The Gcorn plant database was designed for the retrieval of information on homology and evolution of a plant gene of interest based on 2, 682, 261 amino acid sequences of 73 species.
Abstract
Gene homology helps us understand gene function and speciation. The number of plant genes and species registered in public databanks is continuously increasing. It is useful to associate homologous genes of various plants to better understand plant speciation. We designed the Gcorn plant database for the retrieval of information on homology and evolution of a plant gene of interest. Amino acid sequences of 73 species (62 land plants and 11 green algae), containing 2,682,261 sequences, were obtained from the National Center for Biotechnology Information (NCBI) Reference Sequence database. Based on NCBI BLAST searches between these sequences, homologous genes were grouped at various thresholds of homology indices devised by the authors. To show functional and evolutionary traits of a gene of interest, a phylogenetic tree, connecting genes with high homology indices, and line charts of the numbers of genes with various homology indices, are depicted. In addition, such indices are projected on a network graph in which species studied are connected based on the ratios of homologous genes, and on a phylogenetic tree for species based on NCBI Taxonomy. Gcorn plant provides information on homologous genes at various virtual time points along with speciation in plants.
Land plants originated from charophytes, came ashore as bryophytes, and then evolved as vascular plants including gymnosperms and angiosperms (Harrison, 2017). To adjust to the rather severe aspects of land environments relative to water environments, plant gene copy numbers have amplified over time, and genes have diversified after duplication (Zhang, 2003; Rensing et al., 2008; Hori et al., 2014). Tracking such diversification is useful for understanding evolutionary pathways. Genes originating from a common ancestor that diverged after a speciation event are named “orthologs” (Ambrosino and Chiusano, 2017). Conversely, genes that are advantageous to the survival of a certain species and were fortuitously duplicated then adapted to various biological events are named “paralogs.” Although paralogous genes initially shared the same function, they are more likely to have evolved different functions because of purifying selection (Ullah et al., 2015). Therefore, orthologous and paralogous events help us understand the functionality of plant genes as well as the phylogenetic timing of speciation. By comparing a phylogenetic tree for each gene with a phylogenetic tree for species, we can estimate relationships and/or divergence between speciation and gene selection more precisely. Wang et al. (2018) revealed functional divergence in a particular gene family. If such information is provided at the genome level, it is useful as a hypothetical basis for further experimental/theoretical studies to elucidate the evolutionary contexts of gene function and speciation.
There are several databases of plant homologous genes at the genome level, such as PLAZA (containing 3,065,012 genes and 71 plants; Van Bel et al., 2018), PlantOrDB (1,291,670 genes and 41 plants; Li et al., 2015), HomoloGene of the National Center for Biotechnology Information (NCBI; Arabidopsis [Arabidopsis thaliana] and rice [Oryza sativa] as model plants; NCBI Resource Coordinators, 2016), Protein Clusters of NCBI (23 plants; Klimke et al., 2009; NCBI Resource Coordinators, 2016), OrthoDB (31 plants; Zdobnov et al., 2017), and InParanoid (19 plants; Sonnhammer and Östlund, 2015). Although these databases are periodically updated, the number of plant species they contain is fewer than those covered by our preliminary search in 2016 (62 land plants and 11 green algae) except for PLAZA. PLAZA provides many types of information on plant genes, and contains a function to depict a phylogenetic tree for a gene group that contains a plant gene of interest. Although the tree is adjustable in its size and types of information and is helpful for collecting a group of genes homologous to the gene of interest, it is difficult to understand evolutionary and functional aspects of a gene of interest because of the size of the gene group; i.e. in some cases, the trees are very large (e.g. “AT2G37040,” Arabidopsis gene), and in other cases, trees for genes of interests are not depicted (e.g. “AT1G56650,” Arabidopsis gene). PlantOrDB provides two types of phylogenetic trees, a type for genes and a type for species, which show composite aspects of gene evolution. In the database, however, one gene is contained in a single (static) gene group, which can only represent a certain grouping at a single time point along with the evolutionary history of plants. For a deeper grasp of relationships of gene function and evolution, information on gene group dynamics at various time points is useful.
Here, we developed the Gcorn plant database for retrieving functional and evolutionary traits of plant genes. Amino acid sequences and functional descriptions of plant genes from the Reference Sequence (RefSeq) database (O’Leary et al., 2016) of NCBI were obtained and gene homology was analyzed on the basis of BLASTp analysis between these genes. The database is designed to provide gene group dynamics that vary at implicit time points along with plant speciation by using various thresholds of homology indices. Gcorn plant is one of the leading databases for the Gcorn project, which aims to reveal relationships and/or divergence in evolution between genes and species for whole organisms.
