Pinstatic acid is an inactive auxin analog for SCFTIR1/AFB core auxin regulatory pathway and positively modulates auxin efflux transport by inhibiting PIN internalization.
Abstract
Polar auxin transport plays a pivotal role in plant growth and development. PIN-FORMED (PIN) auxin efflux carriers regulate directional auxin movement by establishing local auxin maxima, minima, and gradients that drive multiple developmental processes and responses to environmental signals. Auxin has been proposed to modulate its own transport by regulating subcellular PIN trafficking via processes such as clathrin-mediated PIN endocytosis and constitutive recycling. Here, we further investigated the mechanisms by which auxin affects PIN trafficking by screening auxin analogs and identified pinstatic acid (PISA) as a positive modulator of polar auxin transport in Arabidopsis (Arabidopsis thaliana). PISA had an auxin-like effect on hypocotyl elongation and adventitious root formation via positive regulation of auxin transport. PISA did not activate SCFTIR1/AFB signaling and yet induced PIN accumulation at the cell surface by inhibiting PIN internalization from the plasma membrane. This work demonstrates PISA to be a promising chemical tool to dissect the regulatory mechanisms behind subcellular PIN trafficking and auxin transport.
The plant hormone auxin is a master regulator of plant growth and development. Indole 3-acetic acid (IAA), the predominant natural auxin, regulates numerous and diverse developmental processes such as establishment of embryo polarity, vascular differentiation, apical dominance, and tropic responses to light and gravity (Hayashi, 2012). The auxin responses regulating these diverse developmental events can be modulated at three major steps: auxin metabolism (Korasick et al., 2013; Kasahara, 2016), directional auxin transport (Adamowski and Friml, 2015), and signal transduction (Leyser, 2018).
Polar auxin transport plays a crucial role in auxin-regulated development by influencing local auxin maxima and gradients and is mediated principally by three families of membrane proteins, the Auxin1/Like Aux1 (AUX1/LAX) auxin influx carriers, the PIN-FORMED (PIN) auxin efflux facilitators, and several members of the ATP-binding cassette group B auxin transporters (Adamowski and Friml, 2015).
The polar subcellular localization of the auxin efflux machinery determines the directionality of auxin flow. The spatiotemporal regulation of auxin gradients also depends on the cell-specific expression and subcellular localization of plasma membrane (PM)-localized PIN proteins (PIN1–PIN4 and PIN7), the latter often being responsive to environmental and developmental cues (Adamowski and Friml, 2015). PIN proteins are often asymmetrically distributed within the cell and are constantly recycled between endosomal compartments and the PM. The dynamics of polar localization of PIN proteins regulates the rate and direction of cellular auxin export and this ultimately determines auxin gradients in the tissue. Therefore, the regulatory machinery of the polarity and abundance of PM-localized PIN proteins is crucial for diverse developmental processes and morphogenesis including embryogenesis, initiation of lateral organs, and tropic responses (Robert et al., 2013; Adamowski and Friml, 2015; Rakusová et al., 2015).
The exocytosis and endocytosis of PIN proteins at the PM can be modulated by ADP RIBOSYLATION FACTOR-GUANINE NUCLEOTIDE EXCHANGE FACTORS (ARF-GEFs) including GNOM (Naramoto et al., 2010). PIN proteins are internalized from the PM to the trans-Golgi network/early endosome compartments after which PINs can then proceed along the recycling route to the PM (Adamowski and Friml, 2015). An important tool for investigating exocytic protein sorting is Brefeldin A (BFA), which is a reversible inhibitor of ARF-GEFs including GNOM (Geldner et al., 2001, 2003). BFA treatment leads to accumulation of the endocytosed PINs in artificial intracellular aggregates called “BFA bodies,” the formation of which can be reversed by washing out the BFA (Geldner et al., 2001).
Clathrin-mediated endocytosis is also involved in the internalization of PIN proteins from the PM (Kitakura et al., 2011; Adamowski et al., 2018) and is modulated by the Rho guanidine triphosphate hydrolases of plants (ROP) family of Rho-like GTPases and their associated ROP interactive CRIB motif-containing proteins (Lin et al., 2012; Nagawa et al., 2012). Genetic analysis has revealed that MACCHI-BOU4/ENHANCER OF PID/NAKED PINS IN YUCCA-like1 (MAB4/NPY1), a gene encoding NON-PHOTOTROPIC HYPOCOTYL3-like proteins and homologous MAB4/NPY1–like, regulates PIN abundance at the PM (Furutani et al., 2014). The internalization and trafficking of PIN proteins is dynamically regulated by developmental and environmental cues, such as plant hormones, gravity, and light (Ding et al., 2011; Rakusová et al., 2016). Short-term auxin treatments, in particular using synthetic auxin analogs, blocks clathrin-mediated internalization of PIN proteins from the PM and consequently enhances PIN abundance at the PM and increases auxin efflux (Paciorek et al., 2005; Robert et al., 2010). Auxin also induces PIN1 relocalization from basal to the inner lateral PM of root endodermal and pericycle cells (Prát et al., 2018). Similarly, auxin mediates PIN3 relocalization during gravitropic responses to terminate gravitropic bending (Rakusová et al., 2016). Prolonged auxin treatment induces PIN2 vacuolar targeting and degradation, and this is mediated by the SCFTIR1/AFB (SKP-Cullin-F box [SCF], TRANSPORT INHIBITOR RESISTANT1/AUXIN SIGNALING F-BOX [TIR1/AFB]) pathway (Abas et al., 2006; Baster et al., 2013), which presumably also explains the SCFTIR1/AFBs-dependent auxin effect on PIN2-GFP accumulation in BFA bodies (Pan et al., 2009). In addition, auxin has been reported to reduce the abundance of photoconvertible PIN2-Dendra at the PM by repressing the translocation of newly synthesized PIN2 to the PM (Jásik et al., 2016). Besides an auxin effect on PIN trafficking, other hormones can influence different aspect of PIN trafficking, including cytokinin (Marhavý et al., 2011), salicylic acid (Du et al., 2013), and gibberellic acid (Salanenka et al., 2018), thus providing a possible entry point for crosstalk of these signaling pathways with the auxin distribution network.
Given these different and sometimes contradictory observations for different PINs resulting from investigations in different cells and using different approaches, the underlying cellular and molecular mechanisms for the targeting and recycling of PIN proteins, and in particular for their regulation by auxin, remain largely unknown.
To develop a useful chemical tool for dissecting the regulatory mechanism of PIN trafficking, we have screened phenylacetic acid (PAA) derivatives for selective modulation of PIN trafficking in Arabidopsis (Arabidopsis thaliana). We identified 4-ethoxyphenylacetic acid, which was designated as pinstatic acid (PISA) due to its activity on PIN-mediated polar auxin transport. PISA has an auxin-like effect on hypocotyl elongation and adventitious root formation by positively modulating auxin transport. Similar to conventional auxins, PISA blocks the internalization of PIN proteins from the PM and consequently induces PIN protein accumulation at the PM. PISA is notably different from other known auxin chemical tools, like auxin transport inhibitors 2,3,5‐triiodobenzoic acid (TIBA) and n-1-naphthylphthalamic acid (NPA). Therefore, PISA represents a promising chemical tool for dissecting the complicated regulations of PIN trafficking by auxin.
RESULTS
Pinstatic Acid Is an Inactive PAA Analog on TIR1/AFB-Aux/IAA Coreceptor Complex
Auxins modulate the expression and degradation of PIN proteins via the SCFTIR1/AFB signaling pathway (Baster et al., 2013; Ren and Gray, 2015). On the other hand, clathrin-mediated endocytosis of PIN is inhibited by auxin via a nontranscriptional pathway (Robert et al., 2010). These positive and negative effects of auxin on PIN trafficking hinder access to the regulatory components in PIN trafficking using conventional genetic approaches. Therefore, we searched for an auxin transport modulator that would make PIN trafficking more amenable to experimentation. To this end, we initially screened PAA derivatives according to the following criteria: (1) The derivative should be inactive within the SCFTIR1/AFB pathway and (2) derivative treatment should induce auxin-related phenotypes that are different from the phenotypes typical of auxins or auxin transport inhibitors, such as TIBA and NPA.
In the course of screening, we found that 4-ethoxyphenylacetic acid (later denoted “PISA”) promoted hypocotyl elongation but did not induce auxin-responsive DR5::GUS reporter gene expression, which is mediated by the SCFTIR1/AFB pathway (Figs. 1, A and C, and 2). Thus, PISA was selected as the most promising candidate from a series of 4-alkyloxy-PAA derivatives and further characterized in detail.
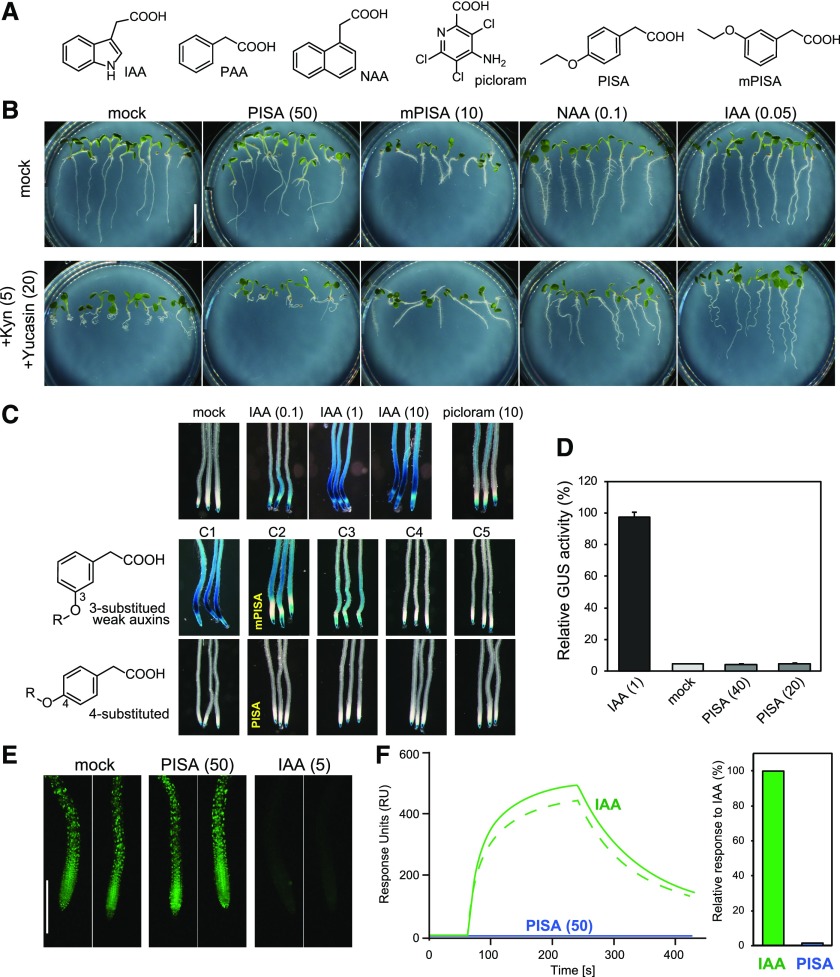
Figure 1.
