A rice MADS-box transcription factor, OsMADS57, plays a major role in nitrate translocation from roots to shoots in low-nitrate supplied rice plants.
Abstract
Root nitrate uptake adjusts to the plant’s nitrogen demand for growth. Here, we report that OsMADS57, a MADS-box transcription factor, modulates nitrate translocation from rice (Oryza sativa) roots to shoots under low-nitrate conditions. OsMADS57 is abundantly expressed in xylem parenchyma cells of root stele and is induced by nitrate. Compared with wild-type rice plants supplied with 0.2 mM nitrate, osmads57 mutants had 31% less xylem loading of nitrate, while overexpression lines had 2-fold higher levels. Shoot-root 15N content ratios were 40% lower in the mutants and 76% higher in the overexpression lines. Rapid NO3− root influx experiments showed that mutation of OsMADS57 did not affect root nitrate uptake. Reverse transcription quantitative PCR analysis of OsNRT2 nitrate transporter genes showed that after 5 min in 0.2 mM nitrate, only OsNRT2.3a (a vascular-specific high-affinity nitrate transporter) had reduced (by two-thirds) expression levels. At 60 min of nitrate treatment, lower expression levels were also observed for three additional NRT2 genes (OsNRT2.1/2.2/2.4). Conversely, in the overexpression lines, four NRT2 genes had much higher expression profiles at all time points tested. As previously reported, OsNRT2.3a functions in nitrate translocation, indicating the possible interaction between OsMADS57 and OsNRT2.3a. Yeast one-hybrid and transient expression assays demonstrated that OsMADS57 binds to the CArG motif (CATTTTATAG) within the OsNRT2.3a promoter. Moreover, seminal root elongation was inhibited in osmads57 mutants, which may be associated with higher auxin levels in and auxin polar transport to root tips of mutant plants. Taken together, these results suggest that OsMADS57 has a role in regulating nitrate translocation from root to shoot via OsNRT2.3a.
Inorganic nitrogen (N) is available for plants as nitrate in aerobic uplands and as ammonium in flooded wetland or acidic soils. For many plants, nitrate acquired by roots is transported to the shoot before being assimilated (Smirnoff and Stewart, 1985; Xu et al., 2012). By contrast, ammonium derived from nitrate reduction or directly from ammonium uptake is preferentially assimilated in the root and then transported in an organic form to the shoot (Xu et al., 2012). Nitrate levels can vary by 3 to 4 orders of magnitude from micromolar to millimolar concentrations, and nitrate is highly mobile in agricultural soils due to its negative charge and solubility (Crawford and Glass, 1998; Miller et al., 2007). In response to external fluctuations of nitrate, plants have evolved at least three nitrate uptake systems, two high-affinity transport systems (HATS) and one low-affinity transport system (LATS), which are responsible for the acquisition of nitrate (Crawford and Glass, 1998).
The constitutive HATS and nitrate-inducible HATS operate to take up nitrate at low nitrate concentrations in external medium, with saturation in the range of 0.2 to 0.5 mM. In contrast, the LATS predominates at high soil nitrate concentrations. Nitrate LATS and HATS uptake systems have been linked to the NPF (originally named as Nitrate Transporter1/Peptide Transporter [NRT1/PTR]) and NRT2 gene families, respectively (Miller et al., 2007; Léran et al., 2014; O’Brien et al., 2016). In Arabidopsis (Arabidopsis thaliana), there are at least 53 and seven members belonging to the NPF (NRT1) and NRT2 families, respectively (Miller et al., 2007; Tsay et al., 2007; O’Brien et al., 2016). The most extensively studied nitrate transporter gene, AtNPF6.3/AtCHL1/AtNRT1.1, which is predominantly expressed in the nascent organs, especially the root tip (Guo et al., 2001), is described as a nitrate transceptor (transporter and receptor), playing multiple roles as a dual-affinity nitrate transporter and a sensor of external nitrate concentration (Liu and Tsay, 2003; Ho et al., 2009; Wang et al., 2009; Gojon et al., 2011; Forde, 2014) and auxin transport at low nitrate concentrations (Krouk et al., 2010a). Several nitrate transporters, mainly from the NRT1/NPF family, involved in nitrate transport in the vascular tissue of roots and shoots have been characterized. AtNRT1.5 is involved in nitrate loading into the root stele and translocating from root to shoot, whose expression is regulated by the transcription factor MYB59 (Lin et al., 2008; Li et al., 2017; Du et al., 2019). AtNRT1.7/AtNRT1.8/AtNRT1.11/AtNRT1.12 have been reported to be involved in nitrate unloading from xylem or loading into phloem (Fan et al., 2009; Li et al., 2010a; Hsu and Tsay, 2013).
In submerged rice (Oryza sativa), nitrate uptake could be comparable with that of ammonium (Kronzucker et al., 2000; Kirk and Kronzucker, 2005) due to oxygen transported by abundant parenchyma in roots into the rhizosphere, resulting in ammonium nitrification by bacteria on the root surface (Kirk, 2003; Li et al., 2008). A few proteins associated with nitrate uptake or transport have been identified (Cai et al., 2008; Feng et al., 2011; Yan et al., 2011; Tang et al., 2012; Hu et al., 2015, 2016; Li et al., 2015; Xia et al., 2015; Fan et al., 2016; Wang et al., 2018a; Wei et al., 2018). Since the nitrate concentration in the rhizosphere of paddy fields is estimated to be less than 10 µm (Kirk and Kronzucker, 2005), NRT2 family members play a major role in nitrate uptake for rice plants (Araki and Hasegawa, 2006; Feng et al., 2011; Yan et al., 2011). OsNRT2.1, OsNRT2.2, and OsNRT2.3a, belonging to the OsNRT2 family, are transcriptionally up-regulated by nitrate supply and require a partner protein, OsNAR2.1, for nitrate uptake (Feng et al., 2011; Yan et al., 2011). OsNRT2.3a is expressed predominantly in xylem parenchyma cells of the root stele and has been demonstrated to play a role in transporting nitrate from the root to the shoot under low-nitrate conditions (Tang et al., 2012). OsNRT2.3b is expressed in the phloem and is suggested to be involved in nitrate transport within the shoot (Fan et al., 2016). OsNRT2.4 encodes a dual-affinity nitrate transporter and functions in nitrate-regulated root growth and nitrate distribution in rice plants (Wei et al., 2018), and several members of the NRT1/NPF family, such as OsNPF2.4, OsNPF2.2, and OsNPF7.2, involved in nitrate transport have also been identified (Li et al., 2015; Xia et al., 2015; Hu et al., 2016).
Knowledge of nitrate transporters has been increasing substantially in the past 30 years. To further illustrate the nitrate uptake mechanism in plants, the regulatory systems controlling the transporter function need to be elucidated (Plett et al., 2018). Among several controlling mechanisms, transcriptional control of nitrate uptake is well documented in which transcription factors (TFs) act as master switches for regulatory networks (Xuan et al., 2017; Kant, 2018; Plett et al., 2018; Wang et al., 2018b). The first TF identified to play a role in N-responsive signaling is the Arabidopsis MADS-box TF (ANR1), which regulates the proliferation of lateral roots in NO3−-rich patches (Zhang and Forde, 1998; Gan et al., 2005) but also exists in the signaling pathway of the transceptor NRT1.1 (Remans et al., 2006). Thereafter, more s involved in complex regulatory networks and nitrate-dependent signaling pathways, such as LATERAL ORGAN BOUNDARY DOMAIN (LBD) family genes (LBD37/38/39; Rubin et al., 2009), NIN-like protein (NLP) family genes (NLP6/7/NLP8; Castaings et al., 2009; Konishi and Yanagisawa, 2013; Marchive et al., 2013; Yan et al., 2016), SBP-box family gene (SPL9; Krouk et al., 2010b), bZIP transcription factors (TGA1/4; Alvarez et al., 2014), Bric-a-Brac/Tramtrack/Broad family genes (BT1/2; Araus et al., 2016), NITRATE REGULATORY GENE2 (NRG2; Xu et al., 2016), and teosinte branched1/cycloidea/proliferating cell factor1-20 gene (TCP20; Guan et al., 2017), have been identified and intensively investigated in Arabidopsis.
In rice, orthologs of the above TFs (e.g. OsMADS) are also found to be regulated under different N supplies (Puig et al., 2013; Yu et al., 2014a; Yang et al., 2015). However, few of them have been demonstrated to regulate developmental responses to nitrate. For instance, OsMADS genes show diverse responses to nitrate supply (Puig et al., 2013; Yu et al., 2014a). Unlike AtANR1, which acts downstream of AtNPF6.3 (Remans et al., 2006), overexpression of nitrate-inducible OsMADS25 significantly increases the expression of nitrate transporter genes and promotes nitrate accumulation and lateral root and shoot growth, suggesting positive effects of OsMADS25 on nitrate uptake via nitrate transporters (Yu et al., 2015). Interestingly, OsMADS genes are targeted by monocot-specific MicroRNA444s (Li et al., 2010b). OsmiR444a overexpression reduces OsMADS expression and root and shoot growth under nitrate supply (Yan et al., 2014). Thus, OsmiR444a/OsMADS may act as a regulatory module for nitrate-dependent signaling and growth responses in rice. However, the specific gene in the regulatory system controlling nitrate uptake and translocation in rice remains unclear. In this study, we report that a rice MADS-box transcription factor, OsMADS57, is involved in the regulation of long-distance translocation of nitrate from the root to the shoot through regulating OsNRT2.3a expression under low nitrate concentration.
RESULTS
OsMADS57 Is Mainly Expressed in Rice Root Stelar Cells and Is Sensitive to Supplies of Nitrate and Auxin
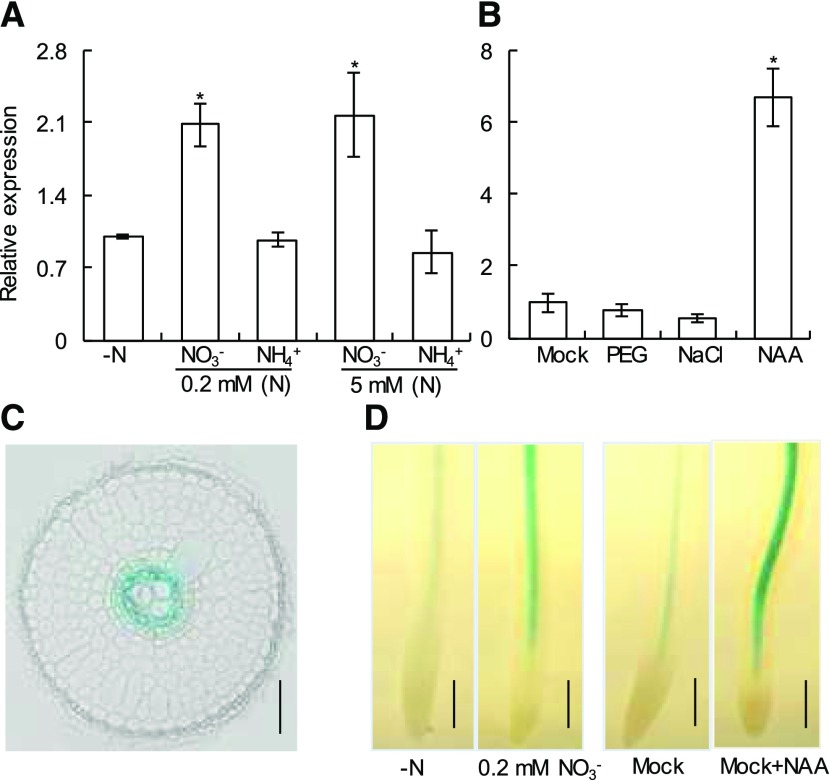
Arabidopsis AtANR1 was identified as a key gene controlling lateral root growth through nitrate signaling (Zhang and Forde, 1998). To understand the biological functions of the ANR1-like-related gene OsMADS57, we performed reverse transcription quantitative PCR (RT-qPCR) analysis and GUS assay to examine its expression pattern. A previous study and the Rice Expression Profile Database (http://ricexpro.dna.affrc.go.jp/GGEP/gene-search.php) have shown that OsMADS57 is abundantly expressed in roots and leaf blades during vegetative stages compared with other stages in rice (Guo et al., 2013). Interestingly, the expression of OsMADS57 was significantly up-regulated by nitrate treatments, irrespective of nitrate concentrations, rather than by ammonium (Fig. 1A). Transgenic rice plants expressing the GUS gene driven by the OsMADS57 promoter (∼3 kb) exhibited GUS activity in the root central cylinder, particularly in the xylem parenchyma cells (Fig. 1, C and D), consistent with previous reports (Puig et al., 2013; Yu et al., 2014a). Stronger GUS staining in the seminal root central cylinder caused by nitrate treatments further demonstrated the above result.
