Abstract
In acute myeloid leukemia (AML), therapy resistance occurs frequently and leads to high mortality of the disease. However, the mechanisms that render leukemic cells drug resistant remain largely undefined. Here, we identified loss of the histone methyltransferase EZH2 and subsequent reduction of histone H3K27 trimethylation as a novel pathway of acquired drug resistance towards tyrosine kinase inhibitors (TKI) and cytotoxic drugs in AML. Low EZH2 protein levels correlated with poor prognosis in AML patients. Suppression of EZH2 protein expression induced chemoresistance of AML cell lines and primary cells as well as in NSG mouse models. Low EZH2 levels resulted in derepression of HOX genes and knockdown of HOXB7 and HOXA9 in resistant cells was sufficient to improve drug sensitivity to TKIs and cytotoxic drugs. The endogenous loss of EZH2 expression in resistant cells and primary blasts from a subset of relapsed AML patients resulted from enhanced CDK1-dependent phosphorylation of EZH2 at T487. This interaction was stabilized by heat shock protein 90 (HSP90) and followed by proteasomal degradation of EZH2 in drug-resistant cells. Accordingly, inhibitors of HSP90, CDK1 and the proteasome prevented EZH2 degradation, decreased HOX gene expression and reinstated drug sensitivity. Finally, patients with reduced EZH2 levels at progression to standard therapy responded to the addition of bortezomib to cytarabine with reestablishment of EZH2 expression and blast clearance. These data suggest restoration of EZH2 protein as a viable approach to overcome treatment resistance in this AML patient population.
Introduction
Chemotherapy with cytarabine (AraC) and an anthracycline remains the standard of care in AML1 despite recent attempts for novel approaches2. Chemotherapy alone cures less than 40% of all adults, with elderly patients having an even worse prognosis mainly due to therapy resistance1, 3. Recently, epigenetic changes have been identified as contributors to chemoresistance4, 5. Epigenetic mechanisms that drive therapy resistance might result from underlying genetic aberrations. Alterations of epigenetic modifiers can determine outcome in hematological malignancies as shown for DNMT3A, ASXL1, TET2 or EZH2 6–10. The Enhancer of Zeste Homologue 2 (EZH2) is a member of the Polycomb Repressive Complex 2 (PRC2) which mediates transcriptional silencing through di- and trimethylation of lysine 27 of histone H3 (H3K27me2/3)11 and plays a crucial role during embryonic development12. Recent studies suggest that EZH2 may have a dual role in cancer pathogenesis acting as oncogene or as tumor suppressor depending on cancer type. EZH2 inactivation in myeloid malignancies such as MDS and MDS/MPN is associated with a poor prognosis9, 13, 14 and can contribute to disease pathogenesis, however entire loss of EZH2 due to genetic deletion in a syngeneic mouse model is reported to prevent the transformation of AML from MDS15. On the other hand, EZH2 is overexpressed in numerous solid tumors such as prostate cancer, melanoma or ovarian cancer16–18 and EZH2 mutations with enhanced methyltransferase activity have been described in non-Hodgkin lymphomas19. In solid tumors, targeting EZH2 can cause regression of tumor cell growth18.
Here we show that loss of EZH2 induces chemoresistance towards multiple drugs in AML. We describe a post-translational mechanism of EZH2 suppression frequently occurring in relapsed AML. Our findings indicate that inhibition of EZH2 degradation in combination with chemotherapy and/or TKIs may be a promising therapy concept in drug-resistant AML.
Results
Loss of EZH2 associates with poor prognosis and chemoresistance in AML
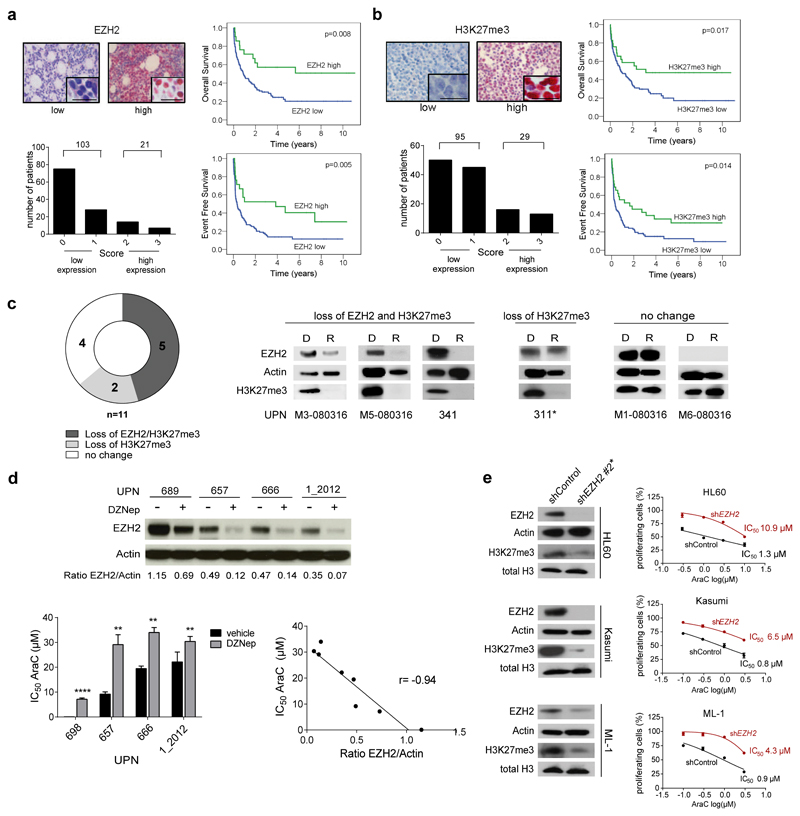
To evaluate the prognostic impact of EZH2 in AML we analyzed EZH2 protein levels in 124 AML patients (Suppl. Table 1) using Tissue Microarrays (TMA). The absence of EZH2 protein expression as analyzed by immunohistochemistry staining significantly correlated with poor overall survival (median 9.6 vs. 47.6 months, p= 0.008, Fig. 1a), poor event-free survival (median 4.06 vs. 46 months, p= 0.005, Fig. 1a) and poor relapse-free survival (median 11.3 versus 55.3 months, p= 0.047, data not shown). Similar findings were obtained for H3K27me3 levels which result from low EZH2 activity (Fig. 1b). H3K27me3 intensity and EZH2 protein expression were closely associated in AML patient samples (p< 0.0001, Suppl. Fig. 1a). In a Cox regression analysis, a combined score of low EZH2 protein expression and low H3K27me3 level remained an independent prognostic parameter in multivariate analysis (p= 0.018, Suppl. Fig. 1b). Low EZH2 mRNA expression as analyzed by qRT-PCR also associated with inferior event free survival (EFS) (p= 0.04, Suppl. Fig. 1c, Suppl. Table 2). A trend towards worse overall survival (OS) was also observed (p= 0.096, Suppl. Fig. 1c). Similar findings were observed in published microarray and RNA-sequencing (RNA-seq) datasets (Suppl. Fig. 1d and e). EZH2 is located on chromosome 7q31.6 and loss of 7q correlates with poor prognosis in myeloid malignancies20. Deletion of chr 7 or 7q in AML blasts was associated with decreased EZH2 mRNA and protein levels (Suppl. Fig. 1f).
Figure 1. Loss of EZH2 associates with poor prognosis and chemoresistance in AML.
(a) EZH2 and b) H3K27me3 immunohistochemistry staining of bone marrow biopsies from 124 AML patients at time of diagnosis. Clinical data are provided in Supplementary Table 1. Nuclear staining of AML blasts was assessed using Remmele´s Immunoreactive Score (IRS). Representative positive and negative stainings are shown. Scale bars indicate 20 µm. Insets show high-magnification images (top left). The number of patients with low or high EZH2 or H3K27me3 protein expression, respectively, is given (bottom left). Kaplan-Meyer Plots for overall survival (OS) and event free survival (EFS) are given for patients with low and high EZH2 or H3K27me3 protein levels (log-rank test) (right).
(c) Frequency of EZH2 and H3K27me3 loss at relapse. Protein extracts were prepared from matched patients blasts at diagnosis and subsequent relapse (n=11 pairs). Immunoblots were performed probing membranes with anti-EZH2, anti-beta Actin and anti-H3K27me3 antibodies. Representative Western Blots for each group are given. For remaining diagnosis-relapse pairs see Suppl. Fig. 1g. UPN= unique patient number, D= Diagnosis, R= Relapse. The asterisk indicates samples with ASXL1 mutation at relapse. Mutation data of 54 genes of a myeloid panel are provided for diagnosis and relapse samples in Suppl. Table 3. For all western blot images full length blots have been cropped for better presentation of results. For full length blots see Supplementary Information.
(d) Primary AML cells from patients with normal karyotype (NK) were exposed to vehicle or 1 µM of the methyltransferase inhibitor DZNep for 24 hours. EZH2 protein levels were analyzed by western blot (top). AML blasts were treated with vehicle (-) or DZNep (+) for 24 hours and subsequently exposed to increasing concentrations of cytarabine (AraC). IC50 values for AraC were determined. Means ± s.d. are shown for three technical replicates (bottom left). EZH2/Actin ratios as calculated from western blots using densitometry (see top panel) and AraC IC50 values (see bottom left panel) determined for blasts from all four AML patients with and without DZNep exposure were plotted. An inverse correlation was observed (r= -0.94, p= 0.0005) (bottom right). UPN= unique patient number. Patients characteristics are given in Suppl. Table 4.
(e) EZH2 suppression by shRNAs. Three different shRNAs were initially tested of which shEZH2 #2 conferred the strongest knockdown (see Fig. 2b) and was chosen for further experiments. EZH2 and H3K27me3 protein expression was analyzed in HL60, Kasumi-1 and ML-1 cells transduced with either shControl or shEZH2 #2 (left). Data are representative for two independent experiments. HL60-, Kasumi- and ML-1 shControl and shEZH2 #2 cells were exposed to increasing concentrations of AraC for 72 hours. MTS assays were performed to determine the percentage of proliferating cells. Means ± s.d. are shown for three independent experiments (right).
In matched diagnosis-relapse AML specimens, a reduction of EZH2 protein and H3K27me3 levels was observed in 45% of the relapse samples (Fig. 1c and Suppl. Fig. 1g). Two patients without EZH2 loss but with reduced H3K27me3 contained an ASXL1 mutation at relapse also known to be associated with reduced H3K27me321 (Suppl. Table 3). Since loss of EZH2 protein was acquired for a subset of AML patients during disease progression, we investigated a functional connection between low EZH2 levels and chemoresistance. We exposed primary AML cells with normal karyotype (Suppl. Table 4) to DZNep, a H3K27 methyltransferase inhibitor reported to reduce EZH2 protein levels in AML cells22. DZNep pretreatment as well as simultaneous application induced resistance to the standard chemotherapeutic agent cytarabine (AraC) (Fig. 1d and Suppl. Fig. 1h). An inverse correlation between EZH2 protein levels and AraC IC50 values was observed (r= -0.94, p= 0.0005, Fig. 1d). In line, knockdown of EZH2 in several AML cell lines consistently induced AraC resistance, with an average 5-fold increase in IC50 values compared to control cells (Fig. 1e).
