Abstract
The sialoglycoprotein podocalyxin is absent in normal pancreas but is overexpressed in pancreatic cancer and is associated with poor clinical outcome. Here we investigate the role of podocalyxin in migration and metastasis of pancreatic adenocarcinomas using SW1990 and Pa03c as cell models. Although ezrin is regarded as a cytoplasmic binding partner of podocalyxin that regulates actin polymerization via Rac1 or RhoA, we did not detect podocalyxin-ezrin association in pancreatic cancer cells. Moreover, depletion of podocalyxin did not alter actin dynamics or modulate Rac1 and RhoA activity in pancreatic cancer cells. Using mass spectrometry, bioinformatics analysis, co-immunoprecipitation, and pulldown assays, we discovered a novel, direct binding interaction between the cytoplasmic tail of podocalyxin and the large GTPase dynamin-2 at its GTPase, middle, and pleckstrin homology domains. This podocalyxin-dynamin-2 interaction regulated microtubule growth rate, which in turn modulated focal adhesion dynamics and ultimately promoted efficient pancreatic cancer cell migration via microtubule- and Src-dependent pathways. Depletion of podocalyxin in a hemispleen mouse model of pancreatic cancer diminished liver metastasis without altering primary tumor size. Collectively, these findings reveal a novel mechanism by which podocalyxin facilitates pancreatic cancer cell migration and metastasis.
Keywords: Podocalyxin, Dynamin-2, Pancreatic Cancer, Cell Migration, Metastasis
Introduction
Podocalyxin (PODXL) is a type 1 transmembrane sialomucin glycoprotein belonging to the CD34 protein subfamily that is endogenously expressed by kidney podocytes (1), hematopoietic progenitor stem cells (2) and vascular endothelium (3). First discovered in adult kidney, the negatively charged ectodomain of PODXL is instrumental for the maintenance of glomerular filtration slits via an anti-adhesive charge repulsion effect (1). In contrast, PODXL expressed on high endothelial venules is instead pro-adhesive, capable of mediating binding to L-selectin on lymphocytes, thereby facilitating lymphocytes tethering, rolling and recruitment to secondary lymphoid organs during inflammation (3).
PODXL plays a pivotal role in regulating cell adhesion, a fundamental aspect of many (patho)physiological processes, including embryonic development, inflammatory and immune responses and cancer metastasis. Importantly, PODXL is also overexpressed by cancer cells, including lung (4), breast (5), ovarian (6), bladder (7), renal (8), colorectal (9) and pancreatic cancers (10). Overexpression of PODXL in cancer cells is associated with aggressive clinicopathological characteristics and poor clinical outcomes (4–10). Both the anti- and pro- adhesive roles of the PODXL ectodomain have been implicated in cancer dissemination. For instance, PODXL has been reported to disrupt cell-cell contact and induce tumor shedding from monolayers of ovarian carcinoma (6), and breast cancer cells (5). Conversely, PODXL, expressed on colon carcinoma (11) and pancreatic cancer cells (12), binds to E- and L-selectin, thereby facilitating circulating cancer cell rolling on vascular endothelium and subsequent extravasation to secondary tumor sites. The structure of PODXL consists of an N-terminal extracellular domain with extensive N- and O-glycosylation and sialylation, and a cytoplasmic intracellular tail with a C-terminal DTHL consensus sequence capable of interacting with PDZ binding domain (13). The cytoplasmic tail of PODXL, despite its small size has been reported to interact with proteins such as ezrin (8,14) and Na(+)/H(+) exchange regulatory cofactor (NHERF) 1 or 2 (15,16) to activate downstream intracellular signaling, primarily via the PI3K (17), RhoA (15) and Rac1 (8) pathways, leading to epithelial-mesenchymal transition (18), migration (19) and invasion (20).
While typically absent in normal pancreas and non-malignant pancreatic epithelial cells, approximately 30–69% of pancreatic ductal adenocarcinomas express PODXL, with the strongest expression exhibited by the invasive front of primary tumors (12,21,22). Moreover, high PODXL expression is associated with poor clinicopathological characteristics and the development of distant metastasis (23,24). PODXL has also been shown to be an independent predictor of poor prognosis and is linked to higher risk of death and reduced survival (10,22–24). These effects are hypothesized to be at least partially due to the ability of PODXL to regulate cell migration and invasion (22,24). Although ezrin is regarded as the main intracellular binding partner of PODXL, our data reveal the lack of any association between PODXL and ezrin in pancreatic cancer cells. Consequently, the mechanism by which PODXL exerts its downstream effects to promote pancreatic cancer cell migration remains unknown. Furthermore, the functional significance of PODXL in promoting pancreatic cancer cell metastasis in vivo has yet to be delineated.
In this study, we examined the roles of PODXL in migration and metastasis of pancreatic cancer cells in vitro and in vivo, using two metastatic pancreatic cancer cell lines, SW1990 and Pa03c. To identify the cytoplasmic binding partner of PODXL in pancreatic cancer cells, we immunoprecipitated PODXL from SW1990 cells, and subjected the specimens to tandem mass spectrometry. Bioinformatics analysis identified dynamin-2 as a potential binding partner of PODXL. Reciprocal co-immunoprecipitation (co-IP) verified the association of PODXL and dynamin-2. This direct binding interaction was further confirmed by in vitro His-tag binding assays, which identified the GTPase, middle and pleckstrin homology domains of dynamin-2 as critical for binding to PODXL. Of note, co-IP assays failed to demonstrate any PODXL-ezrin association. The novel PODXL-dynamin-2 interaction modulates microtubule dynamics, which in turn modulates focal adhesion (FA) assembly/disassembly. Dynamin-2 also regulates FA turnover via Src kinase-dependent pathway. As a result, inhibition or downregulation of dynamin-2, microtubule or Src kinase reverses the pro-migratory phenotype of PODXL in both two-dimensional (2D) and microchannel migration assays. Along these lines, knockdown of PODXL significantly impairs unconfined and confined migration by decreasing microtubule dynamics and increasing FA density. The functional role of PODXL in promoting metastasis is demonstrated using a preclinical murine hepatic metastasis model via a hemispleen injection technique (25).
Materials and Methods
Cell culture and drug treatment.
SW1990 pancreatic cancer cells and MDA-MB-231 breast cancer cells were purchased from the American Type Culture Collection (Manassas, VA), while Pa03c pancreatic cancer cells were obtained as previously described (26). All cell lines were cultured in standard Dulbecco’s Modified Eagle Medium (DMEM, Gibco) supplemented with 10% heat-inactivated fetal bovine serum (FBS, Gibco) and 1% penicillin/streptomycin (Gibco). All cell lines were used for 10 passages after thawing from the frozen vials and were tested routinely for mycoplasma via quantitative polymerase chain reaction. The cell lines were not further authenticated. In select experiments involving drug treatments, cells were incubated with culture media containing 40 μM dynasore (Sigma-Aldrich), 1.2 μM taxol (Sigma-Aldrich), 10 nM dasatinib (Cell Signaling), 10 μM PP2 (EMD Millipore), or the corresponding vehicle control.
shRNA and siRNA knockdown.
Stable cell lines of scramble control and PODXL knockdown (PODXL-KD) SW1990 were generated with short hairpin RNA (shRNA) as previously described (12). Additional PODXL-KD cell lines with SW1990, Pa03c and MDA-MB-231, and NHERF2-KD cell lines with SW1990 were generated using two different lentiviral shRNA sequences as detailed in Supplementary Methods. Transient dynamin-2 knockdown was established by transfecting cells with Dynamin-2 siRNA (Santa Cruz Biotechnology, sc-35236) using Lipofectamine RNAiMAX (Invitrogen) following the manufacturer’s protocol. As a control, cells were transfected with scramble control siRNA (Santa Cruz Biotechnology, sc-35236). Cells were incubated with the lipid complex for 72 h before they were used for subsequent experiments.
Western blot and antibodies.
Standard western blot techniques were performed as previously described (27) using NuPAGE 4–12% Bis-Tris Protein Minigels (Invitrogen). The antibodies used are listed below. Primary antibodies: 1) Podocalyxin-like 1 (3D3) mouse monoclonal antibody (Santa Cruz Biotechnology, sc-23904, 1:500). 2) PODXL (EPR9518) rabbit monoclonal antibody (Abcam, ab150358, 1:1000). 3) Dynamin-2 (DYN2–11) mouse monoclonal antibody (Sigma Aldrich, SAB4200661, 1:500). 4) Dynamin-2 rabbit polyclonal antibody (Abcam, ab3457, 1:1000). 5) Ezrin (3C12) mouse monoclonal antibody (Abcam, ab4069, 1:500). 6) NHERF2 (D3A5) rabbit monoclonal antibody (Cell Signaling, 9568, 1:1000). 7) GST (26H1) mouse monoclonal antibody (Cell Signaling, 2624, 1:2000) 8) 6x-His tag (4E3D10H2/E3) mouse monoclonal antibody (ThermoFisher Scientific, MA1–135, 1:2000). 9) RhoA (7F1.E5) mouse monoclonal antibody (Cytoskeleton, ARH04, 1:500). 10) Rac1 mouse monoclonal antibody (Cytoskeleton, ARC03, 1:500). 11) Actin (C4) mouse monoclonal antibody (BD Transduction, 612656, 1:10000). Secondary antibodies: 1) Anti-mouse IgG, HRP-linked antibody (Cell Signaling, 7076S, 1:2000). 2) Anti-rabbit IgG, HRP-linked antibody (Cell Signaling, 7074S, 1:2000).
Random 2D migration assay.
