Abstract
Combinatorial clinical trials of PARP inhibitors with immunotherapies are ongoing, yet the immunomodulatory effects of PARP inhibition have been incompletely studied. Here, we sought to dissect the mechanisms underlying PARP inhibitor-induced changes in the tumor microenvironment of BRCA1-deficient triple-negative breast cancer (TNBC). We demonstrate that the PARP inhibitor olaparib induces CD8+ T cell infiltration and activation in vivo, and that CD8+ T cell depletion severely compromises anti-tumor efficacy. Olaparib-induced T cell recruitment is mediated through activation of the cGAS/STING pathway in tumor cells with paracrine activation of dendritic cells and is more pronounced in HR-deficient compared to HR-proficient TNBC cells and in vivo models. CRISPR-knockout of STING in cancer cells prevents proinflammatory signaling and is sufficient to abolish olaparib-induced T cell infiltration in vivo. These findings elucidate an additional mechanism of action of PARP inhibitors and provide rationale for combining PARP inhibition with immunotherapies for the treatment of TNBC.
Keywords: PARP inhibitor, CD8+ T cells, STING pathway, BRCA-associated breast cancer, Type I interferon response
Introduction
Triple-negative breast cancer (TNBC) is a highly aggressive and heterogeneous disease that constitutes 10–20% of breast cancer cases and is associated with poor overall survival and a high probability of distant recurrence and death (1,2). Conventional chemotherapy has been the standard-of-care, but initial remissions are often short, and advanced disease remains incurable (2).
A subset of TNBCs are BRCA-associated and are deficient in homologous recombination (HR) repair (2). HR deficiency and inhibition of poly(ADP-ribose) polymerase (PARP) have been shown to produce synthetic lethality by a variety of mechanisms related to catalytic inhibition of the PARP enzyme and trapping of PARP-DNA complexes (3). Consequently, among BRCA-associated breast cancers, agents such as olaparib and talazoparib have improved the rates of response and progression-free survival compared to standard chemotherapy (4, 5). Despite this promising therapeutic efficacy, treatment with PARP inhibitors have been complicated by both de novo and acquired resistance, which has led to the development of combinations with several HR-disrupting strategies in order to sensitize or re-sensitize cancer cells (3).
There has also been substantial interest in the combination of PARP inhibition with immune checkpoint blockade, with combinatorial clinical trials ongoing in breast and other cancer types (6). Reports on the interaction of PARP inhibition with the immune microenvironment have shown variable results in preclinical breast cancer models. In the syngeneic model EMT6, PARP inhibition was shown to decrease T cell infiltration and increase PD-L1 expression via GSK3β inactivation, contributing to immunosuppression that was reversed by addition of an anti-PD-L1 antibody. Consequently, the combination of PARP inhibitor therapy with anti-PD-L1 blockade led to tumor growth inhibition (7). In contrast, in a BRCA1-deficient TNBC humanized mouse xenograft model PARP inhibition was associated with an increased T cell infiltrate and activated interferon signaling (8). Of note, long-term PARP inhibition in cell line and tumor xenograft models has not been associated with an increase in mutational load, suggesting alternative mechanisms for immuno-modulatory effects (9). To this end, in DNA damage response-deficient TNBC cells, endogenous S-phase damage was shown to activate the cyclic GMP-AMP synthase (cGAS)/Stimulator of interferon genes (STING) pathway of cytosolic DNA sensing, leading to proinflammatory cytokine production (10).
We hypothesized that PARP inhibition might activate STING-dependent signaling in BRCA-associated TNBC, leading to an anti-tumor immune response. Here, we show that treatment with the PARP inhibitor olaparib results in the recruitment of CD8+ T cells in a BRCA1- and TP53-deficient genetically-engineered mouse model (GEMM) of TNBC, and that CD8+ T cell depletion diminishes the anti-tumor efficacy. Olaparib-induced pro-inflammatory cytokine production and subsequent T cell infiltration is dependent on cGAS/STING pathway activation exclusively in tumor cells and is more prominent in HR-deficient compared to HR-proficient TNBC cells and in vivo models. Our findings uncover a novel mechanism of action of PARP inhibitors and provide additional mechanistic rationale for combining PARP inhibition with immunotherapies for the treatment of BRCA-associated TNBC.
Results
Efficacy of PARP inhibition depends on recruitment of CD8+ T cells
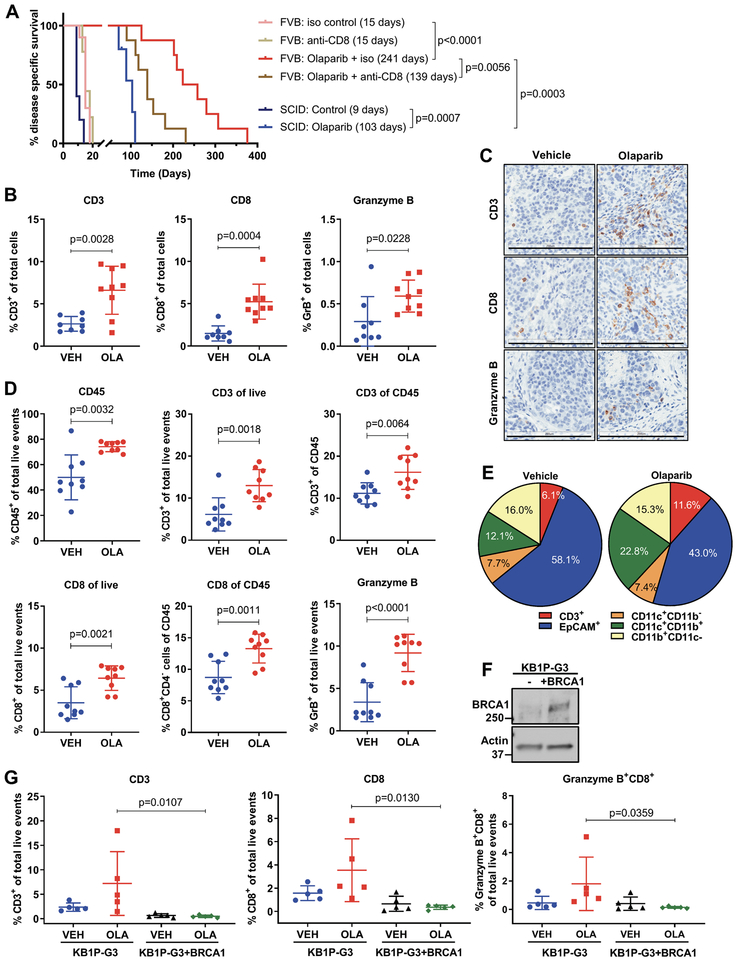
We evaluated the efficacy of olaparib in tumors derived from the K14-Cre-Brca1f/f;Tp53f/f immunocompetent GEMM of TNBC, where spontaneous mammary carcinomas develop after approximately 7 months (11). Individual tumors from this model were transplanted to immunocompetent FVB/129P2 syngeneic mice or to severe combined immunodeficient (SCID) mice and were treated with vehicle or olaparib. In immunocompetent mice, olaparib-treated tumors rapidly regressed, and in some mice completely cleared. Although resistance to olaparib, evidenced by tumor progression, developed between 100–300 days (Supplementary Fig. S1A), olaparib promoted long-term survival, that was increased 16-fold compared to vehicle (Fig. 1A). Notably, the median survival of olaparib-treated SCID mice was significantly lower (103 days) than the median survival of similarly treated immunocompetent mice (241 days) (Fig. 1A), suggesting that an intact immune system is required for an optimal response. To confirm the requirement of an immune response for the anti-tumor efficacy of olaparib, we treated immunocompetent mice with olaparib in the presence of an anti-CD8 antibody. CD8+ T cell depletion, as verified by flow-cytometric analysis (Supplementary Fig. S1B), markedly accelerated tumor growth (Supplementary Fig. S1A) and significantly reduced the median survival of olaparib-treated mice from 241 to 139 days (Fig. 1A). These findings corroborate that CD8+ T cells contribute to the therapeutic efficacy of PARP inhibition.
Figure 1. Efficacy of PARP inhibition depends on recruitment of CD8+ T cells.
(A) Tumor chunks from the BRCA GEMM were transplanted in syngeneic FVB/129P mice (8–10/group), which were treated with vehicle or olaparib along with an isotype (iso) control or an anti-CD8 antibody. Median survivals are shown in parentheses. Tumors were also transplanted in SCID mice (5–6/group) and treated with vehicle or olaparib. Statistical analysis was performed using the Log-rank (Mantel-Cox) test. (B-C) Vehicle (VEH) and olaparib (OLA)-treated tumors were harvested 5 days post-treatment, fixed and subjected to immunohistochemical analysis for CD3, CD8 and granzyme B expression. Staining was quantified using Aperio algorithms. Error bars represent standard deviation (SD). Statistical analyses were performed using unpaired t-tests or unpaired t-tests with Welch’s correction if variances were significantly different. Representative images are shown at 20X magnification. Scale Bar, 200 μm. (D-E) Tumors from vehicle or olaparib-treated mice (n=9) were harvested 5 days post-treatment and analyzed by flow cytometry. Scatter plots show significant increases in CD45+, CD3+, CD8+ and granzyme B+ cells in olaparib-treated animals. Pie charts show the proportions of different cell types (as % of total live events) in the tumor microenvironment. Error bars represent SD. Statistical analyses were performed using unpaired t-tests or unpaired t-tests with Welch’s correction. (F) Immunoblotting for BRCA1 expression in KB1P-G3 and KB1P-G3+BRCA1 cells. (G) KB1P-G3 and KB1P-G3+BRCA1 tumors from 5 vehicle- or olaparib-treated mice were harvested 5 days post-treatment and analyzed by flow cytometry. Scatter plots show % CD3+, CD8+ and granzyme B+ cells. Error bars represent SD. Statistical analyses were performed using two-way ANOVA with Sidak’s post-hoc test.
