’s serine 33, C-terminal CAAX, and a SUMO E3-like domain. Electron microscopy analysis revealed that TNT-like Rhes tunnels appear continuous, cell–cell connections, and <200 nm in diameter. Live-cell imaging shows that Rhes tunnels establish contact with the neighboring cell and deliver Rhes-positive cargoes, which travel across the plasma membrane of the neighboring cell before entering it. The Rhes tunnels carry Rab5a/Lyso 20-positive vesicles and transport mHTT, but not normal HTT, mTOR, or wtTau proteins. SUMOylation-defective mHTT, Rhes C263S (cannot SUMOylate mHTT), or CRISPR/Cas9-mediated depletion of three isoforms of SUMO diminishes Rhes-mediated mHTT transport. Thus, Rhes promotes the biogenesis of TNT-like cellular protrusions and facilitates the cell–cell transport of mHTT involving SUMO-mediated mechanisms.
Introduction
Cell–cell communications, such as synaptic connections, gap junctions, and exosomes, are fundamental to living organisms (Lloyd and McIntyre, 1955; Farquhar and Palade, 1965; Johnstone et al., 1987; Beier et al., 2018; Cervera et al., 2018; Stahl and Raposo, 2018). The tunneling nanotubes (TNTs), the fragile and inconspicuous membranous tunnel-like structures ranging 50 to 200 nm in diameter and 5 to 125 µm in length connecting two cells, have been reported in diverse cell types (Rustom et al., 2004; Gerdes et al., 2007; Hase et al., 2009; Lou et al., 2012; Gousset et al., 2013; Schiller et al., 2013; Austefjord et al., 2014; Burtey et al., 2015; Polak et al., 2015; Wang and Gerdes, 2015; Delage et al., 2016; Desir et al., 2016; Zhu et al., 2016; Keller et al., 2017; Vignais et al., 2017; Dupont et al., 2018; Panasiuk et al., 2018). TNTs lack specific markers, and they are often indistinguishable from a long, filopodia-like protrusion. Thus, their detection in a complex microenvironment in vivo remains a challenge. But elongated protrusions similar to TNTs, termed cytonemes, which contain vesicles on their tip, have been demonstrated in Drosophila melanogaster embryos, and in diverse cell types in vivo (Miller et al., 1995; Ramírez-Weber and Kornberg, 1999; Salas-Vidal and Lomelí, 2004; Teddy and Kulesa, 2004; Chinnery et al., 2008; Pyrgaki et al., 2010; Caneparo et al., 2011). TNTs have been implicated in the transfer of cellular components, such as RNA, calcium signals, proteins, and organelles, and in the formation of electrical and mechanical coupling between cells, as well as transport of viruses and spreading of neurodegenerative disease–linked proteins (Sowinski et al., 2008; Wittig et al., 2012; Gerdes et al., 2013; Abounit et al., 2016; Hashimoto et al., 2016; Jansens et al., 2017; Kumar et al., 2017; Guo et al., 2018; Panasiuk et al., 2018).
Huntington disease (HD) is a monogenic disorder attributable to polyglutamine (>36Q) expansion in Huntingtin (mHTT), a ubiquitously expressed protein. But it is unclear how mHTT promotes the degeneration of the brain’s striatum, a region that controls motor, cognitive, and psychiatric functions (Vonsattel et al., 1985; Reiner et al., 1988; Subramaniam and Snyder, 2011; McColgan and Tabrizi, 2018). Multiple studies have suggested a neuron-to-neuron migration of mHTT both in HD animal models and in human HD patients. The mHTT aggregates were found in healthy striatal cell transplants in the striatum of HD patients (Cicchetti et al., 2014). Healthy human neurons were found to contain mHTT when co-cultured with HD mouse brain slices (Pecho-Vrieseling et al., 2014). In Drosophila, mHTT was found to spread from olfactory receptor neurons to various parts of the brain (Babcock and Ganetzky, 2015). Similarly, human mHTT was found in the striatum of normal mice that had received intraventricular placement of human HD neurons (Jeon et al., 2016). Although the exact mechanisms are unclear, exosomes, i.e., secretory, vesicle-mediated pathways, have been proposed for the transfer of mHTT between cells (Jeon et al., 2016). But the mHTT appears not present in exosomes derived from astrocytic culture (Hong et al., 2017). One study showed that mHTT can be transported via TNTs in CAD cells (Costanzo et al., 2013). Thus, a precise molecular mechanism that underlies the potential transportation of mHTT, and, more important, how it relates to the massive loss of neuronal cells in the striatum in HD, remains unclear (Vonsattel et al., 1985; Kassubek et al., 2004; Subramaniam and Snyder, 2011).
Previously, we linked Rhes, a small GTPase highly enriched in the striatum, to striatal cell loss in HD (Subramaniam and Snyder, 2011). We found that Rhes, a physiological regulator of SUMOylation, interacts with mHTT and promotes its small ubiquitin-like modifier (SUMO) modification, which increases soluble forms of mHTT and promotes cellular toxicity (Subramaniam et al., 2009, 2010). In animal models, we found that the deletion of Rhes prevented HD-related motor and striatal damage, and Rhes overexpression worsened HD-related deficits (Swarnkar et al., 2015). The toxic role of Rhes was demonstrated in several other HD models, including human embryonic stem cell–derived striatal neuronal cells (Okamoto et al., 2009; Seredenina et al., 2011; Baiamonte et al., 2013; Lu and Palacino, 2013; Sbodio et al., 2013; Argenti, 2014). Despite this, the downstream mechanisms by which Rhes and mHTT promote striatal cell toxicity have yet to be resolved.
Here, we report for the first time a novel role for Rhes in the formation of TNT-like intercellular communication, which serves as “highways” for cell–cell transport of Rhes and mHTT, thus providing new insights into Rhes signaling in the striatum and its vulnerability in HD.
Results
Rhes promotes filopodia-like cellular protrusions in striatal neuronal cells
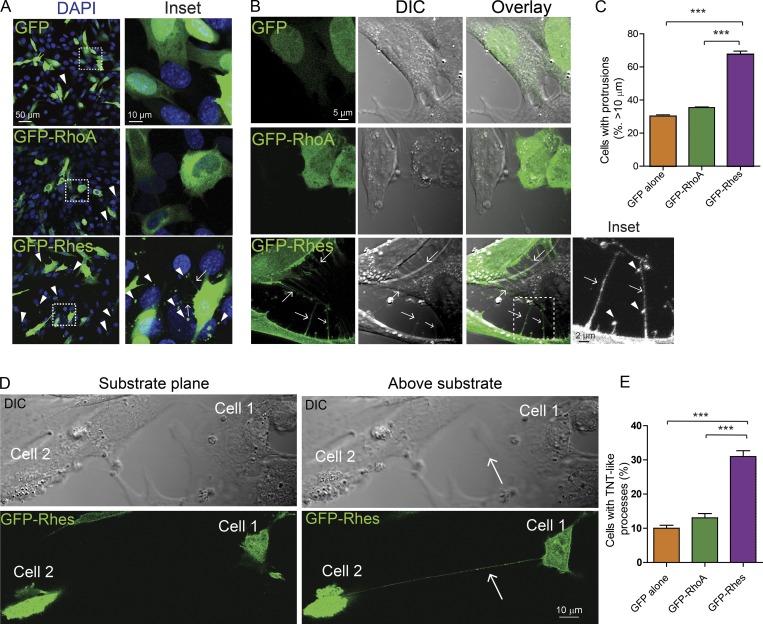
To gain deeper insights into the intracellular distribution of Rhes, we expressed the GFP-tagged Rhes in STHdhQ7/Q7 cells (Trettel et al., 2000), and included controls, GFP alone or GFP-RhoA (Rho family of GTPase). We found numerous GFP-Rhes puncta in neighboring cells that were not transfected with GFP-Rhes (Fig. 1 A, arrowhead). While ∼22% of untransfected cells showed GFP-Rhes puncta per field, that number was ∼4% and ∼9% in GFP alone and in GFP-RhoA–transfected control cells, respectively (Fig. S1 A). Cells expressing GFP-Rhes, but not GFP or GFP-RhoA, revealed conspicuous filopodia-like cellular protrusions that appear connected to the adjacent cells (Fig. 1 A, inset, arrow). Analysis of images captured using differential interference contrast (DIC) microscopy (Fig. 1, B and C) revealed ∼70% of GFP-Rhes–positive cells showed the filopodia-like cellular protrusions (Fig. 1 B, inset, arrow) with vesicle-like puncta (Fig. 1 B, inset, arrowhead), compared with ∼30% in GFP alone or ~35% in GFP-RhoA–expressing cells (Fig. 1 C). In summary, Rhes induces the cellular protrusions that resemble the TNTs in PC12 cells (Rustom et al., 2004) or the “cytonemes” in Drosophila (Ramírez-Weber and Kornberg, 1999; Fig. S1 B, arrowhead). Currently, there are no cellular markers that distinguish cytonemes from TNTs. However, cytonemes appear do not attach to target cells, while TNTs form an open-ended connection between two cells, often hovering above the substratum (Dupont et al., 2018). We found that Rhes-induced protrusions are above the substratum connecting two cells, similar to TNT (Fig. 1 D, arrow). Next, we found ∼30% of GFP-Rhes cells showed TNT-like structures (connecting two cells), compared with ∼10% and ∼13% in GFP alone and GFP-RhoA cells, respectively (Fig. 1 E). Thus, Rhes is a potent inducer of filopodia-like protrusions, resembling TNTs in striatal neuronal cells.
Figure 1.
Rhes promotes filopodia-like cellular protrusions in striatal neuronal cells. (A) Striatal neuronal cells (STHdhQ7/Q7) expressing GFP alone or GFP-RhoA or GFP-Rhes. Inset: Arrowheads show GFP puncta in the untransfected cells, and arrows show the filopodia-like process. DAPI indicates nuclei. (B) Bright field images (DIC) of striatal neuronal cells expressing GFP alone or GFP-RhoA or GFP-Rhes. Arrows point to filopodia-like protrusions. Inset, arrowheads indicate vesicle-like structures. (C) Bar graph shows data mean ± SEM; one-way ANOVA (***, P < 0.001). GFP alone (30.16 ± 1.68, n = 317), GFP-RhoA (35.44 ± 0.67, n = 301), and GFP-Rhes (67.56 ± 3.43, n = 300). (D) Confocal and DIC image of striatal neuronal cell expressing GFP-Rhes shows two different planes (substrate plane or above substrate). Arrow points to TNT-like cellular protrusion visible above the substrate plane from cell 1 to cell 2. (E) Bar graph shows data mean ± SEM; one-way ANOVA (***, P < 0.001). GFP alone (10.00 ± 1.52, n = 317), GFP-RhoA (13.05 ± 2.15, n = 301), and GFP-Rhes (30.98 ± 2.92, n = 300).
Rhes-induced protrusions are membranous structures and show “kiss and run” properties
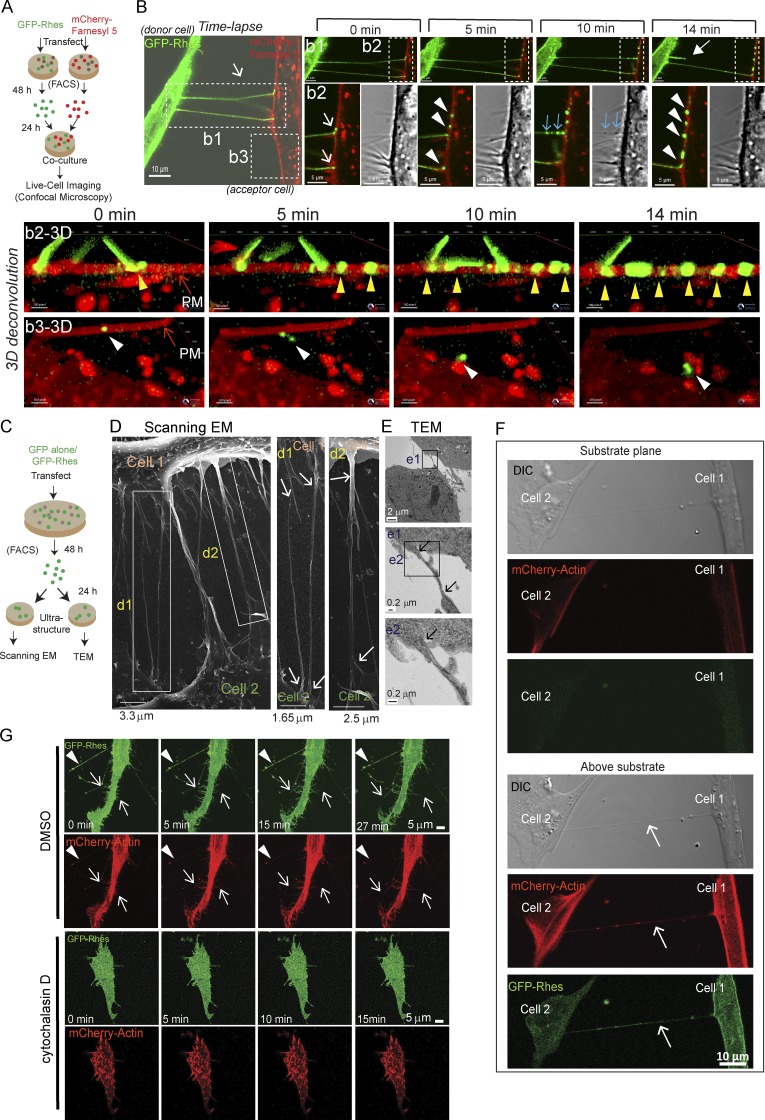
Next, we performed co-culture experiments with FACS, GFP-Rhes, and a membrane marker–expressing cell, followed by live-cell imaging using confocal microscopy (see experimental design, Fig. 2 A). Fig. 2 B and Video 1 shows time-lapse images, where GFP-Rhes–positive cellular protrusions interact with membrane marker (mCherry-Farnesyl 5) in the neighboring cell and deliver Rhes-positive vesicles (Video 1). Inset b1 in Fig. 2 shows, at 0 min, two Rhes-positive cellular protrusions present, and by 14 min (closed arrow), one was retracted, analogous to the “kiss and run” properties (Video 1, see b1 and b2) described for TNT-like structures (Tardivel et al., 2016). Inset b2 in Fig. 2 shows, at 0 min, a clear colocalization (yellow) of a Rhes-induced cellular protrusion (green) with a membrane marker (red)–positive cell (Fig. 2 B, white arrow; and Video 1, b1 and b2). At 5 min (Fig. 2, inset b2), three GFP-Rhes vesicular puncta were delivered to membrane marker–positive cells (Fig. 2 B, b2, arrowhead; and Video 1, see b1 and b2). At 10 min, there are GFP-Rhes–positive puncta lined up in one of the protrusions (blue arrow). Notably, these puncta can be seen in the DIC image (Fig. 2 B, 10 min, blue arrow; and Video 1, b1 and b2). At 14 min, multiple GFP-Rhes vesicular puncta from the protrusion were delivered to the adjacent cell (Fig. 2, b2, arrowhead; and Video 1, see b1 and b2). Deconvolved 3D reconstruction of inset b2 (Fig. 2, b2-3D) shows GFP-Rhes vesicular puncta lined up (arrowhead) and traveling on the plasma membrane (PM) of the adjacent cell, and one of them appears eventually delivered into the lumen (Fig. 2, b3-3D, arrowhead; and Video 1, see b3). We confirmed that GFP-Rhes–induced protrusions are indeed positive for membranes by cotransfecting GFP-Rhes with mCherry-Farnesyl 5 (Fig. S1 C, inset, arrow). Such structures were not apparent in GFP alone–expressing cells (Fig. S1 D). Thus, Rhes-induced cellular protrusions are membrane structures that deliver vesicular GFP-Rhes puncta to the PM of the adjacent cells, and display “kiss and run” properties, resembling TNTs.
