Abstract
Isocitrate dehydrogenase 1 (IDH1) is important for reductive carboxylation in cancer cells, and IDH1 R132H mutant plays a pathogenic role in cancers including acute myeloid leukemia (AML). However, the regulatory mechanisms modulating IDH1 mutant and/or wild-type (WT) function remain unknown. Here we show that two groups of tyrosine kinases (TKs) enhance the activation of IDH1 mutant and WT through preferential Y42 or Y391 phosphorylation. Mechanistically, Y42 phosphorylation occurs in IDH1 monomers, which promotes dimer formation with enhanced substrate (isocitrate or α-ketoglutarate) binding, while Y42-phosphorylated dimers show attenuated disruption to monomers. Y391 phosphorylation occurs in both monomeric and dimeric IDH1, which enhances cofactor (NADP+ or NADPH) binding. Diverse oncogenic TKs activate Src to achieve Y42 and Y391 phosphorylation of IDH1, respectively, which contributes to reductive carboxylation and tumor growth, while FLT3 or FLT3-ITD mutant activate JAK2 to enhance IDH1 mutant activity through phosphorylation of Y391 and Y42, respectively, in AML cells.
INTRODUCTION
The terms metabolic “reprogramming” and “rewiring” have emerged to describe the increasingly better understood metabolic changes observed in cancer cells (1,2). From a definitional perspective, “metabolic reprogramming” represents “software changes” in cancer cells and describes metabolic alterations that are normally induced by growth factors in proliferating cells but are hijacked by oncogenic signals; while “metabolic rewiring” represents “hardware changes” and describes metabolic alterations due to neo-functions of oncogenic mutants, which are not found in normal cells (3). For example, oncogenic signals reprogram cancer cells in an acute manner involving diverse post-translational modifications of metabolic enzymes that also exist in proliferating normal cells (4). The identification of mutations in isocitrate dehydrogenase (IDH) 1 and 2 in glioma and acute myeloid leukemia (AML) represents a rewiring because the mutations confer a neo-function to IDH1/2 to produce the oncometabolite 2-hydroxyglutamate (2-HG) to regulate cancer epigenetics, which is not found in normal cells harboring wild type (WT) IDH1/2 (5–8). We previously reported that oncogenic BRAF V600E rewires the ketogenic pathway to allow cancer cells to benefit from ketone body acetoacetate-promoted BRAF V600E-MEK1 binding, which is not found in cells expressing BRAF WT (3). Thus, clearly distinguishing and characterizing metabolic “reprogramming” and “rewiring” in cancer cells offers apparent advantages to inform therapy development because targeting rewiring (e.g. IDH mutant inhibitors) in cancer cells will have minimal toxicity to normal cells.
IDH1 and IDH2 are two highly homologous members of the IDH family of metabolic enzymes, and are located in the cytoplasm and mitochondria, respectively. IDH1/2 form homodimers and convert isocitrate to α-ketoglutarate (αKG) with the reduction of NADP+ to NADPH (9). αKG is a key intermediate in the Krebs cycle and glutaminolysis, an important nitrogen transporter, and a ligand for αKG-dependent enzymes including histone demethylases such as Jhd1 and methylcytosine dioxygenase enzyme TET2 (10). NADPH not only fuels macromolecular biosynthesis such as lipogenesis but also functions as a crucial antioxidant to quench the reactive oxygen species (ROS) produced during rapid proliferation of cancer cells, which is important for the maintenance of cellular redox homeostasis to protect against toxicity of ROS and oxidative DNA damage (11). Thus, IDH1/2 are important for many metabolic processes in cells including bioenergetics, biosynthesis, and redox homeostasis. Moreover, recent evidence demonstrates that IDH1/2 play an important role in reductive carboxylation that is enhanced in cells under hypoxia, allowing the generation of isocitrate/citrate from αKG and glutamine, which is in particular important in cancer cells for producing citrate and acetyl-CoA that are essential for lipid synthesis during tumorigenesis, as well as reducing mitochondrial ROS to sustain redox homeostasis during anchorage-independent growth (12,13).
Missense mutations of R132 in the enzyme active site of IDH1 were identified in patients with glioblastoma (GBM) and AML cases (5–7,14,15), and corresponding IDH2 R172 mutations as well as a novel R140Q mutant repeatedly occur in AML patients (14,16,17). Overall, IDH1/2 mutations are identified in >75% of grade 2/3 glioma and secondary GBM cases and >20% of AML cases. IDH mutations were also identified in other cancer types such as chondrosarcoma and cholangiocarcinoma (9). IDH mutations are heterozygous events, resulting in loss-of-function of wild type IDH1 enzyme activity but a gain-of-function to mutant IDH1, allowing NADPH-dependent reduction of αKG to produce the oncometabolite 2-HG. 2-HG competitively inhibits the function of αKG-dependent enzymes such as TET2, which in turn causes epigenetic dysregulation including DNA hypermethylation in both GBM and AML, and consequent block of differentiation (18). In AML, IDH1 R132, IDH2 R172, and IDH2 R140 mutations are mutually exclusive, and although IDH1 R132 mutations were detected in AML patients with mutations in NPM1, Fms-related tyrosine kinase 3 (FLT3), CEBPA, or NRAS, concurrence of IDH1 mutations and FLT3-internal tandem duplication (FLT3-ITD) is less common (19).
The pathogenic role of IDH mutations suggested mutant IDH proteins as promising therapeutic targets. Small-molecule inhibitors, including AG-120, enasidenib (AG-221/CC-90007), AG-881, IDH305, and FT-2102, that selectively target mutant IDH1 or IDH2 have been developed and evaluated in preclinical and clinical studies as single agents and in combination with other anti-cancer agents. These inhibitors were designed to bind to the catalytic site of mutant IDH proteins and inhibit the enzyme activity of mutant IDH to convert αKG to 2-HG (20). In addition, recent reports demonstrate the ability of single agent IDH2 R140 mutant inhibitor AG-221 to reverse the DNA hypermethylation and differentiation block induced by IDH2 mutant in leukemia cells, and improve efficacy when given in combination with a FLT3 tyrosine kinase inhibitor in IDH2 mutant expressing leukemia cells (21,22). It remains unknown how oncogenic signals such as oncogenic tyrosine kinases, including FLT3, regulate IDH1 WT and/or mutant IDH1, what the regulatory mechanism is, and whether WT and mutant IDH1 share the same mechanism in the corresponding related cancers.
RESULTS
An intrinsic link between FLT3 and IDH1 in AML
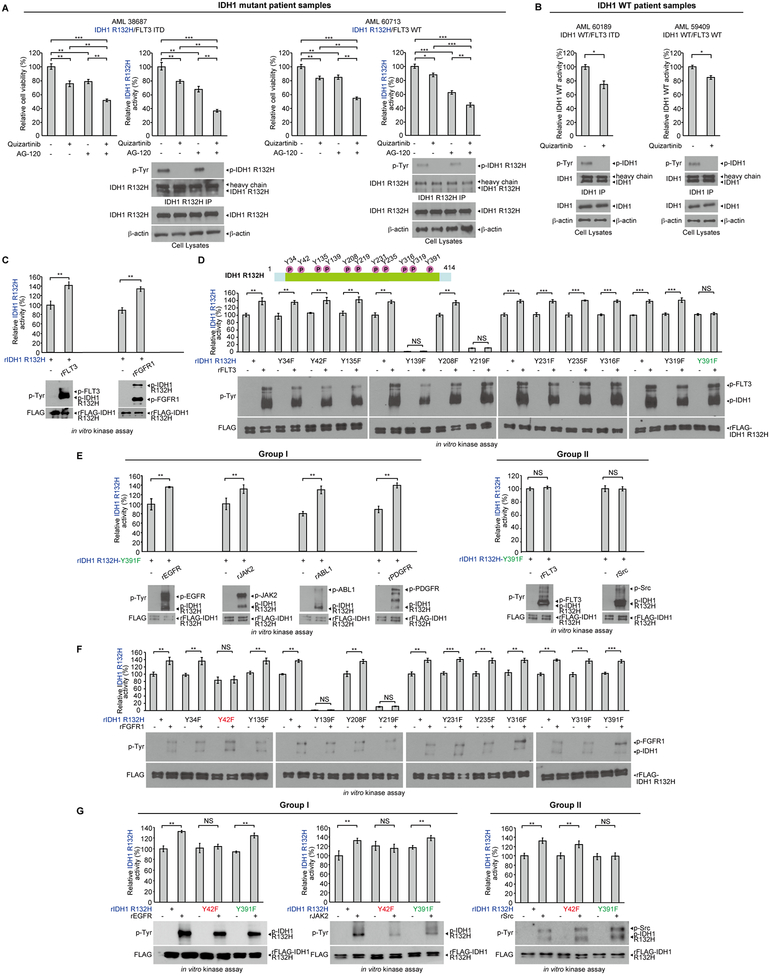
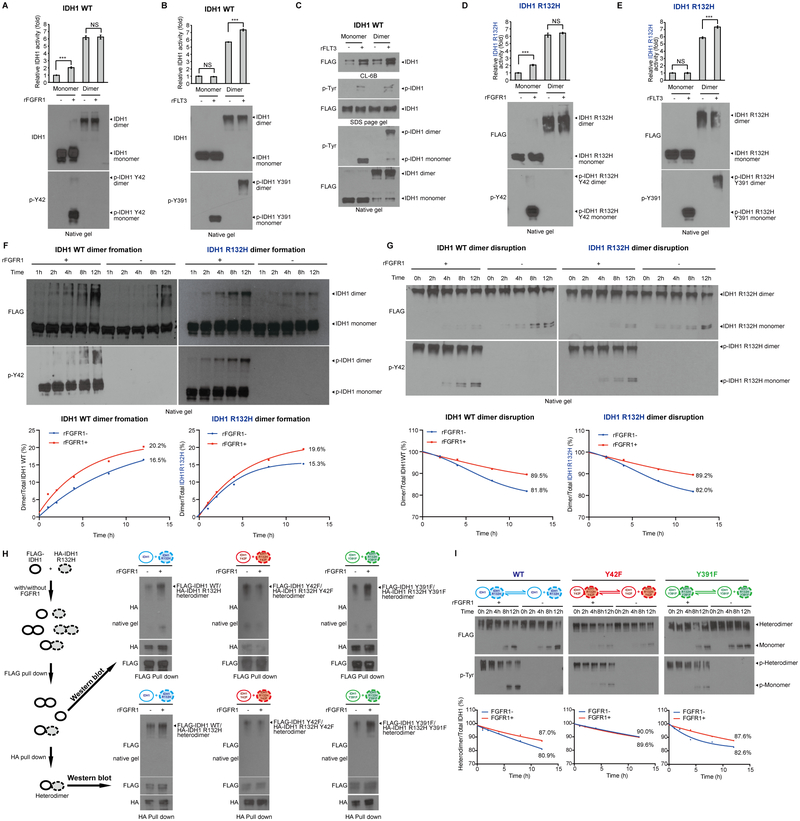
To understand the pathogenic link between mutant FLT3 and IDH1, we treated primary leukemia cells from representative AML patients with the FLT3 inhibitor quizartinib and/or IDH1 R132 mutant inhibitor AG120. We found that single agent treatment for 24 hours resulted in decreased cell viability in IDH1 R132H mutant expressing primary leukemia cells harboring concurrent FLT3-ITD mutation (left left panels, Fig. 1A and S1A), or expressing wild type (WT) FLT3 (left right panels, Fig. 1A and S1A), while combined treatment resulted in further decreased cell viability in these cells. Surprisingly, treatment with either AG120 or quizartinib resulted in decreased mutant IDH1 activity, which was further reduced upon combined treatment in all of the tested primary AML cells, accompanied by decreased tyrosine phosphorylation levels of IDH1 proteins (Fig. 1A and S1A). Similar results were obtained using primary leukemia cells from AML patients expressing IDH1 WT and FLT3-ITD or FLT3 WT, where treatment with quizartinib resulted in decreased IDH1 enzyme activity (Fig. 1B). These data together suggest an intrinsic link between FLT3 and IDH1 regardless of the mutational state of both proteins, likely involving FLT3-dependent tyrosine phosphorylation and activation of IDH1.
Figure 1.
An intrinsic link between FLT3 and IDH1 involves tyrosine phosphorylation in AML and diverse tyrosine kinases phosphorylate and enhance activation of mutant IDH1. A, Human primary AML cells harvested from patients 38687 expressing IDH1 R132H/FLT3-ITD (left panels) and 60713 expressing IDH1 R132H/FLT3 WT (right panels) were treated with FLT3 inhibitor quizartinib (200nM), IDH mutant inhibitor AG120 (1μM), or combination of quizartinib and AG120, followed by cell viability assay (48h) and IDH1 R132H enzyme activity assay (12h) (upper left and right panels). Tyrosine phosphorylation (p-Tyr-100) of IDH1 R132H in each treated group was determined by Western blotting (lower panels). B, Human primary AML cells harvested from patients 60189 expressing IDH1 WT/FLT3-ITD (left panels) and 59409 expressing IDH1 WT/FLT3 WT (right panels) were treated with or without FLT3 inhibitor quizartinib, followed by wild type IDH1 enzyme activity assay (upper panels). Tyrosine phosphorylation of IDH1 WT in each treated group was determined by Western Blotting (lower panels). C, Purified recombinant IDH1 R132H proteins were incubated with recombinant active form of tyrosine kinases including rFLT3 and rFGFR1 in an in vitro kinase assay. IDH1 R132H enzyme activity assay (upper panels) was assessed; tyrosine phosphorylation of IDH1 was detected by Western blotting (lower panels). D, Upper panel: Schematic representation of identified phosphorylated tyrosine residues in IDH1 R132H. Lower panels: Purified FLAG-IDH1 variants were incubated with recombinant active form of FLT3, followed by IDH1 R132H enzyme activity assay (lower upper panels) and Western blotting showing tyrosine phosphorylation of IDH1 R132H and IDH1 R132H YF mutants (lower lower panels). E, Purified FLAG-IDH1 R132H-Y391F mutant was incubated with recombinant active form of Group I tyrosine kinases including rEGFR, rJAK2, rABL1 and rPDGFR (left panels) and Group II tyrosine kinases including rFLT3 and rSrc (right panels), respectively, followed by IDH1 R132H enzyme activity assay (upper panels). Tyrosine phosphorylation of IDH1 R132H in each treated group was determined by Western blotting (lower panels). F, Purified FLAG-IDH1 R132H and FLAG-IDH1 R132H YF mutants were incubated with or without active rFGFR1, followed by IDH1 R132H enzyme activity assay (upper panels). Tyrosine phosphorylation of IDH1 R132H and IDH1 R132H YF mutants was assessed by Western blotting (lower panels). G. Purified FLAG-IDH1 R132H-Y42F and FLAG-IDH1 R132H-Y391F mutants were incubated with or without active rEGFR, rJAK2, or rSrc, followed by IDH1 R132H enzyme activity assay (upper panels). Tyrosine phosphorylation of IDH1 R132H and IDH1 R132H YF mutants was assessed by Western blotting (lower panels).
