The developmentally programmed polarity of the auxin response underlies thermo-induced leaf hyponasty
Abstract
Plants exhibit diverse polar behaviors in response to directional and nondirectional environmental signals, termed tropic and nastic movements, respectively. The ways in which plants incorporate directional information into tropic behaviors is well understood, but it is less well understood how nondirectional stimuli, such as ambient temperatures, specify the polarity of nastic behaviors. Here, we demonstrate that a developmentally programmed polarity of auxin flow underlies thermo-induced leaf hyponasty in Arabidopsis (Arabidopsis thaliana). In warm environments, PHYTOCHROME-INTERACTING FACTOR4 (PIF4) stimulates auxin production in the leaf. This results in the accumulation of auxin in leaf petioles, where PIF4 directly activates a gene encoding the PINOID (PID) protein kinase. PID is involved in polarization of the auxin transporter PIN-FORMED3 to the outer membranes of petiole cells. Notably, the leaf polarity-determining ASYMMETRIC LEAVES1 (AS1) directs the induction of PID to occur predominantly in the abaxial petiole region. These observations indicate that the integration of PIF4-mediated auxin biosynthesis and polar transport, and the AS1-mediated developmental shaping of polar auxin flow, coordinate leaf thermonasty, which facilitates leaf cooling in warm environments. We believe that leaf thermonasty is a suitable model system for studying the developmental programming of environmental adaptation in plants.
Plants actively adjust their growth and architecture to adapt to changing environments, in which the roles of auxin are extensively studied. Both biosynthesis and cellular and organismal distribution of auxin are critical for its function (Petrásek and Friml, 2009; Zhao, 2010; Huang et al., 2017). In particular, it is known that the polar flow of auxin produces its gradients in different plant tissues, leading to asymmetric cell elongation (Ding et al., 2011). Molecular events leading to polar auxin transport are fairly well understood in terms of the tropic behaviors of plant organs, which occur in response to directional stimuli, such as light and gravity (Ding et al., 2011; Rakusová et al., 2011).
Vesicle-to-membrane trafficking of the auxin efflux transporter PIN-FORMED3 (PIN3) determines the polarity of auxin flow (Friml et al., 2002; Ding et al., 2011). It has been reported that localized distribution of PIN proteins at different sides of the cell is regulated by developmental pathways and environmental stimuli (Friml et al., 2002; Zádníková et al., 2010). Under unilateral light conditions, PIN3 is localized to the inner membranes of hypocotyl cells on the illuminated side, while it moves to both the inner and outer membranes of hypocotyl cells at the shaded side (Ding et al., 2011; Rakusová et al., 2011). Accordingly, auxin accumulates on the shaded side, resulting in hypocotyl bending toward light. The polar movement of PIN3 is also important for the gravitropic responses of hypocotyls. It is known that PIN3 is polarized to the outer membranes of hypocotyl cells at the lower side, in response to gravity stimuli (Rakusová et al., 2011).
PINOID (PID) is a protein kinase that is known to phosphorylate PIN3 (Ding et al., 2011). The functional significance of the PID-mediated PIN phosphorylation has been explored (Kleine-Vehn et al., 2009; Zourelidou et al., 2014). It is known that PID-dependent phosphorylation of PIN proteins regulates their polarity (Kleine-Vehn et al., 2009). It also activates the PIN-mediated auxin efflux (Zourelidou et al., 2014). However, the conventional model depicting the PID-mediated regulation of PIN3 polarization through protein phosphorylation might be oversimplified (Weller et al., 2017). It has been suggested that, in addition to the PID-mediated protein phosphorylation, transcriptional regulation of the PIN3 gene, its protein turnover, and as yet unidentified cellular trafficking systems would also contribute to the PIN3-mediated polar auxin distribution (Willige et al., 2011; Wang et al., 2015; Weller et al., 2017).
There is another type of plant movement responses, termed nastic movements, in which nondirectional stimuli, such as temperature, light irradiance, and flooding, trigger directional movements of specific plant organs (Forterre et al., 2005; van Zanten et al., 2009; Keuskamp et al., 2010; Sasidharan and Voesenek, 2015). Unlike tropic movements, nastic movements are not affected by the direction of stimuli but instead are modulated by the quality of stimuli (Forterre et al., 2005). A well-known example is leaf hyponasty, in which nondirectional warm temperature signals stimulate the upward bending of leaf petioles (Koini et al., 2009; van Zanten et al., 2009). The thermally induced nastic leaf behaviors are often termed leaf thermonasty and suggested to play a role in protecting the thermolabile tissues from the radiant heat of the soil surface (Crawford et al., 2012). However, it is unknown how nondirectional temperature signals drive directional leaf movement.
In plants, it has been suggested that the direction of nastic movements is established by certain developmental programs, which determine the polarity of organ patterning (Nick and Schafer, 1989; Polko et al., 2011). In recent years, the molecular mechanisms and signaling schemes specifying leaf polarity have been studied extensively. The adaxial-abaxial polarity of leaves is determined through complicated signaling networks comprising several proteins and RNA molecules (Kerstetter et al., 2001; Iwasaki et al., 2013; Merelo et al., 2016). The Arabidopsis (Arabidopsis thaliana) epigenetic repressors ASYMMETRIC LEAVES1 (AS1) and AS2 act as major upstream regulators of the leaf polarity-determining genes (Iwasaki et al., 2013). They are also required for the polar distribution of auxin at the leaf tip to promote asymmetric differentiation (Zgurski et al., 2005), further supporting the role of AS proteins in specifying the polarity of plant organs.
In this work, we demonstrated that hyponastic leaf movement at warm temperatures is modulated by a developmentally programmed auxin gradient in the petiole. Thermo-activated PHYTOCHROME-INTERACTING FACTOR4 (PIF4) induces the PID gene, which is known to regulate the auxin transporter PIN3. Notably, PIF4 preferentially binds to the PID promoter primarily in the abaxial petiole region at 28°C. Auxin produced via the PIF4-YUCCA8 (YUC8) pathway in the leaf is transported to the petiole and distributed toward the abaxial side of petioles via the polarized action of PID. We conclude that the PIF4-mediated auxin biosynthesis and polar transport and the AS1-directed acceleration of PID expression in the abaxial petiole region constitute a thermal pathway that triggers the upward bending of leaf petioles in response to nondirectional warm temperature signals. We also found that PIF4-governed leaf thermonasty enhances leaf cooling under warm temperature conditions, as has been shown previously (Crawford et al., 2012).
RESULTS
Polarization of Leaf Thermonasty Is Independent of Light Direction and Gravity
The leaves of many plant species bend upward at warm temperatures (Lippincott and Lippincott, 1971; van Zanten et al., 2009). Kinetic analysis of Arabidopsis petiole bending revealed that the hyponastic response was rapidly initiated within several hours following exposure to 28°C (Fig. 1A). We then asked how directional leaf movement occurs in response to ambient temperatures, typical of nondirectional environmental signals.
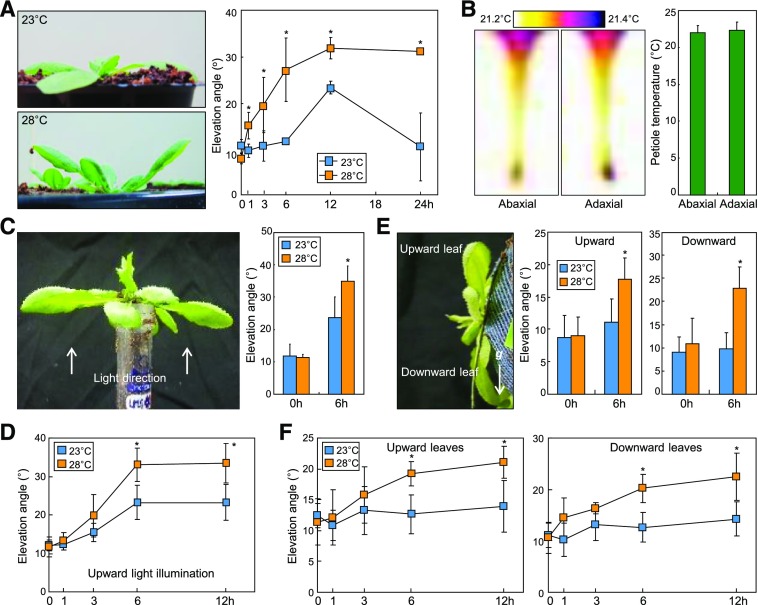
Figure 1.
