Abstract
Additional variant interpretation tools are required to effectively harness genomic sequencing for clinical applications. The American College of Medical Genetics and Genomics (ACMG) and Association for Molecular Pathology (AMP) published guidelines for clinical sequence variant interpretation, incorporating different types of data that lend varying levels of support towards a benign or pathogenic interpretation. Variants of uncertain significance (VUS) are those with either contradictory or insufficient evidence, and their uncertainty complicates patient counseling and management. Functional assays may provide a solution to evidence gaps relegating variants to the VUS category, but the impact of functional evidence in this framework has not been assessed. We employ an algorithmic analysis of the ACMG/AMP combining rules to assess how the availability of strong functional evidence could theoretically improve the ability to make a benign or pathogenic assertion. We follow this with analysis of actual evidence combinations met by variants through expert curations as part of the Clinical Genome Resource (ClinGen). We also examine the impact of functional evidence in a Bayesian adaptation of the ACMG/AMP framework. This lays the groundwork for an evidence-based prioritization of assay development and variant assessment by identifying genes and variants that may benefit the most from functional data.
Keywords: functional assay, Bayesian, VUS, variant interpretation, ACMG/AMP guidelines
INTRODUCTION
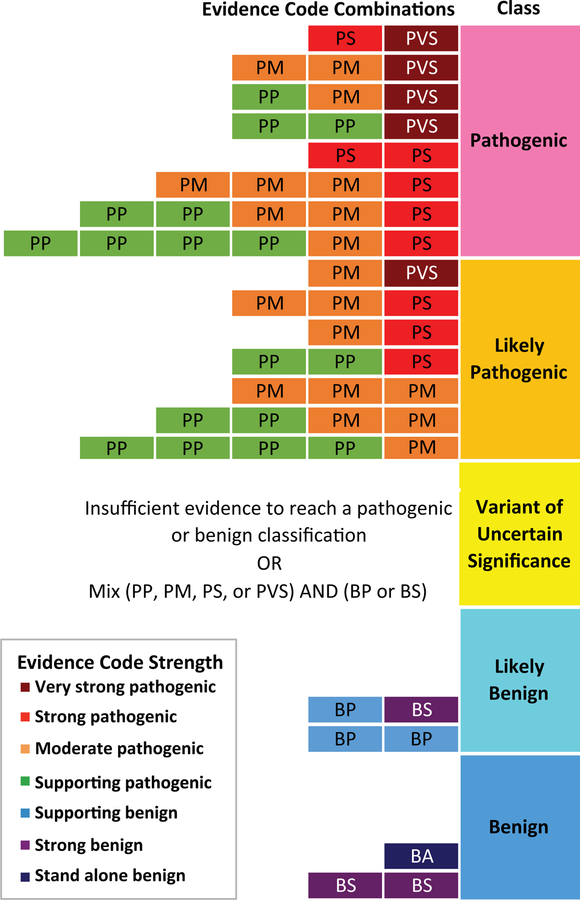
Genomic sequencing is primed to advance screening, diagnosis, and management of disease for improved patient care (Green & Guyer, 2011). To achieve these goals, additional tools are required to understand the clinical significance of human genetic variation. Professional organizations, including the American College of Medical Genetics and Genomics (ACMG) and Association for Molecular Pathology (AMP), have developed variant interpretation guidelines to aid in this process (Richards et al., 2015). Variant interpretation involves an examination of patient phenotype, population data, and an appraisal of the literature for each variant (Evans, Powell, & Berg, 2017). The ACMG/AMP guidelines assign a code to each piece of evidence by type (e.g. population, computational prediction, functional, or segregation data) and strength: benign supporting (BP1–6), strong (BS1–4), or stand-alone (BA1), or pathogenic supporting (PP1–5), moderate (PM1–6), strong (PS1–4), or very strong (PVS1). Then, evidence codes are evaluated in combination using a categorical system to assign a variant’s final interpretation along a 5-tiered spectrum from benign to pathogenic (Richards et al., 2015), as represented in Figure 1. The “variant of uncertain significance” (VUS) classification is applied in two situations: variants with conflicting evidence, or variants with insufficient evidence, as is often the case for novel missense variants (Richards et al., 2015; Starita et al., 2017).
Figure 1. ACMG/AMP categorical combining rules for variant classification.
Possible evidence combinations are represented alongside the corresponding variant classification according to Richards et al. 2015 combining rules. The first letter of each evidence code indicates support towards a pathogenic (P) or benign (B) classification and the second letter indicates the assigned evidence strength: supporting (P), moderate (M), strong (S), very strong (VS), or stand-alone (A). Evidence code boxes are colored by evidence strength. Final classifications are determined by combinations of evidence codes resulting in the following: pathogenic, likely pathogenic, variant of uncertain significance, likely benign, or benign.
Increased application of genetic testing has identified millions of individual variants, but lacking sufficient evidence for a definitive classification, the majority of these remain in the category of VUS (Evans et al., 2017; Starita et al., 2017). Clinical practice guidelines caution against using VUS for risk assessment and decision-making, which complicates patient counseling and management (Domchek, Bradbury, Garber, Offit, & Robson, 2013; Gaba et al., 2016; Richards et al., 2015; Stadler, Schrader, Vijai, Robson, & Offit, 2014; Starita et al., 2017).
Data from well-established in vivo or in vitro functional studies are considered strong evidence in the ACMG/AMP framework (Richards et al., 2015). Functional assays can probe a genetic variant’s effect on the gene or protein product, even in the absence of available patient or segregation data, providing an attractive approach to reclassify VUS (Couch, 2008; Goldgar et al., 2008; Millot et al., 2012). However, the impact of functional data on variant classification within the ACMG/AMP interpretation framework has not been assessed.
