Functional analysis of a previously identified C17 cellulose-inhibitor-revealed that it operates differently from other known inhibitors, enabling the development of a dual herbicide-resistant line.
Abstract
The rapid appearance of herbicide-resistant weeds combined with a lack of novel herbicides being brought to market reduces crop production, thereby threatening food security worldwide. Here, we report on the use of the previously identified cellulose biosynthesis-inhibiting chemical compound C17 as a potential herbicide. Toxicity tests showed that C17 efficiently inhibits the growth of various weeds and widely cultivated dicotyledonous crops, whereas only slight or no growth inhibition was observed for monocotyledonous crops. Surprisingly, when exposed to a mixture of C17 and one of two well-known cellulose biosynthesis inhibitors (CBIs), isoxaben and indaziflam, an additive growth inhibition was observed, demonstrating that C17 has a different mode of action that can be used to sensitize plants toward known CBIs. Moreover, we demonstrate that a C17-resistant CESA3 allele can be used as a positive transformation selection marker and that C17 resistance can be obtained through genome engineering of the wild-type CESA3 allele using clustered regularly interspaced short palindromic repeats-mediated base editing. This editing system allowed us to engineer C17 tolerance in an isoxaben-resistant line, resulting in double herbicide-resistant plants.
Herbicides, chemical substances that are used to control undesirable vegetation, have become an essential part of modern agriculture. Weeds can lead to dramatic crop losses, sometimes even above 90% (Cobb and Reade, 2010; Quareshy et al., 2018). In such situations, herbicides have become crucial for ensuring food security, particularly in the current situation of an ever-increasing demand on global food, driven by rapid population growth and dietary changes. However, despite the need for herbicides, to date only 17 classes of herbicides representing diverse ways to control undesirable vegetation have been found according to the Herbicide Resistance Action Committee classification (Délye et al., 2013). Currently available herbicides disturb the synthesis of biological molecules, such as fatty acids, amino acids, tetrahydrofolate, and ATP, or interfere with essential biological activities, including photosynthesis, cell wall synthesis, microtubule organization, hormone transport, and hormone-based gene regulation (Cobb and Reade, 2010; Délye et al., 2013).
In addition to a limited variety of different herbicides with distinct modes of action, a significant agricultural problem is arising with the increased occurrence of resistance due to natural selection. As of 2014, herbicide tolerance was identified in 220 weed species, covering 21 of the 25 known herbicide sites of action (Heap, 2014). Herbicide-resistant weeds appear or are selected quickly, in as few as six years after a new herbicide is applied (Heap, 2014; Quareshy et al., 2018). It has been reported that 32 2,4-d herbicide-resistant species were generated or selected over only 59 years, between 1957 and 2016 (Quareshy et al., 2018). This rapid generation of herbicide resistance impels the discovery of novel herbicides or strategies to apply to them.
Some herbicides suffer less from the appearance of herbicide resistance development compared to others, including the cellulose biosynthesis inhibitors (CBIs; Heap, 2014). CBIs target the cellulose synthase complexes (CSCs), containing six cellulose synthase subunits (CESAs), which processively catalyze the conversion of UDP-Glc to cellulose (Kimura et al., 1999). Synthesized cellulose can crystallize into cellulose microfibrils through inter- and intramolecular hydrogen bonds and Van der Waals forces and serve as the structural reinforcement of the cell wall (McFarlane et al., 2014). Arabidopsis (Arabidopsis thaliana) holds ten distinct CESA proteins (Somerville, 2006). At least three different CESAs are present in a CSC (Lei et al., 2012; McFarlane et al., 2014), with the CSC of the primary cell wall being mainly composed of CESA1, CESA3, and CESA6 (Lei et al., 2012). Several CBIs, such as isoxaben, flupoxam, dichlobenil, and indaziflam have been developed as herbicides. These herbicides not only structurally belong to different classes of chemicals, but they also appear to operate through different modes of action (Brabham and Debolt, 2013; Tateno et al., 2016). Isoxaben causes the clearance of CSCs from the plasma membrane (PM) focal plane, while indaziflam and dichlobenil increase CSC accumulation but reduce their velocity in the PM (Paredez et al., 2006; DeBolt et al., 2007; Brabham et al., 2014). Although flupoxam’s mode of action on the CSC is still unknown, the amino acid mutation sites in CESA proteins that confer resistance to flupoxam are completely different from those conferring resistance to isoxaben (Tateno et al., 2016). In addition, some new CBIs, such as CESTRIN, acetobixan and quinoxyphen, and new quinoxyphen resistance alleles (cesa1A903V, cesa1R292C, and cesa1G620E) have been identified (Harris et al., 2012; Xia et al., 2014; Worden et al., 2015).
Through a compound screening approach, we recently identified a novel growth inhibitor named C17 (5-(4-chlorophenyl)-7-(2-methoxyphenyl)-1,5,6,7-tetrahydro-[1,2,4]triazolo[1,5-a] pyrimidine). C17 administration was described to result in CESA depletion from the PM. Furthermore, its application results in a weakened cell wall and reduced hypocotyl elongation, correlated with a reduction in cellulose content (Hu et al., 2016). A putative direct interaction between the compound and CESA subunits was suggested by a mutagenesis suppressor screen that identified C17-tolerant mutants carrying single-nucleotide missense changes at either CESA1 or CESA3 (Hu et al., 2016). Here, we demonstrate that C17 might be developed as a potential herbicide operating differently from other known CBIs, whereas its resistance alleles can be used as positive transformation selection markers. Moreover, the ability to mimic C17 resistance through clustered regularly interspaced short palindromic repeats (CRISPR)-mediated base editing suggests the possibility to transfer CBI multiresistance to any crop of interest.
RESULTS
C17 Resistance Protects against CESA PM Depletion
We have previously demonstrated that C17 depletes CSCs from the PM, which resulted in cellulose deficiency, and identified 12 different C17-resistant alleles, being mutant in either CESA1 or CESA3 (Hu et al., 2016). Cellulose is crucial for structural support of the cell, and its deficiency generally induces lignin production (Hamann et al., 2009). C17 administration resulted in an obvious lignin accumulation in the root of wild-type (Col-0) seedlings, but not in those of the C17-resistant CESA3 (CESA3S1037F) 8P mutants (Supplemental Fig. S1). To further validate C17’s disruption of cellulose, we analyzed the cellulose organization with a fluorescent cellulose dye, Pontamine Fast Scarlet 4B (Anderson et al., 2010). As expected, a dramatic reduction of cellulose in the root was observed following C17 treatment (Supplemental Fig. S2), which did not appear in the 8P mutant, supporting the idea that C17 is a cellulose synthesis inhibitor and the C17-resistant allele abolishes C17 function.
