Seedling organs show autonomy as well as interdependency in their ability to respond to elevated temperatures.
Abstract
Plants have a remarkable capacity to adjust their growth and development to elevated ambient temperatures. Increased elongation growth of roots, hypocotyls, and petioles in warm temperatures are hallmarks of seedling thermomorphogenesis. In the last decade, significant progress has been made to identify the molecular signaling components regulating these growth responses. Increased ambient temperature utilizes diverse components of the light sensing and signal transduction network to trigger growth adjustments. However, it remains unknown whether temperature sensing and responses are universal processes that occur uniformly in all plant organs. Alternatively, temperature sensing may be confined to specific tissues or organs, which would require a systemic signal that mediates responses in distal parts of the plant. Here, we show that Arabidopsis (Arabidopsis thaliana) seedlings show organ-specific transcriptome responses to elevated temperatures and that thermomorphogenesis involves both autonomous and organ-interdependent temperature sensing and signaling. Seedling roots can sense and respond to temperature in a shoot-independent manner, whereas shoot temperature responses require both local and systemic processes. The induction of cell elongation in hypocotyls requires temperature sensing in cotyledons, followed by the generation of a mobile auxin signal. Subsequently, auxin travels to the hypocotyl, where it triggers local brassinosteroid-induced cell elongation in seedling stems, which depends upon a distinct, permissive temperature sensor in the hypocotyl.
Increases in ambient temperature affect numerous growth and developmental traits in flowering plants (Ibañez et al., 2017; for review, see Quint et al., 2016; Casal and Balasubramanian, 2019). Among these changes, collectively referred to as thermomorphogenesis (Delker et al., 2014), temperature-induced elongation of hypocotyls, petioles, and roots are hallmark responses (Fig. 1A). Molecular mechanisms involved in thermomorphogenesis have primarily been studied using model phenotypes such as temperature-induced hypocotyl elongation. Temperature changes can be sensed by the inactivation of photoreceptors such as phytochrome B (phyB). phyB function in thermoregulation operates via PHYTOCHROME-INTERACTING FACTOR4 (PIF4), which serves as the major thermomorphogenesis-signaling hub (Koini et al., 2009; Stavang et al., 2009). While light-activated phyB in its Pfr form inhibits PIF4, warmth converts phyB to the inactive Pr conformation (Jung et al., 2016; Legris et al., 2016; Delker et al., 2017). This thermal reversion (or dark reversion) seems to be most prominent in the dark or under low fluence rates, but it can also occur at higher fluence rates if the temperature is high enough (Legris et al., 2016). Other light sensors, such as UV-B RESISTANCE8 (Hayes et al., 2017), phototropins (Fujii et al., 2017), and cryptochromes (Ma et al., 2016), have also been implicated in the regulation of temperature responses, and a cooperative action can be assumed to occur under natural, complex light conditions. The inactivation of phyB (and other light sensors) ultimately results in a derepression of PIF4, which initiates auxin responses (Franklin et al., 2011). Auxin could theoretically induce elongation growth without additional players. However, based on genetic, biochemical, and pharmacological experiments, it was recently reported that auxin action in thermomorphogenesis depends on the brassinosteroid (BR)-activated transcription factor BRASSINAZOLE-RESISTANT1 (BZR1) and its homolog BRI1-EMS-SUPPRESSOR1, which activate elongation growth downstream of auxin (Ibañez et al., 2018; Martínez et al., 2018). BZR1 function may involve heteromerization with other transcription factors such as AUXIN RESPONSE FACTORs and PIF4 (Oh et al., 2012, 2014a), which can affect preferences for specific cis-element-binding sites (Martínez et al., 2018).
Figure 1.
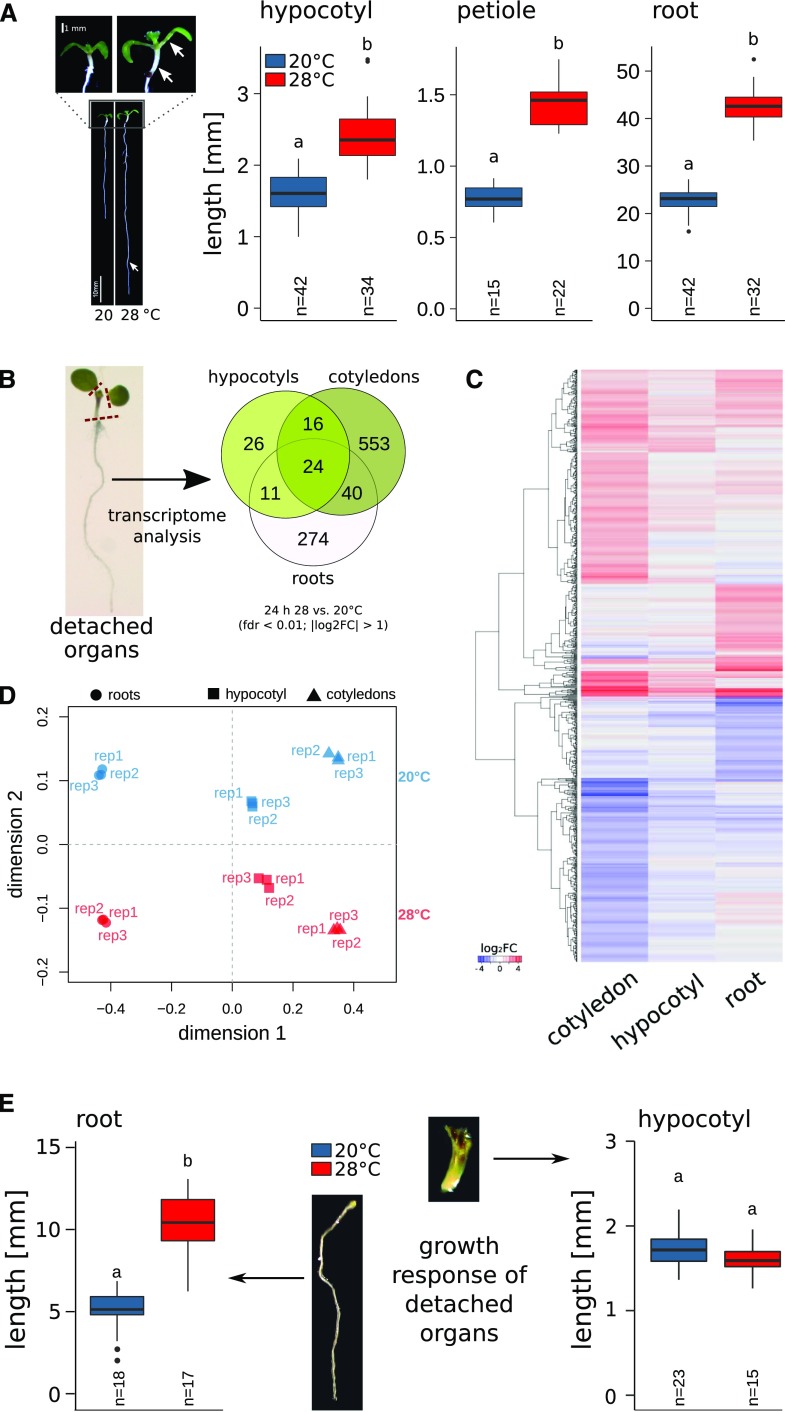
Thermomorphogenesis in seedling organs is autonomous or interdependent. A, Temperature-induced elongation of hypocotyls, roots, and petioles (indicated by white arrows) in 8-d-old seedlings grown at 20°C or 28°C. B, Temperature-induced transcriptome responses in 6-d-old seedlings. Venn diagrams show overlap in differential gene expression after 24 h at 28°C versus control seedlings at 20°C (|log2 fold change [FC]| > 1, false discovery rate [fdr] < 0.01). C, Hierarchical clustering of DEGs using Euclidian distances of log2 FC data. D, MDS of DEGs (shown in C) based on the pairwise Pearson correlation (1 − cor) among all individual biological replicates. E, Elongation responses of detached seedling organs. Petioles and cotyledons or whole shoots were removed from 4-d-old seedlings grown at 20°C. Subsequently, detached organs were placed on growth medium and cultivated for an additional 4 d at 20°C or 28°C. Experiments were performed in long-day (16/8 h) conditions under 90 µmol m−2 s−1 white fluorescent light. Box plots show medians and interquartile ranges of total organ lengths; outliers (greater than 1.5× interquartile range) are shown as black dots. Different letters denote statistical differences at P < 0.05 as assessed by one-way ANOVA and Tukey’s honestly significant difference (HSD) posthoc test.
