Identification of circular RNAs and development of an overexpression method in grape revealed that Vv-circATS1 positively regulates cold tolerance
Abstract
Circular RNAs (circRNAs) are widely distributed and play essential roles in a series of developmental processes, although none have been identified or characterized in grapevines (Vitis vinifera). In this study, we characterized the function of grape circRNA and uncovered thousands of putative back-splicing sites by global transcriptome analysis. Our results indicated that several reported circRNA prediction algorithms should be used simultaneously to obtain comprehensive and reliable circRNA predictions in plants. Furthermore, the length of introns flanking grape circRNAs was closely related to exon circularization. Although the longer introns flanking grape circRNAs appeared to circularize more efficiently, a 20- to 50-nt region seemed large enough to drive grape circRNA biogenesis. In addition, the endogenous introns flanking circularized exon(s) in conjunction with reverse complementary sequences could support the accurate and efficient circularization of various exons in grape, which constitutes a new tool for exploring the functional consequences caused by circRNA expression. Finally, we identified 475 differentially expressed circRNAs in grape leaves under cold stress. Overexpression of Vv-circATS1, a circRNA derived from glycerol-3-P acyltransferase, improved cold tolerance in Arabidopsis (Arabidopsis thaliana), while the linear RNA derived from the same sequence cannot. These results indicate the functional difference between circRNA and linear RNA, and provide new insight into plant abiotic stress resistance.
Noncoding transcripts have become increasingly crucial for various plant functions, including normal growth and development as well as physiological and stress responses (Ariel et al., 2015; Wang et al., 2017a). Circular RNAs (circRNAs) are single-stranded RNAs that are joined head to tail (also referred to as back-spliced); in these circRNAs a downstream 5′ splicing donor site joins to an upstream 3′ splicing acceptor site (Ebbesen et al., 2017). Owing to the development of sequencing technology and associated analytical methods, research has shown that circRNAs are widespread in eukaryotes. Thousands of circRNAs have been investigated in a variety of eukaryotes, including humans (Homo sapiens), mice (Mus musculus), fruit flies (Drosophila), fission yeast (Schizosaccharomyces pombe), nematodes (Caenorhabditis elegans), and archaebacteria (Archaea; Wilusz, 2017; Cooper et al., 2018). At present, circRNAs have been reported in 12 plant species and a total of 95,143 circRNAs have been deposited in PlantcircBase (Chu et al., 2018a).
Several circRNA features seem to be shared in both animals and plants. For instance, both types of circRNAs can originate from exons, untranslated regions (UTRs), introns, noncoding RNA (ncRNA) loci, intergenic regions, and even antisense sequences of known transcripts (Memczak et al., 2013; Lu et al., 2015; Ye et al., 2015). Recent work has revealed that some emerging characteristics in plants, such as alternative circularization (Tan et al., 2017; Wang et al., 2017d) and tissue-specific expression profiles (Chen et al., 2017b) are shared with animal circRNAs (Jeck et al., 2013; Salzman et al., 2013; Westholm et al., 2014; Zhang et al., 2014, 2016b; Gao et al., 2016; Xia et al., 2017). However, some different features exist between animals and plants and even between different plant species. The abundance of circRNAs can be 10 times greater than that of linear RNAs in animals (Salzman et al., 2012; Jeck et al., 2013) but is relatively low in plants (Darbani et al., 2016; Chen et al., 2017b). Furthermore, reverse complementary sequences are enriched in the introns flanking circularized exons in animals (Jeck et al., 2013; Nitsche et al., 2014; Zhang et al., 2014; Ivanov et al., 2015); however, similar repetitive elements and reverse complementary sequences in the flanking introns of exonic circRNAs in plants are scarce (Lu et al., 2015; Ye et al., 2015). Nevertheless, long interspersed nuclear element 1-like elements and their reverse complementary pairs are significantly enriched in the flanking regions of circRNAs in maize (Zea mays; Chen et al., 2017b), and limited complementary sequences exist in both intron regions and flanking splice sites in Arabidopsis (Arabidopsis thaliana; Sun et al., 2016a). Although some studies have been conducted, information on circRNAs in plants is still very limited. Therefore, additional research is highly needed.
Compared with those of animal circRNAs, the biogenesis, regulation, and function of plant circRNAs have remained largely unknown until recently. In animals, the biogenesis of circRNAs is regulated by both cis elements and transacting factors (Li et al., 2018; Wilusz, 2018). Although circRNAs produced from exons that are not bracketed by complementary sequences exist (Westholm et al., 2014; Barrett et al., 2015), flanking complementary intronic sequences contribute to most exon circularization (Liang and Wilusz, 2014; Zhang et al., 2014; Ivanov et al., 2015; Kramer et al., 2015). Flanking intronic repeats as short as <40 nucleotides (nt) are sufficient for exon circularization (Liang and Wilusz, 2014; Kramer et al., 2015). Several works have also identified transacting factors that regulate exon circularization, such as the splicing factor Muscleblind in flies and humans (Ashwal-Fluss et al., 2014), the RNA-binding protein Quaking in humans (Conn et al., 2015), the double-strand RNA-editing enzyme adenosine deaminase 1 acting on RNA in nematodes and humans (Ivanov et al., 2015; Shi et al., 2017), heterogeneous nuclear ribonucleoproteins, and Ser-Arg proteins in flies (Kramer et al., 2015). In plants, however, the results of bioinformatic analyses have suggested that circRNA biogenesis is not driven by base-pairing interactions; instead, the length of the flanking intron of the circularized exon is much longer than that of randomly selected introns (Ye et al., 2015; Chen et al., 2017b; Zhao et al., 2017). However, direct experimental evidence supporting this model has not yet been directly addressed in plants.
The function of plant circRNAs remains largely unknown. Animal circRNAs have been postulated to function as microRNA (miRNA) sponges (Hansen et al., 2013; Memczak et al., 2013); as enhancers of the transcription of their host genes (Zhang et al., 2013; Li et al., 2015); in cell-to-cell information transfer (Lasda and Parker, 2016); as templates for translation (Legnini et al., 2017; Pamudurti et al., 2017; Yang et al., 2017a); or as memory modules, due to their extraordinary stability (Fischer and Leung, 2017). However, little is known about the regulation and function of circRNAs in plants. Until now, only four studies have provided direct evidence via transgenic technology that circRNAs function in plants. In Arabidopsis, overexpression (OE) of a lariat-derived circRNA derived from the first intron of the Ally of AML-1 and LEF-1 gene (ALY4; At5g37720) resulted in pleiotropic phenotypes, including curly and clustered leaves, delayed flowering, and reduced fertility (Li et al., 2016; Cheng et al., 2018). circRNA from exon 6 of the SEPALLATA3 gene is capable of forming an R-loop by direct interaction with its own genomic locus. Overexpression of this circRNA resulted in the production of flowers that had altered floral organ numbers, e.g. increased numbers of petals but fewer stamens (Conn et al., 2017). However, expression of circRNA derived from Phytoene Synthase 1 resulted in different phenotypes of transgenic tomato (Solanum lycopersicum) plants; some lines produced red fruits, while others produced yellow fruits (Tan et al., 2017).
Grape (Vitis vinifera) is one of the most economically important fruit crops worldwide, and it is the first fruit tree species to have its genome sequenced (Jaillon et al., 2007). The investigation into grape circRNAs has recently been suggested due to its complex genome (Sablok et al., 2016, 2017). Cold stress is the most important abiotic stress that influences the regional distribution, normal growth and production of grapes. Several studies have shown that plant circRNAs are differentially expressed under abiotic (Ye et al., 2015; Zuo et al., 2016; Wang et al., 2017b) and biotic stress (Wang et al., 2017d, 2018). However, little is known about how cold stress affects the expression patterns of circRNAs and their functions in stress responses. Accumulating evidence indicates that ncRNAs, especially miRNAs, are key regulators of plant cold stress responses (Wang et al., 2017a; Megha et al., 2018). In particular, we have demonstrated that miRNA 408 (miR408) OE increases the cold stress response in Arabidopsis (Ma et al., 2015) and that miR319 is involved in the cold response in grape (Luo et al., 2018). Robust tests have proven that circRNAs can serve as miRNA sponges to attenuate miRNA-mediated regulation in animals. However, whether and how circRNAs also regulate the cold stress response have not been elucidated, and information about the biological function of circRNAs in grape is lacking.
In this project, we used three widely used circRNA prediction methods, find_circ (Memczak et al., 2013), CIRCexplorer (Zhang et al., 2014), and CIRI (Gao et al., 2015), to identify circRNAs in grape. We selected hundreds of circRNAs for validation via three prediction methods and investigated the features of predicted circRNAs. In addition, we tested flanking intronic sequences that regulate circularization in grape by modulating intron lengths for back splicing. Moreover, we attempted to improve circRNA expression methods in plants. We also recognized and characterized low temperature-induced circRNAs. We ultimately identified and functionally verified a differentially expressed circRNA that is potentially associated with grape responses and adaptability to low temperatures. Our study involved the first genome-wide identification of circRNAs in grape and uncovered a novel functional role of circRNAs in plant responses to cold stress.
RESULTS
Identification and Characterization of Grape circRNAs
To explore circRNAs in grape, we used a mixed sample consisting of five grape tissues (root, stem, leaf, flower, and berry tissues) for RNA sequencing (RNA-seq). After trimming the adaptor sequences and filtering low-quality reads, we generated totals of 117,551,686, 117,541,122, and 117,521,496 reads from three biologic replications. We used three circRNA detection tools (CIRCexplorer, CIRI, and find_circ) with their default settings to identify circRNAs. Based on the back-spliced junction reads, 8,354 unique circRNAs were predicted in five grape tissues (Fig. 1A; Supplemental Fig. 1A; Supplemental Tables S1 and S2). CIRCexplorer (6,293) detected the most circRNA candidates, followed by find_circ (3,509) and CIRI (3,181). Among the circRNA candidates, only 1,432 circRNAs (17.1%) were detected by all three algorithms (Fig. 1A). Fifty circRNAs detected by the three algorithms were all identified successfully by Sanger sequencing (Fig. 1A; Supplemental Fig. 2A). Among the circRNAs predicted by only one algorithm, find_circ exhibited the highest success rate (19 out of 20), followed by CIRI (21 out of 32) and CIRCexplorer (10 out of 40; Fig. 1A; Supplemental Fig. S2, B–D). In addition, we designed 20 pairs of divergent and convergent primers to amplify each circRNA from the total RNA, complementary DNA (cDNA), and genomic DNA (Fig. 1E; Supplemental Fig. S3).
Figure 1.
Grape circRNAs are identified and characterized . A, The circRNAs were identified by three algorithms (find_circ, CIRI, and CIRCexplorer). The Illumina sequencing reads were generated from a mixed sample consisting of five grape tissues (root, stem, leaf, flower, and berry tissues). White boxes show the verification rate of circRNAs according to RT-PCR. B, Shown is the distribution of circRNA lengths in grape. C, A sketch map of different types of circRNAs is shown. The colored rectangles represent exons and the black lines represent introns. D, The genomic origin of grape circRNAs is shown. E, Validation of circRNAs by PCR in conjunction with divergent primers. Upper left, A model showing the convergent and divergent primers used to amplify the linear RNAs and circRNAs, respectively. Lower left, A Sanger sequencing example showing how circRNA_0708 was derived from exon back splicing from VIT_02s0012g02160. Right: An example showing that a pair of divergent primers amplified the circRNA within the cDNA but not within the genomic DNA; a pair of convergent primers was used as a control.
We normalized the content of circRNA in each sample to the number of back-spliced reads per million raw reads (RPM; Supplemental Fig. S1B; Supplemental Table S2). The correlation coefficient analysis revealed that the expression of replicates was divided by different detection tools, indicating essential dissimilarity among the three circRNA detection tools (Supplemental Fig. S1C). Find_circ separated from the other tools mostly in the principal component 1 dimension, while the CIRI and CIRCexplorer tools separated mostly in the principal component 2 dimension, implying better repeatability (Supplemental Fig. S1D).
We extracted 1,432 circRNAs derived by all three algorithms as high-confidence circRNAs to analyze the grape circRNA features. Most circRNAs contained multiple exons (Supplemental Fig. S1E). The majority of back splices (86.94%) spanned one to five exons, and the length ranged from 200 to 700 bp (Fig. 1B). Analysis of the chromosome distribution showed that circRNAs are widely and unevenly transcribed from grape chromosomes (Supplemental Fig. S1F). The predominant circularizing events involved coding DNA sequence (CDS)-CDS-originating (1,306), CDS-5′ UTR (94), and CDS-3′ UTR (28) circRNAs (Fig. 1D). We also analyzed the circRNAs using CIRI_AS (Gao et al., 2016), and 98 circRNAs presented alternative splicing (AS) events (Supplemental Table S3). Excluding those in intron retentions, three types of AS events could be found within circRNAs in all three biological replications (Supplemental Table S3).
In general, we found weak or positive correlations between circRNA levels and the linear-transcript levels of their host genes across different tissues (Fig. 2A). For all tested circRNAs, the circular transcripts were less abundant than were the linear isoforms (Fig. 2A). The results obtained from reverse transcription quantitative PCR (RT-qPCR) further confirmed the tissue-specific expression of circRNAs (Fig. 2A).
