Abstract
Cholesterol sulfotransferase, SULT2B1b, has been demonstrated to modulate both androgen receptor activity and cell growth properties. However, the mechanism(s) by which SULT2B1b alters these properties within prostate cancer cells has not been described. Furthermore, specific advantages of SULT2B1b expression in prostate cancer cells is not understood. In these studies, single-cell mRNA sequencing (scRNA-seq) was conducted to compare the transcriptomes of SULT2B1b knockdown (KD) versus Control KD LNCaP cells. Over 2,000 differentially expressed (DE) genes were identified along with alterations in numerous canonical pathways, including the death receptor signaling pathway. The studies herein demonstrate that SULT2B1b KD increases tumor necrosis factor alpha (TNF) expression in prostate cancer cells and results in NF-κB activation in a TNF-dependent manner. More importantly, SULT2B1b KD significantly enhances TNF-mediated apoptosis in both TNF-sensitive LNCaP cells and TNF-resistant C4–2 cells. Overexpression of SULT2B1b in LNCaP cells also decreases sensitivity to TNF-mediated cell death, suggesting that SULT2B1b modulates pathways dictating the TNF sensitivity capacity of prostate cancer cells. Probing human prostate cancer patient datasets further support this work by providing evidence that SULT2B1b expression is inversely correlated with TNF-related genes, including TNF, CD40LG, FADD, and NFKB1. Together, these data provide evidence that SULT2B1b expression in prostate cancer cells enhances resistance to TNF and may provide a growth advantage. In addition, targeting SULT2B1b may induce an enhanced therapeutic response to TNF treatment in advanced prostate cancer.
Keywords: SULT2B1b, prostate cancer, TNFα, death receptor signaling, scRNA-seq
INTRODUCTION
Prostate cancer is the most commonly diagnosed and second leading cause of non-cutaneous cancer death in American men.(1) Death receptor-targeted therapies, notably Fas, tumor necrosis factor alpha (TNF), and TNF-related apoptosis-inducing ligand (TRAIL), have been studied over several decades as potential therapies for this disease.(2–4) TNF was identified over 40 years ago and demonstrated to induce necrosis of tumor cells.(5) Subsequent to the initial observation, TNF has been defined to have seemingly paradoxical roles in prostate cancer cells. TNF has been shown to induce apoptosis in some prostate cancer cells, such as LNCaP cells, while others, such as the castration resistant prostate cancer (CRPC) cell line C4–2, are resistant to TNF-mediated cell death.(6, 7) TNF studies in prostate cancer provide evidence for both pro-tumorigenic and anti-tumorigenic responses resulting from TNF signaling through TNF-receptors (TNFR1 and TNFR2) in prostate cancer cells.(8–14) However, understanding the mechanisms by which intracellular environmental factors contribute to the paradoxical observations have yet to be elucidated. The mechanism(s) by which the intracellular environment may influence TNF responsiveness in prostate cancer is an understudied field that could impact therapeutic approaches, especially in prostate cancer cells characterized as resistant to TNF-mediated apoptosis.
Prostate cancer cells also demonstrate a dysregulation of cholesterol.(15, 16) We recently reported that RNA interference (RNAi)-mediated knockdown (KD) of SULT2B1b, the cholesterol sulfotransferase, induces apoptosis in a variety of prostate cancer cell lines including C4–2 cells, a model of CRPC.(17) To better understand the mechanism(s) by which SULT2B1b induces cell death in prostate cancer cells, we conducted a single-cell mRNA sequencing (scRNA-seq) analysis comparing SULT2B1b KD to non-targeting (Control) KD LNCaP cells. A single-cell approach was selected for this analysis as it allows for the determination of the quality of siRNA KD at the cellular level, as well as the correlation of the expression of specific genes for identification of SULT2B1b-regulated pathways. Using LNCaP cells as a model limited the heterogeneity in these experiments, yet led to the discovery of many possible avenues of SULT2B1b-mediated regulation. Understanding the role of SULT2B1b in prostate cancer cells and how this enzyme influences cell growth properties will provide a more in-depth understanding of the biology of prostate cancer cells and could lead to identification of alternative therapeutic options for CRPC. Herein, we provide evidence that SULT2B1b regulates TNF-mediated cell signaling and that targeting SULT2B1b by knockdown enhances TNF-induced cell death in both LNCaP cells and TNF-resistant CRPC C4–2 cells.
MATERIALS AND METHODS
Cell Lines
LNCaP cells were purchased from the American Tissue Culture Collection (ATCC; Manassas, VA) and maintained in media conditions identical to those recommended by ATCC. C4–2 cells were purchased from MD Anderson (Houston, TX) and were maintained in T-medium (Invitrogen) supplemented with 10% fetal bovine serum (FBS) and 1% streptomycin/penicillin. LNCaP-SULT2B1b cells, which induce overexpression of SULT2B1b with the addition of doxycycline, were produced by consecutive lentiviral transductions as described previously.(17) All experiments using LNCaP and C4–2 cells were completed within 20 passages of the cells authenticated previously by DDC Medical (Fairfield, OH) through the generous support of the Prostate Cancer Foundation.(17)
Antibodies and Reagents
Primary antibodies against the following proteins were used for these studies: TNFα (Thermo, P300A) and TRAIL/TNFSF10 (R&D Systems, MAB375) for neutralization; β-actin (Cell Signaling, 3700), phospho-IkB (Cell Signaling, 9246), IkBα (Cell Signaling, 4814), TRADD (BD, 610572), and SULT2B1 (Abcam, ab88085) for western blotting; and TNFR1 (Abcam, ab19139) and TNFR2 (Abcam, ab109322) for flow cytometry. Secondary antibodies include goat anti-mouse IgG-HRP (Santa Cruz, sc-2005) for western blotting and donkey anti-rabbit IgG-AlexaFluor647 (Biolegend, 406414) for flow cytometry.