Figure 2.
Rhes-induced filopodia-like protrusions are membranous structures, show “kiss and run” properties, and are abrogated by actin polymerization inhibitor. (A) Experimental design for B. (B) Live-cell imaging of co-cultured striatal neuronal cells expressing GFP-Rhes or mCherry-Farnesyl 5 (membrane marker). Inset b1: Closed arrow shows retraction event at 14 min. Inset b2: Arrows point to TNT-like cellular protrusions interacting with membrane of mCherry-Farnesyl 5. Arrowheads (white) indicate vesicle delivery to mCherry-Farnesyl 5 cells. Blue arrows indicate newly arriving vesicles (also seen in DIC). See related Video 1. b2-3D shows 3D deconvoluted images of inset b2, and arrowheads (yellow) indicate GFP-Rhes vesicular puncta. b3-3D shows 3D convoluted images of inset b3, and arrowheads (white) indicate GFP-Rhes puncta on the membrane and its internalization. See related Video 1. (C) An experimental design for D. (D) Representative scanning EM image of FACS-sorted GFP-Rhes expressing striatal neuronal cells. d1 and d2 inset arrows point to seamless transition of TNT-like cellular protrusions between cells. (E) TEM images of FACS-sorted GFP-Rhes expressing striatal neuronal cells. Insets from e1 and e2 show arrows pointing to TNT-like cellular protrusions. (F) Representative confocal and DIC images of striatal neuronal cells coexpressing GFP-Rhes and mCherry-actin at two different planes (substrate plane or above substrate). Arrows point to TNT-like hovering structures. (G) A snapshot of different time points of confocal live-cell time-lapse images of striatal cells coexpressing GFP-Rhes and mCherry-actin in vehicle (0.1% DMSO; 0, 5, 15, or 27 min) or cytochalasin D (2 µg/ml, 8 h; 0, 5, 10, or 15 min). Arrowheads show the formation of vesicles in TNT-like protrusions; arrows indicate the formation of new protrusions positive for actin. See related Videos 2 and 3.
Next, we enriched the GFP or GFP-Rhes–positive striatal neuronal cells using FACS, which helped to enrich pure GFP alone or the GFP-Rhes cell population (Fig. S2 A), and cultured them for scanning EM or transmission EM (TEM) analysis (see experimental design, Fig. 2 C). In scanning EM, Rhes-induced protrusions appeared to connect two cells, and their surface showed a seamless transition with the surface of connected cells (Fig. 2 D, insets d1 and d2, arrow). In scanning EM, we found <2% of GFP alone–sorted cells showed very few protrusions of ∼450 nm in diameter, but ∼20% of GFP-Rhes–sorted cells showed numerous protrusions, <200 nm in diameter (Fig. S2, B–D). In TEM analysis, Rhes-induced protrusions appeared continuous with the membranes of the connected cell (Fig. 2 E, insets e1 and e2, arrow), and were <150 nm in diameter, compared with GFP alone, whose filopodia were >200 nm in diameter (Fig. S2, E and F). Thus, ultrastructural analysis indicates that Rhes-induced protrusions are thin membranous structures that connect two cells, resembling certain properties of TNTs.
Rhes-induced cellular protrusions are positive for actin and abrogated by cytochalasin D
We found Rhes-induced floating protrusions are positive for actin (Fig. 2 F, arrow) and Exo70 (Fig. S3 A), a known regulator of membrane protrusions and actin polymerization (Liu et al., 2012; Zhao et al., 2013), but negative for tubulin (Fig. S2 G) or for the focal adhesion protein vinculin (Fig. S2 H). Overexpression of Exo70 or actin with control plasmids did not promote the formation of cellular protrusions (Fig. S3, A and B), indicating Rhes may coordinate with actin and/or Exo70 to promote cellular protrusions. Consistent with this notion, live-cell imaging revealed that Rhes-induced, filopodia-like protrusions were readily formed in the vehicle (DMSO)-treated cells (Fig. 2 G, arrow) with vesicular structures (arrowhead) and such dynamic structures were completely abolished in the cytochalasin D (actin polymerization inhibitor)–treated cells (Fig. 2 G and Videos 2 and 3). Collectively, these data indicate that Rhes-induced cellular protrusions are floating structures, connect two cells, and require actin polymerization for their formation resembling TNTs (Dupont et al., 2018). Based on these data, it is unclear whether Rhes-induced TNTs are open-ended/closed-ended connections into the adjacent cell, but live-cell imaging suggests GFP-Rhes puncta that travel on the PM of the acceptor cell are eventually delivered (Fig. 2 B and Video 1). Thus, it is possible that Rhes-induced cellular protrusion may be using a mechanism that involves transiently closed- and open-ended states. Thus, from here onward, we will refer to Rhes-induced cellular protrusions as “TNT-like” cellular protrusions or simply “Rhes tunnels.”
Rhes is transported to neighboring cell/neuron via Rhes tunnels
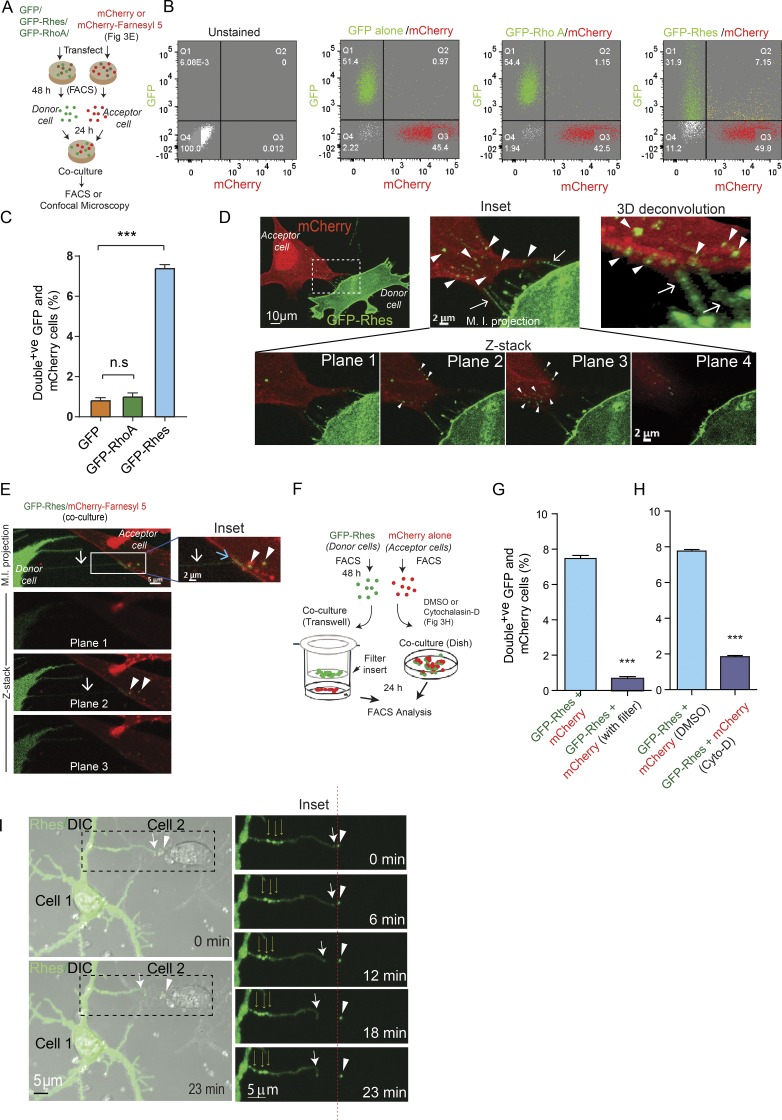
We tested whether the transportation of Rhes occurs between striatal neuronal cells, using co-culture/FACS and confocal analysis (Fig. 3 A). We labeled donor cells with GFP-Rhes or controls (GFP or GFP-RhoA) and acceptor cells with mCherry. We found ∼7% of acceptor cells were double-positive for GFP-Rhes and mCherry, compared with <1% in controls (Fig. 3, B and C). In confocal analysis (Fig. 3 D), multiple TNT-like protrusions from GFP-Rhes donor cells (arrow) connecting mCherry-positive acceptor cells and numerous GFP-Rhes puncta were within mCherry cells (arrowhead, plane 2 and 3). Deconvolved 3D reconstruction further magnified the presence of Rhes tunnels and numerous GFP-Rhes puncta inside mCherry cells (Fig. 3 D, arrowhead; and Video 4). Fig. 3 E (maximum intensity projection) show that Rhes tunnels (white arrow) were connected to the membrane of the mCherry-Farnesyl 5 labeled acceptor cell (inset, blue arrow), and the GFP-Rhes vesicle-like puncta were localized inside the mCherry-Farnesyl 5 cell (inset, arrowhead). Co-culture experiments with GFP or GFP-RhoA did not reveal TNT-like cellular protrusions or GFP puncta (Fig. S3, C and D).
Figure 3.
Rhes is transported to neighboring cells via TNT-like Rhes tunnels. (A) Experimental design for B and C. (B) Representative FACS analysis of co-cultured (20,000 cells) striatal neuronal cells. (C) Bar graph shows data mean ± SEM; one-way ANOVA (***, P < 0.001), n = 3. Quantification of GFP/mCherry double-positive cells (%) for GFP alone, GFP-RhoA, and GFP-Rhes. (D) Representative confocal image of co-cultured GFP-Rhes (green) and mCherry (red)–expressing cells. Arrows indicate TNT-like protrusions, and arrowheads point to vesicular structures in inset and 3D deconvoluted image (see Video 4). (E) Confocal image of FACS-sorted and co-cultured GFP-Rhes or mCherry-Farnesyl 5 (membrane marker)–expressing striatal cells. Images shown at different Z-planes (see Results). M.I. projection, maximum intensity projection. (F) Experimental design for G. (G and H) Bar graph shows data mean ± SEM; one-way ANOVA (***, P < 0.001), n = 3. Quantification of GFP/mCherry double-positive cells (%) in GFP-Rhes control (no Transwell), GFP-Rhes with Transwell, GFP-Rhes vehicle-treated (0.1% DMSO), or GFP-Rhes with cytochalasin D (Cyto-D; 2 µg/ml). (I) Time-lapse images of primary striatal neurons expressing GFP-Rhes. Thick arrows indicate TNT-like process from cell 1, thin arrows show multiple puncta in TNT, and arrowheads indicate GFP-Rhes vesicle in cell 2. Insets show the magnified images. Red dashed line indicates the movement of vesicle comparing its original position at 0 min to 13 min. See related Video 5 and its inset Video 6.
Next, using a Transwell plate containing a 0.4-µm filter and FACS/co-culture (see experimental design, Fig. 3 F), we found that Rhes is transported exclusively by means of physical contact between cells, not by secretory pathways, such as exosomes, which can travel through the filter (Thayanithy et al., 2017). Fig. 3 G shows ∼8% of mCherry cells were positive for GFP-Rhes in the co-culture experiment (without filter), compared with ∼0.8% in a Transwell culture plate (with filter). Next, we tested the effect of cytochalasin D in the cell–cell movement of Rhes. Cytochalasin D completely prevented the transportation of GFP-Rhes to the acceptor cells (Fig. 3 H). These data suggest that a physical cell–cell connection and actin polymerization are necessary for the efficient intercellular transportation of Rhes (Fig. 3, G and H).
Next, we transfected primary striatal neurons with GFP-Rhes or GFP alone and performed live-cell imaging. Fig. 3 I and related Videos 5 and 6 show that at 0 min, the primary striatal neurons transfected with GFP-Rhes (cell 1) formed a long TNT-like cellular protrusion (thick arrow), which at 12 min appeared to be delivering a GFP-Rhes puncta to the neighboring nontransfected cell (cell 2, arrowhead). The inset shows that the Rhes-induced TNT-like cellular protrusion containing multiple puncta (Fig. 3 I, thin arrows) from cell 1 after delivering a GFP-Rhes puncta to cell 2 gets retracted (inset, thick arrow). The red dashed line in the inset demonstrates that the GFP-Rhes puncta was slightly moved, at least 1 µm, into cell 2 at 12 min, compared with 0 min. This suggests that the GFP-Rhes puncta is able to move into cell 2 (Videos 5 and 6). We also found numerous GFP-Rhes puncta in the neighboring untransfected primary striatal neuron, but no such puncta were observed in GFP alone–transfected control cells (Fig. S3, E and F, arrowhead). Thus, Rhes promotes TNT-like Rhes tunnels, as well as travel to neighboring cells through Rhes tunnels in a striatal neuron cell line and in primary cultured striatal neurons.
Rhes SUMO E3-like domain promotes TNT-like Rhes tunnels, but only a full-length Rhes WT can be efficiently transported from cell to cell
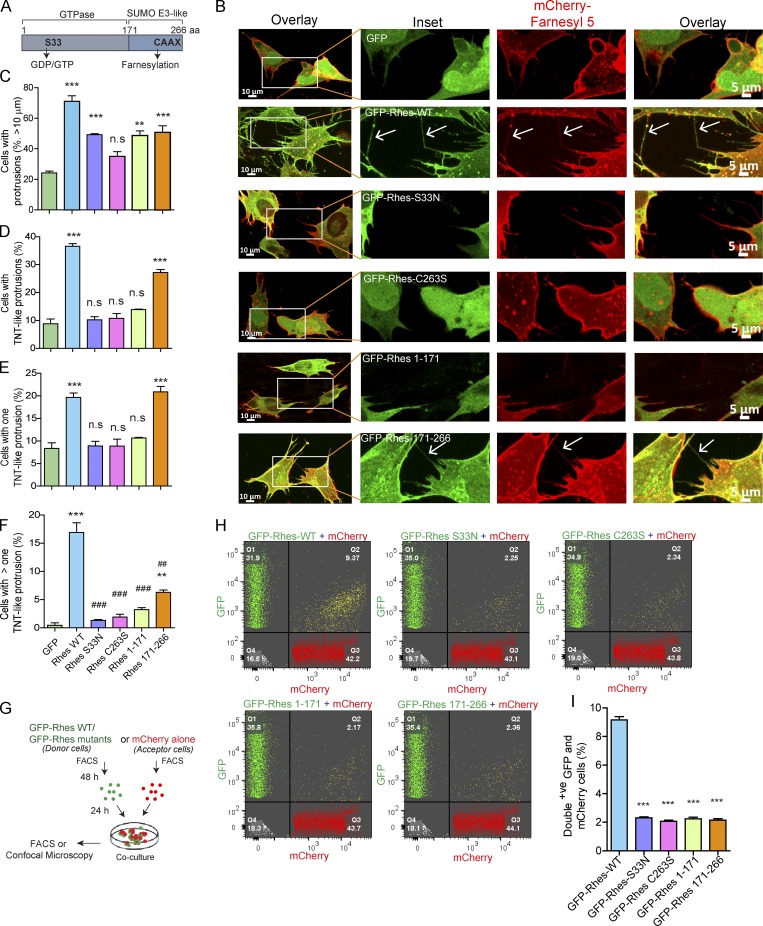
We investigated the effect of various mutants of Rhes (Fig. 4 A and Fig. S4 A), whose expressions were comparable (Fig. S4 B), on the formation of TNT-like Rhes tunnels, using confocal images (Fig. 4 B). We found ∼75% of GFP-Rhes WT–transfected cells had protrusions (all cells with filopodia-like processes), compared with ∼22% in GFP alone or ∼33–50% in Rhes mutants (Fig. 4 C). Cells with all TNT-like cellular protrusions were ∼40% in Rhes WT and 30% in SUMO E3-like domain (Rhes 171–266 aa; Fig. 4 D). Around 20% of cells with at least one TNT-like cellular protrusion were found both in Rhes WT and SUMO E3-like domain, compared with ∼10% in GFP alone and other mutants (Fig. 4 E). But we found ∼16% of Rhes WT–expressing cells showed more than one TNT-like cellular protrusion per cell compared with ∼7% in SUMO E3-like domain and ∼3% or less in control and other mutant-expressing cells (Fig. 4 F). Table 1 shows the mean and SEM for the data plotted in Fig. 4, C–F. Together this study indicates the SUMO E3 ligase domain can promote TNT-like Rhes tunnels, but not the N-terminal GTPase domain. However, serine 33, which is a part of the N-terminal domain, is also crucial for the formation of TNT-like Rhes tunnels (Fig. 4, D and F). Thus, both the GTPase domain and the SUMO E3 ligase domain of Rhes coordinate to induce TNT-like Rhes tunnels. Next, we introduced a mutation in the CAAX domain of 171–266 (Rhes 171–266-C263S), which, like Rhes-FL C263S, was mislocalized to the nucleus, and showed only ∼11% of Rhes tunnels compared with 25% in the intact 171–266-aa domain (Fig. S4, C and D). Thus, membrane anchoring and the SUMO E3 ligase domain of Rhes are necessary for the formation of TNT-like Rhes tunnels. Next, we introduced CAAX to the C-terminal of 1–171 aa, but 1–171 CAAX did not promote TNT-like protrusion, indicating CAAX addition alone is insufficient to promote TNT-like Rhes tunnels (Fig. S4, C and D).
