Abstract
Background
The brown planthopper (Nilaparvata lugens Stål; BPH), one of the most destructive pests of rice, has proven to be a substantial threat, conferring enormous production losses in Asia and becoming a difficult challenge to manipulate and control under field conditions. The continuous use of insecticides promotes the resurgence of BPH, which results in resistant varieties adapting through the upgrading of new BPH biotypes. To overcome resistance acquired by BPH against resistance varieties, different forms of novel resistant gene fusions act as functional domains for breeding to enhance insect resistance.
Results
The current study reports on the novel BPH resistance gene Bph36 derived from two introgression lines (RBPH16 and RBPH17) developed from wild rice GX2183 which was previously reported to be resistant to BPH. Using two F2 crossing populations (Kangwenqizhan × RBPH16 and Huanghuazhan × RBPH17) in a bulked segregant analysis (BSA) for identification of resistant genes and QTL analysis, two QTLs for BPH resistance were generated on the long and short arms of chromosome 4, which was further confirmed by developing BC1F2:3 populations by backcrossing via marker assisted selection (MAS) approach. One BPH resistance locus on the short arm of chromosome 4 was mapped to a 38-kb interval flanked by InDel markers S13 and X48, and then was named Bph36, whereas another locus on the long arm of chromosome 4 was also detected in an interval flanked by RM16766 and RM17033, which was the same as that of Bph27. An evaluation analysis based on four parameters (BPH host selection, honeydew weight, BPH survival rate and BPH population growth rate) shows that Bph36 conferred high levels antibiosis and antixenosis to BPH. Moreover, Bph36 pyramided with Bph3, Bph27, and Bph29 through MAS into elite cultivars 9311 and MH511 (harbored Xa23), creating different background breeding lines that also exhibited strong resistance to BPH in the seedling or tillering stage.
Conclusion
Bph36 can be utilized in BPH resistance breeding programs to develop high resistant rice lines and the high-resolution fine mapping will facilitate further map-based cloning and marker-assisted gene pyramiding of resistant gene. MAS exploited to pyramid with Bph3, Bph27, Bph29, and Xa23 was confirmed the effectiveness for BPH resistance breeding in rice and provided insights into the molecular mechanism of defense to control this devastating insect.
Electronic supplementary material
The online version of this article (10.1186/s12284-019-0289-7) contains supplementary material, which is available to authorized users.
Keywords: Oryza rufipogon Griff, Brown planthopper (Nilaparvata lugens Stål), Resistance gene, Gene mapping, Marker-assisted selection, Near isogenic line, Pyramided line
Background
The brown planthopper (Nilaparvata lugens Stål; BPH) is the most destructive insect pest of rice found throughout tropical Asia where rice is widely planted (Cha et al. 2008). The BPH causes direct damage to rice by sucking the plant’s sap and indirect damage by transmitting viral diseases such as grassy and ragged stunt viruses, resulting in severe yield reduction or complete crop losses referred as a result of the “hopper burn” phenomenon (Rivera et al. 1966; Ling et al. 1978; Sōgawa 1982; Normile 2008). According to the Statistics of China Agriculture Yearbook, over 25 million hectares of rice were infested with rice planthoppers in each year of 2005–2007. To manage the pest, the extensive use of chemical pesticides has generally worked well over the past decades, but the long term application of pesticides resulted in increased BPH resistance to chemical insecticides and presents the risk of destroying the environment (Heinrichs et al. 1982; Tanaka et al. 2000). However, ensuring the genetic resistance of host plants is the most effective and environmentally-friendly approach for the BPH management. Therefore, it is very necessary and imperative to detect more novel resistant genes and to then elucidate the resistance mechanism.
Thirty- five major BPH-resistance genes have been identified from cultivated rice and wild Oryza species (Wang et al. 2018). Wild rice species and their introgression lines have been progressively used as resistance harbors, and almost half of BPH resistance genes are derived from them (Ali and Chowdhury, 2014; Hu et al., 2016a, b). For example, resistance genes Bph10 and Bph18(t) were detected from introgression lines of O. australiensis with the EE genome (Ishii et al. 1994; Jena et al. 2006). Resistance genes bph11(t), bph12(t), Bph13(t), Bph14, Bph15, and QBph4.1 were identified from introgression lines of O. officinalis with the CC genome (Hirabayashi et al., 1998, 1999; Renganayaki et al. 2002; Du et al. 2009; Lv et al. 2014; Hu et al. 2015). Resistance gene Bph12 was derived from the introgression line B14 of O. latifolia with the CCDD genome (Qiu et al. 2012). Resistance genes Bph20(t) and Bph21(t) were identified from O. minuta with the CCDD genome and were mapped to chromosomes 4 and 12, respectively (Rahman et al. 2009). Finally, Bph27 and Bph29 were identified from O. rufipogon with the AA genome (Huang et al. 2013; Wang et al. 2015). Considerable efforts have been made to clone major BPH-resistance genes, and a few successes have been achieved over the past ten years. For instance, Bph14, Bph9, and Bph6 were successfully map-based cloned and to encode one type of NB-LRR protein (Du et al. 2009; Zhao et al. 2016; Guo et al. 2018). Resistance gene Bph3, a cluster of three repeated genes, encodes plasma membrane–localized lectin receptor kinases, confers broad-spectrum insect resistance and serves as a reference for the molecular breeding of insect-resistant cultivars (Liu et al., 2015). Bph29 is a recessive gene derived from O. rufipogon and has been found to contain a B3 DNA-binding domain (Wang et al. 2015).
The interaction of plants and insects is complex and can be described in relation to physical, chemical, cellular, or molecular aspects. To limit damage from insects to plants, one may employ antixenosis, antibiosis, or insect tolerance through physiological functions (Kennedy et al. 1987; Alam and Cohen, 1998). Several BPH resistance genes have been demonstrated to confer one or more types of resistance mechanisms. For instance, Panda and Heinrichs (1983) determined that rice varieties carrying the Bph1 gene exhibit antibiosis and tolerance and observed moderate resistance to the BPH. Moreover, resistance genes Bph6 and Bph14 have been characterized to have antibiotic and antixenotic effects on insects (Qiu et al. 2010; Du et al. 2009). Together, different BPH resistance genes may possess different resistance mechanisms. Thus, it is still necessary to demonstrate resistance mechanisms and to identify resistance effects observed when one novel BPH resistance gene is detected, which should facilitate resistant rice breeding.
The identified BPH resistance genes provide a wealth of resistance resources for rice breeding and improvement. However, it must be noted that the BPH insects can adapt to rice varieties with single resistance gene and then develop new biotypes for practical rice cultivation. For example, two BPH resistance varieties, IR26 and IR36, harboring BPH resistance genes Bph1 and bph2, respectively, were widely cultivated in the 1970s and 1980s, but they have become susceptible in the last few years due to changes in BPH biotypes and infestation patterns (Claridge and Hollander, 1980; Pathak et al. 1969). Previous studies have indicated that rice lines pyramiding two or more BPH resistance genes via marker-assisted selection (MAS) can efficiently improve resistance levels (Qiu et al. 2012; Hu et al., 2016a, b; Jena et al. 2017). Thus, there is still an urgent need to pyramid multiple BPH resistance genes and to develop stronger and more durable varieties that are resistant to BPH.
GX2183, a wild species of O. rufipogon with the AA genome, shows broad spectrum resistance to BPH biotypes, including biotypes 1 and 2, Bangladesh (collected from Bangladesh), Cuu Long (collected from Vietnam), and Pantnagar (collected from India) (Li et al. 2006). Major resistance gene Bph27 has been finely mapped to an 86.3-kb region harbored by markers RM16846 and RM16853 (Huang et al. 2013). Meanwhile, this study reveals the presence of more than one major BPH resistance gene in the rice species of GX2183. In the present study, introgression lines of GX2183 were used to develop the mapping population, near isogenic lines (NILs), and pyramiding lines (PYLs) to map and apply the BPH resistance gene. We identified and finely mapped a new BPH resistance gene Bph36 flanked by InDel markers S13 and X48 on the short arm of rice chromosome 4 and incorporated this gene into the elite rice cultivars via MAS.
Materials and methods
Mapping population, NIL, and PYL development
RBPH16 and RBPH17, two introgression lines derived from O. rufipogon accession GX2183, are highly resistant to BPH and were used as the donor. Rice varieties Kangwenqingzhan (abbreviated as KW) and Huanghuazhan (abbreviated as HHZ) are widely cultivated in southern China and are highly susceptible to BPH. RBPH16 and RBPH17 were respectively crossed with KW and HHZ to develop F2:3 mapping populations. Based on the QTL analysis, the positive F1 (KW/RBPH16) individuals were continuously backcrossed with KW to developed BC1F1, BC2F1, BC3F1, and BC4F1 populations, and each backcrossed individual was verified with tightly linked markers. From the developed populations, single-gene individuals carrying the novel resistance gene were self-crossed twice to develop a BC1F2 population, and then they were used to verify the resistance gene and for fine mapping. Positive BC4F1 individuals were then self-crossed twice to obtain BC4F2 lines homozygous to the resistance genes and taken as NILs.