We next sought to define the effects of olaparib on the immune microenvironment of TNBC. Immunohistochemical (IHC) analysis of tumors from immunocompetent mice showed that olaparib significantly increases CD3+ cells and CD8+ T cells as early as 3 days after treatment (Supplementary Fig. S1C), as well as at 5 (Fig. 1B and C) and 10 days (Supplementary Fig. S1C). Importantly, CD8+ T cell recruitment was accompanied by an increase in granzyme B-positive CD8+ cells (Fig. 1B and C), indicative of cytolytic function. Flow cytometric analysis of immune cell subsets (see Supplementary Fig. S2A and B for gating strategy) in tumors treated with olaparib for 5 days showed that PARP inhibition significantly increases total hematopoietic cell and T cell counts, as measured by CD45 and CD3 staining, respectively (Fig. 1D). Consistent with our IHC analysis, the proportion of CD8+ T cells among CD45+ and total live events, as well as granzyme B expression, were significantly increased in response to olaparib (Fig. 1D). While olaparib also increased CD4+ T cells, the proportion of T regulatory FoxP3+CD4+ T cells (Tregs) was not significantly affected (Supplementary Fig. S1D), implying that olaparib might be increasing CD4+ T-helper cells, which could further contribute to an effective immune response. T cell recruitment and activation in response to olaparib was reproduced in 2 additional independent experiments (Supplementary Fig. S1E). Olaparib not only augmented granzyme B expression in CD8+ T cells but also in natural killer (NK) cells (Supplementary Fig. S1E), suggesting activation of both innate and adaptive immune responses. In addition, as the proportion of tumor cells (EpCAM+) decreased in response to PARP inhibition, the microenvironment became populated with T cells (CD3+) and myeloid cells (CD11b+CD11c+) (Fig. 1E and Supplementary Fig. S1D). These results demonstrate that PARP inhibition can activate a productive anti-tumor immune response in BRCA-deficient breast tumors and that recruitment of cytotoxic CD8+ T cells is required for maximal therapeutic response.
To assess whether T cell infiltration in response to PARP inhibition also occurs in BRCA-proficient TNBC we established tumors using the KB1P-G3 cell line, derived from our GEMM, and a BRCA-reconstituted isogenic cell line (12,13) (Fig. 1F). Treatment with olaparib caused accumulation of significantly higher CD3+, CD8+ and granzyme B-positive CD8+ T cell proportions in BRCA-deficient KB1P-G3 compared to KB1P-G3+BRCA1 tumors (Fig. 1G). Therefore, PARP inhibition potently induces cytotoxic T cell recruitment and activation in BRCA-deficient but not BRCA-proficient TNBC models.
Olaparib activates the STING pathway in the K14-Cre-Brca1f/fTp53f/f TNBC GEMM
The cGAS/STING pathway, triggered by cytosolic DNA, has been established as a critical activator of anti-tumor immune responses (14). To determine whether olaparib-induced cytotoxic T cell recruitment is mediated through cGAS/STING signaling, we examined STING pathway activation in the KB1P-G3−/+BRCA1 isogenic paired cell lines. We assessed the presence of cytosolic DNA and observed that the DNA sensor cGAS, which displayed both nuclear and cytoplasmic localization, was found associated with cytoplasmic micronuclei (Fig. 2A), previously shown to precede cGAS/STING activation (15). Treatment with olaparib significantly increased formation of cGAS-bound micronuclei in KB1P-G3 but not KB1P-G3+BRCA1 cells (Fig. 2A). Next, we assessed the activation of the STING pathway effector TANK-binding kinase 1 (TBK1) by analysis of its activating Ser172 phosphorylation (16) and found that olaparib induced TBK1 phosphorylation in KB1P-G3 but not KB1P-G3+BRCA1 cells (Fig. 2B). We also measured expression of IFN regulatory factor 3 (IRF3), activated downstream of TBK1 by phosphorylation on Ser396 (17). pIRF3 fluorescence intensity was significantly higher in BRCA1-deficient compared to proficient KB1P-G3 cells treated with olaparib (Fig. 2C). We then asked whether TBK1/IRF3 activation correlates with DNA damage accumulation. Formation of γ-H2AX foci, indicative of DNA strand breaks, was induced in both olaparib-treated KB1P-G3 and KB1P-G3+BRCA1 cells (Fig. 2C). However, compared to KB1P-G3+BRCA1, KB1P-G3 cells displayed significantly more γ-H2AX foci (Fig. 2C). To assess whether olaparib-induced cGAS/STING activation results in proinflammatory cytokine production, we measured expression of IFNβ, known to be primarily induced by STING-dependent signaling (14,18). Olaparib significantly upregulated the mRNA levels of IFNβ in KB1P-G3 but not KB1P-G3+BRCA1 cells (Fig. 2D). We also examined the expression of the proinflammatory chemokines CCL5 and CXCL10 that have been associated with STING/TBK1/IRF3-dependent signaling and T cell infiltration in TNBC (10,19–21). Similar to IFNβ, the mRNA levels of CCL5 and CXCL10 were significantly higher in olaparib-treated KB1P-G3 cells compared to their BRCA1-reconstituted pair (Fig. 2D).
Figure 2. Olaparib activates the cGAS/STING pathway in cells derived from the K14-Cre-Brca1f/fTp53f/f GEMM.
The murine isogenic cell line pair KB1P-G3−/+BRCA1 derived from the GEMM was treated with DMSO (0 μM Olaparib) or the indicated doses of olaparib. (A) At 72h post-treatment, cells were stained with picogreen, fixed and subjected to immunofluorescence for cGAS. (Left panel). Representative images of picogreen- (green) and cGAS (red)-stained micronuclei are shown (60x magnification). Scale bar, 8 μm. (Right panel). The number of cGAS-colocalized micronuclei was quantified and expressed as % of total nuclei. Error bars represent standard error of the mean (SEM) of 2 independent experiments. Statistical analysis was performed using two-way ANOVA with Tukey’s post-hoc test. (B) At 72h post-treatment, cells were subjected to immunoblotting for phosphorylated (pTBK1Ser172) and total TBK1 expression with vinculin as a loading control. Markers indicate molecular weight (MW) (kDa). Numbers below the blots represent normalized pTBK1 protein levels quantified by densitometric analysis. (C) At 24h post-treatment, cells were fixed and subjected to immunofluorescence for γ-H2AX (pH2AX Ser139) and pIRF3Ser396. (Left panel) Representative images of DAPI- (blue), γ-H2AX- (green) and pIRF3 (red)- stained cells are shown (20x magnification); scale bar, 8 μm. (Right panels). pIRF3 corrected integrated density/cell was expressed as fold change versus KB1P-G3 DMSO. Statistical analysis was performed using Kruskal-Wallis test with Dunn’s post-hoc test. Error bars represent SEM of 3 independent experiments. The number of cells displaying >5 γ-H2AX foci was quantified, and statistical analysis was performed using two-way ANOVA with Tukey’s post-hoc test. (D) At 72h post-treatment, RNA was extracted and qPCR was performed. IFNβ, CCL5 and CXCL10 mRNA levels were normalized to GAPDH internal control. Error bars represent SEM of 3–4 independent experiments. Statistical analysis was performed using Kruskal-Wallis test with Dunn’s post-hoc test.
We further assessed STING/TBK1/IRF3 pathway activation in K14 cells, another cell line derived from the BRCA1-deficient GEMM (22). We observed a 1500-fold increase in cGAS-synthesized cGAMP levels following olaparib treatment of K14 cells (Supplementary Fig. S3A). Flow-cytometric analysis of phospho-TBK1Ser172, phospho-IRF3Ser396 and γ-H2AX (phospho-H2AXSer139) expression in olaparib-treated K14 cells demonstrated a dose-dependent significant increase, comparable to that of the STING agonist DMXAA (Supplementary Fig. S3B) (gating strategy shown in Supplementary Fig. S3C). Increased phosphorylation of IRF3 and H2AX in response to olaparib was confirmed by immunofluorescence microscopy analysis (Supplementary Fig. S3D). Similar to KB1P-G3 cells, olaparib significantly upregulated the mRNA levels of IFNβ, CCL5 and CXCL10 (Supplementary Fig. S3E). Taken together, the data in the KB1P-G3−/+BRCA1 and K14 cells indicate that PARP inhibition potently activates the cGAS/STING pathway in BRCA1-deficient but not BRCA1-proficient murine tumor cells, leading to type I IFN and proinflammatory chemokine production, known to recruit CD8+ T cells (20,21,23).
