Abstract
Osteosarcoma (OS) is the most common primary bone malignancy, predominately affecting children and adolescents. Due to the introduction of chemotherapy, the 5-year survival rate of OS patients has dramatically improved to 60-70%. Unfortunately, OS patients with recurrence or metastatic disease have less than a 20% chance of long-term survival, despite aggressive therapies. In this study, we aimed to identify gene expression patterns associated with metastasis and recurrence in order to identify potential biomarkers with prognostic power. We found that high expression of polyglutamine tract-binding protein 1 (PQBP1) and low expression of phosphoenolpyruvate carboxykinase 2 (PCK2) were related to a high probability of recurrence and metastasis in OS patients and also predicted shorter recurrence-free survival (RFS) and metastasis-free survival (MFS) after adjustment for other clinical variables. Prediction models based on the combination of PQBP1 and PCK2 expression had good and robust predictive power for recurrence and metastasis. A PQBP1 and PCK2-centered protein interaction network was built, and the hypothetical regulatory path between them was identified and termed the PQBP1-SF3A2-UBA52-PCK2 axis. Gene enrichment analysis indicated that aberrations of metabolism might play an important role in recurrence and metastasis in OS patients. Accordingly, PQBP1 and PCK2 are crucial for recurrence and metastasis in OS, and these findings provide a molecular basis for the exploitation of diagnostic and therapeutic strategies for overcoming recurrence and metastasis in OS.
Keywords: osteosarcoma, recurrence, metastasis, PQBP1, PCK2
Introduction
Osteosarcoma (OS) is the most common primary bone malignancy worldwide, accounting for approximately 6% of all cancers and 56% of malignant bone cancer cases in children and adolescents 1. Although OS predominantly occurs in individuals aged 0 to 24 years old, a second peak of incidence is observed in the elderly 2. Due to the introduction of chemotherapy and surgical removal of the tumor, the 5-year survival rate of patients with localized OS has dramatically improved over the past decades, increasing to nearly 60-70% 3; however, no significant improvement in survival has been reported since the 1970s 4, 5. Unfortunately, due to local recurrence 6 or distant metastasis 7, the current treatments for OS do not always yield satisfactory clinical responses. Approximately 20% of OS patients present with pulmonary metastasis at diagnosis; furthermore, despite intensive chemotherapy and aggressive surgery, 30-40% of patients initially diagnosed without metastatic disease develop metastasis or recurrence 8, 9. OS patients with recurrent or metastatic disease have less than a 20% chance of long-term survival, despite the use of aggressive therapies 5, 10.
Recurrence and metastasis are critical clinical episodes that have significant impacts on OS prognosis. However, it is difficult to predict which patients will develop recurrence or metastasis after therapy using only clinical indicators, such as age, gender, tumor site, and tumor stage. Although magnetic resonance imaging or computed tomography is usually applied, inflammation and recurrent/metastatic tumors are sometimes difficult to distinguish. For the past few years, with the development of high-throughput techniques, many researchers have attempted to identify molecular signatures of OS metastasis and recurrence 11-15. However, OS is a heterogeneous tumor, as confirmed by whole genome sequencing approaches 16, 17; therefore, the lack of consistency in the biomarkers generated from different studies remains one of the major obstacles to their clinical application. Thus, identifying potential key molecules associated with OS metastasis and recurrence that are consistent across various data sources would be of great academic and clinical significance and may lead to the development of novel prediction models and targeted therapies for OS patients with recurrent or metastatic disease, thereby improving patient prognosis.
In this study, we aimed to identify gene expression patterns associated with metastasis and recurrence in order to identify potential biomarkers with prognostic power. We analyzed three OS datasets generated using different microarray platforms and identified two genes associated with metastasis and recurrence in multiple datasets.
Methods
OS patient datasets
We searched the National Center for Biotechnology Information Gene Expression Omnibus database (GEO; https://www.ncbi.nlm.nih.gov/geo/) to identify expression microarray data sets from OS patients with clinically well-defined recurrence or metastasis information. Ultimately, a total of three datasets generated using different platforms were enrolled in our study, including 27 patients with information on metastatic events from a study by Kobayashi et al. (accession number: GSE14827, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE14827)18, 37 patients with information on recurrence events and recurrence times from a study by Kelly et al. (accession number: GSE39055, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE39055)19, and 53 patients with information on metastatic events, metastasis times, survival events, and survival times from a study by Buddingh et al. (accession number: GSE21257, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE21257) 20. Detailed clinical information for the patients in the three datasets included in our study is listed in Table 1.
Table 1.
Summary of the clinical and pathological characteristics of OS patients in 3 included datasets.
| Characteristic | GSE14827 | GSE39055 | GSE21257 |
|---|---|---|---|
| Platform | hgu133plus2 | Illumina HumanHT-12 WG-DASL V4.0 R2 | Illumina Human-6 v2.0 |
| Sample number | 27 | 37 | 53 |
| Age | |||
| median (range) | 15(8- 38) | 11(4 - 71) | 16(3 - 79) |
| Metastasis/Recurrence status | |||
| no metastasis/no recurrence | 18 | 19 | 19 |
| metastasis/recurrence | 9 | 18 | 34 |
| Month to metastasis/recurrence | |||
| median (range) | / | 25.5(2.7 - 151) | 9(1-44) |
| Gender | |||
| male | 17 | 20 | 34 |
| female | 10 | 17 | 19 |
| Histologicalsubtype | |||
| chondroblastic | 3 | / | 6 |
| fibroblastic | 1 | / | 5 |
| osteoblastic | 21 | / | 32 |
| other | 2 | / | 10 |
| Chemotherapeutic response | |||
| good | 11 | / | / |
| poor | 16 | / | / |
| Tumor site | |||
| femur | 15 | / | 27 |
| tibia | 10 | / | 15 |
| humerus | 2 | / | 8 |
| fibula | 0 | / | 2 |
| unknown | 0 | / | 1 |
| Tumor grade | |||
| 1/2/3/4/unkown | / | / | 13/16/13/5/6 |
| Necrosis percentage | |||
| median (range) | / | 65% (0.65%-0.99%) | / |
Acquisition and processing of all expression profiles
The standardized gene expression profile datasets were directly downloaded from the GEO database through the Bioconductor 21 package GEOquery 22. In the case of more than one probe set mapping to the same gene, the average value of the probe sets was used as the gene expression value 23. In consideration of the heterogeneity across the three microarray datasets in terms of systematic measurements, a z-score transformation was performed independently in each dataset 24.
