Abstract
TP63 is frequently amplified or overexpressed in primary Head and Neck Squamous Cell Carcinomas (HNSCCs). Nevertheless, the role of TP63 in the initiation and progression of HNSCCs is not known. Using archival HNSCC tissue sections, we found that TP63 expression is often downregulated in late-stage human HNSCCs. To establish a causal link between TP63 loss and HNSCC tumorigenesis, we developed a genetically engineered mouse model in which Trp63 (the mouse homologue of human TP63) was ablated from head and neck epithelia. Upon exposure of the mice to a chemical carcinogen, we found that Trp63 ablation accelerated HNSCC initiation and progression. To determine whether these findings are relevant for human HNSCCs, we generated TP63 knockdown HNSCC cell lines. These cells were implanted into the tongue of athymic nude mice to generate orthotopic xenografts. We found that loss of TP63 promoted HNSCC progression and metastasis. Further, we determined that tumor metastasis is dependent on MAPK activation in TP63 knockdown HNSCCs. The significance of these findings is underscored by our finding that pharmacological inhibition of MAPK activity by Trametinib drastically impaired HNSCC metastasis mediated by TP63 loss. In conclusion, our data provide novel mechanistic insights into the role of TP63 loss in HNSCC initiation and progression, and provide a rationale for the development of new therapeutic approaches specifically targeting TP63-dependent tumor pathways.
Keywords: Head and Neck Squamous Cell Carcinoma (HNSCC), TP63, MAPK, STAT3, Metastasis
Introduction
Head and Neck Squamous Cell Carcinoma (HNSCC) is the sixth most common cancer worldwide, resulting in approximately 350,000 deaths every year [1]. HNSCCs develop in the mucosal lining of the oral cavity, nasopharynx, hypopharynx, or larynx [2]. Tobacco usage, alcohol consumption, and HPV infection represent the major risk factors for HNSCC development [3]. There are various treatment options for HNSCC including surgical resection, chemotherapy, targeted therapy, immunotherapy, and radiation [4]. These different therapies, alone or in combination, are often successful in eliminating the primary tumor, but treating HNSCC metastases has remained a major challenge [5]. For this reason, the 5-year survival rate for HNSCC is only 40–50% and has not improved over the last decade [6, 7]. To achieve a better clinical outcome, studies focused on understanding the genetic and molecular mechanisms that contribute to HNSCC progression and metastasis are necessary.
TP63 (termed TRP63 in mice), an important regulator of stratified epithelial development and homeostasis [8], has been suggested to play a role in the development and progression of HNSCCs [9]. However, whether TP63 functions as a tumor promoter or a tumor suppressor has remained controversial. A role for TP63 in tumor promotion has been suggested based on findings that the TP63 gene is amplified in a subset of HNSCCs, leading to an overexpression of the ΔNp63α isoform of TP63 [10–12]. Further, forced overexpression of ΔNp63α in SCC cell lines leads to increased survival and proliferation [10, 13]. Although TP63 is often overexpressed in primary tumors, TP63 loss has been associated with tumor progression and metastasis in some tumor types [14–17]. However, whether this also occurs during HNSCC progression and metastasis has not been systematically investigated. Using archival paraffin-embedded HNSCC samples, we found that TP63 is lost in late-stage, but not in early-stage, HNSCCs. Further, using genetically engineered mice as well as human orthotopic xenografts, we determined that loss of TRP63/TP63 promotes HNSCC progression and metastasis through activation of MAPK signaling. In the presence of TP63, this transcription factor directly induces expression of DUSP6, a suppressor of MAPK signaling. Upon TP63 loss, this suppression is relieved, leading to activation of MAPK signaling and subsequent STAT3 activation and MMP15 expression. The importance of our findings is underscored by our observation that pharmacological inhibition of MAPK signaling prevents progression and metastasis of xenograft HNSCCs with low TP63 expression. Collectively, our study provides novel mechanistic insights into TP63 function in HNSCCs in vivo. Further, we have identified key signaling pathways regulated by TP63 that can be therapeutically targeted to prevent HNSCC metastasis.
Materials and Methods
Mouse studies
Krt14.CrePR1 (Jackson laboratory, Bar Harbor, ME) transgenic mice were crossed with Trp63fl/fl mice [18] to obtain Krt14.CrePR1/Trp63fl/+ (control) and Krt14.CrePR1/Trp63fl/fl (Trp63-ablated) mice. To ablate Trp63, mice received RU486 (1mg/ml) via oral gavage from P23 to P33. 4-NQO (Sigma-Aldrich, St. Louis, MO) was added to the drinking water of control and Trp63-ablated mice at a final concentration of 100μg/ml. After 16 weeks, mice were returned to normal drinking water and monitored weekly. Softened food was provided to mice with signs of food-intake difficulty. Mice were euthanized if there was a significant loss of body weight (>15% of original body weight). After euthanasia, tongue, cheek, and cervical lymph nodes were collected for histology, immunostaining, and RNA and protein extraction. Histopathology of all tumors generated was analyzed by Dr. Whitney A. High, a board-certified pathologist. All animal experiments were approved by The University of Colorado Institutional Animal Care and Use Committee (IACUC).
Cell lines and EGF treatment
FaDu and CAL27 cells were maintained in DMEM (Thermo Fisher Scientific, Waltham, MA) supplemented with 10% FBS (Atlanta biologicals, Research triangle park, NC), penicillin, and streptomycin (Thermo Fisher Scientific, Waltham, MA). UMSCCI, 584A2, CCL30 cells were maintained in DMEM-F12 (Thermo Fisher Scientific, Waltham, MA) supplemented with 10% FBS, penicillin and streptomycin. All cells were maintained at 37°C in 5% CO2. FaDu and CAL27 cell lines were authenticated using STR analysis at Molecular Biology Service center (UCD-AMC). The cell lines were routinely tested for mycoplasma contamination using Venor TM GEM Mycoplasma detection kit (Sigma-Aldrich, St. Louis, MO). To activate MAPK signaling, FaDu and CAL27 cells were maintained as described above and treated with 50 ng/ml of EGF (Sigma-Aldrich, St. Louis, MO) for 3 hrs in serum free DMEM.
Plasmids
PHIV-Luc-Zsgreen was a gift from Bryan Welm (Addgene, Cambridge, MA, plasmid #39196). Doxycyline inducible TP63 shRNA (1–3) and Non-silencing shRNA (NS shRNA) were obtained from Dharmacon, inc, Lafayette, CO.
Lentiviral production and infection
PHIV-Luc-Zsgreen lentiviral vector was co-transfected with packaging vectors [pUVMC (plasmid #8449) and pMD2.G (plasmid #12259) from Addgene, Cambridge, MA] into 293T cells. For lentiviral transduction, FaDu and CAL27 cells were seeded at 3 × 105 cells/well of a 6-well plate for 24hrs prior to infection. One ml of viral supernatant was added to 1ml of fresh complete medium plus polybrene (8μg/ml; Sigma-Aldrich, St. Louis, MO) and cells were incubated overnight. The cells were then washed, fed with fresh medium and cultured as above. FaDu and CAL27 cells were stably transduced with NS shRNA and TP63 shRNA plasmids as described above.
qRT-PCR
RNA was extracted using RNeasy mini kit (Qiagen, Germantown, MD). cDNA was synthesized using Verso cDNA synthesis kit (Thermo Fisher Scientific, Waltham, MA). TP63 and β-actin probes were obtained from Thermo Fisher Scientific. Data was analyzed using LC480 thermal cycler (Roche, Basel, Switzerland) and for relative quantification, the comparative cycle threshold (ΔΔCT) method was used.
Cell cycle analysis
FaDu and CAL27 cells transduced with either NS shRNA or TP63 shRNA were fixed and stained with propidium iodide (Thermo Fisher Scientific, Waltham, MA) at room temperature for 20 minutes. The fixed and stained cells were stored at 4°C until further analysis. The cell cycle distribution and data analysis were performed at the University of Colorado-Anschutz medical campus (UCD-AMC) flow cytometry core.
Cell migration assays
FaDu and CAL27 cells were grown to 80–90% confluency in 12-well tissue culture plates. Confluent cells were treated with mitomycin C (10 μg/ml) (Sigma-Aldrich, St. Louis, MO) for 1 hour at 37°C to inhibit cell proliferation. To evaluate cell migration, a scratch was made using 20μl pipette tip. Using a phase contrast inverted microscope (Nikon eclipse TE2000), images were obtained immediately after scratch (Time 0) until the closure of scratch area. Processing of images was done with the NIS Elements 3.2 imaging software (Nikon Instruments Inc, Melville, NY). The percentage of the scratch closure area was quantified using image J.
Cell invasion assays
Cell invasion was analyzed using matrigel coated 8.0 μm-pore polycarbonate membrane transwell inserts (BD Biosciences, San Jose, CA). HNSCC cells (2.5 × 105) were suspended in DMEM media and added to the insert. DMEM containing 10% fetal bovine serum and penicillin and streptomycin was added to the BD Falcon TC companion plate and used as a chemoattractant. HNSCC cells were incubated for 36 hours and non-migrating cells were carefully removed with a cotton swab. Migrating cells were stained with 0.5% crystal violet and counted under 40X magnification using Nikon Eclipse 90I microscope system. Image processing was done with the NIS Elements 3.2 imaging software (Nikon Instruments Inc, Melville, NY). For each condition, five random images of invading cells were selected for quantification, and the percentage of invading cells/field was calculated.