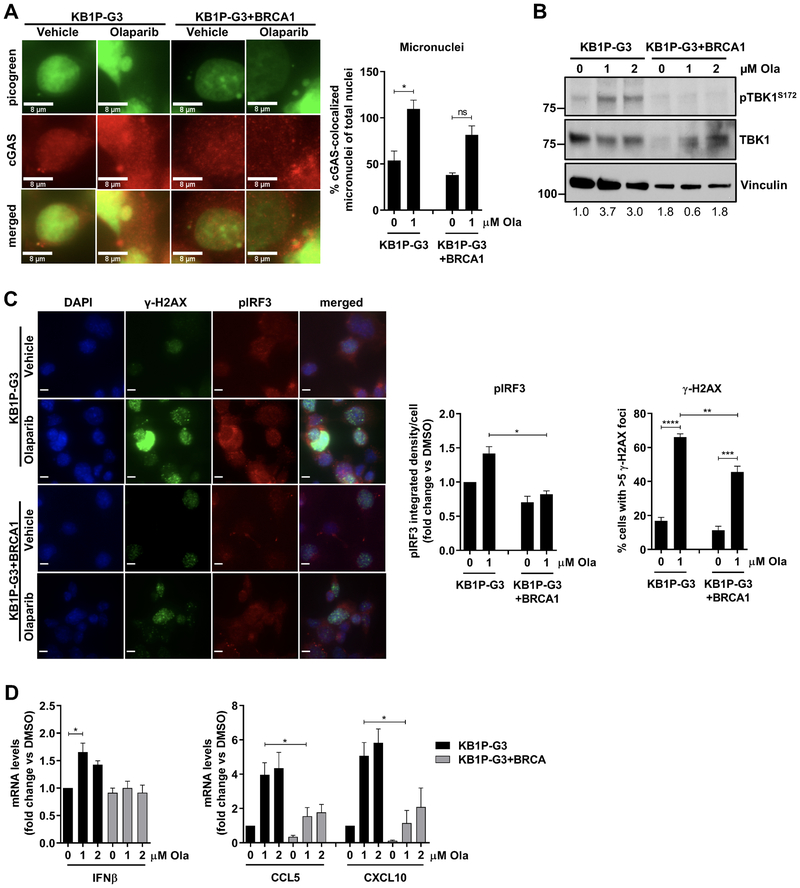
Since the cGAS/STING pathway has previously been shown to be activated in DCs (14), we assessed whether olaparib activates the pathway in both tumor and DCs in vivo by flow-cytometric analysis of harvested tumors treated with vehicle or olaparib (gating strategy shown in Supplementary Fig. S4A and B). PARP inhibition significantly increased the proportion of EpCAM+pIRF3+ cells out of total live events and produced a trend toward increased EpCAM+pTBK1+ cells, demonstrating activation of STING/TBK1/IRF3 signaling in tumor cells in vivo (Fig. 3A). In addition, olaparib significantly increased the proportion of CD11c+CD11b− DCs expressing pTBK1 and pIRF3 (Fig. 3B). pTBK1 and pIRF3 levels were also significantly upregulated in DCs expressing major histocompatibility complex (MHC) class II, indicative of DC maturation and antigen presentation ability (Fig. 3B). The total proportion of mature DCs also increased significantly in response to olaparib (Fig. 3B). Consistent with STING/TBK1/IRF3 pathway activation, mRNA expression analysis of these tumors showed that olaparib increases IFNβ and CCL5 expression (Fig. 3C). Assessment of TBK1/IRF3 signaling in the KB1P-G3−/+BRCA1 isogenic tumor model revealed significantly higher proportions of EpCAM+pTBK1+ and EpCAM+pIRF3+ tumor cells in olaparib-treated KB1P-G3 tumors compared to KB1P-G3+BRCA1 (Fig. 3D), consistent with our in vitro observations.
Figure 3. Olaparib induces pTBK1/pIRF3 signaling in tumor and dendritic cells from the K14-Cre-Brca1f/fTp53f/f GEMM of TNBC.
(A-B) Tumors from 9 vehicle- or olaparib-treated mice were harvested 5 days post-treatment and subjected to flow cytometry for analysis of pTBK1 and pIRF3 expression. Scatter plots demonstrate increases in (A) pTBK1+ and pIRF3+ epithelial (EpCAM+) cells, (B) DCs (CD11b−CD11c+) and mature (CD11c+MHCII+) DCs in olaparib-treated animals. Error bars represent SD. Statistical analyses were performed using unpaired t-tests or unpaired t-tests with Welch’s correction. (C) Tumors from 9 vehicle- or olaparib-treated mice were harvested at 5 days and analyzed for mRNA expression of IFNβ and CCL5 by qPCR. Error bars represent SD. Statistical analyses were performed using an unpaired t-test with Welch’s correction (left panel) or an unpaired t-test (right panel). (D) KB1P-G3 and KB1P-G3+BRCA1 tumors from 5–6 vehicle or olaparib-treated mice were harvested at 5 days post-treatment and subjected to flow cytometry. Scatter plots demonstrate % pTBK1+ and pIRF3+ epithelial (EpCAM+) cells. Error bars represent SD. Statistical analyses were performed using two-way ANOVA with Tukey’s post-hoc test.
To determine whether the observed activation of TBK1/IRF3 signaling in DCs results from a direct effect by PARP inhibitors, we differentiated dendritic cells from bone marrow and cultured them in the presence of olaparib. While the STING agonist DMXAA potently induced TBK1 phosphorylation in DCs resulting in their activation (CD40+) and maturation (MHCII+), olaparib did not affect the expression of these markers (Supplementary Fig. S5A and B). These findings suggest that PARP inhibition potently activates the cGAS/STING pathway in BRCA1-deficient but not proficient tumor cells in vivo, resulting in subsequent activation of TBK1/IRF3 signaling in DCs that stimulates antigen presentation and consequently CD8+ T cell infiltration and activation.
Olaparib activates the STING pathway more potently in HR-deficient human TNBC cells compared to HR-proficient cells
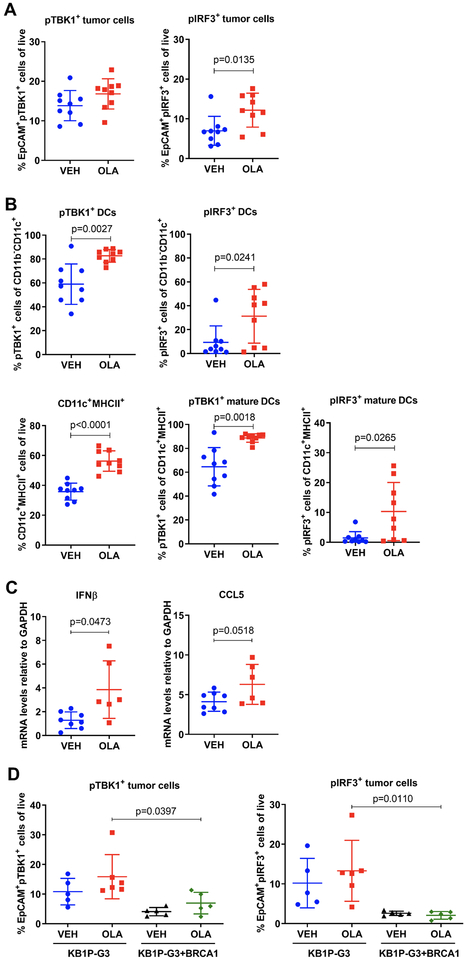
We next assessed the ability of olaparib to activate the cGAS/STING pathway in human TNBC cells. We utilized the BRCA1-deficient MDA-MB-436 cells and its isogenic BRCA1-reconstituted pair (Supplementary Fig. S6A and B), as well as MDA-MB-436 cells with acquired olaparib resistance, generated after prolonged PARP inhibitor exposure (24) (Supplementary Fig. S6B). The mechanism of resistance was shown to be associated with TP53BP1 mutation, facilitating BRCA1-independent end resection, as well as HSP90-mediated stabilization of the mutant form of BRCA1 capable of interacting with PALB2-BRCA2, facilitating RAD51 loading and subsequently HR repair (24). Olaparib significantly increased the number of cGAS-bound micronunclei in MDA-MB-436 control cells, but not in BRCA1-reconstituted or PARP inhibitor-resistant cells (Fig. 4A). Consistent with cGAS activation, cGAMP synthesis was significantly induced in olaparib-treated MDA-MB-436 control cells but not BRCA1-reconstituted or PARP inhibitor-resistant cells (Fig. 4B). Flow cytometric analysis showed that TBK1 phosphorylation in response to olaparib is impaired in PARP inhibitor-resistant and BRCA1-repleted MDA-MB-436 cells, as compared to parental and control cells (Fig. 4C). The same results were obtained when pTBK1 expression was measured by immunoblotting (Fig. 4D). To confirm these findings, we treated a panel of human TNBC cell lines with olaparib according to their 7-day IC50 values (Supplementary Fig. S6B) and assessed pTBK1 expression. In line with our previous observations, olaparib potently induced TBK1 phosphorylation in the HR-deficient MDA-MB-436 cells but not in the BRCA1 wild type (wt) HR-proficient MDA-MB-231 cells (Fig. 4E). Less robust TBK1 phosphorylation occurred following olaparib treatment in BRCA1-mutant HCC1937 cells, which have previously been shown to retain functional HR proficiency (25) (Fig. 4E). The same pattern of pTBK1 activation was observed when these 3 cell lines were treated with the same doses of olaparib (Supplementary Fig. S6C). Of note, STING protein levels did not correlate with pTBK1 activation in these cells (Supplementary Fig. S6C). In fact, STING was overexpressed in PARP inhibitor-resistant MDA-MB-436 cells, suggesting that the inability of PARP inhibition to activate TBK1/IRF3 signaling is not a result of reduced STING expression (Supplementary Fig. S6D).
Figure 4. Olaparib activates the STING pathway more potently in HR-deficient TNBC cells compared to HR-proficient cells.
