Abstract
Background:
Given the scarcity of cell lines from underrepresented populations, it is imperative that genetic ancestry for these cell lines is characterized. Consequences of cell line mischaracterization include squandered resources and publication retractions.
Methods:
We calculated genetic ancestry proportions for 15 cell lines to assess the accuracy of previous race/ethnicity classification and determine previously unknown estimates. DNA was extracted from cell lines and genotyped for ancestry informative markers (AIMs) representing West African (WA), Native American (NA), and European (EUR) ancestry.
Results:
Of the cell lines tested, all previously classified as White/Caucasian were accurately described with mean EUR ancestry proportions of 97%. Cell lines previously classified as Black/African American were not always accurately described. For instance, the 22Rv1 prostate cancer cell line was recently found to carry mixed genetic ancestry using a much smaller panel of markers. However, our more comprehensive analysis determined the 22Rv1 cell line carries 99% EUR ancestry. Most notably, the E006AA-hT prostate cancer cell line, classified as African American, was found to carry 92% EUR ancestry. We also determined the MDA-MB-468 breast cancer cell line carries 23% NA ancestry suggesting possible Afro-Hispanic/Latina ancestry.
Conclusions:
Our results suggest predominantly EUR ancestry for the White/Caucasian-designated cell lines, yet high variance in ancestry for the Black/African American-designated cell lines. Additionally, we revealed an extreme misclassification of the E006AA-hT cell line.
Impact:
Genetic ancestry estimates offer more sophisticated characterization leading to better contextualization of findings. Ancestry estimates should be provided for all cell lines to avoid erroneous conclusions in disparities literature.
Keywords: cell lines, ancestry informative markers, genetic ancestry, E006AA-hT, 22Rv1
INTRODUCTION
In cancer research, biological model systems are used to understand the contribution of cellular mechanisms to tissue function, disease pathogenesis, and drug efficacy (1–4). Human cell lines representing various tissues in normal and disease states are vital tools necessary to establish preclinical research models. Effects and mechanisms of genetic, epigenetic, and chemical perturbations on cellular viability are explored in vitro and in vivo through gene knockdown, knockout, and transfection assays using human cell lines (4–7). Cellular assays employing human cell lines have proven essential to determine the impact of intra- and intergenetic polymorphisms on gene expression and cellular viability (8,9). While impactful mutations altering gene expression, cell function, or cell viability have been observed, there is significant variability in reproducibility among varying cell lines (10,11). A portion of this variability is attributable to differences in genetic background – (e.g. varying polymorphisms, epigenetic signatures, and gene expression patterns) – among cell lines of the same ethnicity as well as between cell lines representing different ethnicities (12–16).
The examination of experimental outcomes using a racial/ethnic variety of cell lines exposed to the same conditions is a useful tool to elucidate genes or pathways driving the biological contribution to disease disparities (15,17–19). Since genetic tapestries differ within and between populations, it is important to include a variety of cell lines derived from individuals within and between populations in an effort to provide an accurate representation of the genetic variability (20–22). Currently, there are many commercially available cell lines representing various tissues and disease stage. However, the majority of these cell lines are from patients of European descent, and there remains a critical need to diversify the selection (23,24). As an example of the existing disparity in cellular model diversity, a recent search of the American Type Culture Collection (ATCC) website for cell lines derived from normal and malignant breast tissue revealed 59 specimens designated as Caucasian/White. Conversely, there were only 14 non-Caucasian/White cell lines consisting of 11 designated as Black, 1 designated as Hispanic, and 1 designated as East Indian.(24) Of note, 10 additional breast tissue cell lines lacked any ethnicity assignment (24).
The lack of adequate genetic population representation among human cell lines leads to a cascade of downstream consequences. It is reasonable to assume that numerous variants, genes, and pathways involved in disease pathogenesis or drug efficacy remain undiscovered when the majority of cell lines used for research are derived from patients of European descent. Since there are lower levels of variation within the European population, we miss potential genetic indicators that steer molecular mechanisms, disease outcomes, and drug response (22,25–27). Also, SNPs identified in European populations do not completely capture the genetic variation contributing to a phenotype in other racial groups (28). More disturbingly, entire demographics are ignored in key mechanistic studies hindered by the restraints monoethnic cellular models afford (29,30). This is problematic as underrepresented racial/ethnic groups, who are more likely to suffer disproportionate disease incidence and mortality, remain also underrepresented in biospecimens available for research (31–34).
Complementary to the aspiration of health disparity researchers to diversify commercially available biospecimens, the National Institutes of Health has currently established guidelines concerning the authentication of key biological resources to ensure identity and validity (35). In response to this recommendation, a recent publication has incorporated ancestry analysis using single nucleotide polymorphism (SNP) genotyping to validate previously reported ethnicities of cell lines as well as elucidate the ancestral proportions of racially unidentified cell lines (23). These efforts are important and provide the scientific community with the 1) confirmation or rejection of the ethnicity designations assigned to these cell lines; 2) prevention of wasted time and money; 3) prevention of misleading publications leading to retractions; and 3) confirmed ethnicity of previously unidentified cell lines; 4) increased variation among cell lines.
