The mitochondrial PDC E1 component contributes to respiratory metabolism and PIN-driven polar auxin transport in Arabidopsis organ development.
Abstract
Pyruvate dehydrogenase is the first enzyme (E1) of the PDH complex (PDC). This multienzyme complex contains E1, E2, and E3 components and controls the entry of carbon into the mitochondrial tricarboxylic acid cycle to enable cellular energy production. The E1 component of the PDC is composed of an E1α catalytic subunit and an E1β regulatory subunit. In Arabidopsis (Arabidopsis thaliana), there are two mitochondrial E1α homologs encoded by IAA-CONJUGATE-RESISTANT 4 (IAR4) and IAR4-LIKE (IAR4L), and one mitochondrial E1β homolog. Although IAR4 was reported to be involved in auxin conjugate sensitivity and auxin homeostasis in root development, its precise role remains unknown. Here, we provide experimental evidence that mitochondrial PDC E1 contributes to polar auxin transport during organ development. We performed genetic screens for factors involved in cotyledon development and identified an uncharacterized mutant, macchi-bou 1 (mab1). MAB1 encodes a mitochondrial PDC E1β subunit that can form both a homodimer and a heterodimer with IAR4. The mab1 mutation impaired MAB1 homodimerization, reduced the abundance of IAR4 and IAR4L, weakened PDC enzymatic activity, and diminished mitochondrial respiration. A metabolomics analysis showed significant changes in metabolites including amino acids in mab1 and, in particular, identified an accumulation of Ala. These results suggest that MAB1 is a component of the Arabidopsis mitochondrial PDC E1. Furthermore, in mab1 mutants and seedlings where the TCA cycle was pharmacologically blocked, we found reduced abundance of the PIN-FORMED (PIN) auxin efflux carriers, possibly due to impaired PIN recycling and enhanced PIN degradation in vacuoles. Therefore, we suggest that mab1 induces defective polar auxin transport via metabolic abnormalities.
In higher plants, the development of organs such as leaves, flowers, and roots is mediated by the phytohormone auxin. The local accumulation of auxin is crucial to aerial and underground organ formation (Vanneste and Friml, 2009). The asymmetric distribution of auxin is established by polar auxin transport and by local auxin biosynthesis during organ development (Wisniewska et al., 2006). PIN-FORMED (PIN) auxin efflux carriers drive polar auxin transport. PIN polarity is arranged to produce an auxin peak at the tips of organs (Benková et al., 2003). The control of PIN localization and auxin level is important for local auxin accumulation during organ development (Geldner et al., 2003; Jaillais et al., 2006; Dhonukshe et al., 2007; Kleine-Vehn et al., 2008). PIN proteins are continually internalized from the plasma membrane into endosomes (Dhonukshe et al., 2007). Some PIN proteins are recycled from endosomes to the plasma membrane and other PIN proteins are targeted to vacuoles where they are degraded (Geldner et al., 2003; Jaillais et al., 2006; Kleine-Vehn et al., 2008). Interference with PIN trafficking causes disruption of PIN polarity and of PIN levels to a variable degree.
Several specific regulators of PIN localization have been identified. For example, Ser/Thr kinase PINOID (PID) controls PIN polarity by phosphorylating PIN proteins (Benjamins et al., 2001; Friml et al., 2004; Michniewicz et al., 2007). In addition, MACCHI-BOU 4/ENHANCER OF PINOID/NAKED PINS IN YUC MUTANTS 1 (MAB4/ENP/NPY1), which encodes a NON-PHOTOTROPIC HYPOCOTYL 3-like protein, is altered in an enhancer mutant that fails to initiate organ development in a pid or yucca1 (yuc1) yuc4 double-mutant genetic background, which partially disrupts auxin biosynthesis (Treml et al., 2005; Cheng et al., 2007b; Furutani et al., 2007). The MAB4/ENP/NPY1 gene and its homologs regulate the PIN level and polarity through a block on PIN internalization (Treml et al., 2005; Furutani et al., 2007, 2011). Mutation of these genes causes defective PIN localization, resulting in defective organ development. YUC genes encode flavin monooxygenases that function as key auxin biosynthesis enzymes and are required for the establishment of a local auxin gradient in many developmental events, including embryogenesis, organ development, and vascular differentiation (Zhao et al., 2001; Cheng et al., 2006, 2007a; Chen et al., 2014). In addition, the TRP AMINOTRANSFERASE OF ARABIDOPSIS (TAA) family of amino transferases functions in the same auxin biosynthetic pathway and contributes to auxin-regulated organ formation (Stepanova et al., 2008; Tao et al., 2008; Mashiguchi et al., 2011; Won et al., 2011;). TAAs convert Trp to indole-3-pyruvate, whereas YUCs play an important role in the conversion of indole-3-pyruvate to indole-3-acetic acid (IAA), the main auxin in plants. IAA-CONJUGATE-RESISTANT4 (IAR4) is a different kind of regulator of local auxin accumulation during organ development. This gene encodes a putative mitochondrial pyruvate dehydrogenase (PDH) E1α subunit that was originally identified in a genetic screen for IAA-amino acid conjugate-resistant mutants (LeClere et al., 2004); subsequently, IAR4 was identified as the gene responsible for the enhancer mutant phenotype in tir1-1 auxin resistance (Quint et al., 2009). PDH E1 is the first component of the three-component PDH complex (PDC) that oxidatively decarboxylates pyruvate to form NADH and acetyl-CoA. E1 contains a catalytic E1α subunit and a regulatory E1β subunit in a heterodimer. Although the precise role of IAR4 in organ formation remains unknown, iar4 mutants display auxin-related phenotypes that can be rescued by increasing IAA levels in the plants. These data suggest that mitochondrial PDH E1α (IAR4) functions in auxin homeostasis and is required for organ development.
Several key regulators of auxin accumulation in organ development have been identified; however, their molecular details and interactions remain poorly understood. To further elucidate the mechanism of auxin-regulated organogenesis, we performed a forward genetic screen to identify mutants lacking cotyledons in plants with a pid genetic background. Here, we report the characterization of an enhancer of the pid mutant, macchi-bou 1 (mab1), that displays defective organ development. We found that a mutation of MAB1(At5g50850), which encodes the Arabidopsis (Arabidopsis thaliana) mitochondrial PDH E1β subunit, causes a decrease in PDC enzymatic activity and metabolic abnormalities. In addition, detailed analyses of PIN trafficking and pharmacological investigations showed that impaired recycling to the plasma membrane and accelerated targeting to the vacuole of PIN2 leads to defective polar auxin transport in the mab1 mutant. Our results indicate that mitochondrial PDH contributes to PIN-dependent auxin transport during organ development, possibly via metabolic regulation.
RESULTS
Identification of Arabidopsis mab1 Mutant Seedlings Exhibiting Defective Organ Formation
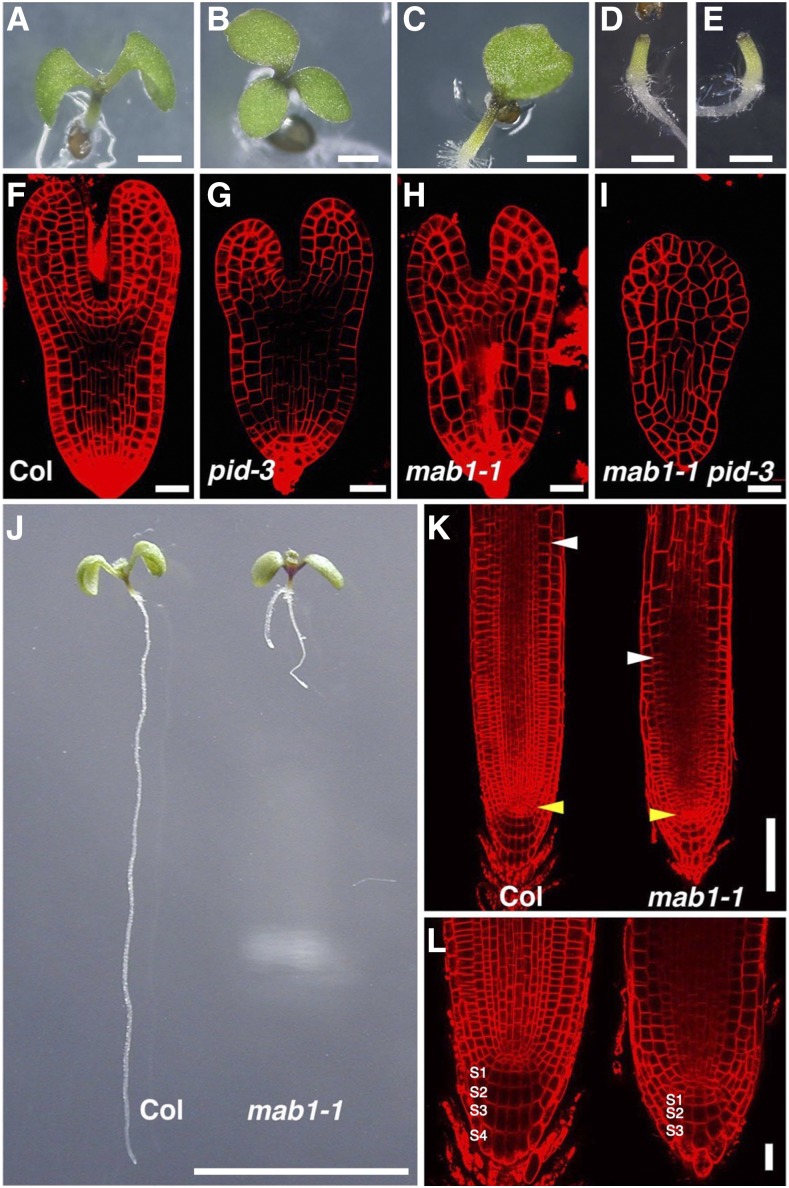
We searched for genes involved in organ development by performing a genetic screen for pid enhancers related to cotyledon development. Single pid mutants often display defects in cotyledon number and position compared to wild-type seedlings (Fig. 1, A and B; Benjamins et al., 2001; Friml et al., 2004). The screen focused on mutations that affected cotyledon formation, causing their absence in the pid-3 background. We isolated a pid enhancer, named mab1-1, from the M2 population of EMS-mutagenized Arabidopsis. Seedlings carrying the mab1-1 pid-3 double mutation lacked cotyledons, whereas mab1-1 single mutant seedlings exhibited aberrant cotyledon numbers and separation of cotyledons (Fig. 1, C and D). Cotyledon development is initiated during embryogenesis. In a wild-type embryo, two cotyledon primordia are formed in the apical region of the embryo after the globular stage (Supplemental Fig. S1, A–C and G). mab1-1 embryos in early embryogenesis appeared almost normal; however, after the globular stage, they displayed aberrant cotyledon development, aberrant cell division in the hypophysis, and reduced cell proliferation throughout the embryo (Supplemental Fig. S1, D–F and H). mab1-1 embryos sometimes produced a single cotyledon or fused cotyledons, and even in mutant embryos that formed two separate cotyledons, we observed defective growth of cotyledon primordia compared to that in wild-type embryos (Fig. 1, F and H). When combined with pid-3 that also caused defective cotyledon development, double mutant embryos completely lacked cotyledon primordia (Fig. 1, G and I). These results indicated that MAB1 functions in cotyledon development together with PID. To investigate the role of MAB1 in postembryonic organ formation, we analyzed flower development in single- and double-mutant plants. Single mab1-1 mutants produced flowers similar to that in the wild type (Supplemental Fig. S1, K and L). However, pid-3 mutants had pin-shaped inflorescences with several defective flowers (Supplemental Fig. S1M). The mab1-1 pid-3 double mutant plants produced no flowers (Supplemental Fig. S1N), indicating that the mab1 mutation enhanced the pid phenotype not only in cotyledon development but also in postembryonic organ development. In addition, mab1-1 mutants had short roots with a minor gravitropic response defect (Fig. 1, J–K; Supplemental Figs. S1, I and J, and S2, A–C): wild-type roots grew in the direction of gravity, whereas mab1-1 mutant roots often elongated in an irregular direction. Mutant roots had a normal cell layer structure, but possessed a reduced meristem zone and a decreased number of layers of columella cells with amyloplasts compared to that in wild-type roots. Root hair development was also repressed in mab1-1 (Supplemental Fig. S2D). These results indicate that MAB1 controls organ growth in both above-ground and underground parts.