Polarization of leaf thermonasty is independent of light direction and gravity. Elevation angles of the fifth and sixth rosette leaves relative to the horizontal plane were measured using 3-week-old plants exposed to 28°C. Three independent measurements, each consisting of 16 individual plants grown under identical conditions, were statistically analyzed using Student’s t test (*, P < 0.01, difference from 23°C). Error bars indicate se. A, Kinetic effects of warm temperatures on petiole bending. Elevation angles were measured in a time course following exposure to 28°C. B, Leaf petiole temperatures. Temperatures on the abaxial and adaxial surfaces of leaf petioles were measured by infrared thermography 6 h following exposure to 28°C. C, Effects of light direction on leaf thermonasty. Plants were exposed to 28°C for 6 h with upward light illumination. D, Kinetic effects of warm temperatures and light directions on petiole bending. Elevation angles were measured in a time course following exposure to 28°C with upward light illumination. E, Effects of gravity on leaf thermonasty. Plants were subjected to horizontal gravitropic stimulation (g) at 28°C for 6 h in the light. F, Kinetic effects of warm temperatures and gravity on petiole bending. Elevation angles were measured in a time course following exposure to 28°C with horizontal gravitropic stimulation, as depicted in E.
We suspected that the polarity of leaf thermonasty might be influenced by directional environmental cues. Infrared thermography showed that temperatures on the adaxial and abaxial surfaces of leaf blades and petioles were identical during temperature treatments (Fig. 1B). We next examined the effects of light direction on leaf thermonasty using upward light illumination. While the extent of hyponastic movement was somewhat different from that observed under downward light illumination, its polarity was not altered by upward light illumination (Fig. 1, C and D). Gravity is another unilateral cue (Rakusová et al., 2011). We were not able to detect any effects of horizontal gravitropic stimulation on the polarity of leaf thermonasty (Fig. 1, E and F). These observations show that the polarity of leaf thermonasty is not influenced by light direction and gravity.
Auxin Biosynthesis and Its Polar Transport Mediate Leaf Thermonasty
The plant growth hormone auxin has been reported to be tightly associated with thermomorphogenesis (Gray et al., 1998; Franklin et al., 2011; Park et al., 2017). To investigate the potential linkage between leaf thermonasty and auxin, we examined localized expression patterns of auxin-responsive genes encoding SMALL AUXIN UP RNA19 (SAUR19) and SAUR22 in the abaxial and adaxial halves of leaf petioles at 28°C (Supplemental Fig. S1A). Thermal induction of SAUR genes was larger in the abaxial samples compared with that in the adaxial samples (Fig. 2A). In addition, a mutation in the gene encoding the auxin receptor TRANSPORT INHIBITOR RESPONSE1 (TIR1) caused reduced thermonastic leaf bending (Fig. 2B). These observations suggest that auxin is involved in the thermal regulation of leaf hyponasty.
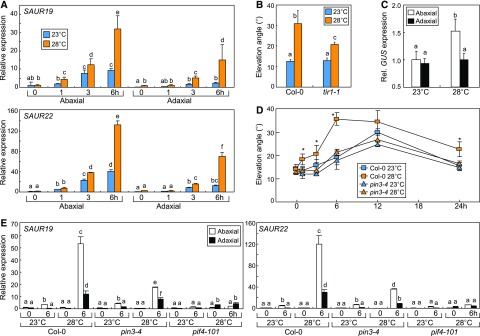
Figure 2.
Expression of auxin response genes is elevated in the abaxial petiole region during leaf thermonasty. Three independent measurements, each consisting of 16 individual plants grown under identical conditions, were subjected to statistical analysis. Different letters represent significant differences (P < 0.01) determined by one-way ANOVA with posthoc Tukey’s test. Error bars indicate se. A, Transcription of SAUR genes. Three-week-old Col-0 plants were exposed to 28°C before preparing dissected petiole samples for total RNA extraction. Transcript levels were analyzed by reverse transcription-mediated quantitative real-time PCR (RT-qPCR). B, Leaf thermonasty in the tir1-1 mutant. Plants were temperature treated as described above. C, Transcription of the GUS reporter. DR5:GUS plants were exposed to 28°C for 6 h, and petiole sampling and RT-qPCR were performed as described above. D, Leaf thermonasty in the pin3-4 mutant. Statistical analysis was performed using Student’s t test (*, P < 0.01, difference from 23°C). E, Transcription of SAUR genes in pin3-4 and pif4-101 mutants. Temperature treatments, preparation of petiole samples, and RT-qPCR were performed as described above.
To investigate the association between polar auxin accumulation and leaf thermonasty, we employed the DR5:GUS auxin reporter plants. Gene expression analysis revealed that the expression of the GUS reporter gene was elevated by ∼2-fold in the abaxial petiole region but was not discernibly altered in the adaxial petiole region in 28°C-treated plants (Fig. 2C). Utilization of the fluorescent auxin system, the DII-VENUS reporter, revealed that fluorescence intensity was significantly lower in the abaxial epidermal regions than in adaxial epidermal regions at 28°C (Supplemental Fig. S1B). Consistent with this, inhibition of auxin transport by 1-N-naphthylphthalamic acid significantly abolished leaf thermonasty (Supplemental Fig. S2A). In addition, exogenous application of indole acetic acid (IAA) to the adaxial petiole region significantly reduced leaf elevation at warm temperatures (Supplemental Fig. S2B). These data suggest that preferential auxin accumulation in the abaxial petiole region is closely linked with leaf thermonasty.
Meanwhile, it is well known that warm temperatures trigger auxin biosynthesis through the PIF4-mediated induction of the YUC8 gene (Gray et al., 1998; Franklin et al., 2011; Park et al., 2017), raising the possibility that PIF4-mediated auxin biosynthesis would be functionally associated with leaf thermonasty. We first examined whether thermal induction of leaf elevation occurs in the yuc8 mutant. As anticipated, leaf thermonastic movement was largely impaired in the mutant (Supplemental Fig. S2C). In addition, pretreatments with yucasin, a potent inhibitor of YUC enzymes (Nishimura et al., 2014), abolished leaf thermonasty in Columbia-0 (Col-0) plants (Supplemental Fig. S2D), supporting that thermo-induced auxin biosynthesis is required for leaf thermonasty. Auxin is produced mainly in leaf blade in response to environmental stimuli (Michaud et al., 2017). Accordingly, when leaf blade was excised, upward petiole bending did not occur at warm temperatures (Supplemental Fig. S2E). In addition, the thermal induction of YUC8 expression was only marginal in the leaf petioles (Supplemental Fig. S2F), further supporting the notion that auxin biosynthesis occurs mainly in leaf blade. Taken together, these observations clearly show that both auxin biosynthesis and its polar transport are important for leaf thermonasty.
On the basis of the functional association between polar auxin transport and leaf hyponasty (Pantazopoulou et al., 2017), we examined any potential roles of the PIN3 transporter during leaf thermonasty. Notably, leaf thermonasty did not occur in the PIN3-deficient pin3-4 mutant (Fig. 2D), consistent with the reduced thermal induction of SAUR genes in the pin3-4 mutant (Fig. 2E). Thermal induction of SAUR genes was also compromised in the pif4-101 mutant, which exhibits disturbed leaf thermonasty (Koini et al., 2009). These observations support that PIN3-mediated polar auxin transport is critical for leaf thermonasty.
PID Is Associated with Leaf Thermonasty
Cellular functions of PIN proteins are regulated at multiple levels, such as gene transcription, intracellular trafficking, and protein stability (Willige et al., 2011; Wang et al., 2015; Weller et al., 2017). Gene expression analysis using dissected leaf petiole samples revealed that the transcription of the PIN3 gene was elevated more than 2-fold on both the abaxial and adaxial sides at 28°C (Supplemental Fig. S3A). Since thermal induction of PIN3 expression occurs on both sides of the petioles, other regulatory mechanisms rather than transcriptional control would play a major role in regulating the asymmetric PIN3 function during leaf thermonasty.
Intracellular distribution of PIN3 is a critical event in establishing polar auxin flow (Ding et al., 2011; Rakusová et al., 2011). To investigate whether intracellular distribution of PIN3 is associated with leaf thermonasty, we expressed a PIN3-GFP gene fusion driven by the endogenous PIN3 promoter in Col-0 plants, and the distribution patterns of GFP signals were examined in the leaf petiole cells. It is known that PIN3 is produced mainly in the endodermal cells of the shoots, which are a major barrier of auxin flow between vasculature and outer cell layers (Ding et al., 2011). Considering the notion that chlorophyll autofluorescence is relatively weaker in the endodermal cells compared with that in the epidermal cells (Keuskamp et al., 2010), we identified the endodermal cells for the examination of the intracellular distribution of PIN3-GFP proteins.
