Abstract
APEX is an engineered peroxidase that catalyzes the oxidation of a wide range of substrates, facilitating its use in a variety of applications, from subcellular staining for electron microscopy to proximity biotinylation for spatial proteomics and transcriptomics. To further advance the capabilities of APEX, we used directed evolution to engineer a split APEX tool (sAPEX). Twenty rounds of FACS-based selections from yeast-displayed fragment libraries, using three different surface display configurations, produced a 200-amino acid N-terminal fragment (with 9 mutations relative to APEX2) called “AP,” and a 50-amino acid C-terminal fragment called “EX”. AP and EX fragments were each inactive on their own but reconstituted to give peroxidase activity when driven together by a molecular interaction. We demonstrate sAPEX reconstitution in the mammalian cytosol, on engineered RNA motifs within a noncoding RNA scaffold, and at mitochondria-endoplasmic reticulum contact sites.
Introduction
APEX2 is an engineered variant of soybean ascorbate peroxidase that, unlike the commonly used horseradish peroxidase (HRP), retains activity when expressed in the cytosol, mitochondria, and other reducing environments within the cell1,2. This feature of APEX2, in addition to its versatile ability to catalyze the H2O2-dependent one-electron oxidation of a wide array of small molecule substrates, has led to its widespread use for a variety of applications, including proteomic mapping of organelles2–6, proximity tagging of protein interactomes7–9, spatial mapping of cellular RNA10, electron microscopy1,11–16, H2O2 sensing17, and protein topology determination1,2,16.
In general, the use of APEX2 begins with fusing it to a protein or peptide in order to target it to a subcellular region or macromolecular complex of interest. For instance, we have targeted APEX2 to the outer mitochondrial membrane (OMM) and the endoplasmic reticulum membrane (ERM) of mammalian cells by fusing the APEX2 gene to transmembrane domains of proteins native to these subcellular locations4,16. These constructs were used for both EM16 and proteomic analysis4 of the OMM and ERM. While this APEX2 fusion strategy has enabled the study of many cellular regions and organelles, there are numerous compartments and structures to which APEX cannot be selectively targeted. For example, there is great interest in the biology of organelle-organelle contact sites, such as the junctions between mitochondria and ER, which participate in calcium signaling18,19, lipid synthesis20–23, and mitochondrial fission24,25. Yet all candidate protein fusions that could potentially target APEX2 to these contact sites, such as to the proteins Drp124, Mfn226–28, SYNJ2BP14, and PDZD829, would also target the peroxidase to locations outside of mito-ER contacts, such as throughout the cytosol30, along the cytoskeleton31, or over the entire OMM4.
Another application for which the conventional APEX2 genetic fusion strategy may be unsuitable is profiling the interactomes of specific cellular RNAs. While several robust methods can identify RNAs that interact with specific proteins of interest32–34, the converse problem—identifying proteins that interact with a particular RNA—is much more challenging using existing methods. One could envision fusing APEX2 to a high-affinity RNA-binding protein (RBP; for example, the bacteriophage MS2 coat protein35), allowing the peroxidase to be ectopically targeted to transcripts that are tagged with that RBP’s cognate RNA motif. However, a major concern would be the excess pool of catalytically active APEX2-RBP fusion protein that is not docked to the tagged RNA and can therefore produce off-target labeling that masks the specific signal.
A general solution to both of these, and related, problems could be a split form of APEX2, in which two inactive fragments of APEX2 reconstitute to give an active peroxidase only when they are physically co-localized (Figure 1A). One could apply this intersectional approach to restrict APEX2 activity specifically to sites of interest –such as mito-ER contacts, or specific RNA binding sites – thus eliminating the background labeling from off-target peroxidase activity.
Figure 1. Split APEX design and screening of potential sAPEX cut sites.

(A) Schematic overview of split APEX (sAPEX). Two inactive fragments (grey) can reconstitute to give active peroxidase (green) when driven together by a protein-protein interaction (PPI). The yellow diamond represents a ligand that can induce dimerization (B) The first screen tested 24 different cut sites. Their locations in the APEX2 protein sequence are indicated by the red vertical lines. Squiggles denote alpha helices. Grey arrows denote beta sheets. Areas shaded green are part of the heme-binding pocket. See Figure S1 for a similarly annotated 3D model. (C) N- and C-terminal sAPEX fragments selected for testing were fused to FRB and FKBP, respectively. (D) Initial screen of cut sites; split occurs after the indicated amino acid. For instance, cut site 7 splits APEX2 between residues 7 and 8. Pairs of constructs were introduced into HEK 293T cells by transient transfection, along with a CFP-NLS (nuclear localization signal) co-transfection marker. Cells were either treated with rapamycin for 24 h (left) or remained untreated (right). Subsequently Amplex UltraRed, a fluorogenic small-molecule peroxidase substrate, and H2O2 were added for 25 minutes, after which cells were fixed and imaged. Resorufin is the fluorescent product of Amplex UltraRed oxidation and indicates peroxidase activity. Scale bars, 20 μm. Three biological replicates were performed. (E) Second cut site screen, focused on residues surrounding G50, and E200. Same assay as in (D). Two biological replicates were performed; representative images shown.
Although split protein reporters have been developed from green fluorescent protein36,37, HRP38, dihydrofolate reductase39, ubiquitin40, luciferase41–44, beta-galactosidase45–48, TEV protease49, Cas950–53, and BioID54, splitting APEX2 presents new challenges. First, APEX2 requires a heme cofactor for its activity, and many cut sites would divide the heme-binding pocket across the two fragments (Figure 1B and Figure S1). Second, in order for split APEX2 to be useful for a broad range of applications, the inactive fragments should have relatively low affinity for one another, such that reconstitution only occurs when the fragments are driven together by a molecular interaction. Not many known split proteins have low-affinity fragments44, and it is challenging to engineer such a property in conjunction with high activity upon reconstitution.
To address the need for an interaction-dependent proximity labeling tool, we describe here the development of an evolved split APEX2 (sAPEX) system. Using a novel yeast display-based directed evolution approach that incorporates positive and negative selection steps, we have attempted to minimize interaction-independent association of the peptide fragments while maintaining high peroxidase activity upon reconstitution. Our resulting sAPEX fragment pair diverges substantially from its parental sequence and shows interaction-dependent reconstitution in multiple contexts in living mammalian cells. Our sAPEX tool adds to the proximity labeling toolkit, and in the future, should extend the utility of APEX-based approaches to new areas of biology at higher spatiotemporal resolution.
Results
Structure-guided screening of potential APEX2 cut sites
We first sought to identify promising cut sites in the APEX2 enzyme using a chemically-inducible protein association system as a test platform. We selected 24 potential cut sites within solvent-exposed loops and turns between secondary structural elements (alpha helices and beta sheets) based on the crystal structure of wild-type ascorbate peroxidase55 (Figure 1B and Figure S1). Cut sites were also selected to avoid the creation of hydrophobic termini that might destabilize the fragments. All fragments were expected to lack a complete heme-binding pocket, with the exception of the large fragments produced from cut sites 7/8 and 29/30. We cloned each of the 24 fragment pairs as fusions to FKBP and FRB, whose interaction can be induced with the small molecule rapamycin (Figure 1C). We introduced the fragment pairs into HEK 293T mammalian cells using transient transfection and evaluated peroxidase activity in the presence or absence of rapamycin. Catalytic activity was detected using an established assay based on the membrane-permeable fluorogenic probe Amplex UltraRed, which is colorless but is converted into the red fluorophore resorufin upon peroxidase-catalyzed oxidization56,57.
Of the 24 fragment pairs tested, seven produced significant resorufin product, indicative of reconstituted activity (Figure 1D). For all cut sites except 7/8, much stronger activity was detected in the presence of rapamycin compared to when rapamycin was omitted, indicating that reconstitution was dependent on a protein-protein interaction. In the case of cut site 7/8, resorufin fluorescence was observed not only when rapamycin was omitted, but also when the large C-terminal fragment (amino acids 8–250) was expressed on its own (Figure S2), making this cut site unsuitable for our purposes. Cut sites within the heme binding cavity (130/131, 135/136, 146/147, 170/171, 173/174, and 181/182) produced fragments that completely failed to reconstitute activity (see Figure 1B and Figure S1 for recolored full length APEX2 (PDB:1OAG)).
To more finely map the optimal cut sites, we performed a second round of screens, evaluating cut sites 1–3 residues away from promising sites identified in our initial screen (Figures 1E and S2). Some cut sites, such as 123/124, were not pursued because, while they may have shown comparable activity to promising sites such as 89/90, they demonstrated poorer expression which led to less consistent biotinylation signal. For cut site 123/124, it is possible this lack of stability may be due to a C-terminal proline formed with the split. Similarly, while cut site 198/199 showed similar activity to cut site 200/201, cut site 200/201 is more distal from secondary structures, i.e. in the middle of a flexible loop region, which we hypothesized could limit structural perturbations. We ultimately identified three optimal cut sites—51/52, 89/90, and 200/201—all of which fell in solvent-exposed loops distal (>15 angstroms away) from the APEX2 active site and heme-binding pocket. For each cut site pair, controls in which either the N- or C-terminal fragments were expressed alone lacked any detectable peroxidase activity (Figure S2).
To assess the efficacy of our split APEX2 fragment pairs for applications in EM and proximity biotinylation, we evaluated their activity using the small molecule substrates diaminobenzidine (DAB)1,16 and biotin phenol (BP)2–4, respectively. Despite positive results in the highly sensitive Amplex UltraRed assay (Figure 1D–E), all three split APEX2 pairs produced far weaker DAB and BP staining than full-length APEX2 (Figure S3 and data not shown). For example, HEK 293T cells expressing full-length APEX2 exhibited very strong DAB staining after 15 minutes, while DAB staining by cut site 200/201 constructs was hardly detectable under identical conditions (Figure S3A). We found that the recently reported58 neighboring cut site, 20½02, exhibited activity similar to that of 200/201 (Figure S3C).
A yeast display-based platform for split APEX2 evolution
The low activity of split APEX2 could be caused by a variety of factors, including poor stability, misfolding, aggregation of the fragments, or distorted geometry of the reconstituted active site and heme binding pocket, which could lead to low catalytic efficiency and/or poor heme recruitment. It is difficult to assess each of these potential problems, and even more difficult to fix them in a rational manner. Hence, we turned to directed evolution, which we and many others have harnessed to improve or alter the properties of enzymes16,38,59–63. We selected yeast display-based directed evolution (Figure 2) because, in contrast to other strategies such as high-throughput screening and phage display, yeast display allows processing of large mutant libraries (>107) with large dynamic range: enrichment based on Fluorescence-Activated Cell Sorting (FACS) enables separation of highly active catalysts from moderately active ones, not only from inactive catalysts.
Figure 2. Yeast-display directed evolution to improve split APEX2.
