Exploration of GAST-like genes across the plant kingdom for their structure and evolution through acquisition of novel motifs and functional conservation uncovers possible roles in hormone signaling.
Abstract
Gibberellic Acid Stimulated Transcript (GAST)-like genes encode small polypeptides, some of which have been implicated in diverse biological processes regulating plant growth and development. However, the occurrence of GASTs among plants, their protein structures, and the mechanisms by which they evolved remain elusive. Here, using a customized workflow, we report genes encoding GAST proteins, identify novel motifs and evolutionary patterns contributing to the subfunctionalization of GAST domains, and explore functional conservation across diverse plant groups. We show that GAST-like sequences evolved initially in the vascular plant Selaginella moellendorffii, after the divergence from bryophytes, and later emerged in gymnosperms and angiosperms. GASTs in angiosperms are characterized by four conserved novel motifs; however, relatively fewer conserved motifs exist in pteridophytes and gymnosperms. Phylogenetic analysis revealed that the GAST-Cysteine Rich1 motif evolved early in the S. moellendorffii GAST, which further acquired subfunctionalization through successive conjugation of other motifs and remained conserved across plants, as supported by their collinearity. Functional characterization of two orthologs from the dicot Arabidopsis (Arabidopsis thaliana; Gibberellic Acid-Stimulated Arabidopsis 10) and the monocot rice (Oryza sativa; Gibberellic Acid Stimulated Transcript-Related 9) suggests hormonal regulation, novel roles in seed germination, and functional conservation among diverse plant groups. Computational modeling predicts that these GAST genes are regulated by several factors, including the phytohormones gibberellin and abscisic acid, through conserved cis-motifs present in their promoters, and that they might act as signaling molecules in a complex feedback loop. Thus, our study identifies GASTs and their encoded proteins, uncovers their structure, novel motifs, and evolutionary pattern among plants, and suggests their functional conservation.
Gibberellins (GAs) are a class of tetracyclic diterpenoid plant hormones involved in the development of roots, trichomes, flowers, fruits, leaves, and seeds, as well as stem elongation and leaf expansion (Griffiths et al., 2006). GA signaling functions as a derepressible system. It is perceived by GA receptor GA-INSENSITIVE DWARF1 (GID1) and interacts with the DELLA (conserved aspartate-glutamate-leucine-leucine-alanine motif containing protein) repressor protein, triggering the degradation of DELLA by the ubiquitin-proteasome pathway, resulting in the liberation of downstream genes (Ueguchi-Tanaka et al., 2005).
Members of Gibberellic Acid Stimulated Transcript (GAST) gene family are regulated by GA and encode small polypeptides that have a C-terminal domain of ∼60 amino acids. The first member of this gene family, GAST1, was identified as a GA-stimulated gene in tomato (Solanum lycopersicum; Shi et al., 1992). To date, 13 homologs of GAST have been identified in Arabidopsis (Arabidopsis thaliana;Gibberellic Acid-Stimulated Arabidopsis [AtGASAs]), and most of them are regulated by phytohormones like GA (Roxrud et al., 2007). Members of the GAST gene family were found to be involved in various biological processes including shoot elongation and vegetative-to-reproductive phase transition (GIP1 [Gibberellin-induced Gene from petunia (Petunia hybrida)], GIP2, GIP4, and GIP5 ); cell elongation during fruit ripening in strawberry (Fragaria x ananassa; FaGASAs); wounding and pathogen infection in potato (Solanum tuberosum; Snakin-1, Snakin-2, and Snakin-3); cell expansion in Gerbera (Gerbera Homolog of GAST); panicle differentiation and cross talk of GA and brassinosteroid signaling in rice (Oryza sativa; OsGASR1, OsGASR2, and OsGSR1); lateral root formation in maize (Zea mays; ZmGSL1); determination of flowering time (AtGASA4 and AtGASA5); and abiotic stress resistance (AtGASA14; Kotilainen et al., 1999; Berrocal-Lobo et al., 2002; Ben-Nissan et al., 2004; de la Fuente et al., 2006; Furukawa et al., 2006; Wang et al., 2009; Zhang et al., 2009; Rubinovich and Weiss, 2010; Zimmermann et al., 2010; Sun et al., 2013; Nahirñak et al., 2016). Since GASTs are GA inducible, they may also be involved in GA signaling and biosynthesis, which remains to be studied.
GA biosynthesis pathways were first studied in the fungus Gibberella fujikuroi and subsequently in diverse plant species (Hedden et al., 2001). The biochemical origins of different bioactive GAs are found to be from geranylgeranyl diphosphate (GGPP), a common C20 precursor for diterpenoids. Though GGPP has not been studied elaborately, GGPP-derived metabolites (bioactive GAs) and their associated enzymes encoded by respective genes are well characterized (Yamaguchi, 2008). Like seed plants, fungi (e.g. G. fujikuroi and Phaeosphaeria) also produce GA; however, GA biosynthesis and signaling vary distinctly from fungi to angiosperms. In fungi, a single bifunctional terpene cyclase is involved in conversion of GGPP to ent-kaurene (a key step in GA biosynthesis). However, the physiological role of GA in fungi has not been identified (Hedden et al., 2001; Hedden and Sponsel, 2015). Physcomitrella patens (a model bryophyte and early non-vascular land plant/moss), also encodes a single bifunctional terpene cyclase gene, similar to fungi. In the due course of evolution, GA-GID-DELLA signaling components that arose in bryophytes (Hirano et al., 2007) showed no interaction in P. patens, but interacted in early vascular lycophytes/pteridophytes (like Selaginella moellendorffii and Selaginella kraussiana; Hirano et al., 2007). Subsequently, the GID-DELLA interaction became responsive to GA stimulus and plants started to respond to DELLA-mediated growth arrest (Yasumura et al., 2007). During the evolution of land plants, GA-mediated signaling was refined and seemed to have evolved in parallel with the GA-DELLA system for regulating growth and development. However, our understanding of the regulatory mechanism acting in or downstream of the GA-GID-DELLA signaling remains largely unexplored and enigmatic. Despite the wide presence of the GA-GID-DELLA signaling module and the GGPP-derived GA biosynthesis pathway, very little is known about the evolution of downstream/upstream components of GA signaling and their conservation/divergence across the plant kingdom.
In this study, we identify the origin of GASTs, explore the evolution of their orthologs across the plant kingdom, and hypothesize their putative role in the GA signaling pathway. Besides their evolutionary conservation, we also highlight their functional conservation. Here, we outline a workflow for the identification of GAST-like genes across the plant kingdom (from algae to angiosperms). We found that GAST-like genes evolved in vascular plants after the bryophyte divergence, whereas pteridophytes and very few gymnosperms have unannotated GAST orthologs (GORs). GORs have 11 conserved Cys residues (distributed in three conserved motifs) at their C-terminus along with a Leu-rich N-terminal motif in some of the sequences. Phylogenetic studies predicted that GASTs evolved initially with motif1 and acquired subfunctionalization in successive conjugation with other motifs through the activity of transposable elements in S. moellendorffii. The presence of conserved motifs in the GAST domain is responsible for the discrete evolution of GORs, which are highly conserved across plant species. Differential expression analysis indicated that Ath-GASA10 and Osa-GASR9 (Gibberellic Acid Stimulated Transcript-Related9) play a role in seed germination, and mutational studies validated that Ath-GASA10 and Osa-GASR9 are functionally conserved in dicots and monocots. In silico annotation analysis suggests that Ath-GASA10 and Osa-GASR9 act as signal molecules for the regulation of the GA biosynthetic pathway either by GA induction or in a negative feedback loop.
RESULTS AND DISCUSSION
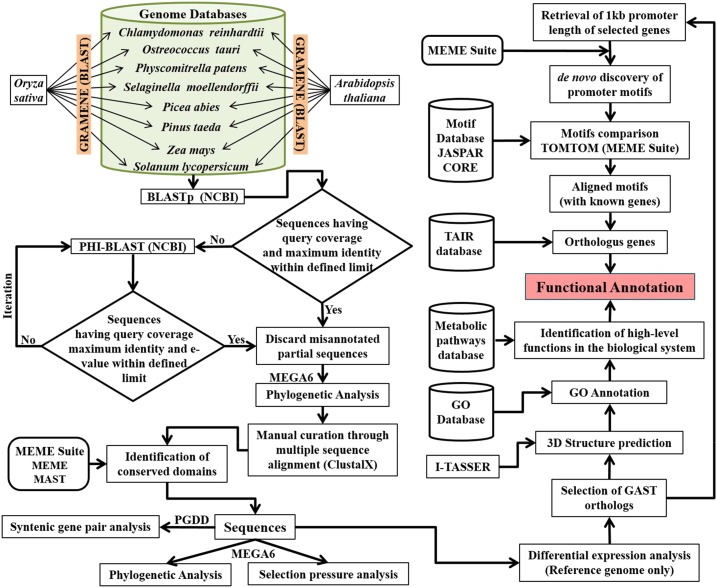
Although the GA signaling pathway is known and extensively studied in angiosperms, the origin, distribution, and evolution of GA-regulated GASTs remains largely unknown (Zhong et al., 2015). We customized a workflow for the identification of GORs across the plant kingdom. Further, we predicted and characterized novel conserved motifs, uncovered their evolutionary pattern, and demonstrated functional conservation (Fig. 1).