Although our understanding of temperature sensing and signaling has greatly improved over the past decade, we tend to generalize findings based on localized responses, primarily in specific shoot tissues. It is, however, unknown whether all elongating organs are regulated similarly and whether all thermoresponsive tissues have the capacity to sense ambient temperature changes. Alternatively, sensing, signaling, and growth responses may be distinct processes that are spatially separated and tissue- or organ-specific. Such spatiotemporal specificities have been demonstrated previously for other regulatory contexts, including light responses mediated by phytochromes (for review, see Montgomery, 2016).
Using a combination of genetic, transcriptomic, physiological, and pharmacological approaches, we here show that elongation responses in the root seem largely independent of shoot-derived signals. In contrast, the hypocotyl is largely unable to trigger temperature responses by itself. Hypocotyl cell elongation requires temperature sensing in the cotyledons, where a mobile auxin signal is generated that delivers the growth stimulus to the hypocotyl. However, the cotyledon-derived auxin signal seems to be gated in hypocotyls by a second permissive thermosensor that determines the capacity for the induction of cell elongation.
RESULTS AND DISCUSSION
Elevated ambient temperature triggers the elongation of petioles, hypocotyls, and roots in Arabidopsis (Arabidopsis thaliana) seedlings (Fig. 1A). First indications for putative specificities in these responses were obtained from organ-specific transcriptome analyses 24 h after a shift to higher ambient temperature. Here, considerable differences in the identity of differentially expressed genes (DEGs) in cotyledons, hypocotyls, and roots to elevated temperature were detected (Fig. 1B; Supplemental Data Sets S1 and S2), indicating considerable spatial differences in temperature responses. Similar spatial specificities have previously been detected in the comparison of shade avoidance transcriptome responses among cotyledons and shoot apices.
Gene Ontology (GO) terms overrepresented in genes with differential temperature responses in all three tissues primarily comprised stress-relevant categories (e.g. reactive oxygen species and response to heat or temperature stimulus; Supplemental Data Set S2) and included genes encoding heat shock proteins (e.g. AT3G12580, AT5G52640, and AT5G12020). While the organ-specific gene sets were partially enriched in similar GO terms, each organ contained also a variety of specifically enriched GO terms (Supplemental Data Set S2). Interestingly, the response to hormone category was significantly enriched only in genes specifically responsive in cotyledons but not in hypocotyl-specific or root-specific genes (Supplemental Data Set S2), suggesting the existence of differences at the signaling level. Overall, the GO term analysis yielded considerable differences in the transcriptional responses to elevated temperatures. However, the classification of genes was made using strict cutoff values (|log2 FC| > 1 and fdr < 0.01) and may thus exclude genes that only marginally fail to meet these criteria. Therefore, we performed a hierarchical cluster analysis of all genes that showed differential expression in at least one tissue (n = 946; Supplemental Data Set S1). Most genes clustered because of their distinct expression response in either roots or cotyledons, whereas the hypocotyl expression response seems generally less pronounced (Fig. 1C), which is in line with the generally low number of DEGs in this organ (Fig. 1B). We further assessed the pattern of the organ-specific expression using a multidimensional scaling (MDS) approach on the DEG set (n = 946). We found that two dimensions were sufficient to separate the individual biological replicates into distinct groups (Fig. 1D). Dimensions 1 and 2 separated the samples according to tissue type and temperature, respectively. Furthermore, dimension 1 clearly separated the root samples from the two shoot organs (Fig. 1D). While these results indicated a high degree of organ specificity in the temperature response of root and shoot tissues, analysis of the response 24 h after the stimulus is likely too late to assess the overlap among initial signaling events.
Based on these global observations, we next inspected the capacity for temperature-induced elongation growth in detached seedling organs. We excised hypocotyls or roots of 4-d-old seedlings and cultivated the excised organs at 20°C or 28°C for an additional 4 d. Generally, detached organs grew well if provided with sucrose as an external energy source. In the absence of any shoot tissue, detached roots were still able to elongate at 28°C proportionally more than at 20°C (Fig. 1E). In contrast, albeit hypocotyls continued to elongate, they did not show a thermomorphogenic response when cotyledons and petioles were removed from the seedling regardless of the presence or absence of the root (Figs. 1E and 2C). This indicated that (1) roots can autonomously sense and respond to temperature and (2) temperature-induced growth in hypocotyls depends on the presence of cotyledons.
Figure 2.
