ERF11 directly activates BT4 in Arabidopsis response to Pst DC3000 infection and depends on the salicylic acid and ethylene signaling pathways
Abstract
Pseudomonas syringae, a major hemibiotrophic bacterial pathogen, causes many devastating plant diseases. However, the transcriptional regulation of plant defense responses to P. syringae remains largely unknown. Here, we found that gain-of-function of BTB AND TAZ DOMAIN PROTEIN 4 (BT4) enhanced the resistance of Arabidopsis (Arabidopsis thaliana) to Pst DC3000 (Pseudomonas syringae pv. tomato DC3000). Disruption of BT4 also weakened the salicylic acid (SA)-induced defense response to Pst DC3000 in bt4 mutants. Further investigation indicated that, under Pst infection, transcription of BT4 is modulated by components of both the SA and ethylene (ET) signaling pathways. Intriguingly, the specific binding elements of ETHYLENE RESPONSE FACTOR (ERF) proteins, including dehydration responsive/C-repeat elements and the GCC box, were found in the putative promoter of BT4. Based on publicly available microarray data and transcriptional confirmation, we determined that ERF11 is inducible by salicylic acid and Pst DC3000 and is modulated by the SA and ET signaling pathways. Consistent with the function of BT4, loss-of-function of ERF11 weakened Arabidopsis resistance to Pst DC3000 and the SA-induced defense response. Biochemical and molecular assays revealed that ERF11 binds specifically to the GCC box of the BT4 promoter to activate its transcription. Genetic studies further revealed that the BT4-regulated Arabidopsis defense response to Pst DC3000 functions directly downstream of ERF11. Our findings indicate that transcriptional activation of BT4 by ERF11 is a key step in SA/ET-regulated plant resistance against Pst DC3000, enhancing our understanding of plant defense responses to hemibiotrophic bacterial pathogens.
Plants are constantly exposed to a wide variety of pathogens; however, few pathogens are capable of successfully colonizing a specific host plant, suggesting the existence of recognition and defense mechanisms (Birkenbihl et al., 2012). In nature, there are two types of microbial pathogens, which differ in how they assimilate nutrition from the host: necrotrophic and biotrophic pathogens (Glazebrook, 2005). Necrotrophic pathogens need to kill living host cells to utilize decayed plant tissue as nutrients for growth and for completion of their life styles, whereas biotrophic pathogens parasitize living host cells for growth and reproduction (Pel and Pieterse, 2013). One general defense strategy of host plants against biotrophic pathogens is to kill infected cells by activating programmed cell death, whereas maintenance of host cell vitality is the main defense response to necrotrophic pathogens (Spoel et al., 2007). Despite this binary classification, most microbial pathogens employ a hemibiotrophic habit to parasitize living host plants, including Magnaporthe grisea and Pseudomonas syringae (Perfect and Green, 2001).
Upon pathogen infection, plants distinguish and resist distinctive pathogens via different phytohormone signaling pathways (Pieterse et al., 2009). In general, the literature links the salicylic acid (SA) pathway to defense responses against biotrophic/hemibiotrophic pathogens and the jasmonic acid (JA) pathway to necrotroph responses, and the SA and JA pathways are considered antagonistic in plant defense responses (Farmer et al., 2003; Vlot et al., 2009; Rivas-San Vicente and Plasencia, 2011; Fu and Dong, 2013; Yang et al., 2015; Zhang et al., 2017). The SA pathway involves defense signaling that increases in response to biotrophic pathogen infection, and this increase often coincides with accumulation of reactive oxygen species (ROS) and induced expression of antimicrobial pathogenesis-related (PR) genes (Delaney et al., 1994; Lawton et al., 1995). However, mutants and transgenic plants with diminished SA synthesis and accumulation, such as sid2 (salicylic acid induction deficient2) and transgenic NahG (bacterial salicylate hydroxylase) plants, fail to trigger plant defense responses and are susceptible to pathogen infection (Gaffney et al., 1993; Nawrath and Métraux, 1999; Wildermuth et al., 2001).
The accumulation of SA and the change in the cellular redox state activate the defense regulator NONEXPRESSOR OF PATHOGENESIS GENES1 (NPR1), a Bric-a-brac, Tramtrack and Broad Complex/Pox virus and Zinc finger (BTB/POZ) domain protein, to translocate to the nucleus and interact with TGACG-motif binding (TGA) transcription factors (TFs), inducing defense responses (Zhang et al., 1999; Després et al., 2000; Zhou et al., 2000; Fan and Dong, 2002; Wang et al., 2005). The core function of NPR1 as a positive regulator in plant defense against biotrophic pathogens has been documented in many species, including rice (Orzya sativa), soybean (Glycine max), orchid (Phalaenopsis aphrodite), mustard (Brassica juncea), and Arabidopsis (Arabidopsis thaliana); Sandhu et al., 2009; Fabro et al., 2011; Chen et al., 2013; Sadumpati et al., 2013; Liu et al., 2017). Exogenous application of SA also activates expression of PR genes and hypersensitive responses to promote cell death, resulting in resistance against virulent and avirulent pathogens (Yalpani et al., 1991; Vlot et al., 2009).
In addition, increasing evidence indicates that the ethylene (ET) signaling pathway is involved in the plant defense response to biotrophic and necrotrophic pathogens (Pieterse et al., 2012). The ET and JA signaling pathways have been shown to act synergistically, which gives plants a potent defense against attack by necrotrophic pathogens. Intriguingly, antagonistic and synergistic interactions between SA and ET have been reported (Pieterse et al., 2012; Guan et al., 2015). The ethylene insensitive2 (ein2) mutants exhibited a diametrically opposite response to Pst DC3000 (Pseudomonas syringae pv. tomato DC3000) in previous reports (Bent et al., 1992; Lawton et al., 1995; Pieterse et al., 1998; Wubben et al., 2001). Overall, our understanding of plant defense against biotrophic pathogens remains limited.
TFs play pivotal roles in the regulation of cross talk between diverse hormone signaling pathways, as well as in signal transduction to mediate defense gene expression. The ET response factor (ERF) proteins belonging to the APETALA2 (AP2)/ERF superfamily, one of the biggest TF families that contain 122 members in Arabidopsis, are plant-specific TFs, and specifically bind to dehydration responsive/C-repeat (DRE/CRT) elements and the GCCGCC motif (GCC) box at the promoter of downstream target genes (Ohme-Takagi and Shinshi, 1995; Li et al., 2011).
Downstream of the ET signaling pathway, most of the ERF genes integrate diverse resistance-related hormone stimuli, such as SA, JA, and ET, and different plant defense signaling pathways (McGrath et al., 2005; Oñate-Sánchez et al., 2007; Pré et al., 2008). Moreover, ERF proteins are crucial integrators of cross talk with different phytohormones (Cheng et al., 2013; Zander et al., 2014; Catinot et al., 2015). Although the SA signaling pathway functions antagonistically with the JA/ET signaling pathways, some ERFs are synergistically induced by SA, JA, and ET, indicating that ERFs can coordinately integrate the SA and the ET/JA signaling pathways, but not antagonize them, to finely modulate the defense response to pathogens (Xu et al., 2007; Zhang et al., 2009, 2016; Seo et al., 2010; Zarei et al., 2011; Chen et al., 2012; Deokar et al., 2015). Moreover, overexpression or disruption of several ERFs enhances the resistance of transgenic Arabidopsis against necrotrophic and biotrophic pathogen challenge (Moffat et al., 2012; Meng et al., 2013). Typically, constitutive overexpression of AtERF1 has been observed to activate the expression of several defense-related genes, including Plant Defensin 1.2 (PDF1.2) and Basic Chitinase (ChiB), and enhance Arabidopsis resistance to necrotrophic pathogens such as Botrytis cinerea, Fusarium oxysporum, and Plectospherella cucumerina but reduce Arabidopsis tolerance to hemibiotrophic Pst DC3000 (Berrocal-Lobo et al., 2002; Lorenzo et al., 2003). In contrast, the ERF protein AtERF4 can negatively regulate expression of PDF1.2 to compromise Arabidopsis tolerance to necrotrophic pathogens (McGrath et al., 2005). These findings suggest that ERF proteins can act as transcriptional activators or repressors to regulate plant defense. For example, in Arabidopsis, AtERF1, AtERF2, and AtERF5 are activators, but AtERF3, AtERF4, AtERF7, and AtERF11 always act as repressors of transcription (Fujimoto et al., 2000).
The BTB AND TAZ domain (BT) proteins, which comprise five members, are plant-specific BTB/POZ domain proteins and regulate transcription (Ren et al., 2007; Robert et al., 2009). Moreover, all five BT proteins can act as calmodulin-binding proteins in response to Ca2+ and are induced by hydrogen peroxide (H2O2) and SA. Following stimulation with Ca2+, H2O2, and SA, BT proteins interact with AtBET10 or AtGET9 to activate transcription of downstream target genes, indicating that BTs play a core integrator role in Ca2+, H2O2, and SA signaling (Du and Poovaiah, 2004; Misra et al., 2018). Increasing amounts of research have demonstrated that transcription regulators are involved in the plant defense response (Spoel et al., 2003; Hao et al., 2013; Liu et al., 2017). BT4 was reported to have a positive function in Arabidopsis defense against the necrotrophic pathogen B. cinerea (Hao et al., 2013). NPR1, a BTB/POZ domain protein, is the core of the SA signaling pathway and acts as a transcription regulator to interact with the TGA TF triggering expression of defense genes (Spoel et al., 2003). The BTB/POZ domain proteins often function in ubiquitination/degradation, contributing to plant defense against pathogen challenge. The E3 ligase OsCRL3 is composed of Cullin3, RBX1, and BTB/POZ proteins and negatively regulates cell death and defense against Magnaporthe oryzae by Cullin-mediated degrading of OsNPR1 in rice (Liu et al., 2017). Moreover, the BTB/POZ-MATH domain proteins BPM1 and BPM3 directly interact with the AP2/ERF transcriptional factor RAP2 to regulate the stress response, indicating that BTB proteins can directly interact with ERF proteins (Weber and Hellmann, 2009). Although the ERF TFs can bind to specific elements of target genes, the function of ERFs in mediating the transcription of BTs is largely unknown.
In this study, we describe the functions of BT4 in plant defense against the hemibiotrophic pathogen Pst DC3000 and the underlying molecular mechanisms. Gain of function of BT4 enhanced resistance of Arabidopsis against Pst DC3000 challenge. Disruption of BT4 weakened the SA-induced defense response to Pst DC3000 in bt4 mutants. Further investigation indicated that transcription of BT4 was associated with SA and ET signaling pathways under Pst infection and was especially dependent on NPR1, EIN2, and EIN3. Bioinformatic assays showed that the putative promoter of BT4 contained DRE/CRT elements and the GCC-box, which specifically target ERF proteins. We confirmed that ERF11 was SA- and Pst DC3000-inducible and was modulated by SA and ET signaling pathways under Pst infection. Moreover, ERF11 loss of function weakened Arabidopsis resistance to Pst DC3000 and the SA-induced defense response. Using the transient expression assay and yeast one-hybrid assay (Y1H), BT4 was identified as a direct target gene of ERF11 in vitro and in vivo. Using an EMSA, we revealed that ERF11 interacted with the GCC-box of the BT4 promoter. In addition, genetic studies further revealed that the BT4-regulated Arabidopsis defense response to Pst DC3000 directly functioned downstream of ERF11. These results suggest that transcriptional activation of BT4 by ERF11 is a key step in SA/ET-regulated plant resistance against Pst DC3000.
RESULTS
BT4 Positively Mediates Plant Defense against Pst DC3000 and Affects the SA-Induced Defense Response
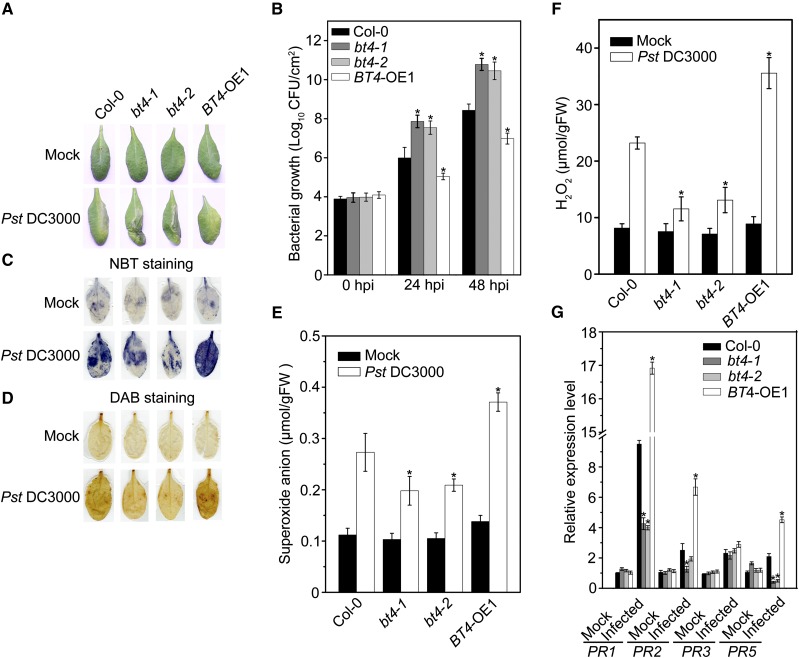
In our previous study, the loss-of-function bt4 mutant exhibited attenuated expression of defense-related genes and resulted in susceptibility to B. cinerea (Hao et al., 2013). There is a strong relationship between plant defenses against necrotrophic pathogens with those against biotrophic/hemibiotrophic pathogens. Therefore, BT4 might function in plant resistance to the hemibiotrophic pathogen Pst DC3000. To confirm our speculation that BT4 functions in defense against Pst DC3000 in Arabidopsis, we used two bt4 mutants (bt4-1 and bt4-2) and one overexpression transgenic plant, BT4-Overexpression1 (BT4-OE1), as described in our previous research (Hao et al., 2013). We determined the responses of 4-week-old wild-type (Col-0), bt4-1, bt4-2, and BT4-OE1 plants to Pst DC3000. At 48 h postinoculation (hpi), leaves presented typical chlorotic symptoms; disease symptoms increased more rapidly in Pst-infected bt4-1 and bt4-2 mutants than in Pst-infected BT4-OE1 plants (Fig. 1A). Moreover, higher bacterial counts were found at 24 and 48 hpi in the two bt4 mutants compared to BT4-OE1 plants (Fig. 1B).
Figure 1.
