Abstract
Acute myeloid leukemia (AML) is a neoplastic disease characterized by the uncontrolled proliferation and accumulation of immature myeloid cells. A common mutation in AML is the inversion of chromosome 16 [inv(16)], which generates a fusion between the genes for core binding factor beta (CBFB) and smooth muscle myosin heavy chain gene (MYH11), forming the oncogene CBFB-MYH11. The expressed protein, CBFβ-SMMHC, forms a heterodimer with the key hematopoietic transcription factor RUNX1. Although CBFβ-SMMHC was previously thought to dominantly repress RUNX1, recent work suggests that CBFβ-SMMHC functions together with RUNX1 to activate transcription of specific target genes. However, the mechanism of this activity or a requirement for additional cofactors is not known. Here, we show that the epigenetic regulator histone deacetylase 1 (HDAC1) forms a complex with CBFβ-SMMHC, co-localizes with RUNX1 and CBFβ-SMMHC on the promoters of known fusion protein target genes, and that Hdac1 is required for expression of these genes. These results imply that HDAC1 is an important component of the CBFβ-SMMHC transcriptional complex, and that leukemia cells expressing the fusion protein may be sensitive to treatment with HDAC1 inhibitors. Using a knock-in mouse model expressing CBFβ-SMMHC, we found that in vivo treatment with the HDAC1 inhibitor entinostat decreased leukemic burden, and induced differentiation and apoptosis of leukemia cells. Together, these results demonstrate that HDAC1 is an important cofactor of CBFβ-SMMHC and a potential therapeutic target in inv(16) AML.
Implications: This report describes a novel role for HDAC1 as a cofactor for the leukemogenic fusion protein CBFβ-SMMHC and shows that inhibitors of HDAC1 effectively target leukemia cells expressing the fusion protein in vivo.
Keywords: Inversion 16, acute myeloid leukemia, CBFβ-SMMHC, HDAC1, RUNX1
Introduction
Acute myeloid leukemia (AML) with inv(16)(p13.1q22) or the related translocation t(16;16)(p13.1;q22) represents 8–10% of all AML cases and usually shows monocytic/ granulocytic differentiation and abnormal eosinophils (1–4). The chromosomal breakpoints for inv(16) occur within the genes CBFB and MYH11, which encode core binding factor beta (CBFβ) and smooth muscle myosin heavy chain (SMMHC), respectively (3,5). The inverted chromosome results in an in-frame fusion between CBFB and the C-terminal coiled-coil region of MYH11 to generate the oncogene CBFB-MYH11. Expression of this oncogene, which produces the protein CBFβ-SMMHC, is the initiating event in inv(16) AML, but additional cooperating mutations are required for transformation to a frank leukemia (6,7).
Core binding factor (CBF) is a heterodimeric transcription factor consisting of one CBFα subunit, which binds DNA, in a complex with CBFβ, which stabilizes the CBFα/DNA interaction. CBFα can be any of the three members of the Runt-related transcription factor family, which includes RUNX1 (AML1, CBFα2), RUNX2 and RUNX3. While the roles of RUNX2 and RUNX3 in blood cells are currently poorly understood, RUNX1 is a well-established, critical regulator of hematopoiesis (8–11). CBFβ-SMMHC retains the RUNX binding site in CBFβ and gains a second high affinity binding domain (HABD) within the SMMHC region (12,13).
Initial models of CBFβ-SMMHC activity proposed that the fusion protein acts by dominantly repressing normal RUNX1 activity. If this model fully described the fusion protein’s activity, one would predict that loss of RUNX1 would be equivalent to expression of the fusion protein. However, knock-in mice expressing Cbfb-MYH11 from the endogenous Cbfb locus (Cbfb+/MYH11) have a more severe block in hematopoietic differentiation and show deregulated expression of a unique set of genes as compared to mice homozygous for a null allele of Runx1 (Runx1−/− ) (14). These findings imply that CBFβ-SMMHC activity is not solely based on RUNX1 repression and raises the possibility that RUNX1 may be dispensable for the fusion protein’s effect. To test this possibility, we generated mice expressing Cbfb-MYH11, but with significantly reduced RUNX1 activity (15). We found that loss of RUNX1 activity impaired Cbfb-MYH11 induced changes in gene expression and myeloid differentiation. Collectively these findings support a new model of CBFβ-SMMHC activity in which the fusion protein doesn’t repress RUNX1, but alters its activity, resulting in the changes in gene expression that lead to leukemogenesis. In support of this model, chromatin immunoprecipitation experiments show that RUNX1 and CBFβ-SMMHC co-localize in the inv(16) cell line, ME-1. Interestingly, the epigenetic modifier histone deacetylase I (HDAC1) was also found to co-localize with RUNX1 and CBFβ-SMMHC, raising the possibility that HDAC1 may contribute to the fusion protein’s transcriptional activity (16).
HDAC1 is a known binding partner of RUNX1 and a member of the class I HDAC family, which also includes HDAC2, HDAC3, and HDAC8. These family members are classified together based on their homology to yeast RPD3, with HDAC1 and 2 being the most similar, and HDAC8 the most divergent (17,18). Class I HDACs’ canonical roles are as epigenetic modifiers associated with transcriptional repression. By removing acetyl groups from lysine residues in histone tails, HDACs create a closed chromatin structure which is inaccessible to the transcriptional machinery (19,20). More recently, Class I HDACs have been shown to have additional roles including participation in transcriptional activation and deacetylation of non-histone proteins (21,22).
Based on these findings, we hypothesize that HDAC1 is part of the RUNX1:CBFβ-SMMHC complex and contributes to the gene expression changes associated with inv(16) AML. In this report, we show that HDAC1 binds to CBFβ-SMMHC and contributes to gene expression changes, maintenance of the differentiation block, and colony growth. In addition, we show that pharmacological inhibition of HDAC1 impairs the growth of CBFβ-SMMHC-expressing leukemia cells in vitro and in vivo, implying that HDAC1 inhibitors may be effective for the treatment of inv(16) AML.
Materials and Methods
Mice
Cbfb+/56M, Mx1-Cre+ or Cbfb+/56M, Mx1-Cre+, Gt(ROSA)26Sortm4(ACTB-tdTomato, -EGFP/Luo/J) (Rosa26tdT/GFP) (Jackson Laboratory, Bar Harbor, ME) mice were maintained on a mixed C57Bl6/129S6 background, genotyped, and treated to develop leukemia, as previously described (15,23–25). Leukemia cells from primary mice were expanded by transplantation into congenic C57Bl6/129S6 F1 6–10 week old mice (Taconic, Hudson, NY), as previously described (14). For in vivo studies, 1×105 – 1×106 cells from Cbfb+/56M, Mx1-Cre+, Rosa26tdT/GFP mice were transplanted into sub-lethally irradiated congenic mice. When GFP in peripheral blood averaged 10–20%, mice were treated by IP injection or oral gavage with 10 mg/kg/day entinostat (Cayman Chemical, Ann Arbor, MI) prepared in PBS with 2.5% DMSO,1% Tween-80 and 5.1% PEG-400, or vehicle alone. All procedures were performed in accordance with guidelines and protocols approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Nebraska Medical Center.
Histology
Tissues were fixed in 4% paraformaldehyde, embedded in paraffin, sectioned, and stained with hematoxylin and eosin. Slides and were examined using a Leica DM4000 B LED microscope at 20X magnification. Cytospins were prepared by centrifugation of cells in a Shandon Cytospin 3 (Thermo Fisher), staining with Wright-Giemsa (Protocol Hema 3 kit, Thermo Fisher), and examined using an Olympus BX51 microscope at 100X magnification.
Cell culture
Cbfb+/56M, Mx1-Cre+ cells were cultured in RPMI-1640 (ATCC, Manassas, Virginia) supplemented with 20% ES cell qualified fetal bovine serum (Thermo Fisher Scientific, Waltham, MA) 1% penicillin/streptomycin, 1% L-glutamine, 10 ng/mL IL-3, 10 ng/mL IL-6, 20 ng/mL SCF (Peprotech, Rocky Hill, NJ) and cryopreserved in RPMI-1640 supplemented with 50% FBS and 10% DMSO. COS-7 cells (ATCC) and HEK293T cells (ATCC) were maintained in DMEM supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin, and 1% L-glutamine. ME-1 cells (kindly provided by P. Liu, NHGRI/NIH) were maintained in RPMI-1640 supplemented with 20% fetal bovine serum, 2.5% of a 10% (w/v) glucose solution, 1% penicillin/streptomycin, 1% L-glutamine, 1% sodium pyruvate, and 2.5% 1M HEPES. Kasumi-1 cells (ATCC) were maintained according to ATCC recommended protocol. Leukemia cell lines were validated by karyotype analysis by the UNMC Human Genetics Laboratory and/or western blot analysis. Cells were maintained in culture for less than 3 months at 37°C, 5% CO2 and routinely monitored for Mycoplasma contamination using MycoAlert PLUS Mycoplasma Detection Kit (Lonza).
