Summary
The early detection of cancers has the potential to save many lives. A recent attempt has been demonstrated successful. However, we note several critical limitations. Given the central importance and broad impact of early cancer detection, we aspire to address those limitations. We explore different supervised learning approaches for multiple cancer type detection and observe significant improvements; for instance, one of our approaches (i.e., CancerA1DE) can double the existing sensitivity from 38% to 77% for the earliest cancer detection (i.e., Stage I) at the 99% specificity level. For Stage II, it can even reach up to about 90% across multiple cancer types. In addition, CancerA1DE can also double the existing sensitivity from 30% to 70% for detecting breast cancers at the 99% specificity level. Data and model analysis are conducted to reveal the underlying reasons. A website is built at http://cancer.cs.cityu.edu.hk/.
Subject Areas: Biological Sciences, Cancer Systems Biology, Cancer, Algorithms, Bioinformatics
Graphical Abstract
Highlights
-
•
We propose an approach (CancerA1DE) to detect early cancers from blood
-
•
CancerA1DE doubles the existing sensitivity for the stage I cancer detection
-
•
For stage II cancers, it can reach up to 90% across multiple cancer types
-
•
The related software is opened and released for future follow-up works
Biological Sciences; Cancer Systems Biology; Cancer; Algorithms; Bioinformatics
Introduction
Cancers are prevalent across the globe (Torre et al., 2015). Millions of deaths could be found due to various cancer types every year (Chen et al., 2016). Unfortunately, the number of deaths is still projected to be increasing even in developed countries (Rahib et al., 2014). Therefore, it is critical to address those cancers in a timely manner.
In particular, the works in cancer protein marker discovery have been fruitful in the past years (Stoeva et al., 2006); for instance, four analytes (leptin, prolactin, osteopontin, and insulin-like growth factor-II) have been discovered to be predictive in the early detection of ovarian cancers (Mor et al., 2005). Three years later, macrophage inhibitory factor and CA-125 have been proposed on top of the previous four proteins to improve the early detection further (Visintin et al., 2008). The combinatorial expression patterns among HSP-27, GST, Annexin II, and L-FABP are also associated with the lymph node metastasis in colorectal cancer (Pei et al., 2007). Blood and fecal protein markers have also been implicated for colorectal cancer diagnosis (Karl et al., 2008). Multiple markers have also been reported for breast cancers (Harbeck et al., 2014), pancreatic cancers (Takadate et al., 2013), lung cancers (Buszewski et al., 2012), gastric cancers (Rugge et al., 2015), liver cancers (Bertino et al., 2012), esophageal cancers (Napier et al., 2014), and others (Zheng et al., 2005). For a list of well-proved markers, one can refer to the past survey (Polanski and Anderson, 2006).
Given the valuable marker information, it is obvious that one can harness the existing advancement in artificial intelligence to comprehend, digest, and combine those information into a comprehensive and accurate early cancer prediction tool. In particular, there are two major modeling paradigms for such a cancer prediction task under the context of binary classification in machine learning: discriminative learning and generative modeling. For discriminative learning, we seek to explore if we can learn any high-dimensional boundary to discriminate cancer cases from normal cases in the available marker space. For generative modeling, we seek to explore if we can build a model to capture the marker distributions of cancer cases and another model to capture the marker distributions of normal cases. Once the two models are ready, we can compute a sample probability belonging to cancer or normal cases as the early cancer prediction.
A recent method called CancerSEEK takes advantages of multiple protein markers in blood for cancer detection and localization across different cancer types and stages using discriminative learning (Cohen et al., 2018). However, we notice that the CancerSEEK method has several limitations; for instance, its front-line cancer detection component is based on logistic regression, whereby linear assumption on different markers is hardly realistic. Its second-line cancer type localization component is based on random forest, a modeling known to be difficult for interpretations. From the user perspective, its lack of public Web service also limits its potential impacts.
To address those limitations altogether, we seek to explore different approaches to solve the multiple cancer type detection problem. A public Web server with open programs is also provided for scientific reproducibility and impact at http://cancer.cs.cityu.edu.hk/.
Results
Data Collection
We have collected the multianalyte blood test data from Cohen et al. (2018). Those data have been processed according to the supplementary guideline provided, resulting in two datasets.
The first dataset has 1,817 patient blood test records, which are designed and adopted to build models to detect cancers as the front-line detector in a binary manner (i.e., cancer or normal). Therefore, to be scalable and economical, it has the minimal number of input feature information involving eight circulating protein marker concentrations and one cell-free DNA mutation score (OmegaScore) as listed in Table S1.
The second dataset has 626 patient blood test records, which are designed and adopted to build models to localize cancer types as the second-line diagnosis (i.e., Breast, Colorectum, Upper GI, Liver, Lung, Ovary, or Pancreas). Therefore, its input feature set covers the previous nine features and includes additional 31 protein markers and patient gender as listed in Table 1.
Table 1.
