Abstract
Background
Diagnostic delays are common for multiple sclerosis (MS) since diagnosis typically depends on the presentation of nonspecific clinical symptoms together with radiologically-determined central nervous system (CNS) lesions. It is important to reduce diagnostic delays as earlier initiation of disease modifying therapies mitigates long-term disability. Developing a metabolomic blood-based MS biomarker is attractive, but prior efforts have largely focused on specific subsets of metabolite classes or analytical platforms. Thus, there are opportunities to interrogate metabolite profiles using more expansive and comprehensive approaches for developing MS biomarkers and for advancing our understanding of MS pathogenesis.
Methods
To identify putative blood-based MS biomarkers, we comprehensively interrogated the metabolite profiles in 12 non-Hispanic white, non-smoking, male MS cases who were drug naïve for 3 months prior to biospecimen collection and 13 non-Hispanic white, non-smoking male controls who were frequency matched to cases by age and BMI. We performed untargeted two-dimensional gas chromatography and time-of-flight mass spectrometry (GCxGC-TOFMS) and targeted lipidomic and amino acid analysis on serum. 325 metabolites met quality control and supervised machine learning was used to identify metabolites most informative for MS status. The discrimination potential of these select metabolites were assessed using receiver operator characteristic curves based on logistic models; top candidate metabolites were defined as having area under the curves (AUC) >80%. The associations between whole-genome expression data and the top candidate metabolites were examined, followed by pathway enrichment analyses. Similar associations were examined for 175 putative MS risk variants and the top candidate metabolites.
Results
12 metabolites were determined to be informative for MS status, of which 6 had AUCs >80%: pyroglutamate, laurate, acylcarnitine C14:1, N-methylmaleimide, and 2 phosphatidylcholines (PC ae 40:5, PC ae 42:5). These metabolites participate in glutathione metabolism, fatty acid metabolism/oxidation, cellular membrane composition, and transient receptor potential channel signaling. Pathway analyses based on the gene expression association for each metabolite suggested enrichment for pathways associated with apoptosis and mitochondrial dysfunction. Interestingly, the predominant MS genetic risk allele HLA-DRB1*15:01 was associated with one of the 6 top metabolites.
Conclusion
Our analysis represents the most comprehensive description of metabolic changes associated with MS to date with the inclusion of genomic and genetic information. We identified atypical metabolic processes that differed between MS patients and controls, which may lead to the development of biological targets for diagnosis and treatment.
Keywords: Multiple sclerosis, metabolomics, serum, gene expression, genotype, biomarker
Introduction
Multiple sclerosis (MS) is an idiopathic autoimmune disease typified by inflammatory events in the central nervous system that trigger demyelination, exacerbating remyelination failure, and subsequent neurodegeneration. Most affected individuals will steadily accrue irreversible neurological disability over the disease course, but there is mounting evidence that earlier initiation of disease modifying therapies (DMTs) may mitigate long-term disability outcomes.1 Unfortunately, MS currently requires a clinical diagnosis that largely depends on neurological symptoms and/or radiologic lesions that are disseminating in time and/or space. As a result, the initiation of DMTs may be delayed. Therefore, there is a need to develop non-invasive diagnostic tools to accelerate MS diagnosis. There is also significant need to further elucidate the patho-etiological mechanisms contributing to MS onset and accrual of disability in order to advance the development of novel therapeutics.
Multiple efforts to identify biological markers (biomarkers) for MS have been made, but most of these potential biomarkers are not MS-specific and many are only discriminatory in cerebrospinal fluid (CSF).2, 3 While CSF is among the most pertinent biological samples for the study of MS, the invasiveness and inherent risk of lumbar punctures makes CSF collection impractical for recurrent testing.4 Instead, biomarkers readily detected in the blood would be ideal for diagnostic and prognostic evaluation. While there are promising candidates for diagnosis, such as neurofilament light protein, the only clinically useful blood biomarkers for MS are antibodies against natalizumab and interferon β drugs; these inform drug efficacy and risk for adverse side-effects.5-7
Increasingly, metabolomic assays are being used to develop disease biomarkers and to identify patho-etiologic processes. Improvements in instrumentation, analytical sensitivity, and consistency have advanced metabolomics-based biomarker development for diseases such as Type II diabetes and cardiovascular disease.8, 9 Most prior metabolomics work in MS has relied on a single assay/platform or had a scope limited to a particular metabolite class.10-18 Only one prior study applied a global untargeted metabolomic approach (ultra-high performance liquid chromatography-tandem mass spectrometry) to MS, which allowed for interrogating a much larger number of metabolites.19 Therefore, by combining targeted and untargeted approaches there are opportunities to conduct more expansive and comprehensive metabolomic investigation in MS, particularly within the multiomic framework.
We profiled serum metabolites in MS cases and controls using untargeted two-dimensional gas chromatography and time-of-flight mass spectrometry (GCxGC-TOFMS) with targeted metabolomic approaches (i.e. lipid and amino acid profiles). GCxGC-TOFMS has been shown to discriminate and identify up to three times more serum metabolites than one-dimensional GC-TOFMS.20, 21 We then identified metabolites predictive of MS status using supervised machine learning and multivariable logistic regression. To explore possible etiologic processes, we integrated whole-genome expression data and genetic data for alleles encoded by human leukocyte antigen (HLA) genes (HLA-DRB1*15:01, HLA-A*02) and 175 putative MS risk single nucleotide polymorphisms (SNPs) outside the major histocompatibility complex (MHC).22 We identified 6 metabolites predictive of MS status and found evidence that pathways involved in apoptosis and mitochondrial dysfunction may be contributing to the altered metabolic profile in MS.
Materials and Methods
Study subjects
The Duke University Measurement to Understand the Reclassification of Disease of Cabarrus/Kannapolis (MURDOCK) longitudinal health study recruitment has been previously described.23 MS cases (n=12) were participants in both the MURDOCK Study Community Registry and Biorepository (Pro00011196) and MURDOCK MS Cohort Study (Pro00023791), while controls (n=13) were drawn from the MURDOCK Study Community Registry and Biorepository (Supplementary Table 1). Participant recruitment and sample collection protocols were approved by the Duke University Institutional Review Board; written informed consent was obtained from all participants. The study population for the current analysis was non-Hispanic white, non-smoking males. MS cases were drug naïve for at least three months prior to biospecimen collection, and medical records were used to confirm a diagnosis of MS and disease-modifying therapy (DMT) history. Controls were age and body mass index (BMI) frequency matched to cases.
Sample collection and storage
The MURDOCK Study collected samples for extraction of serum, RNA, and DNA using standard collection criteria implemented by the MURDOCK Study.23 Serum samples were stored at −80 °C by LabCorp and/or BioStorage Technologies until use in this project. Total RNA was extracted from PAXgene tubes using Qiagen PAXgene Blood RNA Kit (#762164) and stored at −80 °C. Genomic DNA was extracted from whole blood using QIAGEN Gentra Puregene Blood Kits (#158445) and stored at −20 °C.
Sample preparation and data generation
Metabolomic data
For GCxGC-TOFMS, serum aliquots were extracted with methanol/chloroform containing heptadecanoic acid and norleucine as internal standards, as previously described.21 Equal sample aliquots were pooled to create for quality control (QC) samples. Samples were dried under N2, derivatized with methoxyamine in pyridine and then N-Methyl-N-(trimethylsilyl) trifluoroacetamide with 1% trimethylchlorosilane. The samples were analyzed on a LECO Pegasus 4D GCxGC-TOFMS in electron ionization mode. An Agilent (Agilent Technologies, Santa Clara, CA) DB-5ms UI GC column was the primary column and an Agilent DB-17ms was the secondary column. Pooled QC samples were analyzed after every nine samples to assess overall reproducibility and to correct for any observed variations. An alkane retention index standard (C10 – C40) was run at the beginning, middle and end of sample runs. GCxGC-TOFMS Peak lists were generated with LECO ChromaTOF software version 4.50.8.0 (LECO Corporation, St. Joseph, MI) using vendor-recommended parameters. NIST/EPA/NIH Mass Spectral Library 2011 (NIST11), LECO/Fiehn Metabolomics Library, and an in-house library were used as reference libraries for compound identification. MetPP software was used for peak merging and peak list alignment, and retention index (RI) matching was performed using iMatch algorithms with p-value threshold ≤ 0.001.20, 21
For the targeted assay, we used the Biocrates AbsoluteIDQ p150 kit (Biocrates Life Sciences AG, Austria) which profiles specific amino acids, acylcarnitines, hexoses, and phospho- and sphingolipids. For the Biocrates kit, 10 μl serum aliquots were processed and analyzed using the Biocrates AbsoluteIDQ p150 kit protocol.