Altered disease resistance of bt4 and BT4-OE plants against Pseudomonas syringae pv. tomato (Pst) DC3000. A, Typical Pst DC3000 infection symptoms in Col-0, bt4-1, bt4-2, and BT4-OE1 plants. Four-week-old plants were inoculated by Pst DC3000 bacterial suspension or 10 mmol/L MgCl2 and kept at high humidity. Photographs of representative leaves were taken 48 h (hpi. The experiments were repeated three times with similar results. B, Bacterial growth in the inoculated leaves was detected in planta. Bacteria were isolated from plants at 24 and 48 hpi and quantified with gradient dilution assays. The P values (bacterial count of each genotype versus Col-0 under Pst treatment at the same time point) were determined by two-tailed Student’s test assuming equal variance (P < 0.05). C, In situ and (E) quantitative analysis of superoxide anion accumulation in Pst DC3000-inoculated leaves by NBT staining and biochemical testing, respectively. Four-week-old wild-type (Col-0), bt4-1, bt4-2, and BT4-OE1 plants were inoculated with Pst DC3000 or 10 mmol/L MgCl2 and kept in high humidity. Leaf samples were collected at 24 hpi. The P values (superoxide anion of each genotype vs Col-0 under Pst-treatment at the same time point) were determined by two-tailed Student’s test assuming equal variance (P < 0.05). D, In situ and (F) quantitative analysis of H2O2 accumulation in Pst DC3000-inoculated leaves by 3,3-diaminobenzidine (DAB) staining and biochemical testing, respectively. Four-week-old wild-type (Col-0), bt4-1, bt4-2, and BT4-OE1 plants were inoculated with Pst DC3000 or 10 mmol/L MgCl2 and kept in high humidity. Leaf samples were collected at 24 hpi. The P values (H2O2 of each genotype versus Col-0 under Pst treatment at the same time point) were determined by two-tailed Student’s test assuming equal variance (P < 0.05). G, Relative expression levels of PR1, PR2, PR3, and PR5 in the leaves of 4-week-old wild-type (Col-0), bt4-1, bt4-2, and BT4-OE1 plants after Pst DC3000 treatment for 24 h. Relative expression is indicated as folds of the transcript level of an internal AtTub4 gene. The P values (PR expressions of each genotype versus Col-0 under Pst-treatment at the same time point) were determined by two-tailed Student’s test assuming equal variance (P < 0.05). Data presented are the means ± sd from three independent experiments and asterisks indicate significant differences at P < 0.05 between bt4-1/bt4-2/BT4-OE1 and Col-0 plants.
We also compared the patterns for accumulation of ROS and expression levels of defense genes among Col-0, bt4-1, bt4-2, and BT4-OE1 plants at 24 hpi. Accumulations of superoxide anion and H2O2 in leaves were analyzed by nitro-blue tetrazolium (NBT) and 3,3-diaminobenzidine staining (DAB) staining and quantified by biochemical testing. There was no significant difference in accumulation of superoxide anion and H2O2 in unchallenged Col-0, bt4-1, bt4-2, and BT4-OE1 plants (Fig. 1, C–F). Upon Pst DC3000 infection, superoxide anion and H2O2 were accumulated in inoculated leaves at 24 hpi. Superoxide anion and H2O2 accumulation were lower in inoculated leaves of bt4-1 and bt4-2 mutants and higher in BT4-OE1 plants, compared to those in Col-0 (Fig. 1, C–F).
In addition, we quantified the relative expression levels of defense-related genes (PR1, PR2, PR3, and PR5) in response to Pst DC3000 infection. The expression levels of PR genes in unchallenged bt4-1, bt4-2, and BT4-OE1 plants were similar to those in Col-0 (Fig. 1G), suggesting that overexpression and disruption of ERF11 did not affect the basal expression of PR genes. In contrast, higher expression levels of PR1, PR2, and PR5 in the BT4-OE1 plants than in Col-0 at 24 hpi, and especially higher than in bt4-1 and bt4-2 mutants, further supported these phenotypes (Fig. 1G). Furthermore, there were no significant differences in pathogen-induced expression of PR3 among Col-0, bt4-1, bt4-2, and BT4-OE1 plants (Fig. 1G). These results confirmed that disruption of BT4 resulted in Arabidopsis being susceptible to this hemibiotrophic pathogen and that BT4 played a positive role in defense against Pst DC3000.
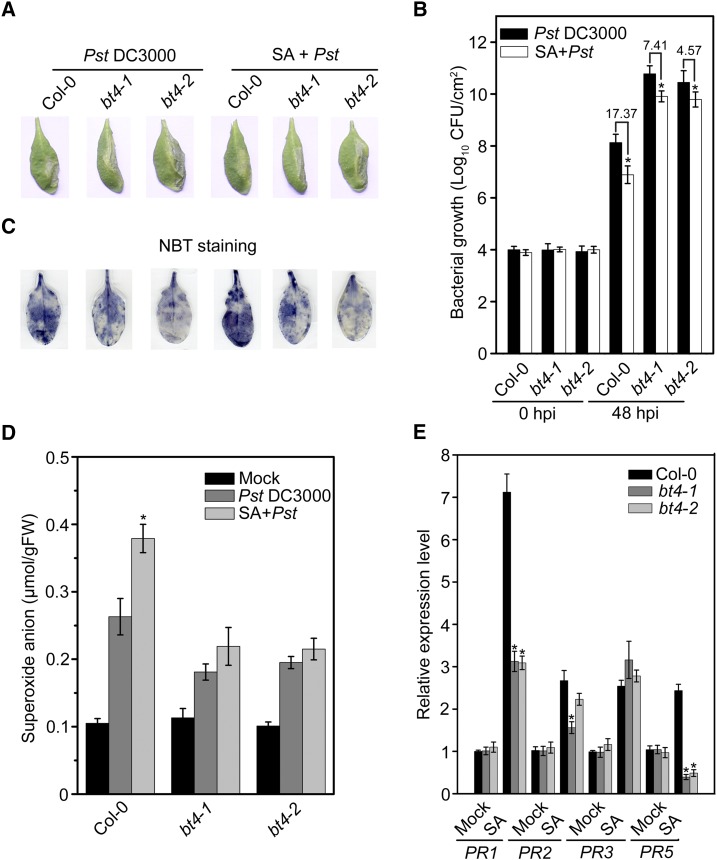
Direct application of SA increases ROS accumulation, activates various PR genes, and enhances resistance to virulent biotrophic pathogens (Mur et al., 2008; Shah, 2009; Coll et al., 2011). To confirm the ability of SA to enhance Arabidopsis resistance to Pst DC3000, we performed infection experiments in four kinds of wild-type Arabidopsis. Plants were sprayed with 1 mm SA or 0.1% ethanol solution (as a control) and inoculated with Pst DC3000 at 24 h after pretreatment. Significantly increased ROS accumulation and a protection effect were observed in SA-pretreated leaves, and most leaves pretreated with 0.1% ethanol solution showed weakened ROS accumulation and extensive chlorosis (Supplemental Fig. S1). To explore whether BT4 is required for the SA-induced defense response, we analyzed and compared the capacity for SA-induced resistance in bt4 mutants. At 48 hpi, disease symptoms were significantly reduced in SA-pretreated Col-0 leaves but nonsignificantly in bt4-1 and bt4-2 mutants compared with the control (Fig. 2A). The bacterial count was significantly decreased in SA-pretreated Col-0 leaves, about 17.37-fold lower compared with the control, but was only 7.41- and 4.57-fold lower in SA-pretreated bt4-1 and bt4-2 mutants compared to the control (Fig. 2B). SA pretreatment did not significantly affect ROS accumulation in bt4 mutants (Fig. 2, C and D). In addition, we also evaluated the expression levels of PR genes in SA-induced Col-0 and bt4 plants by real-time quantitative PCR (qPCR). The relative expression levels of PR1, PR2, PR3, and PR5 were enhanced in Col-0 and bt4 plants after SA treatment for 24 h. However, the SA-induced expression levels of PR1 and PR5 in bt4-1 and bt4-2 mutants as well as induction of PR2 in bt4-1 were significantly lower than those in Col-0 (Fig. 2F). These results indicate that disruption of BT4 impairs the SA-induced defense response to Pst DC3000 in bt4 mutants.
Figure 2.
Attenuated salicylic-acid-induced defense response in bt4 mutants. A, Typical Pst DC3000 infected disease symptoms in wild-type (Col-0), bt4-1, and bt4-2 plants at 48 hpi with or without SA treatment. Four-week-old plants were sprayed with 1 mm SA or 0.1% (v/v) ethanol solution and then inoculated with Pst DC3000 at 24 h after SA treatment. Photographs of representative leaves were taken 48 hpi. B, Bacterial growth in the inoculated leaves of Col-0, bt4-1, and bt4-2 plants in planta with or without SA treatment. Four-week-old plants were sprayed with 1 mm SA or 0.1% (v/v) ethanol solution and then inoculated with Pst DC3000 at 24 h after SA treatment. Bacteria were isolated from plants at 48 hpi and quantified with gradient dilution technique. The P values (bacterial mount of each genotype with SA-pretreatment versus each genotype with mock pretreatment at the same time point) were determined by two-tailed Student’s test assuming equal variance (P < 0.05). C, In situ and (D) quantitative analysis of superoxide anion accumulation in inoculated leaves of Col-0, bt4-1, and bt4-2 plants with or without SA treatment by NBT staining and biochemical testing, respectively. The 4-week-old plants were sprayed with 1 mm SA or 0.1% (v/v) ethanol solution and then inoculated with Pst DC3000 at 24 h after SA treatment. Leaf samples were collected at 24 hpi. The P values (superoxide anion of each genotype with SA-pretreatment versus each genotype without SA pretreatment under Pst-infected at the same time point) were determined by two-tailed Student’s test assuming equal variance (P < 0.05). E, Partial suppression of SA-induced expression of defense genes in bt4 plants. Four-week-old wild-type (Col-0), bt4-1, and bt4-2 plants were sprayed with 1 mm SA or 0.1% (v/v) ethanol solution for 24 h, and then inoculated leaves were collected for RNA isolation. Relative expression is indicated as folds of the transcript level of an internal AtTub4 gene. The P values (PR expressions of each genotype versus Col-0 under SA treatment at the same time point) were determined by two-tailed Student’s test assuming equal variance (P < 0.05). Data presented are the means ± sd from three independent experiments, and asterisks indicate significant differences at P < 0.05 between bt4-1/bt4-2 and Col-0 plants.
BT4 Transcription Is Modulated by the SA and ET Signaling Pathways under Pst DC3000 Treatment
To investigate the relationship between BT4 and plant defense signaling pathways, we first checked its putative promoter sequence (−2500 bp) using a plant cis-acting regulatory DNA element database (https://sogo.dna.affrc.go.jp/cgi-bin/sogo.cgi?lang=en&pj=640&action=page&page=newplace; Higo et al., 1999). As expected, hormone-responsive elements and defense/stress-responsive elements, including JARE, ABRE, SARE, EtRE, DRE/CRT, and GCC-box, were found in the putative promoter of BT4 (Supplemental Fig. S2A). Then, BT4 expression was further analyzed with phytohormone and stress treatment in Col-0. Indeed, the qPCR results showed that BT4 expression was moderately induced by hormone and stress treatment, including JA, SA, 1-aminocyclopropane-1-carboxylic acid (ACC; an ET precursor), abscisic acid (ABA), gibberellin, B. cinerea, Pst DC3000, salt, and drought (Supplemental Fig. S2, B and C).
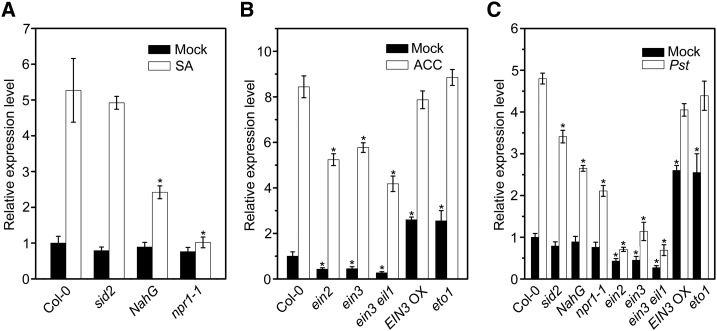
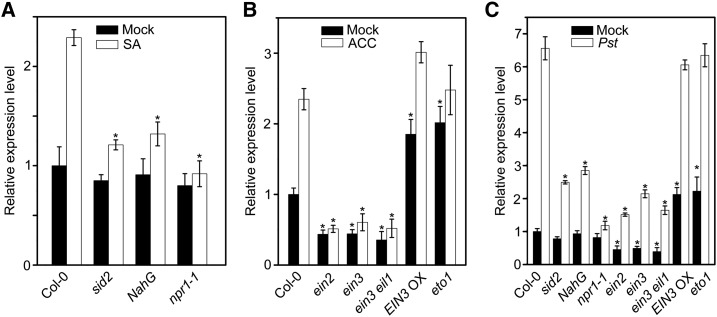
Most of the hormone-responsive elements in the BT4 promoter sequence were related to plant defense signaling pathways, e.g. SA, ET, and JA. Increasing amounts of research have revealed that SA and ET play crucial roles in the plant defense process against Pst DC3000 (Laluk et al., 2011; Guan et al., 2015; Zhang et al., 2016). To investigate whether BT4 transcription is modulated by the SA and ET signaling pathways, we measured BT4 expression in SA/ET synthesis and signaling mutants (e.g. sid2, NahG, npr1-1, ein2, ein3, ein3 eil1, EIN3 Overexpression [EIN3 OX], and eto1) treated with or without hormones and Pst DC3000. Under normal growth conditions, BT4 expression was decreased in sid2, NahG, and npr1-1 plants compared with Col-0 plants (Fig. 3A). Although SA significantly increased BT4 expression in different genotypes, BT4 induction was significantly lower in NahG and npr1-1 plants compared with Col-0 plants treated with 50 μm SA (Fig. 3A). We also measured BT4 expression in ein2, ein3, ein3 eil1, EIN3 OX, and eto1 plants with or without 10 μm ACC. Under normal growth conditions, BT4 expression significantly decreased in ein2, ein3, and ein3 eil1 plants but increased more than 2-fold in EIN3 OX and eto1 plants. Under 10 μm ACC treatment, induction of BT4 was significantly lower in ein2, ein3, and ein3 eil1 compared with Col-0 plants (Fig. 3B). Similar to SA and ACC treatments, Pst DC3000 infection significantly induced expression of BT4 in different genotypes, but induction of BT4 was compromised in sid2, NahG, npr1-1, ein3, ein3 eil1, and ein2 plants compared with Col-0 (Fig. 3C). These results confirm that BT4 functions in the defense process against Pst DC3000 and is modulated by the SA/ET signaling pathway.
Figure 3.
