ABSTRACT
Much of our current knowledge of Rho GTPase networks and the regulation by Rho guanine exchange factors (Rho GEFs) and Rho GTPase activating proteins (Rho GAPs) is based on population-based techniques. Over the last decades, technologies that enable single cell analysis with high spatial and temporal resolution have revealed that Rho GTPase activity in cells is regulated on second timescales and at submicrometer length scales. Therefore, perturbation methods with matching spatial and temporal resolution are crucial to further our understanding of Rho GTPase signaling. Here, we give a brief overview of the components of Rho GTPase signaling networks and review a range of existing perturbation strategies that target a specific component of the Rho GTPase signaling module. The advantages and limitations of each perturbation method are discussed. Several recommendations are formulated to guide future studies aimed at addressing spatiotemporal aspects of Rho GEF and Rho GTPase signaling.
Rho GTPases
Rho GTPases are master regulators of the cytoskeleton, influencing cell migration, neuronal development and trafficking.1 They function as molecular switches that transition between active GTP- and inactive GDP loaded forms. This cycling is regulated by guanine exchange factors (GEF) and GTPase activating proteins (GAP) proteins,2 respectively. Inactive, GDP-bound Rho GTPases are bound to Rho GDP dissociation inhibitors (Rho GDI) and sequestered in the cytoplasm. GTP-loaded Rho GTPases activate downstream effectors to control specific cytoskeletal outputs.3
The human Rho GTPase family consists of ∼20 genes, which can be subdivided in 8 subfamilies.4 An immense amount of experimental work has been conducted on the well-characterized and evolutionary conserved subfamilies of Rac, RhoA and Cdc42, and the identification of their specific functions in terms of cell and molecular biology has gone through several phases of understanding.
After initial experiments with Rac1, RhoA and Cdc42 dominant negative (DN), and constitutively active variants (CA), a picture arose that active Rac1 caused lamellipodia formation and plasma membrane ruffling, RhoA caused formation of stress fibers and focal adhesion complexes and Cdc42 was responsible for filopodia formation at the leading edge.5 It was also shown that Rac1 and Cdc42 caused neurite outgrowth and growth cone expansion, while RhoA impaired these processes and even caused neurite retraction.6 Initially, in terms of cell motility, it was found that Rac1 activity was highest at the leading edge of migrating cells, inducing actin polymerization and adhesion complex formation, leading to membrane protrusions and cell movement,7 while inhibition of Rac1 impaired cell migration in several cell types.5 Cdc42 was shown to be involved in cell polarity based on observations in macrophages that lost their directed movement when lacking Cdc42 while RhoA was thought to be responsible for acto-myosin mediated retraction at the back of cells and thought to be excluded from the leading edge.1
Although it still holds true that overexpression or constitutive activation of these subfamilies of Rho GTPases can lead to this kind of ‘classic’ phenotypes in cells, we now know that the regulation and activation of these signaling molecules is organized at a much more detailed spatiotemporal level.8 The development and constant improvement of Förster Resonance Energy Transfer (FRET) based biosensors for the Rho GTPases9,10 has allowed us to inspect Rho GTPase function in living cells with great spatiotemporal detail. With the use of these techniques, all 3 main Rho GTPase subfamilies have been shown to be activated at the leading edge during distinct phases of plasma membrane protrusion and retraction11 and to have very distinct spatiotemporal activation patterns both at the leading edge and the tail during cell migration in motile fibroblasts.12,13 Moreover, specific spatiotemporal activity patterns have been observed during neural growth cone protrusion and relapse,9 macropinocytosis,14 invadopodia (dis)assembly,14,15 and oocyte wound repair.16,17
The precise molecules and mechanisms involved in the regulation of local Rho GTPase activation and deactivation cycles,18 as well as the interplay between Rho GTPases during complex cell behavior, is a topic of intense ongoing research.19,20
Regulation of Rho GTPase activation
Rho GEFs catalyze the release of GDP from Rho GTPases, thereby converting Rho GTPases into the active, GTP-bound state. There are more than 80 different Rho GEFs found in mammals, which means that they outnumber the Rho GTPases they target by 4 to 1. As Rho GEFs control the activation of Rho GTPase targets, which are involved in virtually every physiologic process in the human body, their specific deregulation or mutants are often linked to developmental problems, cancer progression and several other pathologies.21
Rho GEFs are divided in 2 unrelated families; they either contain a ∼200 amino acid (aa) Dbl-homology domain (DH)22 or a ∼400 aa Dock homology region (DHR).23 The largest and best studied Rho GEF family is the Dbl-homology superfamily (at least 70 members), characterized by one or more DH domains, which are almost always accompanied by a C-terminal ∼100 aa long Pleckstrin Homology (PH) domain. It is well established that the DH domain interacts directly with the Rho GTPase and is responsible for the catalytic activity that accelerates the release of GDP. Since the concentration of GTP is higher than GDP in cells, this results in loading of the Rho GTPase with GTP. The role of the PH domain seems to be less well-defined for this family of Rho GEFs, and their conformation and structural interaction with the DH domain can differ significantly between different Rho GEFs.22 Some PH domains are thought to participate in Rho GTPase binding and possibly assist in the nucleotide exchange reaction. The PH domain may also have a role as plasma membrane (PM) anchor or mediator of allosteric control of GEF activity through its interactions with phospholipids. Furthermore, PH domains have been postulated to function as docking sites for other proteins associated with Rho GTPase signaling cascades.2
The question why there are so many GEFs compared with their Rho GTPase targets has not yet been answered with satisfaction. However there are several possible explanations suggested for this interesting phenomenon.
