How mitochondria damaged in distal axons are cleared is not understood. Zheng et al. find that axonal mitochondria return to neuronal soma for mitophagy after ischemic insult. These spatial features of neuronal mitophagy provide insight into how neurons control mitochondrial quality under pathological conditions.
Abstract
Mitophagy protects against ischemic neuronal injury by eliminating damaged mitochondria, but it is unclear how mitochondria in distal axons are cleared. We find that oxygen and glucose deprivation-reperfusion reduces mitochondrial content in both cell bodies and axons. Axonal mitochondria elimination was not abolished in Atg7fl/fl;nes-Cre neurons, suggesting the absence of direct mitophagy in axons. Instead, axonal mitochondria were enwrapped by autophagosomes in soma and axon-derived mitochondria prioritized for elimination by autophagy. Intriguingly, axonal mitochondria showed prompt loss of anterograde motility but increased retrograde movement upon reperfusion. Anchoring of axonal mitochondria by syntaphilin blocked neuronal mitophagy and aggravated injury. Conversely, induced binding of mitochondria to dynein reinforced retrograde transport and enhanced mitophagy to prevent mitochondrial dysfunction and attenuate neuronal injury. Therefore, we reveal somatic autophagy of axonal mitochondria in ischemic neurons and establish a direct link of retrograde mitochondrial movement with mitophagy. Our findings may provide a new concept for reducing ischemic neuronal injury by correcting mitochondrial motility.
Introduction
Cerebral ischemia is a neurological disorder characterized by interruption of blood supply to the brain. Mitochondrial quality is critical for neuronal survival after ischemic insults (Li and Gao, 2017). Neurons monitor mitochondrial quality either by replenishing mitochondria (Yin et al., 2008; Hayakawa et al., 2016) or, more importantly, by eliminating dysfunctional mitochondria through mitophagy (Rugarli and Langer, 2012; Wang et al., 2018). Mitophagy is the sole mechanism allowing for the degradation of entire mitochondria, and mitophagy deficiency has been linked to a variety of neurological disorders (Karbowski and Neutzner, 2012). In particular, recent evidence indicated that compromised mitophagy exacerbates ischemic brain injury and, conversely, enhanced mitophagy confers neuroprotection (Zhang et al., 2013, 2014; Yuan et al., 2017), indicating the critical contribution of mitophagy to ischemic neuronal survival (Galluzzi et al., 2016).
Neurons are highly differentiated morphologically. More than half of total neuronal mitochondria are located in distal neuron compartments (Nafstad and Blackstad, 1966), where they show higher vulnerability to environmental stresses (Naga et al., 2007; Du et al., 2010). In contrast, autophagosomes and lysosomes were more concentrated in soma (Sagné et al., 2001; Maday et al., 2012; Farías et al., 2017), raising a question as to how neuronal mitophagy is spatially regulated (Ashrafi et al., 2014; Lu, 2014). Under basal conditions, autophagosomes are generated at axonal tips (Maday et al., 2012; Maday and Holzbaur, 2014) and local acute stresses induce axonal mitophagy (Ashrafi et al., 2014; Ashrafi and Schwarz, 2015). On the other hand, soma-restricted mitophagy was suggested in several systems (Cai et al., 2012; Devireddy et al., 2015; Sung et al., 2016). Moreover, mitophagosomes are not readily found in axons under stress conditions (Cai et al., 2012; Maday et al., 2012; Maday and Holzbaur, 2014). It is unclear whether local mitophagy is enough to meet the greater demand for mitochondrial turnover under intensive stress such as ischemia. In addition, mitochondria undergo dynamic movement along with microtubules in physiological and pathological conditions (Schwarz, 2013), and mitochondrial mobility may affect axonal mitophagy. Specifically, immobilized mitochondria are recognized by autophagosomes in axons after stress (Wang et al., 2011; Ashrafi et al., 2014). By contrast, a previous study identified that by budding out the anchoring protein Syntaphilin (SNPH) from mitochondria, axonal mitochondria undergo a selective bidirectional transport. Distinct from the healthy mitochondria undergoing anterogradely, damaged mitochondria go retrogradely under milder stress, serving as a first line defense of mitochondrial quality control (Lin et al., 2017a). However, whether those mitochondria undergo autophagic clearance or fusion and fission during trafficking is still unclear. Taken together, the association of axonal mitochondria motility and mitophagy needs further characterization, especially under stressful conditions like ischemia.
Therefore, we determined the spatial features of mitophagy in neurons that underwent oxygen–glucose deprivation-reperfusion, which mimicked neuronal ischemia. We found that axonal mitochondria were not eliminated locally in axons but retrogradely transported to neuronal soma for mitophagy. Importantly, enhanced retrograde transportation promoted the somatic autophagy of axonal mitochondria and attenuated mitochondrial dysfunction as well as ischemic neuronal injury. The present study identified the somatic autophagy of axonal mitochondria and its regulation by mitochondrial retrograde movement.
Results
Oxygen and glucose deprivation and reperfusion–induced neuronal mitophagy
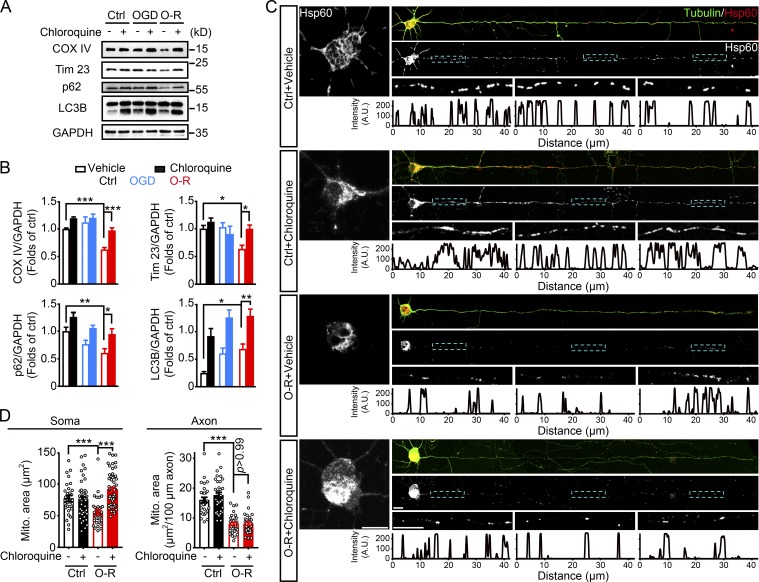
Primary cultured cortical neurons from embryonic mice brains were treated with 20 min of oxygen and glucose deprivation (OGD) plus 40 min of reperfusion. OGD-reperfusion (OGD-Rep) reduced the mitochondrial proteins cytochrome c oxidase subunit IV (COX IV) and translocase of inner mitochondrial membrane 23 (Tim23) and the autophagic adaptor protein p62 in the whole-cell lysis, but this was reversed by the lysosome inhibitor chloroquine, suggesting autophagy-dependent mitochondrial elimination. OGD alone activated neuronal autophagy (accumulation of LC3-II) but failed to induce mitophagy (Fig. 1, A and B), in line with a previous study (Zhang et al., 2013). Hsp60 was immunostained to visualize mitochondrial loss in greater detail (Fig. 1 C). Mitochondrial mass in both neuronal soma and axons was quantified by measuring mitochondrial area. Compared with control, OGD-Rep induced significant parallel mitochondrial loss in neuronal soma and axons (77.2 ± 4.6 vs. 55.6 ± 3.6 µm2 in soma; 16.0 ± 0.9 vs. 8.0 ± 0.5 µm2 per 100 µm in axons; Fig. 1, C and D). Intriguingly, chloroquine reversed mitochondrial loss only in soma but not in axons after OGD-Rep (90.7 ± 3.3 µm2 in soma [P < 0.001] vs. OGD-Rep; 8.0 ± 0.5 µm2 per 100 µm in axons [P > 0.99] vs. OGD-Rep; Fig. 1, C and D), indicating mitophagy may only occur in neuronal soma after ischemic stress.
Figure 1.
OGD-Rep induces global mitochondrial loss in neurons. (A and B) Primary cultured neurons were treated with PBS as a vehicle control or 50 µM chloroquine for 4 h and then subjected to either 1 h of OGD or 20 min of OGD plus 40 min of reperfusion (O-R) in DIV8. (A) The COX IV, Tim23, p62, LC3B, and GAPDH levels in cultured neurons were determined by Western blot. (B) Semiquantitative analysis of COX IV, Tim23, p62, and LC3B-II bands is shown. (C and D) Primary cultured neurons were subjected to 20 min of OGD in DIV8. The mitochondrial marker Hsp60 and neuronal marker β-tubulin III were stained by immunocytochemistry at 40 min after reperfusion. (C) Images show the representative straightened cultured neurons and the line-scan analysis of axonal mitochondria (lower panel). (D) Columns represent the somatic mitochondrial area (left panel) and axonal mitochondrial area per 100-µm axon (right panel). n = 30–60 cells in each group from three independent experiments were included. The data are expressed as means ± SEM. Statistical comparisons were performed with one-way ANOVA with Sidak’s multiple-comparisons test. *P < 0.05, **P < 0.01, ***P < 0.001 versus the indicated group. Scale bar, 10 µm. A.U., arbitrary units; Ctrl, control; Mito, mitochondrial.
OGD-Rep–induced axonal mitochondrial loss is independent of local mitophagy
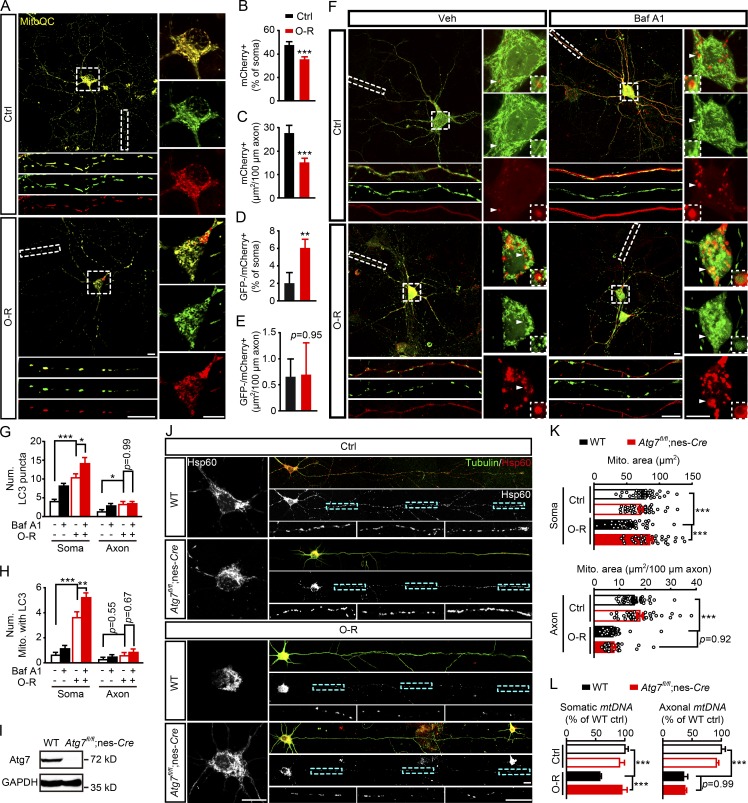
Autophagy can be activated in both neuronal soma and axons in ischemic or excitotoxic models (Wang et al., 2006; Zhang et al., 2013). To further investigate the neuronal compartments where mitophagy occurred after OGD-Rep, a tandem GFP-mCherry–tagged mitochondria-targeting probe (MitoQC) was used (Allen et al., 2013). OGD-Rep reduced the mitochondrial mass (mCherry+) in soma (Fig. 2, A and B) and axons (Fig. 2 C), which confirmed mitochondrial loss. However, OGD-Rep increased the acidic mitochondria (GFP−/mCherry+) in soma (Fig. 2 D) but not in axons (Fig. 2 E), indicating absence of mitophagy in axons. Since axonal autophagosomes may not be mature enough to quench GFP, the colocalization of mitochondria (Mito-GFP) and autophagosomes (mCherry-LC3) was further monitored in neurons with plasmid transfection. After OGD-Rep treatment, mCherry-LC3 puncta increased in both neuronal soma and axons (Fig. 2, F and G), while the number of mitochondria engulfed by autophagosomes increased significantly only in soma but not in axons (Fig. 2 H). In addition, autophagic flux blockage by Bafilomycin A1 (Baf A1) accumulated mCherry-LC3 puncta (Fig. 2, F and G) and the number of mitophagosomes in soma but not in axons (Fig. 2, F and H). These observations indicated that local mitophagy might not occur in ischemic axons and implied that cell bodies were the major cellular compartments for autophagosome biogenesis rather than axons (Maday and Holzbaur, 2014), at least under ischemic conditions.