At 23°C, the PIN3 proteins were distributed equally in the outer membranes of both adaxial and abaxial petiole cells (Fig. 3, A and B; Supplemental Fig. S3B). In contrast, at 28°C, more PIN3 proteins were localized in the outer membranes of abaxial petiole cells than adaxial petiole cells, indicating that warm temperatures stimulate the polarization of PIN3 to the outer membranes of the abaxial petiole cells. It is known that polar PIN3 localization under unilateral light or gravity stimulation is efficiently blocked by brefeldin A (BFA), a potent inhibitor of vesicle trafficking (Ding et al., 2011; Weller et al., 2017). We found that leaf thermonasty was discernibly suppressed in the presence of BFA (Fig. 3C). Polarization of PIN3 to the abaxial petiole cells was also blocked by BFA treatments in warm temperature conditions (Supplemental Fig. S4, A and B). This indicates that, during leaf thermonasty, BFA-sensitive vesicular trafficking drives the thermo-induced polarization of PIN3. Intracellular aggregates, termed BFA bodies, are frequently observed in plant cells, mostly root cells, when treated with BFA (Jásik et al., 2016). We did not observe such BFA bodies in our assay conditions, similar to what was observed with the fluorescence images of hypocotyl cells (Ding et al., 2011). It is likely that BFA bodies may not be readily visible in all tissue cells.
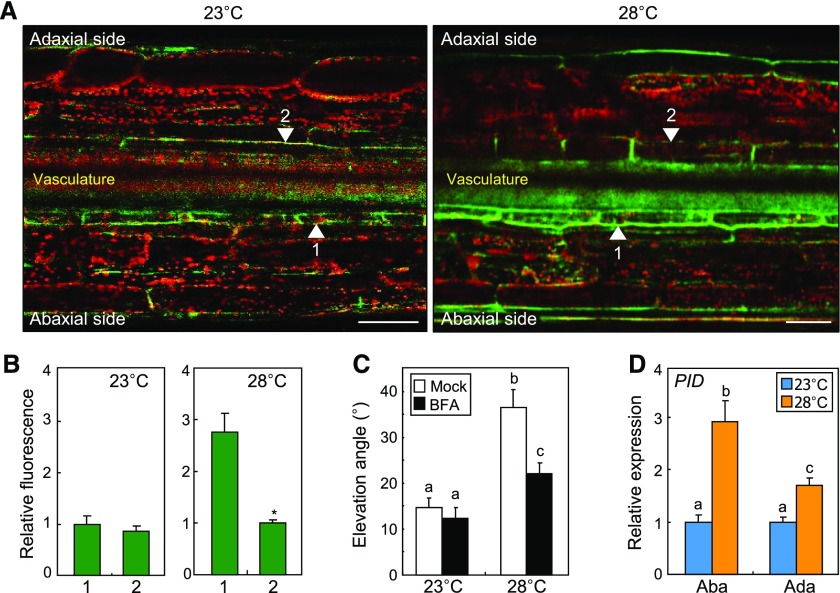
Figure 3.
PIN3 is polarized to the outer membranes of abaxial endodermal cells in leaf petioles at warm temperatures. Three independent measurements, each consisting of eight individual plants grown under identical conditions, were statistically analyzed. Error bars indicate se. In C and D, different letters represent significant differences (P < 0.01) determined by one-way ANOVA with posthoc Tukey’s test. A and B, Polar distribution of PIN3. A, Three-week-old plants expressing a PIN3-GFP fusion driven by the endogenous PIN3 promoter were exposed to 28°C for 6 h before fluorescence microscopy of the fifth leaf petioles. Green and red signals indicate PIN3-GFP and chlorophyll autofluorescence, respectively. Arrowheads indicate the outer membranes of petiole endodermal cells. Bars = 100 μm. B, PIN3-GFP signals were quantitated (Student’s t test, *, P < 0.01). C, Effects of BFA on leaf thermonasty. A 10 μm BFA solution was sprayed on the petioles before exposure to 28°C. Elevation angles were measured and statistically analyzed. D, Transcription of the PID gene. Leaf petioles of Col-0 plants were dissected into abaxial (Aba) and adaxial (Ada) halves. Transcript levels were analyzed by RT-qPCR.
We next analyzed the transcription of genes encoding regulators of PIN3 polarization at warm temperatures. Notably, the transcription of PID and WAG2 genes, which encode Ser/Thr protein kinases that phosphorylate PIN3 (Dhonukshe et al., 2010), was elevated to higher levels in the abaxial petiole cells than in the adaxial petiole cells (Fig. 3D; Supplemental Fig. S4C). The transcription of ARABIDOPSIS H+-ATPase (AHA), SERINE/THREONINE PROTEIN PHOSPHATASE2A (PP2A), and D6 PROTEIN KINASE (D6PK) genes was also elevated slightly in the adaxial petiole regions (Supplemental Fig. S4C). Since polar auxin transport is more active in the abaxial petiole regions, it seems likely that altered transcript levels of AHA, PP2A, and D6PK genes play a minor role during leaf thermonasty. Considering that PID function is regulated primarily at the transcriptional level (Ding et al., 2011), we hypothesized that differential production of PID in the abaxial and adaxial petiole cells would be functionally linked with PIN3 polarization at warm temperatures.
To examine the functional linkage between the differential PID transcription in petiole cells and leaf thermonasty, we analyzed the effects of warm temperatures on the transcription of auxin-responsive genes in transgenic plants overexpressing the PID gene driven by the strong Cauliflower mosaic virus 35S promoter. Because of the molecular nature of the promoter used, it was thought that differential expression of the PID gene on the abaxial and adaxial sides would be disturbed in the transgenic plants. As expected, the polarization patterns of PIN3 to the outer membranes were similar in the abaxial and adaxial petiole cells at both 23°C and 28°C (Fig. 4, A and B; Supplemental Fig. S5, A and B). Leaf thermonasty was also impaired in the PID-overexpressing plants (Fig. 4C), and thermal induction of SAUR22 gene expression was largely reduced in the transgenic plants (Fig. 4D), supporting that PID is important for leaf thermonasty.
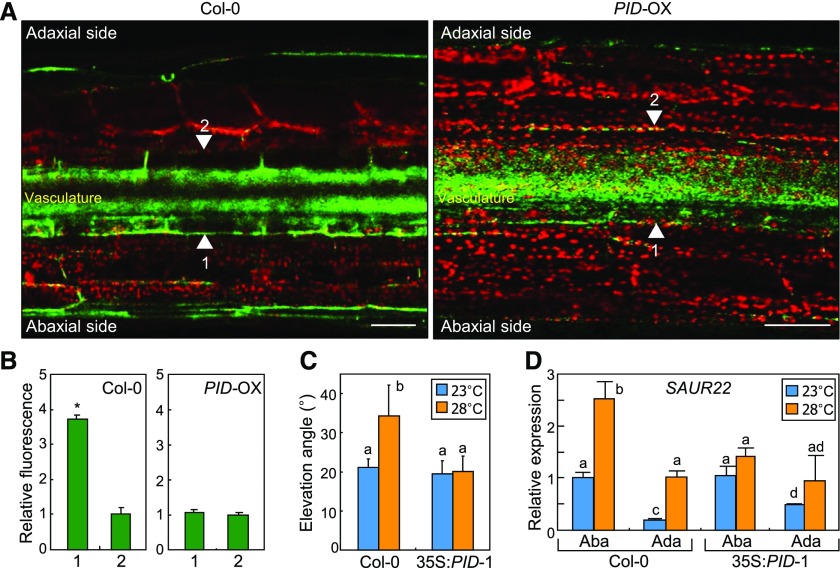
Figure 4.
PID-directed PIN3 polarization underlies leaf thermonasty. Three independent measurements, each consisting of 16 individual plants grown under identical conditions, were statistically analyzed. In C and D, different letters represent significant differences (P < 0.01) determined by one-way ANOVA with posthoc Tukey’s test. Error bars indicate se. A and B, Polar distribution of PIN3. A, Three-week-old plants expressing a PIN3-GFP fusion driven by the endogenous PIN3 promoter were exposed to 28°C for 6 h before fluorescence microscopy of the fifth leaf petioles. Arrowheads indicate the outer membranes of petiole endodermal cells. Bars = 100 μm. B, PIN3-GFP signals were quantitated (Student’s t test, *, P < 0.01). C, Leaf thermonasty in 35S:PID plants. D, Transcription of the SAUR22 gene in 35S:PID plants. Leaf petioles were dissected into abaxial (Aba) and adaxial (Ada) halves. Transcript levels were analyzed by RT-qPCR.