Figure 1.
A flowchart was created for the identification of GORs and functional annotation. The existence of GORs was screened across the plant kingdom. The rice and Arabidopsis genomes were taken as references. 3D structures of GAST proteins (I-TASSER) and promoter motifs (MEME and TOMTOM) were predicted for functional annotation (MEME suite and the KEGG pathway).
GORs Evolved in Pteridophytes After the Divergence from Bryophytes
Among four angiosperm species studied, we identified 12 and 13 GASTs in maize and tomato, respectively. Among 11 GASTs identified in rice, we found one additional sequence, Os10g02625 (Osa-GASR11), and 10 GASTs reported previously (Zimmermann et al., 2010; Supplemental Table S1). Similarly, 15 GORs were identified in Arabidopsis, including 13 previously known members (Roxrud et al., 2007) and two additional GASTs—Ath-GASA13 (AT3G10185) and Ath-GASA15 (AT1G10588; Supplemental Table S1). In this study, GASTs from Arabidopsis and rice were used as references. Surprisingly, we did not find any GASTs in two gymnosperm species (Picea abies and Pinus taeda). However, we have identified a total of 18 GASTs in four other gymnosperms (Pinus pinaster, Pinus tabuliformis, Picea sitchensis, and Picea mariana; Supplemental Table S2) from the National Center for Biotechnology Information database. Though some of the gymnosperms have orthologs of the GA receptor (GID1 and SLEEPY1) and DELLA proteins (Vandenbussche et al., 2007), the complete absence of these orthologs in some gymnosperms might be due to successful adaptive modifications which arose in lineages of gymnosperms that survived after the Eocene epoch, where extreme weather conditions (sharp cooling and drying) caused the diversification and extinction of many lineages of gymnosperms, and led to frequent parallel or convergent evolution (Wang et al., 2000; Ran et al., 2006; Pittermann et al., 2012; Wang and Ran, 2014). However, we cannot rule out the possibility that the absence of GORs in some gymnosperms might be due to the lack of proper annotation. Interestingly, we identified 11 GORs in S. moellendorffii (query coverage >80% and maximum identity >90%) that have not been characterized and annotated previously (Supplemental Table S2). Further, in species like Chlamydomonas reinhardtii and Ostreococcus tauri (unicellular algae), and P. patens (bryophyte), we found that some sequences showed less similarity to references GASTs (Supplemental Table S2).
An unrooted maximum-likelihood (ML) tree was reconstructed using 174 protein sequences retrieved from databases. The branches and sub-branches showed a very low bootstrap value (<30); however, similar sequences were grouped hierarchically (Supplemental Fig. S1). In the tree, GOR sequences were distributed in two clusters, with cluster I having sequences highly similar to reference GASTs and cluster II, divided into subclusters A and B, having sequences both highly similar and less similar to reference GASTs. Subcluster A consisted of sequences with less similarity to references and subcluster B consisted of highly similar sequences from gymnosperms and angiosperms along with pteridophytic S. moellendorffii (Supplemental Fig. S1). The rest of the less similar sequences, from C. reinhardtii, O. tauri (algae), P. patens (bryophytes), P. abies, and P. taeda (gymnosperms) were grouped separately in cluster III and had a very low bootstrap value and no GAST domain, suggesting their unrelatedness to the reference (Supplemental Fig. S1; Supplemental Table S2). Further, these 94 less similar sequences were eliminated, and the remaining 80 sequences were used for analysis.
The presence of GAST domains and their clustering pattern in the phylogeny (Supplemental Fig. S1) suggests that GORs first evolved in the vascular land plant S. moellendorffii, a pteridophyte, after the divergence from bryophytes, and simultaneously in the gymnosperms and angiosperms. However, pteridophytic S. moellendorffii and S. kraussiana possess functional GID1 and DELLA homologs capable of forming a complex with GA, whereas GID1- and DELLA-like proteins are unable to form GA-mediated complexes in P. patens (Yasumura et al., 2007; Yamaguchi, 2008). Unlike vascular plants, non-vascular bryophytes (P. patens) lack GA-mediated growth responses due to the absence of a functional GA-GID1-DELLA signaling module (Yasumura et al., 2007), which could be the reason for the absence of GORs in bryophytes and algae or vice-versa.
Positional Conservation of Cys Residues in Identified Novel Conserved GAST Motifs in GORs
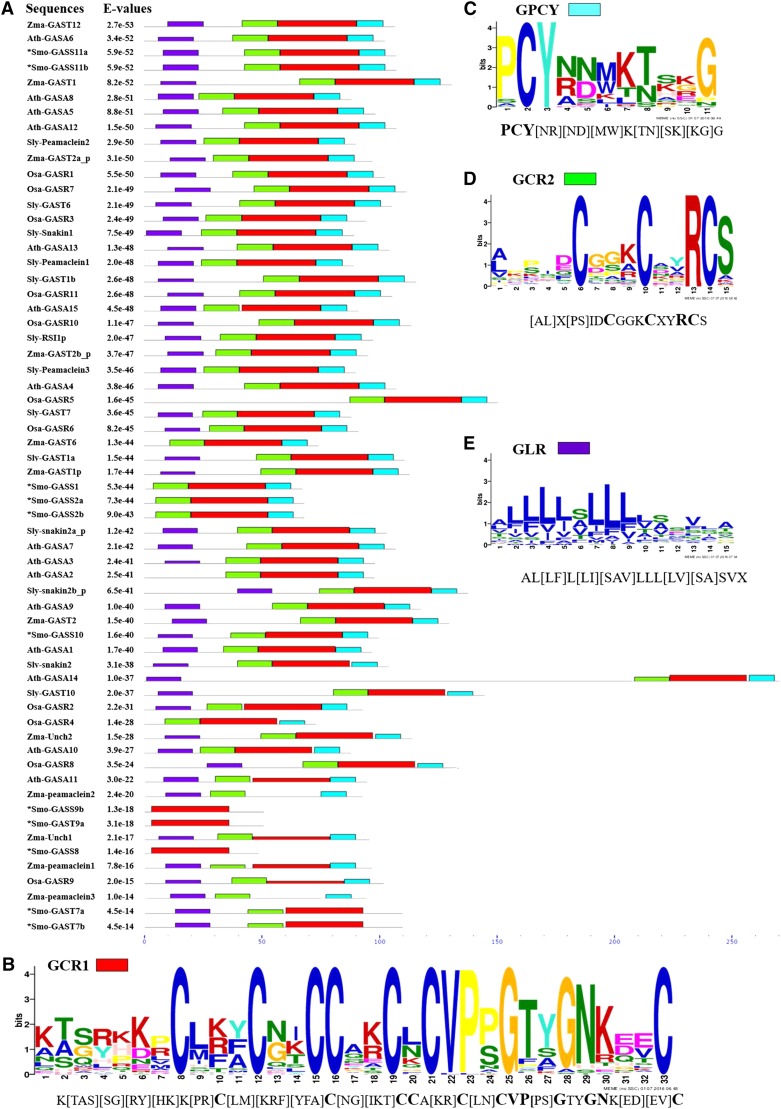
The screened 80 GORs with GAST domains were used for identification of conserved motifs. Using the MEME (Multiple Expectation Maximization for motif elicitation) suite (Bailey et al., 2009) and multiple-sequence alignment (Thompson et al., 1997), we predicted four conserved motifs in 62 highly similar GORs (excluding gymnosperms; Fig. 2; Supplemental Table S2). Based on the bits value of conserved amino acids, motifs were named GAST-Cysteine Rich1 (GCR1; x7Cx3Cx2CCx2CxCVPxGx7), GAST-PCY (GPCY; PCYx3K[TN]x2G), GCR2 (x5Cx3Cx2RCS), and GAST-Leucine Rich (GLR; Fig. 2, B–E). Of the four motifs, three were present consecutively at the C-terminus of GAST sequences (Fig. 2A). GLR has maximum occurrence at the N-terminus, with highly conserved Leu residues (Fig. 2E). Though all four conserved motifs were prevalent among GORs, some of the sequences lacked one, two, or three conserved motifs. The GORs from S. moellendorffii also consist of these conserved motifs (Fig. 2A). Smo-GASS10 (S. moellendorffii Gibberellic Acid-Stimulated Seleginella10), Smo-GASS11a, and Smo-GASS11b have all four motifs; Smo-GASS2a, Smo-GASS2b, and Smo-GASS1 lack the GLR motif; Smo-GASS8, Smo-GASS9a, and Smo-GASS9b have only the GCR1 motif; Smo-GASS7a and Smo-GASS7b lack the GPCY motif (Fig. 2A).
Figure 2.
The distribution of conserved motifs in highly similar GORs is shown. A, Distribution of predicted motifs in 62 highly similar GAST sequences. Identified novel GORs are indicated by asterisks. B to E, Conserved motifs are individually represented as GCR1 (B), GPCY (C), GCR2 (D), and GLR (E). MEME suite v 4.11.2 was used for prediction of the motifs.