The Clinical Genome Resource (ClinGen) consortium (Rehm et al., 2015) established a Sequence Variant Interpretation (SVI) Work Group to refine and enhance the ACMG/AMP guidelines, providing additional recommendations for specific criteria, supporting gene and disease-specific adaptations through Expert Panel groups, and developing quantitative approaches to variant interpretation. The categorical 2015 ACMG/AMP guidelines for clinical sequence variant interpretation (Richards et al.) were recently adapted to a quantitative, Bayesian framework described by Tavtigian and colleagues (2018). Bayesian analysis calculates how the probability of a hypothesis (e.g. pathogenicity) changes with the addition of evidence from multiple sources (Ogino & Wilson, 2004), which can be utilized in variant classification schemes based on data-driven estimates of weights for each piece of evidence.
In this manuscript, we first used an algorithmic approach to identify the theoretical contribution of well-validated functional assays towards reclassification of missense VUS globally. We examined the categorical ACMG/AMP rule combinations (Richards et al., 2015) and assessed how the availability of strong functional evidence could theoretically improve the ability to make a benign or pathogenic assertion. Next, we examined the potential for VUS reclassification in the recent Bayesian adaptation of the ACMG/AMP guidelines (Tavtigian et al., 2018). We followed this with analysis of actual evidence combinations met by missense variants curated through ClinGen’s Inherited Cardiomyopathy Expert Panel (CMP-EP) curation of MYH7-associated cardiomyopathies and RASopathies Expert Panel (RAS-EP) curations of variants in ten established RASopathy-associated genes. These variants served as a case study of functional assays in the context of actual evidence combinations applied in expert variant curation. Importantly, these analyses clarify the utility of functional evidence for clinical variant interpretation in both the categorical and quantitative frameworks. Finally, this analysis lays the groundwork for identifying genes and variants that stand to benefit the most from functional data and thus an evidence-based method for prioritizing assay development and validation.
METHODS
Applicable Rules by Variant Type
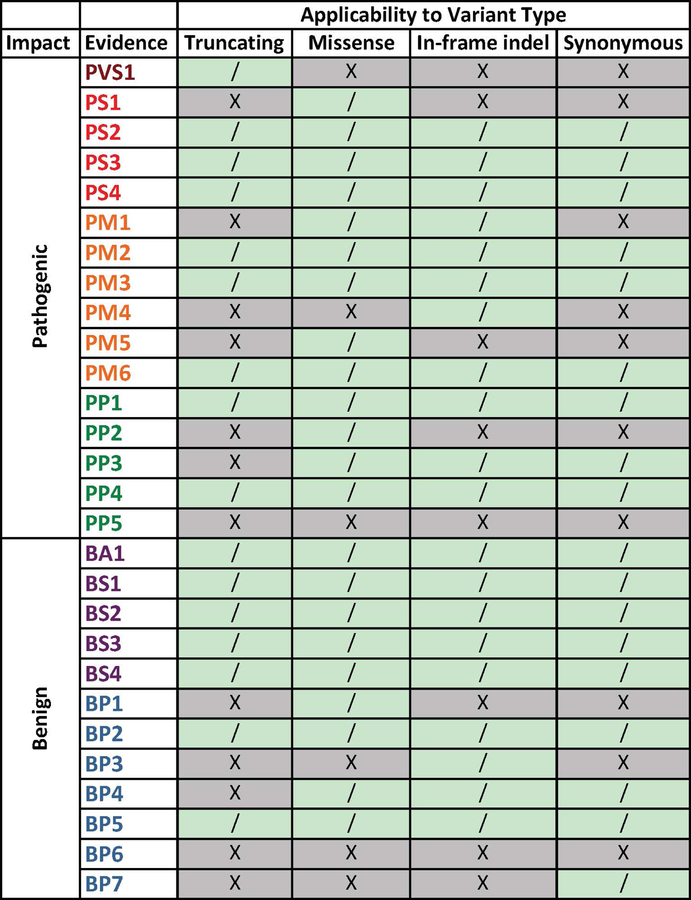
We analyzed the ACMG/AMP variant interpretation guidelines to determine the evidence codes applicable to variants grouped by molecular consequence (missense, truncation, in-frame insertion/deletion, or synonymous) (Figure 2), in order to focus on the typical scenario of a rare missense VUS. PVS1 applies only to loss-of-function or truncating variants, PM4 refers to variants that affect protein length, BP3 is specific to in-frame insertions or deletions, and BP7 is specific to synonymous variants (Richards et al., 2015). We also excluded PP5 and BP6 (reputable source reports) for all variant types, following the recent recommendation from the ClinGen Sequence Variant Interpretation (SVI) Working Group to discontinue their use (Biesecker & Harrison, 2018).
Figure 2. ACMG/AMP evidence codes for classifying sequence variants as applicable to variant molecular consequence.
The first letter of each evidence code indicates support towards a pathogenic (P) or benign (B) variant classification, and is followed by an indicated weight of evidence: supporting (P), moderate (M), strong (S), very strong (VS) or stand-alone (A). See Richards et al. 2015 for a complete description of each code. Variant type refers to the molecular consequence (truncating, missense, in-frame indel, synonymous). If evidence applies to the specific variant type, it is indicated with a “/”, while evidence that does not apply is indicated with an “X”.
Algorithm for Combination Rule Assessment
We wrote a Python script that iteratively calculates the resulting variant interpretation from combinations of satisfied evidence codes, using the ACMG/AMP combining rules outlined by Richards and colleagues (2015). Richards et al. described 30 criteria for variant classification, which includes 28 uniquely named evidence codes and two that account for the ability to increase the strength of pathogenic segregation evidence (PP1) to moderate or strong. For this purpose, we assumed that each piece of evidence considered was independent and met or not met (coded as one or zero). We set up the guidelines as a permutation problem, defining all possible combinations of evidence as: Comball = 230 = 1,073,741,824.
