Abstract
The first high resolution structural views of group II intron maturases illuminate the architectural and functional roles of these multidomain proteins in splicing and DNA invasion. The maturases show striking structural and functional homology to a central protein involved in spliceosomal premessenger RNA splicing, solidifying the idea that group II introns and the spliceosome descended from a common ancestor, an ancient retroelement.
Group II introns comprise an ancient class of mobile genetic elements found in bacteria and eukaryotic organelles1. These elements mobilize as a catalytic RNA intron and an intron encoded protein (IEP), or maturase, that together form a ribonucleoprotein (RNP) complex that mediates splicing and DNA invasion. The RNA intron acts as a ribozyme that catalyzes its own excision from precursor RNA transcripts through a branching pathway analogous to that used for removal of spliceosomal introns, yielding spliced exons and an excised intron lariat. The IEP assists splicing by directly binding the catalytically active form of the RNA intron2. Following splicing, the intronic RNP invades DNA in a remarkable process called retrohoming. During retrohoming, the RNA lariat intron, again aided by the IEP, invades a DNA strand directly by reverse splicing, and then the IEP cleaves the opposite strand and, utilizing a reverse transcriptase domain with homology to non-LTR-retrotransposons, reverse transcribes the inserted RNA intron back into DNA. The first structure of a group II RNA intron was solved nearly ten years ago3 and revealed a catalytic mechanism that has proven indistinguishable from the catalytic mechanism of the spliceosome4. However, the first structure of an IEP has remained elusive, obscuring its mechanism in splicing and DNA invasion. Now, on pages XXX and YYY of this issue, two reports provide the first views of group II intron IEPs. Zhao and Pyle report crystal structures of reverse transcriptase (RT) domains from two IEPs found in Roseburia intestinalis (R.i.) and Eubacterium rectale at 1.2 Å and 2.1 Å, respectively, and Qu et al. report the cryo-EM structure of an RNP complex comprising an endogenously spliced group II intron bound to its IEP from Lactococcus lactis at 3.8 Å. The structures not only reveal fundamental features of group II IEPs and their role in splicing and DNA invasion but also extend the parallels between group II and spliceosomal introns beyond RNA to include key protein cofactors, all but cementing their relationship as evolutionary cousins.
Structures of IEPs have remained elusive, due to the low solubility and stability of these proteins, which Zhao and Pyle attributed to excessive positive charge. To circumvent this limitation, Zhao and Pyle invoked a clever strategy to evaluate alternative IEPs for crystallization, based on their isoelectric point, arginine content, and predicted secondary structure. Remarkably, the top two candidates crystallized. The solved structures reveal a hand-like configuration of the RT module that is characteristic of polymerases and includes the finger and palm domains, although the associated thumb domain was lost during expression due to proteolysis (Figs. 1 and 2).
Figure 1.
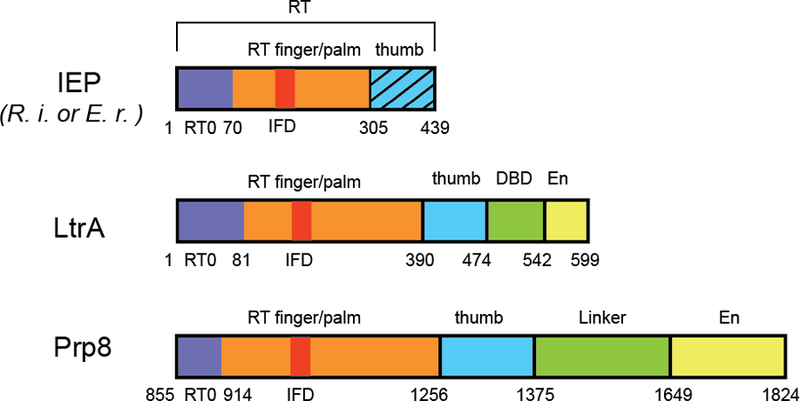
Comparison of domain architecture of group II IEPs and Prp8. RT, reverse transcriptase domain; DBD, DNA binding domain; EN, endonuclease domain. The hatched pattern in the IEP represents the thumb domain missing in the structures of Zhao and Pyle. Image courtesy of Yaming Shao.
