Abstract
Indolizines are heteroaromatic compounds, and their synthetic analogues have reportedly showed promising pharmacological properties. In this study, a series of synthetic 7-methoxy-indolizine derivatives were synthesised, characterised and evaluated for in vitro whole-cell anti-tuberculosis (TB) screening against susceptible (H37Rv) and multi-drug-resistant (MDR) strains of Mycobacterium tuberculosis (MTB) using the resazurin microplate assay method. The cytotoxicity was evaluated using the MTT assay. In silico molecular-docking study was conducted for compounds 5a-j against enoyl-[acyl-carrier] protein reductase, a key enzyme of the type II fatty acid synthesis that has attracted much interest for the development of novel anti-TB compounds. Thereafter, molecular dynamic (MD) simulation was undertaken for the most active inhibitors. Compounds 5i and 5j with the methoxy functional group at the meta position of the benzoyl group, which was at the third position of the indolizine nucleus, demonstrated encouraging anti-TB activity against MDR strains of MTB at 16 μg/mL. In silico studies showed binding affinity within the range of 7.07–8.57 kcal/mol, with 5i showing the highest binding affinity. Hydrogen bonding, π-π- interactions, and electrostatic interactions were common with the active site. Most of these interactions occurred with the catalytic amino acids (Pro193, Tyr158, Phe149, and Lys165). MD simulation showed that 5j possessed the highest binding affinity toward the enzyme, according to the two calculation methods (MM/PBSA and MM/GBSA). The single-crystal X-ray studies of compounds 5c and 5d revealed that the molecular arrangements in these two structures were mostly guided by C-H···O hydrogen-bonded dimeric motifs and C-H···N hydrogen bonds, while various secondary interactions (such as π···π and C-H···F) also contributed to crystal formation. Compounds 5a, 5c, 5i, and 5j exhibited no toxicity up to 500 μg/mL. In conclusion, 5i and 5j are promising anti-TB compounds that have shown high affinity based on docking and MD simulation results.
Introduction
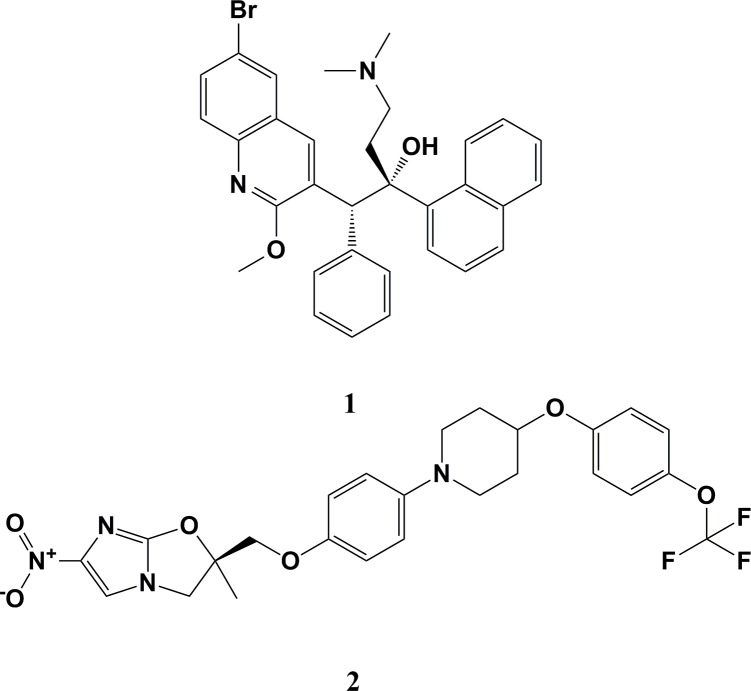
Mycobacterium tuberculosis (MTB) is the bacterial pathogen that underlies the infectious disease known as tuberculosis (TB). This disease affects the lungs and a number of other body systems and structures. According to WHO 2018 report, TB resulted in nearly 1.3 million deaths in those who are HIV-negative, and in 300,000 deaths among those who are HIV-positive [1]. Every year, new TB cases are reported worldwide and human immunodeficiency virus (HIV)-infected persons are up to 37 times more vulnerable to developing TB [2]. The development of multi-drug-resistant (MDR)-TB, extensively drug-resistant (XDR)-TB, and totally drug-resistant (TDR)-TB [3], as well as co-infections with acquired immunodeficiency syndrome (AIDS) and the risks involved in cases of TB among patients with diabetes mellitus [4], has resulted in a grave situation worldwide. Treating MDR-TB and XDR-TB is difficult, as second-line drugs have become far less effective [5]. This problem has been made worse by the evolution of TDR MTB strains [6] that are untreatable using the existing arsenal of anti-TB drugs. Based on the last 40 years of academic and pharmaceutical industry inventions, only bedaquiline (1) was the first novel anti-TB drug permitted by the United States Food and Drug Administration (US FDA) authority in December 2012 for the treatment of MDR-TB [7], while delamanid (2) was the second anti-TB agent to be approved by the European Medicines Agency (EMA) in late 2013 [8] (Fig 1).
Fig 1. Chemical structure of clinically approved anti-TB drugs bedaquiline (1) and delamanid (2).
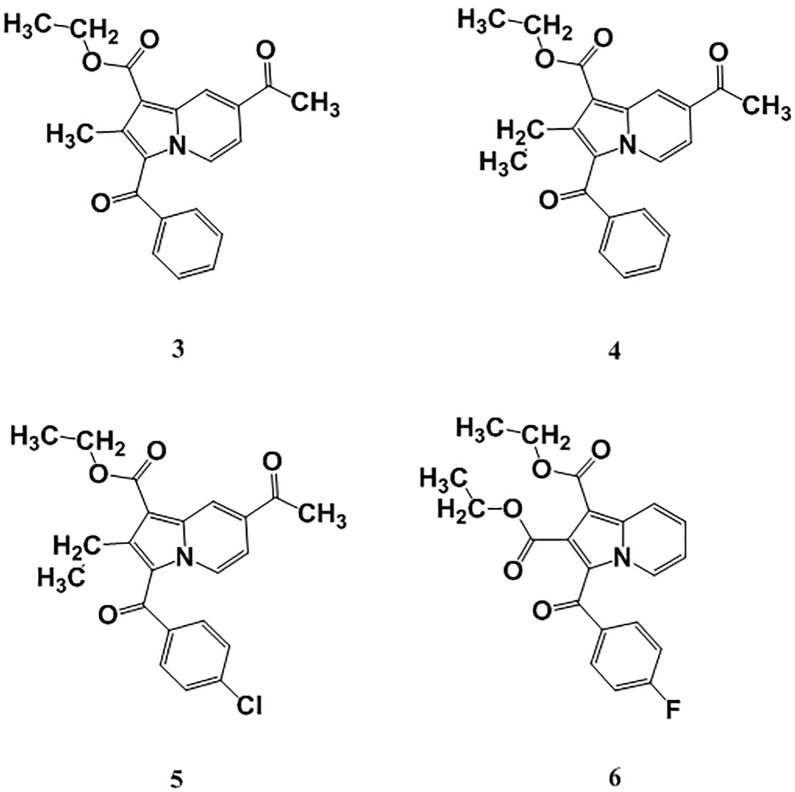
Fatty-acid biosynthesis is critical in the synthesis of the mycobacterial cell wall. The enoyl-[acyl-carrier] protein reductase enzyme elongates fatty-acid chains. Further, it acts as a catalyst to reduce α- and β-unsaturated fatty acids that are in complex with the enzyme, and has become of great interest when developing synthetic indolizine compounds that demonstrate anti-TB activity [9, 10]. Indolizines are heteroaromatic compounds, and their synthetic analogues have reportedly demonstrated promising pharmacological properties [11]. Specifically, they have exhibited analgesic [12], anticancer [13, 14], antidiabetic [15], antihistaminic [16], anti-inflammatory [17, 18], antileishmanial [19], antimicrobial [20], antimutagenic [21], antioxidant [22], antitubercular [10, 23], antiviral [24], larvicidal [25], and herbicidal activities [26]. In continuation of our previous work aimed at developing such synthetic indolizine analogues as enoyl-[acyl-carrier] protein reductase enzyme inhibitors (Fig 2), we undertake the screening of substituted 7-methoxy-indolizine analogues (Fig 3) to determine their whole-cell anti-TB properties against H3Rv and MDR strains of MTB using the resazurin microplate assay (REMA) plate method.
Fig 2. Molecular structure of compounds ethyl 7-acetyl-3-benzoyl-2-methyl-indolizine-1-carboxylate (3), ethyl 7-acetyl-3-benzoyl-2-ethyl-indolizine-1-carboxylate (4), ethyl 7-acetyl-3-(4-chlorobenzoyl)-2-ethyl-indolizine-1-carboxylate (5), and diethyl 3-(4-fluorobenzoyl)indolizine-1,2-dicarboxylate (6) for their anti-TB activity against MDR strains of MTB [10].