Leaf thermonasty was also disturbed in the pid wag1 wag2 triple mutant (Supplemental Fig. S5C). However, the phenotype of the triple mutant should be considered with caution because the lack of leaf thermonasty might be attributed to other developmental defects in the mutant. It has been reported that the triple mutant is defective in leaf organogenesis (Dhonukshe et al., 2010). It is known that PID trafficking to the cellular membranes is associated with PIN function (Kleine-Vehn et al., 2009). Transient expression of the GFP-PID fusion in Arabidopsis protoplasts revealed that PID proteins are predominantly localized to the membranes at both 23°C and 28°C (Supplemental Fig. S5D), which is also consistent with the roles of PID at the cell periphery in the previous report (Kleine-Vehn et al., 2009). Together, these observations indicate that PIN3-mediated polar auxin flow is mediated by PID, which is differentially produced in the abaxial and adaxial petiole cells during leaf thermonasty.
PIF4 Activates PID Transcription
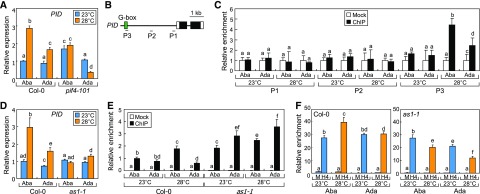
A next question was how PID transcription is differentiated in the abaxial and adaxial petiole cells at warm temperatures. We found that leaf thermonasty and SAUR gene expression were disrupted in the pif4-101 mutant (Fig. 2E; Koini et al., 2009), raising the possibility that PIF4 would regulate PID transcription in response to warm temperatures. Gene expression analysis revealed that PID transcription was somewhat higher in the abaxial petiole cells of pif4-101 at 23°C (Fig. 5A). Notably, thermal induction of PID transcription did not occur in the abaxial petiole cells of the mutant. In addition, PID transcription was even reduced in the adaxial petiole cells of the mutant at warm temperatures. It seems that additional factor(s), in addition to PIF4, might also be involved in the thermal regulation of PID transcription.
Figure 5.
PIF4- and AS1-mediated developmental signals activate PID transcription in the abaxial petiole region at warm temperatures. Three independent measurements, each consisting of 16 individual plants grown under identical conditions, were statistically analyzed. Different letters represent significant differences (P < 0.01) determined by one-way ANOVA with posthoc Tukey’s test. Error bars indicate se (A and D) or sd (C, E, and F). A, PID transcription in the pif4-101 mutant. Three-week-old plants were exposed to 28°C for 6 h. Transcript levels were analyzed by RT-qPCR. B, Genomic structure of the PID locus. Black boxes are exons, and white boxes are 5′ and 3′ untranslated regions. The P1 to P3 sequences were analyzed in ChIP assays. C, PIF4 binding to the PID promoter. Three-week-old plants expressing a PIF4-FLAG fusion driven by the endogenous PIF4 promoter were exposed to 28°C for 6 h. ChIP assays were performed using an anti-FLAG antibody. D, Transcription of the PID gene in as1-1 leaf petioles. Transcript levels were analyzed by RT-qPCR. E, PIF4 binding to the PID promoter in the as1-1 mutant. A PIF4-FLAG fusion was expressed driven by the endogenous PIF4 promoter in Col-0 plants and the as1-1 mutant. The P3 sequence was used in the assay. F, H4 acetylation in PID chromatin. ChIP assays were performed using either Col-0 or as1-1 leaf petioles. An anti-H4Ac antibody was used for immunoprecipitation. H4 acetylation was analyzed by ChIP-qPCR. M, Mock.
To investigate PIF4 binding to PID chromatin in vivo, we conducted chromatin immunoprecipitation (ChIP) assays using transgenic plants expressing a PIF4-FLAG gene fusion driven by the endogenous PIF4 promoter. While PIF4 binding to PID chromatin was not detected at 23°C, PIF4 evidently bound to the P3 sequence region harboring a G-box motif in PID chromatin at 28°C (Fig. 5, B and C), which is known as the PIF4-binding motif (Oh et al., 2012). Notably, thermo-induced DNA binding of PIF4 was more prominent in the abaxial petiole cells (Fig. 5C), which is in good agreement with the higher induction of the PID gene in these cells at 28°C.
We next examined PIN3 distribution in the pif4-101 mutant, which expresses a PIN3-GFP gene fusion driven by the endogenous PIN3 promoter. It was found that the thermally induced polarization of PIN3 to the abaxial endodermal cells was not observed in the pif4-101 mutant (Supplemental Fig. S6), indicating that PIF4 is linked with PIN3 polarization during leaf thermonasty.
AS1 Modulates the PID-Mediated Polar Transport of PIN3
A critical issue was how the polarity of leaf thermonasty is established in response to nondirectional temperature signals. It has been suggested that hyponastic leaf movement would be related to developmental programs that determine leaf polarity (Nick and Schafer, 1989; Polko et al., 2011). AS1 is the epigenetic repressor that functions upstream of leaf polarity-specifying genes (Zgurski et al., 2005; Iwasaki et al., 2013). Thus, we analyzed possible roles of AS1 during leaf thermonasty. It was revealed that the AS1-deficient as1-1 mutant did not exhibit any symptoms of leaf thermonasty (Supplemental Fig. S7A). Different leaf angles in the mutants at 23°C would be attributed to a developmental disruption of abaxial-adaxial polarity, rather than a constitutive thermomorphogenic response, as has been observed in related mutants exhibiting abnormal leaf polarity (Izhaki and Bowman, 2007; Pérez-Pérez et al., 2010). Consistently, it was found that the thermal induction of SAUR and PID genes was abolished in the abaxial petiole cells of the as1-1 mutant (Fig. 5D; Supplemental Fig. S7B). These observations indicate that AS1 plays a role in modulating PID-mediated auxin response during leaf thermonasty.
It has been reported that AS1 modulates AUXIN RESPONSE FACTOR3 (ARF3) expression during the adaxial-abaxial partitioning of a leaf (Iwasaki et al., 2013). In order to examine whether the auxin response is altered in the as1-1 mutant, we analyzed PID transcription in leaf petioles treated with IAA. Consistent with the previous report (Bai and Demason, 2008), exogenous application of IAA promoted PID transcription in the leaf petioles of Col-0 plants (Supplemental Fig. S7C). In contrast, the inductive effects of IAA disappeared in the tir1 mutant. Notably, the IAA-mediated induction of PID transcription still occurred in the as1-1 mutant, suggesting that the as1-1 mutant retains the capacity of auxin responsiveness in gene expression. On the other hand, polar localization of PIN3 to the outer membranes of the abaxial petiole cells was compromised in the mutant at 28°C (Supplemental Fig. S8). It is thus evident that AS1 plays a role in the polar transport of PIN3 during leaf thermonasty.
AS1 Specifies the Polarity of Leaf Thermonasty
The next question was how AS1 modulates PID expression in the petiole cells. Yeast two-hybrid assay revealed that AS1 does not interact with PIF4, the regulator of PID transcription (Supplemental Fig. S7D). In addition, the transcript levels of the PIF4 gene were similar in the petioles of Col-0 plants and the as1-1 mutant (Supplemental Fig. S7E). Considering the signaling linkage between PID-mediated auxin response and AS1, we hypothesized that AS1 would modulate the binding of PIF4 to the promoter of the PID gene at 28°C. ChIP assays revealed that PIF4 binding to the PID promoter was more prominent in the abaxial regions than in the adaxial regions of Col-0 leaf petioles at warm temperatures (Fig. 5E). In contrast, the DNA binding of PIF4 was higher in the adaxial region of as1-1 leaf petioles at both 23°C and 28°C (Fig. 5E). These observations indicate that the DNA-binding affinity of PIF4 is differentially affected in the as1-1 mutant.
To explore molecular events mediating PIF4 binding to the PID promoter, we investigated the chromatin modification patterns in the P3 sequence region of PID chromatin (Fig. 5B). Interestingly, histone 4 (H4) acetylation, an active transcriptional marker (Akhtar and Becker, 2000), was significantly elevated in the abaxial petiole cells but not in the adaxial petiole cells at warm temperatures (Fig. 5F). In contrast, the thermal induction of H4 acetylation in the P3 sequence was even reduced in the as1-1 mutant (Fig. 5F).
We next examined whether H4 acetylation is functionally important for leaf thermonasty by employing chemical inhibitors of histone deacetylases. Leaf thermonasty was found to be disrupted in Col-0 plants treated with trichostatin A and 4-(dimethylamino)-N-[6-(hydroxyamino)-6-oxohexyl]-benzamide (Supplemental Fig. S9A), which strongly inhibit histone deacetylases in plants (Ueno et al., 2007; Supplemental Fig. S9B). While the levels of global H4 acetylation were unaltered at warm temperatures (Supplemental Fig. S9B), the pattern of H4 acetylation was discernibly altered in the PID promoter (Fig. 5F), supporting the notion that the thermal regulation of H4 acetylation is specific to the PID promoter during leaf thermonasty. Together, these observations illustrate that warm temperatures induce H4 acetylation in the PID promoter predominantly in the abaxial petiole cells to facilitate the DNA binding of PIF4.
