Summary:
Metabolic and innate immune signaling pathways have co-evolved to elicit coordinated responses. However, dissecting the integration of these ancient signaling mechanisms remains a challenge. Using Drosophila, we uncovered a role for the innate immune transcription factor NF-κB/Relish in governing lipid metabolism during metabolic adaptation to fasting. We found that Relish is required to restrain fasting-induced lipolysis, and thus conserve cellular triglyceride levels during metabolic adaptation, through specific repression of ATGL/Brummer lipase gene expression in adipose (fat body). Fasting-induced changes in Brummer expression and, consequently, triglyceride metabolism are adjusted by Relish-dependent attenuation of Foxo transcriptional activation function, a critical metabolic transcription factor. Relish limits Foxo function by influencing fasting-dependent histone deacetylation and subsequent chromatin modifications within the Bmm locus. These results highlight that the antagonism of Relish and Foxo functions are crucial in the regulation of lipid metabolism during metabolic adaptation, which may further influence the coordination of innate immune-metabolic responses.
Graphical Abstract
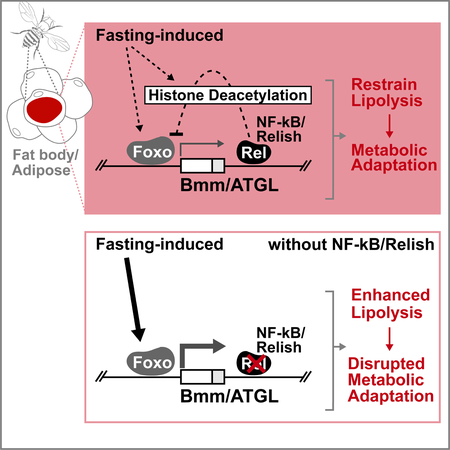
eTOC Blurb:
Molaei et al. show that the Drosophila homolog of NF-κB, Relish, can control lipid homeostasis in response to metabolic adaptation by antagonizing the metabolic transcription factor Foxo. Relish is required to restrain fasting-induced lipolysis by modulating ATGL/Bmm gene expression via the attenuation of Foxo transcription activation function.
Introduction:
The ability to endure bouts of starvation and infection is a primitive challenge in multi-cellular organisms. In order to combat these ancient stressors, organisms have developed tightly regulated and highly integrated adaptive metabolic and innate immune signaling mechanisms. The coupling of metabolic and innate immune responses to regulate metabolism specifically can occur through various mechanisms, such as the interaction between innate immune cells and metabolic tissues (Odegaard and Chawla, 2013; Rosen and Spiegelman, 2014; Schaffler and Scholmerich, 2010). More directly, innate immune signaling pathways (either cell-autonomously or through systemic inflammation) can alter metabolic responses (Becker et al., 2010; Clark et al., 2013; Diangelo et al., 2009; Nunn et al., 2007; Odegaard and Chawla, 2013). To this end, specific tissues such as adipose have coevolved immunological and metabolic function. Adipose tissue plays an important role in the regulation of metabolic homeostasis as an energy and lipid storage organ (Schaffler and Scholmerich, 2010). The majority of stored energy in many metazoans is in the form of lipids, more specifically triglycerides (TAG). Through the coordination of de novo lipid synthesis and lipolysis, adipose (and the liver) mobilize stored TAGs from lipid droplets that are consumed by other tissues in response to shifting energy demands (Arner et al., 2011; Kuhnlein, 2012; Zhao and Karpac, 2017). Animals thus have distinct cell types that act as both nutrient and pathogen sensing systems, allowing for bidirectional and coordinated communication between signaling pathways that respond to diverse stimuli (such as pathogen and diet) in order to adapt metabolic physiology.
NF-κB transcription factors, evolutionarily conserved regulators of innate immunity (Hetru and Hoffmann, 2009; Oeckinghaus et al., 2011), have emerged as a critical node in the bidirectional communication between metabolic and innate immune signaling pathway interactions. Beyond pathogen-dependent innate immunity, these transcription factors also play a diverse role in dictating stress responses to a variety of stimuli through governing assorted gene expression networks. This crosstalk between NF-κB and a number of other signaling pathways helps shape the diverse biological functions of the transcription factor into unique and/or specific responses (reviewed in (Oeckinghaus et al., 2011)). Related to metabolism and independent of infection, NF-κB activation in various metabolic tissues plays an integral part in the breakdown of metabolic homeostasis associated with over-nutrition. However, this is usually driven by its role as an immune regulator through chronic/systemic inflammation (Hotamisligil, 2006; Tornatore et al., 2012). Other findings have revealed an even more intimate, cell-autonomous relationship between NF-κB and metabolism (Mauro et al., 2011). Thus, there is a critical need to characterize the mechanistic connection between NF-κB and various metabolic control networks.
Invertebrate models provide unique advantages to explore the mechanistic underpinnings and the complex integration of these primitive responses, including the incorporation of nutrient and innate immune sensing mechanisms in a distinct tissue (the insect fat body) before these systems evolved into more complex organ types in vertebrates. Here, we leverage the fruit fly Drosophila melanogaster as a model to uncover a potentially ancestral role for NF-κB in governing lipid metabolism.
Results and Discussion:
Relish Function in Fat body Directs Lipid Metabolism in Response to Metabolic Adaptation
In order to explore mechanistic connections between NF-κB and various metabolic control networks, we first assessed lipid homeostasis in Drosophila lacking functional Relish (utilizing the relE20 allele) independent of pathogenic infection. Relish is similar to mammalian p100/p105 NF-κB proteins and contains a Rel-homology domain, as well as ankyrin repeats (found in mammalian inhibitory IκBs) (Buchon et al., 2014; Hetru and Hoffmann, 2009). During ad libitum feeding, NF-κB/Rel mutant adult female flies (relE20 / relE20) have significantly less organismal triglycerides (TAG) compared to genetically matched controls (either OreR or relE20 / + heterozygote flies, 7 days old post-eclosion; Fig. 1A and (Rynes et al., 2012)). However, these changes in TAG correlated with decreases in acute and chronic feeding, and can be rescued by high-calorie (sugar) diets, suggesting that steady-state differences in lipid homeostasis are potentially driven by changes in feeding behavior (Fig. S1A, D). Assaying the major fat storage tissues, we also found that TAG level reduction in mutant animals correlates with strong, but variable, decreases in neutral lipid content in fat body/adipose (Fig. S1B), but not in the intestine (Fig. S1C, E and (Kamareddine et al., 2018)).
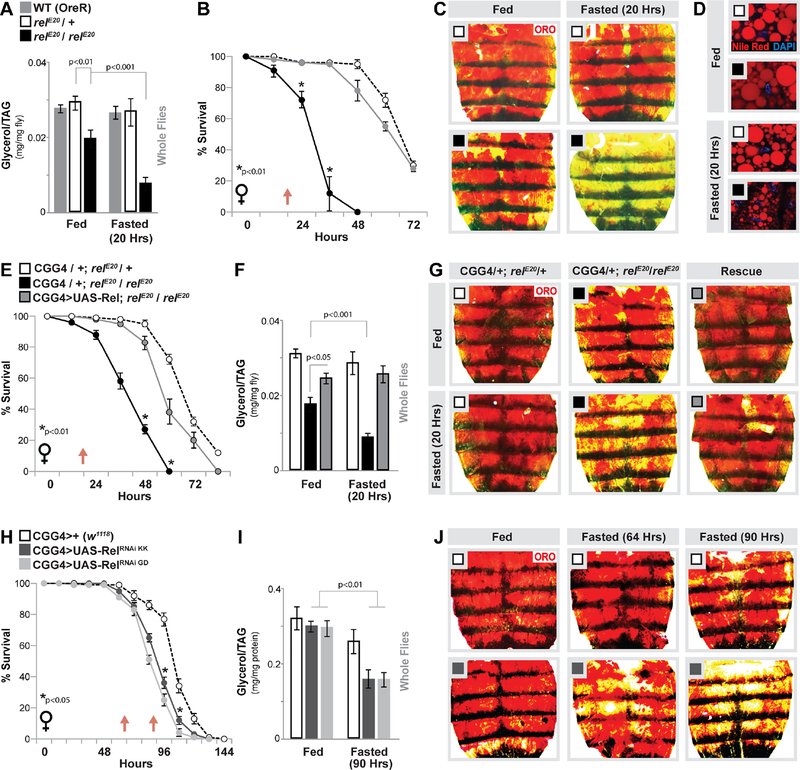
Figure 1: Relish Function in Fat body Directs Lipid Metabolism in Response to Metabolic Adaptation.