In this study, we expand upon this recent focus and include 15 human cell lines representative of various cancer types for ancestry analysis. Using a set of 105 established Ancestry Informative Markers (AIMs) validated as SNP genotypes for population structure analyses, we assign respective West African (WA), Native American (NA), and European (EUR) ancestral proportions (36,37). Key findings of our study include the high NA proportion found within the MDA-MB-468 breast cancer cell line suggesting this patient may have been an Afro-Hispanic/Latina female, although currently simply designated as Black. We also identified the 22Rv1 cell line, currently racially unidentified yet recently categorized as carrying mixed genetic ancestry in a study using 29 AIMs, as a predominantly EUR prostate cancer cell line (23). Furthermore, we conclusively establish that the E006AA-hT prostate cancer cell line, designated as African American, in fact carries majority EUR ancestry.
MATERIALS AND METHODS
Cell Culture
DU145 (ATCC, Cat. # HTB‐81), 22Rv1 (ATCC, Cat. # CRL‐2505), and HeLa (ATCC, Cat. # CRM-CCL-2) cell lines were purchased from American Type Culture Collection (ATCC) and grown in a humidified incubator with 5% CO2 at 37°C in R.A. Kittles’ laboratory. Cells were routinely tested for mycoplasma contamination using PCR Mycoplasma Detection Kit (ABM, Cat. # G238, Richmond, BC). Cell lines were cultured in RPMI 1640 medium (ATCC, Cat. # 30–2001, Manassas, VA) supplemented with 10% fetal bovine serum (ATCC, Cat. # 30–2020, Manassas, VA), penicillin‐streptomycin (Gibco, Cat. # 15140122, Gaithersburg, MD), and gentamicin (Fisher Scientific, Cat. # 15710064, Carlsbad, CA) as recommended by the supplier. 0.2% Normocin (Invivogen, Cat. # ANT‐NR‐1, San Diego, CA) was added to the medium to prevent contamination by mycoplasma, bacteria, or fungi. In accordance with NIH guidelines concerning the authentication of key biological resources and in order to ensure the identity and validity of the resource, cell lines cultured in this study were purchased from ATCC that performs cell line characterizations. DNA was extracted from these cell lines within 2 weeks from receipt and not exceeding 3 passages. DNA extracted from the RWPE1 cell line was provided by L. Nonn. DNA extracted from the E006AA-hT cell line were tested from 3 separate sources, including an original purchase from ATCC, and were kindly provided by S. Ambs, A. Sreekumar, and S. Patierno. DNA extracted from the HCC1500, HCC1806, MCF-10A, MDA-MB-453, MDA-MB-468, T-47D cell lines were provided by S. Kimbro. DNA extracted from the MDA-PCa-2b cell line was provided by S. Lloyd. DNA extracted from the RC-77T/E, LNCaP, and PC3 cell lines were provided by R. Mitra.
SNP Genotyping
Single nucleotide polymorphisms (SNPs) that were previously identified and validated for estimating continental ancestry information in admixed populations were selected to identify the Ancestry Informative Markers (AIMs) (36–38). The AIMs panel consisted of 105 SNPs and were genotyped using the Sequenom MassARRAY genotyping platform with iPLEX chemistry according to manufacturer’s recommendations. iPLEX assays were designed utilizing the Sequenom Assay Design software, allowing for single base extension designs used for multiplexing. DNA was isolated from the cell lines and multiplex assays were performed to amplify 10 ng of genomic DNA by polymerase chain reaction (PCR). PCR reactions were treated with shrimp alkaline phosphatase (SAP) enzyme to neutralize the unincorporated deoxyribonucleotide triphosphate (dNTPs). A post-PCR single base extension reaction was performed for each multiplex reaction using concentrations of 0.625 µM for low mass primers and 1.25 µM for high mass primers. Reactions were diluted with 16 µl of H2O and fragments were purified with resin, spotted onto Sequenom SpectroCHIP microarrays (Agena Bioscience, Product 10500, San Diego, CA) and scanned by MALDI-TOF mass spectrometry. Individual SNP genotype calls were generated using Sequenom TYPER software, which automatically calls allele specific peaks according to their expected masses. A genotype concordance rate of 99% was observed for all markers. Genotyping call rates that exceeded 98.5% were included in the analyses.
DNA Ancestry Analysis
Individual admixture estimates for each cell line were calculated using a model-based clustering method as implemented in the program STRUCTURE v2.3 (39). STRUCTURE 2.3 was run using parental population genotypes from West Africans, Europeans, and Native Americans (36) under the Admixture model using the Bayesian Markov chain Monte Carlo method and a burn-in length of 30,000 for 70,000 repetitions. Since we are unsure about the ancestries of our cell line samples we used the admixture model to determine which estimation of K (number of sub populations) is the best fit for the data. We set K from 2 to 5 and ran 100 iterations. We determined that K=3 had the best fit. We ran used the K=3 estimates for our analyses.
Multidimensional Scaling Analysis
Multidimensional scaling (MDS) analysis was performed to visualize the genetic similarity of the cell lines to worldwide populations. 1000 genomes variant data was downloaded and 824 individuals representing 8 groups were selected to represent worldwide populations--Mende in Sierra Leone (MSL); Luhya in Webuye, Kenya (LWK); Toscani in Italia (TSI); British in England and Scotland (GBR); Indian Telugu in the UK (ITU); Han Chinese in Beijing, China (CHB); Chinese Dai in Xishuangbanna, China (CDX); and Gujarati Indian in Houston, TX (GIH) (40). Markers matching the 105 AIMs were then selected for the MDS analysis. Individuals were removed with missingness > 0.05. Markers were removed with missing genotypes > 0.05 or minor allele frequency < 0.05. Though sparsely located across the genome, PLINK software was still used to remove markers in linkage disequilibrium using an r2 greater than 0.5 in a 50 SNP window with a 5 SNP sliding window in the combined cell line and 1KG variant data (41). PLINK software was also used to perform the MDS analysis on the remaining 87 markers.