Loss of EZH2 induces drug resistance in vitro and in vivo
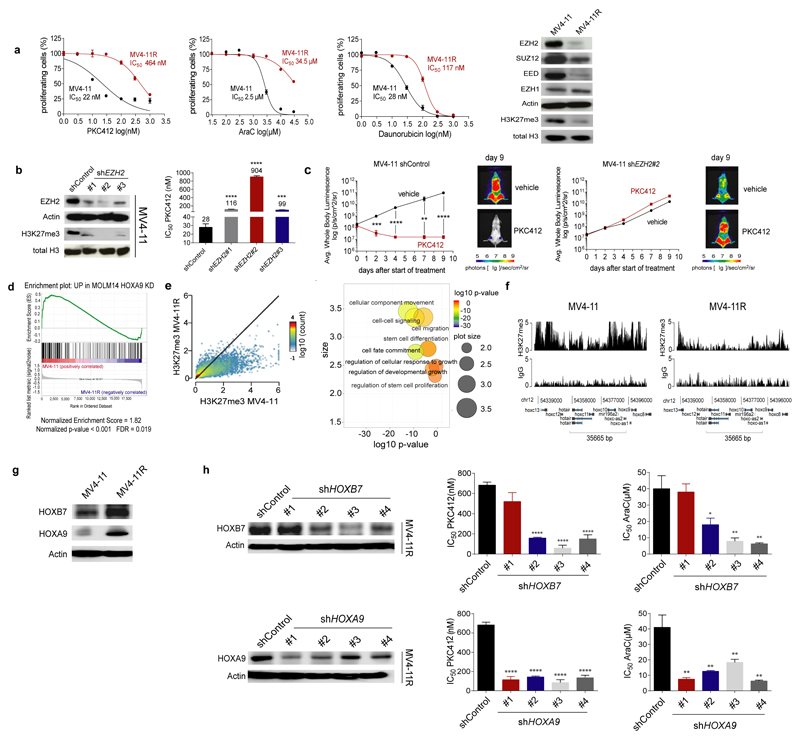
In a complementary line of research we investigated the mechanisms of tyrosine kinase inhibitor (TKI) resistance in the FLT3-ITD positive AML cell line MV4-11 which was generated by prolonged exposure to increasing doses of PKC41223. The PKC412 resistant MV4-11R cells were cross-resistant to other TKIs such as AC220 but also to the standard chemotherapeutic agents cytarabine (AraC) and daunorubicine (Fig. 2a and Suppl. Fig. 2a). These findings pointed towards a generalized drug resistance mechanism. Notably, EZH2 protein levels were lower in MV4-11R compared to parental MV4-11 cells (Fig. 2a). The simultaneous decrease of SUZ12 and EED, the two other main components of the PRC2 complex, suggested destabilization of the complex (Fig. 2a). Accordingly, a strong decrease of H3K27me3 was observed in resistant cells (Fig. 2a). Exome sequencing did not reveal mutations of EZH2 in resistant cells, which could explain the reduced protein levels. Instead the FLT3 N676K mutation as well as TP53 mutations were identified in MV4-11R cells which are known to affect drug sensitivity24, 25 (Suppl. Table 5). But neither introduction of the FLT3 N676K mutation nor shRNA-mediated knockdown of TP53 in parental MV4-11 cells increased drug resistance to the same extent as knockdown of EZH2 which induced a generalized drug resistance (Fig. 2b and Suppl. Fig. 2b-d) although simultaneous knockdown of EZH2 and TP53 showed additive effects on chemoresistance to AraC (Suppl. Fig. 2e). Exposure to EZH2 inhibitors (GSK126, SAH-EZH2, DZNep) also induced drug resistance in sensitive parental MV4-11 (Suppl. Fig. 2f) as well as in FLT3-ITD-positive primary AML blasts (Suppl. Fig. 2g). Of note, drug resistance induced by shRNA-mediated EZH2 knockdown could be reverted by overexpression of a codon-usage modified EZH2 mRNA (Suppl. Fig. 2h). We transplanted MV4-11 control and EZH2 knockdown cells expressing firefly luciferase into NOD-SCID-IL2Rγnull (NSG) mice by intravenous injection. Mice were treated with PKC412 (75mg/kg/d) or vehicle daily for 9 days. PKC412 suppressed growth of MV4-11 control cells whereas EZH2 knockdown cells were resistant to PKC412 treatment in vivo as monitored by Bioluminscence Imaging (BLI) (Fig. 2c and Suppl. Fig. 2i, ** p= 0.0022, *** p= 0.0001, **** p< 0.0001). Further, intravenous transplantation of MV4-11 control and EZH2 knockdown cells in NSG mice and treatment with AC220 or vehicle for 10 days demonstrated a significantly prolonged survival of AC220-treated mice transplanted with control cells compared to mice transplanted with EZH2 knockdown cells (Suppl. Fig. 2j, p= 0.0003).
Figure 2. Loss of EZH2 induces dysregulation of HOX genes in resistant AML cells.
(a) Sensitive and drug-resistant MV4-11 (termed MV4-11R) cells were exposed to increasing concentrations of PKC412, AraC or daunorubicin for 72 hours. MTS assays were performed to determine the percentage of viable, proliferating cells. Means are given for three independent experiments ± s.d. IC50 values were calculated by nonlinear-regression analysis of inhibitor versus normalized response and are indicated (left). Immunoblotting of protein extracts from MV4-11 and MV4-11R was performed to detect PRC2 proteins and H3K27me3. Blots are representative for three independent experiments (right).
(b) Sensitive parental MV4-11 cells were lentivirally transduced with shRNAs targeting EZH2. Protein extracts of MV4-11 shControl and shEZH2 cells were analyzed for EZH2 and H3K27me3 protein expression. Data are representative for two independent experiments (left). MV4-11 shControl and shEZH2 cells were exposed to increasing concentrations of PKC412 for 72 hours. MTS assays were performed to determine the percentage of viable, proliferating cells and IC50 values were calculated as described. Means are indicated for three independent experiments ± s.d. (*** p= 0.0002, **** p< 0.0001) (right).
(c) 5×106 MV4-11 shControl-luc or MV4-11 shEZH2 #2-luc cells were injected i.v. into NSG mice. Three days after tumour cell injection mice were treated with vehicle or PKC412 (75 mg/kg/d) by oral gavaging once daily for 9 consecutive days starting at day 3 post-transplantation. Leukemic bone marrow infiltration was monitored by noninvasive luciferase imaging. Means ± s.d. are given for each group (** p=0.0022, *** p=0.001, **** p< 0.001) (n=6 for PKC412 treatment groups, n=4 for vehicle groups).
(d) Gene Set Enrichment Analysis (GSEA) showed significant enrichment of upregulated genes in sensitive MV4-11 with genes upregulated in Molm14 HOXA9 knockdown cells60 (Normalized Enrichment Score (NES) of 1.82 (p <0.001, false discovery rate [FDR] = 0.019).
(e) ChIP-Seq revealed significant loss of H3K27me3 at multiple loci in MV4-11R cells. Depicted are H3K27me3 positive loci in sensitive and resistant cells (n=11472, r=0.584) (left). Gene Ontology (GO)- pathways analysis was performed for all genes with reduced H3K27me3 in MV4-11R cells. GO- IDs were visualized using RVIGO (Reduce and Visualize Gene Ontology). Representative, significantly overrepresented biological process pathways are shown (right).
(f) ChIP-Seq of H3K27me3 levels at HOX gene loci in MV4-11 and MV4-11R cells are shown. ChIP DNA from three independent experiments was pooled and sequenced.
(g) HOXB7 and HOXA9 protein expression was determined in MV4-11 and MV4-11R cells by western blotting. Blots are representative for two independent experiments.
(h) MV4-11R cells were lentivirally transduced with different shRNAs targeting HOXB7 or HOXA9, respectively, and protein extracts were analyzed for HOXB7 and HOXA9 expression. Data are representative for two independent replicates (left). Control cells, HOXB7- and HOXA9- knockdown cells were exposed to increasing concentrations of PKC412 and AraC respectively, for 72 hours. MTS assays were performed to determine the percentage of viable, proliferating cells. Means of IC50 values ± s.d. are shown for three independent experiments (* p= 0.0131, ** p < 0.009, **** p< 0.0001) (right).
HOX genes are dysregulated in resistant AML cells with loss of EZH2
We next explored the downstream consequences of reduced EZH2 protein levels. In mRNA microarray analyses we identified 202 probes corresponding to 155 genes as being differentially expressed between MV4-11 and MV4-11R cells and a HOXA9 signature emerged in Gene Set Enrichment Analysis (GSEA)26 (Fig. 2d). HOX genes are a family of transcription factors that are highly expressed in hematopoietic stem and progenitor cells whereas HOX gene expression decreases in more differentiated bone marrow cells27. Notably, HOX genes are targets of EZH2 as well as of MLL and MLL fusion proteins28, 29. Classical target genes activated by HOXA9 such as CYBB, CDK6, JUNB, FOS, CDKN1A or IGF1R were upregulated in MV4-11R cells (data not shown). We performed chromatin immunoprecipitation followed by high-throughput DNA sequencing (ChIP-seq) for H3K27me3 to identify genomic regions affected by loss of EZH2 in MV4-11R cells. Gene ontology (GO) analysis revealed that genes affected by loss of H3K27me3 were involved in pathways such as cell fate commitment or stem cell development (Fig. 2e). In line with the microarray expression data, promoter regions of HOX genes from different clusters revealed drastically reduced H3K27me3 levels in the MV4-11R cells (Fig. 2f). Several HOX genes including HOXB7 and HOXA9 were upregulated in resistant cells on mRNA and protein level (Fig. 2g and Suppl. Fig. 3a). In an integrated analysis based on our gene expression and ChIP-seq data, H3K27me3 deposition levels inversely correlated with gene expression whereas H3K4me3 and H3K27ac levels, known as markers for active genes, positively correlated with gene expression (Suppl. Fig. 3b and c). In total, 58 genes, including HOXB7, were differentially regulated in resistant vs. sensitive cells when we combined mRNA expression data with H3K27me3, H3K4me3 and H3K27ac ChIP-seq data30 (Suppl. Fig. 3c and Suppl. Table 6).
Differentially regulated genes were involved in hematopoiesis, myeloid cell differentiation, nucleoside- and ATP- binding and MHC class II pathways (Suppl. Table 7). H3K27me3 levels were also decreased at the promoter of the drug transporter MRP1 (encoded by the ABCC1 gene) in MV4-11R cells with concomitantly increased mRNA and protein levels (Suppl. Fig. 3d and e). In line, increased drug efflux, a typical sign of drug-resistant leukemic (stem) cells, was observed in the resistant cell line (Suppl. Fig. 3f). High HOXB7 and HOXA9 mRNA expression levels were consistently observed in primary AML (Suppl. Fig. 4a) and high mRNA expression levels correlated with poor survival in the TCGA AML data set31 (Suppl. Fig. 4b). Further, EZH2 and HOXB7 or HOXA9 mRNA expression inversely correlated in AML (Suppl. Fig. 4c). EZH2 knockdown in parental sensitive MV4-11 cells as well as in the MLL-translocation negative cell lines HL60, Kasumi-1 and ML-1 induced overexpression of HOXB7 and HOXA9 protein which suggested their regulation by EZH2 in the leukemic cells (Suppl. Fig. 4d and e). Importantly, knockdown of HOXB7 and HOXA9, respectively, restored drug sensitivity in MV4-11R cells and in MLL–translocation negative AML cell lines. (Fig. 2h and Suppl. Fig. 4f and g). The dysregulation of HOX genes as well as an increased transporter efflux point towards a stem-cell-like phenotype of the resistant cells.