Orthotopic xenografts
Orthotopic xenograft SCCs were generated by implanting 5X105 cells into the tongue of 4–5-week-old female athymic nude mice. For bioluminescence imaging of tumors, mice were anaesthetized and injected intraperitoneally with 100μl of D-luciferin (30mg/ml in PBS). Mice were imaged using an IVIS-100 imaging system (Xenogen Corporation, Alameda, CA) and images were captured once weekly. For Trametinib treatment, mice were treated once daily for 5 days by oral gavage with Trametinib (1mg/kg in 4% DMSO/Corn oil) or vehicle (4% DMSO/Corn oil). Mice were monitored daily and euthanized if there was a significant loss in body weight (>15% of original body weight). After euthanasia and necropsy, tumors were visualized using fluorescence microscopy. Tumors were excised for histology, immunostaining, immunohistochemistry, and western blot analysis. Histopathology of all tumors was analyzed by Dr. Whitney A. High, a board-certified pathologist. All animal experiments were approved by The University of Colorado Institutional Animal Care and Use Committee (IACUC).
RNA-sequencing
RNA was extracted using the miRNeasy mini kit (Qiagen). FaDu-TP63 NS shRNA (n=3) and FaDu-TP63 shRNA3 (n=3) SCCs were used for this analysis. Upon library generation, libraries were sequenced (paired end read 150 cycles) using an Illumina 4000 HT sequencer at the microarray and genomics core in UCD-AMC. Raw sequencing reads were converted to FASTQ files and the quality of the reads was determined using the FASTQC tool. Low-quality reads and Illumina sequencing adapters from the 3’ end of the reads were removed using cutadapt tool. High-quality reads were further mapped onto the human genome hg19 (NCBI 37) using STAR (version 2.5.1a). Gene counts were determined using HTSeq tool. Normalization of reads, differential expression analyses, and principal component analyses (PCA) were carried out using the DESeq2 Bioconductor package within the R statistical programming environment. Genes that were differentially expressed (adjusted p-value <0.1) in TP63 shRNA3 SCCs compared to NS shRNA SCCs were used for pathway identification and Gene Ontology (GO) term analysis. DAVID bioinformatics resources [19] and Gene set enrichment analysis (GSEA) [20] were used for pathway identification and GO analysis. The raw unprocessed RNA-seq data is deposited in gene expression omnibus (GEO) under accession number GSE120689.
Western blotting
Flash-frozen HNSCCs were broken down into small pieces on dry ice, and RIPA lysis buffer (Sigma-Aldrich, St. Louis, MO) was added. Tumors were homogenized and sonicated for 30 seconds at low setting on ice. After lysis, tumor lysates were spun at 13,000 rpm for 10 minutes and the supernatant was transferred to a new tube. Protein concentration was determined using BCA method, and equal amounts of protein were loaded for immunoblot analysis. Immunoblots were probed with antibodies against TP63 (mouse monoclonal 4A4, 1:1000, Santa Cruz Biotechnology, Dallas, TX), DUSP6, and MMP15 (mouse monoclonal, 1:1000, Santa Cruz Biotechnology, Dallas, TX) at 4°C overnight followed by incubation for 2 hours at room temperature with goat anti-mouse secondary antibody conjugated to horseradish peroxidase (Vector labs, Burlingame, CA). GAPDH (mouse monoclonal, 1:10,000, EMD Millipore, Burlington, MA) was used as a loading control. Detection was performed using the Supersignal west pico plus chemiluminescent substrate (Thermo Fisher Scientific, Waltham, MA), and images were acquired using Odyssey Imaging Systems (LI-COR Biosciences, Lincoln, NE).
Chromatin Immunoprecipitation
At 70–80% confluency, cells were washed with PBS and crosslinked with 1% formaldehyde for 10 minutes at room temperature. The crosslinking reaction was stopped with 2.5M glycine and the cells were washed twice with ice-cold 1X PBS. After the second wash, cells were lysed using ChIP-SDS lysis buffer (1% SDS, 10mM EDTS, 50mM Tris-HCl, pH 8.1, protease inhibitor cocktail (Roche, Basel, Switzerland)). Lysates were centrifuged, and the supernatant was discarded. The cell pellet containing 2 X106 cells was resuspended in 100 μl of ChIP lysis buffer and sonicated using Bioruptor sonicator (Diagenode, Denville, NJ) using a refrigerated water bath for 10 cycles at high power, 30s ON, and 30s OFF. One ml of 1mg/ml protein lysate was used for immunoprecipitation. ChIP was performed with TP63 (11H1) and IgG antibodies using magnetic beads (Magna ChIP protein A+G beads, Millipore, Burlington, MA) according to the instructions from the manufacturer. Primers (P1-forward (5’-ACTATCTGGGCAGCTTCATTG-3’) and reverse (5’-TAAAGCCGGAGGTTCTCTCT-3’), P2- forward (5’-CGGCGCGTCCCAGCACACCT-3’) and reverse (5’-TAAAACAATGTAGAGTGCAGA-3’), P3- forward (5’- GCGGCTGCTGCTGATGGA-3’) and reverse (5’-CAGCAGCCCGAGCACCGACTC-3’)) spanning the upstream, downstream, and putative TP63 binding motif [21] within the DUSP6 promoter were used for ChIP-qPCR.
Immunofluorescence (IF) and Immunohistochemistry (IHC)
Formalin-fixed paraffin-embedded tumors were sectioned at 4 μm by the UC Denver Morphology and Phenotyping Core. The sections were deparaffinized, subjected to antigen retrieval, and incubated with following primary antibodies: TP63 (mouse monoclonal (4A4), 1:100, Santa Cruz Biotechnology, Dallas, TX, and rabbit polyclonal, 1:100, Cell Signaling Technology inc. Danvers, MA), KRT14, KRT1, KRT13 (guinea pig polyclonal, 1:1000, rabbit polyclonal 1:100, rabbit polyclonal 1:5000, kind gifts from Dennis Roop, UC Denver), P-STA3 (Tyr705 and Ser727) (rabbit monoclonal, 1:100, Cell Signaling Technology inc. Danvers) and Ki67 (mouse monoclonal, 1:100, Cell Signaling Technology inc. Danvers). Visualization was performed using alexa-fluor conjugated secondary antibodies (1:200, Invitrogen, Carlsbad, CA) followed by mounting in DAPI-containing mounting media (Vector labs, Burlingame, CA). Images were acquired using Nikon Eclipse 90I microscope system and processing was done with the NIS Elements 3.10 imaging software (Nikon Instruments Inc, Melville, NY). IHC for P-EK1/2 signaling was performed as described previously using a rabbit anti-P-ERK1/2 antibody (rabbit monoclonal 1:100, Cell Signaling Technology inc. Danvers) [16].
Image quantification
To quantify TRP63/TP63-, Ki67-, KRT1-, KRT13, P-ERK1/2, P-STAT3 (Ser727)-expressing cells, five different images from five different HNSCCs (each image contains approximately 200 cells) were counted. Nuclear staining with DAPI was used to determine the total number of cells. Data is expressed as mean ± standard deviation of the mean.
Statistical analysis
Graph pad (Prism 7, La Jolla, CA) was used to perform statistical analysis. The overall survival for 4-NQO treated mice and xenograft SCCs was estimated using the Kaplan-Meier analysis, and the statistical significance between groups was determined by performing log-rank test. Mann-Whitney U-test was used to compare the number of tumors between groups. To compare among different groups, two-tailed unpaired Student t-tests were performed. In all analyses, P < 0.05 was considered statistically significant.
Results
TP63 expression is reduced in late-stage human HNSCCs
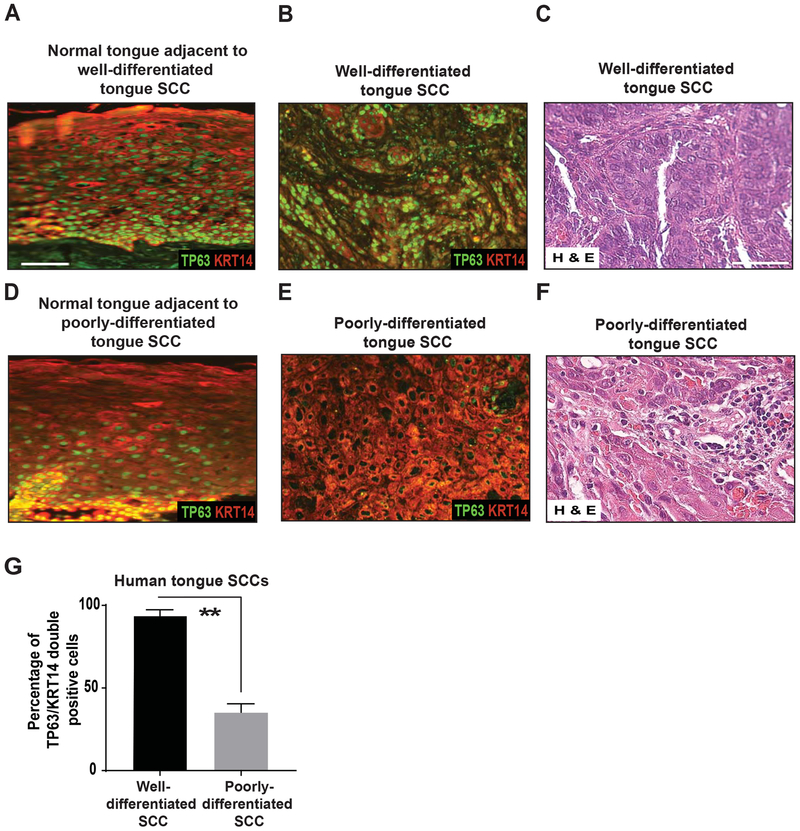
To analyze TP63 expression levels in human HNSCCs, we performed immunofluorescence (IF) staining for TP63 and KRT14 on human archival HNSCC sections [22]. Specifically, we analyzed HNSCCs originating from the tongue and larynx. KRT14 is expressed in stratified epithelia, including head and neck mucosa, and is used to identify the epithelial portion of the tumors. We only quantified expression of TP63 expression, and other markers, in cells expressing KRT14. We found that TP63 expression was dramatically downregulated in poorly differentiated tongue and laryngeal HNSCCs, but not in well-differentiated HNSCCs of the same origin (Figure 1A–G and Supplementary Figure 1A–G), suggesting a general role for TP63 loss in HNSCC tumor progression.