Leaf Thermonasty Lowers Leaf Temperatures
Thermomorphogenic modifications of morphology and architecture are considered to help plants to enhance body-cooling capacity by facilitating heat dissipation (Crawford et al., 2012). Leaf thermonasty is a representative thermomorphogenic event occurring at warm temperatures, indicating that it would be functionally associated with leaf-cooling capacity.
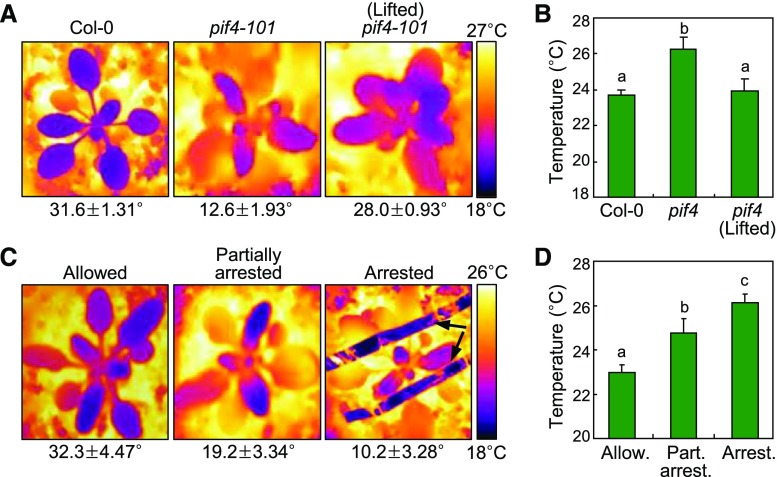
We employed infrared thermography to monitor leaf temperatures under warm temperature conditions. For comparison, the rosette leaves of the pif4-101 mutant were physically lifted so that elevation angles of the mutant leaves were similar to those of Col-0 leaves at warm temperatures (Fig. 6A). It was found that leaf temperature was significantly higher in the pif4-101 mutant compared with that in Col-0 leaves (Fig. 6, A and B). Notably, leaf temperatures were similar in Col-0 plants and in the pif4-101 mutant having physically lifted rosette leaves, supporting that leaf hyponasty is directly related to leaf temperatures.
Figure 6.
Leaf cooling is associated with hyponastic leaf movement. Three-week-old plants were exposed to 28°C for 6 h before taking infrared thermographs (A and C). Elevation angles are given below the thermographs. Temperatures at the central blade areas of the sixth rosette leaves were measured (B and D). Three independent measurements, each consisting of eight individual plants grown under identical conditions, were statistically analyzed. Different letters represent significant differences (P < 0.01) determined by one-way ANOVA with posthoc Tukey’s test. Error bars indicate se. A and B, Leaf temperatures in the pif4-101 mutant. For comparison, the mutant rosette leaves were physically lifted to mimic the increased leaf hyponasty observed in Col-0 plants. C and D, Leaf temperatures in Col-0 plants having physically arrested leaf hyponasty. The rosette leaves were arrested physically to the soil so that leaf hyponasty is not elevated at 28°C. Arrows marks arresting wires.
To further examine the effects of thermonastic leaf movement on leaf cooling, the rosette leaves of Col-0 plants were physically arrested to the soil surface so that the leaves are not able to bend upward at warm temperatures, mimicking those having disrupted thermonastic movement. Leaf cooling was proportional to the elevation angles of rosette leaves (Fig. 6, C and D), verifying that leaf hyponasty is critical for leaf cooling under warm environments.
DISCUSSION
Thermomorphogenesis refers to a suite of plant morphological and architectural modifications, such as hypocotyl elongation, increase of leaf hyponasty, small, thin leaves, and leaf petiole elongation, that occur in response to changes in ambient temperatures (Koini et al., 2009; Franklin et al., 2011; Crawford et al., 2012; Park et al., 2017).
The PIF4 transcription factor, which had been originally identified as a key signaling component of plant photomorphogenesis (Lorrain et al., 2008), plays a central role in warm temperature-mediated morphogenic responses. Accordingly, a wide array of thermomorphogenic responses are impaired in PIF4-defective mutants (Koini et al., 2009). PIF4 directly activates the transcription of the YUC8 gene encoding an auxin biosynthetic enzyme, triggering a complex network of thermal responses (Sun et al., 2012). Recently, it has been reported that BRASSINAZOLE-RESISTANT1, a transcriptional factor that mediates brassinosteroid signaling (Ryu et al., 2007), is involved in PIF4-mediated thermomorphogenesis (Ibañez et al., 2018), further extending the complexity of PIF4 signaling.
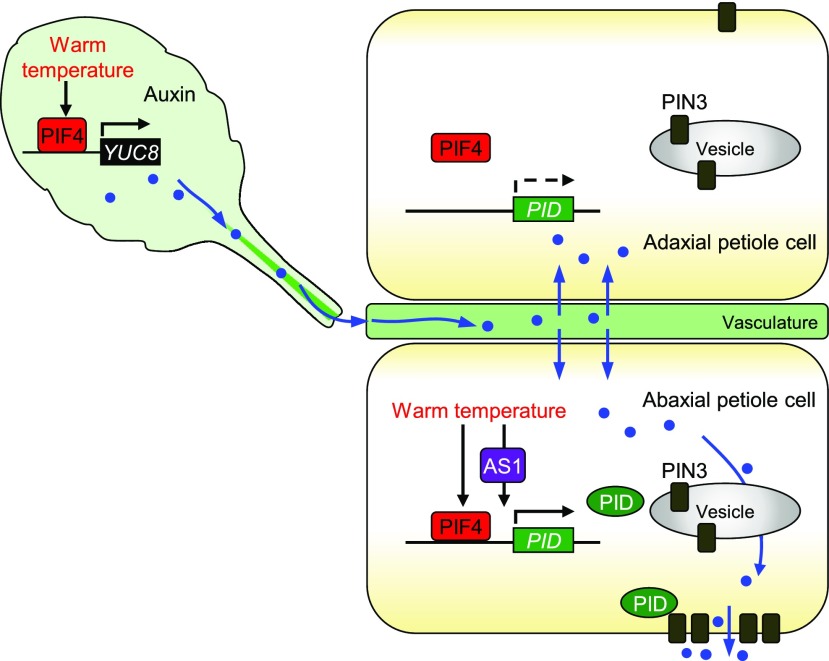
In this work, we demonstrated that the central thermomorphogenic regulator PIF4 constitutes a distinct, two-branched auxin signaling pathway that modulates hyponastic leaf movement under warm temperature conditions. Thermo-activated PIF4 directly stimulates PID transcription in petiole cells, resulting in polar auxin accumulation. In another route, the PIF4-YUC8 branch promotes auxin production in leaf blade, which is transported to the petiole and functions as the substrate for PIN3 machinery. The PIF4-mediated leaf thermonasty also requires AS1-mediated developmental cues that direct PIF4-mediated PID transcription to occur mostly in the abaxial petiole region (Fig. 7). The working scheme of PIF4 and AS1 in triggering leaf thermonasty illustrates a seminal mode of developmental programming of environmental adaptation, which would also be applicable to other nastic movements in plants.
Figure 7.
Schematic model for developmental shaping of polar auxin flow during leaf thermonasty. Thermo-activated PIF4 triggers auxin production in the leaf blade. Auxin is then transported to the petiole, where it is distributed toward the epidermis via PIN3. PIF4 also activates PID transcription in the petiole. The leaf polarity determinant AS1 directs PID transcription to occur predominantly in the abaxial petiole region. The PID-mediated PIN3 polarization to the outer membrane of abaxial petiole cells determines the direction of leaf bending. Blue arrows marks the paths of auxin flow.
Our observations imply that some additional regulators other than PID might also be involved in the thermal activation of PIN3 during leaf thermonasty. For example, PID transcription was differentially regulated in the abaxial and adaxial cells of the pif4-101 mutant, while PIN3 proteins were equally distributed in the petiole cells. In addition, gene expression analysis showed that some additional transcriptional regulators other than PIF4 might be linked with the transcription of PID. It was also found that exogenous application of auxin induced PID transcription, consistent with the previous report (Bai and Demason, 2008), further supporting the notion that PID transcription is modulated by multiple factors. It is probable that auxin-responsive transcription factors, including ARFs, would contribute to PID transcription, directly or indirectly, during leaf thermonasty.
