Biochemical analysis of the aromatic amino acid decarboxylase family in poplar revealed two enzymes involved in the herbivore induced formation of 2-phenylethanol and 2-phenylethylamine.
Abstract
Upon herbivory, the tree species western balsam poplar (Populus trichocarpa) produces a variety of Phe-derived metabolites, including 2-phenylethylamine, 2-phenylethanol, and 2-phenylethyl-β-d-glucopyranoside. To investigate the formation of these potential defense compounds, we functionally characterized aromatic l-amino acid decarboxylases (AADCs) and aromatic aldehyde synthases (AASs), which play important roles in the biosynthesis of specialized aromatic metabolites in other plants. Heterologous expression in Escherichia coli and Nicotiana benthamiana showed that all five AADC/AAS genes identified in the P. trichocarpa genome encode active enzymes. However, only two genes, PtAADC1 and PtAAS1, were significantly upregulated after leaf herbivory. Despite a sequence similarity of ∼96%, PtAADC1 and PtAAS1 showed different enzymatic functions and converted Phe into 2-phenylethylamine and 2-phenylacetaldehyde, respectively. The activities of both enzymes were interconvertible by switching a single amino acid residue in their active sites. A survey of putative AADC/AAS gene pairs in the genomes of other plants suggests an independent evolution of this function-determining residue in different plant families. RNA interference -mediated–downregulation of AADC1 in gray poplar (Populus × canescens) resulted in decreased accumulation of 2-phenylethylamine and 2-phenylethyl-β-d-glucopyranoside, whereas the emission of 2-phenylethanol was not influenced. To investigate the last step of 2-phenylethanol formation, we identified and characterized two P. trichocarpa short-chain dehydrogenases, PtPAR1 and PtPAR2, which were able to reduce 2-phenylacetaldehyde to 2-phenylethanol in vitro. In summary, 2-phenylethanol and its glucoside may be formed in multiple ways in poplar. Our data indicate that PtAADC1 controls the herbivore-induced formation of 2-phenylethylamine and 2-phenylethyl-β-d-glucopyranoside in planta, whereas PtAAS1 likely contributes to the herbivore-induced emission of 2-phenylethanol.
Many plant secondary or specialized metabolites play major roles in plant defense against herbivores. Some accumulate as deterrents or toxins as part of direct plant defense, whereas others are released as volatiles and can attract parasitoids or predators of the herbivores, a phenomenon that has been termed “indirect defense” (Unsicker et al., 2009). Still others may serve as internal signals to activate the formation of both direct and indirect defenses (Maag et al., 2015). Plant-specialized metabolites are highly diverse and belong to different compound classes, including terpenoids, phenylpropanoids, benzenoids, amino acid derivatives, and fatty acid derivatives. Although some of these metabolites, as for example certain terpenes and phenylpropanoids, are ubiquitous among plants, others are exclusively produced in specific families or even a single genus (Fahey et al., 2001; Bieri et al., 2006).
The alcohol 2-phenylethanol and its glucoside 2-phenylethyl-β-d-glucopyranoside are amino acid-derived specialized compounds that are widespread among plants. The free alcohol is well known for its pleasant “rose-like” aroma and has been reported to be a flower, fruit, and vegetative volatile released from a multitude of plant species of more than 50 families (Knudsen et al., 2006). As a bioactive compound, 2-phenylethanol plays diverse roles in plant-insect interactions. It acts, for example, as an attractant for pollinators such as butterflies, bees, and beetles (Roy and Raguso, 1997; Honda et al., 1998; Imai et al., 1998) and mediates direct and indirect plant defenses (Zhu et al., 2005; Galen et al., 2011).
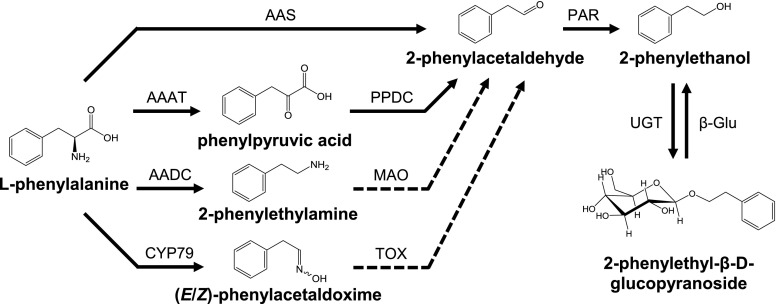
The biosynthesis of 2-phenylethanol from Phe was first elucidated in the yeast Saccharomyces cerevisiae, where it proceeds via the Ehrlich pathway (for review, see Hazelwood et al., 2008). This pathway starts with the transamination of an aromatic or branched-chain amino acid followed by a decarboxylation of the resulting α-keto acid. The aldehyde formed is finally reduced to yield 2-phenylethanol. In plants, the biosynthesis of 2-phenylethanol has been extensively investigated in rose (Rosa spp.; Sakai et al., 2007), petunia (Petunia hybrida; Kaminaga et al., 2006), and tomato (Solanum lycopersicum; Tieman et al., 2006, 2007). The pathways described in these species also start from Phe and involve the formation and subsequent reduction of 2-phenylacetaldehyde; however, the initial reactions are different from those of the Ehrlich pathway (Fig. 1). Tomato, for example, contains several aromatic amino acid decarboxylases (AADCs) that catalyze the formation of 2-phenylethylamine, which could be further converted to 2-phenylacetaldehyde by a monoamine oxidase (Tieman et al., 2006). In contrast, rose and petunia possess a bifunctional aromatic aldehyde synthase (AAS) able to catalyze both Phe decarboxylation and the subsequent oxidative transamination (Kaminaga et al., 2006; Sakai et al., 2007; Farhi et al., 2010). The recent identification of an aromatic amino acid transaminase (AAAT) involved in aroma compound formation in melon (Cucumis melo) as well as the characterization of an AAAT and a phenylpyruvate decarboxylase in rose indicate that the Ehrlich pathway might also contribute to the formation of 2-phenylethanol and other related alcohols in plants (Gonda et al., 2010; Hirata et al., 2016; Sheng et al., 2018). Moreover, recent research on poplar suggests another potential route to 2-phenylethanol that involves the P450-catalyzed production of (E/Z)-phenylacetaldoxime from Phe (Fig. 1; Irmisch et al., 2013, 2014a).
Figure 1.
Proposed pathways for the biosynthesis of 2-phenylethanol in poplar. CYP79, cytochrome P450 family 79 enzyme; PPDC, phenylpyruvic acid decarboxylase; MAO, monoamine oxidase; TOX, transoximase; UGT, UDP-glycosyltransferase; β-Glu, β-glucosidase. Solid lines indicate well-established reactions/enzymes; dashed lines indicate reactions hypothesized in the literature.
AADC and AAS both belong to group II pyridoxal-5′-phosphate (PLP)-dependent enzymes that encompass a large family referred to as the plant aromatic amino acid decarboxylase family (AAAD; Facchini et al., 2000; Torrens-Spence et al., 2018a). AAADs can be assigned to three major groups depending on their substrate specificity for Trp, Tyr, or Phe, respectively. They form mid-sized gene families with an average number of ∼10 members per plant genome, and have been identified in all plant lineages including algae, nonvascular land plants, gymnosperms, and angiosperms (Kumar, 2016). Due to their relatively high sequence identities, a reliable functional prediction of single AAAD members has been difficult in the past. However, two recent studies identified amino acid residues that determine the substrate selectivity and catalytic activity of AAAD enzymes. The different substrate preferences of Trp and Tyr decarboxylases, for example, are indicated by a conserved Gly-Ser polymorphism in the active site of these enzymes (Torrens-Spence et al., 2014a). The difference between AADC and AAS activity is determined by the presence of either a Tyr or Phe (or other apolar residue) on a catalytic loop proximal to the active site (Bertoldi et al., 2002; Torrens-Spence et al., 2013, 2018b).
The last step in the formation of 2-phenylethanol common to all of these pathways is the reduction of 2-phenylacetaldehyde. This reaction is catalyzed by 2-phenylacetaldehyde reductase (PAR), which belongs to the large family of NADPH-dependent short-chain dehydrogenase/reductases (Tieman et al., 2007; Chen et al., 2011). Tomato, for example, possesses two PARs (LePAR1 and LePAR2) that accept 2-phenylacetaldehyde as substrate. Although LePAR1 is highly substrate-specific, LePAR2 can convert a variety of aromatic and aliphatic aldehydes into the respective alcohols (Tieman et al., 2007). Such substrate promiscuity has also been reported for another PAR that is involved in 2-phenylethanol formation in roses (Chen et al., 2011).
In recent years, poplar has been established as a model system for studying plant defenses against herbivores in long-living, woody plant species (Peters and Constabel, 2002; Arimura et al., 2004; Ralph et al., 2006; Frost et al., 2007; Philippe and Bohlmann, 2007; Major and Constabel, 2008; Danner et al., 2011; Irmisch et al., 2014b). After leaf herbivory, the western balsam poplar (Populus trichocarpa) accumulates various defense compounds and releases a complex volatile blend that mediates direct and indirect defense reactions (Irmisch et al., 2013; McCormick et al., 2014). The volatile blend is dominated by terpenes, green leaf volatiles, and nitrogen-containing compounds and contains 2-phenylethanol as one of its main components (Irmisch et al., 2013). RNA interference (RNAi) and labeling experiments indicated that a significant proportion of herbivore-induced 2-phenylethanol seems to be produced via an oxime-dependent pathway (Irmisch et al., 2013, 2014a); however, whether poplar AAAD enzymes may also contribute to the formation of 2-phenylethanol and related compounds was unclear.
To elucidate the possible contribution of AAAD-dependent pathways to the formation of 2-phenylethanol in poplar, we measured the accumulation of pathway intermediates in herbivore-damaged P. trichocarpa leaves using liquid chromatography-tandem mass spectrometry (LC-MS/MS). RNA sequencing (RNA-Seq) experiments and reverse transcription quantitative PCR (RT-qPCR) were used to identify herbivore-induced AAAD candidate genes. Characterization of heterologously produced and purified AAAD proteins, overexpression in Nicotiana benthamiana, and RNAi-mediated downregulation revealed that two candidate enzymes, PtAADC1 and PtAAS1, are involved in herbivore-induced Phe metabolism and 2-phenylethanol formation in poplar.
RESULTS
Potential Precursors and Metabolites of 2-Phenylethanol Accumulate in Herbivore-Damaged P. trichocarpa Leaves
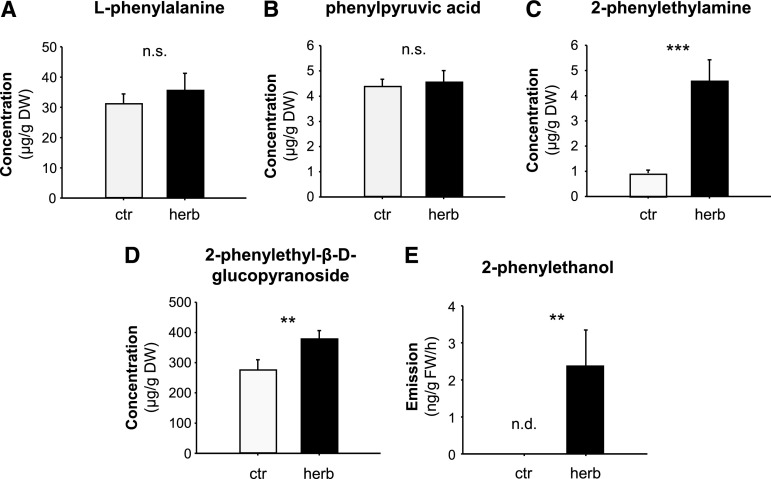
To elucidate the metabolic processes leading to 2-phenylethanol formation in poplar, we measured potential precursors and pathway intermediates in undamaged and gypsy moth (Lymantria dispar) caterpillar-damaged P. trichocarpa leaves (Fig. 2). Phe and phenylpyruvic acid were present in both undamaged and herbivore-damaged leaves and showed no changes in response to herbivory (Fig. 2, A and B). The accumulation of 2-phenylethylamine, however, was significantly increased upon herbivory and mirrored the herbivore-induced emission of 2-phenylethanol (Fig. 2, C and E). 2-Phenylacetaldehyde could not be detected, suggesting a rapid reduction to 2-phenylethanol in poplar.
Figure 2.
Herbivore-damaged leaves of P. trichocarpa accumulate and release Phe-derived metabolites. A–E, The accumulation of l-Phe (A), phenylpyruvic acid (B), 2-phenylethylamine (C), and 2-phenylethyl-β-d-glucopyranoside (D), and the emission of 2-phenylethanol (E) were analyzed in L. dispar damaged (herb) and undamaged control (ctr) leaves. Plant material was extracted with methanol and analyzed via LC-MS/MS (A–D). Volatiles were collected for 8 h and analyzed via GC-FID (E). Means ± se are shown (A–D, n = 10; E, n = 9). Asterisks indicate statistical significance as assessed by Student’s t test or Mann-Whitney Rank Sum tests. l-Phe (P = 0.530, t = −0.64); phenylpyruvic acid (P = 0.772, t = −0.294); 2-phenylethylamine (P < 0.001, T = 55); 2-phenylethyl-β-d-glucopyranoside (P = 0.011, T = 71); 2-phenylethanol (P = 0.005, T = 58.5). DW, dry weight; FW, fresh weight; n.s., not significant; n.d., not detected.