(A) The human TNBC isogenic cell line pair MDA-MB-436 control and BRCA1-reconstituted (+BRCA1) and a PARP inhibitor-resistant MDA-MB-436 clone were treated with DMSO (0 μM olaparib) or the indicated doses of olaparib for 72h and subjected to immunofluorescence with picogreen dye and anti-cGAS antibody. (Left panel) Quantification of micronuclei. Error bars represent SEM of 3 independent experiments. Statistical analysis was performed using one-way ANOVA with Sidak’s post-hoc test. (Right panel) Representative images of picogreen (green) and cGAS (red) stained micronuclei are shown (60x magnification). Scale bar, 8 μm; (B) At 72h post-treatment with DMSO or olaparib, lysates from parental and PARP inhibitor-resistant MDA-MB-436 cells, as well as MDA-MB-436 control and BRCA1-reconstituted cells (+BRCA1) were subjected to ELISA for cGAMP production. cGAMP levels were expressed as fold-change versus DMSO. Error bars represent SEM of 2 independent experiments. Statistical analysis was performed using Kruskal-Wallis test with Dunn’s post-hoc test; (C) The same cell lines were treated as in (B) and subjected to flow cytometric analysis of pTBK1Ser172 expression. Cisplatin (Cis; 1 μM) was used as positive control. Error bars represent SEM of 2–4 independent experiments. Statistical analysis was performed using one-way ANOVA with Sidak’s post-hoc test (left panel) or two-way ANOVA with Tukey’s post-hoc (right panel); (D) Cell lines were treated with DMSO or the indicated concentrations of olaparib for 72h and lysates were subjected to immunoblotting for pTBK1Ser172 and total TBK1 expression. Vertical black lines indicate points of blot cropping. (E) MDA-MB-436, HCC1937 and MDA-MB-231 human TNBC cell lines were treated with olaparib according to their 7-day IC50 values and subjected to immunoblotting for pTBK1Ser172 and total TBK1 expression. (F-G) Parental, PARP inhibitor-resistant, control and BRCA1-reconstituted MDA-MB-436 cells were subjected to flow cytometric analysis for pIRF3Ser396 and pH2AXSer139 expression. Cisplatin (Cis; 1 μM) was used as a positive control. Error bars represent SEM of 2–4 independent experiments. Statistical analysis was performed using one-way ANOVA with Sidak’s post-hoc test or two-way ANOVA with Tukey’s post-hoc. (H) qPCR analysis of IFNβ, CCL5 and CXCL10 mRNA levels in parental, PARP inhibitor-resistant, control and BRCA1-reconstituted MDA-MB-436 cells, normalized to GAPDH internal control. Error bars represent SEM of 4 independent experiments. Statistical analyses were performed using Kruskal-Wallis test Dunn’s post-hoc test.
Similar to pTBK1, olaparib significantly increased the proportion of pIRF3-positive MDA-MB-436 parental and control cells, but not of BRCA1-reconstituted or PARP inhibitor-resistant cells (Fig. 4F). Cisplatin, used as a positive control, induced pIRF3 to the same extent as olaparib (Fig. 4F). Additionally, pIRF3 fluorescence intensity as measured by immunofluorescence microscopy was significantly reduced in the presence of BRCA expression following treatment with olaparib (Supplementary Fig. S6E). TBK1/IRF3 activation correlated with the induction of DNA damage as measured by flow cytometry (Fig. 4G) and immunofluorescence microscopy (Supplementary Fig. S6E). In agreement with these observations, olaparib significantly stimulated IFNβ production in MDA-MB-436 control, but not BRCA1-reconstituted or PARP inhibitor-resistant cells (Fig. 4H). While IFNβ expression was exclusively induced in the HR-deficient parental MDA-MB- 436 cells, there was a trend toward increased CCL5 and CXCL10 production in the BRCA1-reconstituted HR-proficient cells (Fig. 4H), suggesting that these chemokines are possibly induced by a STING-independent mechanism. IRF3 activation and proinflammatory cytokine production in response to olaparib was also evident in HCC1937 cells (Supplementary Fig. S6F).
Moreover, we confirmed that cGAS/STING pathway activation is not specific to olaparib and that the PARP inhibitors veliparib and talazoparib also induce cGAMP synthesis, TBK1 phosphorylation and proinflammatory cytokine production more potently in BRCA-deficient compared to proficient cells (Supplementary Fig. S7A–D). In summary, these data suggest that PARP inhibitors activate the STING/TBK1/IRF3 pathway and subsequent type I IFN production more potently in HR-deficient compared to HR-proficient human TNBC cells, and this correlates with induction of DNA damage more than STING protein expression.
Intratumoral STING pathway activation is required for olaparib-induced T cell recruitment and anti-tumor efficacy
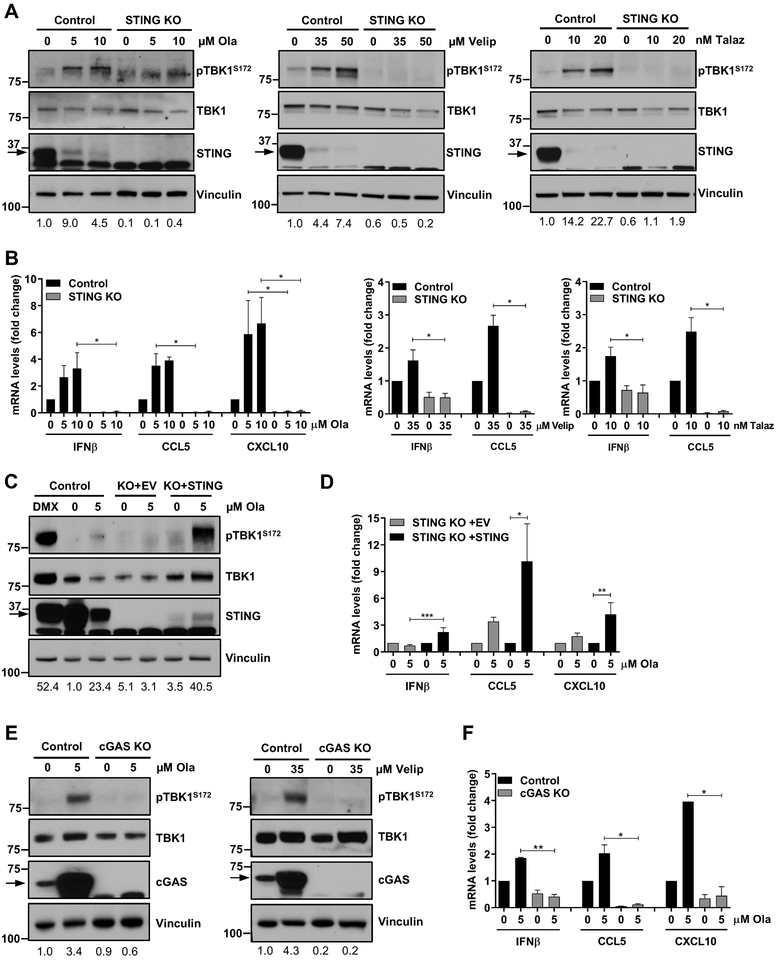
In order to assess the requirement of STING pathway activation in tumor cells for the production of proinflammatory cytokines in response to PARP inhibition, we used CRISPR/Cas9-mediated knockout (KO) of STING in the K14 cell line derived from the BRCA GEMM. CRISPR-mediated KO of STING (STING KO), confirmed by immunoblot analysis (Fig. 5A), did not affect the efficacy of olaparib in vitro (Supplementary Fig. S7E). However, olaparib-induced TBK1 phosphorylation was completely abolished in STING-depleted cells compared to the CRISPR/Cas9 control cells (Fig. 5A). The same result was obtained in response to veliparib and talazoparib (Fig. 5A) (IC50 values are shown in Supplementary Fig. S7E). Consequently, IFNβ, CCL5 and CXCL10 production in response to olaparib, veliparib and talazoparib (Fig. 5B) was blocked in cells lacking STING, indicating that PARP inhibitor-induced proinflammatory cytokine production is mediated through the STING/TBK1/IRF3 pathway. Of note, total STING protein levels were decreased in response to PARP inhibition, likely due to a negative feedback loop leading to STING degradation, as previously reported (26–28).
Figure 5. STING or cGAS depletion abolishes olaparib-induced proinflammatory signaling.
(A-B) Murine K14 CRISPR/Cas9 control or STING knockout (STING KO) cells were treated with DMSO (0 μM) or the indicated doses of olaparib, veliparib or talazoparib for 72h and subjected to immunoblotting or qPCR. STING depletion, as shown by immunoblotting for total STING levels, abolishes PARP inhibitor-induced TBK1 phosphorylation and upregulation of IFNβ, CCL5 and CXCL10 mRNA levels, as measured by immunoblotting and qPCR, respectively. Arrows on the blots indicate specific bands and markers indicate MW (kDa). Error bars represent SEM of 3 independent experiments. Statistical analyses were performed using Kruskal-Wallis test with Dunn’s post-hoc test. (C-D) K14 CRISPR/Cas9 control, STING KO with empty vector (KO+EV) or STING KO with STING-repletion (KO+STING) cells were treated with DMSO or olaparib for 72h and subjected to immunoblotting or qPCR. DMXAA (DMX; 10 μM) was used as a positive control. STING repletion in KO cells, as shown by immunoblotting for total STING levels, rescues olaparib-induced TBK1 phosphorylation and induction of IFNβ, CCL5 and CXCL10 mRNA expression. Error bars represent SEM of 3 independent experiments. Statistical analyses were performed using Kruskal-Wallis test with Dunn’s post-hoc test. (E-F) K14 CRISPR/Cas9 control or cGAS KO cells were treated with the indicated doses of olaparib or veliparib for 72h and subjected to immunoblotting or qPCR. cGAS depletion, as shown by immunoblotting for cGAS levels, abolishes PARP inhibitor-induced TBK1 phosphorylation and upregulation of IFNβ, CCL5 and CXCL10 mRNA levels, as measured by immunoblotting and qPCR, respectively. Error bars represent SEM of 3 independent experiments. Statistical analyses were performed using Kruskal-Wallis test with Dunn’s post-hoc test.
