A glucose-TOR-E2Fa module regulates thermotolerance and thermomemory via regulation of HLP1.
Abstract
Induction of heat shock proteins (HSPs) in response to heat stress (HS) is indispensable for conferring thermotolerance. Glc, a fundamental signaling and metabolic molecule, provides energy to stressed seedlings to combat stress. The recovery of stressed plants from detrimental HS in response to Glc is largely mediated by HSPs, but the mechanistic basis of this thermotolerance is not well defined. In this study, we show that Glc has a prominent role in providing thermotolerance. Glc-mediated thermotolerance involves HSP induction via the TARGET OF RAPAMYCIN (TOR)-E2Fa signaling module. Apart from HSPs, TOR-E2Fa also regulates the Arabidopsis (Arabidopsis thaliana) ortholog of human Hikeshi, named HIKESHI-LIKE PROTEIN1 (HLP1). Expression of proHLP1::GUS in the shoot apical meristem (SAM) after HS coincides with TOR-E2Fa expression, substantiating a role for TOR-E2Fa-HLP1 in providing thermotolerance. We also demonstrate that Glc along with heat could induce proliferation activity in the SAM after HS recovery, which was arrested by the TOR inhibitor AZD-8055. Molecular and physiological studies suggest that HS-activated heat stress transcription factor A1s also positively regulate HLP1 transcription, suggesting convergence of the Glc and HS signaling pathways. Loss of functional HLP1 causes HS hypersensitivity, whereas HLP1 overexpressors display increased thermotolerance. HLP1 binds to the promoters of Glc-regulated HS-responsive genes and promotes chromatin acetylation. In addition, Glc modifies the chromatin landscape at thermomemory-related loci by promoting H3K4 trimethylation (H3K4me3). Glc-primed accumulation of H3K4me3 at thermomemory-associated loci is mediated through HLP1. These findings reveal the novel function of Glc-regulated HLP1 in mediating thermotolerance/thermomemory response.
Coordination of complex networks during cell division and expansion results in growth and development of an organism. These networks keep adapting to an everchanging environment (Gonzalez et al., 2012; Powell and Lenhard, 2012) and involve plant signaling machinery to regulate growth and development. Sugars also coordinate a variety of processes involved in plant growth and respond appropriately to changing environments by altering metabolic and energy demands (Ramon et al., 2008). Among sugars, Glc is a major signaling molecule that affects almost all the processes involved in plant growth and development. Exogenous Glc is perceived by the Glc sensor HEXOKINASE1 (HXK1) or the HXK1-independent cellular receptor REGULATOR OF G-PROTEIN SIGNALING (Chen et al., 2003; Cho et al., 2006, 2009; Huang et al., 2006; Li and Sheen, 2016). Arabidopsis (Arabidopsis thaliana) HXK1 mediates several functions in Glc repression and promotion of transcription and growth (Moore et al., 2003; Cho et al., 2006, 2009).
Arabidopsis TARGET OF RAPAMYCIN (TOR) kinase is activated by energy/nutrient surplus and is essential for transcriptome reprogramming, meristem activation, and plant growth (Schepetilnikov et al., 2013, 2017; Xiong et al., 2013, 2017; Dobrenel et al., 2016; Li and Sheen, 2016; Dong et al., 2017; Li et al., 2017). Glc-activated TOR is an evolutionarily conserved kinase that phosphorylates the E2Fa/b transcription factor for activation of s-phase genes in Arabidopsis (Xiong et al., 2013; Li et al., 2017). E2Fa binds directly to TTTCCCGCC or other similar motifs in the promoter of cell cycle genes and regulates their expression (Chabouté et al., 2000; Xiong et al., 2013). In addition, genes belonging to transcription, stress, defense response, and signaling also contain E2Fa binding motifs in their promoters (Chabouté et al., 2000). Various reports suggested the interaction of Arabidopsis TOR with phytohormones in controlling plant growth and development. Sugar-induced TOR regulates the brassinosteroid signaling transcription factor BZR1, which allows carbon availability to control plant growth (Zhang et al., 2016). Arabidopsis TOR target S6K2 is required for direct phosphorylation of BRASSINOSTEROID INSENSITIVE2 in controlling the photoautotrophic transition (Xiong et al., 2017). In response to auxin, the small GTPase RHO-RELATED PROTEIN FROM PLANTS 2 activates TOR and governs translational reinitiation of upstream open reading frame (uORF)-containing mRNAs (Schepetilnikov et al., 2017). Plants experience various biotic and abiotic stresses that often lead to changes in the internal energy status. Besides interaction with various signaling networks, sugar (Glc) signaling also interacts with pathways of different environmental stresses (Loreti et al., 2005; Mahfouz et al., 2006; Baena-González et al., 2007; Deprost et al., 2007; Baena-González and Sheen, 2008; Wang et al., 2017).
Among the various abiotic stresses studied, temperature is a major factor involved in controlling plant metabolism and growth. Exposure to stress signals results in dynamic reprogramming of the transcriptional networks that enhance stress tolerance. Responses to several types of abiotic stresses in plants are activated and integrated by the expression of thousands of genes encoding proteins involved in numerous biological processes (Seki et al., 2002; Tran et al., 2007; Hua, 2009). In heat shock response (HSR), many of these proteins are involved in preventing or repairing the damage and thus confer increased thermotolerance (Vierling, 1991). It is already known that HSFs of Arabidopsis and other plants bind directly to heat shock elements (HSEs). These HSEs comprise tandem inverted repeats of the pentameric consensus sequence nGAAn (nTTCnnGAAnnTTCn; known as “perfect HSE”) and the AGGGG motifs (Swindell et al., 2007; von Koskull-Döring et al., 2007). Recent reports have shown that heat stress transcription factor A1a (HSFA1a) directly senses HS and becomes activated through alterations in its redox state (Liu et al., 2013). HSFA2 is highly induced under HS and is essential for extending the thermotolerance response (Charng et al., 2007). Heat shock proteins (HSPs) are downstream targets of HSFs and function as molecular chaperones involved in the restoration of protein homeostasis and maintenance of the thermotolerance response. In natural environments, plants experience chronic or recurring stress conditions and have evolved mechanisms to remember past experiences to cope with future stresses. Stress memory refers to changes in the chromatin epigenetic landscape for induced expression of memory-related loci (Lämke et al., 2016). HSFA2 is known to be involved in the maintenance of memory gene induction by binding directly to their promoters and modifying their chromatin through epigenetic modifications (Lämke et al., 2016). HSP101-promoted accumulation of HEAT-STRESS (HS)-ASSOCIATED 32 is required for the maintenance of thermotolerance (Wu et al., 2013). In plants, H3K4 trimethylation (H3K4me3) marks are associated with high induction of gene expression (Guenther et al., 2007). Arabidopsis FORGETTER1 interacts with chromatin remodelers of the CHROMATIN REMODELING BY IMITATION SWITCH family in association with BRAHMA to provide the thermomemory response (Brzezinka et al., 2016).
Crosstalk between the temperature and other stress-response mechanisms in plants has already been proposed (Wang et al., 2004), but the mechanism of adaptive tolerance to temperature stress, when plants have surplus Glc/energy, has not yet been adequately explored. It has already been proposed that human Hikeshi interacts with HSP70 to provides thermotolerance in humans (Kose et al., 2012). The Arabidopsis genome contains a single Hikeshi homolog, which we named HIKESHI-LIKE PROTEIN1 (HLP1). Koizumi et al. (2014) showed that Arabidopsis HLP1 interacts with HSP70 and overexpression of this gene confers thermotolerance to Arabidopsis plants. In our study, we show that Arabidopsis HLP1 plays a crucial role in providing thermotolerance, mediated by Glc. Glc-mediated thermotolerance involves HLP1 induction via the energy sensor TOR-E2Fa signaling module, suggesting a novel crosstalk between Glc/TOR-E2Fa signaling and HS response. We also show that Arabidopsis HLP1 is indispensable for plants to maintain Glc-mediated thermomemory response. This study aims to explore the interaction of Glc signaling and temperature stress in Arabidopsis to provide mechanistic insights into the control of temperature stress by Glc.
RESULTS
Glc Regulates Heat Stress Response in Arabidopsis
Glc regulates a large number of genes involved in a broad range of cellular responses such as signaling during stress, metabolism, growth, and development. We performed a comparative data analysis to find overlap between Glc and HS–regulated genes. According to previous microarray reports (Mishra et al., 2009; Gupta et al., 2015) and recent annotations by The Arabidopsis Information Resource (https://www.arabidopsis.org/), a significant overlap of genes was observed between Glc and HS (Supplemental Fig. S1A; Supplemental Table S1). This led to the hypothesis that Glc may provide thermotolerance to Arabidopsis plants.
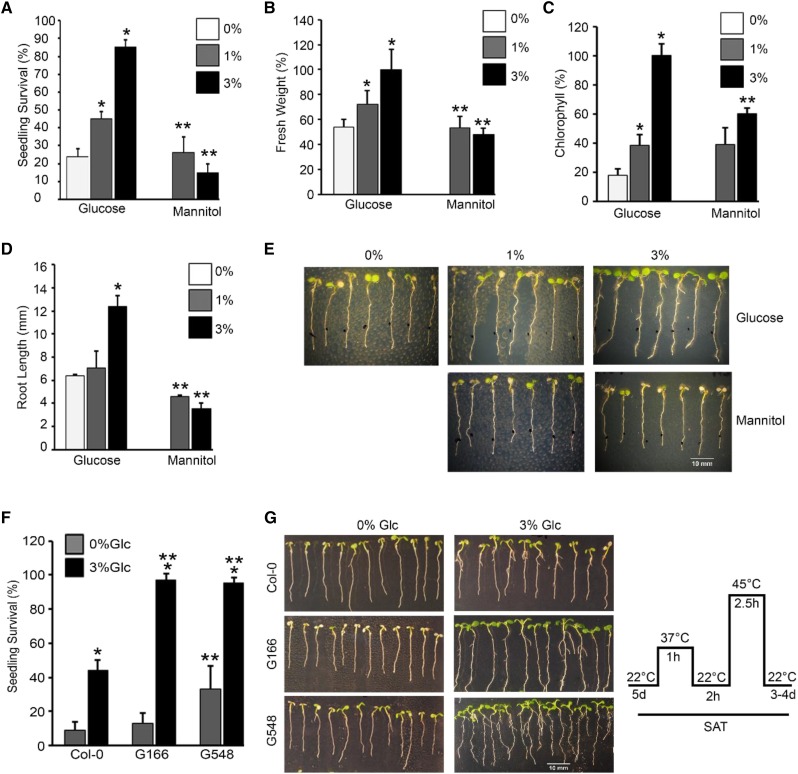
To study Glc-mediated thermotolerance, Arabidopsis Col-0 seedlings were treated with Glc and the effect of HS was analyzed. Five-d–old Arabidopsis Col-0 seedlings were transferred to Murashige and Skoog (MS) media containing increasing concentrations of Glc (0%, 1%, and 3% Glc) for 24 h followed by HS (HS was applied as 1 h at 37°C, 2 h at 22°C, 2.5 h at 45°C, and 3–4 d at 22°C). Arabidopsis Col-0 seedlings supplied with optimum (3% Glc) Glc survived better as compared to seedlings on low Glc (1% Glc) or without Glc (0% Glc; Fig. 1, A–E). To test whether Glc-induced thermotolerance is specific and is not mediated by any other osmotic sugars, Arabidopsis Col-0 seedlings were supplemented with Glc analogs (mannitol and 3-o-methyl-d-Glc [3-OMG]) and the effect of HS was analyzed. Arabidopsis Col-0 seedlings supplied with Glc (1% and 3% Glc) survived better (had higher fresh weight, longer root length, and increased chlorophyll [Chl] content) than seedlings treated with mannitol and 3-OMG (Fig. 1, A–E; Supplemental Fig. S1, B–D). Taken together, these results indicate that a plausible crosstalk exists between Glc and HS, suggesting that Glc may provide thermotolerance to Arabidopsis plants.
Figure 1.
Glc regulates thermotolerance response in Arabidopsis. A–D, Percentage seedling survival, fresh weight, Chl content, and root-length measurement of 5-d–old MS-grown Col-0 seedlings treated without (0% Glc) or with increasing Glc (1% Glc, 3% Glc) and mannitol followed by HS. HS was applied as 1 h at 37°C, 2 h at 22°C, 2.5 h at 45°C, and 3–4 d at 22°C. Data shown are representative of one biological replicate having 25 seedlings. Experiment was repeated three times with similar results. E, Seedling survival phenotype of Col-0 seedlings without or with increasing Glc and mannitol. Images shown are representative of one biological replicate having at least 30 seedlings. Experiments were repeated three times with similar results. F, Percent seedling survival of 5-d–old MS-grown Col-0 and TOR overexpression (G166 and G548) seedlings treated without (0% Glc) or with Glc (3% Glc) followed by HS. Data shown are average of four biological replicates having at least 30 seedlings. G, Seedling survival phenotype of Col-0, G166, and G548 seedlings treated without or with Glc and subjected to HS. Images shown are representative of four biological replicates having at least 30 seedlings. Right shows schematic representation of HS treatment experimental setup. Experiments were repeated four times with similar results. Error bars = sd (Student’s t test, P < 0.05; *control versus treatment; **Glc versus mannitol/wild type versus overexpression).