The expression of BT4 is modulated by SA and ethylene signaling components. A, Relative expression level of BT4 in Col-0, sid2, NahG, and npr1-1 plants with or without 50 µm SA treatment. Seven-day-old seedlings were treated with 50 μm SA or 0.1% (v/v) ethanol solution for 1 h, and plant samples were collected to quantify the relative expression level of BT4 by qPCR. Expression level of BT4 in sid2, NahG, and npr1-1 plants are shown relative to that in mock-treated Col-0. The P values (each genotype versus Col-0 under SA treatment) were determined by two-tailed Student’s test assuming equal variance (P < 0.05). B, Relative expression level of BT4 in Col-0, ein2, ein3, ein3 eil1, EIN3 OX, and eto1 plants with or without 10 µm ACC treatment. Seven-day-old seedlings were treated with 10 μm ACC or H2O for 1 h, and plant samples were collected to quantify the relative expression level of BT4 by qPCR. Expression level of BT4 in ein2, ein3, ein3 eil1, EIN3 OX, and eto1 plants are shown relative to that in mock-treated Col-0. The P values (each genotype versus Col-0 under ACC treatment) were determined by two-tailed Student’s test assuming equal variance (P < 0.05). C, Relative expression level of BT4 in Col-0, sid2, NahG, npr1-1, ein2, ein3, ein3 eil1, EIN3 OX, and eto1 plants without or with Pst DC3000 treatment. Four-week-old plants were treated with Pst DC3000 or 10 mmol/L MgCl2 for 6 h and plant samples were collected to quantify the relative expression level of BT4 by qPCR. Expression level of BT4 in sid2, NahG, npr1-1, ein2, ein3, ein3 eil1, EIN3 OX, and eto1 plants are shown relative to that in mock-treated Col-0. The P values (each genotype vs Col-0 under Pst treatment) were determined by two-tailed Student’s test assuming equal variance (P < 0.05). Data presented are the means ± sd from three independent experiments, and asterisks indicate significant difference at P < 0.05 between inoculated/treated plants and control plants.
ERF11 Is a SA- and Pst-Inducible ERF Gene That Is Transcriptionally Modulated by the SA and ET Signaling Pathways
ERFs, the TFs containing an AP2 DNA-binding domain, are located downstream of the ET signaling pathway and function in cross talk with diverse phytohormones (Zander et al., 2014; Liu et al., 2018). Increasing evidence indicates that ERFs play important roles in abiotic and biotic responses, especially functioning in the Pst-stress response, when Arabidopsis is stimulated in a complex environment (Zhang et al., 2011, 2015, 2016; Mao et al., 2016). The putative promoter of BT4 contained DRE/CRT elements and the GCC-box, which were the specific binding elements of ERF proteins (Supplemental Fig. S2A). Furthermore, BT4 functioned in the defense process against Pst DC3000 and was modulated by the SA and ET signaling pathways (Fig. 3C). Therefore, we assumed that BT4 was modulated by the ET signaling pathway and depended on ERF proteins.
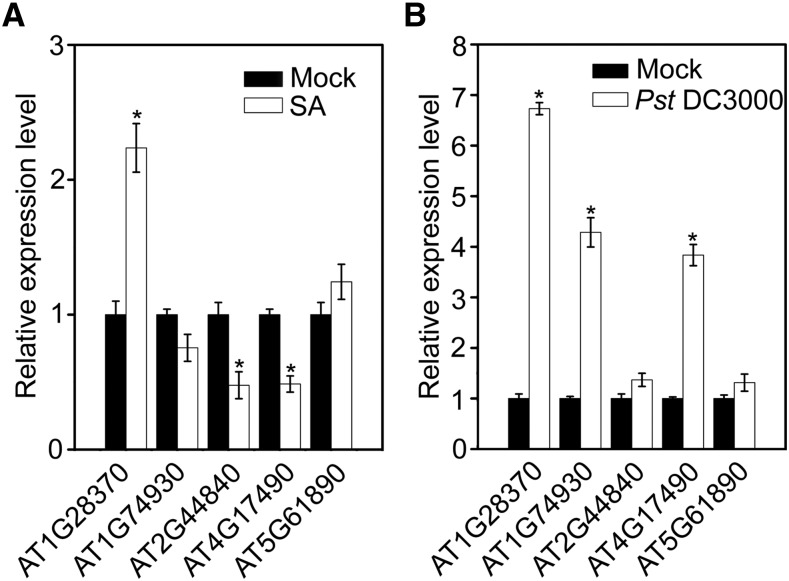
First, using the Gene Expression Omnibus (GEO) database, we performed a genome-wide analysis of ERF genes in the SA and Pst DC3000 responses to determine which ERF gene might function in plant defense against Pst DC3000 and be regulated by the SA signaling pathway. From these putative ERF genes, only five candidates were altered more than 2-fold by SA, Pst DC3000, and null mutation of isochorismic acid synthase (ICS1) gene in three independent transcriptome databases: AT1G28370, AT1G74930, AT2G44840, AT4G17490, and AT5G61890 (Supplemental Fig. S3; Supplemental Table S2). Subsequently, identification using qPCR analysis of Col-0 without (as a control) or with 50 μm SA and Pst DC3000 confirmed that expression levels of AT1G28370, AT2G44840, and AT4G17490 were altered by more than 2-fold after SA treatment for 1 h (Fig. 4A), and expression levels of AT1G28370, AT1G74930, and AT4G17490 were increased more than 2-fold after Pst DC3000 infection for 6 h (Fig. 4B), indicating that AtERF11 (AT1G28370) and AtERF6 (AT4G17490) were simultaneously affected by SA and Pst DC3000. ERF6 is known to function in defense against B. cinerea (Dubois et al., 2015), but regulation of ERF11 has not previously been reported in plant defense against pathogens. Therefore, we investigated the potential function of ERF11 in Arabidopsis defense against Pst DC3000.
Figure 4.
Identification of ERF11 as a SA- and Pst-inducible ERF gene. A, Expression patterns of AT1G28370, AT1G74930, AT2G44840, AT4G17490, and AT5G61890, which were screened from three independent GEO databases (Supplemental Fig. S1; Supplemental Table S2) induced by defense signaling hormones such as SA. Seven-day-old wild-type (Col-0) seedlings were treated with 50 μm SA or 0.1% (v/v) ethanol solution (mock) for 1 h, and plant samples were collected to quantify the relative expression level of AT1G28370, AT1G74930, AT2G44840, AT4G17490, and AT5G61890 by qPCR. Expression levels of these genes are shown relative to that in mock-treated Col-0. The P values (each gene expression with mock-treated versus the expression under SA treatment) were determined by two-tailed Student’s test assuming equal variance (P < 0.05) B, Expression patterns of AT1G28370, AT1G74930, AT2G44840, AT4G17490, and AT5G61890 induced by Pst DC3000. Four-week-old wild-type (Col-0) Arabidopsis were treated with Pst DC3000 or 10 mmol/L MgCl2 (mock) for 6 h, and leaf samples were collected to quantify the relative expression level of AT1G28370, AT1G74930, AT2G44840, AT4G17490, and AT5G61890 by qPCR. Expression levels of these genes are shown relative to that in mock-treated Col-0. The P values (each gene expression with mock-treated versus the expression under Pst treatment) were determined by two-tailed Student’s test assuming equal variance (P < 0.05). Data presented are the means ± sd from three independent experiments, and asterisks indicate significant difference at P < 0.05 between inoculated/treated plants and control plants.
To explore the role of ERF11 in plant defense, we examined whether ERF11 could be induced by pathogen infection and defense signaling hormones such as SA. The expression of ERF11 increased and peaked rapidly to 2.8-fold at 0.5 hours post treatment (hpt), remained at the higher level until 1 hpt, decreased at 3 hpt, and rose once again at 12 hpt (Supplemental Fig. S4). Unlike the pattern following SA treatment, ERF11 expression was moderately increased and peaked up 12-fold at 12 hpi.
To investigate whether transcription of ERF11 is modulated by the SA and ET signaling pathways, we measured ERF11 expression in SA/ET synthesis and signaling mutants, e.g. sid2, NahG, npr1-1, ein2, ein3, ein3 eil1, EIN3 OX, and eto1, treated with or without hormones and Pst DC3000. Under normal growth conditions, ERF11 expression decreased in sid2, NahG, and npr1-1 compared with Col-0 plants (Fig. 5A). Consistent with results in Supplemental Figure S3B and Supplemental Table S2, the transcriptome data from GSE9955 showed that ERF11 expression was lower in sid2/ics1 mutants compared with Col-0 plants. Although SA significantly induced expression of ERF11 in different genotypes, ERF11 induction was significantly lower in sid2, NahG, and npr1-1 compared with Col-0 plants treated with 50 μm SA (Fig. 5A). We also determined the ERF11 expression in ein2, ein3, ein3 eil1, EIN3 OX, and eto1 plants. Under normal growth conditions, ERF11 expression significantly decreased in ein2, ein3, and ein3 eil1 plants but increased almost 2-fold in EIN3 OX and eto1 plants. With 10 μm ACC treatment, ERF11 induction was significantly lower in ein2, ein3, and ein3 eil1 but enhanced in EIX3 OX plants, compared with ACC-treated Col-0 (Fig. 5B). Similar to SA and ACC treatments, Pst DC3000 infection significantly induced ERF11 expression in different genotypes, but ERF11 induction was compromised in sid2, NahG, npr1-1, ein2, ein3, and ein3 eil1 plants compared with Col-0 (Fig. 5C). These results are consistent with expression of BT4 in SA/ET synthesis and signaling mutants. These results led us to speculate that ERF11 functioned in the defense process against Pst DC3000 and was modulated by the SA/ET signaling pathways.
Figure 5.
The expression of ERF11 is modulated by SA and ET signaling components. A, Relative expression level of ERF11 in Col-0, sid2, NahG, and npr1-1 plants with or without 50 µm SA treatment. Seven-day-old seedlings were treated with 50 μm SA or 0.1% (v/v) ethanol solution for 1 h, and plant samples were collected to quantify the relative expression level of ERF11 by qPCR. Expression level of ERF11 in sid2, NahG, and npr1-1 plants are shown relative to that in mock-treated Col-0. The P values (each genotype versus Col-0 under SA treatment) were determined by two-tailed Student’s test assuming equal variance (P < 0.05). B, Relative expression level of ERF11 in Col-0, ein2, ein3, ein3 eil1, EIN3 OX, and eto1 plants with or without 10 µm ACC treatment. Seven-day-old seedlings were treated with 10 μm ACC or H2O for 1 h and plant samples were collected to quantify the relative expression level of ERF11 by qPCR. Expression level of ERF11 in ein2, ein3, ein3 eil1, EIN3 OX, and eto1 plants are shown relative to that in mock-treated Col-0. The P values (each genotype versus Col-0 under ACC treatment) were determined by two-tailed Student’s test assuming equal variance (P < 0.05). C, Relative expression level of ERF11 in Col-0, sid2, NahG, npr1-1, ein2, ein3, ein3 eil1, EIN3 OX, and eto1 plants without or with Pst DC3000 treatment. Four-week-old plants were treated with Pst DC3000 or 10 mmol/L MgCl2 for 6 h, and plant samples were collected to quantify the relative expression level of ERF11 by qPCR. Expression levels of ERF11 in Col-0, sid2, NahG, npr1-1, ein2, ein3, ein3 eil1, EIN3 OX, and eto1 plants are shown relative to that in mock-treated Col-0. The P values (each genotype versus Col-0 under Pst treatment) were determined by two-tailed Student’s test assuming equal variance (P < 0.05). Data presented are the means ± sd from three independent experiments, and asterisks indicate significant difference at P < 0.05 between inoculated/treated plants and control plants.
ERF11 Loss of Function Weakens Arabidopsis Resistance against the Pst DC3000- and SA-Induced Defense Responses
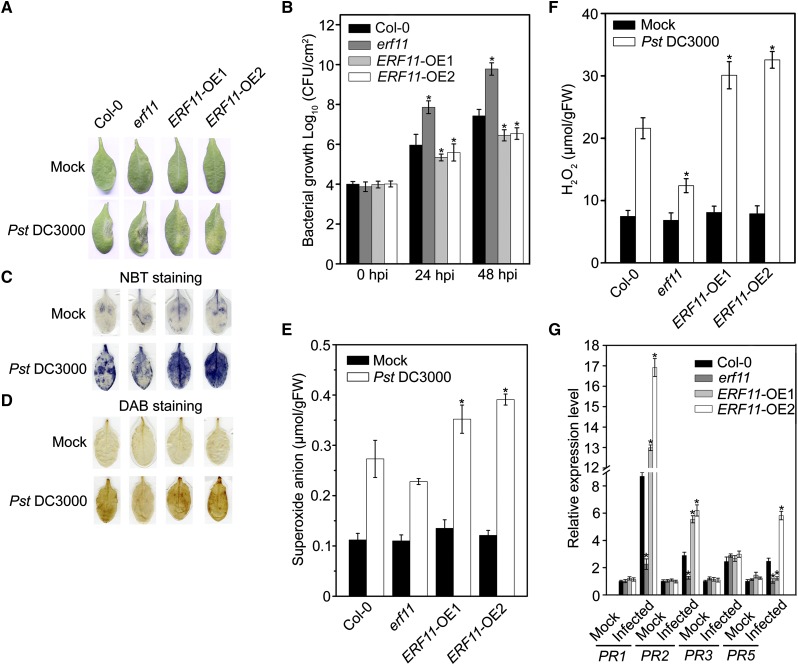
To verify our speculation that ERF11 functions in the defense response against Pst DC3000, the erf11 mutant and two overexpression lines (ERF11-OE1 and ERF11-OE2) described in our previous study (Li et al., 2011) were used for further analysis. We determined differences in response of the Col-0, erf11, and ERF11-OE lines to Pst DC3000 inoculation. At 48 hpi, plants exhibited typical symptoms stimulated by Pst DC3000. Symptom development was significantly reduced in ERF11-OE1 and ERF11-OE2 plants but rapidly increased in erf11 plants, compared with Col-0 (Fig. 6A). Furthermore, a lower level of bacterial growth of Pst DC3000 was evident in ERF11-OE1 and ERF11-OE2 compared with Col-0 and erf11 plants at 24 and 48 hpi (Fig. 6B). We also compared the patterns of ROS content and expression level of defense genes among Col-0, erf11, and ERF11-OE plants at 24 hpi. There were nonsignificant differences in accumulation of superoxide anion and H2O2 in unchallenged Col-0, erf11, ERF11-OE1, and ERF11-OE2 plants (Fig. 6, C–F). Upon Pst DC3000 infection, superoxide anion and H2O2 accumulated in inoculated leaves at 24 hpi. Superoxide anion and H2O2 accumulation was lower in inoculated leaves of the erf11 mutant and significantly higher in ERF11-OE1 and ERF11-OE2 plants, compared with Col-0 (Fig. 6, C–F). Next, we quantified the expression levels of defense-related genes (PR1, PR2, PR3, and PR5) in response to Pst DC3000 infection. The expression levels of PR genes in unchallenged erf11, ERF11-OE1, and ERF11-OE2 plants were similar to those in Col-0, suggesting that overexpression and disruption of ERF11 did not affect basal expression of PR genes (Fig. 6G). In contrast, expression levels of PR1 and PR2 in the ERF11-OE1 and ERF11-OE2 plants, as well as PR5 in ERF11-OE2 plants, were significantly higher than in Col-0 at 24 hpi, and especially higher than those of erf11 mutants (Fig. 6G). Furthermore, there were no significant differences in pathogen-induced expression of PR3 among Col-0, erf11, ERF11-OE1, and ERF11-OE2 plants (Fig. 6G). Taken together, these results indicate that disruption of ERF11 significantly weakens resistance to Pst DC3000 and that ERF11 plays a positive role in defense.