The error bars represent mean values ±SD from three replicates of each sample (*: 0.01<p<0.05; **: 0.01<p<0.001; ***: p<0.001; ns: not significant); Data are mean ±SD; p values were obtained by a two-tailed Student’s test.
Distinct groups of tyrosine kinases enhance activation of WT and mutant IDH1 through preferential phosphorylation of Y42 or Y391
We next performed in vitro kinase assays coupled with mutant IDH1 activity assays using purified, recombinant IDH1 R132H mutant protein (rIDH1 R132H) incubated with recombinant active forms of diverse tyrosine kinases including rFLT3, rFGFR1, rSrc, rPDGFR, rEGFR, rMET, rKit, or rJAK2. We found that purified rIDH1 R132H mutant protein demonstrated basal level enzyme activity, while all of the tested tyrosine kinases were able to directly phosphorylate purified rIDH1 R132H mutant, leading to increased mutant IDH1 activity (Fig. 1C and S1B). We next performed mutational analysis and generated diverse phospho-deficient Y➔F mutants of IDH1 R132H (top, Fig. 1D) based on publicly available data (https://www.phosphosite.org/proteinAction.action?id=10630&showAllSites=true), which identified phosphorylated tyrosine residues of IDH1 in human cancer cells. We found that mutations at Y139 and Y219 intrinsically abolished mutant IDH1 activity, whereas substitution of Y391 with phenylalanine resulted in the abolishment of FLT3-enhanced activation of IDH1 R132H mutant (Fig. 1D).
However, we found that a group of tyrosine kinases (which we termed Group I) including EGFR, JAK2, ABL1, PDGFR, and FGFR1 were still able to phosphorylate and enhance activation of IDH1 R132H mutant with Y391F mutation (Fig. 1E, left and 1F), while FLT3 or Src (termed Group II)-enhanced activation of IDH1 R132H/Y391F mutant was abolished (Fig. 1E, right). Only the Y42F mutation abolished FGFR1-enhanced activation of mutant IDH1 in the coupled kinase and mutant IDH1 activity assays (Fig. 1F). Similar results were obtained using purified rIDH1 WT proteins incubated with rFLT3 (Group II) or rFGFR1 (Group I), where Y391F or Y42F mutations exclusively abolished FLT3 or FGFR1-enhanced activation of IDH1 WT, respectively (Supplementary Fig. S1C). Further studies confirmed that Group I tyrosine kinases including EGFR and JAK2 (Fig. 1G, left) as well as ABL1, FGFR3, c-MET, and PDGFR (Supplementary Fig. S2A) enhanced activation of IDH1 R132H mutant by phosphorylating Y42, whereas Group II tyrosine kinase Src enhanced activation of mutant IDH1 through Y391 phosphorylation (Fig. 1G, right). Similar results were obtained using IDH1 WT proteins incubated with diverse tyrosine kinases (Supplementary Fig. S2B–S2C). These data demonstrate that two groups of tyrosine kinases enhance activation of IDH1 WT or R132H mutant in the same manner through preferential phosphorylation of Y42 or Y391 of IDH1 despite mutational status, suggesting that WT and mutant IDH1 might share the same regulatory mechanisms involving tyrosine phosphorylation.
Y42 phosphorylation promotes IDH1 dimerization and subsequent substrate binding
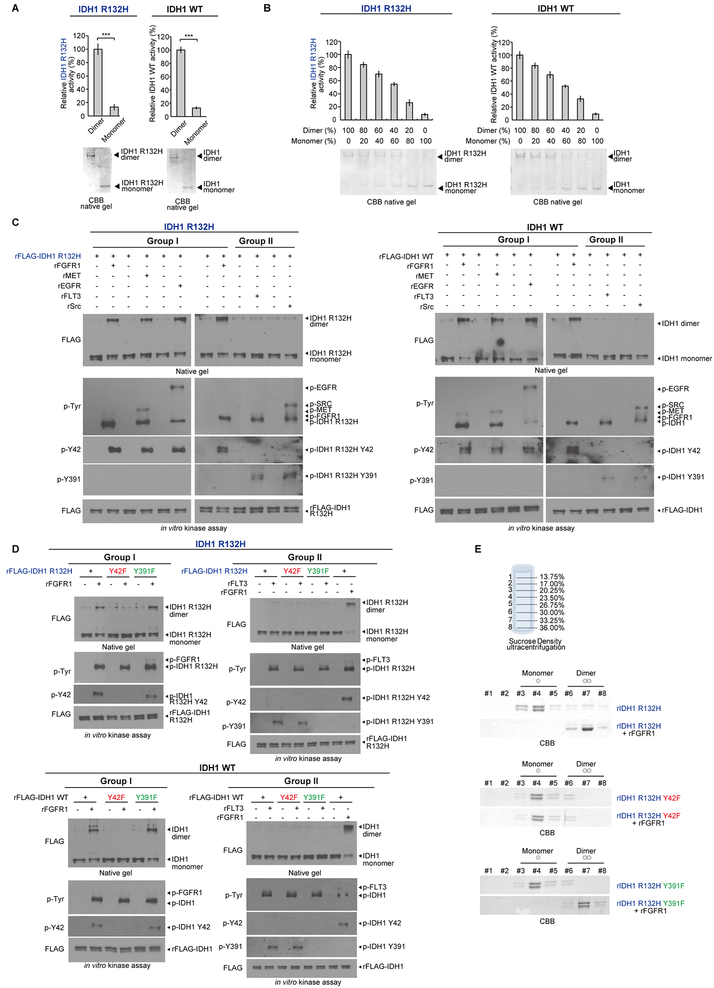
We next sought to elucidate the distinct mechanisms underlying Y42 and Y391 phosphorylation-dependent enhancement of IDH1 activation. Further studies confirmed that purified dimeric and monomeric IDH1 R132H mutant or WT proteins represented active and inactive forms of IDH1, respectively (Fig. 2A, left and right, respectively), and IDH1 R132H mutant or WT enzyme activity correlated with increasing levels of dimer in different compositions of purified monomeric and dimeric IDH1 proteins (Fig. 2B, left and right, respectively). In addition, incubation with the Group I tyrosine kinases rFGFR1, rMET, or rEGFR but not Group II kinases rFLT3 or rSrc resulted in increased dimer formation of IDH1 R132H mutant or WT proteins assessed by Western blot after crosslinking (Fig. 2C, left and right, respectively), suggesting that Y42 phosphorylation by Group I tyrosine kinases may contribute to IDH1 dimer formation. This was further confirmed by incubation of diverse Group I or II kinases with IDH1 R132H mutant proteins with or without Y42F or Y391F mutation, where FGFR1 (Group I) but not FLT3 (Group II) (Fig. 2D, upper), as well as EGFR, and MET (Group I) but not Src (Group II) (Supplementary Fig. S3A) were able to promote dimer formation of IDH1 R132H mutant and R132H/Y391F but not R132H/Y42F proteins through tyrosine phosphorylation. Similar results were obtained using IDH1 WT, Y42F, and Y391F proteins incubated with diverse Group I and II tyrosine kinases (Fig. 2D, bottom and Supplementary Fig. S3B).
Figure 2.
Y42 phosphorylation of IDH1 by Group I tyrosine kinases promotes protein dimerization. A, Purified dimers and monomers of FLAG-IDH1 R132H mutant or FLAG-IDH1 wild type (IDH1 WT) by sucrose density ultracentrifugation were applied to IDH1 R132H or WT enzyme activity assays, respectively (upper panels). Dimeric and monomeric forms of IDH1 were applied to native PAGE and detected by Coomassie brilliant blue (CBB) staining (lower panels). B, FLAG-IDH1 R132H mutant (left panels) and FLAG-IDH1 WT (right panels) protein samples containing different compositions of purified dimeric and monomeric IDH1 by sucrose density ultracentrifugation were applied to IDH1 R132H or WT enzyme activity assays, respectively (upper panels). Dimeric and monomeric forms of IDH1 were applied to native PAGE and detected by Coomassie brilliant blue (CBB) staining (lower panels). C, Purified FLAG-IDH1 R132H mutant (left panels) and FLAG-IDH1 WT (right panels) proteins were incubated with recombinant active form of Group I tyrosine kinases including rFGFR1, rMET and rEGFR and Group II tyrosine kinases including rFLT3 and rSrc, respectively. Dimeric and monomeric IDH1 proteins as well as tyrosine phosphorylation of IDH1 in each treated group were determined by Western blotting. D, Purified FLAG-IDH1 R132H variants (upper panels) and FLAG-IDH1 variants (lower panels) were incubated with recombinant active form of Group I tyrosine kinase rFGFR1 or Group II tyrosine kinase rFLT3 in an in vitro kinase assay. Dimeric and monomeric IDH1 proteins as well as tyrosine phosphorylation of IDH1 proteins in each treated group were determined by Western blotting (left upper and lower and right upper and lower, respectively). In Group II kinases treatment, purified IDH1 and IDH1 R132H proteins incubated with or without Group I kinase FGFR1 were used as positive controls. E, IDH1 R132H (top), IDH1 R132H-Y42F (middle) and IDH1 R132H-Y391F (bottom) proteins were incubated with active rFGFR1 prior to sucrose density ultracentrifugation. Collected fractions were applied to PAGE, followed by CBB staining.
The error bars represent mean values ±SD from three replicates of each sample (***: p<0.001); Data are mean ± SD; p values were obtained by a two-tailed Student’s test.
We purified monomeric and dimeric IDH1 proteins using a sucrose density ultracentrifugation approach (Fig. 2E, top). We found that the majority of recombinant IDH1 R132H proteins were monomers, while phosphorylation by FGFR1 (Group I) resulted in a shift to dimer formation (Fig.2E, middle). This shift was abolished when using rIDH1 R132H/Y42F mutant but retained using rIDH1 R132H/Y391F mutant (Fig. 2E, bottom two panels). Similar results were obtained using rIDH1 WT, Y42F, and Y391F proteins incubated with FGFR1 (Supplementary Fig. S3C).
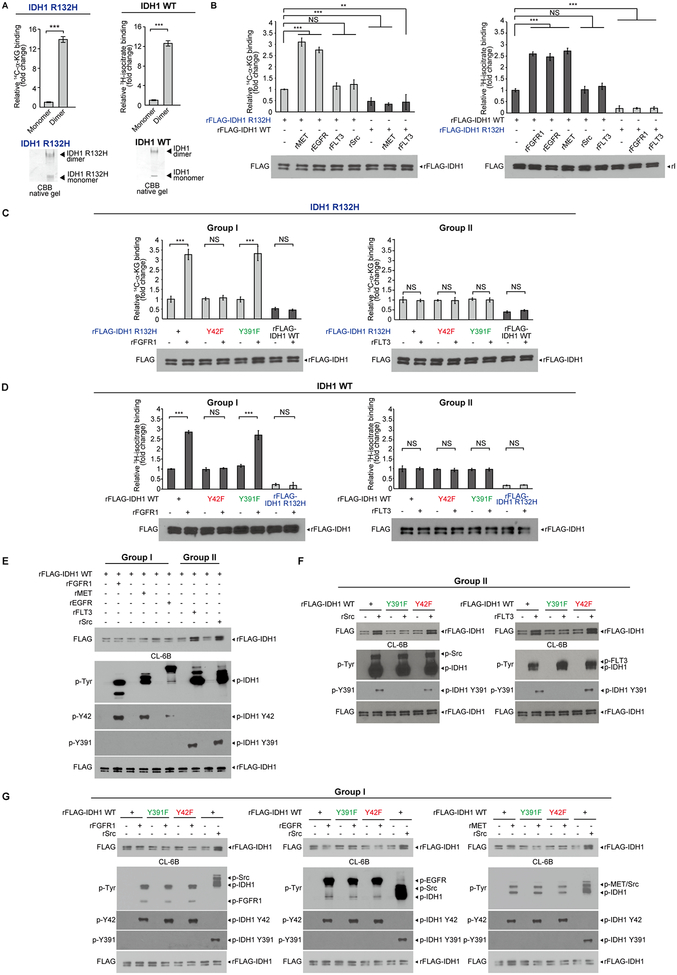
We next determined the underlying mechanism by which Y42 phosphorylation-enhanced dimer formation contributes to IDH1 activation. We separated monomeric and dimeric IDH1 R132H mutant or WT proteins on native gels (Fig. 3A, bottom panels). The gels loaded with IDH1 R132H mutant or WT proteins were incubated with 14C-labeled αKG or 3H-labeled isocitrate, respectively. The binding ability of 14C-αKG or 3H-isocitrate to monomeric/dimeric IDH1 R132H mutant or IDH1 WT proteins, respectively, was assessed by scintillation counting of excised gel bands containing radio-labeled IDH1 monomers or dimers (Fig. 3A). The results of this “overlay” assay suggested that αKG or isocitrate as substrates demonstrated higher binding ability to dimeric IDH1 R132H mutant or IDH1 WT proteins, respectively, compared to corresponding monomers of IDH1 proteins (Fig. 3A, left and right, respectively), suggesting IDH1 dimer formation promotes substrate binding.
Figure 3.
Mutant and WT IDH1 dimer formation contributes to the binding ability to its substrates αKG and isocitrate, respectively. A, Dimeric and monomeric IDH1 R132H or IDH1 WT proteins were separated on native gel and detected by CBB staining (lower panels). Bands containing dimeric or monomeric IDH1 R132H proteins were excised and applied to labeled αKG (14C-αKG) binding assay. The amount of mutant IDH1 protein-bound 14C-αKG were assessed by scintillation counting followed by quantitative analysis (upper left panel). Bands containing dimeric or monomeric IDH1 WT proteins were excised and applied to labeled isocitrate (3H-isocitrate) binding assay. The amount of IDH1 WT-bound 14H-isocitrate were assessed by scintillation counting followed by quantitative analysis (upper right panel). B, Left panels: purified IDH1 R132H proteins were incubated with or without recombinant active form of Group I kinases rMET and rEGFR or Group II kinases rFLT3 and rSrc, followed by 14C-αKG binding assay (Purified IDH1WT proteins incubated with or without Group I kinase rMET or Group II kinase rFLT3 were used as negative controls). Right panels: purified IDH1 WT proteins were incubated with or without purified recombinant active Group I kinases rFGFR1, rEGFR and rMET or Group II kinases Src and FLT3, followed by isocitrate (3H-isocitrate) binding assay. Purified IDH1 R132H proteins incubated with or without Group I kinase rFGFR1 or Group II kinase rFLT3 were used as negative controls. C, Purified FLAG-IDH1 R132H variants were incubated with recombinant active form of Group I tyrosine kinase rFGFR1 and Group II tyrosine kinase rFLT3, respectively, followed by labeled αKG (14C-αKG) binding assays (left and right panels, respectively). Purified IDH1 WT protein was used as a negative control. D, Purified FLAG-IDH1 variants were incubated with recombinant active form of Group I tyrosine kinase rFGFR1 and Group II tyrosine kinase rFLT3, respectively, followed by 3H-isocitrate binding assays (left and right panels, respectively). Purified IDH1 R132H protein was used as a negative control. E, Purified FLAG-IDH1 WT proteins were incubated with recombinant active Group I tyrosine kinases including rFGFR1, rMET and rEGFR and Group II tyrosine kinases including rFLT3 and rSrc, respectively, followed by incubation with Blue Sepharose CL-6B beads that mimic NADP+ binding. Bound IDH1 proteins and tyrosine phosphorylation of IDH1 were determined by Western blotting. F-G, Purified FLAG-IDH1 WT, FLAG-IDH1 Y391F and FLAG-IDH1 Y42F mutant proteins were incubated with recombinant active form of Group II tyrosine kinases including rSrc and rFLT3 (F) and Group I tyrosine kinases rFGFR1, rEGFR and rMET (G), followed by incubation with CL-6B beads that mimic NADP+ binding. Bound IDH1 proteins and tyrosine phosphorylation of IDH1 were determined by Western blotting. Phosphorylation of FLAG IDH1 WT by rSrc was performed in (G) as a positive control.