Figure 2.
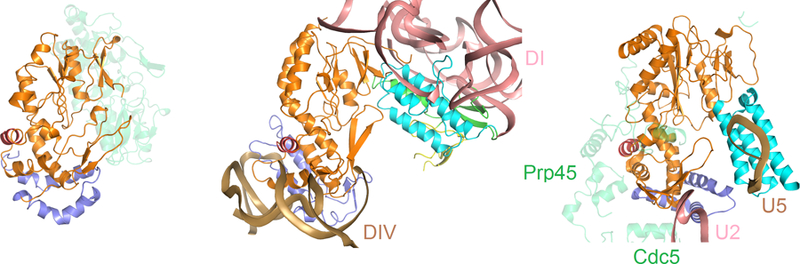
Comparison of RT and RT-like domains from R.i., LtrA and Prp8. Protein domains and motifs are color coded as in Fig. 1. Left: R.i. RT dimer with one protomer colored transparent green. Middle: RNP containing LtrA IEP and group II intron. DI and DIV are colored in salmon and sand, respectively. Right: The RT-like fragment of Prp8 (PDB ID: 3JB9). Interacting proteins are labeled and shown as transparent green; U2, U5 spliceosomal RNA components are shown in salmon and sand, respectively. Image courtesy of Yaming Shao.
The core of the R.i. RT domain, the focus of the study, shows significant structural similarity to telomerase RT (TERT), the RNA-dependent RNA polymerase of hepatitis C virus (HCV), and the RT-like domain of the core splicing factor Prp8, which resides at the heart of the spliceosome5,6. In addition to the core RT domain, group II intron and non-LTR retrotransposon RTs conserve an N-terminal extension (RT0) and an insertion in the finger domain (IFD). Strikingly, when these characteristic domains are included in structural comparisons, the R.i. RT showed the strongest similarity to Prp8 (Fig. 2). While sequence and structural analysis of Prp8 has suggested similarity with group II intron IEPs6,7, the R.i. structure leaves no doubt about their uncanny resemblance and solidifies their evolutionary ties.
Surprisingly, the R.i. RT exhibits almost as strong structural similarity with the RNA-dependent RNA polymerase of hepatitis C virus (HCV) – a similarity that is much stronger than the similarity with other RTs, such as TERT and HIV RT. Zhao and Pyle suggest that this similarity, first recognized phylogenetically8, indicates that the RT of group II introns evolved from a flavivirus RNA polymerase and that more distally related RTs evolved along a distinct lineage.
Curiously, the crystal structure also reveals that the R.i. RT dimerizes through an extensive interface that buries over 1500 Å of surface area, and the authors establish that the dimer is functional for binding to the anchor point on group II intron RNA. Intriguingly, dimerization of the R.i. RT parallels dimerization of LtrA, as implicated by biochemical studies9,10, suggesting a conserved role for dimerization in splicing and/or DNA invasion.
In the second study, Qu et al. circumvented the challenges associated with expression of the LtrA IEP by isolating the protein from its native lactococcal host – and bound to its cognate group II intron RNA, to boot. They isolated this RNP by relocating the open reading frame for LtrA from its usual position within the intron to a downstream location fused to an affinity tag via an intein11. Affinity purification from extracts followed by intein-mediated cleavage yielded an RNP containing the 902-nt lariat RNA intron and 599-amino acid IEP. These particles exhibited activity in splicing and several steps associated with retrohoming, including binding, invasion and cleavage of target DNA. 3D reconstruction by cryo-EM analysis elucidated the global architecture of the RNA alone and for the first time in complex with the IEP.