Feature List for Cancer Type Localization ranked by Information Gain (InfoG)
| InfoG | Input Features | Feature Description |
|---|---|---|
| 1.0389 | TGFa (pg/mL) | Circulating Transforming Growth Factor α Concentration in pg/mL |
| 0.8301 | HE4 (pg/mL) | Circulating Human Epididymis Protein 4 Concentration in pg/mL |
| 0.6135 | sFas (pg/mL) | Circulating soluble Fas Cell Surface Death Receptor Concentration in pg/mL |
| 0.5372 | Thrombospondin-2 (pg/mL) | Circulating Thrombospondin-2 Concentration in pg/mL |
| 0.5073 | AFP (pg/mL) | Circulating Alpha Fetoprotein Precursor Concentration in pg/mL |
| 0.3759 | G-CSF (pg/mL) | Circulating Granulocyte-Colony Stimulating Factor Concentration in pg/mL |
| 0.3633 | IL-6 (pg/mL) | Circulating Interleukin-6 Concentration in pg/mL |
| 0.3597 | CA-125 (U/mL) | Circulating Cancer Antigen 125 Concentration in U/mL |
| 0.2568 | Sex | Patient Gender Information (Male or Female) |
| 0.2352 | sHER2/sEGFR2/sErbB2 (pg/mL) | Circulating sHER2/sEGFR2/sErbB2 Concentration in pg/mL |
| 0.2259 | TIMP-2 (pg/mL) | Circulating Tissue Inhibitor of Metalloproteinases 2 Concentration in pg/mL |
| 0.2231 | CD44 (ng/mL) | Circulating CD44 Concentration in pg/mL |
| 0.183 | CA19-9 (U/mL) | Circulating Cancer Antigen 19-9 Concentration in U/mL |
| 0.1805 | IL-8 (pg/mL) | Circulating Interleukin-8 Concentration in pg/mL |
| 0.164 | CA 15-3 (U/mL) | Circulating Cancer Antigen 15-3 Concentration in U/mL |
| 0.1448 | HGF (pg/mL) | Circulating Hepatocyte Growth Factor Concentration in pg/mL |
| 0.1431 | OPG (ng/mL) | Circulating Osteopontin Concentration in pg/mL |
| 0.1414 | GDF15 (ng/mL) | Circulating Growth Differentiation Factor 15 Concentration in ng/mL |
| 0.1384 | Leptin (pg/mL) | Circulating Leptin Concentration in pg/mL |
| 0.1271 | Myeloperoxidase (ng/mL) | Circulating Myeloperoxidase Concentration in ng/mL |
| 0.125 | Kallikrein-6 (pg/mL) | Circulating Kallikrein-6 Concentration in pg/mL |
| 0.1173 | TIMP-1 (pg/mL) | Circulating Tissue Inhibitor of Metalloproteinases 1 Concentration in pg/mL |
| 0.1122 | Midkine (pg/mL) | Circulating Midkine Concentration in pg/mL |
| 0.1095 | Prolactin (pg/mL) | Circulating Prolactin Concentration in pg/mL |
| 0.1032 | Mesothelin (ng/mL) | Circulating Mesothelin Concentration in ng/mL |
| 0.103 | Galectin-3 (ng/mL) | Circulating Galectin-3 Concentration in ng/mL |
| 0.096 | OPN (pg/mL) | Circulating Osteopontin Concentration in pg/mL |
| 0.0956 | NSE (ng/mL) | Circulating Neuron-Specific Enolase Concentration in ng/mL |
| 0.0901 | sEGFR (pg/mL) | Circulating Soluble Epidermal Growth Factor Receptor Concentration in pg/mL |
| 0.0901 | CEA (pg/mL) | Circulating Carcinoembryonic Antigen Concentration in pg/mL |
| 0.085 | AXL (pg/mL) | Circulating AXL Receptor Tyrosine Kinase Concentration in pg/mL |
| 0.0771 | sPECAM-1 (pg/mL) | Circulating Soluble Platelet and Endothelial Cell Adhesion Molecule 1 Concentration in pg/mL |
| 0.0637 | SHBG (nM) | Circulating Sex Hormone-Binding Globulin Concentration in nM |
| 0.0635 | OmegaScore | Omega Score for Mutations in Circulating Cell-Free DNA [Cohen et al. (2018)] |
| 0 | Angiopoietin-2 (pg/mL) | Circulating Angiopoietin-2 Concentration in pg/mL |
| 0 | DKK1 (ng/mL) | Circulating Dickkopf WNT Signaling Pathway Inhibitor 1 Concentration in ng/mL |
| 0 | CYFRA 21-1 (pg/mL) | Circulating Cytokeratin-19 Fragment Concentration in pg/mL |
| 0 | PAR (pg/mL) | Circulating Protease-Activated Receptor Concentration in pg/mL |
| 0 | Endoglin (pg/mL) | Circulating Endoglin Concentration in pg/mL |
| 0 | FGF2 (pg/mL) | Circulating Fibroblast Growth Factor 2 Concentration in pg/mL |
| 0 | Follistatin (pg/mL) | Circulating Follistatin Concentration in pg/mL |
To visualize the first and second datasets, we have adopted a series of dimensional reduction techniques to project the datasets onto two-dimensional spaces as visualized in Figures S1 and S9, respectively. Linear discriminant analysis was also conducted as depicted in Figure S2. Unfortunately, it can be observed that the datasets are not easily separated even on the full datasets, necessitating advanced algorithmic development under the cross-validations with isolated separations of training and testing data in subsequent sections.
Model Descriptions
To build cancer detection models from the aforementioned datasets, we consider it as a supervised learning task from the machine learning perspective. Therefore, we have selected a range of multiclass supervised learning algorithms: AODE (Webb et al., 2005), deep learning (Angermueller et al., 2016), decision tree (Bhargava et al., 2013), and naive Bayes (NB) (Lewis, 1998). We note that logistic regression and random forest have already been adopted and encapsulated in CancerSEEK (Cohen et al., 2018).
Model Parameter Setting
For AODE, we have adopted A1DE (Webb et al., 2005) and A2DE (Webb et al., 2012) as our classifiers (namely, CancerA1DE and CancerA2DE). The minimum description length (MDL) principle is adopted for continuous marker feature discretization (see the Supplemental Information). For deep learning, since the problem here is a very standard supervised learning task with well-crafted input features, we do not need to increase any model complexity for deep feature learning. Therefore, we have adopted the deep feedforward neural networks with one hidden layer, two hidden layers, and three hidden layers (namely, DeepLearning1, DeepLearning2, and DeepLearning3, respectively) (Angermueller et al., 2016). The remaining training setting follows the default settings of WEKA (Hall et al., 2009). For decision tree, J48 tree building method is adopted (Bhargava et al., 2013). For NB, a Gaussian distribution is assumed for each continuous feature under each class (Lewis, 1998). The remaining parameter setting follows the default parameter values of WEKA (Hall et al., 2009).
