Abstract
Triple-negative breast cancers (TNBCs) represent a heterogeneous disease characterized by several molecular subtypes with different prognoses and responses to therapy. For a correct clinical management of TNBC patients the knowledge of the gene regulation mechanisms related to tumor progression and drug response has become fundamental.
LncRNAs regulate gene expression through various processes, including chromatin modification, transcription and post-transcription and they are emerging as important cancer biomarkers being involved in tumor pathogenesis, metastatic progression and drug resistance.
In this study we aimed to analyze the expression of the lncRNA HOTAIR, mainly involved in breast cancer disease, in a large case series of TNBC patients. We used ISH methods by a RNA probe to better define its staining in tumor tissues and its relation with clinical-pathological parameters and outcomes of patients.
Our results show that high HOTAIR expression in tumor tissues is strongly correlated with lymph nodes metastasis (LNM) (p=0.039), as reported also for other tumor types, and has a direct strong association with Androgen Receptor (AR) expression (p= 0.019).
These data confirm the prognostic role of HOTAIR in TNBC, and, its involvement in the regulation of AR pathway, suggests the possibility to establish new therapeutic strategies for AR+TNBC patients.
Keywords: TNBC, HOTAIR, Androgen Receptor
Introduction
Triple-negative breast cancers (TNBCs) identify a sub-group of breast tumors characterized by the lack of expression of estrogen receptor (ER), progesterone receptor (PR) and the absence of HER2 gene amplification. TNBCs account for approximately 15%-25% approximately of all breast cancers, frequently associated with a younger age and a poor prognosis. For the lack of specific targeted therapies, cytotoxic chemotherapy remains the first choice of medical treatment for TNBC and the median survival is about 13 months 1.
Based on gene expression profiles TNBC appears as a very heterogeneous disease 2. Over the years, several molecular classifications were proposed, from Perou 3 and Lehmann 4, that distinguished seven sub-molecular classes up to that of Jézéquel et al, that suggested 3 distinct molecular subtypes 5. The current molecular stratification based on differences in diagnosis age, grade, local and distant disease progression and histopathology identifies four TNBCs subtypes 6: a luminal androgen receptor (LAR) subtype with the overexpression of Androgen Receptor (AR); a basal-like 1 (BL1) subtype with higher expression of cell cycle, DNA repair, and proliferation genes; a basal-like 2 (BL2) subtype with the upregulation of genes related to growth factor signaling pathway; and a mesenchymal (M) subtype enriched for epithelial-mesenchymal transition and immune-cells related genes 6.
Many studies carried out on large case series showed that TNBC subtypes have a very different prognosis 7 and respond in a completely different way to chemotherapies, both adjuvants and neoadjuvant 8,9. At the moment, this extreme biological and molecular heterogeneity does not allow a correct clinical management of TNBC patients. For this reason, the deepening of knowledge on the gene regulation mechanisms mainly related to tumor progression and drug response has become essential.
LncRNAs are an emerging class of regulatory RNAs abnormally expressed in many tumor diseases. Recently many studies have reported that drug resistance is strongly modulated by lncRNAs through changing genomic stability and promoting the translation of genes involved in cell survival, proliferation, and drug metabolism 10.
The aberrant expression of several lncRNAs has been also reported in TNBC 11. HOTAIR was the first lncRNA able to promote tumor progression and associated with poor prognosis in breast cancer 12. HOTAIR plays significant roles in tumor development and metastatic progression, particularly in lymph node metastatic disease 13. Moreover, aberrant HOTAIR expression was strongly related to drug resistance, both to chemotherapy and biological drug 14-20, in different solid tumors, including breast cancer 21.
Recent in vitro studies have showed that HOTAIR was mainly involved in the regulation of malignant biological behavior of TNBC cells 22. Since the expression of HOTAIR has never been analyzed on large TNBC patient series, in this study we performed its in situ detection on selected TNBC tumor samples, included in prognostic TMAs, to investigate its association with the clinical-pathological parameters and outcome of patients.
Materials and methods
Patients and specimens
From 2003 to 2013, 163 patients who underwent a mastectomy, quadrantectomy or metastectomy at the National Cancer Institute “Giovanni Pascale Foundation” of Naples, Italy, were enrolled into this study.
In our institution, the percentage of tumors classified as Triple Negative is approximately 15-19% of the total number of breast cancer surgical samples. All cases of Triple Negative and non-Triple Negative breast samples were reviewed according to WHO classification criteria, using standard tissue sections and appropriate immunohistochemical slides.