It is notable that the leaf polarity-specifying AS1 incorporates developmental cues into PID expression. AS1 specifies the polarity of lateral organs, in particular, rosette leaves in Arabidopsis (Zgurski et al., 2005; Iwasaki et al., 2013). It was observed that H4 acetylation in PID chromatin and the binding of PIF4 to PID chromatin were discernibly affected in the as1-1 mutant. However, the interpretation of thermomorphogenic phenotypes and biochemical events in as1-1 should be considered with caution in that as1-1 might exhibit stunted growth and pleiotropic effects caused by distorted leaf polarity. It is currently unclear whether the alterations in DNA binding of PIF4 and H4 acetylation in PID chromatin of the as1-1 mutant are functionally interrelated or not. In addition, while PIF4 binds to the PID promoter irrespective of the as1-1 mutation, the thermal induction of PID expression was impaired in the mutant. Thus, it remains to be elucidated whether and how AS1 collaborates with PIF4 in inducing PID transcription during leaf thermonasty.
Our observations demonstrate that the PIN3-mediated polar auxin transport constitutes an important biochemical event during leaf thermonasty. It is notable that PIN proteins are intimately associated with directional movements of plant organs. Especially, PIN3, PIN4, and PIN7 proteins are required for the gravitropic and phototropic responses of hypocotyls (Ding et al., 2011; Rakusová et al., 2011). Meanwhile, the PIN2 protein contributes to root gravitropism (Rahman et al., 2010). It is possible that multiple PIN proteins are modulated by diverse environmental stimuli, such as ambient temperature, gravity, and unilateral light, in order to optimize auxin distribution in different plant organs. It will be interesting to examine whether PIN members other than PIN3 are also functionally associated with leaf thermonasty.
Our data define a distinct signaling network that mediates the developmental shaping of a hyponastic response occurring in leaf petioles at warm temperatures. It is evident that the polarity of leaf thermonasty is not determined by temperature differences and directional light and gravity stimuli but instead is established by the AS1-mediated developmental program. Plants exhibit various types of nastic movements (Forterre et al., 2005; van Zanten et al., 2009; Sasidharan and Voesenek, 2015). In mimosa (Albizia julibrissin) and Venus flytrap (Dionaea muscipula), simple touching promotes water transport to the specific sites of plant organs, resulting in rapid and directional nastic movements (Forterre et al., 2005). In Arabidopsis, flooding triggers ethylene accumulation and asymmetric growth in leaf petioles, causing upward hyponastic bending (Sasidharan and Voesenek, 2015). It is worthy of investigation whether AS1 is also involved in these nastic movements in plants.
MATERIALS AND METHODS
Plant Materials and Culture Conditions
All Arabidopsis (Arabidopsis thaliana) lines used were in the Col-0 background. The pin3-4 (SALK-038609), as1-1 (CS146), yuc8 (SALK-096110), and tir1-1 (CS3798) mutants were obtained from a pool of mutant lines deposited in the Arabidopsis Biological Resource Center (Ohio State University). The pif4-101 mutant (Garlic-114-G06) has been described previously (Lorrain et al., 2008). DII-VENUS plants were obtained from Jae-Yean Kim. The pid wag1 wag2 triple mutant was obtained from Remko Offringa (Dhonukshe et al., 2010).
Expression vectors harboring the pPIN3:PIN3-GFP gene fusions have been described previously (Ganguly et al., 2012). The expression vectors were transformed into pif4-101 and as1-1 mutants (Supplemental Fig. S10). To generate 35S:PID transgenic plants, a PID-coding sequence was amplified from Col-0 cDNA using a pair of primers, MYC-PID F (5′-ACCCGGGTTATGTTACGAGAATCAGACGGT-3′) and MYC-PID R (5′-AATGGATCCTCAAAAGTAATCGAACGCCG-3′), with XmaI and BamHI sites, respectively. The amplified PCR products were fused in frame to the 5′ end of a MYC-coding sequence in the myc-PBA vector. The vector construct was then transformed into transgenic plants expressing the pPIN3:PIN3-GFP fusion in the Col-0 background. The pPIF4:PIF4-FLAG transgenic plants were generated by transforming the pPIF4:PIF4-FLAG-containing vector, which has been described previously (Lee et al., 2014), into the as1-1 mutant.
Sterilized Arabidopsis seeds were cold imbibed at 4°C for 3 d in complete darkness and allowed to germinate either in soil or on one-half-strength Murashige and Skoog-agar plates under long days (16 h of light and 8 h of dark) with white light illumination (120 µmol m−2 s−1) provided by fluorescent FLR40D/A tubes (Osram) in a controlled growth chamber set at 23°C. Three-week-old plants were subjected to temperature treatments at Zeitgeber time 2 (ZT2) for 6 h, unless otherwise mentioned.
Measurement of Leaf Angles
Leaf hyponasty was analyzed using digital images of 3-week-old plants exposed to different temperatures at ZT2. Leaf angles were measured at ZT8 otherwise mentioned. Quantification of elevation angles was performed using the ImageJ software (http://imagej.nih.gov/ij/).
Infrared Thermography
Thermal images of 3-week-old plants were recorded using the thermal imaging camera T420 (FLIR). The thermal images were analyzed using FLIR Tools (http://www.flirkorea.com/home/), and leaf temperatures were recorded in the central area of the sixth rosette leaves.
Sample Preparation for Abaxial and Adaxial Segments
For gene expression analysis, ChIP assay, western-blot assay, and confocal analysis, petioles were divided into abaxial and adaxial halves using razors. Samples were prepared on the basis of previous reports (Polko et al., 2013) with slight modifications.
Gene Expression Analysis
Transcript levels were analyzed by RT-qPCR according to the guidelines that have been proposed to ensure reproducible and accurate measurements (Udvardi et al., 2008). RT-qPCR was conducted in 384-well blocks with the Applied Biosystems QuantStudio 6 Flex using the SYBR Green I master mix in a volume of 10 µL (Han et al., 2019). The two-step thermal cycling profile employed was 15 s at 95°C for denaturation and 1 min at 60°C to 65°C, depending on the calculated melting temperatures of PCR primers, for annealing and polymerization. PCR primers used are listed in Supplemental Table S1. The eIF4A gene (At3g13920) was included as an internal control in PCR to normalize the variations in the amounts of primary cDNAs used.
RT-qPCR was performed using three independent RNA samples, each of which was prepared from a pool of 16 independent plant materials. The comparative ΔΔCT method was employed to evaluate relative quantities of each amplified product in the samples. The threshold cycle was automatically determined for each reaction by the system set with default parameters.
Confocal Microscopy
Transgenic plants expressing the PIN3 gene driven by the endogenous PIN3 promoter have been frequently employed in confocal imaging assays on nastic and tropic responses, such as shade avoidance and hypocotyl phototropism and gravitropism (Keuskamp et al., 2010; Ding et al., 2011; Rakusová et al., 2011).
Three-week-old pPIN3:PIN3-GFP transgenic plants were transferred to 28°C for 6 h. Following temperature treatments, the petioles of the fifth and sixth rosette leaves were dissected so that the longitudinal sections were placed on cover glasses. The dissected petiole samples were subjected to fluorescence imaging using the SP8 X confocal microscope (Leica).
It is known that chlorophyll autofluorescence is relatively weaker in endodermal cells than in epidermal cells (Keuskamp et al., 2010). We visualized PIN3-GFP distribution in the endodermal cells of the petioles using a Leica SP8 X microscope with the following laser and filter setup: white light laser, 488 nm for excitation, 490 to 600 nm for emission to detect GFP, and 561 nm for excitation, and 650 to 750 nm for emission to detect chlorophyll autofluorescence. The magnification value was set to 10. Fluorescence signals from 10 endodermal cells per sample were analyzed using the Leica Application Suite X and ImageJ software. GFP and autofluorescence signals of outer membranes were quantified per 1,500-µm2 sample.
To examine auxin accumulation, 3-week-old DII-VENUS plants were temperature treated, and epidermal regions were subjected to fluorescence imaging using an Olympus BX53 microscope with the following laser and filter setup: Olympus U-HGLGPS laser, 480 to 500 nm for excitation, and 510 to 560 nm for emission to detect VENUS. The magnification value was set to 4. VENUS signals were counted per 2-mm2 sample area to quantify DII-VENUS fluorescence.
Pharmacological Treatment
For BFA treatments, a 10 µm BFA solution (Sigma-Aldrich) was sprayed onto 3-week-old plants prior to temperature treatments. For treatments with histone deacetylase inhibitors, 3 µm trichostatin A (Sigma-Aldrich) and 30 µm 4-(dimethylamino)-N-[6-(hydroxyamino)-6-oxohexyl]-benzamide (Cayman Chemical) solutions were sprayed onto 3-week-old plants prior to temperature treatments. For treatments with auxin biosynthesis and transport inhibitors, 250 µm yucasin (Nishimura et al., 2014) and 10 µm 1-N-naphthylphthalamic acid (Sigma-Aldrich) solutions were sprayed onto 3-week-old plants before exposure to 28°C.