Figure 1.
Defective organ development in mab1 mutants. A to E, Five-day-old seedlings: Wild type (Col; A), pid-3 (B), mab1-1 (C), mab1-1 pid-3 (D), and mab1-2 pid-3 (E). F to I, Comparison of wild-type (Col; F), pid-3 (G), mab1-1 (H), and mab1-1 pid-3 (I) embryos at the late heart stage. J, Root development in 5-d-old wild-type (Col; left) and mab1-1 (right) seedlings. mab1-1 mutants show a short root phenotype. K and L, Comparison of wild-type (Col; left) and mab1-1 (right) root meristems. S1 to S4 indicate columella cell tiers. Yellow arrowheads indicate quiescent center cells, whereas white arrowheads represent the cortex transition zones (K). Scale bars = 1 mm (A to E), 20 µm (F to I, L), 1 cm (J), and 100 µm (K).
MAB1 Encodes a Mitochondrial PDH E1β Subunit
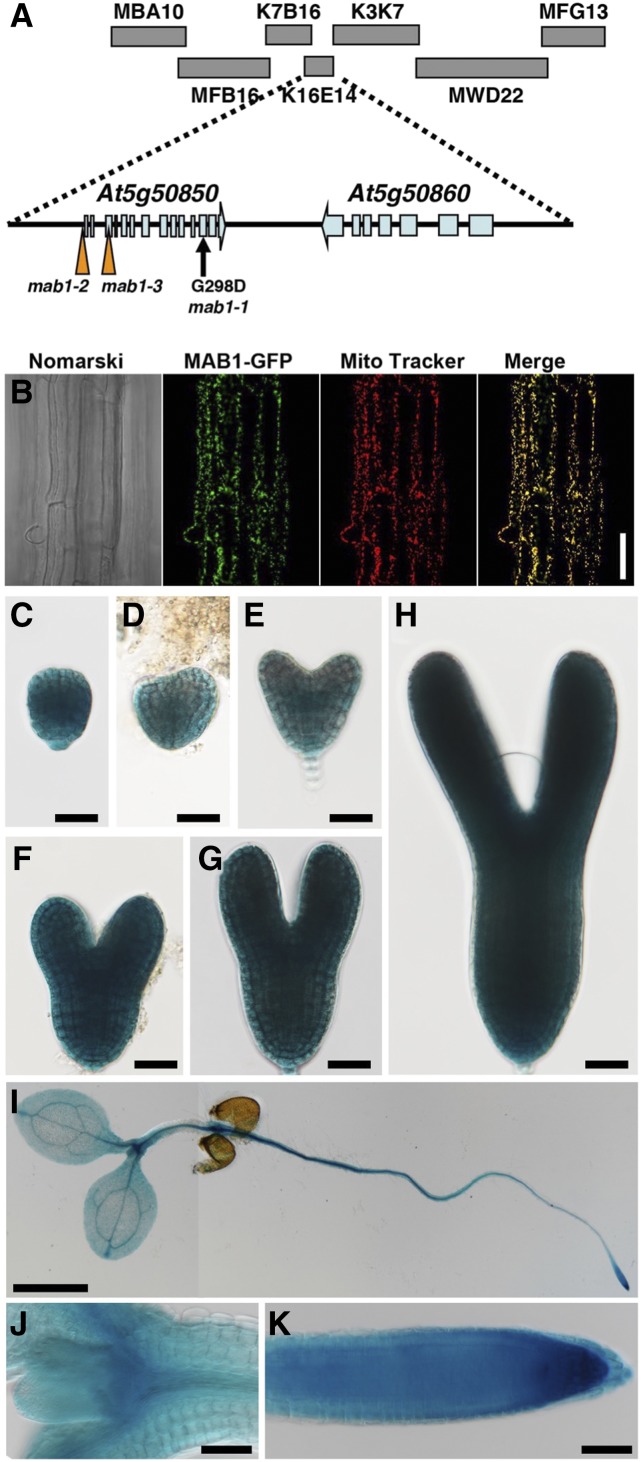
We performed map-based cloning of MAB1 and localized the gene between bacterial artificial chromosomes MBA10 and MFG13 (Fig. 2A). Sequence analysis of several genes within this region identified a G-to-A base substitution in the open reading frame (ORF) of At5g50850; this base change caused a nonconservative Gly-298 to Asp-298 substitution (Fig. 2A). To confirm that the ORF is MAB1, the ORF translationally fused with monomeric red fluorescent protein-encoding gene mCherry was expressed under the control of its own promoter in mab1-1 mutants. The transformants showed complete rescue of the mab1-1 phenotypes regarding both cotyledon and root development, demonstrating that At5g50850 is the MAB1 gene (Supplemental Fig. S3A). In addition, we obtained two other alleles, mab1-2 and mab1-3, that have transfer DNA (T-DNA) insertions located ∼310 bp upstream of the translation start site and in the third exon of At5g50850, respectively (Fig. 2A; Supplemental Fig. S3B). In mab1-2 seedlings where expression of MAB1 was reduced (Supplemental Fig. S3C), root growth and root hair development were severely repressed as in mab1-1 (Supplemental Fig. S2, A, B, and D). As to cotyledon formation, though the mab1-2 single mutant developed two almost normal cotyledons, the mab1-2 mutation also caused failure of cotyledon formation in the pid-3 mutant background (Fig. 1E; Supplemental Fig. S2E). In addition, no mab1-3 homozygotes were found in the progeny of plants heterozygous for mab1-3, indicating that mab1-3 is lethal to gametophytes or embryos. These results indicate that mab1-1 and mab1-2 are partial loss-of-function alleles that cause relatively mild or weak phenotypes and that mab1-3 is a knockout mutation.
Figure 2.
MAB1 encodes a mitochondrial PDC E1β. A, Map position of the MAB1 locus. MAB1 was mapped between the MBA10 and MFG13 bacterial artificial chromosomes on chromosome 5 by map-based cloning. mab1-1 is a point mutation in At5g50850 that causes the amino acid change G298D. In mab1-2 and mab1-3 mutants, T-DNA is inserted ∼310 bp upstream of the translation start site and at the third exon of the gene, respectively. B, Localization of MAB1-GFP and Mito Tracker fluorescence in the epidermis of Arabidopsis roots. The Nomarski image (left), and GFP (second from left) and Mito Tracker (second from right) fluorescence images were obtained using an epifluorescence microscope. At right is a merge of the MAB1-GFP and Mito Tracker images. C to K, Expression patterns of MAB1p:GUS in the embryo at the globular (C), transition (D), heart (E), late heart (F), and torpedo (G, H) stages, and in the 4-d-old seedling (I), around shoot apical meristem (J) and root meristem (K), respectively. GUS staining was performed at 37°C overnight (C to H) and for one hours (I to K). Scale bars = 50 mm (B, J, K), 1 cm (I), and 20 mm (C to H).
At5g50850 encodes a putative mitochondrial PDH E1β subunit of PDH E1 that oxidatively decarboxylates pyruvate. The remaining acetyl group of pyruvate is transferred to CoA by the PDC E2 component to form acetyl-CoA that enters the tricarboxylic acid (TCA) cycle. To confirm that MAB1 is a mitochondrial protein, we fused GFP to its C terminus and expressed this fusion protein under the control of the Cauliflower mosaic virus 35S promoter. The MAB1-GFP protein colocalized with the mitochondrial marker Mito Tracker in root cells (Fig. 2B). The protein has also been previously identified in purified mitochondrial preparations from Arabidopsis as a mitochondrial matrix enzyme (Millar et al., 2001). To examine where MAB1 functions, we used MAB1p:GUS transgenic plants to detect the spatial patterns of MAB1 expression. In embryogenesis, GUS signal was found in whole embryos at the globular, heart, and torpedo stages (Fig. 2, C–G). In MAB1p:GUS plants, GUS activity was detected all over the seedling (Fig. 2, I–K) and strongly in young inflorescences containing inflorescence meristems and juvenile floral organs (Supplemental Fig. S4, A–E). Furthermore, in situ hybridization was performed using an antisense probe against MAB1 mRNA. MAB1 signals were detected throughout embryos (Supplemental Fig. S4, F–I). Expression of MAB1 appeared to decrease in later embryonic stages (Supplemental Fig. S4J); no signals were detected using a MAB1 sense probe in the heart-stage embryo (Supplemental Fig. S4K). This MAB1 expression pattern was almost correlated with the phenotypes of mab1 mutants.