Screening and validation of metastasis- and recurrence-related genes
GSE14827 and GSE39055 were used as the discovery sets to explore gene signatures related to metastasis and recurrence in OS patients, while GSE21257 was used as an independent validation dataset. Differences in the gene expression profiles of OS patients in GSE14827 who did and did not develop metastasis were compared, and similarly, differences in the gene expression profiles of OS patients in GSE39055 who did and did not develop recurrence were analyzed. Using two-tailed t-tests, genes with a p-value < 0.05 and fold change (FC) > 1.5 were considered as differentially expressed genes (DEGs) in these two discovery datasets. Visualization of DEGs in the two discovery datasets was performed using Venn diagrams created by the VennDiagram 25 package in R and heatmap plots based on the Euclidean method created using ComplexHeatmap 26 in Bioconductor. After verification in GSE21257 using two-tailed t-tests, polyglutamine tract-binding protein 1 (PQBP1) and phosphoenolpyruvate carboxykinase 2 (PCK2) were identified as candidates for further analysis.
Survival analysis
Based on the identification of two genes (PQBP1 and PCK2) related to metastasis and recurrence in OS patients, we next evaluated their predictive ability for metastasis and recurrence. We specified PQBP1 and PCK2 mRNA expression cutoff values in GSE14827 to define a binary expression status variable on the basis of the MinPvalue criterion from the Optimal Cutpoints 27 package in R(cutoff values: PQBP1, 0.221; PCK2, 0.771). PQBP1 and PCK2 mRNA expression levels were then classified into high (+) and low (-) expression status in the other two datasets (GSE39055 and GSE21257) according to their cutoff values established in GSE14827. The odds ratio (OR), corresponding 95% confidence intervals (95% CIs), and p-values for the correlations between metastasis/recurrence and PQBP1/PCK2 expression status, individually and in combination, were estimated using Fisher's exact test. Kaplan-Meier survival analysis was conducted to establish survival curves using the survminer 28 package for metastasis-free survival (MFS) or recurrence-free survival (RFS) according to the univariate gene expression status as well as the combined expression status of the two genes. The statistical significance was assessed by log-rank test. In addition, we also performed Kaplan-Meier survival analysis and log-rank tests on overall survival for the purpose of checking the predictive power of the PQBP1 and PCK2 mRNA expression status for late prognosis.
Using the chi-square test via the R package coin 29, we also explored the associations between the PQBP1/PCK2 mRNA expression status and various clinical characteristics that were available in each dataset, such as age, gender, histological subtype, tumor site, and chemotherapeutic response. Univariate analysis with the Cox proportional hazards regression model was used to assess the correlations between the PQBP1/PCK2 mRNA expression status and MFS in GSE21257 or RFS in GSE39055. Multivariate analyses with the Cox proportional hazards regression model was used to test whether the PQBP1 and PCK2 expression statuses were predictors of metastasis and recurrence independent of other clinical characteristics in GSE21257 and GSE39055. Additionally, univariate and multivariate Cox regression analyses for overall survival were conducted in GSE21257. However, due to the lack of information on metastasis times, analysis of the MFS in GSE14827 could not be performed.
Construction and validation of prediction models for metastasis and recurrence
Using the caret 30 package, we constructed prediction models for metastasis in the GSE14827 dataset based on the combination of PQBP1 and PCK2 mRNA expression values using three different machine learning algorithms: random forest, linear kernel support vector machine (SVM), and artificial neural network. These training prediction models were built through five rounds of 10-fold cross validations coupled with an internal parameter debugging process in GSE14827, and then the optimal model for each machine learning algorithm was selected. Subsequently, the three best models were tested for their predictive ability for metastasis in GSE21257 and for recurrence in GSE39055. Using the pROC 31 package in R, the prediction abilities of these models were assessed by receiver operating characteristic (ROC) curves and the corresponding area under the ROC curve (AUC) values.
PQBP1 and PCK2-centered interaction network
To explore the potential regulatory patterns between PQBP1 and PCK2, we built a PQBP1 and PCK2-centered protein interaction network derived from the Search Tool for the Retrieval of Interacting Genes (STRING) database with a combined score above 600 and k nearest neighbors = 1 32. We adopted the fast greedy searching community detection algorithm on this network to find functional modules. We further investigated the shortest path between PQBP1 and PCK2 in this network, and then we detected pairwise associations between the expression values of the genes encoding the proteins involved in this path in GSE14827, GSE39055, and GSE21257. Functional enrichment analysis for all genes encoding proteins in this PQBP1 and PCK2-centered interaction network was performed using hypergeometric tests. Using the complete human genome as the background, terms with an adjusted p-value < 0.05 were considered significantly enriched, and the top ten most significantly enriched terms for each annotation system were visualized as word clouds 33.
All statistical analyses and data mining procedures performed in the present study were conducted with R (version: 3.2.2) and Bioconductor software.