Fig 3. Chemical structures of substituted 7-methoxy-indolizine analogues tested for their anti-TB activity against H37Rv and MDR MTB strains.
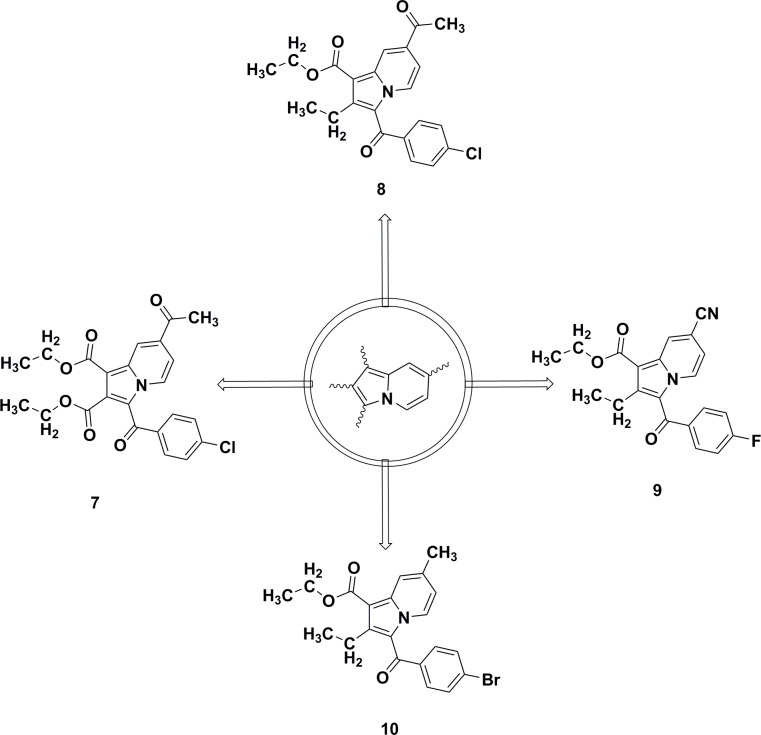
Our group recently investigated various substituted indolizine scaffolds for their synthesis, crystallography, and pharmacological properties, including their anticancer properties [14], their larvicidal activity against Anopheles arabiensis [25, 27], and their cyclooxygenase-2 (COX-2) inhibition properties (Fig 4) [18, 28].
Fig 4. Indolizine lead compounds identified for their anticancer (7) [14] and anti-tubercular (8) properties [10] against MDR strains of MTB, as well as for their COX-2 inhibition (9) [18] and larvicidal activity (10) [25] against Anopheles arabiensis.
Owing to an urgent call for the development of novel scaffold as anti-TB agents, we recently launched a medicinal chemistry program aimed at developing novel, natural, cyclic depsi-peptides [29] and heterocyclic scaffolds as potential anti-TB agents [10, 30–32]. We previously reported the anti-TB activity of indolizines [10], where a series of tri-substituted indolizines were identified as promising anti-TB agents. Among them, indolizine 8 (Fig 4) was found to be potent against H37Rv and MDR strains of MTB. Based on this observation we envisaged to synthesize and test 7-methoxy-indolizine analogues (5a-j) for anti-tubercular properties against H37Rv and MDR MTB strains.
Materials and methods
Materials
All chemicals reported here were obtained from Sigma-Aldrich Co. (St. Louis, MO, USA), while the solvents were obtained from MilliporeSigma (Burlington, MA, USA). Thin-layer chromatography (TLC) was employed to observe chemical reactions, and this process was performed on silica gel (Sigma-Aldrich Co.) on aluminum foil; n-hexane and ethyl acetate (4:6) were used as the solvent. The reactions were visualized under an ultraviolet (UV)-light/iodine chamber. A Büchi melting point B-545 apparatus was used to measure the melting points (Büchi, Labortechnik, Flawil, Switzerland). Infrared (IR) spectra were recorded on a Nicolet 6700 Fourier-transform infrared (FT-IR) spectrometer. Further, 1H and 13C-NMR spectra were recorded using Bruker AVANCE III 400 MHz (Bruker Corporation, Billerica, MA, USA) with CDCl3 (solvent). Chemical shifts (δ) were indicated in ppm, with tetramethylsilane (TMS) as a reference; coupling constants (J) were recorded (Hz). The splitting pattern was documented as follows: s, singlet; d, doublet; q, quartet; and m, multiplet. Liquid chromatography (LC)-mass spectrometry (MS) (Agilent 1100 series) was used to measure the mass spectra, in conjunction with MSD and 0.1% aqueous trifluoroacetic acid in an acetonitrile system on the C18-BDS column. Elemental analysis was conducted using a FLASH EA 1112 CHN analyzer (Thermo Finnigan LLC, New York, NY, USA).
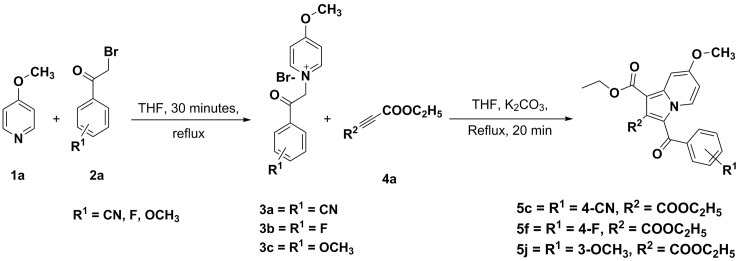
Synthesis of 1-(2-(4-substituted phenyl)-2-oxoethyl)-4-methoxypyridin-1-ium bromide (3a–3c)
4-Substituedphenacylbromide (1a) (0.0091 mol) was added to a well-stirred solution of 4-methoxypyridine (2a) (0.0091 mol) in dry tetrahydrofuran (12 mL) at ambient temperature; and the resulting reaction mixture was refluxed for 30 minutes (Fig 5) and monitored on TLC. The separated product was filtered, recrystallized using ethanol as a solvent, and dried at room temperature to afford a 98%–99% yield of 1-(2-(4-substitutedphenyl)-2-oxoethyl)-4-methoxypyridin-1-ium bromide (3a–3c).
Fig 5. Synthetic scheme for the construction of substituted 7-methoxy-indolizine analogues.
1-(2-(4-cyanophenyl)-2-oxoethyl)-4-methoxypyridin-1-ium bromide (3a)
Appearance: Yellow-colored product. Yield, 98%. 1H-NMR (400MHz, DMSO-d6) δ = 8.96–8.94 (d, J = 7.2 Hz, 2H), 8.52–8.50 (d, J = 7.0 Hz, 2H), 7.90–7.88 (d, J = 7.2 Hz, 2H), 7.58–7.56 (d, J = 8 Hz, 2H), 6.29 (s, 2H), 4.14 (s, 3H); LC-MS (ESI, Positive): m/z: (M+H)+: 253.2.
1-(2-(4-fluorophenyl)-2-oxoethyl)-4-methoxypyridin-1-ium bromide (3b)
Appearance: White-colored product. Yield, 99%. 1H-NMR (400MHz, DMSO-d6) δ = 8.72–8.70 (d, J = 7.0 Hz, 2H), 8.75–8.67 (d, J = 7.2 Hz, 2H), 8.30–8.27 (m, 2H), 8.13–8.10 (t, J = 8.8 Hz, 2H), 6.28 (s, 2H), 4.19 (s, 3H); LC-MS (ESI, Positive): m/z: (M+H)+: 246.12.
4-Methoxy-1-(2-(3-methoxyphenyl)-2-oxoethyl) pyridin-1-ium bromide (3c)
Appearance: Yellow-colored product. Yield, 99%. 1H-NMR (400MHz, DMSO-d6) δ = 9.02–9.00 (d, J = 7.2 Hz, 2H), 8.54–8.52 (d, J = 7.2 Hz, 2H), 7.49–7.47 (d, J = 7.2 Hz, 1H), 7.36–7.31 (t, J = 7.2 Hz, 1H), 7.15 (s, 1H), 7.10–7.08 (d, J = 7.2 Hz, 1H), 6.27 (s, 2H), 4.10 (s, 3H), 3.88 (s, 3H); LC-MS (ESI, Positive): m/z: (M+H)+: 258.2.
Synthetic procedure for the preparation of diethyl 3-(4-cyanobenzoyl)-7-methoxyindolizine-1,2-dicarboxylate (5c) [28]
A mixture of diethyl but-2-ynedioate (4a) (0.1 mmol) and K2CO3 (0.1 mmol) was added to a stirred solution of 1-(2-(4-cyanophenyl)-2-oxoethyl)-4-methoxypyridin-1-ium bromide (3a) (0.1 mmol) in dry tetrahydrofuran (15 mL). The reaction medium was refluxed for 20 minutes and reaction completion was monitored on TLC. Once the reaction was complete, the solvent was removed and diluted with ethyl acetate. The organic layer was rinsed with water and brine and dried over sodium sulfate. The obtained residue was purified by column chromatography to afford a 79% yield of diethyl 3-(4-cyanobenzoyl)-7-methoxyindolizine-1,2-dicarboxylate (5c). Title compounds 5f and 5j were synthesized following the same procedure; Table 1 outlines the physicochemical constants of the characterized title compounds.