Site Directed Mutagenesis
pMIG-CBFB-MYH11Δ179–221, (provided by P. Liu, NHGRI/NIH) (26), was mutated to CBFB-MYH11N63K, N104K, Δ179–221 using the QuikChangeII Site Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA) according to the manufacturer’s recommendations. Primer sequences used in the mutagenesis reaction are available upon request.
Immunoprecipitation and Western Blot
Nuclear lysates were prepared from cells for IP as follows:10mM HEPES pH 7.5, 1.5mM MgCl2,10mM KCl, 0.5mM DTT, and protease inhibitors (Sigma, St. Louis, MO) were added to the cell pellet and incubated on ice for 15 minutes. Cells were centrifuged for 30 seconds at 12,000 rpm and supernatant removed. Next, the previous buffer with the addition of 0.05% NP-40 was added to the cell pellet, vortexed, and centrifuged again. The pellet was resuspended in 20mM HEPES pH 7.5, 25% glycerol, 0.42M NaCl, 0.2mM EDTA, 0.5mM DTT, and protease inhibitors. The samples were alternately incubated on ice for 5 minutes and vortexed a total of five times, then centrifuged for 15 minutes at 13,000 rpm. The nuclear extract was removed for IP. 1 µg (transfected cells) or 2 µg (CM+, ME-1 cells) of the pulldown antibody was added to each sample and incubated with the lysates overnight with rotation. Lysates were incubated for 40 minutes with protein A Dynabeads (Thermo Fisher Scientific), washed five times with 150mM NaCl, 20mM HEPES pH 7.5, 0.2% NP-40, 0.1% Tween and protease inhibitors. Beads were resuspended in 2x Laemmli buffer and boiled at 95°C for 5 minutes. Western blotting was performed as previously described (14). A list of primary and secondary antibodies used can be found in Supplemental Table 1.
Chromatin Immunoprecipitation
Chromatin Immunoprecipitation (ChIP) was performed using MagnaChip A Chromatin Immunoprecipitation Kit (EMD Millipore, Billerica, MA) with some modifications. 10×106 cells were crosslinked with 1.5 mM final concentration of ethylene glycol-bis (succinimidylsuccinate) (EGS) for 30 minutes followed by 10 minutes crosslinking with 1% final concentration paraformaldehyde. Chromatin was sheared using a Bioruptor Plus (Diagenode, Denville, NJ) for 30 total cycles of 30 seconds on/30 seconds off. 5 µg of antibody was used in each pulldown with lysate from approximately 2×106 cells and incubated overnight with 20 µL protein A magnetic beads. The following day, beads were washed as indicated in kit instructions. Reverse crosslinking was achieved with a 5 hour incubation at 62°C. ChIP was followed by qRT-PCR with primers for CDKN1A, MPO, CSF1R, CEBPD, and a negative control gene desert region, as described previously (27,28).
shRNA Knockdown
HEK293 cells were transfected with third generation lentiviral plasmids and Sigma Mission 3xLacO-IPTG plasmid engineered to contain GFP for selection and either HDAC1 shRNA or control with no known target (NTshRNA) (29). HDAC1 and control shRNA were a gift from Saverio Minucci, University of Milan (30). For transduction, CM+ cells were cultured as above with the addition of 57 μM beta-mercaptoethanol and 8 μg/mL polybrene. Cells were spinfected at 2,000 rpm for 90 minutes, followed by a six hour incubation and a second spinfection. 24 hours after the start of transduction, Isopropyl β-D-1-thiogalactopyranoside (IPTG) was added to a final concentration of 1 mM in each well. 48 hours after the start of transduction, cells were sorted on a BD FacsAria (BD Biosciences, Franklin Lakes, NJ).
Quantitative Real-time PCR
RNA was extracted from cells using TRIzol Reagent (Thermo Fisher Scientific) according to manufacturer’s instructions. First strand cDNA synthesis was accomplished using EcoDry Premix (Clontech, Mountain View, CA) according to manufacturer’s instructions. Quantitative real-time PCR (qRT-PCR) was performed on an ABI-PRISM 7000 (Applied Biosystems, Foster City, CA) using SybrGreen 2x Mastermix (Thermo Fisher Scientific) according to manufacturer’s instructions. Primers sequences for Cdkn1a, Mpo, Csf1r, and Cebpd and Actb were described previously (27). Hdac1 primer sequences: forward-TGAAGCCTCACCGAATCCG, reverse-GGGCGAATAGAACGCAGGA.
Flow Cytometry
Cells were stained with the indicated fluorophore-conjugated antibody or dye according to manufacturer’s recommendations. Antibodies used in flow cytometry experiments can be found in Supplemental Table 1. Prior to flow cytometry analysis, mouse peripheral blood was incubated in ACK buffer (Gibco) and bone marrow was lineage depleted with the EasySep Mouse Hematopoietic Progenitor Cell Isolation Kit (StemCell Technologies) according to manufacturer protocols. Flow cytometry analysis or sorting was performed on a BD LSRII or FACSAria (BD Biosciences), respectively. Data was analyzed in FlowJo v.10.0.8 (FlowJo, LLC, Ashland, OR).
Colony-forming Assays
Colony-forming assays (CFA) were performed using MethoCult GF M3434 and SmartDish meniscus-free plates (Stemcell Technologies, Vancouver, Canada). Cells were plated in triplicate in MethoCult mixed with a final concentration of 1 µM entinostat (Cayman Chemical) vorinostat (Active Motif, Carlsbad, CA), RGFP966 (Selleck Chemicals, Houston, TX) or Ro5–3335 (EMD Millipore) or equivalent DMSO control. Cells were incubated at 37°C, 5% CO2, for 14 days and colonies were counted or stained as indicated.
Viability Assay
Cells were treated with increasing doses of Entinostat or RGFP966 and viability was assessed using PrestoBlue Cell Viability Reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions after 72 hours in culture. Fluorescence was detected on a Tecan Infinite M200 (Tecan, Mannedorf, Switzerland). EC50 was calculated using GraphPad Prism 7 (GraphPad Software, La Jolla, CA).
Statistics
All experiments were performed at least three times. Data was analyzed using either the Student’s t-test or ANOVA with Tukey post-hoc test, as appropriate and indicated in the figure legends. Sample size for in vivo experiments was determined by Power Analysis. All statistical tests were performed in GraphPad Prism 7. Data was considered statistically significant at a p-value ≤ 0.05.
Results
HDAC1 is a member of the CBFβ-SMMHC:RUNX1 complex
Because HDAC1 co-localizes with RUNX1 and CBFβ-SMMHC on gene promoters, it is possible that HDAC1 is part of the RUNX1:CBFβ-SMMHC complex. Before testing this, we confirmed that HDAC1 is expressed in leukemia cells from knock-in mice with a conditional Cbfb-MYH11 allele (Cbfb+/56M) under the control of the Mx1-Cre Recombinase (Mx1- Cre+) transgene, (hereafter CM+ cells) and in the human inv(16) cell line, ME-1 (14,15,23,31). We detected increased levels of HDAC1 in three different CM+ mouse samples as compared to bone marrow from wild type mice. HDAC1 was also readily detectable in ME-1 cells (Figure 1A). As HDAC1 and HDAC2 are known to have overlapping functions in normal hematopoiesis, we also analyzed the expression of HDAC2 in CM+ and ME-1 cells (32). HDAC2 was also expressed highly in all three CM+ leukemia samples and in ME-1 cells, similar to HDAC1 (Figure 1B).