Medical records for all cases of Triple Negative breast samples were reviewed for clinical information, including histologic parameters that were determined from the H&E slides. The following clinical and pathological parameters were evaluated for each tumor included in the study: patient age at initial diagnosis, tumor size, histologic subtype, nuclear grade, nodal status, tumor recurrence or distant metastasis.
In addition, all specimens were characterized for all routinely diagnostic immunophenotypic parameters.
Immunohistochemical analysis and evaluation
Immunohistochemical staining (IHC) was done on slides from formalin-fixed, paraffin embedded tissues (FFPE) from TNBC Tissue Micro Arrays (TMAs), to evaluate the expression of ER, PgR, HER2, Ki67 and AR markers. All details of TMA building and IHC analysis was previously reported 23.
Antigen expression was evaluated independently by a pathologist using a light microscopy. Observer was unaware of the clinical outcome. For each sample, at least five fields (inside the tumor and in the area exhibiting tumor invasion) and >500 cells were analyzed. Using a semi-quantitative scoring system microscopically and referring to each antigen scoring method in other studies, an observer evaluated the intensity, extent and subcellular distribution.
For nuclear AR expression we used as the cutoff “low” AR expression if AR positive tumor cells were ≤ 5% and “high” AR expression if AR positive tumor cells were ≥5%.
RNA In Situ Hybridization Assay (RNA ISH)
In situ detection for HOTAIR was performed using the RNAscope (RNAscope® 2.5 HD Detection Reagent - BROWN User Manual) according to the manufacturer's instructions. Briefly, TMAs were cut in 5 mm thick sections. Details of procedure are previously reported 24. The tissue sections were boiled at 95°C for 30 minutes in Target Retrieval solution. Protease treatment was then applied at 40°C for 30 minutes. A provided positive control probe PPIB was used. For negative control, the enclosed negative control probe was applied. The slides were independently evaluated by two separate observers (MDB/GB). Positive staining was indicated by signals as brown dots present in the nucleus and/or cytoplasm. The number of signal staining was counted in 60 tumor cells. The study was performed on tissue microarrays (TMAs) which had 5 cores of 1mm for each case.
The expression of HOTAIR was analyzed in every core of the TMAs. HOTAIR has an expression level varying between 0 to > 10 copies per cell. We used a semi-quantitative scoring utilizing the estimated number of dots present within each cell boundary. We have categorized staining into 4 scores: 0, 1+, 2+, 3+. Staining Score 0: No staining or less than 1 dot to every 10 cells, Score 1: average 2-3 dots/cell, Score 2: average 4-6 dots/cell, Score 3: when more than 10% positive cells have dot clusters >6 dots/cell. For statistical analysis we divided the samples in 2 scoring groups, one representative of the low expression (Score 0-1) and the second of the high expression (Score 2-3).
Statistical analysis
The association between HOTAIR expression and clinical-pathological parameters was conducted using the χ2, Fisher's exact test and T Student test. The level of significance was defined as P<0.05. Overall survival (OS) and disease-free survival (DFS) curves were was plotted using the Kaplan-Meier method with significance valuated using the Mantel-Cox log-rank test. All the statistical analyses were carried out using the Statistical Package for Social Science v. 20 software (SPSS Inc., Chicago, IL, USA).
OS was defined as the time from diagnosis (first surgery) to death by any cause or until the most recent follow-up. DFS was measured as the time from diagnosis to the occurrence of progression, relapse after complete remission, or death from any cause. DFS had a value of zero for patients who did not achieve complete remission. The follow-up duration was five years at least.
Results
Clinical-pathological characteristics of TNBC patients
In our cohort, we have included 163 TNBC samples of breast cancers. The age of patients ranged from 24-91 years, with an average age of 57 years. The percentages of tumor grading were: 89% grade III (145/163), 9.8%grade II (16/163) and 1.2% grade I (2/163). Tumor sizes were: lower than 2 cm in 40% of the samples (62/155), between 2 and 5 cm in 50.3% (78/155) and larger than 5 cm in 9.7% (15/155). For 8 cases we were not able to retrieve that information. Metastatic lymph nodes (LNM) were found in 43.8% (70/160) of patients at surgery. This information was lost for 3 patients. Distant metastases were found in 40.5% (30/74). For 89 cases we were unable to recover this information.