The error bars represent mean values ±SD from three replicates of each sample (**: 0.01<p<0.001; ***: p<0.001; ns: not significant); Data are mean ± SD; p values were obtained by a two-tailed Student’s test.
Moreover, incubation with Group I kinases including rMET and rEGFR but not Group II kinases including rFLT3 and rSrc promoted 14C-αKG binding to IDH1 R132H mutant, while control IDH1 WT showed minimal binding ability to 14C-αKG, which was not altered when phosphorylated by Group I rMET or Group II rFLT3 (Fig. 3B, left). In contrast, incubation with Group I kinases including rFGFR1, rEGFR, rMET and rPDGFR but not Group II kinases including rFLT3 and rSrc promoted 3H-isocitrate binding to IDH1 WT, while control IDH1 R132H mutant showed minimal binding ability to 3H-isocitrate, which was not altered when phosphorylated by Group I rFGFR1 or Group II rFLT3 (Fig. 3B, right).
This was further confirmed by incubation of diverse Group I or II kinases with IDH1 R132H mutant proteins with or without Y42F or Y391F mutation, where FGFR1 (Fig. 3C, left), EGFR, MET and PDGFR (Supplementary Fig. S4A, upper panels) (Group I) but not FLT3 (Fig. 3C, right) or Src (Supplementary Fig. S4A, lower) (Group II) were able to promote 14C-αKG binding to IDH1 R132H mutant and R132H/Y391F but not R132H/Y42F proteins through tyrosine phosphorylation, while IDH1 WT as a negative control showed minimal binding ability to 14C-αKG, which was not altered when phosphorylated by Group I or II tyrosine kinases (Fig. 3C and Supplementary Fig. S4A). Similar results were obtained using IDH1 WT, Y42F, and Y391F proteins incubated with diverse Group I and II tyrosine kinases in the presence of 3H-isocitate (Fig. 3D and Supplementary Fig. S4B).
Y391 phosphorylation enhances cofactor NADP+ binding to IDH1
In multiple structures of IDH1, the location of Y391 is proximal to the NADP+ binding site (approximately 10Å); we thus hypothesized that Y391 phosphorylation may enhance IDH1 activation by impacting cofactor NADP+ binding. To test this hypothesis, we performed a NADP+ binding experiment using Blue Sepharose® CL-6B (Sigma-Aldrich). CL-6B mimics NADP+ and is a pseudo-affinity ligand of many dehydrogenases using NADP+ as a substrate. We found that incubation of Group II kinases including rFLT3 or rSrc but not Group I tyrosine kinases including rFGFR1, rMET or rEGFR resulted in a significant increase in the amount of IDH1 protein bound to the CL-6B agarose beads, indicating increased binding between IDH1 and NADP+ (Fig. 3E), suggesting the Y391 phosphorylation by Group II tyrosine kinases may contribute to cofactor NADP+ binding to IDH1. This was further confirmed by incubation of diverse Group II or I kinases with IDH1 WT, Y42F, or Y391F mutant proteins, where Src and FLT3 (Group II; Fig. 3F) but not FGFR1, EGFR, or MET (Group I; Fig. 3G) were able to promote binding of IDH1 WT and Y42F mutant but not Y391F mutant proteins to CL-6B agarose beads.
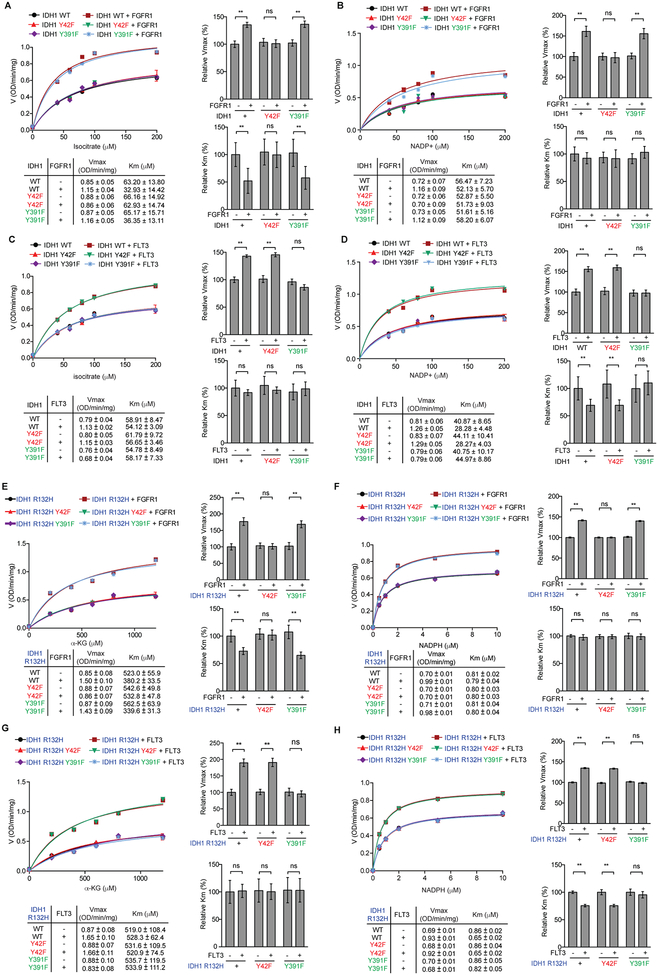
Further kinetics studies confirmed that, in the presence of increasing concentrations of isocitrate as a substrate, phosphorylation of IDH1 WT and Y391F mutant but not Y42F mutant by FGFR1 (Group I) resulted in increased Vmax values (Fig. 4A, top two panels) with decreased Km values for isocitrate (Fig. 4A, bottom two panels), suggesting that Y42 phosphorylation by Group I kinases enhances IDH1 WT activation by promoting substrate binding. In contrast, in the presence of increasing concentrations of NADP+ as a cofactor, although phosphorylation of IDH1 WT and Y391F mutant but not Y42F mutant by FGFR1 resulted in increased Vmax values (Fig. 4B, top two panels), Km values for NADP+ as a cofactor was not altered among IDH1 variants (Fig. 4B, bottom two panels), suggesting that Y42 phosphorylation does not affect cofactor binding. Furthermore, in the presence of increasing concentrations of isocitrate as a substrate, despite the increased Vmax values due to phosphorylation of IDH1 WT and Y42F mutant but not Y391F mutant by FLT3 (Group II) (Fig. 4C, top two panels), the Km values for isocitrate remained unaltered (Fig. 4C, bottom two panels), whereas in the presence of increasing concentrations of NADP+ as a cofactor, phosphorylation of IDH1 WT and Y42F mutant but not Y391F mutant by FLT3 resulted in increased Vmax values (Fig. 4D, top two panels) with decreased Km values for NADP+ as a cofactor (Fig. 4D, bottom two panels). These data suggest that Y391 phosphorylation by Group II kinases enhances IDH1 WT activation by promoting cofactor but not substrate binding.
Figure 4.
Y42 phosphorylation promotes substrate binding while Y391 phosphorylation promotes cofactor binding to enhance IDH1 WT and IDH1 R132H activation. A-D, Vmax and Km of IDH1 WT were measured using purified FLAG-IDH1 and variants incubated with recombinant active form of Group I tyrosine kinase rFGFR1 (A and B) or Group II tyrosine kinase rFLT3 (C and D) in the presence of increasing concentrations of isocitrate (A and C) or NADP+ (B and D), respectively, followed by IDH1 WT enzyme activity assay. Vmax and Km values of each treated group were calculated (lower left) and plotted (upper left). Right 2 panels represent the relative value of Vmax and Km in each treatment. E-H, Vmax and Km of IDH1 R132H were measured using purified FLAG-IDH1 R132H and variants incubated with recombinant active form of Group I tyrosine kinase rFGFR1 (E and F) or Group II tyrosine kinase rFLT3 (G and H) in the presence of increasing concentrations of αKG (E and G) or NADPH (F and H), respectively, followed by IDH1 R132H enzyme activity assay. Vmax and Km values of each treated group were calculated (lower left) and plotted (upper left). Right 2 panels represent the relative value of Vmax and Km in each treatment.
The error bars represent mean values ±SD from three replicates of each sample (**: 0.01<p<0.001; ns: not significant); Data are mean ± SD; p values were obtained by a two-tailed Student’s test.
Similar results were obtained using IDH1 R132H mutant proteins phosphorylated by FGFR1 or FLT3 in the presence of increasing concentrations of αKG or NADPH (Fig. 4E–4H). These results together also demonstrate that the Y42F and Y391F mutations do not affect intrinsic enzyme properties of IDH1 WT or IDH1 R132H mutant.
Y42 phosphorylation occurs in IDH1 monomers, promoting dimer formation but attenuates dimer disruption, while Y391 phosphorylation occurs in both monomeric and dimeric IDH1, enhancing cofactor binding.
In order to confirm whether TKs enhance IDH1 activation predominantly through tyrosine phosphorylation, we performed an in vitro kinase assay followed by protein Tyr phosphatase (PTP) treatment. We found that rIDH1 WT but not Y42F phosphorylated by FGFR1 (Group I) showed enhanced enzyme activity, which was abolished by PTP treatment (Supplementary Fig. S5A, left). In addition, rIDH1 WT but not Y391F phosphorylated by FLT3 (Group II) also showed enhanced activation, which was eliminated by PTP (Supplementary Fig. S5A, right). Consistent with these findings, treatment of IDH1 immunoprecipitates from MOLM-14 cells with PTP resulted in decreased IDH1 enzyme activity with reduced tyrosine phosphorylation (Supplementary Fig. S5B, upper left), and PTP treatment of MOLM-14 cell lysates resulted in a decrease in the amount of dimeric IDH1 proteins (Supplementary Fig. S5B, lower left). Similar results were obtained using IDH1 immunoprecipitates from MOLM-14 cells treated with FLT3 inhibitor TKI258 (Supplementary Fig. S5B, right).
We sought to determine whether Y42 and Y391 can be phosphorylated in monomeric and/or dimeric IDH1. Interestingly, we found that Y42 phosphorylation by FGFR1 (Group I) only occurred in purified monomeric IDH1 WT protein, leading to increased dimer formation from IDH1 monomers accompanied with enhanced enzyme activity, whereas purified dimeric IDH1 WT protein treated with FGFR1 was not phosphorylated at Y42 with unaltered dimer amount and enzyme activity (Fig. 5A). In contrast, Y391 phosphorylation by FLT3 (Group II) occurred in both purified monomeric and dimeric IDH1 WT proteins (Fig. 5B). However, Y391 phosphorylation of IDH1 monomers did not promote dimer formation or enhance enzyme activity, whereas Y391 phosphorylation of IDH1 dimers enhanced enzyme activity despite unaltered dimer amount, which is likely due to increased cofactor NADP+ binding. Indeed, incubation of rFLT3 resulted in increased Y391 phosphorylation of both monomeric and dimeric IDH1 WT proteins with a significant increase in the amount of both monomeric and dimeric IDH1 proteins bound to the CL-6B agarose beads, indicating increased binding between Y391-phosphorylated monomeric and dimeric IDH1 and NADP+ (Fig. 5C). Similar results were obtained using purified monomeric and dimeric IDH1 R132H proteins incubated with rFGFR1 or rFLT3 (Fig. 5D–5E).
Figure 5.
Activation of IDH1 is enhanced by distinct oncogenic tyrosine kinase cascades through direct and indirect phosphorylation. A-B, Purified IDH1 WT monomer and dimer were treated with recombinant active Group I tyrosine kinase FGFR1 (A) or Group II tyrosine kinase FLT3 (B), respectively, followed by IDH1 WT enzyme activity (upper panels). Monomeric and dimeric IDH1 protein levels and tyrosine phosphorylation at Y42 and Y391 of IDH1 were detected by Western blotting using native PAGE (lower panels). C, Purified IDH1 WT monomer and dimer were treated with or without recombinant active Group II tyrosine kinase FLT3, followed by incubation with Blue Sepharose CL-6B beads that mimic NADP+ binding. Bound monomeric and dimeric IDH1 proteins and tyrosine phosphorylation of IDH1 were determined by Western blotting using native PAGE. D-E, Purified IDH1 R132H monomer and dimer were treated with recombinant active Group I tyrosine kinase FGFR1 (D) or Group II tyrosine kinase FLT3 (E), respectively, followed by IDH1 R132H enzyme activity (upper panels). Monomeric and dimeric IDH1 R132H protein levels and tyrosine phosphorylation at Y42 and Y391 of IDH1 R132H were detected by Western blotting using native PAGE (lower panels). F-G, Purified monomeric IDH1 and IDH1 R132H (F) or purified dimeric IDH1 and IDH1 R132H (G) were treated with recombinant active Group I tyrosine kinase FGFR1 in a time dependent manner, followed by native PAGE. Spontaneous dimer formation (F), monomer conversion (G), and tyrosine phosphorylation levels at Y42 of IDH1 and IDH1 R132H were detected by Western blotting. Lower panels show density analysis of corresponding bands to assess dimer formation and monomer conversion in Western blotting. The ratio between homodimers and total IDH1 WT or mutant proteins were quantitatively determined based on density analyses of the Western blotting. H, Left panels: Purified monomers of FLAG-tagged IDH1 and HA-tagged IDH1 R132H were mixed in the presence and absence of recombinant FGFR1, and different monomers and dimers in the mixture were indicated (top two panels, respectively). FLAG pull down was performed to enrich FLAG-IDH1 containing monomers and homo- and hetero-dimers, (middle panel), which were either applied to native gel followed by Western blotting (Figure 5H, upper right), or to HA pull down to further purify the heterodimers (bottom left). Upper right: FLAG-IDH1/HA-R132H heterodimers were detected by HA blotting in native gels. Lower right: FLAG-IDH1/HA-R132H heterodimers purified by sequential FLAG-HA pull downs were detected by FLAG blotting in native gels. I, Purified FLAG-IDH1/HA-IDH1 R132H heterodimers form by non-phosphorylated or FGFR1-phosphorylated monomers of FLAG-IDH1 and HA-IDH1 R132H variants spontaneously disrupt to form monomers in a time dependent manner. Heterodimers and monomer conversions as well as tyrosine phosphorylation levels of dimeric and monomeric proteins were detected by Western blotting. The ratio between heterodimers and total IDH1 proteins were quantitatively determined based on density analyses of the Western blotting.