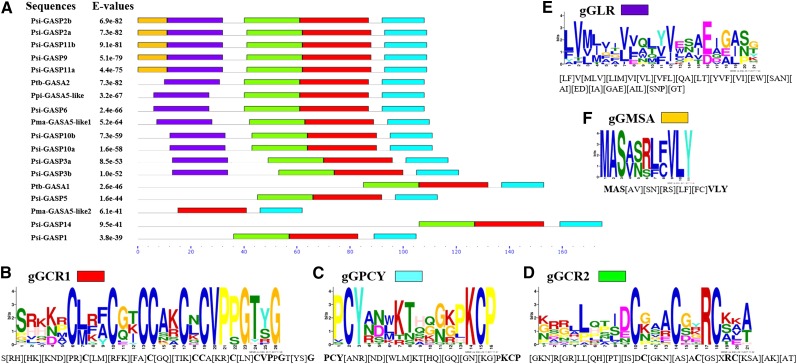
Similarly, we identified five conserved motifs in 18 GORs from gymnosperms (P. pinaster, P. tabuliformis, P. sitchensis, and P. mariana; Fig. 3; Supplemental Table S2). Despite important sequence variation, motifs were very similar, as described above, and were named gymnosperm GAST-Cysteine Rich1 (gGCR1; x5Cx3Cx2CCx2CxCVPxGx2G), gymnosperm GAST-PCY (gGPCY; PCYx3KTx2[GN]xPKCP), gGCR2 (x4Lx4Cx3Cx2RC[KSA]x2), and gymnosperm GAST-Leucine Rich (gGLR; Fig. 3, B–F). GLR has maximum occurrence at the N-terminus, with highly conserved Leu residues (Fig. 3E), which is similar to the case for GORs excluding gymnosperms (Fig. 2E). The fifth novel motif, GAST-MSA (gGMSA; MSAx5VLY), was also predicted by MEME in some of the gymnosperm GORs, namely, Psi-GASP2a (P. sitchensis Gibberellic Acid-Stimulated in Pinaceae2a), Psi-GASP2b, Psi-GASP9, Psi-GASP11a, and Psi-GASP11b (Fig. 3, A and F). The three conserved motifs GCR1, GPCY, and GCR2 (Fig. 2, B–D), and their gymnosperm counterparts gGCR1, gGPCY, and gGCR2 (Fig. 3, B–D), have 11 highly positionally conserved Cys residues at the C-terminus (Figs. 2 and 3), though their sequences are not identical. Based on the variable size of the GAST domain, some level of functional variation in GASTs was previously assumed (Roxrud et al., 2007), but our results, which show tight conservation of Cys residues in GAST motifs, rather suggest functional conservation of GORs, especially those with complete motifs.
Figure 3.
The distribution of conserved motifs in GORs from gymnosperm is shown. A, Distribution of the predicted motifs in 18 GAST sequences from P. pinaster, P. tabuliformis, P. sitchensis, and P. mariana. B to F, Conserved motifs were individually represented as gGCR1 (B), gGPCY (C), gGCR2 (D), gGLR (E), and gGMSA (F). MEME suite v4.11.2. was used for the prediction of motifs.
Conserved GASTs Evolved through Subfunctionalization of Motifs Which Originated in Pteridophytes
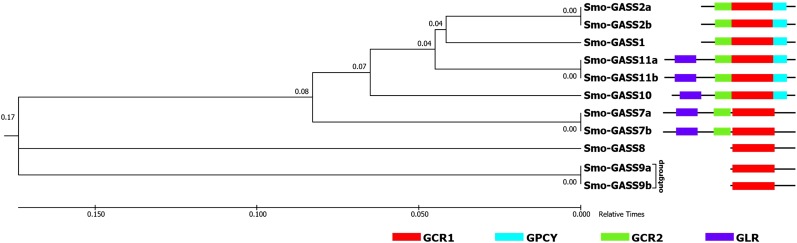
The phylogeny, conserved motifs, and multiple-sequence alignment revealed that the GORs originated in S. moellendorffii. Moreover, our observations indicated that the S. moellendorffii sequences have different combinations of motifs (Fig. 2A). We here show the origin and evolution of GAST domain architecture using a TimeTree of S. moellendorffii GASTs. A TimeTree was reconstructed using the RelTime method (Tamura et al., 2012) inferred from the neighbor-joining method with sequences of S. moellendorffii GAST domains. We observed that GCR1 evolved initially in the sequences of Smo-GASS8, Smo-GASS9a, and Smo-GASS9b, suggesting that GCR1 is the most ancient (Fig. 4). Further, in the relative time frame, Smo-GASS7a and Smo-GASS7b evolved with GCR1, GCR2, and GLR, followed by the evolution of Smo-GASS10, which had acquired GPCY and was the first to possess four complete motifs (Fig. 4). In the subsequent evolution, Smo-GASS11a and Smo-GASS11b acquired all four motifs. Unexpectedly, Smo-GASS1 lost GLR and remained with GCR1, GCR2, and GPCY in the course of evolution (Fig. 4). The sequences Smo-GASS2a and Smo-GASS2b evolved later from Smo-GASS1 with the same combination of motifs. The GAST domain (comprised of GCR1, GCR2, and GPCY) and GLR evolved simultaneously in S. moellendorffii with different combinations of motifs (Fig. 4). We found that the GAST domain motifs are not identical or recurring sequences, suggesting that they are not the repeats of ancient GCR1. Thus, the evolution and rearrangement of motifs did not arise from a duplication event, as S. moellendorffii is an ancient organism with a small genome and a low rate of evolution (Baniaga et al., 2016) and did not arise from either whole-genome duplication or polyploidy events (Banks et al., 2011). Interestingly, it was also found that the S. moellendorffii genome has transposable elements (TEs) in high proportion (37.5%; Banks et al., 2011). It is plausible that the GASTs evolved initially with GCR1 and later acquired subfunctionalization through successive conjugation of other motifs through the activity of TEs in S. moellendorffii.
Figure 4.
The reconstructed TimeTree was used to predict the evolution of S. moellendorffii GASTs containing conserved motifs in a relative time frame. A TimeTree was reconstructed from S. moellendorffii GAST orthologs using the RelTime method inferred from neighbour joining in MEGA7 (Kumar et al., 2016). The TimeTree indicated the evolution of GASTs according to the presence of conserved motifs. Smo-GASS9a and Smo-GASS9b were taken as the outgroup and the distance was calculated in a relative time of 1. On the right, the GAST sequences are shown with their respective motifs arranged according to their evolution. The Smo-GASS sequences and length of the motifs do not represent actual scale.
Evolution of the GAST Domain from Ancient GCR1 through Subfunctionalization is Responsible for the Discrete Evolution of GORs
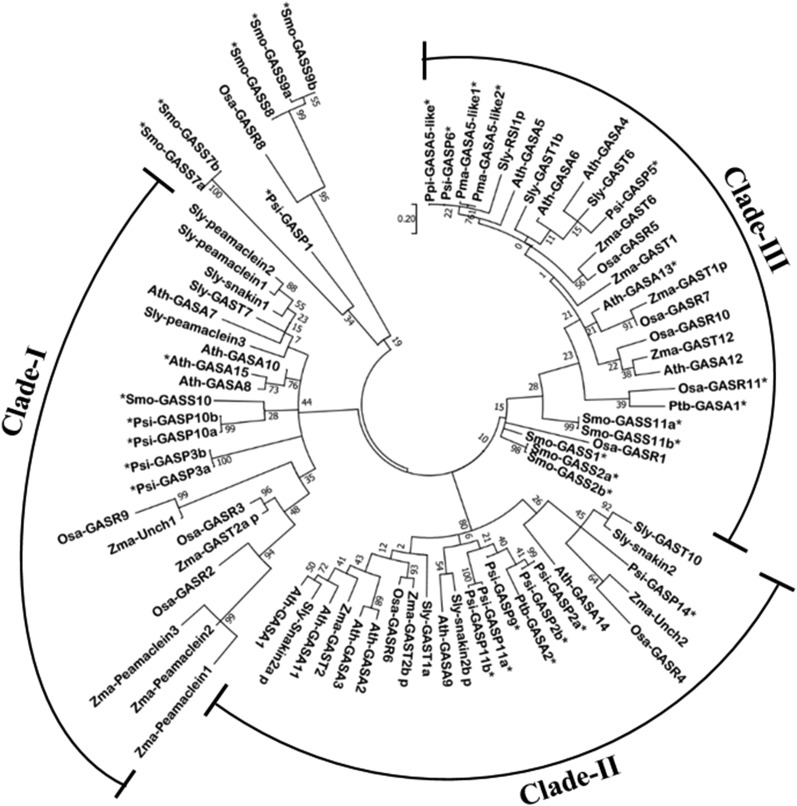
To understand the relatedness among the GORs, we reconstructed one unrooted ML tree using GAST domain sequences of 80 short-listed GORs (Fig. 5). In the ML tree, GASTs diverged from two clusters, one containing sequences of Osa-GASR8, Smo-GASS8, Smo-GASS9a, and Smo-GASS9b, and the other cluster containing Smo-GASS7a and Smo-GASS7b along with Psi-GASP1. Interestingly, only rice Osa-GASR8, with a low rate of substitution, was clustered with Smo- GASS8, Smo-GASS9a, and Smo-GASS9b, and Psi-GASP1, also with a very low rate of substitution, diverged from Smo-GASS7a and Smo-GASS7b (Fig. 5). The rest of the GASTs were diverged and grouped into three clades labeled I, II, and III. Clades I and III have very diverse sequences with highly variable rates of substitution, in comparison to clade II (Fig. 5). In clade I, Osa-GASR9 and Zma-Unch1 (Z. mays-Uncharacterized protein), with high rates of substitution, grouped discretely and evolved faster. Zma-Peamaclein1, Zma-Peamaclein2, and Zma-Peamaclein3, with very high substitution rates, clustered together and diverged from Osa-GASR2. Psi-GASP3a and Psi-GASP3b, with similar sequences, were placed discretely in clade I. However, Psi-GASP10a and Psi-GASP10b, with similar sequences and high rates of substitution, clustered together and diverged from Smo-GASS10. Among four Arabidopsis sequences of clade I, Ath-GASA7 separated with a higher substitution rate from Ath-GASA8, Ath-GASA10, and Ath-GASA15, which grouped together (Fig. 5). The clustering of GORs in this clade might be due to the similarity of the motif sequence to Smo-GASS10, which eventually evolved from Smo-GASS7a and Smo-GASS7b (Fig. 5).