(A-D) Relish-dependent changes in lipid metabolism and survival in response to fasting (A) Total triglyceride (TAG) levels of whole flies (OreR (WT-wild type) control, relE20/+ (heterozygote control), or relE20/relE20 (mutant) genotypes) before and after fasting (20 hours). n = 4–5 samples. (B) Starvation resistance of female flies. n = 5 cohorts (total 87–95 flies). The red arrow indicates time-point of fasting assays. (C) Oil Red O (ORO) and (D) Nile red stain of dissected carcass/ fat body before and after fasting (20 hours). Nile red (neutral lipids; red) and DAPI (DNA; blue) detected by fluorescent histochemistry.
(E-G) Re-expressing Relish (UAS-Rel) in fat body (CGGal4) of Relish-deficient flies restores metabolic adaptation responses. (E) Starvation resistance of female flies (CGGal4/+; relE20/+ (control), CGGal4/+; relE20/relE20 (mutant), or CGGal4/UAS-Rel; relE20/relE20 (Rescue)). n = 5 cohorts (total 79–98 flies). The red arrow indicates time-point of fasting assays. (F) Total TAG levels of whole flies (n = 4–5 samples) and (G) ORO stain of dissected carcass/ fat body before and after fasting (20 hours).
(H-J) Changes in lipid metabolism and survival upon Relish depletion (RNAi lines v108469-KK and v49413-GD) in fat body (CGGal4). (H) Starvation resistance of female flies. n = 6 cohorts (total 139–149 flies). The red arrow indicates time-point of fasting assays. (I) Total TAG levels of whole flies (n = 4–5 samples) and (J) ORO stain of dissected carcass/ fat body (only RNAi v108469-KK shown) before and after fasting (64 and/or 90 hours).
Bars and line graph markers represent mean ± SE. All flies were 7 days old post-eclosion. See also Figures S1–S3.
Since ad libitum effects on lipid homeostasis appear to correspond with feeding deficits (i.e. are potentially indirect), we next assayed changes in fat metabolism in Relish mutant animals during metabolic adaptation to fasting. relE20 / relE20 mutant flies are sensitive to starvation (compared to controls, Fig. 1B). Furthermore, Relish-deficient animals display accelerated decreases in organismal TAG levels during acute fasting (at time-points before significant death occurs), while controls flies show little to no change at these same time-points (always comparing within sibling genotypes, Fig. 1A). These changes in TAG levels correlate with a strong reduction of stored neutral lipids/lipid droplets in carcass fat body (Fig. 1C–D), suggesting that there is enhanced or accelerated lipid breakdown during metabolic adaptation in these animals.
The insect fat body acts as a key sensor to link nutrient status and energy expenditure, and as such is the major lipid depository (mainly triglycerides) that combines energy storage, de novo synthesis, and breakdown functions of vertebrate adipose and hepatic tissues (Arrese and Soulages, 2010; Canavoso et al., 2001; Kuhnlein, 2012). This tissue is also essential for Toll and Relish mediated innate immune responses to bacterial infection (Buchon et al., 2014; Hetru and Hoffmann, 2009; Lemaitre and Hoffmann, 2007). Critically, fat body is integral to properly balance lipid catabolism and anabolism in order to modulate organismal energy homeostasis (through lipid supply to other tissues) in response to metabolic or dietary adaptation (Arrese and Soulages, 2010; Kuhnlein, 2012). Expression of full-length Relish in fat body (CGGal4>UAS-Rel) can rescue reduced starvation survival rates and the accelerated loss of lipid storage in relE20 / relE20 mutant flies during fasting (Fig. 1E–G). These data suggest that Relish function in fat body is required to acutely maintain lipid homeostasis throughout the course of metabolic adaptation.
To further confirm an autonomous and potentially direct role for Relish in the regulation of fasting-mediated changes in lipid metabolism, we inhibited Relish specifically in fat body (using multiple, independent RNAi lines; UAS-RelRNAi KK and GD). Attenuating Relish in fat body of female flies (CGGal4>UAS-RelRNAi) leads to starvation sensitivity, as well as accelerated loss of organismal TAG levels and fat body lipid storage in response to fasting (compared to control flies (CGGal4>w1118), Fig. 1H–J; additional RNAi controls experiments can be found in Fig. S1F–J). As expected, fasting-induced changes in fat body lipid storage occur before significant decreases in total TAG levels of whole animals are observed (Fig. 1H–J). Phenotypes were confirmed with an independent fat body driver (PplGal4; Fig. S2A–C), and similar results were found utilizing males (Fig. S2D–E) but not when utilizing another immune cell (hemocyte) driver (HmlGal4; Fig. S2F–G). Conversely, over-expressing full-length Relish (CGGal4>UAS-Rel) or a constitutively active N-terminal fragment (CGGal4>UAS-Rel.68) in fat body significantly limits fasting-mediated decreases in lipids compared to controls (Fig. S2 H–I).
Furthermore, attenuation of upstream components of the Relish signaling pathway phenocopies these Relish loss-of-function effects on lipid metabolism during metabolic adaption. Relish is governed by conserved regulators TAK1 and the IKK (IκB Kinase) signalosome (which consists of homologs of both IKKβ (Drosophila Ird5) and IKKϒ/NEMO (Drosophila Kenny (key)) (Ganesan et al., 2011; Hetru and Hoffmann, 2009)), while the apical caspase DREDD is required for the proteolytic cleavage of the IkB domain, allowing for nuclear translocation (Hetru and Hoffmann, 2009). Inhibiting Kenny or DREDD in fat body of female flies (CGGal4>UAS-DREDDRNAi or KeyRNAi) leads to starvation sensitivity, as well as accelerated loss of organismal TAG levels and fat body lipid storage in response to fasting (compared to control flies (CGGal4>w1118, Fig. S3A–E)). Similarly, attenuating upstream receptors usually required for NF-κB/Relish activation (PGRP family members PGRP-LC (trans-membrane) or PGRP-LE (cytoplasmic)) also leads to decreased lipid storage in fat body after starvation (Fig. S3F), suggesting that at least part of the canonical innate immune pathway is required for these metabolic phenotypes.
Taken together, these data show that Relish can autonomously regulate lipid metabolism in fat body during metabolic adaptation, and suggest that Relish may direct specific metabolic responses to control the breakdown of triglycerides.
Relish Controls Fasting-induced Lipolysis and Bmm Triglyceride Lipase Gene Expression
Properly balancing energy homeostasis in response to metabolic adaptation depends on the ability to coordinate storage, breakdown, and mobilization of lipids, primarily TAG. This coordination requires precise control of metabolic response networks, including changes in metabolic gene expression. To determine potential mechanisms by which the Relish transcription factor could direct cellular triglyceride metabolism during fasting, we assayed transcriptional changes of various metabolic genes related to lipid catabolism or anabolism in Relish-deficient animals (a subset are shown in Fig. S4A). Specifically, we identified the lipase Brummer (Bmm) as being regulated by Relish (Fig. 2A and Fig. S4A). Bmm is the Drosophila homolog of mammalian adipose triglyceride lipase (ATGL), an enzyme that is critical for lipolysis (Gronke et al., 2005). Bmm plays an essential and conserved role in TAG breakdown, and subsequently fatty acid mobilization, from lipid droplets in fat storage tissues during metabolic adaptation (Gronke et al., 2007). In control flies, bmm transcription is mildly induced during acute fasting, but in relE20 / relE20 mutant flies bmm expression is strongly up-regulated (from whole flies). These Relish-dependent changes in bmm transcription appear unique, as Relish-deficiency does not impact fasting-induced changes in other lipases such as Drosophila hormone-sensitive lipase (dHSL), Drosophila lipase 4 (dlip4) or CG5966 (Fig. 2A). Similar results were found in dissected fat body with specific attenuation of NF-κB/Relish in this same tissue (CGGal4>UAS-RelRNAi KK, Fig. 2B–C). These results suggest that Relish function is required to repress or limit Bmm expression in response to metabolic adaptation, and subsequently restrain triglyceride breakdown.