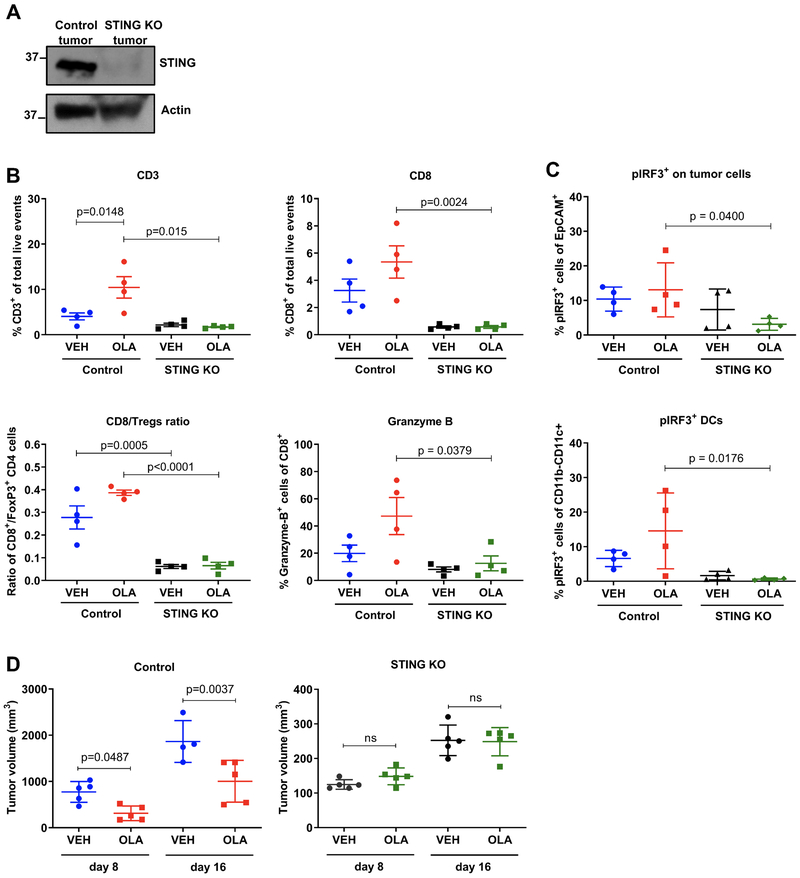
STING repletion in the KO cells (KO+STING) rescued olaparib-induced TBK1 phosphorylation as compared to STING KO cells with empty vector (KO+EV) (Fig. 5C). pTBK1 induction in olaparib-treated STING-repleted cells resembled pTBK1 levels in DMXAA-treated control cells (Fig. 5C). Reconstitution of STING expression in the KO cells also rescued type I IFN and proinflammatory chemokine production (Fig. 5D). To determine whether the DNA sensor cGAS is necessary for PARP inhibitor-induced TBK1/IRF3 signaling, we depleted cGAS from K14 cells. As seen with STING depletion, cGAS KO completely impaired olaparib- and veliparib-induced TBK1 activation (Fig. 5E) and increase in expression of proinflammatory cytokines (Fig. 5F), confirming that the cGAS/STING pathway is required for PARP inhibitor-induced proinflammatory signaling. To assess the requirement of STING pathway activation for PARP inhibitor-induced CD8+ T cell recruitment in vivo, we generated tumors from CRISPR/Cas9 control and STING KO cells in syngeneic mice. The tumors maintained depleted levels of STING (Fig. 6A). We next performed flow cytometric analysis of harvested control and STING KO tumors treated with vehicle or olaparib for 5 days (gating strategies same as in Supplementary Fig. S2 and S4). Consistent with our previous results, olaparib significantly increased the proportion of infiltrating T cells in the CRISPR/Cas9 control tumors with increased CD8+ T cells and granzyme B production (Fig. 6B). In contrast, olaparib-induced CD8+ T cell recruitment and granzyme B expression was abolished in the STING KO tumors (Fig. 6B). Importantly, olaparib-induced IRF3 activation in both tumor and dendritic cells was also impaired when STING was depleted, supporting the notion of cancer cell-mediated DC activation (Fig. 6C).
Figure 6. Intratumoral STING depletion abolishes olaparib-induced T cell recruitment and anti-tumor efficacy.
(A) Immunoblotting for total STING protein levels in K14 control or STING KO tumors. (B-C) Control and STING KO tumors from 4 mice treated with vehicle or olaparib for 5 days were subjected to flow cytometry. Scatter plots demonstrate increases in T cells (CD3+, CD8+ and granzyme B+) and pIRF3+ tumor (EpCAM+) and dendritic (CD11C+CD11B−) cells in olaparib-treated control but not STING KO tumors. Error bars represent SD. Statistical analyses were performed using one-way ANOVA with Sidak’s post-hoc test. (D) K14 CRISPR/Cas9 control or STING KO tumors were transplanted in syngeneic FVB/129P2 mice (5/group), which were treated with vehicle or olaparib. Tumor volumes (mm3) were measured at days 8 and 16. Error bars represent SD. Statistical analysis was performed using one-way ANOVA with Sidak’s post-hoc test.
To directly examine the requirement of STING in the anti-tumor efficacy of olaparib we treated CRSIPR/Cas9 control and STING-depleted tumors with olaparib and measured tumor volume. We observed that the growth of STING KO tumors was delayed compared to control tumors, implying that STING pathway activation might have a pro-tumorigenic role as some studies have suggested (29). Nonetheless, tumor burden in response to olaparib was significantly reduced in mice bearing control tumors, but not STING-depleted tumors, suggesting that STING depletion impairs the efficacy of PARP inhibition (Fig. 6D). These findings demonstrate that tumor cell activation of the STING/TBK1/IRF3 pathway and subsequent production of type I IFNs in response to PARP inhibition is necessary for recruitment and activation of cytotoxic CD8+ T cells and consequent anti-tumor efficacy.
Discussion
In this work, we provide evidence that PARP inhibition triggers an anti-tumor immune response in a BRCA1- and TP53-deficient model of TNBC through activation of the cGAS/STING pathway of cytosolic DNA sensing, and that the anti-tumor efficacy of PARP inhibitors is critically dependent on the recruitment of granzyme B-positive CD8+ T cells. We demonstrate that the STING/TBK1/IRF3 pathway is activated in vivo in BRCA1-deficient but not proficient tumor cells with subsequent paracrine activation in DCs, and that depletion of STING exclusively in tumor cells is sufficient to abolish PARP inhibitor-induced cytotoxic CD8+ T cell infiltration and anti-tumor efficacy.
In other model systems, the PARP inhibitor talazoparib has been associated with an increase in Tregs (30). Despite a substantial increase in total CD4+ T cells, we did not observe a significant change in the numbers of immunosuppressive CD4+FoxP3+ Tregs in response to olaparib in our model. Further studies will be required to determine the precise effects of PARP inhibition on CD4+ T cell subsets and the role of CD4+ T cell infiltration in the anti-tumor efficacy of PARP inhibitors.
In contrast to our findings, it has recently been reported that olaparib decreases cytotoxic CD8+ T cell fractions, as measured by Cytometry by Time of Flight (CyTOF), in the EMT6 model of TNBC, and that addition of an anti-PD-L1 antibody restores CD8+ T cell numbers (7). The failure of olaparib to recruit T cells in this model might account for the limited anti-tumor efficacy that was observed and is likely to be attributed to the BRCA proficiency of this model, where STING pathway induction is substantially less robust than in a BRCA-deficient background. The more pronounced STING pathway activation in BRCA1-deficient tumors may explain the naturally higher lymphocyte infiltration of this subset when compared to other types of breast cancer (10).
While the predominant cell type that produces type I IFNs in the tumor microenvironment are DCs, other cell types, including tumor cells themselves, are emerging as type I IFN producers (14). It has previously been demonstrated that DNA damage response-deficient breast tumor cells that display endogenous S-phase specific DNA damage activate the STING/TBK1/IF3 pathway, resulting in proinflammatory cytokine production and T cell recruitment (10). These findings are consistent with our observations that PARP inhibitor-induced DNA damage, which occurs primarily in S phase, correlates with activation of the cGAS/STING pathway and that this response is more pronounced in HR-deficient compared to HR-proficient TNBC models. This translates to more robust T cell recruitment and activation in vivo. While PARP inhibition activates the STING pathway in both tumor and DCs in vivo, we did not find that olaparib directly induces TBK1/IRF3 signaling in bone marrow-isolated DCs ex vivo. These results are indicative of tumor cell-mediated paracrine activation of the TBK1/IRF3 pathway in DCs that stimulates antigen presentation and are consistent with recent findings in BRCA-deficient ovarian cancer syngeneic and GEMM models (31). Consequently, we find that STING-mediated signaling exclusively within tumor cells is necessary and sufficient for cytotoxic CD8+ T cell infiltration of olaparib-treated tumors and therapeutic efficacy.
Beyond TNBC, the effects of PARP inhibition on the immune microenvironment and cGAS/STING pathway activation have been shown to extend to other tumor types as well. For example, in the BRCA-deficient ovarian cancer model BR5, talazoparib and veliparib have resulted in increased or unchanged CD8+ T cell infiltration, respectively (6,7), with synergistic immune-mediated tumor clearance afforded by the addition of CTLA-4 blockade (10). The survival benefit produced by the CTLA-4 combination was T cell-mediated and dependent on increases in IFN-γ production (10). Additionally, in the BRCA-proficient ID8 ovarian cancer model, PARP inhibition was shown to activate TBK1/IRF3 pro-inflammatory signaling with subsequent CD8+ T cell recruitment. PARP inhibitor efficacy was very modest, although these effects were associated with synergism of PARP inhibition with immune checkpoint blockade (32). In contrast, while potent STING pathway activation in response to olaparib was observed in dendritic cells derived from ID8/BRCA1-null tumors and from a Tp53–/–,BRCA1–/–,Myc overexpressing ovarian GEMM, leading to T cell recruitment, these events did not occur in BRCA-proficient models (31). Taken together, these reports raise the question of the dependence on BRCA deficiency of PARP inhibitor-mediated immune effects specifically in ovarian cancer. In fact, in the clinical study of combined niraparib and pembrolizumab, responses occurred in ovarian cancer patients harboring BRCA wild-type tumors (33). In the breast cancer models examined here, STING pathway activation and immune responses were clearly more pronounced in the BRCA-deficient setting. Notably, the majority of TNBC clinical responses to niraparib/pembrolizumab occurred among patients harboring BRCA mutations (6).