MAB1 Forms a PDH E1 Complex with IAR4 and IAR4L
The E1 component of the PDH complex is a heterotetramer composed of two α- and two β-subunits. In Arabidopsis, IAR4 and its homolog IAR4-LIKE (IAR4L) encode PDH E1α subunit homologs (Luethy et al., 1995; LeClere et al., 2004; Quint et al., 2009). The mutant of IAR4 was reported to show resistance to some IAA-amino acid conjugates (LeClere et al., 2004), suggesting that the mab1 mutants also exhibit less sensitivity to IAA-Ala regarding root growth. As shown in Supplemental Figure S5, A and B, the treatment with IAA-Ala repressed primary root growth in the wild type in a dose-dependent manner, whereas IAA-Ala induced little inhibition of root growth in the mab1 mutants. Furthermore, IAA-Ala enhanced root hair development in wild-type seedlings, whereas in the mab1 mutants it caused no visible differences in root hair development (Supplemental Fig. S5C). These results suggest that MAB1 functions in the same way as IAR4 with regard to root growth.
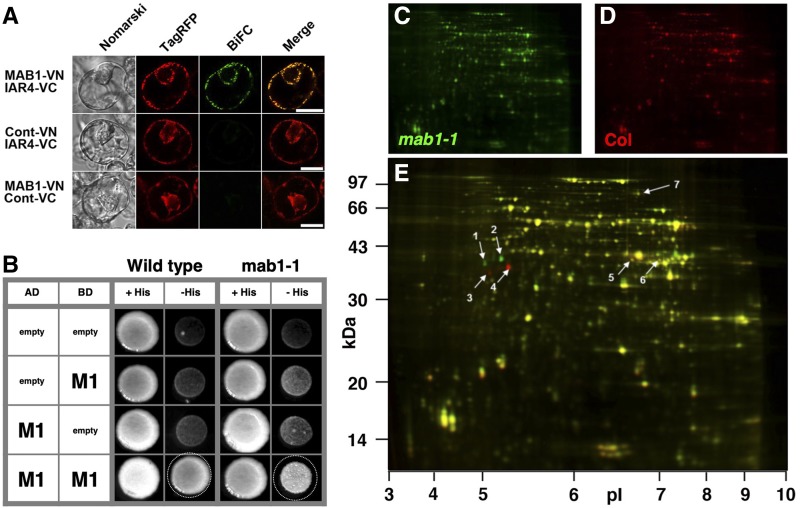
E1β/MAB1 has been shown to interact in heterodimers with both IAR4 and IAR4L in yeast-two hybrid studies (Arabidopsis Interactome Mapping Consortium, 2011). We further tested the interaction of MAB1 with IAR4 using bimolecular fluorescence complementation (BiFC) in Arabidopsis protoplasts. Interaction between MAB1-VN (the N-terminal half of Venus) and IAR4-VC (the C-terminal half of Venus) was observed in mitochondria, whereas no interaction was found between either fusion protein and individual mitochondria-targeted corresponding VN or VC (Fig. 3A). Homodimerization of MAB1 was also found in a yeast two-hybrid experiment (Fig. 3B). The crystal structures of the PDC of other organisms suggest that Gly-298, which is mutated in mab1-1, is located in the putative MAB1 homodimerization domain (Ciszak et al., 2003). We examined the impact of the mab1-1 mutation on MAB1 homodimerization in a yeast two-hybrid assay using the G298D-mutated MAB1 protein. The mutated MAB1 protein showed less frequent homodimerization than did the wild-type MAB1 (Fig. 3B). To further investigate the impact of the mab1-1 mutation in the PDH E1β subunit on mitochondrial protein abundance, we performed difference gel electrophoresis (DIGE) using fluorescence cyanine dyes to compare the mitochondrial proteomes of wild type and mab1-1 (Fig. 3, C and D). DIGE identified seven protein spots that showed significant changes in abundance, including four that were identified as the PDH E1β subunit MAB1 (Fig. 3E; Table 1). The single amino acid mutation of mab1-1 in the PDH E1β subunit shifted the pI and Mr of two MAB1 spots (Fig. 3E). We also observed a decrease of IAR4L (1.6-fold) and IAR4 (2-fold) abundance in mab1-1 mutants (Fig. 3E; Table 1). These results are consistent with MAB1 forming a complex with IAR4 and IAR4L.
Figure 3.
Arabidopsis PDH E1 complex. A, BiFC assay in Arabidopsis protoplast cells. The appropriate BiFC constructs were introduced into protoplasts via PEG-mediated transformation. After 12 h incubation in the dark, Nomarski (left), TagRFP (second from left; mitochondria-targeted marker), and BiFC images (second from right) were captured using an epifluorescence microscope. At right is a merge of the TagRFP and BiFC images. Cont-VN and Cont-VC demonstrate N- and C-terminal halves, respectively, of the Venus fluorescent protein fused with the At2g25140 mitochondrial presequence. TagRFP was also fused with the mitochondrial presequence. Scale bars = 20 µm. B, Yeast two-hybrid assay. MAB1 and mutated MAB1 were expressed as fusion proteins with a transcriptional activator domain and a DNA binding domain. Interaction between the two proteins was tested using the HIS3 reporter gene. C to E, DIGE analysis of a mitochondrial proteome from 3-week-old mab1-1 (C; green) and wild-type (Col; D; red) plants grown hydroponically. Wild-type and mab1-1 proteins with similar abundance are yellow in the overlay (E). Spots indicated by arrows are significantly different and are listed in Table 1.
Table 1. Selected protein spots identified from the DIGE two-dimensional gels shown in Figure 3.
The mab1/wild type column shows the ratios of mab1-1 protein abundance to wild type (Col), with significant changes shown in the t test column; the experiment was performed with three replicates. Proteins whose spot numbers are indicated in Figure 3, C to E, were identified by matrix-assisted laser-desorption ionization time of flight tandem mass spectrometry. The predicted molecular mass and pI of the match are shown along with the molecular weight search score (P < 0.05, when the score is >37), number of peptides matched to tandem mass spectra, and percentage coverage of the matched sequence.
| Spot No. | Accession | Description | mab1/Wild Type | t Test | Score | Matched Peptides | Coverage (%) | Mass | pI |
|---|---|---|---|---|---|---|---|---|---|
| 1 | At5g50850 | MAB1 | 7.28 | 0.0021 | 188 | 5 | 14.3 | 39436 | 5.67 |
| 2 | At5g50850 | MAB1 | 35.41 | 0.0003 | 290 | 5 | 14.3 | 39436 | 5.67 |
| 3 | At5g50850 | MAB1 | −15.30 | 0.0017 | 138 | 3 | 11.3 | 39436 | 5.67 |
| 4 | At5g50850 | MAB1 | −25.15 | 0.0015 | 298 | 6 | 14.3 | 39436 | 5.67 |
| 5 | At1g59900 | IAR4L | −1.63 | 0.0011 | 183 | 3 | 8.5 | 43488 | 7.16 |
| 6 | At1g24180 | IAR4 | −1.98 | 0.0034 | 175 | 4 | 12.2 | 43787 | 8.03 |
| 7 | At1g03090 | MCCA | 2.33 | 0.0051 | 64 | 1 | 2.9 | 78776 | 6.31 |
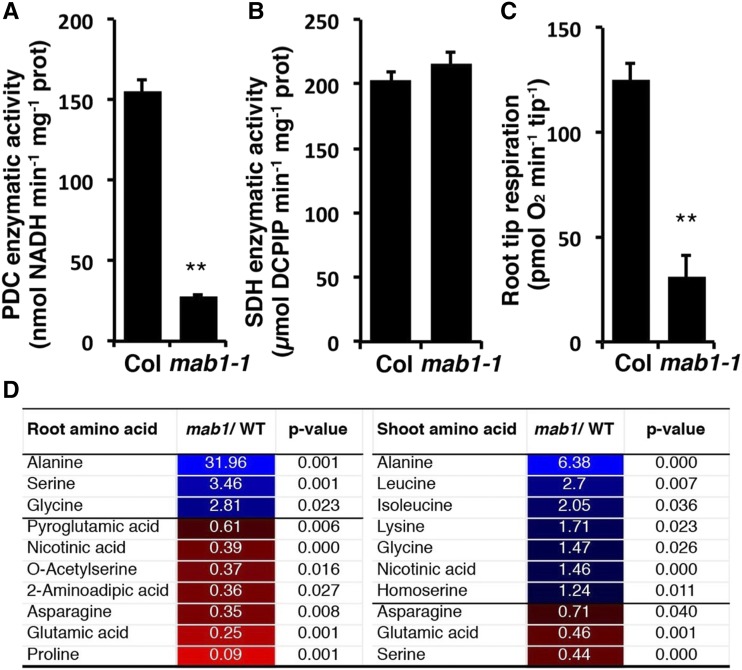
To investigate the effect of the mab1-1 mutation on the function of the PDC, we measured PDC enzymatic activity in isolated mitochondria. PDC activity in mab1-1 mutants was only 17% of that in the wild type, suggesting that the point mutation caused dysfunction of the enzyme (Fig. 4A). To determine whether succinate dehydrogenase (SDH), another mitochondrial dehydrogenase and a key TCA enzyme, was influenced by mab1-1, we also measured its enzymatic activity. As shown in Figure 4B, SDH enzymatic activity was similar in mab1-1 and the wild type, indicating that the mutation specifically affected the PDC. As mab1-1 displayed a short-root phenotype with dysfunctional activity of the mitochondrial PDC, we measured root tip oxygen consumption by a multiplex microrespiratory analysis (Sew et al., 2013). The rate of oxygen consumption in mab1-1 root tips was only 25% of that in wild-type root tips (Fig. 4C), indicating a deficiency in mitochondrial energy production in the former. These results show that MAB1 is a component of the Arabidopsis PDC.
Figure 4.