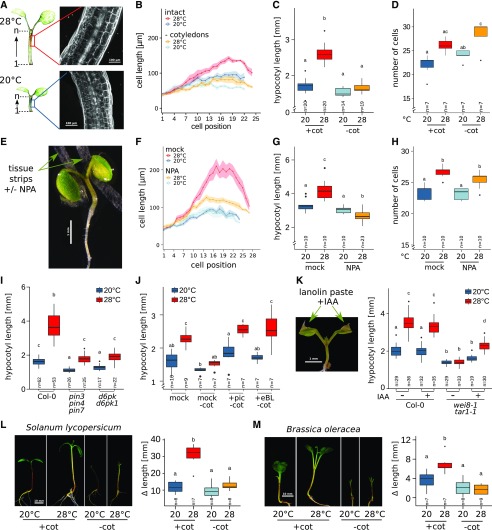
Hypocotyl cell elongation requires cotyledon-derived auxin. A, Lengths of individual cells in a consecutive hypocotyl cortex cell file were determined via confocal microscopy of propidium iodide-stained seedlings. B to D, Effects of cotyledon detachment on hypocotyl cell length (B), total hypocotyl length (C), and hypocotyl cell number in one consecutive cortex cell file (D). Seedlings were grown at 20°C for 4 d. Petioles and cotyledons were removed or seedlings were left intact prior to a shift to 28°C for an additional 3 d. E, Localized block of auxin transport by application of thin tissue strips soaked in medium with or without 100 µm NPA. F to H, Cell length (F), total hypocotyl length (G), and cell number (H) were determined, similar to A to D. I, Total hypocotyl lengths in 8-d-old auxin transport mutants. Seedlings grown at 20°C for 4 d were shifted to 28°C for an additional 4 d. J, Total hypocotyl length of intact seedlings (+cot) or with detached cotyledons (−cot) in the presence or absence of 1 µm picloram (pic) or 100 nm epi-brassinolide (eBL). Four-day-old seedlings grown at 20°C were transferred to medium containing the respective hormones and cultivated at 20°C or 28°C for an additional 3 d. K, Total hypocotyl lengths of 10-d-old seedlings with or without localized application of 1 mm IAA in lanolin paste to cotyledons. Seven-day-old seedlings grown at 20°C were treated with lanolin paste and transferred to 28°C for an additional 3 d. L and M, Temperature-induced changes in hypocotyl lengths of 11-d-old tomato (L) and cabbage (M) seedlings. Plants were initially grown for 8 d at 20°C. Cotyledons were detached or seedlings remained intact prior to the shift to 28°C for an additional 3 d. Δlength was calculated as the hypocotyl length on day 11 minus the length prior to shift. Experiments were performed in long-day conditions (16/8 h) under 30 µmol m−2 s−1 (E–H) or 90 µmol m−2 s−1 (A–D and I–M) white fluorescent light. Control plants in all experiments were treated similarly but were grown at 20°C for the whole time instead of shifting to 28°C. Bold lines in ribbon plots (B and F) show mean lengths of individual cells in a consecutive cortex cell file from the first cell after the root-shoot junction (1) upward to the shoot apex. The shadowed ribbon denotes the se. Box plots show medians and interquartile ranges; outliers (greater than 1.5× interquartile range) are shown as black dots. Different letters denote statistical differences at P < 0.05 as assessed by one-way ANOVA and Tukey’s HSD posthoc test.
A possible explanation for the dependency of the hypocotyl response on the presence of cotyledons would be the spatial separation of temperature sensing from the actual growth response. We hypothesized that hypocotyls may be unable to sense differences in temperature themselves. Instead, they may rely on a long-distance signal triggered by thermosensing events taking place in the cotyledons. We therefore inspected the elongation of hypocotyl cells in intact seedlings and seedlings with detached cotyledons.
Confocal imaging of hypocotyl cells (Fig. 2A) showed that temperature-induced cell elongation in wild-type plants occurred when seedlings were intact. However, this reaction was considerably reduced in seedlings with detached cotyledons, confirming the necessity of a cotyledon-derived signal to induce cell elongation (Fig. 2B) that ultimately results in longer hypocotyls (Fig. 2C). Hypocotyls also showed an increase in cell number, indicating that warm ambient temperature stimulates a moderate increase also in cell division (Fig. 2D). As this response was not affected by the removal of cotyledons, temperature-induced cell division in hypocotyls seems to occur independent of a cotyledon-derived signal. However, the increased cell number did not affect the total hypocotyl length (Fig. 2C) within the analyzed time frame, indicating that cell elongation is the main determinant of hypocotyl thermomorphogenesis.
PIF4 was shown to induce auxin biosynthesis genes in response to elevated temperature (Franklin et al., 2011). As such, auxin, which is an essential regulator of temperature-induced hypocotyl elongation (Gray et al., 1998), would be a likely candidate for a mobile signal. Indeed, the auxin biosynthesis mutant wei8-1 tar1-1 failed to elongate hypocotyl cells, which phenocopied the pifQ mutant that lacks PIF1,3,4,5 activity (Supplemental Fig. S1, A and B). Similarly, seedlings that express a constitutively active phyB variant (YHB) have shorter hypocotyl cells than the corresponding wild type, and YHB hypocotyl cells fail to elongate in response to temperature (Supplemental Fig. S1, C and D), confirming that the derepression of PIF4 via inactivation of phyB is a prerequisite for hypocotyl thermomorphogenesis. Accordingly, the phyABCDE mutant shows hyperelongated hypocotyl cells regardless of the cultivation temperature (Supplemental Fig. S1E). Finally, the yuc1-D gain-of-function mutant, which produces an excess of auxin, also shows a hyperelongation of hypocotyl cells. yuc1-D seedlings retain temperature responsiveness to a certain extent also in the absence of cotyledons (Supplemental Fig. S1F), indicating that an ectopic generation of auxin can overcome the lack of the cotyledon-derived signal.
These results are in accordance the findings of Zheng et al. (2016), who demonstrated that seedlings grown in the presence of the polar auxin transport inhibitor 1-N-naphthylphthalamic acid (NPA; Zhu et al., 2016) showed reduced hypocotyl elongation in response to shade and high temperature under low constant light, which can be counteracted by a local increase of indole-3-acetic acid (IAA) in the hypocotyl. However, Zheng et al. (2016) applied NPA to whole seedlings, which would affect polar auxin transport in all tissues, whereas we were interested in the specific role of cotyledon-derived auxin in hypocotyl thermomorphogenesis. To investigate this, we interrupted auxin transport from cotyledons to the hypocotyl by local application of NPA to petioles of 7-d-old intact seedlings, which were subsequently shifted to elevated temperatures (Fig. 2E). NPA application essentially phenocopied the physical detachment of cotyledons and inhibited temperature-induced cell elongation (Fig. 2F) as well as total hypocotyl elongation (Fig. 2G), while cell division was not affected (Fig. 2H). Genetic data of mutants defective in polar auxin transport further substantiate these findings. Both pin-formed (pin) auxin transport mutants as well as D6PK kinase mutants that regulate the phosphorylation status of PIN proteins (Zourelidou et al., 2009; Zhang et al., 2010; Adamowski and Friml, 2015) showed significant reductions in temperature-induced hypocotyl elongation (Fig. 2I). While illustrating a general requirement for polar auxin transport in hypocotyl temperature responses, potential spatial specificities in PIN action are not resolved here. Collectively, these data indicate that auxin is the sought-for mobile signal that links temperature sensing in the cotyledons with elongation responses in the hypocotyls.
