Summary
Expression of tumor suppressor p53 is regulated at multiple levels, disruption of which often leads to cancer. We have adopted an approach combining computational systems modeling with experimental validation to elucidate the translation regulatory network that controls p53 expression post DNA damage. The RNA-binding protein HuR activates p53 mRNA translation in response to UVC-induced DNA damage in breast carcinoma cells. p53 and HuR levels show pulsatile change post UV irradiation. The computed model fitted with the observed pulse of p53 and HuR only when hypothetical regulators of synthesis and degradation of HuR were incorporated. miR-125b, a UV-responsive microRNA, was found to represses the translation of HuR mRNA. Furthermore, UV irradiation triggered proteasomal degradation of HuR mediated by an E3-ubiquitin ligase tripartite motif-containing 21 (TRIM21). The integrated action of miR-125b and TRIM21 constitutes an intricate control system that regulates pulsatile expression of HuR and p53 and determines cell viability in response to DNA damage.
Subject Areas: Bioinformatics, Biological Sciences, Computational Bioinformatics, Molecular Biology
Graphical Abstract

Highlights
-
•
Systems modeling with experimental validation of p53 translation regulatory network
-
•
Includes miR-125b and HuR in the network as upstream regulators of p53 expression
-
•
Discovers miR-125b and the E3 ligase TRIM21 as new regulators of HuR in the network
-
•
miR-125b and TRIM21 affect cell viability via p53 in response to UV-induced DNA damage
Bioinformatics; Biological Sciences; Computational Bioinformatics; Molecular Biology
Introduction
The protein p53 is the hub of a complex regulatory network of incoming stress signals and outgoing effector pathways that plays a crucial tumor suppressor role (Horn and Vousden, 2007). Different stresses such as DNA damage, hypoxia, and oncogene activation upregulate and activate the p53 protein, mainly by inhibiting the interaction between p53 and the E3 ubiquitin ligase Mdm2 (Horn and Vousden, 2007, Michael and Oren, 2003). The enhanced p53 level causes increased Mdm2 expression, which in turn reduces p53 to a low steady-state level, thereby forming a negative feedback loop post DNA damage (Lahav et al., 2004).
Although the core of the p53 regulatory network consists of the p53-Mdm2 negative feedback loop, it is, nevertheless, clear that other mechanisms for regulating p53 protein levels in response to DNA damage are also important. The observation that the protein synthesis inhibitor cycloheximide (CHX) is capable of blocking p53 induction and partially inhibiting G1 arrest in response to DNA damage provided evidence that the control of p53 mRNA translation plays a key role in p53 induction (Fu et al., 1996, Kastan et al., 1991). Translational regulation of p53 is mediated by the interaction of RNA-binding proteins (RBPs) and microRNAs (miRNAs) with both the 5′ and 3′ UTRs of p53 mRNA (Vilborg et al., 2010). The RBP HuR is a major regulator of p53 mRNA stability and translation in response to DNA damage. HuR undergoes nuclear-cytoplasmic translocation and binds to the p53 mRNA 3′ UTR, enhancing p53 protein synthesis in response to DNA-damaging UVC irradiation (Ahuja et al., 2016, Mazan-Mamczarz et al., 2003).
The first report of an miRNA directly regulating p53 was miR-125b, which is an important negative regulator of p53 protein synthesis and p53-mediated apoptosis (Le et al., 2009). A homolog of miR-125b, miR-125a, also interacts with p53 mRNA 3′ UTR and inhibits translation (Zhang et al., 2009). Recent work has shown that cross talk between miR-125b- and HuR-mediated regulation of p53 mRNA translation controls p53 protein synthesis in response to UV radiation (Ahuja et al., 2016).
Previous studies have shown that the p53 level increases dramatically after exposure to DNA-damaging radiation and then declines in a pulsatile manner (Lev Bar-Or et al., 2000). Interestingly, DNA double-strand breaks, induced by ionizing γ-radiation or radiomimetic drugs such as neocarzinostatin, trigger a series of p53 oscillations with fixed amplitude and duration (Batchelor et al., 2011). In contrast, DNA single-strand breaks, induced by non-ionizing UV radiation, causes a single pulse of p53, with an amplitude dependent on the UV dose (Batchelor et al., 2011, Collister et al., 1998, Gaglia and Lahav, 2014, Purvis et al., 2012). Moreover, quantitative studies examining p53 expression in individual living cells in response to DNA damage have showed that there is heterogeneity of pulsatile expression of p53 among individual cells in a population (Geva-Zatorsky et al., 2006, Lahav et al., 2004). These observations have introduced new questions regarding the mechanism and function of p53 oscillatory dynamics. A number of studies have employed mathematical modeling approaches to elucidate this behavior (Batchelor et al., 2008, Ma et al., 2005, Proctor and Gray, 2008). All the modeling approaches have only considered the p53-Mdm2 negative feedback loop and its upstream regulators, which regulate p53 protein stability, to explain the oscillatory dynamics of p53 (Ciliberto et al., 2005, Geva-Zatorsky et al., 2006, Lahav, 2008, Lev Bar-Or et al., 2000, Ma et al., 2005, Wagner et al., 2005). However, it is important to consider the regulation of p53 protein synthesis to realistically explain the pulsatile expression pattern of p53 in response to DNA damage. This necessitates the inclusion of regulators of p53 mRNA translation that are induced by DNA damage, such as HuR and miR-125b, in the regulatory model of p53 expression in response to genotoxic stress.
Therefore, to understand the translation regulatory network that controls pulsatile p53 expression in response to DNA damage, we have adopted an approach combining computational modeling and experimental validation in a reiterative manner. We have found that both p53 and HuR levels show a pulse over a 12-h time period after exposure to UV irradiation. However, modeling of the HuR-p53-Mdm2 network failed to show the expression pattern obtained experimentally. Further modeling that fitted the experimental observations suggested the presence of a translation inhibitor and a degradation inducer of HuR. The modeling was validated by the discovery of an miRNA inhibiting translation of HuR mRNA and an E3 ubiquitin ligase inducing HuR protein degradation in response to DNA damage. Together these observations have provided the basis of an approach combining computational modeling and biochemical experimentation, which has allowed the discovery of hitherto unknown regulators of p53 expression in the cellular DNA damage response.
Results
Dynamic Modeling of the Minimal p53-Mdm2 Negative Feedback Circuit Shows a Pulsatile Change in p53 Level
The p53-Mdm2 negative feedback circuit was selected as the minimal network, and a mathematical model of the system dynamics containing the key processes that regulate gene expression was developed. The rate equations representing the expression of p53 and Mdm2 were represented by non-linear differential equations consisting of synthesis and degradation terms. Complete model details, model parameters, and parameter testing are described in Modeling Procedures in Materials and Methods. The numerical integration of the set of differential equations with the parametric values (Table S1) showed a pulsatile change in the p53 level (Figure S1). This suggested that the minimal p53-Mdm2 negative feedback loop was sufficient to explain the oscillatory behavior of p53 expression post DNA damage, as in previous studies (Lahav, 2008, Proctor and Gray, 2008).
Inclusion of HuR and miR-125b as Regulators of p53 Expression Fails to Obtain a Fit between the Model and Experimental Data
The pulsatile change in p53 level corresponded to our experimental observation when we exposed MCF7 breast carcinoma cells to a pulse of UVC radiation and observed a single broad pulse of p53 protein approximately over a period of 12 h post UV exposure (Figure 1A). There was a high positive correlation (Pearson R = 0.68) between the result of the simulation (solid purple line) and the experimental data (bar graphs) obtained from seven independent experiments (Figure S1). However, this feedback circuit only considers the stabilization of p53 and does not consider changes in p53 protein synthesis in response to DNA damage. Therefore HuR and miR-125b were introduced in the model as positive and negative regulators, respectively, of p53 protein synthesis (Figure 1B). Terms representing the effect of HuR and miR-125b were included in the differential rate equation representing p53 expression (Figure 1C). However, in the absence of known regulatory processes for miR-125b expression in response to UV, a curve-fitting equation describing the experimentally observed biphasic miR-125b expression pattern in response to UV was included (Figure 1D) in the model. Remarkably, the simulation of these equations now failed to produce a fit (Pearson R = −0.15) with the experimentally observed pulsatile change in p53 expression (Figure 1E). Moreover, the simulation predicted a linear change in HuR level that did not show any correlation (Pearson R = 0.15) with the experimentally observed change in the cytoplasmic level of HuR, which also showed a broad pulse similar to that of p53 (bar graphs in Figures 1E and 1F). This indicated the potential presence of other, hitherto unknown, regulatory factors induced by DNA damage in the regulatory network regulating p53 expression.
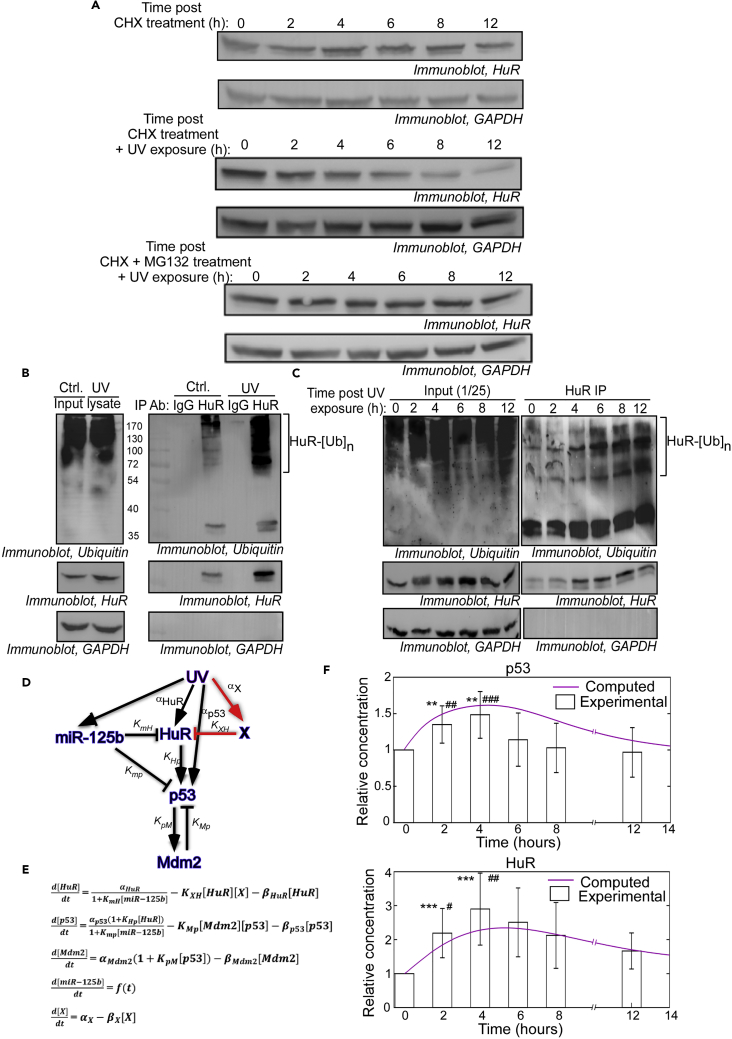
Figure 1.