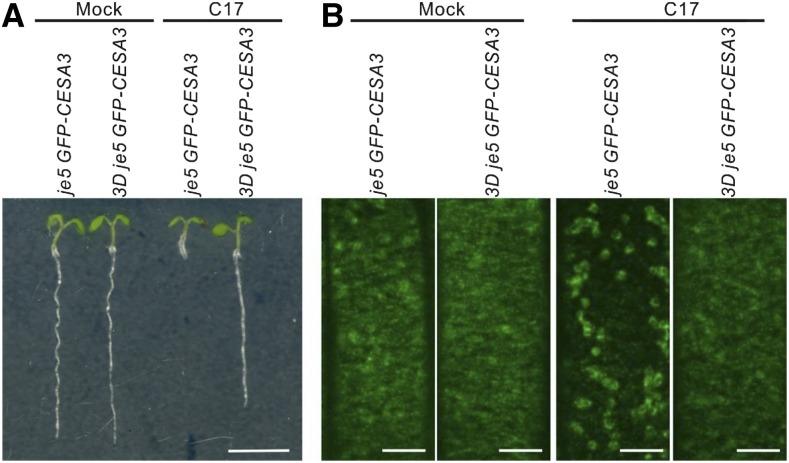
Because we speculated that C17 resistance might be the result of the identified point mutations blocking the depletion of the CESA complexes from the PM, we crossed the CESA3 mutant je5 harboring a GFP-CESA3 reporter (je5 GFP-CESA3) with the C17-tolerant CESA1 (CESA1V297M) 3D mutant, creating the 3D je5 GFP-CESA3 mutant (Desprez et al., 2007; Hu et al., 2016). Compared to je5 GFP-CESA3, 3D je5 GFP-CESA3 plants showed resistance to C17, seen by the absence of a stunted swollen root following compound application (Fig. 1A). Through live cell imaging, we analyzed the abundance of CESA3-containing CSCs associated with the PM as indicated by the GFP-CESA3 signal. Consistent with previous findings (Hu et al., 2016), 200 nm C17 rapidly depleted CESA3s in the je5 GFP-CESA3 membrane (Fig. 1B), whereas no significant change in the GFP-CESA3 signal in the PM was observed when 3D je5 GFP-CESA3 seedlings were exposed to 200 nm C17 (Fig. 1B).
Figure 1.
The 3D point mutation in CESA1 inhibits root growth and abolishes C17-induced CESA3 depletion in the plasma membrane. A, Root growth of 5-d-old je5 GFP-CESA3 and 3D je5 GFP-CESA3 plants grown in the absence (mock) or presence of 200 nm C17. Scale bar, 5 mm. B, Representative spinning confocal microscopy images of 4-d-old je5 GFP-CESA3 and 3D je5 GFP-CESA3 plants treated with 0.1% (v/v) dimethyl sulfoxide (DMSO; mock) and 200 nm C17. The images were taken 20 min after treatment. Scale bars, 5 µm.
C17 Operates Differently from Indaziflam and Isoxaben
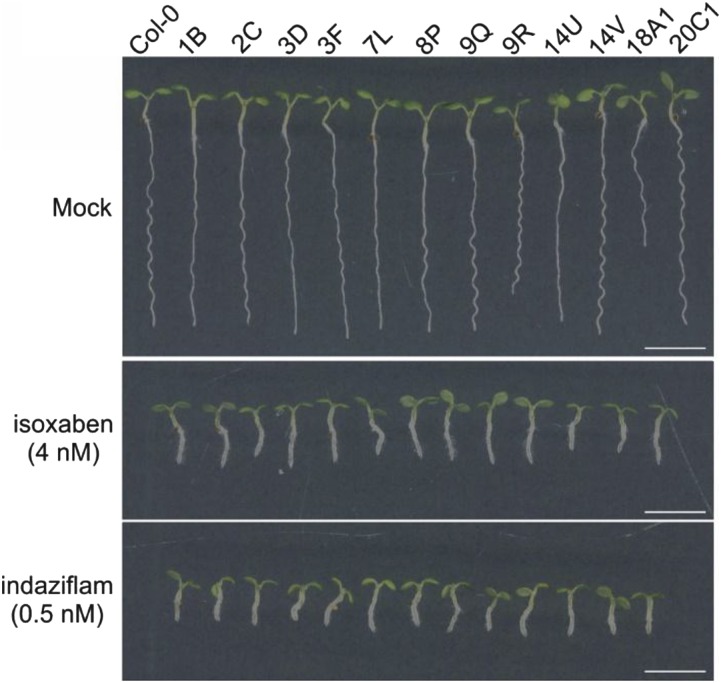
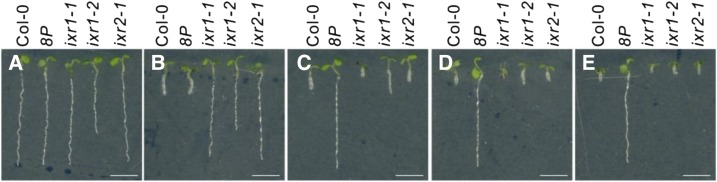
Isoxaben and idaziflam are commercial inhibitors of cellulose biosynthesis. Because C17 displays a chemical structure that is totally different from these two CBIs, we tested whether C17 has a different mode of action by evaluating the sensitivity of 12 C17-tolerant mutants to indaziflam and isoxaben. The C17-tolerant mutants carry a single-nucleotide missense mutation at either CESA1 or CESA3 (Hu et al., 2016). Based on the evaluation of root growth of seedlings germinated on medium containing the CBIs, all C17-tolerant mutants had the same sensitivity as wild-type plants to either isoxaben or idaziflam (Fig. 2), indicating that C17 acts through another mechanism. Conversely, isoxaben-resistant mutants (ixr1-1, ixr1-2, and ixr2-1), corresponding to mutant alleles of CESA3 and CESA6 (Scheible et al., 2001; Desprez et al., 2002), are sensitive to C17 (Fig. 3).
Figure 2.
C17-tolerant mutants are sensitive to isoxaben and idaziflam. Five-day-old wild type (Col-0) and 12 C17-tolerant mutants (1B, 2C, 3D, 3F, 7L, 8P, 9Q, 9R, 14U, 14V, 18A1, and 20C1) grown on one-half Murashige and Skoog (1/2 MS) medium (mock, upper) or 1/2 MS medium supplemented with isoxaben (4 nm, middle) or idaziflam (0.5 nm, bottom). Scale bars, 5 mm.
Figure 3.
Isoxaben-resistant mutants are sensitive to C17. A to E, Root growth of 5-d-old wild type (Col-0), a C17-tolerant mutant (8P), and three isoxaben-tolerant mutants (ixr1-1, ixr1-2, and ixr2-1) grown in the absence (A) or presence of 4 nm isoxaben (B), 200 nm C17 (C), 500 nm C17 (D), and 1 µm C17 (E). Scale bars, 5 mm.
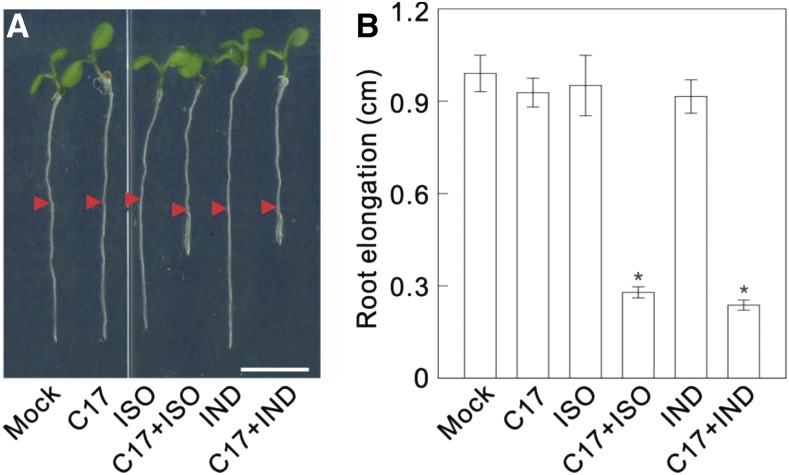
At low concentrations of C17 (50 nm), isoxaben (1 nm), or indaziflam (100 pm), no growth inhibition occurred (Fig. 4A). Strikingly, a dramatic growth inhibition was observed when wild-type plants were treated with a mixture of these low concentrations of C17 and isoxaben or indaziflam, because root elongation only reached 28% or 24% of the control, respectively (Fig. 4), indicating that C17 administration increased the sensitivity to indaziflam and isoxaben.