Figure 2.
Axonal mitophagy is not responsible for OGD-Rep–induced axonal mitochondrial loss. (A–E) Primary cultured cortical neurons expressing MitoQC were subjected to 20 min of OGD followed by 40 min of reperfusion (O-R) in DIV8. (B) Columns present the proportion of mCherry-positive (mCherry+) mitochondrial area to soma area. (C) Columns present mCherry-positive (mCherry+) mitochondrial area per 100-µm axon. (D) Columns present the proportion of GFP-negative–mCherry-positive (GFP-/mCherry+) mitochondrial area to soma area. (E) Columns present GFP-negative–mCherry-positive (GFP-/mCherry+) mitochondrial area per 100-µm axon. (F–H) Primary cultured cortical neurons were previously transfected with mCherry-LC3B and Mito-GFP by electroporation. The cultured neurons were treated with PBS as a vehicle control or 20 µM Baf A1 for 2 h and then subjected to 20 min of OGD. After 40 min of reperfusion, fluorescent images were captured by confocal microscopy. (F) Images show representative examples from three independent experiments. (G and H) Columns represent the number of mCherry-LC3B–positive puncta (G) and the number of mitochondria colocalized with LC3B puncta (H). n = 25–37 cells from three independent experiments for each group. (I–K) Primary cultured cortical neurons from the indicated germline mice were subjected to 20 min of OGD in DIV8. The mitochondrial marker Hsp60 and neuronal marker β-tubulin III were stained by immunocytochemistry at 40 min after reperfusion. (I) The ATG7 level in primary cultured neurons of WT and Atg7fl/fl;nes-Cre mice was measured by Western blot. (J) Images show the representative straightened cultured neurons. (K) Columns represent the somatic mitochondrial (upper panel) and axonal mitochondrial area per 100-µm axon (lower panel), respectively. n = 30–46 neurons from three independent experiments. (L) Cortical neurons from the indicated germline mice were cultured in a microfluidic platform and subjected to 20 min of OGD in DIV8. DNA and RNA in soma and axon compartments were extracted separately at 40 min after reperfusion and amplified by quantitative PCR. Columns represent the relative somatic (left panel) and axonal (right panel) mitochondrial DNA (mtDNA) levels, which are indicated, respectively, by the mt-Atp6 (mitochondria-encoded DNA) to Rpl13 (nucleus-encoded DNA) ratio and mt-Atp6 to Csf1 (axonal housekeeping) ratio. The experiments were repeated three times and the data are expressed as means ± SEM. Statistical comparisons were performed with unpaired Student t tests (B–E) or one-way ANOVA with Sidak’s multiple-comparisons test (G, H, K, and L). *P < 0.05, **P < 0.01, ***P < 0.001 versus the indicated group. Scale bar, 10 µm. Ctrl, control; Mito, mitochondrial/mitochondria; Num, number; Veh, vehicle.
To clarify axonal mitophagy, autophagy-related 7 (Atg7) knockout neurons from Atg7fl/fl crossed with nestin-Cre mice were used (Atg7fl/fl;nes-Cre; verification in Fig. 2 I; Komatsu et al., 2005). Atg7 knockout abolished mitochondrial loss in neuronal soma but not in axons after OGD-Rep (Fig. 2, J and K), which supported the absence of axonal mitophagy. To precisely identify the mitochondrial content in axons, primary cortical neurons were cultured in a microfluidic chamber to isolate axons from neuronal soma (Taylor et al., 2005). We isolated a pure axonal fraction as a PCR assay demonstrated no contamination of neuronal somatic mRNA in H1 histone family member 0 (H1F0; Fig. S1 D). Mitochondrial content was determined by the mitochondrial DNA–encoded gene Atpase 6 (Schips et al., 2011; Zhang et al., 2013). Consistent with the imaging results, Atg7 knockout counteracted the somatic but not axonal mitochondrial DNA loss induced by OGD-Rep (Fig. 2 L), indicating that axonal mitochondria were not eliminated directly by autophagy. Parkin was demonstrated to participate in mitophagy in ischemic neurons (Zhang et al., 2014; Shen et al., 2017) and to be essential for distal mitophagy (Ashrafi et al., 2014). Likewise, Parkin−/− neurons also lost axonal mitochondria after OGD-Rep to a similar extent as WT neurons (Fig. S1). Taken together, these results strongly suggested the absence of direct mitophagy in axons in OGD-Rep neurons.
Somatic autophagy of axonal mitochondria in OGD-Rep neurons
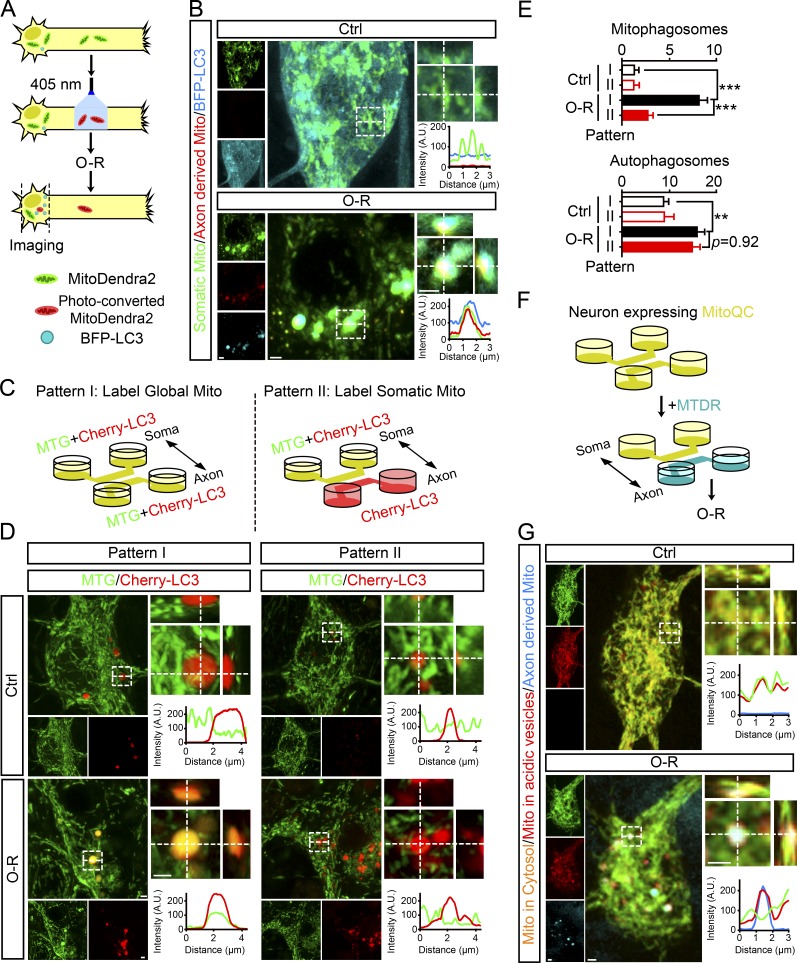
To trace the axonal mitochondria that were lost during OGD-Rep, we transfected neurons with tagBFP-LC3 and MitoDendra2. MitoDendra2 is a mitochondria-located protein that shifts its fluorescence from Ex490 nm/Em507 nm to Ex553 nm/Em573 nm after photo-conversion (Pham et al., 2012). The axonal mitochondria were prelabeled by photo-conversion (Fig. 3 A and Fig. S2 A). After OGD-Rep treatment, axon-derived mitochondria can be found in neuronal soma. Moreover, a remarkable overlap of tagBFP-LC3 and axonal mitochondria was observed, indicated by both orthogonal view display and line scan analysis (Fig. 3 B). To further confirm somatic autophagy of axonal mitochondria, the axonal and somatic mitochondria were separately labeled with different dyes by taking advantage of microfluidic chambers as described in the Materials and methods. Briefly, dyes in the compartment with less medium were not able to diffuse into the other compartment as confirmed by compartment-restricted distribution of Dextran-Rhodamine 123 (Fig. S2 B). Using this strategy, somatic mitochondria of neurons expressing mCherry-LC3 were specifically labeled with MitoTracker Green (MTG; Pattern II in Fig. 3 C). As a control, both chambers were evenly loaded with MTG to label global mitochondria (Pattern I in Fig. 3 C). After OGD-Rep, mitophagy was significantly activated in neuronal soma shown by colocalization of mitochondria and autophagosomes in Pattern I. However, the extent of overlap was dramatically reduced in Pattern II (Fig. 3, D and E). This unexpected result implied axonal mitochondria, compared with somatic and dendritic mitochondria, made up the majority of mitochondria that underwent mitophagy. To further confirm autophagic clearance of axonal mitochondria after OGD-Rep, neurons expressing MitoQC were loaded with MitoTracker Deep Red (MTDR) only in axonal chambers, as described above (Fig. 3 F). In intact neurons, few axonal mitochondria can be detected in soma. However, after OGD-Rep, axon-derived mitochondria appeared in soma and showed the feature of being resided in acidic vesicles (GFP−/mCherry+; Fig. 3 G). Overall, these data revealed that axonal mitochondria were not locally eliminated by mitophagy after OGD-Rep; instead, they appeared in neuronal soma for degradation. It is likely that clearing axon-derived mitochondria was prioritized by autophagy in ischemic neuronal soma.
Figure 3.
Somatic mitophagy preferentially removes axon-derived mitochondria after reperfusion. (A and B) Primary cultured cortical neurons were previously transfected with MitoDendra2 and tagBFP-LC3B by electroporation. After photo-conversion of MitoDendra2 in axons, the cultured neurons were subjected to 20 min of OGD and 20 min of reperfusion (O-R) as shown in A in DIV8. (B) Images show representative examples from three independent experiments. Orthogonal view and line-scan analysis of indicated regions are presented in the right panel. (C–E) Primary cultured cortical neurons expressing mCherry-LC3B were stained with MTG (green) in two different patterns as shown in C in DIV8. After 20 min of OGD followed by 20 min of reperfusion, fluorescent images were captured by confocal microscopy. (D) Images show representative examples from three independent experiments. Orthogonal view and line-scan analysis of indicated regions are presented in the right panel. (E) Columns represent the number of mitophagosomes (upper panel) and autophagosomes (lower panel) in neuronal soma. n = 17–22 cells from three independent experiments for each group. The data are expressed as means ± SEM. Statistical comparisons were performed with one-way ANOVA with Sidak’s multiple-comparisons test. **P < 0.01, ***P < 0.001 versus the indicated group. (F–G) Neurons expressing MitoQC were stained with MTDR (cyan) and then subjected to 20 min of OGD and 20 min of reperfusion (O-R) as shown in F in DIV8. (G) Images show representative examples from three independent experiments. Orthogonal view and line-scan analysis of indicated regions are presented in the right panel. Scale bar, 1 µm. A.U., arbitrary units; Ctrl, control; Mito, mitochondria.