Table 1. Physicochemical parameters of ethyl 7-methoxy-3-(substituted benzoyl)-indolizine-1-carboxylate 5a–5j.
| Compound code | Mol formulae (Mol Mass) | R1 | R2 | Yield (%)a | m.p (°C) reported | m.p (°C) found | cLogPb |
|---|---|---|---|---|---|---|---|
| 5a | C20H16N2O4 (348) | 4-CN | H | 81 | 165 | 165 | 3.9570 |
| 5b | C21H18N2O4 (362) | 4-CN | CH3 | 74 | 191 | 192 | 4.4560 |
| 5c | C23H20N2O6 (420) | 4-CN | COOC2H5 | 79 | - | 171 | 3.4454 |
| 5d | C19H16FNO4 (341) | 4-F | H | 80 | 118 | 118 | 4.5293 |
| 5e | C20H18FNO4 (355) | 4-F | CH3 | 73 | 137 | 138 | 5.0283 |
| 5f | C22H20FNO6 (413) | 4-F | COOC2H5 | 79 | - | 147 | 4.0199 |
| 5g | C19H16BrNO4 (402) | 4-Br | H | 75 | 183 | 182 | 5.2493 |
| 5h | C20H18BrNO4 (416) | 4-Br | CH3 | 77 | 148 | 148 | 5.7483 |
| 5i | C20H19NO5 (353) | 3-OCH3 | H | 85 | 116 | 116 | 4.4986 |
| 5j | C23H23NO7 (425) | 3-OCH3 | COOC2H5 | 76 | - | 142 | 4.0294 |
a Yields calculated after purification by column chromatography.
b ChemDraw Professional 16 was used to calculate cLogP of the title compounds.
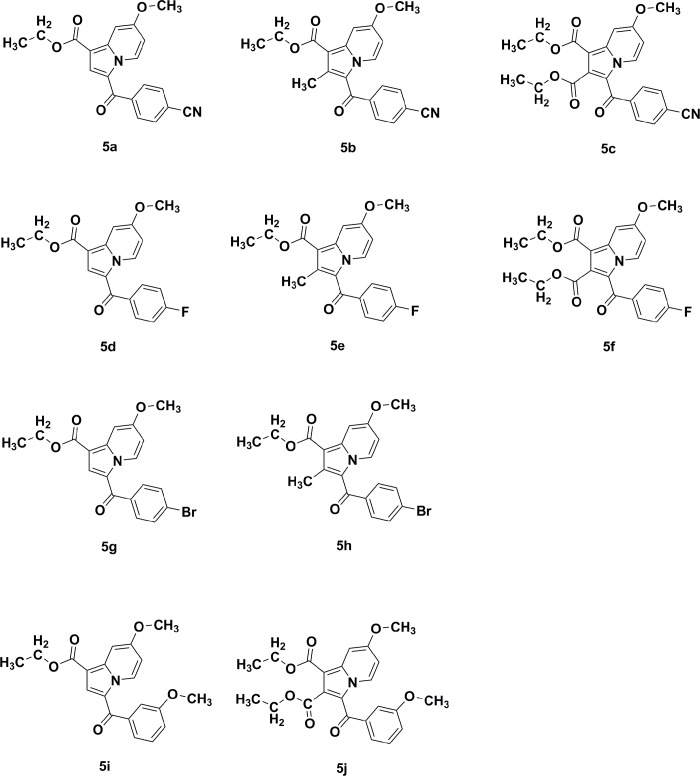
Title compounds 5a, 5b, 5d, 5e, 5g, 5h, and 5i were prepared and the physicochemical constants were compared with a previous report [18] and tabulated in Table 1. The molecular structures of the test compounds are illustrated in Fig 3.
Diethyl 3-(4-cyanobenzoyl)-7-methoxyindolizine-1,2-dicarboxylate (5c)
Appearance: Brown crystalline compound; Fourier transform (FT)-IR (KBr) cm-1 = 2987, 2229, 1737, 1693, 1647, 1596. 1H-NMR (400 MHz CDCl3) δ = 9.55–9.53 (d, J = 7.2Hz, 1H), 7.78 (s, 1H), 7.61–7.55 (m, 4H), 6.82–6.79 (m, 1H), 4.36–4.31 (q, J = 7.2Hz, 2H), 3.99 (s, 3H), 3.76–3.70 (q, J = 7.2Hz, 2H), 1.36–1.33 (t, J = 7.2Hz, 3H), 1.14–1.10 (t, J = 7.2Hz, 3H). 13C-NMR (100 MHz CDCl3) δ = 184.89, 164.98, 163.21, 160.09, 141.57, 138.57, 132.53, 131.21, 130.23, 130.05, 126.28, 119.40, 110.24, 102.64, 97.66, 61.77, 60.28, 55.83, 14.23, 13.60. LC-MS (ESI, Positive): m/z = (M+H)+: 421.2; Anal. calculated for C23H20N2O6; C, 65.71; H, 4.79; N, 6.66; Found; C, 65.70; H, 4.81; N, 6.62.
Diethyl 3-(4-fluorobenzoyl)-7-methoxyindolizine-1,2-dicarboxylate (5f)
Appearance: Yellow amorphous compound; FT-IR (KBr) cm-1 = 2981, 1737, 1699, 1647, 1607, 1230. 1H-NMR (400 MHz CDCl3) δ = 9.50–9.48 (d, J = 7.2Hz, 1H), 7.77–7.71 (m, 3H), 7.15–7.11 (m, 2H), 6.81–6.78 (m, 1H), 4.36–4.31 (q, J = 7.2Hz, 2H), 3.98 (s, 3H), 3.75–3.69 (q, J = 7.2Hz, 2H), 1.36–1.33 (t, J = 7.2Hz, 3H), 1.13–1.10 (t, J = 7.2Hz, 3H). 13C-NMR (100 MHz CDCl3) δ = 184.77, 166.11, 165.02, 163.60, 163.27, 159.96, 141.47, 136.04, 136.00, 132.25, 131.23, 131.14, 129.96, 119.58, 115.59, 114.97, 110.15, 102.44, 97.59, 61.68, 60.25, 55.81, 14.23, 13.62. LC-MS (ESI Positive): m/z = (M+H)+: 414.12; Anal. calculated for C22H20FNO6: C, 63.92; H, 4.88; N, 3,39; Found; C, 63.98; H, 4.85; N, 3.40.
Diethyl 7-methoxy-3-(3-methoxybenzoyl)indolizine-1,2-dicarboxylate (5j)
Appearance: Light-yellow crystalline compound; FT-IR (KBr) cm-1 = 2981, 1738, 1693, 1647, 1608. 1H-NMR (400 MHz CDCl3) δ = 9.56–9.54 (d, J = 7.2Hz, 1H), 7.78 (s, 1H), 7.38–7.36 (m, 1H), 7.29–7.28 (m, 1H), 7.21 (s, 1H), 7.10–7.08 (m, 1H), 6.80–6.78 (m, 1H), 4.35–4.30 (q, J = 7.2Hz, 2H), 3.99 (s, 3H), 3.81 (s, 3H), 3.73–3.68 (q, J = 7.2Hz, 2H), 1.36–1.32 (t, J = 7.2Hz, 3H), 1.11–1.08 (t, J = 7.2Hz, 3H). 13C-NMR (100 MHz CDCl3) δ = 185.90, 165.08, 163.35, 159.91, 159.09, 141.46, 141.00, 132.45, 130.10, 129.14, 121.14, 119.73, 118.38, 112.99, 110.08, 102.45, 97.57, 61.63, 60.21, 55.80, 55.36, 14.22, 13.54. LC-MS (ESI Positive): m/z = (M+H)+: 426.14. Anal. calculated for C23H23NO7; C, 64.93, H, 5.45, N, 3,29; Found; C, 64.95, H, 5.42, N, 3.32.
Crystallography
Crystal growth, single-crystal data collection, and refinement details
Suitable single crystals of compounds 5c and 5d were grown individually from the slow evaporation of toluene at ambient conditions. Single-crystal X-ray diffraction of 5c and 5d was performed using a Bruker SMART APEX II diffractometer with Mo-Kα radiation (χ = 0.71073 Å). Data collection was performed when the temperature reached 110 (2) K using an Oxford Cryostream cooling system (Bruker Apex II software) [33]. The Bruker SAINT software program was used to conduct cell refinement and data reduction [34]. SADABS was used for absorption correction [35], while the structures were solved using SHELXS-97 [36] and refined using full-matrix least-squares methods based on F2 (SHELXL-2018) [37] (WinGX software program, version 2014.1) [38]. The hydrogen atoms were refined using a riding model (Uiso(H) = 1.2Ueq [Caromatic] and Uiso(H) = 1.5Ueq [methyl groups]). One of the–OC2H5 groups (attached to the C19 atom) for the 5c structure was refined as a two-component positional disorder with the occupancy of 83:17. Geometric calculations were carried out using PLATON [39]. ORTEP and packing diagrams were created using the Mercury 3.5.1 (CCDC) program [40]. The crystal data and the structure refinement parameters are given in Table 2.