To create all possible permutations, we used the itertools package product function for Python. Then, each combination was run through a filter to exclude rule combinations that exceeded a maximum of one rule in each of the following categories: population or allele frequency (BA1, BS1, BS2, PS4, PM2), segregation (PP1_Strong, PP1_Moderate, PP1), and de novo variant (PS3 or PM6). Additionally, we grouped inverse rules such that only one of each type could be met (i.e. functional—PS3 or BS3; computational—PP3 or BP4; segregation—PP1_Strong/PP1_Moderate/PP1 or BS4; allelic—PM3 or BP2).
Next, we evaluated and filtered each combination by its applicability to variant type (truncating, synonymous, missense, in-frame insertion/deletion) according to Figure 2. For example, combinations including PVS1 (truncating variant in genes where loss of function is a known mechanism of disease) are binned as “truncating” only, while combinations with criteria that are not specific to a particular molecular consequence were included in all four variant type classes.
As the 2015 ACMG/AMP guidelines (Richards et al., 2015) do not provide a method to resolve conflicting evidence, combinations with both benign and pathogenic evidence were excluded from the initial analysis based on the combining rules. Non-conflicting evidence combinations were then assigned a final classification (pathogenic, likely pathogenic, VUS, likely benign, or benign) using the combining criteria described in Table 5 of Richards et al. (2015).
Algorithmic Analysis of Functional Evidence Impact
We then used this tool to accomplish an analysis of the theoretical utility of a well-validated functional assay, if it were to exist and if the variant were to demonstrate either a damaging effect or no effect on the gene or gene product (PS3 or BS3 in Figure 1 of Richards et al., 2015). This script generated combinations of the ACMG/AMP criteria for missense variants resulting in a VUS interpretation due to insufficient, not contradictory, evidence. We then examined which of these possible evidence combinations were missing functional criteria (PS3/BS3) and how adding this type of strong evidence would affect the ability to assert a benign or pathogenic interpretation. We modified the combinations to include PS3 or BS3 and reanalyzed them using the Richards et al. combining criteria (2015) to obtain a variant classification. We counted the number of combination scenarios that moved from VUS to other categories.
Bayesian Adaptation of the Combining Rules Algorithm
We next asked how this theoretical permutation analysis would change when adapted to the quantitative Bayesian framework recently described by Tavtigian and colleagues (2018). We modified the previous script to include contradictory evidence combinations, as the Bayesian framework provides a method for considering both pathogenic and benign criteria simultaneously. For each evidence combination previously described, we tallied the included evidence by weighted pathogenicity: supporting, moderate, strong, or very strong for pathogenic; and supporting or strong for benign. We did not analyze any combinations including stand-alone benign evidence, as meeting this criterion classifies a variant as benign regardless of other evidence, which is contrary to Bayesian reasoning (Tavtigian et al., 2018). We used the tallies (N) of each category (PP, PM, PS, PVS, BP, BS) to calculate the combined odds of pathogenicity (OddsPath):
This then factored into the calculation of the posterior probability of pathogenicity (Post_P) for each combination, using a default prior probability (Prior_P) of 0.10 as previously described (Tavtigian et al., 2018):
Each Post_P was then assigned a variant classification as follows: benign <0.001; 0.001 ≤ likely benign < 0.1; 0.1 ≤ VUS < 0.90; 0.90 ≤ likely pathogenic < 0.99; 0.99 ≤ pathogenic. As an example, if the algorithm generated a combination of evidence including PM2, PP3, and BP1, the OddsPath calculation would be:
The Post_P would then be 0.325 and the combination classified as VUS.
As before, we then analyzed the impact of adding strong functional criteria to evidence classifications that did not include PS3 or BS3 criteria, recalculating the Post_P and reassigning a variant classification. Using the previous example, the Post_P would be 0.900 (likely pathogenic) with PS3, or 0.025 (likely benign) with BS3. We then counted the number of combinations that moved from VUS to another class.
Curated Variant Analysis
Recently, two ClinGen Expert Panels (EPs) published recommendations for adapting the ACMG/AMP variant classification framework for specific genes and diseases (Gelb et al., 2018; Kelly et al., 2018). The ClinGen Inherited Cardiomyopathy Expert Panel (CMP-EP) curated MYH7 variants using reference transcripts LRG_384t1 and NM_000257.3 (Kelly et al., 2018). The ClinGen RASopathies Expert Panel (RAS-EP) curated variants in nine genes using the indicated reference transcripts: BRAF (LRG_299t1, NM_004333.4), HRAS (NM_005343.2), KRAS (NM_004985.4, NC_000012.12(NM_004985.4)), MAP2K1 (LRG_725t1, NM_002755.3), MAP2K2 (LRG_750t1, NM_030662.3), PTPN11 (LRG_614t1, NM_002834.3), RAF1 (LRG_413t1, NM_002880.3), SHOC2 (LRG_753t1, NM_007373.3), and SOS1 (NM_005633.3) (Gelb et al., 2018). Both groups submitted variants to ClinVar as 3-star expert panel-reviewed interpretations.
We examined their published variant curations, including the ACMG/AMP evidence codes satisfied for each variant, to determine the role of strong functional evidence in the application of the ACMG/AMP combining rules for a final variant classification. For each variant, we extracted the evidence codes applied in expert curation from the respective publications and inserted them in a tabular file for analysis.
We asked how many curated variant interpretations included evidence from a well-validated functional assay demonstrating a damaging effect (PS3) or no effect (BS3) on the gene or gene product, and how this evidence affected the overall classification. We sorted variants by satisfaction of PS3 or BS3 criteria and then grouped them by expert panel-assigned classification. We modeled the impact of strong functional evidence (if it were to become available) on variant reclassification by tabulating the curated evidence plus BS3 or PS3 criteria using the combining rules approach described in Table 5 of Richards et al. (2015).