The expression of proliferation factor Ki67 was high (>20%) in 88%(139/158), and low (≤20%) in 12% of cases (19/158). This information for 5 patients was lost. All clinical-pathological characteristics of TNBC patients are summarized in Table 1.
Table 1.
Relation between HOTAIR expression and clinic pathological features of TNBC patients.
| HOTAIR (n=163) | ||||
|---|---|---|---|---|
| Age | Low | High | P value | R Pearson |
| ≤60 >60 |
65 (69.9%) 51 (72.8%) |
28 (30.1%) 19 (27.2%) |
0,679 | -0,032 |
| Histotype | ||||
| Ductal No Ductal |
95 (68.3%) 21 (87.5%) |
44 (31.7%) 3 (22.5%) |
0,056 | - 0,150 |
| Tumor size | ||||
| ≤2 cm >2≤5 N/A |
42 (67.7%) 69 (68.8%) 5 (62.5%) |
20 (32.3%) 24 (31.2%) 3 (37.5%) |
0,383 | -0,070 |
| Limph Node Metastasis | ||||
| Negative Positive N/A |
70 (77.7%) 44 (62.8%) 2 (66.7%) |
20 (22.3%) 26 (37,2%) 1 (33.3%) |
0,039** | 0,164 |
| Grade | ||||
| G1/G2 G3 |
12 (66.7%) 104 (71.7%) |
6 (33.3%) 41 (28.3%) |
0,655 | -0,035 |
| Metastases | ||||
| Negative Positive N/A |
32 (71.1%) 22 (73.3%) 62 (70.4%) |
13 (28.9%) 8 (24.7%) 26 (29.6%) |
0,834 | -0,024 |
| Ki-67 | ||||
| ≤20% >20% N/A |
16 (84.2%) 97 (69.8%) 3 (60%) |
3 (15.8%) 42 (30.2%) 2 (40%) |
0,191 | 0,104 |
| Status | ||||
| Alive Dead N/A |
37 (67.3%) 24 (77.4%) 55 (71.4%) |
18 (32.7%) 7 (22.6%) 22 (28.6%) |
0,320 | -0,107 |
HOTAIR ISH expression in TNBCs samples
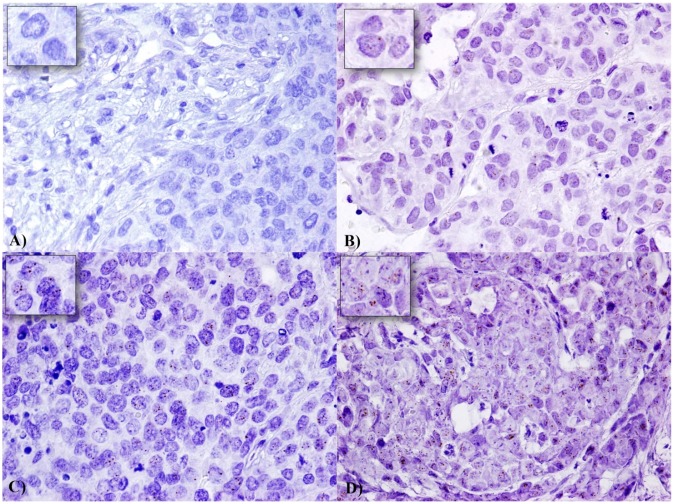
We analysed by in situ RNA hybridization HOTAIR expression on TNBC TMAs. We included only cases which presented at least 60 tumor cell nuclei. HOTAIR showed a tissue-specific distribution in our cases and there was no signals both in non tumor and stromal cells were detected (Figure 1). In detail, 47 TNBC cases showed an high and 116 cases a low expression. HOTAIR expression does not correlate with tumor size, grading, proliferation index, and distant metastases, while shows a trend of statistical association with ductal histotype (pvalue=0.056), and a direct strong statistical association with lymph node metastases (p=0.039) (Table 1).
Figure 1.
Expression of HOTAIR in a series of TNBC samples: Expression levels were depicted as A) No staining or less than 1 dot to every 10 cells (Score 0) (60X magnification), B) average 2-3 dots/cell (Score 1) (60X magnification), C) average 4-6 dots/cell (Score 2) (60X magnification), and D) dot clusters >6 dots/cell (Score 3) (visible at 20-40X magnification). A staining detail is shown in the upper left square.
Representative images for each scoring category of HOTAIR signals are showed in Figure 1.