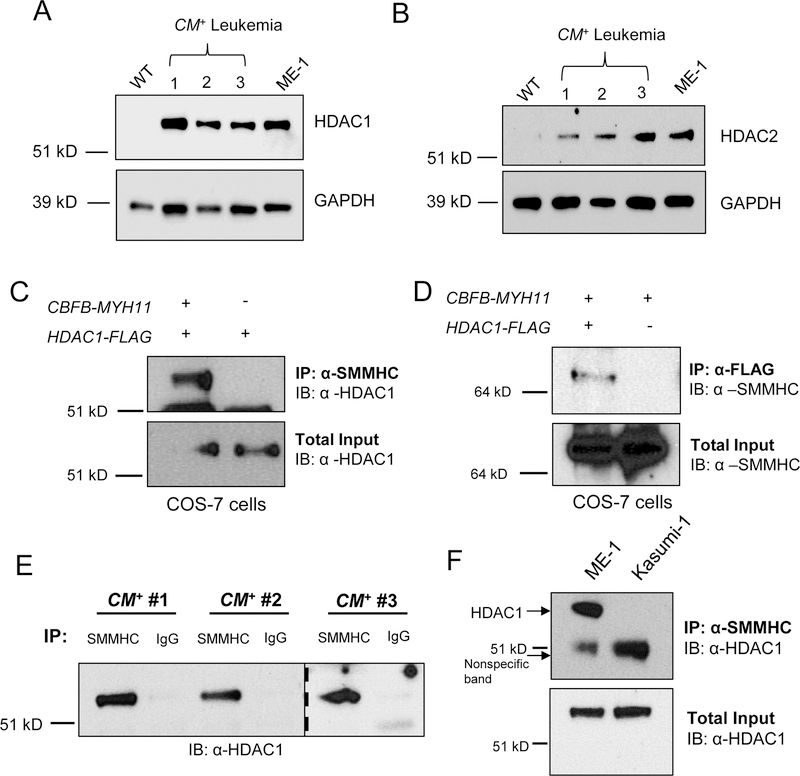
Figure 1. HDAC1 binds to CBFβ-SMMHC.
(A) HDAC1, (B) HDAC2, or GAPDH protein expression was probed in wild-type mouse bone marrow, CM+ mouse cells, and ME-1 cells by western blot. (C) COS-7 cells were transfected with plasmids expressing CBFB-MYH11 or HDAC1-FLAG and IP’s were performed on the lysates with anti-SMMHC or (D) anti-FLAG, followed by western blot. Total inputs are shown below. (E) Lysates from three independent CM+ mice were separated into two equal fractions and incubated with either anti-SMMHC or anti-IgG, followed by western blot to probe for HDAC1. The dotted line indicates separation between two different gels. (F) Lysates from ME-1 cells or Kasumi-1 cells were subjected to IP with anti-SMMHC, followed by western blot for HDAC1. Arrows indicate HDAC1 at its expected size and a non-specific band observed in both lanes. Total input is shown below.
We next tested if HDAC1 and 2 can interact with CBFβ-SMMHC in COS-7 cells transfected with plasmids containing either HDAC1 or HDAC2 fused with a FLAG tag (HDAC1-FLAG, HDAC2-FLAG) and CBFB-MYH11. Using nuclear lysates, we performed co-immunoprecipitations (co-IP’s). IP with an anti-SMMHC antibody resulted in the pulldown of HDAC1-FLAG in cells expressing both CBFβ-SMMHC and HDAC1-FLAG, but not in cells expressing HDAC1-FLAG alone (Figure 1C). In a reciprocal experiment, pulldown with an antibody against FLAG immunoprecipitated CBFβ-SMMHC in cells expressing HDAC1-FLAG and CBFβ-SMMHC, but not in cells expressing HDAC1-FLAG only (Figure 1D). In contrast, immunoprecipitation with anti-SMMHC did not pull down HDAC2 (Supplemental Figure 1). These results suggest that CBFβ-SMMHC can interact with HDAC1 but not HDAC2.
We next tested if endogenous CBFβ-SMMHC and HDAC1 form a complex. Nuclear lysates from leukemic cells from three independent CM+ mice were incubated with either anti-SMMHC or normal rabbit IgG. We observed HDAC1 was immunoprecipitated with anti-SMMHC, but not with IgG, indicating that endogenous CBFβ-SMMHC and HDAC1 interact in mouse leukemia cells (Figure 1E). To confirm this interaction in human leukemia cells, we performed co-IPs using lysates from ME-1 cells and Kasumi-1 cells, a leukemia cell line which expresses HDAC1 but not CBFβ-SMMHC. HDAC1 and CBFβ-SMMHC co-IP’d in ME-1 cells but not in Kasumi-1 cells (Figure 1F). Together, these results indicate that endogenous CBFβ-SMMHC and HDAC1 interact in mouse and human leukemia cells.
HDAC1 is known to bind RUNX1, raising the possibility that RUNX1 mediates the interaction between HDAC1 and CBFβ-SMMHC (33). To test this, we performed IP’s with mutant constructs of CBFB-MYH11 lacking the RUNX1 High Affinity Binding Domain (HABD) in the MYH11 tail (CBFB-MYH11Δ179–221), or with point mutations in the CBFβ domain as well as deletion of the HABD (CBFB-MYH11N63K, N104K, Δ179–221) (Supplemental Figure 2) (34,35). In transfected cells, IP with anti-FLAG was able to pull down both CBFβ-SMMHC mutants, even though the double mutant was not able to pull down RUNX1 (Figure 2A). This indicates that RUNX1 is not required for the interaction between HDAC1 and CBFβ-SMMHC.
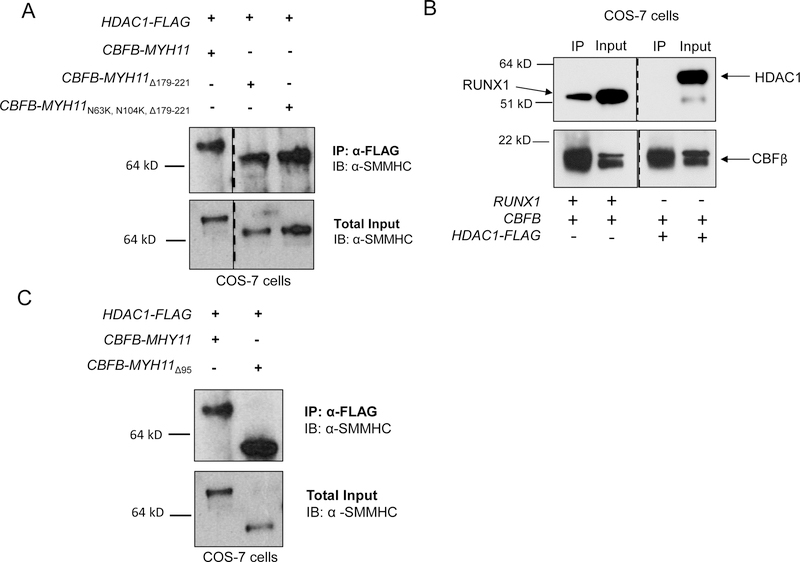
Figure 2. HDAC1 binds to a central SMMHC region in a RUNX1-independent manner.
(A) COS-7 cells were transfected with plasmids containing the indicated construct and the lysates were subjected to IP’s with anti-FLAG, followed by western blot for SMMHC. The dotted line indicates a division between two different regions of the same gel. Total input is shown below. (B) COS-7 cells were transfected with plasmids containing HDAC1-FLAG, CBFB, or RUNX1, and the lysates were subjected to IP with anti-CBFβ antibody followed by western blot for RUNX1 (left side top), HDAC1 (right side top) or CBFβ (bottom). The dotted line indicates where the membrane was cut. (C) COS-7 cells were transfected with plasmids containing HDAC1-FLAG, CBFB-MYH11 or CBFB-MYH11Δ95 and the lysates were subjected to IP with anti-FLAG followed by western blot for SMMHC. Total inputs are shown below.
We next tested the ability of HDAC1 to interact with two other important regions of the fusion protein: the CBFβ region and the c-terminal 95 amino acids, a part of the co-repressor domain. In nuclear lysates from cells expressing HDAC1-FLAG and wild-type CBFβ, IP with anti-FLAG did not co-precipitate detectable CBFβ, and neither did the reciprocal pulldown with anti-CBFβ, although the precipitation of the known CBFβ binding partner RUNX1 was readily apparent (Figure 2B and Supplemental Figure 3). These findings indicate that HDAC1 and wild-type CBFβ do not form a complex. We next tested whether CBFβ-SMMHC’s c-terminus is required for interaction with HDAC1. Nuclear lysates from cells expressing HDAC1-FLAG and a c-terminal deletion mutant of CBFβ-SMMHC (CBFβ-SMMHCΔ95) were immunoprecipitated with anti-FLAG. We found that IP of HDAC1 was able to pulldown CBFβ-SMMHCΔ95 (Figure 2C), indicating that the final 95 residues of SMMHC are not required to form a complex with HDAC1. This is in contrast to what has been shown for the interaction between CBFβ-SMMHC and HDAC8, implying that the fusion protein interacts with HDAC1 and HDAC8 through distinct domains (36).