Enhanced retrograde mitochondrial transport in axons after OGD-Rep
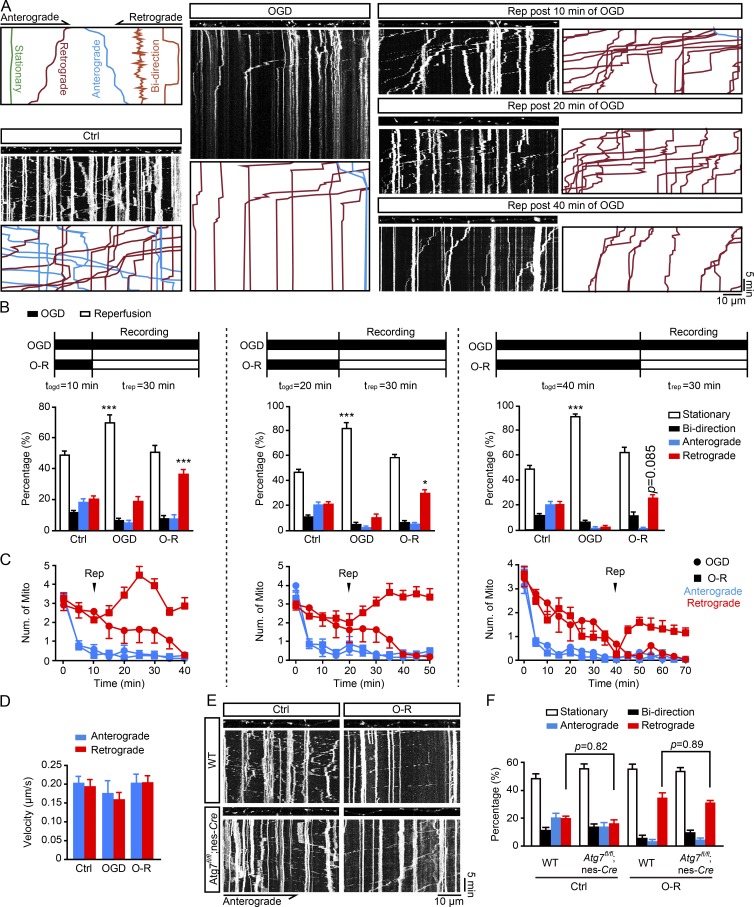
The appearance of somatic autophagy of axonal mitochondria forced us to hypothesize that axonal mitochondria underwent transportation during OGD-Rep. Previous studies reported inconsistent conflicting results of axonal mitochondria movement after neuronal insult (Wang et al., 2011; Cai et al., 2012; Ashrafi et al., 2014; Lin et al., 2017a). We therefore monitored the motility of axonal mitochondria by time-lapse confocal microscopy during the period of OGD-Rep. Mitochondria in the axonal compartment were loaded with MTG, and their motility (defined as stationary, retrograde, anterograde, or bidirectional movement) was recorded by a kymograph (Fig. 4 A, Video 1, Video 2, and Video 3). In basal conditions, axonal mitochondria were stationary (49.1% ± 10.8%), bidirectional (11.0% ± 5.0%), anterograde (19.5% ± 9.3%), or retrograde (20.3% ± 6.9%), all of which were in line with previous descriptions (Cai et al., 2012; Lewis et al., 2016; Zhang et al., 2017). After the onset of OGD, the ratio of immobile mitochondria increased while both anterograde and retrograde movement decreased along with the duration of OGD. Mitochondrial anterograde movement was more vulnerable than retrograde movement to the OGD insult as we documented a complete loss of anterograde and retrograde movement after 10 and 40 min of OGD, respectively. We found a prompt recovery of retrograde, but not anterograde, movement of axonal mitochondria after the onset of reperfusion. The number of retrogradely moved mitochondria decreased along with the increased duration of OGD. Reperfusion after a 40-min OGD treatment led to slight, yet not significant restoration of retrograde mitochondrial movement (Fig. 4, B and C). In addition, mitochondrial velocity did not change significantly during the whole procedure (Fig. 4 D). A recent study showed that axonal mitochondrial mobility reduced in the more mature neurons (Lewis et al., 2016). We determined mitochondrial content and mobility after OGD-Rep in day-in-vitro (DIV) 14 neurons. Similar to the results obtained from neurons in DIV8, OGD-Rep induced mitochondrial loss in both axons and soma (Fig. S3, A–C) and significantly increased mitochondrial retrograde transport (Fig. S3, D and E) in DIV14 neurons. These data suggested that axonal mitochondria were transported to neuronal soma for degradation. In addition, neurons were administrated with carbonyl cyanide 3-chlorophenylhydrazone (CCCP), which induces mitophagy. CCCP, OGD alone, and OGD-Rep all reduced axonal mitochondrial length and membrane potential, indicating mitochondrial dysfunction (Fig. S4, A–D). However, CCCP and OGD alone caused axonal mitochondrial movement arrest, and only OGD-Rep increased the retrograde trafficking of axonal mitochondria (Fig. S4, E and F). These observations indicated that impairment of axonal mitochondria was not sufficient to induce their retrograde transport to soma. Increased retrograde motility of axonal mitochondria in reperfusion injury is consistent with a recent report showing enhanced retrograde transport of axonal mitochondria under mild stress conditions, a condition more relevant to pathophysiological status (Lin et al., 2017a).
Figure 4.
OGD-Rep increases retrograde transport of axonal mitochondria. (A–D) Primary cultured cortical neurons were cultured in a microfluidic platform. After staining with 10 nM MTG, the cultured neurons were subjected to OGD alone for 1 h or OGD-Rep for various time periods as indicated in DIV8. Live cell fluorescent images in axon compartments were captured by confocal microscopy. (A) Images show representative kymographs from three independent experiments. (B) Columns represent the proportion of mitochondria showing different transportation patterns. (C) Curves represent the number of mitochondria either anterogradely or retrogradely transported per 100-µm axon in 5 min. (D) Columns represent the velocity of mitochondria anterogradely or retrogradely transported in axons. (B–D) n = 18 (control, OGD alone, and reperfusion after 10-min OGD), n = 17 (reperfusion after 20-min OGD), and n = 14 (reperfusion after 40-min OGD) cells from three independent experiments for each group. (E and F) Primary cultured cortical neurons from the indicated germline mice were cultured in a microfluidic platform. After staining with 10 nM MTG, the cultured neurons were subjected to 20 min of OGD plus 40 min of reperfusion. Live cell fluorescent images in axon compartments were captured by confocal microscopy. (E) Images show representative kymographs from three independent experiments. (F) Columns represent the proportion of mitochondria showing different transportation patterns. n = 13 cells from three independent experiments for each group. The data are expressed as means ± SEM. Statistical comparisons were performed with one-way ANOVA with Sidak’s multiple-comparisons test. *P < 0.05, ***P < 0.001 versus the indicated group. Ctrl, control; Mito, mitochondria; Num, number; O-R, OGD-Rep.
To clarify whether autophagy has an impact on axonal mitochondrial trafficking, we recorded the axonal mitochondrial movement in Atg7-deficient neurons. Atg7 deletion had no impact on the recovery of mitochondrial retrograde movement (Fig. 4, E and F). Likewise, the Parkin deficiency did not alter the axonal mitochondrial mobility after OGD-Rep (Fig. S4, G and H). These data emphasized that axonal mitochondria were transported rather than degraded directly in axons after OGD-Rep. Furthermore, autophagy may act downstream of mitochondrial movement in ischemic conditions.
Retrograde movement of axonal mitochondria was required for their mitophagy in soma
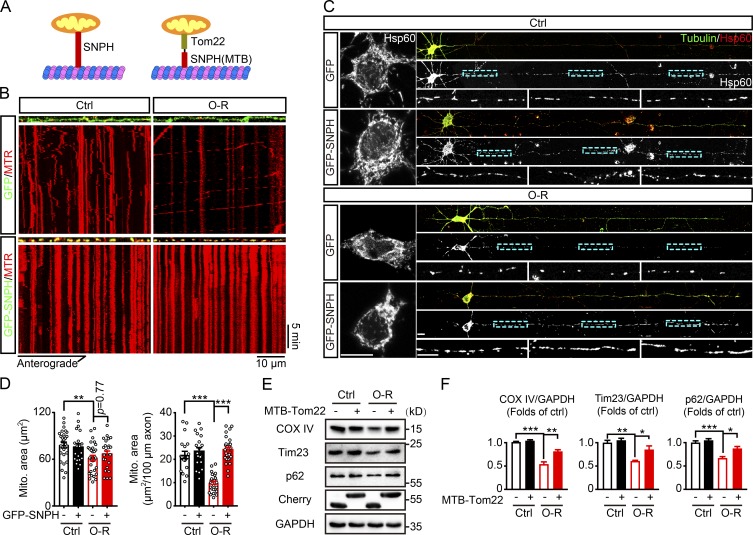
To identify the association of retrograde movement of axonal mitochondria with somatic mitophagy, the axonal mitochondrial movement was arrested by overexpression of SNPH, which anchored axonal mitochondria (Fig. 5 A; Kang et al., 2008). As expected, GFP-SNPH localized to axonal mitochondria and decreased their motility in both intact and OGD-Rep–treated neurons (Fig. 5 B, Video 4, and Video 5). We observed that GFP-SNPH significantly abolished axonal, but not somatic, mitochondrial loss (immunostained by Hsp60) after OGD-Rep treatment (Fig. 5, C and D). These data indicated that mitochondrial movement was essential for axonal mitochondrial loss. It has been recently reported that SNPH could bud out of mitochondria under stress, thus increasing mitochondrial mobility (Lin et al., 2017a). To fully arrest mitochondria, a mitochondrial outer-membrane protein (Tom22) was fused with the microtubule-binding domain (MTB) of SNPH (MTB-Tom22; Kang et al., 2008), thus binding mitochondria with microtubules (Fig. 5 A and Fig. S5 B). Remarkably, the overall mitophagy induced by OGD-Rep was counteracted by MTB-Tom22 expression as reflected by accumulation of mitochondrial proteins (Fig. 5, E and F), revealing that axonal mitochondrial movement was required for ischemic neuronal mitophagy.
Figure 5.
Inhibition of mitochondrial mobility by overexpression of SNPH reverses OGD-Rep–induced mitophagy. (A) Schematic diagrams of GFP-SNPH and mCherry-MTB-Tom22. (B) Primary cultured cortical neurons were previously transfected with GFP or GFP-SNPH by electroporation. After staining with 10 nM MitoTracker Red, the cultured neurons were subjected to 20 min of OGD in DIV14. At reperfusion (O-R), live cell fluorescent images of axonal mitochondria were captured by confocal microscopy for 30 min. Images show representative kymographs from three independent experiments. (C and D) Primary cultured cortical neurons were previously transfected with GFP or GFP-SNPH by electroporation and subjected to 20 min of OGD in DIV14. The mitochondrial marker Hsp60 was stained by immunocytochemistry at 40 min of reperfusion. (C) Images show representative straightened cultured neurons. (D) Columns represent the somatic mitochondrial area (left panel) and axonal mitochondrial area per 100-µm axon (right panel). n = 20–28 cells from three independent experiments for each group. (E) Primary cultured cortical neurons were previously transfected with mCherry-Tom22 or mCherry-MTB-Tom22 by electroporation and subjected to 20 min of OGD followed by 40 min of reperfusion DIV14. The COX IV, Tim23, p62, and GAPDH levels in cultured neurons were determined by Western blot. (F) Semiquantitative analysis of COX IV, Tim23, and p62 bands are shown. The data are expressed as means ± SEM. Statistical comparisons were performed with one-way ANOVA with Sidak’s multiple-comparisons test. *P < 0.05, **P < 0.01, ***P < 0.001 versus the indicated group. Scale bar, 10 µm. Ctrl, control; Mito, mitochondrial.
