Phenotypic screening has identified small-molecule modulators of aging, but the mechanism of compound action often remains opaque due to the complexities of mapping protein targets in whole organisms. Here, we combine a library of covalent inhibitors with activity-based protein profiling to coordinately discover bioactive compounds and protein targets that extend lifespan in Caenorhabditis (C.) elegans. We identify JZL184 – an inhibitor of the mammalian endocannabinoid (eCB) hydrolase monoacylglycerol lipase (MAGL or MGLL) – as a potent inducer of longevity, an initially perplexing result as C. elegans does not possess an MAGL orthologue. We instead identify FAAH-4 as a principal target of JZL184 and show that this enzyme, despite lacking homology with MAGL, performs the equivalent metabolic function of degrading eCB-related monoacylglycerides in C. elegans. Small-molecule phenotypic screening thus illuminates pure pharmacological connections marking convergent metabolic functions in distantly related organisms, implicating the FAAH-4/monoacylglyceride pathway as a regulator of lifespan in C. elegans.
Strict temporal and spatial control of metabolic processes is required for cellular homeostasis and organismal development, and dysregulated metabolism results in age-related pathophysiologies, including cancer, diabetes and neurodegeneration1–3. Metabolic enzymes have accordingly emerged as key regulatory nodes and therapeutic targets in aging-related diseases2–4. Nonetheless, our understanding of the metabolic pathways that can impact physiological lifespan remains incomplete, likely reflecting the methodological challenges associated with studying aging in mammals. Model organisms, such as the roundworm Caenorhabditis elegans, provide an attractive alternative to investigate aging due to their short natural lifespan (approximately 2–3 weeks) and conduciveness to phenotypic screening and genetic analysis. Several aging-related genetic screens have been performed in C. elegans, leading to the discovery of fundamental signaling pathways that regulate lifespan and are conserved in humans5–7.
Phenotypic screening has also identified small molecules that extend lifespan in C. elegans8,9, but elucidating the mechanism of action of these compounds and, in particular, their molecular targets, remains problematic. Conventional small-molecule libraries are mostly composed of compounds that reversibly bind to proteins, and preserving these reversible interactions during biochemical target enrichment protocols is challenging, especially for the lower potency hit compounds that typically emerge from phenotypic screens. More recently, electrophilic compound libraries have been introduced that irreversibly bind to proteins10,11, generating stable interactions that can be isolated and characterized from native biological systems. So far, such electrophilic compound collections have mainly been screened for phenotypic effects in cellular systems12,13.
Among the classes of small-molecule electrophiles potentially suitable for more advanced, whole organism screening, compounds targeting serine hydrolases (SH) are particularly attractive, as these chemotypes (e.g., carbamates, activated ureas) have shown robust in vivo-activity in mammals14,15. Additionally, SHs represent one of the largest and most diverse enzyme families in nature that can be near-universally characterized for activity and small-molecule interactions in native biological systems by activity-based protein profiling (ABPP)16,17. Select members of this 200+-member enzyme class, including fatty acid amide hydrolase (faah-1)18, diacylglycerol lipase (dagl-2)19, and lysosomal lipase (lipl-4)20,21 have been found to affect C. elegans lifespan by impinging on the nutrient-sensing, target of rapamycin (TOR), and nuclear hormone pathways. These findings, however, were obtained using focused genetic methods and identified enzymatic pathways for which overexpression (as opposed to inactivation) promoted longevity. Here, we instead describe a global and systematic assessment of the chemical inhibition of SHs and its impact on longevity in C. elegans. From a structurally diverse library of SH-directed electrophilic compounds, we discovered the aryloxy carbamate JZL184 (1) as a robust lifespan-extending compound. Surprisingly, however, C. elegans lacks an orthologue of the principle target of JZL184 in mammals – the endocannabinoid (eCB)-metabolizing enzyme monoacylglycerol lipase (MAGL or MGLL). Using ABPP, we instead identify FAAH-4 as a major target of JZL184 and related lifespan-extending compounds in C. elegans. We proceed to show that FAAH-4, but not other FAAH enzymes in C. elegans, hydrolyzes 2-arachidonoylgycerol (2-AG), the endogenous eCB substrate of mammalian MAGL. We finally demonstrate that genetic disruption of faah-4 elevates 2-AG, extends lifespan, and protects from oxidative stress in C. elegans. These results thus identify FAAH-4 as a primary 2-AG hydrolase that regulates lifespan and stress responsivity in C. elegans. That the shared metabolic function of C. elegans FAAH-4 and mammalian MAGL is reflected in joint sensitivity to an active site-directed irreversible inhibitor, rather than sequence- or fold-relatedness, underscores the value of small-molecule screening in model organisms as a way to discover functionally analogous druggable pathways that regulate complex biological processes like aging.
Results
A global inventory of C. elegans serine hydrolases.
Prior to embarking on a small-molecule phenotypic screen in C. elegans, we first generated a functional map of SHs in this organism using ABPP combined with quantitative mass spectrometry (MS). 14N- (light) or 15N- (heavy) metabolically labeled animals were lysed, the light-labeled sample heat-denatured, and then both samples treated with a biotinylated fluorophosphonate (FP) probe, which reacts with the conserved serine nucleophile in SH active sites10. We identified 92 SHs that were selectively enriched by the FP probe in native (heavy) compared to heat-denatured control (light) proteomes (Fig. 1a, Supplementary Fig. 1a, and Supplementary Dataset 1). This coverage accounted for ~40% of the predicted C. elegans SHs and included members from all phylogenetic clades of the SH family (Fig. 1a). Both high and low expression SHs, as estimated by public RNA-Seq data22, were found in the FP-enriched data set, indicating that the ABPP technology could broadly and deeply survey SH activities in C. elegans (Supplementary Fig. 1a). The predicted SHs that were not detected by ABPP were mostly found in the low expression group (Supplementary Fig. 1a and Supplementary Dataset 1), suggesting that they may be temporally or spatially restricted in C. elegans. We also cannot exclude the possibility that some of the predicted SHs in C. elegans react poorly with the FP probe under the specific conditions employed or do not produce functional enzymes (e.g., pseudogenes or genes that encode for non-enzymatic proteins).
Figure 1. A chemical proteomic map of serine hydrolase (SH) activities and their chemical inhibition in C. elegans.
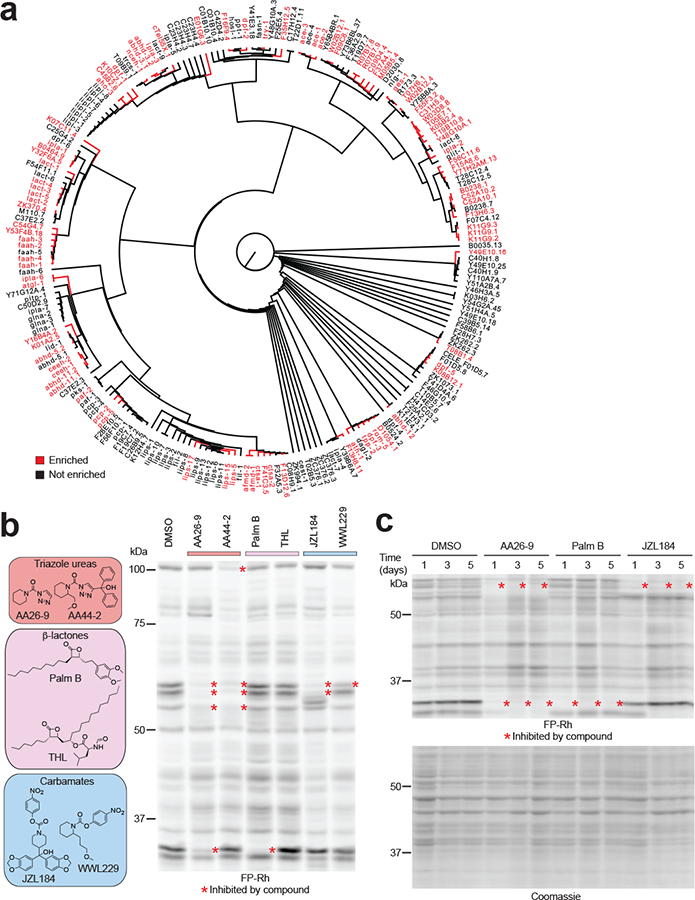
(a) Dendrogram of predicted C. elegans SHs, where blue and black designate enzymes that were enriched or not enriched, respectively, in MS-based ABPP experiments using the SH-directed probe FP-biotin. The dendrogram was constructed by performing a sequence alignment using the ClustalW algorithm, and branch length represents sequence relatedness. (b) Representative classes of SH-directed inhibitors and their activity in C. elegans proteins in vivo as measured by ABPP with FP-rhodamine. Animals were treated with inhibitors (50 μM) for 24 h prior to ABPP. (c) SH targets remain inhibited by test compounds for up to 5 days. C. elegans were dosed with compounds on day 1 of treatment and harvested 1, 3 or 5 days after the initial treatment (soluble fraction) for analysis by ABPP. For b and c, results are representative of 2 independent experiments.
Successful chemical screening in whole organisms requires that compounds engage proteins in vivo, a feature that, for irreversible inhibitors, can be conveniently assessed by competitive ABPP (Supplementary Fig. 1b)17. Considering that our SH-directed small-molecule library, which contains diverse electrophilic chemotypes, including activated ureas23, carbamates15,24, and β-lactones25, has furnished compounds that show good in vivo activity against mammalian SHs, we next asked whether these compounds are also taken up by and engage SHs in C. elegans. We assayed compounds (50 μM, 24 h) in liquid culture with irradiated Op50 bacteria to mitigate the impact of bacterial metabolism on inhibitor activity. Competitive gel-based ABPP17 revealed that each SH-reactive chemotype engaged diverse targets in C. elegans (Fig. 1b and Supplementary Fig. 1c) and these inhibitory events were maintained for select protein targets for up to 5 days (Fig. 1c), underscoring an advantageous feature of irreversible inhibitors, which can furnish sustained target inactivation that is only lost upon re-synthesis of new protein. Having confirmed that members of our inhibitor library engage SHs in C. elegans, we next set out to screen this compound collection for effects on lifespan.
SH-directed inhibitors that extend C. elegans lifespan.