Figure 2.
Expression analysis of circRNAs in different tissues and conservation of predicted 1,432 circRNAs against reported circRNAs from six plants. A, The expression levels of circRNAs differ across tissues and show swing correlation with the linear-transcript expression levels of their host genes. RT-qPCR was applied to target the circRNA junctions (red circle) and the host genes (black line) via divergent and convergent primer pairs, respectively. Three biological replicates per sample were pooled in the experiment. The error bars represent the sd (n = 3). Root, stem, leaf, flower, and fruit samples are shown as Ro, St, Le, Fl, and Fr, respectively. The Pearson correlations (r) between circRNAs and their host genes were calculated. B, Conservation of circRNAs in grape (vvi), Arabidopsis (ath), soybean (gma), rice (osa), trifoliate orange (ptr), tomato (sly), and maize (zma) is analyzed. Upper, The corresponding numbers of circRNAs in grape, Arabidopsis, soybean, rice, trifoliate orange, tomato, and maize and the circRNA numbers for Arabidopsis, soybean, rice, trifoliate orange, tomato, and maize were obtained from PlantcircBase (Release 3; http://ibi.zju.edu.cn/plantcircbase/). Lower, The corresponding ratios of conserved circRNAs between grape and other plant species are shown. The numbers of conserved circRNAs are displayed above each bar.
The results of comparisons between the circRNAs of grape and those of Arabidopsis, soybean (Glycine max), rice (Oryza sativa), trifoliate orange (Poncirus trifoliata), tomato, and maize showed that 31 (2.2%), 7 (0.49%), 19 (1.3%), 12 (0.84%), 8 (0.56%), and 3 (0.21%) grape circRNAs were homologous with Arabidopsis circRNAs, soybean circRNAs, rice circRNAs, trifoliate orange circRNAs, tomato circRNAs and maize circRNAs, respectively (Fig. 2B). Over all, about 5.2% of the grape circRNAs were homologous to the current collection of plant circRNA sequences in the PlantcircBase database.
Grape circRNAs Are Flanked by Long Introns
Analysis of the splice signal of 1,432 grape circRNAs revealed that the majority (98.3%) of circRNAs contain a canonical GT/AG splicing signal (Supplemental Fig. S1G). We therefore extracted the flanking intron sequences of exonic circRNAs from the grape genome for further analysis. No significant positive correlation occurred between the flanking intron length and expression level of all 1,432 circRNAs (Supplemental Fig. S1H). The alignment results between the upstream and downstream flanking intron sequences of the exonic circRNAs revealed that only 85 (5.9%) intronic sequences contained reverse complementary sequences in grape. Furthermore, we compared the flanking intron sequences of genes without or with detectable circRNAs with grape miniature inverted repeat transposable elements (MITEs; Chen et al., 2014). Notably, there was a small difference (17% versus 18%) in the enrichment of MITEs between genes with and those without detectable circRNAs, suggesting that the existence of MITEs might not be involved in circRNA generation.
A one-sample statistical t test analysis revealed that circularized exons were more likely to be flanked by longer introns (Fig. 3A), which led to the speculation that flanking introns might play an important role in exon circularization. To verify this speculation, we used Nicotiana benthamiana plants to study the circularization of grape circRNA; the use of N. benthamiana avoided the potentially detrimental effects of endogenous grape circRNA OE. We cloned two full-length genes (the host genes of circRNA_4363 and circRNA_1975) from grape DNA and inserted them into the split the enhanced GFP (eGFP) gene in a pHB vector. The grape circRNAs were successfully detected by divergent PCR in N. benthamiana leaves (Supplemental Figs. S4 and S5). We then cloned seven circularized exons as well as their flanking introns (circRNA_1218, circRNA_1975, circRNA_4328, circRNA_4363, circRNA_4473, circRNA_5664, and circRNA_7172) from the grape DNA and inserted them into the middle of the split eGFP gene within the pHB vector. The circRNAs with flanking introns were detectable after injection (Fig. 3C). In contrast, circRNAs were not detected in N. benthamiana leaves if the flanking introns were removed (Fig. 3B).
Figure 3.
Identification of flanking introns and their functional analysis in circRNA expression. A, Comparison of flanking intron lengths between the linear transcripts and the 1,432 identified circRNAs commonly identified by all three algorithms (CIRCexplorer, CIRI, and find_circ) in grape. The y axis indicates the log10 value of intron length (bp). Significant difference between groups: **P < 0.001. B to D, The genomic circRNA-forming region for circRNA_1218, circRNA_1975, circRNA_4328, circRNA_4363, circRNA_4473, circRNA_5664, and circRNA_7172 with (C) and without (B) its endogenous flanking intron was analyzed by N. benthamiana leaf transient expression. The expression levels of the circRNAs were quantified by RT-PCR (B and C) and RT-qPCR (D). Different lowercase and capital letters in D indicate significant differences in the expression of circRNAs (black bars) and linear RNAs (gray bars), respectively, at P < 0.05, as determined by Duncan’s multiple-range test. Data are averages ± sd from three independent experiments.
Having shown that the intronic regions immediately flanking circularized exon(s) were sufficient to produce circRNA, we chose two circRNAs (circRNA_7172 and circRNA_1975), which were characterized by relatively high expression (Fig. 3D), to identify the minimal sequence length for circRNA production (Fig. 4; Supplemental Fig. S5). Highly complementary sequences were present in the flanking introns of circRNA_7172 (Fig. 4A, gray rectangles) but not circRNA_1975 (Supplemental Fig. S5A). A basal level of circularization can be accomplished irrespective of the length of the upstream and downstream intronic sequences, provided that they are present. The length is instead relevant for regulating the expression level of circRNAs as longer flanking intronic sequences appear to circularize more efficiently. However, removing the entire flanking intronic sequences of both the 5′ and 3′ ends produced no circRNA at all (Fig. 4, C and D; Supplemental Fig. 5, C and D). Completely deleting either of the complementary sequences of circRNA_7172 largely eliminated circRNA production (Fig. 4, B–D). Interestingly, removing the entire downstream flanking intronic sequence of the circularized exons of circRNA_1975 produced two new circRNAs after the GT/AG splicing signals (Supplemental Fig. S5B). Further analysis revealed that ∼20–50 nt of flanking intronic sequence could support circularization (Fig. 4B; Supplemental Fig. S5B). Since these minimal regions barely contain complementary sequences, we concluded that base pairing between the flanking intronic sequences is unnecessary for circRNA production and that short sequences (20–50 nt) are sufficient to promote back splicing and exon circularization in grape.
Figure 4.
The VIT_218s0001g07710 gene generates a circRNA, and long intronic sequences facilitate circRNA_7172 production. A, The locus and structure of the VIT_218s0001g07710 gene highlight a 6,037-nt region that includes exons 29–31 of this gene. circRNA_7172 is formed when the 5′ splice site at the end of exon 31 is linked to the 3′ splice site at the beginning of exon 29. Black arrows indicate the locations of the divergent and convergent primers. Complementary elements in the designated orientations are shown as gray rectangles. B, circRNA_7172 expression plasmids containing deletions at their 5′ or 3′ ends were transfected into N. benthamiana leaves. Upper, circRNA_7172-1, encompassing 2,584 nt upstream of exon 29 to 2,537 nt downstream of exon 31, characterized the initiation of the complementary sequence partly between the upstream and downstream regions. circRNA_7172-2 characterized the initiation of the complementary sequence perfectly between the upstream and downstream regions, and circRNA_7172-10 characterized the termination of the complementary sequence perfectly between the upstream and downstream regions. The brightness change represents the expression level of the circRNA and linear RNA. No circRNA was identified in circRNA_7172-13 or circRNA_7172-14. Lower, a Sanger sequencing example showing how circRNA_7172 was derived from exon back splicing. C and D, RT-qPCR (C) and RT-PCR (D) were performed to examine circRNA expression. The numbers shown at the bottom of C and D correspond to the column of numbers shown in the left portion of B. The relative amount of RT-PCR products detectable in each lane was determined using gel analysis software (Image Lab). Data are averages ± sd from three independent experiments. Different lowercase and capital letters indicate significant differences in the expression of circRNAs and linear RNAs, respectively, at P < 0.05, as determined by Duncan’s multiple-range test.
Precise and Efficient Expression of circRNAs in Plants
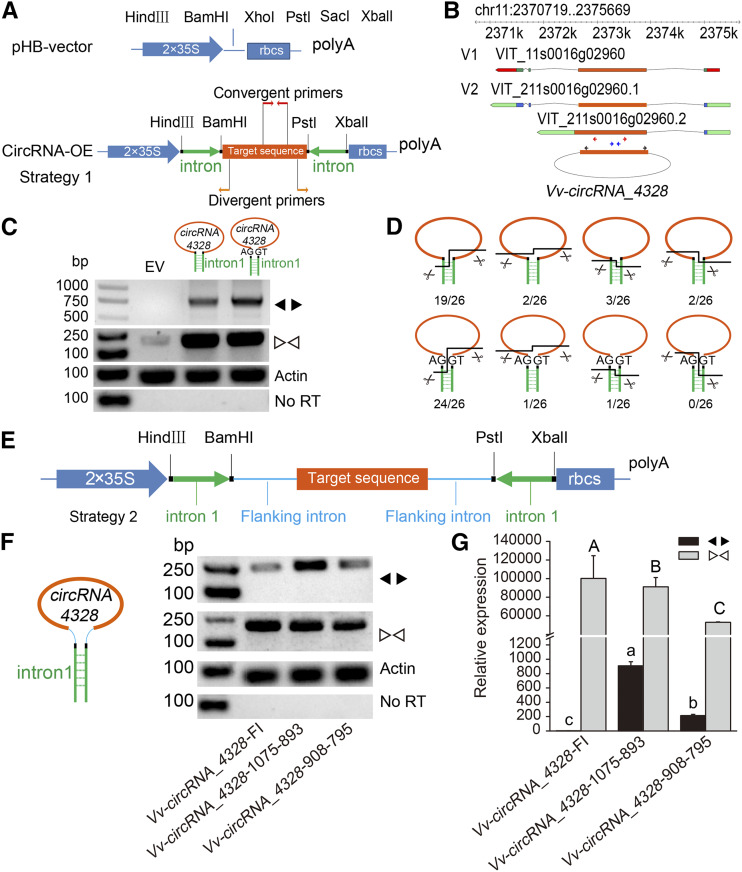
As mentioned above, grape exon circularization was heterologously induced in N. benthamiana by the expression of plasmids that drive exon circularization and that contain the associated flanking sequences. However, the induced expression of some circRNAs, such as circRNA_4328 and circRNA_4363, in N. benthamiana leaves was low (Fig. 3D). Here, we improved the expression levels via a vector construction strategy. We introduced reverse complementary sequences into the OE vector; this approach represents a well-known method for facilitating RNA circularization, and we refer to it as a reverse complement strategy (Strategy 1) in this study (Fig. 5A). We used two randomly selected grape intronic fragments (intron 1, 416 bp; intron 2, 461 bp) and their reverse complementary sequences to promote back splicing of circRNA_4328 and circRNA_4363. Exogenous circRNA_4328 and circRNA_4363 were not detected by convergent primers or divergent primers in empty-vector control N. benthamiana plants, whereas a large amount of transcripts were detected in plants at 4 d after infiltration (Fig. 5C; Supplemental Fig. S6B). Nonetheless, the detected transcripts contained several circular forms (Fig. 5D; Supplemental Figs. S6C and S7). Compared with transcripts in perfect circular form, those in imperfect form were constructed by adding or removing sequences from circularization exons or reverse complementary sequences that aid circularization (Fig. 5D; Supplemental Figs. S6C and S7). We added AG/GT splice sites that we expected might support proper circRNA formation to the flanking regions of the target circularized exon (Starke et al., 2015). Unfortunately, the circularization patterns detected for the AG/GT splice sites were similar to those detected for the non-AG/GT splice sites, and sometimes the splice sites were included in the imperfectly circularized transcripts (Fig. 5D; Supplemental Figs. S6C and S7).
Figure 5.