Analogous to previously solved structures of group II introns3,12–14, conserved motifs and some distinct substructures organize the six characteristic domains (DI-DVI) into an RNA tertiary structure that brings the splice site recognition elements within DI and the branch point (BP) residue within DVI into juxtaposition with the catalytic center, a two-metal-ion motif located within DV that bears striking resemblance to the catalytic RNA core of the spliceosome3. LtrA, the best-characterized IEP9,15, consists of four domains16: a palm/fingers domain and thumb (X) domain of the RT; a DNA binding domain (DBD), and DNA endonuclease (EN) domain (Fig. 1) that together form an extensive bipartite RNA-binding surface consistent with biochemical studies (Figure 2). On one side, LtrA anchoring involves interactions between intron DIV and the RT fingers/palm domain, especially via the aforementioned RT0 extension. On the other side, the IEP interacts with DI, including its exon recognition motifs via an interaction surface that spans EN, DBD, the thumb domain and the C-terminal half of the fingers/palm domain. In addition to the structural homology with other RT’s as noted for the R.i. protein, the LtrA structure extends the parallels to spliceosomal Prp8 even further. Not only do both proteins present a similar overall domain organization (Fig. 1) and RT fold (Fig. 2), but the thumb domains both appear to adopt a similar topological arrangement with respect to the catalytic center and the substrate binding elements of the group II intron and spliceosome, respectively (Fig. 3). Most notably, the LtrA and Prp8 thumb domains interact with the EBS1 loop and U5 loop 1, respectively, functionally equivalent components involved in direct recognition of the 5’ splice site (Fig 3).
Figure 3.
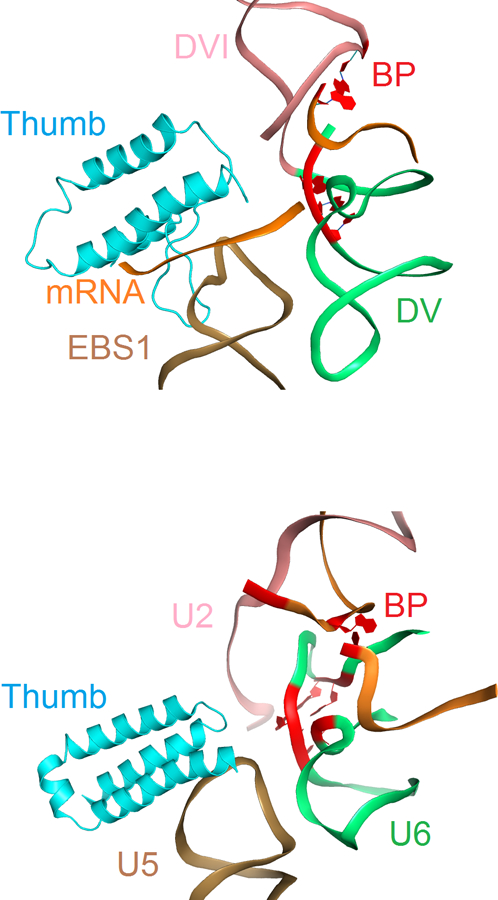
Spatial relationship of thumb domain and RNA active site in group II intron RNP and spliceosome. Top: Intron RNP showing thumb domain interaction with exon binding site 1 (EBS1) relative to the DV helix and branch point adenosine in DVI. Bottom: Spliceosome from Schizosaccharomyces pombe (PDB ID: 3JB9) showing thumb domain interaction with U5 loop 1 relative to U6 helix and branch point adenosine. Thumb domains of LtrA and Prp8 are shown in cyan and other parts of the proteins are omitted. RNAs are shown as ribbons. EBS1 and U5 are shown in sand; domain V and U6 RNA are shown in green; domain VI and U2 RNA are colored in salmon; mRNA, observed in a fraction of the group II is colored in orange. Nucleotides that react to form the branch point (BP) and nucleotides involved in the group II intron and spliceosome catalysis are shown in red. Image courtesy of Yaming Shao.