We tested a set of ~100 compounds (Supplementary Table 1 and Supplementary Dataset 2) containing diverse SH-directed electrophilic groups, including carbamates, ureas, and β-lactones, for effects on C. elegans lifespan (most compounds were tested at 50 μM except for solubility-limited compounds, which were screened at 5–25 μM). To avoid the confounding effects of developmental pathways on lifespan regulation, we administered inhibitors on day 1 of adulthood. While most inhibitors did not substantially alter lifespan, a handful of compounds extended lifespan > 15% (red bars, Fig. 2a), with a single compound – the carbamate JZL184 – increasing lifespan by ~45% (Fig. 2a, b, and Supplementary Dataset 2). Other compounds that shortened lifespan (Fig. 2a) may impair aging processes (see Discussion) or produce general cytotoxic effects at the tested concentrations. Follow-up studies revealed that JZL184 achieved a maximal effect on lifespan at concentrations ranging from 25–50 μM and half-maximal extension at ~6 μM (Fig. 2c).
Figure 2. Phenotypic screening identifies SH-directed inhibitors that extend lifespan in C. elegans.
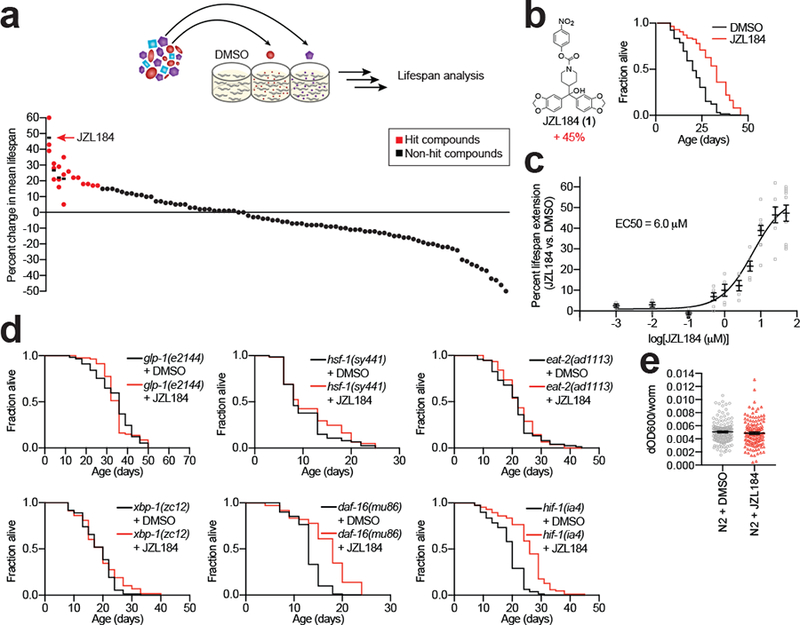
(a) 95 SH-directed inhibitors were screened for their effect on C. elegans lifespan. Screening was done in 96-well plates and compounds were generally screened at 50 μM with a few exceptions that were screened from 5–25 μM due to lower solubility. Approximately 10 animals were incubated per well in liquid culture throughout the assay. Data for representative hit compounds (red dots) are shown as average values from multiple replicates (n = 3 independent experiments, shown on same vertical line, where black horizontal dash marks the average lifespan change for each hit compound). (b) The SH-directed inhibitor JZL184 (1) substantially extends lifespan. Shown is a representative Kaplan-Meier plot revealing ~45% lifespan extension by JZL184 (50 μM) (DMSO n = 80 worms, JZL184 n = 97 worms, P = 1.64E-10, compared to DMSO control). (c) Dose-dependent lifespan increase caused by JZL184. Half-maximal effect (EC50) of JZL184 was calculated to be 6 μM. Data represents mean ± s.e.m. (n = 11 independent experiments). (d) Representative lifespan curves demonstrating the interaction of JZL184 (50 μM) with known aging pathways: glp-1 (DMSO n = 57 worms, JZL184 n = 80 worms, P = 0.8359 compared to glp-1 DMSO control), hsf-1 (DMSO n = 74 worms, JZL184 n = 61 worms, P = 0.2471 compared to hsf-1 DMSO control), eat-2 (DMSO n = 75 worms, JZL184 n = 76 worms, P = 0.5207 compared to eat-2 DMSO control), xbp-1 (DMSO n = 73 worms, JZL184 n = 58 worms, P = 0.2121 compared to xbp-1 DMSO control), daf-16 (DMSO n = 81 worms, JZL184 n = 72 worms, P = 3.79E-11 compared to daf-16 DMSO control) and hif-1 (DMSO n = 79 worms, JZL184 n = 85 worms, P = 4.53E-13 compared to hif-1 DMSO control). For b and d, results are representative of 2 (glp-1) or 3 (all other conditions) independent experiments. Two-sided Mantel-Haenszel version of the log-rank test performed relative to DMSO control for b and d. Statistics for b-d are provided in Supplementary Dataset 2. (e) Food intake per worm in animals treated with DMSO or JZL184 (50 μM). Data represents mean ± s.e.m. (n = 144 wells per condition, P = 0.3752, two-sided unpaired t test, from 6 independent experiments).
JZL184 was able to extend worm lifespan when administered at any tested age prior to day 2 of adulthood; however, when animals were treated with JZL184 at day 2 or later, we observed a sharp decrease in lifespan effect of the compound (Supplementary Fig. 2a, b, and Supplementary Dataset 2). We confirmed that C. elegans brood size was unaltered by JZL184 treatment (Supplementary Fig. 2c), indicating that the compound did not perturb reproductive capacity. Notably, however, we found that JZL184 did not further extend the lifespan of sterile glp-1(e1244) mutant animals (Fig. 2d, Supplementary Fig. 2d, and Supplementary Dataset 2), which are long-lived due to defective germ-cell proliferation and germline signaling26. We tested JZL184 in mutants of several additional canonical longevity pathways, revealing that the compound increased the lifespan of insulin (daf-16(mu86)) and hypoxia (hif-1(ia4)) mutants, but not in animals with mutations in the unfolded protein response (UPR) (xbp-1(zc12)), heat shock factor (hsf-1(sy441)), or dietary restriction pathways (eat-2(ad1113)) (Fig. 2d and Supplementary Dataset 2). JZL184 did not, however, induce endoplasmic reticulum UPR, as tested in the transcriptional GFP-reporter strain zcls4 [hsp-4::gfp] (Supplementary Fig. 3), or alter the food intake of animals, as measured by the amount of bacteria consumed through changes in optical density of the media Fig. 2e). These chemical epistasis experiments suggest that JZL184 acts independently of the insulin/IGF-1 signaling and hypoxia signaling pathways and instead impinges on multiple stress pathways, possibly through a common regulatory node that involves germ-line signaling. To gain further molecular insights into the mechanism of lifespan extension by JZL184, we next sought to identify the SH target(s) of this compound in C. elegans.
FAAH-4 is a primary target of JZL184 in C. elegans.
JZL184 was originally developed as a potent and selective irreversible inhibitor of the mammalian monoacylglycerol lipase (MAGL or MGLL) enzyme27, which is principally responsible for degrading monoacylglycerides (MAGs) including the endocannabinoid 2-arachidonoylglycerol (2-AG)28. Curiously, however, reciprocal BLAST searches revealed that C. elegans lacks an orthologous enzyme to human MAGL (Supplementary Fig. 4). Using quantitative, mass spectrometry-based competitive ABPP methods (Supplementary Fig. 1b), we instead found that JZL184 (50 μM, 24 hr, day 1 adulthood) targeted a handful of the more than 75 quantified SHs in C. elegans – F13H6.3, F15A8.6, Y71H2AM.13, K11G9.2, and fatty acid amide hydrolase-4 (FAAH-4), (Fig. 3a and Supplementary Datasets 3–5). To narrow down the pharmacologically relevant target(s) for the lifespan effects of JZL184, we compared the target profiles of structurally related analogues that showed differential effects on lifespan. We first noted that less electrophilic urea analogues of JZL184 (ALC3 (2) and ALC6 (3)) did not extend C. elegans lifespan (Fig. 3b and Supplementary Dataset 2) and showed, as expected, much lower reactivity with SHs in vivo (Fig. 3c and Supplementary Datasets 3–5). In contrast, WWL154 (4), an analog of JZL184 that maintains the SH-reactive p-nitrophenyl carbamate group, extended lifespan (Fig. 3b and Supplementary Dataset 2) and cross-reacted with three of the five SH targets of JZL184 (FAAH4, F15A8.6, and Y71H2AM.13) in vivo (Fig. 3c and Supplementary Datasets 3–5). Finally, we found that ALC1 (5), another p-nitrophenyl carbamate analog of JZL184, and the human FAAH-1 inhibitor URB597 (6)29 did not extend lifespan (Fig. 3b and Supplementary Dataset 2), despite engaging several of the SH targets of JZL184 with the exception of FAAH-4 in vivo (Fig. 3c, Supplementary Fig. 5, and Supplementary Datasets 3–5). URB597 also strongly engaged C. elegans FAAH-1 in vivo (Fig. 3c and Supplementary Datasets 3–5). By correlating the SH targets and lifespan effects of JZL184 and analogues, we found that FAAH-4 was the only SH that: 1) represented a major target of the compounds that extended lifespan (JZL184, WWL154), and 2) was not inhibited by compounds that did not increase lifespan (ACL1, ACL3, ACL6, URB597). These data designated FAAH-4 as a potential target involved in the lifespan-extending activity of JZL184.
Figure 3. Identification of FAAH-4 as a principal target of JZL184 in C. elegans.