Figure 1. TP63 expression is downregulated in late-stage human tongue HNSCCs.
Immunostaining with antibodies against TP63 (green) and KRT14 (red) on sections of (A, D) normal tongue, (B) well-differentiated tongue SCC (n=5), and (E) poorly differentiated tongue SCC (n=7). TP63 staining studies were performed using the p63 (4A4) antibody. Scale bars = 50 μm. (E, F) Hematoxylin and eosin staining of well-differentiated tongue SCC and poorly differentiated tongue SCC. Scale bars = 50 μm. (G) Quantification of TP63/KRT14 double-positive cells from D and E (two-tailed unpaired Student t-test, **: P < 0.01; Mean ± SD).
Trp63 ablation from head and neck epithelia promotes HNSCC initiation and progression
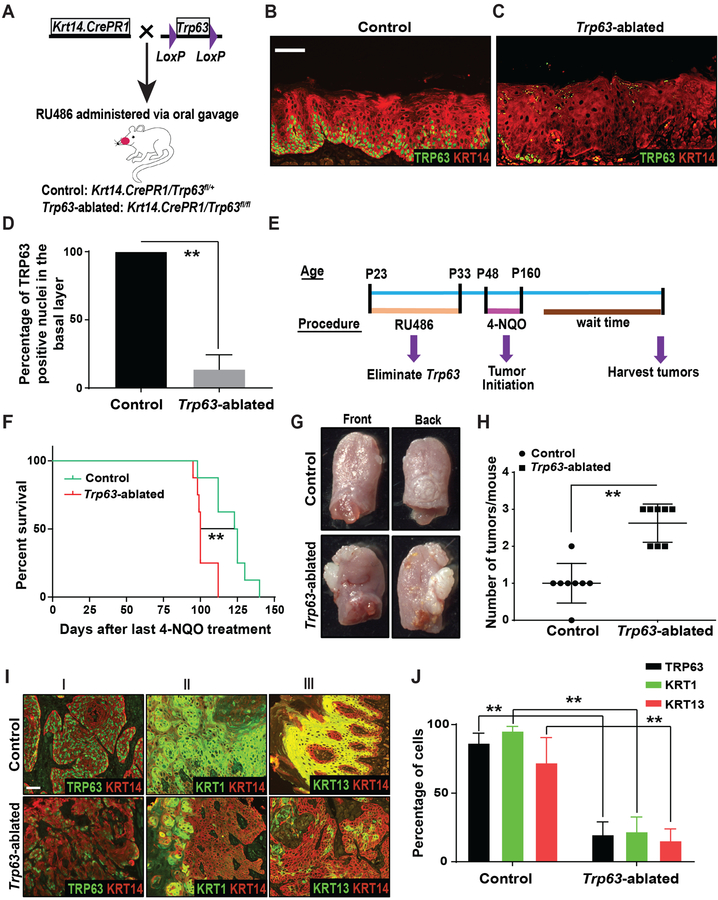
To determine the functional consequences of TP63 loss in HNSCCs, we initially generated inducible, epithelial-specific Trp63 knockout mice (Figure 2A). Briefly, we bred Krt14.CrePR1 mice (Jackson laboratory, Bar Harbor, ME) with Trp63 floxed mice (Trp63fl/fl) [18] to obtain control (Krt14.CrePR1/Trp63fl/+) and Trp63-ablated mice (Krt14.CrePR1/Trp63fl/fl). In the Trp63fl/fl mice, the DNA binding domain, common to all Trp63 isoforms, is flanked by LoxP sites. Thus, Cre-mediated recombination in these mice leads to elimination of all Trp63 isoforms. To ablate Trp63 from head and neck epithelia, mouse tongues were treated with RU486. Wild type mouse tongue only expresses ΔNp63 isoforms (data not shown). IF staining for TRP63 and KRT14 on tongue sections obtained two weeks after completion of RU486 treatment demonstrated the efficient ablation of TRP63 from tongue epithelia in Trp63-ablated mice (Figure 2B–D).
Figure 2. Ablation of Trp63 promotes oral tumorigenesis.
(A) Schematic illustrating the generation of control and Trp63-ablated mice. Immunostaining with antibodies against TRP63 (green) and KRT14 (red) on tongue sections from (B) control and (C) Trp63-ablated mice. TP63 staining studies were performed using the p63α antibody. (D) Quantification of TRP63 positive nuclei from B and C (two-tailed unpaired Student t-test, **: P < 0.01; Mean ± SD). (E) Experimental approach for ablation of Trp63 and generation of oral SCCs by 4-NQO treatment. (F) Graph depicting the survival of control and Trp63-ablated mice (n=8/group) (log-rank test, **: P < 0.01). (G) Representative photographs of SCCs on the front and back of the tongue from control and Trp63-ablated mice. (H) Graph depicting the number of tumors per mouse in control and Trp63-ablated mice (Mann-Whitney U-test, **: P < 0.01) (n=8/group). (I) Immunostaining with antibodies against (I) TRP63 (green) and KRT14 (red), (II) KRT1 (green) and KRT14 (red), and (III) KRT13 (green) and KRT14 (red), on SCCs from control and Trp63-ablated mice. Scale bars = 50 μm. (J) Quantification of TRP63, KRT1 and KRT13 expression from I (two-tailed unpaired Student t-test, **: P < 0.01; Mean ± SD).
As spontaneous tumors did not develop in response to Trp63 ablation, we next treated control and Trp63-ablated mice with 4-Nitroquinoline-1-oxide (4-NQO), a chemical carcinogen that is widely used as a tobacco surrogate in mouse models [23]. Importantly, 4-NQO treatment leads to oral SCC development by faithfully recapitulating human oral carcinogenesis, and the resulting oral tumors display the molecular and histological features of human oral SCCs [24, 25]. Mice were given 4-NQO in their drinking water for 16 weeks after which they were returned to normal drinking water for the remainder of the study (Figure 2E). We found that Trp63-ablated mice developed more tumors than control mice, and had a significantly shorter survival time (Figure 2F–H). We confirmed that TRP63 expression was significantly downregulated in SCCs fromTrp63-ablated mice (Figure 2I, 2J). This downregulation was not associated with an increase in proliferation in these tumors (Supplementary Figure 2A–C). Further, 62.5% of Trp63-ablated tumors were classified as poorly differentiated SCCs, versus only 37.5% of control tumors (Supplementary Figure 2D–H). The poor differentiation grade was further confirmed by the reduced expression of keratin 1 (KRT1) and keratin 13 (KRT13), keratins that are expressed in differentiated cells of normal oral epithelium, but not in malignant oral SCCs [26, 27] (Figure 2I, 2J). Collectively, these data demonstrate that loss of Trp63 leads to the development of more aggressive tumors. We could not analyze the metastatic potential of these tumors in vivo, as the mice had to be euthanized for humane reasons.