Evaluation of PISA for an auxin-like effect in the SCFTIR1/AFB pathway. A, The structures of auxins and PISA. B, Effects of PISA on auxin-deficient root phenotypes. Arabidopsis plants were cultured for 5 d on vertical agar plate containing chemicals with or without auxin biosynthesis inhibitors yucasin DF and L-kynurenine (Kyn). The values in parentheses represent the concentration of chemicals (micromolar). Bar = 5 mm. C, Effects of alkyloxy-PAA on auxin-responsive DR5::GUS expression. Five-d–old DR5::GUS seedlings were incubated with chemicals for 6 h. Methoxy (C1) to pentoxy (C5) PAA derivatives including mPISA and PISA were assessed at 50 μm. D, Quantitative analysis of GUS enzyme activity in the DR5::GUS line treated with IAA and PISA. Values are the means ± sd (n = 9). E, DII-VENUS seedlings were incubated with 10-μm yucasin DF for 3 h and then washed with medium. The seedling was incubated with PISA and IAA for another 60 min. Bar = 500 μm. F, SPR analysis of the auxin-induced interaction between TIR1 and IAA7 degron peptide. The sensorgram shows the effect of 50-μm IAA (green) and 50-μm PISA (blue) on TIR1-DII peptide association and dissociation. The bars show the relative response of PISA to IAA (100%).
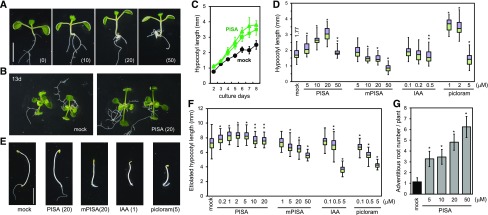
Figure 2.
Effects of PISA on hypocotyl elongation and adventitious root formation. A, Arabidopsis seedlings cultured for 7 d with PISA. The values in parentheses represent the concentration of chemicals (micromolar). Bar = 5 mm. B, 13-d–old plants grown with PISA. C, Time course of hypocotyl length of seedlings cultured with PISA (solid square = 10 μm; solid triangle = 20 μM). Values are the means ± sd (n = 15–20). D, Hypocotyl lengths of seedlings cultured for 7 d with PISA and auxins. The hypocotyl length (millimeters) of the mock-treated seedlings is indicated. Box-and-whisker plots show a median (centerline), upper/lower quartiles (box limits), and maximum/minimum (whiskers; n = 30–38). Statistical significance assessed by Welch’s two-sample t test. Asterisks indicate significant differences (**P < 0.05, *P < 0.01). E, Etiolated seedlings cultured for 5 d in dark with PISA and auxins. F, Hypocotyl lengths of etiolated seedling cultured for 3 d in dark with PISA and auxins. Statistical significance assessed by Welch’s two-sample t test. Asterisks indicate significant differences (n = 50–72, **P < 0.05, *P < 0.01). G, Adventitious root production induced by PISA. Arabidopsis seedlings were cultured for 7 d with PISA and the adventitious root number at shoot and root junction was counted. Asterisks indicate significant differences (n = 30, **P < 0.05, *P < 0.01).
Auxin is biosynthesized by two enzymes, namely TAA1 and YUC in the indole 3-pyruvic acid pathway (Kasahara, 2016). The inhibition of this pathway by l-kynurenine, a TAA1 inhibitor, and yucasin DF, a YUC inhibitor, caused short and curled roots that are typical auxin-deficient phenotypes (Fig. 1B; He et al., 2011; Tsugafune et al., 2017). A quintuple yuc 3 5 7 8 9 mutant showed a similar auxin-deficient root phenotype (Supplemental Fig. S1A; Chen et al., 2014). IAA and 1-naphthylacetic acid (NAA) at 50–100 nm recovered these auxin-deficient root defects in root elongation and gravitropism (Fig. 1B; Supplemental Fig. S1A). An analog of PISA, 3-Ethoxyphenylacetic acid (meta-substituted PISA: mPISA; Fig. 1A), that retains weak auxin activity in DR5::GUS expression (Fig. 1C) also rescued the auxin-deficient curled root phenotype (Fig. 1B). In contrast, PISA did not rescue these root defects caused by auxin deficiency, clearly indicating PISA does not directly act as a typical auxin like IAA or NAA in planta (Fig. 1B; Supplemental Fig. S1A).
The tobacco (Nicotiana tabacum) BY-2 cell suspension culture requires auxin for cell proliferation (Winicur et al., 1998). BY-2 cells proliferated in the presence of IAA and NAA (Supplemental Fig. S1B), but PISA failed to maintain this cell culture (Supplemental Fig. S1B). The cell morphology of the culture treated with PISA showed swollen cell shapes that are a hallmark of auxin-depletion (Supplemental Fig. S1C), further suggesting that PISA does not have the effect as an auxin on cell division (Winicur et al., 1998). Auxin-induced rapid cell elongation in etiolated hypocotyls was demonstrated to be mediated by TR1/AFB receptors (Fendrych et al., 2016). However, PISA failed to induce this rapid cell elongation (Supplemental Fig. S2), suggesting that PISA does not act as a conventional auxin to directly activate the TR1/AFB receptors in the hypocotyl.
IAA and the synthetic auxin picloram cause potent induction of auxin-responsive reporter genes such as DR5 (Fig. 1C). In contrast, PISA did not induce any auxin-responsive DR5::GUS and BA3::GUS reporter expression, again suggesting that it is inactive as a ligand for the SCFTIR1/AFB pathway (Fig. 1, C and D; Supplemental Fig. S3A). DII-VENUS protein is a translational fusion of the TIR1-interacting domain of Aux/IAA proteins and the fluorescent reporter VENUS (Brunoud et al., 2012). IAA promotes the interaction between DII-VENUS and TIR1 receptor to induce the DII-VENUS degradation and loss of the VENUS signal (Fig. 1E). In contrast, PISA did not induce degradation of DII-VENUS, once again suggesting that PISA does not directly modulate TIR1/AFB-Aux/IAA auxin coreceptor complex formation. Additionally, PISA showed no activity in the yeast auxin-inducible degron system (Supplemental Fig. S3B; Nishimura et al., 2009). In this system, the minichromosome maintenance (MCM) complex is essential for DNA replication in yeast and lines in which MCM is deficient fail to grow (Nishimura et al., 2009). The auxins IAA and NAA, and analog mPISA, all repressed the growth of yeast expressing rice OsTIR1 and Aux/IAA-fused MCM4 protein by promoting the degradation of the fused MCM4 protein (Supplemental Fig. S3B; Nishimura et al., 2009). In contrast, PISA did not repress yeast growth in this auxin-inducible degron system, indicating again that PISA is not an active ligand for TIR1.
These findings were further supported by biochemical assays using Surface Plasmon Resonance (SPR) analysis (Fig. 1F) and a pull-down assay (Supplemental Fig. S3C; Lee et al., 2014). IAA promotes assembly of the coreceptor complex of TIR1 and Aux/IAA (domain II) in both assays. In contrast, PISA did not promote the interaction between TIR1 and Aux/IAA in either system (Fig. 1F; Supplemental Fig. S3C). Additionally, the SPR assay also showed that there was no binding of PISA with AFB5 (Supplemental Fig. S3C), and using the SPR assay to test for antiauxin activity by mixing 50-μm PISA with 5-μm IAA showed that PISA did not bind and block the TIR1 auxin-binding site (Supplemental Fig. S3E) whereas the known TIR1/AFB auxin receptor blocker auxinole (Hayashi et al., 2012) reduced the IAA signal dramatically. Thus, in these direct binding assays, PISA does not bind to TIR1/AFB coreceptors. In summary, PISA is completely inactive as a classical auxin that induces the Aux/IAA degradation via TIR1/AFB auxin receptors.
PISA Promotes Hypocotyl Elongation by Positively Modulating Polar Auxin Transport
PISA promotes hypocotyl elongation in a manner that is typical for auxin effects in Arabidopsis seedlings (Fig. 2A). Because PISA did not activate DR5-monitored auxin response, we carefully examined its effects on auxin-related phenotypes in planta to address possible modes of PISA action. In light-grown seedlings, PISA at 5–20-μm promoted hypocotyl elongation (Fig. 2, A–D). In contrast, IAA and mPISA inhibited growth at 0.5 and 20 μm, respectively, whereas the AFB5-selective synthetic auxin picloram strongly promoted hypocotyl elongation (Fig. 2D). In the dark, PISA at 2 μm slightly promoted the elongation of etiolated hypocotyls (Fig. 2, E and F), but did not inhibit their elongation at 20 μm. In contrast, exogenously applied IAA, picloram, and mPISA inhibited the elongation of etiolated hypocotyls (Fig. 2F).
Having explored a set of physiological responses, we made use of genetic and pharmacological tools to gain insight into the mechanism of PISA action. The auxin signaling mutants axr1-3 and tir1-1 afb2-1 showed high resistance to mPISA (Supplemental Fig. S4A), implying that mPISA targets auxin signaling in planta (Hayashi, 2012). In contrast, the hypocotyl of axr1-3 elongated to a similar extent as that of the wild type when treated with PISA (Fig. 3A). Importantly, neither wild type nor axr1-3 responded to PISA after the inhibition of SCFTIR1 auxin signaling by the auxin antagonist auxinole (Fig. 3A; Supplemental Fig. S4B). PISA also failed to promote hypocotyl elongation in the presence of the auxin biosynthesis inhibitor l-kynurenine (Fig. 3A; Supplemental Fig. S4C). These observations indicate that auxin-like effects of PISA on hypocotyl growth require the SCFTIR1/AFB auxin signaling to be activated by endogenous IAA.