It has been shown that 2-phenylethanol can be converted to 2-phenylethyl-β-d-glucopyranoside in roses (Hayashi et al., 2004). Moreover, transgenic poplars overexpressing rose RhPAAS and PAR accumulated large amounts of this glucoside (Costa et al., 2013). However, whether 2-phenylethyl-β-d-glucopyranoside also occurs in wild type poplars was unclear. We could detect 2-phenylethyl-β-d-glucopyranoside both in undamaged and herbivore-damaged leaves and the accumulation of this compound was slightly but significantly induced upon herbivory (Fig. 2D).
Identification of Putative AAAD Family Genes in P. trichocarpa
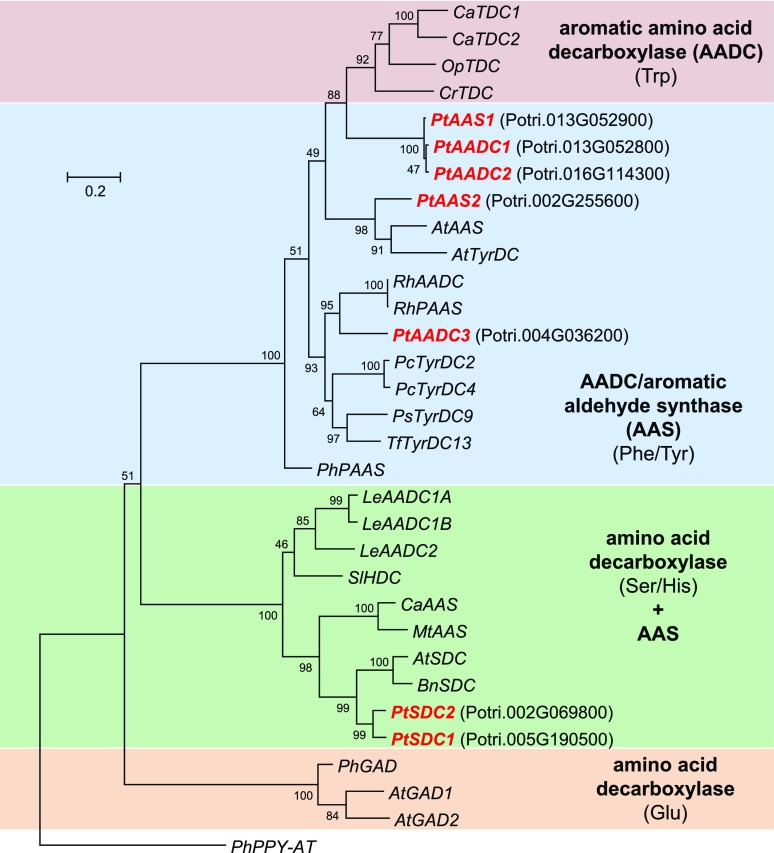
A Basic Local Alignment Search Tool (BLAST; National Center for Biotechnology Information [NCBI]) analysis with RhPAAS (Farhi et al., 2010) as query revealed five putative AAAD genes in the genome of P. trichocarpa (Tuskan et al., 2006; Fig. 3). Considering the function-determining Tyr-Phe polymorphism in the active sites of their encoded proteins, the genes were named PtAADC1 (Potri.013G052800), PtAADC2 (Potri.016G114300), PtAADC3 (Potri.004G036200), PtAAS1 (Potri.013G052900), and PtAAS2 (Potri.002G255600; Supplemental Fig. S1). Two genes, PtSDC1 (Potri.005G190500) and PtSDC2 (Potri.002G069800), which also appeared in the BLAST search, were similar to histidine decarboxylase and serine decarboxylase (SDC) genes (Fig. 3). However, because SDC-like proteins from barrelclover (Medicago truncatula) and chickpea (Cicer arietinum) have recently been characterized as AAS enzymes (Torrens-Spence et al., 2014a), poplar PtSDC1 and PtSDC2 were also chosen as potential AAS gene candidates for further characterization.
Figure 3.
Dendrogram analysis of AAAD family genes from P. trichocarpa and characterized AADC, AAS, and Glu/Ser/His-decarboxylase genes from other plants. Substrates of each clade are indicated in parentheses. The tree was inferred by using the maximum likelihood method and n = 1,000 replicates for bootstrapping. Bootstrap values are shown next to each node. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. Genes described in this study are shown in red and bold with corresponding gene identifiers (italics). Accession numbers are given in Supplemental Table S1.
Biochemical Characterization of Poplar AAAD and SDC Enzymes
The complete open reading frames (ORFs) of the candidate genes were amplified from complementary DNA made from herbivore-damaged leaves of P. trichocarpa and cloned into the vector pET100/D-TOPO. After heterologous expression in Escherichia coli, recombinant proteins carrying an N-terminal His6-Tag were affinity-purified and incubated with Phe, Tyr, or Trp as potential substrates and PLP as cofactor. Both PtAAS1 and PtAAS2 had aldehyde synthase activity and catalyzed the conversion of Phe to 2-phenylacetaldehyde (Table 1). Formation of 2-phenylethylamine, however, could not be detected (Supplemental Fig. S2). Whereas PtAAS1 was highly substrate-specific and exclusively accepted Phe, PtAAS2 showed activity with all tested aromatic amino acids producing the respective aldehydes (Supplemental Fig. S3). Notably, the amino acid residue that has been reported to determine the substrate specificity of Tyr and Trp decarboxylases (Torrens-Spence et al., 2014b) differed between PtAAS1 (Ser-360) and PtAAS2 (Gly-358; Supplemental Fig. S1). This amino acid change might explain the observed differences in substrate promiscuity. In contrast to PtAAS1 and PtAAS2, the enzymes PtAADC1, PtAADC2, and PtAADC3 showed no aldehyde synthase activity but were able to catalyze the decarboxylation of different aromatic amino acids. The Km and kcat values revealed that PtAADC1 and PtAADC2 most likely act as Phe decarboxylases in planta, whereas PtAADC3 is likely a Tyr decarboxylase (Table 1). To test for potential AAAT activity of poplar AAAD candidates, we incubated the enzymes with Phe in the presence of the ammonia acceptor α-ketoglutarate. However, no formation of phenylpyruvic acid could be observed.
Table 1. Kinetic parameters of P. trichocarpa PtAAS1, PtAAS2, PtAADC1, PtAADC2, and PtAADC3.
Enzymes were heterologously expressed in E. coli, purified, and incubated with Phe, Tyr, or Trp. Kinetic parameters are shown as means ± se (n = 3). Volatile phenylacetaldehyde was analyzed via TDU-GC-MS; all other compounds were measured using LC-MS/MS. PHA, phenylacetaldehyde; PEA, 2-phenylethylamine; 4-OH-PHA, 4-hydroxy phenylacetaldehyde; IAAld, indole-3-acetaldehyde; TyrA, tyramine; TrpA, tryptamine.
| Enzyme | Substrate | Product | Km (mm) | Vmax (nmola µg−1 a min−1) | kcat (s−1) | kcat /Km (s−1 mm−1) |
|---|---|---|---|---|---|---|
| PtAAS1 | Phe | PHA | 0.46 ± 0.02 | — | — | — |
| Phe | PEA | — | — | — | — | |
| Tyr | 4-OH-PHA | — | — | — | — | |
| Trp | IAAld | — | — | — | — | |
| PtAAS2 | Phe | PHA | 0.69 ± 0.39 | — | — | — |
| Phe | PEA | — | — | — | — | |
| Tyr | 4-OH-PHAa | detected | — | — | — | |
| Trp | IAAlda | detected | — | — | — | |
| PtAADC1 | Phe | PHA | — | — | — | — |
| Phe | PEA | 0.17 ± 0.03 | 430 ± 12 | 0.39 | 2.3 | |
| Tyr | TyrA | 2.0 ± 0.2 | 85.4 ± 3.9 | 0.08 | 0.04 | |
| Trp | TrpA | 3.0 ± 0.4 | 7.7 ± 0.5 | 0.007 | 0.002 | |
| PtAADC2 | Phe | PHA | — | — | — | — |
| Phe | PEA | 0.19 ± 0.01 | 0.89 ± 0.01 | 0.81 | 4.2 | |
| Tyr | TyrA | 1.2 ± 0.2 | 148 ± 7 | 0.14 | 0.11 | |
| Trp | TrpA | — | — | — | — | |
| PtAADC3 | Phe | PHA | — | — | — | — |
| Phe | PEA | 2.3 ± 0.2 | 6.7 ± 0.2 | 0.14 | 0.003 | |
| Tyr | TyrA | 0.043 ± 0.004 | 3.61 ± 0.06 | 0.003 | 0.08 | |
| Trp | TrpA | — | — | — | — |
Compounds were chemically converted into the corresponding aldoximes before LC-MS/MS analysis (see “Materials and Methods”).
To determine if the putative SDCs PtSDC1 and PtSDC2 exhibit decarboxylase activity toward aromatic amino acids as previously described for SDC-like enzymes in M. truncatula and C. arietinum (Torrens-Spence et al., 2014a), the enzymes were heterologously expressed and tested with Phe, Tyr, Trp, His, and Ser as potential substrates. Both enzymes converted Ser into ethanolamine, but showed no activity with aromatic amino acids and His (Supplemental Fig. S4), indicating that PtSDC1 and PtSDC2 likely function as SDCs in planta. Thus, they were not further considered in this study.
PtAAS1 and PtAADC1 Are Interconvertible by the Change of One Amino Acid
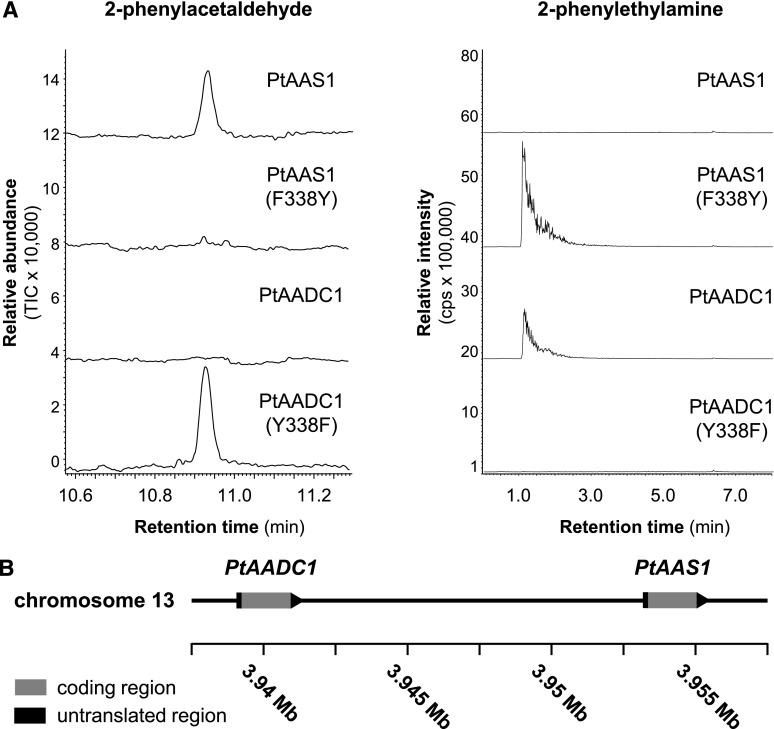
PtAAS1 and PtAADC1 differ in a characteristic amino acid residue that has been described as a function-dictating residue in AADC and AAS enzymes from other plants (Bertoldi et al., 2002; Torrens-Spence et al., 2013, 2018b). To demonstrate that this Tyr–Phe polymorphism also determines the decarboxylase and aldehyde synthase activity of PtAADC1 and PtAAS1, respectively, we constructed the mutant enzymes PtAADC1 (Y338F) and PtAAS1 (F338Y) by in vitro mutagenesis and incubated them with Phe as substrate. As expected, both mutations led to a complete interconversion of the wild-type activities. Whereas PtAADC1 (Y338F) produced exclusively 2-phenylacetaldehyde, PtAAS1 (F338Y) had only decarboxylase activity and produced 2-phenylethylamine (Fig. 4A).
Figure 4.
PtAAS1 and PtAADC1 are interconvertible by the mutation of a single amino acid and are likely derived by tandem gene duplication. A, The recombinant proteins PtAAS1, PtAADC1, and the corresponding mutant enzymes PtAAS1 (F338Y) and PtAADC1 (Y338F) were incubated with l-Phe in the presence of PLP. Enzyme products were analyzed using GC-MS (2-phenylacetaldehyde) and LC-MS/MS (2-phenylethylamine). B, PtAADC1 and PtAAS1 form a gene cluster on chromosome 13. Both genes are oriented in the same direction and there are no coding regions between them. Sequence data were retrieved from the P. trichocarpa Genome Assembly (version 3.0; www.phytozome.net; Tuskan et al., 2006).