Mitochondrial enzymatic activities and root tip respiration in wild type (WT) and mab1-1. A to C, Enzymatic activity of the PDC (A) and SDH (B), and root tip respiration rate (C). Data shown are mean values of three (A, B) and six (C) independent samples, with error representing se. **P < 0.01. D, Selected amino acids showing significantly lower or higher abundance in roots and shoots of mab1-1 compared to the wild type (Col). Plants were grown on plates for 11 d under long-day conditions. Shoots and roots (six biological replicates) were harvested for gas chromatography-mass spectrometry (GS-MS) analysis. Other amino acids and metabolites detected in roots and shoots of mab1-1 and wild-type plants are shown in Supplemental Table S1. Blue boxes and red boxes indicate significantly higher or lower abundance, respectively, in mab1-1.
Metabolomic Changes in the mab1 Mutant
PDC converts pyruvate to acetyl-CoA to provide substrate for the TCA cycle. To determine whether the decrease in PDC enzymatic activity affected pyruvate-related metabolism, we carried out a metabolomic analysis of root and shoot tissues from 11-d-old seedlings grown on agar plates under long-day conditions. Significant differences in abundance of amino acids and other metabolites were identified by comparing these quantities in mutant and wild-type seedlings (Fig. 4D; Supplemental Table S1). The comparison indicated that Ala accumulated to 32-fold and 6.4-fold higher levels in root and shoot, respectively, in the mutant (Fig. 4D). There were some variations in the changes of other amino acids between root and shoot, presumably due to tissue differences. Ala is effectively in equilibrium with pyruvate levels in plant tissues due to the kinetics of conversion of pyruvate to Ala through Ala aminotransferase coupled to the conversion of Glu to 2-oxoglutarate. Hence, the increase in Ala level observed in mab1-1 is highly indicative of a rise in pyruvate concentration. This interpretation is also supported by the observation that Glu was significantly decreased in both root and shoot tissues (Fig. 4D).
Defective PIN-Dependent Auxin Transport in the mab1 Mutant
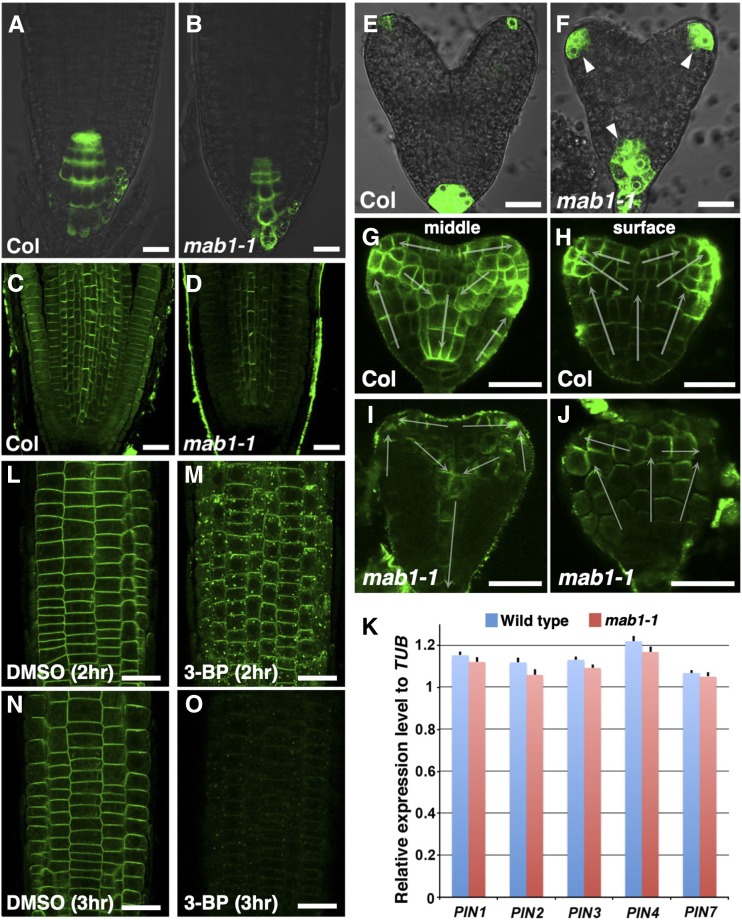
IAR4 was reported to modulate auxin response through the control of auxin homeostasis in root development (Quint et al., 2009). The auxin phenotypes of iar4 roots are believed to arise from altered auxin homeostasis. To clarify the function of MAB1 in auxin-regulated organ formation, we analyzed the expression of an auxin-responsive marker, DR5rev:GFP, in the mab1-1 mutant (Friml et al., 2003). DR5rev:GFP was expressed in the tips of wild-type roots (Fig. 5A). In mab1-1 mutants, the marker gene showed reduced expression in the root tips (Fig. 5B), as previously reported for DR5:GUS expression in iar4 roots (Quint et al., 2009). In contrast, up-regulated GFP signals were found in the vascular tissue of the maturation zone of mab1-1 primary roots (Supplemental Fig. S6, A and B). This analysis indicates that the conflicting findings on DR5 expression cannot simply be explained in terms of MAB1 function in auxin homeostasis. Next, we analyzed the localization of PIN auxin efflux carriers that drive polar auxin transport in the mab1-1 mutant. PINs were localized in the basal side of the plasma membrane in the stele and apically in the epidermis of wild-type roots (Fig. 5C). The amount of PIN proteins (PIN1 and PIN2) appeared to be lower in mab1-1 roots than in wild-type roots, although no defect in PIN polarity was found in mab1-1 (Fig. 5D; Supplemental Fig. S6, C–E). In addition, exogenous auxin treatment did not rescue the short-root phenotypes and decreased PIN2 abundance in mab1-1 mutants, and mab1-1 roots displayed the same auxin response observed in wild-type roots (Supplemental Fig. S6, F–L), indicating that mab1 phenotypes do not result from defective auxin homeostasis and auxin response but rather from defective polar auxin transport in root development. In embryogenesis, we observed enhanced and expanded expression of DR5rev:GFP at the tips of cotyledon primordia and radicles of mab1-1 embryos compared to that in wild-type embryos (Fig. 5, E and F). In wild-type embryos, PIN1 was localized to the upper side of the plasma membrane in the protodermal cells toward the tips of the cotyledon primordia and basally in inner cells (Fig. 5, G and H; Supplemental Fig. S6M). Almost normal PIN1 polarity was found in the protodermal cell layer and inner cells of mab1-1 embryos (Fig. 5, G–J). By contrast, the level of PIN1 was severely reduced in mutant embryos, especially in provascular tissues. The number of PIN1-expressing provascular cells was severely reduced, and discontinuity of the PIN1-expressing provascular cells was often found in mab1-1 embryos (Supplemental Fig. S6, M–O). These results indicate that MAB1 modulates polar auxin transport in organ development through control of the level of PIN. We investigated the cause of the decrease in PIN levels in mab1 mutants by comparing transcription levels of PIN genes in roots and whole seedlings of wild type and mab1-1 mutants. We did not find any evidence of a change in PIN transcription levels in the mutant (Fig. 5K; Supplemental Fig. S7), suggesting that regulation of PIN protein levels via MAB1 might be posttranscriptional.
Figure 5.
Auxin-regulated organ development in wild type and mab1-1. A and B, DR5rev:GFP expression in roots of wild type (Col; A) and mab1-1 (B). Arrowheads indicate expanded GFP signals. C and D, Immunolocalization of PIN1 and PIN2 in wild-type (Col; C) and mab1-1 (D) roots. E and F, DR5rev:GFP expression in heart-stage embryos of wild type (Col; E) and mab1-1 (F). G to J, PIN1 localization in the longitudinal middle (G and I) and surface (H and J) sections of wild-type (Col; G and H) and mab1-1 (I and J) embryos at the midheart stage. The arrow indicates the predicted direction of auxin transport by the polarity of PIN1 localization. K, Reverse transcription quantitative PCR analysis of cDNA from wild-type and mab1-1 roots in 5-d-old seedlings. Expression levels of PIN genes were unaffected by the mab1-1 mutation. Data shown are means of four independent experiments, with error bars representing the sd. L to O, PIN2-GFP localization in PIN2p:PIN2-GFP roots incubated with DMSO (L and N) or 500 mm 3-BP (M and O) for the indicated times. Scale bars = 20 µm.
Furthermore, to examine whether pharmacological inhibition of the TCA cycle also affects PIN localization as the mab1-1 mutation did, we treated PIN2-GFP-expressing wild-type seedlings with 3-bromopyruvate (3-BP), one of the pyruvate mimetics known to function as an inhibitor of several kinds of enzymes in the TCA cycle (Shoshan, 2012). Treatment with 3-BP induced PIN2-GFP aggregations in the cytoplasm and a subsequent severe reduction of PIN2-GFP in the plasma membrane (Fig. 5, L–O; Supplemental Fig. S8). These results imply that the decreased level of PIN proteins in the mab1-1 mutant resulted from the deficiency of the TCA cycle caused by PDC dysfunction.
