Abstract
Background
Histo-blood group antigens (HBGAs) such as fucosyltransferase (FUT)2 and 3 may act as innate host factors that differentially influence susceptibility of individuals and their offspring to pediatric enteric infections.
Methods
In 3 community-based birth cohorts, FUT2 and FUT3 statuses were ascertained for mother-child dyads. Quantitative polymerase chain reaction panels tested 3663 diarrheal and 18 148 asymptomatic stool samples for 29 enteropathogens. Cumulative diarrhea and infection incidence were compared by child (n = 520) and mothers’ (n = 519) HBGA status and hazard ratios (HRs) derived for all-cause diarrhea and specific enteropathogens.
Results
Children of secretor (FUT2 positive) mothers had a 38% increased adjusted risk of all-cause diarrhea (HR = 1.38; 95% confidence interval (CI), 1.15–1.66) and significantly reduced time to first diarrheal episode. Child FUT2 and FUT3 positivity reduced the risk for all-cause diarrhea by 29% (HR = 0.81; 95% CI, 0.71–0.93) and 27% (HR = 0.83; 95% CI, 0.74–0.92), respectively. Strong associations between HBGAs and pathogen-specific infection and diarrhea were observed, particularly for noroviruses, rotaviruses, enterotoxigenic Escherichia coli, and Campylobacter jejuni/coli.
Conclusions
Histo-blood group antigens affect incidence of all-cause diarrhea and enteric infections at magnitudes comparable to many common disease control interventions. Studies measuring impacts of interventions on childhood enteric disease should account for both child and mothers’ HBGA status.
Keywords: randomized controlled clinical trial; controlled human infection model; Escherichia coli infections; diarrhea, prevention and control; fimbriae proteins; colonization factor antigens; antibodies, bacterial; milk proteins, immunology; immunization, passive; bacterial vaccines
The histo-blood group antigens of children and their mothers are determinants of diarrhea in early childhood. This study estimates the innate immunity provided by HBGA for principal etiologies of diarrhea and demonstrates altered risk for bacterial and viral enteropathogens.
Diarrheal disease has been a major cause of illness and premature death throughout history [1]. Despite improvements in water and sanitation, oral rehydration therapy, and the rotavirus vaccine, diarrhea illness still claims 500 000 lives of children a year with only modest declines in morbidity [2, 3]. The variability observed in the effectiveness of preventive interventions [4, 5] and in pathogen-specific diarrhea incidence between populations suggests that inherited host factors may differentially influence susceptibility to certain enteric infections [6–10], and a cluster of fucosyltransferase (FUT) genes—primarily the FUT2 and FUT3 genes—have emerged as candidates [11]. Furthermore, population-based genetic analyses reveal widespread selection and nonneutral evolution of the histo-blood group antigen (HBGA) genes similar only to genes involved in antigen recognition [12].
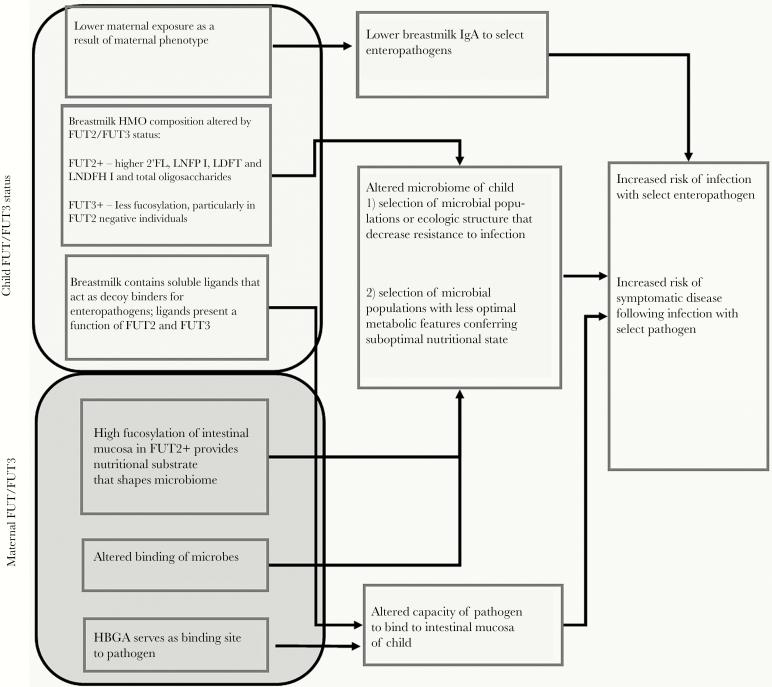
There are multiple hypothesized pathways by which both an infant’s FUT2 and FUT3 genes and those of their nursing mother may alter susceptibility to enteric infection (Figure 1). The FUT2 gene encodes a 2-α-l-fucosyltransferase, an enzyme required for the surface expression of ABO HBGAs on mucosal membranes and in secreted body fluids (Figure 2). HBGA expression on the intestinal epithelia they may serve as host receptors to which enteric viruses and bacteria attach to establish infection [12–16], and individuals expressing at least 1 functional copy of the FUT2 gene are called secretors. Individuals that are homozygous for nonfunctional FUT2 alleles (nonsecretors) do not produce these HBGA on the intestinal epithelia, which confers a degree of innate protection against enteric infection with some noroviruses [8, 13, 15, 17, 18], P[4] and P[8]-type rotavirus [14, 19], enterotoxigenic Escherichia coli (ETEC) [20], and Campylobacter [21] and altering the efficacy of the oral rotavirus vaccine [22]. Fucosyltransferase 3, along with other glycosyltransferases, also modifies HBGAs in mucosal secretions and epithelial cell surfaces [16]. In addition, FUT2 and FUT3 phenotype combinations in nursing mothers influence the distribution and concentration of human milk oligosaccharides (HMOs) expressed in breastmilk. These changes in breastmilk composition can durably alter the child’s microbiome [23, 24], which in turn may alter resistance to enteric infections. Furthermore, these glycans have been shown to bind to certain enteropathogens, such as rotavirus, thereby serving as decoy receptors that prevent infections from becoming patent or symptomatic [11, 21, 25].
Figure 1.
Hypothesized pathways through which maternal and child fucosyltransferase (FUT)2 and FUT3 expression alter susceptibility to enteric infection. 2’-FL, 2’-fucosyllactose; LDFT, lactodifucotetraose; LNDFH I, lacto-N-difucohexaose I; LNFP I, lacto-N-fucopentaose I [38, 43–45].