Results
High expression of PQBP1 and low expression of PCK2 correlated with OS metastasis and recurrence
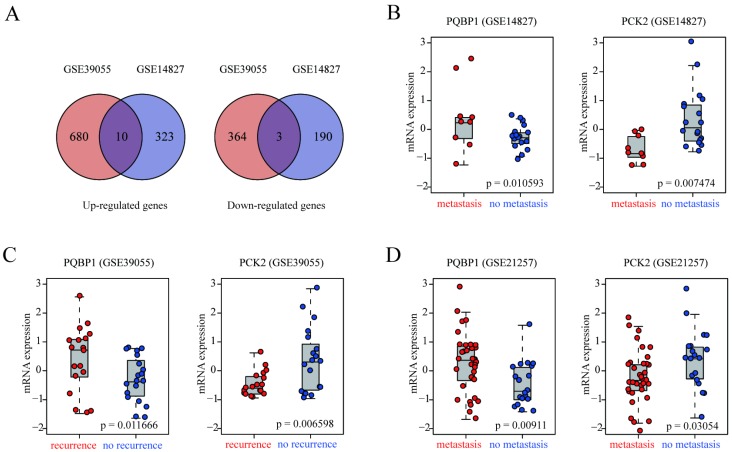
In the present study, we used two OS patient cohorts (GSE14827 and GSE39055) with clinically well-defined metastasis or recurrence information as discovery datasets to identify gene expression alterations associated with OS metastasis and recurrence. GSE14827 included nine patients that developed metastasis and 18 metastasis-free patients, while GSE39055 consisted of 18 patients with recurrence and 19 recurrence-free patients. In GSE14827, we identified 526 DEGs, 333 of which were up-regulated and 193 of which were down-regulated in patients with metastasis compared with levels in non-metastasis patients. In GSE39055, we obtained 1057 DEGs, 690 of which were up-regulated and 367 of which were down-regulated in patients with recurrence compared with levels in recurrence-free patients. Heat maps based on DEGs derived from each discovery dataset demonstrated a clear distinction between the different groups (Figure S1A-B). The complete lists of DEGs from the analyses of GSE14827 and GSE39055 are shown in Tables S1 and S2, respectively. As shown in Figure 1A, the intersecting DEGs derived from the two datasets consisted of ten genes that were up-regulated and three that were down-regulated in patients with metastasis and recurrence (Table S3).
Figure 1.
Identification of genes related to recurrence and metastasis in OS patients. (A) Venn diagrams of DEGs derived from GSE39055 and GSE14827. Ten upregulated and three downregulated were common between the two datasets. (B-D) box plots of PQBP1 and PCK2 mRNA expression levels in patients that did and did not develop recurrence/metastasis in the discovery (GSE39055, GSE14827) and validation (GSE21257) datasets. Red and blue dots represent patients with and without recurrence/metastasis, respectively.
To demonstrate the reproducibility and robustness of these DEGs in an independent dataset, GSE21257, which was generated using a different platform, was selected as a validation cohort. Because PQBP1 was the most significantly up-regulated gene and PCK2 was the most significantly down-regulated gene in the two discovery datasets (Figure 1B-C), we concentrated on these two genes for further verification in GSE21257. Consistent with the results in the discovery datasets, PQBP1 expression was markedly elevated (p = 0.00911) while PCK2 expression was significantly reduced (p = 0.03054) in patients who developed metastasis in GSE21257 (Figure 1D).
PQBP1 and PCK2 act as independent predictors of metastasis and recurrence
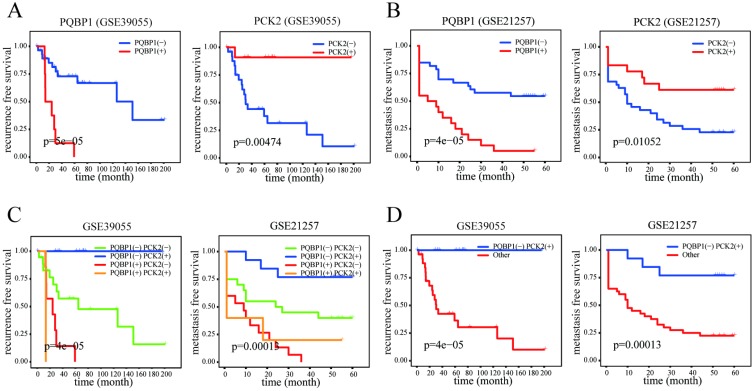
The PQBP1 and PCK2 mRNA expression levels of patients in GSE39055 and GSE21257 were divided into “high” (+) and “low” (-) expression according to their respective cutoff values derived from GSE14827. As shown in Table 2, high expression of PQBP1 and low expression of PCK2 were significantly correlated with a high rate of metastasis in GSE14827 and GSE21257 and a high probability of recurrence in GSE39055. Furthermore, in general, more significant correlations with the incidence of OS metastasis and recurrence were found by combining the expression statuses of these two genes to form four combined expression groups: PCK2(-)PQBP1(-), PCK2(+)PQBP1(+), PCK2(+)PQBP1(-), and PCK2(-)PQBP1(+). In addition, PCK2(+)PQBP1(-) was compared with the other three groups for a binary comparison of combined expression profiles. Kaplan-Meier curves and log-rank tests revealed that both the PQBP1(+) expression group and the PCK2(-) expression group exhibited significantly poorer RFS and MFS in GSE39055 and GSE21257, respectively (Figure 2A-B). When comparing all four combined expression profiles or the PCK2(+)PQBP1(-) profile vs. the other three combined expression profiles, we observed significantly different RFS values in GSE39055 and MFS values in GSE21257 (Figure 2C-D). These findings emphasize the ability of the PQBP1 and PCK2 mRNA expression statuses to predict metastasis and recurrence.
Table 2.
Expression status differences of PQBP1and PCK2 on OS metastasis/recurrence.
| Dataset | PCK2(high) | PQBP1(high) | PCK2+PQBP1 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| OR | 95%CI | P | OR | 95%CI | P | OR | 95%CI | Pa | Pb | |
| GSE14827 | 0.050 | 0.001 -0.544 | 3.639e-03 | 5.755 | 0.754 -55.752 | 0.072 | 0.031 | 0.001 -0.347 | 5.750e-04 | 2.768e-03 |
| GSE39055 | 0.047 | 0.001 -0.417 | 1.098e-03 | 15.311 | 1.739 - 748.736 | 3.626e-03 | 0.043 | 0.001- 0.368 | 4.997e-04 | 5.116e-04 |
| GSE21257 | 0.196 | 0.046 -0.758 | 0.014 | 21.618 | 2.796 -993.201 | 2.640e-04 | 0.092 | 0.013- 0.455 | 7.095e-04 | 6.263e-04 |
a. This comparison were done between PCK2(+)PQBP1(-)and PCK2(+)PQBP1(+)/PCK2(-)PQBP1(-)/PCK2(-)PQBP1(+).
b. Comparisons among 4 groups such as PCK2(+)PQBP1(-)/PCK2(+)PQBP1(+)/PCK2(-)PQBP1(-) and PCK2(-)PQBP1(+).
Figure 2.