RBPH54, an introgression line derived from O. rufipogon, carries BPH-resistance gene Bph29 (Wang et al. 2015). Rice variety PTB33 has been indicated to be highly resistant to BPH and includes resistance gene Bph3 in the short arm of chromosome 4 (Liu et al., 2015). Both were used to develop PYLs with BPH resistance genes. MH511, carrying the Xa23 resistance gene, is a bacterial blight resistant line. Rice line 9311 is a merit restoring line that is highly susceptible to BPH. To develop PYLs with different BPH resistance genes, RBPH16 was first crossed with RBPH54 and PTB33, and then the positive individuals were backcrossed with RBPH16 twice. Next, BC2F1 individuals derived from RBPH16/RBPH54 or RBPH16/PTB33 were crossed and self-crossed twice. Then, the BC2F2 lines were evaluated based on BPH resistance levels, and highly BPH- resistant lines were crossed with MH511 or 9311. Susceptible lines MH511 and9311 were taken as recurrent parents to develop BC3F1, and each backcrossed individual was verified with the tightly linked markers (Additional file 1: Figure S1).
Wild rice accession GX2183 was collected from Guangxi province of China and was stored at Guangxi University. Rice lines RBPH16, RBPH17, RBPH54, and MH511 were developed in our laboratory. Rice varieties 9311, KW, and HHZ were collected and stored in our laboratory as germplasm.
BPH insects and evaluation of BPH resistance
The BPH insects were collected from rice fields in Nanning, Guangxi province of China, and were maintained on Taichung Native 1 (TN1, a susceptible indica variety) under natural conditions in a greenhouse at Guangxi University. A bulk seedling test was conducted to evaluate BPH resistance as described by Qiu et al. (2010). The pre-germinated seeds were sown in rows spaced 3.5 cm apart with 20 seeds in each line in a plastic box (58 × 38 × 7 cm) with a seed bed of 3-cm thick fine puddled soil. Susceptible control TN1, resistant control PTB33 and parents of the mapping population were sown in thecenter. Seedlings of the third-leaf stage (13 or 14 days old) were infested with second- to third-instar hopper nymphs at a density of eight insects per seedling. Temperature and humidity were respectively maintained at 28 ± 2 °C and 75 ± 5% for evaluation. Phenotypic values for the individual plants were recorded on a scale on 0–9 when all plants of susceptible control TN1 were killed. This was done following the Standard Evaluation System (SES) for rice (IRRI, 1996) based on the following scale: 0 = no visible damage; 1 = partial yellowing in first leaf; 3 = first and second leaves have become partially yellow; 5 = pronounced yellowing or minor stunting; 7 = wilting while, the plant is still alive; and 9 = the plant has completely wilted or died. Three replicates of each line were carried out, and all resistance tests were repeated twice.
Marker analysis, QTL detection and gene mapping
Total genomic DNA of the plant leaves was extracted by the CTAB method (Murray and Thompson, 1980). PCR was performed as described by Qiu et al. (2010), and the PCR products were separated on 7% denaturing polyacrylamide gels and visualized by silver staining. Simple sequence repeat (SSR) primer sequences were obtained from the Gramene database (www.gramene.org/archive),and InDel makers were developed through a comparison of genome sequences of japonica cv. Nipponbare and indica cv. 9311.
A bulked segregant analysis (BSA) was performed to identify markers linked to BPH resistance (Michelmore et al. 1991). According to the results of our BPH resistance evaluation, ten extremely resistant and ten extremely susceptible individuals were used to construct two contrasting DNA pools. SSR and InDel markers were screened for polymorphism between the two pools. All polymorphic markers within the study region were used to genotype the F2 individuals. Genetic linkage maps of SSR and InDel markers with BPH resistance loci were constructed using JoinMap 3.0. Genetic linkage maps and phenotypes of the primary mapping population were used for QTL analysis via inclusive composite interval mapping (ICIM) with MapQTL 5.0. QTLs were identified when LOD scores exceeded 3.0.
BPH host selection test
To perform the BPH host selection test, each rice line with a single gene (NIL-Bph36), two genes (NIL-Bph36 + Bph27), and KW were sown into one row with 15 healthy seedlings (4 weeks old) in a plastic bucket containing rice field soil. The second- to third-instar BPH nymphs were released into an average of eight heads per plant. The number of BPH insects attached to each line was counted every day after infestation. After each observation, the base of each rice plant was gently tapped to allow BPH insects to be evenly distributed throughout the water and to select the host plant once again. Three replications were conducted each time and the test was repeated twice.
BPH honeydew excretion measurement
The parafilm sachets method was used to measure BPH honeydew excretion (Pathak et al. 1982). One fifth-instar BPH insect was enclosed in a rectangular (3.5 cm × 3 cm) parafilm bag with a pipette, and then was fastened to the test plants (4 weeks old). Each plant had one or two bags and eight bags for each line of NIL-Bph36, NIL-Bph36 + Bph27, or KW wereused. After 2 days of insect infestation, the bags were removed and the total weight of the bag enclosing the honeydew was weighed as W1. Then, the honeydew was wiped with absorbent cotton and the bag was weighed again (W2). Therefore, the weight of honeydew excretion was calculated as W1 – W2. The test was repeated three times.
BPH survival and growth measurement
The BPH survival rate was determined according to the method described by Qiu et al. (2010). The 5-week-old seedlings were only composed of main stems and were covered with a plastic cup with a hole in the base. Then, each plant was infested with ten third-instar nymphs. The surviving insects were counted 1 d, 2 d, 3 d, 4 d, and 5 d after BPH release. Six plants were arranged for each line of NIL-Bph36, NIL-Bph36 + Bph27, and KW, and the experiment was repeated three times.
To measure the population growth rate (PGR) of the BPH, the rice seedlings were treated as they were for the BPH survival test. The 5-week-old seedlings were infested with 15 pre-weighed second- to third-instar nymphs and then covered with a plastic cup with a hole sealed with absorbent cotton. The weight of the surviving BPH was measured after 4 days of insect infestation. The PGRs of BPH were calculated following the method described by Qiu et al. (2010). Six plants were used for each line of NIL-Bph36, NIL-Bph36 + Bph27, and KW, and the test was repeated three times.
Bacterial blight resistance test
To perform the bacterial blight resistance test, Xanthomonas oryzae pv. oryzae GX1070 was collected from rice plants from Guangxi province of China and was cultured on nutrient agar (NA) media at 28 °C for 4 days. The bacteria were then collected and dissolved with ddH2O to an optical density concentration of OD600 = 1.0. The 8-week-old plants were then inoculated with bacteria using the leaf-clipping method, and then each leaf was measured and scored after 3 weeks of inoculation (Kauffman et al. 1973). Six plants were used for each line, and the test was repeated twice.
MAS for the resistance genes
According to the above study, molecular markers RHD9, RH007, and RHC10 were co-segregated with Bph3 (Liu et al., 2015), and BYL8, BID2, BID3, and BYL7 were tightly linked to Bph29 (Wang et al. 2015). Bph27 was found in the region flanked by SSR markers RM16846 and RM16888, which were used for MAS (Huang et al. 2013). These markers were verified for RBPH16, RBPH54, PTB33, 9311, and MH511 to select suitable markers for MAS.
Agronomic trait measurement
To detect agronomic traits of the developed BPH-resistance lines, each line was composed of five rows and each row included ten plants at a planting density of 20 × 30 cm (20-cm spacing between plants and 30-cm spacing between rows distance). Together with the recurrent parents, three repeats of each line were randomly distributed across the rice field. Then, 20 plants from each repeat were subjected to agronomic trait measurements, (i.e., plant height, effective panicle number, the number of grains per panicle, 1000-grain weight, and seed setting percentage). Field management followed normal agronomic practices. The test was repeated over two seasons.
Data analysis
The Chi-square test for goodness-of-fit was performed with GraphPad Prism 7; and the resistance data were analyzed using one-way ANOVA and comparing the LSD test at a 5% or 1% significance level. The survival rates of insects (%) were arcsine transformed prior to analysis.
Results
Genetic analysis of BPH resistance
A previous study showed that wild rice accession GX2183 (O. rufipogon) contains other BPH resistance genes except for Bph27 (Huang et al. 2013). As it is relatively difficult to develop mapping populations directly crossing with GX2183, we obtained highly BPH-resistant introgression lines and then constructed the associated mapping population. As a result, introgression lines RBPH16 and RBPH17 were derived from GX2183 and were found to be highly resistant to BPH. Furthermore, indica rice cultivars KW and HHZ were found to be highly susceptible to BPH and were applied to develop mapping populations (Fig. 1). Then, F2 lines developed from two different mapping populations were surveyed based on resistance levels via SES. Consequently, the resistance scores of 140 individuals of the F2 population of the crossing KW/RBPH16 varied continuously from 2.0 to 9.0 with some lines showing super-progeny phenomena (Fig. 1a). According to the IRRI scoring criteria for seedling tests and previous studies conducted by Qiu et al. (2010), we considered resistance scores of 0–7.0 to denote resistance and of 7.1–9.0 to denote susceptibility. Therefore, the BPH resistant assay showed that the segregation of resistance to susceptibility in 140 F2 lines disagreed with 3:1 (113:27; χ2 C = 7.02 > χ2 0.05 = 3.84) (Fig. 1a). The same trend of phenotypic variance was observed for the other F2 population derived from HHZ/RBPH17. The segregation of resistance to susceptibility for 120 F2 lines also disagreed with 3:1 (75:45; χ2 C = 9.34 > χ2 0.05 = 3.84) (Fig. 1b). These results indicated that more than one major gene conferred BPH resistance in the introgression lines.