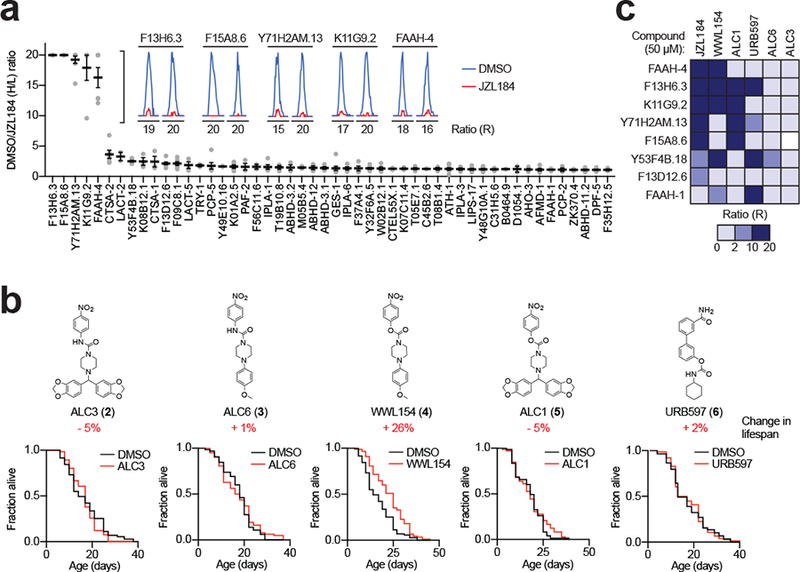
(a) FP-biotin enrichment and identification of SHs endogenously inhibited by JZL184 in C. elegans. Isotopically labeled 15N or light 14N animals were treated with DMSO or JZL184 (50 μM) for 24 h in vivo, respectively. After homogenization, heavy and light proteomes were combined, incubated with FP-biotin, enriched by avidin, tryptically digested and identified and quantified by MS. MS1 chromatograms shown for SHs that were substantially inhibited (> 5-fold relative to DMSO control) by JZL184 (50 μM, 24 h). Top 50 proteins shown. Data represent the mean of median ratios + s.e.m. for peptides quantified for each protein (n = 4 independent experiments). (b) Structures and lifespan effects of analogues of JZL184 (ALC3 (2): DMSO n = 65 worms, ALC3 n = 109 worms, P = 0.3172; ALC6 (3): DMSO n = 65 worms, ALC6 n = 62 worms, P = 0.5191 for ALC6; WWL154 (4): DMSO n = 71 worms, WWL154 n = 88 worms, P = 6.17E-05 for WWL154; ALC1 (5): DMSO n = 73 worms, ALC1 n = 72 worms, P = 0.3744 for ALC1; URB597 (6): DMSO n = 59 worms, URB597 n = 76 worms, P = 0.9948 for URB597). Two-sided Mantel-Haenszel version of the log-rank test performed relative to DMSO control. Results are representative of 4 (ALC6, URB597), 5 (WWL154), 6 (ALC3), or 7 (ALC1) independent experiments. (c) Heatmap depicting competitive MS-ABPP data for the indicated proteins from worms treated with lifespan-extending (JZL184, WWL154) or inactive control (ALC1, URB597, ALC6, ALC3) compounds. Values represent the mean from 2 (ALC1 and ALC3) or 3 (all other conditions) independent experiments. Statistics for b are provided in Supplementary Dataset 2.
We also considered the possibility that JZL184 may covalently inhibit proteins outside the SH family and addressed this question by synthesizing an alkyne analog of JZL184, CL-01 (7), that extends lifespan by ~40% (Supplementary Fig. 6a and Supplementary Dataset 2). Proteomic profiling of N2 animals treated with CL-01, followed by copper-catalyzed azide-alkyne cycloaddition (CuAAC or click) chemistry conjugation to an azide-biotin reporter tag, confirmed the enrichment and JZL184-mediated inhibition of FAAH-4, as well as other SH targets shown in Fig. 3a (Supplementary Fig. 6b and Supplementary Datasets 3, 4). One additional SH – the putative carboxypeptidase F13D12.6 – was enriched by CL-01 and blocked by JZL184 (Supplementary Fig. 6b and Supplementary Datasets 3, 4). This SH was also partially inhibited (~50%) by JZL184 in FP-biotin enrichment experiments (Fig. 3a and Supplementary Datasets 3–5), but was not inhibited by the lifespan-extending analogue WWL154 (Fig. 3c and Supplementary Datasets 3–5). The chemical proteomic experiments with CL-01 thus supported that the covalent target profile of JZL184 did not extend beyond FAAH-4 and the other C. elegans SHs mapped by competitive ABPP experiments with FP-biotin (Fig. 3a, c and Supplementary Datasets 3–5).
FAAH-4 displays monoacylglycerol lipase activity.
FAAH-4 is a member of the amidase signature family, which are atypical SHs that possess a unique structural fold and catalytic machinery (Ser-Ser-Lys catalytic triad, in contrast to the Ser-His-Asp triad found in the more common α/β-hydrolase family)30. Prominent members include mammalian FAAH-1, which degrades the N-acylethanolamine (NAE) class of signaling lipids, including the endocannabinoid anandamide31,32, and the gatA enzyme involved in transamidating misacylated Glu-tRNA(Gln)33. C. elegans has eight predicted amidase signature enzymes, of which five were observed by MS-based competitive ABPP and, among them, only FAAH-4 was substantially inhibited (> 90%) by JZL184 in vivo (Fig. 4a and Supplementary Datasets 3–5). One additional amidase Y53F4B.18 was partly inhibited (~50–60%) by JZL184 in vivo, while the activity of other amidases was unaltered in JZL184-treated worms (Fig. 4a and Supplementary Datasets 3–5). We confirmed and extended the selectivity profile for JZL184 against recombinantly expressed C. elegans amidases, of which six enzymes showed detectable signals in gel-based ABPP assays (FAAH-1–5 and Y53F4B.18; Fig. 4b). We also verified that endogenous and recombinant FAAH-4 showed similar structure-activity relationships with JZL184 analogues (compare Fig. 3c and Fig. 4c) and determined the potency of inhibition of recombinant FAAH-4 by JZL184 (IC50 value of 11 μM; Fig. 4d, e).
Figure 4. FAAH-4 is inhibited by JZL184.
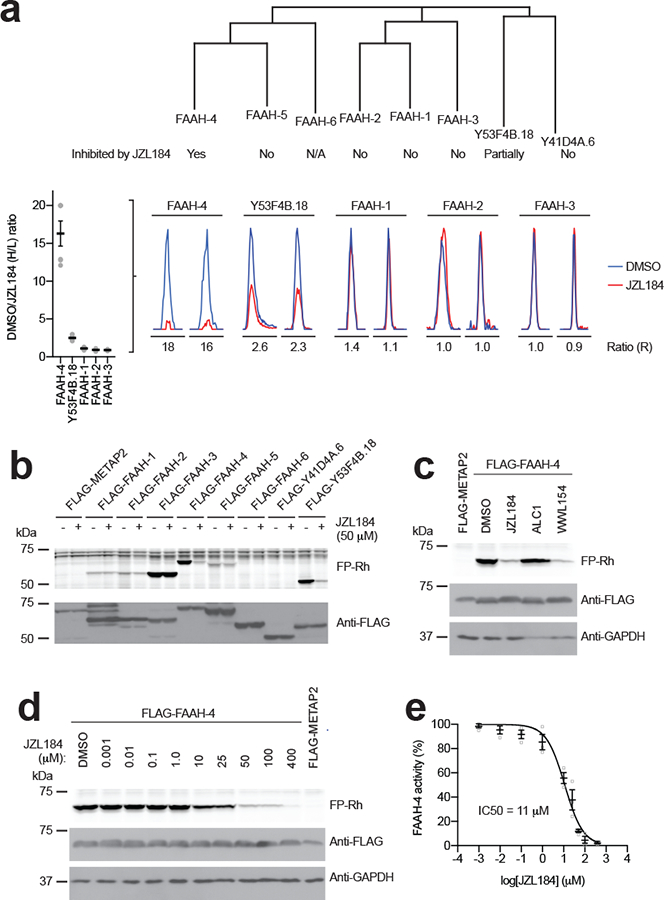
(a) Dendrogram showing the C. elegans amidase family and designating the sensitivity of individual amidases to JZL184. FAAH-6 is marked as N/A because this protein is missing a conserved serine in the amidase signature Ser-Ser-Lys catalytic triad and is therefore considered an inactive member of the family. Images below the dendrogram show representative MS1 traces for inhibition of the indicated amidases in JZL184-treated C. elegans (as determined by MS-based competitive ABPP). Data represent the mean of median ratios + s.e.m. for peptides quantified for each protein (n = 4 independent experiments). (b) Gel-based competitive ABPP showing cross-reactivity of recombinant amidases with JZL184. Only recombinant FAAH-4 and Y53F4B.18 showed evidence of inhibition by JZL184, consistent with the MS-based competitive ABPP analysis of JZL184-treated C. elegans (Fig. 3a). Note that FAAH-6 is missing a conserved serine in the amidase signature Ser-Ser-Lys catalytic triad and is therefore considered an inactive member of the family. (c) Gel-based competitive ABPP results showing inhibition of recombinant FAAH-4 by JZL184 and WWL154 but not ALC1 (50 μM of each compound, 30 min treatment). Full-length gels containing cropped gel data are shown in Supplementary Fig. 7. (d, e) Concentration-dependent inhibition of recombinant FAAH-4 by JZL184 as measured by gel-based ABPP. For e, data represents mean ± s.e.m. (n = 3 independent experiments). Full-length gels containing cropped gel data are shown in Supplementary Fig. 7. For b-d, C. elegans SHs or control protein (METAP2) were recombinantly expressed by transient transfection in HEK-293T cells. For b-d, data are representative of 3 independent experiments.
We were initially surprised to identify FAAH-4 as a unique target of JZL184 among C. elegans amidases, as the amidase signature family does not share any sequence or structural fold homology with mammalian MAGL, which is an α/β-hydrolase, nor is FAAH-4 more closely related to mammalian FAAH-1 (a weak off-target of JZL18434) compared to other C. elegans amidases (e.g., FAAH1–3, 5; Supplementary Fig. 8). Recalling further that C. elegans lacks a sequence-related MAGL protein, we wondered if FAAH-4 might share not only a pharmacological profile, but also metabolic function with MAGL. Consistent with this hypothesis, we found that FAAH-4 was unique among the eight C. elegans amidases in robustly hydrolyzing 2-AG and that this activity was blocked by JZL184 (Fig. 5a and Supplementary Table 2). In contrast, several C. elegans amidases hydrolyzed the amidated substrate AEA (Fig. 5b and Supplementary Table 2). While FAAH-4 was one of the several amidases capable of hydrolyzing AEA, it did so with much lower turnover (20 pmol/min/mg) compared to 2-AG hydrolysis (1200 pmol/min/mg). Finally, we confirmed that other SH targets of JZL184 discovered by MS-ABPP did not exhibit MAG hydrolytic activity in vitro (Supplementary Fig. 9 and Supplementary Table 2). These data, taken together, indicate that, among C. elegans amidases, FAAH-4 displays two special properties: 1) sensitivity to inhibition by JZL184; and 2) robust 2-AG hydrolytic activity. We therefore next wondered whether FAAH-4 acts as a C. elegans MAGL enzyme in vivo.