Modeling of the p53 Regulatory Network, Including miR-125b and HuR as Regulators of p53 mRNA Translation, Fails to Obtain a Fit between the Model and Experimental Observations
(A) Representative immunoblot of cytoplasmic lysates of MCF7 cells exposed to a 10 J/m2 pulse of UVC radiation and collected at indicated time points post UV exposure, probed with p53, and GAPDH antibodies.
(B) Network diagram of the p53 translation regulation network in response to UV irradiation. The edges in red represent newly added regulatory processes.
(C) Rate equations representing the p53 translation regulation network in response to UV irradiation. The rate equation for miR-125b expression consists of a curve-fitting equation with a decreasing exponential function and a Hill equation-like function describing the experimentally observed biphasic miR-125b expression pattern in (C).
(D) Best-fit curve for miR-125b expression over 12-h period post UV irradiation as obtained previously (Ahuja et al., 2016). The curve-fitting equation representing the biphasic expression pattern is represented. The r2 value for curve fitting of miR-125b expression is 0.918695.
(E) Plots representing simulation and experimental data of change of p53 and HuR levels over a 12-h period post exposure to a 10 J/m2 pulse of UVC irradiation. The simulation plots (purple lines) represent the numerical integration of the rate equations in (B). The experimental plots (bar graphs) are composed of intensity values of p53 and HuR bands obtained from cell lysates collected at the designated time points post UV exposure. Experimental data represent mean ± SD values from seven independent immunoblots, normalized to corresponding GAPDH band intensities. The normalized band intensities are scaled to the 0-h time point band intensity, taken as 1. * Represents significant difference from values at 0 h, and # represents significant difference from values at 12-h time points. ∗ or # signifies a p-value ≤ 0.05, ∗∗ or ## signifies a p-value ≤ 0.01, ∗∗∗ or ### signifies a p-value ≤ 0.005.
(F) Representative immunoblot of cytoplasmic lysates of MCF7 cells exposed to UVC radiation and collected at indicated time points post UV exposure and probed with HuR and GAPDH antibodies.
See also Figures S1 and S13 and Table S1.
Incorporation of miR-125b as a Negative Regulator of HuR Improves the Model but Does Not Fit with the Experimental Data
The pulsatile change in HuR level in response to UV irradiation suggested the presence of negative regulator(s) of HuR induced by UV radiation. As miR-125b is induced by UV and represses p53 mRNA translation in the same timescale, we considered the possibility of HuR being a target of miR-125b-mediated translation repression. Interestingly, a target site of miR-125b, with a completely complementary seed sequence, was predicted at the 671–692 nucleotide position in the HuR mRNA 3′ UTR by miRNA target prediction algorithms (Figure 2A). miR-125b levels were negatively correlated with HuR levels in the breast carcinoma cell lines MCF7 and MDA-MB-231, and HuR level in MDA-MB-231 cell line was enhanced in a dose-dependent manner by transfection with an antagomiR to miR-125b (Figure S2). Overexpression of miR-125b in MCF7 cells resulted in a dose-dependent decrease in HuR protein level but not in the HuR mRNA level (Figure 2B, quantification in Figure S3A). miR-125b overexpression (Figure S3B) showed a significant dose-dependent decrease in luciferase activity from a reporter gene construct containing the wild-type (WT) HuR 3′ UTR but not from a reporter gene containing the HuR 3′ UTR with a mutant miR-125b target site (Figure 2C). There was no change in firefly luciferase mRNA level on miR-125b overexpression (Figure S3C). Also, transfection of cells overexpressing miR-125b with a miR-125b antagomiR restored luciferase expression from the reporter gene construct containing the HuR 3′ UTR (Figure 2C). UV irradiation of cells transfected with the luciferase reporter constructs showed significant increase of luciferase activity from reporter gene constructs with HuR WT 3′ UTR and HuR 3′ UTR with mutant miR-125b target site at 2 h post UV irradiation, which coincides with the lowest level of miR-125b. However, luciferase activity from the WT 3′ UTR decreased significantly compared with miR-125b target site mutant 3′ UTR at subsequent time points, which coincides with the increase in miR-125b level (Figure S4). Analysis of ribosomal fractions from cells overexpressing miR-125b showed that HuR mRNA was mostly present in the non-translating mRNA-protein complex (mRNP) fractions compared with that in control cells (Figure 2D). HuR mRNA and miR-125b were found to be associated with Ago2, the major component of the RNA-induced silencing complex, by RNA immunoprecipitation of lysates from cells expressing miR-125b (Figure S5). Together, these data established miR-125b as an UV-induced repressor of HuR protein synthesis.
Figure 2.
Inclusion of miR-125b As a Negative Regulator of HuR Expression Fails to Obtain a Fit between the Model and Experimental Observations
(A) Schematic representation of the region of the HuR 3′ UTR containing the putative miR-125b target site and Homo sapiens miR-125b sequence.
(B) Immunoblots of lysates of MCF7 cells transfected with three increasing concentrations of pSUPER-EGFP-miR-125b probed with HuR and GAPDH antibodies. Semiquantitative RT-PCR of total RNA isolated from the cell lysates using HuR- and GAPDH-specific primers (lower panels).
(C) MCF7 cells transfected with firefly luciferase constructs without 3′ UTR sequence or containing HuR-WT mRNA 3′ UTR or miR-125b-binding-site-mutated HuR mRNA 3′ UTR were cotransfected with three increasing concentrations of pSUPER-miR-125b (left panel). MCF7 cells transfected with firefly luciferase constructs without 3′ UTR sequence or containing HuR-WT mRNA 3′ UTR were cotransfected with the highest concentration of pSUPER-miR-125b and three increasing concentrations of antagomiR-125b (right panel). Fluc values are normalized to Rluc values as transfection control. Data represent mean ± SD values from three independent experiments, each with two technical replicates. The normalized band intensities are scaled to the control lacking miR-125b, taken as 1. ∗ signifies a p-value ≤ 0.05, ∗∗ signifies a p-value ≤ 0.01.
(D) Ribosomal fractions from MCF7 cells, either mock transfected or transfected with pSUPER-miR-125b, were analyzed by sucrose density gradient fractionation. Ribosomal RNA content, measured at 254 nm, is plotted against fraction numbers. RNA isolated from selected fractions was analyzed by semi-quantitative RT-PCR using HuR and GAPDH primers.
(E) Network diagram of the p53 translation regulation network in response to UV irradiation. The edge in red represents newly added regulatory process.
(F) Rate equations representing the p53 translation regulation network in response to UV irradiation. The rate equation for HuR expression contains a term representing the effect of miR-125b (KmH) on HuR synthesis. Other equations are as earlier.
(G) Plots representing simulation and experimental data of change of p53 and HuR levels over a 12-h period post exposure to a 10-J/m2 pulse of UVC irradiation. The simulation plot represents the numerical integration of the rate equations in (F). The experimental graphs are as earlier. * Represents significant difference from values at 0 h, and # represents significant difference from values at 12-h time point. ∗ or # signifies a p-value ≤ 0.05, ∗∗ or ## signifies a p-value ≤ 0.01, ∗∗∗ or ### signifies a p-value ≤ 0.005.
See also Figures S2–S5 and S14 and Table S1.
Therefore, we incorporated the miR-125b mediated downregulation of HuR expression in the model of the p53 regulatory network (Figure 2E) with kmH as the regulatory constant representing the effect of miR-125b on HuR expression (Figure 2F). Simulation of the rate equations showed that p53 protein level attained a plateau after the initial increase. Moreover, HuR expression also exhibited a non-linear increase compared with the continuous linear increase as seen in the previous model. However, in neither case the simulation (solid purple line) matched with the experimentally observed change (bar graphs) in protein levels (Pearson R = −0.10 for p53 and 0.21 for HuR) (Figure 2G). Hence inclusion of miR-125b as a negative regulator of HuR protein synthesis improved the model but failed to match the observed pulsatile change in either HuR or p53. This suggested the presence of yet unknown regulatory factor(s) in the network.
Incorporation of a Hypothetical Protein Degradation Factor as a Negative Regulator of HuR in the Model Obtains a Fit with the Experimental Data
Incorporation of miR-125b as an inhibitor of HuR protein synthesis in the model only modified the nature of the expression kinetics but failed to match the pulsatile expression pattern of HuR. We therefore investigated whether the degradation of HuR protein contributed to the pulsatile change in HuR level. HuR protein level was found to remain nearly unaltered for 12 h in cells treated with CHX, an inhibitor of global protein synthesis, demonstrating HuR to be a highly stable protein (Figure 3A, quantification in Figure S6A). However, UV irradiation with CHX treatment led to a 70% reduction in HuR protein by 12 h, suggesting a rapid degradation of HuR protein induced by UV exposure (Figure 3A, quantification in Figure S6B). Therefore we investigated whether UV induced proteasomal degradation of HuR. Treatment of cells with CHX and MG-132, a proteasome inhibitor, prevented the degradation of HuR post UV exposure (Figure 3A, quantification in Figure S6C). Also, immunoprecipitation of HuR, followed by western blot with anti-ubiquitin antibody, showed enhanced poly-ubiquitination of HuR in UV-treated cells when compared with cells not exposed to UV (Figure 3B). The poly-ubiquitination of HuR increased in a time-dependent manner and showed a maximum between 6 and 12 h post UV exposure, coinciding with the decrease in HuR level (Figure 3C). Together, these observations demonstrated that HuR undergoes ubiquitination and proteasomal degradation induced by UV irradiation.
Figure 3.
Inclusion of a Hypothetical Protein Degradation Factor As a Negative Regulator of HuR Obtains a Fit between the Model and Experimental Observations
(A) Immunoblots of lysates of MCF7 cells treated with cycloheximide (CHX) (upper panels), CHX and UV irradiation (middle panels), or CHX, MG132, and UV irradiation (lower panels) and collected at designated time points over a 12-h period. Lysates were probed with HuR and GAPDH antibodies.
(B) Lysates of MCF7 cells not exposed (Ctrl.) or exposed to UV irradiation and treated with MG132 were immunoprecipitated with HuR antibody or non-immune IgG, and the immunoprecipitates probed with ubiquitin (upper panel), HuR (middle panel), and GAPDH (lower panel) antibodies. Left panels represent the input lysates probed with the same antibodies.