RESULTS
105 AIMs were selected from a larger previously validated set of markers to define critical genome candidate regions and characterize samples from diverse population groups (36,37). This subset of AIMs contains specific SNPs capable of distinguishing WA, NA, and EUR genetic ancestry. Our ancestry estimates suggest cell lines previously classified as White/Caucasian by ATCC were accurately described, with mean EUR ancestry proportions of 97% (range 92–99%) (Table 1). MCF-10A and MDA-MB-453 breast cancer cell lines were found to carry 97% and 99% EUR ancestry, respectively. PC3, DU145, LNCaP, and RWPE1 prostate cancer cell lines were found to carry 98%, 99%, 92%, and 96% EUR ancestry, respectively.
Table 1.
Ancestral proportions of commonly used human cancer cell lines
| Cell Line | ATCC Catalog No. | Gender | Age | Tissue | Designated Race/Ethnicity | WA | NA | EUR | Putative Ancestry |
|---|---|---|---|---|---|---|---|---|---|
| PC3 | CRL-1435 | M | 62 | Prostate Cancer | White/EA | 0.3% | 0.4% | 99.3% | EA |
| DU145 | HTB-81 | M | 69 | Prostate Cancer | White/EA | 0.4% | 0.5% | 99.1% | EA |
| LNCaP | CRL-1740 | M | 50 | Prostate Cancer | White/EA | 6.5% | 1.9% | 91.7% | EA |
| RWPE1 | CRL-11609 | M | 54 | Prostate Cancer | White/EA | 2.2% | 1.7% | 96.1% | EA |
| 22Rv1 | CRL-2505 | M | N/A | Prostate Cancer | N/A | 0.3% | 0.4% | 99.3% | EA |
| E006AA-hT | CRL-3277 | M | 50 | Prostate Cancer* | Black/AA | 1.3% | 7.4% | 91.3% | EA |
| MDA-PCa-2b | CRL-2422 | M | 63 | Prostate Cancer | Black/AA | 86.5% | 3.0% | 10.6% | AA |
| RC-77T/E | N/A | M | 63 | Prostate Cancer | Black/AA | 88.9% | 0.4% | 10.7% | AA |
| HeLa | CCL-2 | F | 31 | Cervical Cancer | Black/AA | 66.5% | 0.4% | 33.1% | AA |
| MCF-10A | CRL-10317 | F | 36 | Breast Disease | White/EA | 0.9% | 2.4% | 96.7% | EA |
| MDA-MB-453 | HBT-131 | F | 48 | Breast Cancer | White/EA | 0.2% | 0.4% | 99.4% | EA |
| HCC1500 | CRL-2329 | F | 32 | Breast Cancer | Black/AA | 78.8% | 0.5% | 20.7% | AA |
| HCC1806 | CRL-2335 | F | 60 | Breast Cancer | Black/AA | 80.2% | 4.4% | 15.4% | AA |
| MDA-MB-468 | HTB-132 | F | 51 | Breast Cancer | Black/AA | 77.1% | 22.7% | 0.2% | AA/HA |
| T-47D | HTB-133 | F | 54 | Breast Cancer | N/A | 0.1% | 0.3% | 99.7% | EA |
E006AA-hT is sold by ATCC as a prostate cancer cell line; however, ATCC acknowledges that the STR profile of this cell line is an 86% match to the 786-O renal cell carcinoma cell line (44).
F, female; M, male; AA, African American; EA, European American; HA, Hispanic American; N/A, not available; No., number; WA, West African; NA, Native American; EUR, European
Similarly, results of some ancestry estimates for cell lines previously classified as Black/African American were accurately described. For example, the MDA-PCa-2b prostate cancer cell line was found to carry 86% WA ancestry (Table 1). Likewise, the HCC1500, HCC1806, and MDA-MB-468 breast cancer cell lines were found to carry 79%, 80%, and 77% WA ancestry, respectively. Interestingly, the MDA-MB-468 breast cancer cell line was also found to carry an appreciable amount of NA ancestry (23%) suggesting possible Afro-Hispanic/Latina ancestry. While the HeLa cervical cancer cell line was found to carry majority WA ancestry (66%), the WA proportion falls below the mean of ~80% WA ancestry typically observed in U.S.-born African Americans.
There are several cell lines that were included in our study that do not have racial identifiers specified by ATCC. For example, T-47D is a breast cancer cell line previously racially unidentified. Our ancestry analysis revealed that T-47D carries 100% EUR ancestry (Table 1). Similarly, the racial identification of the 22Rv1 prostate cancer cell line was never included within the biological characteristics specified when originally derived (42,43). For this reason, no racial identifier has been available through ATCC (23,24). While a recent publication found this cell line to carry mixed genetic ancestry, our ancestry analysis revealed a majority EUR ancestry (99%) (23).