mab1 Alters Intracellular Distribution of PIN Auxin Efflux Carriers
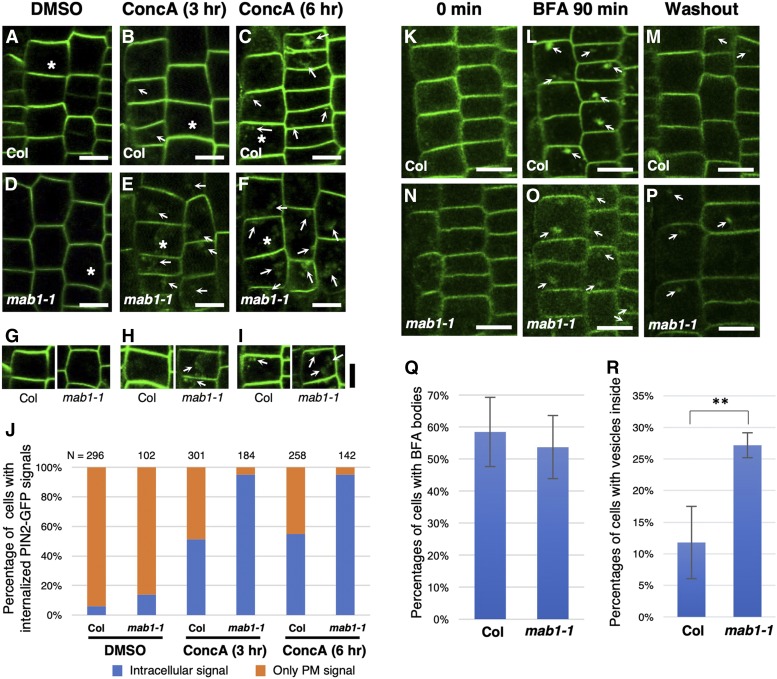
PIN proteins are continuously internalized from the plasma membrane and recycled between the plasma membrane and endosomes (Geldner et al., 2003; Dhonukshe et al., 2007). Furthermore, PIN proteins are targeted to the lytic vacuolar compartment and degraded there (Jaillais et al., 2006, 2007). This raises several possible ways in which PIN targeting to the vacuole could be accelerated and PIN recycling reduced in mab1 mutants. First, we analyzed the effects of the mab1 mutation on PIN targeting to the vacuole using concanamycin A, a specific inhibitor of vacuolar H+-ATPase required for acidification of lytic compartments and protein degradation. We examined the accumulation and trafficking of PIN2-GFP to the vacuole for degradation after concanamycin A treatment. In wild-type roots, a small increase in GFP aggregation was detectable 3 h after concanamycin A treatment (Fig. 6, A–C and G–J). At 6 h after the start of treatment, the cells showed induced accumulation and aggregation of PIN2-GFP in vacuoles in wild-type roots, predominantly in cells soon after division. In contrast, PIN2-GFP aggregation and accumulation in lytic vacuoles was detectable 3 h after concanamycin A treatment in most epidermal cells of mab1-1 roots (Fig. 6, D–F). These results indicate that PIN2 degradation in vacuoles was accelerated in mab1-1. Next, treatment with the phosphatidylinositol-3-kinase inhibitor wortmannin, which alters prevacuolar compartment identity and causes swelling of the compartment, induced much more swelling of intracellular PIN2-GFP signals in mutant roots than in wild-type roots (Supplemental Fig. S9). At the same time, the abundance of PIN2-GFP in the plasma membrane was increased by wortmannin treatment in mab1-1 root epidermis. These results indicate that mab1 results in vacuolar trafficking of PIN2, leading to severe reduction of the PIN2 level in the plasma membrane. Next, we examined whether a reduced PIN2 level in the plasma membrane is a result of defective PIN2 recycling. PIN2-GFP-expressing seedlings were treated with brefeldin A (BFA) for 90 min and then permitted to recover in liquid medium for 120 min. BFA bodies disappeared in most of the epidermal cells of the wild type (Fig. 6, K–M, Q, and R), whereas BFA bodies were often found in the mab1-1 epidermis (Fig. 6, N–R), indicating that mab1 also reduced PIN2 recycling via endosomes.
Figure 6.
PIN2-GFP degradation and recycling in wild-type and mab1-1 roots. A to F, PIN2-GFP localization in epidermis of wild-type (Col; A to C) and mab1-1 (D to F) roots treated with dimethyl sulfoxide (A and D) and with 1 µm concanamycin A (concA) for 3 h (B and E) and 6 h (C and F). G to I, The brightness of the GFP fluorescence was increased equally in both wild-type (Col; left) and mab1-1 (right) epidermal cells harboring PIN2p:PIN2-GFP, marked by an asterisk in A to F. Arrows indicate GFP accumulation in vacuoles and GFP aggregation in the cytosol. J, Percentage of epidermal cells of 5-d-old seedlings with internalized PIN2-GFP signals, treated with dimethyl sulfoxide, and with 1 µm concA for 3 h and 6 h. K to P, PIN2-GFP localization in epidermis of the primary roots of 4-d-old wild-type (Col; K to M) and mab1-1 (N to P) seedlings before (K and N), after treatment with 50 mm BFA for 90 min (L and O), and 120 min after BFA washout (M and P). Q, Percentages of epidermal cells harboring BFA bodies after treatment with BFA for 90 min. n = 992, 571 cells from seven roots in the wild type and eight in mab1-1. Data shown are mean values, with error bars representing the sd. P = 0.392, Student’s t test. R, Percentages of epidermal cells harboring small vesicles within 120 min after BFA washout. n = 716, 510 cells from six roots in the wild type and nine in mab1-1. Data shown are mean values, with error bars representing the sd. ** P < 0.01. Scale bars = 10 µm.
DISCUSSION
The mitochondrial PDC plays a central role in controlling the entry of carbon into the TCA cycle. The enzyme complex, whose biochemical functions underpin energy production, respiration, and primary metabolism, has a remarkable effect on the growth and development of organisms. The PDC consists of three enzyme components: E1, E2, and E3. These components have distinct enzymatic activities that act sequentially to convert pyruvate to acetyl-CoA and to reduce NAD+ to NADH. Eukaryotic mitochondrial PDH consists of 20–30 E1 heterotetramers (E1α and E1β), one E2 60 mer, and six E3 dimers. In Arabidopsis, the components of the mitochondrial PDC are encoded by two E1α genes (IAR4 and IAR4L; Luethy et al., 1995; Quint et al., 2009), one E1β gene (MAB1; Luethy et al., 1994), three E2 genes (mtE2-1, mtE2-2, and mtE2-3; Guan et al., 1995; Thelen et al., 1999; Taylor et al., 2004), and two E3 genes (At1g48030 and At3g17240; Lutziger and Oliver, 2001). However, while the biochemical function of each component is known, the role of PDC activity as a whole, and/or each enzyme component individually, in plant growth and development is not well understood. In our studies, the mab1-1 mutation caused a partial reduction of enzymatic activity and respiration, and showed mild auxin-related phenotypes. The MAB1 knockout mutant, mab1-3, did not produce homozygous plants, suggesting that MAB1 functions as a mitochondrial PDH E1β and plays an essential role in plant development. In addition, the physical interaction between MAB1 and IAR4 (Fig. 3A), specifically the decreased abundance of IAR4 and IAR4L in mab1-1 (Fig. 3, C–E; Table 1) and the phenotypic similarities between mab1 and iar4 (Supplemental Figs. S2 and S5), suggests that IAR4 and IAR4L contribute to PDC activity as E1α components. Previously, IAR4 was identified as a gene for auxin conjugate sensitivity and auxin homeostasis. Most IAA is conjugated to peptides, amino acids, or sugars, while active free IAA occurs in minute amounts (Ludwig-Müller, 2011). This suggests that auxin homeostasis could be regulated through the formation and hydrolysis of auxin conjugates. Many IAA-amino acid conjugates inhibit root elongation in a similar manner to IAA, possibly through hydrolysis by an endogenous auxin conjugate hydrolase. Mutant iar4 plants have reduced sensitivity to several IAA-amino acid conjugates, such as IAA-Ala and IAA-Gly. Interestingly, we found overaccumulation of the corresponding amino acids, especially those derived from pyruvate such as Ala and Gly, in mab1-1 mutants that also have reduced sensitivity to IAA-Ala (Fig. 4D; Supplemental Fig. S5). A mutation in mtE2-1 has been reported to cause overaccumulation of amino acids, especially Ala, and of intermediary metabolites of the TCA cycle (Yu et al., 2012). This suggests that iar4 mutants possibly accumulate the same amino acids as PDH-deficient mutants, leading to an inhibitory effect on the hydrolysis of IAA-amino acids. There appears to be a close correspondence between the specificity of IAA-amino acid conjugate response defects in iar4 and that of the amino acids accumulated in mab1 (Fig. 4D; LeClere et al., 2004).
Role of PDC E1β in Auxin-Regulated Organ Development
Local auxin accumulation promotes aerial and underground organ development. Mutations in Arabidopsis genes encoding subunits of the mitochondrial PDC have been reported to cause defective organ development; we report a similar effect here in mab1-1. The mutant m132, which has a defective mtE2-1 gene, and T-DNA insertion alleles of mtE2-1 and mtE2-3 showed reduction in organ size (Yu et al., 2012; Song and Liu, 2015), whereas iar4 has defects in root elongation (LeClere et al., 2004; Quint et al., 2009). Increasing the levels of auxin can rescue the defects in iar4, suggesting that IAR4 is required for root development through the control of auxin homeostasis. However, the inconsistent auxin response in mab1 mutants cannot simply be explained in terms of auxin levels. Although DR5rev:GFP was reduced in mab1-1 root tips as in iar4 (Fig. 5B; Quint et al., 2009), the auxin response marker was up-regulated in the vasculature of the maturation zone of mab1-1 roots (Fig. 5D; Supplemental Fig. S6, A and B). We suggest that a deficiency of basipetal auxin transport in the vasculature could account for this inconsistent auxin response. This is consistent with the severe reduction of PIN1 expression in mab1-1 roots. In addition, the results of our PIN1 localization analysis indicated that in mab1 embryos auxin is transported to the tips of cotyledon primordia via the protoderm, but does not flow down via provascular tissues due to severe reduction of PIN1 and discontinuity of PIN1-expressing provascular cells in inner cells (Fig. 5; Supplemental Fig. S6). In addition, exogenous auxin treatment failed to rescue impaired PIN2 localization in mab1 roots, indicating that auxin homeostasis is not involved in the control of PIN localization in the mab1 background (Supplemental Fig. S6). Our results suggest that MAB1 is required for organ development by affecting PIN-dependent polar auxin transport. Additionally, we showed that the mab1 mutation accelerated PIN2 trafficking to vacuoles and impaired PIN2 recycling (Fig. 6; Supplemental Fig. S9). Similar reduced PIN2 recycling and enhanced PIN2 targeting to the vacuole have been observed in seedlings treated with inhibitors of exocytosis (Zhang et al., 2016), suggesting that MAB1 is required for PIN exocytosis to the plasma membrane. In addition, our results of pharmacological analysis using 3-BP, the inhibitor of the TCA cycle, suggest that the effect of the mab1 mutation on PIN localization is caused by a dysfunctional TCA cycle. The exact mechanisms by which mitochondrial PDH and/or the TCA cycle affect PIN trafficking remain to be determined. Considering the fact that TCA cycle plays a key role in ATP production, it is possible that ATP depletion affects PIN trafficking. ATP depletion has been shown to induce marked internalization of polarly localized membrane proteins in rat cholangiocytes (Doctor et al., 2000). Although ATP depletion has been shown to inhibit clathrin-mediated endocytosis slightly in Arabidopsis seedlings (Dejonghe et al., 2016), which seems opposite to the effect of mab1 and 3-BP treatment, its effect on PIN trafficking remains obscure. Further investigation of the effect of mab1 and 3-BP treatment on PIN trafficking will be necessary. Another possibility is the generation of reactive oxygen species, which is generally associated with respiration in nongreen tissues. Recently, the importance of redox homeostasis in auxin-dependent organ development has been clarified. A mutation in the ROOT MERISTEMLESS1/CADMIUM SENSITIVE2 (RML1/CAD2) gene, which encodes the first enzyme of glutathione biosynthesis, abolishes postembryonic root development (Vernoux et al., 2000). Additional mutations in the NADPH-dependent thioredoxin reductases, NTRA and NTRB, interfere with developmental processes of aerial organs through modulation of auxin action in the cad2 mutant background (Bashandy et al., 2010). Furthermore, nitric oxide (NO), which regulates redox reactions in plant defense mechanisms, has also been reported to control the PIN1 level in organ development (Fernández-Marcos et al., 2011). The NO-overproducing mutant chlorophyll a/b binding protein underexpressed 1/NO overproducer 1 (cue1/nox1) displays a severe reduction in PIN1 levels in addition to reduced root meristem activity. Thus, redox reactions have been shown to have several roles in PIN regulation. Further analyses of the mab1 mutant will undoubtedly provide insight into the association between mitochondrial PDH enzymatic activity and polar auxin transport.