(C) Lysates of MCF7 cells exposed to UV irradiation and treated with MG132 were collected at designated time point over a 12-h period post UV exposure and immunoprecipitated with HuR antibody and the immunoprecipitates probed with ubiquitin (upper panel), HuR (middle panel), and GAPDH (lower panel) antibodies. Left panels represent the input lysates probed with the same antibodies.
(D) Network diagram of the p53 translation regulation network in response to UV irradiation. The edges in red represent newly added regulatory processes.
(E) Rate equations representing the p53 translation regulation network in response to UV irradiation. The rate equation for HuR expression contains a term representing the effect of X (KXH) on HuR degradation. αX and βX are assumed parameters representing rates of synthesis and degradation, respectively, of X. Other equations are as earlier.
(F) Plots representing simulation and experimental data of change of p53 and HuR levels over a 12-h period post exposure to a 10-J/m2 pulse of UVC irradiation. The simulation plot (purple line) represents the numerical integration of the rate equations in (E). The experimental graphs are as earlier. * Represents significant difference from values at 0 h, and # represents significant difference from values at 12-h time points. ∗ or # signifies a p-value ≤ 0.05, ∗∗ or ## signifies a p-value ≤ 0.01, ∗∗∗ or ### signifies a p-value ≤ 0.005.
See also Figures S6 and S15 and Table S1.
As no regulator of HuR degradation in response to UV irradiation was known, we postulated “X” as a novel negative regulator of HuR that is induced as a result of UV irradiation and leads to degradation of HuR protein (Figure 3D). X was incorporated as a negative regulator of HuR in the differential equation describing the expression of HuR (Figure 3E). Remarkably, simulation of the rate equations generated a pulsatile expression pattern for both p53 and HuR (solid purple lines), which matched satisfactorily (Pearson R = 0.76 for p53 and R = 0.94 for HuR) with the experimentally observed change in HuR and p53 levels (bar graphs) post DNA damage (Figure 3F). This suggested that the pulsatile change in HuR level, and its effect on p53, may be a result of the combination of translation repression of HuR mRNA by miR-125b and degradation of HuR protein by an unknown factor(s) induced by UV radiation.
The Hypothetical HuR Degradation Factor “X” Is the E3 Ubiquitin Ligase TRIM21
To identify the hypothetical HuR protein degradation factor “X,” myc-tagged HuR was overexpressed in MCF7 cells. Cells exposed to UV irradiation and treated with MG132 were lysed 6 h after UV exposure, and lysates were immunoprecipitated with anti-HuR antibody and IgG. The immunoprecipitate was analyzed by mass spectrometry for proteins specifically co-immunoprecipitated with HuR when compared with IgG (Table S2). Two proteins, Serpin B3 and the E3 ubiquitin protein ligase tripartite motif-containing 21 (TRIM21), along with HuR itself, were found to be specifically present in the HuR immunoprecipitate. Serpin B3 was not considered to play a role in HuR degradation as it is a serine protease inhibitor. However, as HuR was found to undergo proteasomal degradation in response to UV irradiation, interaction with an E3 ubiquitin ligase was highly significant. Therefore the E3 ubiquitin ligase TRIM21 was considered as a potential candidate for the unknown factor “X.”
TRIM21 Interacts with HuR in Response to UV Irradiation and Causes Poly-ubiquitination and Degradation of HuR
To confirm the interaction between TRIM21 and HuR, TRIM21 was immunoprecipitated from MCF7 cells treated with MG132, and HuR was found to associate with TRIM21 by immunoblotting with HuR antibody, and vice versa (Figure 4A). This interaction between HuR and TRIM21 was found to be enhanced in UV-exposed cells when compared with cells not exposed to UV (Figure 4B) and was independent of RNA binding (Figure S7). Also, the interaction between TRIM21 and HuR was found to increase in a time-dependent manner over the period of 12 h post UV exposure and was maximum between 4 and 12 h post UV exposure, which coincided with the previously observed decrease in HuR level (Figure 4C). We also checked whether the knockdown of TRIM21 affected the poly-ubiquitination of HuR post UV irradiation. Cells transfected with a small interfering RNA (siRNA) against TRIM21 (Figure 4D, lower panel) showed reduced poly-ubiquitination of HuR post UV exposure, compared with cells transfected with a control siRNA, and the level of poly-ubiquitination of HuR in siTRIM21-transfected cells exposed to UV was similar to that in cells not exposed to UV (Figure 4D, right panel, lanes 1 and 4). Overexpression of TRIM21 in MCF7 cells caused the degradation of HuR upon UV irradiation only, confirming that UV irradiation induced the interaction between TRIM21 and HuR and the subsequent degradation of the latter (Figure 4E). The breast carcinoma cell line MDA-MB-231, which has lower level of HuR compared with MCF7, was found to have higher level of TRIM21, and siRNA-mediated knockdown of TRIM21 in MDA-MB-231 could enhance the level of HuR in these cells (Figure 4F). Together, these observations showed TRIM21 as the E3 ubiquitin ligase responsible for the UV-induced poly-ubiquitination and degradation of HuR.
Figure 4.
TRIM21 Causes Poly-ubiquitination and Degradation of HuR in Response to UV Irradiation
(A) MCF7 cell lysates, treated with MG132, were immunoprecipitated with HuR antibody and immunoblotted with TRIM21 antibody, and vice versa. Non-immune IgG was used as control antibody for immunoprecipitation.
(B) Lysates of MCF7 cells not exposed (Ctrl.) or exposed to UV irradiation, and treated with MG132, were immunoprecipitated with HuR antibody or non-immune IgG and the immunoprecipitates probed with TRIM21 (upper panel), HuR (middle panel), and GAPDH (lower panel) antibodies. Left panels represent the input lysates probed with the same antibodies.
(C) Lysates of MCF7 cells exposed to UV irradiation and treated with MG132 were collected at designated time points over a 12-h period post UV exposure and immunoprecipitated with HuR antibody, and the immunoprecipitates probed with TRIM21 (upper panel), HuR (middle panel), and GAPDH (lower panel) antibodies. Left panels represent the input lysates probed with the same antibodies. The ratio between TRIM21 and HuR band intensities are shown in boxes below left and right panels.
(D) MCF7 cells transfected with control siRNA or siRNA against TRIM21 were not exposed (Ctrl.) or exposed to UV irradiation and treated with MG132. Cell lysates were immunoprecipitated with HuR antibody, and the immunoprecipitates probed with ubiquitin (upper panel), HuR (middle panel), and GAPDH (lower panel) antibodies. Left panels represent the input lysates probed with the same antibodies. The bottom panel represents the input lysates (transfected with Ctrl. siRNA and TRIM21 siRNA) probed with TRIM21 antibody.
(E) MCF7 cells transfected with three increasing concentrations of a TRIM21-expressing vector were treated with CHX and not exposed or exposed to UV irradiation. Cells were collected 6 h after UV exposure, and lysates were immunoblotted with TRIM21 (upper panel), HuR (middle panel), and GAPDH (lower panel) antibodies.
(F) Lysates from MCF7 and MDA-MB-231 cells were immunoblotted with TRIM21 (upper panel), HuR (middle panel), and β-actin (lower panel) antibodies (left panels). Lysates from MDA-MB-231 cells transfected with increasing concentrations of TRIM21 siRNA were immunoblotted with TRIM21 (upper panel), HuR (middle panel), and GAPDH (lower panel) antibodies (right panels).
UV-Induced Ubiquitination of Lys-182 by TRIM21 Causes HuR Degradation
Previously ubiquitination of the Lys-182 (K182) residue in the RNA recognition motif 2 (RRM2) of HuR (Figure 5A) has been shown to be responsible for HuR degradation induced by heat shock, but the E3 ubiquitin ligase that ubiquitinates the K182 residue was not demonstrated (Abdelmohsen et al., 2009). Therefore we investigated whether the UV-induced degradation of HuR involved ubiquitination of K182 and TRIM21 was the E3 ubiquitin ligase responsible. Exogenous expression of Myc-tagged wild-type HuR (HuR-WT) and HuR in which the Lys-182 was mutated to Arg (K182R) in MCF7 cells followed by UV irradiation led to degradation of HuR-WT during the 12-h period post UV irradiation, whereas HuR-K182R was refractory to degradation (Figure 5B). Determination of the half-life of the WT and K182R mutant HuR post UV-irradiation showed that HuR-WT had a half-life of around 7 h, whereas the half-life of the K182R mutant was greater than 12 h (Figure 5C). Overexpression of hemagglutinin (HA)-tagged ubiquitin in cells showed a 2-fold decrease in HuR level compared with cells not overexpressing ubiquitin 6 h post UV exposure (Figure S8). Cells expressing HA-tagged ubiquitin, when transfected with constructs expressing HuR-WT or K182R mutant HuR and exposed to UV, showed significantly higher ubiquitination of HuR-WT compared with K182R mutant HuR (Figure 5D). The overall level of ubiquitination remained nearly unchanged in cells expressing HuR-WT or K182R mutant HuR. Overexpression of TRIM21 in cells expressing wild-type or K182R mutant HuR showed reduced interaction of TRIM21 with K182R mutant HuR post UV irradiation (Figure 5E). Also, overexpression of TRIM21 showed reduced degradation of K182R mutant HuR compared with HuR-WT in a dose-dependent manner 6 h post UV irradiation (Figure 5F, quantification in Figure S9A). Finally, the K182R mutant HuR failed to show the decrease in level 12 h post UV irradiation after the increase at 4 h, as observed for HuR-WT (Figure 5G, quantification in Figure S9B). Together, these observations demonstrate Lys-182 residue of HuR as the target of TRIM21-mediated degradation of HuR post UV irradiation.
Figure 5.
UV-Induced Ubiquitination of Lys-182 by TRIM21 Causes HuR Degradation
(A) Schematic diagram of the HuR wild-type (HuR WT) and Lys-182 to Arg mutant HuR (HuR K182R). * Indicates the position of K182 to R mutation.
(B) Immunoblots of lysates of MCF7 cells transfected with Myc-tagged HuR WT (upper panel) and HuR K182R (lower panel) expression constructs, treated with cycloheximide and UV irradiated, and collected at designated time points over a 12-h period post UV irradiation. Lysates were probed with Myc and β-actin antibodies.
(C) Quantification of Myc-tagged HuR levels from cells expressing HuR WT and HuR K182R over a 12-h period post UV irradiation. Data represent mean ± SD values from three independent experiments. *p ≤ 0.05, **p ≤ 0.01 between corresponding time points.
