Abstract
The metabolic reprogramming associated with characteristic increases in glucose and glutamine metabolism common in advanced cancer is often ascribed to answering a higher demand for metabolic intermediates required for rapid tumor cell growth. Instead, recent discoveries have pointed to an alternative role for glucose and glutamine metabolites as cofactors for chromatin modifiers and other protein post-translational modification enzymes in cancer cells. Beyond epigenetic mechanisms regulating gene expression, many chromatin modifiers also modulate DNA repair, raising the question whether cancer metabolic reprogramming may mediate resistance to genotoxic therapy and genomic instability. Our prior work had implicated N-acetyl-glucosamine (GlcNAc) formation by the hexosamine biosynthetic pathway (HBP) and resulting protein O-GlcNAcylation as a common means by which increased glucose and glutamine metabolism can drive double strand break (DSB) repair and resistance to therapy-induced senescence in cancer cells. We have examined the effects of modulating O-GlcNAcylation on the DNA damage response in MCF7 human mammary carcinoma in vitro and in xenograft tumors. Proteomic profiling revealed deregulated DNA-damage response pathways in cells with altered O-GlcNAcylation. Promoting protein O-GlcNAc modification by targeting O-GlcNAcase (OGA) or simply treating animals with GlcNAc, protected tumor xenografts against radiation. In turn, suppressing protein O-GlcNAcylation by blocking O-GlcNAc transferase (OGT) activity led to delayed DSB repair, reduced cell proliferation, and increased cell senescence in vivo. Taken together, these findings confirm critical connections between cancer metabolic reprogramming, DNA damage response, and senescence and provide a rationale to evaluate agents targeting O-GlcNAcylation in patients as a means to restore tumor sensitivity to radiotherapy.
INTRODUCTION
Along with immortality and genomic instability, metabolic reprogramming is now recognized as a hallmark of cancer (1). Conventional wisdom holds that the increased glucose and glutamine metabolism in cancer cells reflects the greater demand for precursors to feed macromolecular biosynthesis in proliferating cells. However, recent studies have pointed to alternative roles for metabolic intermediates such as acetyl-CoA, S-adenosyl methionine, and α-ketoglutarate as cofactors of chromatin modifying enzymes, deregulating gene expression and driving epigenetic reprogramming (2–8). Often ignored is that along with their well-studied roles in epigenetic regulation of transcription, chromatin-modifying enzymes such as histone acetyltransferases (HATs) and histone methyltransferases (HMTs) also regulate the DNA damage response (DDR). Thereby, cancer metabolic reprogramming may be able to influence DNA repair, genomic instability, and recovery after genotoxic stress.
In our prior work, we probed the influence of cancer metabolism on DNA repair by using cell-permeable metabolic intermediates, small molecule inhibitors, and siRNA to perturb metabolic pathways in MCF7 human mammary adenocarcinoma cells responding to DNA damage after ionizing radiation (IR) (9). Exposure to IR leads to DNA double strand breaks (DSBs) and triggers the DDR pathway. Ionizing radiation-induced foci (IRIF), the multi-kilobase domains of modified chromatin that form at DSBs and then resolve upon repair, can be readily detected by accumulation of γH2AX, the phospho-S139 form of histone H2AX, or other characteristic proteins and modifications, providing mechanistic reporters for DNA damage and repair (10–16). Using delayed IRIF resolution in irradiated cells as a screen for defects in DSB repair, we uncovered distinct roles for glycolysis and glutaminolysis as well as their joint influence via the hexosamine biosynthesis pathway (HBP) in regulating DSB repair and therapy-induced senescence (9).
Beyond roles in secretion, uridine diphosphate-N-acetyl glucosamine (UDP-GlcNAc) is synthesized by the HBP to serve as a cofactor for O-GlcNAc transferase (OGT), which transfers GlcNAc moieties to form O-glycosidic linkages to serine and threonine residues on intracellular proteins (Fig. 1A). Unlike the diversity of Ser/Thr protein kinases, there is only a single OGT gene and just two isoforms, responsible for nucleocytoplasmic and mitochondrial O-GlcNAcylation respectively. Protein O-GlcNAc modifications are highly dynamic and can be rapidly removed by the hexosaminidase O-GlcNAcase (OGA). Together, OGT and OGA regulate O-GlcNAcylation of thousands of cytosolic, mitochondrial, and nuclear proteins, affecting protein folding, stability, localization, and function (17–21). In many OGT substrates, residues modified by O-GlcNAcylation are also subject to phosphorylation by Ser/Thr kinases, providing a link between metabolism and diverse types of cell signaling (22–24). OGT has been implicated in coupling epigenetic regulation to cell metabolism via altered histone modifications (25–27), regulation of Polycomb group proteins and other chromatin modifiers (21,28,29) and by its binding to and modification of TET DNA demethylases (19,26,30,31).
Figure 1.
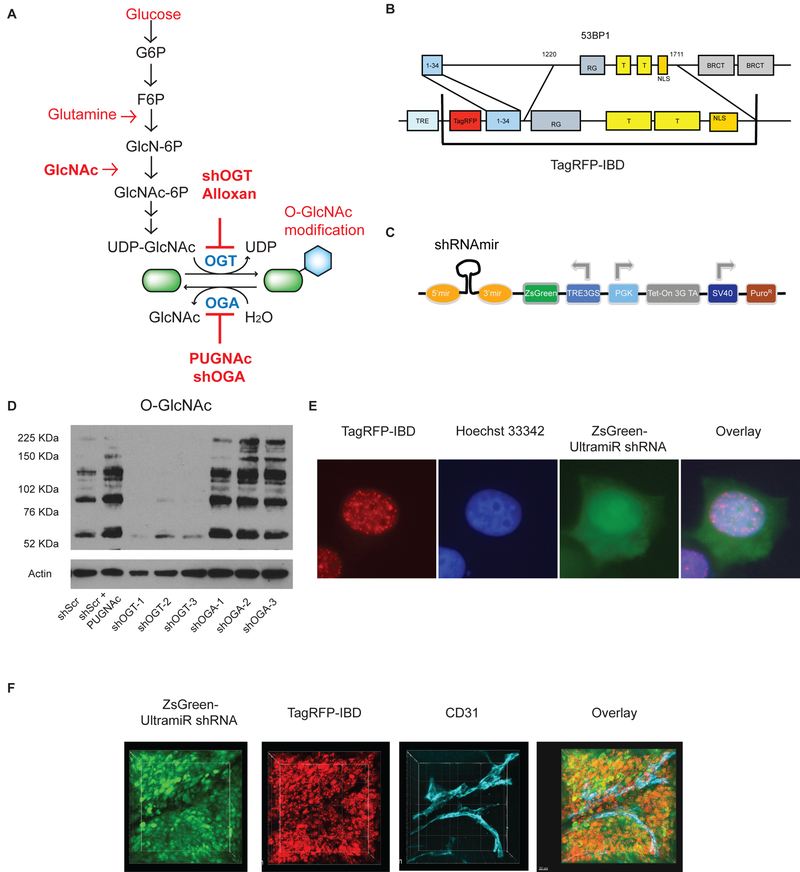
Tools to target protein O-GlcNAcylation and track impacts on DNA damage response in vitro and in vivo. A, The rate limiting step of the hexosamine biosynthetic pathway (HBP) is the transfer of an amine from glutamine to glucose as fructose 6-phosphate (F6P) to form glucosamine 6-phosphate (GlcN-6P) and glutamate. Once acetylated, N-acetylglucosamine 6-phosphate (GlcNAc-6P) is linked to uridine diphosphate to form UDP-GlcNAc. Exogenous GlcNAc is readily phosphorylated and can drive the pathway without feeding glycolysis or glutaminolysis. Although UDP-GlcNAc is often considered as a key metabolite for secretory glycosylation, the enzyme O-GlcNAc transferase (OGT) modifies intracellular protein serines and threonines with the O-GlcNAc post-translational modification. Typically, O-GlcNAcase (OGA) rapidly removes the GlcNAc, restoring the hydroxyl form. Alloxan and PUGNAc are small molecule OGT and OGA inhibitors, respectively. B, The TagRFP-IBD reporter expresses TagRFP fused to residues 1–34 and 1220–1711 of human 53BP1. C, The ZsGreen-shRNA-miR reporter construct for silencing OGT or OGA. D, Western blot analysis of the O-GlcNAc protein modification in MCF7TagRFP-IBD cell lines with shRNAs targeting OGT (shOGT) or OGA (shOGA). Three gene-specific shRNA-miRs were tested for each target. Random scrambled control shRNA-miR (shScr) and the OGA inhibitor PUGNAc were used as negative and positive controls, respectively, for O-GlcNAc protein modification. E, Representative cell image with TagRFP-IBD and ZsGreen-shRNA-miR (Scr) fluorescence examined 2 h after irradiation with 6 Gy (TagRFP, red; ZsGreen, green; Hoechst 33342, blue). F, Intratumoral imaging of MCF7 cells with dual fluorescent reporters for IRIF (TagRFP, red) and shRNA-miR expression (ZsGreen, green) after irradiation with 6 Gy. Anti-CD31 antibody was used for vascular staining (endothelium, cyan).