Detailed information is given on the circRNA OE vector construct used in plants. A, A schematic diagram of the OE circRNA construct (Strategy 1) is shown. A pHB vector was used to generate the new circRNA OE vector. An intron DNA fragment of the grape VIT_13s0074g00100 gene (intron 1, 416 bp) was ligated into both upstream and downstream flanking target sequences (orange rectangle) in an opposite orientation pattern. The convergent primers (red arrows) and divergent primers (orange arrows) used for RT-PCR are indicated. B, The gene structure of circRNA_4328 and its host gene VIT_211s0016g02960 are shown. Red arrows indicate the locations of the divergent primers used in C. Blue arrows indicate the locations of the convergent primers used in C, F, and G. Black arrows indicate the locations of the divergent primers used in F and G. C, RT-PCR products were generated via divergent primers (circRNAs) and convergent primers (linear RNAs) to examine circRNA expression in N. benthamiana leaves transfected with different constructs after 4 d. AG/GT splice sites were added to both flanks of the target sequence. D, The circRNA structures detected experimentally are shown. The orange lines and green boxes represent circRNAs and intron sequences, respectively. The black lines indicate the true dissection sites. The number of experimentally detected circRNA structures was determined. The Sanger sequencing results of the junction reads of the circRNAs are shown in Supplemental Figure S7. E, Schematic diagram of the circRNA OE construct (Strategy 2). Intron 1 (416 bp; green arrow) was introduced in the opposite orientation, and the sequence that was expected to circularize along with the flanking intron by itself (cyan line) was introduced into the vector (BamHI and PstI sites). The inserted sequence for circRNA expression includes the circRNA-forming sequence and its endogenous flanking genomic sequence, which contains the splicing sites. F and G, RT-PCR (F) and RT-qPCR (G) were performed to identify the target gene expression in N. benthamiana leaves transfected with different constructs. circRNA_4328-flanking intron, the endogenous flanking sequence, was used to promote circRNA circularization. circRNA_4328-1075-893, which involves 1075 nt upstream of circRNA_4328 to 893 nt downstream, was used along with reverse complementary sequences (intron 1, 416 bp) to express the circRNA, and circRNA_4328-908-795, which involves the 908 nt upstream of circRNA_4328 to 795 nt downstream, was also used along with reverse complementary sequences (intron 1) to express the circRNA. The different lowercase and capital letters indicate significant differences in the expression of circRNAs and linear RNAs, respectively, at P < 0.05, as determined by Duncan’s multiple range test. Data are averages ± sd from three independent experiments.
With respect to the construction of the circRNA OE vector, Strategy 1 can guarantee only the efficiency of circularization and not the accuracy of circularization (Fig. 5, C and D; Supplemental Fig. S6, B and C). Although few reverse complementary sequences exist between introns bracketing circRNAs in plants, the endogenous sequences flanking the back-splice site contribute to the precision of back splicing (Figs. 3 and 4). Therefore, we proposed a construction method referred to as the reverse complement and flanking intron strategy (Strategy 2), which aimed to increase the accuracy of circularization by flanking introns (Fig. 5E). With respect to the OE of circRNA_4328, circRNA_7172, circRNA_1975, and circRNA_4363, the forming exons of these circRNAs were flanked by their specific upstream and downstream intronic sequences that were cloned from DNA and ligated into pHB vectors, which contained a reverse complementary sequence of intron 1 (Figs. 5F and 6A; Supplemental Figs. S6D and S8A). The results showed that these plasmids could drive the intervening exon(s) to be back spliced accurately, and the circRNA expression levels were tens or hundreds of times greater than those of the vectors driving only the exon(s) that circularize, as well as their immediate flanking sequences (Figs. 5G and 6C; Supplemental Figs. S6E and S8C).
Figure 6.
The endogenous flanking genomic sequence of the circRNA_7172-forming sequence, which can be cloned between the two complementary intron fragments, influenced the accuracy and efficiency of its mature circRNA. A, Upper left, Intron 1 (416 bp; green arrow) was introduced in the opposite orientation, after which the circRNA_7172-forming sequence along with the different lengths of the flanking intron (cyan line) cloned from grape DNA was introduced into the vector. Lower left and right, Circular products were detected after transfection into N. benthamiana leaves. B, Upper left, An intron DNA fragment of the grape VIT_13s0074g00100 (416 bp or 1000 bp; green arrow) gene was introduced in the opposite orientation. The mature circRNA_7172 sequence was then cloned from grape cDNA along with the different lengths of its endogenous flanking intron (cyan line) and introduced into the vector. Lower left and right, Circular products were detected after transfection into N. benthamiana leaves. C and D, RT-qPCR (C) and RT-PCR (D) were performed to detect the target gene expression level in N. benthamiana leaves at 4 d after transfection. circRNA_7172-FI represents the endogenous flanking intronic sequence of the circRNA_7172 sequence and was used to promote circRNA circularization. The numbers shown in the bottom of C and D correspond to the numbered lines shown in the top portions of A and B. Data are averages ± sd from three independent experiments (C). The different lowercase and capital letters indicate significant differences in the expression of circRNAs and linear RNAs, respectively, at P < 0.05, as determined by Duncan’s multiple-range test.
We further tested the circularization efficiency by intercepting the flanking intron sequences based on Strategy 2 (Fig. 6A; Supplemental Fig. S8A). We determined that 50 nt of flanking intronic sequence is required for circularization both in circRNA_1975 and circRNA_7172 and that short intron sequences can severely affect the expression level of the circRNA (Fig. 6C; Supplemental Fig. S8C). Additionally, we cloned a mature circRNA_7172 sequence (29–31 exons of VIT_18s0001g07710) from cDNA amended with endogenous flanking introns. In these situations, 100 nt of flanking intronic sequence is required for circularization (Fig. 6B). In addition, the expression level of the circRNA failed to increase when the length of the reverse complementary intron 1 was extended to 1000 bp (Fig. 6, B and C). Therefore, adding flanking intron sequences (>100 bp upstream and downstream) along with reverse complementary sequences was the best way to overexpress circRNAs in all of our attempts.
Cold Stress Alters the Genome-Wide Profiles of circRNAs in Grape
To perform cold stress treatments, we subjected 1-year-old grape cuttings to 4°C for 0, 2, 4, 8, 12, 24, 48, and 72 h. The cuttings appeared to wilt and lose turgor after cold stress (Supplemental Fig. S9, A and B). Electrolyte leakage (EL) gradually increased under cold stress (Supplemental Fig. S9C). H2O2, malonaldehyde (MDA), peroxidase, and superoxide dismutase production increased once the cold stress began, peaked at 48 h, and then decreased after 72 h (Supplemental Fig. S9, D–G). The transcript levels of VvCBF1 and VvCBF2 slightly decreased at 2 h after cold treatment but gradually accumulated between 4 h and 12 h, after which they decreased drastically (Supplemental Fig. S9, H and I). The transcript levels of VvERF057 rapidly increased once the cold stress began and peaked at both 12 h and 48 h (Supplemental Fig. S9J). Combining the indicators together, we determined that the ideal sampling time points for the identification of differentially expressed circRNAs under low temperature were 0, 4, and 12 h. Using three circRNA detection tools (find_circ, CIRI, and CIRCexplorer), we detected 6,145, 4,406 and 12,322 circRNAs, respectively, in these three samples (Supplemental Tables S4–S6). The coefficient matrices of the independent biological repeats at the same time points were significantly closely correlated, which indicated that the independent biological repeats had similar gene expression profiles (Supplemental Fig. S10). CIRCexplorer detected the most differentially expressed circRNA candidates (289), followed by CIRI (169) and find_circ (120; Fig. 7A; Supplemental Fig. S11; Supplemental Tables S7–S9). When mixed together, we identified 475 differentially expressed circRNAs in grape leaves under cold treatment and used these for further analysis (Fig. 7A; Supplemental Fig. S11; Supplemental Tables S7–S9). We successfully identified 178, 49, and 77 upregulated differentially expressed circRNAs and 151, 63, and 56 downregulated differentially expressed circRNAs in the comparisons of 4 h versus 0 h, 12 h versus 0 h, and 12 h versus 4 h, respectively (Fig. 7B; Supplemental Fig. S11; Supplemental Tables S7–S9).
Figure 7.
Shown is the statistical analysis of differentially expressed circRNAs in grape leaves under control and low-temperature conditions. A, The Venn diagram of differentially expressed grape circRNAs was identified by three algorithms (find_circ, CIRI, and CIRCexplorer) during cold stress (0, 4, and 12 h). B, The histogram shows differentially expressed grape circRNAs identified by three algorithms (find_circ, CIRI, and CIRCexplorer) during cold stress (0, 4, and 12 h). The numbers of upregulated (red) and downregulated (blue) circRNAs are displayed above each bar.
We performed gene ontology (GO) category enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses on the host genes of circRNAs that were differentially expressed under cold stress. With respect to biological processes, the circRNA host genes were involved mainly in response to cold, photosynthesis, unsaturated fatty acid biosynthetic process, and photosynthetic electron transport in photosystem II (Supplemental Fig. S12, A–F). With respect to molecular function, the enriched GO terms included sequence-specific DNA binding, DNA binding, ATP binding, electron transporter, and poly(U) RNA binding (Supplemental Fig. S12, A–F). The significantly enriched KEGG pathways were mainly carbon metabolism, photosynthesis, glycerolipid metabolism, glycerophospholipid metabolism, and carbon fixation in photosynthetic organisms (Supplemental Fig. S12, G–K).
miRNAs participate in the regulation of cold stress in plants (Megha et al., 2018). To detect whether differentially expressed circRNAs in grape under cold stress could affect gene expression via posttranscriptional regulation by binding to miRNAs, we identified the circRNAs predicted to be sponges for differentially expressed miRNAs. As shown in Supplemental Table S10, we identified 1,020 miRNAs along with their corresponding 456 circRNAs (among the 475 differentially expressed circRNAs). Interestingly, several miRNAs, including miR156, miR164, miR167, miR171, miR394, miR395, miR396, miR397, miR398, and miR408, were proven to participate in the cold stress response (Supplemental Fig. S13; Megha et al., 2018).
Overexpression of a circRNA from VvATS1 Enhances Cold Tolerance in Arabidopsis
Of all the differentially expressed circRNAs, circRNA_0708, which is derived from exons 5–8 of the ATS1 gene (referred to as circATS1), was particularly abundant and presented a high fragments per kilobase of exon per million fragments mapped (FPKM) value (Supplemental Table S5). circATS1’s host gene is involved in the glycerolipid metabolism pathway (Supplemental Fig. S12H). Phylogenetic analysis revealed that VvATS1 was most closely related to AtATS1 (Supplemental Fig. S14). We designed a pair of back-to-back divergent primers and used these to verify circATS1 (333 bp; Fig. 8A). Although grape ATS1 protein exhibited 73% sequence identity with the Arabidopsis ATS1 protein, the sequence of the junction area of circATS1 is different. It appears that circATS1 is not present in Arabidopsis (Fig. 8B). The Vv-circATS1 flanking introns were much longer (mean ∼39-fold) than flanking introns in the same region of ATS1 gene in Arabidopsis. Vv-circATS1 was enriched in grape leaf and flower tissues (Fig. 2A). Further, fluorescence in situ hybridization (FISH) revealed that Vv-circATS1 localizes to the cytoplasm, as well as to small amounts of nucleic regions (Fig. 8C). However, Vv-ATS1 mRNA is localized exclusively in the cytosol. As expected, we detected no signal hybridized with the corresponding sense probes (Fig. 8C). Cell fractionation experiments showed that Vv-circATS1 was localized both in the cytoplasm and nucleus, and there existed a number of Vv-circATS1 molecules in the cytoplasm (Fig. 8D). The transcript levels of Vv-circATS1 slightly increased at 2 h of cold stress; decreased at 4, 8, and 12 h; and then increased gradually thereafter (Fig. 8E). In addition, there was no significant correlation between the expression of Vv-ATS1 mRNA and that of Vv-ATS1 circRNA under cold stress; the correlation coefficient was −0.399 (Fig. 8E).
Figure 8.
Functional analysis of Vv-circATS1. A, Validation of full-length Vv-circATS1 sequences by divergent RT-PCR. Left: The diagram shows Vv-circATS1 and the positions of divergent primers used to amplify the circRNAs. Forward and reverse primers were designed to be near each other to ensure that the PCR products were the full size of the circRNAs. Right: RT-PCR products resulting from divergent primers were 333 bp in size. B, Conservation analysis of circATS1 in grape and Arabidopsis. C, FISH images of Vv-circATS1 in grape leaves, in which localization occurred in both the cytosol and nucleus (I, II, V, and VI). FISH images of Vv-ATS1 mRNA are shown for comparison (III and VII). Control sense probes used on grape leaves (IX–XII). Scale bars = 50 μm (I–IV and IX–XII), 10 μm (V–VIII). D, Detection of Vv-circATS1 and Vv-ATS1 mRNA in the cytoplasmic and nuclear fractions by RT-PCR. E, RT-qPCR-based expression analysis of Vv-circATS1 under cold stress. Grape plants were exposed to cold (4°C) treatment for 0–72 h. The relative abundance of circRNAs and linear RNAs was normalized with respect to the abundance of the grape Actin gene. The bars represent standard deviations (sds) for three biological replications. Different lowercase and capital letters indicate significant differences in the expression of circRNAs and linear RNAs, respectively, at P < 0.05, as determined by Duncan’s multiple-range test. F, A schematic diagram of the Vv-circATS1-OE and Vv-circATS1-linear constructs is shown. The upper diagram represents the gene structure of the VIT_202s0012g02160 locus, highlighting a 2023 nt region that includes exons 5 to 8. Black arrows indicate the locations of the divergent and convergent primers. The middle diagram shows that the intronic fragment intron 1 (green arrow) was assembled in the opposite orientation, after which the sequence that was expected to circularize along with the endogenous flanking intron (cyan line, 500 bp upstream and downstream flanking intron sequences of Vv-circATS1) was introduced into the vector. In the lower diagram, Vv-circATS1-linear represented the forming exons of the Vv-circATS1, driven only by the 2× 35S promoter. rbcS, genes encoding the ribulose-1,5-bisphosphate carboxylase small subunit. G, RT-qPCR via divergent (dark gray bars) and convergent (light gray bars) primer pairs was used to target the circularized sequence in wild-type (WT) and four transgenic lines. The Arabidopsis ACTIN2 served as a reference gene. Data are averages ± sd from three independent experiments. Different letters indicate significant differences at P < 0.05, as determined by Duncan’s multiple-range test. H, The phenotype of the Vv-circATS1-OE line is shown. After 30 d at 4°C, only the Vv-circATS1-OE Arabidopsis plants remained green. I, Shown is the chlorophyll content in seedlings treated with 4°C for 30 d. J, The graph shows EL in seedlings treated with 4°C for 30 d. K, The graph shows MDA content in seedlings treated with 4°C for 30 d. Data are averages ± sd from three independent experiments. Different letters indicate significant differences at P < 0.05, as determined by Duncan’s multiple-range test. L, Left, PCR amplification of ES AS mRNAs predicted from Vv-circATS1 is shown. The primers were designed to encompass exons 4 and 9. Right, PCR amplification of AS mRNAs of AtATS1 in Arabidopsis is shown. The primers were designed to encompass exons 4 and 9. M, The expression profile of the AtATS1 gene in transgenic Arabidopsis plants is shown. Different letters indicate significant differences at P < 0.05, as determined by Duncan’s multiple-range test.