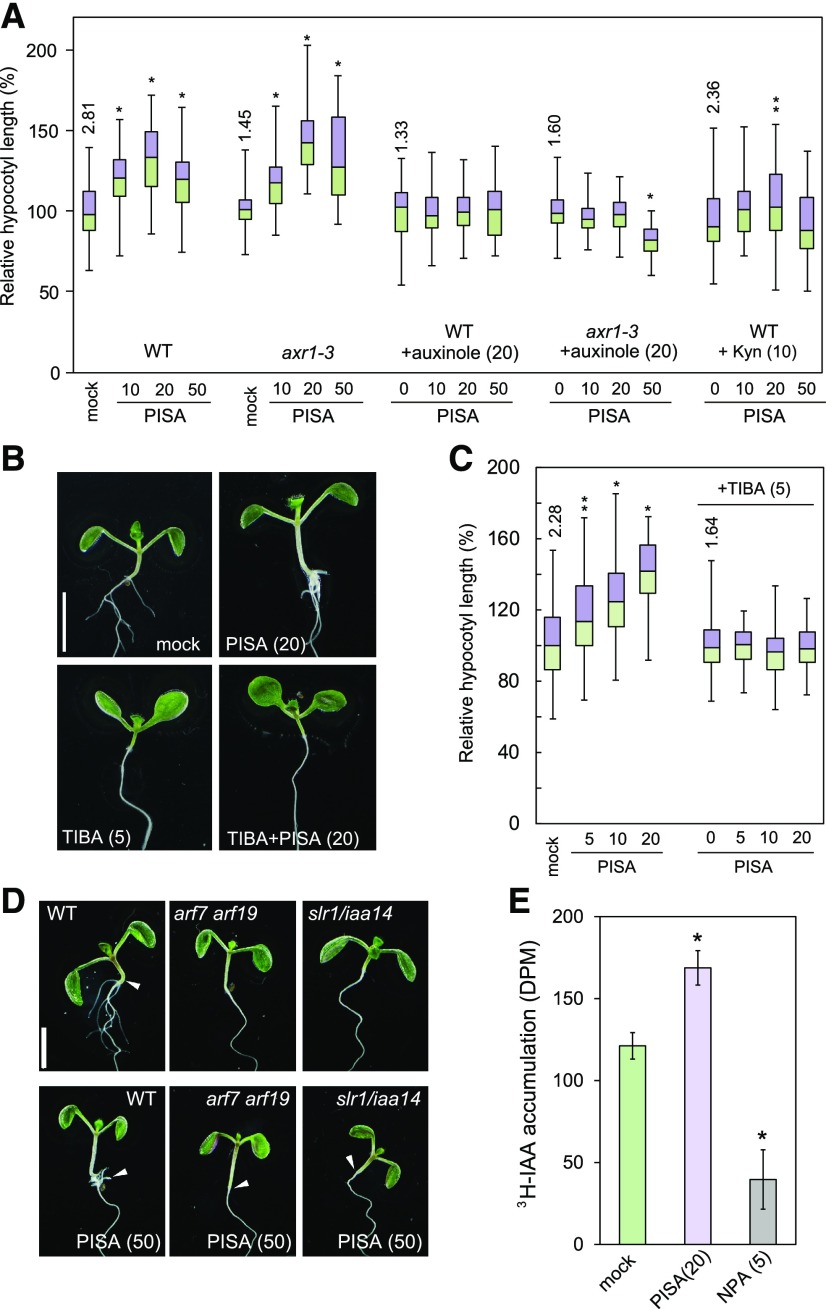
Figure 3.
Auxin signaling and transport inhibitors repress PISA-induced hypocotyl phenotypes and PISA promotes basipetal auxin transport in the hypocotyl. A, The hypocotyl length of Arabidopsis wild-type and axr1-3 mutant seedlings cultured for 7 d with chemicals. Relative hypocotyl length is shown as the percentage of that in mock-treated plants (100%). The actual length (millimeter) of mock-treated hypocotyls are indicated (n = 40–48). B, Seedlings cultured for 7 d with PISA and auxin transport inhibitor, TIBA. C, Hypocotyl length in seedlings cultured with or without TIBA and PISA. Relative hypocotyl length is shown as the percentage of that in mock-treated plants (100%). The actual length (millimeter) of mock-treated hypocotyls is indicated as box-and-whisker plots (n = 40–45). Statistical significance assessed by Welch’s two-sample t test. Asterisks indicate significant differences (**P < 0.05, *P < 0.01). D, Seedlings of wild type, arf7 arf19, and slr1/iaa14 mutants cultured for 7 d with or without PISA. The values in parentheses represents the concentration of chemicals (micromolar). Bar = 5 mm. E, Rootward transport of radiolabeled 3H-IAA in decapitated hypocotyls. NPA, an auxin transport inhibitor, was used as the negative control. (*P < 0.01, n = 9).
To examine the effects of PISA on polar auxin transport, seedlings were cotreated with auxin efflux transport inhibitors and PISA. The promotion of elongation by PISA on hypocotyls was blocked by three auxin efflux transport inhibitors, TIBA, 2-(4-[diethylamino]-2-hydroxybenzoyl)benzoic acid (BUM), and NPA (Fig. 3, B and C; Supplemental Fig. S5A; Fukui and Hayashi, 2018). In addition, treatments with the synthetic auxin picloram and the auxin overproduction line 35S::YUC1 exhibited longer hypocotyls as a high auxin phenotype (Supplemental Fig. S5B), but in these lines TIBA and NPA did not suppress the elongation (Supplemental Fig. S5B). The data suggest that PISA could positively modulate polar auxin transport in hypocotyls. To examine further the effects of PISA on basipetal auxin transport, rootward movement of 3H-IAA was analyzed (Fig. 3E). In this assay, NPA reduced the basipetal movement of 3H-IAA in hypocotyls, whereas PISA enhanced it (Fig. 3E). These results collectively show that PISA positively modulates basipetal auxin transport in hypocotyls. Another possible target of PISA could be the regulation of endogenous auxin concentrations, such as via auxin biosynthesis or catabolism. Analysis of endogenous IAA levels in Arabidopsis seedlings showed that they were not affected by PISA treatment (Supplemental Fig. S6). Together, these results indicate that PISA likely acts by affecting polar auxin transport.
PISA Inhibits Root Growth by Accumulating IAA at the Root Tip
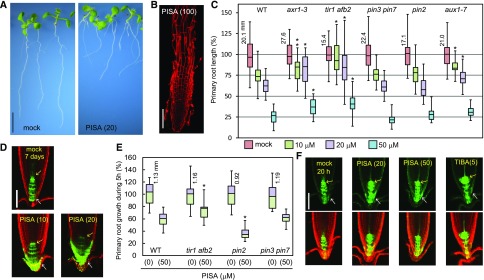
PISA inhibited primary root growth in a manner that is similar to conventional auxins. The seedlings were cultured on vertical plates containing PISA for 7 d (Fig. 4, A–C). The auxin signaling mutants, axr1-3 and tir1 afb2, were insensitive to PISA. Additionally, auxin influx transport mutant aux1-7 was also less sensitive to PISA regarding root growth (Fig. 4C). Taken together with the effects of PISA on auxin transport in the hypocotyl, these results suggest that PISA inhibits primary root growth by modulating auxin transport to affect auxin distribution and maxima. Further, the roots treated with PISA at 100 μm showed severe defects in root cell morphology (Fig. 4B). To examine the effects of PISA on auxin distribution in roots, DR5::GFP seedlings were cultured with PISA for 7 d (Fig. 4D). PISA significantly induced GFP expression in the lateral root cap cell, indicating PISA accumulates IAA in the lateral root cap and root growth is inhibited as a consequence. In contrast to auxin signaling mutants, the sensitivity of pin2 and pin3 pin7 mutants was comparable to that of the wild type (Fig. 4C). To investigate the short-term effects of PISA, seedlings were treated with PISA for 5 h (Fig. 4E). PISA inhibited root elongation within this 5 h incubation. The tir1 afb2 mutant was insensitive to PISA, but the pin2 mutant was more sensitive than the wild type to PISA. Perhaps, in the pin2 mutant, the accumulated IAA is not efficiently transported from the lateral root cap. Consistent with root elongation responses (Fig. 4E), PISA induced DR5::GFP expression in the lateral root cap after 20-h treatment suggesting enhanced accumulation of endogenous IAA (Fig. 4F). The auxin transport inhibitor TIBA blocks IAA efflux and inhibits root elongation by accumulating IAA (Supplemental Fig. S5). TIBA highly induced DR5::GFP expression near the quiescent center (QC) where IAA is biosynthesized (Fig. 4F; Brumos et al., 2018). Taken together, these results indicate that PISA promotes the auxin transport rate leading to accumulations of IAA at the lateral root cap, resulting in the inhibition of root elongation.
Figure 4.
The effects of PISA on root elongation and auxin distribution in the root tip. A, Wild-type (WT) seedlings cultured for 7 d with PISA. Bar = 5 mm. B, Wild-type root cultured with 100-μm PISA . Root was counterstained with propidium iodide. Bar = 100 μm. C, The primary root length of Arabidopsis wild-type and auxin mutants (axr1-3, tir1 afb2, pin3 pin7, pin2/eir1-1, and aux1-7) cultured for 7 d on vertical plate containing PISA. Relative root length is shown as the percentage of that in mock-treated plants (100%). The actual length (millimeter) of mock-treated roots is indicated. Statistical significance was assessed by Welch’s two-sample t test between wild type and mutants. Asterisks indicate significant differences (n = 32–40, **P < 0.05, *P < 0.01). D, The GFP expression of DR5::GFP in roots cultured vertically with PISA for 7 d. Arrows indicate QC (yellow) and lateral root cap (white). Bar = 100 μm. E, The primary root growth of Arabidopsis wild type and auxin mutants over 5 h on vertical plates containing PISA. The actual length (mm) of mock-treated roots is indicated, which was set to 100%. Asterisks indicate significant differences (n = 14–17, *P < 0.01). F, The GFP expression of DR5::GFP cultured vertically with PISA and TIBA for 20 h. The values in parentheses represents the concentration of chemicals (micromolar). Bar = 100 μm.