Gene expression data
Whole-genome expression data were generated from three Illumina HumanHT-12 v4.0 Gene Expression BeadChips for 24 of the 25 study participants (Supplementary Table 1). Biotin-labeled cDNA were generated and hybridized following the Illumina Whole Genome Gene Expression Direct Hybridization Assay. Case and control samples were randomized and sample duplicates included as assay replicate controls. Arrays were scanned with the Illumina iScan Microarray Scanner system. Sample replicates (1 per chip) and array assay controls (1 per assay) confirmed assay integrity and ruled out contamination with identical calls and predicted genotypes.
Genotypic data
Sequenom platform Iplex Gold Reagent Kit multiplexed genotyping assays were designed in ~25 SNP pools using the Assay Design Suite (Agena Biosciences) for 186 of 200 putative non-MHC MS risk variants.22 Linked discovery SNPs or tagging SNPs were substituted for effect SNPs refractory to multiplex design. Samples were spotted to Sequenom SpectroChip(s) and scanned with MALDI-TOFMS. Non-template negative controls, sample duplicates and control DNAs were included. HLA-DRB1*15:01 alleles were determined via TaqMan genotyping of SNP rs9271366 (assay ID: C__33416976_20). HLA-A*02 alleles were determined via TaqMan genotyping of the SNP rs2975033 (assay ID: C__15962692_20). Each SNP assay was reviewed for the genotype call quality based on PCR/Extension yield, proximity to cluster, and raw mass spectra.
Statistical analyses
There were 400 metabolite variables captured across the GCxGC-TOFMS and Biocrates p150 approaches (Supplementary Table 2). 325 metabolites with <30% missing observations were retained for analyses implemented in MetaboAnalyst (http://www.metaboanalyst.ca/).24 ComBat was used to adjust for batch effects (Supplementary Figure 1A), and missing values were replaced using the default setting (by half of the minimum positive values detected in the data) in MetaboAnalyst.25 Random forests, a supervised machine learning (non-parametric) algorithm that is well adapted for prediction and variable importance, fairly robust to the setting of tuning parameters, and capable of analyzing data where predictors outnumber sample size, was used to identify metabolites important (in contrast to a p-value) for MS classification.26 The forest consisted of 5,000 trees and 100 randomly selected metabolites were used to determine classification at each node in a tree. A scree plot of the ranked variable importance scores was used to identify important (top-ranking) metabolites for MS classification (Supplementary Figure 2). Similar results were obtained for quantile normalized and pareto scaled metabolomics data (data not shown). Logistic regression models were used to assess the relationship between each important metabolite and MS status, including a random effect to account for any residual batch effects using the xtlogit function in STATA v13.1 (StataCorp, TX). The area under the curve (AUC) and 95% Bamber and Hanley confidence intervals were determined by non-parametric receiver operating characteristic (ROC) analyses using the roctab function in STATA for each metabolite. Metabolites with AUC>80% were considered our top metabolites.
Gene expression data were background subtracted and quantile normalized within GenomeStudio Software Gene Expression Module (Illumina). Expression values with detection p-values >0.05 and genes with at least 70% of observations were retained; 9067 genes passed QC. ComBat, as implemented in MetaboAnalyst, was applied to remove any chip effects (Supplementary Figure 1B) and data were quantile normalized and pareto scaled. The expression of CD19 (B lymphocyte marker); CD14 (monocyte marker); FCGR3A and NCAM1 (natural killer cell markers); and CD3E, CD3D, CD4, and CD8A (T lymphocyte markers) did not differ between cases and controls via Wilcoxon rank-sum tests, indicating no blood cellular composition differences between cases and controls (data not shown).27 Therefore, we conducted the following multivariable linear mixed-effects regression models: each top metabolite was the outcome and each gene expression value was a predictor, as well MS status, and a random effect to capture residual chip effects using the xtreg function in STATA. The top 2.5% (p-value ranked) of genes associated with each metabolite were analyzed via the Ingenuity Pathway Analysis (IPA) for biological pathway enrichment. The Illumina HumanHT-12 v4.0 Gene Expression BeadChip gene set was used as the reference set. Interaction networks, including endogenous chemicals, and all node types were included in the analysis (35 molecules per network, 25 networks per analysis). Data sources were experimentally observed from mammalian species.
Genetic data were generated along with another 107 samples. QC was applied to the entire genetic data set and included removing samples with >50% missing genotypes; SNPs with >20% missing genotypes; samples with >10% genotypes; and SNPs with Hardy Weinberg Equilibrium p<0.01. After QC, the final genotype dataset included 19 subjects (Supplementary Table 1) and 175 putative non-HLA MS risk SNPs (Supplementary Table 3). Multivariable regression models with the top metabolites at the outcome and each SNP as a predictor, adjusting for MS status and a random effect capturing possible plate effects were conducted. Two HLA alleles were genotyped in all 25 samples, however genotypes for the HLA-DRB1*15:01 and HLA-A*02 tagging SNPs were successfully determined in 22 and 24 samples respectively. Similar regression models were conducted as described above.
A two-sided alpha<0.05 was considered statistically significant for all statistical comparisons.
Results
Identifying top metabolites associated with MS
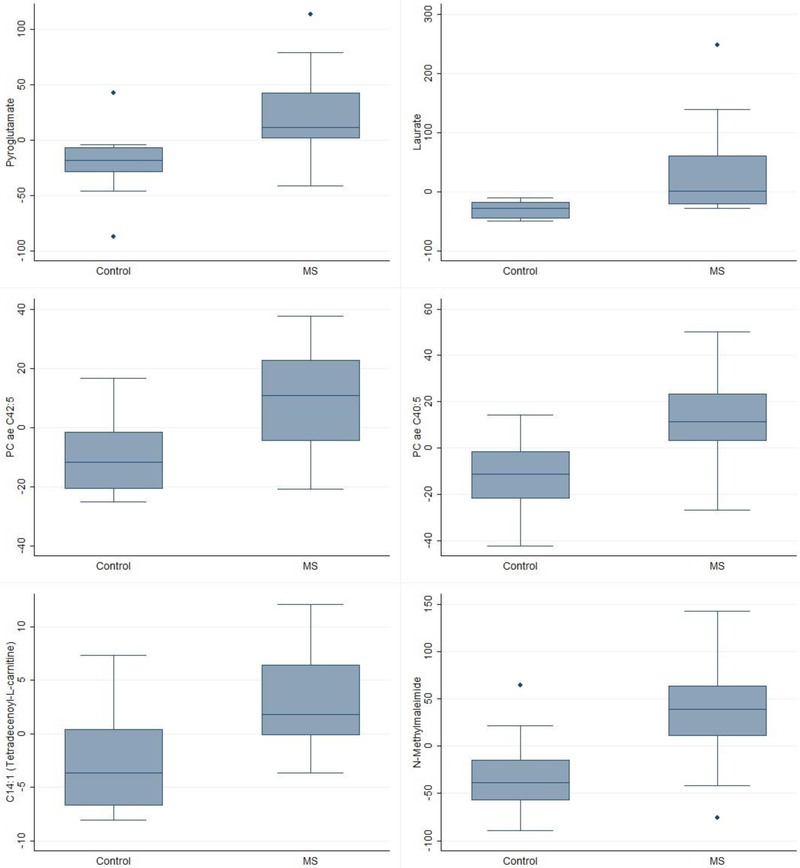
Random forests identified 12 metabolites as informative for MS classification (Table 1, Supplementary Figure 2). The random forests’ out-of-bag error for classifying the 25 samples by MS status was 0.32 (0.42 for cases and 0.23 for controls; Supplementary Figure 3). Eight of these metabolites were associated with MS in logistic regression models (p<0.05), and 6 of them had an AUC >80% (Supplementary Figure 4). The top 6 were two glycerophospholipids, two metabolites associated with fatty acid metabolism, an amino acid, and a biothiol scavenger. All had higher levels in MS cases than controls (Figure 1).