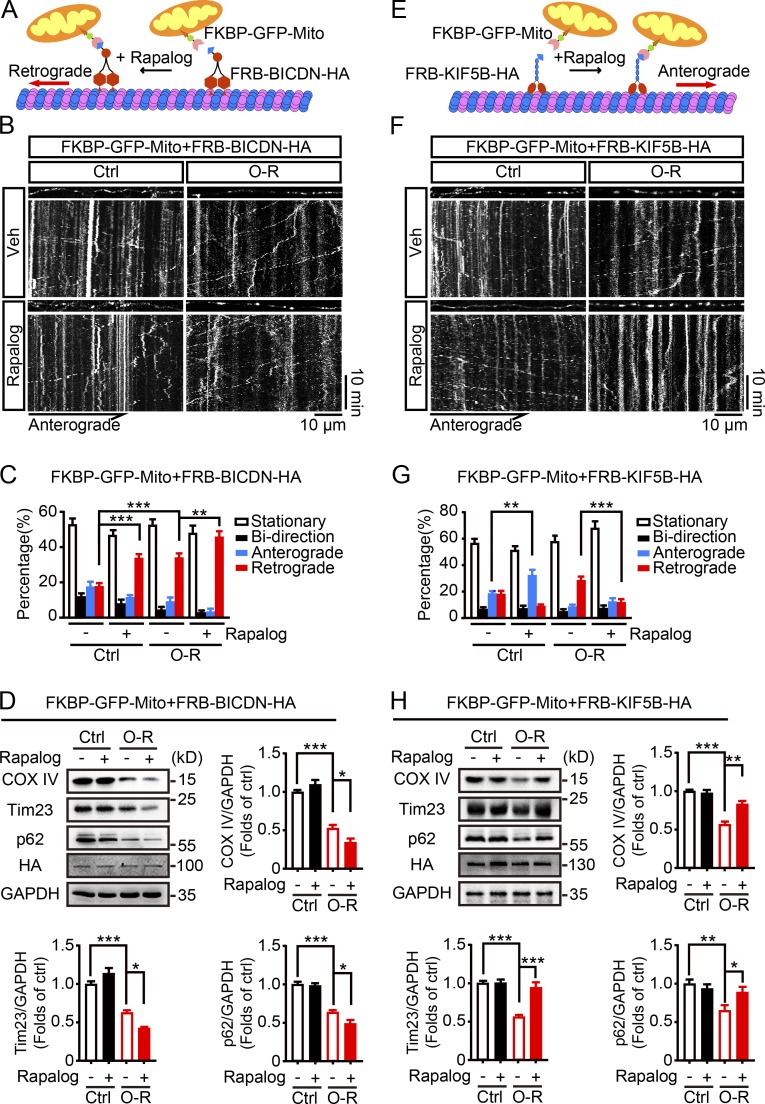
We next asked whether OGD-Rep–induced neuronal mitophagy can be reinforced by promoting retrograde movement of axonal mitochondria. To this end, a protein dimerization tool was used (van Spronsen et al., 2013). The binding of FKBP2 with FRB can be induced in the presence of rapalog. FKBP2 was fused with GFP and mitochondria-targeting sequence. FRB (tagged with HA) was fused with BICDN, the N-terminal domain of the dynein/dynactin interacting protein BICD2, which is capable of inducing selective dynein-mediated transport (Hoogenraad et al., 2003; Fig. 6 A). Indeed, retrograde movement of axonal mitochondria was reinforced by rapalog after OGD-Rep (Fig. 6, B and C; Video 6; and Video 7). Importantly, overall mitophagy was further activated by rapalog treatment since mitochondrial markers COX IV and Tim23 reduced (Fig. 6 D). These data clearly indicated that enhanced retrograde movement of axonal mitochondria was sufficient to reinforce the OGD-Rep–induced mitophagy.
Figure 6.
Mitochondrial retrograde transport is required for OGD-Rep–induced mitophagy. (A and E) Schematic diagrams of rapalog-induced mitochondrial transportation assay. Rapalog induced the recruitment of a fusion protein of FRB with the truncated motor construct of dynein adaptor bicaudal D2 (FRB-BICDN-HA; A) or kinesin-1 (FRB-KIF5B-HA; E) to FKBP-GFP-Mito. (B–D and F–H) Primary cultured cortical neurons were previously transfected with FRB-BICDN-HA (B–D) or FRB-KIF5B-HA (F–H) and FKBP-GFP-Mito by electroporation and subjected to 20 min of OGD in DIV8. At reperfusion, ethanol (vehicle control) or 500 nM rapalog was added. (B and F) Live cell fluorescent images of axonal mitochondria were captured by confocal microscopy. Images show representative kymographs from three independent experiments. (C and G) Columns represent the proportion of mitochondria showing different transportation patterns. Seven cells from three independent experiments were included for each group. (D and H) Neurons were harvested after 40 min of reperfusion. The COX IV, Tim23, p62, HA, and GAPDH levels in cultured neurons were determined by Western blot. Semiquantitative analysis of COX IV, Tim23, and p62 bands is shown. The data are expressed as means ± SEM. Statistical comparisons were performed with one-way ANOVA with Sidak’s multiple-comparisons test. *P < 0.05, **P < 0.01, ***P < 0.001 versus the indicated group. Ctrl, control; Mito, mitochondrial/mitochondria; O-R, reperfusion; Veh, vehicle.
Conversely, we tried to rescue anterograde movement of axonal mitochondria by replacing BICDN with the motor domain and coiled-coil dimerization region of KIF5B, a motor protein for anterograde transport of mitochondria (Fig. 6 E). KIF5B did not rescue anterograde motility loss of axonal mitochondria after OGD-Rep, although it worked in intact neurons; mitochondrial retrograde movement was significantly reduced by this manipulation (Fig. 6, F and G; Video 8; and Video 9). Similar to the results of SNPH expression, in the presence of rapalog, FRB-KIF5B-HA compromised OGD-Rep–induced neuronal mitophagy (Fig. 6 H). Taken together, these results linked the retrograde transportation of axonal mitochondria with their mitophagy in neuronal soma. We provided the first evidence of manipulating neuronal mitophagy by controlling axonal mitochondrial movement.
Retrograde axonal mitochondrial transport rescued ischemic neuron injury by promoting mitophagy
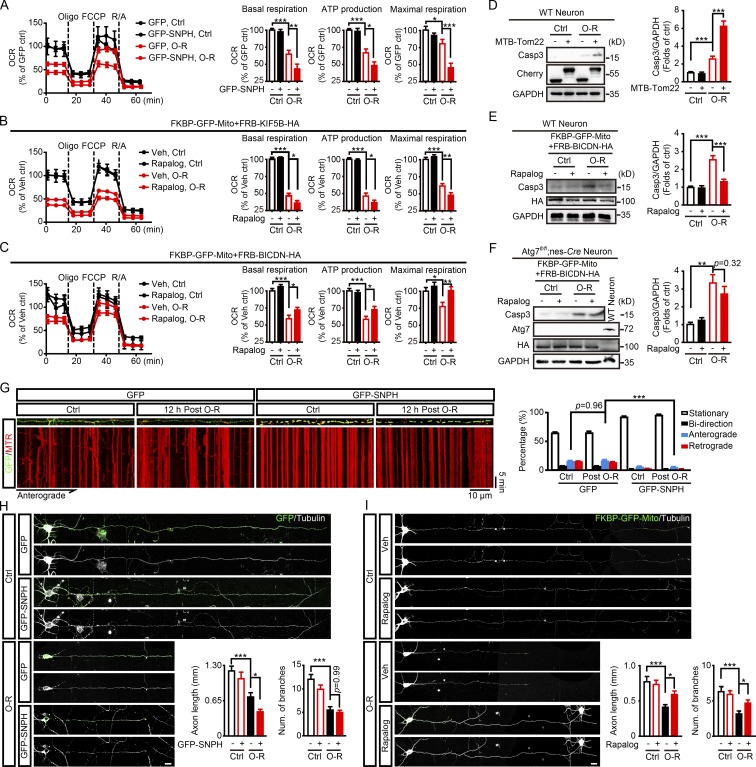
Mitophagy activation protects against ischemic neuronal injury by alleviating mitochondria-dependent apoptosis (Zhang et al., 2013; Yuan et al., 2017). Here we showed somatic autophagy of axonal mitochondria following their retrograde transportation. We next determined whether axonal mitochondrial trafficking was able to affect mitochondrial quality. By measuring mitochondrial oxygen consumption (oxygen consumption rate [OCR]), we found OGD-Rep dramatically compromised mitochondrial quality reflected by reduced mitochondrial basal respiration, ATP production, and maximal respiration (Fig. 7 A). Inhibition of mitochondrial retrograde transport in axons either by SNPH or FRB-KIF5B-HA aggravated OGD-Rep–induced mitochondrial dysfunction (Fig. 7, A and B). Conversely, mitochondrial quality was significantly improved after OGD-Rep when retrograde movement of axonal mitochondrial was reinforced as previously mentioned (Fig. 7 C). OGD-Rep–activated Caspase 3 (Casp3) further increased by MTB-Tom22 expression (Fig. 7 D), suggesting that arrest of axonal mitochondrial mobility aggravated ischemia-induced neuronal injury. Conversely, enhanced retrograde transport attenuated Casp3 activation (Fig. 7 E). By comparison, forced retrograde transport did not reduce neuronal apoptosis in Atg7fl/fl;nes-Cre neurons (Fig. 7 F), demonstrating the requirement of autophagy for the neuroprotection of enhanced mitochondrial retrograde transport. Given the fundamental role of mitochondrial anterograde transport in axon regeneration after injury (Zhou et al., 2016), we next investigated mitochondrial anterograde transport in a later phase of OGD-Rep. After 12 h of reperfusion, mitochondrial anterograde transport recovered to the similar extent as the control group, which was abolished by GFP-SNPH expression (Fig. 7 G). Accordingly, neurons transfected with GFP-SNPH showed greater morphological impairment following OGD-Rep, as reflected by shortened axons after 24 h of reperfusion (Fig. 7 H). Conversely, facilitated mitochondrial retrograde transport attenuated axonal impair and branch loss, suggesting promoted neuronal recovery (Fig. 7 I). Overall, these results highlighted the neuroprotective effects of retrograde movement of axonal mitochondria by promoting neuronal mitophagy.
Figure 7.
Mitochondrial retrograde transport protects against OGD-Rep injuries. (A–C) OCR was assessed by a Seahorse XFe96 analyzer with sequential injections of oligomycin (Oligo), FCCP, and rotenone/antimycin A (R/A). Each value was normalized to the first data-point measurement of control. n = 6–7 repeats were included for each group. The basal respiration, mitochondrial ATP production, and the maximal respiration were calculated as described in the Materials and methods. Primary cultured cortical neurons expressing GFP or GFP-SNPH were subjected to 20 min of OGD-Rep (O-R) in DIV14 (A). Neurons expressing FKBP-GFP-Mito with either FRB-KIF5B-HA (B) or FRB-BICBN-HA (C) were subjected to 20 min of OGD plus 40 min of reperfusion in DIV8 and ethanol (vehicle control) or 500 nM rapalog was added at reperfusion. (D) Primary cultured cortical neurons expressing mCherry-Tom22 or mCherry-MTB-Tom22 were subjected to 20 min of OGD plus 40 min of reperfusion in DIV14. The cleaved Casp3, mCherry, and GAPDH levels in cultured neurons were determined by Western blot. Semiquantitative analysis of Casp3 bands are shown. (E and F) Primary cultured cortical neurons from the indicated germline mice were previously transfected with FRB-BICDN-HA and FKBP-GFP-Mito by electroporation and subjected to 20 min of OGD in DIV8. Ethanol (vehicle control) or 500 nM rapalog was added at reperfusion and the neurons were harvested after 40 min of reperfusion. The cleaved Casp3, HA, and GAPDH levels in cultured neurons were determined by Western blot. Semiquantitative analysis of Casp3 bands is shown. (G) Primary cultured cortical neurons were previously transfected with GFP or GFP-SNPH by electroporation. The cultured neurons were subjected to 20 min of OGD followed by 12 h of reperfusion in DIV14. After staining with 10 nM MitoTracker Red, live cell fluorescent images of axonal mitochondria were captured by confocal microscopy for 30 min. Images show representative kymographs from three independent experiments. Columns represent the proportion of mitochondria showing different transportation patterns. n = 16 (GFP, Ctrl, and 12 h post O-R) and n = 11 (GFP-SNPH, Ctrl, and 12 h post O-R) cells from three independent experiments for each group. (H) Primary cultured cortical neurons expressing GFP or GFP-SNPH were subjected to 20 min of OGD followed by 24 h of reperfusion in DIV14. Columns represent axonal length (left panel) and number of branches (right panel). n = 25–35 cells from three independent experiments for each group. (I) Neurons expressing FKBP-GFP-Mito with FRB-BICBN-HA were subjected to 20 min of OGD followed by 24 h of reperfusion in DIV8. Columns represent axonal length (left panel) and number of branches (right panel). n = 29–31 cells from three independent experiments for each group. The data are expressed as means ± SEM. Statistical comparisons were performed with one-way ANOVA with Sidak’s multiple-comparisons test. *P < 0.05, **P < 0.01, ***P < 0.001 versus the indicated group. Scale bar, 10 µm. Ctrl, control; Mito, mitochondrial/mitochondria; Num, number; Veh, vehicle.