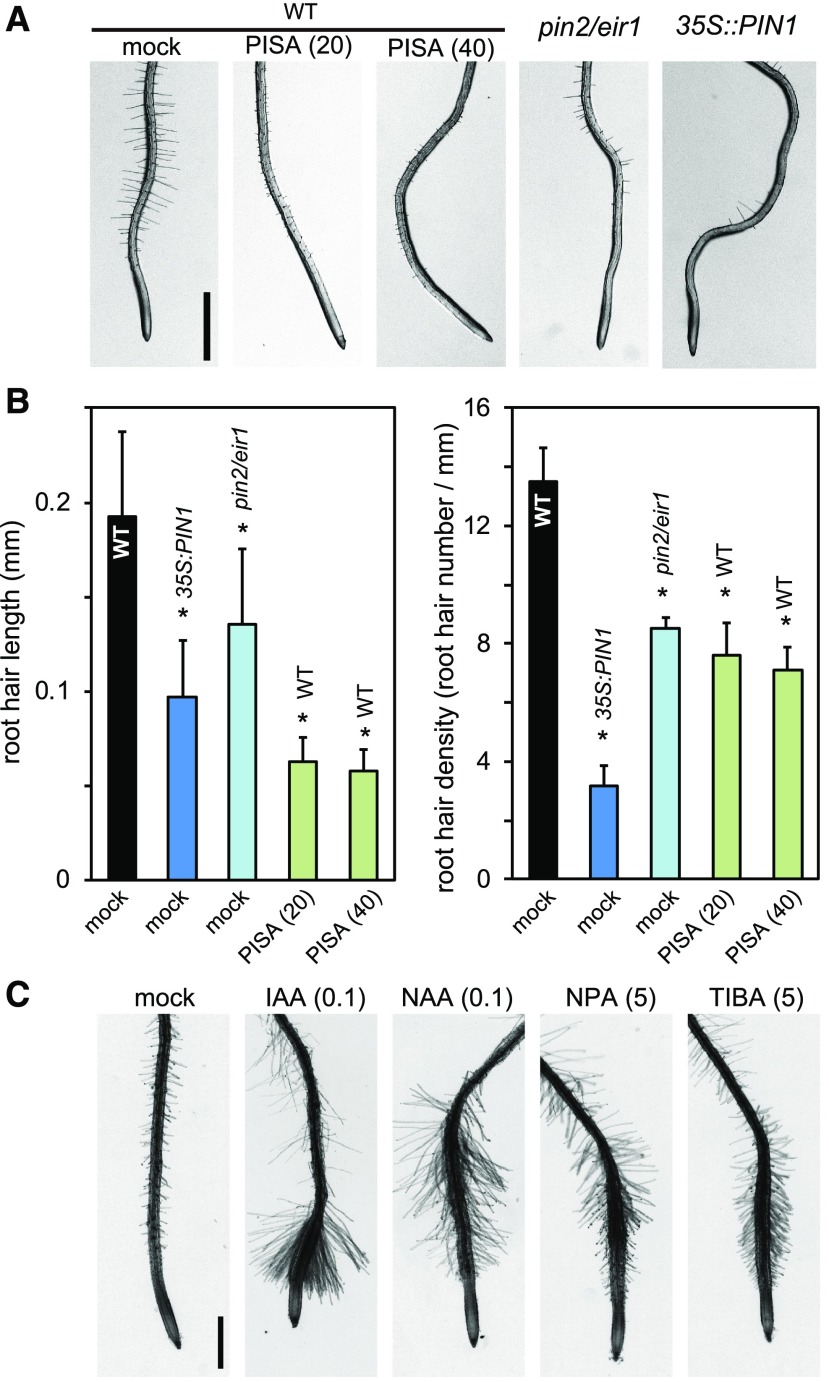
PISA Blocked Root Hair Formation by Positively Modulating Auxin Transport
PISA displayed auxin-like activity in its effects on hypocotyl elongation, primary root inhibition, and adventitious root formation (Fig. 2). Typical auxin efflux transport inhibitors commonly inhibit the elongation of both primary root and hypocotyl, supporting that PISA is not an inhibitor of auxin efflux transport. The effects of PISA on auxin-related phenotypes can be explained if it works by increasing auxin efflux. To further examine the effects of PISA on auxin efflux transport, the root hair phenotype was analyzed. This process involves the PIN2 proteins, which are localized at the apical side of root epidermal cells and mainly contribute to basipetal (shootward) auxin transport (Abas et al., 2006). The loss of function pin2/eir1 mutant displays impaired root hair formation (Fig. 5, A and B). The ectopic overexpression of PIN1 in 35S::PIN1 roots also interferes with this shootward auxin transport, and, consequently, 35S::PIN1 seedlings also show defects in root hair formation (Fig. 5, A and B; Ganguly et al., 2010), suggesting shootward auxin flow is important for root hair formation (Rigas et al., 2013). In contrast, auxin efflux transport inhibitors TIBA and NPA promote root hair formation (Ganguly et al., 2010), probably by increasing the accumulation of endogenous IAA (Fig. 5C). Importantly, PISA inhibits root hair formation, implying PISA has an opposite effects to auxin efflux inhibitors.
Figure 5.
PISA inhibits root hair formation. A, Root hairs of pin2/eir1, 35S::PIN1, and wild-type (WT) plants treated with PISA. Five-d–old seedlings were cultured for 2 d on vertical agar plates with or without PISA. The values in parentheses represent the concentration of chemicals (micromolar). B, The root hair length and density of pin2/eir1, 35S::PIN1, and wild-type plants treated with PISA. The length and density of root hairs within the 2–4-mm region from root tip were measured. Values are the means ± sd. Asterisks indicate significant differences (n = 8–11, *P < 0.01). C, The root hair formation of wild-type seedlings grown with auxins and auxin transport inhibitors. The values in parentheses represent the concentration of chemicals (micromolar). Bar = 1 mm.
PISA Affects Adventitious and Lateral Root Formation by Positively Modulating Auxin Transport
PISA induces adventitious root formation at the shoot/root junction as shown in Figure 2A. Importantly, auxin signaling mutants slr/iaa14 and arf7 arf19 show severe defects in lateral root formation (Fig. 3D; Table 1; Okushima et al., 2007). In these mutants, PISA did not promote adventitious root formation at the shoot/root junction and this is consistent with the auxin-like effects of PISA on hypocotyls (Fig. 3D; Table 1). This suggests that adventitious root formation in response to PISA treatment depends on auxin signaling downstream of SCFTIR1/AFB. In such a situation, auxin efflux transport inhibitors BUM, NPA, and TIBA would reduce polar auxin transport in hypocotyls, resulting in the inhibition of the adventitious root formation and this is indeed what we observed, as shown in Table 1 and Supplemental Figure S5A. Taken together, these results suggest that PISA positively modulates the polar auxin transport system, thereby leading to the accumulation of auxin at the shoot/root junction and promoting adventitious root formation.
Table 1. Effect of PISA on adventitious root formation at shoot/root junction.
| Modulator | Wild Tpe (Col) | arf7 arf19 | slr/iaa14 | TIBA (5 μm) | NPA (5 μm) |
|---|---|---|---|---|---|
| Mock | 1.57 ± 0.65a | 0 | 0 | 0 | 0 |
| PISA (20 μm) | 3.21 ± 0.70 | 0 | 0 | 0 | 0 |
| PISA (50 μm) | 5.07 ± 1.03 | 0 | 0 | 0 | 0 |
Adventitious root number at shoot/root junction for each of the 6-d–old seedlings.
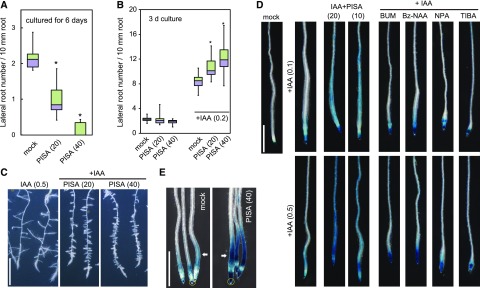
In contrast to the promotion of adventitious roots at the shoot/root junction (Fig. 2A; Table 1), PISA alone repressed lateral root formation in primary roots (Fig. 6A). In contrast, PISA strongly promoted lateral root numbers when coincubated with exogenous IAA (Fig. 6, B and C; Supplemental Fig. S7A). TIBA and NPA did not affect the lateral root number induced by exogenous IAA (Supplemental Fig. S8A), suggesting that inhibition of auxin efflux does not enhance IAA-induced lateral root formation. This was further investigated using the cell cycle reporter CYCB1;1::GUS, which is induced strongly by IAA and NAA in initiating lateral roots. In this assay, PISA enhanced CYCB1;1::GUS expression when in the presence of auxins, IAA and NAA (Supplemental Fig. S7B). Similarly, auxin-induced DR5::GUS expression was dramatically enhanced by pretreatments with PISA (12 h; Fig. 6D). In this experiment, IAA treatment for 6 h at 100 and 500 nm induced DR5::GUS expression in elongation zones only (Fig. 6D). This expression pattern was extended along the entire root by the coincubation of IAA and PISA (Supplemental Fig. S8B). In contrast, cotreatment with IAA and auxin transport inhibitors (NPA, TIBA, 5-benzykoxy IAA, and BUM; Fukui and Hayashi, 2018) activated DR5::GUS expression only at the root tips (Fig. 6D). To examine the effects of PISA on basipetal auxin transport, shootward movement of IAA from the root tip was evaluated by DR5::GUS assay (Fig. 6E; Buer and Muday, 2004; Lewis and Muday, 2009). In this shootward auxin transport assay, the DR5::GUS seedlings were placed on vertical plates containing PISA and then an agar block containing IAA was placed onto the root tips. The seedlings were then incubated for 10 h. PISA promoted DR5::GUS induction derived from root tip IAA (Fig. 6E), suggesting that PISA enhances shootward auxin transport from the root tip. Taken together, these results indicate that PISA increases the net flow of auxin in the roots by positively modulating auxin transport.
Figure 6.
Effects of PISA on IAA-induced lateral root formation and shootward IAA transport. A, Effects of PISA on the lateral root formation. Arabidopsis seedlings were cultured for 6 d with PISA. The number of lateral roots were counted and the density of lateral roots are shown as box-and-whisker plots (n = 14–16). B and C, Effects of PISA on IAA-induced lateral root formation. Five-d–old seedlings were cultured for additional 3 d with PISA in the presence of IAA. The density of lateral roots are shown as box-and-whisker plots (B, n = 14–16) and representative images are shown (C). Bar = 5 mm. D, Effects of PISA on IAA-induced DR5::GUS expression. Five-d–old DR5::GUS seedlings were incubated for 12 h in liquid GM medium with or without PISA or auxin transport inhibitors. IAA was added to the GM medium and the seedlings were further incubated for additional 6 h. The IAA-induced GUS activity was visualized by X-Gluc. Bar = 1 mm. E, Effects of PISA on shootward IAA transport. An agar block containing IAA was applied to DR5::GUS root tips (yellow ring) and the seedlings were incubated on vertical plates containing 40-μm PISA for 10 h. Arrows show the IAA-induced GUS activity. Bar = 1 mm. Statistical significance assessed by Welch’s two-sample t test. Asterisks in (A) and (B) indicate significant differences (*P < 0.01). The values in parentheses represent the concentration of chemicals (micromolar).
Other possible targets for PISA are the AUX1/LAX auxin influx transporters. PISA might promote IAA-induced lateral root formation by increasing the uptake of exogenous IAA. To test the effects of PISA on IAA influx transport, seedlings were cotreated with PISA and the membrane-permeable IAA prodrugs, IAA methyl ester and IAA octyl ester (Supplemental Fig. S9). These lipophilic IAA esters and NAA (Supplemental Fig. S7B) can be incorporated into cells by passive diffusion, but not by the AUX1/LAX transporters. PISA enhanced lateral root formation to the same extent with the two IAA esters, NAA and IAA (Supplemental Fig. S7B and S9), indicating that IAA influx transport is not required for the activity of PISA on lateral root promotion.