In addition, we tallied the evidence for and against pathogenicity for each variant according to strength and entered this into the recent Bayesian adapted guidelines (Tavtigian et al., 2018). We again modeled how the addition of PS3 and BS3 criteria affected variant classification by recalculating the Post_P in this quantitative system.
RESULTS
Algorithmic Analysis of Theoretical Missense Evidence Combinations
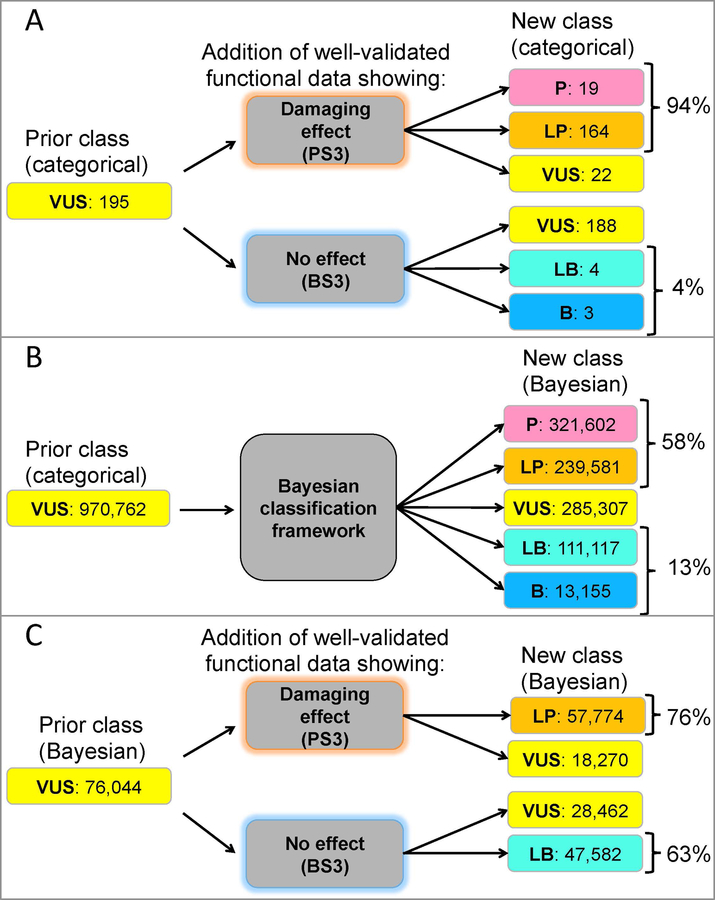
We first used our categorical algorithm to identify the theoretical contribution of well-validated functional assays toward reclassification of missense VUS globally. The categorical rules called 970,762 missense-applicable evidence combinations “VUS”. Of these, 195 missense evidence combinations returned a VUS classification due to limited, not conflicting, evidence and did not include “strong” pathogenic or benign functional evidence codes (PS3 or BS3). We found that 97% could be reclassified with the addition of strong functional data according to the categorical ACMG/AMP combining rules, suggesting that such assays could substantially improve missense VUS interpretations (Figure 3A). Interestingly, this analysis found that satisfaction of the PS3 criterion would be able to reclassify more non-conflicting missense VUS in the categorical framework than the BS3 criterion.
Figure 3. Reclassifying missense VUS evidence combinations in the categorical ACMG/AMP framework and the adapted Bayesian framework.
(A) Modeling the addition of well-validated functional evidence showing a damaging effect (PS3) or no effect (BS3) to non-conflicting missense variant evidence combinations in the categorical ACMG/AMP framework described by Richards et al. (2015) finds 97% of combinations would be reclassified with functional data. (B) Theoretical conflicting and non-conflicting missense evidence combinations resulting in a VUS classification in the categorical ACMG/AMP combining rules (Prior class categorical) were entered into the Bayesian adaptation described by Tavtigian et al. (2018) and the corresponding variant classifications are shown (New class Bayesian). (C) From the theoretical missense evidence combinations resulting in a VUS classification in (B), we examined those that did not include PS3 or BS3 criteria (Prior Class Bayesian). Modeling the addition of well-validated functional evidence (BS3 or PS3) reclassified all combinations (New class Bayesian) by posterior probability calculations in the Bayesian framework described by Tavtigian et al. (2018). ACMG/AMP, American College of Medical Genetics and Genomics/Association for Molecular Pathology; B, benign; BS3, well-established functional studies show no deleterious effect; LB, likely benign; LP, likely pathogenic; P, pathogenic; PS3, well established functional studies show a deleterious effect; VUS, variant of uncertain significance
In the categorical framework, adding evidence (of any type) to conflicting VUS does not change variant interpretation, as there are no specifications for how much evidence is required to outweigh conflicting criteria (Richards et al., 2015). This can be modeled in the Bayesian quantitative framework, however, so we addressed the issue of conflicting evidence in the quantitative adaptation of our Python tool, which contains a formula for calculating the posterior probability of pathogenicity if both pathogenic and benign evidence apply (Tavtigian et al., 2018). Even without additional evidence, when considering both conflicting and non-conflicting missense VUS evidence combinations in the Bayesian-adapted framework, less than 30% remained VUS (Figure 3B). In other words, the quantitative system reclassified about 70% of all missense VUS evidence combinations, likely due to its ability to evaluate a mix of benign and pathogenic evidence. Of the remaining missense VUS combinations that did not include PS3 or BS3, adding PS3 reclassified 76%, while adding BS3 reclassified 63% (Figure 3C). 100% of VUS could be reclassified as likely pathogenic or likely benign in the quantitative framework if the functional assay results are in the same direction as the initial probability of pathogenicity.