Our genetic ancestry analysis of the E006AA-hT prostate cancer cell line revealed an extreme misclassification of racial identity compared to what has been reported in the literature as well as what is described by ATCC (24,44). While this cell line is sold commercially as an African American prostate cancer cell line, ATCC provides a disclaimer within the “Characteristics” section stating “during the accessioning of this line ATCC ran an STR profile for the original starting material. The results match the characterization data in the cited references, however the STR profile was also found to match the STR profile (an 86% match) of another ATCC cell line, 786-O, a cell line derived from a renal cell carcinoma. The originating laboratory did not use the ATCC cell line, 786-O” (45). Due to the perplexity surrounding the true classification of this cell line, we included 3 separate DNA extraction samples provided by 3 separate laboratory groups of the E006AA-hT cell line. The SNP genotyping was found to be identical in all 3 E006AA-hT samples, revealing this cell line carries 91% EUR ancestry (Table 1).
We also included samples in our study of cell lines that are commonly used for research but are not commercially available. For example, the RC-77T/E prostate cancer cell line was developed from an African American prostate cancer patient and we confirmed the WA ancestral proportions of this cell lines at 89% (46).
As an additional method to verify the accuracy of cell line genetic ancestry characterization, MDS analysis was performed to visualize the genetic similarity of the cell lines against worldwide populations from the 1000 genomes project (Figure 1). As expected, when plotting the cell lines and 1KG groups using the first two MDS dimensions, cell lines with predominantly EUR ancestry clustered with individuals from the British in England and Scotland (GBR) and Toscani in Italia (TSI) groups and admixed cell lines with predominantly WA ancestry clustered near the Mende in Sierra Leone (MSL) and Luhya in Webuye, Kenya (LWK) groups. The first two MDS dimensions provide evidence that E006AA-hT is a cell line of predominantly European descent, due to its clustering with the European 1KG individuals, and the MDA-MB-468 cell line as possibly of Afro-Hispanic/Latina descent, due to its position on the axis between the West African and East Asian groups (a proxy for NA ancestry). As the MDA-MB-468 and HeLa cell lines are more heavily admixed their genetic similarity lies between the East Asian to WA axis and between the EUR to WA axis, respectively.
Figure 1. Genetic similarity of cell lines with worldwide populations.
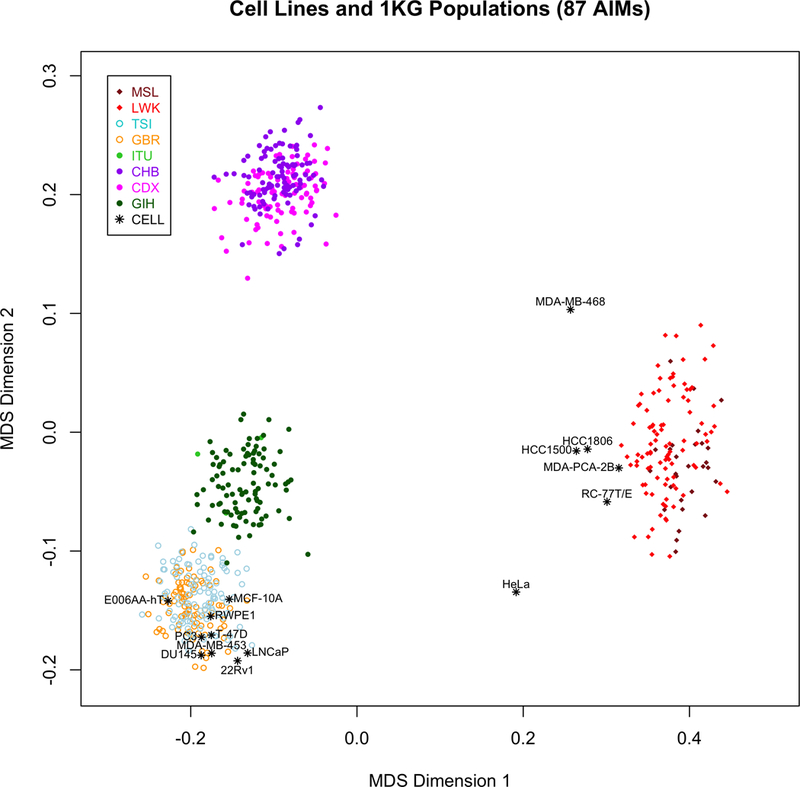
Multidimensional scaling (MDS) analysis was used to determine the genetic similarity of the cell lines to groups in the 1000 genomes (1KG) project. 87 markers were used to generate MDS dimensions for 15 cell lines and 824 1KG individuals. Cell lines and 1KG groups are color-coded. Here, the East Asian group serves as a proxy for Native American groups. Abbreviations: MSL, Mende in Sierra Leone; LWK, Luhya in Webuye, Kenya; TSI, Toscani in Italia; GBR, British in England and Scotland; ITU, Indian Telugu in the UK; CHB, Han Chinese in Beijing, China; CDX, Chinese Dai in Xishuangbanna, China; GIH, Gujarati Indian in Houston, TX; and CELL, a human cancer cell line.
DISCUSSION
In order to promote meaningful and quality health disparities research, there has been a recent interest in incorporating increased racial diversity among human cell lines (17,19,23). Racial classification remains extremely useful for describing general patterns of health as most data are reported by self-identified race (47). However, we recognize that race also embodies social and cultural constructs, and most commonly used human cell lines were developed at a time when self-reported race was considered a sufficient demographic detail (47). As the role of biological determinants in disease acquisition and progression becomes better defined, we cannot undercut the importance of individual genetic background (48–50). Since the use of cell lines is necessary to elucidate genes and pathways driving disease disparities, it is imperative to ascertain the accurate genetic ancestry of cell lines used in pre-clinical research in order to adequately explore the impact of genetic contributions on incidence and progression. Recognizing the importance of precise genetic assignment for research biospecimens, we sought to confirm or negate the current racial identification of commonly used cell lines, provide accurate and robust global ancestry estimates for cell lines from admixed individuals, while also revealing the genetic ancestry of previously racially unidentified cell lines.