CONSTRUCTION
Schema
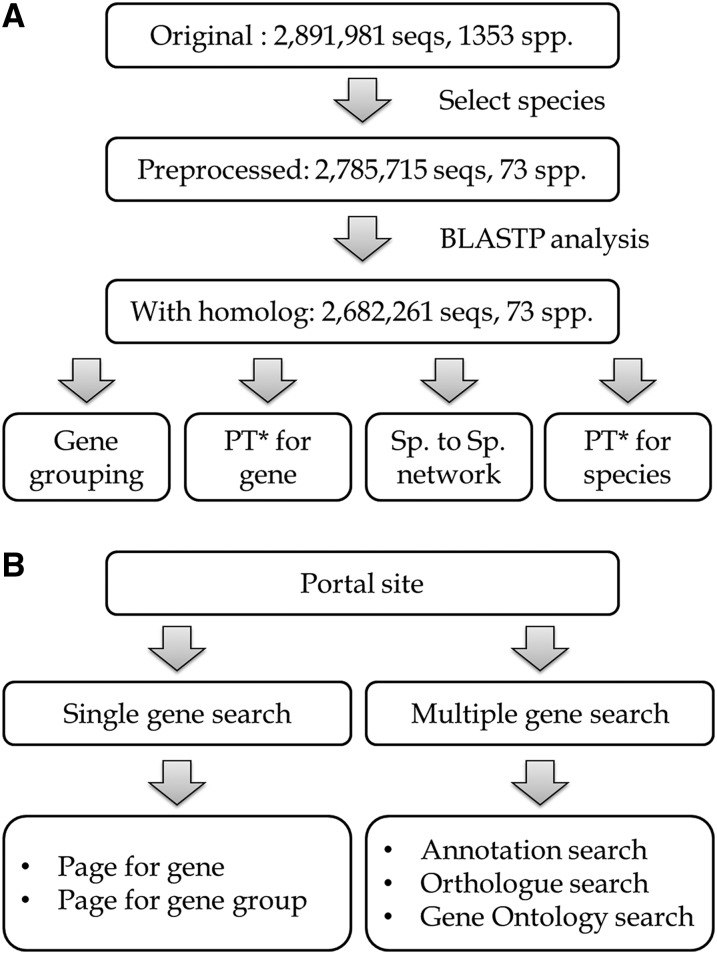
The Gcorn plant database was constructed according to the flowchart in Figure 1. Amino acid sequences of plant genes were obtained from the RefSeq database of NCBI and then used for BLASTp analysis (Fig. 1A). Groups of genes showing a high homology index were selected with various homology indices (see the “Quality Control of Gene Homology”). Several phylogenetic charts can be depicted for each gene (Fig. 1B). Information on the phylogeny of each gene was published in an on-line database.
Figure 1.
A and B, Flowchart of the Gcorn plant database construction (A) and database retrieval of gene(s) of interest (B). PT*, phylogenetic tree.
Implementation
Homology analysis between genes was performed by using the Protein BLAST (BLASTp) program provided by NCBI, and other data processing was executed by the Perl (www.perl.org) programs scripted by the authors. In the Gcorn plant database, Common Gateway Interface scripts based on Perl were adopted for displaying a webpage for a gene or a gene group.
Data Source
FASTA- and GPFF-formatted files of plant genes were obtained from RefSeq (O’Leary et al., 2016) in NCBI in September 2016. At the time of downloading these files, 108 files had been published in each file type. These files contain 2,891,981 sequences from 1353 plants (mainly species). However, there was variation in the numbers of genes contained in individual plants. Therefore, 73 plants for which genome-level genes (i.e. >5,000 genes) were registered were selected for the Gcorn plant database (Table 1). In these plants, 2,785,715 sequences were included (96.3% of the original sequences).
Table 1. Summary of plants used for the Gcorn plant database.
| Species | Sequences | Species | Sequences | Species | Sequences |
|---|---|---|---|---|---|
| Amborella trichopoda | 20382 | Erythranthe guttata | 31862 | Phoenix dactylifera | 38570 |
| Arabidopsis lyrata subsp. lyrata | 32549 | Eucalyptus grandis | 47427 | Physcomitrella patens | 35934 |
| Arabidopsis thaliana | 35374 | Eutrema salsugineum | 29572 | Populus euphratica | 49778 |
| Arachis duranensis | 42563 | Fragaria vesca subsp. vesca | 31354 | Populus trichocarpa | 45942 |
| Arachis ipaensis | 46410 | Glycine max | 71677 | Prunus mume | 29712 |
| Auxenochlorella protothecoides | 7131 | Gossypium arboreum | 47582 | Prunus persica | 29012 |
| Bathycoccus prasinos | 7892 | Gossypium hirsutum | 91198 | Pyrus x bretschneideri | 46190 |
| Beta vulgaris subsp. vulgaris | 31285 | Gossypium raimondii | 59097 | Ricinus communis | 28059 |
| Brachypodium distachyon | 33950 | Jatropha curcas | 28857 | Selaginella moellendorffii | 34817 |
| Brassica napus | 113061 | Malus domestica | 60650 | Sesamum indicum | 33095 |
| Brassica oleracea var oleracea | 56610 | Medicago truncatula | 57693 | Setaria italica | 32964 |
| Brassica rapa | 51063 | Micromonas commoda | 10140 | Solanum lycopersicum | 36213 |
| Camelina sativa | 106361 | Micromonas pusilla CCMP1545 | 10242 | Solanum pennellii | 35077 |
| Capsella rubella | 28797 | Monoraphidium neglectum | 16755 | Solanum tuberosum | 38059 |
| Capsicum annuum | 45478 | Morus notabilis | 27048 | Sorghum bicolor | 33005 |
| Chlamydomonas reinhardtii | 14489 | Musa acuminata subsp. malaccensis | 41734 | Tarenaya hassleriana | 40658 |
| Chlorella variabilis | 9892 | Nelumbo nucifera | 39014 | Theobroma cacao | 44263 |
| Cicer arietinum | 33117 | Nicotiana sylvestris | 48210 | Vigna angularis | 37771 |
| Citrus clementina | 34557 | Nicotiana tabacum | 84630 | Vigna radiata var radiata | 34973 |
| Citrus sinensis | 35654 | Nicotiana tomentosiformis | 45611 | Vitis vinifera | 38136 |
| Coccomyxa subellipsoidea C-169 | 9950 | Oryza brachyantha | 26886 | Volvox carteri f. nagariensis | 14436 |
| Cucumis melo | 29717 | Oryza sativa Japonica Group | 41070 | Zea mays | 58565 |
| Cucumis sativus | 25711 | Ostreococcus lucimarinus CCE9901 | 7603 | Ziziphus jujuba | 37666 |
| Daucus carota subsp. sativus | 44575 | Ostreococcus tauri | 7994 | ||
| Elaeis guineensis | 39543 | Phaseolus vulgaris | 32803 |
BLASTp Analysis
Using the FASTA files of plants studied, pairwise gene homologies were analyzed using the BLASTp program published by NCBI (Camacho et al., 2009). In the analysis, version 2.2.30 of the BLAST+ program was used, and the default E value was used because the index mentioned in the next section was used in the cutoff for determining gene homology. For stable execution of the analyses, each BLAST search was performed by species.
