Abstract
During amphibian development, neural patterning occurs via a two‐step process. Spemann's organizer secretes BMP antagonists that induce anterior neural tissue. A subsequent caudalizing step re‐specifies anterior fated cells to posterior fates such as hindbrain and spinal cord. The neural patterning paradigm suggests that a canonical Wnt‐signaling gradient acts along the anteroposterior axis to pattern the nervous system. Wnt activity is highest in the posterior, inducing spinal cord, at intermediate levels in the trunk, inducing hindbrain, and is lowest in anterior fated forebrain, while BMP‐antagonist levels are constant along the axis. Our results in Xenopus laevis challenge this paradigm. We find that inhibition of canonical Wnt signaling or its downstream transcription factors eliminates hindbrain, but not spinal cord fates, an observation not compatible with a simple high‐to‐low Wnt gradient specifying all fates along the neural anteroposterior axis. Additionally, we find that BMP activity promotes posterior spinal cord cell fate formation in an FGF‐dependent manner, while inhibiting hindbrain fates. These results suggest a need to re‐evaluate the paradigms of neural anteroposterior pattern formation during vertebrate development.
Keywords: BMP signaling, canonical Wnt signaling, hindbrain and spinal cord, neural anteroposterior patterning, Xenopus
Subject Categories: Development & Differentiation, Signal Transduction
Introduction
In vertebrates, the nervous system has a distinct anteroposterior (AP) pattern. After initial neural induction, the nervous system is subdividing into the forebrain, midbrain, hindbrain, and spinal cord regions [rev. in 1, 2]. Historically, much of our knowledge of these earliest patterning events has been elucidated in amphibian and chick model systems [rev. in 3, 4]. In Xenopus, BMP antagonists secreted from the Spemann organizer initially induce anterior neural tissue [5, 6, rev. in 7, 8]. A subsequent caudalizing step specifies these neural induced cells to posterior hindbrain and spinal cord fates 5, 9, 10. These classic experiments suggested that a morphogen gradient could act to caudalize the posterior nervous system. Morphogens are secreted diffusible molecules that can induce changes in cell fates over distance in a concentration‐dependent manner. Morphogen gradients have been suggested to regulate body axis formation during early vertebrate development [rev. in 11].
A secreted morphogen gradient could explain the induction of specific neural cell fates in a concentration‐dependent manner along the vertebrate AP axis. The signaling molecules that caudalize the vertebrate nervous system are retinoic acid, FGF, and Wnt [rev. in 12, 13, 14]. These signaling molecules could all be candidates for morphogens regulating neural AP axis formation. Experimental evidence in Xenopus and chick embryos supports a paradigm in which canonical Wnt signaling acts as a morphogen to form a “low to high” gradient along the AP axis that concomitantly acts with BMP antagonism (neural induction) to pattern the developing nervous system 15, 16. This paradigm suggests that Wnt activity is at its highest levels in the posterior, inducing the spinal cord, at intermediate levels to induce the hindbrain and midbrain in the trunk, and at low activity in the most anterior forebrain regions. Our present results suggest that this paradigm needs to be re‐evaluated.
We show that Xenopus laevis embryos knocked down for zygotic canonical Wnt activity by ectopic Dkk1 protein surprisingly express normal levels of spinal cord markers, and these embryos have remarkably normal spinal cord morphology. These embryos display the typical Wnt knockdown phenotype, having expanded anterior neural‐ectodermal tissues such as forebrain and cement gland, while simultaneously losing hindbrain fates. Thus, while the forebrain is expanded and the hindbrain is lost, the more posterior spinal cord is still intact. If a gradient of high Wnt activity is required to induce the most caudal embryonic regions, then the posterior spinal cord region should have the highest sensitivity to the loss of Wnt activity and not the lowest. In a complementary manner, we show that the knockdown of Zic transcription factor activity gives a similar phenotype. Zic transcription factors mediate Wnt signaling to induce different posterior neural cell fates 17, 18, 19, 20, 21, 22, 23. We show that knockdown of the Wnt‐mediating downstream Zic1 transcription factor protein inhibits hindbrain fates, while expanding the forebrain. However, similar to the inhibition of canonical Wnt signaling, expression of the most posterior spinal cord markers is not decreased, but increased by Zic1 protein knockdown.
In the “classic” paradigm for neural patterning, BMP antagonists and neural caudalizers were proposed to jointly act in specifying posterior cell fates along the vertebrate AP axis. We show in cultured ectodermal ex vivo explants that the more posterior spinal cord markers are not induced in the presence of BMP antagonists and neural caudalizers, while hindbrain markers are robustly induced. Counterintuitively, BMP alone is an efficient inducer of spinal cord marker expression and an inhibitor of hindbrain marker expression in embryos and ectodermal explants. We further found that BMP induction of spinal cord markers is FGF‐dependent.
We propose that the interplay between canonical Wnt activity, BMP activation, and BMP antagonism needs to be re‐assessed concerning neural AP patterning. A simple high‐to‐low gradient of Wnt morphogen activity appears incompatible for determining the AP differences in spinal cord versus hindbrain cell fate specification. Low–moderate BMP signaling levels also appear crucial for spinal cord induction. These findings challenge the present paradigm of neural AP pattern formation in the developing vertebrate nervous system.
Results
Canonical Wnt signaling is required for hindbrain, but not spinal cord formation
It has been a generally accepted paradigm that canonical Wnt signaling acts in a morphogen gradient to pattern neural tissue [rev. in 3] from the most posterior spinal cord (high Wnt) to the most anterior forebrain (low Wnt). Our previous studies showed that Wnt3a in the dorsal–lateral mesoderm induces hindbrain, neural crest, and primary neuron cell fates in the adjacent neural plate 24. Wnt induces these cell fates via direct transcriptional activation of the TALE‐class homeobox protein, Meis3 24, yet Meis3 knockdown has only minor effects on spinal cord development 25. This observation does not necessarily support the idea of a Wnt gradient in neural AP patterning. To rigorously address the role of Wnt signaling in posterior neural patterning, the canonical Wnt‐inhibitor Dkk1 protein was ectopically expressed in one‐cell stage embryos. As seen by sqRT–PCR in neurula stage embryos (Fig 1A), expression of anterior forebrain and cement gland markers (xanf1, otx2, xag1) is increased, but expression of hindbrain markers is reduced (krox20, hoxb3). In an interesting and surprising manner, expression levels of the more posterior spinal cord markers (hoxb9, cdx1, cdx2, cdx4, hoxd10) are barely modulated by Wnt activity inhibition. The hoxb4 gene that is expressed in the hindbrain/spinal cord border region has an intermediate level of inhibition. Similar results were also seen by zygotic ectopic expression of the Nxfz8 protein (Fig 1B), a dominant‐inhibitory Frizzled‐receptor protein that inhibits canonical Wnt signaling 26. When examining embryos by in situ hybridization, the differences between spinal cord and hindbrain markers are striking (Fig 1C). Expression of krox20 in the hindbrain is highly perturbed, but expression of the spinal cord‐specific hoxb9 and cdx4 genes looks normal. Even hoxd10 expression in the most posterior spinal cord region looks quite normal. Mesodermal (dorsal and ventral) markers are expressed at normal levels in gastrula and neurula stage embryos injected with dkk1 mRNA into one blastomere at the one‐cell stage (Appendix Fig S1A–D) or one blastomere/two‐cell stage (Appendix Fig S2A and C). Mesoderm fates do not seem to be perturbed at gastrula or neurula stages. At gastrula stages (Appendix Fig S1A and C), the expression of ventral (vent1, vent2, wnt8), lateral (osr1, osr2), dorsal (xnot, chordin‐chd), and pan‐mesodermal (xbra) markers is at normal levels. At later neurula stages, dorsal‐notochord (xnot, chd, xbra) and dorsal–lateral skeletal muscle (myod, muscle actin‐MA) marker expression is normal (Appendix Figs S1B and D, and S2A and C). In these embryos, anterior neural markers are expanded, hindbrain markers are inhibited, and spinal cord markers are expressed normally (Appendix Figs S1D, and S2A and C). In situ hybridization was performed on embryos that were injected with dkk1 mRNA into one blastomere at the two‐cell stage. These embryos had asymmetric expression of the hindbrain‐specific krox20 marker, but symmetric expression of spinal cord markers (hoxb9, cdx4) and dorsal mesoderm (chd, MA) markers (Appendix Fig S2A and C). We also sectioned the trunk regions of later tailbud stage embryos that were injected with dkk1 mRNA into one blastomere at the two‐cell stage; these embryos had normal spinal cord, notochord, and somite formation (Appendix Fig S2E). These results strongly show that the effects of Wnt inhibition appear specific to the neural plate and not a result of mesoderm perturbation.
Figure 1. Inhibition of canonical Wnt signaling perturbs hindbrain, but not spinal cord cell fates.