The Arabidopsis TOR-E2Fa Module Is Involved in Providing Thermotolerance
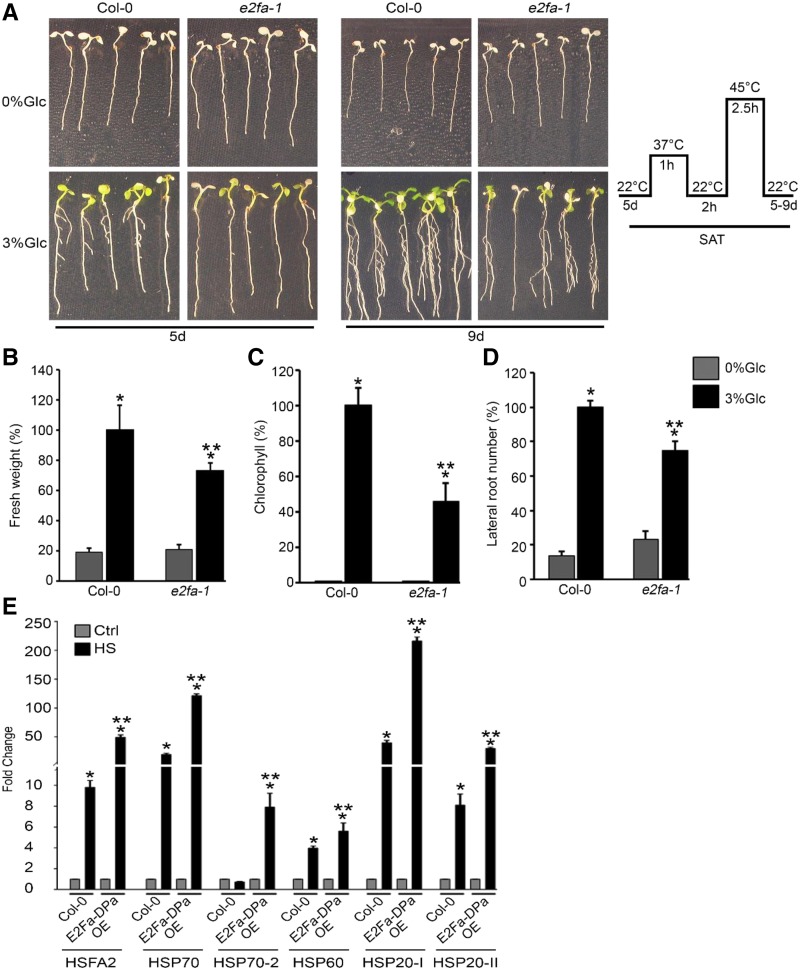
Arabidopsis TOR kinase is known to provide tolerance to various stresses (Deprost et al., 2007; Bakshi et al., 2017; Dong et al., 2017). There are studies that document the inhibition of TOR kinase activity by various stresses (Mahfouz et al., 2006; Wang et al., 2017). However, TOR overexpression lines exhibited increased susceptibility to both bacterial and fungal pathogens (De Vleesschauwer et al., 2017). We therefore investigated the effect of HS on Arabidopsis TOR overexpression lines G166 and G548 (Deprost et al., 2007). Five-d–old Arabidopsis Col-0, G166, and G548 seedlings were treated without or with Glc followed by HS. HS was applied as 1 h at 37°C, 2 h at 22°C, 2.5 h at 45°C, and 3–4 d at 22°C. Arabidopsis G166 and G548 lines exhibited higher seedling survival as compared to Col-0 plants in the presence of Glc (Fig. 1, F and G). Both G166 and G548 lines displayed higher Chl retention and increased lateral root number compared to Col-0 plants due to Glc (Supplemental Fig. S1, E and F). Further, we analyzed the thermosensitive phenotype in tor 35-7 RNA interference (RNAi) lines under HS. Arabidopsis tor 35-7 RNAi showed less seedling survival both at 0% Glc and 3% Glc than Col-0 seedlings (Supplemental Fig. S2, A and B). Moreover, we analyzed the temperature-responsive phenotype in Col-0 seedlings supplemented with the TOR kinase ATP-competitive inhibitors Torin 1 and AZD-8055. Five-d–old MS-grown Arabidopsis Col-0 seedlings were transferred to Glc (3% Glc) without or with TOR inhibitors Torin 1 (10 µM) and AZD-8055 (10 µM) for 24 h followed by HS. Arabidopsis Col-0 seedlings treated with Torin 1 (Cayman Chemical Company) and AZD-8055 displayed less seedling survival with a decrease in expression of HSP genes after HS (Supplemental Fig. S2, C–E). Further, we analyzed the downstream components of TOR signaling for HS phenotypes. As reported earlier, E2Fa is a TOR phosphorylation substrate that governs transcriptional reprogramming of a myriad genes involved in various biological processes (Vandepoele et al., 2005; Naouar et al., 2009; Xiong et al., 2013). We next checked the HS phenotype of the e2fa-1 mutant. Arabidopsis e2fa-1 plants showed reduced fresh weight, lower Chl content, and fewer lateral roots after HS (Fig. 2, A–D). In addition, emergence of true leaves was severely compromised in plants lacking E2Fa, indicating reduced HS recovery (Fig. 2A). Further, increased expression of HS genes was observed in Arabidopsis Col-0 plants transiently overexpressing E2Fa-DPa after HS (Fig. 2E). The reverse transcription quantitative PCR (RT-qPCR) expression of HS genes was also significantly downregulated in the e2fa-1 mutant under HS (Supplemental Fig. S2F). Together, these results suggest a role for the TOR-E2Fa signaling module in sugar-mediated thermotolerance.
Figure 2.
Mutation in E2Fa affects plant tolerance to HS. A, Seedling survival phenotype of Col-0 and e2fa-1 seedlings treated without or with Glc and subjected to HS. Images shown are representative of four biological replicates having at least 30 seedlings. Right shows schematic representation of HS treatment experimental setup. B, Percent fresh weight of 5-d–old MS grown Col-0 and e2fa-1 seedlings treated without (0% Glc) or with Glc (3% Glc) followed by HS. Data shown are average of four technical replicates having at least 20 seedlings. Experiments were repeated two times with similar results. C, Percent Chl content in Arabidopsis Col-0 and e2fa-1 seedlings in presence or absence of Glc and HS. Data shown are average of three biological replicates having at least 30 seedlings. Experiments were repeated three times with similar results. D, Percent lateral root number of Col-0 and e2fa-1 seedlings in the presence of Glc and HS. Data shown are average of three biological replicates having at least 20 seedlings. E, RT-qPCR expression of HS-related genes in Arabidopsis Col-0 and E2Fa-DPa overexpressing (E2Fa-DPa) plants. Four-week–old Col-0 plants were transiently coinfiltrated with 35SCaMV::E2Fa and 35SCaMV::DPa constructs followed by HS. HS was applied as 3 h at 37°C and harvested immediately. DPa transcription factor is known to activate E2Fa activity. Experiments were repeated two times with similar results. Ctrl, control. Error bars = sd (Student’s t test, P < 0.05; *control versus treatment; **wild type versus mutant/overexpression).
HLP1 Transcription Is Regulated by TOR-E2Fa Signaling
Previously reported microarrays revealed that a large number of Glc-regulated genes encode expressed proteins with unknown functions (Mishra et al., 2009; Gupta et al., 2015). These proteins did not contain any known characterized domains. Among them, At1g66080 was reported to be an expressed protein containing a domain of unknown function 775. Sequence homology revealed that this gene encoding domain of unknown function 775 is an ortholog of human Hikeshi, which functions as an HSP70-interacting protein and imports HSP70 into the nucleus to provide thermotolerance (Kose et al., 2012; Koizumi et al., 2014). We named it “HLP1.” Because HLP1 is a single copy gene in Arabidopsis and its ortholog has been shown to play an important role in regulating HS in humans, we selected this gene for further characterization in plants. Arabidopsis HLP1 has been found to be 21-fold–upregulated by Glc and 63-fold–upregulated by HS (Supplemental Fig. S3A; Mishra et al., 2009; Liu et al., 2011). To examine the induction specificity of HLP1 in response to various sugars, Col-0 seedlings were treated with different sugar analogs and RT-qPCR expression was analyzed. The gene expression pattern revealed that HLP1 was induced in response to Glc and Suc, whereas no significant change could be observed in response to different sugar analogs such as mannose, mannitol, and 3-OMG (Supplemental Fig. S3B).
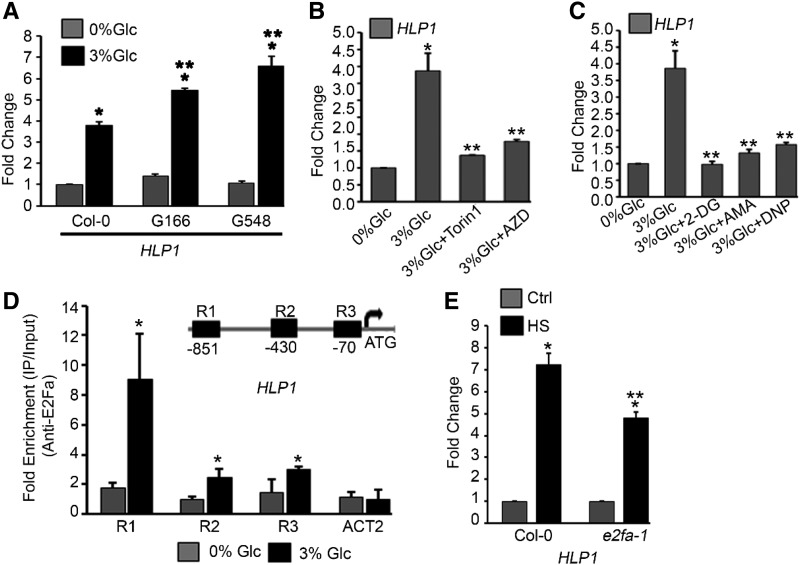
Photosynthesis-controlled TOR signaling is predominantly stimulated by Glc through glycolysis and mitochondrial bioenergetics relay (Xiong et al., 2013; Li et al., 2017). To investigate the dependence of HLP1 on Glc energy signaling, transcript levels of HLP1 were analyzed in seedlings overexpressing TOR. In TOR overexpression lines (G166 and G548), HLP1 mRNA levels were significantly increased in response to Glc compared to Col-0 (Fig. 3A). We also used the estradiol-inducible tor RNAi line (tor es-1) for HLP1 expression. Five-d–old Arabidopsis tor es-1 were transferred to MS medium with or without estradiol for 4 d followed by 24 h starvation at 0% Glc, then treated with Glc (3% Glc for 3 h). Estradiol-induced tor es-1 showed reduced HLP1 expression compared to DMSO-treated plants in response to Glc (Supplemental Fig. S3C). PCNA1 is a known TOR target and was used as a positive control (Supplemental Fig. S3C). In addition, transcript profiling of HLP1 under Torin 1 and AZD-8055 treatment exhibited significantly reduced expression levels (Fig. 3B). Torin 1 and AZD-8055 are ATP-competitive TOR kinase inhibitors, which specifically inhibit TOR phosphorylation of downstream genes (Montané and Menand, 2013; Xiong et al., 2013). These findings indicate that abolition of TOR phosphorylation by biochemical inhibitors results in inhibition of HLP1 expression as similarly reported for E2Fa target genes (Xiong et al., 2013). Furthermore, to check the effect of starvation and energy surplus in HLP1 regulation, we performed a starvation and replenishment assay. Transcript levels of HLP1 decreased consistently at each starvation point, while a sequential increase in HLP1 expression was observed after sugar replenishment (Supplemental Fig. S3D). These results suggest the role of energy metabolism in regulation of HLP1. Moreover, to unravel the involvement of HLP1 in energy signaling, we used chemical inhibitors of glycolysis and mitochondrial bioenergetics relay. HLP1 transcripts were significantly reduced by all metabolic inhibitors used, such as 2-deoxy-Glc, antimycin A, and the mitochondrial uncoupler 2,4-dinitrophenol (Fig. 3C). Taken together, these findings suggest that activation of HLP1 requires specific Glc/Suc energy metabolism provided by mitochondrial electron transport and is regulated by direct energy sensing through a TOR-dependent signaling pathway.
Figure 3.
Glc regulates HLP1 transcription through TOR signaling. A, RT-qPCR expression of HLP1 in Col-0 and TOR overexpression lines (G166 and G548). Five-d–old Arabidopsis plants were first starved with 0% Glc for 24 h followed by Glc treatment (3% Glc for 3 h). Data shown are average of two biological replicates. Experiments were repeated two times with similar results. B, Effect of TOR kinase inhibitors on HLP1 expression. Five-d–old light-grown Arabidopsis seedlings were starved without Glc-containing MS media for 24 h under dark-grown conditions. After starvation, seedlings were treated with TOR inhibitors (Torin 1 and AZD-8055; 10 μM for 4 h in presence of 3% Glc). Data shown are average of two biological replicates. Experiments were repeated two times with similar results. C, Effect of glycolysis and mitochondrial inhibitors on HLP1 expression. Five-d–old light-grown Arabidopsis seedlings were starved in MS media without Glc for 24 h under dark-grown conditions. After starvation, seedlings were treated with glycolysis inhibitor (2-deoxy-Glc, 3%), mitochondrial electron transport inhibitor (antimycin A, 5 µM) and mitochondrial uncoupler (2,4-dinitrophenol 50 µM) for 4 h in presence of 3% Glc. Data shown are average of two biological replicates. Experiments were repeated two times with similar results. D, qPCR showing enrichment of HLP1 promoter fragments in all three regions after ChIP assays with or without Glc treatment (5 mM for 4 h). Protoplasts were harvested from 4-week–old Col-0 plants and subjected to Glc treatment. Fold enrichment of promoter fragments were calculated by comparing samples treated without or with E2Fa-specific antibody. Data shown are the average of four biological replicates. Experiments were repeated four times with similar results. ACT2 was taken as a control for the ChIP assay. E, RT-qPCR PCR showing expression of HLP1 in Col-0 and e2fa-1 mutant under HS. Arabidopsis Col-0 and e2fa-1 seedlings were grown in 1/2-strength MS medium containing 1% Suc for 5 d and then subjected to HS. HS was applied at 37°C for 3 h and the seedlings were harvested immediately. Data shown are the average of two biological replicates. Experiments were repeated two times with similar results. Ctrl, control. Error bars = sd (Student’s t test, P < 0.05; *control versus treatment; **wild type versus overexpression or mutant/3% Glc versus 3% Glc + inhibitors).
Because many genes involved in stress and energy signaling have E2Fa binding sites in their promoters, we scanned the HLP1 promoter for putative transcription factor binding sites (TFBSs). Intriguingly, an ∼1-kb HLP1 promoter contains putative E2Fa TFBSs (region 1–3). To confirm E2Fa binding on the HLP1 promoter, we performed in planta chromatin immunoprecipitation (ChIP) using Arabidopsis protoplasts. Protoplasts were harvested from 4-week–old Col-0 plants and subjected to 3-h Glc treatment (5 mM). ChIP was performed using anti-E2Fa serum to immunoprecipitate E2Fa-bound chromatin and ∼200-bp promoter fragments comprising E2Fa binding sites were analyzed using qPCR. Interestingly, Glc-treated samples exhibited significantly higher enrichment in all three regions checked (region 1–3; Fig. 3D). Furthermore, transcript levels of HLP1 were significantly reduced in e2fa-1 under control as well as HS conditions (Fig. 3E; Supplemental Fig. S3E). Moreover, HLP1 transcript levels were also significantly upregulated in Arabidopsis plants transiently overexpressing E2Fa-DPa under HS (Supplemental Fig. S3F). Altogether, these findings suggest that E2Fa binds to the HLP1 promoter and positively regulates its transcription. These results also suggest that HLP1 works downstream of E2Fa in the Glc/TOR signaling pathway.