22mm×22mm square glass coverslips glued to the bottom a 6-well plate were coated for 1 h with 20 μg/ml of rat tail type I collagen (Gibco). 5×104 cells were seeded onto the coverslips with 2 ml of culture media. The cells were imaged via a 10x Ph1 objective every 10 min for 10 h using stage automation on a Nikon Inverted microscope with a stage top incubator (Tokai Hit Co., Shizuoka, Japan) maintained at 37°C with 5% CO2 and humidity. At least 5 individual migrating single cells from 5 different locations per sample were analyzed using ImageJ (National Institutes of Health, Bethesda, MD). The cell outlines were tracked at every time point to record their spatial position and morphology parameters including projected area, circularity and solidity as previously described (28). A rose plot with the cell migration trajectories and their average mean square displacement were also calculated as described in (28).
Polydimethylsiloxane (PDMS)-based microchannel migration assay.
PDMS-based microchannels devices were fabricated using a photolithography and standard replica molding technique as previously described (29,30). Each device comprises a series of parallel 200 μm-long and 10 μm-high microchannels of prescribed widths varying from 6, 10, 20 to 50μm arrayed perpendicularly between a cell and chemoattractant inlet lines. The microchannels devices were coated with 20 μg/ml of collagen type I to facilitate cell adhesion. Cell migration was visualized and recorded via time-lapse live microscopy in an enclosed, humidified microscope stage maintained at 37°C and 5% CO2 using stage automation and a Nikon Inverted microscope. Phase contrast time-lapse images were taken at every 10 min for 24 h with a 10x Ph1 objective. The spatial x and y positions of all non-dividing and viable cells that entered and migrated in the microchannels were tracked overtime with the Manual Tracking plugin in ImageJ. Motility parameters, namely velocity, speed and persistence, were computed using a custom-written MATLAB code as previously described (29,30).
Cytoskeletal and adhesion dynamics assays.
Actin dynamics was quantified via fluorescence recovery after photobleaching (FRAP) on cells transfected with LifeAct-GFP (31). Microtubule dynamics was assessed by measuring the rate of microtubule growth with time-lapse confocal imaging of EB1-GFP-transfected cells (30). FAs were visualized and quantified via total internal reflectance fluorescence (TIRF) imaging of cells immunostained with phospho-paxillin (28). A comprehensive description of the experimental protocols is provided in Supplementary Methods.
RhoA and Rac active pulldown assays.
RhoA Pulldown Activation Assay (Cytoskeleton, BK036) and Rac1 Pulldown Activation Assay (Cytoskeleton, BK035) were conducted according to manufacturer’s protocols.
Co-IP and mass spectrometry.
Co-IP was performed to identify the binding partner(s) of PODXL in pancreatic cancer cells with Pierce co-IP Kit (Thermo Scientific). The co-IP assays were performed by incubating pre-cleared cell lysates containing 500 μg of total protein with 25 μl of resin immobilized with 10 μg of antibody overnight at 4°C. PODXL (EPR9518) rabbit monoclonal antibody (Abcam, ab150358) was used to immunoprecipitate PODXL. A rabbit IgG monoclonal isotype (EPR25A, Abcam, ab172730) or beads-only controls were included to account for non-specific binding. Unbound proteins were washed away from the antibody-immobilized resin by centrifugation with IP lysis buffer supplemented with 1M NaCl for 10 times. Bound proteins were eluted with 1% SDS and concentrated. The co-IP elution was submitted to the Johns Hopkins Mass Spectrometry and Proteomics Core Facility (Baltimore, MD, USA) for protein identification or analyzed with standard western blot protocol once the candidate protein was identified. In subsequent reciprocal co-IP experiments, Dynamin-2 rabbit polyclonal antibody (Abcam, ab3457) or NHERF2 (D3A5) rabbit monoclonal antibody (Cell Signaling, 9568) were used to immunoprecipitate dynamin-2 or NHERF2, respectively.
In-Vitro His-tag pulldown assay.
In-vitro His-tag pulldown assays were conducted using a recombinant purified His-DNM2 probe (Gift from Dr. Mark McNiven) (32). Additional His-DNM2 constructs consisting of select Dynamin-2 domains, and the His-tagged PODXL cytoplasmic tail (PCT, aa484 to aa558) were created using standard molecular cloning techniques with the same pQE-80L vector as the full length His-DNM2 probe. His-tagged constructs were transformed into and produced with BL21 Competent E.coli under 0.1 mM Isopropyl β-D-1-thiogalactopyranoside induction at 16°C overnight. The recombinant His-tagged proteins were purified using HisPur™ Ni-NTA Spin Purification Kit (Thermo Scientific). The in vitro pulldown assay was performed using Pierce™ His Protein Interaction Pull-Down Kit (Thermo Scientific). For experiments involving purified glutathione S-transferase (GST)-tagged PCT, GST-PCT is cloned into pGEX-2T vector via standard molecular cloning techniques. The recombinant GST-PCT proteins were induced and purified from E.coli using glutathione–agarose beads (Pierce).
Hemispleen mice model.
A preclinical murine model of hepatic metastasis was performed via a hemispleen injection technique (25) to assess the metastatic potential of PODXL-expressing and PODXL-KD pancreatic cancer cells. All animal work was approved and performed in compliance with the Institutional Animal Care and Use Committee (IACUC) at the Johns Hopkins University. Prior to surgery, trypsinized cells were incubated in DMEM with 10% FBS at 37°C with intermittent mixing to restore surface glycoprotein expression. Next, the cells were washed with PBS 3× and resuspended in anti-clumping buffer diluted 1:1000 in Hanks’ balanced salt solution (Gibco) to single cell suspensions of 1×107/ml for Pa03c and 2.5×106/ml for SW1990. The cells were maintained on ice at all times for subsequent steps. 6 to 8 weeks old female NOD-SCID mice were purchased from Johns Hopkins Research Animal Resources (Baltimore, MD, USA) and maintained in accordance with Institutional Animal Care and Use Committee guidelines. At surgery, the spleen of a fully anesthetized mouse was eviscerated, clipped and excised into 2 halves. 100 μl of cell suspension was injected into one half of the excised spleen, followed by a flush with 150 μl of ice-cold PBS. The cells were allowed to flow into the liver via the splenic vessels for 2 min, after which the splenic vessels were clipped and removed, followed by suturing. All mice were monitored regularly and sacrificed when they display morbid characteristics. At necropsy, livers were harvested and examined macroscopically and microscopically with haematoxylin and eosin staining for evidence of metastases. Additionally, any visible splenic tumors formed at the injection sites were collected and measured for size with the formula, V=(LxWxW)/2, where V is the tumor volume, L is the tumor length and W is the tumor width. The primary splenic tumors were also stained for PODXL with immunohistochemistry as detailed in Supplementary Methods.
In vitro and in vivo tumor growth assays.
The proliferative potential of scramble control and PODXL-KD cells were assessed in vitro and in vivo via a subcutaneous implantation model as described in Supplementary Methods.
Statistical analysis.
All data are presented as mean±S.E.M. from 3 independent experiments unless otherwise stated. Graphing and statistical analyses were performed with GraphPad Prism. Statistical significance was determined between pairs of data with a t-test, or between groups of data with one-way ANOVA and a Tukey’s Multiple Comparison post-hoc test.
Results
PODXL knockdown impairs pancreatic cancer cell migration in vitro.
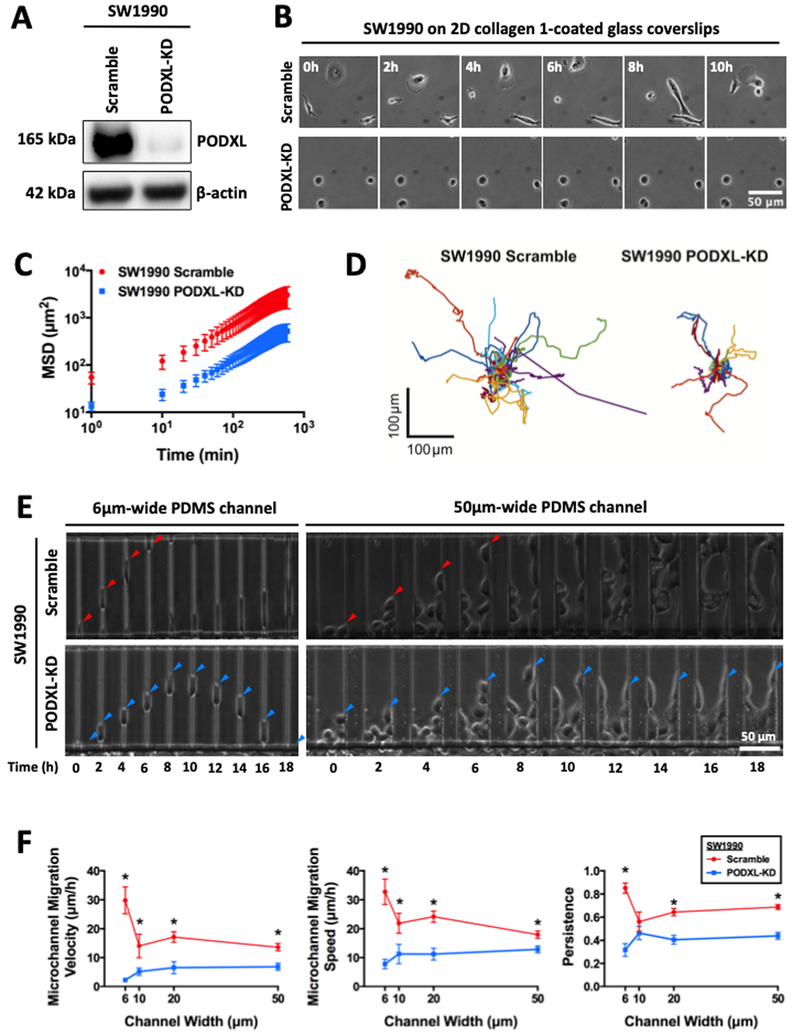
We generated scramble control and stable PODXL-KD SW1990 cells via lipofectamine transfection followed by clonal selection. PODXL knockdown was verified via immunoblotting (Fig. 1A). We first investigated the potential role of PODXL in regulating random 2D migration of pancreatic cancer cells on collagen I-coated glass coverslips. Scramble control relative to PODXL-KD SW1990 cells were more motile (Fig. 1B, Supplementary Video 1), as evidenced by their higher mean squared displacement (Fig. 1C), and as such, were able to explore a larger surrounding area (Fig. 1D). Interestingly, morphometric analysis revealed that PODXL knockdown renders cells smaller, more circular and less protrusive (Supplementary Fig. S1A). Depletion of PODXL also diminishes cell morphodynamics, as evidenced by smaller changes of their various morphological indices (Supplementary Fig. S1B). In line with random 2D migration data, PODXL knockdown significantly suppressed chemotactically-driven cell migration inside both unconfined and confined PDMS-based microchannels (Fig. 1E, Supplementary Video 2), which resulted in lower velocity, speed and persistence relative to scramble control cells (Fig. 1F).