The error bars represent mean values ±SD from three replicates of each sample (***: p<0.001; ns: not significant); Data are mean ± SD; p values were obtained by a two-tailed Student’s test.
We next determined whether Y42 phosphorylation affects dimer formation from monomers and/or dimer disruption to monomers. We found that purified, recombinant monomeric IDH1 WT or R132H mutant proteins spontaneously form dimers in a time dependent manner (Fig. 5F, right halves of left and right panels, respectively), which explains the basal levels of enzyme activity of non-phosphorylated IDH1 WT or R132H mutant proteins, while Y42 phosphorylation of monomeric IDH1 WT or R132H mutant proteins by FGFR1 resulted in enhanced dimer formation (Fig. 5F, left halves of left and right panels, respectively). In addition, purified, recombinant dimeric IDH1 WT or R132H mutant proteins spontaneously disrupt to form inactive monomers in a time dependent manner (Fig. 5G, right halves of left and right panels, respectively), while dimeric IDH1 WT or R132H mutant proteins purified from IDH1 proteins incubated with rFGFR1, which contained Y42-phosphorylated dimers that were likely formed from Y42-phosphorylated monomers, showed attenuated dimer disruption to form monomeric IDH1 proteins (Fig. 5G, left halves of left and right panels, respectively). These data together suggest that Y42 phosphorylation occurs in IDH1 monomers, which promotes dimer formation, and once Y42-phosphorylated monomers form dimers, Y42 phosphorylation attenuates dimer disruption.
IDH1 mutations exist as heterozygous mutations in AML (5–7,14,15), and R132H/WT heterodimers were suggested to be more efficient than R132H homodimers in 2-HG (23). To determine the effect of Y42 phosphorylation on IDH1 heterodimers, we purified monomers of FLAG-tagged IDH1 and HA-tagged IDH1 R132H. The monomers were mixed in the presence and absence of recombinant FGFR1. Theoretically, the mixture contains FLAG-IDH1 and HA-IDH1 R132H monomers, FLAG-IDH1 and HA-IDH1 R132H homodimers, and heterodimers of FLAG-IDH1/HA-IDH1 R132H (Fig. 5H; top of left panel). FLAG pull down enriched FLAG-IDH1 monomers, FLAG-IDH1 homodimers, and FLAG-IDH1/HA-IDH1 R132H heterodimers (Fig. 5H; middle of left panel). The FLAG pull down samples were either applied to native gel followed by Western blotting (Fig. 5H, upper right), or to HA pull down to further purify the heterodimers (Fig. 5H; bottom left). As shown in Fig. 5H, upper right, HA blotting detected the FLAG-IDH1/HA-IDH1 R132H heterodimers, and the results suggested that phosphorylation by rFGFR1 resulted in increased heterodimer formation of IDH1/R132H (left panel) and IDH1 Y391F/R132H Y391F (right panel), but not IDH1 Y42F/R132H Y42F (middle panel). Similar results were obtained using FLAG-IDH1/HA-IDH1 R132H heterodimers purified by sequential FLAG-HA pull downs detected by FLAG blotting (Fig. 5H, lower right). Moreover, as shown in Fig. 5I, purified FLAG-IDH1/HA-IDH1 R132H heterodimers spontaneously disrupt to form monomers in a time dependent manner, while heterodimers formed by FGFR1-phosphorylated monomers of FLAG-IDH1 and HA-IDH1 R132H (left panel) or monomers of FLAG-IDH1 Y391F and HA-IDH1 R132H Y391F (right panel), but not monomers of FLAG-IDH1 Y42F and HA-IDH1 R132H Y42F (middle panel), showed attenuated dimer disruption to monomers.
Distinct oncogenic tyrosine kinase cascades enhance activation of IDH1 through direct and indirect phosphorylation
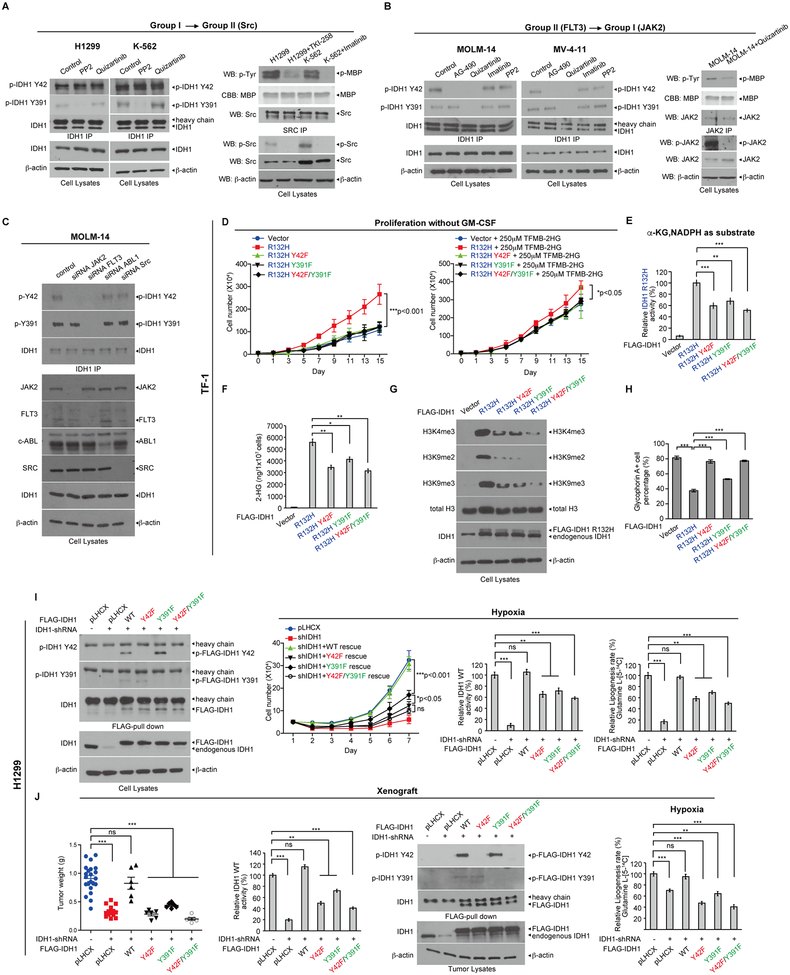
We next sought to determine whether IDH1 R132H mutant and IDH1 WT may be phosphorylated at both or either of the Y42 and Y391 sites in diverse cancer cells expressing different oncogenic tyrosine kinases. We found that both Y42 and Y391 were phosphorylated in lung cancer H1299 cells expressing FGFR1, and leukemia cell lines including K562 cells expressing BCR-ABL, HEL cells expressing JAK2 V617F mutant, and MOLM-14 cells expressing FLT3-ITD (Supplementary Fig. S5C and S5D). Interestingly, treatment with distinct tyrosine kinase inhibitors to target Group I kinases including FGFR1 (TKI258), BCR/ABL (imatinib), and JAK2-V617F (AG490) (Supplementary Fig. S5C), or Group II kinase FLT3-ITD (quizartinib) (Supplementary Fig. S5D) in corresponding cancer/leukemia cells resulted in decreased phosphorylation levels of both Y42 and Y391, suggesting these oncogenic/leukemogenic Group I or II tyrosine kinases activate partner kinases in Group II or I, respectively, to achieve both Y42 and Y391 phosphorylation to further activate IDH1. Indeed, we found that treatment with Group II kinase inhibitors PP2 (Src) but not quizartinib (FLT3) reduced Y391 phosphorylation of IDH1 in H1299 and K562 cells expressing FGFR1 and BCR-ABL, respectively, which are Group I kinases (Fig. 6A, left), suggesting that Group I kinases may activate Src but not FLT3 to achieve Y391 phosphorylation. This was further confirmed by results showing that treatment with Group I kinase inhibitors TKI258 (FGFR1) or imatinib (BCR-ABL) resulted in decreased Src phosphorylation levels in H1299 and K562 cells, respectively (Fig. 6A, bottom right), and reduced activity of Src kinases in an in vitro kinase assay using immunoprecipitated Src incubated with myelin basic protein (MBP) as an exogenous substrate, where Src activity was assessed by the amount of phospho-MBP (Fig. 6A, upper right).
Figure 6.
Abolishment of tyrosine phosphorylation of IDH1 WT and R132H mutant attenuates proliferative and tumor growth potential. A, H1299 and K562 cells were treated with PP2 (Src) or quizartinib (FLT3), followed by endogenous IDH1 immunoprecipitation. Tyrosine phosphorylation at Y42 and Y391 of IDH1 was detected in cell lysates by Western blotting (left panels). Right: H1299 and K562 cells were treated with or without TKI258 and imatinib, respectively. Endogenous Src protein was then immunoprecipitated from cell lysates, followed by in vitro Src kinase activity assay using myelin basic protein (MBP) as an exogenous substrate. Phosphorylation of endogenous Src was tested by Western blotting (right). B, MOLM-14 and MV4–11 cells were treated with AG490, quizartinib, imatinib and PP2, followed by endogenous IDH1 immunoprecipitation. Tyrosine phosphorylation at Y42 and Y391 of IDH1 in cell lysates were detected by Western blotting (left 2 panels). Right: Endogenous JAK2 protein was immunoprecipitated from MOLM-14 cells treated with or without quizartinib, followed by in vitro JAK2 kinase activity assay using MBP as substrate. Phosphorylation of endogenous JAK2 was tested by Western blotting. C, MOLM-14 cells were transfected with siRNAs targeting diverse endogenous tyrosine kinases including JAK2, FLT-3, ABL1 and SRC, followed by endogenous IDH1 immunoprecipitation. Tyrosine phosphorylation at Y42 and Y391 of endogenous IDH1 and knock down efficacy of each tyrosine kinase were detected by Western blotting. D, TF-1 cell lines stably expressing Flag-tagged IDH1 R132H or R132H-Y42F, R132H-Y391F and R132H-Y42F/Y391F mutants were generated, followed by cell proliferation assay in the absence or presence of TFMB-2HG treatment (left and right panel, respectively) in the absence of GM-CSF. E, Flag-tagged IDH1 protein was pulled down from TF-1 stable cell lines expressing IDH1 R132H and variants, followed by IDH1 R132H enzyme activity assay using αKG and NADPH as substrates. F-H, 2-HG production (F), histone methylation (G) and resistance to EPO-induced differentiation (H) of TF-1 stable cell lines expressing IDH1 R132H and variants were measured. I, Generation of H1299 cells with stable knockdown of endogenous human IDH1 and stable “rescue” expression of FLAG-IDH1 WT, Y42F, Y391F or Y42F/Y391F. Rescued IDH1 protein levels were detected by Western blotting (left). Cell proliferation assay (middle left), IDH1 WT enzyme activity assay (middle right) and lipogenesis rate using labeled glutamine (Glutamine L-[5-14C]) as a carbon source (right) were performed using H1299 stable rescue cells under hypoxia. J, From left to right: Tumor weight, IDH1 WT enzyme activity assay, phosphorylation at Y42 and Y391 of IDH1 and lipogenesis rate under hypoxia were detected in xenograft tumors derived from diverse “rescue” H1299 cells expressing IDH1 variants.
The error bars represent mean values ±SD from three replicates of each sample except (J) left (*: 0.01<p<0.05; **: 0.01<p<0.001; ***: p<0.001; ns: not significant); Data are mean ± SD; p values were obtained by a two-tailed Student’s test except a 2-way ANOVA was used for cell proliferation assay.
Similar results were obtained using two FLT3-ITD-expressing leukemia cell lines including MOLM-14 and MV4–11, where treatment with FLT3 inhibitor quizartinib resulted in decreased phosphorylation levels of both Y42 and Y391 of IDH1, whereas JAK2 inhibitors including AG490 and ruxolitinib only reduced Y42 phosphorylation (Fig. 6B, left, and Supplementary Fig. S5E). In contrast, control inhibitors including imatinib and PP2 did not affect Y42 or Y391 phosphorylation of IDH1 (Fig 6B, left). These data suggest that Group II kinase FLT3 may activate Group I kinase JAK2 to achieve Y42 phosphorylation of IDH1. This is consistent with results showing that treatment with FLT3 inhibitor quizartinib resulted in decreased JAK2 phosphorylation levels in MOLM-14 cells (Fig. 6B, bottom right), and reduced JAK2 kinase activity in an in vitro kinase assay using immunoprecipitated JAK2 incubated with MBP as an exogenous substrate (Fig. 6B, upper right). Similar results were obtained using siRNAs to specifically knock down different tyrosine kinases in MOLM-14 cells, where FLT3 siRNA resulted in reduced phosphorylation levels of both Y42 and Y391 of IDH1 while JAK2 siRNA only decreased Y42 phosphorylation, but both siRNAs targeting ABL and Src did not alter IDH1 phosphorylation (Fig. 6C). Further in vitro kinase assays using immunoprecipitated Group I kinase JAK2 or Group II kinase Src (Supplementary Fig. S5F, left and right, respectively) pre-treated with protein tyrosine phosphatase PTP1B to remove tyrosine phosphorylation prior to incubation with Group II kinase rFLT3 or Group I kinases rABL, rFGFR1 or rJAK2, respectively, showed that FLT3 directly phosphorylated JAK2, while ABL, FGFR1 and JAK2 phosphorylated Src. These data supported the proposed distinct Group II➔I and I➔II tyrosine kinase cascades, respectively, which achieve phosphorylation of both Y42 and Y391 of IDH1.