First of all, there is some evidence for the tissue or even cell-specific expression of certain Rho GEFs, for example certain Trio isoforms in the brain.24 Moreover it is possible that certain Rho GEFs are only expressed during particular stages in development, e.g. during the extensive cell migration and neurite network development in embryogenesis, where they perform specific tasks at specific time points. Knockout studies have shown that several Rho GEFs for RhoA are essential for development, and that their function cannot be compensated for by any of the other RhoA specific Rho GEFs.21
Second, almost all Rho GEFs contain additional domains outside their characteristic DH-PH cassette(s). These domains, identified by sequence homology, vary in their functionality and include domains with catalytic activity (Ras GEF / kinase / phosphatase), subcellular localization signals, or modules for protein-protein or protein-membrane interactions.22 These additional domains are likely to contain signaling modules that define context dependent local activity profiles for each Rho GEF in time and space, giving each individual Rho GEF unique signaling properties. A related third option is the possibility that some of these additional domains provide a scaffold function, which sequesters Rho GTPases into specific protein complexes or signaling hubs.
Spatiotemporal aspects of Rho GTPase signaling networks
Rho GTPases regulate complex cellular behavior such as cell migration, neuronal outgrowth and cell polarity, which requires spatial and temporal confinement of signaling networks in cells. Complex cell behavior can only occur when concentrations of signaling proteins are not homogeneous. Moreover, these local gradients must be timed correctly with internal (e.g., cell cycle status) and external signals (e.g., matrix stiffness, chemical cues) to produce relevant cell behavior.
The importance of endomembranes in the generation of activity gradients or local signals, with a special role for the plasma membrane and the vesicular system as integrators of extracellular and intracellular signals, is increasingly recognized.25,26 By localizing proteins on the plasma membrane, the concentration of those molecules is increased up to a 1000-fold due to their spatial confinement in an approximate 2D environment.27 This local concentration effect, which increases the number of active complexes, enables the plasma membrane environment to (temporarily) enhance signaling efficiency. All Rho GTPases contain prenylated CAAX boxes in their C-terminal hypervariable regions, usually in combination with other membrane targeting signals.28 Upon activation they are relocalized to the plasma membrane and other endomembranes where they can exert effects on downstream targets. This highly localized and well-defined Rho GTPase activity is orchestrated by Rho GEFs and Rho GAPs. Rho GEFs and Rho GAPs are often large, multidomain proteins, offering a vast number of possibilities to control their location and activity. By positioning the Rho GEF or Rho GAP near a membrane, scaffold or other cellular structure that is sampled by Rho GTPases, the activity of the Rho GTPase can be effectively increased or decreased respectively. The timing of the activity can be controlled by phosphorylation, allosteric activation or interaction with accessory proteins.2
A clear example of exquisite spatiotemporal control over Rho GTPase activity is provided by p63RhoGEF. The location of the p63RhoGEF is controlled by lipidation of the N-terminus, targeting p63RhoGEF to the plasma membrane. The timing of p63RhoGEF activity is controlled via allosteric activation by a heterotrimeric G-protein (Gaq). Together, these 2 regulatory mechanisms control the location of p63RhoGEF at a micrometer length scale (plasma membrane) and timing of Rho GEF activity at a seconds timescale (heterotrimeric G-protein activation), enabling exquisite control of RhoA activity in cells.29
To advance our knowledge about the complex Rho GEF/Rho GTPase signaling networks that fluctuate in activity over minutes and micrometers, there is a need for technology that perturbs and measures these networks at the right timescales. Currently there are several molecular perturbation techniques available to investigate Rho GTPase signaling networks, which we will review below.
Molecular perturbation technologies
Our current knowledge of Rho GTPases and their regulation by Rho GEFs and Rho GAPs is the result of different experimental approaches from a variety of fields, including genetics, biochemistry, structural biology and cell biology. In cell biology, several molecular perturbation technologies are available that specifically abolish or generate the activity of a single component of the Rho GTPase signaling module. These technologies are used to understand the role of a specific Rho GTPase in a cellular process. Here, we will discuss several perturbation technologies (summarized in Table 1) and we will highlight the benefits and limitations of each strategy. We will evaluate which of the techniques are suitable for disentangling spatiotemporal aspects of Rho GTPase signaling and provide recommendations for future experiments.
Table 1.
Summary of strategies for the perturbation of Rho GEF and Rho GTPase activities in living cells.
| Perturbation strategy | Time scale | Subcellular resolution? | Advantage | Limitation |
|---|---|---|---|---|
| Knock-down (RNAi) | Days | No | Useful in screening | Adaptation, False negatives in case of redundancy |
| Knock-out | Days | No | True null | Adaptation, False negatives in case of redundancy |
| Overexpression | Days | Yes | Simplicity | Adaptation |
| Inhibitor | Minutes-hours | No | Simplicity | Side-effects |
| Adaptation | ||||
| Microinjection | Minutes | No | Single cell kinetics | Labor intensive |
| Heterodimerization–Rapamycin (CID) | Seconds | Yes | Compatible with (FRET) imaging | Irreversible, Side-effects of rapamycin |
| Heterodimerization-Light mediated (LID) | Seconds | Yes | Reversible | Low-throughput |
| Non-invasive |
Micro-injection of constitutive active and dominant negative Rho GTPase variants
A series of seminal publications led by Alan Hall used purified proteins that were introduced into mammalian cells.30–32 First, Rho GTPases that were mutated to either prevent GTPase activity (constitutive active, CA) or GTP binding (dominant negative, DN) were purified from bacteria. Next, the proteins were injected into single Swiss 3T3 fibroblasts and the response of the cells was evaluated by observing their morphology and cytoskeleton by actin staining. Strikingly, several minutes after microinjection, the cells showed distinct changes in shape and cytoskeletal organization. These key studies led to the model that RhoA activity was responsible for stress fiber and focal adhesion formation, Rac1 activity induced pinocytosis and plasma membrane ruffles and Cdc42 activity was involved in the formation of filopodia. There are several important lessons that can be learned from these studies. The 3 Rho GTPases Rho, Rac1 and Cdc42 each have a very characteristic effect on cell morphology. These responses have similarities to the effect of adding growth hormones (EGF, PDGF) and serum (or components thereof, e.g., LPA). The cellular response to the microinjected Rho GTPase is rapid and can be observed after 5–15 minutes. Finally, single cell observations are necessary to appreciate the cellular response to the activated Rho GTPases.