Heat-Induced HLP1 Expression Is Dependent on TOR, and Heat-Triggered HSFA1A Binds to the HLP1 Promoter to Activate Expression
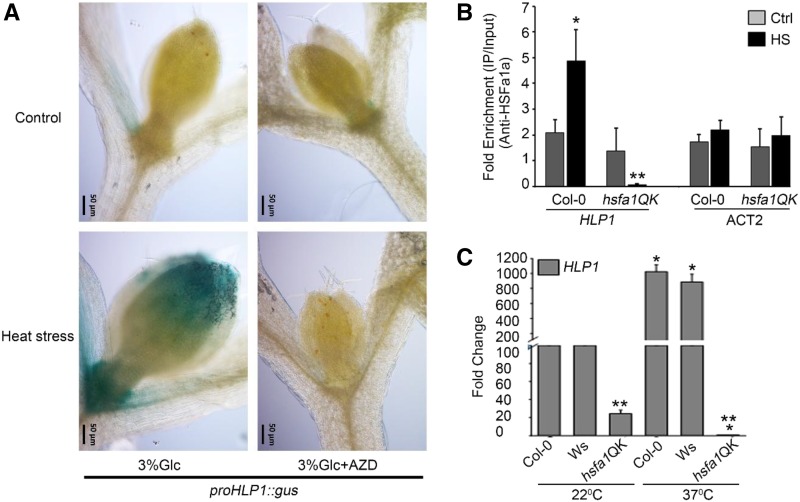
To investigate the heat responsiveness of HLP1 (Supplemental Fig. S3A), proHLP1::GUS seedlings were grown for 7 d in 1/2-strength MS media under 16 h-/8 h-long–day light condition and transferred to Glc-containing MS media (3% Glc) without or with AZD-8055 (1 μM) for 24 h followed by heat treatment. HS was applied as 1 h at 37°C, 2 h at 22°C, and 2.5 h at 45°C. Immediately after HS treatment, the proHLP1::GUS line exhibited strong GUS induction in proliferation-competent cells at the shoot apical meristem (SAM) and leaf tip. Heat strongly induced GUS expression in the leaf primordium at the shoot apex (Fig. 4A). Heat-induLikeced proHLP1::GUS expression in the shoot apex was abolished in seedlings treated with the specific TOR inhibitor AZD-8055 (Fig. 4A). These findings indicate that heat requires a TOR-dependent signal to activate proHLP1::GUS expression in mitotic-competent cells at the shoot apex. Earlier microarrays revealed that HLP1 expression is dependent on HSFA1 but not HSFA2 (Liu and Charng, 2013). Because genes induced by HS or targets of HSFs contain HSEs in their promoters, we scanned ∼1 kb of the HLP1 promoter for putative HSEs using MatInspector Professional Tool (Genomatrix) and the Transcription Factor Database (Wingender et al., 1996; Cartharius et al., 2005). Several putative TFBSs implicated in stress response were found in the HLP1 promoter. The HLP1 promoter has three HSEs along with CCAAT box (Supplemental Table S2). To confirm the presence of putative Cis-acting HSEs, we performed a ChIP experiment. Nine-d–old light-grown Arabidopsis Col-0 and hsfa1QK (hsfa1a/hsfa1b/hsfa1d/hsfa1e) seedlings were subjected to HS at 37°C for 3h. HSFA1A-bound chromatin was immunoprecipitated using an anti-HSFA1a polyclonal antibody and the 166-bp HLP1 promoter harboring HSEs that coimmunoprecipitated with HSFA1a was analyzed with qPCR. We observed a significant enrichment of HSFA1a on the HLP1 promoter due to HS, which was abrogated in hsfa1QK lacking HSFA1s (Fig. 4B). These in vivo interaction studies suggest that HSFA1a binding to the HLP1 promoter increases under HS. To verify the ChIP results, mRNA levels of HLP1 were analyzed in hsfa1QK mutants under HS. In the absence of HS, HLP1 transcripts were significantly reduced in hsfa1QK, whereas HLP1 expression was completely abolished in hsfa1QK under HS (Fig. 4C). These results thus suggest that HSFA1a positively regulates HLP1 transcription by binding to its promoter and thereby modulating thermotolerance response.
Figure 4.
Glc and heat induce HLP1 expression in the SAM in a TOR-dependent manner and heat-induced HsfA1a binds directly to HLP1 promoter. A, proHLP1::GUS expressing seedlings were grown for 7 d in 16-/8-long–day light condition and transferred to MS media containing Glc (3% Glc) without or with AZD-8055 (1 μM) for 24 h followed by heat treatment. HS was applied as 1 h at 37°C, 2 h at 22°C, and 2.5 h at 45°C. After heat treatment, seedlings were transferred immediately (0 h) to GUS buffer and images were captured using a fluorescence microscope (Zeiss). Scale bar = 50 µm. Images shown are representative of two biological replicates each containing at least five seedlings. B, ChIP-qPCR of 166-bp promoter fragment containing HSE after ChIP assay with or without HS (37°C for 3 h). ACT2 was taken as a control for the ChIP assay. Arabidopsis Col-0 and hsfa1QK seedlings were grown in 1/2-strength MS medium containing 1% Suc for 9 d and then subjected to HS as mentioned earlier. Fold enrichment of promoter fragments was calculated by comparing samples treated without or with HsfA1a-specific antibody. CT values without and with anti-HsfA1a antibodies were normalized by input control. Data shown are an average of four biological replicates. Experiments were repeated four times (without or with hsfa1QK/ACT2) with similar results. Ctrl, control. C, RT-qPCR showing expression profile of HLP1 in hsfa1QK (hsfa1a/hsfa1b/hsfa1d/hsfa1e) under control and HS conditions. Arabidopsis Col-0, Wassilewskija-2 (Ws-2), and hsfa1QK seedlings were grown in 1/2-strength MS medium containing 1% Suc for 9 d and then subjected to HS. HS was applied at 37°C for 3 h and the seedlings were harvested immediately. Data shown are an average of two biological replicates. Error bars = sd (Student’s t test, P < 0.05; *control versus treatment; **wild type versus mutant).
Plant Growth after HS Requires Glc/TOR-Dependent Proliferation in the SAM
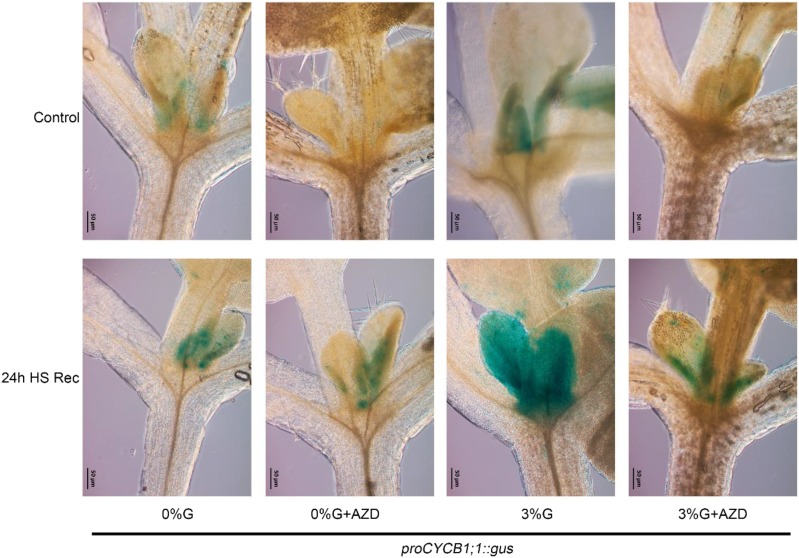
Because cell proliferation and division is required for plant growth once the stress signal has vanished, we examined cell proliferation activity in mitotic-competent cells in the shoot apex using the transcriptional mitotic reporter proCYCLINB (CYCB)1;1::GUS. The proCYCB1;1::GUS-expressing seedlings were grown for 8 d in long-day light conditions and transferred to MS media without (0% Glc) or with Glc (3% Glc) and without or with AZD-8055 (1 µM) for 24 h followed by heat treatment. HS was applied as 1 h at 37°C, 2 h at 22°C, 2.5 h at 45°C, 24 h at 22°C. After 24 h of HS recovery, seedlings were subjected to GUS treatment. As reported earlier, TOR is most abundant in the cell division zones of primary meristems (Menand et al., 2002). This is in good agreement with proCYCB1;1::GUS expression in the SAM after 24-h HS recovery with a much stronger induction in the presence of Glc (Fig. 5). Absence of Glc induced less proCYCB1;1::GUS expression after HS recovery, suggesting less proliferation activity in the SAM and therefore, less survival after HS (Fig. 5). This Glc- and heat-mediated proliferation of mitotically competent cells was abolished in proCYCB1;1::GUS seedlings treated with the TOR inhibitor AZD-8055, suggesting a crucial role of TOR after HS recovery (Fig. 5). Altogether, Glc-mediated proliferation in the SAM is dependent on TOR-mediated signaling for plant growth after HS.
Figure 5.
Cell proliferation in the SAM after HS requires Glc/TOR. proCYCB1;1::GUS-expressing seedlings were grown for 8 d in 16-/8-long–day light conditions and transferred to MS media without (0% Glc) or with Glc (3% Glc), and without or with AZD-8055 (1 μM) for 24 h followed by heat treatment. HS was applied as 1 h at 37°C, 2 h at 22°C, and 2.5 h at 45°C. After 24 h of HS recovery, plants were transferred to GUS buffer and images were captured using a fluorescence microscope (Zeiss). Scale bar = 50 µm. Images shown are representative of two biological replicates each containing at least five seedlings. Experiment was repeated two times with similar results. Rec, recovery after HS.
HLP1 Confers Resistance toward HS
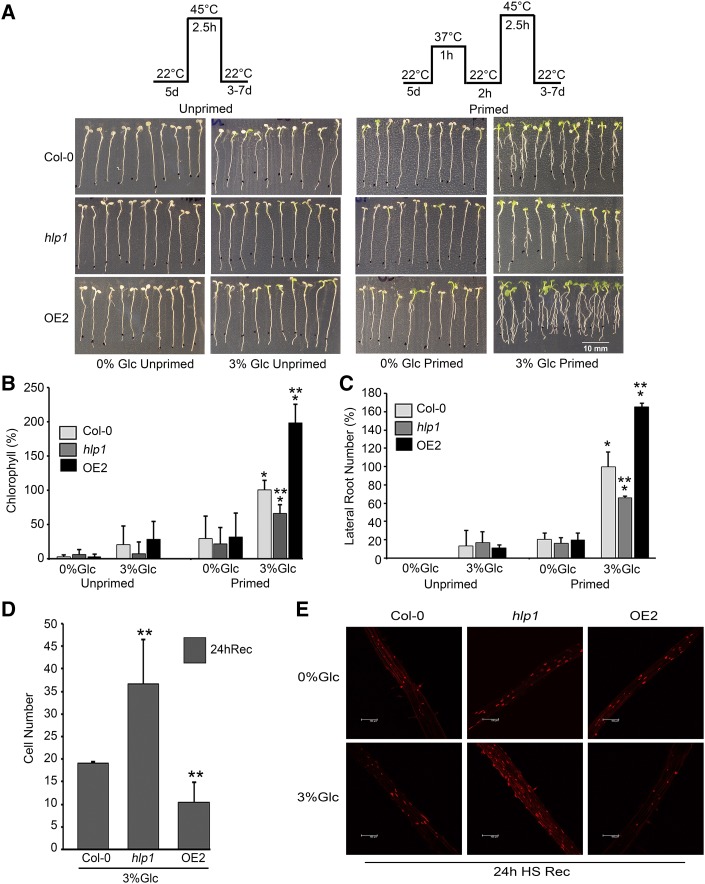
To evaluate the role of HLP1 in thermotolerance, we generated transgenic plants overexpressing HLP1 (OE1 and OE2) under the 35SCaMV promoter. Transcript profiling of OE1 and OE2 lines showed markedly higher expression of HLP1 in the absence of HS (Supplemental Fig. S4A). However, HLP1 overexpressors (OEs) did not show any increase in mRNA expression when exposed to HS (Supplemental Fig. S4B). This suggests that high levels of HLP1 mRNA in OEs under unstimulated conditions might be adequate to impart heat tolerance. To study the effect of HS, Glc-treated OE seedlings were subjected to HS. Both OE lines displayed increased thermotolerance under HS (Supplemental Fig. S4, C–E). In addition, a homozygous line of the hlp1 mutant having a transfer DNA insertion in the 5′UTR region was screened by PCR. RT-qPCR results showed the loss of mRNA accumulation in the hlp1 mutant (Supplemental Fig. S4F). To test short-term acquired thermotolerance (SAT), 5-d–old Arabidopsis Col-0, hlp1 and OE2 seedlings were treated without or with Glc for 24 h followed by HS. HS was applied as follows: Unprimed plants were subjected to HS directly at 45°C for 2.5 h followed by recovery. While in priming, Glc-treated plants were acclimatized at 37°C for 1 h followed by 2-h recovery at 22°C and then subjected to lethal temperature of 45°C for 2.5 h followed by recovery for 3–7 d. No observable changes were found in Col-0, hlp1, and OE2 without priming. Interestingly, Arabidopsis Col-0 and OE2 plants survived better than hlp1 mutants after priming HS (Fig. 6A). After priming, Arabidopsis Col-0 and OE2 seedlings showed faster recovery as observed by the emergence of new leaves, increased Chl content, and increased number of lateral roots (Fig. 6, A–C). In contrast, the hlp1 mutant showed reduced recovery growth, less Chl, and fewer lateral roots (Fig. 6, A–C). In addition, Arabidopsis seedlings overexpressing HLP1 did not show any observable thermotolerance when starved (Fig. 6A; Supplemental Fig. S5, A and C). However, OE2 seedlings supplemented with optimum Glc (3% Glc) displayed enhanced thermotolerance. On the contrary, hlp1 mutants showed a significant decrease in seedling survival under optimum (3% Glc) and higher Glc (5% Glc; Supplemental Fig. S5, A and C). Moreover, a higher degree of Chl retention was observed in OE2 seedlings whereas hlp1 displayed significantly lower Chl content than Col-0 at optimum or higher Glc concentrations (Fig. 6B; Supplemental Fig. S5B).
Figure 6.
Loss of HLP1 function causes susceptibility to HS. A, Seedling survival phenotype of Col-0, hlp1, and OE2 seedlings treated without or with Glc and with or without priming HS. B, Percent Chl content in Arabidopsis Col-0, hlp1, and OE2 seedlings in the presence of Glc and HS. Data shown are an average of three biological replicates having at least 30 seedlings. C, Percent lateral root number of Col-0, hlp1, and OE2 in presence of Glc and HS. Data shown are an average of three biological replicates having at least 20 seedlings. Experiments were repeated three times with similar results. D, Cell number showing cell nuclear fluorescence in Arabidopsis Col-0, hlp1, and OE2 roots stained with SYTOX orange. E, Micrograph showing root cell death after incubation with SYTOX orange. Scale bar = 100 μm. Five-d–old Col-0, hlp1, and OE2 were first treated without or with Glc-containing MS medium and then transferred to HS. HS was applied as 1 h at 37°C, 2 h at 22°C, and 2.5 h at 45°C. After HS, seedlings were recovered at 22°C for 24 h. Roots were stained with SYTOX Orange (1 μM) for 10 min and visualized by confocal microscope. Images shown are representatives of two biological replicates. Experiments were repeated two times with similar results. Rec, recovery after HS. Error bars = sd (Student’s t test, P < 0.05; *control versus treatment; **wild type versus mutant/overexpression).
Furthermore, to test the extent of root cell death in hlp1 and OE2, we used SYTOX orange nucleic acid stain (Thermo Fisher Scientific). This measures loss of cell membrane permeability, reflecting cell death, under HS. No significant difference was observed in Col-0 and hlp1 seedlings without Glc treatment. However, a higher degree of cell death was observed in hlp1 mutants under Glc treatment (Fig. 6, D and E). In contrast, OE2 seedlings supplemented with Glc showed much less fluorescence, suggesting significantly less root cell death (Fig. 6, D and E). In addition, HS plants were 3,3′-Diaminobenzidine (DAB)-stained to measure HS-induced reactive oxygen species production. DAB binds to H2O2 and is often used to quantitate cell death (Ren et al., 2002). The hlp1 mutant displayed higher cell death than Col-0 seedlings even in the 3% Glc (Supplemental Fig. S6, A and B). However, no significant difference was observed in Col-0 and OE2 plants. We speculate that recovery of heat-stressed plants require proliferation activity in mitotic-competent cells in the shoot apex and generation of new leaves from the SAM (Fig. 5), even after shedding off the cotyledons (Fig. 6A, right) and therefore, no difference in Col-0 and OE2 could be observed by DAB staining.