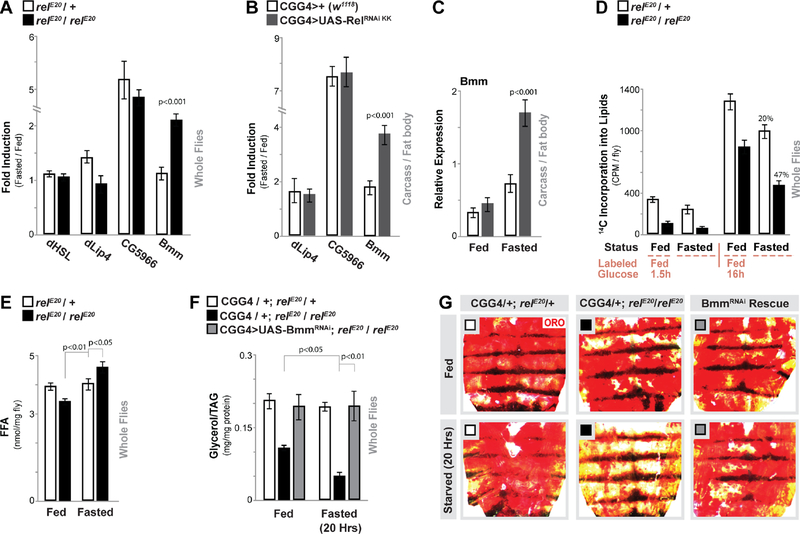
Figure 2: Relish Controls Fasting-induced Triglyceride Lipase Bmm Transcription and Lipolysis.
(A) Drosophila HSL, lip4, CG5966, and bmm transcription (measured by qRTPCR in whole flies, plotted as fold induction (20 hours fasted/fed) of relative expression). relE20/+ (heterozygote control), or relE20/relE20 (mutant) genotypes. n = 3 samples.
(B-C) Changes in lip4, CG5966, and bmm transcription (measured by qRT-PCR in dissected carcass / fat body, plotted as fold induction (64 hours fasted/fed) of relative expression) upon Relish depletion (RNAi line v108469-KK) in fat body (CGGal4). (C) Relative expression values (from (B)) for bmm transcription. n = 3–4 samples.
(D) Quantification of lipid breakdown. Incorporation of 14C-labeled glucose into total lipids (from whole flies) from labeled-glucose fed (1.5 hours or 16 hours) or fasted (20 hours) flies are shown. Percent change in loss of 14C-labeled lipids after fasting is also shown. n = 3–4 samples.
(E) Free fatty acid (FFA) levels measured in whole flies before and after fasting (20 hours). n = 4 samples.
(F-G) Attenuating Bmm (RNAi line v37877) in fat body (CGGal4) of Relish-deficient flies restores metabolic adaptation responses. (F) Total TAG levels of whole flies (n = 3–4 samples) and (G) Oil Red O stain of dissected carcass/ fat body before and after fasting (20 hours, CGGal4/+; relE20/+ (control), CGGal4/+; relE20/relE20 (mutant), or CGGal4/UAS-Bmm RNAi; relE20/relE20 (Rescue)).
Bars represent mean ± SE. All flies were 7 days old post-eclosion. See also Figure S4.
To correlate this difference in gene expression to differences in lipolysis, we next employed an assay to measure dynamic changes in lipid content based on the incorporation of radiolabeled glucose (14C-glucose) into lipids during fatty acid synthesis in vivo. After acute feeding (1.5 hours) of a diet containing 14C-glucose, Relish mutant flies show drastic changes in glucose-incorporation (synthesis) that is likely due to changes in feeding behavior (Fig. 2D and Fig. S1 A). 16 hours of feeding minimized these differences in synthesis, and subsequent analysis of newly synthesized 14C-labeled lipids during fasting showed an increased rate of breakdown in relE20 / relE20 mutant flies (47% in mutants compared to 20% in controls, Fig. 2D). This change in the rate of breakdown correlated with increases in free fatty acids (Fig. 2E). Finally, genetically attenuating Bmm lipase in fat body (CGGal4>UAS-BmmRNAi, (Baumbach et al., 2014)) can rescue the accelerated loss of lipid storage/triglycerides in relE20 / relE20 mutant flies during fasting (Fig. 2F–G).
These data collectively reveal that Relish function is required to limit fasting-induced Bmm gene expression and subsequently restrain triglyceride lipolysis during metabolic adaptation.
Following these results, we wanted to further explore the mechanism by which the Relish transcription factor can context-dependently attenuate Bmm expression. Utilizing Clover (Cis-element OVERrepresentation) software (Frith et al., 2004), we identified conserved NF-κB DNA binding motifs (κB sequence sites identified as GGG R N YYYYY, (Busse et al., 2007)) throughout the first intron of the Bmm locus (Fig. 3A). To assess binding, we used a previously characterized Relish antibody to perform chromatin immunoprecipitation (ChIP)-qPCR experiments (Ji et al., 2016). Relish binding in fed or fasted wild type flies is significantly enriched (compared to IP’s using serum controls) at binding motif(s) approximately 1 kB downstream from the transcriptional start site (R1, Fig. 3A). We also cloned this putative Bmm regulatory region upstream of RFP in order to generate in vivo expression reporters (individual transgenic flies carrying either a wild type reporter (endogenous locus, Bmm_Int_WT-RFP) or a reporter with a deletion in the Relish DNA binding site (Bmm_Int_Δ1-RFP), Fig. 3B). While the unaltered region only slightly influenced RFP reporter activity in fed or fasted conditions, eliminating the Relish binding site leads to minimal enhanced reporter activity under fed conditions and strong increases in RFP activity during fasting (primarily in fat body of carcass and head, Fig. 3B). Thus, this Relish binding site within the first Bmm intron acts as an important regulatory region to limit induced gene expression.
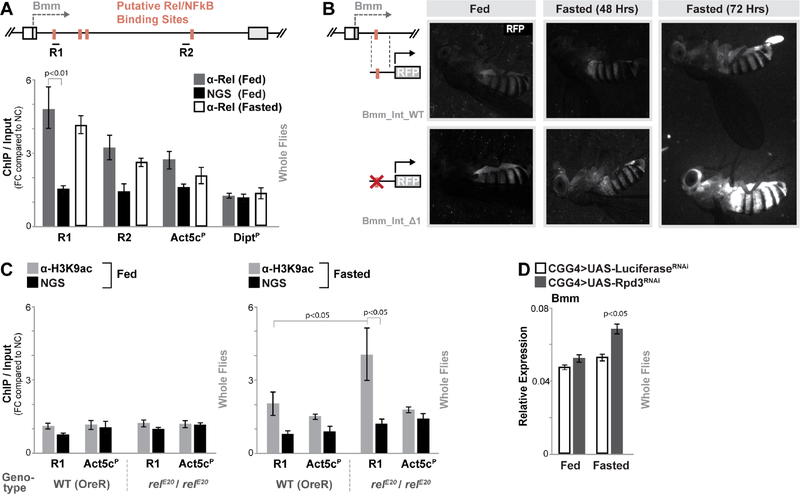
Figure 3: Relish binds to a Regulatory Region in the Bmm Locus.
(A) Schematic shows Bmm locus (focusing on first intron proximal to transcription start site) and putative NF-κB/Rel binding motifs (identified by Clover). R1 and R2 represent regional target sites (and corresponding primer sets) tested in ChIP-qPCR analysis. The histogram represents ChIP-qPCR analysis of Relish binding to the Bmm locus (compared to the Actin5c promoter (Act5cP) and the Diptericin promoter (DiptP) in fed or fasted (20 hours) conditions. ChIP-qPCR analysis with normal goat serum (NGS) is included as a control. Plotted as fold change (FC) of indicated PCR primer sets compared to a negative control (NC) primer set. n = 3 biological replicates.
(B) Requirement of Bmm locus Rel binding site in limiting induced gene expression measured by RFP fluorescence in transgenic flies carrying indicated reporters (during fed and fasted (48 or 72 hours) conditions).