Fig. 1.
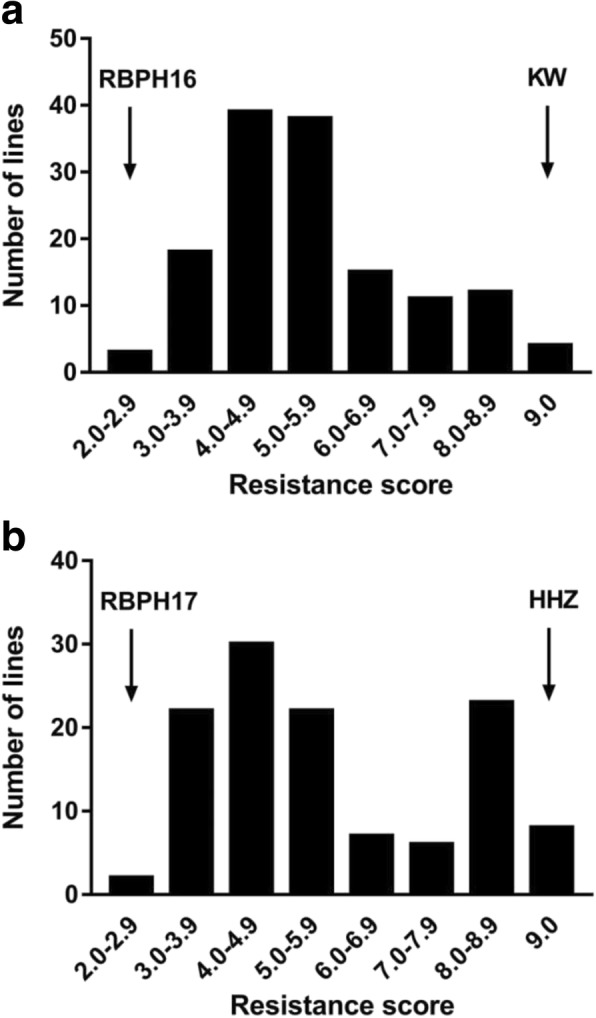
Frequency distribution of the BPH-resistance score in F2 population. a Population derived from cross of KW × RBPH16; b Population derived from cross of HHZ × RBPH17. Rice (Oryza sativa) seedlings were infested with eight BPHs per plant for 9–10 days. RBPH16 and RBPH17 were the resistance parents; KW (Kangwenqingzhan) and HHZ (Huanghuazhan) were the susceptible parents
Primary mapping of resistance genes
A BSA was performed to identify markers linked to the BPH resistance loci. According to the resistance score for F2 individuals of KW/RBPH16 population, a total of 968 molecular markers distributed across 12 rice chromosomes were used to screen polymorphism between extremely resistant and susceptible DNA bulks. As a result, six markers were found to be polymorphic, of which markers RM471, RM17033, and RM16874 and markers RM16502, RM518, and RM335 were identified from the contiguous region of the long and short arms of chromosome 4, respectively. The same analysis was conducted on the F2:3 mapping population of HHZ/RBPH17. A total of 952 markers were surveyed with the two DNA bulks. Markers RM17033, RM16888, RM16874, RM16858, and RM16766 from the long arm exhibited polymorphism, and markers RM335, RM518, RM16465, and RM16502 from the short arm exhibited polymorphism. We found two major resistance loci on chromosome 4. We then used the polymorphic markers to assay the F2 individuals and acquired a local linkage map developed by JoinMap3.0 (Table 1). Based on genotypes and phenotypes of detected F2 individuals within the two mapping populations, we analyzed the resistance loci using MapQTL5.0. As a result, two major loci with large LOD scores and with phenotypic variation were detected on chromosome 4. For mapping population KW/RBPH16, the locus on the long arm of chromosome 4 was detected with an interval flanked by RM16888 and RM16786 with a maximum LOD score of 14.4, contributing 38.2% of the phenotypic variation. The locus on the short arm of chromosome 4 was detected with an interval flanked by RM16465 and RM6659 with a maximum LOD score of 20.3, contributing 49.2% of the phenotypic variation (Table 1). For mapping population HHZ/RBPH17, the locus on the long arm of chromosome 4 was detected with an interval flanked by RM17033 and RM16786 with a maximum LOD score of 11.3, contributing 36.1% of the phenotypic variation. The locus on the short arm of chromosome 4 was detected with an interval flanked by RM16465 and RM16502 with a maximum LOD score of 31.1, contributing 86.8% of the phenotypic variation (Table 1). Taken together, these mapping results suggest that the same chromosome regions harbored BPH resistance loci according to an analysis of different F2 populations. According to a study conducted by Huang et al. (2013), the locus on the long arm of chromosome 4 is generally identical to resistant gene Bph27, while the locus on the short arm differs from the reported genes according to an analysis of genome information on the associate markers. Therefore, the locus harbored by RM16465 and RM16502 (or RM6659) was taken as a novel gene and was tentatively designated as Bph36.
Table 1.
QTL scanning results using 140 lines (KW/RBPH16) and 120 lines (HHZ/RBPH17) resistance scores and marker genotypes of F2 population by MapQTL 5
| Mapping population | Locus | Position (cM) | LOD | PEV (%) | A |
|---|---|---|---|---|---|
| KW/RBPH16-Bph36 | RM335 | 0 | 0.6 | 1 | − 0.2 |
| RM16465 | 9.1 | 0.9 | 0.7 | −1.2 | |
| Bph36 | 9.2 | 20.3 | 49.2 | −1.8 | |
| RM6659 | 10.2 | 20.0 | 48.4 | −1.8 | |
| RM16502 | 15.4 | 1.35 | 2.3 | −0.2 | |
| RM5412 | 18.7 | 0.4 | 0.7 | −0.3 | |
| KW/RBPH16-Bph27 | RM16766 | 0 | 1.7 | 3.4 | 0.6 |
| RM471 | 16.9 | 0.1 | 0.1 | 0 | |
| RM16852 | 17.1 | 0 | 0 | 0.3 | |
| RM16853 | 17.5 | 0.9 | 1.9 | 0.8 | |
| Bph27 | 17.7 | 14.4 | 38.2 | −1.5 | |
| RM16858 | 18.7 | 13.5 | 35.8 | −1.4 | |
| RM16888 | 19.4 | 0.26 | 0.5 | 0.6 | |
| HHZ/RBPH17-Bph36 | RM335 | 0 | 2.5 | 3.6 | 0.2 |
| RM518 | 5 | 2.1 | 3.3 | 0.1 | |
| RM16465 | 13.9 | 4.5 | 54.3 | −1.9 | |
| Bph36 | 17.6 | 31.1 | 86.8 | −2.2 | |
| RM16502 | 20.6 | 19.7 | 9.1 | −0.8 | |
| HHZ/RBPH17-Bph27 | RM16766 | 0 | 0.8 | 2.2 | −0.6 |
| RM16852 | 17 | 9.2 | 1.7 | 0.5 | |
| RM16853 | 19.2 | 11.1 | 36.1 | −1.6 | |
| Bph27 | 20.2 | 11.8 | 36.6 | −1.5 | |
| RM16858 | 23.3 | 0.6 | 30.7 | −1.5 | |
| RM16874 | 24.7 | 0.1 | 0.2 | 0.3 | |
| RM16888 | 25.5 | 0.2 | 0.4 | 0.4 |
PEV (%) Percentage of total phenotypic variance explained by the locus, A additive effect on the resistance allele
Verification and fine mapping of Bph36
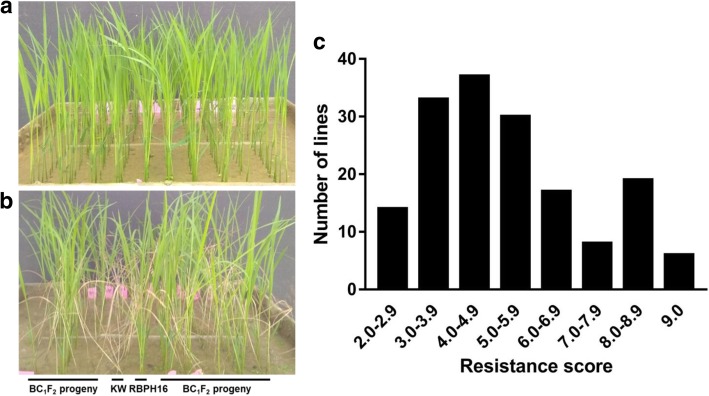
To confirm the genetic segregation of the chromosome region with the target gene, we only used BC1F1 individuals carrying Bph36 to self-cross. Then the developed BC1F2 population derived from KW/RBPH16 was surveyed with a BPH seedling bulk test (Fig. 2a, b). Consequently, the BPH-resistance scores of 161 BC1F2 lines showed a continuous distribution of 2.0 to 9.0, and the segregation of resistant (0–7.0) and susceptible lines (7.1–9.0) presented an acceptable fit to a 3:1 ratio (123:38; χ2 C = 0.168 < χ2 0.05 = 3.84) (Fig. 2c). This result denotes the presence of a single gene determining BPH resistance in the selected mapping population.
