Abstract
When climatic or environmental conditions change, plant populations must either adapt to these new conditions, or track their niche via seed dispersal. Adaptation of plants to different abiotic environments has mostly been discussed with respect to physiological and demographic parameters that allow local persistence. However, rapid modifications in response to changing environmental conditions can also affect seed dispersal, both via plant traits and via their dispersal agents. Studying such changes empirically is challenging, due to the high variability in dispersal success, resulting from environmental heterogeneity, and substantial phenotypic variability of dispersal-related traits of seeds and their dispersers. The exact mechanisms that drive rapid changes are often not well understood, but the ecological implications of these processes are essential determinants of dispersal success, and deserve more attention from ecologists, especially in the context of adaptation to global change. We outline the evidence for rapid changes in seed dispersal traits by discussing variability due to plasticity or genetics broadly, and describe the specific traits and biological systems in which variability in dispersal is being studied, before discussing some of the potential underlying mechanisms. We then address future research needs and propose a simulation model that incorporates phenotypic plasticity in seed dispersal. We close with a call to action and encourage ecologists and biologist to embrace the challenge of better understanding rapid changes in seed dispersal and their consequences for the reaction of plant populations to global change.
Global ecological change is causing plant populations to either adapt or move in response to new environmental conditions. In many species it is thought that seed dispersal may not allow populations to move at a rate that tracks the shifting climate. However, phenotypic plasticity and rapid genetic changes in traits associated with seed dispersal may modify the response capabilities of plants and allow them to responds more quickly. We explore the evidence for rapid modification of seed dispersal traits and propose a path forward to better understand the ways in which seed dispersal may buffer the effects of global ecological change.
Introduction
Global environmental change, in all its forms, is considered one of society’s greatest challenges today. When climatic envelopes shift (Loarie et al. 2009; Mahony et al. 2017), or when frugivore communities change due to invasion or over-exploitation (Bleher and Böhning-Gaese 2001), plant populations can react by either adapting locally to the new environmental conditions (Cochrane et al. 2015) or by dispersing, attempting to track their climatic envelope or ecological niche (Corlett and Westcott 2013; Johnson et al. 2016; Graae et al. 2018). If their dispersal abilities do not match the shift of their ecological niche, a rapid change in, or dependence on, dispersal ability could allow them to respond. Whilst rapid changes in plant traits, such as phenology, mortality and metabolic processes to changed environmental or biotic conditions have been addressed relatively widely (Nicotra et al. 2010; Bardgett et al. 2014; Gratani 2014; Graae et al. 2018), rapid changes in traits affecting plant movement have received considerably less attention, and few studies report evolutionary responses on these traits.
A possible interpretation of this lack of empirical evidence for adaptation of seed dispersal is that selection in plants acts on the immediate factors regulating local survival rather than a process that determines the location of seeds, which then influences survival. However, given the importance of dispersal for the inclusive fitness of a plant, we contend that adaptive changes in seed dispersal do occur. The more plausible explanation for the lack of evidence is that plant movement (dispersal) is notoriously difficult to study (Rogers et al., this issue-a). We hardly understand dispersal as it is, making it challenging to detect changes in dispersal processes due to global change. Moreover, one might conjecture that dispersal becomes more important as climate change progresses and increasing mismatches between current plant distributions and their environmental optima intensify.
For dispersal traits to change rapidly in response to changes in the biotic and abiotic environment, phenotypic diversity associated with dispersal must be present or phenotypes must respond in a plastic manner when they encounter novel environments. Dispersal, defined as the movement of a propagule from its natal source with consequences for gene flow through space (Ronce 2007), has a complex, mostly polygenic, genetic architecture. The speed at which dispersal traits, or suites of traits, can change is associated with the underlying genetic sequence, phenotypic variation in dispersal traits and their covariance with other traits under selection (Saastamoinen et al. 2018), and/or epigenetic mechanisms leading to plastic responses to environmental variability. Whether the mechanisms of rapid changes of dispersal traits are the product of genetic or plastic variation, the outcomes are similar because improved dispersal ability will similarly increase population spread potential.
Our goal in this perspective is to call attention to current research on the potential of rapid changes in seed dispersal as a response to global change. We highlight that, despite some progress, our understanding of the variability regulating seed dispersal remains poor. This is primarily because of the high intrinsic variability of dispersal-related traits of seeds and their dispersers coupled with the high variability in effective dispersal resulting from environmental heterogeneity. Moreover, the logistical challenges, such as long lifespan and the large number of seeds produced, make studying rapid changes in seed dispersal difficult, and few studies to date have tackled these questions. We organize our essay by first discussing the specific traits and biological systems in which changes in dispersal are being studied, along with the ways in which these questions are currently being addressed. Then we discuss potential mechanisms that could result in rapid changes in seed dispersal. We close with recommendations of future research needs and propose a model that incorporates phenotypic plasticity while stressing the need for more empirical studies to untangle the role of genetic and plastic contributions to adaptation of seed dispersal. We show that there is evidence for rapid changes in seed dispersal through multiple mechanisms and that they may allow plants to respond quickly to changing environments (Fig. 1).
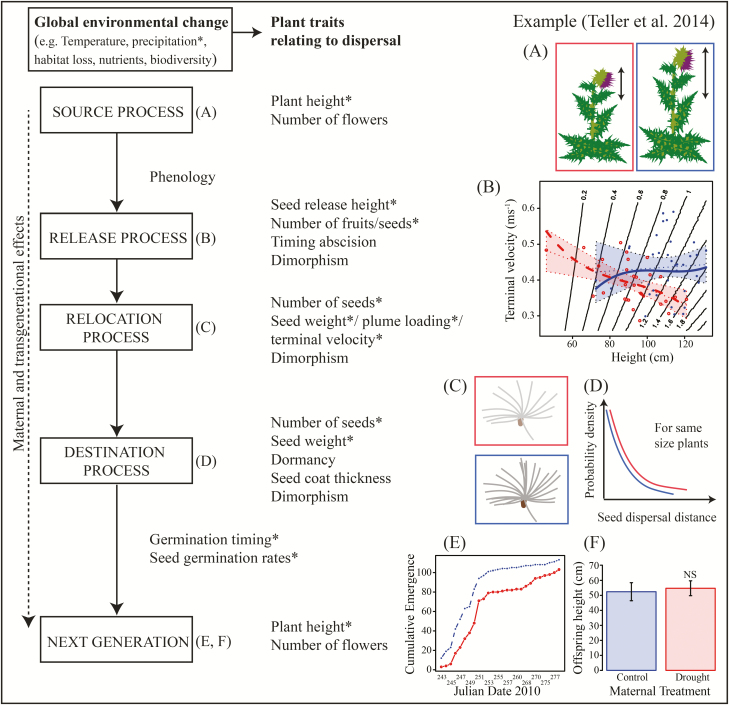
Figure 1.
Dispersal stages, and possibilities of their modification via rapid responses of plant dispersal traits to global environmental change. Global environmental change includes changes in temperature, precipitation, habitat availability, biodiversity and nutrient availability, which in turn can elicit rapid responses in plant traits affecting their dispersal. These responses may affect traits of the maternal plant up to seed release (Source processes), traits that directly influence the initiation of dispersal (maternal or seed traits) (Release processes) and the dispersal process itself (seed traits) (Relocation process), traits that are important after the dispersal process (seed traits) (Destination process) and might even affect the performance of the next generation (maternal and transgenerational effects) (Process stages Jongejans et al. 2015). Potential consequences of a rapid response of dispersal traits to climate change are exemplified with a study by Teller et al. (2014). They studied the effect of drought on varying dispersal traits during different stages of the dispersal process (traits with *) in Carduus nutans. Drought conditions (red figures) reduced plant and seed release height, whereas well-watered plants (blue figures) showed more variability in their height (A). Interestingly, seeds from taller C. nutans plants in the drought treatment should disperse at least as far as same-sized individuals under well-watered conditions due to a decrease in seed terminal velocity in the drought treatment (B). Under drought stress, phenotypic plasticity of maternal plant and seed traits (C) could hence favour longer distance dispersal (D) but with the cost of fewer seeds that might also germinate later (E). Maternal effects for the next generation could not be observed in this example (F). Panels (B), (E) and (F) are reproduced with permission from Teller et al. (2014).
Traits Important to Seed Dispersal
Several traits are important contributors to patterns of seed dispersal. These traits or suite of traits are variable at the subindividual level in their range of phenotypes and plant fitness can be both influenced and mediated by environmental variation (Schupp, this issue; Snell et al. 2019) through different pathways. Though specific traits vary depending on dispersal mode (e.g. anemochory vs. epizoochory) and life-history strategies (Beckman et al. 2018), here we have grouped them into three primary types: traits related to movement, phenology and longevity. Plasticity of movement-related traits—e.g. those associated with attractiveness to frugivores, attachment to dispersers or aerodynamics—is the most obvious, but plants may also vary in phenology/timing of propagule production, or in the length of dormancy on the plant or the ground, with consequences for seed dispersal and hence impacts on plant fitness.
Traits that influence abiotic seed dispersal are largely related to the structure of the seed or plant (see Fig. 1). For wind-dispersed species, seed weight affects falling velocity (Teller et al. 2014), plant height affects distance (Thomson et al. 2011; Zhang et al. 2012) and seed morphology influences the dispersal kernel (Augspurger and Franson 1987). In Arabidopsis thaliana, plant density influences dispersal distance (Wender et al. 2005). Both phenotypic plasticity and genetic variation are responsible for reduced dispersal at the range limits of Cakile edentula (LaRue et al. 2018). For epizoochorous species, the type of attachment affects the likelihood of hitching a ride on a passing animal. Some wind-dispersed and epizoochorous species have heteromorphic seeds (e.g. Martorell and López 2014) which allows them to utilize multiple dispersal vectors, and rapid evolution of a second seed type with a different dispersal mode has been shown to occur within 5–12 generations of selection (Cheptou et al. 2008).
In endozoochorous systems, a complex suite of traits may influence movement, as plants typically respond to a dynamic community of frugivores and may use a variety of traits to attract frugivores. These include pulp:seed ratio, colour, volatiles, fruit size, fruit shape, seed size and shape. There is evidence that fruits respond to the environment in a plastic manner. For example, nutritional quality changed with temperature in cherry tomato (Gautier et al. 2005), fruit and seed size, nutritional content, and pulp:seed ratio responded to soil water availability and nutrients in a desert shrub (Lotan and Izhaki 2013). Fruit weight and seed number were related to rainfall and temperature in a small Mediterranean tree (Chiarucci et al. 1993). In a study of intraspecific variation of 63 fleshy-fruited plant species, fruit size was associated with rainfall, and carbohydrate and lipid content decreased with latitude (Hampe 2003). While there is evidence for plasticity in fruit and seed traits in response to their abiotic environment, there are limited examples of rapid changes in fruit traits to changing biotic environments except for one study finding rapid change in seed size in response to a shifting frugivore community (Galetti et al. 2013). However, rapid change in fruit size, through artificial selection, is a key component of domestication of agricultural crops, and therefore is likely more common than it appears in the literature. For example, in tomatoes, a single loci, fw2.2, has a large impact on fruit size and was important during domestication (Frary et al. 2000).