(C) ChIP-PCR analysis of H3K9ac enrichment in R1/Relish-binding region of the Bmm locus in wild type (WT; OreR) and relE20/relE20 (mutant) genotypes before and after fasting (20 hours). n = 3 biological replicates.
(D) Changes bmm transcription (measured by qRT-PCR in whole flies) before and after fasting (20 hours) upon Rpd3 depletion (RNAi line TRiP 36800) in fat body (CGGal4). Controls are a genetically matched RNAi targeting luciferase. n = 4–6 samples.
Bars represent mean ± SE. All flies were 7 days old post-eclosion. See also Figure S3 and S4.
Relish binding at this region is similar in fed and fasted states (Fig. 3A). We also did not find any evidence of classical Relish transcriptional activation function during acute fasting. First, innate immune target gene expression (Drosomycin and Diptericin) and Relish DNA binding to innate immune gene promoters (Diptericin) were not changed during fasting (Fig. S4B and Fig. 3A). Second, metabolic adaptation did not significantly alter nuclear localization of Relish in fat body (Fig. S4C). Thus, in order to explore how Relish limits or represses fasting-induced Bmm expression, despite its constitutive binding to DNA and distinct from its transcriptional activation function, we assessed histone/chromatin changes in Relish-deficient flies. Histone deacetylases (HDACs) have been shown to accumulate in the nucleus during metabolic adaptation, influencing gene expression in a fasting-dependent manner through chromatin regulation and transcription factor deacetylation (Mihaylova et al., 2011; Nakajima et al., 2016; Wang et al., 2011). Furthermore, previous studies have linked interactions of NF-κB transcription factors and HDACs with NF-κB-dependent transcriptional repression (Ashburner et al., 2001; Dong et al., 2008; Morris et al., 2016). We thus hypothesized that Relish might repress Bmm gene expression through influencing histone modifications during fasting, when histone modifiers (such as HDACs) in the nucleus are elevated. Using ChIP-qPCR, we monitored histone 3 lysine 9 acetylation (H3K9ac, a post-translational modification generally associated with transcriptional activation) at this Bmm regulatory region in Relish-deficient animals and controls. During feeding, there is no change in H3K9ac enrichment at this locus between genotypes (Fig. 3C). However, during fasting relE20 / relE20 mutant flies display a significant enrichment (compared to controls) of H3K9ac at the site of Relish binding (Fig. 3C), indicative of promoter or enhancer activation. Analysis of modEncode ChIP-Seq. databases associated with histone modifications (in adult female flies) also revealed that this site is generally enriched for other modifications linked to gene expression regulation (such as H3K27ac, H3K4me3, and H3K4me1), further indicating that this locus is an important regulatory region (Contrino et al., 2012). Additionally, inhibiting a single HDAC in fat body (Rpd3 (Drosophila HDAC1), CGGal4>UAS-Rpd3RNAi) can drive small, but significant, increases in fasting-induced Bmm transcription (from whole flies, Fig. 3D) and accelerate fat body lipid usage (Fig. S4D).
Taken together, these data show that Relish can bind to a putative regulatory region within the Bmm locus during both feeding and fasting. In response to fasting, the presence of Relish can influence fasting-dependent histone acetylation and chromatin changes that are consistent with transcriptional repression.
Foxo and Relish Antagonism Dictates Fasting-induced Bmm Transcription and Lipolysis
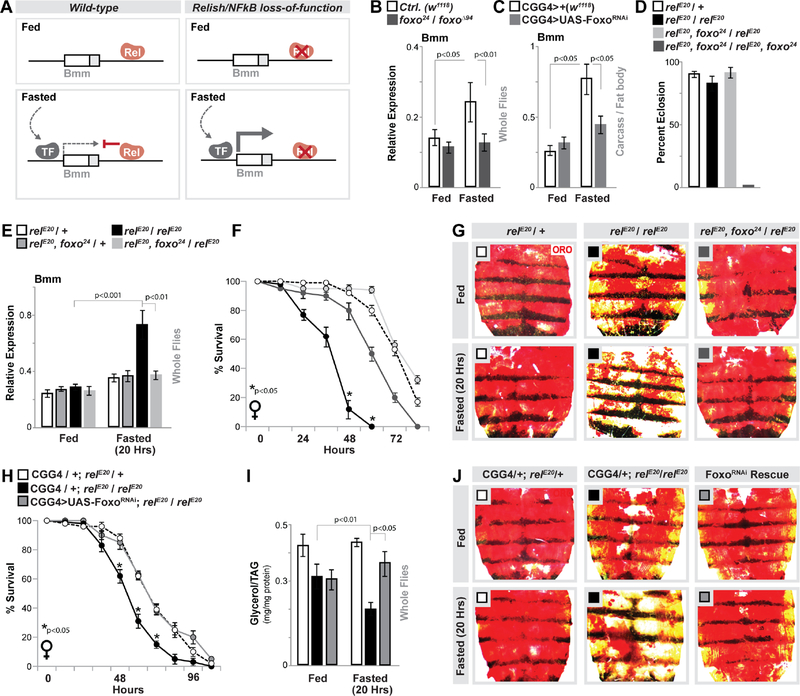
The unique ability of Relish to limit or repress fasting-induced Bmm transcription correlates with attenuation of H3K9ac at Bmm regulatory regions. We thus hypothesized that Relish binding to the Bmm locus leads to fasting-dependent chromatin changes, which subsequently limit transcription activation function of other factors that are induced during metabolic adaptation (Fig. 4A). We assessed various metabolic transcription factors and found that Foxo, a critical regulator of lipolysis and catabolism in general, is required for Relish-dependent changes in ATGL/Bmm expression during metabolic adaptation (Fig. S4E–F). Firstly, Foxo (of which there is a single ortholog in Drosophila) is activated during metabolic adaptation and required for fasting-induced ATGL/Bmm expression across taxa, including in the fly fat body (Fig. 4B–C (from dissected fat body) and (Chakrabarti and Kandror, 2009; Kang et al., 2017; Wang et al., 2011)). Full Relish/Foxo double mutant animals (using the foxo24 allele) are synthetic lethal during metamorphosis (relE20, foxo24 / relE20, foxo24, Fig. 4D). However, simply reducing Foxo gene-dose in NF-κB/Relish mutant flies (relE20, foxo24 / relE20) completely rescues fasting-dependent increases in Bmm expression (from whole flies, Fig. 4E), starvation survival rates, and increases in lipolysis (accelerated loss of lipid storage) in Relish-deficient flies during metabolic adaptation (Fig. 4F–G). Molecular analysis of Foxo transcription activation function also showed that Foxo binding to the Bmm promoter is slightly, but significantly, elevated in Relish-deficient flies only during fasting (Fig. S4G–H). Furthermore, attenuating Foxo specifically in fat body (CGGal4>UAS-FoxoRNAi, (Zhao and Karpac, 2017)) rescues the enhanced depletion of triglycerides/lipid storage and starvation sensitivity associated with relE20 / relE20 mutant flies during fasting (Fig. 4H–J). Foxo transcription activation function is thus required for Relish-dependent changes in lipid metabolism, highlighting that Relish/Foxo integration and antagonism is critical to maintain triglyceride metabolism throughout the course of metabolic adaptation.
Figure 4: Foxo and Relish Antagonism Dictate Fasting-induced Bmm Transcription and Lipolysis.
(A) Putative model highlighting the integration of Relish (Rel) and other fasting-induced transcription factors (TF).
(B-C) Changes in bmm transcription (measured by qRT-PCR in whole flies or dissected fat body) before and after fasting (64 hours) in Foxo mutant (w1118;; foxo24/foxoΔ94) and control (w1118) genotypes, as well as upon Foxo depletion (RNAi line v106097) in fat body (CGGal4). n = 4 samples.
(D) Percent eclosion of adult animals of indicated mutants / double mutants. n = 67–90 animals.
(E) Changes in bmm transcription (measured by qRT-PCR in whole flies) before and after fasting (20 hours) in controls (relE20/+ and relE20, foxo24/+), mutant (relE20/relE20) and mutant with reduction in Foxo gene dose (relE20, foxo24/relE20) genotypes. n = 3 samples.
(F) Starvation resistance of Relish-deficient female flies with reduction in Foxo gene dose (n = 4 cohorts (total 68–78 flies)), and (G) ORO stain of dissected carcass/ fat body before and after fasting (20 hours).