Table 2. Single crystal data and structure refinement parameters for 5c and 5d.
| Identification code | 5c | 5d |
|---|---|---|
| CCDC number | 1873349 | 1873348 |
| Empirical formula | C23 H20 N2 O6 | C19 H16 F N O4 |
| Formula weight | 420.41 | 341.33 |
| Temperature | 110(2) K | 110(2) K |
| Wavelength | 0.71073 Å | 0.71073 Å |
| Crystal system | Monoclinic | Triclinic |
| Space group | P21/n | P-1 |
| Unit cell dimensions | a = 12.1530(12) Å. | a = 4.1368(3) Å |
| b = 17.6845(15) Å. | b = 11.9682(8) Å | |
| c = 19.1511(19) Å. | c = 16.5416(10) Å | |
| α = 90° | α = 74.912(4)° | |
| β = 99.051(4)° | β = 88.675(5)° | |
| γ = 90° | γ = 80.110(5)° | |
| Volume | 4064.7(7) Å3 | 778.79(9) Å3 |
| Z | 8 | 2 |
| Density (calculated) | 1.374 Mg/m3 | 1.456 Mg/m3 |
| Absorption coefficient | 0.101 mm-1 | 0.110 mm-1 |
| F(000) | 1760 | 356 |
| Crystal size | 0.280 x 0.160 x 0.060 mm3 | 0.320 x 0.120 x 0.060 mm3 |
| Theta range for data collection | 1.861 to 27.875°. | 5.017 to 29.575°. |
| Index ranges | -15< = h< = 13, -23< = k< = 23, -25< = l< = 21 | -5< = h< = 5, -15< = k< = 16, -22< = l< = 21 |
| Reflections collected | 30715 | 10474 |
| Independent reflections | 9438 [R(int) = 0.0794] | 4218 [R(int) = 0.0601] |
| Completeness to theta = 25.242° | 98.6% | 97.3% |
| Absorption correction | Semi-empirical from equivalents | Semi-empirical from equivalents |
| Max. and min. transmission | 0.7460 and 0.6507 | 0.7460 and 0.6647 |
| Refinement method | Full-matrix least-squares on F2 | Full-matrix least-squares on F2 |
| Data / restraints / parameters | 9438 / 3 / 577 | 4218 / 0 / 228 |
| Goodness-of-fit on F2 | 1.010 | 1.051 |
| Final R indices [I>2sigma(I)] | R1 = 0.0715, wR2 = 0.1684 | R1 = 0.0580, wR2 = 0.1404 |
| R indices (all data) | R1 = 0.1296, wR2 = 0.1958 | R1 = 0.1010, wR2 = 0.1627 |
| Largest diff. peak and hole | 0.318 and -0.266 e.Å-3 | 0.267 and -0.293 e.Å-3 |
Computational studies
Molecular-docking study
The three-dimensional (3D) molecular structures of the studied compounds (5a–5j) were built using Gaussview and optimized using an AM1 semi-empirical method to their ground state level using Gaussian09 [41]. The crystal structure of the enoyl-[acyl-carrier] protein reductase enzyme was downloaded from the Protein Databank RCSB (PDB; PDB code entry: 1ZID), in which the enzyme was crystalized with an isonicotinic acyl NADH inhibitor. To prepare the protein for docking, water molecules, inhibitor molecules, and any co-crystalized molecules were removed. Autodock 4 [42] was used to dock the studied compounds at the active site of the enzyme. First, the crystalized inhibitor was docked to verify the docking procedure and to confirm the position of the active site; the remaining compounds used the same procedure, in which Kollman-united atom charges neutralized the enzyme with a grid box of 60×60×60 with 0.375 Å distance between points. Then, 250 runs for each inhibitor were carried out using a Lamarckian genetic algorithm. The docked conformations were clustered and ranked according the binding free energy. Discovery Studio 5.0 visualizer was used to visualize the best docked poses and to elucidate the intra-molecular interactions at the enzyme’s active site.
Molecular-dynamics simulation
Amber14 was used to perform all simulations for the enzyme-inhibitor complex immersed in a water box using a TIP3P explicit solvent; an ff14SB force field was employed at a temperature equal to 300 K [43]. Four sodium ions were added to neutralize this system. The system was minimized in two steps, followed by 2.0 fs time step simulations with a cutoff of 10 Å for non-bonded interactions. Short simulation with a constant-volume periodic boundary was performed to increase the temperature from 0 K to 300 K, followed by 3.5 ns of a constant-pressure periodic boundary MD at 300 K using the Langevin thermostat. The same procedure was performed for the enzyme complex and the most active ligands (5i and 5j), as well as for the enzyme that did not have any inhibitor (for comparative purposes).
Antitubercular activity
Resazurin microplate assay (REMA)
Anti-TB screening of test compounds 5a–j was performed using the colorimetric REMA plate method [31, 44].
Determining the minimum inhibitory concentration (MIC)
All test compounds (5a–j) were further evaluated by the agar incorporation method, which was performed three times, and which targeted an H37Rv strain and an MDR-TB strain (isoniazid = 0.2 μg/mL and rifampicin >1.0 μg/mL). MIC determination was performed [45], with some modifications. A Level II Biosafety laboratory was used to carry out this experiment. MTB reference strain H37Rv (American Type Culture Collection [ATCC], Manassas, VA, USA: 25177) and MDR-TB were cultured in Middlebrook 7H11 medium for a total of 3 weeks [46]. The strain was supplemented with OADC (0.005%, v/v, oleic acid; 0.2%, w/v, glucose; 0.085%, w/v, NaCl; 0.02%, v/v, catalase; and 0.5%, 171 w/v, bovine serum albumin [BSA]), and incubated at a temperature of 37°C. Fresh cultures were used to in the preparation of a standardized inoculum in a sterile tube containing 0.05% Tween 80 and 4.5 mL of phosphate buffer; (5 mm in diameter) were used for vortexing. The bacterial supernatant was then standardized to McFarland Number 1 with water, yielding a bacterial concentration of ~1×107cfu/mL. The bacterial suspension was diluted with water; then, a total of 100 μL of the dilution was placed onto Middlebrook 7H10 agar plates containing drug doses ranging from 8–0.125 μg/mL (to begin, 8 μg/mL of the drug was dissolved in distilled water and then diluted twofold to reach the desired concentration before being added to the agar medium). The MICs of the drugs (i.e., that inhibited >1% of the organism’s growth when compared with controls) were obtained 3 weeks following incubation. Table 3 presents the anti-TB results when compared with H37Rv (ATCC: 25177), MDR-MTB, and XDR-MTB.
Table 3. In vitro whole-cell anti-TB activity of 7-methoxy-indolizine analogues (5a–j) against H37RV and MDR-MTB isolates.
| Compound Code | Anti-TB activity—MIC (μg/mL) | |
|---|---|---|
| H37RV isolate | MDR-MTB isolate* | |
| 5a | 8 | 32 |
| 5b | NA | NA |
| 5c | 32 | 64 |
| 5d | 8 | NA |
| 5e | 32 | NA |
| 5f | 32 | NA |
| 5g | NA | NA |
| 5h | NA | NA |
| 5i | 8 | 16 |
| 5j | 8 | 16 |
MIC, minimum inhibitory concentration.
*These isolates were found to be resistant to the first-line antibiotics rifampicin (1 μg/mL), and isoniazid (0.2 μg/mL).
NA: not active
Safety studies- cytotoxicity assay
Title compounds 5a, 5c, 5i, and 5j which exhibited anti-TB activity against MDR strains of MTB, were subjected to safety studies by 3-(4,5-dimethylthiazol-2-yl) -2,5-diphenyltetrazolium bromide (MTT) assay. The MTT cytotoxicity assay is used to evaluate the cytotoxic effects of the most promising compounds against peripheral blood mononuclear cells (PBMCs)–i.e., 5a, 5c, 5i, and 5j –according to the described protocol [47].