Fig. 1. PODXL knockdown in pancreatic cancer cells suppresses random 2D migration and chemotactic migration inside unconfined and confined channels.
(A) Western blot showing efficient knockdown of PODXL in SW1990 pancreatic cancer cells using shRNA lipofectamine transfection followed by stable clonal selection. (B) Representative time-lapse micrographs of scramble control and PODXL-KD SW1990 cells migrating on 2D collagen I-coated glass coverslips. The images are spaced at 2 h intervals. (C) Average mean squared displacement (MSD) over time of migrating scramble control and PODXL-KD SW1990 cells. Data represent the mean±S.E.M from 3 independent experiments. (D) Superimposed images of individual trajectories of scramble control and PODXL-KD SW1990 cells migrating on 2D collagen I-coated surfaces. (E) Representative time-lapse micrographs of scramble control (top row) and PODXL-KD (bottom row) SW1990 cells migrating inside 6 μm-wide, confined microchannels (left panels) and 50 μm-wide, unconfined microchannels (right panels). The images are spaced at 2 h intervals. Arrowheads indicate the leading edge of a migrating cell. (F) Migration velocity (left panel), speed (middle panel) and persistence (right panel) of scramble control and PODXL-KD SW1990 cells in PDMS-based microchannels of 10 μm in height, 200 μm in length, and either 6, 10, 20 or 50 μm in width. Data represent the mean±S.E.M from at least 3 independent experiments. * represents p<0.05 between scramble control and PODXL-KD cells.
To validate our observations, we knocked down PODXL on both SW1990 and Pa03c pancreatic cancer cells using two different shRNA lentivirus sequences. Both sequences were able to knockdown PODXL (Supplementary Fig. S1C) and suppress migration of SW1990 (Supplementary Fig. S1D) and Pa03c (Supplementary Fig. S1E) cells through both unconfined and confined microchannels. To further generalize our findings and to extend their significance to other cancer cell types, we also knocked down PODXL in the widely used human breast cancer cell line, MDA-MB-231 (Supplementary Fig. S1C), and observed a similar inhibition in cell migration through both unconfined and confined microchannels (Supplementary Fig. S1F).
PODXL knockdown alters microtubule and adhesion dynamics in pancreatic cancer cells.
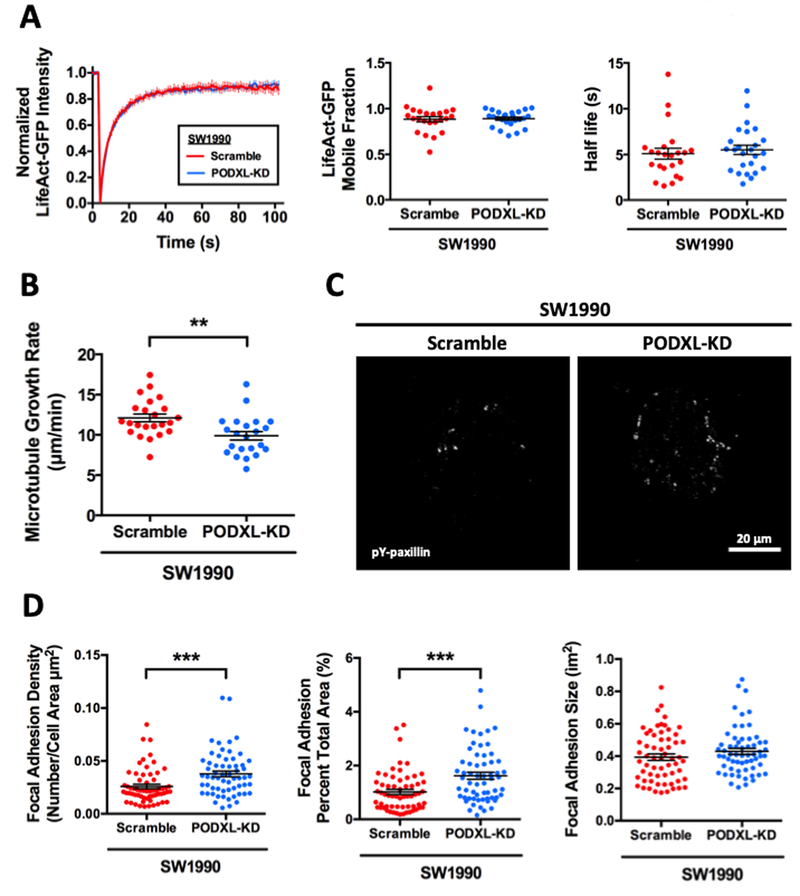
Given the pronounced effect of PODXL knockdown in altering cell morphology and slowing down cell migration, we hypothesized that PODXL mediates these changes by regulating cytoskeletal and/or adhesion dynamics in pancreatic cancer cells. We first quantified actin dynamics in scramble control and PODXL-KD LifeAct-GFP-transfected SW1990 cells using FRAP. Both control and PODXL-KD cells exhibited nearly identical mobile fractions of LifeAct-GFP molecules and half-lives of recovery (Fig. 2A), suggesting that PODXL knockdown does not alter actin dynamics. We next investigated the effects of PODXL knockdown on microtubule dynamics by imaging and quantifying the rate of EB1-GFP incorporation to the positive growing ends of microtubules. Interestingly, SW1990 PODXL-KD cells demonstrated a significantly lower microtubule growth rate compared to the scramble control cells (Fig. 2B), thereby indicating that PODXL depletion slows down microtubule dynamics.
Fig. 2. PODXL knockdown decreases microtubule growth rate and increases FA density without altering actin dynamics.
(A) Actin dynamics was assessed using FRAP at cell-cell junctions of scramble control and PODXL-KD SW1990 cells transiently transfected with LifeAct-GFP. Representative FRAP curves of the fraction of initial fluorescence intensity as a function of time for scramble control (red) and PODXL-KD (blue) cells (left panel). Mobile fraction (middle panel) and recovery half-life (right panel) of LifeAct-GFP molecules in scramble control and PODXL-KD SW1990 cells. Data represent the mean±S.E.M. from 3 independent experiments. (B) Average microtubule growth rate of scramble control and PODXL-KD SW1990 cells, as determined from the life history plots and kymographs of EB1 comets in cells transiently transfected with EB1-GFP. Data represent the mean±S.E.M from 3 independent experiments. (C) Representative TIRF microscopy images of immunostained phospho-paxillin in scramble control and PODXL-KD SW1990 cells. (D) Quantification of the average number of discrete FAs per cell area (left panel), FA area per total cell area (middle panel) and FA size (right panel) of scramble control and PODXL-KD SW1990 cells. Data represent the mean±S.E.M from 3 independent experiments. ** represents p<0.01 and *** represents p<0.001 between scramble control and PODXL-KD cells.
To investigate the potential role of PODXL in regulating cell adhesion, we visualized FAs by staining scramble control and PODXL-KD SW1990 cells for phosphorylated paxillin and imaging by TIRF microscopy (Fig. 2C). PODXL-KD cells relative to scramble controls displayed a higher FA density, quantified either as number of individual FAs over total cell area or as percentage of total cell area covered by FAs (Fig. 2D). Interestingly, the sizes of the individual FAs were not different between scramble control and PODXL-KD cells.
Most cytoskeletal and adhesion dynamics changes are regulated by the activity of small GTPase proteins, such as RhoA and Rac1 (33). However, neither active RhoA (Supplementary Fig. S2A) nor active Rac1 (Supplementary Fig. S2B) were differentially modulated following PODXL knockdown, indicating that the microtubule and adhesion changes induced by PODXL are mediated by a distinct mechanism that is independent of both Rac1 and Rho.
PODXL associates with dynamin-2 in pancreatic cancer cells.