HOTAIR relation with AR expression and survival of TNBCs patients
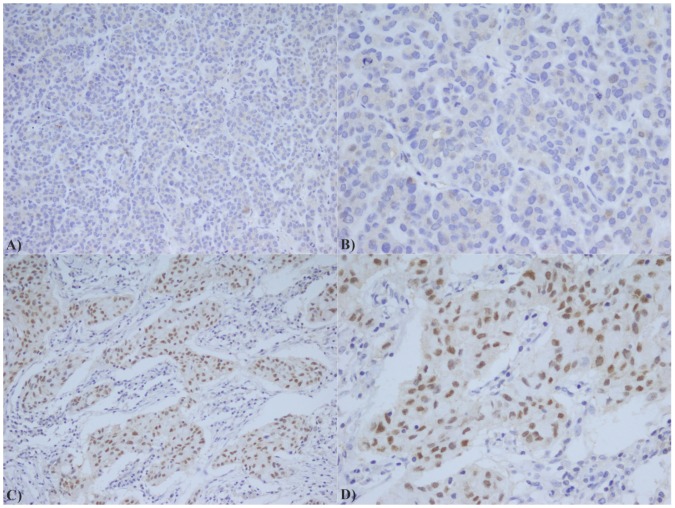
Based on statistical elaboration of HOTAIR with AR staining in TNBC patients (Figure 2), we showed a strong direct association between their combined expression (p value =0.019) as shown in Table 2.
Figure 2.
AR immunostaining in TNBC: A) Low expression of AR (20X); B) Low expression of AR (40X); C) High expression of AR (20X); D) High expression of AR (40X);
Table 2.
Relation between HOTAIR and AR expression.
| HOTAIR (n=163) | ||||
|---|---|---|---|---|
| AR | Low | High | P value | R Pearson |
| Low High N/A |
83 (78.3%) 17 (53.1%) 16 (80%) |
28 (21.7%) 15 (46.7%) 4 (20%) |
0.019 | 0.197 |
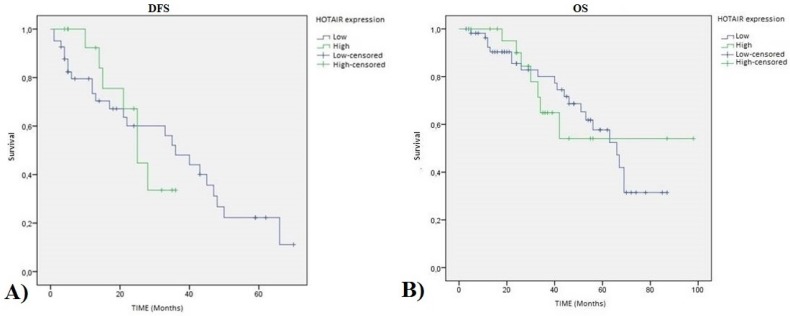
Regarding the relation with patient survival, we showed that the statistical association of HOTAIR expression with disease free survival (DFS) (p value=0.872) and overall survival (OS) (p value=0.990) are not significant as shown in Figure 3.
Figure 3.
HOTAIR high expression TNBC patients Kaplan-Meier curves: A) Disease Free Survival (DFS) (pvalue=0.872 ); and B) Overall Survival (OS) (pvalue=0.990).
Discussion
The recent molecular classifications of TNBC, and the identification of distinct TNBC subtypes with different prognoses and responses to standard chemotherapeutic schemes 1, have opened a new scenario in the therapy of these tumor subtypes, highlighting the possibility of using biological targets, such as AR inhibitors, in LAR subtype, or PARP, CDK and Growth factor inhibitors in basal subtypes 25.
However, the molecular heterogeneity of TNBCs on the one hand would allow to carry out a better prognostic and therapeutic definition, on the other it suggests also to investigate the complex mechanisms that underlie the regulation of gene expression.
In the present study we decided to investigate the role of lncRNA HOTAIR on a large series of TNBCs to better define its prognostic value in this subtype of BC. For the detection of HOTAIR we have optimized an in situ method, through the use of an RNA probe as previously reported 26. Our data have highlighted that 29% of TNBC cases showed an high and 71% cases a low or absent expression of HOTAIR.