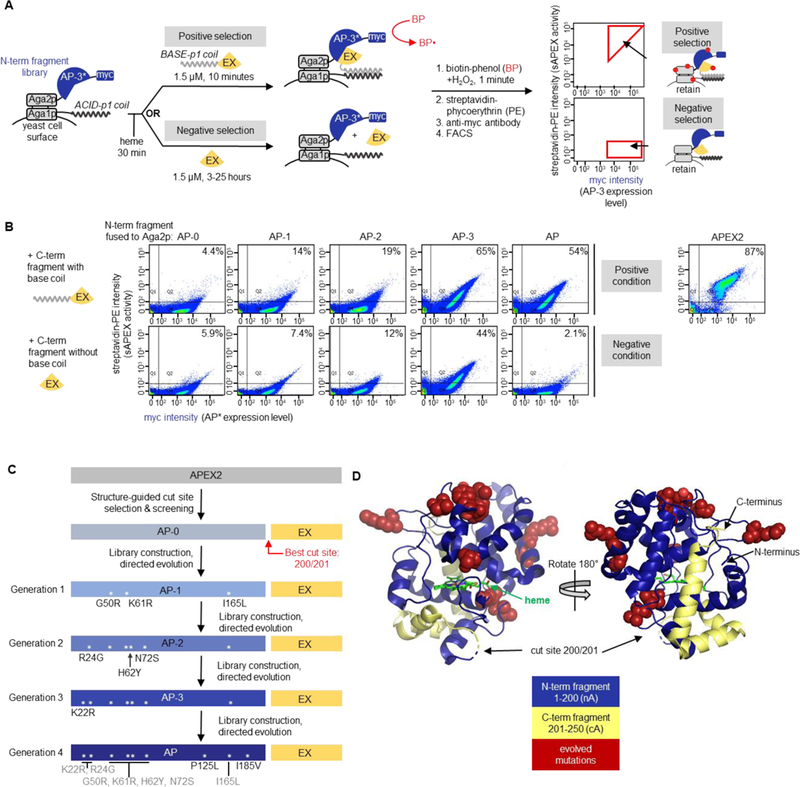
(A) Yeast display-based directed evolution scheme. The scheme shown here was used for Generation 4 selections, but the setup for other generations was similar (detailed in Figures S4–S7). A library of N-terminal fragments based on “AP-3” (the winning clone from Generation 3 selections) was displayed on the yeast cell surface via fusion to the Aga2p mating protein. An acid coil was co-displayed, via fusion to Aga1p, to recruit base coil-fused C-terminal fragment (“EX”, amino acids 201–250 of APEX2). In the positive selection for high sAPEX activity, base coil-EX-GFP was incubated with the AP-3 yeast library for 10 minutes, then reconstituted peroxidase activity was detected by treating the cell mixture with biotin-phenol (BP) and H2O2. Cells with high peroxidase activity label themselves with biotin to a high extent16, enabling their enrichment via FACS after streptavidin-phycoerythryin (PE) staining. In the negative selection to deplete the sAPEX library of AP-3 fragments with excessively high affinity for EX, we incubated the AP-3 yeast library with EX-GFP protein lacking base coil for increasingly long time periods (see Figure S7A), then performed BP labeling. Cells with low streptavidin-PE signal were retained via FACS. Myc staining on the x-axis provides a readout of AP expression level. Note that the fluorescence from GFP was not measured during FACS; GFP was included to increase the solubility of EX. Hypothetical data are shown to illustrate the strategy for gate selection. (B) Summary of improvement of sAPEX activity and PPI-dependence throughout generations of selections in yeast. Full length APEX2 fusion to Aga2p was used as a benchmark for desired activity range. Yeast were prepared as in (A), with the indicated N-terminal fragment of sAPEX expressed on the yeast surface as a fusion to Aga2p. Purified C-terminal fragment (EX), either with or without acid coil, was added to the cells for 10 minutes or 7 hours, respectively. Then BP labeling, streptavidin-PE staining, and FACS were performed as in (A). AP-1, AP-2, and AP-3 are the first, second, and third generation N-terminal fragment clones, whose mutations are shown in (C). The percentage of myc-positive cells in the top right quadrant (Q2) is indicated in the top right corner of each FACS plot. Data are shown for one out of two biological replicates. (C) Summary of split APEX protein engineering. The name of the best N-terminal fragment clone to emerge from Generation 1 selection is “AP-1”, and so on, as indicated. The best clone to emerge from the Generation 4 selection is “AP.” Asterisks depict the locations of mutations within the protein sequence. (D) sAPEX split site and mutations in sAPEX relative to full-length APEX22. The split site is between E200 and G201, and the N-terminal fragment (“AP”) and C-terminal fragment (“EX”) of sAPEX are colored blue and yellow, respectively. The nine residues mutated through directed evolution are colored red and rendered in space-filling mode; the original residues from the parent protein are depicted. Structures based on PBD ID: 1OAG55.
Our initial yeast display selection scheme exploited the yeast mating proteins Aga1p and Aga2p, which are displayed on the yeast cell surface and are joined by two disulfide bridges (Figures 2A and S4A). A library of yeast cells was generated, with each cell displaying on its surface a different mutant of the N-terminal split APEX2 fragment via fusion to the mating protein Aga2p. All cells concomitantly displayed the same C-terminal split APEX2 fragment as a fusion to Aga1p. The fragments were allowed to reconstitute for ~20 hours post-induction of protein expression. To read out the resulting peroxidase activity, APEX2-mediated biotinylation was initiated with biotin-phenol and H2O2, using standard conditions16. Yeast cells that display active reconstituted peroxidase under these conditions should promiscuously biotinylate proteins on their cell surface, which can be quantified using FACS (Figures 2A and S4A).
To establish this selection platform, we first created yeast Aga1p/Aga2p fusion constructs using the three promising split APEX2 pairs identified in the above screen. Because the 51/52 and 89/90 fragment pairs expressed poorly in yeast and gave no detectable activity on the yeast surface (data not shown), we proceeded with directed evolution of the 200/201 fragment pair. The C-terminal fragment (amino acids 201– 250 of APEX2; henceforth called “EX”) was held constant while the N-terminal fragment (amino acids 1–200 of APEX2, called “AP-0”) was mutagenized using error prone PCR. Sequencing showed that our AP-0 library contained an average of 1.4 amino acid mutations per clone (Methods).
We performed four rounds of selection and observed that the activity of the yeast pool (measured by FACS) progressively increased (Figure S4B). We isolated 12 individual clones, characterized their activity on yeast using FACS and in the mammalian cytosol using microscopy, and combined mutations that appeared to be beneficial. The result was “AP-1”, which contains 3 mutations relative to the original APEX2 (Figure 2C). In a side-by-side comparison to the original split APEX2 fragment pair (AP-0 + EX) in HEK 293T cells, AP-1 shows noticeably improved activity in both DAB and BP labeling assays (Figure 3A–C).
Figure 3. Comparing generations of evolved split APEX clones in mammalian cells.

(A) Depiction of protein sequences for FKBP and FRB fusions to sAPEX for analysis in mammalian cells. N-terminal Halotag was fused to the N-terminus of FRB-EX to increase protein solubility. (B) Comparison of sAPEX variants in the mammalian cytosol, assayed by DAB (diaminobenzidine) polymerization activity. In the bright field images, dark regions indicate peroxidase activity. The indicated N-terminal variants of sAPEX were introduced by transient transfection into HEK 293T cells stably expressing FRB-EX, which were incubated with rapamycin for 30 minutes (left) or left untreated (right). The first row shows HEK 293T cells stably expressing full-length APEX2-NES for comparison. Cells were fixed and incubated with DAB and H2O2 for 15 minutes, as previously described16, to allow peroxidase-catalyzed polymerization of DAB. Four separate fields of view are shown per condition. Scale bar, 20 μm. Two biological replicates were performed. (C) Same assay as in (B) except FKBP-AP was introduced by lentiviral infection, and live BP labeling was used to detect peroxidase activity. Infected HEK 293T cells were treated with BP in the presence of H2O2 for 1 minute, then fixed and stained with neutravidin-AlexaFluor647 to visualize peroxidase-catalyzed promiscuous biotinylation2. Scale bar 20 μm. Four biological replicates performed. Additional fields of view shown in Figure S8. Again, the comparison to full-length APEX is with stable HEK 293T cells. (D) Same assay as in (C) but with streptavidin blot readout. Anti-V5 and anti-HA blots detect expression of N-terminal and C-terminal fragments, respectively. Two biological replicates performed.
To further improve the reconstituted activity of split APEX2, we performed another set of yeast selections in which the C-terminal EX fragment was not co-displayed on Aga1p but instead supplied as a purified, soluble protein (Figure S5A). This configuration allowed us to precisely control the concentration of EX added and the time of incubation, and to select for mutations that improved AP-1 expression and stability in the absence of EX co-translation. However, because the fragments only encountered each other on the yeast surface, where endogenous heme is not available, it was necessary to supply exogenous heme to the cells both during and prior to EX fragment addition. Four rounds of selection using this scheme produced the clone “AP-2”, which has three mutations beyond those in AP-1 (Figure 2C).
In a third-generation effort, to drive the fragments together using a protein-protein interaction (PPI) we again added EX as a soluble protein, but we used an artificially designed coiled-coil system, ACID-p1 and BASE-p164, to recruit EX to the proximity of the N-terminal fragment (AP-2) (Figure S6). This configuration mimics the split APEX2 reconstitution that would occur in our target biological applications. Four rounds of selection with gradual reduction of EX concentration and incubation time produced clone “AP-3”, which incorporates one additional mutation relative to “AP-2” and has noticeably higher activity than preceding clones in both DAB and BP labeling assays in HEK 293T cells (Figure 2C and Figure 3).
FACS-based negative selections for reduced intrinsic fragment affinity
An ideal split APEX2 fragment pair would have high catalytic activity when reconstituted, but low intrinsic binding affinity between the fragments, such that reconstitution occurs only when the fragments are driven together by a PPI (Figure 1A). The clone AP-3, obtained after three generations of directed evolution, has much greater reconstituted activity with EX than does the original template (AP-0), but this activity was not dependent on PPI–induced co-proximation. Using FRB and FKBP fusion proteins in HEK 293T cells, for example, we observed considerable DAB and BP signal even in the absence of rapamycin (Figure 3).
Hence we devised a new yeast display selection scheme that alternates between positive and negative selection rounds (Figure 2A). For the positive selection, we supplied the purified EX protein fused to the BASE-p1 helix that facilitates recruitment to Aga1p-ACID-p1. We performed BP labeling followed by streptavidin-phycoerythrin and anti-myc antibody staining. FACS was used to enrich cells with high SA/myc intensity ratios, as above. For the negative selection, we incubated the yeast with EX protein lacking BASE-p1 coil for extended periods of time (3 to 25 hours). AP-3 mutants that reconstituted with EX under these PPI-independent conditions were eliminated by FACS.
Starting with a library of 4.8 × 108 AP-3 variants, we performed two rounds of positive selection and six rounds of negative selection (Figure S7A). By round 8, the yeast population was substantially depleted of cells that could reconstitute APEX2 activity upon addition of EX lacking the BASE-p1 coil (Figure S7B). We isolated 4 unique clones, characterized them by FACS, combined beneficial mutations, and re-tested in the mammalian cytosol. These experiments resulted in AP, the final split APEX2 (sAPEX) N-terminal fragment, which contains 9 mutations compared to the original APEX2 sequence (Figure 2C). Mapping the positions of these 9 mutations (G50R, K61R, I165L from Generation 1; R24G, H62Y, N72S from Generation 2; K22R from Generation 3; and P125L, I185V from Generation 4) onto the structure of wild-type ascorbate peroxidase, we observe that all lie in solvent-exposed regions, and none are at the interface between AP and EX (Figure 2D). Interestingly, half of the mutations are adjacent to cut sites we screened in Figure 1D–E, and many of the mutations were clustered in specific regions of the protein structure (Figure 2D).
Characterization of enriched clones
Having completed 20 rounds of selection using three different yeast display configurations, we characterized key clones side-by-side. First, we prepared yeast displaying AP-0 (the starting template), AP (N-terminal fragment of final sAPEX), full-length APEX2, and AP-1 to AP-3 mutants, as fusions to Aga2p. Aga1p on these cells contained the ACID-p1 coil. We then supplied EX protein, either fused to (Figure 2A, top row) or lacking (bottom row) the BASE-p1 coil for proximity-dependent reconstitution. Figure 2B shows that the streptavidin-phycoerythrin (PE) staining (indicating reconstituted peroxidase activity) progressively increases from the template AP-0 to the finalized AP. However, the signal in the bottom row, reflecting proximity-independent reconstitution with EX (lacking the BASE-p1 coil), also increases, which is undesirable. After the implementation of negative selections in Generation 4, however, the untagged EX signal decreases for the AP clone. The EX+BASE-p1 coil signal remains high for AP, although not quite as high as that seen for AP-3. These observations on yeast indicate that the selections worked as desired and that our final clone AP combines the features of high reconstitution activity with low proximity-independent reconstitution.
We next tested whether these trends would hold in the mammalian cell cytosol. This environment is different from the yeast cell surface: it is 37 °C instead of 30 °C, and reducing rather than oxidizing. We also wished to test the sAPEX clones as soluble proteins rather than membrane-anchored constructs with restricted geometry. Hence, we expressed the N- and C-terminal fragments from each stage of directed evolution as fusions to FKBP and FRB, respectively (Figure 3A). We first compared peroxidase activity with or without rapamycin using our DAB assay (relevant for EM applications1,16), which is much less sensitive than both Amplex UltraRed and BP assays, enabling us to more rigorously compare the activity of our sAPEX fragment pairs (Figure 3B). The original sAPEX template, AP-0, gave barely detectable DAB staining, while AP-1 was dramatically improved, and AP-3 gave the strongest staining. However, as observed in yeast, AP-3 also gave significant signal in the absence of rapamycin, indicating high PPI-independent reconstitution. In contrast, the final AP + EX clones displayed high DAB staining in the presence of rapamycin, but nearly undetectable staining in the majority of cells in the absence of rapamycin.