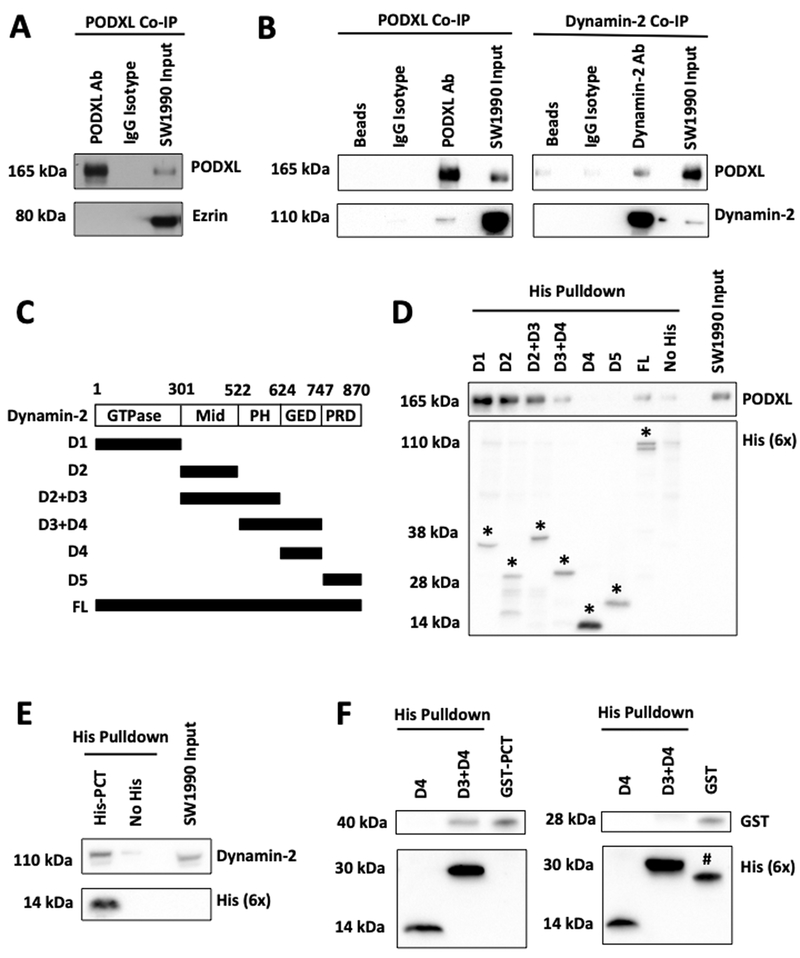
Ezrin is one of the most commonly reported cytoplasmic binding partners of PODXL, where it serves as a scaffold that links the plasma membrane to actin cytoskeleton (8,17). PODXL-ezrin interaction has also been reported to promote migration by activating Rac1 and RhoA (8,15,20). Given that PODXL knockdown fails to alter both actin dynamics and the activities of RhoA and Rac1 (Fig. 2A, Supplementary Fig. S2A,B), we questioned the validity of PODXL-ezrin interaction in pancreatic cancer cells. Interestingly, we failed to detect ezrin in immunoprecipitated PODXL specimens from SW1990 cell lysates (Fig. 3A), thereby indicating that PODXL does not associate with ezrin in pancreatic cancer cells.
Fig. 3. PODXL interacts with dynamin2 in pancreatic cancer cells.
(A) Ezrin was not detected in the immunoprecipitate fraction generated from SW1990 pancreatic cancer cells using a PODXL specific antibody. An IgG isotype antibody was used as a control. The right lane shows the presence of both PODXL and ezrin in SW1990 cell lysate input. (B) Co-IP experiments from SW1990 cell lysates were performed using either a PODXL (left panels) or a dynamin-2 (right panels) specific antibody along with matched isotype controls. The resultant immunoprecipitates were subsequently resolved with SDS-PAGE and blotted with antibodies against PODXL and dynamin-2. Dynamin-2 was detected in the immunoprecipitates of PODXL and vice versa. The right lanes in both panels reveal the presence of both PODXL and dynamin-2 in SW1990 cell lysate inputs. The left lanes in both panels show the beads-only (no antibody) controls. (C) His-tagged constructs encompassing different domains of dynamin-2 were used for the in vitro His-tag pulldown assay. Mid, middle domain; PH, pleckstrin homology domain; GED, GTPase effector domain; PRD, proline-rich domain. (C) In-vitro His-tag pulldown assays demonstrating that the GTPase, middle and pleckstrin homology domains of dynamin-2 are responsible for binding PODXL from SW1990 cell lysates. * denotes the respective His constructs. (E) In-vitro His-tag pulldown assay showing that His-tagged PODXL cytoplasmic tail (PCT) is sufficient to bind to endogenous dynamin-2 from SW1990 cell lysates. (F) In-vitro His-tag pulldown assay demonstrating that purified GST-tagged PCT binds to recombinant His-tagged dynamin-2 D3+D4 domains but not to D4 alone (left panel). Purified GST alone does not bind to either His-tagged constructs. # denotes the protein band of GST (28kDa) from previous immunoblotting with anti-GST antibody.
To identify the cytoplasmic binding partner of PODXL in pancreatic cancer cells, we immunoprecipitated PODXL using a PODXL-specific antibody, EPR9518, along with an appropriate IgG isotype and beads-only (no antibody) controls. Specimens were subjected to mass spectrometry followed by bioinformatics analysis. Only proteins enriched by at least two-fold in PODXL immunoprecipitated samples relative to those of isotype or beads-only control specimens were selected for further analysis (Supplementary Table 1).
Among the listed proteins, dynamin-2 emerged as a promising candidate for the following reasons. First, dynamin-2 is a large GTPase protein that associates with microtubules and promotes their dynamic instability (34). Second, dynamin-2 triggers FA disassembly (32). Third, dynamin-2 is overexpressed in pancreatic ductal carcinoma cells and potentiates migration and invasion (35). As such, we hypothesized that dynamin-2 could serve as a novel cytoplasmic binding partner of PODXL and function as its effector to induce cytoskeletal and migratory changes in pancreatic cancer cells.
By performing reciprocal co-IP experiments using specific antibodies against PODXL and dynamin-2, we demonstrated their physical association in SW1990 pancreatic cancer specimens. Specifically, in PODXL co-IP samples, dynamin-2 was also pulled down and detected only in the PODXL antibody lane, but not in the beads-only or isotype control lanes (Fig. 3B). Similarly, in dynamin-2 immunoprecipitated specimens, PODXL was also found to be co-IPed (Fig. 3B). These findings were validated and extended using Pa03c pancreatic cancer cells (Supplementary Fig. S3A) as well as MDA-MB-231 breast cancer cells (Supplementary Fig. S3B).
Immunofluorescence followed by confocal microscopy revealed the colocalization of PODXL and dynamin-2 around the cell membrane and the thin cytoplasmic area surrounding the nucleus in SW1990 cells (Supplementary Fig. S3C). While PODXL localizes apically in primary cilium (36), embryonic and stem cells (37), normal endothelial and epithelial cells (38,39), and is crucial for the formation of apical lumen in blood vessels (40,41), its spatial localization is not as extensively characterized in cancer cells. Although ectopically expressed PODXL has been reported to localize to the apical surface of OVCAR-3 ovarian and MCF7 breast cancer cells (6,42), confocal imaging disclosed the presence of endogenous PODXL not only at the apical but also at the basal surfaces of SW1990 pancreatic cancer cells (Supplementary Fig. S3C). This spatial localization pattern of PODXL at the basal surfaces enables it to interact and ultimately affect focal adhesion dynamics in pancreatic cancer cells.
To identify the essential domains of dynamin-2 responsible for binding to PODXL, probes consisting of 6x His tags fused to either full-length or truncated constructs encompassing different domains of dynamin-2 were generated and used in in vitro His-tag pulldown assays (Fig. 3C). His-tagged full-length dynamin-2 pulled down PODXL following incubation with SW1990 cell lysate (Fig. 3D). These assays further revealed that the GTPase, middle and pleckstrin homology domains of dynamin-2 are responsible for binding PODXL, as His-constructs containing either of these domains individually or in combination with others were able to pull down PODXL (Fig. 3D). On the other hand, the GTPase effector domain and the proline-rich domain of dynamin-2 were found to be dispensable for this binding interaction (Fig. 3D). Moreover, recombinant His-tagged PCT (aa484 to aa558) was sufficient to bind to endogenous dynamin-2 from SW1990 cell lysate (Fig. 3E). Importantly, we herein demonstrate the direct and specific binding interaction between PODXL and dynamin-2 via in vitro His-tag pulldown assays using both purified recombinant dynamin-2 domains and PCT. Specifically, we show that purified GST-tagged PCT binds to recombinant His-tagged dynamin-2 D3+D4 domains, but not to D4 alone (Fig. 3F). To further demonstrate the specificity of this binding interaction, we show that purified GST alone does not bind to either His-tagged constructs (Fig. 3F). Taken together, mass spectrometry followed by bioinformatics analysis, co-IP, colocalization staining and in vitro His-tag pulldown assays disclose that PODXL and dynamin-2 interact directly to form a complex in pancreatic cancer cells.
Pro-migratory phenotypes of PODXL-expressing cells are coupled to dynamin-2 function.