The CDK1-EZH2 complex is stabilized by HSP90 in drug resistant AML cells with phosphorylation of EZH2 at Threonine 487
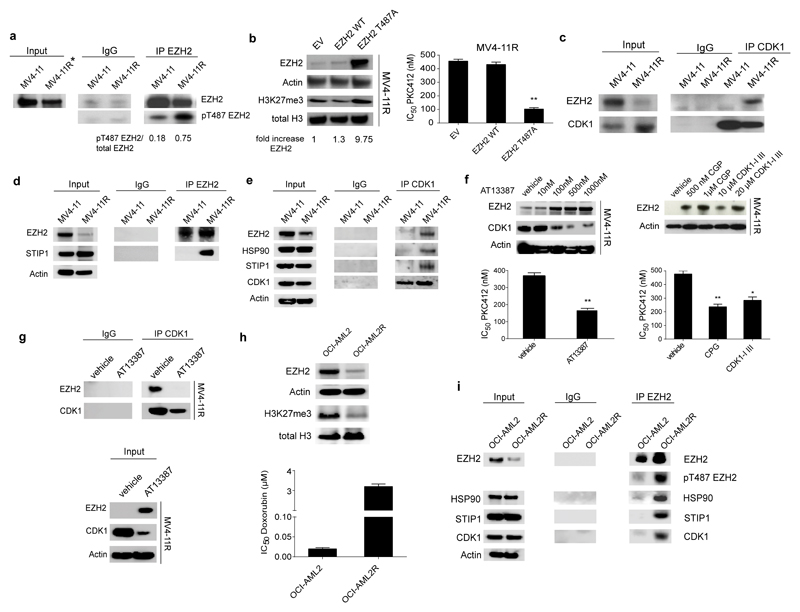
To revert the resistance phenotype we aimed to overexpress EZH2 in MV4-11R cells. However, lentiviral delivery of EZH2 mRNA did not restore EZH2 protein levels in the drug-resistant leukemia cells although the same construct induced EZH2 overexpression in HEK293T cells (Suppl. Fig. 5a). EZH2 mRNA expression, promoter DNA methylation and expression of known regulating miRNAs did not differ significantly between MV4-11 and MV4-11R cells (Suppl. Fig. 5b-d). These data suggested regulation of EZH2 levels by post-translational mechanisms in resistant cells. A label-free quantitative mass spectrometry analysis of immunoprecipitated EZH2 revealed a 2.6-fold increased phosphorylation of the EZH2-residue T487 in resistant cells (p= 0.014, Suppl. Fig. 5e). Resistance-associated phosphorylation at T487 was confirmed by EZH2 immunoprecipitation and western blotting (Fig. 3a). Interestingly, increased phospho-T487-EZH2 levels were also found in relapse samples with reduced total EZH2 from the matched diagnosis-relapse AML pairs (Suppl. Fig. 5f). Expression of a phosphorylation-resistant T487A-EZH2 mutant in MV4-11R cells rescued EZH2 protein expression and increased EZH2 levels by almost 10-fold, whereas wildtype EZH2 protein again could not be expressed (Fig. 3b). The mutant T487A-EZH2 sensitized the formerly resistant cells to PKC412 (Fig. 3b). Analysis of the amino acids in the proximity of T487 revealed the presence of a cyclin dependent kinase 1 (CDK1) consensus sequence [pS/pT]PX[R/K]. Notably, EZH2 protein levels have been shown to be regulated by CDK1-dependent phosphorylation at T487 in HeLa cells and human MSCs32,33, priming EZH2 for ubiquitination and degradation32. A physical interaction between CDK1 and EZH2 was only detected in MV4-11R cells as identified by Co-IP and western blotting. (Fig. 3c). To identify the mechanism of EZH2 phosphorylation and degradation we characterized the EZH2-CDK1 complex by SILAC (Stable Isotope Labeling by Amino Acids in Cell Culture) -based mass spectrometry. IPs with antibodies against phopho-T487-EZH2 and total EZH2 were performed in empty vector (EV) and in mutant T487A-EZH2 overexpressing MV4-11R cells. STIP1 was identified as specific interaction partner of phosphorylated EZH2 in MV4-11R cells, while it was not detected in the phosphorylation-resistant T487A-EZH2 complex (Suppl. Tables 8-11).
Figure 3. CDK1- mediated T487 phosphorylation of EZH2 associates with drug resistance in AML cells.
(a) Protein extracts from MV4-11 and MV4-11R cells were immunoprecipitated using anti-EZH2 antibody. Precipitated proteins were immunoblotted and analyzed with an EZH2 antibody recognizing a different epitope of EZH2 as well as anti-phosphoT487 EZH2 antibody. Results are representative for two independent experiments. For IP experiments a 5-fold excess of resistant IP-lysate was used to normalize IPs for reduced EZH2 levels in resistant cells. The asterisk marks a 5-fold excess of lysate in Input. IP using an IgG antibody served as negative control.
(b) MV4-11R cells were lentivirally transduced with Empty Vector (EV), EZH2 WT or EZH2 T487A mutant. Protein lysates were analyzed for expression of EZH2 and H3K27me3. Results represent data from two independent experiments (left). Resistant cells with EV, EZH2 WT or EZH2 T487A, respectively, were treated with increasing concentrations of PKC412 for 72 hours. MTS assays were performed to determine the percentage of viable, proliferating cells. IC50 values are indicated. Means ± s.d. are shown for three independent experiments (right).
(c) Protein lysates from MV4-11 and MV4-11R cells were prepared and CDK1-bound proteins were pulled-down by anti-CDK1 antibody using 500 µg of protein. Enriched proteins were immunoblotted and probed with anti-CDK1 and anti-EZH2 antibody. Results are representative for two independent experiments.
(d) Protein extracts from MV4-11 and MV4-11R cells were immunoprecipitated using anti-EZH2 antibody. A 5-fold excess of resistant IP-lysate was used to normalize IPs for reduced EZH2 levels in resistant cells. Precipitated proteins were immunoblotted and analyzed with anti-STIP1 antibody. Results are representative for two independent experiments.
(e) Protein extracts from MV4-11 and MV4-11R cells were immunoprecipitated using anti-CDK1 antibody. Precipitated proteins were immunoblotted and analyzed with anti-STIP1, anti-HSP90, anti-EZH2 and anti-CDK1 antibody. Results are representative for two independent experiments.
(f) MV4-11R cells were treated with indicated concentrations of the HSP90 inhibitor AT13387 (left) or with the CDK1 inhibitors CGP74514A (CGP) and CDK1-Inhibitor III (CDK1-I III) (right) for 24 hours. Protein lysates were analyzed for expression of EZH2, CDK1 and Actin by western blot. Blots are representative for two independent experiments (top). Resistant cells were pretreated with 500 nM AT13387, 1µM CGP or 20µM CDK1-I III for 6 hours and proliferation of cells was analyzed in the presence of increasing concentrations of PKC412 after 48 hours by MTS assay. IC50 values are given. Means ± s.d. are shown for three independent experiments (** p< 0.008, * p=0.015) (bottom).
(g) MV4-11R cells were either treated with vehicle or 500nM of the HSP90 inhibitor AT13387 for 24 hours. Protein lysates were prepared and 1000 µg of total protein was immunoprecipitated using anti-CDK1 antibody. Precipitated proteins were immunoblotted and analyzed with anti-EZH2 and anti-CDK1 antibody. IP using an IgG antibody is provided as negative control (top). Input is shown using 20 µg of total protein (bottom). Results are representative for two independent experiments.
(h) Immunoblotting of protein extracts from OCI-AML2 and OCI-AML2R was performed to detect EZH2, Actin, H3K27me3 and total H3. Blots are representative for two independent experiments (top). Sensitive OCI-AML2 and doxorubicin-resistant OCI-AML2R cells were exposed to increasing concentrations of doxorubicin for 72 hours. MTS assays were performed to determine the IC50 of doxorubicin. Means are given for three biological replicates ± s.d. (bottom).
(i) Protein extracts from OCI-AML2 and OCI-AML2R cells were immunoprecipitated using anti-CDK1 antibody. Precipitated proteins were immunoblotted and analyzed with anti-STIP1, anti-HSP90, anti-EZH2, anti-pT487 EZH2 and anti-CDK1 antibodies. Results are representative for two independent experiments.
STIP1 (HOP) is an adaptor protein known to interact with heat shock proteins HSP90 and HSP7034 and we confirmed by Co-IP and western blotting that the interaction between STIP1 and EZH2 occurred in resistant but not in sensitive MV4-11 cells (Fig. 3d). Notably, CDK1 is a known client protein stabilized by HSP9035. STIP1 and HSP90 specifically co-immunoprecipated with CDK1 and EZH2 in MV4-11R cells as analyzed by Co-IP and western blotting (Fig. 3e). These findings suggested that the CDK1-EZH2 interaction could be stabilized by HSP90 in the drug-resistant MV4-11R cells. In line with this hypothesis, treatment with the HSP90 inhibitor AT13387 decreased CDK1 levels and increased EZH2 protein levels as well as drug sensitivity in formerly resistant MV4-11R cells (Fig. 3f) whereas EZH2 levels and drug sensitivity were not significantly affected in the sensitive parental MV4-11 cells (Suppl. Fig. 5g). Also, treatment of MV4-11R cells with CDK1 inhibitors increased EZH2 protein levels and drug sensitivity to PKC412 (Fig. 3f). Inhibition of HSP90 also destabilized the CDK1-EZH2 interaction in MV4-11R cells (Fig. 3g). Notably, in an independent drug resistance model of doxorubicin-resistant OCI-AML2 cells we again identified loss of EZH2 in the resistant cells (Fig. 3h) Here, EZH2 also interacted with STIP1, HSP90 and CDK1 only in the resistant cells (Fig. 3i).
HSP90-mediated stabilization of the CDK1-EZH2 complex appeared to be the predominant mechanism for increased EZH2 phosphorylation in resistant cells. Neither proliferative index, cell cycle distribution, total CDK1 activity nor protein levels of Cyclin B differed between sensitive and resistant cells (Suppl. Fig. 5h and i). Nonetheless, a small contribution of cell cycle alteration cannot be entirely ruled out.
EZH2 undergoes proteasomal degradation in resistant AML and proteasome inhibitors restore EZH2 protein levels and drug sensitivity
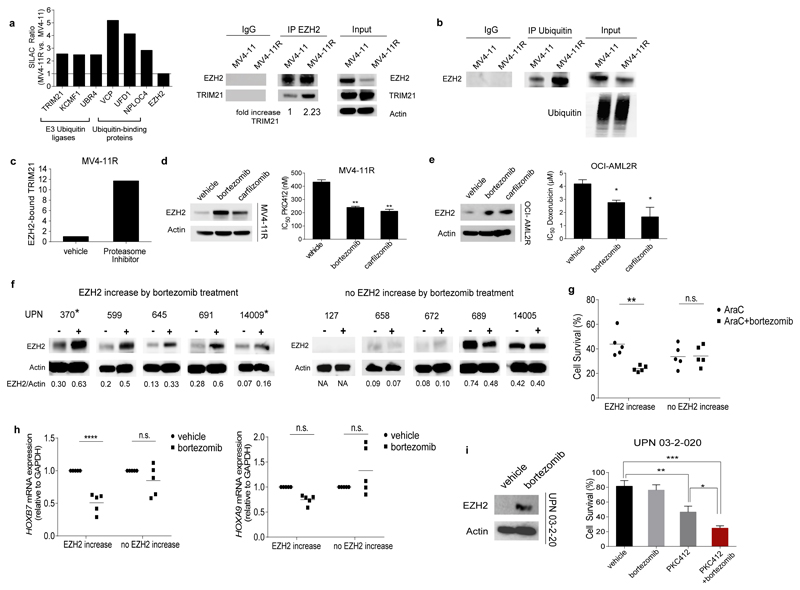
Further SILAC-based mass spectrometry experiments identified enhanced binding of E3 ubiquitin-protein ligases and associated ubiquitin-binding proteins to EZH2 in resistant cells (Fig. 4a and Suppl. Tables 12 and 13). The activity of E3 ubiquitin-protein ligases has been reported to be augmented by phosphorylation of their target proteins36 potentially connecting CDK1-mediated T487 phosphorylation of EZH2 with increased E3 ligase binding and activity. Accordingly, EZH2 protein was strongly ubiquitinated in resistant but not in sensitive MV4-11 cells (Fig. 4b). TRIM21 is an E3-ubiquitin ligase that we identified as an EZH2 binding partner with increased binding to EZH2 in resistant cells (Fig. 4a). In line with this finding, the MS-based protein interaction analysis using an anti-phosphoT487 EZH2 antibody for pull-down of phosphorylated EZH2 confirmed TRIM21 binding to be specific for the phosphoT487-EZH2 in resistant cells (Suppl. Table 10). Furthermore, treatment with proteasome inhibitors and thereby stabilization of EZH2 led to accumulation of TRIM21-EZH2 complexes in MV4-11R cells (Suppl. Table 14 and Fig. 4c). These data suggest that TRIM21 may induce ubiquitination with subsequent proteasomal degradation of phosphoT487-EZH2. Notably, treatment with proteasome inhibitors such as bortezomib, a licensed drug for treatment of multiple myeloma and mantle cell lymphoma, partially restored EZH2 protein levels and drug sensitivity in resistant MV4-11R as well as in resistant OCI-AML2R but not in sensitive parental cells (Fig. 4d, e and Suppl. Fig. 5j and k). In MV4-11R cells HOXB7 and HOXA9 mRNA and protein expression were suppressed by bortezomib treatment (Suppl. Fig. 5l and m). NFkB did not play a role in the resistance mechanism or the bortezomib response of MV4-11R cells (Suppl. Fig. 5n).