Table 1.
Top ranked metabolites from Random forests analyses and logistic regression associations for MS status.
| Metabolite | Random Forest Rank |
Logistic regression p-value |
AUC (95% CI) | Classa | Biological processesa |
Sourcea |
|---|---|---|---|---|---|---|
| Pyroglutamate | 1 | 0.033 | 0.85 (0.68, 1) | Amino acid derivative/Amino acid | Glutathione metabolism/Unknown | Endogenous, food |
| Laurate | 2 | 0.035 | 0.86 (0.72, 1) | Medium chain fatty acid | Lipid and fatty acid metabolism | Endogenous, food |
| UnknownZ11 | 3 | 0.123 | 0.20 (0, 0.41) | Unknown | Unknown | Unknown |
| N-methoxy carbonylproline, methyl ester | 4 | 0.261 | 0.26 (0.03, 0.5) | Amino acid derivative | Unknown | Unknown |
| Phosphatidylcholine PC ae C42:5 | 5 | 0.018 | 0.81 (0.64, 0.99) | Glycerophospholipid | Cellular membranes | Endogenous, food |
| C14:1 (Tetradecenoyl-L-carnitine) | 6 | 0.018 | 0.82 (0.65, 0.99) | Acylcarnitine | Long-chain fatty acid oxidation | Endogenous, food |
| Unknown056 | 7 | 0.048 | 0.21 (0.01, 0.4) | Unknown | Unknown | Unknown |
| Phosphatidylcholine PC ae C40:5 | 8 | 0.016 | 0.82 (0.64, 1) | Glycerophospholipid | Cellular membranes | Endogenous, food |
| N-Methylmaleimide | 9 | 0.015 | 0.82 (0.64, 1) | Electrophile | Biothiol scavenger | Unknown |
| Unknown022 | 10 | 0.71 | 0.61 (0.35, 0.87) | Unknown | Unknown | Unknown |
| Myo-Inositol | 11 | 0.034 | 0.21 (0.03, 0.39) | Cyclohexanol | Galactose, inositol, and phosphatidylinositol metabolism | Endogenous, food |
| Linolic acid | 12 | 0.051 | 0.70 (0.46, 0.94) | Long chain fatty acid | Lipid and fatty acid metabolism | Endogenous, food, environment |
Abbreviations: 95% CI = 95% confidence interval; AUC = area under the curve
Class, gene ontology, and source derived from the Human Metabolome Database (HMDB) and PubChem (NCBI).
Figure 1.
Box plots of the top metabolites by MS status.
Gene expression associations with top metabolites
We explored the relationships between expression for 9,067 genes and each of the top metabolites (Supplementary Table 4). Among MS-related genes, HLA-DRB1 was not associated with any metabolite, however the expression of several other HLA genes were associated with acylcarnitine C14:1 including HLA-DMA, HLA-DMB, HLA-DOA, HLA-DPA1, HLA-DPB1, HLA-DRA, HLA-DRB3, and HLA-DRB6. The expression of multiple genes proximal (+/− 25 kilobases) to the 200 putative non-MHC MS risk variants were associated with the metabolites (Supplementary Table 5).22 Of note are the associations for KPNB1, CLEC16A, PIK3R2, IKZF1, TXK and PHGDH, which were associated with at least three of the 6 metabolites (p<0.1; Supplementary Table 5).
Pathway enrichment analyses of the top 2.5% p-value ranked genes associated with each metabolite, after adjustment for MS status, were conducted and multiple pathways were enriched for each metabolite (Supplementary Table 6). The top-ranked pathways included iron homeostasis signaling (pyroglutamate), ceramide degradation (pyroglutamate), antigen presentation (acylcarnitine C14:1), vitamin D biosynthesis (PC ae 42:5), and mitochondrial dysfunction (PC ae 40:5 and N-Methylmaleimide) (Table 2). We cross-referenced all metabolite-associated enriched pathways with pathways enriched among genes associated with the putative 200 non-MHC risk variants by the International MS Genetics Consortium (Supplementary Table 6).22 A third of the metabolite-associated enriched pathways overlapped with the MS-related pathways. There were also several pathways enriched among the associated genes across metabolites (Supplementary Table 7), specifically apoptosis related processes (death receptor signaling, calcium-induced T lymphocyte apoptosis, lymphotoxin β receptor signaling, and TNFR1 signaling) and altered mitochondrial function (mitochondrial dysfunction, oxidative phosphorylation, and sirtuin signaling pathway).
Table 2.
Top 5 pathways enriched per metabolite based on gene expression analyses.
| Metabolite | Biological Pathways | P-value | Genes |
|---|---|---|---|
| Pyroglutamate | Iron homeostasis signaling pathway | 1.3E-04 | HBZ, FTL, HSCB, CIAO1, BMP6, CUL1, SLC48A1 |
| Ceramide Degradation | 1.5E-03 | ACER3, NAAA | |
| Sphingosine and Sphingosine-1-phosphate Metabolism | 2.6E-03 | ACER3, NAAA | |
| Biotin-carboxyl Carrier Protein Assembly | 2.6E-02 | ACACB | |
| Glutathione Biosynthesis | 2.6E-02 | GCLM | |
| Laurate | Ephrin B Signaling | 7.2E-04 | HNRNPK, GNB1, PXN, RAC2, EPHB1 |
| Inflammasome pathway | 9.1E-04 | MYD88, NLRP1, PYCARD | |
| Germ Cell-Sertoli Cell Junction Signaling | 1.3E-03 | IQGAP1, PAK2, PIK3CD, RHOB, PXN, TUBA4A, RAC2 | |
| Melanocyte Development and Pigmentation Signaling | 2.6E-03 | PIK3CD, PLCG1, RPS6KA4, RPS6KC1, PRKAR1A | |
| SAPK/JNK Signaling | 2.7E-03 | PIK3CD, MAP4K2, HNRNPK, GNB1, RAC2 | |
| Phosphatidylcholine PC ae C42:5 | Asparagine Biosynthesis I | 1.1E-02 | ASNS |
| Role of Oct4 in Mammalian Embryonic Stem Cell Pluripotency | 1.4E-02 | FAM208A, IGF2BP1, NR2F6 | |
| Dolichol and Dolichyl Phosphate Biosynthesis | 2.2E-02 | DOLK | |
| 1,25-dihydroxyvitamin D3 Biosynthesis | 3.3E-02 | CYP2R1 | |
| S-adenosyl-L-methionine Biosynthesis | 3.3E-02 | MAT2A | |
| Phosphatidylcholine PC ae C40:5 | Oxidative Phosphorylation | 2.8E-05 | NDUFB6, COX17, NDUFA7, NDUFB7, ATP5PF, COX5B, VPS9D1 |
| Mitochondrial Dysfunction | 1.1E-04 | CYB5R3, NDUFB6, COX17, NDUFA7, NDUFB7, ATP5PF, COX5B, VPS9D1 | |
| Sirtuin Signaling Pathway | 1.3E-03 | NDUFB6, PARP1, NDUFA7, NDUFB7, ATP5PF, GADD45A, POLR1C, SLC2A1, ATG16L2 | |
| Death Receptor Signaling | 1.9E-03 | PARP12, PARP1, DFFA, PARP14, BIRC3 | |