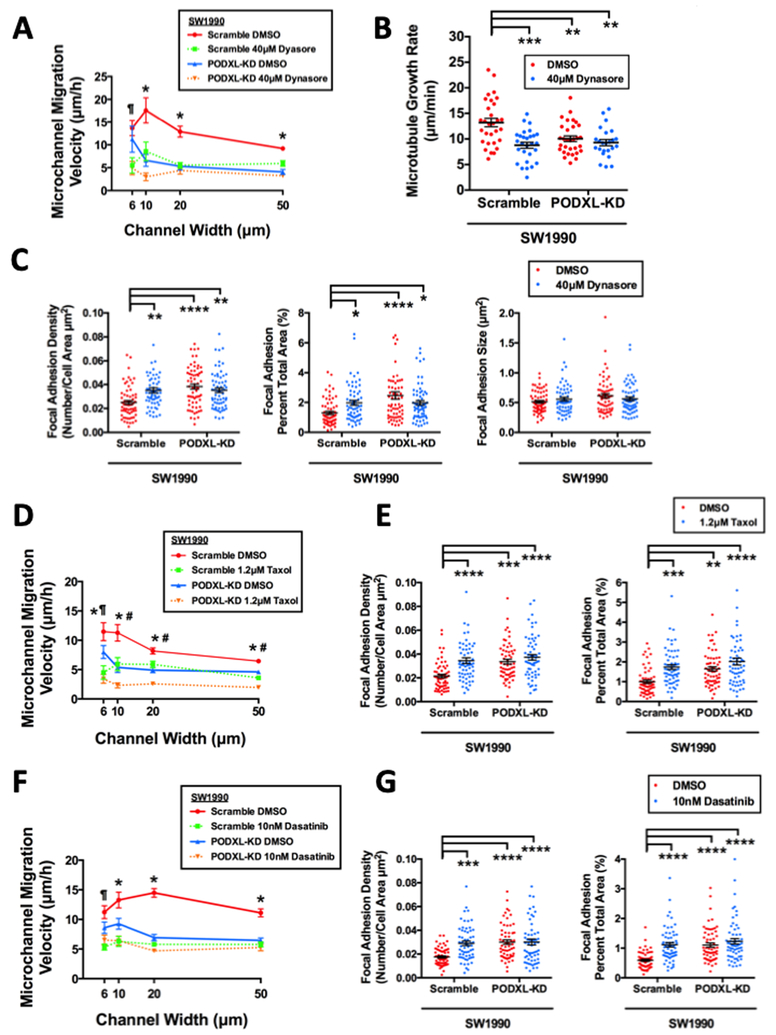
To assess the functional significance of PODXL-dynamin-2 interaction, a dynamin-2 inhibitor, dynasore, or siRNA (siDNM2) were used in both scramble control and PODXL-KD SW1990 cells. Dynasore (40 μM) significantly reduced the migration velocities of scramble control SW1990 cells to the identical levels of the DMSO-treated PODXL-KD cells in both unconfined and confined microchannels (Fig. 4A). Interestingly, dynasore had no additional inhibitory effect on PODXL-KD SW1990 cells. Similar phenotypic mimicry was also observed in the microtubule growth rate using EB1-GFP (Fig. 4B) and FA density (Fig. 4C).
Fig. 4. Inhibiting dynamin-2, microtubules or Src kinase abrogates the migratory potentiation and cytoskeletal remodeling effects of PODXL.
(A) Migration velocity of scramble control and PODXL-KD SW1990 cells inside microchannels of prescribed width after treatment with dynasore (40 μM). Data represent the mean±S.E.M. from 3 independent experiments. * represents p<0.05 between scramble DMSO control and all three other conditions, ¶ represents p<0.05 between scramble DMSO control versus scramble plus dynasore or PODXL-KD plus dynasore. (B) Microtubule growth rate of scramble control and PODXL-KD SW1990 cells following dynasore (40 μM) treatment as measured by an EB1-GFP assay. Data represent the mean±S.E.M from 3 independent experiments. ** represents p<0.01, *** represents p<0.001. (C) FA density quantified as the number of discrete FAs per cell area (left panel) or FA area per total cell area (middle panel) and FA size (right panel) of scramble control and PODXL-KD SW1990 cells following dynasore (40μM) treatment, as assessed by TIRF imaging of phospho-paxillin immunostaining. Data represent the mean±S.E.M, * represents p< 0.05, ** represents p<0.01, *** represents p<0.001, **** represents p<0.0001. (D) Migration velocity of scramble control and PODXL-KD SW1990 cells inside microchannels of prescribed width after taxol (1.2 μM) treatment. Data represent the mean±S.E.M. from 3 independent experiments. * represents p<0.05 between scramble DMSO control and all three other conditions. # represents p<0.05 between PODXL-KD plus taxol versus scramble plus taxol or PODXL-KD plus DMSO control. ¶ represents p<0.05 between PODXL-KD plus taxol versus PODXL-KD plus DMSO control. (E) FA density quantified as described in (C) using scramble control and PODXL-KD SW1990 cells after taxol (1.2μM) treatment, as assessed by TIRF imaging of phospho-paxillin immunostaining. Data represent the mean±S.E.M. from 3 independent experiments. ** represents p<0.01, *** represents p<0.001, **** represents p<0.0001. (F) Migration velocity of scramble control and PODXL-KD SW1990 cells inside microchannels of prescribed width after dasatinib (10 nM) treatment. Data represent the mean±S.E.M. from 3 independent experiments. * represents p<0.05 between scramble DMSO control and all three other conditions, ¶ represent p<0.05 between scramble DMSO control versus scramble control plus dasatinib or PODXL-KD plus dasatinib. (G) FA density quantified as described in (C) using scramble control and PODXL-KD SW1990 cells after dasatinib (10 nM) treatment, as assessed by TIRF imaging of phospho-paxillin immunostaining. Data represent the mean±S.E.M from 3 independent experiments. *** represents p<0.001, **** represents p<0.0001.
To eliminate any potential off-target effects of dynasore, siDNM2 was used to knock down dynamin-2 expression in both scramble control and PODXL-KD SW1990 cells (Supplementary Fig. S4A). Of note, PODXL knockdown did not affect dynamin-2 expression (Supplementary Fig. S4A). Similar to the data acquired using dynasore, dynamin-2 depletion decreased the migration of the scramble control SW1990 cells to the levels of PODXL-KD, without altering migration in PODXL-KD cells (Supplementary Fig. S4B). Cumulatively, these data disclose the functional role of dynamin-2 in mediating the cytoskeletal remodeling and pro-migratory effects of PODXL on pancreatic cancer cells.
According to mass spectrometry results, NHERF2 was also enriched in the immunoprecipitate fraction of PODXL (Supplementary Table 1). Since NHERF2 has been reported to act as a scaffolding protein that bridges the interaction between PODXL and ezrin in glomerular foot processes (16), we sought to delineate the potential role of NHERF2 in PODXL-dependent migration. Although NHERF2 interacts with PODXL in SW1990 pancreatic cancer cells as evidenced by co-IP assays, dynamin-2 was not detected in the NHERF2 immunoprecipitate fraction (Supplementary Fig. S4C). Also, NHERF2 was not detected in the dynamin-2 immunoprecipitate fraction (Supplementary Fig. S4D). We next generated NHERF2-KD SW1990 cells using shRNA lentivirus (Supplementary Fig. S4E), and compared their migratory potential to that of scramble controls. In distinct contrast to PODXL-KD and dynamin-2-KD cells, which displayed a markedly reduced migration velocity, NHERF2-KD cells moved much faster than scramble control cells (Supplementary Fig. S4F). Of note, NHERF2-KD cells exhibit a clear mesenchymal/protrusive phenotype in contrast to the epithelial/rounded morphology of scramble control or PODXL-KD cells. In view of these findings and given that PODXL and/or dynamin-2 knockdown suppress migration to an equivalent extent without any additive effect, we conclude that dynamin-2 regulates PODXL-mediated migration of pancreatic cancer cells via its direct binding interaction with PODXL (Fig. 3F). The precise role of NHERF2 in pancreatic cancer cell migration, which may extend beyond PODXL, deserves further investigation that goes beyond the scope of this current study.
Microtubule and Src kinase are downstream effectors of PODXL-dynamin-2 complex.
Because PODXL knockdown (Fig. 2B) and dynamin-2 inhibition (Fig. 4B) slow down microtubule growth with similar efficiency, we reasoned that microtubules represent a downstream target of the PODXL-dynamin-2 signaling cascade. In light of prior work showing that dynamin-2 regulates microtubule dynamic instability (34), we examined the effects of taxol, which stabilizes microtubules by preventing their depolymerization. Treatment of scramble control SW1990 cells with taxol (1.2 μM) had the same effect as dynamin-2 inhibition/knockdown or PODXL knockdown. Specifically, taxol suppressed the migration velocities of scramble control cells to the levels of untreated PODXL-KD cells (Fig. 4D). Of note, PODXL-KD SW1990 cells treated with taxol displayed a further, albeit moderate, reduction of migratory potential in microchannels with a width ≥10 μm, thereby indicating a potential mild synergistic effect.
Microtubule polymerization towards FAs has been reported to induce FA disassembly, leading to more dynamic FAs (43). Along these lines, stabilizing microtubules via cell treatment with taxol increased both FA density and the percentage of total cell area covered by FAs in scramble control, but not PODXL-KD, SW1990 cells (Fig. 4E). Taken together, our data are in concert with the notion that PODXL, through its binding to dynamin-2, regulates microtubule dynamics, which in turn modulates FA assembly and disassembly.
Evidence suggests that dynamin-2 forms a trimeric complex with FAK and Src kinase to induce FA disassembly by activating integrin endocytosis (32). Interestingly, treatment of scramble control SW1990 cells with the Src kinase inhibitor dasatinib (10 nM) recapitulated the phenotypic responses of PODXL knockdown. Specifically, dasatinib concurrently reduced the migration velocities (Fig. 4F) and increased the FA density and the percentage of total area covered by FAs (Fig. 4G) in scramble control cells. In marked contrast, Src kinase inhibition had a negligible effect on PODXL-KD SW1990 cells (Figs. 4F–4G). Interestingly, dasatinib failed to modulate microtubule dynamics (Supplementary Fig. S4G), suggesting that Src kinase regulates migration and FA dynamics downstream of PODXL without involving microtubules. To validate the role of Src kinase in PODXL-dynamin-2-dependent migration of pancreatic cancer cells, we also assessed the effect of PP2, a more specific Src kinase inhibitor, on migration of both SW1990 scramble control and PODXL-KD cells. Similar to Dasatinib, PP2 inhibition reduced the velocity of the scramble control cells down to the same level as the PODXL-KD cells, without affecting the velocity of PODXL-KD cells (Supplementary Fig. S4H).
PODXL knockdown decreases liver metastasis in vivo.