Overexpression of Rho GTPases
Subsequent studies that addressed the effect of Rho GTPases often use CA and DN Rho GTPase variants.33 To circumvent the labor-intensive protein purification and microinjection, the Rho GTPases are encoded by cDNA on a plasmid, which is introduced in cells by transfection. The host cells produce the protein, and evaluation is performed after sufficient time has passed (typically an overnight incubation) for protein expression to take place. Unlike microinjection, this approach does not reveal the cellular response to an acute effect of the Rho GTPase, since the Rho GTPase is slowly produced over time. In fact, compensatory mechanisms may be triggered when the Rho GTPase is synthesized and the cells may adapt to its presence. For instance, it has been shown that overexpressed Rho GTPase perturbs the levels of endogenous Rho GTPases due to competition for a limited and shared pool of Rho GDIs.34 Therefore, experiments that use overexpression of CA or DN Rho GTPase variants should be interpreted with care. As an alternative, constitutive active Rho GEFs can be used. These will activate endogenous Rho GTPases, without affecting the Rho GTPase/Rho GDI balance. It should be noted that Rho GEFs can be identified that are specific for a certain subfamily (e.g., Cdc42 versus Rac1) but not for specific isoforms35 (e.g., RhoA vs. RhoB). Additionally, cellular adaptation to the ectopically expressed Rho GEF may still occur, for example by compensatory expression of additional Rho GAPs over time.
Knock-down and knockout approaches
The advent of RNAi has spurred a sizeable amount of studies that use this technique to knock down a specific Rho GTPase or Rho GEF36 or perform large scale screening studies.37,38 The RNAi (or siRNA/shRNA) methodology promotes the degradation of mRNA in the host cell, which in turn will reduce protein levels. The efficiency of this method depends on protein stability and will be more successful for proteins that have a high turnover due to rapid degradation. On the other hand, genetic knockouts (by gene-targeting or genome editing) will result in complete absence of the protein. Historically, cells were isolated from knockout animals to study the effects of Rho GTPases.4,39 Nowadays, gene-editing techniques, particularly the generation of indels by CRISPR/Cas9, is a straightforward way to generate knockout of Rho GTPases directly in the cell type of interest.
A recent study highlights the striking adaptation-based difference in signaling outcome between a short-term RNAi knockout and a long-term genetic knockout of the Rac1/Cdc42 GEF Dock6.40 Short-term RNAi mediated depletion of Dock6 triggered actin cytoskeleton collapse and cell rounding during interphase, while long-term genetic knockout approaches did not show obvious actin defects and cells were spread out well during interphase.
Even though siRNA is a powerful tool to perform initial large-scale screenings based on phenotype, acute perturbation is impossible. Therefore, adaptation may occur and similar concerns exist as for plasmid based overexpression studies.
Drugs
Small molecule based drugs represent an important class of reagents that can be used to study protein function. Moreover, these compounds can potentially be used in the clinic to treat diseases. Thus far, several drugs have been reported that target Rho GTPases (e.g., statins41, A1642), Rho GEFs (e.g., ITX343, Y1644) or the interaction interface between Rho GTPases and Rho GEFs (e.g., Rhosin,45 NSC2376646). The kinetics of the response to the drug depends on the properties of the drug (solubility, cell permeability, stability) and its mechanism of action. Optimal drug effects require the optimization of the dose and the incubation time. Once optimized, drugs can be used to perturb signaling at minutes to hours timescales.
A general concern with drugs is their specificity. A clear-cut example is provided by statins. Statins reduce the membrane affinity of Rho GTPases by inhibiting prenylation, but they act on any prenyated protein, e.g., Gγ.47 Another issue is that each Rho GTPase has numerous roles in cell functioning. Hence, even specific inhibition of a Rho GTPase will affect many cellular processes. To increase the specificity of drug action for a specific cellular process, it has been suggested to target the Rho GEF-Rho GTPase interface.48
Small molecule induced activation
Chemical-induced dimerization (CID) is a powerful system to control protein location. Currently, the most popular system is based on the high affinity interaction between rapamycin and 2 of its naturally occurring binding proteins, the rapamycin binding domain of mTOR (FRB) and FK506 binding protein 12 (FKBP12). Rapamycin induces heterodimerization of FRB and FKBP12 within seconds. To circumvent side-effects due to binding of rapamycin to mTOR, rapamycin analogs have been generated that cannot bind mTOR.49
The FRB domain is frequently used as ‘anchor’ for the dimerization system and fused to a protein or amino acid sequence that targets it to a specific location in the cell, e.g., the plasma membrane, the Golgi apparatus or mitochondria.50,51 The FKBP12 domain can be fused to a protein of interest. The subsequent addition of rapamycin can then be used to gain spatiotemporal control over the subcellular location of FKBP12-bound protein during live cell imaging experiments (Fig. 1 and supplemental movie 1).
Figure 1.