Results and discussion
Chemistry
To explore the role of various functional groups on the indolizine nucleus, a series of indolizine scaffolds were synthesized using a greener synthetic approach and the yield was found to be in the range of 73%–85% following purification by column chromatography. The synthetic scheme for the construction of the title compounds (5a–j) is illustrated in Fig 5, and the physicochemical characteristic details are tabulated in Table 1. The intermediates required to develop novel title compounds 5c, 5f, and 5j were synthesized between 92%–99% yield and the characterization details are listed under the experimental section. Title compounds 5a–c, 5d–f, and 5g–h were prepared with nitrile, fluoro, and bromo functional groups, respectively, at the para position of the benzoyl group at the third position of the indolizine nucleus. Compounds 5i and 5j were prepared by having a methoxy functional group at the meta position of the benzoyl group, which is at the third position of the indolizine nucleus. Compounds 5a, 5d, 5g, and 5i were unsubstituted at the second position of the indolizine nucleus, whereas compounds 5b, 5e, and 5h had a methyl substituent at the second position of the indolizine nucleus. Conversely, compounds 5c, 5f, and 5j had an ethyl ester functional group at the second position of the indolizine nucleus. The molecular structure of the resynthesized compounds 5a, 5b, 5d, 5e, 5g, 5h, and 5i was confirmed by LC-MS and melting-point determination. Novel compounds 5c, 5f, and 5j were prepared using a green chemistry approach, and their molecular structures were confirmed by FT-IR, NMR (1H and 13C), LC-MS, and elemental analysis. FT-IR spectra of the title compounds 5c, 5f, and 5j exhibited carbonyl stretching at the 1737–1738 cm–1 range. In the case of proton nuclear magnetic resonance (NMR) spectra, the singlet peak for the methoxy group was observed at 3.98–3.99 ppm. During 13C NMR, carbonyl carbon stretching for compounds 5c, 5f, and 5j was observed in the range of δ = 184.77–184.89. The molecular ion peaks of the compounds observed on LC-MS were in alignment with their molecular mass, whereby the results were within ±0.4% of the calculated theoretical values. cLogP of the title compounds was calculated using ChemDraw Prof version 16.0 and the calculated results were 3.9570–5.7483. The bromo group at the para position of the benzoyl group, which is at the third position of the indolizine nucleus, exhibited the highest cLogP value at 5.7483. Selected title compounds 5c and 5d were subjected to single-crystal X-ray studies and crystal data were deposited into the Cambridge Crystallographic Data Centre (CCDC; numbers 1873349 and 1873348, respectively).
Crystallography
Analysis of the crystal structures of compounds 5c and 5d
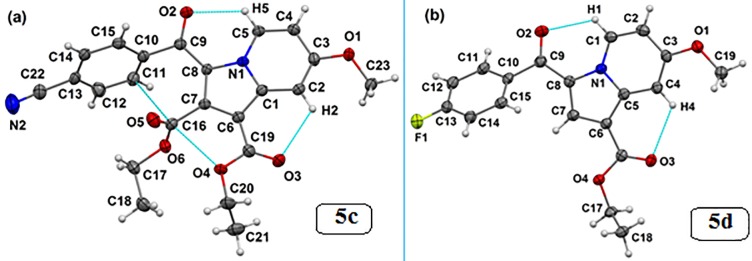
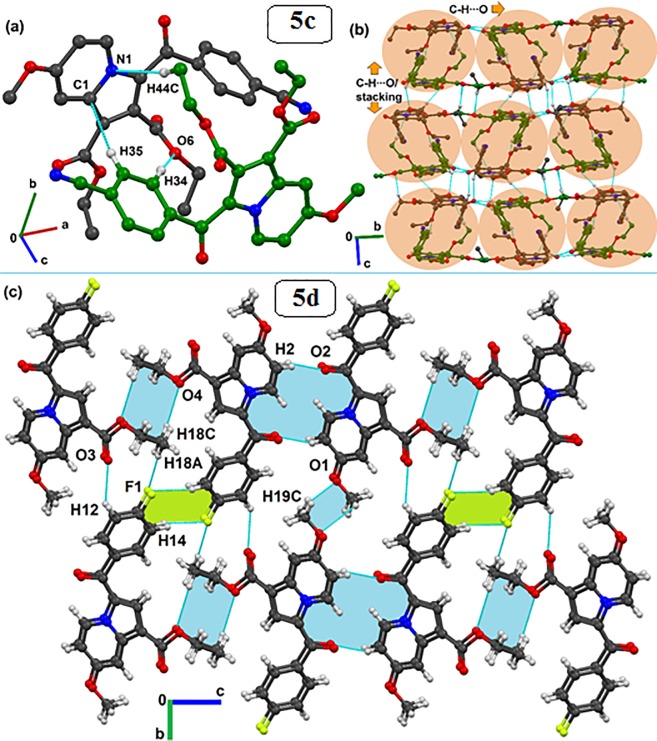
The single-crystal X-ray diffraction study for the title compound revealed that 5c crystallizes in the monoclinic P21/n space group with two symmetrical free molecules (Z′ = 2), while the 5d crystallizes in the P-1 space group of the triclinic crystal system, consisting of one molecule (Z′ = 1) in the asymmetric unit (Table 3). The molecular structures of both 5c and 5d are shown in Fig 6, which depicts that the molecular conformation in the crystal of 5c is primarily stabilized via intramolecular C-H···O, C···O, and C···C(π) contacts. Similarly, intramolecular C-H···O contacts lock the crystal conformation in molecule 5d. Looking for the supramolecular structure of 5c, the presence of two symmetry-independent molecules forms a dimeric motif, which is stabilized by C-H···O, C-H···N and C-H···π hydrogen bonds (see molecules in black and green; Fig 7A). Such hydrogen-bonded dimers (light-red shaded circle in Fig 7B) further extend along the b-crystallographic direction via a C-H···O hydrogen bond, forming a layer of molecules. These molecular layers are assembled through C-H···O and π···π stacking interactions in a parallel manner (see shaded circles) along the crystallographic c-direction to complete the two-dimensional crystal structure of 5c. Conversely, the crystal structure formed by 5d molecules is primarily governed by C-H···O hydrogen-bonded dimers (light-blue shade, Fig 7C).
Fig 6.
Thermal ellipsoidal plots drawn at the 50% probability level for the crystal structures of (a) 5c (second symmetry independent molecule has been omitted for clarity, Z' = 2) and (b) 5d. Dotted lines indicate intramolecular interactions.
Fig 7.
Formation of (a) dimer by two symmetry-independent molecules (black and green) in the asymmetric unit of 5c utilizing C-H···O, C-H···N and C-H···π interactions. (b) Crystal packing for 5c molecules via the association of dimeric motifs (light red) through C-H···O and π···π stacking interactions. (c) Packing arrangement of 5d molecules stabilized via various strong to weak C-H···O (light blue) and C-H···F (light green) dimers. Different-colored carbon atoms indicate different symmetry-independent molecules. Non-interacting hydrogen atoms were removed in the case of 5d to clarify the packing view.
In addition, weak C-H···F (light-green) dimers also provide further support in the formation of molecular sheet-like supramolecular constructs down the bc-crystallographic plane. These molecular sheets are further stacked along the a-direction (Fig 7C). The geometrical parameters of all possible intra- and intermolecular interactions for both 5c and 5d crystal structures are provided in Table 4.
Table 4. List of intra- and intermolecular interactions present in compounds 5c and 5d.