Figure 5.
Unrooted maximum-likelihood phylogenetic tree of GORs using MEGA6. The maximum-likelihood tree was reconstructed using the conserved C-terminal domains of 80 GORs. The bootstrap value was estimated from 1000 replicates. The scale bar represents the amino acid substitution rate. Novel GORs are indicated by asterisks.
Clade II consisted of GORs with less variable rates of substitution and divergence and lacked any S. moellendorffii GORs. Osa-GASR4 and Zma-Unch2, with high substitution rates, diverged hierarchically from Sly-GAST10, Sly-snakin2, Psi-GASP14, and Ath-GASA14 (Fig. 5). Ath-GASA2 and Ath-GASA3 evolved faster than other Arabidopsis GASA sequences in clade II. Psi-GASP2a, Psi-GASP2b, Psi-GASP9, Psi-GASP11a, Psi-GASP 11b, and Ptb-GASA2 grouped together and diverged from Ath-GASA9 and Sly-snakin2b_p (Fig. 5). Clustering of only these gymnosperm GORs, which have a fifth motif, suggests that the relative similarity between their GAST domains or motifs is responsible for their specific clustering in the ML tree and evolutionary pattern.
Clade III consisted of the rest of the S. moellendorffii sequences and some other GORs (Fig. 5). Smo-GASS1, Smo-GASS2a, Smo-GASS2b, and Osa-GASR were the first to diverge in clade III. The subsequent divergence has shown hierarchal separation of the cluster of identical sequences including Smo-GASS11a and Smo-GASS11b and the cluster including Osa-GASR11 and Ptb-GASA1 (Fig. 5). These Smo GORs evolved gradually (Fig. 4). Further, Sly-RSI1p, with a high rate of substitution, grouped together with four gymnosperm GASTs, which had diverged from Ath-GASA5. Ath-GASA4 showed higher rates of substitution in clade III (Fig. 5). The clustering of GORs in these three clades was due to their similarity with the gradually evolved conserved motifs (Fig. 4). Furthermore, these GORs were discretely distributed in the phylogeny due to the variability in occurrence of motifs and distance between motifs, which shows their discrete evolutionary pattern.
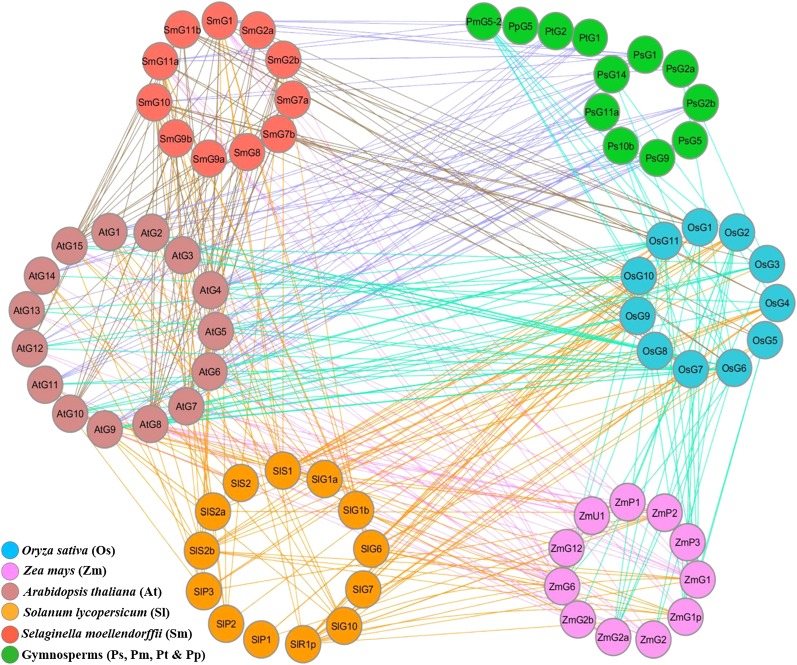
Despite Variable Coverage of Collinearity, Most GORs Show Conservation
To understand the evolution of GASTs and their retention across plant species, we identified duplicated gene pairs in genomic regions that were syntenic using the Plant Genome Duplication Database (PGDD; Lee et al., 2013). Further, the syntenic pairs were shown as a network (Fig. 6) from the log scores provided by PGDD (Hammoudi et al., 2016). Most of the identified S. moellendorffii GORs showed variable coverage of collinearity with GORs from other species. We observed that 45% of S. moellendorffii GORs were collinear with 22% of GORs from gymnosperm (P. mariana-GASA5-like, P. pinaster [Ppi]-GASA5-like, Ppi-Unch8, and Ppi-Unch13; Fig. 6). All S. moellendorffii GORs except Smo-GASS7b showed collinearity with 60% of Arabidopsis GORs and 81% of rice GORs. Smo-GASS7b showed collinearity only with Osa-GASR1, Osa-GASR10, and Osa-GASR11. Similarly, 38% of GORs from tomato and 54% of GORs from maize also showed collinearity with 36% of S. moellendorffii GORs (Fig. 6). All of the primitive S. moellendorffii GORs (Smo-GASS8, Smo-GASS9a, and Smo-GASS9b), with only the GCR1 motif, and the gradually evolved Smo-GASS7a, with GCR1, GCR2, and GLR motifs, shared collinearity only with Arabidopsis GORs (Fig. 6). A possible reason for the maximal syntenic pairing of GORs between these two species is that both genomes have striking similarity in terms of genome size, gene density, and occurrence of TEs (Banks et al., 2011). Several GORs from rice, maize, tomato, and gymnosperms shared collinearity with Smo-GASS10, which has all four motifs, and they clustered together in clade I, indicating their conservation (Fig. 5). The Smo-GASSs (Smo-GASS1, Smo-GASS2, Smo-GASS6, Smo-GASS4, and Smo-GASS3), gradually evolved after Smo-GASS10, shared collinearity in various capacity with GORs from other species, and clustered together in clade III, indicating their evolutionary conservation (Fig. 6). Interestingly, the most recent pteridophytic GOR, Smo-GASS2a, shared collinearity with the next evolved GORs from gymnosperms (Psi-GASP1), Arabidopsis (dicot), and rice (monocot), indicating their conservation and evolutionary lineage.
Figure 6.
Collinearity network among the GORs to understand conservation using Cytoscape v3.6.0. The collinearity network was drawn between syntenic gene pairs using scores provided by PGDD. The “Organic Layout” in Cytoscape v3.6.0 was used for the network representation and optimized manually in species-specific clusters. Specific colors were used to represent each species (colored nodes) in the collinearity network and the colored edges represent specific syntenic gene pairs across the species used for this study. The names of the GORs were shortened to fit in the nodes of the network. For “AtG1,” for example, the first two letters represent the genus and species, the next letter represents the gene name, and the number indicates the numeric value for the the gene family member (Supplemental Table S1).
Though Selaginella evolved many million years ago, prior to the angiosperms, the sequences of GORs remain unperturbed. The whole genome duplication (WGD) event resulted in a higher ploidy level, which was responsible for the evolution and speciation of vascular plants (Barker et al., 2016). Even though all seed and flowering plants have undergone at least one round of polyploidy (Li et al., 2015), it did not strongly affect the sequences of conserved GORs. A possible reason is that unlike more evolved monocots (rice and maize), in wild Arabidopsis weed, selection was not influenced by domestication.