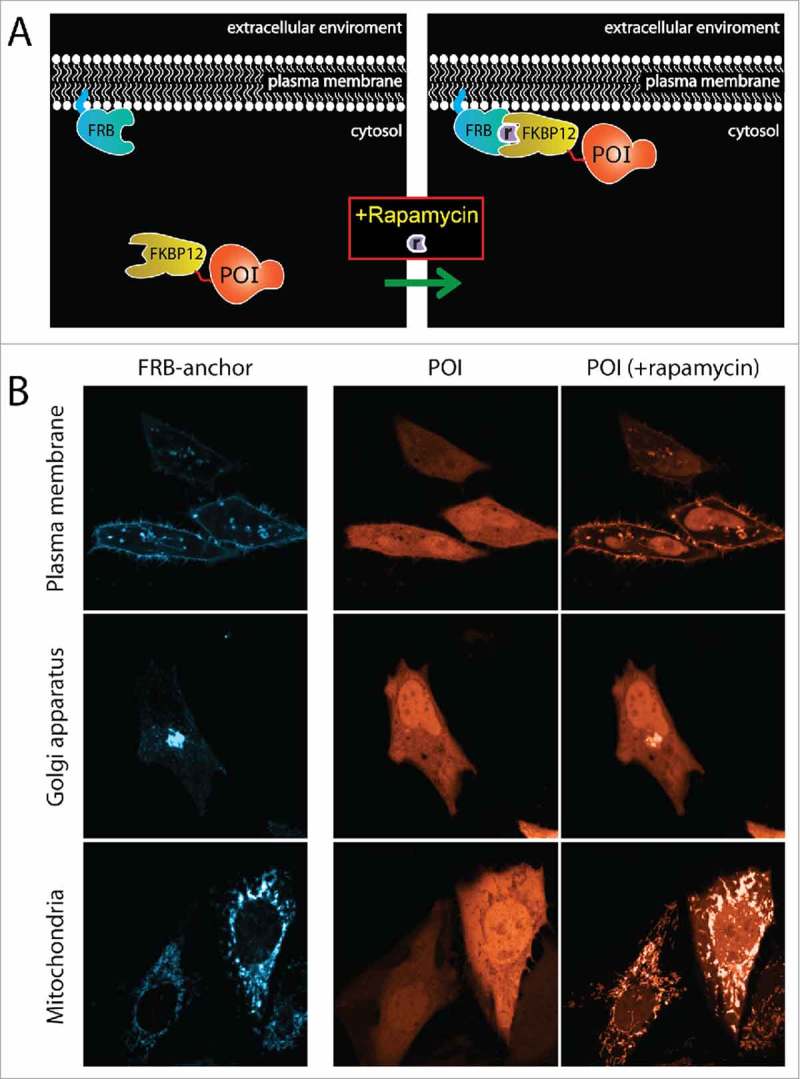
Basic principles of the rapamycin hetero-dimerization system. (A) The rapamycin system consists of 2 small protein domains, FRB and FKBP12, which dimerize upon addition of the small molecule rapamycin. In this example the FRB domain is fused to a plasma membrane targeting sequence. The FKBP12 molecule is fused to a protein of interest (POI), which binds FRB when rapamycin is added, thereby recruiting it to the plasma membrane. (B) The POI (in this case a red fluorescent protein (RFP) was fused to FKBP12) can be targeted to several subcellular locations; e.g., the plasma membrane (top), the Golgi apparatus (middle) and mitochondria (bottom). Left column depicts the location of the FRB anchor (cyan), right columns show the POI before and after addition of rapamycin to the sample (orange). Marten Postma generously provided the LUTs used in (B).
It has been shown that recruiting a Rho GTPase or Rho GEF from the cytosol to the plasma membrane is sufficient to induce profound cell shape changes.52 A follow-up study revealed that recruiting a Rho GEF to the plasma membrane increases RhoA activity, but that relocating the same GEF to either mitochondria or the Golgi apparatus did not alter RhoA activity.51 Thus, demonstrating that activation of a Rho GTPase depends on the subcellular location of the GEF.
One of the challenges is to switch from a low activity state, i.e. cytosolic location, to a fully active state. To achieve this the basal activity of the non-localized Rho GTPase or Rho GEF should be low. Basal activity may be reduced by tethering the protein domain to a subcellular compartment where it is not active.53 Although the CID method is often used to induce activation of signaling it can also be used to inactivate proteins.54 The recruitment of GAP activity, thus far not demonstrated, may be used to inactivate Rho GTPase signaling at specific locations.
To further improve this system, caged (locally inducible), light-inducible and reversible variants of the rapamycin system are currently in development.49 Several alternative CID systems exist that use other small molecules.55 A dimerization system based on the small molecule MeNV-HaXS was recently described.56 This CID system is based on the dimerization between a SNAP and a Halo-tag by MeNV-HaXS, which can be reversed by the blue-light mediated photo cleavage of the dimerizer. Another reversible CID system reports the use of trimethoprim (TMR)-based ligands to induce the dimerization between an E. coli dihydrofolate reductase (eDHFR) anchored to an organelle of interest and protein of interest fused to a mutated version of FKBP (FKBP').57 In this system a synthetic ligand of FKBP' (SLF') -TMR is used to induce dimerization, while regular TMR can act as a competitor for eDHFR, effectively releasing the FKBP'-bound protein of interest. Because FKBP' can still bind rapamycin, this system can be used orthogonally with a rapamycin based FRB anchor, providing means to translocate a protein of interest twice within a living cell.
