Abstract
Immune checkpoint inhibitors (ICIs) are effective in treating a variety of malignancies, including metastatic bladder cancer. A generally accepted hypothesis suggests that ICIs induce tumor regressions by reactivating a population of endogenous tumor-infiltrating lymphocytes (TILs) that recognize cancer neoantigens. Although previous studies have identified neoantigen-reactive TILs from several types of cancer, no study to date has shown whether or not neoantigen-reactive TILs can be found in bladder tumors. To address this, we generated TIL cultures from patients with primary bladder cancer and tested their ability to recognize tumor-specific mutations. We found that CD4+ TILs from one patient recognized mutated C-terminal binding protein 1 (CTBP1Q277R) in an MHC class II-restricted manner. This finding suggests that neoantigen-reactive TILs reside in bladder cancer, which may help explain the effectiveness of immune checkpoint blockade in this disease, and also provides a rationale for the future use of adoptive T-cell therapy targeting neoantigens in bladder cancer.
Introduction
Urothelial carcinoma of the bladder is among the ten most common malignancies worldwide, with an estimated 81,190 new cases and 17,240 deaths per year in the United States (1). Although the early stage disease, which constitutes the majority of newly diagnosed cases, is curable with surgery, there have been no curative treatments for patients with metastases, whose 5-year overall survival remains around 15% (2).
Muscle-invasive bladder cancer (MIBC) is managed with radical cystectomy with neoadjuvant cisplatin-based chemotherapy in selected patients. For patients with metastatic disease, systemic chemotherapy is the standard of care, with excellent but short-lived response rates (2, 3). In addition to these modalities, other therapies have been successfully used in treatment of bladder cancer. In high grade non-muscle-invasive bladder cancer (NMIBC), intravesical instillation of Bacillus Calmette–Guérin (BCG), an attenuated strain of Mycobacterium bovis, was found effective in preventing relapse and progression of localized disease after transurethral resection of the bladder tumor (TURBT) (4, 5). Although its exact mechanism of action is still unclear, BCG was shown to elicit sustained inflammation and recruitment of T lymphocytes to the tumors (6–8).
In recent years, immune checkpoint inhibitors (ICIs) have emerged as an effective immunotherapy for several cancer types, including metastatic bladder cancer (9). ICIs are monoclonal antibodies that block the function of inhibitory molecules, such as PD-1 (programmed cell death protein 1) expressed on the surface of T cells, or its inhibitory ligand PD-L1 (programmed cell death protein 1 ligand 1) expressed on the surface of tumor cells or antigen presenting cells (APCs). Blocking this inhibitory signaling can reactivate T cells to induce anti-tumor immune responses and subsequent tumor regression (10). When administered to patients with metastatic bladder cancer whose disease progressed after standard platinum-based chemotherapy, or who could not tolerate it, ICIs led to durable tumor regressions in 15–20% of cases, with up to 11% complete responses in unselected patients (11–19). These findings established a pivotal role for ICIs in the treatment of metastatic bladder cancer and provided valuable insight into its immunogenicity.
Studies in several cancer types demonstrated a positive correlation between the number of tumor mutations and the responses to ICIs (20–23). Next to melanoma and selected types of lung cancer, bladder carcinoma exhibits the fourth highest mutation burden among all common malignancies, with a median of 155 to 219 mutations per tumor sample across several studies (24–27). Among patients with metastatic bladder cancer treated with atezolizumab, an anti-PD-L1 antibody, the median tumor mutation number was significantly higher in patients who responded to therapy than in those who did not (11, 12, 28). A generally accepted hypothesis suggests that malignancies with a higher mutation burden are more immunogenic, as they are predicted to present a larger number of neoantigens in the MHC-restricted context. Accordingly, ICIs are thought to reactivate tumor-infiltrating lymphocytes (TILs), which can recognize these neoantigens and induce tumor regressions (20, 21, 29–32).
We have previously shown that neoantigen-reactive TILs can be isolated from metastatic melanoma and gastrointestinal cancers, and can lead to durable tumor regressions once in vitro expanded and transferred back into the patients (33–37). Although TILs could be successfully grown from bladder tumors in our previous study (38), no study to date has shown whether or not TILs from bladder tumors can recognize neoantigens. To explore this, we first generated polyclonal TIL cultures from five patients with primary bladder tumors and co-cultured them with autologous APCs presenting the products of cancer mutations. In this study, we describe the isolation and characterization of a neoantigen-reactive TIL population from a patient with primary localized urothelial carcinoma of the bladder.
Materials and Methods
Patients
Five patients with primary localized urothelial carcinoma of the bladder were evaluated and treated at the Urologic Oncology Branch at the National Cancer Institute (NCI). All patients were enrolled on protocols approved by the NCI Institutional Review Board, and they had provided their written informed consent for this study.
Tumor infiltrating lymphocytes
Tumor samples were obtained via TURBT or bladder diverticulectomy. TILs were cultured from tumor fragments following a previously described approach (39). Briefly, tumor tissue was dissected free of hemorrhagic and necrotic areas and cut into approximately 1×1 mm fragments (N=12 or 24), which were then plated individually in 24-well plates and cultured in 2 mL of RPMI medium supplemented with 2 mM L-glutamine, 25 mM HEPES, 10 μg/ml gentamicin (all from Life Technologies, Carlsbad, CA), 10% human AB serum and 6000 IU/ml of IL-2 (Prometheus, San Diego, CA) for 6–8 weeks. Medium was replenished twice weekly; the wells were split in 1:2 fashion when fully confluent and cryopreserved until further use.
Whole exome sequencing
Cancer-specific mutations were identified from tumor samples using whole exome sequencing (WES), as described previously (35). Briefly, genomic DNA was first extracted from tumors and matched normal blood using a Maxwell instrument (Promega, Madison, WI). Next, WES libraries were prepared from genomic DNA (3 μg/sample) using SureSelectXT Target Enrichment System coupled with Human All Exon V4 target bait (Agilent Technologies, Santa Clara, CA). Libraries from Patient 2 (1st resection) and Patient 5 were prepared and sequenced on an Illumina HiSeq2000 sequencer (Axeq/Macrogen USA, Rockville, MD). Libraries from Patient 1, 3 and 4 were prepared and sequenced in-house on a NextSeq 500 desktop sequencer following the manufacturer’s instructions (Illumina, San Diego, CA). Sequencing reads were aligned to human genome build 19 using Novoalign MPI (http://www.novocraft.com/). Duplicates were marked using Picard’s MarkDuplicates tool; in/del realignment and base recalibration was carried out according to the GATK best practices workflow (https://www.broadinstitute.org/gatk/). After the data cleanup, pileup files were created using samtools mpileup (http://samtools.sourceforge.net). Somatic variants were called using Varscan2 (http://varscan.sourceforge.net) according to the following criteria: tumor and normal read counts of 10 or greater, variant allele frequency of 10% or greater, and tumor variant reads of 4 or more. Finally, variants were annotated using Annovar (http://annovar.openbioinformatics.org). Tumor-specific mutations for each patient are listed in Supplementary Table 1.
Tandem minigene and peptide libraries
TMGs were constructed as described previously (40). For non-synonymous point mutations, each mutated amino acid was flanked bilaterally by a sequence encoding 12 wild type (WT) amino acids to generate an individual minigene. For each frameshift mutation, a minigene was designed to contain preceding 12 WT amino acids followed by mutated amino acids in the new reading frame, which terminated at the new stop codon. Next, up to twelve minigenes were linked together to generate tandem minigenes, which were codon-optimized, synthesized and ligated into a pcDNA3.1 vector using an In-Fusion HD EcoDry Cloning Kit (Clontech/Takara, Mountain View, CA). TMG RNA was made by in vitro transcription using a HiScribe T7 Quick High Yield RNA Synthesis Kit (New England BioLabs, Ipswich, MA). RNA samples were purified using RNeasy Kit (Qiagen, Germantown, MD), quantified by spectrophotometry, and stored at −80°C until further use. The amino acid sequences of each TMG used in this study are shown in Supplementary Table 1.
Crude or HPLC-purified (>90% purity) 25-mer peptides, each encoding a point mutation flanked on both sides with 12 WT amino acids, were synthesized by GenScript (Piscataway, NJ) as lyophilized power, resuspended in DMSO and stored at −20°C until use.