HDAC1 is required for CBFβ-SMMHC target gene expression
In the inv(16) AML cell line ME-1, HDAC1 co-localizes with RUNX1 and CBFβ-SMMHC in the promoter regions of target genes (16). To confirm that HDAC1, CBFβ-SMMHC and RUNX1 co-localize in primary CM+ mouse leukemia cells, we performed chromatin immunoprecipitation (ChIP) followed by quantitative real-time PCR for four known target genes: cyclin-dependent kinase inhibitor 1A (Cdkn1a) which encodes p21Waf1/Cip1, myeloperoxidase (Mpo), colony-stimulating factor 1 receptor (Csf1r), and CCAAT/enhancer binding protein delta (Cebpd) (27). We found that HDAC1, CBFβ-SMMHC, and RUNX1 were each significantly enriched at the promoters of Mpo, Csf1r, and Cebpd as compared to control. On the promoter of Cdkn1a, RUNX1 and HDAC1 were significantly enriched, and CBFβ-SMMHC showed a trend towards enrichment, although it did not reach the level of statistical significance (p=0.06) (Figure 3A). To confirm specificity, we tested a gene desert region as a negative control and did not observe enrichment for RUNX1, CBFβ-SMMHC, or HDAC1 (Figure 3A) (28).
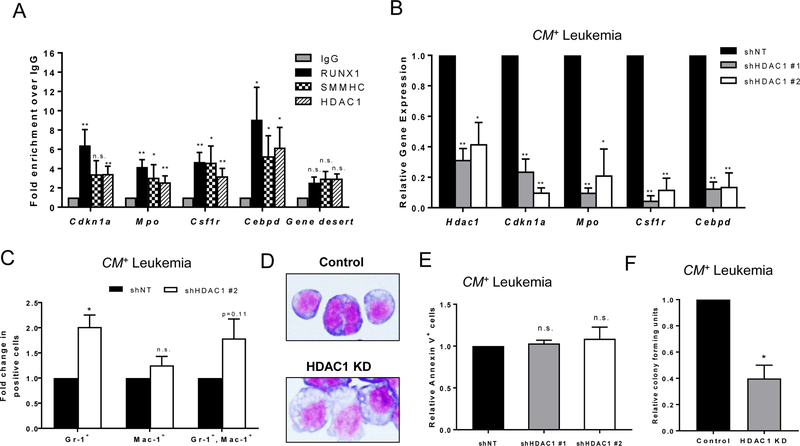
Figure 3. HDAC1 co-localizes with CBFβ-SMMHC and RUNX1 and regulates target gene expression.
(A) Chromatin immunoprecipitation (ChIP) was performed on cell lysates from at least three independent CM+ mice with antibodies against normal rabbit IgG, RUNX1, SMMHC, or HDAC1. Quantitative real-time PCR was used to detect transcript levels of the indicated genes using Actb as a reference control. Data is plotted as fold enrichment compared to IgG. (B) Cells from three independent CM+ mice were transduced with either a control shRNA with no target (shNT) or one of two different shRNA constructs targeting HDAC1 (shHDAC1). RNA/cDNA expression from sorted cells was analyzed using quantitative real-time PCR using Actb as a reference control. Data is plotted as relative gene expression compared to the control shRNA. (C) CM+ cells were lentivirally transduced with control or HDAC1 shRNA constructs, and shRNA expression was induced after sorting. Twenty-four hours later, cells were analyzed for cell surface expression of Gr-1 and Mac-1 by flow cytometry. Data is plotted as fold change in the percentage of total Gr-1 positive, total Mac-1 positive, or Gr-1,Mac-1 double positive cells, compared to control shRNA. (D) Cells from (C) were adhered to slides using a cytospin, stained with Wright-Giemsa and imaged at 100x magnification. (E) Cells from (C) were also stained with an antibody against annexin V, analyzed by flow cytometry, and plotted as fold change compared to control shRNA. (F) CM+ cells were transduced with an shRNA construct targeting HDAC1, induced with IPTG and sorted for live, transduced cells 24 hours later. Cells were plated in triplicate in MethoCult mixed with either IPTG or PBS. Colonies were manually counted 14 days later and plotted as relative colony forming units compared to DMSO control. Data is from two independent experiments. Error bars represent the standard error of the mean (SEM). ANOVA (A,B) or Student’s t-test (C,D) was used to calculate statistical significance. * = p≤0.05, ** = p≤0.01, n.s. = not significant.
To test if HDAC1 activity is required for CBFβ-SMMHC-induced expression of these target genes, we used two different short hairpin RNA (shRNAs) to knockdown HDAC1 in CM+ mouse leukemia cells. Cells were transduced with lentiviral vectors expressing control or HDAC1 shRNAs under an IPTG inducible promoter and GFP from an internal ribosomal entry site (IRES). The cells were treated with IPTG to induce shRNA expression, and twenty-four hours later were sorted for GFP expression. Both shRNAs against HDAC1 caused significant knockdown of Hdac1 as compared to cells transduced with the control shRNA. (Figure 3B). Both shRNAs against HDAC1 also resulted in significant decreases in Cdkn1a, Mpo, Csf1r, and Cebpd expression (Figure 3B). Furthermore, expression of Mpo, Csf1r, and Cebpd appeared to show an HDAC1 dose dependency. This suggests that HDAC1 is required for expression of CBFβ-SMMHC target genes.
To determine if HDAC1 is required for the CBFβ-SMMHC induced block in differentiation, we stained Hdac1 knockdown and control CM+ leukemia cells for expression of Gr-1 (Ly-6G) and Mac-1 (CD11b), which are both markers of mature myeloid cells. Hdac1 knockdown in CM+ cells showed increased expression of Gr-1 and to a lesser extent Mac-1, implying that HDAC1 is required for the CBFβ-SMMHC induced block in differentiation (Figure 3C, Supplemental Figure 4). Cytospins of these cells showed a smaller nuclei to cytoplasmic ratio with more condensed chromatin in the HDAC1 KD cells compared to control, indicating that loss of Hdac1 increased morphological differentiation (Figure 3D). To test if loss of HDAC1 affects the survival of CM+ leukemia cells, we performed staining with annexin V, a marker of apoptosis. In Hdac1 knockdown cells, we did not observe a difference in annexin V staining compared to control, indicating that HDAC1 is likely not regulating cell survival in CM+ leukemia cells (Figure 3E). To test if knockdown of HDAC1 affected colony forming ability, we induced Hdac1 KD in transduced CM+ leukemia cells for 24 hours, then sorted for live GFP positive cells, and plated equal numbers of cells in methylcellulose containing vehicle or IPTG. After 14 days, we observed significantly fewer colonies in the Hdac1 KD plates compared to control (Figure 3F), suggesting that HDAC1 is important for leukemia stem cell activity.
HDAC1 inhibitors impair growth of CBFβ-SMMHC+ leukemia cells in vitro
Our results indicate that HDAC1 is important for CBFβ-SMMHC activity, implying that inv(16) AML cells may be particularly sensitive to treatment with an HDAC1 inhibitor. To test this possibility, we performed colony assays in the presence of entinostat (MS-275), an HDAC1 selective inhibitor (37,38). Equal numbers of cells were plated in the presence of 1 µM entinostat or vehicle and cultured for 14 days. We observed significantly fewer colonies in entinostat treated plates compared to control plates (Figure 4A). The individual colonies also appeared smaller and more diffuse (Figure 4B). After culture, the cells were stained for Gr-1 and Mac-1 expression. There was a large increase in Mac-1+Gr-1+ staining, indicating a more differentiated phenotype (Figure 4C). Cytospins of these cells after colony assay confirmed morphological differentiation with entinostat treated cells exhibiting a greater number of cells with high granularity and convoluted nuclei. (Figure 4D). Similar results were obtained using the class I and II HDAC inhibitor vorinostat (Suberoylanilide Hydroxamic Acid, SAHA) (Supplemental Figure 5A,B).
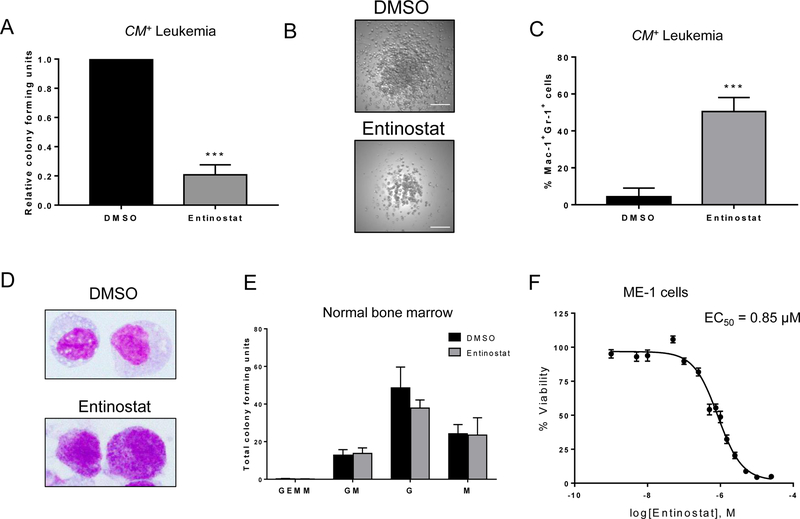
Figure 4. HDAC1 inhibitors reduce growth of CM+ cells in vitro.