To determine the role of Vv-circATS1, we generated transgenic Arabidopsis plants by overexpressing circATS1 along with 500 bp flanking intronic sequences in accordance with Strategy 2 (Fig. 8F). As expected, the expression of Vv-circATS1 significantly increased in the Vv-circATS1-OE lines (Fig. 8G). The RNA expression levels detected by divergent primers and convergent primers were almost the same in all transgenic lines (Fig. 8G). Under cold stress (4°C for 1 month), the leaves of the Vv-circATS1-OE lines maintained their green color, whereas the leaves of the linear control and wild-type lines were brown and necrotic (Fig. 8H). No significant difference in total chlorophyll content, EL, or MDA was detected among the control and the transgenic lines at normal temperature (Fig. 8, I–K). The anthocyanin, EL, and MDA content was significantly lower in the two independent Vv-circATS1-OE lines than in the linear control and wild-type plants treated with 4°C for 30 d. In addition, the chlorophyll content was much higher in the Vv-circATS1-OE plants than in the linear control and wild-type plants (Fig. 8, H and I–K). Further examination revealed no link between circATS1 and the exon skipping (ES) AS variants in both grape and transgenic Arabidopsis lines (Fig. 8L). Additionally, AtATS1 mRNA abundance did not significantly differ between the transgenic Arabidopsis and wild-type lines (Fig. 8M).
Vv-circATS1 Regulates Several miRNAs Associated with Cold Tolerance and Genes Related to Stimulus in Arabidopsis at 4°C
To gain more insight into the role of Vv-circATS1 in response to cold stress, Arabidopsis rosette leaves from wild-type and Vv-circATS1-OE plants (OE4) were collected, after which the total RNA was isolated for miRNA and mRNA-seq analysis after treatment at 4°C for 24 h. Principal component analysis separated the wild-type plants and Vv-circATS1-OE transgenic plants into two clusters (Supplemental Figs. S15A and S16A). Among the miRNAs, 30 were previously unknown, and the sequences of 263 had already been deposited in the miRBase (http://www.mirbase.org/index.shtml; Supplemental Table S11). Interestingly, more miRNAs were detected in the OE plants than in the wild-type plants. Among the 293 identified miRNAs, 23 significantly differed between the OE plants and the wild-type plants: 12 downregulated genes and 11 upregulated genes in the Vv-circATS1-OE plants (Supplemental Fig. S15, B and C; Supplemental Table S12). mRNA-seq results showed that the expression of 212 genes (61.3% upregulated and 38.7% downregulated) was significantly changed by the OE of Vv-circATS1 under cold-stress conditions (Supplemental Fig. S16, B and C; Supplemental Table S13). Thirty-five GO terms were significantly enriched in the Vv-circATS1-induced genes (Supplemental Table S14). Of the enriched GO terms, we noted that Vv-circATS1-induced genes are primarily involved in stress response, and the numbers of genes related to response to stimulus, toxin catabolic process, photosystem II assembly, and proline transport were 76, 9, 7, and 6, respectively (Supplemental Fig. S16D). Interestingly, 56 genes involved in the response to stimulus, seven genes involved in the toxin catabolic process, and five genes involved in proline transport were upregulated by OE of Vv-circATS1. However, all genes involved in photosystem II assembly were downregulated by OE of Vv-circATS1.
DISCUSSION
In animals, circRNAs are becoming a hot research topic due to their substantial regulatory functions. Recent studies have also revealed widespread and various circular ncRNAs in plants (Chu et al., 2018b). However, the characterization of circRNAs is much less known in perennial woody vines, which differ from herbaceous plants in terms of growth and development (Carmona et al., 2007). In this study, we revealed the widespread expression of circRNAs in grape, with some circRNAs exhibiting distinct expression profiles under cold stress. In addition, we demonstrated that flanking introns are critical for the circularization of intervening exons and the length of the upstream and downstream intronic sequences can severely affect the expression level of circRNA production. We also provided new methods to overexpress circRNA in plants in this study. Compared with control plants, transgenic plants overexpressing circRNA derived from the Vv-ATS1 gene were more cold resistant. Our comprehensive analysis of circRNAs in grape provides a basis for improving the knowledge base of woody vine plants on a global scale and elevates grape to the status of a model fruit-bearing species.
Several circRNA Detection Tools Should Be Combined to Achieve Reliable Predictions in Plants
There are three major challenges for circRNA detection in RNA-seq, including discrimination between circRNAs and other types of non-colinear RNAs such as transspliced RNAs and genetic rearrangements; various types of false positive (e.g. sequencing errors, alignment errors, and in vitro artifacts); and biased identification results arising from different circRNA-detecting methods or sequencing data generated from different RNA treatments (Chen et al., 2015). Some approaches have been developed to solve the problem, e.g. computational and experimental strategies, such as RNase R digestion, have been combined to identify circRNAs, but more work is needed (Szabo and Salzman, 2016; Chu et al., 2018a). Computational identification of circRNAs from the total RNA-seq data, which received significant attention, serves as the foundation for investigating the role of these molecules. Approximately 11 circRNA prediction methods have been developed for animal datasets, and attempts have been made to compare these packages (Hansen et al., 2016; Zeng et al., 2017a; Hansen, 2018). Dramatic differences between the algorithms and different conclusions have been drawn with respect to their performance (Hansen et al., 2016; Song et al., 2016; Zeng et al., 2017a; Gao and Zhao, 2018). The UROBORUS pipeline is an efficient tool for detecting circRNAs expressed at low levels within total RNA-seq data (Song et al., 2016). When considering precision and sensitivity, compared with UROBORUS, circRNA_finder, find_circ, MapSplice, NCLScan, PTESFinder, DCC, and Segemehl, CIRI, CIRCexplorer, and KNIFE achieve better balanced performance (Zeng et al., 2017a). The software program PcircRNA_finder was developed to predict specific circRNAs in plants (Chen et al., 2016). However, PcircRNA_finder is limited to exonic circRNA prediction, and its reliability and accuracy have not been reported (Chen et al., 2016). Thus, most studies still use animal circRNA prediction software to predict plant circRNAs (Chu et al., 2018b). In the current study, to improve circRNA identification, we used three tools, find_circ, CIRI, and CIRCexplorer, for genome-wide identification of circRNAs in grape. CIRCexplorer identified the most circRNAs, followed by find_circ and CIRI. These results differed from those in humans, in which, among the three methods, CIRI identified the most circRNAs (Yang et al., 2012; Hansen et al., 2016; Zeng et al., 2017a). In addition, circRNAs predicted by all three methods accounted for a small proportion of the total predicted circRNAs, which indicated tremendous differences between the algorithms when they were applied to grape circRNA libraries. The success rates of circRNAs predicted by a single algorithm differed between cloning and sequencing. Find circ yielded the highest success rate, followed by CIRI and CIRCexplorer. However, in humans, CIRCexplorer and MapSplice generated the most reliable lists of circRNAs (Yang et al., 2012; Hansen et al., 2016). The large differences between mammal and plant genomes may influence the prediction accuracy and sensitivity of detecting circRNAs (Chen et al., 2016). Thus, before more comprehensive performance tools are available, several algorithms should ideally be combined to achieve reliable predictions in both animal and plant species (Hansen et al., 2016, 2018; Gao and Zhao, 2018). New detection methods with more comprehensive ability and higher precision for predicting circRNAs will contribute to revealing the complexity of the circRNA world in plants.
Characteristics and Biogenesis of Grape circRNAs
Recent studies have revealed that circRNAs are widespread in both animals and plants. We found that many interesting characteristics of grape circRNAs were similar to those in other species, including a high proportion of exons, a bias for long flanking introns, and tissue-specific expression patterns. Additionally, grape circRNAs exist as different isoforms, including those that undergo alternative back splicing and display AS circularization patterns. However, the highest proportion of intergenic circRNAs occur in kiwifruit (Actinidia chinensis Planchon; Wang et al., 2017d) and potato (Solanum tuberosum; Zhou et al., 2017), suggesting that variations in circRNAs occur between different species. Previous studies have revealed that circRNAs are conserved between the dicots/monocots divide (Ye et al., 2015; Chen et al., 2017a; Zhao et al., 2017), and some grape circRNAs are also conserved by comparing other plant circRNA sequences, which suggests a common biogenesis mechanism in circRNAs. The relatively low conservation of grape circRNAs compared with those in other plants may be due either to different tissues used in different plants, or to different circRNA-detecting methods (Chu et al., 2018b). In addition, a nonconserved circRNA, circAmotl1, specifically expressed in humans, but not in mice or rats, has a protective effect in Doxorubicin-induced cardiomyopathy (Zeng et al., 2017b).
Revealing circRNA biogenesis gives rise to investigations of possible circRNA functions in plants. Perhaps the current study provides further evidence that sequence determinants are sufficient for triggering circRNA biogenesis irrespective of the plant of origin. Based on our results, concerning circRNA_1975, circRNA_4328, circRNA_4363, and circRNA_7172, and those of previous studies on plants, we have drawn five conclusions with respect to the generation of circRNAs in plants. (1) Instead of a low proportion in rice (206 of 2806; Ye et al., 2017), the majority of grape circRNAs were flanked by canonical GT/AG splicing signals, which is in agreement with results in cotton (Gossypium hirsutum; Zhao et al., 2017) and Arabidopsis (Sun et al., 2016a), indicating that the canonical spliceosomal machinery is involved in back-splicing reactions in most plant species. (2) Few reverse complementary sequences in immediately flanking introns of exonic circRNAs were found in grape, which also is in agreement with results in rice (Lu et al., 2015; Ye et al., 2015), Arabidopsis (Ye et al., 2015), and cotton (Zhao et al., 2017). After constructing vectors that contain different lengths of flanking introns, we demonstrated that the flanking introns were critical for exon circularization. However, we observed that small introns (20–50 nt) that harbored splice sites were sufficient to support circRNA production. Most strikingly, these small introns contain hardly any limited complementary sequences, which indicates that intron pairing-driven circularization does not seem to be the main mechanism of circRNA formation in grape. (3) Generation of Vv-circRNA_0708 was not accompanied by ES events, suggesting that not all circRNAs were generated by ES in grape. This finding is in accordance with a recent report showing that ∼90% of cardiac circRNAs in humans and mice are produced from constitutive exons, whereas fewer (∼10%) are produced from ES events (Aufiero et al., 2018). In addition, it has been speculated that a subset of circRNAs in Arabidopsis likely contribute to the increased splicing efficiency of their cognate ES mRNAs (Conn et al., 2017). These findings indicate that, in addition to the production of an exon-containing lariat, other factors contribute to plant circRNA biogenesis. (4) Interestingly, the sequence length of the flanking introns of circularized exon(s) can significantly alter circularization efficiency (Fig. 4; Supplemental Fig. S5), and it might be that RNA secondary structures or RNA-binding proteins play substantial roles in circRNA biogenesis (Conn et al., 2015; Kramer et al., 2015). (5) In terms of expression levels, circRNAs that are more abundant than their linear counterparts have not been observed in grape or other plant species, suggesting that the efficiency of back-splicing is far less than that of canonical splicing.
Construction of Efficient and Accurate circRNA OE Vectors
Several plasmid construction methods have been used to circularize RNAs in animals, including complementary sequence-mediated circularization (Hansen et al., 2013; Zhang et al., 2014), flanking sequence-mediated circularization (Kramer et al., 2015; Li et al., 2015), and circularization of transfer RNA introns (Noto et al., 2017). Moreover, the Drosophila Laccase2 and human zinc finger protein with KRAB and SCAN domains1 flanking intronic sequences have been optimized so that they can efficiently express “designer” exonic circRNAs in human and fly cells (Kramer et al., 2015; Wilusz, 2017). In addition, a highly efficient and accurate strategy based on the classic complementary sequence-mediated circRNA circularization was developed for overexpressing circRNAs in mammals (Liu et al., 2018). circRNAs in various plant species have been identified, but their functions are currently mostly unknown. This lack of knowledge is due in part to the lack of clear methods available for generating circRNAs efficiently and accurately in plants. Two studies (Lu et al., 2015; Conn et al., 2017) have exploited the feature by which pairing complementary reverse sequences can promote circRNA biogenesis in animals (Hansen et al., 2013; Zhang et al., 2014). However, OE of plasmids can lead to the generation of many undesirable circRNAs in rice (Lu et al., 2015), and no Sanger sequencing results concerning back-splicing sites are available for Arabidopsis (Conn et al., 2017). Therefore, an efficient and accurate circRNA expression strategy is still urgently needed in plants.