Figure 1.
OsMADS57 gene expression pattern. Rice seedlings were grown in International Rice Research Institute (IRRI) solution supplied by N with/without exogenous 10% (w/v) PEG, 100 mM NaCl, and 10 nm 1-naphthaleneacetic acid (NAA) for 7 d. A, Relative OsMADS57 expression in response to N supplies. B, Relative OsMADS57 expression in response to PEG, NaCl, and NAA treatments. Relative mRNA levels of OsMADS57 were normalized relative to OsActin. Data are means of five replications ± se. *, P < 0.05 (Student’s t test) relative to the −N (0 mM N) treatment (A) or the mock (1.25 mM NH4NO3) treatment (B). C, Expression of MADS57 in a seminal root cross section of a MADS57pro::GUS transgenic line. Bar = 50 μm. D, Expression of MADS57 in seminal roots of MADS57pro::GUS transgenic lines. Bars = 0.5 mm.
Furthermore, we performed RT-qPCR to test the relative expression of OsMADS57 in response to external stresses and hormones. The results showed that, compared with the mock treatment, the expression of OsMADS57 was up-regulated by exogenous application of NAA but not by application of polyethylene glycol (PEG) and NaCl (Fig. 1B). Similarly, NAA-induced expression of GUS staining was also observed in the root central cylinder in transgenic lines (Fig. 1D).
OsMADS57 Is Involved in Nitrate Translocation from Roots to Shoots
To determine the specific role of OsMADS57 in rice plants, two independent mutant lines (the transfer-DNA [T-DNA]-inserted osmads57 mutants m1 and m2) and three independent 35S::OsMADS57 overexpression transgenic lines (Ox1–Ox3) were used (Supplemental Fig. S1). Given that OsMADS57 expression was affected only by NO3− supplies, we first analyzed plant growth and N accumulation in rice plants grown under NO3− supplies of 0.2 and 5 mM (Supplemental Fig. S2). Surprisingly, less plant dry weight was observed in the two osmads57 mutants and the three overexpression lines only under low NO3− supply in comparison with wild-type plants. Furthermore, under low NO3− supply, higher N contents were recorded in roots of the mutants and in roots and shoots of the overexpression lines, with shoot accumulation being most pronounced. Although slight decreases were observed in the shoots of the two mutants, they did not show statistical significance.
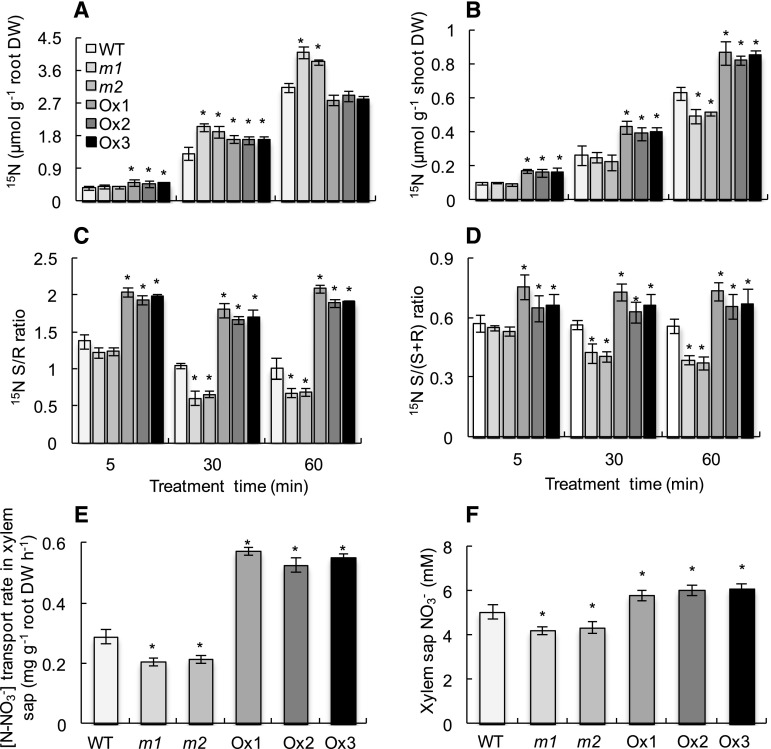
Next, time-course 15N-labeled NO3−-supply experiments were conducted to understand the role of OsMADS57 under low nitrate supply (Fig. 2, A and B). The overexpression lines showed substantially higher 15N accumulation in shoots at all time points (5, 30, and 60 min). The mutant lines showed substantially higher 15N accumulation in roots at 30 and 60 min and lower shoot 15N accumulation at 60 min. These phenotypes resulted in dramatic changes in the shoot-root 15N ratios.
Figure 2.
15N content, 15N distribution ratio, and xylem sap NO3− in the wild type (WT), mutants (m1 and m2), and overexpression lines (Ox1–Ox3). Rice seedlings were grown in IRRI nutrient solution containing 0.2 mM NO3− for 2 weeks and then transferred to IRRI nutrient solution containing 0.2 mM [15N]NO3− for 5, 30, and 60 min. A and B, 15N content in rice roots (A) and shoots (B). C, Shoot-root ratio of 15N content. D, Shoot-(shoot+root) ratio of 15N accumulation. E and F, [15N]NO3− transport rate and concentration in xylem sap of rice plants. Rice seedlings were grown in IRRI solution containing 0.2 mM NO3− for 4 weeks and then were cut 4 cm above the roots for a 1-h xylem sap collection. Data are means of five replications ± se. *, P < 0.05 (Student’s t test) comparing the wild type and other lines at the same experimental period. DW, Dry weight.
Compared with wild-type plants, the shoot-root 15N ratios were 42% to 76% higher in the three overexpression lines, while shoot-root 15N ratios were approximately 40% lower in the two mutants at 30 and 60 min (Fig. 2C), and shoot-(shoot+root) 15N ratios had the same tendency (Fig. 2D). These data suggest that OsMADS57 might be involved in nitrate distribution in rice plants. To confirm if OsMADS57 plays an important role in long-distance nitrate transport, 1-h xylem exudates of rice plants grown in hydroponics were collected, from the cut surface since 4 cm above the ground level was removed, and analyzed (Fig. 2, E and F). The nitrate concentration and nitrate transport rate decreased by 16% and 31% in xylem sap of the two mutant lines in comparison with wild-type plants; conversely, 20% and 2-fold higher nitrate concentrations were recorded in the three overexpression lines. These results illustrated that OsMADS57 might participate in nitrate transport from roots to shoots.
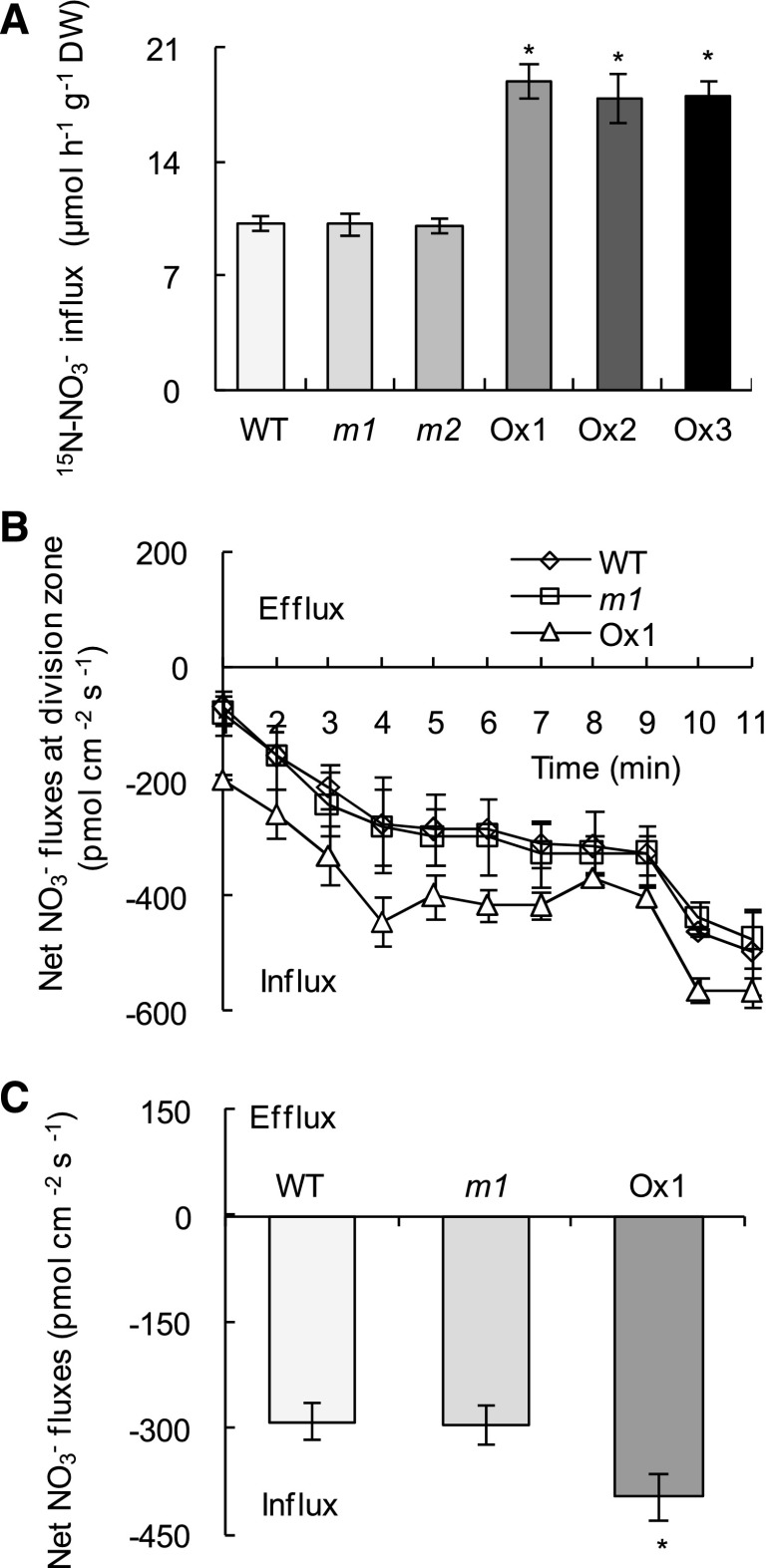
Five-minute influx experiments of 15N-labeled NO3− indicated that NO3− influx rates in OsMADS57 overexpression lines were increased by approximately 80%, whereas no significant difference was recorded between osmads57 mutant lines and wild-type plants (Fig. 3A). Net NO3− fluxes in seminal root tips, using the high-resolution scanning ion-selective electrode technique (SIET), further supported the above results (Fig. 3, B and C). During the entire 11-min experiment, a 38% increase in net nitrate influx was recorded in the overexpression lines in comparison with wild-type plants (Fig. 3C). Thus, higher influx rates by the Ox lines could explain why there is more 15N in the roots of Ox lines.