Given the evidence for dimerization of both the R.i. and LtrA RTs, it is surprising that the cryo-EM structure reveals only a monomer of LtrA bound to the group II intron. Conceivably, dimerization may serve a specific function at a stage of retrohoming distinct from that captured by the endogenous LtrA RNP particle. In this regard, the cryo-EM structure reveals that the RT and endonuclease catalytic cores are 45 Å away from one another, which would require a substantial conformational rearrangement of LtrA or the RNA-DNA substrate to initiate reverse transcription at the site of DNA cleavage. Curiously, dimerization of LtrA according to the R.i. RT structure juxtaposes the RT and endonuclease domains, hinting that the dimer may function at the stage of reverse transcription.
Together, this pair of landmark studies has raised new questions and established a structural framework that is certain to inspire deep mechanistic studies of group II intron splicing and retrohoming. Further, given strong sequence similarity between group II intron RTs and the structurally uncharacterized RTs of non-LTR retroelements, the group II intron RT structures shed new light on these ubiquitous elements that have sculpted over 15% of the human genome. Perhaps most significantly, though, the structures extend previously recognized parallels between group II intron and spliceosomal RNA structure to include not only the structures of central protein cofactors but also the interactions between these cofactors and catalytic RNA (Fig. 3). As such, the structures illuminate the final piece of an evolutionary puzzle and define an ancestral RNP from which group II introns and the spliceosome evolved. Indeed, the work supports the view that spliceosomal small nuclear RNAs derived from fragmentation17 of an ancient, group II intron ribozyme but also that a central RT cofactor co-evolved to maintain splicing activity, while retrohoming activities degenerated.
References
- 1.Lambowitz AM & Belfort M Mobile Bacterial Group II Introns at the Crux of Eukaryotic Evolution. Microbiol Spectr 3, MDNA3-0050-2014 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matsuura M, Noah JW & Lambowitz AM Mechanism of maturase-promoted group II intron splicing. EMBO J 20, 7259–70 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Toor N, Keating KS, Taylor SD & Pyle AM Crystal structure of a self-spliced group II intron. Science 320, 77–82 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fica SM et al. RNA catalyses nuclear pre-mRNA splicing. Nature 503, 229–34 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grainger RJ & Beggs JD Prp8 protein: at the heart of the spliceosome. RNA 11, 533–57 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Galej WP, Oubridge C, Newman AJ & Nagai K Crystal structure of Prp8 reveals active site cavity of the spliceosome. Nature 493, 638–43 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dlakic M & Mushegian A Prp8, the pivotal protein of the spliceosomal catalytic center, evolved from a retroelement-encoded reverse transcriptase. RNA 17, 799–808 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiong Y & Eickbush TH Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J 9, 3353–62 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saldanha R et al. RNA and protein catalysis in group II intron splicing and mobility reactions using purified components. Biochemistry 38, 9069–83 (1999). [DOI] [PubMed] [Google Scholar]
- 10.Rambo RP & Doudna JA Assembly of an active group II intron-maturase complex by protein dimerization. Biochemistry 43, 6486–97 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Gupta K et al. Quaternary arrangement of an active, native group II intron ribonucleoprotein complex revealed by small-angle X-ray scattering. Nucleic Acids Res 42, 5347–60 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marcia M & Pyle AM Visualizing group II intron catalysis through the stages of splicing. Cell 151, 497–507 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robart AR, Chan RT, Peters JK, Rajashankar KR & Toor N Crystal structure of a eukaryotic group II intron lariat. Nature 514, 193–7 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Toor N, Rajashankar K, Keating KS & Pyle AM Structural basis for exon recognition by a group II intron. Nat Struct Mol Biol 15, 1221–2 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wank H, SanFilippo J, Singh RN, Matsuura M & Lambowitz AM A reverse transcriptase/maturase promotes splicing by binding at its own coding segment in a group II intron RNA. Mol Cell 4, 239–50 (1999). [DOI] [PubMed] [Google Scholar]
- 16.Blocker FJ et al. Domain structure and three-dimensional model of a group II intron-encoded reverse transcriptase. RNA 11, 14–28 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharp PA “Five easy pieces”. Science 254, 663 (1991). [DOI] [PubMed] [Google Scholar]