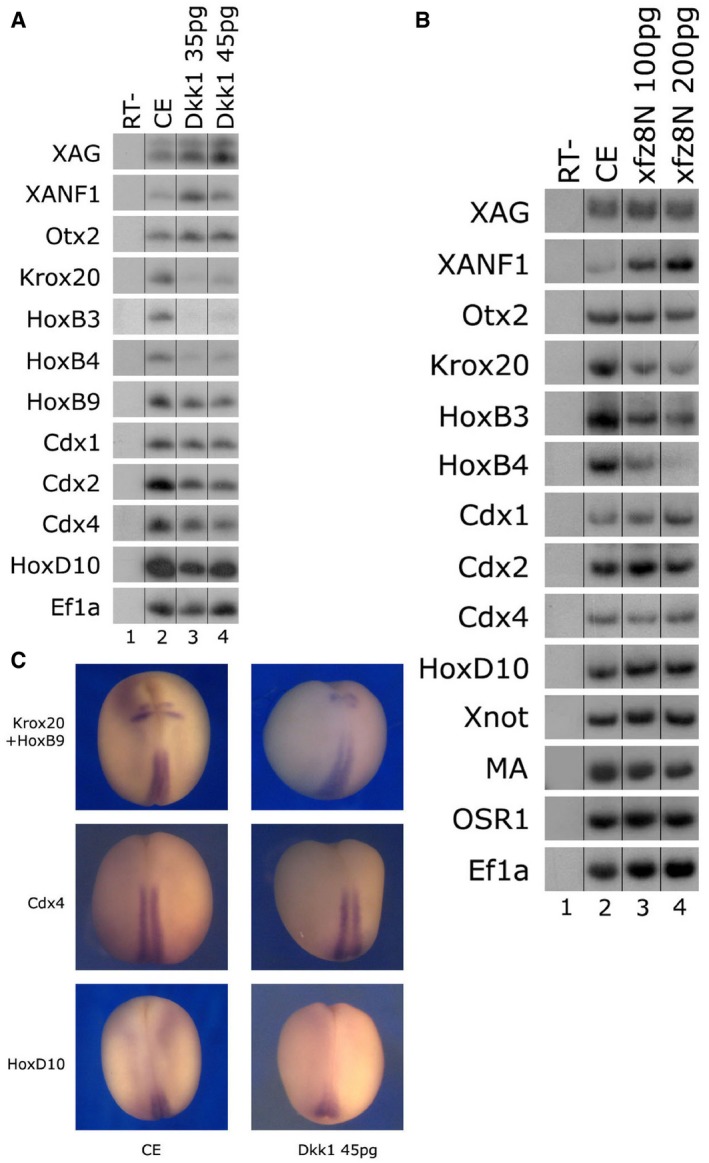
- One‐cell stage embryos were injected animally with mRNA encoding dkk1 (35/45 pg) protein. Total RNA was isolated from 10 control embryos at neurula stage 17 (lane 2) and 10 embryos from each dkk1‐injected group (lanes 3–4). Various neural AP markers were examined by sqRT–PCR: cement gland and forebrain (xag1, xanf1, otx2), hindbrain (krox20, hoxb3), hindbrain/spinal cord border (hoxb4), and spinal cord (hoxb9, cdx1‐2‐4, hoxd10). Ef1α serves as a control for quantitating RNA levels. ‐RT–PCR was performed on total RNA isolated from control embryos (lane 1).
- One‐cell stage embryos were injected animally with the zygotic‐expressing Nxfz8/pCS2 encoding plasmid (100/200 pg). Total RNA was isolated from 10 control embryos at neurula stage 17 (lane 2) and 10 embryos from each Nxfz8‐injected group (lanes 3–4). Various neural AP markers were examined by sqRT–PCR: cement gland and forebrain (xag1, xanf1, otx2), hindbrain (krox20, hoxb3), hindbrain/spinal cord border (hoxb4), and spinal cord (hoxb9, cdx1‐2‐4, hoxd10). Ef1α serves as a control for quantitating RNA levels. ‐RT–PCR was performed on total RNA isolated from control embryos (lane 1).
- Embryos from the same experiment as in (A) underwent in situ hybridization. Expression of the hindbrain marker krox20 (upper two panels) was reduced by Dkk1 (45 pg) in 83% of the embryos (n = 18). Expression of spinal cord markers (hoxb9, cdx4, hox10) was normal, being unchanged in 89% of the embryos (n = 18 for each marker). Control embryos (CE).
To further extend this point, four of the animal blastomeres fated for ectoderm were injected with dkk1 mRNA at the eight‐cell stage. These embryos also expressed lower levels of hindbrain markers (krox20, hoxb3) with normal levels of forebrain (xanf1), spinal cord (hoxb9, cdx2, cdx4), and dorsal mesoderm (chd—notochord, myod—muscle) markers (Appendix Fig S3A). As shown in previous studies 24, we lineage traced embryos injected animally at either the one‐ or eight‐cell stage, and β‐gal expression was localized to the neural plate at stages 17–18 (Appendix Fig S3C). If Wnt was working in a high‐to‐low gradient from posterior to anterior, we would not expect this observed robust expression of spinal cord markers. We would expect spinal cord marker genes to be more sensitive to the loss of Wnt activity than hindbrain markers, but the opposite result is seen.
The Zic1 protein has been previously shown to act with Wnt signaling to mediate induction of neural crest, hindbrain, and primary neuron cell fates 17, 18, 19, 20, 21, 22, 23. We determined how dose‐dependent Zic1‐MO knockdown modulated neural AP cell fates. As can be seen by sqRT–PCR in neurula stage embryos (Fig 2A), forebrain marker expression (xanf1, otx2) is increased and hindbrain markers are reduced (krox20, hoxb1, hoxb3). Expression levels of spinal cord markers (hoxb9, cdx1, cdx4) are strongly increased by Zic1 protein knockdown. The panneural soxd1 marker is unaffected by the loss of Zic1 protein activity. Similar to the Zic1 morphant embryos, ectopic expression of the Zic5 dominant‐negative protein (Fig 2B) inhibited hindbrain marker expression (krox20), while enhancing spinal cord marker expression (cdx1, cdx4). When examining embryos by in situ hybridization, marker expression differences along the neural AP axis are clear (Fig 2C). Expression of the sox2 panneural marker is unaltered by Zic1 knockdown, but forebrain‐specific xanf1 expression is expanded versus the control (Fig 2C). Expression of a representative hindbrain marker such as krox20 is eliminated in comparison with the control (Fig 2C). Interestingly, expression of a representative spinal cord marker, cdx4, is laterally expanded and increased (Fig 2C; similar to Fig 2A and B). It is not clear if the spinal cord markers are anteriorly expanded, but expression of these genes is not perturbed by the knockdown of the Wnt mediator, Zic1. These observations complement the results seen with Dkk1 protein (Fig 1), suggesting that Wnt signaling is required for hindbrain formation, but is not crucial for the specification of more posterior spinal cord fates.
Figure 2. Inhibition of Zic protein activity differentially affects spinal cord and hindbrain cell fates.
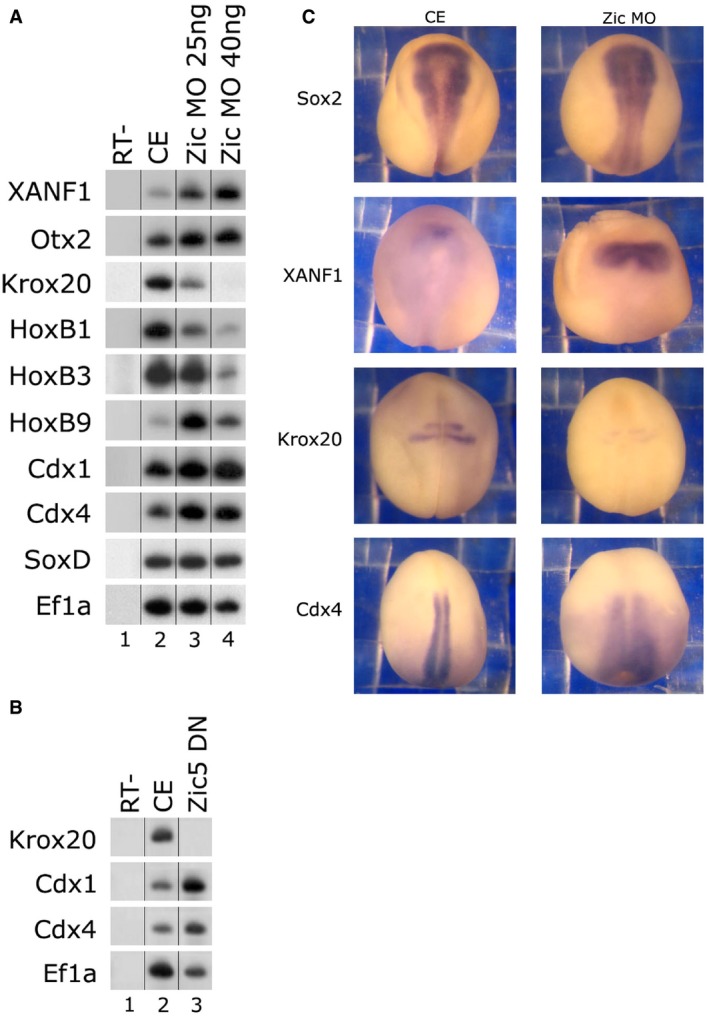
- One‐cell stage embryos were injected animally with the Zic1‐MO (25/40 ng). Total RNA was isolated from 10 control embryos at neurula stage 18 (lane 2) and 10 embryos from each Zic1‐MO‐injected group (lanes 3–4). Neural AP markers were examined by sqRT–PCR: forebrain (xanf1, otx2), hindbrain (krox20, hoxb1, hoxb3), spinal cord (hoxb9, cdx1‐4), and panneural (soxd). Ef1α is the control for quantitating RNA levels. ‐RT–PCR was performed on total RNA isolated from control embryos (lane 1).