RNAi studies were completed using control/non-targeting siRNA complexes (Dharmacon) and siRNA targeting SULT2B1 (IDT, HSC.RNAI.N004605.12.2), TNF (IDT, hs.Ri.TNF.13.2), TL1 (IDT, hs.Ri.TNFSF15.13.3), or DAXX (IDT, hs.Ri.DAXX.13.1) using Lipofectamine RNAiMax (ThermoFisher), according the manufacturer’s instructions. RNA isolation and cDNA synthesis were completed as previously described(17), derived from the EZNA Total RNA Kit I (Omega Bio-tek). PrimeTime ® qRT-PCR gene probes (IDT) used for these studies include: AR (Hs.PT.56a.38770693), KLK3 (PSA) (Hs.PT.56a.38546086), SULT2B1 (Hs.PT.56a.25562421.g), TNF (Hs.PT.58.45380900), and SULT2B1b (Hs.PT.58.22608626).
Additionally, Recombinant human TNFα (Peprotech, 300–01A), human TRADD cell-based ELISA kit (Abnova, KA3564), and human TNFα ELISA kit (Sigma-Aldrich, RAB1089) for cell lysates were used for these studies. The TRADD ELISA kit was used per the manufacturer’s instructions by fixing the adherent cells 72 hours after siRNA transfection and normalizing to crystal violet absorbance, while the TNFα ELISA was performed using cell lysates from samples 72 or 96 hours after siRNA transfection in LNCaP and C4–2 cells, respectively. To assess cell viability, the Cell Counting Kit-8 (Dojindo Molecular Technologies, Inc.) was used for these studies according to the manufacturer’s instructions.
Luciferase Assays
Luciferase assays were conducted by transfecting the pNF-κB-luciferase reporter plasmid (Stratagene) and Renilla luciferase plasmid (pRL-TK) using FuGENE HD transfection reagent (Promega, E2311), followed by assessing luciferase activity using the Dual Luciferase Reporter Assay kit (Promega, E1910).(18) Relative luciferase activity (RLU=Firefly/Renilla) of the NF-κB reporter construct is shown as the mean of the fold change (over samples without siRNA transfection) +/− SEM from at least three independent experiments performed in triplicate. Additional details may be found in Supplemental Methods.
Flow Cytometry
For cell-cycle analysis, fixed cells were stained with propidium iodide (Biolegend #421301), according to the manufacturer’s instructions, and analyzed on a FACSCanto II (BD Biosciences). Further analysis of DNA content was conducted using FlowJo software, version 9 (TreeStar Inc.). For protein expression analysis, cells were analyzed on a BD Fortessa LSR (BD Biosciences) and mean fluorescence intensity (MFI) of experimental samples was compared to a secondary only control sample using FlowJo software.
Cell Preparation and Loading into C1 System
Forty-eight hours following siRNA transfection, LNCaP cells were harvested with trypsin and stained with Zombie Violet viability dye (Biolegend, 423114) before sorting of viable cells on a FACS ARIA III (BD Biosciences). Cells were loaded and stained in a 17–25 µm IFC plate (Fluidigm, 100–5761) followed by exclusion of capture sites containing doublets or dead cells. This procedure was completed on consecutive days for Control and SULT2B1b KD cells and repeated three times for biological replicates. In total, 209 Control siRNA and 190 SULT2B1b siRNA treated cells were used for analysis. Additional details regarding C1 loading and subsequent library preparation is located in Supplemental Methods.
cDNA Harvest, Quantitation, Library Preparation, and Sequencing
Diluted cDNA samples were quantified using a Qubit fluorimeter with the dsDNA HS Assay Kit (Life Technologies, Q32851) and normalized to a concentration of 0.1–0.3 ng/µL for library preparation. Libraries were prepared using the Nextera® XT DNA Sample Preparation Kit and Index Kit (Illumina, FC-131–1096 and FC-131–1002) followed by quantification according to the Fluidgm C1 mRNA sequencing protocol. Unstranded, 2×100bp reads were sequenced using a HiSeq 2500 System (Illumina) on rapid run mode. Three separate sequencing runs were conducted, each with pooled Control KD cell libraries in one lane and pooled SULT2B1b KD cell libraries in the other. cBot Duo (Illumina) was added to allow for distinct sample pools to be added to individual rapid flow cells.
Sequence Trimming and Alignment
Sequences were quality-trimmed and adaptors removed via FastX-Toolkit version 0.0.13.2(19). A FastX trimscore of 30 and a trim length of 50 were used. Maximum length was set to 151 bases. FastQC version 0.11.2(20) was run to check data quality both before and after quality trimming and adaptor removal.
Reads were aligned to the Ensembl Genome Reference Consortium Human Build 38 (GRCh38.p3) using Tophat2.(21, 22) Tophat2 version 2.1.0 was run with default settings with two exceptions: one mismatch was allowed and the library type was set to “fr-unstranded,” due to the non-strand-specificity of the sequencing library. HTSeq v.0.6.1(23) was used in “intersection-nonempty” mode to count reads mapping to features; Biopython v.2.7.3 was used in this analysis.
Statistical Analysis and Pathway Analysis
Statistical analysis of in vitro studies utilized Student’s t-test or analysis of variance (ANOVA) with or without a multiple comparisons test as appropriate, using GraphPad Prism 5 software. Statistically significant values are indicated in figures as *p<0.05, **p<0.01, ***p<0.001, or ****p<0.0001 (no significance is indicated by n.s.). All scRNA-seq data is available in GEO with accession number GSE117410. In total, 36,135 genes were sequenced, many of which exhibited low expression levels. Genes with an average count of less than 5 across all samples were removed and then differential expression was tested using the Bioconductor R package edgeR v.3.2.2,. Controlling for a false discovery rate (FDR) of 5% using the Benjamini-Hochberg procedure yielded 2,029 differentially expressed (DE) genes. DE genes, FDR, log(fold-change), and log(counts per million) were uploaded to Ingenuity Pathway Analysis (IPA) software (Qiagen) and a canonical pathway analysis and upstream regulator analysis were performed. Upstream regulators were predicted in IPA based on the input DE genes and p-values were determined using a one-sided Fisher’s exact test.
Human Prostate Cancer Database Correlations
RNA-seq data from Robinson, et. al including 20 bone marrow-derived metastatic prostate cancer samples without prior treatment and 16 lymph node-derived CRPC samples with prior taxane and abiraterone or enzalutamide treatment were retrieved from cBioPortal database.(24) The data were normalized by log(RPKM+1). Gene co-expression correlations between SULT2B1 and 55 tumor necrosis factor and receptor-related genes were computed using Pearson Correlation Coefficients and were assessed for statistical significance by using a permutation test with 10,000 rounds of random simulation.
RESULTS
scRNA-seq analysis successfully identified altered pathways and DE genes.