(D) Cells-expressing Myc-tagged HuR WT or HuR K182R were transfected with a construct expressing HA-ubiquitin. At 36 h post transfection cells were treated with MG132 and UV irradiated. Lysates from cells collected 6 h post UV irradiation were immunoprecipitated with anti-hemagglutinin (HA) or control IgG antibodies. Immunoprecipitates and input lysates were immunoblotted with anti-Myc, anti-HA, and anti-β-actin antibodies.
(E) Cells expressing Myc-tagged HuR WT or HuR K182R were transfected with a construct expressing TRIM21. At 48 h post transfection cells were treated with MG132 and UV irradiated. Lysates from cells collected 6 h post UV irradiation were immunoprecipitated with anti-TRIM21 or control IgG antibodies. Immunoprecipitates and input lysates were immunoblotted with anti-Myc, anti-TRIM21, and anti-β-actin antibodies.
(F) Cells expressing Myc-tagged HuR WT or HuR K182R were transfected with three increasing concentrations of a construct expressing TRIM21. At 48 h post transfection cells were treated with CHX for 30 min and UV irradiated. Lysates from cells collected 6 h post UV irradiation were immunoblotted with anti-Myc, anti-TRIM21, and anti-β-actin antibodies.
(G) Cells expressing Myc-tagged HuR WT or HuR K182R were UV irradiated. Cytoplasmic lysates from cells collected 4 and 12 h after UV irradiation were immunoblotted with anti-Myc and anti-β-actin antibodies.
See also Figures S8 and S9.
The UV-Induced Pulse of HuR and p53 Is Abolished with Knockdown of TRIM21
To validate TRIM21 as the E3 ubiquitin ligase contributing to the pulsatile change in HuR and p53 levels in response to UV irradiation, cells were transfected with an siRNA against TRIM21 to repress TRIM21 expression. Cells transfected with control siRNA followed by UV exposure showed a pulsatile change in cytoplasmic HuR level as observed earlier (Figure 6A). HuR level became highest at 4 h and gradually decreased thereafter till 12 h. However, cytoplasmic HuR showed an initial increase at 2 h in TRIM21 knockdown lysates and remained constant thereafter till 8–12 h post UV exposure. p53 also showed a pulsatile expression pattern in control siRNA-treated cells, which was abolished with TRIM21 knockdown resulting in a sustained high level of p53 (Figure 6A). To investigate the effect of TRIM21 on HuR degradation, cells transfected with siRNA against TRIM21 or control siRNA were treated with CHX followed by UV irradiation. Immunoblotting of the whole-cell lysate with anti-HuR antibody showed that the HuR level started to decrease from 2 h onward and showed a significant reduction by 12 h in the control siRNA-transfected cells (Figure S10). However, HuR levels remained nearly constant post UV exposure in TRIM21 knockdown cells. These observations demonstrated that the pulsatile change of both HuR and p53 in response to UV irradiation is abolished with knockdown of TRIM21, hence confirming TRIM21 as the E3 ubiquitin ligase involved in HuR degradation and consequently regulating the pulsatile expression of p53.
Figure 6.
Integrated Regulation of HuR by miR-125b and TRIM21 Contributes to the Pulsatile Change in HuR and p53 Levels in Response to UV Irradiation
(A) Cytoplasmic lysates of MCF7 cells transfected with control siRNA or TRIM21 siRNA and exposed to UV irradiation were collected at designated time points post UV exposure and immunoblotted with TRIM21, HuR, p53, and GAPDH antibodies.
(B) Cytoplasmic lysates of MCF7 cells transfected with control oligo, antagomiR-125b, TRIM21 siRNA, or antagomiR-125b and TRIM21 siRNA and exposed to UV irradiation were collected at designated time points post UV exposure and immunoblotted with HuR and GAPDH antibodies (left panels). Similarly treated cell lysates were also immunoblotted with p53 and GAPDH antibodies (right panels).
(C) Plots representing HuR and p53 band intensities from three independent experiments as described in (B) and corresponding best-fit curves. The band intensities of HuR and p53 were determined by densitometry and are normalized to corresponding GAPDH band intensities.
See also Figures S10 and S11.
Integrated Regulation of HuR by miR-125b and TRIM21 Gives Rise to the Pulsatile Change in HuR and p53 Levels and Regulates Cell Viability in Response to UV
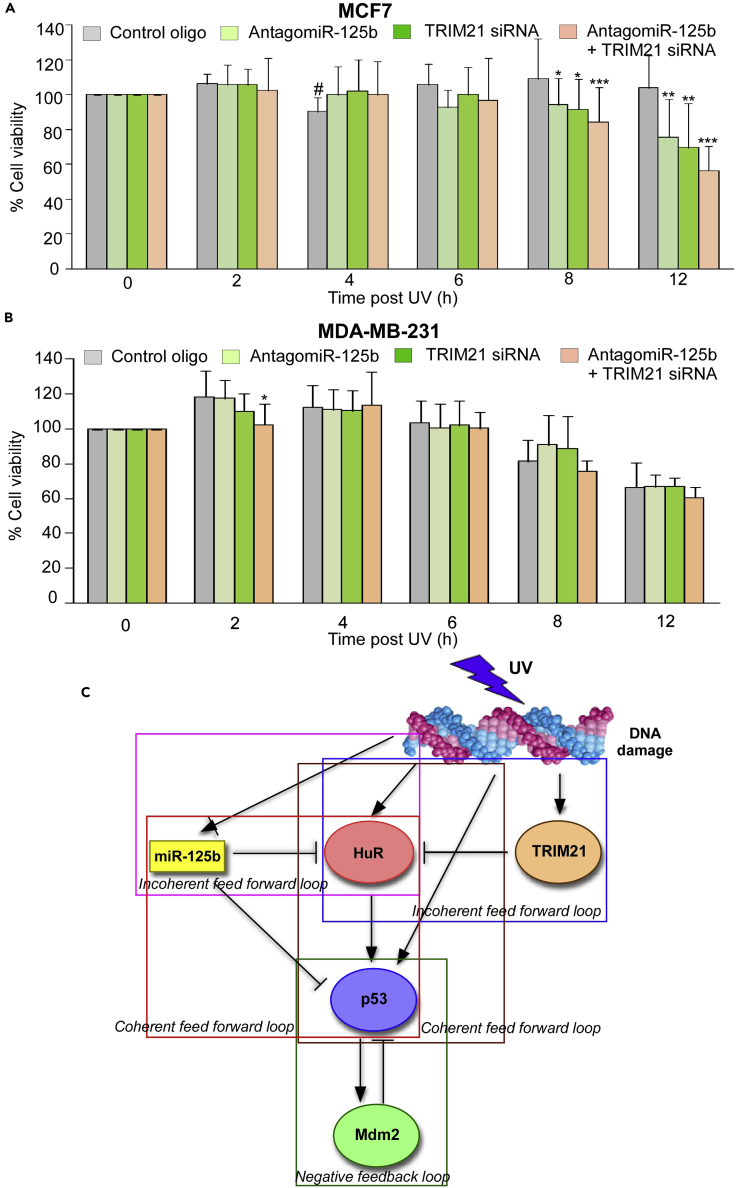
The modeling of p53 and HuR expression post UV-mediated DNA damage led to the discovery of miR-125b and TRIM21 as regulators of HuR synthesis and degradation, respectively. We therefore investigated the combined contributions of miR-125b and TRIM21 in generating the pulsatile change of cytoplasmic HuR and p53 levels in response to UV irradiation. MCF7 cells treated with a control oligo showed the pulsatile pattern of cytoplasmic HuR during the 12-h period post UV irradiation with a peak at around 4 h post UV exposure (Figure 6B). However, treatment with an antagomiR against miR-125b abolished the pulse and resulted in a plateau in HuR level from around 6 h post UV exposure (Figure 6B and best-fit curve in Figure 6C). This demonstrated that antagonizing miR-125b function resulted in de-repression of HuR protein synthesis. Similarly, inhibition of HuR degradation by siRNA-mediated knockdown of TRIM21 resulted in a plateau in HuR level from around 6 h post UV exposure (Figure 6B and best-fit curve in Figure 6C). Remarkably, the combination of antagomiR against miR-125b and the siRNA against TRIM21 resulted in a linear increase in HuR level during the 12 h post UV irradiation (Figure 6B and best-fit curve in Figure 6C). This demonstrated that the combination of translation de-repression and inhibition of degradation converted the pulsatile expression pattern of HuR post DNA damage to a linear increase, as seen in our initial model (Figure 1E). Similar effects of antagomiR-125b and TRIM21 siRNA were observed on p53 levels, except that upon treatment with combination of antagomiR-125b and TRIM21 siRNA, p53 level attained a plateau unlike HuR level (Figure 6B and best-fit curve in Figure 6C). This is likely due to the effect of Mdm2-mediated degradation of p53, which continues to operate during this time period and counteracts the de-repression of p53 protein synthesis. Also, removal of miR-125b from the model, with the retention of the degradation factor, resulted in both p53 and HuR levels attaining plateaux (Figure S11), corresponding to the experimental observations on antagomiR-125b transfection. This shows that the abrogation of either translation repression or protein degradation will result in the loss of the pulsatile change in HuR and p53 levels.
To investigate the effect of the miR-125b-TRIM21-HuR-p53 regulatory network on cell behavior in response to DNA damage, we checked the viability of the UV-treated cells, which have been subjected to inhibition of miR-125b or TRIM21 function, or both. Prolonged increase in p53 level in response to DNA damage will block the cell cycle and decrease cell viability, whereas the pulsatile decrease in p53 level will allow the cells to re-enter the cell cycle and resume cell proliferation. Analysis of the viability of UV-treated MCF7 cells showed that cells transfected with a control oligo did not show a significant change in cell viability, except at 4 h post UV treatment, when the p53 level is the highest (Figure 7A). However, cells that have been transfected with either antagomiR-125b or TRIM21 siRNA or both, in all of which cases p53 levels attains a high plateau from 8–12 h post UV treatment, showed significant reduction in cell viability when compared with control oligo-transfected cells (Figure 7A). We also checked whether the change in cell viability on treatment with antagomiR-125b or TRIM21 siRNA was due to the effect of changes in p53 level. MDA-MB-231 cells, which have non-functional p53, were similarly transfected with a control oligo, antagomiR-125b or TRIM21 siRNA, or a combination of both. Interestingly, MDA-MB-231 cells transfected with control oligo showed significantly reduced viability from 8–12 h post UV irradiation, suggesting that the absence of DNA repair activity of p53 in these cells led to reduced viability (Figure 7B). Also, there was no difference in viability between cells treated with the control oligo and with antagomiR-125b or TRIM21 siRNA or both, unlike in MCF7 cells, indicating that the absence of functional p53 resulted in no effect of inactivation of miR-125b and TRIM21 (Figure 7B). These observations demonstrated that the crucial effect of the miR-125b-TRIM21-HuR regulatory network on cell survival is mediated by its effect on the regulation of p53 expression.