The Tyr-Phe–determined Shift in AADC/AAS Enzyme Activity Evolved Independently in Different Plant Families
Within the poplar AADC/AAS clade, PtAAS1, PtAADC1, and PtAADC2 form a small subfamily with high nucleotide sequence identity of ∼96% to 98% (Fig. 3). Whereas PtAADC2 was found to be located on chromosome 16, PtAADC1 and PtAAS1 were located close to each other on chromosome 13 (Fig. 4B). This and the fact that PtAADC1 and PtAAS1 have no other ORFs between them in the genomic sequence suggest a recent gene duplication that was subsequently followed by neofunctionalization through the mutation of the function-determining amino acid. A survey of putative AADC/AAS sequences in other available plant genomes revealed the presence of comparable AADC/AAS pairs in at least 10 species from diverse plant families, including monocotyledonous and dicotyledonous plants (Supplemental Figs. S5 and S6). The putative AADC and AAS genes in each of these pairs were highly similar to each other and grouped in a species-specific manner, indicating an independent evolution of the Tyr–Phe shift in different plant families or even genera.
Functional Analysis of PtAADC1 and PtAAS1 In Planta
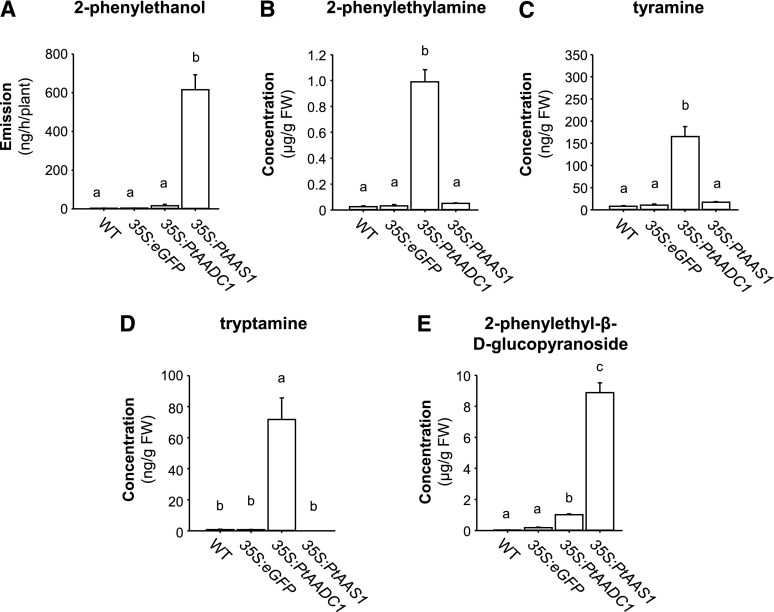
To verify the different enzymatic functions of the characterized poplar Phe-using AAAD enzymes in a heterologous plant system, PtAADC1 and PtAAS1 were chosen for transient expression in N. benthamiana. Plants overexpressing PtAAS1 showed a significant emission of 2-phenylethanol, whereas wild-type plants and plants expressing either eGFP or PtAADC1 produced only trace amounts of this compound, if any (Fig. 5A). In addition, PtAAS1 plants accumulated 2-phenylethyl-β-d-glucopyranoside (Fig. 5E). The overexpression of PtAADC1, however, resulted in the accumulation of 2-phenylethylamine, tyramine, tryptamine, and 2-phenylethyl-β-d-glucopyranoside (Fig. 5, B–E). The aromatic aldehydes 2-phenylacetaldehyde, 4-hydroxy-phenylacetaldehyde, and indole-3-acetaldehyde could not be detected in the tested lines. Altogether, the exclusive emission of 2-phenylethanol from PtAAS1 plants and the different accumulation levels of the aromatic amines in PtAADC1-overexpressing plants corresponded well to the enzymatic activities and kinetic parameters that were determined in vitro (Table 1; Fig. 5). Moreover, the results suggest that the reactive aldehydes can be rapidly converted to less reactive alcohols or other compounds in vivo, even in a heterologous plant system.
Figure 5.
A–E, Overexpression of PtAAS1 and PtAADC1 in N. benthamiana alters the emission of 2-phenylethanol (A) and the accumulation of 2-phenylethylamine (B), tyramine (C), tryptamine (D), and 2-phenylethanol-β-d-glucopyranoside (E). N. benthamiana plants were infiltrated with A. tumefaciens carrying either PtAAS1 or PtAADC1. At 5 d after infiltration, the headspaces of plants were collected and analyzed via GC-MS and GC-FID. Additionally, plants were harvested, extracted with methanol, and metabolite accumulation was analyzed via LC-MS/MS. Different letters above each bar indicate statistically significant differences as assessed by Kruskal-Wallis one-way analysis of variance and Tukey tests. 2-phenylethanol (H = 19.867, P ≤ 0.001); 2-phenylethylamine (H = 16.94, P ≤ 0.001); tyramine (H = 18.267, P ≤ 0.001); tryptamine (H = 18.645, P ≤ 0.001); 2-phenylethyl-β-d-glucopyranoside (H = 21.6, P ≤ 0.001). Means ± se are shown (n = 6). WT, wild type; FW, fresh weight.
Expression of PtAADC1 and PtAAS1 Is Induced in Response to Herbivory in P. trichocarpa Leaves
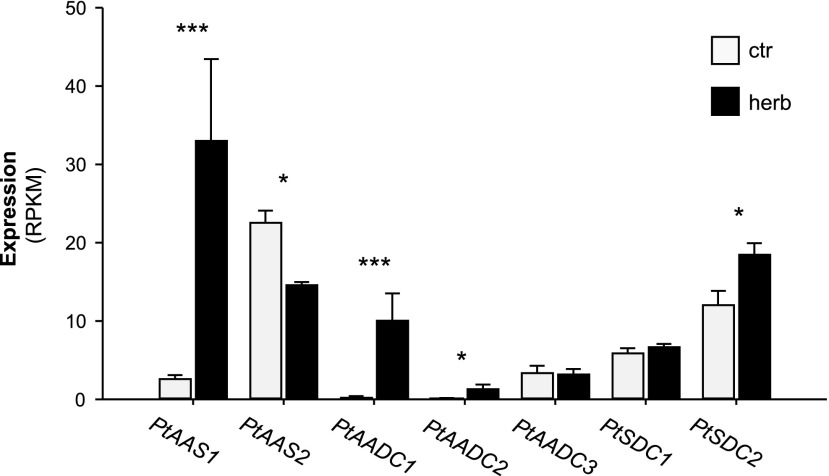
To study the expression of poplar AAAD genes, we sequenced the transcriptomes of herbivore-damaged and undamaged P. trichocarpa leaves and mapped the quality-trimmed sequence reads to the P. trichocarpa Genome (version v3.0; https://phytozome.jgi.doe.gov; Tuskan et al., 2006). All AAAD genes were found to be expressed in poplar leaves (Fig. 6). However, the transcript accumulation of PtAADC1 and PtAAS1 was highly upregulated in herbivore-damaged leaves in comparison to undamaged control leaves with fold changes of 29.4 and 12.4, respectively (Fig. 6). Expression of PtAADC2 and PtSDC2 was also increased in response to the herbivore treatment but showed lower fold increases (6.4 and 1.6, respectively). PtAAS2 showed a significant reduction in gene expression upon herbivory (fold change, −1.5), whereas expression of PtAADC3 and PtSDC1 were not influenced by the treatment (Fig. 6). The RNA-Seq results for poplar AAAD genes were confirmed by RT-qPCR (Supplemental Fig. S7). The highly similar genes PtAAS1, PtAADC1, and PtAADC2 could not be targeted by specific primers and were thus amplified using a universal primer pair. Extensive sequencing of cloned amplicons that encompassed several polymorphisms including the function-determining codon allowed a reliable assignment of the amplicons to specific genes. The RT-qPCR results confirmed the upregulation of PtAAS1 and PtAADC1 gene expression in response to herbivory (Supplemental Table S2). Transcripts of PtAADC2, however, could not be detected in control and herbivore-damaged leaves. Notably, total expression of PtAAS1 in herbivore-damaged leaves (92.5% of sequenced amplicons) was >10 times higher than PtAADC1 (7.5% of sequenced amplicons).
Figure 6.
Transcript accumulation of AAAD and SDC genes in L. dispar-damaged and undamaged P. trichocarpa leaves. Gene expression in herbivore-damaged (herb) and undamaged (ctr) leaves was analyzed by Illumina sequencing and mapping the reads to the transcripts of the P. trichocarpa Genome Assembly (version 3.0; www.phytozome.net; Tuskan et al., 2006). Expression was normalized to reads per kilo base per million mapped reads (RPKM). Significant differences in EDGE tests are visualized by asterisks. Means ± se are shown (n = 4). PtAAS1 (P = 8.23213E-12, weighted difference [WD] = 4.86618E-05); PtAAS2 (P = 0.039452261, WD = −1.11092E-05); PtAADC1 (P = 8.99025E-09, WD = 1.56932E-05); PtAADC2 (P = 0.031269756, WD = 2.02404E-06); PtAADC3 (P = 1, WD = −4.35895E-08); PtSDC1 (P = 0.576261262, WD = 1.61294E-06); PtSDC2 (P = 0.015936012, WD = 1.08813E-05).
RNAi-mediated Downregulation of PtAADC1 in Populus × canescens Affects the Formation of 2-Phenylethylamine and 2-Phenylethyl-β-d-Glucopyranoside
To investigate the function of herbivore-induced AAAD genes in poplar, we aimed to generate RNAi lines with reduced AADC1/AAS1 gene expression by transformation of a DNA fragment complementary to AADC1, AADC2, and AAS1. Because there is no established transformation method for P. trichocarpa, we chose gray poplar (P. × canescens) for transformation, a species commonly used for the generation of transgenic poplar trees (Leple et al., 1992). Gray poplar is a hybrid species derived from a cross between silver poplar (Populus alba) and quaking aspen (Populus tremula) and BLAST searches with the different P. trichocarpa AAAD genes as query and the genomes of P. × canescens, P. alba, and P. tremula (http://aspendb.uga.edu/index.php/databases/spta-717-genome; Xue et al., 2015; Zhou et al., 2015) as templates revealed that all three species possess putative AADC1, AADC2, and AAS1 orthologs (Supplemental Fig. S8). Interestingly, the function-dictating Tyr of P. trichocarpa PtAADC2 was changed into Phe in P. × canescens PcanAADC2 and in P. tremula PtremAADC2, suggesting that both orthologs likely function as aldehyde synthases. However, sequencing of cloned qPCR fragments that were amplified with a primer pair specific for AADC1, AADC2, and AAS1 from cDNA made from herbivore-damaged P. × canescens leaves revealed exclusively PcanAADC1 amplicons (Supplemental Fig. S9). This indicates that the two aldehyde synthase genes PcanAAS1 and PcanAADC2 are likely not expressed in herbivore-damaged leaves of P. × canescens.
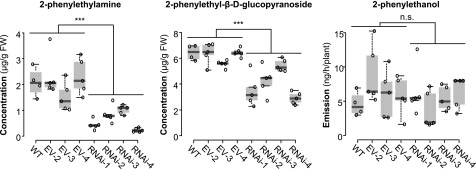
P. × canescens PcanAADC1 RNAi lines as well as wild-type trees and trees carrying an empty vector were subjected to L. dispar herbivory and the emission of 2-phenylethanol and the accumulation of 2-phenylethylamine and 2-phenylethyl-β-d-glucopyranoside in herbivory-induced leaves was measured. Downregulation of PcanAADC1 in the transgenic lines led to a significantly reduced accumulation of herbivore-induced 2-phenylethylamine and 2-phenylethyl-β-d-glucopyranoside (Fig. 7; Supplemental Fig. S10). The emission of 2-phenylethanol, however, remained constant in comparison to wild type and empty vector trees. Expression levels of PcanAAS2 were not affected by the RNAi approach (Supplemental Fig. S10). Thus, the observed metabolic changes in the transgenic lines are most likely caused by the downregulation of PcanAADC1 gene expression.
Figure 7.
RNAi-mediated knockdown of AADC1 in P. × canescens. Sterile P. × canescens calli were transformed via A. tumefaciens. Nontransgenic trees (WT, wild type), empty vector lines (EV-2-4), and RNAi lines (RNAi-1-4) were subjected to L. dispar feeding. Volatile 2-phenylethanol was collected from the headspace and analyzed using GC-MS/FID. Phenylethylamine and 2-phenylethyl-β-d-glucopyranoside were extracted with methanol from ground plant material and analyzed via LC-MS/MS. Biological replicates (nb) and technical replicates (nt) of EV lines and RNAi lines were used to test for statistical differences. Wild type, nb = 4; EV, nb = 3, nt = 5; RNAi, nb = 4, nt = 5. Asterisks indicate statistical significance as assessed by Student’s t test or Mann-Whitney Rank Sum tests. Phenylethylamine (P < 0.001, t = 7.940); 2-phenylethyl-β-d-glucopyranoside (P = 0.011, T = 547); 2-phenylethanol (P = 0.509, T = 404). Medians ± quartiles and outliers are shown. Each data point is represented by a circle. FW, fresh weight; n.s., not significant.