Rather than changing physical traits, plants may adjust the timing of propagule production or ripening in response to changing environments. For example, variation in temperature and precipitation has been show to alter flowering time (Jordan et al. 2015) and some epigenetically regulated (discussed below) hypomethylated regions have been associated with earlier flowering (Fieldes et al. 2005) or seed production (Yakovlev et al. 2012). The timing of fruit/seed production and abscission in plants may respond to environmental factors as well. Ethylene plays a key role in the timing of ripening for many agricultural species (Barry and Giovannoni 2007), and may be related to the timing of ripening and abscission in wild species closely related to tomato (Grumet et al. 1981), yet the role of ethylene is largely unexplored in wild plants. In Costa Rica, the ripening rate of fruit was linked to frugivore visitation rate, such that fruit ripened more rapidly when frugivore visitation was higher (Levey 1987).
Finally, plants may respond to a changing biotic or abiotic environment by relying less on dispersal through space and more on dispersal through time. Traits related to seed longevity in the canopy or soil seed bank may allow plants to persist longer in situations of reduced dispersal. Traits such as seed hardness, physical protection, chemical protection, flesh quantity and likelihood of microbial attack tend to trade-off with traits related to movement (e.g. attractiveness to dispersers). For example, the annual grass Aegilops triuncialis has dimorphic seeds and late season precipitation induces extended seed dormancy in one of the seed types (Dyer 2017).
Rapid Changes in Dispersal in Different Systems
To date, changes in dispersal (or more broadly movement) have been studied in both terrestrial and aquatic systems, though few studies have addressed rapid changes. We specifically highlight responses to effects of global environmental change (Table 1). Most of the studies on dispersal have addressed phenotypic variation across a range of external conditions empirically without investigating the degree to which the phenotypic trait variation stems from genetic or epigenetic factors. In light of rapid genetic responses to environmental changes in other traits (Hoffmann and Sgrò 2011), the heritability of various dispersal traits and their genetic architecture should be of high importance, but heritability of dispersal traits is scarcely known outside of a few traits in annual crops and model plant systems (see the review by Saastamoinen et al. 2018 and citations therein).
Table 1.
Environmental conditions affected by global environmental change, which have been shown to elicit a rapid response in dispersal ability. Given are examples of studies, with conclusions drawn about the main consequence for the dispersal ability of the respective study organisms.
| Global environmental threat | Variables | Exemplary studies | Main consequence for dispersal ability |
|---|---|---|---|
| Plants | |||
| Climate change | Temperature increase | Zhang et al. (2012) | Increase in dispersal distance |
| Teller et al. (2016) | Increase in probability of seed release | ||
| Li et al. (2018) | Increase in fruit dehiscence—phenology of dispersal | ||
| Drought | Teller et al. (2014) | Increase in dispersal distance | |
| Decrease in water availability | Martorell and López (2014) | Increase in highly dispersible seeds | |
| Habitat fragmentation | Increase in distance of suitable habitat patches | Cheptou et al. (2008) | Increase of proportion of non-dispersing seeds in highly fragmented habitat |
| Nutrient cycling | Nutrient depletion | Imbert and Ronce (2001) | Increase in seeds with dispersal structures |
| Biodiversity loss | Loss of dispersal vectors | Galetti et al. (2013) | Decrease in seed size—negative effects on population fitness |
| Animals | |||
| Climate change | Temperature increase | ||
| Drought | Rozen-Rechels et al. (2018) | Increased anxiety (maternal effects) and greater exploration behaviour | |
| Precipitation, extreme weather events | Loe et al. (2016) | Behavioral plasticity leads to increased movement to favourable habitats | |
| Ocean acidification | Rossi et al. (2018) | Maladaption to environmental cues to unsuitable habitat in larval fish | |
| Winter weather conditions | Gurarie et al. (2017) | ||
| Wind | Vansteelant et al. (2017) | ||
| Habitat fragmentation | Moraes et al. (2018) | ||
| Comparison extant metapopulation with fragmented extinct population | Fountain et al. (2016) | Selection for genotypes with higher colonization capacity in fragmented landscapes in butterflies |
To illustrate some of the population and community level impacts that changes in dispersal traits can have, we discuss changes in plant traits, changes in external conditions and changes in animal disperser traits. Including animal dispersers not only gives a more complete picture of the systems in which rapid responses in dispersal are addressed (even if the mechanisms may be different), but also demonstrates how seed dispersal can be affected when biotic dispersal vectors respond to environmental changes and thus influence the total dispersal kernel (Rogers et al., this issue-b). To be comparable with plant dispersal, we consider animal movement broadly, including such topics as natal dispersal, seasonal migration, foraging movement, settlement behaviour and movements within home ranges.
Change in plant traits
Changes in seed dispersal have been mostly studied experimentally within the context of phenotypic variability of dispersal traits (Fig. 1). A common approach has focused on environmental influences on heteromorphic species that produce two or more distinct seed morphs that differ in dispersal potential. Environmental influences on the proportion of more dispersible morphs include herbivory (Fukano et al. 2014) or environmental stress (Mandák and Pyšek 1999; Imbert and Ronce 2001; Cheptou et al. 2008; Martorell and López 2014). Other studies have considered environmental influences on continuous variation in dispersal traits, again, addressing the effects of herbivory (de la Pena and Bonte 2014; Marchetto et al. 2014) and environmental stress (Zhang et al. 2012; Teller et al. 2014) on traits related to wind dispersal such as pappus length, plume loading and plant height. Moreover, the timing of seed release can change in response to the growth conditions experienced by maternal plants (Teller et al. 2016; Li et al. 2018). Similarly, effects of water and soil nutrients on traits associated with endozoochorous seed dispersal such as fruit and seed size, pulp-to-seed ratio and pulp nutrients have been experimentally investigated (Lotan and Izhaki 2013).
Less work has explicitly addressed rapid changes in seed dispersal, although studies have considered within- and among-population genetic variation in traits associated with dispersal (see Saastamoinen et al. 2018 for a review of genetics in seed dispersal) which demonstrate the long-term potential for evolution of dispersal (e.g. Riba et al. 2005). Perhaps the most elegant demonstration of rapid change in dispersal is the reduction in Euterpe edulis fruit size and increased dispersal distance, associated with the selective defaunation of large dispersers in the Atlantic forest of Brazil (Galetti et al. 2013; Carvalho et al. 2016). Studies have also demonstrated directional selection by seed dispersers on a variety of fruit display traits relevant to seed dispersal such as fruit sugar concentration (Palacio et al. 2014), fruit/seed size (e.g. Wheelwright 1993; Hernández 2008) and ripening phenology (e.g. Alcantara et al. 1997); however, these studies have not demonstrated an evolutionary response (but see Heydel and Tackenberg 2016 for a discussion in crops). Other studies have identified differential effectiveness of seed dispersal by animal vectors in different conditions (Martorell and López 2014). As an example, somatic seed polymorphisms in Picris echioides have been found to lead to differential dispersal success by small mammals (Sorensen 1978).
Change in external conditions
It is worth noting that global environmental change could mediate rapid changes in seed dispersal without changes in dispersal traits. In a seed release experiment, Cabra-Rivas et al. (2014) found that stream structure could affect the seed dispersal of the hydrochorous, invasive tree Ailanthus altissima. In particular, dispersal distances increased in degraded reaches because of a loss of complexity and a lack of potential retention zones. Similarly, Greene and Quesada (2011) showed that dispersal distances in the wind-dispersed weed Tragopogon dubius were highly influenced by wind speeds and turbulences. Marchetto et al. (2010) show that the structure and height of surrounding vegetation strongly affects dispersal. For zoochorous transport, seed dispersal could be affected simply by the presence and density of dispersers. This is most apparent if the absence of the animal disperser leads to dispersal network disruptions (Rogers et al. 2017). In a theoretical study, Jones et al. (2017) showed that animal traits such as movement distance, activity level and gut retention time interacted with habitat loss such that seed dispersal distances were largest under intermediate habitat loss. Garcia et al. (2016) used an isotope-based technique to track bird-mediated seed dispersal and found that small differences in seed and animal traits, for example phenology patterns, could lead to large differences of seed dispersal in response to landscape structure. Morales et al. (2013) developed a multispecies mechanistic model of seed dispersal based on frugivore behavioural responses to landscape heterogeneity. Their results indicated that seed dispersal was strongly affected by the frugivore species composition, landscape heterogeneity and the presence of congeneric plant species. If an adult plant can perceive the changed environment, there is potential for plastic responses in fruit or seed traits via epigenetic regulation (Karban and Orrock 2018). If seeds or diaspores with particular traits have higher fitness in these changed abiotic or biotic environments, then there is potential for natural selection and rapid adaptation.
Change in animal disperser traits
Plant seed dispersal could also change in response to rapid changes in the animal disperser traits. The interaction between changes in plant dispersal and animal dispersal has rarely been studied explicitly. However, density-dependent dispersal in animals has been widely addressed, either alone (Ventura et al. 2017; Morton et al. 2018) or in interaction with habitat heterogeneity (Zhang et al. 2018), personality (Cooper et al. 2017) or gender (Paris et al. 2016). Many biotic drivers have been studied including the presence of predators (Payo-Payo et al. 2018) and parasites (Terui et al. 2017), competition (Katz et al. 2017), resource availability (Powell and Bentz 2014; Pope and Jha 2017) and quality (Pepi et al. 2016; Christy et al. 2017; Katz et al. 2017), and habitat fragmentation (Moraes et al. 2018). Other studies have focused on abiotic drivers affecting animal movement such as ocean acidification (Rossi et al. 2018), and climatic conditions including drought (Rozen-Rechels et al. 2018), winter weather conditions (Gurarie et al. 2017) and wind during migration (Vansteelant et al. 2017). Other factors contributing to variation in animal movement include ontogenetic shifts in dispersal (Dahirel et al. 2017; Bombieri et al. 2018) and individual personality (Cote et al. 2013; Michelangeli et al. 2017) though the lability of personality is uncertain (Ochocki and Miller 2017; Szűcs et al. 2017).
From an evolutionary perspective, changes in animal dispersal have been addressed in the context of delayed dispersal in the evolution of cooperative breeding (Wild and Korb 2017), evolution of density-dependent dispersal with range expansion (Fronhofer et al. 2017), the single gene pleiotropic effect on local dispersal (Edelsparre et al. 2014), morphological changes and specialization for limited resources (e.g. Darwin’s Finches) (Grant and Grant 2006) and the quantitative genetic variation underlying local dispersal and the rate of exploration of novel environments (Korsten et al. 2013). However, it is unclear how rapidly these changes occur, but experimental studies in guppies have shown rapid evolution (in several traits including escape) in only a few generations as a response to predator presence (Reznick et al. 2008). In fact, much of our current understanding of rapid evolution comes from animal studies (Hairston et al. 2005; Ellner 2013) including a couple of replicated experiments in natural systems (Losos et al. 1997; Reznick et al. 1997) demonstrating its potential.
Potential Mechanisms Driving Rapid Response of Dispersal Traits
Having reviewed the evidence for rapid changes in seed dispersal traits in response to changing environments, a second question is how this occurs. Research on the mechanisms for rapid change in seed dispersal traits first aims to identify if the trait change is due to plasticity or genotypic change. For plastic traits, the next steps are to understand the mechanism leading to trait change and determine if the change is heritable. For genetic trait changes, the next step is to identify the region under selection and the strength of selection. Studies approach these topics from different directions in observations, experiments and models, some of which we highlight in this section. Though the exact mechanisms contributing to variability and rapid changes may result in similar outcomes for ecology, at least in the first instance, the issue of heritability is important, and the long-term effects of evolutionary and plastic responses may differ. For example, natural selection on standing genetic variation may evolve rapidly in response to environmental changes (Ellner 2013), and is a widely accepted mode of rapid evolution. Epigenetic research is starting to provide a mechanistic bridge between genetic and plastic variation (Moler et al. 2018) and can also occur rapidly. Whether epigenetic inheritance can contribute to rapid adaptive evolution remains a highly contested topic in evolutionary ecology and we hence discuss the potential/debated importance of phenotypic plasticity, the role of epigenetics and how these two processes are involved in non-heritable rapid changes as well as heritable adaptive evolution with implications for seed dispersal.