Figure 4.
C17 shows an additive growth inhibition with isoxaben/indaziflam. A, Root growth of wild type (Col-0) in the absence (mock) or presence of 50 nm C17 (C17), 1 nm isoxaben (ISO), a mixture of 50 nm C17 and 1 nm isoxaben (C17 + ISO), 100 pM indaziflam (IND), and a mixture of 50 nm C17 and 100 pM indaziflam (C17 + IND). Three-day-old seedlings grown on 1/2 MS medium were transferred for 2 days to control medium (control) or medium containing indicated CBIs. Arrowheads indicate the root tip position at the moment of transfer. Scale bar, 5 mm. B, Quantification of root length of plants in A. Data represent mean ± sd (n > 10 plants). Statistically significant differences compared to mock are indicated; *P < 0.01 (two-sided Student’s t test).
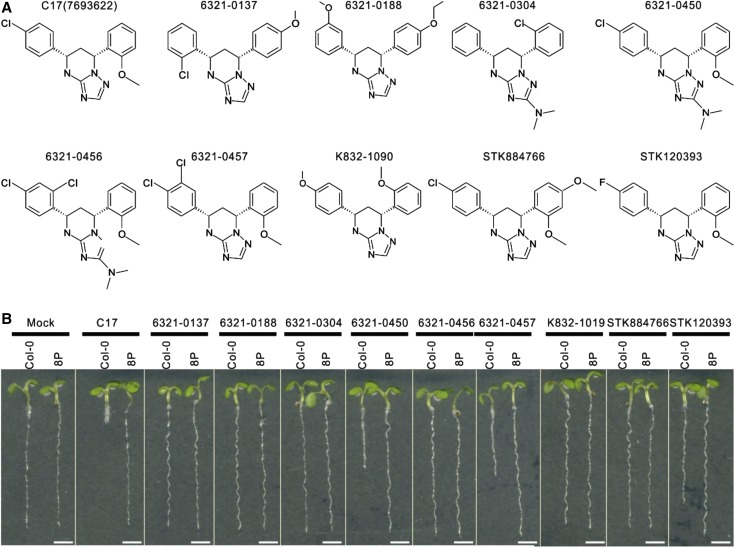
Structure-Activity Analysis of C17-Related Molecules
The C17 molecule is a triazolopyrimidine derivative containing a chlorophenyl and methoxyphenyl side chain (Hu et al., 2016). To identify the pharamocophore that is essential for the inhibition of plant growth, nine commercially available chemical analogs of C17 (Fig. 5A) were examined for their root growth inhibitory activity. A 200 nm concentration was selected, because at this moderate concentration of C17, wild-type root growth was severely inhibited (Fig. 5B). Three tested C17 analogs (6321-0450, 6321-0457, and STK120393) inhibited root growth of the wild type, albeit to a lesser degree than observed for C17 (Fig. 5B), as indicated by the higher compound concentration required to reduce root growth by 50% compared to untreated roots (IC50 value; Supplemental Fig. S3). Growth of the previously identified C17-tolerant mutant 8P (CESA3S1037F) was not affected, indicating that the growth inhibitory effect was linked to CESA3 activity. Six structural analogs did not display growth inhibitory activity in both wild-type and 8P plants. Comparing the different structures indicated that both the chloro group at position 4 of the chlorophenyl side chain and the methoxy group at position 2 of the methoxyphenyl side group are the important factors influencing C17 toxicity, because any substitution and/or position change of these groups in the C17 analogs resulted in a reduced or even complete loss of growth inhibitory activity (Fig. 5). In addition, group addition in C17 (e.g. 6321-0450, 6321-0456, 6321-0457, and STK884766) also affected C17 growth inhibitory activity (Fig. 5).
Figure 5.
C17 structure-function analysis. A, Chemical structures of C17 and its analogs. B, Five-day-old wild type (Col-0) and C17-tolerant mutant 8P grown on 1/2 MS medium (mock) or 1/2 MS medium supplemented with 200 nm of C17 or its analogs. Scale bars, 2.5 mm.
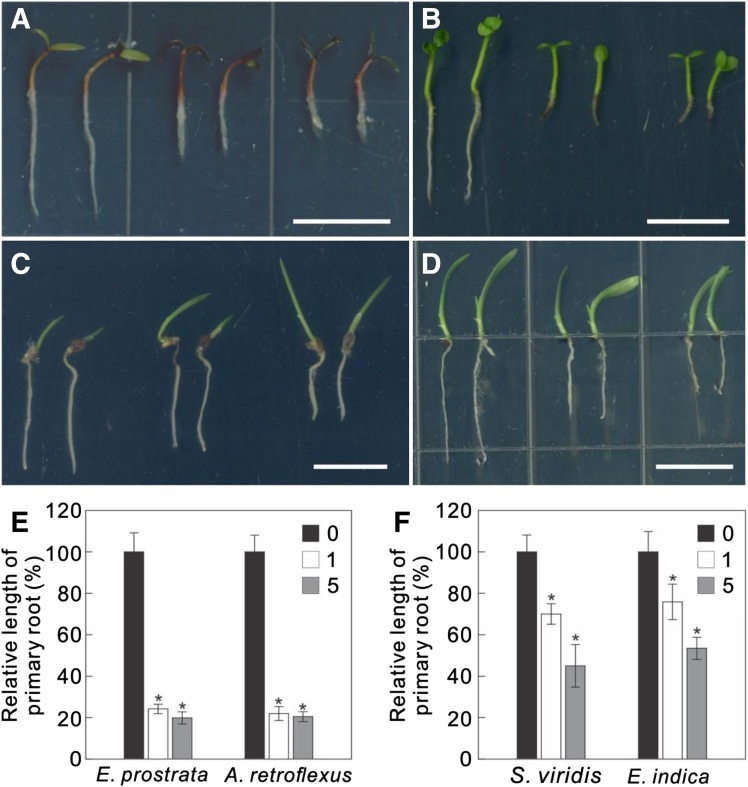
C17 Effectively Inhibits Weed Growth
Because C17 inhibits plant growth, making it a potentially useful herbicide, we tested C17 toxicity for various weeds in vitro. As expected, all tested weeds including Eclipta prostrata, Amaranthus retroflexus, Setaria viridis, and Eleusine indica were susceptible to C17 and their growth was inhibited in a C17 dose-dependent manner (Fig. 6). The dose required to inhibit the growth of monocotyledonous weeds (Fig. 6, C and D) was higher than that for dicotyledonous weeds (Fig. 6, A and B). For instance, root elongation of E. prostrata and A. retroflexus was completely inhibited by application of 1 µm C17, whereas around 50% root elongation of S. viridis and E. indica was still observed when grown at a concentration as high as 5 µm C17 (Fig. 6, E and F).
Figure 6.
C17 effectively inhibits the growth of weeds. A to D, Root growth of E. prostrata (A), A. retroflexus (B), S. viridis (C), and E. indica (D) in the absence (left) or presence of 1 µm (middle) and 5 µm C17 (right). Germinated weeds grown on 1/2 MS medium were transferred for 2 days to control medium or medium containing 1 µm or 5 µm C17. Scale bars, 1 cm. E, Root length quantification of plants in A and B. F, Root length quantification of plants in C and D. Data represent mean ± sd (n > 4 plants). Statistically significant differences compared to plants grown in the absence of C17 are indicated; *P < 0.01 (two-sided Student's t test).