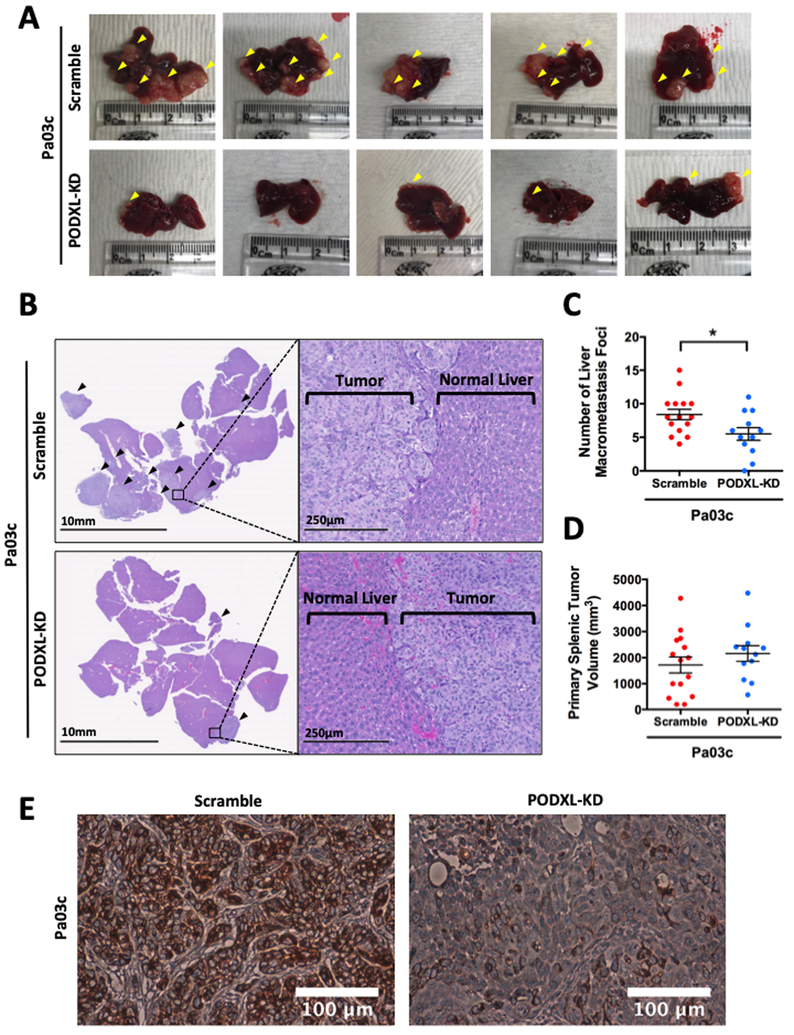
To test whether the enhanced migratory propensity of PODXL-expressing cells observed in vitro facilitates metastasis in vivo, we employed the well-established preclinical model of pancreatic cancer metastasis to the liver following a hemispleen injection technique (25). To this end, we injected 1×106 scramble control or PODXL-KD Pa03c pancreatic cancer cells into the spleen of mice (n=15 for scramble, n=12 for PODXL-KD). Knockdown of PODXL significantly decreased liver metastasis, as evidenced by the gross anatomy photographs (Fig. 5A) and further confirmed microscopically via histology (Fig. 5B). PODXL knockdown generated significantly smaller number of visible liver macrometastasis foci in mice (Fig. 5C). It is noteworthy that the size of the primary splenic tumor at the injection site was similar between scramble control and PODXL-KD Pa03c cells (Fig. 5D). Of note, immunohistochemistry staining for PODXL in primary splenic tumors derived from mice injected with PODXL-KD Pa03c cells showed a dramatic reduction in PODXL staining intensity relative to scramble control specimens (Fig. 5E), indicating that PODXL knockdown was maintained throughout the entire duration of the in vivo hemispleen experiment.
Fig. 5. PODXL knockdown decreases metastasis in vivo in a preclinical murine model of pancreatic cancer metastasis using a hemispleen injection technique.
(A) Gross anatomical pictures of representative livers harvested from mice injected with scramble control (top row) or PODXL-KD (bottom row) Pa03c cells via the hemispleen technique. Yellow arrowheads indicate visible liver macrometastases. (B) Representative histological sections of livers harvested from mice injected with scramble control (top row) or PODXL-KD (bottom row) Pa03c cells. Black arrowheads indicate areas of metastasis. (C) Quantification of the number of visible liver macrometastasis foci observed on the harvested livers and (D) the volume of the primary splenic tumor of mice injected with scramble control or PODXL-KD Pa03c cells. Data represent the mean±S.E.M from 15 and 12 mice for the scramble control and PODXL-KD groups, respectively. * represents p<0.05. (E) Representative immunohistochemistry sections of primary splenic tumors derived from scramble control (left) or PODXL-KD (right) Pa03c cells stained for PODXL reveal that PODXL knockdown is maintained in vivo.
We replicated the aforementioned findings with a second pancreatic cancer cell line, SW1990. Because of the larger size of SW1990 than Pa03c cells, injection of the 1×106 cells resulted in high post-operative mortality approximating 90%, possibly due to clotting and thrombosis. Another potential reason for the differential prothrombotic profile of the two pancreatic cancer cell types could stem from inherent differential surface expression of procoagulant molecules, such as tissue factor that can initiate the coagulation cascade (44). In breast cancer cells, clotting time was determined to be both tissue factor- and cell number- dependent (45). As such, a lower number of cells (2×105) was attempted for injection into the spleen of mice to mitigate this issue. While the lower cell number did improve postoperative survival, about 40% of the mice still died after surgery. Despite the lower sample size (n=7 for scramble, n=5 for PODXL-KD), the inhibitory effect of PODXL knockdown on liver metastasis was even more pronounced, as almost all mice injected with the scramble control SW1990 cells developed liver metastasis, whereas only one out of five mice injected with PODXL-KD SW1990 cells developed metastasis (Supplementary Fig. S5A–C). In concert with our findings using Pa03c cells, PODXL also failed to alter primary splenic tumors size (Supplementary Fig. S5D). Moreover, PODXL knockdown was preserved in vivo for SW1990 tumors as evidenced by immunohistochemistry (Supplementary Fig. S5E).
The growth of scramble control versus PODXL-KD cells was unaltered in vitro and in a subcutaneous injection model in vivo as quantified by the weight of tumors harvested post-mortem, using Pa03c (Supplementary Fig. S5F,G) and SW1990 (Supplementary Fig. S5H,I) cells. Collectively, these data reveal that the increased metastatic potential of PODXL-expressing cells is attributed to their elevated capacity to migrate and metastasize in the liver rather than to proliferate at higher levels at the primary tumor site per se.
Discussion
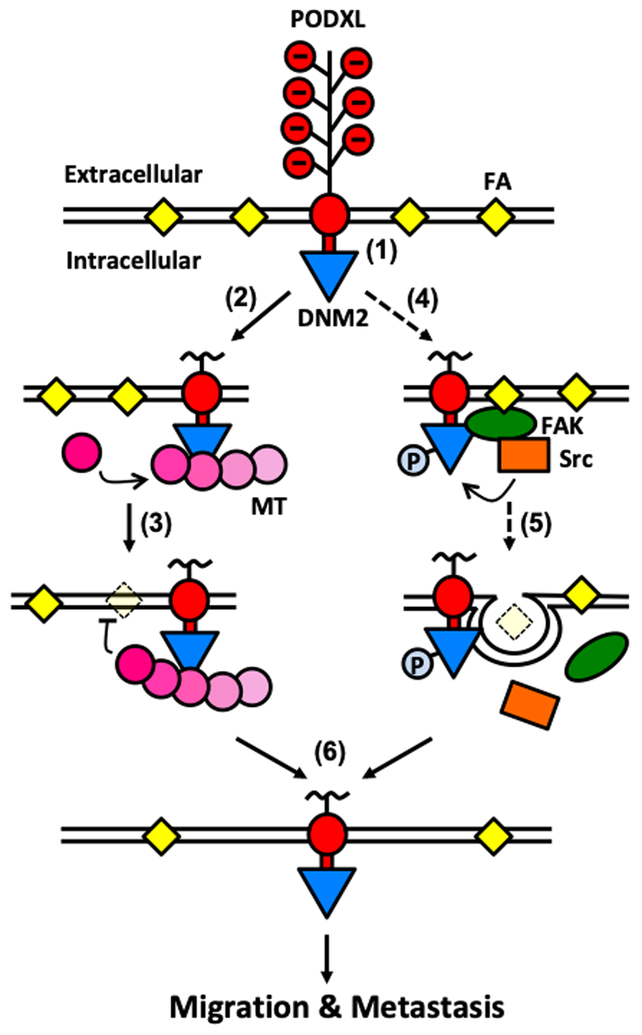
By employing mass spectrometry followed by bioinformatics analysis, co-immunoprecipitation and in vitro His-tag pulldown assays, we herein demonstrate a novel direct binding interaction between the cytoplasmic tail of PODXL and the large GTPase dynamin-2. Given that PODXL knockdown and dynamin-2 inhibition suppress with similar efficiency microtubule growth rate, and that dynamin-2 is a microtubule-associated large GTPase, we reasoned that microtubules are a downstream target of the PODXL-dynamin-2 signaling cascade. This is further substantiated by a previous study showing that dynamin-2 regulates dynamic instability of microtubules (34). Because PODXL knockdown or dynamin-2 inhibition or stabilization of microtubules via cell treatment with taxol increase FA density and the percentage of total cell area covered by FAs, we propose a model by which PODXL, through its binding to dynamin-2, regulates microtubule dynamics, which in turn modulates FA assembly and disassembly (Fig. 6). Specifically, we postulate that PODXL binds to and activates dynamin-2 at the cell periphery, where dynamin-2 could enhance the dynamic instability of microtubules, causing microtubules to polymerize more towards existing FA and promote FA disassembly. Alternatively, dynamin-2, by forming a trimer with FAK and Src kinase (32), could also regulate FA turnover via a Src kinase pathway (Fig. 6). Along these lines, our data reveal that Src kinase inhibition concurrently reduces cell migration and increases the FA density in scramble control, but not PODXL-KD, cells. We further postulate that both the microtubule- and Src kinase-dependent pathways, which are downstream of the PODXL-dynamin-2 signaling axis, regulate FA dynamics, thereby ultimately facilitating cell migration and metastasis (Fig. 6). This proposed model was rigorously tested by inhibiting dynamin-2, microtubules and Src kinase. Interfering with any of these downstream targets causes the scramble cells to phenotypically mimic the PODXL-KD cells, as evidenced by their slower cell migration, decreased microtubule growth rate and increased FA density, thus validating their functional significance in propagating the downstream effects of PODXL.