Complementary sequence-mediated RNA circularization produced a large number of circular transcripts of different lengths, which is in agreement with previous results (Fig. 5D; Supplemental Fig. S6C; Lu et al., 2015). The addition of splice sites (GT/AG) flanking the desired exons could not eliminate this handicap (Fig. 5D; Supplemental Fig. S6C). Here, using flanking intron sequences supplemented by reverse complementary sequences, we improved the expression of circRNAs to levels higher than those resulting from flanking sequence-mediated circularization and ensured the accuracy of circularization. A similar strategy for overexpressing circRNAs in mammalian cell lines has been reported (Liu et al., 2018). The length of the endogenous flanking genomic sequence should not be <100 bp for the accurate expression of circRNAs. Such sequence length has been recommended to be ∼200 bp in mammals (Liu et al., 2018). However, three (circRNA_1975, circRNA_4328, and circRNA_4363) of five (circRNA_1975, circRNA_4328, circRNA_4363, circRNA_7172, and circRNA_0708) circRNAs produced a large number of undesirable linear-form transcripts, in which case the counterpart linear OE constructs are needed to define circRNA functions. Nevertheless, on the basis of our several attempts, this method was the most feasible for overexpressing circRNAs. Of course, relatively more universal and efficient OE vectors remain to be investigated in plants.
Function of Vv-circATS1 in the Cold Stress Response
To date, several reports have described the involvement of plant circRNAs in the stress response (Ye et al., 2015; Zuo et al., 2016; Wang et al., 2017b). In addition, previous research has provided evidence that the functions of circRNAs are closely related to their host genes. Such functions include promoting the transcription of host genes via binding with RNA polymerase II in the nucleus (Li et al., 2015), decreasing the expression of linear RNA (Tan et al., 2017), and promoting ES (Conn et al., 2017). Thus, the cognate linear genes of circRNAs are always used for KEGG and GO functional enrichment analyses to determine the potential function of circRNAs (Wang et al., 2017b; Zhou et al., 2017; Pan et al., 2018; Zeng et al., 2018). In this study, cold response represented differentially enriched GO terms under cold stress, implying that circRNA host genes participate in the cold response (Supplemental Fig. S12, A and C).
Interestingly, our results revealed positive and weak correlations between the expression of grape circRNAs and that of their cognate linear genes, which was in accordance with previous works in Arabidopsis, rice, and barley (Hordeum vulgare; Fig. 2A; Ye et al., 2015; Darbani et al., 2016; Wang et al., 2017d). The relationship between the expression of circRNAs and their cognate linear genes might mean that specific spliceosome-associated proteins may play an important role in regulating the formation of circRNAs. In our case, during cold stress, Vv-circATS1 did not affect the expression of its cognate mRNA (Fig. 8E). The expression of Vv-circATS1 circRNA decreased in the early stage of cold stress. After 24 h of cold treatment, the grape plants were cold, but the leaves started to recover. At this time, the host gene tended to generate more circRNAs for participation in cold resistance (Fig. 8E). This result indicates that the splicing of the circRNA and linear RNA was controlled by different spliceosome-associated proteins. The circRNA formation mechanism involving splicing factors and the regulatory mechanism in response to cold stress would be notable research areas. Unlike the results of two reports by Conn et al. (2017) and Tan et al. (2017), Vv-circATS1 did not seem to be involved in the regulation of its linear isoforms. Genetic evidence suggested that Vv-circATS1-OE lines are insensitive to low temperatures, which is similar to the results for lines overexpressing ATS1 in Arabidopsis (Ariizumi et al., 2002) and tomato (Sui et al., 2007). These findings suggest that, similar to animal circRNAs, plant circRNAs may also regulate gene expression at multiple levels (Lee et al., 2017; Li et al., 2017).
A number of reports have shown that circRNAs can function as miRNA sponges in animals. Several studies have reported that plant circRNAs may be competing endogenous RNAs (Zuo et al., 2016; Liu et al., 2017a; Wang et al., 2017b, 2017c). In plants, miRNAs match their target gene in accordance with a strict complementary base-pairing method (usually a 2- to 4-bp mismatch), and they do not have a seeding region. These properties suggest that plant circRNAs seem to lack basic features in terms of their potential as miRNA sponges. The low abundance of plant circRNAs may have occurred because target cleavage is the predominant functional mode of miRNAs in plants. miRNA-mediated cleavage may be one of the ways in which circRNAs are degraded in plants (Li et al., 2017). In this study, many miRNAs in Vv-circATS1-OE lines still exhibited specific expression patterns under cold stress. With the exception of the downregulation of miR408 in the Vv-circATS1-OE plants compared with the wild-type plants under cold stress (Ma et al., 2015), the altered expression of several miRNAs may help explain the tolerance of Vv-circATS1 transgenic plants to cold stress. miR156 and miR396, which positively regulate cold tolerance (Zhang et al., 2016a; Yang et al., 2017b), were more abundant in Vv-circATS1-OE plants when compared with wild-type plants under cold stress, and miR165 and miR398, which have been shown to negatively regulate cold tolerance in Arabidopsis (Chen et al., 2013; Yan et al., 2016), were significantly reduced in the Vv-circATS1-OE plants. In addition, miR164 regulates senescence and cell death in Arabidopsis leaves, and its expression levels were greater in the Vv-circATS1-OE plants than in the wild-type plants (Kim et al., 2009; Li et al., 2013b; Qiu et al., 2015).
mRNA-seq analysis was employed to uncover how Vv-circATS1 works together with other genes to enhance the cold tolerance. We found that a multitude of stimulus-responsive genes, such as CSD2, PRXCA, PME41, LOX3, and WRKY48, were upregulated in the Vv-circATS1-OE plants. However, it is worth noting that Vv-circATS1 was not able to activate expression of CBF genes under cold conditions, indicating its roles in cold responses through a CBF-independent pathway. Moreover, seven genes involved in the toxin catabolic process, including WRKY28, UGT72E2, ChlADR, GSTU3, CSD2, CXE6, and FH8, were upregulated in the Vv-circATS1-OE plants. In addition, our results also revealed that Vv-circATS1 could activate the transcript of the Pro transport genes (WRKY48, PME41, FH8, CHrC1, and PMEI11) in transgenic Arabidopsis. Regulation of Pro transport is essential for stress tolerance (Kavi Kishor et al., 2005; Lehmann et al., 2010; Kavi Kishor and Sreenivasulu, 2014). Under low temperature and illumination, downregulation of functional PSII is an important protective mechanism (Savitch et al., 2002; Ensminger et al., 2004). The downregulation of PSII may contribute to match the energy requirements for CO2 fixation in the Vv-circATS1-OE plants under low temperature (Huner et al., 1993). It therefore could be speculated that Vv-circATS1 regulates plant cold responses by regulating the expression of miRNAs and stimulus-responsive genes (Supplemental Fig. S17).
MATERIALS AND METHODS
Plant Material and Treatments
For genome-wide identification of circRNAs, plants of the grape (Vitis vinifera) cultivar ‘Muscat Hamburg’ were grown in a vineyard at Shanghai Jiao Tong University, Shanghai, China (31°11′N, 121°29′E). The cultivation medium was a mixture of loam and perlite (1:1). All vines were spaced 1.5 × 2.0 m in north-south oriented rows, and the shoots were trained vertically, with two shoots per vine. The vines were maintained under rain-shelter cultivation. Nutrition and irrigation management was performed as described previously (Wang et al., 2012). Tissues of grape berries, leaves, young apical shoot internodes (stems), flowers, and roots were sampled from the ‘Muscat Hamburg’ plants (three replications) during the 2016 season.
For cold stress treatment, 1-year-old self-rooted ‘Muscat Hamburg’ cuttings were planted in a mixture of loam and manure (1:1, v/v) in plastic tubs in a greenhouse. Cold treatments were performed in a refrigerated chamber maintained at 4°C. The cold treatment was started at 10:00 am under constant light at an intensity of 1000 μmol m−2 s−1. Cuttings with four well-developed leaves were treated, and the well-developed leaves were collected from three independent replications after 0 (used as a control), 2, 4, 8, 12, 24, 48 and 72 h. All the leaf samples were quickly frozen in liquid nitrogen and then stored at –80°C for further analysis. The H2O2 content, MDA content, superoxide dismutase activity, and peroxidase activity were measured by various ready kits from the Nanjing Jiancheng Bioengineering. EL assays were performed immediately with fresh samples as previously described (Ozden et al., 2009). The relative expression of VvCBF1, VvCBF2 (Karimi et al., 2015), and VvERF057 (Sun et al., 2016b) was measured to verify the effects of low-temperature treatment on the cold response of grape at the transcriptional level.
Arabidopsis (Arabidopsis thaliana) and Nicotiana benthamiana plants were cultured in controlled-environment chambers under long-day (16 h light/8 h dark) conditions and a light intensity of 600 μmol m−2 s−1 (produced by cool-white fluorescent lamps) at a temperature of 22°C.
circRNA-Seq on ‘Muscat Hamburg’ Plants
RNA was extracted from various grape tissues using a mirVana miRNA Isolation Kit (AM1561, ThermoFisher Scientific). For genome-wide identification, the total RNA from the grape berries, leaves, young apical shoot internodes (stems), flowers, and roots was mixed in equal proportions. Approximately 10 μg of the total RNA was used to deplete ribosomal RNA (rRNA) using a TruSeq Stranded Total RNA LT with the Ribo-Zero Plant kit (Cat. No. RS-122-2401, Illumina). The rRNA-depleted RNAs were further incubated at 37°C for 1 h with 10 U μg−1 RNase R (Epicentre). Sequencing and bioinformatic analysis was conducted by Shanghai Oe Biotech. Sequencing libraries were constructed as described previously (Zuo et al., 2016). Paired-end sequencing was performed on an Illumina HiSeq 2500 platform (Illumina) by Shanghai Oe Biotech). Clean reads were obtained by removing reads containing adapters using the NGS QC Toolkit (v 2.3.3; Patel and Jain, 2012) and by removing both low-quality reads and those containing poly-N from the raw data. The resulting high-quality clean reads were mapped to the reference grape genome sequence (v 2.1; http://genomes.cribi.unipd.it/DATA/V2/) by BWA-MEM (v 0.7.5a; Li and Durbin, 2009). For genome-wide identification, the software find_circ (v 1.2), CIRCexplorer (v 1), and CIRI (v 2.0.3) were used to extract potential back-splice sites, and CIRI-AS (v 1.2) was used to detect AS events within the circRNAs. The RPM value was used to determine the relative expression levels of the circRNAs. find_circ (v 1.2), CIRCexplorer (v 1), and CIRI (v 2.0.3) were used to identify circRNAs that respond to low temperatures in grape leaves with three biological repeats. The expression level of the circular transcripts was determined as the FPKM. Differentially expressed circRNAs between the two treatment samples were assessed using the DESeq R package (Anders and Huber, 2012). Genes were considered differentially expressed when they met the following criterion: absolute value of log2 (fold change) >1.
Analysis of circRNA Characteristics in Grape
The intron lengths within the circular/linear transcripts was calculated according to current annotations (v 2.1; Vitulo et al., 2014).
With respect to the conservation analysis of circRNAs, the circRNA information was collected from PlantcircBase (http://ibi.zju.edu.cn/plantcircbase/; Release 1) and then Nucleotide Blast was used (E ≤ 1e-5) to align the circRNAs identified in this study against those from Arabidopsis, soybean (Glycine max), rice (Oryza sativa), trifoliate orange (Poncirus trifoliata), tomato (Solanum lycopersicum), and maize (Zea mays).
The upstream flanking intronic sequences were used to BLAST the downstream flanking intronic sequences to detect reverse complementary sequences; the BLAST parameters -ungapped, -word_size 6 and -penalty -1 were used.
Prediction of the miRNA Targets of circRNAs and Functional Annotation Analysis of their Host Genes
Grapevine miRNAs were collected by merging the miRBase (Kozomara and Griffiths-Jones, 2014) and miRVIT databases (Chitarra et al., 2018). The miRNA-targeted circRNAs were predicted by psRNATarget with an expectation ≤5 (http://plantgrn.noble.org/psRNATarget/; Dai et al., 2018). Using hypergeometric distribution, the GO term enrichment and KEGG pathway enrichment analyses of the host genes of all the differentially expressed circRNAs were examined in the comparisons 4 h versus 0 h, 12 h versus 0 h, and 12 h versus 4 h, respectively. All p-values were corrected for multiple testing using Benjamini and Hochberg’s false discovery rate. GO terms were considered significant if their p-value was <0.05 after correcting for multiple testing.