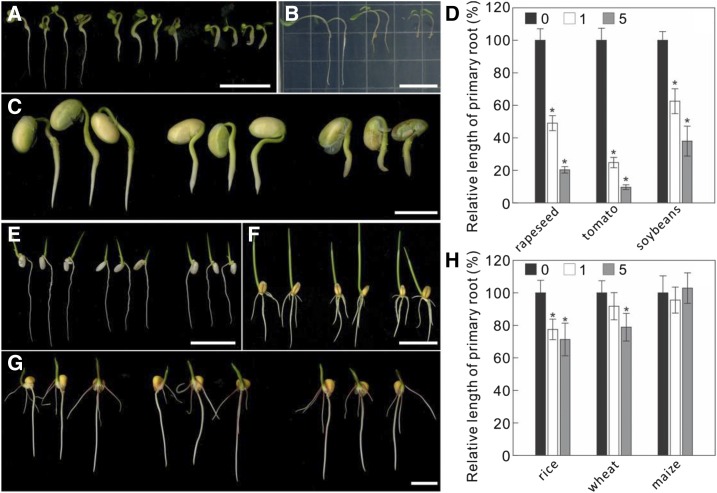
C17 Is Growth Inhibitory for Dicotyledonous Crops
Next, we evaluated the application possibility of C17 in crops. The sensitivity to C17 of six widely cultivated crops was tested, including three dicotyledonous crops (rapeseed [Brassica napus], soybean [Glycine max], tomato [Solanum lycopersicum]) and three monocotyledonous crops (maize [Zea mays], rice [Oryza sativa], and wheat [Triticum aestivum]). As shown in Figure 7, A–D, C17 was able to effectively inhibit root elongation of dicotyledonous crops. However, no significant growth inhibition was observed for maize, and only slight growth inhibition for rice and wheat, which was only significant for wheat when 5 µm C17 was applied (Fig. 7, E–H).
Figure 7.
Crop sensitivity to C17. A to C, Root growth of rapeseed (A), tomato (B), and soybean (C) in the absence (left) or presence of 1 µm (middle) and 5 µm C17 (right). Germinated dicotyledonous crops grown on 1/2 MS medium were transferred for 2 days to control medium or medium containing 1 µm or 5 µm C17. Scale bars, 2 cm. D, Root length quantification of plants in A, B, and C. E to G, Root growth of rice (E), wheat (F), and maize (G) in the absence (left) or presence of 1 µm (middle) and 5 µm C17 (right). Germinated monocotyledonous crops grown on 1/2 MS medium were transferred for 2 days to control medium or medium containing 1 µm or 5 µm C17. Scale bars, 2 cm. H, Root length quantification of plants in E, F, and G. Data represent mean ± sd (n > 4 plants). Statistically significant differences compared to plants in absence of C17 are indicated; *P < 0.01 (two-sided Student's t test).
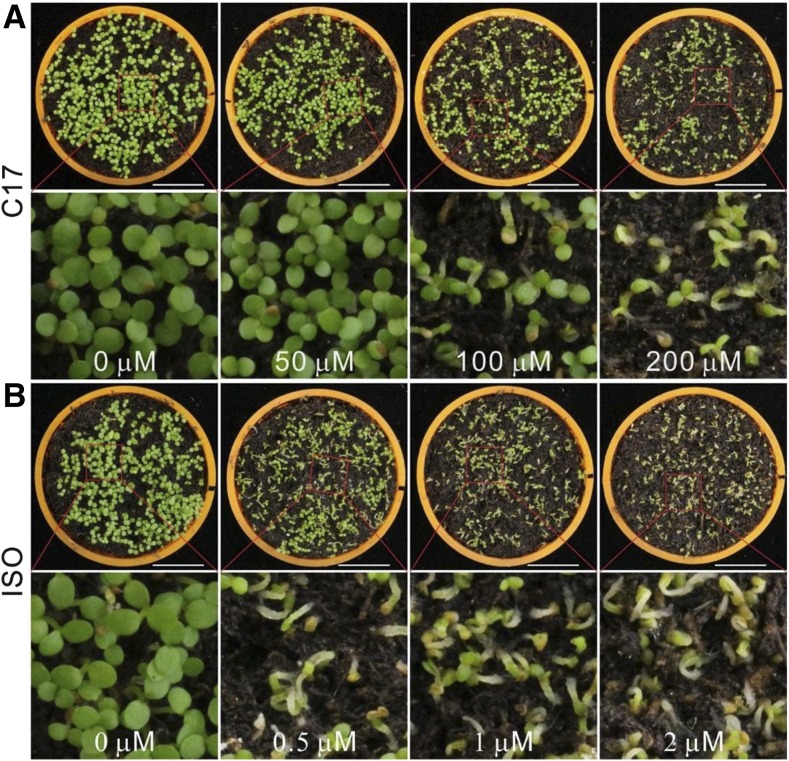
C17 Can be Used as a Spray-Herbicide on Soil
To test the potential use of C17 under non-lab conditions, seeds were sown on soil in the greenhouse and either treated by spraying with C17-containing solution or solvent-containing water only. As the positive control, plants were treated with isoxaben. Six days after sowing, the plants treated with C17 (≥100 μm) displayed a clear growth inhibition, with the seedlings displaying swollen hypocotyls and bleached cotyledons (Fig. 8A). Similar growth inhibition was observed when plants were treated with isoxaben (≥0.5 μm; Fig. 8B). These data demonstrate the potential use of C17 under field conditions.
Figure 8.
C17 effectively inhibits the growth of Arabidopsis on soil. A and B, Six-day-old Arabidopsis seedlings were grown on soil sprayed with a solution containing C17 (50 μm, 100 μm, and 200 μm; A) or isoxaben (0.5 μm, 1 μm, and 2 μm; B) or water containing solvent (0 μm). Scale bars, 1 cm.
Introducing a Mutant CESA3 Allele Confers C17 Tolerance in Arabidopsis
As an herbicide, the generation of C17-resistant plants helps to broaden its application, thereby promoting agricultural value of the compound. To achieve this purpose, the cesa3 mutant allele 2C (CESA3S983F) driven by the constitutive Cauliflower mosaic virus 35S promoter (35S-2C) was transformed into Col-0 wild-type Arabidopsis plants. Surprisingly, we were unable to obtain any 35S-2C transformants out of 20,000 T1 seeds. In contrast, a control construct (35S-GFP) yielded 38 transformants from 20,000 T1 seeds. These data indicated that high (mutant) CESA3 levels in a wild-type background might be lethal. Therefore, to lower CESA3 abundance and activity in transgenic plants, we expressed the 2C mutant allele from its own promoter (pCESA3-2C) and transformed it into the CESA3 mutant je5. Twenty-eight pCESA3-2C transgenic je5 plants were obtained from 20,000 transformed T1 seeds using the transfer DNA (T-DNA) selection marker hygromycin, which were all tolerant to C17, supporting the observation that the 2C mutation confers C17 resistance. Because of the 100% correlation between hygromycin and C17 resistance, we speculated that pCESA3-2C transgenic je5 plants might as well be directly selected on medium supplied with C17. To test this possibility, around 20,000 pCESA3-2C transformed T1 seeds in the je5 background were plated on MS supplemented with 500 nm C17. Thirty C17-tolerant T1 plants were identified, which were all confirmed to harbor the T-DNA by PCR-based genotyping. These data demonstrate that introducing a CESA mutant allele can bring C17 tolerance to plants.