Fig. 2.
Overview of the bulk seedling test and frequency distribution of the BPH-resistance score in BC1F2:3 population carrying Bph36. a Before insect infestation of BC1F2 lines. b After insect infestation of BC1F2 lines. c Frequency distribution of the BPH-resistance score in BC1F2 population carrying Bph36. Rice seedlings were treated with eight BPHs per plant for 9–10 days. RBPH16 and KW were the resistance and susceptible parents, respectively
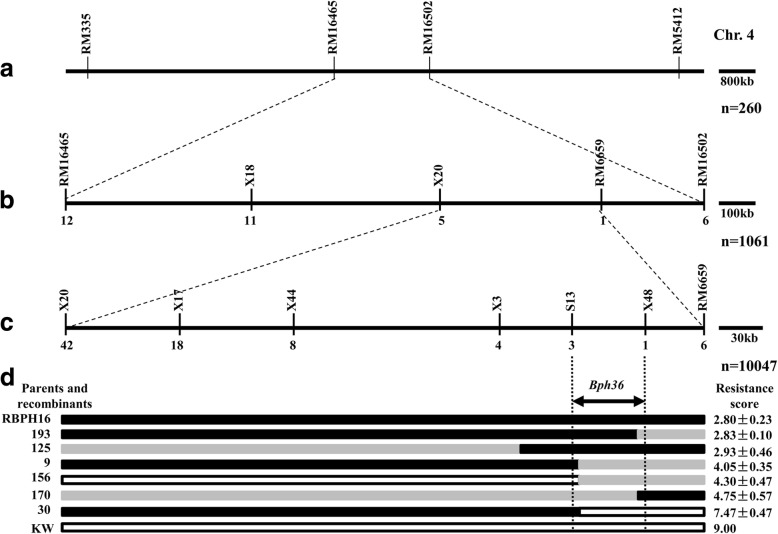
For the high-resolution mapping of the resistance gene, we conducted two rounds of fine mapping. We first developed polymorphic SSR and InDel markers X18, X20, RM6659, RM16465, and RM16502 between RM5412 and RM335 (Fig. 3a,b; Table 2). Then, a total of 1061 BC1F3 individuals from monogenic lines carrying Bph36 were subjected to analysis and 18 recombinant lines were identified from RM16465 and RM6659. According to our analysis of genotypes and resistance scores of the recombinants, the resistance gene was identified in the region between X20 and RM6659 (Fig. 3b). Second, markers X20 and RM6659 were used to screen the genotypes of 10,047 BC2F2 seedlings derived from monogenic lines carrying Bph36. As a result, 68 recombinant individuals were identified from the selected markers (X20 and RM6659). At the same time, polymorphic InDel markers X3, X17, X44, X48, and S13 were developed in the region of interest (Table 2). Based on our analysis of genotypes and resistance scores of the recombinants, recombinants 170 and 193 could delimit the left margin of the gene with X48 and recombinants 9, 30, and 156 could delimit the right margin with S13. Eventually, we determined the candidate genomic region of Bph36 for a 38-kb region flanked by markers S13 and X48 based on the genome of Nipponbare (Fig. 3c, d).
Fig. 3.
High-resolution mapping of Bph36. a Primary mapping of Bph36 on the short arm of chromosome 4; b Fine mapping of Bph36; c High-resolution mapping of Bph36. The number below the lines denotes the number of recombinants between the lateral markers; d Graphical genotypes and resistance phenotypes of the recombinants. The black, white, and gray bars denote the marker genotypes of RBPH16 homozygotes, KW homozygotes, and heterozygotes, respectively
Table 2.
Polymorphic InDel molecular markers used for fine mapping
| ID | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|
| X3 | ATCCGTCCTTATCGTGTGCC | CCCTGTAGCCGATGGAGTTC |
| X17 | GAGCGCAGGGAGTGGATTAG | GTCCAACATGATAGCGACCG |
| X18 | CAGTCGGCCCAGTCATTGAT | GCATAAGTGAACAGAAGTGACCG |
| X20 | TGCATACATCGATGAACAACCTC | ATTTGGCTTTTTCGTAGGGGT |
| X44 | ACAACACAAACCTCAAGGAG | TCCCTTATTGCATTTACAGT |
| X48 | CAAGGGAAACCAAACAGACAA | AAGACGACGGTTCTGGATTG |
| S13 | AGCCCAAATGGAACAAACAA | GCTCACGGTTAAGCAATGGT |
In consideration of the target region, four candidate genes were contained in the corresponding genomic region of Nipponbare. They are hypothetical protein (LOC_Os04g11800), acyltransferase family protein (LOC_Os04g11810), white-brown complex homolog protein (LOC_Os04g11820), and TCP family transcription factor (LOC_Os04g11830). Acyltransferase is a large family of proteins that play an important role in the biosynthesis of some plant secondary metabolites for defensing against insect (Chedgy et al. 2015). White-brown complex (WBC) family protein, is a part of ABC superfamily protein, has been reported to participate in a multitude of physiological processes that allow the plant to adapt to changing environments and cope with biotic and abiotic stresses (Kretzschmar et al. 2011). TCP family transcription factor participate in plant developmental processes, such as development of leaf and flower (Koyama et al. 2017; Yin et al. 2018).
Characterization of the BPH resistance of NILs
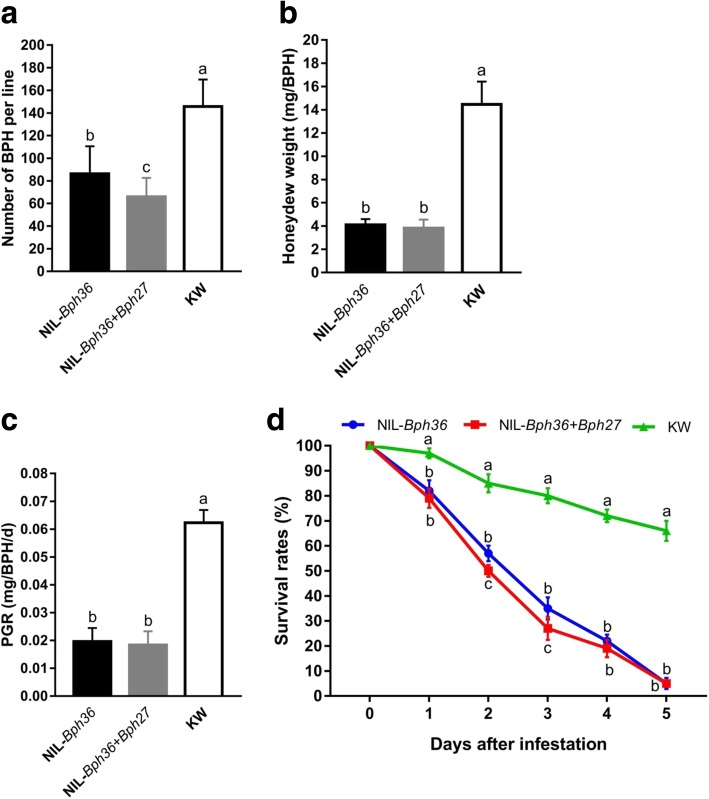
We characterized the BPH resistance of NIL-Bph36 and NIL-Bph36 + Bph27. In the BPH host choice test, the average number of BPHs landing on KW was measured as 145.7, while BPHs landing on NIL-Bph36 + Bph27 and NIL-Bph36 were measured as 66.0 and 86.3, respectively. We observed a significant reduction in the number of settled BPHs between the resistant and susceptible lines (P < 0.001 for NIL-Bph36 and KW or NIL-Bph36 + Bph27 and KW). Moreover, the number of BPHs settled on NIL-Bph36 + Bph27 was found to be significantly lower than that of NIL-Bph36 (P < 0.05) (Fig. 4a). These results indicate that the resistance lines exhibited considerable antixienosis to BPH, and that the lines with two resistance genes exhibited stronger resistance to BPH.
Fig. 4.
Characterization of the BPH resistance of NILs. a BPH host choice among NIL-Bph36, NIL-Bph36 + Bph27, and KW. b Weight of honeydew secreted by BPH on NIL-Bph36, NIL-Bph36 + Bph27, and KW. c Effect of NIL-Bph36, NIL-Bph36 + Bph27, and KW on the BPH PGR (mg/BPH/d). d Survival rates of BPH on NIL-Bph36, NIL-Bph36 + Bph27, and KW. Error bars represent SEs. Means labeled with the same letter are not significantly different at the level of P = 0.05 for A. means labeled with asterisks are significantly different (P < 0.05) for b and c. Statistical analysis was conducted with one-way ANOVA with LSD test
According to our BPH honeydew excretion measurements for the different genotypic plants, we observed 14.46, 4.10, and 3.82 mg/BPH in KW, NIL-Bph36, and NIL-Bph36 + Bph27, respectively (Fig. 4b). To measure resistance effects on the insects, the PGR of BPH was calculated from changes in weight observed before and after feeding the resistant and susceptible plants. Consequently, the PGRs of resistant plants NIL-Bph36 and NIL-Bph36 + Bph27 and of susceptible plants KW were measured as 0.0196, 0.0184, and 0.0623 mg/BPH/d, respectively (Fig. 4c). Significantly less honeydew excretion levels and PGRs were detected from the resistant plants than from the susceptible plants (P < 0.001 for NIL-Bph36 and KW or NIL-Bph36 + Bph27 and KW).