Based on the recently proposed epistastic interaction of BRs and auxin in downstream thermomorphogenesis signaling (Ibañez et al., 2018), exogenous addition of either auxin or BRs should at least partially compensate for the lack of cotyledon-derived auxin in the hypocotyl. To test this hypothesis, 4-d-old plants grown at 20°C on unsupplemented medium were transferred either as intact wild-type seedlings or as seedlings with detached cotyledons to medium containing the synthetic auxin picloram or epi-brassinolide. Either hormone was able to restore temperature responsiveness in hypocotyls even in the absence of cotyledons (Fig. 2J). Further substantiating these observations, local application of IAA (dissolved in lanolin paste) to cotyledons of the wei8-1 tar1-1 auxin biosynthesis mutant was sufficient to partially restore the elongation response to elevated temperature (Fig. 2K). Together, these experiments strongly suggest that auxin is the mobile signal that connects thermosensing in the cotyledons with growth responses in the hypocotyl. This mechanism seems to be of general biological relevance, as the effect of cotyledon removal observed in Arabidopsis was also detected in other flowering plant species. Both tomato (Solanum lycopersicum; Fig. 2L) and cabbage (Brassica oleracea; Fig. 2M) seedlings failed to show hypocotyl thermomorphogenesis when cotyledons were removed prior to the exposure to elevated temperatures.
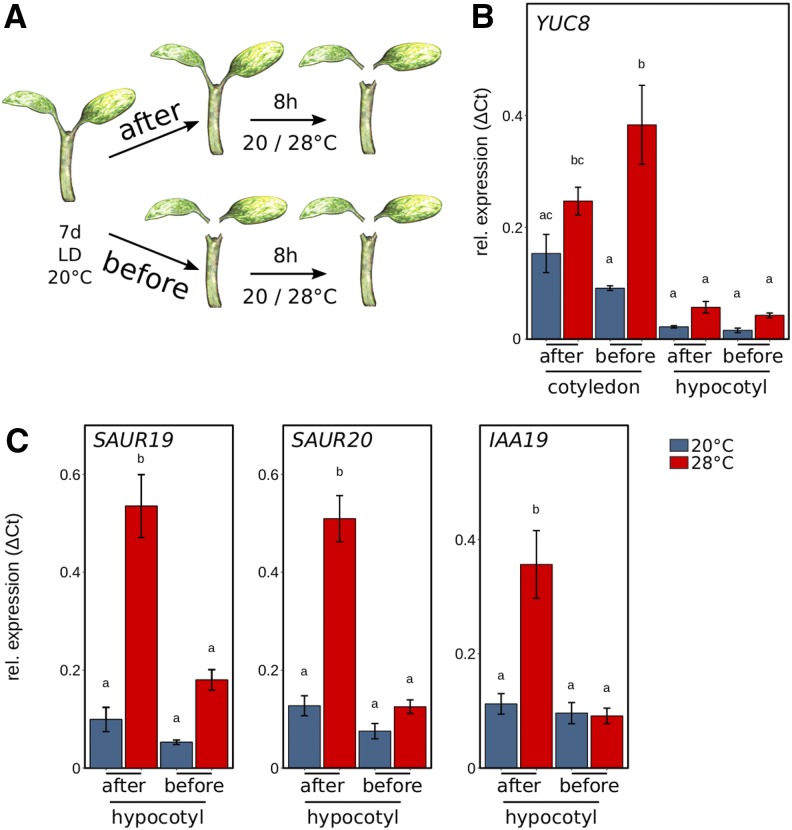
Based on the physiological observations, we next investigated the potential effects of a mobile auxin signal on the transcriptional activation of thermoresponsive genes. Seedlings were dissected in cotyledons and hypocotyls either before or after an 8-h temperature stimulus of 28°C to seedlings previously grown at 20°C (Fig. 3A). In this experimental setup, seedlings dissected after the temperature shift were able to send a mobile auxin signal from the cotyledons to the hypocotyl. Obviously, this was not possible in seedlings that were dissected before temperature treatment. To first test the relevance of temperature-induced auxin biosynthesis in cotyledons versus hypocotyls, we quantified transcript levels of the thermoresponsive auxin biosynthesis gene YUCCA8 (YUC8). As shown in Figure 3B, YUC8 was thermoresponsive in cotyledons but not in hypocotyls. Furthermore, expression levels of YUC8 at elevated temperature were several fold higher in cotyledons in comparison with hypocotyls (Fig. 3B), suggesting that thermoresponsive induction of auxin biosynthesis is most relevant in cotyledons rather than in hypocotyls.
Figure 3.
Organ-specific expression analysis of auxin-related and growth-related genes. A, Seedlings cultivated for 7 d at 20°C were transferred to 20°C or 28°C at zeitgeber time 16 (ZT16) for 8 h prior to harvesting cotyledons and hypocotyls for expression analysis. Seedlings were either dissected after or before the temperature shift, including removal of the root. B, Reverse transcription quantitative PCR (RT-qPCR) expression analysis of the auxin biosynthesis gene YUC8 was assessed for cotyledons and hypocotyls. C, Genes relevant for hypocotyl elongation (SAUR19 and SAUR20) and an auxin response gene (IAA19) were assessed in hypocotyl samples. Seedlings were grown in long-day conditions (16/8 h) under 30 µmol m−2 s−1 white fluorescent light. RT-qPCR analyses were performed on three independent biological replicates. Bar plots show mean values, and error bars denote se. Different letters denote statistical differences at P < 0.05 as assessed by one-way ANOVA and Tukey’s HSD posthoc test.
We then inspected the need for a mobile auxin signal in the induction of thermoresponsive auxin (IAA19) and auxin/BR response genes that are relevant for cell elongation (SAUR19 and SAUR20; Spartz et al., 2012). Hypocotyls that were excised from seedlings after a temperature stimulus showed a strong temperature response in SAUR19, SAUR20, and IAA19 (Fig. 3C), indicative of a successfully transmitted auxin signal from the cotyledons to the hypocotyl. In contrast, the induction was absent if cotyledons were detached prior to the shift to higher temperatures, confirming that auxin has to be translocated to the hypocotyl to induce growth-relevant genes in elevated temperatures.
This mode of regulation strongly resembles processes involved in the regulation of hypocotyl elongation in response to shade (i.e. reduced blue light) or vegetative shade (Keuskamp et al., 2011; Procko et al., 2014, 2016; Nito et al., 2015). Whether temperature perception also shows local spatial preferences within cotyledon and leaf areas similar to the sensing of vegetative shade (Michaud et al., 2017; Pantazopoulou et al., 2017) remains to be clarified.