Importantly, DNA damage response factors are prominent among proteins that accumulate O-GlcNAc modifications under cell stress (32) and loss of O-GlcNAcylation after inhibition of OGT results in increased phosphorylation of DNA repair proteins, including activation of the DNA damage signaling kinase ATM (33). Strikingly, OGT also modifies H2AX on the ATM phosphorylation site S139, leading to decreased γH2AX formation at sites of DNA damage (34). In turn, our prior studies revealed functional links between cellular GlcNAc levels, O-GlcNAcylation, DNA damage response and DSB repair after irradiation (9). Feeding cells GlcNAc or blocking OGA accelerated DSB repair kinetics while targeting OGT resulting in unrepaired breaks, persistent IRIF and increased cellular senescence. Rather than conventional DNA repair factors, we identified the Polycomb Related Complex 2 (PRC2) histone methyltransferase catalytic subunit Ezh2 as a potential link between cancer metabolism and DNA damage response. This built on related studies showing that Ezh2 is O-GlcNAcylated, stabilizing the protein and increasing histone methylation (28), and that its activity promotes DSB repair (35) and blocks cellular senescence (36). Here, we provide direct evidence that protein O-GlcNAcylation is a key regulator of multiple DNA damage response pathways and modulates DSB repair and accelerated senescence not only in vitro but also in tumors, suggesting that targeting cancer metabolism may be a selective means to sensitize cancer to radiation and other genotoxic therapies. Modeling therapy by blocking OGT with a small molecule inhibitor during radiation treatment induced dramatic phenotypes in tumors, suggesting feasibility for this approach.
MATERIALS AND METHODS
Cell line development
MCF7Tet-On Advanced and Lenti-X 293T cell lines (both from Clontech, Mountain View, CA, USA) were grown in high-glucose DMEM with 1% penicillin and streptomycin (Life Technologies, Carlsbad, CA, USA) and 10% Tet system-approved fetal bovine serum (Clontech-Takara Bio, Mountain View, CA, USA). TagRFP (Evrogen-Axxora, Farmingdale, NY, USA) (37) fused to the human 53BP1 IRIF binding domain (IBD) (gift from Halazonetis T.D.) was cloned into the pLVX-Tight-Puro lentiviral vector (Clontech-Takara). Sets of 3 gene-specific shERWOOD-UltramiR lentiviral inducible short hairpin RNA (shRNA) targeting expression of OGT or OGA (MGEA5) with untargeted scrambled control in pZIP-TRE3GS vector were obtained from transOMIC technologies (Huntsville, AL, USA). Lentiviruses were produced in the Lenti-X 293T cell line using a 3rd generation packaging system (Clontech-Takara). Plasmid transfections were performed using FuGENE HD reagent (Promega, Madison, WI, USA). The MCF7Tet-On Advanced cell line was transduced with pLVX-Tight-Puro TagRFP-IBD lentiviruses following transduction with individual pZIP-TRE3GS shRNA-miRs lentiviruses targeting OGT (shOGT), MGEA5 (shOGA) or scrambled control (shScr). Transduced cell lines were selected and cultured in media supplemented with 0.7 μg/mL puromycin. In total, 7 cell lines were developed for this study. Following 48 h of induction with 1 μg/mL doxycycline (Sigma-Aldrich, St. Louis, MO, USA), most cells expressed both TagRFP-IBD as a reporter for DSB repair and ZsGreen fluorescent protein as a reporter for shRNA-miR expression. The cells were tested for mycoplasma and authenticated by short tandem repeat profile (IDEXX BioResearch, Columbia, MO, USA) prior to performing experiments. All experiments were performed within 3 to 10 passages after cell line development.
The shRNA-miR sequences, with targeting sequence in lowercase, used in this study were:
Scrambled control
TGCTGTTGACAGTGAGCGaaggcagaagtatgcaaagcatTAGTGAAGCCACAGATGTAatgctttgcatacttctgcctgTGCCTACTGCCTCGGA
OGT(1)
TGCTGTTGACAGTGAGCGactgaagcagaagattgttataTAGTGAAGCCACAGATGTAtataacaatcttctgcttcagcTGCCTACTGCCTCGGA
OGT(2)
TGCTGTTGACAGTGAGCGcaaccgaggacagattcaaataTAGTGAAGCCACAGATGTAtatttgaatctgtcctcggttaTGCCTACTGCCTCGGA
OGT(3)
TGCTGTTGACAGTGAGCGcccgtatcattttttcacctgaTAGTGAAGCCACAGATGTAtcaggtgaaaaaatgatacggtTGCCTACTGCCTCGGA
MGEA5(1)
TGCTGTTGACAGTGAGCGcaagatggacattcacaaaaaaTAGTGAAGCCACAGATGTAttttttgtgaatgtccatctttTGCCTACTGCCTCGGA
MGEA5(2)
TGCTGTTGACAGTGAGCGcagagagcatagctgaatcaaaTAGTGAAGCCACAGATGTAtttgattcagctatgctctcttTGCCTACTGCCTCGGA
MGEA5(3)
TGCTGTTGACAGTGAGCGctaggatgttttgaaattgcaaTAGTGAAGCCACAGATGTAttgcaatttcaaaacatcctaaTGCCTACTGCCTCGGA
Cell line validation and Western blotting
To evaluate the targeting of OGT or OGA in MCF7TagRFP-IBD cells via shRNA, we examined O-GlcNAc protein modifications in the cell lines. MCF7TagRFP-IBD1 cells with inducible shRNA-miR targeting OGT, OGA or scrambled control were grown with 1 μg/mL doxycycline for 48–72 h. Before harvesting, scrambled control cells were treated overnight with DMSO or the OGA inhibitor PUGNAc. Cells were lysed in M-PER mammalian protein extraction reagent (Thermo Fisher Scientific, Waltham, MA, USA) with protease and phosphatase inhibitors and 50 μM PUGNAc (Sigma-Aldrich, St. Louis, MO, USA). O-linked GlcNAcylation was assessed in cell lysates by Western blotting using an anti-O-GlcNAc antibody (ab2739, 1:1000 dilution, Abcam, Cambridge, MA, USA) and anti-mouse secondary antibody (1:3000, Jackson ImmunoResearch, West Grove, PA, USA). Equal loading was verified by anti-actin-HRP detection.
Protein sample preparation for proteomic analysis
2 × 106 shOGT or shOGA cells were seeded in 150 mm Petri dishes in duplicate and were grown with 1 μg/mL doxycycline for 48 h. Two sets of samples were irradiated with 6 Gy and samples taken after 1 h, along with controls. Cells were scraped from the dishes, washed in ice cold PBS with 50 μM of PUGNAc, pelleted, snap frozen in liquid nitrogen and stored at −80° C.
All steps were performed on ice or in 4° C cold room. Pellets of 6 × 105 - 7 × 106 cells were lysed with 500 μl cytoplasmic lysis buffer (10 mM TrisCl pH 8.0, 10 mM NaCl, 0.2% NP-40) in the presence of protease and phosphatase inhibitors and 50 μM PUGNAc, and passed through a 27 gauge needle 10 times. Lysates were kept on ice for 10 min, then centrifuged at 2000 × g for 5 min at 4° C. The supernatant was transferred to a new 1.5 mL tube, snap frozen in liquid nitrogen and stored at −80° C.
Nuclear lysate was prepared by resuspending the pellet in 300 μL nuclear lysis buffer (50 mM TrisCl pH 8.0, 10 mM EDTA, 1% SDS) supplemented with (300 U MNase and 15 μL 100 mM CaCl2 per sample). MNase digestion was carried out at 37° C for 10 min. The sample was centrifuged at 14,000 × g for 30 min at 4° C. The supernatant was transferred to a new 1.5 mL vial, snap frozen in liquid nitrogen and stored at −80° C.
In-gel trypsinization, HPLC for mass spectrometry and LC-MS/MS data acquisition and analysis were performed as described before (38,39) with modifications as detailed in Supplementary Methods.
IRIF imaging
For IRIF imaging, MCF7TagRFP-IBD cells were seeded on sterile cover glass in 24 well plates at 2.5 × 104 cells per well. Expression of TagRFP-IBD and ZsGreen shRNA-miR were induced with 1 μg/mL doxycycline for 48 h prior to irradiation. After 24 h, cells were fixed, stained with 5 μg/mL Hoechst 33342 and mounted using ProLong Gold (Life Technologies).