One of the most characteristic volatiles released from herbivore-damaged poplars is the nitrile benzyl cyanide (McCormick et al., 2014). Previous studies showed that this compound is mainly formed from Phe via (E/Z)-phenylacetaldoxime through the action of two P450s from the CYP79 and CYP71 families (Irmisch et al., 2013, 2014a). Because 2-phenylethylamine has been proposed as alternative substrate for the formation of benzyl cyanide (Tieman et al., 2006), we measured the emission of the volatile nitrile in herbivore-damaged PcanAADC1 RNAi lines. Downregulation of PcanAADC1 expression and the resulting decrease in 2-phenylethylamine accumulation did not influence benzyl cyanide emission (Supplemental Fig. S11), suggesting that this nitrile is exclusively produced via the oxime-dependent pathway in poplar.
Poplar Aldehyde Reductases PtPAR1 and PtPAR2 Convert 2-Phenylacetaldehyde into 2-Phenylethanol In Vitro
The findings that 2-phenylacetaldehyde could neither be detected in herbivore-damaged poplar leaves nor in PtAAS1-overexpressing N. benthamiana plants suggest a rapid and highly efficient conversion of the aldehyde into the corresponding alcohol in planta, likely catalyzed by a NADPH-dependent reductase (PAR) as already reported in rose and tomato (Tieman et al., 2007; Chen et al., 2011). To identify putative poplar PARs, we performed a “TBLASTN” analysis with the tomato gene LePAR1 (Tieman et al., 2007) as query and the P. trichocarpa genome as template. A dendrogram of putative poplar reductases and characterized reductases from other plants was generated and the five poplar genes that grouped together with either LePAR1 (PtPAR1, PtPAR2) or aliphatic aldehyde reductases (PtAAR1, PtAAR2, PtAAR3) were considered for further biochemical characterization (Supplemental Fig. S12). Heterologously expressed and purified enzymes were tested with a variety of aromatic and aliphatic aldehydes as potential substrates in the presence of the cosubstrate NADPH. PtPAR1 and PtPAR2 exhibited enzyme activity with 2-phenylacetaldehyde and produced 2-phenylethanol (Table 2; Supplemental Fig. S13). However, they were also able to reduce other aromatic and aliphatic aldehyde substrates. In contrast, PtAAR1, PtAAR2, and PtAAR3 were only active with a few aliphatic aldehydes and produced no 2-phenylethanol (Table 2; Supplemental Fig. S13). Gene expression analysis revealed that all five aldehyde reductase genes were constitutively expressed in P. trichocarpa leaves (Supplemental Fig. S14). Their transcript accumulation was not influenced by the herbivore treatment.
Table 2. Enzymatic activity of poplar aldehyde reductases.
Enzymes were heterologously expressed in E. coli, purified, and tested with different aldehydes as potential substrates in the presence of NADPH. Enzyme products were extracted with hexane and analyzed using GC-MS. Symbols: −, no activity detected; + activity detected.
| Substrate | Product | PtPAR1 | PtPAR2 | PtAAR1 | PtAAR2 | PtAAR3 |
|---|---|---|---|---|---|---|
| Citronellal | Citronellol | + | + | + | + | + |
| Geranial | Geraniol | + | + | − | − | − |
| Nonanal | Nonanol | + | + | + | − | − |
| Decanal | Decanol | + | + | + | − | + |
| Hexanal | Hexanol | + | + | − | − | − |
| (E)-2-hexenal | (E)-2-hexenol | + | + | − | − | − |
| Benzaldehyde | Benzyl alcohol | + | + | − | − | − |
| 2-phenylacetaldehyde | 2-phenylethanol | + | + | − | − | − |
DISCUSSION
In response to herbivore feeding, poplar produces several aromatic metabolites including 2-phenylethanol, 2-phenylethyl-β-d-glucopyranoside, and 2-phenylethylamine that may function in direct and indirect plant defense and defense signaling. AADCs and AASs have been shown to be involved in the biosynthesis of Phe-derived specialized compounds in other plants (Kaminaga et al., 2006; Tieman et al., 2007; Hirata et al., 2012). We aimed to study AADC/AAS enzymes in P. trichocarpa to elucidate their roles in the herbivore-induced metabolism of Phe.
PtAAS1 Likely Contributes to the Formation of Herbivore-Induced 2-Phenylethanol in P. trichocarpa
Aromatic aldehyde synthases are bifunctional enzymes that catalyze the decarboxylation and subsequent oxidative deamination of aromatic amino acids. In roses and petunia, AAS enzymes have been shown to produce 2-phenylacetaldehyde as substrate for the formation of the corresponding alcohol 2-phenylethanol (Kaminaga et al., 2006; Sakai et al., 2007; Farhi et al., 2010). P. trichocarpa contains two putative AAS (Fig. 3) and biochemical characterization of the recombinant enzymes confirmed their aldehyde synthase activity (Table 1). Whereas PtAAS1 accepted exclusively Phe as a substrate, PtAAS2 showed activity with Phe, Tyr, and Trp. However, the similar Km values of PtAAS1 and PtAAS2 for Phe suggest that both enzymes catalyze the formation of 2-phenylacetaldehyde in vivo (Table 1). In contrast to PtAAS2, which was constitutively expressed, transcript accumulation of PtAAS1 was highly upregulated in herbivore-damaged leaves of P. trichocarpa and mirrored the herbivore-induced emission of 2-phenylethanol (Figs. 2 and 6). This indicates that PtAAS1 likely contributes to the formation of 2-phenylethanol upon herbivory by providing the precursor 2-phenylacetaldehyde.
Recent studies demonstrated that the herbivore-induced production of 2-phenylethanol in poplar is also mediated by an alternative pathway that involves the formation of (E/Z)-phenylacetaldoxime (Irmisch et al., 2013, 2014a; Fig. 1). This oxime is biosynthesized from Phe only upon herbivory by the P450 enzyme CYP79D6v3. Downregulation of CYP79D6v3 in P. × canescens led to decreased emission of herbivore-induced 2-phenylethanol, whereas N. benthamiana plants overexpressing CYP79D6v3 produced the alcohol, suggesting a role of (E/Z)-phenylacetaldoxime as an alternative precursor for 2-phenylethanol in planta (Irmisch et al., 2013). Indeed, it has been proposed that (E/Z)-phenylacetaldoxime can be converted to 2-phenylacetaldehyde by a transoximase activity (Sørensen et al., 2018) or even directly to 2-phenylethanol by an alcohol dehydrogenase (Ferreira-Silva et al., 2010). The enzymatic machinery behind these reactions, however, is still not known. Because the Km value of CYP79D6v3 for Phe (Km = 744 µM) was similar to that of PtAAS1 (Km = 460 µM), we hypothesize that both enzymes and pathways contribute to the herbivore-induced formation of 2-phenylethanol in P. trichocarpa. Notably, the related poplar species P. × canescens lacked expression of AAS1. It is thus likely that P. × canescens produces the alcohol exclusively via the oxime-dependent pathway upon herbivory. This assumption is strengthened by the fact that the knockdown of AADC1 in P. × canescens had no effect on the herbivore-induced emission of 2-phenylethanol (Fig. 7), ruling out a potential contribution of this enzyme to the formation of volatile 2-phenylethanol.
PtAADC1 Generates 2-Phenylethylamine, Which Can Be Metabolized into 2-Phenylethyl-β-d-Glucopyranoside In Vivo
Plant aromatic amines and derived metabolites play important roles as signaling and defense compounds and are usually produced by AADC enzymes (for a review, see Facchini et al., 2000). We identified and characterized three AADCs in P. trichocarpa that accepted various aromatic amino acids as substrates and converted them into the corresponding aromatic amines in vitro. Kinetic characterization revealed that PtAADC1 and PtAADC2 preferred Phe as substrate, whereas PtAADC3 preferred Tyr (Table 1). N. benthamiana plants overexpressing PtAADC1 produced 2-phenylethylamine and minor amounts of tyramine and tryptamine, confirming the kinetic parameters determined in vitro. RNAi-mediated knockdown of AADC1 in P. × canescens led to decreased levels of herbivore-induced 2-phenylethylamine and 2-phenylethyl-β-d-glucopyranoside, while the emission of 2-phenylethanol was not influenced (Fig. 7). These results indicate (1) that PtAADC1 produces 2-phenylethylamine upon herbivory in planta, and (2) that this compound is further metabolized most likely via 2-phenylacetaldehyde and 2-phenylethanol into 2-phenylethyl-β-d-glucopyranoside. The presence of separate pathways to herbivore-induced 2-phenylethanol and 2-phenylethyl-β-d-glucopyranoside (both constitutive and induced) suggests that these two metabolites have different functions in the plant, even if the metabolic fate of 2-phenylethyl-β-d-glucopyranoside is eventually to be hydrolyzed to 2-phenylethanol. Because emission of the free alcohol was not influenced by the AADC1 knockdown, we propose that the AADC-dependent 2-phenylethanol pathway, in contrast to the AAS-mediated route, is tightly associated with a glucosyltransferase as part of a protein complex. The formation of such complexes or metabolons, which can prevent the release of unstable or toxic pathway intermediates, has been described for the cyanogenic glycoside pathway in sorghum (Sorghum bicolor) where a glucosyltransferase is also included (Nielsen et al., 2008; Laursen et al., 2016).
Constitutive 2-Phenylethyl-β-d-Glucopyranoside Might Be Produced Via PtAAS2 or the Ehrlich Pathway
Undamaged leaves of P. trichocarpa were found to accumulate significant amounts of 2-phenylethyl-β-d-glucopyranoside (Fig. 2). Because PtAAS2 was the only AAAD gene with a considerable constitutive expression, we hypothesize that PtAAS2 provides 2-phenylacetaldehyde as substrate for the formation of 2-phenylethyl-β-d-glucopyranoside in undamaged poplar leaves (Fig. 2). However, the accumulation of phenylpyruvic acid in these leaves indicates that the Ehrlich pathway might also contribute to the formation of 2-phenylethyl-β-d-glucopyranoside. An AAAT has recently been reported in melon. Its characterization and gene expression pattern suggest that the catabolism of aromatic amino acids into aroma volatiles likely proceeds via an initial transamination rather than decarboxylation or decarboxylation/deamination in this species (Gonda et al., 2010). Moreover, roses were shown to possess both the AADC-dependent pathway and the phenylpyruvic acid-dependent pathway for the formation of floral 2-phenylethanol. Whereas the former pathway is active throughout the whole year, the latter is induced exclusively during hot seasons and has been discussed as an adaptation to summer environmental conditions (Hirata et al., 2016). The presence of multiple 2-phenylethanol pathways in poplar may also enable the tree to react to different environmental stresses by producing either 2-phenylethanol or 2-phenylethyl-β-d-glucopyranoside independently.
PtAAS1 and PtAADC1 Have Evolved from a Common Ancestor by Gene Duplication and Neofunctionalization
The emergence of large gene families, such as terpene synthases, cytochrome P450 monooxygenases, UDP-glycosyltransferases, methyltransferases, and acyl transferases, that encode enzymes with diverse substrate specificity, regiospecificity, or catalytic activity, created the basis for the chemical diversity of specialized metabolism in plants (Leong and Last, 2017). These gene families evolved through repeated duplications of single genes, chromosomes, or even whole genomes, followed by neofunctionalization of resulting gene copies (Moghe and Kruse, 2018). It has been shown that single amino acid changes can dramatically alter the specificity or activity of enzymes involved in specialized metabolism (e.g. Johnson et al., 2001; Köllner et al., 2004; Junker et al., 2013; Irmisch et al., 2015; Fan et al., 2017), suggesting that novel enzyme functions may arise in relatively short time periods by the accumulation of only a few nucleotide mutations. PtAADC1 and PtAAS1 provide an example of rapid evolutionary changes. They form a gene cluster on chromosome 13 of the P. trichocarpa genome (Tuskan et al., 2006) and their high similarity to each other indicates a recent tandem gene duplication event. However, PtAADC1 and PtAAS1 have already come to encode different enzymatic activities, catalyzing the decarboxylation and decarboxylation/oxidative transamination of Phe, respectively. Recent studies reported a Tyr/Phe switch that determines the catalytic activity of AADC and AAS enzymes, respectively, in different species (Bertoldi et al., 2002; Torrens-Spence et al., 2013). In vitro mutagenesis experiments performed in our study confirmed the role of this residue as a function-dictating element in PtAADC1 and PtAAS1 (Fig. 4). Interestingly, a survey of putative AADC/AAS genes in a set of sequenced vascular plants (http://www.phytozome.net) revealed similar AADC/AAS gene pairs that differed in the function-dictating residue in at least 10 species from monocotyledonous and dicotyledonous plants (Supplemental Fig. S5). Although none of these enzymes has been characterized so far, their sequence similarities with already published AADC/AAS from other plants suggest similar catalytic functions (Supplemental Fig. S6). The presence of the putative AADC/AAS gene pairs indicates an independent occurrence of the Tyr/Phe-mediated activity switch of AADC/AAS enzymes in single genera or even single species of the angiosperms. The multiple evolution of this trait as well as its conservation in numerous plant lineages suggest different but important functions of AADC/AAS enzyme products in plant metabolism.