Phenotypic plasticity
Phenotypic plasticity is classically defined as the range of phenotypes that a single genotype can express as a function of its environment (Nicotra et al. 2010). Years of common garden studies and norms of reaction analyses have shown that plants have a wide range of phenotypes under different environmental conditions (Pigliucci 2005; Futuyma 2017). A variety of manipulations of environmental conditions and selections of plant individuals from different provenances or genotypes have been combined in common garden experiments or in in vivo settings to measure their effects on different plant traits and distinguish between plastic and genetic responses. Effects of climate changes on dispersal are either simulated with different temperature, moisture or precipitation treatments (see, for example Teller et al. 2014, 2016) or tested along environmental (Martín-Forés et al. 2017) or geographic gradients (LaRue et al. 2018). They are also investigated in transplant experiments with plant individuals from across their ranges or from different provenances (Leiblein-Wild and Tackenberg 2014). In the field of invasion biology, rapid responses to environmental changes are primarily addressed to identify invasion success of exotic species (Davidson et al. 2011; Matesanz et al. 2012; Huang et al. 2015). Here, studies focus on rapid changes in dispersal traits to explain invasion success by either assessing trait responses in the native vs. non-native range (Martín-Forés et al. 2017); at the edges of non-native ranges (Pichancourt and van Klinken 2012) or through the comparison of invasive and closely related, non-invasive species (Sendek et al. 2015; Doudová et al. 2017).
Empirical studies of rapid changes in plant traits are now increasingly used in meta-analyses to generalize responses and identify traits that are most affected (i.e. Davidson et al. 2011; Cochrane et al. 2015). While empirical studies (specifically experimental manipulations) convincingly show plasticity in phenotypic responses to changes in environmental conditions, the mechanisms behind the responses remained elusive for a long time. In recent years, trait measurements are increasingly combined with different molecular methods and hence allow a more detailed interpretation of observed variation (e.g. Molina-Montenegro et al. 2018).
Epigenetics
Though the definition of epigenetics is hotly debated at the moment (Greally 2018), we use the definition of Moler et al. (2018) where epigenetics is defined as the study of changes in gene expression that are not strictly due to changes in the underlying DNA sequence; some of the changes may be heritable. At the molecular level, epigenetics is a regulatory mechanism of phenotypic plasticity controlled by environmental cues resulting in differential gene expression. Not only is epigenetics the explanation for the unexplainable, but it may be the main contributor to phenotypic plasticity (Richards et al. 2017; Moler et al. 2018). There are several mechanisms of epigenetic regulation that may be important to seed dispersal, including chemically controlled DNA histone modification and transcriptional and translational interference via non-coding RNAs (e.g. siRNAs) (Moler et al. 2018), but the majority of epigenetic regulation studies of plant traits tend to focus on the role of DNA methylation of cytosine (5mC) (Birney et al. 2016; Quadrana and Colot 2016; Richards et al. 2017; and reviewed in Moler et al. 2018). This is in part because CG, CHG and CHH (where H can be A, C or T) nucleotide methylation patterns can affect the silencing of genes, including transposable elements, and ultimately gene expression of ecologically important traits (Law and Jacobsen 2010) and because epigenetic controls such as histone modification are not thought to be strictly heritable (but see Nightingale et al. 2006) from one generation to the next (Eichten et al. 2014). In plants, there is an increasing body of evidence illustrating that epigenetic variation (5mC methylation) is widespread in natural populations (Niederhuth et al. 2016). The take-home message is that the different forms of epigenetic modifications affect gene expression by differentially providing access to the underlying genetic code (Richards 2006).
Heritable or transmissible (intergenerational (F0 to F1), multigenerational (F1 to F2) and transgenerational (>F2)) epigenetic marks (e.g. 5mC) are often associated with stable alterations in gene expression associated with environmental cues (Heard and Martienssen 2014; Taudt et al. 2016). Most epigenetic studies in plants are associated with intergenerational inheritance of methylation patterns during cell proliferation (Quadrana and Colot 2016). For example leaf shape (prickly vs. non-prickly) is associated with DNA methylation profile in Ilex aquifolium (Herrera and Bazaga 2013). Epigenetic regulation of plant height has been documented in association with drought and temperature (Mirouze and Paszkowski 2011; Kim et al. 2015). Variation in seed size was found to be associated with 5mC in Lavandula latifolia (Alonso et al. 2018). Additionally, bud-burst timing associated with temperature during embryogenesis contributes to seed production in Picea abies (Yakovlev et al. 2014, 2016) and has been shown to be plastic and under epigenetic control. Recent work has highlighted the role of epigenetic regulation of fruit ripening illustrating the potential for rapid changes in fruit traits (Giovannoni et al. 2017). Epigenetic controlled plasticity may be an important adaptive response when shifts in allele frequency are slow relative to the pace of global change, thus providing an alternative rapid response strategy (Braeutigam et al. 2013).
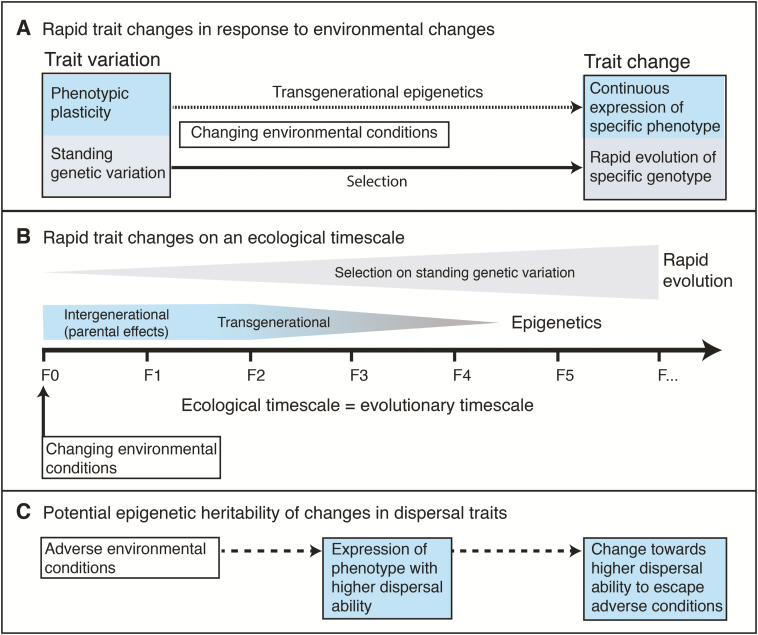
Transmission of epialleles from one generation to the next is far less certain (Quadrana and Colot 2016), and we still do not know how common it is in nature. However, multigenerational (Schmitz et al. 2011), and transgenerational epigenetic inheritance (Richards 2006; Hagmann et al. 2015) has been documented in some model plant systems and could provide a framework for testing the transmission of phenotypic plasticity (Herman et al. 2014; Whipple and Holeski 2016). Epigenetic regulation is a mechanism that enables changes in the environment to switch gene expression on or off, providing a potential means of rapidly altering traits important for dispersal (within one generation) and if stable and heritable may contribute to adaptive evolution (Fig. 2).
Figure 2.
Rapid trait changes in response to environmental change (A) in relation to the eco-evolutionary timescale (B), exemplified for potential epigenetic heritability of changes in dispersal traits (C). (A) Variation of traits in a population can be caused by phenotypic plasticity of one genotype or the standing genetic variation (several genotypes). Changes in environmental conditions could trigger epigenetic responses, with the expression of a specific phenotype. Environmental changes could also select for a specific phenotype based on a specific genotype. If epigenetic effects are transgenerational, they might lead to the continuous expression of a specific phenotype, potentially affecting rapid evolution. Continuous selection for a specific trait from the standing genetic variation can lead to rapid evolution in response to changing environmental conditions. (B) For rapid trait changes, the evolutionary timescale matches the ecological timescale, i.e. trait changes happen in direct response to environmental changes. Epigenetic parental effects might affect only one or two generations, but transgenerational epigenetic effects can span multiple generations. Rapid evolution can occur over a low number of generations, for example by selection on the standing genetic variation. (C) Epigenetic effects in response to environmental changes might be heritable. In this example, epigenetics lead to the expression of a phenotype with higher dispersal ability in response to adverse environmental conditions to escape these conditions. If the epigenetic effect is heritable, the population might change towards having higher dispersal ability and hence be able to escape adverse environmental conditions better.
Rapid evolution
In many organisms, evolution can occur on timescales equivalent to a few generations leading to rapid adaptation to changing environments (Ellner 2013) (Fig. 2B). Rapid adaptive evolution may be the product of selection on standing genetic diversity, or a product of other heritable variability such as epigenetically regulated phenotypic plasticity. Recent advances have been made in the study of invasive plants to identify mechanisms that facilitate their rapid spread using different genotypes or transplants from populations along expansion ranges in common garden experimental settings (Huang et al. 2015; Williams et al. 2016). For example, Huang et al. (2015) tested whether changes in the plume loading of the invasive Mikania micrantha in China was a result of rapid evolution due to selection or phenotypic plasticity by measuring seed traits along the range expansion distance. Seeds along this expansion range were also grown in common garden experiments and measurements of their F1 generation were compared to the field measurements. The comparison showed that dispersal traits in the common garden experiment were not significantly related to the expansion distances. However, the dispersal ability at the edge of the range was higher than in populations towards the source of the spread, indicating rapid and adaptive evolution through selection for higher dispersal ability rather than the plasticity of the dispersal ability (in this case plume load). Additionally, the role of natural selection and rapid evolution (due to spatial sorting of favourable dispersal-related alleles) may be an important factor in sweeping beneficial seed dispersal traits to high frequencies within expanding populations (Ochocki and Miller 2017).
Studies of rapid evolution of dispersal traits focus almost exclusively on genetic components, be it the observations or measurements of heritable traits in experiments (e.g. Cheptou et al. 2008) or measurements of traits combined with genetic or genomic approaches (Zenni et al. 2014; Williams et al. 2016). For example, Cheptou et al. (2008) sampled dispersing and non-dispersing seeds of Crepis sancta in highly fragmented, small habitat patches in an urban area. Observing that the proportion of non-dispersing seeds was much higher in fragmented small patches compared to unfragmented populations and knowing that the ratio of non-dispersing to dispersing seeds is heritable, they concluded that this pattern is evidence for rapid evolution over a few generations due to higher costs of dispersal in fragmented urban populations. Costs of evolution of seed dispersal traits in fragmented landscapes could lead to fitness advantages, but also evolutionary suicide in cases where reduced dispersal evolves and eventually leads to small isolated populations and the accumulation of deleterious alleles (Bonte et al. 2012; Corlett and Westcott 2013).