Base Editing for C17 Resistance
Because a C17-resistant allele could not be introduced into wild-type plants using classical transformation, we applied the recently described CRISPR base-editing system, referred to as BE3 (Komor et al., 2016). This system makes use of apolipoprotein B mRNA editing enzyme, catalytic polypeptide 1, a cytidine deaminase that catalyzes the deamination of cytosine (C) to uracil (U), which is repaired as a thymine (T; Conticello, 2008). Two BE3 vectors were constructed to precisely generate C-to-T transitions to recapitulate the ethyl methanesulfonate mutants, CESA3S983F (2C) and CESA3S1037F (8P). The vectors were transformed into Arabidopsis cell suspension cultures to check the efficiency of targeted base editing in plant cells. Two weeks after transformation with the BE3-CESA3S983F vector, the population of cells showed no edits as determined by Sanger sequencing, but 5 and 7 weeks after transformation, C-to-T mutations were apparent (Supplemental Fig. S4A), demonstrating that this base-editing vector works in Arabidopsis cells.
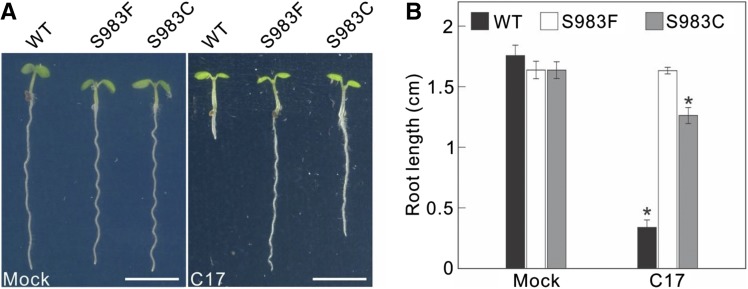
Subsequently, the two vectors were transformed into Arabidopsis by floral dip. A subset of T1 seeds was selected on medium with kanamycin. Of the 23 BE3-CESA3S983F and 19 BE3-CESA3S1037F kanamycin-resistant T1 plants, eight BE3-CESA3S983F T1 plants had clear C-to-T mutations in the sequencing chromatograms (Supplemental Fig. S4B), whereas none of the BE3-CESA3S1037F T1 plants showed any signs of editing. Having established that base editing was occurring in Arabidopsis T1 plants transformed with one of our base-editing vectors, additional T1 seeds were directly sown on medium supplemented with 1 μm C17. Approximately 2,000 T1 BE3-CESA3S983F seedlings were screened and nine C17-tolerant plants were identified (Supplemental Fig. S5A). Sanger sequencing confirmed that four of the C17-tolerant plants had a C-to-T mutation at the BE3-CESA3S983F target. In agreement with the lack of base editing in cell suspensions and kanamycin-resistant T1 plants, none of the ∼1,800 BE3-CESA3S1037F plants screened on C17 showed resistance (Supplemental Fig. S5B). Homozygous, T-DNA-free plants were derived from two independent T1 lines. One contained the intended C-to-T transversion, BE3-CESA3S983F, and the second contained a C-to-G transition, BE3-CESA3S983C. These lines were further used to analyze the inheritance of C17 tolerance. Both editing events could be inherited, with T4 progeny showing C17 tolerance (Fig. 9). BE3-CESA3S983F plants were slightly more tolerant to C17 than CESA3S983C plants, showing no significant difference in growth in the absence of C17 (Fig. 9B).
Figure 9.
C17 tolerance induced by the BE3-CESA3S983F vector could be inherited. A, Root growth of 5-d-old wild type (WT) and T3 progenies of BE3-CESA3S983F (S983F) and BE3-CESA3S983C (S983C) in the absence (mock) or presence of 200 nm C17. Scale bars, 5 mm. B, Root length quantification of plants in A. Data represent mean ± sd (n > 4 plants). Statistically significant differences compared with plants grown in the absence of C17 are indicated; *P < 0.01 (two-sided Student's t test).
Stacked Mutations for Simultaneous Resistance to C17 and Isoxaben
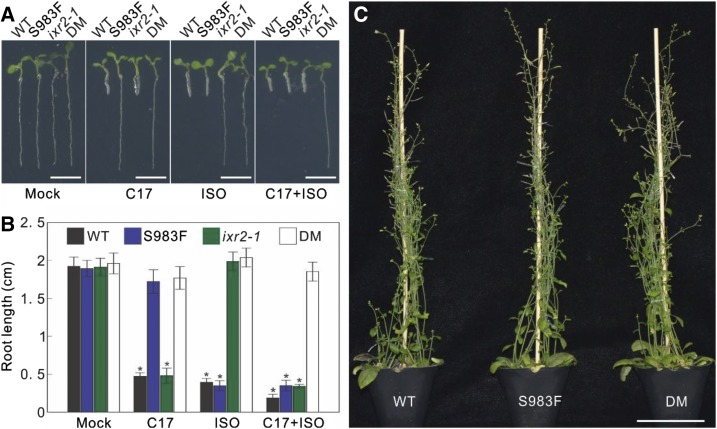
The successful BE3-CESA3S983F vector was transformed into the isoxaben-resistant genotype ixr2-1 and screened for resistance to C17. Because only three C17-resistant T1 plants were initially identified, additional lines were selected on kanamycin. In total, seven ixr2-1 kanamycin-resistant lines were isolated. Seeds from these primary transformants were collected, and C17 resistance was observed in all lines. Offspring plants displayed resistance to both C17 and isoxaben (Fig. 10A). Root lengths of BE-CESA3S983F ixr2-1 mutants grown on medium supplemented with both C17 and isoxaben were similar to those of wild type grown on normal medium without C17 and isoxaben (Fig. 10, A and B). In addition, there was no obvious growth defect in BE-CESA3S983F ixr2-1 that occurred during the whole life cycle in normal medium, compared to the wild type (Fig. 10C).
Figure 10.
Stacked mutations for simultaneous resistance to C17 and isoxaben. A, Root growth of 5-d-old wild type (WT) and progenies of BE3-CESA3S983F (S983F), ixr2-1, and double mutant ixr2-1 BE3-CESA3S983F (DM) in the absence (mock) or presence of 200 nm C17, 3 nm isoxaben (ISO), and the combination of 200 nm C17 and 3 nm isoxaben (C17 + ISO). Scale bars, 5 mm. B, Root length quantification of plants in A. Data represent mean ± sd (n > 4 plants). Statistically significant differences compared with plants in absence of C17 are indicated, *P < 0.01 (two-sided Student's t test). C, Morphology of 60-d-old wild type, S983F, and DM. Scale bar, 5 cm.
DISCUSSION
CBIs and their resistant genes are very valuable, not only to dissect the functions of cellulose and the CESA proteins, but also for the development and application of herbicides (Desprez et al., 2002; Harris et al., 2012; Sethaphong et al., 2013). Generally, CBIs are classified according to their mode of action, triggering either a depletion of the CSCs from the PM, CESA immobilization and accumulation in foci, or resulting in incorrect CESA movement and localization (Brabham and Debolt, 2013). Our results here and before (Hu et al., 2016) show that C17 triggers CESA depletion from the PM, which is prevented in a C17-resistant mutant. The mode of action of C17 appears to be unique or at least different from two well-known commercial herbicides, indaziflam and isoxaben. Previously, isoxaben-resistant plants have shown no cross resistance to indaziflam (Brabham et al., 2014), which might not be surprising given the observations that isoxaben results in CESA depletion from the PM, whereas indaziflam treatment results in a reduced velocity and an increased number of fluorescently labeled CSCs at the PM, clearly demonstrating that both drugs work differently (Paredez et al., 2006; Brabham et al., 2014). More surprisingly, we found that isoxaben- and C17-resistant plants also do not display cross resistance, despite the fact that both drugs affect CESAs in a similar way. This lack of cross resistance suggests that C17 and isoxaben promote CESA depletion differently, probably by targeting distinct contact sites. Indeed, not only do both drugs lack any structural homology, none of the 12 different previously identified CESA alleles that confer C17 resistance are identical to any of the nine known isoxaben-resistant alleles (Hu et al., 2016; Tateno et al., 2016). Moreover, whereas most of the isoxaben-resistance-conferring mutations reside in the predicted CESA cytoplasmic and extracellular loop domains, almost all C17-resistance mutations map to the transmembrane domains (Hu et al., 2016; Tateno et al., 2016).