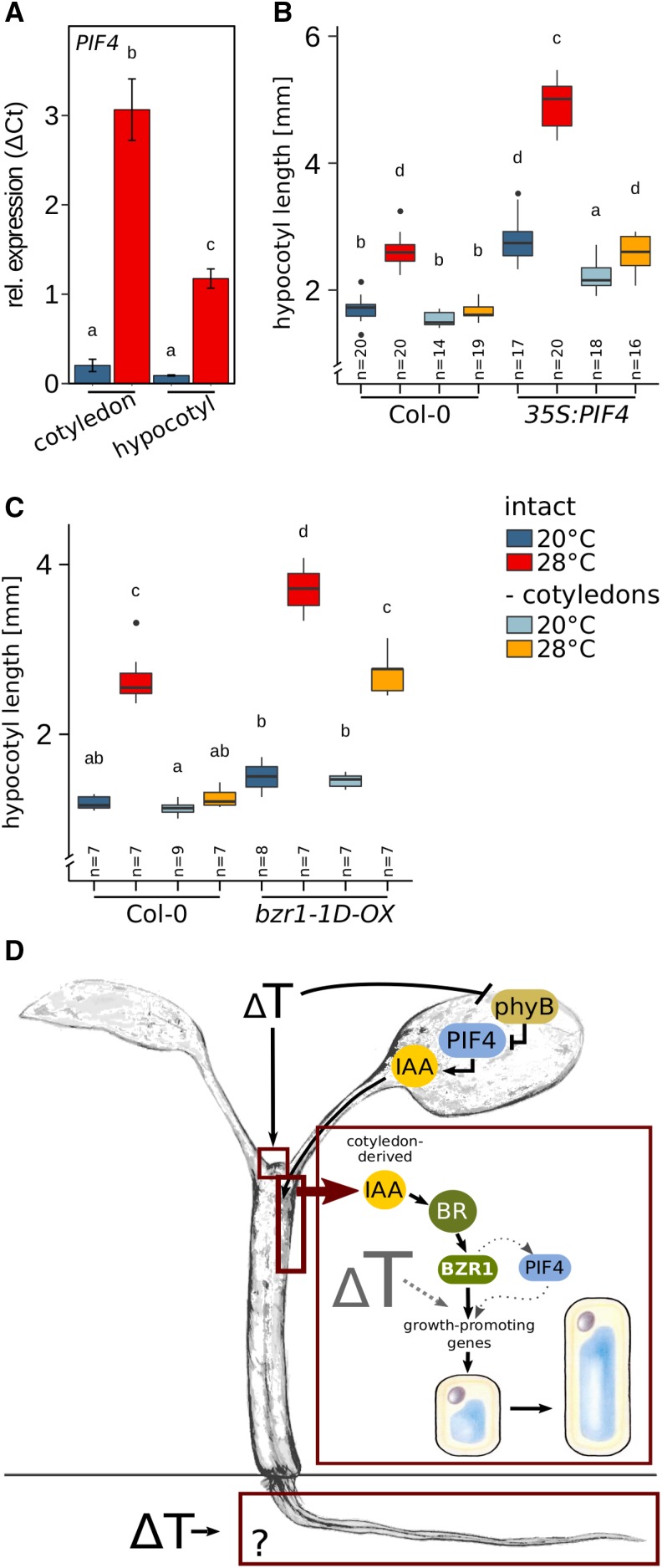
Collectively, these results imply that the predominant function of PIF4 in thermomorphogenesis may be the induction of auxin biosynthesis genes in the cotyledons. Auxin synthesized in the cotyledons then travels to the hypocotyl, where it triggers BR-dependent cell elongation. If it were that simple, there would be no need for thermoresponsive induction of PIF4 transcription in hypocotyls. We observed, however, that in response to temperature, PIF4 was significantly induced in both cotyledons and hypocotyls (Fig. 4A). While the absolute expression level was higher in cotyledons, the induction of expression in hypocotyls may be a consequence of the proposed BZR1-mediated feed-forward regulation (Ibañez et al., 2018) to enable a cooperative interaction of BZR1 and PIF4 in the transcriptional regulation of growth-promoting genes in hypocotyls. The presence of PIF4 may be relevant to fully express BZR1 function, as the interaction with PIF4 seems to affect the specificity for different target cis-elements (Martínez et al., 2018).
Figure 4.
BZR1-mediated hypocotyl thermomorphogenesis requires local permissive temperature sensing. A, PIF4 RT-qPCR expression analysis in cotyledons and hypocotyls dissected after the temperature treatment. The experimental setup was as described in Figure 3. B and C, Effects of cotyledon detachment on hypocotyl length in 35S:PIF4 (B), and bzr1-1D-OX (C) seedlings. Seedlings were initially grown in long-day conditions (16/8 h) under 90 µmol m−2 s−1 white fluorescent light at 20°C for 4 d. Petioles and cotyledons were removed or seedlings were left intact prior to a shift to 28°C for an additional 3 d. Control plants were treated similarly but grown at 20°C. Box plots show medians and interquartile ranges of total organ length; outliers (greater than 1.5× interquartile range) are shown as black dots. Different letters denote statistical differences at P < 0.05 as assessed by one-way ANOVA and Tukey’s HSD posthoc test. D, Model of spatial sensing and signaling specificities in seedling thermomorphogenesis.
To further assess the potential spatial specificities of PIF4 and BZR1, we inspected the effect of cotyledon detachment on the temperature response in PIF4 and BZR1 gain-of-function mutants. 35S:PIF4 hypocotyls hyperelongated in seedlings with intact cotyledons (Fig. 4B) at both temperatures, with an exaggerated response at 28°C. If cotyledons were removed prior to the temperature shift, hypocotyls of seedlings grown at 20°C were still longer than in the wild type but the hypersensitive temperature response was abolished (Fig. 4B), indicating that at 20°C PIF4 can serve as a general regulator of elongation growth if ectopically expressed in relevant tissues. Interestingly, temperature hypersensitivity at 28°C is lost in the absence of cotyledons. Hence, the primary role of PIF4 in elevated temperatures seems to indeed reside in the cotyledons, where its activity is derepressed by the inactivation of photoreceptors (e.g. phyB).
Hypersensitivity of bzr1-1D-OX gain-of-function lines can be largely attributed to hyperelongated hypocotyls at 28°C (Fig. 4C). In dissected seedlings, bzr1-1D-OX still displayed temperature-induced hypocotyl elongation comparable to the wild type. Since detached bzr1-1D-OX hypocotyls are deprived of cotyledon-derived thermosensing via phytochromes and PIF4, this suggests the presence of a second independent thermosensor. This unknown sensor possibly provides a permissive signal that gates cotyledon-derived signaling in the hypocotyls.
Numerous molecular mechanisms have the capacity to serve as thermosensors (for review, see Vu et al., 2019). One possible candidate for a permissive temperature sensor gating BZR1 function in the hypocotyl could be constituted by changes in the chromatin structure. Thermomorphogenesis has been shown to require chromatin remodeling that involves histone deacetylation (Tasset et al., 2018) and the eviction of the H2A.Z histone variants (Kumar and Wigge, 2010). However, at this stage, the nature of the permissive temperature-sensing mechanism remains elusive.
CONCLUSION
Our results imply a physiological model describing temperature-induced elongation of seedling organs as a result of distinct processes in thermosensing, signaling, and elongation growth, which, in some cases, are spatially separated (Fig. 4D). Some organs seem to be able to autonomously sense and respond to temperature, for example roots.
Whether similar sensing and signaling components are involved in the different organs remains to be analyzed, but we are inclined to speculate that thermomorphogenesis signaling differs substantially between roots and shoots. Temperature-induced elongation of hypocotyl cells requires sensing of temperature changes in the cotyledons. Here, derepression of PIF4 can induce the production of auxin, which then serves as a mobile long-distance signal promoting BR biosynthesis and signaling in the hypocotyl. The presence of BR-activated BZR1 in hypocotyl cells subsequently controls transcriptional activity of growth-promoting genes, resulting in cell elongation (Fig. 4D). However, the capacity of BZR1 to induce cell elongation relies on a local, permissive temperature-sensing mechanism that gates BZR1’s capacity to induce elongation at elevated temperatures.
