Abstract
We sought to identify susceptibility genes for high-grade serous ovarian cancer (HGSOC) by performing a transcriptome-wide association study (TWAS) of gene expression and splice junction usage in HGSOC-relevant tissue types (N = 2,169) and the largest GWAS available for HGSOC (N = 13,037 cases/40,941 controls). We identified 25 TWAS significant genes, 7 at the junction level only, including LRRC46 at 19q21.32, (P = 1 × 10−9), CHMP4C at 8q21 (P = 2 × 10−11), and a PRC1 junction at 15q26 (P = 7 × 10−9). In vitro assays for CHMP4C showed the associated variant induces allele specific exon inclusion (P = 0.0024). Functional screens in HGSOC cell lines found evidence of essentiality for three of the novel genes we identified: HAUS6, KANSL1 and PRC1, with the latter comparable to CMYC. Our study implicated at least one target gene for 6/13 distinct GWAS regions, identifying 23 novel candidate susceptibility genes for HGSOC.
Keywords: epithelial ovarian cancer (EOC), high-grade serous ovarian cancer (HGSOC), RNA-sequencing (RNA-seq), expression quantitative trait locus analysis (eQTL), splice site quantitative trait locus analysis (spQTL), ovarian surface epithelial cells (OSECs), fallopian tube secretory epithelial cells (FTSECs), Genome wide association studies (GWAS), The Cancer Genome Atlas (TCGA), Transcriptome-wide association study (TWAS), splice-Transcriptome-wide association study (spTWAS)
INTRODUCTION
Invasive epithelial ovarian cancer (EOC) is a heterogeneous disease with a major heritable component 1. There are several histological subtypes of invasive EOC, each associated with different genetic and epidemiological risk factors, clinical features and likely cells of origin. High grade serous ovarian cancer (HGSOC) is the most common histotype, representing about two-thirds of cases. Highly penetrant germline mutations in the homology directed repair genes including BRCA1 and BRCA2 are the most significant genetic risk factors for HGSOC, but only account for about 10% of cases 1. A major fraction of the remaining EOC risk is due to common, low penetrance risk alleles, and over the last few years, genome-wide association studies (GWASs) have identified 39 different regions of the genome associated with EOC risk, mainly in European populations 2–13.
Typically, SNPs associated with disease risk are located in the non-protein coding genome suggesting they function by altering the activity of non-coding biofeatures (e.g. DNA enhancers, non-coding RNAs) that regulate gene expression. Expression quantitative trait locus (eQTL) analysis can be used to identify associations between risk genotypes and gene expression and several studies have successfully used this approach to identify putative susceptibility genes at GWAS risk loci 2,3,14,15, including in epithelial ovarian cancer 2,3,6–9,14,15. Recently, transcriptome-wide association studies (TWASs) have been proposed as a principled approach to integrate eQTL analyses with GWAS to identify genes whose genetically regulated expression is associated to disease risk 16–20. For a given gene, TWASs use eQTL data to ‘impute’ the total expression across a large cohort of genotyped individuals followed by a test of association with disease risk. TWAS may additionally increase power versus single SNP association testing either by reducing the multiple testing burden or aggregating multiple expression-altering variants into a single test. However, TWASs may also identify significant associations due to pleiotropy between the expression-altering and risk-altering variants or variants they tag 16,21. A TWAS is therefore a first step to prioritize putative target genes, with experimental validation needed to establish causality.
In the current study we established the most comprehensive genome-wide genotype-gene expression datasets available, with >2,000 eQTL samples in primary HGSOCs, EOC precursor tissues (ovarian and fallopian epithelial cells) and other hormonal-related cancers (breast and prostate cancer), to perform multi-tissue TWASs in the largest ovarian cancer GWAS available (N > 50,000). In addition to performing traditional TWAS that test for association at total abundance levels, we extend the TWAS methodology to integrate splice-QTLs by also testing exon junction levels for association to EOC (‘spTWAS’). We identify 25 genes whose expression is significantly associated to EOC risk at genome-wide significance, 7 of which are only significant in the spTWAS, thus underscoring the utility of incorporating splicing association analyses. We use in vitro assays to validate the functional significance of splice-QTL associations for CHMP4C, and evaluate a gene knockout screen in HGSOC 22 to establish the functional essentiality for three of the novel spTWAS genes we identified: HAUS6, KANSL1 and PRC1.
RESULTS
Genetic control of gene expression after tumorigenesis
We first investigated the genetic control of gene expression in EOC precursor tissues and HGSOCs. We assayed genotype, gene expression, and quantified splicing data for 115 primary normal ovarian surface epithelial cells (OSECs), 70 primary normal fallopian tube epithelial cells (FTSECs), and 394 primary HGSOCs profiled by The Cancer Genome Atlas (TCGA). FTSECs are a likely precursor cell type for the majority of HGSOCs 23–27 while OSECs are also a postulated cellular origin for the disease 28–33. We quantified the SNP-heritability (h2g) and genetic correlation (rg) of gene expression and splicing between pairs of tissues (see Methods). For a given tissue, cis-h2g is defined as the fraction of phenotypic variance explained by SNPs within 500 kb of the gene boundary. For a pair of tissues, cis-rg is defined as the correlation of causal genetic effects on expression across all SNPs within 500 kb of the gene boundary. The average cis-h2g was significant in all tissues with an average of 0.026 for overall expression and 0.021 for splice variation, similar to previous observations across different tissues (Supplementary Table 1). There was a higher mean cis-rg between FTSECs and HGSOCs (rg = 0.071, standard error [s.e.] = 0.031; Table 1) than between OSECs and HGSOCs (rg = −0.022, s.e. = 0.029) consistent with the hypothesis that FTSECs are the more likely precursor cell type for HGSOC. We observed a similar, albeit non-significant, trend for heritable splicing events, with a genetic correlation of 0.024 (s.e. = 0.016) between HGSOCs and FTSECs, and −0.018 (s.e. = 0.013) between OSECs and HGSOC. There was a greater genetic correlation for both overall gene expression and splicing events between OSECs and FTSECs (rg = 0.359, s.e. = 0.046 for overall expression; rg = 0.302, s.e. = 0.023 for splicing) indicating that genetic control of gene expression is altered during tumorigenesis. Lastly, we evaluated rg between four molecular subtypes of HGSOCs characterized by gene expression signatures by TCGA 34 but observed no significant divergence from 1.0 and few individually significant genes (Supplementary Table 2, 3). We therefore treated all HGSOCs as a single group for subsequent analyses.
Table 1. Genetic correlation of expression and splicing in ovarian tissues.
Genetic variance/covariance was estimated across all significantly heritable genes (in any panel) using HE-regression, averaged, and transformed to genetic correlation (rg). Standard error shown in parentheses. Sample size: 201 tumor HGSOC, 70 normal FTSEC, 115 normal OSEC.
| Expression | Overall rg | (se) | |
|---|---|---|---|
| Tumor HGSOC | Normal OSEC | −0.022 | (0.029) |
| Tumor HGSOC | Normal FTSEC | 0.071 | (0.031) |
| Normal OSEC | Normal FTSEC | 0.359 | (0.046) |
Cross-cohort validation of TWAS models
We investigated the utility of the expression data assayed in OSECs, FTSECs and HGSOC and other hormonally-regulated cancers (breast and prostate cancers) in building prediction models for TWAS (Figure 1). In addition to OSEC, FTSEC and HGSOC data, we included RNA-seq data from 1,027 primary breast tumors and 84 matched normal precursor tissues, and 483 primary prostate cancers from TCGA 2,9,35. For TCGA cohorts, we also used exon junction events as a measure of alternative splicing to identify predictors that may not be observed through total expression (see Methods). We defined a “panel” as a tissue-state-phenotype triplet (e.g. prostate-tumor-splicing) and performed normalization, correction, and model building within each panel separately so as not to induce confounding due to cross-panel differences. A strength of the TWAS approach is that it is immune to reverse-causal effects of disease on gene expression (independent of genetics), and we show both theoretically and by simulation that this holds for case-only study design (Supplementary Note, Supplementary Figure 1). Each panel underwent stringent quality control and included as covariates genetic and gene expression/splicing principal components, local somatic structural variation, and relevant clinical factors (Supplementary Note, Supplementary Table 4). Accounting for local structural variants significantly increased gene expression heritability for ovarian and breast tumors (Supplementary Figure 2), but tumor/pathology features such as purity, grade/stage, and hormone receptor status did not show substantial genetic heterogeneity except in the case of ovarian tumor expression from BRCA1/2 somatic mutation carriers (Supplementary Note, Supplementary Figure 2, Supplementary Table 3, 5).