Figure 4. EZH2 is degraded by the proteasome in resistant cells and proteasome inhibitors restore EZH2 protein levels and drug sensitivity.
(a) Ubiquitin degradation pathway-associated proteins bind preferentially to EZH2 in resistant cells. Enrichment is represented by the SILAC Ratio ((H/L normalized MV4-11R) / (H/L normalized MV4-11)). H= heavy isotope, L= light isotope. Protein names are indicated at bottom of bars (left). Protein extracts from MV4-11 and MV4-11R cells were immunoprecipitated using anti-EZH2 antibody. A 5-fold excess of resistant IP-lysate was used to normalize IPs for reduced EZH2 levels in resistant cells. Precipitated proteins were immunoblotted and analyzed with anti-TRIM21 and anti-EZH2 antibody. Results are representative for two independent experiments. IP using an IgG antibody is provided as negative control (right).
(b) Immunoprecipitation using anti-Ubiquitin antibody and subsequent immunoblot for EZH2 in MV4-11 and MV4-11R cells. A 5-fold excess of resistant IP-lysate was used to normalize IPs for reduced EZH2 levels in resistant cells. Results are representative for two independent experiments
(c) Enrichment of EZH2-bound TRIM21 in MV4-11R and MV4-11R treated with Proteasome Inhibitors was calculated from the SILAC Ratio ((H/L normalized MV4-11R+Proteasome Inhibitor) / (H/L normalized MV4-11R)) and standardized to EZH2 levels.
(d) MV4-11R cells were exposed to 10 nM bortezomib or carfilzomib for 24 hours. Protein lysates were analyzed for expression of EZH2 by western blot. Data represent two independent experiments (left). MV4-11R cells were pretreated with 10 nM bortezomib or carfilzomib, respectively, for 6 hours and proliferation of cells was analyzed in the presence of increasing concentrations of PKC412 after 48 hours by MTS assay. Means of IC50 values ± s.d. are shown for three independent experiments (** p< 0.01) (right).
(e) OCI-AML2R cells were incubated with 10nM bortezomib or carfilzomib for 24 hours. Protein lysates were analyzed for the expression of EZH2. Data are representative for two independent experiments (left). OCI-AML2R cells were pretreated for 6 hours with 10nM bortezomib or carfilzomib and proliferation of cells was analyzed in the presence of increasing concentrations of doxorubicin after 72 hours by MTS assay. Means of IC50 values ± s.d. are shown for three independent experiments (* < 0.05) (right).
(f) Samples from AML patients with normal karyotype and low to intermediate EZH2 levels were cultured in vitro and exposed to 10nM bortezomib for 6 hours. EZH2/Actin ratios are indicated. Samples from relapsed patients are marked by asterisks. (-) indicates exposure to vehicle, (+) indicates exposure to bortezomib. Clinical characteristics for all patients are listed in Suppl. Table 15. Patients 370, 599 and 689 possess FLT3-ITD mutation.
(g) Treatment response of AML patients blasts with and without EZH2 increase after bortezomib exposure. Blasts were pretreated for 6 hours with 10nM of bortezomib and subsequently exposed to AraC for 48 hours. Data are analyzed by two-way ANOVA with Sidaks post-hoc test (** p< 0.005).
(h) HOXB7 (left) and HOXA9 (right) mRNA expression of AML patients with and without EZH2 increase after bortezomib exposure. The expression of vehicle-treated samples was set at 1 as reference. Data are analyzed by two-way ANOVA with Sidaks post-hoc test (**** p<0.0001).
(i) Primary AML cells from a relapsed AML patient with FLT3-ITD mutation (UPN 03-2-020) were exposed to vehicle or 10 nM bortezomib for 6 hours. EZH2 protein levels were analyzed by western blot (left). Vehicle- or bortezomib-pretreated AML blasts were exposed to 10nM of PKC412. Cell survival was determined by Acridin Orange/PI staining and cell counting from three technical replicates (* p= 0.0122, ** p= 0.0056, *** p= 0.0003) (right).
Restoration of EZH2 protein improves drug sensitivity in AML patient samples
We hypothesized that bortezomib treatment could overcome drug resistance in AML by inhibition of EZH2 degradation and thus analyzed the bortezomib response of primary AML cells in vitro. Low dose bortezomib treatment increased EZH2 protein levels in 5 out of 10 AML patient samples from diagnosis and relapse (Fig. 4f). Increased cytotoxic efficacy of the combination of AraC and bortezomib was predominantly restricted to blasts from patients with increased EZH2 levels after bortezomib exposure (Fig. 4g and Suppl. Fig. 6a and 7a). These blasts also showed decreased HOX gene expression after bortezomib treatment (Fig. 4h and Suppl. Fig. 6b and 7b). Clinical characteristics and cytogenetic findings did not differ between bortezomib responsive and non-responsive patients (Suppl. Table 15). But, non-responsive patients showed a trend for higher number of mutations in a myeloid gene panel (average 4.8 mutations per patient) compared to patients with blasts that did respond to bortezomib exposure (average 2.6 mutations per patient) (p= 0.073, Mann-Whitney test). In FLT3-ITD- positive AML blasts, bortezomib-pretreatment did increase EZH2 levels, sensitivity to AraC (Fig. 4f and Suppl. Fig. 6a) and also to PKC412 (Fig. 4i).
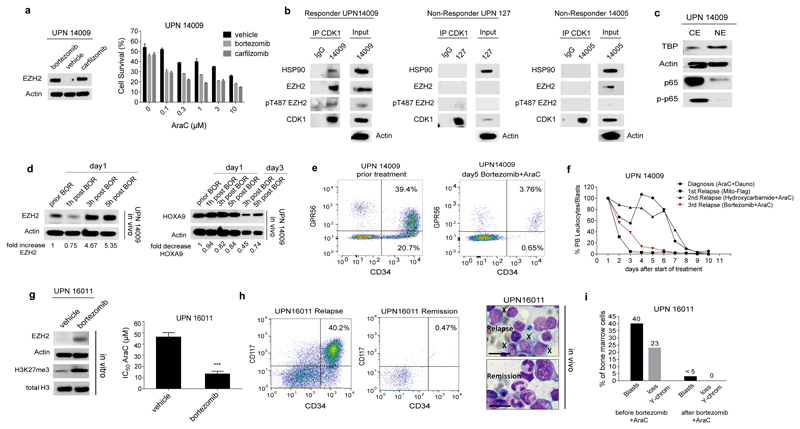
We then focused on one of the tested AML patients (UPN14009) who meanwhile suffered from a third relapse after 2nd allogeneic hematopoietic stem cell transplantation (alloHSCT) and for whom treatment options had been exhausted. Ex vivo treatment with either bortezomib or the second generation proteasome inhibitor carfilzomib induced an increase of EZH2 protein and a solid response to proteasome inhibitors in combination with AraC was observed (Fig. 5a). Primary AML blasts from this patient contained a complex of CDK1-phospho-T487-EZH2-HSP90 as analyzed by Co-IP and western blotting (Fig. 5b). In contrast, the complex was not observed in primary AML blasts that did not show a bortezomib-induced increase of EZH2 protein (Fig. 5b). Nuclear phosphorylated p65, a marker for active NFkB signaling, was not detectable in the blasts of patient 14009 (Fig. 5c). Based on the observed ex vivo responses, the patient was treated with a therapeutic dose of bortezomib (1.3 mg/m2 s.c.) at days 1 and 5 plus 3 g/m2 AraC twice daily at days 1, 3 and 5. Three hours after the initial bortezomib dose, EZH2 protein levels increased in the patient`s blasts (Fig. 5d) with a subsequent decrease of HOXA9 protein levels (Fig. 5d) indicating bortezomib effects on EZH2 stability in an AML patient in vivo. The treatment resulted in a clearance of blasts from the peripheral blood and a significant reduction of CD34+/GPR56+ blasts, representing the more immature blast population37 (Fig. 5e). In addition, the treatment with bortezomib plus AraC induced a more rapid decrease of leukocytes/blasts than most of the conventional chemotherapies applied at diagnosis and former relapses of the patient (Fig. 5f). Blasts from a second AML patient with early relapse on day 240 after allogeneic transplant showed an increase in EZH2 protein levels after in vitro exposure to bortezomib with increased sensitivity to AraC (Fig. 5g). Upon treatment with bortezomib and AraC the patient achieved a second remission with complete blast clearance and 100% donor chimerism (Fig. 5h and i). The patient then underwent a second allogeneic transplant and continued to be in complete remission until the patient died due to graft versus host disease (GVHD) on day 174 after transplantation.
Figure 5. Bortezomib- induced EZH2 increase in AML patients and therapy response.
(a) In vitro treatment of primary cells from the 3rd relapse of patient UPN14009 with bortezomib or the 2nd generation proteasome inhibitor carfilzomib. EZH2 protein levels after 6-hour- treatment are shown (left). Pretreated cells were exposed to increasing concentrations of AraC and cell survival was determined by trypan blue exclusion and cell counting from three technical replicates (right).
(b) Co-IPs were performed for patient samples from which sufficient material was available (UPN 14009, 127 and 14005). Protein extracts were prepared from 5 x107 blast cells and immunoprecipitated using anti-CDK1 antibody. Precipitated proteins were immunoblotted and analyzed with anti-CDK1, anti-EZH2, anti-pT487EZH2 and anti-HSP90 antibodies. Bortezomib Responders and Non-Responders are indicated.
(c) Cytoplasmic (CE) and nuclear extracts (NE) were prepared from Ficoll enriched bone marrow (BM) blasts of patient UPN14009 and analyzed for protein expression of total and phosphorylated p65. Expression of Actin was used as loading control for cytoplasmic extracts. Expression of TATA-Box Binding Protein (TBP) was used as loading control for nuclear extracts. Data are representative for two independent experiments.
(d) Subsequent to in vitro efficacy testing AML relapse patient UPN14009 was treated with bortezomib and AraC. Ficoll-enriched blasts from peripheral blood (PB) were analyzed at indicated time points for EZH2 protein expression (left) and HOXA9 protein expression (right). EZH2 and HOXA9 levels, respectively, were normalized to Actin using densitometry and EZH2 fold change was calculated for different time points after bortezomib application. The EZH2 protein level before bortezomib application was set at 1 as reference. BOR= bortezomib
(e) Flow Cytometry analysis of CD34 and GPR56 expression was performed on blasts of patient UPN14009 during the course of treatment. 10.000 CD45-positive events were analyzed per time point.
(f) Decline of leukocytes is indicated during treatment course of diagnosis, first, second and third relapse of patient UPN14009. Bortezomib+AraC treatment is marked in red. Dosage of each treatment is provided in Suppl. Table 16.
(g) Blasts of patient 16011 were cultured in vitro and exposed to 10nM bortezomib for 24 hours. EZH2 protein levels were analyzed by western blot (left). Blasts were pretreated with 10nM bortezomib for 6 hours and proliferation of cells was analyzed in the presence of increasing concentrations of AraC after 48 hours by MTS assay and IC50 values were calculated. Data represent mean ± s.d. from three technical replicates (*** p= 0.0002) (right).