While this approach reveals the theoretical combinations of evidence that would be amenable to reclassification due to additional functional evidence, it does not evaluate gene- or disease-specific evidence, or indicate the fraction of actual variants that meet those combinations. As such, we followed this theoretical analysis with an examination of publicly available, expertly curated sequence variants for MYH7-associated cardiomyopathies and RASopathies.
Functional Assay Utility in Expert Panel Curation
ClinGen’s RAS-EP and CMP-EP curated evidence for a total of 163 variants in 10 different genes. Of these 163 variants, only 39 met PS3 criteria and expert panels assigned a final classification of pathogenic to all 39. Only one variant (PTPN11 p.Leu560Phe) met BS3 criteria, which in combination with one piece of supporting benign evidence was sufficient to push the final classification from VUS to likely benign (Supp.Table S1). It is more difficult to experimentally demonstrate that a variant has no effect on the gene or gene product, as few assays capture all relevant protein functions. Thus, it is appropriate that only one variant met the BS3 criterion.
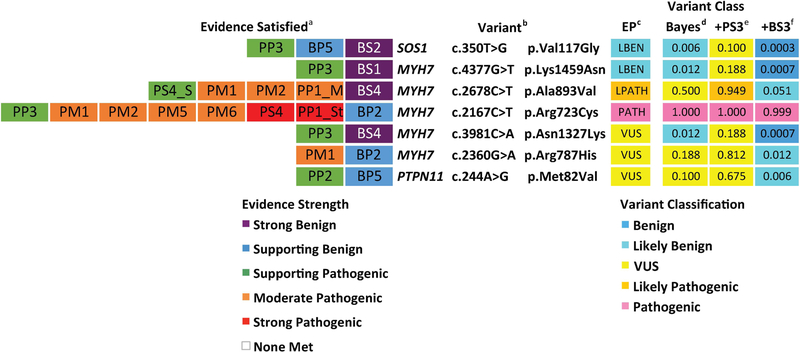
Of 137 missense variants curated, expert panels classified 20 as VUS across six genes (MYH7, HRAS, MAP2K2, PTPN11, RAF1, and SHOC2) and none of these VUS met strong, functional evidence criteria (PS3/BS3). We modeled how adding either PS3 or BS3 criteria would affect the final variant classification, first using the categorical ACMG/AMP combining rules (Richards et al., 2015) and then the quantitative system. Eight non-conflicting VUS reached a likely pathogenic classification with the addition of PS3, and two reached a likely benign classification with the addition of BS3 (Figure 4). The addition of strong functional evidence did not affect the classification of seven non-conflicting VUS, as they either met only one supporting pathogenic criteria or none at all. In all, 10 out of 17 non-conflicting VUS (~59%) would be resolved with strong functional evidence using the categorical combining rules. It should be noted that this number does not include 3 missense variants that were VUS due to conflicting evidence.
Figure 4. Non-conflicting missense variants classified as VUS by expert panels.
aEvidence Satisfied, as determined by expert panels in Gelb et al. (2018) and Kelly et al. (2018), where abbreviations indicate specific criteria described in Richards et al. (2015). Boxes are colored according to evidence strength. bVariant listed by gene, cDNA, and protein. Variants were curated by expert panels using the following reference transcripts: HRAS (NM_005343.2), MAP2K2 (NM_030662.3), RAF1 (NM_002880.3), SHOC2 (NM_007373.3), and MYH7 (NM_000257.3). c Variant class assessed according to categorical ACMG/AMP combining rules (Table 5 in Richards et al., 2015), with Expert Panel (EP) variant classifications considering only evidence satisfied in (a). dVariant class with the addition of either PS3 or BS3 criteria in the categorical interpretation framework (Table 5 in Richards et al.). eVariant class in Bayesian-adapted framework was determined by posterior probability of pathogenicity (Post_P). Post_P was calculated by entering evidence totals by strength in Supplemental Table 1 from Tavtigian et al. (2018), using the default settings: Prior Probability (Prior_P) = 0.100; Odds Path Very Strong = 350; Exponential progression (X) = 2.000. fThese column shows the Post_P value if a strong pathogenic criteria (PS3) or strong benign criteria (BS3) were to be added within the quantitative framework as described in (e). Box colors in (c-f) are coded by variant classification. BS3, well-established functional studies show no deleterious effect; EP, expert panel; LBEN, likely benign; LPATH, likely pathogenic; N/A, none applied; PS3, well established functional studies show a deleterious effect; VUS, variant of uncertain significance
In the Bayesian framework, two non-conflicting missense VUS (MAP2K2 p.Val262Ile and RAF1 p.Val312Ala) reached a likely benign classification simply based on the evidence applied by expert panels (one supporting benign, BP4, in both cases). Modeling the addition of PS3, eight variants reached a likely pathogenic classification, consistent with the categorical assessment. Adding BS3 in the quantitative framework, 11 out of 17 non-conflicting missense VUS reached a likely benign classification, including the two reclassified before adding functional criteria. In sum, 100% of the variants reached a different interpretation in at least one scenario (with either BS3 or PS3).
Using the Bayesian adapted framework, we were also able to model how conflicting variant interpretations might change with the addition of strong functional evidence. We found that for conflicting missense variants with Post_P values from 0.100 to 0.500 (interpreted as VUS in the Bayesian adaptation), adding BS3 reclassified them as likely benign, while the reciprocal (adding PS3) only returned likely pathogenic for one variant (Figure 5).