Downregulation of TP63 in human HNSCC cell lines leads to increased migration and invasion in vitro
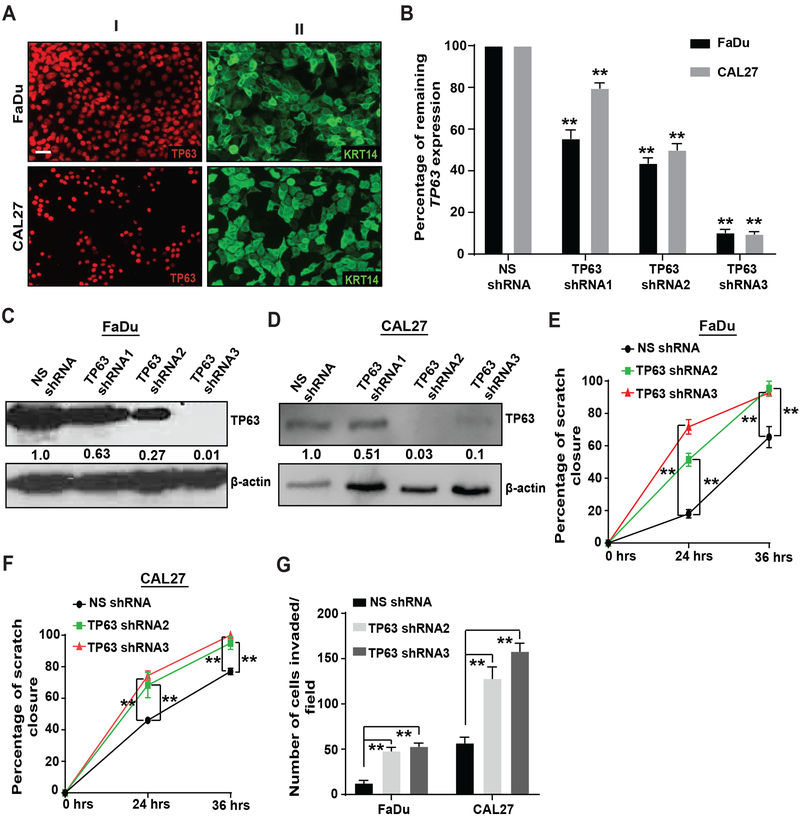
To determine the relevance of our findings for human HNSCC tumorigenesis, we analyzed the consequences of TP63 downregulation in human HNSCC cell lines. FaDu and CAL27 cells were selected as they expressed both TP63, specifically the ΔNp63α isoform, and KRT14 (Figure 3A, Supplementary Figure 3A–C and data not shown). TP63 was downregulated through lentiviral transduction of a doxycycline-inducible TP63 shRNA (three shRNAs were tested [1–3]). A non-silencing shRNA (NS shRNA) was used as a control. All viral constructs also encode RFP to enable the localization of tumors by fluorescence microscopy. After quantification of remaining TP63 expression, we selected FaDu and CAL27 cells transduced with TP63 shRNA2 and TP63 shRNA3 for subsequent studies as they were most efficient at downregulating TP63 expression (Figure 3B–D). These cells will be referred as FaDu-NS shRNA, FaDu-TP63 shRNA2, FaDu-TP63 shRNA3, CAL27-NS shRNA, CAL27-TP63 shRNA2 and CAL27-TP63 shRNA3. Using cell cycle analysis and Ki67 staining of the SCCs, we did not observe differences in proliferation in response to TP63 downregulation (Supplementary Figure 4A–H).
Figure 3. TP63 downregulation increases migration and invasion in vitro.
(A) Immunostaining with antibodies against (I) TP63 and (II) KRT14 on HNSCC cell lines, FaDu and CAL27. Scale bars = 50 μm. FaDu and CAL27 cells were transduced with a recombinant lentivirus encoding TP63 shRNAs (TP63 shRNA1–3) or a non-silencing shRNA as a control (NS shRNA), and TP63 levels were analyzed by (B) qRT-PCR, (two-tailed unpaired Student t-test, **: P < 0.01; Mean ± SD) and (C, D) Western blot analysis using the p63 4A4 antibody. Quantification of relative TP63 levels after normalizing to β-actin is shown at the bottom. (E, F) Effects of TP63 downregulation on migration of FaDu and CAL27 cells were measured using a scratch assay, and the percentage closure was quantified (two-tailed unpaired Student t-test, **: P < 0.01; Mean ± SD). (G) Effects of TP63 downregulation on invasion of FaDu and CAL27 cells was measured using matrigel-coated transwell inserts, and the number of cells invaded/field was quantified (two-tailed unpaired Student t-test, **: P < 0.01; Mean ± SD). Results are combined data from three independent experiments.
Next, we assessed the effects of TP63 loss on migration and invasion of HNSCC cell lines in vitro. For these experiments, cells were treated with mitomycin-C to block proliferation. Using a scratch assay, we observed that TP63 downregulation increased the migration of FaDu and CAL27 cells (Figure 3E, 3F, Supplementary Figure 5A, 5B). Further, TP63 downregulation increased the invasion of FaDu and CAL27 cells in a transwell assay (Figure 3G and Supplementary Figure 5C, 5D). Taken together, these findings demonstrate that downregulation of TP63 increases the migratory and invasive properties of two HNSCC cell lines without affecting proliferation.
Downregulation of TP63 in human HNSCC xenografts promotes cervical lymph node metastasis
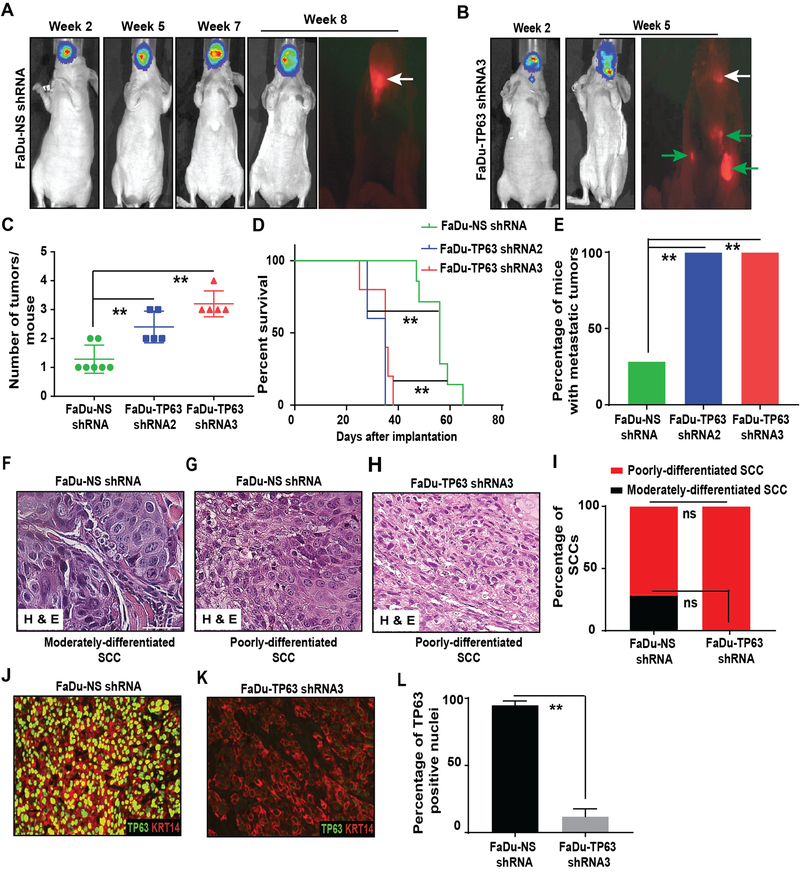
To enable in vivo imaging, we introduced a luciferase-expressing lentivirus into all cell lines used in this study. We then generated orthotopic xenograft HNSCCs by implanting these cells into the tongue of athymic nude mice. Such orthotopic grafts are a well-established method for studying HNSCCs and these models are considered highly physiologically relevant [28]. Mice were fed a doxycycline diet to achieve TP63 knockdown. Strikingly, within 2–3 weeks after implantation, all TP63 knockdown SCCs had metastasized to the cervical lymph nodes (Figure 4A–E, Supplementary Figure 6A–F and Supplementary Figure 7A–E). Further, all mice carrying TP63 knockdown SCCs developed multiple tumors leading to rapid weight loss. In contrast, even after 8–9 weeks most FaDu-NS shRNA (5/7) and CAL27-NS shRNA (7/7) SCCs remained restricted to the tongue (Figure 4A–E, Supplementary Figure 6A–F and Supplementary Figure 7A–E).
Figure 4. Inhibition of TP63 promotes cervical lymph node metastasis.
(A, B) FaDu cells transduced with NS shRNA (n=7) and TP63 shRNA3 (n=5) were implanted into the tongue of athymic female nude and the tumor burden at indicated time points was monitored using bioluminescence imaging. At the end of study, necropsy was performed, and SCCs were identified using fluorescence microscopy. Representative in vivo bioluminescence and RFP images are shown. White arrows indicate primary tongue tumors and green arrows indicate metastatic tumors. Graphs depicting (C) the average number of tumors/mouse, (Mann-Whitney U-test, **: P < 0.01), (D) survival (log-rank test, **: P < 0.01), and (E) percentage of mice with metastatic tumors in mice implanted with FaDu-NS shRNA (n=7), FaDu-TP63 shRNA2 (n=5), and FaDu-TP63 shRNA3 (n=5) (Fisher’s exact test, **: P < 0.05). (F, G, and H) Hematoxylin and eosin staining of moderately and poorly-differentiated SCCs obtained from mice implanted with FaDu-NS shRNA and FaDu-TP63 shRNA3. (I) Graph depicting the percentage of moderately and poorly-differentiated SCCs obtained after implanting mice with FaDu-NS shRNA and FaDU-TP63 shRNAs (Fisher’s exact test, “ns” indicates not significant; P = 0.4615). (J, K) Immunostaining with antibodies against TP63 (red) and KRT14 (green) on SCCs obtained from mice implanted with FaDu-NS shRNA and FaDU-TP63 shRNA3. TP63 staining studies were performed using the p63α antibody. (L) Quantification of TP63 positive nuclei from J and K (two-tailed unpaired Student t-test, **: P < 0.01; Mean ± SD).