Moreover, to test whether HLP1 could sustain priming by mild stress (37°C for 1 h) for longer duration, we transferred primed Arabidopsis Col-0, hlp1, and OE2 plants for a longer recovery period (22°C for 2 d) and then treated with lethal dose (45°C for 2.5 h) of HS. Arabidopsis hlp1 plants showed reduced leaf greening and Chl content than Col-0 plants, whereas OE2 plants better survived the lethal dose of HS after longer recovery (Supplemental Fig. S6, C and D). Altogether, these results suggest that Glc-regulated HLP1 provides thermotolerance to Arabidopsis plants.
HLP1 Binds to the Promoters of HS Genes
Because HSFA1 acts upstream in the HS signaling pathway, we were interested to know whether Glc-regulated thermotolerance is transduced through HSFA1 or other signaling machinery. We analyzed the overlap between Glc- and HSFA1-regulated HS genes (Liu et al., 2011; Gupta et al., 2015). A significant overlap was observed between Glc and HSFA1 (Supplemental Fig. S7A; Supplemental Table S3).
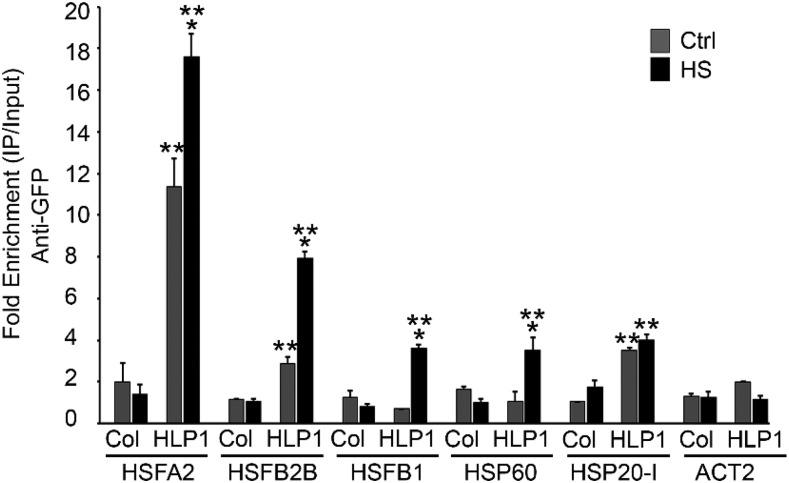
It has previously been reported in humans that Hikeshi functions as a nuclear carrier protein of HSP70 (Kose et al., 2012). Because HLP1 localizes into the nucleus (Koizumi et al., 2014) and is a direct target of E2Fa as well as HSFA1a, it may therefore work downstream of Glc and temperature signaling. We were interested to know how precisely HLP1 transduces Glc-mediated thermotolerance. In this regard, we checked whether HLP1 binds to the promoter of HSP genes. Four-week-old Col-0 plants were infiltrated with Agrobacterium harboring a 35SCAMV::HLP1-YFP (yellow fluorescent protein) construct together with the host RNAi suppression system (pBINR HC-pRO) and then subjected to HS (37°C for 3 h; Supplemental Fig. S8, B and C) followed by ChIP assay. Anti-GFP antibodies were used for immunoprecipitation of HLP1-YFP/chromatin complexes. Approximately 200–250 bp of HSF and HSP promoters harboring HSEs (nGAAnnTTCnnGAAn) that coimmunoprecipitated with HLP1-YFP were analyzed by qPCR. The enrichment of HLP1 at the promoters of HSF/HSP genes was significantly higher under HS in plants overexpressing HLP1-YFP than in uninfiltrated Col-0 plants (Fig. 7). These results suggest that HLP1 binds to the promoters of many heat-responsive genes and may influence their expression.
Figure 7.
HLP1 positively regulates Glc-induced HSP genes by binding to their promoters. ChIP assay of HSP genes after transient expression of 35SCaMV::HLP1-YFP. Four-week–old Col-0 plants were infiltrated with Agrobacterium strain GV3101 containing 35SCaMV::HLP1-YFP construct and kept for 48–72 h under light-grown conditions. ChIP-qPCR of immunoprecipitated promoter fragments containing Cis-acting HSE was performed. ACT2 serves as a control. Amount of immunoprecipitated promoter DNA was calculated by comparing samples treated without or with anti-GFP antibody. CT values without and with anti-GFP antibody were normalized by input control. Data shown are representative of one biological replicate. Experiments were repeated four times (without or with untransformed Col-0/ACT2) with similar results. Ctrl, control. Error bars = sd (Student’s t test, P < 0.05; *control versus treatment; **uninfiltrated versus infiltrated).
Loss-of-Functional HLP1 Affects H3K Acetylation at the Promoters of HS Genes To Activate Expression
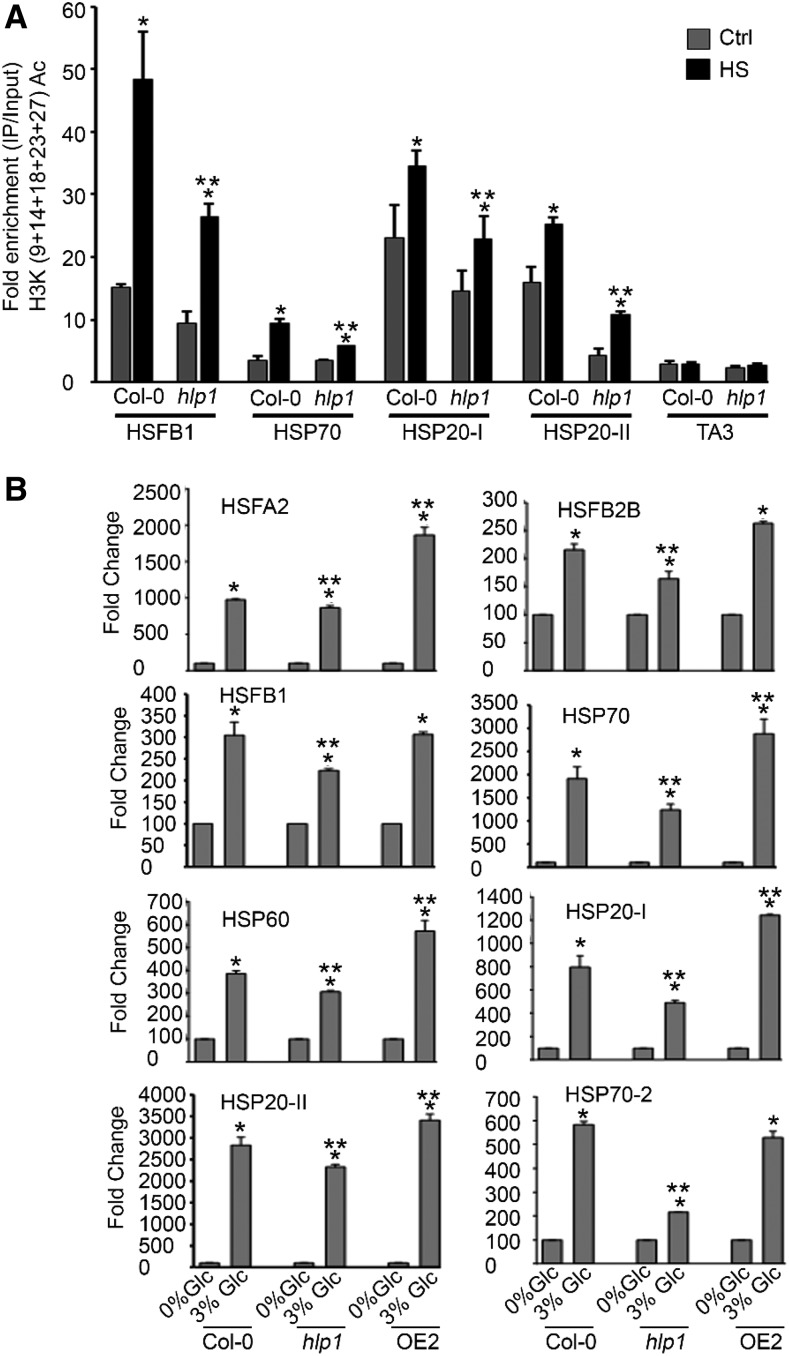
As HLP1 binds to the promoters of HSF/HSP genes, we wanted to know whether HLP1 modulates the epigenetic modifications at these promoters. Histone acetylation as well as H3K4me3 is required to activate gene expression. Earlier reports suggested the role of histone H3 acetylation in gene transcriptional activation, but not in maintenance of memory response (Lämke et al., 2016). We therefore compared the level of H3K (9+14+18+23+27) acetylation at the HSP promoter with or without HS using ChIP-qPCR. We observed a consistent increase in chromatin-activating H3K acetylation marks at the promoters of the tested HS loci in Arabidopsis Col-0 plants under HS. In contrast, hlp1 mutants showed reduced accumulation in chromatin-activating H3K acetylation at these loci (Fig. 8A). We next analyzed whether increased accumulation of H3K acetylation at HS loci in Col-0 and hlp1 correlates with high induction of gene expression. To this end, we compared the RT-qPCR expression of these HS loci in Col-0, hlp1, and OE2 plants. The RT-qPCR expression of these HS genes in OE2 was remarkably higher than in Col-0 plants, whereas hlp1 exhibited significant downregulation of HS-responsive genes (Fig. 8B). These findings showed that HSP levels in OE2 were significantly higher and sufficient to confer thermotolerance. (Figs. 6 and 8B). In contrast, hlp1 mutants showed reduced expression of HSPs, thereby causing hypersensitivity toward HS (Figs. 6 and 8B). Taken together, these results suggest that HLP1 binds to the promoters of heat shock genes and modulates histone H3K acetylation at these promoters to activate transcription.
Figure 8.
HLP1 is required for epigenetic modifications at the promoters of HS genes and positively regulates their transcription. A, ChIP assays of HSF and HSP genes. Seven-d–old MS-grown Arabidopsis Col-0 and hlp1 seedlings were treated with HS at 37°C for 3 h. ChIP-qPCR of immunoprecipitated promoter fragments containing Cis-acting HSEs was performed. Amount of immunoprecipitated promoter DNA was calculated by comparing samples treated without or with anti-Histone H3K (9+14+18+23+27) acetyl antibody. CT values without and with anti-Histone H3K (9+14+18+23+27) acetyl antibody were normalized by input control. Copia-like retrotransposon (TA3) with highly heterochromatinized DNA has been used as a control in the ChIP assay. Data shown are representative of two biological replicates. Experiments were repeated two times with similar results. Ctrl, control. B, Comparison of RT-qPCR showing expression profile of HS genes in Col-0, hlp1 mutant, and 35SCaMV::HLP1 (OE2) transgenic seedlings in the presence of Glc. Five-d–old Col-0, hlp1, and OE2 seedlings were starved for 24 h and then transferred to Glc-free (0% Glc) and Glc-containing (3% Glc) liquid MS medium for 3 h. Data shown are representative of two biological replicates. Experiments were repeated two times with similar results. Error bars = sd (Student’s t test, P < 0.05; *control versus treatment; **wild type versus mutant/overexpression).
Glc Modulates Thermomemory in Arabidopsis
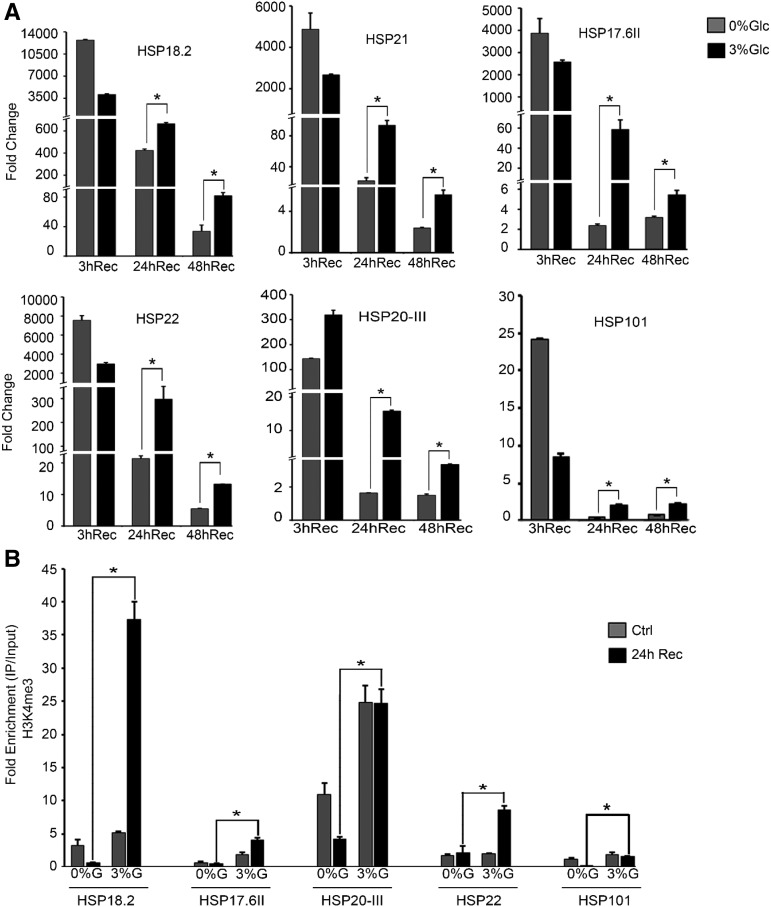
To check whether absence of Glc could induce HS genes, we analyzed the transcript levels of HS genes without Glc (0% Glc). Five-d–old Arabidopsis Col-0 seedlings were starved for 24 h followed by HS (37°C, 3 h) and RT-qPCR was performed. In the absence of Glc, all of these HSP genes were highly induced (Supplemental Fig. S9A). However, Arabidopsis plants supplemented with no Glc (0% Glc) could not exhibit tolerance to HS (Figs. 1, A–D, and 6A). To determine why high levels of HSP induction under 0% Glc are not sufficient to provide thermotolerance, we analyzed genome-wide transcript abundance using an Affymetrix microarray to compare Col-0 plants treated without or with Glc and HS. There was no major change in expression of HSP genes when seedlings treated with or without Glc recovered immediately after HS. The maintenance of acquired thermotolerance over time in the absence of intervening stresses is mediated by thermomemory genes (Charng et al., 2007). Interestingly, expression levels of thermomemory-related genes (HSP17.6II, HSP18.2, HSP22, and HSP20-III) were significantly increased or sustained in the presence of Glc (3% Glc) after a short recovery period (Supplemental Fig. S9B). Microarray reports have shown a number of thermomemory-associated genes, based on their sustained expression, including but not limited to small HSPs such as HSP17.6A, HSP18.2, HSP21, and HSP22 (Stief et al., 2014; Sedaghatmehr et al., 2016). These genes maintained their elevated expression after HS over a period of several days.