Further, 50% and 22% of identified gymnosperm GASTs showed collinearity with 93% of Arabidopsis and 72% of rice GORs, respectively, besides S. moellendorffii (Fig. 6). Arabidopsis GORs, except Ath-GASA2, shared the maximum number of syntenic pairs with gymnosperm GORs (Fig. 6). Interestingly, gymnosperm GORs did not show any synteny with GORs from tomato and maize, which are evolutionarily advanced, partly because of the influence of domestication. We also observed that the slight sequence variation in GAST motifs gGCR1 and gGLR, and the presence of gGMSA in gymnosperms, may cause some level of functional diversification, despite overall strong conservation (Fig. 5). Additionally, the small number of sequenced gymnosperms and poor annotation may affect their collinearity with other species. It is possible that more refined sequencing and annotation of diverse gymnosperms may provide better synteny or collinearity of GORs.
Additionally, 72%, 27%, and 9% of rice GORs showed collinearity with all of the Arabidopsis GORs, 84% of tomato GORs, and 50% of maize GORs (Fig. 6). Similarly, only 7% of Arabidopsis GORs did not show collinearity with any of the Z. mays GORs, while Ath-GASA5, Ath-GASA12, and Ath-GASA13 (20%) did not show collinearity with any of the tomato GORs (Fig. 6). Likewise, 50% of maize GORs have shown collinearity with 77% of tomato GORs (Fig. 6). The maximum number of collinear pairs between rice, maize, and tomato have shared collinearity according to the evolutionary pattern of conserved motifs following their conservation (Fig. 6). Also, the collinearity network suggests that GCR1, along with GPCY and three of GAST domain are highly conserved among the syntenic gene pairs across the plant species used in the current study.
Ath-GASA10 and Osa-GASR9 Act as Positive Regulators of Seed Germination
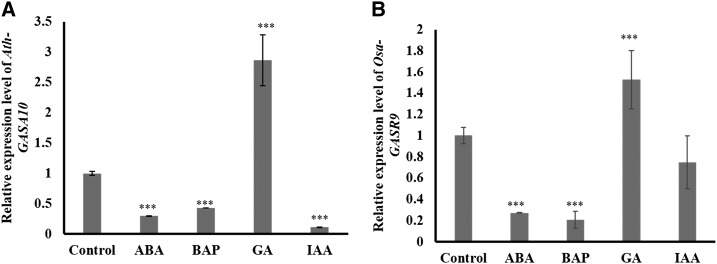
Earlier reports have suggested that GASTs were induced by hormonal treatment (Shi et al., 1992; Wang et al., 2009; Sun et al., 2013). We selected a pair of related orthologs, Ath-GASA10 (a dicot) and Osa-GASR9 (a monocot), from clade I (Fig. 5) to experimentally validate their functional conservation and their response to phytohormones. We observed that GA and ABA induced and reduced the expression level of both Ath-GASA10 and Osa-GASR9 in Arabidopsis and rice seedlings, respectively (Fig. 7, A and B). We also observed similar response of Ath-GASA10 to GA and ABA treatments in germinating Arabidopsis seeds (Supplemental Fig. S2). This indicates their potential role in seed germination (Supplemental Fig. S2), as phytohormones GA and ABA play antagonistic roles during seed germination. Earlier, some members of the Arabidopsis GASA family (AtGASA4 and AtGASA6) were also found to be involved in seed germination (Olszewski et al., 2002; Roxrud et al., 2007; Zhong et al., 2015).
Figure 7.
Expression patterns of Ath-GASA10 and Osa-GASR9 upon treatment with phytohormones. The relative expression patterns of Ath-GASA10 and Osa-GASR9 upon treatment with 10 µm of ABA, BAP, GA, and IAA for 12 h are shown. A, Expression of Ath-GASA10 in Arabidopsis seedling is shown at 5 d after germination. B, Expression of Osa-GASR9 in rice seedlings is shown at 7 d after germination. Error bars indicate the se of three independent experiments. One-way ANOVA was performed. Asterisks indicate significant statistical differences: ***P < 0.001, **P < 0.01, and *P < 0.05.
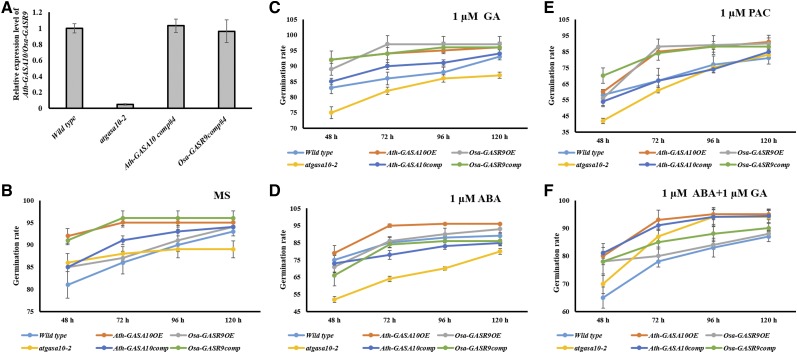
Further, to investigate the potential role of Ath-GASA10 and Osa-GASR9 in seed germination, we analyzed the loss-of-function mutant of Ath-GASA10 (atgasa10-2) and the overexpression line of Ath-GASA10 and Osa-GASR9 (Ath-GASA10OE and Osa-GASR9OE, respectively) in Arabidopsis (Fig. 8). In control conditions, the atgasa10-2 mutant (89%) showed a slower rate of seed germination than wild type (93%) after 120 h (Fig. 8B). Ath-GASA10OE seeds (92%) showed a slightly faster seed germination rate than wild type (81%) after 48 h (Fig. 8B). Upon treatment with 1 µm ABA after 48 h, the difference in the rate of seed germination between wild type (75%) and atgasa10-2 (53%) increased further, whereas it showed less severe impact on germination of Ath-GASA10OE seeds (79%; Fig. 8E). Treatment with GA increased the seed germination rate of Ath-GASA10OE (92% at 48 h) more than that of wild type (83% at 48 h), indicating that Ath-GASA10 plays a role in seed germination in a GA-dependent manner (Fig. 8C). This observation was further strengthened by analyzing the germination rate of transgenic seeds after treatment with paclobutrazol (PAC), whose effect on seed germination rate was in contrast to that of GA (Zhong et al., 2015; Fig. 8D). Since GA and ABA treatment inversely altered the expression of Ath-GASA10, we asked if the lower seed germination rate of atgasa10-2 upon ABA treatment could be rescued by GA treatment. We observed that application of GA with ABA was able to reduce the difference in seed germination between atgasa10-2 and wild type. Upon ABA treatment, only 52% of atgasa10-2 seeds germinated after 48 h, but GA+ABA treatment improved seed germination of atgasa10-2 (70%; Fig. 8F). Osa-GASR9OE seeds showed a germination rate similar to that of Ath-GASA10OE upon treatment with ABA, GA, and PAC, which indicates the functional conservation of Osa-GASR9 and Ath-GASA10 in monocots and dicots, respectively (Fig. 8; Supplemental Table S3).
Figure 8.
Ath-GASA10 and Osa-GASR9 are functionally conserved in the regulation of seed germination. A, The relative expression level of Ath-GASA10 in wild type (Col-0), atgasa10-2, and T1 plants of pAth-GASA10:Ath-GASA10 (Ath-GASA10comp) is shown; expression of OsaGASR9 in T1 plants of pAth-GASA10:OsaGASR9 (Osa-GASR9comp) was analyzed by real-time quantitative PCR. B to F, Arabidopsis seed germination was assayed; seeds of wild type, atgasa10-2, Ath-GASA10comp, Osa-GASR9comp, Ath-GASA10OE, and Osa-GASR9OE were sown on one-half strength MS media with or without phytohormones (PAC, GA, or ABA individually or a combination of GA and ABA). Average germination rates are shown for seeds in one-half strength MS medium (%) without hormone treatment (B), and in one-half strength MS medium supplemented with 1 µm GA (C). 1 µm PAC (D), 1 µm ABA (E), and 1 µm GA + 1 µm ABA (F). The germination experiment was performed in four replicates, and 25 seeds were used in each experiment. Error bars indicate the se.
Rice Osa-GASR9 Can Rescue the Seed Germination Phenotype of Arabidopsis atgasa10-2
To genetically confirm the functional conservation of Ath-GASA10 and Osa-GASR9, we transformed pAth-GASA10:Ath-GASA10 and pAth-GASA10:Osa-GASR9 binary constructs into the atgasa10-2 background. Lines showing expression levels near to those of wild type plants were selected for further analysis (Fig. 8A). Seed germination assay of pAth-GASA10:Ath-GASA10 (Ath-GASA10comp) and pAth-GASA10:Osa-GASR9 (Osa-GASR9comp) transgenic lines showed that 65% were able to rescue the low germination efficiency of atgasa10-2. Seed germination rates of Ath-GASA10comp and Osa-GASR9comp transgenic lines in atgasa10-2 background were comparable to that of wild type plants (Fig. 8). These results indicate that both Ath-GAS10 and Osa-GASR9 are involved in the GA and ABA signaling pathway-mediated seed germination, suggesting their functional conservation, which is also evident from their collinearity (Fig. 6).