Our results may harbor implications for other temperature responses, such as growth responses in later vegetative stages or the induction of flowering. Here, autonomous sensing in specific tissues or organs may be involved and should be considered in approaches to analyze the underlying molecular mechanisms.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) seeds were surface sterilized and stratified for 3 d at 4°C before sowing in all experiments. Genotypes used in this study have been described previously or were obtained from the Nottingham Arabidopsis Stock Centre (NASC; http://arabidopsis.info): yuc1-D (Zhao et al., 2001), wei8-1 tar1-1 (N16412; Stepanova et al., 2008), tir1-1 afb2-3 (N69691; Parry et al., 2009), 35S:PIF4-HA (Nozue et al., 2007), pifQ (N66049; Leivar et al., 2008), bzr1-1D-OX (Oh et al., 2014b), bri1-4 (N3953; Noguchi et al., 1999), phyABCDE (Hu et al., 2013), YHB (p35S:AtPHYBY276H in phyA-201 phyB-5; Su and Lagarias, 2007), and d6pk d6pkl1 (Zourelidou et al., 2009). The pin3 pin4 pin7 mutant was generated by successive crosses of pin3-5 (Friml et al., 2003), pin4-3 (Friml et al., 2002), and pin7-1 (Friml et al., 2003). Wild-type strains were Col-0 (N1092), Ws-2 (N28827), Rrs-7 (N22688), and Ler-0 (NW20). Tomato (Solanum lycopersicum var cerasiforme ‘WV 106’) was kindly provided by Stefan Bennewitz (IPB), and cabbage (Brassica oleracea; N29002) was obtained from NASC. Unless stated otherwise, seedlings were grown on solid Arabidopsis solution (ATS) nutrient medium including 1% (w/v) Suc (Lincoln et al., 1990) on vertically oriented plates under long-day conditions (16 h of light/8 h of dark) with 90 µmol m−2 s−1 photosynthetically active radiation (PAR) from white fluorescent lamps (T5 4000K).
Temperature Response of Hypocotyls, Petioles, or Roots
Organ-specific temperature responses were determined in 8-d-old seedlings grown at 20°C or 28°C. For detached organ growth, 4-d-old seedlings grown at 20°C were dissected to obtain roots or hypocotyls. Isolated organs were placed on ATS medium and cultivated at 20°C or 28°C for an additional 3 or 4 d. All measurements were made from digital photographs of plates using RootDetection (www.labutils.de) and depict the total length of the analyzed organ. Hypocotyl elongation in tomato and cabbage was assessed in 11-d-old plants. Seedlings were cultivated for 8 d at 20°C prior to a shift at 28°C for an additional 3 d. Cotyledons were either detached or seedlings were left intact at the time of the shift. Control plants remained at 20°C. Because of high variability in the germination and growth of tomato and cabbage individuals in the first 8 d, hypocotyl length for each seedling was calculated as the increase in length after transfer to higher temperature (Δlength = length at 11 d – length at 8 d).
IAA and NPA Treatment Assays
Seedlings were grown in long-day conditions at 90 µmol m−2 s−1 for IAA application and at 30 µmol m−2 s−1 for NPA treatments to allow for longer petiole growth for application of NPA plasters. In both experiments, seedlings were initially grown at 20°C for 7 d prior to the pharmacological treatment. For IAA application, 1 mm IAA (Duchefa) in lanolin paste (Sigma-Aldrich) was applied to cotyledons. For NPA treatments, thin strips of tissue were soaked in lukewarm ATS medium with or without the addition of 100 µm NPA (Duchefa) and carefully placed across petioles. For both experiments, seedlings were subsequently cultivated for an additional 3 d at 20°C or 28°C in the respective light conditions.
Cell Measurements
Hypocotyl cell lengths were determined by staining seedlings with 10 µg mL−1 propidium iodide (Sigma-Aldrich) for 5 min. Subsequent microscopic analyses were performed using a Zeiss LSM 700 AxioObserver (Laser 555 nm, Plan-Neofluar 20x/0.50 Ph2) or an ApoTome Zeiss Axio Z1 imager microscope (EC Plan-Neofluar 20x/0.30 M27). Measurements were performed on all individual cells of a consecutive cortex cell file using the LSM 700 ZEN software for seven to 10 independent seedlings per experiment. Experiments were carried out two to three times with similar results, of which one representative experiment is shown.
Statistical Analyses
Statistical differences were assessed by one-way ANOVA and Tukey’s HSD posthoc test, using built-in functions of the statistical environment R (R Development Core Team, 2018). Different letters in graphs denote statistical differences at P < 0.05. Graphs were generated using the ggplot2 R package.
Transcriptome Profiling
Arabidopsis seedlings (Rrs-7; N22688) were cultivated for 5 d at 20°C in long-day conditions with 120 µmol m−2 s−1 PAR. Seedlings were either kept at 20°C or shifted to 28°C for 24 h prior to dissection of the plant material into cotyledons, hypocotyls, and roots. For each seedling organ sample, material for three biological replicates was harvested. RNA was extracted using the RNeasy Plant Mini Kit (Qiagen). RNA samples were further processed and hybridized to the ATH1-121501 microarray by the NASC microarray hybridization service. Raw data were processed with the simpleaffy R package to obtain robust multichip average-normalized log2 expression levels using default settings (Supplemental Data Set S3). The eBayes function of the limma R package (Supplemental Data Set S4) was used to compute log2 FCs, t values, and P values and to correct P values for multiple testing. Genes were considered to be differentially regulated if the 28°C expression values showed a |log2 FC| > 1 and an fdr < 0.01 compared with expression levels at 20°C (Supplemental Data Set S1).
Hierarchical Clustering and MDS
The log2 FC data of DEGs with significant expression responses in at least one organ were subjected to hierarchical clustering in R using the hclust function of the gplots package with Euclidean distances and complete linkage. The heat map was generated using the heatmap.2 function of the gplots package.
MDS was performed in R using the build-in cmdscale function with k = 2 dimensions. Pairwise Pearson correlations (cor) among all individual array samples were computed using the normalized log2 expression levels of DEGs, and 1 − cor served as a distance measure in the MDS.
GO Term Analysis
GO enrichment of DEGs was assessed using PANTHER version 14.0 (Mi et al., 2017). Arabidopsis Genome Initiative (AGI) codes of genes with |log2 FC| > 1 and fdr < 0.01 were analyzed for different gene sets (Supplemental Data Set S2). Enrichment of GO terms was tested using default test settings (Fisher’s exact test and false discovery rate correction) for the PANTHER Biological Process Data Set.
RT-qPCR Expression Analyses
Col-0 seedlings were cultivated at 20°C for 7 d in long-day photoperiods (16/8 h) in 30 µmol m−2 s−1 PAR. Seedlings were shifted to 28°C at Zeitgeber time (ZT) 16 and harvested after 8 h at ZT24. Seedlings were dissected by cutting off cotyledons (with petioles) and roots to allow organ-specific expression analysis. Dissection was performed either before or after the temperature shift at ZT16 or ZT24, respectively. Control seedlings remained at 20°C and were harvested and dissected at the same time points.