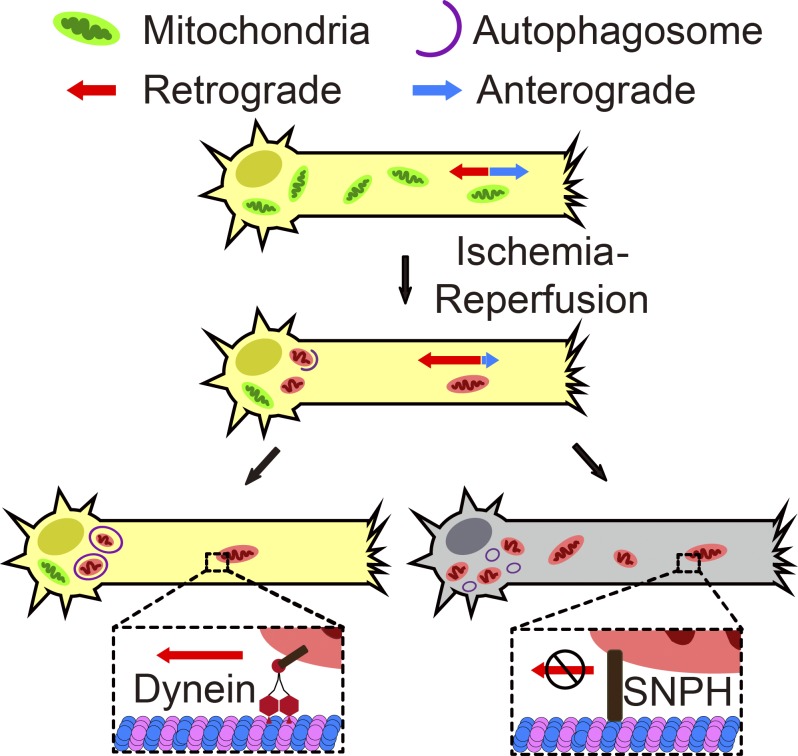
Taken together, the present study revealed the spatial features of mitophagy in ischemic neurons. In particular, axonal mitochondria were retrogradely transported to neuronal soma for autophagic degradation. Mitochondrial mobility arrest aggravated ischemic neuronal injury while reinforced retrograde mitochondrial motility was neuroprotective (Fig. 8).
Figure 8.
Schematic diagram shows the contribution of mitochondrial retrograde transportation to ischemia-reperfusion–induced mitophagy and its neuroprotection. Ischemia-reperfusion increased axonal mitochondrial retrograde transportation specifically, and axonal mitochondria underwent autophagic clearance in neuronal soma. Facilitated retrograde transportation promoted ischemia-reperfusion–induced mitophagy and further neuronal survival. Conversely, inhibition of axonal mitochondrial mobility compromised mitophagy and aggravated ischemic injury.
Discussion
The spatial characteristics of mitophagy in ischemic neurons remain unclear. For the first time, we determined that OGD-Rep led to loss of mitochondria both in neuronal soma and axons. However, mitophagosomes were hardly found in axons and mitochondrial loss in axons was still observed in Atg7fl/fl;nes-Cre and Parkin−/− neurons (Fig. 2 and Fig. S1). These results strongly indicated that axonal mitochondria were not directly degraded by autophagy in axons with ischemic stress. Instead, the axonal mitochondria were retrogradely transported immediately after OGD-Rep. By tracing the axonal mitochondria either with MitoDendra2 or MTDR, we clearly showed that axon-derived mitochondria appeared in neuronal soma and underwent autophagic clearance. Thus, we provide direct evidence indicating the somatic autophagy of axonal mitochondria in ischemic neurons. Although lysosomes were reported to be presented in axons, they are most concentrated in neuronal soma (Farías et al., 2017). Mitophagy in neuronal soma may facilitate mitochondrial elimination by taking advantage of abundant lysosomes in cell bodies to meet the greater demand for mitochondrial clearance after ischemia.
Previous studies found that axonal mitochondria with dissipated mitochondrial membrane potential (ΔΨm) moved retrogradely in intact neurons (Miller and Sheetz, 2004) and mitochondria with Parkin translocation predominantly resided in neuronal cell bodies (Cai et al., 2012). Cell body–restricted mitophagy was also suggested in other in vivo models (Devireddy et al., 2015; Sung et al., 2016). Hence, it was postulated that mitochondria were retrogradely transported for degradation (Yang et al., 2013); however, direct evidence supporting this assumption has been lacking and the consequences of these transported mitochondria remain incompletely understood (Course and Wang, 2016). OGD-Rep increased mitochondrial retrograde transportation (Fig. 4) and those axon-derived mitochondria mainly colocalized with autophagosomes in soma (Fig. 3). More intriguingly, inhibition of mitochondrial movement significantly attenuated OGD-Rep–induced mitophagy (Fig. 5 and Fig. 6, E–H). Conversely, increased retrograde movement of axonal mitochondria was sufficient to reinforce OGD-Rep–induced mitophagy. The current study fills the missing gap between mitochondria trafficking and degradation in a disease-relevant model, and further emphasizes that retrograde movement of axonal mitochondria is essential for ischemic neuronal mitophagy. Several studies also suggested that axonal mitochondrial mobility was arrested by stress and axonal mitophagy occurred locally (Wang et al., 2011; Lovas and Wang, 2013; Ashrafi et al., 2014; Hsieh et al., 2016). Therefore, the associations of mitochondrial mobility with mitophagy may vary depending on distinct stresses. Indeed, compared with OGD-Rep, CCCP arrested mitochondrial mobility rapidly (Fig. S4, E and F) but induced delayed mitophagy (Cai et al., 2012). Given the abundant lysosome content in soma, retrograde transport might be able to facilitate mitochondrial turnover under intensive stress such as ischemia.
After OGD-Rep, nearly 70% of total mitochondria engulfed by somatic autophagosomes were derived from axons (Fig. 3 E) and arresting axonal mitochondria almost totally abolished mitophagy (Fig. 5, E and F). These results indicate that the axonal mitochondria have priority for autophagic degradation among overall mitochondrial content in ischemic neurons. The balance of forces between kinesin and dynein decides the direction of mitochondrial movement in axons (Course and Wang, 2016). We assumed dynein might predominantly occupy the retrogradely transported mitochondria because forced translocation of KIF5B to mitochondria failed to drive anterograde transport during reperfusion (Fig. 6, F and G). Interestingly, several components of the dynein complex contain an LC3 interacting region (LIR) motif, which can be recognized by autophagosomes and thus may confer the prior recognition of retrogradely moved mitochondria. Overall, the present findings reveal the procedures and significance of mitochondrial retrograde transportation-mediated somatic autophagy of axonal mitochondria for the general mitochondrial quality control in ischemic neurons.
Misregulation of mitochondria mobility in ischemic neurons was undetermined. Interestingly, we found that retrograde movement was restored after the recovery of glucose and oxygen. Of note, forced mitochondrial retrograde transport reduced mitochondrial mass by mitophagy but improved the mitochondrial function and further promoted neuronal survival. Conversely, mitochondrial accumulation resulting from inhibition of mitochondrial movement even compromised mitochondrial quality and aggravated neuronal injury (Fig. 5 and Fig. 7). These results imply that mitochondrial quality, rather than mitochondrial mass, is more important for neuronal survival. In this context, mitochondrial retrograde transport may serve as a significant strategy to improve mitochondrial quality control during ischemia–reperfusion. Mitochondrial immobilization in axons may restrict damaged mitochondria from fusing with the healthy mitochondrial network (Wang et al., 2011; Ashrafi et al., 2014), whereas we found that axon-derived mitochondria preferentially underwent degradation without further mitochondrial fusion (Fig. 3, B and G). It is thus less likely that these mitochondria would spread their damage, at least in ischemic neurons. Moreover, reinforced retrograde movement of axonal mitochondria attenuated OGD-Rep–induced neuronal apoptosis and promoted neuronal recovery from ischemic injury. Ischemia–reperfusion also induced axonal mitochondrial loss and increased mitochondrial retrograde transport in more mature neurons (Fig. S3). These results verify the concept of attenuated neuronal injury by precisely manipulating mitochondrial movement (Lovas and Wang, 2013). Corrected mitochondrial motility supports neuronal survival by providing energy at proper sites, and our data further linked the benefits of corrected mitochondrial mobility with efficient mitophagy. Axonal mitochondria took the most, if not all, of the total number of mitochondria that underwent mitophagy. Therefore, our findings highlight the potential value of clearing axonal mitochondria for ischemic therapy.
The mechanisms controlling the selective retrograde movement of axonal mitochondria after ischemic insult are still unclear. A recent study revealed a novel mechanism that the release of SNPH from mitochondria promoted damaged mitochondria that escaped from axons at the early stage of neuronal injury (Lin et al., 2017a). Here we found that mitochondria underwent mitophagy in neuronal soma after they were released from axons. Consistently, the overexpression of SNPH blocked ischemia-induced axonal mitochondrial loss and aggravated neuronal injury. Thus, it might be a universal strategy for neurons to start the hierarchy of the axonal mitochondrial quality control with the release of SNPH from mitochondria during stress (Lin et al., 2017b). Given the significance of axonal mitochondrial health for neuronal homeostasis (Sheng and Cai, 2012), this “transport-and-mitophagy” strategy might apply to a variety of neurological diseases in which SNPH might be a promising target for therapies.
Taken together, we identified, for the first time, somatic autophagy of axonal mitochondria at the reperfusion stage after ischemia. These axonal mitochondria were retrogradely transported back to neuronal cell bodies for degradation. Reinforced retrograde transport of axonal mitochondria promoted mitophagy and attenuated mitochondrial dysfunction and ischemic neuronal injury. These spatial features of mitophagy may provide critical insight into how neurons control mitochondrial quality under pathological conditions.
Materials and methods
Animals
The Parkin−/− mice were kindly provided by Zhuohua Zhang (Central South China University, Changsha, China); the Atg7fl/fl mice were kindly provided by Masaaki Komatsu (Tokyo Metropolitan Institute of Medical Science, Tokyo, Japan). Nestin-Cre transgenic mice were purchased from the Jackson Laboratory. Atg7fl/fl mice were bred with nestin-Cre transgenic mice to produce Atg7fl/fl;nes-Cre mice. For the culture of primary cortical neurons, pregnant mice with embryonic fetuses (embryonic day 18) were used. All experiments were approved by and conducted in accordance with the ethical guidelines of the Zhejiang University Animal Experimentation Committee.