Antigen presenting cells
To generate Epstein–Barr virus-transformed B (EBV-B) cells, peripheral blood samples were collected in Vacutainer CPT (Cell Preparation Tubes with sodium heparin) (BD Bioscience, San Jose, CA), followed by isolation of peripheral blood mononuclear cells (PBMCs) according to the manufacturer’s instructions. Next, 1 × 107 PBMCs were cultured in 4 ml of complete RPMI medium [RPMI 1640 with 10% fetal bovine serum (SAFC, St. Louis, MO) and Antibiotic-Antimycotic (Life Technologies)] with addition 1 ml of B95–8 culture supernatant containing EBV (ATCC, Manassas, VA) and 0.5 μg/ml of cyclosporine A (Sigma Aldrich, St. Louis, MO) for approximately 4–6 weeks. Medium with cyclosporine A was replenished as needed. The cells were further expanded in cyclosporine-free medium and cryopreserved until future use.
For Patient 2, sufficient PBMCs were obtained via leukapheresis, which enabled us to generate autologous dendritic cells (DCs). CD14+ monocytes from PBMC were purified using magnetic anti-CD14 microbeads (BD Biosciences, San Jose, CA) and cultured in Petri dishes (1 × 107 cells / dish) with complete RPMI medium supplemented with 50 ng/ml GM-CSF and 20 ng/ml IL- 4 (PeproTech, Rocky Hill, NJ). 5 ml of fresh cytokine-supplemented medium was added on day 3. Non-adherent and loosely adherent cells were harvested and used for experiments on day 5 or 6.
Library Screening
Recognition of putative neoantigens was assessed by performing overnight co-cultures of T cells and APCs, followed by measuring interferon gamma (IFN-γ) production, or upregulation of activation markers 4–1BB (CD137) and OX40 (CD134) on the surface of T cells.
For peptide library screening, APCs were first incubated with pools of up to 12 individual peptides (final concentration ~1 μM per peptide) for 24 hours at 37 °C and then washed twice prior to the co-culture. For TMG library screening, APCs were electroporated with TMG RNA using Neon Transfection System (Life Technologies) according to the manufacturer’s instructions. Briefly, APCs were first washed with PBS and resuspended in R electroporation buffer at 1 × 107 cells/ml. Next, 10 μl of cellular suspension was mixed with 1 μg of TMG RNA, and electroporated using 1500 V × 30 ms × 1 pulse for DCs or 1600 V × 10 ms × 3 pulses for EBV-B cells. Electroporated cells were rested in Opti-MEM (Life Technologies) for 1 hour at 37 °C prior to the co-culture.
Cryopreserved TILs or TCR-transduced T cells were first rested overnight in complete AIM V [AIM V CTS medium (Life Technologies) with 5% human AB serum (Valley Biomedical, Winchester, VA) and Antibiotic-Antimycotic] supplemented with IL-2 (6000 IU/ml for TILs and 1200 IU/ml for TCR-transduced T cells). The following day, cells were washed twice to remove excess IL-2 and plated (1 × 105 cells/well) with equal number of APCs into U-bottom 96-well plates (for subsequent ELISA and flow cytometry) or MultiScreen-IP filter plates (for ELISPOT). Co-cultures were incubated overnight at 37 °C and 5% CO2. Cell Stimulation Cocktail [phorbol 12-myristate 13-acetate (PMA) and ionomycin] (Affymetrix, San Diego, CA) was used as a positive control in 1:1000 v/v ratio.
Detection of cytokines in lymphocyte co-culture assays
The secretion of IFN-γ, GM-CSF, TNF-α, IL-2 and IL-4 from T cells after overnight co-cultures was measured by enzyme-linked immunosorbent assays (ELISA). Briefly, co-culture plates were first spun at 300 × g for 2 minutes at room temperature, followed by measuring cytokine concentration in the co-culture supernatants using respective ELISA kits (all from ThermoFisher Scientific, Waltham, MA). Cell pellets were resuspended in PBS with 0.5% FBS and set aside for flow cytometric analysis. ELISA plates were read on Spectramax 190 microplate spectrophotometer (Molecular Devices, Sunnyvale, CA) and analyzed using SoftMax Pro 6.2.2 software (Molecular Devices).
Flow cytometric analysis
For all experiments, cells were stained with antibodies diluted in PBS/0.5% FBS in 1:50 V/V ratio at 4°C for 30 minutes. The following antibodies were used: CD4 (clone SK3), CD8 (clone SK1), CD134 (OX40, clone ACT35), CD137 (4-1BB, clone 4B4-1), and anti-mouse TCRβ Chain (clone H57-597) (BD Biosciences, San Jose, CA). Flow cytometric analysis was performed on FACS Canto I cell analyzer (BD Biosciences). Data was analyzed using FlowJo 10.2 software (TreeStar, Ashland, OR).
Identification and synthesis of T-cell receptors (TCRs)
To isolate TCR sequences, a recently described single-cell approach was used, with some modifications (41). Briefly, 1 × 106 TILs were co-cultured overnight with 1 × 106 EBV-B cells pulsed with HPLC-purified CTBP1Q277R 25-mer peptide. The following morning, stimulated T-cells were sorted based on 4–1BB upregulation using the FACS Aria Cell Sorter (BD Biosciences). Sorted 4–1BB+ T cells were subjected to automated single-cell RNA-sequencing sample preparation using the Fluidigm C1 platform (Fluidigm, San Francisco, CA), following the manufacturer’s instruction. Single-cell RNA-seq was performed using Illumina MiSeq system (Illumina). The paired full-length TCRα/β sequences were identified using an in-house software.
To synthesize the identified TCR, TCRα/β constant regions were replaced with modified mouse TCRα/β constant regions to enhance TCR pairing and surface expression (42–44). TCRα and β chains were linked with a furin SGSG P2A linker, and then synthesized and cloned into a MSGV retroviral vector (45).
Generation of TCR-transduced T cells
Retroviral transduction of TCR into donor T cells has been described previously (46). Briefly, on day 1, 293GP cells were plated (1 × 106/well) in poly-D-lysine-coated 6-well plate (Corning, Tewksbury, MA). On day 2, each well was transfected with 1.5 μg pMSGV8-TCR and 0.75 μg pRD114 (VSV-G) using Lipofectamine 2000 Transfection Reagent (Life Technologies). Simultaneously, ~1–3 × 108 donor PBMCs were stimulated with 50 ng/ml anti-CD3 (eBioscience, San Diego, CA) and 1200 IU/ml of IL-2 in a tissue culture flask. Where indicated, PBMCs were enriched for CD4+ T cells by depleting CD8+ T cells using anti-CD8 magnetic beads (BD Biosciences, San Jose, CA) and then processed in the same way. On day 3, retrovirus-containing supernatants from 293GP cells were harvested and spinoculated onto a RetroNectin (Takara Bio USA, Mountain View, CA)-coated 6-well plate at 2000 × g for 2 hours at room temperature, while the 293GP cells were re-fed with fresh media and set aside for an additional 24-hour incubation. Following the removal of unbound supernatant, spinoculated plate was seeded with donor PBMCs (1 × 106 cells/well), centrifuged at 1000 × g for 10 minutes and incubated overnight at 37°C. On day 4, supernatants from 293GP were re-harvested and spinoculated onto a new RetroNectin-coated plate, which was used to re-transduce PBMCs from the previous day following the same protocol. Transduced cells were then cultured in complete AIM V medium supplemented with 1200 IU/ml of IL-2 for additional 5 days. The TCR-transduced T cells were cryopreserved until further use. When using CD8-depleted PBMCs, pure CD4+ transduced T cells were obtained at the end of the process, as described previously (47).
Determination of HLA restriction element
Relevant HLA alleles identified from tumor WES were synthesized and cloned into pcDNA3.1 vector (GeneOracle, Santa Clara, CA). Next, COS-7 cells were plated (2.5 × 104 cells/well) in a flat-bottom 96-well plate and co-transfected the following day with combinations of individual MHC plasmids (150 ng/well each) using Lipofectamine 2000 transfection reagent (0.5 μl/well). The next day, transfected cells were pulsed with WT and mutant 25-mer peptide for 2 hours, washed twice in complete RPMI, and co-cultured overnight with 1 × 105/well of TCR-transduced T cells. IFN-γ production was assessed by ELISA, as described above.
Multi-cytokine analysis
Supernatants from overnight TIL or TCR-transduced donor T cell co-cultures were analyzed using a multiplex sandwich immunoassay named U-PLEX (Meso Scale Diagnostics, Rockville, MD), in accordance with the manufacturer’s instructions. The assay was customized to allow simultaneous detection of IFN-γ, TNF-α, IL-2, IL-4, IL-5, IL-10, IL-13 and GM-CSF.