For immunofluorescence imaging of γH2AX and endogenous 53BP1, MCF7 cells with inducible shRNA were seeded on cover glass at 2.5 × 104 per well in 24 well plates. Expression of the shRNA was induced with 1 μg/mL doxycycline treatment for 48 h. After treatment with PUGNAc or alloxan, cells were fixed with 4% PFA at 24 h after irradiation with 6 Gy and permeabilized with 10% Triton-X 100 for 10 min. After blocking with 5% BSA, primary antibodies for γH2AX (Millipore, 05–636, 1:1000) or 53BP1 (Novus, NB100–304, 1:1000) were then incubated on cell slides overnight at 4° C. Following PBS washes, fluorescent secondary antibodies (Jackson ImmunoReserch) were applied for 1 h at room temperature. After PBS washes, coverslips were mounted with ProLong Gold (Invitrogen) and 1 μg/mL DAPI. Images were captured on a Zeiss Axiovert 40 CFL microscope with a 40X Plan-Neofluar objective and Axiocam digital camera controlled by AxioVision 4.8 software and pseudo-colored using ImageJ or Adobe Photoshop. TagRFP-IBD foci number was counted in ZsGreen shRNA-miR positive cells. Two or more replicates were performed.
Comet assay
For neutral comet assays, MCF7TagRFP-IBD1 cells with or without OGT or OGA shRNA-miR were seeded at 1 × 105 cells per well in 6-well plates with 1 μg/mL doxycycline for 48 h prior to irradiation. After 24 h, cells were mixed with Comet LM agarose and single cell electrophoresis was performed on Comet Slides (Trevigen, Gaithersburg, MD, USA) according to the manufacturer’s protocol. Slides were imaged on an Axiovert 40 with a 20X Plan-NeoFluar objective and AxioCam camera. Images were analyzed using an ImageJ comet assay macro (http://www.med.unc.edu/microscopy/resources/imagej-plugins-and-macros/comet-assay).
Clonogenic assays
MCF7TagRFP-IBD1 cells expressing an shRNA-mir (shScr, shOGT and shOGA) were plated at 500 cells per p100 plate in triplicate in corresponding medium supplemented with 1 μg/mL doxycycline for 48 h prior to irradiation. Radiation was delivered using a GammaCell 60Co source (MDS Nordion) with dose rate 9.2 cGy/sec. Cells remained in culture for 3 weeks and colonies of at least 50 cells were counted.
Immunostaining
Tumors were dissected from mice immediately following humane sacrifice and were fixed in 10% formalin. Tissue processing, embedding, and sectioning were performed by the Human Tissue Resource Center at the University of Chicago. Embedded tissues were cut into 5 μm sections using a microtome (Leica Microsystems RM2125). After deparaffinization and serial rehydration, antigen retrieval was performed in 10 mM sodium citrate buffer pH 6.0, or 10 mM Tris base, 1 mM EDTA, 0.05% Tween 20 buffer pH 9.0, in a tissue steamer for 40 min at 96° C. Slides were then allowed to cool to room temperature for immunostaining.
For immunostaining, blocking was performed using 5% normal goat serum + 5% bovine serum albumin (BSA) in PBS for 30 min at room temperature. Primary antibodies detecting γH2AX (EMD Millipore, ab05–636, 1:100) and O-GlcNAc (Abcam, ab2739, 1:100) were diluted in 1% BSA in 1X PBS were then applied on tissue sections overnight at 4° C in a humidity chamber. Slides were washed 3X briefly in 1X PBS-Tween 20 (0.05%), incubated in biotinylated anti-mouse secondary antibody (Vector Laboratories, MKB-2225, 1:250) diluted in 1% BSA in 1X PBS for 30 min at room temperature, followed by 3X brief washes in 1X PBS-T. Tissue sections were incubated in streptavidin:DyLight 594 (Vector Laboratories, SA-5594, 1:100) diluted in 1X PBS for 30 min at room temperature in the dark. Slides were rinsed with 1X PBS, then stained with 1 μg/mL 4′,6-diamidino-2-phenyl-indole (DAPI) for 20 min at room temperature in the dark. Tissue sections were briefly rinsed with dH2O, mounted using Aqua-Poly/Mount (Polysciences), and coverslipped.
For Ki-67 and EZH2 immunostaining, endogenous peroxidase activity was quenched through incubation in 1% H2O2 in methanol (Sigma-Aldrich) for 20 min. Slides were washed 3X briefly in 1X PBS-T (0.05% Tween-20), then incubated with ImmPRESS HRP polymer anti-rabbit IgG for 30 min at room temperature. Antigen-antibody binding was detected using the ImmPACT AMEC Red peroxidase substrate kit (Vector Laboratories, SK-4285) for Ki-67 or ImmPACT DAB peroxidase (Vector Laboratories, SK-4105) for EZH2. Ki-67 and EZH2 stained tissue sections were briefly immersed in hematoxylin for counterstaining, washed 3X in dH2O, mounted using Aqua-Poly/Mount and covered with cover glasses.
Imaging was performed at the University of Chicago Integrated Light Microscopy Facility. Digital image files were created with a 3D Histech Pannoramic Scan whole slide scanner (Perkin Elmer, Waltham, MA, USA) using either a Zeiss AxioCam MRm CCD camera (immunofluorescence) or a Stingray F146C color camera (chromogenic IHC, Allied Vision Technologies, Stadtroda, Germany). Individual images were created with the 3D Histech Pannoramic Viewer software (Perkin Elmer).
Animals and tumor models
Mice were maintained according to guidelines of the Institutional Animal Care and Use Committee and irradiated using a RadSource RS-2000 X-Ray generator operating at 160 kV and 25 mA at a dose rate of 2.20 Gy/minute, calibrated by NIST traceable dosimetry. 17β-Estradiol pellets (1.7 mg, Innovative Research of America) were implanted s.c. dorsally in female athymic nude mice (Envigo, Indianapolis, IN, USA) 5 days before s.c. injection of 1×107 MCF7TagRFP-IBD cells in 100 μL PBS into the animal’s flank. Once hindlimb tumors grew to a volume of 300 mm3, 2 mg/mL doxycycline was added to the drinking water supplemented with 1% sucrose, for 72 h before irradiation with 6 Gy. Mice were treated by oral gavage with N-acetyl-D-glucosamine (200 mg/kg), or alloxan (50 mg/kg) or PUGNAc (30 mg/kg) (all from Sigma-Aldrich) 2 days before, the day of, and for 7 days after irradiation.
IRIF analysis in tumors
For two-photon confocal imaging of IRIF in tumors, tissue was excised, counterstained with Hoechst 33342 and imaged as described (9).
Transparent Tissue Tomography (T3) (40) was applied for multiplex three-dimensional analysis of IRIF in tumors. In brief, tumors were harvested at 24 h after 6 Gy irradiation, washed with cold PBS, fixed with 2% paraformaldehyde in PBS for 10 min at room temperature, and washed with PBS. The washed tumors were cast in 2% agarose gel (LE Quick Dissolve Agarose, GeneMate, dissolved in distilled water) in 24 well plates. The gel plugs containing tumors were mounted on a vibrating microtome (VT1200S, Leica) equipped with a buffer tray. Sections cut at 0.4 mm thickness were collected in order in cold PBS. The macrosections were stained with DyLight 633-anti-CD31 (BioLegend, clone MEC13.3) antibody in staining buffer (SB, RPMI 1640 media with 10 mg/mL BSA and 0.1% Triton X-100) at 4° C overnight. Then, the macrosections were incubated in 10 ml of 80% (w/v) D-fructose solution for 1 h at 25° C with gentle agitation. The stained sections were imaged by a Leica SP5 AOBS II tandem confocal laser scanning microscope using a HC PL APO 40X/1.25 NA oil objective (0.24 mm working distance) or HCX PL APO 10X/0.4 NA dry objective (2.2 mm working distance) with 488 nm (Argon) excitation and 495–528 nm emission filter for ZsGreen, 561 nm (DPSS) excitation and 585–614 nm filters for TagRFP, and 633 nm excitation and 637–655 nm filter for DyLight 633.
SA-β-Gal assay
Cells were seeded at 2.5 × 104 per well in 6 well plates with 1 μg/mL doxycycline for 48 h prior to irradiation. Cells were fixed 5 days after irradiation and assayed for SA-β-Gal as described (41). SA-β-Gal positive and negative cells were counted in multiple fields, yielding an average percent SA-β-Gal positive staining, indicated on each image as mean ± SEM.
To evaluate senescence in vivo, frozen sections of tumors excised 5 days after irradiation were fixed in 2% paraformaldehyde, stained for SA-β-Gal activity, counterstained with Nuclear Fast Red (Vector Laboratories), dehydrated, mounted and imaged.
Statistical analysis
Statistical significance for IRIF counting and comet assays was determined using the nonparametric Mann-Whitney test. Calculations were performed using Prism software (GraphPad). P value ≤ 0.05 was considered statistically significant.