Overall, our ancestry analysis mostly confirmed the racial classifications previously assigned to cell lines used in this study. In other words, most cell lines classified as “Caucasian/White” carried majority EUR genetic ancestry while most cell lines classified as “African American/Black” carried majority WA genetic ancestry. However, a few key findings from our study refuted what has been reported in the literature and/or by ATCC. Most erroneously, we found E006AA-hT to carry 91% EUR genetic ancestry. This finding is problematic as E006AA-hT is currently marketed and commercially available as an “African American” prostate cancer cell line (24). While ATCC provides a disclaimer that E006AA-hT matches the STR profiling of a renal cell carcinoma cell line, 786-O, there is no indication provided that the racial identifier of “African American” is not accurate (45). Because of the increasing mistrust of the E006AA-hT cell line we have observed in our interactions with others within the prostate cancer research community, we sought to include multiple samples of this cell line from 3 different laboratories located in distant geographical locations to firmly establish the true genetic ancestry. The implications of our finding are considerable as many laboratories have published or are in the process of publishing manuscripts exploring prostate cancer health disparities incorporating E006AA-hT as an African American prostate cancer cell line when, in fact, the cell line is neither African American nor likely derived from the prostate. The revelation of E006AA-hT as carrying predominant EUR ancestry is further disappointing as the field of prostate cancer health disparities research is already hindered by a lack of commercially available African American/Black cell lines, and this new EUR ancestral assignment leaves MDA-PCa-2b as the sole commercially available Black prostate cell line (23,24).
Our study also highlights the advantage of utilizing an ample number of AIMs when conducting ancestry analysis. Recently, the 22Rv1 prostate cancer cell was found to carry mixed genetic ancestry using a smaller subset of 29 AIMs (23,36). While subsets of AIMs as small as 24 have been shown to be useful tools for ascertaining the origin of subjects from particular continents and correct for population stratification in admixed population sample sets, we suggest a more comprehensive subset of at least 100 AIMs as a large decrease in EUR performance has been observed in marker sets smaller than 64 (36). Using a validated set of 105 AIMs, we found 22Rv1 to carry 99% EUR ancestry. Since no racial identifier has ever been assigned to the 22Rv1 cell line, our genetic ancestry results clarify the ambiguity.
Given the high heterogeneity of African Americans, it is imperative to tease out the WA ancestral proportions of individual cell lines (51). We are the first to report that the commonly used HeLa cell line derived from African American cervical cancer patient Henrietta Lacks, carries 66% WA ancestry (52). The average African American carries approximately 80% WA genetic ancestry, yet historically, African Americans have been likely to self-identify as Black regardless of how much or little European background they may possess (53). This self-identification as Black has followed the rule of hypodescent, under which any amount of Black ancestry warrants an association as African American (54,55). While the social and cultural influences of race on disease are undeniable, the role of genetics cannot be ignored. For example, an admixed individual with 20% EUR genetic ancestry may be at greater or lesser risk of certain diseases than an individual with 40% EUR genetic ancestry (56–60). In addition, the implications for pharmacogenomics exist as the nuances of drug effects or mechanisms may not be generalizable in the African American population due to high WA genetic variance (28,61). For these reasons, there remains a burden on the scientific community to expand the collection of currently available human cell lines by incorporating more non-European options to capture the complete picture of genetic contribution to disease incidence, aggressiveness, progression, and response to treatment.
The need for increased diversity in research biospecimens is glaringly obvious when noting the near complete lack of commercially available Hispanic/Latino cell lines (24). In our own ancestry analysis of the cell lines included in this study, we observed very low proportions of NA ancestry in all cell lines except the MDA-MB-468 breast cancer cell line. While the MDA-MB468 cell line is reported as Black, we found that it carries 77% WA ancestry and 23% NA ancestry (24). Based on these genetic ancestry proportions, it is reasonable to presume that the breast cancer patient from whom this cell line was derived may have been Afro-Hispanic/Latina as recent studies have highlighted this admixture proportion within a Hispanic-Caribbean population (62,63). Thus, our genetic ancestry analysis may have uncovered an additional cell line with which to measure the impact of NA ancestry on breast cancer disease incidence, progression, and treatment.
As we progress further into an era of personalized medicine, the importance of racially diverse cell lines will grow clearer. While it is imperative that research biospecimens are designated accurately in terms of race/ethnicity, it is crucial that they be characterized globally and locally for their genetic ancestry so that findings can be properly contextualized for the representative populations. In the future, it would be ideal for commercial companies to report these global and local findings. Additionally, genetic distance mapping of cell lines with the 1000 genomes or Human Genome Diversity Panel populations, using dimensional reduction techniques such as MDS or principal component analysis, should be performed to further determine the race/ethnicity of cell lines through their clustering with worldwide populations (40,64). These techniques should further avoid misclassification that could occur with relying solely on AIMs designed to discern genetic ancestry proportions for a few discrete ancestral populations. We intend for the results of this study to encourage the scientific community to pursue ancestry analysis of additional cell lines as well as develop a wider range of diverse biospecimens.