- One‐cell stage embryos were injected animally with mRNA encoding Zic5 dominant‐negative (0.5 ng) protein. Total RNA was isolated from five control embryos at neurula stage 18 (lane 1) and five embryos from the injected group (lanes 2). Neural AP markers were examined by sqRT–PCR: hindbrain (krox20) and spinal cord (cdx1‐4). Ef1α is the control for quantitating RNA levels. ‐RT–PCR was performed on total RNA isolated from control embryos (lane 3).
- One‐cell stage embryos were injected animally with the Zic1‐MO (25 ng). Embryos underwent in situ hybridization. Expression of the panneural sox2 marker was normal, the forebrain marker xanf1 was expanded in 80% of the embryos, the hindbrain representative marker krox20 was reduced in 80% of the embryos, and the representative spinal cord marker cdx4 was expanded in 90% of the embryos. N = 23 embryos were per marker.
BMP can caudalize neural tissue to spinal cord fates
Our interest in hindbrain patterning led us to perform experiments on the induction of posterior neural markers in animal cap (AC) explants co‐expressing noggin and the Meis3 neural caudalizing protein 24, 25, 27. Meis3 typically induces expression of an array of posterior markers in ACs in the absence of neural induction (Fig 3A, lane 4). When co‐injecting noggin and Meis3 in ACs, neural pattern is altered in a number of ways. The caudalizing Meis3 protein inhibits noggin induction of anterior neural marker 27 expression (Fig 3A, lane 5 versus 6). In these same explants, the combination of noggin and Meis3 enhances hindbrain marker expression as might be expected if a neural inducer and caudalizer are co‐expressed (Fig 3A, lane 4 versus 5). However for spinal cord markers, the opposite is true; noggin represses spinal cord marker expression (Fig 3A, lane 4 versus 5), despite Meis3 protein significantly repressing noggin anteriorizing activity in the same ACs (Fig 3A, lane 5 versus 6). This observation suggests that BMP activity may be required for Meis3 to induce spinal cord markers in the ACs. To further address this point, Meis3 was co‐expressed with a fourfold less noggin RNA concentration (Fig 3A, lane 7). In these explants, anterior markers were still repressed by Meis3, but spinal cord markers were re‐expressed (Fig 3A, lane 5 versus 7). This observation suggests that at the lower noggin concentration, higher endogenous BMP levels are required for the activation or maintenance of spinal cord marker expression in ACs.
Figure 3. BMP activity regulates neural AP patterning.
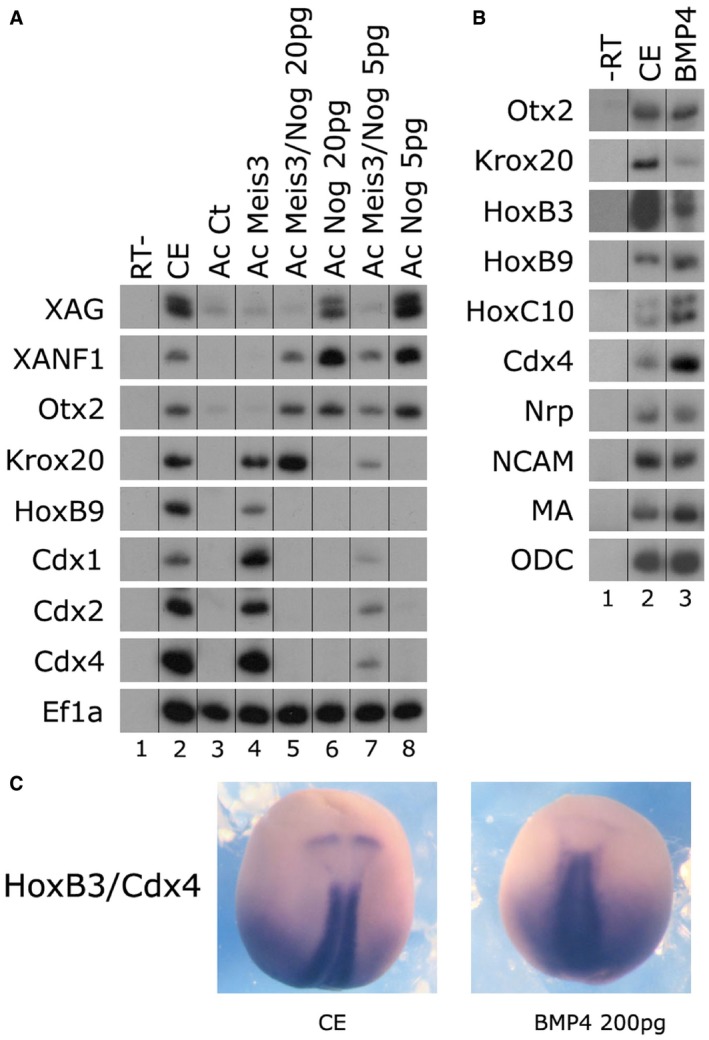
- One‐cell stage embryos were injected animally with mRNAs encoding the noggin (5/20 pg) and/or meis3 (0.5 ng) proteins. AC explants were removed from control and injected embryos at blastula stage 9, and explants were cultured to neurula stage 17. Total RNA was isolated from seven control embryos (lane 2), and from eighteen ACs from each group (lanes 3–8). Neural AP markers were examined by sqRT–PCR: forebrain (xanf1, otx2), hindbrain (krox20, hoxb1, hoxb3), spinal cord (hoxb9, cdx1‐4), and panneural (soxd). Ef1α is the control for quantitating RNA levels. ‐RT–PCR was performed on total RNA isolated from control embryos (lane 1).
- One‐cell stage embryos were injected animally with mRNA encoding bmp4 (0.2 ng) protein. Total RNA was isolated from five control embryos at neurula stage 17 (lane 1) and five embryos from the injected group (lanes 2). Neural and mesodermal markers were examined by sqRT–PCR: forebrain (otx2), hindbrain (krox20, hoxb3), spinal cord (hoxb9, hoxc10, cdx4), panneural (nrp1, ncam), and mesodermal (muscle actin—MA). ODC is the control for quantitating RNA levels. ‐RT–PCR was performed on total RNA isolated from control embryos (lane 1).
- One‐cell stage embryos were injected animally with mRNA encoding bmp4 (0.2 ng) protein Embryos underwent in situ hybridization. Expression of the hindbrain marker hoxb3 was reduced/eliminated by bmp4 (0.2 ng) in 68% of the embryos (n = 19). Expression of the spinal cord marker cdx4 was slightly increased/normal, in 100% of the embryos (n = 19).
We next addressed this same question in vivo. One‐cell (Fig 3B) and eight‐cell (Appendix Fig S3B) stage embryos were injected with RNA encoding BMP4 protein in the animal hemisphere (fated for ectoderm). Previous studies have shown that ectopic BMP ventralizes embryonic mesoderm 28, 29. However, in these present experiments, the RNA was injected at much lower concentrations (five‐ to 10‐fold lower) and was targeted to the ectoderm, and not the mesoderm. As can be seen by sqRT–PCR in neurula stage embryos (Fig 3B), panneural and forebrain marker expression (ncam, nrp1, otx2) is unaltered by the ectopic BMP levels; thus, the dose is not high enough to inhibit neural induction. However, hindbrain markers are strongly reduced (krox20, hoxb3), while spinal cord marker expression is increased (hoxb9, cdx4, hoxd10). Dorsal mesoderm fate does not seem to be perturbed by these low BMP levels, since muscle actin (MA) expression is normal in these embryos. When examining embryos by in situ hybridization, the differences between spinal cord and hindbrain markers are clear (Fig 3C); hoxb3 expression is eliminated, while cdx4 expression looks normal or expanded. Embryos injected with BMP4 into four animal blastomeres at the eight‐cell stage expressed a similar pattern of marker expression like Fig 3A (Appendix Fig S3B), with strongly reduced expression of hindbrain markers (krox20, hoxb3), but normal expression levels of spinal cord markers (hoxb9, hoxc10, cdx2, cdx4), dorsal mesoderm markers (chd, myod), and the anterior cement gland marker xag. In situ hybridization was performed on embryos that were injected with bmp4 mRNA into one blastomere at the two‐cell stage. These embryos had asymmetric expression of the hindbrain‐specific krox20 marker, but symmetric expression of spinal cord markers (hoxb9, cdx4) and dorsal mesoderm (chd, MA) markers (Appendix Fig S2B and D). We also sectioned the trunk regions of later tailbud stage embryos that were injected with bmp mRNA into one blastomere at the two‐cell stage; these embryos had normal spinal cord, notochord, and somite formation (Appendix Fig S2E). Thus, the effects of ectopic BMP levels appear specific to the neural plate and not a result of mesoderm perturbation. These results show that BMP acts to promote spinal cord formation, while repressing hindbrain cell fates, suggesting that BMP is acting to balance caudal cell fates along the neural AP axis.