Quality Control of Gene Homology
In the resultant files from the BLAST searches, homology indices (HIs) between genes (e.g. genes A and B) were calculated using the equation
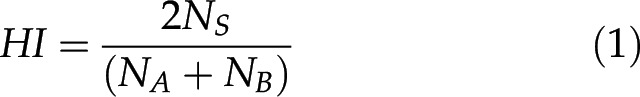 |
where NS represents the number of bases that are shared in both genes, and NA and NB represent the numbers of bases of genes A and B, respectively. This index is coincident to F-measure (harmonic mean of precision and recall indices) in the field of information retrieval. Based on our preliminary research, results with HIs <0.3 were eliminated for containing a considerable amount of random noise.
After the calculation, there were 2,682,261 sequences (92.7% of the original sequences) that were homologous to other genes. Genes with no result from the BLAST search were determined as singletons in the meaning of homology.
Detection of Homologous Gene Groups
Homologous gene groups with various thresholds of HIs were detected. In general, a gene belongs to multiple gene groups detected with different thresholds of HIs; i.e. each gene group hypothetically represents a gene state of a common progenitor as HIs decrease. For each group, the numbers of genes, species, and families were counted. In total, 2,358,763 gene groups were detected and their types of homology were determined; i.e. paralogous or orthologous.
Construction of Phylogenetic Trees for Genes
Phylogenetic trees of individual genes were constructed based on the HI values in a bottom-up manner; i.e. gene pairs with greater HIs were connected before the other pairs. Although this procedure is somewhat similar to the Unweighted Pair Group Method with Arithmetic Mean (UPGMA; Sokal and Michener, 1958), there is a difference when connecting between a single gene and a group of genes; our procedure adopts a strategy in which the single gene was connected to the group on the basis of the maximum HI value, whereas UPGMA uses the average value in that situation. We compared the methods to construct a phylogenetic tree in the Gcorn plant database with UPGMA using a particular gene (“NP_176057,” an Arabidopsis gene), and as a result, this method is equivalent (or slightly better in this case) to that with UPGMA by calculating Robinson-Foulds distances (Robinson and Foulds, 1981) of the trees depicted using the software ClustalW (Supplemental Tables S1 and S2). Genes analyzed were vertically aligned, based on the hypothesis that the mutation rate of each gene pair correlates to evolutionary time lapse.
Construction of the Species-Species Network Based on Gene Homology
Correlation indices (CIs) of gene homology between plants (e.g. plants “C” and “D”) were calculated using the equation
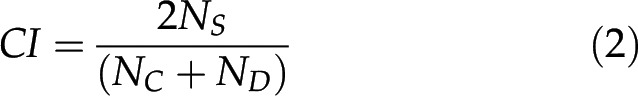 |
where NS represents the number of genes homologous to each other, and NC and ND represent the numbers of genes contained in their genomes, respectively. This index is also coincident with F-measure, similar to HI. A correlation network was depicted based on a threshold of the CI index, which was empirically set at 0.56 so that the plants studied were well separated into clusters and multiple species in one family were not divided into different clusters.
Construction of the Phylogenetic Tree for Species
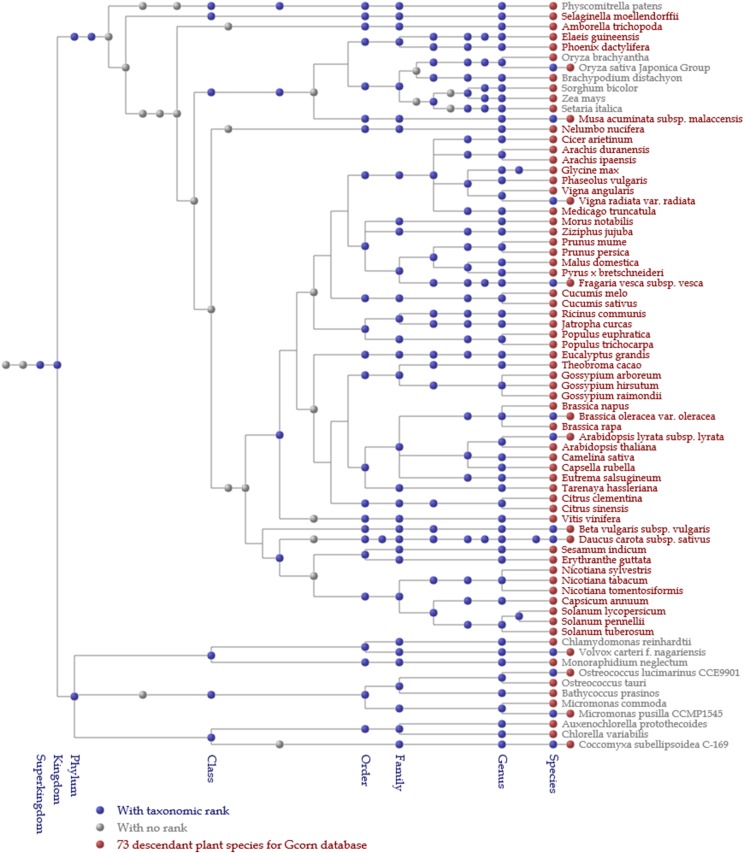
A phylogenetic tree for 73 plants was constructed based on relationships provided by the NCBI Taxonomy database (“nodes.dmp”). When a twig was trifurcated or more, the branching was alleviated using information on the taxonomy of the Angiosperm Phylogeny Group (2009). The top (leftmost) stratum of the phylogenetic tree corresponds to “Eucaryota,” a superkingdom level, whereas the bottom (rightmost) stratum of the tree is the subspecies, varietas, or forma level. There are 30 taxonomic levels such as kingdom, phylum, class, order, family, genus, and species.