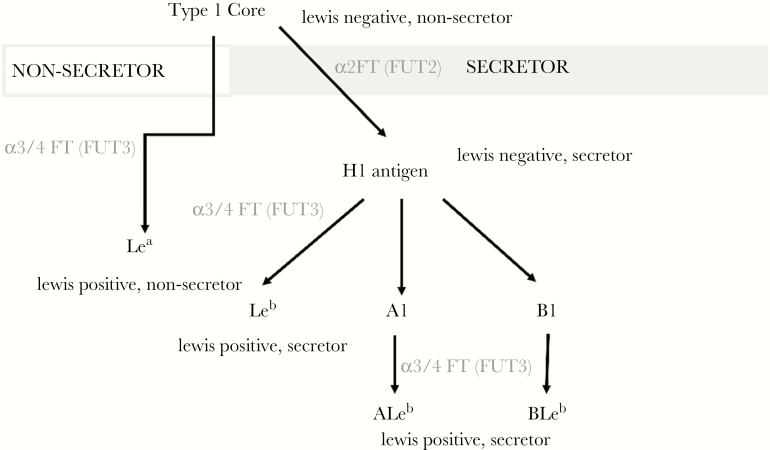
Figure 2.
Pathway of histo-blood group antigen synthesis with assignment of phenotype (Lewis and secretor status) in accordance with the presence of a functional phenotype or allele for the FUT2 gene (encoding α (1,2) fucosyltransferase) and FUT3 gene (encoding enzyme with α(1,3) and α(1,4) fucosyltransferase activities).
The analysis reported here assesses these hypotheses by quantifying and modeling associations between infant and maternal secretor (FUT2) status and Lewis (FUT3) type and episodes of infection with the 5 enteric viruses and 5 enteric bacteria most strongly associated with diarrhea: adenovirus, astrovirus, norovirus, rotavirus, sapovirus, and Campylobacter, enteroaggregative E coli (EAEC), enteropathogenic E coli ([EPEC] typical and atypical), heat-labile enterotoxigenic E coli (LT-ETEC) and heat-stable ETEC (ST-ETEC), and Shigella [10]. The study was done in 3 community-based birth cohorts and used high-resolution diagnostic panels.
METHODS
Study Population
The Etiology Risk Factors and Interactions of Enteric Infections and Malnutrition and the Consequences for Child Health and Development (MAL-ED) project investigates risk factors for enteric infection, diarrheal disease, undernutrition, and related outcomes [26]. Birth cohorts were recruited from communities in 8 low- and middle-income countries, 3 of which sites were included in this substudy to capture differences in distributions of genetic polymorphisms in South Asian, African, and South American contexts: Dhaka, Bangladesh (BGD), Loreto, Peru (PEL), and Haydom, Tanzania (TZH), described in previous publications [27–29]. Subjects were enrolled and monitored continuously over their first 2 years of life from November 2009 to March 2014. Written informed consent was obtained from the caregiver of all participating children.
Ethical Standards
Ethical approval for MAL-ED was given by the Johns Hopkins Institutional Review Board as well as from the respective partner institutions for each site including the following: (BGD) the Institutional Review Board for Health Science Research of the University of Virginia and the Ethical Review Committee of ICDDR,B; (PEL) the Institutional Review Board of the Johns Hopkins School of Public Health, the Ethics Committee of Asociacion Benefica PRISMA, and the Regional Health Department of Loreto; (TZH) the Institutional Review Board for Health Sciences Research of the University of Virginia and the Medical Research Coordinating Committee of the National Institute for Medical Research, and the Ministry of Health and Social Welfare of Tanzania. Written consent was obtained from all participants.
Outcome Variables
Subjects’ stool samples were collected monthly (surveillance samples) and upon reporting of diarrheal episodes by the caregiver. Subjects and their caregivers were visited twice weekly from enrollment (within 17 days of birth) until 2 years of age. Enteropathogen infection status was ascertained by several methods. Samples were assessed prospectively (1) for adenovirus, astrovirus, rotavirus, and Campylobacter by enzyme-linked immunosorbent assay, for Shigella by culture, (2) for E coli pathotypes by culture with polymerase chain reaction (PCR) confirmation, and (3) for norovirus genogroups I and II by reverse-transcription PCR, as described elsewhere [26, 30]. Samples from children who completed 24 months of follow-up were retrospectively tested for 29 enteropathogens (including sapovirus) and, where possible, strains (throughout this article, we use the generic term “strain” to collectively refer to genotypes of rotavirus, genogroups of norovirus, and species of Campylobacter) using probe-based quantitative PCR (qPCR) assays on custom-developed TaqMan Array Cards (Thermo Fisher) [31]. Results from qPCR (75.7% of total observations) were preferentially used in these analyses where available; otherwise, results from the prospective assessment were substituted. To ensure that a single infection episode was not counted multiple times, Campylobacter- and norovirus-positive samples were excluded if they were collected within 30 days of a previous sample that was positive for the same pathogen strain without being separated by an intermediate negative sample. For all other pathogens, a 14-day period was used. Because the TaqMan Array uses the same gene target for Shigella and enteroinvasive E coli (EIEC), these 2 related pathogens were also collapsed into 1 outcome (Shigella).
Covariates
The main exposures of interest in this analysis were secretor (FUT2) status and Lewis (FUT3) type ascertained for study subjects and their mothers from saliva samples using a phenotyping assay [32] in PEL and BGD and sequencing of the FUT2 and FUT3 genes in TZH where specimen collection methods did not allow for the assay to be performed (as detailed in Appendix 1). Phenotypes were classified according to the criteria in Supplemental Table 1a and b. Subjects whose phenotype could not be ascertained were genotyped according to methods described by Reeck et al [32]. In a secondary analysis, blood type was treated as an exposure and categorized as a binary variable comparing group O with groups A, B, and AB (the reference category). In addition, the following a priori-selected covariates were used in the analysis: (1) age: the infants’ age in continuous months at the time of sample collection was treated as the analysis-time variable in all survival analyses; (2) sample type: multivariate analyses were stratified by surveillance and diarrheal stool samples to separately model effects on symptomatic and asymptomatic episodes; (3) study site: a categorical variable indicating whether the subject was enrolled in the BGD (reference category), PEL, or TZH cohort included to adjust for between-site variation in both prevalence of gene expressions and pathogen transmission rates; (4) breastfeeding status: a categorical variable indicating whether, on the day before stool sample collection, subjects were breastfed exclusively, partially (reference category), predominantly, or not at all. (A small number of missing values for breastfeeding status were imputed based on the most common status for that 6-month age group and site.)
Statistical Methods
Using survival analysis, which allowed for multiple failures per subject, Kaplan-Meier estimates were plotted comparing cumulative incidence of diarrhea of any etiology (“all-cause diarrhea”) and individual pathogens and strains between FUT phenotype-positive and negative subjects and subjects’ mother across the first 2 years of life. Hazard ratios (HRs) from univariate, unstratified Cox proportional hazard models were also reported. Multivariate Cox regression models were then fitted for all-cause diarrhea and individual pathogens adjusting mothers’ and infants’ FUT2 and FUT3 status for each other and for covariates and stratifying by sample type (diarrheal or surveillance stools). Separately, stratified Cox models were fitted comparing O with A/B/AB blood groups for those sites where such information was available (BGD and PEL) among secretors adjusting for covariates but not FUT statuses. For 3 pathogen strains with reported binding sites for H Type I antigens, hypothesized to be present at greatest concentration in FUT2+/FUT3− individuals—P[4]/P[8] rotavirus, CFA/I+ ETEC, and Campylobacter jejuni/coli—and for all-cause diarrhea, separate Cox models were fitted comparing HRs of Lewis normal to null type among secretor children (see Figure 2). Analyses were carried out using Stata 13.1 [33].