Figure 7.
The p53 Translation Regulation Network Regulates Cell Viability in Response to UV-Induced DNA Damage
(A) MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay of MCF7 cells transfected with control oligo, antagomiR-125b, TRIM21 siRNA, and both antagomiR-125b and TRIM21 siRNA and collected at designated time points post UV irradiation. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.005 of difference of cell viability from corresponding control oligo-transfected cells at the same time point. #p ≤ 0.05 of difference of cell viability between control oligo-transfected cells between 0- and 4-h time points.
(B) MTT assay of MDA-MB-231 cells transfected with control oligo, antagomiR-125b, TRIM21 siRNA, and both antagomiR-125b and TRIM21 siRNA and collected at designated time points post UV irradiation. *p ≤ 0.05 of difference from corresponding control oligo-transfected cells at the same time point.
(C) Network diagram representing the regulation of p53 mediated by HuR, miR-125b, TRIM21, and Mdm2 in response to UV-induced DNA damage. The network consists of overlapping incoherent and coherent feedforward loops and negative feedback loop with HuR and p53 as the central axis.
See also Figure S16.
Discussion
Dynamic modeling, coupled with experimental validation, of the regulatory system controlling p53 mRNA translation in response to UV-induced DNA damage revealed the role of miR-125b and TRIM21 as regulators of protein synthesis and degradation, respectively, of the RNA-binding protein HuR. HuR undergoes nuclear-cytoplasmic translocation in response to a variety of stress stimuli such as UV, bacterial lipopolysaccharide, and heat shock and regulates the stability or translation of multiple mRNAs (Ahuja et al., 2016, Gallouzi et al., 2001, Mazan-Mamczarz et al., 2003, Poria et al., 2016, Wang et al., 2000). The enhancement of p53 protein synthesis mediated by HuR appears to be crucial in attaining an increased steady-state level of p53, together with the stabilization of p53 protein by inhibition of its interaction with Mdm2. HuR increases p53 protein synthesis not only by directly enhancing p53 mRNA translation but also by preventing the repression of p53 translation by the UV-induced miRNA, miR-125b (Ahuja et al., 2016). The biphasic expression of miR-125b in response to UV allows an enhancement of p53 expression at early time points after UV exposure, but causes repression of p53 mRNA translation at later time points, thereby contributing to the pulsatile nature of p53 expression (Ahuja et al., 2016).
Such complex regulatory networks being amenable to computational modeling, we adopted an approach combining dynamic systems modeling with experimental validation to both understand the systems-level functioning of the p53 regulatory network and find novel regulatory components and interactions that may regulate p53 protein synthesis in response to UV-induced DNA damage. Failure of the network model to match with the experimental data might be due to errors in estimating the parameters used in simulating the model or might indicate the presence of hitherto unknown components or interactions in the network. Parametrization tests, by varying the parameters over a broad range of values, have indicated the absence of major deviations in estimating the parameters used for numerical simulation of the model (Figures S12–S15). Therefore the possibility of involvement of novel regulators of p53 expression was systematically investigated. The dynamics of p53 expression post DNA damage has been the subject of extensive investigation, both experimentally and computationally (Batchelor et al., 2008, Ciliberto et al., 2005, Geva-Zatorsky et al., 2006, Lahav et al., 2004, Lev Bar-Or et al., 2000, Zhang et al., 2007). The dynamics of p53 has been shown to vary substantially across cell lines and in response to different types of DNA-damaging stimuli (Batchelor et al., 2011, Stewart-Ornstein and Lahav, 2017). In our study in MCF7 cells, p53 showed a single broad pulse in response to non-ionizing UV radiation, as observed previously (Batchelor et al., 2011). We have investigated the role of the p53 mRNA translation regulatory network, specifically the translation regulators HuR and miR-125b, in generating this pulsatile change in p53 abundance in response to UV radiation. Among the few known regulators of HuR, miRNAs such as miR-519, miR-16, and miR-34a are reported to downregulate HuR levels (Abdelmohsen et al., 2010, Kojima et al., 2010, Xu et al., 2010). miR-192 has been shown to mediate positive feedback loops that regulate p53 oscillations in response to DNA double-strand breaks (Moore et al., 2015). miR-125a, a member of the miR-125 family, has been shown to be an inhibitor of HuR expression in breast cancer cells, although the same study found that miR-125b, another miR-125 family member, was much less effective in repressing HuR synthesis (Guo et al., 2009). Very recently, miR-125b has been shown to regulate HuR expression in hepatitis C virus-infected liver carcinoma cells (Shwetha et al., 2018). Our computational modeling and experimental observations have demonstrated miR-125b as a negative regulator of HuR protein synthesis in response to DNA damage. miR-125b has been found to target multiple genes in the p53 regulatory network, which includes regulators of both cell proliferation and apoptosis (Le et al., 2011). The demonstration of miR-125b as a regulator of HuR, together with its direct regulation of p53 mRNA translation, establishes it as a major player in the regulation of p53 expression.
The decrease in HuR and p53 levels in the later time period post UV exposure is partially due to translation repression by miR-125b. However, as inclusion of miR-125b as a repressor of HuR protein synthesis failed to generate the pulsatile behavior of HuR and p53, we considered degradation of HuR protein contributing to oscillatory behavior of p53. Despite its stability, HuR protein destabilization by ubiquitin-mediated proteolysis or caspase-mediated cleavage is observed in response to stresses such as moderate heat shock, glycolysis inhibition, DNA damage, and CoCl2-induced hypoxia (Abdelmohsen et al., 2009, Chu et al., 2012, Lucchesi et al., 2016, Talwar et al., 2011). Postulating an unknown factor as mediating the degradation of HuR in response to UV, we found the E3 ubiquitin ligase TRIM21 to be responsible for ubiquitinating HuR leading to its proteasomal degradation. TRIM21, a 52-kD ribonucleoprotein, also denoted as Ro-SSA, is an autoantigen in autoimmune diseases such as systemic lupus erythematosus, rheumatoid arthritis, and Sjögren's syndrome (Ben-Chetrit et al., 1990). TRIM21 functions as a cytosolic IgG receptor known to bind the antibody Fc region via its C-terminal PRYSPRY domain and as an E3 ubiquitin ligase via its N-terminal RING domain (Ikeda and Inoue, 2012, James et al., 2007). TRIM21 negatively regulates the innate immune response to intracellular double-stranded DNA and DNA viruses by ubiquitinating DDX41 and targeting it for degradation (Zhang et al., 2012). It is also an important player in limiting the interferon response by targeting IRF3, IRF5, and other interferon response factors for proteasomal degradation (Higgs et al., 2008, Lazzari et al., 2014). Observations from TRIM21 knockout mice suggest that TRIM21 is a negative regulator of proinflammatory cytokine production and interferon signaling (Espinosa et al., 2009, Yoshimi et al., 2009). TRIM21 is also reported to reduce cell proliferation and acts as a pro-apoptotic molecule by suppressing Bcl-2 expression (Espinosa et al., 2006, Jauharoh et al., 2012). Therefore TRIM21 might play an important role as a link in limiting the inflammatory response and suppressing tumorigenesis. TRIM21 has been connected to regulation of p53 by its role in ubiquitinating guanosine 5′-monophosphate synthase and causing its nuclear localization, which contributes to dissociation of p53 from Mdm2 and its stabilization (Reddy et al., 2014). Our observations now demonstrate a new role for TRIM21 in fine-tuning p53 protein synthesis by modulating HuR protein level in response to UV radiation. This adds a new layer of regulation in the p53 control network activated by DNA damage.
Together, these observations have provided the basis of a systematic approach consisting of computational modeling and biochemical experimentation, which has allowed further elucidation of the intricate regulatory network controlling p53 expression and cell behavior in response to genotoxic stress. The translation regulatory network of p53 in response to UV-induced DNA damage consists of two overlapping incoherent feedforward loops (I-FFL) and two overlapping coherent feedforward loops (C-FFL) with HuR-p53 as the central axis (Figure 7C). Type1 I-FFLs function as pulse generators and response time accelerators, whereas C-FFLs constitute sign-sensitive delay elements that act as persistence detectors, which cause changes in gene expression only in response to persistent signals and protect against brief input fluctuations (Mangan and Alon, 2003). The type 1 I-FFLs in the p53 regulatory network involve damaged DNA, miR-125b, and TRIM21, which act in combination to generate a pulse of HuR (Figure S16). This pulse of HuR, combined with the pulse of miR-125b, which is opposite in phase, allows p53 mRNA translation to oscillate in response to DNA damage. The C-FFLs in the p53 regulatory network include a type 1 C-FFL, which involves damaged DNA, HuR, and p53, and a type 2 C-FFL consisting of miR-125b, HuR, and p53 (Figure S16). These two C-FFLs act in concert to enhance cellular p53 level only in response to persistent DNA damage. The integration of these incoherent and coherent feedforward loops with the p53-Mdm2 negative feedback loops causes the pulsatile change in p53 level in response to genotoxic stress and gives a window of opportunity to the cells to re-enter the cell cycle and resume cell division.
Limitations of the Study
This study develops and validates a mathematical model for the regulation of p53 expression in response to DNA damage that includes the regulators of p53 protein synthesis and stabilization. However, the model excludes the dynamics of the upstream regulators of p53 activation, such as the sensor and effector kinases (Maréchal and Zou, 2013), assuming these to be reflected in the dynamics of Mdm2, their downstream target. The proposed mathematical model can be said to capture the major contributing factors of the modeled phenomena if the model results are close to the experimental observations. However, it is a partial model, and the regulatory system, owing to its high level of complexity, will have aspects that are unaccounted for in the model (Aris and Penn, 1980). Explicit inclusion of the dynamics of the upstream regulators of p53 activation would lead to further refinement of the model and may lead to the discovery of other novel regulators and mediators of the cellular DNA damage response.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
We thank S. Das (IISc. Bangalore, India), S. N. Bhattacharyya (CSIR-IICB, Kolkata, India), and S. K. Singh (BHU, Varanasi, India) for providing reagents and Paul L. Fox (LRI, Cleveland Clinic, Cleveland, USA) for providing laboratory facilities. This work was supported by a Wellcome Trust-DBT India Alliance intermediate fellowship (WT500139/Z/09/Z) to P.S.R., UGC senior research fellowship to D.A., IISER Kolkata senior research fellowship to A. Guha, and CSIR senior research fellowship to S.D.M. The Orbitrap Elite instrument (Proteomics and Metabolomics Core, LRI, Cleveland Clinic) was purchased via an NIH shared instrument grant, 1S10RR031537-01.