Total RNA was extracted from three biological replicates using the NucleoSpin RNA Plant Kit (Macherey-Nagel). First-strand cDNA was synthesized using the PrimeScript RT Reagent Kit (Perfect Real Time) from Takara Bio. qPCR analyses were performed on an AriaMx Real-Time PCR System (Agilent) using Absolute Blue Low Rox Mix (Thermo Fisher Scientific). At1g13320 was used as a reference gene (Czechowski et al., 2005) to calculate relative expression values (2ΔCt values). Oligonucleotide primers used in the analysis are listed in Supplemental Table S1.
Accession Numbers
Expression data from this article can be found in the National Center for Biotechnology Information Gene Expression Omnibus (Edgar et al., 2002) and are accessible under reference number GSE126373. Accession numbers of genes associated with this study are as follows: BZR1 (AT1G75080), D6PK-1 (AT5G556910), D6PKL-1 (AT4G26610), IAA19 (AT3G15540), phyA (AT1G09570), phyB (AT2G18790), phyC (AT5G35840), phyD (AT4G16250), phyE (AT4G18130), PIF1 (AT2G20180), PIF3 (AT1G09530), PIF4 (AT2G43010), PIF5 (AT3G59060), PIN3 (AT1G70940), PIN4 (AT2G01420), PIN7 (AT1G23080), SAUR19 (AT5G18010), SAUR20 (AT5G18020), TAR1 (AT1G23320), WEI8 (AT1G70560), YUC8 (AT4G28720), and PP2AA3 (AT1G13320).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Cell and hypocotyl elongation responses in genotypes affected in auxin, BR, phytochrome, and PIF function.
Supplemental Table S1. Oligonucleotide primers used in qPCR analyses.
Supplemental Data Set S1. Expression data for DEGs.
Supplemental Data Set S2. Gene lists and GO term analysis results.
Supplemental Data Set S3. R script used for microarray normalization.
Supplemental Data Set S4. R script used for microarray data processing.
Acknowledgments
We thank all members of the Department of Molecular Signal Processing at the Leibniz Institute of Plant Biochemistry who helped in collecting the material for the organ-specific transcriptome profiling. We thank Steve Babben and Zihao Zhu for aiding in the collection of plant material for qPCR analyses and Kathrin Denk for technical assistance. We thank Claus Schwechheimer, Stefan Bennewitz, J. Clark Lagarias, and Zhiyong Wang for sharing plant material.
Footnotes
This study was supported by the Rosa Luxemburg Foundation Ph.D. fellowship grant to J.B. and by the Deutsche Forschungsgemeinschaft (grants Qu 141/3-1 and Qu 141/3-2 to M.Q.). The Deutsche Forschungsgemeinschaft provided funding for the German Centre for Integrative Biodiversity Research Halle-Jena-Leipzig (FZT 118).
Articles can be viewed without a subscription.
References
- Adamowski M, Friml J (2015) PIN-dependent auxin transport: Action, regulation, and evolution. Plant Cell 27: 20–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal JJ, Balasubramanian S (2019) Thermomorphogenesis. Annu Rev Plant Biol 70: [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delker C, Sonntag L, James GV, Janitza P, Ibañez C, Ziermann H, Peterson T, Denk K, Mull S, Ziegler J, et al. (2014) The DET1-COP1-HY5 pathway constitutes a multipurpose signaling module regulating plant photomorphogenesis and thermomorphogenesis. Cell Rep 9: 1983–1989 [DOI] [PubMed] [Google Scholar]
- Delker C, van Zanten M, Quint M (2017) Thermosensing enlightened. Trends Plant Sci 22: 185–187 [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30: 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KA, Lee SH, Patel D, Kumar SV, Spartz AK, Gu C, Ye S, Yu P, Breen G, Cohen JD, et al. (2011) Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc Natl Acad Sci USA 108: 20231–20235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J, Benková E, Blilou I, Wisniewska J, Hamann T, Ljung K, Woody S, Sandberg G, Scheres B, Jürgens G, et al. (2002) AtPIN4 mediates sink-driven auxin gradients and root patterning in Arabidopsis. Cell 108: 661–673 [DOI] [PubMed] [Google Scholar]
- Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, Offringa R, Jürgens G (2003) Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426: 147–153 [DOI] [PubMed] [Google Scholar]
- Fujii Y, Tanaka H, Konno N, Ogasawara Y, Hamashima N, Tamura S, Hasegawa S, Hayasaki Y, Okajima K, Kodama Y (2017) Phototropin perceives temperature based on the lifetime of its photoactivated state. Proc Natl Acad Sci USA 114: 9206–9211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Östin A, Sandberg G, Romano CP, Estelle M (1998) High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proc Natl Acad Sci USA 95: 7197–7202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes S, Sharma A, Fraser DP, Trevisan M, Cragg-Barber CK, Tavridou E, Fankhauser C, Jenkins GI, Franklin KA (2017) UV-B perceived by the UVR8 photoreceptor inhibits plant thermomorphogenesis. Curr Biol 27: 120–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Franklin KA, Sharrock RA, Jones MA, Harmer SL, Lagarias JC (2013) Unanticipated regulatory roles for Arabidopsis phytochromes revealed by null mutant analysis. Proc Natl Acad Sci USA 110: 1542–1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibañez C, Poeschl Y, Peterson T, Bellstädt J, Denk K, Gogol-Döring A, Quint M, Delker C (2017) Ambient temperature and genotype differentially affect developmental and phenotypic plasticity in Arabidopsis thaliana. BMC Plant Biol 17: 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibañez C, Delker C, Martinez C, Bürstenbinder K, Janitza P, Lippmann R, Ludwig W, Sun H, James GV, Klecker M, et al. (2018) Brassinosteroids dominate hormonal regulation of plant thermomorphogenesis via BZR1. Curr Biol 28: 303–310.e3 [DOI] [PubMed] [Google Scholar]
- Jung JH, Domijan M, Klose C, Biswas S, Ezer D, Gao M, Khattak AK, Box MS, Charoensawan V, Cortijo S, et al. (2016) Phytochromes function as thermosensors in Arabidopsis. Science 354: 886–889 [DOI] [PubMed] [Google Scholar]
- Keuskamp DH, Sasidharan R, Vos I, Peeters AJM, Voesenek LACJ, Pierik R (2011) Blue-light-mediated shade avoidance requires combined auxin and brassinosteroid action in Arabidopsis seedlings. Plant J 67: 208–217 [DOI] [PubMed] [Google Scholar]
- Koini MA, Alvey L, Allen T, Tilley CA, Harberd NP, Whitelam GC, Franklin KA (2009) High temperature-mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Curr Biol 19: 408–413 [DOI] [PubMed] [Google Scholar]
- Kumar SV, Wigge PA (2010) H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell 140: 136–147 [DOI] [PubMed] [Google Scholar]
- Legris M, Klose C, Burgie ES, Rojas CC, Neme M, Hiltbrunner A, Wigge PA, Schäfer E, Vierstra RD, Casal JJ (2016) Phytochrome B integrates light and temperature signals in Arabidopsis. Science 354: 897–900 [DOI] [PubMed] [Google Scholar]