Immunohistochemistry (IHC)
Tissue sections from formalin-fixed, paraffin-embedded (FFPE) TURBT specimens were stained with H&E and immune-stained with the following antibodies: CD3 (clone 2GV6), CD4 (clone SP35), CD8 (clone SP57) (Ventana, Tucson, AZ), MHC class I (clone HC-10) (kindly provided by Dr. Soldano Ferrone), and HLA-DR (clone TAL.1B5) (Dako, Carpinteria, CA). Staining was performed at the Laboratory of Pathology (NCI); stained sections were examined by a certified pathologist who was blinded to the results of neoantigen screening.
Statistical analysis
Statistical analyses were performed on GraphPad Prism 7.0 software (GraphPad Software, La Jolla, CA). When applicable, data were expressed as mean ± standard deviation (SD).
Results
In this study, we tested TILs from five patients who had primary localized urothelial carcinoma of the bladder, including low- and high-grade NMIBC and high-grade MIBC, and who underwent stage-based treatment with TURBT ± BCG at the NIH Clinical Center. The size of resected tumor specimens, obtained mostly via TURBT that yielded small slices of tumors, was only several mm3 for all five patients. TIL growth occurred slowly and was seen only in a fraction of cultured tumor fragments. The number of tumor-specific mutations on WES ranged between 120 and 232, which was consistent with previous reports in bladder cancer (24, 25). Patient information, tumor characteristics and IHC results are summarized in Table 1; CD8 vs CD4 composition of individual TIL cultures is shown in Supplementary Figure 1.
Table 1.
Patient and tumor characteristics
| Patient ID | Age | Sex | Prior Treatments | Tumor Procurement Method | Tumor Grade | Muscle Invasion | MHC-positive tumor cells (%) | Number of Tumor-Specific Mutations | Tumor CD3+/CD8+/CD4+ infiltration |
Number of TIL Cultures / Plated Tumor Fragments | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Class I | Class II | ||||||||||
| 1 | 75 | M | TURBT, BCG × 2 |
TURBT | High | No | ND | ND | 120 | 0–1 / 0–1 / 0 | 4 / 12 |
| 2 | 39 | M | TURBT | R1: TURBT | High | No | 5–25% | ND | 232 | 0 / 1+ / 0–1 | 7 / 12 |
| R2: TURBT | >50% | 5–50% | 1+ / 0–1 / 0–1 | 4 / 12 | |||||||
| 3 | 65 | M | TURBT | TURBT | High | Yes | >50% | >5% | 194 | 0–1 / 0–1 / 1+ | 8 / 12 |
| 4 | 64 | M | None | Bladder diverticulectomy | Low | No | >50% | >50% | 142 | 1+ / 2+ / 3+ | 4 / 24 |
| 5 | 76 | M | TURBT | TURBT | High | No | 5–50% | ND | 230 | 2–3 / 2–3 / 2–3 | 2 / 24 |
Abbreviations:
BCG, Bacillus Calmette-Guérin; TURBT, transurethral resection of bladder tumor; R1/2, resection #1/2;
ND, not detected; Tumor CD3+/CD8+/CD4+ infiltration, 0 = absent, 1+ = weak/scattered, 2+ = moderate, 3+ = strong
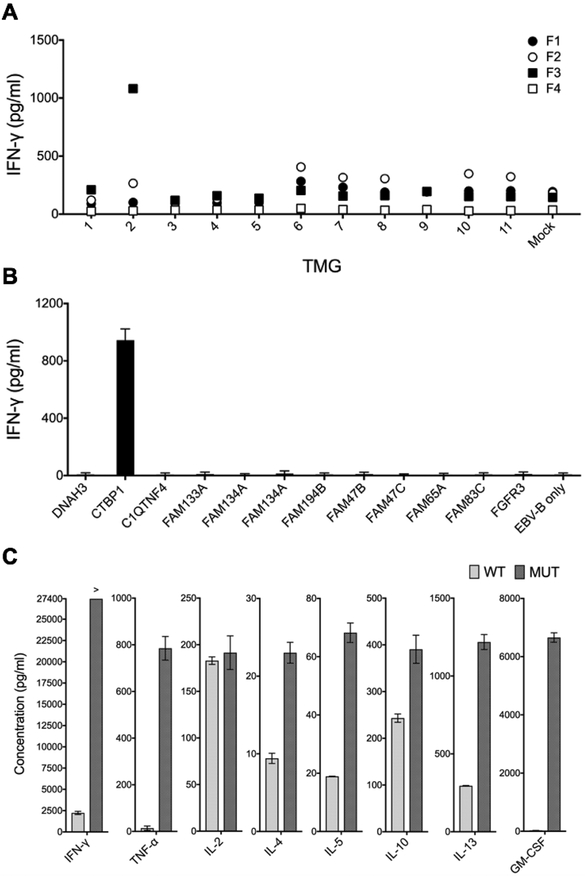
Patient 1, a 75-year-old man whose cancer was previously treated with TURBT and intravesical BCG at an outside institution, presented to the NIH with relapsed localized disease and underwent another TURBT. WES of the fresh tumor sample led to identification of 120 unique mutations, which were incorporated into 11 individual TMGs (Supplementary Table 1). Simultaneously, four TIL cultures (F1 - F4) grew out of 12 plated tumor fragments. We performed overnight co-cultures with TILs and autologous EBV-B cells electroporated with 11 individual TMGs, and then assessed T cell reactivity by performing IFN-γ ELISA. As indicated in Figure 1A, marked IFN-γ production was detected when TIL fragment 3 (F3) was co-cultured with TMG2. To determine which TMG2-encoded mutation was the candidate neoantigen, F3 TILs were co-cultured with EBV-B cells pulsed with individual mutated 25-mer peptides encoded in TMG2. As indicated in Figure 1B, only a mutated peptide harboring a single glutamine-to-arginine substitution in C-terminal binding protein-1 (CTBP1Q277R) elicited IFN-γ production from F3 TILs. Moreover, when compared to its wild type counterpart, HPLC-purified mutant CTBP1 elicited significantly increased production of IFN-γ, TNF-α, IL-5, IL-13 and GM-CSF, and slightly increased production of IL-4 and IL-10, as measured by a multiplex assay (Figure 1C). The increased production of IFN-γ, GM-CSF and TNF-α was further confirmed by ELISA (Supplementary Figure 2).
Figure 1. TILs grown from Patient 1 recognized tumor-specific CTBP1Q277R mutation.
(A) Four TILs (F1–4) were co-cultured with autologous EBV-B cells electroporated with 11 TMGs encoding tumor-specific mutations, or with sterile water alone (Mock). Following an overnight incubation, IFN-γ concentration in co-culture supernatants was determined by IFN-γ ELISA. (B and C) F3 TILs were co-cultured with EBV-B cells pulsed overnight with individual 25-mer peptides encoded in TMG2, followed by IFN-γ ELISA (B) and with wild-type and mutant CTBP1 25-mer, followed by a multiplex assay for IFN-γ, TNF-α, IL-4, IL-5, IL-10, IL-13 and GM-CSF (C). All the experiments were performed once owing to a limited number of available TILs. Bars represent average reads from two duplicate wells; error bars represent SD.
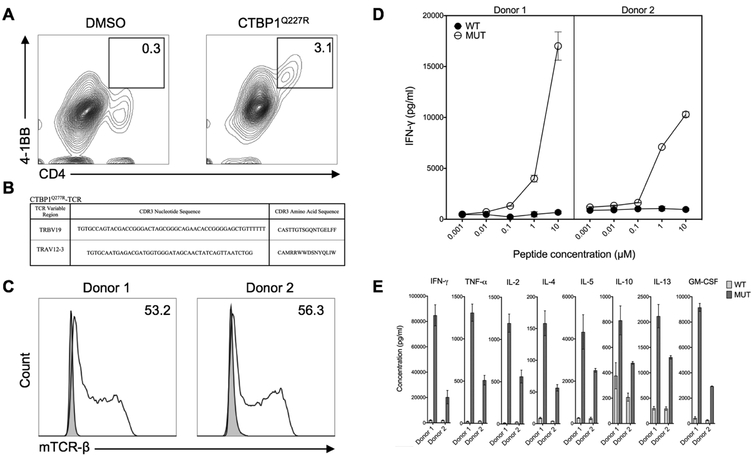
To isolate TCRs that recognized the CTBP1Q277R epitope, F3 TILs were co-cultured overnight with autologous EBV-B cells pulsed with CTBP1Q277R 25-mer peptide, and then analyzed by FACS for upregulation of 4–1BB. As indicated in Figure 2A, 3.1% of cells, all phenotypically CD4+ T lymphocytes, upregulated 4–1BB when co-cultured with CTBP1Q277R peptide, in comparison to 0.3% in the DMSO control. These CD4+ 4–1BB+ TILs were sorted and subjected to single-cell RNA-seq analysis, which identified a single TCR sequence (Figure 2B). This TCR was then synthesized, cloned into a MSGV plasmid and transduced into PBMCs from two unrelated donors with greater than 50% efficiency (Figure 2C). The TCR-transduced T cells were then co-cultured with autologous EBV-B cells pulsed with either CTBP1Q277R 25-mer peptide or the corresponding WT control, followed by assessment of IFN-γ production by ELISA. As indicated in Figure 2D, CTBP1Q277R -TCR-transduced T cells from both donors recognized CTBP1Q277R peptide in a dose-dependent manner, whereas there was no reactivity against the WT peptide. Moreover, in a multiplex assay using CTBP1Q277R -TCR transduced CD4+ T cells, stimulation with mutated 25-mer peptide elicited increased production of TNF-α, IL-2, IL-4, IL-5, IL-10, IL-13 and GM-CSF in comparison to the wild type peptide (Figure 2E). The increased production of GM-CSF, TNF-α, IL-2 and IL-4 was further confirmed by ELISA (Supplementary Figure 3).