| Calcium-induced T Lymphocyte Apoptosis | 1.9E-03 | HLA-DQB1, EP300, CD4, PPP3CC | |
| Tetradecenoyl-L-carnitine (C14:1) | Antigen Presentation Pathway | 4.3E-08 | HLA-DPB1, HLA-DOA, HLA-DMA, HLA-DPA1, HLA-DMB, HLA-DRA, CIITA |
| Allograft Rejection Signaling | 1.5E-06 | HLA-DPB1, HLA-DOA, HLA-DMA, HLA-DPA1, HLA-DMB, HLA-DRA | |
| OX40 Signaling Pathway | 6.6E-06 | HLA-DPB1, HLA-DOA, HLA-DMA, HLA-DPA1, HLA-DMB, HLA-DRA | |
| Cdc42 Signaling | 1.9E-05 | HLA-DPB1, HLA-DOA, HLA-DMA, HLA-DPA1, HLA-DMB, HLA-DRA, ARPC1A, VAV2 | |
| B Cell Development | 7.8E-05 | HLA-DOA, HLA-DMA, HLA-DMB, HLA-DRA | |
| N-Methylmaleimide | Oxidative Phosphorylation | 6.0E-06 | ATP5S, NDUFA4, COX7B, NDUFA9, NDUFA6, ATP5F1C, UQCRB, NDUFAB1 |
| Sirtuin Signaling Pathway | 7.6E-06 | NFKB1, GOT2, CLOCK, SP1, NDUFAB1, RELA, NDUFA4, PARP1, ATG3, NDUFA9, NDUFA6, ATP5F1C, GTF3C2 | |
| Superpathway of Geranylgeranyldiphosphate Biosynthesis I (via Mevalonate) | 2.0E-05 | IDI1, HMGCR, FNTB, ACAT1 | |
| Mitochondrial Dysfunction | 3.6E-05 | ATP5S, NDUFA4, COX7B, NDUFA9, NDUFA6, CASP3, ATP5F1C, UQCRB, NDUFAB1 | |
| Mevalonate Pathway I | 2.4E-04 | IDI1, HMGCR, ACAT1 |
MS risk alleles associations with top metabolites
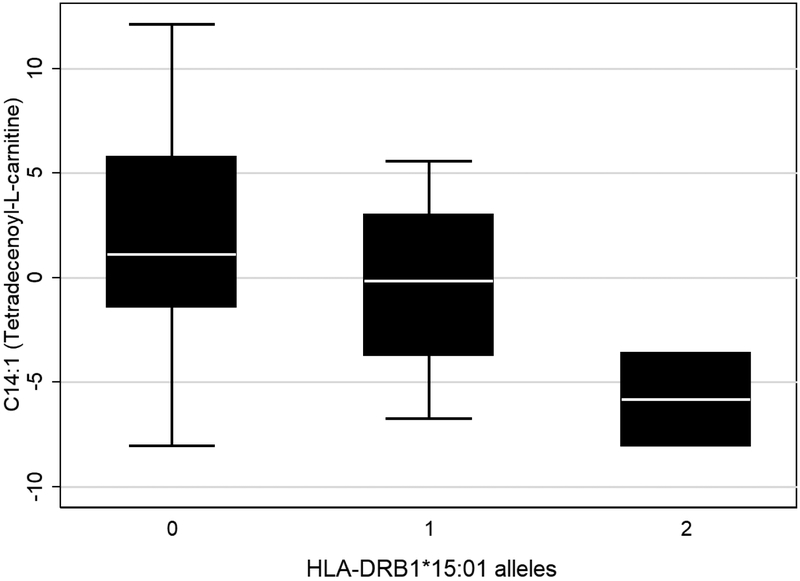
We assessed the relationship between genetic risk and MS-predictive metabolite levels. While no association was observed for HLA-A*02, we identified an association between HLA-DRB1*15:01 and acylcarnitine C14:1 (Table 3; Figure 2). We also observed multiple associations between metabolite levels and the 175 putative non-MHC risk variants that we genotyped (Table 4). Of note were five SNPs associated with both phosphatidylcholines PC ae 40:5 and PC ae 42:5, four of which were genic variants within ETS1, IL2RA and AFF1.
Table 3.
Genetic associations between HLA variants and top metabolites.
| Metabolite | HLA-DRB1*15:01 | HLA-A*02 | ||
|---|---|---|---|---|
| Effect Direction | P-value | Effect Direction | P-value | |
| Pyroglutamate | − | 0.66 | − | 0.31 |
| Laurate | − | 0.31 | + | 0.79 |
| Phosphatidylcholine PC ae C42:5 | + | 0.80 | + | 0.68 |
| Phosphatidylcholine PC ae C40:5 | − | 0.53 | + | 0.21 |
| C14:1 (Tetradecenoyl-L-carnitine) | − | 0.02 | + | 0.75 |
| N-Methylmaleimide | − | 0.10 | + | 0.29 |
Figure 2.
Box plot of C14:1 Tetradecemoyl-L-carnitine by HLA*DRB1*15:01 genotype.
Table 4.
Genic non-MHC MS risk variant associations with the top metabolites.
| Metabolite | Associated SNPs | Location (GRCh38/hg38) |
Minor Allele |
Effect Direction |
P-value | SNP Proximal Genes |
|---|---|---|---|---|---|---|
| Pyroglutamate | rs531612 | chr11:65937961 | C | − | 0.013 | DRAP1, TSGA10IP |
| rs10230723 | chr7:50200284 | T | + | 0.019 | C7orf72, IKZF1 | |
| rs2150879 | chr17:59781849 | A | − | 0.024 | VMP1 | |
| rs34681760 | chr5:6712721 | T | + | 0.033 | LOC100505625, PAPD7 | |
| rs9282641 | chr3:122077921 | A | + | 0.047 | CD86 | |
| Laurate | rs4766578 | chr12:111466567 | T | − | 0.017 | SH2B3 |
| rs2317231 | chr1:157716547 | T | + | 0.019 | FCRL3, FCRL2 | |
| rs9913257 | chr17:75324812 | C | + | 0.023 | GRB2 | |
| rs6032662 | chr20:46105671 | C | + | 0.027 | CD40, NCOA5 | |
| rs2269434 | chr11:47338861 | C | + | 0.034 | MYBPC3 | |
| rs12925972 | chr16:79077400 | T | − | 0.043 | WWOX | |
| rs17780429 | chr6:137901451 | A | + | 0.045 | LOC100130476 | |
| Phosphatidylcholine PC ae 42:5 | rs4262739 | chr11:128551280 | A | − | 0.003 | ETS1 |
| rs12373588 | chr2:111708688 | G | + | 0.007 | ANAPC1, MIR4435-1HG | |
| rs12622670 | chr2:68419404 | C | + | 0.014 | FBXO48, PLEK | |
| rs2705616 | chr4:86941244 | G | + | 0.018 | AFF1 | |
| rs2705618 | chr4:86916393 | T | + | 0.018 | AFF1 | |
| rs62013236 | chr15:78955140 | T | + | 0.020 | CTSH, RASGRF1 | |
| rs6589939 | chr11:122647817 | G | − | 0.020 | MIR100HG, UBASH3B | |
| rs116899835 | chr14:88057144 | T | + | 0.021 | LINC01146 | |
| rs6738544 | chr2:191124630 | A | − | 0.029 | STAT4 | |
| rs12722559 | chr10:6028310 | A | + | 0.033 | IL2RA | |
| rs2269434 | chr11:47338861 | C | − | 0.035 | MYBPC3 | |
| rs9913257 | chr17:75324812 | C | − | 0.047 | GRB2 | |
| rs6952809 | chr7:2408858 | T | + | 0.048 | CHST12 | |
| Phosphatidylcholine PC ae 40:5 | rs1076928 | chr6:36380912 | C | + | 0.001 | ETV7 |
| rs4262739 | chr11:128551280 | A | − | 0.004 | ETS1 | |
| rs12373588 | chr2:111708688 | G | + | 0.007 | ANAPC1, MIR4435-1HG | |
| rs1534422 | chr2:12500615 | A | − | 0.016 | LOC100506457 | |
| rs883871 | chr17:40096407 | A | − | 0.026 | NR1D1 | |
| rs719219 | chr20:49818871 | A | − | 0.028 | B4GALT5, SLC9A8 | |
| rs12722559 | chr10:6028310 | A | + | 0.034 | IL2RA | |
| rs11852059 | chr14:51839373 | C | + | 0.035 | FRMD6, GNG2 | |
| rs137956 | chr22:39897459 | C | + | 0.036 | ENTHD1, GRAP2 | |
| rs9878602 | chr3:71486187 | G | + | 0.038 | FOXP1 | |
| rs140522 | chr22:50532837 | T | − | 0.041 | ODF3B | |
| rs9992763 | chr4:108137562 | T | − | 0.042 | LEF1 | |
| rs2705616 | chr4:86941244 | G | + | 0.044 | AFF1 | |
| rs2705618 | chr4:86916393 | T | + | 0.044 | AFF1 | |
| rs34695601 | chr14:75547955 | C | + | 0.047 | BATF | |
| Acylcarnitine C14:1 | rs2705616 | chr4:86941244 | G | − | 0.001 | AFF1 |
| rs2705618 | chr4:86916393 | T | − | 0.001 | AFF1 | |
| rs12614091 | chr2:203768138 | T | + | 0.006 | CD28, CTLA4 | |
| rs1177228 | chr2:61015275 | A | + | 0.016 | PUS10 | |
| rs2269434 | chr11:47338861 | C | + | 0.02 | MYBPC3 | |
| rs9909593 | chr17:39813896 | G | − | 0.025 | IKZF3 | |
| rs2331964 | chr3:121824051 | T | − | 0.035 | IQCB1 | |
| rs983494 | chr1:160734175 | A | + | 0.045 | CD48, SLAMF7 | |