PISA Perturbed Asymmetric Auxin Distribution and Gravitropism in Root
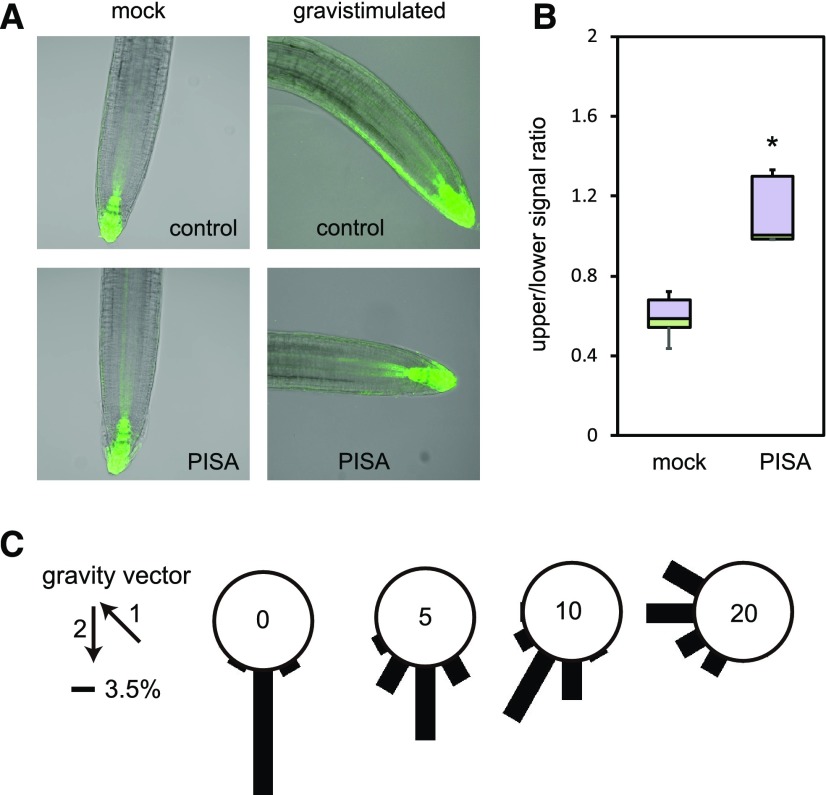
Gravistimulation rapidly induces asymmetric auxin distributions in roots and thereby changes the DR5 reporter expression pattern (Fig. 7A). This gravistimulated asymmetric auxin distribution is driven by PIN-mediated shootward auxin movement in the root epidermis (Wisniewska et al., 2006; Baster et al., 2013). After 4-h gravistimulation, the DR5::GFP signal increased at the lower side of gravistimulated roots. PISA treatment completely diminished this asymmetric expression of DR5::GFP (Fig. 7, A and B) and concomitantly blocked root gravitropic responses (Fig. 7C). These observations show that PISA not only modulates polar auxin transport but specifically affects PIN-mediated asymmetric auxin distribution in gravistimulated roots.
Figure 7.
PISA inhibits auxin distribution and root gravitropism. A, Effect of PISA on auxin asymmetric distribution. Four-d–old DR5::GFP seedlings were transferred to 20-μm PISA and control medium for 1 h. After 1 h, seedlings were gravistimulated for 4 h and imaged. PISA pretreatment abolished auxin asymmetric distribution and seedlings did not respond to gravity stimuli. B, Quantitative evaluation of (A), showing a mean ratio of the signal intensity of the upper-/lower-half of the root. (*P < 0.01). C, Effect of PISA on root gravitropic response. Five-d–old wild-type seedlings were placed on vertical GM agar plates containing PISA and then cultured for 3 h in the dark. The plates were further incubated for 16 h after rotating plates at 135° angle against vertical direction. The arrows indicate the vector of gravity before (1) and after (2) the initiation of gravistimulation. The angles were grouped into 30° classes and plotted as circular histograms.
PISA Blocked the Internalization of PIN Proteins and Promoted Their Accumulation at the PM
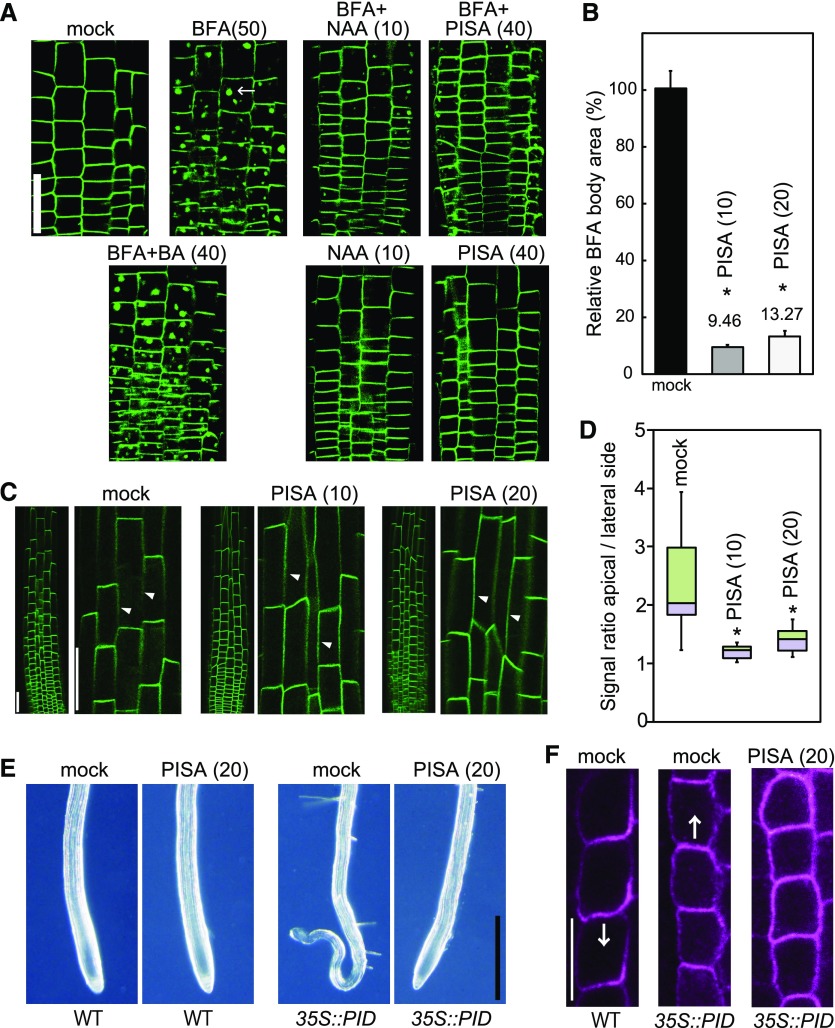
All the phenotypic effects of PISA can be explained by the positive modulation of auxin transport by PISA. PISA treatment did not affect the expression profiles of proPIN1::GFP, proPIN2::GUS and proPIN7::GUS (Supplemental Fig. S10), indicating that the primary target of PISA in auxin transport is not the regulation of PIN transcription. To address the mechanism of positive effects of PISA on auxin efflux, we examined the effects of PISA on the recycling of PIN proteins in roots. BFA induces the formation of BFA bodies that incorporate PIN2-GFP protein in proPIN2::PIN2-GFP line (Geldner et al., 2003). Auxin (NAA) was shown to inhibit BFA body formation by blocking the endocytosis of PIN2 protein (Fig. 8A; Paciorek et al., 2005). The negative control compound benzoic acid (BA) did not affect BFA body formation (Fig. 8A), but PISA inhibited BFA body formation to the same extent as NAA (Fig. 8, A and B). Additionally, BFA body formation with both PIN1-GFP fusion and PIN1 native protein was also blocked by NAA and PISA (Supplemental Fig. S11). These observations suggest that PISA interferes with PIN recycling or vacuolar targeting, and as a consequence promotes the accumulation of PIN proteins at the PM. Because constitutive PIN recycling has been linked to maintenance of its asymmetric, polar distribution, we tested PISA effect on PIN polarity. Indeed, PISA treatment diminished PIN2 polarity at the PM. PIN2 showed pronounced accumulation at the lateral cell sides (Fig. 8, C and D; Supplemental Fig. S12) and PIN1 showed almost no polarity after treatment with PISA (Supplemental Fig. S13). Furthermore, PISA at 100 μm disrupted the root architectures and PIN2 polar localization (Supplemental Fig. S14).
Figure 8.
Effects of PISA on PIN internalization from the PM. A and B, Effect of PISA on the BFA body formation of PIN2-GFP. Five-d–old proPIN2::PIN2-GFP seedlings were incubated for 30 min in liquid GM medium containing PISA, benzoic acid (BA) and NAA, and then BFA was added to the medium. Seedlings were then incubated for additional 60 min. BFA induced PIN2-GFP-marked BFA bodies. The area of BFA body was measured and the area in BFA-treated seedlings (n = 25–40, *P < 0.01) was adjusted to 100%. The value of the area is shown the means ± sd in (B). Bar = 50 μm. C and D, Effect of PISA on the internalization of PIN2-GFP. Five-d–old proPIN2::PIN2-GFP seedlings were incubated for 12 h with PISA. The fluorescence intensity of the apical and lateral sides of cells in the root (n = 18–20, *P < 0.01) were quantified and the fluorescent signal rate (apical side/lateral side) is shown as the means ± sd in (D). The values in parentheses represent the concentration of chemicals (micromolar). Bar = 50 μm. Arrowheads show lateral cell side. E, Effects of PISA on a collapse of the primary root meristem. Five-d–old root tips of wild-type (WT) and 35S::PID plants grown vertically on agar plates containing PISA. Bar = 500 μm. F, Effects of PISA on PIN1 localization in the endodermis of wild-type and 35S::PID roots. Immunolocalization of PIN1 after treatment with PISA for 4 h. Bar = 10 μm. Arrows indicate the direction of IAA transport by PIN1.
This change in the localization of PIN proteins was further investigated using PINOID (PID), a Ser Thr kinase of the AGC kinase family that is known to regulate PIN localization on the cellular membranes (Adamowski and Friml, 2015). Overexpression of PID triggers a basal to apical shift in PIN1 localization, thereby perturbing the auxin gradient in the root tip, depleting auxin from the root tip maxima and leading to meristem collapse (Benjamins et al., 2001; Friml et al., 2004). Consistently, PIN1 was localized at apical side in the endodermis of 35S::PID roots (Fig. 8F). Intriguingly, PISA rescued collapsed root meristems in 35S::PID roots (Fig. 8E) and the apical polarity of PIN1 in 35S::PID was lost and switched to an apolar pattern in endodermal cells (Fig. 8F). Thus, PISA appears to repress IAA depletion from the 35S::PID apical meristem by diminishing shootward IAA transport. This is fully consistent with the PISA effect on the polar localization of PIN proteins.