Figure 4.
Rhes’s SUMO E3-like domain promotes TNT-like Rhes tunnels, but only a Rhes WT travels efficiently from cell to cell. (A) Diagram of Rhes domains. (B) Representative confocal image of striatal neuronal cells coexpressing either GFP-tagged Rhes WT or its mutants with mCherry-Farnesyl 5. Arrows indicate TNT-like process. (C–F) Bar graphs show the quantification of all protrusions (C), all TNT-like protrusions (D), single TNT-like process/cell (E), and more than one TNT-like process/cell (F). Data mean ± SEM are shown in Table 1; one-way ANOVA, **, P < 0.05; ***, P < 0.001 (compared with GFP); ##, P < 0.01; ###, P < 0.001 (compared with Rhes WT); n = 210–230 cells/group, from three independent experiments. (G) Experimental design for H. (H) FACS plots for GFP-tagged Rhes WT and its mutant or mCherry alone expressing striatal neuronal cells (20,000 cells were recorded). (I) Bar graph shows ± SEM; one-way ANOVA (***, P < 0.001, compared with Rhes WT), n = 3. Quantification of GFP and mCherry double-positive population (%) for Rhes-FL, Rhes S33N, Rhes C263S, Rhes 1–171, and Rhes 171–266.
Table 1. Mean and SEM for the data plotted in Fig. 4.
| Sample (n = 3) | Cells with >10-μm protrusions (%) | Cells with TNT-like protrusions (%) | Cells with one TNT-like protrusion (%) | Cells with more than one TNT-like protrusion (%) |
|---|---|---|---|---|
| GFP | 24.23 ± 1.93 | 8.81 ± 2.92 | 8.36 ± 2.14 | 0.45 ± 0.78 |
| Rhes WT | 71.01 ± 6.49 | 36.29 ± 1.64 | 19.66 ± 1.72 | 16.93 ± 2.97 |
| Rhes S33N | 49.21 ± 1.16 | 10.25 ± 1.97 | 8.91 ± 1.76 | 1.33 ± 0.23 |
| Rhes C263S | 34.94 ± 5.48 | 10.78 ± 2.93 | 8.85 ± 2.68 | 1.92 ± 0.82 |
| Rhes 1–171 | 48.64 ± 5.35 | 13.87 ± 0.75 | 10.65 ± 0.28 | 3.22 ± 0.60 |
| Rhes 171–266 | 50.63 ± 7 0.69 | 27.29 ± 1.75 | 20.88 ± 2.07 | 6.30 ± 0.72 |
We next asked whether the SUMO E3 ligase domain (171–266 aa), similar to Rhes WT, can be transported to acceptor cells (see experimental design, Fig. 4 G). Confocal images show that GFP-Rhes WT, but not the SUMO E3 ligase domain or other mutants of Rhes, were found in the mCherry acceptor cells (Fig. S4, E and F, arrowhead). In FACS, around 9% of acceptor cells showed GFP-Rhes WT, but only ∼2% for all other mutants, including SUMO E3 ligase domain (Fig. 4, H and I). Thus, SUMO E3 ligase-like domain of Rhes can promotes the biogenesis of TNT-like Rhes tunnels, but only a full-length Rhes WT can be efficiently transported from cell to cell.
Rhes promotes cell–cell transportation of membranous vesicles via SUMO-E3 ligase domain
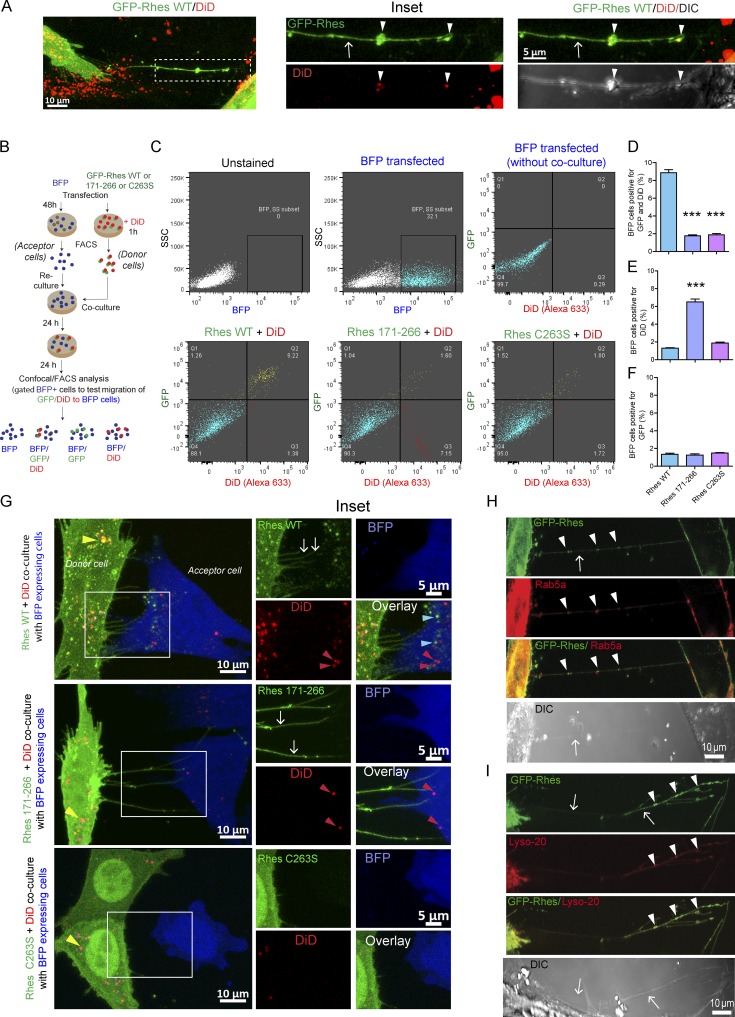
Next, we investigated whether Rhes WT or its mutant modulates the cell–cell transport of vesicles using DiD (1,1'-Dioctadecyl-3,3,3',3'-tetramethylindodicarbocyanine, 4-chlorobenzenesulfonate), which binds to all kinds of vesicles. First, we found TNT-like Rhes tunnels were positive for DiD (Fig. 5 A). Using FACS/co-culture and confocal analysis (see experimental design, Fig. 5 B), we found that ∼9% of GFP-Rhes WT and DiD vesicles were transported to the BFP acceptor cells (Fig. 5, C and D). GFP-Rhes 171–266 was less efficiently transported to acceptor cell (∼2%), but this mutant was able to transport DiD vesicles (∼7%; Fig. 5, C–E). As observed before (Fig. 4), GFP-Rhes C263S showed a diminished effect on its own transport (Fig. 5, C, D, and F) and also the transport of DiD-positive vesicles (<2%, Fig. 5, C–E). Confocal microscopy, in Fig. 5 G, further corroborated that GFP-Rhes WT and GFP-Rhes 171–266 induced TNT-like cellular protrusions (arrows), but only GFP-Rhes WT puncta were seen in the BFP acceptor cell (blue arrowhead). DiD-labeled vesicles were seen in BFP acceptor cells both in WT and 171–266 (red arrowhead), but not in GFP-Rhes C263S. Notably, in donor cells, GFP-Rhes WT vesicular structures were also positive for DiD (Pearson’s coefficient, 0.86), and this property appears to be diminished in the mutants (Pearson’s coefficient is 0.30 and 0.18 for GFP-Rhes 171–266/DiD and GFP-Rhes C263S/DiD, respectively; Fig. 5 G, yellow arrowhead). We also found that Rhes vesicles in TNT-like cellular protrusions (arrow) were positive for an endosome marker, Rab5a (Fig. 5 H, arrowhead), and for a lysosome marker, Lyso-20 (Fig. 5 I, arrowhead), but negative for the ER, Golgi, autophagosome, nucleolus, peroxisome, or mitochondria markers (Fig. S4 G). The Pearson’s correlation coefficients for GFP-Rhes with Rab5a and Lys20 were 0.59 and 0.64, respectively.
Figure 5.
Rhes-induced TNT-like Rhes tunnels contain DiD-, Rab5a-, or Lyso-20–positive vesicles. (A) Representative confocal image of striatal neuronal cells expresing GFP-Rhes and treated with DiD-Red (DiD, vesicle labeling dye). Arrows point to TNT-like protrusions, and arrowheads point to vesicles in TNT-like cellular protrusions. (B) Experimental design for C. (C) FACS analysis of striatal neuronal cells expressing GFP-Rhes WT, GFP-Rhes 171–266, or GFP-Rhes C263S treated with DiD and co-cultured with FACS-sorted BFP-transfected striatal neuronal cells. The BFP population was gated and analyzed for the presence of GFP and/or DiD (Alexa Fluor 633). (D–F) Bar graphs show data mean ± SEM; one-way ANOVA test (***, P < 0.001), n = 3. The quantification of BFP cells positive for both GFP and DiD fluorescence (D), only DiD (E), or only GFP (F). (G) Representative confocal images for co-cultured experiments as indicated in B (see Results). (H and I) Representative confocal and DIC images of striatal neuronal cells cotransfected with GFP-Rhes and Rab5a (H) or GFP-Rhes and Lyso-20 (I). Arrows indicate TNT-like Rhes tunnels, and arrowheads show vesicles in Rhes tunnels.
Collectively, these data suggest that Rhes WT can travel from cell to cell and transport DiD vesicles, Rab5a, and Lyso-20, whereas Rhes 171–266 mutant cannot travel from cell to cell but can deliver DiD vesicles via Rhes tunnels.
Rhes promotes cell–cell transportation of mHTT via TNT-like Rhes tunnels
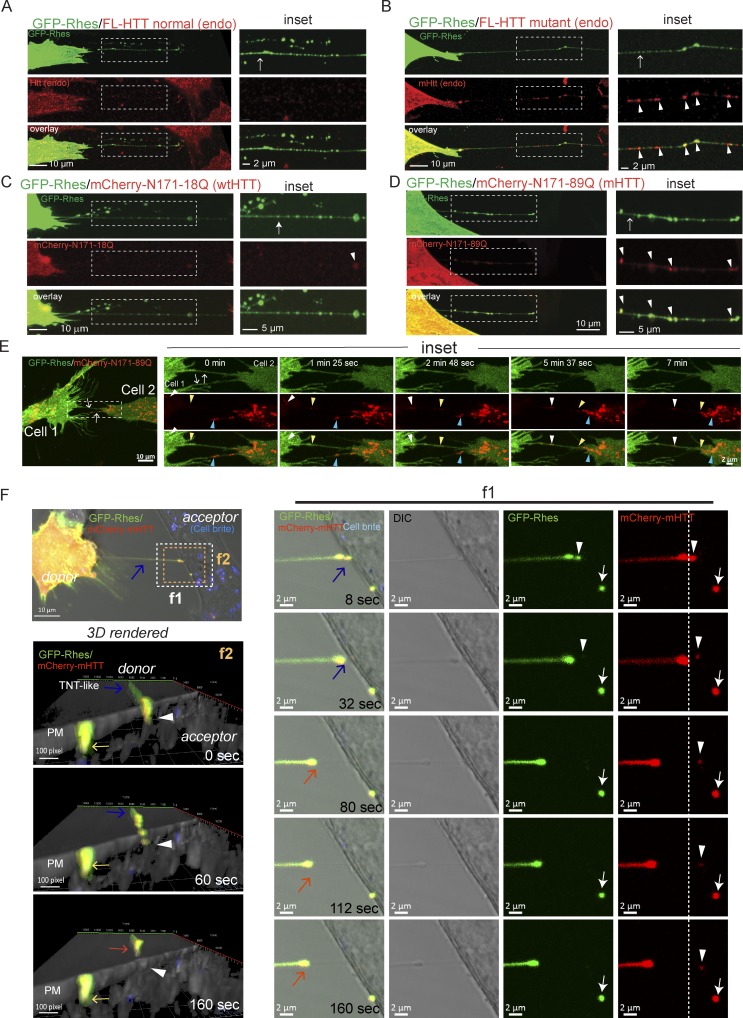
We wondered whether Rhes could transport mHTT from cell to cell via TNT-like Rhes tunnels. We found Rhes tunnels formation both in WT striatal neuronal cells used in this study (STHdhQ7/Q7, which express endogenous full-length wtHTT with seven glutamines) and in mutant striatal neuronal cells (STHdhQ111/Q111, which express full-length mHTT, with humanized 111 polyglutamine repeats; Trettel et al., 2000; Fig. S5, A and B). However, only mHTT in STHdhQ111/Q111, but not wtHTT in STHdhQ7/Q7 cells, was found in Rhes tunnels (Fig. 6, A and B, inset, arrow). Many mHTT puncta in Rhes tunnels appeared as vesicles, and they were partially colocalized with GFP-Rhes (Fig. 6, A and B, inset, arrowhead). Similarly, the N-terminal toxic fragment of mHTT, 1–171-aa–containing 89Q (mCherry-N171-89Q), but not the nontoxic form mCherry-N171-18Q, was found in Rhes tunnels (Fig. 6, C and D, arrow) and colocalized with GFP-Rhes (inset, arrowhead). In GFP alone–expressing cells, we did not see TNT-like cellular protrusions or mHTT transfer (Fig. S5, C and D). Fig. 6 E shows different colors of arrowheads to display, in a time-lapse fashion, the tracking of corresponding mHTT vesicles in Rhes tunnels from cell 1 to cell 2 (Fig. 6 E, inset; and Video 7). Another example of live-cell imaging shows a “leading” Rhes and mHTT puncta that appear paused over the PM of the acceptor cells (Fig. 6 F, inset, f1, white arrow), and a “just approaching” Rhes and mHTT puncta are about to be delivered into the PM of acceptor cells (inset, f1, arrowhead). By 160 s, Rhes tunnels appear retracted (Fig. 6 F, red arrow). The 3D reconstruction of Fig. 6 F (inset, f2, arrow) events shows Rhes tunnels interact with the PM of the acceptor cells, where the leading Rhes and mHTT puncta (yellow arrow) and the just approaching Rhes and mHTT puncta (arrowhead) can be seen on the PM, followed by the retraction of Rhes tunnels (red arrow). Representative time-lapse videos of these events are shown in Video 8. Together this suggests Rhes can transport mHTT from one cell to another via TNT-like Rhes tunnels.