Analogous experiments using the BP assay—relevant for spatial proteomics2,3 and transcriptomics65—showed a similar trend (Figure 3C and Figure S8). While AP-3 showed high activity following rapamycin treatment, it also revealed background labeling in the –rapamycin condition. In contrast, the final sAPEX pair, AP + EX, displayed rapamycin-stimulated activity comparable to that of AP-3, but reduced activity in the majority of cells in the absence of rapamycin. We also characterized BP labeling in HEK 293T cells (Figure 3D) and COS7 cells (Figure S9B) by lysing the cells and blotting the cell lysate with streptavidin-HRP. The same trends were apparent, and we were furthermore able to estimate that reconstituted sAPEX has ~33% of the activity of full-length APEX2 (Figure S9C).
sAPEX reconstitution on a target RNA
As APEX-catalyzed proximity biotinylation has demonstrated utility for mapping protein interactomes7–9, there has been interest in using APEX2 to also map the interactomes of target nucleic acids – RNAs and individual genomic loci. In theory, this could be accomplished by fusing full-length APEX2 to programmable RNA- or DNA-binding proteins (such as CRISPR-Cas13d or Cas9, respectively)66–68 that specifically target the RNA or genomic locus under investigation. Alternatively, target RNAs or DNA loci could themselves be tagged with motifs that specifically bind APEX2 fusion proteins69. However, each of these approaches would be plagued by pools of excess catalytically active APEX2 that is unbound to the target of interest. This is exemplified by recent studies that used dCas9 to recruit APEX2 to specific genomic loci67,68, and a separate study that used the bacteriophage MS2 RNA coat protein (MCP) to recruit promiscuous biotin ligase variants to RNAs appended with the cognate MS2 RNA motif69. An analogous APEX2-MCP fusion system would most likely also suffer from background biotinylation that overwhelms the specific signal from RNA-bound APEX-MCP.
We reasoned that our sAPEX system could potentially be used to alleviate this problem by fusing the evolved AP and EX fragments to orthogonal RNA-binding proteins, and selectively reconstituting APEX2 peroxidase activity on target RNAs that are appended with both of the cognate protein-binding RNA motifs (Figure 4A–B). As proof-of-principle, we applied this strategy to TERC, the noncoding RNA component of the telomerase ribonucleoprotein (RNP), which synthesizes the ends of chromosomes in many clades of eukaryotes70. In addition to providing the template for telomere synthesis, the TERC ncRNA is also thought to serve as a structural “scaffold” onto which the other holoenzyme components assemble71. Critically, proper biogenesis of functional telomerase RNPs can be recapitulated when TERC RNA is transiently expressed from a plasmid, even if the RNA is appended with exogenous sequences at its 5´ end72. We therefore designed a series of variants in which the TERC 5´ terminus is appended by cassettes of motifs recognized by RNA-binding proteins. For this purpose, we chose to express AP and EX as fusions to the MS2 and PP7 bacteriophage nucleocapsid proteins—respectively termed MCP and PCP—which recognize disparate RNA motifs with exquisitely high affinity and specificity (cognate RNA KD ~1nM, non-cognate RNA KD ~1mM)35,73,74,75 (Figure 4A–C). Anticipating that sAPEX reconstitution might be sensitive to the specific geometry with which the AP and EX fragments are co-proximated, we designed a series of TERC variants that positioned MS2 and PP7 stem loops in different orientations (Figure S10).
Figure 4. Split APEX2 reconstitution on RNA binding sites and at mitochondria-ER contacts.

(A) Constructs used to test sAPEX targeting to RNA. MCP and PCP are the bacteriophage coat proteins that bind the MS2 and PP7 RNA stem-loops, respectively. Protein fusions are targeted to the nucleus by an N-terminal nuclear localization signal (NLS). (B) Schematic overview of sAPEX applied to interaction mapping of specific cellular RNAs. Cells expressing sAPEX (with N and C-terminal fragments fused to MCP and PCP, respectively) would localize peroxidase activity and labeling only to hTERC (human telomerase RNA component) RNA sites with adjoining MS2 and PP7 stem-loops. (C) Initial testing of sAPEX targeting to RNAs. HEK 293T cells stably expressing the MCP-AP construct shown in (A) were transfected with PCP-EX expression plasmid and the indicated RNA constructs. Twenty-two hours later, cells were subjected to in situ proximity biotinylation with BP and H2O2, fixed, and then stained with neutravidin-AlexaFluor647 to visualize reconstituted peroxidase activity, anti-HA antibody to visualize EX expression, and anti-V5 antibody to visualize AP expression. Two fields of view are shown per condition. The last two columns show zooms of the boxed regions. Scale bars, 10 μm. Representative fields of view from three biological replicates are shown. Additional fields of view are shown in Figure S11. (D) Constructs used for targeting AP and EX fragments of sAPEX to the outer mitochondrial membrane (OMM) and ER membrane (ERM), respectively. (E) Schematic overview of sAPEX applied at mito-ER contacts. Inactive fragments (grey) fused to FKBP and FRB can reconstitute to give active peroxidase (green) when driven together by an inducible protein-protein interaction (PPI) with the addition of rapamycin (yellow diamond). (F) sAPEX reconstitution in COS7 mammalian cells. COS7 cells stably expressing EX-FRB-ERM from (D) were infected with lentivirus containing OMM-FKBP-AP. Forty-six hours later, cells were incubated with heme for 90 minutes prior to incubation with BP in heme-free media in which rapamycin was added or omitted, as indicated. BP labeling was initiated by the addition of H2O2, and after 1 minute, cells were fixed and stained with neutravidin-AlexaFluor647 to visualize reconstituted sAPEX activity. HA and V5 staining show the localizations of total EX and total AP, respectively. The top row is a negative control with rapamycin omitted. Scale bar, 10 μm. Additional fields of view are shown in Figure S10. This experiment has four biological replicates. (G) Reconstituted sAPEX has reactivity towards DAB in COS7 mammalian cells. COS7 cells stably expressing EX-FRB-ERM from (D) were infected with lentivirus containing OMM-FKBP-AP. After 48 hours, cells were incubated with heme for 90 minutes prior to the rapamycin incubation in heme-free media for 0 or 30 minutes. Cells were then fixed, and DAB polymerization was performed for 15 minutes. In the bright field images, dark regions indicate peroxidase activity. Scale bar, 10 μm. Representative fields of view from three biological replicates are shown. (H) sAPEX can be used as genetically-encoded reporter for EM. Samples from (G) with 30-minute rapamycin incubation were analyzed by EM. Dark staining from osmium tetroxide recruitment to sAPEX-generated DAB polymer is observed exclusively at sites where ER is in close proximity to mitochondria (M). Scale bars, 250 nm. Additional field of view shown in Figure S14A. This experiment represents a single biological replicate.
Expression of MCP–AP and PCP–EX resulted in biotinylation that was dependent on the presence of a TERC RNA bearing both of the cognate MS2 and PP7 motifs (Figure 4C and Figure S11). Critically, we observed no such biotinylation when individual MS2 and PP7 hairpins were localized on separate TERC RNA constructs. Moreover, the labeling pattern appeared punctate, as would be predicted from localizing peroxidase activity to the discrete subnuclear foci characteristic of interphase telomerase76. Consistent with these results, streptavidin blotting of whole cell lysates also shows that expression of MCP–AP and PCP–EX results in biotinylation activity only when they are co-expressed with TERC RNA bearing both MS2 and PP7 hairpins (Figure S12). Together, our data suggest that sAPEX activity can be specifically reconstituted on a target RNA by nucleating protein fragment assembly on a structured RNA cassette.
sAPEX reconstitution at mitochondria-ER contacts
In mammalian cells, an estimated 5–20% of the mitochondrial outer membrane makes intimate contact (<70 nm gap) with the membrane of the endoplasmic reticulum (ER)19. These mito-ER contacts are thought to be important for a variety of functions and signaling processes, from mitochondrial fission to lipid synthesis18,19,20–23,24,25. Recent work has identified proteins that reside at mito-ER contacts and may play a role in tethering the membranes together4,29. A major goal of this field is to comprehensively characterize the molecular composition of these contact sites to better understand how they mediate important cellular processes. To pave the way for such future efforts, we tested whether sAPEX activity could be reconstituted at mito-ER contact sites.
Mito-ER contacts are delicate and easily perturbed structures. Overexpression of various proteins, such as SYNJ2BP or even green fluorescent protein, can lead to dramatic distortion of one or both organellar membranes4,77. Because our optimized sAPEX fragment pair, AP + EX, does not exhibit PPI-independent reconstitution, we reasoned that it might be capable of reconstitution at mito-ER contacts without major perturbation of organellar structure. 4D–F and S13 show AP-FKBP targeted to the OMM and EX-FRB targeted to the ERM in COS7 cells, which are flat and thin, facilitating visualization of mitochondria and ER structures. We observed BP/streptavidin staining in cells treated with rapamycin for 30 minutes. Consistent with the PPI-dependence of sAPEX reconstitution, no BP labeling was observed when rapamycin was omitted. We also stained the COS7 cells for mitochondrial and ER markers and observed minimal disruption of organellar morphology, both before and after rapamycin addition (Figure S13B–C).
To examine sAPEX at mito-ER contacts at higher resolution, we took advantage of APEX’s ability to generate contrast for electron microscopy (EM)1,16. We fixed COS7 cells prepared as above, and overlaid them with DAB and H2O2 to allow reconstituted sAPEX to catalyze the oxidative polymerization and local deposition of DAB1. Bright field imaging showed dark threads corresponding to stained mitochondria in the 30-minute rapamycin-treated samples, but not in the untreated samples (Figure 4G). We next stained the rapamycin-treated samples with OsO4 to deposit electron-dense osmium on the DAB polymer, then embedded and sectioned the samples for EM. EM imaging revealed a dark stain, corresponding to regions of reconstituted sAPEX activity, exclusively at contact sites between mitochondria and ER (Figure 4H and Figure S14A). Two zoomed views show that DAB staining filled the ~25 nm gap between the OMM and ERM in samples that were treated with rapamycin, but was absent from isolated ER and mitochondrial membranes. Furthermore, DAB staining was also absent from samples that were not treated with rapamycin (Figure S14B).
Discussion
Using a combination of rational design and yeast-display directed evolution, we have engineered a low-affinity sAPEX protein complementation assay (PCA) with robust activity upon reconstitution driven by co-proximation. After three generations of evolution, our intermediate AP(−3) fragment was highly active, but gave PPI-independent background reconstitution. To overcome this, our final directed evolution strategy implemented a negative selection that eliminated yeast clones with high fragment affinity. The final engineered sAPEX fragments, AP and EX, possess a total of nine mutations relative to the starting APEX2 template, clustered at solvent exposed regions, that collectively improve the PPI dependence of this system while maintaining high catalytic activity of the reconstituted form. In principle, our yeast-display platform could be extended to engineer other PCA systems. High-affinity fragment pairs such as those of split GFP result in spontaneous and irreversible reconstitution78; similarly, split HRP irreversibly reconstitutes in the ER without a PPI38. Split YFP utilizes fragments that reconstitute in a more PPI-dependent manner79,80, but it still suffers from background fluorescence, especially at high expression levels, and from irreversible YFP self-assembly81,82.
Given the purportedly broad array of novel RNAs and RNA-mediated processes that have eluded mechanistic dissection83, and the growing number of diseases now thought to be mediated by aberrant RNA-protein interactions84,85, there is a great need for identifying the interaction partners of specific RNAs. Conventional approaches address this problem by capturing and enriching a target RNA along with its bound interaction partners, either using affinity-tagged antisense oligonucleotides that isolate endogenous transcripts86–89, or by affinity tagging the transcript of interest itself90,91. However, these approaches are often confounded by the abundance of the target transcript, the unpredictable performance of RNA-based affinity tags92, and nonspecific RNA-protein interactions formed during lysis and enrichments93.
Using RNA scaffolding motifs to target the reconstitution of sAPEX eliminates high off-target background signal from protein overexpression. sAPEX demonstrates promise for BP labeling around specific RNAs, which indicates potential future applications to elucidate interactomes and interaction partners of an RNA of interest. Moreover, since the RNA domain we used to recruit AP and EX was designed to be functionally independent of the RNA to which it is appended, it should in theory be adaptable to other RNA targets94. A downside to this approach, however, is the need to express the transgenic, tagged RNAs that may not properly fold, localize, or function. As an alternative strategy, one might use RNA-targeting CRISPR-Cas systems to recruit AP and EX to endogenously expressed RNAs66. This approach may require substantial optimization to juxtapose the AP and EX fragments in the proper orientation, and may be influenced by the sequence, structure, or proteome of the targeted RNA.
The ability to reconstitute APEX2 activity at an organellar junction represents a promising step towards gaining greater understanding of subcellular locales that were previously intractable to genetic targeting. We found that the reconstitution of sAPEX at mito-ER contact sites was PPI-dependent, despite multiple days of co-translation of the sAPEX fragments on the membranes of these organelles that are known to come into contact. Importantly, sAPEX fragments did not perturb the morphology of these organelles, as shown by fluorescence and electron microscopy. We note that some optimization was required to attain PPI-dependence in the EM experiment; for instance, we found it necessary to reduce overall protein expression by changing the promoter of OMM-FKBP-EX from CMV to human ubiquitin promoter (hUBC).