(A) CM+ cells from three independent mice were plated in triplicate in MethoCult mixed with either 1 µM entinostat or DMSO. Colonies were manually counted 14 days later and plotted as relative colony forming units compared to DMSO control. (B) Representative images of colonies from DMSO treated (top) or entinostat (bottom) plates. Scale bar represents 200 μm. (C) CM+ cells were stained following the colony-forming assay for myeloid differentiation markers Gr-1 and Mac-1 and analyzed by flow cytometry. (D) Cells from (C) were adhered to slides using a cytospin, stained with Wright-Giemsa and imaged at 100x magnification. (E) Wild-type mouse bone marrow was plated as in (A), and colonies were counted and classified according to their constituent cells. Data is plotted as total colony forming units (CFU) for each type of colony. All entinostat bars are not significant (n.s.) compared to DMSO. (F) ME-1 cells were treated with increasing doses of entinostat and cell viability was analyzed with PrestoBlue viability reagent. EC50 was calculated using GraphPad Prism. Error bars represent SEM. Student’s t-test was used to calculate statistical significance. *** = p≤0.001. Abbreviations: GEMM, granulocyte-erythrocyte-monocyte-megakaryocyte; GM, granulocyte-macrophage; G, granulocyte; M, macrophage.
To test the effect of HDACi’s on normal hematopoiesis, we performed colony-forming assays with bone marrow cells from wild type mice. Importantly, there was no significant difference in the growth of any type of colony in the presence of either entinostat or vorinostat, as compared to DMSO (Figure 4E, Supplemental Figure 5C). These results indicate CBFβ-SMMHC-expressing leukemia cells are more sensitive to the effects of HDAC inhibitors than normal hematopoietic cells.
To test if HDACi had a similar effect on gene expression as HDAC1 KD, we treated CM+ mouse leukemia cells with entinostat. Similar to the HDAC1 knockdown, we saw a trend of decreased gene expression for Mpo, Csf1r, and Cebpd, although not for Cdkn1a (Supplemental Figure 6). We tested additional myeloid differentiation genes to determine if some genes were upregulated by HDAC1 inhibition, and found that Cebpe, Cebpa, Gr-1 (Ly6g), and Mac-1 (Itgam) mRNA expression showed a trend towards upregulation in entinostat treated cells, although not to levels of statistical significance, while the early granulopoiesis marker Csf3r (Colony stimulating factor 3 receptor), was significantly downregulated (Supplemental Figure 6). This data demonstrates that entinostat causes changes in gene expression similar to our results above with Hdac1 knockdown, and to previous findings with loss of CBFβ-SMMHC (16).
To test the effect of HDACi on human inv(16) cells, we treated ME-1 cells with increasing doses of entinostat and assayed for cell viability. ME-1 cells showed a dose-dependent decrease in viability over the range of concentrations tested, with an EC50 of 0.85 μM (Figure 4F). This indicates that human CBFβ-SMMHC-expressing cells are also sensitive to treatment with HDACi. Entinostat can also target HDAC3 activity, although less effectively that HDAC1, raising the possibility that inhibition of HDAC3 may contribute to the entinostat’s effect (39). To test this, we treated ME-1 and CM+ cells with the HDAC3 specific inhibitor RGFP966 (40). ME-1 cells were much less sensitive to HDAC3 inhibition, with an EC50 of 7.1 µM (Supplemental Figure 7A). In addition, RGFP966 did not have an effect on the colony-forming ability of CM+ cells (Supplemental Figure 7B). While we have not ruled out a role for HDAC3 in CM+ leukemia, these results indicate that any effect on HDAC3 activity by entinostat is likely minor and that the anti-leukemic effect in CM+ leukemia cells is primarily due to HDAC1 inhibition.
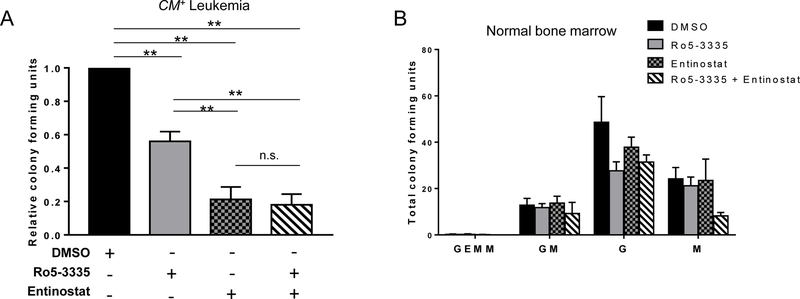
Our data indicates that HDAC1 is required for CBFβ-SMMHC induced gene expression, implying that HDAC1 and RUNX1 are acting in the same pathway. To test this, we treated leukemia cells with either entinostat, the RUNX1 inhibitor Ro5–3335, or both entinostat and Ro5–3335 (41). Either Ro5–3335 or entinostat alone significantly reduced colony growth, as compared to control (Figure 5A). The combination of Ro5–3335 and entinostat significantly reduced the number of colonies compared to DMSO and Ro5–3335 alone but did not further inhibit colony growth compared to entinostat alone, suggesting that these drugs are inhibiting the same pathway (Figure 5A). Neither drug, alone or in combination, had any significant effect on the colony growth of normal bone marrow cells, indicating that CBFβ-SMMHC-expressing leukemia cells are more sensitive to loss of either RUNX1 or HDAC1 activity than normal blood cells (Figure 5B).
Figure 5. RUNX1 or HDAC1 inhibition likely target the same pathway in CM+ cells.
(A) CM+ cells from three independent mice were plated in triplicate in MethoCult mixed with 1 µM of the indicated combinations of Ro5–3335, entinostat, or DMSO. Colonies were manually counted 14 days later and plotted as relative colony forming units (CFU) compared to DMSO control. (B) Wild-type mouse bone marrow was plated as in (A) and colonies were counted and classified according to their constituent cells. Data is plotted as total CFU’s for each type of colony. All bars are not significant (n.s.) compared to DMSO. Error bars represent SEM. ANOVA was used to calculate statistical significance. ** = p≤0.01, n.s = not significant. Abbreviations: GEMM, granulocyte-erythrocyte-monocyte-megakaryocyte; GM, granulocyte-macrophage; G, granulocyte; M, macrophage.
Entinostat decreases leukemic burden in vivo
Entinostat treatment reduced CM+ colony growth in vitro, suggesting that it may be effective against CM+ leukemia in vivo. To test this, we transplanted CM+ primary mouse leukemia samples that also express GFP from the Rosa26 locus (Cbfb+/56M, Mx1-Cre+, Rosa26tdT/GFP) into wild-type recipient mice (25). This system allows us to analyze the effects of drug treatment on both the transplanted, GFP+ leukemia cells, and the recipient mouse’s GFP- normal blood cells.