To validate the microarray data, we performed expression profiling of these thermomemory-associated genes using RT-qPCR. Seven d–old Col-0 seedlings were treated without (0% Glc) or with Glc (3% Glc) for 24 h followed by HS and recovered for various time points (3 h, 24 h, and 48 h). HS was applied as follows: 1 h at 37°C, 90 min at 22°C, and 45 min at 45°C, and then recovered at 22°C for 3 h, 24 h, and 48 h. Thermomemory-related genes were expressed at higher levels in seedlings treated without (0% Glc) or with Glc (3% Glc), when recovered for 3 h. Surprisingly, seedlings treated with Glc showed a more sustained expression of thermomemory-associated genes even under extended recovery periods (24 h and 48 h) than seedlings treated without Glc (Fig. 9A). We also checked the expression of the non-memory gene HSP101 under Glc treatment. The expression levels of HSP101 were comparatively less at 24 h and 48 h recovery than those of the thermomemory genes (Fig. 9A). We also examined the thermomemory HSP gene expression in Arabidopsis Col-0 seedlings treated without (0% Glc) or with low Glc (1% Glc). Arabidopsis plants supplemented with 1% Glc showed more sustained expression of thermomemory genes after 24 h and 48 h of recovery than seedlings treated without Glc (0% Glc; Supplemental Fig. S10). To confirm whether increased or sustained induction of thermomemory genes is due to Glc-mediated thermomemory response and is not due to high basal transcript levels, we compared the basal transcript levels of thermomemory genes in the absence and presence of Glc after 24 h and 48 h. In the absence of HS, Glc alone could not induce the basal expression of thermomemory-associated genes (Supplemental Fig. S9C).
Figure 9.
Glc mediates thermomemory response. A, RT-qPCR expression of thermomemory-related genes in Arabidopsis Col-0 seedlings treated without (0% Glc) and with Glc (3% Glc) followed by HS. HS was applied at 1 h at 37°C, 90 min at 22°C, and 45 min at 45°C. After HS, seedlings were recovered for various time points (3 h, 24 h, and 48 h) at 22°C. Data shown are representative of one biological replicate. Experiments were repeated two times with similar results. B, ChIP-qPCR of thermomemory-related genes in Col-0 seedlings treated without or with Glc followed by HS. HS was applied as 1 h at 37°C, 90 min at 22°C, and 45 min at 45°C. After HS, seedlings were recovered for 24 h at 22°C. Amount of immunoprecipitated promoter DNA was calculated by comparing samples treated without or with anti-Histone H3K4me3 antibody. Data shown are representative of one biological replicate. Experiments were repeated two times with similar results. CT values without and with anti-Histone H3K4me3 antibody were normalized by input control. Rec, recovery after HS; Ctrl, control. Error bars = sd (Student’s t test, P < 0.05; *control versus treatment).
Histone H3K4 trimethylation is known to activate transcription and efficient RNA polymerase II elongation (Ding et al., 2012). H3K4me3 marks are specifically involved in chromatin-based transcriptional memory after drought and HS (Ding et al., 2012; Lämke et al., 2016). In yeast and in mammals, H3K4me3 marks are associated with transcriptional memory of recent exposure to stress (Ng et al., 2003; Guenther et al., 2007). Lämke et al. (2016) showed that histone H3 di- and trimethylation increased toward later phases of HS recovery, when transcription and histone H3 acetylation went down. We hypothesized that increased or sustained expression of memory-related genes might be associated with changes in the epigenetic landscape of their promoters. It is already reported that H3K4me3 chromatin marks are associated with plant memory of recurring HS (Lämke et al., 2016). To this end, we analyzed H3K4me3 marks at HSP17.6II, HSP18.2, HSP21, HSP22, and HSP20-III loci. Seven-d–old Arabidopsis Col-0 plants were acclimatized without (0% Glc) or with Glc (3% Glc) for 24 h followed by HS and recovered for 24 h. HS was applied as follows: 1 h at 37°C, 90 min at 22°C, 45 min at 45°C, and 24 h at 22°C. After a 24-h recovery period, H3K4me3 levels were significantly increased at thermomemory-related loci when treated with 3% Glc (Fig. 9B). In contrast, the non-memory gene HSP101 showed a lesser, although significant, increase in H3K4me3 marks after 24-h recovery under Glc. These results suggest that high induction of non-memory loci at 0% Glc is not sufficient to provide thermotolerance (Fig. 1, A and C; Supplemental Fig. S9A). We, therefore, surmise that Glc facilitates plants to memorize initial HS in the form of H3K4me3 accumulation at thermomemory loci to sustain their expression and provide thermotolerance.
HLP1 Is Required for Sustained Accumulation of H3K4me3 on Thermomemory-Related Loci
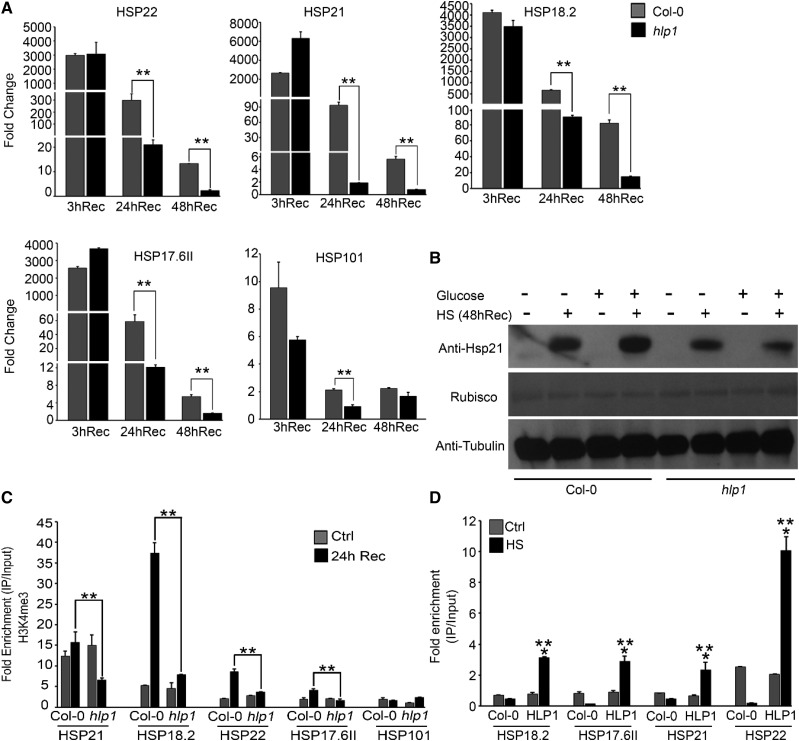
To check whether Glc-primed sustained accumulation of H3K4me3 at thermomemory-related loci is dependent on HLP1, we analyzed the expression levels of thermomemory genes in Arabidopsis Col-0 and hlp1 plants after extended recovery periods (3 h, 24 h, and 48 h). Seven-d-old Col-0 and hlp1 seedlings were transferred to MS media containing Glc (3% Glc) for 24 h followed by HS and recovered for various time points (3 h, 24 h, and 48 h). HS was applied as follows: 1 h at 37°C, 90 min at 22°C, and 45 min at 45°C, and then recovered at 22°C for 3 h, 24 h, and 48 h. No changes were observed in the expression levels of thermomemory genes in Col-0 and hlp1 plants after 3 h of recovery. Interestingly, Arabidopsis Col-0 plants showed sustained expression of thermomemory loci after longer recovery periods of 24 h and 48 h (Fig. 10A). On the other hand, hlp1 plants failed to display sustained expression of thermomemory loci at these time points (Fig. 10A). We also compared the expression levels of the non-memory gene HSP101 in Col-0 and hlp1 after 24-h and 48-h recovery. Non-memory HSP101 did not show sustained expression at these extended recovery time points in Col-0 and the hlp1 mutant. We also compared thermomemory HSP gene expression in Col-0 and hlp1 seedlings treated with low Glc (1% Glc). Arabidopsis Col-0 showed sustained expression of thermomemory genes after extended recovery, whereas hlp1 mutants failed to do so (Supplemental Fig. S11). Further, we also compared the transcript and protein levels of thermomemory-associated HSP21 without or with Glc treatment in Col-0 and hlp1 plants after HS recovery. Thermomemory-associated HSP21 transcripts, as well as protein, accumulated more in the presence of Glc in Col-0 than hlp1 plants (Fig. 10B; Supplemental Fig. S12). However, there was no significant difference in HSP21 transcripts in Col-0 and hlp1 seedlings treated without Glc (0% Glc) after 24 h and 48 h of HS recovery (Supplemental Fig. S12). Moreover, hlp1 mutants did not show sustained accumulation of HSP21 protein at these longer recovery time points as compared to Col-0 plants (Fig. 10B). Importantly, Arabidopsis plants treated with Glc under non-HS conditions showed null levels of HSP21, further substantiating that Glc alone could not induce basal levels of HSP21 protein (Fig. 10B).
Figure 10.
Glc provides thermomemory through HLP1. A, RT-qPCR expression of thermomemory-related genes in Arabidopsis Col-0 and hlp1 seedlings treated with 3% Glc for 24 h followed by HS. HS was applied as 1 h at 37°C, 90 min at 22°C, and 45 min at 45°C. After HS, seedlings were recovered for various time points (3 h, 24 h, and 48 h) at 22°C. Data shown are representative of one biological replicate. Experiments were repeated two times with similar results. B, Immuno-blot detection of HSP21 in Col-0 and hlp1 seedlings treated without or with Glc and HS. After HS, seedlings were recovered for 48 h at 22°C. HSP21 protein was detected using anti-HSP21–specific antibody. Ponceau S-stained Rubisco and Anti-TUBULIN-detected TUBULIN were used as loading controls. C, ChIP-qPCR of thermomemory-related genes in Col-0 and hlp1 seedlings treated without or with HS. After HS, seedlings were recovered for 24 h at 22°C. Amount of immunoprecipitated promoter DNA was calculated by comparing samples treated without or with anti-Histone H3K4me3 antibody. CT values without and with anti-Histone H3K4me3 antibody were normalized by input control. Data shown are representative of one biological replicate. Experiments were repeated two times with similar results. Rec, recovery after HS; Ctrl, control. D, ChIP-qPCR showing binding of HLP1-YFP at thermomemory gene promoters. Approximately 4-week–old Arabidopsis Col-0 plants were infiltrated with 35SCaMV::HLP1-YFP followed by HS at 37°C for 3 h and harvested immediately. Amount of immunoprecipitated promoter DNA was calculated by comparing samples treated without or with anti-GFP antibody. CT values without and with anti-GFP antibody were normalized by input control. Data shown are representative of one biological replicate. Experiments were repeated two times with similar results. Error bars = sd (Student’s t test, P < 0.05; *control versus treatment; **wild type versus mutant/overexpression).
We next examined whether sustained induction of thermomemory genes in Col-0 plants is mediated through accumulation of H3K4me3 marks. To investigate this, we compared the H3K4me3 accumulation in Col-0 and hlp1 plants after 24-h recovery. Arabidopsis Col-0 plants exhibited increased or sustained accumulation of H3K4me3 marks on thermomemory-related loci whereas hlp1 mutants failed to do so (Fig. 10C). Furthermore, Col-0 and hlp1 plants did not show sustained accumulation of H3K4me3 at the non-memory locus HSP101 after 24-h recovery (Fig. 10C). Moreover, we checked whether HLP1 binds to the promoters of thermomemory-associated genes. To this end, we performed in vivo ChIP assays using Arabidopsis Col-0 plants transiently overexpressing HLP1-YFP. Four-week–old Arabidopsis Col-0 plants were infiltrated with HLP1-YFP followed by HS at 37°C for 3 h and harvested immediately. Approximately 200 bp of upstream promoter harboring HSEs were selected for ChIP-qPCR. HLP1-YFP was enriched at the promoters of all four thermomemory-associated genes under HS, compared to no heat (Control) and Col-0 plants without HLP-YFP (Fig. 10D). Next, to test whether complementation of hlp1 with proHLP1::HLP1 restores the thermomemory response, we transformed Arabidopsis hlp1 plants with proHLP1::HLP1 and checked the expression of thermomemory genes after 24 h and 48 h of recovery. Arabidopsis hlp1 plants complemented with proHLP1::HLP1 (proHLP1::HLP1-4 and proHLP1::HLP1-7) showed more sustained expression of thermomemory-related genes after extended recovery than hlp1 plants (Supplemental Fig. S8A). Taken together, these results suggest that HLP1 is required for plants to memorize previous exposure to HS by maintaining chromatin H3K4me3 marks on thermomemory loci for poised induction of thermomemory genes.
DISCUSSION
Our comprehensive molecular, genetic, and physiological study of Arabidopsis revealed an intimate link between Glc- and HS-mediated signaling. Glc transcriptionally regulates a large number of genes involved in a variety of processes such as cell cycle, growth, and stress response. In the literature, there are various reports of crosstalk between Glc-mediated signaling and other signaling pathways such as nutrient-, light-, and hormone-response signaling. Although there are several reports about Glc and phytohormone crosstalk (Szekeres et al., 1996; Gibson, 2004; Mishra et al., 2009; Gupta et al., 2015; Schepetilnikov et al., 2017; Schepetilnikov and Ryabova, 2017; Song et al., 2017; Xiong et al., 2017), only a few reports of a possible interaction between Glc-mediated signaling and stress responses exist (Deprost et al., 2007; Bakshi et al., 2017; De Vleesschauwer et al., 2017; Dong et al., 2017; Wang et al., 2018). A previous microarray experiment identified a significant overlap of Glc-upregulated genes with HS-regulated genes (Gupta et al., 2015). Interestingly, among HS genes, ∼37% of Glc-upregulated genes encoded HSPs. This indicates that Glc regulates HSPs levels in plants and Glc signaling is required for plant survival under HSR. There are a few reports that document the role played by Glc-mediated signaling in stress tolerance. For example, Glc-activated Arabidopsis TOR kinase displays resistance to various stresses (Mahfouz et al., 2006; Deprost et al., 2007; Bakshi et al., 2017; Dong et al., 2017; Wang et al., 2017) and Glc-repressed SnRK1 exhibits plant survival under extreme starvation, darkness, and hypoxia (Baena-González et al., 2007; Baena-González and Sheen, 2008). However, in response to cold and osmotic stress, activity of TOR and its target S6K1 was inhibited (Mahfouz et al., 2006; Wang et al., 2017). Recently, Wang et al. (2018) showed that TOR signaling represses abscisic acid (ABA) signaling, preventing stress activation through phosphorylation of the PYRABACTIN-LIKE ABA receptor in unstressed plants, whereas ABA represses TOR signaling to prevent growth under stress. Nonetheless, no systematic studies have been done that explored the role of Glc-mediated signaling machinery in providing thermotolerance. In this study, we have found that Glc-regulated Arabidopsis TOR mediates enhanced tolerance to HS. Arabidopsis plants overexpressing TOR (G166 and G548) exhibited better thermotolerance, whereas tor 35-7 RNAi plants showed reduced thermotolerance response. The contrasting characteristics (as shown by Wang et al., 2018) in stress responses might be due to differences in stress and recovery conditions, as both TOR G166 and G548 lines showed better stress mitigation when plants recovered after HS, suggesting activation of growth signaling through high Glc/TOR levels at the time of recovery. The fine-tuning of growth and stress signaling under various stress and recovery conditions needs to be explored further. We also compared the overlap between Glc- and TOR-regulated HS genes (Xiong et al., 2013; Gupta et al., 2015). A large overlap of HS genes was observed between Glc and TOR (Supplemental Fig. S7B; Supplemental Table S4), suggesting a common pathway for HS tolerance. We also found that e2fa-1 mutants displayed reduced thermotolerance with a decrease in the expression of HS genes. These several lines of evidence suggest a role for the TOR-E2Fa module in mediating thermotolerance response.