Consistent with the higher rate of metastasis, primary TP63 knockdown SCCs displayed a poorer differentiation grade (Figure 4F–I and Supplementary Figure 7F–J). Further, IF staining for TP63 and KRT14 on FaDu-TP63 shRNA3 and CAL27-TP63 shRNA3 SCCs confirmed significant downregulation of TP63 (Figure 4J–L and Supplementary Figure 7K–M). Collectively, these findings demonstrate that loss of TP63 promotes the progression and metastasis of human xenograft HNSCCs in vivo.
Biological processes regulated by TP63 in HNSCCs
It has been suggested that TP63 downregulation leads to epithelial to mesenchymal transition (EMT) [29]. As SCCs that developed in our models retain KRT14 expression, EMT is unlikely to have contributed to tumor progression. This is further supported by our observation that TP63 knockdown SCCs did not exhibit other hallmarks of EMT including loss of E-cadherin or gain of vimentin (Supplementary Figure 8A–F and data not shown) [30].
To investigate the mechanism by which loss of TP63 promotes metastasis, we performed RNA-Sequencing (RNA-Seq) on FaDu-NS shRNA and FaDu-TP63 shRNA3 SCCs. We identified 176 differently expressed genes (P-value <0.001 and FDR ≤0.1; Supplementary Table 1) in FaDu-TP63 shRNA3 compared to FaDu-NS shRNA SCCs. Pathway analysis using NIH DAVID bioinformatics resources and Gene Set Enrichment Analysis (GSEA) indicated an upregulation of genes associated with EGFR/RAS/MAPK and JAK-STAT3 signaling in TP63 knockdown SCCs (Figure 5A–B and Supplementary Figure 9A, 9B). Further, gene ontology (GO) term functional analysis revealed changes in biological processes related to proliferation, migration, and adhesion (Supplementary Figure 9C, 9D). These data suggest that genes previously associated with migration, invasion, and metastasis in other tumor types [31–33], might be under the control of TP63 in HNSCCs.
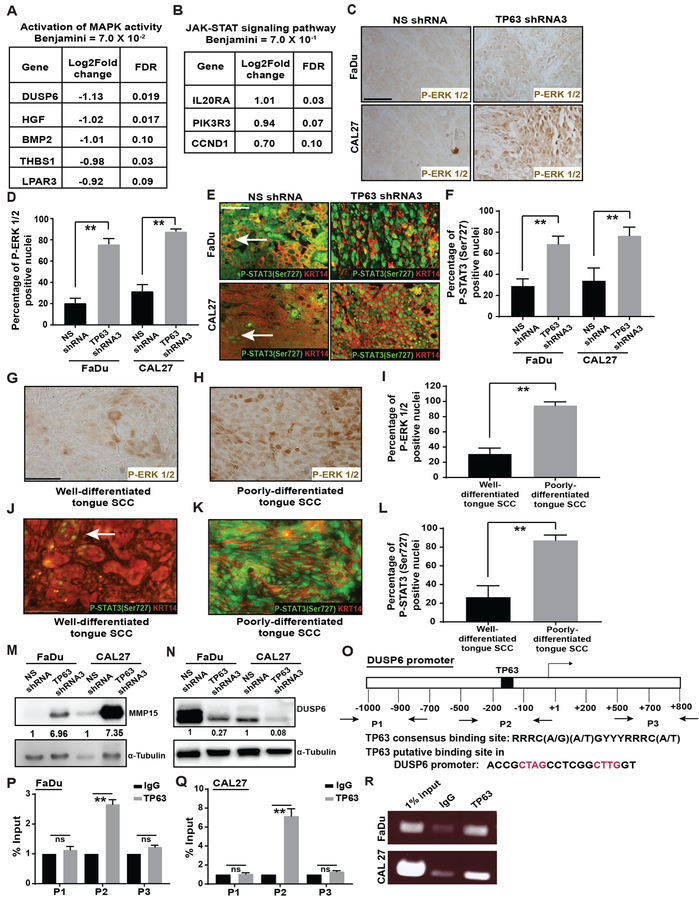
Figure 5. Inhibition of TP63 leads to activation of MAPK and STAT3 signaling in HNSCCs.
(A, B) Significantly differentially expressed genes in TP63 knockdown xenograft HNSCCs that contributed to the identification of indicated pathways (P-value < 0.001 and FDR < 0.1). (C) P-ERK1/2 levels in SCCs obtained from mice implanted with FaDu and CAL27 cells transduced with either NS shRNA or TP63 shRNA3 were analyzed by immunohistochemistry. (D) Quantification of P-ERK 1/2 positive nuclei from C (two-tailed unpaired Student t-test, **: P < 0.01; Mean ± SD). (E) Immunostaining with antibodies against P-STAT3 (Ser727) (green) and KRT14 (red) on SCCs obtained from mice implanted with FaDu cells transduced with either NS shRNA or TP63 shRNA3. (F) Quantification of P-STAT3 (ser727) positive nuclei from E (two-tailed unpaired Student t-test, **: P < 0.01; Mean ± SD). (G, H) P-ERK1/2 levels in well- and poorly-differentiated human tongue SCCs were analyzed by immunohistochemistry. (I) Quantification of P-ERK 1/2 positive nuclei from G and H (two-tailed unpaired Student t-test, **: P < 0.01; Mean ± SD). (J, K) Immunostaining with antibodies against P-STAT3 (Ser727) (green) and KRT14 (red) on well- and poorly-differentiated human tongue SCCs. (L) Quantification of P-STAT3 (Ser727) levels from J and K (two-tailed unpaired Student t-test, **: P < 0.01; Mean ± SD). (M, N) MMP15 and DUSP6 protein levels in SCCs obtained from mice implanted with FaDu cells and CAL27 cells transduced with either NS shRNA or TP63 shRNA3 was measured using western blotting. Quantification of relative MMP15 and DUSP6 levels after normalizing to α-tubulin is shown at the bottom. (O) Schematic illustrating the putative TP63 binding site in the DUSP6 promoter and the primers (P1, P2 and P3) used for ChIP-qPCR. (P-R) Binding of TP63 to the DUSP6 promoter in FaDu and CAL27 cells was assessed by performing ChIP assays using the p63 (11H1) antibody. 1% of total cross-linked, reversed chromatin before immunoprecipitation was considered as input. The enrichment of immunoprecipitated DNA at the DUSP6 promoter was quantified by qPCR followed by agarose gel electrophoresis.
TP63 loss leads to upregulation of MAPK and STAT3 signaling in HNSCCs
To validate the RNA-Seq results, we determined the levels of P-ERK1/2, indicative of activate MAPK signaling. We observed upregulation of P-ERK1/2 in TP63 knockdown SCCs (FaDu and CAL27) compared to their respective controls (Figure 5C, 5D). Further, we performed IF staining for two active forms of STAT3, P-STAT3 (Tyr705) and P-STAT3 (Ser727) [34]. Interestingly, we observed upregulation of P-STAT3 (Ser727), but not P-STAT3 (Tyr705), in TP63 knockdown SCCs (Figure 5E, 5F and Supplementary Figure 10A–D). Lastly, we assessed expression of P-ERK1/2 and P-STAT3 (Ser727) in primary human HNSCCs. In agreement with our data on the xenograft SCCs, we found that loss of TP63 correlated with an upregulation of P-ERK1/2 and P-STAT3 (Ser727). Specifically, poorly differentiated SCCs exhibited TP63 loss and induction of P-ERK1/2 and P-STAT3 (Ser727) whereas well-differentiated SCCs expressed TP63 but exhibited low levels of P-ERK1/2 and P-STAT3 (Ser727) expression (Figure 5G–L, Supplementary Figure 11A–F).
Finally, consistent with findings that MAPK activation promotes tumor progression and metastasis in part by activating MMPs (matrix metalloproteinases) [31], our RNA-seq studies identified significant upregulation of MMP15 (MT2-MMP) in FaDu-TP63 shRNA3 SCCs (Supplementary Table 1). Immunoblot analysis confirmed the upregulation of MMP15 in FaDu-TP63 shRNA3 and CAL27-TP63 shRNA3 SCCs compared to their respective controls (Figure 5M). Taken together, these findings indicate that loss of TP63 upregulates MAPK and STAT3 signaling leading to induction of MMP15 in HNSCCs.
TP63 regulates MAPK signaling in HNSCC by directly binding to the DUSP6 promoter
Our RNA-seq data, validated by Western blot analysis, indicated that Dual Specificity Phosphatase 6 (DUSP6), a negative regulator of MAPK signaling [35], is downregulated in TP63 knockdown SCCs (Figure 5A, 5N). Further, using chromatin immunoprecipitation (ChIP), we found that TP63 directly interacts with a consensus TP63 binding motif [21] within the DUSP6 promoter (P2), but not with upstream or downstream sequences (P1 and P3), suggesting that TP63 suppresses MAPK signaling by inducing DUSP6 expression (Figure 5O–R).
STAT3 serine phosphorylation and MMP15 upregulation in HNSCC are MAPK dependent
As we observed both activation of MAPK and JAK-STAT3 signaling in our system, we wanted to determine whether cross-talk between these two signaling pathways occurs. To this end, we treated FaDu and CAL27 cells with EGF, the upstream ligand for MAPK signaling, and observed a robust activation of P-ERK1/2 and P-STAT3 (Ser727), but not P-STAT3 (Tyr705), in FaDu and CAL27 cells (Supplementary Figure 12A–D). EGF treatment also led to an upregulation of MMP15 (Supplementary Figure 12E, 12F). These data provide a direct link between MAPK activation, STAT3 signaling, and MMP15 expression in HNSCC.