(H-J) Attenuating Foxo (RNAi line v106097) in fat body (CGGal4) of Relish-deficient flies restores metabolic adaptation responses. (H) Starvation resistance of female flies (CGGal4/+; relE20/+ (control), CGGal4/+; relE20/relE20 (mutant), or CGGal4/UAS-Foxo RNAi; relE20/relE20 (Rescue)). n = 6 cohorts (total 117–140 flies). The red arrow indicates time-point of fasting assays. (I) Total TAG levels of whole flies (n = 5 samples) and (J) ORO stain of dissected carcass/ fat body before and after fasting (20 hours).
Bars and line graph markers represent mean ± SE. All flies were 7 days old post-eclosion. See also Figure S4.
In summary, we uncovered a role for the innate immune transcription factor Relish in governing lipid metabolism during metabolic adaptation to fasting utilizing Drosophila. Relish is required to restrain fasting-induced lipolysis, and thus conserve cellular triglyceride levels and promote survival during metabolic adaptation, through specific repression of Bmm lipase gene induction in fat body/adipose. Fasting-induced changes in Bmm expression and triglyceride metabolism are adjusted by Relish-dependent attenuation of Foxo transcriptional activation function, likely through regulation of histone acetylation.
These findings thus suggest that association of Relish with histone modifiers (and subsequent changes in chromatin accessibility at gene regulatory regions) functions to control or limit the induced level of certain metabolic genes. Indeed, a few previous studies have highlighted that mammalian p65/RelA or Drosophila Relish can negatively regulate gene expression, including innate immune targets, through changes in chromatin (Ashburner et al., 2001; Campbell et al., 2004; Dong et al., 2008; Morris et al., 2016). This modification of chromatin and repression of gene expression likely occurs through recruitment of HDACs (HDAC 1/2) to promoter/enhancer regions of target genes, thus influencing histone acetylation, chromatin remodeling, and function of NF-κB itself or other transcriptional activators. We show here that Relish can bind a metabolic target gene during both feeding and fasting, but that uniquely during fasting, it can influence H3K9 acetylation levels at this target gene and subsequently limit transcriptional activation function of positive regulators (such as Foxo). This allows NF-κB transcription factors precise control of specific metabolic gene expression, potentially through metabolic changes in HDAC nuclear localization, in a context-dependent manner. To this end, we have uncovered other metabolic target genes of Relish (in unique tissues with unique metabolic functions) regulated through similar mechanisms (Fig. S4I).
It remains unclear if Relish binding to metabolic target genes in fed states, independent of metabolic stress, can influence gene expression and physiology. However, this may represent constitutive or steady-state function of NF-κB (described in mammals (Baratin et al., 2015; Birbach et al., 2002; O’Dea et al., 2007)), and thus could act as a priming mechanism that promotes the maintenance of metabolic homeostasis during acute stress (such as fasting). While our data show that NF-κB can direct lipid metabolism independent of infection, Foxo (as well as histone deacetylases) are integrated with innate immune responses through a variety of mechanisms (Becker et al., 2010; Karpac et al., 2011; Shakespear et al., 2011). Additional experimental evidence suggests that Relish, and perhaps Relish-Foxo antagonism and Bmm gene regulation, manipulate triglyceride metabolism after chronic systemic bacterial infection (Fig. S4J–M). It is thus possible that NF-κB-dependent regulation of triglyceride catabolism, through limiting Foxo-mediated lipolysis, also play a role in governing innate immune homeostasis in response to pathogenic infections.
STAR★METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jason Karpac (karpac@tamhsc.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Drosophila Husbandry and Strains
A detailed list of fly strains used for these studies is provided in Table S1. All flies were reared on standard yeast and cornmeal-based diet at 25°C and 65% humidity on a 12 hr light/dark cycle, unless otherwise indicated. A standard lab diet (cornmeal-based) for rearing was made with the following protocol: 14g Agar/165.4g Malt Extract/ 41.4g Dry yeast/ 78.2g Cornmeal/ 4.7ml propionic acid/ 3g Methyl 4-Hydroxybenzoate/ 1.5L water.
In order to standardize metabolic results, 2–3 days after eclosion, mated adult flies were placed on a simple sugar-yeast (SY) diet for 5 days. The standard SY diet was made with the following protocol: 1.0g agar/10g sucrose/ 10g yeast/ 0.3 ml propionic acid/ 100 ml water/ 1.5 mL Methyl 4-Hydroxybenzoate. Ingredients were combined, heated to at least 102°C, and cooled before pouring. The high sugar diet (Fig. S1D) was prepared as follows: 1.5 g agar/30 g sucrose/10 g yeast/0.3 ml propionic acid/100 ml water.
For RU486 food, RU486 or vehicle (ethanol 80%) was mixed with food (SY diet), resulting in a 200 uM concentration of RU486 in the food, unless otherwise indicated.
All experiments presented in the results were done utilizing female flies 7 days old post-eclosion (following dietary protocol referenced above) with the exception of data from males (also 7 days old post-eclosion) presented in Figure S2D–E. The UAS-RelishRNAi (both trangenic lines), UAS-FoxoRNAi, UAS-KennyRNAi, UASDreddRNAi, UAS-BmmRNAi, UAS-PGRP-LCRNAi, UAS-PGRP-LERNAi, CGGal4, PplGal4, and HmlGal4 lines used throughout the paper were backcrossed 8–10X into the w1118 background that was used as a control strain. The ebony mutation/marker eS was removed from the relE20 mutant background, with the relE20 mutation finally outcrossed into a wild-type (OreR) background. The efficiency of transgenic RNAi lines UAS-RelishRNAi (VDRC: 108469 (KK) and VDRC: 49413 (GD)), UAS-KennyRNAi (VDRC: 7723 (GD)) and UAS-DreddRNAi (VDRC: 104726 (KK)) were confirmed in this study. The efficiency of transgenic RNAi lines UAS-FoxoRNAi (VDRC: 106097 (KK)), UAS-BmmRNAi (VDRC: 37877 (GD)), UAS-PGRP-LCRNAi (VDRC: 101636 (KK)), UAS-PGRP-LERNAi (VDRC: 23664 (GD)), and UAS-Rpd3RNAi (TRiP: 36800) were confirmed in previous studies.
Generation of Transgenic Flies
To create transgenic flies carrying Brummer/NF-κB expression reporters (Bmm_Int_WT-RFP and Bmm_Int_Δ1-RFP), 300 bp of Bmm/ATGL locus containing κB binding site was selected. The DNA fragment carrying wild-type or mutated (6 bp deletion) κB binding site were synthesized using gBlock technology (Integrated DNA Technology) and cloned into a ϕ31-based DsRed.T4 plasmid reporter system. Finally, the constructs were injected into w−, attP40 embryos (Rainbow Transgenic Flies, Inc).
Bmm/ATGL intronic region containing wild type κB binding site:
GCATGCACGCATTGAATTGAATTTTATTGATAAGCTTGTTTGCGTTTGTAGGT CGCTAGGAAGTCAATGGGGATCTTTCATAATTGACTGCGATAGTGTGTGTGT GTTTTTGGGCGTGTTTGTCCAATTTCGAAGGGGGCTCGTCCCATCCGCTCA AAAGAAAACTGCGGCGCAGTTGAAAAACCTTACGAAAACAGAAAAACAAGTT TCGTATGCCCGGGACAACGCACTTTTGTAAAGCGGCACCCGAATATATGGG CAAATGGTTGGGCACAGCGGTGGGTATATGAATAGCAACGCAGTCCGAAAA CATTTCATCAAACTCGAG
Bmm/ATGL intronic region containing mutant κB binding site:
GCATGCACGCATTGAATTGAATTTTATTGATAAGCTTGTTTGCGTTTGTAGGT CGCTAGGAAGTCAATGTTTCATAATTGACTGCGATAGTGTGTGTGTGTTTTTGGGCGTGTTTGTCCAATTTCGAAGGGGGCTCGTCCCATCCGCTCAAAAGAA AACTGCGGCGCAGTTGAAAAACCTTACGAAAACAGAAAAACAAGTTTCGTAT GCCCGGGACAACGCACTTTTGTAAAGCGGCACCCGAATATATGGGCAAATG GTTGGGCACAGCGGTGGGTATATGAATAGCAACGCAGTCCGAAAACATTTC ATCAAACTCGAG
METHOD DETAILS
de novo Lipid Synthesis Analysis
After 5 days of feeding on standard diet, 200 female flies were transferred to a bottle containing standard diet with 2μCi of 14C-labeled glucose (thatched into the top of food). After 1.5 hrs or 16 hrs feeding, total lipid was extracted from 5 flies for each sample (Fed samples). The other half were transferred to starvation vials (water only) and total lipids were extracted immediately after 12 hrs of fasting (Fasted samples). For extraction of total lipids, 5 flies for each sample were homogenized in 2 ml Folch reagent (CHCl3: MeOH 1:1 v/v). Then, 0.4 mL of cold 0.1 M KCl was added, thoroughly mixed by 1 min vortexing and then spun down at 3000 rpm, 4°C for 5 min. The lower phase wa s transferred to a glass tube and dried down. Dried lipids were re-suspended in 3 ml of scintillation fluid and CPM was counted using a liquid scintillation analyzer (Packard- 2500 TR). Fed samples were indicative of the rate of incorporation of glucose into lipids and Fasted samples were indicative of the breakdown of the labeled lipids.