Unlike most other PCAs, sAPEX possesses the versatile ability to react with many different substrates, enabling its use for a wide array of applications. Because sAPEX activity can be reconstituted in highly specific subcellular regions that are intractable for single-gene constructs, it expands the toolkit of proximity labeling technologies.
Methods
Cloning
See Supplementary Table 1 for a list of genetic constructs used in this study, with annotated epitope tags, promoters, resistance genes, vector name, linkers sequences, etc. For cloning, PCR fragments were amplified using Q5 polymerase (New England BioLabs (NEB)) or PfuUltra II Fusion HS DNA polymerase (Agilent Technologies). The vectors were double-digested and ligated to gel-purified PCR products by T4 ligation or Gibson assembly. Ligated plasmid products were introduced by heat shock transformation into competent XL1-Blue bacteria. The APEX2 gene used for initial cut site screening was amplified from vimentin-APEX2 with codons optimized for mammalian expression. Mutants of AP were generated either using QuikChange mutagenesis (Stratagene) or isolated from individual yeast clones and transferred to mammalian expression vectors using standard cloning techniques.
To generate tagged RNA expression constructs (Figure 4A–C; Figure S10), the TERC ncRNA and the hTR500 3´–processing block72 were amplified as a single product from HEK 293T genomic DNA, via nested PCR. DNA fragments encoding the structured RNA cassettes (Figure S10) were synthesized by primer extension from overlapping oligonucleotides, or as gBlocks (Integrated DNA Technologies, Inc.). TERC and cassette inserts were ligated into the ncRNA transient expression vector pCMV–SV40pA95, from which the existing polyadenylation site had been removed, via standard two- or three-piece restriction-digest cloning.
MCP– and PCP–tagged constructs (Figure 4A–C; Figure S11) were generated as follows. Genes encoding single protomers of the phage coat proteins were amplified from plasmids pHA_MS2-VP64 and pHA_PP7-VP6495. Upstream fragments encoding the Kozak sequence, Nuclear Localization Sequence (NLS), and V5 (MCP) or Flag (PCP) tags were synthesized by primer extension from overlapping oligos (IDT, Inc.). Full length APEX2 was amplified from mito-V5-APEX22. Three-fragment pools (upstream fragments, phage coat protein, APEX2) were restriction digested, then ligated using T4 DNA ligase (New England Biolabs). This ligation mixture was then used as template for a subsequent round of PCR, to isolate the full chimeric insert. These full-length PCR products were then inserted into pCMV–SV40pA by standard restriction-digest cloning. For cloning MCP-AP and PCP-EX, similar methods were employed.
Mammalian cell culture and transfection
HEK 293T and COS7 cells from ATCC (passages <20) were cultured as monolayers in complete growth media consisting of 50% DMEM (Dulbecco’s Modified Eagle medium, Gibco) and 50% MEM (Minimum Essential medium, Gibco) supplemented with 10% w/v FBS (Fetal Bovine Serum, Sigma), 50 units/mL penicillin, and 50 μg/mL streptomycin at 37 °C under 5% CO2. For confocal imaging experiments, cells were grown on 7 × 7 mm glass coverslips in 48-well plates (Corning). To improve the adherence of HEK 293T cells, the glass coverslips were pretreated with 50 μg/mL fibronectin (Millipore) in Dulbecco’s phosphate-buffered saline (DPBS), pH 7.4, for 20 min at 37 °C, followed by two washes with growth media prior to cell plating. Cells were transfected at 70–85% confluence using Lipofectamine 2000 (Life Technologies), with 1.0 μL Lipofectamine 2000 and 200 ng of each plasmid per 300,000 cells in serum-free media for 3–4 h, after which transfection media was replaced with fresh complete growth media. Cells were labeled and/or fixed 18–24 h after transfection. The cell line was not authenticated or tested for mycoplasma.
Lentivirus generation and mammalian cell infection
To generate viruses, HEK 293T cells plated in a T25 flask (Corning) were transfected at ~75–90% confluency with 5 mL of serum-free MEM containing the lentiviral plasmid encoding the gene of interest (2500 ng), the lentiviral packaging plasmids dR8.91 (2250 ng) and pVSV-G (250 ng), and 30 μL of 1 mg/mL Polyethylenimine “Max” (Polysciences) pH 7.1 for 3–4 h, followed by media change with complete MEM. 48 h after transfection, the cell medium containing lentivirus was harvested, filtered through a 0.45-μm filter, and added to other fresh cell cultures to induce expression of the gene of interest. Typically, for both HEK 293T and COS7 cultures plated in wells of a 48-well plate (0.95 cm2 per well), cells were plated approximately 12–16 h prior to lentiviral transduction in 1:1 DMEM:MEM complete growth media. HEK 293T and COS7 cells, at ~70% and ~85% confluency, respectively, were then infected by exchanging the medium with 150–200 μL of complete MEM and 50–100 μL of viral medium. Cells were then incubated for 48 h at 37 °C with 5% CO2 prior to heme incubation, labeling, and imaging.
In the experiment shown in Figure 3D, the N-terminal AP fragment of sAPEX was transiently expressed via lentiviral infection and the full-length APEX2 was stably expressed. This procedure resulted in a reduced number of comparable HEK 293T cells that would express both fragments compared to cells expressing full-length APEX2. Therefore, when comparing our final sAPEX to full-length APEX2 in the cytosol of COS7 cells, both the N-terminal AP fragment of sAPEX and full-length APEX2 were lentivirally introduced with similar infection efficiencies (C-terminal EX fragment of sAPEX was stably expressed in cells infected with FKBP-AP lentivirus, Figure S9).
Generation of stable cell lines
To generate stable cell lines, constructs of interest were cloned into the lentiviral vector pLX304 to make virus, as described above. Low passage HEK 293T cells (<20 passages) plated in 6-well plates (Corning) were infected with 800–1000uL of lentiviral medium for APEX2-NES, FRB-EX, or MCP-AP (Figure 3B–C, Figure 4A–C, Figure S3, and Figure S9A). After 2 days at 37˚C, 5% CO2, each well was trypsinized and replated into two T25 flasks in full growth media containing 8 mg/ml blasticidin S HCl (Calbiochem, CAS no. 3513_03_9). Media was changed to fresh 8 mg/ml blasticidin S HCl media every 24 h. Cells were trypsinized and re-plated before reaching full confluency within the first 7 days. After 7–10 days of selection, surviving cells were used for experiments. To make cells stocks for long-term storage at –80 ˚C, cells from a ~90% confluent T75 flask were harvested, pelleted, resuspended in 5 mL complete media with 5% DMSO, and cooled to –80 ˚C overnight using a Mr. Frosty Freezing Container (Thermo Scientific™). COS7 cells stably expressing EX-FRB-ERM from Figure 3D, Figure 4D–H, Figure S9B and Figure S13 were prepared similarly, with a selection in media containing 10 mg/mL blasticidin S HCl.
Heme supplementation
Where indicated in both yeast and mammalian culture, heme was supplemented by dilution from a thawed 300 μM heme stock solution, which was prepared by dissolving hemin-Cl (Sigma) in 10 mM NaOH with sonication for 3 min, aliquoting, flash freezing in liquid N2, and storing at −80 °C. Heme stocks were used within 1 h after thawing and never reused. Heme stock was diluted to 5 μM and 6 μM in complete growth media and added to HEK 293T and COS7 cells, respectively, for 90 min prior to BP incubation (without heme) or DAB labeling. For yeast heme supplementation in Generations 2–4, washed yeast cells were pelleted at 7000 × g for 1 min and resuspended to a concentration of 1.5 × 107 yeast cells/mL in DPBS-B with 0.1% BSA at the designated heme concentrations (Figure S3B, S4B, S5A) immediately prior to EX incubation.
Amplex UltraRed assay (Figures 1D–E; S2)
HEK 293T cells plated on glass coverslips were transfected as described above with 200 ng of each plasmid encoding a peroxidase fragment and 100 ng of plasmid encoding CFP-NLS in tandem as a transfection marker for 3 h. Transfection media was then replaced with complete medium with or without 400 nM rapamycin (Calbiochem), which was delivered by 1000-fold dilution of a 400 μM stock in DMSO maintained at −20 °C for months and thawed as needed. After 18–22 h, cells were moved to ice, washed three times with DPBS, and treated with a solution of 50 μM Amplex UltraRed (Life Technologies) with 0.02% (6.7 mM) H2O2 in DPBS according to previously published protocols1,16,38. Cells were labeled for 25 min on ice. The reaction was quenched by removal of the substrate solution by aspiration, and cells were subsequently washed three times with DPBS, fixed with 4.0% formaldehyde in DPBS (v/v) on ice for 30 min, and washed 2 more times with DPBS before being imaged by confocal microscopy.
Biotin-phenol labeling (Figures 3C; 4C; 4F; S3B; S3C; S8; S9B; S11; S13)
Genes were introduced into HEK 293T or COS7 cells through either transient transfection with Lipofectamine 2000 or lentiviral infection. After 18–24 h (transfection) or 48 h (lentivirus), BP labeling was initiated by changing the medium to 200 μL of fresh growth medium containing 500 μM BP that was sonicated for at least 5 min to ensure BP was fully dissolved. Cells were incubated at 37 °C under 5% CO2 for 30 min according to previously published protocols. To initiate labeling, 2 μL of 100 mM H2O2 was added to each well, yielding a final concentration of 1 mM H2O2, and the plate was immediately gently agitated. To quench the reaction after 1 min, the BP solution was aspirated, and cells were immediately fixed with 4% formaldehyde in PBS (v/v) with 5 mM Trolox at room temperature for 5 min before continuing fixation on ice for an additional 25 min. Cells were then washed with chilled DPBS three times and permeabilized with pre-chilled methanol at –20 °C for 10 min. Cells were washed again three times with DPBS and blocked with 3% BSA in DPBS for 1 h to overnight with rocking at 4 °C. To detect the expression of sAPEX fusions, cells were incubated with primary antibodies mouse anti-V5 (Life Technologies, Cat. No. R96025, 1:1500 dilution) and rabbit anti-HA (Cell Signaling, 1:1000 dilution) in 1% BSA in DPBS for 1 h to overnight at 4 °C followed by 4 × 5 min washes with chilled DPBS. Cells were then incubated with a 1% BSA in DPBS (v/v) solution containing secondary Alexa Fluor 488-goat anti-rabbit IgG (Life Technologies, Cat. No. A-11001, 1:1000 dilution), Alexa Fluor 568-goat anti-mouse IgG (Life Technologies, Cat. No. A-11004, 1:1000 dilution), and homemade streptavidin–Alexa Fluor 647 (1:1000) for 25–45min at 4 °C with rocking. Cells were then washed 4 × 5 min with chilled DPBS and imaged by confocal microscopy.
Western blotting (Figures 3D; S9A; S12)
For streptavidin blotting, cells were grown on polystyrene 6-well plates (Corning) and labeled under the same conditions described above (see “Biotin-phenol labeling”). After 1 min of labeling, the cells were washed three times with room temperature quencher solution (10 mM sodium azide, 10 mM sodium ascorbate, and 5 mM Trolox in DPBS) and then scraped and pelleted by centrifugation at 1400 rpm for 3 min. The cell pellet was then lysed by gentle resuspension into peroxidase-quencher containing RIPA lysis buffer (50 mM Tris, 150 mM NaCl, 0.1% SDS, 0.5% sodium deoxycholate, 1% Triton X-100, 1× protease cocktail (Sigma Aldrich, catalog no. P8849), 1 mM PMSF (phenylmethylsulfonyl fluoride), 10 mM sodium azide, 10 mM sodium ascorbate, and 5 mM Trolox). After 2 minutes on ice, the lysates were clarified by centrifugation at 15000 × g for 10 min at 4 °C before separation on a 10% SDS-PAGE gel. For blotting analysis, gels were transferred to a nitrocellulose membrane, stained for 10 min using Ponceau S (0.1% (w/v) Ponceau S in 5% (v/v) acetic acid/water), and blocked with “blocking buffer” (3% (w/v) BSA or 5% (w/v) milk in 0.1% Tween-20 in Tris-buffered saline) for 1 h at room temp or 4 °C overnight. The blots were immersed in streptavidin-HRP in blocking buffer (Thermo Scientific, cat. no. 21126, 1:3000 dilution) at room temperature for 1 h and then rinsed with blocking buffer 4 × 5 min. For assessing comparative fragment expression level, identical gels and blots were prepared in parallel and immersed in blocking buffer containing either mouse anti–V5 (Life Technologies, R96025, 1:10,000 dilution), mouse anti-HA (Santa Cruz Biotechnology, G1817, 1:3000 dilution), or mouse anti-FLAG (Sigma Aldrich, F1804, 1:5000 dilution) for 1 h to overnight at 4°C. The blots were rinsed with blocking buffer 4 × 5 minutes before being immersed in anti-Mouse-HRP (Bio-Rad, 1:3000) in blocking buffer at room temp for 1 h, then rinsed with blocking buffer 4 × 5 min again. All blots were developed using Clarity reagent (Bio-Rad) for 5 min and imaged using an Alpha Innotech gel imaging system. Quantitation of Western blots (Figure 3D) was performed using ImageJ on the streptavidin-HRP blot shown. Total band intensity of the entire lane, subtracting endogenously biotinylated protein bands at ~150 kD and ~90 kD (visible in negative controls) and possible self-labeling bands at ~58 kD and ~46 kD, was captured using ImageJ, then normalized to the amount of protein loaded (determined by intensity of lane in the Ponceau-stained blot). Because all three fragments have different epitope tags, bands were not normalized to expression of either fragment.