CancerA1DE Modeling
CancerA1DE has demonstrated the best performance in the subsequent sections. Therefore, we briefly describe CancerA1DE in this section. CancerA1DE is based on the A1DE framework (Webb et al., 2005), which is a variant of NB. Weakening the attribute (or feature) independence assumption on the NB classifier has been proved to achieve significant improvement by various approaches. Nevertheless, the techniques such as the Lazy Bayesian Rules and Tree Augmented Naive Bayes weaken the assumption with the cost of intensive computational power because the optimal subset selection procedure from all attributes or parent attributes could be computationally intensive. In addition, the model selection tends to over-fit the training data and thus increase the estimation variance. To speed it up, Aggregating One-Dependence Estimators (A1DE) was developed; A1DE is designed to avoid any model selection by enumerating all possible 1-dependence classifiers in each of which there is an attribute as the parent of all others (Webb et al., 2005). The mathematical details can be found in the Supplemental Information.
The advantage of A1DE is its relatively low complexities as tabulated in Table S2. We can observe that, although A1DE has significant performance gain over the original NB, its training time complexity is nether quadratic to the number of samples nor cubic to the number of feature attributes. Its training time complexity is just O(tn2) where t is the number of samples and n is the number of feature attributes. Such a property guarantees that it can scale with the number of samples in a linear manner. Its quadratic complexity to the number of feature attributes (i.e., markers here) should not be a big problem since the number of blood test markers is usually finite and limited. In addition, its model formulation is incremental; it means that the CancerA1DE model can be easily updated with new blood marker samples, unlike random forest (i.e. CancerSEEK).
Cancer Detection Results
As ranked in Table S1, it is not surprising that the cancer antigen markers are the most informative features for cancer detection. To act as a control, we have also trained random forests on the features using the Python scikit-learn package (Pedregosa et al., 2011) and explored the feature ranking based on three different measurements: decrease in purity, decrease in accuracy, and recursive feature elimination. The feature ranking results are illustrated in Figure S3. Contrary to the information gain ranking on Table S1, the cancer antigen markers are no longer the top predictive features. Instead, we observe the opposite trend for the purity and accuracy measurements; such a phenomenon exemplifies the underlying complex behavior for cancer detection. It also necessitates our subsequent machine learning approaches.
To explore those features (also known as protein biomarkers) further, we have computed their correlation matrix as visualized in Figure S4. Congruent with our general belief, the cancer antigen markers are positively correlated with statistical significance (p < 0.001). Interestingly, it can be observed that TIMP-1, Myeloperoxidase (or MPO), OPN (or SPP1), and HGF form a positively correlated feature cluster. To explore their relationships, we have mapped the protein markers into STRING network analysis (Szklarczyk et al., 2016) as demonstrated in Figure S5. From the figure, it is now clear that those four proteins did demonstrate different levels of evidences for their interactions; it justifies their positive correlations. On the other hand, we also observe an enriched pathway from the STRING results: PI3K-Akt signaling pathway (ID: 04151) with the supporting proteins (HGF, PRL, and SPP1) at FPR = 0.0243; it indicates that such a feature subset serves as proxy measurements for that pathway in the following cancer detection tasks.
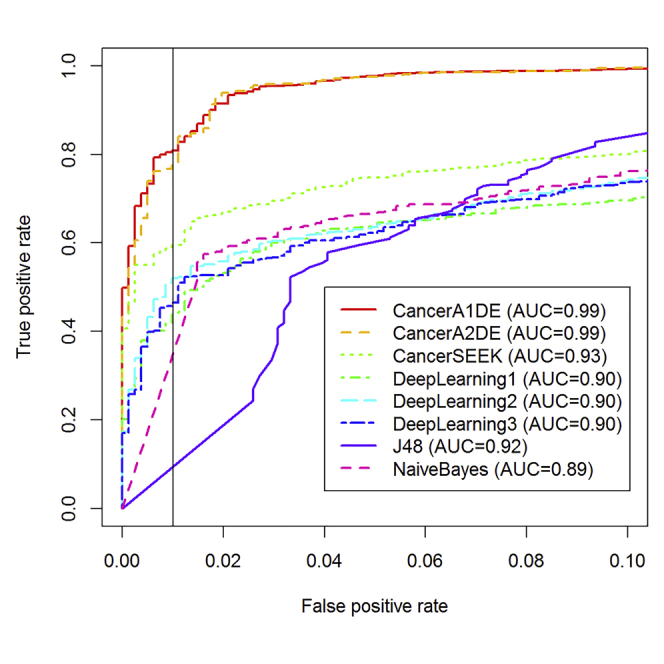
Given those features, we have benchmarked different supervised learning models on the 1,817-patient dataset for cancer detection (i.e., cancer or normal) under 10-fold cross-validations (i.e., 10 randomly divided held-out data subsets for testing, whereas the remaining are for training in 10 rounds) as visualized in Figure 1.
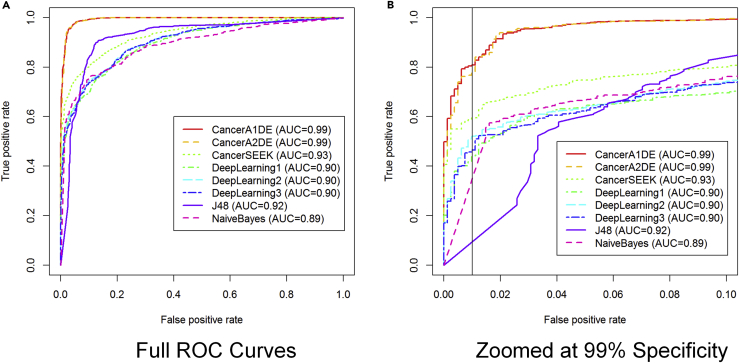
Figure 1.
Receiver Operating Characteristic (ROC) curves for Cancer Detection
Different methods have different colors and line styles. The curves are generated under 10-fold cross-validations. The vertical black line on the right panel is drawn at the 99% specificity level.
(A) Full Scale ROC Curves.
(B) ROC Curves Zoomed to FPR<=0.1.