Isoxaben and indaziflam have been developed as commercial herbicides. Isoxaben is commercially available as Gallery and in combination with trifluralin as Snapshot, while indaziflam is sold under the names Marengo and Specticle (McCullough et al., 2013; Saha et al., 2017). They are widely used for weed control in agriculture, particularly as pre-emergent herbicides in golf courses, vineyards, recreational lawns, orchards, and railroad tracks (Tateno et al., 2016). As a new member of the CBI family, C17 has the potential to be developed as a new herbicide as well. Indeed, our greenhouse experiments demonstrate that C17 can be administered on soil, although currently requiring a relatively high dose. This dose can, however, be significantly decreased with additives, helping the herbicide to remain in the topsoil and attach to the plant. Moreover, our structure-function analysis demonstrates the potential possibility to improve C17’s activity through targeted chemistry.
Our data have shown that C17 can effectively inhibit the growth of dicotyledonous and monocotyledonous weeds, although monocotyledonous weed growth is only inhibited at a higher dose, consistent with previous observations that the growth inhibition triggered by CBIs is more pronounced in dicots than monocots (Corio-Costet et al., 1991; Sabba et al., 1999). Such differences might be explained by the presence of a different cell wall composition or differences in uptake of the CBIs. Only slight or no growth inhibition was observed in monocotyledonous crops, including rice, wheat, and maize, indicating that C17 might be applicable to use in these monocot fields. Additionally, the additive growth inhibition of C17 with isoxaben or indaziflam could be used to, at least partially, overcome the ever-increasing occurrence of herbicide-resistant weeds by making a mixture of C17 with isoxaben or indaziflam.
Next to the potential use as an herbicide, we suggest the use of C17-resistant alleles as a positive selection marker in transformation experiments. In support of such application, we have successfully generated C17-resistant plants by introducing pCESA3-2C into the je5 background. Strikingly, ectopic 2C expression in wild type resulted in no 35S-2C transformants, suggesting that plants might not tolerate a strong increase in CESA3 abundance. Possibly, high levels of CESAs disturb the stoichiometry of the CSCs. The observed inability to introduce an overexpression construct of mutated CESA3 into the genome would limit the transformation with C17-resistant alleles to those plant species for which a CESA knockout or knockdown line is available. To circumvent this restriction, we have successfully applied the CRISPR base editor system BE3 (Komor et al., 2016). Of the nine C17-tolerant plants generated by the CRISPR base editor system that were selected directly on C17-containing medium, the CESA3S983F mutation was confirmed in only four plants. The lack of an absolute linkage between C17 resistance and the presence of the targeted C-to-T mutation can be attributed to the chimeric nature of mutations in somatic cells of T1 Arabidopsis CRISPR plants, which has been demonstrated in several studies (Feng et al., 2013; Fauser et al., 2014). Although the BE3-CESA3S983F vector could efficiently make C-to-T editing, the BE3-CESA3S1037F vector could not. Editing efficiency is dependent on the target sequence context (Komor et al., 2016), where TC and CC sequences are preferred. However, this would not explain the failure here, because both targets are TC. There may be unknown sequence contexts that explain the low efficiency at this site.
As mentioned above, C17 may be applicable as a component of an herbicide mixture with other CBIs to slow down the incidence of herbicide-resistant weeds. This scenario requires the combination of two CESA3 alleles conferring herbicide resistance. Whereas the combination of two such mutant alleles is practically impossible to obtain through recombination in classical breeding programs, we successfully obtained such plants through CRISPR base editing with the BE-CESA3S983F vector in the isoxaben-resistant genotype ixr2-1. Previously, Harris et al. (2012) produced double drug-resistant mutants to quinoxyphen and isoxaben by classic crossing to combine the mutant CESA1 and CESA3 alleles. However, these double drug-resistant plants displayed a pronounced dwarf phenotype, which possibly results from a dramatic loss of CSC activity. In this study, the double drug-resistant plants did not show any growth phenotype, at least under the conditions tested, indicating their potential value in crop design. The possibility to obtain multiherbicide resistance without an obvious growth penalty, combined with the possible use of C17 as an herbicide and its resistant alleles as novel transformation selection markers, earmarks C17 as a novel fundamental and applied research tool.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) Col-0 plants were grown under long-day conditions (16 h of light/8 h of darkness) at 22°C on 1/2 MS germination medium (Murashige and Skoog, 1962). Twelve C17-tolerant mutants (1B, 2C, 3D, 3F, 7L, 8P, 9R, 9Q, 14U, 14V, 18A1, and 20C1), je5, je5 GFP-CESA3, ixr1-1, ixr1-2, and ixr2-1 have been described previously (Scheible et al., 2001; Desprez et al., 2002; Hu et al., 2016). For greenhouse experiments, Col-0 seeds were sown on the surface of the soil and then sprayed with a water solution containing C17 (50 μm, 100 μm, and 200 μm) or isoxaben (0.5 μm, 1 μm, and 2 μm) or solvent (0 μm).
Staining of Lignin and Cellulose
Lignin staining was performed as described by Caño-Delgado et al. (2003), using 7-d-old seedlings that were transferred to liquid 1/2 MS medium supplemented with C17 (2 μm) or solvent only (DMSO). Seedlings were harvested 42 h later. Cellulose observations in the roots were performed as described previously with slight modification as follows (Anderson et al., 2010). Seedlings were collected from the plate and placed in a 1.5-mL centrifuge tube containing 0.01% (w/v) Pontamine Fast Scarlet 4B in liquid 1/2 MS medium for 1 h, washed one time with water, and mounted on slides for observation using a 100× oil immersion objective on a Zeiss710 microscope.
Chemical Treatments
C17 (catalog no. 7693622) and its analogs (catalog no. 6321-0137, 6321-0188, 6321-0304, 6321-0450, 6321-0456, 6321-0457, K832-1019, STK884766, and STK120393) were purchased from ChemBridge, and isoxaben and idaziflam were purchased from Sigma-Aldrich. Chemical compounds were dissolved in DMSO for 50-mm stock solutions and subsequently diluted to the final concentrations described in the text for application. An equal amount of DMSO was used as control. For the sensitivity tests of single compounds in Arabidopsis, seeds were directly plated on 1/2 MS medium with or without the compounds. For analysis of Arabidopsis resistance to a CBI mixture, plants were grown on 1/2 MS medium for 3 d and then transferred to medium with or without the two drugs for 2 d. Note that for easy comparison of the different treatments, seedlings were transferred at the end of the experiments to a single plate for taking pictures. For C17 toxicity tests, weeds and crops germinated on 1/2 MS medium were transferred after 2 d to medium with or without 1 µm or 5 µm C17, and the root elongation after transfer was measured for 2 d. For IC50 estimation, Arabidopsis seeds were grown in liquid 1/2 MS medium containing the tested compounds or DMSO as control. The root length of 5-d-old seedlings was quantified and nonlinear regression analysis performed with GraphPad Prism (version 7.04).