Inhibition of MAPK and P-STAT3 (Ser727) prevents the migration and invasion of TP63 knockdown cells
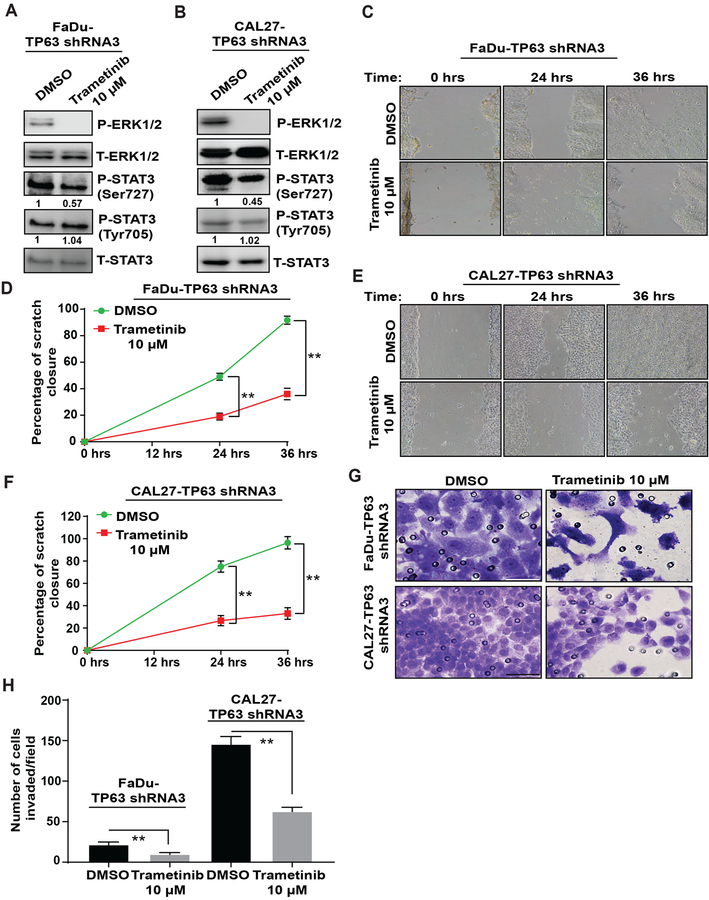
To determine the significance of MAPK activation in TP63 knockdown SCCs, we treated TP63 knockdown cells with the MEK inhibitor Trametinib [36]. The IC50 values of FaDu-TP63 shRNA3 and CAL27-TP63 shRNA3 cells treated with Trametinib are shown in Supplementary Figure 13A, 13B. Trametinib treatment led to inhibition of P-ERK1/2 in FaDu-TP63 shRNA3 and CAL27-TP63 shRNA3 cells (Supplementary Figure 13C, 13D). Consistent with our finding that MAPK functions upstream of P-STAT3 (Ser727) in HNSCC cells, Trametinib treatment also led to inhibition of P-STAT3 (Ser727), but not of P-STAT3 (Tyr705) (Figure 6A, B). Further, Trametinib treatment prevented the migration and invasion of FaDu-TP63 shRNA3 and CAL27-TP63 shRNA3 cells (Figure 6C–H). Thus, the consequences of TP63 loss in HNSCC cells are reversed by inhibition of MAPK signaling.
Figure 6. Trametinib treatment prevents the migration and invasion of TP63 knockdown HNSCC cell lines.
P-ERK1/2, T-ERK1/2, P-STAT3 (Tyr705), P-STA3 (Ser 727), and T-STAT3 protein levels were analyzed in (A) FaDu-TP63 shRNA3 cells or (B) CAL27-TP63 shRNA3 cells treated with DMSO (vehicle) or 10 μM Trametinib by western blotting. (C-F) Effects of Trametinib treatment on migration of FaDu-TP63 shRNA3 and CAL27-TP63 shRNA3 cells were measured using scratch assay, and the percentage closure was quantified (two-tailed unpaired Student t-test, **: P < 0.01; Mean ± SD). (G, H) Effects of Trametinib treatment on invasion of FaDu-TP63 shRNA3 and CAL27-TP63 shRNA3 cells was measured using matrigel-coated transwell inserts, and the number of cells invaded/field was quantified (two-tailed unpaired Student t-test, **: P < 0.01; Mean ± SD).
Trametinib treatment prevents the cervical lymph node metastasis of TP63 knockdown xenografts
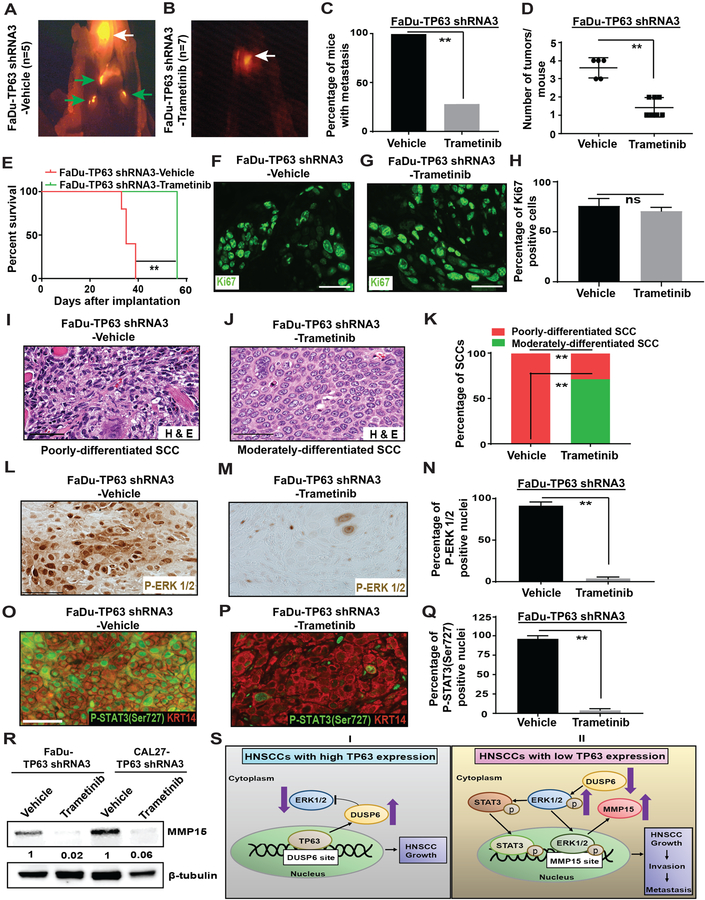
We next determined if Trametinib treatment could prevent metastasis of HNSCCs that lack TP63 expression. To this end, we engrafted mice with FaDu-TP63 shRNA3 or CAL27-TP63 shRNA3 cells into the tongue. Two weeks after grafting, mice were treated with either vehicle or Trametinib once daily. They were also fed a doxycycline diet to downregulate TP63 expression. Whereas all vehicle-treated mice developed cervical lymph node metastases, only 2/7 Trametinib-treated FaDu-TP63 shRNA3 mice and 3/7 Trametinib-treated CAL27-TP63 shRNA3 mice developed cervical lymph node metastases (Figure 7A–C, Supplementary Figure 14A–D, Supplementary Figure 15A–C). Moreover, the tumor burden was lower in Trametinib-treated mice compared to vehicle-treated mice (Figure 7D and Supplementary Figure 15D). Most importantly, Trametinib treatment significantly improved the survival of mice compared to vehicle treatment (Figure 7E and Supplementary Figure 15E). The reduced metastasis was not caused by a possible anti-proliferation effect of Trametinib as we did not observe a difference in proliferation between Trametinib- and vehicle- treated tumors (Figure 7F–H and Supplementary Figure 15F–H).
Figure 7. Inhibition of MAPK signaling by Trametinib treatment prevents the metastasis of TP63 knockdown xenograft HNSCCs.