RESULTS
In order to examine the effect of the HBP pathway and reversible protein O-GlcNAcylation (Fig. 1A) on DNA damage response, we developed dual reporter MCF7 cell lines. As a reporter for formation and resolution of ionizing radiation induced foci (IRIF), we adapted the previously reported doxycycline inducible GFP fusion to the 53BP1 IRIF binding domain (GFP-IBD) (9) by exchanging in the orange-red fluorescent protein TagRFP (λex 555nm, λem 584 nm (37)) to form TagRFP-IBD (Fig. 1B). TagRFP-IBD was stably integrated by lentiviral transduction of MCF7 Tet-On Advanced cells that constitutively express a tetracycline transactivator (tTA) protein, yielding MCF7TagRFP-IBD. Directly comparable to our prior studies with the GFP-IBD reporter, treating MCF7TagRFP-IBD cells or xenograft tumors with doxycycline resulted in pan-nuclear red fluorescence. Upon irradiation, the TagRFP-IBD relocalized to form IRIF at presumptive DSBs. The TagRFP-IBD foci resolved over the following two days with the kinetics of DSB repair (Supplementary Fig. S1). When MCF7 cells expressing TagRFP-IBD and GFP-IBD were coinjected to form flank tumors, they displayed similar numbers of basal foci in unirradiated tumors and comparable foci formation and resolution at 3 and 24 h after irradiation (Supplementary Movies 1, 2 and 3, respectively).
For knockdown of OGT or OGA expression in the MCF7TagRFP-IBD cells, we used sets of three gene-specific shERWOOD-UltramiR lentiviral short hairpin RNAs (shRNA-miR) targeting OGT or OGA (MGEA5) along with an untargeted scrambled control (Scr). Here, each shRNA is encoded in the hairpin of a microRNA scaffold embedded at the 3’ end of an inducible ZsGreen fluorescent marker gene, leading to coexpression of ZsGreen and the shRNA-miR upon treatment with doxycycline (Fig. 1C). To examine effects of shRNA-miR-mediated knockdown in MCF7TagRFP-IBD cells, we assayed protein O-GlcNAcylation by Western blot (Fig. 1D). Compared to shScr control, all three OGT-targeted shRNA-miRs decreased O-GlcNAc protein modification. In turn, protein O-GlcNAcylation was similarly increased by treating shScr cells with the OGA inhibitor PUGNAc or expression of each of the three OGA-targeted shRNA-miRs. The resulting dual reporter cell lines, hereafter denoted as shOGT, shOGA and shScr cells, allowed us to simultaneously visualize IRIF (TagRFP-IBD) and shRNA-miR expression (ZsGreen) in individual cells in vitro (Fig. 1E) or in tumors (Fig. 1F, Supplementary Movie 4) following doxycycline induction.
O-GlcNAc modification modulates DNA-damage response pathways in response to irradiation
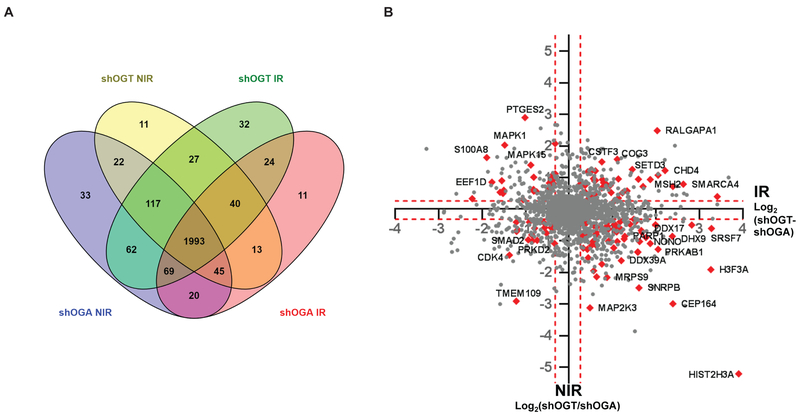
To uncover the effect of O-GlcNAc modification on DNA damage response, we performed mass spectrometry analysis on nuclear extracts of shOGT and shOGA cells that had been treated with 0 or 6 Gy irradiation. A total of 2518 proteins were identified at 1% FDR. Of these, shOGT6 Gy yielded 2263, shOGT0 Gy, 2267, shOGA6 Gy, 2214, and shOGA0 Gy, 2361, with 1993 in common among all four samples. For quantification, we applied a replicate filter of n ≥ 2 reducing the total to 2195 proteins with shOGT6 Gy yielding 2080, shOGT0 Gy, 1958, shOGA6 Gy, 1837, and shOGA0 Gy, 2055 (Fig. 2A).
Figure 2.
Label-free quantitation (LFQ) of tandem mass spectrometry analysis of the nuclear proteomes of shOGT and shOGA cells treated with 0 or 6 Gy. A, Venn diagram showing distribution of the 2518 proteins identified in either shOGT or shOGA cells, with or without irradiation. Of these, 2214 proteins were identified in shOGT0 Gy, 2361 in shOGA0 Gy, 2263 in shOGT6 Gy, and 2214 in shOGA6 Gy, of which 1993 were identified in all four samples. B, XY scatter plot of LFQ intensity ratios of shOGT6 Gy/shOGA6 Gy plotted against shOGT0 Gy/shOGA0 Gy, shown on Log2 scale. Statistically significant cutoffs of ≥1.2-fold change up (Log2, 0.26) and ≤0.8-fold change down (Log2, −0.32) are shown in dashed lines. Protein hits that fall in the DNA Damage and Chromatin pathways are highlighted in red and marked as indicated.
We performed differential expression analysis, using a significance cutoff of ≥ 1.2 fold change (Log2 ≥ 0.26) “Up” and ≤ 0.8 fold change “Down” on the following comparisons: shOGT0 Gy/shOGA0 Gy, shOGT6 Gy/shOGT0 Gy, shOGA6 Gy/shOGA0 Gy, and shOGT6 Gy/shOGA6 Gy. A quadrant plot of the 1993 proteins detected in all four samples, where the X axis represents fold change for shOGT6 Gy/shOGT0 Gy and the Y axis displays fold change for shOGT6 Gy/shOGA6 Gy, yielded several hundred candidates with significant change in abundance after irradiation that fell off the diagonal, potentially identifying proteins whose up- or down-regulation might be O-GlcNAc dependent (Fig. 2B). Considering the effects of the knockdowns alone without irradiation (shOGT0 Gy/shOGA0 Gy), we observed 435 proteins significantly “up” in shOGT cell lines, and 533 “down” in shOGA cell lines. Upon irradiation, we observed upregulation of 499 proteins and downregulation of 599 proteins in shOGA cells (shOGA6 Gy/shOGA0 Gy) and upregulation of 604 proteins and downregulation of 389 proteins in shOGT cells (shOGT6 Gy/shOGT0 Gy). Finally, looking at differential effects of irradiation on the two knockdown cell lines (shOGT6 Gy/shOGA6 Gy), we observed 689 proteins significantly “up” in shOGT but 463 significantly “down” in shOGA (Supplementary Table S1).
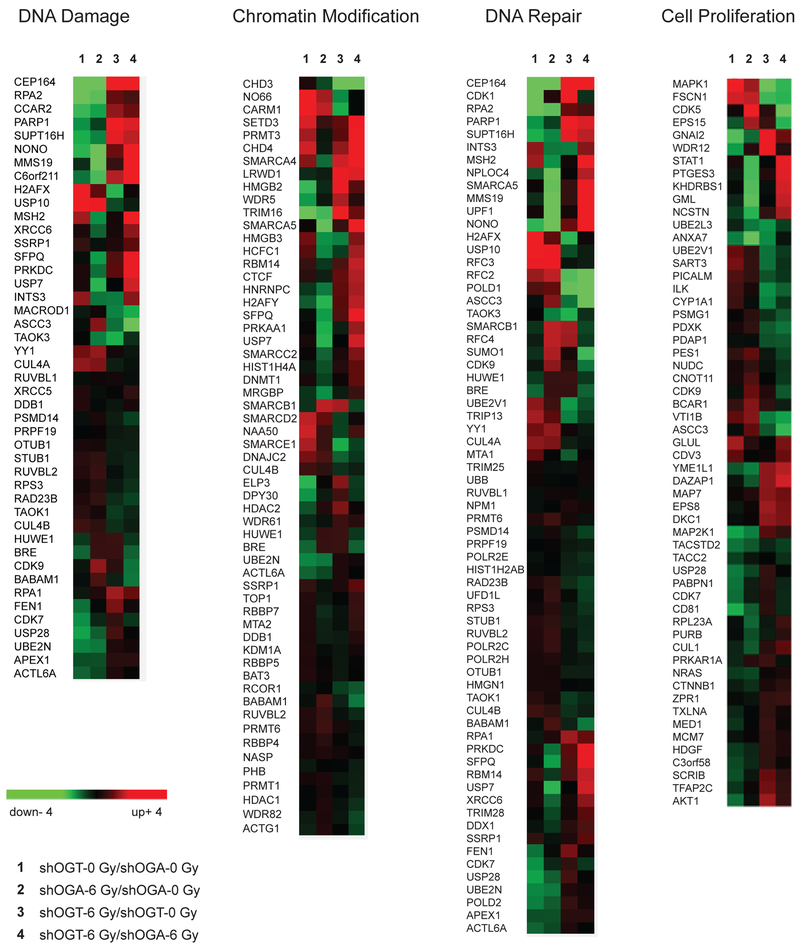
To identify pathways and biological processes that are differentially expressed in shOGT vs. shOGA cells, we performed Reactome pathway analysis (42) via Panther (43). The Reactome analysis identified multiple pathways including cell cycle (R-HSA-1640170), cell cycle checkpoints (R-HSA-69620), metabolism (R-HSA-1430728), NF-κB (R-HSA-5676590), p53 (R-HSA-69541) p38 MAPK signaling (R-HSA-171007), SUMOylation of ubiquitylation proteins (R-HSA-3232142), apoptosis (R-HSA-109581), senescence (R-HSA-2559586), proliferation (GO:0008283), and others. Additionally, this analysis highlighted multiple pathways that contribute in DNA damage response and DNA repair, including chromatin organization (R-HSA-4839726), chromatin modification (R-HSA-3247509), sensing of DNA double strand breaks (R-HSA-5693548), DNA double strand break response (R-HSA-5693606), DNA double strand break repair (R-HSA-5693532), PRC2 methylation of histones and DNA (R-HSA-212300), and DNA methylation (R-HSA-5334118) (Supplementary Table S2). Many of the proteins most differentially expressed (farthest off the diagonal) between shOGT and shOGA were chromatin regulation and DNA-damage response-associated factors (red carets, Fig. 2B). Overall, irradiated shOGT cells displayed marked upregulation of multiple pathways linked to DNA damage response while unirradiated shOGT displayed basal activation of these same pathways. Indeed, hierarchical clustering of the differential expression of proteins in the chromatin modification, DNA-damage DNA-repair and cell proliferation pathways revealed a shared pattern placing the fold change between unirradiated shOGT and shOGA closest to the fold change between irradiated shOGA and unirradiated shOGA (Fig. 3). One interpretation is that inducing shOGT may result in cellular stress, resulting in constitutive activation of DNA damage responses.