Previous studies carried out on TNBC cell models had highlighted that HOTAIR expression was strongly increased by Estrogen in MDA‐MB‐231 and BT549 cells with a consequent increase in their migration capacity 13. Moreover, lapatinib and imatinib are able to repress HOTAIR expression in TNBC cells through inhibition of β‐catenin 22. The analysis of HOTAIR in our TNBC case series showed no statistically significant association with clinical pathological parameters, such as grade and stage but a strong association with lymph node metastases (LNM). This data is in line with the previous studies carried out on different solid tumors, in which HOTAIR primarily appeared as marker of lymph node metastasis 12, 27, 28. In support of this hypothesis, 2 meta-analysis studies have analyzed the large amount of data produced in recent years, showing that high HOTAIR expression in tumor tissue is strongly correlated with LNM 29, 30. In addition, in BC patients high circulating HOTAIR level is also associated with lymph node metastasis 31.
On the contrary, our data showed no significant statistical correlation with patient survival, both DFS and OS. This is in contrast with the literature data, where HOTAIR upregulation appears strongly related to poor survival rate in different tumor types 32. However, the significant biological heterogeneity of TNBC could explain this difference 33.
Our data also draw attention to a strong association between HOTAIR and AR expression in TNBC samples suggesting their potential interaction. A more recent study has described the direct interaction between HOTAIR and AR in prostatic cells 34. Zhang et al, showed that HOTAIR binds to AR protein to block its interaction with E3 ubiquitin ligase MDM2, preventing AR ubiquitination and protein degradation. Consequently, HOTAIR expression is able to induce androgen-independent AR pathway activation in the absence of androgen 34.
TNBC LAR subtype, in addition to having a different biology and molecular profile, also shows a distinct prognosis and a different therapy response compared to other molecular TNBC subtypes. In fact, although AR expression has a positive prognostic role in all different histotypes of breast cancer 35-37, LAR TNBC shows a delayed recurrence compared to the other groups and about the 75% of distant metastasis in LAR subtype occurred more than 3 years after diagnosis 9. In addition, LAR subtypes have the lowest pathological complete response (pCR) rate to neoadjuvant anthracycline and cyclophosphamide followed by taxane (ACT) therapy 9.
In recent years, the possibility of having a biological target has suggested the use of androgens inhibitors also for TNBC therapy. In particular a phase II trial of single-agent enzalutamide in advanced AR+ TNBC has been completed with satisfactory results 38. However, although AR inhibition alone is well tolerated compared to chemotherapy, for the potential development of therapeutic resistance, the best results have been obtained with AR inhibition combined with other agents 39. For this reason it is necessary not only to identify the subtype LAR in TNBC patients but also to understand the regulation mechanisms of AR pathway for a better therapeutic approach. The ability of HOTAIR to modulate AR expression should also be investigated in BC cell models to understand if, as in prostate cancer cells, this interaction can be responsible of anti-androgen drug response.
Numerous mechanisms have clarified the molecular basis of drug resistance, highlighting in particular the contribution of non-mutational regulation of gene expression. LncRNAs are the major modulators of non-mutational gene regulation, and it has been hypothesized that their dynamic changes in response to various drugs, could influence genes involved in cell cycle arrest, inhibition of apoptosis and repair of DNA damage 40. The role of HOTAIR in the modulation of drug resistance mechanisms has been extensively described for several solid tumors 13-19.
In BC HOTAIR aberrant expression increases cell proliferation, and contributes to tamoxifen resistance. In fact, Xue et al. found that HOTAIR accumulated in nuclei and its expression was increased in tamoxifen-resistant breast tumor cells compared to primary, hormone-naïve tumor cells 21.
However, although HOTAIR overexpression is strongly associated with therapeutic resistance, the specific molecular mechanisms through which it regulates drug sensitivity in cancer cells are not yet clear. Additional information on the response of TNBC patients to treatment should be integrated to our data, to evaluate the role of HOTAIR as therapeutic response biomarkers also in this tumor type. Other studies to better understand these mechanisms would offer promise for the development of more effective cancer therapies.
Finally, due to high stability of lncRNAs, as well as all ncRNAs, in biological fluids such as blood, saliva, urine 40-43, the detection of HOTAIR in the blood of TNBC patients could represent a useful prognostic tool especially for the monitoring of therapeutic response.
Acknowledgments
This study was supported by the Italian Ministry of Health and bio-materials for in vivo studies were provided by Institutional BioBank (BBI) of INT Fondazione Pascale.
Ethics approval and consent
This study was approved by the Ethics Committee of INT Fondazione Pascale.