Possible Regulation and Function of Ath-GASA10 and Osa-GASR9 As Putative Signal Molecules
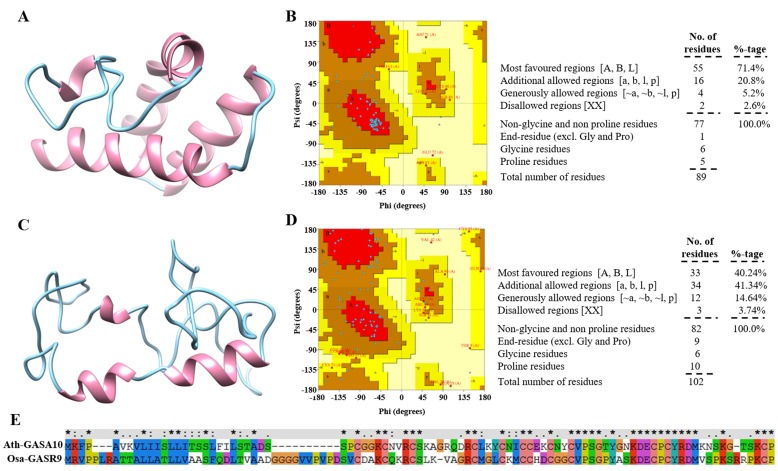
To understand the functional conservation and regulation of Ath-GASA10 and Osa-GASR9, we predicted three-dimensional (3D) structures of Ath-GASA10 (Fig. 9A) and Osa-GASR9 (Fig. 9C) and their stereochemical quality showed 97.4% and 96.3% residues in allowed region of Ramachandran plots, respectively (Fig. 9, B and D), confirming their stable structures. Both proteins have conserved Cys residues and show 42% sequence (Fig. 9E) and 47% structural similarity, which might be due to the presence of conserved motifs. Some of the members of GAST-like genes in Arabidopsis, tomato, rice, and petunia are regulated by GA (Shi et al., 1992; Aubert et al., 1998; Ben-Nissan et al., 2004; Furukawa et al., 2006; Roxrud et al., 2007). Our analysis showed that Ath-GASA10 and Osa-GASR9 expression is antagonistically regulated by GA and ABA (Fig. 7, A and B), indicating their complex regulation, which remains elusive. Therefore, we analyzed the Enzyme Commission (EC) numbers and gene ontology (GO) terms of both Ath-GASA10 and Osa-GASR9 and annotated their molecular function in metabolic pathways (Supplemental Table S4) using 3D structure modeling, which indicates some overlapping functions (Supplemental Table S4).
Figure 9.
Modeled 3D structure of Ath-GASA10 and Osa-GASR9 and its quality check. The protein 3D structures of Ath-GASA10 (A) and Osa-GASR9 (C) were predicted and stereochemical quality was checked using I-TASSER and PROCHECK, respectively. B and D, Ramachandran plot analysis was performed on Ath-GASA10 (B) and Osa-GASR9 (D). 3D structure was visualized through Chimera v 1.6.2. The α-helix is represented in magenta and the coiled coil in cyan. E, Multiple-sequence alignment between Ath-GASA10 and Osa-GASR9 is shown.
Further, we identified cis-motifs and their distribution in upstream promoter sequences of Ath-GASA10 and Osa-GASR9, and we predicted their regulatory function (Supplemental Fig. S3; Supplemental Table S5). We observed the presence of at least five conserved cis-motifs (Supplemental Fig. S3), which may allow the binding of different factors/regulators that have different biological functions such as ABA response, GA signaling/biosynthesis, lateral root development, etc. (Supplemental Fig. S3; Supplemental Table S5).
The best consensus putative function derived for Ath-GASA10 indicates its regulatory role in geranylgeranylation (GO:2000112). The geranylgeranylation process implicates the attachment of one or two 20-carbon lipophilic geranylgeranyl isoprene units from GGPP to one or two Cys residue(s) at the C-terminus of specific proteins (GASTs have Cys-rich C-termini). The geranylgeranylated protein acts as a membrane anchor (Jiang et al., 1995), also supported by the predicted enzymatic activity through Sphingomyelin phosphodiesterase (EC3.1.4.12), which is involved in a membrane lipid metabolic process (GO:0006643; Supplemental Table S4). The GGPP is the precursor of GA, ABA, and the brassinosteroid biosynthetic pathway (Supplemental Fig. S4). Subsequently, these findings are also supported by the presence of promoter motif4, which is responsive to ABA (Supplemental Table S5).
Similarly, the best consensus putative function assigned for Osa-GASR9 indicates its role in the carboxylic acid biosynthetic process (GO:0046394) and the GA biosynthetic process (GO:0009686), and it is well supported by the predicted enzymes EC4.1.1.37 and EC1.13.11.12, indicating its involvement in the formation of the plant hormones jasmonic acid and GA (Supplemental Fig. S4; Supplemental Table S4). The presence of promoter motifs involved in the regulation of lateral root formation, brassinosteroid signaling, seed germination, and GA biosynthesis (promoter motif5) further support this regulation (Supplemental Fig. S3; Supplemental Table S5).
Together, our findings suggest that the functionally conserved Ath-GASA10 and Osa-GASR9 might act as signal molecules for regulation of the GA biosynthetic pathway by GA-mediated induction and/or in a negative feedback loop through a complex regulatory network involving hormone signaling (Supplemental Fig. S5). Our results show GA- and ABA-mediated regulation of Ath-GASA10 and Osa-GASR9 expression. These computational analysis results provide a hypothetical framework for regulation and function of Ath-GASTs and Osa-GASTs, which opens up new avenues for their further characterization and hormonal regulation, as well as their molecular function in different plant physiology and developmental processes.
CONCLUSION
In vascular plants, GA signaling is mediated by the functional GA-GID1-DELLA signaling module, which is non-functional in non-vascular bryophytes (Yasumura et al., 2007), possibly due to the absence of some unknown signaling component. Poorly studied GASTs may be one such component, which have evolved in pteridophytes and are missing in bryophytes. Our results suggest that GASTs are conserved among vascular plants due to the presence of conserved motifs. Phylogenetic studies indicate that GASTs with the GCR1 motif initially evolved in S. moellendorffii, where GASTs with four motifs evolved through successive conjugation of other motifs and subfunctionalization.
The syntenic pair analysis suggests strong conservation of GASTs across vascular plants, which is acquired through moderate reshuffling of sequences. Two closest orthologs from Arabidopsis (Ath-GASA10) and rice (Osa-GAST9) showed structural similarity and functional conservation between dicot and monocot plants, indicating their novel role in seed germination. We also hypothesize that both Ath-GASA10 and Osa-GASR9 are regulated by phytohormones, through conserved motifs in their promoters, and may act as signal molecules in a possible feedback loop in GA- or ABA-mediated regulation.
MATERIALS AND METHODS
Identification of GORs and Sequence Retrieval
The devised flowchart (Fig. 1) was used for the identification of GORs from selected representative species with completely sequenced genomes from each phylum except the gymnosperms (Supplemental Table S1). Initially, two gymnosperm species (P. abies and P. taeda) were taken; later, GAST orthologs were searched in all sequenced gymnosperm species (Supplemental Table S1). First, we retrieved the sequences of all GAST family members from rice (Oryza sativa) and Arabidopsis (Arabidopsis thaiana), and used them as references for the identification of ortholog sequences in other selected genomes using protein BLAST in Gramene (http://ensembl.gramene.org/Tools/Blast?db=core) and the National Center for Biotechnology Information database (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastp&PAGETYPE=BlastSearch&LINKLOC=blasthome). The blast parameters used were: query coverage >80% and maximum identity >90%. For the sequences that did not comply with these criteria, reference GASTs domain were searched through iteration using Position-specific Iterated-BLAST (PHI-BLAST) with altered parameters (query coverage >30%, maximum identity >30% and e-value <2) and partial sequences were discarded. The identification of conserved domains in the GAST sequences were manually curated through multiple-sequence analysis using MEME suite v4.11.2 (http://meme-suite.org/) and MAST (http://meme-suite.org/tools/mast; Bailey et al., 2009). The nomenclature used in the current study to specify GAST family members was of the form “Ath-GASA1” to indicate Arabidopsis GASA1, and a similar pattern was followed for other species.
Phylogenetic Analysis
Phylogenetic analysis was done using MEGA6 (Tamura et al., 2013). Separate trees were reconstructed using full protein sequences and with GAST domains. The ML method was used with the General Time Reversible Substitution model, which has discrete Gamma distribution among sites (four categories) and invariant sites (G + I) as site rates. The bootstrap value was estimated from 1000 replicates. The ML tree was inferred through BioNJ as the initial tree using the heuristic method (Barik et al., 2014, 2015). The identified novel GAST sequence/s were assigned to the GAST family and annotated according to their closeness to or relatedness with reference sequence/s as found in the ML tree. The S. moellendorffii GORs were termed “GASS” (GA-stimulated in Seleginella) and gymnosperm GORs were “GASP” (GA-stimulated in the Pinaceae family; Supplemental Table S1).