Prognostic significance of PQBP1 and PCK2 expression status in OS. (A) Kaplan-Meier plots of RFS with different PQBP1 or PCK2 expression statuses in GSE39055. (B) Kaplan-Meier plots of MFS with different PQBP1 or PCK2 expression statuses in GSE21257. (C) Kaplan-Meier plots of RFS and MFS with four different combined expression profiles in GSE39055 and GSE21257, respectively. (D) Kaplan-Meier plots of RFS and MFS in binary comparison of combined expression profiles in GSE39055 and GSE21257, respectively. For both PQBP1 and PCK2, (-) represents low expression status and (+) represents high expression status.
We further statistically assessed the relationships between the expression levels of the two genes and various clinical characteristics that were available in each dataset, such as age, gender, histological subtype, tumor site, chemotherapeutic response, tumor grade, and necrosis percentage. The only significant correlation observed was the relationship between gender and PCK2 mRNA expression status in GSE21257 (Table S4).
We next conducted a univariate Cox regression analysis to further evaluate the prognostic performance of these two genes and other clinical factors for metastasis and recurrence in each dataset. The results showed that high expression levels of PQBP1 and low expression levels of PCK2 predicted poor MFS in GSE21257 and RFS in GSE39055 (Table 3-4). Multivariate Cox regression analysis further revealed that PQBP1 and PCK2 were significantly and independently associated with the probability of metastasis and recurrence in patients with OS in GSE21257 and GSE39055, after adjusting for other clinical factors.
Table 3.
Univariate and multivariate Cox regression analyses for RFS of OS patients in GSE39055.
| Variables | Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|---|
| HR | 95%CI | p value | HR | 95%CI | p value | ||
| Gender | male vs. female | 1.283 | 0.499 -3.297 | 0.605 | 2.367 | 0.802- 6.986 | 0.119 |
| Age | Low vs. high | 0.449 | 0.128-1.582 | 0.213 | 0.199 | 0.040- 1.001 | 0.050 |
| Necrosis status | low vs. high | 3.808 | 1.093-13.26 | 0.036 | 1.581 | 0.417- 5.985 | 0.500 |
| PCK2 | high vs. low | 0.096 | 0.013 -0.722 | 0.023 | 0.120 | 0.015-0.985 | 0.048 |
| PQBP1 | high vs. low | 6.918 | 2.396 -19.98 | 3.508e-04 | 5.934 | 1.661-21.206 | 0.006 |
Bold text denotes P<0.05
Table 4.
Univariate and multivariate Cox regression analyses for MFS of OS patients in GSE21257.
| Variables | Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|---|
| HR | 95%CI | P value | HR | 95%CI | P value | ||
| Gender | male vs. female | 2.179 | 1.013 - 4.685 | 0.046 | 1.801 | 0.716- 4.529 | 0.211 |
| Age | low vs. high | 2.446 | 1.137 - 5.262 | 0.022 | 1.686 | 0.625- 4.546 | 0.302 |
| Tumor site | femur | 1(reference) | 1(reference) | ||||
| fibula | 0.467 | 0.062 - 3.506 | 0.459 | 0.170 | 0.015- 1.953 | 0.155 | |
| humerus | 0.805 | 0.299 - 2.172 | 0.669 | 0.400 | 0.113- 1.410 | 0.154 | |
| tibia | 0.667 | 0.299 - 1.488 | 0.323 | 0.439 | 0.157- 1.226 | 0.116 | |
| unknown | 0.862 | 0.115 -6.484 | 0.885 | 2.835 | 0.108- 74.743 | 0.532 | |
| Histological subtype | chondroblastic | 1(reference) | 1(reference) | ||||
| fibroblastic | 0.296 | 0.057 -1.531 | 0.146 | 0.158 | 0.012- 1.998 | 0.154 | |
| osteoblastic | 0.707 | 0.266 - 1.878 | 0.487 | 1.592 | 0.381- 6.647 | 0.524 | |
| other | 0.615 | 0.187 -2.019 | 0.423 | 1.700 | 0.341- 8.463 | 0.517 | |
| Tumor grade | G1 | 1(reference) | 1(reference) | ||||
| G2 | 0.454 | 0.186 -1.108 | 0.083 | 0.222 | 0.069- 0.715 | 0.017 | |
| G3 | 0.416 | 0.160 -1.085 | 0.073 | 0.205 | 0.060- 0.699 | 0.011 | |
| G4 | 0.699 | 0.194 -2.529 | 0.586 | 0.398 | 0.090- 1.750 | 0.223 | |
| unknown | 0.601 | 0.190 -1.903 | 0.387 | 0.219 | 0.050- 0.951 | 0.043 | |
| PCK2 | high vs. low | 0.358 | 0.155 -0.826 | 0.016 | 0.254 | 0.085- 0.763 | 0.014 |
| PQBP1 | high vs. low | 3.927 | 1.948 -7.917 | 1.31e-04 | 6.977 | 2.377- 20.478 | 4.07e-04 |
Bold text denotes P<0.05
We further wondered whether the expression levels of PQBP1 and PCK2 were related to the late prognosis of OS patients. To determine this, we created dot and density plots for the mRNA expression distributions of PQBP1and PCK2 based on survival status in the three datasets. The expression distributions of the two genes were not significantly different between living and deceased cases (Figure S2A-B). We also conducted Kaplan-Meier and Cox regression analyses for overall survival based on the expression statuses of the two genes in GSE21257. High expression levels of PQBP1 predicted poor overall survival, while the expression of PCK2 was not significantly related to overall survival (Figure S2C, Table S5). Due to the lack of information on survival times in GSE14827 and GSE39055, statistical analysis for overall survival could not be conducted in these datasets.
Based on the above results, we concluded that PQBP1 and PCK2 can be employed as predictors for metastasis and recurrence in OS patients but are not suitable for the prediction of late prognosis.