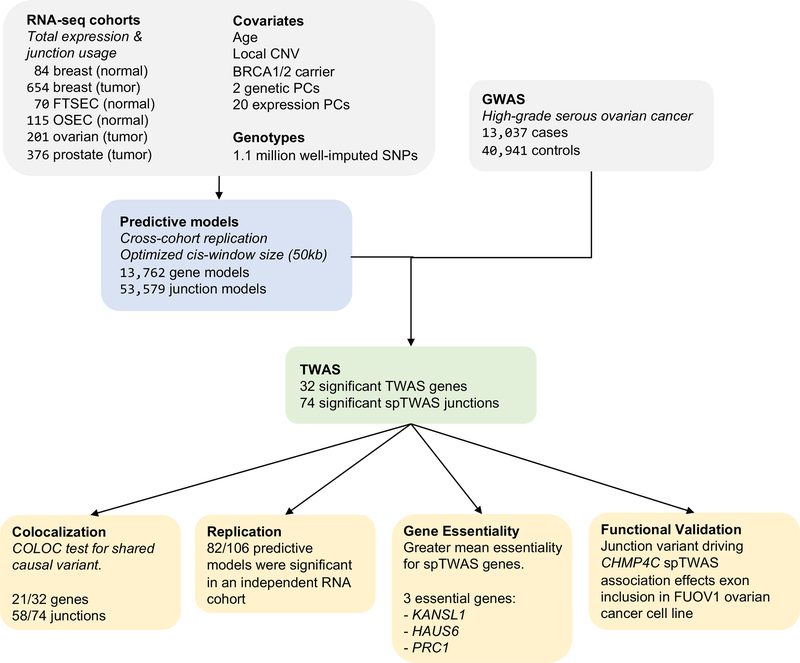
Figure 1. Study schema.
Overview of the analytic workflow. CNV, copy number variation; FTSEC, fallopian tube secretory epithelial cell; OSEC, ovarian surface epithelial cell; PC, principal component; spTWAS, splice transcriptome-wide association study; TWAS, transcriptome-wide association study.
A total of 13,762 significantly heritable genes and 53,579 significantly cis-heritable exon junction events were identified across all cohorts (cis-h2g P < 0.01, for detailed evaluation of cis locus and heritability parameters see Supplementary Note, Supplementary Table 6, Supplementary Figure 3, 4). We then trained multiple penalized predictive models using all SNPs in a locus and evaluated predictive accuracy by five-fold cross-validation against the actual measured expression (see Methods). Mean cross-validated predictor R2 was 0.066 (s.e. 4 × 10−4) and highly significant (median predictor cross-validation P = 4.6 × 10−4), consistent with previous findings that low average heritability of gene expression can be compensated for by sample size sufficient to produce reliable genetic predictors (Supplementary Table 7). We then leveraged the multiple independent cohorts analyzed here to assess replication rates for our predictive models with out-of-sample gene expression (Supplementary Note, Supplementary Table 8, 9). Both out-of-sample correlation and model significance were high across all cohorts and comparable to previous multi-tissue studies 36. Interestingly, predictive models built in tumors had the highest out-of-sample accuracy in the breast and prostate normal panels, consistent with tumor expression capturing genetic effects that are present in normal tissues. Predicting into the normal FTSEC and OSEC samples generally yielded the lowest replication (Supplementary Table 8, 9), likely due to the small size and heterogeneity of these cohorts (notably, predictors constructed in the normal FTSEC and OSEC samples still achieved high out-of-sample accuracy). Similar trends were observed when predicting into healthy samples from the Genotype Tissue Expression (GTEx) cohort (Supplementary Note, Supplementary Table 10, 11; Supplementary Figure 5 for complete details of our validation procedure).
TWAS for EOC identifies candidate susceptibility genes
We performed a transcriptome-wide association study (TWAS) using the GWAS data from 13,037 HGSOC cases and 40,941 controls estimated by the Ovarian Cancer Association Consortium (OCAC) 7 and our trained gene expression / splicing models (Figure 1). The genetically predicted expression of 32 gene-level models (18 unique genes) and 74 junction-level models (17 unique genes) were significantly associated with risk after Bonferroni correction for 66,764 total tests (Supplementary Figure 6). TWASs may identify co-incidental genetic associations due to partial tagging between the expression and disease causing variants, and so we performed additional conditioning and colocalization analyses on a locus-by-locus basis (see Methods).
We validated the expression models for each of the significant TWAS associations by predicting into the independent cohorts from other tissues/states (Supplementary Table 12–14). 82/106 associated models were significantly correlated with expression/splicing measured in at least one independent cohort (after Bonferroni correction for 388 model-by-cohort pairs tested, with 92/106 nominally significant after Bonferroni correction for 4 cohorts tested). Mean replication R2 was 0.11 for overall expression and 0.10 for splicing and we observed no significant differences between the target datasets. 16/32 gene-level models were significantly correlated with gene expression (after Bonferroni correction) in the matching tissue in healthy GTEx samples and mean replication R2 was 0.12, demonstrating little average loss in predictive accuracy in healthy independent samples relative to independent TCGA samples (Supplementary Table 15). Overall we found that more heritable and better cross-validating genes were more likely to show up as significant TWAS associations (Supplementary Note, Supplementary Table 16, Supplementary Figure 7–11).
Novel genes implicated through TWAS
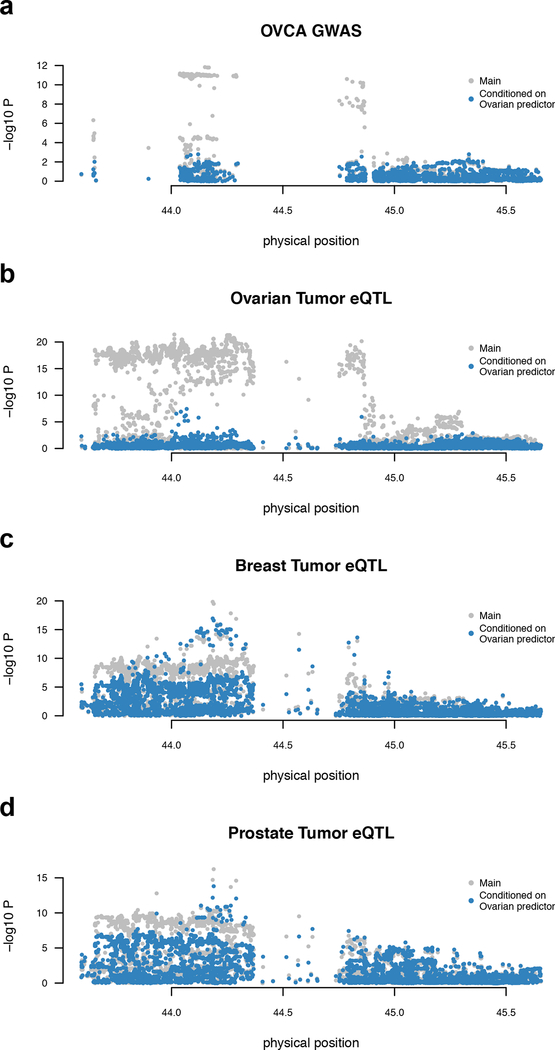
We first characterized gene-level events across the six tissue types, identifying 32 TWAS associations for 18 unique genes after Bonferroni correction (Table 2, Supplementary Table 17, Supplementary Figure 6). A single association was detected in FTSECs: the non-coding RNA TIPARP-AS1 at 3q25.31 (TWAS P = 2.2 × 10−25). Seven genes were associated with risk in HGSOCs; CHMP4C 6 at 8q21.13, and six genes located within an inversion at chromosome 17q21.31 37. ARL17A was a notable example where ovarian-specific eQTLs explained the local GWAS signal, but significant eQTLs observed in breast and prostate were independent (Figure 2, Supplementary Figure 12, 13, Supplementary Table 18). After conditioning on the ARL17A ovarian tumor model (see Methods), the most significant conditional GWAS association in this locus was P = 0.002, whereas after conditioning on the ARL17A breast tumor model, the most significant conditional GWAS association was P =1.2 × 10−05, further supporting this as an ovary-specific association. ARL17A has not been previously implicated in ovarian cancer, although KANSL1-ARL17A gene fusions have been implicated in pancreatic cancer 38. Additionally, RCCD1 at 15q26, which had previously been implicated in a meta-analysis of breast and ovarian cancer 9 was here transcriptome-wide significant (TWAS P = 1.5 × 10−7 in prostate tumor). A follow-up colocalization analysis showed that 21/32 TWAS associations exhibited strong evidence of a single shared causal variant (PP4 > 0.8) and only 4/32 had evidence of joint causal variants (PP3 > 0.2). The number of significant associations was strongly correlated with the number of tested genes (R2 = 0.86) suggesting that these findings are driven by the size and the quality of the expression reference dataset, rather than tissue specificity (Supplementary Figure 14).