To gain further insight into the mechanism by which PISA induces PIN accumulation at the PM, the effects of PISA on PIN2-GFP accumulation were examined in a tir1 afb1 afb2 afb3 quadruple mutant line (Supplemental Fig. S15). As in wild-type roots, PIN2-GFP protein was found to be predominantly located at the apical cell sides and not at lateral cell sides despite the severe growth defects in these roots. PISA promoted the accumulation of PIN2-GFP at lateral cell sides in the quadruple mutant, the same as in the wild-type root. This observation strongly suggests that PISA leads to increases in PIN protein accumulation at the PM without activating the SCFTIR1/AFB pathway.
DISCUSSION
Pinstatic Acid Is an Inert Molecule for the TIR1/AFB-Aux/IAA Coreceptor Complex
In the screening for the auxin transport modulators from the PAA analogs, PISA was found to be the most promising candidate. PISA does not bind to the SCFTIR1/AFB complex. The classical structure activity relationships of monosubstituted PAAs demonstrated that 4-substituted PAA is less or inactive as an auxin (Muir et al., 1967). Consistent with these early structure activity relationship studies of PAA derivatives, our results clearly demonstrated that PISA is not a classical auxin directly modulating the SCFTIR1/AFB machinery (Fig. 1). Consistent with this, a docking study using the auxin binding cavity of TIR1 showed that the 4-ethoxy chain in PISA would prevent stable binding of this compound (Supplemental Fig. S16).
In analogy to PISA, the introduction of alkyloxy chains into IAA and NAA at the 5- or 6-positions diminished their TIR1 binding activity (Tsuda et al., 2011). However, it appears that these alkoxy-IAA and -NAAs are still recognized by PIN efflux proteins to inhibit polar IAA transport in competition with endogenous IAA (Tsuda et al., 2011), suggesting alkoxy-IAAs and alkoxy-NAAs could act as auxin transport inhibitors. On the other hand, PAA is not actively and directionally transported in response to gravitropic stimuli and the distribution of PAA is not inhibited by NPA, suggesting that PAA is distributed by passive diffusion (Sugawara et al., 2015). As for PAA, it seems unlikely that PISA itself would be recognized by PINs in planta.
PISA Positively Modulates Polar Auxin Transport to Induce Auxin-Like Activity
PISA showed characteristic auxin-like activity on primary root and shoot responses. PISA inhibited primary root elongation and induced adventitious root formation at the shoot/root junction (Fig. 2). The auxin signaling mutants axr1-3, tir1 afb2, slr1-1, and arf7 arf19 were resistant to PISA in primary root inhibition and adventitious root formation, suggesting that some PISA-induced responses might be mediated by the SCFTIR1/AFB signaling pathway. However, these responses can also be well explained by the accumulation of endogenous IAA at root tip and the shoot/root junction after elevated IAA efflux (Fig. 4D). Auxin efflux inhibitors completely repressed adventitious root formation induced by PISA (Table 1), suggesting that IAA movement is required for PISA activity on adventitious root formation. In the primary root, IAA is biosynthesized near the QC, where TAA1 is strongly expressed (Brumos et al., 2018). Auxin efflux inhibitors TIBA and NPA are considered to have repressed IAA efflux, leading to induction of DR5::GFP expression near the QC (Fig. 4D) and then results in the inhibition of root elongation (Brumos et al., 2018). In contrast, PISA would promote auxin efflux from the QC to lateral root cap, thereby DR5::GFP signal was induced at that place (Fig. 4D). Thus, PISA inhibits root elongation by distinct mechanism of auxin efflux inhibitors.
Furthermore, PISA promoted hypocotyl elongation. Auxin efflux transport inhibitors, TIBA, NPA, and BUM completely suppressed hypocotyl elongation (Figs. 3, B and C; Supplemental Fig. S5A). Hypocotyl elongation by synthetic auxin picloram or YUC1 overexpression could not be cancelled by auxin efflux transport inhibitors (Supplemental Fig. S5B). These evidences suggest that PISA positively modulated auxin transport to show auxin-like activity in the hypocotyl. This was further confirmed by 3H-IAA transport assays in hypocotyl segments (Fig. 3E). Importantly, no auxin analog has been reported to be positive modulator of auxin transport.
PISA Affects Root Auxin Responses by Positively Modulating Shootward Auxin Transport
In contrast to auxin-like effects on primary root growth and shoot elongation, PISA-treated roots showed typical auxin-repressed phenotypes: reduced root hair formation, fewer lateral roots, and reduced gravitropic response. Auxin transport inhibitors promoted root hair formation (Fig. 5B) by accumulating endogenous IAA, but blocked lateral root formation and gravitropic responses by perturbing auxin distribution. The impaired root phenotypes by PISA resemble the root defects in PIN1 overexpressing roots (Rigas et al., 2013), supporting the hypothesis that PISA represses auxin-regulated phenotypes by enhancing auxin efflux. Intriguingly, PISA dramatically enhanced IAA-induced lateral root formation and PISA also promoted IAA-induced DR5::GUS expression in entire roots when auxin transport inhibitors did not (Fig. 6, B–D). Additionally, PISA enhanced shootward auxin movement from the root tip in basipetal auxin transport assays (Fig. 6E). PISA did not increase the endogenous IAA (Supplemental Fig. S6). Thus, it is unlikely that PISA would elevate endogenous IAA in the shoot by upregulating TAA1 and YUC expression in the IAA biosynthesis pathway or by inhibiting the IAA inactivation pathway involving GH3 and DAO1 (Korasick et al., 2013). These observations suggest that PISA positively modulates shootward IAA transport in the root.
PISA Blocks PIN Internalization to Accumulate PIN at the PM in Arabidopsis
The localization and trafficking of PIN1 and PIN2 proteins have been extensively investigated by Adamowski and Friml (2015) and Rakusová et al. (2015). ROP GTPases-RIC signaling have been shown to inhibit the PIN internalization (Lin et al., 2012; Nagawa et al., 2012), PID kinase and D6 Protein Kinase could directly phosphorylate PIN at the PM to regulate the PIN trafficking in a GNOM-dependent manner (Adamowski and Friml, 2015). However, the molecular mechanism for the regulation of PIN trafficking, especially PIN internalization, by auxin has been unclear. Our results show that PISA inhibited the formation of BFA bodies containing PIN1 and PIN2 proteins (Fig. 8; Supplemental Fig. S11). Furthermore, PISA promoted the accumulation of PIN1 and PIN2 proteins at the lateral side of cells. These observations, together with phenotypic data, clearly indicate that by inhibiting PIN internalization PISA would increase PM-localized PIN content, leading to characteristic phenotypes caused by enhanced auxin efflux.
The target of PISA remains an open and intriguing question. PISA is completely inert for transcriptional auxin signaling modulated by SCF TIR1/AFB–Aux/IAA machinery. PISA enhanced PIN2 accumulation at the PM in tir1 afb1 afb2 afb3 quadruple mutant (Supplemental Fig. S15; Pan et al., 2009), implying TIR1/AFB receptors are not a prerequisite for the inhibition of PIN2 internalization by PISA. Modulation of PIN localization and trafficking are influenced by many regulatory steps (Adamowski and Friml, 2015) and it is likely that auxin could coordinately modulate pathways involving recycling rate, biosynthesis, and degradation of PINs in response to environmental and hormonal stimuli.
Many questions still remain as to the mode of action of PISA. It has been reported that auxin reduced formation of BFA bodies by inhibiting delivery of newly synthesized protein rather than by inhibition of PIN internalization (Jásik et al., 2016). On the other hand, PISA inhibited BFA body formation of PIN2-GFP, but enhanced amounts of PIN2-GFP on the PM, suggesting that delivery is not impaired and internalization is reduced. Given this, we have no reason to believe that PISA would target the regulatory component of PIN internalization to which endogenous auxin would bind. We anticipate that PISA will become a very useful chemical tool to dissect the regulatory mechanism of auxin transport.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Columbia (Col-0) was used for all experiments. The following transgenic and mutant lines were in the Col-0 ecotype: axr1-3 [CS3075], tir1-1 afb2-3 [CS69691], iaa14/slr1-1 (Okushima et al., 2007; Chae et al., 2012; Spartz et al., 2012), arf7 arf19 (Okushima et al., 2007), DII-VENUS (Brunoud et al., 2012), yuc3 5 7 8 9 (Chen et al., 2014), proPIN1::PIN1-GFP (Vieten et al., 2005), proPIN2::PIN2-GFP (Vieten et al., 2005), pin2/eir1-1 [CS16706], 35S::PIN1 [CS9375], 35S::PID (Benjamins et al., 2001), and pPIN2::PIN2-GFP / tir1 afb1 afb2 afb3 (Pan et al., 2009). Seeds were surface-sterilized and grown on germination medium (GM; one-half strength Murashige and Skoog salts [Gibco-BRL], 12 g/L of Suc, 1× B5 vitamins, and 0.2 g/L of MES containing 4 g/L of agar for a horizontal agar plate or 14 g/L of agar for a vertical agar plate, at pH 5.8) containing the indicated hormone and/or chemicals. The length of hypocotyl and lateral root number was measured using the software ImageJ (National Institutes of Health; http://rsb.info.nih.gov/ij).
Chemicals
PISA, and mPISA were synthesized from 4-hydroxyphenylacetic acid methyl ester and 3-hydroxyphenylacetic acid methyl ester, respectively. PISA is commercially available from some chemical suppliers (CAS Registry Number: 4919-33-9, Alfa Aesar, Santa Cruz Biotechnology, and Acros Organics).
Histochemical and Quantitative GUS Measurements
For GUS histochemical analysis, the seedlings were washed with a GUS-staining buffer (100 mm of sodium P at pH 7.0, 10 mm of EDTA, 0.5 mm of K4Fe[CN]6, 0.5 mm of K3Fep[CN]6, and 0.1% [w/v] Triton X-100) and transferred to the GUS-staining buffer containing 1 mm of 5-bromo-4-chloro-3-indolyl-β-d-glucuronide (X-Gluc), the substrate for histochemical staining, and incubated at 37°C until sufficient staining developed. For quantitative measurement, seedlings or the excised roots (n = 15–20) were homogenized in an extraction buffer as described in Hayashi et al. (2012). After centrifugation to remove cell debris, GUS activity was measured with 1 mm of 4-methyl umbelliferyl-β-d-glucuronide as a fluorogenic substrate at 37°C. The protein concentration was determined by Bradford protein assay (Bio-Rad). The experiments were repeated at least three times with four replications.
DII-VENUS Assay
Six-d–old DII-VENUS seedlings (Brunoud et al., 2012) were incubated in GM liquid medium containing 10-μm yucasin DF for 3 h at 24°C. The DII-VENUS seedlings were washed out well with fresh medium and incubated in fresh GM liquid medium for 5 min. Exogenous IAA and PISA was added to this medium and fluorescent images of roots were recorded after 60 min.