Prediction models for metastasis and recurrence based on combined PQBP1 and PCK2 expression had robust predictive power
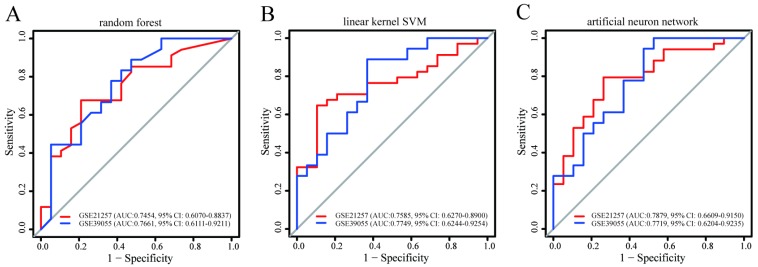
Using three machine learning algorithms, prediction models for metastasis based on a combination of PQBP1 and PCK2 mRNA expression values were built in GSE14827 and subsequently validated in the other two datasets. As shown in Figure 3, the AUC and 95% CI values of these three models indicated good performance for metastasis in GSE21757 and for recurrence in GSE39055 (maximum AUC: 0.7879, 95% CI: 0.6609-0.9150; minimum AUC: 0.7454, 95% CI: 0.6070-0.8837). Among these three models, the model constructed using the artificial neural network algorithm seemed to exhibit the best performance (GSE21257: AUC: 0.7879, 95% CI: 0.6609-0.9150; GSE39055: AUC: 0.7719, 95% CI: 0.6204-0.9235). Despite some differences in performance, however, models based on the combination of PQBP1 and PCK2 from all three machine learning algorithms exhibited robust predictive power for metastasis and recurrence in OS patients. These results further indicate that the combination of the expression statuses of the two genes is able to identify OS patients with high or low risk of metastasis and recurrence.
Figure 3.
The predictive power of the prediction models for recurrence and metastasis built in GSE14827 and applied to the other two datasets. The models were built using (A) random forest, (B) linear kernel support vector machine, and (C) artificial neural network. For each model, the AUCs and 95% CIs are also shown.
Potential regulatory relationship between PQBP1 and PCK2
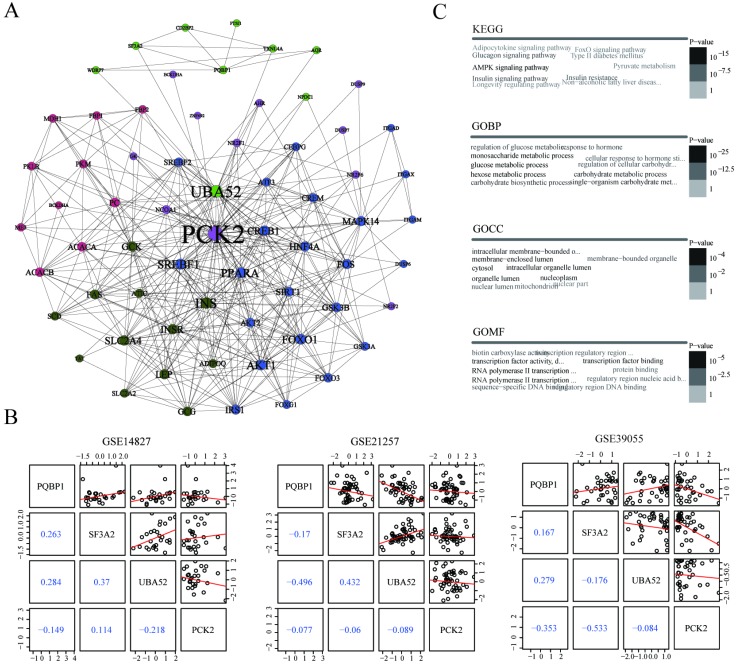
To further explore the interaction relationships between PQBP1 and PCK2, we extracted the minimal PQBP1 and PCK2-centered undirected interaction network (Figure 4A). The network contained five distinct communities with 65 nodes and 400 edges in total. PQBP1 and PCK2 belonged to different communities. We found that PCK2 was the protein most connected with other proteins in the network. Furthermore, we explored the shortest path between PQBP1 and PCK2 in this network, and we found that PQBP1 and PCK2 were connected by two other proteins, SF3A2 and UBA52. Thus, the shortest path was defined as the PQBP1-SF3A2-UBA52-PCK2 axis. The four proteins in this axis belonged to two communities, with PQBP1, SF3A2, and UBA52 in same community and PCK2 belonging to another community. UBA52 and PCK2 were hub genes (genes with an extremely high connectivity) 34 in their own communities. Then, we measured the pairwise expression correlations of the four axis genes in the three datasets (Figure 4B). The expression of UBA52 and PQBP1 was always negatively correlated with that of PCK2. This axis may represent the potential regulatory path between PQBP1 and PCK2.
Figure 4.
Potential regulatory pattern between PQBP1 and PCK2. (A) The minimal PQBP1 and PCK2-centered undirected protein interaction network from the STRING database. Nodes represent proteins, and node size is proportional to the degree of connectivity with other proteins. Edges represent interactions between proteins. Five communities were detected and are colored-coded. (B) Pairwise correlations between expression profiles of the four axis genes in GSE14827, GSE21257, and GSE39055. In the upper triangle, black dots represent pairwise expression values in all samples in each dataset, and red lines represent linear regression values. In the lower triangle, blue numbers show Pearson's correlation coefficients between two genes. (C) Word cloud plots of top ten most significantly enriched terms among genes in the network from KEGG, GOBP, GOCC and GOMF on genes in the network. The font size and gray scale of each term were proportional to the adjusted p value.
To gain insight into the biological roles of the 65 proteins involved in the PQBP1 and PCK2-centered protein interaction network, we performed gene enrichment analyses using the signaling pathways included in the Kyoto Encyclopedia of Genes and Genomes (KEGG) and the cellular component (GOCC), molecular function (GOMF), and biological process (GOBP) categories in the Gene Ontology (GO). The top ten most significantly enriched terms according to each annotation system were visualized as word clouds (Figure 4C). Notably, the top three most significantly enriched GOBP terms were glucose metabolic process, hexose metabolic process, and monosaccharide metabolic process. According to KEGG pathway analysis, the AMPK signaling pathway, insulin signaling pathway, and glucagon signaling pathway were the three most significantly enriched pathways. Thus, both GO and KEGG pathway enrichment analyses identified genes related to PQBP1 and PCK2 that are associated with metabolic dysfunction.