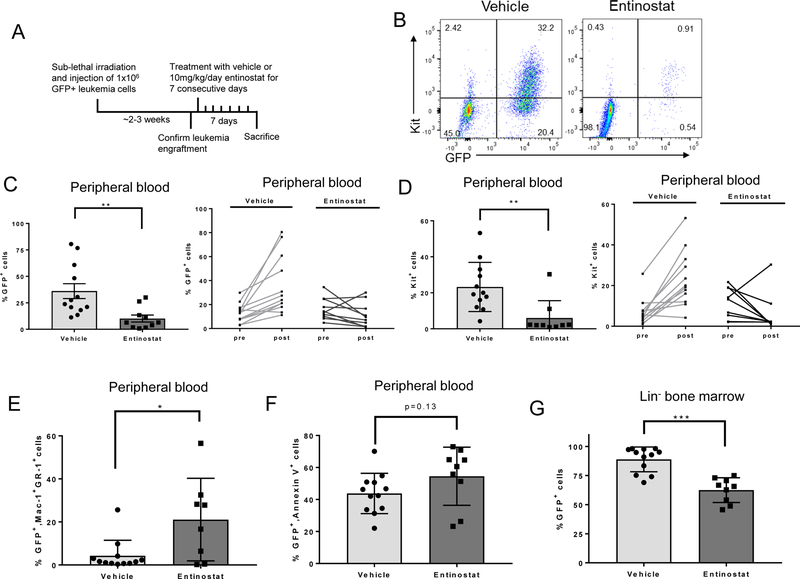
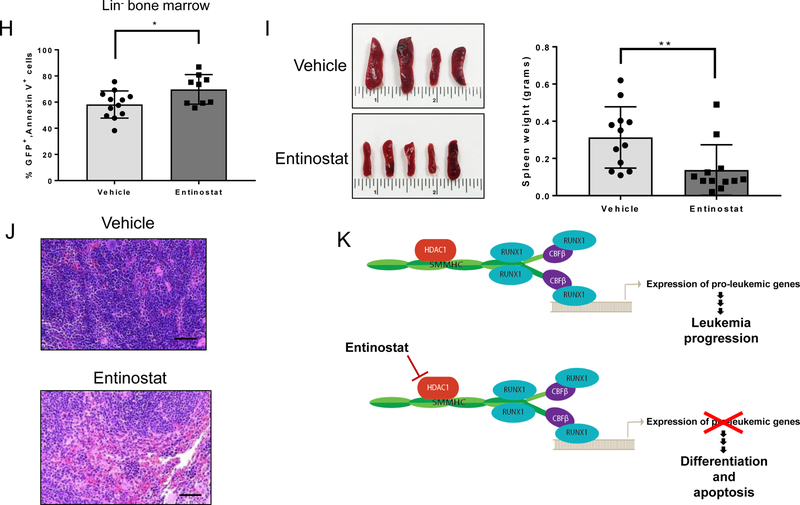
Leukemia cells from 3 independent Cbfb+/56M, Mx1-Cre+, Rosa26tdT/GFP mice were transplanted into congenic recipient mice. Approximately 2–3 weeks later, peripheral blood was analyzed to confirm leukemia engraftment, and mice were treated for 7 days with 10 mg/kg/day entinostat or vehicle. Twenty-four hours after the last treatment, mice were sacrificed, and blood and tissue were harvested (Figure 6A). Mice treated with entinostat showed significant reductions in the number of leukemia cells in the peripheral blood, as determined by GFP or Kit expression (Figure 6B-D). In the entinostat treated mice, the remaining GFP+ leukemia cells in the peripheral blood showed increased expression of both Mac-1 and Gr-1, consistent with our in vitro data (Figure 6E). There was a trend towards increased annexin V+ staining in the GFP+ leukemia cells in the peripheral blood, although this difference did not achieve statistical significance (Figure 6F). There was a parallel decrease in GFP+ cells in the lineage-depleted (lin-) bone marrow of entinostat treated mice (Figure 6G). The remaining GFP+ cells in the bone marrow showed a small, but statistically significant increase in annexin V+ staining (Figure 6H). Entinostat treated mice also had smaller spleens and significantly decreased spleen weights (Figure 6I). Histological examination showed decreased leukemic infiltration in the spleen (Figure 6J). Entinostat treatment did not cause an increase in annexin V, Mac-1, or Gr-1 staining in the GFP- cells, indicating that entinostat does not induce apoptosis or differentiation of normal blood cells (Supplemental Figure 8A,B). These findings indicate that entinostat specifically targets CBFβ-SMMHC-expressing leukemia cells and promotes their differentiation in vivo.
Figure 6. Entinostat treatment decreases leukemic burden in mice with CM+ leukemia.
(A) Schematic of treatment protocol. (B) Representative plots from flow cytometry analysis of Kit and GFP in peripheral blood. (C) Flow cytometry analysis of GFP+ cells in the peripheral blood after treatment with vehicle or entinostat (left) and in each mouse pre-treatment (pre) and post-treatment (post) (right). Each line represents one individual mouse. (D) Percentage of Kit+ cells in peripheral blood (left) and percentage of Kit+ cells pre- and post-treatment (right). Each line represents one individual mouse. (E) Flow cytometry analysis of the percentage of Mac-1+Gr-1+ cells within the GFP+ cell compartment. (F) Flow cytometry analysis of the percentage of annexin V+ cells within the GFP+ cell compartment. (G) Flow cytometry analysis of the percentage of GFP+ cells in lin- bone marrow. (H) Flow cytometry analysis of the percentage of annexin V+ cells in the GFP+ compartment of the lin- bone marrow. (I) Representative images of vehicle or entinostat treated spleens (left) and quantification of spleen weights (right). (J) Representative H&E stained images of spleen sections after treatment taken at 20x magnification. Scale bar = 50 μm. (K) Proposed model of the activity of HDAC1 in CBFβ-SMMHC-expressing cells before (top) and after treatment with entinostat (bottom). Each dot on bar graphs represents one individual mouse. Error bars represent SEM. Student’s t-test was used to calculate statistical significance. * = p≤0.05, ** = p≤0.01, *** = p≤0.001, n.s. = not significant.
In this study, we show that HDAC1 co-localizes with CBFβ-SMMHC and RUNX1, helps to regulate CBFβ-SMMHC target genes, and that either knockdown or pharmacological inhibition of HDAC1 prevents leukemia cell growth and promotes differentiation. Based on these observations, we propose that HDAC1 is a required cofactor for CBFβ-SMMHC induced changes in gene expression and block in differentiation (Figure 6K). Taken together, this data indicates that inhibition of HDAC1 indirectly blocks key leukemogenic activities of CBFβ-SMMHC and is a potential therapeutic strategy for patients with inv(16) AML.
Discussion
CBFβ-SMMHC expression is known to be the initiating event in inv(16) AML, but it is less clear what role the fusion protein has after leukemic transformation. Early models suggested that CBFβ-SMMHC acts as a repressor of RUNX1 by outcompeting CBFβ for binding (13,42–44). More recent work indicates that CBFβ-SMMHC has a direct role in gene expression, likely acting as part of a transcription factor complex requiring RUNX1 (14–16). This raises the possibility that other transcriptional regulators may be recruited to the RUNX1:CBFβ-SMMHC complex and be required for the gene expression changes associated with inv(16) AML. Indeed, the chromatin remodeling factor Chromodomain Helicase DNA Binding Protein 7 (CHD7) is recruited to the RUNX1:CBFβ-SMMHC complex through an interaction with RUNX1 and plays a role in the transactivation activity of the complex in the context of leukemia initiation (45). Our work expands this model, demonstrating that the RUNX1:CBFβ-SMMHC complex includes the epigenetic modifier HDAC1.
We show here a previously unrecognized interaction between HDAC1 and the fusion protein CBFβ-SMMHC in mouse and human leukemia cells. Previous work by others failed to detect this interaction, likely due to the use of whole cell lysates rather than nuclear extracts, as were used in this study (33,36). In fact, we were unable to detect co-IP of HDAC1 and CBFβ-SMMHC from whole cell extracts (data not shown), possibly because HDAC1 is only expressed in the nucleus, whereas CBFβ-SMMHC can localize to the cytoplasm when overexpressed (44). Surprisingly, this interaction was not mediated solely by RUNX1, as evidenced by the retention of HDAC1 binding to CBFβ-SMMHC upon deletion of the high affinity binding domain alone (CBFB-MYH11Δ179–221) or in combination with the CBFβ binding domain (CBFB-MYH11N63K, N104K, Δ179–221). Because the related HDAC family member, HDAC8, can bind to CBFβ-SMMHC on the c-terminal 95 amino acids, we specifically tested this region for HDAC1 binding (33,36). We show that this region is not required for the interaction between HDAC1 and CBFβ-SMMHC, implying that HDAC1 and HDAC8 interact with the fusion protein through distinct domains. We also show that HDAC1 does not form a complex with wild-type CBFβ. Based on these observations it is tempting to conclude that HDAC1 is interacting with the central region of the SMMHC tail. However, it is also possible that HDAC1 interacts with multiple regions of the CBFβ-SMMHC protein or is recruited by multiple CBFβ-SMMHC cofactors. Sin3A, a transcriptional cofactor typically associated with repression, is a known binding partner of both CBFβ-SMMHC and HDAC1, so it could mediate the association between HDAC1 to the fusion protein complex (19,33).