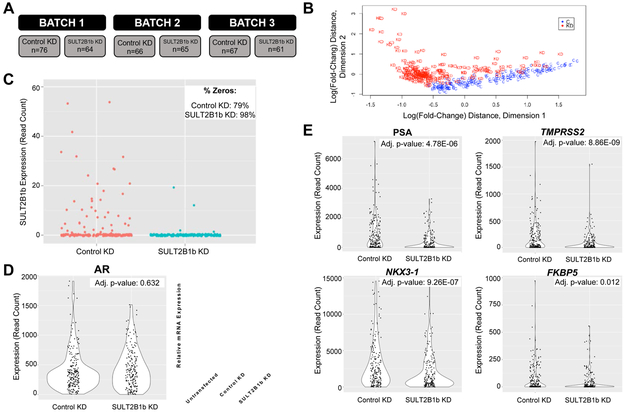
In these studies, scRNA-seq was performed to identify significantly altered genes and pathways in SULT2B1b KD versus Control KD prostate cancer cells. Our previous studies indicate that SULT2B1b KD induces apoptosis in LNCaP cells by 72 hours. Since scRNA-seq requires viable cells, LNCaP were harvested 48 hours after non-targeting or SULT2B1 siRNA (Control KD or SULT2B1b KD, respectively) transfection and then subjected to viable cell sorting prior to single-cell isolation on the Fluidigm C1 Single-Cell Auto Prep System (Supplementary Figure 1A). Viable cell sorting did not impact the efficiency of SULT2B1b KD (Supplementary Figure 1B). Three independent experiments (batches 1–3) were completed each for Control or SULT2B1b KD and the resulting sequencing data were pooled for quality control and analysis, giving a total of 209 Control KD and 190 SULT2B1b KD cells, respectively (Figure 1A). Sequenced reads were determined to be of high quality and minimal batch effects were identified during analysis (Supplementary Figure 1C-D).
Figure 1. scRNA-seq of SULT2B1b KD versus Control KD cells verifies decreased AR activity.
(A) Overview of the number of single cells sequenced. (B) Multidimensional scaling (MDS) plot highlighting the differences due to treatment between single cell groups. “C” indicates Control KD and “KD” indicates SULT2B1b KD. (C) Number of SULT2B1 reads indicating SULT2B1b expression at the single cell level is indicated for the Control KD (Control) or SULT2B1b KD (Knockdown) groups. The upper right-hand corner indicates the percentage of cells in each group with a zero read count. (D) Left: Violin plot representing expression (read count) of AR in scRNA-seq groups; Right: qRT-PCR expression of AR from bulk samples 48 hours after transfection. (E) Violin plots of the indicated AR target genes. Expression (read count) is indicated with adjusted p-values for each gene.
A multidimensional scaling plot demonstrates a degree of separation between cells originating from Control and SULT2B1b KD conditions (Figure 1B). Statistical analysis using a negative binomial distribution followed by the Benjamini-Hochberg procedure identified 2,029 DE genes between the Control and SULT2B1b KD groups. Our analysis determined a reduction, though not significant, in SULT2B1b read counts in SULT2B1b KD versus Control KD treated cells (high number of 0 read counts; Figure 1C) and a significant reduction in AR activity, as seen by reduction in expression of AR target genes (Figure 1D-E). It should be noted that 0 read counts indicate transcript levels below the limit of detection for this assay, rather than no expression. It may also be important to note that AR gene expression at 48 hours remained unchanged in SULT2B1b KD cells compared to Control KD cells in the scRNA-seq dataset, while bulk AR expression by qRT-PCR is reduced after 72 hours of SULT2B1b KD, as we previously reported (Figure 1D,(17)).
Analysis of this scRNA-seq data using Ingenuity Pathway Analysis (IPA) identified numerous significantly altered canonical pathways (Supplementary Figure 2) and putative upstream regulators of the DE genes. Even though the top significantly altered pathways demonstrated a stressed intracellular environment (apoptosis was described in (17)), upregulation of the death receptor signaling pathway was identified as a surprising possible mechanism of SULT2B1b KD-mediated apoptosis (Supplementary Figure 2). Death receptor activation has known implications in prostate cancer and death receptors have been demonstrated to localize in cholesterol-rich domains, but death receptor activation has not been shown to be activated with knockdown of a gene related to cholesterol modification, such as sulfonation.(25, 26) Due to this pathway’s direct association with cell death, we aimed to validate whether SULT2B1b is involved in the regulation of the death receptor signaling pathway and to define the mechanism(s) by which SULT2B1b KD may lead to activation of death receptor signaling.
NF-κB activation is induced by TNF expression within SULT2B1b KD-treated LNCaP cells.
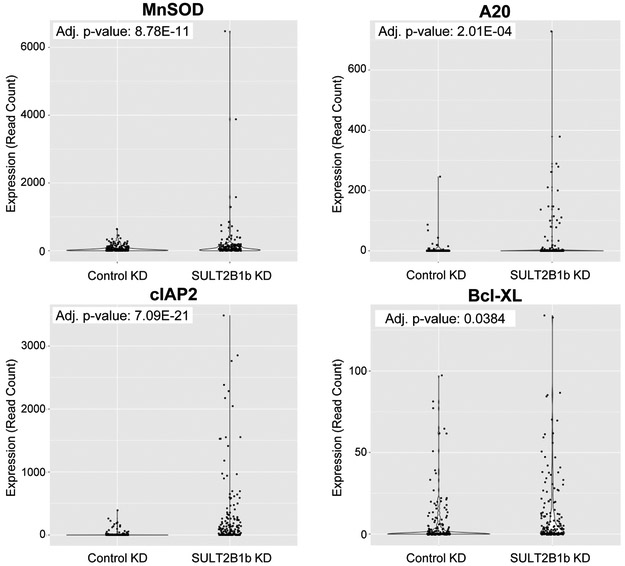
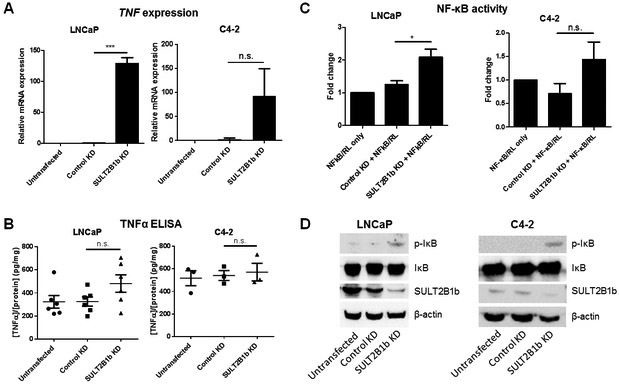
As a contributor to and participant in death receptor signaling, TNF and NF-κB were investigated in the scRNA-seq dataset. Both TNF and the NF-κB complex were predicted to be activated (z-score=4.552, p=1.68E-08 and z-score=4.383, p=2.75E-06, respectively) upstream regulators of many DE genes. Furthermore, four TNF-induced, NF-κB-regulated genes were significantly upregulated in SULT2B1b KD cells, including MnSOD, A20, cIAP2, and Bcl-XL (Figure 2). Further studies using LNCaP and C4–2 cells were conducted to verify the regulation of this pathway by SULT2B1b. SULT2B1b KD in LNCaP cells induces TNF expression and activation of NF-κB, supporting the scRNA-seq data (Figure 3A-D). TNF protein expression is not significantly elevated in LNCaP cells with SULT2B1b KD, but it does trend upward compared to Control KD cells (Figure 3B). In C4–2 cells with SULT2B1b KD, neither TNF expression nor NF-κB activity is significantly increased, but SULT2B1b KD does activate phosphorylation of IκB (Figure 3A-D).