Figure 3.
NO3− acquisition in the wild type (WT), mutants (m1 and m2), and overexpression lines (Ox1–Ox3). Rice seedlings were grown in IRRI nutrient solution containing 0.2 mM NO3− for 2 weeks and then deprived of N for 3 d. The plants were transferred to IRRI nutrient solution containing 0.2 mM [15N]NO3− for 5 min. A, Nitrate influx rates. B, Net NO3− fluxes in the seminal root meristem of rice plants supplied with 0.2 mM NO3− for 11 min. C, Mean rate of NO3− fluxes during the entire 11 min. Data are means of five replications ± se. *, P < 0.05 (Student’s t test) comparing the wild type and other lines. DW, Dry weight.
Root Expression Profiles of Nitrate Reductase and OsNRT2s in the Wild Type, osmads57 Mutants, and Overexpression Lines
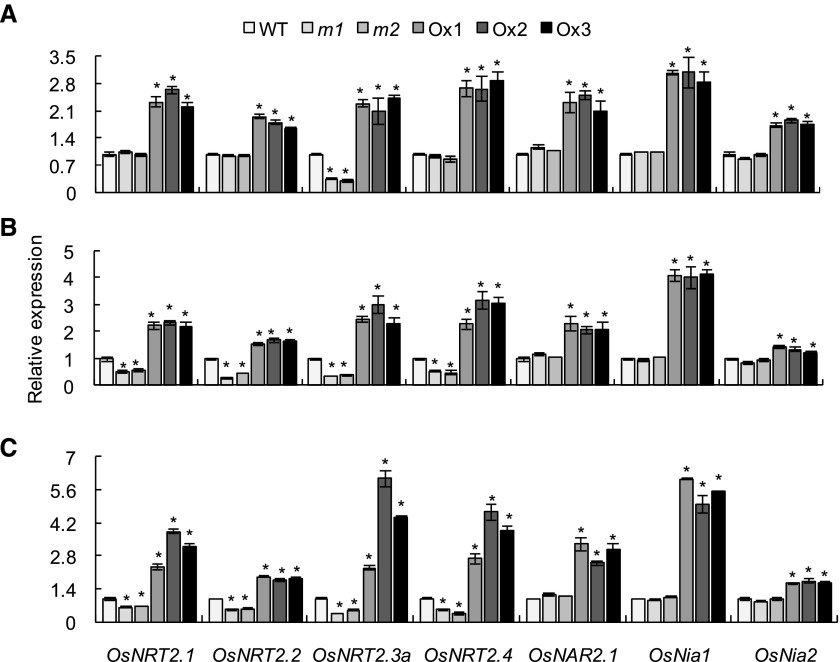
As OsNRT2s, encoding high-affinity nitrate transporters, play a critical role in nitrate uptake and translocation, we performed time-course expression analyses in rice plants supplied with 0.2 mM NO3−. Compared with the wild type, OsNRT2.3a transcripts were significantly decreased in the osmads57 mutants throughout the entire 60-min experimental period (Fig. 4). Decreases in transcript levels were also found for three additional OsNRT2 genes (OsNRT2.1/OsNRT 2.2/OsNRT2.4) in the mutants; however, the time course was delayed: at 5 min, no difference was found between the wild type and mutants (Fig. 4A), but at 30 and 60 min, transcript levels were significantly decreased in the mutants. We also analyzed the expression of OsNAR2.1, which interacts with OsNRT2.1/OsNRT2.2/OsNRT2.3a to produce functional transporters (Xu et al., 2012; Liu et al., 2014), and nitrate reductase (OsNia1/2). The expression of OsNAR2.1 and OsNia1/2 in roots was not affected in the osmads57 mutants, but overexpression of OsMADS57 significantly up-regulated all tested genes (Fig. 4). Because OsNRT2.3b was mainly expressed in shoots, we did not detect its expression in rice roots.
Figure 4.
RT-qPCR analysis of root mRNA expression patterns of nitrate transporter genes (NRT2s) and nitrate reductase genes (Nia1-2) in the wild type (WT), mutants (m1 and m2), and overexpression lines (Ox1–Ox3). Rice seedlings were grown in IRRI nutrient solution containing 0.2 mM NO3− for 2 weeks and then deprived of N for 3 d. The plants were transferred to IRRI nutrient solution containing 0.2 mM NO3− for 5 min (A), 30 min (B), and 60 min (C) before sampling. Relative mRNA levels for individual genes were normalized relative to OsActin. Data are means of five replications ± se. *, P < 0.05 (Student’s t test) comparing the wild type and other lines at the same gene.
OsMADS57 Directly Binds to the OsNRT2.3a Promoter
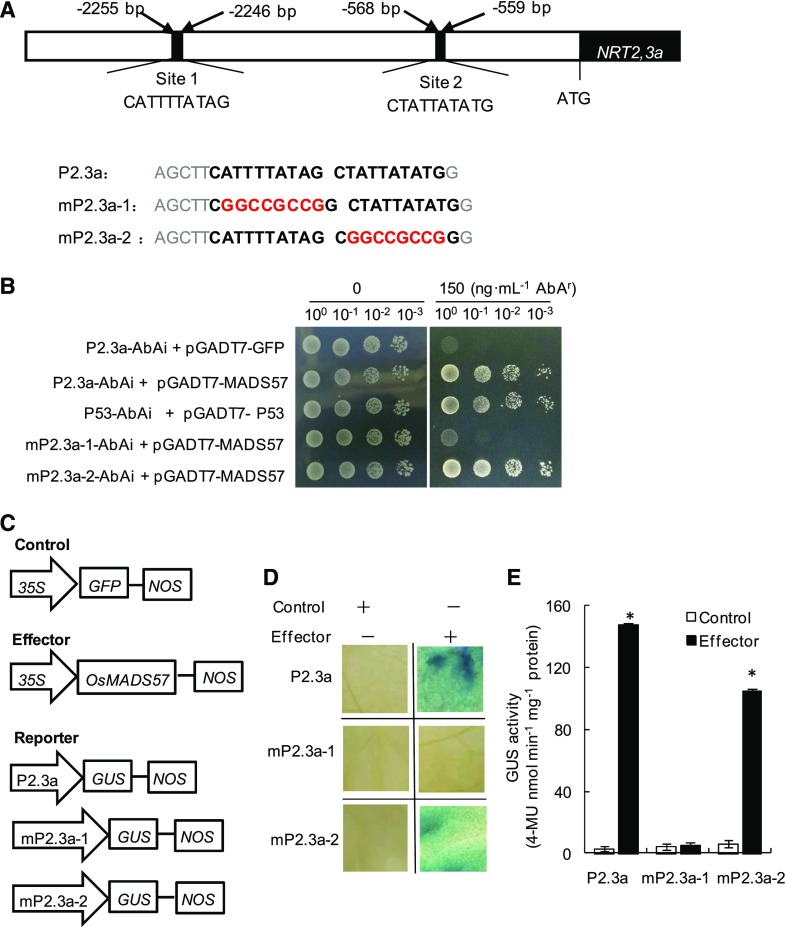
The MADS-box transcription factors can recognize the CArG motif present in the promoters of target genes (Guo et al., 2013). To reveal if OsMADS57 is capable of directly binding to the promoter sequences of OsNRT2.3a, we first examined the distribution of CArG elements in the promoter of OsNRT2.3a. Two putative OsMADS57-binding CArG-box sites, site 1 (CATTTTATAG) and site 2 (CTATTATATG), were found at −2,255 to −2,246 bp and −568 to −559 bp upstream of the translation start site ATG of OsNRT2.3a, respectively (Fig. 5A). Thus, we speculated that OsMADS57 may directly bind to the two CArG boxes in the promoter of OsNRT2.3a. To verify this hypothesis, we first performed a yeast one-hybrid assay using a full-length coding sequence of OsMADS57. One region about 30 bp in length containing the two tandem CArG boxes (P2.3a), an independent mutation of two CArG-box sites in tandem (mP2.3a-1 and mP2.3a-2), and another region about 50 bp in length containing three tandem target DNA sequences of interest designated as P53 were used as the baits for binding assays in the yeast one-hybrid system (Fig. 5B). The interactions between OsMADS57 and the three promoter fragments were tested by growth on medium lacking Ura and Leu. Increasing concentrations of aureobasidin A (AbA) were added to the medium to suppress background activation and assess the strength of the interaction. When the AbA concentration was increased up to 150 nm, the GoldY1H yeast stain containing the P2.3a-AbAi bait and the pGADT7-OsMADS57 prey grew normally on the selective medium, as did the positive control (containing P53-AbAi bait and pGADT7-P53; Fig. 5B), while the construct containing the mP2.3a-1-AbAi bait almost did not grow, similar to the negative control (containing P2.3a-AbAi bait and pGADT7-GFP). However, another mutation of the CArG box (mP2.3a-2) did not affect growth, suggesting that OsMADS57 binds to the CArG box in site 1 rather than in site 2 of the OsNRT2.3a promoter.
Figure 5.
OsMADS57 binds to the OsNRT2.3a promoter. A, Two CArG boxes (sites 1 and 2) in the OsNRT2.3a promoter. Letters in boldface indicate a tandem repeat of two potential CArG boxes in the promoter of OsNRT2.3a (P2.3a), and letters in red are the mutations of site 1 (mP2.3a-1) and site 2 (mP2.3a-2). B, Yeast cells were cotransformed with a bait vector containing a promoter fragment in A fused to an AbAi reporter gene and a prey vector containing MADS57 fused to a GAL4 activation domain. Cells were grown in liquid medium to an OD600 of 1 diluted to 0.1 and diluted in a 10× dilution series (from 10−1 to 10−3). From each dilution, 6 μL was spotted on medium selecting for both plasmids and selecting for interaction (synthetic dropout, –Ura, –Leu), supplemented with 150 nm AbA to suppress background growth and to test the strength of the interaction. C, Schematic diagram of the effector and reporter used for transactivation studies. The plasmid 35S::OsMADS57 was used as the effector, the plasmid P2.3a::GUS and its mutant versions mP2.3a-1::GUS and mP2.3a-2::GUS were used as the reporter, and 35S::GFP was used as an internal control. D and E, Transactivation activity was detected by GUS staining (D) and quantitative analysis of the GUS activity (E) after reporter and effector plasmids were coinfiltrated into N. benthamiana. Data in E are means of five replications ± se. *, P < 0.05 (Student’s t test) compared with the control. 4-MU, 4-Methylumbelliferone.