BMP can activate spinal cord markers independently of mesoderm in ectodermal explants
To address a more direct role for BMP signaling in activating spinal cord markers, we examined whether it can activate spinal cord marker expression in AC explants. We found that ectopic BMP4 can specifically activate expression of spinal cord markers such as hoxb9, cdx1‐2‐4, and hoxc10‐d10 in a dose‐dependent manner (Fig 4A). Hindbrain (krox20) markers are never induced (Fig 4A). Induction of these neural markers appears to be direct in the ectoderm and uncoupled from mesoderm induction. Expression of the lateral mesoderm (kidney) BMP‐target markers osr1‐2 30 is not induced by BMP4 at these concentrations (Fig 4A). Additionally, muscle actin expression is not induced (Fig 4A). These results suggest that BMP does not indirectly mediate expression of spinal cord markers via mesoderm induction, but directly activates their expression in the ectoderm. Supporting this conclusion, ectodermal cytokeratin (E13) expression is not reduced, but constant in the explants (Fig 4A).
Figure 4. BMP activity caudalizes ectoderm to spinal cord fates.
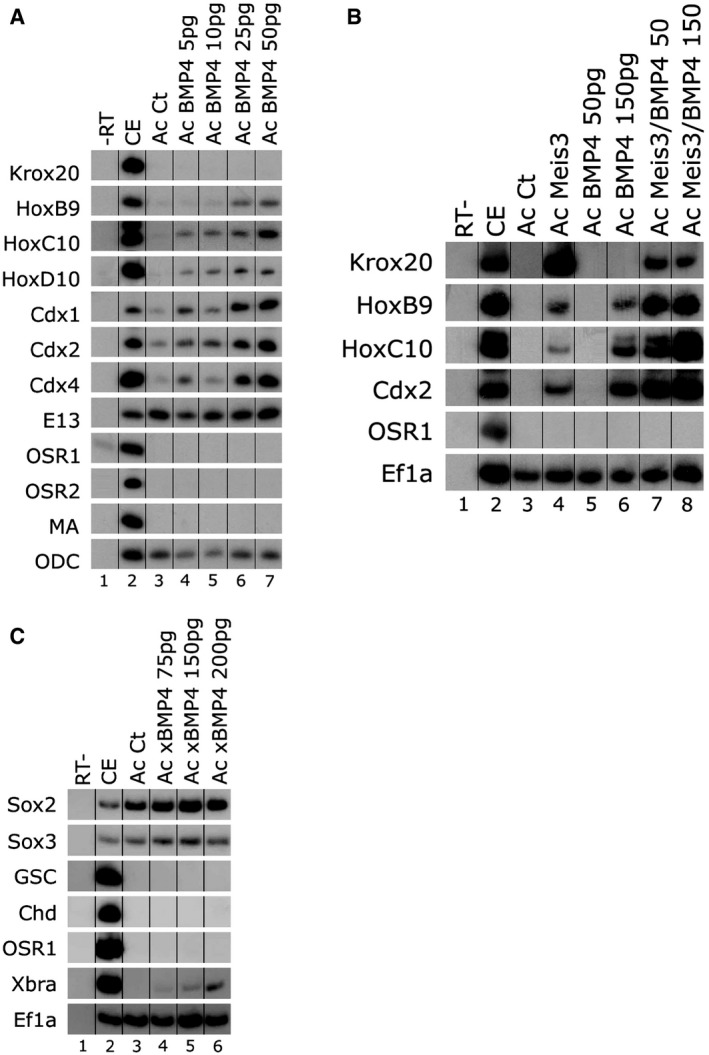
- One‐cell stage embryos were injected animally with increasing concentrations of bmp4 encoding mRNA (5–50 pg). AC explants were removed from control and injected embryos at blastula stage 9, and explants were cultured to neurula stage 16. Total RNA was isolated from five control embryos (lane 2), and from eighteen ACs from each group (lanes 3–7). Neural, epidermal, and mesodermal markers were examined by sqRT–PCR: hindbrain (krox20), spinal cord (hoxb9, hoxc10/d10, cdx1‐2‐4), epidermal cytokeratin (E13), lateral mesoderm (osr1‐2), and dorsal–lateral mesoderm muscle actin (MA). ODC is the control for quantitating the RNA levels. ‐RT–PCR was performed on total RNA isolated from control embryos (lane 1).
- One‐cell stage embryos were injected animally with mRNAs encoding the bmp4 (50/150 pg) and/or meis3 (0.7 ng) proteins. AC explants were removed from control and injected embryos at blastula stage 9, and explants were cultured to neurula stage 16. Total RNA was isolated from five control embryos (lane 2), and from eighteen ACs from each group (lanes 3–8). Neural and mesodermal markers were examined by sqRT–PCR: hindbrain (krox20), spinal cord (hoxb9, hoxc10, cdx2), and lateral mesoderm (osr1). Ef1α is the control for quantitating the RNA levels. ‐RT–PCR was performed on total RNA isolated from control embryos (lane 1).
- One‐cell stage embryos were injected animally with increasing concentrations of bmp4 encoding mRNA (75–200 pg). AC explants were removed from control and injected embryos at blastula stage 9, and explants were cultured to gastrula stage 11–11.5. Total RNA was isolated from five control embryos (lane 2), and from eighteen ACs from each group (lanes 3–6). Marker expression was determined by sqRT–PCR. Xenopus brachyury (xbra) expression was induced by BMP4 in a dose‐dependent manner. Marker expression was determined by sqRT–PCR. Neither dorsal (chd, goosecoid‐gsc) nor ventral–lateral (osr1) mesoderm markers were induced by BMP4. Sox2/3 neural markers are expressed in the ACs and are unchanged by BMP expression. EF1α is the control for quantitating RNA levels. ‐RT–PCR was performed on total RNA isolated from control embryos (lane 1).
In a reciprocal experiment to Fig 3A (noggin/Meis3 co‐expression), Meis3 is co‐expressed with BMP4 in ACs. In these ACs, krox20 expression (hindbrain) is inhibited by BMP (Fig 4B, compare lanes 4–8), but spinal cord marker expression (hoxb9, cdx2, hoxc10) is highly enhanced by BMP (Fig 4B, compare lanes 4–8). Lower concentrations of BMP4 that alone do not induce spinal cord expression (Fig 4B, lane 5) act additively with Meis3 to enhance spinal cord marker expression (Fig 4B, compare lanes 4–5, 7). Higher concentrations of BMP that can induce spinal cord marker expression also additively enhance spinal cord marker expression with Meis3 (Fig 4B, compare lanes 4, 6, 8). Mesoderm is not induced in these explants, as determined by osr1 expression. Studies in other vertebrate species suggested that co‐expression of the brachyury (Bra) and Sox proteins suffices to induce neural precursor cells that will form the spinal cord [31, rev. in 32]. A number of studies suggest that neuromesodermal precursors (NMPs) are present in the vertebrate embryo posterior that will give rise to somite or spinal cord tissues 31, 33, 34, 35, 36. The co‐expression of Bra/Sox proteins may be a transition state in these NMPs on their way to forming spinal cord. We show that BMP indeed induces a transient expression of bra in early neurula stage AC cells (Fig 4C). Interestingly, naive and BMP expressing ACs express similar levels of the sox2 and sox3 genes (Fig 4C). This AC expression of bra is transient, and no other dorsal/lateral (goosecoid‐gsc, chd, osr1, osr2, MA) mesoderm markers are detected in BMP expressing ACs at either gastrula or neurula stages (Fig 4A–C). Previous studies showed that Bra+ NMPs are driven to mesoderm fates by Wnt signaling, but promoted to neural precursor fates by Bra/Sox expression in the absence of signaling 31.
BMP activation of spinal cord markers is FGF‐dependent
FGF activity has been shown to act downstream to Wnt/Meis3 to caudalize the nervous system 24, 37, 38, 39. We thus determined whether BMP caudalizing activity is dependent on FGF signaling activity. Both FGF3 and FGF8 have neural in vivo caudalizing activity in many vertebrate systems [rev. in 12]. Ectopic BMP expression induces fgf3 and fgf8 gene expression in ACs in a dose‐dependent manner (Fig 5A). When co‐expressing BMP4 and the FGF dominant‐negative receptor in ACs, BMP activation of spinal cord markers is highly compromised (Fig 5B, lanes 5–6). FGF gene expression is also inhibited (Fig 5B, lanes 5–6). BMP spinal cord caudalizing activity acts upstream to FGF signaling to activate ligand gene expression, and in the absence of FGF signaling, BMP cannot induce expression of spinal cord markers in AC explants.
Figure 5. BMP4 caudalizes in an FGF‐dependent manner.
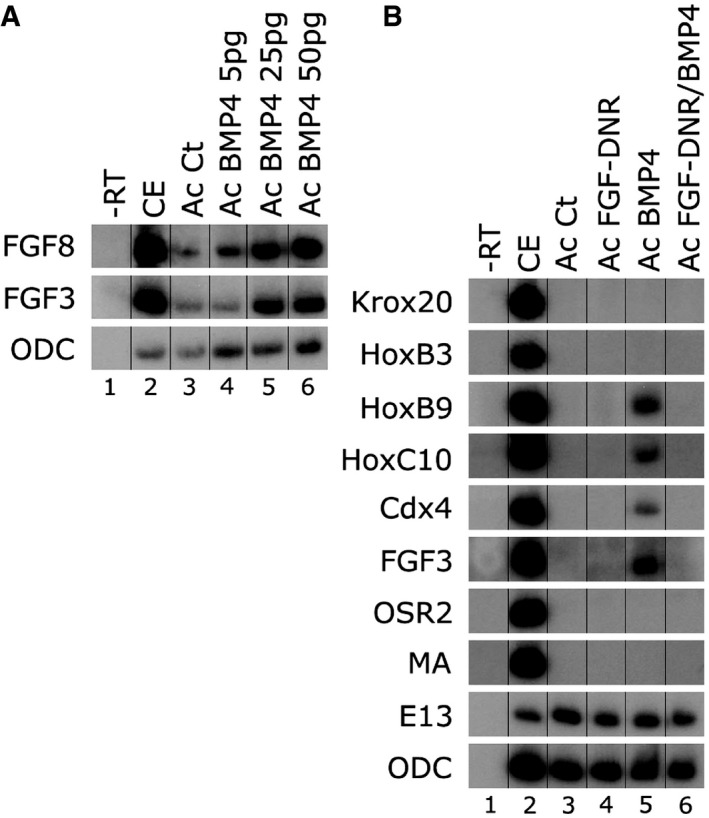
- One‐cell stage embryos were injected animally with increasing concentrations of bmp4 encoding mRNA (5–50 pg). AC explants were removed from control and injected embryos at blastula stage 9, and explants were cultured to neurula stage 16. Total RNA was isolated from five control embryos (lane 2), and from eighteen ACs from each group (lanes 3–6). FGF3‐4‐8 expression was determined by sqRT–PCR. ODC is the control for quantitating RNA levels. ‐RT–PCR was performed on total RNA isolated from control embryos (lane 1).