DAB labeling for light microscopy experiments (Figures 3B; 4G)
Transfected HEK 239T cells plated on glass coverslips were fixed with 4% formaldehyde in PBS (v/v) for 20 min at room temperature. Cells were subsequently washed twice with room temperature DPBS, incubated for 10 min in buffer containing 20 mM glycine to quench unreacted formaldehyde, and washed 3 more times with DPBS. A fresh solution containing 0.5 mg/mL (1.4 mM) DAB (5.4 mg/mL of 3,3′-diaminobenzidine, Sigma, dissolved in 0.1 M HCl) and 0.03% (v/v) (10 mM) H2O2 in DPBS was added to cells for 15 min. To quench polymerization, the DAB solution was removed, and cells were placed on ice and washed 4 × 5 min with chilled DPBS, before imaging.
Labeling the mitochondria and endoplasmic reticulum in COS7 cells for imaging by confocal microscopy (Figures S13B–C)
Mitochondrial morphology was visualized using MitoTracker Red CMXROS (ThermoFisher, M7512); it was diluted to a final concentration of 200 nM and added during the 30 min BP incubation. This dye is retained after aldehyde-fixation. After following the standard primary immunostaining protocol (see biotin-phenol labeling protocol for Figure 4F), cells were then incubated with a 1% BSA in DPBS (w/v) solution containing the secondary antibodies Alexa Fluor 488-goat anti-rabbit IgG (Life Technologies, Cat. No. A-11001, 1:1000 dilution), Alexa Fluor 405-goat anti-mouse IgG (Life Technologies, Cat. No. A-31553, 1:1000 dilution), and homemade streptavidin–Alexa Fluor 647 (1:1000) for 25–45min at 4 °C with rocking.
For visualizing the morphology of the endoplasmic reticulum, COS7 samples were prepared following the procedure for the Figure 4F samples, except that two rounds of immunostaining were required. After the first round of secondary immunostaining (same antibody conditions as the MitoTracker sample), cells were washed 5 × 5 min prior to a second fixation with 4% formaldehyde for 25 min on ice. Cells were then blocked with 2% BSA in DPBS (w/v) for 1 h at 4 °C. Afterwards, a second round of primary immunostaining was performed using an antibody (Santa Cruz Biotechnology, sc-33806) against the KDEL receptor, a resident ER protein, for 1 h at 4 °C. After washing, and secondary immunostaining with Alexa Fluor 568-goat anti-rabbit IgG (Life Technologies, Cat. No. A-11011, 1:2000 dilution in 2% BSA) for 30 min at 4 °C, cells were washed 5 × 5 min each before imaging.
Fluorescence microscopy
Confocal imaging was performed on a Zeiss AxioObserver inverted confocal microscope with a 63× oil-immersion objective, outfitted with a Yokogawa spinning disk confocal head, a Quad-band notch dichroic mirror (405/488/568/647), and 405 (diode), 491 (DPSS), 561 (DPSS) and 640-nm (diode) lasers (all 50 mW). CFP (405 laser excitation; 445/40 emission), Alexa Fluor 488 (491 laser excitation; 528/38 emission), Alexa Fluor 568/resorufin (561 laser excitation; 617/73 emission), Alexa Fluor 647 (647 excitation; 680/30 emission), and differential interference contrast (DIC) images were collected using Slidebook 5.0 or 6.0 software (Intelligent Imaging Innovations). Acquisition times ranged from 100 to 1000 ms. DAB labeled cells were imaged by bright field; acquisition time was 200 ms. All image analysis was performed in Slidebook. Imaging conditions and intensity scales were matched for each data set presented together.
RNA structural modeling
The Docked Fourway Junction construct (“D4WJ”, Figure S10B) was designed based on analogous teco-RNA devices96, supplemented by three-dimensional modeling as performed in COOT97. Briefly, parallel helices of the tetraloop–tetraloop-receptor interaction in the P4–P6 domain of the T. thermophila Group I intron98 (PDB: 1GID) were used as structural templates on which regular A–form RNA duplexes (generated automatically in COOT) were aligned. These duplexes then served as templates on which the structures of RNA-bound MCP and PCP proteins74,99 (PDB IDs: 2BU1, 2QUX, respectively) were aligned. The register of alignment (i.e., the number of base pairs between the MS2 and PP7 loops and the core four-way junction) were systematically altered so as to co-proximate the N- and C-termini of the coat protomers, while minimizing the apparent overall steric clash between the proteins. Images of the final model (Figure S10C) were rendered in PyMol (Schrödinger, LLC).
RNA expression analysis
Quantitative RT–PCR analysis of TERC constructs (Figure S10D) was performed in a manner similar to that described previously95. Briefly, HEK 293T cells were grown in 6-well dishes, as above, and at 80% confluency were transiently transfected with each of the TERC expression constructs, using lipofectamine 2000 (Thermo Fisher Scientific, LLC). Two days thereafter, cell media was aspirated, cells were washed once briefly with warmed PBS, and total cellular RNA was harvested by extraction with Trizol (Thermo Fisher) followed by precipitation with isopropanol, using Glyco Blue (Thermo Fisher) as a carrier. Pellets were resuspended in water and further purified using RNEasy mini-prep columns (QIAgen), following the manufacturer’s “RNA cleanup” protocol with on-column DNAse treatment. 500 ng of purified RNA was used as template for reverse transcription, using Super Script III Reverse Transcriptase (Thermo Fisher), primed with random hexamers (Thermo Fisher). Transcripts were quantified in quadruplicate on a 7900HT Fast Real-Time PCR System (Applied Biosystems) using Rox-normalized Universal SYBR Green Master Mix (Roche) and the following primers:
GAPDH_F: TTCGACAGTCAGCCGCATCTTCTT
GAPDH_R: GCCCAATACGACCAAATCCGTTGA
TERC_F: CGCCTTCCACCGTTCATTC
TERC_R: GGCCAGCAGCTGACATTTT
Data were analyzed using Realtime qPCR Miner100, as described previously95.
Electron microscopy sample preparation and imaging (Figures 4H and Figure S14)
COS7 cells stably expressing EX-ERM under the CMV promoter were plated onto glass bottom dishes (MatTek, part no. P35GC-0–14-C). At 70% confluence, cells were infected with lentivirus to express OMM-AP under the hUBC promoter. After 48 h, media was aspirated and replaced with 2 mL of 6 μM heme-containing cell culture media (see heme supplementation section and mammalian cell culture for more details). After 90 min of heme incubation, heme-containing media was aspirated and replaced with 2 mL of regular cell culture media containing or omitting 400 nM rapamycin. After 30 min of rapamycin incubation, media was aspirated and replaced with 250 μL of pre-warmed, 37 °C 2% (v/v) glutaraldehyde (Electron Microscopy Sciences, cat. no. 16019) in sodium cacodylate buffer (100 mM sodium cacodylate, Electron Microscopy Sciences, cat. no. 12300), pH 7.4, with 2 mM calcium chloride (VWR International, cat. no. 0556–500G)) to commence fixation. After 30 seconds, the glutaraldehyde solution was aspirated and replaced with 1.5–2 mL of fresh room temperature 2% glutaraldehyde solution and placed on ice for 60 minutes. Note, all subsequent steps prior to dehydration were performed on ice. Afterwards, fixation solution was removed, and cells were washed with 2 mL of cold (4 °C) sodium cacodylate buffer 5 times. Cells were subsequently incubated for 5 min with 2 mL of cold 20 mM glycine (VWR International, cat. no. 470301–176) in sodium cacodylate buffer to quench unreacted glutaraldehyde, and then washed with 2 mL of cold sodium cacodylate buffer 5 more times. After the fifth wash was removed, 2 mL of cold fresh DAB labeling solution was added (0.5 mg/mL (1.4 mM) DAB (5.4 mg/mL of 3,3′-diaminobenzidine, Sigma, dissolved in 0.1 M HCl) combined with 0.03% (v/v) (10 mM) H2O2 in cold sodium cacodylate buffer) for 15 min. Extent of DAB polymerization was monitored by bright field microscopy (Figures 4G). Cells were then washed with 2 mL of cold sodium cacodylate buffer 5 times and incubated with 2 mL of freshly prepared OsO4 (Electron Microscopy Sciences, cat. no. 19150) in cold sodium cacodylate buffer for 30 min. OsO4 solution was removed and transferred into sodium sulfite quenching solution (freshly prepared 0.5 M sodium sulfite (Sigma-Aldrich, cat. no. 239321)). Cells were washed five times with 2 mL of ice-cold cell culture grade water. Cells were then incubated with 1.5 mL of cold freshly prepared 2% (w/v) uranyl acetate (Electron Microscopy Sciences, cat. no. 22400) solution for 3 h in the dark at 4 °C. Cells were then washed five times with 2 mL of ice-cold cell culture grade water prior to dehydration via incubation with serially concentrated ethanol solutions (20%, 50%, 70%, 90%, 100% ethanol, then 100% ethanol at room temperature). Cells were then embedded using Durcupan resin exactly as previously described101. Ultrathin sections were cut with a diamond knife and a Leica ultramicrotome at a thickness of 70–80 nm and examined using a FEI Technai 12 (Spirit) transmission EM operated at 120 kV equipped with a Tietz 2k by 2k CCD camera.
Yeast strains, transformation, and culturing
See Supplementary Table 1 for details on all plasmids and primers used in this study. Aga2p-APEX2 (full length) yeast and Generation 2 selection yeast were generated by transformation of the yeast display plasmid pCTCON2 into the Saccharomyces cerevisiae strain EBY100. As previously reported38,102, EBY100, propagated at 30 °C in yeast extract peptone dextrose (YPD) complete medium, was made competent and transformed using the Frozen E-Z Yeast Transformation II Kit (Zymoprep) according to the manufacturer’s protocols. Individual colonies of transformed cells containing the TRP1 gene were selected on synthetic dextrose plus casein amino acid (SDCAA) plates and propagated in SDCAA medium at 30 °C. Protein expression was induced by inoculating 4.5 mL of SGCAA (SDCAA medium with dextrose replaced by an equivalent amount of galactose) with 500 μL of overnight SDCAA yeast culture and incubating at 30 °C for 18–24 h. Successful incorporation was confirmed by sequencing of DNA isolated by Zymoprep (Zymoprep™ Yeast Plasmid Miniprep I, Zymo Research, see modified procedure below) and FACS analysis of immunostained yeast (see procedure below).
All other yeast used in this study were prepared using a two-step transformation of S. cerevisiae strain BJ5465, based on a previously described protocol38,102. BJ5465 yeast, propagated at 30 °C in YPD complete medium, were transformed first with a linearized Yeast Integrating Plasmid (YIP) encoding Aga1p fusion constructs, plated and selected on SDCAA supplemented with 40 μg/mL tryptophan (lacking uracil). FACS analysis and DNA sequencing of PCR products amplified from extracted genomic DNA38,102 were used to confirm homologous recombination of the desired construct. Aga1p was constitutively expressed and immunostaining for the V5 epitope tag was detected in both SDCAA and SGCAA induction media (see Figure S3E for example of yeast surface protein and epitope tags). Next, to introduce individual Aga2p constructs to generate non-library yeast (such as starting templates and individual clones), Aga1p fusion expressing yeast were transformed with the applicable pCTCON2 plasmid and selected on SDCAA plates.