Abolishment of tyrosine phosphorylation of IDH1 mutant and WT attenuates cytokine-independent growth ability with reduced production of oncometabolite 2-HG in human hematopoietic TF-1 erythroleukemia cells
Proliferation of TF-1 erythroleukemia cells depends on the cytokine granulocyte-macrophage colony-stimulating factor (GM-CSF), and when transformed by expression of IDH1 R132H, TF-1 cells can grow in the absence of GM-CSF and are resistant to erythropoietin (EPO)-induced cell differentiation (24,25). We generated TF-1 cell lines with stable expression of FLAG-tagged IDH1 R132H or R132H/Y42F, R132H/Y391F and R132H/Y42F/Y391F mutants by lentiviral transduction. We found that expression of IDH1 R132H resulted in increased GM-CSF-independent cell proliferation, while cells expressing R132H/Y42F, R132H/Y391F and R132H/Y42F/Y391F mutants showed attenuated potential for GM-CSF-independent cell proliferation (Fig. 6D, left), which was partially “rescued” by adding cell permeable TFMB-(R)-2-HG (24) in the culture media (Fig. 6D, right). Consistent with these findings, compared to TF-1 cells expressing IDH1 R132H, cells expressing diverse phosphor-deficient R132H mutants showed attenuated IDH1 mutant enzyme activity (Fig. 6E), 2-HG production (Fig. 6F), histone hypermethylation (Fig. 6G), and resistance to EPO-induced differentiation of TF-1 cells that was assessed using staining with glycophorin A (24) (Fig. 6H and Supplementary Fig. S6A).
Abolishment of tyrosine phosphorylation of IDH1 WT reduces subsequent proliferative and tumor growth potential of H1299 lung cancer cells
We next generated “rescue” H1299 cells with stable knockdown of endogenous IDH1 and stable rescue expression of FLAG-IDH1 WT, Y42F, Y391F, or Y42F/Y391F mutants (Fig. 6I, left). Expression of IDH1 WT reversed the decreased cell proliferation and IDH1 activity of H1299 cells under both normoxia (Supplementary Fig. S6B, left and middle, respectively) and hypoxia (Fig. 6I, left and middle of right panels, respectively), while Y➔F mutants demonstrated attenuated “rescue” of reduced cell proliferation due to decreased IDH1 activity compared to IDH1 WT. In addition, knockdown of endogenous IDH1 or “rescue” expression of IDH1 variants had no effect on lipogenesis under normoxia (Supplementary Fig. S6B, right), whereas IDH1 knockdown resulted in decreased lipogenesis under hypoxia, which was completely or partially rescued by IDH1 WT or IDH1 Y➔F mutants, respectively (Fig. 6I, right of right panel). Consistently, in a xenograft experiment, rescue expression of IDH1 WT reversed the decreased tumor growth rate and masses (Supplementary Fig. S6C, left, and 6J, left, respectively) of injected H1299 cells with stable knockdown of endogenous IDH1, while IDH1 Y➔F mutants demonstrated attenuated rescue ability with reduced IDH1 activity due to abolishment of Y42 and/or Y391 phosphorylation (Fig. 6J, middle two panels, respectively). In tumor cells, IDH1 or “rescue” expression of IDH1 variants had no effect on lipogenesis under normoxia (Supplementary Fig. S6C, middle), whereas IDH1 knockdown resulted in decreased lipogenesis under hypoxia, which was completely or partially rescued by IDH1 WT or IDH1 Y➔F mutants, respectively (Fig. 6J, right). In addition, these tumor growth results correlate with tumor cell proliferation potential assessed by IHC staining of Ki-67 (Supplementary Fig. S6C, right two panels). Similar results were obtained using “rescue” MOLM-14 cells expressing Y➔F mutants of IDH1 with decreased cell proliferation under both normoxia and hypoxia with reduced lipogenesis only under hypoxia (Supplementary Fig. S6D–S6F).
FLT3 WT and ITD mutant enhance activation of WT and mutant IDH1 through direct phosphorylation of Y391 and indirect phosphorylation of Y42 by activating JAK2 in AML cells
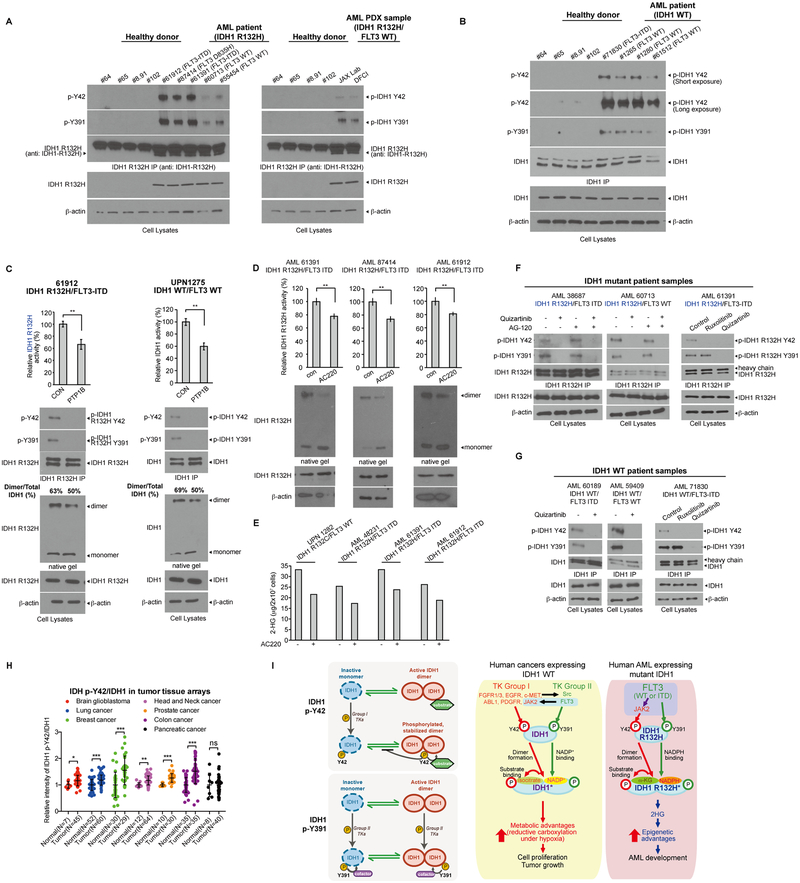
Phosphorylation of Y42 and Y391 was detected in human primary leukemia cells from AML patients harboring IDH1 R132H mutation despite different FLT3 mutational state (Fig. 7A), while upregulated phosphorylation levels of Y42 and Y391 of IDH1 WT were detected in primary leukemia cells from representative AML patients compared to control peripheral blood cells from healthy donors (Fig. 7B). We next confirmed that PTP treatment resulted in decreased enzyme activities of immunoprecipitated IDH1 R132H (Fig. 7C, upper left) or IDH1 WT (Fig. 7C and S7A, upper right) with reduced phosphorylation levels of Y42 and Y391 (Fig. 7C and S7A, middle panels), and that PTP treatment of primary AML cell lysates reduced the amount of dimeric IDH1 proteins (Fig. 7C, lower panels). Moreover, treatment with FLT3 inhibitor quizartinib resulted in decreased amounts of IDH1 R132H dimers but increased monomers (Fig. 7D) with reduced 2-HG production (Fig. 7E) in primary leukemia cells from AML patients expressing mutant IDH1 with either FLT3-ITD or WT. In addition, treatment with quizartinib resulted in decreased Y42 and Y391 phosphorylation of IDH1 R132H mutant (Fig. 7F and S7B) or IDH1 WT (Fig. 7G) in primary leukemia cells from AML patients expressing either FLT3-ITD or WT, while treatment with JAK2 inhibitor ruxolitinib only decreased Y42 phosphorylation of IDH1 R132H or WT (Fig. 7F and 7G, right panels, respectively). Furthermore, IHC staining results showed that relative Y42-phosphorylated IDH1 levels compared to overall IDH1 protein levels were commonly upregulated in diverse primary tumor tissue samples from human patients with GBM, lung, breast, head and neck, prostate, and colon cancers, but not pancreatic cancer, compared to corresponding normal control tissue samples (Fig. 7H and Supplementary Fig. S7C).
Figure 7.
FLT3 WT and ITD mutant enhance activation of WT and mutant IDH1 through direct phosphorylation of Y391 and indirect phosphorylation of Y42 by JAK2 activation in primary AML cells. A, Endogenous IDH1 protein was immunoprecipitated from primary leukemia cells of representative AML patients (left panel) and AML PDX samples (right panel) with IDH1 R132H mutation. Tyrosine phosphorylation of Y42 and Y391 were detected by Western blotting. Peripheral blood samples from four healthy donors were included as controls. B, Endogenous IDH1 protein was immunoprecipitated from primary leukemia cells of representative AML patients with IDH1 WT. Tyrosine phosphorylation of Y42 and Y391 were detected by Western blotting. Peripheral blood samples from four healthy donors were included as controls. C, Primary leukemia cell lysates with IDH1 R132H/FLT3-ITD or IDH1 WT/FLT3 WT from AML patients (left and right panels, respectively) were treated with PTP1B, followed by endogenous IDH1 protein immunoprecipitation and IDH1 R132H and IDH1 WT enzyme activity assay, respectively (upper panels). Tyrosine phosphorylation of Y42 and Y391 of IDH1 was detected by Western blotting (middle panels). Dimeric and monomeric IDH1 protein levels were measured by Western blotting using native PAGE (lower panels). D, Primary leukemia cells from representative AML patients harboring IDH1 R132H and FLT3-ITD mutations were treated with quizartinib, followed by mutant IDH1 enzyme activity assay (upper) and Western blotting to detect dimers and monomers of IDH1 R132H (lower). E, Primary leukemia cells from representative mutant IDH1-expressing AML patients expressing FLT3 WT (left one) or FLT3-ITD (right three) were treated with quizartinib, followed by detection of cellular 2-HG levels by NMR analysis. F, Primary leukemia cells from representative IDH1 R132H AML patients with FLT3-ITD (left) and FLT3 WT (middle) were treated with quizartinib and/or AG120, followed by IDH1 R132H pull down. Right: Primary leukemia cells from representative IDH1 R132H AML patients with FLT3-ITD were treated with JAK2 inhibitor ruxolitinib or FLT3 inhibitor quizartinib, followed by IDH1 R132H immunoprecipitation. Y42 and Y391 phosphorylation of IDH1 R132H was detected by Western blotting. G, Primary leukemia cells from representative wild type IDH1 AML patients with FLT3-ITD (left) and FLT3 WT (middle) were treated with quizartinib, followed by IDH1 immunoprecipitation. Right: Primary leukemia cells from a wild type IDH1 AML patient with FLT3-ITD were treated with JAK2 inhibitor ruxolitinib or FLT3 inhibitor quizartinib, followed by IDH1 pull down. Y42 and Y391 phosphorylation of IDH1 was detected by Western blotting. H, IHC staining of phosphorylated IDH1 (p-Y42) and total IDH1 in normal or tumor tissues in diverse tumor tissue arrays. Relative intensities of p-IDH1 Y42/IDH1 were calculated. I, Working model (detailed description is included in Discussion).
The error bars represent mean values ±SD from three replicates of each sample except (H) (*: 0.01<p<0.05; **: 0.01<p<0.001; ***: p<0.001; ns: not significant); Data are mean ± SD; p values were obtained by a two-tailed Student’s test.
DISCUSSION
Our findings reveal a molecular mechanism shared by WT and mutant IDH1 in related human cancers, which involves distinct tyrosine kinase cascades containing Group I and II tyrosine kinases that preferentially phosphorylate Y42 and Y391, respectively, to fulfill activation of WT and mutant IDH1. Moreover, our finding of tyrosine phosphorylation-dependent IDH1 activation represents a novel isoform-specific activating mechanism for IDH1 in AML, because the functional phosphorylation sites of IDH1 reported in our manuscript, Y42 and Y391 are replaced with F82 and F431, respectively, in IDH2. Our results suggest that spontaneous formation of active IDH1 dimers from monomers and disruption of dimers to form monomers coexist and eventually reach an equilibrium, such that IDH1 dimers but not monomers are able to recruit substrate (isocitrate for WT and α-ketoglutarate for R132H mutant) and thus represent the active portion of total IDH1 proteins (Fig. 7I, upper left). Group I TKs preferentially phosphorylate Y42 on monomeric but not dimeric IDH1, and Y42-phosphorylated IDH1 monomers exhibit enhanced ability to form active dimers, leading to increased substrate binding and enzyme activity (Fig. 7I, upper left). Moreover, active dimers formed by Y42-phosphorylated monomers showed attenuated disruption to form monomers, suggesting that Y42 phosphorylation has a dual role in enhancing IDH1 activation by promoting dimer formation from Y42-phosphrylated monomers and preventing disruption of Y42-phosphorylated dimers, which represents a mechanism to sustain IDH1 activation in cells (Fig. 7I, upper left). In contrast, Y391 phosphorylation by Group II TKs occurs in both monomeric and dimeric IDH1 proteins, which is able to promote cofactor (NADP+ for WT and NADPH for R132H mutant) binding to IDH1 monomers and dimers, respectively (Fig. 7I, lower left). Future structural studies of IDH1 monomers and dimers are warranted to decipher the underlying mechanisms by which Y42 phosphorylation contributes to monomer-dimer equilibrium and subsequent substrate binding, while Y391 phosphorylation promotes cofactor binding to IDH1 despite monomeric or dimeric status. Moreover, it will be interesting to determine whether one or both molecules of an active dimer need to be phosphorylated at Y42 and/or Y391, or both Y42 and Y391 need to be phosphorylated in one molecule in an active dimer to functionally contribute to IDH1 activation.
In human cancers expressing IDH1 WT, diverse Group I tyrosine kinases activate Src (Group II), or FLT3 (Group II) activates JAK2 (Group I) to achieve phosphorylation of both Y42 and Y391, leading to sustained levels of active dimers and enhanced substrate and cofactor NADP+. Phosphorylation-enhanced IDH1 activation is important to provide metabolic advantages to cancer cell proliferation and tumor growth, including reductive carboxylation for lipogenesis under hypoxia (Fig. 7I, middle). In human cancers harboring IDH1 mutations, mutant IDH1 retains such an enhancing mechanism. For example, in AML, FLT3 (Group II) activates JAK2 (Group I) to achieve phosphorylation at both sites and subsequently full activation of mutant IDH1, leading to 2-HG production that provides an epigenetic advantage to tumorigenesis and tumor growth (Fig. 7I, right). However, since IDH1 mutations exist as heterozygous mutations in AML and glioma cases (5–7,14,15), it is believed that homodimers of IDH1 WT and mutant, as well as WT/mutant heterodimers, co-exist. It has been suggested that, although WT:R132H heterodimers exhibit almost abolished IDH1 WT enzyme activity (26), IDH1 mutants require IDH1 WT activity to produce 2-HG (27). Moreover, R132H/WT heterodimers demonstrate a significantly lower Km for αKG as substrate compared to R132H homodimers (23). These studies suggest an overall advantage of R132H/WT heterodimers that favors the R132H mutant-driven conversion of αKG to 2-HG. Our results demonstrate that phosphorylation of Y42 similarly enhances the IDH1 mutant/WT heterodimer formation from monomers and attenuates disruption of heterodimers to monomers as Y42 phosphorylation does to homodimers of IDH1 WT and mutant, which further strengthens the importance of IDH1 tyrosine phosphorylation in mutant IDH1-expressing human cancers.