It has already been proposed that Arabidopsis HLP1 interacts with HSP70 to import it into the nucleus and thus provide thermotolerance (Koizumi et al., 2014). However, no efficient studies have been done to date that suggest a role for Glc-mediated HLP1 activation in thermotolerance. In this study, we have found that Glc-induced HLP1 activation is primarily regulated by the energy sensor TOR kinase. These results were further substantiated by proHLP1::GUS studies under Glc and heat treatment, which showed Glc-heat-induced HLP1 expression in cell division zones of primary meristem. We have found that Glc synergistically works with heat in activating proHLP1::GUS expression and this Glc-heat regulation of HLP1 is dependent on TOR. Previous studies showed the role of TOR in photosynthesis-derived Glc/energy signaling in controlling proliferation of progenitor stem cells in root meristem activation (Xiong et al., 2013). Arabidopsis TOR kinase phosphorylates E2Fa/E2Fb transcription factors in controlling s-phase gene expression in mitotic-competent cells in the shoot and root apical meristem (Li et al., 2017). ProHLP1::GUS expression in the cell division zones of the shoot apex overlap with TOR and E2Fa expression and is abolished by AZD-8055, substantiating the role of Glc-mediated TOR-E2Fa signaling in HLP1-mediated thermotolerance. Furthermore, chemical inhibitors of Glc metabolism partially or completely eliminate HLP1 transcripts, suggesting a plausible role of mitochondrial bioenergetics in Glc-mediated HLP1 regulation.
Characterization of promoter elements containing the E2Fa target motif TTTCCCGCC have been extensively studied (Vandepoele et al., 2005; Naouar et al., 2009; Xiong et al., 2013). Ramirez-Parra et al. (2003) have also shown that genes involved in cell cycle, defense responses, and signaling comprise E2Fa target motifs. However, in addition to TTTCCCGCC, other closely related sequences are also recognized by plant E2Fa. We observed enrichment of TTTCCCGCC and similar E2Fa Cis-acting elements in the promoter of HLP1, which was confirmed by ChIP assay. We found that E2Fa binds directly to the HLP1 promoter and positively regulates its expression. Interestingly, enrichment of E2Fa in the HLP1 promoter was higher under Glc treatment. This further supports the involvement of Glc-mediated HLP1 activation. Mutation in E2Fa significantly inhibited transcriptional activation of HLP1 and HSPs genes that are required to establish thermotolerance, whereas overexpressing E2Fa increased their activation. Moreover, Arabidopsis seedlings treated with Torin 1 and AZD-8055, which prevent TOR-mediated phosphorylation of E2Fa, exhibited perturbation in HLP1 and HSPs transcript synthesis. These evidences suggest a direct involvement of TOR-E2Fa and HLP1 in mediating thermotolerance in plants.
Growth is a highly expensive process in terms of resource consumption and is tightly regulated by internal and external stimuli. When plants sense a stress signal in their environment, they stop or decelerate their growth and once the stress signal has vanished, they restore their growth machinery for organ development. Cell proliferation and division are required for proper growth of the organism and are directed by signals from progenitor stem cells in the root and shoot apex. Notably, expression of the proCYCB1;1::GUS transcriptional reporter in the mitotically active cells was greatly enhanced in response to Glc after 24 h of HS recovery, both at the tip and the base of young leaves. Because cells at the leaf base under normal developmental conditions have exited the proliferation phase, the presence of proCYCB1;1::GUS at the leaf base suggests that during HS recovery, cells might re-enter the cell proliferation phase (Andriankaja et al., 2012). Arabidopsis plants treated without Glc (0% Glc) exhibited less proCYCB1;1::GUS expression in the proliferating zone at the shoot apex, suggesting that Glc is required for plants to maintain their growth after HS. This Glc-regulated proliferation of mitotic cells is dependent on TOR, further demonstrating the role of Arabidopsis TOR kinase in post-HS plant growth.
The HSFA1 and HSFA2 heat shock transcription factors are the major regulators of HSR in plants, involved in initiating and extending the thermotolerance response (Charng et al., 2007; Liu et al., 2011; Liu and Charng, 2013). A previous microarray report showed dependence of HLP1 gene expression on HSFA1s (Liu and Charng, 2013). However, no organized study has so far been done that suggests a possible link between HSFA1s and HLP1. Our study establishes a novel link between HSFA1s and HLP1-mediated thermotolerance. We found that HSFA1a binds to HSEs present in the HLP1 promoter and positively regulates HLP1 expression. This was further confirmed by expression of HLP1 in hsfa1QK mutants. These lines of evidence suggest that HLP1 is a direct downstream target of HSFA1s in regulating thermotolerance.
Several groups have identified the genetic components involved in the regulation of duration of acquired thermotolerance. Different components of HS signaling are important for tolerance against HS regimes of different magnitudes. For example, HSFA3 and HSFA7A are involved in maintaining SAT, whereas HSFA2 and HSA32 are implicated in long-term–acquired thermotolerance (LAT; Yeh et al., 2012). Some of them are also involved in maintaining basal thermotolerance. However, some HS regulators, such as HSFA1, HSFA2, HSFB1/B2B, and HSP101, are involved in regulating basal thermotolerance as well as SAT and LAT (Charng et al., 2007; Ogawa et al., 2007; Ikeda et al., 2011; Liu et al., 2011; Hu et al., 2012). Arabidopsis HLP1 also exhibited regulation of SAT and LAT. This HLP1-mediated SAT and LAT response is dependent on the Glc/energy status of the plant. Arabidopsis seedlings overexpressing HLP1 displayed enhanced thermotolerance in response to Glc. In contrast, hlp1 mutants exhibited hypersensitivity to HS. Growth of the plant is essential after HS and mediated by proliferation in the proliferation zone. Proliferating cells in the shoot and root apex coordinate with the internal and external cues to maintain growth. Arabidopsis hlp1 mutant plants showed reduced growth from the primary meristem as evidenced by compromised growth of the apical meristem and emergence of new leaves (Fig. 6A, right). In contrast, Col-0 and OE2 plants displayed enhanced growth and accelerated emergence of new leaves from the shoot apex (Fig. 6A, right). The reduced growth recovery phenotype of hlp1 mutants from the meristematic zone is in good agreement with the role of TOR-E2Fa-HLP1 signaling in providing thermotolerance, as these regulators are most abundant in meristematic cells (Menand et al., 2002).
Guertin and Lis (2010) have shown that HSEs are the sites of active chromatin, associated with histone acetylation and H3K4 trimethylation (Guertin and Lis, 2010). Mutation in gcn5 decreases H3K9 and H3K14 acetylation in the core promoters of HSFA3 and UVH6 under HS, which results in reduced thermotolerance of gcn5 (Hu et al., 2015). Arabidopsis ASF1A/B transcriptionally activates some of the HSF and HSP genes through accumulation of H3K56 acetylation under HS (Weng et al., 2014). In our study, we have found that HLP1 binds HSF/HSP gene promoters containing HSEs in response to HS. Binding of HLP1 to HSP gene promoters increases H3K (9+14+18+23+27) acetylation in plants. Transgenic lines overexpressing HLP1 exhibit increased expression of HS genes under Glc stimulus, whereas loss of functional HLP1 results in reduced HSP induction. The requisite role of HLP1 in the induction of HSFA2 and other secondary regulators of HSR such as HSPs suggests that these regulators act downstream of HLP1 in HSR.
Priming is a phenomenon in which plants are pre-exposed to mild stress, which provides heightened response to subsequent stresses. Unlike naïve plants, primed plants are alert and show quick response to future stresses. In cases where thermomemory induces transcriptional changes, a possible explanation is alteration in chromatin modifications (Lämke et al., 2016; Liu et al., 2018). Among HSFs, HSFA2 is required for maintenance of thermotolerance. Similarly, the level of HSA32 is maintained over a period of at least 3 d and is required for HS memory (Wu et al., 2013). A number of HS-memory–related genes have been identified by microarray analysis. These genes have sustained expression after primary HS, which lasts several days (Stief et al., 2014). The HS memory is maintained for several days, even in the absence of recurring temperature stress (Charng et al., 2006, 2007; Yeh et al., 2012). Many small HSPs (HSP17.6II, HSP22, HSP18.2) are HS-memory–related genes. Unlike HS-memory–related genes, the expression pattern of HS non-memory genes such as HSP101 peaked immediately after HS and rapidly declined thereafter. Elevation of H3K4me3 levels is required for rapid activation of genes induced by environmental stimuli and acts to prevent them from being silenced (Ng et al., 2003). Brzezinka et al. (2016) showed that nucleosome remodelers of the CHROMATIN REMODELING BY IMITATION SWITCH family together with BRAHMA remodeling complex interact with FORGETTER1 for proper nucleosome assembly at memory-related genes. Chromatin protein BRUSHY1 is also required for sustained induction of thermomemory-associated genes for longer durations (Brzezinka et al., 2018). HSP21 levels are regulated by FtsH6 for thermomemory in plants (Sedaghatmehr et al., 2016). Here, we have shown that Glc induces the accumulation of H3K4me3 on thermomemory-associated genes. H3K4me3 changes the chromatin landscape at thermomemory-related loci to activate sustained expression of thermomemory genes. We also showed that binding of HLP1 to the promoters of thermomemory-associated loci is required for the accumulation of Glc-induced H3K4me3 marks at those loci. These results suggest an epigenetic mode of HLP1-mediated thermomemory response. Taken together, HLP1 acts as a hub of TOR-E2Fa- and HSFA1s-mediated thermotolerance. Our studies demonstrate that HLP1 coordinates heat and sugar signaling by modulating expression of both the memory- and non-memory–HSP genes to provide thermotolerance. In summary, molecular evidences such as histone H3K (9+14+18+23+27) acetylation, expression of non-memory and memory genes, histone H3K4me3 marks, and protein blot of HSP21 are in agreement with the phenotypes exhibited by hlp1 mutants such as compromised growth from the SAM, emergence of new leaves, less Chl, and fewer lateral roots, and strongly support that HLP1 provides thermotolerance response to Arabidopsis plants. Furthermore, expression of proHLP1::GUS and proCYCB1;1::GUS in the SAM further support the role of Glc-regulated HLP1 and -TOR-E2Fa in thermotolerance response. It would, however, be of interest to further examine the role of HLP1 under various abiotic stresses and corelate its functions in diverse environmental stimuli. The work done in this study so far would be useful for forthcoming studies, aimed at gaining functional insights into HS responses in plants.
MATERIALS AND METHODS
Plant Materials and Growth Condition
The Arabidopsis (Arabidopsis thaliana) mutant lines hlp1 (AT1G66080, SALK-135622), e2fa line (AT2G36010, WiscDsLox434F1), the estradiol-inducible TOR RNAi lines (CS69829; es-1), TOR overexpression lines (AT1G50030, GK-548G07-020632), and proCYCB1;1::GUS line were obtained from The Arabidopsis Biological Resource Center at Ohio State University. All of these lines were in the Col-0 background. The TOR (AT1G50030, G166) line was obtained from the Nottingham Arabidopsis Stock Center. The following lines were obtained from the published sources: tor 35-7 RNAi (Col-0 background; Deprost et al., 2007) and HSFA1 quadruple mutant (QK-hsfa1a/hsfa1b/hsfa1d/hsfa1e; Col-0 and Wassilewskija-2 background; Liu and Charng, 2013). Seeds of wild type and mutants were surface-sterilized and imbibed at 4°C for 48 h. Seeds were sown on square petri plates (120 × 120 mm) containing 0.5× MS medium with 1% Suc (29 mM) and 0.8% agar (24 mM), under sterile conditions. Seed germination was carried out in a climate-controlled growth room under long-day conditions (16-h light and 8-h dark, 60 μm m−2 s−1 light intensity) at 22°C ± 2°C temperature.
Transgenic Plants
The full-length coding sequences of HLP1 were amplified from 5-d–old Arabidopsis wild-type seedlings (Col-0) and cloned into the Gateway-based vector pEARLYGATE 201 containing 35SCaMV cassettes. The 35SCAMV::HLP1 construct was then transformed into Agrobacterium strain GV3101. Agrobacterium containing 35SCAMV::HLP1 were transformed into Arabidopsis plants (Col-0) by floral dip. For selection of transgenic plants, 2-week–old plants were sprayed with 120 mg/L of the herbicide BASTA (BASF). Seedlings exhibiting resistance to BASTA were grown at 22°C and seeds were harvested to check homozygosity. Similarly, the first 942-bp upstream of the transcription start site of HLP1 were amplified from Col-0 genomic DNA and cloned into the Gateway-based vector pMDC164 containing the β-glucuronidase reporter. The pro::HLP1-GUS construct was transformed into Arabidopsis plants through Agrobacterium-mediated floral dip transformation and screening of transgenic plants was done by kanamycin selection. Additionally, hlp1 mutants were complemented by proHLP1::HLP1. The proHLP1::HLP1 construct was introduced into hlp1 mutants through Agrobacterium-mediated floral dip and plants were screened through BASTA selection. T1 generation plants were then confirmed by restoration of HLP1 transcript and used for thermomemory gene expression. All primers used are listed in the Supplemental Table S5.
GUS Assay
GUS assay was performed using 7-d–old proHLP1::GUS and proCYCB1;1::GUS promoter lines treated without or with Glc (0% Glc and 3% Glc) followed by HS. HS was applied as 1 h at 37°C, 2 h at 22°C, and 2.5 h at 45°C. After HS, proHLP1::GUS seedlings were transferred immediately (0 h) to GUS buffer, or for the proCYCB1;1::GUS line, transferred to GUS buffer after 24 h of HS recovery (GUS treatment was described by Mishra et al., 2009).
Cell Death Assay
Roots of 5-d–old Arabidopsis seedlings were treated without or with Glc containing MS media followed by HS as described above for HS phenotype. After 24-h recovery at 22°C, seedlings were immersed in 1 μM of SYTOX Orange (Thermo Fisher Scientific) for 5 min, and fluorescence was observed under a confocal microscope (TCS SP2 AOBS System; Leica).
DAB Staining
Seedlings of 5-d–old Arabidopsis Col-0, OE2, and hlp1 mutants were supplemented with Glc and subjected to HS at 37°C for 1 h followed by 2 h of recovery at 22°C and then 45°C for 2.5 h. After HS, seedlings were recovered at 22°C for 24 h and subjected to DAB staining as described in Daudi and O’Brien (2012).