Generation of yeast libraries
For each generation of directed evolution, libraries of AP variants were generated using error-prone PCR according to published protocols16,38,62. In brief, for Generation 1 library, 150 ng of the template sAPEX AP-0 gene in pCTCON2 vector was amplified for 20 rounds with 0.4 μM forward and reverse primers, 2 mM MgCl2, 5 units of Taq polymerase (NEB), and 2 μM each of the mutagenic nucleotide analogs 8-oxo-2′-deoxyguanosine-5′triphosphate (8-oxo-dGTP) and 2′-deoxy-p-nucleoside-5′-triphosphate (dPTP). The PCR product was then gel-purified and amplified through three standard PCR reactions of 30 cycles with primers that introduce overlap to the vector, 100 μM regular dNTPs (VWR) and 5 units of Taq polymerase in 50 μL volume. In parallel, fresh electrocompetent BJ5465 yeast constitutively expressing EX-Aga1p were prepared. BJ5465 yeast expressing EX-Aga1p were passaged at least two times before this procedure to ensure that the cells were healthy. Fresh saturated culture (3 mL) was used to inoculate 100 mL of fresh YPD media, and the cells were cultured with shaking at 220 rpm at 30 °C for 6–8 h until the OD600 reached 1.5–1.8. The cells were then harvested by centrifugation for 3 min at 3000 rpm and resuspended in 50 mL of sterile 100 mM lithium acetate in water, by vigorous shaking. Fresh sterile DTT (1 M stock solution, made on the same day) was added to the yeast cells to a final concentration of 10 mM. The cells were incubated with shaking at 220 rpm for 12 min at 30 °C. Cells were then pelleted by centrifugation at 3000 rpm for 3 min at 4 °C and washed once with 25 mL ice-cold sterile water, pelleted again, and resuspended in 1 mL ice-cold sterile water. The amplified error-prone PCR inserts were electroporated with BamHI-NheI linearized pCTCON2 vector (10 μg insert:1 μg vector) backbone into electrocompetent yeast expressing EX-Aga1p. Electroporation was performed using a Bio-Rad Gene pulser XCell with the following settings: 500-V, 15-ms pulse duration, one pulse only, 2-mm cuvette. The electroporated cells were immediately rescued with 2 mL pre-warmed YPD media and then incubated at 30 °C for 1 h without shaking. Transformation efficiency was determined using 10 μL of this suspension, and the remainder was pelleted to remove the YDP media and resuspended in 100 mL SDCAA media supplemented with 50 units/mL penicillin and 50 μg/mL streptomycin. The culture was grown at 30 °C with shaking at 220 rpm for 1 d, before induction of protein expression for analysis by FACS and Zymoprep. Transformation efficiency was 2.9 × 107. DNA sequencing of 21 distinct colonies showed an average of 1.4 nucleotides changed per clone with a range of 0–4. For Generation 2 AP-1 library, the same procedure was employed except electroporation was performed on S. cerevisiae EBY100. Transformation efficiency was 3.5 × 107. DNA sequencing of 24 distinct colonies showed an average of 1.2 nucleotides changed per clone with a range of 0–5. For Generation 3 AP-2 library, the same procedure was employed except that the rate of mutagenesis was higher; ten rounds of error-prone PCR were performed with 15 μM of each of the mutagenic nucleotide analogs. Electroporation was performed on BJ5465 cells stably expressing acid coil-Aga1p. Transformation efficiency was 3.1 × 107. DNA sequencing of 22 distinct colonies showed an average of 7.3 nucleotides changed per clone with a range of 0–12. For Generation 4 selections, two libraries derived from AP-3 were combined: one generated by ten rounds of error-prone PCR performed with 10 μM each of the mutagenic nucleotide analogs and another generated by 20 rounds of error-prone PCR performed with 4 μM each of the mutagenic nucleotide analogs. Transformation efficiencies of the respective libraries were 2.0 × 107 and 2.8 × 107. DNA sequencing of 24 distinct colonies from each library showed an average of 5.6 and 2.9 nucleotides changed per clone, with a range of 0–9 and 0–4, respectively.
Yeast display selections
For each generation of selections, yeast libraries (as described above) were induced by transferring them to 1:9 SDCAA/SGCAA media and growing the cells for 18–24 h at 30 °C with shaking at 220 rpm. Overnight yeast cultures were pelleted by centrifugation (3000 rpm, room temperature, 3 min), washed with room temperature 0.1% BSA (w/v) DPBS (DPBS-B) twice, and resuspended in an equivalent volume of DPBS-B. The OD600 for a 1/100 dilution of the cell suspension was measured (1 OD600 ~ 1 × 107 yeast cells). An aliquot of yeast with at least a tenfold coverage above the diversity of the yeast pool was pelleted at 7000 × g for 1 min. This oversampling was employed for all passaging and labeling steps except for the first-round sort of each generation, in which we aimed for threefold oversampling during labeling. This lower sampling in the first round was a result of the very large diversity size and the practical difficulties and expense of labeling large numbers of yeast cells. Prior to the first round of sorting, “diversity” was considered to be the number of transformants in the initial library. In subsequent rounds, the “diversity” was the number of yeast cells collected in the previous round of sorting. Yeast libraries from Generations 2–4 were first incubated with heme (see details above); to implement both the positive and negative selections, we added concentrated EX fragment (expressed and purified from E. coli as described below) to the heme-containing yeast DPBS-B culture. For more details regarding the presence of base coil, concentrations, and time of protein and heme incubations, see Figures S5B, and S6B. Yeast were then washed twice with DPBS-B and resuspended in a room temperature solution consisting of 100 μM biotin-phenol and 1 μM H2O2 at a density of 2.5 × 106 cells/mL (7.5 × 106 to 1.0 × 107 cells/mL for first round sorts) by rapid vortexing. After 1 min, the yeast suspension was quenched by addition of an equal volume of 20 mM Trolox and 10 mM sodium ascorbate in DPBS-B, followed by rapid vortexing. Minimal intercellular labeling was observed under these conditions. The cells were pelleted at 14,000 × g for 1 min and washed twice with DPBS-B. For two-dimensional FACS sorting, samples were then resuspended in 50 μL of chicken anti–c-Myc (Invitrogen, Cat. No. A21281, 1:400) and incubated at 4 °C with rotation for 1 h. Samples were washed twice with DPBS-B and resuspended in 50 μL of streptavidin-phycoerythrin (Jackson ImmunoResearch, Cat. No. 016–110-084, 1:250) and Alexa Fluor 647 goat anti-chicken IgG (Life Technologies, 1:300) After 1 h of incubation with rotation at 4 °C, samples were washed twice, and the labeled and fluorophore-stained yeast cells were resuspended in DPBS-B to a concentration of 4.5 × 107 to 1.2 × 108 cells/mL and sorted by FACS into a collection tube containing 5 mL SDCAA with 50 units/mL penicillin and 50 μg/mL streptomycin. Collected yeast cells were immediately placed back into a 30 °C incubator and cultured with shaking at 220 rpm. Yeast cells were grown until saturation and passaged once more in SDCAA; depending on the number of cells collected, it took 1–2 days to reach initial saturation. Fresh saturated yeast culture (0.5 mL) was added to 4.5 mL of SGCAA for induction, labeling, and subsequent rounds of sorting or analysis. For positive selections, trapezoidal gates were drawn to collect yeast cells positive for AF647 signal that also had high PE signal; these were the cells with a high activity to expression ratio. During Generation 4 negative selections, generous rectangular gates were drawn to collect all AF647-positive yeast that were negative for PE signal, representative of cells with no activity despite robust expression (see Figure 2A).
We used a BD FACS Aria II cell sorter with 640nm (for AF 647) and 488 nm or 561(for PE) lasers and appropriate emission filters (780/60 for AF 647), 575/26 for PE).
Sequence analysis of individual yeast clones
Plasmid DNA from yeast cultures was harvested using the Zymoprep Yeast Plasmid Miniprep II Kit (Zymo Research) according to the manufacturer’s instructions for liquid culture except for the following. Zymolyase (6 μL) was added for a total incubation time of 1–3 h, rotating at 37 °C. After the addition of the manufacturer’s Solution 2, the mixture was briefly vortexed. DNA was eluted with 10 μL of warm water, and all eluted DNA was transformed into XL1Blue E. coli cells by heat-shock transformation and rescued for 1 h at 37 °C. The entire culture was then plated on an LB-ampicillin plate, and 18–36 colonies were picked for sequencing of individual clones.
FACS analysis
Yeast samples taken from picked colonies of transformed clones or from samples regrown after sorting were passaged in SDCAA overnight. Expression was induced by inoculating 4.5 mL of SGCAA with 500 μL of fresh saturated SDCAA yeast culture and incubating at 30 °C with shaking at 220 rpm for 18–24 h. Labeling was performed as described above, and yeast populations and clones were analyzed by FACS (BD LSR II flow cytometer, BD Biosciences) with 488-nm and 640-nm lasers and 582/42 (for phycoerythrin) and 670/30 (for AlexaFluor647) emission filters.
Expression and purification of C-terminal sAPEX fragments (EX) from E. coli
For positive selections employed in Generation 2 and negative selections in Generation 4, we cloned N-terminal His6-tagged EX-flag-GFP into pYFJ16 vector for Escherichia coli (E. coli) expression. For positive selections employed in Generations 3 and 4, we added a base coil between the His6-tag and EX. Homemade competent BL21 cells, derived from BL21-CodonPlus(DE3)-RIPL E. coli, were transformed with pYFJ16 constructs and plated on LB-amp plates. Individual colonies were amplified in two separate 5 mL cultures containing B (Luria-Bertani, Miller) media with 100 mg/L ampicillin at 37 °C overnight with shaking. The 10 mL saturated culture was then used to inoculate 1 liter LB with 100 mg/L ampicillin, which was grown at 25 °C with shaking until OD600 = 0.8 at 25 °C. Protein expression was induced with 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG, EMD Millipore), and cultures were grown with shaking at 220 rpm at 25 °C for another 20 h. Bacteria were pelleted by centrifugation at 6000 rpm for 10 min at room temperature and immediately lysed with B-PER buffer (Thermo Fisher Scientific) supplemented with 1× protease cocktail and 1 mM PMSF at 20 mL per 1 liter of original culture. All subsequent steps for protein purification were performed on ice or at 4 °C. The lysate was clarified by centrifugation at 10000 rpm for 15 min, and the supernatant was incubated with 1 mL of Ni-NTA agarose beads suspension (Qiagen) in binding buffer (50 mM Tris, 300 mM NaCl, pH = 7.8), in a total volume of 45 mL, for 20 min with rotation and then transferred to a gravity column. The beads were washed with 10 mL binding buffer followed by 30 mL washing buffer (30 mM imidazole, 50 mM Tris, 300 mM NaCl, 1 mM DTT, pH = 7.8), then the protein was eluted with 7 mL of elution buffer (200 mM imidazole, 50 mM Tris, 300 mM NaCl, pH = 7.8). Fractions were collected and transferred to a centrifugal filter Amicon Ultra-15 and exchanged 3 times into ice-cold DPBS (Dulbecco’s Phosphate Buffer Saline) and concentrated. Protein samples were flash frozen in liquid N2 and stored in aliquots at −80 °C. Purity was checked using SDS-PAGE. Concentrations of total protein content were determined using the bicinchoninic acid (BCA) Protein Assay Kit (Pierce).
Supplementary Material
Acknowledgements
FACS was performed at the Koch Institute Flow Cytometry Core (MIT) and Stanford Shared FACS Facility. S. Han (Stanford) synthesized neutravidin AlexaFluor647. This work was supported by NIH grants to A.Y.T. (R01-CA186568), to D.B. (R01-GM086197), and to M.H.E. (P41-GM103412) for support of the National Center for Microscopy and Imaging Research, in addition to the Chan Zuckerberg Biohub. D.M.S. was supported by HHMI/Jane Coffin Childs Fellowship and by J. Rinn (U. C. Boulder). T.C.B. was supported by Dow Graduate Research and Lester Wolfe Fellowships.
Footnotes
Supporting Information
Supplementary figures are available free of charge via the internet at http://pubs.acs.org.
Conflict of interest disclosure
The authors declare no competing financial interest.