Figure 2. A TCR isolated from CTBP1Q277R-reactive TILs recognized CTBP1Q277R.
(A) F3 TILs were co-cultured overnight with EBV-B cells pulsed with CTBP1Q277R 25-mer peptide, followed by FACS sorting based on 4–1BB upregulation. Contour plots indicate the percentage of CD4+ 4–1BB+ lymphocytes in the respective co-cultures; graphs were gated on all live lymphocytes. (B) CTBP1Q277R-TCR CDR3 sequence, as identified by single-cell RNA sequencing. (C) Transduction efficiencies for CTBP1Q277R-TCR, as assessed by flow cytometry for mouse TCR-β constant chain (mTCR-β) expression. Histograms depict mTCRβ staining on untransduced (shaded) and transduced (unshaded) cells from two donors; numbers (%) indicate the estimated transduction efficiency. Graphs were gated on all live lymphocytes. (D) CTBP1Q277R-TCR-transduced T cells from (C) were co-cultured with autologous EBV-B cells pulsed overnight with serial dilutions of either wild type (WT) or CTBP1Q277R peptide (MUT). IFN-γ concentration was measured in co-culture supernatants by ELISA. A representative of two independently performed experiments is shown. (E) CTBP1Q277R-TCR transduced CD4+ cells from the same donors were co-cultured overnight with EBV-B cells stimulated with 10 μM WT or MUT peptide; concentrations of multiple cytokines in the co-culture supernatants were analyzed for a multiplex assay. Data represents average reads from two duplicate co-culture wells; error bars represent SD.
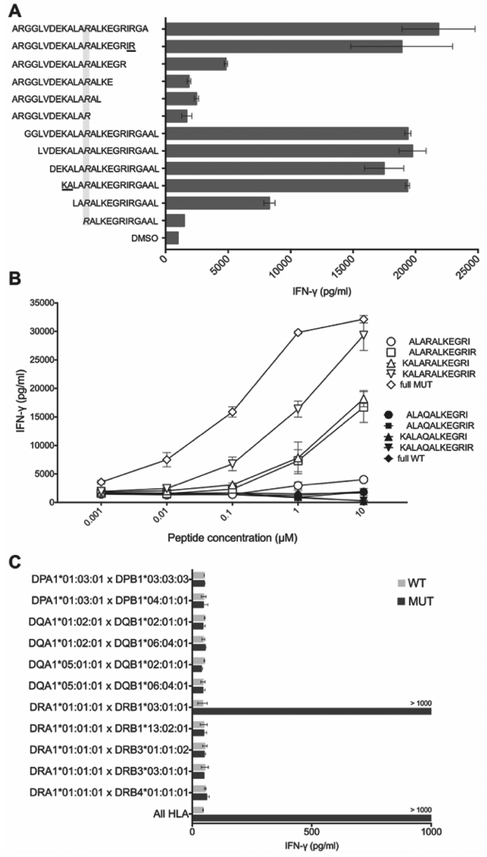
To determine the minimal epitope recognized by CTBP1Q277R-TCR, TCR-transduced T cells were co-cultured with autologous EBV-B cells pulsed with serial truncations of CTBP1Q277R 25-mer peptide. As indicated in Figure 3A, the loss of amino acids delineating the 13-mer (KALARALKEGRIR) resulted in significantly decreased IFN-γ production. We then synthetized this mutated 13-mer peptide and its WT counterpart, alongside several serial truncations of each peptide, and used them in co-cultures with transduced T cells. As shown in Figure 3B, the strongest IFN-γ production was detected when T cells were co-cultured with the mutated 13-mer, with no response to any of the WT peptides, suggesting that this peptide was likely the minimal epitope recognized by CTBP1Q277R-TCR.
Figure 3. Determination of minimal epitope and HLA restriction element for CTBP1Q277R-TCR.
(A) CTBP1Q277R-TCR-transduced T cells were co-cultured with autologous EBV-B cells pulsed with serial two-amino acid truncations of CTBP1Q277R 25-mer. IFN-γ concentration was determined by ELISA. Top 6 sequences on the y axis represent truncations from the C-terminus, and the bottom 6 from the N-terminus of CTBP1Q277R 25-mer. Grayed area highlights the Q277R mutation; underlined amino acids delineate the potential minimal epitope. (B) Transduced T cells were co-cultured with autologous EBV-B cells pulsed with CTBP1Q277R 25-mer, 13-mer identified in (A), and three additional truncations of the latter (white dots). Corresponding WT peptides were used as a control (black dots). IFN-γ concentration was determined by ELISA. (C) COS-7 cells, transfected with combinations of plasmids encoding all paired MHC class II molecules identified in Patient 1, were pulsed for 2 hours with WT or CTBP1Q277R (MUT) peptide and co-cultured with CTBP1Q277R-TCR-transduced T cells. Results of IFN-γ ELISA are shown. For (A-C), a representative of two independent experiments is shown. Data represents average reads from two duplicate co-culture wells; error bars represent SD.
To determine the MHC class II-restriction element for CTBP1Q277R-TCR, COS-7 cells were transfected with pairs of plasmids encoding the following MHC class II molecules identified from this patient’s tumor WES data: DPA1*01:03:01, DPB1*03:01:01, DPB1*04:01:01, DQA1*01:02:01, DQA1*05:01:01, DQB1*02:01:01, DQB1*06:04:01, DRB1*03:01:01, DRB1*13:02:01, DBR3*01:01:02, DRB3*03:01:01, DRB4*01:01:01. Next, transfected cells were pulsed for 2 hours with WT or CTBP1Q277R 25-mer peptide, followed by an overnight co-culture with CTBP1Q277R -TCR-transduced T cells. As indicated in Figure 3C, IFN-γ production was detected only when T cells were co-cultured with COS-7 cells transfected with HLA-DRA1*01:01:01 and HLA-DRB1*03:01:01, thus identifying them as the HLA restriction element for CTBP1Q277R-TCR.
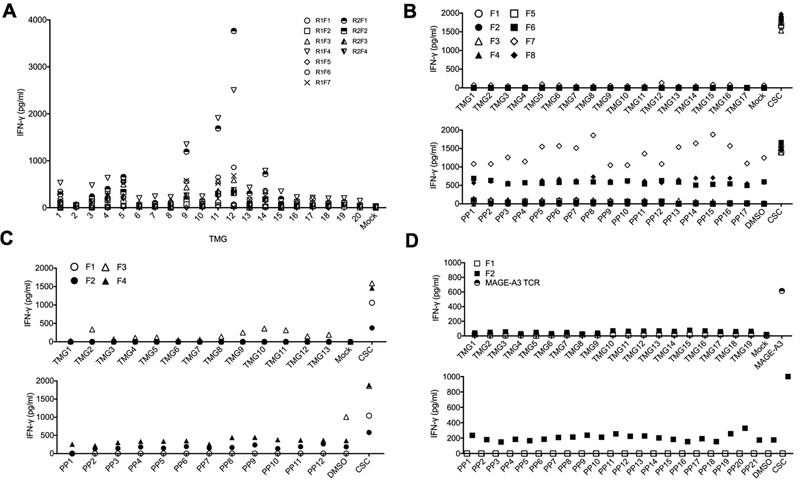
Patient 2, a 39-year-old man with primary bladder cancer, had a single TURBT before presenting to the NIH with relapsed disease. Two consecutive tumor resections were performed, and TILs grew from only four out of 12 fragments obtained with the first procedure, and seven out of 12 fragments obtained with the second. A total of 232 unique mutations were identified by WES, which were then incorporated into 20 individual TMGs. After a co-culture with TMG-electroporated autologous DCs, two TILs from separate resections (R1F4 and R2F1) exhibited marked IFN-γ production against TMG9, TMG11 and TMG12 (Figure 4A). However, co-cultured TILs exhibited no significant 4–1BB upregulation in response to the cognate TMGs (data not shown), indicating that the frequency of reactive T cells was likely very low. Unfortunately, limited availability of samples precluded further experimentation and identification of the neoantigen-specific TCRs.