User Interface
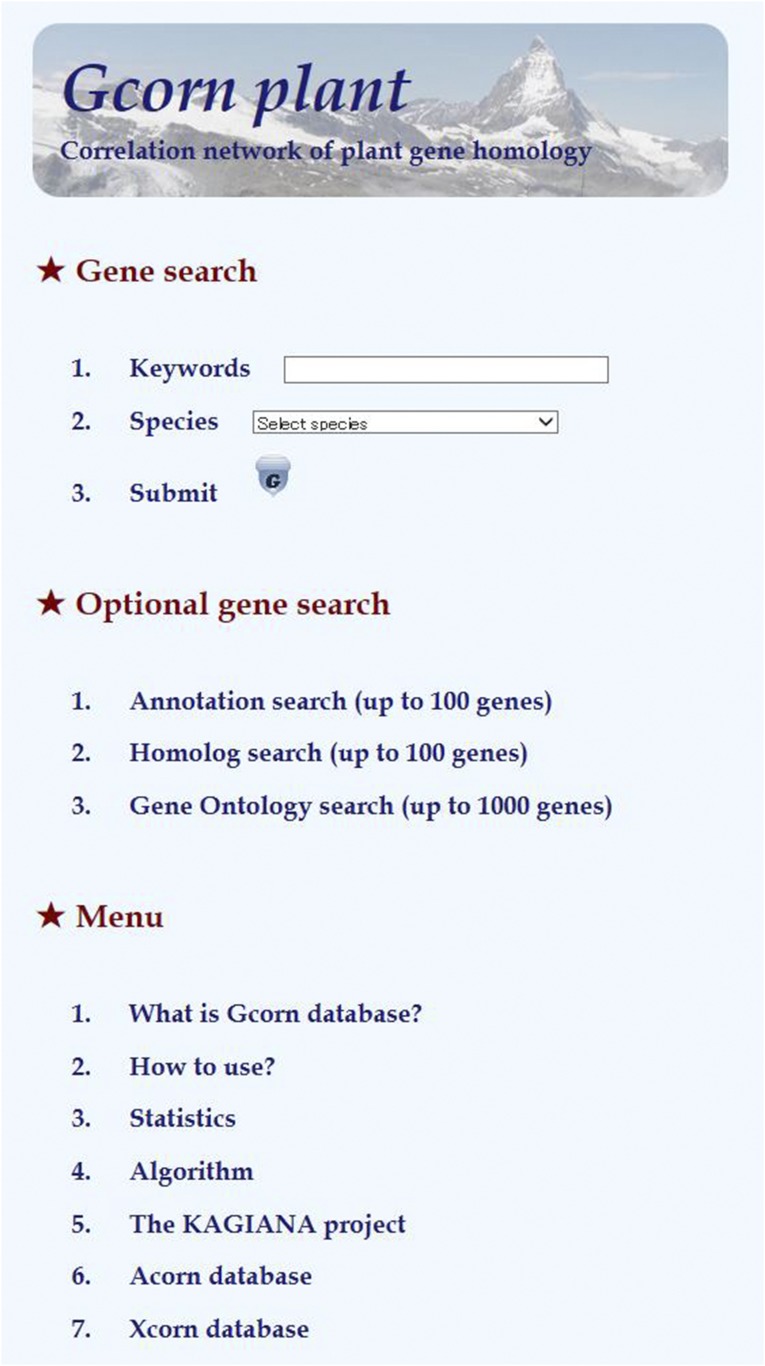
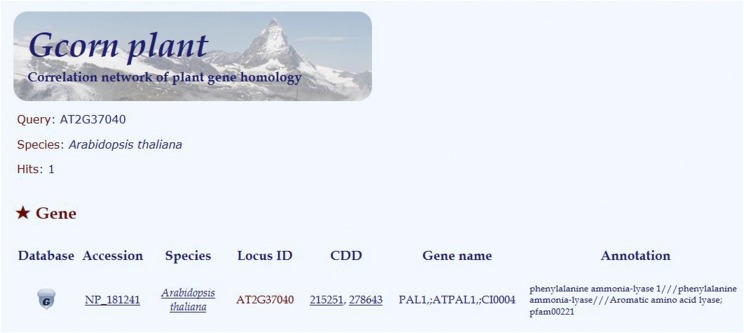
The Gcorn plant database is web-based , available on common internet browsers such as Microsoft Internet Explorer, Mozilla Firefox, and Google Chrome. In the portal, for retrieving information on a single gene, a user is required just to input a term such as a gene identifier or a gene name, selecting a plant using the pull-down menu of a list of plants studied, and then clicking the submit button (Fig. 2). In the subsequent display (Fig. 3), the user is required only to select a gene of interest from the table of candidate genes and then click on the “G” button on the line of the selected gene.