RESULTS
The FUT2 status was ascertained for 534 of the 827 children enrolled at the 3 sites and for 521 of their mothers, whereas 520 children and 519 mothers had FUT3 status ascertained (Table 1). No nonsecretor children and only 1 nonsecretor mother was identified in PEL; however, Lewis null prevalence was highest there of the 3 sites with 21.2% of children and 25.0% of mothers with ascertained FUT3 status. Among subjects for whom FUT2 status was ascertained, prevalence of nonsecretor children was similar in BGD and TZH (22.5% and 19.1%, respectively) but differed substantially among mothers (14.5% in BGD, 22.4% in TZH).
Table 1.
Distribution of Child and Maternal FUT Status Combinations in 3 Birth Cohorts
| Country | Child’s Status | Mother’s Status | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| FUT2+ | FUT2− | FUT3+ | FUT3− | FUT2+ | FUT2− | FUT3+ | FUT3− | ||||
| Bangladesh | Child’s Status | FUT3+ | Number | 120 | 38 | ||||||
| Percentage | (65.2) | (20.7) | |||||||||
| FUT3− | Number | 23 | 3 | ||||||||
| Percentage | (12.5) | (1.6) | |||||||||
| Mother’s Status | FUT2+ | Number | 119 | 28 | 123 | 22 | |||||
| Percentage | (69.2) | (16.3) | (72.4) | (12.9) | |||||||
| FUT2− | Number | 13 | 12 | 24 | 1 | ||||||
| Percentage | (7.6) | (7.0) | (14.1) | (0.6) | |||||||
| FUT3+ | Number | 119 | 37 | 137 | 18 | 131 | 25 | ||||
| Percentage | (69.2) | (21.5) | (80.6) | (10.6) | (76.2) | (14.5) | |||||
| FUT3− | Number | 13 | 3 | 10 | 5 | 16 | 0 | ||||
| Percentage | (7.6) | (1.7) | (5.9) | (2.9) | (9.3) | (0.0) | |||||
| Total | Number | 145 | 42 | 158 | 26 | 147 | 25 | 156 | 16 | ||
| Percentage | (77.5) | (22.5) | (85.9) | (14.1) | (85.5) | (14.5) | (90.1) | (9.9) | |||
| Peru | Child’s status | FUT3+ | Number | 152 | 0 | ||||||
| Percentage | (78.8) | (0.0) | |||||||||
| FUT3− | Number | 41 | 0 | ||||||||
| Percentage | (21.2) | (0.0) | |||||||||
| Mother’s Status | FUT2+ | Number | 189 | 0 | 148 | 39 | |||||
| Percentage | (99.5) | (0.0) | (78.7) | (20.7) | |||||||
| FUT2− | Number | 1 | 0 | 1 | 0 | ||||||
| Percentage | (0.5) | (0.0) | (0.5) | (0.0) | |||||||
| FUT3+ | Number | 142 | 0 | 124 | 18 | 143 | 1 | ||||
| Percentage | (75.1) | (0.0) | (66.3) | (9.6) | (74.5) | (0.5) | |||||
| FUT3− | Number | 47 | 0 | 24 | 21 | 48 | 0 | ||||
| Percentage | (24.9) | (0.0) | (12.8) | (11.2) | (25.0) | (0.0) | |||||
| Total | Number | 195 | 0 | 152 | 41 | 192 | 1 | 144 | 48 | ||
| Percentage | (100.0) | (0.0) | (78.8) | (21.2) | (99.5) | (0.5) | (75.0) | (25.0) | |||
| Tanzania | Child’s Status | FUT3+ | Number | 101 | 22 | ||||||
| Percentage | (71.6) | (15.6) | |||||||||
| FUT3− | Number | 13 | 5 | ||||||||
| Percentage | (9.2) | (3.5) | |||||||||
| Mother’s Status | FUT2+ | Number | 98 | 19 | 95 | 14 | |||||
| Percentage | (64.9) | (12.6) | (66.9) | (9.9) | |||||||
| FUT2− | Number | 24 | 10 | 29 | 4 | ||||||
| Percentage | (15.9) | (6.6) | (20.4) | (2.8) | |||||||
| FUT3+ | Number | 114 | 27 | 118 | 16 | 115 | 31 | ||||
| Percentage | (76.0) | (18.0) | (83.7) | (11.3) | (74.2) | (20.0) | |||||
| FUT3− | Number | 7 | 2 | 5 | 2 | 5 | 4 | ||||
| Percentage | (4.7) | (1.3) | (3.5) | (1.4) | (3.2) | (2.6) | |||||
| Total | Number | 123 | 29 | 125 | 18 | 121 | 35 | 146 | 9 | ||
| Percentage | (80.1) | (19.9) | (87.4) | (12.6) | (77.6) | (22.4) | (94.2) | (5.8) | |||
| Total | 463 | 71 | 435 | 85 | 460 | 61 | 446 | 73 | |||
| Not ascertained | 293 | 307 | 306 | 308 | |||||||
Abbreviations: FUT, fucosyltransferase.
Enteroaggregative E coli was the most prevalent pathogen in both diarrheal and surveillance stools overall and within each of the sites followed by Campylobacter (Table 2). Of the 10 pathogens analyzed, typical EPEC had the lowest prevalence in diarrheal stools, whereas among asymptomatic samples, rotavirus prevalence was lowest. GII norovirus was more prevalent than GI in all sites, and both stool types and atypical GII strains were more common than GII.4. The most commonly isolated rotavirus G-types in diarrheal stools overall were G9 and G12; however, this was mainly due to their high prevalence in the BGD site, where they had isolation rates of 4.2% and 4.0%, respectively. In TZH, G1 rotavirus was by far the most commonly isolated G-strain in both stool types. In PEL, G2 was most common in diarrheal, and G1 was most common in surveillance samples. Among rotavirus P-types, P[8]—the Rotarix vaccine target—was the most commonly isolated in all sites and both sample types, although diarrheal P[4] isolates were much more prevalent in BGD than in the other 2 sites (none were recorded in TZH). Supplemental Figure 1 shows incidence of each outcome before 6 months of age by FUT status and in participants for whom status was not ascertained. There was no evidence of infection incidence differing by ascertainment status. Figure 3 shows Kaplan-Meier plots of the cumulative incidence of all-cause diarrhea by age comparing child and mother FUT2 and FUT3 phenotypes along with HRs from univariate, unstratified Cox models. The FUT2-positive (secretor) infants, and those with secretor mothers, had a statistically significantly increased risk of diarrhea and reduced time to first diarrheal episode. This was especially marked for maternal FUT2 status. At 9 months of age, a cumulative incidence of 85.2% (95% confidence interval [CI], 83.2%–87.1%) was observed in children of secretor mothers compared with 52% (95% CI, 42.3%–63.2%) in children of nonsecretors (log-rank equality test, P ≤ .0001). Overall, children of secretors had 2.24 times (95% CI, 1.89–2.66) the risk of diarrhea episodes than those of nonsecretors. A reduced risk of all-cause diarrhea was seen in nonsecretor compared with secretor children before approximately 5 months of age, but not subsequently. Both maternal and child’s Lewis status had large and highly statistically significant protective effects against diarrhea. Lewis-positive infants had a 27% lower risk of all-cause diarrhea (HR = 0.73; 95% CI, 0.66–0.80) compared with Lewis null infants, and the cumulative incidences for the 2 groups at 9 months of age were 80.1% (95% CI, 78.3%–83.1%) and 92.9% (95% CI, 90.0%–95.2%), respectively. For children of Lewis normal and Lewis null mothers, the equivalent statistics were 70% (HR = 0.70; 95% CI, 0.63–0.77), 81.3% (95% CI, 78.9%–83.6%), and 91.0% (95% CI, 87.0%–93.9%). Equivalent Kaplan-Meier estimates and HRs for each pathogen and strain are in Appendix 2.