To confirm the role of OsMADS57 in the regulation of OsNRT2.3a expression, we performed transient GUS assays in Nicotiana benthamiana as reported by Shim et al. (2013). OsMADS57, with the CaMV35S promoter, was used as the effector. One region about 30 bp in length containing the two tandem CArG boxes (P2.3a) or their mutated versions (mP2.3a-1 and mP2.3a-2) fused to the GUS gene were used as reporters (Fig. 5, A and C). The reporter and effector plasmids were coinfiltrated into N. benthamiana leaves. The GUS reporter gene was activated by coexpressing OsMADS57 with the wild-type promoter P2.3a. However, the mutant reporter containing mP2.3a-1 was not activated but mP2.3a-2 was activated (Fig. 5, D and E). These results demonstrate that OsMADS57 can activate promoter transcription by interacting with the CArG box in site 1 rather than in site 2.
Furthermore, we analyzed time-course 15N translocation and the transcript levels of two high-affinity nitrate transporters (NRT2.1/NRT2.2) in the two osnrt2.3a mutants under IRRI nutrient solution containing 0.2 mM NO3− (Supplemental Fig. S3). The two osnrt2.3a mutant lines showed substantially higher 15N accumulation in roots at 30 and 60 min and lower shoot 15N accumulation at 60 min (Supplemental Fig. S3, A and B), which was consistent with the phenotype of the osmads57 mutants. Furthermore, the relative expression of OsNRT2.1 and OsNRT2.2 in the osnrt2.3a mutants was similar to that of the osmads57 mutants (Supplemental Fig. S3, C–E).
Knockdown of OsMADS57 Inhibited Elongation of Seminal and Adventitious Roots under Low Nitrate
Because root growth is significantly affected in the osmads57 mutants supplied with low nitrate (Supplemental Fig. S2), root architecture was further analyzed in the wild type, mutants, and overexpression lines (Supplemental Fig. S4). Compared with the wild-type plants, total root length was markedly reduced in the mutant plants but increased in the overexpression plants. Interestingly, decreased total root length in the osmads57 mutants was mainly attributed to shorter seminal and adventitious roots rather than to changes in lateral root lengths. The time-course measurement of seminal root growth is shown in Figure 6A. Treatment of 0.2 mM NO3− for 14 d resulted in 20% to 40% shorter seminal roots in the mutant plants compared with the wild-type and overexpression line plants.
Figure 6.
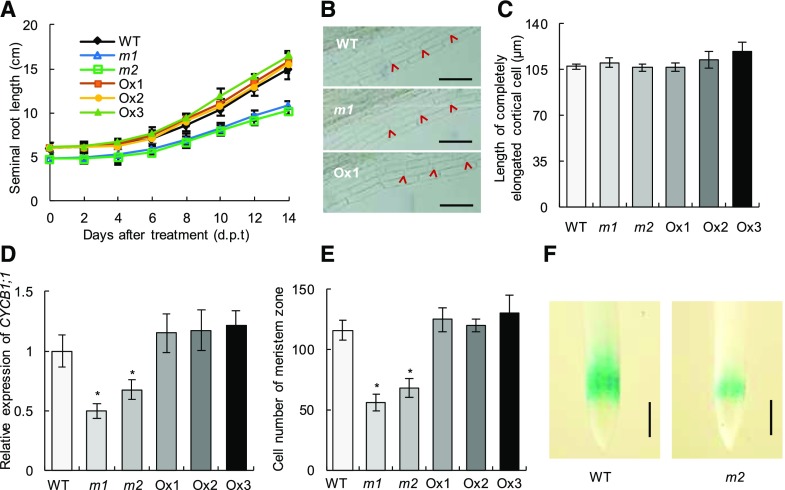
Seminal root elongation of the wild type (WT), mutants (m1 and m2), and overexpression lines (Ox1–Ox3). Rice seedlings were grown in IRRI nutrient solution containing 0.2 mM NO3− for 14 d. A, Seminal root elongation. B, Photographs of fully elongated cells in the seminal root (red arrowheads; n = 15) at 10 d post treatment. Bars = 100 μm. C, Cell length of fully elongated cells. Measurements were obtained directly by measuring 10 cortical cells from 10 plants. D, CYCB1;1 expression in roots at 10 d post treatment. E, Cell numbers of the meristem zone in the seminal root. F, Meristem size and root cell cycle activity of wild-type and m2 plants were measured after treatments for 10 d, as monitored by the proCycB1;1::GUS reporter. Bars = 500 μm. Data in A and C to E are means of five replications ± se. *, P < 0.05 (Student’s t test) comparing the wild type and other lines.
Root elongation is determined by two successive processes: cell division in the root apical meristem and elongation of cells outside the root meristem (Beemster et al., 2003). To investigate defects in root elongation at the cellular level, we observed and measured cortical cell length in the maturity zone and cell proliferation in the meristem zone. osmads57 mutants had little change in the length of cortical cells in the maturity zone (Fig. 6, B and C). The expression of CYCB1;1, responsible for meristematic activity of cells, was markedly weaker in osmads57 mutant plants, whereas in the overexpression lines no difference was observed compared with that in wild-type plants (Fig. 6D). Furthermore, cell numbers in the meristem zone had a similar pattern to that of CYCB1;1 expression (Fig. 6E), suggesting that osmads57 mutations inhibit cell division in the root meristem. Consistently, in the transgenic plant CYCB1;1::GUS, an osmads57 mutation repressed the GUS activity compared with the wild-type plants (Fig. 6F). Taken together, these data suggest that osmads57 mutations inhibit seminal root elongation under the condition of 0.2 mM NO3− by repressing meristematic cell proliferation.
Induced Auxin Distribution Was Observed in Seminal Root Tips of osmads57 Mutants
Auxin plays an important role in the regulation of root system architecture and is required for normal cell proliferation and elongation (Grieneisen et al., 2007). Given the root growth defects of the osmads57 mutants, we hypothesized that auxin may participate in modulating root growth in these mutants. Visualization of the auxin-responsive DR5 promoter (Ulmasov et al., 1997) reporter suggests that auxin levels or responses are elevated in the root tip (0–0.5 cm) of osmads57 mutant plants and are slightly reduced in the overexpression lines compared with wild-type seedlings (Fig. 7A). Exogenous application of NAA increased pDR5::GFP expression throughout the roots and decreased seminal root length of wild-type and overexpression plants to a similar level to that in the osmads57 mutant line (Fig. 7, A and B). These results imply that higher auxin levels in the osmads57 mutants may play an important role in the inhibition of seminal root elongation. Next, endogenous indole-3-acetic acid (IAA) contents in the root tip and lateral root area were analyzed (Fig. 7C). IAA contents in the root tips (0–0.5 cm) of the osmads57 mutants were elevated by approximately 63% compared with the wild type and overexpression lines; no differences were found in lateral root area (2–8 cm) among the rice plants (Fig. 7C). Next, we performed a [3H]IAA transport assay to investigate whether the auxin distribution in root tips is attributed to auxin polar transport. A significant increase in rootward [3H]IAA transport was observed in root tips of the osmads57 mutant in comparison with the wild-type and overexpression plants; no difference was observed in shootward transport from the root apex among the rice plants (Fig. 7D). These results indicated that auxin polar transport toward and within the root was induced in osmads57 mutant plants supplied with 0.2 mM NO3−.
Figure 7.
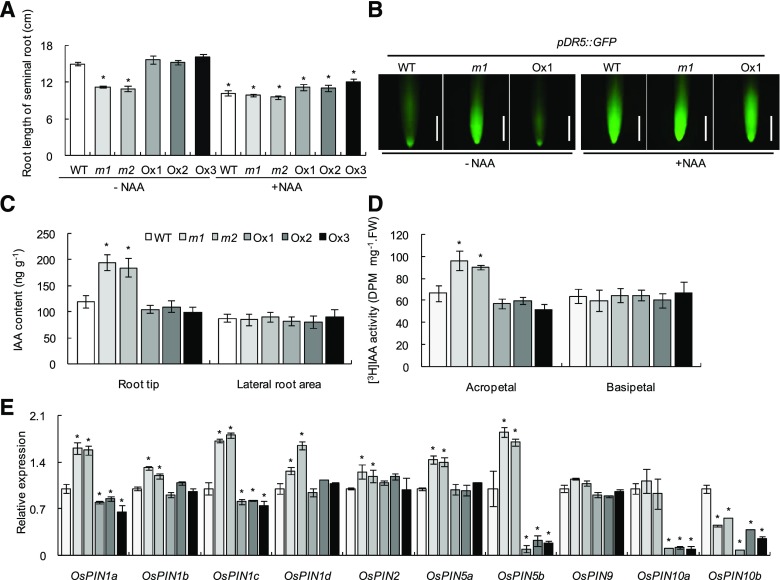
Auxin distribution in root tips of the wild type (WT), mutants (m1 and m2), and overexpression lines (Ox1–Ox3). Seedlings were grown in nutrient solution containing 0.2 mM NO3− with or without 10 nm NAA application. A, Seminal root length. Plants were grown hydroponically for 14 d in nutrient solution containing 0.2 mM NO3− with/without 10 nm NAA application. B, pDR5::GFP (n = 30) expression in seminal root tips. Seedlings were grown for 7 d in nutrient solution containing 0.2 mM NO3− with or without 10 nm NAA application in agar medium. Bars = 1 mm. C to E, Plants were grown hydroponically for 14 d in nutrient solution containing 0.2 mM NO3−. C, IAA content in root tips of 0 to 0.5 cm and lateral root area of 2 to 8 cm adventitious root from the root tip (n = 7). D, [3H]IAA transport (n = 7). E, Relative expression levels of OsPINs. Relative mRNA levels for individual genes were normalized relative to OsActin (n = 3). Data in A and C to E are means ± se. *, P < 0.05 (Student’s t test) comparing the wild type and other lines. FW, Fresh weight.
Auxin transport mostly occurs via the polar transport stream, which is facilitated by proteins of the PIN family (Friml et al., 2003). Our RT-qPCR analyses showed that the expression levels of seven PIN genes (PIN1a–PIN1d, PIN2, and PIN5a and PIN5b) were significantly increased in osmads57 mutant plants, whereas we observed decreased expression of PIN10b in the osmads57 mutants and PIN5b, PIN10a, and PIN10b in the overexpression lines in comparison with wild-type rice seedlings (Fig. 7E).