Author Contributions
Conceptualization, P.S.R. and B.P.; Methodology, P.S.R., B.P., S.D.M., and A. Ghosh; Investigation, A. Guha, D.A., B.P., S.D.M., K.D., and B.W.; Resources, D.R. and V.S., Writing, P.S.R., A. Guha, and S.D.M.; Supervision, P.S.R.; Funding Acquisition, P.S.R.
Declaration of Interests
The authors declare no competing interests.
Published: May 31, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2019.05.002.
Supplemental Information
References
- Abdelmohsen K., Srikantan S., Yang X., Lal A., Kim H.H., Kuwano Y., Galban S., Becker K.G., Kamara D., de Cabo R. Ubiquitin-mediated proteolysis of HuR by heat shock. EMBO J. 2009;28:1271–1282. doi: 10.1038/emboj.2009.67. [DOI] [PMC free article] [PubMed] [Google Scholar]; Abdelmohsen, K., Srikantan, S., Yang, X., Lal, A., Kim, H.H., Kuwano, Y., Galban, S., Becker, K.G., Kamara, D., de Cabo, R., et al. (2009). Ubiquitin-mediated proteolysis of HuR by heat shock. EMBO J. 28, 1271-1282. [DOI] [PMC free article] [PubMed]
- Abdelmohsen K., Kim M.M., Srikantan S., Mercken E.M., Brennan S.E., Wilson G.M., de Cabo R., Gorospe M. miR-519 suppresses tumor growth by reducing HuR levels. Cell Cycle. 2010;9:1354–1359. doi: 10.4161/cc.9.7.11164. [DOI] [PMC free article] [PubMed] [Google Scholar]; Abdelmohsen, K., Kim, M.M., Srikantan, S., Mercken, E.M., Brennan, S.E., Wilson, G.M., de Cabo, R., and Gorospe, M. (2010). miR-519 suppresses tumor growth by reducing HuR levels. Cell Cycle 9, 1354-1359. [DOI] [PMC free article] [PubMed]
- Ahuja D., Goyal A., Ray P.S. Interplay between RNA-binding protein HuR and microRNA-125b regulates p53 mRNA translation in response to genotoxic stress. RNA Biol. 2016;13:1152–1165. doi: 10.1080/15476286.2016.1229734. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ahuja, D., Goyal, A., and Ray, P.S. (2016). Interplay between RNA-binding protein HuR and microRNA-125b regulates p53 mRNA translation in response to genotoxic stress. RNA Biol. 13, 1152-1165. [DOI] [PMC free article] [PubMed]
- Aris R., Penn M. The mere notion of a model. Math. Model. 1980;1:1–12. [Google Scholar]; Aris, R., and Penn, M. (1980). The mere notion of a model. Math. Model. 1, 1-12.
- Batchelor E., Mock C.S., Bhan I., Loewer A., Lahav G. Recurrent initiation: a mechanism for triggering p53 pulses in response to DNA damage. Mol. Cell. 2008;30:277–289. doi: 10.1016/j.molcel.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]; Batchelor, E., Mock, C.S., Bhan, I., Loewer, A., and Lahav, G. (2008). Recurrent initiation: a mechanism for triggering p53 pulses in response to DNA damage. Mol. Cell 30, 277-289. [DOI] [PMC free article] [PubMed]
- Batchelor E., Loewer A., Mock C., Lahav G. Stimulus-dependent dynamics of p53 in single cells. Mol. Syst. Biol. 2011;7:1–8. doi: 10.1038/msb.2011.20. [DOI] [PMC free article] [PubMed] [Google Scholar]; Batchelor, E., Loewer, A., Mock, C., and Lahav, G. (2011). Stimulus-dependent dynamics of p53 in single cells. Mol. Syst. Biol. 7, 1-8. [DOI] [PMC free article] [PubMed]
- Ben-Chetrit E., Fox R.I., Tan E.M. Dissociation of immune responses to the SS-A (Ro) 52-kd and 60-kd polypeptides in systemic lupus erythematosus and Sjögren’s syndrome. Arthritis Rheum. 1990;33:349–355. doi: 10.1002/art.1780330307. [DOI] [PubMed] [Google Scholar]; Ben-Chetrit, E., Fox, R.I., and Tan, E.M. (1990). Dissociation of immune responses to the SS-A (Ro) 52-kd and 60-kd polypeptides in systemic lupus erythematosus and Sjogren’s syndrome. Arthritis Rheum. 33, 349-355. [DOI] [PubMed]
- Chu P.-C., Chuang H.-C., Kulp S.K., Chen C.-S. The mRNA-stabilizing factor HuR protein is targeted by β-TrCP protein for degradation in response to glycolysis inhibition. J. Biol. Chem. 2012;287:43639–43650. doi: 10.1074/jbc.M112.393678. [DOI] [PMC free article] [PubMed] [Google Scholar]; Chu, P.-C., Chuang, H.-C., Kulp, S.K., and Chen, C.-S. (2012). The mRNA-stabilizing factor HuR protein is targeted by β-TrCP protein for degradation in response to glycolysis inhibition. J. Biol. Chem. 287, 43639-43650. [DOI] [PMC free article] [PubMed]
- Ciliberto A., Novak B., Tyson J.J. Steady states and oscillations in the p53/Mdm2 network. Cell Cycle. 2005;4:488–493. doi: 10.4161/cc.4.3.1548. [DOI] [PubMed] [Google Scholar]; Ciliberto, A., Novak, B., and Tyson, J.J. (2005). Steady states and oscillations in the p53/Mdm2 network. Cell Cycle 4, 488-493. [DOI] [PubMed]
- Collister M., Lane D.P., Kuehl B.L. Differential expression of p53, p21(waf1/cip1) and hdm2 dependent on DNA damage in Bloom’s syndrome fibroblasts. Carcinogenesis. 1998;19:2115–2120. doi: 10.1093/carcin/19.12.2115. [DOI] [PubMed] [Google Scholar]; Collister, M., Lane, D.P., and Kuehl, B.L. (1998). Differential expression of p53, p21(waf1/cip1) and hdm2 dependent on DNA damage in Bloom’s syndrome fibroblasts. Carcinogenesis 19, 2115-2120. [DOI] [PubMed]
- Espinosa A., Zhou W., Ek M., Hedlund M., Brauner S., Popovic K., Horvath L., Wallerskog T., Oukka M., Nyberg F. The Sjogren’s syndrome-associated autoantigen Ro52 is an E3 ligase that regulates proliferation and cell death. J. Immunol. 2006;176:6277–6285. doi: 10.4049/jimmunol.176.10.6277. [DOI] [PubMed] [Google Scholar]; Espinosa, A., Zhou, W., Ek, M., Hedlund, M., Brauner, S., Popovic, K., Horvath, L., Wallerskog, T., Oukka, M., Nyberg, F., et al. (2006). The Sjogren’s syndrome-associated autoantigen Ro52 is an E3 ligase that regulates proliferation and cell death. J. Immunol. 176, 6277-6285. [DOI] [PubMed]
- Espinosa A., Dardalhon V., Brauner S., Ambrosi A., Higgs R., Quintana F.J., Sjöstrand M., Eloranta M.-L., Ní Gabhann J., Winqvist O. Loss of the lupus autoantigen Ro52/Trim21 induces tissue inflammation and systemic autoimmunity by disregulating the IL-23-Th17 pathway. J. Exp. Med. 2009;206:1661–1671. doi: 10.1084/jem.20090585. [DOI] [PMC free article] [PubMed] [Google Scholar]; Espinosa, A., Dardalhon, V., Brauner, S., Ambrosi, A., Higgs, R., Quintana, F.J., Sjostrand, M., Eloranta, M.-L., Ni Gabhann, J., Winqvist, O., et al. (2009). Loss of the lupus autoantigen Ro52/Trim21 induces tissue inflammation and systemic autoimmunity by disregulating the IL-23-Th17 pathway. J. Exp. Med. 206, 1661-1671. [DOI] [PMC free article] [PubMed]
- Fu L., Minden M.D., Benchimol S. Translational regulation of human p53 gene expression. EMBO J. 1996;15:4392–4401. [PMC free article] [PubMed] [Google Scholar]; Fu, L., Minden, M.D., and Benchimol, S. (1996). Translational regulation of human p53 gene expression. EMBO J. 15, 4392-4401. [PMC free article] [PubMed]
- Gaglia G., Lahav G. Constant rate of p53 tetramerization in response to DNA damage controls the p53 response. Mol. Syst. Biol. 2014;10:1–8. doi: 10.15252/msb.20145168. [DOI] [PMC free article] [PubMed] [Google Scholar]; Gaglia, G., and Lahav, G. (2014). Constant rate of p53 tetramerization in response to DNA damage controls the p53 response. Mol. Syst. Biol. 10, 1-8. [DOI] [PMC free article] [PubMed]
- Gallouzi I.E., Brennan C.M., Steitz J.A. Protein ligands mediate the CRM1-dependent export of HuR in response to heat shock. RNA. 2001;7:1348–1361. doi: 10.1017/s1355838201016089. [DOI] [PMC free article] [PubMed] [Google Scholar]; Gallouzi, I.E., Brennan, C.M., and Steitz, J.A. (2001). Protein ligands mediate the CRM1-dependent export of HuR in response to heat shock. RNA 7, 1348-1361. [DOI] [PMC free article] [PubMed]
- Geva-Zatorsky N., Rosenfeld N., Itzkovitz S., Milo R., Sigal A., Dekel E., Yarnitzky T., Liron Y., Polak P., Lahav G. Oscillations and variability in the p53 system. Mol. Syst. Biol. 2006;2:1–13. doi: 10.1038/msb4100068. [DOI] [PMC free article] [PubMed] [Google Scholar]; Geva-Zatorsky, N., Rosenfeld, N., Itzkovitz, S., Milo, R., Sigal, A., Dekel, E., Yarnitzky, T., Liron, Y., Polak, P., Lahav, G., et al. (2006). Oscillations and variability in the p53 system. Mol. Syst. Biol. 2, 1-13. [DOI] [PMC free article] [PubMed]
- Guo X., Wu Y., Hartley R.S. MicroRNA-125a represses cell growth by targeting HuR in breast cancer. RNA Biol. 2009;6:575–583. doi: 10.4161/rna.6.5.10079. [DOI] [PMC free article] [PubMed] [Google Scholar]; Guo, X., Wu, Y., and Hartley, R.S. (2009). MicroRNA-125a represses cell growth by targeting HuR in breast cancer. RNA Biol. 6, 575-583. [DOI] [PMC free article] [PubMed]