| Motifs | D–H…A | Symmetry | Geometry | ||
|---|---|---|---|---|---|
| D…A/Å | H…A/ Å | ∠D–H…A/° | |||
| 5c [C1 > C23 –first molecule; C24 > C46 –second molecule] | |||||
| I | C2-H2···O3 | x, y, z (intra) | 3.028(2) | 2.45 | 112 |
| C5-H5···O2 | 2.862(2) | 2.21 | 117 | ||
| C16···O4 | 2.724(2) | - | - | ||
| C16···C11(π) | 3.073(2) | - | - | ||
| C24-H24···O8 | 2.868(2) | 2.21 | 117 | ||
| C27-H27···O9 | 3.007(2) | 2.41 | 113 | ||
| C42···O10 | 2.793(2) | - | - | ||
| C42···C34(π) | 3.026(2) | ||||
| II | C11-H11···O12 | x, y, z | 3.739(3) | 2.66 | 175 |
| C12-H12···O9 | 3.528(3) | 2.71 | 132 | ||
| C23-H23C···N4 | 3.761(3) | 2.71 | 164 | ||
| C17-H17A···N3 | 3.565(3) | 2.77 | 137 | ||
| C34-H34···O6 | 3.535(3) | 2.51 | 157 | ||
| C44-H44C···N1 | 3.672(3) | 2.61 | 168 | ||
| C35-H35···C2(π) | 3.927(3) | 2.87 | 168 | ||
| C12-H12···C27(π) | 4.008(3) | 2.96 | 164 | ||
| III | C25-H25···O2 | -x+1/2, y-1/2, -z+1/2 | 3.393(2) | 2.31 | 178 |
| C14-H14···O8 | 3.212(2) | 2.61 | 115 | ||
| C12-H12···C14(π) | 3.746(2) | 2.70 | 162 | ||
| IV | C23-H23A···O11 | x-1/2, -y+1/2, z-1/2 | 3.356(2) | 2.32 | 159 |
| C20-H20B···O9 | 3.265(2) | 2.53 | 161 | ||
| π ···π (molecular stacking) | 3.994(3) | - | - | ||
| V | C23-H23B···O5 | x-1, y, z | 3.530(3) | 2.51 | 157 |
| C43-H43B···N2 | 3.554(3) | 2.76 | 131 | ||
| VI | C4-H4···O8 | -x-1/2, y+1/2, -z+1/2 | 3.801(2) | 2.81 | 153 |
| C37-H37···O2 | 3.455(2) | 2.45 | 154 | ||
| VII | C45-H45B···O11 | x+1, y, z | 3.563(2) | 2.56 | 154 |
| O7···C37(π) | 3.003(2) | - | - | ||
| VIII | C15-H15···O2 | -x, -y+1, -z | 3.489(3) | 2.48 | 155 |
| C15-H15···C5(π) | 3.728(3) | 2.88 | 135 | ||
| IX | C38-H38···O1 | x+1/2, -y+1/2, z+1/2 | 3.441(2) | 2.58 | 136 |
| X | C41-H41C···O10 | -x,-y+1,-z+1 | 3.616(2) | 2.61 | 155 |
| 5d | |||||
| I | C1-H1···O2 | x, y, z (intra) | 2.930(2) | 2.30 | 115 |
| C4-H4···O3 | 3.066(2) | 2.48 | 113 | ||
| II | C18-H18···F1 | x+1, y-1, z | 3.618(3) | 2.55 | 172 |
| C12-H12···O3 | 4.664(3) | 2.61 | 145 | ||
| III | C17-H17A···F1 | x, y+1, z | 3.295(3) | 2.47 | 133 |
| IV | C17-H17B···O4 | -x+2, -y+1, -z+1 | 3.546(2) | 2.88 | 119 |
| C18-H18C···O4 | 2.492(2) | 2.84 | 119 | ||
| V | C17-H17A···O3 | x+1, y, z | 3.552(3) | 2.70 | 136 |
| C17-H17B···O4 | 3.548(3) | 2.88 | 120 | ||
| VI | C14-H14···F1 | -x+2, -y, -z+1 | 3.333(2) | 2.43 | 140 |
| VII | C19-H19A···O3 | x-1, y, z | 3.782(3) | 2.80 | 152 |
| C17-H17A···O3 | 3.552(3) | 2.70 | 136 | ||
| π ···π (molecular stacking) | 3.632- 4.137(3) |
- | - | ||
| VIII | C19-H19B···O1 | - x-1, -y+2, -z | 3.364(2) | 2.43 | 143 |
| IX | C2-H2···O2 | -x, -y+1, -z | 3.377(2) | 2.33 | 162 |
Computational studies
Docking calculations
To investigate the binding affinity of the studied compounds and to establish potential correlations between the experimental results, a docking study was performed at the active site of the enzyme. The binding affinity and predicted inhibition constant are summarized in Table 5. Indeed, the 10 derivatives showed binding affinity within the range of 7.07–8.57 kcal/mol, with 5i showing the highest. Obviously, the non-bonding interactions between the ligands and amino acids at the active site are responsible for the formation of a stable enzyme-inhibitor complex. For that, the formed complexes with the best binding affinity were visualized and the interactions with the active site were investigated (the interactions are detailed in Fig 8). Hydrogen bonding, pi-pi interactions, and electrostatic interactions are common at the active site. In addition, most of these interactions occurred with the active site residues Pro193, Tyr158, Phe149, and Lys165.
Table 5. Docking free energy and estimated inhibition constant (Ki) of the docked indolizine analogues 5a–j.
| Indolizine analogues | Docking free energy | Inhibition constant |
|---|---|---|
| 5a | –8.46 kcal/mol | 633.99 nM |
| 5b | –8.39 kcal/mol | 703.54 nM |
| 5c | –7.86 kcal/mol | 1.72 uM |
| 5d | –7.90 kcal/mol | 1.62 uM |
| 5e | –8.47 kcal/mol | 617.21 nM |
| 5f | –7.07 kcal/mol | 6.57 uM |
| 5g | –8.54 kcal/mol | 547.97 nM |
| 5h | –8.53 kcal/mol | 563.15 nM |
| 5i | –8.57 kcal/mol | 525.81 nM |
| 5j | –7.36 kcal/mol | 4.01 uM |
Fig 8. Intermolecular interactions of docked 7-methoxy-indolizine analogues 5a–5j at the active site of the enoyl-[acyl-carrier] protein-reductase enzyme.
Molecular dynamic simulations
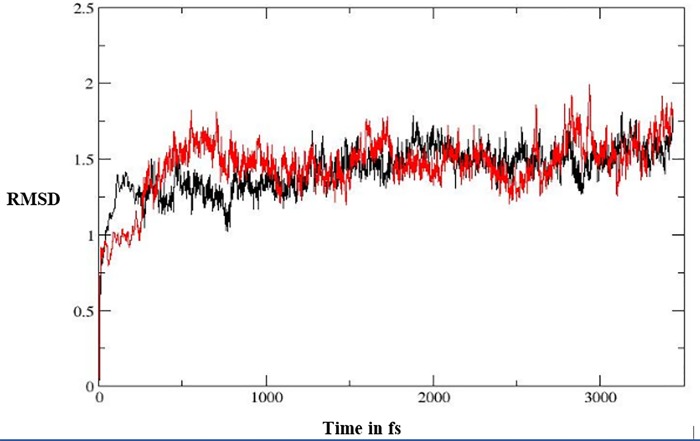
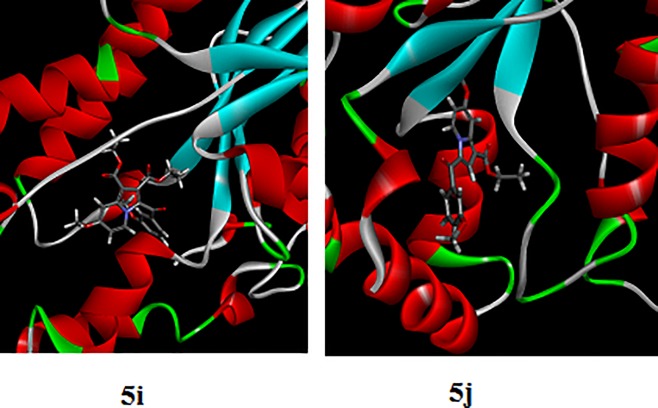
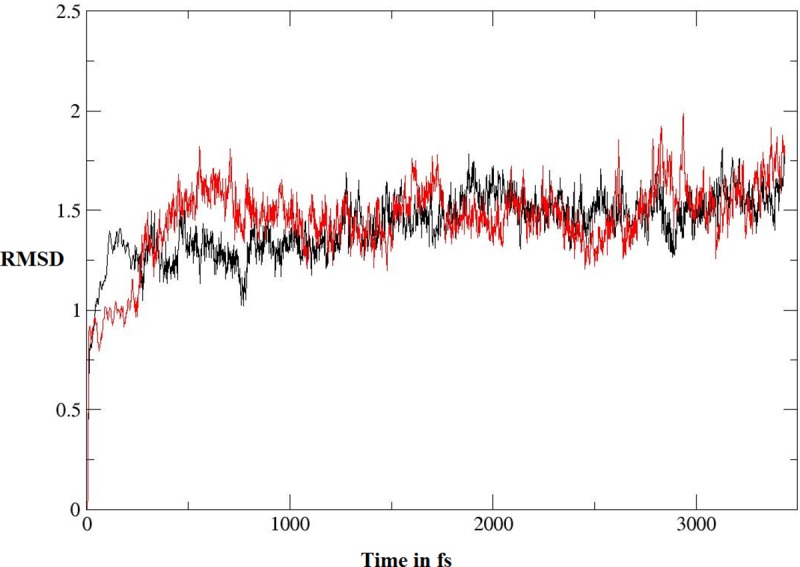
As shown in Table 2, 5I and 5J showed the highest anti-TB biological activity, which was supported by the results of the docking study. To study the stability of the complexes of these compounds with the active site, molecular-dynamic simulation was performed for 3.5 ns (Fig 9). At the beginning, the complexes were optimized, relaxed, and equilibrated followed by long simulation for around 3.0 ns. Following the stability of the simulated complexes, molecular mechanics/Poisson–Boltzmann surface area (MM/PBSA) and molecular mechanics/generalized born surface area (MM/GBSA) were computed; these are widely used to estimate the free binding energy, as shown in Table 6. It was clear that 5j showed higher binding affinity toward the enzyme, according to the two methods of calculation (MM/PBSA and MM/GBSA), which is in contrast with the docking results. It is well known that the docking process is performed while the structure of the enzyme is fixed. In contrast, as the MM/PBSA and MM/GBSA calculations are based on the molecular dynamic simulation, the enzyme-inhibitor complex is flexible which may enhance the reliability of results. Fig 10 illustrates the interaction of ligands 5i and 5j at the active site of the enoyl-[acyl-carrier] protein reductase enzyme following the simulation.