(h) Flow Cytometry analysis of CD34 and CD17 expression was performed on blasts of patient UPN16011 at relapse and upon remission after bortezomib+AraC treatment (left). About 40% blast cells (marked by X) were present in bone marrow at the time of relapse. After therapy with bortezomib and AraC, remission was achieved with differentiating hematopoiesis and absence of leukemic blasts as demonstrated by hematoxylin-eosin staining. Scale bar indicates 10 µm (right).
(i) Results of the bone marrow analysis before and after relapse treatment with bortezomib and AraC. Shown are the fraction of blast cells (<5% indicates remission) and the fraction of bone marrow cells with loss of Y-chromosome in Fluorescence in situ hybridization (500 cells counted). Loss of Y-chromosome was used as the cytogenetic marker for the leukemia cells of this patient.
Discussion
The histone methyltransferase EZH2 plays a complex role in cancer development, acting either as oncogene or tumor suppressor depending on cancer type. Although loss of EZH2 activity by inactivating mutations has been described to lead to poor prognosis in MDS and MDS/MPN not much is known about EZH2`s role in AML. We here identify that low EZH2 protein levels are associated with poor survival in AML and that loss of EZH2 induces resistance towards multiple drugs including TKIs. We show that loss of EZH2 protein occurs in about 45% of relapsed AML samples. Here, decreased levels of EZH2 protein frequently depend on posttranslational dysregulation of EZH2 protein. The loss of EZH2 mediated via CDK1/HSP90 followed by proteasomal degradation constitutes an epigenetic pathway of drug resistance in AML via deregulation of HOX gene expression (Fig. 6). Several other mechanisms have been reported to decrease EZH2 mRNA and/or protein levels in AML such as 7/7q chromosomal deletions since EZH2 is located on chromosome 7q36.1. Further, splicing dysfunction by mutations in spliceosomal genes such as U2AF1 or SRSF2 has been shown to decrease EZH2 mRNA levels in about 10-25% of AML patients38,39. In AML and MDS, patients with -7/del7q are largely refractory to chemotherapy and possess a particularly poor prognosis20, 40. U2AF1 and SRSF2 mutations also have been shown to be associated with an adverse outcome41, 42 and U2AF1 and inactivating EZH2 mutations are mutually exclusive38. The results from our study that loss of EZH2 induces drug resistance may provide a possible explanation for the poor prognosis of patients with −7/del7q or spliceosomal mutations.
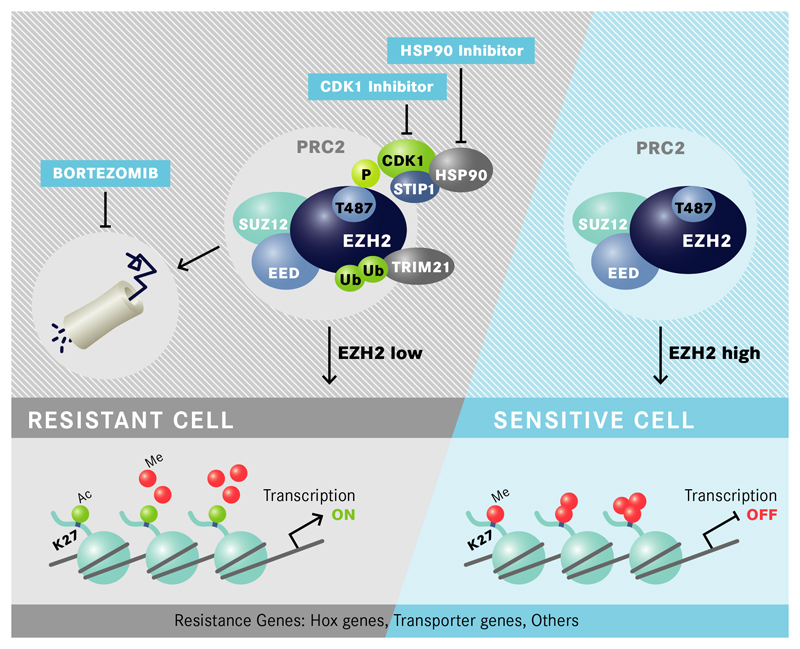
Figure 6. Proposed Model for EZH2- controlled drug resistance in AML cells.
In resistant cells EZH2 is phosphorylated at T487 by CDK1 which is stabilized by STIP1/HSP90 proteins. Phosphorylation of T487 leads to enhanced binding/activity of E3 ubiquitin-protein ligases such as TRIM21, ubiquitination of EZH2 and degradation by the proteasome. EZH2 does no longer introduce H3K27me3 at promoters of resistance genes such as HOXB7, HOXA9 or ABCC1 which become transcriptionally active. EZH2 degradation can be inhibited by proteasome inhibitors as well as HSP90 or CDK1 inhibitors which increase EZH2 protein levels, inactivate expression of resistance genes and restore drug sensitivity (P= phosphorylation, Ub= ubiquitination).
The epigenetic drug resistance mechanism we describe here can be overcome by inhibitors either of CDK1, HSP90 or the proteasome. Notably, in a recent clinical trial the addition of bortezomib to AraC and daunorubicin in induction therapy has been shown to be feasible with a remission frequency of 65% in unselected older patients43. Accordingly, inhibition of EZH2 degradation in combination with chemotherapy and/or TKIs may be a promising therapy concept in drug resistant AML.
Online Methods
Cell Lines and Chemicals
PKC412-resistant MV4-11 cells (termed MV4-11R) were generated by continuous culture of parental MV4-11 with increasing concentrations of PKC412 for 3 months23. Sensitive parental MV4-11, MV4-11R and ML-1 cells were cultured in IMDM (Life Technologies, Darmstadt, Germany) supplemented with 10% fetal bovine serum (Biochrom, Berlin, Germany) at a density of 2 to 10 x105 cells/mL in a humid incubator with 5% CO2 at 37°C. MV4-11R cells were grown in regular medium in the absence of PKC412 for at least 96 hours before start of individual experiments. HL60, Kasumi-1, K562 and U937 cells were cultured in RPMI (Life Technologies, Darmstadt, Germany) supplemented with 10% fetal bovine serum (Biochrom, Berlin, Germany) at a density of 2 to 10 x105 cells/mL in a humid incubator with 5% CO2 at 37°C. Doxorubicin-sensitive and resistant OCI-AML2 (termed OCI-AML2R) were kindly provided by Drs. Ju Han Song and Tae Sung Kim (Korea University, Seoul, Republic of Korea) and cultured in MEM alpha (Life Technologies, Darmstadt, Germany) supplemented with 10% fetal bovine serum (Biochrom, Berlin, Germany) in a humid incubator with 5% CO2 at 37°C. Unless stated otherwise, cell lines were purchased from Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (DSMZ). Cell lines have been tested for mycoplasma contamination.
PKC412 (Midostaurin), CEP-701 (Lestaurtinib), bortezomib (Velcade) and carfilzomib (Kyprolis) were purchased from LC Laboratories (Woburn, MA). ABT-869 (Linifanib) and DZNep were purchased from Cayman Chemicals (Ann Arbor, MI). Cytarabine (AraC) and daunorubicin were obtained from Sigma Aldrich (Taufkirchen, Germany). CGP74514A, CDK1-Inhibitor III and SAH-EZH2 were purchased from Merck Millipore (Schwalbach, Germany). AC220 (Quizartinib) and AT13387 (Onalespib) were purchased from SelleckChem (Houston, TX). GSK126 was obtained from Biovision (Milpitas, CA).
Patient samples
Primary AML samples were obtained from patient peripheral blood (PB) or bone marrow (BM) at the time of diagnosis or relapse with informed consent in accordance with the Declaration of Helsinki. The study was approved by the ethics committees of the Universities of Halle, Münster, Dresden (IRB00001473) and Munich. Mononuclear cells were derived from PB or BM by Ficoll densitiy gradient centrifugation.
Treatment of patients with bortezomib was performed after informed consent on the off-label use following the principles of Helsinki.
Cultivation of primary cells
Primary AML cells were cultured as published44.
Treatment with EZH2 inhibitors
Drug-sensitive MV4-11 cells were treated for 7 days with 10µM of SAH-EZH2 or 3µM GSK126, respectively, as described previously45. The SAH-EZH2 peptide disrupts the interaction of EZH2 and EED thereby reducing EZH2 protein levels whereas GSK126 inhibits the Methyltransferase activity of EZH245. Exposure of sensitive MV4-11 cells and patient samples to 1µM of DZNep was conducted for 24 hours.
Cell viability assays
Leukemic cell lines were seeded in 96-well culture plates at a density of 2.5×104 viable cells/100 µl/well in triplicates and were treated with FLT3-inhibitors or standard chemotherapeutics at the indicated concentrations. Colorimetric CellTiter 96 AQueous One Solution Cell Proliferation assay (Promega, Madison, WI) was used to determine the cytotoxicity. IC50 values were calculated with GraphPad Prism software (San Diego, CA). Experiments were performed in triplicate.
Viability of primary AML cells was determined by trypan blue staining and cell counting or Acridin Orange/PI staining and cell counting, respectively.
Western Blot Analysis
Cells were lysed in RIPA lysis buffer (50 mM Tris-HCL [pH 8], 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, protease inhibitors) for 30 min at 4°C. Protein extracts were resolved by SDS-PAGE and blotted to Nitrocellulose membranes and probed with the following antibodies: anti-beta Actin (Sigma-Aldrich, A5441), anti–EZH2 (AC22, Cell Signaling Technology, 3147S), anti-EZH2 (D2C9, Cell Signaling, 5246), anti-pT487 EZH2 (Abcam, ab109398), anti–SUZ12 (Cell Signaling Technology, 3737S), anti–EED (Millipore, 09-774), anti-EZH1 (Abcam, ab13665), anti-total H3 (Abcam, ab39763), anti-H3K27me3 (Cell Signaling Technologies, 9733), anti-V5 (Abcam, ab9116), anti-total CDK1 (Cell Signaling Technologies, 9112S), anti-pCDK1 (Cell Signaling Technologies, 9111S), anti-MRP1/ABCC1 (Santa Cruz, sc-18835), anti-Ubiquitin (Abcam, ab7780), anti-HOXB7 (Abcam, ab51237), anti-HOXA9 (Abcam, ab140631), anti-TRIM21 antibody (Abcam, ab91432), anti-HSP90 antibody (Abcam, ab13495) and anti-STIP1 antibody (Abcam, ab126724).
Densitometric analysis was performed using ImageJ software (NIH, Bethesda, MD).
Immunoprecipitation and Co-Immunoprecipitation
For EZH2-IPs a 5-fold excess of resistant IP-lysate was used to normalize IPs for reduced EZH2 levels in resistant cells. Immunoprecipitation of EZH2 or Ubiquitin was performed using Dynabeads® Protein G Immunoprecipitation Kit (Life Technologies, Darmstadt, Germany) according to manufacturer`s instructions using following antibodies: rabbit IgG control (Cell Signaling), anti-EZH2 (DC29, Cell Signaling) and anti-Ubiquitin antibody (Abcam, ab7780).
Co-Immunoprecipitation of the CDK1/EZH2/STIP1/HSP90 interaction was performed using Dynabeads® Co-Immunoprecipitation Kit (Life Technologies, Darmstadt, Germany).
Analysis of post-translational modifications of EZH2 by Mass Spectrometry (MS)
Immunoprecipitation of EZH2 using cell lysates from sensitive and resistant cells was performed using Sepharose Beads Immunoprecipitation Kit (Biovision, Milpitas, CA) according to manufacturer`s instructions using following antibodies: anti-EZH2 (DC29, Cell Signaling) and anti-pT487 EZH2 (Abcam, ab109398). The corresponding protein bands on the SDS-PAGE containing EZH2 were cut off and sliced into small gel pieces for following in-gel digestion. After two-times of washes with 25 mM ammonia bicarbonate buffer, gel slices were reduced with dithiothreitol, alkylated with 2-iodoacetamide and digested with trypsin overnight. The resulting peptide mixtures were then extracted and dried in a SpeedVac as described previously46.