Figure 5. Expert-curated missense variants with conflicting evidence.
aEvidence Satisfied, as determined by expert panels in Gelb et al. (2018) and Kelly et al. (2018), where abbreviations indicate specific criteria described in Richards et al. (2015). Boxes are colored according to evidence strength. bVariant listed by gene, cDNA, and protein. Variants were curated by expert panels using the following reference transcripts: SOS1 (NM_005633.3), PTPN11 (NM_002834.3), and MYH7 (NM_000257.3). cExpert Panel (EP) Class as determined by CMP-EP or RAS-EP using modified categorical ACMG/AMP combining rules. dVariant class in Bayesian-adapted framework was determined by posterior probability of pathogenicity (Post_P). Post_P was calculated by entering evidence totals by strength in Supplemental Table 1 from Tavtigian et al. (2018), using the default settings: Prior Probability (Prior_P) = 0.100; Odds Path Very Strong = 350; Exponential progression (X) = 2.000. e,fThese column shows the Post_P value if a strong pathogenic criteria (e) or strong benign criteria (f) were to be added within the quantitative framework as described in (d). Box colors in (c-f) are coded by variant classification. BS3, well-established functional studies show no deleterious effect; EP, expert panel; LBEN, likely benign; LPATH, likely pathogenic; N/A, none applied; PATH, pathogenic; PS3, well established functional studies show a deleterious effect; VUS, variant of uncertain significance
Expert panel and quantitative approaches to conflicting evidence
Expert panels documented evidence in both pathogenic and benign categories for seven missense variants, with five in MYH7 and one each in SOS1 and PTPN11 (Figure 5). Of these seven conflicting variants, the expert panels interpreted two as likely benign, one as likely pathogenic, one as pathogenic, and three as VUS. The quantitative Bayesian framework is congruent with these classifications with two exceptions. First, the CMP-EP interpreted MYH7 p.Ala893Val as likely pathogenic, but the combined evidence in the quantitative framework only reached a Post_P of 0.5, and would be considered a VUS. Second, MYH7 variant p.Asn1327Lys was interpreted as VUS by the CMP-EP, but reached a Post_P of 0.012 and would be considered likely benign in the Bayesian adaptation of the combining rules. Interestingly, another MYH7 variant, p.Lys1459Asn, also met one supporting pathogenic (PP3) and one strong benign (BS1 instead of BS4) and reached a Post_P of 0.012. In this instance, the CMP-EP interpreted it as likely benign, indicating that they permitted BS1 to outweigh PP3 (thus over-riding the “conflicting evidence” guidance in the ACMG/AMP combining rules). Overall, expert panels determined a final, clinically significant variant interpretation for most variants with mixed evidence, and the quantitative approach to conflicting evidence generally agreed with these classifications. Interestingly, we also found that variant MYH7 p.Arg723Cys met pathogenic criteria regardless, indicating that with a Post_P of 1.000, even strongly weighted benign evidence would not be sufficient to alter its classification.
DISCUSSION
The Untapped Potential of Functional Evidence
Functional assays are powerful tools for clinical variant interpretation as demonstrated here. Functional data would reclassify 97% of non-conflicting missense VUS evidence combinations globally in the categorical framework (Figure 3A). Interestingly, 94% of those combinations would be reclassified if the data supports a pathogenic classification (PS3), while only 4% would be reclassified if the data supports a benign classification (BS3). In the quantitative evidence framework analysis, which also included conflicting missense VUS combinations, PS3 would reclassify 76% and BS3 would reclassify 63% of theoretical evidence combinations (Figure 3C).
We found that of the missense variants in the ten genes curated by expert panels, 17 non-conflicting VUS met nine unique combinations of evidence. Six unique combinations met by a total of ten variants (59%) could be reclassified with strong functional evidence in the categorical framework, while 100% could be reclassified in the quantitative analysis. Additionally, all non-conflicting missense variants deemed likely benign by expert panels would be benign with the addition of BS3, while the majority of variants deemed likely pathogenic would be reclassified as pathogenic with the addition of PS3 (Supp. Table S2). The study of real world genetic variants was limited by the number of variants with publicly available clinical interpretations that include the ACMG/AMP evidence codes used in curation. As such, we were unable to perform a gene-by-gene analysis, which may ultimately reveal gene-specific characteristics relevant to functional assay utility. Even so, this preliminary study does support the assertion that strong functional data has great potential to resolve variants relegated to VUS classification due to insufficient evidence (typically rare, missense variants).
Bayesian quantitative analysis is important to resolve conflicting VUS
No standardized method exists within the current ACMG/AMP variant interpretation guidelines to classify variants meeting “conflicting” benign and pathogenic evidence as anything other than VUS (Richards et al., 2015). Further, it is unclear what amount of evidence should be considered “conflicting” on either the pathogenic or the benign side. Meeting one strong pathogenic criterion (e.g. PS3) and one supporting benign criterion (e.g. BP4) is not the same as a variant meeting two strong, four moderate, and one supporting pathogenic criteria in “conflict” with only one supporting benign criterion (e.g. MYH7 p.Arg723Cys, Figure 5). There is lack of consensus on this issue, with different groups approaching conflicting evidence in their own ways. One clinical laboratory developed a hierarchical approach to conflicting variants that places more weight on clinical evidence than functional evidence (Nykamp et al., 2017). An examination of the conflicting missense variants in Figure 5 identifies inconsistencies in consideration of evidence strength for an overall classification, even at the level of disease experts. As previously noted, the CMP-EP weighed BS1 criteria more strongly than BS4 criteria, though Richards et al. (2015) and Tavtigian et al. (2018) treat them equally.
The recently described Bayesian framework provides a standardized, quantitative method for addressing these issues (Tavtigian et al., 2018). Evidence of the same type and strength (e.g. PS3 and PS4) are weighted equally, and a formula is provided that permits inclusion of evidence on both ends of the classification spectrum. As demonstrated in Figure 5, unlike the categorical framework, the Bayesian adaptation can model variant classification in the presence of both benign and pathogenic evidence. This has the potential to reclassify variants restricted to the VUS category based on “conflicting” evidence. Another useful application of the quantitative approach is identification of the types or strengths of evidence required to reach a particular classification.