In the ovarian models, the efficacy of olaparib alone or in combination with immune checkpoint blockade was compromised in STING –/– mice, attributable to a loss of pathway activation in DCs (31,32). Our results highlight the importance of intratumoral STING in the anti-tumor effects of olaparib, which stimulates pathway activation in host dendritic cells in a paracrine manner.
Previously, PARP inhibition was shown to be a potential strategy for ERCC1-deficient non-small cell lung cancer (NSCLC) (34). Consistent with the role of the immune system in anti-tumor activity, PARP inhibition has also been shown to activate a STING-mediated IFN response in ERCC1-deficient NSCLC cells (35). In small cell lung cancer (SCLC) models, olaparib also activates the STING/TBK1/IRF3 pathway. Although olaparib monotherapy did not lead to T cell recruitment or anti-tumor efficacy in vivo, these effects were reversed by the addition of PD-L1 blockade, and the efficacy of combined olaparib and immune checkpoint blockade was abolished by STING or cGAS knockdown in tumor cells (36). These results further support the importance of intratumoral STING for the activity of PARP inhibitor-based treatment. Similar findings were observed with the CHK1 inhibitor prexasertib in SCLC models (36), suggesting that STING pathway activation may be a common mechanism among DNA damage repair inhibitors.
In summary, we have identified CD8+ T cell recruitment via cancer cell STING pathway activation as a critical determinant of the therapeutic efficacy of PARP inhibition in TNBC. These findings suggest that PARP inhibitors can enhance the anti-tumor immune response of TNBCs, which has been associated with improved prognosis (37–39). Importantly, these results provide a further mechanistic rationale for combining PARP inhibition with immunotherapy. In a BRCA-deficient background, PARP inhibitor-mediated DNA damage may leverage the STING pathway to convert immunologically cold into hot tumors and sensitize to immune checkpoint blockade (30,40). In contrast, in BRCA-proficient tumors, where PARP inhibitor-induced DNA damage and STING-dependent type I IFN responses are limited, a direct STING agonist (41) may be the preferred combinatorial partner with immune checkpoint blockade.
Methods
Cell culture.
Human cell lines were purchased from American Type Culture Collection (ATCC) between 2015 and 2017, verified with STR profiling, maintained in the recommended medium and passaged up to 15 times. MDA-MB-436 cells with reconstituted BRCA1 expression and with acquired PARP inhibitor resistance have been previously described (24, 42). The K14 cell line was generated by removing breast tumors from K14-Cre Brca1f/fTrp53f/f females, seeding single-cell suspensions in DMEM (Corning) supplemented with 10% FBS and passaging until phenotypical and growth rate stability were achieved (22). The murine isogenic cell line pair KB1P-G3−/+BRCA1 was generated and maintained as previously described (12,13). All cell lines were routinely tested for the presence of mycoplasma using the MycoAlert Mycoplasma Detection Kit (Lonza). For flow cytometry (FC), trypsinized cells were incubated in Zombie Aqua Fixable Viability dye (Biolegend) and processed according to the antibody (Supplementary Table S1) manufacturer’s protocol ID 407 and 404, for phospho-TBK1 and phospho-IRF3 analysis, respectively.
Viability assays.
Cell viability was assessed by CellTiter-Glo (Promega) according to manufacturer’s guidelines. Viability was normalized to DMSO control and expressed as percentage. Survival curves were generated and IC50 values were derived using non-linear regression with the equation “[Inhibitor] vs. normalized response - Variable slope” (GraphPad Prism v7).
Compounds.
Olaparib (AZD-2281; Selleckchem) and talazoparib (BMN673; Selleckchem) were reconstituted in DMSO (100mM and 50mM, respectively). Veliparib (ABT-888; 10mM) and cisplatin (1mg/ml) were from Selleckchem and Fresenius Kabi, respectively. DMXAA (S1537; Selleckchem) was reconstituted in DMSO (50mM).
In vivo studies.
All animal experiments were conducted in accordance with Institutional Animal Care and Use Committee-approved protocols at Beth Israel Deaconess Medical Center (071–2017) and Dana-Farber Cancer Institute (10–067 and 17–032). Pieces from breast tumors generated in K14-Cre Brca1f/fTrp53f/f female mice were transplanted into the mammary pad of FVB/129P2 or SCID/BEIGE (CB17.Cg-PrkdcscidLystbg-J/Crl; Charles River) recipient females that were at least 6 weeks old. FVB/129P2 recipients were generated by breeding FVB females (Jackson Laboratories) and 129P2 males (Envigo) and using the first-generation litters for experimentation. For KB1P-G3−/+BRCA1 tumor generation 5 million cells mixed with Matrigel (Corning) were injected in the mammary fat pad of FVB/129P2 female mice. When tumors reached 15mm, mice were sacrificed, tumors were harvested and transplanted into recipient FVB/129P2 females. For efficacy studies, treatments were started once the tumors reached 7mm in diameter and continued until tumors reached 20mm in any direction or 1000mm3 in volume, at which point mice were euthanized. For flow cytometry studies, mice bearing tumors of 150–300mm3 in volume were randomized in treatment groups, so that the average tumor volume in each group was the same. DMSO-reconstituted olaparib was diluted in PBS (Corning) immediately before intraperitoneal injection and administered at 50mg/kg daily. Anti-CD8 and IgG2b isotype control antibodies (BioXCell; #BE0117 and BE0090) were dissolved in PBS (Corning) and administered intraperitoneally at 0.2mg/dose twice per week. Tumors were measured every 3–4 days using electronic calipers and tumor volumes calculated by using the ellipsoid formula for volume (L*W*W*pi/6).
Immunohistochemistry.
IHC was performed on the Leica Bond automated staining platform. The CD3 antibody (Cell Signaling Technologies (CST), #99940 clone D4V8L) and CD8 antibody (CST, #98941 clone D4W2Z) were run at 1:150 and 1:400 dilutions, respectively, using the Leica Biosystems Refine Detection Kit with EDTA antigen retrieval. The granzyme B antibody (R&D #AF1865 polyclonal) was run at 1:100 dilution using the Leica Biosystems Refine Detection Kit with citrate antigen retrieval. Appropriate control samples were stained using the respective optimized staining protocols and slides were scanned into the Aperio image analysis platform. Parameters for image analysis macros were developed on the control samples by adapting the membrane v9 or cytoplasmic v2 algorithms as appropriate and used to analyze the CD3, CD8 or granzyme B stains, respectively. IHC analysis of CD8+ cells in Supplementary Fig. 1C was performed by manual counting of CD8+ and total cells in 10 fields/tumor. Images (20x magnification) were acquired using Zeiss Axioimager M1 microscope with Plan-Apochromat 20x/0.8 air.
Tumor digestion and flow cytometry.
Mice were sacrificed at the indicated times and cardiac perfusion was performed. Tumors were extracted, finely minced, blended with the gentleMACS Dissociator (Miltenyi Biotec) and digested with MACS Miltenyi Tumor Dissociation Kit (Miltenyi Biotec #130-096-730) according to manufacturer’s instructions. Dissociated tumor cells were washed with RPMI-1640 medium and lysed with RBC Lysis Solution (Qiagen). Cells were resuspended in FACS buffer; PBS (Life Technologies) containing 0.5% BSA and 2mM EDTA (Sigma-Aldrich). Zombie Aqua Fixable Viability Kit was applied to cells in combination with anti-mouse CD16/CD32 Fcγ receptor II/III blocking antibody (Affymetrix #14-0161-85) for 20min at RT, prior to incubation with primary antibodies (Supplementary Table S1) for 1h at 4°C. Cells were fixed and permeabilized using the FoxP3/Transcription Factor Staining Buffer Set (Affymetrix #00-5523-00), according to the manufacturer’s guidelines, and incubated with antibodies for intracellular antigens overnight at 4°C. Next day cells were washed, resuspended in PBS and analyzed using a BD LSRFortessa flow cytometer. Compensation was performed manually on BD FACS Diva using single color and isotype controls, in line with our previous work (43). Signal threshold definition was defined using all-stain, unstained and isotype controls. Analysis was performed on FlowJo V10. In each experiment a T cell panel and a pTBK1-pIRF3 panel were setup; gating strategies for each panel are provided in figure S2 and S4, respectively.
CRISPR/Cas9-mediated knockout, STING repletion and tumor generation.
The gRNA sequences GTCCAAGTTCGTGCGAGGCTAGG or ATATTCTTGTAGCTCAATCC targeting murine STING or cGAS, respectively, were cloned into the lenti-CRISPR/Cas9v2 vector (Addgene, #52961) according to the Zhang lab protocol (25). The scrambled gRNA sequence CACCGCGTGATGGTCTCGATTGAGT was used as a negative control. Viral infection was performed as described by the RNAi consortium (TRC, Broad Institute) laboratory protocol “Lentivirus production of shRNA or ORF-pLX clones” and single clones were isolated following puromycin selection (2μg/ml; Sigma-Aldrich #P8833) for 3 weeks. For generation of STING-repleted cells, STING KO cells were transfected with pUNO1-mSTINGwt-HA3x (InvivoGen) or pUNO1 empty vector (EV) using FuGENE 6 transfection reagent (Promega). Plasmid-incorporated cells were enriched by blasticidin selection (30 μg/ml; Life Technologies #A1113903) for 3 weeks and polyclonal populations were used in experiments. Tumor generation was performed as stated above for the KB1P-G3 model.
Immunofluorescence microscopy.