Written informed consent was obtained from the patient for publication of this manuscript and any accompanying images. A copy of the written consent is available for review by the Editor-in-Chief of this journal.
References
- 1.Schmadeka R, Harmon BE, Singh M. Triple-negative breast cancer. Am J Pathol. 2014;141:462–477. doi: 10.1309/AJCPQN8GZ8SILKGN. [DOI] [PubMed] [Google Scholar]
- 2.Marotti JD, de Abreu FB, Wells WA, Tsongalis GJ. Triple-Negative Breast Cancer: Next-Generation Sequencing for Target Identification. Am J Pathol. 2017;187:2133–2138. doi: 10.1016/j.ajpath.2017.05.018. [DOI] [PubMed] [Google Scholar]
- 3.Perou CM. Molecular stratification of triple-negative breast cancers. Oncologist. 2010;5:39–48. doi: 10.1634/theoncologist.2010-S5-39. [DOI] [PubMed] [Google Scholar]
- 4.Lehmann BD, Pietenpol JA. Identification and use of biomarkers in treatment strategies for triple-negative breast cancer subtypes. J Pathol. 2014;232:142–50. doi: 10.1002/path.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jézéquel P, Loussouarn D, Guérin-Charbonnel C, Campion L, Vanier A, Gouraud W, Lasla H, Guette C, Valo I, Verrièle V, Campone M. Gene-expression molecular subtyping of triple-negative breast cancer tumours: importance of immune response. Breast Cancer Res. 2015;17:43. doi: 10.1186/s13058-015-0550-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lehmann BD, Jovanović B, Chen X, Estrada MV, Johnson KN, Shyr Y, Moses HL, Sanders ME, Pietenpol JA. Refinement of Triple-Negative Breast Cancer Molecular Subtypes: Implications for Neoadjuvant Chemotherapy Selection. PLoS One. 2016;11:e0157368. doi: 10.1371/journal.pone.0157368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burstein MD, Tsimelzon A, Poage GM, Covington KR, Contreras A, Fuqua SA, Savage MI, Osborne CK, Hilsenbeck SG, Chang JC, Mills GB, Lau CC, Brown PH. Comprehensive genomic analysis identifies novel subtypes and targets of triple-negative breast cancer. Clin Cancer Res. 2015;21:1688–98. doi: 10.1158/1078-0432.CCR-14-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharma P. Biology and Management of Patients With Triple-Negative Breast Cancer. Oncologist. 2016;21:1050–62. doi: 10.1634/theoncologist.2016-0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Masuda H, Baggerly KA, Wang Y, Zhang Y, Gonzalez-Angulo AM, Meric-Bernstam F, Valero V, Lehmann BD, Pietenpol JA, Hortobagyi GN, Symmans WF, Ueno NT. Differential response to neoadjuvant chemotherapy among 7 triple-negative breast cancer molecular subtypes. Clin Cancer Res. 2013;19:5533–5540. doi: 10.1158/1078-0432.CCR-13-0799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhan A, Soleimani M, Mandal SS. Long Noncoding RNA and Cancer: A New Paradigm. Cancer Res. 2017;77:3965–3981. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lv M, Xu P, Wu Y, Huang L, Li W, Lv S, Wu X, Zeng X, Shen R, Jia X, Yin Y, Gu Y, Yuan H, Xie H, Fu Z. LncRNAs as new biomarkers to differentiate triple negative breast cancer from non-triple negative breast cancer. Oncotarget. 2016;7:13047–59. doi: 10.18632/oncotarget.7509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–6. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tao S, He H, Chen Q. Estradiol induces HOTAIR levels via GPER-mediated miR-148a inhibition in breast cancer. J Transl Med. 2015;13:131. doi: 10.1186/s12967-015-0489-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Z, Sun M, Lu K, Liu J, Zhang M, Wu W, De W, Wang Z, Wang R. The long noncoding RNA HOTAIR contributes to cisplatin resistance of human lung adenocarcinoma cells via downregualtion of p21(WAF1/CIP1) expression. PLoS One. 2013;8:e77293. doi: 10.1371/journal.pone.0077293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li J, Yang S, Su N, Wang Y, Yu J, Qiu H, He X. Overexpression of long non-coding RNA HOTAIR leads to chemoresistance by activating the Wnt/β-catenin pathway in human ovarian cancer. Tumour Biol. 2016;37:2057–65. doi: 10.1007/s13277-015-3998-6. [DOI] [PubMed] [Google Scholar]