Sugar Starvation, Replenishment Assay, Sugar Sensitivity Assay, and Treatment with Chemical Inhibitors
Sugar starvation and replenishment assay was done as previously described in Jamsheer and Laxmi (2015) with 0% Suc and 3% (87.6 mM) Suc for various time points . Five-d–old Col-0 seedlings grown under standard growth conditions were used. Sugar sensitivity assay was performed as described in Mishra et al. (2009) using 3% of either Glc (166 mM), Suc (87.6 mM), 3-o-methylglucose (154 mM), mannose (166 mM), or mannitol (164 mM). For sugar sensitivity assay, 5-d-old Col-0 seedlings were used. Sugar sensitivity assay with metabolic inhibitors was done as described in Jamsheer and Laxmi (2015) using 5-d-old Col-0 seedlings under standard growth conditions. For Torin 1 (10 μM) and AZD-8055 (10 μM) treatment, 5-d–old Col-0 seedlings were starved without Glc for 24 h in the dark followed by Glc or Glc + inhibitors treatment for 3 h.
ChIP Assays
ChIP assays were performed by combining a ChIP assay kit (Millipore, Cat. no. 17-295) following the manufacturer’s protocol and that of Saleh et al. (2008) with minor modifications. Protoplasts were harvested from Col-0 plants and treated with Glc (5 mM) for 3 h followed by chromatin isolation. For ChIP in seedlings, 7-d-old Arabidopsis seedlings were transferred to MS media without or with Glc for 24 h followed by HS at 37°C for 3 h and chromatin was isolated. The resuspended chromatin was sonicated in a 4°C water sonicator (Diagenode Bioruptor Plus). Serum containing anti-E2Fa antibodies was obtained from Prof. Lieven De Veylder, VIB Department of Plant Systems Biology, Ghent University (Takahashi et al., 2008). Customized antibodies against HSFA1a were obtained from Abgenex (Cat. no. S10050-A; Project ID no. CP-139-15). Anti-GFP antibodies against HLP1-YFP were obtained from Thermo Fisher Scientific (Cat. no. A-11122). Antibodies against H3K (9+14+18+23+27) acetylation were purchased from Abcam (Cat. no. ab47915). Antibodies against H3K4me3 were purchased from Millipore (Cat. no. 07-473). All primers used are mentioned in the Supplemental Table S5.
Immunoblot Assay
The total plant protein was extracted in cold extraction buffer (137 mM of NaCl, 2.7 mM of KCl, 4.3 mM of Na2HPO4, 1.47 mM of KH2PO4, 10% glycerol [1.1 M], and 1 mM of phenylmethylsulfonyl fluoride) supplemented with plant protease inhibitor cocktail (Sigma-Aldrich, http://www.sigmaald rich.com/). Protein concentration was estimated by Quick Start Bradford reagent (Bio-Rad). Equal amounts of protein samples were loaded. The antibody for HSP21 (Anti-Hsp21; chloroplastic heat shock protein Cat. no. AS08 285) was purchased from Agrisera and monoclonal anti-tubulintyrosine antibody was purchased from Sigma-Aldrich. After detection of HSP21 and tubulin, membranes were subjected to staining with Ponceau S (0.1%; 1.5 mM).
Thermotolerance Test
Arabidopsis seedlings were grown under maintained culture conditions. For temperature stress treatment, all the seedlings were grown initially for 5 d in MS medium with 1% Suc (29 mM). To test the effect of Glc on temperature stress, 5-d–old plants were transferred to 0% Glc and 3% Glc (166 mM) for 24 h and grown at 22°C in the light. For SAT under light-grown condition, seedlings were transferred to the incubator at 37°C for 1 h, allowed to recover for 2 h at 22°C and then transferred to 45°C for 2.5 h, followed by recovery in the culture room for 3–4 d. For LAT, seedlings were transferred to the incubator at 37°C for 1 h, allowed to recover for 2 d at 22°C and then transferred to 45°C for 2.5 h, followed by recovery in the culture room for 3–4 d. For RT-qPCR under HS, seedlings were treated at 37°C for 3 h followed by recovery. To study the effect of Torin 1 and AZD-8055 on the phenotype of Arabidopsis seedlings under HS, 5-d–old Col-0 seedlings were treated with Torin 1 (10 μM) and AZD-8055 (10 µM) for 24 h and then subjected to HS, followed by recovery under maintained culture room conditions. The thermotolerance experiments were performed in an oven preset at the required temperature and recovery of the plants in between the acclimation and severe HS challenge was performed in the standard growth room condition (22°C) in the Arabidopsis culture room.
Thermomemory Test
Arabidopsis seedlings were grown under maintained culture conditions. For thermomemory, RT-qPCR, and ChIP-qPCR, 7-d–old Arabidopsis seedlings were grown on MS media without or with Glc for 24 h followed by HS. For memory, real-time HS was applied as follows: 24 h at 0% and 3% Glc, 1 h at 37°C, 90 min at 22°C, and 45 min at 45°C and then recovered at 22°C for 3 h, 24 h, and 48 h. For ChIP-qPCR, Glc- and HS-treated seedlings were recovered for 24 h at 22°C. For immuno-blotting, Glc and HS-treated seedlings were recovered for 48 h at 22°C. To check the phenotype without priming, 5-d–old Arabidopsis seedlings were transferred to Glc medium followed by HS. HS was applied directly as 45°C for 2.5 h followed by recovery for 3–7 d. For priming, Glc-treated seedlings were transferred to the HS regime as follows: 37°C for 1 h followed by 2-h recovery at 22°C and then treated with 45°C for 2.5 h. After HS, seedlings were recovered at 22°C for 3–7 d.
Measurement of Lateral Root Number and Seedling Survival Assay
For the lateral root number, HS-treated Arabidopsis seedlings were recovered for 3–4 d. For seedling survival assays, 5-d–old Arabidopsis seedlings were transferred to Glc-free (0% Glc) or Glc-containing (3% Glc; 166 mM) 0.5× MS medium and the effect of HS was analyzed. Seedlings showing survival and death were counted manually at 2–7 d after HS and digital images were taken using a Coolpix digital camera (Nikon).
Gene Expression Analysis
For gene expression analysis, RT-qPCR was performed. Arabidopsis seedlings were germinated and grown vertically in square petri plates with 1% Suc containing 0.5× MS medium buffered with 0.5 g/L MES and solidified with 0.8% agar at 24°C in the 16-h day/8-h night conditions. For Glc treatment, 5-d–old Arabidopsis seedlings were transferred to 0% Glc containing medium for 24 h followed by 3% Glc containing medium. For HS, 5-d–old Arabidopsis seedlings were transferred to the incubator at 37°C for 3 h. After the stress treatment, whole seedlings were harvested and frozen in liquid N. Total RNA was isolated from whole seedlings and mRNA was prepared using the RNAeasy Plant Mini Kit (Qiagen). Quantification of RNA was done and tested for quality before subsequent use. First-strand cDNA was synthesized by reverse transcription using 2 μg of total RNA with a high-capacity cDNA Reverse Transcription Kit (Applied Biosystems). Primers were designed by identifying a sequence stretch with a unique sequence. All candidate gene primers were designed using the software Primer Express (version 3.0; Applied Biosystems). An ABI Prism 7900 HP fast real-time PCR system (Applied Biosystems) was used. For the normalization of variance among samples, UBIQUITIN10 and/or 18S rRNA was used as a reference control. The fold-change for each candidate gene in different experimental conditions was determined using the quantitative ΔΔCT method. All primers used are mentioned in the Supplemental Table S5.
Chl Estimation
Chl estimation was performed with seedlings directly placed on square petri plates containing 1/2-strength MS medium with 1% Suc, or 0% and 3% Glc-containing MS medium without Suc. Chl was extracted by keeping seedlings in the dark at 4°C overnight in 2 mL 80% (v/v) acetone. Cell debris was pelleted by centrifugation at 8,000 rpm for 10 min and the amount of Chl was determined at 663 and 646 nm using a SmartSpec Plus spectrophotometer (Bio-Rad). The amount of Chl a (12.21 × l,663 − 2.81 × l,646) and Chl b (20.13 × l,646 − 5.03 × l,663) was quantified and the total amount of Chl (Chl a + Chl b) was normalized per seedling.
Microarray Analysis
For microarray of Arabidopsis Col-0 seedlings under Glc and HS, 5-d–old seedlings were first starved under 0% Glc for 24 h in the dark. After starvation, seedlings were treated without or with Glc for 24 h followed by HS. HS was applied at 37°C for 3 h. Samples were collected both immediately as well as after 3-h recovery. Harvested samples were outsourced for transcriptome profiling. The data were analyzed by the Transcriptome Analysis Console (v3.0; Affymetrix) with default parameters. Microarray has been submitted to ArrayExpress with accession no. E-MTAB-6980. Three biological replicates were used for the microarray experiment.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers: AT1G66080 (HLP1), AT1G50030 (TOR), AT2G36010 (E2FA), AT4G17750 (HSFA1A), AT2G26150 (HSFA2), AT4G11660 (HSFB2B), AT4G36990 (HSFB1), AT3G12580 (HSP70), AT5G02490 (HSP70-2), AT3G23990 (HSP60), AT1G54050 (HSP20-LIKE), AT1G52560 (HSP20-LIKE), AT2G29500 (HSP20-LIKE), AT4G27670 (HSP21), AT5G59720 (HSP18.2), AT5G12020 (HSP17.6II), AT4G10250 (HSP22), AT1G74310 (HSP101), AT3G18780 (ACT2), AT4G37490 (CYCB1;1).
Supplemental Data
The following supplementary data are available.
Supplemental Figure S1. Glc modulates thermotolerance response.
Supplemental Figure S2. Inhibition in TOR signaling causes weaker thermotolerance.
Supplemental Figure S3. HLP1 expression is regulated by Glc energy signaling.
Supplemental Figure S4. Overexpression of HLP1 provides tolerance to HS.
Supplemental Figure S5. Arabidopsis plants defective in HLP1 cause susceptibility to HS.
Supplemental Figure S6. hlp1 mutant showed reduced SAT and LAT.
Supplemental Figure S7. Comparison of Glc-TOR- and Glc-HSFA1-regulated HS genes in microarray.
Supplemental Figure S8. Complementation of hlp1 with proHLP1::HLP1 restores the expression of thermomemory HSPs.
Supplemental Figure S9. Comparison of Glc- and thermomemory-associated genes in microarray.
Supplemental Figure S10. Glc is required for thermomemory response.
Supplemental Figure S11. HLP1 is required in maintaining thermomemory.
Supplemental Figure S12. hlp1 mutant exhibits reduced sustenance of HSP21 transcript after HS recovery under Glc treatment.
Supplemental Table S1. HS genes upregulated by Glc.
Supplemental Table S2. Stress-responsive Cis-elements in the HLP1 promoter.
Supplemental Table S3. List of HS genes coregulated by both Glc and HSFA1.
Supplemental Table S4. HS genes coregulated by Glc and TOR.
Supplemental Table S5. List of primers used in the study.
Acknowledgments
We thank Yee-yung Charng (Agricultural Biotechnology Research Center, Taiwan) for KO-hsfa1a/hsfa1b/hsfa1d/hsfa1e seeds. We also thank Lieven De Veylder (Flemish Institute Biotechnology [VIB], Department of Plant Systems Biology, Ghent University, Belgium) for anti-E2Fa serum. We thank Christian Meyer (French National Institute for Agricultural Research, Institut National de la Recherche Agronomique, Institut Jean-Pierre Bourgin) for providing tor 35-7 RNAi lines. We thank the Central Instrumental Facility, National Institute of Plant Genome Research (NIPGR) for all the required assistance. We thank Muhammed Jamsheer K. (Amity University, Noida), Sandhya Verma (NIPGR), and Harmeet Kaur (National Bureau of Plant Genetic Resources) for advice and valuable discussions. We also thank Sandhya Verma, Harshita B. Saksena (NIPGR), and Manvi Sharma (NIPGR) for critical reading of the article and Aanchal Choudhary (NIPGR) for providing Arabidopsis Col-0 plants for transient infiltration. The authors also acknowledge DBT-eLibrary Consortium (DeLCON) for providing access to e-resources.
Footnotes
This work was supported by the National Institute of Plant Genome Research (Core Grant) and the Department of Biotechnology, Ministry of Science and Technology (DBT grant no. BT/PR8001/BRB/10/1211/2013).