PtPAR1 and PtPAR2 May Participate in the Biosynthesis of 2-Phenylethanol
The last reaction of the various 2-phenylethanol pathways is the reduction of 2-phenylacetaldehyde catalyzed by PAR (Watanabe et al., 2002; Hirata et al., 2012; Fig. 1). We identified a group of five genes in the genome of P. trichocarpa that showed significant sequence similarity to the recently reported PAR genes from tomato and damask rose (Tieman et al., 2007; Chen et al., 2011). Heterologous expression and enzyme characterization revealed PAR activity for two candidates, PtPAR1 and PtPAR2. However, both enzymes also accepted a variety of other aromatic and aliphatic aldehydes as substrates and converted them into the corresponding alcohols (Table 2; Supplemental Fig. S12). Substrate promiscuity has been described for tomato LePAR2 and rose PAR1 (Tieman et al., 2007; Chen et al., 2011) as well as for short-chain dehydrogenases/reductases involved in other metabolic processes (Oberschall et al., 2000; Yamauchi et al., 2011; Sengupta et al., 2015; Jain et al., 2016). Because PtPAR1 and PtPAR2 showed high constitutive expression that remained constant upon herbivory (Supplemental Fig. S12), they might contribute to the constitutive as well as herbivore-induced formation of 2-phenylethanol and its glucoside in planta. However, we cannot rule out that other members of the large short-chain dehydrogenase/reductase family of poplar (Supplemental Fig. S10) are also involved in the biosynthesis of these compounds.
Herbivory-Induced Emission of 2-Phenylethanol and Accumulation of 2-Phenylethylamine Might Serve as Defensive Weapons or Signals in P. trichocarpa
Herbivore-induced volatiles play important roles in plant defense. They can negatively impact the attacking herbivore directly by toxicity or deterrence. Or, they can act as indirect defenses by functioning as cues for natural enemies of the herbivore. The volatile 2-phenylethanol has been shown to be involved in both processes. Florivorous ants, for example, that feed on flowers of the alpine skypilot (Polemonium viscosum) are strongly repelled by higher concentrations of 2-phenylethanol (Galen et al., 2011). On the other hand, the aphid-feeding lacewing Chrysoperla carnea is attracted by 2-phenylethanol released from host plants of the aphid (Zhu et al., 2005), suggesting a function of this compound in indirect defense. We propose that 2-phenylethanol might also be involved in direct or indirect defense in poplar. The volatile blend emitted from L. dispar-infested poplar leaves has been shown to attract Glyptapanteles liparidis parasitoids that oviposit on L. dispar caterpillars (McCormick et al., 2014) and 2-phenylethanol, one of the prominent blend components, could influence the behavior of the parasitic wasp. Additionally, 2-phenylethanol has been shown to act as antimicrobial agent (Zhu et al., 2011; Liu et al., 2014) and thus might protect poplar plants against pathogens associated with herbivores (Chung et al., 2013; Zhu et al., 2014). Alternatively, 2-phenylethanol could also act as an internal plant signal to indicate the presence of herbivore damage to other organs. Other herbivore-induced natural products, especially volatiles, have been shown to play roles as defense signals (Maag et al., 2015). In woody plants, such as poplar where transport through the vascular system involves long distances between branches, volatile signals may be important in providing more rapid information about the location of herbivory (Frost et al., 2007).
Aromatic amines and related condensation products such as hydroxycinnamic acid amides are known to be of central importance in defense responses upon wounding and pathogen attack (Facchini et al., 2002; Macoy et al., 2015). They have been shown to accumulate after tissue damage in a variety of plants including for example Arabidopsis, tomato, bell pepper (Capsicum annuum), and potato (Solanum tuberosum; Schmidt et al., 1998; Newman et al., 2001; Muroi et al., 2009; Campos et al., 2014). In potato, hydroxycinnamic acid amides are induced by Phytophthora infestans infection and elicitor-treated potato cell cultures also accumulate significant amounts of these compounds (Schmidt et al., 1998, 1999). Similar effects have been observed in chitosan-treated potato suspension cultures, suggesting that insect feeding might also trigger related plant responses (Villegas et al., 1990). Whether or not aromatic amines biosynthesized by the herbivore-induced enzyme PtAADC1 might also act as substrates for other defense compounds such as hydroxycinnamic acid amides in poplar should be investigated in future research.
MATERIALS AND METHODS
Plant Treatment
Western balsam poplar (Populus trichocarpa; genotype Muhle Larsen) trees were grown from monoclonal stem cuttings in a greenhouse (24°C, 60% relative humidity, 16-h/8-h light/dark cycle) in a 1:1 mixture of sand and soil (Klasmann potting substrate; Klasmann-Deilmann), until they reached 1 m in height. Wild-type and transgenic gray poplar (Populus × canescens, clone INRA 717-1B4) plants were amplified by micropropagation as described by Behnke et al. (2007). Saplings of ∼10-cm high were repotted to soil (Klasmann potting substrate) and propagated in a controlled environment chamber for 1 month (day, 22°C; night, 18°C; 65% relative humidity; 16-h/8-h light/dark cycle) before they were transferred to the greenhouse.
For herbivore treatment of poplar, plants were enclosed in a polyethylene terephthalate bag (“Bratschlauch”; Toppits) with (herbivory) or without (control) 10 gypsy moth (Lymantria dispar) caterpillars (larval stage, L4) for 24 h (start of the treatment, ∼4 pm of d 1; end of the treatment, ∼4 pm of d 2).
The L. dispar egg batches were provided by Hannah Nadel, US Department of Agriculture, Animal and Plant Health Inspection Service. After hatching, the caterpillars were reared on an artificial diet (gypsy moth diet; MP Biomedicals) until L4 stage. Caterpillars were starved in single cups for 1 d before the experiment.
RNA Extraction and cDNA Synthesis
Poplar leaf material was harvested immediately after the herbivore treatment, flash-frozen in liquid N, and stored at −80°C until further processing. After grinding the frozen leaf material in liquid N to a fine powder with a Vibratory Micro Mill (Pulverisette-0; Fritsch), total RNA was isolated using an InviTrap Spin Plant RNA Kit (Stratec) according to manufacturer’s instructions. RNA concentration was assessed using a spectrophotometer (NanoDrop 2000c; Thermo Fisher Scientific). Before cDNA synthesis, 1 µg of RNA was DNase-treated using 1 U of DNase (Thermo Fisher Scientific). Single-stranded cDNA was prepared using SuperScript III Reverse Transcriptase and Oligo (dT20) primers (Thermo Fisher Scientific).
Identification and Cloning of Putative AAAD Genes
Putative AAAD genes were identified using a BLAST search against the P. trichocarpa genome (Tuskan et al., 2006; http://www.phytozome.net/poplar) with rose (Rosa spp.) RhPAAS (GenBank Accession: ABB04522) as template. The complete ORF of the resulting candidates PtAAS1, PtAAS2, PtAADC1, PtAADC2, and PtAADC3 were amplified from cDNA made from L. dispar-damaged P. trichocarpa leaves using a Phusion High Fidelity DNA Polymerase (New England Biolabs) and inserted into the pCR-Blunt II-TOPO vector (Thermo Fisher Scientific). Primer sequence information is available in Supplemental Table S3.
Identification and Cloning of Putative PAR Genes
For the identification of putative PAR and AAR genes in the P. trichocarpa genome (Tuskan et al., 2006), a BLAST search was carried out with LePAR1 (GenBank Accession: EF613492.1) as query. The complete ORFs of the resulting candidates PtAAR1, PtAAR2, PtAAR3, PtPAR1, and PtPAR2 were amplified from cDNA made from L. dispar-damaged P. trichocarpa leaves using a Phusion High Fidelity DNA Polymerase (New England Biolabs) and inserted into the pET100/D-TOPO vector (Thermo Fisher Scientific). Primer sequence information is available in Supplemental Table S3.
Phylogenetic Analysis and Amino Acid Alignment
Nucleotide sequence alignments were constructed with the Muscle (codon) algorithm (gap open, −2.9; gap extend, 0; hydrophobicity multiplier, 1.5; clustering method, upgmb) implemented in the software MEGA6 (Tamura et al., 2013). Phylogenetic trees were generated with MEGA6 using the Maximum Likelihood method (model/method, Kimura-2-parameter model; substitutions type, nucleotide; rates among sites, uniform rates; gaps/missing data treatment, partial deletion; site coverage cutoff, 80%). An amino acid alignment of poplar AAAD enzymes characterized in this study together with Arabidopsis (Arabidopsis thaliana) AtAAS and AtTyrDC (Lehmann and Pollmann, 2009; Gutensohn et al., 2011) was constructed and visualized with BioEdit (http://www.mbio.ncsu.edu/bioedit/bioedit.html) and the ClustalW algorithm.
RNA-Seq and RT-qPCR Analysis
Total RNA was extracted from leaf material as described above, TruSeq RNA-compatible libraries were prepared, and PolyA enrichment was performed before sequencing the eight transcriptomes of P. trichocarpa (four trees [biological replicates] for each with the control and herbivory treatment) on an IlluminaHiSeq 2500 sequencer (Max Planck Genome Centre, Cologne, Germany) with 18 Mio reads per library, 100 bp, single end. Trimming of the obtained Illumina reads and mapping to the poplar gene model version 3.0 (https://phytozome.jgi.doe.gov/pz/portal.html) were performed with the program CLC Genomics Workbench (Qiagen Bioinformatics; mapping parameter: length fraction, 0.7; similarity fraction, 0.9; maximum number of hits, 25). Empirical analysis of digital gene expression (EDGE) implemented in the program CLC Genomics Workbench was used for gene expression analysis.
For validation of the RNA-Seq results, quantitative real-time PCR was carried out with specific primers for each gene. Therefore, cDNA was prepared as already described here and diluted 1:10 with sterile water. For the amplification of gene fragments with a length of ∼150–300 bp, primer pairs were designed having a Tm ≥ 60°C, a GC content between 40% and 55%, and a primer length in the range of 20–25 nucleotides (see Supplemental Table S3 for primer information). Primer pair efficiency was tested by measuring Ct values in stepwise-diluted cDNA. Amplicon identity was validated via sequencing of cloned fragments. Samples were run in triplicate using Brilliant III SYBR Green qPCR Master Mix (Agilent). The following PCR conditions were applied for all reactions: initial incubation at 95°C for 3 min followed by 40 cycles of amplification (95°C for 20 s and 60°C for 20 s). SYBR green fluorescence (Thermo Fisher Scientific) was measured at the end of each amplification cycle. Melting curves were recorded within a range of 55°C to 95°C at the end of a total of 40 cycles. All samples were run on a Bio-Rad CFX Connect Real-Time PCR Detection System (Bio-Rad) in 96-well Hard-Shell PCR plates with six biological replicates. The relative normalized expression compared to the housekeeping gene Ubiquitin (Irmisch et al., 2013) was analyzed using CFX Manager 3.1 (Bio-Rad).
Biochemical Characterization of Poplar AAAD Enzymes
For heterologous expression in Escherichia coli, the complete ORFs of the candidate AAAD genes were subcloned into the pET100/D-TOPO vector and expressed in E. coli BL21(DE3) cells (Thermo Fisher Scientific) as N-terminal His6-Tag fusion proteins. Cells were grown in TB medium (1.2% [w/v] tryptone, 2.4% [w/v] yeast extract, 0.5% [v/v] glycerol adjusted to pH 7.2 via potassium buffer, and 50 µg/mL of carbenicillin) at 32°C and 220 rpm until an OD600 of 0.4 was reached. Expression was induced by adding 0.4 mm of isopropyl β-D-1-thiogalactopyranoside and the cultures were grown overnight at 18°C under continuous shaking at 220 rpm. Cells were harvested by centrifugation (5000g, 10 min, 4°C), the supernatant was decanted, and the pellet was resuspended in chilled lysis buffer (20 mm of imidazole at pH 7.2, 100 mm of NaCl, 50 mm of KH2PO4, 2% [v/v] glycerol, 10 mm of MgCl2, 0.3 mg/mL of lysozyme, 100 u/L of Benzonase [Thermo Fisher Scientific], and 1 g/L proteinase inhibitor HP; Serva). The cells were frozen in liquid N and thawed in a water bath at 25°C a total of five times and additionally disrupted by a 3 × 30 s treatment with a sonicator (70% intensity, Cat. No. UW2070; Bandelin). The resulting slurry was pelleted via centrifugation (12,000g, 20 min, 4°C) and the supernatant was transferred to a new reaction tube. Soluble protein was purified from the supernatant using a Poly-Prep Chromatography Gravity Flow Column (Bio-Rad) with 500 µL of HisPur Cobalt Resin (Thermo Fischer Scientific) according to the manufacturer’s instructions (equilibration buffer: 100 mm of NaCl, 50 mm of Tris-HCl, 2% [v/v] glycerol, and 20 mm of imidazole at pH 7.2; washing buffer: 100 mm of NaCl, 50 mm of Tris-HCl, 2% [v/v] glycerol, 40 mm of imidazole at pH 7.2; elution buffer: 100 mm of NaCl, 50 mm of Tris-HCl, and 2% [v/v] glycerol, and 150 mm of imidazole at pH 7.2). The enzyme-containing eluate was desalted via an Illustra NAP-10 gravity flow column (GE Healthcare) into a storage buffer (50 mm of Tris-HCl and 0.1 mm of EDTA at pH 8.0) and immediately mixed 1:1 with a PLP-containing buffer (50 mm of Tris-HCl, 0.2 mm of PLP, and 0.1 mm of EDTA at pH 8.0). Protein concentration was determined via a PCA-enhanced Biuret assay as stated by the manufacturer (Roti-Quant Universal; Karl Roth). Protein purity and identity was confirmed via sodium dodecyl sulfate polyacrylamide gel electrophoresis and colloidal Coomassie staining (Roti-Blue; Karl Roth). The relative abundance of the enzymes defined by densitometry (GS-900 Calibrated Densitometer; Bio-Rad) was calculated for the determination of kcat values.