Acknowledgments
Financial Support: National Institutes of Health, Grant Numbers: 1R01MD007105 (RAK); 1T32CA186895 (LWB); U01CA167234 (SL); U54MD012392–02 and P20MD000175–15 (KSK).
Footnotes
Conflict of Interest Disclosure Statement: The authors have no conflicts of interest to declare.
REFERENCES
- 1.Grandori C, Kemp CJ. Personalized Cancer Models for Target Discovery and Precision Medicine. Trends in cancer 2018;4(9):634–42 doi 10.1016/j.trecan.2018.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Galuschka C, Proynova R, Roth B, Augustin HG, Muller-Decker K. Models in Translational Oncology: A Public Resource Database for Preclinical Cancer Research. Cancer research 2017;77(10):2557–63 doi 10.1158/0008-5472.Can-16-3099. [DOI] [PubMed] [Google Scholar]
- 3.Kim HS, Sung YJ, Paik S. Cancer Cell Line Panels Empower Genomics-Based Discovery of Precision Cancer Medicine. Yonsei Med J 2015;56(5):1186–98 doi 10.3349/ymj.2015.56.5.1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144(5):646–74 doi 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 5.Shearer RF, Saunders DN. Experimental design for stable genetic manipulation in mammalian cell lines: lentivirus and alternatives. Genes to cells : devoted to molecular & cellular mechanisms 2015;20(1):1–10 doi 10.1111/gtc.12183. [DOI] [PubMed] [Google Scholar]
- 6.Miest T, Saenz D, Meehan A, Llano M, Poeschla EM. Intensive RNAi with lentiviral vectors in mammalian cells. Methods (San Diego, Calif) 2009;47(4):298–303 doi 10.1016/j.ymeth.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim TK, Eberwine JH. Mammalian cell transfection: the present and the future. Analytical and bioanalytical chemistry 2010;397(8):3173–8 doi 10.1007/s00216-010-3821-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fang X, Li X, Yin Z, Xia L, Quan X, Zhao Y, et al. Genetic variation at the microRNA binding site of CAV1 gene is associated with lung cancer susceptibility. Oncotarget 2017;8(54):92943–54 doi 10.18632/oncotarget.21687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dansonka-Mieszkowska A, Szafron LM, Moes-Sosnowska J, Kulinczak M, Balcerak A, Konopka B, et al. Clinical importance of the EMSY gene expression and polymorphisms in ovarian cancer. Oncotarget 2018;9(25):17735–55 doi 10.18632/oncotarget.24878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freedman LP, Gibson MC, Ethier SP, Soule HR, Neve RM, Reid YA. Reproducibility: changing the policies and culture of cell line authentication. Nature methods 2015;12(6):493–7 doi 10.1038/nmeth.3403. [DOI] [PubMed] [Google Scholar]
- 11.Eisner DA. Reproducibility of science: Fraud, impact factors and carelessness. Journal of molecular and cellular cardiology 2018;114:364–8 doi 10.1016/j.yjmcc.2017.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cajigas-Du Ross CK, Martinez SR, Woods-Burnham L, Duran AM, Roy S, Basu A, et al. RNA sequencing reveals upregulation of a transcriptomic program associated with stemness in metastatic prostate cancer cells selected for taxane resistance. Oncotarget 2018;9(54):30363–84 doi 10.18632/oncotarget.25744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barrett CS, Millena AC, Khan SA. TGF-beta Effects on Prostate Cancer Cell Migration and Invasion Require FosB. The Prostate 2017;77(1):72–81 doi 10.1002/pros.23250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu X, Chen X, Rycaj K, Chao HP, Deng Q, Jeter C, et al. Systematic dissection of phenotypic, functional, and tumorigenic heterogeneity of human prostate cancer cells. Oncotarget 2015;6(27):23959–86 doi 10.18632/oncotarget.4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shiina M, Hashimoto Y, Kato T, Yamamura S, Tanaka Y, Majid S, et al. Differential expression of miR-34b and androgen receptor pathway regulate prostate cancer aggressiveness between African-Americans and Caucasians. Oncotarget 2017;8(5):8356–68 doi 10.18632/oncotarget.14198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Teslow EA, Bao B, Dyson G, Legendre C, Mitrea C, Sakr W, et al. Exogenous IL-6 induces mRNA splice variant MBD2_v2 to promote stemness in TP53 wild-type, African American PCa cells. Molecular oncology 2018;12(7):1138–52 doi 10.1002/1878-0261.12316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Panigrahi GK, Praharaj PP, Peak TC, Long J, Singh R, Rhim JS, et al. Hypoxia-induced exosome secretion promotes survival of African-American and Caucasian prostate cancer cells. Sci Rep 2018;8(1):3853 doi 10.1038/s41598-018-22068-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanchez TW, Zhang G, Li J, Dai L, Mirshahidi S, Wall NR, et al. Immunoseroproteomic Profiling in African American Men with Prostate Cancer: Evidence for an Autoantibody Response to Glycolysis and Plasminogen-Associated Proteins. Mol Cell Proteomics 2016;15(12):3564–80 doi 10.1074/mcp.M116.060244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woods-Burnham L, Cajigas-Du Ross CK, Love A, Basu A, Sanchez-Hernandez ES, Martinez SR, et al. Glucocorticoids induce stress oncoproteins associated with therapy-resistance in African American and European American prostate cancer cells. Sci Rep 2018. doi 10.1038/s41598-018-33150-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shriver MD, Mei R, Parra EJ, Sonpar V, Halder I, Tishkoff SA, et al. Large-scale SNP analysis reveals clustered and continuous patterns of human genetic variation. Human genomics 2005;2(2):81–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manojlovic Z, Christofferson A, Liang WS, Aldrich J, Washington M, Wong S, et al. Comprehensive molecular profiling of 718 Multiple Myelomas reveals significant differences in mutation frequencies between African and European descent cases. PLoS genetics 2017;13(11):e1007087 doi 10.1371/journal.pgen.1007087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Campbell MC, Tishkoff SA. African genetic diversity: implications for human demographic history, modern human origins, and complex disease mapping. Annual review of genomics and human genetics 2008;9:403–33 doi 10.1146/annurev.genom.9.081307.164258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Woods-Burnham L, Basu A, Cajigas-Du Ross CK, Love A, Yates C, De Leon M, et al. The 22Rv1 prostate cancer cell line carries mixed genetic ancestry: Implications for prostate cancer health disparities research using pre-clinical models. The Prostate 2017;77(16):1601–8 doi 10.1002/pros.23437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.American Type Culture Collection 2018.