Fig. 6. Proposed mechanism by which PODXL facilitates pancreatic cancer cell migration and metastasis.
(1) PODXL is a transmembrane protein expressed on pancreatic cancer cells. The cytoplasmic tail of PODXL binds to dynamin-2, thereby recruiting dynamin-2 close to the plasma membrane. (2) The local enrichment of dynamin-2 causes increased microtubule growth rates and dynamics. (3) Polymerization of microtubules towards FA sites induces FA disassembly. (4) Alternatively, dynamin-2 proximal to the membrane could also interact with FAK and Src kinase to form a trimeric complex next to FAs. (5) Src kinase then phosphorylates and activates dynamin-2, leading to endocytic internalization of integrin and FA disassembly. (6) All these events lead to the cells having less FAs, ultimately resulting in increased migration in vitro and enhanced metastasis in vivo. Dashed lines denote data from prior work while solid lines represent findings from this study.
The cytoplasmic domain of PODXL, despite its short size, has been reported to associate with proteins, such as ezrin and NHERF1/2 to mediate downstream signaling involved in cytoskeletal remodeling and cell migration (8,14). PODXL is considered to form a stable complex with ezrin, which in turns connects to actin filaments, leading to redistribution of actin towards the apical membrane in kidney podocytes (14). NHERF1 and 2, which are two highly homologous scaffold proteins with two PDZ domains and an ERM binding domain, interact directly with PODXL and serve as a linker between PODXL and ezrin (16). Interestingly, the N-terminus of ezrin has also been reported to bind directly to the HQRIS sequence in the juxtamembrane region of PODXL independent of NHERF1/2 (15). Binding of ezrin to PODXL, either directly or indirectly, activates ezrin, which in turn activates RhoA through sequestration of RhoGDI by activated ezrin (15). With its second PDZ domain binding to PODXL, the first PDZ domain of NHERF1 can also bind to Rac1 guanine nucleotide exchange factor ARHGEF7 to activate Rac1, promoting lamellipodia formation and cell migration (8).
Although ezrin has emerged as a widely accepted binding partner of PODXL, there are reports showing that PODXL and ezrin may have non-related or even opposing effects in certain cancer types, thereby suggesting that the PODXL-ezrin association may not be as universal as one is led to believe. In urothelial bladder cancer, for instance, while high membranous PODXL expression correlates with poor prognosis (7), low membranous expression ezrin was instead an independent marker of progression and disease-specific survival in another study (46). In our study, we consistently and repeatedly failed to identify the presence of ezrin in immunoprecipitated PODXL specimens either via immunoblotting or mass spectrometry. Moreover, the inability of PODXL knockdown to alter actin dynamics and RhoA or Rac1 activity in pancreatic cancer cells further suggests that the pro-migratory role of PODXL is independent of the actin cytoskeleton and the classical Rac1 and/or RhoA signaling induced by ezrin following its binding to PODXL. Collectively, these previous reports showing that PODXL and ezrin have opposing effects on tumor cells, coupled with our data showing the absence of PODXL-ezrin association and the inability of PODXL knockdown to alter actin polymerization, strongly support the existence of a novel cytoplasmic binding partner, which we demonstrated to be dynamin-2.
While there have been several in vitro studies on PODXL and its involvement in cancer, there is no preclinical animal model study aimed to evaluate the potential oncogenic role of PODXL in pancreatic cancer. To the best of our knowledge, the well-established preclinical model of pancreatic cancer metastasis to the liver via an injection using the hemispleen technique represents the first ever-reported in vivo model that directly assesses the effect of PODXL on pancreatic cancer metastasis. Prior in vivo models associated with PODXL have been performed primarily with breast cancer cells, where impairment in the formation of lung metastases was demonstrated with tail vein injection, subcutaneous injection and orthotopic implantation into mammary fat pad for PODXL-KD cells (20,47). In our work, we demonstrate that PODXL knockdown exerts a pronounced inhibitory effect on liver metastasis using two distinct pancreatic cancer cell lines without affecting primary splenic tumor growth.
The KRAS oncogene undergoes mutational activation in 95% of PDAC patients, and plays a crucial role in initiating and driving pancreatic cancer pathogenesis (48). In addition to KRAS, signal transducer and activator of transcription 3 (STAT3) has been implicated in malignant transformation of pancreatic cancer (49). Interestingly, both KRAS and STAT3 interact with tubulin and microtubules (50,51). Moreover, oncogenic KRAS upregulates Src in pancreatic cancer cells (52). Because 69% of PDACs stain positively for PODXL (12), and given that MTs and Src regulate PODXL-dependent pancreatic cancer cell migration, the potential link among oncogenic KRAS, STAT3 and PODXL deserves to be investigated.
In conclusion, we discovered a novel direct binding interaction between PODXL and dynamin-2 that is critical for promoting the efficient migration in vitro and metastasis in vivo of pancreatic cancer cells. PODXL, through its binding to dynamin-2, regulates microtubule dynamics and Src activation, which in turn modulate FA assembly and disassembly, and ultimately regulate migration. The functional role of PODXL in promoting pancreatic cancer metastasis in vivo was demonstrated using a physiologically relevant hemispleen-based hepatic metastasis model.
Supplementary Material
Statement of Significance.
Findings reveal that a novel interaction between podocalyxin and dynamin-2 promotes migration and metastasis of pancreatic cancer cells by regulating microtubule and focal adhesion dynamics.
Acknowledgments
The authors acknowledge funding support from the US National Institutes of Health (Grant R01CA186286). The authors declare no potential conflicts of interest.
References
- 1.Kerjaschki D, Sharkey DJ, Farquhar MG. Identification and characterization of podocalyxin--the major sialoprotein of the renal glomerular epithelial cell. J Cell Biol 1984;98:1591–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kerosuo L, Juvonen E, Alitalo R, Gylling M, Kerjaschki D, Miettinen A. Podocalyxin in human haematopoietic cells. Br J Haematol 2004;124:809–18 [DOI] [PubMed] [Google Scholar]
- 3.Sassetti C, Tangemann K, Singer MS, Kershaw DB, Rosen SD. Identification of podocalyxin-like protein as a high endothelial venule ligand for L-selectin: parallels to CD34. J Exp Med 1998;187:1965–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kusumoto H, Shintani Y, Kanzaki R, Kawamura T, Funaki S, Minami M, et al. Podocalyxin influences malignant potential by controlling epithelial-mesenchymal transition in lung adenocarcinoma. Cancer Sci 2017;108:528–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Somasiri A, Nielsen JS, Makretsov N, McCoy ML, Prentice L, Gilks CB, et al. Overexpression of the anti-adhesin podocalyxin is an independent predictor of breast cancer progression. Cancer Res 2004;64:5068–73 [DOI] [PubMed] [Google Scholar]
- 6.Cipollone JA, Graves ML, Kobel M, Kalloger SE, Poon T, Gilks CB, et al. The anti-adhesive mucin podocalyxin may help initiate the transperitoneal metastasis of high grade serous ovarian carcinoma. Clin Exp Metastasis 2012;29:239–52 [DOI] [PubMed] [Google Scholar]
- 7.Boman K, Larsson AH, Segersten U, Kuteeva E, Johannesson H, Nodin B, et al. Membranous expression of podocalyxin-like protein is an independent factor of poor prognosis in urothelial bladder cancer. Br J Cancer 2013;108:2321–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsu YH, Lin WL, Hou YT, Pu YS, Shun CT, Chen CL, et al. Podocalyxin EBP50 ezrin molecular complex enhances the metastatic potential of renal cell carcinoma through recruiting Rac1 guanine nucleotide exchange factor ARHGEF7. Am J Pathol 2010;176:3050–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Larsson A, Johansson ME, Wangefjord S, Gaber A, Nodin B, Kucharzewska P, et al. Overexpression of podocalyxin-like protein is an independent factor of poor prognosis in colorectal cancer. Br J Cancer 2011;105:666–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heby M, Elebro J, Nodin B, Jirstrom K, Eberhard J. Prognostic and predictive significance of podocalyxin-like protein expression in pancreatic and periampullary adenocarcinoma. BMC Clin Pathol 2015;15:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thomas SN, Schnaar RL, Konstantopoulos K. Podocalyxin-like protein is an E-/L-selectin ligand on colon carcinoma cells: comparative biochemical properties of selectin ligands in host and tumor cells. Am J Physiol Cell Physiol 2009;296:C505–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dallas MR, Chen SH, Streppel MM, Sharma S, Maitra A, Konstantopoulos K. Sialofucosylated podocalyxin is a functional E- and L-selectin ligand expressed by metastatic pancreatic cancer cells. Am J Physiol Cell Physiol 2012;303:C616–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nielsen JS, McNagny KM. The role of podocalyxin in health and disease. J Am Soc Nephrol 2009;20:1669–76 [DOI] [PubMed] [Google Scholar]