For phosphopeptide enrichment, digested peptides were re-suspended in loading buffer (80% acetonitrile (ACN), 5% trifluoroacetic acid (TFA), 5% glycerol) and loaded onto a pre-equilibrated self-packed TiO2 spin column (5 µm, GL Science, Japan) twice by centrifugation. The TiO2 beads were then sequentially washed with three times of loading buffer, three times of 80% ACN, 5% TFA and one time with 60% ACN, 0.1% TFA. Phosphopeptides were eluted with 0.5 N NH4OH (pH ≥10.5) and dried in a SpeedVac (Thermo Fisher Scientific) as described previously46.
LC-MS/MS analysis was performed on a Q Exactive HF mass spectrometer (Thermo Fisher Scientific, USA) coupled with an Ultimate 3000 RSLC system (Dionex, USA). Phosphopeptides were first enriched on a self-packed precolumn (100 µm × 3 cm, Reprosil-Pur120 C18-AQ 3 µm, Dr. Maisch GmbH, Germany) and then separated on an analytical column (75 µm × 35 cm, ReproSil-Pur 120 C18-AQ, 1.9 µm, Dr. Maisch GmbH) with a 40-min linear gradient of 2%-40% buffer B (80% ACN, 0.1% formic acid (FA)) mixed with corresponding composition of buffer A (0.1% FA in water) at a constant flow rate of 300 nl/min. The Q Exactive HF was operated in a data-dependent acquisition mode where one full MS scan across the 350-1600 m/z range was acquired at a resolution setting of 120,000 FWHM (full width, half maximum) to select up to 20 most abundant peptide precursors of charge states 2 to 6 above a 3×104 intensity threshold, at an isolation width of 1.4 m/z. Precursors were fragmented by Higher Collision Energy Dissociation (HCD) with nitrogen at a normalized collision energy setting of 28%, and their product ion spectra were recorded with a start mass of 110 m/z at resolution of 30,000 FWHM. Automatic gain control target value and maximum ion injection times for MS and MS/MS were 1 × 106 in 50 ms, and 1 × 105 in 64 ms, respectively. Selected precursor m/z values were then excluded for the following 30 s.
The MS raw files were processed by MaxQuant47 (version 1.5.2.8) and MS/MS spectra were searched against UniProt human database (downloaded on Feb, 2015; 89,796 entries) via the Andromeda search engine48. Mass tolerance after recalibration of precursor mass and fragment ion mass were set as 6 and 20 ppm, respectively. Allowed variable modifications included protein deamidation (N), oxidation (M) and phosphorylation (STY). Cysteine carbamidomethylation was defined as a fixed modification. Minimal peptide length was set to 7 amino acids with the maximum of two enzymatic mis-cleavages. The false discovery rate (FDR) was set to 1% for both peptide and protein identifications. Intensities of all identified peptides were determined by the MaxQuant with the option of “match between runs” on.
Identification of Interaction Partners of EZH2 using SILAC-immunoprecipitation Quantitative Proteomics
MV4-11 and MV4-11R cells were each cultured in medium containing arginine and lysine labeled with heavy (H) or light (L) isotopes (SILAC medium, Thermo Scientific, Rockford, USA), respectively, for a total of 5 cell passages. Protein extracts were prepared and immunoprecipitation was performed using either anti-total EZH2/anti-pT487 EZH2 antibody (heavy-labeled extracts) or beads only (light-labeled extracts) for sensitive and resistant cells, respectively. A 5-fold excess of resistant IP-lysate was used to normalize IPs for reduced EZH2 leves in resistant cells. IPs were performed from two cell culture replicates. The beads were resuspended in 50 µl NuPAGE LDS Sample buffer (Invitrogen, Carlsbad, CA, USA) supplemented with 0.1 M DTT. After denaturation for 10 min at 95°C, the denaturated H- and L-labeled proteins were pooled in equimolar amounts and subsequently the samples were separated by size using an 1D-PAGE (Invitrogen, Carlsbad, CA, USA) followed by coomassie blue staining. Each gel lane was cut into 23 gel slices. Proteins from each slice were in-gel digested with trypsin (Promega, Madison, WI) according to the protocol described49. MS measurements were carried out using a nanoflow HPLC (Agilent, Boeblingen, Germany) coupled to a nanoelectrospray Q exactive HF mass spectrometer (Thermo Fisher Scientific, Waltham, MA) as previously described50.
Cloning
For knockdown experiments oligonucleotides corresponding to gene specific small hairpin RNA (shRNA) sequences targeting EZH2 as well as a scrambled control shRNA were cloned into pGreen Puro shRNA Cloning and Expression Lentivector (System Biosciences, Mountain View, CA) following manufacturer`s instructions.
shEZH2 #1 Oligos
fwd: GATCCAAGCTAAGGCAGCTGTTTCAGCTTCCTGTCAGACTGAAACAGCTGCCTTAGCTTTTTTTG
rev: AATTCAAAAAAAGCTAAGGCAGCTGTTTCAGTCTGACAGGAAGCTGAAACAGCTGCCTTAGCTTG
shEZH2 #2 Oligos
fwd: GATCCCGGAAATCTTAAACCAAGAATCTTCCTGTCAGA ATTCTTGGTTTAAGATTTCCGTTTTTG
rev: AATTCAAAAACGGAAATCTTAAACCAAGAATTCTGACAGGAAG ATTCTTGGTTTAAGATTTCCGG
shEZH2#3 Oligos
fwd: GATCCGCTAGGTTAATTGGGACCAAACTTCCTGTCAGATTTGGTCCCAATTAACCTAGCTTTTTG
rev: AATTCAAAAAGCTAGGTTAATTGGGACCAAATCTGACAGGAAGTTTGGTCCCAATTAACCTAGCG
shControl Oligos
fwd: GATCCGTTGACAGTAAGCGATCTCCTTCCTGTCAGAGAGATCGCTTACTGTCAACTTTTTG
rev: AATTCAAAAAGTTGACAGTAAGCGATCTCTCTGACAGGAAGGAGATCGCTTACTGTCAACG
A lentiviral expression vector containing a shRNA against TP53 was purchased from Addgene (shp53 pLKO.1 puro, Plasmid 19119). The pLKO.1 scrambled control was also ordered from Addgene (Plasmid 1864).
For overexpression of WT and mutant T487A EZH2, pTYF Vectors containing WT EZH2, and EZH2 T487A coding sequence were kindly donated by the lab of Yi Zhang (Chapel Hill, NC) and coding regions were subcloned into pCDH lentiviral expression vector (System Biosciences, Mountain View, CA) according to manufacturer`s instructions introducing a V5 tag at the 3` end of the EZH2 sequence. All constructs were verified by DNA sequencing.
For generation of the FLT3-ITD-N676K overexpression lentivector, RNA was isolated from sensitive MV4-11 and reverse transcribed using SuperScript (Life Technologies, Darmstadt, Germany). The coding region of FLT3, harboring the ITD mutation, was amplified from the cDNA introducing a V5 tag at the 3` end. The amplified sequence was introduced into TOPO Cloning Vector and site directed mutagenesis was performed using Quick Change Kit (Promega, Mannheim, Germany). The construct was verified by DNA sequencing for existence of N676K mutation before PCR-based cloning into pCDH lentiviral expression vector.
Lentiviral vectors expressing shRNAs directed against HOXB7 and HOXA9 were purchased from abm Good (Richmond, Canada).
EZH2 codon usage adaption was performed using GENEius and the resulting sequence was synthesized and cloned into pCDH at Eurofins (Penzberg, Germany).
Lentiviral Transfection and Transduction
HEK293T cells were transfected with 10 µg of lentiviral plasmid and ViraPower helper plasmids (Life Technologies, Darmstadt, Germany) using TurboFect transfection Reagent (Thermo Fisher Scientific, Schwerte, Germany). Supernatants containing viral particles were harvested 48 and 72 hours after transfection and virus particles were precipitated by ultracentrifugation. Lentiviral transduction of human cell lines was performed in the presence of 8 μg/ml polybrene (Sigma-Aldrich, Taufkirchen, Germany).
FACS-based assays
Drug Efflux Assay
Activity of drug transporters P-gp, MRP1 and BCRP was flow-cytometrically analyzed by use of eFLUXX-ID Multidrug Resistance Assay Kit (Enzo Life Sciences, Lörrach, Germany).
Cell Cycle Analysis via BrDU
For Cell Cycle Analysis AML cells were synchronized by serum starvation for 24 hours. Upon release cells were incubated with 10µM BrDU for 1 hour, washed in PBS and stained with anti-BrdU-FITC and Propidium Iodid (PI) according to the manufacturer’s instructions (BD Pharmingen, San Diego, CA). Analysis was performed using a BD LSR Fortessa cytometer (BD Biosciences, San Jose, CA).
Ki67 staining
To determine the proliferative index AML cell lines were stained using the PE Mouse Anti-Ki-67 Set (BD Pharmingen, San Diego, CA) according the the manufacturer`s instructions. Analysis was performed using a BD LSR Fortessa cytometer (BD Biosciences, San Jose, CA).
CDK1-Kinase Activity Assay
Analysis of CDK1 activity was performed by using MESACUP® Cdc2/Cdk1 Kinase Assay Kit according to manufacturer`s instructions (MBL International, Woburn, MA)
Immunohistochemistry
For tissue microarray (TMA) preparations, bone marrow trephines were obtained from 136 patients with newly diagnosed AML. Bone marrow trephines were formalin-fixed and paraffin-embedded. Written informed consent was obtained from all patients and the study was approved by the local Institutional Review Board. Only patients treated at the University Hospital of Münster and with available clinical follow-up data were included. The study was in compliance with all applicable national and local ethics guidelines. Only patients receiving intensive treatment were included in further analysis (n=124). Preparation of TMAs and staining was performed as previously described51. For staining following antibodies were used: EZH2 (Cell Signaling Technologies, #3147) and H3K27me3 (Active Motif, #39155). As the range of the blast percentage in individual bone marrow trephines was diverse (ranging from 20-99%) EZH2 expression was only scored in the blast cells, not in normal hematopoietic cells. The staining intensity of blasts was scored 0 (no staining), 1 (weak staining), 2 (moderate staining) or 3 (strong staining) according to Remmele`s Immunoreactive Score (IRS)52. To discriminate between low and high EZH2 expression scores 0 and 1 were considered as low, scores 2 and 3 were considered as high expression. Clinical data of patients are given in Supplementary Table 1.
Xenograft Mouse Models
NOD.Cg-Prkdcscid IL2rgtm1Wjl/SzJ (NSG; Charles River Laboratories, Sulzfeld, Germany) were maintained under specific pathogen-free conditions in the research animal facilities of the Helmholtz Center Munich, Germany and of the University Hospital Halle (Saale), Germany. Animals had free access to food and water, and were housed with a 12-hour light–dark cycle and constant temperature. NSG animal trials were performed in accordance with the current ethical standards of the official committee on animal experimentation (written approval by Regierung von Oberbayern and Landesverwaltungsamt Sachsen-Anhalt, Germany). Animals that lost more than 15% of their body weight during treatment course were euthanized as per institutional policy. For all animal studies no blinding was performed. No statistical method was used to predetermine sample size.
5×106 MV4-11 shControl and MV4-11 shEZH2#2 cells expressing a recombinant codon-optimized form of firefly luciferase were transplanted via tail-vein injection into sublethally irradiated 16-week-old female NSG mice. Tumor burden was assessed by bioluminescence imaging (BLI) using an IVIS Lumina II Imaging system (Caliper Life Sciences, Mainz, Germany) as previously described53. Three days after tumor cell injection randomization was done by assessing leukemia establishment using BLI and confirming that engraftment was equivalent in all drug and vehicle groups before start of treatment. Mice were treated by gavage either with vehicle control (VitE-PEG-CornOil, n=4) or PKC412 at 75 mg/kg/day (n=6) daily for 9 days. Treatment efficacy was monitored by BLI every 2–3 days.