We surveyed the survival number of BPHs left 1 d, 2 d, 3 d, 4 d, and 5 d after treatment. BPH survival rates declined quickly for resistant plants NIL-Bph36 and NIL-Bph36 + Bph27 compared to those of the susceptible plants. After 2 d of infestation, BPH survival rates for the resistant plants were less than 60%while remaining at above 80% for the susceptible plants (P < 0.05 for NIL-Bph36 and KW or NIL-Bph36 + Bph27 and KW). After 5 d of infestation, BPH survival rates of the resistant plants were only measured at 5%, but a value of 62% was measured for the susceptible plants (P < 0.01 for NIL-Bph36 and KW or NIL-Bph36 + Bph27 and KW, Fig. 4d). We must also note that survival rates of NIL-Bph36 + Bph27 were still lower than those of NIL-Bph36 during the observation period, and a statistical difference was detected 2 d and 3 d after infestation (P < 0.05). Taken together the resistant plants had a strong antibiotic effect on the insects and the pyramided line had a stronger effect than the single gene line.
BPH resistance evaluation for PYLs
To select PYLs for advanced breeding application, the developed lines were primarily evaluated through a BPH resistance test and from agronomic trait observations. Highly resistant PYLs (BC3F3) derived from RBPH16/RBPH54 × RBPH16/PTB33 with normal seed settings and heading dates were chosen for BPH resistance gene transformation. A total of seven PYLs were selected and applied to screen with the linked markers and to evaluate BPH resistance levels. Of these, lines VP1720, VP1728, VP1731, VP1732, and VP1733 were found to carry four resistant genes. Specifically, VP1720 and VP1728 were homozygous for the four resistance genes with average resistance scores of 1.76 and 1.59, respectively. VP1732 and VP1733 scored 2.73 and 2.18, respectively, and were found to be homozygous for three resistance genes and heterozygous for one gene. VP1731 was homozygous for Bph36 and Bph27 and heterozygous for the other two genes with an average BPH resistance score of 2.53. VP1722 and VP1734 were homozygous for three resistance genes and also exhibited considerable resistance to the BPH insects (Table 3). Therefore, all of the selected PYLs can be used for BPH resistance transfers for breeding application, especially for lines VP1720 and VP1728.
Table 3.
Selected PYLs carring multiple BPH resistance genes
| ID | X17a | RM16846 | BYL18 | RH007 | Resistant score |
|---|---|---|---|---|---|
| VP1728 | Ab | A | A | A | 1.59 ± 0.41 |
| VP1720 | A | A | A | A | 1.76 ± 0.32 |
| VP1733 | A | A | H | A | 2.18 ± 0.26 |
| VP1722 | A | A | A | B | 2.53 ± 0.68 |
| VP1731 | A | A | H | H | 2.53 ± 0.81 |
| VP1732 | A | A | H | A | 2.73 ± 0.22 |
| VP1734 | A | A | B | A | 2.95 ± 0.18 |
aX17, RM16846, BYL18, and RH007 were the linked molecular markers of Bph36, Bph27, Bph29, and Bph3, respectively. b A, B, and H denoted that the lines were homozygous for resistance and susceptible parents or heterozygous at the resistance gene region, respectively
Breeding applications of resistance genes
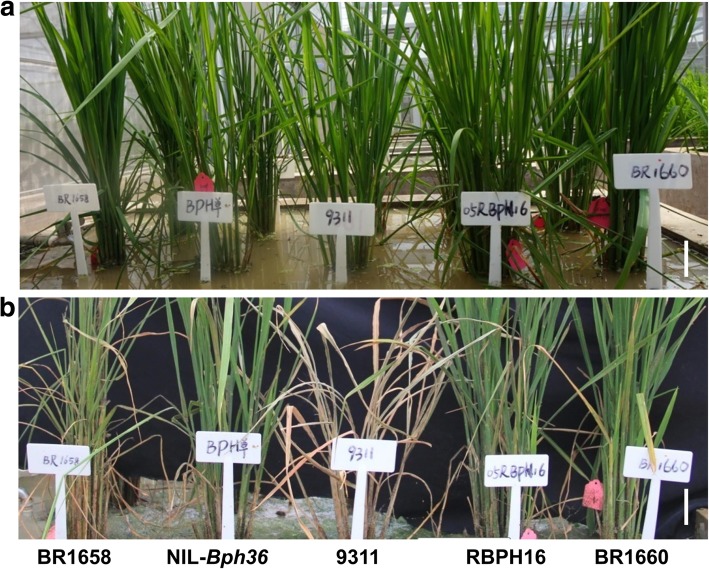
To develop breeding lines with strong BPH resistance and merit agronomic traits, the selected PYL of VP1728 was continuously backcrossed with 9311 and MH511. As a result, rice lines BR1658 and BR1660 carrying Bph36, Bph27, Bph29, Bph3, and Xa23 with an MH511 genetic background were found to be highly resistant to BPH with average resistance scores of 2.93 and 2.12 at the seedling stage, respectively. They also exhibited strong resistance to BPH insects in the tillering stage (Fig. 5). Both lines were then also found to be highly resistant to bacterial blight in the tillering stage (Fig. 6). Average disease infection lengths were measured as 0.6, 0.5, and 10.2 cm for BR1658, BR1660, and KW, respectively (P < 0.001 for BR1658 and BR1660 relative to KW). Rice line BR1693 carrying Bph36, Bph27, Bph3 and Bph29 with the 9311 genetic background also exhibited strong resistance to BPH insects with an average resistance score of 2.14 for the seedling stage.
Fig. 5.
BPH resistance test at tillering stage. a Before infestation with BPH, b After infestation with BPH. RBPH16 and 9311 were the resistance and susceptible controls, respectively. BR1658 and BR1660 had Bph36, Bph27, Bph29, Bph3, and Xa23 with MH511 genetic background and were indicated to be highly resist to BPH. NIL-Bph36 carried single gene Bph36 with KW genetic background. Scale bars, 15 cm
Fig. 6.
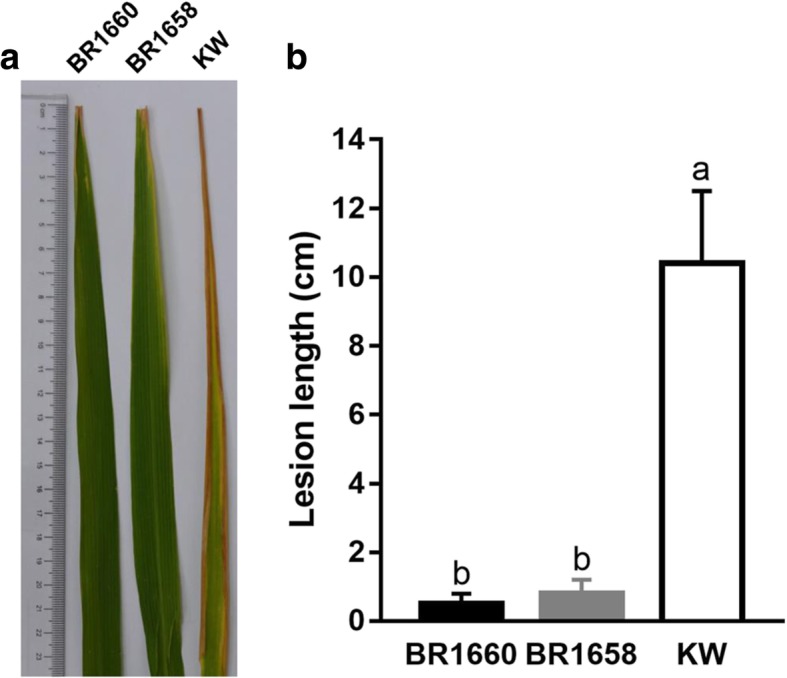
Test for bacterial blight resistance of PYLs at tillering stage. a Bacterial blight resistance of BR1658, BR1660 and KW to GX1070 using the leaf-clipping method. Leaf lesions 2 weeks post inoculation are indicated by representative leaves. b Lesion length measurement. Error bars represent SEs. Means labeled with the same letter are not significantly different at the level of P = 0.05. Statistical analysis was conducted with one-way ANOVA with LSD test
To evaluate the developed breeding lines, we surveyed agronomic traits of BR1658, BR1660, and BR1693 and of recurrent parents MH511 and 9311. The plant height (106.9 cm), number of effective panicles (8.5), and setting rate (84.0) of BR1693 were found to be less than those of 9311, and the number of effective panicles (176.8) was found to be greater than that of 9311. Howerer, we found that no statistical difference between BR1693 and 9311 (P > 0.05). Interestingly, the average 1000-grain weight of BR1693 was measured as 25.4 g, which is lower than that found for 9311 (27.8 g) (F = 16.4, P = 0.042, Table 4). As for rice lines BR1658 and BR1660, all the detected traits were identical to those of MH511 (P > 0.05). Although the setting rates of BR1658 (85.1%) and BR1660 (84.8%) decreased relative to those of MH511 (86.4%) (F = 5.6, P = 0.06 for BR1658 and MH511; F = 4.8 P = 0.046 for BR1660 and MH511), no statistical difference was observed between them (Table 4). These results indicate that the developed breeding lines acquired high BPH and disease resistance values and exhibited the same agronomic traits as their recurrent parents.