Figure 2.
Portal site of the Gcorn plant database. For a single gene search, there are just three steps: inputting a keyword (or two keywords), selecting a species from the pulldown menu, and pushing the submit button. For a multiple gene search, the database provides three items: searches for gene annotation, orthologous genes, and Gene Ontology terms.
Figure 3.
Webpage for selecting a candidate gene from the table named “Gene.” After selecting a gene of interest, just click the “G” button on the line of the gene.
DISCUSSION
Case Study
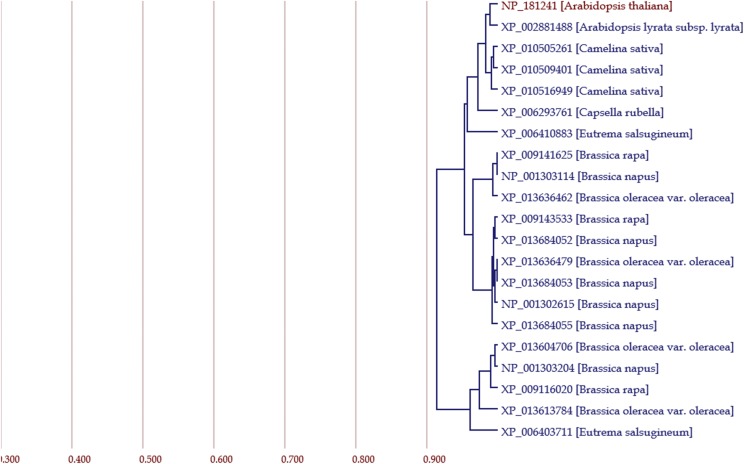
Figures 4–7 show the results of an Arabidopsis gene named “AT2G37040” as its locus code (Arabidopsis Genome Initiative, 2000).
Figure 4.
A phylogenetic tree for AT2G37040, an Arabidopsis gene. The tree contains a gene of interest and 20 genes with the highest HIs. The horizontal axis = HI. A RefSeq identifier of each gene is hyperlinked to its webpage in RefSeq of NCBI.
Figure 7.
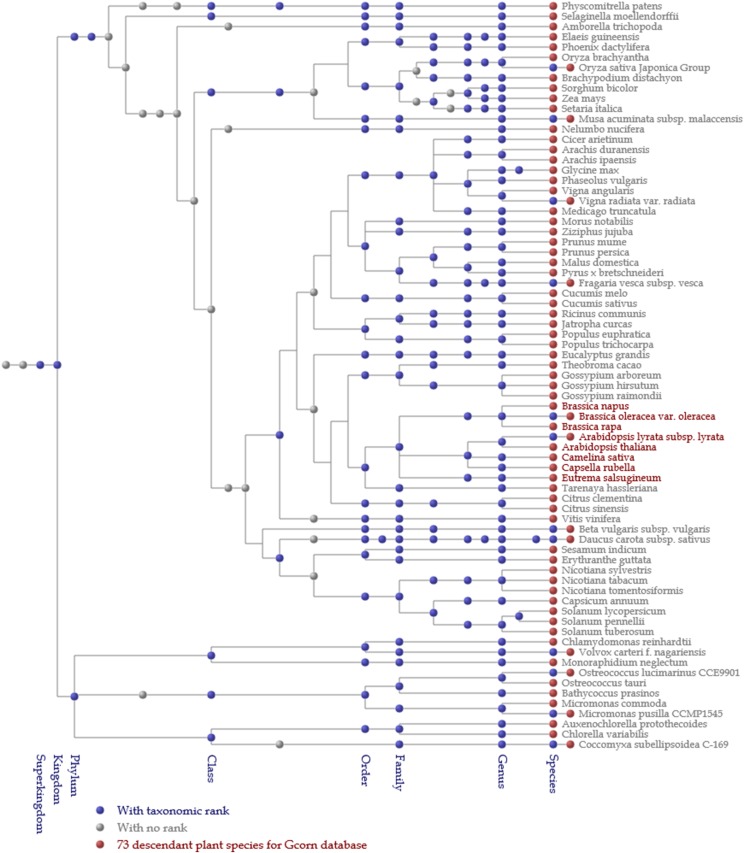
A phylogenetic tree for species for AT2G37040, an Arabidopsis gene. The tree was constructed mainly based on the NCBI Taxonomy database and, in the case of trifurcation or higher branching, twigs were branched based on the Angiosperm Phylogeny Group. Species containing genes homologous to the gene of interest are described in red, otherwise in gray at a threshold of 0.8 of HI. Each node is hyperlinked to its webpage in the NCBI Taxonomy page.
A phylogenetic tree of genes was depicted for the gene of interest with the other 20 genes showing the highest HIs to the gene (Fig. 4). According to the tree, there were three paralogous events that presumably occurred in a group of three genes (Camelina sativaXP_010505261, XP_010509401, and XP_010516949), between two groups of Brassica genes (XP_009141625, XP_001303114, and XP_013636462 for one group and XP_009143533, XP_013684052, XP_013636479, XP_013684053, NP_001302615, and XP_013684055 for another group), and between a group of the upper 16 genes and another group of the lower five genes. In the tree, other homologous events are estimated as orthologous. The HI values between all pairs in the tree were all >0.9, indicating that the amino acid sequences of these genes are quite similar to each other, and thus their functions are also conserved.