| N-methylmaleimide | rs28703878 | chr8:78504987 | G | + | 0.001 | LOC102724874, PKIA |
| rs3923387 | chr8:143912625 | T | + | 0.002 | EPPK1, PLEC | |
| rs12708716 | chr16:11086016 | G | − | 0.004 | CLEC16A | |
| rs11852059 | chr14:51839373 | C | − | 0.005 | FRMD6, GNG2 | |
| rs6837324 | chr4:48125245 | G | + | 0.007 | TXK | |
| rs10245867 | chr7:28102567 | T | + | 0.012 | JAZF1 | |
| rs719316 | chr6:16672529 | C | − | 0.015 | ATXN1 | |
| rs438613 | chr3:28030595 | C | + | 0.026 | CMC1, EOMES | |
| rs35716097 | chr5:177379635 | T | + | 0.031 | RGS14 | |
| rs483180 | chr1:119724882 | G | + | 0.032 | PHGDH | |
| rs12614091 | chr2:203768138 | T | − | 0.041 | CD28, CTLA4 |
Discussion
We combined exploratory open platform metabolomics with transcriptomics and risk variant genotyping to gain a multifaceted multiomic perspective on metabolic changes associated with MS. Our analyses identified 6 metabolites as promising MS predictive candidates involved in glutathione metabolism (pyroglutamate), fatty acid metabolism/oxidation (laurate, C14:1 acylcarnitine), cellular membrane composition (glycerophospholipids PC ae 42:5 and PC ae 40:5), and a transient receptor potential A1 (TRPA1) channel agonism (N-Methylmaleimide). Two of these metabolites have been previously associated with MS (pyroglutamate, laurate), while the remaining four have strong biologically plausible relevance.16, 17, 19, 28, 29 Integrated analyses with genomic and genetic data suggest distinct yet overlapping biological processes may be contributing to elevated levels of these metabolites in MS, including apoptotic processes and mitochondrial dysfunction.
Pyroglutamate (also known as 5-oxoproline) was the most important metabolite identified by random forest analysis. A natural cyclized derivative of glutamate that is generated during biosynthesis of the antioxidant glutathione, it has previously been associated with MS in other untargeted metabolomic analyses of CSF and plasma.16, 17, 19 Elevated pyroglutamate, as detected in our MS cohort, is associated with several rare metabolic disorders, including glutathione synthetase deficiency and 5-oxoprolinase deficiency. These disorders manifest neurological symptoms, including intellectual deficiency, microcephaly, and seizures. Murine experiments suggest that pyroglutamate may directly contribute to neurological symptoms, with peripheral pyroglutamate capable of crossing the blood brain barrier and triggering oxidative damage.30, 31 Pyroglutamate also stimulates sodium-dependent abluminal transport of amino acids across the blood brain barrier.32 As a marker of changes to glutathione metabolism and a source of oxidative damage, pyroglutamate may be both a marker of, and a contributor to, the environment of increased oxidative stress in MS.33 Among the top-ranked pathways enriched among genes associated with this glutathione intermediate were glutathione biosynthesis, iron hemostasis signaling, and ceramide/sphigosine metabolism. The latter two pathways both contribute to apoptosis. Perturbed iron levels promote neuronal death, and abnormal iron deposition in the brain may facilitate neurodegeneration.34 Interestingly, elevated ceramide levels have been observed near MS plaques, and sphingosine metabolism, which is essential for myelin synthesis and maintenance, has been reported to be abnormal in the MS brain.35
The second most important metabolite identified by random forests was laurate (lauric acid), a medium-chain fatty acid that is elevated in the “Western Diet”.6 There is evidence supporting a role for this saturated fatty acid in MS pathology. In vitro treatment of mouse CD4+ T cells with laurate stimulated Th1 and Th17 differentiation and decreased Treg differentiation, while treatment of human CD4+ T cells promoted CD4+interferon-γ+ differentiation.28, 29 In a study of experimental autoimmune encephalomyelitis mice (a murine model of MS), increased dietary laurate promoted differentiation of Th1 and Th17 cells, which contributed to more severe disease.29 Multiple pathways were associated with laurate, including Tec kinase signaling (essential for B and T cell development and activation), lymphotoxin β receptor signaling (apoptotic trigger that can result in the release of interleukin 8 [IL-8], and IL-8 contributes to Th17 development37), IL-8 signaling, sphigosine-1-phosphate signaling, and axonal guidance (Supplementary Table 6).
Two phosphatidylcholines, the primary components of cellular membranes and myelin, were predictive of MS status: PC ae 40:5 and PC ae 42:5. The partial hydrolysis of phosphatidylcholines by phospholipase A2 enzymes generates an array of bioactive products, including eicosanoid pro-inflammatory signaling molecules and myelin-damaging lysophophatidylcholines. In our data, increased expression of a group IV phospholipase A2 enzyme, PLA2G4C, was significantly associated with increased PC ae 40:5 and PC ae 42:5 levels (Supplementary Table 4). PLA2G4C is one of the least well-characterized enzymes in the family, though it is important for mitochondrial function and genetic variants have been associated with schizophrenia and autism spectrum disorder.38, 39 PLA2G4C is expressed throughout the brain and protein levels are highest in nervous system tissues.40,41,42 There is evidence PLA2G4C facilitates macrophage differentiation and polarization and CD4+ T cell transcripts have been found to be restricted to only PLA2G4A and PLA2G4C members of the family, suggesting this PLA2 enzyme may have particular importance in MS-relevant immune cells 43,44.