Validation of circRNAs in Grape
The total RNA was isolated from grape leaves by an RNAprep Pure Plant Kit (DP441, TIANGEN Biotech) in accordance with the manufacturer’s instructions and then reverse transcribed to cDNA using random primers, which were then used as templates for RT-PCR. To confirm the grape circRNAs that were predicted by the software, a set of divergent primers was designed using Primer Express 3.0.1 software (Supplemental Table S15). PCR was carried out using these divergent primers and cDNA templates. The PCR procedure was as follows: 94°C for 3 min; 42 cycles at 94°C for 20 s, 57°C for 20 s, and 72°C for 20 s; and then 1 cycle at 72°C for 5 min. For PCR, 2× Taq master mix (Novoprotein) was used. The PCR products were separated using agarose gel electrophoresis and then purified with a TaKaRa Bio MiniBEST Agarose Gel DNA Extraction Kit (v 4.0). Sanger sequencing was performed to further confirm the presence of the back-spliced junction sites.
RT-qPCR Analysis
With respect to semiquantitative PCR, divergent PCR products were amplified for 38 cycles, while N. benthamiana NbActin products were amplified for 24 cycles. RT-qPCR detection was performed to evaluate the expression levels of circRNAs and circRNA host genes using PrimeScript RT master mix (TaKaRa Bio) with a CFX connect Real-Time PCR Detection System (Bio-Rad). The amplified primers and internal controls used are listed in Supplemental Table S15. The RT-qPCR procedure for the circRNAs and target genes was as follows: 95°C for 30 s followed by 40 cycles of 95°C for 5 s and then 60°C for 10 s. After RT-qPCR amplification, the melting curve and amplification curve were examined to evaluate specific amplification. The relative expression levels were analyzed by the 2−ΔΔct method. Actin was used as an internal control for the circRNAs and target genes. All RT-qPCRs were performed in triplicate.
Construction of Expression Plasmids
All constructs were cloned into pHB (2× 35S promoter) binary plasmids. The genomic region of circRNA_1218, circRNA_1975, circRNA_4328, circRNA_4363, circRNA_4473, circRNA_5664, and circRNA_7172 with or without flanking introns was amplified from grape and then cloned into a pHB vector (Fig. 3, B and C) to obtain a construct for the wild-type expression of circRNA. A series of deletions at the 5′ or 3′ end or at both the 5′ and 3′ ends of the flanking introns of circRNA_1975 and circRNA_7172 were further obtained with PCR amplification, after which they were inserted separately into a pHB vector (Fig. 4; Supplemental Fig. S5). The strategy for generating the circRNA OE constructs is shown in Figures 5 and 6 and in Supplemental Figures S6 and S8. Partial sequences from the grape VIT_13s0074g00100 (intron 1, 416 bp) or VIT_201s0011g00550 (intron 2, 461 bp) genes were cloned into a pHB vector in reverse complementary order with the HindIII/BamHI and PstI/XbalI sites, respectively. The circRNA locus was then amplified from the cDNA and inserted into the middle of the intron (Fig. 5; Supplemental Fig. S6). Furthermore, the circRNA locus amended with a flanking intron was amplified from the DNA and ligated into the middle of intron 1 (Fig. 6; Supplemental Fig. S8). All recombinant plasmids were generated using a ClonExpress II One Step Cloning Kit (Vazyme Biotech). All the primers used for vector construction are listed in Supplemental Table S15.
Transient Agrobacterium tumefaciens-mediated Expression Assays with N. benthamiana
A. tumefaciens strain GV3101 containing recombinant plasmids was grown at 28°C in Luria-Bertani medium that was supplemented with kanamycin and rifampin. When the cells reached an optical density at 600 nm of ∼1.0, the cells were harvested and resuspended in infection buffer [10 mm MgCl2, 10 mm 2-(N-morpholino) ethanesulfonic acid (MES; pH 5.8), and 100 µm acetosyringone], which was then shaken for 4 h at 28°C before the cells were used for infiltration. A. tumefaciens-mediated infiltration was performed as previously described (Dong et al., 2007; Chai et al., 2011; Li et al., 2013a). To examine the expression of the circRNAs and their corresponding linear RNAs, the leaves were collected at 4 d after infiltration. Divergent primers detected circular RNAs. Convergent primers detected both linear and circular transcripts. As a control experiment, no-RT samples were used to prove that circRNAs were not amplified from RNA.
Arabidopsis Transformation
The sense vector was genetically introduced into Arabidopsis using A. tumefaciens-mediated transformation as described in a previous report (Zhang et al., 2006). Transformants were selected based on their resistance to the herbicide Basta. Positive transgenic seedlings were grown in pots containing a mixture of peat and vermiculite (1:1, v/v) to select for T2 and T3 generations.
FISH
In situ hybridization was performed using specific probes of Vv-circATS1 (Supplemental Table S15). Probe for the back-splice region of Vv-circATS1 (antisense-Vv-circATS1) was labeled with fluorescein isothiocyanate dye in the 5′ terminal, probe for the linear region only was labeled with trimethine cyanine dye in the 5′ terminal (antisense-Vv-ATS1 mRNA). The FISH experiment was performed as previously described (Iglesias-Fernández et al., 2013; Hernández-Castellano et al., 2017). Grape leaves were fixed in 5% (v/v) formaldehyde, 5% (v/v) acetic acid, and 63% (v/v) alcohol. After dehydrating, dissecting, and deparaffinizing the samples, tissue sections were permeabilized with proteinase K in Tris-EDTA buffer at 37°C for 1 h. Slides were then rinsed three times in phosphate-buffered saline for 5 min. After prehybridization at 37°C for 1 h, tissues and RNA probes (6 ng/μL) were incubated at 37°C overnight. Slides were washed with 2× standard sodium citrate (SSC) at 37°C for 10 min, twice with 1× SSC at 37°C for 2 min, and 0.5× SSC at 37°C for 10 min. Nuclei were counterstained with 4,6-diamidino-2-phenylindole for 8 min. The images were acquired by digital fluorescence microscopy (Eclipse Ti-SR; Nikon).
Nuclear-Cytoplasmic Fractionation
Cytoplasmic and nuclear fragments were isolated from grape leaves as described previously (Wang et al., 2011). RT-PCR was performed to examine the levels of Vv-circATS1, nuclear control transcript (U6), and cytoplasmic control transcript (Actin) in these fractions.
Cold Tolerance Assays for Transgenic Arabidopsis
Cold tolerance assays for transgenic Arabidopsis were performed on Murashige and Skoog (MS) medium. For the MS medium assay, after they were surface sterilized, 25 seeds of both wild-type and transgenic Arabidopsis plants were sown on one-half strength MS media that were supplemented with 1.5% (w/v) Suc and 0.7% (w/v) agar. The seeds were refrigerated at 4°C for 3 d and then incubated in a growth chamber at 22°C with a 16 h light/8 h dark photoperiod. Four-day-old plants were treated with low temperature (4°C). Four-day-old plants grown on MS medium under continuous light were treated with low temperature (4°C) for 1 month. Chlorophyll was extracted using 95% (v/v) ethanol and analyzed using UV spectrophotometry (Choi et al., 2014). The EL assays were performed as described (Liu et al., 2017b). The content of MDA was measured using kits that were provided by the Nanjing Jiancheng Bioengineering Institute.
miRNA Transcriptomic Analysis and mRNA Seq Analysis in Transgenic Arabidopsis
Twenty-day-old wild-type and transgenic Arabidopsis seedlings were kept at 4°C until harvest. Mature leaves were harvested at 24 h after treatment. For miRNA transcriptomic analysis, isolation of small RNAs and preparation of small RNA libraries were performed based on a previously described procedure (Liang et al., 2015; Feng et al., 2017) with two biological replicates. Then, sequencing was performed on an Illumina Hiseq2500 based on the manufacturer’s protocol. Bioinformatic analysis of sequencing data and miRNA prediction were processed as described by Feng et al. (2017). Briefly, after sequencing, raw reads were filtered to remove low-quality reads, adapter sequences, reads >41 nt and <18 nt in length and ‘N’ contaminations. The filtered clean reads were mapped to Repbase and Rfam database (v 10.1) to exclude ncRNAs (rRNA, small nuclear RNA, small nucleolar RNA, and tRNA) and repeat gene sequences. To identify known miRNAs, all the unmapped sequences were blasted against Arabidopsis miRNAs present in the miRBase database. The remaining unmatched sequences were analyzed by the miRDeep2 algorithm (Friedländer et al., 2012) to identify novel miRNAs. The miRNA expression level was analyzed using transcripts per million (Sun et al., 2014). DESeq library in the R statistical software package was then used to identify the differentially expressed miRNAs (Anders and Huber, 2012).
For mRNA-seq analysis, RNA-seq libraries were generated using the Truseq Stranded mRNA LT sample prep kit (Illumina). The Illumina HiSeq2500 paired-end sequencing was conducted by Shanghai Oe Biotech. The gene expression values were normalized as FPKM. Differential expression analysis between wild-type and transgenic Arabidopsis seedlings (two biological replicates for each group) was performed using the DESeq R package. Genes with at least 1.5-fold change in expression between the two groups and a p-value <0.05 were considered differentially expressed. GO enrichment was performed using the web-based tool Singular Enrichment Analysis of agriGO with the default parameters (Tian et al., 2017).
Statistical Analysis
RT-qPCR and physiological data were analyzed using ANOVA, and the averages were compared by the Duncan’s New Multiple Range test (P < 0.05) using SPSS Statistics 17.0. Pearson’s correlation analysis was applied to measure the significance of the correlation of the expressions between circRNAs and their host genes.
Accession Numbers
Genes and accession numbers are as follows: Vv-ATS1, VIT_202s0012g02160; AtATS1, AT1G32200; AtGPAT1, At1g06520; AtGPAT2, At1g02390; AtGPAT3, At4g01950; AtGPAT4, At1g01610; AtGPAT5, At3g11430; AtGPAT6, At2g38110; AtGPAT7, At5g06090; AtGPAT8, AT4G00400, and AtGPAT9, AT5G60620. The data sets of circRNA-Seq for genome-wide identification on ‘Muscat Hamburg’ Plants (SRP152999), circRNA-Seq on grape leaves under cold treatment (SRP151394), miRNA-Seq on Arabidopsis (SRP151733), and mRNA-Seq on Arabidopsis (SRP173029) were generated and deposited in the NCBI Sequence Read Archive.
Supplemental Data
The following supplemental information is available:
Supplemental Figure S1.Features of grape circRNAs.
Supplemental Figure S2. Experimental testing of grape circRNA predictions by three algorithms (find_circ, CIRI, and CIRCexplorer).
Supplemental Figure S3. Experimental analysis of circRNAs in grape.
Supplemental Figure S4. The full-length grape VIT_211s0016g03830 gene (the host gene of circRNA_4363) can induce back splicing and exon circularization via heterologous expression in N. benthamiana.
Supplemental Figure S5. The VIT_05s0124g00390 gene generates a circRNA, and long intronic sequences facilitate circRNA_1975 production.
Supplemental Figure S6. The duplication test of the strategy for overexpressing circRNAs is shown in Figure 5; the target sequence was circRNA_4363.
Supplemental Figure S7. Sanger sequencing results of the circular sequences of PCR products generated by divergent primers.
Supplemental Figure S8. The endogenous flanking genomic sequence of the circRNA_1975-forming sequence, which can be cloned between the two complementary intron fragments, influenced the accuracy and efficiency of its mature circRNA.
Supplemental Figure S9. The physiological response and expression of marker genes of grape leaves under cold stress.
Supplemental Figure S10. The coefficient matrices of the independent biological repeats identified by three tools, find_circ, CIRI, and CIRCexplorer.
Supplemental Figure S11. Hierarchical clustering analysis of differentially expressed circRNAs.
Supplemental Figure S12. Pathway enrichment analyses.
Supplemental Figure S13. Prediction of circRNA-miRNA networks in the leaves of grape cuttings under cold stress (4°C).
Supplemental Figure S14. Phylogenetic tree showing the relationships between Vv-ATS1 in grape and GPAT family proteins in Arabidopsis.
Supplemental Figure S15. Cold stress altered the expression patterns of low temperature-related miRNAs in Vv-circATS1-OE lines.
Supplemental Figure S16. Differentially expressed genes affected by OE of Vv-circATS1 under cold conditions.
Supplemental Figure S17. VvcircATS1 biogenesis and its function in the modulation of cold stress.
Supplemental Table S1. The annotation of circRNAs in grape.
Supplemental Table S2. The expression profiles of circRNAs in grape; expression values were determined by the spliced reads per million (RPM).
Supplemental Table S3. Alternative splicing detection in CIRI-AS.
Supplemental Table S4. The relative expression levels (FPKM) and annotation of circRNAs detected by find_circ from control and cold sample in grape leaves.
Supplemental Table S5. The relative expression levels (FPKM) and annotation of circRNAs detected by CIRI from control and cold sample in grape leaves.
Supplemental Table S6. The relative expression levels (FPKM) and annotation of circRNAs detected by CIRCexplorer from control and cold sample in grape leaves.
Supplemental Table S7. The differentially expressed circRNAs detected by find_circ in grape leaves under low temperature stress.
Supplemental Table S8. The differentially expressed circRNAs detected by CIRI in grape leaves under low temperature stress.
Supplemental Table S9. The differentially expressed circRNAs detected by CIRCexplorer in grape leaves under low temperature stress.
Supplemental Table S10. Predicted circRNA-miRNA connection for differentially expressed circRNAs in grape leaves under low temperature stress.