- One‐cell stage embryos were injected animally with mRNAs encoding the bmp4 (50 pg) and/or FGF dominant‐negative receptor (FGF‐DNR, 1.5 ng) proteins. AC explants were removed from control and injected embryos at blastula stage 9, and explants were cultured to neurula stage 16. Total RNA was isolated from five control embryos (lane 2), and from eighteen ACs from each group (lanes 3–6). In addition to fgf3, neural, epidermal, and mesodermal markers were examined by sqRT–PCR: hindbrain (krox20, hoxb3), spinal cord (hoxb9, hoxc10, cdx4), lateral mesoderm (osr1), dorsal–lateral mesoderm muscle actin (MA), and epidermal cytokeratin (E13). ODC is the control for quantitating RNA levels. ‐RT–PCR was performed on total RNA isolated from control embryos (lane 1).
Discussion
Classic experimental embryology studies suggested a two‐step model where a caudalizing factor gradient confers posterior identity to tissue which has initially been induced as anterior neuroectoderm (6, 9, 10). To address the relevance of such a gradient for canonical Wnt/β‐catenin signaling activity, frog and chick embryos and explants were treated with differing doses of Wnt inhibitors 15, 16. Wnt‐inhibitor treatment of chick neural plate explants suggested a progressive requirement for Wnt activity, at least for repressing anterior neural cell fates 16. Similar observations were seen in Xenopus embryos expressing increasing levels of Wnt‐inhibitor proteins 15. Low levels of these Wnt inhibitors expanded anterior marker expression, while posterior marker expression was shifted more caudally. Recombinant explant experiments support a Wnt ligand–morphogen model; Wnt3a caudalizes cells non‐autonomously, but β‐catenin cannot 15. Luciferase Wnt reporters in posterior slices of late gastrula embryos had higher luciferase activity than anterior slices. Moreover, endogenous β‐catenin staining revealed nuclear localization in posterior neural ectoderm versus decreased nuclear β‐catenin in anterior regions 15. These studies show the necessity of Wnt activity for correct hindbrain and forebrain AP patterning 15, 16. However, regulation of hindbrain versus spinal cord induction was not addressed, since more posterior markers were not examined 15, 16.
The specification of more posterior spinal cord cell fates may reflect interactions of different signaling pathways and Hox proteins, rather than a graded activity of a single one. Our results and those of others suggest that inducing all axial neural cell identities is more complex than just a simplified single gradient of “low to high” Wnt activity to delineate all cell fates from the forebrain to the most posterior spinal cord. The response of hindbrain versus spinal cord hox genes to changes in Wnt signaling levels does not fit a gradient model. Kiecker and Niehrs 15 showed that inhibition of Wnt signaling by Dkk1 strongly modulated hindbrain formation during Xenopus development, and these observations have been further shown in different vertebrate species [rev. in 40]. Our previous studies and those of others in Xenopus have shown that perturbation of Wnt signaling prevents transcriptional activation of the earliest expressed hindbrain regulating homeobox genes, such as meis3, hoxd1, hoxa2, and gbx2 15, 24, 41, 42, without disrupting mesoderm formation 24. Loss of activity of these early neural plate expressed homeoproteins significantly compromises hindbrain formation in different vertebrates [rev. in 40], without significantly altering the spinal cord 24; this study). Meis3, hoxd1, hoxa1, and gbx2 are all Wnt direct‐target genes, directly activated by β‐catenin/TCF transcription factors, and their expression is activated by Wnt signaling in the neural plate at the earliest developmental stages crucial for neural patterning and specification 24, 41, 42, 43, 44.
Moreover, the Wnt8‐MO inhibited expression of Paralogous group 1 (PG‐1) hox genes that are Wnt/β‐catenin direct targets expressed in the hindbrain, while surprisingly up‐regulating and expanding spinal cord‐specific hoxc6 expression 41. This result contradicts a simple posterior to anterior gradient model for patterning the posterior nervous system. This study 41 also demonstrated a differential expression response intensity of the various PG‐1 hox genes to modulation of Wnt8, which are all expressed at a similar AP level, while the more posterior hoxb4 remained unaltered 41, 45. These findings are not compatible with a gradient, but suggest that upon initial induction of the PG‐1 hox gene expression, more complex interactions, with the Wnt, RA, and FGF pathways, are integrated to specify their expression. Hoxd1 expression is a good example; it is a Wnt/β‐catenin‐ and RA‐direct‐target gene, as well as a direct target of the Wnt/β‐catenin mediator Meis3 24, 41, 46, 47. Alternatively, differential Wnt activity could be achieved by changing the time periods of neuroectoderm exposure to the Wnt signal. The most anterior prechordal plate mesoderm expresses several secreted and autonomous Wnt inhibitors, while the paraxial‐fated mesoderm is required in vivo to induce posterior neural tissue via the expression and secretion of Wnt3a protein to the overlying posterior neural plate 24, 48, 49.
While Kiecker and Niehrs 15 show graded nuclear‐localized β‐catenin staining in sagittal sections that included involuted mesoderm and neuroectoderm, comparison of sagittal and parasagittal sections reveals a different picture 50. In parasagittal sections, the most intense staining is in the posterior involuted mesoderm, but staining in the neural plate region was fairly uniform along the AP axis 50. This neural plate region corresponds to the cells expressing the highest levels of the earliest hindbrain and spinal cord promoting genes (meis3, gbx2, hox, and cdx), in zebrafish, Xenopus, and chick embryos at late gastrula stages. Some of these genes are induced by the underlying paraxial‐fated mesoderm via secretion of Wnt3a 24, 44, but this region does not appear to have a β‐catenin gradient 50. Further supporting a role for neural‐specific Wnt activity in hindbrain formation, a recent mapping of endogenous in vivo Wnt activity utilizing a GFP‐reporter gene in Xenopus tropicalis shows extensive GFP expression in posterior neural plate tissue at late gastrula to early neurula stages 51. There is no apparent GFP gradient in these transgenic frogs. This coincides with the temporal window of posterior neural patterning that is the obvious spatial target of Wnt3a‐MO or Dkk1 inhibition in Xenopus embryos, in which either the lack of endogenous secreted Wnt3a from the dorsal–lateral mesoderm or the presence of Dkk1 in the neural plate significantly inhibit hindbrain formation 15, 24.
The question of whether a spatial, or temporal Wnt signal gradient, or even different intrinsic responses to a constant Wnt signal mediate these different inductive properties is still open. Graded expression of Wnt pathway components or inhibitors has not been convincingly observed in the posterior nervous system. These observations could also argue for differential threshold responses to a constant signal, rather than a graded one. Our results do not contradict these previous results 15, 16. Wnt is required to repress forebrain and induce the hindbrain; there is no doubt about this observation. However, the elimination of Wnt activity and/or downstream transcription factors mediating Wnt activity strongly disrupted hindbrain formation, while only minimally modulating spinal cord formation. Thus, a Wnt gradient cannot explain the specification of all cell fates along the entire neural AP axis. Whether low levels of Wnt are required for spinal cord formation still needs to be determined, but we clearly show that the most posterior hox and cdx genes of the spinal cord are expressed robustly when Wnt signaling is highly compromised and hindbrain formation is perturbed.