Cells were seeded on chamber slides (Thermo Scientific™ Nunc™ Lab-Tek™) and treated as specified. For picogreen-cGAS staining cells were incubated with pico488 DNA quantification reagent (Lumiprobe Life Science Solutions; 1:500) for 2h at 37°C. For all immunofluorescence staining, cells were washed in PBS, fixed in 4% formaldehyde/PBS (10min, RT) and permeabilized in 0.25% Triton/PBS (20min, RT). Blocking was performed in 5% BSA/0.1% Triton/PBS (30min, RT), followed by incubation with primary antibodies (Supplementary Table S1) in blocking buffer (1h, 37°C). Cells were washed in PBS and incubated with secondary antibodies (Supplementary Table S1) in blocking buffer (1h, RT), followed by washing and mounting on coverslips using ProLong Gold antifade reagent with DAPI (#P36935, Life Technologies). Cells were imaged using Nikon widefield confocal microscope at the specified magnification and images were analyzed using ImageJ/Fiji software. For micronuclei and γ-H2AX foci quantification, at least 50 and 150 nuclei were counted in 5–10 fields, respectively. For quantification of fluorescence intensity, the area and integrated density of equally exposed images were measured. Background fluorescence was calculated by measuring the mean grey value from 3 different areas on the image containing no cells. The corrected integrated density was calculated by multiplying the area of the image by the background fluorescence and subtracting the product from the integrated density of the entire image. This was divided by the number of cells in the image to give the corrected integrated density per cell (44).
cGAMP ELISA.
cGAMP ELISA (Cayman chemical) was performed on cell lysates according to manufacturer’s instructions. Briefly, cells were lysed and protein concentration was determined by Bio-rad protein assay. Titration was performed to determine ideal concentration of protein, which was 25 μg for human and 50 μg for mouse lysates. Two technical replicates were used per biological replicate.
Quantitative PCR.
RNA was isolated using the PureLink™ RNA kit (Invitrogen) and reversed transcribed using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems), according to the manufacturer’s guidelines. qPCR was performed using Power SYBR Green PCR MasterMix (Applied Biosystems) in a QuantStudio 3 Real-Time PCR System (Applied Biosystems). Primer sequences (5’−3’):
human IFNβ forward AACTTGCTTGGATTCCTACAAAG and reverse TATTCAAGCCTCCCATTCAATTG,
mouse IFNβ forward CCAGCTCCAAGAAAGGACGA and reverse CGCCCTGTAGGTGAGGTTGAT,
human CCL5 forward TGCCCACATCAAGGAGTATTT and reverse CTTTCGGGTGACAAAGACG,
mouse CCL5 forward GCTCCAATCTTGCAGTCGTG and reverse GCTCCAATCTTGCAGTCGTG,
human CXCL10 forward GGCCATCAAGAATTTACTGAAAGCA and reverse TCTGTGTGGTCCATCCTTGGAA,
mouse CXCL10 forward CCAAGTGCTGCCGTCATTTT and reverse CTCAACACGTGGGCAGGATA,
human GAPDH forward GAGTCAACGGATTTGGTCGT and reverse TTGATTTTGGAGGGATCTCG,
mouse GAPDH forward ACCACAGTCCATGCCATCAC and reverse TCCACCACCCTGTTGCTGTA.
Bone marrow derived dendritic cells.
Dendritic cells were derived from FVB/129P2 F1 mice as follows: Using a 25G 5/8” needled syringe bone marrow was flushed out of femurs with HBSS (Gibco) into a 50ml conical tube. Flushed bone marrow was filtered through a 70μM cell strainer (Fisherbrand) into another 50ml tube. Cells were spun down (5min, 1500rpm, 4°C) and the pellet was resuspended in ACK buffer (Gibco) to lyse red blood cells. Cells were washed and resuspended in growing media (RPMI/1% Penicillin-Streptomycin/0.05 mM 2-Mercaptoethanol/10% heat inactivated FBS; all from Gibco) supplemented with 200 ng/ml flt3l (Peprotech) for 7 days. At day 7, cells were collected, counted and seeded in the presence of DMSO, DMXAA (5 μg/ml) or olaparib (5 μM). After 20h, cells were collected, resuspended in FACS buffer and stained for flow cytometry as detailed above (“tumor digestion and FC” section).
Immunoblotting.
Cells were lysed in RIPA buffer (Boston Bioproducts) supplemented with protease and phosphatase inhibitors (Calbiochem), quantitated for protein using the BCA Protein Assay Kit (Pierce) and equal amounts of protein were resolved by SDS-PAGE. Membranes were blocked in 5% milk/TBS-T (Boston Bioproducts) and incubated with antibodies (Supplementary Table S1). Immunodetection was performed using SuperSignal West Pico and Femto Chemiluminescent Substrate (ThermoFisher Scientific). Blot stripping was performed using Restore PLUS Western Blot Stripping Buffer (Life Technologies) according to manufacturer’s guidelines. Protein levels were quantified by densitometric analysis using ImageJ/Fiji. Phosphorylated protein levels were normalized to total proteinband then to loading control and expressed as fold-change versus control DMSO. Immunoblots shown are representative of 3 independent experiments.
Statistical analyses.
Statistical analyses were performed using GraphPad Prism v7 and v8. For comparison of two sets of measurements, an unpaired t-test was performed. Unpaired t test with Welch’s correction was used when sample variances were not equal, as defined by the Brown-Forsythe test. For two sets of fold-change measurements, a one sample t-test was used (measurements were compared to the value of 1). For comparison of three or more sets of unpaired measurements, one-way ANOVA was performed with Tukey’s post-hoc test if all sets were analyzed, or Sidak’s post-hoc test if selected relevant pairs were analyzed (as recommended by GraphPad Prism). When the Brown-Forsythe test indicated that sample variances were not equal the Brown-Forsythe ANOVA test was performed with Games-Howell post-hoc test. When two variables were present two-way ANOVA with Tukey’s post-hoc test was performed. For comparison of three or more sets of measurements that did not follow a normal distribution e.g. fold-change measurements, the Kruskal-Wallis test with Dunn’s post-hoc test was performed. Normality was examined using the Kolmogorov-Smirnov test. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Supplementary Material
Significance.
This work demonstrates cross-talk between PARP inhibition and the tumor microenvironment related to STING/TBK1/IRF3 pathway activation in cancer cells that governs CD8+ T cell recruitment and anti-tumor efficacy. The data provide novel insight into the mechanism of action PARP inhibitors in BRCA-associated breast cancer.
Acknowledgments
This work was supported by the Dana-Farber/Harvard Cancer Center (DF/HCC) Specialized Program of Research Excellence (SPORE) in Breast Cancer, P50 CA168504 (G.I.S., G.M.W.), a P50 CA168504 Career Enhancement Award (J.L.G.), NIH R01 CA090687 (G.I.S.), Susan Komen Career Catalyst Award CCR18547597 (J.L.G.), Merck Investigator Studies Program (MISP) Grant LKR163726 (G.I.S., G.M.W.), as well as the Breast Cancer Research Foundation (G.M.W.), the Ludwig Center at Harvard (G.M.W.), NIH R01 CA226776 (G.M.W.) and ERC CoG-681572 (S.R.).
We thank Shawn Johnson for support with preliminary experiments, Brett Gross for technical help, Liam Cornell for help with CRISPR technology and Bose Kochupurakkal for assistance with immunohistochemical analyses. We thank Dana-Farber/Harvard Cancer Center in Boston, MA, for the use of the Specialized Histopathology Core, which provided histology and immunohistochemistry services. Dana-Farber/Harvard Cancer Center is supported in part by an NCI Cancer Center Support Grant NIH 5 P30 CA06516.
Disclosure of potential conflicts of interest
J.L.G. is a consultant for Glaxo-Smith Kline (GSK) and receives sponsored research support from GSK and Eli Lilly. G.M.W. reports grants from Merck & Co. during the conduct of the study. G.M.W. has a patent Application 14/348810, Compositions and Methods for the Treatment of proliferative diseases pending, and a patent US 20090258352 A1, Pin1 as a marker for abnormal cell growth licensed to Cell Signaling; R&D Systems. G.I.S. has received research funding from Eli Lilly, Merck KGaA/EMD-Serono, Merck & Co., and Sierra Oncology. He has served on advisory boards for Pfizer, Eli Lilly, G1 Therapeutics, Roche, Merck KGaA/EMD-Serono, Sierra Oncology, Bicycle Therapeutics, Fusion Pharmaceuticals, Cybrexa Therapeutics, Astex, Almac, Ipsen, Bayer, Angiex, and Daiichi Sankyo. The Dana-Farber Cancer Institute has received funding from Pfizer and Array BioPharma for the conduct of investigator-initiated clinical trials led by Dr. Shapiro.