We note that our current understanding of the role of epigenetics and phenotypic plasticity of seed dispersal is progressing, and more work is needed. One way that these mechanisms could be combined to contribute to rapid changes in seed dispersal is if epigenetics (whether intergenerational or transgenerational) acts as interpretive machinery of the underlying genetic diversity and thus regulates phenotypic plasticity. As phenotypic plasticity can create diversity in phenotypes increasing the options for selection to act upon, it is likely in some cases to be adaptive when the expressed phenotype provides the highest level of fitness. Then natural selection on the adaptive phenotypes leads to evolution of these genes. This means that phenotypic plasticity and epigenetics may provide a means for rapid evolution of seed dispersal traits. We tend to agree with Price et al. (2003) and Haig (2007) and argue that epigenetic and genetic components probably both contribute to population differentiation and influence adaptive genetic evolution on ecological timescales. In sum, epigenetic mechanisms regulating phenotypic plasticity could contribute to rapid evolution of seed dispersal.
What Is the Consequence of Rapid Change in Seed Dispersal for Population Persistence and Spread?
A final question is how important the reviewed changes in dispersal parameters are for population persistence and spread under global change (Snell et al. 2019). The question is challenging to answer, because few studies have explicitly linked seed dispersal to successful establishment of adult plants, and thus demonstrated demographic consequences to changes in dispersal (Traveset et al. 2014), despite the widespread acceptance of dispersal as an important factor in population dynamics and spread. One notable exception is the example of the wind-dispersed thistle. Skarpaas and Shea (2007) estimated the spread rate of two invasive thistles (Carduus nutans and C. acanthoides) in North America, and showed that plant traits such as seed release height and seed weight affect dispersal distances. In subsequent studies, it was demonstrated that warming led to higher seed release probabilities in C. nutans (via changes in the plastic traits of seed release height and seed weight, Fig. 2) (Teller et al. 2014) as well as higher seed release probability in wind tunnels (Teller et al. 2016), effectively increasing the spread rate significantly under warmer climate scenarios. Zhang et al. (2012) found parental effects of warming on the early life stages of C. nutans might also be heritable. Which mechanisms cause the phenotypic plasticity in those dispersal traits is still unknown.
It is important to note that predictive models rarely take into account variation in dispersal traits (see Box 1 for an example of how this can be achieved) although inclusion of such variation could greatly alter predictions under global change (Travis and Dytham 2002; Clobert et al. 2009; Burton et al. 2010; Jongejans et al. 2011; Travis et al. 2013; Bourne et al. 2014). There is some evidence that the ability to rapidly change dispersal traits can influence the success of biological invasions and extend the range of populations to include suboptimal environments (Pichancourt and van Klinken 2012). This suggests that the ability to rapidly change dispersal traits may be relevant to the spread rates of species and to how effectively populations can be expected to adapt to climate change. We ran numerical experiments with our model (Box 1) where we considered the effects of plasticity in seed dispersal traits on the rate of invasion of a population into a periodically fragmented one-dimensional environment with alternating regions of more or less favourable habitat. We found that in some cases the model predicted that trait plasticity could lead to increased invasion speed, but that depends on the details of the environment and the specific traits and plastic responses of the population (for details, see the Supporting Information—File S1). We expect that more detailed study based on the model will lead to additional insights on when and why plasticity in dispersal traits might be advantageous. In addition to our model, some models addressing invasive species have looked at the evolution of dispersal (Hastings et al. 2005; Perkins et al. 2013) in the context of spatial spread and these models could provide a starting point for further work. Models that have looked at whether a species can keep up with a moving suitable habitat (e.g. Berestycki et al. 2008) have not previously included plastic responses or genetic or epigenetic adaptation (or details of dispersal) but could and should be modified to do so.
Box 1.
We use an integro-difference equation framework to model total seed dispersal pathways that allows us to examine and compare plasticity strategies (see Supporting Information for details of the model). Here the plant produces seeds of various types, where the seed type i corresponds to seeds produced by the plant whose dispersal pathway is governed by the composite integro-difference kernel ki. Here the index i may be discrete or continuous, and the proportion of seeds at of type i is non-negative and satisfies in the discrete case and in the continuous case. Summing over i in the discrete case or integrating over i in the continuous case and carefully tracking seed production, dispersal and establishment, the model will take the adult plant density at location x in spatial region Ω and time t to the next generation adult plant density . The model can be expanded to compare the evolution of different plasticity strategies by dividing the overall population into different subclasses, which produce different proportions of the possible seed types. This is done by indexing the subclasses (e.g. by the index j, for with density for the jth subclass) letting the proportion of seeds of dispersal type i vary with subclass, so that in subclass j is replaced with . One then elaborates the model into a competition model for in the spirit of adaptive dynamics. In the elaborated model one may track the transient and/or asymptotic distributions of .
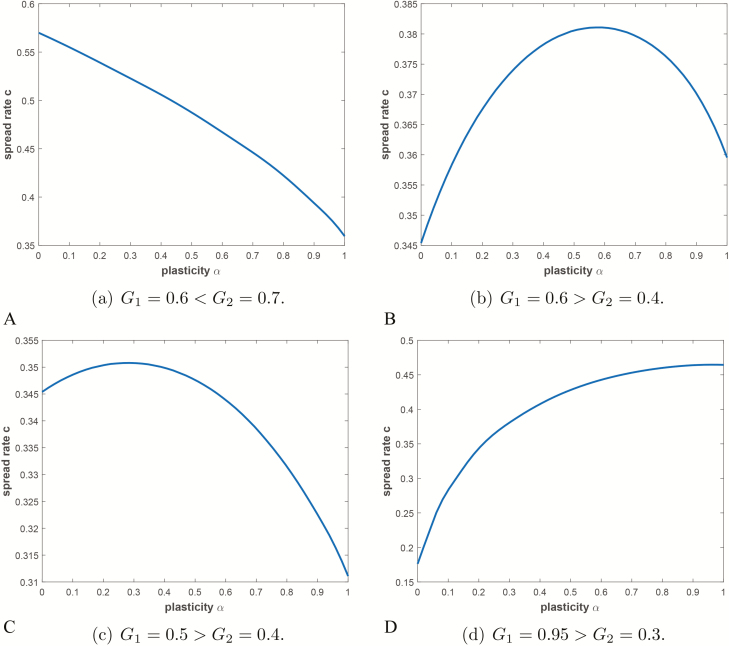
We carry out numerical simulations of the model under the simplifying assumption that the plant is an annual that produces two types of seeds with distinct dispersal pathways corresponding to the same underlying probability distribution but with different parameters. We consider both Gaussian and Laplace distributions. In this case . We also assume that the landscape is one dimensional and spatially periodic with alternating patches of constant poorer resource availability and constant richer resource availability. Our simulations examine the rate of spread of the population as a function of the degree of plasticity on . We show that a mixed plasticity strategy can sometimes maximize the spread rate depending upon the overall survivability of each seed type. We illustrate this below for the case of Gaussian kernels (Fig. 3). Details of the model and the simulations are given in the Supporting Information.
Figure 3.
The figure shows the spread rate of the plant species in question as a function of the plasticity parameter α. Here G1 and G2 represent overall survival rates associated to the two seed dispersal pathways.
Previous theoretical work acknowledges the existence of both random and non-random reasons for trait variability, but most mathematical or simulation approaches then concentrated on heritable or non-heritable random variability. For example, Albert et al. (2011) showed in a simple simulation of wind dispersal that variability in the terminal falling velocity of seeds increases the probability of long-distance dispersal. Thus, dispersal distances would be underestimated when considering only the average falling velocity in the model. This is a particular example of Jensen’s inequality, which states that if the relationship between variable traits and a response is non-linear, then predictions based on the mean trait (rather than the full distribution of trait values) could overestimate or underestimate the response (Bolnick et al. 2011; Moran et al. 2016). Such non-linear averaging effects induced by random trait variability, as well as the tendency of trait variability to spread risks (portfolio effect), are relatively well understood.
Comparatively less attention has focused on theoretical models to understand the effect of non-random non-heritable or heritable (via hard or soft selection, see, e.g. Messer and Petrov 2013) variation. In general, such variation should improve the performance and adaptive potential of plants to environmental variation (see Malanson (2018) for a simulation study of the role of intraspecific variability and climatic volatility). Imbert and Ronce (2001) showed that heteromorphic Asteraceae increase the dispersal of their progeny under environmental stress by varying the relative proportion of wind- and animal-dispersed seed morphs. More interestingly, the plastic increase in dispersal could only be observed under nutrient depletion as a typically longer-lasting environmental stress but not for herbivore pressure as a more unpredictable stress, which corroborates theoretical expectations.
Predicting potential effects of variability in seed dispersal traits on community dynamics is even more complicated than predicting population effects. Despite its complexity, biotic and abiotic interactions and their evolutionary dynamics must be considered to avoid incomplete or erroneous conclusions regarding the mechanisms influencing patterns of seed dispersal (Hand et al. 2015; Luikart et al. 2018). Concurrently, the effect of phenotypic plasticity on coexistence of competing plants remains largely unclear (Turcotte and Levine 2016). Hart et al. (2016) used a theoretical model to test the effects of random variation in demographic rates and competitive parameters (through non-linear averaging) as well as demographic stochasticity. In contrast to common expectations, they found that intraspecific variation could destabilize rather than stabilize coexistence. However, their model concentrated only on random variation and ignored spatial dynamics while dispersal plays a crucial role in metacommunity dynamics and can affect local and regional diversity in a number of ways (Leibold et al. 2004). The competition–colonization trade-off predicts that high colonization ability in competitively inferior species could promote coexistence with dominant competitors that are poorer dispersers. The mass effect perspective predicts that species could be rescued from local competitive exclusion by immigration from other source communities. From these theoretical expectations, we could conceive of different ways in which variability in seed dispersal could promote coexistence. For example, plastic increases in dispersal could not only help escape from high intraspecific competition, but also escape from stressful environments. If plant species differ in their adaptive potential, species with a lower adaptive potential could be at a relative disadvantage.
As a complicating factor, seed dispersal in many species is also affected by mutualistic interactions between plant species and their animal seed dispersers, and these interactions can help to stabilize or destabilize coexistence (Jeltsch et al. 2013). Differences in the adaptive potential of plants and animal dispersers (e.g. phenological response) may lead to disruption of the seed disperser network or establishment of novel seed dispersal interactions with unclear effect on population and community dynamics (Warren et al. 2011; McConkey et al. 2012). In the future, more concerted effort is needed to link empirical and theoretical approaches as well as targeted experiments to explore the impact of rapid changes in seed dispersal plasticity in complex community dynamics (Miner et al. 2005; Turcotte and Levine 2016).
Summary, Challenges and Future Directions
Seed dispersal is an essential life-history stage for plants, yet our understanding about the details of this process remains limited. This is both because of the high variability in effective dispersal due to exogenous factors (climate, wind, terrain), but also because of the high intrinsic variability of dispersal-related traits of seeds as well as their dispersers, associated with phenotypic variation (Herrera 2017). Moreover, challenges to studying rapid changes in dispersal exist because there are extensive logistical constraints to untangling these processes. In this perspective essay, we have argued that the variability in dispersal-related traits plays an essential role in regulating dispersal processes, in particular in non-equilibrium situations, and thus deserves closer attention by ecologists studying the role of seed dispersal, in particular in connection with the adaptation to global environmental change.
We have shown that there is evidence and potential for rapid regulation of seed dispersal processes via heritable or non-heritable variation, both in plants, and in seed dispersers. The effects of this variation for plant fitness and plant community dynamics are less clear. While the induced variation in plants may be adaptive (i.e. fitness enhancing), it is very difficult to demonstrate this, due to the necessity of understanding the entire life cycle of a plant to determine fitness. The effect of behavioural changes of seed dispersers is even more elusive, in the sense that they will optimize their own survival, an objective that is not necessarily aligned with the fitness of their associated seed plants. Together, these processes engender substantial uncertainty about the stability of seed dispersal processes and thus plant reproduction under different environmental conditions.