Enzyme assays were performed in closed glass tubes containing ∼1 µg of purified enzyme and 1 mm of substrate in a total volume of 200 µL of PLP buffer. The reaction was carried out for 30 min under shaking at 300 rpm and 32°C and was stopped by adding 800 µL of methanol. After placing on ice for 30 min, the denatured enzymes were removed by centrifugation (4.000g, 4°C, 10 min) and the supernatant was transferred into a glass vial or a 96-Deep–Well 41-mm plate (Agilent) for LC-MS/MS analysis.
For the determination of the kinetic parameters, assays were carried out in triplicate and with varying substrate concentrations. The enzyme concentration and the incubation times were chosen so that the reaction velocity was linear during the incubation time.
Biochemical Characterization of Poplar PAR Enzymes
The complete ORFs of the candidate AAR and PAR genes were cloned into the pET100/D-TOPO vector and expressed in E. coli BL21(DE3) cells (Thermo Fisher Scientific) as N-terminal His6-Tag fusion proteins, as described for AADC and AAS enzymes. Soluble protein was purified from the supernatant as described by Irmisch et al. (2015). Protein purity and identity was confirmed via SDS-PAGE and colloidal Coomassie staining (Roti-Blue; Karl Roth). Enzyme assays were performed in closed glass tubes containing ∼1 µg of purified enzyme, 1 mm of substrate, and 1 mm of NADPH in a total volume of 100 µL of reaction buffer (10% [v/v] glycerol, 10 mm of TrisHCl at pH 7.5, and 1 mm of dithiothreitol) and were carefully overlaid with 150 µL of hexane and incubated for 1.5 h. After incubation, enzyme assays were thoroughly mixed, the hexane phase was transferred to a new glass vessel, and 1 µL was analyzed as described in Irmisch et al. (2015).
Site-Directed Mutagenesis of PtAAS1 and PtAADC1
For site-directed mutagenesis of PtAAS1 and PtAADC1, specific primers possessing the desired mutated codon were designed after the method described by Ho et al., 1989 (Supplemental Table S3). The vectors pET100/D-TOPO:PtAADC1 and pET100/D-TOPO:PtAAS1 were used as templates for a Phusion High Fidelity DNA PCR (New England BioLabs). The elongation time was adapted to the total length of the template vector according to the instructions of the supplier. The double-nicked vector was directly transformed into chemically competent E. coli cells (TOP10 One Shot; Thermo Fisher Scientific) and screened for successful mutagenesis by sequencing of plasmids purified from single colonies. Plasmids containing the desired mutations were transformed into E. coli BL21(DE3) cells (Thermo Fisher Scientific) and heterologous expression, protein purification, and activity tests were performed.
Transient Expression of PtAADC1 and PtAAS1 in Nicotiana benthamiana
For expression in N. benthamiana, the coding regions of PtAADC1 and PtAAS1 were amplified from the respective pET100/D-TOPO vectors and inserted into the pCAMBiA2300U vector as described in Irmisch et al. (2013). The pCAMBiA 2300U vector and vectors carrying eGFP and p19 were kindly provided by the group of D. Werck-Reichhart, Strasbourg, France. After verification of the sequence integrity, pCAMBiA vectors were separately transferred into Agrobacterium tumefaciens strain C58p90. Protein expression was confirmed by fluorescence analysis (microscope) of N. benthamiana plants transformed with eGFP. Five milliliters of an overnight culture (220 rpm, 28°C) were used to inoculate 200 mL of Luria-Bertani medium (50 µg/mL of kanamycin, 25 μg/mL of rifampicin, and 25 μg/mL of gentamicin) for overnight growth. The following day, the cultures were centrifuged (4000g, 5 min) and the cells were resuspended in infiltration buffer (10 mm of 2-ethanesulfonic acid, 10 mm of MgCl2, and 100 μM of acetosyringone at pH 5.6) to reach a final OD600 of 0.5. After shaking for at least 2 h at room temperature, the cultures carrying PtAADC1, PtAAS1, or eGFP were mixed with an equal volume of cultures carrying pBIN:p19. For transformation, 3- to 4-week–old N. benthamiana plants were dipped upside-down in an Agrobacterium solution and vacuum was applied to infiltrate the leaves. Infiltrated plants were placed in a room without direct sunlight. At 3 d after transformation, plants were placed under low direct light (light-emitting diode intensity 40%) for an additional 3 d. Volatiles were measured on the 6th d after transformation. For volatile collection, plants were separately placed in gas-tight 3-L glass desiccators. Charcoal-filtered air was pumped into the desiccators at a flow rate of 1 L/min and the air left the desiccators through a filter packed with 30 mg of Porapak Q (Sigma-Aldrich). Volatiles were collected for 6 h (10 am to 4 pm). The volatile compounds were desorbed from the filters and analyzed by gas chromatography-mass spectrometry (GC-MS) and gas chromatography–flame ionization detection (GC-FID). Plants were harvested after the volatile collection, ground in liquid N, and stored at −80°C until further analysis.
Vector Construction and Transformation of Poplar
The construction of the binary vector was described by Levée et al. (2009). The transformation of the P. × canescens clone INRA 7171-B4 followed a protocol published by Meilan and Ma (2007). To target only AADC1/AAS1 mRNA, a fragment between position 455 and 765 of the coding sequence of PtAADC1 was selected. Transgenic RNAi poplar plants were amplified by micropropagation as described by Behnke et al. (2007). To test the level of transgenicity, RT-qPCR analysis was done on wild-type, vector control (pCAMBiA), and RNAi plants.
Chemical Conversion of PtAAS Enzyme Products into Aldoximes
For the identification of reactive aromatic aldehydes, PtAAS enzymes were incubated with single amino acids as substrates, overlaid with 400 µL of pure toluol and incubated for 1 h at 32°C. The aromatic aldehydes produced by the enzymes were extracted into the toluol phase and transferred to a new glass vial. The toluol phase was incubated with an aqueous solution of 100 mm of hydroxylamine (pH 7.0) at 25°C for 1 h as described in Irmisch et al. (2013) and Hofmann et al. (1981) to form the corresponding aldoxime. The toluol phase was then transferred to a fresh glass vial and dried-down under a continuous flow of N. The pellet was dissolved in 200 µL of methanol and analyzed via LC-MS/MS as described in Irmisch et al. (2013).
LC-MS/MS Analysis of Plant Methanol Extracts
Metabolites were extracted from ground plant material (poplar or N. benthamiana) with methanol (10:1 v/w). Analytes were separated using a 1200 HPLC system (Agilent) on a Zorbax Eclipse XDB-C18 column (50 × 4.6 mm, 1.8 µm; Agilent). Mobile phase consisted of formic acid (0.05% [v/v]) in water (A) and acetonitrile (B). The column temperature was maintained at 25°C. HPLC parameters are given in Supplemental Table S4.
The liquid chromatography was coupled to an API-5000 Tandem Mass Spectrometer (Applied Biosystems) equipped with a Turbospray ion source (ion spray voltage, 5500 eV; turbo gas temperature, 700°C; nebulizing gas, 70 pressure per square inch [p.s.i.]; curtain gas, 35 p.s.i.; heating gas, 70 p.s.i.; collision gas, 2 p.s.i.). Multiple reaction monitoring was used to monitor a parent ion → product ion reaction for each analyte. Detailed parameters are given in Supplemental Table S5. Identification and quantification of compounds were performed using standard curves made from authentic and commercially available standards of 2-phenylethylamine, tyramine, tryptamine, 2-phenylethyl-β-d-glucopyranoside, phenylpyruvic acid, and Phe (Supplemental Table S6). Relative quantification of ethanolamine (Supplemental Fig. S4) was carried out under the conditions mentioned in Supplemental Tables S4 and S5. Ethanolamine was identified via an authentic standard (Supplemental Table S6).
GC-MS/FID Analysis of Plant Volatiles, PAR, and AAR Products
A gas chromatograph (model No. 6890; Hewlett-Packard) was employed with helium (MS) or H2 (FID) as carrier gas at 2 mL/min, splitless injection (injector temperature, 230°C; injection volume, 1 μL), and a DB-5MS column (30 m × 0.25 mm × 0.25 µm; Agilent). The GC oven temperature was held for 2 min and then increased to 225°C with a gradient of 5°C/min, held for another 2 min, and then further increased to 250°C with 100°C/min and held for 1 min. A coupled mass spectrometer (model no. 5973; Hewlett-Packard) with a quadrupole mass selective detector (transfer line temperature, 230°C; source temperature, 230°C; quadrupole temperature, 150°C; ionization potential, 70 eV; scan range of 40–400 atomic mass units) was then used. Quantification was performed with the trace of a FID operated at 300°C. For analysis of PAR and AAR reaction products, 1 μL of the hexane phase covering the enzyme assays was injected into GC-MS. The compounds 2-phenylethanol and 2-phenylacetaldehyde were identified via authentic standards (Supplemental Table S6).
Thermal Desorption Unit-GC-MS Analysis of PtAAS Enzyme Products
PtAAS enzyme products were collected from the headspace of the assays with polydimethylsiloxane (PDMS) silicone tubes as described by Kallenbach et al. (2014). For the determination of Km values, enzyme assays as described above were incubated with preconditioned PDMS silicone tubes in an air-tight vial for 30 min. After the incubation, PDMS tubes were immediately analyzed using thermal desorption GC-MS. For preparing a standard curve, 2-phenylacetaldehyde was dissolved in 100% ethanol and added in specific amounts to the buffer system used for the enzyme assays. As an enzyme control, 1 μg of bovine serum albumin was added per assay and total volume of the assay was set to 200 μL.
A thermal desorption unit (TDU; model no. TD-20; Shimadzu) combined with an ultra gas chromatograph (model no. GC-MS-QP2010; Shimadzu) was employed with He as carrier gas. Thermal desorption of PDMS tubes was carried out with a sample flow of 60 mL/min, a desorption temperature of 200°C, a valve temperature of 250°C, and a transfer line temperature of 230°C. The trap tube was cooled (−20°C) and, upon completed desorption, heated to 230°C to release the sample onto the GC column. The gas chromatograph was operated at a column flow rate of 1.5 mL/min (He), split injection (split ratio: 20), and a model no. ZB-5MS column (30 m × 0.25 mm × 0.25 µm; Zebron; Phenomenex). The GC oven temperature was set to 45°C, held for 3 min, then increased to 180°C with a gradient of 6°C/min, finally increased to 300°C with a 100°C/min increment, and held for another 2.3 min. The coupled mass spectrometer with a quadrupole mass selective detector was operated in “Scan mode selective” for 33–350 atomic mass units (interface temperature, 250°C; source temperature, 230°C; ionization potential, 70 eV).
Statistical Analysis
Statistical analysis was carried out as described in the figure legends for the respective experiments. Student’s t tests, Mann-Whitney Rank Sum tests, Kruskal-Wallis one-way analysis of variance, and Tukey tests were performed with the software SigmaPlot 12.0 (Systat Software). EDGE tests were performed with CLC Genomics Workbench (Qiagen Informatics). Data visualization was done with the software SigmaPlot 12.0 and R Version 3.3.1 (R Development Core Team; http://www.R-project.org).
Accession Numbers
Sequence data for PtAAS1 (MK440562), PtAAS2 (MK440561), PtAADC1 (MK440568), PtAADC2 (MK440567), PtAADC3 (MK440566), PtSDC1 (MK440558), PtSDC2 (MK440569), PtPAR1 (MK440560), PtPAR2 (MK440559), PtAAR1 (MK440565), PtAAR2 (MK440564), and PtAAR3 (MK440563) can be found in the NCBI GenBank (https://www.ncbi.nlm.nih.gov/genbank/) under the corresponding identifiers. Raw reads of the RNA-Seq experiment were deposited in the NCBI Sequence Read Archive under the accession PRJNA516861.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Amino acid alignment of PtAAS and PtAADC enzymes from P. trichocarpa and characterized AtAAS and AtTyrDC from Arabidopsis.