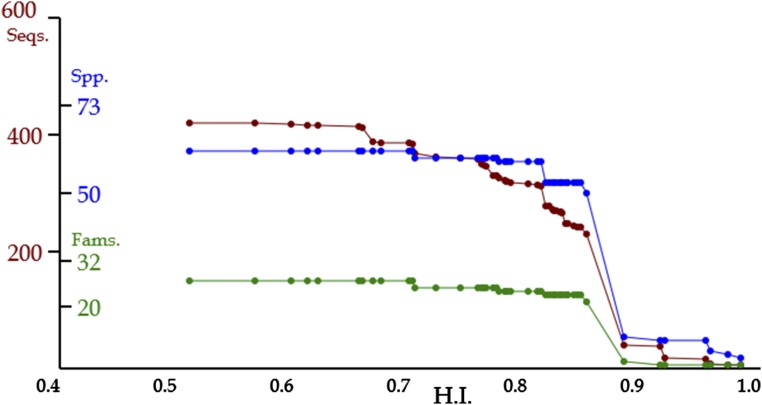
In Figure 5, the red line represents the number of genes (sequences) with various thresholds of HIs along with evolutionary time and the blue and green lines represent the numbers of species and families containing these genes, respectively. From the left to the right along with evolutionary time, the numbers of genes were 229 at 0.851 of HI, 37 at 0.914, 16 at 0.953, and then five at 0.983. Because the decrease of the gene number represents the occurrence of a homologous event, the timing after the gene group with 0.851 of HI represents a major homologous event. With that timing, the blue line shows such a strict decrease that this event is presumably orthologous. On the other hand, in the timing between 0.668 and 0.656 of HIs, the species number showed no decrease despite the decrease of the gene number. This indicates that the event is estimated as paralogous. In the case of this event, the number of species was 62, in accordance with the number of all land plants studied. Therefore, the paralogous event hypothetically occurred in the last common ancestor of all land plants.
Figure 5.
Line charts for a lineage of gene groups for AT2G37040, an Arabidopsis gene. Red, blue, and green lines represent the numbers of genes (sequences), species, and families, respectively, contained in individual gene groups. The horizontal axis = HI. At the time of an orthologous event, the numbers of genes and species decrease. At the time of a paralogous event, in contrast, the number of genes decreases, but the number of species is retained.
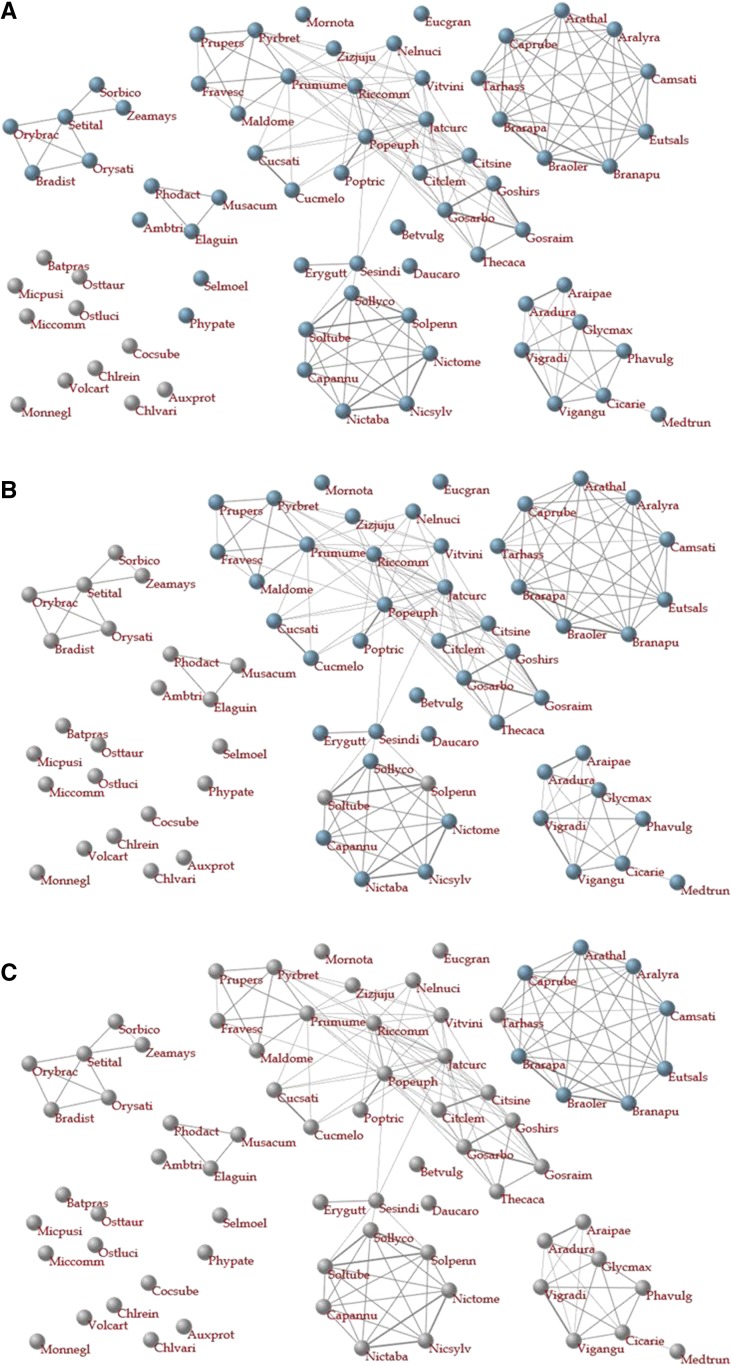
Figure 6 shows a network in which nodes represent species and are connected to other species based on a threshold of CIs. In the network, blue nodes represent species containing gene(s) homologous to the gene of interest (i.e. AT2G37040) at thresholds of 0.6 (Fig. 6A), 0.8 (Fig. 6B), and 0.9 (Fig. 6C), respectively. According to these projections of HI values on the network, genes homologous to the gene of interest are shared in all land plants studied, and genes with relatively greater HIs to the gene are concentrated in Brassicaceae.