CONCLUSIONS
Here, we show that MAB1, which encodes a mitochondrial PDH E1β, is a novel molecular player in auxin-regulated organ formation in Arabidopsis. The mab1 mutation affected PDC enzymatic activity, respiration, and amino acid metabolism. Interestingly, in the mab1 mutant, the level of PIN auxin efflux carrier was decreased, possibly by impaired PIN recycling and accelerated targeting to lytic vacuoles. Our findings suggest a functional link between this mitochondrial metabolic pathway and PIN trafficking in organ formation.
MATERIALS AND METHODS
Plant Materials and Growth Condition
Arabidopsis (Arabidopsis thaliana) accession Columbia (Col) was used as the wild type. The following mutant alleles were used: pid-3 (Col; Bennett et al., 1995) and pid-2 (Lansberg erecta [Ler]; Christensen et al., 2000). mab1-1 was isolated from ethylmethane sulfonate-mutagenized M2 Col seeds obtained from LEHLE SEEDS (http://www.arabidopsis.com/). mab1-2 and mab1-3 carry T-DNA insertions at ∼310 bp upstream of the translation start site and in the third exon of MAB1, respectively. mab1-2 (SALK_111681) and mab1-3 (GABI_550G12) were obtained from the Arabidopsis Biological Resource Center and Nottingham Arabidopsis Stock Center (Alonso et al., 2003; Rosso et al., 2003). Plants carrying the mutant alleles were backcrossed three times to Col prior to phenotypic analysis. The plants were grown on soil as previously described (Fukaki et al., 1996). Siliques were collected for analyses of embryo phenotype, immunolocalization, and in situ hybridization. For analysis of seedling phenotypes, seeds were surface sterilized and germinated on Murashige and Skoog plates as previously described (Furutani et al., 2004).
Mapping and Cloning of MAB1 Genes
The mab1-1 mutant was crossed with pid-2 (Ler) plants to map the MAB1 locus. Approximately 200 F2 seedlings lacking cotyledons and exhibiting short roots were screened, and the MAB1 locus was mapped between the bacterial artificial chromosome contigs MBA10 and MFG13 on chromosome V. A 5.0-kb DNA fragment, which included the 1.0 kb upstream region and the 0.7 kb downstream region of the At5g50850 gene, was cloned into the binary vector pBIN19. In addition, the MAB1-coding region was inserted into pGWB5 between the cauliflower mosaic virus 35S promoter and the GFP-coding region (Nakagawa et al., 2007a). For MAB1p:MAB1-mCherry transformants, the genome fragment of the 1.0-kb sequence upstream of the translation start site of MAB1 (At5g50850), the coding sequence of MAB1, and the mCherry sequence were amplified and cloned into pGWB501 vector (Nakagawa et al., 2007b). These constructs were transformed into Agrobacterium tumefaciens strain MP90, which was then transformed into mab1-1 plants using the floral dip method (Clough and Bent, 1998). Transformants were selected by germinating seeds on medium containing 30 µg/mL kanamycin. Homozygous lines were obtained in the T3 generation, and T3 or T4 homozygous lines were used for complementation testing and reporter analysis.
Microscopy
For histological analysis, embryos were stained with 30 µg/ml of FM4-64 and fluorescence was imaged using a confocal laser-scanning microscopy (FV1000; Olympus). Whole-mount immunofluorescence was performed using a previously described protocol (Sauer et al., 2006). Antibodies were diluted as follows: 1:150 for goat anti-PIN1, 1:400 for sheep anti-PIN2, 1:600 for Alexa488- and Alexa546-conjugated anti-rabbit, anti-goat, and anti-sheep secondary antibodies. For visualizing mitochondria, seedlings were incubated in 1 µm of MitoTracker Red580 (Molecular Probes) for 15 min in the dark and then washed three times for 15 min. Images for all staining procedures were obtained using an Olympus FV1000 confocal microscope.
BiFC
The MAB1 and IAR4 open reading frames and the At2g25140/CLPB4 (CASEIN LYTIC PROTEINASE B4) mitochondrial presequence, which corresponds to the region spanning amino acids 1–90, were cloned into BiFC vectors or a fluorescent protein expression vector (Lee et al., 2007; Taoka et al., 2011). For the BiFC experiments, 3 µg each of Venus-N-terminal- and Venus-C-terminal-tagged protein expression vector (MAB1-VN and IAR4-VC, CLPB4-VN and IAR4-VC, and MAB1-VN and CLPB4-VC), and 3 µg of mitochondria-targeted TagRFP expression plasmid (CLPB4-TagRFP), were cotransformed into Arabidopsis protoplasts as previously described (Takeuchi et al., 2000). The CLPB4-TagRFP expression plasmid was introduced as a marker for transformation efficiency. After incubation at 23°C for 12 h in the dark, Venus (BiFC) and TagRFP (transformation marker) fluorescence was analyzed by confocal microscopy.
Yeast Two-Hybrid Assay
In order to eliminate mitochondrial transit peptides from MAB1, we performed a PCR amplification of a truncated 1005-bp fragment of MAB1 complementary DNA (cDNA) that corresponded to the region spanning amino acids 30–363. The fragment was subcloned into pAD-GAL4-GWRFC and pBD-GAL4-GWRFC (Yamaguchi et al., 2008). The mutation that causes the amino acid change G284E, found in the mab1-1 mutant background, was introduced into the truncated MAB1 gene. The plasmids were introduced into the AH109 strain using a Fast-Yeast Transformation kit (G-Biosciences). Empty vectors were used as negative controls. Interaction between the bait and target proteins was detected by the expression of the HIS3 reporter gene. To distinguish between leaky expression of the HIS3 gene and the specific interaction of proteins, detection of a second reporter gene (lacZ) was determined using a filter lift assay.
In Situ Hybridization
In situ hybridization was performed as previously described (Furutani et al., 2004). Hybridization was carried out at 45°C. The template for transcription of a MAB1 antisense probe was derived from a 606-bp fragment amplified using the primers pdhe1-b-for in situ-fw and pdhe1-b-for in situ-rv, listed in Supplemental Table S2.
GUS Staining
For MAB1p:GUS reporter lines, the genome fragment of the 1.0-kb sequence upstream of the translation start site of MAB1 (At5g50850) was amplified and cloned into pBI101.3 vector (Clontech). These constructs were transformed into Agrobacterium tumefaciens strain GV3101, which was then transformed into Col plants using the floral dip method (Clough and Bent, 1998).
Tissues were fixed in 90% (v/v) ice-cold acetone and then incubated in GUS staining solution (100 mm sodium phosphate, pH 7.0, 10 mm EDTA, 10 mm ferricyanide, 10 mm ferrocyanide, 0.1% [v/v] Triton X-100, and 0.5 mg/mL 5-bromo-4-chloro-3-indolyl-β-d-GlcA) at 37°C. After the GUS reaction, samples were destained with 70% (v/v) ethanol, cleared with 8:1:2 (w/v/v) mixture of 2,2,2-trichloroethane-1,1-diol, glycerol, and water, and observed under a microscope equipped with Nomarski optics (Ni-U; Nikon).
Isolation of Arabidopsis Mitochondria
Mitochondria were isolated from 3-week-old wild-type or mab1-1 seedlings grown hydroponically under long-day conditions using differential centrifugation and samples were collected and washed (Lee et al., 2008). Aliquots were frozen at −80°C for further analysis of enzymatic activity and differential in gel electrophoresis.
PDC Enzymatic Activity Assay
PDC activity was assayed spectrophotometrically by measuring NADH formation at 340 nm in a medium containing 50 mm Tes-NaOH (pH 7.6); 0.2% (v/v) Triton X-100; 1 mm MgCl2; 2 mm β-NAD; 0.2 mm thiamine pyrophosphate; 0.12 mm lithium -CoA, 2 mm l-Cys, and 1 mm sodium pyruvate (Huang et al., 2015). Assays were initiated by the addition of 100-µL aliquots of mitochondria into a cuvette (containing 40–60 µg of protein). An extinction coefficient of 6229 m−1 cm−1 at 340 nm for NADH was used for calculations. Succinate dehydrogenase enzymatic activity was determined based on the method described previously (Huang et al., 2010). The mitochondrial SDH activity was assayed spectrophotometrically by monitoring A600, at 25°C, in 1 ml of a reaction medium (50 mm potassium phosphate, pH 7.4, 10 mm sodium succinate, 0.1 mm EDTA, 0.1% (w/v) BSA, 10 mm potassium cyanide, 0.12 mm dichlorophenolindophenol, and 1.6 mm phenazine methosulfate). The reaction was initialized by adding isolated mitochondria. An extinction coefficient of 21 mm−1 cm−1 at 600 nm for reduced dichlorophenolindophenol was used for calculations.