ChIP
ChIP assays were performed essentially as described previously (Lee et al., 2014). Briefly, 3-week-old plants grown on Murashige and Skoog-agar plates were harvested at ZT24 and vacuum infiltrated with 1% (v/v) formaldehyde for cross-linking. The plant materials were then ground in liquid nitrogen after quenching the cross-linking process and resuspended in 30 mL of nuclear extraction buffer (1.7 m Suc, 10 mm Tris-Cl, pH 7.5, 2 mm MgCl2, 0.15% [v/v] Triton X-100, 5 mm β-mercaptoethanol, and 0.1 mm phenylmethylsulfonyl fluoride) containing protease inhibitors (Sigma-Aldrich) and filtered through Miracloth filters (Millipore). The filtered mixture was centrifuged at 4,300g for 20 min at 4°C, and nuclear fractions were isolated by the Suc cushion method. The nuclear fractions were lysed with lysis buffer (50 mm Tris-Cl, pH 8, 0.5 m EDTA, and 1% [w/v] SDS) containing protease inhibitors and sonicated to obtain chromatin fragments of 400 to 700 bp.
Five micrograms of anti-FLAG (Sigma-Aldrich) or anti-H4Ac (Millipore) antibody was added to the chromatin solution and incubated for 16 h at 4°C. The Protein-G or Protein-A agarose beads (Millipore) were then added to the solution and incubated for 1 h. The incubated mixture was centrifuged at 4,000g for 2 min at 4°C. Following reverse cross-linking of the precipitates, residual proteins were removed by treatments with proteinase K. DNA fragments were purified using a silica membrane spin column (Promega). To determine the amounts of DNA enriched in chromatin preparations, qPCR was performed, and the values were normalized to the amount of input in each sample.
Immunological Assay
Plant materials were ground in liquid nitrogen. The ground plant materials were resuspended in protein extraction buffer (100 mm Tris-Cl, pH 6.8, 4% SDS, 0.2% Bromophenol Blue, 20% glycerol, and 200 mm β-mercaptoethanol). The mixtures were boiled for 10 min and then centrifuged at 16,000g for 10 min at 4°C. The supernatants were analyzed by SDS-PAGE. The separated proteins were transferred to polyvinylidene difluoride membranes. Anti-H4Ac (Millipore), anti-tubulin (Sigma-Aldrich), and anti-H3 (Millipore) antibodies were used for the immunological detection of H4Ac, tubulin, and H3 proteins, respectively. Anti-rabbit and anti-mouse IgG-peroxidase antibodies (Santa Cruz Biotechnology) were used as secondary antibodies for the immunoblot assays with anti-H4Ac, anti-H3, and anti-tubulin primary antibodies, respectively.
Statistical Analysis
The statistical significance between two means of measurements was determined using a two-sided Student’s t test with P < 0.01 or P < 0.05. To determine statistical significance for more than two populations, one-way ANOVA with post hoc Tukey’s test (P < 0.01) was used. Statistical analyses were performed using the Rstudio software (https://www.rstudio.com/). Three independent measurements were statistically analyzed for phenotypic assays and gene expression analysis unless otherwise mentioned.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: PIF4, AT2G43010; PID, AT2G34650; PIN3, AT1G70940; AS1, AT2G37630; SAUR19, AT5G18010; SAUR22, AT5G18050; TIR1, AT3G62980; AHA1, AT2G18960; AHA2, AT4G30190; PP2A, AT1G69960; D6PK, AT5G55910; WAG1, AT1G53700; WAG2, AT3G14370; YUC8, AT4G28720; and eIF4A, AT3G13920.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Auxin distribution in the abaxial and adaxial petiole regions during leaf thermonasty.
Supplemental Figure S2. Both auxin biosynthesis and its polar transport are involved in leaf thermonasty.
Supplemental Figure S3. PIN3 is associated with leaf thermonasty.
Supplemental Figure S4. Thermo-induced polarization of PIN3 is mediated by vesicular trafficking.
Supplemental Figure S5. PID is associated with leaf thermonasty.
Supplemental Figure S6. PIF4 mediates PIN3 polarization during leaf thermonasty.
Supplemental Figure S7. Thermonastic leaf movements in the as1-1 mutant.
Supplemental Figure S8. Thermo-induced PIN3 polarization is disrupted in the as1-1 mutant.
Supplemental Figure S9. H4 acetylation is related to leaf thermonasty.
Supplemental Figure S10. Transcription of PIN3 and PID genes in different genetic backgrounds.
Supplemental Table S1. Primers used in this work.
Acknowledgments
We thank Jae-Yean Kim for the DII-VENUS reporter plants and Tomokazu Koshiba for providing yucasin. The pid wag1 wag2 mutant was obtained from Remko Offringa.
Footnotes
This work was supported by the Leaping Research Program (NRF-2018R1A2A1A19020840) provided by the National Research Foundation of Korea and the Next-Generation BioGreen 21 Program (PJ013134) provided by the Rural Development Administration of Korea. Y.-J.P. was partially supported by the Global Ph.D. Fellowship Program through the National Research Foundation of Korea (NRF-2016H1A2A1906534).
References
- Akhtar A, Becker PB (2000) Activation of transcription through histone H4 acetylation by MOF, an acetyltransferase essential for dosage compensation in Drosophila. Mol Cell 5: 367–375 [DOI] [PubMed] [Google Scholar]
- Bai F, Demason DA (2008) Hormone interactions and regulation of PsPK2:GUS compared with DR5:GUS and PID:GUS in Arabidopsis thaliana. Am J Bot 95: 133–145 [DOI] [PubMed] [Google Scholar]
- Crawford AJ, McLachlan DH, Hetherington AM, Franklin KA (2012) High temperature exposure increases plant cooling capacity. Curr Biol 22: R396–R397 [DOI] [PubMed] [Google Scholar]
- Dhonukshe P, Huang F, Galvan-Ampudia CS, Mähönen AP, Kleine-Vehn J, Xu J, Quint A, Prasad K, Friml J, Scheres B, et al. (2010) Plasma membrane-bound AGC3 kinases phosphorylate PIN auxin carriers at TPRXS(N/S) motifs to direct apical PIN recycling. Development 137: 3245–3255 [DOI] [PubMed] [Google Scholar]
- Ding Z, Galván-Ampudia CS, Demarsy E, Łangowski Ł, Kleine-Vehn J, Fan Y, Morita MT, Tasaka M, Fankhauser C, Offringa R, et al. (2011) Light-mediated polarization of the PIN3 auxin transporter for the phototropic response in Arabidopsis. Nat Cell Biol 13: 447–452 [DOI] [PubMed] [Google Scholar]
- Forterre Y, Skotheim JM, Dumais J, Mahadevan L (2005) How the Venus flytrap snaps. Nature 433: 421–425 [DOI] [PubMed] [Google Scholar]
- Franklin KA, Lee SH, Patel D, Kumar SV, Spartz AK, Gu C, Ye S, Yu P, Breen G, Cohen JD, et al. (2011) Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc Natl Acad Sci USA 108: 20231–20235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J, Wiśniewska J, Benková E, Mendgen K, Palme K (2002) Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 415: 806–809 [DOI] [PubMed] [Google Scholar]
- Ganguly A, Lee SH, Cho HT (2012) Functional identification of the phosphorylation sites of Arabidopsis PIN-FORMED3 for its subcellular localization and biological role. Plant J 71: 810–823 [DOI] [PubMed] [Google Scholar]
- Gray WM, Ostin A, Sandberg G, Romano CP, Estelle M (1998) High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proc Natl Acad Sci USA 95: 7197–7202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han SH, Park YJ, Park CM (2019) Light primes the thermally induced detoxification of reactive oxygen species during development of thermotolerance in Arabidopsis. Plant Cell Physiol 60: 230–241 [DOI] [PubMed] [Google Scholar]
- Huang CF, Yu CP, Wu YH, Lu MJ, Tu SL, Wu SH, Shiu SH, Ku MSB, Li WH (2017) Elevated auxin biosynthesis and transport underlie high vein density in C4 leaves. Proc Natl Acad Sci USA 114: E6884–E6891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibañez C, Delker C, Martinez C, Bürstenbinder K, Janitza P, Lippmann R, Ludwig W, Sun H, James GV, Klecker M, et al. (2018) Brassinosteroids dominate hormonal regulation of plant thermomorphogenesis via BZR1. Curr Biol 28: 303–310.e3 [DOI] [PubMed] [Google Scholar]
- Iwasaki M, Takahashi H, Iwakawa H, Nakagawa A, Ishikawa T, Tanaka H, Matsumura Y, Pekker I, Eshed Y, Vial-Pradel S, et al. (2013) Dual regulation of ETTIN (ARF3) gene expression by AS1-AS2, which maintains the DNA methylation level, is involved in stabilization of leaf adaxial-abaxial partitioning in Arabidopsis. Development 140: 1958–1969 [DOI] [PubMed] [Google Scholar]