Table 2.
Numbers and Percentages of Stool Samples That Were Positive for Enteropathogens in 3 MAL-ED Study Sites and Overall
| Enteropathogen | BGD | PEL | TZH | Total | ||||
|---|---|---|---|---|---|---|---|---|
| n | (%) | n | (%) | n | (%) | n | (%) | |
| Adenovirus 40/41 | ||||||||
| Asymptomatic infections | 927 | (16.2) | 1140 | (16.6) | 462 | (8.3) | 2529 | (13.9) |
| Diarrheal episodes | 651 | (40.9) | 458 | (23.3) | 14 | (13.0) | 1123 | (30.7) |
| Astrovirus | ||||||||
| Asymptomatic infections | 926 | (16.1) | 827 | (12.0) | 334 | (6.0) | 2087 | (11.5) |
| Diarrheal episodes | 420 | (26.4) | 449 | (22.8) | 8 | (7.4) | 877 | (23.9) |
| Norovirus—Either Genogroup | ||||||||
| Asymptomatic infections | 871 | (15.2) | 945 | (13.8) | 806 | (14.5) | 2622 | (14.4) |
| Diarrheal episodes | 356 | (22.4) | 607 | (30.9) | 23 | (21.3) | 986 | (26.9) |
| GI Norovirus | ||||||||
| Asymptomatic infections | 302 | (5.3) | 281 | (4.1) | 216 | (3.9) | 799 | (4.4) |
| Diarrheal episodes | 122 | (7.7) | 181 | (9.2) | 5 | (4.6) | 308 | (8.4) |
| GII.4 Norovirus | ||||||||
| Asymptomatic infections | 70 | (1.2) | 125 | (1.8) | 129 | (2.3) | 324 | (1.8) |
| Diarrheal episodes | 35 | (2.2) | 64 | (3.3) | 2 | (1.9) | 101 | (2.8) |
| Other GII Norovirus | ||||||||
| Asymptomatic infections | 564 | (9.8) | 519 | (7.6) | 465 | (8.4) | 1548 | (8.5) |
| Diarrheal episodes | 169 | (10.6) | 219 | (11.1) | 12 | (11.1) | 400 | (10.9) |
| Rotavirus—Any Genotype | ||||||||
| Asymptomatic infections | 270 | (4.7) | 180 | (2.6) | 246 | (4.4) | 696 | (3.8) |
| Diarrheal episodes | 351 | (22.1) | 105 | (5.3) | 17 | (15.7) | 473 | (12.9) |
| G1 Rotavirus | ||||||||
| Asymptomatic infections | 29 | (0.5) | 27 | (0.4) | 70 | (1.3) | 126 | (0.7) |
| Diarrheal episodes | 15 | (0.9) | 4 | (0.2) | 6 | (5.6) | 25 | (0.7) |
| G2 Rotavirus | ||||||||
| Asymptomatic infections | 5 | (0.1) | 23 | (0.3) | 7 | (0.1) | 35 | (0.2) |
| Diarrheal episodes | 11 | (0.7) | 25 | (1.3) | 0 | (0.0) | 36 | (1.0) |
| G3 Rotavirus | ||||||||
| Asymptomatic infections | 0 | (0.0) | 20 | (0.3) | 2 | (0.0) | 22 | (0.1) |
| Diarrheal episodes | 0 | (0.0) | 20 | (1.0) | 0 | (0.0) | 20 | (0.5) |
| G4 Rotavirus | ||||||||
| Asymptomatic infections | 0 | (0.0) | 0 | (0.0) | 2 | (0.0) | 2 | (0.0) |
| Diarrheal episodes | 0 | (0.0) | 0 | (0.0) | 0 | (0.0) | 0 | (0.0) |
| G8 Rotavirus | ||||||||
| Asymptomatic infections | 0 | (0.0) | 1 | (0.0) | 5 | (0.1) | 6 | (0.0) |
| Diarrheal episodes | 0 | (0.0) | 0 | (0.0) | 0 | (0.0) | 0 | (0.0) |
| G9 Rotavirus | ||||||||
| Asymptomatic infections | 44 | (0.8) | 7 | (0.1) | 1 | (0.0) | 52 | (0.3) |
| Diarrheal episodes | 67 | (4.2) | 10 | (0.5) | 0 | (0.0) | 77 | (2.1) |
| G12 Rotavirus | ||||||||
| Asymptomatic infections | 30 | (0.5) | 2 | (0.0) | 16 | (0.3) | 48 | (0.3) |
| Diarrheal episodes | 64 | (4.0) | 4 | (0.2) | 0 | (0.0) | 68 | (1.9) |
| P[4] Rotavirus | ||||||||
| Asymptomatic infections | 21 | (0.4) | 26 | (0.4) | 5 | (0.1) | 52 | (0.3) |
| Diarrheal episodes | 47 | (3.0) | 31 | (1.6) | 0 | (0.0) | 78 | (2.1) |
| P[6] Rotavirus | ||||||||
| Asymptomatic infections | 13 | (0.2) | 9 | (0.1) | 8 | (0.1) | 30 | (0.2) |
| Diarrheal episodes | 15 | (0.9) | 10 | (0.5) | 0 | (0.0) | 25 | (0.7) |
| P[8] Rotavirus | ||||||||
| Asymptomatic infections | 45 | (0.8) | 30 | (0.4) | 64 | (1.2) | 139 | (0.8) |
| Diarrheal episodes | 92 | (5.8) | 22 | (1.1) | 5 | (4.6) | 119 | (3.2) |
| Sapovirus | ||||||||
| Asymptomatic infections | 798 | (13.9) | 737 | (10.7) | 470 | (8.5) | 2005 | (11.0) |
| Diarrheal episodes | 358 | (22.5) | 350 | (17.8) | 16 | (14.8) | 724 | (19.8) |
| Campylobacter spp | ||||||||
| Asymptomatic infections | 1763 | (30.7) | 1287 | (18.7) | 2130 | (38.4) | 5180 | (28.5) |
| Diarrheal episodes | 597 | (37.5) | 552 | (28.1) | 41 | (38.0) | 1190 | (32.5) |
| EAEC | ||||||||
| Asymptomatic infections | 3326 | (58.0) | 3361 | (48.9) | 3546 | (64.0) | 10233 | (56.4) |
| Diarrheal episodes | 837 | (52.6) | 1005 | (51.1) | 71 | (65.7) | 1913 | (52.2) |
| Atypical EPEC | ||||||||
| Asymptomatic infections | 1178 | (20.5) | 1419 | (20.7) | 1404 | (25.3) | 4001 | (22.0) |
| Diarrheal episodes | 301 | (18.9) | 404 | (20.6) | 30 | (27.8) | 735 | (20.1) |
| Typical EPEC | ||||||||
| Asymptomatic infections | 966 | (16.8) | 664 | (9.7) | 848 | (15.3) | 2478 | (13.7) |
| Diarrheal episodes | 285 | (17.9) | 235 | (12.0) | 19 | (17.6) | 539 | (14.7) |
| LT-ETEC | ||||||||
| Asymptomatic infections | 803 | (14.0) | 1009 | (14.7) | 1336 | (24.1) | 3148 | (17.3) |
| Diarrheal episodes | 235 | (14.8) | 384 | (19.5) | 29 | (26.9) | 648 | (17.7) |
| ST-ETEC | ||||||||