- 14.Orlando RA, Takeda T, Zak B, Schmieder S, Benoit VM, McQuistan T, et al. The glomerular epithelial cell anti-adhesin podocalyxin associates with the actin cytoskeleton through interactions with ezrin. J Am Soc Nephrol 2001;12:1589–98 [DOI] [PubMed] [Google Scholar]
- 15.Schmieder S, Nagai M, Orlando RA, Takeda T, Farquhar MG. Podocalyxin activates RhoA and induces actin reorganization through NHERF1 and Ezrin in MDCK cells. J Am Soc Nephrol 2004;15:2289–98 [DOI] [PubMed] [Google Scholar]
- 16.Takeda T, McQuistan T, Orlando RA, Farquhar MG. Loss of glomerular foot processes is associated with uncoupling of podocalyxin from the actin cytoskeleton. J Clin Invest 2001;108:289–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sizemore S, Cicek M, Sizemore N, Ng KP, Casey G. Podocalyxin increases the aggressive phenotype of breast and prostate cancer cells in vitro through its interaction with ezrin. Cancer Res 2007;67:6183–91 [DOI] [PubMed] [Google Scholar]
- 18.Meng X, Ezzati P, Wilkins JA. Requirement of podocalyxin in TGF-beta induced epithelial mesenchymal transition. PLoS One 2011;6:e18715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fernandez D, Horrillo A, Alquezar C, Gonzalez-Manchon C, Parrilla R, Ayuso MS. Control of cell adhesion and migration by podocalyxin. Implication of Rac1 and Cdc42. Biochem Biophys Res Commun 2013;432:302–7 [DOI] [PubMed] [Google Scholar]
- 20.Lin CW, Sun MS, Liao MY, Chung CH, Chi YH, Chiou LT, et al. Podocalyxin-like 1 promotes invadopodia formation and metastasis through activation of Rac1/Cdc42/cortactin signaling in breast cancer cells. Carcinogenesis 2014;35:2425–35 [DOI] [PubMed] [Google Scholar]
- 21.Ney JT, Zhou H, Sipos B, Buttner R, Chen X, Kloppel G, et al. Podocalyxin-like protein 1 expression is useful to differentiate pancreatic ductal adenocarcinomas from adenocarcinomas of the biliary and gastrointestinal tracts. Hum Pathol 2007;38:359–64 [DOI] [PubMed] [Google Scholar]
- 22.Taniuchi K, Furihata M, Naganuma S, Dabanaka K, Hanazaki K, Saibara T. Podocalyxin-like protein, linked to poor prognosis of pancreatic cancers, promotes cell invasion by binding to gelsolin. Cancer Sci 2016;107:1430–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saukkonen K, Hagstrom J, Mustonen H, Juuti A, Nordling S, Fermer C, et al. Podocalyxin Is a Marker of Poor Prognosis in Pancreatic Ductal Adenocarcinoma. PLoS One 2015;10:e0129012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chijiiwa Y, Moriyama T, Ohuchida K, Nabae T, Ohtsuka T, Miyasaka Y, et al. Overexpression of microRNA-5100 decreases the aggressive phenotype of pancreatic cancer cells by targeting PODXL. Int J Oncol 2016;48:1688–700 [DOI] [PubMed] [Google Scholar]
- 25.Soares KC, Foley K, Olino K, Leubner A, Mayo SC, Jain A, et al. A preclinical murine model of hepatic metastases. J Vis Exp 2014:51677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 2008;321:1801–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen SH, Hung WC, Wang P, Paul C, Konstantopoulos K. Mesothelin binding to CA125/MUC16 promotes pancreatic cancer cell motility and invasion via MMP-7 activation. Sci Rep 2013;3:1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stroka KM, Wong BS, Shriver M, Phillip JM, Wirtz D, Kontrogianni-Konstantopoulos A, et al. Loss of giant obscurins alters breast epithelial cell mechanosensing of matrix stiffness. Oncotarget 2017;8:54004–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tong Z, Balzer EM, Dallas MR, Hung WC, Stebe KJ, Konstantopoulos K. Chemotaxis of cell populations through confined spaces at single-cell resolution. PLoS One 2012;7:e29211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Balzer EM, Tong Z, Paul CD, Hung WC, Stroka KM, Boggs AE, et al. Physical confinement alters tumor cell adhesion and migration phenotypes. FASEB J 2012;26:4045–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shriver M, Stroka KM, Vitolo MI, Martin S, Huso DL, Konstantopoulos K, et al. Loss of giant obscurins from breast epithelium promotes epithelial-to-mesenchymal transition, tumorigenicity and metastasis. Oncogene 2015;34:4248–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Y, Cao H, Chen J, McNiven MA. A direct interaction between the large GTPase dynamin-2 and FAK regulates focal adhesion dynamics in response to active Src. Mol Biol Cell 2011;22:1529–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Actin Etienne-Manneville S. and microtubules in cell motility: which one is in control? Traffic 2004;5:470–7 [DOI] [PubMed] [Google Scholar]
- 34.Tanabe K, Takei K. Dynamic instability of microtubules requires dynamin 2 and is impaired in a Charcot-Marie-Tooth mutant. J Cell Biol 2009;185:939–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eppinga RD, Krueger EW, Weller SG, Zhang L, Cao H, McNiven MA. Increased expression of the large GTPase dynamin 2 potentiates metastatic migration and invasion of pancreatic ductal carcinoma. Oncogene 2012;31:1228–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Francis SS, Sfakianos J, Lo B, Mellman I. A hierarchy of signals regulates entry of membrane proteins into the ciliary membrane domain in epithelial cells. J Cell Biol 2011;193:219–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Doyonnas R, Nielsen JS, Chelliah S, Drew E, Hara T, Miyajima A, et al. Podocalyxin is a CD34-related marker of murine hematopoietic stem cells and embryonic erythroid cells. Blood 2005;105:4170–8 [DOI] [PubMed] [Google Scholar]
- 38.Mrozowska PS, Fukuda M. Regulation of podocalyxin trafficking by Rab small GTPases in 2D and 3D epithelial cell cultures. J Cell Biol 2016;213:355–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Richards M, Hetheridge C, Mellor H. The Formin FMNL3 Controls Early Apical Specification in Endothelial Cells by Regulating the Polarized Trafficking of Podocalyxin. Curr Biol 2015;25:2325–31 [DOI] [PubMed] [Google Scholar]
- 40.Strilic B, Eglinger J, Krieg M, Zeeb M, Axnick J, Babal P, et al. Electrostatic cell-surface repulsion initiates lumen formation in developing blood vessels. Curr Biol 2010;20:2003–9 [DOI] [PubMed] [Google Scholar]
- 41.Strilic B, Kucera T, Eglinger J, Hughes MR, McNagny KM, Tsukita S, et al. The molecular basis of vascular lumen formation in the developing mouse aorta. Dev Cell 2009;17:505–15 [DOI] [PubMed] [Google Scholar]
- 42.Nielsen JS, Graves ML, Chelliah S, Vogl AW, Roskelley CD, McNagny KM. The CD34-related molecule podocalyxin is a potent inducer of microvillus formation. PLoS One 2007;2:e237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stehbens S, Wittmann T. Targeting and transport: how microtubules control focal adhesion dynamics. J Cell Biol 2012;198:481–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mitrugno A, Tormoen GW, Kuhn P, McCarty OJ. The prothrombotic activity of cancer cells in the circulation. Blood Rev 2016;30:11–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Berny-Lang MA, Aslan JE, Tormoen GW, Patel IA, Bock PE, Gruber A, et al. Promotion of experimental thrombus formation by the procoagulant activity of breast cancer cells. Phys Biol 2011;8:015014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Palou J, Algaba F, Vera I, Rodriguez O, Villavicencio H, Sanchez-Carbayo M. Protein expression patterns of ezrin are predictors of progression in T1G3 bladder tumours treated with nonmaintenance bacillus Calmette-Guerin. Eur Urol 2009;56:829–36 [DOI] [PubMed] [Google Scholar]
- 47.Snyder KA, Hughes MR, Hedberg B, Brandon J, Hernaez DC, Bergqvist P, et al. Podocalyxin enhances breast tumor growth and metastasis and is a target for monoclonal antibody therapy. Breast Cancer Res 2015;17:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zeitouni D, Pylayeva-Gupta Y, Der CJ, Bryant KL. KRAS Mutant Pancreatic Cancer: No Lone Path to an Effective Treatment. Cancers (Basel) 2016;8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scholz A, Heinze S, Detjen KM, Peters M, Welzel M, Hauff P, et al. Activated signal transducer and activator of transcription 3 (STAT3) supports the malignant phenotype of human pancreatic cancer. Gastroenterology 2003;125:891–905 [DOI] [PubMed] [Google Scholar]
- 50.Thissen JA, Gross JM, Subramanian K, Meyer T, Casey PJ. Prenylation-dependent association of Ki-Ras with microtubules. Evidence for a role in subcellular trafficking. J Biol Chem 1997;272:30362–70 [DOI] [PubMed] [Google Scholar]
- 51.Yan B, Xie S, Liu Z, Luo Y, Zhou J, Li D, et al. STAT3 association with microtubules and its activation are independent of HDAC6 activity. DNA Cell Biol 2015;34:290–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kelber JA, Reno T, Kaushal S, Metildi C, Wright T, Stoletov K, et al. KRas induces a Src/PEAK1/ErbB2 kinase amplification loop that drives metastatic growth and therapy resistance in pancreatic cancer. Cancer Res 2012;72:2554–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.