Discussion
The results of this study showed that the gene expression patterns of PQBP1 and PCK2 were associated with metastasis and recurrence, making these genes potential biomarkers with prognostic power. PQBP1, also known as NPW38, encodes a widely expressed 38-kDa protein 35, 36 that contains several protein interaction domains: a WW domain through which PQBP1 interacts with RNA polymerase II 37, a polar amino acid-rich domain that binds to polyglutamine tracts 36, a nuclear localization signal, and a carboxy-terminal domain that interacts with splicing factors 38-40. Mutations in PQBP1 are reportedly associated with several X-linked mental retardation disorders, including Renpenning, Hamel, Sutherland-Haan, Golabi-Ito-Hall, and Porteous syndromes 41-45. However, the biological function of PQBP1 in tumors remains unknown, and thus far, there has been no research into the role of PQBP1 in OS.
PCK2, also known as PEPCK2, is located on chromosome 14q11.2. The protein encoded by PCK2 functions as a rate-limiting enzyme involved in gluconeogenesis and glycolysis 46, 47. Studies have reported potential roles for PCK2 mRNA and protein in tumors, but due to discrepancies among studies, the results remain inconclusive. For example, Vincent et al. 48 showed that PCK2 mRNA was highly expressed in the thyroid, breast, kidney, and bladder and in non-small-cell lung cancer tissue. PCK2 knockdown hampered tumor cell proliferation, reduced tumor size, and increased tumor cell death 49-51. Conversely, another study reported that overexpression of PCK2 hindered tumorigenesis and slowed the growth of tumor-repopulating cells growth 52. Moreover, expression of the PCK2 protein gradually decreased in cisplatin-resistant bladder cancer cells compared to levels in cisplatin-naïve cells, and this decrease was dependent on the cisplatin concentration 53.
In our study, a comprehensive analysis of gene expression profiles in OS patients in the GSE14827 and GSE39055 datasets was conducted, and we identified 13 intersecting genes related to metastasis and recurrence. Among these 13 genes, PQBP1 and PCK2 were successfully validated in an independent dataset (GSE21257). High expression of PQBP1 and low expression of PCK2 were associated with a high probability of metastasis and recurrence in OS patients. Multivariate Cox regression analysis revealed that the predictive power of PQBP1 and PCK2 for RFS and MFS were independent of other clinicopathological covariates, such as gender, necrosis status, and age, in OS patients from GSE39055 and GSE21257. Furthermore, metastasis and recurrence prediction models for OS patients were built based on a combination of PQBP1 and PCK2 mRNA expression values using three machine learning algorithms in GSE14827. We found that these prediction models had robust prognostic power for metastasis and recurrence in GSE39055 and GSE21257, regardless of which machine learning algorithm was adopted. These findings indicate that PQBP1 and PCK2 may allow for the stratification of OS patients in future clinical trials. We also analyzed the PQBP1 and PCK2-centered protein network in an attempt to identify the roles of these proteins in the development of metastasis and recurrence in OS, and we extracted the shortest path between the two proteins, which reflects a possible regulatory axis between PQBP1 and PCK2. Interestingly, we noted that the most significantly enriched GO terms and KEGG pathways for genes encoding proteins in the network were related to metabolic dysfunction, suggesting that aberrations in metabolism may play an important role in OS metastasis and recurrence. Previous research has reported that PCK2 downregulation leads to a reduction in energy metabolism and may subsequently decrease susceptibility to chemotherapeutic drugs 53. This may explain why low expression of PCK2 is associated with increased likelihoods of metastasis and recurrence in OS patients. However, the molecular mechanisms underlying the predictive roles of PQBP1 and PCK2 in OS metastasis and recurrence remain unclear, and thus the above hypothesis should be further explored and confirmed in future experimental research.
Conclusions
In this study, we newly identified PQBP1 and PCK2 as potential predictive biomarkers for metastasis and recurrence in patients with OS, providing a molecular basis for the development of targeted diagnostic and therapeutic strategies for overcoming metastasis and recurrence in OS. It is our hope that these findings will enhance clinical decision-making and strengthen disease surveillance. However, the molecular mechanisms underlying the upregulation of PQBP1 and downregulation of PCK2 in metastatic and recurrent OS patients remain unclear and should be elucidated in future studies.
Supplementary Material
Supplementary figures and tables.
Acknowledgments
This work was generously supported by the National Natural Science Foundation of China (No.81601992), the Natural Science Foundation of Jiangsu Province (No.BK20160343), the Research project of Jiangsu Provincail Health Department (H201417) and the Jiangsu Provincial Clinical Orthopedic Center.
Abbreviations
- OS
osteosarcoma
- PQBP1
polyglutamine tract-binding protein 1
- PCK2
phosphoenolpyruvate carboxykinase 2
- RFS
recurrence free survival
- MFS
metastasis free survival
- GEO
Gene Expression Omnibus database
- DEGs
differentially expressed genes
- OR
odds ratio
- 95% CI
95% confidence interval
- SVM
support vector machine
- ROC
receiver operating characteristic
- AUC
area under the ROC curve
- STRING database
Search Tool for the Retrieval of Interacting Genes database
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- GO
Gene Ontology
- GOCC
Gene Ontology cellular component
- GOMF
Gene Ontology molecular function
- GOBP
Gene Ontology biological process
- HR
hazard ratio
References
- 1.Ottaviani G, Jaffe N. The epidemiology of osteosarcoma. Cancer Treat Res. 2009;152:3–13. doi: 10.1007/978-1-4419-0284-9_1. [DOI] [PubMed] [Google Scholar]