Figure 4. TILs from Patient 2 recognized autologous DCs electroporated with TMG 9, 11 and 12, whereas TILs from the remaining patients did not recognize any mutations.
(A) Eleven TILs from Patient 2 (R1F1-R1F7 from the first and R2F1-R2F4 from second tumor resection) were co-cultured with autologous DCs electroporated with 20 TMGs encoding tumor-specific mutations, or with sterile water alone (Mock). (B-D) TILs from Patient 3 (B), 4 (C) and 5 (D) were co-cultured with autologous EBV-B cells that were either electroporated with TMGs encoding tumor-specific mutations (upper panels) or pulsed with peptide pools (PP) encoding the same mutations (lower panels). IFN-γ concentration was determined in co-culture supernatants using IFN-γ ELISA. In (D), PBMCs transduced with MAGE-A3 TCR were co-cultured with cognate EBV-B cells electroporated with MAGE-A3 RNA and used as a positive control. All the experiments were performed once owing to a limited number of available TILs. CSC = cell stimulation cocktail.
TILs from the remaining three patients exhibited no significant IFN-γ production following an overnight co-culture with neoantigen-loaded autologous EBV-B cells. Briefly, for Patient 3, eight TIL cultures grew from 12 tumor fragments, and none demonstrated specific reactivity against EBV-B cells electroporated with TMGs (N=17) or pulsed with mutated peptide pools (N=17) encoding 194 tumor-specific mutations (Figure 4B). For Patient 4, four TIL cultures grew from 24 tumor fragments; no specific reactivity was observed against any of the TMGs (N=13) or mutated peptide pools (N=12) representing 142 mutations (Figure 4C). Finally, for Patient 5, two TIL cultures grew from 24 tumor fragments; no specific reactivity was seen against any of the TMGs (N=19) or mutated peptide pools (N=20) encoding a total of 230 cancer mutations (Figure 4D).
Discussion
In this study, we grew TILs from five patients with primary localized urothelial carcinoma of the bladder, including low and high-grade NMIBC and high-grade MIBC, and screened them for recognition of tumor-specific mutations. In our previous experience with screening TILs from patients with metastatic melanoma and gastrointestinal cancers, the majority of hits could be obtained from both screening against the mutated peptides and the TMGs. The peptide approach, however, showed a slight bias towards detecting CD4+ TIL responses, whereas the TMG approach slightly biased the detection towards the CD8+ TILs (unpublished data). To avoid such biases in this study, we utilized both approaches when screening TILs, whenever we had sufficient number of cells available.
We found that a CD4+ subset of TILs from Patient 1, who had high-grade NMIBC previously treated with BCG, specifically recognized a point mutation in CTBP1 protein (CTBP1Q277R). Furthermore, we isolated an HLA-DRB1*03:01-restricted, CTBP1Q277R-reactive TCR and confirmed that it could recognize the mutant, but not the WT CTBP1 peptide. CTBP1 is a transcriptional corepressor that plays a role in oncogenesis through suppression of genes that regulate cell cycle progression, DNA repair, apoptosis and intercellular adhesion (48). Although CTBP1 overexpression has previously been detected in several cancers, oncogenic repercussions of its mutations remain largely unknown. In addition, CTBP1 mutation at the amino acid position 277 has not been reported in the COSMIC (Catalogue of Somatic Mutations in Cancer) database, and thus appears to be unique to Patient 1.
Even though ICIs have previously been shown to elicit durable regressions of metastatic bladder tumors, identification of specific neoantigens has been lacking. Our study, conducted in patients with primary disease, shows that neoantigen-reactive TILs can be isolated from a bladder tumor specimen, a finding consistent with the previous evidence that tumors express unique antigens can be recognized by T cells (49). Although a previous study found that circulating CD8+ T cells from a patient with primary bladder cancer recognized a tumor-specific point mutation in LPGAT1 (also known as KIAA0205), a ubiquitously expressed enzyme involved in membrane phospholipid metabolism, this reactivity arose from a circulating T cell clone after in vitro stimulation with irradiated tumor cells (50, 51). Thus, this T-cell clone might not necessarily represent T cells residing within the tumor.
Several technical challenges have limited our ability to discover additional neoantigens. The starting amount of tumor tissue was very modest and had been immersed in urine prior to excision. Tumor infiltration by T cells, as assessed by IHC for CD3, CD8 and CD4, was overall weak (Table 1) and could not readily explain the success rate in raising TIL cultures, likely because of the heterogeneity between the parts of primary tumors used for IHC and TIL culturing. In contrast to our previous studies of melanoma and gastrointestinal tumors, TIL growth was observed in only a fraction of plated tumor fragments (Table 1), which further limited the number and repertoire of cells available for testing. TIL growth occurred slowly, raising concerns that non-specific, bystander T cells may have outgrown the exhausted, neoantigen-reactive T cells in the long-term cultures (52). Lastly, as in the case of Patient 2, whose TILs exhibited significant IFN-γ production in response to three TMGs, the frequency of the potentially reactive T cells was likely too low to allow the detection by flow cytometry based on 4–1BB up-regulation, thereby precluding the possibility of further enrichment and testing of these T cells.
In addition to these limitations, tumor-intrinsic properties could have reduced our ability to identify neoantigen-reactive TILs in bladder cancer. The heterogeneity of primary bladder tumors, which could be more pronounced than in metastases, could have prevented the identification of TILs recognizing mutations unique to the parts of the tumor that were not captured by WES analysis of a limited tumor fragment. Additionally, several previous studies reported a high prevalence of partial or complete MHC class I molecule loss from primary bladder tumors (53–57), which could restrict the presentation of tumor neoantigens.
Tumor MHC expression in our study cohort was variable, as was the T cell infiltration (Table 1). The finding of a CTBP1Q277R-reactive CD4+ T cell clone from a tumor with seemingly undetectable MHC class II expression and sparse T cell infiltration highlights how tumor heterogeneity, well-described in primary bladder tumors (58), can limit the use of a single tumor biopsy in defining the biomarkers that could predict the sensitivity to immunotherapies. Accordingly, our finding could potentially help explain the lack of consistency in correlation between clinical benefits of immunotherapy and the tumor immune features in some circumstances.
It has been shown that CD4+ T cells can induce significant tumor regressions in cancer patients by targeting MHC class II-restricted cancer antigens (36, 59, 60). One important question is whether the tumor cells without detectable MHC class II expression can still be eliminated by the tumor antigen-reactive CD4+ T cells. Although previous work in a murine melanoma model suggested that tumor regressions occur following IFN-γ-mediated in vivo upregulation of MHC class II molecules on tumor cells (61, 62), further research is needed to test whether this applies to human tumors. Alternatively, CD4+ T cells could recognize tumor antigens presented by antigen-presenting cells and then prime the immune responses against additional tumor antigens, some of which could be MHC class I-restricted - a phenomenon called “antigen spreading”(63). Again, further evidence is necessary to support this hypothesis in human cancer.
Due to its small size and the aforementioned technical limitations, our study cannot be used to accurately determine whether the neoantigen-specific T cells are significantly represented in the population of patients with bladder cancer, and whether they truly drive anti-tumor immune responses. To better address these questions, future studies would have to test a larger number of patients, especially patients with more advanced/metastatic disease, from which larger tumor specimens could be obtained. Alternatively, PD-1+ T cells from the peripheral blood could be screened in an attempt to identify cancer neoantigens, as previously reported in patients with metastatic melanoma (64). Further research will also be needed to determine the dynamics of neoantigen responses in patients with metastatic bladder cancer, and to explore the potential differences between those who responded to ICIs and those who were resistant.