Figure 6.
Rhes promotes cell–cell transportation of mHTT via Rhes tunnels. (A–D) Representative confocal images of GFP-Rhes in striatal cells (STHdhQ7/Q7) or mutant striatal cells (STHdhQ111/Q111) with immunocytochemistry for endogenous (endo) normal full-length (FL) HTT (A) or endogenous FL mutant HTT with anti-HTT MAB2166 antibody (B). WT striatal cells expressing GFP-Rhes and mCherry-tagged N-terminal nonpathogenic HTT fragment (N171-18Q; C), or N-terminal pathogenic HTT fragment (N171-89Q; D). Arrows indicate TNT-like Rhes tunnels, and arrowheads indicate the colocalization of mHTT/HTT in the Rhes tunnel. (E) Confocal time-lapse image (∼7 min) of striatal cells coexpressing GFP-Rhes and mCherry mHTT (N171-89Q). Arrows point to TNT-like Rhes tunnel connecting cell 1 and cell 2. Arrowheads in inset point to the movement of mHTT vesicle in the Rhes tunnel. See related Video 7. (F) Confocal time-lapse image (∼160 s) of striatal cells coexpressing GFP-Rhes and mCherry mHTT (N171-89Q). Blue arrow points to TNT-like Rhes tunnel connecting cell 1 and cell 2. Inset f1 points to the delivery of leading Rhes and mHTT puncta (arrow) and just approaching Rhes and mHTT vesicular puncta (arrowhead) into the acceptor cells. Inset f2 shows 3D rendering of the time-lapse image of F. Arrowhead points to the just approaching Rhes and mHTT puncta, yellow arrow indicates the leading Rhes and mHTT puncta, and red arrow indicates the retracting Rhes tunnel.
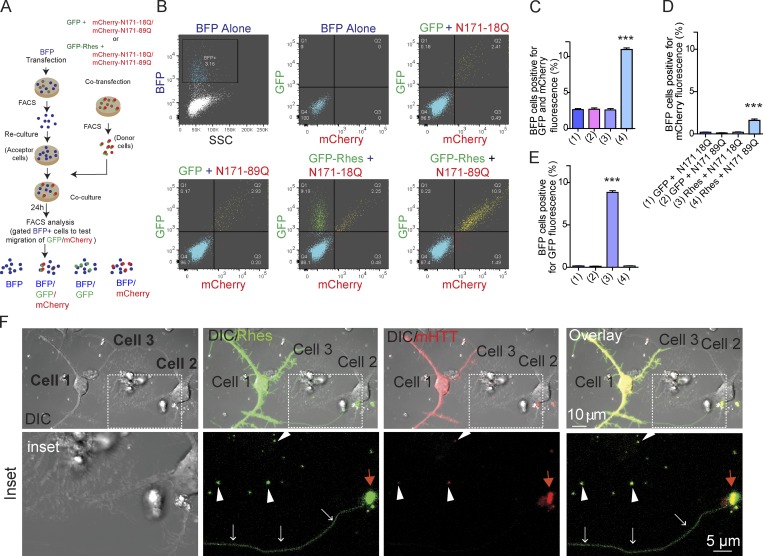
In a co-culture/FACS analysis (see experimental design, Fig. 7 A), we found that ∼12% of acceptor BFP cells were copositive for GFP-Rhes and mHTT, but only ∼2.5% were copositive for GFP-Rhes and wtHTT, which was similar to the GFP control (Fig. 7, B and C), indicating that Rhes and mHTT transported to the same population of BFP+ cells (acceptor cells). The mHTT alone is transported much less efficiently (∼0.3%; Fig. 7, B and D), but with Rhes, mHTT transport is increased by 20-fold (∼12%; Fig. 7, B and C). Note that ∼2% of acceptor cells are positive only for mHTT in the presence of GFP-Rhes compared with ∼0.3% in GFP (Fig. 7 D). This indicates that a small but a significant portion of mHTT can be transported without Rhes, but that transportation is still Rhes dependent. Normal HTT alone cannot be transported with or without Rhes (Fig. 7 D). In addition, ∼10% of the acceptor cells are positive for Rhes alone, but negative for wtHTT, indicating that Rhes transportation is not affected by the presence of wtHTT (Fig. 7 E). In confocal image analysis of FACS-sorted cells, we confirmed that acceptor cells were positive for Rhes and mHTT, but not in the GFP control cells (Fig. S5, E and F). Thus, mHTT, but not wtHTT, is very efficiently transported from one cell to another by Rhes, and Rhes travel is not affected by either wtHTT or mHTT.
Figure 7.
Rhes efficiently transports mHTT between striatal neuronal cells, and in primary striatal neurons via TNT-like Rhes tunnels. (A) Experimental design for B and C. (B) FACS plot and co-culture of striatal cells transfected with Rhes and HTT constructs as indicated. BFP cell population was gated and a total 20,000 cells recorded. (C–E) Bar graphs show data mean ± SEM; one-way ANOVA test (***, P < 0.001), n = 3. Quantification of BFP cells (%) positive for both GFP and mCherry fluorescence (C), only mCherry (D), or only GFP fluorescence (E). (F) Representative confocal and DIC images of primary striatal neurons cotransfected with GFP-Rhes and mCherry mHTT. Arrows (white) indicate TNT-like process from cell 1 connecting cell 2. Arrowheads indicate GFP-Rhes and mHTT vesicular puncta in neighboring cell 3. Red arrows indicate the colocalization of Rhes and mHTT puncta in cell 2.
Next, we found a thin and long (∼55 µm) GFP-Rhes–positive, TNT-like protrusion from primary striatal neurons expressing GFP-Rhes and mCherry mHTT (N171-89Q; cell 1; Fig. 7 K, white arrow, inset), which was colocalized with mHTT puncta in cell 2 (Fig. 7 K, red arrow, inset). Multiple GFP-Rhes and mCherry mHTT double-positive vesicular puncta were found in neighboring cell 3 (Fig. 7 K, arrowhead). In GFP alone and mHTT-expressing primary striatal neurons, we did not detect TNT-like protrusions or obvious vesicular puncta in neighboring cells (Fig. S5 G). Thus, Rhes induces cell–cell transportation of mHTT in primary striatal neurons via a TNT-like cellular protrusion.
Rhes requires physical cell–cell contact to transport mHTT, which associates with vesicles in acceptor cells
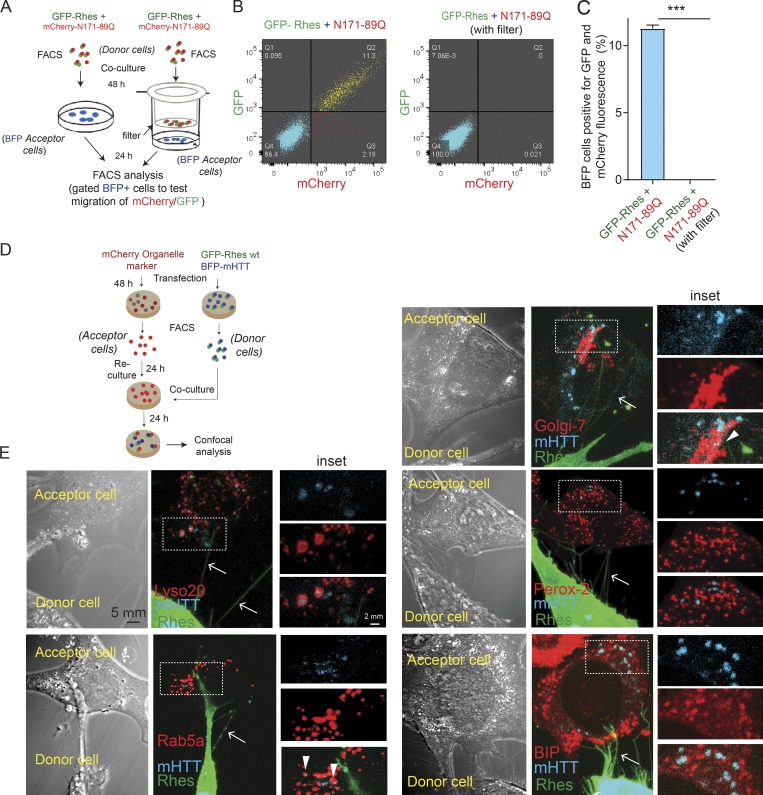
We next investigated whether exosomes or any other diffusible modes of transport might contribute to mHTT transfer by Rhes, using a FACS/co-culture Transwell plate, separated by a 0.4-µm filter (see experimental design, Fig. 8 A). Rhes could transport mHTT to ∼12% of BFP cells in the co-culture experiment (without filter), as in Fig. 6, but this effect by Rhes is completely abolished with a filter (Fig. 8, B and C). This suggests that the absence of diffusible factors, and a necessity of a physical cell–cell contact, in Rhes-mediated transfer of mHTT between cells.
Figure 8.
Rhes requires physical cell–cell contact to transport mHTT, which in acceptor cells associates with lysosome and other vesicles. (A) Experimental design for B and C. (B) FACS plot for the indicated co-cultured striatal neuronal cells. BFP cell population was gated, and 20,000 cells were recorded per sample. (C) Bar graphs show data mean ± SEM; Student’s t test (***, P < 0.001), n = 3, indicating percentage of BFP cells (acceptor cells) positive for Rhes + N171 89Q and Rhes + N171-89Q with Transwell (filter). (D) Experimental design for E. (E) Representative confocal and DIC images and their insets indicating donor cell (GPF-Rhes/BFP-mHTT) and acceptor cell (mCherry-tagged various organelle markers). Green channel shows GFP-Rhes, blue color represents BFP-mHTT, and red channel indicates the mCherry-tagged Lysosome (Lyso 20), Rab5a, Golgi-7, Peroxisome (Perox-2), and BIP individual organelle markers. Arrows indicate colocalization between BFP-mHTT and different organelle markers.
We next wondered where mHTT might associate in the acceptor cells, using FACS/co-culture experiments (see experimental design, Fig. 8 D). Fig. 8 E shows numerous Rhes-positive TNT-like cellular protrusions (arrow) from donor cell interacting with acceptor cell that contained mHTT, which predominantly localized with the lysosomal marker (Lyso-20, Pearson’s coefficient, 0.82), but very little association was found with endocytic markers (Rab5a, Pearson’s coefficient, 0.29), Golgi (Golgi-7, Pearson’s coefficient, 0.37), ER (BiP, Pearson’s coefficient, 0.23), or peroxisome (Perox-2, Pearson’s coefficient, 0.19) in acceptor cells (inset, arrowhead). Thus, mHTT appears to be associated primarily with lysosomes and to some extent other vesicles in acceptor cells.
Rhes transports Ataxin-3 (Atxn3) and poly 72Q protein but not mTOR or wtTau
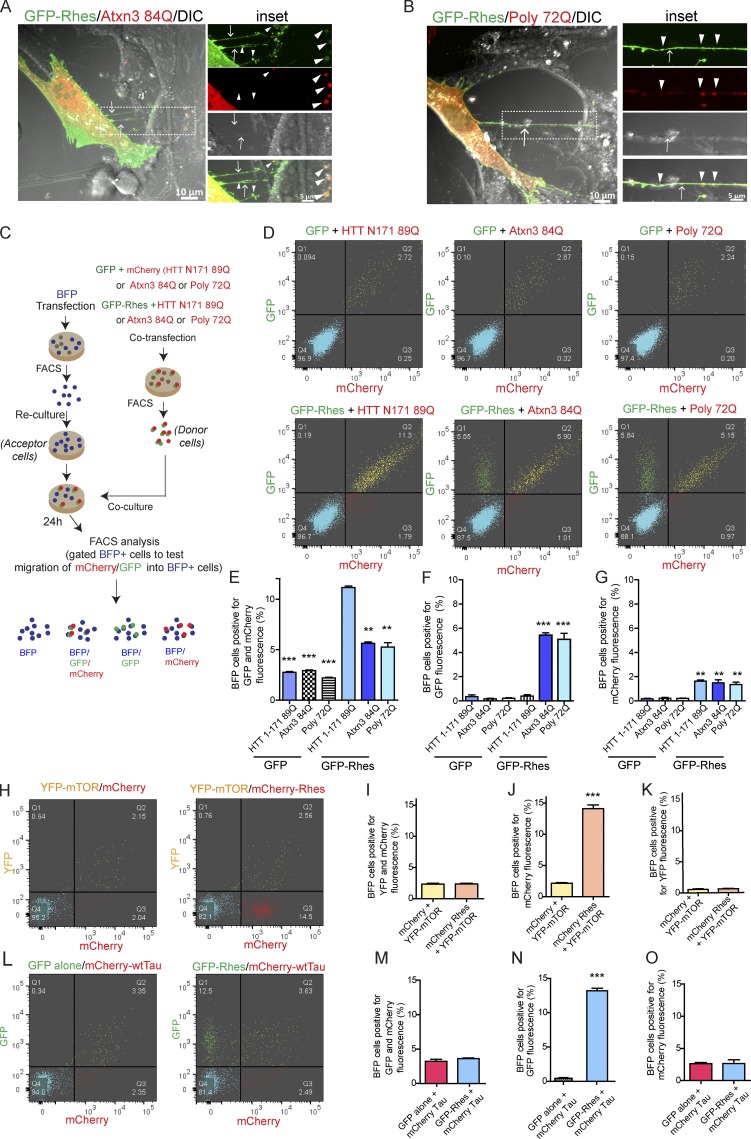
We investigated whether poly-Q expansion is a determining factor for the Rhes-mediated intercellular transportation of mHTT. Thus, we tested whether Atxn3-containing 84Q [(Atxn3 84Q)], which affects the striatum in Machado–Joseph disease (Taniwaki et al., 1997; Yen et al., 2000; Alves et al., 2008; Duarte-Neves et al., 2015), is transported via TNT-like Rhes tunnels. We included a stretch of 72Q (poly 72Q) as additional control. Confocal imaging showed the presence of Atxn3 84Q and poly 72Q in the GFP-Rhes–induced TNT-like cellular protrusions (Fig. 9, A and B). Quantification using co-culture experiments and FACS analysis (see experimental design, Fig. 9 C) showed Rhes could transport ∼5% of Atxn3 84Q or ∼5% of the poly 72Q to BFP acceptor cells, compared with ∼12% of mHTT (Fig. 9, D–G). GFP alone did not induce Atxn3 84Q or poly 72Q transport (Fig. 9, D–G; and Fig. S6, A and B), and the expression level of all three poly-Q proteins appeared comparable in the Western blot (Fig. S6, C and D). Interestingly, as shown before (Fig. 7, C and D), almost all mHTT transported together with Rhes, but only a portion of Atxn3 or poly 72Q transported with Rhes to the BFP acceptor cells (Fig. 9 E). Notably, Rhes transportation, which remained around 12%, was not affected by Atxn3 or poly 72Q protein (Fig. 9 F). This indicates Rhes efficiently promotes cell–cell transportation of mHTT, compared with Atxn3 or poly 72Q only proteins. A small percentage of acceptor cells (∼2%) were positive only for mHTT, Atxn3 84Q, or poly 72Q proteins in the presence of Rhes, compared with ∼0.3% in control (GFP alone; Fig. 9 G), indicating that poly-Q proteins can be cell–cell transported on their own (without Rhes), yet their transportation is facilitated by Rhes. Together, as in Fig. 7, C and D, we found there is a selective transport of mHTT (∼20-fold more) by Rhes, compared with other poly-Q proteins, which are ∼10-fold more transported by Rhes (Fig. 9, E and G). A differential protein stability or interaction of poly-Q–enriched proteins with Rhes might contribute to such differential cell–cell transfer efficiency. To investigate this, we tested the transport of mTOR, a known interactor of Rhes, or Tau, which is known to be transported via TNT (Subramaniam et al., 2011; Tardivel et al., 2016). In confocal analysis (Fig. S6, E and F), we found GFP-Rhes–positive puncta (arrowhead) in TNT-like cellular protrusions (arrow), but they were positive for neither mTOR nor wtTau (MAPTau-C-10). FACS/co-culture experiments showed GFP-Rhes is transported to acceptor cells (∼12%; Fig. 9, H, J, L, and N), but it cannot transport mTOR (Fig. 9, H–K) or Tau (Fig. 9, L–O), which were <2% similar to GFP or mCherry controls.