Interestingly, it can be observed that our proposed CancerA1DE and CancerA2DE outperformed CancerSEEK by a significant margin in terms of the Area Under Curve (AUC) values (0.99 vs 0.93). If the figure is zoomed to the regions with false-positive rates less than 0.1, such a performance gain is even pronounced. At the 99% specificity level, our CancerA1DE and CancerA2DE can even achieve around 80% sensitivity, which is higher than that of CancerSEEK by 20% (Cohen et al., 2018). We attribute such a performance gain to two reasons: (1) the generative modeling approach undertaken by CancerA1DE and CancerA2DE and (2) the feature discretization of CancerA1DE and CancerA2DE based on the MDL principle (Kononenko, 1995). Reason (1) is obvious in that generative modeling can generalize itself well over the discriminative modeling, which is prone to over-fitting, whereas reason (2) is not intuitive but insightful. Its feature discretization performance suggests that we should focus on the protein marker concentration intervals rather than on the actual protein marker concentration magnitudes. As depicted in Figure S6, we can observe that the input features are well separated into different groups based on MDL. In particular, it is interesting that the feature groups are associated with different levels of misclassification risks, which can well inform CancerA1DE and CancerA2DE to make cancer detection decisions in a probabilistically generative manner.
To confirm its performance further, we have conducted a performance sensitivity analysis based on different withheld data amount settings. Specifically, we randomly divided the dataset into two subsets: training set and testing set in different proportions as tabulated in the Table S3. From the table, we can observe that our CancerA1DE and CancerA2DE models can reach the AUC performance of 0.98 as soon as it has about 20% data for model training (i.e., 363 patient samples). Such a rapid learning characteristic is important as patient sample costs are substantial. Its underlying Bayesian generative modeling also prevents it from over-fitting.
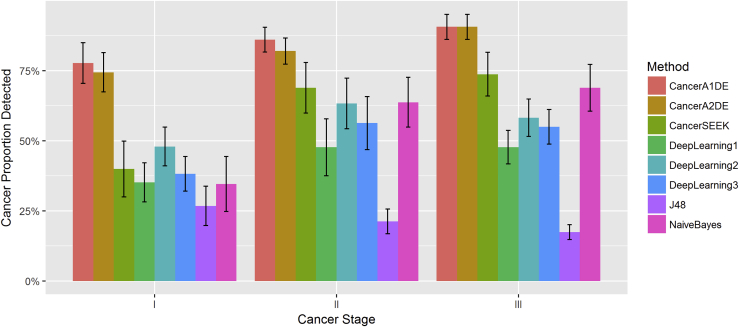
In the context of cancer detection, its early detection performance is the most important task in the clinical setting. Therefore, we proceed to wonder how early different methods can detect cancers for timely follow-ups. On the other hand, we would also like to ensure that the false-positive rate can be minimized. Therefore, we limit our cancer detections to the 99% specificity level where the detected proportions of cancers with different stages are depicted in Figure 2.
Figure 2.
Proportion of Detected Cancers with Different Stages at the 99% Specificity Level
Each color represents a method, and the horizontal axis has been ordered by cancer stages. Each bar represents the median sensitivity of each method on each cancer stage with standard errors.
We can observe that our CancerA1DE and CancerA2DE can detect close to 77% of the early-stage cancers (i.e., Stage I), whereas the others are well below 50%. Such a phenomenon is clinically critical since early cancer treatment intervention can often lead to increased patient survival rates (Miller et al., 2016).
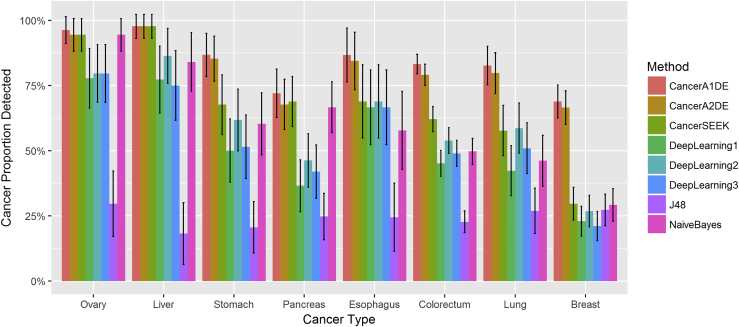
To investigate whether any of the methods is biased to specific cancer types, we have also plotted the detected proportion of different cancer types at the 99% specificity level in Figure 3.
Figure 3.
Detected Proportions of Different Cancer Types at the 99% Specificity Level
Different colors represent different methods. The horizontal axis is ordered by cancer types. Each bar represents the sensitivity of each method on each cancer type with 95% confidence intervals.
Clearly, it can be observed that both CancerA1DE and CancerA2DE have broadly competitive edges over the other state-of-the-art methods across different cancer types. It can achieve at least 60% sensitivity for all tested cancer types at the 99% specificity level. In particular, CancerA1DE and CancerA2DE can double the detected proportions of breast cancer, which is the second most common cancer diagnosed among women in the United States (DeSantis et al., 2014).
To provide further insights, we have plotted the conditional probabilities of CancerA1DE for all possible combinations. An example is depicted in Figure S7, whereas the others are exhaustively enumerated in Figure S8. From those figures, it can be observed that the conditional probabilities are highly specific to normal samples (see the spikes and the monotonically decreasing trends in the figures). This implies that the CancerA1DE modeling places strong pattern recognition emphasis on filtering out the normal samples; therefore, it explains why the CancerA1DE can achieve the highly sensitive detection performance at the 99% specificity level as previously demonstrated.
Cancer Type Localization Results
As ranked in Table 1 and Figure S10, it is interesting to observe that the previous nine features in cancer detection are no longer the top features for cancer type localization. This necessitates our motivation that we have to include additional features (i.e., additional 31 protein markers and patient gender) for cancer type localization.
However, we also need to ensure the nonredundancy of those features. Therefore, we have computed the feature correlation matrix as heatmap in Figure S11. It can be observed that the protein marker features are not highly correlated to each other. Nonetheless, if we zoom in specific regions, we did observe a few weak feature communities here. Therefore, similar to the previous section, we have mapped the protein marker names into STRING network analysis (Szklarczyk et al., 2016) in Figure S12. Unfortunately, most of the interactions are based on text mining, which is not conclusive. Nonetheless, based on the evidence strength, we can still observe few existing communities that are congruent with our observations from the correlation heatmap. Different from the previous cancer detection task, several pathways are enriched as listed in Table S4; this indicates the comprehensiveness and diversity of the protein markers for the following cancer type localization tasks.