Table 2. TWAS analyses in ovarian cancer.
FTSEC, fallopian tube secretory epithelial cell; OSEC, ovarian surface epithelial cell; HGSOC, high-grade serous ovarian cancer.
| Site | Type | Samples | Tested genes | Tested splice variants | Significant genes | Significant splice variants | Significant splice variant genes |
|---|---|---|---|---|---|---|---|
| Breast | Normal tissue | 84 | 1984 | 5391 | 3 | 9 | 3 |
| Breast | Tumor tissue | 654 | 4465 | 21568 | 7 | 23 | 9 |
| Ovary | Normal FTSEC | 70 | 541 | - | 2 | - | - |
| Ovary | Normal OSEC | 115 | 607 | - | 0 | - | - |
| Ovary | Tumor tissue (HGSOC) | 201 | 1744 | 8759 | 7 | 18 | 8 |
| Prostate | Tumor tissue | 376 | 4414 | 17857 | 13 | 24 | 10 |
Figure 2. Ovary-specific TWAS association for ARL17A.
The ARL17A gene is under strong genetic control in multiple tissues but only colocalizes with GWAS in ovarian tumors. Each panel shows Manhattan plot of SNP-phenotype association before (gray) and after (blue) conditioning on the TWAS predictor trained on ovarian tumor expression: a, GWAS associations, with signal fully explained after conditioning on the predictor (N = 53,978); b, ovarian tumor eQTLs (N = 201); c, breast tumor eQTL (N = 654); d, prostate tumor eQTL (N = 376). a and b show that associations are explained by the ovarian TWAS predictor, whereas c and d show that associations are independent of the ovarian TWAS predictor. Two sided p-value was computed from the GWAS summary data (a) or by linear regression (b-d).
We replicated 10/18 unique genes using independent prediction models from the GTEx study after Bonferroni correction (Supplementary Table 19). Only two were significant using GTEx ovary models - the paralogs LRRC37A and LRRC37A2, which were significant in nearly all tissues except for testis and normal prostate tissues (Supplementary Table 15). A TWAS analysis of all 84,064 available GTEx models identified two additional transcriptome-wide significant loci: MLLT10 at 10p12.31 which was significant in leg and spleen; and DNALI1 at 1p34 which was significant in 9 tissues (but not in breast, prostate or ovary). The DNALI1 locus was previously reported as genome-wide significant in serous EOC where the RSPO1 gene was proposed as a putative target gene, but no eQTL association was detected 11. Conditioning on the predicted expression of DNALI1 accounted for all the genome-wide significant signal, consistent with these genes being potential mediators of the association (Supplementary Figure 15). In breast tissue, where models were trained in TCGA tumor and normal tissues and GTEx healthy tissues, genes that were predictable in multiple cohorts produced highly concordant TWAS test statistics, underscoring the consistency of these models between tumor/normal and case/control expression (Supplementary Figure 5).
Novel transcripts implicated through junction spTWAS
Next, we performed a spTWAS across all significantly heritable exon junction events, identifying 74 splice-TWAS associations with EOC risk in 18 unique genes (after Bonferroni correction; Table 2, Supplementary Table 20). This included 7 genes that did not have a significant gene-level TWAS association in the TWAS analysis of overall expression. Colocalization analysis 39 showed that 58/74 associations were consistent with a shared causal variant (posterior on shared > 0.8) and 70/74 were inconsistent with a single distinct causal variant (posterior on distinct < 0.2). Three loci contained only a single significantly associated gene and we investigated these loci in detail.
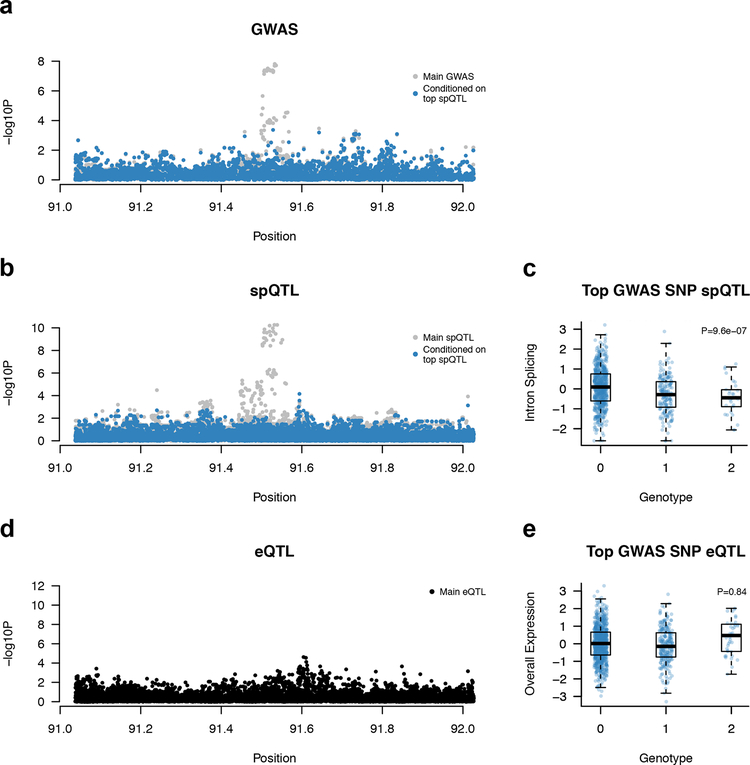
First, we identified a splice-TWAS association for PRC1 (in breast tumors) which fully explains the GWAS signal at the 15q26.1 locus (TWAS P = 8.9 × 10−8, PP4 = 1.0), which is associated with both breast and ovarian cancer risk 9 (Figure 3). This spTWAS model replicated significantly in ovarian tumor tissue (P = 8.6 × 10−4; Supplementary Table 13). Notably, we found no significant eQTLs for overall expression of PRC1, highlighting a genetic effect on splicing that is independent of total expression. We separately identified a significant TWAS association for the nearby RCCD1 gene in prostate tumors which is modestly correlated with PRC1 (Supplementary Figure 9) and was previously implicated as a candidate breast/ovarian cancer susceptibility gene 9. Second, we identified multiple spTWAS associations for CHMP4C (in all tumor panels) which fully explained the 8q21 locus. CHMP4C harbors a missense risk variant and was previously implicated by eQTL analysis 6. Here, the lead spQTL (rs74758321; GWAS P = 1.1 × 10−10) is within 300 bp of the splice junction and in perfect linkage disequilibrium with the top GWAS SNP at this locus, further implicating splicing as the potential causal mechanism (see additional validation below). Third, we identified a splice-TWAS association for HAUS6 (in prostate tumors) at the 9p22.1 locus (TWAS P = 2.8 × 10−7, PP4 = 0.7, PP3 = 0.01) which was not genome-wide significant (GWAS P = 5.9 × 10−6). Although a conditional analysis fully accounted for the local GWAS signal (Supplementary Figure 16), cross-validation accuracy of the predictive model was nominally significant (P = 2.8 × 10−3) and the model did not replicate in other tissues (Supplementary Table 13), necessitating further replication to confirm this locus.