Figure 9.
Rhes transports Ataxin3 and poly 72Q protein but not mTOR or wtTau. (A and B) Representative confocal and DIC images of striatal neuronal cells expressing GFP-Rhes and mCherry-Ataxin 3 (Atxn3 84Q; A) or GFP-Rhes and mCherry-Poly 72Q (B). Arrows point to a TNT-like process. Arrowheads indicate GFP-Rhes colocalization with Atxn 84Q or Poly-72Q. (C) An experimental design for D and E. (D) Representative FACS plot for the indicated co-cultured cells. A total of 20,000 cells was recorded. (E–G) Bar graphs show data mean ± SEM; one-way ANOVA test (***, P < 0.001; **, P < 0.01), n = 3. Quantification of BFP cells (%) positive for both GFP and mCherry fluorescence (E), only GFP (F), and only mCherry (G). (H) FACS-sorted double-positive YFP-mTOR/mCherry alone or YFP-mTOR/mCherry-Rhes striatal neuronal cells (donor cells) were co-cultured with FACS-sorted BFP-expressing cells (acceptor cells). (I–K) Bar graphs show data mean ± SEM; Student’s t test (***, P < 0.001), n = 3. Quantification of BFP cells (%) positive for both YFP and mCherry fluorescence (I), mCherry only (J), or YFP only (K). (L) FACS-sorted double-positive (GFP alone/mCherry-Tau or GFP-Rhes/mCherry-Tau striatal neuronal cells [donor cells]) were co-cultured with FACS-sorted BFP-expressing cells (acceptor cells). (M–O) Bar graphs shows data mean ± SEM; Student’s t test (***, P < 0.001), n = 3. Quantification of BFP cells (%) positive for both GFP and mCherry fluorescence (M), GFP only (N), and mCherry only (O).
Thus, Rhes selectively transports mHTT or poly-Q expanded proteins but not mTOR or Tau, indicating that the transport is regulated and at least partially via poly-Q–dependent mechanisms.
SUMO participates in Rhes-mediated cell–cell transport of mHTT
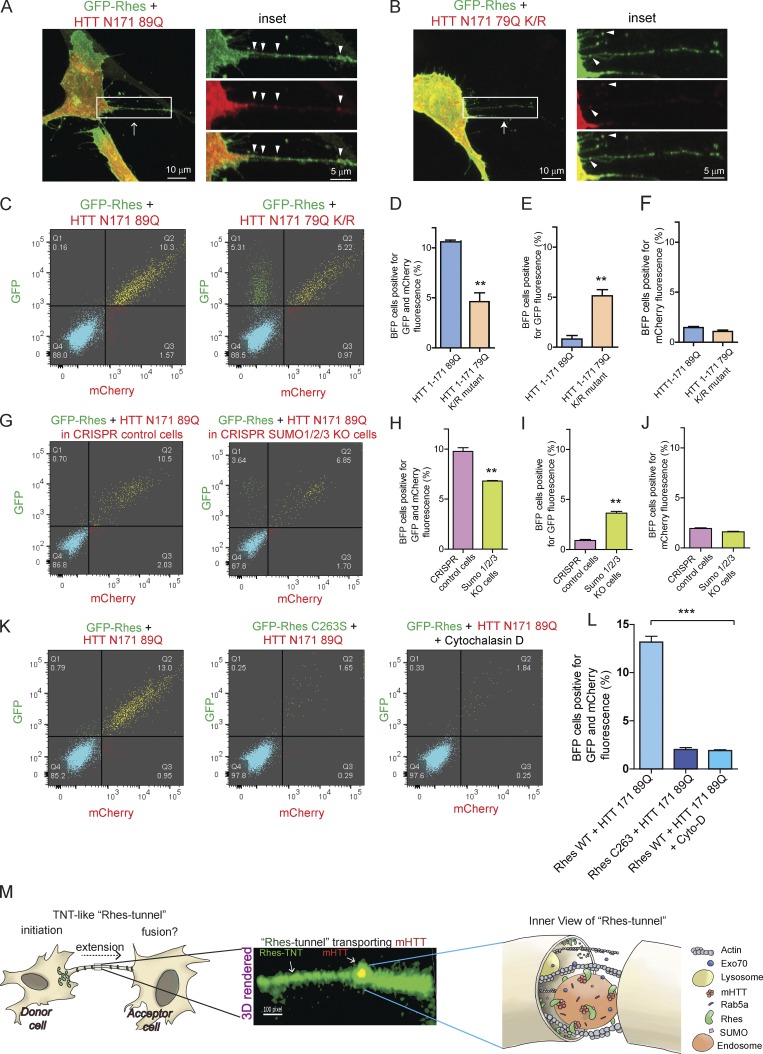
Since the SUMO E3-like domain of Rhes contributes to TNT-like cellular protrusions biogenesis (Fig. 4), and Rhes promotes SUMOylation of mHTT at lysine 6, 9, 15, and 91 and multiple other targets (Subramaniam et al., 2009, 2010; O’Rourke et al., 2013), we wondered whether SUMOylation of mHTT might be an additional contributing factor in the Rhes-mediated cell–cell transport of mHTT. To investigate whether SUMO plays a role in the formation of TNT-like Rhes tunnels and mHTT transport, first we compared the transfer efficiency of SUMOylation-intact mHTT (N171-89Q) versus SUMOylation-defective mHTT that contains mutations at lysine 6, 9, 15, and 91 to arginine (N171-79Q K/R). In confocal analysis, we found that N171-89Q appeared more in TNT-like Rhes tunnels, compared with the N171-79Q K/R mutant (Fig. 10, A and B, inset, arrowhead). In FACS and co-culture experiments (see experimental design, Fig. S7 A), we found ∼10% of BFP acceptor cells that were positive for N171-89Q and GFP-Rhes (Fig. 10, C, and D), as before (Figs. 7 and 8), but only ∼5% of acceptor cells were positive for N171-79Q K/R mutant and GFP-Rhes (Fig. 10, C and D). However, the overall transport of Rhes was not affected by N171-79Q K/R mutant (Fig. 10, C and E), which was altogether ∼10% both in N171-79Q K/R or N171-89Q conditions. Either mHTT or its K/R mutant alone show negligible levels of cell–cell transport (Fig. 10 F). Together these data indicate that the Rhes transports SUMOylation-defective mHTT much less efficiently than SUMOylation-intact mHTT via TNT-like Rhes tunnels.
Figure 10.
SUMO participates in Rhes-mediated cell–cell transportation of mHTT. (A) Representative confocal images of striatal neuronal cells coexpressing GFP-Rhes and mCherry-HTT N171 89Q, or (B) GFP-Rhes and mCherry-HTT N171 79Q K/R mutant (K 6, 9, 15, and 91 R). Arrow indicates TNT-like Rhes tunnel, and arrowheads indicate mHTT in Rhes tunnel. (C) Representative FACS plot of double-positive striatal neuronal cells (GFP-Rhes and mCherry-HTT N171 89Q or GFP-Rhes and mCherry-HTT N171 79Q K/R mutant) co-cultured with FACS-sorted BFP-expressing cells for 24 h (see experimental design in Fig. S7 A). (D–F) Bar graphs show data mean ± SEM; Student’s t test (**, P < 0.01), n = 3. Quantification of percentage of BFP cells positive for GFP and mCherry (D), only GFP (E), or only mCherry fluorescence (F). (G) Representative FACS plot of control CRISPR- or SUMO1/2/3-depleted double-positive striatal neuronal cells (GFP-Rhes and mCherry-HTT N171 89Q) co-cultured with FACS-sorted BFP-expressing cells (see experimental design in Fig. S7 F). (H–J) Bar graphs show data mean ± SEM; Student’s t test (**, P < 0.01), n = 3. Quantification of percentage of BFP cells, positive for both GFP and mCherry (H), for GFP only (I), or for mCherry only (J). (K) Representative FACS plot for the indicated co-cultured cells. (L) Bar graphs show mean ± SEM; one-way ANOVA (***, P < 0.001), n = 3. Quantification percentage of BFP cells positive for Rhes WT + HTT N171-89Q, Rhes C263S + HTT N171-89Q, and Rhes WT + HTT N171-89Q in cytochalasin D. (M) Model depicting the Rhes-induced TNT-like protrusion, Rhes tunnel connecting two cells, shows a 3D-rendered snapshot of a Rhes-TNT–like protrusion transporting mHTT and a view inside the Rhes tunnel.
To further investigate the role of SUMO, we generated striatal neuronal cells depleted for all three SUMO isoforms (Wilkinson and Henley, 2010) using the CRISPR/Cas-9 tool. This method yielded a 60–70% loss of all three SUMO isoforms, as quantified by Western blotting (Fig. S7, B–E). We then compared the transfer efficiency of mHTT N171-89Q between SUMO-intact and SUMO-depleted cells, using FACS and co-culture experiments (see experimental design, Fig. S7 F). We found Rhes could transport mHTT to ∼11% of BFP cells (acceptor cells) from SUMO-intact cells, but ∼7% from SUMO-depleted cells (Fig. 10, G and H). But, as in K/R mutant data above, the overall Rhes transport was not affected in SUMO-depleted cells versus SUMO-intact cells (Fig. 10, G–I). SUMO-depleted cells do not modulate the cell–cell transport of mHTT (Fig. 10 J). Thus, SUMO may not affect the Rhes-induced TNT-like Rhes tunnels formation or transport of Rhes itself, but it might contribute to transport of mHTT, via Rhes tunnels. Next, we investigated the effect of Rhes C263S, which we found had no SUMOylation activity toward mHTT (Subramaniam et al., 2009), as well as cytochalasin D, the actin polymerization inhibitor, on the Rhes-mediated cell–cell transportation of mHTT. The GFP-Rhes C263S failed to promote the cell–cell transfer of mHTT, and cytochalasin D blocked GFP-Rhes WT–induced transfer of mHTT (Fig. 10, K and L).
Altogether, we demonstrate that Rhes promotes TNT-like Rhes tunnels, through which Rhes, endosome, lysosome vesicles, and mHTT are transported to the neighboring cells. This entire process requires actin, SUMO, and intact membrane and GTP binding/SUMO E3-like domains of Rhes (Fig. 10 M).
Discussion
The serendipitous finding that Rhes increased the biogenesis of the TNT-like cellular protrusion and transport to the adjacent cells raises many intriguing questions both at the conceptual and mechanistic levels. One exciting question is, what is the need for such a long-range (up to 110 µm) physical connection between cells? It is plausible that Rhes tunnels may serve as a novel mode of cell–cell communication to maintain cellular homeostasis. In the striatum, which connects to multiple regions of the brain, the potential involvement of a TNT-like cellular protrusion offers an unprecedented complexity by which Rhes may signal within and outside the striatum. The TNT-like cellular protrusion is found in vivo—for example, in Drosophila embryos (cytonemes; Ramírez-Weber and Kornberg, 1999; Hsiung et al., 2005; Bischoff et al., 2013), in immune cells, in major histocompatibility complex class II+ cells, and in several malignant tumors, including astrocytoma in the brain (Osswald et al., 2015; Ariazi et al., 2017), which resemble Rhes tunnels. The presence of TNT-like Rhes tunnels in the striatum or other brain regions remains unknown. This is the first report, to our knowledge, that describes the role of a brain-enriched protein in the biogenesis of TNT-like cellular protrusions, thus raising the prospect that a TNT-like Rhes tunnels may participate in normal striatal functions in vivo.
What are the mechanisms by which Rhes induces TNT-like cellular protrusions? Broadly, there are at least three components in TNT-like protrusion biogenesis: initiation, extension, and fusion (Fig. 10 M). We propose that Rhes-positive vesicles dock the membrane and initiate the membrane protrusion. This step can be facilitated by Exo70, a known regulator of membrane protrusion (Zhao et al., 2013). Actin polymerization then extends the protrusion, forming TNT-like cellular protrusions and allowing vesicles to carry cargo (Fig. 10 M). Vesicle-leading TNTs could then fuse with the target cell via unknown mechanisms and deliver the vesicles. Our EM, membrane labeling, and live imaging studies suggest that Rhes tunnels at the fusion step may be an open-ended structure. However, we cannot rule out the possibility that this step can be transiently open and then rapidly close, as proposed in the “tunnel and toll booth” mechanism (Wang et al., 2010). Alternatively, as shown in Fig. 3 I, in the primary striatal neuron, a portion of the Rhes-induced TNT-like cellular protrusions containing vesicles can be engulfed by the recipient cell, analogous to the “snatch and grab” mechanism (Wang et al., 2010; Smith et al., 2011; Al Heialy et al., 2015). Evidence for open-ended TNT-like cellular protrusions exists in several cell types that displayed some form of membrane/cytoplasmic continuity between cells (Watkins and Salter, 2005; Sartori-Rupp et al., 2019). An active gap junction has also been implicated as a gatekeeper for TNTs at the fusion step in the recipient cells (Lock et al., 2016). Whether the gap junction participates in TNT-like Rhes tunnel fusion with recipient cells remains unknown. One unique feature of TNT-like Rhes tunnels is that the vesicle-like puncta, once delivered, appear to travel along the membrane for a while (e.g., Fig. 2 B and Fig. 6 F) before being delivered into the acceptor cell. This raises a prospect that the vesicles from Rhes tunnels bearing cargoes are delivered in a regulated manner, potentially involving endocytic mechanisms. It is worth noting that Rhes interacts with multiple proteins implicated in endocytic pathways in the striatum (Shahani et al., 2016).