We believe that future research directions focused on understanding how rapid modifications in seed dispersal can change the capacity of species to respond to global ecological change can focus on three points.
First, we urgently need improved data on the range of subindividual variation associated with seed dispersal traits. We must dissect the degree to which the subindividual variability is the product of genetic variability or phenotypic plasticity. This can be accomplished using targeted experiments, molecular approaches and common gardens (Whipple and Holeski 2016; Moler et al. 2018). Further, to the degree possible, the proportion of phenotypic plasticity that is associated with patterns of DNA methylation (epigenetically induced variation) needs to be quantified. If phenotypic plasticity is determined to be under epigenetic control (e.g. 5mC) then the heritability needs to be quantified, this can be done using multigenerational reciprocal common garden experiments. If variation in seed dispersal traits (e.g. seed size or mass) are unexplained by genotype or environment variability, and persist for multiple generations, then this would suggest that epigenetic variation could be adaptive and heritable. This task is complicated by the number of different traits that contribute to seed dispersal (seed, plant and disperser) and which vary at the subindividual scale. The empirical findings must be deposited in comprehensive databases that contain information on mean seed dispersal trait values and standard deviations; ideally with environmental covariates identified.
Second, theoretical models that describe these changes broadly, based on identified plant traits and the adaptive landscape on which dispersal traits operate for each plant, need to be developed so that we can predict how seed traits may change in response to biotic and abiotic factors and stressors. Our model (Box 1) begins to address this research direction but must be expanded upon to include real-world examples which can inform empirical research and decision-making. Similar models should be constructed for key dispersal functional groups of plants. A general theory of trait change in plants in response to changes in the biotic and abiotic environment could both guide further empirical tests and link to plant demographic models, improving models of spatial spread (e.g. Neubert and Caswell 2000; Jongejans et al. 2011) and dynamic vegetation models (e.g. Hartig et al. 2012; Snell et al. 2014).
Lastly, in these times of global change, it is crucial to understand which plant species are under threat because they are unable to keep up with their environmental niche. Including rapid responses in seed dispersal traits will allow us to refine our predictions for various species and inform our decisions about how to respond proactively to global change to minimize population/species loss in the future. If we combine our empirical understanding of heritable and non-heritable mechanisms of seed dispersal with theoretical and mathematical modelling of dispersal pathways, we are likely to improve predictions and improve our ability to predict further into the future for best- and worst-case scenarios.
Sources of Funding
The ideas and discussion for this manuscript stemmed from the authors participation in a National Science Foundation-funded (NSF DEB # 1548194) workshop on seed dispersal held at the National Socio-Environmental Synthesis Center (SESYNC, funded by NSF DBI-1052875) in May 2016. RSC, CC, and XY were supported in part by NSF (DMS # 1514752). DZ received funding from the Swiss National Science Foundation (SNF, grant: PZ00P3_168136/1) and from the German Science Foundation (DFG, grant: ZU 361/1-1).
Contributions by the Authors
J.S.J. and G.P. led the paper, everyone else alphabetical. All authors contributed to discussions, writing and approved the manuscript.
Conflict of Interest
None declared.
Supporting Information
The following additional information is available in the online version of this article—
File S1. Model for plants with Plasticity in seed dispersal.
Acknowledgements
We thank B. Loiselle for useful discussion and comments on the manuscript. We also thank two anonymous reviewers, associate editor and editor for their constructive feedback which improved this paper.
Glossary
| Term | Definition | Citation |
|---|---|---|
| Adaptive trait | A trait that has higher fitness in a specific environment. | Charlesworth et al. (2017) |
| Adaptive and neutral evolution | Neutral evolution is when alleles and their associated phenotypes change without being selected for (neutral drift), while adaptive evolution refers to changes in allele frequencies due to selection. | (Luikart et al. 2018) |
| Adaptive phenotypic plasticity | The part of phenotypic plasticity that improves the fitness of an organism (e.g. adaptation to a spatially or temporally variable environment), and should thus be selected for. | Sensu Dudley and Schmitt (1996) |
| Dispersal | The movement of a propagule from its natal source with consequences for gene flow through space. | Ronce (2007) |
| Epialleles | Loci differing in chromatin state among cells or organisms. | Moler et al. (2018) |
| Epigenetics | The study of changes in gene expression that are not due to changes in the underlying DNA sequence regardless of heritability. | Moler et al. (2018) |
| Epigenetic inheritance | The phenomenon that epigenetic regulation of gene expressions can be maintained during cell proliferation (mitosis) or between generations (meiosis). Intergenerational F0, multigenerational F0–F1 or transgenerational >F2 | Quadrana and Colot (2016) |
| Local adaptation | As a result of genotype by environment interactions, and in the absence of other forces and constraints, divergent selection should cause local populations to evolve traits that provide an advantage under its local environmental conditions. | Kawecki and Ebert (2004) |
| Reaction Norm | The range of phenotypes expressed by a given genotype across some environmental gradient. | Via and Lande (1985) |
| Parental effects | An effect of the parental phenotype on offspring phenotype. Epigenetic inheritance from F0 to F1. | Uller (2008) |
| Phenotypic plasticity | The ability of a single genotype to produce different phenotypes when exposed to different environmental conditions. | Pigliucci (2005) |
| Rapid evolution | A genetic change occurring rapidly enough to have a measurable impact within a few generations. | Hairston et al. (2005) |
| Standing genetic variation | The genetic variability in a population. | Barrett and Schluter (2008) |
Literature Cited
- Albert CH, Grassein F, Vieilledent G, Violle C. 2011. When and how should intraspecific variability be considered in trait-based plant ecology? Perspectives in Plant Ecology Evolution and Systematics 13:217–225. [Google Scholar]
- Alcantara JM, Rey PJ, Valera F, SanchezLafuente AM, Gutierrez JE. 1997. Habitat alteration and plant intra-specific competition for seed dispersers. An example with Olea europaea var. sylvestris. Oikos 79:291–300. [Google Scholar]
- Alonso C, Pérez R, Bazaga P, Medrano M, Herrera CM. 2018. Within-plant variation in seed size and inflorescence fecundity is associated with epigenetic mosaicism in the shrub Lavandula latifolia (Lamiaceae). Annals of Botany 121:153–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augspurger CK, Franson SE. 1987. Wind dispersal of artificial fruits varying in mass, area, and morphology. Ecology 68:27–42. [Google Scholar]
- Bardgett RD, Mommer L, De Vries FT. 2014. Going underground: root traits as drivers of ecosystem processes. Trends in Ecology & Evolution 29:692–699. [DOI] [PubMed] [Google Scholar]
- Barrett RD, Schluter D. 2008. Adaptation from standing genetic variation. Trends in Ecology & Evolution 23:38–44. [DOI] [PubMed] [Google Scholar]
- Barry CS, Giovannoni JJ. 2007. Ethylene and fruit ripening. Journal Plant Growth Regulation 26:143–159. [Google Scholar]
- Beckman NG, Bullock JM, Salguero-Gómez R. 2018. High dispersal ability is related to fast life-history strategies. Journal of Ecology 106:1349–1362. [Google Scholar]
- Berestycki H, Diekmann O, Nagelkerke CJ, Zegeling PA. 2008. Can a species keep pace with a shifting climate? Bulletin of Mathematical Biology 71:399–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Smith GD, Greally JM. 2016. Epigenome-wide association studies and the interpretation of disease-omics. PLoS Genetics 12:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleher B, Böhning-Gaese K. 2001. Consequences of frugivore diversity for seed dispersal, seedling establishment and the spatial pattern of seedlings and trees. Oecologia 129:385–394. [DOI] [PubMed] [Google Scholar]
- Bolnick DI, Amarasekare P, Araújo MS, Bürger R, Levine JM, Novak M, Rudolf VH, Schreiber SJ, Urban MC, Vasseur DA. 2011. Why intraspecific trait variation matters in community ecology. Trends in Ecology & Evolution 26:183–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bombieri G, Fasciolo A, Penteriani V, Carlos Illera J, Chamberlain D, del Mar Delgado M. 2018. Disentangling the effects of genetic and environmental factors on movement behaviour. Ethology 124:139–148. [Google Scholar]
- Bonte D, Van Dyck H, Bullock JM, Coulon A, Delgado M, Gibbs M, Lehouck V, Matthysen E, Mustin K, Saastamoinen M, Schtickzelle N, Stevens VM, Vandewoestijne S, Baguette M, Barton K, Benton TG, Chaput-Bardy A, Clobert J, Dytham C, Hovestadt T, Meier CM, Palmer SC, Turlure C, Travis JM. 2012. Costs of dispersal. Biological Reviews of the Cambridge Philosophical Society 87:290–312. [DOI] [PubMed] [Google Scholar]
- Bourne EC, Bocedi G, Travis JMJ, Pakeman RJ, Brooker RW, Schiffers K. 2014. Between migration load and evolutionary rescue: dispersal, adaptation and the response of spatially structured populations to environmental change. Proceedings of the Royal Society B: Biological Sciences 281:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braeutigam K, Vining KJ, Lafon-Placette C, Fossdal CG, Mirouze M, Gutierrez Marcos J, Fluch S, Fernandez Fraga M, Angeles Guevara M, Abarca D, Johnsen O, Maury S, Strauss SH, Campbell MM, Rohde A, Diaz-Sala C, Cervera M-T. 2013. Epigenetic regulation of adaptive responses of forest tree species to the environment. Ecology and Evolution 3:399–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton OJ, Phillips BL, Travis JM. 2010. Trade-offs and the evolution of life-histories during range expansion. Ecology Letters 13:1210–1220. [DOI] [PubMed] [Google Scholar]
- Cabra-Rivas I, Alonso A, Castro-Diez P. 2014. Does stream structure affect dispersal by water? A case study of the invasive tree Ailanthus altissima in Spain. Management of Biological Invasions 5:179–186. [Google Scholar]