Collinearity Calculation among GORs
To estimate GAST conservation among the genomes, syntenic gene pairs were identified for the calculation of collinearity. These GAST pairs were retrieved from the PGDD (http://chibba.agtec.uga.edu/duplication/index/blast). The PGDD scores of syntenic gene pairs were present in the form of network through Cytoscape v 3.6.0. The “Organic Layout” was used for the network representation and further optimized manually to highlight the relatedness among the GOR syntenic pairs in species-specific clusters.
3D Structure Prediction and Identification of Conserved Motifs in Promoter
Three dimensional structure was determined using the Iterative Threading Assembly Refinement server (Supplemental Table S6; Zhang, 2008; Roy et al., 2010). The functional annotation was predicted on the basis of predicted EC numbers with active sites, GO terms, and ligand binding sites (Zhang, 2008; Roy et al., 2010; Yang et al., 2015). 3D structures of the best predicted model of individual proteins were visualized using Chimera v1.6.2. Further, predicted consensus GO terms and EC numbers for GAST were used for identification of their high-level functions in the biological system through the metabolic pathway database KEGG (http://www.genome.jp/kegg/pathway.html). De novo discovery of promoter motifs was carried out using different tools available in the MEME suite (v4.11.2). Predicted promoter motifs were compared by aligning with the different transcription factor binding sites in eukaryotes (experimentally defined), available on the JASPAR CORE database (http://jaspar.genereg.net/), with the help of TOMTOM (from the MEME Suite). These identified transcription factor binding sites were further used to identify their respective orthologous genes in Arabidopsis (http://www.arabidopsis.org/).
Plant Materials and Growth Conditions
The seeds of Arabidopsis Col-0 (ecotype Columbia-0), and a transfer-DNA insertion mutant atgasa10-2 (Salk_119972) were procured from Arabidopsis Biological Resource Center. atgasa10-2 (AT5G59845) contains a single exon with transfer-DNA insertion in the promoter region and showed 80% reduction in the expression level of Ath-GASA10. Seeds were sterilized, germinated, grown, and maintained as described previously (Singh et al., 2012, 2014). One hundred seeds of each line were sterilized, stratified, and sown on one-half strength Murashige and Skoog (MS) medium supplemented with any of 1 µm ABA, 1 µm GA, or 1 µm PAC, and kept for germination under normal growth condition (22°C). We scored seed germination rate from 48 h to 120 h; emergence of a radical tip was marked as the first sign of seed germination. For rice study, the indica rice cultivar IR64 was used. Seed sterilization and germination assay were performed as described previously (Singh et al., 2016). The germinated seeds were transferred to hydroponic media for their growth and development (Yoshida et al., 1976). Further, we have also generated overexpression lines of Ath-GASA10 and Osa-GASR9 in Arabidopsis and the rate of seed germination was analyzed in Ath-GASA10OE, atgasa10-2, and Osa-GASR9OE.
RNA Isolation and Relative Expression Analysis
RNA was isolated using the TRI-reagent (Sigma-Aldrich)-based method and treated with RNase-free DNase I (Thermo Fisher Scientific); 2 μg RNA was converted to complementary DNA with M-MLV Reverse Transcriptase (Invitrogen) as described previously (Singh et al., 2017). Each sample was analyzed in triplicate by real-time quantitative PCR using 2× Brilliant II SYBR Master Mix (Agilent Technologies). The PCR reaction was placed in an ABI PRISM 7900 HT Fast Real Time PCR System (Applied Biosystems). Actin/GAPDH genes and actin/ubiquitin were used as endogenous controls in the case of rice and Arabidopsis, respectively. For relative quantification of transcript abundance, the ΔΔCt method was used (Livak and Schmittgen, 2001; primers are listed in Supplemental Table S7).
Construction of Transgenic Plants
The Ath-GASA10 promoter was cloned into the pCAMBIA1301 vector between the PstI and NcoI restriction enzyme sites by replacing the Cauliflower mosaic virus (CaMV)35S promoter. The Ath-GASA10 coding region (270 bp) was amplified and cloned into the pJET1.2 cloning vector and subcloned into the pCAMBIA1301 vector between the BglII and BstEII restriction enzyme (New England BioLabs) sites under control of the CaMV35S promoter for overexpression and the Ath-GASA10 native promoter for complementation construct. Similarly, the coding region of Osa-GASR9 (309 bp) was cloned into the pCAMBIA1301 vector between the BglII and BstEII restriction enzyme sites under control of the CaMV35S promoter for overexpression and the Ath-GASA10 native promoter for complementation construct. These constructs were transformed in Arabidopsis plants using the floral dip method (Clough and Bent, 1998). T1 plants were selected on hygromycin and resistant plants were used for expression level analysis. Homozygous T3 seeds were used for further experiments.
Statistical Analysis
For syntenic pair and collinearity network analysis, log scores were provided by PGDD (Hammoudi et al., 2016). Biological experiments were performed in at least three replicates. Error bars indicate the se for three independent experiments. Statistical significance for all the experiments was evaluated using one-way ANOVA in Excel (Microsoft), and statistical differences were indicated by asterisks: ***P < 0.001, **P < 0.01, *P < 0.05.
Accession Numbers
Accession numbers of the genes used in this study are Ath-Actin7, AT5G09810; Ath-Ubiquitin1, AT3G62250; Osa-Actin1, Os03g50885 and Osa-GAPDH, Os08g03290. All other genes are listed in Supplemental Table S1.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Phylogenetic analysis of GORs in different species.
Supplemental Figure S2. Expression analysis of Ath-GASA10 in germinating seeds.
Supplemental Figure S3. Prediction of cis-motifs in the promoter of Ath-GASA10 and Osa-GASR9 using MEME-suite v4.11.2.
Supplemental Figure S4. Biosynthesis pathways of plant hormones retrieved from KEGG database.
Supplemental Figure S5. The putative functional pathway of Ath-GASA10/Osa-GASR9 postulated through in silico analysis.
Supplemental Table S1. List of protein and nucleotide sequences identified across the plant kingdom for the determination of GORs using a reference genome.
Supplemental Table S2. Identified GOR sequences across the selected species from plant kingdom.
Supplemental Table S3. Average germination rates (AGR in %) of wild type and transgenic seeds under hormone treatment and control conditions.
Supplemental Table S4. Predicted consensus GO terms and EC numbers of Osa-GASR9 and Ath-GASA10.
Supplemental Table S5. Features of promoter motifs identified from Osa-GASR9 and Ath-GASA10.
Supplemental Table S6. Best predicted 3D model of Ath-GASA10 and Osa-GASR9 with their C-Score, TM Score, and RMSD value (I-TASSER server) along with predicted Ramachandran Plot statistics of 3D structure.
Supplemental Table S7. List of primers used in the study.
Acknowledgments
We thank the central instrumental facility of the National Institute of Plant Genome research. We thank the Arabidopsis Biological Resource Center for Arabidopsis seeds and Shalini Mukherjee, Swati Verma, Archita Singh, and Sandeep Yadav for critical review of the article.
Footnotes
Funding provided by the Science and Engineering Research Board, Department of Science and Technology, India for a National Post Doctoral Fellowship (NPDF/2015/000232) to A.K., the Council of Scientific and Industrial Research, India for a fellowship to P.K., and the Department of Biotechnology, Government of India for a DBT-Research Associate fellowship (BT/PR3292/AGR/2/811/2011 to A.S.).