There was little overlap in the pathways enriched among the genes whose expression was associated with either phosphatidylcholine, though MS risk variants in IL2RA, ETS1, and AFF1 were associated with both metabolites. ETS1 is a transcription factor that influences a wide array of cellular processes; interestingly, it is predicted to regulate PLA2G4C based on the presence of core consensus sequences in the promoter.45 In our data, EST1 expression was associated with PC ae 42:5 levels (p=0.007; Supplementary Table 5). AFF1 is a core member of the transcription factor positive transcription elongation factor b (P-TEFb), which regulates RNA polymerase II-mediated transcription.46 In our data, AFF1 expression was associated with PC ae 40:5 levels (p=0.017; Supplementary Table 5). Of note, variants near or within AFF1 have been associated with circulating lipid levels in genome-wide association studies, which supports an as yet uncharacterized role for AFF1 in lipid metabolism.47-50
The fourth lipid-related metabolite predictive of MS in our analyses was acylcarnitine C14:1, an intermediate of fatty acid oxidation and a marker for the rare genetic metabolic disorder VCLAD (very long-chain acyl-dehydrogenase deficiency). In general, acylcarnitines are indicative of mitochondrial function and energy metabolism, which are both relevant to, and perturbed by, MS pathogenesis.51, 52 The sirtuin signaling pathway, which is associated with longevity and mitochondrial efficiency, was enriched among genes associated with acylcarnitine C14:1; it was also enriched among genes associated with PC ae 40:5 and N-methylmaleide (Supplementary Table 7).53 Also enriched in association with C14:1 were genes involved with antigen presentation, including the expression of multiple class II HLA genes: HLA-DMA, HLA-DMB, HLA-DOA, HLA-DPA1, HLA-DPB1, HLA-DRA, HLA-DRB3, and HLA-DRB6 (Supplementary Table 4). Carriage of HLA-DRB1*15:01 risk alleles was also associated with C14:1, though not HLA-DRB1 expression (Table 3, Figure 2). The presentation of antigens by antigen presenting cells activates CD4+ helper T cells, which in turn triggers a switch in metabolic programming from fatty acid oxidation to glycolysis.54 We hypothesize that a change in acylcarnitine levels paired with a change in class II HLA gene expression may reflect an activated immune environment promoting a transition in immune cell energy metabolism.
The final metabolite important for predicting MS status in these data was N-methylmaleimide, a transient receptor potential ankyrin 1 (TRPA1) channel agonist. TRPA1 is a nonselective membrane cation channel predominantly expressed in nociceptive sensory neurons but also astrocytes and other non-neuronal cells.55-57 TRPA1 is activated by numerous endogenous and exogenous agonists and participates in hyperalgesia and neurogenic inflammation.56-58 In mice with cuprizone-induced demyelination (a murine MS model that circumvents an autoimmune response), TRPA1 deficiency significantly reduced non-immune mediated demyelination by reducing apoptosis of mature oligodendrocyte.59, 60 As for N-methylmaleimide, little else is known about this electron deficient thiol-blocking agent. We did, however, associate this metabolite with an enrichment for pathways involved in mitochondrial functioning (mitochondrial dysfunction, oxidative phosphorylation, sirtuin signaling), cholesterol/steroid synthesis (mevalonate pathway 1), and apoptosis (Table 2, Supplementary Tables 6 and 7).
There are several strengths to this exploratory investigation of altered metabolic profiles in MS. First, we combined untargeted and targeted metabolomics approaches to sera from a homogeneous cohort of non-Hispanic white, non-smoking males with and without MS, who were frequency matched for age and BMI. Second, we focused on cases who were drug naïve for at least 3 months; thus, we are confident the metabolic signal is not modulated by drug activity. Third, we applied robust non-parametric methods to discern metabolites important for MS status, then used parametric models to determine metabolites most predictive of MS status. And lastly, we integrated multiomic data (transcriptomic and genetic) to explore possible patho-etiologic relationships contributing to variation in the metabolites of interest.
There are a few limitations we must acknowledge. First is the absence of a replication data set. Second, the use of prevalent and not incident MS cases was not ideal; as a result, the identified metabolites may not reflect processes conferring risk but ongoing pathophysiological processes. For example, we identified two phosphatidylcholines which are common components of myelin. Third, our cohort was relatively homogenous for sex, disease type, smoking status, BMI (most are overweight/obese), and disease duration (Supplementary Table 1) – while this was a strength in reducing confounding, it precluded us from investigating associations for these traits in this sample.
In conclusion, we applied targeted and untargeted discovery metabolomics approaches to identify 6 metabolites with elevated levels in MS cases compared with controls. Two of these MS-predictive metabolites have prior evidence for a role in MS, and there is a strong biological rationale for the remaining four peripheral blood metabolites being relevant to MS pathology.
Our integrated analyses suggested that perturbations in mitochondrial functioning, apoptosis, and energy metabolism contributed to the altered MS metabolic profiles. This study has generated a wealth of information that may lead to the development of new metabolic targets for improved MS diagnosis and treatment. Further research is warranted, particularly larger studies including diverse patient populations with considerations for sex, race and MS subtype. It would also be of great interest to investigate the relationships between the metabolites and MS progression (i.e. laurate was associated with disease severity in a murine MS model). And lastly, it would be important to determine if the metabolomics associations are specific to MS or represent inflammatory or neurodegenerative process that may be shared with other autoimmune and neurological disorders.
Supplementary Material
Highlights.
Six metabolites were predictive for MS in males
Results support lipidomic changes in MS
Several MS risk variants were associated with the top predictive metabolites
Pathways analyses suggest processes involved in MS pathology
Acknowledgments
We are grateful to MURDOCK Study participants, leadership, and staff; the MS Society of the Greater Carolinas; and the clinicians and staff of Duke Neurology, Atrium Health Neurosciences Institute - Charlotte, NorthEast Neurology - Concord, Neurology Associates - Greenville, and Raleigh Neurology Associates for providing samples and support. We thank Huiyuan Chen of the DHMRI Analytical Sciences Laboratory and Stephen Siecinski, Elizabeth Burns, and Stephanie Arvai of Duke Molecular Physiology Institute for their contributions. We also thank Xiang Zhang and Xiaoli Wei of the Center for Regulatory and Environmental Analytical Metabolomics (CREAM), University of Louisville for MetPP software. Finally, we are deeply grateful to Herman Stone and family, Eric and Erika Braun, and Jason Cox for their exceedingly generous support of our research.
Funding
This work was supported by gifts from Herman Stone, Eric and Erika Braun, Braun Fundraiser donors, and the Cox Family Foundation. The MURDOCK Study is supported by a gift to Duke University from the David H. Murdock Institute for Business and Culture and Duke University's CTSA grant (UL1TR001117) from the National Institutes of Health’s National Center for Advancing Translational Sciences.