The previous methods are also adopted here for fair benchmark comparisons. In particular, the multiclass supervised learning setting is experimented on the 626-patient dataset for cancer type localization (i.e., Breast, Colorectum, Upper GI, Liver, Lung, Ovary, and Pancreas) under 10-fold cross-validations as visualized in Figure S13.
It can be observed that both CancerA1DE and CancerA2DE can well maintain the receiver operating characteristic curves above the diagonal baseline, whereas the others suffer from several drawbacks; for instance, CancerSEEK appears to be polarized into two extreme performance groups; this indicates that CancerSEEK, which is based on random forest, could have bias towards specific cancer type differentiations, consistent with the observation to be made from Figure 4. On the other hand, the deep learning methods cannot scale to full performance once the specificity level is relaxed. Interestingly, the NB classifier demonstrates surprising classification performance across few cancer types. Congruent with CancerA1DE and CancerA2DE, such a phenomenon reflects that multiclass generative modeling is more suitable than multiclass discriminative modeling for the cancer type localization here.
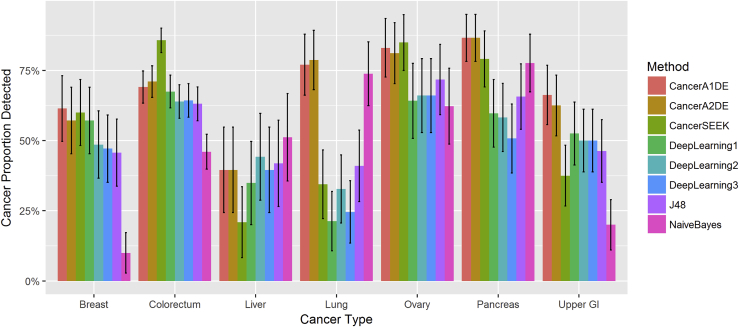
Figure 4.
Localized Proportions of Different Cancer Types using the Top One Prediction Approach
Different colors represent different methods. The horizontal axis is ordered by cancer types. Each bar represents the sensitivity of each method on each cancer type with 95% confidence intervals.
To compare the methods in a more realistic setting than the previous curves, we seek to validate the methods for the top-one predictions. In other words, for each patient record, we allow each method to predict and localize its cancer type once only. Under the 10-fold cross-validation, the results are visualized in Figure 4.
Congruent with the ROC curves, CancerA1DE and CancerA2DE demonstrate stable performance across different cancer types. Although CancerSEEK can score well on colorectal and ovarian cancers, its random forest has sacrificed its performance on other cancer types. In addition, we also observe that most methods cannot perform well on the liver cancer localization. Such a performance deviation can be attributed to the scarce data availability issue here.
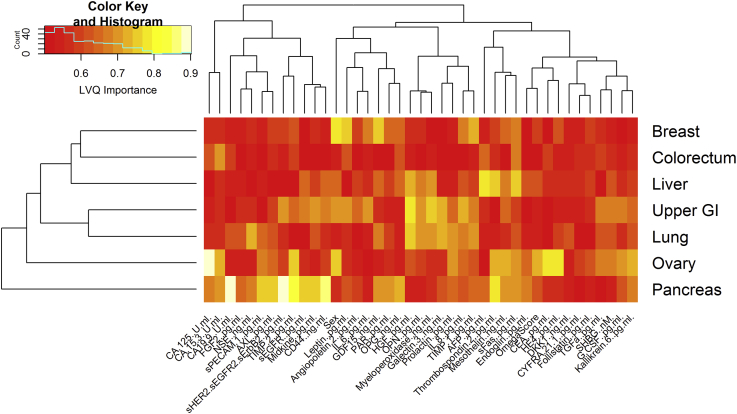
To investigate the reasons further, we have adopted Learning Vector Quantization to perform feature importance ranking under the one-class-versus-all setting with 10-fold cross-validations. The complete results are listed in Figure S14. To summarize the results, we have performed hierarchical clustering and heatmap visualization on the feature importance results as depicted in Figure 5. Interestingly, we observe that the cancer types can be clustered into three groups. The ovarian cancers and pancreatic cancers form independent groups; it explains why most methods can perform well on those two cancers since their important features are well isolated from the others. In contrast, the other five cancer types share significant portions of important features. This forms a machine learning trap that discriminative learning algorithms (e.g., the random forest adopted by CancerSEEK) could be biased towards the cancer type that has the largest data samples (e.g., colorectal cancer here as demonstrated in Figure 4).
Figure 5.
Feature Importance Heatmap for Cancer Type Localization under One-Class-versus-Others Setting
The feature rankings are measured based on the Learning Vector Quantization (LVQ) building under Python caret package (Bischl et al., 2016). Ten-fold cross-validations are run to compute the feature importance values. After that, the function “heatmap.2” in R language is adopted with the default setting to cluster and visualize the feature importance values. Further details can be found in Figure S14.
Discussion
In this study, we have explored different approaches for multiple cancer type detection from multianalyte blood test results. With eight circulating protein markers and one circulating DNA mutation score, the best approach (CancerA1DE) can outperform the existing approach (CancerSEEK) for cancer detection at the 99% specificity level.
Nonetheless, such an observation is based only on the available 1,817 patient blood samples. It is undeniable that different approaches can result in different performance on different datasets. This is further complicated by the fact that we have diverse cancer types within the 1,817 patient blood samples. The main competitiveness of our approach is that it can take into account the nonlinear relationships between the available markers, although the only cost is the slightly increased computational complexity. However, we conceive such a cost as manageable under the current computing environment (e.g., it takes only less than 1 sec for the CancerA1DE model building on our desktop computer using the Weka library).