Figure 2. SULT2B1b KD is predicted to activate TNF and NF-κB in scRNA-seq.
Violin plots representing expression (read count) of indicated NF-κB target genes in Control KD versus SULT2B1b KD groups of the scRNA-seq analysis, with adjusted p-value indicated in the upper left-hand corner.
Figure 3. SULT2B1b KD induces TNF expression and activates NF-κB in LNCaP, but not C4-2 cells.
LNCaP and C4-2 cells were transfected with control or SULT2B1b siRNA and evaluated for (A-B) TNF expression or (C-D) NF-κB activity. (A) qRT-PCR for TNF expression 72 hours after siRNA KD. (B) TNF protein expression was quantified by cell lysate-based ELISA 72 hours after siRNA KD. (C) Luciferase assay completed 48 hours after siRNA transfection. NF-κB luciferase activity was normalized to Renilla luciferase. Samples were further normalized to control wells without siRNA transfection. Bars indicate the mean +/− SEM of at least four independent experiments using a Student’s t-test. (D) Western blot of indicated proteins 72 hours after siRNA KD.
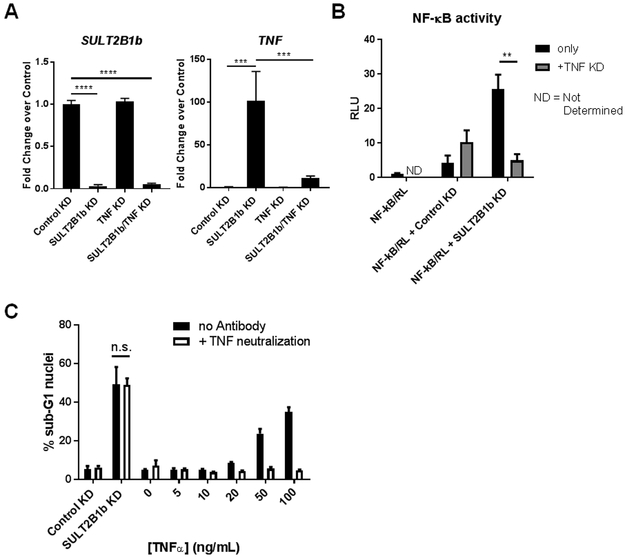
To determine if NF-κB activation by SULT2B1b KD is due to the induction of TNF expression in LNCaP cells, a consecutive KD of TNF in SULT2B1b KD cells was performed. These data demonstrate that the loss of TNF expression in SULT2B1b KD cells abrogates the induction of NF-κB activity via luciferase assay (Figure 4A-B). Understanding that LNCaP cells are sensitive to TNF-mediated cell death, the data presented in Figures 3 and 4B call into question whether the TNF produced as a result of SULT2B1b KD is the cause of apoptosis in these cells. TNF neutralization experiments were conducted to address this question; these studies demonstrate that antibody-based neutralization of TNF in SULT2B1b KD cells has no effect on cell death by analysis of sub-G1 nuclei, while exogenous TNF can be efficiently neutralized in these cells (Figure 4C). Thus, TNF is produced by SULT2B1b KD in LNCaP cells leading to NF-κB activation, but TNF neutralization in these cells cannot block apoptosis induced by SULT2B1b KD.
Figure 4. TNF expression in SULT2B1b KD cells is the cause of NF-κB activation, but not induction of cell death.
(A) pRT-PCR indicates SULT2B1b and TNF expression in LNCaP cells that were consecutively transfected with SULT2B1b siRNA followed by TNF siRNA. Significance was determined with a one-way ANOVA with a multiple comparisons test. (B) NF-κB luciferase assay was completed 48 hours after siRNA transfection and relative luciferase units (RLU) are shown. Bars indicate the mean +/− SEM of at least triplicate samples by two-way ANOVA considering only Control KD and SULT2B1b KD groups and the graph is representative of two independent experiments. (C) LNCaP cells were pre-treated with TNF neutralizing antibody (40 µg/mL) and then transfected with Control or SULT2B1b siRNA or treated with indicated concentrations of exogenous TNFα. Approximately 72 hours post-siRNA transfection, cells were harvested and prepared for cell cycle analysis by flow cytometry. Bars indicate the average percent sub-G1 nuclei +/− SEM of triplicate samples. This graph is representative of at least two independent experiments.
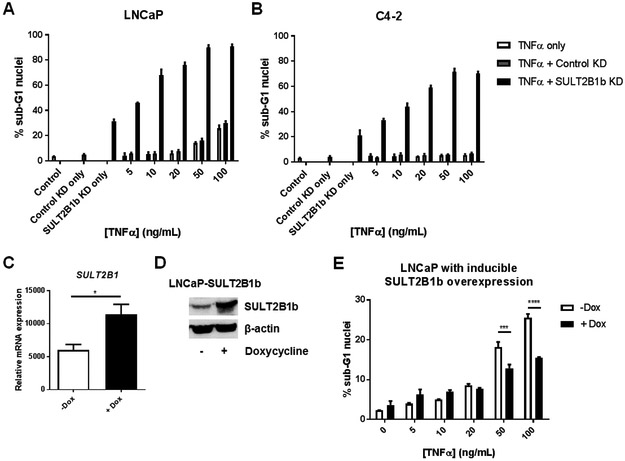
SULT2B1b expression alters sensitivity to TNF-mediated cell death in both LNCaP and C4-2 cells.