Figure 6.
Species-species networks for AT2G37040, an Arabidopsis gene. In these networks, nodes represent species and are connected to other species based on the ratio of genes homologous to each other at a threshold of 0.56, because in networks with this threshold, species belonging to single families are well grouped such as Brassicaceae, Leguminosae, and Poaceae. Blue and gray nodes represent species with and without genes homologous to a gene of interest, respectively. A–C, The networks, in which nodes are colored, are depicted using different thresholds of 0.6 (A), 0.8 (B), and 0.9 (C). In (A), all land plants are in blue and, in (C), only plants of the Brassicaceae are in blue.
The phylogenetic tree for species (Fig. 7) was depicted based on the Taxonomy database of NCBI. In Figure 7, each node corresponds to certain NCBI taxa. Blue and gray nodes represent taxa with and without taxonomic ranks, respectively. Red nodes represent 73 plant species studied for the Gcorn plant database, whereas the species (or subspecies) names shown in red represent species containing genes homologous to the gene of interest. According to Figure 7 (0.8 or greater of HI), genes homologous to the gene of interest emerged only in Brassicaceae. In Figure 8 (the case of AT2G03720 with 0.4 of HI), however, species shown in red appeared in most major (order-level) clades of angiosperms with some exceptions (e.g. Poales and Amborellales), indicating that the amino acids of these homologous genes could be presumably mutated in several lineages such as Poales more frequently than in the other angiosperms, independent of speciation.
Figure 8.
A phylogenetic tree for species for AT2G03720, an Arabidopsis gene. The tree was depicted at a threshold of 0.45 of HI. Dissimilar to that for AT2G37040 (Fig. 7), species in red are dispersed in the range of land plants, indicating the difference in evolutionary time between speciation and amino acid mutation.
The Gcorn plant database allows a user to know several gene (amino acid)-level evolutionary events such as the timing of a homologous gene duplication event and trends among speciation and amino acid sequence diversification of plant genes. Such information promotes the understanding of relationships and/or divergence between gene function and speciation.
Comparison with Other Databases
There are several databases on plant homologous genes at the genome level, such as PlantOrDB (Li et al., 2015), HomoloGene of NCBI (NCBI Resource Coordinators, 2016), Protein Clusters of NCBI (Klimke et al., 2009; NCBI Resource Coordinators, 2016), OrthoDB (Zdobnov et al., 2017), and InParanoid (Sonnhammer and Östlund, 2015). As shown in Table 1, the number of plant species and genes (or subspecies) contained in the Gcorn plant database is greater than those in these databases. For a simple comparison to discuss gene function and evolution with these databases, the gene introduced in the previous section (i.e. AT2G37040) was referred to in these databases.
HomoloGene provides a simple viewer to visualize homologous genes throughout all organisms. In the case of the reference gene, it displayed 13 homologous genes (11 of plants and two of fungi). However, the database contains only two plant species, and thus it is insufficient to discuss relationships in gene function and evolution of plant genes.
Protein Clusters provides a function for multiple alignments of homologous genes of interest. It displayed 91 homologous genes from 12 plants out of 23 species. By selecting genes and clicking the “Multiple Alignment” link, a user can obtain multiple alignments of the genes. The webpages for the reference gene contain information on basic statistics and a table of the homologous genes, buttheviewer introduced by Klimke et al. (2009) is not avalable. Therefore, such information is insufficient for discussing the relationship between gene function and evolution.
OrthoDB provides detailed information on homologous genes throughout the Eucaryota (including 31 plants), bacteria, archaea, and viruses. For the reference gene, it showed 664 genes in 347 species. Although the number of species is sufficient for discussing gene function and evolution, there is no viewer for summarizing a group containing a lot of genes in some properties (e.g. species classification). Therefore, it is difficult to discuss evolutionary traits of gene function.
InParanoid provides a viewer of the functional features of genes of 19 plants. For the reference gene, it displayed 132 pairs of homologous gene clusters between pairs of organisms. Although the results are useful for retrieving homologous genes in other species, it is insufficient for discussing gene function and evolution.
PlantOrDB contains 1,291,670 genes from 41 plants and provides good viewers for discussing gene function and evolution. Two types of phylogenetic trees for species and for genes are provided. For the reference gene, it showed 278 genes (sequences) of 35 plant species. According to the phylogenetic tree for species, genes homologous to the reference gene are found in land plants, but not in green algae. The phylogenetic tree for genes revealed whether homologous events for these genes were paralogous or orthologous, providing information on gene evolution. However, the homologous genes are fixed as a single (static) gene group, and thus it is difficult to grasp gene groups along with evolutionary time. Moreover, because a single (fixed) resolution for gene homology is used for the tree, it is hard to overview the group containing a lot of genes with respect to gene evolution.
Additionally, although these databases provide amino acid sequences of genes, RefSeq protein identifiers such as NP_181241 for AT2G37040 are not available at the databases except for HomoloGene and Protein Clusters provided by NCBI, which makes identification and retrieval of the sequences difficult.
The Gcorn plant database provides two levels of information on gene evolution; i.e. one is a phylogenetic tree for a gene of interest with 20 genes showing the greatest HIs and the other is line charts of the numbers of genes, species, and families contained in homologous gene groups with various thresholds of HI. These viewers are designed for a simpler overview of gene evolution in the relatively near past and remote past, respectively.