Figure 5. FAAH-4 has 2-AG and AEA hydrolytic activity in vitro.
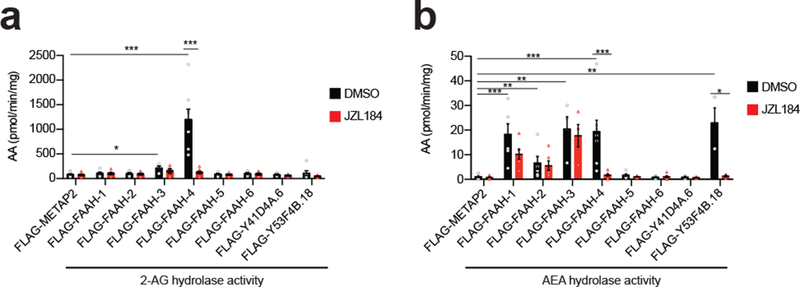
(a, b) 2-AG and AEA hydrolytic activities of recombinant C. elegans amidases or control protein METAP2 (5 μg of lysate) in presence of JZL184 (50 μM, 30 min pre-treatment) or DMSO. Following treatment, AEA or 2-AG (50 μM) was added to the lysates and the reaction proceeded for 20 min at 37 °C prior to quenching. Hydrolytic activities were determined by quantifying arachidonic acid (AA) product relative to a d8-arachidonic acid (AA) internal standard by LC-MS/MS. Data represent the mean ± s.e.m. n = 4 (Y41D4A.6, Y53F4B.18 for AEA), 6 (FAAH-1, 2, 5, 6, Y53F4B.18 for 2-AG), or 8 (METAP2, FAAH-4) independent samples per group. *P < 0.05, **P < 0.01, and ***P < 0.001 (two-sided Mann-Whitney test performed relative to METAP2 or DMSO control). The P values are provided in Supplementary Table 2.
FAAH-4 regulates C. elegans lifespan and MAG content.
To further characterize the function of FAAH-4 in C. elegans, we generated a faah-4 deletion animal (Δfaah-4) using CRISPR/CAS-9 technology and confirmed that this animal model lacks FAAH-4 protein by MS-based ABPP (Supplementary Fig. 10a and Supplementary Datasets 3, 4). Lifespan measurements revealed that Δfaah-4 animals lived substantially longer than wildtype N2 animals (Fig. 6a and b (left panel), and Supplementary Dataset 2). JZL184 did, however, further extend the lifespan increase observed in Δfaah-4 animals (~22% without JZL184 compared to N2 with DMSO; ~25% with JZL184 compared to Δfaah-4 with DMSO) to an absolute magnitude that matched, but did not appreciably exceed the longevity effects observed in JZL184-treated wild type worms (44% increase in lifespan) (Fig. 6b (left panel) and Supplementary Dataset 2). The relative lifespan extension of JZL184 was accordingly smaller in Δfaah-4 animals compared to N2 animals (Fig. 6b, right panel).
Figure 6. FAAH-4 regulates MAG content and lifespan of C. elegans.
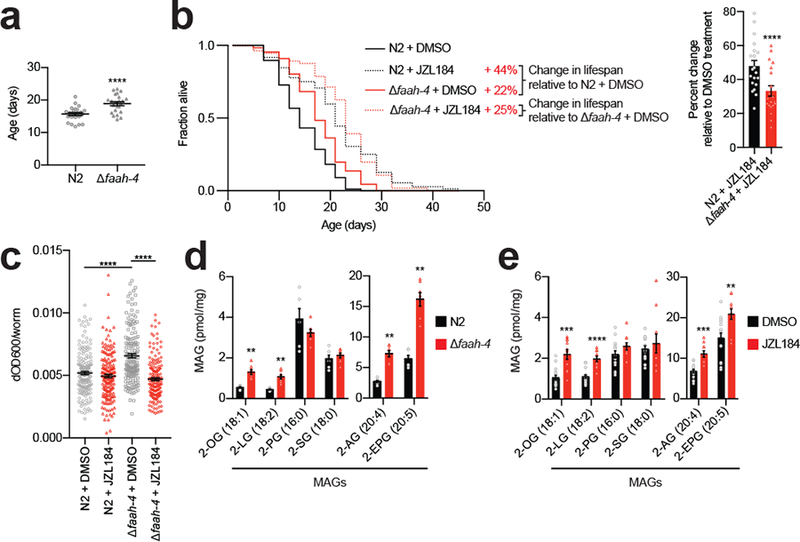
(a) Δfaah-4 animals are long-lived. Each point represents a cohort in an experiment. Data represent the mean lifespan ± s.e.m. from n = 23 independent experiments (at least 43 worms per cohort, 23 cohorts per group; ****P = 6.98E-11, two-sided paired t test). (b) Left, JZL184 further extends lifespan in Δfaah-4 animals. No additional extension in lifespan was seen in Δfaah-4 animals treated with JZL184 compared to N2 animals treated with JZL184. Lifespan curves for N2 and Δfaah-4 animals treated with JZL184 (50 μM) or DMSO (N2 DMSO n = 88, N2 JZL184 n = 72, Δfaah-4 DMSO n = 66, Δfaah-4 JZL184 n = 56). This result is a representative of 20 independent experiments. Right, percent change in mean lifespan of N2 and Δfaah-4 animals treated with JZL184 relative to N2 and Δfaah-4 animals treated with DMSO. Data represent the mean + s.e.m. (n = 20 independent experiments; ****P = 6.69E-07, two-sided paired t test). Statistics are provided in Supplementary Dataset 2. (c) Food intake per worm in N2 or Δfaah-4 animals treated with DMSO or JZL184 (50 μM). Data represent the mean ± s.e.m. (N2 DMSO n = 183, N2 JZL184 n = 178, Δfaah-4 DMSO n = 160, Δfaah-4 JZL184 n = 164, ****P = 1.46E-10 for N2 vs Δfaah-4 DMSO, ****P < 1.00E-15 for Δfaah-4 DMSO vs JZL184, two-sided unpaired t test performed, from 8 independent experiments). (d-e) Mono- and poly-unsaturated MAGs are elevated in (d) Δfaah-4 animals or (e) animals treated with JZL184. Quantification of MAGs from indicated groups of animals was performed by targeted LC-MS/MS. 2-OG, 2-oleoylglycerol (C18:1); 2-LG, 2-linoleoylglycerol (C18:2); 2-PG, 2-palmitoylglycerol (C16:0); 2-SG, 2-stearoylglycerol (C18:0); 2-AG, 2-arachidonoylglycerol (C20:4); 2-EPG, 2-eicosapentaenoylglycerol (C20:5). For d-e, data represent the mean ± s.e.m.; n = 6 (d, N2), 7 (d, Δfaah-4), 10 (e, JZL184), or 15 (e, DMSO) independent samples per group. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 (two-sided Mann-Whitney test performed relative to N2 or DMSO control). The P values for d, e are provided in Supplementary Table 3 and 4, respectively.
In our attempts to further understand the mechanism(s) by which JZL184 and FAAH-4 loss contribute to lifespan, we found that both Δfaah-4 animals and wild-type animals treated with JZL184 showed a similarly greater resistance to paraquat-induced death compared to control animals (Supplementary Fig. 10b), suggesting that the lifespan extension associated with FAAH-4 disruption may involve, at least in part, improved responses to oxidative stress35 (Supplementary Fig. 10b). We also observed that, in contrast to JZL184-treated animals, which did not show altered food intake (Fig. 6c), Δfaah-4 animals displayed a paradoxical increase in food consumption that was reversed by JZL184 treatment (Fig. 6c). As reductions in food intake can promote lifespan extension in C. elegans36, these data suggest that an adaptation to consume more food may suppress the extent of lifespan increase observed in Δfaah-4 animals.
That JZL184 can further extend lifespan of Δfaah-4 animals, as well as reverse their heightened food consumption phenotype, supports the involvement of additional SHs in the action of this compound. To initially explore this concept, we evaluated another amidase Y53F4B.18 that was partially inhibited by JZL184 and the lifespan-extending analogue WWL154 (Fig. 3c) and found that y53f4b.18 deletion animals (Δy53f4b.18) lived ~15% longer than N2 animals (Supplementary Fig. 11, Supplementary Datasets 3, 4 and Supplementary Dataset 2). As was observed in Δfaah-4 animals, JZL184 further extended the lifespan of Δy53f4b.18 animals (Supplementary Fig. 11b and Supplementary Dataset 2). Notably, however, the Δfaah-4;Δy53f4b.18 double knockout mutant exhibited a similar lifespan extension to either single mutant animal (Supplementary Fig. 11b and Supplementary Dataset 2), suggesting that FAAH-4 and Y53F4B.18 may act within the same lifespan-regulating pathway.
Finally, we evaluated the lipid content of Δfaah-4 animals and found that they displayed significantly elevated mono- and poly-unsaturated MAGs, including 2-AG, compared to wild type animals (Fig. 6d and Supplementary Table 3), while NAEs and free fatty acids (FFAs) were mostly unchanged (Supplementary Fig. 12a, b, and Supplementary Table 3). We also found that JZL184-treated worms displayed a similar profile of increased MAGs (Fig. 6e and Supplementary Table 4) with mostly unaltered NAE and FFA content (Supplementary Fig. 12c, d, and Supplementary Table 4). Fewer lipid changes were observed in Δy53f4b.18 animals, but a handful of NAEs and MAGs were also elevated in these animals (Supplementary Fig. 12e–g and Supplementary Table 5).
These results, taken together indicate that JZL184 increases lifespan through a combination of inhibiting FAAH-4 and one or more other SH targets (e.g., Y53F4B.18). Our mechanistic studies further demonstrate that FAAH-4 regulates not only lifespan, but also stress responses in C. elegans, where the enzyme also serves as a hydrolytic enzyme for 2-AG and other MAG lipids. Considering that FAAH-4 also hydrolyzes AEA in vitro (Fig. 5b), it remains possible that the enzyme additionally contributes, along with other amidases, to regulating AEA signaling in vivo.