Light-induced activation
One of the first examples of light-induced Rho GTPase activity is the engineering of a photoactivatable Rac1.58 A photosensitive protein domain sterically hinders Rac1 activity by blocking its interaction with effectors. Upon absorption of light by the photosensitive Light Oxygen Voltage-sensing (LOV) domain, its interaction with Rac1 is reduced and Rac1 activity toward effectors is increased. A more generalizable strategy was recently reported and applied to both Rho GTPases and Rho GEFs.59
Another flexible strategy, similar to CID, is to use light-induced dimerization (LID), similar to the chemical induced dimerization. Several light-induced dimerization systems have been reported60 with different characteristics. Some aspects to consider when using these systems are (i) reversibility, (ii) requirement for addition of co-factor and (iii) activation spectrum (the optimal wavelength to activate the system). These systems have in common that activity can be controlled with high spatial resolution. By guiding the light to a small region in a cell, effects of local activity can be evaluated. Several of these systems have been used to demonstrate that light-induced recruitment of a Rho GEF can be used to induce local cell shape changes.61,62
Conclusion and recommendations
Of all the perturbation techniques discussed here, only microinjection, CID and LID enable acute perturbation. Since microinjection is labor-intensive and relatively invasive we do not consider it further. Both CID and LID allow acute perturbation with high subcellular resolution. In case of CID this is achieved by placing the anchor at specific membranes or structures. This can also be done for LID, but local activation with this system can also be achieved by pointing the activating light at specific cellular locations. The Rho GTPase signaling network in cells is tightly regulated by Rho GEF and Rho GAP activity on a timescale of seconds and micrometers,8,63 and CID and LID are the only methods that can perturb the signaling network on both this time and length scale in single cells (Table 1). The read-out from the other perturbation techniques will suffer from adaptation of the cell based on both biochemical and mechanical feedback.64 It was recently shown that the long-term (genomically altered) perturbation of a Rho GEF results in completely different signaling response compared with short-term perturbations (overnight).40 We believe the same will hold true on the scale of minutes to hours.
One of the challenges is to combine local activation with a quantitative read-out of the cellular response. Shape changes can be quantified51 and may be informative to study effects of local Rho GTPase activity on cell morphology. More detailed information can be obtained by using specific probes that report on local effects of signaling proteins or the cytoskeleton. For instance, fluorescent protein fusions with actin probes can be paired with any of the described systems. Similarly, the location and dynamics of myosin II, microtubules or focal adhesion proteins can be followed over time.
Powerful and direct read-outs can be acquired by using FRET based sensors, providing quantitative information with high spatial and temporal resolution.11 Since most high-contrast based FRET sensors are based on CFP-YFP pairs, light based systems that use blue light are not compatible. However, FRET based reporters can be paired with the rapamycin system or light-based systems that use red light. An alternative solution is to develop red-shifted FRET pairs that do not require blue light for imaging.
To conclude, there are several systems for the local and acute (in)activation of Rho GTPases signaling. These systems match the second timescales and micrometer length-scales at which Rho GTPase activity is controlled in cells. Therefore, these perturbation strategies provide a powerful toolkit to examine effects of local signaling by Rho GTPases in a wide variety of processes. Nature will undoubtedly continue to surprise us, and other genetically encoded modules that enable ways to activate local activities are anticipated. We are convinced that the combination of acute local perturbation and single cell analysis will greatly improve our understanding of Rho GTPase signaling.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- [1].Etienne-Manneville S, Hall A. Rho GTPases in cell biology. Nature 2002; 420:629-35; PMID:12478284; https://doi.org/ 10.1038/nature01148 [DOI] [PubMed] [Google Scholar]
- [2].Cherfils J, Zeghouf M. Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol Rev 2013; 93:269-309; PMID:23303910; https://doi.org/ 10.1152/physrev.00003.2012 [DOI] [PubMed] [Google Scholar]
- [3].Spiering D, Hodgson L. Dynamics of the Rho-family small GTPases in actin regulation and motility. Cell Adh Migr 2011; 5:170-80; PMID:21178402; https://doi.org/ 10.4161/cam.5.2.14403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Heasman SJ, Ridley AJ. Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat Rev Mol Cell Biol 2008; 9:690-701; PMID:18719708; https://doi.org/ 10.1038/nrm2476 [DOI] [PubMed] [Google Scholar]
- [5].Hall A. Rho GTPases and the actin cytoskeleton. Science 1998; 279:509-14; PMID:9438836; https://doi.org/ 10.1126/science.279.5350.509 [DOI] [PubMed] [Google Scholar]