On the other hand, for cancer type localization with additional 31 circulating protein markers and gender information, we observed diverse performance behavior among different approaches. Such performance discrepancy is attributed to several reasons ranging from the cancer-type-specific feature importance to the supervised learning methodology paradigm (i.e., generative or discriminative modeling) as revealed. In particular, we observe the resurge of the generative Bayesian approach (CancerA1DE), which has been demonstrated to be more successful than the state-of-the-art methods such as random forest (CancerSEEK) and deep learning in this study.
With the advancement of edge computing devices (e.g., mobile phones) and biotechnology (e.g., biosensor devices), we envision that the integration between them can provide one-stop blood tests accessible to patients. Our current study can provide a reference to deploy a sensitive and robust blood test model for routine cancer screening at the software level.
In the future, it can be interesting if additional marker features can be explored; however, the related costs are substantial on actual patient cohorts. Comprehensive genomics and pathway analysis on the newly arisen markers are also needed for providing mechanistic insights before incorporating into the current study. Continuous assessment on the proposed approaches is also required for extensive uses. Therefore, with the public Web server and open software programs for scientific reproducibility, we believe that this study can serve as a useful platform for future studies on early cancer detection from blood.
Limitation of Study
The current study is limited by the absence of a truly independent validation set.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
The authors would like to thank the reviewers for their constructive comments. They would also like to thank Prashant Sridhar and Ajay Rajnikanth for their English proofreading. The work described in this paper was substantially supported by three grants from the Research Grants Council of the Hong Kong Special Administrative Region [CityU 21200816], [CityU 11203217], and [CityU 11200218]. We acknowledge the donation support of the Titan Xp GPU from the Nvidia Corporation.
Author Contributions
K.-C.W. conceived and designed the study; K.-C.W., X.L., C.L., C.P., Q.L, S.K., and J.Y. supervised the study; K.-C.W., J.C., J.Z., J.L., S.Y., and S.Z. implemented the study; K.-C.W., J.C., X.L., and J.Y. wrote the manuscript.
Declaration of Interests
The authors declare no competing interests.
Published: May 31, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2019.04.035.
Supplemental Information
References
- Angermueller C., Parnamaa T., Parts L., Stegle O. Deep learning for computational biology. Mol. Syst. Biol. 2016;12:878. doi: 10.15252/msb.20156651. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angermueller, C., Parnamaa, T., Parts, L., and Stegle, O. (2016). Deep learning for computational biology. Molecular systems biology, 12(7):878. [DOI] [PMC free article] [PubMed]
- Bertino G., Ardiri A., Malaguarnera M., Malaguarnera G., Bertino N., Calvagno G.S. Hepatocellualar carcinoma serum markers. In: Fojo A.T., editor. Vol 39. Elsevier; 2012. pp. 410–433. (Seminars in Oncology). [DOI] [PubMed] [Google Scholar]; Bertino, G., Ardiri, A., Malaguarnera, M., Malaguarnera, G., Bertino, N., and Calvagno, G. S. (2012). Hepatocellualar carcinoma serum markers. In Seminars in oncology, Vol 39, pages 410-433. Elsevier. [DOI] [PubMed]
- Bhargava N., Sharma G., Bhargava R., Mathuria M. Decision tree analysis on j48 algorithm for data mining. Proc. Int. J. Adv. Res. Comput. Sci. Softw. Eng. 2013;3 [Google Scholar]; Bhargava, N., Sharma, G., Bhargava, R., and Mathuria, M. (2013). Decision tree analysis on j48 algorithm for data mining. Proceedings of International Journal of Advanced Research in Computer Science and Software Engineering, 3(6).
- Bischl B., Lang M., Kotthoff L., Schiffner J., Richter J., Studerus E., Casalicchio G., Jones Z.M. mlr: Machine learning in r. J. Mach. Learn. Res. 2016;17:1–5. [Google Scholar]; Bischl, B., Lang, M., Kotthoff, L., Schiffner, J., Richter, J., Studerus, E., Casalicchio, G., and Jones, Z. M. (2016). mlr: Machine learning in r. Journal of Machine Learning Research, 17(170):1-5.
- Buszewski B., Ligor T., Jezierski T., Wenda-Piesik A., Walczak M., Rudnicka J. Identification of volatile lung cancer markers by gas chromatography–mass spectrometry: comparison with discrimination by canines. Anal. Bioanal. Chem. 2012;404:141–146. doi: 10.1007/s00216-012-6102-8. [DOI] [PMC free article] [PubMed] [Google Scholar]; Buszewski, B., Ligor, T., Jezierski, T., Wenda-Piesik, A., Walczak, M., and Rudnicka, J. (2012). Identification of volatile lung cancer markers by gas chromatography-mass spectrometry: comparison with discrimination by canines. Analytical and bioanalytical chemistry, 404(1):141-146. [DOI] [PMC free article] [PubMed]
- Chen W., Zheng R., Baade P.D., Zhang S., Zeng H., Bray F., Jemal A., Yu X.Q., He J. Cancer statistics in China, 2015. CA Cancer J. Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]; Chen, W., Zheng, R., Baade, P. D., Zhang, S., Zeng, H., Bray, F., Jemal, A., Yu, X. Q., and He, J. (2016). Cancer statistics in china, 2015. CA: a cancer journal for clinicians, 66(2):115-132. [DOI] [PubMed]
- Cohen J.D., Li L., Wang Y., Thoburn C., Afsari B., Danilova L., Douville C., Javed A.A., Wong F., Mattox A. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science. 2018;359:926–930. doi: 10.1126/science.aar3247. [DOI] [PMC free article] [PubMed] [Google Scholar]; Cohen, J. D., Li, L., Wang, Y., Thoburn, C., Afsari, B., Danilova, L., Douville, C., Javed, A. A., Wong, F., Mattox, A., Hruban, R. H., Wolfgang, C. L., Goggins, M. G., Dal Molin, M., Wang, T. L., Roden, R., Klein, A. P., Ptak, J., Dobbyn, L., Schaefer, J., Silliman, N., Popoli, M., Vogelstein, J. T., Browne, J. D., Schoen, R. E., Brand, R. E., Tie, J., Gibbs, P., Wong, H. L., Mansfield, A. S., Jen, J., Hanash, S. M., Falconi, M., Allen, P. J., Zhou, S., Bettegowda, C., Diaz, L. A., Tomasetti, C., Kinzler, K. W., Vogelstein, B., Lennon, A. M., and Papadopoulos, N. (2018). Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science, 359(6378):926-930. [DOI] [PMC free article] [PubMed]
- DeSantis C., Ma J., Bryan L., Jemal A. Breast cancer statistics, 2013. CA Cancer J. Clin. 2014;64:52–62. doi: 10.3322/caac.21203. [DOI] [PubMed] [Google Scholar]; DeSantis, C., Ma, J., Bryan, L., and Jemal, A. (2014). Breast cancer statistics, 2013. CA: a cancer journal for clinicians, 64(1):52-62. [DOI] [PubMed]
- Hall M., Frank E., Holmes G., Pfahringer B., Reutemann P., Witten I.H. The weka data mining software: an update. ACM SIGKDD Explor. Newslett. 2009;11:10–18. [Google Scholar]; Hall, M., Frank, E., Holmes, G., Pfahringer, B., Reutemann, P., and Witten, I. H. (2009). The weka data mining software: an update. ACM SIGKDD explorations newsletter, 11(1):10-18.