FGF signaling plays a role in both hindbrain and spinal cord formation in vertebrates 24, 37, 39, 52, 53, 54, 55. The FGF3/8 signaling center in r4 is crucial for early hindbrain specification in vertebrates 53, 54, 56. This signaling center appears not to be involved in spinal cord induction. The nature of the spinal cord inducing FGF activity is elusive. It is still unknown how signal timing, germ layer locale, and expression specificity of FGF ligands specify spinal cord fates. FGF3 and FGF8 genes are expressed in the correct time and place to account for this later spinal cord specification, with high transcript levels in the posterior circumblastoporal collar region at stages 11.5–12.5 56, 57. BMP4 gene expression and Smad1 protein localization are also found in the ventral–posterior mesoectodermal regions adjacent to and overlapping the circumblastoporal collar 50, 58, 59. In this study, we show that BMP signaling plays a pivotal role in neural caudalization by acting upstream to or by directly interacting with FGF signaling. Relatively low BMP levels efficiently caudalize embryos and explants in a highly differential manner. We show that BMP inhibits hindbrain, while activating spinal cord marker expression. This activation of spinal cord markers is FGF‐dependent and could be mediated by direct activation of FGF ligand gene expression or by a different mechanism. A study in zebrafish embryos showed that BMP and FGF signaling cooperate to specify spinal cord fates 60. A more recent study in mice showed that BMP/FGF signaling interactions were crucial for caudal neural tube closure, neural crest specification, and posterior axis extension 61. Another work suggests that components of Wnt, BMP, and FGF signaling may interact to control neural AP patterning 62. In Xenopus, the FGF8a splice version protein appears to be an efficient inducer of spinal cord 63, but its precise role in spinal cord induction still needs to be elucidated. At late gastrula–early neurula stages, fgf8 is expressed in the most posterior mesoderm/neural plate regions, adjacent to posterior BMP expressing epidermal regions 63. BMP/FGF interactions could potentially induce spinal cord in the posterior neural plate. A study in differentiating human ES cells also showed that FGF can efficiently induce expression of spinal cord markers in the absence of Wnt signaling 64. Interestingly, at later stages of tailbud formation in the most posterior embryo regions, FGF and BMP were shown to be necessary for spinal cord formation in both zebrafish 31, 65 and Xenopus embryos 66, 67. Canonical Wnt signaling was important for promoting mesoderm progenitor cells, but not neural progenitors in the posterior spinal cord 31. These findings, while at later stages of development, support our observations. Similar to the spinal cord neural fated precursors found in zebrafish and other vertebrates 31, 32, we show that BMP4 induces bra/sox positive NMP‐like cells that will express spinal cord markers. Previous studies showed that Bra expressing NMPs are driven to mesoderm fates by Wnt signaling, but promoted to neural precursor fates by Bra/Sox expression in the absence of Wnt signaling 31. By transiently activating bra gene expression, coupled to endogenous sox gene expression, BMP/FGF signaling could induce posterior neural precursors in AC explants.
Despite hints from these few studies, the concept of BMP acting as a neural caudalizer seems fairly counterintuitive, but its levels in posterior ectoderm or mesoderm could control FGF activity in the posterior nervous system. We show that FGF and BMP activities are coordinated to promote spinal cord cell fates, in which canonical Wnt activity seems expendable. While Wnt signaling is linked to activating expression of many spinal cord promoting transcription factors, FGF also induces many of these same factors. The bioassays used in many systems may not have finely resolved the actual in vivo regulatory mechanisms. One example is the Sall4 protein. The Sall4 protein required for spinal cord formation in Xenopus was initially detected as a Wnt‐target gene, but its function in vivo is controlled by FGF, and not Wnt 68. Many cdx and hox genes expressed along the neural AP axis are induced by both FGF and Wnt signaling in various bioassays. These genes are clearly involved in distinguishing between spinal cord and hindbrain fates. The ultimate goal will be to determine the actual in vivo hierarchy of signaling that specifies the distinct caudal neural regions. We suggest that Wnt signaling is mainly involved in hindbrain formation, by initially activating expression of meis3 and the most anterior hox genes 18, 24, 41, 42, 45, 46. Our previous studies suggest that this initial Wnt‐caudalizing event occurs early in development, in early‐mid gastrula stages 24. FGF seems to play two separate temporally and spatially different roles that regulate hindbrain and spinal cord formation. Early FGF3/8 expression is downstream to Wnt/Meis3 in the hindbrain 18, 37. In contrast, later FGF activity specifies spinal cord and is required for the activation of cdx and the more posterior hox genes 52, 69. FGF induction of cdx gene expression activates spinal cord hox gene expression, while simultaneously repressing more anterior hindbrain hox gene expression 48, 69, 70, 71, 72. A recent study in chick and mouse embryos suggests that regionalization of the spinal cord and activation of cdx gene expression is separated in time and space from the induction of the hindbrain/forebrain regions 73. This study emphasizes that different lineage origins separate these different neural AP regions. In Xenopus, we suggest that the caudalizing forces driving neural AP pattern are the “earlier” Wnt‐Meis3‐FGF activity that induces the hindbrain 24, 25, 37, which is coupled to a “later” BMP‐FGF‐Cdx activity that specifies the spinal cord 48, 71, 72, 74. This separation of time, space, and signaling for the hindbrain and spinal cord 31, 73 does not fit a model in which a single gradient of Wnt signaling demarcates the entire AP character of the nervous system.
Recent findings in the hemichordate, Saccoglossus kowalevskii (Acorn worm) support our observations 75. In the acorn worm, canonical Wnt signaling represses anterior and induces mid‐axial ectodermal fates during early development. Matching our findings in Xenopus, specification of the most posterior fates, as shown by posterior Hox gene expression, was not perturbed by modulation of canonical Wnt levels during early AP axis formation at late blastula to early gastrula stages. We show the first proof for an analogous role of canonical Wnt signaling in vertebrates. Canonical Wnt signaling is crucial for establishing mid‐axial neural structures (hindbrain) versus the more posterior (spinal cord), suggesting that this role for Wnt signaling in neural patterning is evolutionarily conserved between vertebrates and hemichordates.
The pathways regulating neural AP patterning are highly conserved among vertebrates. However, the actual “nuts and bolts” of the process are far from being elucidated. The challenge of the future is to fully understand how the interactions of all the different signaling pathways regulate Hox, Cdx, and Meis gene expression and protein activities to correctly pattern the neural AP axis. This study has examined the fundamental regulation of neural AP patterning during its earliest stages. We have found that two long‐held paradigms: (i) A Wnt‐signaling AP “low to high” gradient drives all posterior neural cell fate formation, and (ii) BMP signaling is incompatible with AP neural cell fate patterning both need to be re‐assessed. Further experimentation will explain how BMP/FGF activities interact to specify spinal cord cell fates in the developing vertebrate nervous system.
Materials and Methods
Xenopus embryos
Ovulation, in vitro fertilization, culture, and explant dissections and treatments were as described 27, 29, 76. Embryos were sectioned in paraffin as described 77.
RNA and DNA injections
Capped sense in vitro transcribed mRNA constructs of bmp4, noggin, dkk1, Zic5‐DN, FGF‐DNR, and meis3 18, 24, 29, 43, 78 were injected into the animal hemisphere of one‐, two‐, or eight‐cell stage embryos. For zygotic expression, the Nxfz8/pCS2 plasmid was injected into the animal hemisphere of one‐cell stage embryos 26. β‐gal RNA was injected and detected as described 24, 76.
In situ hybridization
Whole‐mount in situ hybridization was performed 79 with digoxigenin‐labeled probes to sox2, xanf1, hoxb3, krox20, cdx4, hoxb9, hoxd10, muscle actin, and chordin 18, 25, 68, 74, 80, 81.
Semi‐quantitative (sq) RT–PCR analysis
sqRT–PCR was performed 82. In all sqRT–PCR experiments, three to six independent experimental repeats were typically performed. In all experiments, samples are routinely assayed a minimum of two times for each marker. sqRT–PCR Primers (see Appendix Table S1): efα, odc, nrp1, soxd, ncam, nrp1, xanf1, otx2, xag1, krox20, hoxb1, hoxb3, hoxb4, hoxb9, cdx1, cdx2, cdx4, n‐tub, hoxc10, hoxd10, epidermal cytokeratin (E13), osr1, osr2, fgf3, fgf8, xbra, xnot, chd, gsc, muscle actin (MA), myod, vent1, vent2, and wnt8 18, 24, 80, 81, 83, 84, 85.
Author contributions
DF, HP, YEG and AM conceived and designed the research. HP, YEG, and AM performed most of the experiments, with assistance from YME and YV. YME and YV performed preliminary experiments in this project. DF, HP, YEG, and AM wrote the manuscript with input from YME.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supporting information
Appendix
Review Process File
Acknowledgements
We thank Dr. Aviad Keren for help with embryo sectioning. D.F. was supported by grants from the Israel Science Foundation (658/09, 658/15) and the Israel Cancer Research Fund.