Analysis of Gene Expression
Total RNA from the whole bodies or dissected fat body/carcass (with all of the eggs and intestines removed) of flies were extracted using Trizol and complementary DNAs were synthesized using Superscript III (Invitrogen). Quantitative Real-Time PCR (qRT-PCR) was performed using SYBR Green, the Applied Biosystems StepOnePlus Real-Time PCR system, and the primer sets described in Table S2. Results are the average ± standard error of at least three independent biological samples, and quantification of gene expression levels calculated using the ΔCt method and normalized to actin5C expression levels.
Metabolite Measurements
For triglyceride (TAG) assays, five beheaded adult females were homogenized in 200 μl of PBST (PBS, 0.1% Tween 20) and heated at 70°C for 5 min to inactivate endogenous enzymes. Samples were centrifuged at 4000 rpm for 3 min at 4 °C and 10 uL of the cleared extract were used to measure triglycerides (StanBio Liquicolor Triglycerides Kit) or protein concentrations (Bio-Rad Protein Assay Kit) according to the manufacturer instructions. TAG levels were normalized to weight or protein levels depending on genotype (some genotyped reveal drastic changes in wet weight after starvation that limits interpretation of normalized metabolite data). Note: The kit measures glycerol cleaved from TAG and diacylglycerol (DAG), as well as minimal amounts of free glycerol; the majority of neutral lipids extracted from whole flies are TAG.
The levels of free fatty acids (FFAs) were measured using the Free Fatty Acid Quantification Kit (Sigma-Aldrich), following the manufacturer’s instruction. Metabolite samples preparation was the same as described for TAG measurements.
Oil Red O staining
Intestines and fat body/carcasses (with all of the eggs and intestines removed) of flies were dissected in PBS and fixed in 4% paraformaldehyde for 20 min, then washed twice with PBS, incubated for 20 min in fresh Oil Red O solution (6 ml of 0.1% Oil Red O in isopropanol and 4 ml distilled water, and passed through a 0.45 μm syringe), followed by rinsing with distilled water. Bright-field images were collected using a Leica M165 FluoCombi stereoscope system (utilizing a single focal plane) and processed using Leica software and Adobe Photoshop. Note: Contrast (red – neutral lipids vs. yellow/black – cuticle) was enhanced using Adobe Photoshop (equal for all images) in order to better visualize the red stain.
Nile Red staining
Fat body/carcasses were dissected in PBS (with all of the eggs and intact intestines removed) and fixed in 4% paraformaldehyde for 20 min. Fixed carcasses were then washed twice with PBS, incubated for 30 min in fresh Nile Red solution with DAPI (1μl of 0.004% Nile Red Solution in 500 μl PBS), followed by rinsing with distilled water. Confocal images were collected using a Nikon Eclipse Ti confocal system (utilizing a single focal plane) and processed using Nikon software and Adobe Photoshop.
Immunostaining and Microscopy
Flies were dissected in PBS and fat body/carcasses were fixed with 4% paraformaldehyde containing 0.1% Tween-20 and 0.1% Triton X-100 for 10 min at room temperature, washed 2 times with PBS containing 0.1% Triton X-100 (PBST) and then blocked in blocking buffer (PBST containing 0.1% BSA and 0.0025 Sodium Azide) for 1 h. The primary antibody Rabbit anti-Relish (RayBiotech, RB-14–0004) (1:500) was applied overnight at 4°C. Alexa Fluor 488-conjugated anti-Rabbit IgG antibody (Jackson Immunoresearch, 1:500) was incubated for 2 h at room temperature. Hoechst was used to counterstain DNA. Confocal images were collected using a Nikon Eclipse Ti confocal system (utilizing a single focal plane) and processed using the Nikon software and Adobe Photoshop.
Feeding Behavior
The CAFE assay was performed as follows: Briefly, a single fly was transferred from SY standard food to vials filled with 5 ml of 1.5% agar that maintains internal humidity and serves as a water source. Flies were fed with 5% sucrose solution and maintained in 5 ul capillaries (VWR, #53432–706). After twelve hours habituation, the old capillaries were replaced with a new one at the start of the assay. The amount of liquid food consumed was recorded after 24 hr and corrected on the basis of the evaporation (typically < 10% of ingested volumes) observed the identical vials without flies.
Feeding assays on blue dye-labeled food were performed as follows: 30 flies were transferred from standard food to vials filled with identical medium containing 0.5% brilliant blue. Feeding was interrupted after 1h and 5 flies each were transferred to 50 μl 1 x PBS containing 0.1% Triton X-100 (PBST) and homogenized immediately. Blue dye consumption was quantified by measuring the absorbance of the supernatant at 630 nm (A630). Various amounts of dye-containing food were weighed, homogenized in PBST, and measured (A630) in order to create a standard curve used to quantify blue dye food consumption of flies.
Chromatin Immunoprecipitation (ChIP)
Approximately 100 adult female flies (5–10 days old post-eclosion) were ground in liquid nitrogen then homogenized and cross-linked (10 minutes at RT) in 600 uL of 1xPBS containing 1% formaldehyde, 1mM PMSF and 1x Protease Inhibitor cocktail (Thermo Scientific). The homogenate was centrifuged for 20 min at 12000 x rpm (4° C). The pellet was washed twice by resuspending in 600 uL of 1x PBS containing 1mM PMSF and 1x Protease Inhibitor cocktail and centrifuged at 12000 x rpm for 20 min (4° C). To lyse tissue and cells, the pellet was resuspended in 600 uL of RIPA buffer (10 mM Tris-HCl, pH 7.6, 1 mM EDTA, 0.1% SDS, 0.1% Na-Deoxycholate, 1% Triton X-100, 1mM PMSF and 1x Protease Inhibitor cocktail) then incubated at RT for 30 min.
The chromatin was sheared to 500–1000 bp DNA fragments using a Diagenode sonicator (20 min sonication, highest power, 30 sec sonication, 30 sec rest). After sonication, the sheared chromatin was centrifuged for 20 min at 12000x rpm, 4° C. The supernatant was collected, aliquoted, snap-frozen, and stored at −80° C. For immunoprecipitation, 40 uL of protein A magnetic beads (Thermos Scientific) were conjugated (4 hours incubation at 4° C) with 10 uL of Normal Goat Serum (Rockland, used as control), 10 uL of Rabbit anti-Relish (RayBiotech, RB-14–0004), or 2 uL of anti-Histone H3 (acetyl 9) antibody (abcam, ab4441). After applying beads to the magnet and removing supernatant, 100 uL of chromatin was diluted 1:10 with dilution buffer (20 mM Tris-HCl, pH 8, 2 mM EDTA, pH 8, 150 mM NaCl, 1% Triton X-100) and incubated overnight with beads. Beads were washed with following buffers at 4° C, for 10 min each: 2x with 1mL of RIPA buffer + 1mM PMSF + 1x protease inhibitor; 2x with 1mL of RIPA buffer + 0.3 M NaCl; 2x with 1mL of LiCl buffer (0.25 M LiCl, 0.5% Triton X-100, 0.5% NADOC); 1x with 1 mL of 1x TE + 0.2 Triton X-100; 1x with 1mL of 1x TE.