Fig 9. RMSD graph computed for 3.5 ns; the black line represents compound 5i and the red line represents compound 5j.
Table 6. MM/PBSA and MM/GBSA calculations for compounds 5i and 5j.
| Compound | ΔG (MM/PBSA) | ΔG (MM/GBSA) |
|---|---|---|
| 5i | –5.803 | –18.841 |
| 5j | –7.004 | –21.346 |
Fig 10. The interaction of ligands 5i and 5j at the active site of the enoyl-[acyl-carrier] protein-reductase enzyme following simulation.
Anti-TB activity
Title compounds 5a–5j were evaluated for their MIC against H37Rv and MDR strains of MTB (Table 2). Compounds 5i and 5j have a methoxy group at the meta position of the benzoyl group, which was at the third position of the indolizine nucleus; they exhibited similar anti-TB activity at 8 μg/mL against the H37Rv strain and at 16 μg/mL against the MDR strains of MTB. Compound 5a, which has a nitrile group at the fourth position of the benzoyl group, revealed activity at 8 μg/mL against H37Rv and at 32 μg/mL against MDR strains of MTB. However, di-ethyl ester functional groups on the indolizine nucleus in compound 5c with the nitrile group at the fourth position of the benzoyl group showed anti-TB activity at 8 μg/mL against H37Rv and at 32 μg/mL against MDR strains of MTB. Although compounds 5e and 5f exhibited anti-TB activity against the susceptible H37Rv strain at 32 μg/mL, they failed to show anti-TB activity against MDR strains of MTB; meanwhile, compounds 5b, 5g, and 5h exhibited no activity against either of the anti-TB strains in the experiment.
Safety studies
The anti-TB test compounds 5a, 5c, 5i, and 5j from the series in Fig 5 were evaluated in safety studies by MTT assay. It was found that test compounds 5a, 5c, 5i, and 5j exhibited no toxicity up to 500 μg/mL across PBM cell lines.
Conclusions
Indolizine compounds were previously identified as a class of anti-TB agents against MDR strains of MTB. Here, we presented our medicinal chemistry efforts that were aimed at screening indolizine analogues with various functional groups to determine their anti-TB activity in vitro. We performed computational docking for the compounds 5a to 5j and dynamics simulations for the compounds 5i and 5j, and we also detailed the crystallographic insights of two selected compounds that had different substituents to assess the role of inter- and intra-molecular interactions. Compounds 5i and 5j emerged as promising anti-TB agents against MDR strains of MTB with no toxicity up to 500μg/mL. Docking and MD-simulation results tended to support the corresponding observed biological activity; these data showed that 5j has higher binding affinity when compared with 5i. The findings of the crystallographic analysis clearly suggest that the molecular arrangements of the 5c and 5d structures are mostly guided by C-H···O hydrogen-bonded dimeric motifs and C-H···N hydrogen bonds, while various secondary interactions (including π···π and C-H···F) were also found to contribute to the crystal formation.
Supporting information
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
Acknowledgments
The authors wish to express their gratitude to the Deanship of Scientific Research, King Faisal University, Kingdom of Saudi Arabia for providing support and encouragement. The authors thank Dr. Hong Su, Centre for Supramolecular Chemistry Research, Department of Chemistry, University of Cape Town, Rondebosch 7701 for single crystal X-ray data collection. DC and SB thank IISER Bhopal for providing the research facilities and infrastructure for this study. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Abbreviations
- MDR-TB
multi-drug-resistant tuberculosis
- INH
isoniazid
- TB
tuberculosis
- TDR
totally drug resistant
- XDR-TB
extensively drug resistant
- MM/PBSA
molecular mechanics/Poisson–Boltzmann surface area
- MM/GBSA
Molecular mechanics/Generalized Born surface area
- MIC
minimum inhibitory concentration
- REMA
resazurin microplate assay
- DMF
Dimethylformamide
- MTB
Mycobacterium tuberculosis
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The authors are grateful to the Deanship of Scientific Research, King Faisal University, Kingdom of Saudi Arabia, for providing financial support (grant number: 17122011).
References
- 1.WHO. Executive summary_21Sept2018. http://wwwwhoint/tb/publications/global_report/Exec_summary_21Sept2018v11pdf?ua=1 visited on September 29, 2018. 2017.
- 2.WHO, Executive summary_21Sept2018. http://wwwwhoint/tb/publications/global_report/Exec_summary_21Sept2018v11pdf?ua=1 visited on September 29, 2018. 2018.
- 3.Velayati AA, Masjedi MR, Farnia P, Tabarsi P, Ghanavi J, Ziazarifi AH, et al. Emergence of new forms of totally drug-resistant tuberculosis bacilli: super extensively drug-resistant tuberculosis or totally drug-resistant strains in iran. Chest. 2009;136(2):420–5. 10.1378/chest.08-2427 [DOI] [PubMed] [Google Scholar]
- 4.Baker MA, Lin HH, Chang HY, Murray MB. The risk of tuberculosis disease among persons with diabetes mellitus: a prospective cohort study. Clin Infect Dis. 2012;54(6):818–25. 10.1093/cid/cir939 [DOI] [PubMed] [Google Scholar]
- 5.Hu Y, Xu L, He YL, Pang Y, Lu N, Liu J, et al. Prevalence and molecular characterization of second-line drugs resistance among multidrug-resistant Mycobacterium tuberculosis isolates in Southwest of China. BioMed Research International. 2017;2017:4563826 10.1155/2017/4563826 PMC5536135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parida SK, Axelsson-Robertson R, Rao MV, Singh N, Master I, Lutckii A, et al. Totally drug-resistant tuberculosis and adjunct therapies. J Intern Med. 2015;277(4):388–405. 10.1111/joim.12264 [DOI] [PubMed] [Google Scholar]
- 7.Cox E, Laessig K. FDA Approval of Bedaquiline—The benefit–risk balance for drug-resistant Tuberculosis. N Engl J Med. 2014;371(8):689–91. 10.1056/NEJMp1314385 . [DOI] [PubMed] [Google Scholar]
- 8.Barry Iii CE. Timing is everything for compassionate use of delamanid. Nat Med. 2015;21(3):211–. 10.1038/nm.3823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ling LL, Xian J, Ali S, Geng B, Fan J, Mills DM, et al. Identification and Characterization of Inhibitors of Bacterial Enoyl-Acyl Carrier Protein Reductase. Antimicrob Agents Chemother. 2004;48(5):1541–7. 10.1128/AAC.48.5.1541-1547.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khedr MA, Pillay M, Chandrashekharappa S, Chopra D, Aldhubiab BE, Attimarad M, et al. Molecular modeling studies and anti-TB activity of trisubstituted indolizine analogues; molecular docking and dynamic inputs. J Biomol Struct Dyn. 2018;36(8):2163–78. Epub 2017/06/29. 10.1080/07391102.2017.1345325 . [DOI] [PubMed] [Google Scholar]
- 11.Sandeep C, Venugopala KN, Mohammed AK, Mahesh A, Basavaraj P, Rashmi SK, et al. Review on chemistry of natural and synthetic indolizines with their chemical and pharmacological properties. J Basic Clin Pharm. 2016;8(2):49–61. [Google Scholar]
- 12.Vaught JL, Carson JR, Carmosin RJ, Blum PS, Persico FJ, Hageman WE, et al. Antinociceptive action of McN-5195 in rodents: a structurally novel (indolizine) analgesic with a nonopioid mechanism of action. J Pharmacol Exp Ther. 1990;255(1):1–10. [PubMed] [Google Scholar]
- 13.Butler MS. Natural products to drugs: natural product-derived compounds in clinical trials. Nat Prod Rep. 2008;25(3):475–516. 10.1039/b514294f [DOI] [PubMed] [Google Scholar]
- 14.Sandeep C, Padmashali B, Venugopala KN, Kulkarni RS, Venugopala R, Odhav B. Synthesis and characterization of ethyl 7-acetyl-2-substituted 3-(substituted benzoyl)indolizine-1-carboxylates for in vitro anticancer activity. Asian J Chem. 2016;28(5):1043–8. 10.14233/ajchem.2016.19582. . [DOI] [Google Scholar]
- 15.Mederski W, Beier N, Burgdorf LT, Gericke R, Klein M, Tsaklakidis C. Indolizine derivatives and the use thereof as antidiabetics. US Patent. 2012, Jan 31;8(106,067 B2).