Figure 6.
Altered disease resistance of ERF11-OE and erf11 plants against Pst DC3000. A, Typical Pst DC3000-infected symptoms detected in wild-type (Col-0), erf11, ERF11-OE1, and ERF11-OE2 plants. Four-week-old plants were inoculated by Pst DC3000 bacterial suspension or 10 mmol/L MgCl2 and kept at high humidity. Photographs of representative leaves were taken 48 hpi. The experiments were repeated three times with similar results. B, Bacterial growth in the inoculated leaves detected in planta. Bacteria were isolated from plants at 24 and 48 hpi and quantified with gradient dilution assays. The P values (bacterial count of each genotype versus Col-0 under Pst-treatment at the same time point) were determined by two-tailed Student’s test assuming equal variance (P < 0.05). C, In situ and (E) quantitative analysis of superoxide anion accumulation in Pst DC3000-inoculated leaves by NBT staining and biochemical testing, respectively. Four-week-old plants were inoculated with Pst DC3000 or 10 mmol/L MgCl2 and kept in high humidity. Leaf samples were collected at 24 hpi. The P values (superoxide anion of each genotype versus Col-0 under Pst-treatment at the same time point) were determined by two-tailed Student’s test assuming equal variance (P < 0.05). D, In situ and (F) quantitative analysis of H2O2 accumulation in Pst DC3000-inoculated leaves by DAB staining and biochemical testing, respectively. Four-week-old plants were inoculated with Pst DC3000 or 10 mmol/L MgCl2 and kept in high humidity. Leaf samples were collected at 24 hpi. The P values (H2O2 accumulation of each genotype versus Col-0 under Pst-treatment at the same time point) were determined by two-tailed Student’s test assuming equal variance (P < 0.05). G, Relative expression levels of PR1, PR2, PR3, and PR5 in leaves of 4-week-old wild-type (Col-0), erf11, ERF11-OE1, and ERF11-OE2 plants after Pst DC3000 treatment for 24 h. The P values (PR expressions of each genotype versus Col-0 under Pst-treatment at the same time point) were determined by two-tailed Student’s test assuming equal variance (P < 0.05). Data presented are the means ± sd from three independent experiments, and asterisks indicate significant differences at P < 0.05 between erf11/ERF11-OE1/ERF11-OE2 and Col-0 plants.
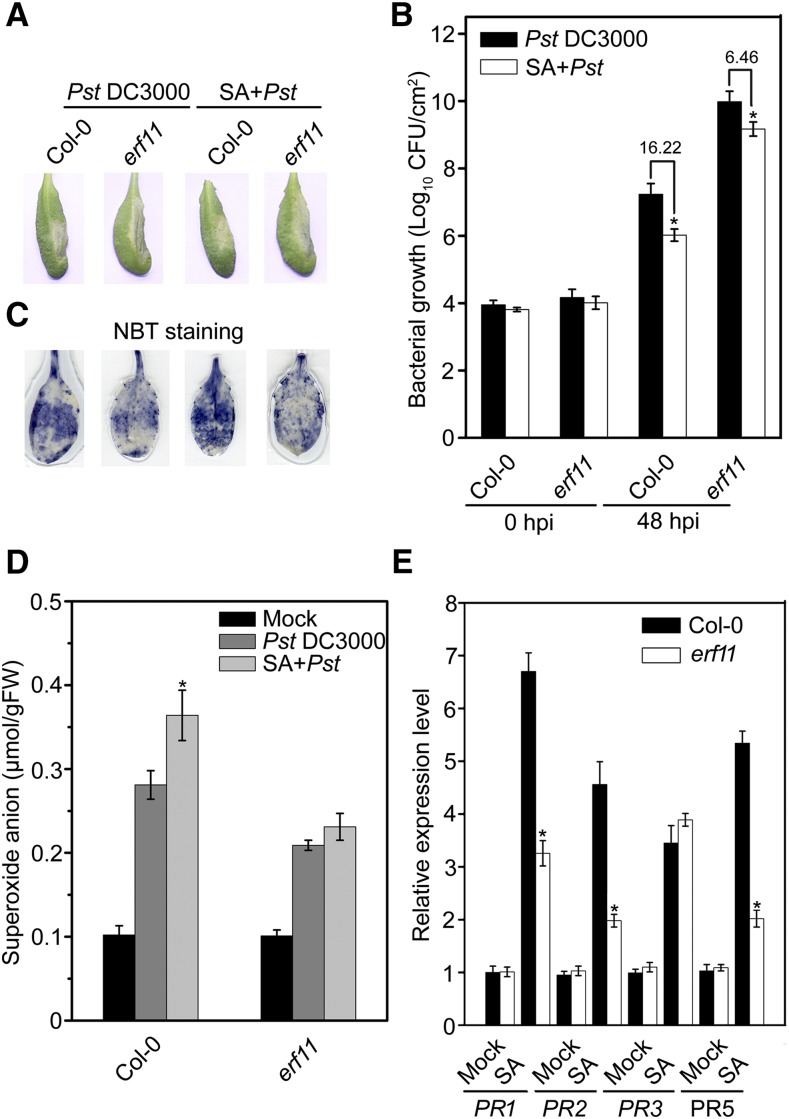
To address the potential roles of ERF11 in the SA-induced defense response, we determined the capacity of SA-enhanced resistance to Pst DC3000 in erf11 mutants. At 48 hpi, disease symptoms were significantly reduced in SA-pretreated Col-0 plants, but nonsignificantly in SA-pretreated erf11 plants, compared with the control (Fig. 7A). The SA pretreatment resulted in a 16.22-fold decrease in bacterial growth in the Pst-inoculated Col-0, but only a 6.46-fold decrease in Pst-inoculated erf11 plants (Fig. 7B). Moreover, SA pretreatment did not significantly affect ROS accumulation in erf11 mutants (Fig. 7, C and D). We also compared the expression levels of PR genes in SA-induced Col-0 and erf11 plants using qPCR. Expressions of PR1, PR2, PR3, and PR5 were induced by SA in Col-0 and erf11 plants (Fig. 7E). However, SA-induced expression of PR1, PR2, and PR5 in erf11 mutants was compromised compared with Col-0 (Fig. 7E). These results indicate that disruption of ERF11 partially weakens the SA-induced defense response to Pst DC3000 in erf11 mutants.
Figure 7.
Attenuated SA-induced defense response in erf11 mutants. A, Typical Pst DC3000-infected disease symptoms in Col-0 and erf11 plants at 48 hpi with or without SA treatment. Four-week-old plants were sprayed with 1 mm SA or 0.1% (v/v) ethanol solution and then inoculated with Pst DC3000 at 24 h after SA treatment. Photographs of representative leaves were taken 48 hpi. B, Bacterial growth in inoculated leaves of Col-0 and erf11 plants in planta with or without SA treatment. Four-week-old plants were sprayed with 1 mm SA or 0.1% (v/v) ethanol solution and then inoculated with Pst DC3000 at 24 h after SA treatment. Bacteria were isolated from the plants at 48 hpi and quantified with gradient dilution assay. The P values (bacterial count of each genotype with SA pretreatment versus each genotype with mock pretreatment at the same time point) were determined by two-tailed Student’s test assuming equal variance (P < 0.05). C, In situ and (D) quantitative analysis of superoxide anion accumulation in inoculated leaves of Col-0 and erf11 plants with or without SA treatment by NBT staining and biochemical testing, respectively. Four-week-old wild-type plants were sprayed with 1 mm SA or 0.1% (v/v) ethanol solution and then inoculated with Pst DC3000 at 24 h after SA treatment. Leaf samples were collected at 24 hpi. The P values (superoxide anion of each genotype with SA pretreatment versus each genotype without SA pretreatment under Pst-infected at the same time point) were determined by two-tailed Student’s test assuming equal variance (P < 0.05). E, Partial suppression of SA-induced expression of defense genes in erf11 plants. Four-week-old wild-type (Col-0) and erf11 plants were sprayed with 1 mm SA or 0.1% (v/v) ethanol solution for 24 h and then inoculated leaves were collected for RNA isolation. Relative expression is shown relative to the transcript levels of an internal AtTub4 gene. The P values (PR expressions of each genotype versus Col-0 under SA treatment at the same time point) were determined by two-tailed Student’s test assuming equal variance (P < 0.05). Data presented are the means ± sd from three independent experiments, and asterisks indicate significant differences at P < 0.05 between erf11 and Col-0 plants.
BT4 Directly Functions Downstream of ERF11 in Arabidopsis Defense against Pst DC3000
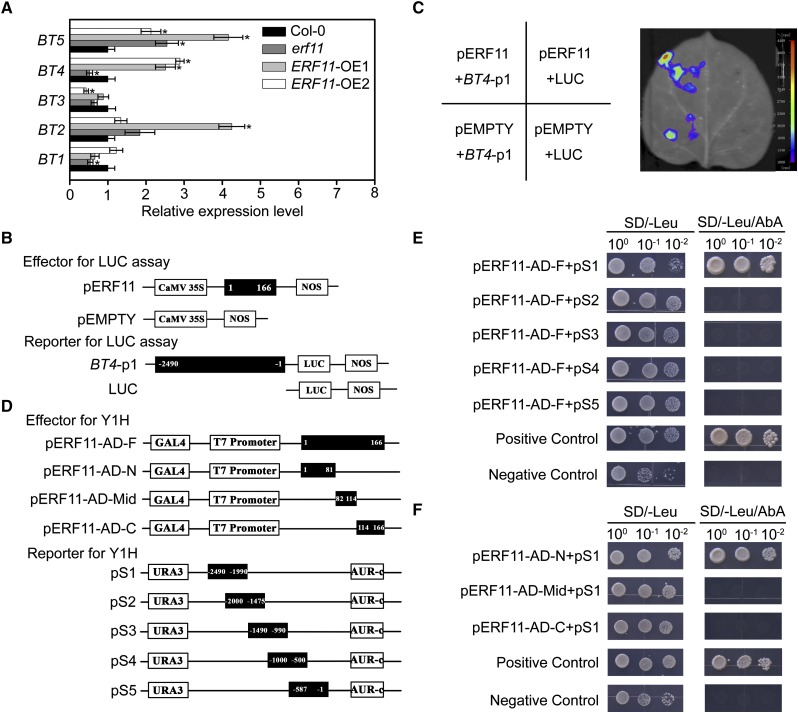
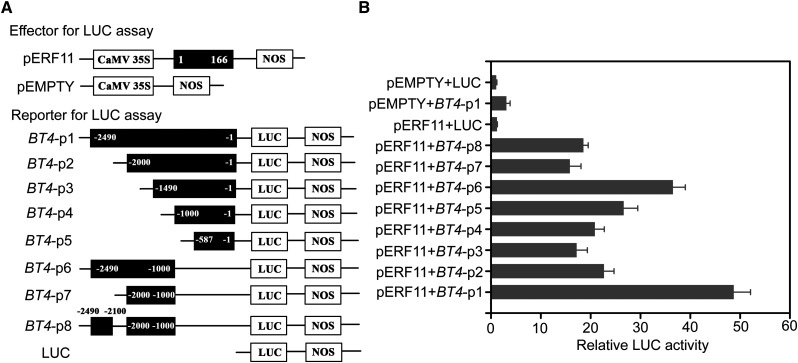
Both BT4 and ERF11 had positive roles in Arabidopsis defense against Pst DC3000 and were modulated by the SA and ET signaling pathways (Figs. 1, 3, 5, and 6). Moreover, DRE/CRT elements and the GCC-box were found in the putative promoter of BT4 (Supplemental Fig. S2A). We then determined whether BT4 transcription was controlled by ERF11. The expression levels of BT genes in Col-0, erf11, ERF11-OE1, and ERF11-OE2 plants were evaluated. Only BT4 transcription showed a close correlation with ERF11 expression (Fig. 8A). BT4 expression was significantly increased in ERF11-OE1 and ERF11-OE2 plants but decreased in erf11 mutants compared with Col-0. To further analyze whether ERF11 could activate BT4 expression, we performed a tobacco transient expression assay—the 2490 bp promoter upstream from the initiation codon of BT4 (BT4-p1) was fused into the luciferase (LUC) reporter gene and cotransfected with the effector of full-length ERF11 protein (pERF11) into tobacco leaves (Fig. 8, B and C). The pERF11 effector coexpressed with BT4-p1 reporter significantly increased LUC activity compared with the control (Fig. 8C). These results demonstrate that ERF11 can activate BT4 transcription.
Figure 8.
ERF11 targets BT4 promoter. A, Expression of BT4 modulated by ERF11. Relative expression of BT1, BT2, BT3, BT4, and BT5 in 4-week-old wild-type (Col-0), erf11, ERF11-OE1, and ERF11-OE2 plants. Relative expression is indicated as folds of the transcript level of an internal AtTub4 gene. The P values (BT expressions of each genotype versus Col-0 at the same time point) were determined by two-tailed Student’s test assuming equal variance (P < 0.05). Data presented are the means ± sd from three independent experiments, and asterisks indicate significant differences at P < 0.05 between erf11/ERF11-OE1/ERF11-OE2 and Col-0 plants. B, Schematic diagram of effector and reporter employed in LUC activity assay. The numbers in fragments (pERF11, BT4-p1) indicate the positions of the nucleotides at the 5′ or 3′ end of each fragment relative to the translation start site in reporter or amino acids in effector. C, Transient expression assays showed that ERF11 activates the transcription of BT4. Luminescence imaging of Nicotiana tabacum leaves is shown 48 h after coinfiltration with reporter and effector. D, Schematic diagram of effector and reporter used in Y1H assay. The numbers in fragments (pERF11-AD-F, pERF11-AD-N, pERF11-AD-Mid, pERF11-AD-C, pS1, pS2, pS3, pS4, and pS5) indicate the positions of the nucleotides at the 5′ or 3′ end of each fragment relative to the translation start site in reporter or amino acids in effector. E, Interaction of full-length ERF11 with different fragments of the BT4 promoter. pS1, pS2, pS3, pS4, and pS5 indicate the reporters carrying different fragments of the BT4 promoter as schematic diagram of reporter for Y1H. Transformed yeast cells containing both effector and reporter were plated on the selective medium (SD/−Leu/AbA). AbA, Aureobasidin A. Cotransformation of pGBKT7-53 and pGADT7-Rec T was employed as positive control. Cotransformation of pGBKT7-lam and pGADT7-Rec T was used as negative control. F, Interaction of the BT4 promoter fragment pS1 with different lengths of ERF11. pERF11-AD-F, pERF11-AD-N, pERF11-AD-Mid, and pERF11-AD-C indicate the effectors carrying full-length protein, N-terminal, middle-region, and C-terminal portions of ERF11, respectively. Transformed yeast cells containing both effector and reporter were plated on the selective medium (SD/−Leu/AbA). Cotransformation of pGBKT7-53 and pGADT7-Rec T was employed as positive control. Cotransformation of pGBKT7-lam and pGADT7-Rec T was used as negative control.