Abbreviations
- biomarkers
biological markers
- GCxGC-TOFMS
two-dimensional gas chromatography and time-of-flight mass spectrometry
- HLA
human leukocyte antigen
- LD
linkage disequilibrium
- MURDOCK
Duke University Measurement to Understand the Reclassification of Disease of Cabarrus/Kannapolis
- QC
quality control
- RR-MS
relapsing-remitting MS
- SNP
single-nucleotide polymorphism
- SP-MS
secondary-progressive MS
Footnotes
Declarations of interest: none
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Comi G Induction vs. escalating therapy in multiple sclerosis: practical implications. Neurol Sci. 2008; 29 Suppl 2: S253–5. [DOI] [PubMed] [Google Scholar]
- 2.Housley WJ, Pitt D and Hafler DA. Biomarkers in multiple sclerosis. Clin Immunol. 2015. [DOI] [PubMed] [Google Scholar]
- 3.Comabella M and Montalban X. Body fluid biomarkers in multiple sclerosis. The Lancet Neurology. 2014; 13: 113–26. [DOI] [PubMed] [Google Scholar]
- 4.Wright BL, Lai JT and Sinclair AJ. Cerebrospinal fluid and lumbar puncture: a practical review. Journal of neurology. 2012; 259: 1530–45. [DOI] [PubMed] [Google Scholar]
- 5.Novakova L, Zetterberg H, Sundstrom P, et al. Monitoring disease activity in multiple sclerosis using serum neurofilament light protein. Neurology. 2017; 89: 2230–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sorensen PS, Jensen PE, Haghikia A, et al. Occurrence of antibodies against natalizumab in relapsing multiple sclerosis patients treated with natalizumab. Multiple sclerosis. 2011; 17: 1074–8. [DOI] [PubMed] [Google Scholar]
- 7.Bertolotto A, Gilli F, Sala A, et al. Persistent neutralizing antibodies abolish the interferon beta bioavailability in MS patients. Neurology. 2003; 60: 634–9. [DOI] [PubMed] [Google Scholar]
- 8.Rebholz CM, Yu B, Zheng Z, et al. Serum metabolomic profile of incident diabetes. Diabetologia. 2018; 61: 1046–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paynter NP, Balasubramanian R, Giulianini F, et al. Metabolic Predictors of Incident Coronary Heart Disease in Women. Circulation. 2018; 137: 841–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lutz NW, Viola A, Malikova I, et al. Inflammatory multiple-sclerosis plaques generate characteristic metabolic profiles in cerebrospinal fluid. PloS one. 2007; 2: e595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mehrpour M, Kyani A, Tafazzoli M, Fathi F and Joghataie MT. A metabonomics investigation of multiple sclerosis by nuclear magnetic resonance. Magn Reson Chem. 2013; 51: 102–9. [DOI] [PubMed] [Google Scholar]
- 12.Moussallieh FM, Elbayed K, Chanson JB, et al. Serum analysis by 1H nuclear magnetic resonance spectroscopy: a new tool for distinguishing neuromyelitis optica from multiple sclerosis. Multiple sclerosis. 2014; 20: 558–65. [DOI] [PubMed] [Google Scholar]
- 13.Pieragostino D, D'Alessandro M, di Ioia M, et al. An integrated metabolomics approach for the research of new cerebrospinal fluid biomarkers of multiple sclerosis. Molecular bioSystems. 2015; 11: 1563–72. [DOI] [PubMed] [Google Scholar]
- 14.Reinke SN, Broadhurst DL, Sykes BD, et al. Metabolomic profiling in multiple sclerosis: insights into biomarkers and pathogenesis. Mult Scler. 2014; 20: 1396–400. [DOI] [PubMed] [Google Scholar]
- 15.Tavazzi B, Batocchi AP, Amorini AM, et al. Serum metabolic profile in multiple sclerosis patients. Mult Scler Int. 2011; 2011: 167156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim HH, Jeong IH, Hyun JS, Kong BS, Kim HJ and Park SJ. Metabolomic profiling of CSF in multiple sclerosis and neuromyelitis optica spectrum disorder by nuclear magnetic resonance. PloS one. 2017; 12: e0181758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poddighe S, Murgia F, Lorefice L, et al. Metabolomic analysis identifies altered metabolic pathways in Multiple Sclerosis. Int J Biochem Cell Biol. 2017; 93: 148–55. [DOI] [PubMed] [Google Scholar]
- 18.Stoessel D, Stellmann JP, Willing A, et al. Metabolomic Profiles for Primary Progressive Multiple Sclerosis Stratification and Disease Course Monitoring. Front Hum Neurosci. 2018; 12: 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhargava P, Fitzgerald KC, Calabresi PA and Mowry EM. Metabolic alterations in multiple sclerosis and the impact of vitamin D supplementation. JCI Insight. 2017; 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wei X, Shi X, Koo I, et al. MetPP: a computational platform for comprehensive two-dimensional gas chromatography time-of-flight mass spectrometry-based metabolomics. Bioinformatics. 2013; 29: 1786–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Winnike JH, Wei X, Knagge KJ, Colman SD, Gregory SG and Zhang X. Comparison of GC-MS and GCxGC-MS in the Analysis of Human Serum Samples for Biomarker Discovery. J Proteome Res. 2015; 14: 1810–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.International Multiple Sclerosis Genetics Consorti NP, Baranzini Sergio E., Santaniello Adam, Shoostari Parisa, Cotsapas Chris, Wong Garrett, Beecham Ashley H., James Tojo, Replogle Joseph, Vlachos Ioannis, McCabe Cristin, Pers Tune, Brandes Aaron, White Charles, Keenan Brendan, Cimpean Maria, Winn Phoebe, Panteliadis Ioannis-Pavlos, Robbins Allison, Andlauer Till F. M., Zarzycki Onigiusz, Dubois Benedicte, Goris An, Sondergaard Helle Bach, Sellebjerg Finn, Sorensen Per Soelberg, Ullum Henrik, Thoerner Lise Wegner, Saarela Janna, Cournu-Rebeix Isabelle, Damotte Vincent, Fontaine Bertrand, Guillot-Noel Lena, Lathrop Mark, Vukusik Sandra, Berthele Achim, Biberacher Viola, Buck Dorothea, Gasperi Christiane, Graetz Christiane, Grummel Verena, Hemmer Bernhard, Hoshi Muni, Knier Benjamin, Korn Thomas, Lill Christina M, Luessi Felix, Muhlau Mark, Zipp Frauke, Dardiotis Efthimios, Agliardi Cristina, Amoroso Antonio, Barizzone Nadia, Benedetti Maria Donata, Bernardinelli Luisa, Cavalla Paola, Clarelli Ferdinando, Comi Giancarlo, Cusi Daniele, Esposito Federica, Ferre Laura, Galimberti Daniela, Guaschino Clara, Leone Maurizio A., Martinelli Vittorio, Moiola Lucia, Salvetti Marco, Sorosina Melissa, Vecchio Domizia, Zauli Andrea, Santoro Silvia, Zuccala Miriam, Mescheriakova Julia, van Duijn Cornelia, Bos Steffan D., Celius Elisabeth G., Spurkland Anne, Comabella Manuel, Montalban Xavier, Alfredsson Lars, Bomfim Izaura L., Gomez-Cabrero David, Hillert Jan, Jagodic Maja, Linden Magdalena, Piehl Fredrik, Jelcic Ilijas, Martin Roland, Sospedra Mireia, Baker Amie, Ban Maria, Hawkins Clive, Hysi Pirro, Kalra Seema, Karpe Fredrik, Khadake Jyoti, Lachance Genevieve, Molyneux Paul, Neville Matthew, Thorpe John, Bradshaw Elizabeth, Caillier Stacy J., Calabresi Peter, Cree Bruce A. C., Cross Anne, Davis Mary F., de Bakker Paul, Delgado Silvia, Dembele Marieme, Edwards Keith, Fitzgerald Kate, Frohlich Irene Y., Gourraud Pierre-Antoine, Haines Jonathan L., Hakonarson Hakon, Kimbrough Dorlan, Isobe Noriko, Konidari Ioanna, Lathi Ellen, Lee Michelle H., Li Taibo, An David, Zimmer Andrew, Lo Albert, Madireddy Lohith, Manrique Clara P., Mitrovic Mitja, Olah Marta, Patrick Ellis, Pericak-Vance Margaret A., Piccio Laura, Schaefer Cathy, Weiner Howard, Lage Kasper, - ANZgene, - IIBDGC, - WTCCC2, Compston Alastair, Hafler David, Harbo Hanne F., Hauser Stephen L., Stewart Graeme, D'Alfonso Sandra, Hadjigeorgiou Georgios, Taylor Bruce, Barcellos Lisa F., Booth David, Hintzen Rogier, Kockum Ingrid, Martinelli-Boneschi Filippo, McCauley Jacob L., Oksenberg Jorge R., Oturai Annette, Sawcer Stephen, Ivinson Adrian J., Olsson Tomas, De Jager Philip L.. The Multiple Sclerosis Genomic Map: Role of peripheral immune cells and resident microglia in susceptibility. bioRxiv. 2017. [Google Scholar]
- 23.Bhattacharya S, Dunham AA, Cornish MA, et al. The Measurement to Understand Reclassification of Disease of Cabarrus/Kannapolis (MURDOCK) Study Community Registry and Biorepository. American journal of translational research. 2012; 4: 458–70. [PMC free article] [PubMed] [Google Scholar]