Figure 3. Splice-TWAS association at PRC1 implicates novel target gene independent of genetic effects on total expression.
Panels a, b, d show Manhattan plot of SNP-phenotype association before (gray) and after (blue) conditioning on the top splice-QTL (a: GWAS, N = 53,978; b,c: breast tumor junction, N = 654; d,e: breast tumor expression, N = 654). Panels c, e show box and scatter plots of normalized junction (top) and overall (bottom) expression, stratified by lead GWAS SNP genotype. Two sided p-value was computed from the GWAS summary data (a) or by linear regression (b-e). c,e: thick line is median, box is the interquartile range (IQR), whiskers are quartiles plus 1.5 IQR.
The majority of associations (51/74) were at the 17q21.31 locus within an inversion polymorphism spanning ~900 kb. This region contains hundreds of variants in high linkage disequilibrium that all represent putative causal alleles and are involved in genetic co-regulation of 9 genes, with evidence of multiple clusters of independent associations (Supplementary Figure 11). We observed a complex co-regulation of >3 unique genes at one other locus - 19p13.11 - with evidence of multiple independent associations. We performed stepwise conditional analysis of all significant TWAS/spTWAS associations in the locus to identify the minimal set of genes that jointly explained the most genome-wide significant signal. The final model contained two splicing events for the BABAM1 gene (chr19:17378336–17379565 in ovarian tumors and chr19:17378336–17379603 in prostate tumors, correlated with R2 = 0.59), reducing the lead GWAS SNP from P = 7.8 × 10−25 to P = 8.2 × 10−6 (Supplementary Figure 11, 17). The 19p13.11 risk locus is also associated with triple-negative breast cancer 2 and BABAM1, a known BRCA1-interacting protein, is therefore a compelling target gene. Our previous gene-level functional studies failed to find strong functional evidence of a role for BABAM1 in ovarian and breast tumorigenesis, but instead implicated the neighboring genes ABHD8 and ANKLE1 2. These new analyses suggest that characterizing the functional significance of BABAM1 splice variants is warranted. Further studies will be needed to understand these apparently contradictory results which suggest either multiple causal variants or complex local haplotype structure.
GWAS variance and pleiotropy explained by TWAS associations
Overall, the GWAS contained 13 contiguous genome-wide significant regions, of which 6 were within 500 kb of a TWAS or splice-TWAS association (Supplementary Table 21). These 6 regions implicated a total of 106 associated features out of a 1,134 tested, demonstrating a substantial number of heritable gene/tissue combinations that have also been ruled out as likely cis targets. All gene expression and splicing models, without thresholding, explained 31% (s.e. 11%; P = 3.5 × 10−3) of EOC SNP-heritability (estimated by a modified LD-score regression, see Methods). This estimate includes any tagged genetic effects that alter expression and risk independently, and thus should be interpreted as an upper bound.
We further tested the 106 transcriptome-wide significant features (74 splice events and 32 genes) for pleiotropic associations with breast cancer risk from a recent breast cancer risk GWAS 40. 70 out of 106 features showed evidence of significant TWAS association (P < 0.05/106), demonstrating extensive pleiotropy between breast and ovarian cancer at these loci that appears to operate through the same genes (Supplementary Table 22). No significant differences were observed in the rate of pleiotropic association using breast, prostate, or ovarian models (Supplementary Table 23). Of the 70 pleiotropic associations, four were genome-wide significant (P < 5 × 10−8) for breast cancer: gene-level association with RCCD1, and exon-level associations with PRC1, LRRC37A, and KANSL1. These results highlight two robust genome-wide significant loci associated with breast and ovarian cancer that also exhibit effects on expression of the same genes. We repeated the same analysis for a recent GWAS for prostate cancer 41 but did not identify any features significant after Bonferroni correction, suggesting that the extensive expression-based pleiotropy we observe between breast and ovarian cancer is not expected by chance.
Functional assays support the CHMP4C splicing association
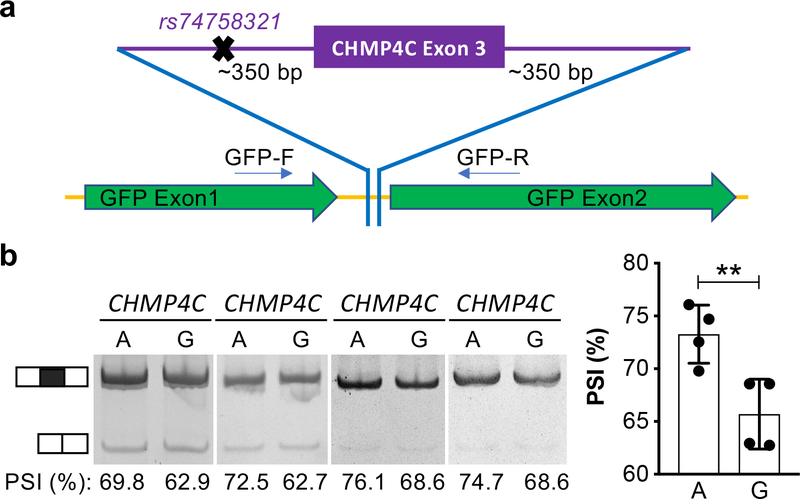
As described above, we identified four spTWAS associations in the CHMP4C gene, the most significant of which was rs74758321, which is in perfect linkage disequilibrium with the top GWAS risk SNPs for ovarian cancer identified in this region. In silico and in vitro functional analyses were performed to established if this is a likely causal SNP at this locus. SNP rs74758321 is most significantly associated with the chr8:82665476:82667605 junction in ovarian, breast, and prostate tumors with similar effect-sizes across all phenotypes (Supplementary Table 24). In a joint regression testing of the association between the SNP and all four splice junctions, this junction was the most significant feature in all tumor cohorts, but was non-significant in the normal tissues, though a significant joint association was observed for other junctions as well (Supplementary Table 25). This was also the only variant identified that fell within the consensus splice site sequence (within 300 bp of a junction). We therefore evaluated the effects of different alleles of rs74758321 on splicing in an in vitro splicing reporter assay performed in FUOV1 ovarian cancer cells. We observed exon inclusion (7.6+/−1.6%) more frequently in cells transfected with the ‘A’ allele compared to the ‘G’ allele (P = 0.0024, two tailed paired Student’s T-test) (Figure 4).
Figure 4. CHMP4C splicing is associated with EOC risk allele.
a. Schematic of the splicing assay, CHMP4C exon 3 with flanking intronic sequence was cloned into a splicing reporter vector. Plasmids were generated to harbor either the ‘G’ or ‘A’ allele of the SNP. b. In FUOV1 ovarian cancer cells the ‘A’ allele is associated with higher rates of exon inclusion. PSI, percent spliced in. Data shown are mean with SD from N = 4 independent experiments, ** P =0.0024, two-tailed paired Student’s T-test.
Importantly, we did not observe functional evidence supporting transcriptional regulatory activity or the previously implicated missense variant. Enhancer scanning assays performed to evaluate allele-specific enhancer activity of ~2 kb genomic tiles containing nine credible (1:100) causal variants identified in GWAS, including rs74758321, at this locus failed to detect any differential regulatory activity (Supplementary Note, Supplementary Figure 18). In addition, we performed a CHMP4C protein stability analysis to determine the effect of the missense variant SNP rs35094336 (Ala232Thr). No difference was observed in protein expression in transiently transfected ovarian (IOSE4CMYC) or 293FT human embryonic kidney cells. Also, no change in the stability of CHMP4C containing either the rs35094336 ‘A’ or ‘G’ allele was detected after cycloheximide treatment (not shown).