| Asymptomatic infections | 1477 | (25.7) | 595 | (8.7) | 1275 | (23.0) | 3347 | (18.4) |
| Diarrheal episodes | 589 | (37.0) | 233 | (11.9) | 29 | (26.9) | 851 | (23.2) |
| Shigella spp/EIEC | ||||||||
| Asymptomatic infections | 587 | (10.2) | 596 | (8.7) | 797 | (14.4) | 1980 | (10.9) |
| Diarrheal episodes | 318 | (20.0) | 243 | (12.4) | 15 | (13.9) | 576 | (15.7) |
| Total stool samples | ||||||||
| Surveillance collections | 5736 | (31.6) | 6871 | (37.9) | 5541 | (30.5) | 18148 | (100.0) |
| Diarrheal collections | 1590 | (43.4) | 1965 | (53.6) | 108 | (2.9) | 3663 | (100.0) |
Abbreviations: BGD, Bangladesh; E coli, Escherichia coli; EAEC, enteroaggregative E coli; EIEC, enteroinvasive E coli; EPEC, enteropathogenic E coli; LT-ETEC, heat-labile enterotoxigenic E coli; MAL-ED, Malnutrition and the Consequences for Child Health and Development; PEL, Peru; ST-ETEC, heat-stable ETEC; TZH, Tanzania.
Figure 3.
Kaplan-Meier plot of cumulative incidence of diarrhea of any etiology by age for phenotype positive and negative subjects by fucosyltransferase (FUT) gene and for child’s and mother’s status with hazard ratios (HRs) comparing phenotype positive to negative status from univariate Cox regression models (with 95% confidence interval [CI] [***, P < .001, **, P = .001–.01, and *, P = .01–.05] and significance level). Maternal FUT2 status consistently had the largest association with the risk of all-cause diarrhea, with risk ratios greatest between 3 and 12 months of age but significant until 24 months of age.
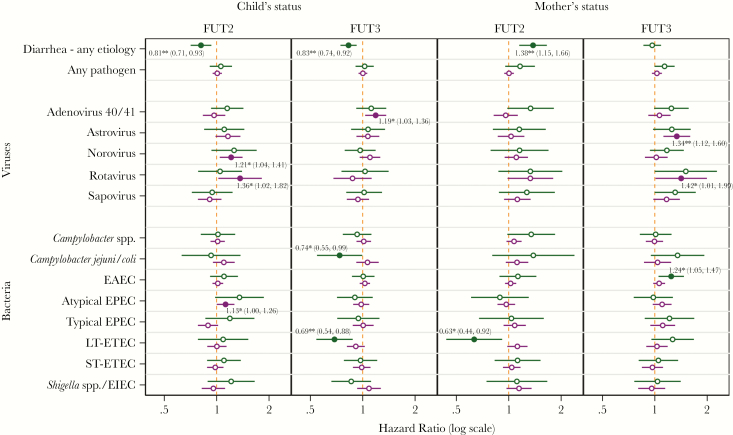
After adjustment for the child’s Lewis type and maternal secretor status and Lewis type, study site, and breastfeeding status, the child’s secretor status was associated with a 19% risk reduction for all-cause diarrhea (HR = 0.81; 95% CI, 0.71–0.93) during the first 2 years of life (Figure 4), a significant change from the univariate model findings. The similarly adjusted association of child’s FUT3 status demonstrated a 27% reduction in all-cause diarrhea risk (HR = 0.83; 95% CI, 0.74–0.92) in Lewis-positive compared with Lewis null children, consistent with the univariate model. Children who were secretors had a statistically significant 21% increased risk of asymptomatic norovirus (HR = 1.21; 95% CI, 1.04–1.41) but not norovirus diarrhea. However, secretor children had a 36% increased risk of asymptomatic rotavirus illness from all genotypes (HR = 1.36; 95% CI, 1.02–1.82) and a 13% increased risk of asymptomatic atypical EPEC (HR = 1.13; 95% CI, 1.00–1.26) relative to nonsecretors. A reduction in C jejuni/coli diarrhea risk was seen in Lewis normal infants (HR = 0.74; 95% CI, 0.55–0.99), but not in asymptomatic infection, or in infection from combined Campylobacter species. Lewis normal infants also experienced a decreased risk (HR = 0.69; 95% CI, 0.54–0.88) of LT-ETEC diarrhea, but not of asymptomatic LT-ETEC infection, and an increased risk of asymptomatic adenovirus (HR = 1.19; 95% CI, 1.03–1.36), but not of adenovirus diarrhea.
Figure 4.
Hazard ratios for diarrhea and for infection with common enteric pathogens comparing phenotype positive to negative status from multivariate Cox models that adjusted for mothers’ fucosyltransferase (FUT)2 and FUT3 status (for child estimates) and child’s FUT2 and FUT3 (for maternal estimates) and covariates stratified by diarrheal (green) and surveillance (purple) stool samples. Statistically significant estimates (P ≤ .05) are represented by solid markers and labeled with the hazard ratio (with 95% confidence interval; ***, P < .001, **, P = .001–.01, and *, P = .01–.05) and significance level.
Maternal secretor-positive status conferred an excess all-cause diarrhea risk of 38% (HR = 1.38; 95% CI, 1.15–1.66) to children after adjusting for other phenotypes and covariates. The only pathogen-specific association with maternal secretor-positive status was seen for LT-ETEC with children of secretors showing a 27% reduced risk of LT-ETEC diarrhea than those of nonsecretors (HR = 0.63; 95% CI, 0.44–0.92). Maternal FUT3 was no longer associated with all-cause diarrhea risk in the multivariate model but conferred a 34% increased risk of astrovirus infection (HR = 1.34; 95% CI, 1.12–1.60), but not astrovirus diarrhea. Children of Lewis normal mothers showed a 42% increased risk of rotavirus diarrhea (HR = 1.42; 95% CI, 1.01–1.99), but no difference in asymptomatic infections, and a 24% increase in EAEC diarrhea (HR = 1.24; 95% CI, 1.05–1.47). No associations were observed between any maternal or child HBGA status and risk of infection or diarrhea with sapovirus, typical EPEC, ST-ETEC, or Shigella.