To reverse crosslink, beads were re-suspended in 100uL of 1x TE + 3 uL 10% SDS + 5 uL of 20mg/mL Proteinase K (VWR) and incubated at 65° C overnight. Beads were applied to the magnet and supernatant was transferred to a PCR purification column (Qiagen PCR purification kit) to purify DNA. To prepare Input (chromatin extract without Immunoprecipitation), 10 uL of chromatin extract were incubated with proteinase K then applied to PCR purification column. For all Immunoprecipitated (IP) and Input samples, DNA was eluted in 30 uL of water, and 2uL of that was used as template for qRT-PCR (see Table S2 for primer sets). The upstream region of the actin5c gene (Act5CP) and Normal Goat Serum were used as controls.
To assess enrichment, %Input was calculated first (between ChIP DNA and input DNA for each primer set). Then the fold change in enrichment was calculated by dividing %Input of each primer set to %Input of a negative control primer set designed for Drosophila (Drosophila Negative Control primer set 1, Active Motif).
Generating Germ-Free Animals
Female adult flies were fed Penicillin/Streptomycin for 5 days to remove bacteria from the gut. Then single flies were washed with 70% ethanol (to remove bacteria from cuticle), dried completely and ground in 200 uL of sterile LB broth and quickly spun down. 10 uL of the supernatant was cultured on Nutrient agar plates. Colonies were counted after 2 days incubation at 29° C.
Starvation Sensitivity Analysis
Adult flies (20–25 flies per vial/cohort) were provided with only water (absolutely no food) on filter paper with a KimWipe, ensuring water was present throughout the analysis. The number of dead flies in each vial was recorded every 12 hours, and data is presented as the mean survival of cohorts.
Septic/Systemic Infection Assay
To induce systemic infection, 5–10 days adult female flies of indicated genotypes were pocked in their thorax with a sterilized tungsten needle dipped into a concentrated overnight culture of Ecc15 (Erwinia carotovora carotovora 15, gram-negative bacteria, OD600 ~ 300). The pocked flies were incubated at 25° C and total RNA and metabolite samples were collected from the whole body of flies at 16 hours and 40 hours after septic infection, as well as from unchallenged flies. Heat-killed bacteria were inactivated by incubating a 1 mL aliquot of the bacterial suspension at 65° C for 20 mins before poking.
QUANTIFICATION AND STATISTICAL ANALYSIS
All p-values were calculated using the Student’s t test with unpaired samples. All error bars represent standard error, and n representations are as follows: Number of whole flies: Figure 1A–B, E–F, H–I; Figure 2A, D, E–F; Figure 3A, C–D; Figure 4B, D, E–F, H–I; Figure S1A, D–J; FigureS2A–B, D–I; Figure S3A–D; Figure S4A–B, E–M.
Number of dissected carcass/fat body: Figure 2B–C; Figure 4C.
Exact values of all n’s can be found in Figure legends.
Supplementary Material
Highlights:
Fat body NF-κB/Relish function controls fasting-induced metabolic adaptation
NF-κB/Relish directs fasting-induced lipolysis and triglyceride metabolism
NF-κB/Relish regulates Foxo-dependent triglyceride lipase gene expression
Foxo and NF-κB/Relish antagonism dictate triglyceride metabolism
Acknowledgments
This work was supported by the National Institute of Diabetes and Digestive and Kidney Diseases (grant R01 DK108930 to J.K) and by the American Heart Association (pre-doctoral fellowship to M.M).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests.
References:
- Arner P, Bernard S, Salehpour M, Possnert G, Liebl J, Steier P, Buchholz BA, Eriksson M, Arner E, Hauner H, et al. (2011). Dynamics of human adipose lipid turnover in health and metabolic disease. Nature 478, 110–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrese EL, and Soulages JL (2010). Insect fat body: energy, metabolism, and regulation. Annual review of entomology 55, 207–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner BP, Westerheide SD, and Baldwin AS Jr. (2001). The p65 (RelA) subunit of NF-kappaB interacts with the histone deacetylase (HDAC) corepressors HDAC1 and HDAC2 to negatively regulate gene expression. Mol Cell Biol 21, 7065–7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baratin M, Foray C, Demaria O, Habbeddine M, Pollet E, Maurizio J, Verthuy C, Davanture S, Azukizawa H, Flores-Langarica A, et al. (2015). Homeostatic NF-kappaB Signaling in Steady-State Migratory Dendritic Cells Regulates Immune Homeostasis and Tolerance. Immunity 42, 627–639. [DOI] [PubMed] [Google Scholar]
- Basset A, Khush RS, Braun A, Gardan L, Boccard F, Hoffmann JA, and Lemaitre B (2000). The phytopathogenic bacteria Erwinia carotovora infects Drosophila and activates an immune response. Proc Natl Acad Sci U S A 97, 3376–3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumbach J, Hummel P, Bickmeyer I, Kowalczyk KM, Frank M, Knorr K, Hildebrandt A, Riedel D, Jackle H, and Kuhnlein RP (2014). A Drosophila in vivo screen identifies store-operated calcium entry as a key regulator of adiposity. Cell Metab 19, 331–343. [DOI] [PubMed] [Google Scholar]
- Becker T, Loch G, Beyer M, Zinke I, Aschenbrenner AC, Carrera P, Inhester T, Schultze JL, and Hoch M (2010). FOXO-dependent regulation of innate immune homeostasis. Nature 463, 369–373. [DOI] [PubMed] [Google Scholar]
- Birbach A, Gold P, Binder BR, Hofer E, de Martin R, and Schmid JA (2002). Signaling molecules of the NF-kappa B pathway shuttle constitutively between cytoplasm and nucleus. J Biol Chem 277, 10842–10851. [DOI] [PubMed] [Google Scholar]
- Buchon N, Silverman N, and Cherry S (2014). Immunity in Drosophila melanogaster--from microbial recognition to whole-organism physiology. Nat Rev Immunol 14, 796–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busse MS, Arnold CP, Towb P, Katrivesis J, and Wasserman SA (2007). A kappaB sequence code for pathway-specific innate immune responses. EMBO J 26, 3826–3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell KJ, Rocha S, and Perkins ND (2004). Active repression of antiapoptotic gene expression by RelA(p65) NF-kappa B. Mol Cell 13, 853–865. [DOI] [PubMed] [Google Scholar]
- Canavoso LE, Jouni ZE, Karnas KJ, Pennington JE, and Wells MA (2001). Fat metabolism in insects. Annu Rev Nutr 21, 23–46. [DOI] [PubMed] [Google Scholar]
- Chakrabarti P, and Kandror KV (2009). FoxO1 controls insulin-dependent adipose triglyceride lipase (ATGL) expression and lipolysis in adipocytes. J Biol Chem 284, 13296–13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee N, and Bohmann D (2012). A versatile PhiC31 based reporter system for measuring AP-1 and Nrf2 signaling in Drosophila and in tissue culture. PLoS One 7, e34063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark RI, Tan SW, Pean CB, Roostalu U, Vivancos V, Bronda K, Pilatova M, Fu J, Walker DW, Berdeaux R, et al. (2013). MEF2 is an in vivo immune-metabolic switch. Cell 155, 435–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contrino S, Smith RN, Butano D, Carr A, Hu F, Lyne R, Rutherford K, Kalderimis A, Sullivan J, Carbon S, et al. (2012). modMine: flexible access to modENCODE data. Nucleic Acids Res 40, D1082–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diangelo JR, Bland ML, Bambina S, Cherry S, and Birnbaum MJ (2009). The immune response attenuates growth and nutrient storage in Drosophila by reducing insulin signaling. Proc Natl Acad Sci U S A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, et al. (2007). A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448, 151–156. [DOI] [PubMed] [Google Scholar]