Genes showing evidence of essentiality in a knockout screen
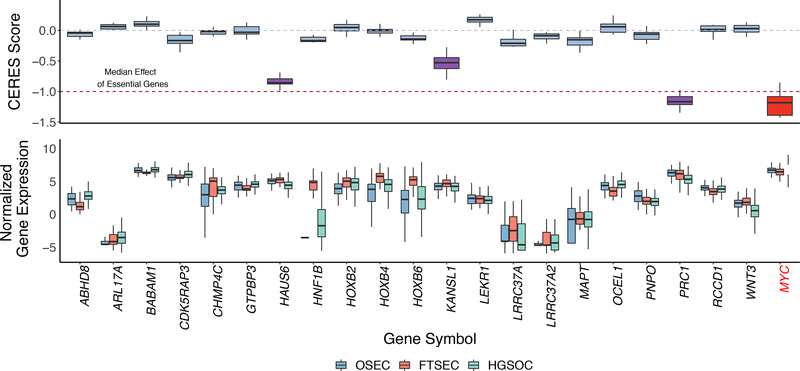
We explored the functional role of the 25 candidate susceptibility genes identified from our TWAS and spTWAS analyses using publicly available data from a gene essentiality screen 22. We utilized gene knockout data for 24 genes in 13 HGSOC cell lines (Figure 5). Three genes showed evidence of essentiality (CERES Score < −0.5) - KAT8 regulatory NSL complex subunit 1 (KANSL1, mean CERES score = −0.53, s.d. = 0.15), HAUS Augmin Like Complex Subunit 6 (HAUS6, mean CERES score = −0.84 s.d. 0.07), and protein regulator of cytokinesis 1 (PRC1, mean CERES score = −1.13, s.d. = 0.14). PRC1 shows similar levels of essentiality as MYC, a key oncogenic transcription factor in many tumor types, including ovarian cancer 42. All three genes were identified only through the splice-TWAS and not previously reported. Indeed, the mean CERES score across significant splice-TWAS genes (−0.21 s.e. 0.03) was significantly lower (i.e. more essential) than that of genes not associated through splicing (−0.01 s.e. 0.02; P = 4.8 × 10−6 for difference by Wilcoxon rank sum test). This significant difference suggests that risk variants affecting splicing, and thus protein structure, may be more likely to target essential genes in ovarian cancer cells than risk variants that apparently only affect transcription (i.e. protein abundance). As the CERES functional screens model complete gene knockouts, further functional assays of specific isoforms and allelic series will be required to validate this hypothesis.
Figure 5. Functional analyses show evidence of essentiality for three TWAS/spTWAS genes.
a. Gene knockout experiments to determine gene essentiality for 13 HGSOC cell lines. CERES Score is a copy number corrected indicator of depletion of gene-targeting guide RNAs; the lower the CERES Score, the more essential the gene. MYC is a known essential gene and is included as a positive control. CERES Score thresholds corresponding to the median score for non-essential and essential genes are indicated with a blue dashed line at 0 and red dashed line at −1, respectively. Genes with a CERES Score ≤ −0.5 (and therefore showing evidence of essentiality) are highlighted with purple boxes. b. Relative expression of each gene in OSECs (N = 120), FTSECs (N = 71) and HGSOCs (N = 394). ARL17B and TIPARP-AS1 were not evaluated in the CERES screen and were therefore excluded from the plots. Boxes in each plot represent the first and third quartiles, and whiskers extend to 1.5 * IQR.
DISCUSSION
In this study we integrated tissue specific gene expression and genotyping data with the largest GWAS dataset available for HGSOC 7 to identify 25 candidate susceptibility genes, one of which was experimentally validated and three which showed promising functional evidence of essentiality. The spTWAS analysis identified 7 genes that were not implicated by the gene-level TWAS, nearly doubling the number of candidate susceptibility genes we identified. This included PRC1 (at 15q26.1) which explained all of the GWAS signal while exhibiting no eQTL association and was not previously identified in a locus-specific eQTL analysis. Notably, PRC1 showed similar levels of essentiality as MYC (a known essential gene and likely GWAS target gene in HGSOC 13) strongly indicating PRC1 plays a functional role in the development of ovarian tumors. In breast, ovarian and prostate tumors we identified an spTWAS association for CHMP4C, at chromosome 8q21.13. In an in vitro splicing assay, the two alleles of rs74758321 were associated with significantly different rates of exon 3 inclusion. We performed comprehensive testing for allele-specific activity for all candidate causal variants in the region, using a set of in vitro assays that are commonly used to evaluate allele-specific activity of risk SNPs, including enhancer scanning, electrophoretic mobility shift assays and protein stability assays 2,40,43–45. Beyond the validated role for rs74758321 in splicing there was no evidence to support a functional role for any of the other candidate causal variants at this locus, indicating rs74758321 is the most likely ‘causal’ variant at this risk locus. Taken together, these findings indicate that alternative splicing should be considered more broadly in post-GWAS functional analyses. CHMP4C expression has been implicated in several cancers and has been proposed as a diagnostic tumor marker and therapeutic target for ovarian cancer 46,47. We observed a striking overlap between significant TWAS genes in GWAS for HGSOC and a recent GWAS of breast cancer 40, including genome-wide significant associations with PRC1 and KANSL1. These findings merit further studies based on TWAS methodologies to identify pleiotropy and common cancer susceptibility genes for these cancers.
It remains likely that our TWAS analysis missed a unquantifiable proportion of true associations, while some associations may represent false positive findings due to chance co-regulation 21. This is emphasized by a recent parallel publication from Lu et al. 48 reporting a TWAS for ovarian cancer using total expression models constructed in the GTEx cohort. The use of distinct transcriptomic data in our study and our focus on splicing variation likely contributes to the differences in the candidate ovarian cancer susceptibility genes identified in each study. In particular, the one novel locus identified by Lu et al., FZD4 at 11q14.2 and a plausible candidate because of its role as a member of the frizzled gene family associated with Wnt signaling, was not heritable in any of the tissues we investigated (h2g < 0.006 in any tissue). We note that selecting the appropriate tissue/cell type for TWAS is critical in avoiding false positives in causal gene identification from TWAS and remains an active research area for TWAS analyses; see ref. 21 for a broader discussion of tissue choice in causal gene identification following TWAS or ref. 7 for a specific discussion of tissue of origin for different histotypes of epithelial ovarian cancer. Future studies to improve the power of TWAS analysis in ovarian cancer will need to establish substantially larger gene expression and genotyped datasets for normal precursor tissues, for HGSOC and for other EOC histotypes that were not evaluated in the current study.
In summary, we have performed a TWAS based on the integration of GWAS data for HGSOC and gene expression data for both normal and tumor tissues associated with HGSOC pathogenesis, to identify candidate susceptibility genes associated with inherited HGSOC risk. Most importantly, this study established spTWAS associations as a major component of HGSOC heritability, a principle that also likely applies to many other phenotypes.
ONLINE METHODS
Data processing and QC
Genotypes.
Germline DNA from normal OSEC and FTSEC samples were genotyped using the Oncoarray platform 7. For TCGA data, SNP genotype calls using Birdsuite were downloaded from the TCGA legacy archive and imputed using the EAGLE pipeline provided by the Michigan imputation server. The following genotype QC was performed across all studies: SNPs were retained if they had imputation INFO>0.9; locus missingness <5%; Hardy-Weinberg equilibrium two-tailed P-value > 5 × 10−6; and minor allele frequency > 1% (thresholds based on GTEx Consortium recommendations). Individuals were excluded if they had more than 5% missing sites. Two genotype principal components were computed to account for ancestry and included as covariates in all subsequent analyses.
Gene/exon expression in HGSOC precursor tissues.
OSECs and FTSECs were harvested from histologically normal ovaries and fallopian tubes removed from women diagnosed with ovarian, uterine or cervical cancer. Short-term cultures were established 49,50. OSECs were harvested using a cytobrush and cultured in NOSE-CM media containing 15% fetal bovine serum (FBS, Hyclone), 34 μg ml−1 bovine pituitary extract, 10 ng ml−1 epidermal growth factor (Life Technologies), 5 μg ml−1 insulin and 500 ng ml−1 hydrocortisone (Sigma-Aldrich). Fallopian epithelia were dissociated from stromal tissues by Pronase/DNase I digestion (Roche and Sigma-Aldrich, respectively) for 48–72 hours at 4°C. Purified epithelia were cultured on collagen I (Sigma-Aldrich) using DMEM/F12 base media supplemented with 2% Ultroser G (Pall Corporation). At ~80% confluency, cells were lysed using the QIAzol reagent and RNA extracted using the RNeasy Mini kit (both QIAgen). RNA sequencing was performed by the University of Southern California Epigenome Core Facility using 50 bp single end reads. All data processing was performed using ‘R’ and ‘Bioconductor’, and packages therein.