For AC220 inhibitor studies, female 8-10-weeks-old NSG mice (Charles River Laboratories, Germany) were sublethally irradiated with 2Gy of total body irradiation 6 hours before injection of leukemic cells. 3×106 MV4-11 -control and -EZH2 knockdown cells were intravenously injected. Before start of treatment mice were randomly split into groups of 7 mice and mice were treated by gavage either with vehicle (22% hydroxypropyl-β-cyclodextrin) or AC220 (SelleckChem, Houston, TX) at 10mg/kg daily for 10 days. Survival of the drug- and vehicle-treated mice was measured as the time from transplantation until moribund state. Survival benefit was assessed by Kaplan–Meier analysis.
Quantitative RT-PCR (qPCR)
TaqMan Gene expression Assays (Applied Biosystems) using primer/probe sets for GAPDH (assay Hs02758991_g1), EZH2 (assay Hs00544833_m1), HOXB7 (Hs04187556_m1), HOXA9 (Hs00365956_m1), HOXD12 (Hs00706957_s1) and ABCC1 (Hs01561502_m1) were performed according to manufacturer`s instructions. Clinical data of patients analyzed by qPCR for EZH2 expression are given in Supplementary Table 2.
Exome Sequencing
Libraries for Exome Sequencing were prepared using the SureSelect XT Human All Exon V5+UTR kit (Agilent, Böblingen, Germany) and 3 µg of gDNA following the manufacturer`s instructions. Briefly, DNA was sheared by sonication, end-repaired and adenylated at 3` ends. Paired end adapters were ligated and DNA was amplified. Libraries were hybridized to exonic probes and captured DNA was amplified and index-tagged for sample pooling. Exome enriched Libraries were subject to 2 × 100 cycles paired-end sequencing on a HiScanSQ instrument (Illumina, San Diego, CA). An average of 77.516.833 reads was obtained per sample with a read length of 101 bps. 98.15% and 98.33% of reads could be mapped for sensitive and resistant MV4-11 cells, respectively, with a mean target coverage of 91.9.
The Burrows-Wheeler alignment algorithm (BWA)54 was applied to map raw reads from the Illumina HiScanSQ to the hg19 genome. Duplicate reads were removed and the Genome Analysis Toolkit (GATK) was used to apply local realignment, base quality score recalibration and SNP and INDEL discovery across all samples simultaneously. Standard filtering parameters were applied to remove low quality mutations. Detected variations were transferred to the Ingenuity© Variant Analysis (CA, USA) online software.
Diagnostic Sequencing
Molecular alterations in patients were evaluated by targeted resequencing using a commercially available TruSight Myeloid assay (Illumina, Chesterford, UK) which covers 54 genes or gene hotspots related to myeloid neoplasms as follows: BCOR, BCORL1, CDKN2A, CEBPA, CUX1,DNMT3A, ETV6, EZH2, IKZF1, KDM6A, PHF6, RAD21, RUNX1, STAG2 and ZRSR2 and oncogenic hotspots of ABL1, ASXL1, ATRX, BRAF, CALR, CBL, CBLB, CBLC, CDKN2A, CSF3R, FBXW7, FLT3, GATA1, GATA2, GNAS, HRAS, IDH1, IDH2, JAK2, JAK3, KIT, KRAS, MLL, MPL, MYD88, NOTCH1, NPM1, NRAS, PDGFRA, PTEN, PTPN11, SETBP1, SF3B1, SMC1A, SMC3, SRSF2, TET2, TP53, U2AF1 and WT1. For preparation of target enrichment libraries, 50 ng of genomic DNA were used and prepared as recommended in the manufactures protocol (TruSight Myeloid Sequencing Panel Reference Guide 15054779 v02, Illumina). Samples were paired-end sequenced (2x225 bp) on a MiSeq sequencer using MiSeq Reagent Kits V3 (Illumina). Sequence data alignment of demultiplexed FastQ files, variant calling and filtering was done using the Sequence Pilot software package (JSI medical systems GmbH, Ettenheim, Germany) with default settings and a 5% variant allele frequency (VAF) mutation calling cut-off. Detection of large insertions was performed using PINDEL algorithm55 following BWA-MEM54 mapping with default settings. Human genome build HG19 was used as reference genome for mapping algorithms.
Reduced Representation Bisulfite Sequencing (RRBS)
A total of 0.3 to 1 μg of DNA was used for RRBS library preparation using published protocols56, 57.
mRNA Expression Arrays
Human Gene 1.0 ST Arrays (Affymetrix, CA, USA) using RNA isolated from sensitive MV4-11 and resistant MV4-11R cells were processed according to the manufacturer`s instructions. Arrays were scanned at 1.56 μm resolution using the Affymetrix GeneChip Scanner 3000. Raw gene expression data were imported to the Affymetrix expression console and subject to robust multi-array average (RMA). Differential gene expression was calculated with RankProd software (Ref6).
Expression array raw data are available at NCBI Gene Expression Omnibus (GSE61786).
mRNA expression data sets
Published mRNA expression data sets of AML patients deposited at The Cancer Genome Atlas (TCGA) (https://tcga-data.nci.nih.gov/tcga/) or Leukemia Gene Atlas (LGA) (www.leukemia-gene-atlas.org) were either directly analyzed or downloaded and analyzed using Graph Pad Prism6 (GraphPad Software, San Diego, CA).
Chromatin IP (ChIP) and High Throughput Sequencing
Cells were fixed for 10 min in culture media containing 1% formaldehyde and were processed for ChIP as previously described58. Briefly, formaldehyde fixed chromatin was sonicated using Bioruptor (Diagnode, Liege, Belgium) in IP buffer (2 volumes SDS Buffer : 1 volume Triton Dilution Buffer; SDS Buffer: 100mM NaCl 50mM Tris-Cl, pH8.1, 5mM EDTA, pH 8.0, 0.2% NaN3, 2% SDS; Triton Dilution Buffer: 100mM Tris-Cl, pH 8.6, 100mM NaCl, 5mM EDTA, pH 8.0, 0.2% NaN3, 5.0% Triton X-100) for 14 cycles (30sec on and 30 sec off). 10ug DNA (sonicated chromatin) was used for each ChIP in IP buffer. Following antibodies were used: Rabbit IgG (DAKO), rabbit mAb against-H3K27me3 (Cell Signaling, #9733), H3K4me3 (Cell Signaling, #9751) and H3K27ac (Abcam, ab4729). Samples were incubated with antibodies overnight at 4° C and immunocomplexes were precipitated with Protein A-sepharose beads with 3 h rotation at 4° C. After subsequent washes the samples were decrosslinked overnight at 68° C with shaking in elution buffer (20mM Tris-HCl pH= 7.5, 5mM EDTA, 50mM NaCl), 1%SDS and 50ug/ml Proteinase K. Finally, ChIP DNA was purified using Qiagen PCR purification kit. ChIP DNA from three parallel ChIPs was pooled and 10 ng was used for preparation of ChIP-seq libraries. The libraries were prepared using ChIP seq DNA sample preparation kit from NEB following manufacturer’s instructions. Samples were multiplexed and run on a HiSeq2000 (Illumina). Sequencing of ChIP-seq samples produced single-end 42bp reads after removal of multiplex index sequence tags. Reads were trimmed for low quality nucleotides and mapped to the hg19 human genome sequence using Bowtie v0.12.7 applying the parameters "-S -m 1". Subsequent handling, analysis and visualization of tracks and scatter plots was done using EaSeq59. Aligned datasets were filtered for multiple reads at the same position occurring after PCR amplication. Where more than one read mapped to the same position and at the same strand duplicates were removed. The reads were extended to a DNA fragment size of 200 bp, and the number of reads overlapping with an area of +/-5 kbp of TSS for H3K27me3 and +/-1 kbp of TSS for H3K27ac and H3K4me3, shown in 2D-histograms or at the positions used in the tracks, was counted and normalized to fragments pr. Mreads pr. kbp. The number of fragments was derived from the count by dividing it with (1 + DNA fragment size / bin size). ChIP-seq raw data are available at NCBI Gene Expression Omnibus (GSE61786). Correlation of mRNA expression data with H3K27me3, H3K4me3 and H3K27ac ChIP-seq data was performed as previously described30.
Statistical Analysis
For statistical analyses, SPSS Version 22 (IBM, Ehningen, Germany) and GraphPad Prism 6 (GraphPad Software, San Diego, CA) were used.
Data were analyzed for normal distribution before statistical analyses. Values are presented as mean ± s.d. of replicates. Two-tailed Student’s test and Mann-Whitney-U test were used to determine statistical significance for bar charts unless stated otherwise. The comparison of multiple groups was carried out by two-way ANOVA with Sidak´s post-hoc test or Tukey´s post-hoc test, respectively. Nonlinear-regression analysis of inhibitor versus normalized response was performed for calculation of IC50 values. For Kaplan-Meier survival analysis, the log-rank test was used to determine statistical significance. P-values < 0.05 were considered to be statistically significant. No statistical method was used to predetermine sample size. The investigators were not blinded to allocation during experiments and outcome assessments.
Supplementary Material
Acknowledgements
We are grateful to Drs. Ju Han Song and Tae Sung Kim from Korea University, Seoul, Republic of Korea and to Dr. Salem Chouaib from INSERM U1186, Gustave Roussy Cancer Campus, Villejuif, France for providing cell lines.
The authors thank Dorte Wendt-Cousin and Anika Küttner for excellent technical assistance, and Konstantin Agelopoulos and Isabel Schulze for assistance with Exome library preparation and processing. We are grateful to the University of Münster core facilities IFG/IZKF for performing the Affymetrix gene expression arrays and the LIFA for Illumina sequencing of Exome libraries. We thank atelier42 for designing the graphical model of chemoresistance. SG and CMT hold the copyright for the graphic art.
The Münster laboratory of W.E.B. was supported by the Deutsche Forschungsgemeinschaft, DFG EXC 1003 Cells in Motion – Cluster of Excellence. This work was further supported by grants from the Deutsche Forschungsgemeinschaft (SPP1463, MU1328/9-2 to C.M.T), the German Cancer Aid Foundation (111286 to C.M.T), the German José-Carreras Leukemia Foundation (DJCLS R 13/04 to C.M.T) and the state of Sachsen-Anhalt.
Footnotes
Accession Codes
Gene Expression Omnibus GSE61786
Data availability
ChIP-Seq and mRNA microarray data are stored at NCBIs Gene Expression Omnibus (GEO) data repository with the Accession Code GSE61786.
Author Contributions
S.G., S.A.-S. and T.S. performed cell culture experiments, ChIP-Seq, quantitative PCR, RNAi, western blotting, immunoprecipitations and flow cytometry. T.S., S.T., G.K. and C.W. performed tissue microarray production, staining and analysis. F.S., G.E. and C.T. performed EZH2 mRNA expression analysis of AML patients and established the MV4-11R cell line. C.T., W.E.B., A.Z., L.P.M and K. S. provided patient samples. H.-U.K., M.D., S.A.-S., K.H., C.R. and M.L. performed bioinformatic analysis of mRNA expression microarrays, ChIP-Seq and Exome-Seq data. T.O., H.S., K.-T.P. and H.U. performed and analyzed label free and SILAC-labeled mass spectrometry of EZH2. C.T. and S.H. performed diagnostic sequencing of primary AML samples. S.G., C.P., B.V. and I.J. performed and analyzed mouse experiments. All authors discussed the results and commented on the manuscript. S.G. and C.M.-T. designed the study, analyzed the data and wrote the paper.