- 16.Fang S, Gao H, Tong Y, Yang J, Tang R, Niu Y, Li M, Guo L. Long noncoding RNA-HOTAIR affects chemoresistance by regulating HOXA1 methylation in small cell lung cancer cells. Lab Invest. 2016;96:60–8. doi: 10.1038/labinvest.2015.123. [DOI] [PubMed] [Google Scholar]
- 17.Yan J, Dang Y, Liu S, Zhang Y, Zhang G. LncRNA HOTAIR promotes cisplatin resistance in gastric cancer by targeting miR-126 to activate the PI3K/AKT/MRP1genes. Tumour Biol. 2016;37:16345–16355. doi: 10.1007/s13277-016-5448-5. [DOI] [PubMed] [Google Scholar]
- 18.Liu MY, Li XQ, Gao TH, Cui Y, Ma N, Zhou Y, Zhang GJ. Elevated HOTAIR expression associated with cisplatin resistance in non-small cell lung cancer patients. J Thorac Dis. 2016;8:3314–3322. doi: 10.21037/jtd.2016.11.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang L, Dong P, Wang W, Huang M, Tian B. Gemcitabine treatment causes resistance and malignancy of pancreatic cancer stem-like cells via induction of lncRNA HOTAIR. Exp Ther Med. 2017;14:4773–4780. doi: 10.3892/etm.2017.5151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang H, Qin R, Guan A, Yao Y, Huang Y, Jia H, Huang W, Gao J. HOTAIR enhanced paclitaxel and doxorubicin resistance in gastric cancer cells partly through inhibiting miR-217 expression. J Cell Biochem; 2018. Jun 1. [DOI] [PubMed] [Google Scholar]
- 21.Xue X, Yang YA, Zhang A, Fong KW, Kim J, Song B, Li S, Zhao JC, Yu J. LncRNA HOTAIR enhances ER signaling and confers tamoxifen resistance in breast cancer. Oncogene. 2016;35:2746–55. doi: 10.1038/onc.2015.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang YL, Overstreet AM, Chen MS, Wang J, Zhao HJ, Ho PC, Smith M, Wang SC. Combined inhibition of EGFR and c-ABL suppresses the growth of triple-negative breast cancer growth through inhibition of HOTAIR. Oncotarget. 2015;6:11150–61. doi: 10.18632/oncotarget.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Collina F, Cerrone M, Peluso V, Laurentiis MD, Caputo R, Cecio RD, Liguori G, Botti G, Cantile M, Di Bonito M. Downregulation of androgen receptor is strongly associated with diabetes in triple negative breast cancer patients. Am J Transl Res. 2016;8:3530–9. [PMC free article] [PubMed] [Google Scholar]
- 24.Grabinski TM(1), Kneynsberg A(1), Manfredsson FP(1), Kanaan NM(1). A method for combining RNAscope in situ hybridization with immunohistochemistry in thick free-floating brain sections and primary neuronal cultures. PLoS One. 2015 Mar 20;10(3):e0120120. doi: 10.1371/journal.pone.0120120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee A, Djamgoz MBA. Triple negative breast cancer: Emerging therapeutic modalities and novel combination therapies. Cancer Treat Rev. 2018;62:110–122. doi: 10.1016/j.ctrv.2017.11.003. [DOI] [PubMed] [Google Scholar]
- 26.Gökmen-Polar Y, Vladislav IT, Neelamraju Y, Janga SC, Badve S. Prognostic impact of HOTAIR expression is restricted to ER-negative breast cancers. Sci Rep; 2015. p. 5. 5:8765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu ZH, Wang XL, Tang HM, Jiang T, Chen J, Lu S, Qiu GQ, Peng ZH, Yan DW. Long non-coding RNA HOTAIR is a powerful predictor of metastasis and poor prognosis and is associated with epithelial-mesenchymal transition in colon cancer. Oncol Rep. 2014;32:395–402. doi: 10.3892/or.2014.3186. [DOI] [PubMed] [Google Scholar]