- Carvalho CS, Galetti M, Colevatti RG, Jordano P. 2016. Defaunation leads to microevolutionary changes in a tropical palm. Scientific Reports 6:31957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D, Barton Nicholas H, Charlesworth B. 2017. The sources of adaptive variation. Proceedings of the Royal Society B: Biological Sciences 284:20162864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheptou PO, Carrue O, Rouifed S, Cantarel A. 2008. Rapid evolution of seed dispersal in an urban environment in the weed Crepis sancta. Proceedings of the National Academy of Sciences of the United States of America 105:3796–3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiarucci A, Pacini E, Loppi S. 1993. Influence of temperature and rainfall on fruit and seed production of Arbutus unedo L. Botanical Journal of the Linnean Society 111:71–82. [Google Scholar]
- Christy MT, Savidge JA, Adams AAY, Gragg JE, Rodda GH. 2017. Experimental landscape reduction of wild rodents increases movements in the invasive brown treesnake (Boiga irregularis). Management of Biological Invasions 8:455–467. [Google Scholar]
- Clobert J, Le Galliard JF, Cote J, Meylan S, Massot M. 2009. Informed dispersal, heterogeneity in animal dispersal syndromes and the dynamics of spatially structured populations. Ecology Letters 12:197–209. [DOI] [PubMed] [Google Scholar]
- Cochrane A, Yates CJ, Hoyle GL, Nicotra AB. 2015. Will among-population variation in seed traits improve the chance of species persistence under climate change? Global Ecology and Biogeography 24:12–24. [Google Scholar]
- Cooper EB, Taylor RW, Kelley AD, Martinig AR, Boutin S, Humphries MM, Ben D, Lane JE, McAdam AG. 2017. Personality is correlated with natal dispersal in North American red squirrels Tamiasciurus hudsonicus. Behaviour 154:939–961. [Google Scholar]
- Corlett RT, Westcott DA. 2013. Will plant movements keep up with climate change? Trends in Ecology & Evolution 28:482–488. [DOI] [PubMed] [Google Scholar]
- Cote J, Fogarty S, Tymen B, Sih A, Brodin T. 2013. Personality-dependent dispersal cancelled under predation risk. Philosophical Transactions of the Royal Society B-Biological Sciences 280:20132349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahirel M, Vong A, Ansart A, Madec L. 2017. Individual boldness is life stage-dependent and linked to dispersal in a hermaphrodite land snail. Ecological Research 32:751–755. [Google Scholar]
- Davidson AM, Jennions M, Nicotra AB. 2011. Do invasive species show higher phenotypic plasticity than native species and, if so, is it adaptive? A meta-analysis. Ecology Letters 14:419–431. [DOI] [PubMed] [Google Scholar]
- de la Pena E, Bonte D. 2014. Above- and belowground herbivory jointly impact defense and seed dispersal traits in Taraxacum officinale. Ecology and Evolution 4:3309–3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudová J, Douda J, Mandák B. 2017. The complexity underlying invasiveness precludes the identification of invasive traits: a comparative study of invasive and non-invasive heterocarpic Atriplex congeners. PLoS One 12:e0176455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley SA, Schmitt J. 1996. Testing the adaptive plasticity hypothesis: density-dependent selection on manipulated stem length in Impatiens capensis. The American Naturalist 147:445–465. [Google Scholar]
- Dyer AR. 2017. The seed ecology of Aegilops triuncialis: linking trait variation to growing conditions. Seed Science Research 27:183–198. [Google Scholar]
- Edelsparre AH, Vesterberg A, Lim JH, Anwari M, Fitzpatrick MJ. 2014. Alleles underlying larval foraging behaviour influence adult dispersal in nature. Ecology Letters 17:333–339. [DOI] [PubMed] [Google Scholar]
- Eichten SR, Schmitz RJ, Springer NM. 2014. Epigenetics: beyond chromatin modifications and complex genetic regulation. Plant Physiology 165:933–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellner SP. 2013. Rapid evolution: from genes to communities, and back again? Functional Ecology 27:1087–1099. [Google Scholar]
- Fieldes MA, Schaeffer SM, Krech MJ, Brown JC. 2005. DNA hypomethylation in 5-azacytidine-induced early-flowering lines of flax. TAG Theoretical and Applied Genetics 111:136–149. [DOI] [PubMed] [Google Scholar]
- Fountain T, Nieminen M, Sirén J, Wong SC, Lehtonen R, Hanski I. 2016. Predictable allele frequency changes due to habitat fragmentation in the Glanville fritillary butterfly. Proceedings of the National Academy of Sciences of the United States of America 113:2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frary A, Nesbitt TC, Grandillo S, Knaap E, Cong B, Liu J, Meller J, Elber R, Alpert KB, Tanksley SD. 2000. Fw2.2: a quantitative trait locus key to the evolution of tomato fruit size. Science 289:85–88. [DOI] [PubMed] [Google Scholar]
- Fronhofer EA, Gut S, Altermatt F. 2017. Evolution of density-dependent movement during experimental range expansions. Journal of Evolutionary Biology 30:2165–2176. [DOI] [PubMed] [Google Scholar]
- Fukano Y, Hirayama H, Tanaka K. 2014. A herbivory-induced increase in the proportion of floating seeds in an invasive plant. Acta Oecologica-International Journal of Ecology 56:27–31. [Google Scholar]
- Futuyma DJ. 2017. Evolutionary biology today and the call for an extended synthesis. Interface Focus 7:20160145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galetti M, Guevara R, Côrtes MC, Fadini R, Von Matter S, Leite AB, Labecca F, Ribeiro T, Carvalho CS, Collevatti RG, Pires MM, Guimarães PR Jr, Brancalion PH, Ribeiro MC, Jordano P. 2013. Functional extinction of birds drives rapid evolutionary changes in seed size. Science 340:1086–1090. [DOI] [PubMed] [Google Scholar]
- Garcia D, Carlo TA, Martínez D. 2016. Differential effect of landscape structure on the large-scale dispersal of co-occurring bird-dispersed trees. Basic and Applied Ecology 17:428–437. [Google Scholar]
- Gautier H, Rocci A, Buret M, Grasselly D, Causse M. 2005. Fruit load or fruit position alters response to temperature and subsequently cherry tomato quality. Journal of the Science of Food and Agriculture 85:1009–1016. [Google Scholar]
- Giovannoni J, Nguyen C, Ampofo B, Zhong S, Fei Z. 2017. The epigenome and transcriptional dynamics of fruit ripening. Annual Review of Plant Biology 68:61–84. [DOI] [PubMed] [Google Scholar]
- Graae BJ, Vandvik V, Armbruster WS, Eiserhardt WL, Svenning J-C, Hylander K, Ehrlen J, Speed JDM, Klanderud K, Brathen KA, Milbau A, Opedal OH, Alsos IG, Ejrnaes R, Bruun HH, Birks HJB, Westergaard KB, Birks HH, Lenoir J. 2018. Stay or go - how topographic complexity influences alpine plant population and community responses to climate change. Perspectives in Plant Ecology Evolution and Systematics 30:41–50. [Google Scholar]
- Grant PR, Grant BR. 2006. Evolution of character displacement in Darwin’s finches. Science 313:224–226. [DOI] [PubMed] [Google Scholar]
- Gratani L. 2014. Plant phenotypic plasticity in response to environmental factors. Advances in Botany 2014:1–17. [Google Scholar]
- Greally JM. 2018. A user’s guide to the ambiguous word ‘epigenetics’. Nature Reviews Molecular Cell Biology 19:207–208. [DOI] [PubMed] [Google Scholar]
- Greene DF, Quesada M. 2011. The differential effect of updrafts, downdrafts and horizontal winds on the seed abscission of Tragopogon dubius. Functional Ecology 25:468–472. [Google Scholar]
- Grumet R, Fobes JF, Herner RC. 1981. Ripening behavior of wild tomato species. Plant Physiology 68:1428–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurarie E, Cagnacci F, Peters W, Fleming CH, Calabrese JM, Mueller T, Fagan WF. 2017. A framework for modelling range shifts and migrations: asking when, whither, whether and will it return. The Journal of Animal Ecology 86:943–959. [DOI] [PubMed] [Google Scholar]
- Hagmann J, Becker C, Müller J, Stegle O, Meyer RC, Wang G, Schneeberger K, Fitz J, Altmann T, Bergelson J, Borgwardt K, Weigel D. 2015. Century-scale methylome stability in a recently diverged Arabidopsis thaliana lineage. PLoS Genetics 11:e1004920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haig D. 2007. Weismann rules! OK? Epigenetics and the Lamarckian temptation. Biology & Philosophy 22:415–428. [Google Scholar]
- Hairston NG, Ellner SP, Geber MA, Yoshida T, Fox JA. 2005. Rapid evolution and the convergence of ecological and evolutionary time. Ecology Letters 8:1114–1127. [Google Scholar]
- Hampe A. 2003. Large-scale geographical trends in fruit traits of vertebrate-dispersed temperate plants. Journal of Biogeography 30:487–496. [Google Scholar]
- Hand BK, Lowe WH, Kovach RP, Muhlfeld CC, Luikart G. 2015. Landscape community genomics: understanding eco-evolutionary processes in complex environments. Trends in Ecology & Evolution 30:161–168. [DOI] [PubMed] [Google Scholar]
- Hart SP, Schreiber SJ, Levine JM. 2016. How variation between individuals affects species coexistence. Ecology Letters 19:825–838. [DOI] [PubMed] [Google Scholar]
- Hartig F, Dyke J, Hickler T, Higgins SI, Ohara R, Scheiter S, Huth A. 2012. Connecting dynamic vegetation models to data - an inverse perspective. Journal of Biogeography 39:2240–2252. [Google Scholar]
- Hastings A, Cuddington K, Davies KF, Dugaw CJ, Elmendorf S, Freestone A, Harrison S, Holland M, Lambrinos J, Malvadkar U, Melbourne BA, Moore K, Taylor C, Thomson D. 2005. The spatial spread of invasions: new developments in theory and evidence. Ecology Letters 8:91–101. [Google Scholar]
- Heard E, Martienssen RA. 2014. Transgenerational epigenetic inheritance: myths and mechanisms. Cell 157:95–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman JJ, Spencer HG, Donohue K, Sultan SE. 2014. How stable ‘should’ epigenetic modifications be? Insights from adaptive plasticity and bet hedging. Evolution 68:632–643. [DOI] [PubMed] [Google Scholar]
- Hernández Á. 2008. Birds and guelder rose Viburnum opulus: selective consumption and dispersal via regurgitation of small-sized fruits and seeds. Plant Ecology 203:111–122. [Google Scholar]
- Herrera CM. 2017. The ecology of subindividual variability in plants: patterns, processes, and prospects. Web Ecology 17:51–64. [Google Scholar]
- Herrera CM, Bazaga P. 2013. Epigenetic correlates of plant phenotypic plasticity: DNA methylation differs between prickly and nonprickly leaves in heterophyllous Ilex aquifolium (Aquifoliaceae) trees. Botanical Journal of the Linnean Society 171:441–452. [Google Scholar]
- Heydel F, Tackenberg O. 2016. How are the phenologies of ripening and seed release affected by species’ ecology and evolution? Oikos 126:738–747. [Google Scholar]
- Hoffmann AA, Sgrò CM. 2011. Climate change and evolutionary adaptation. Nature 470:479–485. [DOI] [PubMed] [Google Scholar]
- Huang F, Peng S, Chen B, Liao H, Huang Q, Lin Z, Liu G. 2015. Rapid evolution of dispersal-related traits during range expansion of an invasive vine Mikania micrantha. Oikos 124:1023–1030. [Google Scholar]
- Imbert E, Ronce O. 2001. Phenotypic plasticity for dispersal ability in the seed heteromorphic Crepis sancta (Asteraceae). Oikos 93:126–134. [Google Scholar]
- Jeltsch F, Bonte D, Pe’er G, Reineking B, Leimgruber P, Balkenhol N, Schröder B, Buchmann CM, Mueller T, Blaum N, Zurell D, Böhning-Gaese K, Wiegand T, Eccard JA, Hofer H, Reeg J, Eggers U, Bauer S. 2013. Integrating movement ecology with biodiversity research - exploring new avenues to address spatiotemporal biodiversity dynamics. Movement Ecology 1:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JS, Gaddis KD, Cairns DM, Lafon CW, Krutovsky KV. 2016. Plant responses to global change: next generation biogeography. Physical Geography 37:93–119. [Google Scholar]