A Y1H assay was performed to investigate whether ERF11 physically interacted with the promoter of BT4. The generation of full-length (pERF11-AD-F) effectors and reporters of BT4 promoter pS1, pS2, pS3, pS4, and pS5 is schematically described in Figure 8D. When effector pERF11-AD-F and reporters pS1, pS2, pS3, pS4, and pS5 were cotransformed into the Y1H gold yeast cell, respectively, pERF11-AD-F significantly activated AbA resistance in pS1, but not the other reporters (Fig. 8E). To further determine which domain of ERF11 protein (pERF11) directly bound to the region from −2490 to −1990 of the BT4 promoter (pS1), we produced diverse effectors, including N-terminal region (pERF11-AD-N), middle region (pERF11-AD-Mid), and C-terminal region (pERF11-AD-C), consulting to pERF11 domains (Supplemental Fig. S5). When the effectors pERF11-AD-N, pERF11-AD-Mid, and pERF11-AD-C were respectively cotransformed with pS1 into the Y1H gold yeast cell, pERF11-AD-N significantly activated AbA resistance in the pS1 reporter (Fig. 8F). Analysis of the pERF11 domain revealed that the AP2 domain was located in the N-terminal region and the ETHYLENE-RESPONSE FACTOR Amphiphilic Repression (EAR) motif was located at the C-terminal region of ERF11 (Supplemental Fig. S5). These results suggest that the N-terminal region of ERF11, possibly the AP2 domain, interacts with the region from −2490 to −1990 bp (pS1) of the BT4 promoter in yeasts.
Analysis of the BT4 promoter sequence revealed that the GCC-box and two DRE elements were located −2065, −2186, and −1890 bp upstream of the initiation codon, respectively (Supplemental Fig. S2). It has been suggested that some ERF proteins impart tolerance to abiotic stress through DRE/CRT elements, while others use the GCC-box element (Wang et al., 2014; Zhu et al., 2014; Phukan et al., 2017). We speculate that the pERF11 directly interacts with the GCC-box of the BT4 promoter. To further confirm the binding of ERF11 to BT4 promoter in vivo, a LUC activity assay was performed using the transient expression assay system in Arabidopsis protoplasts. The construction of effector pERF11 and reporters BT4-p1, BT4-p2, BT4-p3, BT4-p4, BT4-p5, BT4-p6, BT4-p7, and BT4-p8 is illustrated in Figure 9A. Under the activation of effector pERF11, LUC activity significantly increased in reporters that contained the ERF11-binding core sequence such as BT4-p1 and BT4-p6; however, low LUC activity was noted in reporters without the ERF11-binding core sequence: BT4-p2, BT4-p3, BT4-p4, BT4-p5, and BT4-p7. Moreover, LUC activity significantly decreased in the reporter with BT4-p8, in which the fragment from −−2100 to −2000 bp was deleted (Fig. 9B). These results suggest that ERF11 targets the region from −2100 to −2000 bp of the BT4 promoter, probably the GCC-box, to activate its transcription.
Figure 9.
The region from −2100 to −2000 bp of BT4 promoter is recognized by ERF11 in a transient expression assay. A, Schematic diagram of effector and reporter constructs used in protoplast-mediated transient cotransformation expression assay. The coding domain of ERF11 is fused downstream of Cauliflower mosaic virus 35S in pCAMBIA1307. The promoter fragment of BT4 is fused upstream of the LUC gene in pGreenII-0800-LUC. The numbers in fragments indicate the positions of the nucleotides at the 5′ or 3′ end of each fragment relative to the translation start site in reporter or amino acids in effector. B, Relative luciferase activity detected by transient cotransformation with reporter and effector into Arabidopsis protoplasts. To normalize the values obtained for each independent cotransformation, the REN from Renilla spp. was used as an internal control. Luciferase activity is quantified in arbitrary units relative to REN. sd is based on three independent experiments.
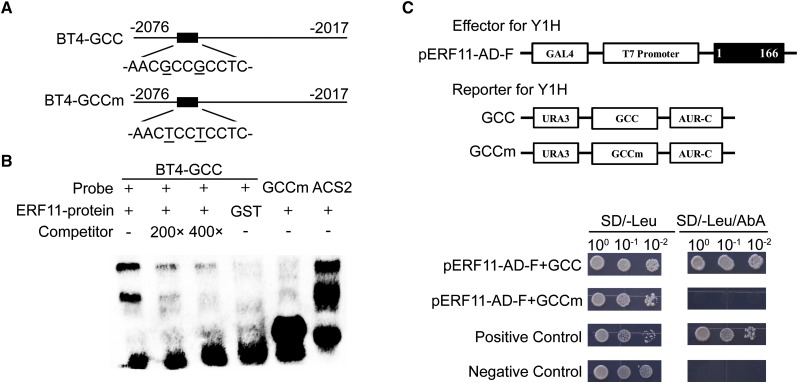
To further confirm whether ERF11 physically binds to the GCC-box of the BT4 promoter, we performed the EMSA and expressed and purified the GST-tagged ERF11 fusion protein in Escherichia coli. The positive control indicated that the GST-ERF11 fusion protein interacted with the DRE probe of ACS2 (Fig. 10B), as previously reported (Li et al., 2011). Similarly, the GST-ERF11 fusion protein was able to bind to the DNA probes containing the GCC-box of the BT4 promoter (BT4-GCC) but failed to bind to the mutated probes (BT4-GCCm). Furthermore, increasing the concentration of unlabeled BT4-GCC probes in the binding reactions led to much weaker combined bands. These results were further confirmed using the Y1H assay, and the effector pERF11-AD-F was found to significantly activate AbA resistance in the GCC reporter, but not in the GCCm reporter (Fig. 10C). These results suggest that ERF11 physically interacts with the GCC-box of the BT4 promoter in vitro.
Figure 10.
Recognition of GCC-box of BT4 promoter by ERF11 in vitro and in yeast cells. A, Schematic diagram shows the positions of the probes or bait employed in EMSA and Y1H assay. The numbers in fragments (BT4-GCC, BT4-GCCm) indicate the positions of the nucleotides at the 5′ or 3′ end of each fragment relative to the translation start site. BT4-GCCm was similar to BT4-GCC except the base G mutated to T. B, EMSA for binding to GCC-box sequence in the promoter of BT4 by ERF11 in vitro. The full length of ERF11 protein fused to GST was used to detect interaction. Biotin-labeled probes were incubated with ERF11-GST protein. GST protein was used as a negative control. ACS2 probe was used as a positive control, a mutated version of BT4-GCC (GCCm) was used as a negative control. Unlabeled DNA was added in 200- and 400-fold molar excess as competitors. “−” and “+” represent absence or presence, respectively. C, Y1H assay for binding to GCC-box region of BT4 promoter by ERF11 in yeast cell. Cotransformation of pERF11-AD-F and GCC or GCCm reporter was used as test group. Transformed yeast cells containing both effector and reporter were plated on the selective medium (SD/−Leu/AbA). AbA, Aureobasidin A. Cotransformation of pGBKT7-53 and pGADT7-Rec T was employed as positive control. Cotransformation of pGBKT7-lam and pGADT7-Rec T was utilized as negative control. The numbers in fragments indicate the positions of the nucleotides at the 5′ or 3′ end of each fragment relative to the translation start site in reporter or amino acids in effector.
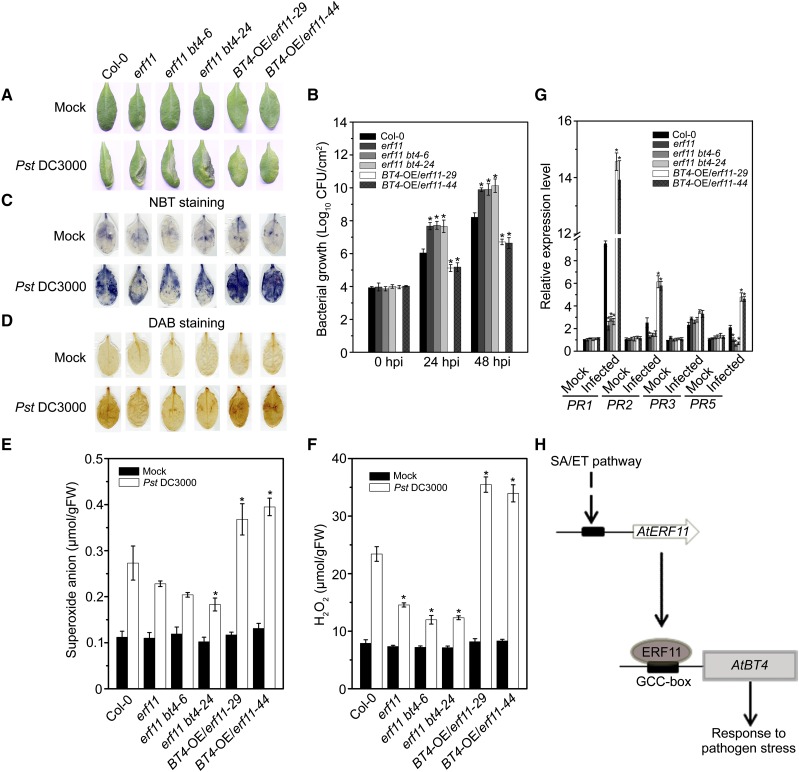
To confirm the genetic relationship between BT4 and ERF11 in Arabidopsis defense against Pst DC3000, we further generated erf11 bt4 and BT4-OE/erf11 plants by crossing. We obtained two double mutants and two complement transgenic plants: erf11 bt4-6, erf11 bt4-24, BT4-OE/erf11-29, and BT4-OE/erf11-44. Morphological phenotypes and expression analysis are shown in Supplemental Figure S6; intriguingly, double mutants and complement transgenic plants showed no obvious morphological abnormalities and were indistinguishable from their parents. We first analyzed the disease symptoms of erf11 bt4-6, erf11 bt4-24, BT4-OE/erf11-29, and BT4-OE/erf11-44 plants following Pst infection. Analysis of disease symptoms in the BT4-OE/erf11-29 and BT4-OE/erf11-44 complement plants as well as erf11 bt4-6 and erf11 bt4-24 double mutants compared with single erf11 mutants revealed that complement lines exhibited conspicuous resistance against the virulent pathogen Pst DC3000, but disease symptoms of the double mutants were similar to those of erf11 mutants (Fig. 11A). In agreement with this finding, the bacterial counts in erf11 bt4-6 and erf11 bt4-24 double mutants did not significantly differ to those of single erf11 mutants. However, there were lower bacterial counts in the BT4-OE/erf11-29 and BT4-OE/erf11-44 complement transgenic plants compared with Col-0, especially lower than those in single and double mutants (Fig. 11B). Accumulation of superoxide anion and H2O2 of BT4-OE/erf11-29 and BT4-OE/erf11-44 inoculated leaves were higher than in Col-0 and significantly higher than in erf11 bt4-6 and erf11 bt4-24 double mutants at 24 hpi, whereas those of erf11 bt4-6 and erf11 bt4-24 double mutants were similar to erf11 mutants (Fig. 11, C–F). Moreover, we quantified the relative expression levels of defense-related genes in double mutants and complement transgenic plants to compare with single erf11 mutants during Pst DC3000 infection. Expression levels of PR1, PR2, and PR5 in the BT4-OE/erf11-29 and BT4-OE/erf11-44 plants were significantly higher than those in the Col-0, especially than those of erf11 mutants, whereas expression levels of these genes in erf11 bt4-6 and erf11 bt4-24 double mutants were similar to erf11 mutants (Fig. 11G). Taken together, these results not only indicate that BT4 is directly downstream of ERF11 and overexpression of BT4 in the erf11 background could rescue the erf11 mutant phenotype during Pst DC3000 inoculation, but also suggest that ERF11 and BT4 genes belong to the same signaling pathway to regulate the Arabidopsis resistance against Pst DC3000.
Figure 11.
Overexpression of BT4 in erf11 background rescues the resistance to Pst DC3000. A, Typical Pst DC3000-infected symptoms in wild-type (Col-0), erf11, erf11 bt4-6, erf11 bt4-24, BT4-OE/erf11-29, and BT4-OE/erf11-44 plants. Four-week-old plants were inoculated by Pst DC3000 bacterial suspension or 10 mmol/L MgCl2 and kept at high humidity. Photographs of representative leaves were taken 48 hpi. The experiments were repeated three times with similar results. B, Bacterial growth in the inoculated leaves detected in planta. Bacteria were isolated from the plants 24 and 48 hpi and quantified with gradient dilution technique. The P values (bacterial count of each genotype versus Col-0 under Pst treatment at the same time point) were determined by two-tailed Student’s test assuming equal variance (P < 0.05). C, In situ and (E) quantitative analysis of superoxide anion accumulation in Pst DC3000-inoculated leaves by NBT staining and biochemical testing, respectively. Four-week-old plants were inoculated with Pst DC3000 or 10 mmol/L MgCl2 and kept in high humidity. Leaf samples were collected at 24 hpi. The P values (superoxide anion of each genotype versus Col-0 under Pst-treatment at the same time point) were determined by two-tailed Student’s test assuming equal variance (P < 0.05). D, In situ and (F) quantitative analysis of H2O2 accumulation in Pst DC3000-inoculated leaves by DAB staining and biochemical testing, respectively. Four-week-old plants were inoculated with Pst DC3000 or 10 mmol/L MgCl2 and kept in high humidity. Leaf samples were collected at 24 hpi. The P values (H2O2 accumulation of each genotype versus Col-0 under Pst-treatment at the same time point) were determined by two-tailed Student’s test assuming equal variance (P < 0.05). G, Relative expression levels of PR1, PR2, PR3, and PR5 in leaves of 4-week-old wild-type (Col-0), erf11, erf11 bt4-6, erf11 bt4-24, BT4-OE/erf11-29, and BT4-OE/erf11-44 plants after Pst DC3000 treatment for 24 h. The P values (PR expressions of each genotype versus Col-0 under Pst-treatment at the same time point) were determined by two-tailed Student’s test assuming equal variance (P < 0.05). Data presented are the means ± sd from three independent experiments andasteriks indicate significant differences at P < 0.05 between erf11, erf11 bt4-6, erf11 bt4-24, BT4-OE/erf11-29, BT4-OE/erf11-44, and Col-0 plants. H, Model of ERF11 transcriptional activates BT4 to modulate SA/ET-regulated plant resistance against Pst DC3000. During Pst infection, ERF11 transcription was modulated by SA and ET signaling pathways. Then, numerous ERF11 TFs accumulate in the nucleus. ERF11, in turn, interacts with the GCC-box of BT4 promoter to activate expression of BT4. Next, BT4 protein mediates transcription of PR genes to regulate plant resistance to Pst DC3000.