- 2.Mirabello L, Troisi RJ, Savage SA. Osteosarcoma incidence and survival rates from 1973 to 2004: data from the Surveillance, Epidemiology, and End Results Program. Cancer. 2009;115:1531–43. doi: 10.1002/cncr.24121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bielack SS, Kempf-Bielack B, Delling G. et al. Prognostic factors in high-grade osteosarcoma of the extremities or trunk: an analysis of 1,702 patients treated on neoadjuvant cooperative osteosarcoma study group protocols. J Clin Oncol. 2002;20:776–90. doi: 10.1200/JCO.2002.20.3.776. [DOI] [PubMed] [Google Scholar]
- 4.Bacci G, Longhi A, Versari M. et al. Prognostic factors for osteosarcoma of the extremity treated with neoadjuvant chemotherapy: 15-year experience in 789 patients treated at a single institution. Cancer. 2006;106:1154–61. doi: 10.1002/cncr.21724. [DOI] [PubMed] [Google Scholar]
- 5.Chou AJ, Geller DS, Gorlick R. Therapy for osteosarcoma: where do we go from here? Paediatr Drugs. 2008;10:315–27. doi: 10.2165/00148581-200810050-00005. [DOI] [PubMed] [Google Scholar]
- 6.Bielack SS, Kempf-Bielack B, Branscheid D. et al. Second and subsequent recurrences of osteosarcoma: presentation, treatment, and outcomes of 249 consecutive cooperative osteosarcoma study group patients. J Clin Oncol. 2009;27:557–65. doi: 10.1200/JCO.2008.16.2305. [DOI] [PubMed] [Google Scholar]
- 7.PosthumaDeBoer J, Witlox MA, Kaspers GJ. et al. Molecular alterations as target for therapy in metastatic osteosarcoma: a review of literature. Clin Exp Metastasis. 2011;28:493–503. doi: 10.1007/s10585-011-9384-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qu L, Liu B. Cyclooxygeanse-2 promotes metastasis in osteosarcoma. Cancer Cell Int. 2015;15:69. doi: 10.1186/s12935-015-0220-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daw NC, Chou AJ, Jaffe N. et al. Recurrent osteosarcoma with a single pulmonary metastasis: a multi-institutional review. Br J Cancer. 2015;112:278–82. doi: 10.1038/bjc.2014.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kempf-Bielack B, Bielack SS, Jurgens H. et al. Osteosarcoma relapse after combined modality therapy: an analysis of unselected patients in the Cooperative Osteosarcoma Study Group (COSS) J Clin Oncol. 2005;23:559–68. doi: 10.1200/JCO.2005.04.063. [DOI] [PubMed] [Google Scholar]
- 11.Hou CH, Lin FL, Tong KB. et al. Transforming growth factor alpha promotes osteosarcoma metastasis by ICAM-1 and PI3K/Akt signaling pathway. Biochem Pharmacol. 2014;89:453–63. doi: 10.1016/j.bcp.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 12.Zhang K, Gao J, Ni Y. Screening of candidate key genes associated with human osteosarcoma using bioinformatics analysis. Oncol Lett. 2017;14:2887–93. doi: 10.3892/ol.2017.6519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu X, Zhang C, Zhang Y. et al. Down regulation of human positive coactivator 4 suppress tumorigenesis and lung metastasis of osteosarcoma. Oncotarget. 2017;8:53210–25. doi: 10.18632/oncotarget.18290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thanapprapasr K, Nartthanarung A, Thanapprapasr D. et al. pFAK-Y397 overexpression as both a prognostic and a predictive biomarker for patients with metastatic osteosarcoma. PLoS One. 2017;12:e0182989. doi: 10.1371/journal.pone.0182989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jin H, Luo S, Wang Y. et al. miR-135b Stimulates Osteosarcoma Recurrence and Lung Metastasis via Notch and Wnt/beta-Catenin Signaling. Mol Ther Nucleic Acids. 2017;8:111–22. doi: 10.1016/j.omtn.2017.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen X, Bahrami A, Pappo A. et al. Recurrent somatic structural variations contribute to tumorigenesis in pediatric osteosarcoma. Cell Rep. 2014;7:104–12. doi: 10.1016/j.celrep.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuijjer ML, Hogendoorn PC, Cleton-Jansen AM. Genome-wide analyses on high-grade osteosarcoma: making sense of a genomically most unstable tumor. Int J Cancer. 2013;133:2512–21. doi: 10.1002/ijc.28124. [DOI] [PubMed] [Google Scholar]
- 18.Kobayashi E, Masuda M, Nakayama R. et al. Reduced argininosuccinate synthetase is a predictive biomarker for the development of pulmonary metastasis in patients with osteosarcoma. Mol Cancer Ther. 2010;9:535–44. doi: 10.1158/1535-7163.MCT-09-0774. [DOI] [PubMed] [Google Scholar]
- 19.Kelly AD, Haibe-Kains B, Janeway KA. et al. MicroRNA paraffin-based studies in osteosarcoma reveal reproducible independent prognostic profiles at 14q32. Genome Med. 2013;5:2. doi: 10.1186/gm406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buddingh EP, Kuijjer ML, Duim RA. et al. Tumor-infiltrating macrophages are associated with metastasis suppression in high-grade osteosarcoma: a rationale for treatment with macrophage activating agents. Clin Cancer Res. 2011;17:2110–9. doi: 10.1158/1078-0432.CCR-10-2047. [DOI] [PubMed] [Google Scholar]
- 21.Gentleman RC, Carey VJ, Bates DM. et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davis S, Meltzer PS. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics. 2007;23:1846–7. doi: 10.1093/bioinformatics/btm254. [DOI] [PubMed] [Google Scholar]
- 23.Li W, Li K, Zhao L. et al. Bioinformatics analysis reveals disturbance mechanism of MAPK signaling pathway and cell cycle in Glioblastoma multiforme. Gene. 2014;547:346–50. doi: 10.1016/j.gene.2014.06.042. [DOI] [PubMed] [Google Scholar]
- 24.Cheadle C, Vawter MP, Freed WJ. et al. Analysis of microarray data using Z score transformation. J Mol Diagn. 2003;5:73–81. doi: 10.1016/S1525-1578(10)60455-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen H, Boutros PC. VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinformatics. 2011;12:35. doi: 10.1186/1471-2105-12-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gu Z. ComplexHeatmap: Making Complex Heatmaps. R package version 1.10.2. 2016. https://github.com/jokergoo/ComplexHeatmap.
- 27.López-Ratón M, Rodríguez-Álvarez MX, Cadarso-Suárez C, OptimalCutpoints: An R Package for Selecting Optimal Cutpoints in Diagnostic Tests. Journal of Statistical Software; 2014. p. 061. [Google Scholar]
- 28.Kassambara A, Kosinski M. survminer: Drawing Survival Curves using 'ggplot2'. R package version 0.3.1. 2017. https://CRAN.R-project.org/package=survminer.
- 29.Hothorn T, Hornik K, Wiel MAVD. et al. A Lego System for Conditional Inference. American Statistician. 2006;60:257–63. [Google Scholar]
- 30.Kuhn M. caret: Classification and Regression Training. R package version 6.0-76. 2017. https://CRAN.R-project.org/package=caret.