References
- (1).Martell JD; Deerinck TJ; Sancak Y; Poulos TL; Mootha VK; Sosinsky GE; Ellisman MH; Ting AY Engineered Ascorbate Peroxidase as a Genetically Encoded Reporter for Electron Microscopy. Nat. Biotechnol. 2012, 30, 1143–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Rhee HW; Zou P; Udeshi ND; Martell JD; Mootha VK; Carr SA; Ting AY Proteomic Mapping of Mitochondria in Living Cells via Spatially Restricted Enzymatic Tagging. Science 2013, 339, 1328–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Hung V; Zou P; Rhee HW; Udeshi ND; Cracan V; Svinkina T; Carr SA; Mootha VK; Ting AY Proteomic Mapping of the Human Mitochondrial Intermembrane Space in Live Cells via Ratiometric APEX Tagging. Mol. Cell 2014, 55, 332–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Hung V; Lam SS; Udeshi ND; Svinkina T; Guzman G; Mootha VK; Carr SA; Ting AY Proteomic Mapping of Cytosol-Facing Outer Mitochondrial and ER Membranes in Living Human Cells by Proximity Biotinylation. elife 2017, 6, e24463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Loh KH; Stawski PS; Draycott AS; Udeshi ND; Lehrman EK; Wilton DK; Svinkina T; Deerinck TJ; Ellisman MH; Stevens B; Carr SA; Ting AY Proteomic Analysis of Unbounded Cellular Compartments: Synaptic Clefts. Cell 2016, 166, 1295–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Chen C-L; Hu Y; Udeshi ND; Lau TY; Wirtz-Peitz F; He L; Ting AY; Carr S. a; Perrimon N Proteomic Mapping in Live Drosophila Tissues Using an Engineered Ascorbate Peroxidase. Proc. Natl. Acad. Sci. U. S. A. 2015, 112, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Paek J; Kalocsay M; Staus DP; Wingler L; Pascolutti R; Paulo JA; Gygi SP; Kruse AC Multidimensional Tracking of GPCR Signaling via Peroxidase-Catalyzed Proximity Labeling. Cell 2017, 169, 338–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Han S; Udeshi ND; Deerinck TJ; Svinkina T; Ellisman MH; Carr SA; Ting AY Proximity Biotinylation as a Method for Mapping Proteins Associated with MtDNA in Living Cells. Cell Chem. Biol. 2017, 24, 404–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Lobingier BT; Hüttenhain R; Eichel K; Miller KB; Ting AY; von Zastrow M; Krogan NJ An Approach to Spatiotemporally Resolve Protein Interaction Networks in Living Cells. Cell 2017, 169, 350–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Kaewsapsak P; Shechner DM; Mallard W; Rinn JL; Ting AY Live-Cell Mapping of Organelle-Associated RNAs via Proximity Biotinylation Combined with Protein-RNA Crosslinking. eLife 2017, 6, e29224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Shvets E; Bitsikas V; Howard G; Hansen CG; Nichols BJ Dynamic Caveolae Exclude Bulk Membrane Proteins and Are Required for Sorting of Excess Glycosphingolipids. Nat. Commun. 2015, 6, 6867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Ludwig A; Nichols BJ; Sandin S Architecture of the Caveolar Coat Complex. J. Cell Sci. 2016, 129, 3077–3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Liu LK; Choudhary V; Toulmay A; Prinz WA An Inducible ER-Golgi Tether Facilitates Ceramide Transport to Alleviate Lipotoxicity. J. Cell Biol. 2017, 216, 131–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Hyenne V; Apaydin A; Rodriguez D; Spiegelhalter C; Hoff-Yoessle S; Diem M; Tak S; Lefebvre O; Schwab Y; Goetz JG; Labouesse M RAL-1 Controls Multivesicular Body Biogenesis and Exosome Secretion. J. Cell Biol. 2015, 211, 27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Joesch M; Mankus D; Yamagata M; Shahbazi A; Schalek R; Suissa-Peleg A; Meister M; Lichtman JW; Scheirer WJ; Sanes JR Reconstruction of Genetically Identified Neurons Imaged by Serial-Section Electron Microscopy. elife 2016, 5, e15015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Lam SS; Martell JD; Kamer KJ; Deerinck TJ; Ellisman MH; Mootha VK; Ting AY Directed Evolution of APEX2 for Electron Microscopy and Proximity Labeling. Nat. Methods 2015, 12, 51–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Dwyer DJ; Belenky PA; Yang JH; MacDonald IC; Martell JD; Takahashi N; Chan CTY; Lobritz MA; Braff D; Schwarz EG; Ye JD; Pati M; Vercruysse M; Ralifo PS; Allison KR; Khalil AS; Ting AY; Walker GC; Collins JJ Antibiotics Induce Redox-Related Physiological Alterations as Part of Their Lethality. Proc. Natl. Acad. Sci. 2014, 111, 2100–2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Szabadkai G; Bianchi K; Várnai P; De Stefani D; Wieckowski MR; Cavagna D; Nagy AI; Balla T; Rizzuto R Chaperone-Mediated Coupling of Endoplasmic Reticulum and Mitochondrial Ca2+ Channels. J. Cell Biol. 2006, 175, 901–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Rizzuto R; Pinton P; Carrington W; Fay FS; Fogarty KE; Lifshitz LM; Tuft RA; Pozzan T Close Contacts with the Endoplasmic Reticulum as Determinants of Mitochondrial Ca2+ Responses. Science 1998, 280, 1763–1766. [DOI] [PubMed] [Google Scholar]
- (20).Kornmann B; Osman C; Walter P The Conserved GTPase Gem1 Regulates Endoplasmic Reticulum-Mitochondria Connections. Proc. Natl. Acad. Sci. 2011, 108, 14151–14156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Kornmann B; Currie E; Collins SR; Schuldiner M; Nunnari J; Weissman JS; Walter P An ER-Mitochondria Tethering Complex Revealed by a Synthetic Biology Screen. Science 2009, 325, 477–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Lewin TM; Van Horn CG; Krisans SK; Coleman RA Rat Liver Acyl-CoA Synthetase 4 Is a Peripheral-Membrane Protein Located in Two Distinct Subcellular Organelles, Peroxisomes, and Mitochondrial-Associated Membrane. Arch. Biochem. Biophys. 2002, 404, 263–270. [DOI] [PubMed] [Google Scholar]
- (23).Rusiñol AE; Cui Z; Chen MH; Vance JE A Unique Mitochondria-Associated Membrane Fraction from Rat Liver Has a High Capacity for Lipid Synthesis and Contains Pre-Golgi Secretory Proteins Including Nascent Lipoproteins. J. Biol. Chem. 1994, 269, 27494–27502. [PubMed] [Google Scholar]
- (24).Friedman JR; Lackner LL; West M; DiBenedetto JR; Nunnari J; Voeltz GK ER Tubules Mark Sites of Mitochondrial Division. Science 2011, 334, 358–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Murley A; Lackner LL; Osman C; West M; Voeltz GK; Walter P; Nunnari J ER-Associated Mitochondrial Division Links the Distribution of Mitochondria and Mitochondrial DNA in Yeast. elife 2013, 2013, e00422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).De Brito OM; Scorrano L Mitofusin 2 Tethers Endoplasmic Reticulum to Mitochondria. Nature 2008, 456, 605–610. [DOI] [PubMed] [Google Scholar]
- (27).Filadi R; Greotti E; Turacchio G; Luini A; Pozzan T; Pizzo P Mitofusin 2 Ablation Increases Endoplasmic Reticulum–mitochondria Coupling. Proc. Natl. Acad. Sci. 2015, 112, E2174–E2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Cosson P; Marchetti A; Ravazzola M; Orci L Mitofusin-2 Independent Juxtaposition of Endoplasmic Reticulum and Mitochondria: An Ultrastructural Study. PLoS One 2012, 7, e46293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Hirabayashi Y; Kwon SK; Paek H; Pernice WM; Paul MA; Lee J; Erfani P; Raczkowski A; Petrey DS; Pon LA; Polleux F ER-Mitochondria Tethering by PDZD8 Regulates Ca2+ Dynamics in Mammalian Neurons. Science 2017, 358, 623–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Smirnova E; Griparic L; Shurland DL; van der Bliek AM Dynamin-Related Protein Drp1 Is Required for Mitochondrial Division in Mammalian Cells. Mol. Biol. Cell 2001, 12, 2245–2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Henning MS; Stiedl P; Barry DS; McMahon R; Morham SG; Walsh D; Naghavi MH PDZD8 Is a Novel Moesin-Interacting Cytoskeletal Regulatory Protein That Suppresses Infection by Herpes Simplex Virus Type 1. Virology 2011, 415, 114–121. [DOI] [PubMed] [Google Scholar]
- (32).Hendrickson D; Kelley DR; Tenen D; Bernstein B; Rinn JL Widespread RNA Binding by Chromatin-Associated Proteins. Genome Biol. 2016, 17, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Sibley CR Individual Nucleotide Resolution UV Cross-Linking and Immunoprecipitation (ICLIP) to Determine Protein–RNA Interactions. Meth. Mol. Biol. 2018, 1649, 427–454. [DOI] [PubMed] [Google Scholar]
- (34).Garzia A; Morozov P; Sajek M; Meyer C; Tuschl T PAR-CLIP for Discovering Target Sites of RNA-Binding Proteins. Methods Mol. Biol. 2018, 1720, 55–75. [DOI] [PubMed] [Google Scholar]
- (35).Peabody DS The RNA Binding Site of Bacteriophage MS2 Coat Protein. EMBO J. 1993, 12, 595–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Cabantous S; Terwilliger TC; Waldo GS Protein Tagging and Detection with Engineered Self-Assembling Fragments of Green Fluorescent Protein. Nat. Biotechnol. 2005, 23, 102–107. [DOI] [PubMed] [Google Scholar]
- (37).Ghosh I; Hamilton AD; Regan L Antiparallel Leucine Zipper-Directed Protein Reassembly: Application to the Green Fluorescent Protein. J. Am. Chem. Soc. 2000, 122, 5658–5659. [Google Scholar]
- (38).Martell JD; Yamagata M; Deerinck TJ; Phan S; Kwa CG; Ellisman MH; Sanes JR; Ting AY A Split Horseradish Peroxidase for the Detection of Intercellular Protein–protein Interactions and Sensitive Visualization of Synapses. Nat. Biotechnol. 2016, 34, 774–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Pelletier JN; Campbell-Valois FX; Michnick SW Oligomerization Domain-Directed Reassembly of Active Dihydrofolate Reductase from Rationally Designed Fragments. Proc. Natl. Acad. Sci. 1998, 95, 12141–12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Johnsson N; Varshavsky A Split Ubiquitin as a Sensor of Protein Interactions in Vivo. Proc. Natl. Acad. Sci. U. S. A. 1994, 91, 10340–10344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Paulmurugant R; Gambhir SS Monitoring Protein-Protein Interactions Using Split Synthetic Renilla Luciferase Protein-Fragment-Assisted Complementation. Anal. Chem. 2003, 75, 1584–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Luker KE; Smith MCP; Luker GD; Gammon ST; Piwnica-Worms H; Piwnica-Worms D Kinetics of Regulated Protein-Protein Interactions Revealed with Firefly Luciferase Complementation Imaging in Cells and Living Animals. Proc. Natl. Acad. Sci. 2004, 101, 12288–12293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Ozawa T; Kaihara A; Sato M; Tachihara K; Umezawa Y Split Luciferase as an Optical Probe for Detecting Protein-Protein Interactions in Mammalian Cells Based on Protein Splicing. Anal. Chem. 2001, 73, 2516–2521. [DOI] [PubMed] [Google Scholar]
- (44).Dixon AS; Schwinn MK; Hall MP; Zimmerman K; Otto P; Lubben TH; Butler BL; Binkowski BF; Machleidt T; Kirkland TA; Wood MG; Eggers CT; Encell LP; Wood KV NanoLuc Complementation Reporter Optimized for Accurate Measurement of Protein Interactions in Cells. ACS Chem. Biol. 2016, 11, 400–408. [DOI] [PubMed] [Google Scholar]
- (45).Olson KR; Eglen RM β Galactosidase Complementation: A Cell-Based Luminescent Assay Platform for Drug Discovery. Assay Drug Dev. Technol. 2007, 5, 137–144. [DOI] [PubMed] [Google Scholar]
- (46).Broome AM; Bhavsar N; Ramamurthy G; Newton G; Basilion JP Expanding the Utility of β-Galactosidase Complementation: Piece by Piece. Mol. Pharm. 2010, 7, 60–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Ullmann A; Jacob F; Monod J Characterization by in Vitro Complementation of a Peptide Corresponding to an Operator-Proximal Segment of the β-Galactosidase Structural Gene of Escherichia Coli. J. Mol. Biol. 1967, 24, 339–343. [DOI] [PubMed] [Google Scholar]
- (48).Rossi F; Charlton CA; Blau HM Monitoring Protein-Protein Interactions in Intact Eukaryotic Cells by Beta-Galactosidase Complementation. Proc. Natl. Acad. Sci. U. S. A. 1997, 94, 8405–8410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Wehr MC; Laage R; Bolz U; Fischer TM; Grünewald S; Scheek S; Bach A; Nave KA; Rossner MJ Monitoring Regulated Protein-Protein Interactions Using Split TEV. Nat. Methods 2006, 3, 985–993. [DOI] [PubMed] [Google Scholar]