(A, B) Athymic nude mice were implanted with FaDu-TP63 shRNA3 and treated with either vehicle or Trametinib. At the end of study, necropsy was performed, and SCCs were identified using fluorescence microscopy. Representative in vivo bioluminescence and RFP images are shown. White arrows indicate primary tongue tumors and green arrows indicate metastatic tumors. Graphs depicting (C) the average number of tumors/mouse (Mann-Whitney U-test, **: P < 0.01), (D) survival (log-rank test, **: P < 0.01), and (E) percentage of mice with metastatic tumors in mice implanted with FaDu-TP63 shRNA3 and treated with either vehicle or Trametinib. (Fisher’s exact test, **: P < 0.05). (F,G) Immunostaining with antibody against Ki67 on SCCs obtained from mice implanted with FaDu-TP63 shRNA 3 cells and treated with either vehicle or Trametinib. (H) Quantification of Ki67-positive cells from f and G (two-tailed unpaired Student t-test, “ns” indicates not significant; P > 0.05). (I, J) Hematoxylin and eosin staining of moderately and poorly-differentiated SCCs obtained from mice implanted with FaDu-TP63 shRNA3 and treated with either Vehicle or Trametinib. (K) Graph depicting the percentage of moderately and poorly-differentiated SCCs obtained from I and J (Fisher’s exact test, **: P < 0.05). (L, M) Immunohistochemistry with antibody against P-ERK1/2 on SCCs obtained from mice implanted with FaDu-TP63 shRNA 3 and treated with either vehicle or Trametinib. (N) Quantification of P-ERK 1/2 positive nuclei from L and M (two-tailed unpaired Student t-test, **: P < 0.01; Mean ± SD). (O, P) Immunostaining with antibody against P-STAT3 (Ser727) on SCCs obtained from mice implanted with FaDu-TP63 shRNA 3 cells and treated with either vehicle or Trametinib. (Q) Quantification of P-STAT3 (Ser727) positive nuclei from O and P (two-tailed unpaired Student t-test, **: P < 0.01; Mean ± SD). (R) MMP15 protein levels in SCCs obtained from mice implanted with FaDu-TP63 shRNA3 treated with vehicle or Trametinib were measured using western blotting. Quantification of relative MMP-15 levels after normalizing to β-actin is shown at the bottom. (S) (I) In HNSCCs with high TP63 expression, TP63 downregulates MAPK signaling by directly inducing the MAPK inhibitor DUSP6. (II) In HNSCCs with low TP63 expression, MAPK signaling is activated through downregulation of DUSP6. MAPK signaling leads to upregulation of P-STAT3 (Ser727) and MMP15 to promote HNSCC growth, invasion, and metastasis.
Histopathological analysis confirmed that Trametinib treatment prevented the progression to poorly differentiated SCCs. Specifically, whereas all vehicle treated SCCs were classified as poorly differentiated, 71.4% of FaDu-TP63 shRNA3 SCCs and 43% of CAL27-TP63 shRNA3 SCCs were classified as moderately differentiated (Figure 7I–K, Supplementary Figure 15I–K). Finally, Trametinib treatment reversed the induction of P-ERK1/2, P-STAT3 (Ser727), and MMP15 in both FaDu-TP63 shRNA3 SCCs and CAL27-TP63 shRNA3 SCCs (Figure 7L–R and Supplementary Figure 15L–Q). Collectively, these findings demonstrate that Trametinib treatment prevents the metastasis of HNSCCs with low TP63 expression through downregulating the levels of P-ERK1/2, P-STAT3 (Ser727), and MMP15.
Discussion
TP63 is frequently overexpressed in primary human SCCs, including HNSCCs, and is often believed to function as an oncogene during early stages of tumorigenesis [10, 11, 13, 17]. Further, loss of TP63 during later stages of tumorigenesis has been linked to tumor aggressiveness and metastasis in skin, breast, prostate, and bladder cancers [14–16, 37]. However, the role of TP63 in metastasis of HNSCCs has not been investigated. In this study, we demonstrate that TP63 expression is specifically downregulated in poorly differentiated HNSCCs (Figure 1 and Supplementary Figure 1). Further, using genetically engineered mice as well as xenograft models, we determined that TRP63/TP63 downregulation accelerates HNSCC progression and metastasis. The role for TP63 in suppressing tumor metastasis is mediated by induction of DUSP6, a negative regulator of RAS-MAPK signaling (Figure 5O–R). Upon TP63 downregulation, activation of MAPK signaling leads to STAT3 phosphorylation and MMP15 induction, providing a mechanistic explanation for the high metastatic potential of TP63 knockdown SCCs (Figure 7S). Consistent with these findings, we previously analyzed publicly available patient datasets (TCGA) and observed a correlation between low TP63 expression and an overall shorter survival time in HNSCC patients [16].
The TP63/TRP63 gene can produce at least six isoforms by using two different promoters and alternative splicing [38]. Consistent with previous reports on TP63/TRP63 expression in stratified epithelia and squamous tumors [8], we found that both the mouse tongue and the human HNSCC lines used in this study only express ΔNp63α. Further, our strategies were designed to eliminate all TRP63/TP63 isoforms. Thus, even if other TRP63/TP63 isoforms are expressed in these cells or tissues at low levels, they would have been eliminated in our experiments.
The mechanism underlying TP63 loss in HNSCCs is not known. Although induction of EMT has been suggested to occur in response to TP63 loss, we did not observe EMT in our tumors. Further, mutations in TP63 gene are rarely found in HNSCCs [39, 40]. However, silencing of TP63 gene expression through promoter methylation has been reported in lung and bladder cancer [41, 42]. Additionally, in some non-squamous tumor types, TP63 expression is downregulated by microRNAs [43, 44]. However, whether these mechanisms contribute to TP63 loss in HNSCCs remains to be investigated.
Our conclusion that TRP63/TP63 functions as a tumor suppressor gene in oral tumorigenesis appears to contradict previously published work that concluded that ablation of TRP63 in established mouse oral SCCs leads to tumor regression [45]. In this study, TRP63 was ablated in SCCs that developed in response to DMBA treatment. Importantly, the oral tumors that developed in these mice were located on the outer lip and facial skin rather than within the oral cavity. Thus, these tumors do not mimic the intra-oral SCCs that developed in our model. Further, the use of DMBA, rather than 4NQO, as a tumor initiator complicates the interpretation of the findings as it is well-established that only 4NQO leads to the development of oral SCCs that mimic human SCCs at the molecular and cellular level [46, 47]. Our analysis of 4NQO-induced oral tumors as well as xenograft tumors clearly establishes a critical role for TRP63/TP63 loss in tumor progression and metastasis.
Using RNA-sequencing followed by functional studies, we determined that upregulation of MAPK and STAT3 in TP63 knockdown HNSCCs is responsible for the high metastatic potential of these tumors. Previous studies have demonstrated that a crosstalk between MAPK and STAT3 signaling pathways occurs in cancer cell lines [48]. However, a crosstalk between these two signaling pathways has not been investigated in HNSCC progression and metastasis in vivo. Here, we report that the serine phosphorylation of STAT3 in HNSCCs is dependent on MAPK activation (Supplementary Figure 12). Further, MAPK inhibition by Trametinib treatment led to a downregulation of P-STAT3 (Ser727) levels and prevention of tumor metastasis (Figure 6, 7). Thus, we have identified a novel functional link between MAPK and P-STAT3 (Ser727) in HNSCC progression.
A potential limitation of this study is the use of HNSCC cell lines harboring TP53 mutations [49]. A previous study using breast cancer cells found that some mutant TP53 proteins can directly interact with TP63 affecting the ability of TP63 to transactivate genes that are involved in suppression of metastasis [50]. It is possible that these interactions also occur in HNSCCs carrying TP53 mutations. However, TP53-mediated inactivation of TP63 does not seem to occur in the HNSCC cell lines used in our study as we observed a clear difference in phenotype between cells and xenografts with or without TP63 expression. Nevertheless, future studies using HNSCC cell lines without TP53 mutations would be informative in providing further insights into the specific role of TP63 in HNSCC progression and metastasis.
In summary, we have identified a novel role for TP63 loss in HNSCC progression and metastasis. Our study has uncovered the upregulation of the MAPK-P-STAT3 (Ser727)-MMP15 axis as the mechanism by which TP63 promotes metastasis. Further, therapeutic inhibition of this axis prevents HNSCC metastasis caused by loss of TP63. Our data demonstrating that metastasis of HNSCCs that have lost TP63 expression can be prevented by inhibiting MAPK-P-STAT3 signaling has a direct translational impact relevant to the treatment of these HNSCCs.
Supplementary Material
Implications:
Our findings uncover a novel functional role for TP63 loss in HNSCC metastasis and identify MAPK signaling as a potential therapeutic target for treating HNSCCs with low TP63 expression.
Acknowledgements
We would like to thank Drs. Peter J. Koch and Shi-Long Lu (University of Colorado Anschutz Medical Campus) for helpful comments on this manuscript. We also would like to thank Dr. James K. Wahl, III (University of Nebraska Medical Center) for generating the monoclonal anti-TP63 antibody (11H1). This work was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) of the National Institutes of Health (NIH) under Award Number R01 AR061506. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. This work was also supported by a research grant from the Cancer League of Colorado.
Financial Support: This work was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) under Award Number R01 AR061506 to MIK. This work was also supported by a research grant from the Cancer League of Colorado to MIK.
Footnotes
Conflict of Interest: The authors state no conflicts of interest.