When identical multivariate models were fitted to evaluate associations between maternal and child HBGA status and risk of strain-specific rotavirus and norovirus infections, no associations were observed for maternal statuses, but numerous strong associations were observed for the child’s status (Figure 5A). The most marked increase in risk among secretor compared with nonsecretor children was for GII.4 norovirus diarrhea (HR = 10.45; 95% CI, 1.40–78.02). Asymptomatic norovirus GII.4 infection was also 3.95 times more common in secretor than nonsecretor children (HR = 3.95; 95% CI, 1.92–8.14). Child secretor status conferred a 24% increased risk (HR = 1.24; 95% CI, 1.02–1.50) of asymptomatic infection of less common GII noroviruses, but not for diarrheal episodes. There were no associations between GI noroviruses and any HBGA status. Child secretor status conferred almost 8 times the risk of P[4] rotavirus diarrhea (HR = 7.60; 95% CI, 1.71–33.88) and 5 times the risk of asymptomatic P[8] rotavirus infection (HR = 4.88; 95% CI, 1.77–13.42). A child’s Lewis-positive status increased the adjusted risk of diarrheal P[8] rotavirus by almost fourfold (HR = 3.82; 95% CI, 1.36–10.70), but not the risk of asymptomatic infection. No diarrhea attributable to rotavirus G1 was seen in Lewis null children, and just 1 such episode was seen in secretor children. Lewis-positive children were at a 68% decreased risk (HR = 0.32; 95% CI, 0.11–0.90) of symptomatic diarrhea from rotavirus P[6], but no difference was observed in the risk of asymptomatic infection. Lewis normal children had a 21% greater risk of infections (HR = 1.21; 95% CI, 1.00–1.45) with atypical GII norovirus, but not diarrhea caused by these infections.
Figure 5.
Hazard ratios (HRs) for infection with common virus strains comparing phenotype positive to negative status from multivariate Cox models that adjusted for child and mothers’ fucosyltransferase (FUT)2 and FUT3 status and covariates stratified by diarrheal (green) and surveillance (purple) stool samples. Statistically significant estimates (P ≤ .05) are represented by solid markers and labeled with the HR (with 95% confidence interval; ***, P < .001, **, P = .001–.01, and *, P = .01–.05) and significance level. Diarrhea was not noted in children who were Lewis positive with G1 rotavirus infections, thus a HR was undefined. (Just 1 case each of G1 and G2 rotavirus diarrhea was observed in nonsecretor infants, and no cases of G1 rotavirus diarrhea were observed in Lewis null infants, so it was not possible to calculate HRs for those associations.)
There was also evidence of differential susceptibility of children to ETEC infections and diarrhea due to ETEC based on the specific identity of colonization antigens/fimbrae as a function of the child’s, but not maternal, secretor status (Appendix 5). Secretor children had 1.79 (95% CI, 1.03–3.12) times the risk of ETEC diarrhea associated with CFA/1, 4.91 (95% CI, 1.12–21.49) times the risk of CS5-producing ETEC diarrhea, and 1.88 (95% CI, 1.14–3.08) times the risk of CS6 compared with nonsecretors. No effect was observed of Lewis type or maternal HBGA status on any individual ETEC fimbria.
In the models comparing blood group O subjects to those of other groups (Appendices 3 and 4), no significant effects on individual pathogens were observed except a 24% reduction in adenovirus diarrhea (HR = 0.76; 95% CI, 0.61–0.95) and a 69% reduction in P[8] rotavirus diarrhea (HR = 0.31; 95% CI, 0.14–0.70). However, a 21% increase in all-cause diarrhea risk was observed for blood group O subjects (HR = 1.21; 95% CI, 1.06–1.38). Because multiple enteric pathogens (rotavirus P[4], P[6], P[8] [34, 35], and C jejuni [21]) have been described as binding to H-Type I HBGA, which would be maximally expressed in Lewis null secretor children. We compared infection and diarrhea from these pathogens between FUT2+/FUT3− and FUT2+/FUT3+ children (see Figure 2). The HR associated with being a Lewis null secretor for P[8] rotavirus diarrhea was 5.65 (95% CI, 1.77–18.10) compared with Lewis normal secretors, whereas the equivalent for P[4] rotavirus diarrhea was 2.45 (95% CI, 1.04–5.82) demonstrating that although binding has been described to both Leb and Type I antigens [7, 36, 37], the epidemiologic data strongly supports the binding to Type I antigen as being of primary importance in determining innate susceptibility to disease. However, no such effect was observed for either C jejuni/coli or other species of Campylobacter, another species with purported binding the Type I antigen [21].
DISCUSSION
In this analysis of a longitudinal study of 827 children on 3 continents, we demonstrate that HBGAs of both the child and the child’s mother were important determinants of all-cause diarrhea, as well as incidence of several important diarrhea-associated enteric pathogens. Although many of these associations are consistent with those described or hypothesized previously, the empirical results of this study yield more precise, population-level effect size estimates than have been available to date, as well as being suggestive of hitherto unidentified associations of HBGAs with less well studied pathogens such as adenovirus, astrovirus, EAEC, and atypical EPEC. The use of a PCR array card with simultaneous detection of a large panel of pathogens and adherence factors is a strength of the current study because it allowed for the relative risk of different microbes and genotypes to be compared with each other in the same populations. The inclusion of 3 populations with relatively large genetic differences is a second strength of the current work.
Although previous studies have examined associations between HBGAs and individual viruses and their strains in vivo [19] and single bacteria species in vitro [21], this is the first study to increase the scope to account for the effect of both secretor and Lewis status in both members of the mother-child dyad on asymptomatic and diarrheal episodes and utilize comprehensive enteric pathogen detection methods across 3 geographically and genetically diverse populations.
An unexpected finding was that maternal secretor status showed a stronger association with a child’s risk of diarrhea than the equivalent status of the child itself. Furthermore, upon adjustment for other HBGA statuses and covariates, this effect remained strong and statistically significant and in the opposing direction to the protective effects of both the child’s own secretor status and their Lewis type. Although the apparent protection conferred by a child’s Lewis normal status against all-cause diarrhea appears to be accounted for by the preventive effect on risk of diarrheal C jejuni/coli, LT-ETEC, and P[6] rotavirus, the similar effect of secretor status is not explained by any particular pathogen species, a finding for which we lack a clear explanation. It is possible that the diagnostics we did, although comprehensive by current standard, is not detecting some etiologic agents. However, these findings do suggest that decoy binding to pathogen receptors does not explain the protective effect of breastmilk (which is certainly multifactorial), because children of secretor mothers had more all-cause diarrhea and higher pathogen carriage then those of nonsecretors. It appears more likely that glycans affect host susceptibility to infection principally by altering the microbiome through the manipulation of Bacteroides species [38].