RNAseq data for 394 HGSOC samples was obtained from The Cancer Genome Atlas (TCGA) data portal as protected data (raw sequencing, fastq files) and downloaded via CGHub’s geneTorrent. Data were aligned to a reference genome (hg19) using STAR. Quality control of aligned samples was performed using RSeQC. GC bias and batch effect corrections were performed using EDASeq and ‘sva’. To adjust for batch effects we used an empirical Bayes framework (comBat), available in ‘sva’.
Gene/exon expression in non-ovarian TCGA samples.
Normalized gene and exon level events were downloaded from the TCGA FireCloud. Exon usage was previously quantified using MapSplice. Finally, all expression/exon measurements were quantile normalized. As with the ovarian data, all expression/exon measurements were quantile normalized and three expression principal components were computed and used as covariates in all subsequent analyses.
Gene expression in GTEx samples.
Processed and normalized expression and genotypes were downloaded from dbGAP and the GTEx Portal as described in 51. For each tissue the following covariates were included in all analyses: three genetic principal components, sex, platform, and 14–35 expression factors 52 as selected by the main GTEx analysis.
Clinical factors for TCGA samples.
We extracted all relevant clinical factors available for TCGA samples for use as covariates and to evaluate expression heterogeneity (Supplementary Table 4, 5). We quantified BRCA1/2 somatic mutation carriers using MutSigCV2 calls from tumor whole-exome sequencing made available by TCGA. We quantified somatic structural variants using BirdSuite CNV calls from tumor/normal genotype array data made available by TCGA. Tumor purity was systematically estimated across the TCGA cohorts by Aran et al. 53.
Heritability and genetic correlation
To evaluate tumor subtype heterogeneity we quantified the genetic correlation of gene expression between subtypes. For each panel and factor we divided the samples into two groups either as carriers/non-carriers for dichotomous factors such as somatic BRCA1/2 mutation, or as low/high for quantitative factors such as age. Mean heritability and genetic correlation were estimated using Haseman-Elston regression 54 as implemented in GCTA 55. All SNPs within 500 kb of the gene boundary were used to define the cis locus and construct the corresponding kinship matrix. Standard errors for genetic correlation across all genes were estimated as in 56.
Construction and validation of gene prediction models
TWAS predictors were computed using the FUSION software (see Web Resources). Briefly, for each gene or exon junction, SNPs from +/−500 kb of the feature boundary were extracted and used to estimated cis-SNP-heritability 55. Clinical features and gene expression principal components were always included as covariates to account for trans variation (see Supplementary Note for detailed analyses). Features that had nominally significant cis-SNP-heritability (likelihood ratio test P<0.01) were retained for model building and TWAS. We elected to use a heritability-based cutoff rather than specify a cutoff on the cross-validation R2 because the former uses all available data, however we report both statistics for all associations. Notably, for the 106 TWAS-significant models, 105/106 has nominal cross-validation P<0.05 and 85/106 had Bonferroni significant cross-validation P<0.05/106. The genotypes were used to train TWAS predictive models using BLUP, elastic net, and LASSO algorithms.
Five-fold cross-validation was performed for each reference panel and gene/splicing model. Gene expression for each fold of the data was hidden in turn; the full prediction model was then trained on the remaining expression and genetic data; and the trained model was then predicted into the held-out fold samples. This procedure was repeated across all folds to compute the overall cross-validated prediction, an adjusted R2 (and corresponding two-tailed P-value) was then computed between the cross-validated prediction and the measured expression by ordinary least squares. The lasso and elastic net models require a penalty parameter that is itself fit by leave-one-out cross-validation and this was performed within each fold (i.e. double cross-validation where the testing data is hidden from all parameter tuning). Across all tissues and features, the top eQTL was the best predictor only 26% of the time. Surprisingly, the BLUP predictor - which has the weakest penalization in favor of sparsity - was the most common best predictor (best 33% of the time), suggesting a greater degree of effect heterogeneity in this data than studies of normals where cis-expression effects are typically sparse 57.
For models trained in GTEx (v6), TWAS expression weights were previously computed as described in 16, and downloaded from the FUSION website.
We investigated the concordance in heritability and R2 estimates between the TCGA tumors and the corresponding healthy GTEx tissues. For each pair of TCGA/GTEx tissues we measured Pearson correlation (and significance) between the estimated cis-heritability across all evaluated genes. We note that the GTEx heritability estimates have greater estimation error than signal (mean cis-h2g estimate 0.042, mean s.e. estimate 0.045) which is expected to greatly deflate this estimate. We therefore additionally estimated the relationship between the two estimates using regression of GTEx estimate on TCGA estimate (Supplementary Table 11). We similarly evaluated the correlation between the TCGA in-sample cross-validation R2 and the TCGA-into-GTEx prediction R2.
Correlation of predictive models between the TCGA panels was estimated in the 1000 Genomes Project European reference samples. First, each model was predicted into the reference samples. Second, for each gene that was modelled in multiple panels and each pair of panels, the correlation between predicted values from panel 1 and panel 2 was computed. The mean correlation across all pairs of panels and shared genes is reported in (Supplementary Table S12).
TWAS analysis
GWAS data.
GWAS data from the Ovarian Cancer Association Consortium as described in 7 were downloaded and aligned to hg19 HapMap3 SNPs (excluding A/T or C/G SNPs due to strand ambiguity). These SNPs are consistently imputed with high accuracy across diverse genotyping platforms and were used to compute all TWAS weights.
TWAS tests.
The FUSION software was used to perform TWAS tests across all predictive models 17. Models were considered “transcriptome-wide significant” if they passed Bonferroni correction for all genes and exon events tested.
Summary-based conditional analyses between TWAS and GWAS associations were performed using FUSION 58. For a given significant TWAS association, the gene/exon expression was predicted into the 1000 Genomes EUR samples to estimate the LD between the predicted model and each SNP in the locus. Each GWAS SNP was then conditioned on the predicted model using the LD estimate to quantify the amount of residual association signal. Stepwise model selection was performed by including each TWAS-associated feature (from most significant to least) into the model until no feature remained conditionally significant.
Summary-based conditional analyses for individual SNPs were performed using GCTA-COJO 58. Colocalization analyses were performed using the COLOC software 39 and the marginal eQTL/spQTL statistics for a given feature.
Conditional and colocalization analyses
First, we condition every GWAS association on the predicted value of each significant TWAS gene to assess how much association signal remains independent of the TWAS association (see Methods). We note that comparing GWAS and TWAS effect-sizes directly poses a challenge because the associations are on different scales: GWAS effect-sizes are on the allelic odds ratio scale, whereas TWAS effect-sizes are on the standardized expression scale as there is no formal “allele” for the expression predictor. Instead, the conditional analysis serves to quantify whether the TWAS association can “explain” the GWAS association after accounting for correlation between the predictive model and GWAS risk SNPs. Residual GWAS signal, after conditioning, is an indicator that the TWAS association is partially tagging the causal variants, or that other independent causal variants are present at the locus (as with SNP-based conditioning).
Second, we perform a “colocalization” analysis using the COLOC software 39, which evaluates the posterior probability that the genetic association to the gene/exon is driven by a single shared causal variant with the GWAS risk association (termed “PP4” in the COLOC notation). This model does not consider colocalization between multiple causal variants, so high PP4 is a more stringent threshold to clear than the TWAS association and may miss true colocalization at loci with heterogeneous effects on expression and disease. COLOC additionally estimates the probability that the expression and GWAS are driven by two distinct causal variants (PP3), and we use low PP3 as a less stringent threshold for evidence of non-independent association signal (though it may still be confounded by multiple causal variants).