Discussion
There remains much interest in deducing protein orthology across distantly related species as a means to uncover conserved pathways and functions throughout organismal evolution37,38. Mapping protein orthologues has largely been pursued using sequence relatedness as a guiding principle37,38, but evolutionarily unrelated proteins can also perform the same biochemical functions in different species, and the identification of such non-homologous isofunctional proteins39 remains technically challenging. Previous strategies include the proteomic characterization of protein interaction networks40 or the comparative analysis of genomic operons41 across species, although the former requires that proteins are part of stable complexes and the latter is largely restricted in applicability to prokaryotes. Our findings that FAAH-4 serves the primary metabolic function in C. elegans performed by the MAGL enzyme in mammals and is also sensitive to the MAGL-directed inhibitor JZL184 illuminate a provocative alternative – that pharmacological convergence, or the property of conserved interaction with chemical probes, offers another powerful way to excavate non-homologous isofunctional proteins39. We anticipate that more detailed mechanistic and structural studies may illuminate at an atomic level how FAAH-4, but not other C. elegans amidases, shares both inhibitor and substrate profiles with mammalian MAGL. We should also note that, while our ABPP experiments support that JZL184 selectively inhibits FAAH-4 over other FAAH enzymes in C. elegans, and conversely URB597 preferentially inhibits FAAH-1, it would be important to confirm these in vivo selectivity profiles with additional assays (e.g., substrate-based) in future studies. The broader application of chemical proteomic screening in whole organisms, both phenotypic as shown herein and correlative42, may facilitate the discovery of additional functionally analogous proteins and pathways that display pharmacological convergence. Future phenotypic screens would also benefit from a more detailed understanding of the physicochemical properties that impart good uptake of compounds into worms.
While our data support that FAAH-4 and Y53F4B.18 make a substantive contribution to longevity in C. elegans, these enzymes only partly accounted for the lifespan-extending effects of JZL184. Our studies with the JZL184-based click probe CL-01 indicate that the covalent targets of JZL184 are confined to FAAH-4 and a handful of additional SHs. We cannot exclude, however, that non-covalent targets may exist for JZL184. Understanding which of the additional SH target(s) of JZL184 contribute to its remarkable effects on lifespan, as well as to the apparent adaptation in feeding behavior caused by loss of FAAH-4 (which was reversed by JZL184 treatment; Fig. 6c) is an important future objective. Regardless, our data support and augment a growing body of evidence that lipid pathways play fundamental roles in aging. Monounsaturated43 and polyunsaturated44 fatty acids have been found to extend lifespan, while lipid peroxides suppress lifespan45 in C. elegans, and members of the NAE lipid class exert variable, possibly acyl-chain dependent effects on aging18,21. Lipolysis in germ cells also regulates longevity in C. elegans20. To what extent these outcomes reflect a role for lipids in metabolism (e.g., serving as sources of energy or building blocks for membranes) versus signaling functions (e.g., acting as ligands for G-protein-coupled46 or nuclear hormone21 receptors) remains less well understood. Within our library of test inhibitors of SHs were compounds that target other enzymes in the endocannabinoid system, and we call attention to DH376 (8) and DO34 (9), two carbamate inhibitors of the mammalian diacylglycerol lipase enzymes DAGLA and DAGLB47 both of which produced substantial lifespan-shortening effects in our primary screen (Supplementary Dataset 2). While further studies are needed to confirm whether DH376 and DO34 inhibit C. elegans DAGL-2 in vivo, it is noteworthy that the genetic loss of this enzyme also leads to reductions in lifespan in C. elegans19. We should also note that FAAH-4’s contributions to lipid metabolism and signaling may extend beyond 2-AG/MAG lipids. Considering, for instance, that FAAH-4 also hydrolyzes AEA in vitro (Fig. 5b), it remains possible that the enzyme contributes, along with other amidases, to regulating AEA signaling in vivo. Finally, when considering the potential translational relevance of our findings, we note that the endocannabinoid system has been proposed to play an important role in aging in mammals48,49, and major components of this system are substantially altered in the brains of older mice, including reductions in 2-AG and elevations in MAGL50. With the recent discovery of endocannabinoid-like receptors in C. elegans that respond to 2-AG46, which is produced by DAGL-2 and degraded by FAAH-4, two enzymes that show opposing impacts on lifespan in this organism19, it seems plausible that both worms and mammals possess a fully isofunctional lipid signaling pathway that regulates longevity and possibly aging-related disorders.
Methods
C. elegans strains and nomenclature
All strains were maintained at 20 °C on standard nematode growth medium (NGM) plates with Escherichia coli (OP50–1, streptomycin-resistant). The following strains were obtained from the CGC: CF1038 (daf-16(mu86) I), DA1113 (eat-2(ad1113) II), ZG31 (hif-1(ia4) V), PS3551 (hsf-1(sy441) I), SJ17 (xbp-1(zc12) III; zcls4 V), CF1903 (glp-1(e2144)III), and RB1668 (C02H7.2(ok2068) X). The faah-4 deletion strain (Δfaah-4) named OD3609 (genotype: faah-4(lt121)III) and y53f4b.18 deletion strain (Δy53f4b.18) named VV213 (genotype: y53f4b.18(vq3)II) were engineered as described51. All experiments were performed using hermaphrodites.
Cell culture methods
HEK293T cells (ATCC CRL-3216) were maintained at 37 °C with 5% CO2. HEK293T cells were grown in DMEM (Gibco) supplemented with 10% (v/v) fetal bovine serum (FBS, Omega Scientific), penicillin (100 U/ml, GE Life Sciences), streptomycin (100 μg/ml, GE Life Sciences) and L-glutamine (2 mM).
Generation of C. elegans database and predicted SH list
A highly non-redundant, gene-centric, protein sequence database was curated from the UniProt C. elegans proteome (UP000001940, accessed 10/17/2017, 26778 proteins). All protein entries with reviewed status were kept by default. Otherwise, the longest protein sequence was chosen for every WormBaseGene identifier in the UniProt database. Redundancy was further reduced using CD-HIT (98% identity) resulting in 19660 protein sequences in the final database. A reverse-concatenated version of the full database was used for MS analysis (ProLuCID). To generate a list of predicted C. elegans SHs, the database above was searched against Pfam-A using PfamScan and filtered using a manually curated mammalian SH list. It should be noted that this list contains primarily metabolic SHs and may not account for C. elegans SHs if they contain unannotated SH domains that are different from mammalian SH domains.
Preparation of C. elegans proteomes for MS-ABPP analysis
To isotopically label C. elegans, isotopically labeled Op50 bacteria were used as the food source. Op50 bacteria were cultured in minimal media enriched with either (14NH4)2SO4 or 15NH4)2SO4 and gamma-irradiated52. C. elegans were fed 14N- (light) or 15N- (heavy) OP50 for three generations to ensure full isotopic incorporation. Per replicate, over 10,000 C. elegans were grown in liquid culture with 6mg/mL Op50. Fluorodeoxyuridine (FUDR, 0.6 mM, Sigma) was added to L4 animals to prevent the development of progeny. For comparative ABPP experiments, on day 1 of adulthood, animals were washed 6X in PBS and flash frozen until homogenization. For competitive ABPP experiments, on day 1 of adulthood, 14N- (light) and 15N- (heavy) animals were treated with compound (50 μM) or DMSO (Sigma), respectively, for 24 h, washed 6X in PBS and flash frozen until homogenization. To harvest proteomes, a small scoop each of 0.5 mm Borosilicate Glass Beads (Next Advance) and 1.4 mm Zirconium Oxide Beads (Precellys) were added to the animals and animals were homogenized in the Bullet Blender (5 min, setting 9, 2X, Next Advance). Cell lysates were then centrifuged (100,000 × g, 45 min) to yield membrane (pellet) and soluble (supernatant) fractions. Membrane pellets were resuspended in PBS by sonication. Protein concentrations were determined using the DC Protein Assay (Bio-Rad) and absorbance read at 750 nm using an Infinite F500 plate reader (Tecan). For FP-enrichment experiments related to Figure 1, the light samples were heat-denatured at 95 °C for 5 min followed by 15 min on ice (3X) prior to FP-labeling.
MS-ABPP sample preparation
Methods were adapted from those previously reported53. In brief, isotopically labeled 14N- (light) or 15N- (heavy) samples (1 mg/mL in 1 mL of PBS) were labeled with FP-biotin (5 μM) or CL-01 (5 μM) for 2 h at 37 °C. Samples treated with CL-01 were conjugated to an azide-biotin reporter tag by copper-catalyzed azide-alkyne cycloaddition (CuAAC or click) chemistry.54 Following labeling, proteomes were denatured and precipitated with the addition of H2O (1 mL), MeOH (2 mL), and CHCl3 (500 μL); the resulting suspension was then centrifuged (4,000 rpm, 10 min) to create a protein disc at the interface between the aqueous and organic phases. The protein disc was washed with once with 4:4:1 MeOH/H2O/CHCl3 and then with MeOH. Light and heavy protein pellets were combined, resuspended in 25 mM ammonium bicarbonate/6 M urea in water, reduced using dithiothreitol (DTT, 10 mM, 15 min) at 65 °C, and then alkylated using iodoacetamide (40 mM, 30 min) at 23°C in the dark. Biotinylated proteins were enriched with pre-washed avidin-agarose beads (Sigma-Aldrich) by rotating at 23 °C in PBS with 0.2% SDS (6 mL, 2 h). The beads were then washed sequentially in PBS with 1% SDS (3X), 10 mL PBS (3X) and H2O (3X). On-bead digestion was performed using sequencing-grade trypsin (2 g; Promega) in 25 mM ammonium bicarbonate/2 M urea in water with 1 mM CaCl2 (200 μL, 12–14 h) at 37 °C. Peptides obtained from this procedure were acidified with formic acid (5%) and pressure-loaded onto a biphasic (strong cation exchange/reverse phase) capillary column for mass spectrometry analysis on a Finnigan LTQ Orbitrap (Thermo Scientific) or LTQ Orbitrap Velos (Thermo Scientific).