Supplemental Figure S2. PtAAS1 and PtAAS2 do not release 2-phenylethylamine.
Supplemental Figure S3. Detection of poplar AAS products via the conversion to corresponding aldoximes.
Supplemental Figure S4. PtSDC1 and PtSDC2 convert l-Ser to ethanolamine.
Supplemental Figure S5. Phylogeny and comparison of plant AAAD gene clusters.
Supplemental Figure S6. Dendrogram analysis of putative AADC/AAS gene pairs and characterized AADC, AAS, and Glu/Ser/His-decarboxylase genes from other plants.
Supplemental Figure S7. Transcript accumulation of PtAAS1, PtAADC1, PtAADC2, PtAAS2, and PtAADC3 in caterpillar-damaged (herb) and undamaged (ctr) leaves of P. trichocarpa.
Supplemental Figure S8. Dendrogram analysis of AAAD genes from P. trichocarpa, P. × canescens, P. tremula, and P. alba.
Supplemental Figure S9. Transcript accumulation of AAS1, AADC1, and AADC2 in gypsy moth (L. dispar) caterpillar-damaged P. × canescens leaves.
Supplemental Figure S10. Transcript accumulation of PcanAADC1 and PcanAAS2 in caterpillar-damaged leaves of control and AADC1/AADC2/AAS1 RNAi lines of P. × canescens.
Supplemental Figure S11. The RNAi-mediated knockdown of PcanAADC1 in P. × canescens does not influence the herbivore-induced emission of benzyl cyanide.
Supplemental Figure S12. Dendrogram analysis of putative reductases from poplar and characterized reductases from other plants.
Supplemental Figure S13. PtPAR1 and PtPAR2 convert 2-phenylacetaldehyde into 2-phenylethanol in vitro.
Supplemental Figure S14. Transcript accumulation of putative aldehyde reductase genes in L. dispar-damaged and undamaged P. trichocarpa leaves.
Supplemental Table S1. Accession numbers of genes used for phylogenetic analysis.
Supplemental Table S2. Expression of PtAAS1, PtAADC1, and PtAADC2 in undamaged and herbivore-damaged P. trichocarpa leaves.
Supplemental Table S3. Oligonucleotides used for isolation and RT-qPCR analysis of poplar AAS, AADC, SDC, PAR, and AAR genes.
Supplemental Table S4. HPLC gradients used for separation and analysis of metabolites.
Supplemental Table S5. MS/MS parameters used for LC-MS/MS analysis.
Supplemental Table S6. Compounds used as substrates for enzyme assays and as standards for LC-MS/MS and GC-MS quantification.
Acknowledgments
We thank Tamara Krügel, Danny Kessler, and all the Max Planck Institute for Chemical Ecology gardeners for their help with rearing the poplar and N. benthamiana plants. D. Werck-Reichhart, Strasbourg, France, is thanked for kindly providing the pCAMBiA vectors and for the nice introduction to the USER cloning system. We thank Tabea Kröber, Daniel Veit, Natascha Rauch, and Marion Stäger for excellent technical assistance and Grit Kunert for statistical advice. Additionally, we thank Sybille B. Unsicker (Max Planck Institute for Chemical Ecology) and her group members for rearing L. dispar caterpillars. Alexander Haverkamp is thanked for help with the data visualization.
Footnotes
This work was supported by the Max-Planck Society.
Articles can be viewed without a subscription.
References
- Arimura G, Huber DPW, Bohlmann J (2004) Forest tent caterpillars (Malacosoma disstria) induce local and systemic diurnal emissions of terpenoid volatiles in hybrid poplar (Populus trichocarpa × deltoides): cDNA cloning, functional characterization, and patterns of gene expression of (−)-germacrene D synthase, PtdTPS1. Plant J 37: 603–616 [DOI] [PubMed] [Google Scholar]
- Behnke K, Ehlting B, Teuber M, Bauerfeind M, Louis S, Hänsch R, Polle A, Bohlmann J, Schnitzler JP (2007) Transgenic, non-isoprene emitting poplars don’t like it hot. Plant J 51: 485–499 [DOI] [PubMed] [Google Scholar]
- Bertoldi M, Gonsalvi M, Contestabile R, Voltattorni CB (2002) Mutation of tyrosine 332 to phenylalanine converts dopa decarboxylase into a decarboxylation-dependent oxidative deaminase. J Biol Chem 277: 36357–36362 [DOI] [PubMed] [Google Scholar]
- Bieri S, Brachet A, Veuthey JL, Christen P (2006) Cocaine distribution in wild Erythroxylum species. J Ethnopharmacol 103: 439–447 [DOI] [PubMed] [Google Scholar]
- Campos L, Lisón P, López-Gresa MP, Rodrigo I, Zacarés L, Conejero V, Bellés JM (2014) Transgenic tomato plants overexpressing tyramine n-hydroxycinnamoyltransferase exhibit elevated hydroxycinnamic acid amide levels and enhanced resistance to Pseudomonas syringae. Mol Plant Microbe Interact 27: 1159–1169 [DOI] [PubMed] [Google Scholar]
- Chen XM, Kobayashi H, Sakai M, Hirata H, Asai T, Ohnishi T, Baldermann S, Watanabe N (2011) Functional characterization of rose phenylacetaldehyde reductase (PAR), an enzyme involved in the biosynthesis of the scent compound 2-phenylethanol. J Plant Physiol 168: 88–95 [DOI] [PubMed] [Google Scholar]
- Chung SH, Rosa C, Scully ED, Peiffer M, Tooker JF, Hoover K, Luthe DS, Felton GW (2013) Herbivore exploits orally secreted bacteria to suppress plant defenses. Proc Natl Acad Sci USA 110: 15728–15733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa MA, Marques JV, Dalisay DS, Herman B, Bedgar DL, Davin LB, Lewis NG (2013) Transgenic hybrid poplar for sustainable and scalable production of the commodity/specialty chemical, 2-phenylethanol. PLoS One 8: e83169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danner H, Boeckler GA, Irmisch S, Yuan JS, Chen F, Gershenzon J, Unsicker SB, Köllner TG (2011) Four terpene synthases produce major compounds of the gypsy moth feeding-induced volatile blend of Populus trichocarpa. Phytochemistry 72: 897–908 [DOI] [PubMed] [Google Scholar]
- Facchini PJ, Huber-Allanach KL, Tari LW (2000) Plant aromatic l-amino acid decarboxylases: Evolution, biochemistry, regulation, and metabolic engineering applications. Phytochemistry 54: 121–138 [DOI] [PubMed] [Google Scholar]
- Facchini PJ, Hagel J, Zulak KG (2002) Hydroxycinnamic acid amide metabolism: Physiology and biochemistry. Can J Bot 80: 577–589 [Google Scholar]
- Fahey JW, Zalcmann AT, Talalay P (2001) The chemical diversity and distribution of glucosinolates and isothiocyanates among plants. Phytochemistry 56: 5–51 [DOI] [PubMed] [Google Scholar]
- Fan P, Miller AM, Liu X, Jones AD, Last RL (2017) Evolution of a flipped pathway creates metabolic innovation in tomato trichomes through BAHD enzyme promiscuity. Nat Commun 8: 2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhi M, Lavie O, Masci T, Hendel-Rahmanim K, Weiss D, Abeliovich H, Vainstein A (2010) Identification of rose phenylacetaldehyde synthase by functional complementation in yeast. Plant Mol Biol 72: 235–245 [DOI] [PubMed] [Google Scholar]
- Ferreira-Silva B, Lavandera I, Kern A, Faber K, Kroutil W (2010) Chemo-promiscuity of alcohol dehydrogenases: Reduction of phenylacetaldoxime to the alcohol. Tetrahedron 66: 3410–3414 [Google Scholar]
- Frost CJ, Appel HM, Carlson JE, De Moraes CM, Mescher MC, Schultz JC (2007) Within-plant signalling via volatiles overcomes vascular constraints on systemic signalling and primes responses against herbivores. Ecol Lett 10: 490–498 [DOI] [PubMed] [Google Scholar]
- Galen C, Kaczorowski R, Todd SL, Geib J, Raguso RA (2011) Dosage-dependent impacts of a floral volatile compound on pollinators, larcenists, and the potential for floral evolution in the alpine skypilot Polemonium viscosum. Am Nat 177: 258–272 [DOI] [PubMed] [Google Scholar]
- Gonda I, Bar E, Portnoy V, Lev S, Burger J, Schaffer AA, Tadmor Y, Gepstein S, Giovannoni JJ, Katzir N, et al. (2010). Branched-chain and aromatic amino acid catabolism into aroma volatiles in Cucumis melo L. fruit. 61: 1111–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutensohn M, Klempien A, Kaminaga Y, Nagegowda DA, Negre-Zakharov F, Huh JH, Luo H, Weizbauer R, Mengiste T, Tholl D, et al. (2011) Role of aromatic aldehyde synthase in wounding/herbivory response and flower scent production in different Arabidopsis ecotypes. Plant J 66: 591–602 [DOI] [PubMed] [Google Scholar]
- Hayashi S, Yagi K, Ishikawa T, Kawasaki M, Asai T, Picone J, Turnbull C, Hiratake J, Sakata K, Takada M, et al. (2004) Emission of 2-phenylethanol from its β-d-glucopyranoside and the biogenesis of these compounds from [2H8] l-phenylalanine in rose flowers. Tetrahedron 60: 7005–7013 [Google Scholar]
- Hazelwood LA, Daran JM, Van Maris AJA, Pronk JT, Dickinson JR (2008) The Ehrlich pathway for fusel alcohol production: A century of research on Saccharomyces cerevisiae metabolism. Appl Environ Microbiol 74: 2259–2266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata H, Ohnishi T, Ishida H, Tomida K, Sakai M, Hara M, Watanabe N (2012) Functional characterization of aromatic amino acid aminotransferase involved in 2-phenylethanol biosynthesis in isolated rose petal protoplasts. J Plant Physiol 169: 444–451 [DOI] [PubMed] [Google Scholar]
- Hirata H, Ohnishi T, Tomida K, Ishida H, Kanda M, Sakai M, Yoshimura J, Suzuki H, Ishikawa T, Dohra H, et al. (2016) Seasonal induction of alternative principal pathway for rose flower scent. Sci Rep 6: 20234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR (1989) Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77: 51–59 [DOI] [PubMed] [Google Scholar]
- Hofmann F, Rausch T, Hilgenberg W (1981) Preparation of radioactively labelled indole-3-acetic acid precursors. J Lab Compd Radiopharm 18: 1491–1495 [Google Scholar]
- Honda K, Ômura H, Hayashi N (1998) Identification of floral volatiles from Ligustrum japonicum that stimulate flower-visiting by cabbage butterfly, Pieris rapae. J Chem Ecol 24: 2167–2180 [Google Scholar]
- Imai T, Maekawa M, Tsuchiya S, Fujimori T (1998) Field attraction of Hoplia communis to 2-phenylethanol, a major volatile component from host flowers, Rosa spp. J Chem Ecol 24: 1491–1497 [Google Scholar]
- Irmisch S, McCormick AC, Boeckler GA, Schmidt A, Reichelt M, Schneider B, Block K, Schnitzler J-P, Gershenzon J, Unsicker SB, Köllner TG (2013) Two herbivore-induced cytochrome P450 enzymes CYP79D6 and CYP79D7 catalyze the formation of volatile aldoximes involved in poplar defense. Plant Cell 25: 4737–4754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irmisch S, Clavijo McCormick A, Günther J, Schmidt A, Boeckler GA, Gershenzon J, Unsicker SB, Köllner TG (2014a) Herbivore-induced poplar cytochrome P450 enzymes of the CYP71 family convert aldoximes to nitriles which repel a generalist caterpillar. Plant J 80: 1095–1107 [DOI] [PubMed] [Google Scholar]
- Irmisch S, Jiang Y, Chen F, Gershenzon J, Köllner TG (2014b) Terpene synthases and their contribution to herbivore-induced volatile emission in western balsam poplar (Populus trichocarpa). BMC Plant Biol 14: 270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irmisch S, Müller AT, Schmidt L, Günther J, Gershenzon J, Köllner TG (2015) One amino acid makes the difference: The formation of ent-kaurene and 16α-hydroxy-ent-kaurane by diterpene synthases in poplar. BMC Plant Biol 15: 262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain D, Khandal H, Khurana JP, Chattopadhyay D (2016) A pathogenesis related-10 protein CaARP functions as aldo/keto reductase to scavenge cytotoxic aldehydes. Plant Mol Biol 90: 171–187 [DOI] [PubMed] [Google Scholar]
- Johnson ET, Ryu S, Yi H, Shin B, Cheong H, Choi G (2001) Alteration of a single amino acid changes the substrate specificity of dihydroflavonol 4-reductase. Plant J 25: 325–333 [DOI] [PubMed] [Google Scholar]