DISCUSSION
Plant roots take up nitrate from the external medium and transport a large portion of nitrate from roots to shoots (Kronzucker et al., 2000; Kirk and Kronzucker, 2005). A number of nitrate transporters have been functionally characterized for their specific functions in the acquisition and distribution of nitrate in plants (Huang et al., 1999; Liu and Tsay, 2003; Chiu et al., 2004; Almagro et al., 2008; Lin et al., 2008; Fan et al., 2009, 2016; Wang et al., 2009, 2018a; Li et al., 2010a, 2015; Wang and Tsay, 2011; Tang et al., 2012; Xu et al., 2012; Hu et al., 2015, 2016; Xia et al., 2015; Xuan et al., 2017; Plett et al., 2018). Improvements of nitrate uptake and transport in crops through manipulation of nitrate transporters has recently been successful (Hu et al., 2015; Fan et al., 2016; Wang et al., 2018a); however, it stands to reason that further improvements will require more complete knowledge of the regulatory system to maximize N uptake and utilization. Although increasing evidence suggests that N uptake involves complex gene regulatory networks, and nitrate-dependent signaling pathways have been intensively investigated in Arabidopsis (Xuan et al., 2017; Plett et al., 2018; Wang et al., 2018b), it remains unclear which gene is involved in modulating nitrate uptake and transport in rice. In this study, we demonstrated that OsMADS57, a rice MADS-box transcription factor whose expression is induced by nitrate, mediates the transport of root-acquired nitrate to the shoots through modulating the expression of OsNRT2.3a.
OsMADS57 Is Involved in Nitrate Transport through Regulating OsNRT2.3a Expression
Our data indicate that OsMADS57 functions as a transcriptional regulator of OsNRT2.3a to modulate rice nitrate translocation from roots to shoots and are supported by the following evidence: (1) the expression of OsMADS57 and OsNRT2.3a under nitrate treatments was significantly higher than under ammonium treatments in roots; (2) OsMADS57 and OsNRT2.3a expression overlapped spatially; (3) osmads57 mutants showed a similar phenotype to the osnrt2.3a mutant by RNA interference, such as long-distance nitrate translocation but not root influx at low nitrate supply; and (4) OsMADS57 bound to the cis-element in the OsNRT2.3a promoter.
The expression of OsMADS57 can be induced by nitrate as previously reported (Puig et al., 2013; Yu et al., 2014a), suggesting a specific role in the nitrate response. Staining of transgenic GUS plants indicated that OsMADS57 is preferentially expressed in the stele of rice roots, and the expression increased after nitrate treatments (Fig. 1, A and D). Feng et al. (2011) reported that the expression of OsNRT2.3a is significantly induced by nitrate rather than ammonium. Tang et al. (2012) further confirmed that OsNRT2.3a is mainly expressed in xylem parenchyma cells in the stele of nitrate-supplied roots. These results indicate that the two proteins may have overlapping localization, mainly in the central cylinder of the root.
Phenotype analyses further showed that osmads57 mutants exhibited a smaller phenotype under a low-nitrate condition compared with wild-type plants, which is similar to that of osnrt2.3a-RNA interference plants (Tang et al., 2012). When plants were exposed to relatively long-term supplies of low nitrate, the two independent osmads57 mutants accumulated much higher nitrate in roots than wild-type plants (Supplemental Fig. S2E). Analyses of NO3− transport rates in xylem sap and the distribution of 15N in the roots and shoots showed a strong decrease in nitrate translocation from roots to shoots in the mutants under low nitrate supply (Fig. 2). Short-term [15N]NO3− influx analyses of both whole plants and the root meristem zone showed that NO3− influx rates in OsMADS57 overexpression lines were significantly increased, whereas no significant difference was recorded in osmads57 mutants compared with wild-type plants. Higher [15N]NO3− influx in overexpression lines may be attributed to negative feedback signals from long-distance transport and/or root storage pools at low external supply (Fig. 3). These results demonstrate that OsMADS57 has a major role in nitrate translocation from roots to shoots. Furthermore, we also analyzed time-course [15N]NO3− translocation in osnrt2.3a mutants and observed higher 15N accumulation in roots at 30 and 60 min and lower shoot 15N accumulation at 60 min (Supplemental Fig. S3, A and B), which was consistent with the phenotype of the osmads57 mutants.
Interestingly, results from time-course expression analyses showed that, compared with the wild type, OsNRT2.3a transcripts were significantly decreased in the osmads57 mutants throughout the entire 60-min experimental period (Fig. 4); however, the time course was delayed for three additional OsNRT2 genes (OsNRT2.1/OsNRT2.2/OsNRT2.4) in the mutants: at 5 min supplied by NO3−, no difference was found between the wild type and the mutants (Fig. 4A), but at 30 and 60 min, transcript levels were significantly decreased (Fig. 4, B and C). Furthermore, we also performed time-course expression analyses in osnrt2.3a mutants. The relative expression of OsNRT2.1 and OsNRT2.2 in the osnrt2.3a mutants was similar to that of the osmads57 mutants (Supplemental Fig. S3, C–E). Since OsNRT2.3a has been reported to mediate nitrate root-to-shoot translocation at low nitrate concentrations, these further suggest that decreased nitrate translocation from roots to shoots in mads57 mutants may be due to the decreased expression level of OsNRT2.3a. Both the yeast one-hybrid assay and transient expression in N. benthamiana suggest that OsMADS57 directly binds to the first specific CArG box in the OsNRT2.3a promoter (Fig. 5), confirming that OsMADS57 may be a direct regulator of OsNRT2.3a.
Reduced Seminal Root Elongation in the osmads57 Mutants Might Be Associated with the Increasing Auxin Accumulation in the Root Tip
The MADS-box gene family has been reported to modulate plant root development. In Arabidopsis, AGL17/AGL21 and ANR1 belong to the AGL17 clade, which is preferentially expressed in roots (Zhang and Forde, 1998; Gan et al., 2005, 2012; Han et al., 2008; Yu et al., 2014b). Among them, AGL21 has been reported to play a crucial role in lateral root formation and elongation in response to N signals (Yu et al., 2014b), and ANR1 is involved in nitrate-stimulated lateral root development (Zhang and Forde, 1998; Gan et al., 2012). In miR444a-overexpressing rice lines, expression of the target ANR1-like genes was down-regulated and lateral root elongation was less responsive to localized nitrate (Yan et al., 2014). And OsMADS25 has been reported to regulate root development and nitrate accumulation in roots (Yu et al., 2015). These results indicated that ANR1-like genes might have a similar role in regulating root development, especially lateral root growth, in response to N in Arabidopsis and rice, despite the evolutionary distance between the two species.
Guo et al. (2013) reported that OsMADS57 interacts with OsTB1 to modulate rice tillering via D14. Our results showed that OsMADS57 is exclusively expressed in the xylem parenchyma cells of roots and is significantly induced by nitrate and NAA (Fig. 1), consistent with previous studies (Puig et al., 2013; Yu et al., 2014a), suggesting its potential role in root development in response to external signals. Under the sufficient-nitrate condition, similar root growth and N content were recorded among wild-type plants, osmads57 mutants, and overexpression lines (Supplemental Fig. S2). Interestingly, under the low-nitrate condition, the mutation of OsMADS57 results in a decrease of seminal and adventitious root elongation through reducing cell numbers in the meristem zone and the expression level of CYCB1;1, but it did not affect lateral root growth (Fig. 6; Supplemental Fig. S4). Although mutants and overexpression plants showed opposite phenotypes in both nitrate translocation (Fig. 2) and transcriptional regulation (Fig. 4), the elongation of seminal and adventitious roots was only affected in mutant plants but not in overexpression plants relative to wild-type plants. Possibly, the increased nitrate uptake and translocation in the overexpression plants can maintain root growth under low-nitrate conditions, but the reduced nitrate uptake and translocation in the mutant plants may result in N deficiency and root growth inhibition. Furthermore, the smaller root diameter in the overexpression lines might result in less dry weight in comparison with wild-type plants (Supplemental Fig. S5).
Plants adjust root growth and development in response to changing environmental conditions through the perception and integration of external signals into the signaling pathways of plant hormones, such as auxin (López-Bucio et al., 2003; Malamy, 2005; Rubio et al., 2009; Krouk et al., 2011; Kazan, 2013; Krouk, 2016). In rice, auxin plays dominant roles in regulating root growth, such as seminal and adventitious root elongation responding to fluctuating N supplies (Sun et al., 2014). This study found that the regulatory role played by auxin in root development of the osmads57 mutants under low-nitrate conditions likely occurs via modulation of auxin accumulation in the root tip. In our study, under the sufficient-nitrate condition, similar auxin content was found among wild-type plants, osmads57 mutants, and overexpression lines (Supplemental Fig. S6). Under the 0.2 mM nitrate condition, compared with wild-type plants, (1) elevated IAA content in the mutant root tips and (2) higher activities of [3H]IAA transport from shoots to roots in the mutants (in comparison with wild-type plants) suggested that auxin polar transport was promoted in the osmads57 mutants. These results were correlated with an increase in the expression of seven PIN genes in mutant roots (relative to wild-type plants). Exogenous NAA application restored seminal root elongation and DR5::GFP expression levels in seminal root tips of wild-type plants to levels similar to those in the osmads57 mutants. Taken together, these results indicate that more auxin accumulation repressed the cell proliferation activity in the root tip of the osmads57 mutants.
CONCLUSION
In conclusion, we found that the transcription factor OsMADS57 may be a positive regulator of high-affinity OsNRT2.3a expression involved in nitrate translocation from roots to shoots. Moreover, inhibition of seminal root elongation in osmads57 mutants may be associated with more auxin accumulation in the root tips, thus repressing the cell proliferation activity.
MATERIALS AND METHODS
Vector Construction and Rice Transformation
For the overexpression construct, the full-length complementary DNA of OsMADS57 was amplified with the primer set MADS57OxF and MADS57OxR (Supplemental Table S1) and then was inserted into the BamHI and SacI sites in the pTCK303 vector. For the fusion construct of the OsMADS57 promoter and the GUS coding sequence (pOsMADS57-GUS), the immediate upstream region of the putative 2.95-kb promoter from the ATG start codon of OsMADS57 was amplified from rice (Oryza sativa ‘Dongjin’) genomic DNA using the primers MADS57Gus-F and MADS57Gus-R (Supplemental Table S1) and inserted into the pS1aGUS-3 vector at the PacI and AscI sites. These constructs were introduced into Agrobacterium tumefaciens (strain EHA105) and transformed into callus derived from mature seeds of cv Dongjin. Selection of transgenic lines (T1 generation) was conducted in the presence of 50 mg L−1 hygromycin (Roche), and GUS staining analysis was performed as previously described (Feng et al., 2011).
Identification of OsMADS57 T-DNA Insertion Mutants and Overexpression Lines
Two independent T-DNA insertion mutants in the OsMADS57 locus, m1 (PFG_3A-15619.R) and m2 (PFG_3A-60459.L), were identified in the SIGnAL database and obtained (http://signal.salk.edu/cgi-bin/RiceGE). Genotyping of the osmads57 segregating population was performed by PCR (Supplemental Fig. S1, A and B). The insertions were confirmed by qPCR using OsMADS57-specific primers (Supplemental Fig. S1C) and T-DNA border primers (LB and RB). The flanking sequences of the T-DNA insertion sites were sequenced by Genescript. The expression levels of OsMADS57 in osmads57 mutants were determined by RT-qPCR (Supplemental Fig. S1D). Transcripts of Actin were amplified as a control, using the primer set Actin-FP and Actin-RP. All primer sequences for the PCR and RT-qPCR experiments are listed in Supplemental Table S1. The independent overexpression transgenic lines with one copy, namely Ox-1, Ox-2, and Ox-3, were obtained by Southern-blot analysis (Jia et al., 2011). And the expression levels of OsMADS57 were determined by RT-qPCR using the primers presented in Supplemental Table S1.