Generation of Transgenic Arabidopsis Plants
The plasmids of 35S-2C (CESA3S983F) and pCESA3-2C were generated by cloning the open reading frame of the mutated CESA3 from the 2C mutant into pK7WG2D and pGWB6-pCESA3, respectively (Karimi et al., 2002; Desprez et al., 2007). Cloning primers are listed in Supplemental Table S1. The resulting plasmids were transformed into Col-0 (for 35S-2C) or the je5 mutant (for pCESA3-2C) by Agrobacterium tumefaciens-mediated floral dipping (Clough and Bent, 1998).
Base Editing Vector Construction
Gateway entry vectors were modified to make them compatible for Golden Gate cloning (GreenGate system; Lampropoulos et al., 2013). The entire region between attP1 and attP2 of pDONR221 (ccdB and chloramphenicol resistant marker) was amplified using forward and reverse primers that contained the attB (4, 1, or 2r)-BsaI site plus the overhang A (ACCT) and attB (1r, 2, or 3)-BsaI site plus overhang G (ATAC), respectively (Supplemental Table S1). The PCR fragments were purified and used for the BP recombination reaction using pDONRP4P1R, pDONOR221, and pDONRP2RP3, resulting in the generation of pEN-L4-AG-R1, pEN-L1-AG-L2, and pEN-R2-AG-L3, respectively.
A set of Golden Gate entry modules was generated as follows: Cas9-D10A (without NLS) was PCR amplified using Q5 polymerase from pDe-CAS9-D10A (a gift from Holger Puchta; Fauser et al., 2014) with oligos 45 and 47 (Fauser et al., 2014; Supplemental Table S1). The amplification product was digested with BsaI and ligated using T4 DNA ligase into the BsaI-digested pGGC000 vector, which was a gift from Jan Lohmann (Addgene plasmid #48858). The resulting plasmid was named pGG-C-CAS9-D10A-D. Synthetic gene fragments for APOBEC-(GGS)5 and UGI-NLS (Invitrogen) were BsaI digested and ligated respectively into the BsaI-digested pGGB000 and pGGD000 vectors to generate pGG-B-APOBEC-(GGS)5-C and pGG-d-UGI-NLS-E. pGGB000 and pGGD000 were a gift from Jan Lohmann (Addgene plasmids #48857 and #48859, respectively). The PcUbi promoter was PCR amplified from pDe-CAS9 (a gift from Holger Puchta; Fauser et al., 2014). The PCR product was cloned into pGGA000 (Lampropoulos et al., 2013) to generate pGG-A-PcUbi-B. The terminator G7T was amplified from pEN-R2-9-L3 (Karimi et al., 2007) and cloned into pGG000E to create pGG-E-G7T-F. All colonies were screened using colony PCR with oligos 81 and 87 for pGG-C-CAS9-D10-A and primers 61 and 62 for pGG-B-APOBEC-(GGS)5-C and pGG-d-UGI-NLS-E (Supplemental Table S1). Plasmid DNA was extracted from positive colonies with the GeneJET plasmid miniprep kit (Thermo Fisher Scientific) and verified with Sanger sequencing at the VIB Genetic Service Facility (Wilrijk, Belgium).
The Golden Gate Entry modules pGG-A-PcUBIP-B, pGG-B-APOBEC-(GGS)5-C, pGG-C-CAS9-D10A-D, pGG-D-UGI-NLS-E, pGG-E-G7T-F, and a linker of annealed oligos (LinkerFG_F and Linker FG_R) were cloned into pEN-L4-AG-R1 via a Golden Gate reaction, resulting in the Gateway entry vector pEN-L4-CytD-R1. The reaction was performed according to Lampropoulos et al. (2013) with some small modifications. In brief, 100 ng of each of the modules was mixed with 150 ng of the destination vector, 1.5 µL CutSmart buffer, 1.5 µL ATP (10 mm), 0.5 µL T4 DNA ligase (400 u/µL), and 0.5 µL BsaI in a total volume of 15 µL. The reaction was performed in a thermocycler with the following conditions: 30 cycles of 37°C for 3 min and 16°C for 3 min, followed by 50°C for 5 min and 80°C for 5 min. Five microliters of the reaction mixture was used for heat-shock transformation of 50 µL DH5α-competent E. coli cells. These cells were recovered in 950 µL super optimal broth with catabolite repression medium and 150 µL was plated on LB medium containing 100 µg/mL spectinomycin. Colonies were screened by colony PCR using primers 23 and 86, and purified plasmids validated by restriction digestion with EcoRI.
The annealed oligo pairs 49/51 and 50/52 were ligated into a BbsI-digested pEN-Chimera vector (a gift from Holger Puchta; Fauser et al., 2014). This generated pCh-gRNA_CESA3S983F and pCh-gRNA_CESA3S1037F, respectively. Clones were screened using oligos 49 or 51 and 19, and the plasmids were sequence validated for the region with the inserted oligos. pEN-L4-CytD-R1 with pCh-gRNA_CESA3S983F or pCh-gRNA_CESA3S1037F were recombined in pK7m24GW,3 by LR multisite reaction (Karimi et al., 2002), resulting in the generation of BE3-CESA3S983F and BE3-CESA3S1037F. The expression clones were screened by colony PCR using primers 85 and 26 and validated with a restriction digest using NheI.
Arabidopsis Cell Suspension Cultures
Arabidopsis cell suspension culture PSB-d was transformed as previously described (Van Leene et al., 2007). Aliquots of cells were harvested 2, 5, and 7 weeks after transformation. DNA was extracted using the Edward’s extraction method (Edwards et al., 1991). PCR was performed using 2×MyTaq mix (Bioline), and Sanger sequencing was performed by Eurofins Scientific using Oligo 122.
Spinning Disk Microscopy and Image Analysis
The 3D je5 GFP-CESA3 line was generated by crossing je5 GFP-CESA3 and 3D je5 GFP-CESA3. The 4-d-old seedlings were treated with 0.1% (v/v) DMSO (mock) and 200 nm C17 (C17). The GFP fluorescence was analyzed using an Ultraview spinning disk microscope (Perkin Elmer).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: CESA1 (AT4G32410) and CESA3 (AT5G05170).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Lignin staining of C17-treated plants.
Supplemental Figure S2. Cellulose visualization in the roots of C17-treated plants.
Supplemental Figure S3. IC50 estimation of C17 and the active analogs.
Supplemental Figure S4. Base editing for C17 resistance in Arabidopsis.
Supplemental Figure S5. Primary Arabidopsis transformants selected on C17 medium.
Supplemental Table S1. Sequences of primers used in this work.
Acknowledgments
The authors thank Annick Bleys for help in preparing the manuscript, Hilde Van den Daele for help in preparing figures, and Dr. Shiguo Chen for providing weed seeds.
Footnotes
This work was supported by a grant of the Research Foundation Flanders (G.023616N) and National Key R&D Program of China (2018YFD1000706).
Articles can be viewed without a subscription.