This report provides new insights into the role of Rhes in cell–cell communication and HD, potentially by spreading mHTT via Rhes tunnels in the striatum. It is important to note that mHTT does not seem to affect Rhes transportation via Rhes tunnels, but Rhes is a critical for the mHTT transportation between striatal cells. In vivo evidence for mHTT transport between striatal cells was obtained from implant studies in HD patients. When implanted, the healthy fetal striatal cells eventually degenerated in the striatum of HD patients, and histopathology revealed the intracellular accumulation of mHTT in the implants and enhanced inflammatory changes around the implant. This patient study suggested that mHTT moves from the affected HD striatum to healthy striatal cells (Cicchetti et al., 2009, 2014). Studies in animal models and human-derived cells support mHTT transportation between cells (Pecho-Vrieseling et al., 2014; Babcock and Ganetzky, 2015; Jeon et al., 2016). But a precise molecular mechanism underlying the potential spread of mHTT, and more important, how it relates to the massive loss of neuronal cells in the striatum in HD, remains unclear (Vonsattel et al., 1985; Kassubek et al., 2004; Subramaniam and Snyder, 2011). Our data support that Rhes may be a critical factor for mHTT intercellular transport in the striatum. Within the striatum, Rhes mRNA is abundantly present in the medium-spiny neuron, which incurs early and massive damage in HD. It raises some critical questions, such as, what are the cell types through which Rhes transports mHTT via Rhes tunnels? This question is counterintuitive because mHTT is a ubiquitously expressed protein, and the need for cell–cell transfer remains obscure. However, mHTT expression may vary between each cell type, and, perhaps, Rhes-induced transportation of mHTT may affect the stoichiometry of mHTT levels in a cell type–specific manner. Another possibility would be that Rhes might promote the transportation of mHTT from medium-spiny neurons to inflammatory cells as a means of removing mHTT, presumably for degradation via the lysosome-mediated pathway. Or, Rhes-mediated transportation of mHTT between medium-spiny neurons and other cells in the striatum is a stochastic process that is randomly determined. A recent study indicates Rhes also mediates Tau pathology in an Alzheimer’s disease mouse model (Hernandez et al., 2019). Although we did not observe wtTau being transported by Rhes tunnels, but Tau mutants may be, a possibility remains to be tested. A future mechanistic investigation of the TNT-like intercellular communication by Rhes and its corroboration in ex vivo and in vivo models is needed. Also, studies on mHTT transport via Rhes tunnels will generate a new perspective in understanding of the mechanisms of striatal vulnerability in HD, which may lead to the novel therapeutic opportunities to block the cell–cell movement of mHTT by Rhes, and thus, HD progression.
Materials and methods
Cell culture and chemicals
Mouse normal striatal neuronal cells (STHdhQ7/Q7) or HD mutants (STHdhQ111/Q111; Trettel et al., 2000) were cultured in growth medium containing Dulbecco’s modified Eagle’s medium (Thermo Fisher Scientific) with 10% FBS and 1% penicillin-streptomycin, as described in our previous works (Subramaniam et al., 2009; Pryor et al., 2014; Shahani et al., 2016).
Primary neuron culture
Animals were cared for in accordance with the guidelines set forth by the National Institutes of Health regarding the proper treatment and use of laboratory animals and with the approval of Institutional Animal Care and Use Committee of The Scripps Research Institute. Striata of postnatal day 1 C57BL/6 mice were removed and digested at 37°C for 15 min in a final concentration of 0.25% papain and resuspended in neuronal plating media (Neurobasal-A media; Thermo Fisher Scientific), with 5% FBS, 0.5 mM glutamax, and 1% penicillin-streptomycin. Tissues were dissociated by trituration with a pipette. Further, cells were plated in 35-mm glass-bottom dishes (D11140H; Matsunami) coated with 100 µg/ml poly-D-lysine at the density of 2 × 105 cells per dish. Dishes were maintained in a 37°C, 5% CO2 incubator. After the cells adhered (1–3 h after plating), plating media were replaced with growth media (Neurobasal-A media, 2% B27, 0.5 mM glutamax, and 1% penicillin-streptomycin).
Antibodies, chemicals, and treatments of cells
Huntingtin mouse monoclonal antibody (MAB 2166) was purchased from Millipore. GFP (sc-9996) and actin monoclonal antibody (sc-47778) were obtained from Santa Cruz Biotechnology. mTOR (# 2983), SUMO1 (# 4930), and SUMO2/3 (# 4974) antibodies were obtained from Cell Signaling Technologies. Alexa Fluor 568 anti-mouse antibody was purchased from Thermo Fisher Scientific. Cytochalasin D was purchased from Tocris Biosciences (# 1233). For the actin polymerization inhibition experiment, cytochalasin D (2 µg/ml) was added for 8 h to the cells after 40 h of the transfection of indicated DNA. Mitotracker (M7511) was purchased from Thermo Fisher Scientific. For mitochondrial staining, mitotracker was dissolved in DMSO and used at 200 nM concentration. Mitotracker was added for 30 min after 48 h of Rhes transfection; cells were washed with D-PBS and fixed with 1% PFA. Vybrant DiD cell-labeling solution was purchased from Thermo Fisher Scientific (V22887) and used as per the manufacturer’s recommendations.
Plasmids and transfection
For GFP-Rhes and GFP-Rhes domains, we amplified their respective cDNA from pCMV-Myc-Rhes (Subramaniam et al., 2009) and cloned it in EGFP-C1 vector. These cDNA were amplified using specific primers and then subcloned at BspEI and Sal1 restriction sites. GFP-Rhes mutants were generated by site-directed mutagenesis. mCherry N171 18Q, mCherry N171 89Q, mCherry poly 72Q, and mHTT N171 K/R mutant cDNA were amplified using the PCR-based method. Further, these cDNA were subcloned in the mCherry-C1 vector at BspEI and BamHI restriction sites. GFP-Atxn3 84Q was obtained from Addgene (22123). It was subcloned in mCherry-C1 vector using SalI and MluI restriction enzymes and confirmed by DNA sequencing. The following plasmids were obtained from Addgene (Table 2). Striatal neuronal cells seeded in 35-mm glass-bottom dishes or other plates were transfected 24 h later with cDNA constructs using PolyFect (Qiagen) as per the manufacturer’s instructions. Primary neurons were transfected with Lipofectamine 2000 (Invitrogen) in the ratio of 1:2 (DNA:Lipofectamine). For a 10-cm dish, we transfected ∼8 µg of DNA, and for 35-mm dishes, ranging from 1 to 2 µg total DNA.
Table 2. Plasmids used in this study obtained from Addgene.
| Plasmids | Provided by | Addgene ID |
|---|---|---|
| GFP-RhoA | Channing Der (The University of North Carolina, Chapel Hill, NC) | 23224 |
| mCherry-alpha-tubulin | Gia Voeltz (University of Colorado Boulder, Boulder, CO) | 49149 |
| mCherry-actin 7 | Michael Davidson (The Florida state University, Tallahassee, FL) | 54966 |
| pEGFP-C3-Exo70 | Channing Der (The University of North Carolina, Chapel Hill, NC) | 53761 |
| Rab5a-pmCherryC1 | Choursistein Merrifield (Institut de biologie integrative de la cellule, Gif-sur-Yvette, France) | 27679 |
| BiP-mCherry | Erik Snapp (Janelia Research Campus, Ashburn, VA) | 62233 |
| mCherry-peroxisome-2 | Michael Davidson (The Florida state University, Tallahassee, FL) | 54520 |
| mCherry-Golgi-7 | Michael Davidson (The Florida state University, Tallahassee, FL) | 55052 |
| mCherry-Lysosomes-20 | Michael Davidson (The Florida state University, Tallahassee, FL) | 55073 |
| mApple-Fibrillarin-7 | Michael Davidson (The Florida state University, Tallahassee, FL) | 54900 |
| pEGFP-C1-Ataxin3Q84 | Henry Paulson (University of Michigan health system, Ann Arbor, MI) | 22123 |
| mCherry-MAPTau-C-10 | Michael Davidson (The Florida state University, Tallahassee, FL) | 55077 |
| mCherry-Farnesyl 5 | Michael Davidson (The Florida state University, Tallahassee, FL) | 55045 |
| peYFP-C1-mTOR | Jie Chen and Taekjip Ha (University of Illinois, Chicago, IL) | 73384 |
| pmCherry-vinculin | Chinten James Lim (University of British coloumbia, Vancouver, Canada) | 80024 |
Flow cytometry
We used different FACS strategies to address Rhes-induced TNT like processes and mHTT transportation. Each of them is discussed here briefly.
In Fig. 3, A–D, striatal neuronal cells were transfected with GFP, GFP-RhoA, GFP-Rhes, or mCherry cDNA constructs in a 10-cm dish. After 48 h, cells were trypsinized and resuspended in FACS buffer (25 mM Hepes, 10 U/ml DNase, 5 mM EDTA, and 2% FBS in Ca2+/Mg2+-free D-PBS). Cells were filtered through a 40-µm nylon filter and sorted in a BD Biosciences FACSAria sorter. After sorting, cells were co-cultured in 12-well plates in equal ratios (1:1) for 24 h. Further cells were trypsinized and washed three times with D-PBS and analyzed by flow cytometry (BD Biosciences; LSR Fortessa cell analyzer). Each experiment was performed three times, and 10,000–20,000 cells were counted for each sample. Data were compensated with single color controls and plotted using FlowJo software.
In Fig. 3, F and G, FACS-sorted GFP-Rhes or mCherry alone–expressing cells were plated together or on 0.4-µm filters (Nunc) placed on top of FACS-sorted mCherry-expressing cells that prevent cell–cell contact. After co-culture for 24 h, the filters were removed, and cells were analyzed by flow cytometry as described above. The indicated FACS-sorted cells in Fig. 3 H were co-cultured for 12 h and then treated with vehicle or cytochalasin D (2 µg/ml) for another 12 h.
In Fig. 4, H and I, striatal neuronal cells were transfected with GFP-Rhes or indicated mutants/domains or mCherry construct in 10-cm dishes (one dish for GFP-tagged protein and four dishes for mCherry). After 48 h, cells were FACS-sorted and co-cultured. After 24 h, cells were analyzed by flow cytometry as described above.
In Fig. 5, B–G, Striatal neuronal cells were transfected with BFP-C1 plasmid (8 µg) in six 10-cm dishes at day 0. At day 1, a different set of striatal neuronal cells was transfected with a GFP-Rhes WT or GFP-Rhes 171–266 or GFP-Rhes C263S plasmid (8 µg) in two 10-cm dishes for each. At day 2, the cells transfected with BFP-C1 (48-h transfection) were sorted to get a BFP+ pure population (acceptor cells) based on blue fluorescence. BFP+ cells were seeded in 12-well plates for 24 h. At day 3, Rhes WT or GFP-Rhes 171–266 or GFP-Rhes C263S–expressing cells (48-h transfection) were incubated with DiD dye according to the manufacturer’s recommendations for 1 h and sorted as a double-positive population (donor cells) using GFP and an Alexa Fluor 633 laser and co-cultured with already seeded BFP+ cells for 24 h. After that, cells were analyzed by FACS or confocal imaging as described above. Data were presented as a BFP+ population that was gated and analyzed for migration of GFP/mCherry-tagged proteins.
In Fig. 7, A–E, striatal neuronal cells were transfected with BFP-C1 plasmid (8 µg) in six 10-cm dishes at day 0. At day 1, a different set of striatal neuronal cells was cotransfected with a combination of GFP + mCherry wtHTT/mHTT or GFP-Rhes + mCherry wtHTT/mHTT (4 µg each) in two 10-cm dishes for each. At day 2, the cells transfected with BFP-C1 (48 h transfection) were sorted to get a BFP+ pure population (acceptor cells) based on blue fluorescence. BFP+ cells were seeded in 12-well plates for 24 h. At day 3, cotransfected cells (GFP + mCherry wtHTT/mHTT or GFP-Rhes + mCherry wtHTT/mHTT) were sorted (48 h transfection) as a double-positive population (donor cells) and co-cultured with already seeded BFP+ cells for 24 h. After that, cells were analyzed by FACS as described above. Data were presented as a BFP+ population that was gated and analyzed for migration of GFP/mCherry-tagged proteins. A similar strategy was used for other co-culture experiments.
Similarly, in the experiment shown in Fig. 8, A–C, FACS-sorted GFP-Rhes and mCherry-mHtt double-positive cells were plated on 0.4-µm filters (Nunc) placed on top of FACS-sorted BFP-expressing cells that prevent cell–cell contact. After co-culture for 24 h, the filters were removed and the BFP-expressing cells were analyzed by flow cytometry as described above.
Immunofluorescence
At the indicated times after transfection, cells were washed in D-PBS and fixed for 5 min in 1% PFA (Electron Microscopy Sciences). Note, 4% PFA or prolonged incubation with PFA disrupts highly fragile TNT-like processes. The cells were permeabilized with 0.1% Triton X-100 and labeled with mouse anti-Huntingtin antibody (1:100 for 18 h at 4°C) or rabbit anti-mTOR antibody (1:400 for 18 h at 4°C). The Alexa Fluor 568 secondary antibodies were used at 1:500 for 1 h at room temperature.
Image processing and TNT-like protrusion quantification
All the fluorescent confocal images were taken in Zeiss 880 microscope using 20× or 63× oil immersion Plan-apochromat objective (1.4 NA). Excitation was via a 405-, 561-, or 633-nm diode-pumped solid-state laser, and the 488-nm line of an argon ion laser. Time-lapse acquisitions were performed using a 63× oil-immersion lens (1.4 NA). Images of striatal neuronal cells used for 3D reconstruction and TNT-like protrusion detection were acquired with an optimal Z-step of 0.27 µm covering the whole cellular volume. Processing was performed with Zen software black/blue edition 2012. 3D analyses and remodeling were done by Zen 2012 black edition software. For protrusion quantification, we counted all cells with filopodia-like structures that were >10 µm in length and <200 nm. For TNT-like process quantification among these protrusions, we used the same parameters except that they had to have been connected to two cells.
Generation of SUMO 1/2/3 (triple) KO striatal cells
SUMO 1/2/3 (triple) KO striatal neuronal cells were generated using SUMO1/2/3 CRISPR/Cas9 plasmids from Santa Cruz Biotechnology. First, we transfected the striatal neuronal cells with SUMO1 CRISPR/Cas9 plasmid (SC-423588) or CRISPR/Cas9 control plasmid (SC-418922) in the 10-cm dish. After 48 h, we sorted the cells based on GFP fluorescence and recultured them. We passaged them three to four times and prepared lysate to confirm the SUMO1 depletion by Western blotting using SUMO1 antibody. Once we established the SUMO1 reduction, we transfected them with SUMO2 CRISPR/Cas9 plasmid (SC-431342) and sorted them after 48 h. Again, cells were passaged three to four times and proceeded for SUMO3 CRISPR/Cas9 (SC-423045) transfection followed by sorting. Finally, cells were blotted for SUMO1 or SUMO2/3 antibody to assess the depletion of SUMO protein.
Protein expression and Western blots
To check the protein expression level of GFP-Rhes or its domains or mutants (Fig. S4 B), striatal neuronal cells (STHdhQ7/Q7) were transfected with an indicated plasmid (1.5 µg per well) using 3 µl Lipofectamine 2000. After 48 h, cells were lysed in buffer containing 1% Triton X-100 in 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, and protease inhibitor cocktail. 40 µg of protein extracts from each sample was separated on polyacrylamide gels and transferred to polyvinylidene fluoride membranes for immunoblotting (Shahani et al., 2016). Where indicated, the band intensities were quantified with ImageJ software.