In conclusion, we demonstrated that a cancer-specific CTBP1Q277R mutation elicited a TCR-mediated, MHC class II-restricted recognition by endogenous TILs. This proof-of-principle study provides preliminary evidence that neoantigen-reactive TILs can be isolated from bladder tumors, which in turn may offer a mechanistic insight into the effectiveness of immune checkpoint inhibition in bladder cancer treatment. Furthermore, our findings provide a preliminary rationale for future use of adoptively transferred neoantigen-reactive T cells in the treatment of patients with bladder cancer. This could be accomplished either by expanding and administering TILs that specifically recognized cancer neoantigens, as previously reported by our group, or by isolating the neoantigen-reactive TCRs and transferring them into autologous PBMCs, which would then be infused back to the patients (32, 65).
Supplementary Material
Key points.
Tumor-infiltrating lymphocytes (TILs) can be grown from primary bladder tumors.
Neoantigen-reactive TILs can be isolated from a bladder tumor specimen.
Acknowledgments
The authors thank John R. Wunderlich, Chyi-Chia Richard Lee, Li Jia, Kenichi Hanada and James C. Yang for suggestions and technical support. Single-cell RNA-seq was conducted at the CCR Genomics Core at the National Cancer Institute. This work utilized the computational resources of the NIH HPC Biowulf cluster (http://hpc.nih.gov).
Grant support: Intramural Research Program of the National Cancer Institute.
References
- 1.Siegel RL, Miller KD, and Jemal A. 2018. Cancer statistics, 2018. CA Cancer J Clin 68: 7–30. [DOI] [PubMed] [Google Scholar]
- 2.von der Maase H, Sengelov L, Roberts JT, Ricci S, Dogliotti L, Oliver T, Moore MJ, Zimmermann A, and Arning M. 2005. Long-term survival results of a randomized trial comparing gemcitabine plus cisplatin, with methotrexate, vinblastine, doxorubicin, plus cisplatin in patients with bladder cancer. J Clin Oncol 23: 4602–4608. [DOI] [PubMed] [Google Scholar]
- 3.Sternberg CN, de Mulder PH, Schornagel JH, Theodore C, Fossa SD, van Oosterom AT, Witjes F, Spina M, van Groeningen CJ, de Balincourt C, Collette L, R. European Organization for, and G. Treatment of Cancer Genitourinary Tract Cancer Cooperative. 2001. Randomized phase III trial of high-dose-intensity methotrexate, vinblastine, doxorubicin, and cisplatin (MVAC) chemotherapy and recombinant human granulocyte colony-stimulating factor versus classic MVAC in advanced urothelial tract tumors: European Organization for Research and Treatment of Cancer Protocol no. 30924. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 19: 2638–2646. [DOI] [PubMed] [Google Scholar]
- 4.Kamat AM, Hahn NM, Efstathiou JA, Lerner SP, Malmstrom PU, Choi W, Guo CC, Lotan Y, and Kassouf W. 2016. Bladder cancer. Lancet 388: 2796–2810. [DOI] [PubMed] [Google Scholar]
- 5.Sylvester RJ, van der Meijden AP, Witjes JA, and Kurth K. 2005. Bacillus calmette-guerin versus chemotherapy for the intravesical treatment of patients with carcinoma in situ of the bladder: a meta-analysis of the published results of randomized clinical trials. J Urol 174: 86–91; discussion 91–82.15947584 [Google Scholar]
- 6.Bohle A, and Brandau S. 2003. Immune mechanisms in bacillus Calmette-Guerin immunotherapy for superficial bladder cancer. J Urol 170: 964–969. [DOI] [PubMed] [Google Scholar]
- 7.Kresowik TP, and Griffith TS. 2009. Bacillus Calmette-Guerin immunotherapy for urothelial carcinoma of the bladder. Immunotherapy 1: 281–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Redelman-Sidi G, Glickman MS, and Bochner BH. 2014. The mechanism of action of BCG therapy for bladder cancer--a current perspective. Nat Rev Urol 11: 153–162. [DOI] [PubMed] [Google Scholar]
- 9.Ribas A, and Wolchok JD. 2018. Cancer immunotherapy using checkpoint blockade. Science 359: 1350–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nguyen LT, and Ohashi PS. 2015. Clinical blockade of PD1 and LAG3--potential mechanisms of action. Nat Rev Immunol 15: 45–56. [DOI] [PubMed] [Google Scholar]
- 11.Rosenberg JE, Hoffman-Censits J, Powles T, van der Heijden MS, Balar AV, Necchi A, Dawson N, O’Donnell PH, Balmanoukian A, Loriot Y, Srinivas S, Retz MM, Grivas P, Joseph RW, Galsky MD, Fleming MT, Petrylak DP, Perez-Gracia JL, Burris HA, Castellano D, Canil C, Bellmunt J, Bajorin D, Nickles D, Bourgon R, Frampton GM, Cui N, Mariathasan S, Abidoye O, Fine GD, and Dreicer R. 2016. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet 387: 1909–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balar AV, Galsky MD, Rosenberg JE, Powles T, Petrylak DP, Bellmunt J, Loriot Y, Necchi A, Hoffman-Censits J, Perez-Gracia JL, Dawson NA, van der Heijden MS, Dreicer R, Srinivas S, Retz MM, Joseph RW, Drakaki A, Vaishampayan UN, Sridhar SS, Quinn DI, Duran I, Shaffer DR, Eigl BJ, Grivas PD, Yu EY, Li S, Kadel EE 3rd, Boyd Z, Bourgon R, Hegde PS, Mariathasan S, Thastrom A, Abidoye OO, Fine GD, Bajorin DF, and I. M. S. Group. 2017. Atezolizumab as first-line treatment in cisplatin-ineligible patients with locally advanced and metastatic urothelial carcinoma: a single-arm, multicentre, phase 2 trial. Lancet 389: 67–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Massard C, Gordon MS, Sharma S, Rafii S, Wainberg ZA, Luke J, Curiel TJ, Colon-Otero G, Hamid O, Sanborn RE, O’Donnell PH, Drakaki A, Tan W, Kurland JF, Rebelatto MC, Jin X, Blake-Haskins JA, Gupta A, and Segal NH. 2016. Safety and Efficacy of Durvalumab (MEDI4736), an Anti-Programmed Cell Death Ligand-1 Immune Checkpoint Inhibitor, in Patients With Advanced Urothelial Bladder Cancer. J Clin Oncol 34: 3119–3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Powles T, O’Donnell PH, Massard C, Arkenau HT, Friedlander TW, Hoimes CJ, Lee JL, Ong M, Sridhar SS, Vogelzang NJ, Fishman MN, Zhang J, Srinivas S, Parikh J, Antal J, Jin X, Gupta AK, Ben Y, and Hahn NM. 2017. Efficacy and Safety of Durvalumab in Locally Advanced or Metastatic Urothelial Carcinoma: Updated Results From a Phase 1/2 Open-label Study. JAMA Oncol 3: e172411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Apolo AB, Infante JR, Balmanoukian A, Patel MR, Wang D, Kelly K, Mega AE, Britten CD, Ravaud A, Mita AC, Safran H, Stinchcombe TE, Srdanov M, Gelb AB, Schlichting M, Chin K, and Gulley JL. 2017. Avelumab, an Anti-Programmed Death-Ligand 1 Antibody, In Patients With Refractory Metastatic Urothelial Carcinoma: Results From a Multicenter, Phase Ib Study. J Clin Oncol 35: 2117–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharma P, Callahan MK, Bono P, Kim J, Spiliopoulou P, Calvo E, Pillai RN, Ott PA, de Braud F, Morse M, Le DT, Jaeger D, Chan E, Harbison C, Lin CS, Tschaika M, Azrilevich A, and Rosenberg JE. 2016. Nivolumab monotherapy in recurrent metastatic urothelial carcinoma (CheckMate 032): a multicentre, open-label, two-stage, multi-arm, phase 1/2 trial. Lancet Oncol 17: 1590–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharma P, Retz M, Siefker-Radtke A, Baron A, Necchi A, Bedke J, Plimack ER, Vaena D, Grimm MO, Bracarda S, Arranz JA, Pal S, Ohyama C, Saci A, Qu X, Lambert A, Krishnan S, Azrilevich A, and Galsky MD. 2017. Nivolumab in metastatic urothelial carcinoma after platinum therapy (CheckMate 275): a multicentre, single-arm, phase 2 trial. Lancet Oncol 18: 312–322. [DOI] [PubMed] [Google Scholar]