Plant Material and Growth Conditions
Two T-DNA insertion mutant lines (m1 and m2) in the rice (ssp. japonica) cv Dongjin background were obtained from RiceGE, the Rice Functional Genomics Express Database. All transgenic lines were developed in the genetic background of cv Dongjin.
Plants were grown in a greenhouse under natural light at day/night temperatures of 30°C/18°C. Rice seeds of the wild type (cv Dongjin), mutants (homologies), and overexpression lines (T2 generation with one copy) were surface sterilized and germinated in one-half-strength Murashige and Skoog (Duchefa) standard medium. Seven-day-old seedlings of uniform size and vigor were selected and then transferred to a tank containing 8 L of IRRI nutrient solution [(mM) 0.3 KH2PO4, 0.35 K2SO4, 1 CaCl2, 1 MgSO4·7H2O, 0.5 Na2SiO3 and (µm) 20 Fe-EDTA, 9 MnCl2, 0.39 (NH4)6Mo7O4·2H2O, 20 H3BO3, 0.77 ZnSO4, 0.32 CuSO4, pH 5.5]. The nutrient solution was replaced with fresh solution daily. Nitrate and ammonium were supplied in the nutrient medium as Ca(NO3)2 and (NH4)2SO4. To exclude the potential effects of Ca2+ on the treatments, the solutions in the same experimental system were supplemented with Ca2+ to the same level as those under the higher NO3− conditions using CaCl2. To inhibit nitrification, 7 μm dicyandiamide (C2H4N4) was added to each tank to prevent ammonium oxidation.
For analysis of gene expression in response to N, osmotic stress, and auxin, 7-d-old rice seedlings were grown in IRRI solution supplied by N with/without exogenous PEG (10%, w/v), NaCl (100 mM), and NAA (10 nm) for 7 d. The roots were collected and frozen in liquid N.
Determination of Total N Content, [15N]NO3− Influx Rate, and 15N Accumulation
Rice seedlings were grown in IRRI solution containing 0.2 or 5 mM NO3− for 2 weeks. At each harvest, rice roots and shoots were separated and washed with 0.1 mM CaSO4 for 1 min, placed in an oven at 105°C for 30 min to inactivate the enzymes, and finally dried to a constant weight at 70°C. The dry weight was recorded. Total N content in plants was determined by the Kjeldahl method (Li et al., 2006).
The influx rate of [15N]NO3− and 15N accumulation were assayed as previously described (Delhon et al., 1995). Rice seedlings were grown in IRRI nutrient solution containing 0.2 mM NO3− for 2 weeks and then deprived of N for 3 d. The plants were transferred first to 0.1 mM CaSO4 for 1 min, then to a complete nutrient solution containing 0.2 mM [15N]NO3− (atom % 15N: 99%) for 5, 30, and 60 min, and finally to 0.1 mM CaSO4 for 1 min. After grinding in liquid N, one aliquot of powder was dried to a constant weight at 70°C. Approximately 6 mg of powder from each sample was analyzed using an Isotope Ratio Mass Spectrometer system (Thermo Fisher Scientific).
Xylem Sap Nitrate Content Analysis
Rice seedlings were grown in IRRI nutrient solution containing 0.2 mM NO3− for 4 weeks and then were cut 4 cm above the ground level, and the roots were immediately transferred to 0.2 mM NO3−. A preweighed absorbent cotton ball was attached to the cut surface and covered with plastic film for 1 h. Next, the collected xylem sap was squeezed from the cotton with a syringe, and the volume of the exudates was calculated from the increase in the weight of the cotton. Nitrate concentration was determined as previously described (Tang et al., 2012).
Measurement of the Net NO3− Flux Rate in Rice Plants with the SIET System
Rice seedlings were grown in the IRRI nutrient solution containing 0.2 mM NO3− for 2 weeks and then deprived of N for 3 d. Net NO3− flux in the root meristem zone was measured using the noninvasive SIET technique as described previously (Tang et al., 2012). The roots of seedlings were equilibrated in the measuring solution for 30 min. The equilibrated seedlings were then transferred to the measuring chamber filled with the solution containing 0.2 mM NO3−. Net NO3− fluxes were measured under the experimental conditions for 11 min to decrease the variability due to fluctuations. Prior to the flux measurements, the ion-selective electrodes were calibrated using NO3− concentrations of 0.05 and 0.5 mM. Each plant was measured once. The final flux values were means of more than five individual plants. The measuring solution was composed of 0.2 mM CaCl2, 0.1 mM NaCl, 0.1 mM MgSO4, and 0.3 mM MES (pH 6, adjusted with 1 m NaOH). The measurements were carried out using the SIET system BIO-003A (Younger USA Science and Technology).
Yeast One-Hybrid Assays
Yeast one-hybrid assays were used to check the binding of OsMADS57 to OsNRT2.3a using the Matchmaker Gold Yeast One-Hybrid Library Screening System kit and the Yeastmaker Transformation System 2 kit (Clontech) following the manufacturer’s instructions. One region about 30 bp in length containing the two tandem CArG boxes in the OsNRT2.3a promoter or a mutation of the CArG boxes was inserted into the vector pAbA. The construct was linearized by BstBI digestion and transformed into a Y1HGold strain to generate a Y1H bait strain. Complementary DNA encoding full-length OsMADS57 was inserted into the pGADT7AD vector to generate pGADT7AD-OsMADS57 using primers M57BamHIF and M57SacIR. The OsMADS57 construct was transformed into the Y1H bait strain and selected on a synthetic dropout-Ura and -Leu plate containing AbA.
Transient Expression in Nicotiana benthamiana Leaves
Transient expression was analyzed according to the method of Yang et al. (2000). Four-week-old N. benthamiana plants were used for infiltration. The constructs were individually transformed into the A. tumefaciens strain EHA105. The A. tumefaciens cells were infiltrated onto the abaxial surface of N. benthamiana leaves using 2-mL needleless syringes. After infiltration, the N. benthamiana plants were grown in a greenhouse under dark conditions for 48 to 60 h.
Histochemical activity of GUS in transgenic plant materials and quantitative analysis of GUS activity in N. benthamiana leaves were detected according to the method of Jefferson et al. (1987).
Measurement of Root System Architecture and Histological Observation
The rice root system is composed of seminal and adventitious roots each bearing lateral roots. Rice seedlings were grown in the IRRI nutrient solution containing 0.2 mM NO3− for 2 weeks before sampling. The length of seminal and adventitious roots was measured using a ruler, and lateral root density was calculated by dividing the lateral root number by root length. Total root length and lateral root length were measured using the WinRhizo scanner-based image-analysis system (Regent Instruments).
To analyze the length of the cortical cells in the maturity zone and cell number in the meristem zone, the root tips of the seminal roots were treated for 1 h with 1.8 m KOH solution heated to 90°C to clear the tissues and then treated with 1% (v/v) HCl solution for 5 min (Sun et al., 2016). The cortical cells of the root tips were observed with a microscope using a color CCD camera (Olympus Optical). The length of mature cells was measured and cell numbers of the root meristem were determined according to Sun et al. (2016).
The pCYCB1;1::GUS Construct
The pCYCB1;1::GUS fusion construct was transformed into wild-type plants (cv Dongjin) and the osmads57 mutant. The construct was kindly provided by Chuanzao Mao. The root tips were used in histochemical GUS staining analyses. The stained tissues were photographed using an Olympus SZX2-ILLK microscope with a color CCD camera (Olympus; Sun et al., 2016).
Determination of IAA
The content of IAA in the root tips and the lateral root area were determined (Song et al., 2011). Two root zones were sampled: the root tip (0–0.5 cm) and the lateral root zone where lateral root initiation, emergence, and elongation (2–8 cm) occurs. The fresh weight of the samples was measured, after which specimens were immediately frozen in liquid N. We performed sample measurement of free IAA by HPLC. A standard IAA sample was obtained from Sigma-Aldrich.
To detect IAA distribution patterns in plants, the pDR5::GFP fusion construct was transformed into wild-type plants, m1 mutants, and Ox-1 lines. The pDR5::GFP vector was constructed from the pDR5::GUS construct provided by Chuanzao Mao and was described by Huang et al. (2015). We analyzed the fluorescence of GFP in the cells using 543-nm helium-neon and 488-nm argon lasers using a confocal laser scanning microscope (LSM410; Carl Zeiss).
[3H]IAA Transport Assay
[3H]IAA polar transport was assayed after the 0.2 mM NO3− treatment for 2 weeks. Ten replicate roots were sampled. The [3H]IAA solution contained 0.5 mM [3H]IAA (20 Ci mmol−1) in 2% (v/v) dimethyl sulfoxide, 25 mM MES (pH 5.2), and 0.25% agar (Song et al., 2013).
Shoot-to-root auxin transport in intact plants was monitored as follows. [3H]IAA solution (20 µL) was applied to the cut surface after rice shoots were removed at 2 cm above the root-shoot junction and then incubated in scintillation solution (4 mL) for 18 h. After an 18-h (overnight) incubation in darkness, rice roots were sampled and weighed. [3H]IAA radioactivity was detected using a multipurpose scintillation counter (LS6500; Beckman-Coulter).
The assay for basipetal auxin transport (transport away from the root tip) was performed using 3-cm-long excised root tip segments. [3H]IAA solution (3 µL) was applied to the root tip placed horizontally on a plastic film. After incubation in a humid, dark environment for 18 h (overnight), root segments were cut into two parts: (1) the distal end 1 cm from the root tip and (2) the remaining 2 cm. [3H]IAA radioactivity was measured in the 2-cm-long segments.
RT-qPCR Analysis
Total RNA was isolated from the roots of rice seedlings. RNA extraction, RT, and qPCR procedures followed the reported procedure (Chen et al., 2012). The primer sets targeting nitrate-related genes and the PIN genes are listed in Supplemental Table S1.
Statistical Analysis
Data from the experiments were pooled for calculation of means and se and analyzed by Student’s t test at P ≤ 0.05 to determine the statistical significance of the differences when comparing the wild type and other lines. All statistical evaluations were conducted using the SPSS (version 11.0) statistical software (SPSS).
Accession Numbers
The sequences of rice data in this article can be downloaded from https://blast.ncbi.nlm.nih.gov/Blast.cgi or http://rice.plantbiology.msu.edu: OsMADS57 (AK108784), OsNRT2.1 (AB008519), OsNRT2.2 (AK109733), OsNRT2.3a (AK109776), OsNRT2.4 (NM_193361), OsNART2.1 (NM_001053852.2), OsNiA1 (AK102178), OsNiA2 (AK102363), OsPIN1a (AK103208), OsPIN1b (AK102343), OsPIN1c (AK103181), OsPIN1d (LOC_Os12g04000) OsPIN2 (AK101191), OsPIN5a (AK066552), OsPIN5b (AK100297), OsPIN9 (AK05922), OsPIN10a (LOC_Os01g45550), OsPIN10b (LOC_Os05g50140), and OsACT (AK100267).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Identification of the osmads57 mutants and overexpression lines.
Supplemental Figure S2. Comparison of growth and total N content of the wild type, osmads57 mutants, and overexpression lines.
Supplemental Figure S3. 15N content and root mRNA expression pattern of nitrate transporter genes in the wild type and osnrt2.3a mutants.