Overall, our results demonstrate that ERF11 directly activates BT4 in the Arabidopsis response to Pst DC3000 infection and is dependent on the SA and ET signaling pathways.
DISCUSSION
Earlier studies showed that BT4 is required for resistance against B. cinerea in Arabidopsis and indicated that it regulated the expression of defense-related genes in response to the SA and JA signaling pathways (Hao et al., 2013). Here, we suggested that BT4 was modulated by the SA and ET signaling pathways to positively regulate Arabidopsis defense against Pst DC3000. Moreover, BT4 loss of function compromised the SA-induced defense response to Pst DC3000. We found that the putative promoter of BT4 contained DRE/CRT elements and the GCC-box, which are specific target elements for ERF TFs. Further analyses focused on screening potential ERF genes involved in the Arabidopsis defense against Pst DC3000 depending on the SA and ET signaling pathways. Through mining the available microarray databases and combined transcriptional confirmation, we observed that ERF11 was induced by SA, ACC, and Pst DC3000 treatment and modulated by the SA and ET signaling pathways. Indeed, ERF11 loss-of-function compromised Arabidopsis resistance against Pst DC3000 and the SA-induced defense response. Next, we focused on the relationship between ERF11 and BT4 to address the idea that an ERF11-BT4 transcriptional cascade was involved in Arabidopsis defense against Pst DC3000. Our data indicated that ERF11 was bound to the promoter of BT4 in vitro and in vivo. Moreover, either ERF11-OE or BT4-OE was sufficient to increase the expression levels of PR genes under Pst infection and enhance defense against Pst DC3000. In addition, BT4 overexpression in the erf11 background also enhanced expression levels of PR genes with Pst inoculation and increased resistance against Pst DC3000. Therefore, this research revealed that the transcriptional activation of BT4 by ERF11 is a key step in SA/ET-regulated plant resistance against Pst DC3000.
In Arabidopsis, both SA and ET signaling are necessary to regulate the defense response (Nawrath and Métraux, 1999; Wildermuth et al., 2001; Berrocal-Lobo et al., 2002). Furthermore, the JA signaling pathway is known to regulate Arabidopsis resistance against necrotrophic pathogens, whereas SA signaling contributes to defense against biotrophic/hemibiotrophic pathogens, and the SA and JA pathways are antagonistic in plant defense responses (Vlot et al., 2009; Fu and Dong, 2013; Zander et al., 2014; Yang et al., 2015; Zhang et al., 2017). Previously, we demonstrated that BT4 had a positive function in resistance against B. cinerea in Arabidopsis and regulated the expression of defense-related genes in response to the SA and JA signaling pathways (Hao et al., 2013). However, in this research, we found that BT4 also had a positive function in defense against the hemibiotrophic pathogen Pst DC3000 and was modulated by the SA and ET pathways (Figs. 1 and 3). These results indicate that ET and the ET signaling pathway are important integrators in the cross talk between SA and JA. An increasing number of studies have revealed that several ERFs are coordinately induced by SA, JA, and ET, indicating that ERFs can synergistically integrate the SA and the ET/JA signaling pathways but not antagonize them (Zarei et al., 2011; Chen et al., 2012; Deokar et al., 2015; Zhang et al., 2015, 2016). We revealed that ERF11 was coordinately induced by SA, ACC, and JA treatment (Fig. 5; Supplemental Fig. S7). Being downstream of the ET signaling pathway, we demonstrated that ERF11 physically interacted with the BT4 promoter (Figs. 9–11). Thus, we confirmed that ERF11 plays a vital role in synergistic cross talk with SA, JA, and ET. Perhaps ERF11 synergistic integration with the SA, JA, and ET signaling pathways leads to BT4 playing a synergistic role in plant defense against necrotrophic and hemibiotrophic pathogens.
SA contributes to plant defense against biotrophic/hemibiotrophic pathogens (Delaney et al., 1994; Lawton et al., 1995). However, mutants and transgenic plants, with diminished SA synthesis and accumulation, are compromised in triggering plant defense responses and are susceptible to pathogen infection. Direct application of SA and its analogs has been reported to increase ROS accumulation, activate expression of PR genes, and enhance resistance to biotrophic pathogens (Mur et al., 2008; Shah, 2009; Coll et al., 2011). Indeed, we revealed that direct application of SA enhanced resistance and ROS accumulation in the wild type: Col-0, No, Ler, and Ws (Supplemental Fig. S1). However, disruption of ERF11 or BT4 compromised SA-induced resistance in erf11 and bt4 mutants (Figs. 2 and 7), indicating that ERF11 and BT4 played critical roles in the SA defense response against Pst DC3000.
Downstream of the multiple interactions of diverse hormone signaling pathways, TFs play very important functions in regulating the expression of PRs and mediating plant defense. The ERF proteins, as plant-specific TFs, are focused in plant defense responses and involved in regulation of PRs. For example, the ERF1 TF triggers transcription of PDF1.2, enhancing resistance against B. cinerea, and this regulation depends on the integral ET signaling pathway, especially on EIN3 (Berrocal-Lobo et al., 2002; Berrocal-Lobo and Molina, 2004). In this study, through screening and identification of available microarray data, we revealed that ERF11 was induced by both SA and Pst DC3000 (Fig. 4; Supplemental Fig. S4). Following further analysis of ERF11, we found that ERF11 had a positive role in Arabidopsis-Pst DC3000 interaction, and transcription of ERF11 was modulated by the SA and ET signaling pathways during Pst infection, such as by NPR1 and EIN3 (Figs. 5 and 6). pERF11 belongs to subfamily VIII-B-1a, a group not reported in plant defense responses, which have vastly different amino acid sequences to ERF1. In addition to different amino acid sequences, ERF11 was not found to directly bind to the promoter of PR genes. Interestingly, we found that BT4 expression significantly increased in ERF11-OE1 and ERF11-OE2 plants but decreased in erf11 mutants (Fig. 8A). Many of the ERF TFs specifically bind to the GCC-box (AGCCGCC) and DRE/CRT elements (TACCGACAT), the core cis-motif present in the promoter of target genes (Yamaguchi-Shinozaki and Shinozaki, 1994; Ohme-Takagi and Shinshi, 1995; Brown et al., 2003; Van der Does et al., 2013). With the help of Y1H and EMSA, these results indicated that ERF11 could bind to the promoters of BT4 to activate transcription of BT4 (Figs. 8–10). The ERF11 belongs to the subfamily VIII-B-1a, and all members of this subfamily (ERF3, ERF4, and ERF7–12) contain a transcription repressor EAR motif near their C terminus (McGrath et al., 2005; Nakano et al., 2006a, 2006b). In earlier research, we found that ERF11 interacts with the DRE motif of the ACS2/5 promoters to repress its transcription, resulting in decreased ET biosynthesis, suggesting that the EAR motif of ERF transcription repressors plays a crucial role in modulating expression of target genes (Li et al., 2011). In this study, the N terminus of ERF11, AP2 domain, was revealed to bind to the GCC-box of the BT4 promoter to activate BT4 expression and mediate resistance against Pst DC3000 (Figs. 8–10). This finding indicates that different domains of the same TF play activation or repression functions in diverse stress responses. However, the mechanisms by which the plant regulates the same TF to activate or suppress target genes remain unclear.
Increasing evidence demonstrates that transcription regulators are involved in the plant defense response (Spoel et al., 2003; Hao et al., 2013; Liu et al., 2017). The NPR1 protein, belonging to BTB/POZ domain proteins, is the core of the SA signaling pathway (Durrant and Dong, 2004; Kesarwani et al., 2007). The NPR1 protein is unable to transcriptionally regulate target genes and acts as a transcription regulator to interact with TGA TFs activating the expression of defense genes (Fan and Dong, 2002). However, we found that BT4 protein possessed transactivation activity in yeast cells and was located in the nucleus. Furthermore, BT4 was observed to play an important role in Arabidopsis defense against B. cinerea and Pst DC3000 by regulating the expression of defense-related genes (Hao et al., 2013; Fig. 1G). These results indicate that transcription regulator BT4 possesses the characteristics of a TF to regulate transcription of defense-related genes. In future studies, we will focus on whether BT4 directly binds to the promoter of defense-related genes to mediate plant defense against pathogen challenge.
The plant defense response is a complex process that involves multiple physiological, pathological, and molecular mechanisms. In such a process, transcriptional regulation is a key step for plant defense against pathogens. Here, we focused on how ERF11 transcriptionally regulated BT4 expression to enhance the Arabidopsis defense response against Pst DC3000. Based on our research, we propose a regulatory model for ERF11 mediation in the transcription of BT4 during Pst DC3000 infection in Arabidopsis (Fig. 11H). During Pst DC3000 infection, ERF11 transcription was modulated by the SA and ET signaling pathways, followed by much accumulation of ERF11 TFs in the nucleus. ERF11, in turn, interacted with the GCC-box of the BT4 promoter to activate BT4 expression. Next, the BT4 protein mediated the transcription of PR genes to enhance plant resistance to Pst DC3000.
MATERIALS AND METHODS
Plant Material and Bacterial Strains
The background of all Arabidopsis (Arabidopsis thaliana) mutants used in this study was Col-0. The Arabidopsis mutants bt4-1 (SALK_015577.54.25.x), bt4-2 (SALK_045370C), erf11 (SALK_116053), sid2 (SALK_045134), NahG, npr1-1 (SALK_046187), ein2 (CS3071), ein3-1(CS8025), and eto1 (CS3072) were obtained from the Arabidopsis Biological Resource Center (http://abrc.osu.edu/), and the ein3 eil1 double mutant was provided by Professor H.W. Guo at Southern University of Science and Technology. Transgenic BT4-OE plants constitutively overexpressing BT4 driven by the Cauliflower mosaic virus 35S promoter and transgenic AtERF11-overexpressing plants (ERF11-OE1 and ERF11-OE2) driven by the 35S promoter labeled with HA (influenza hemagglutinin epitope) were developed in previous studies (Li et al., 2011; Hao et al., 2013). The erf11 bt4-6 and erf11 bt4-24 double mutants were made by crossing erf11 and bt4-1 as well as erf11 and bt4-2 plants. The BT4-OE/erf11 was produced by crossing the erf11 mutant with BT4-OE line. All seeds were first surface sterilized using ethanol, sown on Murashige and Skoog medium plates containing 0.5% (w/v) phytagel, incubated at 4°C in darkness for 3 to 5 d, and then cultivated at 22°C with a 16/8 h light/dark cycle.
Analysis of Available Microarray Data
The expression pattern of ERF11 during biotic and hormone stress in Arabidopsis was carried out using publicly available microarray CEL files in the GEO database (Barrett et al., 2013). GSE5520, GSE51626, and GSE9955 were used for expression analysis (Naseem et al., 2012; Singh et al., 2015). The data were analyzed by GEO2R, an R-based web application, to help identify differentially expressed genes (DEGs; Barrett et al., 2013). The putative DEGs between mutant and wild type or between control (mock) and treatment were identified using a two-step process: (1) genes that were 2-fold up- or down-regulated were selected and (2) Welch’s t test was performed (P < 0.05). Finally, a volcano map illustrating DEGs was constructed using Graphpad Prism 6 software (https://www.graphpad.com).
Pathogen Inoculation and Hormone Treatments
Pst DC3000 (Pseudomonas syringae pv. tomato DC3000) was cultured overnight at 28°C in King’s B medium containing 25 μg/mL rifampicin. When the bacterial cell concentration reached OD600 of 0.8 to 1.0, the cells were centrifuged and resuspended in 10 mm MgCl2 buffer to OD600 of 0.002. Then, bacterial cells were inoculated into rosette leaves by hand infiltration using 1-mL syringes without a needle, and the infected plants kept in a container with high humidity and in darkness for 24 h. To determine the bacterial population in plants, leaf disks were obtained from different inoculated leaves and homogenized with 200 μL of MgCl2 solution. After a series of gradient dilutions, the suspension was plated on King’s B medium supplemented with 25 μg/mL rifampicin, and bacterial colonies were counted at 2 d after incubation at 28°C.
For analysis of gene expression after phytohormone treatment, sterilized Arabidopsis seeds grown in Murashige and Skoog medium for 7 d were transferred to Whatman filter paper containing 50 μm SA or 10 μm ACC. For the control, seeds were transferred onto filter paper containing 0.1% (v/v) ethanol solution or water. To verify SA-induced plant resistance against Pst DC3000, 4-week-old Arabidopsis plants were pretreated with 1 mm SA or 0.1% (v/v) ethanol solution for 24 h and then inoculated with Pst DC3000.
RNA Extraction and Real Time QuantitativePCR Analysis of Gene Expression
Total RNA was extracted from 7-d-old plants or 4-week-old mature plants and treated with hormones or the pathogen using Trizol reagent (Invitrogen, http://www.invitrogen.com/). Then the total RNA was reverse transcribed to complementary DNA (cDNA) using M-MLV reverse transcriptase (Reverse Transcriptase system; Promega, http://www.promega.com/) according to the manufacturer’s instructions. Subsequently, gene expression was measured by real-time qPCR analysis with SYBR Premix (Takara, http://www.takarabiomed.com.cn/) using the IQ5 real-time system (Bio-Red, http://www.bio-rad.com/). All PCR amplifications were performed in 96-well optical reaction plates with 45 cycles of denaturation for 15 s at 95°C, annealing for 20 s at 56°C, and extension for 45 s at 72°C. Expression levels were normalized using AtTUB4. The primers used in qPCR are listed in Supplemental Table S1. Each qPCR was repeated thrice independently.
Measurement of ROS Accumulation
To detect superoxide anion and H2O2 accumulation in situ, NBT staining and DAB were used as described by Zhang et al. (2016). Leaves were transferred to 1 mg/mL DAB solution and vacuum-infiltrated at 37°C for 30 min. Subsequently, pigments from the leaves were removed with 95% ethanol until colorless.
Superoxide anions in leaves were quantified using a superoxide assay kit (Beyotime; http://www.beyotime.com/product/S0063.htm) according to the manufacturer’s instructions. Fluorescence was measured with a Bio-Tek Synergy 4 plate reader (excitation, 370 nm; emission, 420 nm). The superoxide anion concentration in each sample was calculated using a standard curve, which was linear with NaNO2 concentration.