- 18.Bellmunt J, de Wit R, Vaughn DJ, Fradet Y, Lee JL, Fong L, Vogelzang NJ, Climent MA, Petrylak DP, Choueiri TK, Necchi A, Gerritsen W, Gurney H, Quinn DI, Culine S, Sternberg CN, Mai Y, Poehlein CH, Perini RF, Bajorin DF, and K.−. Investigators. 2017. Pembrolizumab as Second-Line Therapy for Advanced Urothelial Carcinoma. N Engl J Med 376: 1015–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Balar AV, Castellano D, O’Donnell PH, Grivas P, Vuky J, Powles T, Plimack ER, Hahn NM, de Wit R, Pang L, Savage MJ, Perini RF, Keefe SM, Bajorin D, and Bellmunt J. 2017. First-line pembrolizumab in cisplatin-ineligible patients with locally advanced and unresectable or metastatic urothelial cancer (KEYNOTE-052): a multicentre, single-arm, phase 2 study. Lancet Oncol. [DOI] [PubMed] [Google Scholar]
- 20.Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, Lee W, Yuan J, Wong P, Ho TS, Miller ML, Rekhtman N, Moreira AL, Ibrahim F, Bruggeman C, Gasmi B, Zappasodi R, Maeda Y, Sander C, Garon EB, Merghoub T, Wolchok JD, Schumacher TN, and Chan TA. 2015. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 348: 124–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Allen EM, Miao D, Schilling B, Shukla SA, Blank C, Zimmer L, Sucker A, Hillen U, Foppen MHG, Goldinger SM, Utikal J, Hassel JC, Weide B, Kaehler KC, Loquai C, Mohr P, Gutzmer R, Dummer R, Gabriel S, Wu CJ, Schadendorf D, and Garraway LA. 2015. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 350: 207–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hugo W, Zaretsky JM, Sun L, Song C, Moreno BH, Hu-Lieskovan S, Berent-Maoz B, Pang J, Chmielowski B, Cherry G, Seja E, Lomeli S, Kong X, Kelley MC, Sosman JA, Johnson DB, Ribas A, and Lo RS. 2016. Genomic and Transcriptomic Features of Response to Anti-PD-1 Therapy in Metastatic Melanoma. Cell 165: 35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yarchoan M, Hopkins A, and Jaffee EM. 2017. Tumor Mutational Burden and Response Rate to PD-1 Inhibition. N Engl J Med 377: 2500–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, Xie M, Zhang Q, McMichael JF, Wyczalkowski MA, Leiserson MDM, Miller CA, Welch JS, Walter MJ, Wendl MC, Ley TJ, Wilson RK, Raphael BJ, and Ding L. 2013. Mutational landscape and significance across 12 major cancer types. Nature 502: 333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cazier JB, Rao SR, McLean CM, Walker AK, Wright BJ, Jaeger EE, Kartsonaki C, Marsden L, Yau C, Camps C, Kaisaki P, Oxford-Illumina WGSC, Taylor J, Catto JW, Tomlinson IP, Kiltie AE, and Hamdy FC. 2014. Whole-genome sequencing of bladder cancers reveals somatic CDKN1A mutations and clinicopathological associations with mutation burden. Nat Commun 5: 3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, Carter SL, Stewart C, Mermel CH, Roberts SA, Kiezun A, Hammerman PS, McKenna A, Drier Y, Zou L, Ramos AH, Pugh TJ, Stransky N, Helman E, Kim J, Sougnez C, Ambrogio L, Nickerson E, Shefler E, Cortes ML, Auclair D, Saksena G, Voet D, Noble M, DiCara D, Lin P, Lichtenstein L, Heiman DI, Fennell T, Imielinski M, Hernandez B, Hodis E, Baca S, Dulak AM, Lohr J, Landau DA, Wu CJ, Melendez-Zajgla J, Hidalgo-Miranda A, Koren A, McCarroll SA, Mora J, Crompton B, Onofrio R, Parkin M, Winckler W, Ardlie K, Gabriel SB, Roberts CWM, Biegel JA, Stegmaier K, Bass AJ, Garraway LA, Meyerson M, Golub TR, Gordenin DA, Sunyaev S, Lander ES, and Getz G. 2013. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499: 214–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Balbas-Martinez C, Sagrera A, Carrillo-de-Santa-Pau E, Earl J, Marquez M, Vazquez M, Lapi E, Castro-Giner F, Beltran S, Bayes M, Carrato A, Cigudosa JC, Dominguez O, Gut M, Herranz J, Juanpere N, Kogevinas M, Langa X, Lopez-Knowles E, Lorente JA, Lloreta J, Pisano DG, Richart L, Rico D, Salgado RN, Tardon A, Chanock S, Heath S, Valencia A, Losada A, Gut I, Malats N, and Real FX. 2013. Recurrent inactivation of STAG2 in bladder cancer is not associated with aneuploidy. Nat Genet 45: 1464–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Powles T, Duran I, van der Heijden MS, Loriot Y, Vogelzang NJ, De Giorgi U, Oudard S, Retz MM, Castellano D, Bamias A, Flechon A, Gravis G, Hussain S, Takano T, Leng N, Kadel EE 3rd, Banchereau R, Hegde PS, Mariathasan S, Cui N, Shen X, Derleth CL, Green MC, and Ravaud A. 2017. Atezolizumab versus chemotherapy in patients with platinum-treated locally advanced or metastatic urothelial carcinoma (IMvigor211): a multicentre, open-label, phase 3 randomised controlled trial. Lancet. [DOI] [PubMed] [Google Scholar]
- 29.Rooney MS, Shukla SA, Wu CJ, Getz G, and Hacohen N. 2015. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 160: 48–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schumacher TN, and Schreiber RD. 2015. Neoantigens in cancer immunotherapy. Science 348: 69–74. [DOI] [PubMed] [Google Scholar]
- 31.Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, Skora AD, Luber BS, Azad NS, Laheru D, Biedrzycki B, Donehower RC, Zaheer A, Fisher GA, Crocenzi TS, Lee JJ, Duffy SM, Goldberg RM, de la Chapelle A, Koshiji M, Bhaijee F, Huebner T, Hruban RH, Wood LD, Cuka N, Pardoll DM, Papadopoulos N, Kinzler KW, Zhou S, Cornish TC, Taube JM, Anders RA, Eshleman JR, Vogelstein B, and Diaz LA Jr. 2015. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med 372: 2509–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tran E, Robbins PF, and Rosenberg SA. 2017. ‘Final common pathway’ of human cancer immunotherapy: targeting random somatic mutations. Nat Immunol 18: 255–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ellebaek E, Iversen TZ, Junker N, Donia M, Engell-Noerregaard L, Met O, Holmich LR, Andersen RS, Hadrup SR, Andersen MH, thor Straten P, and Svane IM. 2012. Adoptive cell therapy with autologous tumor infiltrating lymphocytes and low-dose Interleukin-2 in metastatic melanoma patients. J Transl Med 10: 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rosenberg SA, Yang JC, Sherry RM, Kammula US, Hughes MS, Phan GQ, Citrin DE, Restifo NP, Robbins PF, Wunderlich JR, Morton KE, Laurencot CM, Steinberg SM, White DE, and Dudley ME. 2011. Durable complete responses in heavily pretreated patients with metastatic melanoma using T-cell transfer immunotherapy. Clin Cancer Res 17: 4550–4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tran E, Robbins PF, Lu YC, Prickett TD, Gartner JJ, Jia L, Pasetto A, Zheng Z, Ray S, Groh EM, Kriley IR, and Rosenberg SA. 2016. T-Cell Transfer Therapy Targeting Mutant KRAS in Cancer. N Engl J Med 375: 2255–2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tran E, Turcotte S, Gros A, Robbins PF, Lu YC, Dudley ME, Wunderlich JR, Somerville RP, Hogan K, Hinrichs CS, Parkhurst MR, Yang JC, and Rosenberg SA. 2014. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science 344: 641–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robbins PF, Lu YC, El-Gamil M, Li YF, Gross C, Gartner J, Lin JC, Teer JK, Cliften P, Tycksen E, Samuels Y, and Rosenberg SA. 2013. Mining exomic sequencing data to identify mutated antigens recognized by adoptively transferred tumor-reactive T cells. Nature medicine 19: 747–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haas GP, Solomon D, and Rosenberg SA. 1990. Tumor-infiltrating lymphocytes from nonrenal urological malignancies. Cancer Immunol Immunother 30: 342–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dudley ME, Wunderlich JR, Shelton TE, Even J, and Rosenberg SA. 2003. Generation of tumor-infiltrating lymphocyte cultures for use in adoptive transfer therapy for melanoma patients. J Immunother 26: 332–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu YC, Yao X, Crystal JS, Li YF, El-Gamil M, Gross C, Davis L, Dudley ME, Yang JC, Samuels Y, Rosenberg SA, and Robbins PF. 2014. Efficient identification of mutated cancer antigens recognized by T cells associated with durable tumor regressions. Clin Cancer Res 20: 3401–3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu YC, Zheng Z, Robbins PF, Tran E, Prickett TD, Gartner JJ, Li YF, Ray S, Franco Z, Bliskovsky V, Fitzgerald PC, and Rosenberg SA. 2017. An Efficient Single-Cell RNA-Seq Approach to Identify Neoantigen-Specific T Cell Receptors. Mol Ther. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cohen CJ, Zhao Y, Zheng Z, Rosenberg SA, and Morgan RA. 2006. Enhanced antitumor activity of murine-human hybrid T-cell receptor (TCR) in human lymphocytes is associated with improved pairing and TCR/CD3 stability. Cancer Res 66: 8878–8886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cohen CJ, Li YF, El-Gamil M, Robbins PF, Rosenberg SA, and Morgan RA. 2007. Enhanced antitumor activity of T cells engineered to express T-cell receptors with a second disulfide bond. Cancer Res 67: 3898–3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haga-Friedman A, Horovitz-Fried M, and Cohen CJ. 2012. Incorporation of transmembrane hydrophobic mutations in the TCR enhance its surface expression and T cell functional avidity. J Immunol 188: 5538–5546. [DOI] [PubMed] [Google Scholar]
- 45.Wargo JA, Robbins PF, Li Y, Zhao Y, El-Gamil M, Caragacianu D, Zheng Z, Hong JA, Downey S, Schrump DS, Rosenberg SA, and Morgan RA. 2009. Recognition of NY-ESO-1+ tumor cells by engineered lymphocytes is enhanced by improved vector design and epigenetic modulation of tumor antigen expression. Cancer Immunol Immunother 58: 383–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morgan RA, Dudley ME, Wunderlich JR, Hughes MS, Yang JC, Sherry RM, Royal RE, Topalian SL, Kammula US, Restifo NP, Zheng Z, Nahvi A, de Vries CR, Rogers-Freezer LJ, Mavroukakis SA, and Rosenberg SA. 2006. Cancer regression in patients after transfer of genetically engineered lymphocytes. Science 314: 126–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yao X, Lu YC, Parker LL, Li YF, El-Gamil M, Black MA, Xu H, Feldman SA, van der Bruggen P, Rosenberg SA, and Robbins PF. 2016. Isolation and Characterization of an HLA-DPB1*04: 01-restricted MAGE-A3 T-Cell Receptor for Cancer Immunotherapy. J Immunother 39: 191–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Blevins MA, Huang M, and Zhao R. 2017. The Role of CtBP1 in Oncogenic Processes and Its Potential as a Therapeutic Target. Mol Cancer Ther 16: 981–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parmiani G, De Filippo A, Novellino L, and Castelli C. 2007. Unique human tumor antigens: immunobiology and use in clinical trials. Journal of immunology 178: 1975–1979. [DOI] [PubMed] [Google Scholar]
- 50.Gueguen M, Patard JJ, Gaugler B, Brasseur F, Renauld JC, Van Cangh PJ, Boon T, and Van den Eynde BJ. 1998. An antigen recognized by autologous CTLs on a human bladder carcinoma. J Immunol 160: 6188–6194. [PubMed] [Google Scholar]
- 51.Yang Y, Cao J, and Shi Y. 2004. Identification and characterization of a gene encoding human LPGAT1, an endoplasmic reticulum-associated lysophosphatidylglycerol acyltransferase. J Biol Chem 279: 55866–55874. [DOI] [PubMed] [Google Scholar]
- 52.Simoni Y, Becht E, Fehlings M, Loh CY, Koo SL, Teng KWW, Yeong JPS, Nahar R, Zhang T, Kared H, Duan K, Ang N, Poidinger M, Lee YY, Larbi A, Khng AJ, Tan E, Fu C, Mathew R, Teo M, Lim WT, Toh CK, Ong BH, Koh T, Hillmer AM, Takano A, Lim TKH, Tan EH, Zhai W, Tan DSW, Tan IB, and Newell EW. 2018. Bystander CD8(+) T cells are abundant and phenotypically distinct in human tumour infiltrates. Nature 557: 575–579. [DOI] [PubMed] [Google Scholar]
- 53.Romero JM, Jimenez P, Cabrera T, Cozar JM, Pedrinaci S, Tallada M, Garrido F, and Ruiz-Cabello F. 2005. Coordinated downregulation of the antigen presentation machinery and HLA class I/beta2-microglobulin complex is responsible for HLA-ABC loss in bladder cancer. Int J Cancer 113: 605–610. [DOI] [PubMed] [Google Scholar]
- 54.Maleno I, Romero JM, Cabrera T, Paco L, Aptsiauri N, Cozar JM, Tallada M, Lopez-Nevot MA, and Garrido F. 2006. LOH at 6p21.3 region and HLA class I altered phenotypes in bladder carcinomas. Immunogenetics 58: 503–510. [DOI] [PubMed] [Google Scholar]
- 55.Maleno I, Aptsiauri N, Cabrera T, Gallego A, Paschen A, Lopez-Nevot MA, and Garrido F. 2011. Frequent loss of heterozygosity in the beta2-microglobulin region of chromosome 15 in primary human tumors. Immunogenetics 63: 65–71. [DOI] [PubMed] [Google Scholar]
- 56.Levin I, Klein T, Goldstein J, Kuperman O, Kanetti J, and Klein B. 1991. Expression of class I histocompatibility antigens in transitional cell carcinoma of the urinary bladder in relation to survival. Cancer 68: 2591–2594. [DOI] [PubMed] [Google Scholar]
- 57.Cabrera T, Pedrajas G, Cozar JM, Garrido A, Vicente J, Tallada M, and Garrido F. 2003. HLA class I expression in bladder carcinomas. Tissue Antigens 62: 324–327. [DOI] [PubMed] [Google Scholar]
- 58.Garrido F, Aptsiauri N, Doorduijn EM, Garcia Lora AM, and van Hall T. 2016. The urgent need to recover MHC class I in cancers for effective immunotherapy. Curr Opin Immunol 39: 44–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hunder NN, Wallen H, Cao J, Hendricks DW, Reilly JZ, Rodmyre R, Jungbluth A, Gnjatic S, Thompson JA, and Yee C. 2008. Treatment of metastatic melanoma with autologous CD4+ T cells against NY-ESO-1. N Engl J Med 358: 2698–2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lu YC, Parker LL, Lu T, Zheng Z, Toomey MA, White DE, Yao X, Li YF, Robbins PF, Feldman SA, van der Bruggen P, Klebanoff CA, Goff SL, Sherry RM, Kammula US, Yang JC, and Rosenberg SA. 2017. Treatment of Patients With Metastatic Cancer Using a Major Histocompatibility Complex Class II-Restricted T-Cell Receptor Targeting the Cancer Germline Antigen MAGE-A3. J Clin Oncol 35: 3322–3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Muranski P, Boni A, Antony PA, Cassard L, Irvine KR, Kaiser A, Paulos CM, Palmer DC, Touloukian CE, Ptak K, Gattinoni L, Wrzesinski C, Hinrichs CS, Kerstann KW, Feigenbaum L, Chan CC, and Restifo NP. 2008. Tumor-specific Th17-polarized cells eradicate large established melanoma. Blood 112: 362–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Quezada SA, Simpson TR, Peggs KS, Merghoub T, Vider J, Fan X, Blasberg R, Yagita H, Muranski P, Antony PA, Restifo NP, and Allison JP. 2010. Tumor-reactive CD4(+) T cells develop cytotoxic activity and eradicate large established melanoma after transfer into lymphopenic hosts. J Exp Med 207: 637–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gulley JL, Madan RA, Pachynski R, Mulders P, Sheikh NA, Trager J, and Drake CG. 2017. Role of Antigen Spread and Distinctive Characteristics of Immunotherapy in Cancer Treatment. J Natl Cancer Inst 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gros A, Parkhurst MR, Tran E, Pasetto A, Robbins PF, Ilyas S, Prickett TD, Gartner JJ, Crystal JS, Roberts IM, Trebska-McGowan K, Wunderlich JR, Yang JC, and Rosenberg SA. 2016. Prospective identification of neoantigen-specific lymphocytes in the peripheral blood of melanoma patients. Nat Med 22: 433–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lu YC, and Robbins PF. 2016. Cancer immunotherapy targeting neoantigens. Semin Immunol 28: 22–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.