Oxygen Consumption Assays
O2 consumption by a single root tip (5 mm) from 9-d-old plants grown on plates was measured as previously described (Sew et al., 2013). The respiration medium contains 10 mm HEPES, 10 mm MES, and 2 mm CaCl2 at pH 7.2.
DIGE Analysis
Mitochondrial proteins (50 µg) from wild-type (Col) and mab1-1 plants, as well as a 1:1 mixture of both samples, were acetone-precipitated and resolubilized in lysis buffer as previously described (Huang et al., 2013). Individual proteins were labeled with fluorescent cyanine dyes Cy2, Cy3, or Cy5 and then combined and separated on isoelectric focusing strips (pH 3–10 nonlinear). Two-dimensional gels were run at 50 mA current for 6 h. The fluorescence-labeled proteins were visualized with a Typhoon Trio laser scanner (GE Healthcare) and compared with the DECYDER software package (version 6.5; GE Healthcare). Mass spectrum identification of protein spots was conducted as previously reported (Lee et al., 2008).
Metabolite Extraction and GC-MS Analysis
Plants were grown on plates for 11 d under long-day conditions. Shoots and roots (six biological replicates) were harvested for GC-MS analysis as previously described (Huang et al., 2013). Raw GC-MS data preprocessing and statistical analysis were performed using METABOLOME-EXPRESS software (version 1.0, http://www.metabolome-express.org).
Reverse Transcription Quantitative PCR and Semiquantitative Reverse Transcription Quantitative PCR
Total RNA was isolated from roots or seedlings of 5-d-old seedlings using the RNeasy Plant Mini Kit (Qiagen). First-strand cDNA was synthesized from 2 µg of DNA-treated total RNA with an oligo(dT)24 primer and SuperScriptII reverse transcriptase (Invitrogen). Reverse transcription quantitative PCR was performed with the LightCycler 96 (Roche) with SYBR Premix Ex Taq (TaKaRa) and the gene-specific primer sets according to the manufacturer’s instructions. β-TUBULIN was measured for an internal control and used to normalize the data. All primer sequences are listed in Supplemental Table S2.
For semiquantitative reverse transcription PCR, cDNA was synthesized from 1 µg of DNA-treated total RNA with an oligo(dT)24 primer and SuperScriptII reverse transcriptase (Invitrogen). ACT2 was used for an internal control. All primer sequences are also listed in Supplemental Table S2.
Drug Application
Drugs were exogenously applied by incubation of 5-d-old seedlings in MS liquid medium supplemented with 3-bromopyruvate (250 mm, 500 mm), concanamycin A (1 µm), and wortmannin (33 µm). Control treatments contained an equivalent amount of solvent (dimethyl sulfoxide). For concanamycin A and wortmannin treatment, plants were rinsed twice in half MS liquid medium after treatment. For the brefeldin A (BFA) washout experiment, 4-d-old seedlings were transferred into half MS liquid medium with 50 µm BFA and incubated for 90 min. The washout of BFA was performed by incubating the seedlings in half MS liquid medium without BFA for 120 min after 2× rinsing with half MS liquid media.
Accession Numbers
The accession numbers for DNA sequences used in this study are listed in Supplemental Table S3.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Phenotypic analyses of the mab1-1 mutant.
Supplemental Figure S2. Phenotypic analyses of mab1 mutants in seedling development.
Supplemental Figure S3. Complementation test and characterization of the mab1-2 allele.
Supplemental Figure S4. Expression patterns of MAB1.
Supplemental Figure S5. Effects of IAA-Ala on seedling development in mab1 mutants.
Supplemental Figure S6. Auxin response in the mab1-1 mutant.
Supplemental Figure S7. Expression analyses of PIN genes.
Supplemental Figure S8. Effects of 3-BP on PIN2 internalization.
Supplemental Figure S9. Effects of wortmannin on PIN2 internalization in the mab1-1 mutant.
Supplemental Table S1. Metabolomic analysis of shoot and root tissue.
Supplemental Table S2. Primers used in this study.
Supplemental Table S3. Accession numbers of the genes in this study.
Acknowledgments
We thank Dr. Ben Scheres and Dr. Jiří Friml for providing us with PIN1p:PIN1-GFP-, PIN2p:PIN2-GFP-, and DR5rev:GFP-expressing plants, Dr. Tsuyoshi Nakagawa for providing the pGWB5 binary vector, and Arabidopsis Biological Resource Center and Nottingham Arabidopsis Stock Center for providing materials. We also thank Asami Mori and Keiko Uno for excellent technical assistance.
Footnotes
This work was supported in part by the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan, through Grants-in-Aid for Scientific Research on Priority Areas (14036222 and 19060007 to M.T.), a Grant-in-Aid for Young Scientists (B; 20770034), Grants-in-Aid for Scientific Research on Innovative Areas (26113513 and 16H01244), and a Global Centers of Excellence Program in Nara Institute for Science and Technology (Frontier Biosciences: Strategies for survival and adaptation in a changing global environment) to M.F. S.H. and A.H.M. were funded as Australian Research Council Australian Future Fellows (FT130101338 and FT110100242, respectively) and by the Australian Research Council Centre of Excellence in Plant Energy Biology (CE140100008).
Articles can be viewed without a subscription.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Arabidopsis Interactome Mapping Consortium (2011) Evidence for network evolution in an Arabidopsis interactome map. Science 333: 601–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashandy T, Guilleminot J, Vernoux T, Caparros-Ruiz D, Ljung K, Meyer Y, Reichheld JP (2010) Interplay between the NADP-linked thioredoxin and glutathione systems in Arabidopsis auxin signaling. Plant Cell 22: 376–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamins R, Quint A, Weijers D, Hooykaas P, Offringa R (2001) The PINOID protein kinase regulates organ development in Arabidopsis by enhancing polar auxin transport. Development 128: 4057–4067 [DOI] [PubMed] [Google Scholar]
- Benková E, Michniewicz M, Sauer M, Teichmann T, Seifertová D, Jürgens G, Friml J (2003) Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602 [DOI] [PubMed] [Google Scholar]
- Bennett SRM, Alvarez J, Bossinger G, Smyth DR (1995) Morphogenesis in pinoid mutants of Arabidopsis thaliana. Plant J 8: 505–520 [Google Scholar]
- Chen Q, Dai X, De-Paoli H, Cheng Y, Takebayashi Y, Kasahara H, Kamiya Y, Zhao Y (2014) Auxin overproduction in shoots cannot rescue auxin deficiencies in Arabidopsis roots. Plant Cell Physiol 55: 1072–1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y (2006) Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev 20: 1790–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y (2007a) Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell 19: 2430–2439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Qin G, Dai X, Zhao Y (2007b) NPY1, a BTB-NPH3-like protein, plays a critical role in auxin-regulated organogenesis in Arabidopsis. Proc Natl Acad Sci USA 104: 18825–18829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen SK, Dagenais N, Chory J, Weigel D (2000) Regulation of auxin response by the protein kinase PINOID. Cell 100: 469–478 [DOI] [PubMed] [Google Scholar]
- Ciszak EM, Korotchkina LG, Dominiak PM, Sidhu S, Patel MS (2003) Structural basis for flip-flop action of thiamin pyrophosphate-dependent enzymes revealed by human pyruvate dehydrogenase. J Biol Chem 278: 21240–21246 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dejonghe W, Kuenen S, Mylle E, Vasileva M, Keech O, Viotti C, Swerts J, Fendrych M, Ortiz-Morea FA, Mishev K, et al. (2016) Mitochondrial uncouplers inhibit clathrin-mediated endocytosis largely through cytoplasmic acidification. Nat Commun 7: 11710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhonukshe P, Aniento F, Hwang I, Robinson DG, Mravec J, Stierhof YD, Friml J (2007) Clathrin-mediated constitutive endocytosis of PIN auxin efflux carriers in Arabidopsis. Curr Biol 17: 520–527 [DOI] [PubMed] [Google Scholar]
- Doctor RB, Dahl RH, Salter KD, Fouassier L, Chen J, Fitz JG (2000) ATP depletion in rat cholangiocytes leads to marked internalization of membrane proteins. Hepatology 31: 1045–1054 [DOI] [PubMed] [Google Scholar]
- Fernández-Marcos M, Sanz L, Lewis DR, Muday GK, Lorenzo O (2011) Nitric oxide causes root apical meristem defects and growth inhibition while reducing PIN-FORMED 1 (PIN1)-dependent acropetal auxin transport. Proc Natl Acad Sci USA 108: 18506–18511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, Offringa R, Jürgens G (2003) Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426: 147–153 [DOI] [PubMed] [Google Scholar]
- Friml J, Yang X, Michniewicz M, Weijers D, Quint A, Tietz O, Benjamins R, Ouwerkerk PB, Ljung K, Sandberg G, et al. (2004) A PINOID-dependent binary switch in apical-basal PIN polar targeting directs auxin efflux. Science 306: 862–865 [DOI] [PubMed] [Google Scholar]
- Fukaki H, Fujisawa H, Tasaka M (1996) Gravitropic response of inflorescence stems in Arabidopsis thaliana. Plant Physiol 110: 933–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furutani M, Vernoux T, Traas J, Kato T, Tasaka M, Aida M (2004) PIN-FORMED1 and PINOID regulate boundary formation and cotyledon development in Arabidopsis embryogenesis. Development 131: 5021–5030 [DOI] [PubMed] [Google Scholar]
- Furutani M, Kajiwara T, Kato T, Treml BS, Stockum C, Torres-Ruiz RA, Tasaka M (2007) The gene MACCHI-BOU 4/ENHANCER OF PINOID encodes a NPH3-like protein and reveals similarities between organogenesis and phototropism at the molecular level. Development 134: 3849–3859 [DOI] [PubMed] [Google Scholar]