- Izhaki A, Bowman JL (2007) KANADI and class III HD-Zip gene families regulate embryo patterning and modulate auxin flow during embryogenesis in Arabidopsis. Plant Cell 19: 495–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jásik J, Bokor B, Stuchlík S, Mičieta K, Turňa J, Schmelzer E (2016) Effects of Auxins on PIN-FORMED2 (PIN2) Dynamics Are Not Mediated by Inhibiting PIN2 Endocytosis. Plant Physiol 172: 1019–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerstetter RA, Bollman K, Taylor RA, Bomblies K, Poethig RS (2001) KANADI regulates organ polarity in Arabidopsis. Nature 411: 706–709 [DOI] [PubMed] [Google Scholar]
- Keuskamp DH, Pollmann S, Voesenek LA, Peeters AJ, Pierik R (2010) Auxin transport through PIN-FORMED 3 (PIN3) controls shade avoidance and fitness during competition. Proc Natl Acad Sci USA 107: 22740–22744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Vehn J, Huang F, Naramoto S, Zhang J, Michniewicz M, Offringa R, Friml J (2009) PIN auxin efflux carrier polarity is regulated by PINOID kinase-mediated recruitment into GNOM-independent trafficking in Arabidopsis. Plant Cell 21: 3839–3849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koini MA, Alvey L, Allen T, Tilley CA, Harberd NP, Whitelam GC, Franklin KA (2009) High temperature-mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Curr Biol 19: 408–413 [DOI] [PubMed] [Google Scholar]
- Lee HJ, Jung JH, Cortés Llorca L, Kim SG, Lee S, Baldwin IT, Park CM (2014) FCA mediates thermal adaptation of stem growth by attenuating auxin action in Arabidopsis. Nat Commun 5: 5473. [DOI] [PubMed] [Google Scholar]
- Lippincott BB, Lippincott JA (1971) Auxin-induced hyponasty of the leaf blade of Phaseolus vulgaris. Am J Bot 58: 817–826 [Google Scholar]
- Lorrain S, Allen T, Duek PD, Whitelam GC, Fankhauser C (2008) Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J 53: 312–323 [DOI] [PubMed] [Google Scholar]
- Merelo P, Ram H, Pia Caggiano M, Ohno C, Ott F, Straub D, Graeff M, Cho SK, Yang SW, Wenkel S, et al. (2016) Regulation of MIR165/166 by class II and class III homeodomain leucine zipper proteins establishes leaf polarity. Proc Natl Acad Sci USA 113: 11973–11978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaud O, Fiorucci AS, Xenarios I, Fankhauser C (2017) Local auxin production underlies a spatially restricted neighbor-detection response in Arabidopsis. Proc Natl Acad Sci USA 114: 7444–7449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick P, Schafer E (1989) Nastic response of maize (Zea mays L.) coleoptiles during clinostat rotation. Planta 179: 123–131 [DOI] [PubMed] [Google Scholar]
- Nishimura T, Hayashi K, Suzuki H, Gyohda A, Takaoka C, Sakaguchi Y, Matsumoto S, Kasahara H, Sakai T, Kato J, et al. (2014) Yucasin is a potent inhibitor of YUCCA, a key enzyme in auxin biosynthesis. Plant J 77: 352–366 [DOI] [PubMed] [Google Scholar]
- Oh E, Zhu JY, Wang ZY (2012) Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat Cell Biol 14: 802–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantazopoulou CK, Bongers FJ, Küpers JJ, Reinen E, Das D, Evers JB, Anten NPR, Pierik R (2017) Neighbor detection at the leaf tip adaptively regulates upward leaf movement through spatial auxin dynamics. Proc Natl Acad Sci USA 114: 7450–7455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YJ, Lee HJ, Ha JH, Kim JY, Park CM (2017) COP1 conveys warm temperature information to hypocotyl thermomorphogenesis. New Phytol 215: 269–280 [DOI] [PubMed] [Google Scholar]
- Pérez-Pérez JM, Candela H, Robles P, López-Torrejón G, del Pozo JC, Micol JL (2010) A role for AUXIN RESISTANT3 in the coordination of leaf growth. Plant Cell Physiol 51: 1661–1673 [DOI] [PubMed] [Google Scholar]
- Petrásek J, Friml J (2009) Auxin transport routes in plant development. Development 136: 2675–2688 [DOI] [PubMed] [Google Scholar]
- Polko JK, Voesenek LA, Peeters AJ, Pierik R (2011) Petiole hyponasty: An ethylene-driven, adaptive response to changes in the environment. AoB Plants 2011: plr031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polko JK, Pierik R, van Zanten M, Tarkowská D, Strnad M, Voesenek LA, Peeters AJ (2013) Ethylene promotes hyponastic growth through interaction with ROTUNDIFOLIA3/CYP90C1 in Arabidopsis. J Exp Bot 64: 613–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman A, Takahashi M, Shibasaki K, Wu S, Inaba T, Tsurumi S, Baskin TI (2010) Gravitropism of Arabidopsis thaliana roots requires the polarization of PIN2 toward the root tip in meristematic cortical cells. Plant Cell 22: 1762–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakusová H, Gallego-Bartolomé J, Vanstraelen M, Robert HS, Alabadí D, Blázquez MA, Benková E, Friml J (2011) Polarization of PIN3-dependent auxin transport for hypocotyl gravitropic response in Arabidopsis thaliana. Plant J 67: 817–826 [DOI] [PubMed] [Google Scholar]
- Ryu H, Kim K, Cho H, Park J, Choe S, Hwang I (2007) Nucleocytoplasmic shuttling of BZR1 mediated by phosphorylation is essential in Arabidopsis brassinosteroid signaling. Plant Cell 19: 2749–2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasidharan R, Voesenek LA (2015) Ethylene-mediated acclimations to flooding stress. Plant Physiol 169: 3–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Qi L, Li Y, Chu J, Li C (2012) PIF4-mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating Arabidopsis hypocotyl growth. PLoS Genet 8: e1002594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udvardi MK, Czechowski T, Scheible WR (2008) Eleven golden rules of quantitative RT-PCR. Plant Cell 20: 1736–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno Y, Ishikawa T, Watanabe K, Terakura S, Iwakawa H, Okada K, Machida C, Machida Y (2007) Histone deacetylases and ASYMMETRIC LEAVES2 are involved in the establishment of polarity in leaves of Arabidopsis. Plant Cell 19: 445–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zanten M, Voesenek LA, Peeters AJ, Millenaar FF (2009) Hormone- and light-mediated regulation of heat-induced differential petiole growth in Arabidopsis. Plant Physiol 151: 1446–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HZ, Yang KZ, Zou JJ, Zhu LL, Xie ZD, Morita MT, Tasaka M, Friml J, Grotewold E, Beeckman T, et al. (2015) Transcriptional regulation of PIN genes by FOUR LIPS and MYB88 during Arabidopsis root gravitropism. Nat Commun 6: 8822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller B, Zourelidou M, Frank L, Barbosa IC, Fastner A, Richter S, Jürgens G, Hammes UZ, Schwechheimer C (2017) Dynamic PIN-FORMED auxin efflux carrier phosphorylation at the plasma membrane controls auxin efflux-dependent growth. Proc Natl Acad Sci USA 114: E887–E896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willige BC, Isono E, Richter R, Zourelidou M, Schwechheimer C (2011) Gibberellin regulates PIN-FORMED abundance and is required for auxin transport-dependent growth and development in Arabidopsis thaliana. Plant Cell 23: 2184–2195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zádníková P, Petrásek J, Marhavy P, Raz V, Vandenbussche F, Ding Z, Schwarzerová K, Morita MT, Tasaka M, Hejátko J, et al. (2010) Role of PIN-mediated auxin efflux in apical hook development of Arabidopsis thaliana. Development 137: 607–617 [DOI] [PubMed] [Google Scholar]
- Zgurski JM, Sharma R, Bolokoski DA, Schultz EA (2005) Asymmetric auxin response precedes asymmetric growth and differentiation of asymmetric leaf1 and asymmetric leaf2 Arabidopsis leaves. Plant Cell 17: 77–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. (2010) Auxin biosynthesis and its role in plant development. Annu Rev Plant Biol 61: 49–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zourelidou M, Absmanner B, Weller B, Barbosa IC, Willige BC, Fastner A, Streit V, Port SA, Colcombet J, de la Fuente van Bentem S, et al. (2014) Auxin efflux by PIN-FORMED proteins is activated by two different protein kinases, D6 PROTEIN KINASE and PINOID. eLife 3: e02860. [DOI] [PMC free article] [PubMed] [Google Scholar]