While SULT2B1b KD activates NF-κB through expression of TNF, this does not seem to be a direct mechanism of apoptosis in these cells (Figure 3–4). Data presented in Figure 3 also demonstrate that SULT2B1b KD has differing effects on TNF expression and NF-κB activity in LNCaP and C4–2 cell lines. We postulated that upregulation of genes related to death receptor signaling would sensitize SULT2B1b KD cells to death ligand stimuli. Further investigation of the impact of SULT2B1b KD in LNCaP and C4–2 cells demonstrates that SULT2B1b KD enhances cell death in response to exogenous TNF in both LNCaP and C4–2 cells compared to controls, determined by cell cycle analysis (Figure 5A-B). A dose response showing enhanced cell death can also be seen in SULT2B1b KD cells of both cell lines when increased concentrations of TNF is added (Figure 5A-B).
Figure 5. SULT2B1b regulates sensitivity to TNF-mediated cell death in LNCaP and C4-2 cells.
LNCaP (A) or C4-2 (B) cells were transfected with Control or SULT2B1b siRNA prior to addition of indicated concentrations of TNF. All cells were harvested 72 hours later and prepared for cell cycle analysis by flow cytometry. Bars indicate the average percent sub-G1 nuclei +/− SEM of triplicate wells. For all TNF-treated cells in (A) and (B), SULT2B1b KD significantly enhances sub-G1 nuclei percentage over Control KD cells in both LNCaP and C4-2 cells by two-way ANOVA (p<0.0001). Graphs are representative of three independent experiments. (C-D) SULT2B1b expression was evaluated by (C) qRT-PCR and (D) western blot in LNCaP-SULT2B1b cells. (E) LNCaP-SULT2B1b cells were treated with doxycycline prior to addition of indicated concentrations of TNF. After 72 hours, all cells were harvested and prepared for cell cycle analysis by flow cytometry. Bars indicate the mean of % sub-G1 nuclei +/− SEM of triplicate wells evaluated by two-way ANOVA with multiple comparisons test. The graph is representative of duplicate experiments.
Following our previous report that SULT2B1b KD reduces AR expression and activity in LNCaP cells, we aimed to determine if TNF treatment in LNCaP or C4–2 cells with SULT2B1b KD cells exacerbates the effect on AR activity. In LNCaP cells with SULT2B1b KD, treatment of low-dose TNF resulted in a significant interaction between SULT2B1b KD and TNF treatment in reducing both AR (p=0.0361) and PSA (p=0.0421) expression, while no significant interaction was determined between SULT2B1b KD and TNF treatment in C4–2 cells by two-way ANOVA (Supplemental Figure 3). However, C4–2 cells with SULT2B1b KD and TNF treatment did significantly reduce PSA expression compared to Control KD cells (Supplemental Figure 3B). Thus, while TNF and SULT2B1b KD are independently capable of inhibiting AR activity as previously reported,(6, 17) the combination results in the greatest reduction in AR activity, although interaction between the molecules occurs only in LNCaP cells and not in a model of CRPC.
The enhanced sensitivity to TNF-induced apoptosis does not seem to be due to altered expression of TNF receptor expression, since no significant increase in TNFR1 or TNFR2 can be observed by flow cytometry (Supplemental Figure 4). Previous reports suggested that loss of TNF receptor-associated adaptor protein (TRADD) expression is responsible for decreased sensitivity of advanced prostate cancer cells, such as C4–2, to TNF-mediated cell death.(6) Western blot and ELISA analyses of TRADD expression in SULT2B1b KD versus Control KD cells indicated that TRADD expression is increased with SULT2B1b KD treatment in LNCaP, but not C4–2, cells (Supplemental Figure 5). Thus, it seems that SULT2B1b may modulate TRADD levels and could account for altered sensitivity to TNF in LNCaP cells, but cannot explain the enhanced sensitivity of C4–2 cells to TNF treatment.
Additional studies were completed to determine whether the SULT2B1b KD-mediated increased sensitivity to TNF-induced cell death was simply an artifact of cells that were previously demonstrated to be undergoing apoptosis, or whether SULT2B1b is a regulator of this death receptor pathway. To address this question, LNCaP-SULT2B1b cells were produced to induce stable SULT2B1b overexpression with the addition of doxycycline. Experiments with LNCaP-SULT2B1b demonstrate that SULT2B1b modulates TNF-mediated cell death, as the addition of doxycycline results in significantly reduced sensitivity to exogenous TNF treatment compared to no doxycycline induction (Figure 5C-E). These studies confirm a role for SULT2B1b as a regulator of prostate cancer cell sensitivity to TNF-mediated cell death.
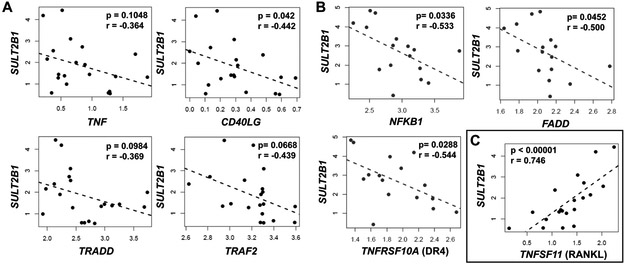
SULT2B1b expression levels inversely correlate with TNF-related genes in human patients.