MS and data analysis
MS-ABPP samples were analyzed on a Finnigan LTQ Orbitrap (Thermo Scientific) or LTQ Orbitrap Velos (Thermo Scientific) following previously reported protocols14. Peptides were eluted during a 5-step multidimensional LC-MS protocol using 0%, 25%, 50%, 80%, and 100% salt bumps of 500 mM aqueous ammonium acetate and followed by an increasing gradient of acetonitrile/0.1% formic acid in each step. Data were collected in data-dependent acquisition mode (400–1,800 m/z). MS2 information was extracted using RawXtract (v 1.9.9.2) and searched on the Integrated Proteomics Pipeline (IP2) using the ProLuCID algorithm. Spectra were searched against a reverse-concatenated non-redundant (gene-centric) database of C. elegans protein sequences that had been assembled from the UniProt database (see methods described above). Heavy isotopic labeling searching was enabled (N15). Peptide modifications were specified on cysteine residues (static, 57.0215 m/z; iodoacetamide alkylation) and methionine (differential, 15.9949; oxidation). Spectral matches were further filtered using DTASelect, where the spectrum false positive rate was restricted to 1% and peptides were required to have at least one tryptic end. Heavy-to-light ratios were quantified using in-house CIMAGE software55. CIMAGE reports a maximum ratio of 20 (such that highly abundant proteins do not get assigned drastically large ratios); this is also the ratio it assigns to singletons—peptides for which MS1 signal was detected solely for the m/z of the heavy ion (but not light). Samples from the membrane or soluble proteome fractions were first analyzed separately, retaining proteins that met these quality control criteria for further analysis: 1) two sequence unique peptides per protein and 2) peptides present in at least two replicates. Proteins that met these criteria with heavy-to-light ratio > 5 and annotated as a predicted SH were designated as FP-enriched proteins (Supplementary Dataset 1). In competition experiments, targets were defined as FP-enriched proteins that had an average ratio >5. For competitive ABPP experiments with analogs of JZL184, the established targets of JZL184 were manually inspected to include high confidence peptides that otherwise did not meet previous requirements. For heatmap analysis, proteins were designated as membrane or soluble proteins based on the FP-enrichment data (Supplementary Dataset 1) (membrane proteins: F15A8.6, FAAH04, Y53F4B.18, Y71H2AM.13 and soluble proteins: F13H6.3 and K11G9.2). The average of ratios was calculated from membrane or soluble replicates. For CL-01 experiments, the maximum ratio was set to 10 and proteins were required to meet the following criteria: 1) two quantified peptides per replicate and 2) two valid ratio values between replicates.
Lifespan assay and analysis
The lifespan assays and analysis was carried out as previously described56. In brief, approximately 10 age-synchronized C. elegans were cultured at 20 °C in 96-well plates containing S. complete media and irradiated Op50 bacteria (6 mg/mL). Animals, except the glp-1 strain, were given FUDR (0.6 mM, Sigma) at the L4 developmental stage to prevent the development of progeny. The glp-1 stain was temperature shifted to 25 °C at the L1 larval stage until day 1, where they were shifted back to 20 °C for the remainder of the assay. On day 1 of adulthood, animals were treated with compound (5–50 μM, dependent on solubility of compound) or DMSO (v/v 0.5%, Sigma) and continuously exposed throughout the assay. On day 5 of adulthood, Op50 (5 μg, irradiated) was added to each well to prevent starvation. Living animals were scored by eye. Statistical analysis was done using STATA software. Percent changes in lifespan were calculated by comparing mean lifespan values. P-values were calculated using two-sided Mantel–Haenszel version of the log-rank test.
Preparation of C. elegans proteomes for gel-based ABPP
Approximately 3,000 N2 C. elegans were grown in liquid culture with irradiated Op50 (6 mg/mL) per replicate. On day 1 of adulthood, animals were treated with compound or DMSO, respectively, for 24 h, washed 6X in PBS and flash frozen until homogenization. To harvest proteomes, a small scoop each of 0.5 mm Borosilicate Glass Beads (Next Advance) and 1.4 mm Zirconium Oxide Beads (Precellys) were added to the animals and animals were homogenized in the Bullet Blender (5 min, setting 9, 2X). Cell lysates were then centrifuged (100,000 × g, 45 min) to yield membrane (pellet) and soluble (supernatant) fractions. Membrane pellets were resuspended in PBS by sonication. Protein concentrations were determined using the DC Protein Assay (Bio-Rad) and absorbance read at 750 nm using an Infinite F500 plate reader (Tecan). Proteomes were prepared at 1 mg/mL in PBS.
Gel-based ABPP analysis
Competitive gel-based ABPP experiments were performed as previously described57. HEK293T and C. elegans proteomes (25 μL, 1 mg/mL) were treated with FP-rhodamine (1 or 5 μM, respectively) at 37 °C (30 min or 1 hr, respectively). The reactions were quenched by the addition of 4X SDS-PAGE loading buffer. Samples were visualized by in-gel fluorescence on a ChemiDoc MP flatbed fluorescence scanner (Bio-Rad) with an exposure time between 10–60 s. Rhodamine fluorescence is presented in gray scale. Relative band intensities were quantified using ImageJ software.
Food intake assay
The assay was carried out as previously described58. In brief, approximately 10 age-synchronized C. elegans were cultured at 20 °C in 96-well plates containing S. complete media and irradiated Op50 bacteria (6 mg/mL). Animals were given FUDR (0.6 mM, Sigma) at the L4 developmental stage to prevent the development of progeny. On day 1 of adulthood, animals were treated with JZL184 (50 μM) or DMSO (v/v 0.5%, Sigma) and continuously exposed throughout the assay. Bacterial clearance (ΔOD600) was determined by the difference between OD600 values on day 1 and day 4 of adulthood. Food intake per worm (ΔOD600/X0) was calculated by normalizing the bacterial clearance (ΔOD600) values to the number of worms per well (X0). Statistical analysis was done in Prism. P-values were calculated by two-sided unpaired t test.
Brood size assay
The assay was carried out as previously described56. In brief, age-synchronized N2 C. elegans were cultured at 20 °C in 96-well plates containing S. complete media and irradiated Op50 bacteria (6 mg/mL). On day 1, C. elegans were treated with DMSO or JZL184 (50 μM). Every 24 hours until day 5 of adulthood, the adult animals were transferred to the next well in the plate and the progeny were scored from the previous well. Brood sizes were around 200 and may be lower due to liquid culture.
UPRER Stress imaging
The assay was carried out as previously described56. In brief, age-synchronized SJ4005 hsp-4::gfp reporter animals were cultured at 20 °C in 96-well plates containing S. complete media and irradiated Op50 bacteria (6 mg/mL). To prevent self-fertilization, 5-fluoro-2’- deoxyuridine (FUDR, 0.12 mM final) (Sigma, cat # 856657) was added at the L4 developmental stage. 24 hours before imaging, JZL184 (between 2 – 50 μM) was added to the worms. On day 1, worms in the positive control group were treated with 5 μg/ml tunicamycin and all worms were imaged 8 hours later. GFP fluorescence was imaged with ImageXpress Micro XL High-Content screening system (Molecular Devices) and a 2× objective.
Oxidative stress response assay
Approximately 10 age-synchronized C. elegans were cultured at 20 °C in 96-well plates containing S. complete media and irradiated Op50 bacteria (6 mg/mL). Animals were given FUDR (0.6 mM, Sigma) at the L4 developmental stage to prevent the development of progeny. On day 1 of adulthood, animals were treated with JZL184 (50 μM) or DMSO (v/v 0.5%, Sigma) and continuously exposed throughout the assay. On day 5 of adulthood, Op50 (5 μg, irradiated) was added to each well and animals were exposed to oxidative stress via a solution of paraquat (50 mM). Living animals were scored by eye 48 hours later. Statistical analysis was done in Prism. P-values were calculated by two-sided unpaired t test.
Plasmid construction for recombinant expression
Full-length C. elegans cDNA encoding for F13H6.3, F15A8.6, FAAH-1, FAAH-2, FAAH-3, FAAH-4, FAAH-5, FAAH-6, K11G9.2, Y41D4A.6, Y53F4B.18, and Y71H2AM.13 were cloned into expression vector pRK5 with a C-terminal FLAG-tag using SalI (N-terminal) and NotI (C-terminal) restriction sites. For gene sequences containing an internal SalI restriction site (faah-2 and faah-6), gene products were first subcloned into the pRK5 vector via NotI (N- and C-terminal). Point mutations for synonymous codons were introduced to remove the SalI restriction site using QuikChange site-directed mutagenesis (Stratagene). Mutant clones were then cloned into expression vector pRK5 with a C-terminal FLAG-tag using SalI (N-terminal) and NotI (C-terminal) restriction sites. The plasmid pRK5:FLAG-METAP259 was used as a control vector. All clone sequences were analyzed and verified. For a list of primers used see Supplementary Table 6. Primers were purchased from Integrated DNA Technologies. All sequencing was performed by Eton Biosciences Inc. The pRK5 vector was a gift from David Sabatini (MIT).
Overexpression of proteins
To recombinantly overexpress proteins, HEK293T cells were grown to 40% confluence in 6 well plates and transiently transfected with 2 μg of the desired construct using polyethyleneimine ‘MAX’ (MW 40,000, PEI, Polysciences, Inc.) following the manufacturer’s protocol. ‘Mock’ transfected cells were transfected with 2 μg of pRK5:FLAG-METAP2. Cells were washed in PBS and harvested 48 hours after transfection and cell pellets were flash-frozen and stored at −80 °C until further use.
Western blotting
Cell proteomes were denatured at 95 °C for 5 min and resolved by SDS-PAGE (10% acrylamide), transferred to nitrocellulose membrane (45 V for 120 min), and blocked by 5% milk in TBS-Tween. The primary antibodies used and dilutions are as follows: anti-FLAG (Rabbit, Sigma-Aldrich, F7425, 1:20,000) and anti-GAPDH (Mouse, Santa Cruz, SC-32233, 1:10,000). The secondary antibodies used and dilutions are as follows: HRP-labeled anti-rabbit (Goat, Santa Cruz, SC-2030, 1:5,000) and HRP-labeled anti-mouse (Goat, Santa Cruz, SC-2005, 1:5,000).
Substrate hydrolysis assay
Methods to measure the hydrolytic activities of recombinantly expressed enzymes were adapted from previous protocols14. HEK293T cells overexpressing recombinant proteins were sonicated to obtain lysate, which were diluted in PBS (5 μg protein, 70 μL). Lysates were then treated with JZL184 (50 μM) or DMSO (30 min, 37 °C). Following treatment, a solution of AEA or 2-AG (30 μL, 0.167 mM, sonicated in PBS; final AEA or 2-AG concentration of 50 μM) was added to the lysates and the reaction proceeded for 20 min at 37 °C. The reaction was quenched by adding 2:1 CHCl3:MeOH with 1 nmol of d8-arachidonic acid (AA) internal standard. Samples were vortexed and centrifuged (1,400 × g, 3 min) to separate organic and aqueous phases. The organic phase was collected and subjected to LC-MS/MS analysis. AA was measured as described below.