EM
TEM and scanning EM were performed in the Emory EM Core facility using a protocol described previously (Kumar et al., 2017).
TEM
Monolayer cell samples close to 90% confluency were fixed with 2.5% glutaraldehyde in 0.1 M cacodylate buffer. Samples were then washed twice with 0.1 M cacodylate buffer and post-fixed with buffered 1% osmium tetroxide at room temperature for 1 h. Following buffer washes, samples were dehydrated, infiltrated, and embedded in Epnate 12 resin (Ted Pella). Ultrathin sections were cut at 70–80 nm and counterstained using uranyl acetate and lead citrate. The examination of the ultrathin sections was performed on a JEOL JEM-1400 transmission electron microscope equipped with a Gatan US1000 CCD camera (Gatan).
Scanning EM
Monolayer cells fixed with 2.5% glutaraldehyde in 0.1 M cacodylate were placed in a 4°C refrigerator overnight. Samples were then rinsed with 0.1 M cacodylate buffer twice before post-fixation with 1% osmium tetroxide in 0.1 M cacodylate for 1 h. Samples were then rinsed in deionized water, dehydrated through an ethanol series, and passed through 100% dry ethanol. The dehydrated monolayer cell samples were placed into labeled microporous specimen capsules and loaded into the sample boat of a chilled Polaron E3000 critical point drying unit. The unit was sealed and filled with liquid CO2 under pressure. The CO2 was allowed to gently wash through the chamber and exchange for the ethanol in the tissue. When the exchange was complete, the CO2 was brought to its critical point of 1,073 psi and 31°C and allowed to gently bleed away. The monolayer cell sample was secured on labeled scanning EM stubs. The stubs were sputter-coated with 15 nm chromium using a Denton DV-602 magnetron sputter coater. The samples were imaged at 10 kV using the upper stage of a Topcon DS130 field emission scanning electron microscope, and images collected using a Quartz PCI digital image collection system.
Statistical analysis
Unless otherwise noted, all experiments were performed in duplicate and repeated at least three times. The statistical comparison was performed between groups using one-way ANOVA and Student’s t test, and significance values were set at P < 0.05, using Graph Pad Prism7.
Online supplemental material
Fig. S1 shows Rhes-induced TNT-like cell protrusions, Rhes tunnels. Fig. S2 shows scanning EM and TEM images for Rhes-induced TNT-like cell protrusions and Rhes transportation to a neighboring cell and GFP-Rhes tunnels negative for tubulin or vinculin. Fig. S3 shows Rhes-induced TNT-like cell protrusions are positive for Exo70 and numerous GFP-Rhes puncta in the neigboring cells of a primary striatal neuron transfected with GFP-Rhes. Fig. S4 represents the role of Rhes domain/mutants in production of Rhes-induced TNT-like cell protrusions. Fig. S5 shows mHTT N171 89Q transportation via Rhes-induced TNT-like Rhes tunnels. Fig. S6 shows that Rhes tunnels are negative for mTOR and wtTau proteins. Fig. S7 shows the experimental design for various experiments and SUMO 1/2/3 protein depletion. Video 1 shows Rhes-induced cellular protrusions, and corresponding b1, b2, and b3 inset videos. Videos 2 and 3 show the effect of DMSO or actin polymerization inhibitor on TNT-like Rhes tunnels. Video 4 is an animation of Fig. 3 D showing Rhes tunnels interacting with mCherry cells and delivering GFP-Rhes. Video 5 shows a GFP-Rhes–positive neuron (cell 1) making contact with cell 2 and delivering GFP-Rhes puncta. Video 6 is an animation of a cellular protrusion in Video 5 showing multiple vesicles in cell 1 (thin arrow) and a delivery of GFP-Rhes puncta (thick arrow) to cell 2. Video 7 shows mHTT transport from cell 1 to cell 2 via Rhes tunnels (GFP). Video 8 is another example that shows Rhes and mHTT transport through a Rhes tunnel (GFP) and their delivery to the neighboring cells. Here, a “leading” Rhes and mHTT puncta, a “just approaching” Rhes and mHTT puncta, and a retracting Rhes tunnel can be observed (see text for details).
Supplementary Material
Acknowledgments
We would like to thank Nicole Galli for artwork of the model; Dr. Neelam Shahani for critically reading and providing suggestions to improve the manuscript; Mary McAllister for editing services; Dr. Yi Hong (Emory University, Atlanta, GA) for EM support; Dr. Long Yan (Max Planck Institute of Neuroscience, Jupiter, FL) for confocal imaging help; and Alta Johnson and Bivian Torres (flow cytometry core, The Scripps Research Institute, Jupiter, FL) for help in cell sorting and data analysis.
This research was partially supported by a training grant in Alzheimer’s drug discovery from the Lottie French Lewis Fund of the Community Foundation for Palm Beach and Martin Counties. This research was supported by funding from National Institutes of Health/National Institute of Neurological Disorders and Stroke grant R01-NS087019-01A1, National Institutes of Health/National Institute of Neurological Disorders and Stroke grant R01-NS094577-01A1, and grants from Cure Huntington Disease Initiative (CHDI) foundation.
The authors declare no competing financial interests.
Author contributions: M. Sharma made initial observations. S. Subramaniam further conceptualized and co-designed the project with M. Sharma. M. Sharma carried out all the experiments. M. Sharma and S. Subramaniam analyzed the data. S. Subramaniam wrote the manuscript with input from M. Sharma.
References
- Abounit S., Wu J.W., Duff K., Victoria G.S., and Zurzolo C.. 2016. Tunneling nanotubes: A possible highway in the spreading of tau and other prion-like proteins in neurodegenerative diseases. Prion. 10:344–351. 10.1080/19336896.2016.1223003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Heialy S., Zeroual M., Farahnak S., McGovern T., Risse P.A., Novali M., Lauzon A.M., Roman H.N., and Martin J.G.. 2015. Nanotubes connect CD4+ T cells to airway smooth muscle cells: novel mechanism of T cell survival. J. Immunol. 194:5626–5634. 10.4049/jimmunol.1401718 [DOI] [PubMed] [Google Scholar]
- Alves S., Régulier E., Nascimento-Ferreira I., Hassig R., Dufour N., Koeppen A., Carvalho A.L., Simões S., de Lima M.C., Brouillet E., et al. . 2008. Striatal and nigral pathology in a lentiviral rat model of Machado-Joseph disease. Hum. Mol. Genet. 17:2071–2083. 10.1093/hmg/ddn106 [DOI] [PubMed] [Google Scholar]
- Argenti M.2014. The role of mitochondrial dysfunction in Huntington’s disease pathogenesis and its relation with striatal Rhes protein. Ph.D Thesis. Università degli Studi di Padova, Padova, Italy. [Google Scholar]
- Ariazi J., Benowitz A., De Biasi V., Den Boer M.L., Cherqui S., Cui H., Douillet N., Eugenin E.A., Favre D., Goodman S., et al. . 2017. Tunneling Nanotubes and Gap Junctions-Their Role in Long-Range Intercellular Communication during Development, Health, and Disease Conditions. Front. Mol. Neurosci. 10:333 10.3389/fnmol.2017.00333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austefjord M.W., Gerdes H.H., and Wang X.. 2014. Tunneling nanotubes: Diversity in morphology and structure. Commun. Integr. Biol. 7:e27934 10.4161/cib.27934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babcock D.T., and Ganetzky B.. 2015. Transcellular spreading of huntingtin aggregates in the Drosophila brain. Proc. Natl. Acad. Sci. USA. 112:E5427–E5433. 10.1073/pnas.1516217112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baiamonte B.A., Lee F.A., Brewer S.T., Spano D., and LaHoste G.J.. 2013. Attenuation of Rhes activity significantly delays the appearance of behavioral symptoms in a mouse model of Huntington’s disease. PLoS One. 8:e53606 10.1371/journal.pone.0053606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beier C., Palanker D., and Sher A.. 2018. Stereotyped Synaptic Connectivity Is Restored during Circuit Repair in the Adult Mammalian Retina. Curr. Biol. 28:1818–1824.e2. 10.1016/j.cub.2018.04.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff M., Gradilla A.C., Seijo I., Andrés G., Rodríguez-Navas C., González-Méndez L., and Guerrero I.. 2013. Cytonemes are required for the establishment of a normal Hedgehog morphogen gradient in Drosophila epithelia. Nat. Cell Biol. 15:1269–1281. 10.1038/ncb2856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burtey A., Wagner M., Hodneland E., Skaftnesmo K.O., Schoelermann J., Mondragon I.R., Espedal H., Golebiewska A., Niclou S.P., Bjerkvig R., et al. . 2015. Intercellular transfer of transferrin receptor by a contact-, Rab8-dependent mechanism involving tunneling nanotubes. FASEB J. 29:4695–4712. 10.1096/fj.14-268615 [DOI] [PubMed] [Google Scholar]
- Caneparo L., Pantazis P., Dempsey W., and Fraser S.E.. 2011. Intercellular bridges in vertebrate gastrulation. PLoS One. 6:e20230 10.1371/journal.pone.0020230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervera J., Pietak A., Levin M., and Mafe S.. 2018. Bioelectrical coupling in multicellular domains regulated by gap junctions: A conceptual approach. Bioelectrochemistry. 123:45–61. 10.1016/j.bioelechem.2018.04.013 [DOI] [PubMed] [Google Scholar]
- Chinnery H.R., Pearlman E., and McMenamin P.G.. 2008. Cutting edge: Membrane nanotubes in vivo: a feature of MHC class II+ cells in the mouse cornea. J. Immunol. 180:5779–5783. 10.4049/jimmunol.180.9.5779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicchetti F., Saporta S., Hauser R.A., Parent M., Saint-Pierre M., Sanberg P.R., Li X.J., Parker J.R., Chu Y., Mufson E.J., et al. . 2009. Neural transplants in patients with Huntington’s disease undergo disease-like neuronal degeneration. Proc. Natl. Acad. Sci. USA. 106:12483–12488. 10.1073/pnas.0904239106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicchetti F., Lacroix S., Cisbani G., Vallières N., Saint-Pierre M., St-Amour I., Tolouei R., Skepper J.N., Hauser R.A., Mantovani D., et al. . 2014. Mutant huntingtin is present in neuronal grafts in Huntington disease patients. Ann. Neurol. 76:31–42. 10.1002/ana.24174 [DOI] [PubMed] [Google Scholar]
- Costanzo M., Abounit S., Marzo L., Danckaert A., Chamoun Z., Roux P., and Zurzolo C.. 2013. Transfer of polyglutamine aggregates in neuronal cells occurs in tunneling nanotubes. J. Cell Sci. 126:3678–3685. 10.1242/jcs.126086 [DOI] [PubMed] [Google Scholar]
- Delage E., Cervantes D.C., Pénard E., Schmitt C., Syan S., Disanza A., Scita G., and Zurzolo C.. 2016. Differential identity of Filopodia and Tunneling Nanotubes revealed by the opposite functions of actin regulatory complexes. Sci. Rep. 6:39632 10.1038/srep39632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desir S., Dickson E.L., Vogel R.I., Thayanithy V., Wong P., Teoh D., Geller M.A., Steer C.J., Subramanian S., and Lou E.. 2016. Tunneling nanotube formation is stimulated by hypoxia in ovarian cancer cells. Oncotarget. 7:43150–43161. 10.18632/oncotarget.9504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte-Neves J., Gonçalves N., Cunha-Santos J., Simões A.T., den Dunnen W.F., Hirai H., Kügler S., Cavadas C., and Pereira de Almeida L.. 2015. Neuropeptide Y mitigates neuropathology and motor deficits in mouse models of Machado-Joseph disease. Hum. Mol. Genet. 24:5451–5463. 10.1093/hmg/ddv271 [DOI] [PubMed] [Google Scholar]
- Dupont M., Souriant S., Lugo-Villarino G., Maridonneau-Parini I., and Vérollet C.. 2018. Tunneling Nanotubes: Intimate Communication between Myeloid Cells. Front. Immunol. 9:43 10.3389/fimmu.2018.00043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farquhar M.G., and Palade G.E.. 1965. Cell junctions in amphibian skin. J. Cell Biol. 26:263–291. 10.1083/jcb.26.1.263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes H.H., Bukoreshtliev N.V., and Barroso J.F.. 2007. Tunneling nanotubes: a new route for the exchange of components between animal cells. FEBS Lett. 581:2194–2201. 10.1016/j.febslet.2007.03.071 [DOI] [PubMed] [Google Scholar]
- Gerdes H.H., Rustom A., and Wang X.. 2013. Tunneling nanotubes, an emerging intercellular communication route in development. Mech. Dev. 130:381–387. 10.1016/j.mod.2012.11.006 [DOI] [PubMed] [Google Scholar]
- Gousset K., Marzo L., Commere P.H., and Zurzolo C.. 2013. Myo10 is a key regulator of TNT formation in neuronal cells. J. Cell Sci. 126:4424–4435. 10.1242/jcs.129239 [DOI] [PubMed] [Google Scholar]
- Guo R., Davis D., and Fang Y.. 2018. Intercellular transfer of mitochondria rescues virus-induced cell death but facilitates cell-to-cell spreading of porcine reproductive and respiratory syndrome virus. Virology. 517:122–134. 10.1016/j.virol.2017.12.018 [DOI] [PubMed] [Google Scholar]
- Hase K., Kimura S., Takatsu H., Ohmae M., Kawano S., Kitamura H., Ito M., Watarai H., Hazelett C.C., Yeaman C., and Ohno H.. 2009. M-Sec promotes membrane nanotube formation by interacting with Ral and the exocyst complex. Nat. Cell Biol. 11:1427–1432. 10.1038/ncb1990 [DOI] [PubMed] [Google Scholar]
- Hashimoto M., Bhuyan F., Hiyoshi M., Noyori O., Nasser H., Miyazaki M., Saito T., Kondoh Y., Osada H., Kimura S., et al. . 2016. Potential Role of the Formation of Tunneling Nanotubes in HIV-1 Spread in Macrophages. J. Immunol. 196:1832–1841. 10.4049/jimmunol.1500845 [DOI] [PubMed] [Google Scholar]
- Hernandez I., Luna G., Rauch J.N., Reis S.A., Giroux M., Karch C.M., Boctor D., Sibih Y.E., Storm N.J., Diaz A., et al. . 2019. A farnesyltransferase inhibitor activates lysosomes and reduces tau pathology in mice with tauopathy. Sci. Transl. Med. 11:eaat3005 10.1126/scitranslmed.aat3005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y., Zhao T., Li X.J., and Li S.. 2017. Mutant Huntingtin Inhibits αB-Crystallin Expression and Impairs Exosome Secretion from Astrocytes. J. Neurosci. 37:9550–9563. 10.1523/JNEUROSCI.1418-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiung F., Ramirez-Weber F.A., Iwaki D.D., and Kornberg T.B.. 2005. Dependence of Drosophila wing imaginal disc cytonemes on Decapentaplegic. Nature. 437:560–563. 10.1038/nature03951 [DOI] [PubMed] [Google Scholar]