- 25.Brockmoller J, Tzvetkov MV. Pharmacogenetics: data, concepts and tools to improve drug discovery and drug treatment. European journal of clinical pharmacology 2008;64(2):133–57 doi 10.1007/s00228-007-0424-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jorde LB, Watkins WS, Bamshad MJ, Dixon ME, Ricker CE, Seielstad MT, et al. The distribution of human genetic diversity: a comparison of mitochondrial, autosomal, and Y-chromosome data. American journal of human genetics 2000;66(3):979–88 doi 10.1086/302825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee SS. Racializing drug design: implications of pharmacogenomics for health disparities. Am J Public Health 2005;95(12):2133–8 doi 10.2105/ajph.2005.068676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hernandez W, Gamazon ER, Aquino-Michaels K, Patel S, O’Brien TJ, Harralson AF, et al. Ethnicity-specific pharmacogenetics: the case of warfarin in African Americans. The pharmacogenomics journal 2014;14(3):223–8 doi 10.1038/tpj.2013.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mosher JT, Pemberton TJ, Harter K, Wang C, Buzbas EO, Dvorak P, et al. Lack of population diversity in commonly used human embryonic stem-cell lines. N Engl J Med 2010;362(2):183–5 doi 10.1056/NEJMc0910371. [DOI] [PubMed] [Google Scholar]
- 30.Tofoli FA, Dasso M, Morato-Marques M, Nunes K, Pereira LA, da Silva GS, et al. Increasing The Genetic Admixture of Available Lines of Human Pluripotent Stem Cells. Sci Rep 2016;6:34699 doi 10.1038/srep34699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hagiwara N, Berry-Bobovski L, Francis C, Ramsey L, Chapman RA, Albrecht TL. Unexpected findings in the exploration of African American underrepresentation in biospecimen collection and biobanks. J Cancer Educ 2014;29(3):580–7 doi 10.1007/s13187-013-0586-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA: a cancer journal for clinicians 2018;68(1):7–30 doi 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 33.Ragin C, Park JY. Biospecimens, biobanking and global cancer research collaborations. Ecancermedicalscience 2014;8:454 doi 10.3332/ecancer.2014.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singh GK, Jemal A. Socioeconomic and Racial/Ethnic Disparities in Cancer Mortality, Incidence, and Survival in the United States, 1950–2014: Over Six Decades of Changing Patterns and Widening Inequalities. J Environ Public Health 2017;2017:2819372 doi 10.1155/2017/2819372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.National Institutes of Health. Implementing rigor and transparency in NIH and AHRQ Research Grant Applications October 9, 2015;NOT-OD-16–011. [Google Scholar]
- 36.Kosoy R, Nassir R, Tian C, White PA, Butler LM, Silva G, et al. Ancestry informative marker sets for determining continental origin and admixture proportions in common populations in America. Human mutation 2009;30(1):69–78 doi 10.1002/humu.20822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nassir R, Kosoy R, Tian C, White PA, Butler LM, Silva G, et al. An ancestry informative marker set for determining continental origin: validation and extension using human genome diversity panels. BMC genetics 2009;10:39 doi 10.1186/1471-2156-10-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Torres JB, Stone AC, Kittles R. An anthropological genetic perspective on Creolization in the Anglophone Caribbean. American journal of physical anthropology 2013;151(1):135–43 doi 10.1002/ajpa.22261. [DOI] [PubMed] [Google Scholar]
- 39.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 2003;164(4):1567–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA, et al. A map of human genome variation from population-scale sequencing. Nature 2010;467(7319):1061–73 doi 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience 2015;4:7 doi 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sramkoski RM, Pretlow TG 2nd, Giaconia JM, Pretlow TPS, Sy MS, et al. A new human prostate carcinoma cell line, 22Rv1. In Vitro Cell Dev Biol Anim 1999;35(7):403–9 doi 10.1007/s11626-999-0115-4. [DOI] [PubMed] [Google Scholar]
- 43.Pretlow TG, Wolman SR, Micale MA, Pelley RJ, Kursh ED, Resnick MI, et al. Xenografts of primary human prostatic carcinoma. Journal of the National Cancer Institute 1993;85(5):394–8. [DOI] [PubMed] [Google Scholar]
- 44.Koochekpour S, Willard SS, Shourideh M, Ali S, Liu C, Azabdaftari G, et al. Establishment and characterization of a highly tumorigenic African American prostate cancer cell line, E006AA-hT. International journal of biological sciences 2014;10(8):834–45 doi 10.7150/ijbs.9406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.American Type Culture Collection E006AA-hT 2018. [Google Scholar]