- (50).Wright AV; Sternberg SH; Taylor DW; Staahl BT; Bardales JA; Kornfeld JE; Doudna JA Rational Design of a Split-Cas9 Enzyme Complex. Proc. Natl. Acad. Sci. 2015, 112, 2984–2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Zetsche B; Volz SE; Zhang F A Split-Cas9 Architecture for Inducible Genome Editing and Transcription Modulation. Nat. Biotech. 2015, 33, 139–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Truong DJ; Kühner K; Kühn R; Werfel S; Engelhardt S; Wurst W; Ortiz O Development of an Intein-Mediated Split-Cas9 System for Gene Therapy. Nucleic Acids Res. 2015, 43, 6450–6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Nihongaki Y; Kawano F; Nakajima T; Sato M Photoactivatable CRISPR-Cas9 for Optogenetic Genome Editing. Nat. Biotechnol. 2015, 33, 755–760. [DOI] [PubMed] [Google Scholar]
- (54).Schopp IM; Amaya Ramirez CC; Debeljak J; Kreibich E; Skribbe M; Wild K; Béthune J Split-BioID a Conditional Proteomics Approach to Monitor the Composition of Spatiotemporally Defined Protein Complexes. Nat. Commun. 2017, 8, 15690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Sharp KH; Mewies M; Moody PCE; Raven EL Crystal Structure of the Ascorbate Peroxidase-Ascorbate Complex. Nat. Struct. Biol. 2003, 10, 303–307. [DOI] [PubMed] [Google Scholar]
- (56).Mohanty JG; Jaffe JS; Schulman ES; Raible DG A Highly Sensitive Fluorescent Micro-Assay of H2O2 Release from Activated Human Leukocytes Using a Dihydroxyphenoxazine Derivative. J. Immunol. Methods 1997, 202, 133–141. [DOI] [PubMed] [Google Scholar]
- (57).Zhou M; Diwu Z; Panchuk-Voloshina N; Haugland RP A Stable Nonfluorescent Derivative of Resorufin for the Fluorometric Determination of Trace Hydrogen Peroxide: Applications in Detecting the Activity of Phagocyte NADPH Oxidase and Other Oxidases. Anal. Biochem. 1997, 253, 162–168. [DOI] [PubMed] [Google Scholar]
- (58).Xue M; Hou J; Wang L; Cheng D; Lu J; Zheng L; Xu T Optimizing the Fragment Complementation of APEX2 for Detection of Specific Protein-Protein Interactions in Live Cells. Sci. Rep. 2017, 7, 9160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Morawski B; Lin Z; Cirino P; Joo H; Bandara G; Arnold FH Functional Expression of Horseradish Peroxidase in Saccharomyces Cerevisiae and Pichia Pastoris. Protein Eng. 2000, 13, 377–384. [DOI] [PubMed] [Google Scholar]
- (60).Arnold FH Design by Directed Evolution. Acc. Chem. Res. 1998, 31, 125–131. [Google Scholar]
- (61).Esvelt KM; Carlson JC; Liu DR A System for the Continuous Directed Evolution of Biomolecules. Nature 2011, 472, 499–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Boder ET; Wittrup KD Yeast Surface Display for Screening Combinatorial Polypeptide Libraries. Nat. Biotechnol. 1997, 15, 553–557. [DOI] [PubMed] [Google Scholar]
- (63).Branon TC; Bosch JA; Sanchez AD; Udeshi ND; Svinkina T; Carr SA; Feldman JL; Perrimon N; Ting AY Efficient Proximity Labeling in Living Cells and Organisms with TurboID. Nat. Biotechnol. 2018, 36, 880–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).O’Shea EK; Lumb KJ; Kim PS Peptide “Velcro”: Design of a Heterodimeric Coiled Coil. Curr. Biol. 1993, 3, 658–667. [DOI] [PubMed] [Google Scholar]
- (65).Kaewsapsak P; Shechner DM; Mallard W; Rinn JL; Ting AY Live-Cell Mapping of Organelle-Associated RNAs via Proximity Biotinylation Combined with Protein-RNA Crosslinking. eLife 2017, 6, e29224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Konermann S; Lotfy P; Brideau NJ; Oki J; Shokhirev MN; Hsu PD Transcriptome Engineering with RNA-Targeting Type VI-D CRISPR Effectors. Cell 2018, 173, 665–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Myers SA; Wright J; Peckner R; Kalish BT; Zhang F; Carr SA Discovery of Proteins Associated with a Predefined Genomic Locus via DCas9-APEX-Mediated Proximity Labeling. Nat. Methods 2018, 15, 437–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Gao XD; Tu LC; Mir A; Rodriguez T; Ding Y; Leszyk J; Dekker J; Shaffer SA; Zhu LJ; Wolfe SA; Sontheimer EJ C-BERST: Defining Subnuclear Proteomic Landscapes at Genomic Elements with DCas9-APEX2. Nat. Methods 2018, 15, 433–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Ramanathan M; Majzoub K; Rao DS; Neela PH; Zarnegar BJ; Mondal S; Roth JG; Gai H; Kovalski JR; Siprashvili Z; Palmer TD; Carette JE; Khavari PA RNA–protein Interaction Detection in Living Cells. Nat. Methods 2018, 15, 207–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Blackburn EH; Greider CW; Szostak JW Telomeres and Telomerase: The Path from Maize, Tetrahymena and Yeast to Human Cancer and Aging. Nat. Med. 2006, 12, 1133–1138. [DOI] [PubMed] [Google Scholar]
- (71).Zappulla DC; Cech TR RNA as a Flexible Scaffold for Proteins: Yeast Telomerase and Beyond. Cold Spring Harb. Symp. Quant. Biol. 2006, 71, 217–224. [DOI] [PubMed] [Google Scholar]
- (72).Fu D; Collins K Distinct Biogenesis Pathways for Human Telomerase RNA and H/ACA Small Nucleolar RNAs. Mol. Cell 2003, 11, 1361–1372. [DOI] [PubMed] [Google Scholar]
- (73).LeCuyer KA; Behlen LS; Uhlenbeck OC Mutants of the Bacteriophage MS2 Coat Protein That Alter Its Cooperative Binding to RNA. Biochemistry 1995, 34, 10600–10606. [DOI] [PubMed] [Google Scholar]
- (74).Chao JA; Patskovsky Y; Almo SC; Singer RH Structural Basis for the Coevolution of a Viral RNA-Protein Complex. Nat. Struct. Mol. Biol. 2008, 15, 103–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Lim F; Peabody DS RNA Recognition Site of PP7 Coat Protein. Nucleic Acids Res. 2002, 30, 4138–4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Theimer CA; Jády BE; Chim N; Richard P; Breece KE; Kiss T; Feigon J Structural and Functional Characterization of Human Telomerase RNA Processing and Cajal Body Localization Signals. Mol. Cell 2007, 27, 869–881. [DOI] [PubMed] [Google Scholar]
- (77).Snapp EL; Hegde RS; Francolini M; Lombardo F; Colombo S; Pedrazzini E; Borgese N; Lippincott-Schwartz J Formation of Stacked ER Cisternae by Low Affinity Protein Interactions. J. Cell Biol. 2003, 163, 257–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Magliery TJ; Wilson CGM; Pan W; Mishler D; Ghosh I; Hamilton AD; Regan L Detecting Protein-Protein Interactions with a Green Fluorescent Protein Fragment Reassembly Trap: Scope and Mechanism. J. Am. Chem. Soc. 2005, 127, 146–157. [DOI] [PubMed] [Google Scholar]
- (79).Nagai T; Ibata K; Park ES; Kubota M; Mikoshiba K; Miyawaki A A Variant of Yellow Fluorescent Protein with Fast and Efficient Maturation for Cell-Biological Applications. Nat. Biotechnol. 2002, 20, 87–90. [DOI] [PubMed] [Google Scholar]
- (80).Hu CD; Chinenov Y; Kerppola TK Visualization of Interactions among BZIP and Rel Family Proteins in Living Cells Using Bimolecular Fluorescence Complementation. Mol. Cell 2002, 9, 789–798. [DOI] [PubMed] [Google Scholar]
- (81).Chumakov SP; Kravchenko YE; Chumakov PM Protein Complementation as Tool for Studying Protein-Protein Interactions in Living Cells. Mol. Biol. 2012, 46, 627–638. [PubMed] [Google Scholar]
- (82).Horstman A; Tonaco IAN; Boutilier K; Immink RG A Cautionary Note on the Use of Split-YFP/BiFC in Plant Protein-Protein Interaction Studies. Int. J. Mol. Sci. 2014, 15, 9628–9643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Kopp F; Mendell JT Leading Edge Review Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 2018, 172, 393–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Pereira B; Billaud M; Almeida R RNA-Binding Proteins in Cancer: Old Players and New Actors. Trends Cancer 2017, 3, 506–528. [DOI] [PubMed] [Google Scholar]
- (85).Osborne RJ; Thornton CA RNA-Dominant Diseases. Hum. Mol. Genet. 2006, 15, 162–169. [DOI] [PubMed] [Google Scholar]
- (86).Lee N; Moss WN; Yario TA; Steitz JA EBV Noncoding RNA Binds Nascent RNA to Drive Host PAX5 to Viral DNA. Cell 2015, 160, 607–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (87).Chu C; Zhang QC; da Rocha ST; Flynn RA; Bharadwaj M; Calabrese JM; Magnuson T; Heard E; Chang HY Systematic Discovery of Xist RNA Binding Proteins. Cell 2015, 161, 404–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (88).McHugh CA; Chen CK; Chow A; Surka CF; Tran C; McDonel P; Pandya-Jones A; Blanco M; Burghard C; Moradian A; Sweredoski MJ, Shishkin AA; Su,; Lander ES; Hess S; Plath K; Guttman M The Xist LncRNA Interacts Directly with SHARP to Silence Transcription through HDAC3. Nature 2015, 521, 232–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (89).Minajigi A; Froberg JE; Wei C; Sunwoo H; Kesner B; Colognori D; Lessing D; Payer B; Boukhali M; Haas W; Lee JT A Comprehensive Xist Interactome Reveals Cohesin Repulsion and an RNA-Directed Chromosome Conformation. Science 2015, 349, 2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (90).Walker SC; Scott FH; Srisawat C; Engelke DR RNA Affinity Tags for the Rapid Purification and Investigation of RNAs and RNA-Protein Complexes. Methods Mol. Biol. 2008, 488, 23–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (91).Jazurek M; Ciesiolka A; Starega-Roslan J; Bilinska K; Krzyzosiak WJ Identifying Proteins That Bind to Specific RNAs - Focus on Simple Repeat Expansion Diseases. Nucleic Acids Res. 2016, 44, 9050–9070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (92).Walker SC; Good PD; Gipson TA; Engelke DR The Dual Use of RNA Aptamer Sequences for Affinity Purification and Localization Studies of RNAs and RNA-Protein Complexes. Methods Mol. Biol. 2011, 714, 423–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (93).Mili S; Steitz JA Evidence for Reassociation of RNA-Binding Proteins after Cell Lysis: Implications for the Interpretation of Immunoprecipitation Analyses. RNA 2004, 10, 1692–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (94).Hogg JR; Collins K RNA-Based Affinity Purification Reveals 7SK RNPs with Distinct Composition and Regulation. RNA 2007, 13, 868–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (95).Shechner DM; Hacisuleyman E; Younger ST; Rinn JL Multiplexable, Locus-Specific Targeting of Long RNAs with CRISPR-Display. Nat. Methods 2015, 12, 664–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (96).Geary C; Chworos A; Jaeger L Promoting RNA Helical Stacking via A-Minor Junctions. Nucleic Acids Res. 2011, 39, 1066–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (97).Emsley P; Lohkamp B; Scott WG; Cowtan K Features and Development of Coot. Acta Crystallogr. .D. Biol. Crystallogr. 2010, 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (98).Cate JH; Gooding AR; Podell E; Zhou K; Golden BL; Kundrot CE; Cech TR; Doudna JA Crystal Structure of a Group I Ribozyme Domain: Principles of RNA Packing. Science 1996, 273, 1678–1685. [DOI] [PubMed] [Google Scholar]
- (99).Grahn E; Moss T; Helgstrand C; Fridborg K; Sundaram M; Tars K; Lago H; Stonehouse NJ; Davis DR; Stockley PG; Lijas L Structural Basis of Pyrimidine Specificity in the MS2 RNA Hairpin-Coat-Protein Complex. RNA 2001, 7, 1616–1627. [PMC free article] [PubMed] [Google Scholar]
- (100).Zhao S; Fernald RD Comprehensive Algorithm for Quantitative Real-Time Polymerase Chain Reaction. J. Comput. Biol. 2005, 12, 1047–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (101).Martell JD; Deerinck TJ; Lam SS; Ellisman MH; Ting AY Electron Microscopy Using the Genetically Encoded APEX2 Tag in Cultured Mammalian Cells. Nat. Protoc. 2017, 12, 1792–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (102).Wang W; Wildes CP; Pattarabanjird T; Sanchez MI; Glober GF; Matthews GA; Tye KM; Ting AY A Light- and Calcium-Gated Transcription Factor for Imaging and Manipulating Activated Neurons. Nat. Biotechnol. 2017, 35, 864–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