- Harbeck N., Sotlar K., Wuerstlein R., Doisneau-Sixou S. Molecular and protein markers for clinical decision making in breast cancer: today and tomorrow. Cancer Treat. Rev. 2014;40:434–444. doi: 10.1016/j.ctrv.2013.09.014. [DOI] [PubMed] [Google Scholar]; Harbeck, N., Sotlar, K., Wuerstlein, R., and Doisneau-Sixou, S. (2014). Molecular and protein markers for clinical decision making in breast cancer: today and tomorrow. Cancer treatment reviews, 40(3):434-444. [DOI] [PubMed]
- Karl J., Wild N., Tacke M., Andres H., Garczarek U., Rollinger W., Zolg W. Improved diagnosis of colorectal cancer using a combination of fecal occult blood and novel fecal protein markers. Clin. Gastroenterol. Hepatol. 2008;6:1122–1128. doi: 10.1016/j.cgh.2008.04.021. [DOI] [PubMed] [Google Scholar]; Karl, J., Wild, N., Tacke, M., Andres, H., Garczarek, U., Rollinger, W., and Zolg, W. (2008). Improved diagnosis of colorectal cancer using a combination of fecal occult blood and novel fecal protein markers. Clinical Gastroenterology and Hepatology, 6(10):1122-1128. [DOI] [PubMed]
- Kononenko, I. (1995). On biases in estimating multi-valued attributes. In 14th International Joint Conference on Artificial Intelligence, pages 1034–1040.
- Lewis D.D. Naive (Bayes) at forty: the independence assumption in information retrieval. In: Nédellec C., Rouveirol C., editors. European Conference on Machine Learning. Springer; 1998. pp. 4–15. [Google Scholar]; Lewis, D. D. (1998). Naive (bayes) at forty: The independence assumption in information retrieval. In European conference on machine learning, pages 4-15. Springer.
- Miller K.D., Siegel R.L., Lin C.C., Mariotto A.B., Kramer J.L., Rowland J.H., Stein K.D., Alteri R., Jemal A. Cancer treatment and survivorship statistics, 2016. CA Cancer J. Clin. 2016;66:271–289. doi: 10.3322/caac.21349. [DOI] [PubMed] [Google Scholar]; Miller, K. D., Siegel, R. L., Lin, C. C., Mariotto, A. B., Kramer, J. L., Rowland, J. H., Stein, K. D., Alteri, R., and Jemal, A. (2016). Cancer treatment and survivorship statistics, 2016. CA: a cancer journal for clinicians, 66(4):271-289. [DOI] [PubMed]
- Mor G., Visintin I., Lai Y., Zhao H., Schwartz P., Rutherford T., Yue L., Bray-Ward P., Ward D.C. Serum protein markers for early detection of ovarian cancer. Proc. Natl. Acad. Sci. U S A. 2005;102:7677–7682. doi: 10.1073/pnas.0502178102. [DOI] [PMC free article] [PubMed] [Google Scholar]; Mor, G., Visintin, I., Lai, Y., Zhao, H., Schwartz, P., Rutherford, T., Yue, L., Bray-Ward, P., and Ward, D. C. (2005). Serum protein markers for early detection of ovarian cancer. Proceedings of the National Academy of Sciences of the United States of America, 102(21):7677-7682. [DOI] [PMC free article] [PubMed]
- Napier K.J., Scheerer M., Misra S. Esophageal cancer: a review of epidemiology, pathogenesis, staging workup and treatment modalities. World J. Gastrointest. Oncol. 2014;6:112. doi: 10.4251/wjgo.v6.i5.112. [DOI] [PMC free article] [PubMed] [Google Scholar]; Napier, K. J., Scheerer, M., and Misra, S. (2014). Esophageal cancer: A review of epidemiology, pathogenesis, staging workup and treatment modalities. World journal of gastrointestinal oncology, 6(5):112. [DOI] [PMC free article] [PubMed]
- Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., Blondel M., Prettenhofer P., Weiss R., Dubourg V. Scikit-learn: machine learning in python. J. Mach. Learn. Res. 2011;12:2825–2830. [Google Scholar]; Pedregosa, F., Varoquaux, G., Gramfort, A., Michel, V., Thirion, B., Grisel, O., Blondel, M., Prettenhofer, P., Weiss, R., Dubourg, V., et al. (2011). Scikit-learn: Machine learning in python. Journal of machine learning research, 12(Oct):2825-2830.