Table 4.
Agronomic traits of recurrent parents and breeding lines
| Linesa | Plant height (cm) | Number of effective panicle per plant | Number of grains per panicle | Setting rate (%) | 1000-grain weight (g) |
|---|---|---|---|---|---|
| 9311 | 108.6 ± 0.70 | 8.7 ± 0.37 | 168.9 ± 5.06 | 84.7 ± 1.53 | 27.8 ± 0.35 |
| BR1693 | 106.9 ± 0.92 | 8.5 ± 0.44 | 176.8 ± 4.15 | 84.0 ± 1.28 | 25.4* ± 0.16 |
| MH511 | 96.4 ± 2.21 | 6.9 ± 0.57 | 141.6 ± 5.06 | 86.4 ± 2.15 | 21.3 ± 0.23 |
| BR1658 | 95.4 ± 1.15 | 7.0 ± 0.42 | 139.7 ± 7.46 | 85.1 ± 1.56 | 21.6 ± 0.37 |
| BR1660 | 94.9 ± 1.76 | 6.8 ± 0.52 | 141.8 ± 5.24 | 84.8 ± 1.72 | 20.9 ± 0.68 |
a9311 and MH511 were the susceptible parents; BR1693 was the highly BPH resistance line with 9311 genetic background; BR1658 and BR1660 were the highly BPH and bacterial blight resistance lines with MH511 genetic background. *P < 0.05 derived from one-way ANOVA with LSD test
Discussion
BPHexerts chronic biotic stress and severely constrains rice production in Asia and in other parts of the world. Accordingly, BPH resistance gene identification, mapping and characterization are valuable in ensuring long-term resistance against diverse predominant biotypes for sustainable rice outputs. Previous reports demonstrate the highly resistant wild rice accession GX2183 contains more than one major gene for BPH resistance. In this study, RBPH16 and RBPH17 originating from GX2183 were employed as donor parents and were respectively crossed to KW and HHZ to develop F2:3 mapping populations. Subsequently, with the exception of the previously detected gene Bph27 (Huang et al. 2013), novel resistance gene Bph36 was identified via high-resolution mapping to accupy a 38-kb region harbored by markers S13 and X48. Rice chromosomes 4 and 12 have been considered as the mainframe of BPH resistance genes (Jena and Kim 2010; Fujita et al. 2013; Hu et al., 2016a, b). For instance, thirteen genes are found in two distinct clusters of chromosome 4. Six of them (Bph6, bph12, Bph27, bph18(t), Bph27(t), and Bph34) are clustered on the long arm of chromosome 4 (Qiu et al. 2010, 2012; Huang et al. 2013; Fujita et al. 2013; Kumar et al. 2018); and seven genes (Bph3, Bph12, Bph15, Bph17, Bph20(t), Bph30, and Bph33) are found on the short arm of chromosome 4 (Liu et al., 2015; Yang et al. 2002; Sun et al. 2005; Yang et al. 2004; Rahman et al. 2009; Wang et al. 2018; Hu et al. 2018). According to the present study Bph36 also occupies this cluster, and via high-resolution mapping, it was successfully identified on the short arm of chromosome 4 roughly 2 Mb downstream from the Bph12, Bph17, Bph15, and Bph20 regions (Yang et al. 2002; Sun et al. 2005; Yang et al. 2004; Rahman et al. 2009) and 0.6 Mb upstream from Bph3 in the Nipponbare genome (Liu et al., 2015). Gene Bph36 presented a large LOD score and accounts for the considerable variance in BPH resistance observed as shown in Table 1. This was also proved through the characterization of NILs and PYLs as shown in Figs. 4 and Fig. 5.
Plants generally apply antixenosis, antibiosis, and tolerance against insect attacks. Previous studies indicate that BPH resistance genes such as Bph6, Bph14, and Bph27 have strong antixenotic and antibiotic effects on the insects (Qiu et al. 2010; Du et al. 2009; Huang et al. 2013), while the resistance gene Bph7 mainly confers tolerance to BPH (Qiu et al. 2014). Moreover, plants pyramiding with two or more genes significantly increase resistance to BPH insects (Qiu et al. 2012; Hu et al. 2012). According to tests on BPH host choices, honeydew excretion, survival rates, and PGR, lines of NIL-Bph36 and NIL-Bph36 + Bph27 show significant antixenotic and antibiotic effects on insects relative to those of susceptible plants, and it must be noted that lines of NIL-Bph36 + Bph27 have significantly stronger resistance effects than NIL-Bph36 as shown in host choice and BPH survival rates (Fig. 4).
Wild rice species offer natural and unique pools of resistance genes with strong resistance to all BPH biotypes (Heinrichs et al. 1982; Hu et al. 2015). At the same time, modern technologies offer means of using molecular markers in molecular breeding approaches to accelerate the identification and application of resistance loci (Ashkani et al. 2015). Recently, Jiang et al. (2018) introduced six dominant BPH-resistance genes into Jin23B using MAS and in turn improved BPH-resistance breeding lines. Moreover, Hu et al. (2016a, b) pyramided Bph3, Bph14, and Bph15 into elite indica cultivar Heimeizhan and obtained high BPH resistance breeding lines. Via high-resolution mapping, Bph36 has been found to include tightly linked markers that are effective and efficient for MAS performance (Additional file 2: Figure S2). We also pyramided Bph36 with Bph27, Bph29, and Bph3 by continuous backcrossing and MAS, and obtained several highly resistant PYLs. Of these, VP1720 and VP1728 were found to be homozygous across all four BPH-resistance genes and exhibited strong resistance to insects (Table 3). Then, VP1728 was backcrossed with MH511 and 9311 to develop approximate resistant breeding lines with diverse backgrounds. Consequently, an agronomic trait survey suggests that the developed breeding lines are almost the same as the recurrent parents (Table 4). Therefore, the developed PYLs may be applied for insect resistance rice breeding.
Regarding agricultural traits of the developed PYLs, setting rates of BR1658 (85.1%) and BR1660 (84.8%) declined relative to those of MH511 (86.4%) (F = 5.6, P = 0.06 for BR1658 and MH511; F = 4.8 P = 0.046 for BR1660 and MH511). The 1000-grain weight of BR1693 (25.4 g) was also lower than that of 9311 (27.8 g) (F = 16.4, P = 0.042, Table 4). This suggests that linkage drag remains a serious problem when pyramiding different genes via MAS. Many previous studies have revealed the same problem. For instance, a previous study shows that Bph3 from Rathu Heenati is positioned very close to the Wxa allele (roughly 380 kb), which confers amylose content in rice (Jairin et al. 2007, 2009). However, the authors acquired breeding lines with Bph3 and without Wxa alleles by screening large backcrossed individuals (Jairin et al. 2009). However, Hu et al. (2016a, b) failed to break up linkages of Wxa while pyramiding Bph3 with Bph14 and Bph15 in Hemeizhan, as the numbers of screened backcrossed individuals was still too low. Therefore, more backcrosses and MAS tests are needed to break up unexpected linkage genes for breeding purposes.
Our results validate the usefulness of the Bph36 gene for the control of BPH and show that applying it to susceptible rice varieties produces resistance effect by suppressing the feeding behavior, growth and longevity of BPH insects. This can in turn facilitate the development of BPH-resistant rice varieties in the future and help limit herbicide use. Tightly linked markers will facilitate the marker-assisted breeding and high-resolution mapping of Bph36, laying the foundation for insect resistance mechanism research, map-based cloning and functional analysis and providing a theoretical basis and intermediate materials to facilitate resistance to BPH.
Additional files
Figure S1. The pedigree of pyramiding lines (PYLs) containing resistance genes Bph36, Bph27, Bph29, Bph3, and Xa23 developed using marker-assisted selection (MAS). (TIF 359 kb)
Figure S2 Amplified bands of BC1F2 individuals derived from KW/RBPH16 with InDel marker X17 and detected by 7% PAGE. M: Marker DL2000, RBPH16: resistance parent, KW: susceptible parent, Lanes 1–22: individuals of BC1F2. (TIF 638 kb)
Table S1 Markers for marker-assisted selection (MAS) of BPH and bacterial blight resistance genes. (DOCX 16 kb)
Acknowledgements
This research was supported by the National Key R&D Program of China (2016YFD0100600), the National Natural Science Foundation of China (31560423), the Guangxi Natural Science Foundation (2013GXNSFGA019009), and Guangxi innovation-driven development special funding project (Guike-AA17204070).