Additional Function
The Gcorn plant database provides information on the functions of multiple genes. Using an annotation search, homolog search, and Gene Ontology search for multiple genes of interest, annotated descriptions of gene function, homologous genes in the other species, and Gene Ontology terms, respectively, are available.
Future Development
In the Gcorn project, analyses of fungi and protozoa have been completed, and their databases are under construction. For the next category, analysis of invertebrates will be performed. At present, the RefSeq database provides information on plant genes in which 94 plants contain genes at the genome level. Therefore, we are ready to reanalyze data of plants and update the Gcorn plant database.
Furthermore, we plan to search homologous genes between multiple categories of organisms such as plants and fungi. For instance, according to the genome of a bat obtained from the Genome database of NCBI, the genome contains two genes showing high homology to those in Brassicaceae. Although these genes can be presumably artificial or contaminated, some pairs of homologous genes can coexist over categories of organisms.
CONCLUSION
The Gcorn plant database is a web-based database for retrieving and grasping relationships in gene function and evolution in 73 plants. It provides information on homologous genes at various virtual time points along with speciation in plants. The Gcorn plant database is available at http://www.plant.osakafu-u.ac.jp/∼kagiana/gcorn/p/.
Supplemental Data
The following supplemental materials are available.
Supplemental Table S1. Genes used for the comparison between phylogenetic trees.
Supplemental Table S2. Comparisons of phylogenetic tress based on the algorithms.
Acknowledgments
We are grateful to Kotaro Ishizaka and the class of Plant Bioscience Data Processing, School of Life and Environmental Sciences, Osaka Prefecture University for bug reporting of Gcorn.
Footnotes
This project was partly supported by the Promotion of Dissimilar Field Collaboration Research at Osaka Prefecture University (grant no. 0203041700) and by the Ministry of Education, Culture, Sports, Science, and Technology of Japan Grants-in-Aid for Scientific Research (MEXT grant no. 17HP8034 to Y.O.).
Articles can be viewed without a subscription.
References
- Ambrosino L, Chiusano ML (2017) Transcriptologs: A transcriptome-based approach to predict orthology relationships. Bioinform Biol Insights 11: 1177932217690136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angiosperm Phylogeny Group (2009) An update of the angiosperm phylogeny group classification for the orders and families of flowering plants: APG III. Bot J Linn Soc 161: 105–121 [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL (2009) BLAST+: architecture and applications. BMC Bioinformatics 10: 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison CJ. (2017) Development and genetics in the evolution of land plant body plans. Philos Trans R Soc Lond B Biol Sci 372: 20150490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori K, Maruyama F, Fujisawa T, Togashi T, Yamamoto N, Seo M, Sato S, Yamada T, Mori H, Tajima N, et al. (2014) Klebsormidium flaccidum genome reveals primary factors for plant terrestrial adaptation. Nat Commun 5: 3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimke W, Agarwala R, Badretdin A, Chetvernin S, Ciufo S, Fedorov B, Kiryutin B, O’Neill K, Resch W, Resenchuk S, et al. (2009) The National Center for Biotechnology Information’s Protein Clusters Database. Nucleic Acids Res 37: D216–D223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Ji G, Ye C, Shu C, Zhang J, Liang C (2015) PlantOrDB: A genome-wide ortholog database for land plants and green algae. BMC Plant Biol 15: 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NCBI Resource Coordinators (2016) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 44(D1): D7–D19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, Rajput B, Robbertse B, Smith-White B, Ako-Adjei D, et al. (2016) Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res 44(D1): D733–D745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing SA, Lang D, Zimmer AD, Terry A, Salamov A, Shapiro H, Nishiyama T, Perroud PF, Lindquist EA, Kamisugi Y, et al. (2008) The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319: 64–69 [DOI] [PubMed] [Google Scholar]
- Robinson DF, Foulds LR (1981) Comparison of phylogenetic trees. Math Biosci 53: 131–147 [Google Scholar]
- Sokal R, Michener C (1958) A statistical method for evaluating systematic relationships. Univ Kans Sci Bull 38: 1409–1438 [Google Scholar]
- Sonnhammer EL, Östlund G (2015) InParanoid 8: Orthology analysis between 273 proteomes, mostly eukaryotic. Nucleic Acids Res 43: D234–D239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah I, Sjöstrand J, Andersson P, Sennblad B, Lagergren J (2015) Integrating sequence evolution into probabilistic orthology analysis. Syst Biol 64: 969–982 [DOI] [PubMed] [Google Scholar]
- Van Bel M, Diels T, Vancaester E, Kreft L, Botzki A, Van de Peer Y, Coppens F, Vandepoele K (2018) PLAZA 4.0: An integrative resource for functional, evolutionary and comparative plant genomics. Nucleic Acids Res 46(D1): D1190–D1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Shi X, Chen S, Ma C, Xu S (2018) Evolutionary origin, gradual accumulation and functional divergence of heat shock factor gene family with plant evolution. Front Plant Sci 9: 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdobnov EM, Tegenfeldt F, Kuznetsov D, Waterhouse RM, Simão FA, Ioannidis P, Seppey M, Loetscher A, Kriventseva EV (2017) OrthoDB v9.1: Cataloging evolutionary and functional annotations for animal, fungal, plant, archaeal, bacterial and viral orthologs. Nucleic Acids Res 45(D1): D744–D749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. (2003) Evolution by gene duplication: An update. Trends Ecol Evol 18: 292–298 [Google Scholar]