SPR Assay
SPR assays were performed as described in Quareshy et al. (2017). A quantity of 50-μm IAA or PISA was used to assay for the formation of the auxin-induced TIR1-IAA7 coreceptor complex, or AFB5-IAA7 complex. For the antiauxin assay, 5-μm IAA and 50-μm PISA (or control compound) were mixed and the sensorgram assessed for a reduced signal to the IAA.
Exogenous IAA-Induced Lateral Root Promotion
For lateral root growth, Arabidopsis seedlings were grown vertically for 5 d in continuous light on a GM agar plate. The seedlings were transferred to liquid GM medium containing the indicated concentration of IAA and PISA. The seedlings were cultivated under continuous light for another 3 d at 24°C and then the lateral root numbers were recorded. Three independent experiments were performed.
Gravitropic Response Assay
Six-d–old seedlings were grown vertically on GM agar plates under continuous light at 24°C. The seedlings were then transferred to agar plates containing chemicals and cultured vertically for 2 h. The plates were rotated 135° in the vertical plane, followed by incubation for 16 h in the dark. Photographs of the roots were recorded with a digital camera.
Auxin Transport Assay
For shoot basipetal transport, 6-d–old Col-0 etiolated seedlings grown on GM agar plates were decapitated to avoid endogenous auxin biosynthesis in cotyledons and a droplet of GM agar (12 g/L agar) with 3H-IAA was applied to apical part of the hypocotyls. The seedlings were preincubated with 20-μm PISA for 1 h on an agar plate containing PISA. After 6 h, all roots were removed, hypocotyls were collected, homogenized using grinder and liquid N, and incubated overnight in Opti-Fluor scintillation solution (Perkin Elmer). The amount of 3H-IAA was measured in a scintillation counter (300SL; Hidex) for 300 s with three technical repetitions. The decapitated seedlings were placed on a GM agar plate containing 5-μm NPA to inhibit auxin transport, and then a 3H-IAA agar droplet was applied to each apical part. The negative control (diffusion) was estimated with seedlings transferred to GM agar containing 5-μm NPA during the 3H-IAA droplets incubation (6 h) to inhibit auxin transport.
The root basipetal transport assay was carried out with slight modifications according to the method of Lewis and Muday (2009). A narrow strip of aluminum foil was vertically embedded in GM agar plate (20 g/L agar) containing 40-μm PISA. Five-d–old DR5::GUS seedlings were placed on the GM agar so that the root tip stepped over the edge of the foil strip. An agar block (10-μm IAA and 40-μm PISA) was placed on the root tip. The aluminum strip blocks the diffusion of IAA into the GM agar plate. The plate was incubated vertically for 10 h and GUS activity was visualized histochemically with X-Gluc.
Asymmetric Auxin Distribution Measurement and PIN Immunolocalization Analysis
All measurements were performed using the software ImageJ. Quantification of auxin asymmetry was performed on maximal intensity projection of Z-scans of root tip by measuring the ratio of signal intensity of upper/lower half of the root. DR5rev-GFP reporter line was imaged before and after gravistimulation. PIN immunolocalizations of primary roots were carried out as described in Sauer et al. (2006) and Robert et al. (2010). The antibodies used in this study were anti-PIN1, 1:1,000; and anti-PIN2, 1:1,000.
Imaging and Image Analysis
Fluorescence images were recorded with a fluorescence microscope (BX-50; Olympus) and a laser scanning confocal microscope (FV-3000; Olympus). Typically, the seedlings were incubated with GM medium containing chemicals for the indicated time at 24°C and fluorescence images were then immediately recorded. For quantification of the fluorescent signal in epidermal cell in proPIN2::PIN2-GFP and proPIN1::PIN1-GFP. The same image acquisition parameters were used for all signal measurements. The regions of the visible BFA bodies in the same number and area of root cell were selected and the BFA body signal area (the area of BFA body/the constant root cell area containing same cell number) were calculated by the software ImageJ. To measure signal intensity of PM-localized PIN2-GFP, the mean pixel intensities were obtained from the apical and lateral sides of the individual cells by ImageJ. The PM-accumulation of PIN2-GFP was shown as the ratio of intensity (the apical side/the lateral side), 50–60 cells were analyzed for 5–7 seedlings in three independent treatments.
Statistical Analysis
Statistically significant differences in the results (**P < 0.05 or *P < 0.01) are based on Welch’s two-sample t test by SigmaPlot (v.14; Systat). The values of mock-treated and PISA-treated samples (Figs. 2, 3, and 5–8) and the values of wild-type and mutant samples treated with PISA at the same concentration (Fig. 4) were statistically tested. Data are means ± sd of independent replicates. Box-and-whisker plots show a median (centerline), upper/lower quartiles (box limits), and maximum/minimum (whiskers).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers: TIR1 (At3g62980), AFB2 (At3g26810), AXR1 (At1g05180), PIN1 (At1g73590), PIN2 (At5g57090), PIN3 (At1g70940), IAA14 (At4g14550), AUX1 (At2g38120), and PID (At2g34650).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Auxin activity in an auxin-deficient Arabidopsis mutant and BY2 tobacco cell culture.
Supplemental Figure S2. Effects of PISA on rapid cell expansion in hypocotyl.
Supplemental Figure S3. Effects of PISA on SCFTIR1 signaling.
Supplemental Figure S4. Effects of mPISA and PISA on the phenotype related to SCFTIR1/AFB pathway.
Supplemental Figure S5. Auxin transport inhibitors blocked PISA-induced high-auxin phenotype, but did not inhibit the high-auxin phenotypes by picloram and YUC1 overexpression.
Supplemental Figure S6. Effects of PISA on endogenous IAA level.
Supplemental Figure S7. Phenotype of Arabidopsis seedlings cocultured with PISA and auxins.
Supplemental Figure S8. Effects of PISA and auxin transport inhibitors on auxin response in root.
Supplemental Figure S9. PISA promoted the lateral root formation induced by membrane-permeable IAA precursors.
Supplemental Figure S10. PISA did not affect the expression of PIN1::GUS, PIN2::GUS and PIN7::GUS reporter expression.
Supplemental Figure S11. Effect of PISA on the BFA body formation of PIN1.
Supplemental Figure S12. Effect of PISA on the internalization of PIN2-GFP.
Supplemental Figure S13. Effect of PISA on the internalization of PIN1.
Supplemental Figure S14. Effect of PISA on the internalization of PIN2 at high concentration.
Supplemental Figure S15. Effects of PISA on PIN2 membrane localization in tir1 afb 1 afb 2 afb3 mutant.
Supplemental Figure S16. Molecular docking study of PAA, mPISA, and PISA with TIR1.
Supplemental Methods.Tobacco BY2 cell culture, pull-down assay, rapid cell elongation assay and the endogenous IAA measurement.
Acknowledgments
We thank Dr. H. Fukaki (University of Kobe), Dr. R. Offringa (Leiden University), Dr. Jianwei Pan (Zhejiang Normal University), and Dr. M. Estelle (University of California at San Diego) for providing mutants and transgenic line seeds.
Footnotes
This work was supported by the Ministry of Education, Culture, Sports, Science, and Technology (Grant-in-Aid for Scientific Research no. JP25114518 to K.H.), the Biotechnology and Biological Sciences Research Council (award no. BB/L009366/1 to R.N. and S.K.), and the European Union’s Horizon2020 program (European Research Council grant agreement no. 742985 to J.F.).
Articles can be viewed without a subscription.
References
- Abas L, Benjamins R, Malenica N, Paciorek T, Wiśniewska J, Moulinier-Anzola JC, Sieberer T, Friml J, Luschnig C (2006) Intracellular trafficking and proteolysis of the Arabidopsis auxin-efflux facilitator PIN2 are involved in root gravitropism. Nat Cell Biol 8: 249–256 [DOI] [PubMed] [Google Scholar]
- Adamowski M, Friml J (2015) PIN-dependent auxin transport: Action, regulation, and evolution. Plant Cell 27: 20–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamowski M, Narasimhan M, Kania U, Glanc M, De Jaeger G, Friml J (2018) A functional study of AUXILIN-LIKE1 and 2, two putative clathrin uncoating factors in Arabidopsis. Plant Cell 30: 700–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baster P, Robert S, Kleine-Vehn J, Vanneste S, Kania U, Grunewald W, De Rybel B, Beeckman T, Friml J (2013) SCF(TIR1/AFB)-auxin signalling regulates PIN vacuolar trafficking and auxin fluxes during root gravitropism. EMBO J 32: 260–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamins R, Quint A, Weijers D, Hooykaas P, Offringa R (2001) The PINOID protein kinase regulates organ development in Arabidopsis by enhancing polar auxin transport. Development 128: 4057–4067 [DOI] [PubMed] [Google Scholar]
- Brumos J, Robles LM, Yun J, Vu TC, Jackson S, Alonso JM, Stepanova AN (2018) Local auxin biosynthesis is a key regulator of plant development. Dev Cell 47: 306–318 e305 [DOI] [PubMed] [Google Scholar]
- Brunoud G, Wells DM, Oliva M, Larrieu A, Mirabet V, Burrow AH, Beeckman T, Kepinski S, Traas J, Bennett MJ, et al. (2012) A novel sensor to map auxin response and distribution at high spatio-temporal resolution. Nature 482: 103–106 [DOI] [PubMed] [Google Scholar]
- Buer CS, Muday GK (2004) The transparent testa4 mutation prevents flavonoid synthesis and alters auxin transport and the response of Arabidopsis roots to gravity and light. Plant Cell 16: 1191–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae K, Isaacs CG, Reeves PH, Maloney GS, Muday GK, Nagpal P, Reed JW (2012) Arabidopsis SMALL AUXIN UP RNA63 promotes hypocotyl and stamen filament elongation. Plant J 71: 684–697 [DOI] [PubMed] [Google Scholar]
- Chen Q, Dai X, De-Paoli H, Cheng Y, Takebayashi Y, Kasahara H, Kamiya Y, Zhao Y (2014) Auxin overproduction in shoots cannot rescue auxin deficiencies in Arabidopsis roots. Plant Cell Physiol 55: 1072–1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z, Galván-Ampudia CS, Demarsy E, Łangowski Ł, Kleine-Vehn J, Fan Y, Morita MT, Tasaka M, Fankhauser C, Offringa R, et al. (2011) Light-mediated polarization of the PIN3 auxin transporter for the phototropic response in Arabidopsis. Nat Cell Biol 13: 447–452 [DOI] [PubMed] [Google Scholar]