- 24.Chong J, Soufan O, Li C, et al. MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018; 46: W486–W94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson WE, Li C and Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007; 8: 118–27. [DOI] [PubMed] [Google Scholar]
- 26.Goldstein BA, Polley EC and Briggs FB. Random forests for genetic association studies. Stat Appl Genet Mol Biol. 2011; 10: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levine ME, Crimmins EM, Weir DR and Cole SW. Contemporaneous Social Environment and the Architecture of Late-Life Gene Expression Profiles. American journal of epidemiology. 2017; 186: 503–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hammer A, Schliep A, Jorg S, et al. Impact of combined sodium chloride and saturated long-chain fatty acid challenge on the differentiation of T helper cells in neuroinflammation. Journal of neuroinflammation. 2017; 14: 184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haghikia A, Jorg S, Duscha A, et al. Dietary Fatty Acids Directly Impact Central Nervous System Autoimmunity via the Small Intestine. Immunity. 2015; 43: 817–29. [DOI] [PubMed] [Google Scholar]
- 30.Pederzolli CD, Sgaravatti AM, Braum CA, et al. 5-Oxoproline reduces non-enzymatic antioxidant defenses in vitro in rat brain. Metab Brain Dis. 2007; 22: 51–65. [DOI] [PubMed] [Google Scholar]
- 31.Pederzolli CD, Mescka CP, Zandona BR, et al. Acute administration of 5-oxoproline induces oxidative damage to lipids and proteins and impairs antioxidant defenses in cerebral cortex and cerebellum of young rats. Metab Brain Dis. 2010; 25: 145–54. [DOI] [PubMed] [Google Scholar]
- 32.Lee WJ, Hawkins RA, Peterson DR and Vina JR. Role of oxoproline in the regulation of neutral amino acid transport across the blood-brain barrier. J Biol Chem. 1996; 271: 19129–33. [DOI] [PubMed] [Google Scholar]
- 33.Fischer MT, Sharma R, Lim JL, et al. NADPH oxidase expression in active multiple sclerosis lesions in relation to oxidative tissue damage and mitochondrial injury. Brain : a journal of neurology. 2012; 135: 886–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stankiewicz JM and Brass SD. Role of iron in neurotoxicity: a cause for concern in the elderly? Curr Opin Clin Nutr Metab Care. 2009; 12: 22–9. [DOI] [PubMed] [Google Scholar]
- 35.Jana A and Pahan K. Sphingolipids in multiple sclerosis. Neuromolecular Med. 2010; 12: 351–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haase S, Haghikia A, Gold R and Linker RA. Dietary fatty acids and susceptibility to multiple sclerosis. Mult Scler. 2018; 24: 12–6. [DOI] [PubMed] [Google Scholar]
- 37.Souwer Y, Groot Kormelink T, Taanman-Kueter EW, et al. Human TH17 cell development requires processing of dendritic cell-derived CXCL8 by neutrophil elastase. J Allergy Clin Immunol. 2018; 141: 2286–9 e5. [DOI] [PubMed] [Google Scholar]
- 38.Liu S, Qiu S, Lu Y, et al. The rs251684 Variant of PLA2G4C Is Associated with Autism Spectrum Disorder in the Northeast Han Chinese Population. Genet Test Mol Biomarkers. 2016; 20: 747–52. [DOI] [PubMed] [Google Scholar]
- 39.Nadalin S and Buretic-Tomljanovic A. An association between PLA2G6 and PLA2G4C gene polymorphisms and schizophrenia risk and illness severity in a Croatian population. Prostaglandins Leukot Essent Fatty Acids. 2017; 121: 57–9. [DOI] [PubMed] [Google Scholar]
- 40.Underwood KW, Song C, Kriz RW, Chang XJ, Knopf JL and Lin LL. A novel calcium-independent phospholipase A2, cPLA2-gamma, that is prenylated and contains homology to cPLA2. J Biol Chem. 1998; 273: 21926–32. [DOI] [PubMed] [Google Scholar]
- 41.Pickard RT, Strifler BA, Kramer RM and Sharp JD. Molecular cloning of two new human paralogs of 85-kDa cytosolic phospholipase A2. J Biol Chem. 1999; 274: 8823–31. [DOI] [PubMed] [Google Scholar]
- 42.Schmidt T, Samaras P, Frejno M, et al. ProteomicsDB. Nucleic Acids Res. 2018; 46: D1271–D81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ishihara K, Kuroda A, Sugihara K, Kanai S, Nabe T and Akiba S. Regulation of macrophage differentiation and polarization by group IVC phospholipase A(2). Biochemical and biophysical research communications. 2011; 416: 325–30. [DOI] [PubMed] [Google Scholar]
- 44.Percher F, Curis C, Peres E, et al. HTLV-1-induced leukotriene B4 secretion by T cells promotes T cell recruitment and virus propagation. Nat Commun. 2017; 8: 15890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fishilevich S, Nudel R, Rappaport N, et al. GeneHancer: genome-wide integration of enhancers and target genes in GeneCards. Database (Oxford). 2017; 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lu H, Li Z, Xue Y, et al. AFF1 is a ubiquitous P-TEFb partner to enable Tat extraction of P-TEFb from 7SK snRNP and formation of SECs for HIV transactivation. Proceedings of the National Academy of Sciences of the United States of America. 2014; 111: E15–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Waterworth DM, Ricketts SL, Song K, et al. Genetic variants influencing circulating lipid levels and risk of coronary artery disease. Arterioscler Thromb Vasc Biol. 2010; 30: 2264–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spracklen CN, Chen P, Kim YJ, et al. Association analyses of East Asian individuals and transancestry analyses with European individuals reveal new loci associated with cholesterol and triglyceride levels. Human molecular genetics. 2017; 26: 1770–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Teslovich TM, Musunuru K, Smith AV, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010; 466: 707–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Willer CJ, Schmidt EM, Sengupta S, et al. Discovery and refinement of loci associated with lipid levels. Nature genetics. 2013; 45: 1274–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Friese MA, Schattling B and Fugger L. Mechanisms of neurodegeneration and axonal dysfunction in multiple sclerosis. Nat Rev Neurol. 2014; 10: 225–38. [DOI] [PubMed] [Google Scholar]
- 52.De Rosa V, Galgani M, Porcellini A, et al. Glycolysis controls the induction of human regulatory T cells by modulating the expression of FOXP3 exon 2 splicing variants. Nature immunology. 2015; 16: 1174–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Verdin E, Hirschey MD, Finley LW and Haigis MC. Sirtuin regulation of mitochondria: energy production, apoptosis, and signaling. Trends Biochem Sci. 2010; 35: 669–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Slack M, Wang T and Wang R. T cell metabolic reprogramming and plasticity. Mol Immunol. 2015; 68: 507–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shigetomi E, Tong X, Kwan KY, Corey DP and Khakh BS. TRPA1 channels regulate astrocyte resting calcium and inhibitory synapse efficacy through GAT-3. Nat Neurosci. 2011; 15: 70–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen J and Hackos DH. TRPA1 as a drug target--promise and challenges. Naunyn Schmiedebergs Arch Pharmacol. 2015; 388: 451–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Paulsen CE, Armache JP, Gao Y, Cheng Y and Julius D. Structure of the TRPA1 ion channel suggests regulatory mechanisms. Nature. 2015; 520: 511–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bautista DM, Jordt SE, Nikai T, et al. TRPA1 mediates the inflammatory actions of environmental irritants and proalgesic agents. Cell. 2006; 124: 1269–82. [DOI] [PubMed] [Google Scholar]
- 59.Saghy E, Sipos E, Acs P, et al. TRPA1 deficiency is protective in cuprizone-induced demyelination-A new target against oligodendrocyte apoptosis. Glia. 2016; 64: 2166–80. [DOI] [PubMed] [Google Scholar]
- 60.Bolcskei K, Kriszta G, Saghy E, et al. Behavioural alterations and morphological changes are attenuated by the lack of TRPA1 receptors in the cuprizone-induced demyelination model in mice. J Neuroimmunol. 2018; 320: 1–10. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.