Sample preparation for metabolomic analysis
C. elegans (20,000) were grown in liquid culture with irradiated Op50 (6 mg/mL). At the L4 developmental stage, animals were washed 6X in PBS and flash frozen until homogenization. For experiments with compound treatment, animals were treated with JZL184 (50 μM) or DMSO from the L2/3 to L4 developmental stage (24 h). To harvest proteomes, a small scoop each of 0.5 mm Borosilicate Glass Beads (Next Advance) and 1.4 mm Zirconium Oxide Beads (Precellys) along with PBS (300 μL) were added to the animals and animals were homogenized in the Bullet Blender (5 min, setting 9, 2X). After homogenization, PBS was added to samples (300 μL) and samples were vortexed and centrifuged (500 × g, 3 min). The supernatant was used to measure protein concentration using the DC Protein Assay (Bio-Rad) and absorbance read at 750 nm using an Infinite F500 plate reader (Tecan). MeOH (600 μL) and lipid standards (1–100 pmol based on endogenous lipid abundance, Cayman Chemical) were added to each sample and samples were vortexed and centrifuged (500 × g, 3 min). Supernatant (1 mL) was added to CHCl3 (1 mL), vortexed and then centrifuged (1,400 × g, 3 min). The organic layer was removed, CHCl3 (1 mL) with formic acid (1:500) was added to the remaining sample, and the extraction was repeated. The combined organic extracts were dried under nitrogen, resuspended in 2:1 v/v CHCl3/MeOH, and stored at −80°C until LC-MS/MS analysis.
Metabolite measurement
Metabolomic analysis was adapted from previous methods14. Metabolites were quantified by targeted metabolomics, using a liquid chromatography with tandem mass spectrometry (LC-MS/MS) based multiple reaction monitoring (MRM) metabolomics platform on the Agilent G6410B Triple-Quad instrument. A Gemini reverse-phase C18 column (50 mm, 4.6 mm with 5 μm diameter particles, Phenomonex) was used to achieve LC separation. Mobile phase A consisted of 95:5 v/v H2O/MeOH and mobile phase B consisted of 60:35:5 v/v/v iPrOH/MeOH/H2O. Ammonium hydroxide (0.1%, negative ionization mode) or formic acid (0.1%, positive ionization mode) were added to mobile phases A and B to assist in ion formation. The flow rate for each positive mode run started at 0.1 mL/min with 100% A and 0% B (5 min). In the next step, the flow rate was increased (0.4 mL/min) and solvent changed to 70% A and 30 % B. The percentage of solvent B increased linearly for 15 min until the solvent system was 0% A and 100% B. Solvent B (100%) continued to flow at 0.5 mL/min for 8 min, followed by equilibration for 3 min with 100% A and 0% B. The negative mode run started with a flow rate of 0.1 mL/min with 100% A and 0% B (5 min). At 5 min, the flow rate was increased (0.4 mL/min) and the percentage of solvent B increased linearly for 15 min until the solvent system was 0% A and 100% B. Following the linear gradient was an isocratic gradient of 100% B at 0.5 mL/min for 8 min. Lastly, the column was equilibrated for 3 min with 100% A and 0% B. During MS analysis, the electrospray ionization source had the following parameters: drying gas temperature = 350 °C, drying gas flow rate = 11 L/min, and the nebulizer pressure = 35 psi. The following parameters were used to measure metabolites by MRM (precursor ion, product ion, collision energy in V, polarity): 2-PG (16:0) (331.2, 239.1, 8, positive), 2-SG (18:0) (359.3, 267.1, 8, positive), 2-OG (18:1) (357.3, 265.1, 8, positive), 2-LG (18:2) (355.3, 263.1, 8, positive), 2-AG (20:4) (379.3, 287.1, 8, positive), 2-EPG (20:5) (377.3, 285.1, 8, positive), 2-AG-d5 (20:4) (384.3, 287.1, 8, positive), PEA (16:0) (300.2, 62, 11, positive), POEA (16:1) (298.3, 62, 11, positive), SEA (18:0) (328.2, 62, 11, positive), OEA (18:1) (326.2, 62, 11, positive), LEA (18:2) (324.2, 62, 11, positive), AEA (20:4) (348.3, 62, 11, positive), EPEA (20:5) (346.3, 62, 11, positive), AEA-d4 (20:4) (352.3, 66, 11, positive), EPEA-d4 (20:5) (350.3, 66, 11, positive), PEA-d4 (16:0) (304.3, 66, 11, positive), PA (16:0) (255, 255, 0, negative), SA (18:0) (283.3, 283.3, 0, negative), OA (18:1) (281.3, 281.3, 0, negative), LA (18:2) (279.2, 279.2, 0, negative), AA (20:4) (303.2, 303.2, 0, negative), DHA (22:6) (327.2, 327.2, 0, negative), and AA-d8 (20:4) (311.3, 311.3, 0, negative). Metabolites were quantified by integrating the area under the peak and comparing values to internal standards.
Generation of CRISPR-mediated FAAH-4 deletion strain
A faah-4 deletion strain (Δfaah-4, OD3609) and y53f4b.18 (Δy53f4b.18, VV213) deletion strain were generated using previously reported methods51. In brief, http://crispr.mit.edu was used to select sgRNA sequences targeting the N- and C- terminus of faah-4 and y53f4b.18 and crRNA was purchased from Integrated DNA Technologies. (faah-4 N terminal sgRNA1: 5´–GAAGCTGTTGATTGAGAAGG–3´, faah-4 C terminal sgRNA2: 5´–TACAGCATTCGGCAACCCGG–3´, y53f4b.18 N terminal sgRNA3: 5’–TACTGTATTTCAGGGGTAAGCGG–3´, y53f4b.18 C terminal sgRNA4: 5’- GGGGTTTAATTTGGGGAGAAAGG–3´) Young N2 adults were injected with a mix of trRNA (28.5 μM), sgRNA1 (14.3 μM), sgRNA2 (14.3 μM) and recombinant Cas9 (28.5 μM). The next day, individual worms were transferred to a new plate. Once progeny (F1) reached the L4 developmental stage, they were transferred to a new plate and genotyped the next day, after eggs were laid. (faah-4 genotyping primers: 5´–CGTGTCGAGACCCAGTAACG -3´, 5´- GCGCCTTAATGGCATGAATA–3´, 5´–CGAAGGAGATTTGCTTCTGAAA–3´, WT: 548 bp, deletion: 794 bp, purchased from Eton Biosciences Inc). (y53f4b.18 genotyping primers: 5´–ACCGACGGTATCCTGCTCTT–3´, 5´–GAGACCTTTGGATATTTGTTGGA–3´, 5´–TAACCGTTGCCTTGCTCTCT–3´, WT: 530 bp, deletion: 943 bp, purchased from Eton Biosciences Inc). The homozygous deletion strain was sequenced and backcrossed to the N2 strain 6 times. For a list of primers see Supplementary Table 6.
Compound synthesis
FP-biotin and FP-rhodamine were synthesized in-house as previously described16,60. Compounds used in the screen that have been published were synthesized in-house. See Supplementary Table 1 for information on the published compounds and Synthetic Methods for synthesis and characterization of unpublished compounds.
Data availability
The data that support the findings of this study are available within the paper [and its supplementary information files] or from the corresponding author upon reasonable request.
Supplementary Material
Acknowledgments
We are grateful to M. Hansen, J. Chang, X. She, and G. Solis for discussions and technical expertise in C. elegans and the Caenorhabditis Genetics Center for strains. This work was supported by the NIH (DA033760, CA215249), the Damon Runyon Cancer Research Foundation, and Abide Therapeutics.
Footnotes
Competing interests.
B.F.C. is a founder and advisor to Abide Therapeutics, a biotechnology company interested in developing serine hydrolase inhibitors and therapeutics.