- Junker A, Fischer J, Sichhart Y, Brandt W, Dräger B (2013) Evolution of the key alkaloid enzyme putrescine n-methyltransferase from spermidine synthase. Front Plant Sci 4: 260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallenbach M, Oh Y, Eilers EJ, Veit D, Baldwin IT, Schuman MC (2014) A robust, simple, high-throughput technique for time-resolved plant volatile analysis in field experiments. Plant J 78: 1060–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminaga Y, Schnepp J, Peel G, Kish CM, Ben-Nissan G, Weiss D, Orlova I, Lavie O, Rhodes D, Wood K, et al. (2006) Plant phenylacetaldehyde synthase is a bifunctional homotetrameric enzyme that catalyzes phenylalanine decarboxylation and oxidation. J Biol Chem 281: 23357–23366 [DOI] [PubMed] [Google Scholar]
- Knudsen JT, Eriksson R, Gershenzon J, Ståhl B (2006) Diversity and distribution of floral scent. Bot Rev 72: 1–120 [Google Scholar]
- Köllner TG, Schnee C, Gershenzon J, and Degenhardt J (2004). The variability of sesquiterpenes emitted from two Zea mays cultivars is controlled by allelic variation of two terpene synthase genes encoding stereoselective multiple product enzymes. Plant Cell 16: 1115–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R. (2016) Evolutionary trails of plant group II pyridoxal phosphate-dependent decarboxylase genes. Front Plant Sci 7: 1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laursen T, Borch J, Knudsen C, Bavishi K, Torta F, Martens HJ, Silvestro D, Hatzakis NS, Wenk MR, Dafforn TR, et al. (2016) Characterization of a dynamic metabolon producing the defense compound dhurrin in sorghum. Science 354: 890–893 [DOI] [PubMed] [Google Scholar]
- Lehmann T, Pollmann S (2009) Gene expression and characterization of a stress-induced tyrosine decarboxylase from Arabidopsis thaliana. FEBS Lett 583: 1895–1900 [DOI] [PubMed] [Google Scholar]
- Leong BJ, Last RL (2017) Promiscuity, impersonation and accommodation: Evolution of plant specialized metabolism. Curr Opin Struct Biol 47: 105–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leple JC, Brasileiro AC, Michel MF, Delmotte F, Jouanin L (1992) Transgenic poplars: Expression of chimeric genes using four different constructs. Plant Cell Rep 11: 137–141 [DOI] [PubMed] [Google Scholar]
- Levée V, Major I, Levasseur C, Tremblay L, MacKay J, Séguin A (2009) Expression profiling and functional analysis of Populus WRKY23 reveals a regulatory role in defense. New Phytol 184: 48–70 [DOI] [PubMed] [Google Scholar]
- Liu P, Cheng Y, Yang M, Liu Y, Chen K, Long CA, Deng X (2014) Mechanisms of action for 2-phenylethanol isolated from Kloeckera apiculata in control of Penicillium molds of citrus fruits. BMC Microbiol 14: 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maag D, Erb M, Köllner TG, Gershenzon J (2015) Defensive weapons and defense signals in plants: Some metabolites serve both roles. BioEssays 37: 167–174 [DOI] [PubMed] [Google Scholar]
- Macoy DM, Kim WY, Lee SY, Kim MG (2015) Biotic stress related functions of hydroxycinnamic acid amide in plants. J Plant Biol 58: 156–163 [Google Scholar]
- Major IT, Constabel CP (2008) Functional analysis of the Kunitz trypsin inhibitor family in poplar reveals biochemical diversity and multiplicity in defense against herbivores. Plant Physiol 146: 888–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick AC, Boeckler GA, Köllner TG, Gershenzon J, Unsicker SB (2014) The timing of herbivore-induced volatile emission in black poplar (Populus nigra) and the influence of herbivore age and identity affect the value of individual volatiles as cues for herbivore enemies. BMC Plant Biol 14: 304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meilan R, Ma C (2007) Poplar (Populus spp.) BT - Agrobacterium Protocols. In Wang K, ed, Humana Press, Vol 2 Totowa, NJ, pp 143–151 [Google Scholar]
- Moghe GD, Kruse LH (2018) The study of plant specialized metabolism: Challenges and prospects in the genomics era. Am J Bot 105: 959–962 [DOI] [PubMed] [Google Scholar]
- Muroi A, Ishihara A, Tanaka C, Ishizuka A, Takabayashi J, Miyoshi H, Nishioka T (2009) Accumulation of hydroxycinnamic acid amides induced by pathogen infection and identification of agmatine coumaroyltransferase in Arabidopsis thaliana. Planta 230: 517–527 [DOI] [PubMed] [Google Scholar]
- Newman MA, von Roepenack-Lahaye E, Parr A, Daniels MJ, Dow JM (2001) Induction of hydroxycinnamoyl-tyramine conjugates in pepper by Xanthomonas campestris, a plant defense response activated by hrp gene-dependent and hrp gene-independent mechanisms. Mol Plant Microbe Interact 14: 785–792 [DOI] [PubMed] [Google Scholar]
- Nielsen KA, Tattersall DB, Jones PR, Møller BL (2008) Metabolon formation in dhurrin biosynthesis. Phytochemistry 69: 88–98 [DOI] [PubMed] [Google Scholar]
- Oberschall A, Deák M, Török K, Sass L, Vass I, Kovács I, Fehér A, Dudits D, Horváth GV (2000) A novel aldose/aldehyde reductase protects transgenic plants against lipid peroxidation under chemical and drought stresses. Plant J 24: 437–446 [DOI] [PubMed] [Google Scholar]
- Peters DJ, Constabel CP (2002) Molecular analysis of herbivore-induced condensed tannin synthesis: Cloning and expression of dihydroflavonol reductase from trembling aspen (Populus tremuloides). Plant J 32: 701–712 [DOI] [PubMed] [Google Scholar]
- Philippe RN, Bohlmann J (2007) Poplar defense against insect herbivores. Can J Bot 85: 1111–1126 [Google Scholar]
- Ralph S, Oddy C, Cooper D, Yueh H, Jancsik S, Kolosova N, Philippe RN, Aeschliman D, White R, Huber D, et al. (2006) Genomics of hybrid poplar (Populus trichocarpa × deltoides) interacting with forest tent caterpillars (Malacosoma disstria): Normalized and full-length cDNA libraries, expressed sequence tags, and a cDNA microarray for the study of insect-induced defences in poplar. Mol Ecol 15: 1275–1297 [DOI] [PubMed] [Google Scholar]
- Roy BA, Raguso RA (1997) Olfactory versus visual cues in a floral mimicry system. Oecologia 109: 414–426 [DOI] [PubMed] [Google Scholar]
- Sakai M, Hirata H, Sayama H, Sekiguchi K, Itano H, Asai T, Dohra H, Hara M, Watanabe N (2007) Production of 2-phenylethanol in roses as the dominant floral scent compound from l-phenylalanine by two key enzymes, a PLP-dependent decarboxylase and a phenylacetaldehyde reductase. Biosci Biotechnol Biochem 71: 2408–2419 [DOI] [PubMed] [Google Scholar]
- Schmidt A, Scheel D, Strack D (1998) Elicitor-stimulated biosynthesis of hydroxycinnamoyltyramines in cell suspension cultures of Solanum tuberosum. Planta 205: 51–55 [Google Scholar]
- Schmidt A, Grimm R, Schmidt J, Scheel D, Strack D, Rosahl S (1999) Cloning and expression of a potato cDNA encoding hydroxycinnamoyl-CoA:tyramine n-(hydroxycinnamoyl)transferase. J Biol Chem 274: 4273–4280 [DOI] [PubMed] [Google Scholar]
- Sengupta D, Naik D, Reddy AR (2015) Plant aldo-keto reductases (AKRs) as multi-tasking soldiers involved in diverse plant metabolic processes and stress defense: A structure-function update. J Plant Physiol 179: 40–55 [DOI] [PubMed] [Google Scholar]
- Sheng L, Zeng Y, Wei T, Zhu M, Fang X, Yuan X, Luo Y, Feng L (2018) Cloning and functional verification of genes related to 2-phenylethanol biosynthesis in Rosa rugosa. Genes (Basel) 9: 576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen M, Neilson EHJ, Møller BL (2018) Oximes: Unrecognized chameleons in general and specialized plant metabolism. Mol Plant 11: 95–117 [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30: 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieman D, Taylor M, Schauer N, Fernie AR, Hanson AD, Klee HJ (2006) Tomato aromatic amino acid decarboxylases participate in synthesis of the flavor volatiles 2-phenylethanol and 2-phenylacetaldehyde. Proc Natl Acad Sci USA 103: 8287–8292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieman DM, Loucas HM, Kim JY, Clark DG, Klee HJ (2007) Tomato phenylacetaldehyde reductases catalyze the last step in the synthesis of the aroma volatile 2-phenylethanol. Phytochemistry 68: 2660–2669 [DOI] [PubMed] [Google Scholar]
- Torrens-Spence MP, Liu P, Ding H, Harich K, Gillaspy G, Li J (2013) Biochemical evaluation of the decarboxylation and decarboxylation–deamination activities of plant aromatic amino acid decarboxylases. J Biol Chem 288: 2376–2387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrens-Spence MP, Lazear M, von Guggenberg R, Ding H, Li J (2014a) Investigation of a substrate-specifying residue within Papaver somniferum and Catharanthus roseus aromatic amino acid decarboxylases. Phytochemistry 106: 37–43 [DOI] [PubMed] [Google Scholar]
- Torrens-Spence MP, von Guggenberg R, Lazear M, Ding H, Li J (2014b) Diverse functional evolution of serine decarboxylases: Identification of two novel acetaldehyde synthases that uses hydrophobic amino acids as substrates. BMC Plant Biol 14: 247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrens-Spence MP, Chiang Y, Smith T, Vicent MA, Wang Y (2018a) Structural basis for independent origins of new catalytic machineries in plant AAAD proteins. bioRxiv 404970 [Google Scholar]
- Torrens-Spence MP, Pluskal T, Li FS, Carballo V, Weng JK (2018b) Complete pathway elucidation and heterologous reconstitution of Rhodiola salidroside biosynthesis. Mol Plant 11: 205–217 [DOI] [PubMed] [Google Scholar]
- Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A, et al. (2006) The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313: 1596–1604 [DOI] [PubMed] [Google Scholar]
- Unsicker SB, Kunert G, Gershenzon J (2009) Protective perfumes: The role of vegetative volatiles in plant defense against herbivores. Curr Opin Plant Biol 12: 479–485 [DOI] [PubMed] [Google Scholar]
- Villegas M, Brodelius PE, Kylin A (1990) Elicitor‐induced hydroxycinnamoyl‐CoA:tyramine hydroxycinnamoyltransferase in plant cell suspension cultures. Physiol Plant 78: 414–420 [Google Scholar]
- Watanabe S, Hayashi K, Yagi K, Asai T, MacTavish H, Picone J, Turnbull C, Watanabe N (2002) Biogenesis of 2-phenylethanol in rose flowers: Incorporation of [2H8]L-phenylalanine into 2-phenylethanol and its β-D-glucopyranoside during the flower opening of Rosa ‘Hoh-Jun’ and Rosa damascena Mill. Biosci Biotechnol Biochem 66: 943–947 [DOI] [PubMed] [Google Scholar]
- Xue LJ, Alabady MS, Mohebbi M, Tsai CJ (2015) Exploiting genome variation to improve next-generation sequencing data analysis and genome editing efficiency in Populus tremula × alba 717-1B4. Tree Genet Genomes 11: 82 [Google Scholar]
- Yamauchi Y, Hasegawa A, Taninaka A, Mizutani M, Sugimoto Y (2011) NADPH-dependent reductases involved in the detoxification of reactive carbonyls in plants. J Biol Chem 286: 6999–7009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Jacobs TB, Xue LJ, Harding SA, Tsai CJ (2015) Exploiting SNPs for biallelic CRISPR mutations in the outcrossing woody perennial Populus reveals 4-coumarate:CoA ligase specificity and redundancy. New Phytol 208: 298–301 [DOI] [PubMed] [Google Scholar]
- Zhu F, Poelman EH, Dicke M (2014) Insect herbivore-associated organisms affect plant responses to herbivory. New Phytol 204: 315–321 [Google Scholar]
- Zhu J, Obrycki JJ, Ochieng SA, Baker TC, Pickett JA, Smiley D (2005) Attraction of two lacewing species to volatiles produced by host plants and aphid prey. Naturwissenschaften 92: 277–281 [DOI] [PubMed] [Google Scholar]
- Zhu YJ, Zhou HT, Hu YH, Tang JY, Su MX, Guo YJ, Chen QX, Liu B (2011) Antityrosinase and antimicrobial activities of 2-phenylethanol, 2-phenylacetaldehyde and 2-phenylacetic acid. Food Chem 124: 298–302 [Google Scholar]