- Higgs R., Ní Gabhann J., Ben Larbi N., Breen E.P., Fitzgerald K.A., Jefferies C.A. The E3 ubiquitin ligase Ro52 negatively regulates IFN-beta production post-pathogen recognition by polyubiquitin-mediated degradation of IRF3. J. Immunol. 2008;181:1780–1786. doi: 10.4049/jimmunol.181.3.1780. [DOI] [PMC free article] [PubMed] [Google Scholar]; Higgs, R., Ni Gabhann, J., Ben Larbi, N., Breen, E.P., Fitzgerald, K.A., and Jefferies, C.A. (2008). The E3 ubiquitin ligase Ro52 negatively regulates IFN-beta production post-pathogen recognition by polyubiquitin-mediated degradation of IRF3. J. Immunol. 181, 1780-1786. [DOI] [PMC free article] [PubMed]
- Horn H.F., Vousden K.H. Coping with stress: multiple ways to activate p53. Oncogene. 2007;26:1306–1316. doi: 10.1038/sj.onc.1210263. [DOI] [PubMed] [Google Scholar]; Horn, H.F., and Vousden, K.H. (2007). Coping with stress: multiple ways to activate p53. Oncogene 26, 1306-1316. [DOI] [PubMed]
- Ikeda K., Inoue S. TRIM proteins as RING finger E3 ubiquitin ligases. Adv. Exp. Med. Biol. 2012;770:27–37. doi: 10.1007/978-1-4614-5398-7_3. [DOI] [PubMed] [Google Scholar]; Ikeda, K., and Inoue, S. (2012). TRIM proteins as RING finger E3 ubiquitin ligases. Adv. Exp. Med. Biol. 770, 27-37. [DOI] [PubMed]
- James L.C., Keeble A.H., Khan Z., Rhodes D.A., Trowsdale J. Structural basis for PRYSPRY-mediated tripartite motif (TRIM) protein function. Proc. Natl. Acad. Sci. U S A. 2007;104:6200–6205. doi: 10.1073/pnas.0609174104. [DOI] [PMC free article] [PubMed] [Google Scholar]; James, L.C., Keeble, A.H., Khan, Z., Rhodes, D.A., and Trowsdale, J. (2007). Structural basis for PRYSPRY-mediated tripartite motif (TRIM) protein function. Proc. Natl. Acad. Sci. U S A. 104, 6200-6205. [DOI] [PMC free article] [PubMed]
- Jauharoh S.N.A., Saegusa J., Sugimoto T., Ardianto B., Kasagi S., Sugiyama D., Kurimoto C., Tokuno O., Nakamachi Y., Kumagai S. SS-A/Ro52 promotes apoptosis by regulating Bcl-2 production. Biochem. Biophys. Res. Commun. 2012;417:582–587. doi: 10.1016/j.bbrc.2011.12.010. [DOI] [PubMed] [Google Scholar]; Jauharoh, S.N.A., Saegusa, J., Sugimoto, T., Ardianto, B., Kasagi, S., Sugiyama, D., Kurimoto, C., Tokuno, O., Nakamachi, Y., Kumagai, S., et al. (2012). SS-A/Ro52 promotes apoptosis by regulating Bcl-2 production. Biochem. Biophys. Res. Commun. 417, 582-587. [DOI] [PubMed]
- Kastan M.B., Onyekwere O., Sidransky D., Vogelstein B., Craig R.W. Participation of p53 protein in the cellular response to DNA damage. Cancer Res. 1991;51:6304–6311. [PubMed] [Google Scholar]; Kastan, M.B., Onyekwere, O., Sidransky, D., Vogelstein, B., and Craig, R.W. (1991). Participation of p53 protein in the cellular response to DNA damage. Cancer Res. 51, 6304-6311. [PubMed]
- Kojima K., Fujita Y., Nozawa Y., Deguchi T., Ito M. MiR-34a attenuates paclitaxel-resistance of hormone-refractory prostate cancer PC3 cells through direct and indirect mechanisms. Prostate. 2010;70:1501–1512. doi: 10.1002/pros.21185. [DOI] [PubMed] [Google Scholar]; Kojima, K., Fujita, Y., Nozawa, Y., Deguchi, T., and Ito, M. (2010). MiR-34a attenuates paclitaxel-resistance of hormone-refractory prostate cancer PC3 cells through direct and indirect mechanisms. Prostate 70, 1501-1512. [DOI] [PubMed]
- Lahav G. Oscillations by the p53-Mdm2 feedback loop. Adv. Exp. Med. Biol. 2008;641:28–38. doi: 10.1007/978-0-387-09794-7_2. [DOI] [PubMed] [Google Scholar]; Lahav, G. (2008). Oscillations by the p53-Mdm2 feedback loop. Adv. Exp. Med. Biol. 641, 28-38. [DOI] [PubMed]
- Lahav G., Rosenfeld N., Sigal A., Geva-Zatorsky N., Levine A.J., Elowitz M.B., Alon U. Dynamics of the p53-Mdm2 feedback loop in individual cells. Nat. Genet. 2004;36:147–150. doi: 10.1038/ng1293. [DOI] [PubMed] [Google Scholar]; Lahav, G., Rosenfeld, N., Sigal, A., Geva-Zatorsky, N., Levine, A.J., Elowitz, M.B., and Alon, U. (2004). Dynamics of the p53-Mdm2 feedback loop in individual cells. Nat. Genet. 36, 147-150. [DOI] [PubMed]
- Lazzari E., Korczeniewska J., Ní Gabhann J., Smith S., Barnes B.J., Jefferies C.A. TRIpartite motif 21 (TRIM21) differentially regulates the stability of interferon regulatory factor 5 (IRF5) isoforms. PLoS One. 2014;9:e103609. doi: 10.1371/journal.pone.0103609. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lazzari, E., Korczeniewska, J., Ni Gabhann, J., Smith, S., Barnes, B.J., and Jefferies, C.A. (2014). TRIpartite motif 21 (TRIM21) differentially regulates the stability of interferon regulatory factor 5 (IRF5) isoforms. PLoS One 9, e103609. [DOI] [PMC free article] [PubMed]
- Le M.T.N., Teh C., Shyh-Chang N., Xie H., Zhou B., Korzh V., Lodish H.F., Lim B. MicroRNA-125b is a novel negative regulator of p53. Genes Dev. 2009;23:862–876. doi: 10.1101/gad.1767609. [DOI] [PMC free article] [PubMed] [Google Scholar]; Le, M.T.N., Teh, C., Shyh-Chang, N., Xie, H., Zhou, B., Korzh, V., Lodish, H.F., and Lim, B. (2009). MicroRNA-125b is a novel negative regulator of p53. Genes Dev. 23, 862-876. [DOI] [PMC free article] [PubMed]
- Le M.T.N., Shyh-Chang N., Khaw S.L., Chin L., Teh C., Tay J., O’Day E., Korzh V., Yang H., Lal A. Conserved regulation of p53 network dosage by microRNA-125b occurs through evolving miRNA-target gene pairs. PLoS Genet. 2011;7:e1002242. doi: 10.1371/journal.pgen.1002242. [DOI] [PMC free article] [PubMed] [Google Scholar]; Le, M.T.N., Shyh-Chang, N., Khaw, S.L., Chin, L., Teh, C., Tay, J., O’Day, E., Korzh, V., Yang, H., Lal, A., et al. (2011). Conserved regulation of p53 network dosage by microRNA-125b occurs through evolving miRNA-target gene pairs. PLoS Genet. 7. [DOI] [PMC free article] [PubMed]
- Lev Bar-Or R., Maya R., Segel L.A., Alon U., Levine A.J., Oren M. Generation of oscillations by the p53-Mdm2 feedback loop: a theoretical and experimental study. Proc. Natl. Acad. Sci. U S A. 2000;97:11250–11255. doi: 10.1073/pnas.210171597. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lev Bar-Or, R., Maya, R., Segel, L.A., Alon, U., Levine, A.J., and Oren, M. (2000). Generation of oscillations by the p53-Mdm2 feedback loop: a theoretical and experimental study. Proc. Natl. Acad. Sci. U S A. 97, 11250-11255. [DOI] [PMC free article] [PubMed]
- Lucchesi C., Sheikh M.S., Huang Y. Negative regulation of RNA-binding protein HuR by tumor-suppressor ECRG2. Oncogene. 2016;35:2565–2573. doi: 10.1038/onc.2015.339. [DOI] [PubMed] [Google Scholar]; Lucchesi, C., Sheikh, M.S., and Huang, Y. (2016). Negative regulation of RNA-binding protein HuR by tumor-suppressor ECRG2. Oncogene 35, 2565-2573. [DOI] [PubMed]
- Ma L., Wagner J., Rice J.J., Hu W., Levine A.J., Stolovitzky G.A. A plausible model for the digital response of p53 to DNA damage. Proc. Natl. Acad. Sci. U S A. 2005;102:14266–14271. doi: 10.1073/pnas.0501352102. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ma, L., Wagner, J., Rice, J.J., Hu, W., Levine, A.J., and Stolovitzky, G.A. (2005). A plausible model for the digital response of p53 to DNA damage. Proc. Natl. Acad. Sci. U S A 102, 14266-14271. [DOI] [PMC free article] [PubMed]
- Mangan S., Alon U. Structure and function of the feed-forward loop network motif. Proc. Natl. Acad. Sci. U S A. 2003;100:11980–11985. doi: 10.1073/pnas.2133841100. [DOI] [PMC free article] [PubMed] [Google Scholar]; Mangan, S., and Alon, U. (2003). Structure and function of the feed-forward loop network motif. Proc. Natl. Acad. Sci. U S A. 100, 11980-11985. [DOI] [PMC free article] [PubMed]
- Maréchal A., Zou L. DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb. Perspect. Biol. 2013;5:a012716. doi: 10.1101/cshperspect.a012716. [DOI] [PMC free article] [PubMed] [Google Scholar]; Marechal, A., and Zou, L. (2013). DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb. Perspect. Biol. 5, a012716. [DOI] [PMC free article] [PubMed]
- Mazan-Mamczarz K., Galbán S., López de Silanes I., Martindale J.L., Atasoy U., Keene J.D., Gorospe M. RNA-binding protein HuR enhances p53 translation in response to ultraviolet light irradiation. Proc. Natl. Acad. Sci. U S A. 2003;100:8354–8359. doi: 10.1073/pnas.1432104100. [DOI] [PMC free article] [PubMed] [Google Scholar]; Mazan-Mamczarz, K., Galban, S., Lopez de Silanes, I., Martindale, J.L., Atasoy, U., Keene, J.D., and Gorospe, M. (2003). RNA-binding protein HuR enhances p53 translation in response to ultraviolet light irradiation. Proc. Natl. Acad. Sci. U S A. 100, 8354-8359. [DOI] [PMC free article] [PubMed]