Previous work demonstrates that HDAC1 co-localizes with RUNX1 and CBFβ-SMMHC on chromatin in ME-1 cells (16). We have extended this finding by showing their co-localization in primary mouse CM+ cells on the promoters of genes that are regulated by CBFβ-SMMHC (16). We also show that knockdown of HDAC1 results in a 2-fold or greater downregulation of Cdkn1a, Mpo, and Cebpd in CM+ leukemia cells, suggesting cooperation between HDAC1 and CBFβ-SMMHC in regulating gene expression changes (16). This data is consistent with the previous finding that knockdown of CBFβ-SMMHC decreased expression of MPO, CDKN1A, and CEBPD in ME-1 cells (16). While a decrease in expression of myeloid genes seems paradoxical to the observed myeloid differentiation induced by HDAC1 knockdown or inhibition, the maturation of myeloid cells consists of multiple phases of gene expression and repression. The MPO, CSF1R, and CSF3R genes are all expressed during the initial steps of myelopoiesis, but their expression decreases with further differentiation. In contrast, CEBPE, GR-1 and MAC-1 expression is restricted to more mature myeloid cells (46). Therefore, it is not necessarily surprising that loss of MPO and CSF1R expression would accompany terminal myeloid differentiation.
Our findings also support the model of the RUNX1:CBFβ-SMMHC complex acting as an active transcription factor complex in inv(16) AML. It is noteworthy that knockdown of HDAC1 resulted in decreased expression of target genes, since HDACs are traditionally thought to function only as transcriptional co-repressors. However, this assumption is challenged in recent reports showing HDAC1 is associated with the promoters of highly expressed genes, and that genetic depletion or inhibition of HDAC activity results in increased gene expression (21,22). These findings suggest that HDACs may have non-canonical roles in transcriptional activation as well. Currently, the mechanism of HDACs’ role in transcriptional activation is unclear, but has been proposed to involve deacetylation of non-histone proteins or the turnover of acetylation marks between rounds of transcription (21,22).
The requirement of HDAC1 for CBFβ-SMMHC activity implies that HDAC inhibitors may be able to inhibit the fusion protein indirectly. In fact, we observed that treatment with either entinostat, which is selective for HDAC1, or the pan-HDAC inhibitor vorinostat significantly reduced the growth of CBFβ-SMMHC-expressing leukemia cells. It is significant that the combination of entinostat and the RUNX1 inhibitor Ro5–3335 did not have an increased effect as compared to treatment with entinostat alone. This is consistent with a model in which HDAC1 and RUNX1 are both required for CBFβ-SMMHC’s ability to regulate gene expression. While much work has focused on finding an inhibitor of the RUNX1:CBFβ or RUNX1:CBFβ-SMMHC interaction, our results indicate that HDAC1 inhibitors, which are already in use clinically, can have a similar effect on CBFβ-SMMHC activity.
As strong support for this hypothesis, entinostat treatment of mice with CM+ leukemia had a strong anti-leukemic effect, reducing the number of leukemic cells and promoting their differentiation. Entinostat was much more potent against leukemia cells than normal blood cells, implying a more stringent requirement for HDAC1 in CBFβ-SMMHC-expressing leukemia cells than in normal hematopoietic cells. This may in part be due to HDAC2, which is known to have overlapping functions with HDAC1 in normal hematopoietic cells so may be able to compensate for HDAC1 inhibition. We did not detect an interaction between HDAC2 and CBFB-SMMHC, implying that HDAC2 does not contribute to the fusion protein’s activity. Currently, we cannot rule out a role for HDAC3 in CBFβ-SMMHC-expressing leukemia cells. While our gene expression, cell morphology, and colony-forming assay data demonstrate that entinostat treatment mirrors HDAC1 knockdown, inhibition of HDAC3 or other yet unknown proteins in addition to HDAC1 may explain the subtle phenotypic differences we observed in HDAC1 knockdown and entinostat treated CM+ cells.
Our data parallels what has been shown in the related CBF leukemia defined by the t(8;21) rearrangement. The resultant fusion protein, AML1-ETO, is known to bind the HDAC1 corepressor complex (47). In addition, treatment with entinostat or the HDACi valproic acid causes differentiation and/or apoptosis in RUNX1-ETO expressing leukemia cells. Entinostat has also been shown to cause differentiation and apopotosis in leukemia cells expressing the fusion gene MLL-AF9. Although this fusion protein is not known to interact with HDAC1, it does require RUNX1 expression for its leukemogenic activity (48–50). These results may imply a common role for HDAC1 in RUNX1-dependent leukemia.
In summary, our results show that HDAC1 forms a complex with CBFβ-SMMHC and plays an important role in the fusion protein’s activity. We also show that pharmacological inhibition of HDAC1 blocks the growth of CBFβ-SMMHC-expressing leukemia cells and promotes their differentiation, indicating that the use of HDAC inhibitors may be useful for the treatment of inv(16) AML.
Supplementary Material
Acknowledgements
This work was supported by funding from the Fred & Pamela Buffett Cancer Center Support Grant from the National Cancer Institute under award number P30 CA036727, Nebraska Department of Health and Human Services, the Cattlemen’s Ball of Nebraska, St. Baldrick’s Foundation, and the UNMC graduate student assistantship. The authors would like to thank the UNMC Department of Biochemistry and Molecular Biology, UNMC Flow Cytometry Research Facility, UNMC Comparative Medicine, and UNMC Tissue Sciences Facility.
Footnotes
Conflict of Interest Statement: The authors have no conflicts of interest to declare.
References
- 1.Kihara R, Nagata Y, Kiyoi H, Kato T, Yamamoto E, Suzuki K, et al. Comprehensive analysis of genetic alterations and their prognostic impacts in adult acute myeloid leukemia patients. Leukemia 2014;28:1586–95. [DOI] [PubMed] [Google Scholar]
- 2.Le Beau MM, Larson RA, Bitter MA, Vardiman JW, Golomb HM, Rowley JD. Association of an inversion of chromosome 16 with abnormal marrow eosinophils in acute myelomonocytic leukemia. A unique cytogenetic-clinicopathological association. N Engl J Med 1983;309:630–6. [DOI] [PubMed] [Google Scholar]
- 3.Liu PP, Hajra a, Wijmenga C, Collins FS. Molecular pathogenesis of the chromosome 16 inversion in the M4Eo subtype of acute myeloid leukemia. Blood 1995;85:2289–302. [PubMed] [Google Scholar]
- 4.Arber DA, Orazi A, Hasserjian R, Borowitz MJ, Beau MM Le, Bloomfield CD, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 2016;127:2391–406. [DOI] [PubMed] [Google Scholar]
- 5.Liu P, Tarlé S a, Hajra a, Claxton DF, Marlton P, Freedman M, et al. Fusion between transcription factor CBF beta/PEBP2 beta and a myosin heavy chain in acute myeloid leukemia. Science 1993;261:1041–4. [DOI] [PubMed] [Google Scholar]
- 6.Castilla LH, Garrett L, Adya N, Orlic D, Dutra A, Anderson S, et al. The fusion gene Cbfb-MYH11 blocks myeloid differentiation and predisposes mice to acute myelomonocytic leukaemia. Nat Genet 1999;23:144–6. [DOI] [PubMed] [Google Scholar]
- 7.Castilla LH, Perrat P, Martinez NJ, Landrette SF, Keys R, Oikemus S, et al. Identification of genes that synergize with Cbfb-MYH11 in the pathogenesis of acute myeloid leukemia. Proc Natl Acad Sci U S A 2004;101:4924–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hart SM, Foroni L. Core binding factor genes and human leukemia. Haematologica 2002;87:1307–23. [PubMed] [Google Scholar]
- 9.Speck N a, Gilliland DG. Core-binding factors in haematopoiesis and leukaemia. Nat Rev Cancer 2002;2:502–13. [DOI] [PubMed] [Google Scholar]
- 10.Speck N a. Core binding factor and its role in normal hematopoietic development. Curr Opin Hematol 2001;8:192–6. [DOI] [PubMed] [Google Scholar]
- 11.de Bruijn MF, Speck NA. Core-binding factors in hematopoiesis and immune function. Oncogene 2004;23:4238–48. [DOI] [PubMed] [Google Scholar]