- 31.Robin X, Turck N, Hainard A. et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Szklarczyk D, Franceschini A, Kuhn M. et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39:D561–8. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kolde R. GOsummaries: Word cloud summaries of GO enrichment analysis. R package version 2.6.0. 2016. https://github.com/raivokolde/GOsummaries.
- 34.Yang Y, Han L, Yuan Y. et al. Gene co-expression network analysis reveals common system-level properties of prognostic genes across cancer types. Nat Commun. 2014;5:3231. doi: 10.1038/ncomms4231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Komuro A, Saeki M, Kato S. Npw38, a novel nuclear protein possessing a WW domain capable of activating basal transcription. Nucleic Acids Res. 1999;27:1957–65. doi: 10.1093/nar/27.9.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Waragai M, Lammers CH, Takeuchi S. et al. PQBP-1, a novel polyglutamine tract-binding protein, inhibits transcription activation by Brn-2 and affects cell survival. Hum Mol Genet. 1999;8:977–87. doi: 10.1093/hmg/8.6.977. [DOI] [PubMed] [Google Scholar]
- 37.Okazawa H, Rich T, Chang A. et al. Interaction between mutant ataxin-1 and PQBP-1 affects transcription and cell death. Neuron. 2002;34:701–13. doi: 10.1016/s0896-6273(02)00697-9. [DOI] [PubMed] [Google Scholar]
- 38.Llorian M, Beullens M, Lesage B. et al. Nucleocytoplasmic shuttling of the splicing factor SIPP1. J Biol Chem. 2005;280:38862–9. doi: 10.1074/jbc.M509185200. [DOI] [PubMed] [Google Scholar]
- 39.Waragai M, Junn E, Kajikawa M. et al. PQBP-1/Npw38, a nuclear protein binding to the polyglutamine tract, interacts with U5-15kD/dim1p via the carboxyl-terminal domain. Biochem Biophys Res Commun. 2000;273:592–5. doi: 10.1006/bbrc.2000.2992. [DOI] [PubMed] [Google Scholar]
- 40.Zhang Y, Lindblom T, Chang A. et al. Evidence that dim1 associates with proteins involved in pre-mRNA splicing, and delineation of residues essential for dim1 interactions with hnRNP F and Npw38/PQBP-1. Gene. 2000;257:33–43. doi: 10.1016/s0378-1119(00)00372-3. [DOI] [PubMed] [Google Scholar]
- 41.Kalscheuer VM, Freude K, Musante L. et al. Mutations in the polyglutamine binding protein 1 gene cause X-linked mental retardation. Nat Genet. 2003;35:313–5. doi: 10.1038/ng1264. [DOI] [PubMed] [Google Scholar]
- 42.Lenski C, Abidi F, Meindl A. et al. Novel truncating mutations in the polyglutamine tract binding protein 1 gene (PQBP1) cause Renpenning syndrome and X-linked mental retardation in another family with microcephaly. Am J Hum Genet. 2004;74:777–80. doi: 10.1086/383205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Germanaud D, Rossi M, Bussy G. et al. The Renpenning syndrome spectrum: new clinical insights supported by 13 new PQBP1-mutated males. Clin Genet. 2011;79:225–35. doi: 10.1111/j.1399-0004.2010.01551.x. [DOI] [PubMed] [Google Scholar]
- 44.Kleefstra T, Franken CE, Arens YH. et al. Genotype-phenotype studies in three families with mutations in the polyglutamine-binding protein 1 gene (PQBP1) Clin Genet. 2004;66:318–26. doi: 10.1111/j.1399-0004.2004.00308.x. [DOI] [PubMed] [Google Scholar]
- 45.Lubs H, Abidi FE, Echeverri R. et al. Golabi-Ito-Hall syndrome results from a missense mutation in the WW domain of the PQBP1 gene. J Med Genet. 2006;43:e30. doi: 10.1136/jmg.2005.037556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beale EG, Harvey BJ, Forest C. PCK1 and PCK2 as candidate diabetes and obesity genes. Cell Biochem Biophys. 2007;48:89–95. doi: 10.1007/s12013-007-0025-6. [DOI] [PubMed] [Google Scholar]
- 47.Zhao C, Dong J, Jiang T. et al. Early second-trimester serum miRNA profiling predicts gestational diabetes mellitus. PLoS One. 2011;6:e23925. doi: 10.1371/journal.pone.0023925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vincent EE, Sergushichev A, Griss T. et al. Mitochondrial Phosphoenolpyruvate Carboxykinase Regulates Metabolic Adaptation and Enables Glucose-Independent Tumor Growth. Mol Cell. 2015;60:195–207. doi: 10.1016/j.molcel.2015.08.013. [DOI] [PubMed] [Google Scholar]
- 49.Balsa-Martinez E, Puigserver P. Cancer Cells Hijack Gluconeogenic Enzymes to Fuel Cell Growth. Mol Cell. 2015;60:509–11. doi: 10.1016/j.molcel.2015.11.005. [DOI] [PubMed] [Google Scholar]
- 50.Mendez-Lucas A, Hyrossova P, Novellasdemunt L. et al. Mitochondrial phosphoenolpyruvate carboxykinase (PEPCK-M) is a pro-survival, endoplasmic reticulum (ER) stress response gene involved in tumor cell adaptation to nutrient availability. J Biol Chem. 2014;289:22090–102. doi: 10.1074/jbc.M114.566927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leithner K, Hrzenjak A, Trotzmuller M. et al. PCK2 activation mediates an adaptive response to glucose depletion in lung cancer. Oncogene. 2015;34:1044–50. doi: 10.1038/onc.2014.47. [DOI] [PubMed] [Google Scholar]
- 52.Luo S, Li Y, Ma R. et al. Downregulation of PCK2 remodels tricarboxylic acid cycle in tumor-repopulating cells of melanoma. Oncogene. 2017;36:3609–17. doi: 10.1038/onc.2016.520. [DOI] [PubMed] [Google Scholar]
- 53.Taoka Y, Matsumoto K, Ohashi K. et al. Protein expression profile related to cisplatin resistance in bladder cancer cell lines detected by two-dimensional gel electrophoresis. Biomed Res. 2015;36:253–61. doi: 10.2220/biomedres.36.253. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures and tables.