- 46.Theodore S, Sharp S, Zhou J, Turner T, Li H, Miki J, et al. Establishment and characterization of a pair of non-malignant and malignant tumor derived cell lines from an African American prostate cancer patient. Int J Oncol 2010;37(6):1477–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.DeSantis CE, Siegel RL, Sauer AG, Miller KD, Fedewa SA, Alcaraz KI, et al. Cancer statistics for African Americans, 2016: Progress and opportunities in reducing racial disparities. CA: a cancer journal for clinicians 2016;66(4):290–308 doi 10.3322/caac.21340. [DOI] [PubMed] [Google Scholar]
- 48.Bird A Genetic determinants of the epigenome in development and cancer. Swiss medical weekly 2017;147:w14523 doi 10.4414/smw.2017.14523. [DOI] [PubMed] [Google Scholar]
- 49.Ozdemir BC, Dotto GP. Racial Differences in Cancer Susceptibility and Survival: More Than the Color of the Skin? Trends in cancer 2017;3(3):181–97 doi 10.1016/j.trecan.2017.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jing L, Su L, Ring BZ. Ethnic background and genetic variation in the evaluation of cancer risk: a systematic review. PLoS One 2014;9(6):e97522 doi 10.1371/journal.pone.0097522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Halder I, Yang BZ, Kranzler HR, Stein MB, Shriver MD, Gelernter J. Measurement of admixture proportions and description of admixture structure in different U.S. populations. Human mutation 2009;30(9):1299–309 doi 10.1002/humu.21045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.C BJ. HeLa (for Henrietta Lacks). Science 1974;184(4143):1268 doi 10.1126/science.184.4143.1268. [DOI] [PubMed] [Google Scholar]
- 53.Zakharia F, Basu A, Absher D, Assimes TL, Go AS, Hlatky MA, et al. Characterizing the admixed African ancestry of African Americans. Genome Biol 2009;10(12):R141 doi 10.1186/gb-2009-10-12-r141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hollinger David A.. The One Drop Rule and the One Hate Rule. American Academy of Arts and Sciences, 2005;134(1):18–28. [Google Scholar]
- 55.National Research Council (US) Panel on Race E, and Health in Later Life,. Critical Perspectives on Racial and Ethnic Differences in Health in Late Life National Academies Press; (US), 2004;Racial and Ethnic Identification. [PubMed] [Google Scholar]
- 56.Fejerman L, John EM, Huntsman S, Beckman K, Choudhry S, Perez-Stable E, et al. Genetic ancestry and risk of breast cancer among U.S. Latinas. Cancer research 2008;68(23):9723–8 doi 10.1158/0008-5472.Can-08-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Al-Alem U, Rauscher G, Shah E, Batai K, Mahmoud A, Beisner E, et al. Association of genetic ancestry with breast cancer in ethnically diverse women from Chicago. PloS one 2014;9(11):e112916 doi 10.1371/journal.pone.0112916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Klimentidis YC, Arora A, Zhou J, Kittles R, Allison DB. The Genetic Contribution of West-African Ancestry to Protection against Central Obesity in African-American Men but Not Women: Results from the ARIC and MESA Studies. Frontiers in genetics 2016;7:89 doi 10.3389/fgene.2016.00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cappetta M, Berdasco M, Hochmann J, Bonilla C, Sans M, Hidalgo PC, et al. Effect of genetic ancestry on leukocyte global DNA methylation in cancer patients. BMC cancer 2015;15:434 doi 10.1186/s12885-015-1461-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bress A, Kittles R, Wing C, Hooker SE, Jr., King A. Genetic ancestry as an effect modifier of naltrexone in smoking cessation among African Americans: an analysis of a randomized controlled trial. Pharmacogenetics and genomics 2015;25(6):305–12 doi 10.1097/fpc.0000000000000138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kaye JB, Schultz LE, Steiner HE, Kittles RA, Cavallari LH, Karnes JH. Warfarin Pharmacogenomics in Diverse Populations. Pharmacotherapy 2017;37(9):1150–63 doi 10.1002/phar.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Irizarry-Ramirez M, Kittles RA, Wang X, Salgado-Montilla J, Nogueras-Gonzalez GM, Sanchez-Ortiz R, et al. Genetic ancestry and prostate cancer susceptibility SNPs in Puerto Rican and African American men. The Prostate 2017;77(10):1118–27 doi 10.1002/pros.23368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Via M, Gignoux CR, Roth LA, Fejerman L, Galanter J, Choudhry S, et al. History shaped the geographic distribution of genomic admixture on the island of Puerto Rico. PLoS One 2011;6(1):e16513 doi 10.1371/journal.pone.0016513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cann HM, de Toma C, Cazes L, Legrand MF, Morel V, Piouffre L, et al. A human genome diversity cell line panel. Science (New York, NY) 2002;296(5566):261–2. [DOI] [PubMed] [Google Scholar]