The H2O2 in leaves was quantified using a hydrogen peroxide assay kit (Beyotime; http://www.beyotime.com/product/S0038.htm/) according to the manufacturer’s instructions. Fluorescence was measured with a BioTek Synergy 4 plate reader (excitation, 530 nm; emission, 590 nm). The H2O2 concentration in each sample was calculated using a standard curve, which was linear with H2O2 concentration.
Y1H Assay
Matchmaker One-Hybrid System (Clontech; http://www.clontech.com/) was used with slight modification to perform the Y1H assay for investigating the interaction of TFs with target gene promoters. The BT4 promoter fragment (−2490 to −1 bp) was divided into five sections: S1 (−2490 to −1990 bp), S2 (−2000 to −1475 bp), S3 (−1490 to −990 bp), S4 (−1000 to −500 bp), and S5 (−587 to −1 bp). Each section was PCR amplified. The obtained PCR-amplified fragments were connected into the pAbAi vector as reporters. The reporter vectors were linearized at the BbsI or BstBI site as described in the user manual and transformed into Y1H gold strain. The full-length cDNA of ERF11 as well as the N-terminal, middle-region, and C-terminal fragments were cloned into the pGADT7 vector containing a GAL4 transcriptional activation domain, yielding effectors pERF11-AD-F, pERF11-AD-N, pERF11-AD-Mid, and pERF11-AD-C, respectively. After confirming integration of the reporter vectors into the yeast strain, the effector vectors were respectively transformed into the Y1H gold strain, which carried a different reporter vector. The cotransformation yeasts were cultivated in SD/−Leu and SD/−Leu/AbA medium. The Y1H assay was performed according to the manufacturer’s protocol (Matchmaker One-Hybrid System; Clontech; http://www.clontech.com/). To confirm ERF11 binding to the GCC-box of AtBT4 promoter, the fragments containing the GCC-box and GCC-box mutation of AtBT4 promoter were obtained from IDOBIO, connected into the pAbAi vector as reporters, and the Y1H assay was performed (Matchmaker One-Hybrid System; Clontech).
Luciferase Activity Assay
To further investigate the ERF11 interaction with BT4 promoter, the LUC activity assay was performed using leaves of tobacco (Nicotiana tabacum) and protoplasts of Arabidopsis. The full-length cDNA of ERF11 was cloned into the pCAMBIA1307 vector containing a 35S Cauliflower mosaic virus promoter to achieve constitutive overexpression of ERF11 as an effector. The BT4 promoter, p1 (−2490 to −1 bp), p2 (−2000 to −1 bp), p3 (−1490 to −1 bp), p4 (−1000 to −1 bp), p5 (−587 to −1 bp), p6 (−2490 to −1000 bp), p7 (−2000 to −1000 bp), and p8 (−2490 to −2100 and −2000 to −1000 bp), were cloned with primers given in Supplemental Table S1 and introduced into the pGreenII-0800-LUC vector containing REN and LUC genes. The reporter and effector plasmids were respectively transformed into Agrobacterium tumefaciens strain GV3101. The strains were incubated in Yeast Mannitol Medium (YEB) overnight and centrifuged to harvest the cells, and the cells resuspended in dilution buffer (10 mm MES, 0.2 mm acetosyringone, and 10 mm MgCl2) to a concentration of OD600 = 1.0. Then, equal volumes of different bacterial suspensions were coinjected into the leaves of 4-week-old tobacco plants with a needleless syringe. After bacterial infection, plants were cultivated in darkness for 12 h and then kept under 16/8 h of light/dark cycle for 48 h at 24°C. The leaves were sprayed with 100 mm luciferin (VivoGlo Luciferin; Promega; https://www.promega.com.cn/) and placed in darkness for 5 min. The LUC activity was observed using a low-light cooled CCD imaging apparatus (iXon; Andor Technology; http://www.andor.com/). The experiments were performed in triplicate.
Analysis of transient expression of LUC activity in protoplasts was performed as described by Zhao et al. (2016). In brief, the leaf debris (0.5-mm width) were cut from the second leaves using a razor blade and soaked in 15 mL of enzyme solution containing 20 mm MES (pH 5.7), 1.5% (w/v) cellulase R10 (Cellulase Onozuka R10; Yakult, http://www.yakult.co.jp/ypi/en/tos.html), 0.4% (w/v) macerozyme R-10 (Macerozyme R-10; Yakult), 0.4 m mannitol, 20 mm KCl, 10 mm CaCl2, 1 mm β-mercaptoethanol, and 0.1% bovine serum albumin (BSA). Subsequently, the leaves were incubated at room temperature and 20 rpm for 4 h in darkness. The cell lysate was filtered with a sieve and washed twice with W5 buffer: 2 mm MES (pH 5.7), 154 mm NaCl, 125 mm CaCl2, and 5 mm KCl. The protoplast suspension was centrifuged at 100g for 3 min to harvest protoplast cells. Then, protoplast cells were resuspended in MMG solution (4 mm MES [pH 5.7], 0.4 m mannitol, and 15 mm MgCl2), mixed with plasmid DNA mixture and 110 μL of PEG solution (40% [w/v] PEG-4000, 0.2 m mannitol, and 100 mm CaCl2), and incubated in darkness for 15 min at 28°C. Subsequently, the protoplasts were washed twice with W5 solution to eliminate PEG solution and incubated in W5 solution in darkness for 12 h at 28°C. The protoplast LUC activity was determined using a multifunction microplate reader (TriStar LB 941; Berthold; https://www.berthold.com/) using a dual luciferase reporter gene assay kit (Beyotime; http://www.beyotime.com/product/RG027.htm). All experiments were performed in triplicate.
EMSA
To construct plasmids for the expression of full-length (1–166 amino acids) pERF11 in Escherichia coli BL21, the cDNA fragments of AtERF11 were obtained by PCR amplification and inserted into the multicloning sites of the pGEX-6p-1 vector. The fusion protein was purified using ProteinIso GST Resin according to the manufacturer’s instructions (TransGen Biotech, http://www.transgen.com.cn). The EMSA was performed using a Light Shift Chemuluminescent EMSA kit according to the manufacturer’s protocol (Thermo Fisher Scientific; http://www.thermofisher.com). The probes were synthesized with oligonucleotides (Supplemental Table S2) and labeled using a biotin 3′ end DNA labeling kit (Thermo Fisher Scientific). Each binding reaction mixture, containing 100 ng of ERF11-GST recombinant protein or GST protein, 20 fmol of labeled DNA probe, 0.05% NP-40, 50 ng of poly(dI-dC), 5 mm MgCl2, 2.5% glycerol, 1× binding buffer, and ultrapure water to a final volume of 20 mL, was incubated at 25°C for 25 min. Unlabeled DNA was added in 200- and 400-fold molar excess as competitors. The reaction mixtures were then loaded onto 5% polyacrylamide gels to separate free and bound DNA. The DNA on the gel was transferred onto nylon membranes (GE Life Sciences; https://www.gelifesciences.com). After UV cross linking, the DNA on the membrane was detected using a chemiluminescent nucleic acid detection module (Thermo Fisher Scientific).
Statistical Analysis
Statistically significant differences (*P < 0.05) were based on Student’s t test computed by SigmaPlot 10.0 (http://sigmaplot.software.informer.com/10.0/). Data presented are means ± sd of three independent experimental replicates.
Accession Numbers
The sequence data from this study can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: ERF11 (At1g28370), BT4 (At5g67480), SID2 (Ag1g74710), NPR1 (At1g64280), EIN2 (At5g03280), EIN3 (At3g20770), ETO1 (At3g51770), ERF6 (AT4g17490), ERF13 (AT2g44840), ERF114 (AT5g61890), ORA47 (AT1g74930), PR1 (AT2g14610), PR2 (AT3g57260), PR3 (AT3g12500), PR5 (AT1g75040), and TUB4 (At5g44340).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Increased disease resistance following application of SA in wild-type Arabidopsis.
Supplemental Figure S2. Expression of BT4 is induced by various treatments.
Supplemental Figure S3. Analysis of differentially expressed ERF genes in three independent GEO databases.
Supplemental Figure S4. Expression pattern of ERF11 induced by SA and Pseudomonas syringae pv. tomato DC3000.
Supplemental Figure S5. SMART analysis reveals that pERF11 includes AP2, a low complexity region, and an EAR domain.
Supplemental Figure S6. Phenotypic analysis of ERF11-related and BT4-related plants.
Supplemental Figure S7. Expression of ERF11 is induced by treatment with jasmonic acid.
Supplemental Table S1. Oligonucleotides and primers used in this study.
Supplemental Table S2. ERF transcriptome database.
Acknowledgments
The authors greatly appreciate the efforts of the editors and anonymous reviewers who improved the text. We thank Dr. Hongwei Guo (Southern University of Science and Technology, Shenzhen, China) for providing Arabidopsis material and members of the J.D. and R.H. laboratory for insightful discussions throughout this work.
Footnotes
This study was supported by the Natural Science Foundation of China (grant no. 31200203), the Research Fund for the Doctoral Program of Higher Education of China (grant no. 20121302120007), the Natural Science Foundation of Hebei Province, China (grant no. C2012204032), and a Starting Grant from Hebei Agricultural University (grant no. 2001023). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Articles can be viewed without a subscription.
References
- Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, et al. (2013) NCBI GEO: Archive for functional genomics data sets—update. Nucleic Acids Res 41: D991–D995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bent AF, Innes RW, Ecker JR, Staskawicz BJ (1992) Disease development in ethylene-insensitive Arabidopsis thaliana infected with virulent and avirulent Pseudomonas and Xanthomonas pathogens. Mol Plant Microbe Interact 5: 372–378 [DOI] [PubMed] [Google Scholar]
- Berrocal-Lobo M, Molina A (2004) Ethylene response factor 1 mediates Arabidopsis resistance to the soilborne fungus Fusarium oxysporum. Mol Plant Microbe Interact 17: 763–770 [DOI] [PubMed] [Google Scholar]
- Berrocal-Lobo M, Molina A, Solano R (2002) Constitutive expression of ETHYLENE-RESPONSE-FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J 29: 23–32 [DOI] [PubMed] [Google Scholar]
- Birkenbihl RP, Diezel C, Somssich IE (2012) Arabidopsis WRKY33 is a key transcriptional regulator of hormonal and metabolic responses toward Botrytis cinerea infection. Plant Physiol 159: 266–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown RL, Kazan K, McGrath KC, Maclean DJ, Manners JM (2003) A role for the GCC-box in jasmonate-mediated activation of the PDF1.2 gene of Arabidopsis. Plant Physiol 132: 1020–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catinot J, Huang JB, Huang PY, Tseng MY, Chen YL, Gu SY, Lo WS, Wang LC, Chen YR, Zimmerli L (2015) ETHYLENE RESPONSE FACTOR 96 positively regulates Arabidopsis resistance to necrotrophic pathogens by direct binding to GCC elements of jasmonate- and ethylene-responsive defence genes. Plant Cell Environ 38: 2721–2734 [DOI] [PubMed] [Google Scholar]
- Chen JC, Lu HC, Chen CE, Hsu HF, Chen HH, Yeh HH (2013) The NPR1 ortholog PhaNPR1 is required for the induction of PhaPR1 in Phalaenopsis aphrodite. Bot Stud (Taipei, Taiwan) 54: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Yang Q, Gruber M, Kang J, Sun Y, Ding W, Zhang T, Zhang X (2012) Expression of an alfalfa (Medicago sativa L.) ethylene response factor gene MsERF8 in tobacco plants enhances resistance to salinity. Mol Biol Rep 39: 6067–6075 [DOI] [PubMed] [Google Scholar]
- Cheng MC, Liao PM, Kuo WW, Lin TP (2013) The Arabidopsis ETHYLENE RESPONSE FACTOR1 regulates abiotic stress-responsive gene expression by binding to different cis-acting elements in response to different stress signals. Plant Physiol 162: 1566–1582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll NS, Epple P, Dangl JL (2011) Programmed cell death in the plant immune system. Cell Death Differ 18: 1247–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney TP, Uknes S, Vernooij B, Friedrich L, Weymann K, Negrotto D, Gaffney T, Gut-Rella M, Kessmann H, Ward E, et al. (1994) A central role of salicylic acid in plant disease resistance. Science 266: 1247–1250 [DOI] [PubMed] [Google Scholar]
- Deokar AA, Kondawar V, Kohli D, Aslam M, Jain PK, Karuppayil SM, Varshney RK, Srinivasan R (2015) The CarERF genes in chickpea (Cicer arietinum L.) and the identification of CarERF116 as abiotic stress responsive transcription factor. Funct Integr Genomics 15: 27–46 [DOI] [PubMed] [Google Scholar]
- Després C, DeLong C, Glaze S, Liu E, Fobert PR (2000) The Arabidopsis NPR1/NIM1 protein enhances the DNA binding activity of a subgroup of the TGA family of bZIP transcription factors. Plant Cell 12: 279–290 [PMC free article] [PubMed] [Google Scholar]
- Du L, Poovaiah BW (2004) A novel family of Ca2+/calmodulin-binding proteins involved in transcriptional regulation: Interaction with fsh/Ring3 class transcription activators. Plant Mol Biol 54: 549–569 [DOI] [PubMed] [Google Scholar]
- Dubois M, Van den Broeck L, Claeys H, Van Vlierberghe K, Matsui M, Inzé D (2015) The ETHYLENE RESPONSE FACTORs ERF6 and ERF11 antagonistically regulate mannitol-induced growth inhibition in Arabidopsis. Plant Physiol 169: 166–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrant WE, Dong X (2004) Systemic acquired resistance. Annu Rev Phytopathol 42: 185–209 [DOI] [PubMed] [Google Scholar]
- Fabro G, Steinbrenner J, Coates M, Ishaque N, Baxter L, Studholme DJ, Körner E, Allen RL, Piquerez SJ, Rougon-Cardoso A, et al. (2011) Multiple candidate effectors from the oomycete pathogen Hyaloperonospora arabidopsidis suppress host plant immunity. PLoS Pathog 7: e1002348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan W, Dong X (2002) In vivo interaction between NPR1 and transcription factor TGA2 leads to salicylic acid-mediated gene activation in Arabidopsis. Plant Cell 14: 1377–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer EE, Alméras E, Krishnamurthy V (2003) Jasmonates and related oxylipins in plant responses to pathogenesis and herbivory. Curr Opin Plant Biol 6: 372–378 [DOI] [PubMed] [Google Scholar]
- Fu ZQ, Dong X (2013) Systemic acquired resistance: Turning local infection into global defense. Annu Rev Plant Biol 64: 839–863 [DOI] [PubMed] [Google Scholar]
- Fujimoto SY, Ohta M, Usui A, Shinshi H, Ohme-Takagi M (2000) Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell 12: 393–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffney T, Friedrich L, Vernooij B, Negrotto D, Nye G, Uknes S, Ward E, Kessmann H, Ryals J (1993) Requirement of salicylic acid for the induction of systemic acquired resistance. Science 261: 754–756 [DOI] [PubMed] [Google Scholar]