- Jones LR, Duke-Sylvester SM, Leberg PL, Johnson DM. 2017. Closing the gaps for animal seed dispersal: separating the effects of habitat loss on dispersal distances and seed aggregation. Ecology and Evolution 7:5410–5425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongejans E, Shea K, Skarpaas O, Kelly D, Ellner SP. 2011. Importance of individual and environmental variation for invasive species spread: a spatial integral projection model. Ecology 92:86–97. [DOI] [PubMed] [Google Scholar]
- Jongejans E, Skarpaas O, Ferrari MJ, Long ES, Dauer JT, Schwarz CM, Jabbour R, Isard SA, Lieb DA, Sezen Z, Hulting AG. 2015. A unifying gravity framework for dispersal. Theoretical Ecology 8:207–223. [Google Scholar]
- Jordan CY, Ally D, Hodgins KA. 2015. When can stress facilitate divergence by altering time to flowering? Ecology and Evolution 5:5962–5973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karban R, Orrock JL. 2018. A judgment and decision-making model for plant behavior. Ecology 99:1909–1919. [DOI] [PubMed] [Google Scholar]
- Katz N, Shavit R, Pruitt JN, Scharf I. 2017. Group dynamics and relocation decisions of a trap-building predator are differentially affected by biotic and abiotic factors. Current Zoology 63:647–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawecki TJ, Ebert D. 2004. Conceptual issues in local adaptation. Ecology Letters 7:1225–1241. [Google Scholar]
- Kim JM, Sasaki T, Ueda M, Sako K, Seki M. 2015. Chromatin changes in response to drought, salinity, heat, and cold stresses in plants. Frontiers in Plant Science 6:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korsten P, van Overveld T, Adriaensen F, Matthysen E. 2013. Genetic integration of local dispersal and exploratory behaviour in a wild bird. Nature Communications 4:2362. [DOI] [PubMed] [Google Scholar]
- LaRue EA, Holland JD, Emery NC. 2018. Environmental predictors of dispersal traits across a species’ geographic range. Ecology 99:1857–1865. [DOI] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE. 2010. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nature Reviews Genetics 11:204–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiblein-Wild MC, Tackenberg O. 2014. Phenotypic variation of 38 European Ambrosia artemisiifolia populations measured in a common garden experiment. Biological Invasions 16:2003–2015. [Google Scholar]
- Leibold MA, Holyoak M, Mouquet N, Amarasekare P, Chase JM, Hoopes MF, Holt RD, Shurin JB, Law R, Tilman D, Loreau M, Gonzalez A. 2004. The metacommunity concept: a framework for multi-scale community ecology. Ecology Letters 7:601–613. [Google Scholar]
- Levey DJ. 1987. Facultative ripening in Hamelia patens (Rubiaceae): effects of fruit removal and rotting. Oecologia 74:203–208. [DOI] [PubMed] [Google Scholar]
- Li XR, Deb J, Kumar SV, Østergaard L. 2018. Temperature modulates tissue-specification program to control fruit dehiscence in Brassicaceae. Molecular Plant 11:598–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loarie SR, Duffy PB, Hamilton H, Asner GP, Field CB, Ackerly DD. 2009. The velocity of climate change. Nature 462:1052–1055. [DOI] [PubMed] [Google Scholar]
- Loe LE, Hansen BB, Stien A, Albon SD, Bischof R, Carlsson A, Irvine RJ, Meland M, Rivrud IM, Ropstad E, Veiberg V, Mysterud A. 2016. Behavioral buffering of extreme weather events in a high-Arctic herbivore. Ecosphere 7:e01374. [Google Scholar]
- Losos JB, Warheitt KI, Schoener TW. 1997. Adaptive differentiation following experimental island colonization in Anolis lizards. Nature 387:70. [Google Scholar]
- Lotan A, Izhaki I. 2013. Could abiotic environment shape fleshy fruit traits? A field study of the desert shrub Ochradenus baccatus. Journal of Arid Environments 92:34–41. [Google Scholar]
- Luikart G, Kardos M, Hand BK, Rajora OP, Aitken SN, Hohenlohe PA. 2018. Population genomics: advancing understanding of nature. In: Rajora OP, ed. Population genomics: concepts, approaches and applications. Cham, Switzerland: Springer, 3–79. [Google Scholar]
- Mahony CR, Cannon AJ, Wang T, Aitken SN. 2017. A closer look at novel climates: new methods and insights at continental to landscape scales. Global Change Biology 23:3934–3955. [DOI] [PubMed] [Google Scholar]
- Malanson GP. 2018. Intraspecific variability may not compensate for increasing climatic volatility. Population Ecology 60:287–295. [Google Scholar]
- Mandák B, Pyšek P. 1999. Effects of plant density and nutrient levels on fruit polymorphism in Atriplex sagittata. Oecologia 119:63–72. [DOI] [PubMed] [Google Scholar]
- Marchetto KM, Jongejans E, Isard SA. 2010. Plant spatial arrangement affects projected invasion speeds of two invasive thistles. Oikos 119:1462–1468. [Google Scholar]
- Marchetto KM, Shea K, Kelly D, Groenteman R, Sezen Z, Jongejans E. 2014. Unrecognized impact of a biocontrol agent on the spread rate of an invasive thistle. Ecological Applications: a Publication of the Ecological Society of America 24:1178–1187. [DOI] [PubMed] [Google Scholar]
- Martín-Forés I, Avilés M, Acosta-Gallo B, Breed MF, Del Pozo A, de Miguel JM, Sánchez-Jardón L, Castro I, Ovalle C, Casado MA. 2017. Ecotypic differentiation and phenotypic plasticity combine to enhance the invasiveness of the most widespread daisy in Chile, Leontodon saxatilis. Scientific Reports 7:1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martorell C, López MM. 2014. Informed dispersal in plants: Heterosperma pinnatum (Asteraceae) adjusts its dispersal mode to escape from competition and water stress. Oikos 123:225–231. [Google Scholar]
- Matesanz S, Horgan-Kobelski T, Sultan SE. 2012. Phenotypic plasticity and population differentiation in an ongoing species invasion. PLoS One 7:e44955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConkey KR, Prasad S, Campos-Arceiz A, Brodie JF, Rogers H, Santamaria L. 2012. Seed dispersal in changing landscapes. Biological Conservation 146:1–13. [Google Scholar]
- Messer PW, Petrov DA. 2013. Population genomics of rapid adaptation by soft selective sweeps. Trends in Ecology & Evolution 28:659–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelangeli M, Smith CR, Wong BBM, Chapple DG. 2017. Aggression mediates dispersal tendency in an invasive lizard Animal Behaviour 133:29–34. [Google Scholar]
- Miner BG, Sultan SE, Morgan SG, Padilla DK, Relyea RA. 2005. Ecological consequences of phenotypic plasticity. Trends in Ecology & Evolution 20:685–692. [DOI] [PubMed] [Google Scholar]
- Mirouze M, Paszkowski J. 2011. Epigenetic contribution to stress adaptation in plants. Current Opinion in Plant Biology 14:267–274. [DOI] [PubMed] [Google Scholar]
- Moler EV, Abakir A, Eleftheriou M, Johnson JS, Krutovsky KV, Lewis LC, Ruzov A, Whipple AV, Rajora OP. 2018. Population epigenomics: advancing understanding of phenotypic plasticity, acclimation, adaptation, and diseases. In: Rajora OP, ed. Population genomics: concepts, approaches and applications. Cham, Switzerland: Springer, 179–260. [Google Scholar]
- Molina-Montenegro MA, Acuña-Rodríguez IS, Flores TSM, Hereme R, Lafon A, Atala C, Torres-Díaz C. 2018. Is the success of plant invasions the result of rapid adaptive evolution in seed traits? Evidence from a latitudinal rainfall gradient. Frontiers in Plant Science 9:208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraes AM, Ruiz-Miranda CR, Galetti PM Jr, Niebuhr BB, Alexandre BR, Muylaert RL, Grativol AD, Ribeiro JW, Ferreira AN, Ribeiro MC. 2018. Landscape resistance influences effective dispersal of endangered golden lion tamarins within the Atlantic forest. Biological Conservation 224:178–187. [Google Scholar]
- Morales JM, García D, Martínez D, Rodriguez-Pérez J, Herrera JM. 2013. Frugivore behavioural details matter for seed dispersal: a multi-species model for Cantabrian thrushes and trees. PLoS One 8:e65216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran EV, Hartig F, Bell DM. 2016. Intraspecific trait variation across scales: implications for understanding global change responses. Global Change Biology 22:137–150. [DOI] [PubMed] [Google Scholar]
- Morton ER, McGrady MJ, Newton I, Rollie CJ, Smith GD, Mearns R, Oli MK. 2018. Dispersal: a matter of scale. Ecology 99:938–946. [DOI] [PubMed] [Google Scholar]
- Neubert MG, Caswell H. 2000. Demography and dispersal: calculation and sensitivity analysis of invasion speed for structured populations. Ecology 81:1613–1628. [Google Scholar]
- Nicotra AB, Atkin OK, Bonser SP, Davidson AM, Finnegan EJ, Mathesius U, Poot P, Purugganan MD, Richards CL, Valladares F, van Kleunen M. 2010. Plant phenotypic plasticity in a changing climate. Trends in Plant Science 15:684–692. [DOI] [PubMed] [Google Scholar]
- Niederhuth CE, Bewick AJ, Ji L, Alabady MS, Kim KD, Li Q, Rohr NA, Rambani A, Burke JM, Udall JA, Egesi C, Schmutz J, Grimwood J, Jackson SA, Springer NM, Schmitz RJ. 2016. Widespread natural variation of DNA methylation within angiosperms. Genome Biology 17:194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nightingale KP, O’Neill LP, Turner BM. 2006. Histone modifications: signalling receptors and potential elements of a heritable epigenetic code. Current Opinion in Genetics & Development 16:125–136. [DOI] [PubMed] [Google Scholar]
- Ochocki BM, Miller TEX. 2017. Rapid evolution of dispersal ability makes biological invasions faster and more variable. Nature Communications 8:14315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacio FX, Lacoretz M, Ordano M. 2014. Bird-mediated selection on fruit display traits in Celtis ehrenbergiana (Cannabaceae). Evolutionary Ecology Research 16:51–62. [Google Scholar]
- Paris D, Nicholls AO, Hall A, Harvey A, Massaro M. 2016. Female-biased dispersal in a spatially restricted endemic island bird. Behavioral Ecology and Sociobiology 70:2061–2069. [Google Scholar]
- Payo-Payo A, Sanz-Aguilar A, Genovart M, Bertolero A, Piccardo J, Camps D, Ruiz-Olmo J, Oro D. 2018. Predator arrival elicits differential dispersal, change in age structure and reproductive performance in a prey population. Scientific Reports 8:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepi AA, Broadley HJ, Elkinton JS. 2016. Density-dependent effects of larval dispersal mediated by host plant quality on populations of an invasive insect. Oecologia 182:499–509. [DOI] [PubMed] [Google Scholar]
- Perkins TA, Phillips BL, Baskett ML, Hastings A. 2013. Evolution of dispersal and life history interact to drive accelerating spread of an invasive species. Ecology Letters 16:1079–1087. [DOI] [PubMed] [Google Scholar]
- Pichancourt JB, van Klinken RD. 2012. Phenotypic plasticity influences the size, shape and dynamics of the geographic distribution of an invasive plant. PLoS One 7:e32323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigliucci M. 2005. Evolution of phenotypic plasticity: where are we going now? Trends in Ecology & Evolution 20:481–486. [DOI] [PubMed] [Google Scholar]