Our studies revealed that a spectrum of tyrosine residues of IDH1 WT and R132H can be phosphorylated. These include the previously reported Y139, which is believed to play a critical role in IDH1 R132H mediated catalysis of αKG by compensating the increased negative charge on the C2 atom of αKG during reduction of αKG to 2-HG (28). IDH1 R132H/Y139A mutant demonstrates <1% activity (28), which is consistent with our finding that Y139F mutant is catalytically deficient. Future structural and mechanistic studies are warranted to determine whether phosphorylation at Y139 also contributes to IDH1 mediated catalysis. In contrast, unlike Y139F or Y219F mutations that almost abolish IDH1 enzyme activity, Y42F and Y391F mutations do not alter the intrinsic enzymatic properties of IDH1 WT or R132H mutant, which was supported by the diverse kinetic studies.
These results also provide an additional rationale supporting the combination of tyrosine kinase and mutant IDH1 inhibitors for the clinical treatment of IDH1 mutant-expressing human cancers. It is plausible to consider combined therapy using inhibitors of mutant IDH1 and FLT3 to treat AML patients harboring IDH1 mutations regardless of the mutational status of FLT3, given both FLT3 WT and ITD mutant are able to phosphorylate IDH1 mutant directly and indirectly through activation of JAK2. It was reported that FLT3-ITD is able to induce STAT5 phosphorylation independent of JAK2, where FLT3 may directly phosphorylate STAT5 (29). Thus, it is likely that FLT3 WT or ITD mutant are able to phosphorylate and activate JAK2, which might be dispensable for STAT5 phosphorylation and activation but is critical for the phosphorylation and subsequently enhanced activation of WT and mutant IDH1 in AML leukemia cells. Taken together, our studies elucidate a novel regulatory mechanism of IDH1, which not only provides insight into how signaling impacts metabolic function but also informs mechanism-based combination therapeutic studies.
METHODS
Primary tissue samples from patients with leukemia and healthy donors.
Approval of use of human specimens was given by the Institutional Review Board of Emory University School of Medicine. The clinical samples numbered with UPN were obtained with informed consent with approval by the Emory University Institutional Review Board. The clinical samples from Memorial Sloan-Kettering Cancer Center were approved from the Institutional Review Board of Memorial Sloan-Kettering Cancer Center. Patients provided written informed consent in all cases at time of enrollment. Only samples from leukemia patients that were not previously treated with chemotherapy or radiation therapy were used. Mononuclear cells of peripheral blood and bone marrow samples from leukemia patients or healthy donors were isolated using lymphocyte separation medium (Cellgro). Cells were then counted and cultured in RPMI 1640 medium supplemented with 10% FBS and P/S for further indicated treatments.
Cell culture
TF-1 cells (#CRL-2003, ATCC; purchased 2018; not authenticated) were cultured in RPMI 1640 medium with 10% FBS and 2 ng/ml recombinant human GM-CSF (R&D Systems). K-562 (#CCL-243, ATCC; purchased 2015; not authenticated; Mycoplasma tested in Nov. 2018), H1299 (#CRL-5803, ATCC; purchased 2015; not authenticated; Mycoplasma tested in Nov. 2018), MOLM-14, MV-4–11 and HEL cells were cultured in RPMI 1640 medium with 10% FBS. HEK293T was cultured in Dulbecco Modified Eagle Medium (DMEM) with 10% FBS. Cells were cultured at 37°C with of 5% CO2. MOLM-14, MV-4–11, HEL and HEK293T cells were obtained from Dr. Gary Gilliland’s laboratory at Brigham and Women’s Hospital in 2004; not authenticated.
RNAi-resistant IDH1 wild type and diverse YF mutant plasmids
The shRNA sequence for wild-type IDH1 is CGAATCATTTGGGAATTGATT. The wild-type IDH1 and diverse YF mutant plasmid plasmids were mutagenized by PCR to generate IDH1 plasmids that contain 3 silent mutations introduced in the 21 bp IDH1 shRNA target sequence (CGTATCATATGGGAGTTGATT). The mutations were confirmed by sequence analysis.
Cell lines
Stable knockdown of endogenous IDH1 was achieved by using lentiviral vectors harboring shRNA constructs. To be specific, each shRNA construct was co-transfected with psPAX2 packaging plasmid and pMD2.G envelope plasmid (Addgene) into HEK293T cells using TransIT®-LT1 Transfection Reagent (Mirus Bio) according to the manufacturer’s instructions. Supernatant of culture medium containing lentivirus was collected 36–48 hours after transfection, filtered by 0.2μm filter, followed by adding to host cell lines. 24 hours after infection, target cells were then subjected to puromycin (2μg/ml) selection for 24–48 hours. Knockdown efficiency of endogenous IDH1 protein was confirmed by Western blotting. Stable overexpression of IDH1 wild type and mutants in MOLM14, H1299 and TF-1 cells was conducted by using retroviral vectors harboring RNAi-resistant FLAG-tagged IDH1 wild type, FLAG-tagged IDH1 R132H, and RNAi-resistant FLAG-tagged IDH1 wild type or R132H plus diverse YF mutants. Briefly, to produce retrovirus, each construct was co-transfected with VSVG, EcoPak packaging plasmid and envelope plasmid (Addgene) into HEK293T cells using FuGENE™ Transfection Reagent (Promega) according to the manufacturer’s instructions. Retrovirus-containing supernatant medium was collected 48 hours after transfection, filtered before adding to the indicated host cell lines (1M HEPES was used to adjust the pH to 7.4 in culture medium). 24 hours after infection, target cells were subjected to hygromycin selection (Invitrogen). The overexpression of proteins was confirmed by Western blotting using antibodies against IDH1.
Antibodies
Antibodies against DYKDDDDK (FLAG) tag, p-Tyrosine (p-Tyr-100), Jak2, p-Jak2 (Tyr1007/1008), di-methyl-histone H3 (Lys9), histone H3, FLT3 and Src were from Cell Signaling Technology (CST). Antibodies against β-actin and p-Src (Tyr418) were from Sigma-Aldrich. Antibody against HA was from Santa Cruz Biotechnology. Antibody against trimethyl (Lys9) was from Abcam. Antibodies against IDH1-R132H and Trimethyl-Histone H3 (Lys4) were from EMD Millipore. Antibody against IDH1 was from R&D SYSTEMS. Antibody against PE-Cy™5 Mouse Anti-Human CD235a was from BD Pharmingen™. Goat anti-Mouse IgG (H+L) secondary antibody and goat anti-rabbit IgG (H+L) secondary antibody were from Thermo Fisher Scientific. Antibodies against p-IDH1 Y42 and p-IDH1 Y391 were custom-made by SHANGHAI GENOMICS, INC. Antibodies against Ki67 and IDH1 for IHC staining studies were from Abcam.
Reagents
Myelin basic protein from bovine, DL-isocitric acid trisodium salt hydrate, β-nicotinamide adenine dinucleotide phosphate hydrate (NADP+), β-nicotinamide adenine dinucleotide 2′-phosphate reduced tetrasodium salt hydrate (NADPH), α-ketoglutaric acid sodium salt, diaphorase from Clostridium kluyveri, resazurin sodium salt, and ATP disodium salt hydrate were purchased from Sigma-Aldrich. Ketoglutaric acid sodium salt, α-[1-14C], 50μCi (1.85MBq) and glucose, D-[U-14C]- were from PerkinElmer. Glutamine L-[5-14C] and isocitric acid[3H(G)] were from ARC. Inhibitors including dovitinib (TKI-258, CHIR-258), imatinib, AG-490, quizartinib, PP2, erlotinib, ruxolitinib, dasatinib and AG-120 were purchased from Selleckchem. Recombinant proteins including FLT3, SRC, PDGFRα, PDGFRβ, EGFR, MET, KIT, JAK2, FGFR3 and ABL1 were purchased from Thermo Fisher.
Cell proliferation and cell viability assay
Cell proliferation assays were performed by seeding 5 × 104 cells in a 6-well plate in normoxia (5% CO2 and 95% air) or hypoxia (5% CO2, 1% O2 and 94% N2) with daily counting of cell numbers. Cell proliferation was determined by cell numbers recorded after being seeded and normalized to that of each cell line at the starting time (T=0 hour) by trypan blue exclusion using TC20 Automated Cell Counter (BioRad). For cell viability assay, 5 × 104 cells were seeded in each well of a 96-well plate and incubated with ivosidenib (AG-120) or quizartinib for 48 hr. Relative cell viability was determined using CellTiter 96 Aqueous One solution proliferation kit (Promega).
Mutant IDH1 enzyme activity assay
An in vitro or in vivo IDH1 R132H activity assay was performed as previously described (30). To be specific, 20 ng purified IDH1 R132H and variants proteins treated with or without recombinant active tyrosine kinases in an in vitro kinase assay were added to 50 μl assay buffer (150 mM NaCl, 20 mM Tris-HCl (pH7.5), 10 mM MgCl2, 0.05% (w/v) bovine serum albumin) containing 15 μM NADPH (Sigma-Aldrich), 1.5 mM α-KG (Sigma-Aldrich). After 1-hour incubation at room temperature, 20 μM resazurin and 10 μg/ml diaphorase were added to the reaction mixture and incubated for another 10 minutes at room temperature. To determine the IDH1 R132H activity in cells, 1×106 cells were harvested and directly lysed using 100 μl 1% NP-40 cell lysis buffer. 25 μl whole cell lysates were then added with 25 μl 2x assay buffer (300 mM NaCl, 40 mM Tris-HCl (pH7.5), 20 mM MgCl2, 0.05% (w/v) bovine serum albumin) containing 30 μM NADPH and 3 mM α-KG. After 1 hour incubation at room temperature, 20 μM resazurin and 10 μg/ml diaphorase were added to the primary reaction and incubated for another 10 minutes at room temperature. Fluorescence was recorded on a Spectramax GEMINIEM plate reader (Molecular Devices) at Ex544/Em590.
Wild type IDH1 enzyme activity assay
An in vitro enzyme activity assay for wild type IDH1 was performed as previously described (8). In brief, 20 ng purified IDH1 and YF variant proteins that were pre-treated with or without recombinant active tyrosine kinases in an in vitro kinase assay were added to 100 μl enzyme activity assay buffer (25mM Tris-HCl (pH7.5), 10 mM MgCl2, 5 mM DTT) containing 0.5 mM NADP+ (Sigma-Aldrich) and 1 mM isocitric acid (Sigma-Aldrich). To determine the wild type IDH1 activity in cells, endogenous IDH1 from 1×107 cells was bound to protein G-Sepharose 4 Fast Flow beads (Sigma-Aldrich) by immunoprecipitation using IDH1 antibody (CST). To test IDH1 activity in xenograft tumor tissues, around 2 mg of total tumor lysates were used. After immunoprecipitation, the IDH1-bound beads were washed 3 times and eluted using IDH1 peptide (CST). Then 10 ul of the supernatant was added to 100μl assay buffer (25mM Tris-HCl, pH7.5, 10 mM MgCl2, 5 mM DTT) containing 0.5 mM NADP+ and 1 mM isocitric acid. IDH1 activity was measured by recording absorbance of 340 nm in kinetic mode every 20 seconds for 15 minutes using a SpectraMax Plus spectrophotometer (Molecular Devices).
Immunoprecipitation
1–2 mg of cells or tumor lysates were incubated with anti-IDH1 antibody (R&D SYSTEMS) or anti-IDH1 antibody (CST) overnight at 4°C. After incubation, protein G-Sepharose was used for precipitation for 2 hours. The beads were then washed 3 times with 1xTBS and eluted by boiling in SDS sample buffer for Western blotting analysis.
Purification of prokaryotic recombinant IDH1 proteins
6xHis-FLAG-IDH1 or 6xHis-HA-IDH1 and variants proteins were purified by sonicating of high expressing BL21(DE3) pLysS cells obtained from a 250 ml culture subjected to IPTG-induction for 16 hours at 30°C. Bacteria cell lysates were obtained by centrifugations and loaded onto a Ni-NTA column within 20 mM imidazole. The bound proteins were eluted with 250 mM imidazole, followed by desalting using a PD-10 column. The purified recombinant IDH1 proteins were examined by Coomassie Brilliant Blue staining and western blotting.
In vitro tyrosine kinase assays
In vitro kinase assays were performed as previously described (31,32). To be specific, 1μg recombinant IDH1, IDH1 R132H and variants proteins were incubated with diverse recombinant active form of tyrosine kinases for 90 min in the presence of 800 uM ATP (Sigma) at 30°C in the following assay buffer, respectively. For FGFR1 assay buffer, 10 mM HEPES (pH 7.5), 10 mM MnCl2, 150 mM NaCl, 5 mM DTT, 0.01% Triton® X-100 was used; For FLT3 assay buffer, 60 mM HEPES (pH 7.5), 3 mM MgCl2, 3 mM MnCl2, 3 μM Na3VO4, 1.2 mM DTT was used; For FGFR3 assay buffer, 60 mM HEPES (pH 7.5), 3 mM MgCl2, 3 mM MnCl2, 3 μM Na3VO4, and 1.2 mM DTT was used; For JAK2 assay buffer, 25 mM HEPES (pH 7.5), 10 mM MgCl2, 0.5 mM EGTA, 0.5 mM Na3VO4, 5 mM β-glycerophosphate, 2.5 mM DTT, and 0.01% Triton® X100 was used; For ABL1 assay buffer, 60 mM HEPES (pH 7.5), 3 mM MgCl2, 3 mM MnCl2, 3 μM Na3VO4, 1.2 mM DTT was used; For SRC buffer, 50 mM HEPES (pH 7.5), 10 mM MgCl2, 10% Glycerol, 2.5 mM DTT, and 0.01% Triton® X-100 was used; For EGFR buffer, 20 mM Tris (pH 7.5), 10 mM MgCl2, 1 mM EGTA, 1 mM Na3VO4, 5 mM β−glycerophosphate, 2 mM DTT, and 0.02% Triton® X-100 was used; For MET assay buffer, 25 mM Tris (pH 7.5), 10 mM MgCl2, 0.5 mM EGTA, 0.5 mM Na3VO4, 5 mM β-glycerophosphate, 2.5 mM DTT, and 0.01% Triton® X-100 was used. The amount of each recombinant active kinases used for the reaction was as follows: 100 ng rFGFR1 (0.1 U), 160 ng rMET (0.06 U), 800 ng rEGFR (0.1 U), 200 ng rJAK2 (0.06 U), 120 ng rPDGFRα (0.066 U), 250 ng rPDGFRβ (0.066 U), 25 ng rFLT3 (0.06 U), 100 ng rSRC (0.2 U) and 120 ng rABL1 (0.06 U). For in vitro kinase assay of IDH1 dimer or monomer, 1 mg IDH1 WT or R132H protein was used for sucrose gradient centrifugation to separate monomer or dimer, then 1 μg monomer or dimer protein was used for in vitro kinase assay with or without recombinant active form of rFGFR1 or rFLT3. Then samples were injected for the Western blotting analysis with native gel.