Figure 3.
Gene ontology analysis of O-GlcNAcylation-dependent protein expression in the DNA damage response. Hierarchal clustering represented with heat maps showing Log2 LFQ intensity ratios of proteins associated with DNA Damage, Chromatin Modification, DNA Repair, and Cell Proliferation pathways for shOGT0 Gy/shOGA0 Gy, shOGA6 Gy/shOGA0 Gy, shOGT6 Gy/shOGT0 Gy, and shOGT6 Gy/shOGA6 Gy.
OGT regulates radiation sensitivity in MCF7 human mammary adenocarcinoma cells
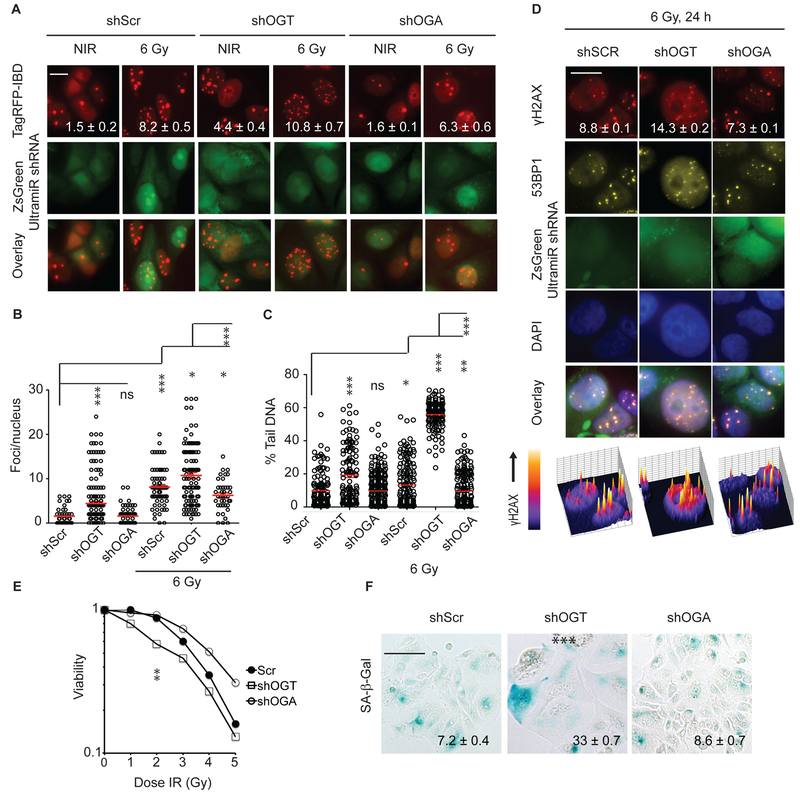
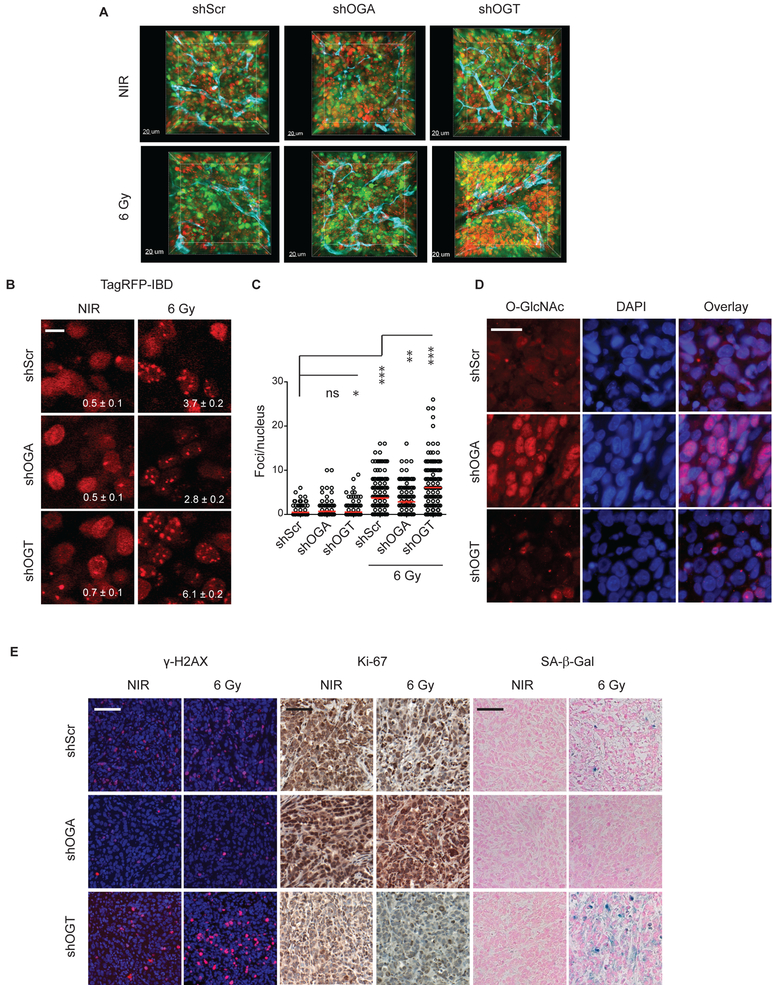
To further explore regulation of DNA damage-related pathways by O-GlcNAcylation, we examined radiation response in shOGT, shOGA and shScr cells. Compared to shScr, shOGT displayed IRIF persistence, marked by increased numbers of TagRFP-IBD foci when cells were examined 24 h after irradiation (Fig. 4A and B). Notably, shOGT also displayed increased basal TagRFP-IBD foci, with some cells displaying ten to twenty IRIF even without irradiation, suggesting high levels and/or lack of normal repair of spontaneous DNA damage. Compared to shScr, shOGA displayed a similar level of basal IRIF but greater IRIF resolution at 24 h. Examining longer times after 6 Gy irradiation, the majority of shScr, shOGT and shOGA cells returned to their basal levels of IRIF by 48 or 72 h. In each cell line, a fraction of cells retained numerous persistent foci (>60 per cell) even at 5 days after irradiation, consistent with the pattern of relative foci persistence at shorter time points, with shScr displaying 16%, shOGT, 24%, and shOGA, 10%, of these characteristic cells.
Figure 4.
Targeting the HBP pathway via silencing of either OGT or OGA alters DSB repair, IRIF resolution, viability and IR-induced senescence in MCF7TagRFP-IBD cells. A, Silencing of OGT or OGA has opposing effects on IRIF resolution in MCF7 cells. TagRFP-IBD foci in representative cells 24 h after 6 Gy are shown. Scale bar 20 μm. B, Plots of IRIF per nucleus in individual cells. Red bar indicates mean ± SEM; *, P <0.05; ***, P < 0.0001, (Mann-Whitney test). C, Effects of OGT or OGA silencing on DSB repair. Cell lines with inducible shRNA-miRs targeting OGT, OGA or with scrambled control were grown in medium containing different concentrations of glucose for 24 h. Following irradiation with 6 Gy, neutral comet assays were performed 24 h post IR. Plots of percent tail DNA are shown. Red line indicates mean ± SEM. ***, P < 0.0001, (Mann-Whitney test). D, Immunofluorescence staining of endogenous 53BP1 and γH2AX in shScr control, shOGA, and shOGT cells 24 h after 6 Gy. Shown are individual channels and overlay of anti-γH2AX (red), anti-53BP1 (yellow), shRNA-miR expression (green), and DAPI (blue) for representative cells. Mean γH2AX foci per cell ± SEM is indicated. Below are perspective plots of γH2AX staining intensity. Scale bar 20 μm. E, Clonogenic survival of cells after irradiation. Normalized survival fractions indicating average of 3 replicates are shown. F, Senescence induction in cells bearing shRNA-miRs targeting OGT or OGA evaluated by SA-β-Gal (blue) staining 5 days after 6 Gy. Representative images are shown. Percent SA-β-Gal positive cells are indicated as mean ± SEM. Scale bar 50 μm.