This study demonstrates that the same FUT phenotype can significantly affect a single outcome in opposing directions depending on whether the child or mother’s status is considered. This is most striking in the qualitatively different effect of maternal (risk greatest in children of secretor mothers) compared with child’s secretor status (risk decreased among secretor children) on all-cause diarrhea. Furthermore, competing effects of a particular HBGA on different strains of the same pathogen were observed. Lewis normal status in a child conferred an increased risk of G1 and P[8] rotavirus but a decreased risk of diarrheal P[6] rotavirus. Such competing effects are likely evidence of balancing selection maintaining the expression of a given HBGA phenotype within particular populations despite their risk for certain individual infections.
Data from a health facility in Dhaka specializing in treatment of moderate to severe pediatric diarrhea from 2015 to 2017 show that, even with high-quality case management, admitted infants aged 0–5 months have a case fatality rate more than 3 times that of those aged 6–11 months—a risk ratio of 3.16 (95% CI, 2.33–4.30) [39]. This suggests that, although cumulative incidence at 2 years may approach 100% for many of the pathogens analyzed here, any factors that prolong the time to first episode until after the critical first 6 months of life—such as differences in breast milk HMO profile—may have the potential to reduce diarrhea-attributable mortality. In contrast, genetic factors that shorten this time may have exerted selective survival pressures on historical populations that universally lacked access to case management. Several of the effects of HBGAs identified here seem to play this role of delaying illness episodes to beyond this critical window for a large proportion of infants, but the effects of FUT2 are most notable. That secretors are more likely but their nonsecretor offspring less likely to survive infancy favors the persistence of nonfunctioning alleles in the population.
CONCLUSIONS
The effects observed in this analysis are comparable in magnitude to those of common interventions such as rotavirus vaccine, which has an effectiveness at preventing severe disease of approximately 50% in high child mortality settings [40] and water and sanitation improvements and hygiene promotion [41, 42]. The HBGA antigen testing is simple, affordable, and available for implementation at the level of regional laboratories. Researchers assessing the impacts of such interventions should adjust for the distribution of HBGA in both the children in study populations and their mothers, which may act as effect modifiers impacting on their ability to influence outcomes. More detailed characterization of the pathogen- and strain-specific effects of HBGAs on enteric infection can inform the development of precision public health and improve the success of regionalized and targeted interventions.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We thank the participants, their families, and the study community for their dedicated time and effort to better the understanding the transmission and more enduring impact of enteric infections in early childhood. We also thank the following: Jan Vinje (Centers for Disease Control and Prevention) for critical input and manuscript review; Dr. Leah Jager for consultation regarding the statistical analysis; Dr. Ben Jann (University of Bern, Switzerland) for guidance in generating the figures; Christine Szymanski for insight and encouragement, particularly regarding Campylobacter infection and disease patency; Chris Damman and Anita Zaidi for input on early iterations of the analysis; and Dick Guerrant for final reflections.
Financial support. This work was funded by the Etiology, Risk Factors, and Interactions of Enteric Infections and Malnutrition and the Consequences for Child Health and Development Project (MAL-ED) is carried out as a collaborative project funded by the Bill & Melinda Gates Foundation (BMGF) (BMGF-47075), the Foundation for the National Institutes of Health, and the National Institutes of Health, Fogarty International Center, whereas additional support was obtained from BMGF for the examination of host innate factors on enteric disease risk and enteropathy (Grants OPP1066146 and OPP1152146; to M. N. K.). Additional funding was obtained from teh Sherrilyn and Ken Fisher Center for Environmental Infectious Diseases, Johns Hopkins School of Medicine (to M. N. K) and the National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institues of health 1UL1TR001079.
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
References
- 1. Kramer B, Kanof A. Diarrhea in children: a historical review. J Pediatr 1960; 57:769–83. [DOI] [PubMed] [Google Scholar]
- 2. Liu L, Oza S, Hogan D, et al. Global, regional, and national causes of under-5 mortality in 2000–15: an updated systematic analysis with implications for the sustainable development goals. Lancet 2016; 388:3027–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kotloff KL, Nataro JP, Blackwelder WC, et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet 2013; 382:209–22. [DOI] [PubMed] [Google Scholar]
- 4. Desselberger U. Differences of rotavirus vaccine effectiveness by country: likely causes and contributing factors. Pathog 2017; 6:E65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Null C, Stewart CP, Pickering AJ, et al. Effects of water quality, sanitation, handwashing, and nutritional interventions on diarrhoea and child growth in rural Kenya: a cluster-randomised controlled trial. Lancet Glob Heal 2018; 6:e316–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mohan VR, Karthikeyan R, Babji S, et al. Rotavirus infection and disease in a multisite birth cohort: results from the MAL-ED study. J Infect Dis 2017; 92:680–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jiang X, Liu Y, Tan M. Histo-blood group antigens as receptors for rotavirus, new understanding on rotavirus epidemiology and vaccine strategy. Emerg Microbes Infect 2017; 6:e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lopman BA, Trivedi T, Vicuña Y, et al. Norovirus infection and disease in an ecuadorian birth cohort: association of certain norovirus genotypes with host FUT2 secretor status. J Infect Dis 2015; 211:1813–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rouhani S, Peñataro Yori P, Paredes Olortegui M, et al. Norovirus infection and acquired immunity in 8 countries: results from the MAL-ED study. Clin Infect Dis 2016; 62:1210–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Platts-Mills JA, Liu J, Rogawski ET, et al. Use of quantitative molecular diagnostic methods to assess the aetiology, burden, and clinical characteristics of diarrhoea in children in low-resource settings: a reanalysis of the MAL-ED cohort study. Lancet Glob Heal 2018; 6:e1309–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Newburg DS, Ruiz-Palacios GM, Morrow AL. Human milk glycans protect infants against enteric pathogens. Annu Rev Nutr 2005; 25:37–58. [DOI] [PubMed] [Google Scholar]
- 12. Fumagalli M, Cagliani R, Pozzoli U, et al. Widespread balancing selection and pathogen-driven selection at blood group antigen genes. Genome 2008; 19:199–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ferrer-Admetlla A, Sikora M, Laayouni H, et al. A natural history of FUT2 polymorphism in humans. Mol Biol Evol 2009; 26:1993–2003. [DOI] [PubMed] [Google Scholar]