References
- 1.Carey L, Winer E, Viale G, Cameron D, Gianni L. Triple-negative breast cancer: disease entity or title of convenience? Nat Rev Clin Oncol 2010;7:683–92. [DOI] [PubMed] [Google Scholar]
- 2.Bianchini G, Balko JM, Mayer IA, Sanders ME, Gianni L. Triple-negative breast cancer: challenges and opportunities of a heterogeneous disease. Nat Rev Clin Oncol 2016;13:674–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lord CJ, Ashworth A. PARP inhibitors: Synthetic lethality in the clinic. Science 2017;355:1152–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robson M, Im S-A, Senkus E, Xu B, mchek S, Masuda N, et al. Olaparib for Metastatic Breast Cancer in Patients with a Germline BRCA Mutation. New Engl J Med 2017;377:523–33. [DOI] [PubMed] [Google Scholar]
- 5.Litton JK, Rugo HS, Ettl J, Hurvitz SA, Gonçalves A, Lee K-HH, et al. Talazoparib in Patients with Advanced Breast Cancer and a Germline BRCA Mutation. New Engl J Med 2018;379:753–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vinayak S, Tolaney SM, Schwartzberg L, Mita MM, McCann G, Tan AR, et al. TOPACIO/Keynote-162: Niraparib + pembrolizumab in patients (pts) with metastatic triple-negative breast cancer (TNBC), a phase 2 trial. J Clin Oncol 2018;36:1011 [abstract]. [Google Scholar]
- 7.Jiao S, Xia W, Yamaguchi H, Wei Y, Chen M-K, Hsu J-M, et al. PARP Inhibitor Upregulates PD-L1 Expression and Enhances Cancer-Associated Immunosuppression. Clin Cancer Res 2017;23:3711–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Z, Sun K, Xiao Y, Feng B, Mikule K, Ma X, et al. Niraparib activates interferon signaling and potentiates anti-PD-1 antibody efficacy in tumor models. Sci Rep 2019;9:1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Póti Á, Berta K, Xiao Y, Pipek O, Klus GT, Ried T, et al. Long-term treatment with the PARP inhibitor niraparib does not increase the mutation load in cell line models and tumour xenografts. Brit J Cancer 2018;119:1392–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parkes EE, Walker SM, Taggart LE, McCabe N, Knight LA, Wilkinson R, et al. Activation of STING-Dependent Innate Immune Signaling By S-Phase-Specific DNA Damage in Breast Cancer. J Natl Cancer Inst 2017;109:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu X, Holstege H, van der Gulden H, Treur-Mulder M, Zevenhoven J, Velds A, et al. Somatic loss of BRCA1 and p53 in mice induces mammary tumors with features of human BRCA1-mutated basal-like breast cancer. Proc Natl Acad Sci USA 2007;104:12111–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jaspers JE, Kersbergen A, Boon U, Sol W, van Deemter L, Zander SA, et al. Loss of 53BP1 Causes PARP Inhibitor Resistance in Brca1-Mutated Mouse Mammary Tumors. Cancer Discov 2013;3:68–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barazas M, Gasparini A, Huang Y, Küçükosmanoğlu A, Annunziato S, Bouwman P, et al. Radiosensitivity is an acquired vulnerability of PARPi-resistant BRCA1-deficient tumors. Cancer Res 2018;79:452–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corrales L, McWhirter SM, Dubensky TW, Gajewski TF. The host STING pathway at the interface of cancer and immunity. The Journal of clinical investigation. J Clin Invest 2016;126:2404–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harding SM, Benci JL, Irianto J, Discher DE, Minn AJ, Greenberg RA. Mitotic progression following DNA damage enables pattern recognition within micronuclei. Nature 2017;548:466–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma X, Helgason E, Phung QT, Quan CL, Iyer RS, Lee MW, et al. Molecular basis of Tank-binding kinase 1 activation by transautophosphorylation. Proc Natl Acad Sci USA 2012;109:9378–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Servant MJ, Grandvaux N, tenOever BR, Duguay D, Lin R, Hiscott J. Identification of the minimal phosphoacceptor site required for in vivo activation of interferon regulatory factor 3 in response to virus and double-stranded RNA. J Biol Chem 2003;278:9441–7. [DOI] [PubMed] [Google Scholar]
- 18.Bose D cGAS/STING Pathway in Cancer: Jekyll and Hyde Story of Cancer Immune Response. Int J Mol Sci 2017;18:2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee H, Lee J-J, Song I, Park I, Kang J, Yu J, et al. Prognostic and predictive value of NanoString-based immune-related gene signatures in a neoadjuvant setting of triple-negative breast cancer: relationship to tumor-infiltrating lymphocytes. Breast Cancer Res Treat 2015;151:619–27. [DOI] [PubMed] [Google Scholar]
- 20.Araujo JM, Gomez AC, Aguilar A, Salgado R, Balko JM, Bravo L, et al. Effect of CCL5 expression in the recruitment of immune cells in triple negative breast cancer. Scientific Rep 2018;8:4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mulligan A, Raitman I, Feeley L, Pinnaduwage D, Nguyen LT, O’Malley FP, et al. Tumoral Lymphocytic Infiltration and Expression of the Chemokine CXCL10 in Breast Cancers from the Ontario Familial Breast Cancer Registry. Clin Cancer Res 2013;19:336–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Juvekar A, Hu H, Yadegarynia S, Lyssiotis CA, Ullas S, Lien EC, et al. Phosphoinositide 3-kinase inhibitors induce DNA damage through nucleoside depletion. Proc Natl Acad Sci USA 2016;113:E4338–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hervas-Stubbs S, Perez-Gracia J, Rouzaut A, Sanmamed MF, Bon A, Melero I. Direct Effects of Type I Interferons on Cells of the Immune System. Clin Cancer Res 2011;17:2619–27. [DOI] [PubMed] [Google Scholar]
- 24.Johnson N, Johnson SF, Yao W, Li Y-C, Choi Y-E, Bernhardy AJ, et al. Stabilization of mutant BRCA1 protein confers PARP inhibitor and platinum resistance. Proc Natl Acad Sci USA 2013;110:17041–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson SF, Cruz C, Greifenberg A, Dust S, Stover DG, Chi D, et al. CDK12 Inhibition Reverses De Novo and Acquired PARP Inhibitor Resistance in BRCA Wild-Type and Mutated Models of Triple-Negative Breast Cancer. Cell Rep 2016;17:2367–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prabakaran T, Bodda C, Krapp C, Zhang B, Christensen MH, Sun C, et al. Attenuation of cGAS-STING signaling is mediated by a p62/SQSTM1-dependent autophagy pathway activated by TBK1. Embo J 2018;37:e97858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Konno H, Konno K, Barber GN. Cyclic Dinucleotides Trigger ULK1 (ATG1) Phosphorylation of STING to Prevent Sustained Innate Immune Signaling. Cell 2013;155:688–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dobbs N, Burnaevskiy N, Chen D, Gonugunta VK, Alto NM, Yan N. STING Activation by Translocation from the ER Is Associated with Infection and Autoinflammatory Disease. Cell Host Microbe 2015;18:157–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khoo L, Chen L. Role of the cGAS–STING pathway in cancer development and oncotherapeutic approaches. EMBO Rep 2018;19:e46935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang J, Wang L, Cong Z, Amoozgar Z, Kiner E, Xing D, et al. The PARP1 inhibitor BMN 673 exhibits immunoregulatory effects in a Brca1−/− murine model of ovarian cancer. Biochem Biophys Res Commun 2015;463:551–6. [DOI] [PubMed] [Google Scholar]
- 31.Ding L, Kim H-J, Wang Q, Kearns M, Jiang T, Ohlson CE, et al. PARP Inhibition Elicits STING-Dependent Antitumor Immunity in Brca1-Deficient Ovarian Cancer. Cell Rep 2018;25:2972–2980.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shen J, Zhao W, Ju Z, Wang L, Peng Y, Labrie M, et al. PARPi triggers the STING-dependent immune response and enhances the therapeutic efficacy of immune checkpoint blockade independent of BRCAness. Cancer Res 2019;79:311–19.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Konstantinopoulos PA, Waggoner SE, Vidal GA, Mita MM, Fleming GF, Holloway RW, et al. TOPACIO/Keynote-162 (NCT02657889): A phase 1/2 study of niraparib + pembrolizumab in patients (pts) with advanced triple-negative breast cancer or recurrent ovarian cancer (ROC)—Results from ROC cohort. J Clin Oncol 2018;36:suppl, A106 [abstract]. [Google Scholar]
- 34.Postel-Vinay S, Bajrami I, Friboulet L, Elliott R, Fontebasso Y, Dorvault N, et al. A high-throughput screen identifies PARP1/2 inhibitors as a potential therapy for ERCC1-deficient non-small cell lung cancer. Oncogene 2013;32:5377–87. [DOI] [PubMed] [Google Scholar]
- 35.Chabanon RM, Muirhead G, Krastev DB, Adam J, Morel D, Garrido M, et al. PARP inhibition enhances tumor cell-intrinsic immunity in ERCC1-deficient non-small cell lung cancer. J Clin Invest 2019;129:1211–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sen T, Rodriguez LB, Chen L, Corte C, Morikawa N, Fujimoto J, et al. Targeting DNA damage response promotes anti-tumor immunity through STING-mediated T-cell activation in small cell lung cancer. Cancer Discov 2019;February 18 [epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loi S, Sirtaine N, Piette F, Salgado R, Viale G, Eenoo F, et al. Prognostic and Predictive Value of Tumor-Infiltrating Lymphocytes in a Phase III Randomized Adjuvant Breast Cancer Trial in Node-Positive Breast Cancer Comparing the Addition of Docetaxel to Doxorubicin With Doxorubicin-Based Chemotherapy: BIG 02–98. J Clin Oncol 2013;31:860–7. [DOI] [PubMed] [Google Scholar]
- 38.Ono M, Tsuda H, Shimizu C, Yamamoto S, Shibata T, Yamamoto H, et al. Tumor-infiltrating lymphocytes are correlated with response to neoadjuvant chemotherapy in triple-negative breast cancer. Breast Cancer Res Treat 2012;132:793–805. [DOI] [PubMed] [Google Scholar]
- 39.Kwa M, Adams S. Prognostic and Predictive Value of Tumor-Infiltrating Lymphocytes in Breast Cancer. Curr Breast Cancer Rep 2016;8:1–13. [Google Scholar]
- 40.Higuchi T, Flies DB, Marjon NA, Mantia-Smaldone G, Ronner L, Gimotty PA, et al. CTLA-4 Blockade Synergizes Therapeutically with PARP Inhibition in BRCA1-Deficient Ovarian Cancer. Cancer Immunology Res 2015;3:1257–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iurescia S, Fioretti D, Rinaldi M. Targeting Cytosolic Nucleic Acid-Sensing Pathways for Cancer Immunotherapies. Frontiers Immunol 2018;9:711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Y, Krais JJ, Bernhardy AJ, Nicolas E, Cai KQ, Harrell MI, et al. RING domain–deficient BRCA1 promotes PARP inhibitor and platinum resistance. J Clin Invest 2016;126:3145–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guerriero JL, Sotayo A, Ponichtera HE, Castrillon JA, Pourzia AL, Schad S, et al. Class IIa HDAC inhibition reduces breast tumours and metastases through anti-tumour macrophages. Nature 2017;543:428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ferreira T and Rasband WS ImageJ User Guide - IJ 1.46. 2010 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.