- Furutani M, Sakamoto N, Yoshida S, Kajiwara T, Robert HS, Friml J, Tasaka M (2011) Polar-localized NPH3-like proteins regulate polarity and endocytosis of PIN-FORMED auxin efflux carriers. Development 138: 2069–2078 [DOI] [PubMed] [Google Scholar]
- Geldner N, Anders N, Wolters H, Keicher J, Kornberger W, Muller P, Delbarre A, Ueda T, Nakano A, Jürgens G (2003) The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell 112: 219–230 [DOI] [PubMed] [Google Scholar]
- Guan Y, Rawsthorne S, Scofield G, Shaw P, Doonan J (1995) Cloning and characterization of a dihydrolipoamide acetyltransferase (E2) subunit of the pyruvate dehydrogenase complex from Arabidopsis thaliana. J Biol Chem 270: 5412–5417 [DOI] [PubMed] [Google Scholar]
- Huang S, Taylor NL, Narsai R, Eubel H, Whelan J, Millar AH (2010) Functional and composition differences between mitochondrial complex II in Arabidopsis and rice are correlated with the complex genetic history of the enzyme. Plant Mol Biol 72: 331–342 [DOI] [PubMed] [Google Scholar]
- Huang S, Taylor NL, Ströher E, Fenske R, Millar AH (2013) Succinate dehydrogenase assembly factor 2 is needed for assembly and activity of mitochondrial complex II and for normal root elongation in Arabidopsis. Plant J 73: 429–441 [DOI] [PubMed] [Google Scholar]
- Huang S, Lee CP, Millar AH (2015) Activity Assay for Plant Mitochondrial Enzymes. In Whelan J and Murcha MW, eds, Plant Mitochondria: Methods and Protocols. Springer New York, New York, : pp 139–149 [DOI] [PubMed] [Google Scholar]
- Jaillais Y, Fobis-Loisy I, Miège C, Rollin C, Gaude T (2006) AtSNX1 defines an endosome for auxin-carrier trafficking in Arabidopsis. Nature 443: 106–109 [DOI] [PubMed] [Google Scholar]
- Jaillais Y, Santambrogio M, Rozier F, Fobis-Loisy I, Miège C, Gaude T (2007) The retromer protein VPS29 links cell polarity and organ initiation in plants. Cell 130: 1057–1070 [DOI] [PubMed] [Google Scholar]
- Kleine-Vehn J, Leitner J, Zwiewka M, Sauer M, Abas L, Luschnig C, Friml J (2008) Differential degradation of PIN2 auxin efflux carrier by retromer-dependent vacuolar targeting. Proc Natl Acad Sci USA 105: 17812–17817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeClere S, Rampey RA, Bartel B (2004) IAR4, a gene required for auxin conjugate sensitivity in Arabidopsis, encodes a pyruvate dehydrogenase E1α homolog. Plant Physiol 135: 989–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CP, Eubel H, O’Toole N, Millar AH (2008) Heterogeneity of the mitochondrial proteome for photosynthetic and non-photosynthetic Arabidopsis metabolism. Mol Cell Proteomics 7: 1297–1316 [DOI] [PubMed] [Google Scholar]
- Lee U, Rioflorido I, Hong SW, Larkindale J, Waters ER, Vierling E (2007) The Arabidopsis ClpB/Hsp100 family of proteins: Chaperones for stress and chloroplast development. Plant J 49: 115–127 [DOI] [PubMed] [Google Scholar]
- Ludwig-Müller J. (2011) Auxin conjugates: Their role for plant development and in the evolution of land plants. J Exp Bot 62: 1757–1773 [DOI] [PubMed] [Google Scholar]
- Luethy MH, Miernyk JA, Randall DD (1994) The nucleotide and deduced amino acid sequences of a cDNA encoding the E1 β-subunit of the Arabidopsis thaliana mitochondrial pyruvate dehydrogenase complex. Biochim Biophys Acta 1187: 95–98 [DOI] [PubMed] [Google Scholar]
- Luethy MH, Miernyk JA, Randall DD (1995) The mitochondrial pyruvate dehydrogenase complex: Nucleotide and deduced amino-acid sequences of a cDNA encoding the Arabidopsis thaliana E1 α-subunit. Gene 164: 251–254 [DOI] [PubMed] [Google Scholar]
- Lutziger I, Oliver DJ (2001) Characterization of two cDNAs encoding mitochondrial lipoamide dehydrogenase from Arabidopsis. Plant Physiol 127: 615–623 [PMC free article] [PubMed] [Google Scholar]
- Mashiguchi K, Tanaka K, Sakai T, Sugawara S, Kawaide H, Natsume M, Hanada A, Yaeno T, Shirasu K, Yao H, et al. (2011) The main auxin biosynthesis pathway in Arabidopsis. Proc Natl Acad Sci USA 108: 18512–18517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michniewicz M, Zago MK, Abas L, Weijers D, Schweighofer A, Meskiene I, Heisler MG, Ohno C, Zhang J, Huang F, et al. (2007) Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell 130: 1044–1056 [DOI] [PubMed] [Google Scholar]
- Millar AH, Sweetlove LJ, Giegé P, Leaver CJ (2001) Analysis of the Arabidopsis mitochondrial proteome. Plant Physiol 127: 1711–1727 [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T (2007a) Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Suzuki T, Murata S, Nakamura S, Hino T, Maeo K, Tabata R, Kawai T, Tanaka K, Niwa Y, et al. (2007b) Improved gateway binary vectors: High-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci Biotechnol Biochem 71: 2095–2100 [DOI] [PubMed] [Google Scholar]
- Quint M, Barkawi LS, Fan KT, Cohen JD, Gray WM (2009) Arabidopsis IAR4 modulates auxin response by regulating auxin homeostasis. Plant Physiol 150: 748–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B (2003) An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol Biol 53: 247–259 [DOI] [PubMed] [Google Scholar]
- Sauer M, Paciorek T, Benková E, Friml J (2006) Immunocytochemical techniques for whole-mount in situ protein localization in plants. Nat Protoc 1: 98–103 [DOI] [PubMed] [Google Scholar]
- Sew YS, Ströher E, Holzmann C, Huang S, Taylor NL, Jordana X, Millar AH (2013) Multiplex micro-respiratory measurements of Arabidopsis tissues. New Phytol 200: 922–932 [DOI] [PubMed] [Google Scholar]
- Shoshan MC. (2012) 3-Bromopyruvate: Targets and outcomes. J Bioenerg Biomembr 44: 7–15 [DOI] [PubMed] [Google Scholar]
- Song L, Liu D (2015) Mutations in the three Arabidopsis genes that encode the E2 subunit of the mitochondrial pyruvate dehydrogenase complex differentially affect enzymatic activity and plant growth. Plant Cell Rep 34: 1919–1926 [DOI] [PubMed] [Google Scholar]
- Stepanova AN, Robertson-Hoyt J, Yun J, Benavente LM, Xie DY, Dolezal K, Schlereth A, Jürgens G, Alonso JM (2008) TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133: 177–191 [DOI] [PubMed] [Google Scholar]
- Takeuchi M, Ueda T, Sato K, Abe H, Nagata T, Nakano A (2000) A dominant negative mutant of sar1 GTPase inhibits protein transport from the endoplasmic reticulum to the Golgi apparatus in tobacco and Arabidopsis cultured cells. Plant J 23: 517–525 [DOI] [PubMed] [Google Scholar]
- Tao Y, Ferrer JL, Ljung K, Pojer F, Hong F, Long JA, Li L, Moreno JE, Bowman ME, Ivans LJ, et al. (2008) Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133: 164–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taoka K, Ohki I, Tsuji H, Furuita K, Hayashi K, Yanase T, Yamaguchi M, Nakashima C, Purwestri YA, Tamaki S, et al. (2011) 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature 476: 332–335 [DOI] [PubMed] [Google Scholar]
- Taylor NL, Heazlewood JL, Day DA, Millar AH (2004) Lipoic acid-dependent oxidative catabolism of α-keto acids in mitochondria provides evidence for branched-chain amino acid catabolism in Arabidopsis. Plant Physiol 134: 838–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelen JJ, Muszynski MG, David NR, Luethy MH, Elthon TE, Miernyk JA, Randall DD (1999) The dihydrolipoamide S-acetyltransferase subunit of the mitochondrial pyruvate dehydrogenase complex from maize contains a single lipoyl domain. J Biol Chem 274: 21769–21775 [DOI] [PubMed] [Google Scholar]
- Treml BS, Winderl S, Radykewicz R, Herz M, Schweizer G, Hutzler P, Glawischnig E, Ruiz RA (2005) The gene ENHANCER OF PINOID controls cotyledon development in the Arabidopsis embryo. Development 132: 4063–4074 [DOI] [PubMed] [Google Scholar]
- Vanneste S, Friml J (2009) Auxin: A trigger for change in plant development. Cell 136: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Vernoux T, Wilson RC, Seeley KA, Reichheld JP, Muroy S, Brown S, Maughan SC, Cobbett CS, Van Montagu M, Inzé D, (2000) The ROOT MERISTEMLESS1/CADMIUM SENSITIVE2 gene defines a glutathione-dependent pathway involved in initiation and maintenance of cell division during postembryonic root development. Plant Cell 12: 97–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewska J, Xu J, Seifertová D, Brewer PB, Ruzicka K, Blilou I, Rouquié D, Benková E, Scheres B, Friml J (2006) Polar PIN localization directs auxin flow in plants. Science 312: 883. [DOI] [PubMed] [Google Scholar]
- Won C, Shen X, Mashiguchi K, Zheng Z, Dai X, Cheng Y, Kasahara H, Kamiya Y, Chory J, Zhao Y (2011) Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proc Natl Acad Sci USA 108: 18518–18523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M, Kubo M, Fukuda H, Demura T (2008) Vascular-related NAC-DOMAIN7 is involved in the differentiation of all types of xylem vessels in Arabidopsis roots and shoots. Plant J 55: 652–664 [DOI] [PubMed] [Google Scholar]
- Yu H, Du X, Zhang F, Zhang F, Hu Y, Liu S, Jiang X, Wang G, Liu D (2012) A mutation in the E2 subunit of the mitochondrial pyruvate dehydrogenase complex in Arabidopsis reduces plant organ size and enhances the accumulation of amino acids and intermediate products of the TCA cycle. Planta 236: 387–399 [DOI] [PubMed] [Google Scholar]
- Zhang C, Brown MQ, van de Ven W, Zhang ZM, Wu B, Young MC, Synek L, Borchardt D, Harrison R, Pan S, et al. (2016) Endosidin2 targets conserved exocyst complex subunit EXO70 to inhibit exocytosis. Proc Natl Acad Sci USA 113: E41–E50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Christensen SK, Fankhauser C, Cashman JR, Cohen JD, Weigel D, Chory J (2001) A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291: 306–309 [DOI] [PubMed] [Google Scholar]