Overall, these data suggested a defect in DSB repair associated with shOGT. Indeed, comet assays confirmed a defect in DSB repair in shOGT cells and lower levels of residual DSBs in shOGA expressing cells as compared to shScr control cells (Fig. 4C).
A potential drawback of TagRFP-IBD is that it is an indirect reporter for IRIF where apparent foci intensity may vary depending on expression level. Thus, we also examined formation of γH2AX and relocalization of endogenous 53BP1 in shSCR, shOGT and shOGA cells by indirect immunofluorescence. As shown in representative images in Fig. 4D, γH2AX and 53BP1 display similar patterns of colocalization in each cell line at 24 h after irradiation. Further, counting the γH2AX foci revealed IRIF persistence in shOGT as observed with the TagRFP-IBD reporter. As indicated by the 3D rendering in Fig. 4D, the γH2AX immunoreactivity of persistent IRIF was comparable in each cell line, though shOGA appeared to display lower total H2AX phosphorylation.
Clonogenic assays revealed that compared with control (surviving fraction at 2 Gy, SF2 = 0.89 ± 0.04) or shOGA cells (SF2 = 0.94 ± 0.04, P = 0.46 (NS)), shOGT cells displayed increased radiation sensitivity (SF2 = 0.58 ± 0.02, P < 0.003, Fig. 4E). However, the suppression of colony formation was not associated with significantly reduced cell viability after irradiation, suggesting only a minor contribution from apoptotic or necrotic cell death (Supplementary Fig. S2A). Instead, while surviving irradiated shScr and shOGT cells formed rapidly growing colonies, a large fraction of irradiated shOGT cells formed abortive microcolonies characterized by senescence-like morphology (Supplementary Fig. S2B). A direct assay for radiation-induced senescence revealed a significantly higher fraction of large, flat shOGT cells expressing senescence-associated beta-galactosidase (SA-β-Gal) at 7 days after irradiation compared to shOGA or shScr cells (Fig. 4F).
A concern is that the pattern of cellular responses observed after irradiation upon suppression of O-GlcNAcylation, characterized by persistent IRIF, slow DSB repair, enhanced senescence and lack of cell death, might be specific to MCF7 cells, a human ER+ breast carcinoma cell line that is p53 wild type but does not express caspase 3, and thus displays an attenuated apoptotic response. We therefore examined multiple human and murine tumor cell lines to assay the effects of the OGA inhibitor PUGNAc and OGT inhibitor alloxan on IRIF resolution, apoptosis induction and cell viability upon irradiation. Consistent with our shOGA results in MCF7, treatment of MDA-MB-231 human breast cancer, MDA-MB-435 human melanoma, B16F10 murine melanoma or Panc02 murine pancreatic ductal adenocarcinoma cells with PUGNAc improved IRIF resolution at 24 h after irradiation (Supplementary Fig. S3). In turn, as observed for shOGT, alloxan-treated cells revealed IRIF persistence at 24 h. Similarly to MCF7, B16F10 and Panc02 cells did not demonstrate significantly increased apoptosis or cell death after irradiation with 6 Gy, alone or with PUGNAc or alloxan treatment (Supplementary Fig. S4, Supplementary Movie 5). Instead, alloxan increased radiation-induced senescence in Panc02 cells in a dose-dependent manner (Supplementary Fig. S5).
Silencing OGT and OGA mediate opposing effects on DNA damage response in irradiated xenograft tumors
To examine the influence of OGT, OGA and O-GlcNAcylation on DNA damage response in vivo, we established MCF7TagRFP-IBD tumor xenografts expressing shOGT, shOGA or shScr (control). Mice were treated with doxycycline for 2 days to induce TagRFP-IBD and shRNA-miR expression prior to irradiation with a single dose of 6 Gy. 3D imaging of intact tumor tissue at 24 h after 0 or 6 Gy revealed similar basal IRIF for unirradiated tumors but foci persistence after irradiation in shOGT (Fig. 5A, Supplementary Movie 3). Examining IRIF persistence at 24 h after irradiation at high resolution revealed that compared to shScr control, shOGA tumors displayed accelerated IRIF resolution while shOGT tumors displayed IRIF persistence (Fig. 5B and C). In a parallel group of animals, tumors were excised 7 days after 6 Gy. Immunofluorescence analysis of total protein O-GlcNAcylation in frozen sections revealed markedly increased immunoreactivity after induction of shOGA and a decrease in shOGT tumors (Fig. 5D). Immunohistochemistry in FFPE thin sections was performed probing for γH2AX to detect persistent DNA damage, Ki-67 to assess cell proliferation, and SA-β-Gal to evaluate radiation-induced senescence. The shOGT tumors displayed persistent DNA damage, decreased proliferation and greater senescence compared to shScr control while the irradiated shOGA tumors were nearly indistinguishable from unirradiated tumors (Fig. 5E).
Figure 5.
Silencing of OGT or OGA alters the DNA damage response in irradiated tumors. A, Three-dimensional imaging of MCF7 tumor xenografts with dual fluorescent reporters for IRIF (TagRFP, red) and shRNA-miR expression (ZsGreen, green). Anti-CD31 antibody was used for vascular staining (endothelium, cyan). B, Intratumoral imaging of the IRIF reporter TagRFP-IBD 24 h after 6 Gy. Number of foci per nucleus is shown (mean ± SEM). Scale bar 10 μm. C, Quantification of IRIF per nucleus in individual cells 24 h after 6 Gy. Red bar indicates mean ± SEM. ***, P < 0.0001; * P < 0.05 (Mann-Whitney test). D, Immunofluorescence of O-GlcNAcylated proteins in xenograft tumors displays the increase in O-GlcNAcylation following shOGA knock down. DAPI (blue) was used as a nuclear stain. Scale bar: 25 μm. E, Thin sections of tumors excised 7 days after 6 Gy immunostained for γH2AX (red), Ki-67 (brown) and SA-β-Gal activity (blue). Representative images are shown. Scale bar: 100 μm.
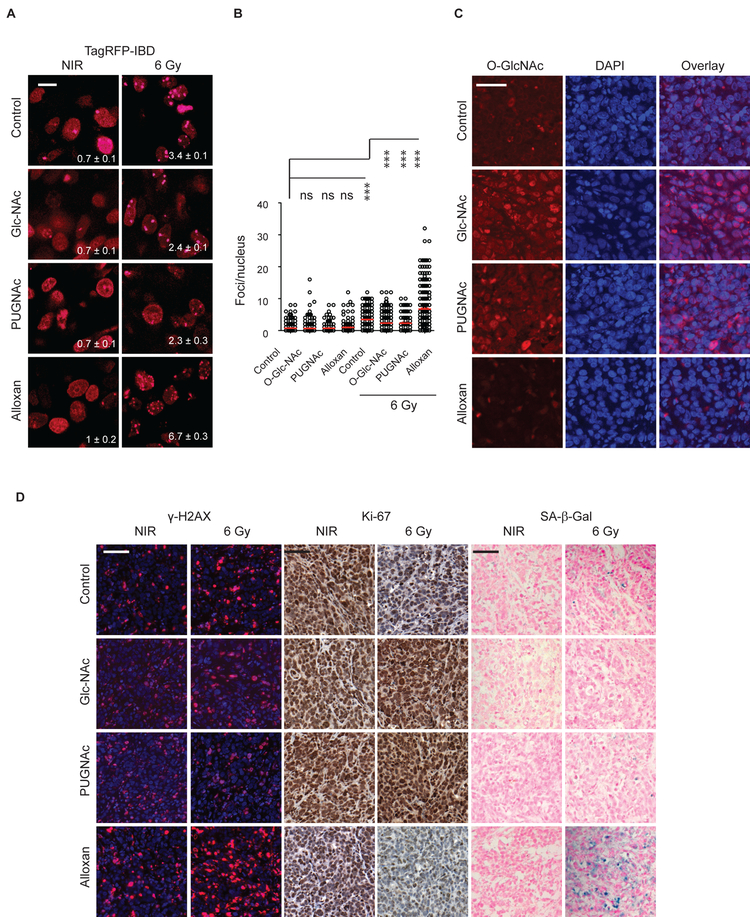
Targeting O-GlcNAcylation with small molecule inhibitors modulates radiation response in irradiated tumors
In order to assess the potential translational impact of our findings, we established shScr MCF7TagRFP-IBD tumors and treated the mice with GlcNAc to drive UDP-GlcNAc biosynthesis, PUGNAc to inhibit OGA or alloxan to block OGT. Much like for shOGA, imaging TagRFP-IBD foci in shScr tumors treated with GlcNAc or PUGNAc revealed increased IRIF resolution at 24 h after 6 Gy irradiation compared to untreated control (Fig. 6A and B). In turn, as observed for shOGT, alloxan-treated tumors revealed IRIF persistence at 24 h. Immunofluorescence analysis in thin sections from tumors excised 7 days after irradiation revealed increased protein O-GlcNAcylation in tumors treated with GlcNAc or PUGNAc tumors treated with alloxan displayed lower O-GlcNAc levels (Fig. 6C). Analysis of γH2AX, Ki-67 and SA-β-Gal showed that treatment with GlcNAc or PUGNAc spared tumors from radiation damage whereas alloxan led to persistent DNA damage, decreased proliferation and increased senescence compared to untreated control (Fig. 6D).