FLAG-pull down assay
200 μg total protein from whole cell lysates were incubated with 30 μl of ANTI-FLAG M2 Affinity Gel (Sigma-Aldrich) for 4 hours at 4°C, followed by washing with phosphate-buffered saline (PBS) for 3 times to remove unbound materials. The bound proteins were then eluted from beads by boiling in SDS buffer (50 mM Tris-Cl (pH 6.8), 2% (w/v) SDS, 0.1% (w/v) bromophenol blue, 100 mM DTT) for 10 minutes and visualized via immunoblotting analysis.
Native PAGE
Native gels were prepared as described (31) using 0.375M Tris-HCl, pH 8.8,10% acrylamide, without SDS. Cell lysates were mixed with 5 × native gel sample buffer (0.05% bromophenol blue, 10% glycerol, 312.5mM Tris-HCl, pH 6.8) before loading. The samples were applied to native PAGE that ran in buffer (25mM Tris-HCl, 192mM glycine), followed by Western blotting.
Sucrose density ultracentrifugation
Sucrose gradient centrifugation was performed as previously described (31). In brief, purified recombinant 6xHis-FLAG-IDH1 or R132H and variant protein was laid on a 13.75%−36% sucrose gradient and spun at 50,000 rpm for 12 hours using a Beckman MLS-50 rotor. Fractions in each section were collected and analyzed by Coomassie blue staining.
Substrate binding assay
Purified recombinant 6xHis-FLAG-IDH1 and variants that are immobilized on anti-FLAG beads were treated with or without diverse recombinant active tyrosine kinases in an in vitro kinase assay. The beads were then incubated with 0.1 mM isocitric acid [3H(G)] (ARC) at room temperature for 1 hours and then washed twice with TBS to remove the unbound isocitric acid [3H(G)] molecule. IDH1 and variant proteins were then eluted with 3x FLAG peptide. Bound isocitric acid molecules to proteins were measured using a scintillation counter. Purified recombinant 6xHis-FLAG-IDH1 R132H and variants that are immobilized on anti-FLAG beads were treated with or without diverse recombinant active tyrosine kinases in an in vitro kinase assay. The beads were incubated with 0.1 mM α-ketoglutaric acid [1-14C] (Perkin Elmer) at room temperature for 1 hour and then washed twice with TBS to remove the unbound α-ketoglutaric acid [1-14C]. The IDH1-R132H and variant proteins were eluted with 3× FLAG peptide. Bound α-ketoglutaric acid molecules to proteins were measured using a scintillation counter.
Substrate binding assay for monomer or dimer of IDH1 or IDH1 R132H
Purified recombinant IDH1 monomers and dimers were separated on a native gel. The gel was incubated with 0.1 mM isocitric acid [3H(G)] (ARC) for 2 hours at 4°C and then washed twice with TBS to remove the unbound isocitric acid [3H(G)]. The IDH1 proteins in monomer and dimer form were cut from the gel and the retained isocitric acid [3H(G)] on both forms of IDH1 proteins were measured using a scintillation counter. Purified recombinant IDH1 R132H monomers and dimers were separated on a native gel, then incubated with 0.1 mM α-ketoglutaric acid [1-14C] (Perkin Elmer) for 2 hours at 4°C and then washed twice with TBS to remove the unbound α-ketoglutaric acid [1-14C]. The IDH1 R132H proteins in monomer and dimer form were cut from the gel and the retained α-ketoglutaric acid [1-14C] on both forms of IDH1 R132H proteins were measured using a scintillation counter.
NADP+ binding assay
NADP+ binding assay was performed as previously described (33). Briefly, 200 ng of 6xHis-FLAG-IDH1 and variant proteins were incubated with 30 μl Blue Sepharose CL-6B beads (Amersham Biosciences) which mimics NADP+ at 4°C for 2 hours, then washed with 20 mM Tris-HCL (pH 8.6) for 3 times. The beads were then subjected to SDS-PAGE, followed by Western blotting analysis. For NADP+ binding assay with IDH1 dimer or monomer, 1 mg IDH1 WT protein was used for sucrose gradient centrifugation to separate monomer or dimer, then 1 ug monomer or dimer protein was used for in vitro kinase assay with or without FGFR1. After kinase assay, 200 ng protein were incubated with 30 μl Blue Sepharose CL-6B beads (Amersham Biosciences) at 4°C for 2 hours, then washed with 20 mM Tris-HCL (pH 8.6) 3 times. The beads were subjected to SDS-PAGE, followed by Western blotting. The same amount of protein was loaded in parallel as loading control.
Kinetic assay
20 ng purified IDH1 and YF variant proteins that pre-treated with or without recombinant active tyrosine kinases in an in vitro kinase assay were incubated with various concentrations of NADP+ (0–200 μM; Sigma-Aldrich) or isocitrate acid (0–200 μM; Sigma-Aldrich) and 1 mM isocitric Acid (Sigma-Aldrich) or 0.5 mM NADP+ (Sigma-Aldrich), respectively at 100 μl enzyme activity assay buffer (25 mM Tris-HCl pH7.5), 10 mM MgCl2, 5 mM DTT). Absorbance at 340 nm was measured every 20 seconds for 5 minutes using a SpectraMax Plus spectrophotometer (Molecular Devices).
20 ng purified IDH1 R132H and YF variants proteins that pre-treated with or without recombinant active tyrosine kinases in an in vitro kinase assay were incubated with various concentrations of NADPH (0–10μM; Sigma-Aldrich) and 1.5 mM α-KG (Sigma-Aldrich) or various concentrations of α-KG (0–1500 μM; Sigma-Aldrich) and 15 μM NADPH (Sigma-Aldrich) at 100 μl enzyme activity assay buffer (25 mM Tris-HCl (pH7.5), 10 mM MgCl2, 5 mM DTT). Absorbance at 340 nm was measured every 20 seconds for 5 minutes using a SpectraMax Plus spectrophotometer (Molecular Devices). Non-linear regression analysis (Michaelis-Menten) was performed in Graphpad Prism 6.0.
IDH1 WT or R132H protein dimer formation
1 mg IDH1 WT or R132H protein was used for Sucrose gradient centrifugation to separate monomer, then 1μg monomer protein was used for in vitro kinase assay with FGFR1 as indicated time and stopped in fridge of minus 80. Then samples were injected for the Western blotting analysis with native gel.
NADP+ competitive binding assay
1 μg of 6xHis-FLAG-IDH1 protein was incubated with 100 μl Blue Sepharose CL-6B beads (Amersham Biosciences) at 4°C for 2 hours, then washed with 20 mM Tris-HCL (pH 8.6) for 3 times and separated to 3 tubes equally. The beads were incubated with indicated concentration of NADP+ at 4°C for 1 hours. Then the samples were centrifuged to separate the supernatant and beads. The supernatant or beads were subjected to SDS-PAGE, followed by Western blotting analysis.
IDH1 heterodimer formation
1 mg FLAG-IDH1 WT and HA-IDH1 R132H protein was used for Sucrose gradient centrifugation to separate monomer, then 100 μg monomer protein was mixed and used for in vitro kinase assay with FGFR1 for 2 hours. The protein was incubated with 50 μl of ANTI-FLAG M2 Affinity Gel (Sigma-Aldrich) for 4 hours at 4°C, followed by washing with phosphate-buffered saline (PBS) for 3 times to remove unbound materials. Then bound protein was elute with 100 μg FLAG peptide (Sigma-Aldrich). Then half of the eluted protein was injected for the Western blotting analysis with native gel. The rest of the eluted protein was incubated with 30 μl of monoclonal Anti-HA Agarose (Sigma-Aldrich) for 4 hours at 4°C, followed by washing with phosphate-buffered saline (PBS) for 3 times to remove unbound materials. The bound protein was elute with 10 μg HA peptide (Sigma-Aldrich), then the samples were applied to native gels for the Western blotting analysis.
IDH1 WT or R132H protein monomer conversion
1 mg IDH1 WT or R132H protein was used for in vitro kinase assay with FGFR1, then the dimer was separated by sucrose gradient centrifugation. 1μg dimer was collected as indicated time and the monomer conversion was stopped by fridge of minus 80, then the samples were loaded on native gels for the Western blotting analysis.
The conversion of IDH1 heterodimer to monomer
For purification of IDH1 heterodimer, 1 mg FLAG-IDH1 WT and HA-IDH1 R132H protein was used for in vitro kinase assay with FGFR1 for 2 hours. The protein was incubated with 100 μl of ANTI-FLAG M2 Affinity Gel (Sigma-Aldrich) for 4 hours at 4°C, followed by washing with PBS for 3 times to remove unbound materials. Then bound protein was elute with 200 μg FLAG peptide (Sigma-Aldrich). The eluted protein was incubated with 1000 μl of monoclonal Anti-HA Agarose (Sigma-Aldrich) for 4 hours at 4°C, followed by washing with PBS for 3 times to remove unbound materials. Then bound protein was eluted with 50 μg HA peptide (Sigma-Aldrich). Then the heterodimer was collected at indicated time and the monomer conversion was stopped by fridge of minus 80. Then the samples were loaded on a native gel for the Western blotting analysis.
Cell culture treatment
Treatments with tyrosine kinase inhibitors were performed by incubating cells with 0.1 μM TKI-258, 1 μM imatinib, 50 μM AG-490, 100 nM erlotinib, 0.1 μM ruxolitinib, 200 nM quizartinib, 50 nM dasatinib or 1 μM AG-120 for 4 hours. PP2 treatment was performed by incubating cells with 10 μM PP2 for 4 hours.
Small interfering RNA-mediated knockdown
The transfection of small interfering RNA (siRNA) into MOLM-14 cells was carried out using Lipofectamine® 2000 transfection reagent (Thermo Fisher), according to the manufacturer’s instructions. Briefly, siRNA and Lipofectamine 2000 reagent were mixed in Opti-MEM medium (Thermo Fisher) and incubated for 30 min at room temperature to allow the complex formation. Then the cells were washed with Opti-MEM medium (Thermo Fisher), and the mixtures were added. Twelve hours after transfection, the culture medium was replaced by fresh complete medium. The cells were harvested 72 h after transfection, followed by further analysis. The following siRNA sequences were used for knockdown: negative control siRNA (nonsilencing; QIAGEN; SI03650325); Hs_ABL1_6 Flexi Tube siRNA (QIAGEN; SI00299089); Hs_SRC_7 Flexi Tube siRNA (QIAGEN; SI02223928); Hs_JAK2_7 Flexi Tube siRNA (QIAGEN; SI02659657); Hs_FLT3_6 Flexi Tube siRNA (QIAGEN; SI02659608).
TF-1 cell differentiation assay
TF-1 stable cell lines were washed four times with plain RPMI medium and cultured for 24 hours without GM-CSF. Then 2U/mL EPO (recombinant human erythropoietin) was added for differentiation. 8 days later, glycophorin A was used for erythroid differentiation assay by flow cytometry. Cell surface staining for glycophorin A was performed as follows: cells were washed once in PBS supplemented with 2% FBS (PBS-FBS) and then resuspended in a 1:200 dilution of PE-Cy5-conjugated mouse anti-human CD235a antibody (BD Pharmingen) in PBS-FBS. The cells were incubated for 20 minutes at 4°C in the dark, then washed once with PBS-FBS. The cells were analyzed on a FACSCanto™ II cytometer (Becton Dickinson). All flow cytometry data was analyzed using FlowJo software (TreeStar, Inc.).
2-HG measurement in cell lines, patient samples and tumor samples
The sample preparation for ex vivo NMR analysis followed a previously described method (34). Briefly, 1 × 107 cells were thawed in 99.996% saline deuterium oxide (D2O, Sigma-Adrich) in the sample holder/rotor (4 mm ZrO2). A 50 μl insert was subsequently placed in the sample holder to stabilize the sample and to provide the balance for the rotor. For NMR experiments, D2O (99.996%) containing 0.75% 3-(trimethylsilyl) propionic acid (TSP) was added to get a frequency-lock signal, as well as to serve as an internal reference for chemical shift and concentration measurements. Each sample added with TSP containing D2O was reweighed for metabolite quantification. All NMR samples were treated rapidly on ice to avoid possible degradation. HRMAS NMR experiments were conducted with a dedicated 4 mm HRMAS probe at 4°C using a Bruker AVANCE 600 WB solid state NMR spectrometer (Bruker Instruments, Inc.). The entire process throughout the experiment was maintained at 4°C (±0.1°C) via a variable temperature control unit and the probe-head was precooled to 4°C before loading the sample. To ensure the spin sidebands did not affect the spectrum, the spinning rates of sample were controlled in the range of 2,800 KHz (±2 Hz) or at the lower spin rate of 800 Hz if the rotor-synchronized delay alternating with nutation for tailored excitation sequence was used. The presaturation of water was achieved with a zqpr sequence before acquisition pulses. A rotor-synchronized Carr-Purcell-Meibom-Gill pulse sequence was used to suppress broad signals from macromolecules. The number of transients was 256. In all experiments, the repetition time was 5.0 s, and the spectral width was 10 kHz. 95% pure 2HG compound (Santa Cruz Biotechnology) in solution was used as 2HG resonance identification and assignment. One-dimensional (1D) NMR spectra of a pure 2HG compound, and a mixture of 2HG and a combination of glutamate (Glu) and glutamine (Gln) compounds (together termed Glx), were obtained at ~10 mM 2HG in D2O, pH 7.0 and collected at 300 MHz at 25°C. J-coupling correlations and patterns of protons in the pure 2HG were then analyzed by a two-dimensional (2D) J-coupled correlated spectroscopy (COSY) method. For the tumor sample analysis, data of 2D COSY were collected at 4°C with 6,000 Hz spectral width and 1.5-s relaxation delay. Thirty-two transients in the time domain t2 were averaged for each of the 512 increments in time domain of t1 with a total acquisition time of ~3 hours.
Lipid biosynthesis assay
For 14C-lipid biosynthesis assay, cells were pre-incubated with 4 μl of glucose, D-[14C(U)] (PerkinElmer) under normoxia (5% CO2 and 95% air) for 2 hours or 4 mM glutamine L-[5-14C] (ARC) under hypoxia (5% CO2, 1% O2 and 94% N2) for 24 hours. Lipids were then extracted by the addition of 500 μl of hexane: isopropanol (3:2 v/v), dried, resuspended in 50 μl of chloroform, and subjected to scintillation counting.