References
- Aubert D, Chevillard M, Dorne AM, Arlaud G, Herzog M (1998) Expression patterns of GASA genes in Arabidopsis thaliana: The GASA4 gene is up-regulated by gibberellins in meristematic regions. Plant Mol Biol 36: 871–883 [DOI] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS (2009) MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res 37: W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baniaga AE, Arrigo N, Barker MS (2016) The small nuclear genomes of Selaginella are associated with a low rate of genome size evolution. Genome Biol Evol 8: 1516–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks JA, Nishiyama T, Hasebe M, Bowman JL, Gribskov M, dePamphilis C, Albert VA, Aono N, Aoyama T, Ambrose BA, et al. (2011) The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science 332: 960–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barik S, SarkarDas S, Singh A, Gautam V, Kumar P, Majee M, Sarkar AK (2014) Phylogenetic analysis reveals conservation and diversification of micro RNA166 genes among diverse plant species. Genomics 103: 114–121 [DOI] [PubMed] [Google Scholar]
- Barik S, Kumar A, Sarkar Das S, Yadav S, Gautam V, Singh A, Singh S, Sarkar AK (2015) Coevolution pattern and functional conservation or divergence of miR167s and their targets across diverse plant species. Sci Rep 5: 14611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker MS, Arrigo N, Baniaga AE, Li Z, Levin DA (2016) On the relative abundance of autopolyploids and allopolyploids. New Phytol 210: 391–398 [DOI] [PubMed] [Google Scholar]
- Ben-Nissan G, Lee JY, Borohov A, Weiss D (2004) GIP, a Petunia hybrida GA-induced cysteine-rich protein: A possible role in shoot elongation and transition to flowering. Plant J 37: 229–238 [DOI] [PubMed] [Google Scholar]
- Berrocal-Lobo M, Segura A, Moreno M, López G, García-Olmedo F, Molina A (2002) Snakin-2, an antimicrobial peptide from potato whose gene is locally induced by wounding and responds to pathogen infection. Plant Physiol 128: 951–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- de la Fuente JI, Amaya I, Castillejo C, Sánchez-Sevilla JF, Quesada MA, Botella MA, Valpuesta V (2006) The strawberry gene FaGAST affects plant growth through inhibition of cell elongation. J Exp Bot 57: 2401–2411 [DOI] [PubMed] [Google Scholar]
- Furukawa T, Sakaguchi N, Shimada H (2006) Two OsGASR genes, rice GAST homologue genes that are abundant in proliferating tissues, show different expression patterns in developing panicles. Genes Genet Syst 81: 171–180 [DOI] [PubMed] [Google Scholar]
- Griffiths J, Murase K, Rieu I, Zentella R, Zhang ZL, Powers SJ, Gong F, Phillips AL, Hedden P, Sun TP, et al. (2006) Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell 18: 3399–3414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammoudi V, Vlachakis G, Schranz ME, van den Burg HA (2016) Whole-genome duplications followed by tandem duplications drive diversification of the protein modifier SUMO in Angiosperms. New Phytol 211: 172–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden P, Sponsel V (2015) A century of gibberellin research. J Plant Growth Regul 34: 740–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden P, Phillips AL, Rojas MC, Carrera E, Tudzynski B (2001) Gibberellin biosynthesis in plants and fungi: A case of convergent evolution? J Plant Growth Regul 20: 319–331 [DOI] [PubMed] [Google Scholar]
- Hirano K, Nakajima M, Asano K, Nishiyama T, Sakakibara H, Kojima M, Katoh E, Xiang H, Tanahashi T, Hasebe M, et al. (2007) The GID1-mediated gibberellin perception mechanism is conserved in the Lycophyte Selaginella moellendorffii but not in the Bryophyte Physcomitrella patens. Plant Cell 19: 3058–3079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Proteau P, Poulter D, Ferro-Novick S (1995) BTS1 encodes a geranylgeranyl diphosphate synthase in Saccharomyces cerevisiae. J Biol Chem 270: 21793–21799 [DOI] [PubMed] [Google Scholar]
- Kotilainen M, Helariutta Y, Mehto M, Pollanen E, Albert VA, Elomaa P, Teeri TH (1999) GEG participates in the regulation of cell and organ shape during corolla and carpel development in Gerbera hybrida. Plant Cell 11: 1093–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33: 1870–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TH, Tang H, Wang X, Paterson AH (2013) PGDD: A database of gene and genome duplication in plants. Nucleic Acids Res 41: D1152–D1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Baniaga AE, Sessa EB, Scascitelli M, Graham SW, Rieseberg LH, Barker MS (2015) Early genome duplications in conifers and other seed plants. Sci Adv 1: e1501084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Nahirñak V, Rivarola M, Gonzalez de Urreta M, Paniego N, Hopp HE, Almasia NI, Vazquez-Rovere C (2016) Genome-wide analysis of the Snakin/GASA gene family in Solanum tuberosum cv. Kennebec. Am J Potato Res 93: 172–188 [Google Scholar]
- Olszewski N, Sun TP, Gubler F (2002) Gibberellin signaling: Biosynthesis, catabolism, and response pathways. Plant Cell 14(Suppl): S61–S80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittermann J, Stuart SA, Dawson TE, Moreau A (2012) Cenozoic climate change shaped the evolutionary ecophysiology of the Cupressaceae conifers. Proc Natl Acad Sci USA 109: 9647–9652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran JH, Wei XX, Wang XQ (2006) Molecular phylogeny and biogeography of Picea (Pinaceae): Implications for phylogeographical studies using cytoplasmic haplotypes. Mol Phylogenet Evol 41: 405–419 [DOI] [PubMed] [Google Scholar]
- Roxrud I, Lid SE, Fletcher JC, Schmidt ED, Opsahl-Sorteberg HG (2007) GASA4, one of the 14-member Arabidopsis GASA family of small polypeptides, regulates flowering and seed development. Plant Cell Physiol 48: 471–483 [DOI] [PubMed] [Google Scholar]
- Roy A, Kucukural A, Zhang Y (2010) I-TASSER: A unified platform for automated protein structure and function prediction. Nat Protoc 5: 725–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinovich L, Weiss D (2010) The Arabidopsis cysteine-rich protein GASA4 promotes GA responses and exhibits redox activity in bacteria and in planta. Plant J 64: 1018–1027 [DOI] [PubMed] [Google Scholar]
- Shi L, Gast RT, Gopalraj M, Olszewski NE (1992) Characterization of a shoot-specific, GA3- and ABA-regulated gene from tomato. Plant J 2: 153–159 [PubMed] [Google Scholar]
- Singh A, Singh S, Panigrahi KC, Reski R, Sarkar AK (2014) Balanced activity of microRNA166/165 and its target transcripts from the class III homeodomain-leucine zipper family regulates root growth in Arabidopsis thaliana. Plant Cell Rep 33: 945–953 [DOI] [PubMed] [Google Scholar]
- Singh A, Kumar P, Gautam V, Rengasamy B, Adhikari B, Udayakumar M, Sarkar AK (2016) Root transcriptome of two contrasting indica rice cultivars uncovers regulators of root development and physiological responses. Sci Rep 6: 39266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S, Singh A, Roy S, Sarkar AK (2012) SWP1 negatively regulates lateral root initiation and elongation in Arabidopsis. Plant Signal Behav 7: 1522–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S, Singh A, Yadav S, Gautam V, Singh A, Sarkar AK (2017) Sirtinol, a Sir2 protein inhibitor, affects stem cell maintenance and root development in Arabidopsis thaliana by modulating auxin-cytokinin signaling components. Sci Rep 7: 42450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Wang H, Yu H, Zhong C, Zhang X, Peng J, Wang X (2013) GASA14 regulates leaf expansion and abiotic stress resistance by modulating reactive oxygen species accumulation. J Exp Bot 64: 1637–1647 [DOI] [PubMed] [Google Scholar]
- Tamura K, Battistuzzi FU, Billing-Ross P, Murillo O, Filipski A, Kumar S (2012) Estimating divergence times in large molecular phylogenies. Proc Natl Acad Sci USA 109: 19333–19338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30: 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Ashikari M, Nakajima M, Itoh H, Katoh E, Kobayashi M, Chow TY, Hsing YI, Kitano H, et al. (2005) GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437: 693–698 [DOI] [PubMed] [Google Scholar]
- Vandenbussche F, Fierro AC, Wiedemann G, Reski R, Van Der Straeten D (2007) Evolutionary conservation of plant gibberellin signalling pathway components. BMC Plant Biol 7: 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XQ, Ran JH (2014) Evolution and biogeography of gymnosperms. Mol Phylogenet Evol 75: 24–40 [DOI] [PubMed] [Google Scholar]
- Wang L, Wang Z, Xu Y, Joo SH, Kim SK, Xue Z, Xu Z, Wang Z, Chong K (2009) OsGSR1 is involved in crosstalk between gibberellins and brassinosteroids in rice. Plant J 57: 498–510 [DOI] [PubMed] [Google Scholar]
- Wang XQ, Tank DC, Sang T (2000) Phylogeny and divergence times in Pinaceae: Evidence from three genomes. Mol Biol Evol 17: 773–781 [DOI] [PubMed] [Google Scholar]
- Yamaguchi S. (2008) Gibberellin metabolism and its regulation. Annu Rev Plant Biol 59: 225–251 [DOI] [PubMed] [Google Scholar]
- Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y (2015) The I-TASSER Suite: Protein structure and function prediction. Nat Methods 12: 7–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasumura Y, Crumpton-Taylor M, Fuentes S, Harberd NP (2007) Step-by-step acquisition of the gibberellin-DELLA growth-regulatory mechanism during land-plant evolution. Curr Biol 17: 1225–1230 [DOI] [PubMed] [Google Scholar]
- Yoshida S, Forno DA, Cock JH, Kwanchai AG (1976) Laboratory Manual for Physiological Studies of Rice. 3rd ed. International Rice Research Institute, Los Baños, the Philippines: pp 61–66 [Google Scholar]
- Zhang Y. (2008) I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Yang C, Peng J, Sun S, Wang X (2009) GASA5, a regulator of flowering time and stem growth in Arabidopsis thaliana. Plant Mol Biol 69: 745–759 [DOI] [PubMed] [Google Scholar]
- Zhong C, Xu H, Ye S, Wang S, Li L, Zhang S, Wang X (2015) Gibberellic Acid-Stimulated Arabidopsis6 serves as an integrator of gibberellin, abscisic acid, and glucose signaling during seed germination in Arabidopsis. Plant Physiol 169: 2288–2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann R, Sakai H, Hochholdinger F (2010) The Gibberellic Acid Stimulated-Like gene family in maize and its role in lateral root development. Plant Physiol 152: 356–365 [DOI] [PMC free article] [PubMed] [Google Scholar]