- 16.Cingolani GM, Claudi F, Massi M, Venturi F. Indolizine derivatives with biological activity VI 1-(2-aminoethyl)-3-benzyl-7-methoxy-2-methylindolizine, benanserin structural analogue. Cingolani. 1990;25(8):709–12. 10.1016/0223-5234(90)90138-S. [DOI] [Google Scholar]
- 17.Hagishita S, Yamada M, Shirahase K, Okada T, Murakami Y, Ito Y, et al. Potent inhibitors of secretory phospholipase A2: synthesis and inhibitory activities of indolizine and indene derivatives. J Med Chem. 1996;39(19):3636–58. Epub 1996/09/13. 10.1021/jm960395q . [DOI] [PubMed] [Google Scholar]
- 18.Sandeep C, Venugopala KN, Khedr MA, Padmashali B, Kulkarni RS, Rashmi V, et al. Design and synthesis of novel indolizine analogues as COX-2 inhibitors: Computational perspective and in vitro screening. Indian J Pharm Educ. 2017;51(3):452–60. [Google Scholar]
- 19.Jaisankar P, Pal B, Manna KN, Pradhan PK, Medda S, Basu MK, et al. Synthesis of antileishmanial (5R)-(-)-5-carbomethoxy-3-formyl-5,6-dihydroindolo-[2,3-a]-indolizine. ARKIVOC. 2003;(9):150–7. [Google Scholar]
- 20.Hazra A, Mondal S, Maity A, Naskar S, Saha P, Paira R, et al. Amberlite-IRA-402 (OH) ion exchange resin mediated synthesis of indolizines, pyrrolo [1,2-a] quinolines and isoquinolines: antibacterial and antifungal evaluation of the products. Eur J Med Chem. 2011;46(6):2132–40. 10.1016/j.ejmech.2011.02.066 [DOI] [PubMed] [Google Scholar]
- 21.Olejnikova P, Birosova L, Svorc L. Antimicrobial and antimutagenic properties of newly synthesized derivatives of indolizine. Sci Pharm. 2009;77:216. [Google Scholar]
- 22.Nasir AI, Gundersen L-L, Rise F, Antonsen Ø, Kristensen T, Langhelle B, et al. Inhibition of lipid peroxidation mediated by indolizines. Bioorg Med Chem Lett. 1998;8(14):1829–32. 10.1016/S0960-894X(98)00313-8. [DOI] [PubMed] [Google Scholar]
- 23.Dannhardt G, Meindl W, Gussmann S, Ajili S, Kappe T. Anti-mycobacterial 7-hydroxy-2,3-dihydro-1H-indolizin-5-ones. Eur J Med Chem. 1987;22(6):505–10. 10.1016/0223-5234(87)90290-X. [DOI] [Google Scholar]
- 24.Mishra BB, Tiwari VK. Natural products in drug discovery: Clinical evaluations and investigations. Opportunity Challenge and Scope of Natural Products in Medicinal Chemistry. 2011:1–61. [Google Scholar]
- 25.Sandeep C, Venugopala KN, Gleiser RM, Chetram A, Padmashali B, Kulkarni RS, et al. Greener synthesis of indolizine analogues using water as a base and solvent: study for larvicidal activity against Anopheles arabiensis. Chem Biol Drug Des. 2016;88(6):899–904. 10.1111/cbdd.12823 [DOI] [PubMed] [Google Scholar]
- 26.Smith SC, Clarke ED, Ridley SM, Bartlett D, Greenhow DT, Glithro H, et al. Herbicidal indolizine-5,8-diones: photosystem I redox mediators. Pest Manag Sci. 2005;61(1):16–24. Epub 2004/12/14. 10.1002/ps.980 . [DOI] [PubMed] [Google Scholar]
- 27.Sandeep C, Venugopala KN, Nayak SK, M. Gleiser R, García DA, Kumalo HM, et al. One-pot microwave assisted synthesis and structural elucidation of novel ethyl 3-substituted-7-methylindolizine-1-carboxylates with larvicidal activity against Anopheles arabiensis. J Mol Struct. 2018;1156:377–84. 10.1016/j.molstruc.2017.11.131 [DOI] [Google Scholar]
- 28.Chandrashekharappa S, Venugopala KN, Tratrat C, Mahomoodally FM, Aldhubiab BE, Haroun M, et al. Efficient synthesis and characterization of novel indolizines: exploration of in vitro COX-2 inhibitory activity and molecular modelling studies. New J Chem. 2018;42(7):4893–901. 10.1039/C7NJ05010K [DOI] [Google Scholar]
- 29.Narayanaswamy VK, Albericio F, Coovadia YM, Kruger HG, Maguire GE, Pillay M, et al. Total synthesis of a depsidomycin analogue by convergent solid-phase peptide synthesis and macrolactonization strategy for antitubercular activity. J Pept Sci. 2011;17(10):683–9. Epub 2011/07/19. 10.1002/psc.1389 . [DOI] [PubMed] [Google Scholar]
- 30.Venugopala KN, Nayak SK, Pillay M, Prasanna R, Coovadia YM, Odhav B. Synthesis and antitubercular activity of 2-(substituted phenyl/benzyl-amino)-6-(4-chlorophenyl)-5-(methoxycarbonyl)-4-methyl-3,6-dihydropyrimidin-1-ium chlorides. Chem Biol Drug Des. 2013;81(2):219–27. 10.1111/cbdd.12065 [DOI] [PubMed] [Google Scholar]
- 31.Venugopala KN, Dharma Rao GB, Bhandary S, Pillay M, Chopra D, Aldhubiab BE, et al. Design, synthesis, and characterization of (1-(4-aryl)- 1H-1,2,3-triazol-4-yl)methyl, substituted phenyl-6-methyl-2-oxo-1,2,3,4-tetrahydropyrimidine-5-carboxylates against Mycobacterium tuberculosis. Drug Des Devel Ther. 2016;10:2681–90. 10.2147/DDDT.S109760 PMC5003518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Venugopala KN, Sandeep C, Pillay M, Bhandary S, Kandeel M, Mahomoodally FM, et al. Synthesis and structural elucidation of novel benzothiazole derivatives as anti-tubercular agents: In-silico screening for possible target identification. Med Chem. 2018;15(3):311–326. Epub 2018/07/04. 10.2174/1573406414666180703121815 . [DOI] [PubMed] [Google Scholar]
- 33.Apex2, Version 2 User Manual, M86-E01078, Bruker Analytical X-ray Systems Madison, WI. 2006. [Google Scholar]
- 34.Siemens, SMART System, Siemens Analytical X-ray Instruments Inc; Madison, MI: 1995. [Google Scholar]
- 35.Sheldrick GM. SADABS; Bruker AXS, Inc.: Madison, WI: 2007. [Google Scholar]
- 36.Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64:112–22. 10.1107/S0108767307043930 [DOI] [PubMed] [Google Scholar]
- 37.Sheldrick G. Crystal structure refinement with SHELXL. Acta Cryst C. 2015;71:3–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Farrugia L. WinGX suite for small-molecule single-crystal crystallography. J Appl Crystallogr. 1999;32(4):837–8. 10.1107/S0021889899006020 [DOI] [Google Scholar]
- 39.Spek A. Single-crystal structure validation with the program PLATON. J Appl Crystallogr. 2003;36(1):7–13. 10.1107/S0021889802022112 [DOI] [Google Scholar]
- 40.Macrae CF, Bruno IJ, Chisholm JA, Edgington PR, McCabe P, Pidcock E, et al. Mercury CSD 2.0—new features for the visualization and investigation of crystal structures. J Appl Crystallogr. 2008;41(2):466–70. 10.1107/S0021889807067908 [DOI] [Google Scholar]
- 41.Frisch MJ, Trucks GW, Schlegel HB, Scuseria G.E., et al. Gaussian 09, Revision B.1, Gaussian Inc, Wallingford, CT: 2009. [Google Scholar]
- 42.Zhang R, Lv K, Wang B, Li L, Wang B, Liu M, et al. Design, synthesis and antitubercular evaluation of benzothiazinones containing an oximido or amino nitrogen heterocycle moiety. RSC Advances. 2017;7(3):1480–3. 10.1039/C6RA25712G [DOI] [Google Scholar]
- 43.Case DA, Berryman JT, Betz RM, Cerutti DS, Cheatham TE III; Darden TA, et al. AMBER 2015. University of California: San Francisco, CA, USA: 2015. [Google Scholar]
- 44.Martin A, Morcillo N, Lemus D, Montoro E, Telles MA, Simboli N, et al. Multicenter study of MTT and resazurin assays for testing susceptibility to first-line anti-tuberculosis drugs. Int J Tuberc Lung Dis. 2005;9(8):901–6. [PubMed] [Google Scholar]
- 45.Yoshikuni O, Mayumi T, Kenichi S. Inhibitory activity of quinolones against DNA gyrase of Mycobacterium tuberculosis. J Antimicrob Chemother. 2001;47:447–50. 10.1093/jac/47.4.447 [DOI] [PubMed] [Google Scholar]
- 46.Middlebrook G, Reggiards Z, Tigertt WD. Automable radiometric detection of growth of Mycobacterium tuberculosis in selective media. Am Rev Respir Dis. 1977;115:1067–9. [DOI] [PubMed] [Google Scholar]
- 47.Mossman T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.