- Glazebrook J. (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 43: 205–227 [DOI] [PubMed] [Google Scholar]
- Guan R, Su J, Meng X, Li S, Liu Y, Xu J, Zhang S (2015) Multilayered regulation of ethylene induction plays a positive role in Arabidopsis resistance against Pseudomonas syringae. Plant Physiol 169: 299–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao CC, Jia J, Chen Z, Xing JH, Dong JG, Han JM (2013) Functional analysis of BT4 of Arabidopsis thaliana in resistance against Botrytis cinerea. Australas Plant Pathol 42: 393–401 [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27: 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesarwani M, Yoo J, Dong X (2007) Genetic interactions of TGA transcription factors in the regulation of pathogenesis-related genes and disease resistance in Arabidopsis. Plant Physiol 144: 336–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laluk K, Luo H, Chai M, Dhawan R, Lai Z, Mengiste T (2011) Biochemical and genetic requirements for function of the immune response regulator BOTRYTIS-INDUCED KINASE1 in plant growth, ethylene signaling, and PAMP-triggered immunity in Arabidopsis. Plant Cell 23: 2831–2849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawton K, Weymann K, Friedrich L, Vernooij B, Uknes S, Ryals J (1995) Systemic acquired resistance in Arabidopsis requires salicylic acid but not ethylene. Mol Plant Microbe Interact 8: 863–870 [DOI] [PubMed] [Google Scholar]
- Li Z, Zhang L, Yu Y, Quan R, Zhang Z, Zhang H, Huang R (2011) The ethylene response factor AtERF11 that is transcriptionally modulated by the bZIP transcription factor HY5 is a crucial repressor for ethylene biosynthesis in Arabidopsis. Plant J 68: 88–99 [DOI] [PubMed] [Google Scholar]
- Liu K, Li Y, Chen X, Li L, Liu K, Zhao H, Wang Y, Han S (2018) ERF72 interacts with ARF6 and BZR1 to regulate hypocotyl elongation in Arabidopsis. J Exp Bot 69: 3933–3947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Ning Y, Zhang Y, Yu N, Zhao C, Zhan X, Wu W, Chen D, Wei X, Wang GL, et al. (2017) OsCUL3a negatively regulates cell death and immunity by degrading OsNPR1 in rice. Plant Cell 29: 345–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O, Piqueras R, Sánchez-Serrano JJ, Solano R (2003) ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15: 165–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao JL, Miao ZQ, Wang Z, Yu LH, Cai XT, Xiang CB (2016) Arabidopsis ERF1 mediates cross-talk between ethylene and auxin biosynthesis during primary root elongation by regulating ASA1 expression. PLoS Genet 12: e1005760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath KC, Dombrecht B, Manners JM, Schenk PM, Edgar CI, Maclean DJ, Scheible WR, Udvardi MK, Kazan K (2005) Repressor- and activator-type ethylene response factors functioning in jasmonate signaling and disease resistance identified via a genome-wide screen of Arabidopsis transcription factor gene expression. Plant Physiol 139: 949–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Xu J, He Y, Yang KY, Mordorski B, Liu Y, Zhang S (2013) Phosphorylation of an ERF transcription factor by Arabidopsis MPK3/MPK6 regulates plant defense gene induction and fungal resistance. Plant Cell 25: 1126–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra A, McKnight TD, Mandadi KK (2018) Bromodomain proteins GTE9 and GTE11 are essential for specific BT2-mediated sugar and ABA responses in Arabidopsis thaliana. Plant Mol Biol 96: 393–402 [DOI] [PubMed] [Google Scholar]
- Moffat CS, Ingle RA, Wathugala DL, Saunders NJ, Knight H, Knight MR (2012) ERF5 and ERF6 play redundant roles as positive regulators of JA/Et-mediated defense against Botrytis cinerea in Arabidopsis. PLoS One 7: e35995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mur LA, Kenton P, Lloyd AJ, Ougham H, Prats E (2008) The hypersensitive response; the centenary is upon us but how much do we know? J Exp Bot 59: 501–520 [DOI] [PubMed] [Google Scholar]
- Nakano T, Suzuki K, Fujimura T, Shinshi H (2006a) Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol 140: 411–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T, Suzuki K, Ohtsuki N, Tsujimoto Y, Fujimura T, Shinshi H (2006b) Identification of genes of the plant-specific transcription-factor families cooperatively regulated by ethylene and jasmonate in Arabidopsis thaliana. J Plant Res 119: 407–413 [DOI] [PubMed] [Google Scholar]
- Naseem M, Philippi N, Hussain A, Wangorsch G, Ahmed N, Dandekar T (2012) Integrated systems view on networking by hormones in Arabidopsis immunity reveals multiple crosstalk for cytokinin. Plant Cell 24: 1793–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath C, Métraux JP (1999) Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell 11: 1393–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohme-Takagi M, Shinshi H (1995) Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell 7: 173–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oñate-Sánchez L, Anderson JP, Young J, Singh KB (2007) AtERF14, a member of the ERF family of transcription factors, plays a nonredundant role in plant defense. Plant Physiol 143: 400–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pel MJ, Pieterse CM (2013) Microbial recognition and evasion of host immunity. J Exp Bot 64: 1237–1248 [DOI] [PubMed] [Google Scholar]
- Perfect SE, Green JR (2001) Infection structures of biotrophic and hemibiotrophic fungal plant pathogens. Mol Plant Pathol 2: 101–108 [DOI] [PubMed] [Google Scholar]
- Phukan UJ, Jeena GS, Tripathi V, Shukla RK (2017) Regulation of Apetala2/Ethylene response factors in plants. Front Plant Sci 8: 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse CM, van Wees SC, van Pelt JA, Knoester M, Laan R, Gerrits H, Weisbeek PJ, van Loon LC (1998) A novel signaling pathway controlling induced systemic resistance in Arabidopsis. Plant Cell 10: 1571–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse CM, Leon-Reyes A, Van der Ent S, Van Wees SC (2009) Networking by small-molecule hormones in plant immunity. Nat Chem Biol 5: 308–316 [DOI] [PubMed] [Google Scholar]
- Pieterse CM, Van der Does D, Zamioudis C, Leon-Reyes A, Van Wees SC (2012) Hormonal modulation of plant immunity. Annu Rev Cell Dev Biol 28: 489–521 [DOI] [PubMed] [Google Scholar]
- Pré M, Atallah M, Champion A, De Vos M, Pieterse CM, Memelink J (2008) The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol 147: 1347–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren S, Mandadi KK, Boedeker AL, Rathore KS, McKnight TD (2007) Regulation of telomerase in Arabidopsis by BT2, an apparent target of TELOMERASE ACTIVATOR1. Plant Cell 19: 23–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas-San Vicente M, Plasencia J (2011) Salicylic acid beyond defence: Its role in plant growth and development. J Exp Bot 62: 3321–3338 [DOI] [PubMed] [Google Scholar]
- Robert HS, Quint A, Brand D, Vivian-Smith A, Offringa R (2009) BTB and TAZ domain scaffold proteins perform a crucial function in Arabidopsis development. Plant J 58: 109–121 [DOI] [PubMed] [Google Scholar]
- Sadumpati V, Kalambur M, Vudem DR, Kirti PB, Khareedu VR (2013) Transgenic indica rice lines, expressing Brassica juncea Nonexpressor of pathogenesis-related genes 1 (BjNPR1), exhibit enhanced resistance to major pathogens. J Biotechnol 166: 114–121 [DOI] [PubMed] [Google Scholar]
- Sandhu D, Tasma IM, Frasch R, Bhattacharyya MK (2009) Systemic acquired resistance in soybean is regulated by two proteins, Orthologous to Arabidopsis NPR1. BMC Plant Biol 9: 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo YJ, Park JB, Cho YJ, Jung C, Seo HS, Park SK, Nahm BH, Song JT (2010) Overexpression of the ethylene-responsive factor gene BrERF4 from Brassica rapa increases tolerance to salt and drought in Arabidopsis plants. Mol Cells 30: 271–277 [DOI] [PubMed] [Google Scholar]
- Shah J. (2009) Plants under attack: Systemic signals in defence. Curr Opin Plant Biol 12: 459–464 [DOI] [PubMed] [Google Scholar]
- Singh M, Bag SK, Bhardwaj A, Ranjan A, Mantri S, Nigam D, Sharma YK, Sawant SV (2015) Global nucleosome positioning regulates salicylic acid mediated transcription in Arabidopsis thaliana. BMC Plant Biol 15: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel SH, Koornneef A, Claessens SM, Korzelius JP, Van Pelt JA, Mueller MJ, Buchala AJ, Métraux JP, Brown R, Kazan K, et al. (2003) NPR1 modulates cross-talk between salicylate- and jasmonate-dependent defense pathways through a novel function in the cytosol. Plant Cell 15: 760–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel SH, Johnson JS, Dong X (2007) Regulation of tradeoffs between plant defenses against pathogens with different lifestyles. Proc Natl Acad Sci USA 104: 18842–18847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Does D, Leon-Reyes A, Koornneef A, Van Verk MC, Rodenburg N, Pauwels L, Goossens A, Körbes AP, Memelink J, Ritsema T, et al. (2013) Salicylic acid suppresses jasmonic acid signaling downstream of SCFCOI1-JAZ by targeting GCC promoter motifs via transcription factor ORA59. Plant Cell 25: 744–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlot AC, Dempsey DA, Klessig DF (2009) Salicylic acid, a multifaceted hormone to combat disease. Annu Rev Phytopathol 47: 177–206 [DOI] [PubMed] [Google Scholar]
- Wang D, Weaver ND, Kesarwani M, Dong X (2005) Induction of protein secretory pathway is required for systemic acquired resistance. Science 308: 1036–1040 [DOI] [PubMed] [Google Scholar]
- Wang L, Qin L, Liu W, Zhang D, Wang Y (2014) A novel ethylene-responsive factor from Tamarix hispida, ThERF1, is a GCC-box- and DRE-motif binding protein that negatively modulates abiotic stress tolerance in Arabidopsis. Physiol Plant 152: 84–97 [DOI] [PubMed] [Google Scholar]
- Weber H, Hellmann H (2009) Arabidopsis thaliana BTB/ POZ-MATH proteins interact with members of the ERF/AP2 transcription factor family. FEBS J 276: 6624–6635 [DOI] [PubMed] [Google Scholar]
- Wildermuth MC, Dewdney J, Wu G, Ausubel FM (2001) Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414: 562–565 [DOI] [PubMed] [Google Scholar]
- Wubben MJ II, Su H, Rodermel SR, Baum TJ (2001) Susceptibility to the sugar beet cyst nematode is modulated by ethylene signal transduction in Arabidopsis thaliana. Mol Plant Microbe Interact 14: 1206–1212 [DOI] [PubMed] [Google Scholar]
- Xu ZS, Xia LQ, Chen M, Cheng XG, Zhang RY, Li LC, Zhao YX, Lu Y, Ni ZY, Liu L, et al. (2007) Isolation and molecular characterization of the Triticum aestivum L. ethylene-responsive factor 1 (TaERF1) that increases multiple stress tolerance. Plant Mol Biol 65: 719–732 [DOI] [PubMed] [Google Scholar]
- Yalpani N, Silverman P, Wilson TM, Kleier DA, Raskin I (1991) Salicylic acid is a systemic signal and an inducer of pathogenesis-related proteins in virus-infected tobacco. Plant Cell 3: 809–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K (1994) A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6: 251–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Li B, Zheng XY, Li J, Yang M, Dong X, He G, An C, Deng XW (2015) Salicylic acid biosynthesis is enhanced and contributes to increased biotrophic pathogen resistance in Arabidopsis hybrids. Nat Commun 6: 7309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zander M, Thurow C, Gatz C (2014) TGA transcription factors activate the salicylic acid-suppressible branch of the ethylene-induced defense program by regulating ORA59 expression. Plant Physiol 165: 1671–1683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarei A, Körbes AP, Younessi P, Montiel G, Champion A, Memelink J (2011) Two GCC boxes and AP2/ERF-domain transcription factor ORA59 in jasmonate/ethylene-mediated activation of the PDF1.2 promoter in Arabidopsis. Plant Mol Biol 75: 321–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Chen M, Li L, Xu Z, Chen X, Guo J, Ma Y (2009) Overexpression of the soybean GmERF3 gene, an AP2/ERF type transcription factor for increased tolerances to salt, drought, and diseases in transgenic tobacco. J Exp Bot 60: 3781–3796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Huang L, Dai Y, Liu S, Hong Y, Tian L, Huang L, Cao Z, Li D, Song F (2015) Arabidopsis AtERF15 positively regulates immunity against Pseudomonas syringae pv. tomato DC3000 and Botrytis cinerea. Front Plant Sci 6: 686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Hong Y, Huang L, Li D, Song F (2016) Arabidopsis AtERF014 acts as a dual regulator that differentially modulates immunity against Pseudomonas syringae pv. tomato and Botrytis cinerea. Sci Rep 6: 30251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Li Z, Quan R, Li G, Wang R, Huang R (2011) An AP2 domain-containing gene, ESE1, targeted by the ethylene signaling component EIN3 is important for the salt response in Arabidopsis. Plant Physiol 157: 854–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Corwin JA, Copeland D, Feusier J, Eshbaugh R, Chen F, Atwell S, Kliebenstein DJ (2017) Plastic transcriptomes stabilize immunity to pathogen diversity: the jasmonic acid and salicylic acid networks within the Arabidopsis/Botrytis pathosystem. Plant Cell 29: 2727–2752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Fan W, Kinkema M, Li X, Dong X (1999) Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene. Proc Natl Acad Sci USA 96: 6523–6528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Chan Z, Gao J, Xing L, Cao M, Yu C, Hu Y, You J, Shi H, Zhu Y, et al. (2016) ABA receptor PYL9 promotes drought resistance and leaf senescence. Proc Natl Acad Sci USA 113: 1949–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou JM, Trifa Y, Silva H, Pontier D, Lam E, Shah J, Klessig DF (2000) NPR1 differentially interacts with members of the TGA/OBF family of transcription factors that bind an element of the PR-1 gene required for induction by salicylic acid. Mol Plant Microbe Interact 13: 191–202 [DOI] [PubMed] [Google Scholar]
- Zhu X, Qi L, Liu X, Cai S, Xu H, Huang R, Li J, Wei X, Zhang Z (2014) The wheat ethylene response factor transcription factor pathogen-induced ERF1 mediates host responses to both the necrotrophic pathogen Rhizoctonia cerealis and freezing stresses. Plant Physiol 164: 1499–1514 [DOI] [PMC free article] [PubMed] [Google Scholar]