Supplemental Table S11. All miRNA discovered in Vv-cirATS1-OE and wild-type Arabidopsis under 4°C for 12 h.
Supplemental Table S12. Significant differentially expressed miRNAs between Vv-cirATS1-OE and wild-type Arabidopsis under 4°C for 12 h.
Supplemental Table S13. Significant differentially expressed mRNAs between Vv-cirATS1-OE and wild-type Arabidopsis under 4°C for 12 h.
Supplemental Table S14. Significant GO terms for differentially expressed genes between Vv-cirATS1-OE and wild-type Arabidopsis under 4°C for 12 h.
Supplemental Table S15. Primers used in this study.
Acknowledgments
We thank Amnon Lers for providing so many useful suggestions.
Footnotes
This work was supported by the National Key Research and Development Program of China (grant no. 2018YFD1000300), the National Natural Science Foundation of China (grant no. 31701888), the Shanghai Natural Science Foundation (grant no. 17ZR1448800) and the Special Funds of Modern Industrial Technology System for Agriculture (CARS-29-zp-7).
References
- Anders S, Huber W (2012) Differential expression of RNA-Seq data at the gene level: The DESeq package. European Molecular Biology Laboratory (EMBL) https://bioconductor.org/packages/release/bioc/vignettes/DESeq/inst/doc/DESeq.pdf [Google Scholar]
- Ariel F, Romero-Barrios N, Jégu T, Benhamed M, Crespi M (2015) Battles and hijacks: Noncoding transcription in plants. Trends Plant Sci 20: 362–371 [DOI] [PubMed] [Google Scholar]
- Ariizumi T, Kishitani S, Inatsugi R, Nishida I, Murata N, Toriyama K (2002) An increase in unsaturation of fatty acids in phosphatidylglycerol from leaves improves the rates of photosynthesis and growth at low temperatures in transgenic rice seedlings. Plant Cell Physiol 43: 751–758 [DOI] [PubMed] [Google Scholar]
- Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S (2014) circRNA biogenesis competes with pre-mRNA splicing. Mol Cell 56: 55–66 [DOI] [PubMed] [Google Scholar]
- Aufiero S, van den Hoogenhof MMG, Reckman YJ, Beqqali A, van der Made I, Kluin J, Khan MAF, Pinto YM, Creemers EE (2018) Cardiac circRNAs arise mainly from constitutive exons rather than alternatively spliced exons. RNA 24: 815–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett SP, Wang PL, Salzman J (2015) Circular RNA biogenesis can proceed through an exon-containing lariat precursor. eLife 4: e07540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmona MJ, Calonje M, Martínez-Zapater JM (2007) The FT/TFL1 gene family in grapevine. Plant Mol Biol 63: 637–650 [DOI] [PubMed] [Google Scholar]
- Chai YM, Jia HF, Li CL, Dong QH, Shen YY (2011) FaPYR1 is involved in strawberry fruit ripening. J Exp Bot 62: 5079–5089 [DOI] [PubMed] [Google Scholar]
- Chen G, Cui J, Wang L, Zhu Y, Lu Z, Jin B (2017a) Genome-Wide Identification of Circular RNAs in Arabidopsis thaliana. Front Plant Sci 8: 1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen I, Chen C-Y, Chuang T-J (2015) Biogenesis, identification, and function of exonic circular RNAs. Wiley Interdiscip Rev RNA 6: 563–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Hu Q, Zhang Y, Lu C, Kuang H (2014) P-MITE: A database for plant miniature inverted-repeat transposable elements. Nucleic Acids Res 42: D1176–D1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Yu Y, Zhang X, Liu C, Ye C, Fan L (2016) PcircRNA_finder: A software for circRNA prediction in plants. Bioinformatics 32: 3528–3529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Zhang P, Fan Y, Lu Q, Li Q, Yan J, Muehlbauer GJ, Schnable PS, Dai M, Li L (2017b) Circular RNAs mediated by transposons are associated with transcriptomic and phenotypic variation in maize. New Phytol 217: 1292–1306 [DOI] [PubMed] [Google Scholar]
- Chen Y, Jiang J, Song A, Chen S, Shan H, Luo H, Gu C, Sun J, Zhu L, Fang W, et al. (2013) Ambient temperature enhanced freezing tolerance of Chrysanthemum dichrum CdICE1Arabidopsis via miR398. BMC Biol 11: 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Zhang Y, Li Z, Wang T, Zhang X, Zheng B (2018) A lariat-derived circular RNA is required for plant development in Arabidopsis. Sci China Life Sci 61:204–213 [DOI] [PubMed] [Google Scholar]
- Chitarra W, Pagliarani C, Abbà S, Boccacci P, Birello G, Rossi M, Palmano S, Marzachì C, Perrone I, Gambino G (2018) miRVIT: A novel miRNA database and its application to uncover Vitis responses to Flavescence dorée infection. Front Plant Sci 9: 1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi W-G, Toyota M, Kim S-H, Hilleary R, Gilroy S (2014) Salt stress-induced Ca2+ waves are associated with rapid, long-distance root-to-shoot signaling in plants. Proc Natl Acad Sci USA 111: 6497–6502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu Q, Bai P, Zhu X, Zhang X, Mao L, Zhu Q-H, Fan L, Ye C-Y (2018a) Characteristics of plant circular RNAs. Brief Bioinform 00: 1–9 [DOI] [PubMed] [Google Scholar]
- Chu QJ, Shen EH, Ye CY, Fan LJ, Zhu QH (2018b) Emerging roles of plant circular RNAs. J Plant Cell Dev 1: 1–14 [Google Scholar]
- Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA, Goodall GJ (2015) The RNA binding protein quaking regulates formation of circRNAs. Cell 160: 1125–1134 [DOI] [PubMed] [Google Scholar]
- Conn VM, Hugouvieux V, Nayak A, Conos SA, Capovilla G, Cildir G, Jourdain A, Tergaonkar V, Schmid M, Zubieta C, et al. (2017) A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nat Plants 3: 17053. [DOI] [PubMed] [Google Scholar]
- Cooper DA, Cortés-López M, Miura P (2018) Genome-wide circRNA profiling from RNA-seq data. In Dieterich C and Papantonis A, eds, Circular RNAs: Methods and Protocols. Springer, New York: pp 27–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, Zhuang Z, Zhao PX (2018) psRNATarget: A plant small RNA target analysis server (2017 release). Nucleic Acids Res 46(W1): W49–W54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darbani B, Noeparvar S, Borg S (2016) Identification of circular RNAs from the parental genes involved in multiple aspects of cellular metabolism in barley. Front Plant Sci 7: 776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Burch-Smith TM, Liu Y, Mamillapalli P, Dinesh-Kumar SP (2007) A ligation-independent cloning tobacco rattle virus vector for high-throughput virus-induced gene silencing identifies roles for NbMADS4-1 and -2 in floral development. Plant Physiol 145: 1161–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebbesen KK, Hansen TB, Kjems J (2017) Insights into circular RNA biology. RNA Biol 14: 1035–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ensminger I, Sveshnikov D, Campbell DA, Funk C, Jansson S, Lloyd J, Shibistova O, Oquist G (2004) Intermittent low temperatures constrain spring recovery of photosynthesis in boreal Scots pine forests. Glob Change Biol 10: 995–1008 [Google Scholar]
- Feng JL, Yang ZJ, Chen SP, El-Kassaby YA, Chen H (2017) High throughput sequencing of small RNAs reveals dynamic microRNAs expression of lipid metabolism during Camellia oleifera and C. meiocarpa seed natural drying. BMC Genomics 18: 546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer JW, Leung AK (2017) CircRNAs: A regulator of cellular stress. Crit Rev Biochem Mol Biol 52: 220–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedländer MR, Mackowiak SD, Li N, Chen W, Rajewsky N (2012) miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res 40: 37–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Zhao F (2018) Computational strategies for exploring circular RNAs. Trends Genet 34: 389–400 [DOI] [PubMed] [Google Scholar]
- Gao Y, Wang J, Zhao F (2015) CIRI: An efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol 16: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Wang J, Zheng Y, Zhang J, Chen S, Zhao F (2016) Comprehensive identification of internal structure and alternative splicing events in circular RNAs. Nat Commun 7: 12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen TB. (2018) Improved circRNA identification by combining prediction algorithms. Front Cell Dev Biol 6: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J (2013) Natural RNA circles function as efficient microRNA sponges. Nature 495: 384–388 [DOI] [PubMed] [Google Scholar]
- Hansen TB, Venø MT, Damgaard CK, Kjems J (2016) Comparison of circular RNA prediction tools. Nucleic Acids Res 44: e58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Castellano S, Nic-Can GI, De-la-Peña C (2017) Localization of miRNAs by in situ hybridization in plants using conventional oligonucleotide probes. In Kovalchuk I, ed, Plant Epigenetics: Methods and Protocols. Springer US, Boston, pp 51–62 [DOI] [PubMed] [Google Scholar]
- Huner NP, Oquist G, Hurry VM, Krol M, Falk S, Griffith M (1993) Photosynthesis, photoinhibition and low temperature acclimation in cold tolerant plants. Photosynth Res 37: 19–39 [DOI] [PubMed] [Google Scholar]
- Iglesias-Fernández R, Barrero-Sicilia C, Carrillo-Barral N, Oñate-Sánchez L, Carbonero P (2013) Arabidopsis thaliana bZIP44: A transcription factor affecting seed germination and expression of the mannanase-encoding gene AtMAN7. Plant J 74: 767–780 [DOI] [PubMed] [Google Scholar]
- Ivanov A, Memczak S, Wyler E, Torti F, Porath HT, Orejuela MR, Piechotta M, Levanon EY, Landthaler M, Dieterich C, et al. (2015) Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Reports 10: 170–177 [DOI] [PubMed] [Google Scholar]
- Jaillon O, Aury JM, Noel B, Policriti A, Clepet C, Casagrande A, Choisne N, Aubourg S, Vitulo N, Jubin C, et al. ; French-Italian Public Consortium for Grapevine Genome Characterization (2007) The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449: 463–467 [DOI] [PubMed] [Google Scholar]
- Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE (2013) Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19: 141–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Ebadi A, Mousavi SA, Salami SA, Zarei A (2015) Comparison of CBF1, CBF2, CBF3 and CBF4 expression in some grapevine cultivars and species under cold stress. Sci Hortic (Amsterdam) 197: 521–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavi Kishor PB, Sreenivasulu N (2014) Is proline accumulation per se correlated with stress tolerance or is proline homeostasis a more critical issue? Plant Cell Environ 37: 300–311 [DOI] [PubMed] [Google Scholar]
- Kavi Kishor PB, Sangam S, Amrutha RN, Sri Laxmi P, Naidu KR, Sambasiva Rao KRS, Rao S, Reddy KJ, Theriappan P, Sreenivasulu N (2005) Regulation of proline biosynthesis, degradation, uptake and transport in higher plants: Its implications in plant growth and abiotic stress tolerance. Curr Sci 88: 424–438 [Google Scholar]
- Kim JH, Woo HR, Kim J, Lim PO, Lee IC, Choi SH, Hwang D, Nam HG (2009) Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis. Science 323: 1053–1057 [DOI] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S (2014) miRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 42: D68–D73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer MC, Liang D, Tatomer DC, Gold B, March ZM, Cherry S, Wilusz JE (2015) Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes Dev 29: 2168–2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasda E, Parker R (2016) Circular RNAs co-precipitate with extracellular vesicles: A possible mechanism for circRNA clearance. PLoS One 11: e0148407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SM, Kong HG, Ryu CM (2017) Are circular RNAs new kids on the block? Trends Plant Sci 22: 357–360 [DOI] [PubMed] [Google Scholar]
- Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Wade M, et al. (2017) Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell 66: 22–37.e9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann S, Funck D, Szabados L, Rentsch D (2010) Proline metabolism and transport in plant development. Amino Acids 39: 949–962 [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Ji K, Sun Y, Luo H, Wang H, Leng P (2013a) The role of FaBG3 in fruit ripening and B. cinerea fungal infection of strawberry. Plant J 76: 24–35 [DOI] [PubMed] [Google Scholar]
- Li QF, Zhang YC, Chen YQ, Yu Y (2017) Circular RNAs roll into the regulatory network of plants. Biochem Biophys Res Commun 488: 382–386 [DOI] [PubMed] [Google Scholar]
- Li X, Yang L, Chen L-L (2018) The biogenesis, functions, and challenges of circular RNAs. Mol Cell 71: 428–442 [DOI] [PubMed] [Google Scholar]
- Li Z, Peng J, Wen X, Guo H (2013b) ETHYLENE-INSENSITIVE3 is a senescence-associated gene that accelerates age-dependent leaf senescence by directly repressing miR164 transcription in Arabidopsis. Plant Cell 25: 3311–3328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, et al. (2015) Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 22: 256–264 [DOI] [PubMed] [Google Scholar]