- Michael D., Oren M. The p53-Mdm2 module and the ubiquitin system. Semin. Cancer Biol. 2003;13:49–58. doi: 10.1016/s1044-579x(02)00099-8. [DOI] [PubMed] [Google Scholar]; Michael, D., and Oren, M. (2003). The p53-Mdm2 module and the ubiquitin system. Semin. Cancer Biol. 13, 49-58. [DOI] [PubMed]
- Moore R., Ooi H.K., Kang T., Bleris L., Ma L. MiR-192-Mediated positive feedback loop controls the robustness of stress-induced p53 oscillations in breast cancer cells. PLoS Comput. Biol. 2015;11:e1004653. doi: 10.1371/journal.pcbi.1004653. [DOI] [PMC free article] [PubMed] [Google Scholar]; Moore, R., Ooi, H.K., Kang, T., Bleris, L., and Ma, L. (2015). MiR-192-Mediated positive feedback loop controls the robustness of stress-induced p53 oscillations in breast cancer cells. PLoS Comput. Biol. 11, e1004653. [DOI] [PMC free article] [PubMed]
- Poria D.K., Guha A., Nandi I., Ray P.S. RNA-binding protein HuR sequesters microRNA-21 to prevent translation repression of proinflammatory tumor suppressor gene programmed cell death 4. Oncogene. 2016;35:1703–1715. doi: 10.1038/onc.2015.235. [DOI] [PMC free article] [PubMed] [Google Scholar]; Poria, D.K., Guha, A., Nandi, I., and Ray, P.S. (2016). RNA-binding protein HuR sequesters microRNA-21 to prevent translation repression of proinflammatory tumor suppressor gene programmed cell death 4. Oncogene 35, 1703-1715. [DOI] [PMC free article] [PubMed]
- Proctor C.J., Gray D.A. Explaining oscillations and variability in the p53-Mdm2 system. BMC Syst. Biol. 2008;2:75. doi: 10.1186/1752-0509-2-75. [DOI] [PMC free article] [PubMed] [Google Scholar]; Proctor, C.J., and Gray, D.A. (2008). Explaining oscillations and variability in the p53-Mdm2 system. BMC Syst. Biol. 2, 75. [DOI] [PMC free article] [PubMed]
- Purvis J.E., Karhohs K.W., Mock C., Batchelor E., Loewer A., Lahav G. p53 dynamics control cell fate. Science. 2012;336:1440–1444. doi: 10.1126/science.1218351. [DOI] [PMC free article] [PubMed] [Google Scholar]; Purvis, J.E., Karhohs, K.W., Mock, C., Batchelor, E., Loewer, A., and Lahav, G. (2012). p53 dynamics control cell fate. Science 336, 1440-1444. [DOI] [PMC free article] [PubMed]
- Reddy B.A., van der Knaap J.A., Bot A.G.M., Mohd-Sarip A., Dekkers D.H.W., Timmermans M.A., Martens J.W.M., Demmers J.A.A., Verrijzer C.P. Nucleotide biosynthetic enzyme GMP synthase is a TRIM21-controlled relay of p53 stabilization. Mol. Cell. 2014;53:458–470. doi: 10.1016/j.molcel.2013.12.017. [DOI] [PubMed] [Google Scholar]; Reddy, B.A., van der Knaap, J.A., Bot, A.G.M., Mohd-Sarip, A., Dekkers, D.H.W., Timmermans, M.A., Martens, J.W.M., Demmers, J.A.A., and Verrijzer, C.P. (2014). Nucleotide biosynthetic enzyme GMP synthase is a TRIM21-controlled relay of p53 stabilization. Mol. Cell 53, 458-470. [DOI] [PubMed]
- Shwetha S., Sharma G., Raheja H., Goel A., Aggarwal R., Das S. Interaction of miR-125b-5p with Human antigen R mRNA: mechanism of controlling HCV replication. Virus Res. 2018;258:1–8. doi: 10.1016/j.virusres.2018.09.006. [DOI] [PubMed] [Google Scholar]; Shwetha, S., Sharma, G., Raheja, H., Goel, A., Aggarwal, R., and Das, S. (2018). Interaction of miR-125b-5p with Human antigen R mRNA: mechanism of controlling HCV replication. Virus Res. 258, 1-8. [DOI] [PubMed]
- Stewart-Ornstein J., Lahav G. P53 dynamics in response to DNA damage vary across cell lines and are shaped by efficiency of DNA repair and activity of the kinase ATM. Sci. Signal. 2017;10:eaah6671. doi: 10.1126/scisignal.aah6671. [DOI] [PMC free article] [PubMed] [Google Scholar]; Stewart-Ornstein, J., and Lahav, G. (2017). P53 dynamics in response to DNA damage vary across cell lines and are shaped by efficiency of DNA repair and activity of the kinase ATM. Sci. Signal. 10, eaah6671. [DOI] [PMC free article] [PubMed]
- Talwar S., Jin J., Carroll B., Liu A., Gillespie M.B., Palanisamy V. Caspase-mediated cleavage of RNA-binding protein HuR regulates c-Myc protein expression after hypoxic stress. J. Biol. Chem. 2011;286:32333–32343. doi: 10.1074/jbc.M111.255927. [DOI] [PMC free article] [PubMed] [Google Scholar]; Talwar, S., Jin, J., Carroll, B., Liu, A., Gillespie, M.B., and Palanisamy, V. (2011). Caspase-mediated cleavage of RNA-binding protein HuR regulates c-Myc protein expression after hypoxic stress. J. Biol. Chem. 286, 32333-32343. [DOI] [PMC free article] [PubMed]
- Vilborg A., Wilhelm M.T., Wiman K.G. Regulation of tumor suppressor p53 at the RNA level. J. Mol. Med. (Berl) 2010;88:645–652. doi: 10.1007/s00109-010-0609-2. [DOI] [PubMed] [Google Scholar]; Vilborg, A., Wilhelm, M.T., and Wiman, K.G. (2010). Regulation of tumor suppressor p53 at the RNA level. J. Mol. Med. (Berl). 88, 645-652. [DOI] [PubMed]
- Wagner J., Ma L., Rice J.J., Hu W., Levine A.J., Stolovitzky G.A. p53-Mdm2 loop controlled by a balance of its feedback strength and effective dampening using ATM and delayed feedback. Syst. Biol. (Stevenage) 2005;152:109–118. doi: 10.1049/ip-syb:20050025. [DOI] [PubMed] [Google Scholar]; Wagner, J., Ma, L., Rice, J.J., Hu, W., Levine, A.J., and Stolovitzky, G.A. (2005). p53-Mdm2 loop controlled by a balance of its feedback strength and effective dampening using ATM and delayed feedback. Syst. Biol. (Stevenage). 152, 109-118. [DOI] [PubMed]
- Wang W., Furneaux H., Cheng H., Caldwell M.C., Hutter D., Liu Y., Holbrook N., Gorospe M. HuR regulates p21 mRNA stabilization by UV light. Mol. Cell. Biol. 2000;20:760–769. doi: 10.1128/mcb.20.3.760-769.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wang, W., Furneaux, H., Cheng, H., Caldwell, M.C., Hutter, D., Liu, Y., Holbrook, N., and Gorospe, M. (2000). HuR regulates p21 mRNA stabilization by UV light. Mol. Cell. Biol. 20, 760-769. [DOI] [PMC free article] [PubMed]
- Xu F., Zhang X., Lei Y., Liu X., Liu Z., Tong T., Wang W. Loss of repression of HuR translation by miR-16 may be responsible for the elevation of HuR in human breast carcinoma. J. Cell. Biochem. 2010;111:727–734. doi: 10.1002/jcb.22762. [DOI] [PubMed] [Google Scholar]; Xu, F., Zhang, X., Lei, Y., Liu, X., Liu, Z., Tong, T., and Wang, W. (2010). Loss of repression of HuR translation by miR-16 may be responsible for the elevation of HuR in human breast carcinoma. J. Cell. Biochem. 111, 727-734. [DOI] [PubMed]
- Yoshimi R., Chang T.-H., Wang H., Atsumi T., Morse H.C., Ozato K., Ozato K. Gene disruption study reveals a nonredundant role for TRIM21/Ro52 in NF-kappaB-dependent cytokine expression in fibroblasts. J. Immunol. 2009;182:7527–7538. doi: 10.4049/jimmunol.0804121. [DOI] [PMC free article] [PubMed] [Google Scholar]; Yoshimi, R., Chang, T.-H., Wang, H., Atsumi, T., Morse, H.C., Ozato, K., and Ozato, K. (2009). Gene disruption study reveals a nonredundant role for TRIM21/Ro52 in NF-kappaB-dependent cytokine expression in fibroblasts. J. Immunol. 182, 7527-7538. [DOI] [PMC free article] [PubMed]
- Zhang T., Brazhnik P., Tyson J.J. Exploring mechanisms of the DNA-damage response: p53 pulses and their possible relevance to apoptosis. Cell Cycle. 2007;6:85–94. doi: 10.4161/cc.6.1.3705. [DOI] [PubMed] [Google Scholar]; Zhang, T., Brazhnik, P., and Tyson, J.J. (2007). Exploring mechanisms of the DNA-damage response: p53 pulses and their possible relevance to apoptosis. Cell Cycle 6, 85-94. [DOI] [PubMed]
- Zhang Y., Gao J.-S., Tang X., Tucker L.D., Quesenberry P., Rigoutsos I., Ramratnam B. MicroRNA 125a and its regulation of the p53 tumor suppressor gene. FEBS Lett. 2009;583:3725–3730. doi: 10.1016/j.febslet.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]; Zhang, Y., Gao, J.-S., Tang, X., Tucker, L.D., Quesenberry, P., Rigoutsos, I., and Ramratnam, B. (2009). MicroRNA 125a and its regulation of the p53 tumor suppressor gene. FEBS Lett. 583, 3725-3730. [DOI] [PMC free article] [PubMed]
- Zhang Z., Bao M., Lu N., Weng L., Yuan B., Liu Y.-J. The E3 ubiquitin ligase TRIM21 negatively regulates the innate immune response to intracellular double-stranded DNA. Nat. Immunol. 2012;14:172–178. doi: 10.1038/ni.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]; Zhang, Z., Bao, M., Lu, N., Weng, L., Yuan, B., and Liu, Y.-J. (2012). The E3 ubiquitin ligase TRIM21 negatively regulates the innate immune response to intracellular double-stranded DNA. Nat. Immunol. 14, 172-178. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.