- Pei H., Zhu H., Zeng S., Li Y., Yang H., Shen L., Chen J., Zeng L., Fan J., Li X. Proteome analysis and tissue microarray for profiling protein markers associated with lymph node metastasis in colorectal cancer. J. Proteome Res. 2007;6:2495–2501. doi: 10.1021/pr060644r. [DOI] [PubMed] [Google Scholar]; Pei, H., Zhu, H., Zeng, S., Li, Y., Yang, H., Shen, L., Chen, J., Zeng, L., Fan, J., Li, X., et al. (2007). Proteome analysis and tissue microarray for profiling protein markers associated with lymph node metastasis in colorectal cancer. Journal of proteome research, 6(7):2495-2501. [DOI] [PubMed]
- Polanski M., Anderson N.L. A list of candidate cancer biomarkers for targeted proteomics. Biomarker Insights. 2006;1:1–48. [PMC free article] [PubMed] [Google Scholar]; Polanski, M. and Anderson, N. L. (2006). A list of candidate cancer biomarkers for targeted proteomics. Biomarker insights, 1:1-48. [PMC free article] [PubMed]
- Rahib L., Smith B.D., Aizenberg R., Rosenzweig A.B., Fleshman J.M., Matrisian L.M. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74:2913–2921. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]; Rahib, L., Smith, B. D., Aizenberg, R., Rosenzweig, A. B., Fleshman, J. M., and Matrisian, L. M. (2014). Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the united states. Cancer research, 74(11):2913-2921. [DOI] [PubMed]
- Rugge M., Fassan M., Graham D.Y. Epidemiology of gastric cancer. In: Strong V.E., editor. Gastric Cancer. Springer; 2015. pp. 23–34. [Google Scholar]; Rugge, M., Fassan, M., and Graham, D. Y. (2015). Epidemiology of gastric cancer. In Gastric Cancer, pages 23-34. Springer.
- Stoeva S.I., Lee J.-S., Smith J.E., Rosen S.T., Mirkin C.A. Multiplexed detection of protein cancer markers with biobarcoded nanoparticle probes. J. Am. Chem. Soc. 2006;128:8378–8379. doi: 10.1021/ja0613106. [DOI] [PubMed] [Google Scholar]; Stoeva, S. I., Lee, J.-S., Smith, J. E., Rosen, S. T., and Mirkin, C. A. (2006). Multiplexed detection of protein cancer markers with biobarcoded nanoparticle probes. Journal of the American Chemical Society, 128(26):8378-8379. [DOI] [PubMed]
- Szklarczyk D., Morris J.H., Cook H., Kuhn M., Wyder S., Simonovic M., Santos A., Doncheva N.T., Roth A., Bork P. The string database in 2017: quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 2016;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]; Szklarczyk, D., Morris, J. H., Cook, H., Kuhn, M., Wyder, S., Simonovic, M., Santos, A., Doncheva, N. T., Roth, A., Bork, P., et al. (2016). The string database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic acids research, page 45:D362-D368. [DOI] [PMC free article] [PubMed]
- Takadate T., Onogawa T., Fukuda T., Motoi F., Suzuki T., Fujii K., Kihara M., Mikami S., Bando Y., Maeda S. Novel prognostic protein markers of resectable pancreatic cancer identified by coupled shotgun and targeted proteomics using formalin-fixed paraffin-embedded tissues. Int. J. Cancer. 2013;132:1368–1382. doi: 10.1002/ijc.27797. [DOI] [PubMed] [Google Scholar]; Takadate, T., Onogawa, T., Fukuda, T., Motoi, F., Suzuki, T., Fujii, K., Kihara, M., Mikami, S., Bando, Y., Maeda, S., et al. (2013). Novel prognostic protein markers of resectable pancreatic cancer identified by coupled shotgun and targeted proteomics using formalin-fixed paraffin-embedded tissues. International journal of cancer, 132(6):1368-1382. [DOI] [PubMed]
- Torre L.A., Bray F., Siegel R.L., Ferlay J., Lortet-Tieulent J., Jemal A. Global cancer statistics, 2012. CA Cancer J. Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]; Torre, L. A., Bray, F., Siegel, R. L., Ferlay, J., Lortet-Tieulent, J., and Jemal, A. (2015). Global cancer statistics, 2012. CA: a cancer journal for clinicians, 65(2):87-108. [DOI] [PubMed]
- Visintin I., Feng Z., Longton G., Ward D.C., Alvero A.B., Lai Y., Tenthorey J., Leiser A., Flores-Saaib R., Yu H. Diagnostic markers for early detection of ovarian cancer. Clin. Cancer Res. 2008;14:1065–1072. doi: 10.1158/1078-0432.CCR-07-1569. [DOI] [PubMed] [Google Scholar]; Visintin, I., Feng, Z., Longton, G., Ward, D. C., Alvero, A. B., Lai, Y., Tenthorey, J., Leiser, A., Flores-Saaib, R., Yu, H., et al. (2008). Diagnostic markers for early detection of ovarian cancer. Clinical Cancer Research, 14(4):1065-1072. [DOI] [PubMed]
- Webb G.I., Boughton J.R., Wang Z. Not so naive Bayes: Aggregating one-dependence estimators. Mach. Learn. 2005;58:5–24. [Google Scholar]; Webb, G. I., Boughton, J. R., and Wang, Z. (2005). Not so naive bayes: Aggregating one-dependence estimators. Machine learning, 58(1):5-24.
- Webb G.I., Boughton J.R., Zheng F., Ting K.M., Salem H. Learning by extrapolation from marginal to full-multivariate probability distributions: decreasingly naive bayesian classification. Mach. Learn. 2012;86:233–272. [Google Scholar]; Webb, G. I., Boughton, J. R., Zheng, F., Ting, K. M., and Salem, H. (2012). Learning by extrapolation from marginal to full-multivariate probability distributions: decreasingly naive bayesian classification. Machine learning, 86(2):233-272.
- Zheng G., Patolsky F., Cui Y., Wang W.U., Lieber C.M. Multiplexed electrical detection of cancer markers with nanowire sensor arrays. Nat. Biotechnol. 2005;23:1294. doi: 10.1038/nbt1138. [DOI] [PubMed] [Google Scholar]; Zheng, G., Patolsky, F., Cui, Y., Wang, W. U., and Lieber, C. M. (2005). Multiplexed electrical detection of cancer markers with nanowire sensor arrays. Nature biotechnology, 23(10):1294. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.