References
- 1.Mourad M, et al. , Epidemiological Trends of Head and Neck Cancer in the United States: A SEER Population Study. Journal of Oral and Maxillofacial Surgery, 2017. 75(12): p. 2562–2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marur S and Forastiere AA, Head and Neck Squamous Cell Carcinoma: Update on Epidemiology, Diagnosis, and Treatment. Mayo Clin Proc, 2016. 91(3): p. 386–96. [DOI] [PubMed] [Google Scholar]
- 3.Galbiatti AL, et al. , Head and neck cancer: causes, prevention and treatment. Braz J Otorhinolaryngol, 2013. 79(2): p. 239–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cognetti DM, Weber RS, and Lai SY, Head and neck cancer: an evolving treatment paradigm. Cancer, 2008. 113(7 Suppl): p. 1911–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zibelman M and Mehra R, Overview of Current Treatment Options and Investigational Targeted Therapies for Locally Advanced Squamous Cell Carcinoma of the Head and Neck. Am J Clin Oncol, 2016. 39(4): p. 396–406. [DOI] [PubMed] [Google Scholar]
- 6.Hauswald H, et al. , Long-term outcome and patterns of failure in patients with advanced head and neck cancer. Radiat Oncol, 2011. 6: p. 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Svahn MF, et al. , Trends in all-cause five-year mortality after head and neck cancers diagnosed over a period of 33 years. Focus on estimated degree of association with human papillomavirus. Acta Oncol, 2016. 55(9–10): p. 1084–1090. [DOI] [PubMed] [Google Scholar]
- 8.Koster MI, p63 in skin development and ectodermal dysplasias. J Invest Dermatol, 2010. 130(10): p. 2352–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rothenberg SM and Ellisen LW, The molecular pathogenesis of head and neck squamous cell carcinoma. J Clin Invest, 2012. 122(6): p. 1951–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saladi SV, et al. , ACTL6A Is Co-Amplified with p63 in Squamous Cell Carcinoma to Drive YAP Activation, Regenerative Proliferation, and Poor Prognosis. Cancer Cell, 2017. 31(1): p. 35–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamaguchi K, et al. , Frequent gain of the p40/p51/p63 gene locus in primary head and neck squamous cell carcinoma. Int J Cancer, 2000. 86(5): p. 684–9. [DOI] [PubMed] [Google Scholar]
- 12.Hibi K, et al. , AIS is an oncogene amplified in squamous cell carcinoma. Proceedings of the National Academy of Sciences of the United States of America, 2000. 97(10): p. 5462–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rocco JW, et al. , p63 mediates survival in squamous cell carcinoma by suppression of p73-dependent apoptosis. Cancer Cell, 2006. 9(1): p. 45–56. [DOI] [PubMed] [Google Scholar]
- 14.Tucci P, et al. , Loss of p63 and its microRNA-205 target results in enhanced cell migration and metastasis in prostate cancer. Proc Natl Acad Sci U S A, 2012. 109(38): p. 15312–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Urist MJ, et al. , Loss of p63 expression is associated with tumor progression in bladder cancer. American Journal of Pathology, 2002. 161(4): p. 1199–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lakshmanachetty S, et al. , TRP63/TP63 loss accelerates skin tumorigenesis through activation of Wnt/beta-catenin signaling. J Dermatol Sci, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sniezek JC, et al. , Dominant negative p63 isoform expression in head and neck squamous cell carcinoma. Laryngoscope, 2004. 114(12): p. 2063–2072. [DOI] [PubMed] [Google Scholar]
- 18.Mills AA, Qi Y, and Bradley A, Conditional inactivation of p63 by Cre-mediated excision. Genesis, 2002. 32(2): p. 138–41. [DOI] [PubMed] [Google Scholar]
- 19.Huang DW, et al. , DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res, 2007. 35(Web Server issue): p. W169–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Subramanian A, et al. , Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A, 2005. 102(43): p. 15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kouwenhoven EN, et al. , Genome-wide profiling of p63 DNA-binding sites identifies an element that regulates gene expression during limb development in the 7q21 SHFM1 locus. PLoS Genet, 2010. 6(8): p. e1001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koster MI, et al. , Reactivation of developmentally expressed p63 isoforms predisposes to tumor development and progression. Cancer Res, 2006. 66(8): p. 3981–6. [DOI] [PubMed] [Google Scholar]
- 23.Vitale-Cross L, et al. , Chemical carcinogenesis models for evaluating molecular-targeted prevention and treatment of oral cancer. Cancer Prev Res (Phila), 2009. 2(5): p. 419–22. [DOI] [PubMed] [Google Scholar]
- 24.Kanojia D and Vaidya MM, 4-nitroquinoline-1-oxide induced experimental oral carcinogenesis. Oral Oncol, 2006. 42(7): p. 655–67. [DOI] [PubMed] [Google Scholar]
- 25.Yang Z, et al. , Comparable molecular alterations in 4-nitroquinoline 1-oxide-induced oral and esophageal cancer in mice and in human esophageal cancer, associated with poor prognosis of patients. In Vivo, 2013. 27(4): p. 473–84. [PMC free article] [PubMed] [Google Scholar]
- 26.Yanagawa T, et al. , Loss of cytokeratin 13 expression in squamous cell carcinoma of the tongue is a possible sign for local recurrence. Journal of Experimental & Clinical Cancer Research, 2007. 26(2): p. 215–220. [PubMed] [Google Scholar]
- 27.Frohwitter G, et al. , Cytokeratin and protein expression patterns in squamous cell carcinoma of the oral cavity provide evidence for two distinct pathogenetic pathways. Oncol Lett, 2016. 12(1): p. 107–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sano D and Myers JN, Xenograft models of head and neck cancers. Head Neck Oncol, 2009. 1: p. 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barbieri CE, et al. , Loss of p63 leads to increased cell migration and up-regulation of genes involved in invasion and metastasis. Cancer Res, 2006. 66(15): p. 7589–97. [DOI] [PubMed] [Google Scholar]
- 30.Higashikawa K, et al. , Snail-induced down-regulation of DeltaNp63alpha acquires invasive phenotype of human squamous cell carcinoma. Cancer Res, 2007. 67(19): p. 9207–13. [DOI] [PubMed] [Google Scholar]
- 31.Reddy KB, Nabha SM, and Atanaskova N, Role of MAP kinase in tumor progression and invasion. Cancer Metastasis Rev, 2003. 22(4): p. 395–403. [DOI] [PubMed] [Google Scholar]
- 32.Peng Q, et al. , Mitogen-activated protein kinase signaling pathway in oral cancer. Oncol Lett, 2018. 15(2): p. 1379–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Teng Y, Ross JL, and Cowell JK, The involvement of JAK-STAT3 in cell motility, invasion, and metastasis. JAKSTAT, 2014. 3(1): p. e28086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Levy DE and Lee CK, What does Stat3 do? Journal of Clinical Investigation, 2002. 109(9): p. 1143–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Z, et al. , Dual specificity phosphatase 6 (DUSP6) is an ETS-regulated negative feedback mediator of oncogenic ERK signaling in lung cancer cells. Carcinogenesis, 2010. 31(4): p. 577–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.King JW and Nathan PD, Role of the MEK inhibitor trametinib in the treatment of metastatic melanoma. Future Oncol, 2014. 10(9): p. 1559–70. [DOI] [PubMed] [Google Scholar]
- 37.Bergholz J, et al. , DeltaNp63alpha regulates Erk signaling via MKP3 to inhibit cancer metastasis. Oncogene, 2014. 33(2): p. 212–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang AN, et al. , p63, a p53 homolog at 3q27–29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Molecular Cell, 1998. 2(3): p. 305–316. [DOI] [PubMed] [Google Scholar]
- 39.Li H, et al. , Genomic analysis of head and neck squamous cell carcinoma cell lines and human tumors: a rational approach to preclinical model selection. Mol Cancer Res, 2014. 12(4): p. 571–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stransky N, et al. , The mutational landscape of head and neck squamous cell carcinoma. Science, 2011. 333(6046): p. 1157–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shi YX, et al. , Genome-wide DNA methylation profiling reveals novel epigenetic signatures in squamous cell lung cancer. BMC Genomics, 2017. 18(1): p. 901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park BJ, et al. , Frequent alteration of p63 expression in human primary bladder carcinomas. Cancer Res, 2000. 60(13): p. 3370–4. [PubMed] [Google Scholar]
- 43.Papagiannakopoulos T, Shapiro A, and Kosik KS, MicroRNA-21 targets a network of key tumor-suppressive pathways in glioblastoma cells. Cancer Res, 2008. 68(19): p. 8164–72. [DOI] [PubMed] [Google Scholar]
- 44.Manni I, et al. , The microRNA miR-92 increases proliferation of myeloid cells and by targeting p63 modulates the abundance of its isoforms. Faseb j, 2009. 23(11): p. 3957–66. [DOI] [PubMed] [Google Scholar]
- 45.Ramsey MR, et al. , FGFR2 signaling underlies p63 oncogenic function in squamous cell carcinoma. J Clin Invest, 2013. 123(8): p. 3525–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ishida K, et al. , Current mouse models of oral squamous cell carcinoma: Genetic and chemically induced models. Oral Oncol, 2017. 73: p. 16–20. [DOI] [PubMed] [Google Scholar]
- 47.Schoop RA, Noteborn MH, and Baatenburg de Jong RJ, A mouse model for oral squamous cell carcinoma. J Mol Histol, 2009. 40(3): p. 177–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gkouveris I, et al. , Erk1/2 activation and modulation of STAT3 signaling in oral cancer. Oncol Rep, 2014. 32(5): p. 2175–82. [DOI] [PubMed] [Google Scholar]
- 49.Nichols AC, et al. , Frequent mutations in TP53 and CDKN2A found by next-generation sequencing of head and neck cancer cell lines. Arch Otolaryngol Head Neck Surg, 2012. 138(8): p. 732–9. [DOI] [PubMed] [Google Scholar]
- 50.Adorno M, et al. , A Mutant-p53/Smad Complex Opposes p63 to Empower TGF beta-Induced Metastasis. Cell, 2009. 137(1): p. 87–98. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.