- Pope NS, Jha S. 2017. Seasonal food scarcity prompts long-distance foraging by a wild social bee. The American Naturalist 191:45–57. [DOI] [PubMed] [Google Scholar]
- Powell JA, Bentz BJ. 2014. Phenology and density-dependent dispersal predict patterns of mountain pine beetle (Dendroctonus ponderosae) impact Ecological Modelling 273:173–185. [Google Scholar]
- Price TD, Qvarnström A, Irwin DE. 2003. The role of phenotypic plasticity in driving genetic evolution. Proceedings of the Royal Society of London. Series B: Biological Sciences 270:1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadrana L, Colot V. 2016. Plant transgenerational epigenetics. Annual Review of Genetics 50:467–491. [DOI] [PubMed] [Google Scholar]
- Reznick DN, Ghalambor CK, Crooks K. 2008. Experimental studies of evolution in guppies: a model for understanding the evolutionary consequences of predator removal in natural communities. Molecular Ecology 17:97–107. [DOI] [PubMed] [Google Scholar]
- Reznick DN, Shaw FH, Rodd FH, Shaw RG. 1997. Evaluation of the rate of evolution in natural populations of guppies (Poecilia reticulata). Science 275:1934–1937. [DOI] [PubMed] [Google Scholar]
- Riba M, Mignot A, Freville H, Colas B, Vile D, Virevaire M. 2005. Variation in dispersal traits in a narrow-endemic plant species, Centaurea corymbosa Pourret. (Asteraceae). Evolutionary Ecology 19:241–254. [Google Scholar]
- Richards EJ. 2006. Inherited epigenetic variation–revisiting soft inheritance. Nature Reviews Genetics 7:395–401. [DOI] [PubMed] [Google Scholar]
- Richards CL, Alonso C, Becker C, Bossdorf O, Bucher E, Colomé-Tatché M, Durka W, Engelhardt J, Gaspar B, Gogol-Döring A, Grosse I, van Gurp TP, Heer K, Kronholm I, Lampei C, Latzel V, Mirouze M, Opgenoorth L, Paun O, Prohaska SJ, Rensing SA, Stadler PF, Trucchi E, Ullrich K, Verhoeven KJF. 2017. Ecological plant epigenetics: evidence from model and non-model species, and the way forward. Ecology Letters 20:1576–1590. [DOI] [PubMed] [Google Scholar]
- Rogers H, Aslan C, Beckman NG, Shea K, HilleRisLambers J, Fricke E, Brodie JF, Bronstein J, Bruna E, Bullock JM, Cantrell S, Decker R, Effiom E, Gurski K, Hartig F, Hastings A, Johnson JS, Kogan O, Loiselle BA, Miriti M, Neubert MG, Pejchar L, Powell JA, Pufal G, Razafindratsima OH, Sandor M, Schreiber SJ, Schupp EW, Snell RS, Strickland C, Zambrano J, Zhou J, Zurell D. this issue-a. Seed dispersal and seed predation-under-appreciated but critically important processes. AoB Plants. [Google Scholar]
- Rogers HS, Buhle ER, HilleRisLambers J, Fricke EC, Miller RH, Tewksbury JJ. 2017. Effects of an invasive predator cascade to plants via mutualism disruption. Nature Communications 8:14557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers H, Pufal G, Johnson JS, Zurell D, Shea K, Bruna EM, Hartig F, Bullock JM, Cantrell S, Beckman NG. this issue-b. The total dispersal kernel: a review and future directions. AoB Plants [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronce O. 2007. How does it feel to be like a rolling stone? Ten questions about dispersal evolution. Annual Review of Ecology, Evolution, and Systematics 38:231–253. [Google Scholar]
- Rossi T, Pistevos JCA, Connell SD, Nagelkerken I. 2018. On the wrong track: ocean acidification attracts larval fish to irrelevant environmental cues. Scientific Reports 8:5840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen-Rechels D, Dupoue A, Meylan S, Decenciere B, Guingand S, Le Galliard J-F. 2018. Water restriction in viviparous lizards causes transgenerational effects on behavioral anxiety and immediate effects on exploration behavior. Behavioral Ecology and Sociobiology 72:23 [Google Scholar]
- Saastamoinen M, Bocedi G, Cote J, Legrand D, Guillaume F, Wheat CW, Fronhofer EA, Garcia C, Henry R, Husby A, Baguette M, Bonte D, Coulon A, Kokko H, Matthysen E, Niitepõld K, Nonaka E, Stevens VM, Travis JMJ, Donohue K, Bullock JM, Del Mar Delgado M. 2018. Genetics of dispersal. Biological Reviews of the Cambridge Philosophical Society 93:574–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz RJ, Schultz MD, Lewsey MG, O’Malley RC, Urich MA, Libiger O, Schork NJ, Ecker JR. 2011. Transgenerational epigenetic instability is a source of novel methylation variants. Science 334:369–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schupp EW. this issue. Trait variation. AoB Plants. [Google Scholar]
- Sendek A, Herz K, Auge H, Hensen I, Klotz S. 2015. Performance and responses to competition in two congeneric annual species: does seed heteromorphism matter? Plant Biology 17:1203–1209. [DOI] [PubMed] [Google Scholar]
- Skarpaas O, Shea K. 2007. Dispersal patterns, dispersal mechanisms, and invasion wave speeds for invasive thistles. The American Naturalist 170:421–430. [DOI] [PubMed] [Google Scholar]
- Snell RS, Huth A, Nabel JEMS, Bocedi G, Travis JMJ, Gravel D, Bugmann H, Gutiérrez AG, Hickler T, Higgins SI, Reineking B, Scherstjanoi M, Zurbriggen N, Lischke H. 2014. Using dynamic vegetation models to simulate plant range shifts. Ecography 37:1184–1197. [Google Scholar]
- Snell RS, Strickland C, Sullivan LL, Giladi I, Hastings A, Holbrook K, Jongejans E, Kogan O, Rudolph J, Zwolak R, Beckman NG, Schupp E, Cavazos BR, Fricke E, Rogers HS, Loiselle BA, Montaño-Centellas F, Carvalho CS, Jones LR, Lichti NI, Lustenhouwer N, Schreiber S. 2019. Consequences of intraspecific variation in seed dispersal for plant demography, communities, evolution, and global change. AoB Plants plz016, doi:10.1093/aobpla/plz016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen AE. 1978. Somatic polymorphism and seed dispersal. Nature 276:174. [Google Scholar]
- Szűcs M, Vahsen ML, Melbourne BA, Hoover C, Weiss-Lehman C, Hufbauer RA. 2017. Rapid adaptive evolution in novel environments acts as an architect of population range expansion. Proceedings of the National Academy of Sciences 114:13501–13506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taudt A, Colomé-Tatché M, Johannes F. 2016. Genetic sources of population epigenomic variation. Nature Reviews Genetics 17:319–332. [DOI] [PubMed] [Google Scholar]
- Teller BJ, Campbell C, Shea K. 2014. Dispersal under duress: can stress enhance the performance of a passively dispersed species? Ecology 95:2694–2698. [Google Scholar]
- Teller BJ, Zhang R, Shea K. 2016. Seed release in a changing climate: initiation of movement increases spread of an invasive species under simulated climate warming. Diversity and Distributions 22:708–716. [Google Scholar]
- Terui A, Ooue K, Urabe H, Nakamura F. 2017. Parasite infection induces size-dependent host dispersal: consequences for parasite persistence. Proceedings of the Royal Society B: Biological Sciences 284:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson FJ, Moles AT, Auld TD, Kingsford RT. 2011. Seed dispersal distance is more strongly correlated with plant height than with seed mass. Journal of Ecology 99:1299–1307. [Google Scholar]
- Traveset A, Heleno R, Nogales M. 2014. The ecology of seed dispersal. In: Gallagher RS, ed. Seeds: the ecology of regeneration in plant communities. Wallingford, UK: CABI, 62–93. [Google Scholar]
- Travis JMJ, Delgado M, Bocedi G, Baguette M, Barton K, Bonte D, Boulangeat I, Hodgson JA, Kubisch A, Penteriani V, Saastamoinen M, Stevens VM, Bullock JM. 2013. Dispersal and species’ responses to climate change. Oikos 122:1532–1540. [Google Scholar]
- Travis JMJ, Dytham C. 2002. Dispersal evolution during invasions. Evolutionary Ecology Research 4:1119–1129. [Google Scholar]
- Turcotte MM, Levine JM. 2016. Phenotypic plasticity and species coexistence. Trends in Ecology & Evolution 31:803–813. [DOI] [PubMed] [Google Scholar]
- Uller T. 2008. Developmental plasticity and the evolution of parental effects. Trends in Ecology & Evolution 23:432–438. [DOI] [PubMed] [Google Scholar]
- Vansteelant WMG, Kekkonen J, Byholm P. 2017. Wind conditions and geography shape the first outbound migration of juvenile honey buzzards and their distribution across sub-Saharan Africa. Proceedings of the Royal Society B: Biological Sciences 284:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura L, Smith DR, Lubin Y. 2017. Crowding leads to fitness benefits and reduced dispersal in a colonial spider. Behavioral Ecology 28:1384–1392. [Google Scholar]
- Via S, Lande R. 1985. Genotype-environment interaction and the evolution of phenotypic plasticity. Evolution 39:505–522. [DOI] [PubMed] [Google Scholar]
- Warren RJ II, Bahn V, Bradford MA. 2011. Temperature cues phenological synchrony in ant-mediated seed dispersal. Global Change Biology 17:2444–2454. [Google Scholar]
- Wender NJ, Polisetty CR, Donohue K. 2005. Density-dependent processes influencing the evolutionary dynamics of dispersal: a functional analysis of seed dispersal in Arabidopsis thaliana (Brassicaceae). American Journal of Botany 92:960–971. [DOI] [PubMed] [Google Scholar]
- Wheelwright NT. 1993. Fruit size in a tropical tree species - variation, preference by birds, and heritability. Vegetatio 108:163–174. [Google Scholar]
- Whipple AV, Holeski LM. 2016. Epigenetic inheritance across the landscape. Frontiers in Genetics 7:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild G, Korb J. 2017. Evolution of delayed dispersal and subsequent emergence of helping, with implications for cooperative breeding. Journal of Theoretical Biology 427:53–64. [DOI] [PubMed] [Google Scholar]
- Williams JL, Kendall BE, Levine JM. 2016. Rapid evolution accelerates plant population spread in fragmented experimental landscapes. Science 353:482–485. [DOI] [PubMed] [Google Scholar]
- Yakovlev IA, Carneros E, Lee Y, Olsen JE, Fossdal CG. 2016. Transcriptional profiling of epigenetic regulators in somatic embryos during temperature induced formation of an epigenetic memory in Norway spruce. Planta 243:1237–1249. [DOI] [PubMed] [Google Scholar]
- Yakovlev I, Fossdal CG, Skrøppa T, Olsen JE, Jahren AH, Johnsen Ø. 2012. An adaptive epigenetic memory in conifers with important implications for seed production. Seed Science Research 22:63–76. [Google Scholar]
- Yakovlev IA, Lee Y, Rotter B, Olsen JE, Skroppa T, Johnsen O, Fossdal CG. 2014. Temperature-dependent differential transcriptomes during formation of an epigenetic memory in Norway spruce embryogenesis. Tree Genetics & Genomes 10:355–366. [Google Scholar]
- Zenni RD, Bailey JK, Simberloff D. 2014. Rapid evolution and range expansion of an invasive plant are driven by provenance-environment interactions. Ecology Letters 17:727–735. [DOI] [PubMed] [Google Scholar]
- Zhang R, Jongejans E, Shea K. 2012. Warming increases the spread of an invasive thistle. PLoS One 6:e21725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Sun T, Xue S, Yang W, Shao D. 2018. Habitat-mediated, density-dependent dispersal strategies affecting spatial dynamics of populations in an anthropogenically-modified landscape. The Science of the Total Environment 625:1510–1517. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.