Figure 6.
Targeting of the HBP pathway with small molecule inhibitors alters the DNA damage response in irradiated tumors. A, Intratumoral imaging of the IRIF reporter TagRFP-IBD 24 h after 6 Gy. Foci numbers per nucleus are shown (mean ± SEM). Scale bar: 10 μm. B, Quantification of IRIF per nucleus in individual cells 24 h after 6 Gy. Red bar indicates mean ± SEM. ***, P < 0.0001; * P < 0.05 (Mann-Whitney test). C, Immunofluorescence of O-GlcNAcylated proteins (red) in tumors treated with GlcNAc, OGA inhibitor PUGNAc or OGT inhibitor alloxan in comparison with control tumor tissue. DAPI (blue) was used as a nuclear stain. Scale bar: 50 μm. D, γH2AX (red), Ki-67 (brown) and SA-β-Gal (blue) in tumor sections at day 7 after 6 Gy. Representative images are shown. Scale bar: 100 μm.
Among pathways highlighted by the proteomic analysis was PRC2 methylation of histones and DNA (R-HSA-212300). We had previously identified the PRC2 histone methyltransferase catalytic subunit Ezh2 as a candidate mediator of O-GlcNAc-dependent regulation of DSB repair (9). Consistent with prior in vitro results that Ezh2 abundance is regulated by OGT (28), immunohistochemistry analysis of Ezh2 expression in the unirradiated and irradiated shOGT or alloxan-treated shScr tumors at 7 days after irradiation displayed decreased immunostaining compared with shOGA or GlcNAc- or PUGNAc-treated shScr tumors or shScr controls (Supplementary Fig. S6A and S6B).
DISCUSSION
Metabolic reprogramming is a hallmark of cancer often described as a necessary adaptation to the increased demands for energy and building blocks for macromolecular biosynthesis to support cancer cell growth and proliferation (1–3). We have explored alternative pathways whereby cancer metabolism may impact other cancer hallmarks such as genomic instability and cell immortality. The recent discovery that 2-hydroxyglutarate (2-HG) acts as an oncometabolite to promote carcinogenesis without being further metabolized has led to the discovery of epigenetic enzymes as primary targets, suggesting mechanisms based on deregulated gene expression (44). Our prior work provided an alternative model, implicating 2-HG along with the hexosamine biosynthetic pathway metabolite N-acetylglucosamine (GlcNAc) in promoting DNA double strand break repair and blocking onset of cellular senescence via their effects on chromatin modification (9).
While high levels of 2-HG are associated with relatively rare IDH mutations (45), increased cellular GlcNAc is likely a far more common feature of cancer (31,46–49). High O-GlcNAcylation may be as simple as the result of increased flux through the HBP due to deregulated glycolysis and the Warburg effect, but other metabolic adaptations, feedback loops, and activating mutations may be implicated. As such, targeting the HBP and/or protein O-GlcNAcylation might represent a relatively general, cancer cell-selective means to sensitize tumors to genotoxic stress. To explore this concept, in this study we examined how modulating protein O-GlcNAcylation by targeting O-GlcNAc transferase or O-GlcNAcase affects radiation response in cells and tumors.
Prior proteomic analysis has focused on discovery of OGT substrates, leading to more than 4,000 O-GlcNAc modified proteins identified to date (50). Many of these proteins have links to DNA damage and repair and their dynamic O-GlcNAcylation is implicated in the cellular response to diverse forms of stress (23,51–53). Despite many leads, functional analysis has lagged considerably, leaving roles for O-GlcNAcylation in DNA damage response poorly defined. Thus, we performed quantitative proteomic differential expression profiling of the shOGT and shOGA cell lines with and without irradiation. Reactome pathway analysis revealed marked changes in regulation of multiple pathways associated with cell proliferation, cellular signaling, and DNA damage responses between shOGT and shOGA. Overall, our pathway analysis suggested that OGT functions in unperturbed cells to suppress genomic instability and decrease cellular stress but becomes critical in irradiated cells for DNA damage repair and recovery from checkpoint arrest.
Given these findings, we examined the radiation response in cells expressing inducible shOGT or shOGA and observed effects on DSB repair, IRIF resolution, cell viability and radiation-induced senescence consistent with a key role for O-GlcNAcylation in DNA damage response. In particular, shOGT delayed DNA repair, decreased clonogenicity and increased senescence after irradiation compared to knockdown of OGA or scrambled control. Taken together, our in vitro data suggest that increased O-GlcNAc modification promotes normal functions of DNA-damage and repair pathways in response to irradiation. As a result, preventing physiological O-GlcNAcylation of cellular proteins induces persistent DNA damage and increased DNA damage signaling upon irradiation. Further, blocking OGT appears to increase basal levels of DSBs even in unperturbed cells, suggesting a role in protection from oxidative stress and/or cell cycle defects.
To evaluate the impact of O-GlcNAc modification on radiation response in vivo, we examined O-GlcNAcylation dependent effects in tumors irradiated 48 h after inducing shRNA-mirs. Consistent with the in vitro experiments, irradiated tumors expressing shRNA-miRs targeting OGT displayed decreased O-GlcNAc modification, persistent DNA damage, decreased cell proliferation and increased senescence. Importantly, treating animals bearing control tumors with alloxan, a non-specific small-molecule OGT inhibitor, recapitulated the phenotype of shRNA-miR targeting OGT.
Perhaps of equal importance, silencing OGA conferred a relatively dramatic radioresistance phenotype in vivo. OGA knockdown spared tumors from most of the deleterious effects of radiation, blocking accumulation of DNA damage, preserving cell proliferation and preventing radiation-induced senescence. Treating mice bearing control tumors with PUGNAc, a non-specific OGA inhibitor, or the HBP metabolite GlcNAc increased O-GlcNAcylation and similarly protected irradiated tumors from persistent DNA damage, loss of proliferation or senescence. Considering that GlcNAc is non-toxic and readily available, these results may suggest a new approach to radiation protection and mitigation.
Taken together, our studies suggest that the hexosamine biosynthetic pathway, via its impact on protein O-GlcNAcylation, is a key determinant of the DNA damage response in cancer, providing a mechanistic link between metabolic reprogramming, genomic instability and therapeutic response. Our findings also implicate the HBP as an important metabolic contributor to therapeutic resistance via promoting DNA repair and protecting cells from cell cycle arrest and senescence. These results provide a strong rationale for targeting O-GlcNAcylation as a route to overcoming resistance to radiation and other genotoxic therapies.
Supplementary Material
IMPLICATIONS.
Finding that the hexosamine biosynthetic pathway, via its impact on protein O-GlcNAcylation, is a key determinant of the DNA damage response in cancer provides a mechanistic link between metabolic reprogramming, genomic instability and therapeutic response and suggests novel therapeutic approaches for tumor radiosensitization.
Acknowledgments
We thank Nicholas Rymut and Joanna Pagacz for technical support, Shirley Bond in the Integrated Light Microscopy Core for advice and assistance, and Julian Lutze for manuscript editing.
Grant Support
These studies were supported by NIH NCI R01s CA164492, CA176843 and CA217182 to S.J.K.
Conflicts of Interest: S.-Y.L., V.P.B. and S.J.K. are founders of Transnostics. S.J.K. is a founder of OncoSenescence and receives funding from AbbVie.