Xenograft studies
Approval of the use of mice and designed experiments was given by the Institutional Animal Care and Use Committee (IACUC) of Emory University. Briefly, NSG mice (NOD scid gamma, female 6-week old, The Jackson Lab) were subcutaneously injected with 1 × 106 H1299 cells stably expressing IDH1 WT, IDH1 Y42F, IDH1 Y391F and IDH1 Y42F/Y391F with stable knockdown of endogenous IDH1 on the left and right flanks, respectively. Tumor growth was recorded every two days from 7–12 days after inoculation by measurement of two perpendicular diameters using the formula 4π/3 × (width/2)2 × (length/2). The masses of tumors (mg) derived from treatments were analyzed. For single cell isolation from tumor tissues, fresh tumors were removed from xenograft mice and were placed in PBS. The tumors were then minced into small pieces using scissors and digested with 5ml digestion buffer (Accumax@ Stemcell) for 1 hour at room temperature on a horizontal shaker. The digestion buffer was then neutralized with equal amount of cell culture media after digestion and xenograft tumor cells were strained through 70 μm cell strainer, followed by washing twice with PBS. Cells were then counted and cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS) and penicillin/streptomycin (P/S, 100 Unit/ml) for further treatment.
PTP1B treatment
For in vivo assay, 1×107 cells were harvested and lysed using 900 μl NP-40 cell lysis buffer with 100 μl 10 × PTP1B buffer (500 mM HEPES, pH 7.2, 10 mM EDTA, 10 mM DTT, 0.5% NP-40) containing 0.5 μg BSA), followed by incubation with 2μg PTP1B protein for 90 min at 30°C. 200 μl reaction mixture was then stored as input samples, while the remaining 800 μl reaction mixture was subjected to IDH1 immunoprecipitation using anti-IDH1 antibody (R&D SYSTEMS) for 6 hours at 4°C. After immunoprecipitation, protein G-Sepharose was added for precipitation for 2 hours. The beads were then washed 3 times with TBS, followed by analysis using PAGE and Western blotting. Input samples were applied to native gel followed by Western blotting. For the in vitro assay, recombinant FLAG-tagged IDH1 and mutant proteins (1 μg each) were incubated with FLAG beads (30 μl) for 3 hours at 4°C. After incubation, the beads were washed with TBS 3 times, followed by in vitro tyrosine kinase assay in the presence or absence of recombinant active form of FGFR1 or FLT3 for 90 min at 30°C. The beads were then washed 3 times with TBS to remove kinases, followed by adding 1 μg PTP1B in 100 μl reaction buffer (50 mM HEPES, pH 7.2, 1 mM EDTA, 1 mM DTT, 0.05% NP-40 containing 0.1 μg of BSA) and incubated for 90 min at 30°C. At the end of the reaction, the beads were washed with TBS 3 times, then IDH1 proteins were eluted by adding 2 μg of FLAG peptide, followed by Western blotting analysis and enzyme activity assay.
Immunohistochemical staining
Immunohistochemical staining for Ki67, phospho-IDH1 Y42 and IDH1 were performed as previously described (35–37). In brief, tumor tissues from xenograft mice were fixed in 10% buffered formalin, embedded in paraffin and mounted on slides. Slides of xenograft tumor tissues or tumor tissue arrays were deparaffinized and rehydrated, followed by incubation in 3% hydrogen peroxide to suppress endogenous peroxidase activity. High-pressure antigen retrieval was achieved in 10 mM sodium citrate (pH6.0). Sections were then blocked by incubation in 10% goat serum. Human Ki67 antibody (Abcam; 1:500), human phospho-IDH1 Y42 antibody (SHANGHAI GENOMICS, INC; 1:200) or human IDH1 antibody (Abcam; 1:500) were applied overnight at 4°C. Dako IHC kit (Agilent technologies) was used for detection. Secondary antibody was applied at room temperature for 1 hour. Slides were then stained with 3,3′-diaminobenzidine, washed, counterstained with hematoxylin, dehydrated, and mounted.
QUANTIFICATION AND STATISTICAL ANALYSIS
In studies in which statistical analyses were performed, a 2-tailed Student’s t test was used to generate p values, except a two-way ANOVA was used for cell proliferation assay and tumor growth. p values less than or equal to 0.05 were considered significant. Data with error bars represent mean ± SD, except for tumor growth curves which represent mean ± SEM. There is no estimate of variation in each group of data and the variance is similar between the groups. No statistical method was used to predetermine sample size. The experiments were not randomized. The investigators were not blinded to allocation during experiments and outcome assessment. All data are expected to have normal distribution. Statistical analysis and graphical presentation were performed using Prism 6.0 (GraphPad) and Microsoft Office Excel 2016.
Supplementary Material
SIGNIFICANCE.
We demonstrated an intrinsic connection between oncogenic TKs and activation of WT and mutant IDH1, which involves distinct tyrosine kinase cascades in related cancers. In particular, these results provide an additional rationale supporting the combination of FLT3 and mutant IDH1 inhibitors as a promising clinical treatment of mutant IDH1-positive AML.
Acknowledgments
We thank Dr. Anthea Hammond for critical reading and editing of the manuscript.
Grant Support
This work was supported in part by NIH grants including CA140515, CA183594, CA174786 (J.C.), R35197594, CA173636, MSKCC Support Grant/Core Grant P30 CA008748 (R.L.L.), CA169937 (H.M.), and K08CA181507–01A1 (A.H.S.), and Joel A. Katz Music Medicine Fund supported by the T.J. Martell Foundation/Winship Cancer Institute (J.C. and R.L.). R.L. and A.H.S. are Special Fellows of the Leukemia and Lymphoma Society. J.C. is the Winship 5K Scholar and the R. Randall Rollins Chair in Oncology.
Conflict of Interest Statement: R.L.L. is on the supervisory board of Qiagen and is a scientific advisor to Loxo, Imago, C4 Therapeutics and Isoplexis, which each include an equity interest. He receives research support from and consulted for Celgene and Roche, he has received research support from Prelude Therapeutics, and he has consulted for Incyte, Novartis, Astellas, Morphosys and Janssen. He has received honoraria from Lilly and Amgen for invited lectures and from Gilead for grant reviews.
Footnotes
SUPPLEMENTARY DATA
Supplementary data includes seven figures.
REFERENCES
- 1.Warburg O On the origin of cancer cells. Science 1956;123(3191):309–14. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144(5):646–74 doi S0092–8674(11)00127–9 [pii] 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Kang HB, Fan J, Lin R, Elf S, Ji Q, Zhao L, et al. Metabolic Rewiring by Oncogenic BRAF V600E Links Ketogenesis Pathway to BRAF-MEK1 Signaling. Molecular cell 2015;59(3):345–58 doi 10.1016/j.molcel.2015.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hitosugi T, Chen J. Post-translational modifications and the Warburg effect. Oncogene 2014;33(34):4279–85 doi 10.1038/onc.2013.406. [DOI] [PubMed] [Google Scholar]
- 5.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, et al. An integrated genomic analysis of human glioblastoma multiforme. Science 2008;321(5897):1807–12 doi 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. The New England journal of medicine 2009;361(11):1058–66 doi 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, et al. IDH1 and IDH2 mutations in gliomas. The New England journal of medicine 2009;360(8):765–73 doi 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 2009;462(7274):739–44 doi 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cairns RA, Mak TW. Oncogenic isocitrate dehydrogenase mutations: mechanisms, models, and clinical opportunities. Cancer Discov 2013;3(7):730–41 doi 10.1158/2159-8290.CD-13-0083. [DOI] [PubMed] [Google Scholar]
- 10.Inoue S, Lemonnier F, Mak TW. Roles of IDH1/2 and TET2 mutations in myeloid disorders. Int J Hematol 2016;103(6):627–33 doi 10.1007/s12185-016-1973-7. [DOI] [PubMed] [Google Scholar]
- 11.Kroemer G, Pouyssegur J. Tumor cell metabolism: cancer’s Achilles’ heel. Cancer Cell 2008;13(6):472–82. [DOI] [PubMed] [Google Scholar]
- 12.Jiang L, Shestov AA, Swain P, Yang C, Parker SJ, Wang QA, et al. Reductive carboxylation supports redox homeostasis during anchorage-independent growth. Nature 2016;532(7598):255–8 doi 10.1038/nature17393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Metallo CM, Gameiro PA, Bell EL, Mattaini KR, Yang J, Hiller K, et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature 2011;481(7381):380–4 doi 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gross S, Cairns RA, Minden MD, Driggers EM, Bittinger MA, Jang HG, et al. Cancer-associated metabolite 2-hydroxyglutarate accumulates in acute myelogenous leukemia with isocitrate dehydrogenase 1 and 2 mutations. J Exp Med 2010;207(2):339–44 doi 10.1084/jem.20092506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marcucci G, Maharry K, Wu YZ, Radmacher MD, Mrozek K, Margeson D, et al. IDH1 and IDH2 gene mutations identify novel molecular subsets within de novo cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 2010;28(14):2348–55 doi 10.1200/JCO.2009.27.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell 2010;17(3):225–34 doi 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Green A, Beer P. Somatic mutations of IDH1 and IDH2 in the leukemic transformation of myeloproliferative neoplasms. N Engl J Med 2010;362(4):369–70 doi 10.1056/NEJMc0910063. [DOI] [PubMed] [Google Scholar]
- 18.McKenney AS, Levine RL. Isocitrate dehydrogenase mutations in leukemia. J Clin Invest 2013;123(9):3672–7 doi 10.1172/JCI67266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patel KP, Ravandi F, Ma D, Paladugu A, Barkoh BA, Medeiros LJ, et al. Acute myeloid leukemia with IDH1 or IDH2 mutation: frequency and clinicopathologic features. Am J Clin Pathol 2011;135(1):35–45 doi 10.1309/AJCPD7NR2RMNQDVF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Medeiros BC, Fathi AT, DiNardo CD, Pollyea DA, Chan SM, Swords R. Isocitrate dehydrogenase mutations in myeloid malignancies. Leukemia 2017;31(2):272–81 doi 10.1038/leu.2016.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yen K, Travins J, Wang F, David MD, Artin E, Straley K, et al. AG-221, a First-in-Class Therapy Targeting Acute Myeloid Leukemia Harboring Oncogenic IDH2 Mutations. Cancer Discov 2017;7(5):478–93 doi 10.1158/2159-8290.CD-16-1034. [DOI] [PubMed] [Google Scholar]
- 22.Shih AH, Meydan C, Shank K, Garrett-Bakelman FE, Ward PS, Intlekofer AM, et al. Combination Targeted Therapy to Disrupt Aberrant Oncogenic Signaling and Reverse Epigenetic Dysfunction in IDH2- and TET2-Mutant Acute Myeloid Leukemia. Cancer Discov 2017;7(5):494–505 doi 10.1158/2159-8290.CD-16-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brooks E, Wu X, Hanel A, Nguyen S, Wang J, Zhang JH, et al. Identification and Characterization of Small-Molecule Inhibitors of the R132H/R132H Mutant Isocitrate Dehydrogenase 1 Homodimer and R132H/Wild-Type Heterodimer. J Biomol Screen 2014;19(8):1193–200 doi 10.1177/1087057114541148. [DOI] [PubMed] [Google Scholar]
- 24.Losman JA, Looper RE, Koivunen P, Lee S, Schneider RK, McMahon C, et al. (R)-2-hydroxyglutarate is sufficient to promote leukemogenesis and its effects are reversible. Science 2013;339(6127):1621–5 doi 10.1126/science.1231677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang F, Travins J, DeLaBarre B, Penard-Lacronique V, Schalm S, Hansen E, et al. Targeted inhibition of mutant IDH2 in leukemia cells induces cellular differentiation. Science 2013;340(6132):622–6 doi 10.1126/science.1234769. [DOI] [PubMed] [Google Scholar]
- 26.Zhao S, Lin Y, Xu W, Jiang W, Zha Z, Wang P, et al. Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1alpha. Science 2009;324(5924):261–5 doi 10.1126/science.1170944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ward PS, Lu C, Cross JR, Abdel-Wahab O, Levine RL, Schwartz GK, et al. The potential for isocitrate dehydrogenase mutations to produce 2-hydroxyglutarate depends on allele specificity and subcellular compartmentalization. J Biol Chem 2013;288(6):3804–15 doi 10.1074/jbc.M112.435495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang B, Zhong C, Peng Y, Lai Z, Ding J. Molecular mechanisms of “off-on switch” of activities of human IDH1 by tumor-associated mutation R132H. Cell Res 2010;20(11):1188–200 doi 10.1038/cr.2010.145. [DOI] [PubMed] [Google Scholar]
- 29.Choudhary C, Brandts C, Schwable J, Tickenbrock L, Sargin B, Ueker A, et al. Activation mechanisms of STAT5 by oncogenic Flt3-ITD. Blood 2007;110(1):370–4 doi 10.1182/blood-2006-05-024018. [DOI] [PubMed] [Google Scholar]
- 30.Davis MI, Gross S, Shen M, Straley KS, Pragani R, Lea WA, et al. Biochemical, cellular, and biophysical characterization of a potent inhibitor of mutant isocitrate dehydrogenase IDH1. The Journal of biological chemistry 2014;289(20):13717–25 doi 10.1074/jbc.M113.511030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fan J, Lin R, Xia S, Chen D, Elf SE, Liu S, et al. Tetrameric Acetyl-CoA Acetyltransferase 1 Is Important for Tumor Growth. Mol Cell 2016;64(5):859–74 doi 10.1016/j.molcel.2016.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fan J, Shan C, Kang HB, Elf S, Xie J, Tucker M, et al. Tyr phosphorylation of PDP1 toggles recruitment between ACAT1 and SIRT3 to regulate the pyruvate dehydrogenase complex. Molecular cell 2014;53(4):534–48 doi 10.1016/j.molcel.2013.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shan C, Elf S, Ji Q, Kang HB, Zhou L, Hitosugi T, et al. Lysine acetylation activates 6-phosphogluconate dehydrogenase to promote tumor growth. Molecular cell 2014;55(4):552–65 doi 10.1016/j.molcel.2014.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kalinina J, Carroll A, Wang L, Yu Q, Mancheno DE, Wu S, et al. Detection of “oncometabolite” 2-hydroxyglutarate by magnetic resonance analysis as a biomarker of IDH1/2 mutations in glioma. Journal of molecular medicine 2012;90(10):1161–71 doi 10.1007/s00109-012-0888-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kang HB, Fan J, Lin R, Elf S, Ji Q, Zhao L, et al. Metabolic Rewiring by Oncogenic BRAF V600E Links Ketogenesis Pathway to BRAF-MEK1 Signaling. Mol Cell 2015;59(3):345–58 doi 10.1016/j.molcel.2015.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin R, Xia S, Shan C, Chen D, Liu Y, Gao X, et al. The Dietary Supplement Chondroitin-4-Sulfate Exhibits Oncogene-Specific Pro-tumor Effects on BRAF V600 E Melanoma Cells. Mol Cell 2018;69(6):923–37 e8 doi 10.1016/j.molcel.2018.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao L, Fan J, Xia S, Pan Y, Liu S, Qian G, et al. HMG-CoA synthase 1 is a synthetic lethal partner of BRAFV600E in human cancers. J Biol Chem 2017;292(24):10142–52 doi 10.1074/jbc.M117.788778. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.