Supplemental Figure S4. Root systems of the wild type, osmads57 mutants, and overexpression lines.
Supplemental Figure S5. Seminal root diameter of the wild type, osmads57 mutant, and overexpression line.
Supplemental Figure S6. Auxin content in roots of the wild type, osmads57 mutants, and overexpression lines.
Supplemental Table S1. Primer sequences used in this study.
Acknowledgments
We thank Nigel Crawford (University of California, San Diego) for polishing the article and Chuanzao Mao (Zhejiang University) for providing the pCYCB1;1::GUS fusion construct.
Footnotes
This work was supported by the National Key R&D Program of China (2017YFD0200100 and 2018YFD0200503), the National Nature Science Foundation of China (31471936 and 31672225), the 111 Project (12009), the Innovative Research Team Development Plan of the Ministry of Education of China, the PAPD project of the Jiangsu Higher Education Institutions, and the Jiangsu Overseas Research & Training Program for University Prominent Young & Middle-Aged Teachers and Presidents.
References
- Almagro A, Lin SH, Tsay YF (2008) Characterization of the Arabidopsis nitrate transporter NRT1.6 reveals a role of nitrate in early embryo development. Plant Cell 20: 3289–3299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez JM, Riveras E, Vidal EA, Gras DE, Contreras-López O, Tamayo KP, Aceituno F, Gómez I, Ruffel S, Lejay L, et al. (2014) Systems approach identifies TGA1 and TGA4 transcription factors as important regulatory components of the nitrate response of Arabidopsis thaliana roots. Plant J 80: 1–13 [DOI] [PubMed] [Google Scholar]
- Araki R, Hasegawa H (2006) Expression of rice (Oryza sativa L.) genes involved in high-affinity nitrate transport during the period of nitrate induction. Breed Sci 56: 295–302 [Google Scholar]
- Araus V, Vidal EA, Puelma T, Alamos S, Mieulet D, Guiderdoni E, Gutiérrez RA (2016) Members of BTB gene family of scaffold proteins suppress nitrate uptake and nitrogen use efficiency. Plant Physiol 171: 1523–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beemster GTS, Fiorani F, Inzé D (2003) Cell cycle: The key to plant growth control? Trends Plant Sci 8: 154–158 [DOI] [PubMed] [Google Scholar]
- Cai C, Wang JY, Zhu YG, Shen QR, Li B, Tong YP, Li ZS (2008) Gene structure and expression of the high-affinity nitrate transport system in rice roots. J Integr Plant Biol 50: 443–451 [DOI] [PubMed] [Google Scholar]
- Castaings L, Camargo A, Pocholle D, Gaudon V, Texier Y, Boutet-Mercey S, Taconnat L, Renou JP, Daniel-Vedele F, Fernandez E, et al. (2009) The nodule inception-like protein 7 modulates nitrate sensing and metabolism in Arabidopsis. Plant J 57: 426–435 [DOI] [PubMed] [Google Scholar]
- Chen Y, Fan X, Song W, Zhang Y, Xu G (2012) Over-expression of OsPIN2 leads to increased tiller numbers, angle and shorter plant height through suppression of OsLAZY1. Plant Biotechnol J 10: 139–149 [DOI] [PubMed] [Google Scholar]
- Chiu CC, Lin CS, Hsia AP, Su RC, Lin HL, Tsay YF (2004) Mutation of a nitrate transporter, AtNRT1:4, results in a reduced petiole nitrate content and altered leaf development. Plant Cell Physiol 45: 1139–1148 [DOI] [PubMed] [Google Scholar]
- Crawford NM, Glass ADM (1998) Molecular and physiological aspects of nitrate uptake in plants. Trends Plant Sci 3: 389–395 [Google Scholar]
- Delhon P, Gojon A, Tillard P, Passama L (1995) Diurnal regulation of NO3− uptake in soybean plants. I. Changes in NO3− influx, efflux, and N utilization in the plant during the day/night cycle. J Exp Bot 46: 1585–1594 [Google Scholar]
- Du XQ, Wang FL, Li H, Jing S, Yu M, Li J, Wu WH, Kudla J, Wang Y (2019) The transcription factor MYB59 regulates K+/NO3− translocation in the Arabidopsis response to low K+ stress. Plant Cell 31: 699–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan SC, Lin CS, Hsu PK, Lin SH, Tsay YF (2009) The Arabidopsis nitrate transporter NRT1.7, expressed in phloem, is responsible for source-to-sink remobilization of nitrate. Plant Cell 21: 2750–2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X, Tang Z, Tan Y, Zhang Y, Luo B, Yang M, Lian X, Shen Q, Miller AJ, Xu G (2016) Overexpression of a pH-sensitive nitrate transporter in rice increases crop yields. Proc Natl Acad Sci USA 113: 7118–7123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H, Yan M, Fan X, Li B, Shen Q, Miller AJ, Xu G (2011) Spatial expression and regulation of rice high-affinity nitrate transporters by nitrogen and carbon status. J Exp Bot 62: 2319–2332 [DOI] [PubMed] [Google Scholar]
- Forde BG. (2014) Nitrogen signalling pathways shaping root system architecture: An update. Curr Opin Plant Biol 21: 30–36 [DOI] [PubMed] [Google Scholar]
- Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, Offringa R, Jürgens G (2003) Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426: 147–153 [DOI] [PubMed] [Google Scholar]
- Gan Y, Filleur S, Rahman A, Gotensparre S, Forde BG (2005) Nutritional regulation of ANR1 and other root-expressed MADS-box genes in Arabidopsis thaliana. Planta 222: 730–742 [DOI] [PubMed] [Google Scholar]
- Gan Y, Bernreiter A, Filleur S, Abram B, Forde BG (2012) Overexpressing the ANR1 MADS-box gene in transgenic plants provides new insights into its role in the nitrate regulation of root development. Plant Cell Physiol 53: 1003–1016 [DOI] [PubMed] [Google Scholar]
- Gojon A, Krouk G, Perrine-Walker F, Laugier E (2011) Nitrate transceptor(s) in plants. J Exp Bot 62: 2299–2308 [DOI] [PubMed] [Google Scholar]
- Grieneisen VA, Xu J, Marée AF, Hogeweg P, Scheres B (2007) Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature 449: 1008–1013 [DOI] [PubMed] [Google Scholar]
- Guan P, Ripoll JJ, Wang R, Vuong L, Bailey-Steinitz LJ, Ye D, Crawford NM (2017) Interacting TCP and NLP transcription factors control plant responses to nitrate availability. Proc Natl Acad Sci USA 114: 2419–2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo FQ, Wang R, Chen M, Crawford NM (2001) The Arabidopsis dual-affinity nitrate transporter gene AtNRT1.1 (CHL1) is activated and functions in nascent organ development during vegetative and reproductive growth. Plant Cell 13: 1761–1777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Xu Y, Liu H, Mao Z, Zhang C, Ma Y, Zhang Q, Meng Z, Chong K (2013) The interaction between OsMADS57 and OsTB1 modulates rice tillering via DWARF14. Nat Commun 4: 1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han P, García-Ponce B, Fonseca-Salazar G, Alvarez-Buylla ER, Yu H (2008) AGAMOUS-LIKE 17, a novel flowering promoter, acts in a FT-independent photoperiod pathway. Plant J 55: 253–265 [DOI] [PubMed] [Google Scholar]
- Ho CH, Lin SH, Hu HC, Tsay YF (2009) CHL1 functions as a nitrate sensor in plants. Cell 138: 1184–1194 [DOI] [PubMed] [Google Scholar]
- Hsu PK, Tsay YF (2013) Two phloem nitrate transporters, NRT1.11 and NRT1.12, are important for redistributing xylem-borne nitrate to enhance plant growth. Plant Physiol 163: 844–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Wang W, Ou S, Tang J, Li H, Che R, Zhang Z, Chai X, Wang H, Wang Y, et al. (2015) Variation in NRT1.1B contributes to nitrate-use divergence between rice subspecies. Nat Genet 47: 834–838 [DOI] [PubMed] [Google Scholar]
- Hu R, Qiu D, Chen Y, Miller AJ, Fan X, Pan X, Zhang M (2016) Knock-down of a tonoplast localized low-affinity nitrate transporter OsNPF7.2 affects rice growth under high nitrate supply. Front Plant Sci 7: 1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang NC, Liu KH, Lo HJ, Tsay YF (1999) Cloning and functional characterization of an Arabidopsis nitrate transporter gene that encodes a constitutive component of low-affinity uptake. Plant Cell 11: 1381–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Chen S, Liang Z, Zhang C, Yan M, Chen J, Xu G, Fan X, Zhang Y (2015) Knockdown of the partner protein OsNAR2.1 for high-affinity nitrate transport represses lateral root formation in a nitrate-dependent manner. Sci Rep 5: 18192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H, Ren H, Gu M, Zhao J, Sun S, Zhang X, Chen J, Wu P, Xu G (2011) The phosphate transporter gene OsPht1;8 is involved in phosphate homeostasis in rice. Plant Physiol 156: 1164–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant S. (2018) Understanding nitrate uptake, signaling and remobilisation for improving plant nitrogen use efficiency. Semin Cell Dev Biol 74: 89–96 [DOI] [PubMed] [Google Scholar]
- Kazan K. (2013) Auxin and the integration of environmental signals into plant root development. Ann Bot 112: 1655–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk GJD. (2003) Rice root properties for internal aeration and efficient nutrient acquisition in submerged soil. New Phytol 159: 185–194 [DOI] [PubMed] [Google Scholar]
- Kirk GJ, Kronzucker HJ (2005) The potential for nitrification and nitrate uptake in the rhizosphere of wetland plants: A modelling study. Ann Bot 96: 639–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi M, Yanagisawa S (2013) Arabidopsis NIN-like transcription factors have a central role in nitrate signalling. Nat Commun 4: 1617. [DOI] [PubMed] [Google Scholar]
- Kronzucker HJ, Glass ADM, Siddiqi MY, Kirk GJD (2000) Comparative kinetic analysis of ammonium and nitrate acquisition by tropical lowland rice: Implications for rice cultivation and yield potential. New Phytol 145: 471–476 [DOI] [PubMed] [Google Scholar]
- Krouk G. (2016) Hormones and nitrate: A two-way connection. Plant Mol Biol 91: 599–606 [DOI] [PubMed] [Google Scholar]
- Krouk G, Lacombe B, Bielach A, Perrine-Walker F, Malinska K, Mounier E, Hoyerova K, Tillard P, Leon S, Ljung K, et al. (2010a) Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev Cell 18: 927–937 [DOI] [PubMed] [Google Scholar]
- Krouk G, Mirowski P, LeCun Y, Shasha DE, Coruzzi GM (2010b) Predictive network modeling of the high-resolution dynamic plant transcriptome in response to nitrate. Genome Biol 11: R123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krouk G, Ruffel S, Gutiérrez RA, Gojon A, Crawford NM, Coruzzi GM, Lacombe B (2011) A framework integrating plant growth with hormones and nutrients. Trends Plant Sci 16: 178–182 [DOI] [PubMed] [Google Scholar]