- Li Z, Wang S, Cheng J, Su C, Zhong S, Liu Q, Fang Y, Yu Y, Lv H, Zheng Y, et al. (2016) Intron lariat RNA inhibits microRNA biogenesis by sequestering the dicing complex in Arabidopsis. PLoS Genet 12: e1006422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang D, Wilusz JE (2014) Short intronic repeat sequences facilitate circular RNA production. Genes Dev 28: 2233–2247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G, Ai Q, Yu D (2015) Uncovering miRNAs involved in crosstalk between nutrient deficiencies in Arabidopsis. Sci Rep 5: 11813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Conn V, Goodall GJ, Conn SJ (2018) A highly efficient strategy for overexpressing circRNAs. Methods Mol Biol 1724: 97–105 [DOI] [PubMed] [Google Scholar]
- Liu T, Zhang L, Chen G, Shi T (2017a) Identifying and characterizing the circular RNAs during the lifespan of Arabidopsis leaves. Front Plant Sci 8: 1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Jia Y, Ding Y, Shi Y, Li Z, Guo Y, Gong Z, Yang S (2017b) Plasma Membrane CRPK1-Mediated Phosphorylation of 14-3-3 Proteins Induces Their Nuclear Import to Fine-Tune CBF Signaling during Cold Response. Mol Cell 66: 117–128.e5 [DOI] [PubMed] [Google Scholar]
- Lu T, Cui L, Zhou Y, Zhu C, Fan D, Gong H, Zhao Q, Zhou C, Zhao Y, Lu D, et al. (2015) Transcriptome-wide investigation of circular RNAs in rice. RNA 21: 2076–2087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Gao Z, Li H, Li Q, Zhang C, Xu W, Song S, Ma C, Wang S (2018) Selection of reference genes for miRNA qRT-PCR under abiotic stress in grapevine. Sci Rep 8: 4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C, Burd S, Lers A (2015) miR408 is involved in abiotic stress responses in Arabidopsis. Plant J 84: 169–187 [DOI] [PubMed] [Google Scholar]
- Megha S, Basu U, Kav NNV (2018) Regulation of low temperature stress in plants by microRNAs. Plant Cell Environ 41: 1–15 [DOI] [PubMed] [Google Scholar]
- Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. (2013) Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495: 333–338 [DOI] [PubMed] [Google Scholar]
- Nitsche A, Doose G, Tafer H, Robinson M, Saha NR, Gerdol M, Canapa A, Hoffmann S, Amemiya CT, Stadler PF (2014) Atypical RNAs in the coelacanth transcriptome. J Exp Zoolog B Mol Dev Evol 322: 342–351 [DOI] [PubMed] [Google Scholar]
- Noto JJ, Schmidt CA, Matera AG (2017) Engineering and expressing circular RNAs via tRNA splicing. RNA Biol 14: 978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozden M, Demirel U, Kahraman A (2009) Effects of proline on antioxidant system in leaves of grapevine (Vitis vinifera L.) exposed to oxidative stress by H2O2. Sci Hortic 119: 163–168 [Google Scholar]
- Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, Hanan M, Wyler E, Perez-Hernandez D, Ramberger E, et al. (2017) Translation of CircRNAs. Mol Cell 66: 9–21.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan T, Sun X, Liu Y, Li H, Deng G, Lin H, Wang S (2018) Correction to: Heat stress alters genome-wide profiles of circular RNAs in Arabidopsis. Plant Mol Biol 96: 231. [DOI] [PubMed] [Google Scholar]
- Patel RK, Jain M (2012) NGS QC Toolkit: A toolkit for quality control of next generation sequencing data. PLoS One 7: e30619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu K, Li Z, Yang Z, Chen J, Wu S, Zhu X, Gao S, Gao J, Ren G, Kuai B, et al. (2015) EIN3 and ORE1 accelerate degreening during ethylene-mediated leaf senescence by directly activating chlorophyll catabolic genes in Arabidopsis. PLoS Genet 11: e1005399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sablok G, Zhao H, Sun X (2016) Plant circular RNAs (circRNAs): Transcriptional regulation beyond miRNAs in plants. Mol Plant 9: 192–194 [DOI] [PubMed] [Google Scholar]
- Sablok G, Powell B, Braessler J, Yu F, Min XJ (2017) Comparative landscape of alternative splicing in fruit plants. Curr Plant Biol 9–10: 29–36 [Google Scholar]
- Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO (2012) Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One 7: e30733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO (2013) Cell-type specific features of circular RNA expression. PLoS Genet 9: e1003777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitch LV, Leonardos ED, Krol M, Jansson S, Grodzinski B, Huner NPA, Öquist G (2002) Two different strategies for light utilization in photosynthesis in relation to growth and cold acclimation. Plant Cell Environ 25: 761–771 [Google Scholar]
- Shi L, Yan P, Liang Y, Sun Y, Shen J, Zhou S, Lin H, Liang X, Cai X (2017) Circular RNA expression is suppressed by androgen receptor (AR)-regulated adenosine deaminase that acts on RNA (ADAR1) in human hepatocellular carcinoma. Cell Death Dis 8: e3171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Zhang N, Han P, Moon BS, Lai RK, Wang K, Lu W (2016) Circular RNA profile in gliomas revealed by identification tool UROBORUS. Nucleic Acids Res 44: e87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starke S, Jost I, Rossbach O, Schneider T, Schreiner S, Hung LH, Bindereif A (2015) Exon circularization requires canonical splice signals. Cell Reports 10: 103–111 [DOI] [PubMed] [Google Scholar]
- Sui N, Li M, Zhao SJ, Li F, Liang H, Meng QW (2007) Overexpression of glycerol-3-phosphate acyltransferase gene improves chilling tolerance in tomato. Planta 226: 1097–1108 [DOI] [PubMed] [Google Scholar]
- Sun J, Wang S, Li C, Ren Y, Wang J (2014) Novel expression profiles of microRNAs suggest that specific miRNAs regulate gene expression for the sexual maturation of female Schistosoma japonicum after pairing. Parasit Vectors 7: 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Wang L, Ding J, Wang Y, Wang J, Zhang X, Che Y, Liu Z, Zhang X, Ye J, et al. (2016a) Integrative analysis of Arabidopsis thaliana transcriptomics reveals intuitive splicing mechanism for circular RNA. FEBS Lett 590: 3510–3516 [DOI] [PubMed] [Google Scholar]
- Sun X, Zhao T, Gan S, Ren X, Fang L, Karungo SK, Wang Y, Chen L, Li S, Xin H (2016b) Ethylene positively regulates cold tolerance in grapevine by modulating the expression of ETHYLENE RESPONSE FACTOR 057. Sci Rep 6: 24066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo L, Salzman J (2016) Detecting circular RNAs: Bioinformatic and experimental challenges. Nat Rev Genet 17: 679–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J, Zhou Z, Niu Y, Sun X, Deng Z (2017) Identification and functional characterization of tomato circRNAs derived from genes involved in fruit pigment accumulation. Sci Rep 7: 8594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian T, Liu Y, Yan H, You Q, Yi X, Du Z, Xu W, Su Z (2017) agriGO v2.0: A GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res 45(W1): W122–W129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitulo N, Forcato C, Carpinelli EC, Telatin A, Campagna D, D’Angelo M, Zimbello R, Corso M, Vannozzi A, Bonghi C, et al. (2014) A deep survey of alternative splicing in grape reveals changes in the splicing machinery related to tissue, stress condition and genotype. BMC Plant Biol 14: 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, He JJ, Duan CQ, Yu XM, Zhu LN, Xie ZS, Zhang CX, Xu WP, Wang SP (2012) Root restriction affects anthocyanin accumulation and composition in berry skin of ‘Kyoho’ grape (Vitis vinifera L. x Vitis labrusca L.) during ripening. Sci Hortic 137: 20–28 [Google Scholar]
- Wang J, Meng X, Dobrovolskaya OB, Orlov YL, Chen M (2017a) Non-coding RNAs and their roles in stress response in plants. Genomics Proteomics Bioinformatics 15: 301–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Yang Y, Jin L, Ling X, Liu T, Chen T, Ji Y, Yu W, Zhang B (2018) Re-analysis of long non-coding RNAs and prediction of circRNAs reveal their novel roles in susceptible tomato following TYLCV infection. BMC Plant Biol 18: 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Ye R, Xin Y, Fang X, Li C, Shi H, Zhou X, Qi Y (2011) An importin β protein negatively regulates microRNA activity in Arabidopsis. Plant Cell 23: 3565–3576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wang Q, Gao L, Zhu B, Luo Y, Deng Z, Zuo J (2017c) Integrative analysis of circRNAs acting as ceRNAs involved in ethylene pathway in tomato. Physiol Plant 161: 311–321 [DOI] [PubMed] [Google Scholar]
- Wang Y, Yang M, Wei S, Qin F, Zhao H, Suo B (2017b) Identification of circular RNAs and their targets in leaves of Triticum aestivum L. under dehydration stress. Front Plant Sci 7: 2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Liu Y, Li D, Li L, Zhang Q, Wang S, Huang H (2017d) Identification of circular RNAs in kiwifruit and their species-specific response to bacterial canker pathogen invasion. Front Plant Sci 8: 413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westholm JO, Miura P, Olson S, Shenker S, Joseph B, Sanfilippo P, Celniker SE, Graveley BR, Lai EC (2014) Genome-wide analysis of Drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Reports 9: 1966–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE. (2017) Circular RNAs: Unexpected outputs of many protein-coding genes. RNA Biol 14: 1007–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE. (2018) A 360° view of circular RNAs: From biogenesis to functions. Wiley Interdiscip Rev RNA 9: e1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S, Feng J, Lei L, Hu J, Xia L, Wang J, Xiang Y, Liu L, Zhong S, Han L, et al. (2017) Comprehensive characterization of tissue-specific circular RNAs in the human and mouse genomes. Brief Bioinform 18: 984–992 [DOI] [PubMed] [Google Scholar]
- Yan J, Zhao C, Zhou J, Yang Y, Wang P, Zhu X, Tang G, Bressan RA, Zhu JK (2016) The miR165/166 mediated regulatory module plays critical roles in ABA homeostasis and response in Arabidopsis thaliana. PLoS Genet 12: e1006416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Simpson JP, Li-Beisson Y, Beisson F, Pollard M, Ohlrogge JB (2012) A land-plant-specific glycerol-3-phosphate acyltransferase family in Arabidopsis: Substrate specificity, sn-2 preference, and evolution. Plant Physiol 160: 638–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, Jin Y, Yang Y, Chen LL, Wang Y, et al. (2017a) Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res 27: 626–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Zhang X, Su Y, Zou J, Wang Z, Xu L, Que Y (2017b) miRNA alteration is an important mechanism in sugarcane response to low-temperature environment. BMC Genomics 18: 833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye CY, Chen L, Liu C, Zhu QH, Fan L (2015) Widespread noncoding circular RNAs in plants. New Phytol 208: 88–95 [DOI] [PubMed] [Google Scholar]
- Ye CY, Zhang X, Chu Q, Liu C, Yu Y, Jiang W, Zhu QH, Fan L, Guo L (2017) Full-length sequence assembly reveals circular RNAs with diverse non-GT/AG splicing signals in rice. RNA Biol 14: 1055–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng rf, Zhou jj, Hu cg, Zhang jz (2018) Transcriptome-wide identification and functional prediction of novel and flowering-related circular RNAs from trifoliate orange (Poncirus trifoliata L. Raf.). Planta 247: 1191–1202 [DOI] [PubMed] [Google Scholar]
- Zeng X, Lin W, Guo M, Zou Q (2017a) A comprehensive overview and evaluation of circular RNA detection tools. PLOS Comput Biol 13: e1005420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Du WW, Wu Y, Yang Z, Awan FM, Li X, Yang W, Zhang C, Yang Q, Yee A, et al. (2017b) A circular RNA binds to and activates AKT phosphorylation and nuclear localizationrReducingaApoptosis andeEnhancing cardiac repair. Theranostics 7: 3842–3855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Henriques R, Lin SS, Niu QW, Chua NH (2006) Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat Protoc 1: 641–646 [DOI] [PubMed] [Google Scholar]
- Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L (2014) Complementary sequence-mediated exon circularization. Cell 159: 134–147 [DOI] [PubMed] [Google Scholar]
- Zhang XO, Dong R, Zhang Y, Zhang JL, Luo Z, Zhang J, Chen LL, Yang L (2016b) Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res 26: 1277–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Wang W, Wang M, Zhang HY, Liu JH (2016a) The miR396b of Poncirus trifoliata functions in cold tolerance by regulating ACC oxidase gene expression and modulating ethylene-polyamine homeostasis. Plant Cell Physiol 57: 1865–1878 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Zhu S, Yang L, Chen LL (2013) Circular intronic long noncoding RNAs. Mol Cell 51: 792–806 [DOI] [PubMed] [Google Scholar]
- Zhao T, Wang L, Li S, Xu M, Guan X, Zhou B (2017) Characterization of conserved circular RNA in polyploid Gossypium species and their ancestors. FEBS Lett 591: 3660–3669 [DOI] [PubMed] [Google Scholar]
- Zhou R, Zhu Y, Zhao J, Fang Z, Wang S, Yin J, Chu Z, Ma D (2017) Transcriptome-wide identification and characterization of potato circular RNAs in response to Pectobacterium carotovorum subspecies brasiliense infection. Int J Mol Sci 19: 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J, Wang Q, Zhu B, Luo Y, Gao L (2016) Deciphering the roles of circRNAs on chilling injury in tomato. Biochem Biophys Res Commun 479: 132–138 [DOI] [PubMed] [Google Scholar]