Primary cortical neuron cultures and OGD procedures
The primary cortical neuronal culture experiments were performed as described previously (Fan et al., 2011). Briefly, the dissected cortex from embryonic day 18 fetal mice was digested with 0.25% trypsin (Invitrogen; 25200-056). Approximately 105 cells/cm2 were seeded onto poly-L-lysine (Sigma; P1399)–coated microscope coveslips (Fisher Scientific; 12-545-80) for immunostaining; six-well plates (Corning) for Western blot and glass-bottom dishes (Cellvis, D35-20-0-N) were used for live cell imaging. For microfluidic chamber (Xona; RD450), 20 µl medium containing 2 × 105 cells was added onto the microscope coverslips (Deckgläser; 25 mm Φ) attached to the microfluidic devices. The neurons were grown in Neurobasal Plus medium (Invitrogen; 35829-01) supplemented with 2% B27 Plus Supplement (Invitrogen; 35828-01) and 0.25% GlutaMAX Supplement (Invitrogen; 35050-061). Cultures were maintained for 8–14 d before treatment.
For OGD treatment, oxygen- and glucose-free DMEM was prepared by bubbling glucose-free DMEM (Invitrogen; 11966-025) with 5% CO2 and 95% N2 for 30 min. DIV8–14 primary cultured neurons were refreshed with oxygen- and glucose-free DMEM. Cells were then immediately placed in a sealed chamber (Billups-Rothenburg; MIC-101) loaded with mixed gas containing 5% CO2 and 95% N2. Control groups were switched to DMEM (Invitrogen; 11995-065) with glucose for the same period time as the OGD group. For reperfusion, neurons were refreshed with normal culture medium. In Fig. 1, neurons were treated with PBS as a vehicle control or 50 µM chloroquine (Sigma; C6628) for 4 h before OGD. PBS took up 0.1% (vol:vol) in the final culture medium. Neurons were cultured in microscope coverslips for immunostaining or six-well plates for Western blot. In Fig. 2 and Fig. S1, neurons were cultured in glass-bottom dishes for live cell imaging, microscope coverslips for immunostaining, or microfluidic chambers for DNA/RNA isolation. OGD-Rep was performed in both somatic and axonal compartments. In Fig. 2, F–H, neurons were treated with PBS as a vehicle control or 20 µM Baf A1 (Abcam; ab120497) for 2 h before OGD. PBS took up 0.1% (vol:vol) in the final culture medium. In Fig. 3, Fig. 4, Fig. S3 D, and Fig. S4, A, E, and G, neurons were seeded on the microfluidic chambers and both somatic and axonal compartments were subjected to OGD-Rep for the indicated time. In Figs. 5, 6, 7, Fig. S3 A, Fig. S4 C, and Fig. S5, neurons were cultured in glass-bottom dishes for live cell imaging, microscope coverslips for immunostaining, or six-well plates for Western blot.
Plasmid construction and transfection
FRB-KIF5B-HA, FRB-BICDN-HA, and FKBP-GFP-mito were gifts from Casper C. Hoogenraad (Utrecht University, Utrecht, Netherlands). MitoDendra2 was purchased from YouBio (V2304). Human SNPH cDNA was amplified from a human cDNA library (YouBio; G111702) by PCR with the forward (Fw) primer 5′-GTAGATCTATGGCCATGTCCCTGCC-3′ and the reverse (Rv) primer 5′-CTGGATCCTCAGAGCTGGGAGCCGC-3′. The PCR product was inserted into the BglII and BamHI site of pEGFP-C1 plasmid to construct the pEGFP-SNPH plasmid. Mouse Tom22 cDNA was amplified from a mouse cDNA library (YouBio; G138812) by PCR with the Fw primer 5′-TTGAATTCTATGGCCGCCGCCGTCGC-3′ and the Rv primer 5′-GGGGATCCCTACATCTTTCCAGGAAGTGGAG-3′. The PCR product was inserted into the EcoRI and BamHI site of pmCherry-C1 plasmid to construct the pmCherry-Tom22 plasmid. MTB of human SNPH was amplified with the Fw primer 5′-CACGAATTCAATGATCCGGCACCTGAAAGCC-3′ and the Rv primer 5′-CTGCAGCGACGGCGGCGGCCATCTTGTCAATCAGGTTGTTCTTG-3′. MTB-Tom22 was amplified with the Fw primer 5′-CACGAATTCAATGATCCGGCACCTGAAAGCC-3′ and the Rv primer 5′-GAGGGATCCCTACATCTTTCCAGGAAGTGGAG-3′. The PCR product was inserted into the EcoRI and BamHI site of pmCherry-C1 plasmid to construct the pmCherry-MTB-Tom22 plasmid. The residues 101–152 of human Fis1 cDNA were amplified from a human cDNA library (YouBio; G111702) by PCR with the Fw primer 5′-GGAGATCTGAGCCCCAGAACAACCAGGCCAAGG-3′ and the Rv primer 5′-TGGAATTCTCAGGATTTGGACTTGGACACAGCAAGTC-3′. The PCR product was inserted into the BglII and EcoRI site of pEGFP-C1 plasmid to construct the pEGFP-FIS1 plasmid. Then EGFP-Fis1 cDNA was amplified by PCR with the Fw primer 5′-TTAAGCTTCGATGGTGAGCAAGGGCGAGGAGCTGTT-3′ and the Rv primer 5′-TGGAATTCTCAGGATTTGGACTTGGACACAGCAAGTC-3′. The PCR product was inserted into the HindIII and EcoRI site of pmCherry-C1 plasmid to construct the MitoQC plasmid. Human microtubule-associated protein 1 light-chain 3β (LC3B) cDNA was amplified from a human cDNA library (YouBio; G102982) by PCR with the Fw primer 5′-TCAGATCTATGCCGTCGGAGAAGACCT-3′ and the Rv primer 5′-GCGAATTCTTACACTGACAATTTCATCCCG-3′. The PCR product was inserted into the BglII and EcoRI site of ptagBFP-C plasmid to construct the ptagBFP-LC3B plasmid.
Primary cortical neurons were transfected with Mouse Neuron Nucleofector Kit (Lonza; VPG-1001) according to the manufacturer’s protocol. Briefly, 5 × 106 cells were collected during primary neuronal culture and then resuspended with 100 µl Nucleofector Solution containing 3 µg DNA. The cell suspension was transferred into the cuvette and electroporated with Nucleofector Program O-005. Approximately 50% of neurons can be transfected.
Axonal DNA/RNA isolation and real-time PCR
The isolation of axonal sample was performed as reported previously (Taylor et al., 2009). Briefly, neurons were cultured in microfluidic chambers, and OGD-Rep was performed in both somatic and axonal compartments in DIV8. At 40 min of reperfusion, the medium of both soma and axonal sides of microfluidic chamber was removed. 100 µl of lysis buffer was added to the axonal well and recollected after 10 s of incubation at room temperature. Continual aspiration is needed in soma compartment to prevent cellular material of soma from leaking into the axonal compartment. After collection of the axonal sample, DNA/RNA from the cell body compartment was isolated with the same procedures, except for aspiration in axonal compartment. A DNA/RNA/Protein isolation kit (Omega; R6734-01) was used to extract both DNA and RNA of one sample at the same time.
After verification of no nuclear gene expression in the axonal sample, aliquots of 3 to ∼10 ng total DNA were analyzed via real-time PCR to evaluate the expression of the axonal mitochondrial gene mt-Atp6 and the axonal housekeeping gene Csf1 (Taylor et al., 2009). Relative expression was presented as mt-Atp6:Csf1. Relative expression of somatic mitochondria was presented as mt-Atp6:Rpl-13. The primer sequences were as follows: mouse Atp6 (Fw: 5′-TCCTAGGCCTTTTACCACATACA-3′; Rv: 5′-TTTGTGTCGGAAGCCTGTAA-3′), mouse Rpl13 (Fw: 5′-CCTGCTGCTCTCAAGGTTGT-3′; Rv: 5′-GGTACTTCCACCCGACCTC-3), mouse Csf1 (Fw: 5′-CAAGGACTATGAGGAGCAGAACAAG-3′; Rv: 5′-GAAGTTCTTGATCTTCTCCAGCA-3′), mouse H1F0 (Fw: 5′-ACCTATTGTGCAAGGACAGC-3′; Rv: 5′-ACCAGGTCTCCAACTTACC-3′).
Immunostaining and imaging analysis
For immunostaining, cells were fixed with PBS containing 4% formaldehyde and permeabilized with 0.3% Triton X-100 in PBS for 15 min. After being blocked with PBS containing 1% BSA and 5% donkey serum for 1 h, cells were incubated at 4°C overnight with primary antibody against Hsp60 (1:400; Cell Signaling Technology; 12165) or β-tubulin III (1:200; Cell Signaling Technology; 4466). Secondary antibodies labeled with Alex Fluor 488, 594, or 647 (Invitrogen; 21202, A21207, and A31571) were subsequently added to the cells. Coverslips were observed on a confocal microscope (Fluoview FV1000; Olympus). Images were collected from six to ten random fields in each slide and the experiments were performed at least three times. Neurons were traced from cell body to the axonal tips during imaging and the axons were recognized by their morphological characteristics: long, thin, and uniform in diameter with sparse branches and growth cones (Banker and Cowan, 1979). The “Segmented Line” and the “Straighten” tool in Image J (National Institutes of Health) were used to show the profile of the entire axon (Kang et al., 2008). The images were transferred to 8-bit format and the somatic mitochondrial area were measured after setting scale and threshold. As for axonal mitochondria, the area and length were measured by Image J using the “Analyze Particles” tool. Neuronal morphology was analyzed with the “Neuron J” plugin. All data were collected and analyzed in a blind manner.
Live neuron imaging analysis
For colocalization assay of autophagosomes and mitochondria, neurons on glass-bottom dishes were observed by a 63× 1.4-NA oil immersion objective on a confocal microscope (Leica TCS SP8) with an incubator (5% CO2, 37°C). Z-stack images were acquired at a 1,024 × 1,024-pixel resolution and step-size was 0.3–1 µm. Images were collected from six to ten random fields in each slide and the experiments were performed at least three times. The number of autophagosomes and mitophagosomes were counted by the experimenters who were blind to the groups. Orthogonal view images and line-scan analysis of indicated regions were acquired by Image J with the “Orthogonal Views” and “Plot Profile” tool.
Photo-converting axonal MitoDendra2 was achieved by bleaching the axonal compartments of microfluidic devices loaded with MitoDendra2 expressed neurons with a 405-nm laser (60% laser power, 10 µs/pixel) for two flames. OGD-Rep was performed in both somatic and axonal compartments of microfluidic devices for the indicated time. Neuronal soma were observed by a 63× 1.4-NA oil-immersion objective on a confocal microscope (Carl Zeiss LSM 880 with Airyscan) with the Airyscan array detector, and the images were processed using the Airyscan processing toolbox in the ZEN software. Images of MitoDendra2 and tagBFP-LC3B fluorescence in each neuronal soma were captured in two consecutive stacks with the same Z-stack settings. Z-stack images were acquired at a 1560 × 1560-pixel resolution and step-size was 0.19–0.22 µm. Images were collected from six to ten random fields in each slide and the experiments were performed at least three times. Orthogonal view images and line profile of indicated region were acquired by Image J with the “Orthogonal Views” and “Plot Profile” tool.