- Jansens R.J.J., Van den Broeck W., De Pelsmaeker S., Lamote J.A.S., Van Waesberghe C., Couck L., and Favoreel H.W.. 2017. Pseudorabies virus US3-induced tunneling nanotubes contain stabilized microtubules, interact with neighbouring cells via cadherins and allow intercellular molecular communication. J. Virol. 10.1128/JVI.00749-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon I., Cicchetti F., Cisbani G., Lee S., Li E., Bae J., Lee N., Li L., Im W., Kim M., et al. . 2016. Human-to-mouse prion-like propagation of mutant huntingtin protein. Acta Neuropathol. 132:577–592. 10.1007/s00401-016-1582-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone R.M., Adam M., Hammond J.R., Orr L., and Turbide C.. 1987. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). J. Biol. Chem. 262:9412–9420. [PubMed] [Google Scholar]
- Kassubek J., Juengling F.D., Kioschies T., Henkel K., Karitzky J., Kramer B., Ecker D., Andrich J., Saft C., Kraus P., et al. . 2004. Topography of cerebral atrophy in early Huntington’s disease: a voxel based morphometric MRI study. J. Neurol. Neurosurg. Psychiatry. 75:213–220. [PMC free article] [PubMed] [Google Scholar]
- Keller K.E., Bradley J.M., Sun Y.Y., Yang Y.F., and Acott T.S.. 2017. Tunneling Nanotubes are Novel Cellular Structures That Communicate Signals Between Trabecular Meshwork Cells. Invest. Ophthalmol. Vis. Sci. 58:5298–5307. 10.1167/iovs.17-22732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., Kim J.H., Ranjan P., Metcalfe M.G., Cao W., Mishina M., Gangappa S., Guo Z., Boyden E.S., Zaki S., et al. . 2017. Influenza virus exploits tunneling nanotubes for cell-to-cell spread. Sci. Rep. 7:40360 10.1038/srep40360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Zhao Y., Sun Y., He B., Yang C., Svitkina T., Goldman Y.E., and Guo W.. 2012. Exo70 stimulates the Arp2/3 complex for lamellipodia formation and directional cell migration. Curr. Biol. 22:1510–1515. 10.1016/j.cub.2012.05.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd D.P., and McIntyre A.K.. 1955. Transmitter potentiality of homonymous and heteronymous monosynaptic reflex connections of individual motoneurons. J. Gen. Physiol. 38:789–799. 10.1085/jgp.38.6.789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lock J.T., Parker I., and Smith I.F.. 2016. Communication of Ca(2+) signals via tunneling membrane nanotubes is mediated by transmission of inositol trisphosphate through gap junctions. Cell Calcium. 60:266–272. 10.1016/j.ceca.2016.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou E., Fujisawa S., Morozov A., Barlas A., Romin Y., Dogan Y., Gholami S., Moreira A.L., Manova-Todorova K., and Moore M.A.. 2012. Tunneling nanotubes provide a unique conduit for intercellular transfer of cellular contents in human malignant pleural mesothelioma. PLoS One. 7:e33093 10.1371/journal.pone.0033093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B., and Palacino J.. 2013. A novel human embryonic stem cell-derived Huntington’s disease neuronal model exhibits mutant huntingtin (mHTT) aggregates and soluble mHTT-dependent neurodegeneration. FASEB J. 27:1820–1829. 10.1096/fj.12-219220 [DOI] [PubMed] [Google Scholar]
- McColgan P., and Tabrizi S.J.. 2018. Huntington’s disease: a clinical review. Eur. J. Neurol. 25:24–34. 10.1111/ene.13413 [DOI] [PubMed] [Google Scholar]
- Miller J., Fraser S.E., and McClay D.. 1995. Dynamics of thin filopodia during sea urchin gastrulation. Development. 121:2501–2511. [DOI] [PubMed] [Google Scholar]
- O’Rourke J.G., Gareau J.R., Ochaba J., Song W., Raskó T., Reverter D., Lee J., Monteys A.M., Pallos J., Mee L., et al. . 2013. SUMO-2 and PIAS1 modulate insoluble mutant huntingtin protein accumulation. Cell Reports. 4:362–375. 10.1016/j.celrep.2013.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto S., Pouladi M.A., Talantova M., Yao D., Xia P., Ehrnhoefer D.E., Zaidi R., Clemente A., Kaul M., Graham R.K., et al. . 2009. Balance between synaptic versus extrasynaptic NMDA receptor activity influences inclusions and neurotoxicity of mutant huntingtin. Nat. Med. 15:1407–1413. 10.1038/nm.2056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osswald M., Jung E., Sahm F., Solecki G., Venkataramani V., Blaes J., Weil S., Horstmann H., Wiestler B., Syed M., et al. . 2015. Brain tumour cells interconnect to a functional and resistant network. Nature. 528:93–98. [DOI] [PubMed] [Google Scholar]
- Panasiuk M., Rychłowski M., Derewońko N., and Bieńkowska-Szewczyk K.. 2018. Tunneling Nanotubes as a Novel Route of Cell-to-Cell Spread of Herpesviruses. J. Virol. 92:e00090-18 10.1128/JVI.00090-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecho-Vrieseling E., Rieker C., Fuchs S., Bleckmann D., Esposito M.S., Botta P., Goldstein C., Bernhard M., Galimberti I., Müller M., et al. . 2014. Transneuronal propagation of mutant huntingtin contributes to non-cell autonomous pathology in neurons. Nat. Neurosci. 17:1064–1072. 10.1038/nn.3761 [DOI] [PubMed] [Google Scholar]
- Polak R., de Rooij B., Pieters R., and den Boer M.L.. 2015. B-cell precursor acute lymphoblastic leukemia cells use tunneling nanotubes to orchestrate their microenvironment. Blood. 126:2404–2414. 10.1182/blood-2015-03-634238 [DOI] [PubMed] [Google Scholar]
- Pryor W.M., Biagioli M., Shahani N., Swarnkar S., Huang W.C., Page D.T., MacDonald M.E., and Subramaniam S.. 2014. Huntingtin promotes mTORC1 signaling in the pathogenesis of Huntington’s disease. Sci. Signal. 7:ra103 10.1126/scisignal.2005633 [DOI] [PubMed] [Google Scholar]
- Pyrgaki C., Trainor P., Hadjantonakis A.K., and Niswander L.. 2010. Dynamic imaging of mammalian neural tube closure. Dev. Biol. 344:941–947. 10.1016/j.ydbio.2010.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez-Weber F.A., and Kornberg T.B.. 1999. Cytonemes: cellular processes that project to the principal signaling center in Drosophila imaginal discs. Cell. 97:599–607. 10.1016/S0092-8674(00)80771-0 [DOI] [PubMed] [Google Scholar]
- Reiner A., Albin R.L., Anderson K.D., D’Amato C.J., Penney J.B., and Young A.B.. 1988. Differential loss of striatal projection neurons in Huntington disease. Proc. Natl. Acad. Sci. USA. 85:5733–5737. 10.1073/pnas.85.15.5733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustom A., Saffrich R., Markovic I., Walther P., and Gerdes H.H.. 2004. Nanotubular highways for intercellular organelle transport. Science. 303:1007–1010. 10.1126/science.1093133 [DOI] [PubMed] [Google Scholar]
- Salas-Vidal E., and Lomelí H.. 2004. Imaging filopodia dynamics in the mouse blastocyst. Dev. Biol. 265:75–89. 10.1016/j.ydbio.2003.09.012 [DOI] [PubMed] [Google Scholar]
- Sartori-Rupp A., Cordero Cervantes D., Pepe A., Gousset K., Delage E., Corroyer-Dulmont S., Schmitt C., Krijnse-Locker J., and Zurzolo C.. 2019. Correlative cryo-electron microscopy reveals the structure of TNTs in neuronal cells. Nat. Commun. 10:342 10.1038/s41467-018-08178-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sbodio J.I., Paul B.D., Machamer C.E., and Snyder S.H.. 2013. Golgi protein ACBD3 mediates neurotoxicity associated with Huntington’s disease. Cell Reports. 4:890–897. 10.1016/j.celrep.2013.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller C., Huber J.E., Diakopoulos K.N., and Weiss E.H.. 2013. Tunneling nanotubes enable intercellular transfer of MHC class I molecules. Hum. Immunol. 74:412–416. 10.1016/j.humimm.2012.11.026 [DOI] [PubMed] [Google Scholar]
- Seredenina T., Gokce O., and Luthi-Carter R.. 2011. Decreased striatal RGS2 expression is neuroprotective in Huntington’s disease (HD) and exemplifies a compensatory aspect of HD-induced gene regulation. PLoS One. 6:e22231 10.1371/journal.pone.0022231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahani N., Swarnkar S., Giovinazzo V., Morgenweck J., Bohn L.M., Scharager-Tapia C., Pascal B., Martinez-Acedo P., Khare K., and Subramaniam S.. 2016. RasGRP1 promotes amphetamine-induced motor behavior through a Rhes interaction network (“Rhesactome”) in the striatum. Sci. Signal. 9:ra111 10.1126/scisignal.aaf6670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith I.F., Shuai J., and Parker I.. 2011. Active generation and propagation of Ca2+ signals within tunneling membrane nanotubes. Biophys. J. 100:L37–L39. 10.1016/j.bpj.2011.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowinski S., Jolly C., Berninghausen O., Purbhoo M.A., Chauveau A., Köhler K., Oddos S., Eissmann P., Brodsky F.M., Hopkins C., et al. . 2008. Membrane nanotubes physically connect T cells over long distances presenting a novel route for HIV-1 transmission. Nat. Cell Biol. 10:211–219. 10.1038/ncb1682 [DOI] [PubMed] [Google Scholar]
- Stahl P.D., and Raposo G.. 2018. Exosomes and extracellular vesicles: the path forward. Essays Biochem. 62:119–124. 10.1042/EBC20170088 [DOI] [PubMed] [Google Scholar]
- Subramaniam S., and Snyder S.H.. 2011. Huntington’s disease is a disorder of the corpus striatum: focus on Rhes (Ras homologue enriched in the striatum). Neuropharmacology. 60:1187–1192. 10.1016/j.neuropharm.2010.10.025 [DOI] [PubMed] [Google Scholar]
- Subramaniam S., Sixt K.M., Barrow R., and Snyder S.H.. 2009. Rhes, a striatal specific protein, mediates mutant-huntingtin cytotoxicity. Science. 324:1327–1330. 10.1126/science.1172871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam S., Mealer R.G., Sixt K.M., Barrow R.K., Usiello A., and Snyder S.H.. 2010. Rhes, a physiologic regulator of sumoylation, enhances cross-sumoylation between the basic sumoylation enzymes E1 and Ubc9. J. Biol. Chem. 285:20428–20432. 10.1074/jbc.C110.127191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam S., Napolitano F., Mealer R.G., Kim S., Errico F., Barrow R., Shahani N., Tyagi R., Snyder S.H., and Usiello A.. 2011. Rhes, a striatal-enriched small G protein, mediates mTOR signaling and L-DOPA-induced dyskinesia. Nat. Neurosci. 15:191–193. 10.1038/nn.2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarnkar S., Chen Y., Pryor W.M., Shahani N., Page D.T., and Subramaniam S.. 2015. Ectopic expression of the striatal-enriched GTPase Rhes elicits cerebellar degeneration and an ataxia phenotype in Huntington’s disease. Neurobiol. Dis. 82:66–77. 10.1016/j.nbd.2015.05.011 [DOI] [PubMed] [Google Scholar]
- Taniwaki T., Sakai T., Kobayashi T., Kuwabara Y., Otsuka M., Ichiya Y., Masuda K., and Goto I.. 1997. Positron emission tomography (PET) in Machado-Joseph disease. J. Neurol. Sci. 145:63–67. 10.1016/S0022-510X(96)00242-0 [DOI] [PubMed] [Google Scholar]
- Tardivel M., Bégard S., Bousset L., Dujardin S., Coens A., Melki R., Buée L., and Colin M.. 2016. Tunneling nanotube (TNT)-mediated neuron-to neuron transfer of pathological Tau protein assemblies. Acta Neuropathol. Commun. 4:117 10.1186/s40478-016-0386-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teddy J.M., and Kulesa P.M.. 2004. In vivo evidence for short- and long-range cell communication in cranial neural crest cells. Development. 131:6141–6151. 10.1242/dev.01534 [DOI] [PubMed] [Google Scholar]
- Thayanithy V., O’Hare P., Wong P., Zhao X., Steer C.J., Subramanian S., and Lou E.. 2017. A transwell assay that excludes exosomes for assessment of tunneling nanotube-mediated intercellular communication. Cell Commun. Signal. 15:46 10.1186/s12964-017-0201-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trettel F., Rigamonti D., Hilditch-Maguire P., Wheeler V.C., Sharp A.H., Persichetti F., Cattaneo E., and MacDonald M.E.. 2000. Dominant phenotypes produced by the HD mutation in STHdh(Q111) striatal cells. Hum. Mol. Genet. 9:2799–2809. 10.1093/hmg/9.19.2799 [DOI] [PubMed] [Google Scholar]
- Vignais M.L., Caicedo A., Brondello J.M., and Jorgensen C.. 2017. Cell Connections by Tunneling Nanotubes: Effects of Mitochondrial Trafficking on Target Cell Metabolism, Homeostasis, and Response to Therapy. Stem Cells Int. 2017:6917941 10.1155/2017/6917941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonsattel J.P., Myers R.H., Stevens T.J., Ferrante R.J., Bird E.D., and Richardson E.P. Jr. 1985. Neuropathological classification of Huntington’s disease. J. Neuropathol. Exp. Neurol. 44:559–577. 10.1097/00005072-198511000-00003 [DOI] [PubMed] [Google Scholar]
- Wang X., and Gerdes H.H.. 2015. Transfer of mitochondria via tunneling nanotubes rescues apoptotic PC12 cells. Cell Death Differ. 22:1181–1191. 10.1038/cdd.2014.211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Veruki M.L., Bukoreshtliev N.V., Hartveit E., and Gerdes H.H.. 2010. Animal cells connected by nanotubes can be electrically coupled through interposed gap-junction channels. Proc. Natl. Acad. Sci. USA. 107:17194–17199. 10.1073/pnas.1006785107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins S.C., and Salter R.D.. 2005. Functional connectivity between immune cells mediated by tunneling nanotubules. Immunity. 23:309–318. 10.1016/j.immuni.2005.08.009 [DOI] [PubMed] [Google Scholar]
- Wilkinson K.A., and Henley J.M.. 2010. Mechanisms, regulation and consequences of protein SUMOylation. Biochem. J. 428:133–145. 10.1042/BJ20100158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittig D., Wang X., Walter C., Gerdes H.H., Funk R.H., and Roehlecke C.. 2012. Multi-level communication of human retinal pigment epithelial cells via tunneling nanotubes. PLoS One. 7:e33195 10.1371/journal.pone.0033195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen T.C., Lu C.S., Tzen K.Y., Wey S.P., Chou Y.H., Weng Y.H., Kao P.F., and Ting G.. 2000. Decreased dopamine transporter binding in Machado-Joseph disease. J. Nucl. Med. 41:994–998. [PubMed] [Google Scholar]
- Zhao Y., Liu J., Yang C., Capraro B.R., Baumgart T., Bradley R.P., Ramakrishnan N., Xu X., Radhakrishnan R., Svitkina T., and Guo W.. 2013. Exo70 generates membrane curvature for morphogenesis and cell migration. Dev. Cell. 26:266–278. 10.1016/j.devcel.2013.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Xue C., Xu X., Guo Y., Li X., Lu J., Ju S., Wang Y., Cao Z., and Gu X.. 2016. Rab8a/Rab11a regulate intercellular communications between neural cells via tunneling nanotubes. Cell Death Dis. 7:e2523 10.1038/cddis.2016.441 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.