- [6].Jalink K, van Corven EJ, Hengeveld T, Morii N, Narumiya S, Moolenaar WH. Inhibition of lysophosphatidate- and thrombin-induced neurite retraction and neuronal cell rounding by ADP ribosylation of the small GTP-binding protein Rho. J Cell Biol 1994; 126:801-10; PMID:8045941; https://doi.org/ 10.1083/jcb.126.3.801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Small JV, Stradal T, Vignal E, Rottner K. The lamellipodium: where motility begins. Trends Cell Biol 2002; 12:112-20; PMID:11859023; https://doi.org/ 10.1016/S0962-8924(01)02237-1 [DOI] [PubMed] [Google Scholar]
- [8].Pertz O. Spatio-temporal Rho GTPase signaling - where are we now? J Cell Sci 2010; 123:1841-50; PMID:20484664; https://doi.org/ 10.1242/jcs.064345 [DOI] [PubMed] [Google Scholar]
- [9].Fritz RD, Letzelter M, Reimann A, Martin K, Fusco L, Ritsma L, Ponsioen B, Fluri E, Schulte-Merker S, van Rheenen J, et al.. A Versatile Toolkit to Produce Sensitive FRET Biosensors to Visualize Signaling in Time and Space. Sci Signal 2013; 6:rs12; PMID:23882122; https://doi.org/ 10.1126/scisignal.2004135 [DOI] [PubMed] [Google Scholar]
- [10].Yoshizaki H. Activity of Rho-family GTPases during cell division as visualized with FRET-based probes. J Cell Biol 2003; 162:223-32; PMID:12860967; https://doi.org/ 10.1083/jcb.200212049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Machacek M, Hodgson L, Welch C, Elliott H, Pertz O, Nalbant P, Abell A, Johnson GL, Hahn KM, Danuser G. Coordination of Rho GTPase activities during cell protrusion. Nature 2009; 461:99-103; PMID:19693013; https://doi.org/ 10.1038/nature08242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Martin K, Vilela M, Jeon NL, Danuser G, Pertz O. A growth factor-induced, spatially organizing cytoskeletal module enables rapid and persistent fibroblast migration. Dev Cell 2014; 30:701-16; PMID:25268172; https://doi.org/ 10.1016/j.devcel.2014.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Martin K, Reimann A, Fritz RD, Ryu H, Jeon NL, Pertz O. Spatio-temporal co-ordination of RhoA, Rac1 and Cdc42 activation during prototypical edge protrusion and retraction dynamics. Sci Rep 2016; 6:21901; PMID:26912264; https://doi.org/ 10.1038/srep21901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Zawistowski JS, Sabouri-Ghomi M, Danuser G, Hahn KM, Hodgson L. A RhoC biosensor reveals differences in the activation kinetics of RhoA and RhoC in migrating cells. PLoS One 2013; 8:e79877; PMID:24224016; https://doi.org/ 10.1371/journal.pone.0079877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Moshfegh Y, Bravo-Cordero JJ, Miskolci V, Condeelis J, Hodgson L. A Trio-Rac1-Pak1 signalling axis drives invadopodia disassembly. Nat Cell Biol 2014; 16:574-86; PMID:24859002; https://doi.org/ 10.1038/ncb2972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Benink HA, Bement WM. Concentric zones of active RhoA and Cdc42 around single cell wounds. J Cell Biol 2005; 168:429-39; PMID:15684032; https://doi.org/ 10.1083/jcb.200411109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Simon CM, Vaughan EM, Bement WM, Edelstein-Keshet L. Pattern formation of Rho GTPases in single cell wound healing. Mol Biol Cell 2013; 24:421-32; PMID:23264464; https://doi.org/ 10.1091/mbc.E12-08-0634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Moissoglu K, Schwartz MA. Spatial and temporal control of Rho GTPase functions. Cell Logist 2014; 4:e943618; PMID:25610718; https://doi.org/ 10.4161/21592780.2014.943618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Fusco L, Lefort R, Smith K, Benmansour F, Gonzalez G, Barillari C, Rinn B, Fleuret F, Fua P, Pertz O. Computer vision profiling of neurite outgrowth dynamics reveals spatiotemporal modularity of Rho GTPase signaling. J Cell Biol 2016; 212:91-111; PMID:26728857; https://doi.org/ 10.1083/jcb.201506018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Yang HW, Collins SR, Meyer T. Locally excitable Cdc42 signals steer cells during chemotaxis. Nat Cell Biol 2015; 18:191-201; PMID:26689677; https://doi.org/ 10.1038/ncb3292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Cook DR, Rossman KL, Der CJ. Rho guanine nucleotide exchange factors: regulators of Rho GTPase activity in development and disease. Oncogene 2013; 33:4021-35; PMID:24037532; https://doi.org/ 10.1038/onc.2013.362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Rossman KL, Der CJ, Sondek J. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat Rev Mol Cell Biol 2005; 6:167-80; PMID:15688002; https://doi.org/ 10.1038/nrm1587 [DOI] [PubMed] [Google Scholar]
- [23].Côté J-F, Vuori K. GEF what? Dock180 and related proteins help Rac to polarize cells in new ways. Trends Cell Biol 2007; 17:383-93; PMID:17765544; https://doi.org/ 10.1016/j.tcb.2007.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Portales-Casamar E, Briançon-Marjollet A, Fromont S, Triboulet R, Debant A. Identification of novel neuronal isoforms of the Rho-GEF Trio. Biol Cell 2006; 98:183-93; PMID:16033331; https://doi.org/ 10.1042/BC20050009 [DOI] [PubMed] [Google Scholar]
- [25].Grecco HE, Schmick M, Bastiaens PIH. Signaling from the living plasma membrane. Cell 2011; 144:897-909; PMID:21414482; https://doi.org/ 10.1016/j.cell.2011.01.029 [DOI] [PubMed] [Google Scholar]