- Dong J, Jimi E, Zhong H, Hayden MS, and Ghosh S (2008). Repression of gene expression by unphosphorylated NF-kappaB p65 through epigenetic mechanisms. Genes Dev 22, 1159–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frith MC, Fu Y, Yu L, Chen JF, Hansen U, and Weng Z (2004). Detection of functional DNA motifs via statistical over-representation. Nucleic Acids Res 32, 1372–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesan S, Aggarwal K, Paquette N, and Silverman N (2011). NF-kappaB/Rel proteins and the humoral immune responses of Drosophila melanogaster. Curr Top Microbiol Immunol 349, 25–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronke S, Mildner A, Fellert S, Tennagels N, Petry S, Muller G, Jackle H, and Kuhnlein RP (2005). Brummer lipase is an evolutionary conserved fat storage regulator in Drosophila. Cell Metab 1, 323–330. [DOI] [PubMed] [Google Scholar]
- Gronke S, Muller G, Hirsch J, Fellert S, Andreou A, Haase T, Jackle H, and Kuhnlein RP (2007). Dual lipolytic control of body fat storage and mobilization in Drosophila. PLoS biology 5, e137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazelrigg T, Levis R, and Rubin GM (1984). Transformation of white locus DNA in drosophila: dosage compensation, zeste interaction, and position effects. Cell 36, 469–481. [DOI] [PubMed] [Google Scholar]
- Hedengren M, Asling B, Dushay MS, Ando I, Ekengren S, Wihlborg M, and Hultmark D (1999). Relish, a central factor in the control of humoral but not cellular immunity in Drosophila. Mol Cell 4, 827–837. [DOI] [PubMed] [Google Scholar]
- Hennig KM, Colombani J, and Neufeld TP (2006). TOR coordinates bulk and targeted endocytosis in the Drosophila melanogaster fat body to regulate cell growth. J Cell Biol 173, 963–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetru C, and Hoffmann JA (2009). NF-kappaB in the immune response of Drosophila. Cold Spring Harb Perspect Biol 1, a000232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotamisligil GS (2006). Inflammation and metabolic disorders. Nature 444, 860–867. [DOI] [PubMed] [Google Scholar]
- Ji Y, Thomas C, Tulin N, Lodhi N, Boamah E, Kolenko V, and Tulin AV (2016). Charon Mediates Immune Deficiency-Driven PARP-1-Dependent Immune Responses in Drosophila. Journal of immunology 197, 2382–2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamareddine L, Robins WP, Berkey CD, Mekalanos JJ, and Watnick PI (2018). The Drosophila Immune Deficiency Pathway Modulates Enteroendocrine Function and Host Metabolism. Cell Metab. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang P, Chang K, Liu Y, Bouska M, Birnbaum A, Karashchuk G, Thakore R, Zheng W, Post S, Brent CS, et al. (2017). Drosophila Kruppel homolog 1 represses lipolysis through interaction with dFOXO. Sci Rep 7, 16369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpac J, Younger A, and Jasper H (2011). Dynamic coordination of innate immune signaling and insulin signaling regulates systemic responses to localized DNA damage. Dev Cell 20, 841–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhnlein RP (2012). Thematic review series: Lipid droplet synthesis and metabolism: from yeast to man. Lipid droplet-based storage fat metabolism in Drosophila. J Lipid Res 53, 1430–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaitre B, and Hoffmann J (2007). The host defense of Drosophila melanogaster. Annu Rev Immunol 25, 697–743. [DOI] [PubMed] [Google Scholar]
- Mauro C, Leow SC, Anso E, Rocha S, Thotakura AK, Tornatore L, Moretti M, De Smaele E, Beg AA, Tergaonkar V, et al. (2011). NF-kappaB controls energy homeostasis and metabolic adaptation by upregulating mitochondrial respiration. Nat Cell Biol 13, 1272–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihaylova MM, Vasquez DS, Ravnskjaer K, Denechaud PD, Yu RT, Alvarez JG, Downes M, Evans RM, Montminy M, and Shaw RJ (2011). Class IIa histone deacetylases are hormone-activated regulators of FOXO and mammalian glucose homeostasis. Cell 145, 607–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris O, Liu X, Domingues C, Runchel C, Chai A, Basith S, Tenev T, Chen H, Choi S, Pennetta G, et al. (2016). Signal Integration by the IkappaB Protein Pickle Shapes Drosophila Innate Host Defense. Cell Host Microbe 20, 283–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima E, Shimaji K, Umegawachi T, Tomida S, Yoshida H, Yoshimoto N, Izawa S, Kimura H, and Yamaguchi M (2016). The Histone Deacetylase Gene Rpd3 Is Required for Starvation Stress Resistance. PLoS One 11, e0167554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunn AV, Bell J, and Barter P (2007). The integration of lipid-sensing and anti-inflammatory effects: how the PPARs play a role in metabolic balance. Nucl Recept 5, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Dea EL, Barken D, Peralta RQ, Tran KT, Werner SL, Kearns JD, Levchenko A, and Hoffmann A (2007). A homeostatic model of IkappaB metabolism to control constitutive NF-kappaB activity. Mol Syst Biol 3, 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odegaard JI, and Chawla A (2013). The immune system as a sensor of the metabolic state. Immunity 38, 644–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeckinghaus A, Hayden MS, and Ghosh S (2011). Crosstalk in NF-kappaB signaling pathways. Nat Immunol 12, 695–708. [DOI] [PubMed] [Google Scholar]
- Perkins LA, Holderbaum L, Tao R, Hu Y, Sopko R, McCall K, Yang-Zhou D, Flockhart I, Binari R, Shim HS, et al. (2015). The Transgenic RNAi Project at Harvard Medical School: Resources and Validation. Genetics 201, 843–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen ED, and Spiegelman BM (2014). What we talk about when we talk about fat. Cell 156, 20–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rynes J, Donohoe CD, Frommolt P, Brodesser S, Jindra M, and Uhlirova M (2012). Activating transcription factor 3 regulates immune and metabolic homeostasis. Mol Cell Biol 32, 3949–3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffler A, and Scholmerich J (2010). Innate immunity and adipose tissue biology. Trends Immunol 31, 228–235. [DOI] [PubMed] [Google Scholar]
- Shakespear MR, Halili MA, Irvine KM, Fairlie DP, and Sweet MJ (2011). Histone deacetylases as regulators of inflammation and immunity. Trends Immunol 32, 335–343. [DOI] [PubMed] [Google Scholar]
- Sinenko SA, and Mathey-Prevot B (2004). Increased expression of Drosophila tetraspanin, Tsp68C, suppresses the abnormal proliferation of ytr-deficient and Ras/Raf-activated hemocytes. Oncogene 23, 9120–9128. [DOI] [PubMed] [Google Scholar]
- Slack C, Giannakou ME, Foley A, Goss M, and Partridge L (2011). dFOXO-independent effects of reduced insulin-like signaling in Drosophila. Aging Cell 10, 735–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykiotis GP, and Bohmann D (2008). Keap1/Nrf2 signaling regulates oxidative stress tolerance and lifespan in Drosophila. Dev Cell 14, 76–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornatore L, Thotakura AK, Bennett J, Moretti M, and Franzoso G (2012). The nuclear factor kappa B signaling pathway: integrating metabolism with inflammation. Trends in cell biology 22, 557–566. [DOI] [PubMed] [Google Scholar]
- Wang B, Moya N, Niessen S, Hoover H, Mihaylova MM, Shaw RJ, Yates JR 3rd, Fischer WH, Thomas JB, and Montminy M (2011). A hormone-dependent module regulating energy balance. Cell 145, 596–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber K, Johnson N, Champlin D, and Patty A (2005). Many P-element insertions affect wing shape in Drosophila melanogaster. Genetics 169, 1461–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiklund ML, Steinert S, Junell A, Hultmark D, and Stoven S (2009). The N-terminal half of the Drosophila Rel/NF-kappaB factor Relish, REL-68, constitutively activates transcription of specific Relish target genes. Dev Comp Immunol 33, 690–696. [DOI] [PubMed] [Google Scholar]
- Zhao X, and Karpac J (2017). Muscle Directs Diurnal Energy Homeostasis through a Myokine-Dependent Hormone Module in Drosophila. Curr Biol 27, 1941–1955 e1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinke I, Kirchner C, Chao LC, Tetzlaff MT, and Pankratz MJ (1999). Suppression of food intake and growth by amino acids in Drosophila: the role of pumpless, a fat body expressed gene with homology to vertebrate glycine cleavage system. Development 126, 5275–5284. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.