- 12.Warren a J, Bravo J, Williams RL, Rabbitts TH. Structural basis for the heterodimeric interaction between the acute leukaemia-associated transcription factors AML1 and CBFbeta. EMBO J 2000;19:3004–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lukasik SM, Zhang L, Corpora T, Tomanicek S, Li Y, Kundu M, et al. Altered affinity of CBF beta-SMMHC for Runx1 explains its role in leukemogenesis. Nat Struct Biol 2002;9:674–9. [DOI] [PubMed] [Google Scholar]
- 14.Hyde RK, Kamikubo Y, Anderson S, Kirby M, Alemu L, Zhao L, et al. Cbfb/Runx1 repression-independent blockage of differentiation and accumulation of Csf2rb-expressing cells by Cbfb-MYH11. Blood 2010;115:1433–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hyde RK, Zhao L, Alemu L, Liu PP. Runx1 is required for hematopoietic defects and leukemogenesis in Cbfb-MYH11 knock-in mice. Leukemia 2015;29:1771–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mandoli a, Singh a a, Jansen PWTC, Wierenga a TJ, Riahi H, Franci G, et al. CBFB-MYH11/RUNX1 together with a compendium of hematopoietic regulators, chromatin modifiers and basal transcription factors occupies self-renewal genes in inv(16) acute myeloid leukemia. Leukemia 2014;28:770–8. [DOI] [PubMed] [Google Scholar]
- 17.Taunton J, Hassig C a, Schreiber SL. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science 1996;272:408–11. [DOI] [PubMed] [Google Scholar]
- 18.Gray SG, Ekström TJ. The human histone deacetylase family. Exp Cell Res 2001;262:75–83. [DOI] [PubMed] [Google Scholar]
- 19.Delcuve GP, Khan DH, Davie JR. Roles of histone deacetylases in epigenetic regulation: emerging paradigms from studies with inhibitors. Clin Epigenetics 2012;4:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Ruijter AJM, van Gennip AH, Caron HN, Kemp S, van Kuilenburg ABP. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem J 2003;370:737–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, et al. Genome-wide Mapping of HATs and HDACs Reveals Distinct Functions in Active and Inactive Genes. Cell 2009;138:1019–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nusinzon I, Horvath CM. Histone Deacetylases as Transcriptional Activators? Role Reversal in Inducible Gene Regulation. Sci Signal 2005;2005:1–7. [DOI] [PubMed] [Google Scholar]
- 23.Kuo Y-H, Landrette SF, Heilman S a, Perrat PN, Garrett L, Liu PP, et al. Cbf beta-SMMHC induces distinct abnormal myeloid progenitors able to develop acute myeloid leukemia. Cancer Cell 2006;9:57–68. [DOI] [PubMed] [Google Scholar]
- 24.Hyde RK, Liu PP. RUNX1 Repression Independent Mechanisms of Leukemogenesis by Fusion Genes CBFB-MYH11 and AML1-ETO (RUNX1-RUNX1T1). J Cell Biochem 2010;110:1039–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis 2007;45:593–605. [DOI] [PubMed] [Google Scholar]
- 26.Kamikubo Y, Zhao L, Wunderlich M, Corpora T, Hyde RK, Paul T a., et al. Accelerated Leukemogenesis by Truncated CBFβ-SMMHC Defective in High-Affinity Binding with RUNX1. Cancer Cell 2010;17:455–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuo Y-H, Zaidi SK, Gornostaeva S, Komori T, Stein GS, Castilla LH. Runx2 induces acute myeloid leukemia in cooperation with Cbfbeta-SMMHC in mice. Blood 2009;113:3323–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giaimo BD, Ferrante F, Borggrefe T. Chromatin Immunoprecipitation (ChIP) in Mouse T-cell Lines. J Vis Exp 2017; [DOI] [PMC free article] [PubMed]
- 29.Dull T, Zufferey R, Kelly M, Mandel RJ, Nguyen M, Trono D, et al. A third-generation lentivirus vector with a conditional packaging system. J Virol 1998;72:8463–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Santoro F, Botrugno O a., Dal Zuffo R, Pallavicini I, Matthews GM, Cluse L, et al. A dual role for Hdac1: oncosuppressor in tumorigenesis, oncogene in tumor maintenance. Blood 2013;121:3459–68. [DOI] [PubMed] [Google Scholar]
- 31.Yanagisawa K, Horiuchi T, Fujita S. Establishment and characterization of a new human leukemia cell line derived from M4E0. Blood 1991;78:451–7. [PubMed] [Google Scholar]
- 32.Wilting RH, Yanover E, Heideman MR, Jacobs H, Horner J, van der Torre J, et al. Overlapping functions of Hdac1 and Hdac2 in cell cycle regulation and haematopoiesis. EMBO J 2010;29:2586–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Durst KL, Lutterbach B, Kummalue T, Friedman AD, Hiebert SW. The inv ( 16 ) Fusion Protein Associates with Corepressors via a Smooth Muscle Myosin Heavy-Chain Domain. Mol Cell Biol 2003;23:607–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang YY, Shi J, Zhang L, Davis A, Bravo J, Warren AJ, et al. Energetic and functional contribution of residues in the core binding factor β (CBFβ) subunit to heterodimerization with CBFα. J Biol Chem 2000;275:39579–88. [DOI] [PubMed] [Google Scholar]
- 35.Nagata T, Werner MH. Functional mutagenesis of AML1/RUNX1 and PEBP2 beta/CBF beta define distinct, non-overlapping sites for DNA recognition and heterodimerization by the Runt domain. J Mol Biol 2001;308:191–203. [DOI] [PubMed] [Google Scholar]
- 36.Qi J, Singh S, Hua W-K, Cai Q, Chao S-W, Li L, et al. HDAC8 Inhibition Specifically Targets Inv(16) Acute Myeloid Leukemic Stem Cells by Restoring p53 Acetylation. Cell Stem Cell 2015;17:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bradner JE, West N, Grachan ML, Greenberg EF, Haggarty SJ, Warnow T, et al. Chemical phylogenetics of histone deacetylases. Nat Chem Biol 2010;6:238–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu WS, Parmigiani RB, Marks P. Histone deacetylase inhibitors: molecular mechanisms of action. Oncogene 2007;26:5541–52. [DOI] [PubMed] [Google Scholar]
- 39.Hu E Identification of Novel Isoform-Selective Inhibitors within Class I Histone Deacetylases. J Pharmacol Exp Ther 2003;307:720–8. [DOI] [PubMed] [Google Scholar]
- 40.Malvaez M, McQuown SC, Rogge GA, Astarabadi M, Jacques V, Carreiro S, et al. HDAC3-selective inhibitor enhances extinction of cocaine-seeking behavior in a persistent manner. Proc Natl Acad Sci 2013;110:2647–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cunningham L, Finckbeiner S, Hyde RK, Southall N, Marugan J, Yedavalli VRK, et al. Identification of benzodiazepine Ro5–3335 as an inhibitor of CBF leukemia through quantitative high throughput screen against RUNX1-CBF interaction. Proc Natl Acad Sci 2012;109:14592–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang G, Shigesada K, Ito K, Wee HJ, Yokomizo T, Ito Y. Dimerization with PEBP2beta protects RUNX1/AML1 from ubiquitin-proteasome-mediated degradation. EMBO J 2001;20:723–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang G, Shigesada K, Wee H-J, Liu PP, Osato M, Ito Y. Molecular basis for a dominant inactivation of RUNX1/AML1 by the leukemogenic inversion 16 chimera. Blood 2004;103:3200–7. [DOI] [PubMed] [Google Scholar]
- 44.Adya N, Stacy T, Speck N a, Liu PP. The leukemic protein core binding factor beta (CBFbeta)-smooth-muscle myosin heavy chain sequesters CBFalpha2 into cytoskeletal filaments and aggregates. Mol Cell Biol 1998;18:7432–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhen T, Kwon EM, Zhao L, Hsu J, Hyde RK, Lu Y, et al. Chd7 deficiency delays leukemogenesis in mice induced by Cbfb-MYH11. Blood 2017;130:2431–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Friedman AD. Transcriptional regulation of granulocyte and monocyte development. Oncogene 2002;21:3377–90. [DOI] [PubMed] [Google Scholar]
- 47.Wang J, Hoshino T, Redner RL, Kajigaya S, Liu JM. ETO, fusion partner in t(8;21) acute myeloid leukemia, represses transcription by interaction with the human N-CoR/mSin3/HDAC1 complex. Proc Natl Acad Sci U S A 1998;95:10860–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou L, Ruvolo VR, McQueen T, Chen W, Samudio IJ, Conneely O, et al. HDAC inhibition by SNDX-275 (Entinostat) restores expression of silenced leukemia-associated transcription factors Nur77 and Nor1 and of key pro-apoptotic proteins in AML. Leukemia 2013;27:1358–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Blagitko-Dorfs N, Jiang Y, Duque-Afonso J, Hiller J, Yalcin A, Greve G, et al. Epigenetic Priming of AML Blasts for All-trans Retinoic Acid-Induced Differentiation by the HDAC Class-I Selective Inhibitor Entinostat. PLoS One 2013;8:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hyde RK, Liu P, Friedman AD. RUNX1 and CBFβ mutations and activities of their wild-type alleles in AML. Adv Exp Med Biol 2017. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.