EMBO Reports (2019) 20: e45842
References
- 1. Gamse J, Sive H (2000) Vertebrate anteroposterior patterning: the Xenopus neurectoderm as a paradigm. BioEssays 22: 976–986 [DOI] [PubMed] [Google Scholar]
- 2. Pera EM, Acosta H, Gouignard N, Climent M, Arregi I (2014) Active signals, gradient formation and regional specificity in neural induction. Exp Cell Res 321: 25–31 [DOI] [PubMed] [Google Scholar]
- 3. Carron C, Shi DL (2016) Specification of anteroposterior axis by combinatorial signaling during Xenopus development. Wiley Interdiscip Rev Dev Biol 5: 150–168 [DOI] [PubMed] [Google Scholar]
- 4. Stern CD (2005) Neural induction: old problem, new findings, yet more questions. Development 132: 2007–2021 [DOI] [PubMed] [Google Scholar]
- 5. Nieuwkoop PD (1952) Activation and organization of the central nervous system in amphibians. III Synthesis of a new working hypothesis. J Exp Zool 120: 83–108 [Google Scholar]
- 6. Spemann H, Mangold H (1924) Uber Induktion von Embryonalanlagen durch Implantation artfremder Organisatoren. Arch Mikr Anat Entw Mech 100: 599–638 [Google Scholar]
- 7. De Robertis EM, Kuroda H (2004) Dorsal‐ventral patterning and neural induction in Xenopus embryos. Annu Rev Cell Dev Biol 20: 285–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Harland RM (2000) Neural induction. Curr Opin Genet Dev 10: 357–362 [DOI] [PubMed] [Google Scholar]
- 9. Eyal‐Giladi H (1954) Dynamic aspects of neural induction. Arch Biol 65: 180–259 [PubMed] [Google Scholar]
- 10. Toivonen S, Saxen L (1968) Morphogenetic interaction of presumptive neural and mesodermal cells mixed in different ratios. Science 159: 539–540 [DOI] [PubMed] [Google Scholar]
- 11. Tuazon FB, Mullins MC (2015) Temporally coordinated signals progressively pattern the anteroposterior and dorsoventral body axes. Semin Cell Dev Biol 42: 118–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dorey K, Amaya E (2010) FGF signaling: diverse roles during early vertebrate embryogenesis. Development 137: 3731–3742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Elkouby YM, Frank D (2010) Wnt‐catenin signaling in vertebrate posterior neural development In Colloquium series on developmental biology, Kessler DS. (ed.), eBook 4. San Rafael, CA: Morgan & Claypool Life Sciences; [PubMed] [Google Scholar]
- 14. White RJ, Schilling TF (2008) How degrading: Cyp26s in hindbrain development. Dev Dyn 237: 2775–2790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kiecker C, Niehrs C (2001) A morphogen gradient of Wnt/beta‐catenin signaling regulates anteroposterior neural patterning in Xenopus . Development 128: 4189–4201 [DOI] [PubMed] [Google Scholar]
- 16. Nordstrom U, Jessell TM, Edlund T (2002) Progressive induction of caudal neural character by graded Wnt signaling. Nat Neurosci 5: 525–532 [DOI] [PubMed] [Google Scholar]
- 17. Bae CJ, Park BY, Lee YH, Tobias JW, Hong CS, Saint‐Jeannet J (2014) Identification of Pax3 and Zic1 targets in the developing neural crest. Dev Biol 386: 473–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gutkovich YE, Ofir R, Elkouby YM, Dibner C, Gefen A, Elias S, Frank D (2010) Xenopus Meis3 protein lies at a nexus downstream to Zic1 and Pax3 proteins, regulating multiple cell‐fates during early nervous system development. Dev Biol 338: 50–62 [DOI] [PubMed] [Google Scholar]
- 19. Merzdorf CS, Sive HL (2006) The zic1 gene is an activator of Wnt signaling. Int J Dev Biol 50: 611–617 [DOI] [PubMed] [Google Scholar]
- 20. Monsoro‐Burq AH, Wang E, Harland RM (2005) Msx1 and Pax3 cooperate to mediate FGF8 and WNT signals during Xenopus neural crest induction. Dev Cell 8: 167–178 [DOI] [PubMed] [Google Scholar]
- 21. Nagai T, Aruga J, Takada S, Günther T, Spörle R, Schughart K, Mikoshiba K (1997) The expression of the mouse Zic1, Zic2, and Zic3 gene suggests an essential role for Zic genes in body pattern formation. Dev Biol 182: 299–313 [DOI] [PubMed] [Google Scholar]
- 22. Plouhinec JL, Roche DD, Pegoraro C, Figueiredo AL, Maczkowiak F, Brunet LJ, Milet C, Vert JP, Pollet N, Harland RM et al (2014) Pax3 and Zic1 trigger the early neural crest gene regulatory network by the direct activation of multiple key neural crest specifiers. Dev Biol 386: 461–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sato T, Sasai N, Sasai Y (2005) Neural crest determination by co‐activation of Pax3 and Zic1 genes in Xenopus ectoderm. Development 132: 2355–2363 [DOI] [PubMed] [Google Scholar]
- 24. Elkouby YM, Elias S, Casey ES, Blythe SA, Tsabar N, Klein PS, Root H, Liu KJ, Frank D (2010) Mesodermal Wnt signaling organizes the neural plate via Meis3. Development 137: 1531–1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dibner C, Elias S, Frank D (2001) XMeis3 protein activity is required for proper hindbrain patterning in Xenopus laevis embryos. Development 128: 3415–3426 [DOI] [PubMed] [Google Scholar]
- 26. Deardorff MA, Tan C, Conrad LJ, Klein PS (1998) Frizzled‐8 is expressed in the Spemann organizer and plays a role in early morphogenesis. Development 125: 2687–2700 [DOI] [PubMed] [Google Scholar]
- 27. Salzberg A, Elias S, Nachaliel N, Bonstein L, Henig C, Frank D (1999) A Meis family protein caudalizes neural cell fates in Xenopus . Mech Dev 80: 3–13 [DOI] [PubMed] [Google Scholar]
- 28. Jones CM, Dale L, Hogan BL, Wright CV, Smith JC (1996) Bone morphogenetic protein‐4 (BMP‐4) acts during gastrula stages to cause ventralization of Xenopus embryos. Development 122: 1545–1554 [DOI] [PubMed] [Google Scholar]
- 29. Re'em‐Kalma Y, Lamb T, Frank D (1995) Competition between noggin and bone morphogenetic protein 4 activities may regulate dorsalization during Xenopus development. Proc Natl Acad Sci USA 92: 12141–12145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. James RG, Schultheiss TM (2005) Bmp signaling promotes intermediate mesoderm gene expression in a dose‐dependent, cell‐autonomous and translation‐dependent manner. Dev Biol 288: 113–125 [DOI] [PubMed] [Google Scholar]
- 31. Martin BL, Kimelman D (2012) Canonical Wnt signaling dynamically controls multiple stem cell fate decisions during vertebrate body formation. Dev Cell 22: 223–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Martin BL (2016) Factors that coordinate mesoderm specification from neuromesodermal progenitors with segmentation during vertebrate axial extension. Semin Cell Dev Biol 49: 59–67 [DOI] [PubMed] [Google Scholar]
- 33. Davis RL, Kirschner MW (2000) The fate of cells in the tailbud of Xenopus laevis . Development 127: 255–267 [DOI] [PubMed] [Google Scholar]
- 34. Gentsch GE, Owens ND, Martin SR, Piccinelli P, Faial T, Trotter MW, Gilchrist MJ, Smith JC (2013) In vivo T‐box transcription factor profiling reveals joint regulation of embryonic neuromesodermal bipotency. Cell Rep 4: 1185–1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Taniguchi Y, Kurth T, Weiche S, Reichelt S, Tazaki A, Perike S, Kappert V, Epperlein HH (2017) The posterior neural plate in axolotl gives rise to neural tube or turns anteriorly to form somites of the tail and posterior trunk. Dev Biol 422: 155–170 [DOI] [PubMed] [Google Scholar]
- 36. Tzouanacou E, Wegener A, Wymeersch FJ, Wilson V, Nicolas JF (2009) Redefining the progression of lineage segregations during mammalian embryogenesis by clonal analysis. Dev Cell 17: 365–376 [DOI] [PubMed] [Google Scholar]
- 37. Aamar E, Frank D (2004) Xenopus Meis3 protein forms a hindbrain inducing center by activating FGF/MAP kinase and PCP pathways. Development 131: 153–163 [DOI] [PubMed] [Google Scholar]
- 38. Domingos PM, Itasaki N, Jones CM, Mercurio S, Sargent MG, Smith JC, Krumlauf R (2001) The Wnt/beta‐catenin pathway posteriorizes neural tissue in Xenopus by an indirect mechanism requiring FGF signaling. Dev Biol 239: 148–160 [DOI] [PubMed] [Google Scholar]
- 39. Ribisi S, Mariani FV, Aamar E, Lamb TM, Frank D, Harland RM (2000) Ras‐mediated FGF signaling is required for the formation of posterior but not anterior neural tissue in Xenopus laevis . Dev Biol 227: 183–196 [DOI] [PubMed] [Google Scholar]
- 40. Frank D, Sela‐Donenfeld D (2018) Hindbrain induction during early vertebrate development. Cell Mol Life Sci 76: 941–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. In der Rieden PM, Vilaspasa FL, Durston AJ (2010) Xwnt8 directly initiates expression of labial Hox genes. Dev Dyn 239: 126–139 [DOI] [PubMed] [Google Scholar]
- 42. Janssens S, Denayer T, Deroo T, van Roy F, Vleminckx K (2010) Direct control of Hoxd1 and Irx3 expression by Wnt/beta catenin signaling during anteroposterior patterning of the neural axis in Xenopus . Int J Dev Biol 54: 1435–1442 [DOI] [PubMed] [Google Scholar]
- 43. Elkouby YM, Polevoy H, Gutkovich YE, Michaelov A, Frank D (2012) A hindbrain‐repressive Wnt3a/Meis3/Tsh1 circuit promotes neuronal differentiation and coordinates tissue maturation. Development 139: 1487–1497 [DOI] [PubMed] [Google Scholar]