References
- Andriankaja M, Dhondt S, De Bodt S, Vanhaeren H, Coppens F, De Milde L, Mu P (2012) Exit from proliferation during leaf development in Arabidopsis thaliana: A not-so-gradual process. 22: 64–78 [DOI] [PubMed] [Google Scholar]
- Baena-González E, Sheen J (2008) Convergent energy and stress signaling. Trends Plant Sci 13: 474–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena-González E, Rolland F, Thevelein JM, Sheen J (2007) A central integrator of transcription networks in plant stress and energy signalling. Nature 448: 938–942 [DOI] [PubMed] [Google Scholar]
- Bakshi A, Moin M, Kumar MU, Reddy ABM, Ren M, Datla R, Siddiq EA, Kirti PB (2017) Ectopic expression of Arabidopsis target of rapamycin (AtTOR) improves water-use efficiency and yield potential in rice. Sci Rep 7: 42835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzezinka K, Altmann S, Czesnick H, Nicolas P, Gorka M, Benke E, Kabelitz T, Jähne F, Graf A, Kappel C, et al. (2016) Arabidopsis FORGETTER1 mediates stress-induced chromatin memory through nucleosome remodeling. eLife 5: e17061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzezinka K, Altmann S, Bäurle I (2018) BRUSHY1/TONSOKU/MGOUN3 is required for heat stress memory. Plant Cell Environ 42: 771-781 [DOI] [PubMed] [Google Scholar]
- Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M, Werner T (2005) MatInspector and beyond: Promoter analysis based on transcription factor binding sites. Bioinformatics 21: 2933–2942 [DOI] [PubMed] [Google Scholar]
- Chabouté ME, Clément B, Sekine M, Philipps G, Chaubet-Gigot N (2000) Cell cycle regulation of the tobacco ribonucleotide reductase small subunit gene is mediated by E2F-like elements. Plant Cell 12: 1987–2000 [PMC free article] [PubMed] [Google Scholar]
- Charng YY, Liu HC, Liu NY, Hsu FC, Ko SS (2006) Arabidopsis Hsa32, a novel heat shock protein, is essential for acquired thermotolerance during long recovery after acclimation. Plant Physiol 140: 1297–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charng YY, Liu HC, Liu NY, Chi WT, Wang CN, Chang SH, Wang TT (2007) A heat-inducible transcription factor, HSFA2, is required for extension of acquired thermotolerance in Arabidopsis. Plant Physiol 143: 251–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J-G, Willard FS, Huang J, Liang J, Chasse SA, Jones AM, Siderovski DP (2003) A seven-transmembrane RGS protein that modulates plant cell proliferation. Science 301: 1728–1731 [DOI] [PubMed] [Google Scholar]
- Cho J-I, Ryoo N, Eom J-S, Lee D-W, Kim H-B, Jeong S-W, Lee Y-H, Kwon Y-K, Cho M-H, Bhoo SH, et al. (2009) Role of the rice hexokinases OsHXK5 and OsHXK6 as glucose sensors. Plant Physiol 149: 745–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YH, Yoo SD, Sheen J (2006) Regulatory functions of nuclear hexokinase1 complex in glucose signaling. Cell 127: 579–589 [DOI] [PubMed] [Google Scholar]
- Daudi A, O’Brien JA (2012) Detection of hydrogen peroxide by DAB staining in Arabidopsis leaves. Bio Protoc 2: e263. [PMC free article] [PubMed] [Google Scholar]
- Deprost D, Yao L, Sormani R, Moreau M, Leterreux G, Nicolaï M, Bedu M, Robaglia C, Meyer C (2007) The Arabidopsis TOR kinase links plant growth, yield, stress resistance and mRNA translation. EMBO Rep 8: 864–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vleesschauwer D, Filipe O, Hoffman G, Seifi HS, Haeck A, Canlas P, Van Bockhaven J, De Waele E, Demeestere K, Ronald P, et al. (2017) TARGET OF RAPAMYCIN signaling orchestrates growth–defense trade-offs in plants. New Phytol 217: 305–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Fromm M, Avramova Z (2012) Multiple exposures to drought “train” transcriptional responses in Arabidopsis. Nat Commun 3: 740–749 [DOI] [PubMed] [Google Scholar]
- Dobrenel T, Caldana C, Hanson J, Robaglia C, Vincentz M, Veit B, Meyer C (2016) TOR signaling and nutrient sensing. Annu Rev Plant Biol 67: 261–285 [DOI] [PubMed] [Google Scholar]
- Dong Y, Silbermann M, Speiser A, Forieri I, Linster E, Poschet G, Allboje Samami A, Wanatabe M, Sticht C, Teleman AA, et al. (2017) Sulfur availability regulates plant growth via glucose-TOR signaling. Nat Commun 8: 1174. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Gibson SI. (2004) Sugar and phytohormone response pathways: Navigating a signalling network. J Exp Bot 55: 253–264 [DOI] [PubMed] [Google Scholar]
- Gonzalez N, Vanhaeren H, Inzé D (2012) Leaf size control: Complex coordination of cell division and expansion. Trends Plant Sci 17: 332–340 [DOI] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA (2007) A chromatin landmark and transcription initiation at most promoters in human cells. Cell 130: 77–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guertin MJ, Lis JT (2010) Chromatin landscape dictates HSF binding to target DNA elements. PLoS Genet 6: e1001114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Singh M, Laxmi A (2015) Multiple interactions between glucose and brassinosteroid signal transduction pathways in Arabidopsis are uncovered by whole-genome transcriptional profiling. Plant Physiol 168: 1091–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C, Lin S-Y, Chi W-T, Charng Y-Y (2012) Recent gene duplication and subfunctionalization produced a mitochondrial GrpE, the nucleotide exchange factor of the Hsp70 complex, specialized in thermotolerance to chronic heat stress in Arabidopsis. Plant Physiol 158: 747-758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Song N, Zheng M, Liu X, Liu Z, Xing J, Ma J, Guo W, Yao Y, Peng H, et al. (2015) Histone acetyltransferase GCN5 is essential for heat stress-responsive gene activation and thermotolerance in Arabidopsis. Plant J 84: 1178–1191 [DOI] [PubMed] [Google Scholar]
- Hua J. (2009) From freezing to scorching, transcriptional responses to temperature variations in plants. Curr Opin Plant Biol 12: 568–573 [DOI] [PubMed] [Google Scholar]
- Huang J, Taylor JP, Chen J-G, Uhrig JF, Schnell DJ, Nakagawa T, Korth KL, Jones AM (2006) The plastid protein THYLAKOID FORMATION1 and the plasma membrane G-protein GPA1 interact in a novel sugar-signaling mechanism in Arabidopsis. Plant Cell 18: 1226–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M, Mitsuda N, Ohme-Takagi M (2011) Arabidopsis HSFB1 and HSFB2b act as repressors of the expression of heat-inducible Hsfs but positively regulate the acquired thermotolerance. Plant Physiol 157: 1243–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamsheer KM, Laxmi A (2015) Expression of Arabidopsis FCS-like zinc finger genes is differentially regulated by sugars, cellular energy level, and abiotic stress. Front Plant Sci 6: 746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi S, Ohama N, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K (2014) Functional analysis of the HIKESHI-like protein and its interaction with HSP70 in Arabidopsis. Biochem Biophys Res Commun 450: 396–400 [DOI] [PubMed] [Google Scholar]
- Kose S, Furuta M, Imamoto N (2012) Hikeshi, a nuclear import carrier for Hsp70s, protects cells from heat shock-induced nuclear damage. Cell 149: 578–589 [DOI] [PubMed] [Google Scholar]
- Lämke J, Brzezinka K, Altmann S, Bäurle I (2016) A hit-and-run heat shock factor governs sustained histone methylation and transcriptional stress memory. EMBO J 35: 162–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Sheen J (2016) Dynamic and diverse sugar signaling. Curr Opin Plant Biol 33: 116–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Cai W, Liu Y, Li H, Fu L, Liu Z, Xu L, Liu H, Xu T, Xiong Y (2017) Differential TOR activation and cell proliferation in Arabidopsis root and shoot apexes. 114: 2765-2770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HC, Charng YY (2013) Common and distinct functions of Arabidopsis class A1 and A2 heat shock factors in diverse abiotic stress responses and development. Plant Physiol 163: 276–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HC, Liao HT, Charng YY (2011) The role of class A1 heat shock factors (HSFA1s) in response to heat and other stresses in Arabidopsis. Plant Cell Environ 34: 738–751 [DOI] [PubMed] [Google Scholar]
- Liu H, Lämke J, Lin S, Hung M-J, Liu K-M, Charng Y, Bäurle I (2018) Distinct heat shock factors and chromatin modifications mediate the organ-autonomous transcriptional memory of heat stress. Plant J 95: 401–413 [DOI] [PubMed] [Google Scholar]
- Liu Y, Zhang C, Chen J, Guo L, Li X, Li W, Yu Z, Deng J, Zhang P, Zhang K, et al. (2013) Arabidopsis heat shock factor HSFA1a directly senses heat stress, pH changes, and hydrogen peroxide via the engagement of redox state. Plant Physiol Biochem 64: 92–98 [DOI] [PubMed] [Google Scholar]
- Loreti E, Poggi A, Novi G, Alpi A, Perata P (2005) A genome-wide analysis of the effects of sucrose on gene expression in Arabidopsis seedlings under anoxia. Plant Physiol 137: 1130–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahfouz MM, Kim S, Delauney AJ, Verma DPS (2006) Arabidopsis Target Of Rapamycin interacts with RAPTOR, which regulates the activity of S6 kinase in response to osmotic stress signals. Plant Cell 18: 477–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menand B, Desnos T, Nussaume L, Berger F, Bouchez D, Meyer C, Robaglia C (2002) Expression and disruption of the Arabidopsis TOR (target of rapamycin) gene. Proc Natl Acad Sci USA 99: 6422–6427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra BS, Singh M, Aggrawal P, Laxmi A (2009) Glucose and auxin signaling interaction in controlling Arabidopsis thaliana seedlings root growth and development. PLoS One 4: e4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montané MH, Menand B (2013) ATP-competitive mTOR kinase inhibitors delay plant growth by triggering early differentiation of meristematic cells but no developmental patterning change. J Exp Bot 64: 4361–4374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore B, Zhou L, Rolland F, Hall Q, Cheng WH, Liu YX, Hwang I, Jones T, Sheen J (2003) Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science 300: 332–336 [DOI] [PubMed] [Google Scholar]
- Naouar N, Vandepoele K, Lammens T, Casneuf T, Zeller G, van Hummelen P, Weigel D, Rätsch G, Inzé D, Kuiper M, et al. (2009) Quantitative RNA expression analysis with Affymetrix Tiling 1.0R arrays identifies new E2F target genes. Plant J 57: 184–194 [DOI] [PubMed] [Google Scholar]
- Ng HH, Robert F, Young RA, Struhl K (2003) Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell 11: 709–719 [DOI] [PubMed] [Google Scholar]
- Ogawa D, Yamaguchi K, Nishiuchi T (2007) High-level overexpression of the Arabidopsis HSFA2 gene confers not only increased thermotolerance but also salt/osmotic stress tolerance and enhanced callus growth. J Exp Bot 58: 3373–3383 [DOI] [PubMed] [Google Scholar]
- Powell AE, Lenhard M (2012) Control of organ size in plants. Curr Biol 22: R360–R367 [DOI] [PubMed] [Google Scholar]
- Ramirez-Parra E, Fründt C, Gutierrez C (2003) A genome-wide identification of E2F-regulated genes in Arabidopsis. Plant J 33: 801–811 [DOI] [PubMed] [Google Scholar]
- Ramon M, Rolland F, Sheen J (2008) Sugar sensing and signaling. Arabidopsis Book 6: e0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D, Yang H, Zhang S (2002) Cell death mediated by MAPK is associated with hydrogen peroxide production in Arabidopsis. J Biol Chem 277: 559–565 [DOI] [PubMed] [Google Scholar]
- Saleh A, Alvarez-Venegas R, Avramova Z (2008) An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat Protoc 3: 1018–1025 [DOI] [PubMed] [Google Scholar]
- Schepetilnikov M, Ryabova LA (2017) Auxin signaling in regulation of plant translation reinitiation. Front Plant Sci 8: 1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepetilnikov M, Dimitrova M, Mancera-Martínez E, Geldreich A, Keller M, Ryabova LA (2013) TOR and S6K1 promote translation reinitiation of uORF-containing mRNAs via phosphorylation of E1F3h. EMBO J 32: 1087–1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepetilnikov M, Makarian J, Srour O, Geldreich A, Yang Z, Chicher J, Hammann P, Ryabova LA (2017) GTPase ROP2 binds and promotes activation of Target Of Rapamycin, TOR, in response to auxin. EMBO J 36: 886–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedaghatmehr M, Mueller-Roeber B, Balazadeh S (2016) The plastid metalloprotease FtsH6 and small heat shock protein HSP21 jointly regulate thermomemory in Arabidopsis. Nat Commun 7: 12439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Ishida J, Narusaka M, Fujita M, Nanjo T, Umezawa T, Kamiya A, Nakajima M, Enju A, Sakurai T, et al. (2002) Monitoring the expression pattern of around 7,000 Arabidopsis genes under ABA treatments using a full-length cDNA microarray. Funct Integr Genomics 2: 282–291 [DOI] [PubMed] [Google Scholar]
- Song Y, Zhao G, Zhang X, Li L, Xiong F, Zhuo F, Zhang C, Yang Z, Datla R, Ren M, et al. (2017) The crosstalk between Target Of Rapamycin (TOR) and jasmonic acid (JA) signaling existing in Arabidopsis and cotton. Sci Rep 7: 45830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stief A, Altmann S, Hoffmann K, Pant BD, Scheible W-R, Bäurle I (2014) Arabidopsis miR156 regulates tolerance to recurring environmental stress through SPL transcription factors. Plant Cell 26: 1792–1807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swindell WR, Huebner M, Weber AP (2007) Transcriptional profiling of Arabidopsis heat shock proteins and transcription factors reveals extensive overlap between heat and non-heat stress response pathways. BMC Genomics 8: 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekeres M, Németh K, Koncz-Kálmán Z, Mathur J, Kauschmann A, Altmann T, Rédei GP, Nagy F, Schell J, Koncz C (1996) Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell 85: 171–182 [DOI] [PubMed] [Google Scholar]
- Takahashi N, Lammens T, Boudolf V, Maes S, Yoshizumi T, De Jaeger G, Witters E, Inzé D, De Veylder L (2008) The DNA replication checkpoint aids survival of plants deficient in the novel replisome factor ETG1. EMBO J 27: 1840–1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran L-SP, Nakashima K, Shinozaki K, Yamaguchi-Shinozaki K (2007) Plant gene networks in osmotic stress response: From genes to regulatory networks. Methods Enzymol 428: 109–128 [DOI] [PubMed] [Google Scholar]
- Vandepoele K, Vlieghe K, Florquin K, Hennig L, Beemster GTS, Gruissem W, Van de Peer Y, Inzé D, De Veylder L, et al. (2005) Genome-wide identification of potential plant E2F target genes. Plant Physiol 139: 316–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierling E. (1991) The roles of heat shock proteins in plants. Annu Rev Plant Physiol Plant Mol Biol 42: 579–620 [Google Scholar]
- von Koskull-Döring P, Scharf KD, Nover L (2007) The diversity of plant heat stress transcription factors. Trends Plant Sci 12: 452–457 [DOI] [PubMed] [Google Scholar]
- Wang L, Li H, Zhao C, Li S, Kong L, Wu W, Kong W, Liu Y, Wei Y, Zhu J-K, et al. (2017) The inhibition of protein translation mediated by AtGCN1 is essential for cold tolerance in Arabidopsis thaliana. Plant Cell Environ 40: 56–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Zhao Y, Li Z, Hsu C-C, Liu X, Fu L, Hou Y-J, Du Y, Xie S, Zhang C, et al. (2018) Reciprocal regulation of the TOR kinase and ABA receptor balances plant growth and stress response. Mol Cell 69: 100–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Vinocur B, Shoseyov O, Altman A (2004) Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci 9: 244–252 [DOI] [PubMed] [Google Scholar]
- Weng M, Yang Y, Feng H, Pan Z, Shen W-H, Zhu Y, Dong A (2014) Histone chaperone ASF1 is involved in gene transcription activation in response to heat stress in Arabidopsis thaliana. Plant Cell Environ 37: 2128–2138 [DOI] [PubMed] [Google Scholar]
- Wingender E, Dietze P, Karas H, Knüppel R (1996) TRANSFAC: A database on transcription factors and their DNA binding sites. Nucleic Acids Res 24: 238–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TY, Juan YT, Hsu YH, Wu SH, Liao HT, Fung RWM, Charng YY (2013) Interplay between heat shock proteins HSP101 and HSA32 prolongs heat acclimation memory posttranscriptionally in Arabidopsis. Plant Physiol 161: 2075–2084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong F, Zhang R, Meng Z, Deng K, Que Y, Zhuo F, Feng L, Guo S, Datla R, Ren M (2017) Brassinosteroid Insensitive 2 (BIN2) acts as a downstream effector of the Target Of Rapamycin (TOR) signaling pathway to regulate photoautotrophic growth in Arabidopsis. New Phytol 213: 233–249 [DOI] [PubMed] [Google Scholar]
- Xiong Y, McCormack M, Li L, Hall Q, Xiang C, Sheen J (2013) Glucose-TOR signalling reprograms the transcriptome and activates meristems. Nature 496: 181–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh CH, Kaplinsky NJ, Hu C, Charng YY (2012) Some like it hot, some like it warm: Phenotyping to explore thermotolerance diversity. Plant Sci 195: 10–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Zhu J-Y, Roh J, Marchive C, Kim S-K, Meyer C, Sun Y, Wang W, Wang Z-Y (2016) TOR signaling promotes accumulation of BZR1 to balance growth with carbon availability in Arabidopsis. Curr Biol 26: 1854–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]