Fluidic isolation in microfluidic chambers was achieved as reported previously (Taylor et al., 2005). Briefly, neurons expressing mCherry-LC3B were cultured in microfluidic chambers and loaded with 10 nM MTG (Invitrogen; M7514) in both somatic and axonal compartments to label global mitochondria. For somatic mitochondrial labeling, 180 µl culture medium containing 10 nM MTG was added to only the somatic compartment, immediately followed by the addition of 200 µl regular culture medium to axonal compartments. After 15 min of staining, both somatic and axonal compartments were washed with regular culture medium and subjected to OGD-Rep. Neuronal soma were observed by a 63× 1.4-NA oil-immersion objective on a confocal microscope (Carl Zeiss LSM 880 with Airyscan) with the Airyscan array detector, and the images were processed using the Airyscan processing toolbox in the ZEN software. Z-stack images were acquired at a 1,560 × 1,560-pixel resolution and step-size was 0.19–0.22 µm. Images were collected from six to ten random fields in each slide and the experiments were performed at least three times. The numbers of autophagosomes and mitophagosomes were counted by the experimenters who were blinded to the groups. Neurons expressing MitoQC were cultured in microfluidic chambers and loaded with 10 nM MTDR (Invitrogen; M22426) in only axonal compartments to label axonal mitochondria similarly as mentioned above. After 15 min of staining, neurons in both somatic and axonal compartments were washed with regular culture medium and subjected to OGD-Rep. Neuronal soma were observed by a 63× 1.4-NA oil-immersion objective on a confocal microscope (Leica TCS SP8) with an incubator (5% CO2, 37°C). Z-stack images were acquired at a 1,024 × 1,024-pixel resolution and step-size was 0.3–1 µm. Images were collected from six to ten random fields in each slide and the experiments were performed at least three times. Orthogonal view images and line-scan analysis of the indicated region were acquired by Image J with the “Orthogonal Views” and “Plot Profile” tool.
To monitor axonal mitochondrial transport, neurons were cultured in microfluidic chambers and loaded with 10 nM MTG in both somatic and axonal compartments. OGD-Rep was performed in both somatic and axonal compartments of microfluidic devices, and the axonal compartments of microfluidic devices were imaged by a 63× 1.4-NA oil-immersion objective on a confocal microscope (Leica TCS SP8) with an incubator (5% CO2, 37°C). Time-lapse images were acquired at a 1,024 × 1,024-pixel resolution every 10 s for 30 min. Axons were straightened by Image J with the “Straighten” tool, and kymographs were generated with the “Kymograph” tool. A mitochondrion was considered stationary if its initial and final position was the same and it showed no movements in the entire recording period. Those mitochondria that moved but returned to the initial position were defined as bidirectional mitochondria. Retrograde transportation is characterized by movements toward soma and displacement >5 µm, and opposite direction movements were defined as anterograde transportation. The velocity of each mitochondrion that is in motion was calculated with the slope of the lines in the kymograph (Wang and Schwarz, 2009).
Neurons expressing GFP, GFP-SNPH, or FKBP-GFP-Mito with either FRB-KIF5B-HA or FRB-BICDN-HA were cultured in glass-bottom dishes and subjected to OGD-Rep for the indicated time. Neurons were observed by a 63× 1.4-NA oil-immersion objective on a confocal microscope (Leica TCS SP8) with an incubator (5% CO2, 37°C). Time-lapse images were acquired and analyzed as described above. The axons were recognized by their morphological characteristics: long, thin, and uniform in diameter with sparse branches and growth cones (Banker and Cowan, 1979).
To detect mitochondrial membrane potential, neurons were cultured in glass-bottom dishes and stained with 100 nM tetramethylrhodamine methyl ester (Biotium; 70017) in DIV8. After 30 min, neurons were washed with regular culture medium and subjected to 1 h of 1 µM CCCP (Sigma; C2759) incubation, 1 h of OGD alone, or 20 min of OGD plus 40 min of reperfusion. Neurons were observed by a 63× 1.4-NA oil-immersion objective on a confocal microscope (Leica TCS SP8) with an incubator (5% CO2, 37°C). Z-stack images were acquired at a 1,024 × 1,024-pixel resolution and step-size was 0.5–1 µm. Images were collected from four to six random fields in each slide and the experiments were performed at least three times. Images of different groups were captured with the same confocal settings and the equal laser power. Fluorescence intensity was measured with Image J by the experimenters who were blind to the groups.
All data were collected and analyzed in a blind manner.
Rapalog-induced mitochondrial transportation
Neurons overexpressing FKBP-GFP-Mito with either FRB-KIF5B-HA or FRB-BICDN-HA were treated with ethanol as a vehicle control or 500 nM rapalog (Clontech; 635057) at the reperfusion after 20 min OGD in DIV8. Then cells were fixed for immunostaining or harvested for Western blot at 40 min of reperfusion. For morphology analysis, rapalog was washed out with regular culture medium after 40 min of incubation. Neurons were fixed for immunostaining at 24 h of reperfusion.
Measurement of mitochondrial respiration
Mitochondrial respiration was measured by detecting OCR using a Seahorse XF96 Extracellular Flux analyzer (Seahorse Bioscience, Agilent). Cortical neurons were seeded into Seahorse XF96-well plates at a density of 3 × 104 cells per well. Sensor cartridges were hydrated in XF calibrant (Agilent; 100840-000) and maintained at 37°C in air without CO2 24 h before OGD treatment. DIV8–14 neurons were subjected to OGD-Rep and the mitochondrial stress test was performed according to the manufacturer’s protocol. In brief, cells were washed once and incubated with XF base medium (Agilent; 103334-100) supplemented with 25 mM glucose (Sigma; G7021), 1 mM pyruvate sodium (Sigma; P5280), and 1% GlutaMAX for 1 h at 37°C in the absence of CO2 before the beginning of the assay. Changes in cellular respiration were assessed over time after sequential injections of oligomycin (Sigma; 75351) in port A, FCCP (Sigma; C2920) in port B, and rotenone (Sigma; R8875)/antimycin A (Sigma; A8674) in port C. The final concentrations of these compounds were 1 µM oligomycin, 0.5 µM FCCP, and 0.5 µM rotenone/2.5 µM antimycin A. Three measurements were performed in each session at 3-min intervals. The OCR values were normalized to protein concentration per well. The basal respiration was calculated by subtracting the average OCR of three measurements following rotenone/antimycin A injection from the average OCR of three measurements preceding oligomycin injection. The mitochondrial ATP production was calculated by subtracting the average OCR of three measurements between oligomycin and FCCP injection from the average OCR of three measurements preceding oligomycin injection. The maximum respiration was calculated by subtracting the average OCR of three measurements following rotenone/antimycin A injection from the average OCR of three measurements between FCCP and rotenone/antimycin A injection.
Western blot
The cells were homogenized in RIPA buffer (Sangon Biotech; C500005). A 40-µg aliquot of protein from each sample was separated using SDS-PAGE. The following primary antibodies were used: p62 (1:1,000; MBL; PM045), cleaved Casp3 (1:1,000; Cell Signaling Technology; 9661), COX IV (1:1,000; Cell Signaling Technology; 4850), Tim23 (1:100; Santa Cruz; sc-514463), RFP (1:1,000; MBL; PM005), HA (1:1,000; Cell Signaling Technology; 3724), LC3 (1:1,000; Sigma; L7543), Parkin (1:1,000; Cell Signaling Technology; P5748), or GAPDH (1:3,000; KangChen; KC-5G4). Secondary antibodies conjugated with HRP against either rabbit or mouse IgG (1:5,000; Cell Signaling Technology; 7071 and 7072) were applied. Digital images were quantified using densitometric measurement with Quantity-One software (Bio-Rad).
Statistical analysis
Statistical comparisons between two groups were performed by an unpaired t test. Comparisons between three or more groups were performed by one-way ANOVA with Sidak’s multiple-comparisons test. P < 0.05 was considered statistically significant.
Online supplemental material
Fig. S1 shows that Parkin deficiency fails to reverse OGD-Rep–induced axonal mitochondrial loss. Fig. S2 shows selective labeling of axonal mitochondria. Fig. S3 shows OGD-Rep–induced axonal mitochondrial loss and retrograde transportation in DIV14 neurons. Fig. S4 shows that reperfusion selectively maintains mitochondrial mobility and Parkin defciency fails to reverse reperfusion-induced mitochondrial retrograde transportation. Fig. S5 shows inhibition of SNPH (MTB-Tom22) on mitochondrial mobility during reperfusion. Video 1 shows axonal mitochondrial motion in the intact neuron. Video 2 shows that OGD arrests axonal mitochondrial mobility. Video 3 shows that reperfusion increases mitochondrial retrograde transportation. Video 4 shows the motion of axonal mitochondria stained with MitoTracker Red during reperfusion. Video 5 demonstrates that overexpression of GFP-SNPH arrests axonal mitochondrial transport during reperfusion. Video 6 shows the motion of axonal mitochondria in the neurons overexpressing FKBP-GFP-Mito and FRB-BICDN-HA during reperfusion. Video 7 shows that rapalog reinforces axonal mitochondrial retrograde transport in the neurons overexpressing FKBP-GFP-Mito and FRB-BICDN-HA during reperfusion. Video 8 shows the motion of axonal mitochondria in the neurons overexpressing FKBP-GFP-Mito and FRB-KIF5B-HA during reperfusion. Video 9 shows that rapalog arrests mitochondrial mobility in the neurons overexpressing FKBP-GFP-Mito and FRB-KIF5B-HA during reperfusion.
Supplementary Material
Acknowledgments
We are grateful to Professor Zhuohua Zhang from Central South China University for offering the Parkin gene knockout mice and Professor Masaaki Komatsu of Tokyo Metropolitan Institute of Medical Science for kindly offering the Atg7fl/fl mice. The plasmids encoding FRB-KIF5B-HA, FRB-BICDN-HA, and FKBP-GFP-Mito were kindly provided by Professor Casper C. Hoogenraad from Utrecht University. We are grateful to Professor Minxin Guan from Zhejiang University for help with the measurement of mitochondrial respiration and to the Imaging Facilities, Zhejiang University School of Medicine, for help with confocal microscopy.
This work was funded by the National Natural Science Foundation of China (grants 81573406, 81630098, 81521062, and 81822044).
The authors declare no competing financial interests.
Author contributions: Y. Zheng observed that reperfusion induced mitochondrial retrograde transport and carried out most of primary neuron culture as well as confocal microscopy imaging; X. Zhang designed the experiments to label somatic and axonal mitochondria separately and discovered that axonal mitochondria had a priority for autophagic clearance; X. Wu. carried out most of the biochemistry experiments with the help of S. Ma; X Zhang, X Wu, and L. Jiang analyzed kymograph data in a blind manner; Y. Zheng designed and constructed the plasmids with the help of A. Ahsan; and Z. Xiao; Y. Zheng, X. Zhang, F. Han, Z. Qin, W. Hu, and Z. Chen organized and interpreted the data. X. Zhang wrote the manuscript with input from all authors.