- 14. Payne DC, Currier RL, Staat MA, et al. Epidemiologic association between FUT2 secretor status and severe rotavirus gastroenteritis in children in the United States. JAMA Pediatr 2015; 169:1040–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Larsson MM, Rydell GE, Grahn A, et al. Antibody prevalence and titer to norovirus (genogroup II) correlate with secretor (FUT2) but not with ABO phenotype or Lewis (FUT3) genotype. J Infect Dis 2006; 194:1422–7. [DOI] [PubMed] [Google Scholar]
- 16. Marionneau S, Airaud F, Bovin NV, Le Pendu J, Ruvoën-Clouet N. Influence of the combined ABO, FUT2, and FUT3 polymorphism on susceptibility to Norwalk virus attachment. J Infect Dis 2005; 192:1071–7. [DOI] [PubMed] [Google Scholar]
- 17. Thorven M, Grahn A, Hedlund KO, et al. A homozygous nonsense mutation (428G–>A) in the human secretor (FUT2) gene provides resistance to symptomatic norovirus (GGII) infections. J Virol 2005; 79:15351–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lindesmith L, Moe C, Marionneau S, et al. Human susceptibility and resistance to Norwalk virus infection. Nat Med 2003; 9:548–53. [DOI] [PubMed] [Google Scholar]
- 19. Zhang XF, Long Y, Tan M, et al. P[8] and P[4] rotavirus infection associated with secretor phenotypes among children in South China. Sci Rep 2016; 6:34591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mottram L, Wiklund G, Larson G, Qadri F, Svennerholm AM. FUT2 non-secretor status is associated with altered susceptibility to symptomatic enterotoxigenic Escherichia coli infection in Bangladeshis. Sci Rep 2017; 7:10649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ruiz-Palacios GM, Cervantes LE, Ramos P, Chavez-Munguia B, Newburg DS. Campylobacter jejuni binds intestinal H(O) antigen (Fuc alpha 1, 2Gal beta 1, 4GlcNAc), and fucosyloligosaccharides of human milk inhibit its binding and infection. J Biol Chem 2003; 278:14112–20. [DOI] [PubMed] [Google Scholar]
- 22. Lee B, Dickson DM, deCamp AC, et al. Histo-blood group antigen phenotype determines susceptibility to genotype-specific rotavirus infections and impacts measures of rotavirus vaccine efficacy. J Infect Dis 2018; 217:1399–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lewis ZT, Totten SM, Smilowitz JT, et al. Maternal fucosyltransferase 2 status affects the gut bifidobacterial communities of breastfed infants. Microbiome 2015; 3:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zivkovic AM, German JB, Lebrilla CB, Mills DA. Human milk glycobiome and its impact on the infant gastrointestinal microbiota. Proc Natl Acad Sci U S A 2011; 108 (Suppl 1):4653–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Morrow AL, Ruiz-Palacios GM, Jiang X, Newburg DS. Human-milk glycans that inhibit pathogen binding protect breast-feeding infants against infectious diarrhea. J Nutr 2005; 135:1304–7. [DOI] [PubMed] [Google Scholar]
- 26. MAL-ED Network Investigators. The MAL-ED study: a multinational and multidisciplinary approach to understand the relationship between enteric pathogens, malnutrition, gut physiology, physical growth, cognitive development, and immune responses in infants and children up to 2 years of age in resource-poor environments. Clin Infect Dis 2014; 59 (Suppl 4):S193–206. [DOI] [PubMed] [Google Scholar]
- 27. Yori PP, Lee G, Olortegui MP, et al. Santa Clara de Nanay: the MAL-ED cohort in Peru. Clin Infect Dis 2014; 59 (Suppl 4):S310–6. [DOI] [PubMed] [Google Scholar]
- 28. Mduma ER, Gratz J, Patil C, et al. The etiology, risk factors, and interactions of enteric infections and malnutrition and the consequences for child health and development study (MAL-ED): description of the Tanzanian site. Clin Infect Dis 2014; 59 (Suppl 4):S325–30. [DOI] [PubMed] [Google Scholar]
- 29. Ahmed T, Mahfuz M, Islam MM, et al. The MAL-ED cohort study in Mirpur, Bangladesh. Clin Infect Dis 2014; 59 (Suppl 4):S280–6. [DOI] [PubMed] [Google Scholar]
- 30. Houpt E, Gratz J, Kosek M, et al. Microbiologic methods utilized in the MAL-ED cohort study. Clin Infect Dis 2014; 59 (Suppl 4):S225–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liu J, Gratz J, Amour C, et al. Optimization of quantitative PCR methods for enteropathogen detection. PLoS One 2016; 11:e0158199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Reeck A, Kavanagh O, Estes MK, et al. Serological correlate of protection against norovirus-induced gastroenteritis. J Infect Dis 2010; 202:1212–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. StataCorp. Stata Statistical Software: Release 15. College Station, TX, 2017. [Google Scholar]
- 34. Liu Y, Ramelot TA, Huang P, et al. Glycan specificity of P[19] rotavirus and comparison with those of related P genotypes. J Virol 2016; 90:9983–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Le Pendu J, Nyström K, Ruvoën-Clouet N. Host-pathogen co-evolution and glycan interactions. Curr Opin Virol 2014; 7:88–94. [DOI] [PubMed] [Google Scholar]
- 36. Ramani S, Hu L, Venkataram Prasad BV, Estes MK. Diversity in rotavirus-host glycan interactions: a “Sweet” spectrum. Cell Mol Gastroenterol Hepatol 2016; 2:263–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Huang P, Xia M, Tan M, et al. Spike protein VP8* of human rotavirus recognizes histo-blood group antigens in a type-specific manner. J Virol 2012; 86:4833–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kashyap PC, Marcobal A, Ursell LK, et al. Genetically dictated change in host mucus carbohydrate landscape exerts a diet-dependent effect on the gut microbiota. Proc Natl Acad Sci U S A 2013; 110:17059–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ahmed T. Case fatality rates for patients admitted to two health facilities for moderate to severe diarrhea in Bangladesh. 2018. [Google Scholar]
- 40. Burnett E, Jonesteller CL, Tate JE, Yen C, Parashar UD. Global impact of rotavirus vaccination on childhood hospitalizations and mortality from diarrhea. J Infect Dis 2017; 215:1666–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fewtrell L, Kaufmann RB, Kay D, et al. Water, sanitation, and hygiene interventions to reduce diarrhoea in less developed countries: a systematic review and meta-analysis. Lancet Infect Dis 2005: 5:42–52. [DOI] [PubMed] [Google Scholar]
- 42. Wolf J, Hunter PR, Freeman MC, et al. Impact of drinking water, sanitation and handwashing with soap on childhood diarrhoeal disease: updated meta-analysis and meta-regression. Trop Med Int Heal 2018; 23:508–25. [DOI] [PubMed] [Google Scholar]
- 43. Yu Y, Lasanajak Y, Song X, et al. Human milk contains novel glycans that are potential decoy receptors for neonatal rotaviruses. Mol Cell Proteomics 2014; 13:2944–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ashline DJ, Yu Y, Lasanajak Y, et al. Structural characterization by multistage mass spectrometry (MSn) of human milk glycans recognized by human rotaviruses. Mol Cell Proteomics 2014; 13:2961–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Magalhães A, Rossez Y, Robbe-Masselot C, et al. Muc5ac gastric mucin glycosylation is shaped by FUT2 activity and functionally impacts Helicobacter pylori binding. Sci Rep 2016; 6:25575. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.