The data presented herein suggest that SULT2B1b KD induces many intracellular changes in prostate cancer cells, specifically increased activation of death receptor signaling and sensitivity to TNF. To investigate whether SULT2B1b may also play a role in TNF sensitivity or death receptor signaling in the clinical setting, we obtained metastatic prostate cancer data from cBioPortal for gene expression correlations. SULT2B1 was negatively correlated with multiple TNF-related genes in bone metastasis samples with no prior abiraterone or enzalutamide treatment, including death receptor ligands TNF (r=−0.364) and CD40LG (r=−0.442), as well as TNFR-associated genes TRADD (r=−0.369) and TRAF2 (r=−0.439) (Figure 6A). Furthermore, SULT2B1b negatively correlates with TNF-related genes in lymph node-derived CRPC samples with prior therapy, including NFKB1 (r=−0.533), apoptotic regulator FADD (r=−0.500), and death receptor 4 (DR4, r=−0.544) (Figure 6B). These data demonstrate that SULT2B1b expression is inversely correlated with TNF and related genes in metastatic prostate cancer and CRPC patient samples, as was suggested by cell line data described herein. A complete list of significant negative correlations between SULT2B1 and TNF-related genes in bone-derived samples is listed in Supplemental Table 1. Interestingly, one gene, TNFSF11, or RANKL, has a strong positive correlation with SULT2B1 (r=0.746) in human prostate cancer bone metastasis specimens (Figure 6C). Elevated expression of RANKL has a known association with bone metastasis niche formation and cancer progression by promoting osteoclast-mediated bone resorption.(27, 28) Together, these data demonstrate that SULT2B1b expression in prostate cancer cells is inversely associated with TNF-related genes, but positively associated with a gene involved with osteoclast activation and cancer cell survival in bone.
Figure 6. SULT2B1b expression correlates with numerous TNF-related genes in human patients.
(A) Gene co-expression indicates SULT2B1 negatively correlates with TNF, CD40LG, TRADD, and TRAF2 in bone marrow-derived metastatic prostate cancer samples from individuals with no prior treatment. (B) Co-expression of SULT2B1 with NFKB1, FADD, and DR4 determines significant negative correlations with each of these genes in lymph node-derived CRPC samples. (C) Gene co-expression of SULT2B1 with RANKL demonstrates a strong positive correlation in bone marrow-derived prostate cancer samples. Pearson correlation coefficients (r) and p-values determined by a permutation test (p) are indicated for each correlation plot. Dots indicate samples obtained from individual patients.
DISCUSSION
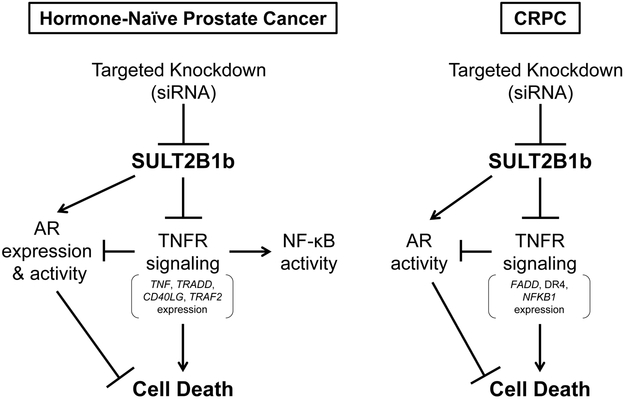
We provide evidence herein demonstrating that siRNA-mediated knockdown of SULT2B1b in LNCaP cells results in vast alterations in multiple signaling pathways, as detected by scRNA-seq analysis. A model illustrating the proposed role of SULT2B1b on death receptor signaling is shown in Figure 7, where SULT2B1b is shown to be a key modulator of cell death via NF-κB and AR.
Figure 7.
Diagram of proposed SULT2B1b influence on death receptor signaling and AR activity in advanced, hormone-naive prostate cancer and CRPC cells.
One goal of this analysis was to understand the heterogeneity of SULT2B1b expression within each treatment group. Interestingly, SULT2B1 expression levels were both low and heterogeneous within cells of both groups (Figure 1C), therefore expression of this gene could not be used for direct correlation with other genes of interest in this single-cell analysis. Analysis of SULT2B1 expression across many tissue types using the GTEx Portal supports that transcription of this gene is low in prostate cells (Supplemental Figure 6) as our data in Figure 1 suggests, but the large number of cells with zero expression may also be due to the high dropout rates observed with single-cell analysis.(29–31) Even though expression of SULT2B1 is relatively low, it is notable that loss of SULT2B1b remains profound with over 2000 DE genes.
We previously demonstrated that SULT2B1b KD induces apoptosis in LNCaP cells and, as expected, intracellular stress- and apoptosis-related pathways were significantly altered including protein ubiquitination, mitochondrial dysfunction, role of BRCA1 in DNA damage response, p53 signaling, and induction of apoptosis by HIV1 (Supplemental Figure 2). It was surprising that SULT2B1b KD cells yield significant alterations in genes of the canonical death receptor signaling pathway compared to Control KD cells, but alterations in genes identified in this pathway, including TNFSF15 (TL1), TNFSF10 (TRAIL), and DAXX genes did not contribute to SULT2B1b KD-induced apoptosis (Supplemental Figure 7).
Further studies in vitro determined that SULT2B1b KD induces TNF expression, causing NF-κB activation in LNCaP cells, and these effects were predicted by the scRNA-seq data. Surprisingly, SULT2B1b KD significantly increased expression of four anti-apoptotic genes illustrating the complex intracellular effects of SULT2B1b (Figure 2). The induction of TNF expression by LNCaP cells by SULT2B1b KD does not appear to be the cause of SULT2B1b KD-mediated apoptosis, since TNF neutralization does not abrogate apoptosis (Figure 4C). Thus, we conclude that the induction of TNF by SULT2B1b KD may not directly induce apoptosis through the death receptor at the protein level, but increased TNF expression does influence downstream signaling in SULT2B1b KD cells. SULT2B1b KD and overexpression studies (Figure 5) indicate that SULT2B1b is a novel regulator of prostate cancer sensitivity to TNF-mediated cell death. This is further supported by data in Figure 6 and Supplemental Table 1 which indicate SULT2B1b expression inversely correlates with expression of numerous TNF-associated genes in metastatic prostate cancer.
It has been demonstrated that TNF signaling can reduce AR expression and activity.(32) We previously showed that SULT2B1b KD decreases AR activity, but this occurs to a lesser extent in CRPC (Supplemental Figure 3,(17)). It is not likely that TNF expression induced by SULT2B1b KD is the cause of AR inhibition, since no significant elevation of TNF protein was detected and neutralization of TNF had no effect on apoptosis (Figures 3–4). Thus, the exact mechanism of AR regulation by SULT2B1b is not understood, but SULT2B1b KD does aid in reducing PSA expression in TNF-treated C4–2 cells. Perhaps SULT2B1b KD causes intracellular changes which enable TNF treatment to have a greater effect on AR activity in CRPC.