- Du Y, Tejos R, Beck M, Himschoot E, Li H, Robatzek S, Vanneste S, Friml J (2013) Salicylic acid interferes with clathrin-mediated endocytic protein trafficking. Proc Natl Acad Sci USA 110: 7946–7951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fendrych M, Leung J, Friml J (2016) TIR1/AFB-Aux/IAA auxin perception mediates rapid cell wall acidification and growth of Arabidopsis hypocotyls. eLife 5: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J, Yang X, Michniewicz M, Weijers D, Quint A, Tietz O, Benjamins R, Ouwerkerk PB, Ljung K, Sandberg G, et al. (2004) A PINOID-dependent binary switch in apical-basal PIN polar targeting directs auxin efflux. Science 306: 862–865 [DOI] [PubMed] [Google Scholar]
- Fukui K, Hayashi KI (2018) Manipulation and sensing of auxin metabolism, transport and signaling. Plant Cell Physiol 59: 1500–1510 [DOI] [PubMed] [Google Scholar]
- Furutani M, Nakano Y, Tasaka M (2014) MAB4-induced auxin sink generates local auxin gradients in Arabidopsis organ formation. Proc Natl Acad Sci USA 111: 1198–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly A, Lee SH, Cho M, Lee OR, Yoo H, Cho HT (2010) Differential auxin-transporting activities of PIN-FORMED proteins in Arabidopsis root hair cells. Plant Physiol 153: 1046–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner N, Friml J, Stierhof YD, Jürgens G, Palme K (2001) Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature 413: 425–428 [DOI] [PubMed] [Google Scholar]
- Geldner N, Anders N, Wolters H, Keicher J, Kornberger W, Muller P, Delbarre A, Ueda T, Nakano A, Jürgens G (2003) The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell 112: 219–230 [DOI] [PubMed] [Google Scholar]
- Hayashi K. (2012) The interaction and integration of auxin signaling components. Plant Cell Physiol 53: 965–975 [DOI] [PubMed] [Google Scholar]
- Hayashi K, Neve J, Hirose M, Kuboki A, Shimada Y, Kepinski S, Nozaki H (2012) Rational design of an auxin antagonist of the SCF(TIR1) auxin receptor complex. ACS Chem Biol 7: 590–598 [DOI] [PubMed] [Google Scholar]
- He W, Brumos J, Li H, Ji Y, Ke M, Gong X, Zeng Q, Li W, Zhang X, An F, et al. (2011) A small-molecule screen identifies l-kynurenine as a competitive inhibitor of TAA1/TAR activity in ethylene-directed auxin biosynthesis and root growth in Arabidopsis. Plant Cell 23: 3944–3960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jásik J, Bokor B, Stuchlík S, Mičieta K, Turňa J, Schmelzer E (2016) Effects of auxins on PIN-FORMED2 (PIN2) dynamics are not mediated by inhibiting PIN2 endocytosis. Plant Physiol 172: 1019–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara H. (2016) Current aspects of auxin biosynthesis in plants. Biosci Biotechnol Biochem 80: 34–42 [DOI] [PubMed] [Google Scholar]
- Kitakura S, Vanneste S, Robert S, Löfke C, Teichmann T, Tanaka H, Friml J (2011) Clathrin mediates endocytosis and polar distribution of PIN auxin transporters in Arabidopsis. Plant Cell 23: 1920–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korasick DA, Enders TA, Strader LC (2013) Auxin biosynthesis and storage forms. J Exp Bot 64: 2541–2555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Sundaram S, Armitage L, Evans JP, Hawkes T, Kepinski S, Ferro N, Napier RM (2014) Defining binding efficiency and specificity of auxins for SCF(TIR1/AFB)-Aux/IAA co-receptor complex formation. ACS Chem Biol 9: 673–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis DR, Muday GK (2009) Measurement of auxin transport in Arabidopsis thaliana. Nat Protoc 4: 437–451 [DOI] [PubMed] [Google Scholar]
- Leyser O. (2018) Auxin signaling. Plant Physiol 176: 465–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D, Nagawa S, Chen J, Cao L, Chen X, Xu T, Li H, Dhonukshe P, Yamamuro C, Friml J, et al. (2012) A ROP GTPase-dependent auxin signaling pathway regulates the subcellular distribution of PIN2 in Arabidopsis roots. Curr Biol 22: 1319–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marhavý P, Bielach A, Abas L, Abuzeineh A, Duclercq J, Tanaka H, Pařezová M, Petrášek J, Friml J, Kleine-Vehn J, et al. (2011) Cytokinin modulates endocytic trafficking of PIN1 auxin efflux carrier to control plant organogenesis. Dev Cell 21: 796–804 [DOI] [PubMed] [Google Scholar]
- Muir RM, Fujita T, Hansch C (1967) Structure-activity relationship in the auxin activity of mono-substituted phenylacetic acids. Plant Physiol 42: 1519–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagawa S, Xu T, Lin D, Dhonukshe P, Zhang X, Friml J, Scheres B, Fu Y, Yang Z (2012) ROP GTPase-dependent actin microfilaments promote PIN1 polarization by localized inhibition of clathrin-dependent endocytosis. PLoS Biol 10: e1001299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naramoto S, Kleine-Vehn J, Robert S, Fujimoto M, Dainobu T, Paciorek T, Ueda T, Nakano A, Van Montagu MC, Fukuda H, et al. (2010) ADP-ribosylation factor machinery mediates endocytosis in plant cells. Proc Natl Acad Sci USA 107: 21890–21895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M (2009) An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Methods 6: 917–922 [DOI] [PubMed] [Google Scholar]
- Okushima Y, Fukaki H, Onoda M, Theologis A, Tasaka M (2007) ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell 19: 118–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paciorek T, Zazímalová E, Ruthardt N, Petrásek J, Stierhof YD, Kleine-Vehn J, Morris DA, Emans N, Jürgens G, Geldner N, et al. (2005) Auxin inhibits endocytosis and promotes its own efflux from cells. Nature 435: 1251–1256 [DOI] [PubMed] [Google Scholar]
- Pan J, Fujioka S, Peng J, Chen J, Li G, Chen R (2009) The E3 ubiquitin ligase SCFTIR1/AFB and membrane sterols play key roles in auxin regulation of endocytosis, recycling, and plasma membrane accumulation of the auxin efflux transporter PIN2 in Arabidopsis thaliana. Plant Cell 21: 568–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prát T, Hajný J, Grunewald W, Vasileva M, Molnár G, Tejos R, Schmid M, Sauer M, Friml J (2018) WRKY23 is a component of the transcriptional network mediating auxin feedback on PIN polarity. PLoS Genet 14: e1007177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quareshy M, Uzunova V, Prusinska JM, Napier RM (2017) Assaying auxin receptor activity using SPR assays with F-Box proteins and Aux/IAA degrons. Methods Mol Biol 1497: 159–191 [DOI] [PubMed] [Google Scholar]
- Rakusová H, Fendrych M, Friml J (2015) Intracellular trafficking and PIN-mediated cell polarity during tropic responses in plants. Curr Opin Plant Biol 23: 116–123 [DOI] [PubMed] [Google Scholar]
- Rakusová H, Abbas M, Han H, Song S, Robert HS, Friml J (2016) Termination of shoot gravitropic responses by auxin feedback on PIN3 polarity. Curr Biol 26: 3026–3032 [DOI] [PubMed] [Google Scholar]
- Ren H, Gray WM (2015) SAUR proteins as effectors of hormonal and environmental signals in plant growth. Mol Plant 8: 1153–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigas S, Ditengou FA, Ljung K, Daras G, Tietz O, Palme K, Hatzopoulos P (2013) Root gravitropism and root hair development constitute coupled developmental responses regulated by auxin homeostasis in the Arabidopsis root apex. New Phytol 197: 1130–1141 [DOI] [PubMed] [Google Scholar]
- Robert HS, Grones P, Stepanova AN, Robles LM, Lokerse AS, Alonso JM, Weijers D, Friml J (2013) Local auxin sources orient the apical-basal axis in Arabidopsis embryos. Curr Biol 23: 2506–2512 [DOI] [PubMed] [Google Scholar]
- Robert S, Kleine-Vehn J, Barbez E, Sauer M, Paciorek T, Baster P, Vanneste S, Zhang J, Simon S, Čovanová M, et al. (2010) ABP1 mediates auxin inhibition of clathrin-dependent endocytosis in Arabidopsis. Cell 143: 111–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salanenka Y, Verstraeten I, Löfke C, Tabata K, Naramoto S, Glanc M, Friml J (2018) Gibberellin DELLA signaling targets the retromer complex to redirect protein trafficking to the plasma membrane. Proc Natl Acad Sci USA 115: 3716–3721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer M, Paciorek T, Benková E, Friml J (2006) Immunocytochemical techniques for whole-mount in situ protein localization in plants. Nat Protoc 1: 98–103 [DOI] [PubMed] [Google Scholar]
- Spartz AK, Lee SH, Wenger JP, Gonzalez N, Itoh H, Inzé D, Peer WA, Murphy AS, Overvoorde PJ, Gray WM (2012) The SAUR19 subfamily of SMALL AUXIN UP RNA genes promote cell expansion. Plant J 70: 978–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara S, Mashiguchi K, Tanaka K, Hishiyama S, Sakai T, Hanada K, Kinoshita-Tsujimura K, Yu H, Dai X, Takebayashi Y, et al. (2015) Distinct characteristics of indole-3-acetic acid and phenylacetic acid, two common auxins in plants. Plant Cell Physiol 56: 1641–1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda E, Yang H, Nishimura T, Uehara Y, Sakai T, Furutani M, Koshiba T, Hirose M, Nozaki H, Murphy AS, et al. (2011) Alkoxy-auxins are selective inhibitors of auxin transport mediated by PIN, ABCB, and AUX1 transporters. J Biol Chem 286: 2354–2364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsugafune S, Mashiguchi K, Fukui K, Takebayashi Y, Nishimura T, Sakai T, Shimada Y, Kasahara H, Koshiba T, Hayashi KI (2017) Yucasin DF, a potent and persistent inhibitor of auxin biosynthesis in plants. Sci Rep 7: 13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieten A, Vanneste S, Wisniewska J, Benková E, Benjamins R, Beeckman T, Luschnig C, Friml J (2005) Functional redundancy of PIN proteins is accompanied by auxin-dependent cross-regulation of PIN expression. Development 132: 4521–4531 [DOI] [PubMed] [Google Scholar]
- Winicur ZM, Zhang GF, Staehelin LA (1998) Auxin deprivation induces synchronous Golgi differentiation in suspension-cultured tobacco BY-2 cells. Plant Physiol 117: 501–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewska J, Xu J, Seifertová D, Brewer PB, Ruzicka K, Blilou I, Rouquié D, Benková E, Scheres B, Friml J (2006) Polar PIN localization directs auxin flow in plants. Science 312: 883. [DOI] [PubMed] [Google Scholar]