- 44. Li B, Kuriyama S, Moreno M, Mayor R (2009) The posteriorizing gene Gbx2 is a direct target of Wnt signaling and the earliest factor in neural crest induction. Development 136: 3267–3278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. McNulty CL, Peres JN, Bardine N, van den Akker WM, Durston AJ (2005) Knockdown of the complete Hox paralogous group 1 leads to dramatic hindbrain and neural crest defects. Development 132: 2861–2871 [DOI] [PubMed] [Google Scholar]
- 46. Dibner C, Elias S, Ofir R, Souopgui J, Kolm PJ, Sive H, Pieler T, Frank D (2004) The Meis3 protein and retinoid signaling interact to pattern the Xenopus hindbrain. Dev Biol 271: 75–86 [DOI] [PubMed] [Google Scholar]
- 47. Kolm PJ, Sive H (1995) Hindbrain patterning requires retinoid signaling. Dev Biol 192: 1–16 [DOI] [PubMed] [Google Scholar]
- 48. Faas L, Isaacs HV (2009) Overlapping functions of Cdx1, Cdx2, and Cdx4 in the development of the amphibian Xenopus tropicalis . Dev Dyn 238: 835–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. McGrew LL, Hoppler S, Moon RT (1997) Wnt and FGF pathways cooperatively pattern anteroposterior neural ectoderm in Xenopus . Mech Dev 69: 105–114 [DOI] [PubMed] [Google Scholar]
- 50. Schohl A, Fagotto F (2002) Beta‐catenin, MAPK and Smad signaling during early Xenopus development. Development 129: 37–52 [DOI] [PubMed] [Google Scholar]
- 51. Borday C, Parain K, Thi Tran H, Vleminckx K, Perron M, Monsoro‐Burq AH (2018) An atlas of Wnt activity during embryogenesis in Xenopus tropicalis . PLoS One 13: e0193606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bel‐Vialar S, Itasaki N, Krumlauf R (2002) Initiating Hox gene expression: in the early chick neural tube differential sensitivity to FGF and RA signaling subdivides the HoxB genes in two distinct groups. Development 129: 5103–5115 [DOI] [PubMed] [Google Scholar]
- 53. Maves L, Jackman W, Kimmel CB (2002) FGF3 and FGF8 mediate a rhombomere 4 signaling activity in the zebrafish hindbrain. Development 129: 3825–3837 [DOI] [PubMed] [Google Scholar]
- 54. Walshe J, Maroon H, McGonnell IM, Dickson C, Mason I (2002) Establishment of hindbrain segmental identity requires signaling by FGF3 and FGF8. Curr Biol 12: 1117–1123 [DOI] [PubMed] [Google Scholar]
- 55. Weisinger K, Wilkinson DG, Sela‐Donenfeld D (2008) Inhibition of BMPs by follistatin is required for FGF3 expression and segmental patterning of the hindbrain. Dev Biol 324: 213–225 [DOI] [PubMed] [Google Scholar]
- 56. Lombardo A, Isaacs HV, Slack JM (1998) Expression and functions of FGF‐3 in Xenopus development. Int J Dev Biol 42: 1101–1107 [PubMed] [Google Scholar]
- 57. Monsoro‐Burq AH, Fletcher RB, Harland RM (2003) Neural crest induction by paraxial mesoderm in Xenopus embryos requires FGF signals. Development 130: 3111–3124 [DOI] [PubMed] [Google Scholar]
- 58. Fainsod A, Steinbeisser H, De Robertis EM (1994) On the function of BMP‐4 in patterning the marginal zone of the Xenopus embryo. EMBO J 13: 5015–5025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Knöchel S, Dillinger K, Köster M, Knöchel W (2001) Structure and expression of Xenopus tropicalis BMP‐2 and BMP‐4 genes. Mech Dev 109: 79–82 [DOI] [PubMed] [Google Scholar]
- 60. Kudoh T, Wilson SW, Dawid IB (2002) Distinct roles for Fgf, Wnt and retinoic acid in posteriorizing the neural ectoderm. Development 129: 4335–4346 [DOI] [PubMed] [Google Scholar]
- 61. Anderson MJ, Schimmang T, Lewandoski M (2016) An FGF3‐BMP signaling axis regulates caudal neural tube closure, neural crest specification and anterior‐posterior axis extension. PLoS Genet 12: e1006018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hashiguchi M, Mullins MC (2013) Anteroposterior and dorsoventral patterning are coordinated by an identical patterning clock. Development 140: 1970–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Fletcher RB, Baker JC, Harland RM (2006) FGF8 spliceforms mediate early mesoderm and posterior neural tissue formation in Xenopus . Development 133: 1703–1714 [DOI] [PubMed] [Google Scholar]
- 64. Lupo G, Novorol C, Smith JR, Vallier L, Miranda E, Alexander M, Biagioni S, Pedersen RA, Harris WA (2013) Multiple roles of Activin/Nodal, bone morphogenetic protein, fibroblast growth factor and Wnt/β‐catenin signaling in the anterior neural patterning of adherent human embryonic stem cell cultures. Open Biol 3: 120167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Goto H, Kimmey SC, Row RH, Matus DQ, Martin BL (2017) FGF and canonical Wnt signaling cooperate to induce paraxial mesoderm from tailbud neuromesodermal progenitors through regulation of a two‐step epithelial to mesenchymal transition. Development 144: 1412–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Beck CW, Whitman M, Slack JM (2001) The role of BMP signaling in outgrowth and patterning of the Xenopus tail bud. Dev Biol 238: 303–314 [DOI] [PubMed] [Google Scholar]
- 67. Beck CW, Christen B, Slack JM (2003) Molecular pathways needed for regeneration of spinal cord and muscle in a vertebrate. Dev Cell 5: 429–439 [DOI] [PubMed] [Google Scholar]
- 68. Young JJ, Kjolby RA, Kong NR, Monica SD, Harland RM (2014) Spalt‐like 4 promotes posterior neural fates via repression of pou5f3 family members in Xenopus . Development 141: 1683–1693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Isaacs HV, Pownall ME, Slack JM (1998) Regulation of Hox gene expression and posterior development by the Xenopus caudal homologue Xcad3. EMBO J 17: 3413–3427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Pownall ME, Isaacs HV, Slack JM (1998) Two phases of Hox gene regulation during early Xenopus development. Curr Biol 8: 673–676 [DOI] [PubMed] [Google Scholar]
- 71. Shimizu T, Bae YK, Hibi M (2006) Cdx‐Hox code controls competence for responding to Fgfs and retinoic acid in zebrafish neural tissue. Development 133: 4709–4719 [DOI] [PubMed] [Google Scholar]
- 72. Skromne I, Thorsen D, Hale M, Prince VE, Ho RK (2007) Repression of the hindbrain developmental program by Cdx factors is required for the specification of the vertebrate spinal cord. Development 134: 2147–2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Metzis V, Steinhauser S, Pakanavicius E, Gouti M, Stamataki D, Ivanovitch K, Watson T, Rayon T, Mousavy Gharavy SN, Lovell‐Badge R et al (2018) Nervous system regionalization entails axial allocation before neural differentiation. Cell 175: 1105–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Keenan ID, Sharrard RM, Isaacs HV (2006) FGF signal transduction and the regulation of Cdx gene expression. Dev Biol 299: 478–488 [DOI] [PubMed] [Google Scholar]
- 75. Darras S, Fritzenwanker JH, Uhlinger KR, Farrelly E, Pani AM, Hurley IA, Norris RP, Osovitz M, Terasaki M, Wu M et al (2018) Anteroposterior axis patterning by early canonical Wnt signaling during hemichordate development. PLoS Biol 16: e2003698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Bonstein L, Elias S, Frank D (1998) Paraxial‐fated mesoderm is required for neural crest induction in Xenopus embryos. Dev Biol 193: 156–168 [DOI] [PubMed] [Google Scholar]
- 77. Keren A, Bengal E, Frank D (2005) p38 MAP kinase regulates the expression of XMyf5 and affects distinct myogenic programs during Xenopus development. Dev Biol 288: 73–86 [DOI] [PubMed] [Google Scholar]
- 78. Henig C, Elias S, Frank D (1998) A POU protein regulates mesoderm competence to FGF in Xenopus . Mech Dev 71: 131–142 [DOI] [PubMed] [Google Scholar]
- 79. Harland RM (1991) In situ hybridization: an improved whole‐mount method for Xenopus embryos. Methods Cell Biol 36: 685–695 [DOI] [PubMed] [Google Scholar]
- 80. Fonar Y, Gutkovich YE, Root H, Malyarova A, Aamar E, Golubovskaya VM, Elias S, Elkouby YM, Frank D (2011) Focal adhesion kinase protein regulates Wnt3a gene expression to control cell fate specification in the developing neural plate. Mol Biol Cell 22: 2409–2421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Polevoy H, Malyarova A, Fonar Y, Elias S, Frank D (2017) FoxD1 protein interacts with Wnt and BMP signaling to differentially pattern mesoderm and neural tissue. Int J Dev Biol 61: 293–302 [DOI] [PubMed] [Google Scholar]
- 82. Snir M, Ofir R, Elias S, Frank D (2006) Xenopus laevis POU91 protein, an Oct3/4 homologue, regulates competence transitions from mesoderm to neural cell‐fates. EMBO J 25: 3664–3674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Bin‐Nun N, Lichtig H, Malyarova A, Levy M, Elias S, Frank D (2014) PTK7 modulates Wnt signaling activity via LRP6. Development 141: 410–421 [DOI] [PubMed] [Google Scholar]
- 84. Hudry B, Thomas‐Chollier M, Volovik Y, Duffraisse M, Dard A, Frank D, Technau U, Merabet S (2014) Molecular insights into the origin of the Hox‐TALE patterning system. Elife 3: e01939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Zetser A, Frank D, Bengal E (2001) MAP kinase converts MyoD into an instructive muscle differentiation factor in Xenopus . Dev Biol 240: 168–181 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Review Process File