- 28.Qiu JJ, Lin YY, Ye LC, Ding JX, Feng WW, Jin HY, Zhang Y, Li Q, Hua KQ. Overexpression of long non-coding RNA HOTAIR predicts poor patient prognosis and promotes tumor metastasis in epithelial ovarian cancer. Gynecol Oncol. 2014;134:121–8. doi: 10.1016/j.ygyno.2014.03.556. [DOI] [PubMed] [Google Scholar]
- 29.Sun Z, Wu XY, Wu CL. The association between LncRNA HOTAIR and cancer lymph node metastasis and distant metastasis: a meta-analysis. Neoplasma. 2018;65:178–184. doi: 10.4149/neo_2018_170114N34. [DOI] [PubMed] [Google Scholar]
- 30.Cai B, Wu Z, Liao K, Zhang S. Long noncoding RNA HOTAIR can serve as a common molecular marker for lymph node metastasis: a meta-analysis. Tumour Biol. 2014;35:8445–50. doi: 10.1007/s13277-014-2311-4. [DOI] [PubMed] [Google Scholar]
- 31.Lv R, Zhang J, Zhang W, Huang Y, Wang N, Zhang Q, Qu S. Circulating HOTAIR expression predicts the clinical response to neoadjuvant chemotherapy in patients with breast cancer. Cancer Biomark; 2018. Mar 30. [DOI] [PubMed] [Google Scholar]
- 32.Serghiou S, Kyriakopoulou A, Ioannidis JP. Long noncoding RNAs as novel predictors of survival in human cancer: a systematic review and meta-analysis. Mol Cancer. 2016;15:50. doi: 10.1186/s12943-016-0535-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hubalek M, Czech T, Müller H. Biological Subtypes of Triple-Negative Breast Cancer. Breast Care (Basel) 2017;12:8–14. doi: 10.1159/000455820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang A, Zhao JC, Kim J, Fong KW, Yang YA, Chakravarti D, Mo YY, Yu J. LncRNA HOTAIR Enhances the Androgen-Receptor Mediated Transcriptional Program and Drives Castration-Resistant Prostate Cancer. Cell Rep. 2015;13:209–221. doi: 10.1016/j.celrep.2015.08.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Basile D, Cinausero M, Iacono D, Pelizzari G, Bonotto M, Vitale MG, Gerratana L, Puglisi F. Androgen receptor in estrogen receptor positive breast cancer: Beyond expression. Cancer Treat Rev. 2017;61:15–22. doi: 10.1016/j.ctrv.2017.09.006. [DOI] [PubMed] [Google Scholar]
- 36.Rahim B, O'Regan R. AR Signaling in Breast Cancer. Cancers (Basel) 2017;9:3. doi: 10.3390/cancers9030021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rampurwala M, Wisinski KB, O'Regan R. Role of the androgen receptor in triple-negative breast cancer. Clin Adv Hematol Oncol. 2016;14:186–93. [PMC free article] [PubMed] [Google Scholar]
- 38.Traina T, Miller K, Yardley D, O'Shaughness, J, Cortes J, Awada A. Results from a phase 2 study of enzalutamide (ENZA), an androgen receptor (AR) inhibitor, in advanced AR+ triple-negative breast cancer (TNBC). J Clin Oncol. 2015; 2015. 33: 1003. [Google Scholar]
- 39.Barton VN, Gordon MA, Richer JK, Elias A. Anti-androgen therapy in triple-negative breast cancer. Ther Adv Med Oncol. 2016;8:305–8. doi: 10.1177/1758834016646735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lipovich L, Johnson R, Lin CY. MacroRNA underdogs in a microRNA world: evolutionary, regulatory, and biomedical significance of mammalian long non-protein-coding RNA. Biochim Biophys Acta. 2010;1799:597–615. doi: 10.1016/j.bbagrm.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 41.Shi T, Gao G, Cao Y. Long Noncoding RNAs as Novel Biomarkers Have a Promising Future in Cancer Diagnostics. Dis Markers. 2016;2016:9085195. doi: 10.1155/2016/9085195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiao ZY, Tian Q, Li N, Wang HB, Li KZ. Plasma long non-coding RNAs (lncRNAs) serve as potential biomarkers for predicting breast cancer. Eur Rev Med Pharmacol Sci. 2018;22:1994–1999. doi: 10.26355/eurrev_201804_14727. [DOI] [PubMed] [Google Scholar]
- 43.Avazpour N, Hajjari M, Tahmasebi Birgani M. HOTAIR: A Promising LongNon-coding RNA with Potential Role in Breast Invasive Carcinoma. Front Genet. 2017;8:170. doi: 10.3389/fgene.2017.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]