Quantifying the categorical combining rules: emerging inconsistencies
We entered each of the criteria met by the EP-curated non-conflicting missense variants into the Bayesian framework using the supplemental table in Tavtigian et al. (2018) and compared the resulting classifications to the EP class. While mostly consistent, we identified two instances that demonstrate the utility of comparison between the categorical and quantitative methods for combining evidence. First, two missense variants reached a likely benign classification using the Bayesian combining rules based on just one supporting benign piece of evidence (BP4 in both cases), while the EP interpreted them as VUS. This might reflect an inconsistency in the Bayesian adaptation of the ACMG/AMP combining rules, as Richards et al. (2015) requires at least two supporting benign criteria for a “likely benign” classification. It is possible that the prior probability assumption (Prior_P) or odds of pathogenicity (Odds_Path) may need to be adjusted so that one supporting benign criterion returns a posterior probability in the range of VUS. Similarly, two variants in RAF1 (p.Ser257Pro and p.Val263Gly) were interpreted as likely pathogenic by the RAS-EP, in agreement with the categorical combining rules. However, calculating the Post_P demonstrates that two supporting and four moderate pathogenic criteria would be nearly sufficient to meet the pathogenic threshold at 0.994. This represents another inconsistency of the ACMG/AMP combining rules when adapted to the Bayesian model, suggesting that the ClinGen SVI should further evaluate combinations of evidence outside of those explicitly outlined by Richards et al. (2015) for future guideline revisions and recommendations.
Challenges of assigning evidence strength to functional data
For reproducible variant interpretations, evidence must be applied consistently. Questions remain about how functional assays should qualify as “well-established” in the interpretation of clinical variants, resulting in inconsistent application of the PS3/BS3 criteria (Harrison et al., 2017). Richards et al. (2015) specify that assays validated in a Clinical Laboratory Improvement Amendments (CLIA) certified laboratory are the most well established. However, such validation does not necessarily indicate that the level of evidence supporting a pathogenic or benign interpretation is “strong” in the Bayesian quantitative sense for each assay or each variant. In addition, should equally compelling evidence from non-CLIA certified labs carry less weight? Further, as it is more difficult to appropriately apply the BS3 criterion in variant interpretation, could curators downgrade the strength of BS3 for less comprehensive studies demonstrating no effect on the gene or gene product? A complete evaluation of a variant’s functional impact may require results from multiple experimental assays (e.g. splicing, protein stability, enzyme activity), though guidelines for integrating these data are vague. The ACMG/AMP guidelines outline strength stratifications for increasing evidence weight with segregation data (PP1), but do not include moderate strength or very strong benign criteria, so modifying BS3 strength poses challenges in the current categorical framework (Richards et al., 2015). One consideration is that the categorical combining rules cannot be applied as written for moderate benign evidence, though by the Bayesian logic outlined by Tavtigian et al. (2018), moderate evidence is equivalent to the combined weight of two supporting criteria.
Robust validation requires benchmarking results with variants of known clinical significance in order to determine the assay’s sensitivity and specificity. Other consortia such as the International Society for Gastrointestinal Hereditary Tumours (InSiGHT), the Evidence-based Network for the Interpretation of Germline Mutant Alleles (ENIGMA), and the International Agency for Research on Cancer (IARC) already established this as a requirement for functional assay application (Couch, 2008; Millot et al., 2012; Thompson et al., 2015). This type of performance benchmarking permits the calculation of likelihood ratios that can then be converted to qualitative strengths (very strong, strong, moderate, or supporting) as described in the categorical ACMG/AMP framework, or directly applied as odds of pathogenicity in the Bayesian framework. This approach also would permit the stratification and consideration of multiple pieces of functional evidence in a single variant’s clinical interpretation, similar to how it is considered in the Sherloc adaptation described by Nykamp and colleagues (2017). The ClinGen SVI should work to address these questions as guidelines evolve and more expert panels adapt the guidelines for specific diseases and genes.
Potential as prioritization method
The analysis presented here lays the groundwork for identifying genes and variants that stand to benefit the most from functional data and thus an evidence-based method for prioritizing assay development and validation. While methods of mass mutagenesis and variant evaluation have advanced substantially, permitting saturated variant assessment for relatively small genes like PPARG (Majithia et al., 2016), the processes involved in developing and validating a high-throughput assay are still costly and time-consuming. As such, an evidence-based method for prioritizing reclassification of the numerous clinically challenging VUS is necessary to best allocate resources and research efforts (Starita et al., 2017). All of the genes examined by the CMP-EP and RAS-EP have at least one in vivo or in vitro assay approved for variant interpretation applications, yet few variants met PS3 or BS3 evidence criteria (Gelb et al., 2018; Kelly et al., 2018). The VUS identified in Figure 4 that would benefit from the addition of strong functional evidence should be prioritized for analysis using these approved methods. Further, as larger and more well annotated datasets become available, mining them for evidence codes applied in variant curation will provide a high-throughput, evidence-based method of prioritizing genes and variants for functional assessment, and will contextualize the importance of experimental evidence for meaningful clinical interpretation.
Finally, there is a strong need for a centralized database of approved functional assays and associated data to support curation efforts as we define what a “well validated” functional assay means. This type of database will be imperative for the next steps in prioritizing the reanalysis and functional assessment of VUS that stand to benefit most. As expert panels and others identify well-validated experimental assays for variant interpretation, they should be submitted to a central repository that links to the ClinGen Variant Curation Interface (https://curation.clinicalgenome.org) and to ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/).