T.O. and S.A.-S. share second authorship of this paper.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Roboz GJ. Novel approaches to the treatment of acute myeloid leukemia. Hematology Am Soc Hematol Educ Program. 2011;2011:43–50. doi: 10.1182/asheducation-2011.1.43. [DOI] [PubMed] [Google Scholar]
- 2.Schenk T, et al. Inhibition of the LSD1 (KDM1A) demethylase reactivates the all-trans-retinoic acid differentiation pathway in acute myeloid leukemia. Nat Med. 2012;18:605–611. doi: 10.1038/nm.2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krug U, Buchner T, Berdel WE, Muller-Tidow C. The treatment of elderly patients with acute myeloid leukemia. Dtsch Arztebl Int. 2011;108:863–870. doi: 10.3238/arztebl.2011.0863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Knoechel B, et al. An epigenetic mechanism of resistance to targeted therapy in T cell acute lymphoblastic leukemia. Nat Genet. 2014;46:364–370. doi: 10.1038/ng.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharma SV, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ribeiro AF, et al. Mutant DNMT3A: a marker of poor prognosis in acute myeloid leukemia. Blood. 2012;119:5824–5831. doi: 10.1182/blood-2011-07-367961. [DOI] [PubMed] [Google Scholar]
- 7.Chou WC, et al. TET2 mutation is an unfavorable prognostic factor in acute myeloid leukemia patients with intermediate-risk cytogenetics. Blood. 2011;118:3803–3810. doi: 10.1182/blood-2011-02-339747. [DOI] [PubMed] [Google Scholar]
- 8.Metzeler KH, et al. ASXL1 mutations identify a high-risk subgroup of older patients with primary cytogenetically normal AML within the ELN Favorable genetic category. Blood. 2011;118:6920–6929. doi: 10.1182/blood-2011-08-368225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ernst T, et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat Genet. 2010;42:722–726. doi: 10.1038/ng.621. [DOI] [PubMed] [Google Scholar]
- 10.Shih AH, Abdel-Wahab O, Patel JP, Levine RL. The role of mutations in epigenetic regulators in myeloid malignancies. Nat Rev Cancer. 2012;12:599–612. doi: 10.1038/nrc3343. [DOI] [PubMed] [Google Scholar]
- 11.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O'Carroll D, et al. The polycomb-group gene Ezh2 is required for early mouse development. Mol Cell Biol. 2001;21:4330–4336. doi: 10.1128/MCB.21.13.4330-4336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nikoloski G, et al. Somatic mutations of the histone methyltransferase gene EZH2 in myelodysplastic syndromes. Nat Genet. 2010;42:665–667. doi: 10.1038/ng.620. [DOI] [PubMed] [Google Scholar]
- 14.Guglielmelli P, et al. EZH2 mutational status predicts poor survival in myelofibrosis. Blood. 2011;118:5227–5234. doi: 10.1182/blood-2011-06-363424. [DOI] [PubMed] [Google Scholar]
- 15.Sashida G, et al. Ezh2 loss promotes development of myelodysplastic syndrome but attenuates its predisposition to leukaemic transformation. Nat Commun. 2014;5:4177. doi: 10.1038/ncomms5177. [DOI] [PubMed] [Google Scholar]
- 16.Varambally S, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 17.Zingg D, et al. The epigenetic modifier EZH2 controls melanoma growth and metastasis through silencing of distinct tumour suppressors. Nat Commun. 2015;6:6051. doi: 10.1038/ncomms7051. [DOI] [PubMed] [Google Scholar]
- 18.Bitler BG, et al. Synthetic lethality by targeting EZH2 methyltransferase activity in ARID1A-mutated cancers. Nat Med. 2015;21:231–238. doi: 10.1038/nm.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morin RD, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet. 2010;42:181–185. doi: 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grimwade D, et al. Refinement of cytogenetic classification in acute myeloid leukemia: determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood. 2010;116:354–365. doi: 10.1182/blood-2009-11-254441. [DOI] [PubMed] [Google Scholar]
- 21.Abdel-Wahab O, et al. ASXL1 mutations promote myeloid transformation through loss of PRC2-mediated gene repression. Cancer Cell. 2012;22:180–193. doi: 10.1016/j.ccr.2012.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fiskus W, et al. Combined epigenetic therapy with the histone methyltransferase EZH2 inhibitor 3-deazaneplanocin A and the histone deacetylase inhibitor panobinostat against human AML cells. Blood. 2009;114:2733–2743. doi: 10.1182/blood-2009-03-213496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stolzel F, et al. Mechanisms of resistance against PKC412 in resistant FLT3-ITD positive human acute myeloid leukemia cells. Ann Hematol. 2010;89:653–662. doi: 10.1007/s00277-009-0889-1. [DOI] [PubMed] [Google Scholar]
- 24.Heidel F, et al. Clinical resistance to the kinase inhibitor PKC412 in acute myeloid leukemia by mutation of Asn-676 in the FLT3 tyrosine kinase domain. Blood. 2006;107:293–300. doi: 10.1182/blood-2005-06-2469. [DOI] [PubMed] [Google Scholar]
- 25.Bunz F, et al. Disruption of p53 in human cancer cells alters the responses to therapeutic agents. J Clin Invest. 1999;104:263–269. doi: 10.1172/JCI6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sauvageau G, et al. Differential expression of homeobox genes in functionally distinct CD34+ subpopulations of human bone marrow cells. Proc Natl Acad Sci U S A. 1994;91:12223–12227. doi: 10.1073/pnas.91.25.12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Armstrong SA, Golub TR, Korsmeyer SJ. MLL-rearranged leukemias: insights from gene expression profiling. Semin Hematol. 2003;40:268–273. doi: 10.1016/s0037-1963(03)00196-3. [DOI] [PubMed] [Google Scholar]
- 29.Hanson RD, et al. Mammalian Trithorax and polycomb-group homologues are antagonistic regulators of homeotic development. Proc Natl Acad Sci U S A. 1999;96:14372–14377. doi: 10.1073/pnas.96.25.14372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klein HU, et al. Integrative analysis of histone ChIP-seq and transcription data using Bayesian mixture models. Bioinformatics. 2014 doi: 10.1093/bioinformatics/btu003. [DOI] [PubMed] [Google Scholar]
- 31.Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu SC, Zhang Y. Cyclin-dependent kinase 1 (CDK1)-mediated phosphorylation of enhancer of zeste 2 (Ezh2) regulates its stability. J Biol Chem. 2011;286:28511–28519. doi: 10.1074/jbc.M111.240515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wei Y, et al. CDK1-dependent phosphorylation of EZH2 suppresses methylation of H3K27 and promotes osteogenic differentiation of human mesenchymal stem cells. Nat Cell Biol. 2011;13:87–94. doi: 10.1038/ncb2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen S, Smith DF. Hop as an adaptor in the heat shock protein 70 (Hsp70) and hsp90 chaperone machinery. J Biol Chem. 1998;273:35194–35200. doi: 10.1074/jbc.273.52.35194. [DOI] [PubMed] [Google Scholar]
- 35.Caldas-Lopes E, et al. Hsp90 inhibitor PU-H71, a multimodal inhibitor of malignancy, induces complete responses in triple-negative breast cancer models. Proc Natl Acad Sci U S A. 2009;106:8368–8373. doi: 10.1073/pnas.0903392106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weissman AM. Themes and variations on ubiquitylation. Nat Rev Mol Cell Biol. 2001;2:169–178. doi: 10.1038/35056563. [DOI] [PubMed] [Google Scholar]
- 37.Pabst C, et al. GPR56 identifies primary human acute myeloid leukemia cells with high repopulating potential in vivo. Blood. 2016;127:2018–2027. doi: 10.1182/blood-2015-11-683649. [DOI] [PubMed] [Google Scholar]
- 38.Khan SN, et al. Multiple mechanisms deregulate EZH2 and histone H3 lysine 27 epigenetic changes in myeloid malignancies. Leukemia. 2013;27:1301–1309. doi: 10.1038/leu.2013.80. [DOI] [PubMed] [Google Scholar]
- 39.Lindsley RC, et al. Acute myeloid leukemia ontogeny is defined by distinct somatic mutations. Blood. 2015;125:1367–1376. doi: 10.1182/blood-2014-11-610543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dohner H, et al. Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood. 2010;115:453–474. doi: 10.1182/blood-2009-07-235358. [DOI] [PubMed] [Google Scholar]
- 41.Makishima H, et al. Mutations in the spliceosome machinery, a novel and ubiquitous pathway in leukemogenesis. Blood. 2012;119:3203–3210. doi: 10.1182/blood-2011-12-399774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang SJ, et al. Genetic analysis of patients with leukemic transformation of myeloproliferative neoplasms shows recurrent SRSF2 mutations that are associated with adverse outcome. Blood. 2012;119:4480–4485. doi: 10.1182/blood-2011-11-390252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Attar EC, et al. Bortezomib added to daunorubicin and cytarabine during induction therapy and to intermediate-dose cytarabine for consolidation in patients with previously untreated acute myeloid leukemia age 60 to 75 years: CALGB (Alliance) study 10502. J Clin Oncol. 2013;31:923–929. doi: 10.1200/JCO.2012.45.2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pabst C, et al. Identification of small molecules that support human leukemia stem cell activity ex vivo. Nat Methods. 2014;11:436–442. doi: 10.1038/nmeth.2847. [DOI] [PubMed] [Google Scholar]
- 45.Kim W, et al. Targeted disruption of the EZH2-EED complex inhibits EZH2-dependent cancer. Nat Chem Biol. 2013;9:643–650. doi: 10.1038/nchembio.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Atanassov I, Urlaub H. Increased proteome coverage by combining PAGE and peptide isoelectric focusing: comparative study of gel-based separation approaches. Proteomics. 2013;13:2947–2955. doi: 10.1002/pmic.201300035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 48.Cox J, et al. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 49.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc. 2006;1:2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 50.Oellerich T, et al. FLT3-ITD and TLR9 use Bruton tyrosine kinase to activate distinct transcriptional programs mediating AML cell survival and proliferation. Blood. 2015;125:1936–1947. doi: 10.1182/blood-2014-06-585216. [DOI] [PubMed] [Google Scholar]
- 51.Homme C, et al. Low SMC1A protein expression predicts poor survival in acute myeloid leukemia. Oncol Rep. 2010;24:47–56. doi: 10.3892/or_00000827. [DOI] [PubMed] [Google Scholar]
- 52.Remmele W, Stegner HE. Recommendation for uniform definition of an immunoreactive score (IRS) for immunohistochemical estrogen receptor detection (ER-ICA) in breast cancer tissue. Pathologe. 1987;8:138–140. [PubMed] [Google Scholar]
- 53.Vick B, et al. An advanced preclinical mouse model for acute myeloid leukemia using patients' cells of various genetic subgroups and in vivo bioluminescence imaging. PLoS One. 2015;10:e0120925. doi: 10.1371/journal.pone.0120925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009;25:2865–2871. doi: 10.1093/bioinformatics/btp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Meissner A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schoofs T, et al. DNA methylation changes are a late event in acute promyelocytic leukemia and coincide with loss of transcription factor binding. Blood. 2013;121:178–187. doi: 10.1182/blood-2012-08-448860. [DOI] [PubMed] [Google Scholar]
- 58.Dietrich N, et al. REST-mediated recruitment of polycomb repressor complexes in mammalian cells. PLoS Genet. 2012;8:e1002494. doi: 10.1371/journal.pgen.1002494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lerdrup M, Johansen JV, Agrawal-Singh S, Hansen K. An interactive environment for agile analysis and visualization of ChIP-sequencing data. Nat Struct Mol Biol. 2016;23:349–357. doi: 10.1038/nsmb.3180. [DOI] [PubMed] [Google Scholar]
- 60.Faber J, et al. HOXA9 is required for survival in human MLL-rearranged acute leukemias. Blood. 2009;113:2375–2385. doi: 10.1182/blood-2007-09-113597. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.