- Leivar P, Monte E, Oka Y, Liu T, Carle C, Castillon A, Huq E, Quail PH (2008) Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr Biol 18: 1815–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln C, Britton JH, Estelle M (1990) Growth and development of the axr1 mutants of Arabidopsis. Plant Cell 2: 1071–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D, Li X, Guo Y, Chu J, Fang S, Yan C, Noel JP, Liu H (2016) Cryptochrome 1 interacts with PIF4 to regulate high temperature-mediated hypocotyl elongation in response to blue light. Proc Natl Acad Sci USA 113: 224–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez C, Espinosa-Ruíz A, de Lucas M, Bernardo-García S, Franco-Zorrilla JM, Prat S (2018) PIF4-induced BR synthesis is critical to diurnal and thermomorphogenic growth. EMBO J 37: 99552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Huang X, Muruganujan A, Tang H, Mills C, Kang D, Thomas PD (2017) PANTHER version 11: Expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res 45: D183–D189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaud O, Fiorucci AS, Xenarios I, Fankhauser C (2017) Local auxin production underlies a spatially restricted neighbor-detection response in Arabidopsis. Proc Natl Acad Sci USA 114: 7444–7449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery BL. (2016) Spatiotemporal phytochrome signaling during photomorphogenesis: From physiology to molecular mechanisms and back. Front Plant Sci 7: 480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nito K, Kajiyama T, Unten-Kobayashi J, Fujii A, Mochizuki N, Kambara H, Nagatani A (2015) Spatial regulation of the gene expression response to shade in Arabidopsis seedlings. Plant Cell Physiol 56: 1306–1319 [DOI] [PubMed] [Google Scholar]
- Noguchi T, Fujioka S, Choe S, Takatsuto S, Yoshida S, Yuan H, Feldmann KA, Tax FE (1999) Brassinosteroid-insensitive dwarf mutants of Arabidopsis accumulate brassinosteroids. Plant Physiol 121: 743–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozue K, Covington MF, Duek PD, Lorrain S, Fankhauser C, Harmer SL, Maloof JN (2007) Rhythmic growth explained by coincidence between internal and external cues. Nature 448: 358–361 [DOI] [PubMed] [Google Scholar]
- Oh E, Zhu JY, Wang ZY (2012) Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat Cell Biol 14: 802–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Zhu JY, Bai MY, Arenhart RA, Sun Y, Wang ZY (2014a) Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl. eLife 3: e03031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Zhu JY, Ryu H, Hwang I, Wang ZY (2014b) TOPLESS mediates brassinosteroid-induced transcriptional repression through interaction with BZR1. Nat Commun 5: 4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantazopoulou CK, Bongers FJ, Küpers JJ, Reinen E, Das D, Evers JB, Anten NPR, Pierik R (2017) Neighbor detection at the leaf tip adaptively regulates upward leaf movement through spatial auxin dynamics. Proc Natl Acad Sci USA 114: 7450–7455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry G, Calderon-Villalobos LI, Prigge M, Peret B, Dharmasiri S, Itoh H, Lechner E, Gray WM, Bennett M, Estelle M (2009) Complex regulation of the TIR1/AFB family of auxin receptors. Proc Natl Acad Sci USA 106: 22540–22545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Procko C, Crenshaw CM, Ljung K, Noel JP, Chory J (2014) Cotyledon-generated auxin is required for shade-induced hypocotyl growth in Brassica rapa. Plant Physiol 165: 1285–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Procko C, Burko Y, Jaillais Y, Ljung K, Long JA, Chory J (2016) The epidermis coordinates auxin-induced stem growth in response to shade. Genes Dev 30: 1529–1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quint M, Delker C, Franklin KA, Wigge PA, Halliday KJ, van Zanten M (2016) Molecular and genetic control of plant thermomorphogenesis. Nat Plants 2: 15190. [DOI] [PubMed] [Google Scholar]
- R Development Core Team (2018) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria, http://www.R-project.org [Google Scholar]
- Spartz AK, Lee SH, Wenger JP, Gonzalez N, Itoh H, Inzé D, Peer WA, Murphy AS, Overvoorde PJ, Gray WM (2012) The SAUR19 subfamily of SMALL AUXIN UP RNA genes promote cell expansion. Plant J 70: 978–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavang JA, Gallego-Bartolomé J, Gómez MD, Yoshida S, Asami T, Olsen JE, García-Martínez JL, Alabadí D, Blázquez MA (2009) Hormonal regulation of temperature-induced growth in Arabidopsis. Plant J 60: 589–601 [DOI] [PubMed] [Google Scholar]
- Stepanova AN, Robertson-Hoyt J, Yun J, Benavente LM, Xie DY, Dolezal K, Schlereth A, Jürgens G, Alonso JM (2008) TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133: 177–191 [DOI] [PubMed] [Google Scholar]
- Su YS, Lagarias JC (2007) Light-independent phytochrome signaling mediated by dominant GAF domain tyrosine mutants of Arabidopsis phytochromes in transgenic plants. Plant Cell 19: 2124–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasset C, Singh Yadav A, Sureshkumar S, Singh R, van der Woude L, Nekrasov M, Tremethick D, van Zanten M, Balasubramanian S (2018) POWERDRESS-mediated histone deacetylation is essential for thermomorphogenesis in Arabidopsis thaliana. PLoS Genet 14: e1007280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu LD, Gevaert K, De Smet I (2019) Feeling the heat: Searching for plant thermosensors. Trends Plant Sci 24: 210–219 [DOI] [PubMed] [Google Scholar]
- Zhang J, Nodzyński T, Pencík A, Rolcík J, Friml J (2010) PIN phosphorylation is sufficient to mediate PIN polarity and direct auxin transport. Proc Natl Acad Sci USA 107: 918–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Christensen SK, Fankhauser C, Cashman JR, Cohen JD, Weigel D, Chory J (2001) A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291: 306–309 [DOI] [PubMed] [Google Scholar]
- Zheng Z, Guo Y, Novák O, Chen W, Ljung K, Noel JP, Chory J (2016) Local auxin metabolism regulates environment-induced hypocotyl elongation. Nat Plants 2: 16025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Bailly A, Zwiewka M, Sovero V, Di Donato M, Ge P, Oehri J, Aryal B, Hao P, Linnert M, et al. (2016) TWISTED DWARF1 mediates the action of auxin transport inhibitors on actin cytoskeleton dynamics. Plant Cell 28: 930–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zourelidou M, Müller I, Willige BC, Nill C, Jikumaru Y, Li H, Schwechheimer C (2009) The polarly localized D6 PROTEIN KINASE is required for efficient auxin transport in Arabidopsis thaliana. Development 136: 627–636 [DOI] [PubMed] [Google Scholar]