Supplementary Material
ACKNOWLEDGEMENTS
This work is primarily funded through National Human Genome Research Institute grants U01 HG007437, U41 HG009650. S.E.B. is also supported in part by National Institute of General Medical Sciences grants 5T32 GM007092 and 5T32 GM008719-6. S.E.B. is also a recipient of support from the University Cancer Research Fund as an MD/PhD Scholar. J.S.B. is a recipient of the Yang Family Biomedical Scholars Award.
Grant numbers: U01 HG007437, U41 HG009650, 5T32 GM008719-6, 5T32 GM007092
REFERENCES
- Biesecker LG, & Harrison SM (2018). The ACMG/AMP reputable source criteria for the interpretation of sequence variants. Genetics in Medicine 10.1038/gim.2018.42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couch FJ (2008). Assesment of Functional Effects of Unclassified Genetic Variants. Hum Mutat, 29(11), 1314–1326. 10.1002/humu.20899.Assessment [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domchek SM, Bradbury A, Garber JE, Offit K, & Robson ME (2013). Multiplex genetic testing for cancer susceptibility: Out on the high wire without a net? Journal of Clinical Oncology, 31(10), 1267–1270. 10.1200/JCO.2012.46.9403 [DOI] [PubMed] [Google Scholar]
- Evans JP, Powell BC, & Berg JS (2017). Finding the rare pathogenic variants in a human genome. JAMA - Journal of the American Medical Association 10.1001/jama.2017.0432 [DOI] [PubMed] [Google Scholar]
- Gaba P, Martijn Bos J, Cannon BC, Cha YM, Friedman PA, Asirvatham SJ, & Ackerman MJ (2016). Implantable cardioverter-defibrillator explantation for overdiagnosed or overtreated congenital long QT syndrome. Heart Rhythm, 13(4), 879–885. 10.1016/j.hrthm.2015.12.008 [DOI] [PubMed] [Google Scholar]
- Gelb BD, Cavé H, Dillon MW, Gripp KW, Lee JA, Mason-Suares H, … Vincent LM (2018). ClinGen’s RASopathy Expert Panel consensus methods for variant interpretation. Genetics in Medicine 10.1038/gim.2018.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldgar DE, Easton DF, Byrnes GB, Spurdle AB, Iversen ES, & Greenblatt MS (2008). Genetic evidence and integration of various data sources for classifying uncertain variants into a single model. Human Mutation, 29(11), 1265–1272. 10.1002/humu.20897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green ED, & Guyer MS (2011). Charting a course for genomic medicine from base pairs to bedside. Nature, 470(7333), 204–213. 10.1038/nature09764 [DOI] [PubMed] [Google Scholar]
- Harrison SM, Dolinsky JS, Knight Johnson AE, Pesaran T, Azzariti DR, Bale S, … Rehm HL (2017). Clinical laboratories collaborate to resolve differences in variant interpretations submitted to ClinVar. Genetics in Medicine, 19(10), 1096–1104. 10.1038/gim.2017.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly MA, Caleshu C, Morales A, Buchan J, Wolf Z, Harrison SM, … Wilde A (2018). Adaptation and validation of the ACMG/AMP variant classification framework for MYH7-associated inherited cardiomyopathies: Recommendations by ClinGen’s Inherited Cardiomyopathy Expert Panel. Genetics in Medicine, 20(3), 351–359. 10.1038/gim.2017.218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majithia AR, Tsuda B, Agostini M, Gnanapradeepan K, Rice R, Peloso G, … Altshuler D (2016). Prospective functional classification of all possible missense variants in PPARG. Nature Genetics, 48(12), 1570–1575. 10.1038/ng.3700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millot GA, Carvalho MA, Caputo SM, Vreeswijk MPG, Brown MA, Webb M, … Monteiro ANA (2012). A guide for functional analysis of BRCA1 variants of uncertain significance. Human Mutation, 33(11), 1526–1537. 10.1002/humu.22150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nykamp K, Anderson M, Powers M, Garcia J, Herrera B, Ho YY, … Topper S (2017). Sherloc: A comprehensive refinement of the ACMG-AMP variant classification criteria. Genetics in Medicine, 19(10), 1105–1117. 10.1038/gim.2017.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino S, & Wilson RB (2004). Bayesian analysis and risk assessment in genetic counseling and testing. Journal of Molecular Diagnostics, 6(1), 1–9. 10.1016/S1525-1578(10)60484-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehm HL, Berg JS, Brooks LD, Bustamante CD, Evans JP, Landrum MJ, … Watson MS (2015). ClinGen — The Clinical Genome Resource. New England Journal of Medicine, 372(23), 2235–2242. 10.1056/NEJMsr1406261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, … Rehm HL (2015). Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genetics in Medicine, 17(5), 405–423. 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler ZK, Schrader KA, Vijai J, Robson ME, & Offit K (2014). Cancer genomics and inherited risk. Journal of Clinical Oncology, 32(7), 687–698. 10.1200/JCO.2013.49.7271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starita LM, Ahituv N, Dunham MJ, Kitzman JO, Roth FP, Seelig G, … Fowler DM (2017). Variant Interpretation: Functional Assays to the Rescue. American Journal of Human Genetics, 101(3), 315–325. 10.1016/j.ajhg.2017.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavtigian SV, Greenblatt MS, Harrison SM, Nussbaum RL, Prabhu SA, Boucher KM, & Biesecker LG (2018). Modeling the ACMG/AMP variant classification guidelines as a Bayesian classification framework. Genetics in Medicine 10.1038/gim.2017.210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson BA, Spurdle A, Plazzer J-P, Greenblatt MS, Akagi K, Al-Mulla F, … Farrell MP (2015). Application of a five-tiered scheme for standardized classification of 2,360 unique mismatch repair gene variants lodged on the InSiGHT locus-specific database. Nature Genetics, 46(2), 107–115. 10.1038/ng.2854.Application [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.