While therapeutic applications of TNF have been diminished in recent years, it remains an important molecule within the tumor microenvironment that could impact therapeutic responses. Circulating TNF levels have been demonstrated to increase in CRPC patients compared to patients with localized prostate cancer.(33) If SULT2B1b expression/activity maintains TNF resistance in prostate cancer cells as suggested by the data herein, then targeting SULT2B1b may sensitize advanced prostate cancer cells to TNF in the microenvironment. However, it should be noted that TNF itself may impact SULT2B1b expression, since low-dose treatment of TNF reduces SULT2B1b expression in C4–2 cells (Supplemental Figures 3B, 5D). Thus, it is possible that elevated TNF in the microenvironment suppresses SULT2B1b expression in CRPC. Understanding the inflammatory status of individual patients with this disease may be critical for determining whether this targeting approach will alter the viability/cytotoxicity balance in cancer cells, since elevated expression of both SULT2B1b and TNF may be necessary for treatment efficacy.
It remains possible that SULT2B1b influences the death receptor signaling pathway through its function in cholesterol metabolism as a regulator of LXR.(17, 34) Cholesterol-rich lipid rafts are known to stabilize intracellular signaling, but are also likely sites for the initiation of death receptor signaling.(16, 25) While total cholesterol levels are not significantly altered by SULT2B1b KD in LNCaP cells (data not shown), SULT2B1b KD does reduce intracellular levels of cholesterol sulfate.(17) It is not known if cholesterol sulfate (versus unmodified cholesterol) is important in lipid raft stability, but perturbations in lipid rafts certainly lead to vast disruptions of many canonical signaling pathways, including TNFR complex formation and signaling.(35)
The data demonstrated herein also provide evidence that SULT2B1b expression inversely correlates with expression of multiple TNF-related genes indicating a protective role of SULT2B1b in metastatic hormone-naïve and castration-resistant prostate cancer, but SULT2B1b also had a strong positive correlation with osteoclast regulator, RANKL (Figure 6C). These data suggest that SULT2B1b may be involved in both evading death ligand-induced apoptosis in prostate cancer cells as well as establishing a prostate cancer bone metastatic niche during progression of the disease. This supports a role for SULT2B1b in establishing metastatic lesions within bone in patients with hormone-naïve prostate cancer. Targeting SULT2B1b would perhaps limit osteoclast formation, but this strategy would likely need to be used in combination with other targeting strategies to offset activating signaling pathways in metastatic disease, such as lipid raft stability with statin drugs.(36–38) In patients with high TNF levels, such as in CRPC, SULT2B1b targeting could aid in sensitizing CRPC cells to exogenous TNF and/or inhibiting AR activity causing cell death. SULT2B1b should be considered as a therapeutic target in advanced prostate cancer and its expression status could be used along with other information (i.e. TNF level, inflammatory status, statin use or other therapeutic history, etc.) to guide patient treatment options.
Supplementary Material
Implications: These data suggest that SULT2B1b expression enhances resistance to TNF and may promote prostate cancer.
ACKNOWLEDGMENTS
These studies were supported by the Department of Defense Prostate Cancer Research Program grant #W81XWH-14–1-0588 (TL Ratliff, AD Mesecar, SA Crist, and RE Vickman) and the Walther Cancer Foundation (TL Ratliff). The authors would like to thank the Prostate Cancer Foundation for generously supporting and DDC Medical for conducting authentication of our cell lines. We gratefully acknowledge the support of the Purdue Genomics Core Facility (specifically Phillip SanMiguel and Paul Parker), Purdue Flow Cytometry and Cell Separation facility, Bioinformatics shared resource, Purdue University Center for Cancer Research (NIH grant P30 CA023168), IU Simon Cancer Center (NIH grant P30 CA082709, NA Lanman), and the Walther Cancer Foundation. We also greatly appreciate Dr. Bennett Elzey and members of the Ratliff lab for continued suggestions and support of this project.
Abbreviations List:
- ANOVA
analysis of variance
- AR
androgen receptor
- CRPC
castration resistant prostate cancer
- DAXX
death domain associated protein
- DE
differentially expressed
- FBS
fetal bovine serum
- FDR
false discovery rate
- IPA
Ingenuity Pathway Analysis
- KD
knockdown
- LXR
liver X receptor
- PSA
prostate specific antigen
- RANKL
TNF ligand superfamily member 11
- RNAi
RNA interference
- RPKM
reads per kilobase per million mapped reads
- scRNA-seq
single-cell mRNA sequencing
- SULT2B1b
sulfotransferase 2B1b; cholesterol sulfotransferase
- TL1
TNF superfamily member 15
- TNF
tumor necrosis factor
- TNFR
TNF receptor
- TRADD
TNF receptor associated death domain
- TRAIL
TNF-related apoptosis-inducing ligand
Footnotes
Conflict of Interest Statement: The authors of this manuscript declare no potential conflicts of interest.