Funding
This research was supported by the National Key R&D Program of China (2016YFD0100600), the National Natural Science Foundation of China (31560423), the Guangxi Natural Science Foundation (2013GXNSFGA019009), and Guangxi innovation-driven development special funding project (Guike-AA17204070).
Availability of data and materials
All relevant data are provided as Additional file 3: Table S1.
Authors’ contributions
ZH L, YF Q, and RB L conceived the idea and wrote the manuscript. ZH L, YX X, and HL Z conducted the experiments and performed data analysis. Y L, B U, XZ J, XY W, F L and BX Q participated in material development and sample preparation. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Rongbai Li, Phone: +86-771-3225260, Email: lirongbai@126.com.
Yongfu Qiu, Phone: +86-771-3225260, Email: yfqiu@126.com.
References
- Alam SN, Cohen MB. Detection and analysis of QTLs for resistance to the brown planthopper, Nilaparvata lugens, in a doubled-haploid rice population. Theor Appl Genet. 1998;97(8):1370–1379. doi: 10.1007/s001220051031. [DOI] [Google Scholar]
- Ali MP, Chowdhury TR. Tagging and mapping of genes and QTLs of Nilaparvata lugens resistance in rice. Euphytica. 2014;195:1–30. doi: 10.1007/s10681-013-0983-2. [DOI] [Google Scholar]
- Ashkani S, Rafii MY, Shabanimofrad M, Miah G, Sahebi M, Azizi P, Tanweer FA, Akhtar MS, Nasehi A. Molecular breeding strategy and challenges towards improvement of blast disease resistance in rice crop. Front Plant Sci. 2015;6(7773):886. doi: 10.3389/fpls.2015.00886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha YS, Ji H, Yun DW, Ahn BO, Lee MC, Suh SC, Lee CS, Ahn EK, Jeon YH, Jin ID, Sohn JK, Koh HJ, Eun MY. Fine mapping of the rice Bph1 gene, which confers resistance to the brown planthopper (Nilaparvata lugens Stål), and development of STS markers for marker-assisted selection. Mol Cells. 2008;26(2):146–151. [PubMed] [Google Scholar]
- Chedgy RJ, KÖllner TG, Constabel CP. Functional characterization of two acyltransferases from Populus trichocarpa capable of synthesizing benzyl benzoate and salicyl benzoate, potential intermediates in salicinoid phenolic glycoside biosynthesis. Phytochemistry. 2015;113:149–159. doi: 10.1016/j.phytochem.2014.10.018. [DOI] [PubMed] [Google Scholar]
- Claridge MF, Hollander J. The ‘biotypes’ of the rice brown planthopper, Nilaparvata lugens. Entomol Exp Appl. 1980;27:23–30. doi: 10.1111/j.1570-7458.1980.tb02942.x. [DOI] [Google Scholar]
- Du B, Zhang WL, Liu BF, Hu J, Wei Z, Shi ZY, He RF, Zhu LL, Chen RZ, Han B, He GC. Identification and characterization of Bph14, a gene conferring resistance to brown planthopper in rice. Proc Natl Acad Sci U S A. 2009;106(52):22163–22168. doi: 10.1073/pnas.0912139106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita D, Kohli A, Horgan FG. Rice resistance to planthoppers and leafhoppers. Crit Rev Plant Sci. 2013;32(3):162–191. doi: 10.1080/07352689.2012.735986. [DOI] [Google Scholar]
- Guo J, Xu C, Wu D, Zhao Y, Qiu Y, Wang X, Ouyang Y, Cai B, Liu X, Jing S, Shangguan X, Wang H, Ma Y, Hu L, Wu Y, Shi S, Wang W, Zhu L, Xu X, Chen R, Du B, He G. Bph6 encodes an exocyst-localized protein and confers broad resistance to planthoppers in rice. Nat Genet. 2018;50:297–306. doi: 10.1038/s41588-018-0039-6. [DOI] [PubMed] [Google Scholar]
- Heinrichs EA, Reissig WH, Valencia S, Chellliah S. Rates and effect of resurgence-inducing insecticides on populations of Nilaparvata lugens (Homoptera: Delphacidae) and its predators. Environ Entomol. 1982;11:1269–1273. doi: 10.1093/ee/11.6.1269. [DOI] [Google Scholar]
- Hirabayashi H, Angeles ER, Kaji R, Ogawa T, Brar DS, Khush GS. Identification of brown planthopper resistance gene derived from O. officinalis using molecular markers in rice. Breed Sci. 1998;48(Suppl):82. [Google Scholar]
- Hirabayashi H, Kaji R, Angeles ER, et al. RFLP analysis of a new gene for resistance to brown planthopper derived from O. officinalis on rice chromosome 4. Breed Sci. 1999;48:48–53. [Google Scholar]
- Hu J, Chang X, Zou L, Tang W, Wu W. Identification and fine mapping of Bph33, a new brown planthopper resistance gene in rice (Oryza sativa L) Rice. 2018;11:55. doi: 10.1186/s12284-018-0249-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Li X, Wu C, Yang C, Hua H, Gao G, Xiao J, He Y. Pyramiding and evaluation of the brown planthopper resistance genes Bph14 and Bph15 in hybrid rice. Mol Breed. 2012;29(1):61–69. doi: 10.1007/s11032-010-9526-x. [DOI] [Google Scholar]
- Hu J, Xiao C, Cheng M, Gao G, Zhang Q, He Y. Fine mapping and pyramiding of brown planthopper resistance genes QBph3 and QBph4 in an introgression line from wild rice O. officinalis. Mol Breed. 2015;35:3. doi: 10.1007/s11032-015-0228-2. [DOI] [Google Scholar]
- Hu J, Xiao C, He Y. Recent progress on the genetics and molecular breeding of brown planthopper resistance in rice. Rice. 2016;9:30. doi: 10.1186/s12284-016-0099-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Xiao H, Hu K, Jiang Y, Zhang Y. Application of marker-assisted backcross to introgress Bph3, Bph14 and Bph15 into an elite indica rice variety for improving its resistance to brown planthopper. Plant Breed. 2016;135:291–300. doi: 10.1111/pbr.12364. [DOI] [Google Scholar]
- Huang D, Qiu Y, Zhang Y, Huang F, Meng J, Wei S, Li R, Chen B. Fine mapping and characterization of BPH27, a brown planthopper resistance gene from wild rice (Oryza rufipogon Griff.) Theor Appl Genet. 2013;126:219–229. doi: 10.1007/s00122-012-1975-7. [DOI] [PubMed] [Google Scholar]
- IRRI . Standard evaluation systems for rice. Phillipines: IRRI; 1996. [Google Scholar]
- Ishii T, Brar DS, Multani DS, Khush GS. Molecular tagging of genes for brown planthopper resistance and earliness introgressed from Oryza australiensis into cultivated rice, O. sativa. Genome. 1994;37:217–221. doi: 10.1139/g94-030. [DOI] [PubMed] [Google Scholar]
- Jairin J, Phengrat K, Teangdeerith S, Vanavichit A, Toojinda T. Mapping of a broad-spectrum brown planthopper resistance gene, Bph3, on rice chromosome 6. Mol Breed. 2007;19(1):35–44. doi: 10.1007/s11032-006-9040-3. [DOI] [Google Scholar]
- Jairin J, Teangdeerith S, Leelagud P, Kothcharerk J, Sansen K, Yi M, Vanavichit A, Toojinda T. Development of rice introgression lines with brown planthopper resistance and KDML105 grain quality characteristics through marker-assisted selection. Field Crop Res. 2009;110(3):263–271. doi: 10.1016/j.fcr.2008.09.009. [DOI] [Google Scholar]
- Jena KK, Hechanova SL, Verdeprado H, Prahalada GD, Kim SR. Development of 25 near-isogenic lines (NILs) with ten BPH resistance genes in rice (Oryza sativa L.): production, resistance spectrum, and molecular analysis. Theor Appl Genet. 2017;130(Suppl):1–16. doi: 10.1007/s00122-017-2963-8. [DOI] [PubMed] [Google Scholar]
- Jena KK, Jeung JU, Lee JH, Choi HC, Brar DS. High-resolution mapping of a new brown planthopper (BPH) resistance gene, Bph18(t), and marker-assisted selection for BPH resistance in rice (Oryza sativa L.) Theor Appl Genet. 2006;112:288–297. doi: 10.1007/s00122-005-0127-8. [DOI] [PubMed] [Google Scholar]
- Jena KK, Kim SM. Current status of brown planthopper (BPH) resistance and genetics. Rice. 2010;3(2–3):161–171. doi: 10.1007/s12284-010-9050-y. [DOI] [Google Scholar]
- Jiang H, Hu J, Li Z, Liu J, Gao G, Zhang Q, Xiao J, He Y. Evaluation and breeding application of six brown planthopper resistance genes in rice maintainer line Jin 23B. Rice. 2018;11(1):22. doi: 10.1186/s12284-018-0215-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman HE, Reddy APK, Hsieh SPY, Merca SD. An improved technique for evaluating resistance to rice varieties of Xanthomonas oryzae pv. oryzae. Plant Dis Rep. 1973;57:537–541. [Google Scholar]