References
- Anderson CT, Carroll A, Akhmetova L, Somerville C (2010) Real-time imaging of cellulose reorientation during cell wall expansion in Arabidopsis roots. Plant Physiol 152: 787–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brabham C, Debolt S (2013) Chemical genetics to examine cellulose biosynthesis. Front Plant Sci 3: 309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brabham C, Lei L, Gu Y, Stork J, Barrett M, DeBolt S (2014) Indaziflam herbicidal action: A potent cellulose biosynthesis inhibitor. Plant Physiol 166: 1177–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caño-Delgado A, Penfield S, Smith C, Catley M, Bevan M (2003) Reduced cellulose synthesis invokes lignification and defense responses in Arabidopsis thaliana. Plant J 34: 351–362 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cobb A, Reade JPH (2010) Herbicides and Plant Physiology, Ed 2. Wiley-Blackwell, Oxford [Google Scholar]
- Conticello SG. (2008) The AID/APOBEC family of nucleic acid mutators. Genome Biol 9: 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corio-Costet M-F, Dall’Agnese M, Scalla R (1991) Effects of isoxaben on sensitive and tolerant plant cell cultures: I. Metabolic fate of isoxaben. Pestic Biochem Physiol 40: 246–254 [Google Scholar]
- DeBolt S, Gutierrez R, Ehrhardt DW, Somerville C (2007) Nonmotile cellulose synthase subunits repeatedly accumulate within localized regions at the plasma membrane in Arabidopsis hypocotyl cells following 2,6-dichlorobenzonitrile treatment. Plant Physiol 145: 334–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Délye C, Jasieniuk M, Le Corre V (2013) Deciphering the evolution of herbicide resistance in weeds. Trends Genet 29: 649–658 [DOI] [PubMed] [Google Scholar]
- Desprez T, Vernhettes S, Fagard M, Refrégier G, Desnos T, Aletti E, Py N, Pelletier S, Höfte H (2002) Resistance against herbicide isoxaben and cellulose deficiency caused by distinct mutations in same cellulose synthase isoform CESA6. Plant Physiol 128: 482–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desprez T, Juraniec M, Crowell EF, Jouy H, Pochylova Z, Parcy F, Höfte H, Gonneau M, Vernhettes S (2007) Organization of cellulose synthase complexes involved in primary cell wall synthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 104: 15572–15577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards K, Johnstone C, Thompson C (1991) A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res 19: 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauser F, Schiml S, Puchta H (2014) Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana. Plant J 79: 348–359 [DOI] [PubMed] [Google Scholar]
- Feng Z, Zhang B, Ding W, Liu X, Yang DL, Wei P, Cao F, Zhu S, Zhang F, Mao Y, Zhu JK (2013) Efficient genome editing in plants using a CRISPR/Cas system. Cell Res 23: 1229–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamann T, Bennett M, Mansfield J, Somerville C (2009) Identification of cell-wall stress as a hexose-dependent and osmosensitive regulator of plant responses. Plant J 57: 1015–1026 [DOI] [PubMed] [Google Scholar]
- Harris DM, Corbin K, Wang T, Gutierrez R, Bertolo AL, Petti C, Smilgies D-M, Estevez JM, Bonetta D, Urbanowicz BR, Ehrhardt DW, Somerville CR, et al. (2012) Cellulose microfibril crystallinity is reduced by mutating C-terminal transmembrane region residues CESA1A903V and CESA3T942I of cellulose synthase. Proc Natl Acad Sci USA 109: 4098–4103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heap I. (2014) Global perspective of herbicide-resistant weeds. Pest Manag Sci 70: 1306–1315 [DOI] [PubMed] [Google Scholar]
- Hu Z, Vanderhaeghen R, Cools T, Wang Y, De Clercq I, Leroux O, Nguyen L, Belt K, Millar AH, Audenaert D, Hilson P, Small I, et al. (2016) Mitochondrial defects confer tolerance against cellulose deficiency. Plant Cell 28: 2276–2290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Karimi M, Bleys A, Vanderhaeghen R, Hilson P (2007) Building blocks for plant gene assembly. Plant Physiol 145: 1183–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S, Laosinchai W, Itoh T, Cui X, Linder CR, Brown RM Jr (1999) Immunogold labeling of rosette terminal cellulose-synthesizing complexes in the vascular plant vigna angularis. Plant Cell 11: 2075–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR (2016) Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533: 420–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampropoulos A, Sutikovic Z, Wenzl C, Maegele I, Lohmann JU, Forner J (2013) GreenGate—a novel, versatile, and efficient cloning system for plant transgenesis. PLoS One 8: e83043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei L, Li S, Gu Y (2012) Cellulose synthase complexes: Composition and regulation. Front Plant Sci 3: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough PE, Yu J, Gómez de Barreda D (2013) Efficacy of preemergence herbicides for controlling a dinitroaniline-resistant goosegrass (Eleusine indica) in Georgia. Weed Technol 27: 639–644 [Google Scholar]
- McFarlane HE, Döring A, Persson S (2014) The cell biology of cellulose synthesis. Annu Rev Plant Biol 65: 69–94 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Paredez AR, Somerville CR, Ehrhardt DW (2006) Visualization of cellulose synthase demonstrates functional association with microtubules. Science 312: 1491–1495 [DOI] [PubMed] [Google Scholar]
- Quareshy M, Prusinska J, Li J, Napier R (2018) A cheminformatics review of auxins as herbicides. J Exp Bot 69: 265–275 [DOI] [PubMed] [Google Scholar]
- Sabba RP, Durso NA, Vaughn KC (1999) Structural and immunocytochemical characterization of the walls of dichlobenil‐habituated BY‐2 tobacco cells. Int J Plant Sci 160: 275–290 [Google Scholar]
- Saha D, Marble SC, Stewart C, Chandler A (2017) Preemergence and postemergence control of artilleryweed (Pilea microphylla) in container nurseries and landscapes. Weed Technol 31: 574–581 [Google Scholar]
- Scheible W-R, Eshed R, Richmond T, Delmer D, Somerville C (2001) Modifications of cellulose synthase confer resistance to isoxaben and thiazolidinone herbicides in Arabidopsis Ixr1 mutants. Proc Natl Acad Sci USA 98: 10079–10084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethaphong L, Haigler CH, Kubicki JD, Zimmer J, Bonetta D, DeBolt S, Yingling YG (2013) Tertiary model of a plant cellulose synthase. Proc Natl Acad Sci USA 110: 7512–7517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville C. (2006) Cellulose synthesis in higher plants. Annu Rev Cell Dev Biol 22: 53–78 [DOI] [PubMed] [Google Scholar]
- Tateno M, Brabham C, DeBolt S (2016) Cellulose biosynthesis inhibitors—a multifunctional toolbox. J Exp Bot 67: 533–542 [DOI] [PubMed] [Google Scholar]
- Van Leene J, Stals H, Eeckhout D, Persiau G, Van De Slijke E, Van Isterdael G, De Clercq A, Bonnet E, Laukens K, Remmerie N, Henderickx K, De Vijlder T, et al. (2007) A tandem affinity purification-based technology platform to study the cell cycle interactome in Arabidopsis thaliana. Mol Cell Proteomics 6: 1226–1238 [DOI] [PubMed] [Google Scholar]
- Worden N, Wilkop TE, Esteve VE, Jeannotte R, Lathe R, Vernhettes S, Weimer B, Hicks G, Alonso J, Labavitch J, Persson S, Ehrhardt D, et al. (2015) CESA TRAFFICKING INHIBITOR inhibits cellulose deposition and interferes with the trafficking of cellulose synthase complexes and their associated proteins KORRIGAN1 and POM2/CELLULOSE SYNTHASE INTERACTIVE PROTEIN1. Plant Physiol 167: 381–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y, Lei L, Brabham C, Stork J, Strickland J, Ladak A, Gu Y, Wallace I, DeBolt S (2014) Acetobixan, an inhibitor of cellulose synthesis identified by microbial bioprospecting. PLoS One 9: e95245. [DOI] [PMC free article] [PubMed] [Google Scholar]