REFERENCES
- 1.Society AC (2018) Cancer Facts & Figures 2018. (Atlanta: American Cancer Society; ). [Google Scholar]
- 2.An J, et al. (2003) Drug interactions between the proteasome inhibitor bortezomib and cytotoxic chemotherapy, tumor necrosis factor (TNF) alpha, and TNF-related apoptosis-inducing ligand in prostate cancer. Clin Cancer Res 9(12):4537–4545. [PubMed] [Google Scholar]
- 3.Norris JS, et al. (2001) The use of Fas Ligand, TRAIL and Bax in gene therapy of prostate cancer. Curr Gene Ther 1(1):123–136. [DOI] [PubMed] [Google Scholar]
- 4.O’Kane HF, et al. (2006) Targeting death receptors in bladder, prostate and renal cancer. J Urol 175(2):432–438. [DOI] [PubMed] [Google Scholar]
- 5.Carswell EA, et al. (1975) An endotoxin-induced serum factor that causes necrosis of tumors. Proc Natl Acad Sci U S A 72(9):3666–3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang D, et al. (2009) Reduced tumor necrosis factor receptor-associated death domain expression is associated with prostate cancer progression. Cancer Res 69(24):9448–9456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chopra DP, Menard RE, Januszewski J, & Mattingly RR (2004) TNF-alpha-mediated apoptosis in normal human prostate epithelial cells and tumor cell lines. Cancer Lett 203(2):145–154. [DOI] [PubMed] [Google Scholar]
- 8.Kimura K, Bowen C, Spiegel S, & Gelmann EP (1999) Tumor necrosis factor-alpha sensitizes prostate cancer cells to gamma-irradiation-induced apoptosis. Cancer Res 59(7):1606–1614. [PubMed] [Google Scholar]
- 9.Chung TD, et al. (1998) Tumor necrosis factor-alpha-based gene therapy enhances radiation cytotoxicity in human prostate cancer. Cancer Gene Ther 5(6):344–349. [PubMed] [Google Scholar]
- 10.Pirtskhalaishvili G, Shurin GV, Esche C, Trump DL, & Shurin MR (2001) TNF-alpha protects dendritic cells from prostate cancer-induced apoptosis. Prostate Cancer Prostatic Dis 4(4):221–227. [DOI] [PubMed] [Google Scholar]
- 11.Harada S, et al. (2001) Long-term exposure of tumor necrosis factor alpha causes hypersensitivity to androgen and anti-androgen withdrawal phenomenon in LNCaP prostate cancer cells. Prostate 46(4):319–326. [DOI] [PubMed] [Google Scholar]
- 12.Mizokami A, Gotoh A, Yamada H, Keller ET, & Matsumoto T (2000) Tumor necrosis factor-alpha represses androgen sensitivity in the LNCaP prostate cancer cell line. J Urol 164(3 Pt 1):800–805. [DOI] [PubMed] [Google Scholar]
- 13.Lu L, et al. (2012) Gambogic acid inhibits TNF-alpha-induced invasion of human prostate cancer PC3 cells in vitro through PI3K/Akt and NF-kappaB signaling pathways. Acta Pharmacol Sin 33(4):531–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Radhakrishnan P, et al. (2011) TNFalpha enhances the motility and invasiveness of prostatic cancer cells by stimulating the expression of selective glycosyl- and sulfotransferase genes involved in the synthesis of selectin ligands. Biochem Biophys Res Commun 409(3):436–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yue S, et al. (2014) Cholesteryl ester accumulation induced by PTEN loss and PI3K/AKT activation underlies human prostate cancer aggressiveness. Cell Metab 19(3):393–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freeman MR & Solomon KR (2004) Cholesterol and prostate cancer. J Cell Biochem 91(1):54–69. [DOI] [PubMed] [Google Scholar]
- 17.Vickman RE, et al. (2016) Cholesterol Sulfonation Enzyme, SULT2B1b, Modulates AR and Cell Growth Properties in Prostate Cancer. Mol Cancer Res 14(9):776–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hsu CC & Hu CD (2012) Critical role of N-terminal end-localized nuclear export signal in regulation of activating transcription factor 2 (ATF2) subcellular localization and transcriptional activity. J Biol Chem 287(11):8621–8632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gordon AaH G (2010) FASTX-Toolkit. [Google Scholar]
- 20.Andrews S (2010) FastQC. [Google Scholar]
- 21.Kim D, et al. (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14(4):R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trapnell C, Pachter L, & Salzberg SL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25(9):1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anders S, Pyl PT, & Huber W (2015) HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31(2):166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robinson D, et al. (2015) Integrative clinical genomics of advanced prostate cancer. Cell 161(5):1215–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Legler DF, Micheau O, Doucey MA, Tschopp J, & Bron C (2003) Recruitment of TNF receptor 1 to lipid rafts is essential for TNFalpha-mediated NF-kappaB activation. Immunity 18(5):655–664. [DOI] [PubMed] [Google Scholar]
- 26.Lewis AK, et al. (2016) Death Receptor 5 Networks Require Membrane Cholesterol for Proper Structure and Function. J Mol Biol 428(24 Pt A):4843–4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones DH, et al. (2006) Regulation of cancer cell migration and bone metastasis by RANKL. Nature 440(7084):692–696. [DOI] [PubMed] [Google Scholar]
- 28.Li X, et al. (2014) Potential role of the OPG/RANK/RANKL axis in prostate cancer invasion and bone metastasis. Oncol Rep 32(6):2605–2611. [DOI] [PubMed] [Google Scholar]
- 29.Li WV & Li JJ (2018) An accurate and robust imputation method scImpute for single-cell RNA-seq data. Nat Commun 9(1):997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ziegenhain C, et al. (2017) Comparative Analysis of Single-Cell RNA Sequencing Methods. Mol Cell 65(4):631–643 e634. [DOI] [PubMed] [Google Scholar]
- 31.Kharchenko PV, Silberstein L, & Scadden DT (2014) Bayesian approach to single-cell differential expression analysis. Nat Methods 11(7):740–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ko S, Shi L, Kim S, Song CS, & Chatterjee B (2008) Interplay of nuclear factor-kappaB and B-myb in the negative regulation of androgen receptor expression by tumor necrosis factor alpha. Mol Endocrinol 22(2):273–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chadha KC, et al. (2014) New serum biomarkers for prostate cancer diagnosis. Clin Cancer Investig J 3(1):72–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen W, Chen G, Head DL, Mangelsdorf DJ, & Russell DW (2007) Enzymatic reduction of oxysterols impairs LXR signaling in cultured cells and the livers of mice. Cell Metab 5(1):73–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muppidi JR, Tschopp J, & Siegel RM (2004) Life and death decisions: secondary complexes and lipid rafts in TNF receptor family signal transduction. Immunity 21(4):461–465. [DOI] [PubMed] [Google Scholar]
- 36.Zhuang L, Kim J, Adam RM, Solomon KR, & Freeman MR (2005) Cholesterol targeting alters lipid raft composition and cell survival in prostate cancer cells and xenografts. J Clin Invest 115(4):959–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhuang L, Lin J, Lu ML, Solomon KR, & Freeman MR (2002) Cholesterol-rich lipid rafts mediate akt-regulated survival in prostate cancer cells. Cancer Res 62(8):2227–2231. [PubMed] [Google Scholar]
- 38.Srivatsav AT, Mishra M, & Kapoor S (2018) Small-Molecule Modulation of Lipid-Dependent Cellular Processes against Cancer: Fats on the Gunpoint. Biomed Res Int 2018:6437371. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.