- Kennedy GG, Gould F, Deponti OMB, Stinner RE. Ecological, agricultural, genetic, and commercial considerations in the deployment of insect-resistant germplasm. Environ Entomol. 1987;16(2):327–338. doi: 10.1093/ee/16.2.327. [DOI] [Google Scholar]
- Koyama T, Sato F, Ohmetakagi M. Roles of miR319 and TCP transcription factors in leaf development. Plant Physiol. 2017;175(2):874–885. doi: 10.1104/pp.17.00732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretzschmar T, Burla B, Lee Y, Martinoia E, Nagy R. Functions of ABC transporters in plants. Essays Biochem. 2011;50:145–160. doi: 10.1042/bse0500145. [DOI] [PubMed] [Google Scholar]
- Kumar K, Sarao PS, Bhatia D, Neelam K, Kaur A, Mangat GS, Brar DS, Singh K. High-resolution genetic mapping of a novel brown planthopper resistance locus, Bph34 in Oryza sativa L. × Oryza nivara (Sharma & Shastry) derived interspecific F2 population. Theor Appl Genet. 2018;131(5):1163–1171. doi: 10.1007/s00122-018-3069-7. [DOI] [PubMed] [Google Scholar]
- Li R, Li L, Wei S, Wei Y, Chen Y, Bai D, Yang L, Huang F, Lu W, Zhang X, Li X, Yang X, Wei Y. The evaluation and utilization of new genes for brown planthopper resistance in common wild rice (Oryza rufipogon Griff.) Mol Plant Breed. 2006;4(3):365–371. [Google Scholar]
- Ling KC, Tiongco ER, Aguiero VM. Rice ragged stunt, a new virus disease. Plant Dis Rep. 1978;62(8):701–705. [Google Scholar]
- Liu Y, Wu H, Chen H, Liu Y, He J, Kang H, Sun Z, Pan G, Wang Q, Hu J, Zhou F, et al. A gene cluster encoding lectin receptor kinases confers broad-spectrum and durable insect resistance in rice. Nat Biotechnol. 2015;33:301–305. doi: 10.1038/nbt.3069. [DOI] [PubMed] [Google Scholar]
- Lv W, Du B, Shangguan X, Zhao Y, Pan Y, Zhu L, He Y, He G. BAC and RNA sequencing reveal the brown planthopper resistance gene BPH15 in a recombination cold spot that mediates a unique defense mechanism. BMC Genomics. 2014;15:674. doi: 10.1186/1471-2164-15-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelmore RW, Paran I, Kesseli RV. Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci U S A. 1991;88(21):9828–9832. doi: 10.1073/pnas.88.21.9828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray MG, Thompson WK. Rapid isolation of high molecular-weight plant DNA. Nucleic Acids Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normile D. Reinventing rice to feed the world. Science. 2008;321(5887):330–333. doi: 10.1126/science.321.5887.330. [DOI] [PubMed] [Google Scholar]
- Panda N, Heinrichs EA. Levels of tolerance and antibiosis in rice varieties having moderate resistance to the brown planthopper, Nilaparvata lugens (Stål) (Hemiptera: Delphacidae) Environ Entomol. 1983;12(4):1204–1214. doi: 10.1093/ee/12.4.1204. [DOI] [Google Scholar]
- Pathak MD, Cheng CH, Fortuno ME. Resistance to Nephotettix impicticeps, and Nilaparvata lugens, in varieties of rice. Nature. 1969;223(5205):502–504. doi: 10.1038/223502a0. [DOI] [Google Scholar]
- Pathak PK, Saxena RC, Heinrichs EA. Parafilm sachet for measuring honeydew excretion by Nilaparvata lugens on rice. J Econ Entomol. 1982;75(2):194–195. doi: 10.1093/jee/75.2.194. [DOI] [Google Scholar]
- Qiu Y, Guo J, Jing S, Zhu L, He G. High-resolution mapping of the brown planthopper resistance gene Bph6 in rice and characterizing its resistance in the 9311 and Nipponbare near isogenic backgrounds. Theor Appl Genet. 2010;121:1601–1611. doi: 10.1007/s00122-010-1413-7. [DOI] [PubMed] [Google Scholar]
- Qiu Y, Guo J, Jing S, Zhu L, He G. Development and characterization of japonica rice lines carrying the brown planthopper-resistance genes BPH12 and BPH6. Theor Appl Genet. 2012;124:485–494. doi: 10.1007/s00122-011-1722-5. [DOI] [PubMed] [Google Scholar]
- Qiu Y, Guo J, Jing S, Zhu L, He G. Fine mapping of the rice brown planthopper resistance gene BPH7 and characterization of its resistance in the 93-11 background. Euphytica. 2014;198:369–379. doi: 10.1007/s10681-014-1112-6. [DOI] [Google Scholar]
- Rahman ML, Jiang W, Chu SH, Qiao Y, Ham TH, et al. High-resolution mapping of two rice brown planthopper resistance genes, Bph20(t) and Bph21(t), originating from Oryza minuta. Theor Appl Genet. 2009;119:1237–1246. doi: 10.1007/s00122-009-1125-z. [DOI] [PubMed] [Google Scholar]
- Renganayaki K, Fritz AK, Sadasivam S, Pammi S, Harrington SE, Mccouch SR, Kumar SM, Reddy AS. Mapping and progress toward map-based cloning of brown planthopper biotype-4 resistance gene introgressed from Oryza officinalis into cultivated rice O. sativa. Crop Sci. 2002;42:2112–2117. doi: 10.2135/cropsci2002.2112. [DOI] [Google Scholar]
- Rivera CT, Ou SH, Iida TT. Grassy stunt disease of rice and its transmission by the planthopper Nilaparvata lugens Stål. Plant Dis Rep. 1966;50:453–456. [Google Scholar]
- Sōgawa K. The rice brown planthopper: feeding physiology and host plant interactions. Annu Rev Entomol. 1982;27(1):49–73. doi: 10.1146/annurev.en.27.010182.000405. [DOI] [Google Scholar]
- Sun L, Su C, Wang C, Zhai HQ, Wan JM. Mapping of a major resistance gene to brown planthopper in the rice cultivar Rathu Heenati. Breed Sci. 2005;55:391–396. doi: 10.1270/jsbbs.55.391. [DOI] [Google Scholar]
- Tanaka K, Endo S, Kazano H. Toxicity of insecticides to predators of rice planthoppers: spiders, the mirid bug and the dryinid wasp. Appl Entomol Zool. 2000;35:177–187. doi: 10.1303/aez.2000.177. [DOI] [Google Scholar]
- Wang H, Shi S, Guo Q, Nie L, Du B, Chen R, Zhu L, He G. High-resolution mapping of a gene conferring strong antibiosis to brown planthopper and developing resistant near-isogenic lines in 9311 background. Mol Breed. 2018;38:107. doi: 10.1007/s11032-018-0859-1. [DOI] [Google Scholar]
- Wang Y, Cao L, Zhang Y, Cao C, Liu F, Huang F, Qiu Y, Li R, Lou X. Map-based cloning and characterization of BPH29, a B3 domain-containing recessive gene conferring brown planthopper resistance in rice. J Exp Bot. 2015;66:6035–6045. doi: 10.1093/jxb/erv318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Ren X, Weng Q, Zhu L, He G. Molecular mapping and genetic analysis of a rice brown planthopper (Nilaparvata lugens Stål) resistance gene. Hereditas. 2002;136:39–43. doi: 10.1034/j.1601-5223.2002.1360106.x. [DOI] [PubMed] [Google Scholar]
- Yang H, You A, Yang Z, Zhang F, He R, Zhu L, He G. High-resolution genetic mapping at the Bph15 locus for brown planthopper resistance in rice (Oryza sativa L.) Theor Appl Genet. 2004;110:182–191. doi: 10.1007/s00122-004-1844-0. [DOI] [PubMed] [Google Scholar]
- Yin D, Liu X, Shi Z, Li D, Zhu L. An AT-hook protein DEPRESSED PALEA1 physically interacts with the TCP family transcription factor RETARDED PALEA1 in rice. Biochem Biophys Res Commun. 2018;495(1):487–492. doi: 10.1016/j.bbrc.2017.11.031. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Huang J, Wang Z, Jing S, Wang Y, et al. Allelic diversity in an NLR gene BPH9 enables rice to combat planthopper variation. Proc Natl Acad Sci U S A. 2016;113:12850–12855. doi: 10.1073/pnas.1614862113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. The pedigree of pyramiding lines (PYLs) containing resistance genes Bph36, Bph27, Bph29, Bph3, and Xa23 developed using marker-assisted selection (MAS). (TIF 359 kb)
Figure S2 Amplified bands of BC1F2 individuals derived from KW/RBPH16 with InDel marker X17 and detected by 7% PAGE. M: Marker DL2000, RBPH16: resistance parent, KW: susceptible parent, Lanes 1–22: individuals of BC1F2. (TIF 638 kb)
Table S1 Markers for marker-assisted selection (MAS) of BPH and bacterial blight resistance genes. (DOCX 16 kb)
Data Availability Statement
All relevant data are provided as Additional file 3: Table S1.