References:
- 1.Finkel T The metabolic regulation of aging. Nat Med 21, 1416–1423, doi: 10.1038/nm.3998 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Imai S & Guarente L NAD+ and sirtuins in aging and disease. Trends Cell Biol 24, 464–471, doi: 10.1016/j.tcb.2014.04.002 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson SC, Rabinovitch PS & Kaeberlein M mTOR is a key modulator of ageing and age-related disease. Nature 493, 338–345, doi: 10.1038/nature11861 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yoshino J, Mills KF, Yoon MJ & Imai S Nicotinamide mononucleotide, a key NAD(+) intermediate, treats the pathophysiology of diet- and age-induced diabetes in mice. Cell Metab 14, 528–536, doi: 10.1016/j.cmet.2011.08.014 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kenyon C, Chang J, Gensch E, Rudner A & Tabtiang R A C. elegans mutant that lives twice as long as wild type. Nature 366, 461–464, doi: 10.1038/366461a0 (1993). [DOI] [PubMed] [Google Scholar]
- 6.Hamilton B et al. A systematic RNAi screen for longevity genes in C. elegans. Genes Dev 19, 1544–1555, doi: 10.1101/gad.1308205 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garigan D et al. Genetic analysis of tissue aging in Caenorhabditis elegans: a role for heat-shock factor and bacterial proliferation. Genetics 161, 1101–1112 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Evason K, Huang C, Yamben I, Covey DF & Kornfeld K Anticonvulsant medications extend worm life-span. Science 307, 258–262, doi: 10.1126/science.1105299 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Petrascheck M, Ye X & Buck LB An antidepressant that extends lifespan in adult Caenorhabditis elegans. Nature 450, 553–556, doi: 10.1038/nature05991 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Bachovchin DA et al. Superfamily-wide portrait of serine hydrolase inhibition achieved by library-versus-library screening. Proc Natl Acad Sci U S A 107, 20941–20946, doi: 10.1073/pnas.1011663107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Backus KM et al. Proteome-wide covalent ligand discovery in native biological systems. Nature 534, 570–574, doi: 10.1038/nature18002 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gruner BM et al. An in vivo multiplexed small-molecule screening platform. Nat Methods 13, 883–889, doi: 10.1038/nmeth.3992 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roberts AM et al. Chemoproteomic Screening of Covalent Ligands Reveals UBA5 As a Novel Pancreatic Cancer Target. ACS Chem Biol 12, 899–904, doi: 10.1021/acschembio.7b00020 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Hsu KL et al. DAGLbeta inhibition perturbs a lipid network involved in macrophage inflammatory responses. Nat Chem Biol 8, 999–1007, doi: 10.1038/nchembio.1105 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cognetta AB 3rd et al. Selective N-Hydroxyhydantoin Carbamate Inhibitors of Mammalian Serine Hydrolases. Chem Biol 22, 928–937, doi: 10.1016/j.chembiol.2015.05.018 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Y, Patricelli MP & Cravatt BF Activity-based protein profiling: the serine hydrolases. Proc Natl Acad Sci U S A 96, 14694–14699 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Niphakis MJ & Cravatt BF Enzyme inhibitor discovery by activity-based protein profiling. Annu Rev Biochem 83, 341–377, doi: 10.1146/annurev-biochem-060713-035708 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Lucanic M et al. N-acylethanolamine signalling mediates the effect of diet on lifespan in Caenorhabditis elegans. Nature 473, 226–229, doi: 10.1038/nature10007 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin YH et al. Diacylglycerol lipase regulates lifespan and oxidative stress response by inversely modulating TOR signaling in Drosophila and C. elegans. Aging Cell 13, 755–764, doi: 10.1111/acel.12232 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang MC, O’Rourke EJ & Ruvkun G Fat metabolism links germline stem cells and longevity in C. elegans. Science 322, 957–960, doi: 10.1126/science.1162011 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Folick A et al. Aging. Lysosomal signaling molecules regulate longevity in Caenorhabditis elegans. Science 347, 83–86, doi: 10.1126/science.1258857 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rangaraju S et al. Suppression of transcriptional drift extends C. elegans lifespan by postponing the onset of mortality. Elife 4, e08833, doi: 10.7554/eLife.08833 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adibekian A et al. Click-generated triazole ureas as ultrapotent in vivo-active serine hydrolase inhibitors. Nat Chem Biol 7, 469–478, doi: 10.1038/nchembio.579 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang JW, Cognetta AB 3rd, Niphakis MJ & Cravatt BF Proteome-wide reactivity profiling identifies diverse carbamate chemotypes tuned for serine hydrolase inhibition. ACS Chem Biol 8, 1590–1599, doi: 10.1021/cb400261h (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kamat SS et al. Immunomodulatory lysophosphatidylserines are regulated by ABHD16A and ABHD12 interplay. Nat Chem Biol 11, 164–171, doi: 10.1038/nchembio.1721 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arantes-Oliveira N, Apfeld J, Dillin A & Kenyon C Regulation of life-span by germ-line stem cells in Caenorhabditis elegans. Science 295, 502–505, doi: 10.1126/science.1065768 (2002). [DOI] [PubMed] [Google Scholar]
- 27.Long JZ et al. Selective blockade of 2-arachidonoylglycerol hydrolysis produces cannabinoid behavioral effects. Nat Chem Biol 5, 37–44, doi: 10.1038/nchembio.129 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grabner GF, Zimmermann R, Schicho R & Taschler U Monoglyceride lipase as a drug target: At the crossroads of arachidonic acid metabolism and endocannabinoid signaling. Pharmacol Ther 175, 35–46, doi: 10.1016/j.pharmthera.2017.02.033 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kathuria S et al. Modulation of anxiety through blockade of anandamide hydrolysis. Nat Med 9, 76–81, doi: 10.1038/nm803 (2003). [DOI] [PubMed] [Google Scholar]
- 30.Shin S et al. Characterization of a novel Ser-cisSer-Lys catalytic triad in comparison with the classical Ser-His-Asp triad. J Biol Chem 278, 24937–24943, doi: 10.1074/jbc.M302156200 (2003). [DOI] [PubMed] [Google Scholar]
- 31.Cravatt BF et al. Molecular characterization of an enzyme that degrades neuromodulatory fatty-acid amides. Nature 384, 83–87, doi: 10.1038/384083a0 (1996). [DOI] [PubMed] [Google Scholar]
- 32.Giang DK & Cravatt BF Molecular characterization of human and mouse fatty acid amide hydrolases. Proc Natl Acad Sci U S A 94, 2238–2242 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Curnow AW et al. Glu-tRNAGln amidotransferase: a novel heterotrimeric enzyme required for correct decoding of glutamine codons during translation. Proc Natl Acad Sci U S A 94, 11819–11826 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chang JW et al. Highly selective inhibitors of monoacylglycerol lipase bearing a reactive group that is bioisosteric with endocannabinoid substrates. Chem Biol 19, 579–588, doi: 10.1016/j.chembiol.2012.03.009 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dai DF, Chiao YA, Marcinek DJ, Szeto HH & Rabinovitch PS Mitochondrial oxidative stress in aging and healthspan. Longev Healthspan 3, 6, doi: 10.1186/2046-2395-3-6 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Greer EL & Brunet A Different dietary restriction regimens extend lifespan by both independent and overlapping genetic pathways in C. elegans. Aging Cell 8, 113–127, doi: 10.1111/j.1474-9726.2009.00459.x (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Altenhoff AM et al. The OMA orthology database in 2018: retrieving evolutionary relationships among all domains of life through richer web and programmatic interfaces. Nucleic Acids Res, doi: 10.1093/nar/gkx1019 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dolinski K & Botstein D Orthology and functional conservation in eukaryotes. Annu Rev Genet 41, 465–507, doi: 10.1146/annurev.genet.40.110405.090439 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Omelchenko MV, Galperin MY, Wolf YI & Koonin EV Non-homologous isofunctional enzymes: a systematic analysis of alternative solutions in enzyme evolution. Biol Direct 5, 31, doi: 10.1186/1745-6150-5-31 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bandyopadhyay S, Sharan R & Ideker T Systematic identification of functional orthologs based on protein network comparison. Genome Res 16, 428–435, doi: 10.1101/gr.4526006 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kurnasov O et al. NAD biosynthesis: identification of the tryptophan to quinolinate pathway in bacteria. Chem Biol 10, 1195–1204 (2003). [DOI] [PubMed] [Google Scholar]
- 42.Martell J et al. Global Cysteine-Reactivity Profiling during Impaired Insulin/IGF-1 Signaling in C. elegans Identifies Uncharacterized Mediators of Longevity. Cell Chem Biol 23, 955–966, doi: 10.1016/j.chembiol.2016.06.015 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Han S et al. Mono-unsaturated fatty acids link H3K4me3 modifiers to C. elegans lifespan. Nature 544, 185–190, doi: 10.1038/nature21686 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O’Rourke EJ, Kuballa P, Xavier R & Ruvkun G omega-6 Polyunsaturated fatty acids extend life span through the activation of autophagy. Genes Dev 27, 429–440, doi: 10.1101/gad.205294.112 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shmookler Reis RJ et al. Modulation of lipid biosynthesis contributes to stress resistance and longevity of C. elegans mutants. Aging (Albany NY) 3, 125–147, doi: 10.18632/aging.100275 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oakes MD, Law WJ, Clark T, Bamber BA & Komuniecki R Cannabinoids Activate Monoaminergic Signaling to Modulate Key C. elegans Behaviors. J Neurosci 37, 2859–2869, doi: 10.1523/JNEUROSCI.3151-16.2017 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ogasawara D et al. Rapid and profound rewiring of brain lipid signaling networks by acute diacylglycerol lipase inhibition. Proc Natl Acad Sci U S A 113, 26–33, doi: 10.1073/pnas.1522364112 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fagan SG & Campbell VA The influence of cannabinoids on generic traits of neurodegeneration. Br J Pharmacol 171, 1347–1360, doi: 10.1111/bph.12492 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bilkei-Gorzo A The endocannabinoid system in normal and pathological brain ageing. Philos Trans R Soc Lond B Biol Sci 367, 3326–3341, doi: 10.1098/rstb.2011.0388 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Piyanova A et al. Age-related changes in the endocannabinoid system in the mouse hippocampus. Mech Ageing Dev 150, 55–64, doi: 10.1016/j.mad.2015.08.005 (2015). [DOI] [PubMed] [Google Scholar]
- 51.Paix A, Folkmann A, Rasoloson D & Seydoux G High Efficiency, Homology-Directed Genome Editing in Caenorhabditis elegans Using CRISPR-Cas9 Ribonucleoprotein Complexes. Genetics 201, 47–54, doi: 10.1534/genetics.115.179382 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rangaraju S, Solis GM & Petrascheck M High-throughput small-molecule screening in Caenorhabditis elegans. Methods Mol Biol 1263, 139–155, doi: 10.1007/978-1-4939-2269-7_11 (2015). [DOI] [PubMed] [Google Scholar]
- 53.Hulce JJ, Cognetta AB, Niphakis MJ, Tully SE & Cravatt BF Proteome-wide mapping of cholesterol-interacting proteins in mammalian cells. Nat Methods 10, 259–264, doi: 10.1038/nmeth.2368 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rostovtsev VV, Green LG, Fokin VV & Sharpless KB A stepwise huisgen cycloaddition process: copper(I)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angew Chem Int Ed Engl 41, 2596–2599, doi: (2002). [DOI] [PubMed] [Google Scholar]
- 55.Weerapana E et al. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature 468, 790–795, doi: 10.1038/nature09472 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Solis GM et al. Translation attenuation by minocycline enhances longevity and proteostasis in old post-stress-responsive organisms. Elife 7, doi: 10.7554/eLife.40314 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jessani N et al. A streamlined platform for high-content functional proteomics of primary human specimens. Nat Methods 2, 691–697, doi: 10.1038/nmeth778 (2005). [DOI] [PubMed] [Google Scholar]
- 58.Gomez-Amaro RL et al. Measuring Food Intake and Nutrient Absorption in Caenorhabditis elegans. Genetics 200, 443–454, doi: 10.1534/genetics.115.175851 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bar-Peled L et al. A Tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science 340, 1100–1106, doi: 10.1126/science.1232044 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Patricelli MP, Giang DK, Stamp LM & Burbaum JJ Direct visualization of serine hydrolase activities in complex proteomes using fluorescent active site-directed probes. Proteomics 1, 1067–1071, doi: (2001). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available within the paper [and its supplementary information files] or from the corresponding author upon reasonable request.