BIBLIOGRAPHY
- 1.Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell 2012;21(3):297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab 2008;7(1):11–20. [DOI] [PubMed] [Google Scholar]
- 3.Koppenol WH, Bounds PL, Dang CV. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat Rev Cancer 2011;11(5):325–37. [DOI] [PubMed] [Google Scholar]
- 4.Lu C, Thompson CB. Metabolic regulation of epigenetics. Cell Metab 2012;16(1):9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaelin WG Jr., McKnight SL. Influence of metabolism on epigenetics and disease. Cell 2013;153(1):56–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu J, Kim J, Oberdoerffer P. Metabolic modulation of chromatin: implications for DNA repair and genomic integrity. Front Genet 2013;4:182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gut P, Verdin E. The nexus of chromatin regulation and intermediary metabolism. Nature 2013;502(7472):489–98. [DOI] [PubMed] [Google Scholar]
- 8.Li X, Egervari G, Wang Y, Berger SL, Lu Z. Regulation of chromatin and gene expression by metabolic enzymes and metabolites. Nat Rev Mol Cell Biol 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Efimova EV, Takahashi S, Shamsi NA, Wu D, Labay E, Ulanovskaya OA, et al. Linking Cancer Metabolism to DNA Repair and Accelerated Senescence. Mol Cancer Res 2016;14(2):173–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bonner WM, Redon CE, Dickey JS, Nakamura AJ, Sedelnikova O, Solier S, et al. GammaH2AX and cancer. Nat Rev Cancer 2008;8(12):957–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Attikum H, Gasser S. Crosstalk between histone modifications during the DNA damage response. Trends in Cell Biology 2009;19(5):207–17. [DOI] [PubMed] [Google Scholar]
- 12.Rossetto D, Truman AW, Kron SJ, Côté J. Epigenetic modifications in double-strand break DNA damage signaling and repair. Clin Cancer Res 2010;16(18):4543–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hunt CR, Ramnarain D, Horikoshi N, Iyengar P, Pandita RK, Shay JW, et al. Histone modifications and DNA double-strand break repair after exposure to ionizing radiations. Radiat Res 2013;179(4):383–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smeenk G, van Attikum H. The chromatin response to DNA breaks: leaving a mark on genome integrity. Annu Rev Biochem 2013;82:55–80. [DOI] [PubMed] [Google Scholar]
- 15.Kao J, Milano MT, Javaheri A, Garofalo MC, Chmura SJ, Weichselbaum RR, et al. gamma-H2AX as a therapeutic target for improving the efficacy of radiation therapy. Curr Cancer Drug Targets 2006;6(3):197–205. [DOI] [PubMed] [Google Scholar]
- 16.Goodarzi A, Jeggo P. Irradiation induced foci (IRIF) as a biomarker for radiosensitivity. Mutat Res 2012;736(1–2):39–47. [DOI] [PubMed] [Google Scholar]
- 17.Hart GW, Housley MP, Slawson C. Cycling of O-linked beta-N-acetylglucosamine on nucleocytoplasmic proteins. Nature 2007;446(7139):1017–22. [DOI] [PubMed] [Google Scholar]
- 18.Hanover JA, Krause MW, Love DC. Bittersweet memories: linking metabolism to epigenetics through O-GlcNAcylation. Nat Rev Mol Cell Biol 2012;13(5):312–21. [DOI] [PubMed] [Google Scholar]
- 19.Dehennaut V, Leprince D, Lefebvre T. O-GlcNAcylation, an Epigenetic Mark. Focus on the Histone Code, TET Family Proteins, and Polycomb Group Proteins. Frontiers in endocrinology 2014;5:155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang X, Qian K. Protein O-GlcNAcylation: emerging mechanisms and functions. Nat Rev Mol Cell Biol 2017;18(7):452–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leturcq M, Lefebvre T, Vercoutter-Edouart AS. O-GlcNAcylation and chromatin remodeling in mammals: an up-to-date overview. Biochem Soc Trans 2017;45(2):323–38. [DOI] [PubMed] [Google Scholar]
- 22.Hart GW, Slawson C, Ramirez-Correa G, Lagerlof O. Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annu Rev Biochem 2011;80:825–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Slawson C, Hart G. O-GlcNAc signalling: implications for cancer cell biology. Nat Rev Cancer 2011;11(9):678–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hardiville S, Hart GW. Nutrient Regulation of Signaling, Transcription, and Cell Physiology by O-GlcNAcylation. Cell Metab 2014;20(2):208–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sakabe K, Wang Z, Hart GW. Beta-N-acetylglucosamine (O-GlcNAc) is part of the histone code. Proc Natl Acad Sci U S A 2010;107(46):19915–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deplus R, Delatte B, Schwinn MK, Defrance M, Mendez J, Murphy N, et al. TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. EMBO J 2013;32(5):645–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hardiville S, Hart GW. Nutrient regulation of gene expression by O-GlcNAcylation of chromatin. Curr Opin Chem Biol 2016;33:88–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chu CS, Lo PW, Yeh YH, Hsu PH, Peng SH, Teng YC, et al. O-GlcNAcylation regulates EZH2 protein stability and function. Proc Natl Acad Sci U S A 2014;111(4):1355–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Y, Wang L, Liu J, Zhang P, An M, Han C, et al. O-GlcNAcylation modulates Bmi-1 protein stability and potential oncogenic function in prostate cancer. Oncogene 2017;36(45):6293–305. [DOI] [PubMed] [Google Scholar]
- 30.Wu D, Cai Y, Jin J. Potential coordination role between O-GlcNAcylation and epigenetics. Protein Cell 2017;8(10):713–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hanover JA, Chen W, Bond MR. O-GlcNAc in cancer: An Oncometabolism-fueled vicious cycle. J Bioenerg Biomembr 2018. [DOI] [PubMed] [Google Scholar]
- 32.Zachara N, Molina H, Wong K, Pandey A, Hart G. The dynamic stress-induced “O-GlcNAc-ome” highlights functions for O-GlcNAc in regulating DNA damage/repair and other cellular pathways. Amino Acids 2011;40(3):793–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhong J, Martinez M, Sengupta S, Lee A, Wu X, Chaerkady R, et al. Quantitative phosphoproteomics reveals crosstalk between phosphorylation and O-GlcNAc in the DNA damage response pathway. Proteomics 2015;15(2–3):591–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Q, Yu X. OGT restrains the expansion of DNA damage signaling. Nucleic Acids Res 2016;44(19):9266–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Campbell S, Ismail IH, Young LC, Poirier GG, Hendzel MJ. Polycomb repressive complex 2 contributes to DNA double-strand break repair. Cell Cycle 2013;12(16):2675–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bracken AP, Kleine-Kohlbrecher D, Dietrich N, Pasini D, Gargiulo G, Beekman C, et al. The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev 2007;21(5):525–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Merzlyak EM, Goedhart J, Shcherbo D, Bulina ME, Shcheglov AS, Fradkov AF, et al. Bright monomeric red fluorescent protein with an extended fluorescence lifetime. Nat Methods 2007;4(7):555–7. [DOI] [PubMed] [Google Scholar]
- 38.Wolfgeher D, Dunn DM, Woodford MR, Bourboulia D, Bratslavsky G, Mollapour M, et al. The dynamic interactome of human Aha1 upon Y223 phosphorylation. Data Brief 2015;5:752–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lyon SM, Mayampurath A, Rogers MR, Wolfgeher DJ, Fisher SM, Volchenboum SL, et al. A method for whole protein isolation from human cranial bone. Anal Biochem 2016;515:33–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee SS, Bindokas VP, Kron SJ. Multiplex three-dimensional optical mapping of tumor immune microenvironment. Sci Rep 2017;7(1):17031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci USA 1995;92(20):9363–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fabregat A, Jupe S, Matthews L, Sidiropoulos K, Gillespie M, Garapati P, et al. The Reactome Pathway Knowledgebase. Nucleic Acids Res 2018;46(D1):D649–D55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res 2003;13(9):2129–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim S, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell 2011;19(1):17–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dang L, Yen K, Attar EC. IDH mutations in cancer and progress toward development of targeted therapeutics. Ann Oncol 2016;27(4):599–608. [DOI] [PubMed] [Google Scholar]
- 46.Slawson C, Copeland RJ, Hart GW. O-GlcNAc signaling: a metabolic link between diabetes and cancer? Trends Biochem Sci 2010;35(10):547–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fardini Y, Dehennaut V, Lefebvre T, Issad T. O-GlcNAcylation: A New Cancer Hallmark? Front Endocrinol 2013;4:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ma Z, Vosseller K. Cancer metabolism and elevated O-GlcNAc in oncogenic signaling. J Biol Chem 2014;289(50):34457–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu Y, Cao Y, Pan X, Shi M, Wu Q, Huang T, et al. O-GlcNAc elevation through activation of the hexosamine biosynthetic pathway enhances cancer cell chemoresistance. Cell Death Dis 2018;9(5):485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ma J, Hart GW. O-GlcNAc profiling: from proteins to proteomes. Clin Proteomics 2014;11(1):8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Groves J, Lee A, Yildirir G, Zachara N. Dynamic O-GlcNAcylation and its roles in the cellular stress response and homeostasis. Cell Stress and Chaperones 2013;18(5):535–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ferrer CM, Lynch TP, Sodi VL, Falcone JN, Schwab LP, Peacock DL, et al. O-GlcNAcylation regulates cancer metabolism and survival stress signaling via regulation of the HIF-1 pathway. Mol Cell 2014;54(5):820–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Groves JA, Maduka AO, O’Meally RN, Cole RN, Zachara NE. Fatty acid synthase inhibits the O-GlcNAcase during oxidative stress. J Biol Chem 2017;292(16):6493–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.