- [26].Groves JT, Kuriyan J. Molecular mechanisms in signal transduction at the membrane. Nat Struct Mol Biol 2010; 17:659-65; PMID:20495561; https://doi.org/ 10.1038/nsmb.1844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kholodenko BN, Hoek JB, Westerhoff HV. Why cytoplasmic signalling proteins should be recruited to cell membranes. Trends Cell Biol 2000; 10:173-8; PMID:10754559; https://doi.org/ 10.1016/S0962-8924(00)01741-4 [DOI] [PubMed] [Google Scholar]
- [28].Michaelson D, Silletti J, Murphy G, D'Eustachio P, Rush M, Philips MR. Differential localization of Rho GTPases in live cells: regulation by hypervariable regions and RhoGDI binding. J Cell Biol 2001; 152:111-26; PMID:11149925; https://doi.org/ 10.1083/jcb.152.1.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].van Unen J, Yin T, Wu YI, Mastop M, Gadella TWJ, Goedhart J. Kinetics of recruitment and allosteric activation of ARHGEF25 isoforms by the heterotrimeric G-protein Gαq. Sci Rep 2016; 6:36825; https://doi.org/ 10.1038/srep36825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Paterson HF, Self AJ, Garrett MD, Just I, Aktories K, Hall A. Microinjection of recombinant p21rho induces rapid changes in cell morphology. J Cell Biol 1990; 111:1001-7; PMID:2118140; https://doi.org/ 10.1083/jcb.111.3.1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Nobes CD, Hall A. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell 1995; 81:53-62; PMID:7536630; https://doi.org/ 10.1016/0092-8674(95)90370-4 [DOI] [PubMed] [Google Scholar]
- [32].Ridley AJ, Hall A. The small GTP-binding protein rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell 1992; 70:389-99; PMID:1643657; https://doi.org/ 10.1016/0092-8674(92)90163-7 [DOI] [PubMed] [Google Scholar]
- [33].Feig LA. Tools of the trade: use of dominant-inhibitory mutants of Ras-family GTPases. Nat Cell Biol 1999; 1:E25-7; PMID:10559887; https://doi.org/ 10.1038/10018 [DOI] [PubMed] [Google Scholar]
- [34].Boulter E, Garcia-Mata R, Guilluy C, Dubash A, Rossi G, Brennwald PJ, Burridge K. Regulation of Rho GTPase crosstalk, degradation and activity by RhoGDI1. Nat Cell Biol 2010; 12:477-83; PMID:20400958; https://doi.org/ 10.1038/ncb2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Jaiswal M, Dvorsky R, Ahmadian MR. Deciphering the molecular and functional basis of Dbl family proteins: a novel systematic approach toward classification of selective activation of the Rho family proteins. J Biol Chem 2013; 288:4486-500; PMID:23255595; https://doi.org/ 10.1074/jbc.M112.429746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Monypenny J, Zicha D, Higashida C, Oceguera-Yanez F, Narumiya S, Watanabe N. Cdc42 and Rac family GTPases regulate mode and speed but not direction of primary fibroblast migration during platelet-derived growth factor-dependent chemotaxis. Mol Cell Biol 2009; 29:2730-47; PMID:19273601; https://doi.org/ 10.1128/MCB.01285-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Vaqué JP, Dorsam RT, Feng X, Iglesias-Bartolome R, Forsthoefel DJ, Chen Q, Debant A, Seeger MA, Ksander BR, Teramoto H, et al.. A Genome-wide RNAi screen reveals a Trio-Regulated Rho GTPase circuitry transducing mitogenic signals initiated by G Protein-coupled receptors. Mol Cell 2012; 49:94-108; PMID:23177739; https://doi.org/ 10.1016/j.molcel.2012.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Sanz-Moreno V, Gadea G, Ahn J, Paterson H, Marra P, Pinner S, Sahai E, Marshall CJ. Rac activation and inactivation control plasticity of tumor cell movement. Cell 2008; 135:510-23; PMID:18984162; https://doi.org/ 10.1016/j.cell.2008.09.043 [DOI] [PubMed] [Google Scholar]
- [39].Wang L, Zheng Y. Cell type-specific functions of Rho GTPases revealed by gene targeting in mice. Trends Cell Biol 2007; 17:58-64; PMID:17161947; https://doi.org/ 10.1016/j.tcb.2006.11.009 [DOI] [PubMed] [Google Scholar]
- [40].Cerikan B, Shaheen R, Colo GP, Gläßer C, Hata S, Knobeloch K-P, Alkuraya FS, Fässler R, Schiebel E. Cell-Intrinsic adaptation arising from chronic ablation of a key Rho GTPase regulator. Dev Cell 2016; 39:28-43; PMID:27693507; https://doi.org/ 10.1016/j.devcel.2016.08.020 [DOI] [PubMed] [Google Scholar]
- [41].Riganti C, Aldieri E, Doublier S, Bosia A, Ghigo D. Statins-mediated inhibition of rho GTPases as a potential tool in anti-tumor therapy. Mini Rev Med Chem 2008; 8:609-18; PMID:18537716; https://doi.org/ 10.2174/138955708784534436 [DOI] [PubMed] [Google Scholar]
- [42].Cromm PM, Spiegel J, Grossmann TN, Waldmann H. Direct Modulation of Small GTPase Activity and Function. Angew Chem Int Ed Engl 2015; 54:13516-37; PMID:26470842; https://doi.org/ 10.1002/anie.201504357 [DOI] [PubMed] [Google Scholar]
- [43].Bouquier N, Vignal E, Charrasse S, Weill M, Schmidt S, Léonetti J-P, Blangy A, Fort P. A cell active chemical GEF inhibitor selectively targets the Trio/RhoG/Rac1 signaling pathway. Chem Biol 2009; 16:657-66; PMID:19549603; https://doi.org/ 10.1016/j.chembiol.2009.04.012 [DOI] [PubMed] [Google Scholar]