- Léran S, Varala K, Boyer JC, Chiurazzi M, Crawford N, Daniel-Vedele F, David L, Dickstein R, Fernandez E, Forde B, et al. (2014) A unified nomenclature of NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER family members in plants. Trends Plant Sci 19: 5–9 [DOI] [PubMed] [Google Scholar]
- Li B, Xin W, Sun S, Shen Q, Xu G (2006) Physiological and molecular responses of nitrogen-starved rice plants to re-supply of different nitrogen sources. Plant Soil 287: 145–159 [Google Scholar]
- Li H, Yu M, Du XQ, Wang ZF, Wu WH, Quintero FJ, Jin XH, Li HD, Wang Y (2017) NRT1.5/NPF7.3 functions as a proton-coupled H+/K+ antiporter for K+ loading into the xylem in Arabidopsis. Plant Cell 29: 2016–2026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JY, Fu YL, Pike SM, Bao J, Tian W, Zhang Y, Chen CZ, Zhang Y, Li HM, Huang J, et al. (2010a) The Arabidopsis nitrate transporter NRT1.8 functions in nitrate removal from the xylem sap and mediates cadmium tolerance. Plant Cell 22: 1633–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YL, Fan XR, Shen QR (2008) The relationship between rhizosphere nitrification and nitrogen-use efficiency in rice plants. Plant Cell Environ 31: 73–85 [DOI] [PubMed] [Google Scholar]
- Li YF, Zheng Y, Addo-Quaye C, Zhang L, Saini A, Jagadeeswaran G, Axtell MJ, Zhang W, Sunkar R (2010b) Transcriptome-wide identification of microRNA targets in rice. Plant J 62: 742–759 [DOI] [PubMed] [Google Scholar]
- Li Y, Ouyang J, Wang YY, Hu R, Xia K, Duan J, Wang Y, Tsay YF, Zhang M (2015) Disruption of the rice nitrate transporter OsNPF2.2 hinders root-to-shoot nitrate transport and vascular development. Sci Rep 5: 9635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SH, Kuo HF, Canivenc G, Lin CS, Lepetit M, Hsu PK, Tillard P, Lin HL, Wang YY, Tsai CB, et al. (2008) Mutation of the Arabidopsis NRT1.5 nitrate transporter causes defective root-to-shoot nitrate transport. Plant Cell 20: 2514–2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu KH, Tsay YF (2003) Switching between the two action modes of the dual-affinity nitrate transporter CHL1 by phosphorylation. EMBO J 22: 1005–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Huang D, Tao J, Miller AJ, Fan X, Xu G (2014) Identification and functional assay of the interaction motifs in the partner protein OsNAR2.1 of the two-component system for high-affinity nitrate transport. New Phytol 204: 74–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Bucio J, Cruz-Ramírez A, Herrera-Estrella L (2003) The role of nutrient availability in regulating root architecture. Curr Opin Plant Biol 6: 280–287 [DOI] [PubMed] [Google Scholar]
- Malamy JE. (2005) Intrinsic and environmental response pathways that regulate root system architecture. Plant Cell Environ 28: 67–77 [DOI] [PubMed] [Google Scholar]
- Marchive C, Roudier F, Castaings L, Bréhaut V, Blondet E, Colot V, Meyer C, Krapp A (2013) Nuclear retention of the transcription factor NLP7 orchestrates the early response to nitrate in plants. Nat Commun 4: 1713. [DOI] [PubMed] [Google Scholar]
- Miller AJ, Fan X, Orsel M, Smith SJ, Wells DM (2007) Nitrate transport and signalling. J Exp Bot 58: 2297–2306 [DOI] [PubMed] [Google Scholar]
- O’Brien JA, Vega A, Bouguyon E, Krouk G, Gojon A, Coruzzi G, Gutiérrez RA (2016) Nitrate transport, sensing, and responses in plants. Mol Plant 9: 837–856 [DOI] [PubMed] [Google Scholar]
- Plett DC, Holtham LR, Okamoto M, Garnett TP (2018) Nitrate uptake and its regulation in relation to improving nitrogen use efficiency in cereals. Semin Cell Dev Biol 74: 97–104 [DOI] [PubMed] [Google Scholar]
- Puig J, Meynard D, Khong GN, Pauluzzi G, Guiderdoni E, Gantet P (2013) Analysis of the expression of the AGL17-like clade of MADS-box transcription factors in rice. Gene Expr Patterns 13: 160–170 [DOI] [PubMed] [Google Scholar]
- Remans T, Nacry P, Pervent M, Filleur S, Diatloff E, Mounier E, Tillard P, Forde BG, Gojon A (2006) The Arabidopsis NRT1.1 transporter participates in the signaling pathway triggering root colonization of nitrate-rich patches. Proc Natl Acad Sci USA 103: 19206–19211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin G, Tohge T, Matsuda F, Saito K, Scheible WR (2009) Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. Plant Cell 21: 3567–3584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio V, Bustos R, Irigoyen ML, Cardona-López X, Rojas-Triana M, Paz-Ares J (2009) Plant hormones and nutrient signaling. Plant Mol Biol 69: 361–373 [DOI] [PubMed] [Google Scholar]
- Shim JS, Jung C, Lee S, Min K, Lee YW, Choi Y, Lee JS, Song JT, Kim JK, Choi YD (2013) AtMYB44 regulates WRKY70 expression and modulates antagonistic interaction between salicylic acid and jasmonic acid signaling. Plant J 73: 483–495 [DOI] [PubMed] [Google Scholar]
- Smirnoff N, Stewart GR (1985) Nitrate assimilation and translocation by higher plants: Comparative physiology and ecological consequences. Physiol Plant 64: 133–140 [Google Scholar]
- Song W, Makeen K, Wang D, Zhang C, Xu Y, Zhao H, Tu E, Zhang Y, Shen Q, Xu G (2011) Nitrate supply affects root growth differentially in two rice cultivars differing in nitrogen use efficiency. Plant Soil 343: 357–368 [Google Scholar]
- Song W, Sun H, Li J, Gong X, Huang S, Zhu X, Zhang Y, Xu G (2013) Auxin distribution is differentially affected by nitrate in roots of two rice cultivars differing in responsiveness to nitrogen. Ann Bot 112: 1383–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Tao J, Liu S, Huang S, Chen S, Xie X, Yoneyama K, Zhang Y, Xu G (2014) Strigolactones are involved in phosphate- and nitrate-deficiency-induced root development and auxin transport in rice. J Exp Bot 65: 6735–6746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Bi Y, Tao J, Huang S, Hou M, Xue R, Liang Z, Gu P, Yoneyama K, Xie X, et al. (2016) Strigolactones are required for nitric oxide to induce root elongation in response to nitrogen and phosphate deficiencies in rice. Plant Cell Environ 39: 1473–1484 [DOI] [PubMed] [Google Scholar]
- Tang Z, Fan X, Li Q, Feng H, Miller AJ, Shen Q, Xu G (2012) Knockdown of a rice stelar nitrate transporter alters long-distance translocation but not root influx. Plant Physiol 160: 2052–2063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsay YF, Chiu CC, Tsai CB, Ho CH, Hsu PK (2007) Nitrate transporters and peptide transporters. FEBS Lett 581: 2290–2300 [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YY, Tsay YF (2011) Arabidopsis nitrate transporter NRT1.9 is important in phloem nitrate transport. Plant Cell 23: 1945–1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Xing X, Wang Y, Tran A, Crawford NM (2009) A genetic screen for nitrate regulatory mutants captures the nitrate transporter gene NRT1.1. Plant Physiol 151: 472–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Hu B, Yuan D, Liu Y, Che R, Hu Y, Ou S, Liu Y, Zhang Z, Wang H, et al. (2018a) Expression of the nitrate transporter gene OsNRT1.1A/OsNPF6.3 confers high yield and early maturation in rice. Plant Cell 30: 638–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YY, Cheng YH, Chen KE, Tsay YF (2018b) Nitrate transport, signaling, and use efficiency. Annu Rev Plant Biol 69: 85–122 [DOI] [PubMed] [Google Scholar]
- Wei J, Zheng Y, Feng H, Qu H, Fan X, Yamaji N, Ma JF, Xu G (2018) OsNRT2.4 encodes a dual-affinity nitrate transporter and functions in nitrate-regulated root growth and nitrate distribution in rice. J Exp Bot 69: 1095–1107 [DOI] [PubMed] [Google Scholar]
- Xia X, Fan X, Wei J, Feng H, Qu H, Xie D, Miller AJ, Xu G (2015) Rice nitrate transporter OsNPF2.4 functions in low-affinity acquisition and long-distance transport. J Exp Bot 66: 317–331 [DOI] [PubMed] [Google Scholar]
- Xu G, Fan X, Miller AJ (2012) Plant nitrogen assimilation and use efficiency. Annu Rev Plant Biol 63: 153–182 [DOI] [PubMed] [Google Scholar]
- Xu N, Wang R, Zhao L, Zhang C, Li Z, Lei Z, Liu F, Guan P, Chu Z, Crawford NM, et al. (2016) The Arabidopsis NRG2 protein mediates nitrate signaling and interacts with and regulates key nitrate regulators. Plant Cell 28: 485–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuan W, Beeckman T, Xu G (2017) Plant nitrogen nutrition: Sensing and signaling. Curr Opin Plant Biol 39: 57–65 [DOI] [PubMed] [Google Scholar]
- Yan D, Easwaran V, Chau V, Okamoto M, Ierullo M, Kimura M, Endo A, Yano R, Pasha A, Gong Y, et al. (2016) NIN-like protein 8 is a master regulator of nitrate-promoted seed germination in Arabidopsis. Nat Commun 7: 13179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan M, Fan X, Feng H, Miller AJ, Shen Q, Xu G (2011) Rice OsNAR2.1 interacts with OsNRT2.1, OsNRT2.2 and OsNRT2.3a nitrate transporters to provide uptake over high and low concentration ranges. Plant Cell Environ 34: 1360–1372 [DOI] [PubMed] [Google Scholar]
- Yan Y, Wang H, Hamera S, Chen X, Fang R (2014) miR444a has multiple functions in the rice nitrate-signaling pathway. Plant J 78: 44–55 [DOI] [PubMed] [Google Scholar]
- Yang W, Yoon J, Choi H, Fan Y, Chen R, An G (2015) Transcriptome analysis of nitrogen-starvation-responsive genes in rice. BMC Plant Biol 15: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Li R, Qi M (2000) In vivo analysis of plant promoters and transcription factors by agroinfiltration of tobacco leaves. Plant J 22: 543–551 [DOI] [PubMed] [Google Scholar]
- Yu C, Su S, Xu Y, Zhao Y, Yan A, Huang L, Ali I, Gan Y (2014a) The effects of fluctuations in the nutrient supply on the expression of five members of the AGL17 clade of MADS-box genes in rice. PLoS ONE 9: e105597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, Liu Y, Zhang A, Su S, Yan A, Huang L, Ali I, Liu Y, Forde BG, Gan Y (2015) MADS-box transcription factor OsMADS25 regulates root development through affection of nitrate accumulation in rice. PLoS ONE 10: e0135196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu LH, Miao ZQ, Qi GF, Wu J, Cai XT, Mao JL, Xiang CB (2014b) MADS-box transcription factor AGL21 regulates lateral root development and responds to multiple external and physiological signals. Mol Plant 7: 1653–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Forde BG (1998) An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science 279: 407–409 [DOI] [PubMed] [Google Scholar]