Functional analyses
CHMP4C spQTL analyses:
We tested rs74758321 for association to CHMP4C junction usage in each of the TCGA cohorts (Supplementary Table 24, 25; Life Sciences Reporting Summary). To keep effect sizes as consistent as possible across the studies (and since these junctions were already shown to be highly heritable with all covariates), we did not incorporate any covariates and only performed simple rescaling of each junction count to mean 0, variance 1. We first tested each junction (in each tissue) in turn for association to the SNP by OLS regression (Supplementary Table 24). We then tested the joint effect of the SNP on junction usage by reversing the regression : SNP ~ junction1 + junction2 + junction3 + junction4 (Supplementary Table 25). While the individual effect-sizes from this multiple regression are difficult to interpret, the significance of each association is an indicator of which junction more strongly drives the genetic association.
For splicing assays, IOSE11 cells were grown in NOSE-CM 59 and FUOV1 ovarian cancer cells were grown in DMEM/F12 (Thermo Fisher Scientific) supplemented with 10% FBS (Sigma-Aldrich) and L-Glutamine (Lonza, Catalogue number:17–605E). A minigene construct was generated to include chr8:82667209–82668009, representing CHMP4C exon 3 +/− ~350 bp (see Supplementary Note for primers used). IOSE11 DNA was used as a template for PCR, as this cell line is heterozygous for rs74758321. PCR products were cloned into the splicing reporter vector pZW1 60 to generate a pair of plasmids containing the two alleles of SNP rs74758321. The constructs were confirmed by double digestion and further verified by Sanger sequencing. Twelve-well FUOV1 ovarian cancer cells were transfected with 2 μg splicing reporter when cells are 80% confluency. Cells were harvested 24 h post transfection and total RNA was subsequently extracted. cDNA was made from 5 μg of total RNA by reverse transcription and one twentieth of cDNA was used as template to amplify both inclusion and skipping form of splicing reporter GFP transcript with or without CHMP4C exon 3 by PCR within 25 cycles using GFP-F and GFP-R (see Supplementary Note). PCR products were subjected to 5% polyacrylamide gel electrophoresis, and the resulting gels were imaged. Expression of each band was quantified using ImageJ software (http://imagej.nih.gov/ij/) and the inclusion rate of the target exon was calculated.
Enhancer scanning assays -
We tested ~2 kb tiles containing all nine (rs137960856, rs11782652, rs74544416, rs78740005, rs78724141, rs111683632, rs74758321, rs76837345, rs35094336) credible (1:100; 10/26/2015 imputation) SNPs in this locus for regulatory enhancer activity using enhancer scanning 61 in IOSE4CMYC cells 62. Tiles were tested in both orientations. Primers are provided in Supplementary Table 27.
Protein stability assays -
SNP rs35094336 missense variant (Ala232Thr) was evaluated for CHMP4C protein stability. CHMP4C cDNA containing each allele were cloned into pCMV6-entry vector. iOSE4CMYC cells were stably transfected with FuGENE-HD and clones selected by G418 (500 μg/ml, Gibco). Stable clones were treated with cicloheximide (Sigma) for up to 48 h and protein levels were assessed. The missense variant (rs35094336) does not impact CHMP4C transcript stability.
Knockout screen data analyses -
Differential gene expression analyses were performed using the OSEC, FTSEC and HGSOC data sets described above. Processed CERES knockout data were downloaded 22 and data for the 13 HGSOC lines included in this study were used in our analyses.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by multiple grants: An NIH/NCI R21 award “The Role of Splice Quantitative Traits in Ovarian Cancer Pathogenesis” (CA22007801); an NIH/NCI U19 award “Follow-up of Ovarian Cancer genetic association and Interaction studies (FOCI)” as part of the Genetic Mechanisms in Oncology (GAME-ON) consortium (CA148112); an NIH/NCI R01 award “Functional Effects of Ovarian Cancer Risk Variants” (CA211707); an NIH/NCI R01 award “Epidemiology and biology of lncRNAs in ovarian cancer” (CA207456); an NIH/NCI R01 award “Identifying causal variants and genes underlying breast cancer risk loci” (CA204954); and an NIH/NCI R01 award “(PQ3) A functional genomic approach to identification and interpretation of germline-tumor genetic interactions “ (CA227237). S.A.G is additionally supported by the Barth Family Chair in Cancer Genetics at Cedar-Sinai Medical Center. K.L. is supported in part by a K99/R00 Pathway to Independence Award from the NIH (R00CA184415) and institutional support from the Samuel Oschin Comprehensive Cancer Institute at Cedars-Sinai Medical Center. H.N. and M.A.S.F. are supported by grants 2015/07925–5 and 2017/08211–1 from Sao Paulo Research Foundation (FAPESP). H.N. is also supported by an institutional grant (Henry Ford Hospital). This work was supported in part by the Ovarian Cancer Research Fund Alliance Program Project Development Grant (373356): Co-Evolution of Epithelial Ovarian Cancer and Tumor Stroma. Additional support for this work came from NIH/NCI grants 1R01CA211707 and 1R01CA207456 and OCRF award 258807. The results shown here are in part based upon data generated by the TCGA Research Network: http://cancergenome.nih.gov/. Some of the normal tissue specimens were collected as part of the USC Jean Richardson Gynecologic Tissue and Fluid Repository, which is supported by a grant from the USC Department of Obstetrics & Gynecology and the NCT Cancer Center Shared Grant award P30 CA014089 (to the Norris Comprehensive Cancer Center).
Footnotes
Conflict of Interest: The authors have no financial conflicts of interest to declare.
DATA AVAILABILITY STATEMENT
Code, documentation for all methods, all trained TWAS models for all genes and splice variants has been made available on the TWAS/FUSION web-site (http://gusevlab.org/projects/fusion/). Full TWAS association statistics have been made available in an interactive database on http:///www.twas-hub.org.
Editorial summary:
A multi-tissue transcriptome-wide association study based on genetic predictors of expression level and alternative splicing in relevant tissues identifies 25 candidate genes associated with high grade serous ovarian cancer.