- [44].Shang X, Marchioni F, Evelyn CR, Sipes N, Zhou X, Seibel W, Wortman M, Zheng Y. Small-molecule inhibitors targeting G-protein-coupled Rho guanine nucleotide exchange factors. Proc Natl Acad Sci USA 2013; 110:3155-60; PMID:23382194; https://doi.org/ 10.1073/pnas.1212324110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Shang X, Marchioni F, Sipes N, Evelyn CR, Jerabek-Willemsen M, Duhr S, Seibel W, Wortman M, Zheng Y. Rational design of small molecule inhibitors targeting RhoA Subfamily Rho GTPases. Chem Biol 2012; 19:699-710; PMID:22726684; https://doi.org/ 10.1016/j.chembiol.2012.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Gao Y, Dickerson JB, Guo F, Zheng J, Zheng Y. Rational design and characterization of a Rac GTPase-specific small molecule inhibitor. Proc Natl Acad Sci USA 2004; 101:7618-23; PMID:15128949; https://doi.org/ 10.1073/pnas.0307512101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Liao JK, Laufs U. Pleiotropic effects of statins. Annu Rev Pharmacol Toxicol 2005; 45:89-118; PMID:15822172; https://doi.org/ 10.1146/annurev.pharmtox.45.120403.095748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Lin Y, Zheng Y. Approaches of targeting Rho GTPases in cancer drug discovery. Expert Opin Drug Discov 2015; 10:991-1010; PMID:26087073; https://doi.org/ 10.1517/17460441.2015.1058775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Putyrski M, Schultz C. Protein translocation as a tool: The current rapamycin story. FEBS Lett 2012; 586:2097-105; PMID:22584056; https://doi.org/ 10.1016/j.febslet.2012.04.061 [DOI] [PubMed] [Google Scholar]
- [50].Komatsu T, Kukelyansky I, McCaffery JM, Ueno T, Varela LC, Inoue T. organelle-specific, rapid induction of molecular activities and membrane tethering. Nat Methods 2010; 7(3):206-8; PMID:20154678; https://doi.org/ 10.1038/nmeth.1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].van Unen J, Reinhard NR, Yin T, Wu YI, Postma M, Gadella TWJ, Goedhart J. Plasma membrane restricted RhoGEF activity is sufficient for RhoA-mediated actin polymerization. Sci Rep 2015; 5:14693; PMID:26435194; https://doi.org/ 10.1038/srep14693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Inoue T, Heo WD, Grimley JS, Wandless TJ, Meyer T. An inducible translocation strategy to rapidly activate and inhibit small GTPase signaling pathways. Nat Methods 2005; 2:415-8; PMID:15908919; https://doi.org/ 10.1038/nmeth763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Phua SC, Pohlmeyer C, Inoue T. Rapidly relocating molecules between organelles to manipulate small GTPase activity. ACS Chem Biol 2012; 7:1950-5; PMID:22999378; https://doi.org/ 10.1021/cb300280k [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Robinson MS, Sahlender DA, Foster SD. Rapid inactivation of proteins by rapamycin-induced rerouting to mitochondria. Dev Cell 2010; 18:324-31; PMID:20159602; https://doi.org/ 10.1016/j.devcel.2009.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Voss S, Klewer L, Wu YW. Chemically induced dimerization: reversible and spatiotemporal control of protein function in cells. Curr Opin Chem Biol 2015; 28:194-201; PMID:26431673; https://doi.org/ 10.1016/j.cbpa.2015.09.003 [DOI] [PubMed] [Google Scholar]
- [56].Zimmermann M, Cal R, Janett E, Hoffmann V, Bochet CG, Constable E, Beaufils F, Wymann MP. Cell-permeant and photocleavable chemical inducer of dimerization. Angew Chem Int Ed Engl 2014; 53:4717-20; PMID:24677313; https://doi.org/ 10.1002/anie.201310969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Liu P, Calderon A, Konstantinidis G, Hou J, Voss S, Chen X, Li F, Banerjee S, Hoffmann J-E, Theiss C, et al.. A bioorthogonal small-molecule-switch system for controlling protein function in live cells. Angew Chem Int Ed Engl 2014; 53:10049-55; PMID:25065762; https://doi.org/ 10.1002/anie.201403463 [DOI] [PubMed] [Google Scholar]
- [58].Wu YI, Frey D, Lungu OI, Jaehrig A, Schlichting I, Kuhlman B, Hahn KM. A genetically encoded photoactivatable Rac controls the motility of living cells. Nature 2009; 461:104-8; PMID:19693014; https://doi.org/ 10.1038/nature08241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Dagliyan O, Tarnawski M, Chu P-H, Shirvanyants D, Schlichting I, Dokholyan NV, Hahn KM. Engineering extrinsic disorder to control protein activity in living cells. Science 2016; 354:1441-4; PMID:27980211; https://doi.org/ 10.1126/science.aah3404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Shcherbakova DM, Shemetov AA, Kaberniuk AA, Verkhusha VV. Natural photoreceptors as a source of fluorescent proteins, biosensors, and optogenetic tools. Annu Rev Biochem 2015; 84:519-50; PMID:25706899; https://doi.org/ 10.1146/annurev-biochem-060614-034411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Guntas G, Hallett RA, Zimmerman SP, Williams T, Yumerefendi H, Bear JE, Kuhlman B. Engineering an improved light-induced dimer (iLID) for controlling the localization and activity of signaling proteins. Proc Natl Acad Sci USA 2015; 112:112-7; PMID:25535392; https://doi.org/ 10.1073/pnas.1417910112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Levskaya A, Weiner OD, Lim WA, Voigt CA. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature 2009; 461:997-1001; PMID:19749742; https://doi.org/ 10.1038/nature08446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Fritz RD, Pertz O. The dynamics of spatio-temporal Rho GTPase signaling: formation of signaling patterns. F1000Res 2016; 5. pii: F1000 Faculty Rev-749; PMID:27158467; https://doi.org/ 10.12688/f1000research.7370.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Artemenko Y, Axiotakis L, Borleis J, Iglesias PA, Devreotes PN. Chemical and mechanical stimuli act on common signal transduction and cytoskeletal networks. Proc Natl Acad Sci USA 2016; 113:E7500-9; PMID:27821730; https://doi.org/ 10.1073/pnas.1608767113 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