References
- Allen G.F., Toth R., James J., and Ganley I.G.. 2013. Loss of iron triggers PINK1/Parkin-independent mitophagy. EMBO Rep. 14:1127–1135. 10.1038/embor.2013.168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashrafi G., and Schwarz T.L.. 2015. PINK1- and PARK2-mediated local mitophagy in distal neuronal axons. Autophagy. 11:187–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashrafi G., Schlehe J.S., LaVoie M.J., and Schwarz T.L.. 2014. Mitophagy of damaged mitochondria occurs locally in distal neuronal axons and requires PINK1 and Parkin. J. Cell Biol. 206:655–670. 10.1083/jcb.201401070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banker G.A., and Cowan W.M.. 1979. Further observations on hippocampal neurons in dispersed cell culture. J. Comp. Neurol. 187:469–493. 10.1002/cne.901870302 [DOI] [PubMed] [Google Scholar]
- Cai Q., Zakaria H.M., Simone A., and Sheng Z.H.. 2012. Spatial parkin translocation and degradation of damaged mitochondria via mitophagy in live cortical neurons. Curr. Biol. 22:545–552. 10.1016/j.cub.2012.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Course M.M., and Wang X.. 2016. Transporting mitochondria in neurons. F1000 Res. 5:1735 10.12688/f1000research.7864.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devireddy S., Liu A., Lampe T., and Hollenbeck P.J.. 2015. The organization of mitochondrial quality control and life cycle in the nervous system in vivo in the absence of PINK1. J. Neurosci. 35:9391–9401. 10.1523/JNEUROSCI.1198-15.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H., Guo L., Yan S., Sosunov A.A., McKhann G.M., and Yan S.S.. 2010. Early deficits in synaptic mitochondria in an Alzheimer’s disease mouse model. Proc. Natl. Acad. Sci. USA. 107:18670–18675. 10.1073/pnas.1006586107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y.Y., Hu W.W., Dai H.B., Zhang J.X., Zhang L.Y., He P., Shen Y., Ohtsu H., Wei E.Q., and Chen Z.. 2011. Activation of the central histaminergic system is involved in hypoxia-induced stroke tolerance in adult mice. J. Cereb. Blood Flow Metab. 31:305–314. 10.1038/jcbfm.2010.94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farías G.G., Guardia C.M., De Pace R., Britt D.J., and Bonifacino J.S.. 2017. BORC/kinesin-1 ensemble drives polarized transport of lysosomes into the axon. Proc. Natl. Acad. Sci. USA. 114:E2955–E2964. 10.1073/pnas.1616363114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L., Bravo-San Pedro J.M., Blomgren K., and Kroemer G.. 2016. Autophagy in acute brain injury. Nat. Rev. Neurosci. 17:467–484. 10.1038/nrn.2016.51 [DOI] [PubMed] [Google Scholar]
- Hayakawa K., Esposito E., Wang X., Terasaki Y., Liu Y., Xing C., Ji X., and Lo E.H.. 2016. Transfer of mitochondria from astrocytes to neurons after stroke. Nature. 535:551–555. 10.1038/nature18928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogenraad C.C., Wulf P., Schiefermeier N., Stepanova T., Galjart N., Small J.V., Grosveld F., de Zeeuw C.I., and Akhmanova A.. 2003. Bicaudal D induces selective dynein-mediated microtubule minus end-directed transport. EMBO J. 22:6004–6015. 10.1093/emboj/cdg592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh C.H., Shaltouki A., Gonzalez A.E., Bettencourt da Cruz A., Burbulla L.F., St Lawrence E., Schüle B., Krainc D., Palmer T.D., and Wang X.. 2016. Functional impairment in Miro degradation and mitophagy is a shared feature in familial and sporadic Parkinson’s disease. Cell Stem Cell. 19:709–724. 10.1016/j.stem.2016.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J.S., Tian J.H., Pan P.Y., Zald P., Li C., Deng C., and Sheng Z.H.. 2008. Docking of axonal mitochondria by syntaphilin controls their mobility and affects short-term facilitation. Cell. 132:137–148. 10.1016/j.cell.2007.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbowski M., and Neutzner A.. 2012. Neurodegeneration as a consequence of failed mitochondrial maintenance. Acta Neuropathol. 123:157–171. 10.1007/s00401-011-0921-0 [DOI] [PubMed] [Google Scholar]
- Komatsu M., Waguri S., Ueno T., Iwata J., Murata S., Tanida I., Ezaki J., Mizushima N., Ohsumi Y., Uchiyama Y., et al. 2005. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 169:425–434. 10.1083/jcb.200412022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis T.L. Jr., Turi G.F., Kwon S.K., Losonczy A., and Polleux F.. 2016. Progressive decrease of mitochondrial motility during maturation of cortical axons in vitro and in vivo. Curr. Biol. 26:2602–2608. 10.1016/j.cub.2016.07.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., and Gao S.. 2017. Mitochondrial dysfunction in ischemic stroke. In Translational Research in Stroke. Lapchak P.A., and Yang G.-Y., editors. Springer Singapore, Singapore: 201–221. 10.1007/978-981-10-5804-2_10 [DOI] [Google Scholar]
- Lin M.Y., Cheng X.T., Tammineni P., Xie Y., Zhou B., Cai Q., and Sheng Z.H.. 2017a Releasing syntaphilin removes stressed mitochondria from axons independent of mitophagy under pathophysiological conditions. Neuron. 94:595–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M.Y., Cheng X.T., Xie Y., Cai Q., and Sheng Z.H.. 2017b Removing dysfunctional mitochondria from axons independent of mitophagy under pathophysiological conditions. Autophagy. 13:1792–1794. 10.1080/15548627.2017.1356552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovas J.R., and Wang X.. 2013. The meaning of mitochondrial movement to a neuron’s life. Biochim. Biophys. Acta. 1833:184–194. 10.1016/j.bbamcr.2012.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B. 2014. Neuronal mitophagy: long-distance delivery or eating locally? Curr. Biol. 24:R1006–R1008. 10.1016/j.cub.2014.09.037 [DOI] [PubMed] [Google Scholar]
- Maday S., and Holzbaur E.L.. 2014. Autophagosome biogenesis in primary neurons follows an ordered and spatially regulated pathway. Dev. Cell. 30:71–85. 10.1016/j.devcel.2014.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maday S., Wallace K.E., and Holzbaur E.L.. 2012. Autophagosomes initiate distally and mature during transport toward the cell soma in primary neurons. J. Cell Biol. 196:407–417. 10.1083/jcb.201106120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller K.E., and Sheetz M.P.. 2004. Axonal mitochondrial transport and potential are correlated. J. Cell Sci. 117:2791–2804. 10.1242/jcs.01130 [DOI] [PubMed] [Google Scholar]
- Nafstad P.H., and Blackstad T.W.. 1966. Distribution of mitochondria in pyramidal cells and boutons in hippocampal cortex. Z. Zellforsch. Mikrosk. Anat. 73:234–245. 10.1007/BF00334866 [DOI] [PubMed] [Google Scholar]
- Naga K.K., Sullivan P.G., and Geddes J.W.. 2007. High cyclophilin D content of synaptic mitochondria results in increased vulnerability to permeability transition. J. Neurosci. 27:7469–7475. 10.1523/JNEUROSCI.0646-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham A.H., McCaffery J.M., and Chan D.C.. 2012. Mouse lines with photo-activatable mitochondria to study mitochondrial dynamics. Genesis. 50:833–843. 10.1002/dvg.22050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rugarli E.I., and Langer T.. 2012. Mitochondrial quality control: a matter of life and death for neurons. EMBO J. 31:1336–1349. 10.1038/emboj.2012.38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagné C., Agulhon C., Ravassard P., Darmon M., Hamon M., El Mestikawy S., Gasnier B., and Giros B.. 2001. Identification and characterization of a lysosomal transporter for small neutral amino acids. Proc. Natl. Acad. Sci. USA. 98:7206–7211. 10.1073/pnas.121183498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schips T.G., Wietelmann A., Höhn K., Schimanski S., Walther P., Braun T., Wirth T., and Maier H.J.. 2011. FoxO3 induces reversible cardiac atrophy and autophagy in a transgenic mouse model. Cardiovasc. Res. 91:587–597. 10.1093/cvr/cvr144 [DOI] [PubMed] [Google Scholar]
- Schwarz T.L. 2013. Mitochondrial trafficking in neurons. Cold Spring Harb. Perspect. Biol. 5:a011304 10.1101/cshperspect.a011304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z., Zheng Y., Wu J., Chen Y., Wu X., Zhou Y., Yuan Y., Lu S., Jiang L., Qin Z., et al. 2017. PARK2-dependent mitophagy induced by acidic postconditioning protects against focal cerebral ischemia and extends the reperfusion window. Autophagy. 13:473–485. 10.1080/15548627.2016.1274596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng Z.H., and Cai Q.. 2012. Mitochondrial transport in neurons: Impact on synaptic homeostasis and neurodegeneration. Nat. Rev. Neurosci. 13:77–93. 10.1038/nrn3156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung H., Tandarich L.C., Nguyen K., and Hollenbeck P.J.. 2016. Compartmentalized Regulation of Parkin-Mediated Mitochondrial Quality Control in the Drosophila Nervous System In Vivo. J. Neurosci. 36:7375–7391. 10.1523/JNEUROSCI.0633-16.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor A.M., Blurton-Jones M., Rhee S.W., Cribbs D.H., Cotman C.W., and Jeon N.L.. 2005. A microfluidic culture platform for CNS axonal injury, regeneration and transport. Nat. Methods. 2:599–605. 10.1038/nmeth777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor A.M., Berchtold N.C., Perreau V.M., Tu C.H., Li Jeon N., and Cotman C.W.. 2009. Axonal mRNA in uninjured and regenerating cortical mammalian axons. J. Neurosci. 29:4697–4707. 10.1523/JNEUROSCI.6130-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Spronsen M., Mikhaylova M., Lipka J., Schlager M.A., van den Heuvel D.J., Kuijpers M., Wulf P.S., Keijzer N., Demmers J., Kapitein L.C., et al. 2013. TRAK/Milton motor-adaptor proteins steer mitochondrial trafficking to axons and dendrites. Neuron. 77:485–502. 10.1016/j.neuron.2012.11.027 [DOI] [PubMed] [Google Scholar]
- Wang X., and Schwarz T.L.. 2009. Imaging axonal transport of mitochondria. Methods Enzymol. 457:319–333. 10.1016/S0076-6879(09)05018-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Shao B.Z., Deng Z., Chen S., Yue Z., and Miao C.Y.. 2018. Autophagy in ischemic stroke. Prog. Neurobiol. 163-164:98–117. 10.1016/j.pneurobio.2018.01.001 [DOI] [PubMed] [Google Scholar]
- Wang Q.J., Ding Y., Kohtz D.S., Mizushima N., Cristea I.M., Rout M.P., Chait B.T., Zhong Y., Heintz N., and Yue Z.. 2006. Induction of autophagy in axonal dystrophy and degeneration [published correction appears in J. Neurosci 2006;26]. J. Neurosci. 26:8057–8068. 10.1523/JNEUROSCI.2261-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Winter D., Ashrafi G., Schlehe J., Wong Y.L., Selkoe D., Rice S., Steen J., LaVoie M.J., and Schwarz T.L.. 2011. PINK1 and Parkin target Miro for phosphorylation and degradation to arrest mitochondrial motility. Cell. 147:893–906. 10.1016/j.cell.2011.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Coleman M., Zhang L., Zheng X., and Yue Z.. 2013. Autophagy in axonal and dendritic degeneration. Trends Neurosci. 36:418–428. 10.1016/j.tins.2013.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin W., Signore A.P., Iwai M., Cao G., Gao Y., and Chen J.. 2008. Rapidly increased neuronal mitochondrial biogenesis after hypoxic-ischemic brain injury. Stroke. 39:3057–3063. 10.1161/STROKEAHA.108.520114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y., Zheng Y., Zhang X., Chen Y., Wu X., Wu J., Shen Z., Jiang L., Wang L., Yang W., et al. 2017. BNIP3L/NIX-mediated mitophagy protects against ischemic brain injury independent of PARK2. Autophagy. 13:1754–1766. 10.1080/15548627.2017.1357792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Zhao C., Lv C., Liu X., Du S., Li Z., Wang Y., and Zhang W.. 2017. Geniposide alleviates amyloid-induced synaptic injury by protecting axonal mitochondrial trafficking. Front. Cell. Neurosci. 10:309 10.3389/fncel.2016.00309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Yan H., Yuan Y., Gao J., Shen Z., Cheng Y., Shen Y., Wang R.R., Wang X., Hu W.W., et al. 2013. Cerebral ischemia-reperfusion-induced autophagy protects against neuronal injury by mitochondrial clearance. Autophagy. 9:1321–1333. 10.4161/auto.25132 [DOI] [PubMed] [Google Scholar]
- Zhang X., Yuan Y., Jiang L., Zhang J., Gao J., Shen Z., Zheng Y., Deng T., Yan H., Li W., et al. 2014. Endoplasmic reticulum stress induced by tunicamycin and thapsigargin protects against transient ischemic brain injury: Involvement of PARK2-dependent mitophagy. Autophagy. 10:1801–1813. 10.4161/auto.32136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B., Yu P., Lin M.Y., Sun T., Chen Y., and Sheng Z.H.. 2016. Facilitation of axon regeneration by enhancing mitochondrial transport and rescuing energy deficits. J. Cell Biol. 214:103–119. 10.1083/jcb.201605101 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.