REFERENCES
- 1.Jones MR, Kamara D, Karlan BY, Pharoah PDP & Gayther SA Genetic epidemiology of ovarian cancer and prospects for polygenic risk prediction. Gynecol. Oncol 147, 705–713 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Lawrenson K et al. Functional mechanisms underlying pleiotropic risk alleles at the 19p13.1 breast-ovarian cancer susceptibility locus. Nat. Commun 7, 12675 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lawrenson K et al. Cis-eQTL analysis and functional validation of candidate susceptibility genes for high-grade serous ovarian cancer. Nat. Commun 6, 8234 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Song H et al. A genome-wide association study identifies a new ovarian cancer susceptibility locus on 9p22.2. Nat. Genet 41, 996–1000 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bolton KL et al. Common variants at 19p13 are associated with susceptibility to ovarian cancer. Nat. Genet 42, 880–884 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pharoah PDP et al. GWAS meta-analysis and replication identifies three new susceptibility loci for ovarian cancer. Nat. Genet 45, 362–70, 370e1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Phelan CM et al. Identification of 12 new susceptibility loci for different histotypes of epithelial ovarian cancer. Nat. Genet 49, 680–691 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kelemen LE et al. Genome-wide significant risk associations for mucinous ovarian carcinoma. Nat. Genet 47, 888–897 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kar SP et al. Genome-Wide Meta-Analyses of Breast, Ovarian, and Prostate Cancer Association Studies Identify Multiple New Susceptibility Loci Shared by at Least Two Cancer Types. Cancer Discov 6, 1052–1067 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen K et al. Genome-wide association study identifies new susceptibility loci for epithelial ovarian cancer in Han Chinese women. Nat. Commun 5, 4682 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Kuchenbaecker KB et al. Identification of six new susceptibility loci for invasive epithelial ovarian cancer. Nat. Genet 47, 164–171 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bojesen SE et al. Multiple independent variants at the TERT locus are associated with telomere length and risks of breast and ovarian cancer. Nat. Genet 45, 371–84, 384e1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goode EL et al. A genome-wide association study identifies susceptibility loci for ovarian cancer at 2q31 and 8q24. Nat. Genet 42, 874–879 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Q et al. Expression QTL-based analyses reveal candidate causal genes and loci across five tumor types. Hum. Mol. Genet 23, 5294–5302 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Q et al. Integrative eQTL-based analyses reveal the biology of breast cancer risk loci. Cell 152, 633–641 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mancuso N et al. Integrating Gene Expression with Summary Association Statistics to Identify Genes Associated with 30 Complex Traits. Am. J. Hum. Genet 100, 473–487 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gusev A et al. Integrative approaches for large-scale transcriptome-wide association studies. Nat. Genet 48, 245–252 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu Z et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet 48, 481–487 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Xu Z, Wu C, Wei P & Pan W A Powerful Framework for Integrating eQTL and GWAS Summary Data. Genetics 207, 893–902 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gamazon ER et al. A gene-based association method for mapping traits using reference transcriptome data. Nat. Genet 47, 1091–1098 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wainberg M et al. Vulnerabilities of transcriptome-wide association studies. BioRxiv (2017). doi: 10.1101/206961 [DOI] [Google Scholar]
- 22.Meyers RM et al. Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat. Genet 49, 1779–1784 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leeper K et al. Pathologic findings in prophylactic oophorectomy specimens in high-risk women. Gynecol. Oncol 87, 52–56 (2002). [DOI] [PubMed] [Google Scholar]
- 24.Paley PJ et al. Occult cancer of the fallopian tube in BRCA-1 germline mutation carriers at prophylactic oophorectomy: a case for recommending hysterectomy at surgical prophylaxis. Gynecol. Oncol 80, 176–180 (2001). [DOI] [PubMed] [Google Scholar]
- 25.Carcangiu ML et al. Atypical epithelial proliferation in fallopian tubes in prophylactic salpingo-oophorectomy specimens from BRCA1 and BRCA2 germline mutation carriers. Int. J. Gynecol. Pathol 23, 35–40 (2004). [DOI] [PubMed] [Google Scholar]
- 26.Callahan MJ et al. Primary fallopian tube malignancies in BRCA-positive women undergoing surgery for ovarian cancer risk reduction. J. Clin. Oncol 25, 3985–3990 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Gilks CB et al. Incidental nonuterine high-grade serous carcinomas arise in the fallopian tube in most cases: further evidence for the tubal origin of high-grade serous carcinomas. Am. J. Surg. Pathol 39, 357–364 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Auersperg N et al. Expression of two mucin antigens in cultured human ovarian surface epithelium: influence of a family history of ovarian cancer. Am. J. Obstet. Gynecol 173, 558–565 (1995). [DOI] [PubMed] [Google Scholar]
- 29.Dyck HG et al. Autonomy of the epithelial phenotype in human ovarian surface epithelium: changes with neoplastic progression and with a family history of ovarian cancer. Int. J. Cancer 69, 429–436 (1996). [DOI] [PubMed] [Google Scholar]
- 30.He Q-Y et al. Proteomic analysis of a preneoplastic phenotype in ovarian surface epithelial cells derived from prophylactic oophorectomies. Gynecol. Oncol 98, 68–76 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Casey MJ et al. Histology of prophylactically removed ovaries from BRCA1 and BRCA2 mutation carriers compared with noncarriers in hereditary breast ovarian cancer syndrome kindreds. Gynecol. Oncol 78, 278–287 (2000). [DOI] [PubMed] [Google Scholar]
- 32.Lu KH et al. Occult ovarian tumors in women with BRCA1 or BRCA2 mutations undergoing prophylactic oophorectomy. J. Clin. Oncol 18, 2728–2732 (2000). [DOI] [PubMed] [Google Scholar]
- 33.Adler E, Mhawech-Fauceglia P, Gayther SA & Lawrenson K PAX8 expression in ovarian surface epithelial cells. Hum. Pathol 46, 948–956 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature 474, 609–615 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ross-Adams H et al. HNF1B variants associate with promoter methylation and regulate gene networks activated in prostate and ovarian cancer. Oncotarget 7, 74734–74746 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Consortium GTEx et al. Genetic effects on gene expression across human tissues. Nature 550, 204–213 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Permuth-Wey J et al. Identification and molecular characterization of a new ovarian cancer susceptibility locus at 17q21.31. Nat. Commun 4, 1627 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goecks J et al. Open pipelines for integrated tumor genome profiles reveal differences between pancreatic cancer tumors and cell lines. Cancer Med 4, 392–403 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Giambartolomei C et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet 10, e1004383 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Michailidou K et al. Association analysis identifies 65 new breast cancer risk loci. Nature 551, 92–94 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schumacher FR et al. Association analyses of more than 140,000 men identify 63 new prostate cancer susceptibility loci. Nat. Genet 50, 928–936 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reyes-González JM et al. Targeting c-MYC in Platinum-Resistant Ovarian Cancer. Mol. Cancer Ther 14, 2260–2269 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baskin R et al. Functional analysis of the 11q23.3 glioma susceptibility locus implicates PHLDB1 and DDX6 in glioma susceptibility. Sci. Rep 5, 17367 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.French JD et al. Functional variants at the 11q13 risk locus for breast cancer regulate cyclin D1 expression through long-range enhancers. Am. J. Hum. Genet 92, 489–503 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pasquali L et al. Pancreatic islet enhancer clusters enriched in type 2 diabetes risk-associated variants. Nat. Genet 46, 136–143 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fujita K et al. Proteomic analysis of urinary extracellular vesicles from high Gleason score prostate cancer. Sci. Rep 7, 42961 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nikolova DN et al. Genome-wide gene expression profiles of ovarian carcinoma: Identification of molecular targets for the treatment of ovarian carcinoma. Mol. Med. Report 2, 365–384 (2009). [DOI] [PubMed] [Google Scholar]
- 48.Lu Y et al. A Transcriptome-Wide Association Study Among 97,898 Women to Identify Candidate Susceptibility Genes for Epithelial Ovarian Cancer Risk. Cancer Res 78, 5419–5430 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lawrenson K et al. In vitro three-dimensional modelling of human ovarian surface epithelial cells. Cell Prolif 42, 385–393 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Karst AM, Levanon K & Drapkin R Modeling high-grade serous ovarian carcinogenesis from the fallopian tube. Proc Natl Acad Sci USA 108, 7547–7552 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.GTEx Consortium. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348, 648–660 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stegle O, Parts L, Piipari M, Winn J & Durbin R Using probabilistic estimation of expression residuals (PEER) to obtain increased power and interpretability of gene expression analyses. Nat. Protoc 7, 500–507 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aran D, Sirota M & Butte AJ Systematic pan-cancer analysis of tumour purity. Nat. Commun 6, 8971 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Haseman JK & Elston RC The investigation of linkage between a quantitative trait and a marker locus. Behav. Genet 2, 3–19 (1972). [DOI] [PubMed] [Google Scholar]
- 55.Yang J, Lee SH, Goddard ME & Visscher PM GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet 88, 76–82 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Falconer DS & Mackay TFC Introduction to Quantitative Genetics (4th Edition). (Pearson, 1996). [Google Scholar]
- 57.Wheeler HE et al. Survey of the Heritability and Sparse Architecture of Gene Expression Traits across Human Tissues. PLoS Genet 12, e1006423 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang J et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet 44, 369–75, S1 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li NF et al. A modified medium that significantly improves the growth of human normal ovarian surface epithelial (OSE) cells in vitro. Lab. Invest 84, 923–931 (2004). [DOI] [PubMed] [Google Scholar]
- 60.Hsiao Y-HE et al. Alternative splicing modulated by genetic variants demonstrates accelerated evolution regulated by highly conserved proteins. Genome Res 26, 440–450 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Buckley M et al. Enhancer scanning to locate regulatory regions in genomic loci. Nat. Protoc 11, 46–60 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lawrenson K et al. Senescent fibroblasts promote neoplastic transformation of partially transformed ovarian epithelial cells in a three-dimensional model of early stage ovarian cancer. Neoplasia 12, 317–325 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.